
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 March 2023
Sec. Plant Pathogen Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1146915
This article is part of the Research Topic Relationships Between Plant Disease and Microbiomes View all 5 articles
Asexual spore serves as essential inoculum of rice blast during the disease cycle, and differentiation of young conidium from conidiophore is intimately regulated by cell cycle. Mih1 encodes a dual-specificity phosphatase that involved in the G2/M transition of the mitotic cell cycle by regulating the Cdk1 activity in eukaryotes. Till now, the roles of Mih1 homologue, however, remain unclear in Magnaporthe oryzae. We here functionally characterized the Mih1 homologue MoMih1 in M. oryzae. MoMih1 is localized to both the cytoplasm and nucleus and can physically interact with the CDK protein MoCdc28 in vivo. Loss of MoMih1 led to delayed nucleus division and a high level of Tyr15 phosphorylation of MoCdc28. The MoMih1 mutants showed retarded mycelial growth with a defective polar growth, less fungal biomass, and shorter distance between diaphragms, compared with the KU80. Asexual reproduction altered in MoMih1 mutants, with both abnormal conidial morphogenesis and decreased conidiation. The MoMih1 mutants severely attenuated the virulence to host plants due to the impaired ability of penetration and biotrophic growth. The incapability of scavenging of host-derived reactive oxygen species, which was possibly ascribed to the severely decreased extracellular enzymes activities, were partially associated with deficiency of pathogenicity. Besides, the MoMih1 mutants displayed also improper localization of retromer protein MoVps26 and polarisome component MoSpa2, and defects of cell wall integrity (CWI), melanin pigmentation, chitin synthesis, and hydrophobicity. In conclusion, our results demonstrate that MoMih1 plays pleiotropic roles during fungal development and plant infection of M. oryzae.
Rice blast, caused by the fungal pathogen Magnaporthe oryzae (synonym to Pyricularia oryzae), is one of the most destructive diseases that seriously threatens the rice production worldwide (Wilson and Talbot, 2009). The blast fungus differentiates three-celled pyriform spores as the primary inoculum, and their dispersal is essential for the epidemiology of this disease (Fernandez and Orth, 2018). The blast disease begins at the attachment of those asexual spores to the susceptible rice leaves, and under permitted condition, they germinate and develop a specialized dome-shape appressorium at the tip of germ tube, which helps to rupture the rice leaf cuticle cells (Sabnam and Barman, 2017). After penetration, the invasive hyphae grow biotrophically in the initially infected plant cells and thereafter followed by a necrotrophic phase, which resulted in cell death and appearance of disease lesions on the plants (Ebbole, 2007; Zhang et al., 2016).
The development of both pyriform spores and appressorium is one of the most complicated morphological and physiological events (Zhou et al., 2009; Liu et al., 2010; Oses-Ruiz and Talbot, 2017; Oses-Ruiz et al., 2021). The switches from conidiophore growth to young conidium differentiation, and germ tube to melanized appressorium are intimately linked to cell cycle regulation in M. oryzae (Liu et al., 2010; Saunders et al., 2010). During conidiation, two rounds of mitosis and cytokinesis have occurred to develop the mature pyriform conidia, in which each cell contains a single nucleus (Liu et al., 2010; Li et al., 2018). Blocking of Dam1, a subunit of DASH complex involved in cell cycle, delayed the mitotic progression and impaired sporulation under permitted conditions (Shah et al., 2019). Cdc14 is a conserved dual-specificity phosphatases that directly accelerates the cytokinesis during cell division (Meitinger et al., 2012). In M. oryzae, the CDC14 homologue is essential for mitosis and cytokinesis during asexual spore differentiation, and its disruption results in defective mitotic exit, and incorrect septum formation during conidium and appressorium development in M. oryzae (Li et al., 2018). Besides, the Pombe Cdc15 homology (PCH) MoCDC15, which is required for cytokinesis by directly regulating the formation of actomyosin ring, has also been identified for asexual spore development and plant infection of M. oryzae (Goh et al., 2011). During infection process, appressorium mediated plant infection, which is crucial for the full virulence of M. oryzae, is tightly regulated by the cell cycle checkpoint (Saunders et al., 2010). Mutation of two checkpoints such as NIM1 and NimA could prevent the fungal infection of rice plants (Saunders et al., 2010). What’s more, the block of cell cycle by prohibiting S-phase checkpoint or mitotic exit also prevents appressorium-mediated plant infection, further highlighting the essential role of cell cycle regulation in determining the full virulence of M. oryzae (Saunders et al., 2010; Oses-Ruiz et al., 2017). Though successful proceeding of cell cycle is critical for the development of asexual spores and infection structures, the precise regulatory mechanism of the cell-cycle check point controlled by the special regulator, have yet to be definitively understood in M. oryzae.
Protein phosphorylation and dephosphorylation are two most important post-translational modifications that regulate the cellular processes, such as cell cycle, DNA replication, and gene transcription, and have mainly been regulated by protein kinase and protein phosphatase in eukaryotes (Zhao et al., 2007; Li et al., 2012). Dual-specificity phosphatases are recognized to dephosphorylate tyrosine and serine/threonine residues within the substrate, and thus control a serial of fundamental physiological processes such as cell growth, differentiation, and apoptosis (Tonks, 2006). Mih1 encodes a dual-specificity phosphatase that activates the enzymes activity of cyclin-dependent protein kinase (Cdk1) by removing the phosphate group on the catalytic subunit Cdc28, then promotes G2/M transition of mitotic cell cycle (Camps et al., 2000; Pal et al., 2008). In fungi like Schizosaccharomyces pombe, Ustilago maydis and Aspergillus nidulans, the Cdc25 encodes a dual-specificity protein phosphatase that is essential for the G2/M phase transition, and the disruption of Cdc25 makes the mutant postpone entry into mitosis and develop abnormally large cell (Lew and Kornbluth, 1996; Sgarlata and Perez-Martin, 2005). However, in budding yeast Saccharomyces cerevisiae, inactivation of Mih1, a homolog of S. pombe Cdc25, could only lead to a moderate delay of mitotic entry, which is inconsistent with the previous findings in other fungi, suggesting a potential exist of redundancy of the dephosphorylation of Cdk1 in different eukaryotes (Pal et al., 2008). In addition, Mih1 has been confirmed to control the phosphorylation state of Vps26, a subunit of retromer subcomplex that directly recognizes the sorting signal in cargo transport, and mutation of Mih1 lead to decreased affinity of retromer to the targets, defective polar growth and pathogenesis of the fungi (Riquelme, 2013; Zheng et al., 2015; Cui et al., 2017; Qu et al., 2022). Taken together, the cell cycle was differentially regulated in the fungi when responses to diverse environmental stimuli, and understanding the roles of dual-specificity phosphatase Cdc25 would provide novel clues to insight of regulatory mechanism of the cell cycle in eukaryotes.
In this study, we report a crucial role for a putative cell division control protein Cdc25 which shows high similarity to the S. cerevisiae Mih1 during fungal development and pathogenesis in M. oryzae. Our results demonstrate that the loss of MoMih1 led to delayed nucleus division and a high level of Tyr15 phosphorylation of MoCdc28. The MoMih1 mutant was severely reduced in polar growth and asexual reproduction. The virulence of MoMih1 mutants were significantly attenuated due to the impaired ability of penetration and biotrophic growth. The MoMih1 mutants displayed also improper localization of retromer protein MoVps26 and polarisome component MoSpa2, and defects of cell wall integrity (CWI), melanin pigmentation, chitin distribution, hydrophobicity, and extracellular enzymes activities. Taken together, our results indicate that MoMih1 is an important regulator required for the fungal development and plant infection of M. oryzae.
The wild-type strain Guy11 was used the parental strain to delete the MoGin4 and MoCyc2 gene, while the KU80 strain was used as the parental strain to disrupt the MoMih1 gene in M. oryzae (Villalba et al., 2008). Mycelia used for microscopic examination, DNA and RNA isolation, protein extraction were cultured in liquid CM at 28°C for 24 to 48 h. To assay vegetative growth, mycelial plugs of KU80, MoMih1-41, MoMih1-44, and MoMih1c were respectively inoculated on complete medium (CM), minimal medium (MM), oatmeal medium (OM), and rice straw decoction and corn medium (RDC) (Qi et al., 2012), and cultured at 28°C in darkness for 5 d. To measure fungal biomass, four mycelial plugs of tested strains were cultured in 150 mL liquid CM at 28°C for 3 d, then froze dry to measure dry weight.
Conidia suspension of KU80, MoMih1-41, MoMih1-44 and MoMih1c were dropped on hydrophilic coverslips to develop mycelium, then stained with CFW to examine septum formation. For melanin pigmentation, all strains were cultured on CM for 5 d and/or in liquid CM for 7 d at 28°C in darkness. To investigate asexual reproduction, all strains were cultured on RDC medium, and the conidia were collected, and counted as the reference described (Guo et al., 2017). Conidial morphology and size, and the average numbers of septa per conidia were recorded using a light microscope. To evaluate the responses to different stressors, all strains were inoculated on CM plates supplemented with or without 5 mM H2O2, 200 μg/mL CFW, 200 μg/mL CongoRed, and 5 mM caffeine, respectively, and then cultured at 28°C for 5 d. To detect CWI, equal amount of mycelia of tested strain was treated with 7.5 mg/mL lysing enzyme, and the release of protoplasts was recorded after enzyme treatment for 30, 60, and 90 min.
To investigate the phylogenetic relationship among the orthologues involved in the cell cycle in different species, the amino acid sequences of each orthologue in S. cerevisiae were used as a prey to search the NCBI database, all the orthologous proteins in diverse species were downloaded, and aligned using the Muscle (Edgar, 2004). The aligned sequences were imported into software IQ-TREE 2, and thereafter phylogenetically analyzed to evaluate the evolutionary relationship among those orthologues using the maximum likelihood method (Minh et al., 2020).
The 1200 bp DNA fragments, which respectively flanked the upstream and downstream of the MoMih1 gene, were amplified from the M. oryzae genomic DNA with primers MoMih1-1F/MoMih1-2R and MoMih1-3F/MoMih1-4R (Table S1). The hygromycin B cassette (HPH) was amplified from plasmid pCB1003 with primers HPH-F/HPH-R (Table S1). The above DNA fragments were fused to the enzymes digested (XbaI and HindIII) pKO1B vector (Lu et al., 2014) using the gap repair system in yeast FY834. The resultant plasmid pKO1B-MoMih1-HPH was transformed into Agrobacterium tumefaciens strain AGL1, and the MoMih1 deletion mutants was generated using the A. tumefaciens-mediated transformation (ATMT) of M. oryzae. The putative MoMih1 mutants were further verified by PCR, RT-PCR, and Southern blot.
To generate complemented strain, a 3030 bp DNA fragment containing a native promoter (1180 bp) and a full length of MoMih1 coding region (1850 bp) was amplified using primers Mih1pF/Mih1pR (Table S1) and transformed into yeast strain XK125 together with XhoI linearized pYF11. The complemental vector pYF11-MoMih1 was introduced into the MoMih1-44 mutant by PEG-mediated transformation of M. oryzae. Both MoGin4 (MGG_02810) and MoCyc2 (MGG_07065) deletion mutants were generated following the above-described methods.
To observe the localization of MoMih1 in M. oryzae, the full length of MoMih1 coding region (1853 bp) was amplified with primers RP27Mih1pF/Mih1pR (Table S1) and ligated with XhoI linearized pYF11. The resultant plasmid pYF11-MoMih1-eGFP was transformed into Guy11. To visualize the colocalization of MoMih1 with the nucleus signal, the plasmid containing H1-mRFP fusion gene was transformed into strain expressing MoMih1-eGFP. The derivative strains, which expressed MoMih1-eGFP or both MoMih1-eGFP and H1-mRFP, were examined under a confocal laser scanning microscope (CLSM).
The full length of MoVps26 coding region (1098 bp) was amplified from genomic DNA of Guy11 with primers Vps26pF/Vps26pR (Table S1), and then ligated to XhoI digested pYF11 following the above-described procedures. The plasmid pYF11-MoVps26-eGFP was respectively introduced in KU80 and MoMih1-44 mutant, and then hyphal tips of each strain expressing MoVps26-eGFP fusion protein were examined under CLSM. To evaluate whether the microtubule-destabilizing drug affected the localization of MoVps26-eGFP, the above transformants were cultured in liquid CM with or without 0.4 μg/mL benomyl, and then hyphal tips of them were examined under CLSM.
To determine the localization of MoSpa2 in MoMih1 mutant, the full length of MoSpa2 coding region (3.0 kb) was cloned with primers Spa2pF/Spa2pR (Table S1) and ligated into pYF11 as the procedures described above. The plasmid pYF11-MoSpa2-eGFP was then introduced into KU80 and MoMih1-44 mutant, respectively, and hyphal tips of the derivative strain expressing MoSpa2-eGFP were examined under CLSM.
To test pathogenicity, conidial suspensions (1 × 104 spores/mL) of KU80, MoMih1-41, MoMih1-44 and MoMih1c were dropped on barley leaves (Hordeum vulgare cv Golden Promise; one-week-old), and/or sprayed on rice seedlings (Oryza sativa cv CO-39; two-week-old), respectively. The fungi-inoculated plants were placed in a moist chamber at 28°C in dark for 24 h, and then shifted to a photoperiod of 12 h light for another 96 h. To examine fungal infection, conidial suspension (1 × 104 spores/mL) was dropped on lower epidermis of barley leaves, cultured at 28°C for 24 h and/or 48 h, and then invasive hyphae were viewed under light microscopy. The DAB staining was performed following the previous methods (Guo et al., 2017).
The chitin contents of KU80, MoMih1-41, MoMih1-44 and MoMih1c were determined following the previous method (Yin et al., 2016). 5 mg freeze-dried fungal powder was mixed with 1 mL 6% KOH solution, water bathed at 80°C for 90 min, and then centrifuged (16 000g, 10 min). The sediment was washed by 1 mL PBS for three times, resuspended in 0.5 mL Mcilvine’s buffer (pH 6), and then added 100 μL chitinase (C6137, Sigma-Aldrich, Shanghai, China) in suspension. After reacting in water bath at 37°C for 16 h, 100 μL sodium borate solution (0.27 M, pH 9) was added in the tubes, then boiled at 100°C for 10 min. 1 mL diluted Ehrlich’s reagent solution (10-folds) was finally added to the reaction tubes, and then incubated at 37°C for 20 min. The absorbance value of each sample was determined at OD585. A standard curve was prepared using the D-GlcNAc (A8625, Sigma-Aldrich, Shanghai, China). This experiment was repeated three times.
The strains of KU80, MoMih1-41, MoMih1-44 and MoMih1c were respectively inoculated on CM plates with or without 0.2 mM ABTS (2,2’-Azinobis-(3-ethylbenzthiazoline-6-sulphonate)), placed in chamber at 28°C for 24 h, and then recorded the purple hydrolysis circle on the media. In liquid CM, extracellular enzyme activities, including the laccases and peroxidases, were determined following the previous procedures (Chi et al., 2009). These experiments were repeated three times.
The total RNA of each sample was extracted and reversely transcribed following the previous procedures (Guo et al., 2016). The expression of target genes involving in melanogenesis, conidiation, hydrophobicity or genes encoding peroxidase, laccase, chitin synthase were detected using the BIO-RAD CFX96 touch qRT-PCR system (Bio-Rad, Hercules, California, USA) according to the established procedures (Guo et al., 2016). All the primers used here are listed in Table S1.
To determine the roles of MoMih1 on cytokinesis, the vector pMoCH245Y-H3-mRFP was respectively transformed in KU80 and MoMih1-44 mutant via ATMT to generate KU80-H3-mRFP and MoMih1-H3-mRFP strains. To visualize nuclear distribution, both mycelia and conidia of each tested strain were cultured as described above, stained with CFW, and then visualized by CLSM.
To determine the roles of MoMih1 during autophagy, the vector pMoCH245Y-eGFP-ATG8 was transformed into KU80 and MoMih1-44 mutant, respectively. The derivative strains of KU80-eGFP-ATG8 and MoMih1-eGFP-ATG8 were respectively grown in liquid CM at 28°C for 36 h, then transferred to nitrogen starvation (SD-N) medium for another 2 or 4 h. The eGFP-MoAtg8 fusion protein and free GFP were detected using the anti-GFP antibodies, and analyzed by the ImageJ software to calculate the ratio of free GFP and fusion eGFP-MoATG8 (Zhu et al., 2018).
The full-length of the coding sequences of MoMih1 and MoCdc28 were cloned, and inserted into the bait construct pGADT7 (630442, Clontech, Dalian, China) and prey construct pGBKT7 (630443, Clontech, Dalian, China), respectively. Both the resultant bait construct pGADT7-MoMih1 and prey construct pGBKT7-MoCdc28 were co-transformed into yeast strain AH109 (MATα, trp1-901, leu2-3, 112, ura3-52, his3-200), and proteins interaction was validated following the described methods (Zheng et al., 2021a).
The MoMih1-CYFP and MoCdc28-NYFP fusion constructs were generated by ligating the coding sequences of MoMih1 and MoCdc28, which were amplified with primers pHZ68Mih1F/pHZ68Mih1R and pHZ65Cdc28F/pHZ65Cdc28, into pHZ68 and pHZ65, respectively. The vector pairs of MoMih1-CYFP and MoCdc28-NYFP, MoMih1-CYFP and NYFP, CYFP and MoCdc28-NYFP, and CYFP and NYFP were simultaneously transformed into Guy11. YFP signals were examined under the CLSM. The primers are listed in Table S1.
To determine the phosphorylation level of Cdc28, total protein of the tested strains was separated by 12% SDS-PAGE and immunoblotted with an anti-phospho-Cdc2 (Tyr15) antibody (Cell Signaling Technology). The anti-Cdc2 antibody (Santa Cruz Biotechnology) was used as a negative control.
The proceeding of cell cycle is essential for the precise initiation of conidiophore and appressorium, and is accurately regulated by the cell cycle-related genes in filamentous fungus (Saunders et al., 2010; Oses-Ruiz and Talbot, 2017; Yue et al., 2017). In this study, a family of cell cycle-related proteins was identified and phylogenetically grouped into 17 clades (Figure 1A). Previously, two cell cycle-related genes, including MoCks1 (Cks1 family) and CHM1 (CLA4 family), were individually deleted and functionally characterized in M. oryzae (Li et al., 2004; Yue et al., 2017). In this study, to investigate the roles of the rest 15 cell cycle-related genes in M. oryzae, a high-throughput gene deletion system (Lu et al., 2014) was used following the homologous gene disruption strategy (Figure S1A). At present, three of them, including MoMih1 (MGG_07734), MoGin4 (MGG_02810) and MoCyc2 (MGG_07065), were individually deleted from the parental strain KU80 or Guy11, respectively, and validated by PCR, RT-PCR and Southern blot (Figures S1B–D; Figures S2A–D). The two mutants (MoGin4 and MoCyc2), except for MoMih1, were dispensable for vegetative growth and pathogenicity, and thus did not perform further evaluations (Figures 1B, C). Subsequent bioinformatic analysis revealed that MoMih1 encodes a dual-specificity phosphatase that contains a conserved DCR motif (aa 392-394) and HCXXXXXR sequence (aa 434-441) (Figure S3) unique to Cdc25 and Mih1 phosphatases in S. pombe and S. cerevisiae, respectively, implying a conserved role of MoMih1 in the cell cycle regulation during the development and plant infection of M. oryzae.
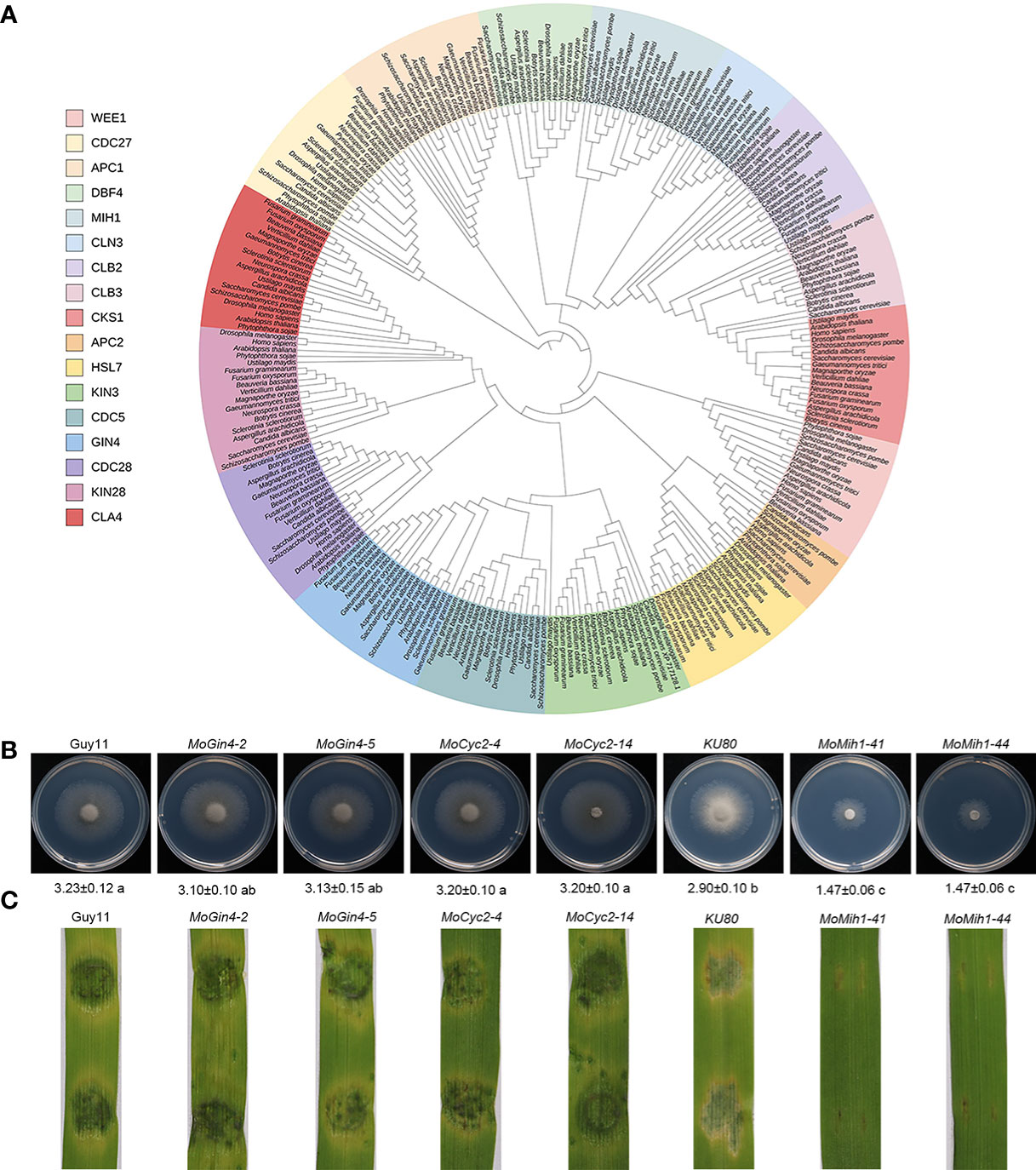
Figure 1 The orthologues of cell cycle and morphogenetic proteins in M. oryzae. (A) Phylogenetic analysis and classification of cell cycle and morphogenetic related genes in diverse species. IQ-TREE 2 was used to establish a phylogenetic tree based on the full length of these proteins using the maximum likelihood method. (B) The mycelial growth of the tested strains. The colony diameters of the tested strains were assayed on MM medium at 5 dpi. (C) The pathogenic assay of the tested strains. Mycelial plugs of the tested strains were inoculated on 7-day-old barley leaves, and the diseased leaves were recorded at 5 dpi.
To ascertain the cellular localization of MoMih1 in M. oryzae, the MoMih1-eGFP fusion protein was transformed into the wild type strain Guy11, and then examined under CLSM. The MoMih1-eGFP fusion protein was mostly distributed in cytoplasm of the conidia (Figures 2A, B; Figure S4A), and mainly concentrated as patches in the vegetative and invasive hyphae (Figures S4B, C). To ascertain whether the patches of MoMih1-eGFP was localized at nucleus in M. oryzae, the red fluorescent fusion protein H1-RFP was co-expressed in the MoMih1-eGFP strain. A co-localization of MoMih1-eGFP and H1-RFP was observed in the vegetative hyphae of M. oryzae (Figures 2C, D).
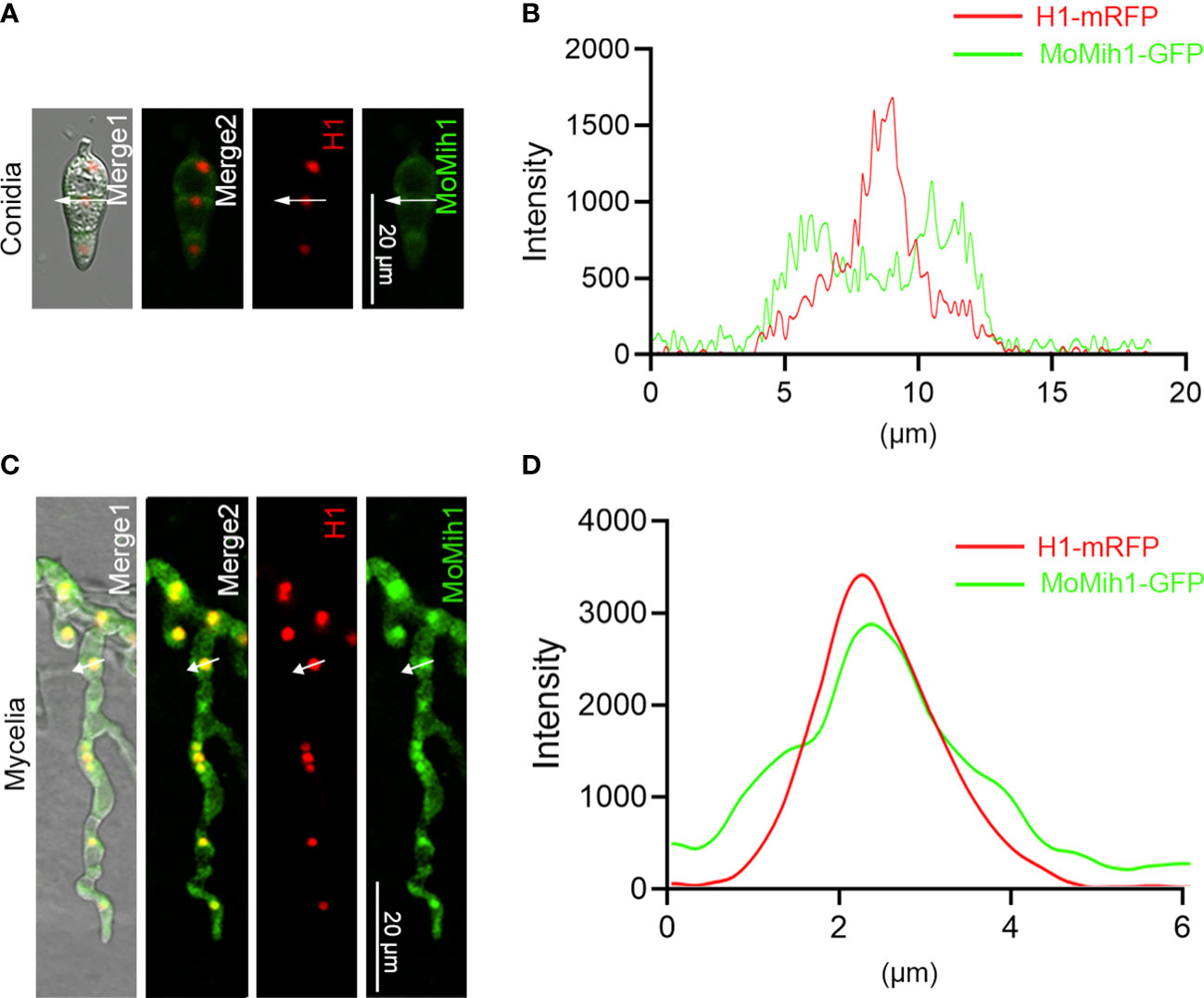
Figure 2 The subcellular localization of MoMih1 in M. oryzae. (A) The conidia of the transformants co-expressing the MoMih1-eGFP and H1-mRFP fusion protein were examined under a CLSM. Bar = 20 μm. (B) Line scan graph analysis of the fluorescence. Line scan graph indicates that MoMih1 don’t colocalized with the histone protein at spore stage. (C) The hyphae of the transformants co-expressing the MoMih1-eGFP and H1-mRFP fusion protein were examined under a CLSM. Bar = 20 μm. (D) Line scan graph analysis of the red and green fluorescence. Line scan graph analysis of fluorescence indicates the co-localization of MoMih1 with histone at mycelia.
To validate the biological role of MoMih1 in M. oryzae, a complemental strain MoMih1c was subsequently generated by reintroducing the full length of MoMih1 into MoMih1-44 mutant, and verified by RT-PCR (Figure S1C). The mycelial growth rate of MoMih1 mutants were significantly reduced in comparison to the KU80 and MoMih1c (Figures 3A, B). In liquid CM, mycelial pellets of MoMih1 mutants were much smaller but denser than that of KU80 and MoMih1c (Figure 3C). The average length of sub-apical cell of the MoMih1 mutants was approximately 50% shorter than that of KU80 and MoMih1c (Figures 3D, E). Besides, the MoMih1 mutants significantly decreased the fungal biomass compared with that of KU80 and MoMih1c (Figure 3F). These results indicate that MoMih1 is required for the vegetative growth of M. oryzae.
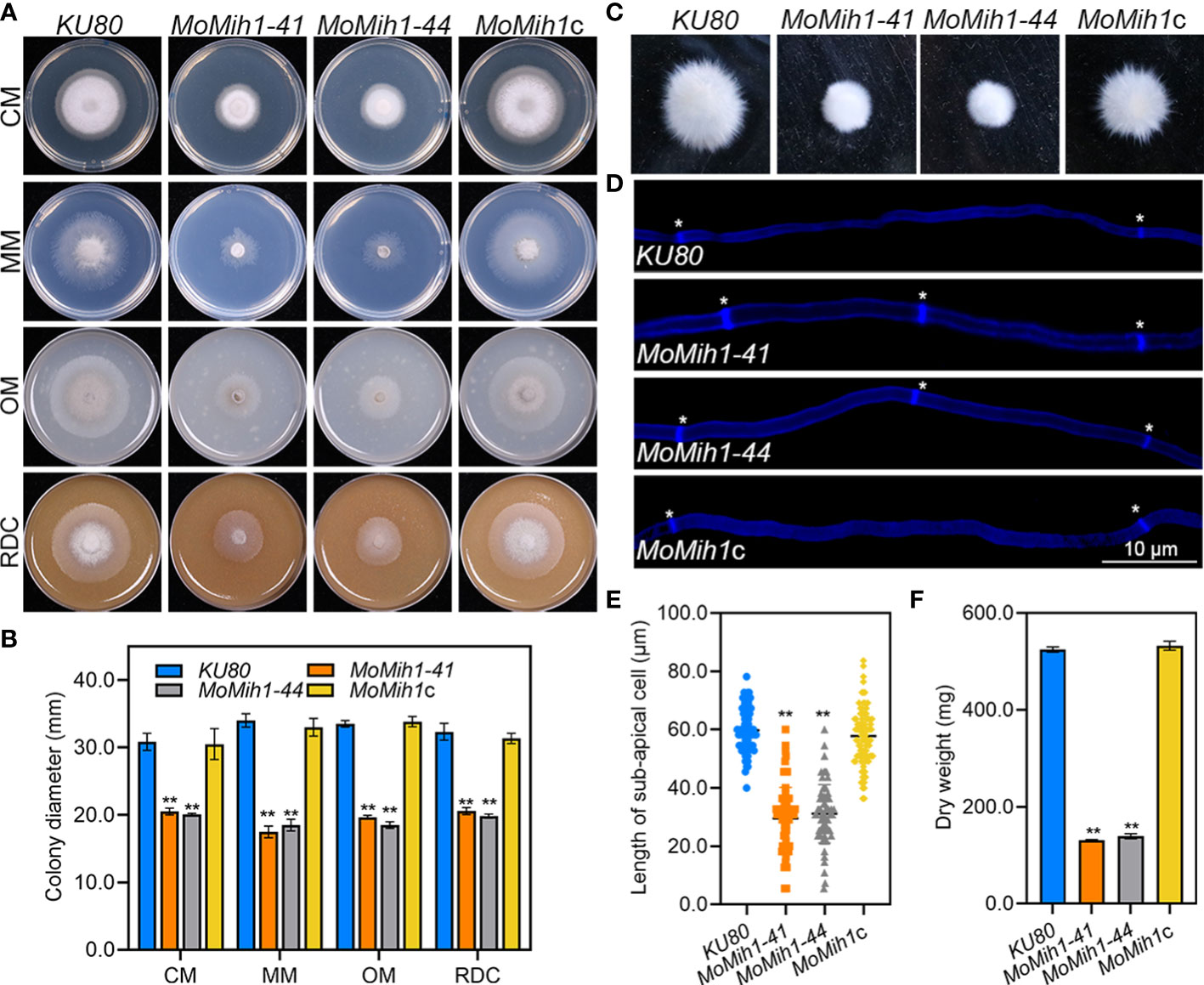
Figure 3 MoMih1 is required for vegetative growth of M. oryzae. (A) MoMih1 mutants displayed retarded growth on four artificial media. The mycelial plugs of KU80, MoMih1-41, MoMih1-44, and MoMih1c were inoculated on CM, MM, OM, and RDC, respectively, and cultured at 28°C in darkness for 5 days. (B) The analysis of colony diameter of the MoMih1 mutants. The colony diameter of the tested strains grown on four artificial media were measured at 5 dpi, and then statistically analyzed. The error bars represent the SD of three replicates, whereas “**” represents a significant difference (p < 0.01). (C) Mycelia growth of the MoMih1 mutants in liquid CM. All strains were inoculated in liquid CM at 28°C for 48 h in darkness and then photographed. (D) Examination of hyphal septa in the MoMih1 mutants. The mycelia of the tested strains were stained with CFW, and then observed under fluorescent microscopy (UV 330-380 nm). Bar = 10 μm. (E) Analysis of the cell length between two diaphragms in vegetative hyphae. The length between two diaphragms of vegetative hyphae of the tested strains were recorded, and statistically analyzed. “**” represents the significant difference between the KU80 and MoMih1 mutants (p < 0.01). (F) Measurement of fungal biomass. Fungal mycelia of KU80, MoMih1-41, MoMih1-44, and MoMih1c was collected at 3 dpi, freezing dried, and statistically analyzed (p < 0.01).
The aerial hyphae growth and conidiophore development were compared, and the MoMih1 mutants developed less fluffy aerial hyphae than that of KU80 and MoMih1c (Figure 4A). Microscopic examination showed that most of the mutants develop abnormal conidiophores, bearing significantly less conidia than that of KU80 and MoMih1c (Figure 4B). The conidial production of MoMih1 mutants was apparently decreased compared with KU80 and MoMih1c (Figure 4C). To further understand the potential role of MoMih1 on conidiation, the expression of sporulation-related genes was investigated, and all of them significantly down-regulated in the MoMih1 mutant, compared with that in KU80 strain (Figure 4D). Besides, the MoMih1 mutants showed abnormal conidial morphology (Figure 4E), with evidently increased conidial length, compared with KU80 and MoMih1c (Figure 4F). Meanwhile, the MoMih1 mutants developed a greater number of one or two-celled conidia, compared with that by KU80 and MoMih1c (Figure 4G).
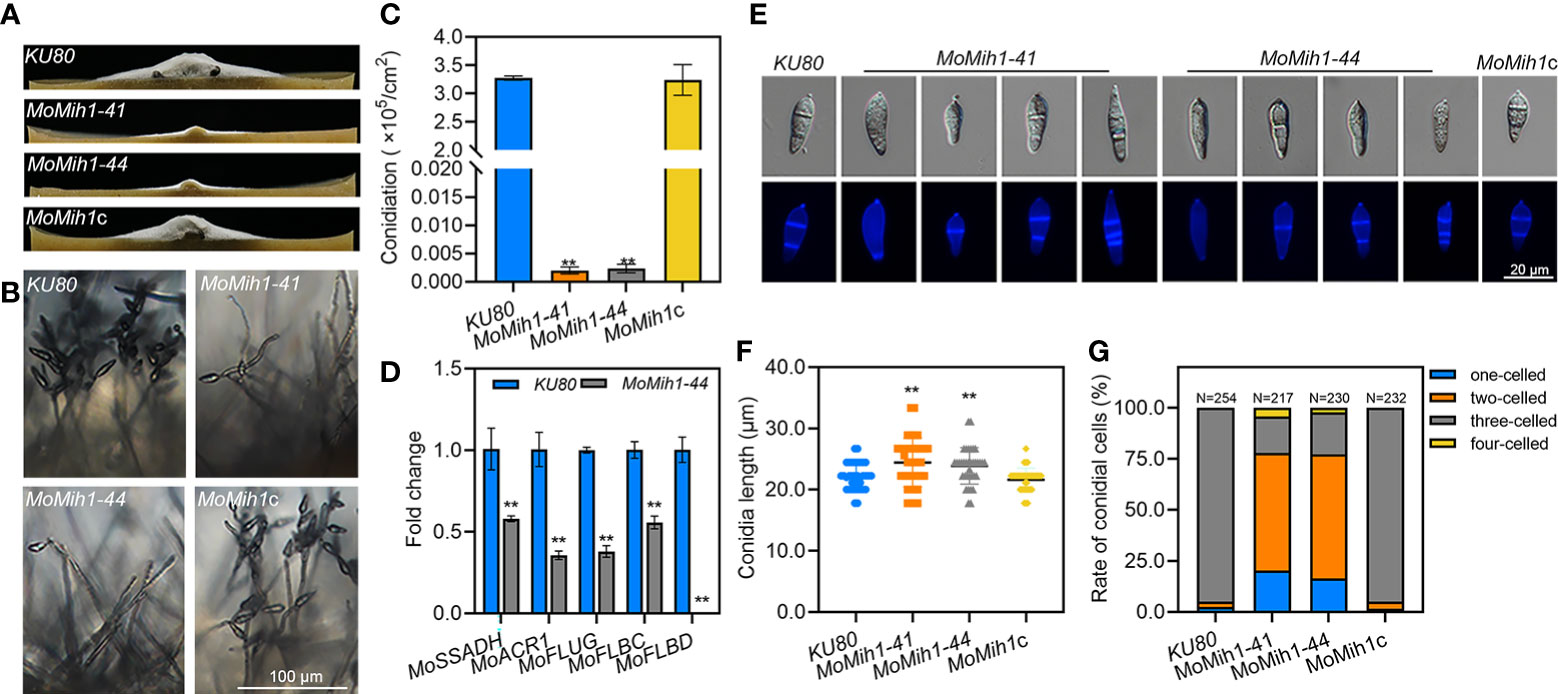
Figure 4 MoMih1 is required for the conidiation and conidial morphology of M. oryzae. (A) Comparison of aerial hyphae growth. All strains were grown on RDC media under darkness, and then photographed at 5 dpi. (B) Conidia formation on the conidiophores. Light microscopic observation was carried out to examine the conidia development on the conidiophores of the MoMih1 mutants that grown on RDC at 7dpi. Bar = 100 μm. (C) Statistical analysis of conidia reproduction. The conidia formed by each tested strain were collected and statically analyzed. “**” means significant difference among the tested strains (p < 0.01), and error bars indicate SD. (D) Transcription of the conidiation-related genes. The expression of conidiation-related genes was examined by qRT-PCR in the KU80 and MoMih1 mutants. “**” means significant differences (p<0.01), and error bars represent SD. (E) Conidial morphology of MoMih1 mutant. Conidia of each tested strain were stained with CFW, and then observed by differential interference contrast (DIC) microscopy and UV fluorescence microscopy. (F) Statistical analysis of conidial length. The conidial length was determined by measuring more than 200 conidia, and then statistically analyzed. “**” means significant difference among the strains (p < 0.01), and error bars represent SD. (G) Ratio of conidia with different number of cells in the MoMih1 mutant. The proportion of conidia with different cell numbers was statistically analyzed from three independent experiments. “**” means significant difference among the tested strains (p < 0.01), and error bars represent SD.
The pathogenicity of KU80, MoMih1 mutants and MoMih1c was conducted on both rice and barley leaves. The MoMih1 mutants significantly attenuated the virulence on host plant leaves, compared with KU80 and MoMih1c (Figures 5A, B). The infection ability of the MoMih1 mutants was impaired, with most of them exhibiting incapability of rupturing the barley leaf epidermis cells, compared with KU80 and MoMih1c (Figure 5C). The penetration rate of MoMih1 mutants was approximately 60% less than that of by KU80 and MoMih1c (Figure 5D). At 48 hpi, the invasive hyphae of the MoMih1 mutants were mostly restricted in the initially invaded cells, in contrast to the free expanding growth to the neighboring cells by the KU80 and MoMih1c (Figures 5C, E), suggesting that MoMih1 plays an important role in the pathogenesis of M. oryzae.
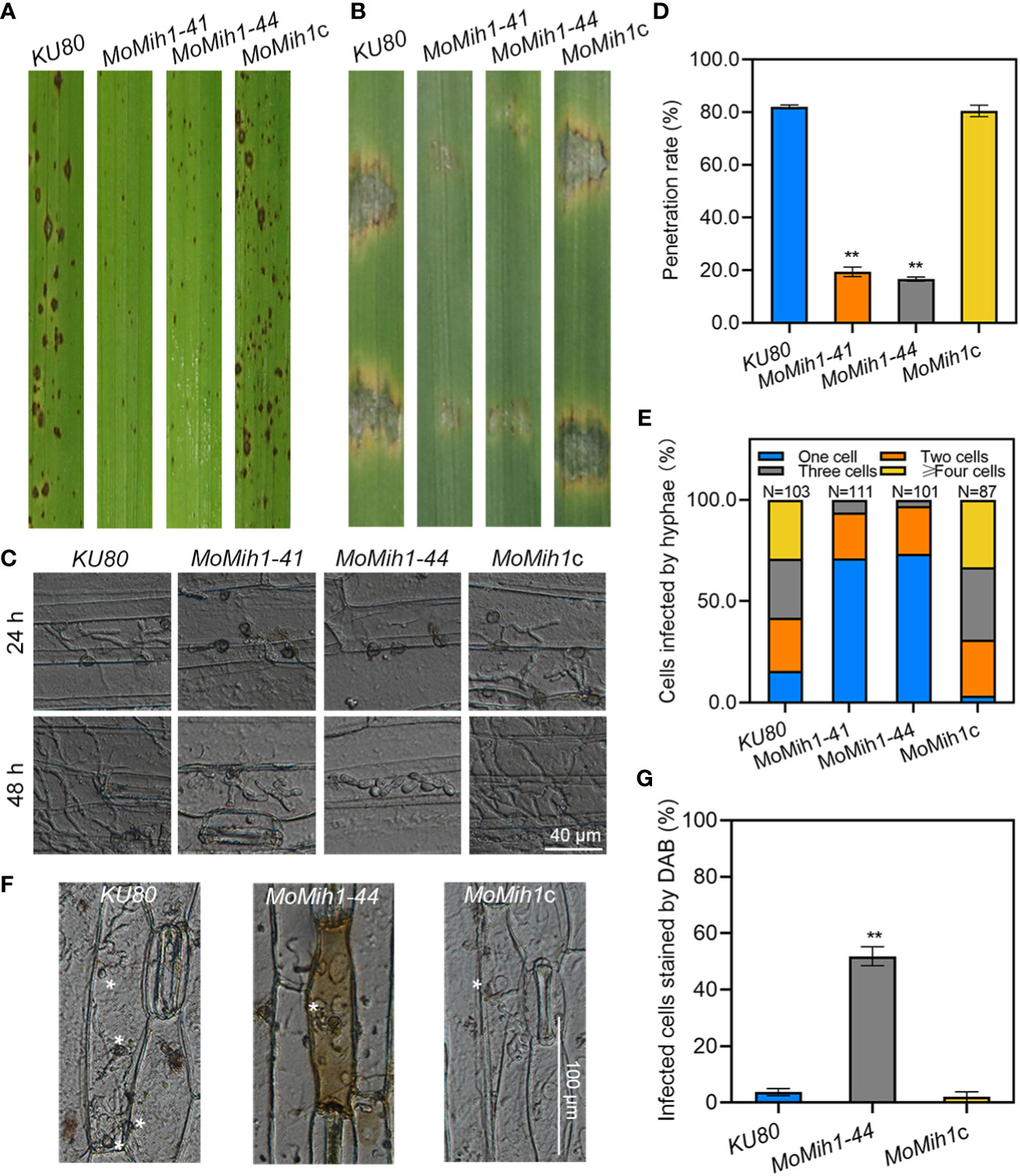
Figure 5 MoMih1 is essential for pathogenicity of M. oryzae. (A) Deletion of MoMih1 affects the virulence of M. oryzae on rice leaves. Conidia suspension of the MoMih1 mutants were sprayed on the rice seedlings, and the diseased leaves were collected at 5 dpi. (B) Pathogenic assays on barley leaves. Conidial suspension of KU80, MoMih1 mutants and MoMih1c was inoculated on barley leaves, and the diseased leaves were collected at 5 dpi. (C) Penetration assay. Conidial suspension of KU80, MoMih1 mutants and MoMih1c was inoculated on the barley leaves, cultured under the condition of 28°C for 24 h or 48 h, and then examined under light microscopy. (D) Statistical analysis of penetration rate. The appressoria mediated penetration of barley epidermal cells were examined, measured at 24 hpi, and then statistically analyzed. “**” represents significant difference between KU80 and MoMih1 mutants (p < 0.01), and error bars represent SD. (E) The MoMih1 deletion affects the invasive hyphae expansion. The percentage of plant cells occupied by the invasive hyphae of each strain was measured and analyzed at 48 hpi. (F) The MoMih1 deletion resulted in the accumulation of ROS at infection sites. Conidial suspensions (1 × 104 spores mL-1) of KU80, MoMih1-44 mutant and MoMih1c were inoculated on barley leaves for 24 h, and then stained with DAB solution. Plant cells infected by the MoMih1 mutant was mostly stained with DAB. Bar = 100 μm. (G) Statistical analysis of the proportion of plant cells stained by DAB. “**” means significant difference between KU80 and MoMih1 mutants (p < 0.01), and error bars represent SD.
Successful removal of host-derived reactive oxygen species (ROS) from plant infection site is an essential prerequisite to ensure the infection of M. oryzae (Guo et al., 2010; Guo et al., 2011; Wang et al., 2017). The sensitivity of the MoMih1 mutants to hydrogen peroxide (H2O2) was firstly verified, with all of them showing incremental sensitivity to H2O2, compared to KU80 and MoMih1c (Figures S5A, B). Since the MoMih1 mutants developed a great amount of tiny necrotic lesions on the rice leaves (Figure 5A), the ROS burst at the infection site was thus monitored among the tested strains. An average 51.80 ± 3.34% of the plant cells infected by MoMih1 mutant could be stained by DAB, compared with 3.71 ± 1.28% and 2.00 ± 1.79% of the KU80 and MoMih1c, respectively (Figures 5F, G), suggesting the critical role of MoMih1 in detoxifying the host-derived ROS during plant infection. The potential reason for decreased ability to scavenge host-derived ROS was thereafter explored, and the extracellular enzyme activity, including peroxidases and laccases, was almost abolished in MoMih1 mutants, compared with KU80 and MoMih1c (Figures S5C, D). The transcriptions of five putative peroxidase and laccase encoding genes were significantly down regulated in the MoMih1 mutants, compared with KU80 (Figure S5E).
Intact cell wall of M. oryzae is a guarantee for the proper infection on rice plants (Feng et al., 2021), the sensitivity of MoMih1 mutants to different cell wall stressors was therefore tested. The MoMih1 mutants are more sensitive to those stressors than that of KU80 and MoMih1c (Figures 6A, B). Besides, the protoplasts released by the MoMih1 mutants were also significantly increased compared with KU80 and MoMih1c at each given time (Figure 6C), implying a severe defect of CWI in MoMih1 mutants.
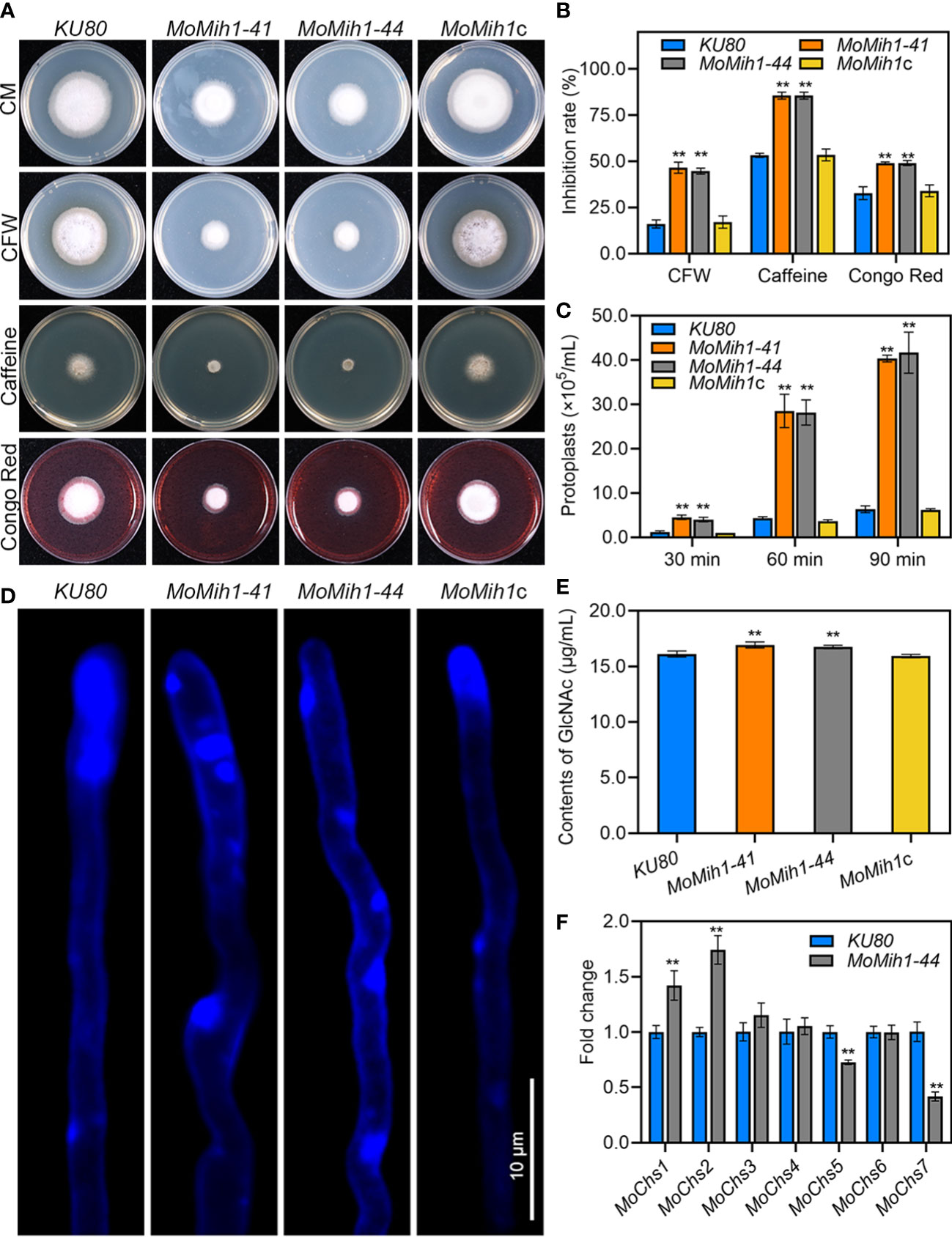
Figure 6 MoMih1 is required for cell wall integrity in M. oryzae (A) Mycelial growth under cell wall stressors. The KU80, MoMih1 mutants and MoMih1c were inoculated on CM with CFW, caffeine, or Congo red, cultured at 28°C for 5 days, and then photographed. (B) Statistical analysis of the inhibition rate under different cell wall stressors. “**” mean significant difference between KU80 and MoMih1 mutants (p < 0.01), and error bars represent SD. (C) Protoplasts released under the treatment of cell wall lysing enzyme. The release of protoplast was quantified at 30 min intervals. “**” means significant difference between KU80 and MoMih1 mutants (p < 0.01), and error bars represent SD. (D) Examination of chitin on the cell wall of MoMih1 mutant. Hypha of KU80, MoMih1 mutants and MoMih1c were stained with CFW for 5 min, and then examined under UV fluorescent microscope. Bar = 10 μm. (E) Measurement of GlcNac. GlcNac was determined in the KU80, MoMih1 mutants and MoMih1c by the fluorimetric Morgan-Elson method. “**” means significant difference between KU80 and MoMih1 mutants (p < 0.01), and error bars represent SD. (F) Expression of chitin synthase encoding genes. Transcription of the chitin synthase encoding genes was compared between KU80 and MoMih1 mutants by qRT-PCR. “**” means significant difference between KU80 and MoMih1 mutants (p < 0.01), and error bars represent SD.
Chitin, one of the main components of fungal cell wall, is essential for the CWI of M. oryzae (Yin et al., 2016; Guo et al., 2017). After staining with CFW, the blue fluorescence was mostly congregated at the hyphal tip of KU80 and MoMih1c, however, patches of bright fluorescence were unevenly distributed on the hyphal wall but not the tips of the MoMih1 mutants (Figure 6D). Quantitative detection of chitin contents showed, interestingly, that chitin contents of the MoMih1 mutant were significantly increased, compared with that of KU80 and MoMih1c (Figure 6E). The expression of seven chitin synthase genes was assayed, with MoChs1 and MoChs2 showing increased expression, but MoChs5 and MoChs7 displaying decreased expression, compared with that of in KU80 (Figure 6F).
The MoMih1 mutants were defective in melanin pigmentation compared with KU80 and MoMih1c (Figure 7A). The melanin biosynthesis related genes, such as MoRSY1, MoALB1, MoBUF1 and MoHNR1, were significantly downregulated in the MoMih1 mutants compared with that in KU80 (Figure 7B). Moreover, the MoMih1 mutants were more sensitive to tricyclazole, a specific inhibitor of melanin biosynthesis (Figures 7C, D), implying that MoMih1 was required for melanin biosynthesis in M. oryzae.
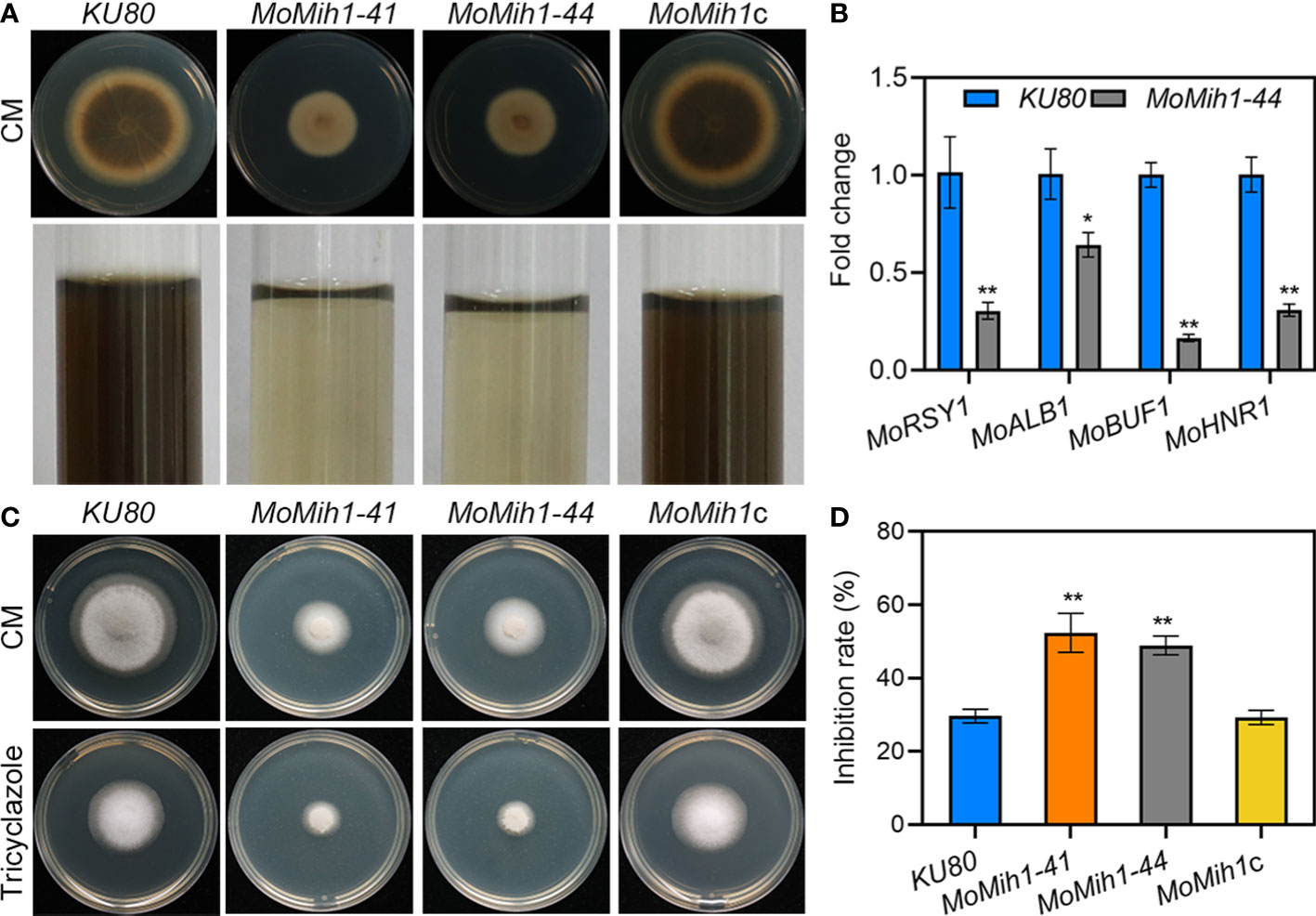
Figure 7 MoMih1 is required for melanin pigmentation of M. oryzae. (A) Colony pigmentation of the MoMih1 mutant. All the strains were inoculated on CM and cultured at 28°C for 5 days. Reduced melanin pigmentation was observed on both CM agar and liquid CM. (B) The transcription of melanin synthesis genes in the hypha of MoMih1 mutant. The expression levels of MoRSY1, MoALB1, MoBUF1 and MoHNR1 was examined by qRT-PCR, with all of them significantly decreased expression in the MoMih1 mutant. “**” mean significant difference in gene expression between two strains (p < 0.01), and error bars represent SD. (C) Sensitivity of the MoMih1 mutant to tricyclazole. The MoMih1 mutant was inoculated on CM amended with tricyclazole, cultured at 28°C for 5 days, and then fungal colony was recorded. (D) Statistical analysis of the inhibition rate under tricyclazole stress. “**” stands for the significant difference between KU80 and MoMih1 mutants (p < 0.01), and error bars represent SD.
The hydrophobicity was determined on the fungal culture of KU80 and MoMih1 mutants, respectively. The water droplets remained on the mycelial culture of KU80 and MoMih1c after 24 hpi, in contrast to the immediate soaking into the mycelial cultures by the MoMih1 mutants (Figure S6A). Due to the wettable phenotype of the mutants, the transcription of hydrophobin-encoding genes including MPG1, MHP1, and two MHP1 homologs (MGG_09134 and MGG_10105) was verified, and all of them showed remarkably down-regulated expression in MoMih1 mutant, compared with that of KU80 (Figure S6B).
In view of the retarded growth of the MoMih1 mutants, the polar growth was thus examined. Approximate 40% mutant mycelia displayed abnormally swollen tips, compared with KU80 (Figures 8A, B). When stained with CFW, the blue fluorescence was mostly converged at the mycelial tips of KU80 and MoMih1c, but not the swollen tips of MoMih1 mutants (Figure 8C), implying an inactive chitin synthesis in the mycelial tips of MoMih1 mutant. Spa2 is a component of the polarisome and functions in regulating hyphal morphology and extension during fungal polar growth (Virag and Harris, 2006), therefore, the MoSpa2-eGFP fusion protein were examined in KU80 and MoMih1-44, respectively. The MoSpa2 can accurately localize to the normal growing hyphal tip of KU80, but not the swollen tips of the MoMih1 mutant (Figure 8D), suggesting MoMih1 is required for polarisome formation during the polarized growth of M. oryzae.

Figure 8 MoMih1 is required for polarized growth of M. oryzae (A) Deletion of MoMih1 resulted in the development of swollen structures at the mycelia tips. All tested strains were inoculated in liquid CM at 28°C for 48 h in darkness, and then swollen structures at the mycelia tips were photographed Bar = 20 μm. (B) Statistical analysis of abnormal structures at the mycelial tips. The rate of abnormal mycelial tips was recorded and statistically analyzed. The error bars represent the SD of three replicates, whereas “**” represents a significant difference (p < 0.01). (C) Chitin distribution on the cell wall of mycelial tips. Hyphal tips of the tested strains were stained by CFW and observed by confocal microscopy. Chitin distribution was abnormal at the swollen tips of the MoMih1 mutant. Bar = 20 μm. (D) Localization of MoSpa2-eGFP in the MoMih1 mutant. Hyphal tips of KU80 and MoMih1-44 that expressed MoSpa2-eGFP were examined by confocal microscopy. Localization of MoSpa2-eGFP to the polarisome was absent in swollen structures of the MoMih1 mutants. Bar = 20 μm.
In S. cerevisiae, Mih1 directly regulates the phosphorylation of Vps26, a subunit of the retromer subcomplex, and thereby modulates the binding affinities of the retromer to specific cargo (Cui et al., 2017), we thus examined the cellular location of MoVps26 in the MoMih1 mutant. A dispersive distribution of the MoVps26-eGFP was observed in the hyphal cytoplasm of MoMih1 mutant, compared with the mostly punctate structures in strain KU80 (Figure 9A). The immunoblotting with anti-GFP showed that the proteolysis of MoVps26-eGFP has increased in the MoMih1 mutant (Figure 9B). The mobility of MoVps26-eGFP depends on microtubules, and could be prevented by the microtubule-destabilizing agent in M. oryzae (Zheng et al., 2015). When treating with benomyl, the MoMih1 mutant showed both obviously increased sensitivity to fungicide and convergent localization of MoVps26-eGFP nearby the hyphal tips, compared with that of KU80 (Figures 9C, D, E). Besides, the ratio of the hyphae containing the swollen tips has remarkably increased in the MoMih1 mutants (Figure 9F), compared with control strain.
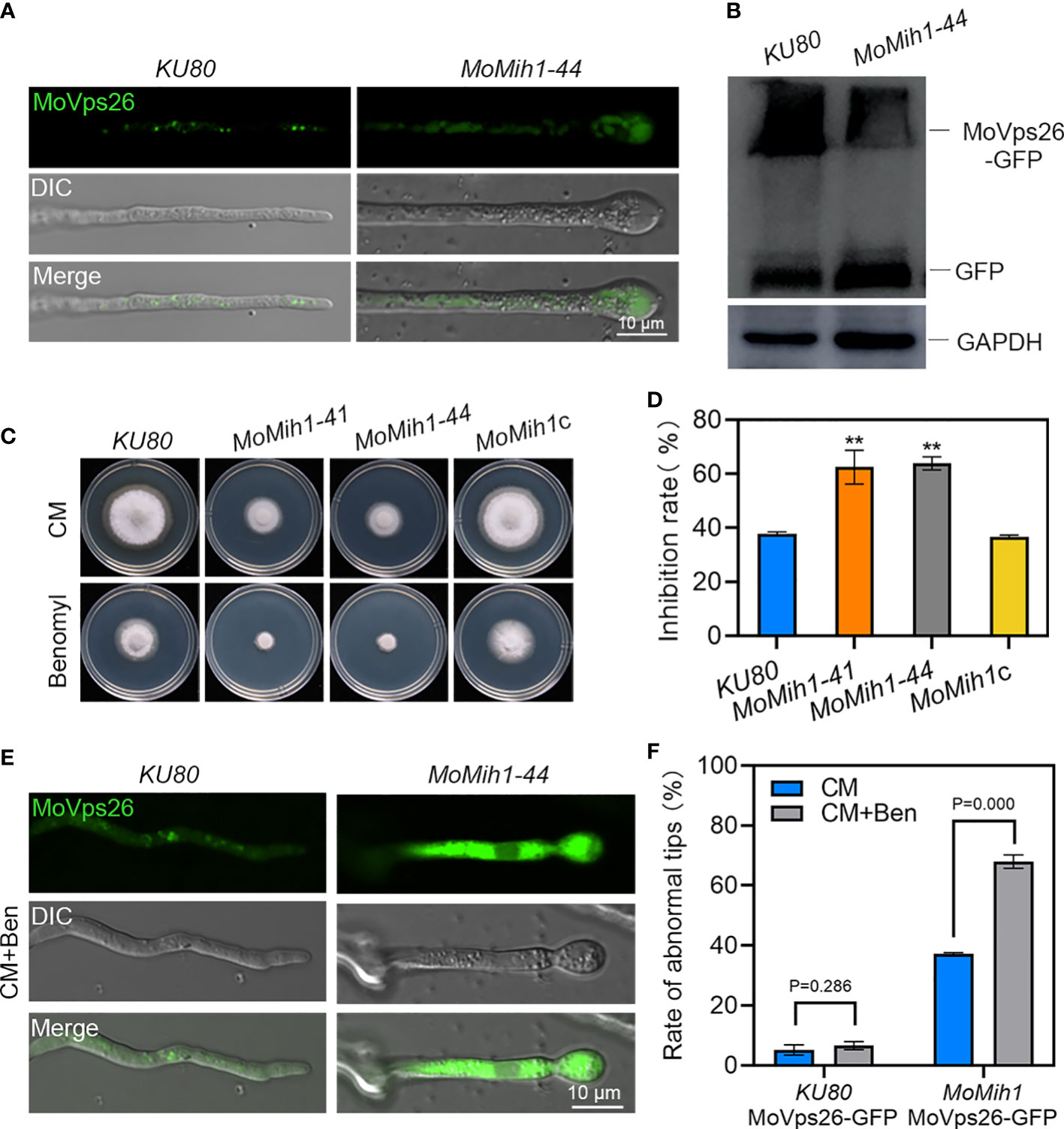
Figure 9 MoMih1 is required for the localization of MoVps26 in M. oryzae. (A) The localization of MoVps26-eGFP in MoMih1 mutants. The strains KU80 and MoMih1-44 were respectively expressed the MoVps26-eGFP fusion protein, and then hyphal tips of resultant transformants were examined under confocal microscopy. The localization of MoVps26-eGFP was abnormal in the swollen tips of the MoMih1 mutants. Bar = 10 μm. (B) The proteolysis of MoVps26-eGFP has increased in the MoMih1 mutant. Mycelia of KU80 and MoMih1-44 expressing MoVps26-eGFP were cultured in liquid CM at 28°C for 36 h, and the MoVps26-eGFP fusion protein was detected by immunoblotting with anti-GFP. The blotting of GAPDH was used as a control. (C) Mycelia growth under benomyl treatment. All tested strains were inoculated on CM supplemented with 0.4 μg/mL benomyl and cultured at 28 °C for 5 days. (D) Statistical analysis of the inhibition rate under benomyl treatment. “**” represents the significant difference between KU80 and MoMih1 mutants (p < 0.01), and Error bars represent SD. (E) Localization of MoVps26-eGFP under the treatment with 0.4 μg/mL benomyl. The strain MoMih1/MoVps26-eGFP was inoculated in liquid CM at 28°C for 24 h in darkness, then added with benomyl and grew at 28°C for 16 h in darkness. Bar = 10 μm. (F) Treatment of benomyl stimulated the formation of swollen structures in the MoMih1 mutants. The swollen structures of the KU80/MoVps26-eGFP and MoMih1/MoVps26-eGFP was recorded by amending with or without benomyl, and the rate of abnormal mycelial tips was statistically analyzed. The error bars represent the SD of three replicates, whereas “**” represents a significant difference (p < 0.01).
In S. cerevisiae, protein tyrosine phosphatase Mih1 control the cell cycle progress through affecting the phosphorylation levels of Cdc28. In M. oryzae, a putative catalytic subunit of Cdk1 protein named as MoCdc28 (MGG_01362) was identified to directly interact with MoMih1 based on yeast two-hybrid (Y2H) and BiFC assay (Figures 10A, B). Moreover, western blot analysis with anti-phospho-Cdk1 antibody also revealed that the Cdk1 phosphorylation signal in the MoMih1 mutant was much stronger than that in the KU80 (Figure 10C), suggesting an essential role of MoMih1 in removing the phosphate group on MoCdc28 during fungal development in M. oryzae.
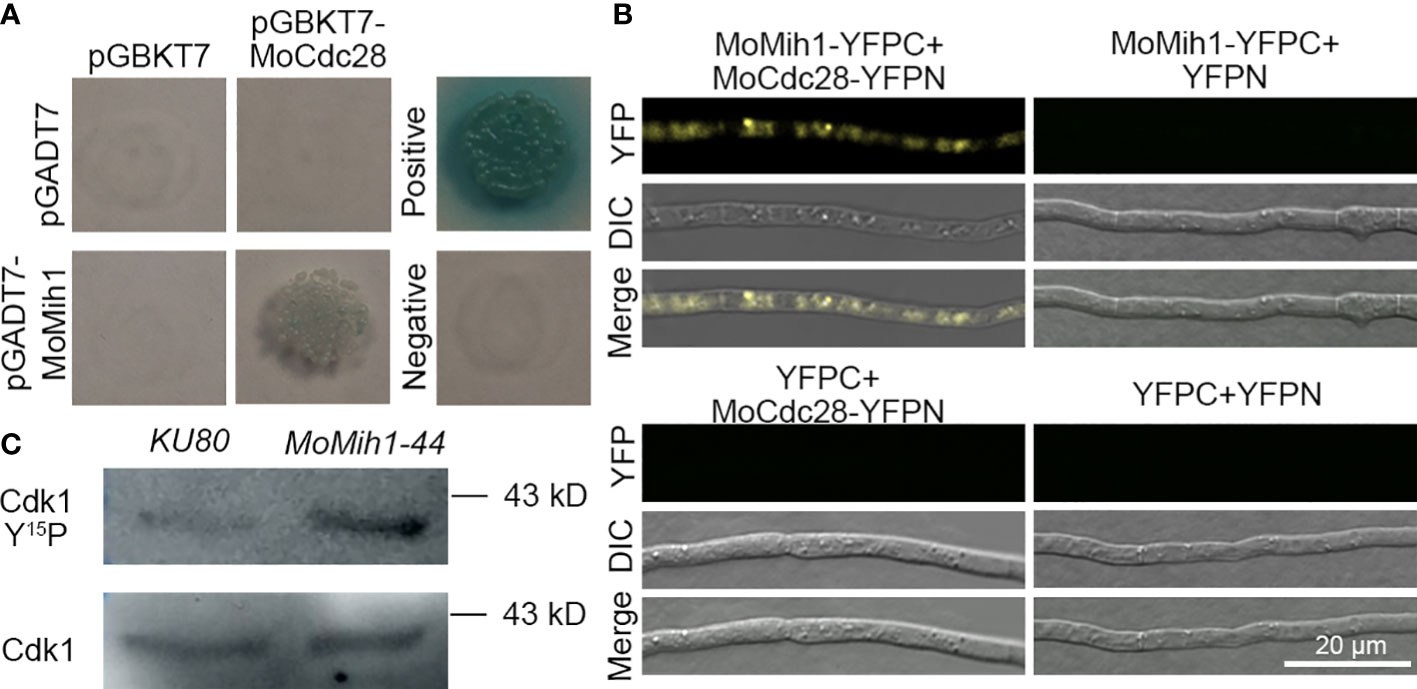
Figure 10 MoMih1 physically interacts with MoCdc28 and regulates the Cdk1 phosphorylation in M. oryzae (A) Yeast two-hybrid assay. The coding sequences of MoMih1 and MoCdc28 were inserted into the vector pGADT7 and pGBKT7, respectively, and then co-transformed into yeast strain AH109. Yeast cells co-transformed with pGADT7/pGBKT7-MoCdc28, pGADT7-MoMih1/pGBKT7, pGADT7-T/pGBKT7-53, and pGADT7-T/pGBKT7-Lam were used as control, respectively. (B) Detection of interaction between MoMih1 and MoCdc28 by bimolecular fluorescence complementation (BiFC) assay. A pair of MoCdc28-NYFP and CYFP-MoMih1 constructs were co-transformed into wild type strain Guy11. Construct pairs of MoCdc28-NYFP and CYFP, CYFP-MoMih1 and NYFP, and NYFP and CYFP were co-transformed into Guy11 to serve as negative controls. YFP signals were detected under the CLSM. (C) Detection of the Cdk1 phosphorylation in the MoMih1 mutant. Western blot was carried out to detect Cdk1 Y15P (for phosphorylation signal) and Cdk1 in protein extracts using the anti-phospho-Cdc2 (Tyr15) and anti-Cdc2 antibodies, respectively. Note that the phosphorylation signal was intensified in MoMih1 deletion mutant.
To determine whether MoMih1 affects the cell cycle of M. oryzae, nuclear segregation was examined in the strains KU80-H3-mRFP and MoMih1-H3-mRFP. Most of the nuclei showed delayed separation in the mycelium of MoMih1 mutant, compared with KU80 (Figure 11A). However, nuclear segregation seems normally proceed in the MoMih1 mutant during the appressorium formation, which is similar with the wild type strain KU80-H3-mRFP (Figure 11B).
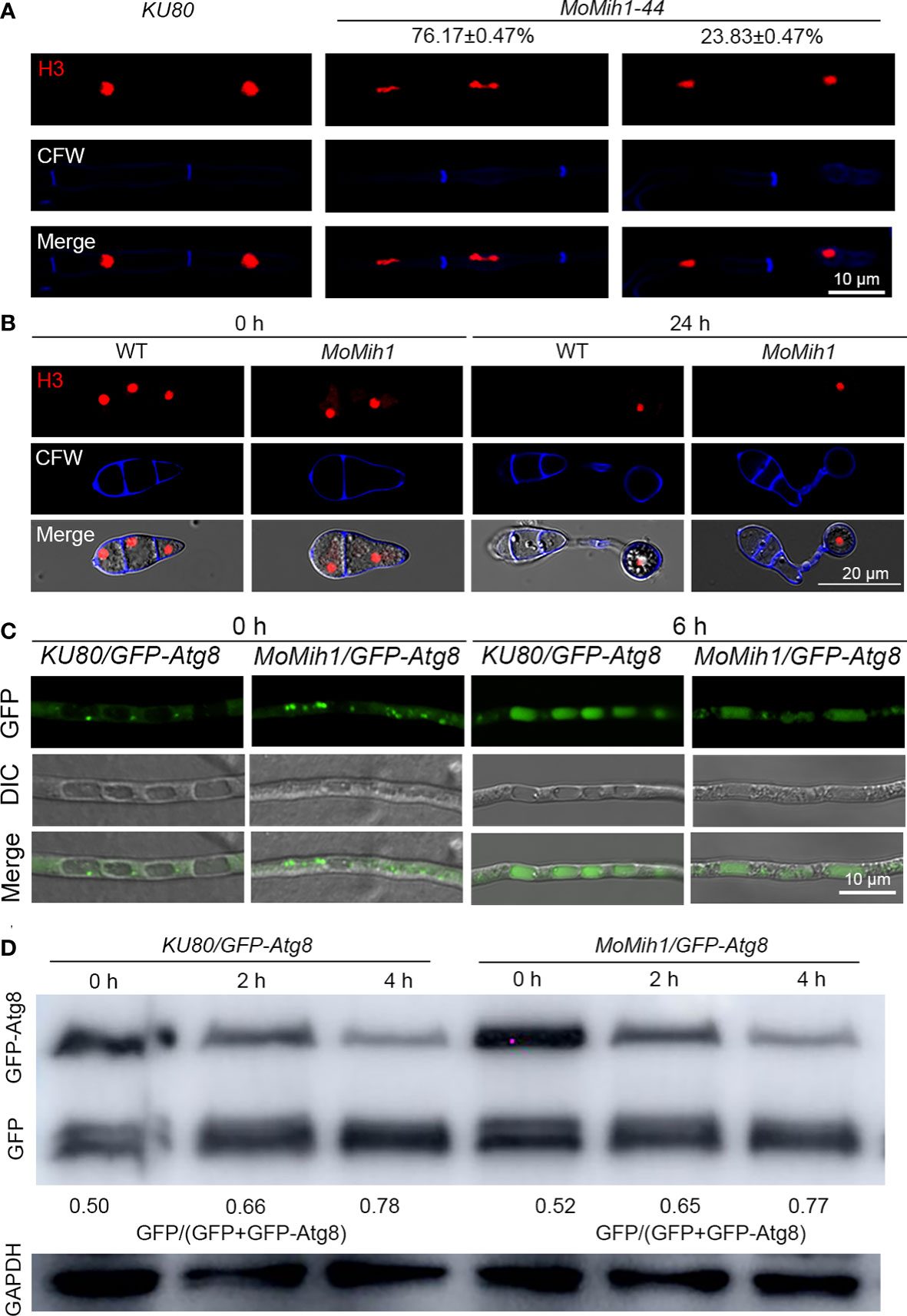
Figure 11 MoMih1 is required for nuclear division but not non-selective autophagy in M. oryzae. (A) Nuclear division in the hyphae of MoMih1 mutant. Hyphae of the KU80 and MoMih1 mutant that respectively expressed H3-RFP were stained with CFW and examined by CLSM. Bar = 10 μm. The proportion of each type of fungal cells was indicated in the figure. (B) Nuclear distribution during the appressorium development in MoMih1 mutant. Conidia of KU80 and MoMih1 mutant that respectively expressed H3-RFP were inoculated on hydrophobic coverslip for 0 or 24 h, stained with CFW, and examined by CLSM. Bar = 20 μm. (C) Localization of eGFP-MoAtg8 in the MoMih1-44 mutant. Both the parental strain KU80 and MoMih1-44 mutant were grown in liquid CM for 36 h, transferred to SD-N medium for 4 h and then examined by CLSM. Bar = 10 μm. (D) Immunoblot to detect the degradation of eGFP-MoAtg8. All tested strain were under the treatment of nitrogen starvation for 2 and 4 h, and then immunoblotted with anti-GFP antibody to analyze the breakdown of eGFP-MoAtg8 in the KU80 and MoMih1 mutant. The ratio of eGFP with eGFP+eGFP-MoAtg8 was calculated.
Autophagy usually synchronized with the cell cycle in M. oryzae, to test whether MoMih1 regulates the cell cycle via affecting the fungal autophagy, the autophagy was compared between the MoMih1 mutant and KU80. When grown in CM, both the KU80-eGFP-Atg8 and MoMih1-eGFP-Atg8 strains, which expressed eGFP-MoAtg8 fusion protein, showed similar amount of autophagosomes within hyphae (Figure 11C). After transferring into minimum medium minus nitrogen (MM-N), interestingly, the MoMih1 mutant accumulated similar autophagosomes to that of in wild-type KU80-eGFP-Atg8 (Figure 11C). Western blot showed that the non-selective autophagy in KU80-eGFP-Atg8 strain was comparable with MoMih1-eGFP-Atg8 strain (Figure 11D), suggesting that MoMih1 is dispensable for non-selective autophagy in M. oryzae.
Successful proceeding of mitosis is necessary for the growth and development of eukaryotes (Hindley et al., 1987). In M. oryzae, cell cycle regulates the appressorium morphology and plant infection, and the completion of mitosis is critical for the full virulence of this fungus (Oses-Ruiz et al., 2017). In M. oryzae genome, a series of cell cycle regulators that is homologous to proteins involved in cell cycle regulation in S. cerevisiae, were identified (Oses-Ruiz and Talbot, 2017), and some of them have been verified to participate in the growth, development, sporulation and pathogenicity of this fungus (Saunders et al., 2010; Yue et al., 2017; Li et al., 2018), suggesting the essential roles of the cell cycle regulation during fungal development and plant infection of M. oryzae. In S. pombe, the dual-specificity phosphatase Cdc25 work together with Wee1 kinase to regulate the phosphorylation of Cdk1 to control the proceeding of mitosis, and the dephosphorylation of Cdk1 by Cdc25 promotes the cells to enter mitosis (Dunphy, 1994). In S. cerevisiae, deletion of Mih1, a homolog of Cdc25 in fission yeast, results in abnormally enlarging cells and moderately delayed mitotic process (Pal et al., 2008). Besides, Cdc25 and its homologs play also important roles in fungal growth and pathogenicity in many other filamentous fungi, and for example, the disruption of Cdc25 could incur retarded growth, diminished sporulation, and attenuated ability to infect the hosts by Beauveria bassiana and U. maydis (Sgarlata and Perez-Martin, 2005; Qiu et al., 2015), implying the critical conserved role of this Cdc25 protein during the growth and pathogenesis of those fungi. In M. oryzae, a dual-specificity phosphatase name as MoMih1 has been identified in the fungal genome, however, the underlying mechanism regulated by this cell cycle protein has yet to be understood till now. In this study, we found that MoMih1 is conserved in structures that are unique to Cdc25 and Mih1 phosphatases in S. pombe and S. cerevisiae, respectively. MoMih1 is required for normal mitosis and fungal development in M. oryzae, and deletion of MoMih1 lead to decreased activity of extracellular enzyme and ability to scavenge active oxygen of M. oryzae. These results indicate that MoMih1 plays an important role in determining the fungal morphogenesis and virulence of M. oryzae.
The hyphae of the filamentous fungal pathogens are highly polarized cells, and their polar growth at the hyphal tip is a prerequisite for vegetative growth, development and plant infection of those pathogens (Steinberg, 2007; Guo et al., 2015). In fungal pathogens, polarized growth depends on a specialized protein subcomplex called polarisome (Harris et al., 2005). The accurate localization of Spa2, a central component of polarisome, is required for establishment of proper hyphae polarized growth in filamentous fungal pathogens (Meyer et al., 2008; Li et al., 2014; Cao et al., 2021; Zheng et al., 2021b; Zhu et al., 2022). In this study, approximate 40% of the vegetative hyphae of the MoMih1 showed abnormally swollen tips, and the central localization of MoSpa2-eGFP observed in the polarisome of KU80 was completely abolished in the swollen hyphae tips or asymmetrically localized in the twisted hyphae tips of MoMih1 mutant. In view of the critical role of MoMih1 in protein dephosphorylation, and improper localization of Spa2 homologs to polarisome resulted in defective hyphal tip growth in M. oryzae and other phytopathogens (Li et al., 2014; Cao et al., 2021; Zhu et al., 2022), we speculate that MoMih1 might regulate polarized growth by dephosphorylation of certain components of the polarisome, such as MoSpa2, during fungal development in M. oryzae. Besides, continuous polarized tip growth also requires the constant delivery of cargo to the cell apex (Pantazopoulou et al., 2014). MoVps26, a subunit of retromer subcomplex that participate in intracellular transport of secretory vesicles cargo (Zheng et al., 2015), showed punctate structures at or near the vacuolar of KU80, but dispersed, cytoplasmic signals in the MoMih1 mutant, implying an essential role of MoMih1 in establishment of cargo selection complex of the retromer in M. oryzae. The treatment of benomyl resulted in remarkably convergent localization of MoVps26-eGFP nearby the hyphal tips and increased ratio of swollen hyphal tips in MoMih1 mutants, further suggesting that MoMih1 might affect the polarized tip growth by destabilizing the microtubule cytoskeleton in the hyphae of M. oryzae. Based above, the abolishment of MoSpa2 and disordered localization of MoVps26 that affected by MoMih1 might be the reason for the abnormal polarized tip growth in M. oryzae.
The massive formation and diffusion of asexual spore directly determines the epidemic degree of the rice blast in the disease cycle (Zhang et al., 2016). Young conidia are produced on the tip of conidiophores differentiated from the fluffy arial hyphae in an axial manner (Liu et al., 2010). The fluffy growth of arial hyphae is proportional to the development of conidiophores, and essential for asexual sporulation of M. oryzae (Jeon et al., 2008; Guo et al., 2017; Matheis et al., 2017). In M. oryzae, deletion of the MoMih1 produced less arial hyphae, and significantly reduced conidiophores and conidia, suggesting a critical role of MoMih1 in determining the asexual development in the blast fungus. The significant downregulation of two transcription factors MoFLBC and MoFLBD, which are essential for aerial hyphae development and spore formation (Matheis et al., 2017), respectively, might be partially responsible for defects of conidiophore development and conidial formation in MoMih1 mutant. The MoACR1, a stage-specific negative regulator for conidiation, is essential for the establishment of sympodial pattern of spore formation, and a mutation of this protein lead to produce head-to-tail conidia in M. oryzae (Lau and Hamer, 1998). Besides, blocking of MoSsadh also completely abolished the conidiation due to the loss of conidiophore development in M. oryzae (Guo et al., 2011). We here found that the transcription level of those two genes significantly decreased in the MoMih1 mutant, further suggesting an important role of MoMih1 in conidiophore development. Taken together, our results implied that MoMih1 might function as a key upstream regulator to stimulate the transcription of conidiation related genes during asexual development of M. oryzae.
The scavenging of ROS at the infection site of the plants is a prerequisite for the successful fungal penetration during plant-microbe interaction (Molina and Kahmann, 2007; Chi et al., 2009; Guo et al., 2010; Guo et al., 2011; Zhang et al., 2021). In M. oryzae, the basic leucine zipper (bZIP) family transcriptional factors MoAP1 and Moatf1 are two important regulators in response to oxidative stressors, and deletion either of them resulted in attenuation of degrading the host-derived ROS, and thus reduced the pathogenicity of the mutants (Guo et al., 2010; Guo et al., 2011). In this study, deletion of MoMih1 attenuated the fungal virulence, with both decreased penetration and retarded invasive hyphae extension during plant infection. The MoMih1 mutant was more sensitive to hydrogen peroxide, and almost lost the secretion of extracellular laccases and peroxidases that help to scavenge the host-derived ROS. Since ROS generally acts as the second messenger that activate the defense pathway during plant microbe interactions (Molina and Kahmann, 2007; Guo et al., 2010; Guo et al., 2011), we speculated that MoMih1 may participate in fungal pathogenicity by regulating the extracellular enzyme activity to detoxify host-derived ROS and then facilitated the fungal penetration and invasive hyphae extension in M. oryzae.
The plant infection of M. oryzae is mediated by development of a functional appressorium (Wilson and Talbot, 2009; Zhang et al., 2011; Ryder et al., 2019). The proceeding of cell cycle is critical for the formation of appressorium and secondary infection hyphae (Saunders et al., 2010; Oses-Ruiz et al., 2017). In S. cerevisiae, the checkpoint that inhibits anaphase onset mainly depends on the activity of Cdk1, and the Mih1 has been proved to remove the phosphate group of Cdk1Y19, which as a result activated the Cdk1 activity, and promote the transition of mitosis in S. cerevisiae (Pal et al., 2008). In this study, MoMih1 directly interacted with the Cdk1 homologue MoCdc28 in vivo, and deletion of MoMih1 resulted in incremental numbers of nucleus in many hyphal cell compartments of the fungus and significantly increased phosphorylation level of MoCdc28Y15 in MoMih1 mutant, confirmed that MoMih1 functioned as important cell cycle regulator to proceed the mitosis during development of M. oryzae. This finding agrees with the role of Cdc25 in S. pombe and U. maydis, in which the deletion of Cdc25 strongly blocked the initiation of M phase, further suggesting a conserved role of Cdc25 tyrosine phosphatases in controlling the cell division in diverse fungi (Sgarlata and Perez-Martin, 2005). Interestingly, nuclear division arrest was not identified during appressorium development, we attributed this to the rapid mitosis whose duration time is too short to be captured during conidia germination in MoMih1 mutant. In S. cerevisiae, Mih1 localized to the nucleus and cytoplasm through the mechanism of nucleocytoplasmic shuttle, and in most situation functioned in the nucleus (Yano et al., 2013). In this study, MoMih1 was localized to both the nucleus and cytoplasm at mycelial stage, whereas only located in the cytoplasm at conidial stage, indicating the conserved role of MoMih1 during fungal development, and this spatial expression difference at diverse stages might reflect a critical role of MoMih1 in determining polar growth, cargo transport and full virulence of M. oryzae.
In conclusion, this study revealed the role of MoMih1 in the development of M. oryzae, which laid a foundation for further study on the role of MoMih1 in the cell cycle of M. oryzae.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
MG conceived the experiments. SL and XG performed the experiments. MG and SL wrote the manuscript. JM and SW helped to analyze the data and revise the manuscript. All authors contributed to the article and approved the submitted version.
This study was granted by the Foundation for the Excellent Talents of Anhui Agricultural University (Grant No: RC2015002 to MG), and the National Natural Science Foundations of China (Grant No: 31671976 to MG).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1146915/full#supplementary-material
Supplementary Figure 1 | Validation of MoMih1 deletion in M. oryzae. (A). The gene deletion strategy for generating the MoMih1 mutants. (B). The M. oryzae MoMih1 mutants were validated by PCR. The genomic DNA was extracted from MoMih1-41 and MoMih1-44, and verfied using the primers of P1/P2 or P3/P4 by PCR. The MoMih1 deletion mutants can amplify 1.485 and 1.486 kb fragments using the primers of P1/P2 or P3/P4. (C). The M. oryzae MoMih1 mutants were validated by RT-PCR. The total RNA was extracted from KU80, MoMih1-41, MoMih1-44, and MoMih1c, reversely transcribed, and verfied using the primer pairs to detect MoMih1 (MGG_07734) and Actin (MGG_03892). (D, E). Southern blot to validate the deletion of MoMih1 mutants (MoMih1-41, MoMih1-44). To validate the deletion of MoMih1 in M. oryzae, genomic DNA of KU80 and MoMih1 deletion mutants were digested with SphI and hybridized with MoMih1-LF probe; To validate the copy number of HPH in MoMih1 deletion mutants, genomic DNA of KU80 and MoMih1 deletion mutants were digested with HindIII, and hybridized with the HPH probe.
Supplementary Figure 2 | The Validation of MoGin4 and MoCyc2 deletion in M. oryzae. (A). The M. oryzae MoGin4 mutants were validated by PCR. The genomic DNA was extracted from MoGin4-2 and MoGin4-5 mutants, and detected using the primers GP1/P2 and P3/GP4 by PCR. The MoGin4 deletion mutants can amplify 1.601and 1.504 kb fragments by the primers GP1/P2 and P3/GP4. (B). The M. oryzae MoGin4 mutants were validated by RT-PCR. The total RNA was extracted from Guy11, MoGin4-2, and MoGin4-5, reversely transcribed, and verfied by the primers MoGin4-RT-1 and MoGin4-RT-2 to detect the deltion of MoGin4 (MGG_02810). (C). The M. oryzae MoCyc2 mutants were validated by PCR. The genomic DNA was extracted from MoCyc2-4 and MoCyc2-14, and validated using the primers CP1/P2 and P3/CP4 by PCR. The MoCyc2 deletion mutants can amplify 1.586 and 1.604 kb fragments, respetively. (D). The MoCyc2 mutants were validated by RT-PCR. The total RNA of Guy11, MoCyc2-4, and MoCyc2-14 was respetively extracted, reversely transcribed, and verfied by using the primer pairs MoCyc2-RT-1 and MoCyc2-RT-2 to detect the deletion of MoCyc2 (MGG_07065).
Supplementary Figure 3 | Bioinformatic analysis of MoMih1 and Cdc25 homologues. Bioinformatic analysis revealed that MoMih1 contains a conserved HCXXXXXR sequence and DCR motif unique to Cdc25 and Mih1 phosphatases in S. pombe and S. cerevisiae, respectively. The conserved DCR motif (aa 392-394) and the HCXXXXXR consensus sequence (aa 434-441) are marked with the black frame.
Supplementary Figure 4 | The subcellular localization of MoMih1 in M. oryzae. The conidia (A), mycelia (B), and invasive hyphae (C) of the transformants expressing the MoMih1-eGFP fusion protein were examined under a CLSM.
Supplementary Figure 5 | The MoMih1 mutants are hypersensitive to hydrogen peroxide in M. oryzae. (A). Mycelia growth under hydrogen peroxide. The KU80, MoMih1 mutants and MoMih1c were inoculated on CM with or without 5 mM H2O2 and cultured at 28 °C for 5 days. (B). Statistical analysis of the inhibition rate under hydrogen peroxide. “**” stands for the significant difference among the tested strains (p < 0.01), and error bars represent SD. (C). Laccase activity test. The laccase activity was monitored in CM supplemented with 0.2 mM 2, 2’-azino-di-3-ethylbenzathiazoline-6-sulfonate (ABTS). (D). Measurement of the laccase and peroxidase activity. (E). Transcription of peroxidases and laccases encoding genes. The expression of putative peroxidases and laccases encoding genes was examined by qRT-PCR in the KU80 and MoMih1 mutants.
Supplementary Figure 6 | MoMih1 is required for the hydrophobicity of M. oryzae. (A). Hydrophobicity assay for the MoMih1 mutants. Drops of water were inoculated on the colony surfaces of the tested strains. An easily-wettable phenotype of MoMih1 mutants was observed and photographed at 24 hpi. (B). The transcriptional levels of hydrophobin genes. The expression of four hydrophobin genes, such as MGG_09134, MGG_10105, MoMPG1, and MoMHP1, was significantly reduced in the MoMih1 mutant. “**” mean significant difference in gene expression (p< 0.01), and error bars represent SD.
Camps, M., Nichols, A., Arkinstall, S. (2000). Dual specificity phosphatases: a gene family for control of MAP kinase function. FASEB J. 14, 6–16. doi: 10.1096/fasebj.14.1.6
Cao, S. L., Li, W., Li, C. H., Wang, G. H., Jiang, W. Q., Sun, H. Y., et al. (2021). The CHY-type zinc finger protein FgChy1 regulates polarized growth, pathogenicity, and microtubule assembly in fusarium graminearum. Mol. Plant-Microbe Interact. 34, 362–375. doi: 10.1094/MPMI-07-20-0206-R
Chi, M.H., Park, S.Y., Kim, S., Lee, Y.H. (2009). A novel pathogenicity gene isrequired in the rice blast fungus to suppress the basal defenses of the host. PloS Pathog. 5 (4), e1000401. doi: 10.1371/journal.ppat.1000401
Cui, T. Z., Peterson, T. A., Burd, C. G. (2017). CDC25 family protein phosphatase gates cargo recognition by the Vps26 retromer subunit. Elife 6, e24126. doi: 10.7554/eLife.24126
Dunphy, W. G. (1994). The decision to enter mitosis. Trends Cell Biol. 4, 202–207. doi: 10.1016/0962-8924(94)90142-2
Ebbole, D. J. (2007). Magnaporthe as a model for understanding host-pathogen interactions. Annu. Rev. Phytopathol. 45, 437–456. doi: 10.1146/annurev.phyto.45.062806.094346
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Feng, W., Yin, Z., Wu, H., Liu, P., Liu, X., Liu, M., et al. (2021). Balancing of the mitotic exit network and cell wall integrity signaling governs the development and pathogenicity in Magnaporthe oryzae. PloS Pathog. 17, e1009080. doi: 10.1371/journal.ppat.1009080
Fernandez, J., Orth, K. (2018). Rise of a cereal killer: The biology of magnaporthe oryzae biotrophic growth. Trends Microbiol. 26, 582–597. doi: 10.1016/j.tim.2017.12.007
Goh, J., Kim, K. S., Park, J., Jeon, J., Park, S. Y., Lee, Y. H. (2011). The cell cycle gene MoCDC15 regulates hyphal growth, asexual development and plant infection in the rice blast pathogen Magnaporthe oryzae. Fungal Genet. Biol. 48, 784–792. doi: 10.1016/j.fgb.2011.05.001
Guo, M., Chen, Y., Du, Y., Dong, Y. H., Guo, W., Zhai, S., et al. (2011). The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PloS Pathog. 7 (2), e24126. doi: 10.1371/journal.ppat.1001302
Guo, M., Guo, W., Chen, Y., Dong, S. M., Zhang, X., Zhang, H. F., et al. (2010). The basic leucine zipper transcription factor Moatf1 mediates oxidative stress responses and is necessary for full virulence of the rice blast fungus Magnaporthe oryzae. Mol. Plant-Microbe Interact. 23, 1053–1068. doi: 10.1094/MPMI-23-8-1053
Guo, M., Kilaru, S., Schuster, M., Latz, M., Steinberg, G. (2015). Fluorescent markers for the spitzenkörper and exocytosis in Zymoseptoria tritici. Fungal Genet. Biol. 79, 158–165. doi: 10.1016/j.fgb.2015.04.014
Guo, M., Tan, L. Y., Nie, X., Zhang, Z. G. (2017). A class-II myosin is required for growth, conidiation, cell wall integrity and pathogenicity of Magnaporthe oryzae. Virulence 8, 1335–1354. doi: 10.1080/21505594.2017.1323156
Guo, M., Tan, L. Y., Nie, X., Zhu, X. L., Pan, Y. M., Gao, Z. M. (2016). The Pmt2p-mediated protein O-mannosylation is required for morphogenesis, adhesive properties, cell wall integrity and full virulence of Magnaporthe oryzae. Front. In Microbiol. 7, 630. doi: 10.3389/fmicb.2016.00630
Harris, S. D., Read, N. D., Roberson, R. W., Shaw, B., Seiler, S., Plamann, M., et al. Polarisome meets spitzenkörper: Microscopy, genetics, and genomicsconverge (2005). Eukaryot Cell 4, 225–229. doi: 10.1128/EC.4.2.225-229.2005
Hindley, J., Phear, G., Stein, M., Beach, D. (1987). Sucl+ encodes a predicted 13-kilodalton protein that is essential for cell viability and is directly involved in the division cycle of Schizosaccharomyces pombe. Mol. Cell. Biol. 7, 504–511. doi: 10.1128/mcb.7.1.504-511.1987
Jeon, J., Goh, J., Yoo, S., Chi, M. H., Choi, J., Rho, H. S., et al. (2008). A putative MAP kinase kinase kinase, MCK1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Mol. Plant-Microbe Interact. MPMI 21, 525. doi: 10.1094/MPMI-21-5-0525
Lau, G. W., Hamer, J. E. (1998). Acropetal: a genetic locus required for conidiophore architecture and pathogenicity in the rice blast fungus. Fungal Genet. Biol. 24, 228–239. doi: 10.1006/fgbi.1998.1053
Lew, D. J., Kornbluth, S. (1996). Regulatory roles of cyclin dependent kinase phosphorylation in cell cycle control. Curr. Opin. Cell Biol. 8, 795–804. doi: 10.1016/S0955-0674(96)80080-9
Li, C. H., Cao, S. L., Zhang, C. K., Zhang, Y. H., Zhang, Q., Xu, J. R., et al. (2018). MoCDC14 is important for septation during conidiation and appressorium formation in Magnaporthe oryzae. Mol. Plant Pathol. 19, 328–340. doi: 10.1111/mpp.12523
Li, L., Xue, C., Bruno, K., Nishimura, M., Xu, J. R. (2004). Two PAK kinase genes, CHM1 and MST20, have distinct functions in Magnaporthe grisea. Mol. Plant Microbe Interact. 17, 547–556. doi: 10.1094/MPMI.2004.17.5.547
Li, C., Yang, J., Zhou, W., Chen, X. L., Huang, J. G., Cheng, Z. H., et al. (2014). A spindle pole antigen gene MoSPA2 is important for polar cell growth of vegetative hyphae and conidia, but is dispensable for pathogenicity in Magnaporthe oryzae. Curr. Genet. 60, 255–263. doi: 10.1007/s00294-014-0431-4
Li, G., Zhou, X., Xu, J. R. (2012). Genetic control of infection-related development in magnaporthe oryzae. Curr. Opin. Microbiol. 15, 678–684. doi: 10.1016/j.mib.2012.09.004
Liu, W. D., Xie, S. Y., Zhao, X. H., Chen, X., Zheng, W. H., Lu, G. D., et al. (2010). A homeobox gene is essential for conidiogenesis of the rice blast fungus Magnaporthe oryzae. Mol. Plant-Microbe Interact. 23, 366–375. doi: 10.1094/MPMI-23-4-0366
Lu, J., Cao, H., Zhang, L., Huang, P., Lin, F. (2014). Systematic analysis of Zn2Cys6 transcription factors required for development and pathogenicity by high-throughput gene knockout in the rice blast fungus. PloS Pathog. 10, e1004432. doi: 10.1371/journal.ppat.1004432
Matheis, S., Yemelin, A., Scheps, D., Andresen, K., Jacob, S., Thines, E., et al. (2017). Functions of the Magnaporthe oryzae Flb3p and Flb4p transcription factors in the regulation of conidiation. Microbiol. Res. 196, 106–117. doi: 10.1016/j.micres.2016.12.010
Meitinger, F., Palani, S., Pereira, G. (2012). The power of MEN in cytokinesis. Cell Cycle 11, 219–228. doi: 10.4161/cc.11.2.18857
Meyer, V., Arentshorst, M., van den Hondel, C. A., Ram, A. F. (2008). The polarisome component SpaA localises to hyphal tips of aspergillus niger and is important for polar growth. Fungal Genet. Biol. 45, 152–164. doi: 10.1016/j.fgb.2007.07.006
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era (vol 37, pg 1530, 2020). Mol. Biol. Evol. 37, 2461–2461.
Molina, L., Kahmann, R. (2007). An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell 19, 2293–2309. doi: 10.1105/tpc.107.052332
Oses-Ruiz, M., Cruz-Mireles, N., Martin-Urdiroz, M., Soanes, D. M., Eseola, A. B., Tang, B. Z., et al. (2021). Appressorium-mediated plant infection by Magnaporthe oryzae is regulated by a Pmk1-dependent hierarchical transcriptional network. Nat. Microbiol. 6, 1383–U1106. doi: 10.1038/s41564-021-00978-w
Oses-Ruiz, M., Sakulkoo, W., Littlejohn, G. R., Martin-Urdiroz, M., Talbot, N. J. (2017). Two independent s-phase checkpoints regulate appressorium-mediated plant infection by the rice blast fungus magnaporthe oryzae. Proc. Natl. Acad. Sci. U.S.A. 114, E237–E244. doi: 10.1073/pnas.1611307114
Oses-Ruiz, M., Talbot, N. J. (2017). Cell cycle-dependent regulation of plant infection by the rice blast fungus Magnaporthe oryzae. Commun. Integr. Biol. 10, e1372067. doi: 10.1080/19420889.2017.1372067
Pal, G., Paraz, M. T., Kellogg, D. R. (2008). Regulation of Mih1/Cdc25 by protein phosphatase 2A and casein kinase 1. J. Cell Biol. 180, 931–945. doi: 10.1083/jcb.200711014
Pantazopoulou, A., Pinar, M., Xiang, X., Penalva, M. A. (2014). Maturation of late golgi cisternae into RabE(RAB11) exocytic post-golgi carriers visualized in vivo. Mol. Biol. Cell 25, 2428–2443. doi: 10.1091/mbc.e14-02-0710
Qi, Z. Q., Wang, Q., Dou, X. Y., Wang, W., Zhao, Q., Lv, R. L., et al. (2012). MoSwi6, an APSES family transcription factor, interacts with MoMps1 and is required for hyphal and conidial morphogenesis, appressorial function and pathogenicity of Magnaporthe oryzae. Mol. Plant Pathol. 13, 677–689. doi: 10.1111/j.1364-3703.2011.00779.x
Qiu, L., Wang, J. J., Ying, S. H., Feng, M. G. (2015). Wee1 and Cdc25 control morphogenesis, virulence and multistress tolerance of Beauveria bassiana by balancing cell cycle-required cyclin-dependent kinase 1 activity. Environ. Microbiol. 17, 1119–1133. doi: 10.1111/1462-2920.12530
Qu, Y. M., Cao, H. J., Huang, P. Y., Wang, J., Liu, X. H., Lu, J. P., et al. (2022). A kelch domain cell end protein, PoTea1, mediates cell polarization during appressorium morphogenesis in Pyricularia oryzae. Microbiological Res. 259. doi: 10.1016/j.micres.2022.126999
Riquelme, M. (2013). Tip growth in filamentous fungi: a road trip to the apex. Annu. Rev. Microbiol. 67, 587–609. doi: 10.1146/annurev-micro-092412-155652
Ryder, L. S., Dagdas, Y. F., Kershaw, M. J., Venkataraman, C., Madzvamuse, A., Yan, X., et al. (2019). A sensor kinase controls turgor-driven plant infection by the rice blast fungus. Nature 574, 423–427. doi: 10.1038/s41586-019-1637-x
Sabnam, N., Barman, S. R. (2017). WISH, a novel CFEM GPCR is indispensable for surface sensing, asexual and pathogenic differentiation in rice blast fungus. Fungal Genet. And Biol. 105, 37–51. doi: 10.1016/j.fgb.2017.05.006
Saunders, D. G. O., Aves, S. J., Talbot, N. J. (2010). Cell cycle-mediated regulation of plant infection by the rice blast fungus. Plant Cell 22, 497–507. doi: 10.1105/tpc.109.072447
Sgarlata, C., Perez-Martin, J. (2005). The cdc25 phosphatase is essential for the G2/M phase transition in the basidiomycete yeast Ustilago maydis. Mol. Microbiol. 58, 1482–1496. doi: 10.1111/j.1365-2958.2005.04925.x
Shah, H., Rawat, K., Ashar, H., Patkar, R., Manjrekar, J. (2019). Dual role for fungal-specific outer kinetochore proteins during cell cycle and development in Magnaporthe oryzae. J. Cell Sci. 132. doi: 10.1242/jcs.224147
Steinberg, G. (2007). Hyphal growth: a tale of motors, lipids, and the spitzenkorper. Eukaryot Cell 6, 351–360. doi: 10.1128/EC.00381-06
Tonks, N. K. (2006). Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 7, 833–846. doi: 10.1038/nrm2039
Villalba, F., Collemare, J., Landraud, P., Lambou, K., Brozek, V., Cirer, B., et al. (2008). Improved gene targeting in Magnaporthe grisea by inactivation of MgKU80 required for non-homologous end joining. Fungal Genet. Biol. 45, 68–75. doi: 10.1016/j.fgb.2007.06.006
Virag, A., Harris, S. D. (2006). Functional characterization of aspergillus nidulans homologues of Saccharomyces cerevisiae Spa2 and Bud6. Eukaryot Cell 5, 881–895. doi: 10.1128/EC.00036-06
Wang, J. Z., Yin, Z. Y., Tang, W., Cai, X. J., Gao, C. Y., Zhang, H. F., et al. (2017). The thioredoxin MoTrx2 protein mediates reactive oxygen species (ROS) balance and controls pathogenicity as a target of the transcription factor MoAP1 in Magnaporthe oryzae. Mol. Plant Pathol. 18, 1199–1209. doi: 10.1111/mpp.12484
Wilson, R. A., Talbot, N. J. (2009). Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 7, 185–195. doi: 10.1038/nrmicro2032
Yano, K., Uesono, Y., Yoshida, S., Kikuchi, A., Kashiwazaki, J., Mabuchi, I., et al. (2013). Mih1/Cdc25 is negatively regulated by Pkc1 in Saccharomyces cerevisiae. Genes to Cells 18, 425–441. doi: 10.1111/gtc.12047
Yin, Z. Y., Tang, W., Wang, J. Z., Liu, X. Y., Yang, L. N., Gao, C. Y., et al. (2016). Phosphodiesterase MoPdeH targets MoMck1 of the conserved mitogen-activated protein (MAP) kinase signalling pathway to regulate cell wall integrity in rice blast fungus Magnaporthe oryzae. Mol. Plant Pathol. 17, 654–668. doi: 10.1111/mpp.12317
Yue, X. F., Que, Y. W., Deng, S. Z., Xu, L., Oses-Ruiz, M., Talbot, N. J., et al. (2017). The cyclin dependent kinase subunit Cks1 is required for infection-associated development of the rice blast fungus Magnaporthe oryzae. Environ. Microbiol. 19, 3959–3981. doi: 10.1111/1462-2920.13796
Zhang, H., Shen, W., Zhang, D., Shen, X., Wang, F., Hsiang, T., et al. (2021). The bZIP transcription factor LtAP1 modulates oxidative stress tolerance and virulence in the peach gummosis fungus Lasiodiplodia theobromae. Front. Microbiol. 12, 741842. doi: 10.3389/fmicb.2021.741842
Zhang, H., Tang, W., Liu, K., Huang, Q., Zhang, X., Yan, X., et al. (2011). Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae. PloS Pathog. 7, e1002450.
Zhang, H. F., Zheng, X. B., Zhang, Z. G. (2016). The Magnaporthe grisea species complex and plant pathogenesis. Mol. Plant Pathol. 17, 796–804.
Zhao, X. H., Mehrabi, R., Xu, J. R. (2007). Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryotic Cell 6, 1701–1714.
Zheng, H. W., Li, L. P., Yu, Z., Yuan, Y. P., Zheng, Q. J., Xie, Q. R., et al. (2021b). FgSpa2 recruits FgMsb3, a Rab8 GAP, to the polarisome to regulate polarized trafficking, growth and pathogenicity in Fusarium graminearum. New Phytol. 229, 1665–1683.
Zheng, C. C., Zhang, W. W., Zhang, S. L., Yang, G. G., Tan, L. Y., Guo, M. (2021a). Class I myosin mediated endocytosis and polarization growth is essential for pathogenicity of Magnaporthe oryzae. Appl. Microbiol. And Biotechnol. 105, 7395–7410.
Zheng, W. H., Zhou, J., He, Y. L., Xie, Q. R., Chen, A. H., Zheng, H. W., et al. (2015). Retromer is essential for autophagy-dependent plant infection by the rice blast fungus. PloS Genet. 11. doi: 10.1371/journal.pgen.1005704
Zhou, Z., Li, G., Lin, C., He, C. (2009). Conidiophore stalk-less1 encodes a putative zinc-finger protein involved in the early stage of conidiation and mycelial infection in Magnaporthe oryzae. Mol. Plant Microbe Interact. 22, 402–410. doi: 10.1094/MPMI-22-4-0402
Zhu, J. D., Hu, D. H., Liu, Q. Q., Hou, R., Xu, J. R., Wang, G. H. (2022). Stage-specific genetic interaction between FgYCK1 and FgBNI4 during vegetative growth and conidiation in Fusarium graminearum. Int. J. Mol. Sci. 23. doi: 10.3390/ijms23169106
Keywords: M. oryzae, dual-specificity phosphatase, MoMih1, conidiogenesis, pathogenicity
Citation: Liu S, Gong X, Ma J, Wang S and Guo M (2023) MoMih1 is indispensable for asexual development, cell wall integrity, and pathogenicity of Magnaporthe oryzae. Front. Plant Sci. 14:1146915. doi: 10.3389/fpls.2023.1146915
Received: 18 January 2023; Accepted: 27 February 2023;
Published: 14 March 2023.
Edited by:
Mingming Yang, Northwest A&F University, ChinaReviewed by:
Jiaoyu Wang, Zhejiang Academy of Agricultural Sciences, ChinaCopyright © 2023 Liu, Gong, Ma, Wang and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Guo, a2FuZHlsZW1vbkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.