
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 February 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1135684
This article is part of the Research Topic Research on Brassicaceae Crops Genomics and Breeding View all 10 articles
Orphan genes are essential to the emergence of species-specific traits and the process of evolution, lacking sequence similarity to any other identified genes. As they lack recognizable domains or functional motifs, however, efforts to characterize these orphan genes are often difficult. Flowering is a key trait in Brassica rapa, as premature bolting can have a pronounced adverse impact on plant quality and yield. Bolting resistance-related orphan genes, however, have yet to be characterized. In this study, an orphan gene designated BOLTING RESISTANCE 1 (BR1) was identified and found through gene structural variation analyses to be more highly conserved in Chinese cabbage than in other available accessions. The expression of BR1 was increased in bolting resistant Chinese cabbage and decreased in bolting non-resistant type, and the expression of some mark genes were consist with bolting resistance phenotype. BR1 is primarily expressed in leaves at the vegetative growth stage, and the highest BR1 expression levels during the flowering stage were observed in the flower buds and silique as compared to other tissue types. The overexpression of BR1 in Arabidopsis was associated with enhanced bolting resistance under long day (LD) conditions, with these transgenic plants exhibiting significant decreases in stem height, rosette radius, and chlorophyll content. Transcriptomic sequencing of WT and BR1OE plants showed the association of BR1 with other bolting resistance genes. Transcriptomic sequencing and qPCR revealed that six flowering integrator genes and one chlorophyll biosynthesis-related gene were downregulated following BR1 overexpression. Six key genes in photoperiodic flowering pathway exhibited downward expression trends in BR1OE plants, while the expression of floral repressor AtFLC gene was upregulated. The transcripts of these key genes were consistent with observed phenotypes in BR1OE plants, and the results indicated that BR1 may function through vernalization and photoperiodic pathway. Instead, the protein encoded by BR1 gene was subsequently found to localize to the nucleus. Taken together, we first propose that orphan gene BR1 functions as a novel regulator of flowering time, and these results suggested that BR1 may represent a promising candidate gene to support the selective breeding of Chinese cabbage cultivars with enhanced bolting resistance.
Orphan genes (OGs) are species- or lineage-specific genes that lack sequence similarity with other known genes expressed by other species (Jiang et al., 2022), often arising as a result of rapid evolutionary activity (Cai et al., 2006). To date, OGs have been reported in many genomic sequencing analyses of species including Aegiceras corniculatum (Ma et al., 2021), Arabidopsis thaliana (Donoghue et al., 2011), Brassica rapa (Jiang et al., 2018), eight Cucurbitaceae species (Ma et al., 2022), Oryza sativa (Cui et al., 2015), and Vigna unguiculata (Li et al., 2019). As OGs do not contain recognizable domains, functional motifs, or folding patterns, their functions are often unclear such that detailed functional characterization is ultimately necessary. Several studies have successfully demonstrated the diverse roles played by specific OGs as mediators of metabolite synthesis (Li et al., 2009; Li and Wurtele, 2015; Li et al., 2015b; O’Conner et al., 2018; Jiang et al., 2020b; Fang et al., 2021; Shen et al., 2021; Tanvir et al., 2022b), biotic stresses response (Jiang et al., 2018; Qi et al., 2019; Brennan et al., 2020; Jiang et al., 2020a; Wang et al., 2021; Moon et al., 2022; Tanvir et al., 2022a), abiotic stresses response (Yadeta et al., 2014; Li et al., 2019; Ma et al., 2020), species-specific traits, or the regulation of growth and development (Chen et al., 2017; Ni et al., 2017; Wang et al., 2017; Zhao et al., 2018; Dossa et al., 2021). These characteristics make OGs important targets for plant breeding efforts aimed at enhancing specific traits of interest to improve plant stress resistance and quality. However, the ability of specific OGs to regulate the timing of flowering has largely been overlooked to date.
Flowering timing is an agronomically important trait that can determine reproductive success and shape crop yields. This timing is thus regulated by a complex network of signaling proteins and processes that are responsive to external stimuli and developmental cues (Wang, 2014; Blümel et al., 2015). Five genetically defined pathways have been identified to date as important regulators of floral transition, including the age, photoperiod, hormone, autonomous, and vernalization pathways (Srikanth and Schmid, 2011; Li et al., 2015a; Freytes et al., 2021). These pathways integrate diverse signaling inputs associated with flowering and ultimately regulate the expression of important genes that govern flowering timing including APETALA1 (AP1, AT1G69120), SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1, AT2G45660), FLOWERING LOCUS T (FT, AT1G65480), and the plant-specific transcription factor (TF) LEAFY (LFY, AT5G61850) (Wang, 2014; Bao et al., 2020). The autonomous pathway can promote flowering in a manner that is independent of the length of the day through the suppression of FLOWERING LOCUS C (FLC, AT5G10140), which is a central repressor of flowering and vernalization (Simpson and Dean, 2002; Jung and Müller, 2009; Cheng et al., 2017). FLC is an important inhibitor of flowering activity that can suppress shoot apical meristem (SAM) floral transition-related TF expression (Cho et al., 2017). In the photoperiod pathway, daily patterns of expression for the floral regulators CONSTANS (CO, AT5G15840) and FT are regulated by a range of positive and negative factors that can influence protein-protein interactions, chromatin structural characteristics, protein stability, and transcriptional activity (Johansson and Staiger, 2015). Circadian rhythms are closely linked to the photoperiod-mediated control of the transition from vegetative to reproductive plant growth (Creux and Harmer, 2019). Gibberellin (GA) pathway signaling serves as a major hormonal mechanism that regulates flowering activity, although other hormones including jasmonate, brassinosteroid, abscisic acid, cytokinins, and ethylene also play regulatory roles in this context (Izawa, 2021). The miR156-SPL (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE) and miR172-AP2 (APETALA2, AT4G36920) modules have been suggested as key regulatory hubs involved in the age pathway that facilitate plant flowering under non-inductive conditions (Wang, 2014; Kinoshita and Richter, 2020). While these results provide important insight into the mechanisms that control the timing of flowering, the mechanistic links among these pathways and the regulatory crosstalk between them have yet to be characterized in detail.
Heading is among the most important agronomic traits for B. rapa ssp. pekinensis (Chinese cabbage) or Brassica oleracea var. capitata (cabbage) and has been the subject of extensive research interest (Zhang et al., 2022). Premature bolting can have a severe adverse impact on Chinese cabbage or cabbage yields and quality, restricting the geographic distribution and planting season for this economically important species (Su et al., 2018). Accordingly, there is a pressing need to breed novel cultivars with enhanced bolting resistance. The histone H4 protein encoded by BrHIS4.A04 (Bra035673) has previously been shown to attenuate photoperiod-related flowering gene expression under drought conditions in Chinese cabbage plants via signaling through the ABA pathway, thus preventing premature bolting (Xin et al., 2020). SET DOMAIN GROUP 8 (BrSDG8, BraA07g040740.3C) serves as an additional regulator of early bolting in B. rapa ssp. pekinensis, with FLC H3K6 methylation activity increasing when the function of this gene is disrupted (Fu et al., 2020). BrFLC5 (Bra022771) was also previously found to be expressed at lower levels than two other BrFLC genes, supporting efforts to breed B. rapa plants resistant to premature bolting (Xi et al., 2018). Cabbage BoFLC4-1 played a similar role to Arabidopsis FLC in regulating flowering time (Lin et al., 2005). The intron I 215-bp indel of BoFLC2 influenced the flowering time of cabbage, which might offer critical information to promote the study of epigenetic gene silencing processes in flowering-related genes (Li et al., 2022). Study also showed that BoFLC1, BoFLC3, and BoFLC5 were within the confidence intervals of known flowering time quantitative trait loci (QTL) (Razi et al., 2008). These prior results thus offer valuable insights that can be leveraged to better facilitate the genetic control of the bolting and flowering processes.
In this study, the OG designated BOLTING RESISTANCE 1 (BR1) was the target of functional characterization efforts exploring its relationship with the timing of flowering. Structural genotypic variations and sequence characteristics in the BR1 gene region were analyzed in 524 B. rapa accessions, and its expressions in bolting resistant or bolting non-resistant Chinese cabbage were detected. Representative mark genes were determined in bolting resistant type inbred lines. And BR1 expression patterns were examined over the course of Chinese cabbage development in different tissue compartments. The impact of BR1 overexpression on flowering time was additionally assessed in Arabidopsis thaliana, while transcriptomic sequencing was used to explore the mechanistic basis for the ability of BR1 to delay A. thaliana flowering time. Subcellular localization of the BR1 protein was additionally analyzed. Together, these analyses identified the B. rapa ssp. pekinensis BR1 gene as a key regulator of delayed flowering time in A. thaliana.
B. rapa ‘Chiifu’ cultivar, Chinese cabbage inbred lines, A. thaliana ecotype ‘Columbia-0’ (Col-0), and the transgenic A. thaliana lines were cultivated as in our prior studies (Jiang et al., 2018; Jiang et al., 2020b). Nicotiana benthamiana was cultivated as in prior study (Zhan et al., 2022).
The CD-Search tool was used to search the NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) for conserved domains in BR1. The SignalP 5.0 server (https://services.healthtech.dtu.dk/service.php?SignalP-5.0) was used to identify predicted signal peptide sequences in BR1. The PROSITE database (https://prosite.expasy.org/) was used to predict motifs in BR1, while TF predictions were made with the Plant Transcription Factor Database (PlantTFDB v5.0, http://planttfdb.gao-lab.org/). Genotypic analyses of structural variations in the BR1 gene region were performed by comparing 524 B. rapa accessions with the Polymorph tool using the Brassicaceae Database (BRAD, http://brassicadb.cn/) as reported previously (Cai et al., 2021), with these 524 different B. rapa accessions being derived from a separate report (Cheng et al., 2016; Su et al., 2018; Cai et al., 2021). Ten orphan genes analyzed in this study were identified in our previous study (Jiang et al., 2018). The B. rapa genome version 3.0 was used for structural variation analyses.
Quantitative real-time PCR (qPCR) analyses were performed as detailed previously (Jiang et al., 2018), using primers compiled in Supplementary Table S1. Sampling of plants at the seedling and flowering stages was performed as published previously (Jiang et al., 2018), while sampling at the rosette and heading stages was performed at the 6th and 8th weeks, respectively. When rosette stage sampling was performed, three leaves were collected from each of nine individual ‘Chiifu’ plants (three biological replicates, three plants per replicate). With the oldest leaf numbered as leaf one, samples were collected from different positions (top, middle, bottom) on three different leaves (outer leaf, first leaf; middle leaf, 10th leaf; inner leaf, 20th leaf), designated as the RL1, RL2, and RL3 from the outer to the inner leaves. In the heading stage, three leaves (outer leaf, first leaf; middle leaf, 20th leaf; inner leaf, 40th leaf) were similarly collected from each of nine ‘Chiifu’ plants, with these samples being respectively designated as HL1, HL2, and HL3. For BR1 expression in bolting resistant or bolting non-resistant Chinese cabbage, ten inbred lines from heading stage were selected from our laboratory, the top point of short stem (GP) and the top point of inner leaf (TP) were sampled, TP was used as control. Three biological replicates with three plants of different lines per replicate were sampled. After collection, samples were snap-frozen with liquid nitrogen and stored at −80°C for subsequent RNA isolation. qPCR primers for the expression analyses of representative mark genes were listed in Supplementary Table S1.
Vector constructs, Arabidopsis transformation, and selection were all performed as detailed in our prior report (Jiang et al., 2020b). Vector construction was performed using primer pairs compiled in Supplementary Table S1. Phenotypic analyses of transgenic Arabidopsis plants were performed as detailed in our prior study (Jiang et al., 2020b).
Three biological replicate samples were collected from the aerial portions of WT and BR1OE mutant plants 25 days post-planting. After snap freezing using liquid nitrogen, these samples were stored at −80°C. RNA extraction was performed as detailed previously (Jiang et al., 2018), and 1% agarose gel electrophoresis was used to detect any RNA contamination or degradation while a NanoPhotometer® instrument (IMPLEN, CA, USA) was used to confirm RNA purity. A Qubit® RNA Assay Kit and a Qubit® 2.0 Fluorometer (Life Technologies, CA, USA) were used to quantify the RNA concentrations in individual samples, while an RNA Nano 6000 Assay Kit and a Bioanalyzer 2100 instrument (Agilent Technologies, CA, USA) were used to confirm RNA integrity. Sequencing libraries were prepared from 1 μg of RNA per sample with a NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) based on provided directions. Library sequencing was then performed with an Illumina Hiseq platform to generate 125 bp/150 bp paired-end reads.
Initial data were filtered with Fastp (v0.19.3) to remove adapter-containing reads, reads containing > 10% N bases, and reads with > 50% low-quality (Q ≤ 20) bases. The clean reads were then compared to the Arabidopsis TAIR10 genome which was downloaded from The Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org/) using HISAT (v2.1.0). New gene predictions were made using StringTie (v1.3.4d), while gene alignment was calculated with FeatureCounts (v1.6.2), and fragments per kilobases of exons per million mapped reads (FPKM) expression values were then calculated for all transcripts. Differentially expressed genes (DEGs) were identified using DESeq2 (v1.22.1) based on Benjamini & Hochberg-corrected p-values, a |log2Fold Change| ≥ 1, and a false discovery rate (FDR) < 0.05. Hypergeometric tests were used for Gene Ontology (GO) term and KEGG pathway enrichment analyses. Gene expression was validated using primer pairs listed in Supplementary Table S1.
The BR1 coding sequence was cloned into the pCAM35-GFP vector without the corresponding stop codons using the KpnI and BamHI cleavage sites. pCAM35::BR1::GFP expression vector construction and N. benthamiana epidermal cell infection was performed as reported previously (Zhang et al., 2021). Nuclei were visualized through the co-expression of a mCherry-labeled nuclear marker. All experiments were independently repeated in triplicate, and a Leica confocal microscope (SP8, Germany) was used to visualize cells at 48 h following agro-infiltration. Primer pairs used in vector construction are compiled in Supplementary Table S1.
SPSS 19.0 was used to compare data through Student’s t-tests or one-way ANOVAs with Duncan’s multiple range test as appropriate.
Genotypic analyses of structural variations of ten orphan genes were randomly selected from our prior study (Jiang et al., 2018) (Supplementary Table S2), which was used to screen potential bolting resistance orphan genes. BRAD was used to perform genotypic analyses of structural variations in this BR1 gene region across 524 B. rapa accessions. There were no any structural variations in four orphan genes (BraA01g024790.3C, BraA02g027570.3C, BraA03g056750.3C, and BraA09g030600.3C). The highest average sequence conservation rates was found in the gene region of BR1 (BraA10g003580.3C), including just four single nucleotide polymorphisms (SNPs) in this region (A10_1866376, A10_1866412, A10_1866527, and A10_1866663) (Figures 1A-D), which accounting for ~80% in Chinese cabbage relative to ~46% in other accessions (Figure 1E). The A10_1866412 SNP exhibited the highest sequence conservation ratio (~98%) in Chinese cabbage, while the average variation ratio in Chinese cabbage was ~20%, with this value being lower than in other accessions (~54%). The A10_1866376 SNP exhibited the highest variation ratio (~76%) in other accessions. Average BR1 sequence conservation rates were approximately 77%, 75%, and 88% in Chinese cabbage spring, summer, and autumn ecotypes, respectively (Figure 1F). No variations in the A10_1866412 SNP were observed among these three Chinese cabbage ecotypes, and the average percentage of variation in these three respective ecotypes was approximately 23%, 25%, and 12%. BR1 located on chromosome A10 at positions 1866284 – 1866805, and this gene contains no introns and consists of 522 bases that encode a protein 173 amino acids in length. The BR1 protein does not contain any known signal peptides, motifs, or conserved domains, and it was not identified as a TF in subsequent analyses. As such, the BR1 gene is more conserved in Chinese cabbage than in other accessions, potentially owing to domestication and associated selection for leafy head development and bolting resistance in this economically important species.
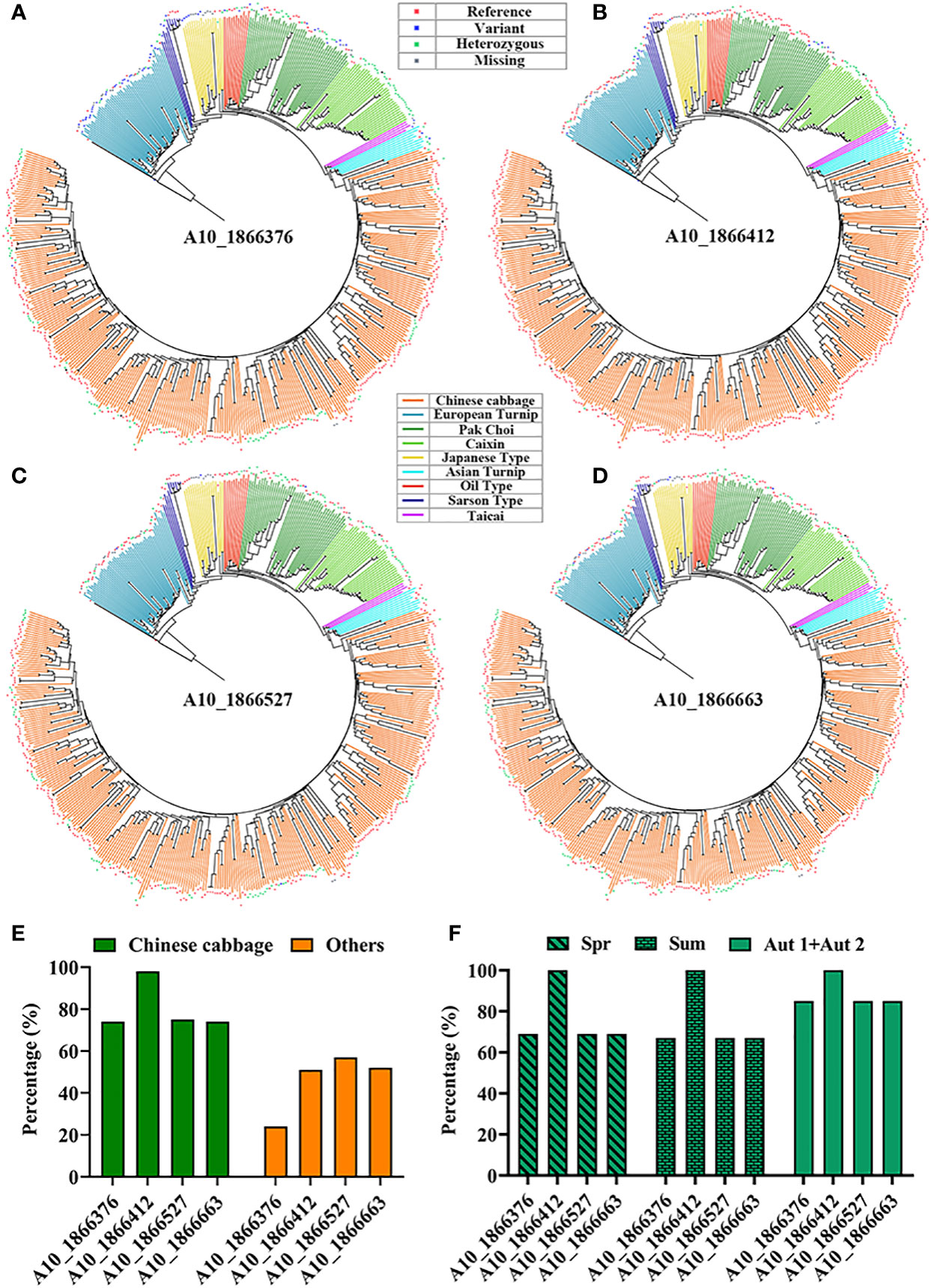
Figure 1 Genotypic analyses of SNPs in the BR1 gene region. (A-D) Different SNP types identified across 524 B rapa accessions in the BR1 gene region. If a particular accession was consistent with the reference genome, this is denoted by a reference designation, whereas the variant designation was used to indicate an accession with a genotype divergent from the reference genome. Deletion events were designated by ‘Missing’, and heterozygous typing was represented by ‘Heterozygous’. (E) The percentage of sequence conservation in Chinese cabbage and other accessions. (F) Percentage of sequence conservation in different Chinese cabbage ecotypes, with Spr and Sum respectively corresponding to the spring and summer ecotypes, while the autumn ecotypes are denoted by Aut1+Aut2.
qPCR was used to analyze BR1 expression patterns in an effort to explore its potential functional roles during different stages of B. rapa development. Significant increases in BR1 expression were observed in both roots and leaves in the seedling stage, with peak expression in the leaves (Figure 2A). During the rosette and heading stages, BR1 expression levels rose significantly in the middle (RL2 and HL2) and inner leaves (RL3 and HL3) relative to the outer leaves (RL1 and HL1) (Figures 2B, C). During the flowering stage, higher BR1 expression levels were detected in the flower buds and silique relative to other analyzed tissues (Figure 2D). These results provide a basis for the further exploration of BR1 as an OG associated with the regulation of different stages of vegetative and reproductive growth in Chinese cabbage plants. Next, the expression levels of BR1 were detected within bolting resistant (BR type) and bolting non-resistant (BN type) Chinese cabbage to explore its functions. Study indicated that Chinese cabbage with the rounded apices of its short stem belonging to BR type, and BN type possessed the pointed apices (Mero and Honma, 1984). Surprisingly, BR1 possessed increase trends in five lines from BR type, but showed suppressed trends in BN type (Figure 3A). Meanwhile, the expression patterns of mark genes were analyzed to confirm the function of controlling bolting resistance in Chinese cabbage. Mark genes including BrFLCs, BrFTs, BrSOC1s, and BrLFYs. BR type inbred lines ‘BR-98’ was selected for further analysis. As expected, four BrFLCs showed increased trends, while BrFTs, BrSOC1s, and BrLFY2 were displayed decreased trends (Figure 3B). The expression of BrFT3 and BrLFY1 was not detected. These expression patterns correspond to the bolting resistance phenotype of inbred line ‘BR-98’. Such results suggested that BR1 may directly or indirectly involve in bolting resistance of Chinese cabbage.
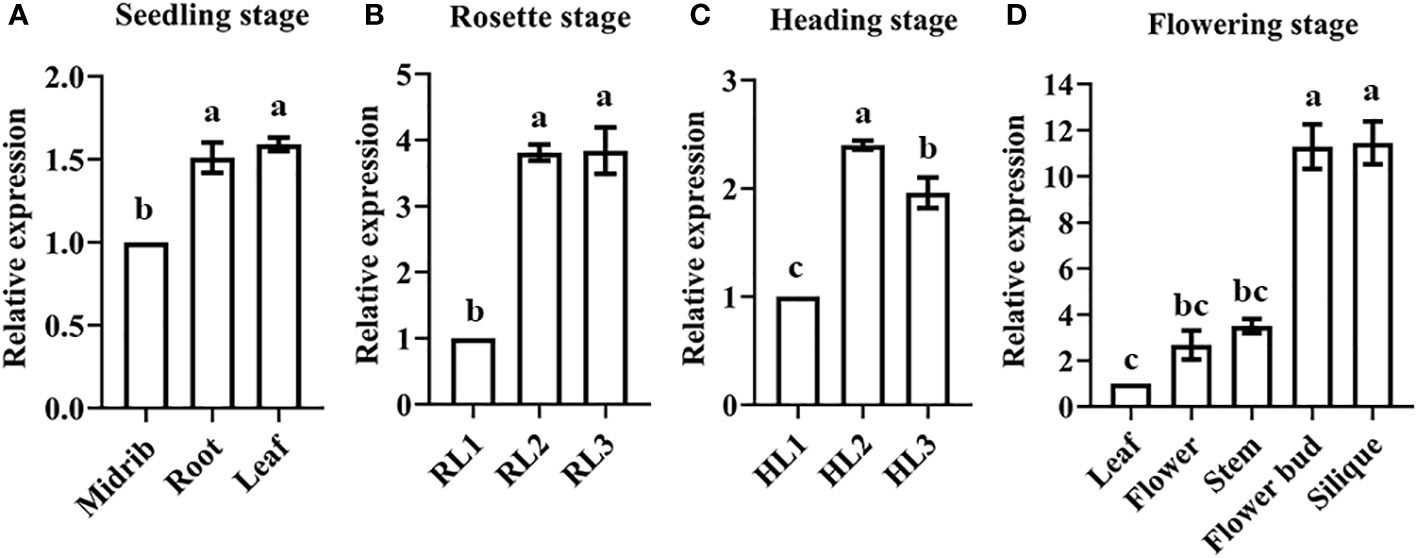
Figure 2 BR1 expression patterns during different developmental stages in Chinese cabbage. BR1 expression was assessed in the (A) seedling, (B) rosette, (C) heading, and (D) flowering stages. Data are means ± SE of three independent measurements. Statistically distinct groups are marked with black letters (one-way ANOVA, p < 0.05).
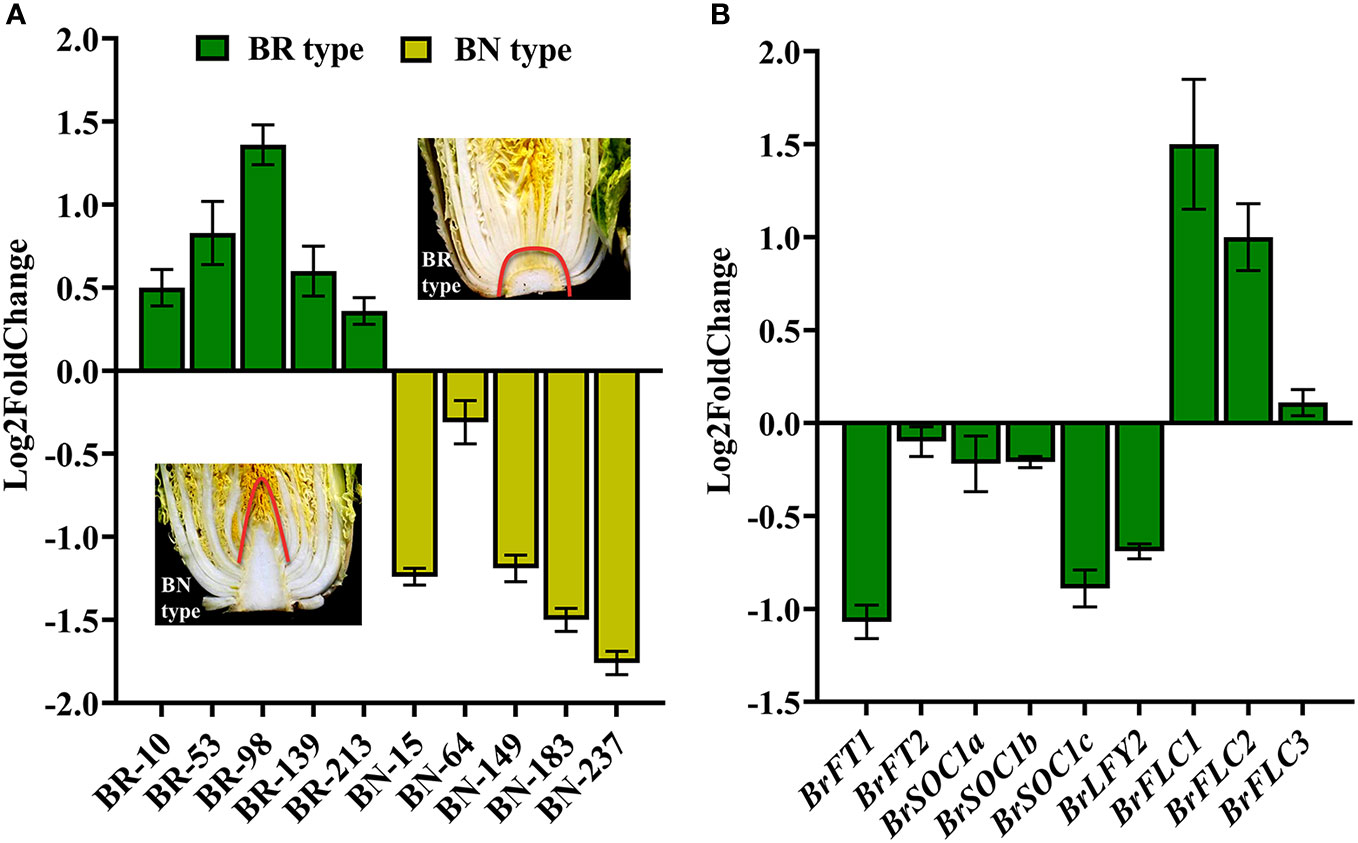
Figure 3 Expression patterns of BR1 (A) and representative mark genes (B) in bolting resistant or non-resistant Chinese cabbage during heading stage. Examples of bolting resistant type (BR type) and bolting non-resistant type (BN type) Chinese cabbage were showed in the figure. Green column represented BR type, yellow column indicated BN type. BR type inbred lines ‘BR-98’ was used for expression analyses.
To better understand the functional roles played by BR1, A. thaliana plants overexpressing this gene were prepared. After floral dip transformation, the DsRed marker was used to select T2 homozygous seeds from different self-pollinated T1 transgenic seed lines, and T3 plants were then planted to assess phenotypes following the harvesting of seeds from T2 plants. Phenotypes were compared between the wild-type (WT) and BR1 overexpressing plants (BR1OE) in the vegetative and reproductive phases of growth under LD conditions (16 h of light/8 h of dark). These analyses revealed that BR1 overexpression strongly delayed Arabidopsis floral transition (Figure 4A), with BR1OE plants exhibiting a 28% delay in flowering time relative to WT controls (Figure 4B). Rosette radius was also reduced by ~26% in these BR1OE mutant plants (Figure 4C), suggesting that BR1 may regulate leaf elongation under standard growth conditions. BR1OE plants also exhibited a decrease in final stem height relative to WT controls (Figure 4D), whereas silique length and numbers of seeds per silique were unchanged (Figures 4E, F). Notably, BR1OE plants exhibited reductions in chlorophyll content relative to WT plants (Figures 4G-I). Accordingly, the overexpression of BR1 enhances Arabidopsis bolting resistance.
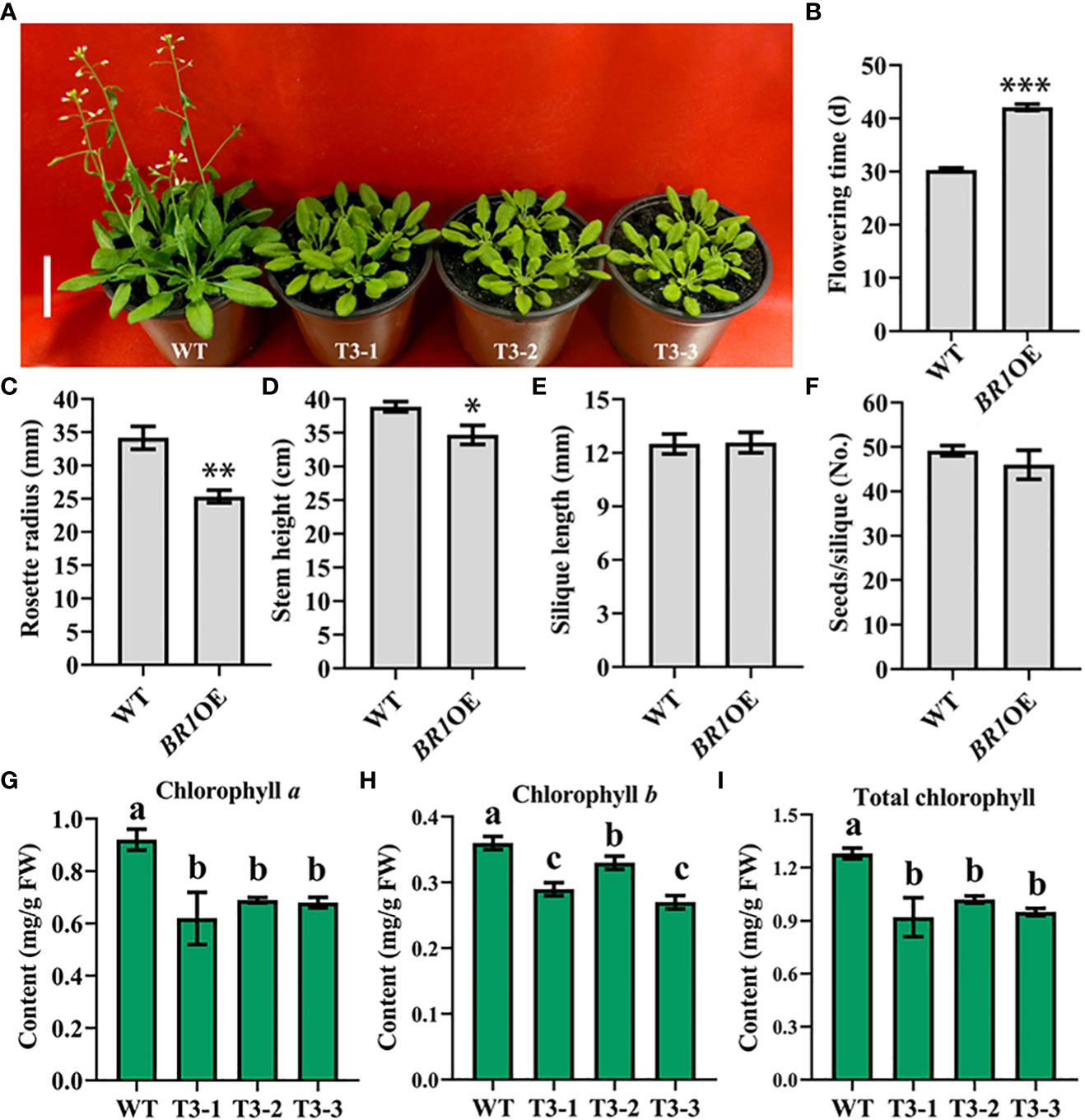
Figure 4 BR1OE mutant phenotypic characterization. (A) Under LD conditions the BR1OE transgenic plant exhibited a late-flowering phenotype. Representative images of 32-day-old transgenic plants from three separate genetic transformation events (T3-1, T3-2, and T3-3) and wild-type Col-0 (WT) are shown. Scale bar: 4 cm. (B) Flowering time. (C) Rosette radius. (D) Stem height. (E) Silique length. (F) Seed number per silique. Data are means ± SE of three independent measurements. Significantly differences were identified when comparing WT and BR1OE plants using Student’s t-tests (*p < 0.05, **p < 0.01, ***p < 0.001). (G) Chlorophyll a content. (H) Chlorophyll b content. (I) Total chlorophyll content. Analyses were performed with transgenic plants from three separate genetic transformation events (T3-1, T3-2, and T3-3) and wild-type Col-0 (WT) Arabidopsis. FW: fresh weight. Data are means ± SE of three independent measurements. Statistically distinct groups are marked with black letters (one-way ANOVA, p < 0.05).
Transcriptomic sequencing was conducted to explore the molecular mechanisms underlying the effects of BR1 on delayed flowering time in A. thaliana by preparing cDNA libraries from the leaves of WT and BR1OE plants. All samples exhibited Q20 values > 97% and Q30 values > 92% (Supplementary Table S3). Raw and clean read numbers for individual samples respectively ranged from 43,611,780 - 48,073,534 and 42,257,488 - 46,814,082, consistent with highly reliable transcriptomic detection results.
Relative to WT plants, the BR1OE mutants exhibited significantly delayed flowering time. To better understand the transcriptomic changes underlying this phenotype, FPKM values for individual unigenes were compared between these two A. thaliana varieties to identify genes that were differentially expressed. In total, 254 DEGs were identified of which 73 and 181 were respectively up- and downregulated (28.74% and 71.26%, respectively) (Figure 5A, Supplementary Table S4). GO enrichment analyses of these DEGs identified 44 significantly enriched GO terms (Figure 5B, Supplementary Table S5). These included enriched biological processes (including cellular process, metabolic process, biological regulation, developmental process, regulation of biological process, multicellular organismal progress, signaling, reproductive process, reproduction, growth, and rhythmic process), cellular component (including cell, cell part, organelle, membrane, membrane part, and extracellular region), and molecular function (including binding, catalytic activity, transcription regulator activity, transporter activity, structural molecule activity, antioxidant activity) terms. KEGG pathway enrichment analysis also identified 47 significantly enriched KEGG pathways (Supplementary Table S6), with the top 20 for each DEG set being presented in Figure 5C. The majority of these DEGs were enriched in pathways including “metabolic pathways and biosynthesis of secondary metabolites (ko01100)”, “plant-pathogen interaction (ko04626)”, “phenylpropanoid biosynthesis (ko00940)”, “plant MAPK signaling pathway (ko04016)”, and “porphyrin and chlorophyll metabolism (ko00860)”.
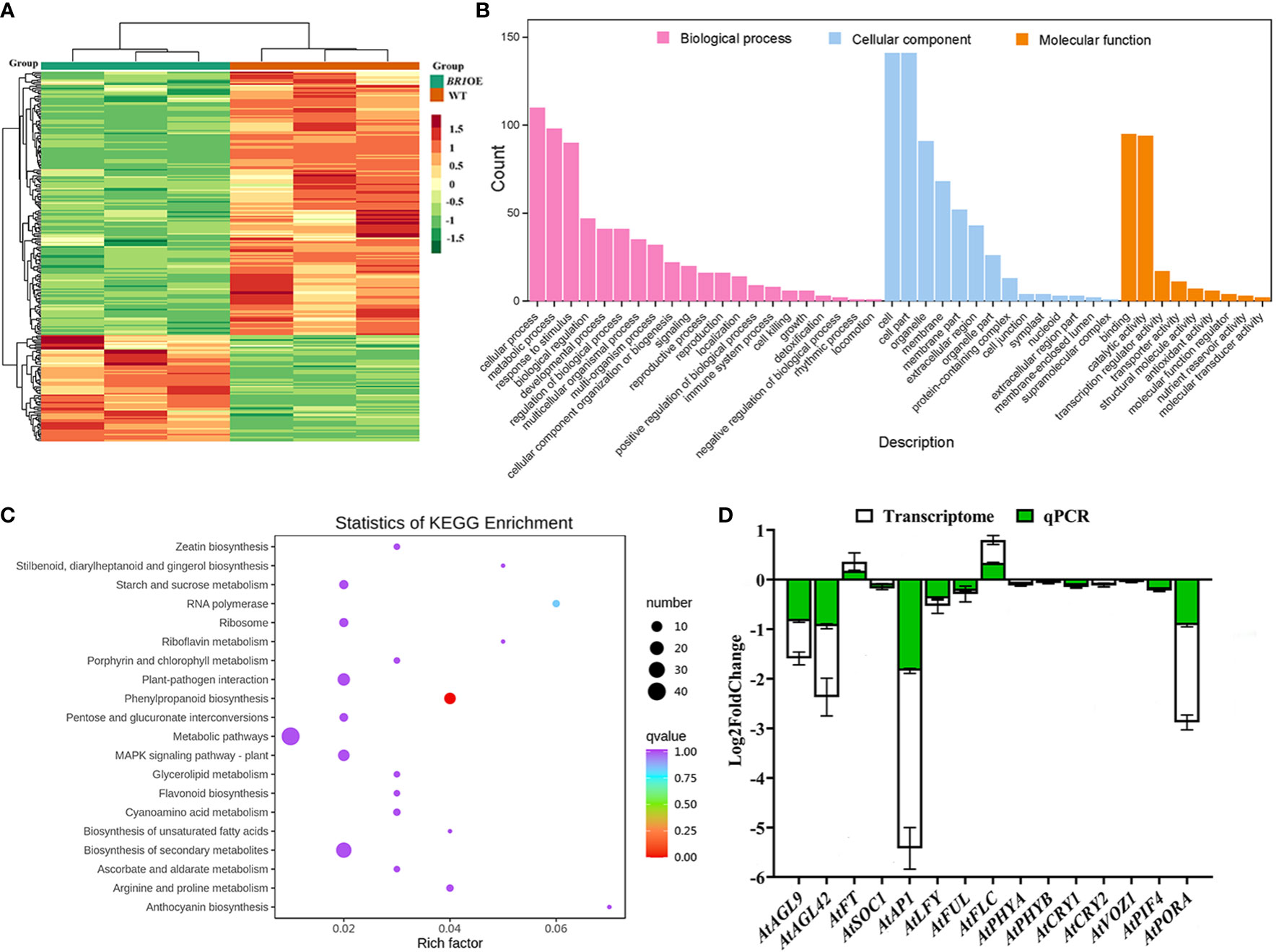
Figure 5 Identification of genes differentially expressed between WT and BR1OE plants. (A) DEG heatmap analysis. (B) DEG Gene Ontology (GO) classification was performed, with individual bars denoting the numbers of DEGs mapped to particular GO categories (pink: biological process; blue: cellular component; orange: molecular function). (C) The top 20 most enriched KEGG pathways associated with identified DEGs. Pathways are provided with the corresponding enrichment factors. (D) qPCR validation of gene expression profiles.
Next, DEGs associated with the floral transition pathway were compared between WT and BR1OE plants, revealing three MADS-box TFs among these DEGs including AGAMOUS-LIKE 9 (AtAGL9, AT1G24260), AtAP1, and AGAMOUS-LIKE 42 (AtAGL42, AT5G62165) (Figure 5D). These TFs were significantly downregulated in BR1OE plants relative to WT controls, in line with the delayed flowering time phenotype (Figure 4A). While not significantly downregulated, other floral integrator genes exhibited downward expression trends in BR1OE plants, including AtSOC1, AtLFY, and FRUITFULL (AtFUL, AT5G60910), whereas AtFT was expressed at low levels in both WT and mutant plants. The expression level of floral repressor AtFLC gene showed upward trends in BR1OE plants as expected. Interestingly, the downward expression trends of six key genes that involved in photoperiodic flowering pathway were observed in BR1OE plants, including PHYTOCHROME A (AtPHYA, AT1G09570), PHYTOCHROME B (AtPHYB, AT2G18790), CRYPTOCHROME 1 (AtCRY1, AT4G08920), CRYPTOCHROME 2 (AtCRY2, AT1G04400), VASCULAR PLANT ONE ZINC FINGER PROTEIN 1 (AtVOZ1, AT1G28520), and PHYTOCHROME INTERACTING FACTOR 4 (AtPIF4, AT2G43010). Moreover, the key chlorophyll biosynthesis-related gene Protochlorophyllide oxidoreductase A (AtPORA, AT5G54190) was also significantly downregulated in BR1OE mutant plants relative to WT controls, explaining the observed reduction in chlorophyll content (Figures 4G-I). These results indicated that BR1 gene may regulate flowering primarily through vernalization and photoperiodic pathway, which further supported a model in which BR1 serves as an important regulator of Arabidopsis floral transition.
A vector encoding the 35S::BR1::GFP fusion protein under the control of the CaMV35S promoter and a control empty 35S::GFP vector were prepared and used to conduct subcellular localization analyses. These assays revealed that BR1-GFP primarily localized to the nucleus, whereas control GFP signal was visible throughout the nuclear and cytosolic compartments (Figure 6). BR1-GRP and nuclear marker co-expression revealed complete overlap between these two signals, confirming that BR1 is a protein that localizes to the nucleus.
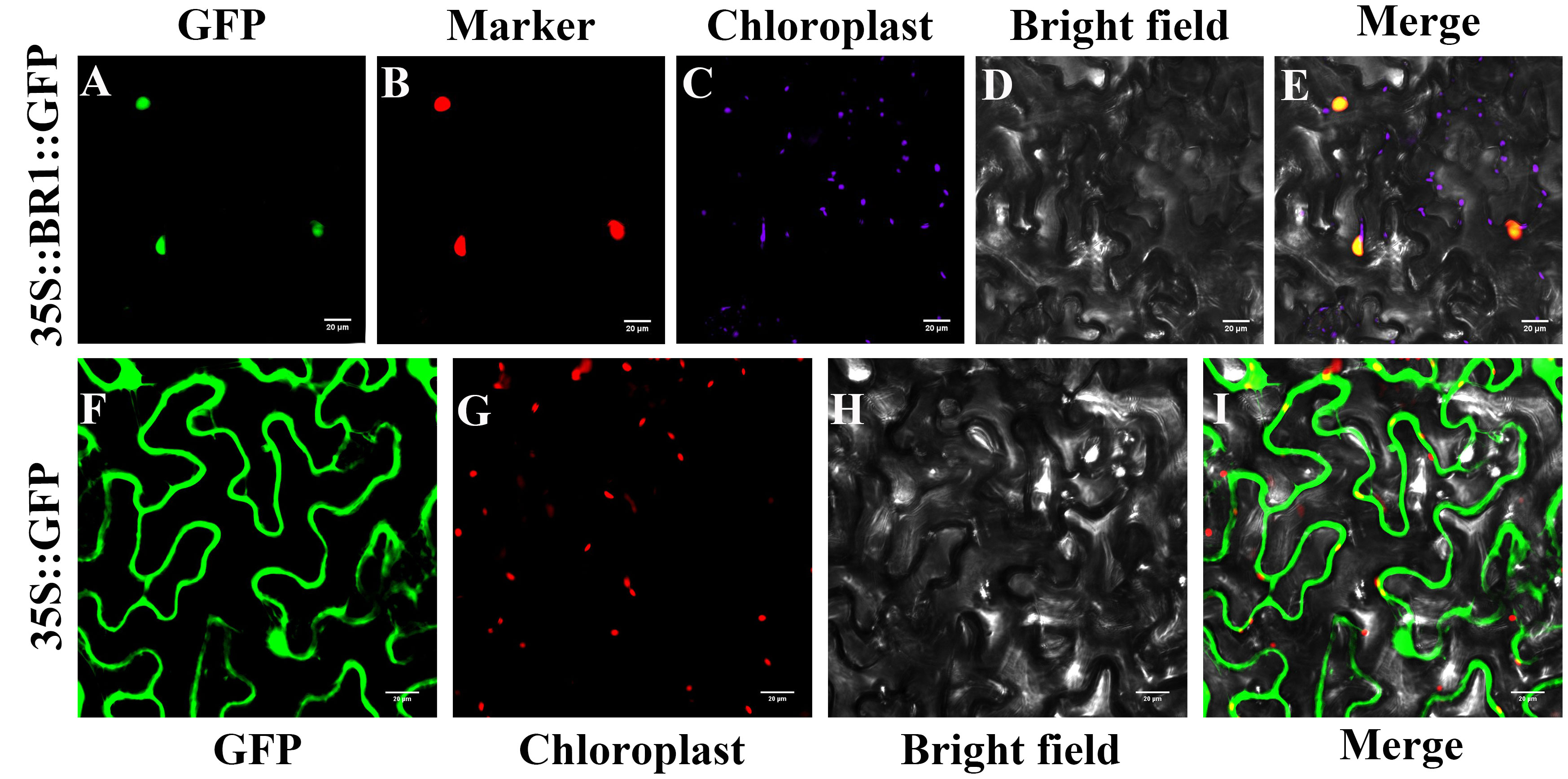
Figure 6 BR1 subcellular localization analyses. (A, F) GFP fluorescence. (B) Nuclear marker fluorescence. (C, G) chloroplast signals. (D, H) Brightfield images. (E, I) Merged images. A Leica confocal microscope was used to collect images at 48 h following agro-infiltration. Control GFP localization was evident throughout these cells. Scale bar: 20 μm.
Genotypic analyses of these OGs across 524 accessions revealed four SNPs in the BR1 gene, consistent with the high levels of BR1 conservation in Chinese cabbage relative to other accessions. This provided preliminary support for the role of this and other OGs as regulators of flowering habits, offering insight into the distinct bolting mechanisms that may have evolved in B. rapa that are absent in Arabidopsis (Jiang et al., 2022). Potential OG and lineage-specific gene family contributions to species-specific phenotypes have previously been characterized in the context of multicellular development in dictyostelids (Luna and Chain, 2021). OGs are thus associated with species-specific traits, in line with the present results. Despite the close relationships between Chinese cabbage and A. thaliana, these plants have evolved certain distinct characteristics over the course of evolution including distinct leaf shape, leaf size, and flowering timing. During the heading stage of growth, Chinese cabbage plants exhibit a leafy head consisting of leaves with extremely inwardly curved blades on their shoot apex, with these leafy heads being composed of many heading leaves that generally curve following the rosette stage (Ren et al., 2018). BR1 is mainly expressed in leaves, especially in the middle and inner leaves during the vegetative stage, suggesting that BR1 may play a role in Chinese cabbage leafy head formation. OGs also exhibit a high degree of tissue-specific expression in different tissues and stages of development (Donoghue et al., 2011; Jiang et al., 2018), which may reflect the putative functions of BR1 in the developmental regulation of Chinese cabbage. Well correspondence with delayed flowering phenotype in ‘BR1OE’, the expression of BR1 was increased in BR type, whereas decreased in BN type, which indicating that BR1 may play a role in bolting resistance of Chinese cabbage. The expression levels of several mark genes were detected in representative bolting resistance inbred line, floral inhibition factor BrFLCs displayed upregulated expression, and other flowering integrators showed downregulated expression. Studies indicated that FLC homologues in Brassica species act similarly to AtFLC that can delay flowering (Yuan et al., 2009; Xi et al., 2018). The expression of integration factors was similar to previous study (Dong et al., 2016). These results further verified the bolting resistance phenotype of ‘BR-98’, and reflected the potential role of BR1 in the regulation of bolting resistance.
Recent years, bolting resistance of Chinese cabbage had attracted more and more attention from scientists. In this study, the OG BR1 was identified as a novel regulator of plant morphological characteristics. Arabidopsis BR1OE mutants exhibited notably delayed flowering time together with significant decreases in stem height, rosette radius, and chlorophyll content. The overexpression of GRAINS NUMBER 2 (GN2) gene unique to the ‘Yuanjiang’ common wild rice genome, was similarly found to promote a later heading date, decreased plant height, and a reduction in grain number relative to WT controls (Chen et al., 2017). Brassica-Specific Genes 1 (BSGs1) has similarly been shown to be specifically expressed in Chinese cabbage during the heading stage, suggesting that it may be related to leafy head formation (Jiang et al., 2018). Flowering time is an agronomic trait that is critically important to Chinese cabbage production, and premature bolting can significantly reduce harvest yields and quality (Yuan et al., 2009). Chinese cabbage bolting and flowering generally necessitate vernalization and photoperiodism (Elers and Wiebe, 1984), and BR1-related flowering time delays may be linked to enhanced bolting resistance, highlighting a promising new pathway that can help prevent premature flowering for these economically important Chinese cabbage plants.
An appropriately timed floral transition is vital to ensuring (Srikanth and Schmid, 2011). By analyzing the transcriptome of the Arabidopsis BR1OE overexpression plants, we identified several flowering-related genes responsive to BR1 overexpression. The transcriptomic sequencing and qPCR analyses performed herein confirmed that BR1 overexpression in Arabidopsis significantly altered the expression of AtAGL9, AtAGL42, and AtAP1, in addition to promoting the downregulation of the floral transition identity genes AtSOC1, AtLFY, and AtFUL. Decreases in the expression of these integrator genes were consistent with observed flowering delays in mutant BR1OE plants. AtSOC1 can interact with FRUITFULL (AtFUL) to promote the activation of AtLFY, triggering the expression of the floral meristem identity gene AtAP1 promoting floral commitment such that flowers develop on the SAM (Jaudal et al., 2018). SPL TFs can promote flowering time and floral fate, with MADS-box TFs including AtAP1, AtFUL, and AtAGL9 serving as regulators of floral fate (Freytes et al., 2021). The SOC1-like gene, AtAGL42, is linked to the control of SAM and axillary meristem (AM) floral transition (Dorca-Fornell et al., 2011). AtAGL9 was shown to bind to and repress AtSOC1, while activating expression of a large number of floral homeotic genes (Srikanth and Schmid, 2011). The expression level of AtFLC gene displayed upward trends in BR1OE plants as expected. As a major inhibitor of flowering, FLC suppressed the expression of transcription factors (TFs) needed for the SAM floral transition (Cho et al., 2017). Low expression level in AtFT was also evident in BR1OE plants relative to WT controls, and this phenomenon still needs further clarification. Eight core regulatory genes in photoperiodic flowering pathway showed downward expression trends, including AtPHYA, AtPHYB, AtCRY1, AtCRY2, AtVOZ1, and AtPIF4. These genes regulated flowering, and both of these mutants flower late under different conditions (Simpson and Dean, 2002; Thines et al., 2014). How the overexpression of BR1 gene affects the expression of these genes still needs further study.
Notably, BR1 overexpression resulted in significant AtPORA downregulation in Arabidopsis, consistent with the observed reductions in chlorophyll levels in BR1OE transgenic plants (Figure 4G, H, I). POR is a key enzyme for the light-induced greening of etiolated angiosperm plants, which is one of three known light-dependent enzymes, catalyses reduction of the photosensitizer and substrate protochlorophyllide to form the pigment chlorophyllide (Reinbothe et al., 2010; Buhr et al., 2017; Zhang et al., 2019). The Arabidopsis porA-1 mutant plants have been shown to exhibit severe photoautotrophic growth defects and decreases in total chlorophyll content (Paddock et al., 2012), confirming the present results. Through comparisons of transcriptomic profiles, most enriched KEGG pathways were metabolic pathways related to the biosynthesis of secondary metabolites. GO enrichment analyses indicated that all DEGs were related to metabolic, developmental, reproductive, and growth-related processes. Based on the above analysis, we speculated that BR1 may regulate flowering time delay through vernalization and photoperiodic pathway. However, the mechanisms through which the BR1 pathway contributes to delayed flowering warrant further research. As BR1 localizes to the nuclear compartment, the identification of upstream regulatory TFs and downstream target proteins can be readily performed through respective yeast one-hybrid and yeast two-hybrid screens. Further knockout and overexpression analyses in Chinese cabbage will thus be essential to fully characterize how BR1 regulates bolting resistance.
The highly conserved nature of BR1 in Chinese cabbage accessions supports its potential role as a regulator of bolting resistance. The expression of BR1 was increased in bolting resistant Chinese cabbage and decreased in bolting non-resistant type. The overexpression of BR1 in Arabidopsis resulted in delayed flowering time and other obvious phenotypes. Both transcriptomic sequencing and qPCR analyses further revealed that nuclear-located orphan gene BR1 may function as a new mediator of flowering time through vernalization and photoperiodic pathway. These results offer novel insight into the links between OGs and flowering time in B. rapa, providing a theoretical basis for future studies aimed at examining the mechanisms governing bolting resistance in this economically important species.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: PRJNA922732 (SRA).
MJ and HL conceived and designed the experiments. MJ, YZ, and HL performed the experiments. MJ, YZ, XY, XL, and HL analyzed the data. MJ and HL drafted the manuscript. All authors contributed to the article and approved the submitted version.
This study was funded by the Doctoral Initiating Fund Project of Jilin Agricultural Science and Technology College (Grant No. 20215019).
We thank Prof. Zhongyun Piao, Dr. Wenxing Pang and Dr. Zongxiang Zhan (College of Horticulture, Shenyang Agricultural University) for their kindly help on this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1135684/full#supplementary-material
Bao, S., Hua, C., Shen, L., Yu, H. (2020). New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 62, 118–131. doi: 10.1111/jipb.12892
Blümel, M., Dally, N., Jung, C. (2015). Flowering time regulation in crops-what did we learn from Arabidopsis? Curr. Opin. Biotechnol. 32, 121–129. doi: 10.1016/j.copbio.2014.11.023
Brennan, C. J., Zhou, B., Benbow, H. R., Ajaz, S., Karki, S. J., Hehir, J. G., et al. (2020). Taxonomically restricted wheat genes interact with small secreted fungal proteins and enhance resistance to septoria tritici blotch disease. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00433
Buhr, F., Lahroussi, A., Springer, A., Rustgi, S., von Wettstein, D., Reinbothe, C., et al. (2017). NADPH:protochlorophyllide oxidoreductase b (PORB) action in Arabidopsis thaliana revisited through transgenic expression of engineered barley PORB mutant proteins. Plant Mol. Biol. 94, 45–59. doi: 10.1007/s11103-017-0592-x
Cai, X., Chang, L., Zhang, T., Chen, H., Zhang, L., Lin, R., et al. (2021). Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa. Genome Biol. 22, 166. doi: 10.1186/s13059-021-02383-2
Cai, J. J., Woo, P. C., Lau, S. K., Smith, D. K., Yuen, K. Y. (2006). Accelerated evolutionary rate may be responsible for the emergence of lineage-specific genes in ascomycota. J. Mol. Evol. 63, 1–11. doi: 10.1007/s00239-004-0372-5
Chen, H., Tang, Y., Liu, J., Tan, L., Jiang, J., Wang, M., et al. (2017). Emergence of a novel chimeric gene underlying grain number in rice. Genetics 205, 993–1002. doi: 10.1534/genetics.116.188201
Cheng, F., Sun, R., Hou, X., Zheng, H., Zhang, F., Zhang, Y., et al. (2016). Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 48, 1218–1224. doi: 10.1038/ng.3634
Cheng, J. Z., Zhou, Y. P., Lv, T. X., Xie, C. P., Tian, C. E. (2017). Research progress on the autonomous flowering time pathway in Arabidopsis. Physiol. Mol. Biol. Plants 23, 477–485. doi: 10.1007/s12298-017-0458-3
Cho, L. H., Yoon, J., An, G. (2017). The control of flowering time by environmental factors. Plant J. 90, 708–719. doi: 10.1111/tpj.13461
Creux, N., Harmer, S. (2019). Circadian rhythms in plants. Cold Spring Harb. Perspect. Biol. 11, a034611. doi: 10.1101/cshperspect.a034611
Cui, X., Lv, Y., Chen, M., Nikoloski, Z., Twell, D., Zhang, D. (2015). Young genes out of the male: an insight from evolutionary age analysis of the pollen transcriptome. Mol. Plant 8, 935–945. doi: 10.1016/j.molp.2014.12.008
Dong, X., Yi, H., Han, C. T., Nou, I. S., Swaraz, A. M., Hur, Y. (2016). Genome-wide analysis of genes associated with bolting in heading type Chinese cabbage. Euphytica 212, 65–82. doi: 10.1007/s10681-016-1759-2
Donoghue, M. T., Keshavaiah, C., Swamidatta, S. H., Spillane, C. (2011). Evolutionary origins of brassicaceae specific genes in Arabidopsis thaliana. BMC Evol. Biol. 11, 47. doi: 10.1186/1471-2148-11-47
Dorca-Fornell, C., Gregis, V., Grandi, V., Coupland, G., Colombo, L., Kater, M. M. (2011). The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. Plant J. 67, 1006–1017. doi: 10.1111/j.1365-313X.2011.04653.x
Dossa, K., Zhou, R., Li, D., Liu, A., Qin, L., Mmadi, M. A., et al. (2021). A novel motif in the 5’-UTR of an orphan gene ‘Big root biomass’ modulates root biomass in sesame. Plant Biotechnol. J. 19, 1065–1079. doi: 10.1111/pbi.13531
Elers, B., Wiebe, H. J. (1984). Flower formation of Chinese cabbage. i. response to vernalization and photoperiods. Sci. Hortic. 22, 219–231. doi: 10.1016/0304-4238(84)90055-4
Fang, H., Shen, S., Wang, D., Zhang, F., Zhang, C., Wang, Z., et al. (2021). A monocot-specific hydroxycinnamoylputrescine gene cluster contributes to immunity and cell death in rice. Sci. Bull. 66, 2381–2393. doi: 10.1016/j.scib.2021.06.014
Freytes, S. N., Canelo, M., Cerdán, P. D. (2021). Regulation of flowering time: When and where? Curr. Opin. Plant Biol. 63, 102049. doi: 10.1016/j.pbi.2021.102049
Fu, W., Huang, S., Gao, Y., Zhang, M., Qu, G., Wang, N., et al. (2020). Role of BrSDG8 on bolting in Chinese cabbage (Brassica rapa). Theor. Appl. Genet. 133, 2937–2948. doi: 10.1007/s00122-020-03647-4
Izawa, T. (2021). What is going on with the hormonal control of flowering in plants? Plant J. 105, 431–445. doi: 10.1111/tpj.15036
Jaudal, M., Zhang, L., Che, C., Li, G., Tang, Y., Wen, J., et al. (2018). A SOC1-like gene MtSOC1a promotes flowering and primary stem elongation in medicago. J. Exp. Bot. 69, 4867–4880. doi: 10.1093/jxb/ery284
Jiang, M., Dong, X., Lang, H., Pang, W., Zhan, Z., Li, X., et al. (2018). Mining of Brassica-specific genes (BSGs) and their induction in different developmental stages and under Plasmodiophora brassicae stress in Brassica rapa. Int. J. Mol. Sci. 19, 2064. doi: 10.3390/ijms19072064
Jiang, C., Hei, R., Yang, Y., Zhang, S., Wang, Q., Wang, W., et al. (2020a). An orphan protein of Fusarium graminearum modulates host immunity by mediating proteasomal degradation of TaSnRK1α. Nat. Commun. 11, 4382. doi: 10.1038/s41467-020-18240-y
Jiang, M., Li, X., Dong, X., Zu, Y., Zhan, Z., Piao, Z., et al. (2022). Research advances and prospects of orphan genes in plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.947129
Jiang, M., Zhan, Z., Li, H., Dong, X., Cheng, F., Piao, Z. (2020b). Brassica rapa orphan genes largely affect soluble sugar metabolism. Hortic. Res. 7, 181. doi: 10.1038/s41438-020-00403-z
Johansson, M., Staiger, D. (2015). Time to flower: Interplay between photoperiod and the circadian clock. J. Exp. Bot. 66, 719–730. doi: 10.1093/jxb/eru441
Jung, C., Müller, A. E. (2009). Flowering time control and applications in plant breeding. Trends Plant Sci. 14, 563–573. doi: 10.1016/j.tplants.2009.07.005
Kinoshita, A., Richter, R. (2020). Genetic and molecular basis of floral induction in Arabidopsis thaliana. J. Exp. Bot. 71, 2490–2504. doi: 10.1093/jxb/eraa057
Li, L., Foster, C. M., Gan, Q., Nettleton, D., James, M. G., Myers, A. M., et al. (2009). Identification of the novel protein QQS as a component of the starch metabolic network in Arabidopsis leaves. Plant J. 58, 485–498. doi: 10.1111/j.1365-313X.2009.03793.x
Li, L., Li, X., Liu, Y., Liu, H. (2015a). Flowering responses to light and temperature. Sci. China Life Sci. 59, 403–408. doi: 10.1007/s11427-015-4910-8
Li, Q., Peng, A., Yang, J., Zheng, S., Li, Z., Mu, Y., et al. (2022). A 215-bp indel at intron I of BoFLC2 affects flowering time in Brassica oleracea var. capitata during vernalization. Theor. Appl. Genet. 135, 2785–2797. doi: 10.1007/s00122-022-04149-1
Li, G., Wu, X., Hu, Y., Muñoz-Amatriaín, M., Luo, J., Zhou, W., et al. (2019). Orphan genes are involved in drought adaptations and ecoclimatic-oriented selections in domesticated cowpea. J. Exp. Bot. 70, 3101–3110. doi: 10.1093/jxb/erz145
Li, L., Wurtele, E. S. (2015). The QQS orphan gene of Arabidopsis modulates carbon and nitrogen allocation in soybean. Plant Biotechnol. J. 13, 177–187. doi: 10.1111/pbi.12238
Li, L., Zheng, W., Zhu, Y., Ye, H., Tang, B., Arendsee, Z. W., et al. (2015b). QQS orphan gene regulates carbon and nitrogen partitioning across species via NF-YC interactions. Proc. Natl. Acad. Sci. U.S.A. 112, 14734–14739. doi: 10.1073/pnas.1514670112
Lin, S. I., Wang, J. G., Poon, S. Y., Su, C. L., Wang, S. S., Chiou, T. J. (2005). Differential regulation of FLOWERING LOCUS c expression by vernalization in cabbage and Arabidopsis. Plant Physiol. 137, 1037–1048. doi: 10.1104/pp.104.058974
Luna, S. K., Chain, F. J. J. (2021). Lineage-specific genes and family expansions in dictyostelid genomes display expression bias and evolutionary diversification during development. Genes (Basel) 12, 1628. doi: 10.3390/genes12101628
Ma, D., Ding, Q., Guo, Z., Zhao, Z., Wei, L., Li, Y., et al. (2021). Identification, characterization and expression analysis of lineage-specific genes within mangrove species Aegiceras corniculatum. Mol. Genet. Genomics 296, 1235–1247. doi: 10.1007/s00438-021-01810-0
Ma, D., Lai, Z., Ding, Q., Zhang, K., Chang, K., Li, S., et al. (2022). Identification, characterization and function of orphan genes among the current cucurbitaceae genomes. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.872137
Ma, S., Yuan, Y., Tao, Y., Jia, H., Ma, Z. (2020). Identification, characterization and expression analysis of lineage-specific genes within Triticeae. Genomics 112, 1343–1350. doi: 10.1016/j.ygeno.2019.08.003
Mero, C. E., Honma, S. (1984). Inheritance of bolt resistance in an interspecific cross of Brassica species: II. chikale (B. campestris l. ssp. pekinensis × B. napus l.) × Chinese cabbage. J. Hered. 75, 6485–6487. doi: 10.1093/oxfordjournals.jhered.a109991
Moon, H., Jeong, A. R., Kwon, O. K., Park, C. J. (2022). Oryza-specific orphan protein triggers enhanced resistance to Xanthomonas oryzae pv. oryzae in rice. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.859375
Ni, F., Qi, J., Hao, Q., Lyu, B., Luo, M. C., Wang, Y., et al. (2017). Wheat Ms2 encodes for an orphan protein that confers male sterility in grass species. Nat. Commun. 8, 15121. doi: 10.1038/ncomms15121
O’Conner, S., Neudorf, A., Zheng, W., Qi, M., Zhao, X., Du, C., et al. (2018). “From arabidopsis to crops: the arabidopsis QQS orphan gene modulates nitrogen allocation across species,” in Engineering nitrogen utilization in crop plants. Eds. Shrawat, A., Zayed, A., Lightfoot, D. A. (Switzerland, AG: Springer Cham), 95–117. doi: 10.1007/978-3-319-92958-3_6
Paddock, T., Lima, D., Mason, M. E., Apel, K., Armstrong, G. A. (2012). Arabidopsis light-dependent protochlorophyllide oxidoreductase a (PORA) is essential for normal plant growth and development. Plant Mol. Biol. 78, 447–460. doi: 10.1007/s11103-012-9873-6
Qi, M., Zheng, W., Zhao, X., Hohenstein, J. D., Kandel, Y., O’Conner, S., et al. (2019). QQS orphan gene and its interactor NF-YC4 reduce susceptibility to pathogens and pests. Plant Biotechnol. J. 17, 252–263. doi: 10.1111/pbi.12961
Razi, H., Howell, E. C., Newbury, H. J., Kearsey, M. J. (2008). Does sequence polymorphism of FLC paralogues underlie flowering time QTL in Brassica oleracea? Theor. Appl. Genet. 116, 179–192. doi: 10.1007/s00122-007-0657-3
Reinbothe, C., El Bakkouri, M., Buhr, F., Muraki, N., Nomata, J., Kurisu, G., et al. (2010). Chlorophyll biosynthesis: Spotlight on protochlorophyllide reduction. Trends Plant Sci. 15, 614–624. doi: 10.1016/j.tplants.2010.07.002
Ren, W., Wang, H., Bai, J., Wu, F., He, Y. (2018). Association of microRNAs with types of leaf curvature in Brassica rapa. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00073
Shen, S., Peng, M., Fang, H., Wang, Z., Zhou, S., Jing, X., et al. (2021). An Oryza-specific hydroxycinnamoyl tyramine gene cluster contributes to enhanced disease resistance. Sci. Bull. 66, 2369–2380. doi: 10.1016/j.scib.2021.03.015
Simpson, G. G., Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289. doi: 10.1126/science.296.5566.285
Srikanth, A., Schmid, M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68, 2013–2037. doi: 10.1007/s00018-011-0673-y
Su, T., Wang, W., Li, P., Zhang, B., Li, P., Xin, X., et al. (2018). A genomic variation map provides insights into the genetic basis of spring Chinese cabbage (Brassica rapa ssp. pekinensis) selection. Mol. Plant 11, 1360–1376. doi: 10.1016/j.molp.2018.08.006
Tanvir, R., Ping, W., Sun, J., Cain, M., Li, X., Li, L. (2022a). AtQQS orphan gene and NtNF-YC4 boost protein accumulation and pest resistance in tobacco (Nicotiana tabacum). Plant Sci. 317, 111198. doi: 10.1016/j.plantsci.2022.111198
Tanvir, R., Wang, L., Zhang, A., Li, L. (2022b). Orphan genes in crop improvement: enhancing potato tuber protein without impacting yield. Plants (Basel) 11, 3076. doi: 10.3390/plants11223076
Thines, B. C., Youn, Y., Duarte, M. I., Harmon, F. G. (2014). The time of day effects of warm temperature on flowering time involve PIF4 and PIF5. J. Exp. Bot. 65, 1141–1151. doi: 10.1093/jxb/ert487
Wang, J. W. (2014). Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 65, 4723–4730. doi: 10.1093/jxb/eru246
Wang, C., Chen, S., Feng, A., Su, J., Wang, W., Feng, J., et al. (2021). Xa7, a small orphan gene harboring promoter trap for AvrXa7, leads to the durable resistance to Xanthomonas oryzae pv. oryzae. Rice 14, 48. doi: 10.1186/s12284-021-00490-z
Wang, Z., Li, J., Chen, S., Heng, Y., Chen, Z., Yang, J., et al. (2017). Poaceae-specific MS1 encodes a phospholipid-binding protein for male fertility in bread wheat. Proc. Natl. Acad. Sci. U.S.A. 114, 12614–12619. doi: 10.1073/pnas.1715570114
Xi, X., Wei, K., Gao, B., Liu, J., Liang, J., Cheng, F., et al. (2018). BrFLC5: a weak regulator of flowering time in Brassica rapa. Theor. Appl. Genet. 131, 2107–2116. doi: 10.1007/s00122-018-3139-x
Xin, X., Su, T., Li, P., Wang, W., Zhao, X., Yu, Y., et al. (2020). A histone H4 gene prevents drought-induced bolting in Chinese cabbage by attenuating the expression of flowering genes. J. Exp. Bot. 72, 623–635. doi: 10.1093/jxb/eraa452
Yadeta, K. A., Valkenburg, D. J., Hanemian, M., Marco, Y., Thomma, B. P. (2014). The brassicaceae-specific EWR1 gene provides resistance to vascular wilt pathogens. PloS One 9, e88230. doi: 10.1371/journal.pone.0088230
Yuan, Y. X., Wu, J., Sun, R. F., Zhang, X. W., Xu, D. H., Bonnema, G., et al. (2009). A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J. Exp. Bot. 60, 1299–1308. doi: 10.1093/jxb/erp010
Zhan, Z., Liu, H., Yang, Y., Liu, S., Li, X., Piao, Z. (2022). Identification and characterization of putative effectors from Plasmodiophora brassicae that suppress or induce cell death in Nicotiana benthamiana. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.881992
Zhang, X., Feng, C., Wang, M., Li, T., Liu, X., Jiang, J. (2021). Plasma membrane-localized SlSWEET7a and SlSWEET14 regulate sugar transport and storage in tomato fruits. Hortic. Res. 8, 186. doi: 10.1038/s41438-021-00624-w
Zhang, S., Heyes, D. J., Feng, L., Sun, W., Johannissen, L. O., Liu, H., et al. (2019). Structural basis for enzymatic photocatalysis in chlorophyll biosynthesis. Nature 574, 722–725. doi: 10.1038/s41586-019-1685-2
Zhang, X., Ma, W., Liu, M., Li, X., Li, J., Lu, Y., et al. (2022). OCTOPUS regulates BIN2 to control leaf curvature in Chinese cabbage. Proc. Natl. Acad. Sci. U.S.A. 119, e2208978119. doi: 10.1073/pnas.2208978119
Keywords: Brassica rapa, orphan gene, BR1, bolting resistance, Arabidopsis
Citation: Jiang M, Zhang Y, Yang X, Li X and Lang H (2023) Brassica rapa orphan gene BR1 delays flowering time in Arabidopsis. Front. Plant Sci. 14:1135684. doi: 10.3389/fpls.2023.1135684
Received: 01 January 2023; Accepted: 15 February 2023;
Published: 24 February 2023.
Edited by:
Yoonkang Hur, Chungnam National University, Republic of KoreaReviewed by:
Zhansheng Li, Insititute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (CAAS), ChinaCopyright © 2023 Jiang, Zhang, Yang, Li and Lang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaonan Li, Z3JhY2VzbGVleG5AMTYzLmNvbQ==; Hong Lang, bGFuZ2hvbmdAamxua3UuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.