
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 12 May 2023
Sec. Plant Cell Biology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1142868
This article is part of the Research Topic Cellular Heterogeneity in Plants View all 8 articles
Oil palm micropropagation through tissue culture is a technique to provide elite oil palms to meet the desired traits. This technique is commonly carried out through somatic embryogenesis. However, the oil palm’s somatic embryogenesis rate is quite low. Several approaches have been made to overcome this problem, including transcriptome profiling through RNA-seq to identify key genes involved in oil palm somatic embryogenesis. RNA sequencing was applied in high- and low-embryogenic ortets of Tenera varieties based on the somatic embryoid rate at the callus, globular, scutellar, and coleoptilar embryoid stages. Cellular analysis of embryoid inductions and proliferations showed that high-embryogenic ortets resulted in higher embryoid proliferation and germinations than low-embryogenic ortets. Transcriptome profiling showed that there are a total of 1,911 differentially expressed genes (DEGs) between high- and low-embryogenic ortets. ABA signaling-related genes such as LEA, DDX28, and vicilin-like protein are upregulated in high-embryogenic ortets. Furthermore, DEGs associated with other hormone signaling, such as HD-ZIP associated with brassinosteroids and NPF associated with auxin, are upregulated in high-embryogenic ortets. This result suggests a physiological difference between high- and low-embryogenic ortets that is connected to their capacity for somatic embryogenesis. These DEGs will be used as potential biomarkers for high-embryogenic ortets and will be validated in further studies.
As the most productive oil-bearing plant in the world (FAO, 2021), improvement of oil palm planting materials is necessary to fulfill the high demand for this vegetable oil. One of the techniques that can be applied to improve oil palm planting material is tissue culture. In vitro clonal seedling techniques can increase yield between 20% and 30% in comparison to conventional seedlings. This increase is due to the use of high-yielding ortets as an explant source (Corley & Tinker, 2003) and true-to-type plants that are genetically identical to their ortets (Shearman et al., 2013). Oil palm is a monocotyledon plant that only has one shoot apical meristem (Weckx et al., 2019). Oil palm in vitro clonal seedling techniques are carried out through somatic embryogenesis. However, the oil palm somatic embryogenesis rate is only 1%–5% (Roowi et al., 2010). Somatic embryogenesis is affected by several factors, including genotype, mother plant, media type, and explant source (Constantin et al., 2015). The plant genotype is a main factor in the oil palm embryogenic competence (Roowi et al., 2010). Somatic embryogenesis involves various physiological, cellular, and molecular activities, including dedifferentiation and reprogramming of gene expression patterns (Karami & Saidi, 2010).
Transcriptomics is an RNA transcript used for the study of gene expression differences and gene pathways in activating or suppressing several genes during somatic embryogenesis (Rajesh et al., 2016). Several genes related to oil palm embryogenesis have been identified using molecular analyses. For example, using expressed sequence tags, two genes, namely, lipid-transfer protein (LTP) and glutathione S-transferase (GST), were differentially expressed between embryogenic and non-embryogenic callus (Low et al., 2008). Moreover, using microarray and quantitative real-time PCR (qPCR), some oil palm somatic embryogenesis genes, i.e., IAA-amino acid hydrolase ILR1-like 1 (ILR1) and late embryogenesis abundant (LEA2), were also detected as biomarkers to differentiate the embryoid stage from the callus stage (Syariyanto et al., 2018). Recently, transcriptome profiling of leaves of high- and low-embryogenic ortets revealed that flowering-related genes such as flowering locus T-interacting protein (FTIP), frigida-like (FRL), and nuclear transcription factor y subunit A-7 (NF-YA) were upregulated in high-embryogenic ortets (Ooi et al., 2021). However, the study of RNA transcripts in the somatic embryoid development of high- and low-embryogenic oil palm is not yet available. Moreover, somatic embryogenesis encompasses three developmental stages consisting of embryonic induction, embryonic, and developmental stages (Elhiti et al., 2013). Therefore, it is important to analyze the molecular mechanisms of somatic embryoids not only in the induction stages but also in the embryonic and developmental stages.
RNA sequencing enables high throughput of RNA transcript analysis through cDNA sequencing and can provide quantitative information on gene expression and differentially expressed genes (DEGs) (Kukurba & Montgomery, 2015). Thus, this technique allows the identification of the key genes involved in oil palm somatic embryogenesis through analysis of DEGs between high- and low-embryogenic ortets. In this study, RNA sequencing was used to obtain a transcriptome profile of the high- and low-embryogenic ortets at the callus and somatic embryoid stages. This study examined the DEGs between the high- and low-embryogenic ortets at the callus and several embryoid stages. Functional gene identification related to embryogenesis is expected to facilitate the decision-making process in improving the efficiency of large-scale oil palm in vitro propagation.
The callus and three somatic embryoid development stages consisting of globular, scutellar, and coleoptilar (Figure 1) from four Tenera (DxP) mother palms from Deli Dura origin with high oil productivity (8–9 tons·ha−1·year−1) were used. Mother palms were coded as ortets 10818-r, 20818-r, 10119-t, and 10319-t. The ortet somatic embryoid rates performances were used as criteria for the ortet selection. The average somatic embryoid rate was 1% and used as an internal benchmark (PT. SMART Tbk, 2020). There were ortets that showed distinct somatic embryoid performance and were selected for this study. Ortets 10119-t and 10319-t were the highest embryogenic ortets, whereas ortets 10818-r and 20818-r were the lowest embryogenic ortets. Callus and somatic embryoid were obtained from immature leaves that were cultured in Murashige and Skoog medium (Murashige & Skoog, 1962) with the addition of 5% sucrose and 0.65% agar. Callus induction was conducted by subculturing immature leaf explants every 3 months during the 12 months of the incubation period, while callus was subcultured every 2 months during the 12 months of the incubation period to induce the somatic embryoid. In total, 3,500 immature leaf explants with a size of 1 × 1 cm from each ortet were cultured in callus induction media. Cellular analyses were conducted to measure the callus formation rate, somatic embryo formation rate, somatic embryo proliferation rate, and germination rate. The callus formation rate was calculated from the number of clump calli per explant, while the somatic embryo rate was calculated from the number of somatic embryoids per clump callus in each ortet. Embryoid proliferations were performed by subculturing the first clump of each somatic embryoid line every 2 months in proliferation and germination media. Embryoids were cultured in a 100-ml Erlenmeyer flask containing 15 clumps of 3–5-mm proliferated embryoid. The proliferation and germination rates of each embryoid line per subculture period were calculated based on the frequency of proliferated and germinated cultures against the number of initial cultures in the first subculture. The germinated embryoid was the embryoid that had formed at least one bud and leaf.
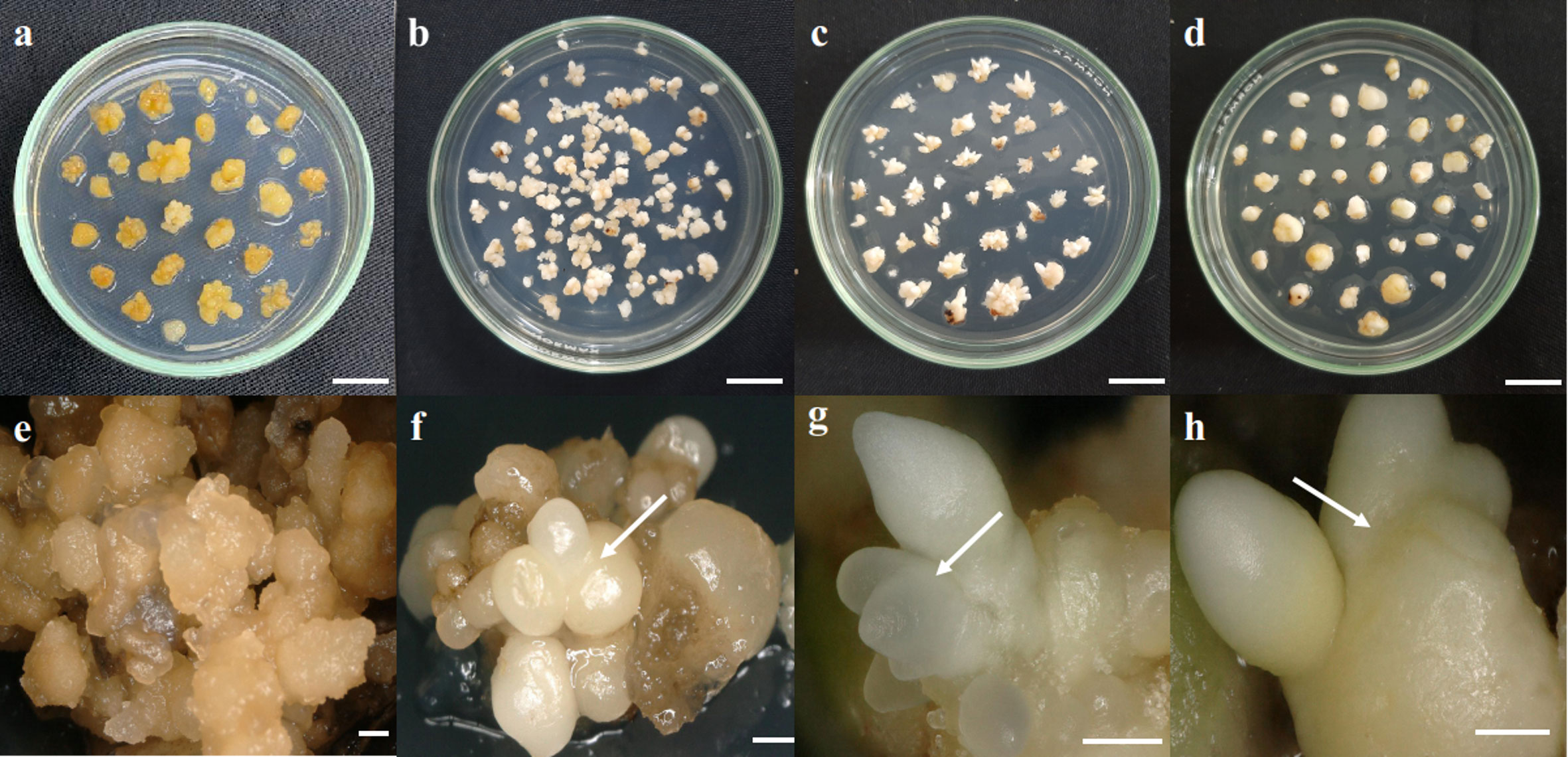
Figure 1 Somatic embryoid development stages. (A, E) Callus, (B, F) globular embryoid, (C, G) scutellar embryoid, and (D, H) coleoptilar embryoid. Scale bars: (A–D), 1 cm; (E), 1 mm; (F–H), 0.5 mm.
The identification of callus and somatic embryoid development was conducted using a Keyence VHX-6000 stereo microscope (Keyence, Osaka, Japan). The somatic embryoid of each stage was separated from each other, consisting of callus, globular, scutellar, and coleoptilar phases. Total RNA was extracted from the callus and three somatic embryoid stages using the RNeasy® Plant Mini Kit (Qiagen, Hilden, Germany) and following the manufacturer’s instructions. The total RNA of the callus was extracted from three biological replicates for each ortet. Each replication was collected by bulking five callus lines from the sixth subculture, resulting in a total of 12 samples. The total RNA of each somatic embryoid development and each ortet was extracted from three biological replicates. The first and second replications were obtained from different embryoid lines, while the third replication was obtained by bulking two embryoid lines of the first and second replications. The embryoids of each stage and ortet were collected from several flasks from the fourth to seventh subcultures derived from the same embryoid line. Thus, the total sample of embryoid stages was 36. In total, there were 48 observation units for RNA-seq. The purity of the RNA was examined using a NanoDrop™2000c Spectrophotometer (Thermo Scientific, Massachusetts, United States), and the RNA integrity number (RIN) was examined using a QC 2100 Bioanalyzer Instrument (Agilent Company, Santa Clara, CA, United States).
RNA sequencing was conducted by Novogene Co., Ltd., Beijing, China. Total RNA was used for cDNA synthesis through reverse transcriptase polymerase chain reaction (RT-PCR) for cDNA library construction. The libraries were sequenced by Illumina HiSeq 4000 (Illumina, San Diego, CA, United States). The reads were mapped to the Elaeis guineensis transcriptome reference (GCF_000442705.1_EG5) in the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov). Transcripts per million (TPM) and the number of expressed genes were counted using Kallisto software (Bray et al., 2016). Identification of DEGs with |log2fold change| > 1 and p-value < 0.05 was performed through pairwise comparisons between high- and low-embryogenic samples at each stage using DESeq2 (Love et al., 2014). DEG enrichment and gene ontology were analyzed by PlantRegMap software (Tian et al., 2020).
The callus induction (CI) rates of four ortets ranged from 11.9% to 17.0% (Figure 2A). Ortets 10119-t and 10319-t showed a lower CI rate than the internal benchmark of 16.8% (PT. SMART Tbk, 2020). Ortets 10818-r and 20818-r showed CI rates of 17.0% and 16.3%, respectively. However, their somatic embryoid rates were only 0.6% and 0.2%, respectively (Figure 2B). Ortets 10119-t and 10319-t showed high somatic embryogenesis rates at 20.1% and 33.8%, respectively, which are higher than the internal benchmark of 1.0% (PT. SMART Tbk, 2020). Based on these performances, ortets 10119-t and 10319-t were categorized as high-embryogenic palms, while ortets 10818-r and 20818-r were categorized as low-embryogenic palms. The callus induction rate of the four ortets did not show a significant difference. Significant differences from the four ortets are shown in the callus differentiation stage to form somatic embryos. These results indicated that there were differences in the callus differentiation process between high- and low-embryogenic ortets that affected embryogenesis. Thus, a molecular analysis of the callus stage was needed to reveal differences in callus differentiation between the two ortet categories.
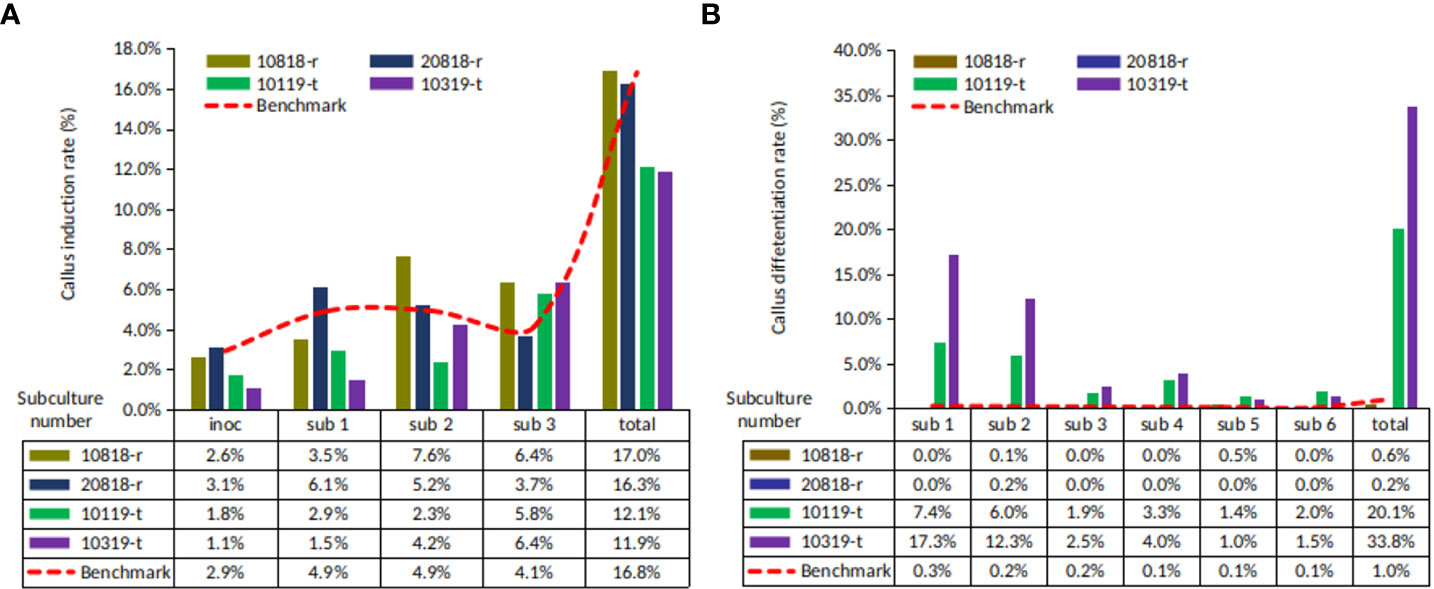
Figure 2 Callus and somatic embryoid rate from oil palm leaf explant of four ortets. (A) Callus induction rate. (B) Somatic embryoid rate. Inoc, inoculation; sub-1, first subculture; sub-2, second subculture; sub-3, third subculture, etc.
Somatic embryoid proliferation growth during subculture formed a sigmoid curve (Figure 3A). The embryoid proliferation of all ortets was increased gradually until the fifth subculture and decreased continually until the 15th subculture. High-embryogenic ortets (ortets 10119-t and 10319-t) showed a higher embryoid proliferation than low-embryogenic ortets (ortets 10818-r and 20818-r). The embryoid of high-embryogenic ortets proliferated until the 15th subculture, whereas the low-embryogenic ortets only proliferated until the 10th subculture. At the fifth subculture, high-embryogenic ortets showed the highest embryoid proliferation, increasing 18-fold from the starting point. Meanwhile, the highest embryoid proliferation of the low-embryogenic ortets was only 12-fold. The somatic embryoids reached the maturation stage and then germinated into shoots at the fourth subculture. The highest germination rates from high- and low-embryogenic ortets were observed in the eighth subculture at 25-fold and 22-fold, respectively (Figure 3B). The differences in embryoid proliferation and germination between high- and low-embryogenic ortets indicated differences in embryoid development. These results indicated the requirement for a molecular analysis of embryoid development to reveal the differences between the two ortet categories.
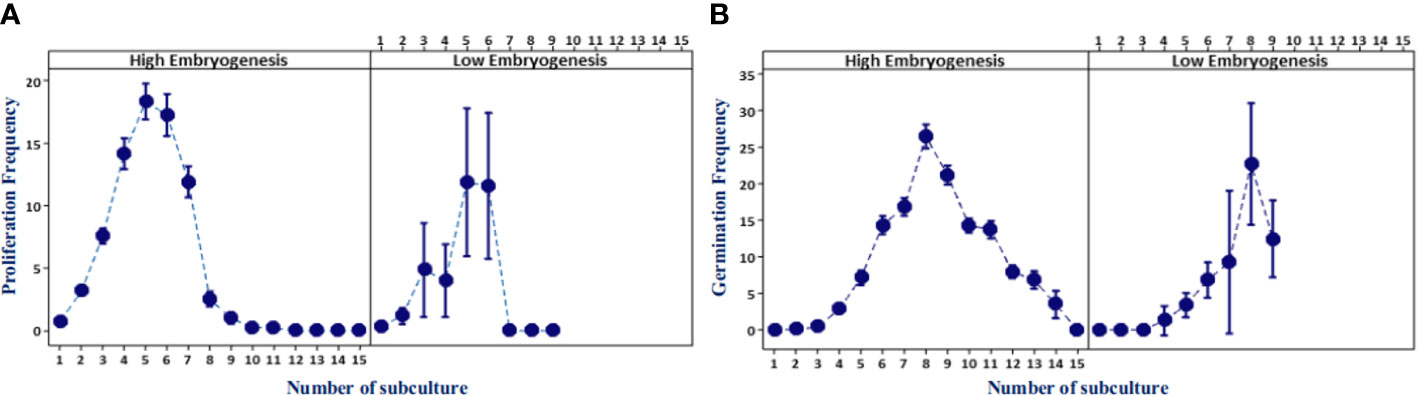
Figure 3 Embryoid proliferation and germination frequency of high- and low-embryogenic ortets. (A) Embryoid proliferation. (B) Embryoid germination. Two ortets represent the high embryogenesis category, ortets 10119-t and 10319-t. Two ortets represent the low embryogenesis category, ortets 10818-r and 20818-r.
In total, 1,911 DEGs were identified between high- and low-embryogenic ortets at the callus and embryoid stages. The DEG distribution was presented in a volcano plot (Figure 4). Gene outer line |log2FC| > 1 and p-value < 0.05 were considered to be differentially expressed. As far as the distance of the DEG distribution from the |log2FC| line, the log2 fold change value will be higher. The p-value showed the significance level of the DEGs transcript to the total transcript of all samples, which described the transcript abundance of DEGs compared to the total transcript. Meanwhile, the log2FC value showed the level of DEG fold change between samples.
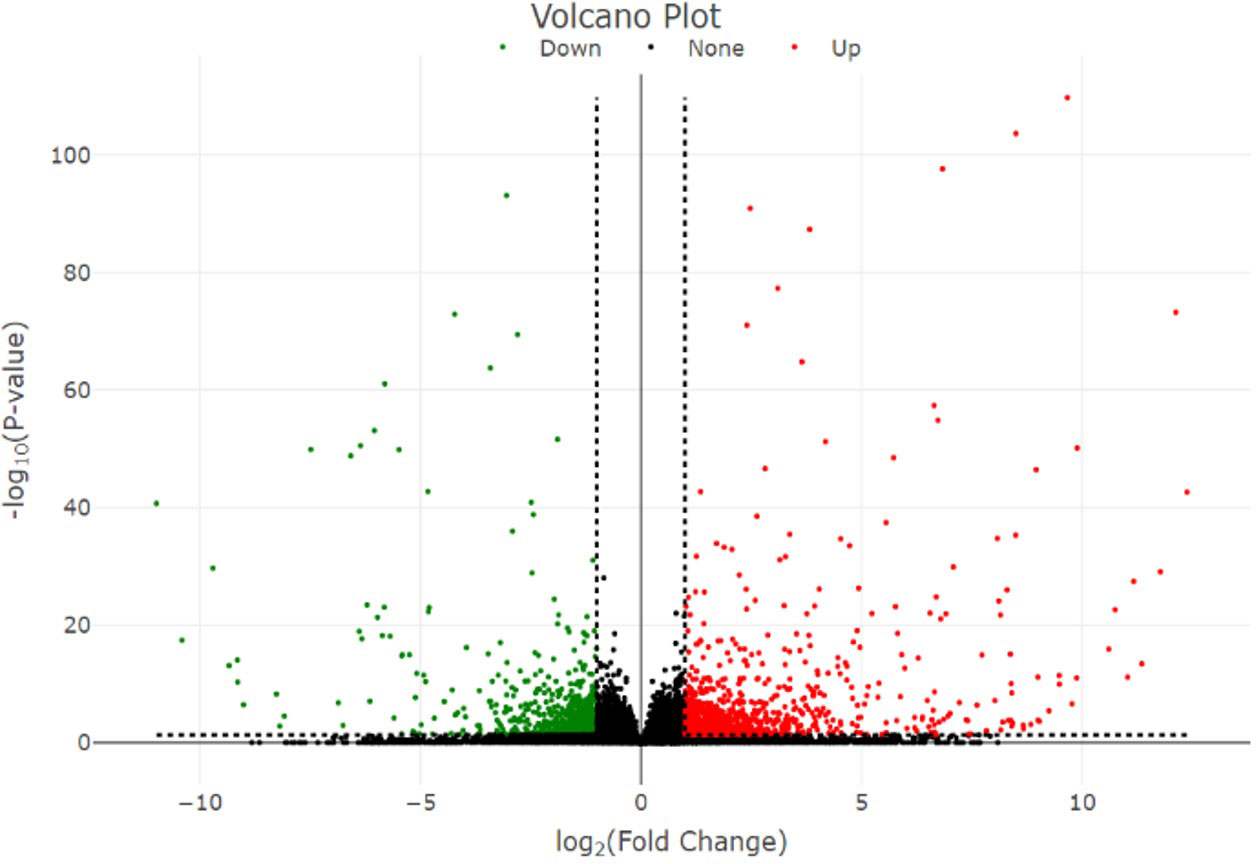
Figure 4 Volcano plot of the DEGs. Volcano plot represents the distribution of significant genes. The y-axis represents the significant level of gene expression between samples measured by the p-value, while the x-axis represents the fold change (log2 fold change) of DEGs between high- and low-embryogenic samples. DEGs, differentially expressed genes.
The gene expression in high- and low-embryogenic ortets is shown in the heatmap (Figure 5). The heatmap displays two major clusters on the x-axis. The first cluster contained low-embryogenic ortets, while the second contained high-embryogenic ortets. One group of DEGs exhibited higher expression in the high-embryogenic ortets but lower expression in the low-embryogenic ortets, while the other group of DEGs showed higher expression in the low-embryogenic ortets but lower expression in the high-embryogenic ortets. This result demonstrated the differences in transcriptome profiles of high- and low-embryogenic ortets at the callus and embryoid stages. Principal component analysis (PCA) also grouped the sample into two major groups, i.e., the high embryogenesis group (red) and the low embryogenesis group (blue), based on the number of transcripts per million DEGs (Figure 6A). Callus was grouped separately from globular, scutellar, and coleoptilar embryoid. This result indicated that transcriptome profiles of embryoid development were more similar to each other and different from the callus transcriptome profile.
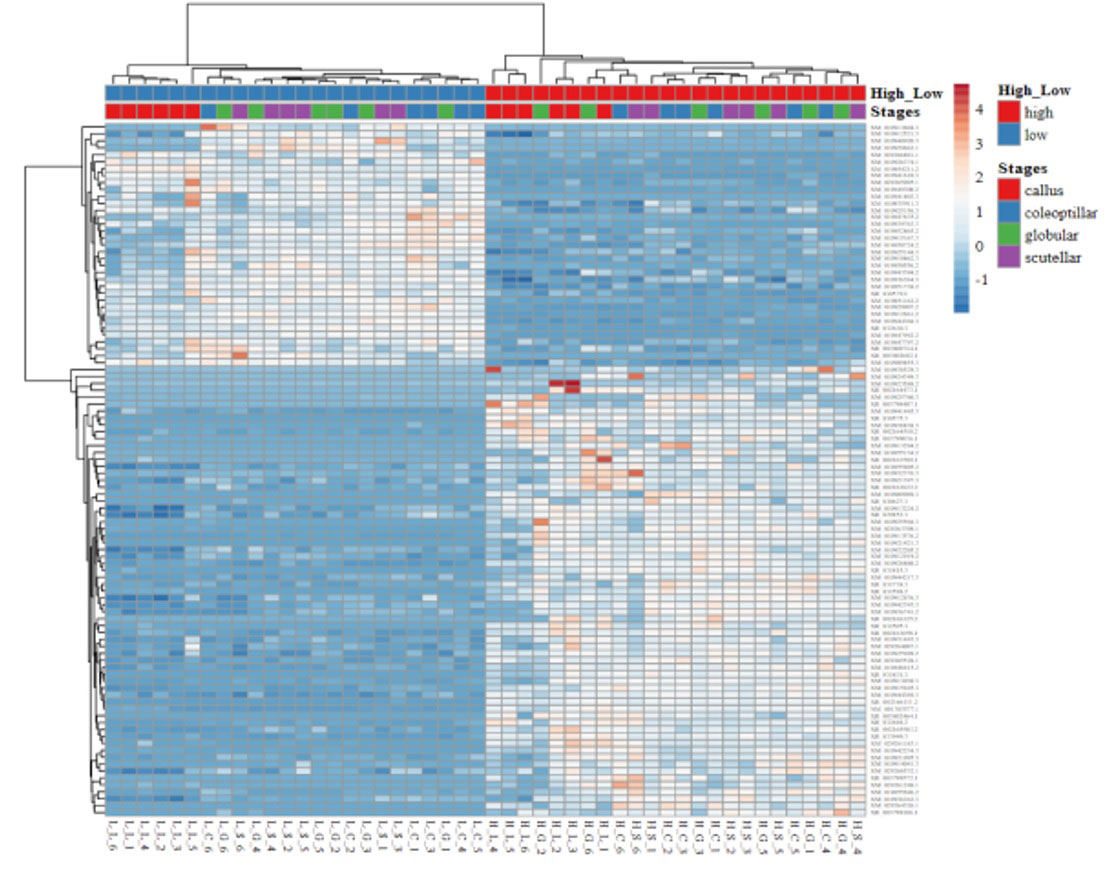
Figure 5 Heatmap of the DEGs. Heatmap of gene expression profiles of high- and low-embryogenic samples based on the transcript per million (TPM) of top 100 DEGs based on p-value. DEGs, differentially expressed genes.
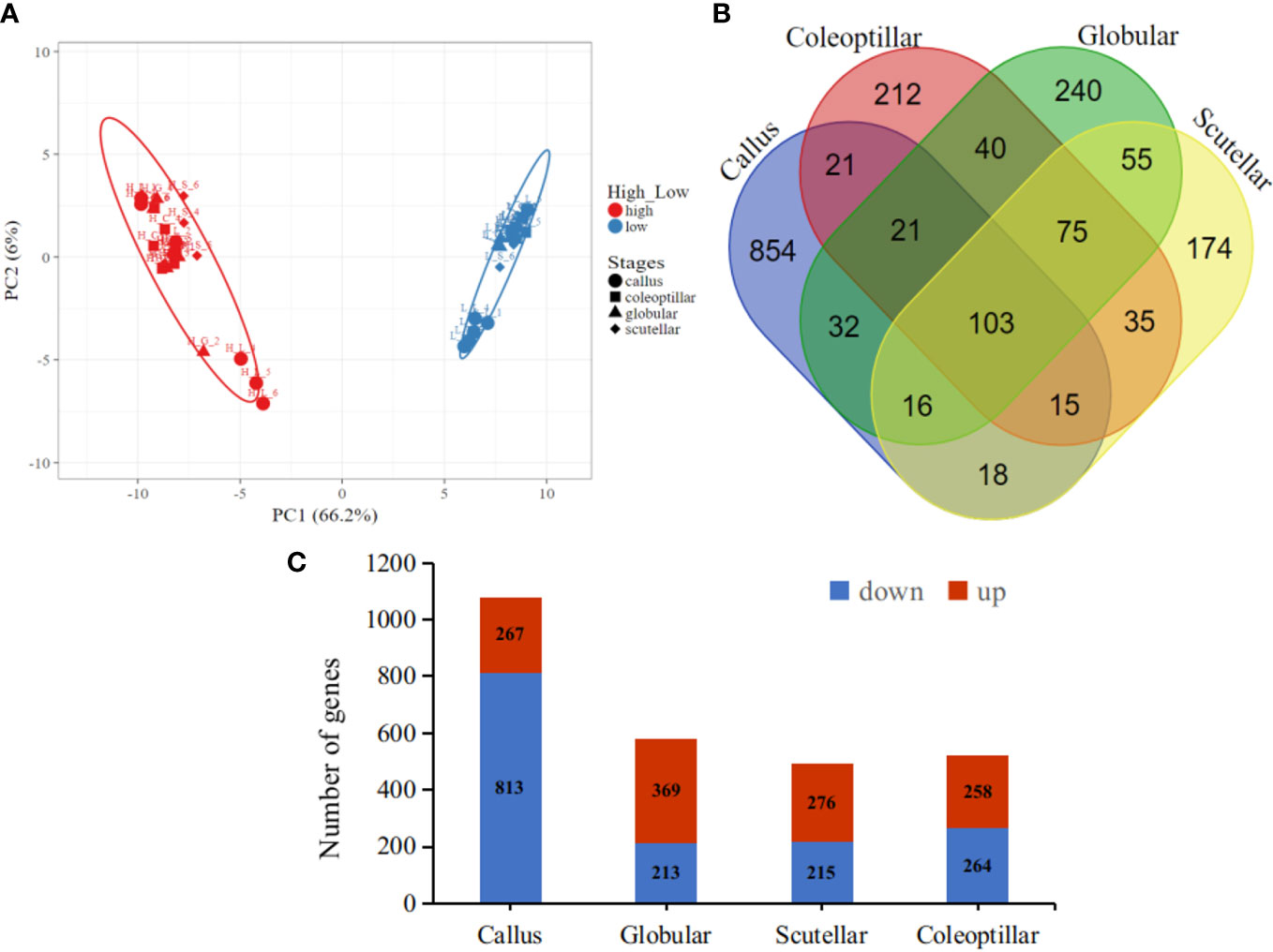
Figure 6 Transcriptome profiles of the DEGs. (A) PCA of the callus and somatic embryoid based on the transcript per million (TPM) of DEG, high embryogenesis (red) and low embryogenesis (blue). (B) Venn DEG diagrams of four somatic embryogenesis stages. (C) The number of upregulated and downregulated DEGs at each embryogenesis stage. DEGs, differentially expressed genes; PCA, principal component analysis.
In total, 103 DEGs were expressed in all somatic embryogenesis stages. Furthermore, 854, 240, 174, and 212 DEGs were specifically expressed in the callus, globular, scutellar, and coleoptilar stages, respectively (Figure 6B). The callus stage had the highest number of DEGs when compared to the other stages. Most DEGs at the callus stage were downregulated in high-embryogenic ortets, while most DEGs at embryoid development stages were upregulated in high-embryogenic ortets (Figure 6C). This result indicated that the transcriptome profile of the somatic embryoid induction stage was different from that of the somatic embryoid development stage. This result showed that more genes were involved at the embryoid induction stages than at the embryoid developmental stages.
The DEGs of high- and low-embryogenic ortets were grouped into 425 gene ontology (GO), consisting of 49 cellular components (11.5%), 131 molecular functions (30.8%), and 245 biological processes (57.6%) (Table 1). The “single organism process” GO term (GO:0044699) showed the highest number of genes in the biological processes category, followed by “single-organism metabolic process” (GO:0044710) and “metabolic process” (GO:0008152). The “response to stimulus” GO term (GO:0050896) was a sub-category that also showed a high number of genes. This pattern suggests that the appearance of “response to stimulus” GO is in accordance with the environmental factors during the tissue culture process that involved various physical and chemical stimuli such as light, temperature, humidity, and media hormone concentration.
The gene candidates involved in oil palm somatic embryogenesis were determined by the highest value of |log2FC| of DEGs and their physiological function (Table 2). The top DEGs based on |log2FC| were elongation factor 1-alpha (EF1a), vicilin-like seed storage protein At2g28490, nuclear pore complex protein (NUP1), uncharacterized LOC105049880, protein NRT1/PTR FAMILY 8.3 (NPF), multiple myeloma tumor-associated protein 2 homolog (MMTAG2), AP2-like ethylene-responsive transcription factor AIL7 (AP2/ERF AIL7), late embryogenesis abundant (LEA), DEAD-box ATP-dependent RNA helicase 28 (DDX28), and homeobox-leucine zipper protein ROC2 (HD-ZIP). The expression of these genes based on the TPM value showed a significant difference between high- and low-embryogenic ortets (Figure 7). The expression level of the top genes was varied, but most of them were upregulated in high-embryogenic ortets, i.e., EF1a, vicilin-like protein, NPF, MMTAG2, AP2/ERF AIL7, LEA, DDX28, and HD-ZIP. In contrast, the two genes were downregulated, i.e., NUP1 and uncharacterized LOC105049880.
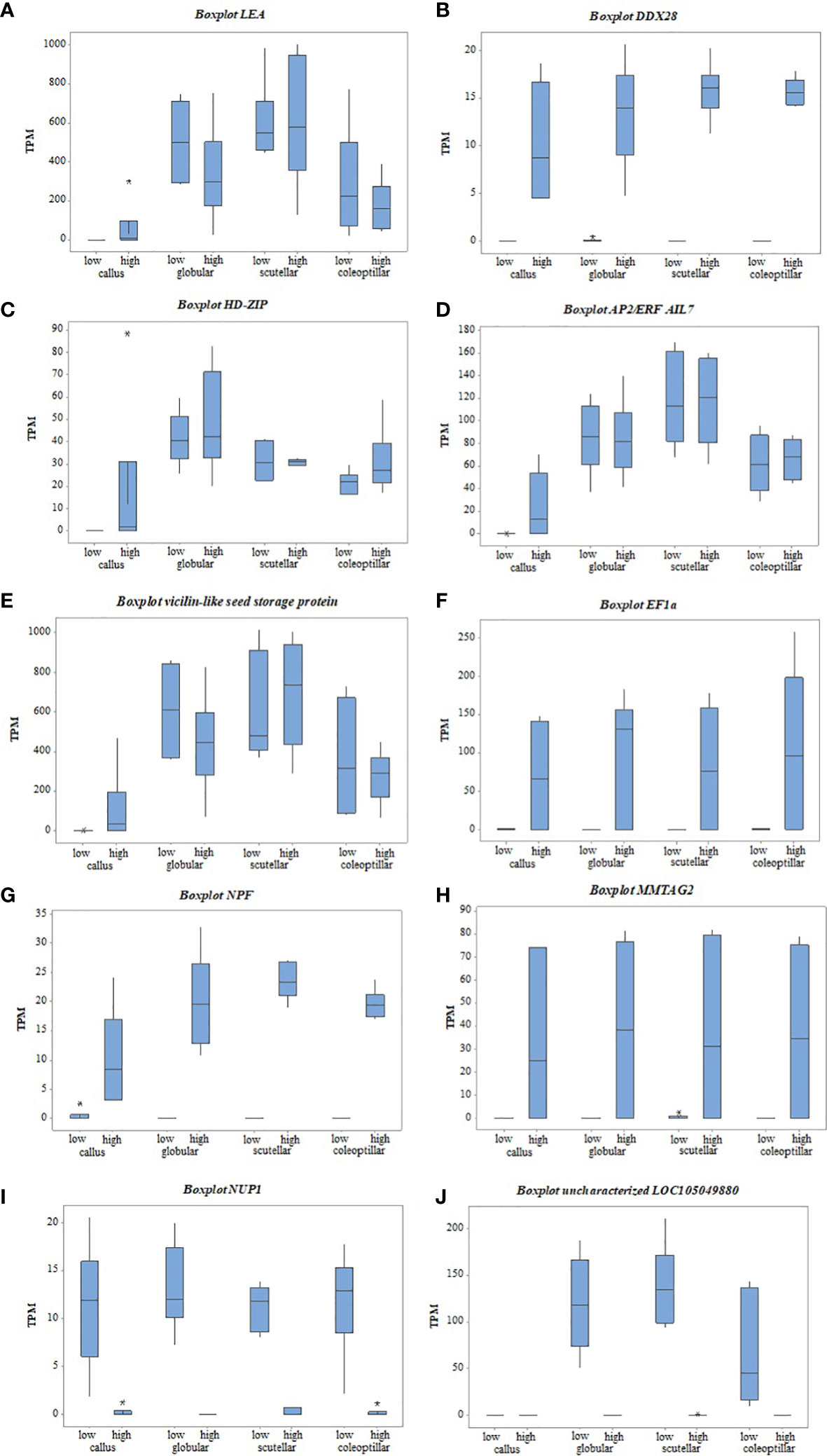
Figure 7 The gene expression of top DEGs between high- and low-embryogenic ortets at callus and embryoid stages based on TPM values. (A) LEA gene, (B) DDX28 gene, (C) HD-ZIP gene, (D) AP2/ERF AIL7 gene, (E) Vicilin-like seed gene, (F) EF1a gene, (G) NPF gene, (H) MMTAG2 gene, (I) NUP1 gene, (J) uncharacterized LOC105049880 gene. DEGs, differentially expressed genes; TPM, transcript per million.
Transcriptome analysis of callus revealed that several top DEGs were upregulated in the high-embryogenic ortets and absent in the low-embryogenic ortets, i.e., LEA, DDX28, and HD-ZIP (Figures 7A-C). The expression of LEA increased gradually from callus to globular, scutellar, and coleoptilar (Figure 7A). It could be suggested that LEA was more involved in somatic embryoid development than in embryoid induction. However, the expression of this gene was lacking in the callus of the low-embryogenic ortets. Hence, it might indicate that this gene is necessary for embryogenic induction. The absence of DDX28 and HD-ZIP expression (Figures 7B, C) in the callus of low-embryogenic ortets may influence the embryogenic potential, but this requires more detailed investigation. In this study, HD-ZIP was expressed in all stages of somatic embryoid development in both embryogenic categories but was absent in the callus of low-embryogenic ortets.
The top DEGs that were upregulated in the high-embryogenic ortets but downregulated in the low-embryogenic ortets were AP2/ERF, vicilin-like protein, EF1, NPF, and MMTAG2. The expression of AP2/ERF and vicilin-like protein continually increased from callus to globular, scutellar, and coleoptilar stages in either high- or low-embryogenic ortets, but the significant difference only occurred in the callus stage (Figures 7D, E). This pattern suggested that AP2/ERF and vicilin-like protein were involved in all stages of the somatic embryoid, but the low expression of this gene in the callus stage might result in low-embryogenic induction.
The identification of the DEGs in the embryoid development stages, i.e., globular, scutellar, and coleoptilar, was necessary to analyze the low proliferation and germination of embryoids in low-embryogenic ortets. EF1a, NPF, and MMTAG2 were identified as DEGs at callus and embryoid development. These genes were upregulated in the high-embryogenic ortets and downregulated in the low-embryogenic ortets at callus and embryoid stages (Figures 7F-H). The downregulation of these genes in the low-embryogenic ortets might influence not only embryoid induction but also embryoid development, i.e., embryoid proliferation and germination. Meanwhile, there were two downregulated DEGs in the high-embryogenic ortets, i.e., NUP1 and uncharacterized LOC105049880 (Figures 7I, J).
Calli were formed from leaf explants through cell dedifferentiation. The callus induction rate of four ortets ranged from 11.9% to 17.0%. The significant difference between the four ortets was shown in callus differentiation to form somatic embryoids. Two ortets showed high embryogenesis performance, i.e., 10119-t and 10319-t; the other two ortets showed low embryogenesis performance, i.e., 10818-r and 20818-r. Callus differentiation involves the transition from callus cells to stem-like cells that will initiate embryoids (Rose et al., 2022). Auxin gradients at the transition stage from callus to embryonic stem-like cells promoted embryonic cell induction (Chen et al., 2022). This process stimulated transcription factors for early embryoid development that induced the activation of somatic embryo-related genes.
The expression of several DEGs was upregulated in the high-embryogenic ortets and absent in the low-embryogenic ortets i.e., LEA, DDX28, and HD-ZIP (Figures 7A-C). LEA was also identified as DEGs between embryogenic and non-embryogenic calli by microarray (Budinarta et al., 2012). The expression of this gene was validated with q-PCR and showed that the LEA expression was higher at the embryoid development stages than at the callus stage (Syariyanto et al., 2018), and we obtained a similar result in this study. Another study revealed that LEA plays an important role in zygotic as well as somatic embryogenesis through ABA treatment (Ikeda et al., 2006). A recent study on oil palm leaf transcriptome profiling reported that DDX28 was also differentially expressed between high- and low-embryogenic ortets (Ooi et al., 2021). Based on in silico analysis in rice, the member of DDX28 interacted with the pathway of ABA phytohormone signaling (Macovei et al., 2012). In addition to auxin, ABA was the predominant phytohormone that affected embryogenesis potential. In wheat and barley, somatic embryogenesis potential is determined by the level of indoleacetic and abscisic acids (Seldimirova et al., 2019). In the oil palm, DDX28 is also involved in inflorescence sex determination (Ho et al., 2016). In another research, HD-ZIP was reported to be highly expressed during early oil palm somatic embryogenesis (Ooi et al., 2016). HD-ZIP is the transcription factor that has a role in various plant growth functions, plant adaptation to several environmental stressors, and plant growth regulator pathways (Hong et al., 2021). In Arabidopsis, HD-ZIP is a transcription factor that controls the regulation of brassinosteroid-related-homeobox 3 (BHB3). Brassinosteroid is a phytohormone that plays an important role as a plant growth regulator (Hasegawa et al., 2022). Based on the results, our study showed that most of the DEGs were associated with phytohormone regulation, especially with ABA.
Some DEGs were expressed in the callus of high-embryogenic ortets but downregulated in the callus of low-embryogenic ortets, i.e., AP2/ERF and vicilin-like protein (Figures 7D, E). AP2/ERF and vicilin-like protein were involved in all stages of somatic embryoids. However, the low expression of this gene in the callus of low-embryogenic ortets might result in low-embryogenic induction. A transcription factor from apetala2 (AP2) family, such as BABY BOOM (BBM), plays an important role in cell proliferation and induction of somatic embryogenesis in Arabidopsis thaliana and Brassica napus (Boutilier et al., 2002). Another study reported that AP2/ERF is involved in various primary metabolic processes as well as secondary metabolic processes, growth and development of the plant, and response to environmental stress (Licausi et al., 2013). Based on Basic Local Alignment Search Toll (BLAST) data, AP2-ERF AIL7 protein in NCBI showed a functional domain of AP2-type that was similar to BBM family in A. thaliana and B. napus with homology of 75.21% and 80.95%, respectively. The involvement of the AP2 family in embryogenic callus and somatic embryo development was also reported in oil palm (Morcillo et al., 2007). Moreover, in Arabidopsis, overexpression of the AINTEGUMENTA-LIKE (AIL) transcription factors promotes embryogenesis and organogenesis (Magnani et al., 2017). A similar pattern with AP2/ERF expression was exhibited by vicilin-like protein gene. Vicilin-like protein encodes a storage protein located in the chloroplast as well as cytoplasm (Van de Vondel et al., 2020) that is found in cereal seeds and nuts (Burrieza et al., 2019). Vicilin-like proteins play roles in seed growth and response to external stress (Ma et al., 2022). Vicilin-like protein is also necessary for LEA protein coding for seed germination that was provided by ABA (Hu & Ma, 2006).
Some DEGs were significant not only in the callus stage but also in embryoid development, i.e., EF1a, NPF, and MMTAG2. The downregulation of these genes in the low-embryogenic ortets might influence not only embryoid induction but also embryoid development, i.e., embryoid proliferation and germination. Another study reported that EF1a was expressed during somatic embryogenesis and germination of the Liriodendron hybrid (Li et al., 2021). The involvement of EF1a in somatic embryo maturation was also reported in Norway spruce (Picea abies) (Wang-Lougheed et al., 2004). EF1a is a member of the GTP binding protein (guanine nucleotide-binding protein) that plays a role in protein synthesis, actin filaments, and microtubules during the cell cycle (Li et al., 2021). The main function of this protein was to transport tRNA to the ribosome. Furthermore, NPF plays a role in the transport of various substrates, including nitrates and several hormones. NPF acts as a nitrate sensor that affects auxin transport. The auxin transport by NPF is inhibited by certain nitrate concentrations (Corratgé-Faillie & Lacombe, 2017). In Arabidopsis, NPF plays a role in the nitrogen content of the somatic embryo and is highly expressed at the maturation stage (Leran et al., 2015). The nitrogen content influences the protein content in the somatic embryoid of Glycine max (L.) (Truong et al., 2013). Then, the protein content determined the quality of the mature somatic embryoid of Medicago sativa (L.) (Lai & Mckersie, 1994). Another study reported that nitrogen supply affected the germination of the somatic embryoid of P. abies (Carlsson et al., 2018). The function of MMTAG2 in oil palm in addition to being related to somatic embryogenesis is not well characterized. MMTAG2 is reported to be involved in the mechanism of resistance to gummy stem blight (GSB) disease and is used as a marker-assisted selection in melon plant breeding (Han et al., 2022). MMTAG2 causes DNA imbalance and encourages the activation of DNA repair pathways caused by the production of reactive oxygen species (ROS) (Saitoh & Oda, 2021). ROS are involved in embryogenesis during the dedifferentiation of somatic cells into callus cells (Rose et al., 2022).
Two DEGs were downregulated in high-embryogenic ortets and upregulated in low-embryogenic ortets, i.e., NUP1 and uncharacterized LOC105049880. NUP is a nuclear membrane protein that regulates ABA in order to respond the abiotic stress (Zhu et al., 2017). NUP1 is necessary for miRNA transport from the nucleus to the cytoplasm. The decrease in NUP1 expression resulted in the inhibition of the miRNA transport (Zhang et al., 2020). NUP1 interacts with some locus in the chromosome that could inhibit, activate, and regulate the expression of several genes (Sumner & Brickner, 2022). The nuclear pore complex (NPC) mutant was reported to inhibit the somatic embryo development in Arabidopsis. It was because the signaling pathway of some phytohormones was sensitive to the interference of NPC (Wu et al., 2022). The function of uncharacterized LOC105049880 gene in addition to being related to somatic embryogenesis is not well characterized.
The interaction of appropriate genotypes and the expression of specific genes from each embryogenesis stage promote embryoid induction. The biological stages of embryogenic induction consisted of dedifferentiation, embryogenic stem cells, and early embryo development (Rose et al., 2022). Each step involved a specific gene that supports somatic embryogenesis. The transition of callus cells to stem-like cells required an auxin gradient and involved genes related to auxin signaling and transport (Chen et al., 2022). In this study, NPF, a gene related to auxin transport, was identified as DEGs between high- and low-embryogenic ortets. The differential expression of auxin transport genes might contribute to the determination of auxin levels between high- and low-embryogenic ortets. In Medicago truncatula, the auxin gradient was described as high in early callus development and proliferation but degraded at the transition of callus to embryogenic stem cells (Chen et al., 2022). Auxin induced the expression of genes that modified the genetic program of somatic cells and regulated the transition of somatic cells to somatic embryo development (Loyola-Vargas & Ochoa-Alejo, 2016). The use of auxins such as 2,4-D was reported to increase the expression of a transcription factor for somatic embryo induction (Shivani et al., 2017).
The degradation of auxin levels in the transition of callus to embryogenic stem cells and the increase of ABA in this stage promoted high somatic embryogenesis (Chen et al., 2022). Several DEGs were related to ABA signaling, which were LEA, DDX28, vicilin-like protein, and NUP1. ABA has an important role in the induction of somatic embryoids. Several plants required exogenous ABA in the medium to enhance the somatic embryoid (Ikeda et al., 2006; Méndez-Hernández et al., 2019). ABA displayed additive effects on promoting cell fate transition from callus cells to embryogenic stem cells (Chen et al., 2022). The application of ABA in oil palm tissue culture is reported to increase the maturation of somatic embryos (Teixeira et al., 1994; Bertossi et al., 2001; Jayanthi et al., 2011). However, its application in somatic embryo induction has not been reported. In this study, several DEGs were associated with ABA signaling in the callus stage. This result indicated that ABA is involved in not only embryoid development but also embryoid induction. The addition of ABA to embryoid induction media and analysis of endogenous content in the callus stage of high- and low-embryogenic ortets could be used to investigate the role of ABA in the induction of oil palm somatic embryos.
The transition of callus to embryogenic required transcription factors for further embryoid development. In this study, transcription factors AP2/ERF and HD-ZIP were identified as DEGs between high and low in the embryoid induction stages. Transcription factors of embryogenesis were very important in the somatic embryoid induction (Chu et al., 2017) and affected the expression of the somatic embryoid-related gene (Méndez-Hernández et al., 2019). The interaction of appropriate hormones and transcription was required for the embryonic stem cells to develop into embryos (Rose et al., 2022).
Cellular analysis at explant and callus stages showed that the embryogenesis rates of high-embryogenic ortets 10119-t and 10319-t were 20.1% and 33.8%, respectively, but the low-embryogenic ortets 10818-r and 20818-r were only 0.6% and 0.2%, respectively. High-embryogenic ortets have higher somatic embryoid proliferation and germination rates at 18-fold and 25-fold, respectively, but the low-embryogenic ortets were only 12-fold and 22-fold, respectively. Transcriptome analysis through RNA sequencing showed that the high-embryogenic ortets had a different transcriptome profile from the low-embryogenic ortets at callus and somatic embryoid stages based on the heatmap and PCA. A total of 103 DEGs were expressed in all embryogenesis stages; 854 DEGs were specifically expressed in the callus stage, 240 DEGs in the globular stage, 174 DEGs in the scutellar stage, and 212 DEGs in the coleoptilar stage. Embryogenesis-related gene candidates were selected based on the top |log2FC| value and their physiological function. Gene candidates were commonly associated with the regulation of hormones such as ABA, auxins, and brassinosteroids. The role of each gene indicates a physiological state associated with the potential for somatic embryogenesis. The top DEGs need to be validated in further studies.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
AS, RR, NW and TL designed the research. AS performed the research and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by PT SMART Tbk under a research project code of 3.4.1.046.
We thank the management of PT SMART Tbk, which financially supported the full project under the research code 3.4.1.046. We also thank Zulfikar Ahmad Tanjung and Hadi Septian Guna Putra for their advice on the bioinformatics and statistical analyses. Credits are also addressed to Dr. Condro Utomo, Dr. Reno Tryono, Dr. Andree SK, Dr. Wisnu Adi Wicaksono, and Victor Aprilyanto for their suggestions for the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bertossi, F., Chabrillange, N., Duval, Y. (2001). Abscisic acid and desiccation tolerance in oil palm (Elaeis guineensis jacq.) somatic embryos. Genet. Selection Evol. 33 (1), 75–84. doi: 10.1186/bf03500874
Boutilier, K., Offringa, R., Sharma, V. K., Kieft, H., Ouellet, T., Zhang, L., et al. (2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14 (8), 1737–1749. doi: 10.1105/tpc.001941
Bray, N. L., Pimentel, H., Melsted, P., Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34 (5), 525–527. doi: 10.1038/nbt.3519
Budinarta, W., Hatorangan, M. R., Joanito, I., Tanjung, Z. A., Toruan-Mathius, N. (2012). “Penapisan gen yang berperan dalam embriogenesis pada kelapa sawit (Elaeis guineensis jacq.),” in Technical annual report of tissue culture and biotechnology 2012. Eds. Nugroho, Y. A., Rohman, R. A., Utomo, C. (Tissue culture and Biotechnology, PT. SMART, Tbk).
Burrieza, H. P., Rizzo, A. J., Moura Vale, E., Silveira, V., Maldonado, S. (2019). Shotgun proteomic analysis of quinoa seeds reveals novel lysine-rich seed storage globulins. Food Chem. 293, 299–306. doi: 10.1016/j.foodchem.2019.04.098
Carlsson, J., Egertsdotter, U., Ganeteg, U., Svennerstam, H. (2018). Nitrogen utilization during germination of somatic embryos of Norway spruce : revealing the importance of supplied glutamine for nitrogen metabolism. Trees 0 (0). doi: 10.1007/s00468-018-1784-y
Chen, Y., Yu, H., Wang, Y., Li, F., Xing, Y., Ge, X. (2022). Uniconazole augments abscisic acid in promoting somatic embryogenesis in cotton (Gossypium hirsutum l.). Front. Plant Sci. 13 (April). doi: 10.3389/fpls.2022.865778
Chu, Z., Chen, J., Sun, J., Dong, Z., Yang, X., Wang, Y., et al. (2017). De novo assembly and comparative analysis of the transcriptome of embryogenic callus formation in bread wheat (Triticum aestivum l.). BMC Plant Biol. 17 (1), 1–22. doi: 10.1186/s12870-017-1204-2
Constantin, M., Nchu, W. A., Godswill, N., Made, N., Wiendi, A., Ebongue, N., et al. (2015). Induction of oil palm (Elaeis guineensis jacq. var. tenera) callogenesis and somatic embryogenesis from young leaf explants. J. Appl. Biol. Biotechnol. 3 (4), 4–10. doi: 10.7324/JABB.2015.3402
Corratgé-Faillie, C., Lacombe, B. (2017). Substrate (un)specificity of arabidopsis NRT1/PTR FAMILY (NPF) proteins. J. Exp. Bot. 68 (12), 3107–3113. doi: 10.1093/jxb/erw499
Elhiti, M., Stasolla, C., Wang, A. (2013). Molecular regulation of plant somatic embryogenesis. Vitro Cell. Dev. Biol. - Plant 49 (6), 631–642. doi: 10.1007/s11627-013-9547-3
FAO (2021) FAOSTAT. Available at: http://www.fao.org/faostat/en/#data/QD/visualize.
Han, J., Dong, S., Shi, Y., Miao, H., Liu, X., Beckles, D. M., et al. (2022). Fine mapping and candidate gene analysis of gummy stem blight resistance in cucumber stem. Theor. Appl. Genet. 135 (9), 3117–3125. doi: 10.1007/s00122-022-04172-2
Hasegawa, R., Arakawa, T., Fujita, K., Tanaka, Y., Ookawa, Z., Sakamoto, S., et al. (2022). Arabidopsis homeobox-leucine zipper transcription factor BRASSINOSTEROID-RELATED HOMEOBOX 3 regulates leaf greenness by suppressing BR signaling. Plant Biotechnol. 39 (2), 209–214. doi: 10.5511/plantbiotechnology.22.0128a
Ho, H., Low, J. Z., Gudimella, R., Tammi, M. T., Harikrishna, J. A. (2016). Expression patterns of inflorescence- and sex-specific transcripts in male and female inflorescences of African oil palm (Elaeis guineensis). Ann. Appl. Biol. 168 (2), 274–289. doi: 10.1111/aab.12263
Hong, Y., Liu, Y., Zhang, Y., Jia, L., Yang, X., Zhang, X., et al. (2021). Genome-wide characterization of homeobox-leucine zipper gene family in tomato (Solanum lycopersicum) and functional analysis of SlHDZ34 (III sub-family member) under salinity stress. Environ. Exp. Bot. 192 (September), 104652. doi: 10.1016/j.envexpbot.2021.104652
Hu, W., Ma, H. (2006). Characterization of a novel putative zinc finger gene MIF1: involvement in multiple hormonal regulation of arabidopsis development. Plant J. 45 (3), 399–422. doi: 10.1111/j.1365-313X.2005.02626.x
Ikeda, M., Umehara, M., Kamada, H. (2006). Embryogenesis-related genes; its expression and roles during somatic and zygotic embryogenesis in carrot and arabidopsis. Plant Biotechnol. 23 (2), 153–161. doi: 10.5511/plantbiotechnology.23.153
Jayanthi, M., Mohan, N. M., Mandal, P. K. (2011). Direct somatic embryogenesis and plantlet regeneration in oil palm. J. Plant Biochem. Biotechnol. 20 (2), 249–251. doi: 10.1007/s13562-011-0053-6
Karami, O., Saidi, A. (2010). The molecular basis for stress-induced acquisition of somatic embryogenesis. Mol. Biol. Rep. 37 (5), 2493–2507. doi: 10.1007/s11033-009-9764-3
Kukurba, K. R., Montgomery, S. B. (2015). RNA Sequencing and analysis. Cold Spring Harbor Protoc. 2015 (11), 951–969. doi: 10.1101/pdb.top084970
Lai, F., Mckersie, B. D. (1994). Regulation of starch and protein accumulation in alfalfa (Medicago sativa l.) somatic embryos. Plant Sci. 100, 211–219. doi: 10.1016/0168-9452(94)90078-7
Leran, S., Garg, B., Boursiac, Y., Corratge-Faillie, C., Brachet, C., Tillard, P., et al. (2015). AtNPF5.5, a nitrate transporter affecting nitrogen accumulation in arabidopsis embryo. Sci. Rep. 5, 1–7. doi: 10.1038/srep07962
Li, T., Yuan, W., Qiu, S., Shi, J. (2021). Selection of reference genes for gene expression analysis in liriodendron hybrids’ somatic embryogenesis and germinative tissues. Sci. Rep. 11 (1), 1–12. doi: 10.1038/s41598-021-84518-w
Licausi, F., Ohme-Takagi, M., Perata, P. (2013). APETALA2/Ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199 (3), 639–649. doi: 10.1111/nph.12291
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 1–21. doi: 10.1186/s13059-014-0550-8
Low, E. T. L., Alias, H., Boon, S. H., Shariff, E. M., Tan, C. Y. A., Ooi, L. C. L., et al. (2008). Oil palm (Elaeis guineensis jacq.) tissue culture ESTs: identifying genes associated with callogenesis and embryogenesis. BMC Plant Biol. 8, 1–19. doi: 10.1186/1471-2229-8-62
Loyola-Vargas, V. M., Ochoa-Alejo, N. (2016). “Somatic embryogenesis. an overview,” in Somatic embryogenesis: fundamental aspects and applications (Cham: Springer), 1–10. doi: 10.1007/978-3-319-33705-0
Ma, J., Pan, C., Chen, H., Chen, W., Chen, W., Zhang, M., et al. (2022). Insight of the functional and biological activities of coconut (Cocos nucifera l.) protein by proteomics analysis and protein-based bioinformatics. Molecules 27 (9), 2987. doi: 10.3390/molecules27092987
Macovei, A., Vaid, N., Tula, S., Tuteja, N. (2012). A new DEAD-box helicase ATP-binding protein (OsABP) from rice is responsive to abiotic stress. Plant Signaling Behav. 7 (9), 1138–1143. doi: 10.4161/psb.21343
Magnani, E., Jiménez-Gómez, J. M., Soubigou-Taconnat, L., Lepiniec, L., Fiume, E. (2017). Profiling the onset of somatic embryogenesis in arabidopsis. BMC Genomics 18 (1), 1–12. doi: 10.1186/s12864-017-4391-1
Méndez-Hernández, H. A., Ledezma-Rodríguez, M., Avilez-Montalvo, R. N., Juárez-Gómez, Y. L., Skeete, A., Avilez-Montalvo, J., et al. (2019). Signaling overview of plant somatic embryogenesis. Front. Plant Sci. 10 (February). doi: 10.3389/fpls.2019.00077
Morcillo, F., Gallard, A., Pillot, M., Jouannic, S., Aberlenc-Bertossi, F., Collin, M., et al. (2007). EgAP2-1, an AINTEGUMENTA-like (AIL) gene expressed in meristematic and proliferating tissues of embryos in oil palm. Planta 226 (6), 1353–1362. doi: 10.1007/s00425-007-0574-3
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarium 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Ooi, S. E., Feshah, I., Nuraziyan, A., Sarpan, N., Ata, N., Lim, C. C., et al. (2021). Leaf transcriptomic signatures for somatic embryogenesis potential of elaeis guineensis. Plant Cell Rep. 40, 1141–1154. doi: 10.1007/s00299-021-02698-1
Ooi, S. E., Ramli, Z., Syed Alwee, S. S. R., Kulaveerasingam, H., Ong-Abdullah, M. (2016). EgHOX1, a HD-zip II gene, is highly expressed during early oil palm (Elaeis guineensis jacq.) somatic embryogenesis. Plant Gene 8, 16–25. doi: 10.1016/j.plgene.2016.09.006
PT. SMART Tbk (2020). Annual progress report: tissue culture department (Plant Production and Biotechnology Division).
Rajesh, M. K., Fayas, T. P., Naganeeswaran, S., Rachana, K. E., Bhavyashree, U., Sajini, K. K., et al. (2016). De novo assembly and characterization of global transcriptome of coconut palm (Cocos nucifera l.) embryogenic calli using illumina paired-end sequencing. Protoplasma 253 (3), 913–928. doi: 10.1007/s00709-015-0856-8
Roowi, S. H., Ho, C. L., Alwee, S. S. R. S., Abdullah, M. O., Napis, S. (2010). Isolation and characterization of differentially expressed transcripts from the suspension cells of oil palm (Elaeis guineensis jacq.) in response to different concentration of auxins. Mol. Biotechnol. 46 (1), 1–19. doi: 10.1007/s12033-010-9262-9
Rose, R. J., Chen, Y., Yu, H., Wang, Y., Li, F., Xing, Y., et al. (2022). Somatic embryogenesis in the medicago truncatula model: cellular and molecular mechanisms. Front. Plant Sci. 10 (March). doi: 10.3389/fpls.2022.865778
Saitoh, T., Oda, T. (2021). Dna damage response in multiple myeloma: the role of the tumor microenvironment. Cancers 13 (3), 1–22. doi: 10.3390/cancers13030504
Seldimirova, O. A., Kudoyarova, G. R., Kruglova, N. N., Galin, I. R., Veselov, D. S. (2019). Somatic embryogenesis in wheat and barley calli in vitro is determined by the level of indoleacetic and abscisic acids. Russ. J. Dev. Biol. 50, 3, 124–135. doi: 10.1134/S1062360419030056
Shearman, J. R., Jantasuriyarat, C., Sangsrakru, D., Yoocha, T., Vannavichit, A., Tragoonrung, S., et al. (2013). Transcriptome analysis of normal and mantled developing oil palm flower and fruit. Genomics 101 (5), 306–312. doi: 10.1016/j.ygeno.2013.02.012
Shivani Awasthi, P, Sharma, V, Kaur, N, Kaur, N, Pandey, P, et al. (2017). Genome-wide analysis of transcription factors during somatic embryogenesis in Banana (Musa Spp.) Cv. Grand Naine. PLoS ONE. 12 (8), 1–19. doi: 10.1371/journal.pone.0182242
Sumner, M. C., Brickner, J. (2022). The nuclear pore complex as a transcription regulator. Cold Spring Harb. Perspect. Biol. 14 (7). doi: 10.1101/cshperspect.a039438
Syariyanto, N., Widyastuti, U., Mathius, N. T. (2018). Gene expression analysis of somatic embryogenesis in oil palm. Int. J. Oil Palm 1 (1), 38–49. doi: 10.35876/ijop.v1i1.5
Teixeira, J. B., Söndahl, M. R., Kirby, E. G. (1994). Somatic embryogenesis from immature inflorescences of oil palm. Plant Cell Rep. 13 (5), 247–250. doi: 10.1007/BF00233313
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J., Gao, G. (2020). PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48 (D1), D1104–D1113. doi: 10.1093/nar/gkz1020
Truong, Q., Koch, K., Yoon, J. M., Everard, J. D., Shanks, J. V. (2013). Influence of carbon to nitrogen ratios on soybean somatic embryo (cv. jack) growth and composition. J. Exp. Bot. 64, 10, 2985–2995. doi: 10.1093/jxb/ert138
Van de Vondel, J., Lambrecht, M. A., Delcour, J. A. (2020). Osborne Extractability and chromatographic separation of protein from quinoa (Chenopodium quinoa willd.) wholemeal. Food Sci. Technol. 126, 1–8. doi: 10.1016/j.lwt.2020.109321
Wang-Lougheed, W., Beardmore, T., Park, Y., Adams, G. (2004). Use of KN1, EF-1a, 2S seed storage protein and LEA genes as early indicators of somatic embryo maturation in Norway spruce (Picea abies). In Vitro Cell. Dev. Biol. 40, 60A.
Weckx, S., Inzé, D., Maene, L. (2019). Tissue culture of oil palm: finding the balance between mass propagation and somaclonal variation. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00722
Wu, X., Han, J., Changkui, G. (2022). Function of nuclear pore complexes in regulation of plant defense signaling. Int. J. Mol. Sci. 23 (6), 1–17. doi: 10.3390/ijms23063031
Zhang, B., You, C., Zhang, Y., Zeng, L., Hu, J., Zhao, M., et al. (2020). Linking key steps of microRNA biogenesis by TREX-2 and the nuclear pore complex in arabidopsis. Nat. Plants 6 (8), 957–969. doi: 10.1038/s41477-020-0726-z
Keywords: differentially expressed genes, globular embryoid, high embryogenic, oil palm, somatic embryogenesis, RNA-seq
Citation: Sahara A, Roberdi R, Wiendi NMA and Liwang T (2023) Transcriptome profiling of high and low somatic embryogenesis rate of oil palm (Elaeis guineensis Jacq. var. Tenera). Front. Plant Sci. 14:1142868. doi: 10.3389/fpls.2023.1142868
Received: 12 January 2023; Accepted: 20 April 2023;
Published: 12 May 2023.
Edited by:
Ying Hua Su, Shandong Agricultural University, ChinaReviewed by:
Kelvin Kamfwa, University of Zambia, ZambiaCopyright © 2023 Sahara, Roberdi, Wiendi and Liwang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Asri Sahara, YmlvdGVjaG5vbG9neUBzaW5hcm1hcy1hZ3JpLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.