- 1Department of Biology, Jeju National University, Jeju, Republic of Korea
- 2Department of Applied Biology, College of Agriculture and Life Sciences, Chonnam National University, Gwangju, Republic of Korea
Introduction
Plant organelles, including chloroplasts and mitochondria, derived from free-living cyanobacteria and α-proteobacteria (Andersson et al., 2003; Timmis et al., 2004). During the evolution, the massive organellar genes were transferred to the nuclear genome, resulting in only 3-209 proteins are encoded from the current chloroplast or mitochondrial genome (Kleine et al., 2009). The organelles-encoded proteins are crucial for organellar gene expression (OGE), photosynthesis, organellar electron transport chain, cellular metabolism, and ATP production (Maier et al., 2013). Notably, OGE is chiefly regulated at post-transcriptional steps, including RNA cleavage, RNA stability, RNA editing (C to U), and RNA splicing (Lee and Kang, 2016; Lee and Kang, 2020). The fine-tuned regulation of organellar post-transcriptional RNA metabolism essentially require hundreds of the nucleus-encoded chloroplast and mitochondrial RNA-binding proteins (nCMRBPs), including pentatricopeptide repeat (PPR) proteins, chloroplast ribosome maturation and splicing domain (CRM) proteins, DEAD-Box RNA helicases (RHs), S1 domain-containing proteins (SDPs), and mitochondrial transcription termination factors (mTERFs), during the entire period of plant growth and development (Hammani and Giegé, 2014; Quesada, 2016; Leister et al., 2017; Lee and Kang, 2020). Many recent studies have demonstrated that the lack of these proteins leads to the defective RNA metabolsim in plants under various environmental stresses (Lee and Kang, 2020; Qin et al., 2021).
PPR proteins are extensively distributed to plant lineages, containing more than 400 family members, and function as organellar-specific RNA-binding proteins (Schmitz-Linneweber and Small, 2008; Barkan and Small, 2014; Brown et al., 2014). Interestingly, a redesign of PPR tracts was shown to recognize programmed RNA targets and to control organellar gene expression (Coquille et al., 2014; Yagi et al., 2014; Colas des Francs-Small et al., 2018; McDowell et al., 2022). Here, we highlight a novel idea to redesign the PPR tracts of PPR4 and PPR19 proteins, which could affect the affinity of cognate RNAs in plant’s response to developmental and environmental cues.
Mechanism of P-type PPR proteins for target RNA recognition
The PPR constitutes a pair of antiparallel α helices of degenerate 35-amino acid tandem repeated-motifs ranging from 2 to 30 tracts (Small and Peeters, 2000; Barkan and Small, 2014). The binding specificity of PPR motifs relies on the basis of 1 repeat to 1 nucleotide binding module, leading to the association of single-stranded RNA (Barkan and Small, 2014; Cheng et al., 2016). P-class PPR proteins mainly consist of pure PPR motifs, which are important for organellar transcript processing, RNA stabilization, and group II intron splicing (Barkan and Small, 2014; Cheng et al., 2016; Wang et al., 2021). The PPR-RNA binding codes show that the amino acid residues at the fifth and thirty-fifth positions are essential for sequence specificity in the association of PPR protein-RNA substrate (Barkan and Small, 2014; Coquille et al., 2014; Shen et al., 2015; Yan et al., 2019). Importantly, amino acid codes that recognize the specific RNA bases harbor side chains playing an important role for hydrogen bond interaction with the similar manner of Watson and Crick base paring (Barkan and Small, 2014; Coquille et al., 2014; Shen et al., 2016), suggesting further possibility to predict and/or customize target RNA sequences via the combinational PPR-RNA binding code.
Redesign of artificial PPR proteins in organelles
To date, the usage of amino acid codes for nucleotide recognition suggests that modifying amino acid of PPR motifs causes a weak or strong ability in perceiving target RNA substrates (Barkan et al., 2012; Yin et al., 2013; Shen et al., 2015; Yan et al., 2019). Indeed, artificial PPRs exhibit the ability of recognizing programmed RNA targets and can modulate chloroplast gene expression (Coquille et al., 2014; Yagi et al., 2014). For instance, ZmPPR10 recognizing 5’ UTR of chloroplast ZmatpH was modified to successfully switch on the nuclear transgene expression in tobacco plastids and potato amyloplasts (Rojas et al., 2019; Yu et al., 2019). The native PPR proteins of dPPRrbcL and dPPRpetL were tailored to be associated with the specific sites in the 5’ UTR of rbcL and petL mRNAs in Arabidopsis chloroplasts, respectively (Manavski et al., 2021). The RPF2, an Arabidopsis mitochondrial PPR protein, specifically binds to two cognate RNA sites located within the 5’ UTRs of cox3 and nad9 genes (Colas des Francs-Small et al., 2018). Interestingly, the reprogrammed PPR tracts of RFP2 protein, which were designed to bind new target RNA sequences within the open reading frame of nad6, recognized the new RNA target site and subsequently led to a cleavage of the RNA molecule in vivo (Colas des Francs-Small et al., 2018). These results demonstrate the specificities and in vivo functionalities of the artificial PPR proteins.
Engineering of PPR4 and PPR19 proteins for organellar gene regulation
Previous studies showed that PPR19, which includes tandem-repeated 19 PPR tracts, is crucial for the stabilization of nad1 intron 3a in Arabidopsis mitochondria and PPR4, which possesses one RNA-recognition motif (RRM) and tandem-repeated 15 PPR tracts, plays an essential role in the splicing of rps12 intron 1b in Arabidopsis chloroplasts (Lee et al., 2017; Lee et al., 2019). Although the RRM and PPR motifs of PPR4 are associated with the strong RNA-binding activity of the PPR4 protein, the PPR motifs alone can strongly bind to the specific sequences of rps12 intron 1b (Lee et al., 2019). Importantly, the designed 11 and 14 PPR tracts showed a similar binding affinity to intended target substrates (Miranda et al., 2018), whereas the longer repeated-tracts tend to promote off-target action and reduce target-binding specificity. Notably, previous researches focused on switching on transgenes in chloroplasts or targeting to new RNA substrates in chloroplasts and mitochondria by combinations of manipulated PPR motifs (Colas des Francs-Small et al., 2018; Miranda et al., 2018; Rojas et al., 2019; Yu et al., 2019; Manavski et al., 2021). However, we suggest that rdPPR4 and rdPPR19 proteins are manipulated in its motifs for promoting the affinity of the proteins to its cognate RNAs (Figure 1). We anticipate that these optimizations will enhance the splicing of chloroplast rps12 intron 1 and the stability of nad1 intron 3 in native target location, which will increase the function of photosystems and mitochondrial electron transfer chains in plants under optimal conditions and environmental cues. The improvement of organellar functions will confer the phenotypic tolerance in plant’s response to abiotic stresses, including high salinity, drought, extreme temperatures, and high light.
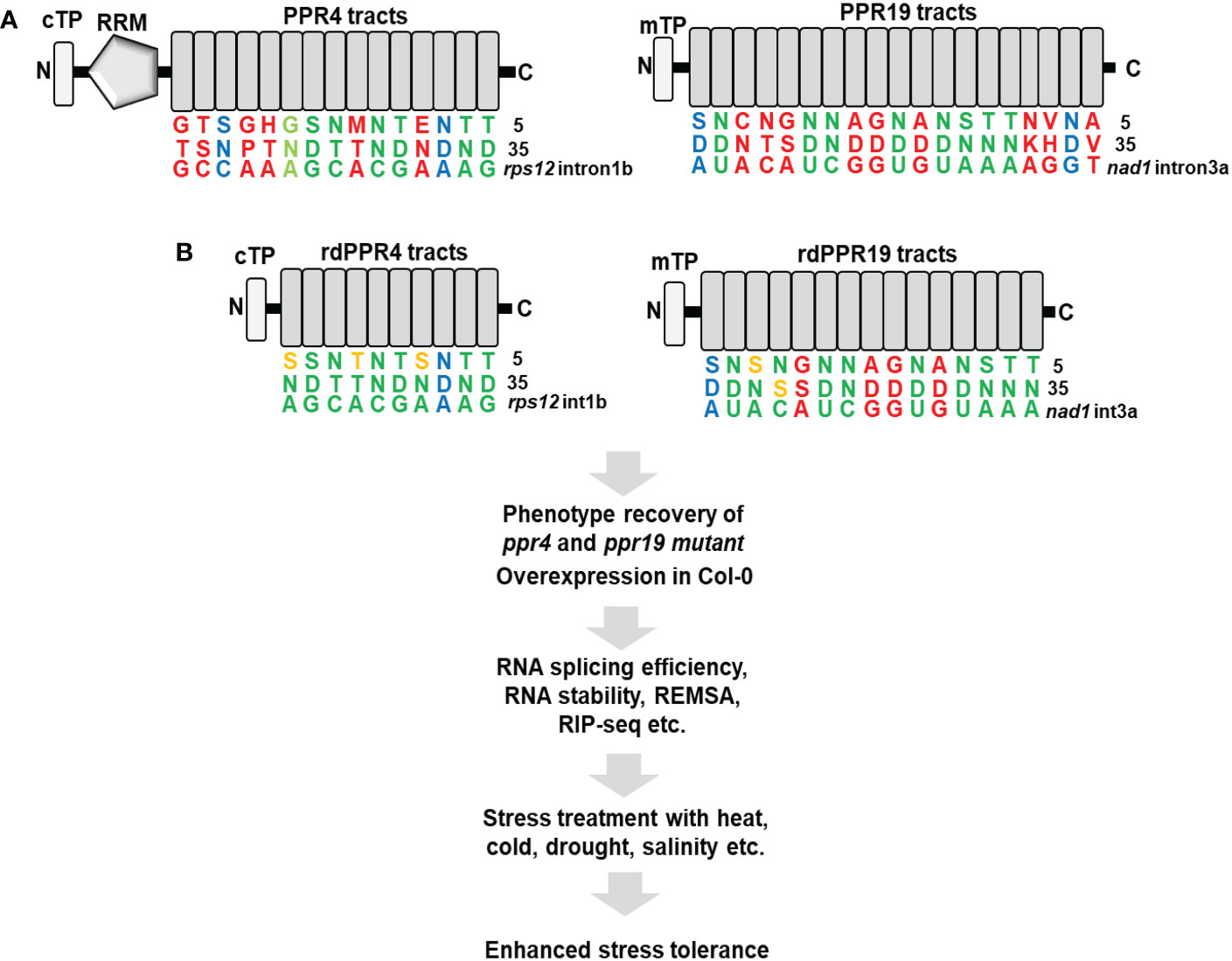
Figure 1 A redesign of Arabidopsis PPR4 and PPR19 proteins targeting to native RNA substrates. (A) The combination of the fifth and thirty-fifth amino acids on the native PPR4 and PPR19 proteins specifically recognizes target RNA sequences indicated with green colors for perfect matches, light green colors for less perfect matches, blue colors for mismatches, and red colors for no predictable matches. (B) The redesigned Arabidopsis PPR4 and PPR19 proteins with reprogrammed PPR tracts, which is named rdPPR4 tracks and rdPPR19 tracks. The RRM and N-terminal PPR tracts of the PPR4 protein and the C-terminal PPR tracts of the PPR19 protein, which do not participate in target RNA recognition, are eliminated. The PPR-RNA binding codes are modified for better matches with its cognate target RNA sequences as shown with orange colors. REMSA, RNA electrophoretic mobility shift assay; RIP-seq, RNA immunoprecipitation-sequencing; cTP, chloroplast transit peptide; mTP, mitochondrial transit peptide.
Conclusion and perspectives
Engineering PPR proteins can be harnessed for the manipulation of intron splicing and/or stabilization of organellar RNA molecules, and these redesigned PPR proteins will be a potential means to improve plant’s fitness to developmental and environmental cues. Engineered PPR proteins can endow the PPR proteins with a scaffolder for RNA targets, which can be potentially applicable for synthetic and RNA biology. Engineering PPR proteins combined with the artificial organelles-trafficking system can be utilized in agricultural crops to produce or restore cytoplasmic male sterility (CMS) which maternally fails to produce fertile pollens due to abnormalities of mitochondrial RNA metabolism.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This research was supported by Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (2019R1A6A1A10072987) to KL and by a grant from the Mid-career Researcher Program through the NRF of Korea funded by the Ministry of Science, ICT and Future Planning (NRF-2021R1A2C1004187) to HK, Republic of Korea.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Andersson, G., Karlberg, O., Canbäck, B., Kurland, C. G. (2003). On the origin of mitochondria: A genomics perspective. Philos. Trans. R. Soc Lond. B Biol. Sci. 358, 165–179. doi: 10.1098/rstb.2002.1193
Barkan, A., Rojas, M., Fujii, S., Yap, A., Chong, Y. S., Bond, C. S., et al. (2012). A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PloS Genet. 8, e1002910. doi: 10.1371/journal.pgen.1002910
Barkan, A., Small, I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442. doi: 10.1146/annurev-arplant-050213-040159
Brown, G. G., Colas des Francs-Small, C., Ostersetzer, O. (2014). Group II intron splicing factors in plant mitochondria. Front. Plant Sci. 5, 35. doi: 10.3389/fpls.2014.00035
Cheng, S., Gutmann, B., Zhong, X., Ye, Y., Fisher, M. F., Bai, F., et al. (2016). Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 85, 532–547. doi: 10.1111/tpj.13121
Colas des Francs-Small, C., Vincis Pereira Sanglard, L., Small, I. (2018). Targeted cleavage of nad6 mRNA induced by a modified pentatricopeptide repeat protein in plant mitochondria. Commun. Biol. 1, 1–7. doi: 10.1038/s42003-018-0166-8
Coquille, S., Filipovska, A., Chia, T., Rajappa, L., Lingford, J. P., Razif, M. F., et al. (2014). An artificial PPR scaffold for programmable RNA recognition. Nat. Commun. 5, 1–9. doi: 10.1038/ncomms6729
Hammani, K., Giegé, P. (2014). RNA Metabolism in plant mitochondria. Trends Plant Sci. 19, 380–389. doi: 10.1016/j.tplants.2013.12.008
Kleine, T., Maier, U. G., Leister, D. (2009). DNA Transfer from organelles to the nucleus: The idiosyncratic genetics of endosymbiosis. Annu. Rev. Plant Biol. 60, 115–138. doi: 10.1146/annurev.arplant.043008.092119
Lee, K., Han, J. H., Park, Y. I., Colas des Francs-Small, C., Small, I., Kang, H. (2017). The mitochondrial pentatricopeptide repeat protein PPR19 is involved in the stabilization of NADH dehydrogenase 1 transcripts and is crucial for mitochondrial function and arabidopsis thaliana development. New Phytol. 215, 202–216. doi: 10.1111/nph.14528
Lee, K., Kang, H. (2016). Emerging roles of RNA-binding proteins in plant growth, development, and stress responses. Mol. Cells 39, 179. doi: 10.14348/molcells.2016.2359
Lee, K., Kang, H. (2020). Roles of organellar RNA-binding proteins in plant growth, development, and abiotic stress responses. Int. J. Mol. Sci. 21, 4548. doi: 10.3390/ijms21124548
Lee, K., Park, S. J., Colas des Francs-Small, C., Whitby, M., Small, I., Kang, H. (2019). The coordinated action of PPR4 and EMB2654 on each intron half mediates trans-splicing of rps12 transcripts in plant chloroplasts. Plant J. 100, 1193–1207. doi: 10.1111/tpj.14509
Leister, D., Wang, L., Kleine, T. (2017). Organellar gene expression and acclimation of plants to environmental stress. Front. Plant Sci. 8, 387. doi: 10.3389/fpls.2017.00387
Maier, U.-G., Zauner, S., Woehle, C., Bolte, K., Hempel, F., Allen, J. F., et al. (2013). Massively convergent evolution for ribosomal protein gene content in plastid and mitochondrial genomes. Genome Biol. Evol. 5, 2318–2329. doi: 10.1093/gbe/evt181
Manavski, N., Mathieu, S., Rojas, M., Méteignier, L.-V., Brachmann, A., Barkan, A., et al. (2021). In vivo stabilization of endogenous chloroplast RNAs by customized artificial pentatricopeptide repeat proteins. Nucleic Acids Res. 49, 5985–5997. doi: 10.1093/nar/gkab390
McDowell, R., Small, I., Williams-Carrier, R., Bond, C. S. (2022). Synthetic PPR proteins as tools for sequence-specific targeting of RNA. Methods 208, 19–26. doi: 10.1016/j.ymeth.2022.10.003
Miranda, R. G., McDermott, J. J., Barkan, A. (2018). RNA-Binding specificity landscapes of designer pentatricopeptide repeat proteins elucidate principles of PPR–RNA interactions. Nucleic Acids Res. 46, 2613–2623. doi: 10.1093/nar/gkx1288
Qin, T., Zhao, P., Sun, J., Zhao, Y., Zhang, Y., Yang, Q., et al. (2021). Research progress of PPR proteins in RNA editing, stress response, plant growth and development. Front. Genet. 12, 765580. doi: 10.3389/fgene.2021.765580
Quesada, V. (2016). The roles of mitochondrial transcription termination factors (MTERFs) in plants. Physiol. Plant 157, 389–399. doi: 10.1111/ppl.12416
Rojas, M., Yu, Q., Williams-Carrier, R., Maliga, P., Barkan, A. (2019). Engineered PPR proteins as inducible switches to activate the expression of chloroplast transgenes. Nat. Plants 5, 505–511. doi: 10.1038/s41477-019-0412-1
Schmitz-Linneweber, C., Small, I. (2008). Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 13, 663–670. doi: 10.1016/j.tplants.2008.10.001
Shen, C., Wang, X., Liu, Y., Li, Q., Yang, Z., Yan, N., et al. (2015). Specific RNA recognition by designer pentatricopeptide repeat protein. Mol. Plant 8, 667–670. doi: 10.1016/j.molp.2015.01.001
Shen, C., Zhang, D., Guan, Z., Liu, Y., Yang, Z., Yang, Y., et al. (2016). Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 7, 11285. doi: 10.1038/ncomms11285
Small, I. D., Peeters, N. (2000). The PPR motif–a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 45–47. doi: 10.1016/S0968-0004(99)01520-0
Timmis, J. N., Ayliffe, M. A., Huang, C. Y., Martin, W. (2004). Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135. doi: 10.1038/nrg1271
Wang, X., An, Y., Xu, P., Xiao, J. (2021). Functioning of PPR proteins in organelle RNA metabolism and chloroplast biogenesis. Front. Plant Sci. 12, 627501. doi: 10.3389/fpls.2021.627501
Yagi, Y., Nakamura, T., Small, I. (2014). The potential for manipulating RNA with pentatricopeptide repeat proteins. Plant J. 78, 772–782. doi: 10.1111/tpj.12377
Yan, J., Yao, Y., Hong, S., Yang, Y., Shen, C., Zhang, Q., et al. (2019). Delineation of pentatricopeptide repeat codes for target RNA prediction. Nucleic Acids Res. 47, 3728–3738. doi: 10.1093/nar/gkz075
Yin, P., Li, Q., Yan, C., Liu, Y., Liu, J., Yu, F., et al. (2013). Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504, 168–171. doi: 10.1038/nature12651
Keywords: engineering PPR proteins, organellar gene expression, organellar RNA metabolism, chloroplast, mitochondria
Citation: Lee K and Kang H (2023) Engineering of pentatricopeptide repeat proteins in organellar gene regulation. Front. Plant Sci. 14:1144298. doi: 10.3389/fpls.2023.1144298
Received: 17 January 2023; Accepted: 21 February 2023;
Published: 01 March 2023.
Edited by:
Jirong Huang, Shanghai Institute for Biological Sciences (CAS), ChinaReviewed by:
Ping Yin, Huazhong Agricultural University, ChinaCopyright © 2023 Lee and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kwanuk Lee, a3VsZWVAamVqdW51LmFjLmty; Hunseung Kang, aHNrYW5nQGpudS5hYy5rcg==
 Kwanuk Lee
Kwanuk Lee Hunseung Kang
Hunseung Kang