
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 February 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1133029
This article is part of the Research Topic Abiotic and Biotic Stress in Horticultural Crops: Insight into Recent Advances in the Underlying Tolerance Mechanism View all 46 articles
 Sandeep Jaiswal1,2
Sandeep Jaiswal1,2 Krishnayan Paul1,2
Krishnayan Paul1,2 K. Venkat Raman1
K. Venkat Raman1 Saurabh Tyagi1
Saurabh Tyagi1 Manjesh Saakre1,2
Manjesh Saakre1,2 Jyotsana Tilgam1,2
Jyotsana Tilgam1,2 Sougata Bhattacharjee1,2
Sougata Bhattacharjee1,2 Joshitha Vijayan1
Joshitha Vijayan1 Kalyan Kumar Mondal3
Kalyan Kumar Mondal3 Rohini Sreevathsa1
Rohini Sreevathsa1 Debasis Pattanayak1*
Debasis Pattanayak1*Cold-induced sweetening (CIS) is an unwanted physiological phenomenon in which reducing sugars (RS) get accumulated in potato (Solanum tuberosum) upon cold storage. High RS content makes potato commercially unsuitable for processing due to the unacceptable brown color in processed products like chips, fries, etc., and the production of a potential carcinogen, acrylamide. UDP-glucose pyrophosphorylase (UGPase) catalyzes the synthesis of UDP-glucose towards the synthesis of sucrose and is also involved in the regulation of CIS in potato. The objective of the present work was RNAi-mediated downregulation of the StUGPase expression level in potato for the development of CIS tolerant potato. Hairpin RNA (hpRNA) gene construct was developed by placing UGPase cDNA fragment in sense and antisense orientation intervened by GBSS intron. Internodal stem explants (cv. Kufri Chipsona-4) were transformed with hpRNA gene construct, and 22 transgenic lines were obtained by PCR screening of putative transformants. Four transgenic lines showed the highest level of RS content reduction following 30 days of cold storage, with reductions in sucrose and RS (glucose & fructose) levels of up to 46% and 57.5%, respectively. Cold stored transgenic potato of these four lines produced acceptable chip colour upon processing. The selected transgenic lines carried two to five copies of the transgene. Northern hybridization revealed an accumulation of siRNA with a concomitant decrease in the StUGPase transcript level in these selected transgenic lines. The present work demonstrates the efficacy of StUGPase silencing in controlling CIS in potato, and the strategy can be employed for the development of CIS tolerant potato varieties.
Cold storage of potato (Solanum tuberosum L.) is an integral part of post-harvest handling of this semi-perishable crop. However, potato becomes sweet in taste due to the accumulation of reducing sugars (RS), a physiological process known as cold-induced sweetening (CIS) (Sowokinos, 1990). CIS occurs due to the cold-induced imbalance between the rate of breakdown of starch and its metabolism, resulting in higher glucose and fructose content in tubers (Sowokinos, 2001a; Blenkinsop et al., 2010). It is an undesirable physiological process as sweetened potato is not preferred for both table and processing purposes. Potato with a high amount of RS content develops brown to a black colour and bitter taste when fried at a higher temperature. Reducing sugars react with the amino group of free amino acids during frying or processing at high temperatures and develop dark pigmentation by a non-enzymatic reaction called Maillard reaction (Murata, 2021). Dark-coloured potato chips, fries, etc. taste bitter and are unsafe due to the presence of a high level of acrylamide, a byproduct of the Maillard reaction (Keijbets, 2008). Acrylamide has been classified as a ‘Probable carcinogen to humans (Group 2A) (Nursten, 2005; Borda and Alexe, 2011; Halford et al., 2011).
Due to a rapid expansion of the potato processing industry in India, the demand for quality raw materials throughout the year is also increasing and it is anticipated to rise from 6 million tonnes in 2025 to 25 million tonnes in 2050 (Singh, 2012). At present nearly 8.9% of the total potato produce is being processed, which is projected to be 10.76% by the year 2025 (CPRI Vision, 2050). However, the lack of supply of processing-grade potatoes throughout the year is still a major constraint for the potato processing industry (Das et al., 2021). Potato is kept in cold storage after harvest for yearlong supply and to avoid problems like price crashes and glut situations (Pradel et al., 2019). However, so far bred Indian potato cultivars including indigenous processing grade potato varieties by ICAR-CPRI, Shimla (i.e. ‘Kufri Chipsona1’, ‘Kufri Chipsona2’, ‘Kufri Chipsona3’, and ‘Kufri Chipsona4’) are not good cold chipper. Hence potato becomes unfit for processing after cold storage due to susceptibility to CIS and accumulation of a high amount of RS during cold storage (Pandey et al., 2009; Marwaha et al., 2010; Kumar, 2011; Raigond et al., 2018; Gupta et al., 2021). Therefore, the development of new cultivars with low RS build-up during cold storage or the improvement of existing genotypes with traits like cold/CIS tolerance is very important for the sustenance of potato processing industry. However, no resistant cultivar has indeed been developed so far by conventional breeding that could be regarded as good “cold chippers (Xiong et al., 2002; Hamernik et al., 2009). The biggest impediments towards the development of CIS resistant potato cultivars through conventional breeding are the lack of suitable germplasm, the genetic complexity of potato, heterozygosity, and polysomic tetraploid inheritance of this crop (Ortiz and Watanabe, 2004; Bradshaw et al., 2006; Hirsch et al., 2013). Therefore, genetic engineering is the most practical alternative for the amelioration of CIS in potato. Numerous efforts have been made to manipulate the expression of genes encoding the key regulatory enzymes, transcriptional factors, and other regulatory molecules that are directly or indirectly involved in CIS with variable degrees of success in reducing RS accumulation during CIS (Schwimmer et al., 1961; Sowokinos, 2001a; Geigenberger et al., 2004; Liu et al., 2010; Brummell et al., 2011; Liu et al., 2013; McKenzie et al., 2013; Zhang et al., 2013; Shivalingamurthy et al., 2018; Shi et al., 2021, Shi et al., 2022). Vacuolar acid invertase (vaINV), catalyzing the penultimate step, has been identified as one of the major factors involved in CIS (Zrenner et al., 1996; Matsuura-Endo et al., 2004; McKenzie et al., 2005; Liu et al., 2021). A few studies reported considerable improvement of CIS by down regulation of the StvaINV expression (Bhaskar et al., 2010; Wiberley-Bradford et al., 2014; Zhu et al., 2014; Hameed et al., 2018). However, it was observed that a high amount of sucrose gets accumulated in cold-stored StvaINV silenced potato, which imparted an undesirable sweet taste to chips (Pattanayak et al., 2009). This necessitates that an alternative strategy be devised to circumvent the problem of sucrose accumulation.
UGPase catalyzes the first reversible and committed step in sucrose synthesis through the formation of high-energy glycosyl nucleotide, UDP-glucose (Rees and Morrell, 1990; Spychalla et al., 1994; Borovkov et al., 1996). Double knockout mutant for UGPase alleles (atugp1 and atugp2), confirmed the essential role of AtUGPase in carbohydrate metabolism in both the vegetative and reproductive phases in Arabidopsis thaliana (Park et al., 2010). During the initiation of CIS in potato tubers, the change in StUGPase expression and the concentration of UDP-Glc were found to be closely parallel to that of sucrose level, indicating the crucial role of UGPase in regulating carbon flux for sucrose biosynthesis in tubers during cold storage (Zrenner et al., 1993; Spychalla et al., 1994; Hill et al., 1996; Bagnaresi et al., 2008; Chen et al., 2008).To date, two UGPase alleles have been reported and designated as UgpA (A-II isozyme; UGP5) and UgpB (A-I isozyme; UGP3) in potato (Katsube et al., 1990; Spychalla et al., 1994; Sowokinos et al., 2018). Interestingly, the allelic ratios of UgpA and UgpB are reported to determine the degree of CIS resistance (Sowokinos, 2001b; Sowokinos et al., 2018). Despite a 96% reduction in UGPase activity through antisense inhibition in transgenic potato, no detrimental effect was observed in the growth and development of genetically engineered lines (Zrenner et al., 1993). While transgenic potato tubers expressing the StUGPase antisense construct were cold-stored, Spychalla et al. (1994) noticed a considerable shift in the sucrose concentration. Therefore, in the current study, we attempted RNAi-mediated downregulation of StUGPase to develop cold-chipping potato transgenic lines. Both sucrose and RS content reduced considerably in UGPase RNAi transgenic potato after cold storage at 4 °C for 30 days.
Virus-free in vitro grown plantlet of potato cultivar Kufri Chipsona-4 (KC4), obtained from the Division of Crop Improvement, ICAR-Central Potato Research Institute, Shimla, was used as plant material for this study. The plantlets were cultured, multiplied, and maintained in vitro under the growth condition of 16h light-8h dark photoperiod cycle and 24 °C temperature. Internodal stem segments were used as explants for Agrobacterium-mediated transformation.
The StUGPase cDNA sequence of 1758 nt was retrieved from the NCBI (Sequence ID: D00667.1). The sequence was divided into four regions/fragments, and a similarity search was performed through the BLAST search in NCBI to identify the region with the least homology with other non-target genes. Moreover, the web tool dsCheck (http://dsCheck.RNAi.jp/) was used to identify the target region with maximum siRNA generation potential with the least off-targeting effect. The ~500 nt long sequences corresponding to the 450-949 nt region of the StUGPase cDNA were selected for the development of the hpRNA construct.
PureLink™ RNA Mini Kit (Invitrogen™) was used to extract total RNA from KC4 leaves following the manufacturer’s protocol. cDNA was synthesized from 1 µg of total RNA using PrimeScript™ 1st strand cDNA Synthesis Kit (Takara Bio ink). DNA fragments of ~500 bp were amplified from KC4 cDNA using specially designed StUGPase sense and antisense specific primers having specific restriction sites incorporated in them (Supplementary Table 1) using a thermal cycler (Eppendorf, Germany). The amplified fragments were ligated, individually onto the pGEM T-Easy vector. Potato Granule Bound Starch Synthase (GBSS) intron of 105 nt corresponding to 3821-3925 nt of GBSS gene sequence (StGBSS; NCBI Accession ID: X58453) was amplified with specific sets of primers (Supplementary Table 2) containing suitable restriction enzyme sites and ligated onto the pGEM T-Easy vector. After sequence confirmation the UGPase sense (UGPase-S) fragment was taken out from the pGEM T- Easy vector by XbaI-SacI digestion and ligated onto XbaI-SacI digested pUC19 vector to get pUC19::UGPase-S. Similarly, 105 bp potato GBSS intron was taken out by XmaI-SacI digestion of pGEM T-Easy vector and cloned onto the pUC19::UGPase-S, linearized by XmaI-SacI digestion, to get pUC19::UGPase-S:GBSS-Int. UGPase antisense (UGPase-AS) fragment was finally subcloned onto BamHI-SacI sites of the pUC19::UGPase-S:GBSS-Int to get pUC19::UGPase-S:GBSSInt:UGPase-AS (designated as pU::hpU; hereby the hpRNA StUGPase construct is abbreviated as hpU). Each step of the assembly was confirmed by restriction digestion and sequencing. Finally, the hpU construct was subcloned onto pBI121 binary vector linearized by XbaI- SacI digestion to get the binary vector cassette pBI121::hpUGPase (hereby, designated as pB:hpU; Figure 1). The constructed recombinant binary vector cassette was confirmed by restriction analysis and sequencing. The binary vector was then transformed into A. tumefaciens strain EHA105 by freeze-thaw method (Höfgen and Willmitzer, 1988). Colonies harboring pB:hpU were confirmed by PCR and then used for potato transformation.
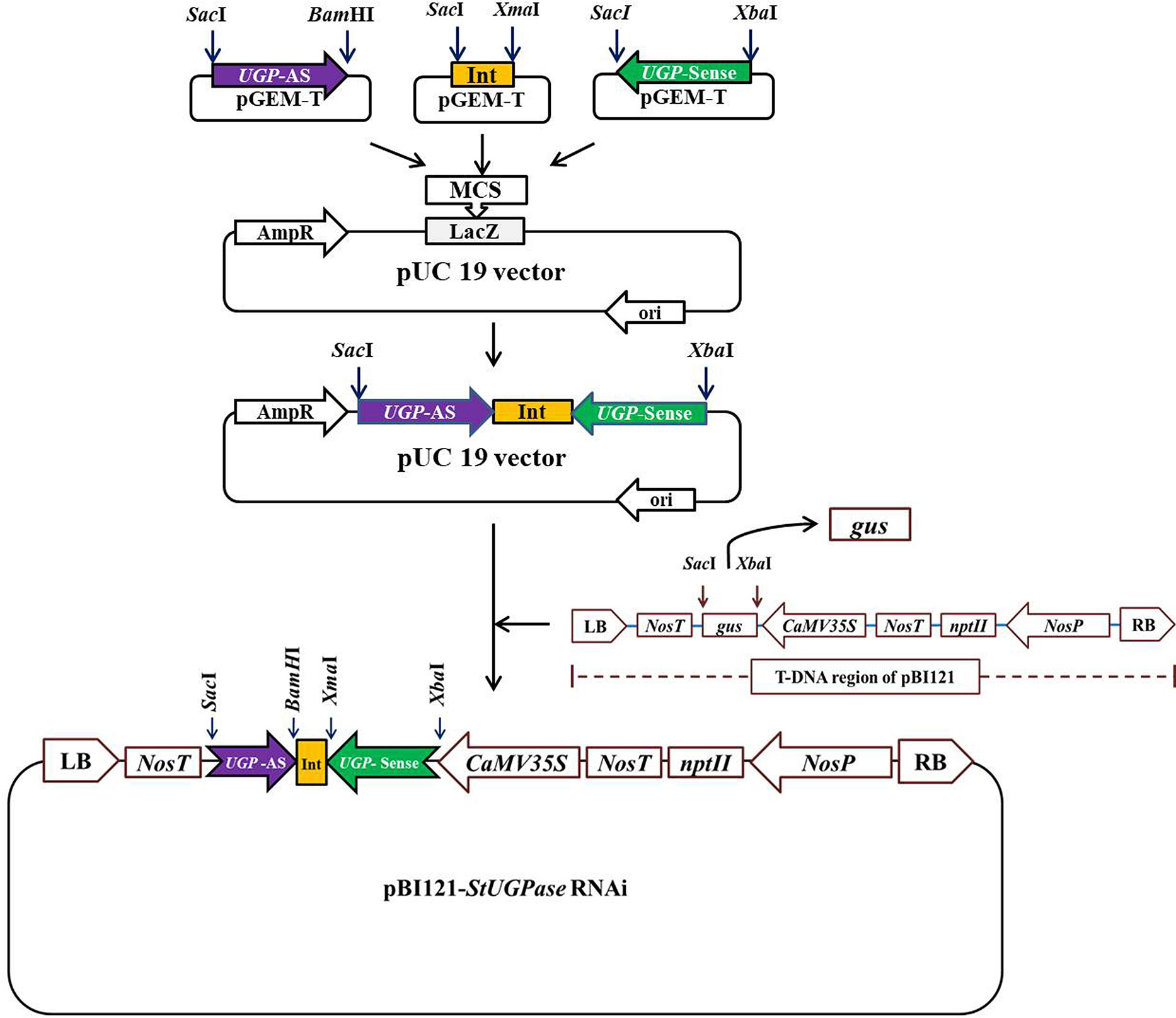
Figure 1 Outline of the strategy for the development of StUGPase hpRNA binary vector cassette, pBI121::UGPase-S:GBSS-Int:UGPase-AS (designated as pB::hpU) for potato transformation. The details of the cloning strategy have been described in Result section 3.1.
in vitro grown potato plantlets (20-23 days old) were used as a source of internodal stem explants. About 100 such explants were placed on Petri plates containing filter paper (90 mm Whatman No.3) placed on pre-culture medium (M.S. medium without any hormone or antibiotics, pH- 5.8) for two days in dark conditions. The pre-cultured internodal stem explants were taken out from Petri plates and dipped in diluted (1:5) Agrobacterium culture, harbouring pB:hpU binary vector cassette, for 1 min in an aseptic Petri plate. The plate containing explant with Agrobacterium culture was shaken slowly in rotational movement. The explants were dried on sterile filter papers and kept on the same pre-culture plates for two days for co-cultivation in the dark. After that, transformed internodal explants were sub-cultured on selection medium [M.S. basal medium (PT021, Himedia) supplemented with 2 mg/l Ca-pantothenate, 3 mg/l GA3, 3 mg/l Trans-Zeatin Riboside, 0.05 mg/l IAA, 50 mg/l kanamycin, 250 mg/l cefotaxime and 250 mg/l carbenicillin, 20g/l sucrose, pH = 5.8], and the plates were kept at 24 °C under 16 hours light and 8 hours darkness. After 6-8 days, the cultures were shifted into fresh regeneration plates to avoid agro-contamination followed by regular sub-culturing after 10-15 days. After 30-40 days of selection, a tiny adventitious shoot/plantlet proliferating from one/both ends of the explant can be visible. The shoots were allowed to grow up to 2-3 cm and then excised and transferred into tubes containing selective propagation medium for a better shoot and root growth and kept at 24 °C. After rooting, the plants are allowed to grow for 3-4 weeks and multiplied at regular intervals.
The putative transformants were screened by PCR for transgene integration using specific sets of primers (UGPase-S F & GBSS-Int R; Supplementary Table 2) to amplify 600 bp fragment encompassing UGPase-S and GBSS-Int. PCR amplification (Supplementary Table 3) was carried out in a thermal cycler (Eppendorf, Germany) programmed with a hot start at 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, 55°C for 1 min and 72°C for 1.5 min and a final extension at 72°C for 7 min. The amplified PCR products were resolved on 1% agarose gel, visualized on a UV- trans-illuminator, and photographed using a gel documentation system. Transgene expression in the PCR-confirmed transgenic lines was confirmed by RT-PCR. Total RNA was extracted from the UGPase RNAi potato lines and wild-type control potato plants using the SpectrumTM Plant Total RNA Extraction kit (Sigma, USA) according to the manufacturer’s protocol. The cDNA was synthesized from 800 ng of total RNA using the PrimeScriptTM single-strand cDNA Synthesis Kit (Takara Bio Ink). RT-PCR was done using the same primer set used for PCR analysis. The amplified RT-PCR products were electrophoresed on 1% agarose gel, visualized on U.V.- transilluminator, and photographed using a gel documentation system.
The RT-PCR positive putative transgenic lines were further multiplied in-vitro and were grown in soil in a transgenic net house under short-day winter conditions. Haulm cutting was done 120 days after planting (DAP), and tubers were collected after 7 days of haulm cutting. The baby tubers so obtained were kept in a cold chamber in March, and taken for sprouting in the first week of October. Sprouted tubers were planted in soil in a transgenic net house in the last week of October under the natural short-day winter condition of Delhi, India. Haulm cutting was done 120 days after planting and harvesting was done 7 days after haulm cutting. Processing attributes were analyzed at fresh harvest, and the rest of the tubers were stored at 4°C in a controlled environment growth chamber (Percival- Scientific, USA). Uniform-size tubers were taken out from the cold chamber after 30 days of cold storage for analysis of cold chipping attributes.
The chipping attributes of the control, and transgenic lines, were assessed at various stages like fresh harvest (without cold treatment), 30 days of cold storage at 4°C, and 21 days of reconditioning at room temperature. Six freshly harvested tubers from each transgenic line were randomly selected for the preparation of chips. Potato chips prepared by deep-frying at 184 °C in groundnut oil were assigned a colour score on a scale of 1-10 based on visual inspection as per the colour score reference chart for potato chips developed by Ezekiel et al. (2003). Chips with a colour score of 1 are the lightest in colour and chips with a score of 10 are the darkest in colour. Chips with colour scores of 1-3 are preferred and are considered acceptable as per the standard norm of the processing industry.
Total sugar was extracted following the protocol of Viola and Davies (1992) with slight modifications. Tuber samples were ground to a fine powder in a pre-chilled pestle and mortar, and about 200 mg of the sample was taken in 2 ml Eppendorf tubes containing 1 ml of 80% ethanol. Sampling was done in replication of three for each transgenic line. Tubes were kept at 4°C overnight with intermittent vortexing. The next morning the mixture was boiled at 80°C for 10 minutes and then allowed to cool at RT. Tubes were centrifuged for a brief period at 3000xg for 2 min, and the clarified supernatant was transferred to fresh tubes. The above step of ethanol extraction was repeated twice with the same sample. The collected clarified supernatant was kept in a vacuum evaporator (CentriVap Centrifugal Concentrator) at 40° C till the entire solution is evaporated. Sugar was dissolved in 200 ul of water and the soluble sugar (glucose, fructose, and sucrose) content was estimated using the Sucrose/D-Fructose/D-Glucose Assay Kit (Product code: K-SUFRG from Megazyme). All the reagents were prepared as per the manufacturer’s instructions (https://www.megazyme.com/documents/Assay_Protocol/K-SUFRG_DATA.pdf) and were stored at the appropriate temperature. The 1/10 diluted sugar sample was used for the assay reaction in triplicate and the absorbance reading during enzymatic assay was taken in a spectrophotometer at 340 nm.
Southern hybridization was done to confirm the transgene integration and the transgene copy number in the genome of UGPase RNAi transgenic potato lines. Approximately 15 µg of genomic DNA per sample was completely digested with 5 µl of EcoRI (EcoRI-HF® 20U/ µl) in a 50 µl reaction and resolved on 0.8 % Agarose gel (without ethidium bromide) in 1X TAE buffer. Southern blot preparation was done according to Russell and Sambrook (2001) using a positively charged nylon membrane (Hybond N+, Amersham, U.K.). The DNA blotting (capillary transfer) was carried out at room temperature for 16 hr. The membrane was carefully removed from the gel, rinsed in 2X SSC (0.3 M NaCl, 0.03 M Trisodium Citrate, pH 7.0), and placed on Whatman 3 paper for air-drying. The membrane was fixed at 22 J/cm for 2 min using Stratalinker UV-crosslinker (Stratagene, U.K.). A 790 bp of DIG-labelled probe was PCR amplified by using nptII gene-specific primers. Probe preparation, prehybridization, hybridization, and detection were performed by following the protocol provided by DIG Luminescent Detection Kit (Roche). The hybridized membrane was exposed to X-ray film (Fujifilm, Kodak) in an intensifying cassette under dark conditions. The cassette was placed for 2 hours, and the exposed X-ray film was developed to visualize the results.
RNA was isolated from the cold stored tubers following the protocol developed by Kumar et al. (2007). For small RNA northern blot analysis, a 5% metaphor gel was prepared and 90 µg total RNA per sample was loaded for separation. RNA samples were separated at 70-75 V at 4°C for 3-4 hr and then transferred to a positively charged nylon membrane (N+, Bright Star-Plus, Applied Biosystem) through capillary blotting setup, same as described for Southern blot transfer in the section 2.7.1. The capillary transfer of RNA was carried out for 7-8 hours while maintaining the transfer buffer temperature at 4˚C. For cross-linking, EDC reagent was used as described by Pall and Hamilton (2008) in place of U.V.cross-linking. A PCR DIG-Probe Synthesis Kit with DIG-dUTP from Roche was used for the PCR labelling of a 500 nt long probe specific to the target sequence using StUGPase-S-specific primers by following the manufacturer’s instructions. Pre-hybridization, hybridization, and band detection were performed following the protocol provided by DIG Luminescent Detection Kit (Roche, Basel, Switzerland).
For detecting the downregulation of StUGPase expression in transgenic lines compared to the control at various treatment conditions (fresh vs. cold treatment), qRT-PCR analysis was performed using StUGPase-specific primers. For tuber tissue, sampling was done at fresh harvest as well as after 30 days of cold storage. qRT-PCR reactions were performed in the Agilent qPCR system under the following conditions, initial denaturation at 95°C for 5 minutes, followed by 40 cycles of 95˚C for 30 sec, 62˚C for 30 sec and 72˚C for 20 seconds. The reactions were carried out using gene-specific real-time primers (Supplementary Table 4) and the StEF1α gene as an internal control. The relative expression levels were calculated using the 2−ΔCt method (Pfaffl, 2001). The specificity of the PCR amplification was checked through melt curve analysis.
The strategy adopted was to develop the StUGPase hpRNA gene construct by cloning the target sequence in an inverted repeat (IR) orientation intervened by potato “granule-bound starch synthase (GBSS)” intron. The complete cDNA sequence of StUGPase (Sequence ID: D00667.1) was retrieved from GenBank (Supplementary Figure 1). For the development of hpRNA construct, a 500 nt sequence corresponding to the 450-949 region of StUGPase cDNA (Supplementary Figure 2) was selected using the web tool dsCheck (http://dsCheck.RNAi.jp/). Potato genomic DNA sequence of 105 nt corresponding to 3821-3925 nt of potato GBSS genomic DNA sequence (Sequence ID: X58453) comprised of 13 nt GBSS exon-13 (5′ exons), 85 nt GBSS intron-13 (last intron), and 6 nt GBSS exon 14 (3′ exons) was used as an intervening sequence for inverted repeat (IR) gene construct (Supplementary Figure 3).
The targeted region of StUGPase was RT-PCR amplified using two sets of primer pairs (Supplementary Table 1), and the 500 bp long amplified DNA fragments, designated as sense (S) and antisense (AS), were ligated separately onto pGEM T-Easy vector through TA-cloning. Similarly, the 105 bp potato GBSS intron was PCR amplified from KC-4 genomic DNA and cloned onto the pGEM T-Easy vector. The StUGPase-S, StGBSS-Int, and StUGPase-AS fragments were confirmed by sequencing, and then sequentially subcloned onto the pUC19 vector to generate UGPase hpRNA construct (Figure 1). At every step of assembly, the clones (pUC19::StUGPase-S, pUC19::StUGPase-S:StGBSS-Int, and pU::hpU) were confirmed by restriction digestion and sequencing (Figure 1). The assembled ~1100 bp long hpU fragment was released from the pU::hpU by XbaI-SacI digestion and then subcloned onto the pBI121 backbone, generated by replacing the GUS coding region by XbaI-SacI digestion to get pB::hpU RNAi binary vector cassette. Development of pB::hpU was confirmed through different combinations of restriction digestion (data not shown) and then introduced into Agrobacterium tumefaciens EHA105 competent cells for potato transformation.
About 800 inter-nodal stem explants of the potato cultivar, KC4, were used for the Agrobacterium-mediated genetic transformation technique and 27 putative transformants were developed. These putative transformants were screened by PCR and RT-PCR and 22 UGPase RNAi lines were obtained (Figure 2). A DNA fragment of ~600 bp corresponding to the cloned sequence of StUGPase-S and StGBSS-Int region was amplified from the 22 UGPase RNAi transgenic lines but not from the non-transgenic control (Supplementary Figures 4, 5). These RNAi transgenic lines were grown in the transgenic net house as short-day winter crop. During the net house trial, normal growth and development were observed in all the UGPase RNAi transgenic lines, and no morphological abnormalities were seen in any of the transgenic lines (Supplementary Figure 6).

Figure 2 Transformation, regeneration, and net house trial of UGPase RNAi potato transgenic lines of KC4. (A) pre-culture, (B) selection, (C) regeneration, (D-F). shoot elongation, (G) transfer into rooting media, (H) rooting, (I) hardening, (J) net house grown plants, (K) harvested tuber.
In vitro multiplied plantlets of the 22 UGPase RNAi transgenic lines and the non-transgenic KC4 control (approx. 20 plants for each line) were grown in the net house as a short-day winter crop. Tubers were harvested in February after 120 days of planting and kept in cold storage for seed purpose as they were small and of varying size. The tubers were taken from cold storage in the first week of October and kept at room temperature for sprouting. Sprouted seed tubers were planted in the last week of October in soil in the net house under natural short-day conditions. Haulm cutting was done after 120 days of planting, and harvesting was done 7 days after haulm cutting. The yield attribute of the RNAi transgenic lines was found variable across the lines and significantly different when compared to non-transgenic control. Out of the total of 22 lines, the highest tuber yield was recorded in UG4 (327.04 g/plant), whereas the lowest was recorded in UG11 (91.60 g/plant) (Supplementary Table 5). After the evaluation of cold chipping abilities, a total of four transgenic lines (UG14, UG18, UG19, and UG21) were selected for further analysis. The average tuber yield of the four selected lines (272.60 gm/plant) was found similar to the average yield of the control line (305.06 gm/plant).
Chips were prepared from both fresh harvest and cold stored tubers. At fresh harvest, all the RNAi transgenic lines and non-transgenic control produced desirable light colour chips with a score of 1.0. However, chips prepared after 30 days of cold storage displayed drastic differences in chip colour scores ranging from 2.5 to 6.5. KC4 control tubers produced the darkest color chips (colour score 6.5) after 30 days of cold storage. In general, there was deterioration in chip colour in all the UGPase RNAi lines after cold storage compared to that of fresh harvest. The cold-chipping performance showed that out of 22 RNAi transgenic lines evaluated, chips produced from the four RNAi lines, UG14, UG18, UG19, and UG21, had acceptable light colour after 30 days of cold storage. The chip colour score for the two UGPase RNAi lines, UG14 and UG19, was 2.5, while for the other two RNAi lines, UG 18 and UG 21, it was 3.0 (Figure 3; Supplementary Table 6). There was considerable improvement in chip colour in all the RNAi lines after reconditioning of cold-stored tubers for 21 days at room temperature. Chips colour for the three best performing RNAi lines, UG14, UG18, and UG19 was 1.5, while for the other best performing RNAi line, UG21, it was 2.0. In contrast, the non-transgenic counterpart produced chips with colour score of 3.5 after reconditioning (Figure 3).
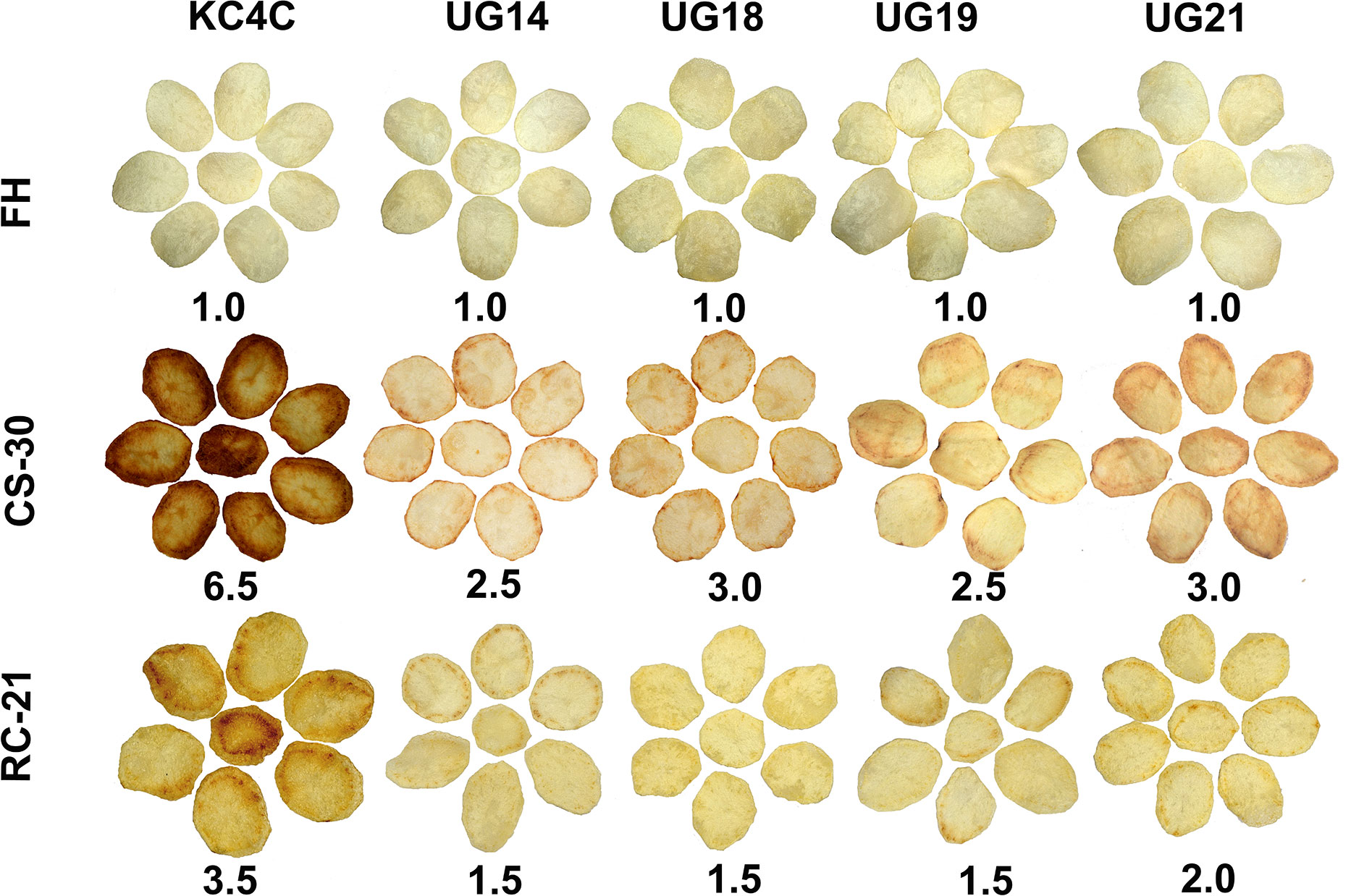
Figure 3 Cold chipping performance of the four best performing UGPase RNAi potato transgenic lines of KC4 at fresh harvest (FH), after 30 days of cold storage (CS) and after 21 days of room temperature reconditioning (RC) of the cold stored tubers. The number below chips of each line indicates chip colour score.
Total soluble sugars [Reducing sugar (RS), glucose and fructose; and sucrose] content of the four best performing UGPase RNAi lines were estimated from tubers at fresh harvest, after cold storage, and after reconditioning. In general, RS and sucrose content was much less in all the four selected RNAi transgenic lines at all three treatment conditions studied. At harvest, RS content in the four selected RNAi lines ranged from 4.9-8.13 mg/100g fresh weight compared to 12mg/100g fresh weight in KC4 non-transformed control. The maximum reduction in RS content was observed in UG18 (59.31%) followed by UG21 (50.12%) and UG19 (37.58%), whereas the least reduction was recorded in UG14 (32.41%) (Figure 4; summarised in the Supplementary Table 6). Around 15-25 times increase in RS level was observed in both the RNAi transgenic and control tubers after cold storage for 30 days. However, the extent of increase in RS content was 54.09% to 57.52% lesser in the four RNAi transgenic lines than that of the control after cold storage. The RS content in cold-stored tubers of these four UGPase RNAi lines ranged from 120 mg to 163 mg per 100 g FW, whereas it was more than 250 mg per 100 g FW in the control tubers (Figure 4). RS level reduced considerably in both the RNAi lines and control after reconditioning. RS level varied from 48.25 mg to 77.7 mg per 100 g FW in the RNAi lines, whereas it was 110.6 mg per 100 g FW in the control line after reconditioning (Figure 4). The reduction in RS content in the RNAi lines was 37.4% to 59.3% over that of 30-day cold storage (Figure 4).
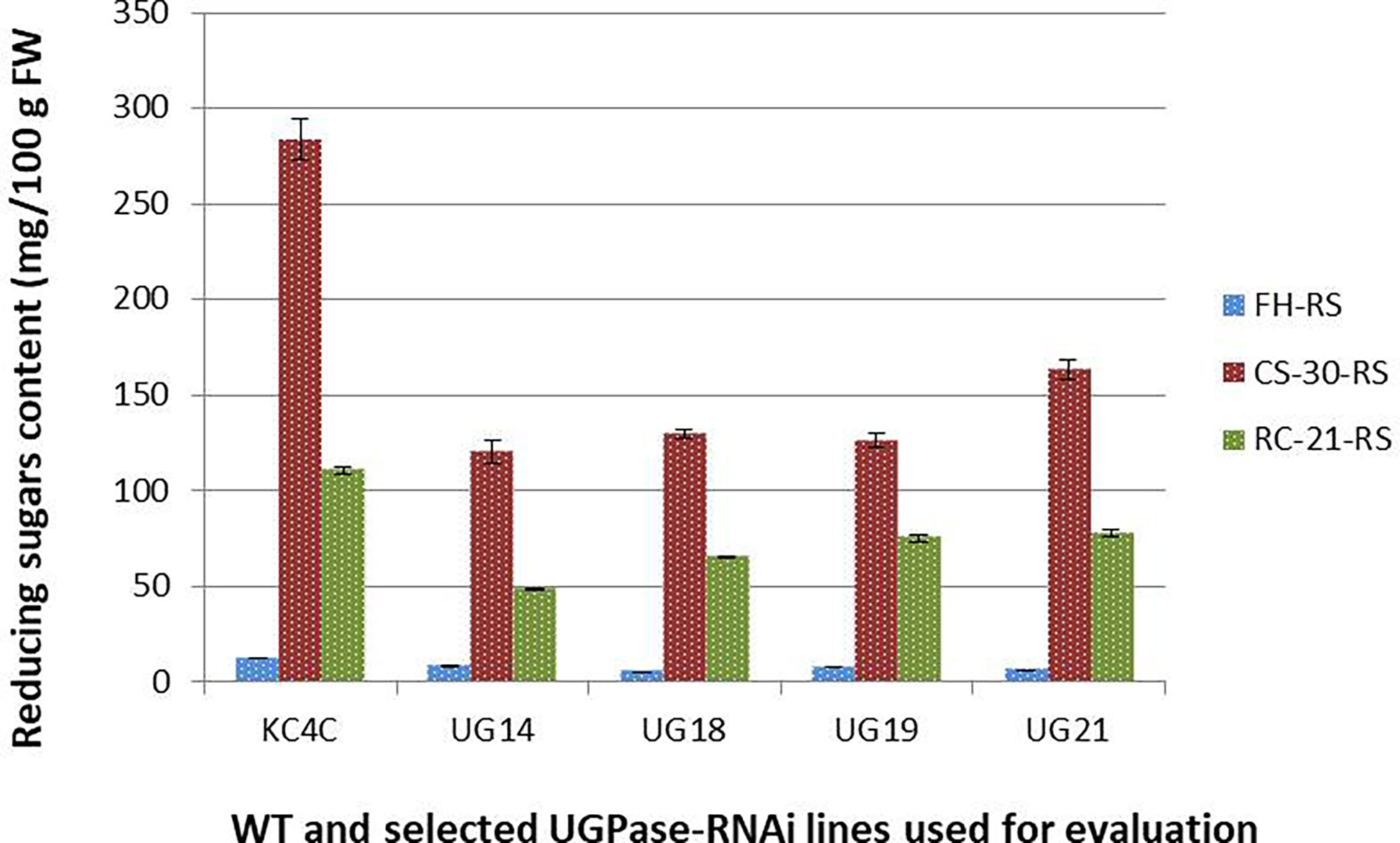
Figure 4 Reducing sugar content in the UGPase RNAi potato transgenic lines of KC4. KC4C, Kufri Chipsona-4 control; UG14, UG18, UG19, and UG21 are the four selected UGPase RNAi transgenic lines of KC4.FH, Fresh harvest; CS-30, cold stored for 30 days; RC-21, 21 days reconditioning at room temp after cold storage.
Similarly, there was a significant reduction in sucrose level in the UGPase RNAi transgenic lines than that of the non-transformed control. At harvest, the control tuber had sucrose content of 320.16 mg/100 g fresh weight, and the average sucrose content of the RNAi lines was 170.23 mg/100 g fresh weight which is around 43% less than that of the control (Figure 5). The highest reduction of sucrose content was observed in line UG21 (50%) followed by UG19 (47.55%), UG14 (46.77%), and UG18 (43.55%). After cold storage at 4° C for 30 days, sucrose content increased upto 910 mg/100g FW in the UGPase RNAi transgenic lines, while it was 1314 mg/100g FW in the control tubers implying a 4-5.4 times increase in sucrose content under cold stored condition than that of fresh harvest (Figure 5). The sucrose level in the RNAi transgenic lines was 31-46% lesser than that of the non-transgenic counterpart. The highest reduction was noticed in line UG14 (46.12%), followed by UG19 (39.9%), UG21 (33.9%), and UG18 (30.7%). When the sucrose content of tubers after reconditioning was compared with that of fresh harvest, the reconditioned tubers were found to possess slightly higher sucrose levels than that of the control tuber. Following the trend, the sucrose content of the transgenic lines was 17-22% lower than that of the non-transgenic counterpart after reconditioning (Figure 5).
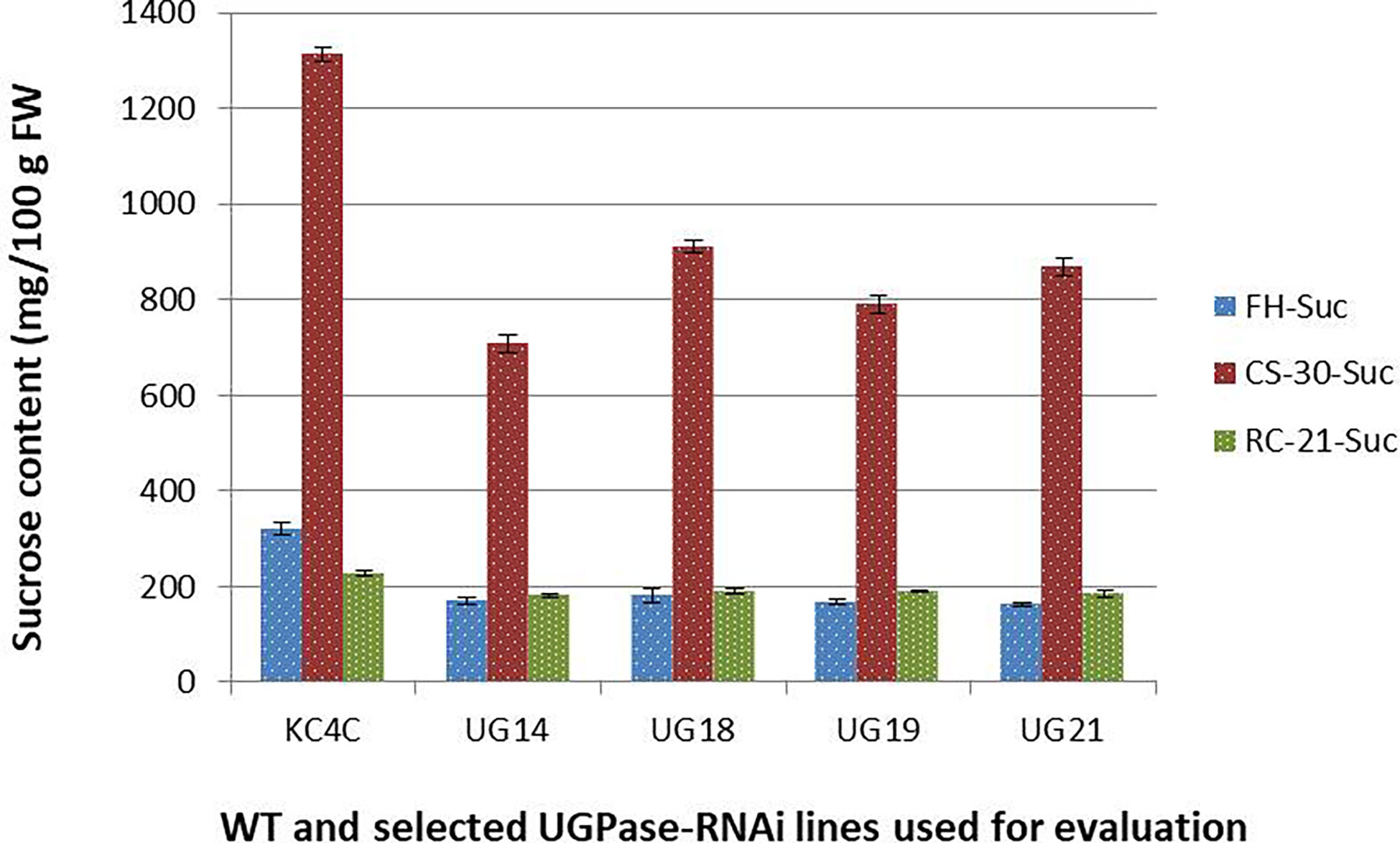
Figure 5 Sucrose content in the UGPase RNAi potato transgenic lines of KC4. KC4C, Kufri Chipsona-4 control; UG14, UG18, UG19, and UG21 are the four selected UGPase RNAi transgenic lines of KC4. FH, Fresh harvest; CS-30, cold stored for 30 days; RC-21, 21 days reconditioning at room temp after cold storage.
The four UGPase RNAi potato lines were analyzed for confirmation of gene integration and copy number by Southern hybridization. Genomic DNA isolated from leaves of the four RNAi transgenic lines and non-transformed KC4 control was digested with EcoRI, which had a single recognition site within the T-DNA region of pB:hpU binary vector. PCR amplified 790 bp nptII fragment was labeled with DIG-dUTP and used as a probe. The selected four UGPase RNAi transgenic potato lines carried 2-5 copies of the transgene in their genome. Out of four selected transgenic potato lines, UG19 had five, UG14 had four, UG18 had three and UG21 had 2 copies of the transgene (Figure 6).
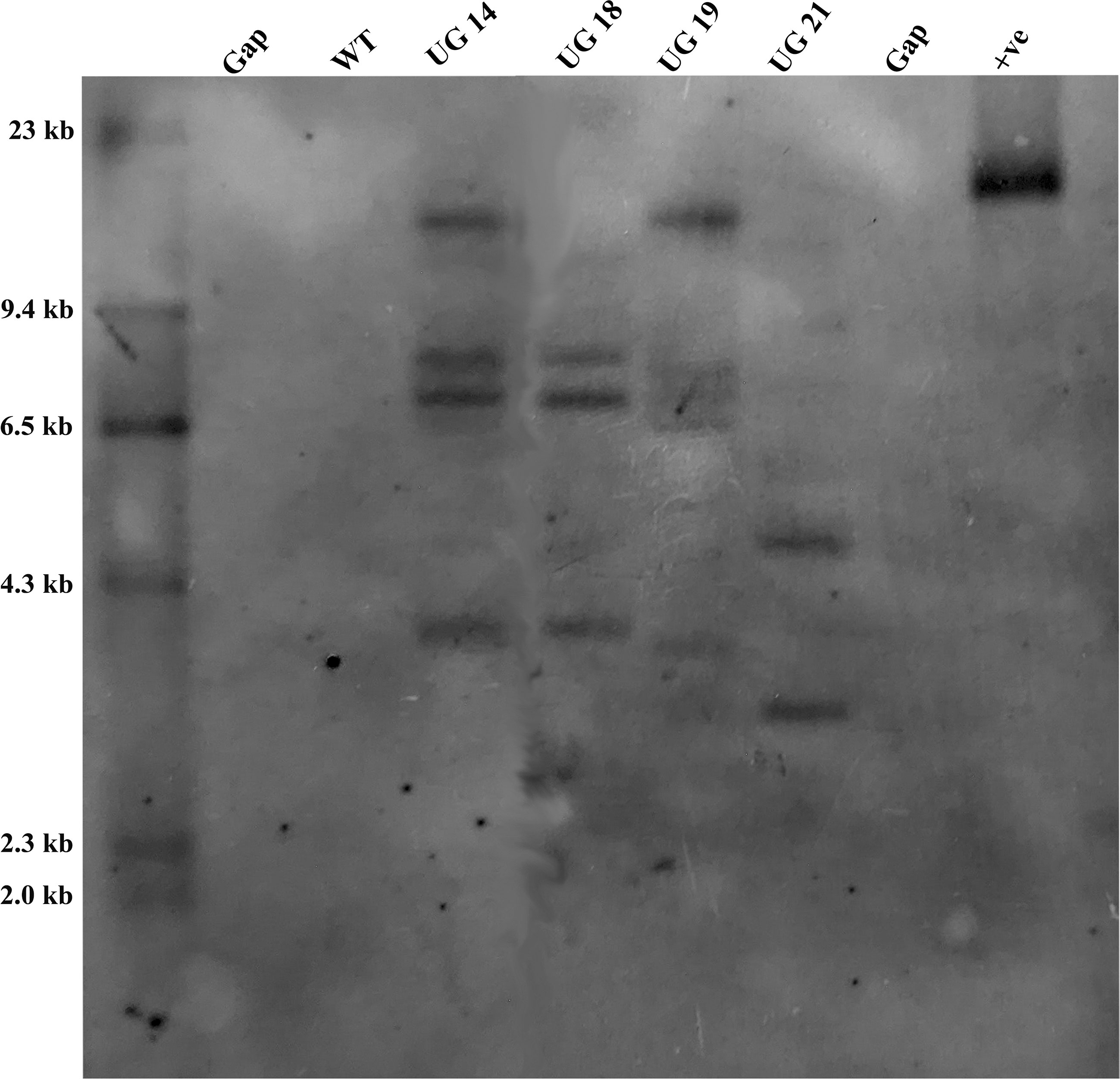
Figure 6 Southern blot analysis of the UGPase RNAi transgenic potato lines of KC4. Lanes: WT, wild type control; UG14, UG18, UG19, UG21 represent transgenic potato lines; +ve, positive control.
Small RNA northern hybridization was done to detect the accumulation of siRNA in the RNAi transgenic lines. Total RNA isolated from the cold stored tubers was resolved on metaphor agarose gel and blotted onto the membrane, which was probed with randomly labeled UGPase-S cDNA. Positive control of 21nt long oligo with sequence similarity with UGPase mRNA target region was used as control. A prominent band of ~21 nt long was detected in all the four RNAi transgenic lines indicating UGPase-specific siRNA accumulation (Figure 7). No UGPase-specific siRNA band was observed in case of the non-transgenic control.
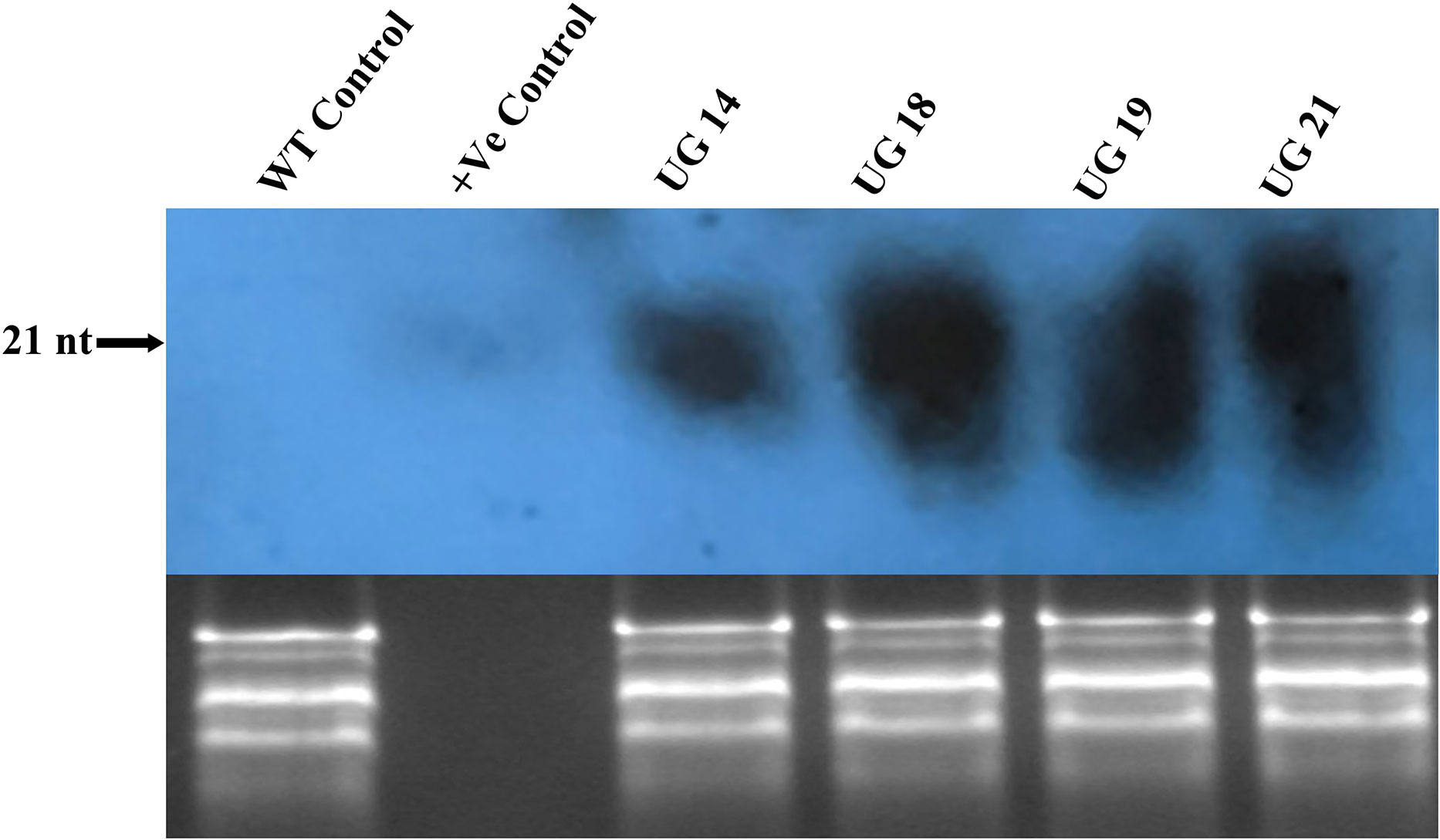
Figure 7 Small RNA northern blot of the UGPase RNAi transgenic potato lines of KC4. siRNA band corresponding to ~21nt long positive control oligo was detected in the fourRNAi transgenic lines. Lanes: Lanes: WT, wild type control; +ve, positive control; UG14, UG18, UG19, UG21 represent transgenic potato lines.
Real-time PCR analysis was conducted to detect the extent of downregulation of the target gene, StUGPase, expression. cDNA was prepared from RNA isolated from both the freshly harvested and cold stored tubers of the four UGPase RNAi transgenic lines and wild-type control and used for real-time analysis. Many-fold reduction in the steady-state StUGPase transcript abundance was recorded in cold-stored tubers of the RNAi potato line was detected (Figure 8). The highest reduction (4.5 fold at FH and 5 fold at CS-30) in transcript level was observed in UG19. The level of reduction in StUGPase transcript in UG14, UG18, and UG21 was 3.5, 2.8, and 2.2 folds, respectively, at fresh harvest (FH) and 3.9, 2.4, and 1.8 folds, respectively, after cold storage for 30 days (CS-30) compared to that of the control. In comparison to the freshly harvested and reconditioned tubers, the level of UGPase expression was 2.51- 3.32 times higher in the cold stored tuber. But still, in both the conditions, the same pattern of downregulation of UGPase expression was observed.
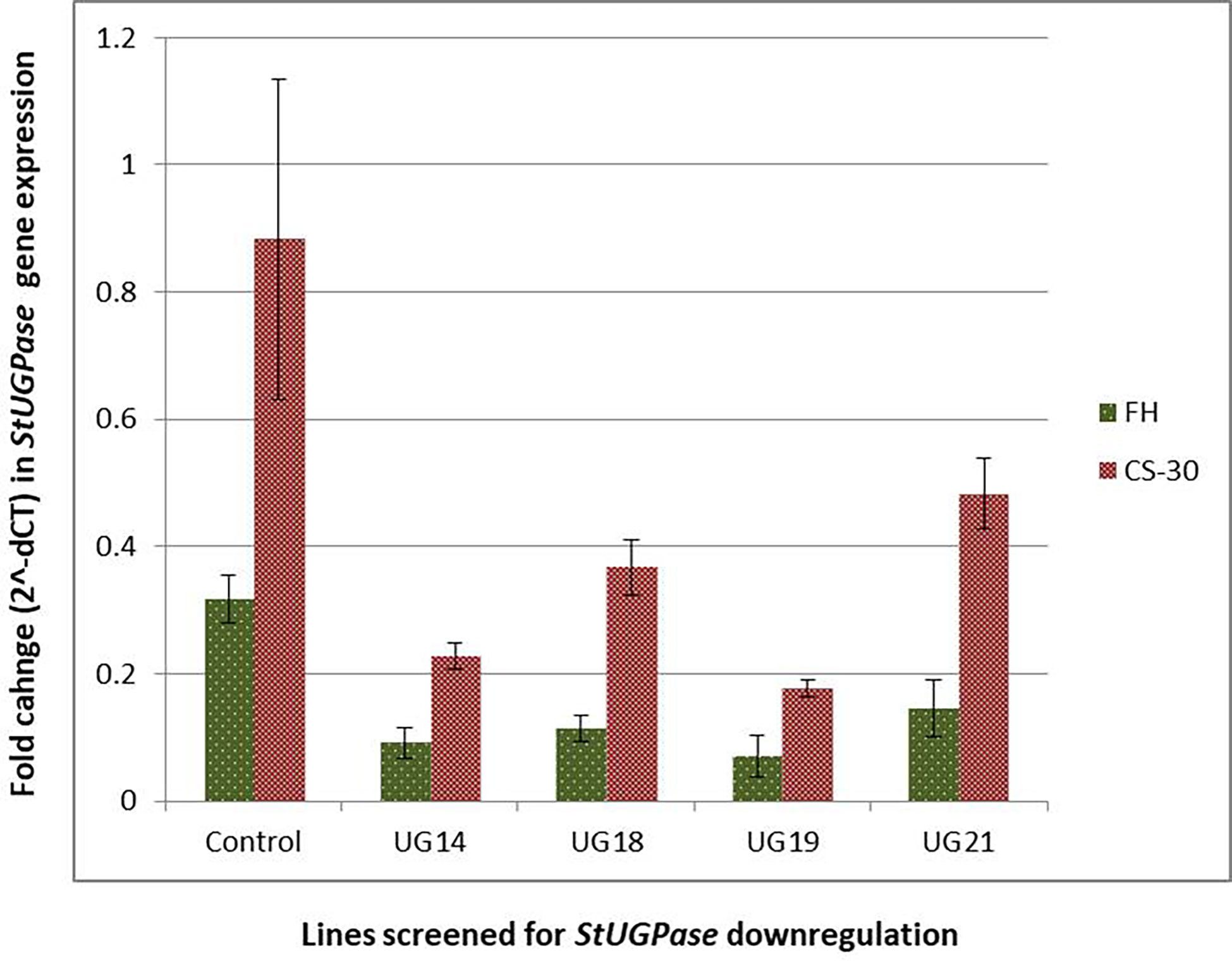
Figure 8 Reduction in StUGPase expression in the four RNAi potato transgenic lines of KC4 as revealed by Real-Time PCR analysis. Control, KC4C plant; UG14, UG18, UG19, and UG21 represents RNAi transgenic lines. FH, fresh harvest; CS-30, cold stored for 30 days.
The data presented in the current study demonstrate that RNAi-mediated silencing of the StUGPase gene is an effective approach of reducing RS accumulation and consequently improving cold chipping attributes in processing potato cultivars. Based on previous studies the two enzymes, namely vaINV and UGPase, have been considered to be the two key regulatory enzymes of CIS. To date, considerable efforts were made to reduce vaINV activity and RS accumulation in potato tubers utilizing a multitude of molecular strategies, including overexpression of invertase inhibitors (Greiner et al., 1999; McKenzie et al., 2013) and silencing of the gene encoding vaINV (Zhang et al., 2008; Pattanayak et al., 2009; Bhaskar et al., 2010; Wiberley-Bradford et al., 2014; Zhu et al., 2014; Clasen et al., 2016; Yasmeen et al., 2022). Since vaINV is associated with the breakdown of sucrose into RS during CIS, the inhibition of vaINV activity could lead to the accumulation of sucrose in the tuber. Although sucrose does not influence potato chip colour, but it imparts an undesirable sweet taste in chips prepared from cold stored vaINV silenced potato (Pattanayak et al., 2009). This sucrose accumulation could further be prevented by manipulating another key enzyme, UGPase, since it has significant control over the flux of carbon toward sucrose biosynthesis and found to play a crucial role during CIS (Zrenner et al., 1993; Spychalla et al., 1994; Hill et al., 1996; Bagnaresi et al., 2008; Chen et al., 2008). Therefore, the present study was designed to suppress StUGPase expression by hpRNA-mediated silencing of StUGPase and consequent amelioration of CIS in potato. As UGPase is involved in many important biological functions it was perceived that complete knockout of the gene might have detrimental effects in growth and development. Hence, RNAi-mediated silencing of StUGPase was adopted considering the ease of hpRNA construct development and high efficacy of the technique (He and Hannon, 2004; Duda et al., 2014; Liang et al., 2015; Swiech et al., 2015). Previous attempts of silencing of StUGPase employing antisense approach demonstrated variable success. Zrenner et al. (1993) could not find any significant change in RS content despite 96% decrease in UGPase activity of antisense line, Spychalla et al. (1994) observed a significant change in the sucrose content of cold stored transgenic potato tubers expressing an antisense construct of StUGPase. In the present study, significant reduction of soluble sugar content (both sucrose and RS content) was observed in the selected four RNAi transgenic lines’ tubers of KC4 after cold storage for 30 days. Reduction of RS content was reflected in the colour of the chips prepared from cold stored transgenic tubers. Chip colour of the cold stored RNAi transgenic tubers was in the acceptable range (score of 2.5 to 3.0) compared to very dark brown colour (score of 6.5) in the cold stored control tubers. It was reported that RS level in cold stored potato should not exceed 160mg/100g tissue fresh weight to get acceptable colour in chips with score of 1-3 (Ezekiel et al., 2003). As both the RS content and chip colour score of cold stored RNAi transgenic tubers were at the border line of acceptance processing attributes of the selected RNAi transgenic potato lines were tested after reconditioning of tubers at room temperature (20-24° C) for 21 days. Room temperature reconditioning of cold stored potatoes is normally followed by the processing industries for improvement in colour and texture in the processed potato products like chips and fries (Ezekiel et al., 2003; Marwaha et al., 2010). In the present study, much improvement in sucrose and RS content and consequently in chip colour were recorded after reconditioning. Sucrose content in all the four RNAi lines was almost to that of the respective fresh harvest level, and RS content was reduced to half of the respective level after cold storage and also almost half compared to that of control after reconditioning. That at each point of the study (fresh harvest, 30 days of cold storage and after reconditioning) soluble sugar level (both sucrose and RS content) was almost 50% less in the RNAi lines compared to that of control indicates the efficacy of the strategy.
Downregulation of the target gene was recorded in all the selected transgenic lines (54-72 % in FH and 45-80% under CS-30 conditions). StUGPase expression was reported to be substantially up-regulated in potato tubers at low temperature (Spychalla et al., 1994; Borovkov et al., 1996) which is also evident in the current study. StUGPase expression was enhanced many folds in untransformed WT plants under cold storage. Despite that, the extent of StUGPase down regulation was almost similar during both at fresh harvest and after cold storage. Detection of StUGPase-specific siRNAs in cold stored RNAi transgenic tubers, but not in cold stored control tubers, confirms that this down regulation of StUGPase expression was mediated by these siRNA mediated degradation of StUGPase mRNA. This indicates that RNAi-mediated silencing was not inhibited in cold, and was effectively operational during cold storage (Szittya et al., 2003). Although we could not find a strict correlation among the extent of down regulation of StUGPase expression, reduction of soluble sugar content in the cold stored RNAi transgenic tubers, and chip colour. This could be due to the influence of some other factors on tuber sugar content and chip colour. It is important to note that the tuber sugar content and chip colour are influenced by many factors like disease stress, temperature and water stress, management practices, and mechanical handling (Kumar et al., 2004).
Amelioration of CIS in transgenic potato developed by RNAi-mediated silencing of StUGPase demonstrates the important role played by the enzyme in the regulation of sucrose biosynthesis and the accumulation of RS in the cold-stored potato tubers. A considerable improvement in the cold-chipping ability of UGPase RNAi transgenic potato in terms of reduction in sucrose and RS content and improvement in chip colour demonstrates the efficacy of the strategy. No growth and developmental abnormalities were observed during the net house trial. The apparent variation in yield was due to the planting of different size baby seed tubers. However, the selected four lines showing a 2 to 5 fold reduction in StUGPase expression had a comparable average yield to that of the control. Considering the vital role UGPase plays in plant growth, development, survival under stress conditions, and also in CIS it can be said that our strategy could strike a balance between induction of a moderate level of silencing of this essential gene and achieving considerable improvement in reduction of soluble sugar content and improvement in chip colour. Our approach, being less disruptive, might be the choice to get commercial success in near future. In the future, modern genome editing tools can also be employed to get any mutant with optimum silencing of StUGPase and development of transgene free CIS tolerant potato lines.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
This article does not contain any studies involving animals or human participants as objects of research.
Conceiving and designing the experiment were done by DP. Data curation and analysis were done by SJ, JT, and SB. The experiment was conducted by SJ, KR, ST, MS, and KP. Supervision of the research work by DP. Writing of the original draft by SJ. Review and editing of the manuscript by DP, KR, KM, JV, and RS. All authors contributed to the article and approved the submitted version.
The study was supported by the PG Fellowship to SJ by ICAR-IARI, New Delhi, and institute research fund to DP by ICAR-NIPB.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1133029/full#supplementary-material
Bagnaresi, P., Moschella, A., Beretta, O., Vitulli, F., Ranalli, P., Perata, P. (2008). Heterologous microarray experiments allow the identification of the early events associated with potato tuber cold sweetening. BMC Genomics 9, 1–23. doi: 10.1186/1471-2164-9-176
Bhaskar, P. B., Wu, L., Busse, J. S., Whitty, B. R., Hamernik, A. J., Jansky, S. H., et al. (2010). Suppression of the vacuolar invertase gene prevents cold-induced sweetening in potato. Plant Physiol. 154, 939–948. doi: 10.1104/pp.110.162545
Blenkinsop, R. W., Yada, R. Y., Marangoni, A. G. (2010). “Metabolic control of low-temperature sweetening in potato tubers during postharvest storage,” in Horticultural reviews (Oxford, UK: John Wiley and Sons, Inc), 317–354. doi: 10.1002/9780470650837.ch8
Borovkov, A. Y., Mcclean, P. E., Sowokinos, J. R., Ruud, S. H., Secor, G. A. (1996). Effect of expression of UDP-glucose pyrophosphorylase ribozyme and antisense RNAs on the enzyme activity and carbohydrate composition of field-grown transgenic potato plants. J. Plant Physiol. 147, 644–652. doi: 10.1016/S0176-1617(11)81473-2
Bradshaw, J. E., Bryan, G. J., Ramsay, G. (2006). Genetic resources (including wild and cultivated solanum species) and progress in their utilisation in potato breeding. Potato Res. 49, 49–65. doi: 10.1007/s11540-006-9002-5
Brummell, D. A., Chen, R. K., Harris, J. C., Zhang, H., Hamiaux, C., Kralicek, A. V., et al. (2011). Induction of vacuolar invertase inhibitor mRNA in potato tubers contributes to cold-induced sweetening resistance and includes spliced hybrid mRNA variants. J. Exp. Bot. 62, 3519–3534. doi: 10.1093/jxb/err043
Chen, S., Hajirezaei, M. R., Zanor, M. I., Hornyik, C., Debast, S., Lacomme, C., et al. (2008). RNA Interference-mediated repression of sucrose-phosphatase in transgenic potato tubers (Solanum tuberosum) strongly affects the hexose-to-sucrose ratio upon cold storage with only minor effects on total soluble carbohydrate accumulation. Plant Cell Environ. 31, 165–176. doi: 10.1111/j.1365-3040.2007.01747.x
Clasen, B. M., Stoddard, T. J., Luo, S., Demorest, Z. L., Li, J., Cedrone, F., et al. (2016). Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol. J. 14, 169–176. doi: 10.1111/pbi.12370
Das, S., Mitra, B., Luthra, S. K., Saha, A., Hassan, M. M., Hossain, A. (2021). Study on morphological, physiological characteristics and yields of twenty-one potato (Solanum tuberosum l.) cultivars grown in eastern sub-himalayan plains of India. Agronomy 11, 335. doi: 10.3390/agronomy11020335
Datir, S. S., Yousf, S., Sharma, S., Kochle, M., Ravikumar, A., Chugh, J. (2020). Cold storage reveals distinct metabolic perturbations in processing and non-processing cultivars of potato (Solanum tuberosum l.). Sci. Rep. 10, 6268. doi: 10.1038/s41598-020-63329-5
Duda, K., Lonowski, L. A., Kofoed-Nielsen, M., Ibarra, A., Delay, C. M., Kang, Q., et al. (2014). High-efficiency genome editing via 2A-coupled co-expression of fluorescent proteins and zinc finger nucleases or CRISPR/Cas9 nickase pairs. Nucleic Acids Res. 42, e84–e84. doi: 10.1093/nar/gku251
Ezekiel, R., Singh, B., Kumar, D. (2003). A reference chart for potato chip colour for use in India. J. Indian Potato Assoc. 30, 259–265.
Geigenberger, P., Stitt, M., Fernie, A. R. (2004). Metabolic control analysis and regulation of the conversion of sucrose to starch in growing potato tubers. Plant Cell Environ. 27, 655–673. doi: 10.1111/j.1365-3040.2004.01183.x
Greiner, S., Rausch, T., Sonnewald, U., Herbers, K. (1999). Ectopic expression of a tobacco invertase inhibitor homolog prevents cold-induced sweetening of potato tubers. Nat. Biotechnol. 17, 708–711. doi: 10.1038/10924
Gupta, V. K., Luthra, S. K., Bhardwaj, V., Rawal, S. (2021). Varieties and Technologies for Processing. Chakrabarti, S. K., Bhardwaj, V. (Eds.) Potato in Sub-tropics: A Saga of Success. (pp 207–223). New Delhi International Books and Periodical Supply Service.
Halford, N. G., Curtis, T. Y., Muttucumaru, N., Postles, J., Mottram, D. S. (2011). Sugars in crop plants. Ann. Appl. Biol. 158, 1–25. doi: 10.1111/j.1744-7348.2010.00443.x
Hameed, A., Zaidi, S. S. E. A., Shakir, S., Mansoor, S. (2018). Applications of new breeding technologies for potato improvement. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00925
Hamernik, A. J., Hanneman, R. E., Jansky, S. H. (2009). Introgression of wild species germplasm with extreme resistance to cold sweetening into the cultivated potato. Crop Sci. 49, 529–542. doi: 10.2135/cropsci2008.04.0209
He, L., Hannon, G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531. doi: 10.1038/nrg1379
Hill, L. M., Reimholz, R., Schröder, R., Nielsen, T. H., Stitt, M. (1996). The onset of sucrose accumulation in cold-stored potato tubers is caused by an increased rate of sucrose synthesis and coincides with low levels of hexose-phosphates, an activation of sucrose phosphate synthase and the appearance of a new form of amylase. Plant Cell Environ. 19, 1223–1237. doi: 10.1111/j.1365-3040.1996.tb00001.x
Hirsch, C. N., Hirsch, C. D., Felcher, K., Coombs, J., Zarka, D., Van Deynze, A., et al. (2013). Retrospective view of north American potato (Solanum tuberosum l.) breeding in the 20th and 21st centuries. G3: Genes Genomes Genet. 3, 1003–1013. doi: 10.1534/g3.113.005595
Höfgen, R., Willmitzer, L. (1988). Storage of competent cells for agrobacterium transformation. Nucleic Acids Res. 16, 9877. doi: 10.1093/nar/16.20.9877
Katsube, T., Kazuta, Y., Mori, H., Nakano, K., Tanizawa, K., Fukui, T. (1990). UDP-Glucose pyrophosphorylase from potato tuber: cDNA cloning and sequencing. J. Biochem. 108, 321–326. doi: 10.1093/oxfordjournals.jbchem.a123200
Keijbets, M. J. H. (2008). Potato processing for the consumer: developments and future challenges. Potato Res. 51, 271–281. doi: 10.1007/s11540-008-9104-3
Kumar, D. (2011) Cold-induced sweetening development in Indian potato (Solanum tuberosum l.) varieties. Available at: http://nopr.niscpr.res.in/handle/123456789/11615.
Kumar, G. M., Iyer, S., Knowles, N. R. (2007). Extraction of RNA from fresh, frozen, and lyophilized tuber and root tissues. J. Agric. Food Chem. 55, 1674–1678. doi: 10.1021/jf062941m
Kumar, D., Singh, B. P., Kumar, P. (2004). An overview of the factors affecting sugar content of potatoes. Ann. Appl. Biol. 145, 247–256. doi: 10.1111/j.1744-7348.2004.tb00380.x
Liang, X., Potter, J., Kumar, S., Zou, Y., Quintanilla, R., Sridharan, M., et al. (2015). Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 208, 44–53. doi: 10.1016/j.jbiotec.2015.04.024
Liu, X., Cheng, S., Liu, J., Ou, Y., Song, B., Zhang, C., et al. (2013). The potato protease inhibitor gene, St-inh, plays roles in the cold-induced sweetening of potato tubers by modulating invertase activity. Postharvest Biol. Technol. 86, 265–271. doi: 10.1111/pbi.12054
Liu, X., Song, B., Zhang, H., Li, X. Q., Xie, C., Liu, J. (2010). Cloning and molecular characterization of putative invertase inhibitor genes and their possible contributions to cold-induced sweetening of potato tubers. Mol. Genet. Genomics 284, 147–159. doi: 10.1007/s00438-010-0554-3
Liu, C., Xi, H., Chen, X., Zhao, Y., Yao, J., Si, J., et al. (2021). Genome-wide identification and expression pattern of alkaline/neutral invertase gene family in dendrobium catenatum. Biotechnol. Biotechnol. Equip. 35, 527–537. doi: 10.1080/13102818.2021.1901610
Marwaha, R. S., Pandey, S. K., Kumar, D., Singh, S. V., Kumar, P. (2010). Potato processing scenario in India: industrial constraints, future projections, challenges ahead and remedies–a review. J. Food Sci. Technol. 47, 137–156. doi: 10.1007/s13197-010-0026-0
Matsuura-Endo, C., Kobayashi, A., Noda, T., Takigawa, S., Yamauchi, H., Mori, M. (2004). Changes in sugar content and activity of vacuolar acid invertase during low-temperature storage of potato tubers from six Japanese cultivars. J. Plant Res. 117, 131–137. doi: 10.1007/s10265-003-0137-z
McKenzie, M. J., Chen, R. K., Harris, J. C., Ashworth, M. J., Brummell, D. A. (2013). Post-translational regulation of acid invertase activity by vacuolar invertase inhibitor affects resistance to cold-induced sweetening of potato tubers. Plant Cell Environ. 36, 176–185. doi: 10.1111/j.1365-3040.2012.02565.x
McKenzie, M. J., Sowokinos, J. R., Shea, I. M., Gupta, S. K., Lindlauf, R. R., Anderson, J. A. (2005). Investigations on the role of acid invertase and UDP-glucose pyrophosphorylase in potato clones with varying resistance to cold-induced sweetening. Am. J. Potato Res. 82, 231–239. doi: 10.1007/BF02853589
Murata, M. (2021). Browning and pigmentation in food through the maillard reaction. Glycoconj. J. 38, 283–292. doi: 10.1007/s10719-020-09943-x
Nursten, H. E. (2005). The maillard reaction: chemistry, biochemistry, and implications (Royal Society of Chemistry).
Ortiz, R., Watanabe, K. N. (2004). Genetic contributions to breeding polyploid crops. Recent Res. Dev. Genet. Breed. 1 (Part II), 269–286.
Pall, G. S., Hamilton, A. J. (2008). Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 3, 1077–1084. doi: 10.1038/nprot.2008.67
Pandey, S. K., Marwaha, R. S., Dinesh, K., Singh, S. K. (2009). Indian potato processing story: Industrial limitations, challenges ahead and vision for the future. Potato Journal 36, 1–13.
Park, J. I., Ishimizu, T., Suwabe, K., Sudo, K., Masuko, H., Hakozaki, H., et al. (2010). UDP-Glucose pyrophosphorylase is rate limiting in vegetative and reproductive phases in Arabidopsis thaliana. Plant Cell Physiol. 51, 981–996. doi: 10.1093/pcp/pcq057
Pattanayak, D., Pandey, S. K., Chamail, A., Verma, A., Huddone, S., Malhotra, B., et al. (2009). Indian Patent No. 305855.
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45–e45. doi: 10.1093/nar/29.9.e45
Pradel, W., Gatto, M., Hareau, G., Pandey, S. K., Bhardway, V. (2019). Adoption of potato varieties and their role for climate change adaptation in India. Climate Risk Manage. 23, 114–123. doi: 10.1016/j.crm.2019.01.001
Raigond, P., Mehta, A., Singh, B. (2018). Sweetening during low-temperature and long-term storage of Indian potatoes. Potato Res. 61, 207–217. doi: 10.1007/s11540-018-9369-0
Rees, T. A., Morrell, S. (1990). Carbohydrate metabolism in developing potatoes. Am. Potato J. 67, 835–847. doi: 10.1007/BF03044296
Russell, D. W., Sambrook, J. (2001). Molecular cloning: A laboratory manual (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory) Vol. 1, p. 112.
Schwimmer, S., Makower, R. U., Rorem, E. S. (1961). Invertase & invertase inhibitor in potato. Plant Physiol. 36, 313. doi: 10.1104/pp.36.3.313
Shi, W., Ma, Q., Yin, W., Liu, T., Song, Y., Chen, Y., et al. (2022). StTINY3 enhances cold-induced sweetening resistance by coordinating starch resynthesis and sucrose hydrolysis in potato. J. Exp. Bot. doi: 10.1093/jxb/erac171
Shi, W., Song, Y., Liu, T., Ma, Q., Yin, W., Shen, Y., et al. (2021). StRAP2. 3, an ERF-VII transcription factor, directly activates StInvInh2 to enhance cold-induced sweetening resistance in potato. Hortic. Res. 8. doi: 10.1038/s41438-021-00522-1
Shivalingamurthy, S. G., Anangi, R., Kalaipandian, S., Glassop, D., King, G. F., Rae, A. L. (2018). Identification and functional characterization of sugarcane invertase inhibitor (ShINH1): A potential candidate for reducing pre-and post-harvest loss of sucrose in sugarcane. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00598
Singh, S. (2012). “Marketing channels and their implications for smallholder farmers in India,” in The transformation of agri-food systems (Routledge), 303–334.
Sowokinos, J. (1990). Stress-induced alterations in carbohydrate metabolism. Mol. Cell. Biol. Potato, 137–158.
Sowokinos, J. R. (2001a). Biochemical and molecular control of cold-induced sweetening in potatoes. Am. J. Potato Res. 78, 221–236. doi: 10.1007/bf02883548
Sowokinos, J. R. (2001b). Allele and isozyme patterns of UDP-glucose pyrophosphorylase as a marker for cold-sweetening resistance in potatoes. Am. J. Potato Res. 78, 57–64. doi: 10.1007/BF02874825
Sowokinos, J. R., Hayes, R. J., Thill, C. A. (2018). Coordinated regulation of cold induced sweetening in tetraploid potato families by isozymes of UDP-glucose pyrophosphorylase and vacuolar acid invertase. Am. J. Potato Res. 95, 487–494. doi: 10.1007/s12230-018-9653-1
Spychalla, J. P., Scheffler, B. E., Sowokinos, J. R., Bevan, M. W. (1994). Cloning, antisense RNA inhibition, and the coordinated expression of UDP-glucose pyrophosphorylase with starch biosynthetic genes in potato tubers. J. Plant Physiol. 144, 444–453. doi: 10.1016/S0176-1617(11)82121-8
Swiech, L., Heidenreich, M., Banerjee, A., Habib, N., Li, Y., Trombetta, J., et al. (2015). In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 33, 102–106. doi: 10.1038/nbt.3055
Szittya, G., Silhavy, D., Molnár, A., Havelda, Z., Lovas, A., Lakatos, L., et al. (2003). Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 22, 633–640. doi: 10.1093/emboj/cdg74
Viola, R., Davies, H. V. (1992). A microplate reader assay for rapid enzymatic quantification of sugars in potato tubers. Potato Res. 35, 55–58. doi: 10.1007/BF02357723
Wiberley-Bradford, A. E., Busse, J. S., Jiang, J., Bethke, P. C. (2014). Sugar metabolism, chip color, invertase activity, and gene expression during long-term cold storage of potato (Solanum tuberosum) tubers from wild-type and vacuolar invertase silencing lines of katahdin. BMC Res. Notes 7, 1–10. doi: 10.1186/1756-0500-7-801
Xiong, X., Tai, G. C. C., Seabrook, J. E. A., Wehling, P. (2002). Effectiveness of selection for quality traits during the early stage in the potato breeding population. Plant Breed. 121, 441–444. doi: 10.1046/j.1439-0523.2002.00699.x
Yasmeen, A., Shakoor, S., Azam, S., Bakhsh, A., Shahid, N., Latif, A., et al. (2022). CRISPR/Cas-mediated knockdown of vacuolar invertase gene expression lowers the cold-induced sweetening in potatoes. Planta 256, 1–5. doi: 10.1007/s00425-022-04022-x
Zhang, L., Häusler, R. E., Greiten, C., Hajirezaei, M. R., Haferkamp, I., Neuhaus, H. E., et al. (2008). Overriding the co-limiting import of carbon and energy into tuber amyloplasts increases the starch content and yield of transgenic potato plants. Plant Biotechnol. J. 6, 453–464. doi: 10.1111/j.1467-7652.2008.00332.x
Zhang, H., Liu, X., Liu, J., Ou, Y., Lin, Y., Li, M., et al. (2013). A novel RING finger gene, SbRFP1, increases resistance to cold-induced sweetening of potato tubers. FEBS Lett. 587, 749–755. doi: 10.1016/j.febslet.2013.01.066
Zhu, X., Richael, C., Chamberlain, P., Busse, J. S., Bussan, A. J., Jiang, J., et al. (2014). Vacuolar invertase gene silencing in potato (Solanum tuberosum l.) improves processing quality by decreasing the frequency of sugar-end defects. PloS One 9, e93381. doi: 10.1371/journal.pone.0093381
Zrenner, R., Schüler, K., Sonnewald, U. (1996). Soluble acid invertase determines the hexose-to-sucrose ratio in cold-stored potato tubers. Planta 198, 246–252. doi: 10.1007/BF00206250
Keywords: cold induced sweetening, hpRNA, potato, reducing sugar, RNAi, UDP-glucose pyrophosphorylase
Citation: Jaiswal S, Paul K, Raman KV, Tyagi S, Saakre M, Tilgam J, Bhattacharjee S, Vijayan J, Mondal KK, Sreevathsa R and Pattanayak D (2023) Amelioration of cold-induced sweetening in potato by RNAi mediated silencing of StUGPase encoding UDP-glucose pyrophosphorylase. Front. Plant Sci. 14:1133029. doi: 10.3389/fpls.2023.1133029
Received: 28 December 2022; Accepted: 30 January 2023;
Published: 17 February 2023.
Edited by:
Muhammad Ahsan Altaf, Hainan University, ChinaReviewed by:
Rahat Sharif, Yangzhou University, ChinaCopyright © 2023 Jaiswal, Paul, Raman, Tyagi, Saakre, Tilgam, Bhattacharjee, Vijayan, Mondal, Sreevathsa and Pattanayak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Debasis Pattanayak, ZGViYXNpc3BhdHRhbmF5YWtAeWFob28uY28uaW4=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.