
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 13 June 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1142211
This article is part of the Research Topic Identification and Functional Analysis of Differentially Expressed Genes in Plant Response to Abiotic Stresses View all 24 articles
Rusty root syndrome is a common and serious disease in the process of Panax ginseng cultivation. This disease greatly decreases the production and quality of P. ginseng and causes a severe threat to the healthy development of the ginseng industry. However, its pathogenic mechanism remains unclear. In this study, Illumina high-throughput sequencing (RNA-seq) technology was used for comparative transcriptome analysis of healthy and rusty root-affected ginseng. The roots of rusty ginseng showed 672 upregulated genes and 526 downregulated genes compared with the healthy ginseng roots. There were significant differences in the expression of genes involved in the biosynthesis of secondary metabolites, plant hormone signal transduction, and plant–pathogen interaction. Further analysis showed that the cell wall synthesis and modification of ginseng has a strong response to rusty root syndrome. Furthermore, the rusty ginseng increased aluminum tolerance by inhibiting Al entering cells through external chelating Al and cell wall-binding Al. The present study establishes a molecular model of the ginseng response to rusty roots. Our findings provide new insights into the occurrence of rusty root syndrome, which will reveal the underlying molecular mechanisms of ginseng response to this disease.
Ginseng (Panax ginseng C. A. Meyer), which commonly refers to the dried root and rhizome of P. ginseng, is known worldwide for its potent pharmacological activities, including its antiaging, antidiabetic, immunomodulatory, anticancer, and neuromodulatory properties (Kiefer and Pantuso, 2003; Zheng et al., 2012; Yang et al., 2014; Wang et al., 2016; Zheng et al., 2018). As a precious and important traditional Chinese medicine, ginseng has been cultivated for more than 4,000 years. Currently, the main cultivation areas of ginseng are in China and South Korea; it is also grown in Japan, North Korea, Canada, and the United States. The northeastern region is the most important ginseng production area in China. In addition, the phylogeny and population genetics of ginseng suggest that Fusong town in Northeast China might be the domestication center of Asian ginseng, as the variety grown there is closest to wild ginseng (Li et al., 2015). As a perennial medicinal plant, the active ingredients of ginseng will accumulate over time. Ginseng takes 4–6 years of consecutive cultivation to obtain high-quality ginseng with good medicinal properties. However, after 4 years of continuous planting, soil-borne diseases increased, resulting in significantly reduced yield and quality (Kim et al., 2017; Farh et al., 2018).
The rusty root is one of the most destructive root diseases of ginseng, characterized by small or large reddish-brown spots on the surface of the ginseng roots. This disease has attracted considerable attention due to its significant yield decline of between 20% and 30% per year and great economic losses (Garbeva et al., 2004). Rusty root-affected ginseng has less fibrous roots and reddish-brown to orange-brown irregular discolored lesions on the surface of the ginseng tap or lateral roots. In seriously diseased fields, sometimes even the whole root is covered with reddish-brown spots. Often, superficial lesions are easily scraped off, revealing the white healthy tissue inside. Presently, the cause of rusty root syndrome is still controversial. Some studies have shown that rusty root was closely related to the rhizosphere soil conditions (such as higher moisture, nitrate concentrations, and active Al species) and are considered a non-infectious physiological disease (Liu et al., 2014). Furthermore, microbes also play a key role in this disease, as suggested in other studies that ginseng root could be infected by putative pathogens, including bacteria (Choi et al., 2005; Lee et al., 2011), fungi (Rahman and Punja, 2005; Reeleder et al., 2006; Guan et al., 2014; Lu et al., 2015; Lu et al., 2020), or a combination of both (Ellouze et al., 2014; Liu et al., 2019), and showed rust symptom after inoculation of the isolates.
In this study, high-throughput sequencing was employed to systematically characterize changes in the healthy and rusty ginseng transcriptome. The results showed that the response of ginseng to the disease involved hormones (BR), Ca2+, HR, and cell wall-related signals. Our findings provide not only a new understanding of the origin of this disease but also a theoretical basis for its prevention and treatment.
P. ginseng samples were collected in Henglu, Ji’an City, Jilin Province (41°34′65″N, 125°81′65″E; 428 m). These were randomly selected from 100 m2 of a 4-year ginseng plantation, and three healthy (CK) and rusty root (RR) ginseng plants were collected from the selected area as three biological replicates. The rusty ginseng samples were covered with red rust-colored lesions throughout the root.
The collected ginseng root tissues were rinsed with RNase-free water and then drained with clean paper. The ginseng root tissue was cut into small pieces ≤0.5 cm2 and quickly placed into the pre-cooled RNase-free threaded cryotube with written numbers. The samples were immediately frozen in liquid nitrogen for 1 h and then transferred to −80°C for long-term storage.
Total RNA Purification Kit, TRK1001 (LC Science, Houston, TX, USA), was used to extract the total RNA of six P. ginseng samples following the operation manual. RNA contamination and degradation were monitored on 1% agarose gels. The total RNA quantity and purity were measured using Bioanalyzer 2100 and RNA 1000 Nano LabChip Kit (Agilent, Santa Clara, CA, USA) with RIN number >7.0. Poly(A) RNA was purified from a total of 5 μg of RNA in two rounds using poly-T oligo-attached magnetic beads. After purification, the mRNA was fragmented into small pieces using divalent cations at elevated temperatures. The cleaved RNA fragments were then reverse transcribed according to the mRNA-seq sample preparation kit (Illumina, San Diego, CA, USA) to create the final cDNA library with an average insert size of 300 bp ( ± 50 bp) for the paired-end libraries. Then, we performed the paired-end sequencing on an Illumina HiSeq 4000 (LC Sciences, Houston, TX, USA) following the supplier’s recommended protocol.
We aligned sequencing reads to the Ginseng Genome Database (http://ginsengdb.snu.ac.kr/) reference genome using Hisat package v.2.0 (Kim et al., 2015). The Hisat software initially removed a part of the reads based on the quality information accompanying each read and then mapped the reads to the reference genome.
Mapped reads of each sample were assembled using StringTie (v.1.3.0) (Pertea et al., 2015). Then, all transcriptomes from samples were merged to reconstruct a comprehensive transcriptome using perl scripts. After the final transcriptome was generated, the expression levels of all transcripts were estimated using StringTie and edgeR. edgeR was used to perform the expression level of mRNAs by calculating FPKM (fragments per kilobase of transcript per million mapped reads). Differential expression analysis of the two groups was performed using the R package (v.3.2.5). The differentially expressed mRNAs and genes were selected using |log2(foldchange)| ≥ 1 (p < 0.05) as selection criteria.
The expression of differential expression genes was verified by quantitative real-time PCR (qRT-PCR) on a Real-Time PCR System (analytikjena-qTOWER2.2) using 2× SYBR® Green Supermix (China). The specific primers of differentially expressed genes (DEGs) were designed by the online website (https://bioinfo.ut.ee/primer3-0.4.0/primer3/), and all of the primers used in this study are listed in Table S2. RNA was extracted from three biological replicates of rusty root-affected and healthy ginseng roots, and cDNA was synthesized using TUREscript 1st Strand cDNA Synthesis Kit (Aidlab). For each reaction system, 0.5 μl of the forward and reverse primers and 1 μl of the cDNA template were added. Conditions of qRT-PCR were as follows: 95°C (3 min), followed by 39 cycles of 95°C (10 s), 60°C (30 s), and 72°C (30 s). The 2−ΔΔCt method was used to calculate the relative gene expression level (Arocho et al., 2006), which was log2 transformed and plotted against the corresponding means data from the RNA-seq and qRT-PCR.
In this study, all of the data are represented as the mean ± SD of the three technical replicates. Student’s t-test was used for statistical comparison between the CK and RR groups. Statistical analysis was performed with Microsoft Excel 2010. We considered p < 0.05 to be statistically significant.
To gain a global overview of the rusty root transcript profile, healthy (CK) and rusty root-affected ginseng (RR) samples were sequenced by Illumina HiSeq 4000. Three biological replicates were designed in two groups. A total of six samples were obtained. In this study, a total of 267,199,488 raw reads were generated by sequencing CK vs. RR samples. After removing adaptor sequences and low-quality sequences, 49,741,788, 37,829,990, 37,235,008, 25,698,520, 46,584,096, and 40,707,226 clean reads were obtained (Table 1). These data were then mapped onto the Ginseng Genome Database (http://ginsengdb.snu.ac.kr/) reference genome using Hisat software, resulting in most of the valid reads being well-matched with the reference genome. It indicated that the transcriptome data were deemed suitable for the subsequent analysis.
The expression levels of the genes from the ginseng transcriptomes were calculated and normalized to FPKM values. Through comparison with healthy ginseng samples, and using |log2(foldchange)| ≥ 1 (p < 0.05) as selection criteria, 1,198 genes (672 upregulated, 526 downregulated) with significant differential expression were identified in the diseased ginseng (Table S1). Compared with healthy ginseng, only external two to three layers of cells were affected and displayed a red to brown color in the rusty root-affected ginseng (Luo et al., 2022). In this study, we found that there was less differential expression of genes, which may be due to the high content of white ginseng root tissue (the unaffected root tissue).
To validate the reliability of the RNA-seq data, eight DEGs were selected to investigate their transcriptional expression in the P. ginseng roots via qRT-PCR using gene-specific premiers (Table S2). To compare the expression data between RNA-seq and qRT-PCR, the relative expression level was transformed to log2(foldchange). The results showed a strong correlation between the RNA-seq and qRT-PCR data (R2 = 0.976; Figure 1), demonstrating the reliability of the RNA-seq expression profile in this study.
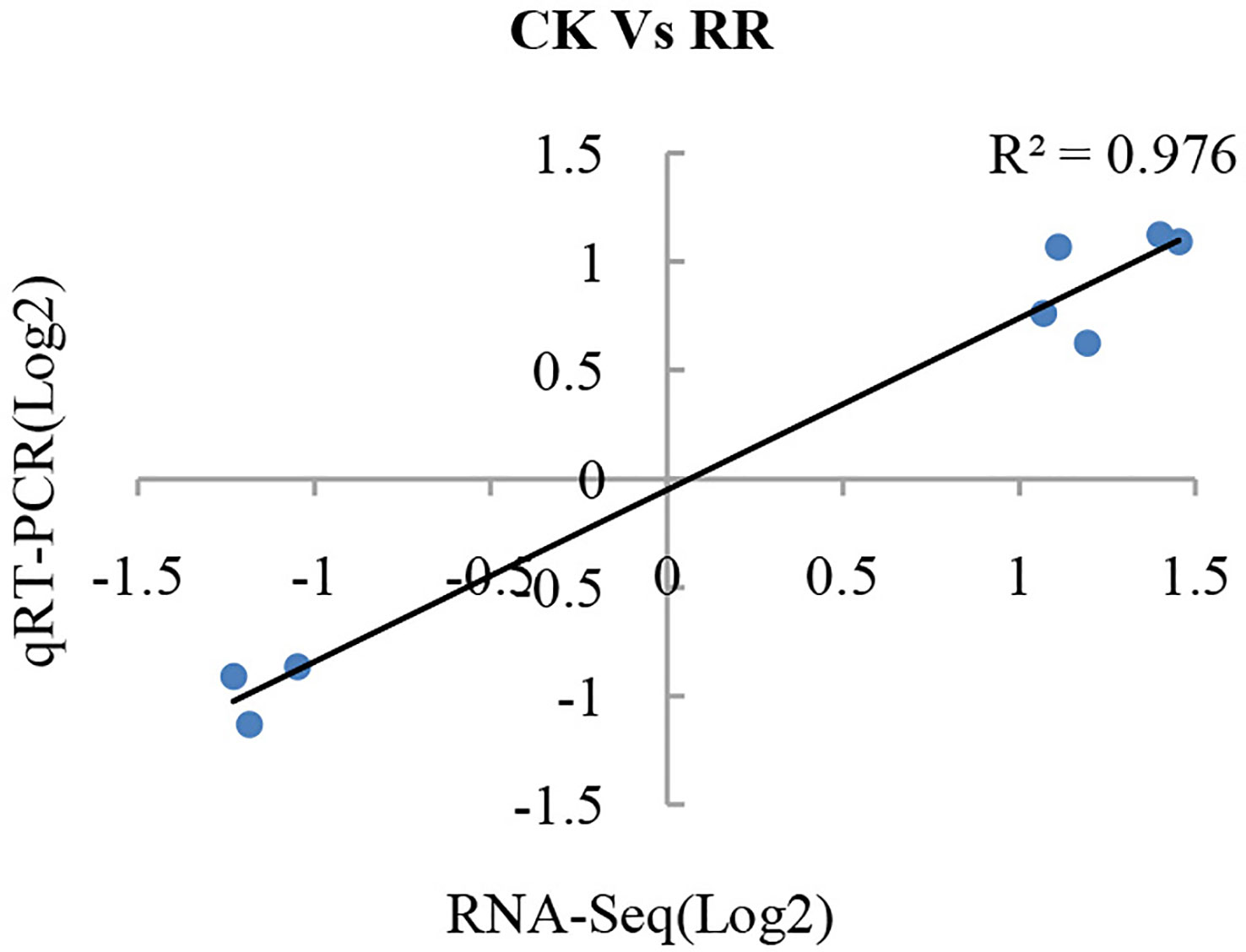
Figure 1 Validation of gene expression from RNA-seq by qRT-PCR analysis. Eight differentially expressed genes were randomly selected and used to examine the expression profiles by qRT-PCR analysis. The values from RNA-seq data were plotted against the corresponding qRT-PCR values and fitted into a linear regression. The R2 represents the correlation coefficient.
To characterize the functions of all these DEGs, we performed gene ontology (GO) enrichment analysis to classify all the DEGs into 107 biological processes (BP), 18 cellular components (CC), and 52 molecular functions (MF) depending on p-value < 0.05 (Table S3). The DEGs in GO-enriched categories of the biological processes were mainly involved in the oxidation–reduction process, the ethylene-activated signaling pathway, the response to cadmium ion, and cell wall organization. In the cellular component, most of the DEGs were significantly mapped to the extracellular region, the membrane, the plasmodesma, and the cell wall. According to molecular function, the DEGs were mainly associated with transcription factor activity and oxidoreductase activity (Table 2). GO enrichment results showed DEGs with functions related to binding activity, oxidoreductase activity, and transferase activity after infection with the rusty root of ginseng. These genes primarily participated in biological processes such as gene function regulation, plant hormone regulation, defense response to microorganisms, response to abiotic stress, secondary metabolite biosynthetic process, and cell wall organization.
In addition, we performed the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis to identify the metabolic pathways involved in the responses of rusty roots in P. ginseng. Based on the rich factors and p-values, the DEGs were significantly enriched in 22 main pathways. Among these, plant hormone signal transduction, pentose and glucuronate interconversions, fatty acid metabolism, biosynthesis of unsaturated fatty acids, and ascorbate and aldarate metabolism were significantly enriched of the DEGs (Figure 2; Table S4). The enrichment analysis results of the GO and KEGG showed the responses of ginseng to rusty root syndrome. Compared with those in healthy ginseng, many DEGs in rusty root-affected ginseng were involved in plant hormone signal transduction, plant–pathogen interaction, and synthesis of cellulose and lignin.
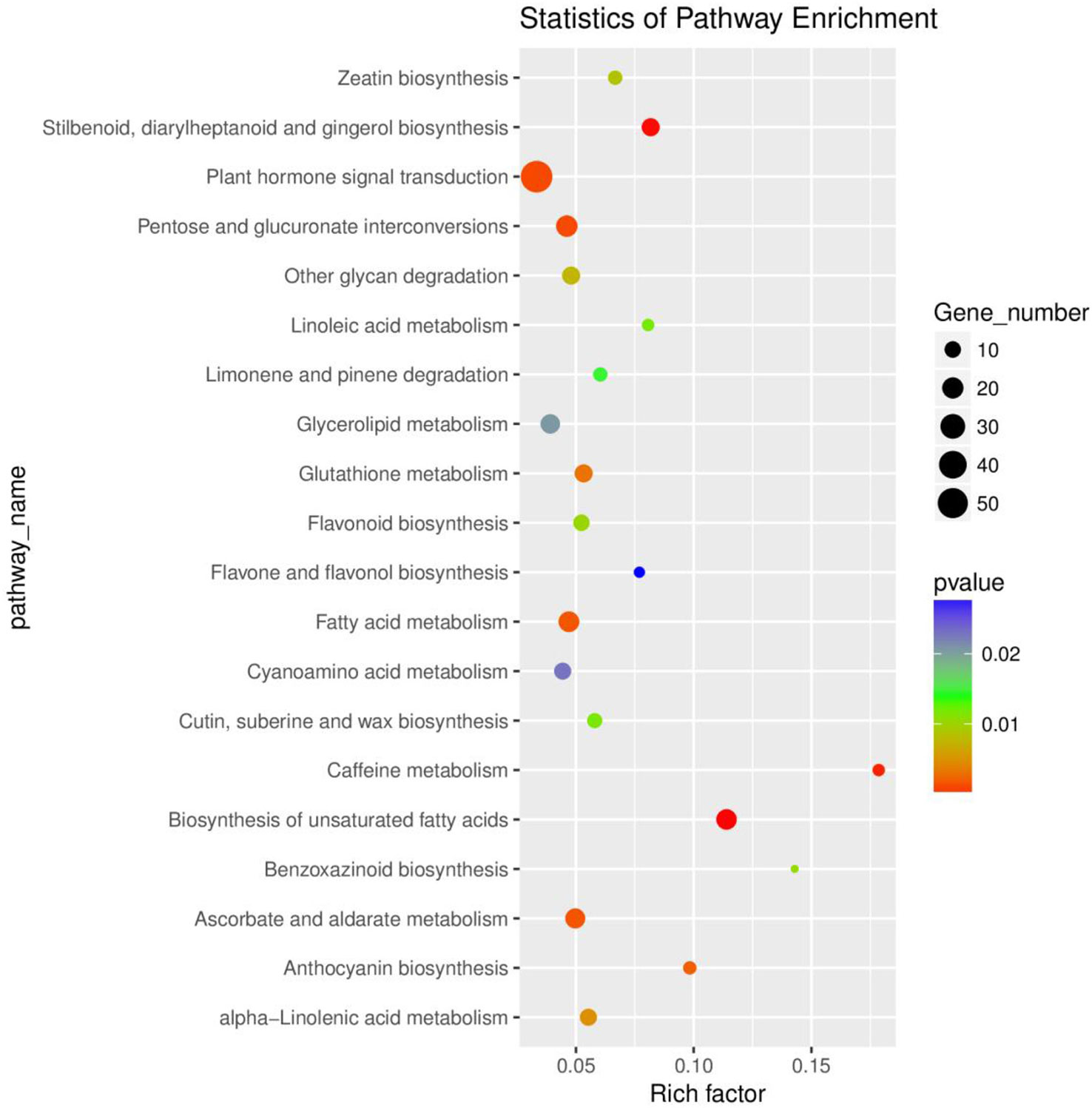
Figure 2 Functional classification of the DEGs via KEGG analysis. The top 20 enriched KEGG pathways are indicated, with dot size representing the ratio of selected genes to total genes in the pathway and dot color illustrating adjusted p-values. The ordinate shows the name of the KEGG terms, and the abscissa shows the ratio of the number of differentially expressed genes annotated to the pathway and the total of genes annotated to the pathway. Gene number and p-value are shown on the right. DEGs, differentially expressed genes; KEGG, Kyoto Encyclopedia of Genes and Genomes.
According to the analysis, these pathways explained the possible roles of these DEGs in the occurrence of ginseng rusty roots. In the “biosynthesis of unsaturated fatty acids” pathway, most DEGs were annotated as omega-6 fatty acid desaturase/acyl-lipid omega-6 desaturase (Delta-12 desaturase) [EC:1.14.19.6 1.14.19.22], which usually played an active role in resisting environmental stresses such as cold, salt, and high temperature (Zhang et al., 2005; Wang et al., 2014), but their expression was mostly (17 of 18) promoted in the root of rusty ginseng (Table S4). The expression trend of these genes in diseased ginseng was contrary to the study of Gong et al. (2020).
Plant hormones are secondary metabolites that regulate plant growth and development at extremely low concentrations. Therefore, we further examine the transcription level of hormone-related genes. The analysis detected 84 DEGs involved in the brassinosteroid (BR), jasmonic acid (JA), auxin (IAA), ethylene (ET), gibberellin (GA), abscisic acid (ABA), cytokine (CK), and salicylic acid (SA) biosynthesis, metabolism, and signal transduction in rusty ginseng (Figures 3; Table S5). It shows that plant hormones play an important role in the response of ginseng to rusty root disease, especially BRs. Among these DEGs, 19 of the 22 BR metabolism and signaling pathway genes in rusty roots were significantly downregulated compared with healthy ginseng roots (Figure 4). All seven TCH4 (xyloglucan: xyloglucosyl transferase), which encode xyloglucan endotransglycosylase (XET), were downregulated in the roots of diseased ginseng (Figure 4). Furthermore, 15 DEGs (nine upregulated/six downregulated) were enriched in rusty root syndrome of ginseng compared with healthy ginseng (Table S5). These were primarily involved in JA metabolism and signaling pathways, including the phospholipase A1, lipoxygenase, 12-oxophytodienoic acid reductase, transcription factor MYC2, and jasmonate ZIM domain-containing protein (Figure 4; Table S5). Nine of the 14 IAA-related genes were upregulated in diseased ginseng, such as those encoding indole-3-pyruvate monooxygenase, N-hydroxythioamide S-beta-glucosyltransferase, auxin influx carrier (AUX1 LAX family), auxin response factor, and SAUR family protein (Figure 4; Table S5). Nine of the 12 ET-related genes were significantly upregulated in rusty roots, including those encoding aminocyclopropanecarboxylate oxidases, serine/threonine-protein kinase CTR1, ethylene-responsive transcription factor 1, and ethylene-responsive transcription factor 2 (Figure 4; Table S5). Furthermore, nine GA-related DEGs (seven upregulated/two downregulated), six ABA-related DEGs (three upregulated/three downregulated), five cytokine-related DEGs (four upregulated/one downregulated), and one SA-related DEG (upregulated) were enriched in diseased ginseng compared with healthy ginseng (Figure 4; Table S5).
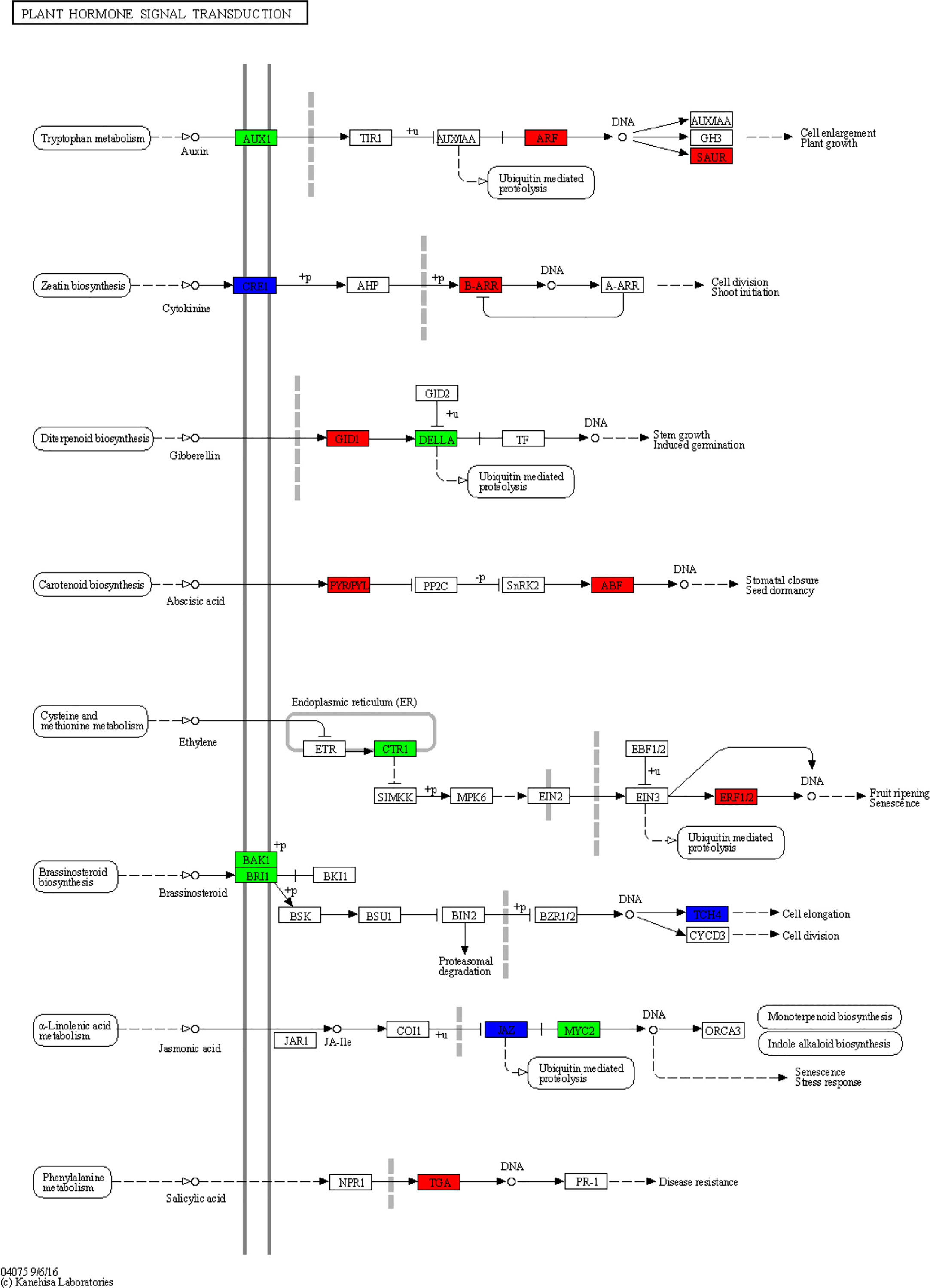
Figure 3 Overview of plant hormone signal transduction pathway. This diagram represents the signaling pathway of eight plant hormones in Panax ginseng. The different colors of the frame indicate the different gene transcription in RR compared with CK (red frame, upregulation; blue frame, downregulation).
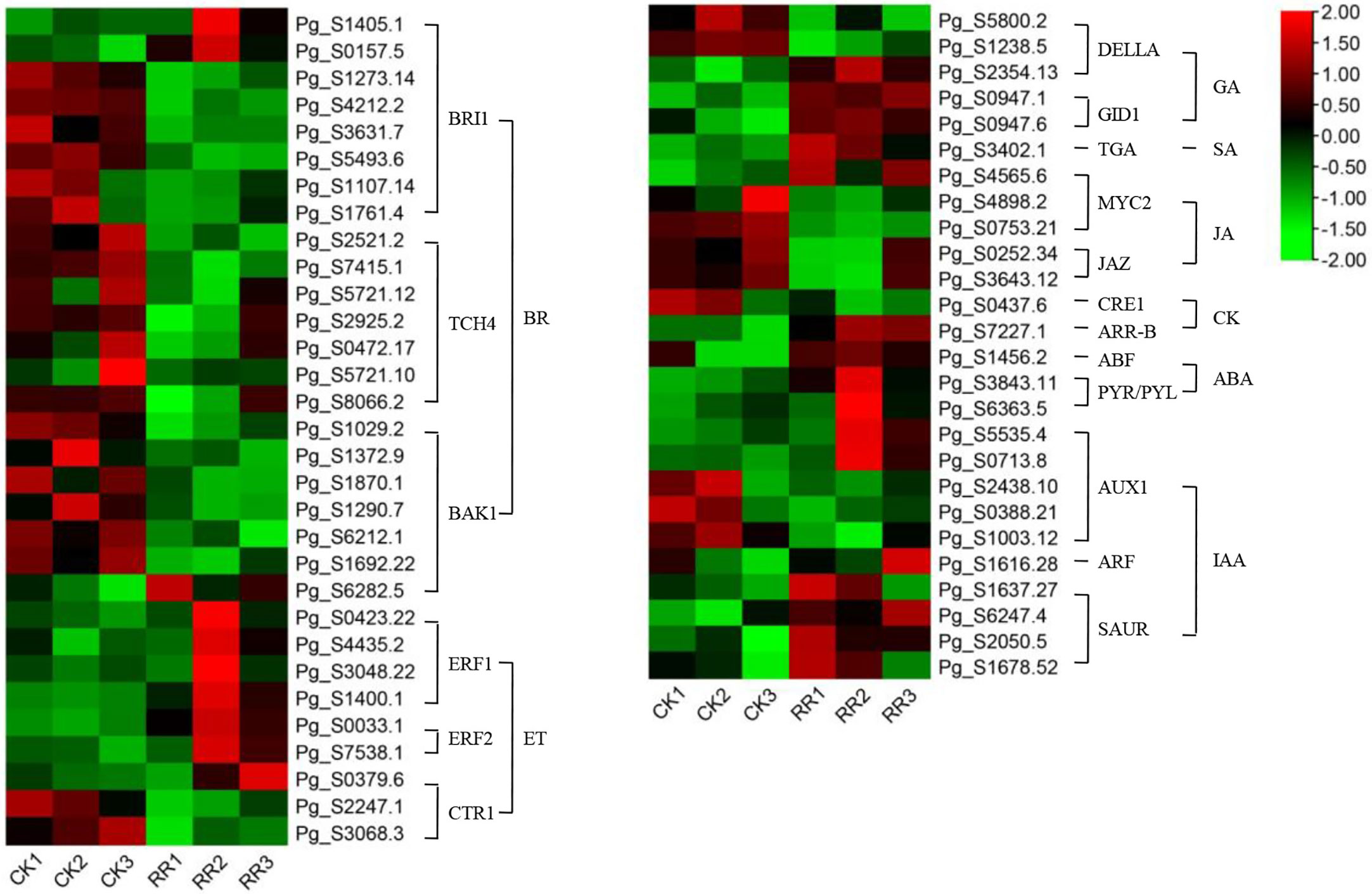
Figure 4 DEGs related to plant hormone signal transduction. The heatmap was constructed according to the expression level of these functional DEGs, and its log2-fold expression limit was 2 (red) to −2 (green). DEGs, differentially expressed genes.
There were 48 DEGs involved in the “plant–pathogen interaction” pathway (Table S1). It is speculated that these genes may play a key role in the response of ginseng to rusty roots. Compared with healthy ginseng, downregulation of Ca2+-dependent protein kinases (CDPKs) and cyclic nucleotide-gated channels (CNGCs), which affected Ca2+ flow and usually led to hypersensitive response (HR), were detected in diseased ginseng. Although FLS2 was downregulated, its downstream induced some transcription factors (TFs) responsible for inducing defense-related genes in rusty ginseng, such as WRKYs. In addition, several genes associated with effector-triggered immunity, including Pti4, Pti6, RPM1, RAR1, RPS2, PBS1, EDS1, Rd19, HCD1, and WRKY1/2, were distinctly differential expressed in rusty roots of ginseng (Figures 5, 6).
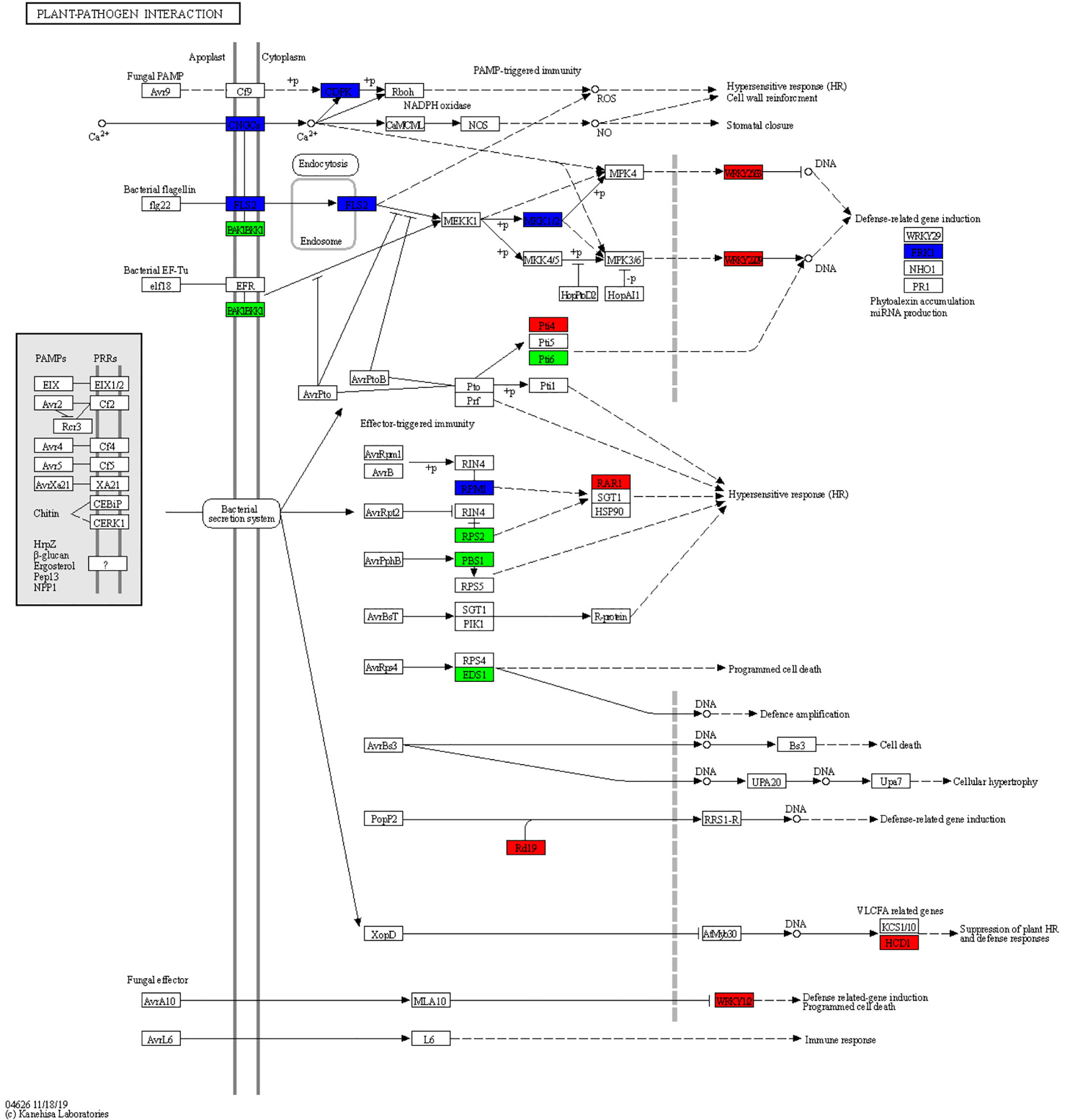
Figure 5 Overview of the plant–pathogen interaction pathway. The different colors of the frame indicate the different gene transcription in RR compared with CK (red frame, upregulation; blue frame, downregulation).
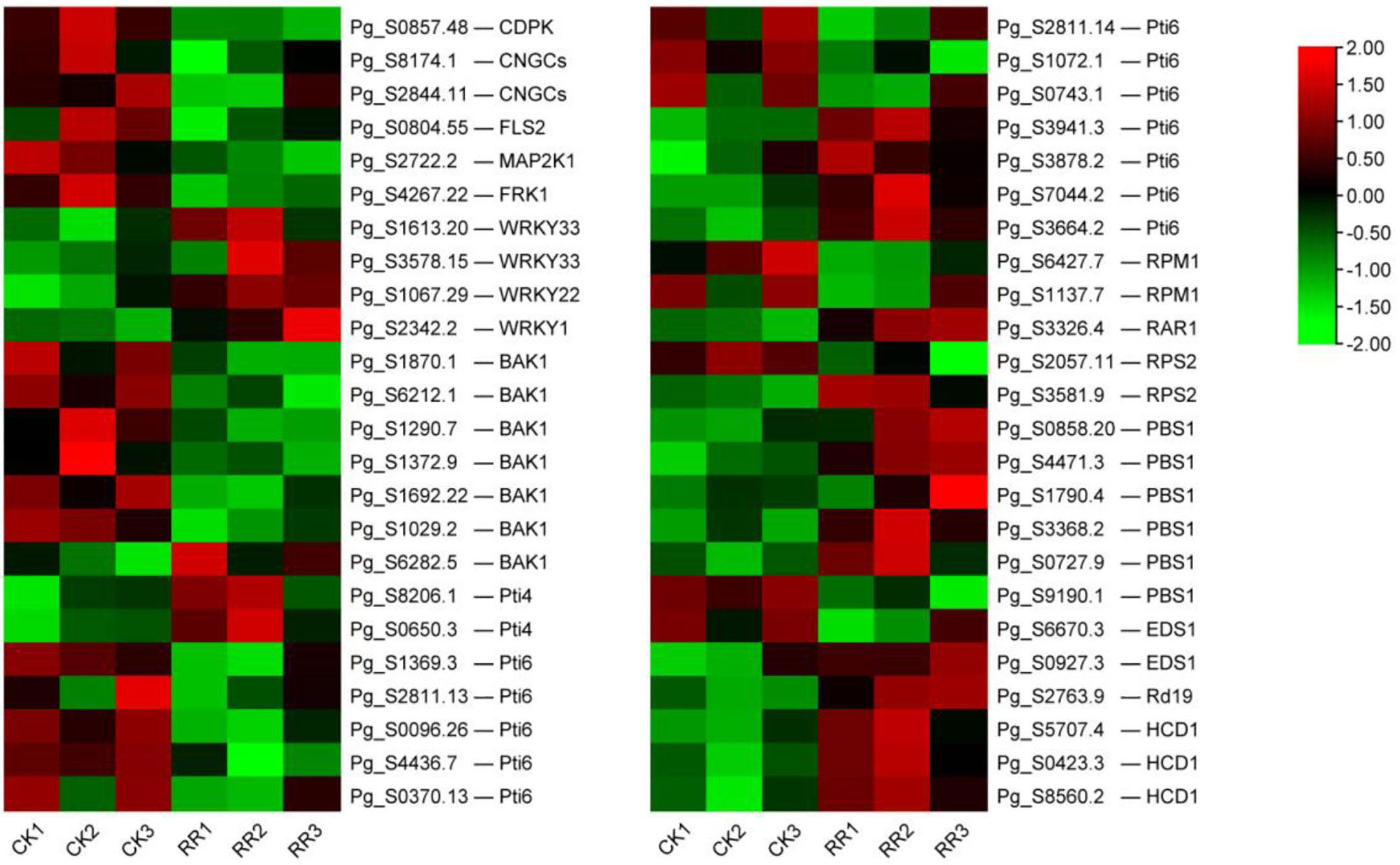
Figure 6 DEGs related to plant–pathogen interaction. The heatmap was constructed according to the expression level of these functional DEGs, and its log2-fold expression limit was 2 (red) to −2 (green). DEGs, differentially expressed genes.
CNGCs, which participate in the plant autoimmune cascade of early hypersensitivity, are one of the important Ca2+-conducting channels (Wang et al., 2022). CDPKs are usually involved in Ca2+ signaling cascades (Boudsocq and Sheen, 2013). As a secondary massager, Ca2+ plays a crucial role in activating the downstream defense reaction. The downregulation of CDPKs and CNGCs may be related to HR. FLS2, a pattern recognition receptor (PRR), can trigger innate immunity in plants. The downregulation of the above genes may indicate that part of PAMP-triggered immunity (PTI) is inhibited in rusty roots. Overall, the immune system of rusty root-affected was inhibited, thus affecting the HR and cell wall reinforcement, being sensitive to external stress, and reducing the survival rate of ginseng.
In addition to maintaining the cell structure, the cell wall also directly affects the response to environmental stresses, acting as a physical barrier (Yan et al., 2021). To confirm the response of cell walls in ginseng roots to rusty root syndrome, the relevant GO terms are listed in Figure 7. The analysis results discovered that many genes related to cell walls were differentially expressed in rusty ginseng, indicating that cell walls played a key role in the response of ginseng to rusty roots. There are two typical cell walls in plants: primary cell wall (PCW) and secondary cell wall (SCW). Their biosynthetic components and cellular locations are clearly distinguished. Each plant cell is surrounded by relatively thin and extensible PCW, which is mainly composed of three polysaccharide-derived polymers: cellulose, hemicelluloses, and pectin. SCW mainly includes cellulose, hemicelluloses, and lignin biopolymers. Cellulose in plants is synthesized by Cellulose Synthase Complex (CSC) containing at least three different cellulose synthases (CESAs) and also requires endo-1,4-beta-d-glucanase (EG) activity (Vain et al., 2014). In diseased ginseng, we found that one cellulose synthase (UDP-forming) gene was upregulated, and two EG genes were differential expression (Figure 8). Xylans are a diverse family of hemicellulosic polysaccharides found in abundance within the cell walls of nearly all flowering plants (Smith et al., 2022). Two transcripts of xylan synthase had inhibited expression in the rusty ginseng root (Figure 8). Pectin is composed of three polysaccharides, homogalacturonan (HG), rhamnogalacturonan I (RG-I), and rhamnogalacturonan II (RG-II). The most abundant pectic polysaccharide is HG, which is synthesized by GAUT gene family of α-1,4-galacturonosyltransferases (Mohnen et al., 2012). The occurrence of rusty root disease inhibits the expression of GAUT gene in ginseng. The biosynthesis of lignin, the main component of SCW, involves the complex regulation of many enzymes, such as cinnamoyl CoA reductase (CCR), caffeoyl-CoA O-methyltransferase (CCoAOMT), shikimate O-hydroxycinnamoyltransferase (HCT), and ferulate-5-hydroxylase (F5H, CYP84A) (Vanholme et al., 2010). The gene encoding CCR is a key gene for lignin biosynthesis, which is induced to express under stress conditions (Srivastava et al., 2015). CCoAOMT is an enzyme involved in monolignol synthesis that affects the efficiency of lignification and lignin composition (Rakoczy et al., 2018). Downregulation of HCT gene, an essential enzyme of the phenylpropanoid metabolism, reduces lignin content in alfalfa (Medicago sativa L.) (Bhattarai et al., 2018). F5H is a cytochrome P450-dependent monooxygenase, which is the enzyme responsible for the last hydroxylation of the syringyl-type lignin precursors (Franke et al., 2000). All DEGs encoding these four lignin synthetases are upregulated (Figure 8). Class III peroxidases (PRXs) promote lignin polymerization by oxidizing lignin monomers (monolignols) (Hoffmann et al., 2020; Zhu et al., 2021), although a beta-glucosidase, which is thought to play a key role in lignification by releasing the monolignol aglycones, inhibits the expression in the root of diseased ginseng (Dharmawardhana et al., 1995). Six of the seven DEGs related to lignin synthesis were upregulated in rusty roots (Figure 8). The transcriptional abundance of lignin synthesis-related transcripts increased, indicating the lignification process of rusty ginseng was enhanced. Lignin deposition is one of the responses to stress.
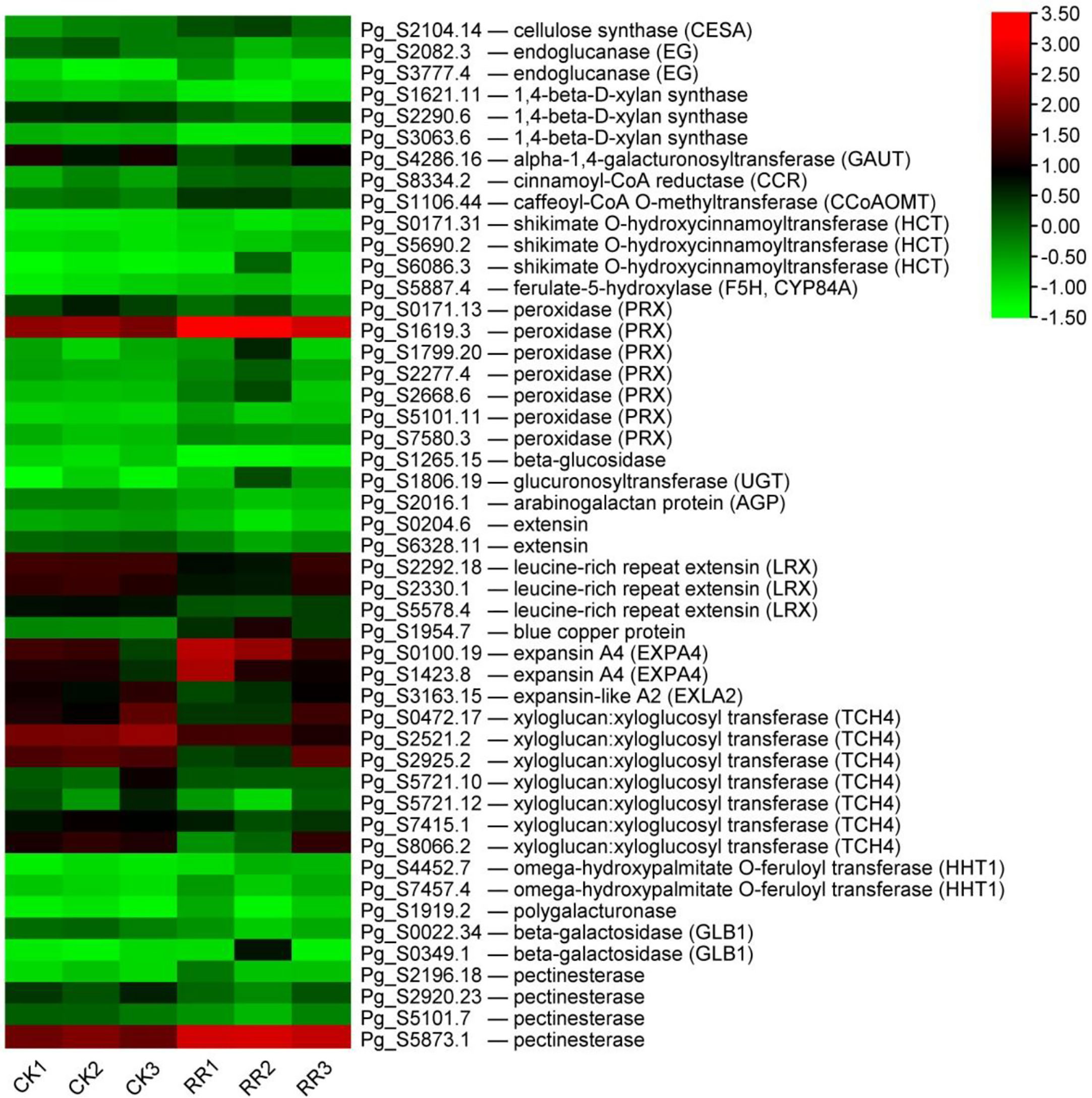
Figure 8 DEGs related to the cell walls. The heatmap was constructed according to the expression level of these functional DEGs, and its log2-fold expression limit was 2 (red) to −2 (green). DEGs, differentially expressed genes.
Arabinogalactan proteins (AGPs) and extensins are major members of the hydroxyproline-rich glycoprotein (HRGP) superfamily, which are unique components of plant cell walls (Tan et al., 2018). Glucuronosyltransferases are involved in the synthesis of the carbohydrate moieties of AGPs (Endo et al., 2013). AGPs in Arabidopsis positively regulate cell wall biosynthesis and root growth by regulating ABA signaling (Seifert et al., 2014; Hromadova et al., 2021). Extensins are involved in cell wall reinforcement in higher plants and defense against pathogen attacks (Castilleux et al., 2021). One AGP and five transcripts putatively coding for extension (including leucine-rich repeat extension, LRX) were all downregulated, while one glucuronosyltransferase was upregulated in the rusty root ginseng (Figure 8). LRXs modify cell wall expansion and also represent a connection between the cell wall and plasma membrane, perceiving extracellular signals and indirectly relaying this information to the cytoplasm (Herger et al., 2019). Additionally, blue copper protein, a gene putatively coding for a protein associated with the cell wall, was upregulated in rusty roots over fourfold (Figure 8). Cell wall modifications or alterations in cell wall structure are a common mechanism of defense against disease, and specific cell wall genes may contribute to plant defense. We found that there were differences in the expression profiles of cell wall-modifying genes. Expansins are plant cell wall-modifying proteins that have four subfamilies: α-expansin (EXPA), β-expansin (EXPB), expansin-like A (EXLA), and expansin-like B (EXLB) (Jin et al., 2020). As an important cell growth regulator, expansin also promoted interfascicular fiber cell elongation and cell wall thickness but did not alter the cellulose content in the cell wall (Li et al., 2017). XETs are most likely to modify the cell wall, which is the fundamental determinant of plant morphology (Xu et al., 1995). Two transcripts encoding EXPA were upregulated, while seven TCH4 and one EXLA were downregulated in the roots of diseased ginseng (Figure 8). Regulation of cell wall-modifying genes may underlie plant morphogenetic responses to the environment. BRs, a specific class of plant steroids, play a role in promoting growth involving cell division and elongation (Mandava, 1988; Hu et al., 2000; Sasse, 2003). Some BR-regulated genes have been proven to be related to cell elongation and cell wall organization (Goda et al., 2002). One of these genes is the TCH4 gene, which encodes the xyloglucan endotransglucosylase/hydrolase (XTH). It has been proposed that these specific XTH isoenzymes play a role in strengthening the lateral wall of root hairs and the cell walls of the root differentiation zone after the completion of cell expansion (Maris et al., 2009). The expression of the TCH4 gene in Arabidopsis is rapidly regulated by environmental variables, and changes in the expression level of the TCH4 gene directly lead to modifications in cell wall properties and structure (Xu et al., 1995). In our study, the mRNA levels of seven TCH4 genes were significantly reduced in the roots of rusty ginseng (Figure 4). It is revealed that the regulation of cell wall modifying enzyme genes is crucial to the regulation of plant morphogenetic response to the environment (Surgun and Burun, 2015). The reason for regulation is to change the properties of the cell walls so that plants can rapidly adapt to environmental conditions (Xu et al., 1995; Surgun and Burun, 2015). Our research results indicated that the occurrence of rusty root disease induces the cell wall modification of ginseng roots, which may also be one of the mechanisms for ginseng to adapt to stress. Omega-hydroxypalmitate O-feruloyl transferase (HHT1) could be involved in suberin biosynthesis. Suberin is a cell wall biopolymer with aliphatic and aromatic domains, which is synthesized in plant wound tissues to restrict water loss and pathogen infection (Molina et al., 2009). It was found that two HHT1 genes were induced to express in diseased ginseng (Figure 8). In addition, several cell wall-degrading enzymes (mainly polygalacturonase, beta-galactosidase, and pectinesterase), leading to cell wall loosening, were differential expressions (Figure 8). The modification in the cell wall may contribute greatly to the occurrence of rusty root syndrome.
Chinese scholars have reported that continuous cultivation of ginseng results in soil acidification, and the content of active Al in soil increases (Liu et al., 2014; You et al., 2015). Aluminum stress is believed to be related to the rusty root syndrome of ginseng (Zhou et al., 2017; Ma et al., 2021). Therefore, this study analyzed the expression of genes related to aluminum stress from the transcriptional level. Organic acid ions (including malate, citrate, and oxalate) secreted by plant roots can directly chelate external Al to prevent aluminum toxicity (Ma et al., 2001; Ryan et al., 2001). The expression of citrate synthetic enzyme CS (citrate synthase) and PEPCK (phosphoenolpyruvate carboxykinase, utilized in the dissimilation of malate/citrate) were decreased, while malate synthase MDH (malate dehydrogenase) was increased in rusty ginseng (Figure 9). Organic acids produced by plants are exuded outside the root to the rhizosphere through membrane transporters. The malate transporter ALMT (aluminum-activated malate transporter) and the citrate transporter MATE (multidrug and toxin extrusion) facilitate root malate and citrate exudation, respectively (Hoekenga et al., 2006). Transcription levels of two ALMTs were significantly upregulated in rusty ginseng roots (Figure 9). This study speculated that the synthesis and outward transportation of malate in ginseng roots were enhanced during the process of ginseng infection with rusty roots, indicating that malate played an important role in the rusty roots. In Arabidopsis thaliana, plant roots secrete both malate and citrate in response to Al stress, with malate being the major player in Al resistance (Liu et al., 2009). Accordingly, the increased malate synthesis and secretion might contribute significantly to the enhanced Al resistance in rusty ginseng, while the negative effect of reduced citrate synthesis on the Al resistance is negligible.
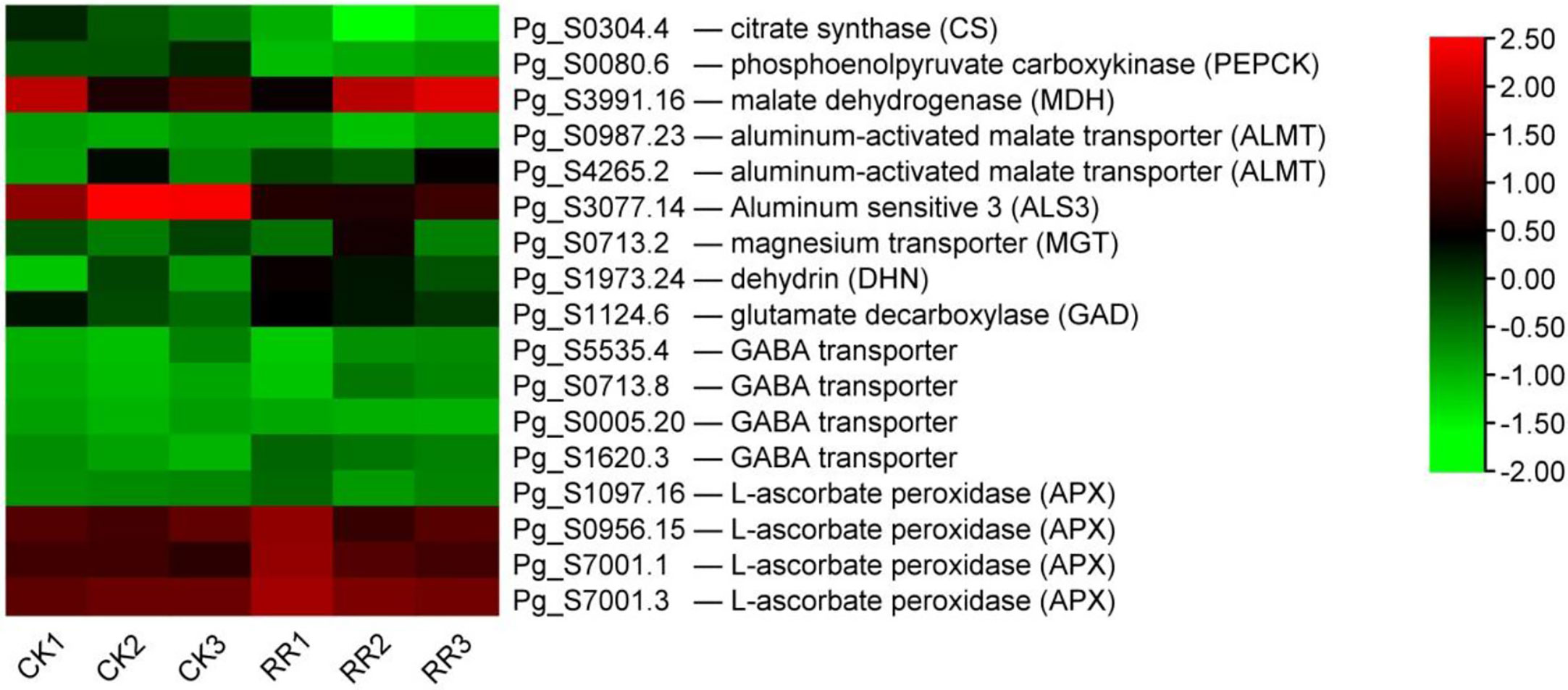
Figure 9 DEGs related to aluminum stress. The heatmap was constructed according to the expression level of these functional DEGs, and its log2-fold expression limit was 2 (red) to −2 (green). DEGs, differentially expressed genes.
In addition to the organic acid exudation-based Al-resistance mechanism, another effective strategy for Al exclusion is the absorption of Al by plant cell wall polysaccharides. At a high Al concentration in the environment, barley may absorb ~85% of the peripheral Al into its root cell walls (Wang et al., 2006), and the giant alga Chara coralline may even absorb up to 99.9% of total Al into its cell walls (Taylor et al., 2000). The total aluminum content in the rusty ginseng was higher, and it mainly accumulated in the diseased root and its periderm (Zhou et al., 2017). The internal detoxification of plants utilizes the ALS (aluminum-sensitive) encoding tonoplast-localized ATP-binding cassette (ABC) transporter to chelate internal Al into the vacuoles (Larsen et al., 2007). The bacterial-type ABC transporter complex STAR1/ALS3 is responsible for the transport of UDP (uridine diphosphate)-glucose that can modify cell walls and therefore masks Al-binding sites (Larsen et al., 2005). The transcription level of ALS3 was inhibited in the rusty ginseng root (Figure 9). It indicated that the Al-binding sites of cell walls were exposed, while the internal detoxification of aluminum was weakened. The results of this study revealed that aluminum tolerance in rusty ginseng roots was mainly through external chelation aluminum and cell wall-binding aluminum to restrain aluminum into cells. Therefore, it is considered that the rusty root syndrome of ginseng is a spontaneous self-protection mechanism of ginseng under aluminum stress for a long time.
Some genes related to aluminum tolerance were induced to express in ginseng root infected with the rusty syndrome. It is reported that magnesium transporter (MGT) is one of the cellular targets of Al toxicity, and overexpression of MGT1 confers Al tolerance on the plant (Deng et al., 2006). Dehydrins (DHNs) are considered molecular protectors. MsDHN1 is involved in increasing the tolerance of alfalfa to Al stress (Lv et al., 2021). GABA has been reported to regulate the malate-transporting plasma membrane channel during Al stress in wheat (Ramesh et al., 2015; Ramesh et al., 2018). GABA synthetase glutamate decarboxylase (GAD) and four GABA transporters are upregulated (Figure 9). Ascorbate (AsA) and glutathione (GSH) are at the heart of the redox hub, and AsA–GSH system plays pivotal roles in Al tolerance. The changes in the levels and status of AsA and GSH were correlated with alterations in AsA–GSH cycle-allied enzymes, including ascorbate peroxidase (APX), dehydroascorbate reductase (DHAR), monodehydroascorbate reductase (MDHAR), and glutathione reductase (GR). Compared with healthy ginseng, the transcriptional levels of four APXs were upregulated, while the expression levels of DHAR, MDHAR, and GR had no significant changes (Figure 9). The above results indicate that aluminum resistance-related genes are induced to express, and aluminum resistance function is activated in the root of rusty ginseng.
Transcriptome analysis provided clues for the molecular mechanism of ginseng-infected rusty roots. In conclusion, the immune system of rusty ginseng was inhibited, and the lignification process and the cell wall were enhanced. The aluminum resistance-related genes were induced, and the synthesis and outward transportation of malate were enhanced in the rusty roots of ginseng. It was indicated that the rusty ginseng increased aluminum resistance, and aluminum resistance mainly inhibited aluminum from entering cells through external chelating aluminum and cell wall-binding aluminum. Therefore, it is considered that ginseng rusty root syndrome is the spontaneous self-protection mechanism of ginseng under long-term aluminum stress.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI Sequence Read Archive (SRA) database (Accession Number: PRJNA941146).
AT and WL contributed to the experimental design and manuscript writing. HW and XL contributed to the data analysis. GX and JZ contributed to the qRT-PCR verification experiment. All authors contributed to the article and approved the submitted version.
This work was supported by the Science and Technology Project of the Jilin Education Department (JJKH20210547KJ, JJKH20170439KJ) and the Science Technology Development Plan Project of Jilin Province (YDZJ202301ZYTS523).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1142211/full#supplementary-material
Arocho, A., Chen, B., Ladanyi, M., Pan, Q. (2006). Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn. Mol. Pathol. 15, 56–61. doi: 10.1097/00019606-200603000-00009
Bhattarai, K., Rajasekar, S., Dixon, R. A., Monteros, M. J. (2018). Agronomic performance and lignin content of HCT down-regulated alfalfa (Medicago sativa l.). BioEnerg. Res. 11, 505–515. doi: 10.1007/s12155-018-9911-6
Boudsocq, M., Sheen, J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18, 30–40. doi: 10.1016/j.tplants.2012.08.008
Castilleux, R., Plancot, B., Vicre, M., Nguema-Ona, E., Driouich, A. (2021). Extensin, an underestimated key component of cell wall defence? Ann. Bot. 127, 709–713. doi: 10.1093/aob/mcab001
Choi, J. E., Ryuk, J. A., Kim, J. H., Choi, C. H., Chun, J., Kim, Y., et al. (2005). Identification of endophytic bacteria isolated from rusty-colored root of Korean ginseng (Panax ginseng) and its induction. Korean J. Medicinal Crop Science 13, 1–5.
Deng, W., Luo, K., Li, D., Zheng, X., Wei, X., Smith, W., et al. (2006). Overexpression of an arabidopsis magnesium transport gene, AtMGT1, in nicotiana benthamiana confers Al tolerance. J. Exp. Bot. 57, 4235–4243. doi: 10.1093/jxb/erl201
Dharmawardhana, D. P., Ellis, B. E., Carlson, J. E. (1995). A beta-glucosidase from lodgepole pine xylem specific for the lignin precursor coniferin. Plant Physiol. 107, 331–339. doi: 10.1104/pp.107.2.331
Ellouze, W., Taheri, A. E., Bainard, L. D., Yang, C., Bazghaleh, N., Navarro-Borrell, A., et al. (2014). Soil fungal resources in annual cropping systems and their potential for management. BioMed. Res. Int. 2014, 531824. doi: 10.1155/2014/531824
Endo, M., Kotake, T., Watanabe, Y., Kimura, K., Tsumuraya, Y. (2013). Biosynthesis of the carbohydrate moieties of arabinogalactan proteins by membrane-bound beta-glucuronosyltransferases from radish primary roots. Planta. 238, 1157–1169. doi: 10.1007/s00425-013-1959-0
Farh, M. E., Kim, Y. J., Kim, Y. J., Yang, D. C. (2018). Cylindrocarpon destructans/Ilyonectria radicicola-species complex: causative agent of ginseng root-rot disease and rusty symptoms. J. Ginseng Res. 42, 9–15. doi: 10.1016/j.jgr.2017.01.004
Franke, R., McMichael, C. M., Meyer, K., Shirley, A. M., Cusumano, J. C., Chapple, C. (2000). Modified lignin in tobacco and poplar plants over-expressing the arabidopsis gene encoding ferulate 5-hydroxylase. Plant J. 22, 223–234. doi: 10.1046/j.1365-313x.2000.00727.x
Garbeva, P., Veen, J. A., Elsas, J. D. (2004). Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 42, 243–270. doi: 10.1146/annurev.phyto.42.012604.135455
Goda, H., Shimada, Y., Asami, T., Fujioka, S., Yoshida, S. (2002). Microarray analysis of brassinosteroid-regulated genes in arabidopsis. Plant Physiol. 130, 1319–1334. doi: 10.1104/pp.011254
Gong, L., Gao, J., Xu, T., Qu, J., Wang, Z., Yang, Z., et al. (2020). Transcriptome analysis of field-grown Asian ginseng provides clues to environmental conditions and developmental mechanisms related to red skin root syndrome. Ind. Crop Prod. 153. doi: 10.1016/j.indcrop.2020.112486
Guan, Y. M., Lu, B. H., Wang, Y., Gao, J., Wu, L. J. (2014). First report of root rot caused by fusarium redolens on ginseng (Panax ginseng) in jilin province of China. Plant Dis. 98, 844. doi: 10.1094/PDIS-09-13-0922-PDN
Herger, A., Dunser, K., Kleine-Vehn, J., Ringli, C. (2019). Leucine-rich repeat extensin proteins and their role in cell wall sensing. Curr. Biol. 29, R851–R858. doi: 10.1016/j.cub.2019.07.039
Hoekenga, O. A., Maron, L. G., Pineros, M. A., Cancado, G. M., Shaff, J., Kobayashi, Y., et al. (2006). AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in arabidopsis. PNAS. 103, 9738–9743. doi: 10.1073/pnas.0602868103
Hoffmann, N., Benske, A., Betz, H., Schuetz, M., Samuels, A. L. (2020). Laccases and peroxidases Co-localize in lignified secondary cell walls throughout stem development. Plant Physiol. 184, 806–822. doi: 10.1104/pp.20.00473
Hromadova, D., Soukup, A., Tylova, E. (2021). Arabinogalactan proteins in plant roots - an update on possible functions. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.674010
Hu, Y., Bao, F., Li, J. (2000). Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in arabidopsis. Plant J. 24, 693–701. doi: 10.1046/j.1365-313x.2000.00915.x
Jin, K. M., Zhuo, R. Y., Xu, D., Wang, Y. J., Fan, H. J., Huang, B. Y., et al. (2020). Genome-wide identification of the expansin gene family and its potential association with drought stress in moso bamboo. Int. J. Mol. Sci. 21. doi: 10.3390/ijms21249491
Kim, Y. S., Balaraju, K., Jeon, Y. H. (2017). Biological characteristics of bacillus amyloliquefaciens AK-0 and suppression of ginseng root rot caused by cylindrocarpon destructans. J. Appl. Microbiol. 122, 166–179. doi: 10.1111/jam.13325
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Larsen, P. B., Cancel, J., Rounds, M., Ochoa, V. (2007). Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta. 225, 1447–1458. doi: 10.1007/s00425-006-0452-4
Larsen, P. B., Geisler, M. J., Jones, C. A., Williams, K. M., Cancel, J. D. (2005). ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in arabidopsis. Plant J. 41, 353–363. doi: 10.1111/j.1365-313X.2004.02306.x
Lee, C., Kwang, Y. K., Jo-Eun, L., Sunghan, K., Dongkul, R., Jae-Eul, C., et al. (2011). Enzymes hydrolyzing structural components and ferrous ion cause rusty-root symptom on ginseng (Panax ginseng). J. Microbiol. Biotechn. 21, 192–196. doi: 10.4014/jmb.1008.08010
Li, J., Hu, X., Huang, X., Huo, H., Li, J., Zhang, D., et al. (2017). Functional identification of an EXPA gene (NcEXPA8) isolated from the tree neolamarckia cadamba. Biotechnol. Biotec. Eq. 31, 1116–1125. doi: 10.1080/13102818.2017.1362960
Li, M. R., Shi, F. X., Zhou, Y. X., Li, Y. L., Wang, X. F., Zhang, C., et al. (2015). Genetic and epigenetic diversities shed light on domestication of cultivated ginseng (Panax ginseng). Mol. Plant 8, 1612–1622. doi: 10.1016/j.molp.2015.07.011
Liu, J., Magalhaes, J. V., Shaff, J., Kochian, L. V. (2009). Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer arabidopsis aluminum tolerance. Plant J. 57, 389–399. doi: 10.1111/j.1365-313X.2008.03696.x
Liu, D., Sun, H., Ma, H. (2019). Deciphering microbiome related to rusty roots of panax ginseng and evaluation of antagonists against pathogenic ilyonectria. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01350
Liu, X., Yang, Z., Gao, L., Xiang, W., Zhang, B., Xie, Z., et al. (2014). Comparison of the characteristics of artificial ginseng bed soils in relation to the incidence of ginseng red skin disease. Exp. Agr. 50, 59–71. doi: 10.1017/S0014479713000367
Lu, X. H., Jiao, X. L., Chen, A. J., Luo, Y., Gao, W. W. (2015). First report of ilyonectria robusta causing rusty root of Asian ginseng in China. Plant Dis. 99, 156. doi: 10.1094/PDIS-06-14-0663-PDN
Lu, X. H., Zhang, X. M., Jiao, X. L., Hao, J. J., Zhang, X. S., Luo, Y., et al. (2020). Taxonomy of fungal complex causing red-skin root of panax ginseng in China. J. Ginseng Res. 44, 506–518. doi: 10.1016/j.jgr.2019.01.006
Luo, M., Li, A., Wang, F., Jiang, J., Wang, Z., You, J. (2022). Integrative analysis of multiple metabolomes and transcriptome revealed color expression mechanism in red skin root syndrome of panax ginseng. Ind. Crop Prod. 177. doi: 10.1016/j.indcrop.2021.114491
Lv, A., Wen, W., Fan, N., Su, L., Zhou, P., An, Y. (2021). Dehydrin MsDHN1 improves aluminum tolerance of alfalfa (Medicago sativa l.) by affecting oxalate exudation from root tips. Plant J. 108, 441–458. doi: 10.1111/tpj.15451
Ma, R., Fu, B., Song, S., Zhang, C., Yuan, X., Zhang, T., et al. (2021). Ginsenoside biosynthesis in panax ginseng with red-skin disease is inhibited by soil characteristics. J. Soil Sci. Plant Nutt. 21, 2264–2273. doi: 10.1007/s42729-021-00519-8
Ma, J. F., Ryan, P. R., Delhaize, E. (2001). Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 6, 273–278. doi: 10.1016/s1360-1385(01)01961-6
Mandava, N. B. (1988). Plant growth-promoting brassinosteroids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 23–52.
Maris, A., Suslov, D., Fry, S. C., Verbelen, J. P., Vissenberg, K. (2009). Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of arabidopsis and their effect on root growth and cell wall extension. J. Exp. Bot. 60, 3959–3972. doi: 10.1093/jxb/erp229
Mohnen, D., Atmodjo, M., Tan, L., Amos, R., Zhua, X., Atwood, J. A., et al. (2012). Synthesis of the plant cell wallˈs most complex glycan: pectin - surprises in glycosyltransferase processing and anchoring in the golgi. FASEB J. 26, 349(3). doi: 10.1096/fasebj.26.1_supplement.349.3
Molina, I., Li-Beisson, Y., Beisson, F., Ohlrogge, J. B., Pollard, M. (2009). Identification of an arabidopsis feruloyl-coenzyme a transferase required for suberin synthesis. Plant Physiol. 151, 1317–1328. doi: 10.1104/pp.109.144907
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Rahman, M., Punja, Z. K. (2005). Factors influencing development of root rot on ginseng caused by cylindrocarpon destructans. Phytopathology. 95, 1381–1390. doi: 10.1094/PHYTO-95-1381
Rakoczy, M., Femiak, I., Alejska, M., Figlerowicz, M., Podkowinski, J. (2018). Sorghum CCoAOMT and CCoAOMT-like gene evolution, structure, expression and the role of conserved amino acids in protein activity. Mol. Genet. Genomics 293, 1077–1089. doi: 10.1007/s00438-018-1441-6
Ramesh, S. A., Kamran, M., Sullivan, W., Chirkova, L., Okamoto, M., Degryse, F., et al. (2018). Aluminum-activated malate transporters can facilitate GABA transport. Plant Cell. 30, 1147–1164. doi: 10.1105/tpc.17.00864
Ramesh, S. A., Tyerman, S. D., Xu, B., Bose, J., Kaur, S., Conn, V., et al. (2015). GABA signalling modulates plant growth by directly regulating the activity of plant-specific anion transporters. Nat. Commun. 6, 7879. doi: 10.1038/ncomms8879
Reeleder, R. D., Hoke, S. M. T., Zhang, Y. (2006). Rusted root of ginseng (Panax quinquefolius) is caused by a species of rhexocercosporidium. Ecol. Epidemiol. 96, 1243–1254. doi: 10.1094/PHYTO-96-1243
Ryan, P., Delhaize, E., Jones, D. (2001). Function and mechanism of organic anion exudation from plant roots. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 527–560. doi: 10.1146/annurev.arplant.52.1.527
Sasse, J. M. (2003). Physiological actions of brassinosteroids: an update. J. Plant Growth Regul. 22, 276–288. doi: 10.1007/s00344-003-0062-3
Seifert, G. J., Xue, H., Acet, T. (2014). The arabidopsis thaliana FASCICLIN LIKE ARABINOGALACTAN PROTEIN 4 gene acts synergistically with abscisic acid signalling to control root growth. Ann. Bot. 114, 1125–1133. doi: 10.1093/aob/mcu010
Smith, P. J., Curry, T. M., Yang, J. Y., Barnes, W. J., Ziegler, S. J., Mittal, A., et al. (2022). Enzymatic synthesis of xylan microparticles with tunable morphologies. ACS Mater. Au. 2, 440–452. doi: 10.1021/acsmaterialsau.2c00006
Srivastava, S., Vishwakarma, R. K., Arafat, Y. A., Gupta, S. K., Khan, B. M. (2015). Abiotic stress induces change in cinnamoyl CoA reductase (CCR) protein abundance and lignin deposition in developing seedlings of leucaena leucocephala. Physiol. Mol. Biol. Plants. 21, 197–205. doi: 10.1007/s12298-015-0289-z
Surgun, Y., Burun, B. (2015). Analysis of effects of 24-epibrassinolide and boron treatments on the expression of CycD3;1, TCH4 and KOR genes in arabidopsis thaliana (L.) heynh seedlings using semi-quantitative RT-PCR. J. Appl. Biol. Sci. 9, 59–65.
Tan, L., Tees, D., Qian, J., Kareem, S., Kieliszewski, M. J. (2018). Intermolecular interactions between glycomodules of plant cell wall arabinogalactan-proteins and extensins. Cell Surf. 1, 25–33. doi: 10.1016/j.tcsw.2018.03.001
Taylor, G. J., McDonald-Stephens, J. L., Hunter, D. B., Bertsch, P. M., Elmore, D., Rengel, Z., et al. (2000). Direct measurement of aluminum uptake and distribution in single cells of chara corallina. Plant Physiol. 123, 987–996. doi: 10.1104/pp.123.3.987
Vain, T., Crowell, E. F., Timpano, H., Biot, E., Desprez, T., Mansoori, N., et al. (2014). The cellulase KORRIGAN is part of the cellulose synthase complex. Plant Physiol. 165, 1521–1532. doi: 10.1104/pp.114.241216
Vanholme, R., Demedts, B., Morreel, K., Ralph, J., Boerjan, W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153, 895–905. doi: 10.1104/pp.110.155119
Wang, C. Z., Anderson, S., Du, W., He, T. C., Yuan, C. S. (2016). Red ginseng and cancer treatment. Chin. J. Nat. Med. 14, 7–16. doi: 10.3724/SP.J.1009.2016.00007
Wang, C., Mao, X., Zhao, D., Yu, H., Duo, H., Sun, E., et al. (2022). Transcriptomic analysis reveals that cell wall- and hypersensitive response (HR)-related genes are involved in the responses of apple to valsa mali. Plant Biotechnol. Rep. 16, 539–551. doi: 10.1007/s11816-022-00763-z
Wang, J. P., Raman, H., Zhang, G. P., Mendham, N., Zhou, M. X. (2006). Aluminium tolerance in barley (Hordeum vulgare l.): physiological mechanisms, genetics and screening methods. J. Zhejiang Univ. Sci. B. 7, 769–787. doi: 10.1631/jzus.2006.B0769
Wang, H. S., Yu, C., Tang, X. F., Zhu, Z. J., Ma, N. N., Meng, Q. W. (2014). A tomato endoplasmic reticulum (ER)-type omega-3 fatty acid desaturase (LeFAD3) functions in early seedling tolerance to salinity stress. Plant Cell Rep. 33, 131–142. doi: 10.1007/s00299-013-1517-z
Xu, W., Purugganan, M. M., Polisensky, D. H., Antosiewicz, D. M., Fry, S. C., Braam, J. (1995). Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell. 7, 1555–1567. doi: 10.1105/tpc.7.10.1555
Yan, L., Riaz, M., Liu, J., Liu, Y., Zeng, Y., Jiang, C. (2021). Boron reduces aluminum deposition in alkali-soluble pectin and cytoplasm to release aluminum toxicity. J. Hazard Mater. 401, 123388. doi: 10.1016/j.jhazmat.2020.123388
Yang, W. Z., Hu, Y., Wu, W. Y., Ye, M., Guo, D. A. (2014). Saponins in the genus panax l. (Araliaceae): a systematic review of their chemical diversity. Phytochemistry. 106, 7–24. doi: 10.1016/j.phytochem.2014.07.012
You, J., Liu, X., Zhang, B., Xie, Z., Hou, Z., Yang, Z. (2015). Seasonal changes in soil acidity and related properties in ginseng artificial bed soils under a plastic shade. J. Ginseng Res. 39, 81–88. doi: 10.1016/j.jgr.2014.08.002
Zhang, M., Barg, R., Yin, M., Gueta-Dahan, Y., Leikin-Frenkel, A., Salts, Y., et al. (2005). Modulated fatty acid desaturation via overexpression of two distinct omega-3 desaturases differentially alters tolerance to various abiotic stresses in transgenic tobacco cells and plants. Plant J. 44, 361–371. doi: 10.1111/j.1365-313X.2005.02536.x
Zheng, S. D., Wu, H. J., Wu, D. L. (2012). Roles and mechanisms of ginseng in protecting heart. Chin. J. Integr. Med. 18, 548–555. doi: 10.1007/s11655-012-1148-1
Zheng, M., Xin, Y., Li, Y., Xu, F., Xi, X., Guo, H., et al. (2018). Ginsenosides: a potential neuroprotective agent. BioMed. Res. Int. 2018, 8174345. doi: 10.1155/2018/8174345
Zhou, Y., Yang, Z., Gao, L., Liu, W., Liu, R., Zhao, J., et al. (2017). Changes in element accumulation, phenolic metabolism, and antioxidative enzyme activities in the red-skin roots of panax ginseng. J. Ginseng Res. 41, 307–315. doi: 10.1016/j.jgr.2016.06.001
Keywords: Panax ginseng, rusty root, transcriptome, cell wall, aluminum tolerance
Citation: Tong A, Liu W, Wang H, Liu X, Xia G and Zhu J (2023) Transcriptome analysis provides insights into the cell wall and aluminum toxicity related to rusty root syndrome of Panax ginseng. Front. Plant Sci. 14:1142211. doi: 10.3389/fpls.2023.1142211
Received: 11 January 2023; Accepted: 02 May 2023;
Published: 13 June 2023.
Edited by:
Waqar Afzal Malik, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Zhengnan Li, Inner Mongolia Agricultural University, ChinaCopyright © 2023 Tong, Liu, Wang, Liu, Xia and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Liu, bHd0b25nQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.