
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 08 March 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1139961
This article is part of the Research Topic Enhancing Salinity Tolerance in Crop Plants through Agronomic, Genetic, Molecular, and Physiological Approaches View all 17 articles
 Lei Lei1,2,3,4
Lei Lei1,2,3,4 Liangzi Cao2,3,4
Liangzi Cao2,3,4 Guohua Ding2,3,4
Guohua Ding2,3,4 Jinsong Zhou2,3,4
Jinsong Zhou2,3,4 Yu Luo2
Yu Luo2 Liangming Bai2,3
Liangming Bai2,3 Tianshu Xia2
Tianshu Xia2 Lei Chen5
Lei Chen5 Jiangxu Wang5
Jiangxu Wang5 Kai Liu5
Kai Liu5 Qingjun Lei6
Qingjun Lei6 Tingting Xie2
Tingting Xie2 Guang Yang2
Guang Yang2 Xueyang Wang5
Xueyang Wang5 Shichen Sun2,3,4*
Shichen Sun2,3,4* Yongcai Lai2,4*
Yongcai Lai2,4*Rice has been reported to be highly sensitive to salt stress at the seedling stage. However, the lack of target genes that can be used for improving salt tolerance has resulted in several saline soils unsuitable for cultivation and planting. To characterize new salt-tolerant genes, we used 1,002 F2:3 populations derived from Teng-Xi144 and Long-Dao19 crosses as the phenotypic source to systematically characterize seedlings’ survival days and ion concentration under salt stress. Utilizing QTL-seq resequencing technology and a high-density linkage map based on 4,326 SNP markers, we identified qSTS4 as a major QTL influencing seedling salt tolerance, which accounted for 33.14% of the phenotypic variation. Through functional annotation, variation detection and qRT-PCR analysis of genes within 46.9 Kb of qSTS4, it was revealed that there was one SNP in the promoter region of OsBBX11, which resulted in a significant response difference between the two parents to salt stress. Transgenic plants using knockout-based technology and demonstrated that Na+ and K+ in the roots of the functional-loss-type OsBBX11 were translocated largely to the leaves under 120 mmol/L NaCl compared with the wild-type, causing osbbx11 leaves to die after 12 days of salt stress due to an imbalance in osmotic pressure. In conclusion, this study identified OsBBX11 as a salt-tolerance gene, and one SNPs in the OsBBX11 promoter region can be used to identify its interacting transcription factors. This provides a theoretical basis for finding the molecular mechanism of OsBBX11 upstream and downstream regulation of salt tolerance and molecular design breeding in the future.
Rice (Oryza sativa L.) is a staple food for much of the global population (Ponce et al., 2021), Approximately 30% of the global rice cultivation area is affected by salinity (Takehisa et al., 2004). According to the data from the United Nations Educational, Scientific and Cultural Organization and the United Nations Food and Agriculture Organization, China has 9913 hectares of saline-alkali land (He and Luo, 2021). When the concentration of soluble salt in the soil reaches 0.3%, the photosynthetic efficiency of rice seedlings decreases, and the organism exhibits ionic imbalance and high osmotic pressure, leading to plant wilting and apoptosis, and reduced rice yield (Akbar et al., 1972; Ma et al., 2017), which severely restricts the distribution of rice planting. Therefore, screening salt-tolerant rice varieties and analyzing the genetic mechanism of salt tolerance in rice are of great significance for ensuring rice production.
Salt tolerance of rice is a typical quantitative trait controlled by multiple genes (Johnson et al., 1992). To date, approximately one thousand QTLs associated with salt tolerance in different growth stages of rice have been reported (Prakash et al., 2022). However, most of the QTLs are difficult to utilize in molecular marker-assisted breeding, leading to only qSKC-1 and Saltol (Lin et al., 2004; Ren et al., 2005) being finely mapped or map-cloned, and the candidate gene for Saltol has yet to be isolated. Therefore, it is highly challenging to clone salt-tolerance QTLs in rice. Reverse genetic approaches have confirmed that more than 300 rice genes, including transcription factors OsbZIP20 (Baoxiang et al., 2022), OsbZIP72 (Baoxiang et al., 2021), OsMYB106 (Wang et al., 2020), ion transporter protein OsHKT2;2 (Kader et al., 2006), OsNHX2 (Fukuda et al., 2011), protein kinase SIT1/SIT2 (Li et al., 2014), and OsMAPKKK63 (Na et al., 2019), regulate salt tolerance in rice. However, transgenic salt-tolerant rice typically displays growth deficiencies and lacks genetic salt-tolerant allele data, making it difficult to utilize for rice molecular breeding. Molecular marker-assisted selection breeding can reduce the duration of salt-tolerant rice breeding by four to seven years, with Saltol being the most widely employed. (Bimpong et al., 2016), only Saltol has been extensively used in salt tolerance breeding (Salam et al., 2007; Singh et al., 2011; Babu et al., 2012; Linh et al., 2012; Gregorio et al., 2013; Vinod et al., 2013). However, its utilization is restricted to India, the Philippines, Bangladesh, Thailand, Vietnam, and Senegal(Linh et al., 2012; Singh et al., 2016), and no salt-tolerant genes suitable for application in the Chinese rice cultivation context have been identified. Consequently, it is imperative to conduct further research and uncover salt-tolerant QTL/genes for potential functional applications in China.
In comparison to traditional QTL mapping, the integration of QTL-Seq technology and high-density linkage maps can expedite the identification of major genes of rice traits(Kodama et al., 2018; Sun et al., 2019). Additionally, resistance genes can be extracted from local cultivated varieties and utilized for molecular breeding, taking advantage of its superior allelic variations without the need to consider the genetic background interference caused by kinship (Xiao et al., 2018; Xu et al., 2020), thus yielding salt-tolerant varieties suitable for production. Nevertheless, the salt-tolerant QTLs reported mainly originate from the varieties developed by the International Rice Research Institute and the indica or japonica rice in southern China, while few are from northern China. In the present study, we used a QTL-Seq and high-density linkage mapping approach with 1002 F2 populations derived from a cross between Teng-Xi144 (TX144, a salt-sensitive variety) and Long-Dao19 (LD19, a strongly salt-tolerant variety) and five traits: survival days of seedlings (SDSs), the shoot Na+ concentration (SNC), shoot K+ concentration (SKC), root Na+ concentration (RNC), and root K+ concentration (RKC) under 120mmol/L NaCl stress, to identify major QTL intervals for salt tolerance at the seedling stage (STS) in rice. The candidate genes of QTL were identified by detecting the variation of sequence and the expression level of genes under salt stress. Next, the salt tolerance function of the gene was characterized by the salt tolerance phenotype of the transgenic plants. This provides a key theory for the molecular breeding of salt tolerance genes and the molecular mechanism of salt tolerance in Longdao 19.
For QTL-sequencing and linkage mapping, the two extreme rice cultivars, namely, TX144 (salt-sensitive) as the female parent and LD19 (salt-tolerant) as the male parent, were crossed to develop a population of 1002 F2:3 by single seed descent at the experimental station of Heilongjiang Academy of Agricultural Sciences (Heilongjiang Province, China; 47°98′ N, 128°08′ E; 128 m above sea level).
Salt tolerance for all 1002 F2:3 and their parents was evaluated in a hydroponics system at the experimental station of Northeast Agricultural University. Briefly, the individuals were harvested and kept at 40°C for 2 days to break seed dormancy. Afterward, the seeds were surface-sterilized with 2% NaClO3 for 10 min, washed thrice with sterile water, and soaked in water in Petri dishes at 30°C for germination. When the seed bud was equal to half the length of the seed, 50 uniformly germinated seeds were selected in each Petri dish and hydroponically grown in the Yoshida nutrient solution. Each experimental treatment was repeated thrice. The climatic cabinet temperature was set at 28°C/25°C day/night cycle with 60% relative humidity. Rice reached the three-leaf stage and was treated with 120 mmol/L NaCl solution in a stressful environment for 7 days, following which the salt solution was replaced with nutrient solution, and the incubation was continued for 5 days. Next, survival days of seedlings (SDSs) were recorded for each plant in days from seeding to death. To observe physiological traits, the second experiment was performed. The procedure and administration of the experiments were the same as those described above. At 7 days after treatment with NaCl, each sample’s shoots were harvested and dried at 100°C for 30 min and 60°C for 1 week. A total of 0.1 g of dried sample was ground and subsequently digested with 0.1 N nitric acid (Fisher Scientific) at 70°C for 8 h (Campbell et al., 2017). The shoot Na+ concentration (SNC), shoot K+ concentration (SKC), root Na+ concentration (RNC), and root K+ concentration (RKC) were measured using a flame photometer (Sherwood 410, Cambridge, UK) to evaluate the salt tolerance at the seedling stage.
To prepare paired-end libraries with an insert size of 500 bp using the paired-end DNA sample prep kit (Illumina, San Diego, CA, USA), the total genomic DNA was extracted from bulked pools, and at least 3 g of the genomic DNA were used to construct genomic libraries. A HiSeq X10 (Illumina) NGS platform was used at Gene Denovo (Guangzhou, China) to sequence these libraries. Quality trimming is an essential step in generating high-confidence variant calling. To achieve high-quality clean reads, we used the following three stringent criteria to filter the raw reads: 1) removing reads with more than 10% unidentified nucleotides; 2) removing reads with > 50% PhRED quality scores of > 20; and 3) aligning reads to barcodes.
Burrows–Wheeler Aligner (v.0.7.16a-r1181) was used to align filtered reads with the Nipponbare reference genome sequence (Matsumoto et al., 2005) using the parameter ‘mem-M’. Using mem-M, shorter split-alignment hits were marked as secondary alignments (Kumar et al., 2019). UnifiedGenotyper (v.3.5) was used for variant calling (http://gatk.broadinstitute.org/hc/en-us/community/posts/36007363711-0-UnifiedGenotyper-in-GATK4). In the ANNOVAR tool (Wang et al., 2010), all mutations were annotated for genes, functions, and genomic regions. SNP-Index (Takagi et al., 2013), G-Statistic (Magwene et al., 2011; Mansfeld and Grumet, 2018), ED (Hill et al., 2013) and two-tailed Fisher’s exact test (Fisher, 1922) based on SNPs were used for association analysis and calculated using a 2000-kb sliding window with a step size of 20 kb and excluding windows with less than 10 SNP/Indel. The results obtained were merged into the next window in case of insufficient SNPs. The final QTL interval was calculated using the overlapping intervals of the four methods. The raw Illumina sequencing data from this study were submitted to the NCBI’s Sequence Read Archive (SRA) database under accession number SRR22671405, SRR22671406, SRR22671407, and SRR22671408.
To extract the genomic DNA, Fresh leaflets 3 to 5 cm in length were collected and ground, and frozen in liquid nitrogen (Promega Corporation, Madison, WI, USA). SNP assays were conducted using DNA samples showing high quality (minimum 10 kb fragments) and an appropriate concentration (10–50 ng/mL) according to agarose gel electrophoresis. Only high-quality DNA samples were used for SNP analysis. Polymorphism in 1002 F2:3 lines and their parents was detected using a 10 K rice genotyping by target sequencing (GBTS) liquid phase chip based on GenioBits technology (Molbreeding Biotechnology Co., Shijiazhuang, China). We first screened SNPs according to parental genotypes, and afterward removed redundant markers from datasets using the IciMapping 4.2 software’s “BIN” computing module. Finally, these BINs were mapped using the JoinMap 4.0 software (Van Ooijen, 2006). QTL analysis was based on the arithmetic mean values of three replicates for each trait, determined by the inclusive composite interval mapping in IciMapping 4.2. LOD ≥ 2.5 was set as the threshold to decide the QTL for each trait, with 1 cM walking speed.
At the two-leaf stage, plants in the control experiment were grown under normal conditions, and seedlings in the stress experiment were exposed to salt stress (120 mM NaCl). Leaf samples were collected and rice leaves from TX144 and LD19 were sampled from both sets of experiments at 0 and 12 h of stress and immediately frozen in liquid nitrogen. Next, 2 g of the total RNA was extracted using the TRIzol reagent, and first-strand cDNA was synthesized using Invitrogen’s Superscript II Reverse Transcriptase kit (Invitrogen). The qRT-PCR was performed using a Roche Light Cycler 2.10 system and the 2Fast qPCR Master Mix. Primer 5.0 software was used to develop gene-specific primer sequences (Supplementary Table 1). Three biological replicates and three technical replicates were used for measurements. Actin-EFα1 (LOC_Os03g08010) was used as the internal control (Livak and Schmittgen, 2001). The relative gene expression was calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
To generate qsts4 knockdown plants, plasmids were introduced into the Agrobacterium tumefaciens strain EHA105. Plasmid construction and plant transformation were performed as described previously (Li et al., 2017). Two target sequences, including point accepted mutation (PAM) (GGTGTGCTGCGCGGACGAGGCGG/GGCGTTCATCTTCTGCGTGGAGG) were selected within the target genes. After PCR-mediated DNA amplification in the knockout lines, the designed target amplicons (300-500 bp) were sequenced directly and identified using degenerate sequence decoding (Ma et al., 2015). Knockout lines were detected by PCR sequencing with primers 5’-TGTGCGGTTTTGAGGGGAGT-3’ and 5’-TAATCTTGTTATCAAGAATG-3’. Primers used for vector construction are listed in Supplementary Table 1.
The descriptive statistical analysis and differences between parental and F2:3 lines for salt tolerance were performed using the SigmaPlot software (v.12.5; Systat Software, San Jose, CA, USA). Data represent the mean ± standard deviation. All graphs were drawn using Origin 2018 (OriginLab, Northampton, MA, USA) or SPSS 18.0 (IBM, Armonk, NY, USA).
We assessed the phenotypic variations under salt stress and control conditions. Out of two parental varieties, TX144 but not LD19, presented a salt-sensitive phenotype, indicating that LD19 had stronger salt tolerance (Figure 1A). In addition, survival days of seedlings (SDS) of 1002 F2:3 progeny (Figure 1B) ranged from 20 to 34 days, of which 50 most and least salt-tolerant progeny were assigned to T-pools and S-pools, respectively, for DNA re-sequencing. To study physiological traits under salt stress, four traits were measured. The F2:3 population showed continuous segregation. The SNC, SKC, RKC, and RNC demonstrated a normal distribution (Figures 1C–F), which was consistent with the inheritance of quantitative traits. Moreover, the absolute values of skewness and kurtosis for all traits were < 1, indicating that the data of these traits were suitable for QTL analysis (Supplementary Table 2).
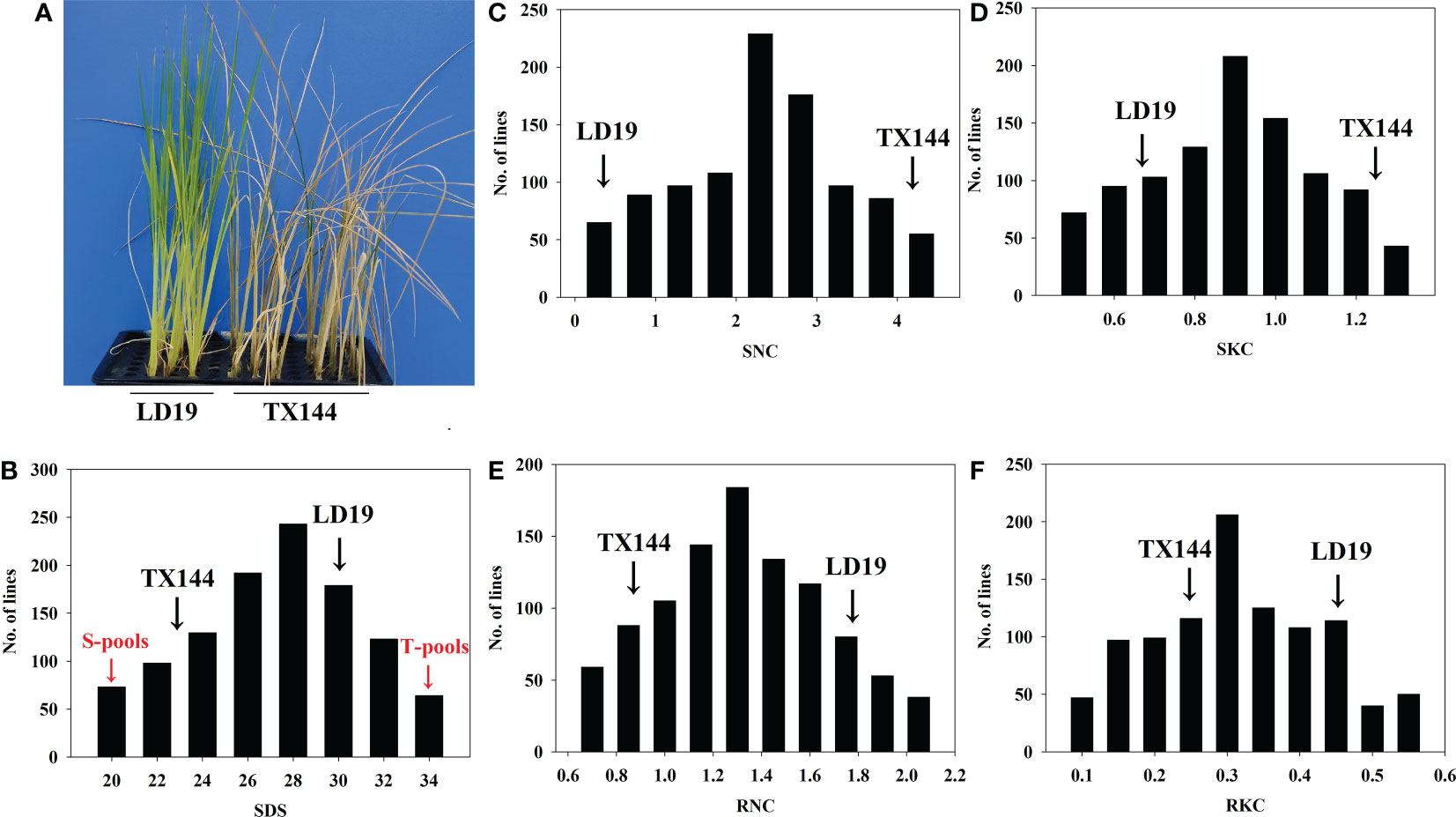
Figure 1 Phenotypic analysis of the parents and their derived populations under salt stress. (A) Comparison of the growth of seedling plants of the two parents under salt stress. (B–F) Frequency distribution of days of seedling survival and four physiological traits of shoots and roots of 1002 F2:3 lines under salt stress.
Pearson correlation coefficients were used to assess the correlation between different traits. Supplementary Table 3 shows a significant correlation among the five traits under salt stress. SDS was only negatively correlated with SNC in the shoots, whereas SNC was negatively correlated with SKC in the shoots. We observed a positive correlation between RNC and between RKC for both traits in seedling roots, suggesting a more significant influence of these traits under salt conditions.
The average sequencing depth of parents and the two pools was 50×. In addition, 1,067,436 SNPs and 177,492 indels were blasted using the ‘Nipponbare’ reference genome, followed by trimming and filtering to finally generate 443,872 SNPs and 72,173 indels. Finally, 516,045 high-quality SNPs/indels were preferentially selected for QTL-sequencing analysis (Supplementary Table 4). Δ (SNP-Index) values (Takagi et al., 2013), Euclidean distance (ED) values (Hill et al., 2013), G values (Magwene et al., 2011; Mansfeld and Grumet, 2018), and Fisher’s exact test P-values (Fisher, 1922) were used to identify the candidate QTL regions contributing to salt tolerance at the seedling stage (STS). Figure 2 and Supplementary Table 5 show that only a significant (P < 0.01) peak in the Δ (SNP-Index), ED, and G values, whereas E distributions spanned the intervals of 14.4-Mb (16.3-30.7 Mb), 4.2-Mb (21.2-25.4 Mb), and 5.2 (20.4-25.6 Mb) on chromosome 4, respectively. We designated this QTL as qSDS4. Altogether, a 4.2-Mb overlapping interval of the three methods resulted in the final qSTS4 interval.
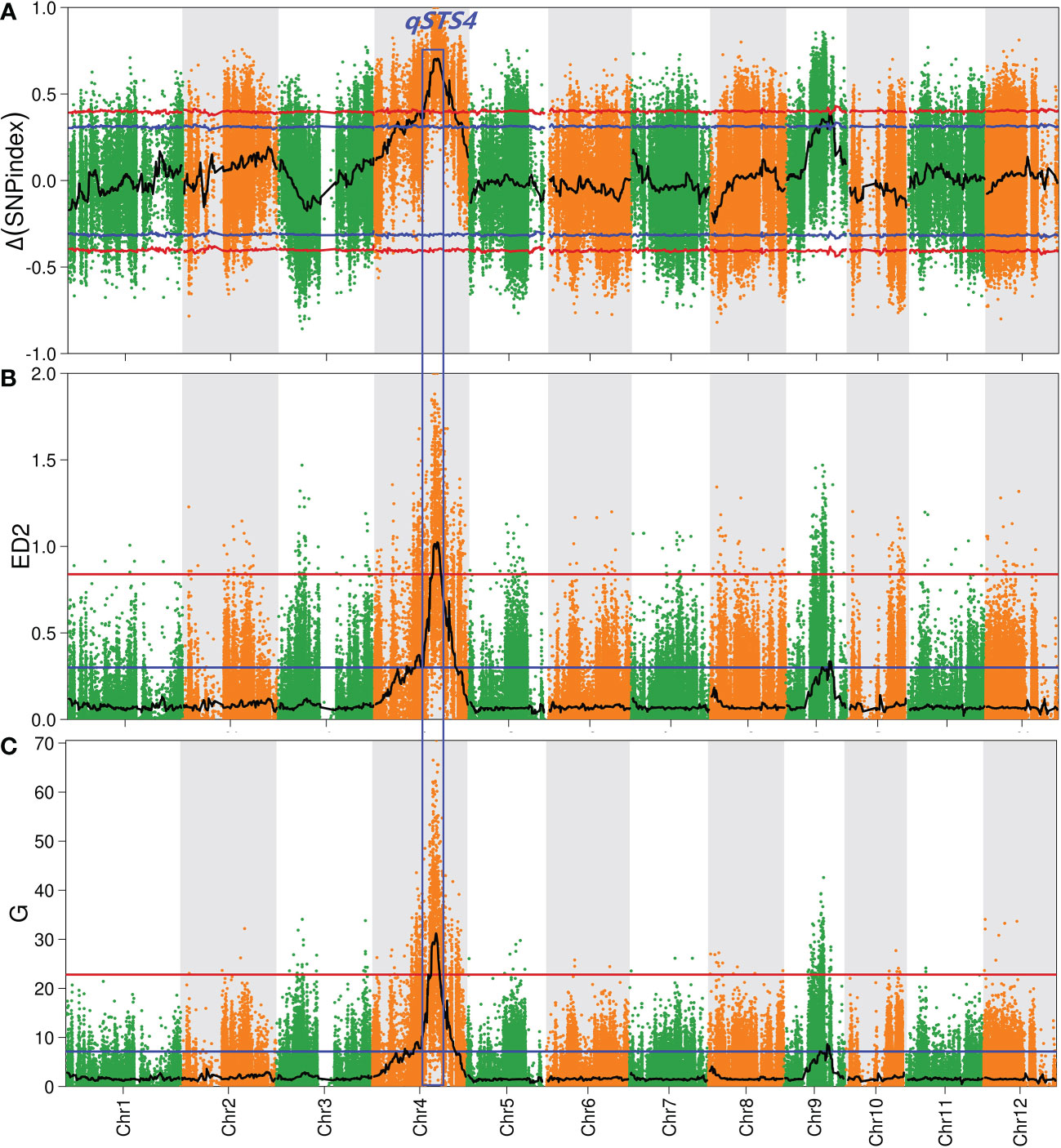
Figure 2 Quantitative trait locus (QTL) analysis of rice salt tolerance at seeding stage by QTL-seq. (A–C) represent Manhattan plots of Δ (SNP-index), Euclidean distance (ED2) and G-value on chromosomes, respectively. The blue and red lines represent the 95% and 99% confidence intervals, respectively, and the black line represents the mean of the four algorithms, plotted using sliding window analysis. The numbers on the horizontal coordinates represent chromosome numbers.
The 10K rice genotyping chip was used to detect 4,326 SNPs in 1002 F2:3 lines and their parents. A total of 1,107 bin markers were evenly distributed on 12 chromosomes and obtained using the “BIN” calculation module of the IciMapping 4.2 software. A linkage map covering a total genetic distance of 2252.63 cM of the rice genome was obtained, and the average distance between Bin markers was 2.03 cM (Figure S1). For SDS, two QTLs were detected on chromosomes 1 and 4, respectively (Supplementary Table 6; Figure S1). qSDS4-1 which exerted the most significant effect contributed to 13.89% of the total phenotypic variance. The additive effect of the LD19 allele increased SDS by 3.20 days. The other QTLs, such as qSDS1, accounted for 6.0% of the total phenotypic variance, and the LD19 allele at qSDS1 increased the SDS (Supplementary Table 6). For SKC, SNC, and RKC, a significant LOD peak interval on chromosome 4 was detected. We designated these QTLs as qSKC4, qSNC4, and qRKC4, respectively. The percentages of phenotypic variance explained by qSKC4, qSNC4, and qRKC4 were 29.21%, 30.40%, and 28.09%, respectively. For RNC, only qRNC was detected on chromosome 4. This QTL had a relatively small effect, which contributed to 8.12% of the total phenotypic variance (Supplementary Table 6). Altogether, qSDS4-1, qSKC4, qSNC4, and qRKC4 were anchored within the 46.9-Kb interval between 24625017-bp and 24671947-bp, which overlapped completely with the qSDS4-1 interval detected by QTL-sequencing. Therefore, this peak interval with one-causal multiple effects was considered the STS QTL and termed qSTS4 (Figure 3A).
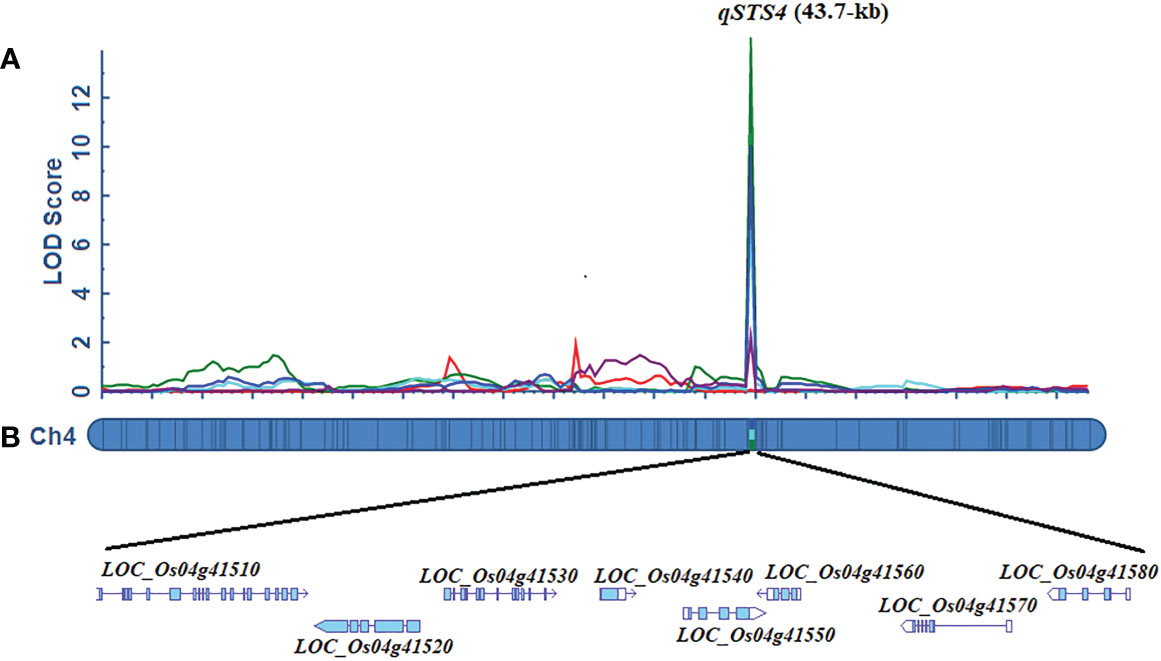
Figure 3 Fine mapping of the qSTS4. (A) Localization of qSTS4 based on high-density mapping by ICIM module of QTL IciMapping 4.2. (B) Putative salt genes identified at qSTS4 using annotation information from the ‘Nipponbare’ reference genome (http://plants.ensembl.org/index.html/).
Based on the Nipponbare genome annotation (http://plants.ensembl.org/index.html/), the annotation information of the eight genes within the qSTS4 interval is shown in Supplemental Table 7 and Figure 3B. These genes include 1 retrotransposon protein, 2 serine/threonine-protein kinase GCN2, 1 CRS1/YhbY domain-containing protein, 1 B-box zinc finger family protein, 1 ethylene-responsive protein-related, 1 glycine-rich protein, and 1 Calmodulin-related calcium sensor protein. Out of the eight annotated genes, only LOC_Os04g41560 was a B-box zinc finger family protein (OsBXX11), which was annotated as the function of “response to abiotic stimulus (GO:0009628, GO:0009791).” Orthologous group of LOC_Os04g41560 in plants, AT2G31380 in Arabidopsis, Sb04g025400 and Sb06g021170 in Sorghum, GRMZM2G018876, GRMZM2G019335, and GRMZM2G095299 in maize, and Bradi3g48180 in Brachypodium were annotated as putative salt tolerance genes using the http://rice.uga.edu/ database. Moreover, two of the eight annotated genes contained two SNP loci in the upstream or CDS, including one SNP in the upstream LOC_Os04g41560 (Figure 4) and one SNP in the CDS of LOC_Os04g41520 (Supplemental Table 8). To verify the expression characteristics of eight genes under salt stress, their expression patterns under salt stress and control (no stress) were determined by real-time reverse transcription PCR (qRT-PCR). Under salt stress, LOC_Os04g41520 and LOC_Os04g41570 were downregulated in both LD19 and TX144, and LOC_Os04g41510, LOC_Os04g41530, LOC_Os04g41540, LOC_Os04g41550, and LOC_Os04g41580 were up-regulated in both LD19 and TX144. Compared to TX144, only LOC_Os04g41560 was upregulated in LD19 under salt stress (Figure 5), implicating LOC_Os04g41560 in regulating salt tolerance in rice.
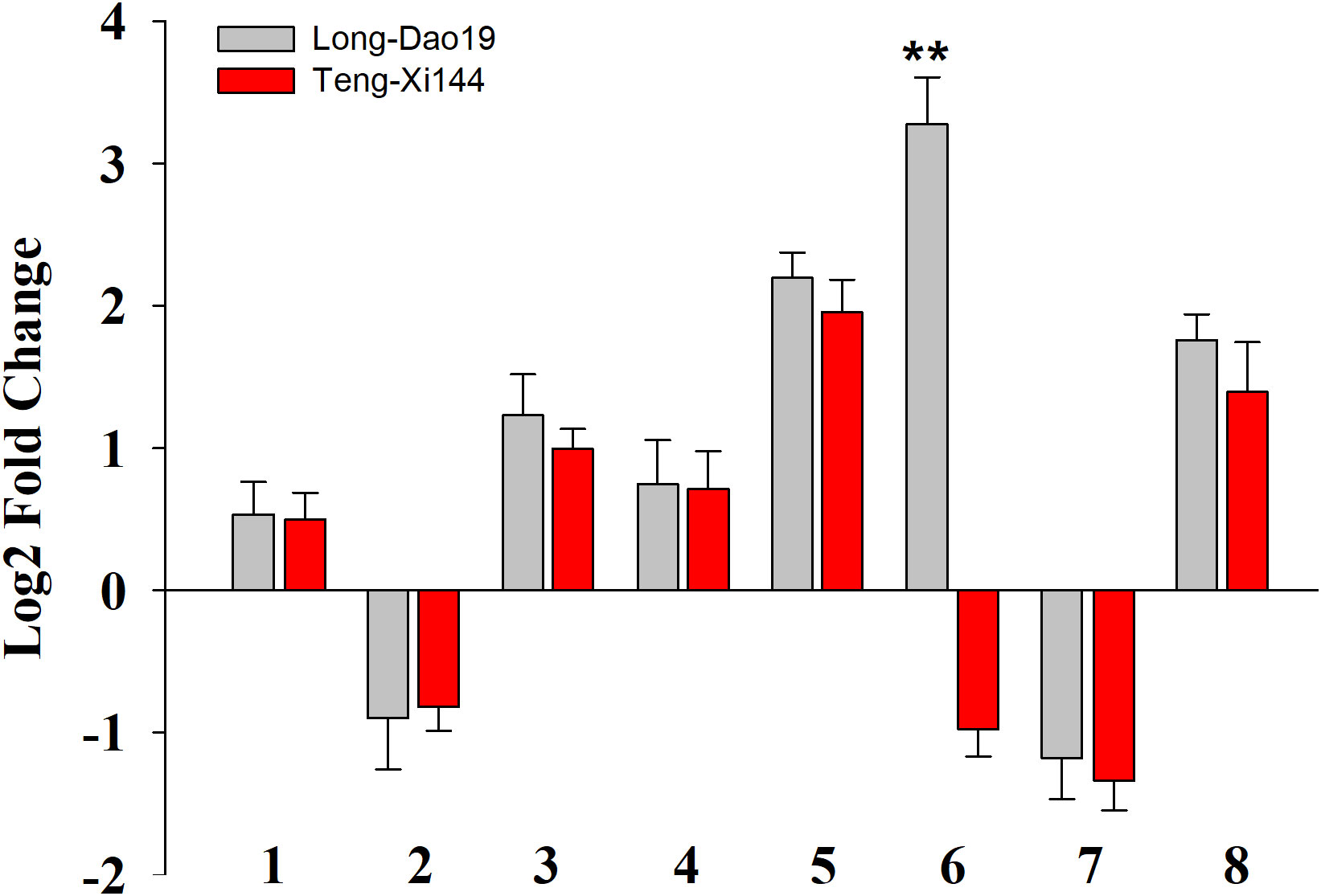
Figure 5 The expression level of selected genes in LD19 and TX144 12h after exposure to salt stress. Log2-fold change was calculated for gene expression under salt stress compared with control. 1–8 represents the genes used for expression analysis. 1-LOC_Os04g41510; 2-LOC_Os04g41520; 3-LOC_Os04g41530; 4-LOC_Os04g41540; 5-LOC_Os04g41550; 6-LOC_Os04g41560; 7-LOC_Os04g41570; 8-LOC_Os04g41580. **P < 0.01; Student’s t-test.
Next, we verified the salt tolerance of LOC_Os04g41560 (OsBXX11) using knockout lines generated by clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated gene editing. As shown in Figure 6A, osbbx11 exhibited a strong loss-of-function phenotype under salt stress, i.e., almost all seedlings died after 10 days of salt stress and 12 days of resuming growth in a non-salt environment (Figure 6B). Subsequently, we collected these rice seedlings and measured the SNC, SKC, RNC, and RKC. The results showed that the SNC and SKC in osbbx11 shoots were more than two times higher than in LD19 shoots. In contrast, the values of RNC and RKC in the roots of osbbx11 were low compared with those of LD19 (Figure 6C). Altogether, OsBBX11 has a positive and significant function in STS.
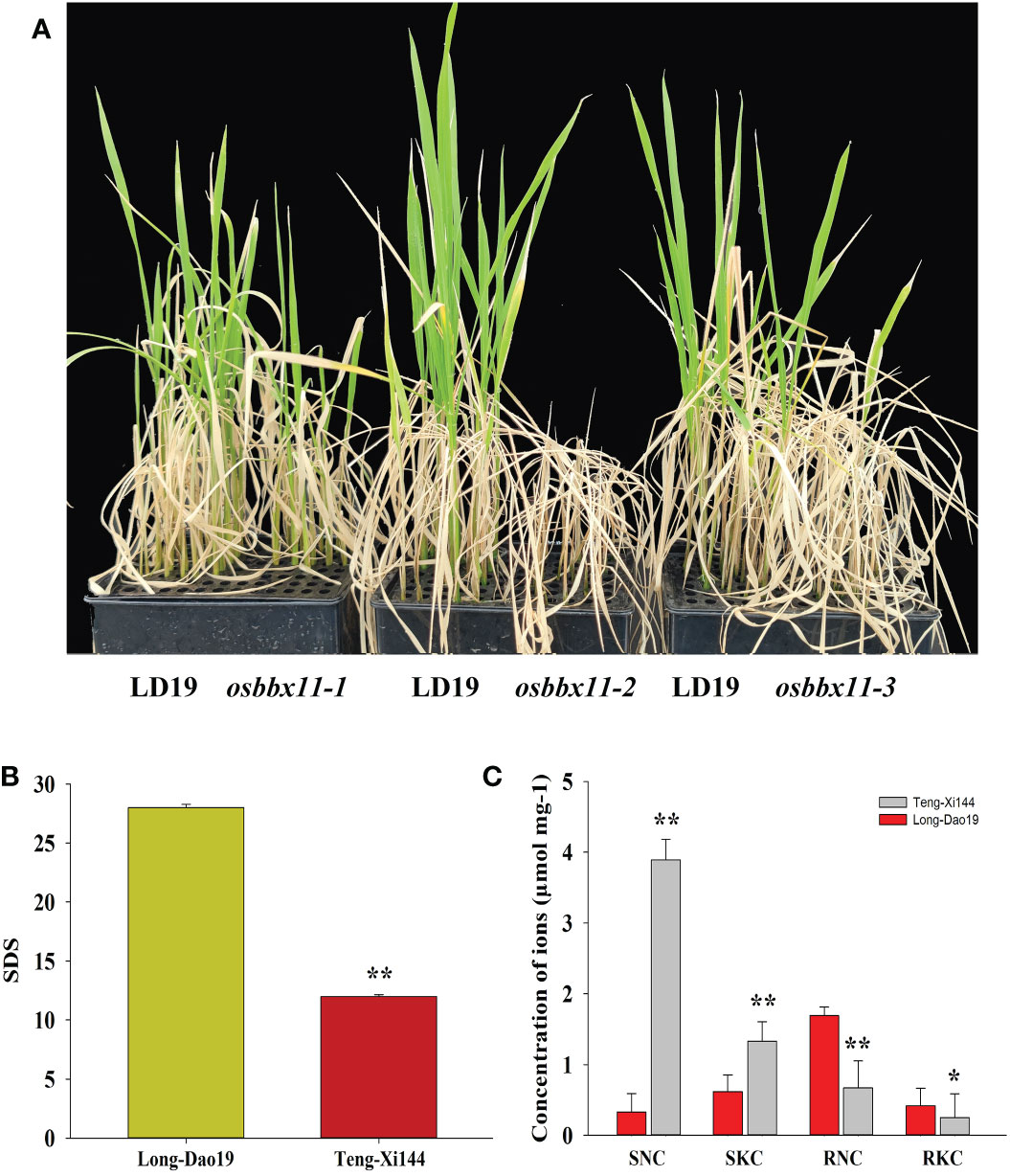
Figure 6 Functional analysis of rice’s B-box zinc finger family protein LOC_Os04g41560 (OsBX11). (A) Salt tolerance phenotype of knockout plants after 10 days of salt stress and 12 days of resuming growth in a non-salt environment. (B) Comparison of seedling survival days after salt stress between knockout and wild-type plants. (C) Comparison of Na+1 and K+1 concentrations in roots and shoots of knockout and wild-type plants after salt stress. **P < 0.01; Student’s t-test *P < 0.05; Student’s t-test.
Salt tolerance at the seedling stage is highly crucial because the effects on initial plant stages decide the production (Liang et al., 2015). Therefore, it is essential to decipher the mechanism of salt tolerance at the seedling stage to improve rice yield. Although several QTLs and candidate genes contributing to seedling salt tolerance have been identified in rice(Nayyeripasand et al., 2021; Yadav et al., 2021; de Ocampo et al., 2022; Yuan et al., 2022). To date, the molecular mechanism of salt tolerance in rice at the seedling stage has not been elucidated. Several factors, such as chlorophyll contents, Na+ uptake, K+ uptake, ionic balance, and ionic compartment contribute to salt tolerance (Nieves-Cordones et al., 2016; Le et al., 2021; Nakhla et al., 2021; Tian et al., 2021).STS was regulated by the balanced relationship between Na+ and K+ in the roots and shoots. As shown in Figure 1, in response to salt stress, the salt-sensitive variety TX144 excessively accumulated Na+ and K+ in the leaves compared with the salt-tolerant variety LD19. However, the ion concentration in the roots of TX144 was reduced, and it could survive for only 23 days. Furthermore, Several studies have demonstrated that the uptake of Na+ and K+ by rice is independent under salt stress (Yeo et al., 1987; Koyama et al., 2001). However, the data in Supplemental Table 3 reveals a negative correlation between SKC and SNC, indicating competition between Na+ and K+ in the shoots. However, in the roots, the relationship between Na+ and K+ was synergistic. For related traits, QTLs were often mapped in the same chromosomal region (Paterson et al., 1991; Lin et al., 2004; Pundir et al., 2021). We found that qSTS4 was linked to SDS, SNC, SKC, and RKC. These results suggest that the correlation of traits could be attributed to polymorphic effects or close linkage of genes. In addition, the results indicate a vital function of qSTS4 in determining salt tolerance in both parental genotypes.
It is highly challenging to develop high-density genetic maps and a series of near-isogenic lines using traditional map-based QTL cloning methods. QTL-sequencing has merged as a powerful tool to rapidly detect major QTLs. Moreover, it has been applied to the genetic study of salt tolerance in crops; however, these studies reveal hundreds of candidate genes in the QTL interval that need to be researched (Tiwari et al., 2016; Shamaya et al., 2017; Kodama et al., 2018; Wu et al., 2020). An alternative method to identify candidate genes is to design molecular markers within the interval obtained by QTL-sequencing and further narrow QTL intervals through linkage maps. (Zhang et al., 2018). However, because salt tolerance is a complex quantitative inheritance in crops, the difference interval of parents on salt tolerance should be mined from all chromosomes. We used QTL-sequencing and linkage analysis to detect an overlapping peak interval (Figure 3A). Linkage analysis additionally detected qSDS1 and qRNC7 on chromosomes 1 and 7, indicating a difference in the allele ratio among the F2:3 populations. Interestingly, OsHsfA7, located in the qSDS1 interval, is a heat shock protein. Under salt treatment, rice overexpressing OsHsfA7 relieved damage symptoms and higher survival rates (Liu et al., 2013). qSTS4 was a major QTL detected on chromosome 7, contributing to 33.14%, 28.09%, 30.40%, and 29.21% of the phenotypic variations for SDS, RKC, SNC, and SKC, respectively (Supplementary Table 6), whereas Na+ concentration exerted only a small effect and accounted for 8.12% of the variance for qRNC7. It is noteworthy that qSTS4 and qRNC7 were not detected in the previous study, probably due to the lack of variations in these two QTLs between the two parents. Therefore, these two loci can be used as new salt-tolerant targets in LD19 for subsequent molecular breeding. Currently, the different functions of alleles of several key genes under salt stress have been confirmed, such as OsCPK17 and OsHKT1;5 (Negrão et al., 2013), HKT1;5 and HKT2;3 (Mishra et al., 2016), as well as the salt-tolerant alleles obtained from 3000 rice resources(Campbell et al., 2017; Liu et al., 2019). However, these alleles were considered for salt-tolerant breeding of northern japonica rice before the separation of japonica rice in northern China was resolved. In our study, both TX144 and LD19 are northern japonica rice. The results of QTL-seq and linkage analysis suggest that the SNP in the confidence interval causes phenotypic separation, indicating that there are alleles that can distinguish salt tolerance in these two cultivars. In our study, it is unclear whether the SNP difference in the promoter of OsBBX11 of TX144 and LD19 is the cause of the differential response to salt stress. To elucidate this issue, we plan to clone the alleles of OsBBX11, construct expression vectors of plants and yeast, and express these vectors in rice protoplasts, yeast, and Arabidopsis to compare the effects of this natural allele variations on gene expression and subcellular localization under salt stress.
The B-box (BBX) gene family, encoding zinc-finger proteins, is an important family of transcription factors in plants. It has several functions, such as response to environmental changes and plant growth and development (Robson et al., 2001; Lira et al., 2020). In rice, the BBX genes such as OsCO3 (Kim et al., 2008), Hd1 (Zong et al., 2021), OsCCT19 (Zhang et al., 2015), DTH2 (Wu et al., 2013), OsCOL15 (Wu et al., 2018), and OsBBX14 (Bai et al., 2016) are largely involved in regulating the heather dying stage and rice response to light. However, none of the BBX genes have been characterized to be associated with salinity tolerance in rice. However, several BBX genes have been reported to be associated with the regulation of salt tolerance in plants. For instance, the overexpressed Malus domestica gene MdBBX10 (Jackson and Linsley, 2004) enhanced salt tolerance in Arabidopsis, as well as AtBBX5 (Min et al., 2015) and AtBBX24 (Nagaoka and Takano, 2003) in Arabidopsis are induced by salt stress. Although A and B genes,.OsBBX4 (Liu et al., 2012) and OsBBX22 (Jain et al., 2007) in rice are induced by salt stress, their salt-tolerance functions have yet to be characterized. Recently, several key genes, such as OsMIR408, OsmiR535, OsNAC3, and BEARI, have been intervened by gene knockout technology to alleviate the damage of salt stress in plants (Yue et al., 2020; Zhang et al., 2021; Zhou et al., 2021; Teng et al., 2022). In our study, OsBBX11 was mapping as a B-box zinc finger family protein (BBX). Although transgenic lines confirmed that OsBBX11 may play a salt-tolerant function by affecting the transport of sodium and potassium ions from underground to aboveground, the underlying molecular mechanism remains elusive. Previous studies have demonstrated that OsBBX11 was down-regulated by including NAA (a member of the auxin family), GA3 (a gibberellin), and KT (a cytokinin) (Huang et al., 2012). Based on this, the study of the salt tolerance mechanism of OsBBX11 should consider the effects of ion transport channel proteins or hormone regulation. As a newly identified salt-tolerance regulatory factor, the physiological mechanism of salt-tolerance regulation of OsBBX11 remains unclear. In addition, the molecular mechanism of promoter components interacting with transcription factors and the protein factors interacting with OsBBX11 also need further investigation. Moreover, whether hormones are involved in regulating the response to salt stress warrants verification by creating multiple types of transgenic material.
Based on QTL-seq and high-density linkage map, we identified QTLs associated with seedling salt-tolerance in rice. qRT-PCR analysis showed that the salt-responsive gene OsBBX11 was a candidate gene in qSTS4 region. Transgenic plant phenotypic detection confirmed that OsBBX11 plays a salt-tolerance function by affecting the transport of Na+ and K+ from underground to aboveground. In addition, its alleles of the salt-specifically induced expression pattern of the Longdao-9 can be an important target for salt-tolerance improvement in rice.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI SRA: SRX18634378, SRR22671406, SRR22671407, and SRR22671408.
LL, LZC and GHD conceived and designed the research. LMC, JXW, JSZ, TSX and LC participated in data analysis. YL, KL, QJL, TTX, GY and XYW performed material development, sample preparation, and data analysis. LL and SCS wrote the manuscript. SCS and YCL corrected the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the Heilongjiang Postdoctoral Foundation (LBH-Z22265), Key R & D Program of Heilongjiang (2022ZX02B04-3), and the earmarked fund for CARS-01.
The authors would like to thank all the reviewers who participated in the review and MJEditor (www.mjeditor.com) for its linguistic assistance during the preparation of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1139961/full#supplementary-material
Supplementary Figure 1 | A linkage map covering a total genetic distance of 2252.63 cM and 4326 SNP markers in the rice genome with an average distance between Bin markers of 2.03 cM. qSDS4-1, qSKC4, qSNC4, qRKC4, qSDS1 and qRNC7 were detected based on a threshold of LOD>3.
Akbar, M., Yabuno, T., Nakao, S. (1972). Breeding for saline-resistant varieties of rice: I. variability for salt tolerance among some rice varietles. Japanese J. Breed. 22, 277–284. doi: 10.1270/jsbbs1951.22.277
Babu, N., Sharma, S., Ellur, R., Singh, V., Pal, M., Pathania, S., et al. (2012). “Marker assisted improvement of pusa basmati 1121 for salinity tolerance,” in International conference on plant biotechnology for food security: New frontiers, national agricultural science centre, New Delhi. 72–73.
Bai, B., Zhao, J., Li, Y., Zhang, F., Zhou, J., Chen, F., et al. (2016). OsBBX14 delays heading date by repressing florigen gene expression under long and short-day conditions in rice. Plant Sci. 247, 25–34. doi: 10.1016/j.plantsci.2016.02.017
Baoxiang, W., Bo, X., Yan, L., Jingfang, L., Zhiguang, S., Ming, C., et al. (2022). A novel mechanisms of the signaling cascade associated with the SAPK10-bZIP20-NHX1 synergistic interaction to enhance tolerance of plant to abiotic stress in rice (Oryza sativa l.). Plant Sci. 323, 111393. doi: 10.1016/j.plantsci.2022.111393
Baoxiang, W., Yan, L., Yifeng, W., Jingfang, L., Zhiguang, S., Ming, C., et al. (2021). OsbZIP72 is involved in transcriptional gene-regulation pathway of abscisic acid signal transduction by activating rice high-affinity potassium transporter OsHKT1; 1. Rice Sci. 28, 257–267. doi: 10.1016/j.rsci.2021.04.005
Bimpong, I. K., Manneh, B., Sock, M., Diaw, F., Amoah, N. K. A., Ismail, A.M., et al. (2016). Improving salt tolerance of lowland rice cultivar ‘Rassi’ through marker-aided backcross breeding in West Africa. Plant Sci. 242, 288–299. doi: 10.1016/j.plantsci.2015.09.020
Campbell, M. T., Bandillo, N., Al Shiblawi, F. R. A., Sharma, S., Liu, K., Du, Q., et al. (2017). Allelic variants of OsHKT1;1 underlie the divergence between indica and japonica subspecies of rice (Oryza sativa) for root sodium content. PloS Genet. 13, e1006823. doi: 10.1371/journal.pgen.1006823
de Ocampo, M. P., Ho, V. T., Thomson, M. J., Mitsuya, S., Yamauchi, A., Ismail, A. M., et al. (2022). QTL mapping under salt stress in rice using a kalarata-azucena population. Euphytica 218, 74. doi: 10.1007/s10681-022-03026-8
Fisher, R. A. (1922). On the interpretation of χ 2 from contingency tables, and the calculation of p. J. R. Stat. Soc. 85, 87–94. doi: 10.1111/j.2397-2335.1922.tb00768.x
Fukuda, A., Nakamura, A., Hara, N., Toki, S., Tanaka, Y. (2011). Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta 233, 175–188. doi: 10.1007/s00425-010-1289-4
Gregorio, G., Islam, M., Vergara, G., Thirumeni, S. (2013). Recent advances in rice science to design salinity and other abiotic stress tolerant rice varieties. Sabrao J. Breed. Genet. 45, 31–41.
He, L., Luo, Y. (2021). Effects of long-term sand mixing ameliorating measures on soil salinity characteristics of saline-alkali land in northern shaanxi. IOP Conf. Series: Earth Environ. Sci. 781, 22084. doi: 10.1088/1755-1315/781/2/022084
Hill, J. T., Demarest, B. L., Bisgrove, B. W., Gorsi, B., Su, Y. C., Yost, H. J., et al. (2013). MMAPPR: Mutation mapping analysis pipeline for pooled RNA-seq. Genome Res. 23, 687–697. doi: 10.1101/gr.146936.112
Huang, J., Zhao, X., Weng, X., Wang, L., Xie, W. (2012). The rice b-box zinc finger gene family: genomic identification, characterization, expression profiling and diurnal analysis. PloS One 7, e48242. doi: 10.1371/journal.pone.0048242
Jackson, A. L., Linsley, P. S. (2004). Noise amidst the silence: Off-target effects of siRNAs? Trends Genet. 20, 521–524. doi: 10.1016/j.tig.2004.08.006
Jain, M., Nijhawan, A., Arora, R., Agarwal, P., Ray, S., Sharma, P., et al. (2007). F-box proteins in rice. genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143, 1467–1483. doi: 10.1104/pp.106.091900
Johnson, D., Smith, S. E., Dobrenz, A. (1992). Genetic and phenotypic relationships in response to NaCl at different developmental stages in alfalfa. Theor. Appl. Genet. 83, 833–838. doi: 10.1007/BF00226705
Kader, M. A., Seidel, T., Golldack, D., Lindberg, S. (2006). Expressions of OsHKT1, OsHKT2, and OsVHA are differentially regulated under NaCl stress in salt-sensitive and salt-tolerant rice (Oryza sativa l.) cultivars. J. Exp. Bot. 57, 4257–4268. doi: 10.1093/jxb/erl199
Kim, S. K., Yun, C. H., Lee, J. H., Jang, Y. H., Park, H. Y., Kim, J. K., et al. (2008). OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta 228, 355–365. doi: 10.1007/s00425-008-0742-0
Kodama, A., Narita, R., Yamaguchi, M., Hisano, H., Adachi, S., Takagi, H., et al. (2018). QTLs maintaining grain fertility under salt stress detected by exome QTL-seq and interval mapping in barley. Breed Sci. 68, 561–570. doi: 10.1270/jsbbs.18082
Koyama, M. L., Levesley, A., Koebner, R. M., Flowers, T. J., Yeo, A. R. (2001). Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiol. 125, 406–422. doi: 10.1104/pp.125.1.406
Kumar, S., Agarwal, S., Ranvijay (2019). Fast and memory efficient approach for mapping NGS reads to a reference genome. J. Bioinf. Comput. Biol. 17. doi: 10.1142/s0219720019500082
Le, T. D., Gathignol, F., Vu, H. T., Nguyen, K. L., Tran, L. H., Vu, H. T. T., et al. (2021). Genome-wide association mapping of salinity tolerance at the seedling stage in a panel of Vietnamese landraces reveals new valuable QTLs for salinity stress tolerance breeding in rice. Plants (Basel) 10. doi: 10.3390/plants10061088
Li, H., Hu, B., Chu, C. (2017). Nitrogen use efficiency in crops: Lessons from arabidopsis and rice. J. Exp. Bot. 68, 2477–2488. doi: 10.1093/jxb/erx101
Li, C. H., Wang, G., Zhao, J. L., Zhang, L. Q., Ai, L. F., Han, Y. F., et al. (2014). The receptor-like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell 26, 2538–2553. doi: 10.1105/tpc.114.125187
Liang, J. L., Qu, Y. P., Yang, C. G., Ma, X. D., Cao, G. L., Zhao, Z. W., et al. (2015). Identification of QTLs associated with salt or alkaline tolerance at the seedling stage in rice under salt or alkaline stress. Euphytica 201, 441–452. doi: 10.1007/s10681-014-1236-8
Lin, H., Zhu, M., Yano, M., Gao, J., Liang, Z., Su, W., et al. (2004). QTLs for na+ and k+ uptake of the shoots and roots controlling rice salt tolerance. Theor. Appl. Genet. 108, 253–260. doi: 10.1007/s00122-003-1421-y
Linh, L. H., Linh, T. H., Xuan, T. D., Ham, L. H., Ismail, A. M., Khanh, T. D., et al. (2012). Molecular breeding to improve salt tolerance of rice (Oryza sativa l.) in the red river delta of Vietnam. Int. J. Plant Genomics 2012, 949038. doi: 10.1155/2012/949038
Lira, B. S., Oliveira, M. J., Shiose, L., Wu, R. T. A., Rosado, D., Lupi, A. C.D., et al. (2020). Light and ripening-regulated BBX protein-encoding genes in solanum lycopersicum. Sci. Rep. 10, 19235. doi: 10.1038/s41598-020-76131-0
Liu, C., Chen, K., Zhao, X., Wang, X., Shen, C., Zhu, Y., et al. (2019). Identification of genes for salt tolerance and yield-related traits in rice plants grown hydroponically and under saline field conditions by genome-wide association study. Rice (N Y) 12, 88. doi: 10.1186/s12284-019-0349-z
Liu, Y., Xing, L., Li, J., Dai, S. (2012). Rice b-box zinc finger protein OsBBX25 is involved in the abiotic response. Chin. Bull. Bot. 47, 366–378. doi: 10.3724/SP.J.1259.2012.00366
Liu, A. L., Zou, J., Liu, C. F., Zhou, X. Y., Zhang, X. W., Luo, G. Y., et al. (2013). Over-expression of OsHsfA7 enhanced salt and drought tolerance in transgenic rice. BMB Rep. 46, 31–36. doi: 10.5483/bmbrep.2013.46.1.090
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, C., Wang, Y., Gu, D., Nan, J., Chen, S., Li, H., et al. (2017). Overexpression of s-Adenosyl-l-Methionine synthetase 2 from sugar beet M14 increased arabidopsis tolerance to salt and oxidative stress. Int. J. Mol. Sci. 18, 847. doi: 10.3390/ijms18040847
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Magwene, P. M., Willis, J. H., Kelly, J. K. (2011). The statistics of bulk segregant analysis using next generation sequencing. PloS Comput. Biol. 7, e1002255. doi: 10.1371/journal.pcbi.1002255
Mansfeld, B. N., Grumet, R. (2018). QTLseqr: An r package for bulk segregant analysis with next-generation sequencing. Plant Genome 11, 180006. doi: 10.3835/plantgenome2018.01.0006
Matsumoto, T., Wu, J. Z., Kanamori, H., Katayose, Y., Fujisawa, M., Namiki, N., et al. (2005). The map-based sequence of the rice genome. Nature 436, 793–800. doi: 10.1038/nature03895
Min, J. H., Chung, J. S., Lee, K. H., Kim, C. S. (2015). The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in arabidopsis. J. Integr. Plant Biol. 57, 313–324. doi: 10.1111/jipb.12246
Mishra, S., Singh, B., Panda, K., Singh, B. P., Singh, N., Misra, P., et al. (2016). Association of SNP haplotypes of HKT family genes with salt tolerance in Indian wild rice germplasm. Rice (N Y) 9, 15. doi: 10.1186/s12284-016-0083-8
Na, Y. J., Choi, H. K., Park, M. Y., Choi, S. W., Xuan Vo, K. T., Jeon, J. S., et al. (2019). OsMAPKKK63 is involved in salt stress response and seed dormancy control. Plant Signal Behav. 14, e1578633. doi: 10.1080/15592324.2019.1578633
Nagaoka, S., Takano, T. (2003). Salt tolerance-related protein STO binds to a myb transcription factor homologue and confers salt tolerance in arabidopsis. J. Exp. Bot. 54, 2231–2237. doi: 10.1093/jxb/erg241
Nakhla, W. R., Sun, W., Fan, K., Yang, K., Zhang, C., Yu, S., et al. (2021). Identification of QTLs for salt tolerance at the germination and seedling stages in rice. Plants (Basel) 10, 428. doi: 10.3390/plants10030428
Nayyeripasand, L., Garoosi, G. A., Ahmadikhah, A. (2021). Genome-wide association study (GWAS) to identify salt-tolerance QTLs carrying novel candidate genes in rice during early vegetative stage. Rice (N Y) 14, 9. doi: 10.1186/s12284-020-00433-0
Negrão, S., Almadanim, M. C., Pires, I. S., Abreu, I. A., Maroco, J., Courtois, B., et al. (2013). New allelic variants found in key rice salt-tolerance genes: an association study. Plant Biotechnol. J. 11, 87–100. doi: 10.1111/pbi.12010
Nieves-Cordones, M., Martínez, V., Benito, B., Rubio, F. (2016). Comparison between arabidopsis and rice for main pathways of k+ and na+ uptake by roots. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00992
Paterson, A. H., Damon, S., Hewitt, J. D., Zamir, D., Rabinowitch, H. D., Lincoln, S. E., et al. (1991). Mendelian factors underlying quantitative traits in tomato: comparison across species, generations, and environments. Genetics 127, 181–197. doi: 10.1093/genetics/127.1.181
Ponce, K. S., Meng, L., Guo, L., Leng, Y., Ye, G. (2021). Advances in sensing, response and regulation mechanism of salt tolerance in rice. Int. J. Mol. Sci. 22, 2254. doi: 10.3390/ijms22052254
Prakash, N. R., Lokeshkumar, B. M., Rathor, S., Warraich, A. S., Yadav, S., Vinaykumar, N. M., et al. (2022). Meta-analysis and validation of genomic loci governing seedling and reproductive stage salinity tolerance in rice. Physiol. Plant 174, e13629. doi: 10.1111/ppl.13629
Pundir, P., Devi, A., Krishnamurthy, S. L., Sharma, P. C., Vinay, K. N. M. (2021). QTLs in salt rice variety CSR10 reveals salinity tolerance at reproductive stage. Acta Physiologiae Plantarum 43, 35. doi: 10.1007/s11738-020-03183-0
Ren, Z. H., Gao, J. P., Li, L. G., Cai, X. L., Huang, W., Chao, D. Y., et al. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37, 1141–1146. doi: 10.1038/ng1643
Robson, F., Costa, M. M., Hepworth, S. R., Vizir, I., Piñeiro, M., Reeves, P. H., et al. (2001). Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28, 619–631. doi: 10.1046/j.1365-313x.2001.01163.x
Salam, M., Rahman, M., Bhuiyan, M., Uddin, K., Sarker, M., Yasmeen, R., et al. (2007). BRRI dhan47: A salt tolerant variety for the boro season. Int. Rice Res. Notes 32, 42–43. doi: 10.3860/irrn.v32i1.1093
Shamaya, N. J., Shavrukov, Y., Langridge, P., Roy, S. J., Tester, M. (2017). Genetics of na(+) exclusion and salinity tolerance in Afghani durum wheat landraces. BMC Plant Biol. 17, 209. doi: 10.1186/s12870-017-1164-6
Singh, A., Gopalakrishnan, S., Singh, V., Prabhu, K., Mohapatra, T., Singh, N., et al. (2011). Marker assisted selection: A paradigm shift in basmati breeding. Indian J. Genet. Plant Breed. 71, 120. doi: 10.1002/pca.1274
Singh, R., Singh, Y., Xalaxo, S., Verulkar, S., Yadav, N., Singh, S., et al. (2016). From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network. Plant Sci. 242, 278–287. doi: 10.1016/j.plantsci.2015.08.008
Sun, B. R., Fu, C. Y., Fan, Z. L., Chen, Y., Chen, W. F., Zhang, J., et al. (2019). Genomic and transcriptomic analysis reveal molecular basis of salinity tolerance in a novel strong salt-tolerant rice landrace changmaogu. Rice (N Y) 12, 99. doi: 10.1186/s12284-019-0360-4
Takagi, H., Abe, A., Yoshida, K., Kosugi, S., Natsume, S., Mitsuoka, C., et al. (2013). QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74, 174–183. doi: 10.1111/tpj.12105
Takehisa, H., Shimodate, T., Fukuta, Y., Ueda, T., Yano, M., Yamaya, T., et al. (2004). Identification of quantitative trait loci for plant growth of rice in paddy field flooded with salt water. Field Crops Res. 89, 85–95. doi: 10.1016/j.fcr.2004.01.026
Teng, Y., Lv, M., Zhang, X., Cai, M., Chen, T. (2022). BEAR1, a bHLH transcription factor, controls salt response genes to regulate rice salt response. J. Plant Biol., 65, 217–230. doi: 10.1007/s12374-022-09347-4
Tian, Q., Shen, L., Luan, J., Zhou, Z., Guo, D., Shen, Y., et al. (2021). Rice shaker potassium channel OsAKT2 positively regulates salt tolerance and grain yield by mediating k+ redistribution. Plant Cell Environ. 44, 2951–2965. doi: 10.1111/pce.14101
Tiwari, S., Sl, K., Kumar, V., Singh, B., Rao, A. R., Mithra Sv., A., et al. (2016). Mapping QTLs for salt tolerance in rice (Oryza sativa l.) by bulked segregant analysis of recombinant inbred lines using 50K SNP chip. PloS One 11, e0153610. doi: 10.1371/journal.pone.0153610
Van Ooijen, J. (2006). JoinMap® 4, software for the calculation of genetic linkage maps in experimental populations. Kyazma BV Wageningen 33.
Vinod, K., Krishnan, S. G., Babu, N. N., Nagarajan, M., Singh, A. (2013). “Improving salt tolerance in rice: Looking beyond the conventional,” in Salt stress in plants (Springer), 219–260. doi: 10.1007/978-1-4614-6108-1_10
Wang, K., Li, M. Y., Hakonarson, H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164. doi: 10.1093/nar/gkq603
Wang, J., Nan, N., Li, N., Liu, Y., Wang, T. J., Hwang, I., et al. (2020). A DNA methylation reader-chaperone regulator-transcription factor complex activates OsHKT1;5 expression during salinity stress. Plant Cell 32, 3535–3558. doi: 10.1105/tpc.20.00301
Wu, F., Yang, J., Yu, D., Xu, P. (2020). Identification and validation a major QTL from “Sea rice 86” seedlings conferred salt tolerance. Agronomy 10, 410. doi: 10.3390/AGRONOMY10030410
Wu, W., Zhang, Y., Zhang, M., Zhan, X., Shen, X., Yu, P., et al. (2018). The rice CONSTANS-like protein OsCOL15 suppresses flowering by promoting Ghd7 and repressing RID1. Biochem. Biophys. Res. Commun. 495, 1349–1355. doi: 10.1016/j.bbrc.2017.11.095
Wu, W., Zheng, X. M., Lu, G., Zhong, Z., Gao, H., Chen, L., et al. (2013). Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl. Acad. Sci. U.S.A. 110, 2775–2780. doi: 10.1073/pnas.1213962110
Xiao, N., Gao, Y., Qian, H., Gao, Q., Wu, Y., Zhang, D., et al. (2018). Identification of genes related to cold tolerance and a functional allele that confers cold tolerance. Plant Physiol. 177, 1108–1123. doi: 10.1104/pp.18.00209
Xu, Y., Wang, R., Wang, Y., Zhang, L., Yao, S. (2020). A point mutation in LTT1 enhances cold tolerance at the booting stage in rice. Plant Cell Environ. 43, 992–1007. doi: 10.1111/pce.13717
Yadav, A. K., Kumar, A., Grover, N., Ellur, R. K., Bollinedi, H., Krishnan, S. G., et al. (2021). Genome-wide association study reveals marker-trait associations for early vegetative stage salinity tolerance in rice. Plants (Basel) 10, 559. doi: 10.3390/plants10030559
Yeo, A., Yeo, M., Flowers, T. (1987). The contribution of an apoplastic pathway to sodium uptake by rice roots in saline conditions. J. Exp. Bot. 38, 1141–1153. doi: 10.1093/jxb/38.7.1141
Yuan, L., Zhang, L., Wei, X., Wang, R., Li, N., Chen, G., et al. (2022). Quantitative trait locus mapping of salt tolerance in wild rice oryza longistaminata. Int. J. Mol. Sci. 23, 2379. doi: 10.3390/ijms23042379
Yue, E., Cao, H., Liu, B. (2020). OsmiR535, a potential genetic editing target for drought and salinity stress tolerance in oryza sativa. Plants (Basel) 9, 1337. doi: 10.3390/plants9101337
Zhang, L., Li, Q., Dong, H., He, Q., Liang, L., Tan, C., et al. (2015). Three CCT domain-containing genes were identified to regulate heading date by candidate gene-based association mapping and transformation in rice. Sci. Rep. 5, 7663. doi: 10.1038/srep07663
Zhang, X., Long, Y., Chen, X., Zhang, B., Xin, Y., Li, L., et al. (2021). A NAC transcription factor OsNAC3 positively regulates ABA response and salt tolerance in rice. BMC Plant Biol. 21, 546. doi: 10.1186/s12870-021-03333-7
Zhang, X., Wang, W., Guo, N., Zhang, Y., Bu, Y., Zhao, J., et al. (2018). Combining QTL-seq and linkage mapping to fine map a wild soybean allele characteristic of greater plant height. BMC Genomics 19, 226. doi: 10.1186/s12864-018-4582-4
Zhou, J., Yuan, M., Zhao, Y., Quan, Q., Yu, D., Yang, H., et al. (2021). Efficient deletion of multiple circle RNA loci by CRISPR-Cas9 reveals Os06circ02797 as a putative sponge for OsMIR408 in rice. Plant Biotechnol. J. 19, 1240–1252. doi: 10.1111/pbi.13544
Keywords: japonica rice, seedling stage, salt tolerance, b-box zinc finger family protein, function characterization
Citation: Lei L, Cao L, Ding G, Zhou J, Luo Y, Bai L, Xia T, Chen L, Wang J, Liu K, Lei Q, Xie T, Yang G, Wang X, Sun S and Lai Y (2023) OsBBX11 on qSTS4 links to salt tolerance at the seeding stage in Oryza sativa L. ssp. Japonica. Front. Plant Sci. 14:1139961. doi: 10.3389/fpls.2023.1139961
Received: 08 January 2023; Accepted: 20 February 2023;
Published: 08 March 2023.
Edited by:
Muhammad Umair Hassan, Jiangxi Agricultural University, ChinaReviewed by:
Farrukh Azeem, Government College University, PakistanCopyright © 2023 Lei, Cao, Ding, Zhou, Luo, Bai, Xia, Chen, Wang, Liu, Lei, Xie, Yang, Wang, Sun and Lai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shichen Sun, c3Vuc2hpY2hlbjE5NzlAMTYzLmNvbQ==; Yongcai Lai, eW9uZ2NhaWxhaTE5NjRAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.