
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 February 2023
Sec. Technical Advances in Plant Science
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1131064
This article is part of the Research Topic Exploring Complex Biosphere Molecular Signaling Networks: Plant-Microbes Symbiosis at Microscopic to Macroscopic Levels View all 8 articles
Magnesium (Mg) is an essential element for plant growth and development. Rice is an important food crop in the world, but there are few studies on the uptake and translocation of Mg2+ in rice. We used a multi-parent advanced generation inter-cross (MAGIC) population constructed using four parental lines and genotyped by a 55 K rice SNP array for association analysis to locate QTLs related to Mg2+ uptake and translocation in rice at the seedling stage. Four QTLs (qRMg1, qRMg2, qRMg7 and qRMg8) were detected for the root Mg2+ concentration, which explained 11.45-13.08% of the phenotypic variation. The Mg2+ transporter gene, OsMGT1, was within the region of qRMg1. Three QTLs (qSMg3, qSMg7 and qSMg10) were detected for the shoot Mg2+ concentration, which explained 4.30-5.46% of the phenotypic variation. Two QTLs (qTrMg3 and qTrMg8) were found to affect the translocation of Mg2+ from the roots to the shoots, and explained 10.91% and 9.63% of phenotypic variation. qSMg3 and qTrMg3 might be the same, since they are very close to each other on chromosome 3. Analysis of candidate genes in the region of qSMg3 and qTrMg3 through qRT-PCR, complementation assay in the yeast Mg2+ transport-defective mutant CM66, and sequence analysis of the parental lines suggested that LOC_Os03g04360 may play important roles in Mg2+ uptake, translocation and accumulation in rice. Overexpression of LOC_Os03g04360 can significantly increase the Mg2+ concentration in rice seedlings, especially under the condition of low Mg2+ supply.
As a key macronutrient, magnesium (Mg) plays an important role in plant growth, development and reproductive success (Verbruggen and Hermans, 2013). The ionic form of magnesium (Mg2+) is the most abundant divalent cation in plant cells, and its most prominent role may be its function as a central atom of chloroplast molecules in photosynthesis (Rissler et al., 2002; Cakmak, 2013). Mg is also a necessary element in carbohydrate partitioning (Cakmak et al., 1994; Cakmak, 2013). Lack of magnesium reduces the rate of photosynthesis and disrupts the distribution of carbohydrates from source to sink in plants, while inhibiting the growth of plant organs, eventually leading to a significant decrease in crop quality and productivity (Aitken et al., 1999; Rissler et al., 2002; Verbruggen and Hermans, 2013). The only effective form of Mg absorption by plants is Mg2+, which has the smallest ionic radius but the largest hydrated ionic radius in cations (Maguire and Cowan, 2002). This unique chemical property makes Mg2+ bind weakly to negative charged soil colloids and root cell walls, which leads to the loss of the exchangeable Mg easily from soil (Aitken et al., 1999; Hermans et al., 2004). Additionally, excessive application of high rates of K+ and fertilizers antagonistically interfere with plant Mg uptake, thus enhances the risk of Mg deficiency (Gransee and Führs, 2013). With the increase of crop yield and multi-cropping, Mg consumption leads to the lack of Mg in soil (Rosanoff, 2013). Therefore, crop production problems caused by magnesium deficiency have gradually become important issues in agriculture.
In view of the unique chemical property and biological significance of Mg2+, more and more studies have been conducted to gain understanding of the genetic and molecular mechanism of Mg2+ uptake, translocation and distribution in plants (Li et al., 2008; Hermans et al., 2013). MHX (Mg2+/H+ Exchanger), HKT (High-Affinity K+ Transport), CNGC (Cyclic Nucleotide-Gated Channel) and MGT (Magnesium Transporter) have been identified as Mg2+ transporters in plants (Shaul et al., 1999; Gebert et al., 2009; Tang and Luan, 2017; Zhang et al., 2017). MHX is a unique vacuolar Mg2+ transporter in Arabidopsis (Shaul et al., 1999). OsHKT2;4 has the function of low-affinity Mg2+ transporter in rice (Zhang et al., 2017). A CNGC family protein, AtCNGC10, has been indicated to mediate Mg2+ influx, particularly in the root meristem and distal elongation zones (Guo et al., 2010). MGT is the best-studied gene family of Mg2+ transporter in plants (Gebert et al., 2009; Saito et al., 2013). Although most of the members of the MGT family have Mg transport activity as proven by functional complementation with yeast and bacteria mutants, their physiological roles in plants are largely different. MGT6 can mediate Mg2+ uptake in the roots of Arabidopsis (Yan et al., 2018). Two tonoplast-localized transporters, MGT2 and MGT3, are involved in the transport of Mg2+ into vacuole in Arabidopsis (Conn et al., 2011). MGT4, MGT5 and MGT9 were found to promote pollen development and male fertility through Mg2+ influx in Arabidopsis (Chen et al., 2009; Li et al., 2015; Xu et al., 2015). MGT10 located on the chloroplast envelope membrane regulates Mg2+ homeostasis in chloroplasts of Arabidopsis (Drummond et al., 2006; Sun et al., 2017). The expression of OsMGT1 was highly induced by Mg2+ deficiency in shoots, and knockout of OsMGT1 resulted in a significant reduction in Mg2+ content and biomass at seedling stage when grown under Mg-limited conditions (Zhang et al., 2019). Knockout of OsMGT1 results in decreased Mg2+ uptake in the roots by a stable isotope 25Mg2+ uptake experiment (Chen et al., 2012). This evidence indicates that OsMGT1 is a transporter for root Mg2+ uptake in rice. Li et al. (2020) found that a chloroplast-localized Mg2+ transporter gene, OsMGT3, which is rhythmically expressed in leaf mesophyll cells, partly modulates Mg fluctuations in rice chloroplasts. OsPRR95 and OsPRR59 in rice are transcriptional repressors to negatively regulate the rhythmic expression of OsMGT3, which encodes a chloroplast-localized Mg2+ transporter (Chen et al., 2022).
As an important food crop in the world, it is of great significance to improve resistance/tolerance to various stresses in rice, including Mg deficiency. Quantitative trait locus (QTL) mapping has been widely used to analyze genetic factors of agronomic traits, including absorption and transport of metal ions (Yang et al., 2018; Gao et al., 2019; Yan et al., 2019). Norton et al. (2010) conducted a multi-element analysis of the leaves and grains of a field-grown rice F2 population, and detected eight QTLs for Mg concentration in leaves and five QTLs for Mg concentration in grains. Yang et al. (2018) identified two QTLs for Mg concentration in shoots at the mature stage through a genome-wide association study (GWAS). So far, no major QTLs have been found, fine-mapped or cloned for Mg2+ uptake and transport in rice. In this study, we aimed to illuminate the genetic basis of Mg2+ uptake and transport at the seedling stage through GWAS with a highly diverse MAGIC population genotyped using a 55 K single nucleotide polymorphisms (SNPs) array.
The MAGIC population used in this experiment was the DC1 population described by Meng et al. (2017). DC1 population was developed by four parental lines A, B, C and D, which came from different countries and had different agronomic characters (Table S1). A random sample of 215 lines formed this population were used in this study.
The paddy rice seeds were surface-sterilized with 10% (v/v) hydrogen peroxide solution for 30 minutes, washed with deionized water and germinated for 48 hours under dark condition and 30°C (Chen et al., 2020). Experiment was laid out according to an augmented randomized complete block design with the four parental lines being replicated in four blocks. A total of 24 seeds per plot were randomly sown in a 96 well PCR plate, with perforations at the bottom of the plate to facilitate the roots to fully contact with the nutrient solution (Naz et al., 2019). The full strength IRRI solution used has the following compositions: 1.0 mM MgSO4·7H2O, 1.25 mM NH4NO3, 0.3 mM KH2PO4, 1.0 mM CaCl2, 0.35 mM K2SO4, 0.5 mM Na2SiO3, 20.0 μM Fe-EDTA, 20.0 μM H3BO3, 9.0 μM MnCl2, 0.77 μM ZnSO4, 0.32 μM CuSO4, and 0.39 μM (NH4)6Mo7O24, pH 5.5. Rice seedlings were first grown in the 1/4 strength solution for two weeks, and then transferred to the full-strength solution with different Mg2+ concentrations for three weeks. The nutrient solution was replaced every three days and the pH was adjusted to 5.5 every day. Plants were grown in a growth room with a 14 h light (30°C) (8:00 – 22:00)/10 h dark (22°C)(22:00 – 8:00) and 60% relative humidity. We explored the Mg2+ concentration of four parental lines under normal growth and Mg2+ deficiency treatment. After 3-week treatment, the plants were divided into two parts: root and shoot. The tissue samples were dried at 70°C, and 24 seedlings of each line were mixed to measure the dry weight and Mg2+ concentration.
The dried samples were crushed, and then wet-digested in concentrated HNO3 at 120°C for 30 min, and then further digested with HClO4 at 180°C until the samples became transparent. Then the samples were diluted with ultrapure water. The Mg2+ concentration was determined by inductively coupled plasma mass spectrometry (ICP-MS).
Meng et al. (2017) genotyped the MAGIC population with a 55K SNPs array. Selection of high-quality SNPs for QTL mapping used a three-step filtering strategy. First, markers monomorphic among the four parents were removed. Second, set all heterozygous genotypes to “deletion” and delete markers with deletion values greater than 10%. Finally, markers with a minor allele frequency of less than 3% were deleted. The number of markers remaining was 22,160.
The MLM (Mixed Linear Model) implemented in TASSEL version 5.2.3 was used to analyze the associations between SNP markers and traits. P < 0.001 was used as the threshold to declare the significance of marker-trait associations. R2 was used to evaluate the percentage of phenotypic variance explained of related loci to phenotypic traits.
To examine the expression response of the candidate genes to Mg2+ deficiency, seedlings of rice variety Nipponbare or the four parental lines were first grown in the 1/4 strength IRRI solution for two weeks, and then cultured in the full strength IRRI solution with 1.0 mM Mg2+ or without Mg2+ for three weeks. The roots were sampled for RNA extraction. A randomized complete block design with three replicates and a plot size of 24 seedling were used to layout the experiment. Samples were taken from all the 24 seedlings of a plot and mixed for RNA extraction.
To investigate the expression pattern of the candidate genes in different organs at different growth stages, 3-week-old seedling of Nipponbare precultured hydroponically were transplanted to the paddy field in the Experimenal Farm in Shenzhen of the Agricultural Genomics Institute in Shenzhen, Chinese Academy of Agricultural Sciences. Tissue samples taken includes roots, basal stem, leaf sheath and leaf blade at the vegetative stage and roots, basal stem, lower leaf sheath, lower leaf blade, flag leaf sheath, flag leaf blade, node I–II, inter node II, peduncle, rachis, spikelet, husk and seed at the reproductive stages. A single plant was regarded as a biological replicate and three biological replicates were used.
The total RNA was extracted by Trizol (Vazyme Biotech Co. Ltd, China). Then the total RNA was reverse- transcripted with the HiScript Q RT SuperMix for qPCR kit (Vazyme Biotech Co). The AceQ Universal SYBR qPCR Master Mix kit (Vazyme Biotech Co) was used for quantitative analysis (Chen et al., 2020). The primers for qRT-PCR were shown in Supplementary Table S2.
The Mg2+ translocation ability of each candidate gene protein was examined by a yeast complementation assay. The yeast mutant CM66, which lacks plasma membrane Mg2+ transporters ALR1 and ALR2, was used (Li et al., 2001). The open reading frames of all candidate genes were amplified from the full-length cDNA of rice cv. Nipponbare, and the primer sequences were shown in Supplementary Table S3. Each candidate gene was ligated into a pYES2 vector with correct direction.
Empty vector pYES2 and candidate genes vectors were introduced into CM66 yeast cells, respectively, according to the manufacturer’s protocol (Yeast Transformation Kit; Beijing Kulaibo Technology Co. Ltd, China), and transformants were selected on synthetic dextrose medium without uracil (SD-U). Positive clones were cultured in SD-U liquid medium until the early logarithmic phase, concentrated and washed three times with sterile distilled water. After sequential 10-fold dilution, 8 μL of the cell suspension were spotted on SD-U plates containing 1, 4, 64 mmol/L MgCl2, respectively. The plates were incubated at 30°C for 3 d before the growth phenotypes were evaluated.
The growth of CM66 yeast strain transformed with various plasmids in liquid SD-U media containing Mg2+ was determined. Overnight yeast cells were prepared and the optical density (OD) at 600 nm was adjusted to 0.5 with sterile distilled water. Then, 20 μL of cell suspensions was added to 20 mL liquid SD-U media containing 4, 64, 128 mmol/L MgCl2 in each bottle. The OD values at 600 nm were determined at indicated time.
Seedlings of the four parental lines were used for DNA extraction by the DNeasy Plant Mini Kit (Qiagen, Germany). For each parental line, a single plant was used and the experiment was conducted in triplicate. The DNA samples of the four parental lines were used as templates to amplify the full-length genomic sequence and promoter of LOC_Os03g04360, LOC_Os03g04430 and LOC_Os03g04480 by the KOD-FX polymerase (Toyobo, Japan) using specific primers (Table S4). After PCR amplification, PCR products were sequenced by the Sangon Biotech Co., Ltd. (Shanghai, China). Sequences of LOC_Os03g04360, LOC_Os03g04430 and LOC_Os03g04480 from the four parental lines were aligned and analyzed using MEGA 7.0.
Using the Gateway (Invitrogen) recombi-nation reaction, the open reading frame sequence of LOC_Os03g04360 was transferred into the destination vector pCAMBIA1300. The vectors were transformed into rice as described previously (Chen et al., 2020).
We evaluated the Mg2+ translocation and accumulation in the four parental lines of the MAGIC DC1 population and its four parental lines. Two-week-old seedlings of the four parental lines with normal growth were treated with 1.0 mM Mg2+ (Figure 1A) or without Mg2+ supply (Figure 1B) for three weeks. Under the condition of 1.0 mM Mg2+ supply, no significant difference among the four parental lines was found for the Mg2+ concentration in roots or shoots (Figure 1C), nor for Mg2+ translocation (the ratio of Mg2+ accumulation in shoots to roots) (Figure 1E). After three weeks of Mg2+-free treatment, the root and shoot Mg2+ concentrations were highest in the parental line B, while the lowest in the parental line D. The root Mg2+ concentration of parental line B was 1.83 times that of the parental line D. The shoot Mg2+ concentration of B was 2.40 times that of parental line D (Figure 1D). The Mg2+ translocation of parental line B was the highest, while that of parental line D was the lowest. The Mg2+ translocation of parental line B was 2.75 times that of the parental line D (Figure 1F). The MAGIC population exhibited significant phenotypic variation for Mg2+ concentration in roots and shoots, and for Mg2+ translocation (Table 1). All the traits displayed normal distributions (Figure 2).
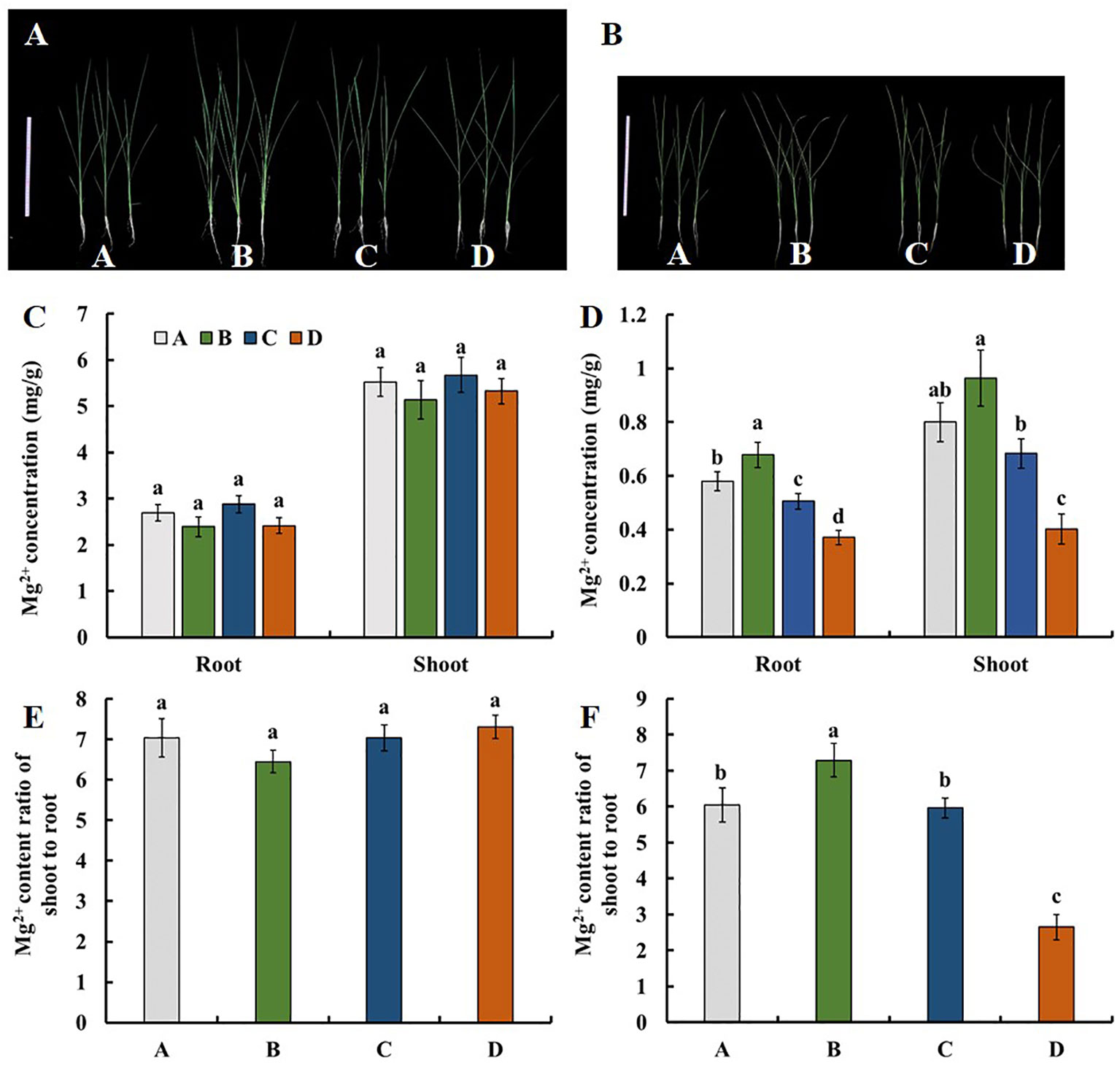
Figure 1 Mg2+ uptake and translocation of the four parental lines of the DC1 population. Rice seedlings cultured in the 1/4 strength IRRI solution for two weeks, and in the full strength IRRI solution with (A) 1 mM Mg2+ or (B) without Mg2+ for three weeks. Bar: 30 cm. The Mg2+ concentration of seedlings treated with (C) 1 mM Mg2+ or (D) without Mg2+. The shoot-to-root ratio of the Mg2+ content (Mg2+ translocation) of seedlings treated with (E) 1 mM Mg2+ or (F) without Mg2+. Values are mean ± SE (n = 4). The different letters above the bars indicate a significant difference between each line (P < 0.01).
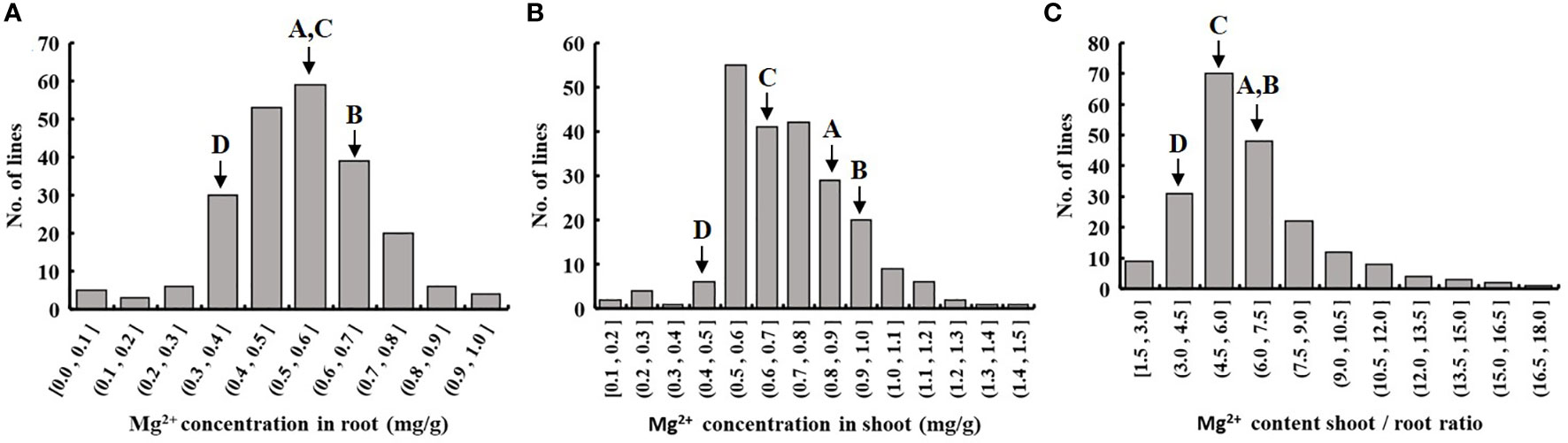
Figure 2 Frequency distribution of Mg2+ uptake and translocation traits in the DC1 population. Rice seedlings of the MAGIC population in the 1/4 strength IRRI solution for two weeks, and in the full strength IRRI solution without Mg2+ for three weeks. Frequency distribution of (A) root Mg2+ concentration, (B) shoot Mg2+ concentration and (C) shoot-to-root ratio of Mg2+ content. A, B, C and D represent the parental lines A, B, C and D.
We performed GWAS analysis for the above traits. Four QTLs for root Mg2+ concentration was located on chromosomes 1, 2, 7 and 8, respectively. They explained 13.08%, 12.32%, 11.61% and 11.45% of the phenotypic variation, respectively (Table 2 and Figures 3A, B). Three QTLs were identified for shoot Mg2+ concentration, which were distributed on chromosomes 3, 7 and 10, and explained 5.57%, 6.30% and 5.46% of phenotypic variation, respectively (Table 2 and Figures 3C, D). Two QTLs were identified for Mg2+ translocation, which were distributed on chromosomes 3 and 8, and explained 10.91% and 9.63% of phenotypic variation, respectively (Table 2 and Figures 3E, F).
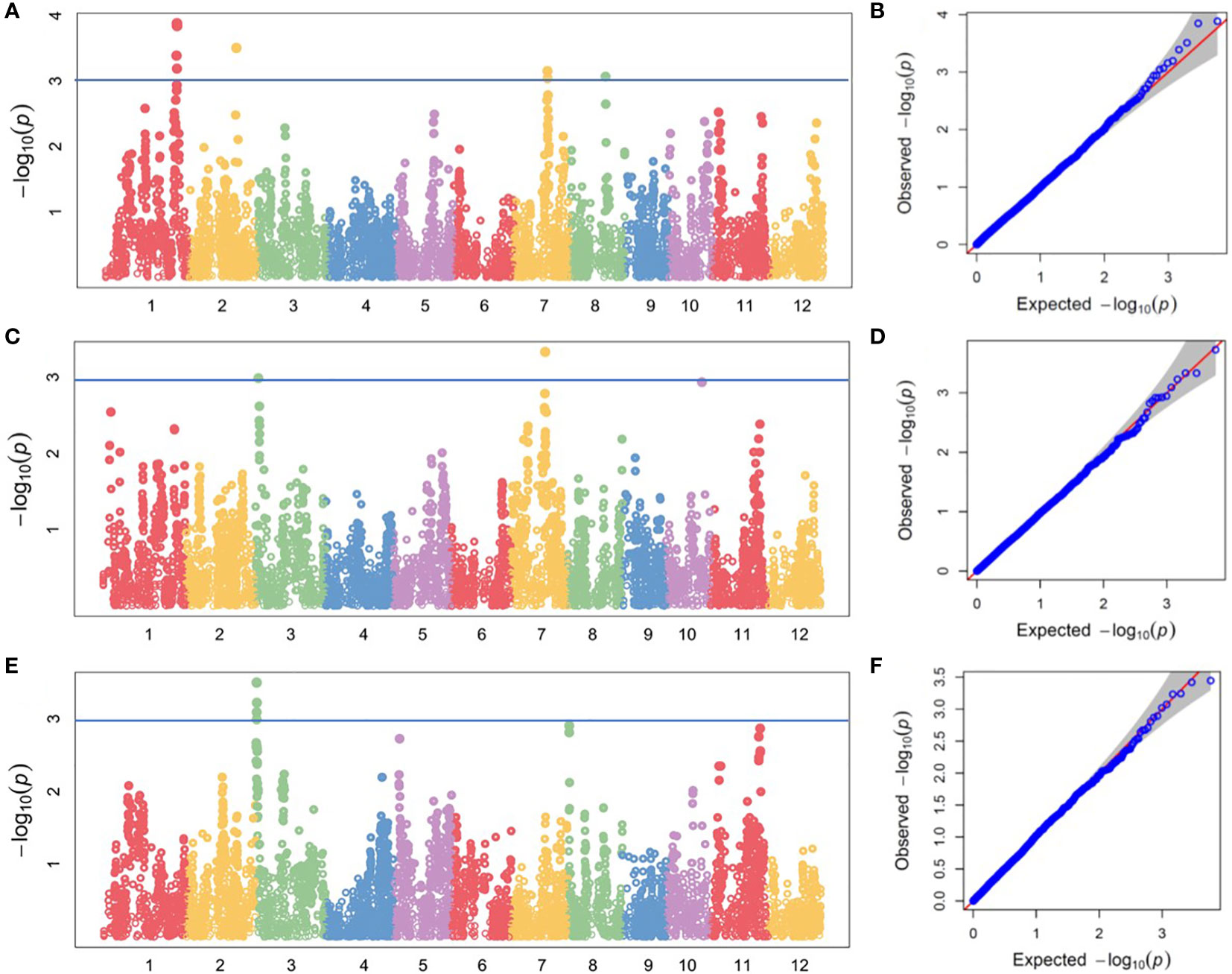
Figure 3 GWAS of magnesium uptake and translocation in the DC1 population. Manhattan plots of for (A) root Mg2+ concentration, (C) shoot Mg2+ concentration and (E) Mg2+ content ratio of shoot to root at seedling stage; Quantile-Quantile plots for (B) root Mg2+ concentration, (D) shoot Mg2+ concentration and (F) shoot-to-root ratio of Mg2+ content.
The chromosome 3 has a region harboring QTLs for two traits. They are qSMg3 for shoot Mg2+ concentration and qTrMg3 for Mg2+ translocation. Through annotation (http://rice.plantbiology.msu.edu/index.shtml) and literature information, 16 genes were chosen as the candidate genes responsible for Mg2+ translocation and accumulation. Among them, eight encode proteins with transmembrane structure (Figure S1), including OsMGT9 (LOC_Os03g04480), a member of MGT family (Table 3). Five genes including LOC_Os03g03590, LOC_Os03g03660, LOC_Os03g04360, LOC_Os03g04430 and LOC_Os03g04480 were found to be induced by Mg2+ deficiency (Figure 4).
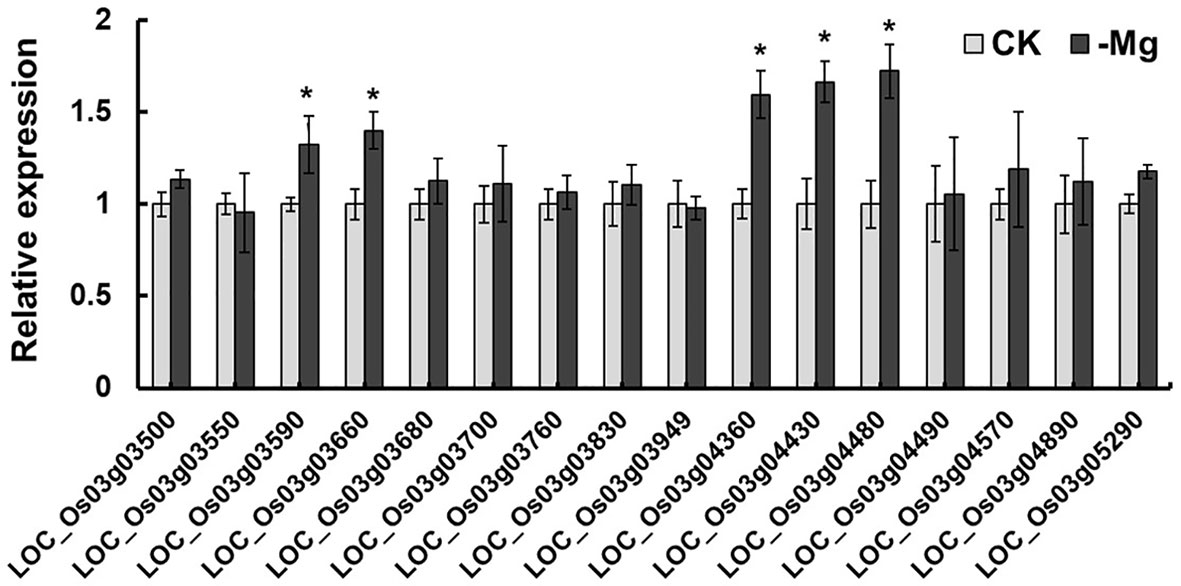
Figure 4 Relative expression of candidate genes for qSMg3 and qTrMg3 under different Mg2+ concentrations. The rice cv. Nipponbare seedlings in the 1/4 strength IRRI solution for two weeks, and in IRRI solution with 1 mM Mg2+ (CK) or without Mg2+ (-Mg) for three weeks. RNA was extracted from rice roots. Values are mean ± SE (n = 3). * A significant difference at the 0.01 level.
The five genes responsive to Mg2+ deficiency and three other genes with complete transmembrane structure (LOC_Os03g03680, LOC_Os03g03700, LOC_Os03g04570) were expressed in the yeast mutant CM66. Under the condition of 1 or 4 mmol/L Mg2+ supply under solid medium for four days, LOC_Os03g04430 increased the growth of CM66 and while LOC_Os03g04360 inhibited (Figure 5). The other seven genes, including OsMGT9 (LOC_Os03g04480), did not show any effects on the growth of CM66 (Figure 5). The experiment of liquid medium with different Mg2+ concentrations confirmed that the growth of CM66 under 4 mmol/L Mg2+ supply was increased by LOC_Os03g04430, inhibited by LOC_Os03g04360 (Figure S2).
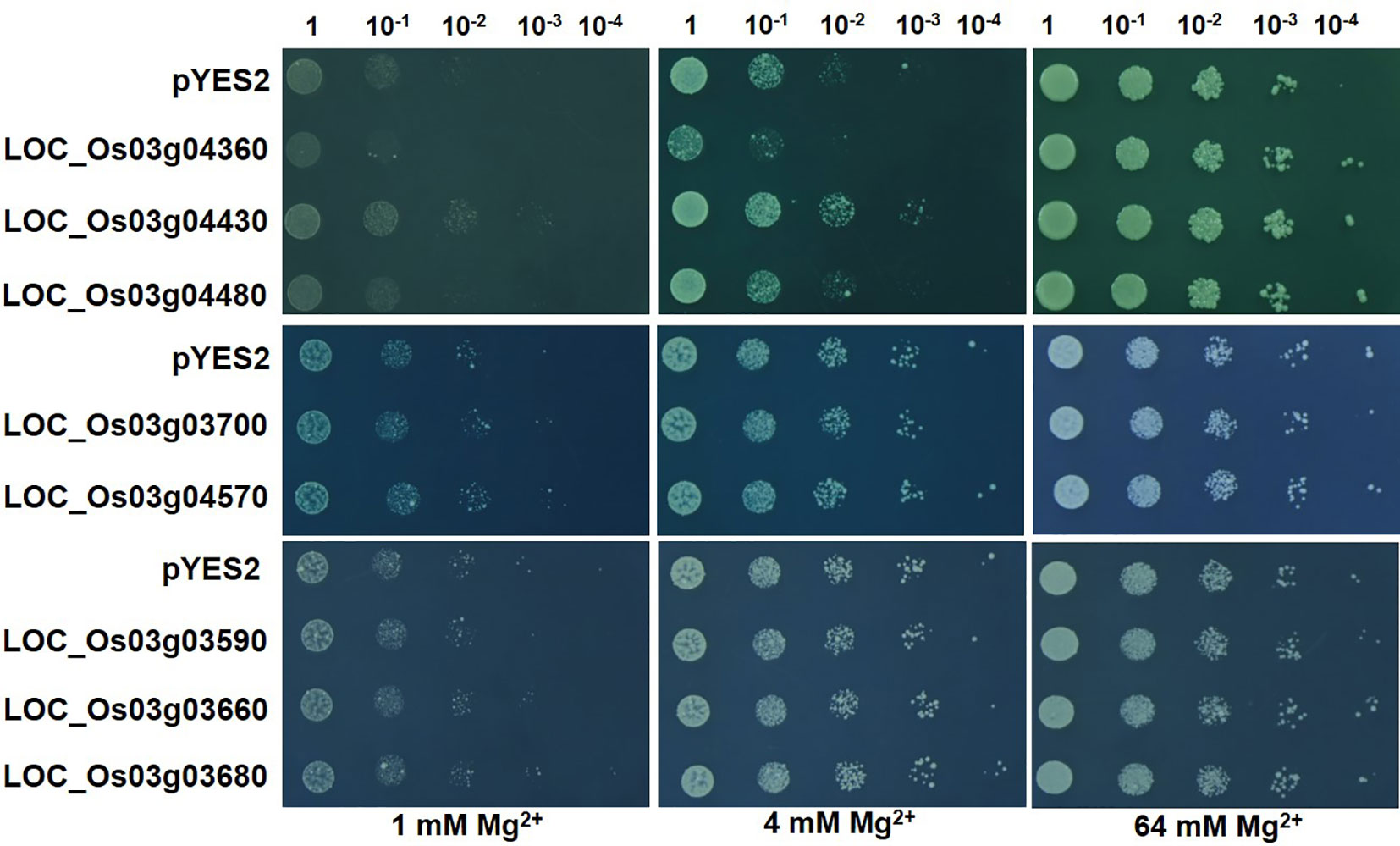
Figure 5 Expression of the candidate genes in yeast mutant CM66. Overnight yeast cell suspension of CM66 transformed with empty vector pYES2 or candidate genes were serially diluted (1:10) and spotted on the solid media containing 1, 4 or 64 mmol/L MgSO4. Pictures were taken after four days growth at 30°C.
To further screen candidate genes to obtain the target genes, the sequences of LOC_Os03g04360, LOC_Os03g04430 and LOC_Os03g04480 of the four parental lines were analyzed by PCR amplification and sequencing. The coding region sequence of LOC_Os03g04360, LOC_Os03g04430 and LOC_Os03g04480, including exon and intron, had no difference among the four parental lines (Figures 6A, S4A, B). There was no difference among A, B and C in the 3000 bp region of the upstream of the ATG and the 3’-UTR sequence after the TGA for both of LOC_Os03g04360 and LOC_Os03g04430 (Figures 6A, S3A). Compared with A, B and C, D has five single-nucleotide mutations in the 3000 bp region of the upstream of the ATG and two single-nucleotide mutations in the 3’-UTR of LOC_Os03g04360 (Figure 6A), plus a single-nucleotide mutation in the 3000 bp upstream of the ATG of LOC_Os03g04430 (Figure S3A). At the same time, we analyzed the gene sequence of LOC_Os03g04480 of the four parental lines, and found no natural variation (Figure S3B).
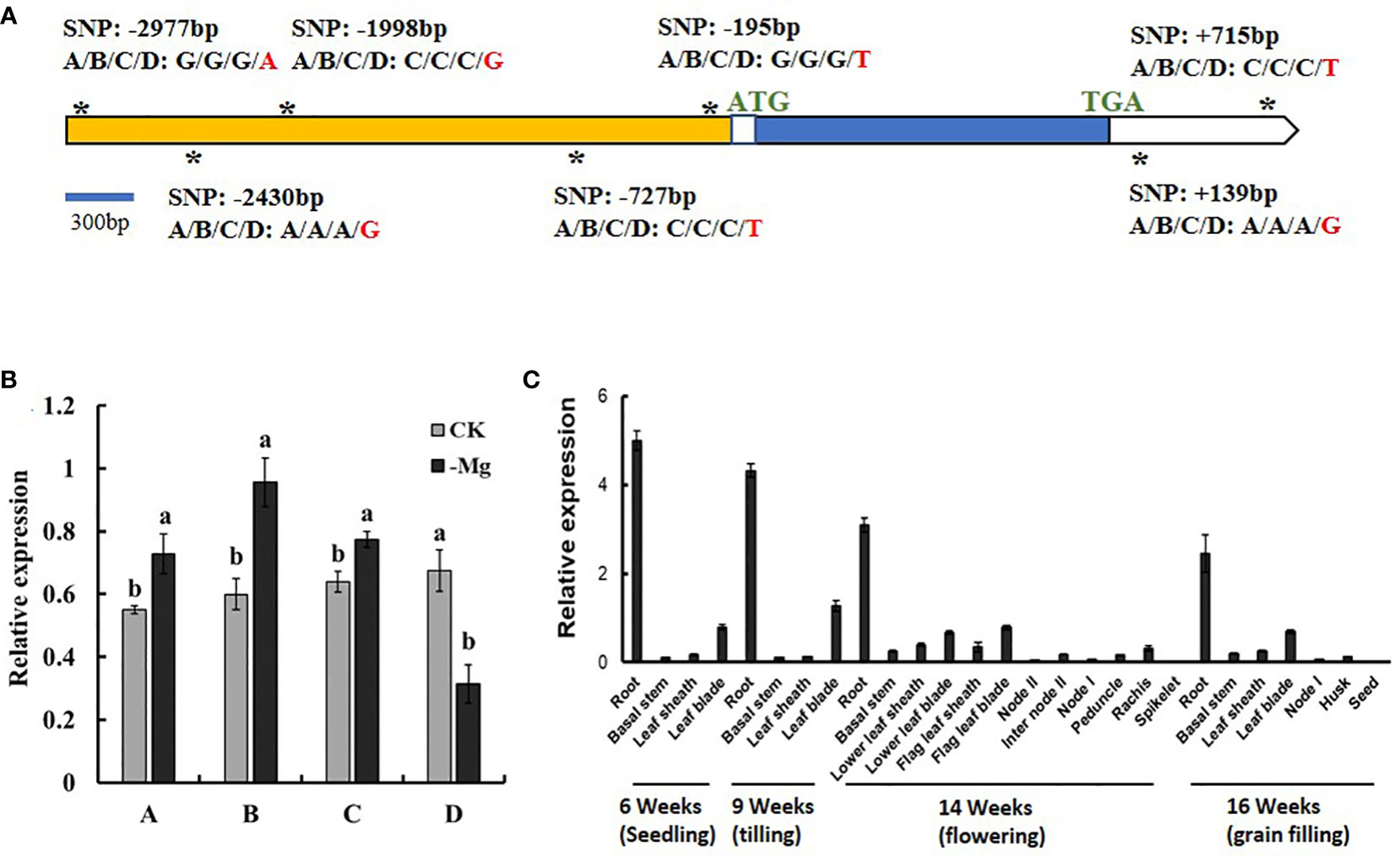
Figure 6 Sequence and expression of LOC_Os03g04360 in the four parental lines of the DC1 population. (A) Gene structure of LOC_Os03g04360 and polymorphism locations (SNPs, asterisks). The SNPs are underlined with asterisks. (B) Relative expression of LOC_Os03g04360 under different Mg2+ concentrations. The rice seedlings grown in the 1/4 strength IRRI solution for two weeks, and in the full strength IRRI solution with 1 mM Mg2+ (CK) or without Mg2+ (-Mg) for three weeks. RNA was extracted from rice roots. (C) Expression of LOC_Os03g04360 in different organs at different growth stages. RNA Samples were taken from cv. Nipponbare grown in a paddy field. Values are mean ± SE (n = 3). The different letters above the bars indicate significant difference between the control and treatments at P < 0.01.
The expression patterns of LOC_Os03g04360, LOC_Os03g04430 and LOC_Os03g04480 in the four parental lines are shown in Figures 6B, S3C, D. Compared with the normal Mg2+ supply, the expression of LOC_Os03g04360 was significantly induced by Mg2+ deficiency in A, B and C while significantly reduced in D (Figure 6B). Compared with the normal Mg2+ supply, the expression level of LOC_Os03g04430 was increased under Mg2+ deficiency in all the four parental lines (Figure S3C). LOC_Os03g04480 was significantly induced by Mg2+ deficiency in all parental lines (Figure S3D). Therefore, LOC_Os03g04430 is not considered as a candidate gene underlying qSMg3 and qTrMg3.
In order to further analyze the biological functions of LOC_Os03g04360, its expression in different organs at different growth stages were investigated. LOC_Os03g04360 strongly expressed in root at different growth stages, but only weakly expressed in other tissues (Figure 6C).
Analysis of candidate genes in the region of qSMg3 and qTrMg3 through qRT-PCR, complementation assay in the yeast Mg2+ transport-defective mutant CM66, and sequence analysis of the parental lines suggested that LOC_Os03g04360 may play important roles in Mg2+ uptake, translocation and accumulation in rice. We introduced pUbi : LOC_Os03g04360 expression construct into cv. Nipponbare (WT) using agrobacterium tumefaciens-mediated transformation. The expression of LOC_Os03g04360 in roots increased 26-42-fold in OE1 and OE2 compared with WT, but only 1-fold in OE3 (Figure 7C).
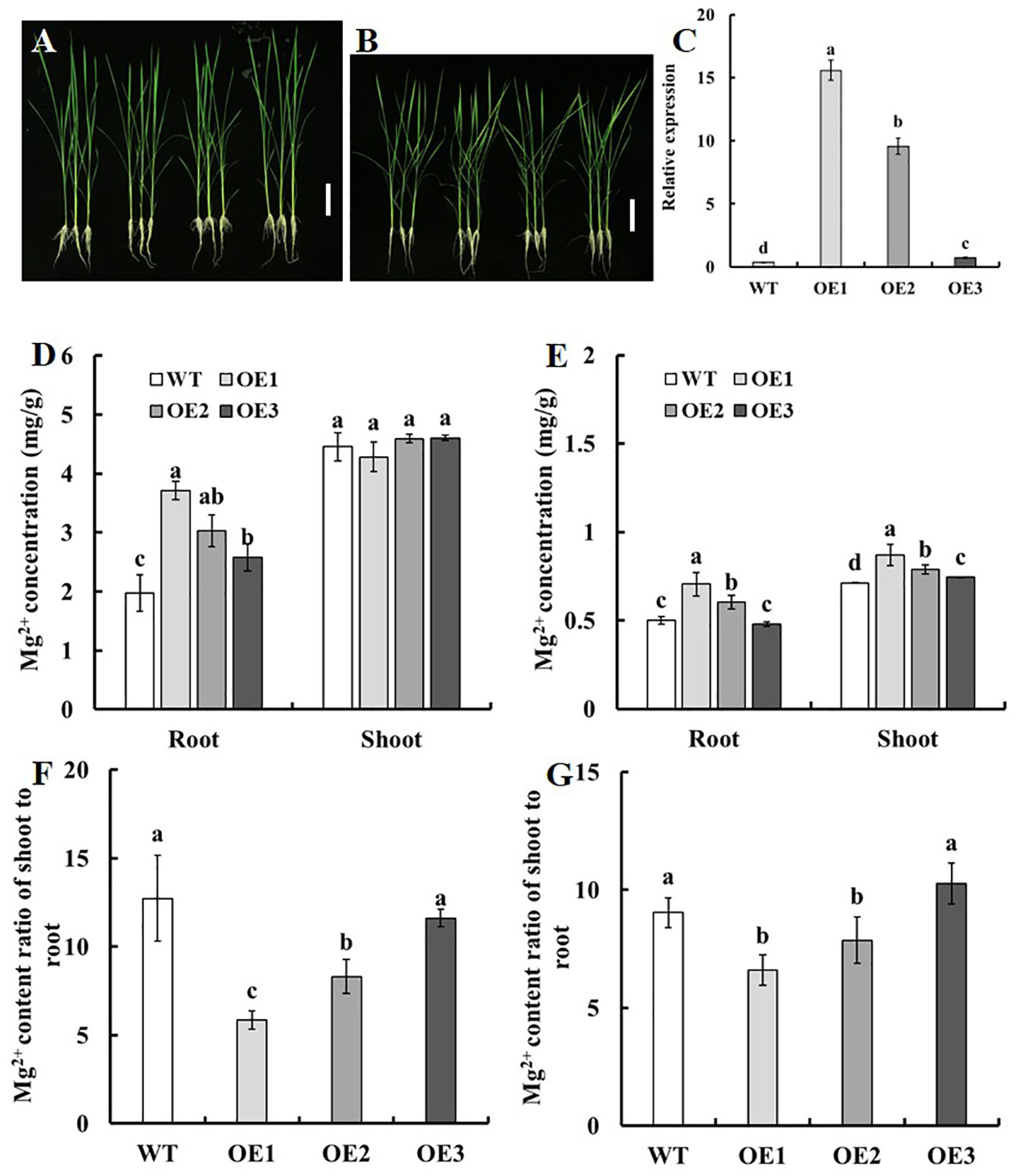
Figure 7 Mg2+ uptake and translocation of the LOC_Os03g04360 overexpression lines. Two-week-old seedlings of the transgenic lines and WT with normal growth were treated with 250 μM Mg2+ or 10 μM Mg2+ supply for two weeks. (A, B) Phenotype of the LOC_Os03g04360 overexpression lines grown with (A) 250 μM Mg2+ and (B) 10 μM Mg2+ supply. (C) Quantitative real-time PCR analysis of endogenous LOC_Os03g04360 expression in various transgenic lines and WT. RNA was collected from root. The Mg2+ concentration in roots and shoots in LOC_Os03g04360 overexpression lines and WT under (D) 250 μM Mg2+ or (E) 10 μM Mg2+ supply. The shoot-to-root ratio of the Mg2+ content of seedlings treated with (F) 250 μM Mg2+ or (G) 10 μM Mg2+. Error bars: SE (n = 3 plants). The different letters above the bars indicate a significant difference between each line (P < 0.01).
We further analyzed the effect of overexpression of LOC_Os03g04360 on Mg2+ accumulation at seedling stage. Two-week-old seedlings of the transgenic lines and WT with normal growth were treated with 250 μM Mg2+ (Figure 7A) or 10 μM Mg2+ supply (Figure 7B) for two weeks. Under the condition of 250 μM Mg2+ supply, no significant difference among the transgenic lines and WT was found for the Mg2+ concentration in shoots (Figure 7D), compared with WT, the Mg2+ concentration in roots of OE1, OE2 and OE3 increased by 87.7%, 53.4% and 30.5% respectively (Figure 7D). Under the condition of 10 μM Mg2+ supply, compared with WT, the Mg2+ concentration in roots of OE1, OE2 and OE3 increased by 40.6%, 20.2% and 0% respectively, and that in shoots increased by 21.9%, 10.5% and 5.4% respectively (Figure 7E). Under the two treatments, the Mg2+ content ratio of shoot to root of OE1 and OE2 were significantly reduced, and there was no difference between OE3 and WT (Figures 7F, G).
High-resolution mapping of rice by using MAGIC populations has been previously reported (Bandillo et al., 2013; Meng et al., 2016a; Meng et al., 2016b; Meng et al., 2017). We previously identified QTLs associated with the toxicity tolerance of rice to three essential metals (Fe, Zn, and Al) by using three MAGIC populations including DC1, DC2 and 8way populations (Meng et al., 2017). The four parents of the MAGIC DC1 population used in this study displayed substantial differences in Mg2+ uptake, translocation and accumulation under Mg2+ deficiency (Figure 1 and Table 1). In this study, we aimed to identify the loci related to Mg2+ uptake and transport in the seedling stage by screening the MAGIC DC1 population in a hydroponic system, using a 55K SNPs array, and using a mixed linear model to conduct association analysis.
In the present study, four QTLs related to the concentration of Mg2+ in roots, three QTLs related to the shoot Mg2+ concentration, and two QTLs related to the Mg2+ translocation were identified (Table 2). These QTLs did not co-locate with the QTLs of Mg concentration in leaves or shoots at the mature stage in the field of Norton et al. (2010) and Yang et al. (2018), which may be caused by differences in growth conditions and growth stages. The qRMg1 (Chr.1: 37.5 Mb) was located 187 kb away from the OsMGT1. OsMGT1 is a plasma membrane-localized transporter, and has high expression in root tips and vascular tissues (Chen et al., 2012), overexpression of OsMGT1 increased Mg2+ content under low-Mg2+ supply (Zhang et al., 2019).
A set of candidate genes for qSMg3 and qTrMg3 located on the same region of chromosome 3 were studied in the present study (Table 2).
LOC_Os03g04360, was found to be responsive to Mg2+ deficiency (Figure 4) and affected the growth of CM66 under low Mg2+ condition (Figures 5, S3). LOC_Os03g04360 inhibited the growth of CM66 under low Mg2+ supply (Figures 5, S3). LOC_Os03g04360 belongs to the phosphate transporter gene family OsPHT1 (Chiou and Lin, 2011; Secco et al., 2013). The PHT1 family promotes both Pi uptake in soil and Pi transfer in plants (Ai et al., 2009). PHT1 family members also participate in other biological processes. PvPht1;4, OsPT4, OsPT8 and PHT1;1 participate in the uptake and accumulation of As (V) by plants (Catarecha et al., 2007; Wang et al., 2016; Cao et al., 2017; Ye et al., 2017; Sun et al., 2020). OsPT8 also regulates disease resistance by regulating rice mitogen-resistant protein interaction factor kinase BWMK1 (Dong et al., 2019). Phosphate transporter OsPHT1;8 was involved in auxin signal transduction in rice roots (Jia et al., 2017). The coding region sequence of LOC_Os03g04360 had no difference among the four parental lines (Figure 6A). Compared with A, B and C, the parental line D showed five single-nucleotide mutations in the 3000 bp region of the upstream of ATG, and two single-nucleotide mutations in the 3’-UTR of LOC_Os03g04360 (Figure 6A). It is well-known that variation of promoter sequence of a gene can change its expression and function. The natural variation of the GSE5 promoter contributes to the grain size diversity of rice (Duan et al., 2017). Natural variation in OsCBL10 promoter can affect flood tolerance during seed germination of rice subspecies (Ye et al., 2018). Natural variation in OsHMA3 promoter contributes to differential grain cadmium accumulation between Indica and Japonica rice (Liu et al., 2020). Our results that Mg2+ deficiency significantly induced the expression of LOC_Os03g04360 in A, B and C but inhibited it in D (Figure 6B) could be caused by the observed variations in the promoter. The protein of LOC_Os03g04360 may have Mg2+ transport activity and participate in the Mg2+ uptake and translocation process in rice.
We introduced pUbi : LOC_Os03g04360 expression construct into Nipponbare using agrobacterium tumefaciens-mediated transformation. It was found that overexpression of LOC_Os03g04360 could significantly increase the Mg2+ concentration in rice roots under different Mg2+ supply at seedling stage (Figures 7D, E), but decreased Mg2+ content ratio of shoot to root (Figures 7F, G). Dai et al. (2022) showed that LOC_Os03g04360 was located in the plasma membrane of cells. LOC_Os03g04360 was strongly expressed in roots at different growth stages, but weakly expressed in other tissues (Figure 6C). Taken together, the present results suggested that LOC_Os03g04360 may play an important role in the uptake of Mg2+ in roots and the translocation of Mg2+ from root to shoot. It means that LOC_Os03g04360 may promote the uptake and accumulation of Mg2+ and inhibit translocation in rice. Further characterizations of LOC_Os03g04360 is needed to elucidate its functional significance in Mg2+ uptake, translocation and accumulation in rice.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Conceived and designed the experiments: JC and GY. Performed the experiments: SZ, JC, WZ, JinyL, LM and JindL. Analyzed the data: SZ, JC and JindL. Wrote and revised the paper: JC, GY and SZ. All authors contributed to the article and approved the submitted version.
This research was financially supported by the National Natural Science Foundation of China (31902103), the Guangdong Basic and Applied Basic Research Foundation (2022A1515012381), the Young Elite Scientists Sponsorship Program by CAST (2020QNRC001) and the Shenzhen Science and Technology Program (JCYJ20210324124409027 and JCYJ20200109150650397).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1131064/full#supplementary-material
Supplementary Table 1 | Description of the four parental lines used for developing the DC1 population. GID: germplasm identification number used in the International Rice Information System (http://irri.org/).
Supplementary Figure 1 | Topological model of proteins encoded by candidate genes. The probability of transmembrane location predicted by the TMHMM algorithm (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0). Red areas indicate predicted TM spans.
Supplementary Figure 2 | Expression of the candidate genes in yeast mutant CM66 in liquid medium. Yeast strains were grown in liquid media with (A) 4mmol/L MgSO4, (B) 64 mmol/L MgSO4, or (B) 128 mmol/L MgSO4 for 75 h. The absorbance at 600 nm (OD600) of cell cultures was measured every 5 h. Values are mean ± SE (n = 3). The asterisks above the bars indicate significant difference between lines at P < 0.01.
Supplementary Figure 3 | Sequence and expression of LOC_Os03g04430 and LOC_Os03g04480 in the four parental lines of the DC1 population. Gene structure of (A) LOC_Os03g04430 and (B) LOC_Os03g04480 and polymorphism locations (SNPs, asterisks). The SNPs are underlined with asterisks. Relative expression of (C) LOC_Os03g04430 and (D) LOC_Os03g04480 under different Mg2+ concentrations. The rice seedlings in the 1/4 strength IRRI solution for two weeks, and in the full strength IRRI solution with 1 mM Mg2+ (CK) or without Mg2+ (-Mg) for three weeks. RNA was extracted from rice roots. Values are mean ± SE (n = 3). The different letters above the bars indicate significant difference between the control and treatments at P < 0.01.
Ai, P., Sun, S., Zhao, J., Fan, X., Xin, W., Guo, Q., et al. (2009). Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 57 (5), 798–809. doi: 10.1111/j.1365-313X.2008.03726.x
Aitken, R. L., Dickson, T., Hailes, K. J., Moody, P. W. (1999). Response of field-grown maize to applied magnesium in acidic soils in north-eastern Australia. Aust. J. Agric. Res. 50 (2), 191–198. doi: 10.1071/A98149
Bandillo, N., Raghavan, C., Muyco, P. A., Sevilla, M. A. L., Lobina, I. T., Dilla-Ermita, C. J., et al. (2013). Multi-parent advanced generation inter-cross (MAGIC) populations in rice: Progress and potential for genetics research and breeding. Rice 6 (1), 11. doi: 10.1186/1939-8433-6-11
Cakmak, I. (2013). Magnesium in crop production, food quality and human health. Plant Soil 368 (1), 1–4. doi: 10.1007/s11104-013-1781-2
Cakmak, I., Hengeler, C., Marschner, H. (1994). Changes in phloem export of sucrose in leaves in response to phosphorus, potassium and magnesium deficiency in bean plants. J. Exp. Bot. 45 (9), 1251–1257. doi: 10.1093/jxb/45.9.1251
Cao, Y., Sun, D., Ai, H., Mei, H., Liu, X., Sun, S., et al. (2017). Knocking out OsPT4 gene decreases arsenate uptake by rice plants and inorganic arsenic accumulation in rice grains. Environ. Sci. Technol. 51 (21), 12131–12138. doi: 10.1021/acs.est.7b03028
Catarecha, P., Segura, M. D., Franco-Zorrilla, J. M., García-Ponce, B., Lanza, M., Solano, R., et al. (2007). A mutant of the arabidopsis phosphate transporter PHT1;1 displays enhanced arsenic accumulation. Plant Cell 19 (3), 1123–1133. doi: 10.1105/tpc.106.041871
Chen, J., Li, L., Liu, Z., Yuan, Y., Guo, L., Mao, D., et al. (2009). Magnesium transporter AtMGT9 is essential for pollen development in arabidopsis. Cell Res. 19 (7), 887–898. doi: 10.1038/cr.2009.58
Chen, J., Liu, X., Liu, S., Fan, X., Zhao, L., Song, M., et al. (2020). Co-Overexpression of OsNAR2.1 and OsNRT2.3a increased agronomic nitrogen use efficiency in transgenic rice plants. Front. Plant Sci. 11, 1245. doi: 10.3389/fpls.2020.01245
Chen, C. Q., Tian, X. Y., Li, J., Bai, S., Zhang, Z. Y., Li, Y., et al. (2022). Two central circadian oscillators OsPRR59 and OsPRR95 modulate magnesium homeostasis and carbon fixation in rice. Mol. Plant 15 (10), 1602–1614. doi: 10.1016/j.molp.2022.09.008
Chen, Z. C., Yamaji, N., Motoyama, R., Nagamura, Y., Ma, J. F. (2012). Up-regulation of a magnesium transporter gene OsMGT1 is required for conferring aluminum tolerance in rice. Plant Physiol. 159 (4), 1624–1633. doi: 10.1104/pp.112.199778
Chiou, T.-J., Lin, S.-I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62 (1), 185–206. doi: 10.1146/annurev-arplant-042110-103849
Conn, S. J., Conn, V., Tyerman, S. D., Kaiser, B. N., Leigh, R. A., Gilliham, M. (2011). Magnesium transporters, MGT2/MRS2-1 and MGT3/MRS2-5, are important for magnesium partitioning within arabidopsis thaliana mesophyll vacuoles. New Phytol. 190 (3), 583–594. doi: 10.1111/j.1469-8137.2010.03619.x
Dai, C., Dai, X., Qu, H., Men, Q., Liu, J., Yu, L., et al. (2022). The rice phosphate transporter OsPHT1;7 plays a dual role in phosphorus redistribution and anther development. Plant Physiol. 188 (4), 2272–2288. doi: 10.1093/plphys/kiac030
Dong, Z., Li, W., Liu, J., Li, L., Pan, S., Liu, S., et al. (2019). The rice phosphate transporter protein OsPT8 regulates disease resistance and plant growth. Sci. Rep. 9 (1), 5408. doi: 10.1038/s41598-019-41718-9
Drummond, R. S. M., Tutone, A., Li, Y. C., Gardner, R. C. (2006). A putative magnesium transporter AtMRS2-11 is localized to the plant chloroplast envelope membrane system. Plant Sci. 170 (1), 78–89. doi: 10.1016/j.plantsci.2005.08.018
Duan, P., Xu, J., Zeng, D., Zhang, B., Geng, M., Zhang, G., et al. (2017). Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant 10 (5), 685–694. doi: 10.1016/j.molp.2017.03.009
Gao, Z., Wang, Y., Chen, G., Zhang, A., Yang, S., Shang, L., et al. (2019). The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat. Commun. 10 (1), 5207. doi: 10.1038/s41467-019-13110-8
Gebert, M., Meschenmoser, K., Svidová, S., Weghuber, J., Schweyen, R., Eifler, K., et al. (2009). A root-expressed magnesium transporter of the MRS2/MGT gene family in arabidopsis thaliana allows for growth in low-Mg2+ environments. Plant Cell. 21 (12), 4018–4030. doi: 10.1105/tpc.109.070557
Gransee, A., Führs, H. (2013). Magnesium mobility in soils as a challenge for soil and plant analysis, magnesium fertilization and root uptake under adverse growth conditions. Plant Soil 368 (1-2), 5–21. doi: 10.1007/s11104-012-1567-y
Guo, K. M., Babourina, O., Christopher, D. A., Borsic, T., Rengel, Z. (2010). The cyclic nucleotide-gated channel AtCNGC10 transports Ca2+ and Mg2+ in arabidopsis. Physiol. Plant 139 (3), 303–312. doi: 10.1111/j.1399-3054.2010.01366.x
Hermans, C., Conn, S., Chen, J., Xiao, Q., Verbruggen, N. (2013). An update on magnesium homeostasis mechanisms in plants. Metallomics 5 (9), 1170–1183. doi: 10.1039/c3mt20223b
Hermans, C., Johnson, G. N., Strasser, R. J., Verbruggen, N. (2004). Physiological characterisation of magnesium deficiency in sugar beet: Acclimation to low magnesium differentially affects photosystems I and II. Planta 220 (2), 344–355. doi: 10.1007/s00425-004-1340-4
Jia, H., Zhang, S., Wang, L., Yang, Y., Zhang, H., Cui, H., et al. (2017). OsPht1;8, a phosphate transporter, is involved in auxin and phosphate starvation response in rice. J. Exp. Bot. 68 (18), 5057–5068. doi: 10.1093/jxb/erx317
Li, J., Huang, Y., Tan, H., Yang, X., Tian, L., Luan, S., et al. (2015). An endoplasmic reticulum magnesium transporter is essential for pollen development in arabidopsis. Plant Sci. 231, 212–220. doi: 10.1016/j.plantsci.2014.12.008
Li, L.-G., Sokolov, L. N., Yang, Y.-H., Li, D.-P., Ting, J., Pandy, G. K., et al. (2008). A mitochondrial magnesium transporter functions in arabidopsis pollen development. Mol. Plant 1 (4), 675–685. doi: 10.1093/mp/ssn031
Li, L., Tutone, A. F., Drummond, R. S., Gardner, R. C., Luan, S. (2001). A novel family of magnesium transport genes in arabidopsis. Plant Cell 13 (12), 2761–2775. doi: 10.1105/tpc.010352
Li, J., Yokosho, K., Liu, S., Cao, H. R., Yamaji, N., Zhu, X. G., et al. (2020). Diel magnesium fluctuations in chloroplasts contribute to photosynthesis in rice. Nat. Plants 6 (7), 848–859. doi: 10.1038/s41477-020-0686-3
Liu, C., Gao, Z., Shang, L., Yang, C., Ruan, B., Zeng, D., et al. (2020). Natural variation in the promoter of OsHMA3 contributes to differential grain cadmium accumulation between indica and japonica rice. J. Integr. Plant Biol. 62 (3), 314–329. doi: 10.1111/jipb.12794
Maguire, M. E., Cowan, J. A. (2002). Magnesium chemistry and biochemistry. Biometals 15 (3), 203–210. doi: 10.1023/A:1016058229972
Meng, L., Guo, L., Ponce, K., Zhao, X., Ye, G. (2016a). Characterization of three indica rice multiparent advanced generation intercross (MAGIC) populations for quantitative trait loci identification. Plant Genome 9, 2. doi: 10.3835/plantgenome2015.10.0109
Meng, L., Wang, B., Zhao, X., Ponce, K., Qian, Q., Ye, G. (2017). Association mapping of ferrous, zinc, and aluminum tolerance at the seedling stage in indica rice using MAGIC populations. Front. Plant Sci. 8, 1822 . doi: 10.3389/fpls.2017.01822
Meng, L., Zhao, X., Ponce, K., Ye, G., Leung, H. (2016b). QTL mapping for agronomic traits using multi-parent advanced generation inter-cross (MAGIC) populations derived from diverse elite indica rice lines. Field Crops Res. 189, 19–42. doi: 10.1016/j.fcr.2016.02.004
Naz, M., Luo, B., Guo, X., Li, B., Chen, J., Fan, X. (2019). Overexpression of nitrate transporter OsNRT2.1 enhances nitrate-dependent root elongation. Genes 10 (4), 290. doi: 10.3390/genes10040290
Norton, G. J., Deacon, C. M., Xiong, L., Huang, S., Meharg, A. A., Price, A. H. (2010). Genetic mapping of the rice ionome in leaves and grain: Identification of QTLs for 17 elements including arsenic, cadmium, iron and selenium. Plant Soil 329 (1), 139–153. doi: 10.1007/s11104-009-0141-8
Rissler, H. M., Collakova, E., DellaPenna, D., Whelan, J., Pogson, B. J. (2002). Chlorophyll biosynthesis. expression of a second chl I gene of magnesium chelatase in arabidopsis supports only limited chlorophyll synthesis. Plant Physiol. 128 (2), 770–779. doi: 10.1104/pp.010625
Rosanoff, A. (2013). Changing crop magnesium concentrations: Impact on human health. Plant Soil 368 (1), 139–153. doi: 10.1007/s11104-012-1471-5
Saito, T., Kobayashi, N. I., Tanoi, K., Iwata, N., Suzuki, H., Iwata, R., et al. (2013). Expression and functional analysis of the CorA-MRS2-ALR-Type magnesium transporter family in rice. Plant Cell Physiol. 54 (10), 1673–1683. doi: 10.1093/pcp/pct112
Secco, D., Jabnoune, M., Walker, H., Shou, H., Wu, P., Poirier, Y., et al. (2013). Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell. 25 (11), 4285–4304. doi: 10.1105/tpc.113.117325
Shaul, O., Hilgemann, D. W., de-Almeida-Engler, J., Van Montagu, M., Inzé, D., Galili, G. (1999). Cloning and characterization of a novel Mg2+/H+ exchanger. EMBO J. 18 (14), 3973–3980. doi: 10.1093/emboj/18.14.3973
Sun, D., Feng, H., Li, X., Ai, H., Sun, S., Chen, Y., et al. (2020). Expression of new pteris vittata phosphate transporter PvPht1;4 reduces arsenic translocation from the roots to shoots in tobacco plants. Environ. Sci. Technol. 54 (2), 1045–1053. doi: 10.1021/acs.est.9b05486
Sun, Y., Yang, R., Li, L., Huang, J. (2017). The magnesium transporter MGT10 is essential for chloroplast development and photosynthesis in arabidopsis thaliana. Mol. Plant 10 (12), 1584–1587. doi: 10.1016/j.molp.2017.09.017
Tang, R.-J., Luan, S. (2017). Regulation of calcium and magnesium homeostasis in plants: from transporters to signaling network. Curr. Opin. Plant Biol. 39, 97–105. doi: 10.1016/j.pbi.2017.06.009
Verbruggen, N., Hermans, C. (2013). Physiological and molecular responses to magnesium nutritional imbalance in plants. Plant Soil 368 (1), 87–99. doi: 10.1007/s11104-013-1589-0
Wang, P., Zhang, W., Mao, C., Xu, G., Zhao, F.-J. (2016). The role of OsPT8 in arsenate uptake and varietal difference in arsenate tolerance in rice. J. Exp. Bot. 67 (21), 6051–6059. doi: 10.1093/jxb/erw362
Xu, X.-F., Wang, B., Lou, Y., Han, W.-J., Lu, J.-Y., Li, D.-D., et al. (2015). Magnesium transporter 5 plays an important role in mg transport for male gametophyte development in arabidopsis. Plant J. 84 (5), 925–936. doi: 10.1111/tpj.13054
Yan, Y.-W., Mao, D.-D., Yang, L., Qi, J.-L., Zhang, X.-X., Tang, Q.-L., et al. (2018). Magnesium transporter MGT6 plays an essential role in maintaining magnesium homeostasis and regulating high magnesium tolerance in arabidopsis. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00274
Yan, H., Xu, W., Xie, J., Gao, Y., Wu, L., Sun, L., et al. (2019). Variation of a major facilitator superfamily gene contributes to differential cadmium accumulation between rice subspecies. Nat. Commun. 10 (1), 2562. doi: 10.1038/s41467-019-10544-y
Yang, M., Lu, K., Zhao, F.-J., Xie, W., Ramakrishna, P., Wang, G., et al. (2018). Genome-wide association studies reveal the genetic basis of ionomic variation in rice. Plant Cell. 30 (11), 2720–2740. doi: 10.1105/tpc.18.00375
Ye, Y., Li, P., Xu, T., Zeng, L., Cheng, D., Yang, M., et al. (2017). OsPT4 contributes to arsenate uptake and transport in rice. Front. Plant Sci. 8, 2197. doi: 10.3389/fpls.2017.02197
Ye, N.-H., Wang, F.-Z., Shi, L., Chen, M.-X., Cao, Y.-Y., Zhu, F.-Y., et al. (2018). Natural variation in the promoter of rice calcineurin b-like protein10 (OsCBL10) affects flooding tolerance during seed germination among rice subspecies. Plant J. 94 (4), 612–625. doi: 10.1111/tpj.13881
Zhang, C., Li, H., Wang, J., Zhang, B., Wang, W., Lin, H., et al. (2017). The rice high-affinity k+ transporter OsHKT2;4 mediates Mg2+ homeostasis under high-Mg2+ conditions in transgenic arabidopsis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01823
Keywords: Mg2+ uptake, Mg2+ translocation, association analysis, quantitative trait loci (QTL), MAGIC population, rice
Citation: Zhi S, Zou W, Li J, Meng L, Liu J, Chen J and Ye G (2023) Mapping QTLs and gene validation studies for Mg2+ uptake and translocation using a MAGIC population in rice. Front. Plant Sci. 14:1131064. doi: 10.3389/fpls.2023.1131064
Received: 24 December 2022; Accepted: 13 February 2023;
Published: 23 February 2023.
Edited by:
Ashok Samuel, Waseda University, JapanReviewed by:
Zhenyu Gao, China National Rice Research Institute (CAAS), ChinaCopyright © 2023 Zhi, Zou, Li, Meng, Liu, Chen and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingguang Chen, Y2hlbmpnMjhAbWFpbC5zeXN1LmVkdS5jbg==; Guoyou Ye, Zy55ZUBpcnJpLm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.