- 1Sanya Nanfan Research Institute & Key Laboratory of Quality Regulation of Tropical Horticultural Crop in Hainan Province, School of Horticulture, Hainan University, Haikou, China
- 2Key Laboratory of Tropical Fruit Biology, Ministry of Agriculture and Rural Affairs, South Subtropical Crops Research Institute, Chinese Academy of Tropical Agricultural Sciences, Zhanjiang, China
Introduction: Flavonoids are important secondary metabolites in plants and light is a crucial environmental factor regulating flavonoids biosynthesis. However, effect of light on the different flavonoids compositions accumulation in mango and the relevant molecular mechanism still need to be clarified.
Methods: In this study, green-mature fruits of red mango cultivar ‘Zill’ were subjected to postharvest light treatment, and fruit peel color, total soluble solids content, total organic acid, and firmness of flesh were measured. The flavonoids metabolites profile, and the expression of flavonoids-related genes and light signal pathway genes were also analyzed.
Results: Results showed that light treatment promoted the red coloration of fruit peel and increased the total soluble solids content and firmness of flesh. The concentration of flavonols, proanthocyanidins and anthocyanins, and expression of key flavonoids biosynthetic genes including MiF3H, MiFLS, MiLAR, MiANS, MiUFGT1, and MiUFGT3 were significantly induced by light. The MYBs regulating flavonols and proanthocyanidins, i.e. MiMYB22 and MiMYB12, as well as the key light signal pathway transcription factors (TFs) MiHY5 and MiHYH, were identified in mango. The transcription of MiMYB1, MiMYB12, MiMYB22, MiHY5 and MiHYH was up-regulated by light.
Discussion: Our results provide a postharvest technology to improve mango fruit appearance quality, and are helpful to reveal the molecular mechanism of light-induced flavonoids biosynthesis in mango.
Introduction
Flavonoids are important secondary metabolites in plants which determine fruit quality due to their essential contribution to fruit color, antioxidation capacity and nutritive value. Flavonoids consist of various classes including flavonols, proanthocyanidins (PAs), and anthocyanins, which are the three main subgroups (Williams and Grayer, 2004). Flavonoids are synthesized via phenylpropanoid and flavonoid pathway (Winkel-Shirley, 2001). The early flavonoids biosynthetic genes (EBGs) include Phenylalanine ammonia-lyase (PAL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), and flavonoid 3’-hydroxylase (F3’H). The late flavonoids biosynthetic genes (LBGs) involve dihydroflavonol reductase (DFR), flavonol synthase (FLS), anthocyanidin synthase (ANS), leucoanthocyanidin reductase (LAR), anthocyanidin reductase (ANR), and UDP-glucose: flavonoid 3-O-glucosyltransferase (UFGT). Among them, FLS, UFGT, and LAR and ANR catalyze the last step of flavonols, anthocyanins, and proanthocyanidins biosynthesis, respectively.
The expression of flavonoids biosynthetic genes is regulated by MYB-bHLH-WD40 (MBW) complex, with the crucial contribution of R2R3-MYB transcription factor (TF) (Broun, 2005). In Arabidopsis, total 125 R2R3-MYB TFs are divided into 25 subgroups, and the 5th, 6th and 7th subgroups participate in the biosynthesis of proanthocyanidins, anthocyanins, and flavonols, with the representative MYB123/TT2, MYB75/PAP1, and MYB12/PEG1, respectively (Stracke et al., 2001; Dubos et al., 2010). The MYBs controlling flavonoids biosynthesis in fruits have also been widely reported. In strawberry, FaMYB9/FaMYB11, the homologues of AtTT2, were found to form the complex with FabHLH3 and FaTTG1 to control proanthocyanidins biosynthesis in strawberry fruits (Schaart et al., 2013). Proanthocyanidins and flavonols accumulation in red-fleshed apple are regulated by MYB12 and MYB22, respectively (Wang et al., 2017). PpMYB17 could activate the expression of PpCHS, PpCHI, PpF3H, and PpFLS to positively regulate flavonols biosynthesis in pear (Premathilake et al., 2020). With the great contribution to the red coloration in fruits, MYBs regulating anthocyanins biosynthesis have been identified in diverse fruit species, including MdMYB1/A/10 in apple (Takos et al., 2006; Ban et al., 2007; Espley et al., 2007), PyMYB10 and PyMYB114 in pear (Feng et al., 2010; Yao et al., 2017), VvMYBA1 in grape (Kobayashi et al., 2004), and CsRuby in citrus (Butelli et al., 2012; Huang et al., 2018; Huang et al., 2019). In mango, the anthocyanins biosynthesis is regulated by MiMYB1 (Kanzaki et al., 2020; Shi et al., 2021), while more MYBs controlling other flavonoids compositions biosynthesis need to be discovered.
Flavonoids biosynthesis in fruits is affected by environmental factors such as light. Postharvest light treatment is widely used to induce flavonoids accumulation in numerous fruit species including apple (Peng et al., 2012), pear (Qian et al., 2013; Sun et al., 2014), peach (Santin et al., 2019), nectarine (Scattino et al., 2014), grape (Sheng et al., 2018), tomato (Liu et al., 2011), and blueberry (Yang et al., 2019). CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1), ELONGATED HYPOCOTYL 5 (HY5), and HY5-HOMOLOG (HYH) are the key proteins regulating photomorphogenesis in plants such as flavonoids accumulation (Podolec et al., 2021). COP1 is an ubiquitin E3 ligase, which negatively regulate light-induced flavonoids biosynthesis by degrading flavonoids-related TFs including MYB (Li et al., 2012), and bHLH (Tao et al., 2020). HY5 and HYH are light-responsive TFs, which positively regulate flavonoids accumulation in plants by activating the expression of flavonoids-related genes including CHS, ANS, FLS, and MYB, through the binding to the G-box or ACE-box in the promoter region of target genes (Holm et al., 2002; Loyola et al., 2016; Henry-Kirk et al., 2018; Tao et al., 2018).
Mango (Mangifera indica L.) is one the most popular tropical fruit species due to its unique aroma, flavor, and enriched nutrition. Following banana, grape, apple and orange, mango is the fifth most produced fruit crop worldwide (http://www.fao.org/faostat/). The effect of light on flavonoids accumulation in mango has been reported. Preharvest bagging treatment inhibited the accumulation of flavonols and anthocyanins but promoted the accumulation of proanthocyanidins in mango (Shi et al., 2021). Postharvest light treatment could increase the total phenols, total flavonoids, and anthocyanins content in the fruit skin of mango (González-Aguilar et al., 2007; Cao et al., 2016; Ni et al., 2022; Shi et al., 2022). However, the effect of postharvest light treatment on the accumulation of different flavonoids components, i.e. flavonols, proanthocyanidins and anthocyanins is still unknown.
In this study, green mature bagged ‘Zill’ mango fruits were subjected to postharvest light treatment, and fruit peel was sampled at 0, 6, 24, 72, 144, and 240 hours of exposure. Fruit quality indexes including firmness, fruit color, total soluble solids content, total organic acid content, and solidity-acid ratio were measured. Metabolomic profiling was established to analyze the concentration of flavonols, proanthocyanidins and anthocyanins in the fruit peel during treatment. The expression of flavonoids biosynthetic and regulatory genes (especially different MYBs), as well as the key light signal pathway genes including COP1, HY5 and HYH, was also measured. This study will enrich our knowledge regarding the mechanism of light-induced flavonoids biosynthesis in mango.
Materials and methods
Plant materials and treatments
Mango (Mangifera indica cv. Zill) fruits were obtained from South Subtropical Crops Research Institute (SSCRI) in Zhanjiang, China. Fruitlets were bagged with double layers yellow-black paper bags (Qingdao Kobayashi Co., Ltd., Qingdao, China) at 20 days after full bloom to block out all the light. Green mature fruits (130 days after full bloom) were harvested with bags, transported to the lab, and debagged for postharvest light treatment in plant growth chambers (Conviron, Adaptis A 1000, Winnipeg, Canada). 180 unblemished fruits with uniformed size were divided into two groups, with half fruits subjected to mimic sunlight treatment (mixture of 4.5 μW•cm-2 UV-B and 16 W•m−2 white light) and the rest retained in darkness as control. The relative humidity and temperature were 80% and 17°C, respectively. All the conditions for treatment were according to our previous patent (Qian et al., 2022). 30 fruits were regarded as one biological replicate. Fruit peel of 5 fruits per replicate was collected at 0, 6, 24, 72, 144, and 240 hours of light exposure for metabolomic and gene expression analyses. For fruit peel sampling, the exposed side was sampled by a peeler for light-treated fruit, and the up-side fruit peel was sampled for control fruit. Fruit peel was sampled as thin as possible to ensure the minimum collection of flesh.
Fruit quality measurement
Firmness was measured by a TA touch texture profile analyzer (Bosin Tech, Shanghai, China). After removing the peel, a 2 mm diameter probe was inserted to the equatorial part of the flesh at a 90 °C angle and depth of 5 mm. The analyzer parameters for operation were set as follows: 2 mm s-1 for pre-test speed; 4 mm s-1 for test speed; 3 mm s-1 post-test speed, and 2 s for intermediate interval. Fruit color indexes in the equatorial part of fruit (L*, a*, and b* values) were measured by a portable colorimeter (LS170, Shenzhen Linshang Technology Co.,Ltd., Shenzhen, China) according to the instruction of the manufacturer. Total soluble solids content, total organic acid, and solidity-acid ratio were measured by a brix-acidity meter (PAL-BX/ACID15, ATAGO, Tokyo, Japan) according to the user manual. Analysis was performed in three biological replicates.
Metabolomic profiling of flavonoids
The details for metabolomic profiling of flavonoids were described in the previous study (Shi et al., 2021), which was conducted by Metware Biotechnology Co. Ltd. (Wuhan, China). In brief, fruit peel was successively freeze-dried, ground, and added to the extraction solution (50% methanol containing 0.1% HCl), and the supernatant was used for high-performance liquid chromatography with tandem mass spectrometry (HPLC−MS/MS) analysis. The identification of flavonoids compounds was based on the Metware Database (MWDB). The quantification of flavonoids was according to the area of the chromatographic peak, and the concentration was calculated by using the linear equation of corresponding standard. Analysis was performed in three biological replicates.
RNA extraction, cDNA synthesis, and Q-PCR analysis from mango peel
Total RNA was extracted using an RNA prep pure plant kit (Tiangen, DP441, Beijing, China). First-strand cDNA was synthesized from 1 µg of total RNA using the HiScript IIQ RT SuperMix (Vazyme, R223-01, Nanjing, China). The Q-PCR primers were designed by Primer 3 (https://bioinfo.ut.ee/primer3-0.4.0/) and synthesized by Sangon Biotech Co. Ltd, Shanghai, China (Table S1). The Q-PCR reactions (15µL) were performed on a real-time PCR machine (qTOWER3G, Jena, Germany) containing 7.5µL SYBR premix ExTaqTMII (Takara, Japan), 5.5µL of cDNA (20 times diluted) and 1µL of both forward and reverse primers (10µM). Relative expression of mRNA was calculated by the cycle threshold (Ct) 2-ΔΔCt method, with the normalization by the mango actin gene.
Identification, multiple sequence alignment and phylogenetic tree construction of MYBs, HY5, and HYH
Apple MdMYB22 (AAZ20438.1), MdMYB12 (XP_008337875.1), MdHY5 (NP_001280752.1), and MdHYH (XP_008369576.1) sequences were used to search for the mango homologs by blasting to mango genome database using TBtools (Chen et al., 2020; Wang et al., 2020a). The amino acid sequences of R2R3 domain of flavonoids-related MYBs, and the whole amino acid sequences of HY5 and HYH from mango and other plant species were used for multiple sequence alignment by Jalview (Waterhouse et al., 2009). Full-length flavonoids-related MYBs, HY5, HYH protein sequences were used for phylogenetic tree construction by MEGA X with the neighbor-joining (NJ) method.
Statistical analysis
Data of fruit quality indexes were subjected to a one-way Analysis of Variance (ANOVA) using SPSS 27.0 (SPSS, Chicago, IL, USA), and mean values were separated by Tukey’s multiple range test. Probability values of <0.05 were considered statistically significant. Metabolic and gene expression data were subjected to a Student’s t-test using SPSS 27.0 to analyze the statistic difference between control and treatment. Probability values of <0.05 and <0.01 were considered statistically significant and highly statistically significant, marked with one asterisk (*) and two asterisks (**), respectively. Correlation analysis was conducted by ChiPlot (https://www.chiplot.online/).
Results
Effect of postharvest light exposure on the fruit quality of mango
During postharvest light exposure, no red coloration was detected in the control fruits which were kept in darkness, while the light-treated fruits started turning red at 6 days of exposure, and the red coloration was enhanced at 10 days (Figure 1A). After 10 days of treatment, the firmness of both light-treated and control fruits was decreased, and control fruits showed significant lower firmness than light-treated fruits (Figure 1B). Red coloration decreased the lightness (L* value), and yellowness (b* value) of light-treated fruits, but tremendously increased the fruit redness (a* value) (Figures 1C–E). Light treatment increased the total soluble solids content, but had no effect on the total organic acid content and solidity-acid ratio (Figures 1F–H).
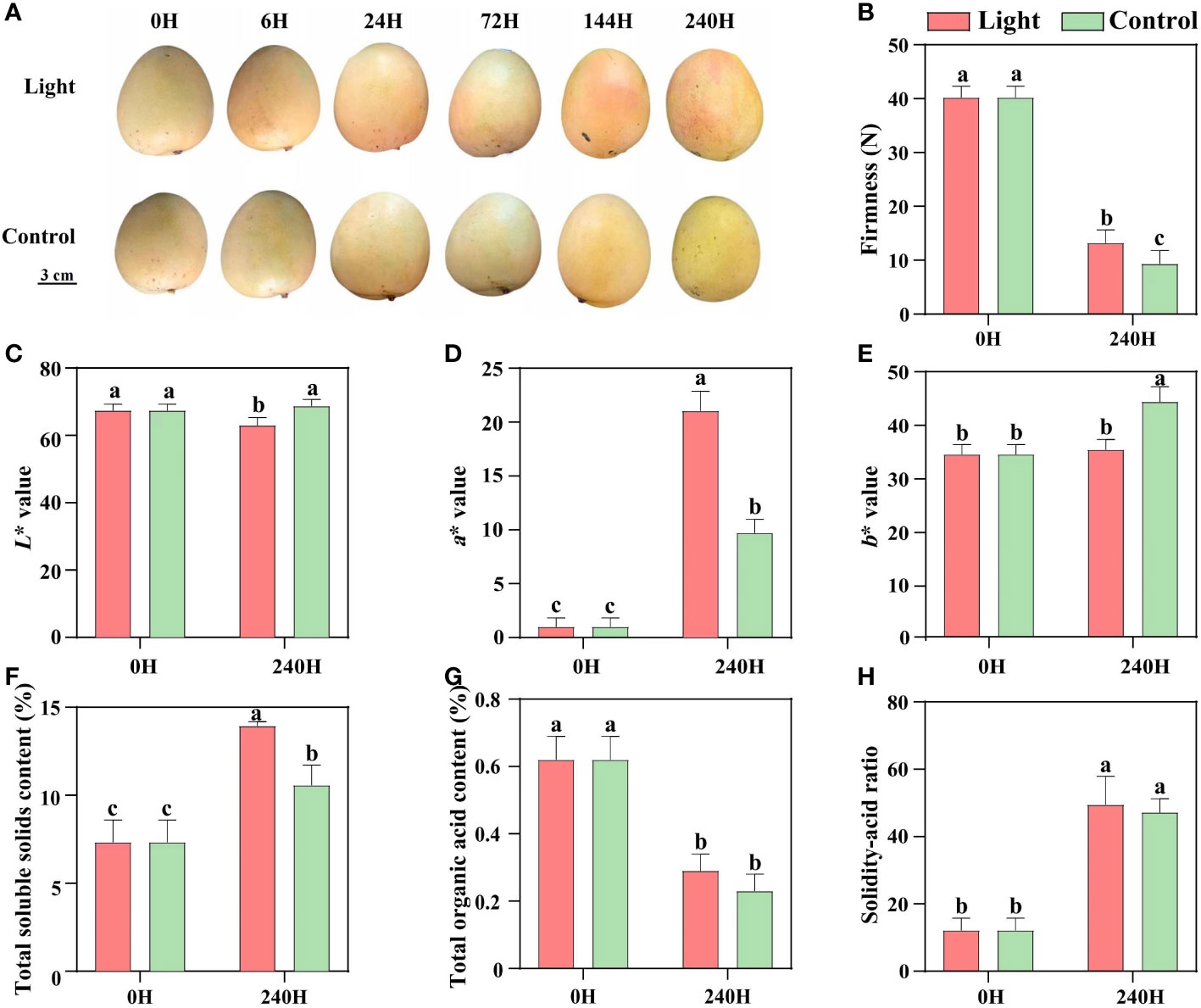
Figure 1 Effect of light on the appearance quality and internal quality of ‘Zill’ mango. (A) Representative images of light-treated and control (darkness) ‘Zill’ mango fruits. (B) Effect of light treatment on flesh firmness. (C–E) Effect of light treatment on fruit color indexes L*, a* and b*. (F–H) Effect of light treatment on the total soluble solids content, total organic acid content and solidity-acid ratio in mango flesh. Each value represents the mean ± standard deviation of three biological replicates. Values without the same letter are significantly different, p < 0.05 according to Tukey tests.
Effect of postharvest light exposure on the concentration of different flavonoids compositions in mango fruit peel
A total of 108 standards include 9 flavonols, 6 proanthocyanidins, and 93 anthocyanins, while most substance could not be detected or exists at a very low level in our sample (Table S2). According to the results of metabolites profile, naringenin-7-O-glucoside and quercetin-3-O-glucoside, procyanidin B1 and procyanidin B3, and cyanidin-3-O-galactoside and peonidin-3-O-glucoside were the main flavonols, proanthocyanidins, and anthocyanins components in ‘Zill’ mango fruit peel, respectively (Figure 2). The increase of flavonoids concentration was detected by light treatment, while accumulation pattern differed among components (Figure 2). The concentration of flavonols and proanthocyanidins in light-treated sample peaked at day 6, and then decreased at day 10 but still with a relatively higher concentration (Figure 2). For anthocyanins, significant accumulation of Cyanidin-3-O-galactoside, and Peonidin-3-O-glucoside were detected at day 6 and day 3, respectively, continued and peaked at day 10 (Figure 2). Control samples showed very low content of anthocyanins, but accumulated certain amount of flavonols and proanthocyanidins (Figure 2). Concentration of most flavonoids compositions was relatively stable in control samples during the whole treatment (Figure 2).
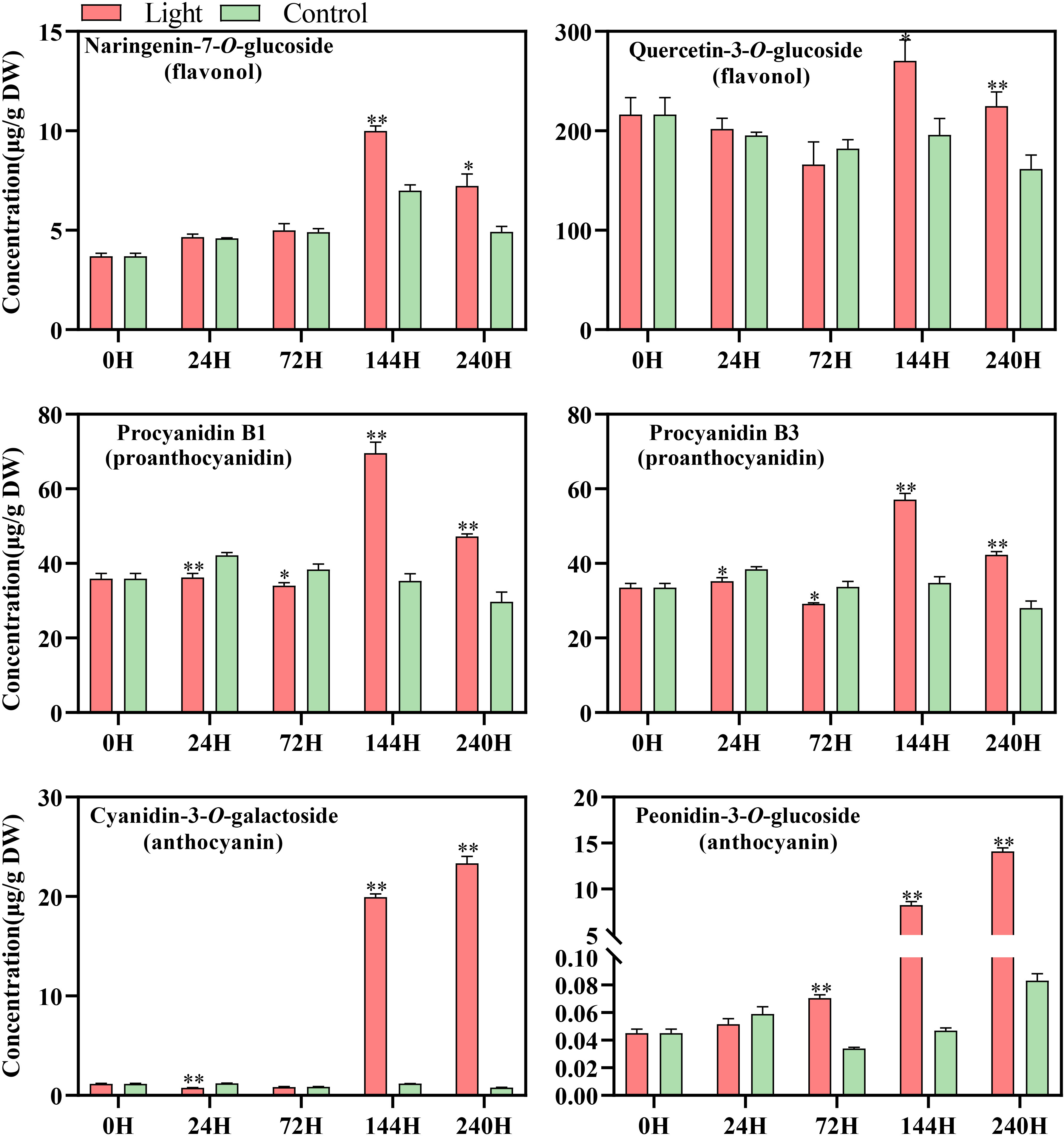
Figure 2 Concentration of flavonoid compounds detected in the fruit skin of light-treated and control (darkness) ‘Zill’ mango fruits during different postharvest light treatment stages. Each value represents the mean ± standard deviation of three biological replicates. * indicates significant difference (p-value < 0.05), and ** indicates very significant difference (p-value < 0.01) between control and treatment, as determined by Student’s t-test.
Effect of postharvest light exposure on the structural genes expression of flavonoids biosynthesis in mango fruit peel
Light up-regulated the expression of all the structural genes of flavonoids biosynthesis, while the expression pattern differed among genes (Figures 3A, B).
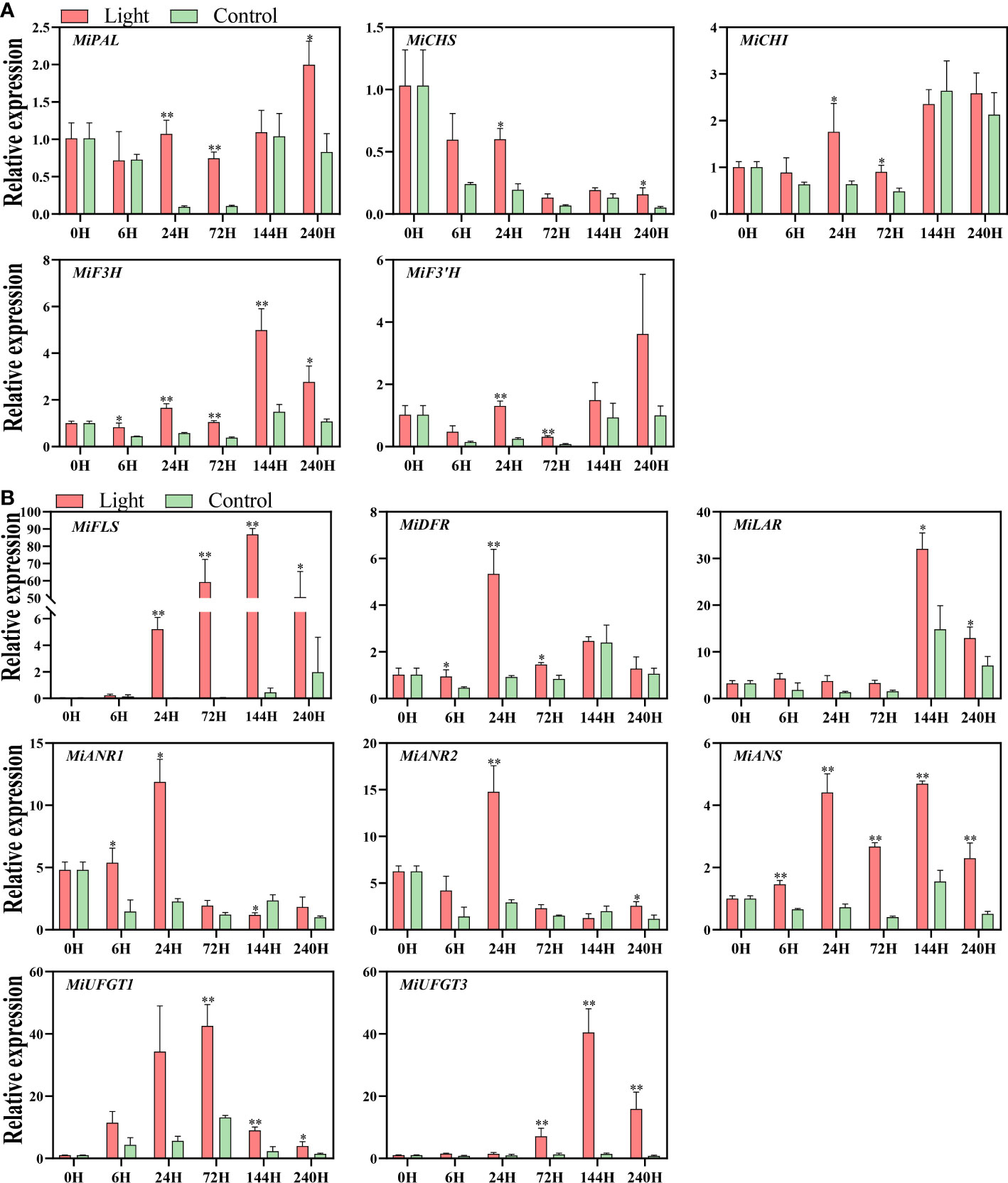
Figure 3 Effect of light treatment on the expression of early flavonoids biosynthetic genes (A) and late flavonoids biosynthetic genes (B) in ‘Zill’ mango peel. Each value represents the mean ± standard deviation of three biological replicates. * indicates significant difference (p-value < 0.05), and ** indicates very significant difference (p-value < 0.01) between control and treatment, as determined by Student’s t-test.
For EBGs, the expression of MiPAL, MiCHI, and MiF3’H was increased at day 1 and day 3 by light treatment, and MiCHS expression was induced at day 1 and day10 (Figure 3A). The expression of MiF3H was increased at 6 hours of light treatment, and stayed at a higher level in the light-treated fruits during the whole experiment, with the expression peak at day 6 (Figure 3A).
For LBGs, the increase of MiDFR expression was detected at 6 hours, 1 day, and 3 days of exposure, and up-regulation of MiANS started from 6 hours of treatment and lasted during the whole treatment (Figure 3B). As a flavonols specific gene, MiFLS expression of light-treated sample was tremendously induced at day 1, peaked almost at day 6, and kept at a high expression level during the treatment (Figure 3B). For proanthocyanidins specific genes, MiLAR transcription in light-treated fruits was up-regulated at day 6 and day 10, with the peak at day 6, which is highly correlated to the proanthocyanidins concentration (Figure 3B). The expression of MiANR1 responded at the early stages of light treatment, i.e. 6 hours and 1 day, while MiANR2 expression was significantly up-regulated in light-treated fruits at day1 and day 10 (Figure 3B). For anthocyanins specific genes, the increased expression of both MiUFGT1 and MiUFGT3 was detected at day 3, day 6, and day10, and the expression peak of MiUFGT1 and MiUFGT3 occurred at day 3 and day 6, respectively (Figure 3B).
Identification of flavonoids-related MYBs and expression of genes encoding MBW complex in response to light
The MYB regulating anthocyanins biosynthesis in mango has already been reported by Kanzaki et al. (2020), named MiMYB1, while the flavonols and proanthocyanidins-related MYBs are still unknown. In this study, we used MdMYB12 and MdMYB22, which have been proven to regulate proanthocyanidins and flavonols biosynthesis in apple by Wang et al. (2017), to search for the mango homologs MYBs in the mango genome database (Wang et al., 2020a). Finally, Mi12g07270.1, and Mi09g04990.1 were defined as the homologs of apple MdMYB12 and MdMYB22 in mango, named MiMYB12 and MiMYB22, respectively. Multiple sequence alignment showed that MiMYB1, MiMYB22, and MiMYB12 shared a very conserved R2R3 domain with the relevant MYBs in other plant species including Arabidopsis, strawberry, grape, and apple (Figure 4A). Phylogenetic analysis based on the full protein sequence showed that MYBs from Subgroups (SGs) 5, 6, and 7 were clustered together, and mango MiMYB1, MiMYB12 and MiMYB22 showed the closest relationship with the homologs from petunia, apple and grape, respectively (Figure 4B).
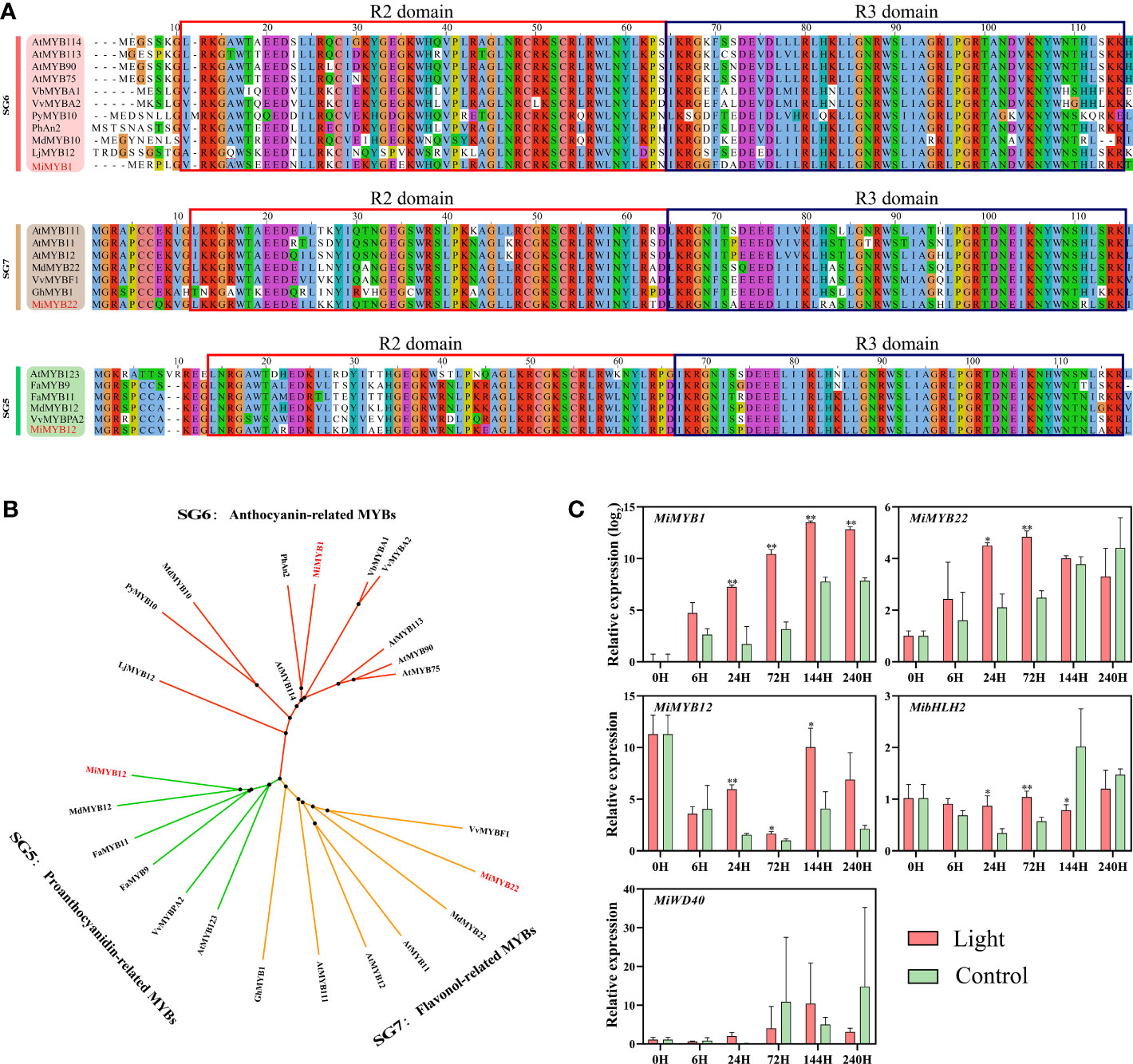
Figure 4 (A) Multiple sequence alignment of flavonoids-related MYBs in mango and other plant species. The R2R3 domain is boxed. The mango proteins are highlighted in red. (B) Phylogenetic tree derived from amino acid sequences of flavonoids-related MYBs in mango and other plant species. The mango proteins are highlighted in red. All sequences were retrieved from NCBI database with accessions as follows: SG6: Arabidopsis thaliana AtMYB114 (Q9FNV8.1), AtMYB113 (Q9FNV9.1), AtMYB90 (Q9ZTC3.1) and AtMYB75 (Q9FE25.1); Vitis betulifolia VbMYBA1 (AGH68552.1); Vitis vinifera VvMYBA2 (BAD18978.1); Pyrus pyrifolia PyMYB10 (ALN66630); Petunia x hybrida PhAN2 (AAF66727); Malus domestica MdMYB10 (ACQ45201.1); Lilium japonicum var. Abeanum LjMYB12 (BAP00661.1). SG7: Arabidopsis thaliana AtMYB111 (NP_199744.1), AtMYB11 (Q9LZK4.1) and AtMYB12 (NP_182268.1); Malus domestica MdMYB22 (AAZ20438.1), Vitis vinifera VvBF1 (NP_001267930.1); Gossypium hirsutum GhMYB1 (NP_001313761.1). SG5: Arabidopsis thaliana AtMYB123 (Q9FJA2.1); Fragaria x ananassa FaMYB9 (AFL02460.1), FaMYB11 (AFL02461.1); Malus domestica MdMYB12 (XP_008337875.1); Vitis vinifera VvMYBPA2 (NP_001267953.1). (C) Effect of light treatment on the expression of flavonoids-related MiMYBs, MibHLH2, and MiWD40 in ‘Zill’ mango peel. Each value represents the mean ± standard deviation of three biological replicates. * indicates significant difference (p-value < 0.05), and ** indicates very significant difference (p-value < 0.01) between control and treatment, as determined by Student’s t-test.
The expression of MiMYB1, MiMYB12, and MiMYB22 was all induced by light, with the most significant response by MiMYB1 (Figure 4C). MiMYB1 transcription in light-treated samples was up-regulated from day 1, and lasted during the whole experiment (Figure 4C). The increased expression of MiMYB12 and MiMYB22 by light treatment was detected at day 1, day 3, and day 6, and day 1 and day 3, respectively (Figure 4C). MibHLH2 expression was slightly increased at day 1 and day 3, and subsequently slightly decreased at day 6, while the expression of MiWD40 showed no response to light (Figure 4C).
Identification of mango HY5 and HYH, and expression of light signal genes in response to light
Using the apple MdHY5 (NP_001280752.1) and MdHYH (XP_008369576.1) sequences, the respective homologs in mango were identified, named MiHY5 (mango009397) and MiHYH (mango023606). Multiple sequence alignment showed that both MiHY5 and MiHYH contain a conserved bZIP domain (Figure 5A). The phylogenetic tree showed that HY5 and HYH were clustered into two groups, and all the HY5 or HYH proteins were clustered together (Figure 5B). Q-PCR analysis showed that MiCOP1 expression was only induced by light at 6 hours of treatment (Figure 5C). The transcription of MiHY5 was mainly up-regulated by light at the early stages of treatment, i.e. from 6 hours to 3 days, while MiHYH expression was significantly in response to light during the whole treatment (Figure 5C).
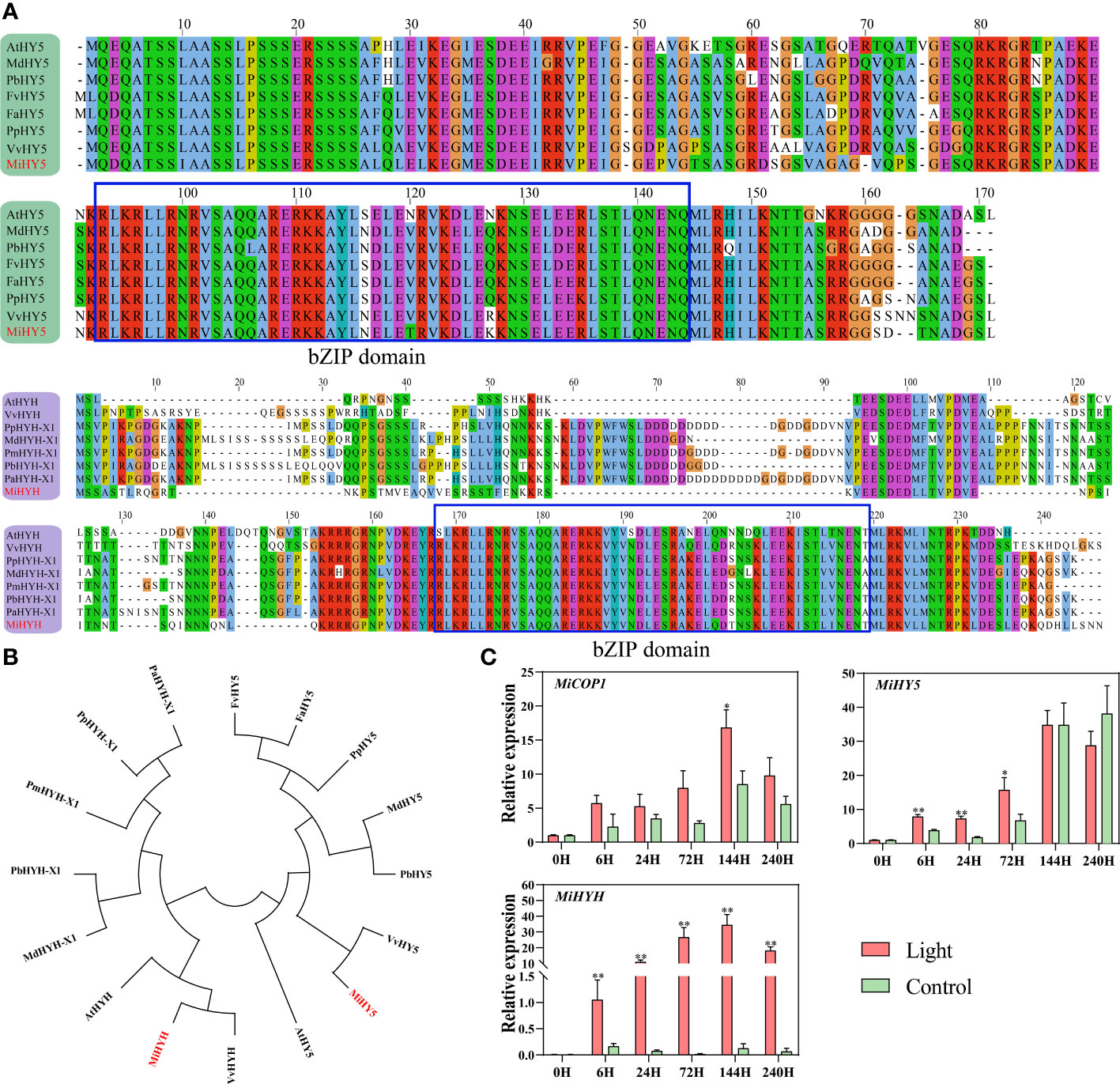
Figure 5 (A) Multiple sequence alignment of HY5 and HYH proteins in mango and other plant species. The bZIP domain is boxed. The mango proteins are highlighted in red. (B) Phylogenetic tree derived from amino acid sequences of HY5 and HYH in mango and other plant species. The mango proteins are highlighted in red. All sequences were retrieved from NCBI database with accessions as follows: Arabidopsis thaliana AtHY5 (O24646.1) and AtHYH (OAP05786.1); Malus domestica MdHY5 (NP_001280752.1) and MdHYH-X1 (XP_008369576.1); Pyrus x bretschneideri PbHY5 (XP_009355719.1) and PbHYH-X1 (XP_009353456.1); Fragaria vesca subsp. Vesca FvHY5 (XP_004291469.1); Fragaria x ananassa FaHY5 (AKG58815.1); Prunus persica PpHY5 (XP_020411091.1) and PpHYH-X1 (XP_020409867.1); Vitis vinifera VvHY5 (XP_010648648.1) and VvHYH (AHX24181.1); Prunus mume PmHYH-X1 (XP_008222315.1); Prunus avium PaHYH-X1 (XP_021812250.1). (C) Effect of light treatment on the expression of flavonoids-related MiCOP1, MiHY5, and MiHYH in ‘Zill’ mango peel. Each value represents the mean ± standard deviation of three biological replicates. * indicates significant difference (p-value < 0.05), and ** indicates very significant difference (p-value < 0.01) between control and treatment, as determined by Student’s t-test.
Correlation analysis between flavonoids contents and gene expression
Correlation analysis was carried out to understand the relationship among flavonoids contents and gene expression. Results showed that 6 flavonoids components exhibited high positive correlation with each other (r > 0.5, p < 0.01) (Figure 6). The most significant correlations were observed between two proanthocyanidins, i.e. procyanidin B1 and procyanidin B3 (r = 0.98, p < 0.001), and two anthocyanins, i.e. cyanidin-3-O-galactoside, and peonidin-3-O-glucoside (r = 0.98, p < 0.001), respectively (Figure 6). For correlations between flavonoids contents and gene expression, MiF3H, MiLAR, MiUFGT3, and MiMYB1 were highly positively correlated with all the flavonoids components (r > 0.6, p < 0.001) (Figure 6). MiFLS was highly correlated to naringenin-7-O-glucoside concentration (r = 0.72, p < 0.001), but lowly correlated to quercetin-3-O-glucoside concentration (r = 0.44, p < 0.05) (Figure 6). Procyanidin B1 and procyanidin B3 showed high correlation with MiLAR, but low correlation with MiANR1 and MiANR2 (Figure 6). Cyanidin-3-O-galactoside, and peonidin-3-O-glucoside showed high correlation with MiUFGT3, but low correlation with MiUFGT1 (Figure 6). MiMYB12 was highly correlated with quercetin-3-O-glucoside (r = 0.67, p < 0.001), while MiMYB22 showed low correlation with flavonoids concentration (Figure 6).
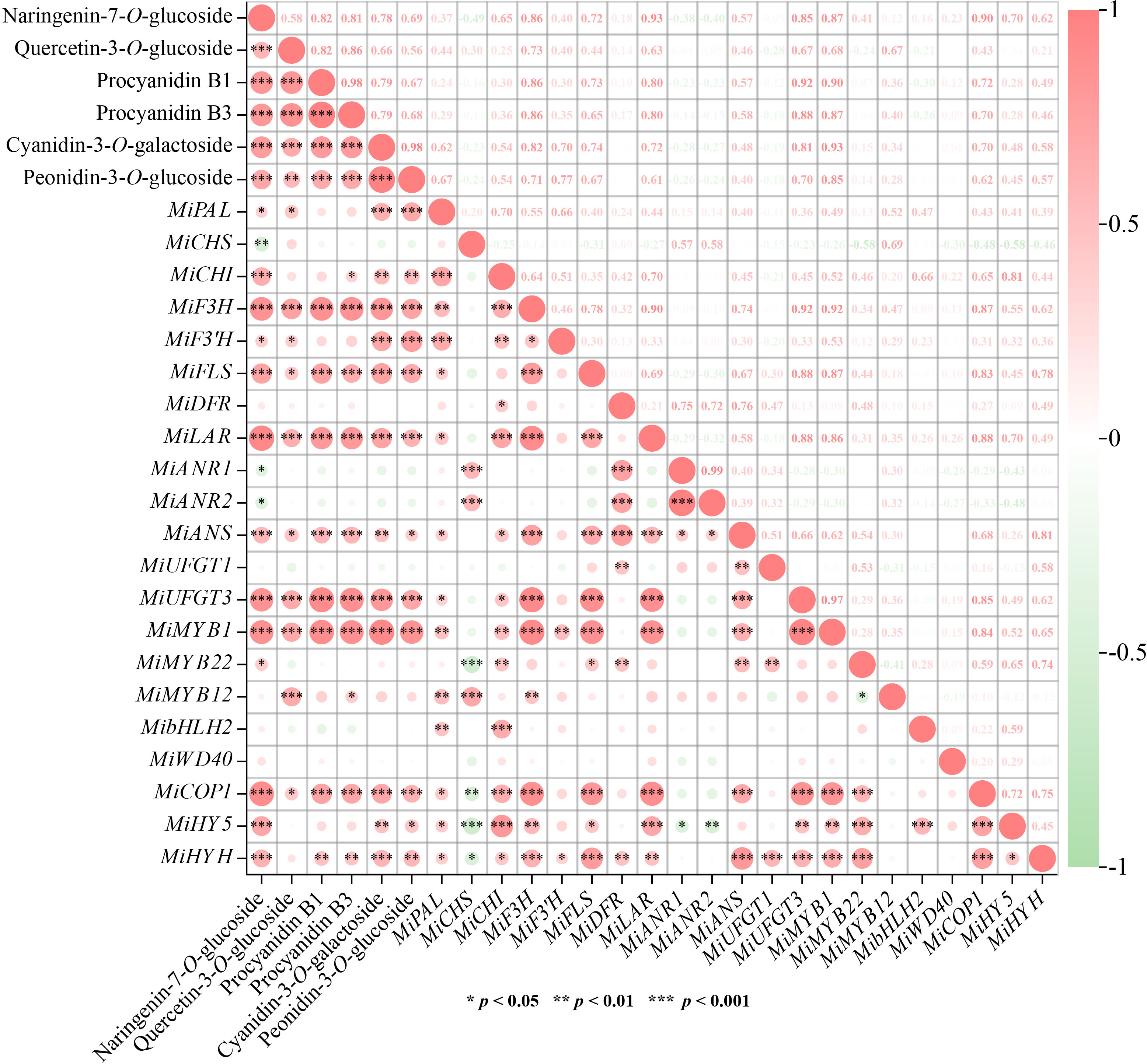
Figure 6 Correlation analysis among flavonoids contents and gene expression. Positive and negative correlations are indicated by colors, with red representing positive and green representing negative (*, p < 0.05, **, p < 0.01 and ***, p < 0.001). The numbers represent the Pearson correlation coefficient between two variables.
Discussion
Light induces the flavonoids accumulation in mango fruit peel but also indicates the competition among different flavonoids compositions
Light is one of the most crucial environmental factors not only regulates plant growth and development, but also induces secondary metabolites accumulation, for instance, flavonoids. Numerous preharvest and postharvest studies have shown that light promotes the total flavonoids concentration in plant (Huang et al., 2009; Qian et al., 2021), as well as different flavonoids components, including flavonols, proanthocyanidins and anthocyanins (Qian et al., 2013; Scattino et al., 2014; Shi et al., 2021). In this study, light induced the accumulation of flavonols, proanthocyanidins and anthocyanins in mango peel (Figure 2), which was similar with the previous studies. In addition, the accumulation of flavonols and proanthocyanidins peaked at day 6 and subsequently decreased towards day 10 (Figure 2), while the obvious red coloration and anthocyanins accumulation started at day 6, continuously increased, and peaked at day 10 (Figure 2), indicating the competition among different flavonoids compositions. Flavonols, proanthocyanidins and anthocyanins are synthesized from different branches which are derived from the same pathway, and after accumulating certain amount of flavonoids, plants fine-tune the pathway to fulfill the demand of growth and development instead of over-accumulating flavonoids. For example, many fruits accumulate flavonols and proanthocyanidins as the main flavonoids components at the early developmental stage to provide astringent taste against early feeding (Jaakola, 2013), while anthocyanins become the dominant composition in ripe fruits to attract insects and animals for seed dispersal (Harborne and Williams, 2000). The competition between proanthocyanidins and anthocyanins is more obvious because they are derived from the same precursor, anthocyanidins. Therefore, the increased content of proanthocyanidins or anthocyanins usually leads to the decrease of the other substance (Xie et al., 2003; Han et al., 2012).
Flavonoids accumulation is associated with the up-regulation of the key structural genes
Among all the EBGs, MiF3H was continuously induced by light (Figure 3A), ensuring the sufficient precursor accumulation for flavonoids biosynthesis, i.e. dihydroflavonols. Dihydroflavonols are important intermediate products in flavonoids pathway, which can be directly catalyzed by FLS to form flavonols or further processed for proanthocyanidins and anthocyanins biosynthesis. MiFLS expression was also tremendously induced by light resulting in the increased accumulation of flavonols (Figures 2, 3B). It has been showed that antisense expression of a FLS gene in petunia could inhibit flavonols accumulation and switched the petal color from light pink to red (Holton et al., 1993), while over-expression of a Camellia nitidissima FLS gene in tobacco promoted flavonols accumulation and change floral color from pink to white or light yellow (Zhou et al., 2013). The enhanced expression of MiANS in light-treated fruit peels was detected from 6 hours to 10 days (Figure 3B). ANS is crucial for both proanthocyanidins and anthocyanins biosynthesis since it catalyzes the formation of anthocyanidins, which can be further catalyzed by ANR to form proanthocyanidins, or by UFGT to form anthocyanins. The up-regulation of MiUFGT1 and MiUFGT3 mainly occurred form day 3 to day10 (Figure 3B), while MiANR1 and MiANR2 responded to light at the early stage (Figure 3B), indicating most anthocyanidins were converted to anthocyanins instead of proanthocyanidins at the late stage of light treatment, which was correlated to the increasing accumulation of anthocyanins from day 6 to day 10 (Figure 2). MiLAR expression was highly correlated with proanthocyanidins concentration (Figures 2, 3B, 6), indicating the conversion from leucoanthocyanins to proanthocyanidins catalyzed by LAR, rather than from anthocyanidins by ANR, was predominant during proanthocyanidins biosynthesis. It has been reported that overexpression of an apple ANR gene in tobacco suppressed the tobacco LAR expression, which also indicated the competition between these two NAPDH-dependent reductases (Han et al., 2012).
MYBs play essential roles in light-induced flavonoids biosynthesis
By regulating the expression of flavonoids biosynthetic genes, MYBs are regarded as the most important TFs mediating flavonoids synthesis, and this process is often in response to light signal. Over-expression of a light-induced Tartary buckwheat FtMYB6 gene in Tartary buckwheat hairy roots and tobacco could significantly increase the accumulation of flavonols (Yao et al., 2020). Over-expression of Arabidopsis AtMYB111 in tobacco promoted flavonols accumulation, which requires light (Pandey et al., 2014). UV-B light responsive MYB134 promotes proanthocyanidins synthesis in poplar by binding to the promoter regions of PA pathway genes including PAL and ANR (Mellway et al., 2009). Compared with flavonols and proanthocyanidins, anthocyanins are more widely studied due to their great contribution to the plants coloration, and MYBs regulating light-induced anthocyanins biosynthesis has been reported in diverse fruit species including apple (Li et al., 2012; Bai et al., 2016), pear (Feng et al., 2010; Ni et al., 2019), blood orange (Huang et al., 2019), Chinese bayberry (Niu et al., 2010), litchi (Lai et al., 2014), and mango (Kanzaki et al., 2020). In this study, the putative MYBs regulating flavonols and proanthocyanidins biosynthesis, i.e. MiMYB22 and MiMYB12, have been identified, respectively (Figures 4A, B), and the expression of MiMYB1, MiMYB22 and MiMYB12 was significantly increased by light treatment (Figure 4C), suggesting these MYBs probably regulate light-induced flavonoids biosynthesis.
COP1, HY5 and HYH are key upstream regulators of light-induced flavonoids biosynthesis
After being sensed by different photoreceptors, light signal pathway is transduced by COP1, HY5, and HYH. Under darkness, COP1 is located in the nucleus to degrade flavonoids-related regulators such as HY5 and MYBs via ubiquitination (Osterlund et al., 2000; Saijo et al., 2003; Li et al., 2012; Maier et al., 2013). Under light conditions, COP1 is translocated to cytoplasm leading to the accumulation of TFs such as HY5 and MYBs and subsequent flavonoids biosynthesis (Jiao et al., 2007; Li et al., 2012). In this study, the expression of MiCOP1 was induced by light at day 6, but showed no difference between light-treated sample and control sample at the other time points (Figure 5C), indicating that COP1 functions mainly through the protein subcellular localization instead of gene expression.
HY5 and HYH have also been widely reported to regulate light-induced flavonoids accumulation. In apple, MdHY5 regulates light-induced anthocyanins biosynthesis via binding to E-box and G-box motifs in the promoter region of MdMYB10 (An et al., 2017). PyHY5 is involved in the light-induced anthocyanins biosynthesis in pear by promoting the expression of PyWD40 and PyMYB10 (Wang et al., 2020b). As the homolog of HY5, HYH has also been shown to contribute to the light-induced flavonoids biosynthesis in peach (Zhao et al., 2022) and Arabidopsis (Holm et al., 2002). In this study, both MiHY5 and MiHYH responded very quickly to light and the increased expression lasted for quite a long time (Figure 5C), indicating the essential roles of HY5 and HYH during the light signal transduction.
Conclusions
Postharvest light treatment promoted the red peel coloration and increased the total soluble solids content and firmness in ‘Zill’ mango fruit. Metabolites profile showed that the accumulation of flavonols, proanthocyanidins and anthocyanins was also induced by light, as well as the key flavonoids biosynthetic genes including MiF3H, MiFLS, MiLAR, MiANS, MiUFGT1, and MiUFGT3. The flavonoids related MYBs, and essential light signal TFs HY5 and HYH, were also identified in mango, and their expression showed a light responsive pattern. Our results provide some molecular clues about light-induced flavonoids accumulation in mango.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author contributions
Conceptualization: WZ, HW, and MQ. Methodology: WZ, HW, CY, BS, BZ, XM, and KZ. Data curation: WZ, HW, CY, BS, BZ, XM, and KZ. Writing—original draft preparation: WZ, HW, CY, BS, BZ, XM, KZ, and MQ. Writing—review and editing: WZ, HW, CY, BS, BZ, XM, KZ, and MQ. Funding acquisition: HW and MQ. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by National Natural Science Foundation of China (grant number: 32160678), the Major Science and Technology Plan of Hainan Province (grant number: ZDKJ2021014), Hainan Provincial Natural Science Foundation of China (grant numbers: 322RC568; 320QN192), Hainan Province Science and Technology Special Fund (grant number: ZDYF2022XDNY255), and the Scientific Research Foundation of Hainan University (grant number: KYQD(ZR)20053).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1136281/full#supplementary-material
References
An, J.-P., Qu, F.-J., Yao, J.-F., Wang, X.-N., You, C.-X., Wang, X.-F., et al. (2017). The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 4 (1), 17023. doi: 10.1038/hortres.2017.23
Bai, S., Tuan, P. A., Saito, T., Honda, C., Hatsuyama, Y., Ito, A., et al. (2016). Epigenetic regulation of MdMYB1 is associated with paper bagging-induced red pigmentation of apples. Planta 244 (3), 573–586. doi: 10.1007/s00425-016-2524-4
Ban, Y., Honda, C., Hatsuyama, Y., Igarashi, M., Bessho, H., Moriguchi, T. (2007). Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 48 (7), 958–970. doi: 10.1093/pcp/pcm066
Broun, P. (2005). Transcriptional control of flavonoid biosynthesis: A complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8 (3), 272–279. doi: 10.1016/j.pbi.2005.03.006
Butelli, E., Licciardello, C., Zhang, Y., Liu, J., Mackay, S., Bailey, P., et al. (2012). Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24 (3), 1242–1255. doi: 10.1105/tpc.111.095232
Cao, S. F., Zhang, Y., Yu, X., Braun, C., Harrison, D., Joyce, D. C., et al. (2016). Postharvest light treatments increase skin blush in mango fruit. Acta Hortic. 1111, 399–404. doi: 10.17660/ActaHortic.2016.1111.57
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15 (10), 573–581. doi: 10.1016/j.tplants.2010.06.005
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49 (3), 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Feng, S., Wang, Y., Yang, S., Xu, Y., Chen, X. (2010). Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 232 (1), 245–255. doi: 10.1007/s00425-010-1170-5
González-Aguilar, G. A., Zavaleta-Gatica, R., Tiznado-Hernández, M. E. (2007). Improving postharvest quality of mango ‘Haden’ by UV-c treatment. Postharvest Biol. Technol. 45 (1), 108–116. doi: 10.1016/j.postharvbio.2007.01.012
Han, Y. P., Vimolmangkang, S., Soria-Guerra, R. E., Korban, S. S. (2012). Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J. Exp. Bot. 63 (7), 2437–2447. doi: 10.1093/jxb/err415
Harborne, J. B., Williams, C. A. (2000). Advances in flavonoid research since 1992. Phytochemistry 55 (6), 481–504. doi: 10.1016/S0031-9422(00)00235-1
Henry-Kirk, R. A., Plunkett, B., Hall, M., McGhie, T., Allan, A. C., Wargent, J. J., et al. (2018). Solar UV light regulates flavonoid metabolism in apple (Malus x domestica). Plant Cell Environ. 41 (3), 675–688. doi: 10.1111/pce.13125
Holm, M., Ma, L.-G., Qu, L.-J., Deng, X.-W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16 (10), 1247–1259. doi: 10.1101/gad.969702
Holton, T. A., Brugliera, F., Tanaka, Y. (1993). Cloning and expression of flavonol synthase from Petunia hybrida. Plant J. 4 (6), 1003–1010. doi: 10.1046/j.1365-313X.1993.04061003.x
Huang, D., Wang, X., Tang, Z., Yuan, Y., Xu, Y., He, J., et al. (2018). Subfunctionalization of the Ruby2–Ruby1 gene cluster during the domestication of citrus. Nat. Plants 4 (11), 930–941. doi: 10.1038/s41477-018-0287-6
Huang, C., Yu, B., Teng, Y., Su, J., Shu, Q., Cheng, Z., et al. (2009). Effects of fruit bagging on coloring and related physiology, and qualities of red Chinese sand pears during fruit maturation. Sci. Hortic. 121 (2), 149–158. doi: 10.1016/j.scienta.2009.01.031
Huang, D., Yuan, Y., Tang, Z., Huang, Y., Kang, C., Deng, X., et al. (2019). Retrotransposon promoter of Ruby1 controls both light- and cold-induced accumulation of anthocyanins in blood orange. Plant Cell Environ. 42 (11), 3092–3104. doi: 10.1111/pce.13609
Jaakola, L. (2013). New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 18 (9), 477–483. doi: 10.1016/j.tplants.2013.06.003
Jiao, Y., Lau, O. S., Deng, X. W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8 (3), 217–230. doi: 10.1038/nrg2049
Kanzaki, S., Ichihi, A., Tanaka, Y., Fujishige, S., Koeda, S., Shimizu, K. (2020). The R2R3-MYB transcription factor MiMYB1 regulates light dependent red coloration of ‘Irwin’ mango fruit skin. Sci. Hortic. 272, 109567. doi: 10.1016/j.scienta.2020.109567
Kobayashi, S., Goto-Yamamoto, N., Hirochika, H. (2004). Retrotransposon-induced mutations in grape skin color. Science 304 (5673), 982–982. doi: 10.1126/science.1095011
Lai, B., Li, X.-J., Hu, B., Qin, Y.-H., Huang, X.-M., Wang, H.-C., et al. (2014). LcMYB1 is a key determinant of differential anthocyanin accumulation among genotypes, tissues, developmental phases and ABA and light stimuli in Litchi chinensis. PloS One 9 (1), e86293. doi: 10.1371/journal.pone.0086293
Li, Y. Y., Mao, K., Zhao, C., Zhao, X. Y., Zhang, H. L., Shu, H. R., et al. (2012). MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 160 (2), 1011–1022. doi: 10.1104/pp.112.199703
Liu, C., Han, X., Cai, L., Lu, X., Ying, T., Jiang, Z. (2011). Postharvest UV-b irradiation maintains sensory qualities and enhances antioxidant capacity in tomato fruit during storage. Postharvest Biol. Technol. 59 (3), 232–237. doi: 10.1016/j.postharvbio.2010.09.003
Loyola, R., Herrera, D., Mas, A., Wong, D. C. J., Höll, J., Cavallini, E., et al. (2016). The photomorphogenic factors UV-b RECEPTOR 1, ELONGATED HYPOCOTYL 5, and HY5 HOMOLOGUE are part of the UV-b signalling pathway in grapevine and mediate flavonol accumulation in response to the environment. J. Exp. Bot. 67 (18), 5429–5445. doi: 10.1093/jxb/erw307
Maier, A., Schrader, A., Kokkelink, L., Falke, C., Welter, B., Iniesto, E., et al. (2013). Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in arabidopsis. Plant J. 74 (4), 638–651. doi: 10.1111/tpj.12153
Mellway, R. D., Tran, L. T., Prouse, M. B., Campbell, M. M., Constabel, C. P. (2009). The wound-, pathogen-, and ultraviolet b-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiol. 150 (2), 924–941. doi: 10.1104/pp.109.139071
Ni, J., Bai, S., Zhao, Y., Qian, M., Tao, R., Yin, L., et al. (2019). Ethylene response factors Pp4ERF24 and Pp12ERF96 regulate blue light-induced anthocyanin biosynthesis in ‘Red zaosu’ pear fruits by interacting with MYB114. Plant Mol. Biol. 99 (1), 67–78. doi: 10.1007/s11103-018-0802-1
Ni, J., Liao, Y., Zhang, M., Pan, C., Yang, Q., Bai, S., et al. (2022). Blue light simultaneously induces peel anthocyanin biosynthesis and flesh Carotenoid/Sucrose biosynthesis in mango fruit. J. Agric. Food Chem. 70 (50), 16021–16035. doi: 10.1021/acs.jafc.2c07137
Niu, S.-S., Xu, C.-J., Zhang, W.-S., Zhang, B., Li, X., Lin-Wang, K., et al. (2010). Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry (Myrica rubra) fruit by a R2R3 MYB transcription factor. Planta 231 (4), 887–899. doi: 10.1007/s00425-009-1095-z
Osterlund, M. T., Hardtke, C. S., Wei, N., Deng, X. W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 (6785), 462–466. doi: 10.1038/35013076
Pandey, A., Misra, P., Bhambhani, S., Bhatia, C., Trivedi, P. K. (2014). Expression of Arabidopsis MYB transcription factor, AtMYB111, in tobacco requires light to modulate flavonol content. Sci. Rep. 4 (1), 5018. doi: 10.1038/srep05018
Peng, T., Saito, T., Honda, C., Ban, Y., Kondo, S., Liu, J.-H., et al. (2012). Screening of UV-b-induced genes from apple peels by SSH: Possible involvement of MdCOP1-mediated signaling cascade genes in anthocyanin accumulation. Physiol. Plant 148, 432–444. doi: 10.1111/j.1399-3054.2012.12002.x
Podolec, R., Demarsy, E., Ulm, R. (2021). Perception and signaling of ultraviolet-b radiation in plants. Annu. Rev. Plant Biol. 72 (1), 793–822. doi: 10.1146/annurev-arplant-050718-095946
Premathilake, A. T., Ni, J., Bai, S., Tao, R., Ahmad, M., Teng, Y. (2020). R2R3-MYB transcription factor PpMYB17 positively regulates flavonoid biosynthesis in pear fruit. Planta 252 (4), 59. doi: 10.1007/s00425-020-03473-4
Qian, M., Rosenqvist, E., Prinsen, E., Pescheck, F., Flygare, A.-M., Kalbina, I., et al. (2021). Downsizing in plants–UV light induces pronounced morphological changes in the absence of stress. Plant Physiol. 187 (1), 378–395. doi: 10.1093/plphys/kiab262
Qian, M. J., Zhang, D., Yue, X. Y., Wang, S. K., Li, X. G., Teng, Y. W. (2013). Analysis of different pigmentation patterns in 'Mantianhong' (Pyrus pyrifolia Nakai) and 'Cascade' (Pyrus communis L.) under bagging treatment and postharvest UV-b/visible irradiation conditions. Sci. Hortic. 151, 75–82. doi: 10.1016/j.scienta.2012.12.020
Qian, M., Zhou, K., Wu, H., Yang, C., Shi, B. (2022). Method for inducing post-harvest red Mangifera indica L. to be colored. south Africa patent no 2022/04464. Pretoria: South Afr. Patent Office.
Saijo, Y., Sullivan, J. A., Wang, H., Yang, J., Shen, Y., Rubio, V., et al. (2003). The COP1–SPA1 interaction defines a critical step in phytochrome a-mediated regulation of HY5 activity. Genes Dev. 17 (21), 2642–2647. doi: 10.1101/gad.1122903
Santin, M., Lucini, L., Castagna, A., Rocchetti, G., Hauser, M.-T., Ranieri, A. (2019). Comparative “phenol-omics” and gene expression analyses in peach (Prunus persica) skin in response to different postharvest UV-b treatments. Plant Physiol. Biochem. 135, 511–519. doi: 10.1016/j.plaphy.2018.11.009
Scattino, C., Castagna, A., Neugart, S., Chan, H. M., Schreiner, M., Crisosto, C. H., et al. (2014). Post-harvest UV-b irradiation induces changes of phenol contents and corresponding biosynthetic gene expression in peaches and nectarines. Food Chem. 163, 51–60. doi: 10.1016/j.foodchem.2014.04.077
Schaart, J. G., Dubos, C., Romero de la Fuente, I., van Houwelingen, A. M. M. L., de Vos, R. C. H., Jonker, H. H., et al. (2013). Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 197 (2), 454–467. doi: 10.1111/nph.12017
Sheng, K., Zheng, H., Shui, S., Yan, L., Liu, C., Zheng, L. (2018). Comparison of postharvest UV-b and UV-c treatments on table grape: Changes in phenolic compounds and their transcription of biosynthetic genes during storage. Postharvest Biol. Technol. 138, 74–81. doi: 10.1016/j.postharvbio.2018.01.002
Shi, B., Wu, H., Zheng, B., Qian, M., Gao, A., Zhou, K. (2021). Analysis of light-independent anthocyanin accumulation in mango (Mangifera indica L.). Horticulturae 7 (11), 423. doi: 10.3390/horticulturae7110423
Shi, B., Wu, H., Zhu, W., Zheng, B., Wang, S., Zhou, K., et al. (2022). Genome-wide identification and expression analysis of WRKY genes during anthocyanin biosynthesis in the mango (Mangifera indica L.). Agriculture 12 (6), 821. doi: 10.3390/agriculture12060821
Stracke, R., Werber, M., Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 (5), 447–456. doi: 10.1016/S1369-5266(00)00199-0
Sun, Y., Qian, M., Wu, R., Niu, Q., Teng, Y., Zhang, D. (2014). Postharvest pigmentation in red Chinese sand pears (Pyrus pyrifolia Nakai) in response to optimum light and temperature. Postharvest Biol. Technol. 91 (0), 64–71. doi: 10.1016/j.postharvbio.2013.12.015
Takos, A. M., Jaffe, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., Walker, A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. doi: 10.1104/pp.106.088104
Tao, R., Bai, S., Ni, J., Yang, Q., Zhao, Y., Teng, Y. (2018). The blue light signal transduction pathway is involved in anthocyanin accumulation in ‘Red zaosu’ pear. Planta 248 (1), 37–48. doi: 10.1007/s00425-018-2877-y
Tao, R., Yu, W., Gao, Y., Ni, J., Yin, L., Zhang, X., et al. (2020). Light-induced Basic/Helix-Loop-Helix64 enhances anthocyanin biosynthesis and undergoes CONSTITUTIVELY PHOTOMORPHOGENIC1-mediated degradation in pear. Plant Physiol. 184 (4), 1684–1701. doi: 10.1104/pp.20.01188
Wang, P., Luo, Y., Huang, J., Gao, S., Zhu, G., Dang, Z., et al. (2020a). The genome evolution and domestication of tropical fruit mango. Genome Biol. Evol. 21 (1), 60. doi: 10.1186/s13059-020-01959-8
Wang, N., Xu, H., Jiang, S., Zhang, Z., Lu, N., Qiu, H., et al. (2017). MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90 (2), 276–292. doi: 10.1111/tpj.13487
Wang, Y., Zhang, X., Zhao, Y., Yang, J., He, Y., Li, G., et al. (2020b). Transcription factor PyHY5 binds to the promoters of PyWD40 and PyMYB10 and regulates its expression in red pear ‘Yunhongli no. 1’. Plant Physiol. Biochem. 154, 665–674. doi: 10.1016/j.plaphy.2020.07.008
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., Barton, G. J. (2009). Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25 (9), 1189–1191. doi: 10.1093/bioinformatics/btp033
Williams, C. A., Grayer, R. J. (2004). Anthocyanins and other flavonoids. Nat. Prod. Rep. 21 (4), 539–573. doi: 10.1039/B311404J
Winkel-Shirley, B. (2001). Flavonoid biosynthesis. a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126 (2), 485–493. doi: 10.1104/pp.126.2.485
Xie, D.-Y., Sharma, S. B., Paiva, N. L., Ferreira, D., Dixon, R. A. (2003). Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299 (5605), 396–399. doi: 10.1126/science.1078540
Yang, J., Shi, W., Li, B., Bai, Y., Hou, Z. (2019). Preharvest and postharvest UV radiation affected flavonoid metabolism and antioxidant capacity differently in developing blueberries (Vaccinium corymbosum L.). Food Chem. 301, 125248. doi: 10.1016/j.foodchem.2019.125248
Yao, P., Huang, Y., Dong, Q., Wan, M., Wang, A., Chen, Y., et al. (2020). FtMYB6, a light-induced SG7 R2R3-MYB transcription factor, promotes flavonol biosynthesis in tartary buckwheat (Fagopyrum tataricum). J. Agric. Food Chem. 68 (47), 13685–13696. doi: 10.1021/acs.jafc.0c03037
Yao, G., Ming, M., Allan, A. C., Gu, C., Li, L., Wu, X., et al. (2017). Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 92 (3), 437–451. doi: 10.1111/tpj.13666
Zhao, L., Sun, J., Cai, Y., Yang, Q., Zhang, Y., Ogutu, C. O., et al. (2022). PpHYH is responsible for light-induced anthocyanin accumulation in fruit peel of Prunus persica. Tree Physiol. 42 (8), 1662–1677. doi: 10.1093/treephys/tpac025
Keywords: mango, flavonoids, light treatment, metabolites profile, gene expression
Citation: Zhu W, Wu H, Yang C, Shi B, Zheng B, Ma X, Zhou K and Qian M (2023) Postharvest light-induced flavonoids accumulation in mango (Mangifera indica L.) peel is associated with the up-regulation of flavonoids-related and light signal pathway genes. Front. Plant Sci. 14:1136281. doi: 10.3389/fpls.2023.1136281
Received: 02 January 2023; Accepted: 28 February 2023;
Published: 13 March 2023.
Edited by:
María Serrano, Miguel Hernández University of Elche, SpainReviewed by:
Yue Wang, Zhejiang University, ChinaHui Liu, Zhengzhou Fruit Research Institute (CAAS), China
Copyright © 2023 Zhu, Wu, Yang, Shi, Zheng, Ma, Zhou and Qian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minjie Qian, bWluamllLnFpYW5AaGFpbmFudS5lZHUuY24=
†These authors have contributed equally to this work
 Wencan Zhu1†
Wencan Zhu1† Bin Shi
Bin Shi Xiaowei Ma
Xiaowei Ma Kaibing Zhou
Kaibing Zhou Minjie Qian
Minjie Qian