- 1Agrotechnology and Rural Development Division, Council of Scientific and Industrial Research (CSIR)-North East Institute of Science and Technology, Jorhat, Assam, India
- 2Advanced Computation and Data Sciences Division, Council of Scientific and Industrial Research (CSIR)-North East Institute of Science and Technology, Jorhat, Assam, India
Solanum khasianum is a medicinally important plant that is a source of steroidal alkaloids ‘solasodine.’ It has various industrial applications, including oral contraceptives and other pharmaceutical uses. The present study was based on 186 germplasm of S. khasianum, which were analyzed for the stability of economically important traits like solasodine content and fruit yield. The collected germplasm was planted during Kharif 2018, 2019, and 2020 in RCBD with three replications at the experimental farm of CSIR-NEIST, Jorhat, Assam, India. A multivariate approach for stability analysis was applied to identify stable germplasm of S. khasianum for economically important traits. The germplasm was analyzed for additive main effects and multiplicative interaction (AMMI), GGE biplot, multi-trait stability index, and Shukla’s variance which were evaluated for three environments. The AMMI ANOVA revealed significant GE interaction for all the studied traits. The stable and high-yielding germplasm was identified from the AMMI biplot, GGE biplot, Shukla’s variance value, and MTSI plot analysis. Lines no. 90, 85, 70, 107, and 62 were identified as highly stable fruit yielders while, lines no. 1, 146, and 68 were identified as stable high solasodine lines. However, considering both traits, i.e., high fruit yield and solasodine content, MTSI analysis was performed which showed that lines 1, 85, 70,155, 71, 114, 65, 86, 62, 116, 32, and 182 could be used in a breeding program. Thus, this identified germplasm can be considered for further varietal development and could be used in a breeding program. The findings of the present study would be beneficial for the S. khasianum breeding program.
1 Introduction
The Solanaceae family with worldwide distribution comprises 90 genera, containing more than 3000 species. The Solanaceae family’s largest genus is Solanum with more than 2000 species (Kaunda and Zhang, 2019). Solanum khasianum (C.B. Clarke), also called Solanum aculeatissimum Jacq., is a member of the Solanaceae family and is cultivated in China, Bhutan, and the Indian states of Assam, Arunachal Pradesh, Meghalaya, Sikkim, Manipur, Orissa, Nilgiri Hill, and West Bengal (Gogoi et al., 2021). Alkaloids, flavonoids, phenols, and numerous other phytoconstituents found in S. khasianum are recognized to have the ability to reduce inflammation, blood sugar levels, and oxidative stress (Gupta, 2019). The potential glycoalkaloids reported to be present in S. khasianum include solasodine, solamargine, solanine, and solasonine (Srivastava et al., 2016; Kaunda and Zhang, 2019). Among the Solanum plants screened for solasodine production, S. khasianum was found to possess the maximum solasodine content (Weissenberg, 2001; Begum et al., 2022; Lal et al., 2022a). It is widely known for its range of therapeutic applications. Solasodine was found to possess many pharmacological properties like anti-inflammatory, antioxidant, antinociceptive, antimicrobial, cytotoxic, anti-obesity, antiandrogenic, and hepatoprotective properties (Srivastava et al., 2016; Kumar et al., 2019; Gogoi et al., 2021). Moreover, previous reports suggested that the berries possess anti-diabetic, anti-inflammatory, anti-cholinesterase, anticancer, and antibacterial properties (Rosangkima and Jagetia, 2015; Gogoi et al., 2021; Pavani and Shasthree, 2021). As an ethnomedicine, the plant has been used for treating diseases like whooping cough, bronchitis, smallpox, trachoma, snake bites, rheumatism, and tooth and skin infections (Schmelzer and Gurib, 2008; Chirumamilla et al., 2021; Lal et al., 2022a).
Solanum khasianum is a sturdy, branched shrub with a height of approximately 0.75 to 1.5 m. The stem and leaves bear spines on both its surfaces with the shape of ovate to lobate. It possesses hermaphrodite flowers and abundant berries and turns yellowish upon maturity (Begum et al., 2022; Lal et al., 2022a). The cultivation area of S. khasianum is more than 4000 ha in India across the states of Assam, Meghalaya, Tripura, Manipur, Maharashtra, and Karnataka (Gogoi et al., 2021). S. khasianum is a storehouse of solasodine, the steroidal alkaloid which has high industrial and medicinal significance (Sanwal et al., 2016; Gogoi et al., 2021). It is the starting base for synthesizing steroid hormones, such as cortisone and oral contraceptives (Begum et al., 2022). Solasodine is a key intermediate and substrate in steroidal drugs and sex hormone production (Sanwal et al., 2016; Lal et al., 2022a). It is used in steroidal hormone production for commercial use in oral pills or injections (Sinani and Eltayeb, 2017; Lal et al., 2022a). It, thus, plays a crucial role in human birth control purposes. The valuable part of the plant is the berries which contain the source of solasodine that can be extracted from ripened berries. The maximum content of solasodine is produced when the berries have ripened and turned yellow in color. The seeds are mucilage in nature, small in size, and are the primary source of propagation (Begum et al., 2022). Increased consumption of this medicine and reckless harvesting of the plant from the wild as a raw material source has led to the depletion of genetic diversity and habitat loss (Canter et al., 2005; Julsing et al., 2007). Due to its rapid growth and low initial culture cost, it is one of the best sources of affordable raw materials for industrially significant solasodine extraction. Therefore, it would be very advantageous to identify and include plant breeding for this neglected plant species.
As for the definition of stability, a genotype that can consistently produce consistent performance regardless of any change in its environmental conditions can be termed a stable genotype. In plant breeding, the stability of yield traits is a significant aspect. However, with ever-changing fluctuations in environmental conditions, the determination of yield factor is challenging. The phenotypic expression of a genotype is a controlled result of both genotypic constitutions of the genotype and the environment. Hence, the genotype may show inconsistent performance under varying environmental conditions. A stable genotype with superior yield in varying environments is considered the mostly desired genotype. This compels the necessary study of the genotype × environment to identify stable genotypes with desirable traits.
A stable genotype is one which produces a stable yield in an environment (Annicchiarico, 2002; Munda et al., 2020). The performance of a genotype in multi-environments is difficult to study when the genotype-environment interaction is stronger (Alwala et al., 2010). The selection of a stable genotype is difficult due to the interaction between genotype × environments (Gupta et al., 2015). The genotypic response to an environment is multivariate; hence, the determination of stability using parametric and nonparametric statistical analysis which is univariate would not be very effective. There are many multivariate models available for the analysis of stability. Among the various multivariate model, additive main effects and multiplicative interaction (AMMI) method and ‘GGE biplot’ are widely used (Lal et al., 2022b). The AMMI model is an amalgamation of the analysis of variance and the principal component analysis. It efficiently illustrates the adaptive responses of genotypes to the environment by providing details on the main and interaction effects along with a biplot. The main effects of G × E are assessed using variance analysis (main additive effects) while the G × E interaction is estimated using the principal analysis components (multiplicative interactions). The GGE biplot was developed to represent graphically the G × E interaction and genotype main effects. Shukla’s variance analysis is effective to partition the total G × E interaction for the estimation of the variance attribution for each of the genotypes in an unbiased manner. For the simultaneous estimation of stability and identification of genotypes with multiple superior traits, the MTSI is useful for exploiting wide adaptability. The plant breeding program is aimed at the identification of superior yielding lines due to varying genotypes’ performance and varying environments. Thus, the results obtained are either for the identification of genotypes with high-yielding traits or different environment-adapted genotypes. However, with the multivariate approach for stability analysis across multilocation, both the issues of varying genotypic and environmental interactions can be addressed. The present stability analysis was verified by AMMI, GGE, MTSI, and Shukla’s variance methods. High fruit-yielding germplasm with high solasodine content, which shows a consistent performance across different locations of NE India, is the major selection criteria for the identification of elite lines.
So far, none of the stability studies in S. khasianum are available in the public domain. Hence the present study was conducted for the first time to assess the performance of large numbers of germplasm of S. khasianum with high solasodine content.
2 Materials and methods
2.1 Materials and methods
Initially, 273 germplasm of S. khasianum were collected from different regions of NE India. Among them, 186 high solasodine content-rich (>0.8%) lines were identified and used in the present study (Begum et al., 2022). The identified 186 germplasm were planted for three years in Kharif in 2018, 2019, and 2020 at the experimental farm of CSIR-North East Institute of Science and Technology (NEIST), Jorhat, Assam, India, and were evaluated for three years. The plant specimens were identified by the plant breeder of CSIR-NEIST, Jorhat, India, and the prepared herbarium was deposited in the departmental record with identification ID: RRLJ-SK-1 to RRLJ-SK-186.
2.2 Morphological characters recorded
The morphological data were recorded for the three years of study. The data included plant spread (cm), plant height (cm), leaf width (cm), leaf length (cm), fruit diameter (cm), number of fruits/plants, fresh fruit weight per plant (kg), and days to maturity. Along with the morphological characters, the solasodine content (%) was also recorded for each year (Table S1). All the data were recorded for ten randomly selected healthy plants for each germplasm.
2.3 Solasodine extraction and quantification
The dried ripe berries of S. khasianum were used for the estimation of solasodine. Solasodine extraction and quantification were done as per the method suggested by Begum et al. (2022).
2.4 Statistical analysis
R software was used for performing all the statistical analyses. The mean-multi-environment trial analysis package in R software was used in the study (Olivoto et al., 2019). The analysis of variance (ANOVA), AMMI, multi-trait stability index (MTSI), GGE, and Shukla’s variance were used to analyze the three-year data of Solanum khasianum for economic traits like solasodine content and fresh fruit weight per plant.
3 Results
The evaluation of germplasm is pivotal to elite germplasm; therefore, the identified or selected germplasm needs to be further assessed for its stability and consistent performance in varied environments. The present study focuses on the identification of stable germplasm of S. khasianum for economically important traits. The pooled data for three years were analyzed with ANOVA and various stability analyses (AMMI, GGE, MTSI, and Shukla’s variance). The coefficient of variation is a significant parameter for the comparison between traits based on phenotypic variation. Concerning the coefficient of variation, a higher variation was found in the fresh fruit weight per plant followed by plant height while a moderate coefficient of variation was observed for solasodine content and days to maturity (Table 1).
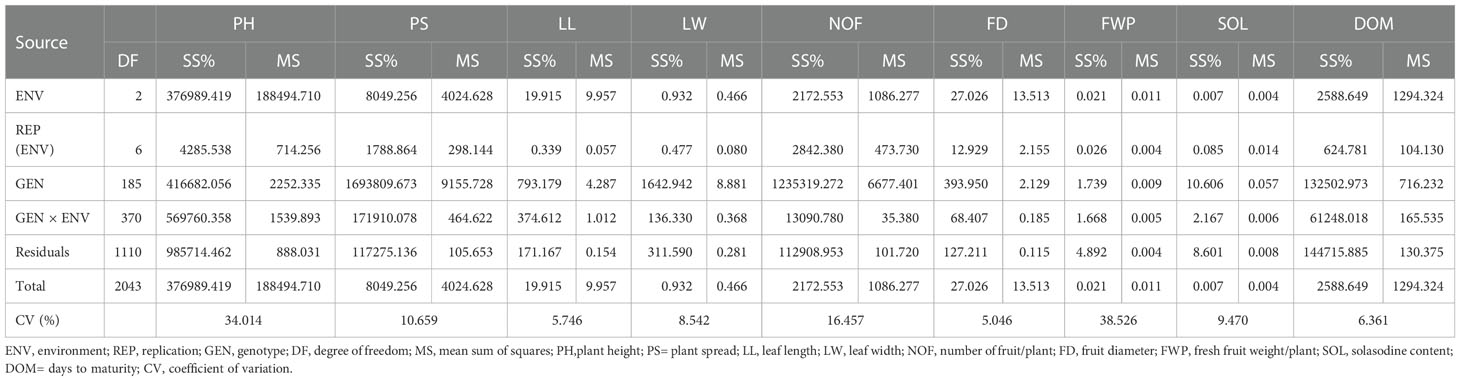
Table 1 The analysis of variance on the three-year data of Solanum khasianum germplasm that was used in the study.
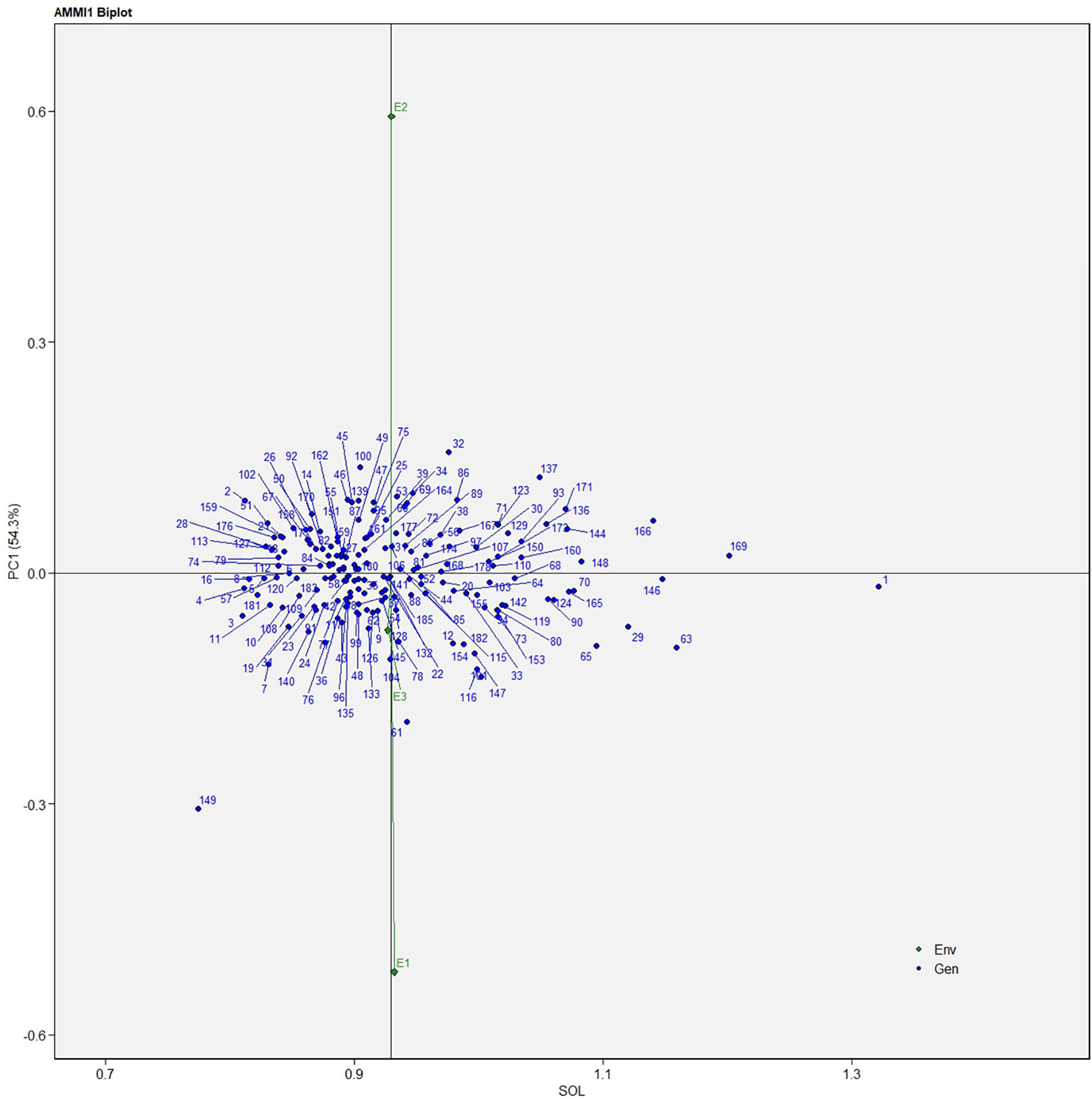
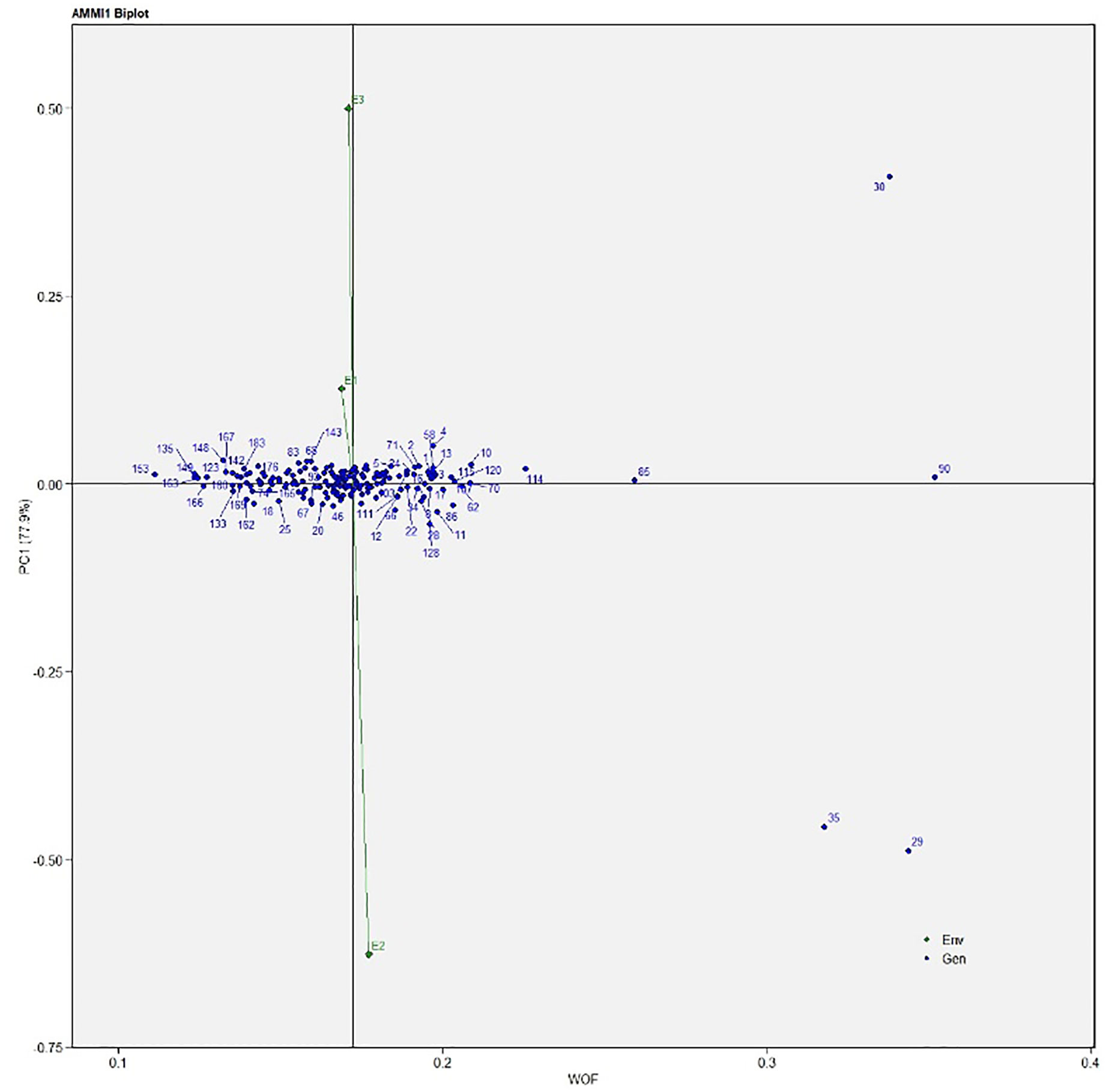
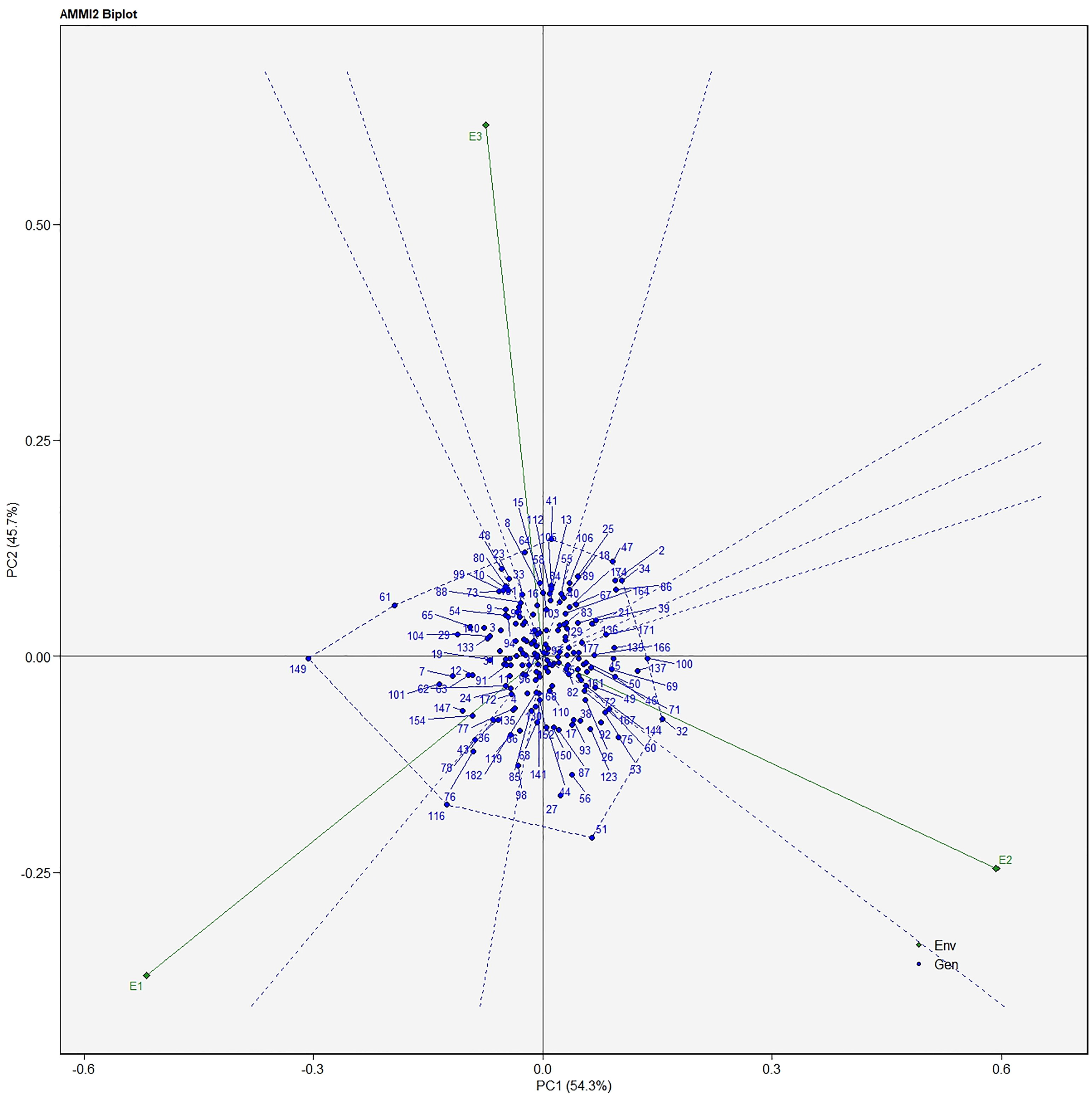
The AMMI analysis of variance (ANOVA) revealed that the environmental effect was significant for all the traits while the genotypic effect was significant for plant height, fruit diameter, fresh fruit weight per plant, solasodine content, and days to maturity. Significant G × E interaction was observed for all the studied traits indicating divergent performances across the different environments. Thus, a precise selection of germplasm with elite traits is essential for a breeding program. The interaction effect was analyzed from the principal component analysis (IPCA) of AMMI ANOVA from which G × E interaction sum of squares percentage value of 48.660 and 16.955 was recorded for fresh fruit weight per plant and solasodine content, respectively (Table 2). A more precise analysis for the selection of germplasm with elite traits from the genotype-environment interaction was identified through the AMMI biplot. The genotype-environment interaction for yield is an important analysis in which the less interacting genotypes are closer to the center of the AMMI biplot and considered more stable than their counterpart for that region. In contrast, the far-centered genotypes have individual stability with each environment which indicates the high-performing germplasm for each environment. The AMMI was performed for the economically important traits i.e., solasodine content and fresh fruit weight per plant for which AMMI biplots were constructed. From the AMMI results, it was found that lines no. 1, 146, 68, 178, 52, 107, and 44 were among the high yielders and were stable for solasodine content (Figure 1). Lines no. 90, 85, 70, 107, and 62 were considered stable and high yielders for fresh fruit weight per plant (Figure 2). The individual competence of the genotypes in the studied environments was evaluated using the AMMI2 model in which the first two principal components (Factor 1 and Factor 2) were used to generate the plot. In the AMMI2 biplot for solasodine content, line no. 116 was considered a high yielder in environment one, line no. 51 and 32 in environment two, and line no. 105 in environment three, respectively (Figure 3).
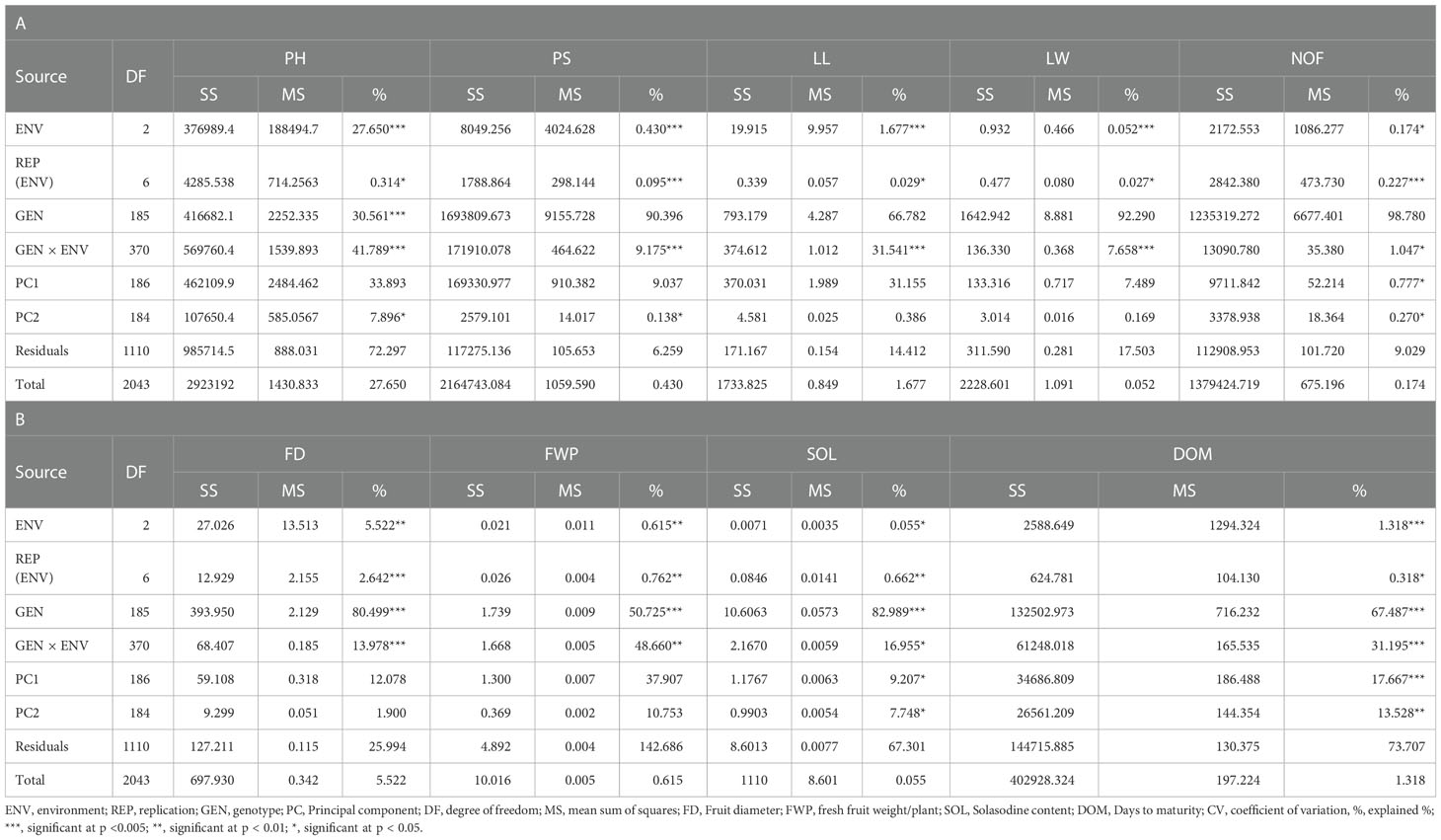
Table 2 The AMMI Analysis of variance on the three-year data of Solanum khasianum germplasm that was used in the study.
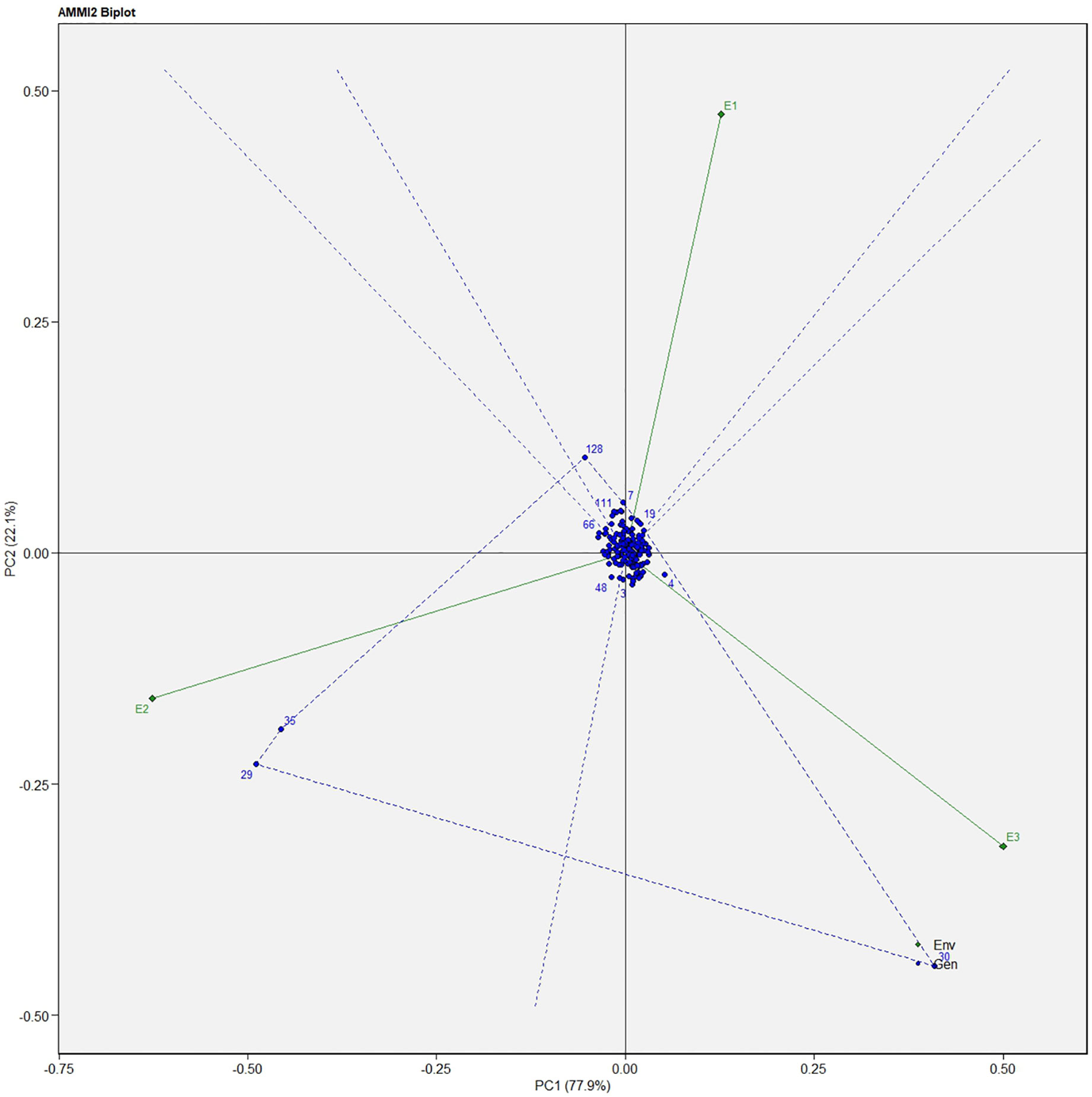
Similarly, for the fresh fruit weight per plant, line no. 128 and 7 were considered stable in environment one, lines no. 29 and 35 in environment two, and line no. 30 in environment three, respectively (Figure 4). The above-identified genotypes showed the best performance in the studied environments and were found to be highly competent.
The genotype in a studied environment that possesses the highest mean performance and absolute stability can be considered the ideal genotype. Such a genotype possessing a superior trait with zero-G × E interaction is ideal. It should be described as the one possessing the most excellent vector length from genotype markers to the origin of the biplot. In the GGE biplot, lines no. 149, 61, and 32 were the most unstable lines. Their vector lengths were longer compared to the other genotypes. Whereas lines no.1, 169, and 146 had high solasodine content and were found to be stable genotypes since their vector lengths were short (Figures 5, 6). While considering the trait of the fresh fruit weight per plant from the GGE biplot, genotypes 29, 30, and 35 had higher weights of fresh fruit/plant than average but were unstable as they exhibited longer vector lengths. Lines no. 90, 85, and 114 had high fresh fruit weight per plant and exhibited short vector lengths which characterize a stable genotype (Figures 7, 8). The discriminating ability is directly proportional to the vector length. Environments E2 and E3 had longer vectors than E1 for the trait fresh fruit weight per plant (Figure 8). While, for the trait solasodine content, E1 and E2 had longer vectors than E3 (Figure 6). Thus, E2 and E3 were the best environments to exhibit germplasm differentiation for fresh fruit yield per plant while environment E1 and E2 were the most discriminating environment for solasodine content.
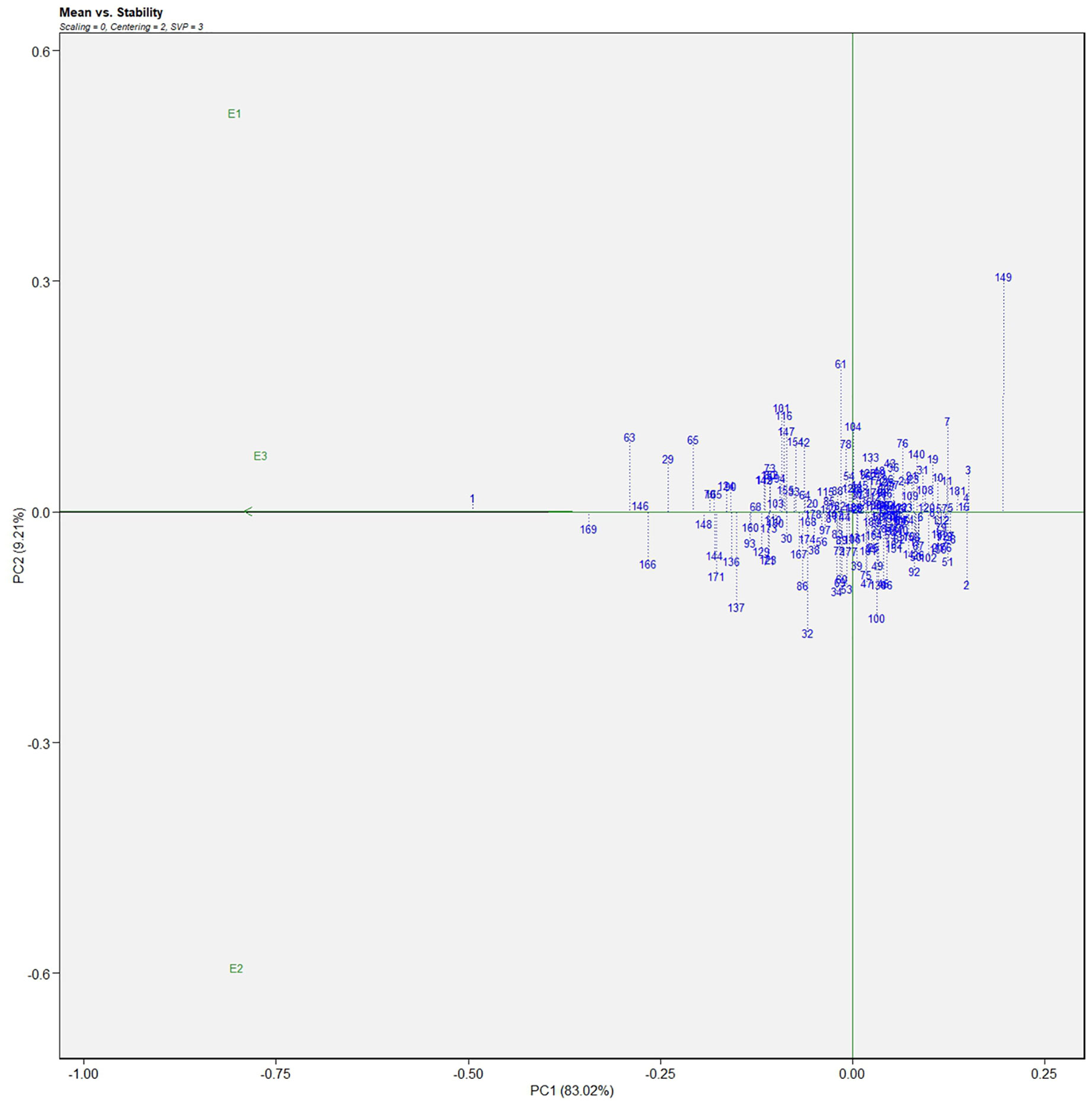
Figure 5 Average environment coordination views of the GGE-biplot based on environment-focused scaling for the mean performance and stability 186 germplasm of S. khasianum for solasodine content.
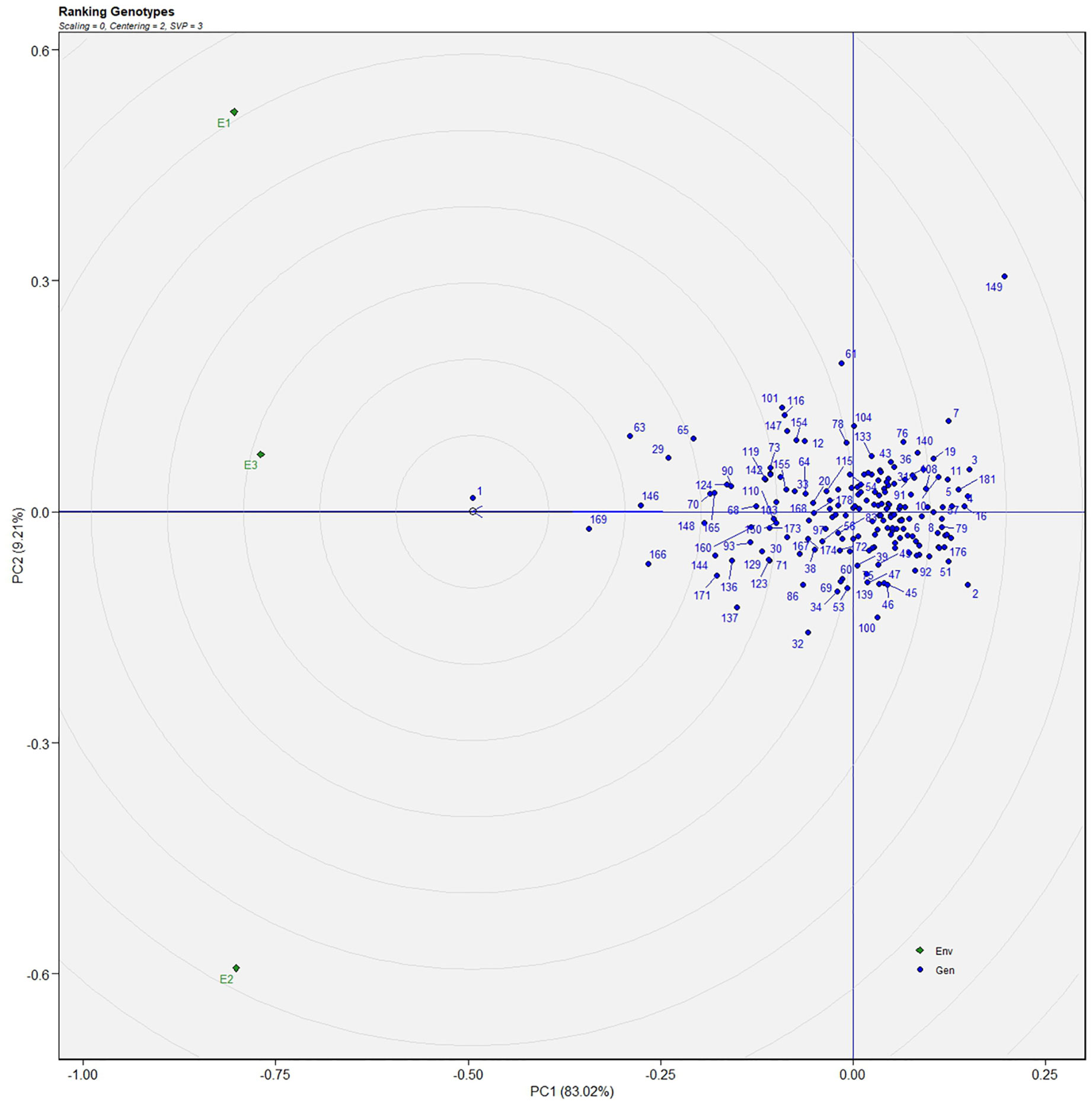
Figure 6 GGE-biplot based on genotype and environment-focused scaling for the comparison of genotype and environments for solasodine content.
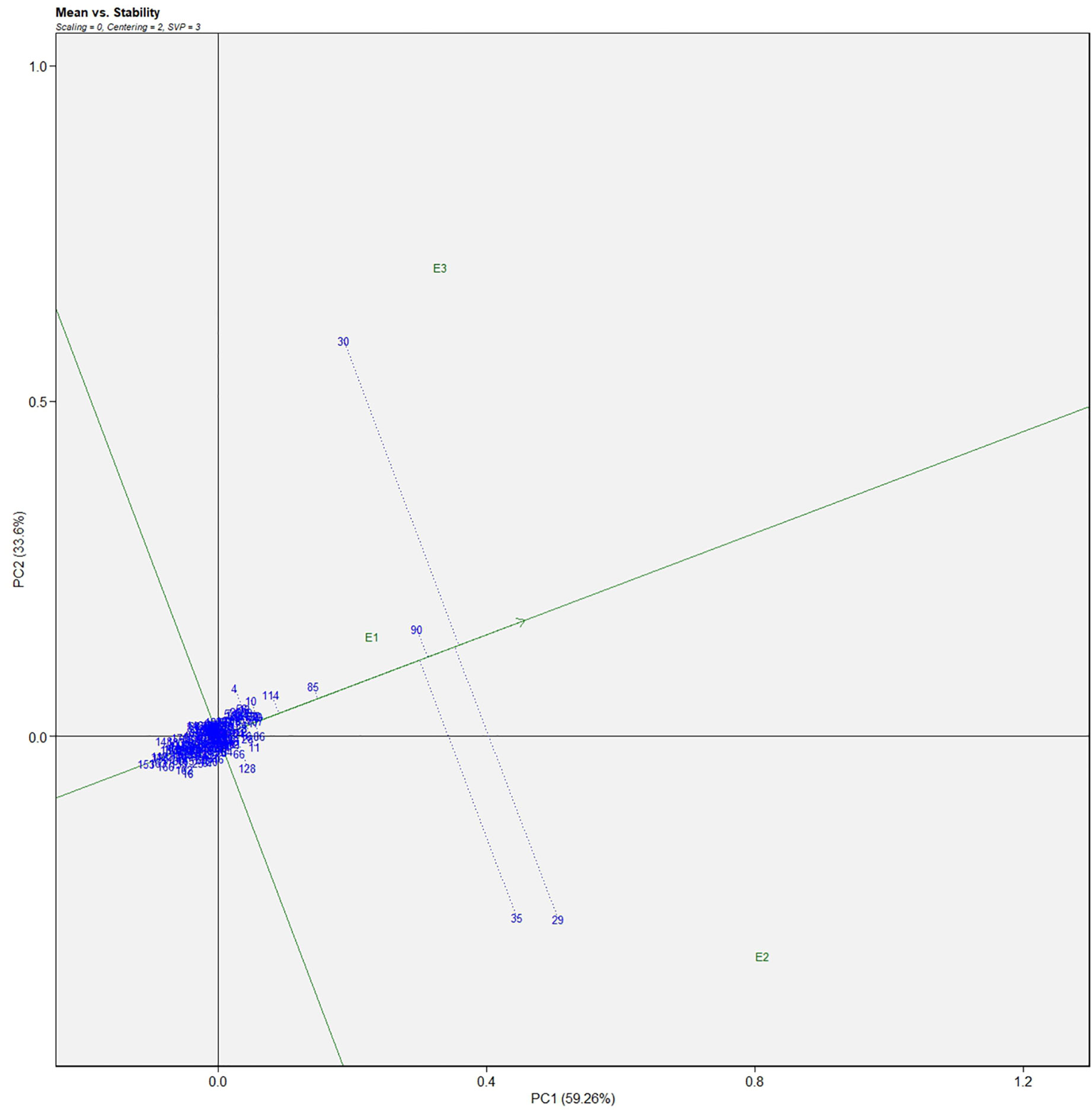
Figure 7 Average environment coordination views of the GGE-biplot based on environment-focused scaling for the mean performance and stability 186 germplasm of S. khasianum for the weight of fruit.

Figure 8 GGE-biplot based on genotype and environment-focused scaling for the comparison of genotype and environments for the weight of fruit.
Shukla’s variance was studied for genotype performance across the environments according to how the germplasm with minimum values is intended to be more stable. On the basis of our present results, lines no. 153, 135, 149, 163, 166, and 123 were considered stable for fresh fruit weight per plant while lines no. 149, 3, 4, 2, 16, 181, 5, and 28 were considered stable for solasodine content (Table 3). The MTSI study revealed that the mean of the selected germplasm (Xs) was higher than the mean of the original (Xo) for fresh fruit weight per plant and solasodine content. The selection gain (SG) was positive for both traits with SG% of 7.022 and 7.610 for the traits of fresh fruit weight per plant and solasodine content, respectively. The selection differential (SD) was favorable for both traits and heritability of 0.520 and 0.873 for fresh fruit weight per plant and solasodine content, respectively (Table 4). As per the MTSI analysis, lines no. 90, 1, 85, 70, 155, 63, 115, 107, 71, 103, 114, 65, 86, 124, 101, 62, 34, 110, 137, 171, 174, 116, 12, 32, 22, 182, 147, and 33 were considered as stable germplasm (Figure 9). Based on the multivariate and univariate approaches used in the study, it can be concluded that lines 90, 85, 107, and 62 were the elite lines with a high yield and stable nature. The study of stability is an important aspect in the identification of stable genotypes with elite traits. The elite germplasm in the study refers to the stable germplasm with a high fruit yield and solasodine content. The study of S. khasianum is significant for commercially cultivating the industrially important solasodine-rich plant.
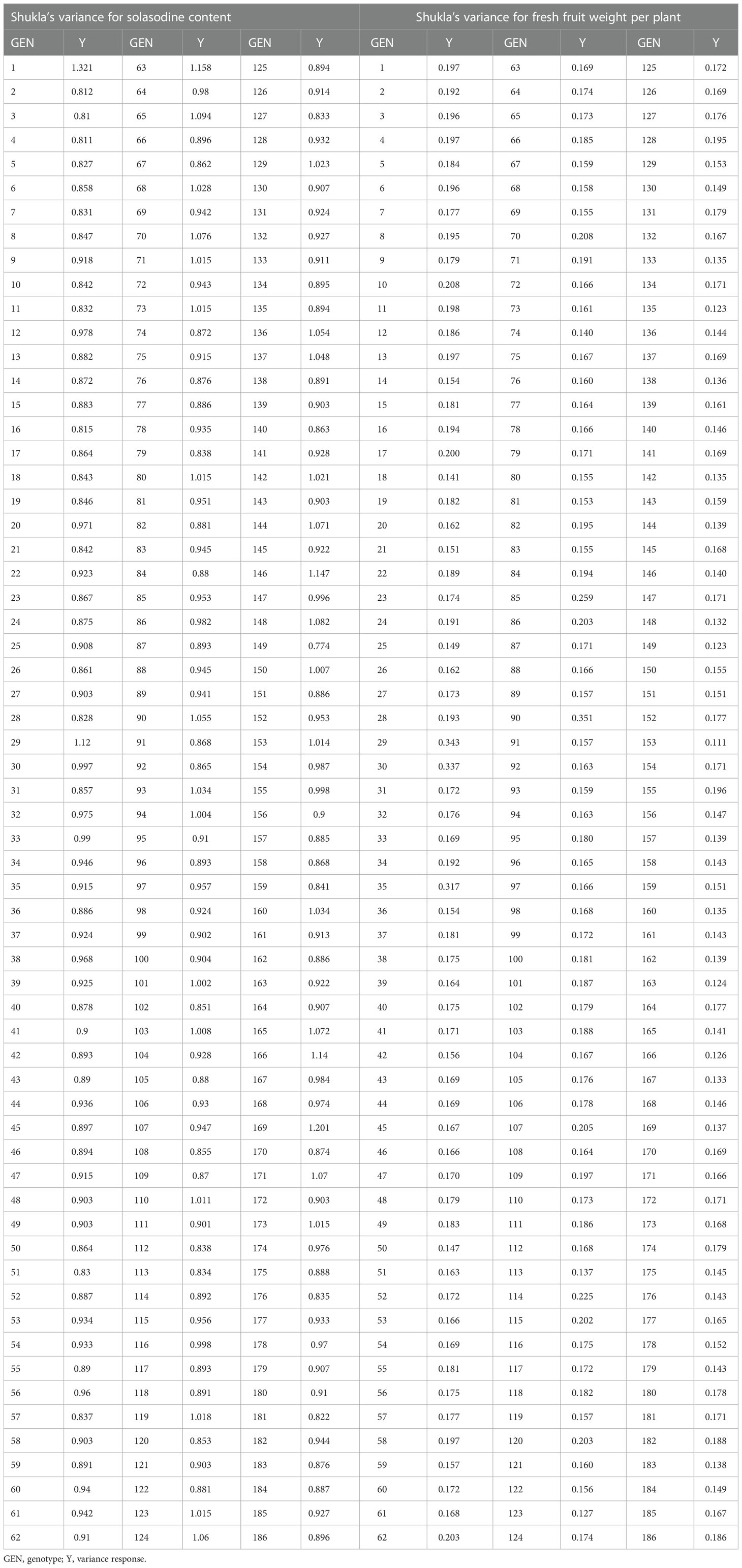
Table 3 Shukla’s variance on the three-year data for solasodine content and weight of fruit for Solanum khasianum germplasm that was used in the study.
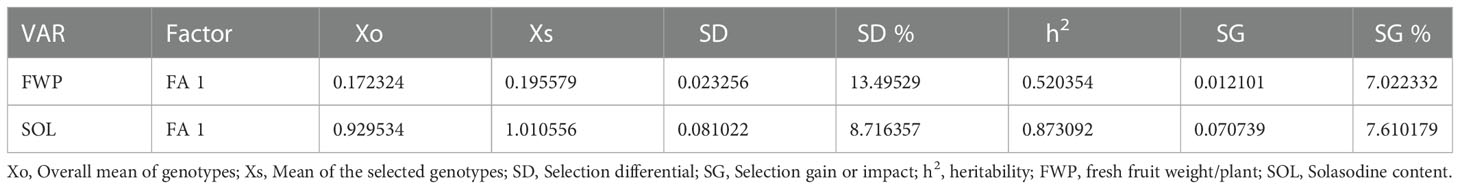
Table 4 Estimates of selection differential, selection gain, and heritability based on MTSI for 186 S. khasianum germplasm across three environments.
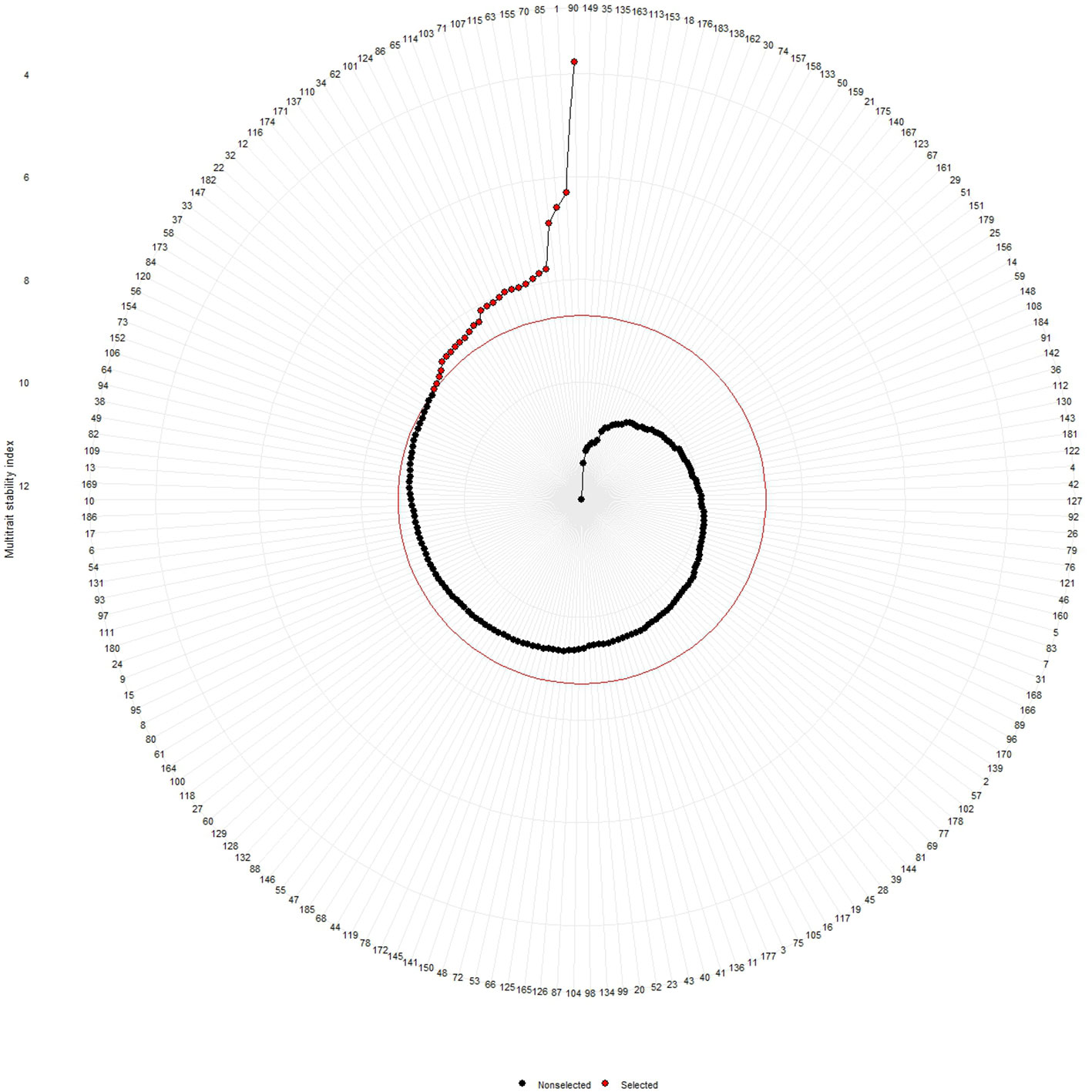
Figure 9 Multi trait stability index (MTSI) of 186 germplasm of S. khasianum based on the traits of solasodine content and weight of fruit.
4 Discussion
The assessment of the different landraces worldwide is crucial for identifying and preserving genetically rich resources. The various landraces of different crops have been the storehouse for significant desirable traits. These are widely used in various breeding programs, which can be exploited to increase genetic variability. Hashemi et al. (2018) stated that different crop genotypes exhibit variable responses under different environmental conditions; hence, to reduce the influence of the environment on the genotypes, the compatibility and stability of the genotype need to be studied for multiple years at the multilocation. The most preferable genotype from the breeding point of view is the one exhibiting stability along with a significantly positive main effect, or in other terms, which has broad adaptability (Lal et al., 2022c; Munda et al., 2023). So far, no stability analysis has been studied to date for S. khasianum; hence, the results were considered with previous reports of the Solanaceae family.
A study on Solanum melongena from Iran was performed by Khankahdani et al. (2022) on fifteen germplasm for stability analysis by using the AMMI model. The AMMI biplot revealed that genotype 7 (AM5) followed by genotypes 12 (Y7), 5 (AM7), and 13 (SA15) were identified and ranked for their stability against yield factor since they revealed the lowest genotype × environment interaction. The least stable genotypes were the ones that exhibited the highest level of genotype × environment interaction, and the most unstable identified genotypes were genotypes 1 (Y), 8 (AM4), and 19 (GHE12), respectively. Similar results were obtained in our study on the basis of which germplasm with commercially important traits and stability were identified for future breeding programs.
A similar study by Bagheri et al. (2016) reported the AMMI stability assay where 22 germplasm of the long eggplant were studied for three years. A report on the stability analysis in tobacco by the AMMI model has been previously reported by Sadeghi and Samizadeh (2011). The AMMI model was also used to study stability in general along with individual stability in different studied environments for production yield in numerous other crops like cotton (Damavandi-Kamali et al., 2011), barley (Koocheki et al., 2012), bread wheat (Bigonah, 2012), Soybean (Soltan Mohamadi et al., 2017), and durum wheat (Sadeghzadeh et al., 2018). Scavo et al. (2023) assessed six genotypes of Solanum tuberosum L. for AMMI and GGE. The study revealed the Salad Blue genotype as the most suitable genotype for total phenolic content, dry matter, FRAP, and vitamin C throughout eight studied environments due to its above-average mean values and stability. Another study by Das et al. (2021) was conducted on 21 potato cultivars in the eastern sub-Himalayan plains of West Bengal, India, to assess their suitability, stability, and yield factors. The study revealed ‘Kufri Pukhraj’ and ‘Kufri Chipsona-3’ as higher yielder cultivars along with ‘Kufri Surya’ as the heat-resistant cultivar. However, two cultivars, namely, ‘Kufri Khyati’and ‘HPS II/67’ were identified as stable for eastern Himalayan conditions. A study on stability analysis for nine genotypes of Elite Irish Potato (Solanum tuberosum L.) was conducted in western Ethiopia. The study revealed CIP384321.30 as the stable genotype for the region on the basis of the mean tuber yield, regression slope identified, and GGE biplot (Fufa and Fufa, 2021). Similar conclusions were drawn from our present study in which stable germplasm was identified across the studied environments along with individual stable germplasm for each environment with the which-won-where pattern approach.
A study on the African tomato landraces was reported by Tembe et al. (2018) in which the morphological traits were assessed. The study revealed that significant variations among the studied samples can be useful as a potential source of genetic diversity for the crop improvement of tomatoes. Similar previous findings on other vegetable crops have also been reported by different workers in tomato (Manzano et al., 2015; Sousaraei et al., 2021), lettuce (Missio et al., 2018), and eggplant fields (Taher et al., 2017; Gramazio et al., 2019), identifying superior genetic resources. Tonk et al. (2011) studied the GGE biplot analysis in maize, revealing that the G16 hybrid was found to have high suitability to the tested environments for grain yield. A study was conducted on 10 genotypes of potatoes in different environments for AMMI by Almohammedi et al. (2019). The percentage of variation of 98.44%, 0.24%, and 1.31% was recorded for genotypes, environments, and GEI, respectively, which revealed that genotypes contributed more towards variation than the other parameters. As per the definition by Bernardo (2002), when the progeny of a genotype shows consistent results for the traits of interest, the genotype is regarded as possessing a good breeding value. Based on our study, lines no.1, 90, 146, 68, 85, 107, 70, and 62 were identified as superior stable germplasm. Germplasm no. 90, 85, 70, 107, and 62 were identified as high yielding and stable for fresh fruit weight per plant. While germplasm no. 1, 146, and 68 were identified for high solasodine stable lines. Thus, this identified germplasm can be considered for further large-scale cultivation on a commercial level or can be used in breeding programs.
5 Conclusions
The natural source of solasodine from S. khasianum is an important medicinal plant species. The identification of high fruit-yielding and solasodine-rich germplasm is significant for the commercial success of the industrially important solasodine. The multivariate stability analysis is a novel approach that provides an easy way to select high-performance and stable lines in S. khasianum that performs better under varied environmental conditions. On the basis of the present study, lines no. 90, 85, 70, 107, and 62 were identified as stable and high-yielding fruit yields per plant. Further, lines no. 1, 146, and 68 were identified as stable and high-solasodine lines. MTSI analysis revealed lines 1, 85, 70, 155, 71, 114, 65, 86, 62, 116, 32, and 182 as stable for both high yielders, high fruit yields, and high solasodine lines. The large-scale cultivation of this germplasm can prove to be a cheap natural source of steroidal alkaloid solasodine. Moreover, the AMMI, GGE, MTSI, and Shukla’s variance models were found to be effective plant breeding models for the evaluation of stability which would maximize the utilization of resources thereby making a significant contribution to the sustainability of breeding programs. The identified genotypes of the study can be utilized for commercial cultivation and can be useful in future breeding programs.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
TB: wrote the original draft, methodology, and edited the manuscript; SM: software analysis; TG: wrote the original draft; RG: editing; VKC: data curation; SC: formal analysis and visualization; HL: data curation and formal analysis; GNS: editing and supervision; ML: conceptualization, methodology, editing, validation, and supervision. All authors contributed to the article and approved the submitted version.
Funding
The present work was supported by NMPB, New Delhi, Ministry of Ayush, govt. of India in the form of project no. R&D/AS-01/2018-19.
Acknowledgments
The authors are thankful to the Director, CSIR-NEIST, Jorhat, for providing the laboratory and field facility of the institute.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1143778/full#supplementary-material
References
Almohammedi, O. H. M., Al-Abdaly, M. M., Mahmod, S. A. (2019). Study of genotype and environmental interaction on yield analysis of tuber of potato (Solanum tuberosum l.) using AMMI in Iraq. Plant Arch. 19 (1), 978–982.
Alwala, S., Kwolek, T., McPherson, M., Pellow, J., Meyer, D. (2010). A comprehensive comparison between Eberhart and Russell joint regression and GGE biplot analyses to identify stable and high yielding maize hybrids. Field Crops Res. 119, 225–230. doi: 10.1016/j.fcr.2010.07.010
Annicchiarico, P. (2002). Genotype × environmental interactions; challenges and opportunities for plant breeding and cultivar recommendations. FAO plant production and protection Paper.174 (Rome: FAO).
Bagheri, M., Keshavarz, S., Kakhki, A. (2016). Evaluation of selected lines from eggplant (Solanum melongena l.) landraces. J. Seed Plant Breed. 32-1 (2), 165–180. doi: 10.22092/SPIJ.2017.111295
Begum, T., Munda, S., Pandey, S. K., Lal, M. (2022). Estimation of selection criteria through multi-year assessment of variability parameters, association studies and genetic diversity of Solanum khasianum CB Clarke. Sci. Hortic. 297, 110923. doi: 10.1016/j.scienta.2022.110923
Bernardo, R. (2002). Reinventing quantitative genetics for plant breeding: something old, something new, something borrowed, something BLUE. Heredity 125, 375–385. doi: 10.1038/s41437-020-0312-1
Bigonah, H. H. (2012). Yield stability of promising lines of winter and facultative wheat in different climate of Iran. Afr. J. Agric. Res. 7 (15), 2304–2311. doi: 10.5897/AJAR11.931
Canter, P. H., Thomas, H., Ernst, E. (2005). Bringing medicinal plants into cultivation: opportunities and challenges for biotechnology. Trend Biotechnol. 23, 180–185. doi: 10.1016/j.tibtech.2005.02.002
Chirumamilla, P., Gopu, C., Jogam, P., Taduri, S. (2021). Highly efficient rapid micropropagation and assessment of genetic fidelity of regenerants by ISSR and SCoT markers of Solanum khasianum Clarke. Plant Cell Tissue Organ Cult. 144 (2), 397–407. doi: 10.1007/s11240-020-01964-6
Damavandi-Kamali, S., Babaeian, J. N., Aalishah, A. (2011). Evaluation of stability and compatibility of the yield of cotton varieties based on one-variable parametric and non-parametric methods and AMMI model. Iranian J. Crop Sci. 42 (2), 397–407.
Das, S., Mitra, B., Luthra, S. K., Saha, A., Hassan, M. M., Hossain, A. (2021). Study on morphological, physiological characteristics and yields of twenty-one potato (Solanum tuberosum l.) cultivars grown in Eastern Sub-Himalayan plains of India. Agron 11, 335. doi: 10.3390/agronomy11020335
Fufa, T. W., Fufa, A. N. (2021). Yield stability analysis of elite Irish potato (Solanum tuberosum l.) varieties in Western Ethiopia. In. J. Biochem. Biophy Mol. Biol. 6 (1), 6–10. doi: 10.11648/j.ijbbmb.20210601.13
Gogoi, R., Sarma, N., Pandey, S. K., Lal, M. (2021). Phytochemical constituents and pharmacological potential of Solanum khasianum C.B. clarke., extracts: special emphasis on its skin whitening, anti-diabetic, acetylcholinesterase, genotoxic activities. Trends Phytochem. Res. 5 (2), 47–61. doi: 10.30495/TPR.2021.1917249.1190
Gramazio, P., Chatziefstratiou, E., Petropoulos, C., Chioti, V., Mylona, P., Kapotis, G., et al. (2019). Multi-level characterization of eggplant accessions from Greek islands and the mainland contributes to the enhancement and conservation of this germplasm and reveals a large diversity and signatures of differentiation between both origins. Agronomy 9 (887), 1–20. doi: 10.3390/agronomy9120887
Gupta, S. (2019). In-vitro anti-inflammatory activity of s. xanthocarpum and A. ofcinarum herb by human red blood cell membrane stabilization method. J. Drug Deliv.Ther. 9 (3), 9663–9666. doi: 10.22270/jddt.v9i3-s2948
Gupta, P., Dhawan, S. S., Lal, R. K. (2015). Adaptability and stability based differentiation and selection in aromatic grasses (Cymbopogon species) germplasm. Ind. Crop Prod. 78, 1–8. doi: 10.1016/j.indcrop.2015.10.018
Hashemi, A., Nematzadeh, G. A., Oladi, M., Afkhami, G. A., Gholizadeh, G. A. (2018). Study of rapeseed (Brassica napus) promising genotypes adaptation in different regions of mazandaran. J. Crop Breed. 10 (28), 119–124. doi: 10.29252/jcb.10.28.119
Julsing, M. K., Quax, W. J., Kysar, O. (2007). The engineering of medicinal plants prospects and limitation of medicinal biotechnology. Med. Plant Biotechnol. 3, 6. doi: 10.1002/9783527619771.ch1
Kaunda, J. S., Zhang, Y. J. (2019). The genus solanum: an ethnopharmacological, phytochemical and biological properties review. Nat. Products Bioprospect 9, 77–137. doi: 10.1007/s13659-019-0201-6
Khankahdani, H. H., Bagheri, M., Khoshkam, S. (2022). Stability and compatibility of some Iranian eggplant (Solanum melongena l.) lines using AMMI method. Int. J. Hortic. Sci. Technol. 9 (1), 52–39. doi: 10.22059/ijhst.2020.301667.364
Koocheki, A. R., Sorkhi, B., Eslamzadeh, H. M. R. (2012). Study on stability of elite barley (Hordeum vulgar l.) genotypes for cold regions of Iran using AMMI method. Cereal Res. 2 (4), 249–261. doi: 20.1001.1.22520163.1391.2.4.1.7
Kumar, R., Khan, M. I., Prasad, M. (2019). Solasodine: A perspective on their roles in health and disease. Res. J. Pharm. Technol. 12 (5), 2571–2576. doi: 10.5958/0974-360X.2019.00432.3
Lal, M., Munda, S., Begum, T., Gupta, T., Paw, M., Chanda, S. K., et al. (2022b). Identification and registration for high-yielding strain through ST and MLT of Curcuma caesia Roxb.(Jor Lab KH-2): A high-value medicinal plant. Genes 13 (10), 1807. doi: 10.3390/genes13101807
Lal, M., Munda, S., Bhandari, S., Saikia, S., Begum, T., Pandey, S. K. (2022a). Molecular genetic diversity analysis using SSR marker amongst high solasodine content lines of Solanum khasianum C.B. Clarke, an industrially important plant. Ind. Crops Prod. 184, 115073. doi: 10.1016/j.indcrop.2022.115073
Lal, M., Munda, S., Gogoi, A., Begum, T., Baruah, J., Chanda, S. K., et al. (2022c). Use of stability statistics in the selection of Clausena heptaphylla (Roxb.) Wight & arn for novel anethole rich strain (Jor Lab CH-2). Front. Plant Sci. 13, 1060492. doi: 10.3389/fpls.2022.1060492
Manzano, S., Navarro, P., Martínez, C., Megías, Z. M., Rebolloso, M. M., Jamilena, M. (2015). Evaluation of fruit quality in tomato landraces under organic greenhouse conditions. Acta Hortic 1099, 645–652. doi: 10.3389/fpls.2018.10491
Missio, J. C., Rivera, A., Figàs, M. R., Casanova, C., Camí, B., Soler, S., et al. (2018). A comparison of landraces vs. modern varieties of lettuce in organic farming during the winter in the Mediterranean area: an approach considering the viewpoints of breeders, consumers, and farmers. Front. Plant Sci. 9 (1491), 1–15.
Munda, S., Paw, M., Saikia, S., Begum, T., Baruah, J., Lal, M. (2023). Stability and selection of trait specific genotypes of Curcuma caesia roxb. using AMMI, BLUP, GGE, WAAS and MTSI model over three years evaluation. JARMAP 32, 100446. doi: 10.1016/j.jarmap.2022.100446
Munda, S., Sarma, N., Lal, M. (2020). GxE interaction of 72 accessions with three year evaluation of Cymbopogon winterianus jowitt. using regression coefficient and additive main effects and multiplicative interaction model (AMMI). Ind. Crop Prod. 146, 112169. doi: 10.1016/j.indcrop.2020.112169
Olivoto, T., L’ucio, A. D. C., da silva, J. A. G., Marchioro, V. S., de Souza, V. Q., Jost, E. (2019). Mean performance and stability in multi-environment trials I: combining features of AMMI and BLUP techniques. Agron. J 111, 2949–2960. doi: 10.2134/agronj2019.03.0220
Pavani, C., Shasthree, T. (2021). Biological activity of green synthesized silver nanoparticles and different plant extracts of Solanum khasianum Clarke. Int. Res. J. Adv. Sci. Hub. 3, 12–17. doi: 10.47392/irjash.2021.103
Rosangkima, G., Jagetia, G. C. (2015). In-vitro anticancer screening of medicinal plants of mizoram state, India, against dalton’s lymphoma, MCF-7 and HELA cells. Int. J. Recent Sci. Res. 6, 5648–5653.
Sadeghi, S. M., Samizadeh, H. (2011). Evaluation of yield stability of Virginia tobacco hybrids using stability parameters and pattern analysis via AMMI model. Electronic J. Crop Prod. 4 (2), 103–119. https://www.sid.ir/paper/134936/en.
Sadeghzadeh, B., Mohammadi, R., Ahmadi, H., Abedi, G., Ahmadi, M. M., Mohammadfam, M., et al. (2018). GGE biplot and AMMI application in the study of adaptability and grain yield stability of durum wheat lines under dryland conditions. Env. Stresses Crop Sci. 11 (2), 241–260. doi: 10.22077/escs.2018.381.1075
Sanwal, C. S., Kumar, R., Anwar, R., Kakade, V., Kerketta, S., Bhardwaj, S. D. (2016). Growth and yield of Solanum khasianum in Pinus roxburghii forest based silvi-medicinal system in mid hills of Indian himalaya. Ecosyst 3, 19. doi: 10.1186/S40663-016-0078-3
Scavo, A., Mauromicale, G., Ierna, A. (2023). Genotype × environment interactions of potato tuber quality characteristics by AMMI and GGE biplot analysis. Scientia Hortic. 310, 111750. doi: 10.1016/j.scienta.2022.111750
Schmelzer, G. H., Gurib, F. A. (2008). “Medicinal plants 1,” in Plant resources of tropical Africa, Netherlands, vol. 11. (Netherlands: PROTA Foundation), 518–520.
Sinani, S. S. S., Eltayeb, E. A. (2017). The steroidal glycoalkaloids solamargine and solasonine in solanum plants. S. Afr. J. Bot. 112, 253–269. doi: 10.1016/j.sajb.2017.06.002
Soltan Mohamadi, S., Peyghambri, S. A., Babaei, H. R. (2017). Study the adaptability and yield sustainability of soybean genotypes in four regions of Iran. Iranian. J. Crop Sci. 48 (2), 389–397. doi: 10.22059/IJFCS.2017.63482
Sousaraei, N., Mashayekhi, K., Mousavizadeh, S. J., Akbarpour, V., Medina, J., Aliniaeifard, S. (2021). Screening of tomato landraces for drought tolerance based on growth and chlorophyll fluorescence analyses. Hortic. Env. Biotech. 62, 521–535. doi: 10.1007/s13580-020-00328-5
Srivastava, M., Sharma, S., Misra, P. (2016). Elicitation based enhancement of secondary metabolites in Rauwolfia serpentina and Solanum khasianum hairy root cultures. Pharmacogn. Mag Suppl. 3, S315–S320. doi: 10.4103/0973-1296.185726
Taher, D., Solberg, S., Prohens, J., Chou, Y., Rakha, M., Wu, T. (2017). World vegetable center eggplant collection: origin, composition, seed dissemination and utilization in breeding. Front. Plant Sci. 8 (1484), 1–12. doi: 10.3389/fpls.2017.01484
Tembe, K. O., Chemining’wa, G., Ambuko, J., Owino, W. (2018). Evaluation of African tomato landraces (Solanum lycopersicum) based on morphological and horticultural traits. Agricu. Nat. Resources 52, 536–542. doi: 10.1016/j.anres.2018.11.014
Tonk, F. A., Ilker, E., Tosun, M. (2011). Evaluation of genotype X environment interactions in maize hybrids using GGE biplot analysis. Crop Breed. App. Biotech. 11, 1–9. doi: 10.1590/S1984-70332011000100001
Keywords: anova, AMMI, GGE biplot, MTSI, Shukla’s variance, solasodine, Solanum khasianum
Citation: Begum T, Munda S, Gupta T, Gogoi R, Choubey VK, Chanda SK, Lekhak H, Sastry GN and Lal M (2023) Stability estimation through multivariate approach among solasodine-rich lines of Solanum khasianum (C.B. Clarke): an important industrial plant. Front. Plant Sci. 14:1143778. doi: 10.3389/fpls.2023.1143778
Received: 13 January 2023; Accepted: 16 March 2023;
Published: 11 May 2023.
Edited by:
Weibiao Liao, Gansu Agricultural University, ChinaReviewed by:
Abdurrahim Yılmaz, Abant Izzet Baysal University, TürkiyeAnanya Kuanar, Siksha O Anusandhan University, India
Copyright © 2023 Begum, Munda, Gupta, Gogoi, Choubey, Chanda, Lekhak, Sastry and Lal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohan Lal, bW9oYW5AbmVpc3QucmVzLmlu
 Twahira Begum
Twahira Begum Sunita Munda
Sunita Munda Tanmita Gupta1
Tanmita Gupta1 Vikash Kumar Choubey
Vikash Kumar Choubey Mohan Lal
Mohan Lal