
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 07 March 2023
Sec. Plant Symbiotic Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1130924
This article is part of the Research Topic Molecular Markers of Plant Adaptation to Multiple Environmental Stressors View all 3 articles
 Yu Gao1
Yu Gao1 Siyuan Huang2
Siyuan Huang2 Yujie Wang1
Yujie Wang1 Hongxin Lin3
Hongxin Lin3 Zhiyong Pan4
Zhiyong Pan4 Shubao Zhang1
Shubao Zhang1 Jie Zhang2
Jie Zhang2 Wenquan Wang2
Wenquan Wang2 Shanhan Cheng1*
Shanhan Cheng1* Yinhua Chen1*
Yinhua Chen1*Introduction: Plants and arbuscular mycorrhizal fungi (AMF) mutualistic interactions are essential for sustainable agriculture production. Although it is shown that AMF inoculation improves cassava physiological performances and yield traits, the molecular mechanisms involved in AM symbiosis remain largely unknown. Herein, we integrated metabolomics and transcriptomics analyses of symbiotic (Ri) and asymbiotic (CK) cassava roots and explored AM-induced biochemical and transcriptional changes.
Results: Three weeks (3w) after AMF inoculations, proliferating fungal hyphae were observable, and plant height and root length were significantly increased. In total, we identified 1,016 metabolites, of which 25 were differentially accumulated (DAMs) at 3w. The most highly induced metabolites were 5-aminolevulinic acid, L-glutamic acid, and lysoPC 18:2. Transcriptome analysis identified 693 and 6,481 differentially expressed genes (DEGs) in the comparison between CK (3w) against Ri at 3w and 6w, respectively. Functional enrichment analyses of DAMs and DEGs unveiled transport, amino acids and sugar metabolisms, biosynthesis of secondary metabolites, plant hormone signal transduction, phenylpropanoid biosynthesis, and plant-pathogen interactions as the most differentially regulated pathways. Potential candidate genes, including nitrogen and phosphate transporters, transcription factors, phytohormone, sugar metabolism-related, and SYM (symbiosis) signaling pathway-related, were identified for future functional studies.
Discussion: Our results provide molecular insights into AM symbiosis and valuable resources for improving cassava production.
Plants’ adaptation to diverse ecological systems worldwide depends on many factors, among which their ability to develop mutualistic interactions with symbionts plays important roles (Parniske, 2008; Chen et al., 2022). Arbuscular mycorrhiza (AM) is the most widespread symbiotic association between AM fungi (AMF) and plant roots (Bapaume and Reinhardt, 2012; Ho-Plágaro and García-Garrido, 2022b). The majority of landplant species interact with AMF from the Glomeromycotina phylum (Pimprikar and Gutjahr, 2018; Ho-Plágaro and García-Garrido, 2022b). This reciprocal relationship is highly beneficial and reposed on a bidirectional nutrient exchange between fungi and host plants (Ho-Plágaro and García-Garrido, 2022a). Host plants provide AMF with carbon (photosynthates and fatty acids), while in return, fungi supply plants with essential mineral nutrients, principally phosphorus (P) and nitrogen (N) (Parniske, 2008; Püschel et al., 2020; Zai et al., 2021; Ma et al., 2022; Ho-Plágaro and García-Garrido, 2022a). Moreover, AM symbiosis improves plant growth, production, and tolerance to various biotic and abiotic stresses (Sabra et al., 2018; Gao et al., 2020; Zhang et al., 2020; Jajoo, 2021; Lin et al., 2021; Jumrani et al., 2022). Accordingly, AMF have become symbionts of several research interests and are recommended for wide use as bio-fertilizers to enhance crops’ quality and productivity under the ever-changing climate and to ensure sustainable agriculture (Igiehon and Babalola, 2017; Begum et al., 2019).
AM symbiosis development can be divided into four distinct stages (Gutjahr, 2014; Diédhiou and Diouf, 2018). The first stage, pre−contact signaling, consists of cross-talk between the two symbionts via diffusible signal molecules (fungi mainly exudate lipo-chitooligosaccharides, while plants strigolactones, STs). The second step, contact between plant roots and fungal hyphae, consists of physical contact between the two partners, followed by the beginning of hyphopodium formation on root surfaces. The third step, the intra−radical proliferation of the fungal hyphae, consists of the penetration and growth of fungal hyphae in the apoplast of the cortex, altering their typical appearance. The last step, arbuscules formation, consists of fungal hyphaes’ penetration and proliferation in the inner cortex, causing colonization surface (Diédhiou and Diouf, 2018). Arbuscules are the main sites of nutrient exchange between the two symbionts, and their development induces de novo synthesis of the peri arbuscular membrane to surround the cytoplasmic membrane (Abdallah et al., 2014; Diédhiou and Diouf, 2018). The AMF recognition by the host plants is mediated by a common symbiosis (SYM) signaling pathway, which is partially shared with Rhizobium-legume symbiosis (Bonfante and Genre, 2010; Genre and Russo, 2016). The SYM signaling pathway and symbionts interactions are mainly regulated by an interplay between TFs and phytohormones (auxin, gibberellin, ABA, STs, and ethylene) (Gutjahr, 2014; Diédhiou and Diouf, 2018; Jiang et al., 2018; Müller and Harrison, 2019; Faizan et al., 2020; Liu et al., 2020; Tominaga et al., 2020; Mitra et al., 2021). Among them, GRAS TFs, specifically the genes NSP1 and RAM1, play essential roles (Floss et al., 2013; Nagae et al., 2014; Hohnjec et al., 2015; Rich et al., 2017; Hartmann et al., 2019; Ho-Plágaro et al., 2019; Ho-Plágaro and García-Garrido, 2022b). Arbuscules’ formation causes changes in the expression patterns of many genes in the AM roots, leading to variation in primary and secondary metabolites and production improvement (Ren et al., 2019; Sakamoto et al., 2019; Shtark et al., 2021; Kaur et al., 2022; Mishra et al., 2022; Zhao et al., 2022). For instance, the expression of plant transporter family genes is significantly induced during AM symbiosis (Porcel et al., 2016; Kameoka et al., 2019; Banasiak et al., 2021; Rui et al., 2022). It is shown that AM-induced molecular mechanisms differ according to the species, genotype, AMF type, and growing conditions (Tsiokanos et al., 2022). Therefore, dissecting the AM-induced mechanisms and regulation in diverse plant species will enable the establishment of an efficient agro-biotechnological approach for using AM in sustainable agriculture and improve the economic and quality values of crops.
Cassava (Manihot esculenta Crantz) is a perennial shrub that belongs to the Euphorbiaceae family (Blagbrough et al., 2010). Also called yucca or manioc, cassava originated in South America, from where it was subsequently introduced to tropical and subtropical regions of Asia and Africa (Blagbrough et al., 2010; Mombo et al., 2016). Its tuberous roots are valuable food sources in developing countries and are extensively used to produce starch, bioethanol, and other bio-based products, such as medicine, feed, biopolymers, and cosmetics (Li et al., 2017). Among carbohydrate food sources, cassava ranks fourth in the tropics after rice, maize, and sugar cane (Blagbrough et al., 2010). In Asia, cassava drives the rural economy of several countries as it is cultivated by over 8 million farmers (Malik et al., 2020). Accordingly, one of the main breeding objectives in cassava is to improve storage root and starch yield to ensure the availability of food supply in the current situation of the growing population (Sonnewald et al., 2020). Previous studies have demonstrated that AMF inoculation improved cassava root fresh weight, mineral nutrition, total biomass, and productivity (Ceballos et al., 2013; Aliyu et al., 2018). However, the molecular mechanisms involved in AM symbiosis in cassava are unknown. With the availability of genome information on cassava (Wang et al., 2014; Hu et al., 2021), deciphering AM-induced molecular changes will offer important resources to optimize the crop production and tolerance abilities to meet food and market demand. Furthermore, it is demonstrated that cassava and fungal genetic variation and genotype × genotype specifications regulate exchanges between the two partners (Mateus et al., 2019; Savary et al., 2020). Hence, identifying key biochemical markers and candidate genes may facilitate the efficient use of AMF in improving cassava production. Knowledge of differentially accumulated metabolites during AM symbiosis in cassava will facilitate the understanding of molecular interactions and provide metabolic markers for discriminating efficient symbiosis. Metabolomics analysis is an efficient and widely used molecular approach to assess the metabolome underlying organisms’ phenotype and investigate the variability of metabolites among different organs, varieties, and species of the same taxa (Scalbert et al., 2011; Dossou et al., 2021). Moreover, it helps understand biological processes and metabolic pathways (Farag et al., 2018; Dossou et al., 2022).
In the present study, we investigated the impact of AMF (Rhizophagus irregularis, DOAM197198) inoculation on cassava growth characteristics. We mainly examined metabolome and transcriptional changes during AM symbiosis in cassava via comparative metabolomics and transcriptomics analyses and unveiled key metabolites, pathways, and potential candidate genes. Our findings provide insights into AM symbiosis and valuable resources for cassava improvement.
Huanan No. 9, an edible Manihot esculenta Crantz variety widely cultivated in China, and the fungus Rhizophagus irregularis (Ri), strain DOAM197198 were used as the host and AMF in this study.
Selected stems of Huanan No. 9, with the same thickness and size, were divided into uniform lengths (15cm). Next, the stem pieces were soaked in a mixed solution of acetamiprid, carbendazim, and acetamiprid to remove bacteria, fungi, eggs, etc. Then they were grown in pots (diameter of 15 cm) filled with a sterilized (high temperature of 121°C and high-pressure steam for 1 h) substrate at 2/3 height of the pot (Figure S1). The substrate was composed of a mixture of river sand and vermiculite at a ratio of 4:1. Prior to sown the pieces of stem in the substrate, about 800 spores (saturated in 1 g of distilled water) were spread on the surface of the substrate of the treatment group (to induce the AM symbiosis) and covered with an amount of the sterilized substrate until the stems could stand upright in the pot. An equal amount of sterilized substrate was added to the control group. In total, three groups, including two treatment groups (Ri, inoculated with the AMF) and one control group (CK, without inoculation), were formed. Each group was composed of 20 replicates. The Ri groups were Ri-3w (allowed to grow up to three weeks after inoculation) and Ri-6w (allowed to grow up to six weeks after inoculation). The pots were maintained in a greenhouse at 28°C and 16 h photoperiod (16 h light and 8 h darkness). All the pots (CK and Ri) were irrigated every week with 100 ml of low-phosphorus Hoagland nutrient solution (phosphorus content of 20 µM).
CK and Ri-3w root samples were collected three weeks after treatment, while Ri-6w roots were sampled after six weeks. The entire root system was cut off with scissors in clean Petri dishes and gently rinsed with tap water to remove the substrate. For each treatment, two groups of samples in three replications were prepared, one for microscopic observations and the other was immediately frozen-dried in liquid nitrogen, followed by storing at -80°C for metabolomics analysis and transcriptome sequencing. From our microscopic observations, the root starts to contact with the soil mycorrhizal layer and symbiosis occurs at 3 weeks, while a stable symbiosis occurs at 6 weeks. Therefore, the comparison between CK and Ri-3w is expected to show the changes of root response at the initial stage of symbiosis. The changes in root response from the initial stage of symbiosis to the stable symbiosis stage will be elucidated by comparing Ri-3w and Ri-6w samples. Samples of 3- weeks after inoculation were selected for metabolomics and transcriptomics analyses based on microscopic observations and to investigate molecular mechanisms at the initial stage of fungal hyphae colonization of cassava roots. The Ri-6w samples were taken to examine dynamic changes in the accumulation of DAMs and expression patterns of DEGs in inoculated plants.
At the sampling time, some physiological performances were evaluated, including plant height, stem thickness, root length, root weight (biomass), and leaf chlorophyll content. For the root biomass, root samples were oven dried at 60 °C for 72 hours. The total leaf chlorophyll content was measured on three fully opened leaves, with a SPAD (single-photon avalanche diode) meter on the plants. Three technical measures per leaf were conducted.
The WGA (wheat germ agglutinin) fluorescent dye staining method was used for the mycorrhizal staining directly after the samples’ collection. The root samples were introduced in a clean 50 ml centrifuge tube containing 10% KOH solution. Then they were fixed in FAA (formaldehyde alcohol acetic acid) solution for 24 h, followed by washing with distilled water. Next, root samples were placed in a water bath at 90°C for 5 minutes and then soaked again in a 10% KOH solution. After removing the KOH by rinsing three to five times with purified water, root samples were subsequently immersed in a 2% HCl solution, gently stretched using tweezers, left at room temperature for 15-30 minutes, and then washed four to five times in distilled water. Following, root samples were soaked in PBS buffer for 30 minutes; gently stretched with tweezers; and then washed to discard the PBS buffer. Finally, the root samples were placed on a clean 2 ml centrifuge tube, and a fresh mixture solution of PBS buffer and WGA488 (V:V = 1000:1) was added. The tubes were wrapped in tin foil paper and kept overnight in the refrigerator at 4°C. The following day, the symbiotic cells were observed using scanning electron microscopy (SEM, S-3000N, Hitachi Co., Ltd., Matsuda, Japan).
The widely targeted metabolomics profiling of root samples was carried out at MetWare Co., Ltd. (Wuhan, China). Briefly, 100 mg of each root sample (beforehand vacuum freeze-dried) was ground into a fine powder and dissolved in 1 mL of methanol (70%), vortex-mixed and extracted overnight at 4°C. Thereafter, extracts were centrifugated at 12,000 g for 10 minutes, and supernatants were collected and filtered with a 0.22 μm microporous membrane. All samples’ final extract was stored in a vial for subsequent metabolomics analysis. All sample extracts were mixed equally to form the quality control (QC) samples. The data acquisition system was a UPLC-MS/MS (ultra-performance liquid chromatography-mass spectrometry) platform composed of a UPLC (SHIMADZU Nexera X2, www.shimadzu.com.cn/) and a tandem mass spectrometry (MS/MS) (Applied Biosystems 4500 QTRAP, http://sciex.com/). The qualitative identification and quantification of metabolites; and multivariate analyses, including orthogonal partial least squares discriminant analysis (OPLS-DA), principal component analysis (PCA), hierarchical clustering analysis (HCA), differentially accumulated metabolites (DAMs) analysis, and functional annotation and enrichment analyses were conducted as per reported methods (Chen et al., 2013; Dossou et al., 2021; Dossou et al., 2022). Significant DAMs were filtered out at the thresholds of VIP (variable importance in projection) ≥ 1 and t-test p < 0.05.
The total RNA extraction from cassava root samples, subsequent integrity and quality checking, sequencing on Illumina Hiseq platform, construction of cDNA library, and de novo assembly were carried out as per recently described methods (Sakamoto et al., 2019; Wang et al., 2020). Then the clean reads were mapped into the cassava reference genome (version 6.1, https://phytozome-next.jgi.doe.gov/info/Mesculenta_v6_1) (Bredeson et al., 2016), with the TopHat2 software (Kim et al., 2013). The parameter of no more than one mismatch was accepted in the alignment (Kim et al., 2013).
The expression level of each gene was normalized to the number of FPKM (Fragments Per Kilobase of transcript per Million reads) using the Cufflinks 2.0 software (Trapnell et al., 2012). The DEGs were identified using the DESeq2 software at thresholds of |log2Fold Change| ≥ 1 and p-value < 0.5 (Love et al., 2014). The functional annotations of DEGs were conducted via GO (gene ontology) and KEGG (Kyoto encyclopedia for genes and genomes) pathway enrichment analyses. The Blast2GO and KOBAS2.0 programs were used for the GO and KEGG analyses, respectively (Kanehisa and Goto, 2000; Conesa et al., 2005).
The establishment of mutualistic interactions between plant roots and beneficial microorganisms is governed by a common pathway (the SYM signaling pathway) which has been well characterized in Medicago truncatula (Bonfante and Genre, 2010; Genre and Russo, 2016). To identify SYM signaling pathway-related genes in cassava, M. truncatula SYM pathway genes were downloaded from the phytozome website and used for blast analysis against the cassava genome (Bredeson et al., 2016). Further, the DEGs related to this pathway were screened out as potential candidate genes for future studies.
To validate the transcriptome data, twelve genes with varying expression patterns were randomly selected for qRT-PCR analysis. The analysis was performed on LightCycler480 (Roche, Switzerland) real-time PCR system, with ChamQ™ SYBR1 qPCR Master Mix (Vazyme Biotech, Nanjing, China) (Song et al., 2021). The EF-1α (elongation factor 1-α) gene was used as the internal control. The specific primers of each gene are listed in Table S1. The expression level of each gene was computed via the 2¯ΔΔCT method (Livak and Schmittgen, 2001).
Statistical analyses and graphing were conducted using GraphPad Prism v9.0.0121 (GraphPad 159 Software Inc., La Jolla, CA, USA). The data are presented as the mean ± SD, and statistical differences were achieved by t-test at P < 0.05. Heatmaps of gene expression patterns were carried out in TBtools software (Chen et al., 2020).
To confirm the establishment of AM symbiosis between the inoculated cassava roots and the fungi, we conducted microscopic observations on root samples at three and six weeks (3w and 6w) after inoculation. The results showed that symbiotic cells were already established at 3w, and proliferating fungal hyphae could be observed (Figures 1A–D). After 6w of inoculation, the fungal colonized a large surface of root cells, and hyphopodium could be observed (Figure 1D). As shown in Figures S1A, S1B, the AM symbiosis improved the physiological performances of inoculated cassava plants (Ri). Compared to the control plants (CK), the plant height and root length of Ri plants were significantly higher (Figures 1E, G). The stem thickness and root biomass (root dry weight) of Ri plants were slightly increased compared to CK (Figures 1F, H); however, the differences were not statistically significant. There was no difference in the chlorophyll contents of CK and Ri plants (Figure 1I).
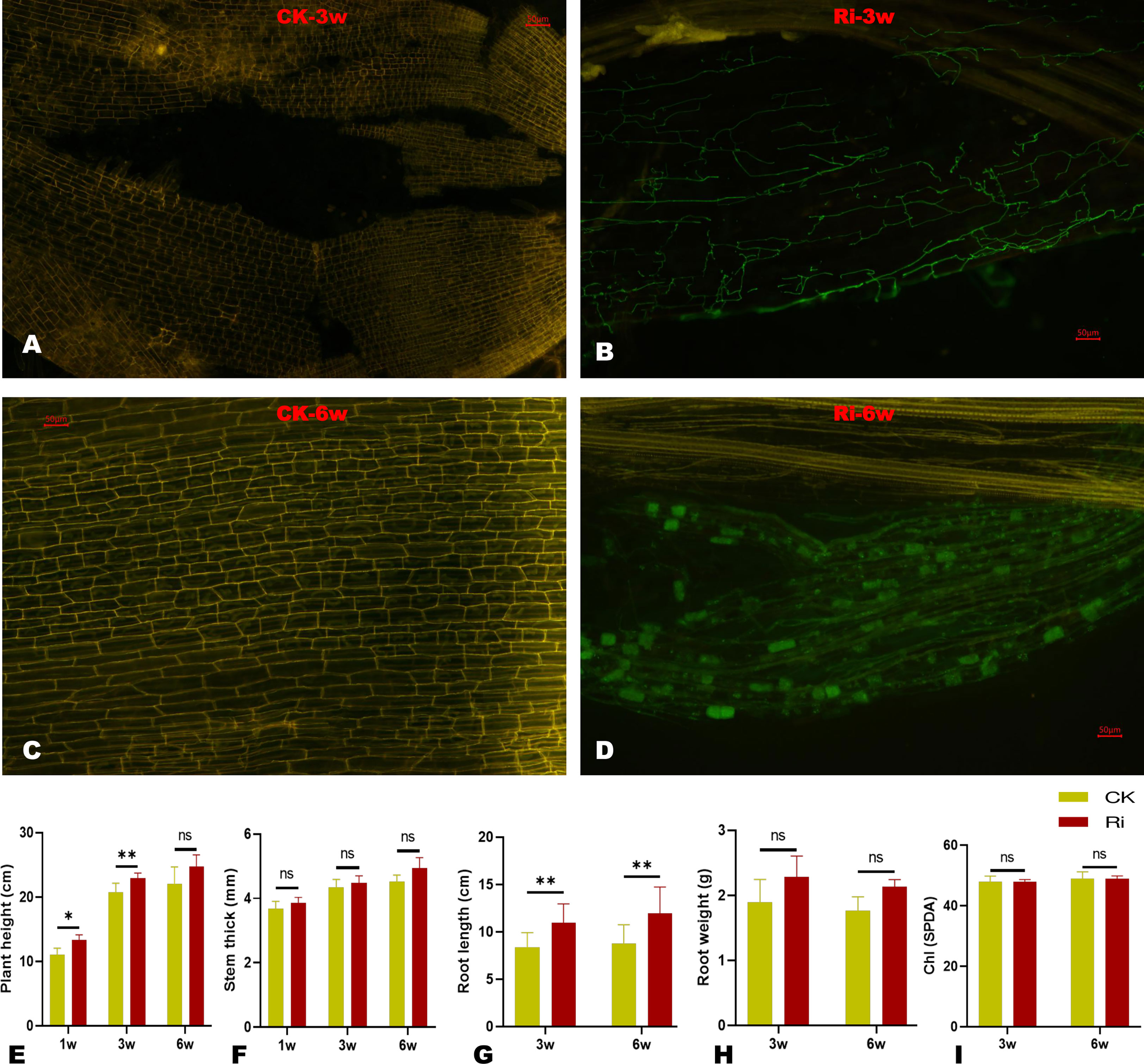
Figure 1 Microscopic observation and impacts of AM symbiosis on some physiological traits after three (3w) and six (6w) weeks of AMF inoculation. (A, B) Microscopic image of symbiotic cells at 3w of control (CK) and inoculated plants (Ri), respectively. (C, D) Microscopic images of symbiotic cells at 6w of control (CK) and inoculated plants (Ri), respectively. The green organelles in D and F indicate proliferating fungal hyphae. Pictures were taken at 10x, and scale bars indicate 50 µm. (E–J) Evaluation of some physiological traits, including plant height, stem thickness, root length, root dry weight, and leaf chlorophyll content, respectively. The data are presented as mean ± SD of three replicates. Comparisons were evaluated between CK and Ri at each time point using t-test at P < 0.05. * and ** indicate significant differences at P < 0.05 and P < 0.01, respectively. “ns” indicate not significantly different.
To examine the influence of AMF inoculation on cassava plants’ metabolism, we carried out metabolomics analysis of CK (3w) and Ri (3w and 6w) root samples. In total, 1016 metabolites, including 529 and 487 at the positive and negative ions, respectively, were identified and classified (Table S2). The main classes of metabolites identified were flavonoids (18.21%), followed by phenolic acids (16.83%), lipids (16.34%), amino acids and derivatives (10.43%), organic acids (8.07%), saccharides and alcohols (6.69%), alkaloids (6.50%), and nucleotides and derivatives (6.40%) (Figure S2).
Hierarchical cluster analysis (HCA) and Principal component analysis (PCA) allow for exploring the variability of metabolites among samples of the same and different groups. As presented in Figure 2, the CK, Ri-3w, and Ri-6w samples were clustered separately on the PCA and HCA plots, indicating that their metabolite profiles were very different. Some metabolites were specifically induced in the roots of Ri plants (Figure 2B). To confirm the observed metabolite variation, we performed an OPLS-DA analysis. The results were supportive (Figure S2). We obtained strong goodness of fit (R2X > 0.643, R2Y> 0.997) and high predictability (Q2 > 0.777) of the models (Figure S3).
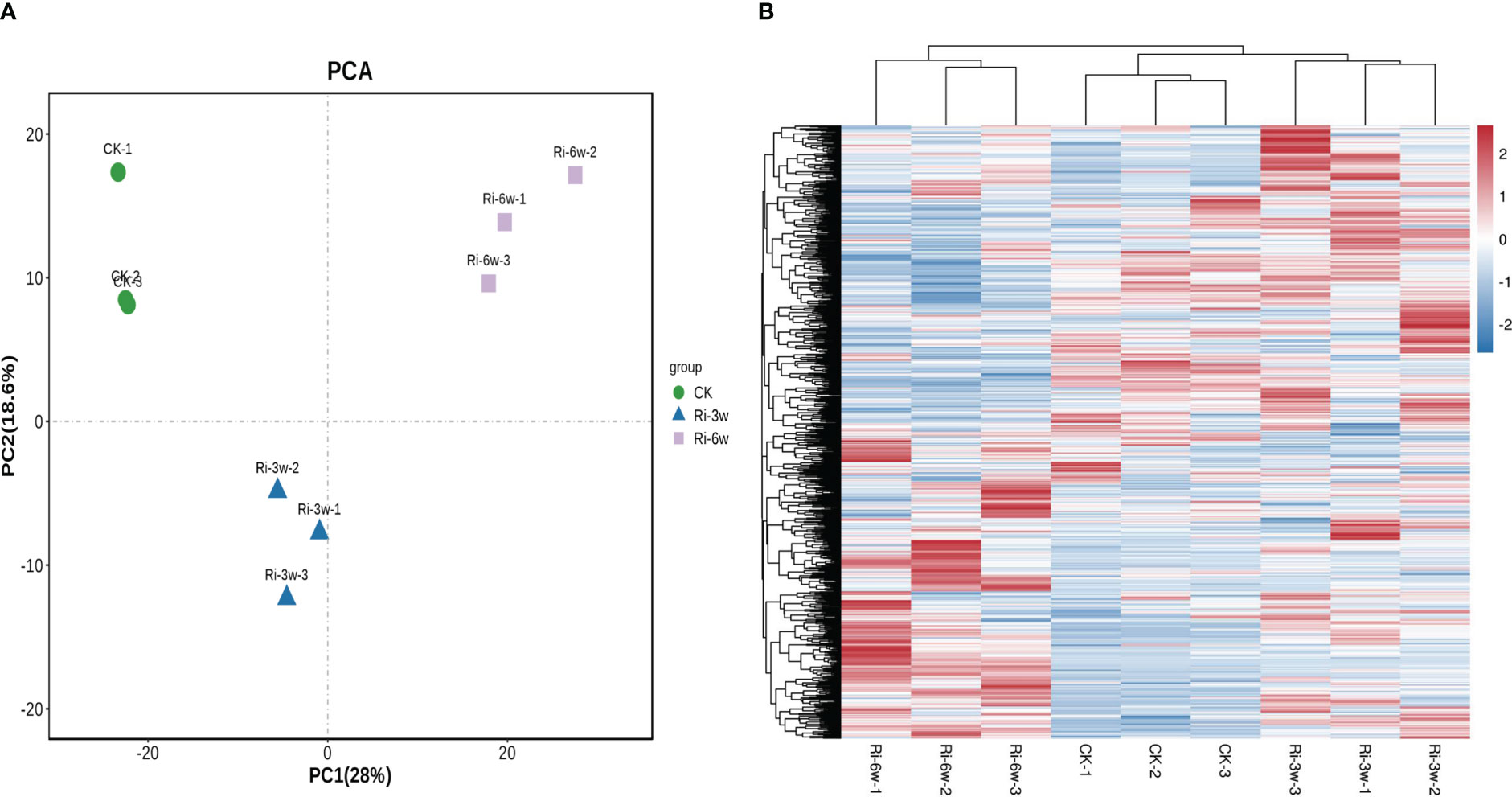
Figure 2 Principal component (A) and hierarchical clustering analyses (B) of asymbiotic (CK) and symbiotic (Ri) cassava roots based on their respective metabolite profiles. 3w and 6w indicate three and six weeks after AMF inoculation, respectively.
In order to identify major metabolites involved in AM symbiosis in cassava, we performed DAMs analysis. Significant DAMs in pairwise comparison between groups were detected at thresholds of VIP ≥ 1 and p-value < 0.05. The volcano plots are shown in Figure S4. The results indicate that there were 25 (22 up-regulated) significantly differential metabolites in pairwise comparison between CK and Ri-3w (Figure 3A; Table S3). Meanwhile, 73 DAMs, including 40 up-regulated and 33 down-regulated, were identified between CK and Ri-6w (Figure 3A; Table S4). Venn diagram analysis showed that only eight DAMs overlapped between the two pairwise comparisons (Figure 3B), indicating a dynamic metabolome change in AM-inoculated cassava roots. Heatmap clustering analysis revealed that 5-aminolevulinic acid, L-glutamic acid, and lysoPC 18:2 were the most significantly induced metabolites in inoculated cassava (Figure 3C). The classification of the DAMs between CK and Ri-6w is shown in Figure 3D. It was noteworthy that 14 (14/18) flavonoids were down-regulated, while 7 (7/9) and 9 (9/10) nucleotides and amino acids, respectively, were up-regulated at 6w (Figure 3D).
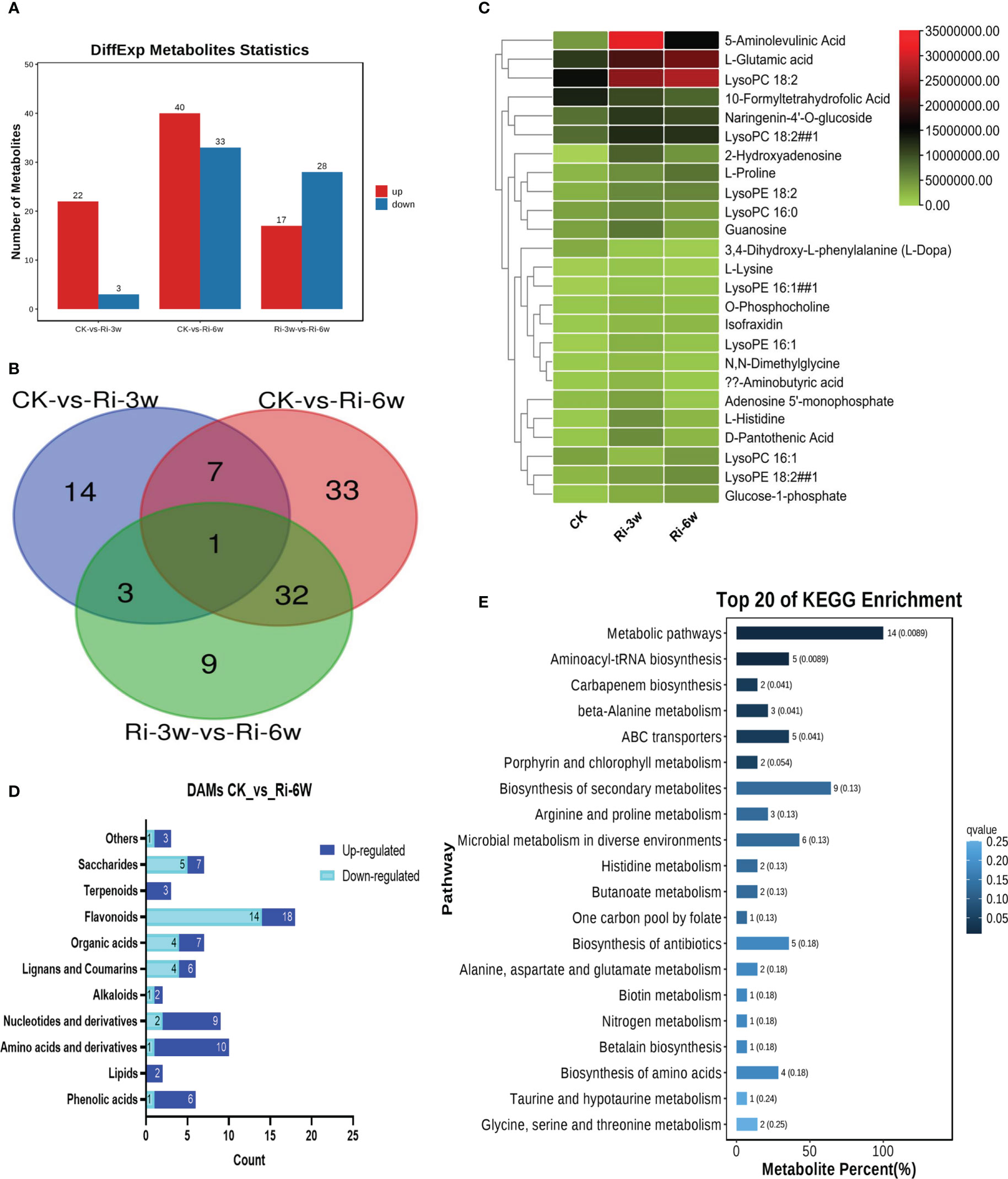
Figure 3 Differentially accumulated metabolites (DAMs) in pairwise comparison between groups and functional analysis. (A) Number of up- and down-regulated DAMs between groups. (B) Venn diagram showing the number of common DAMs shared by groups. (C) Heatmap of DAMs in between CK and Ri-3w. (D) Classification of DAMs between CK and Ri-6w. (E) KEGG annotation and enrichment result of DAMs between CK and Ri-3w.
To explore the molecular mechanisms involved in AM symbiosis in cassava, we carried out functional annotation and enrichment analyses of DAMs. The results showed that the DAMs between CK and Ri-3w were primarily involved in secondary metabolites biosynthesis, microbial metabolism in diverse environments, ABC transporters, aminoacyl-tRNA biosynthesis, biosynthesis of antibiotics, and amino acids metabolism (Figure 3E; Table S5). In addition to these pathways, the DAMs between CK and Ri-6w were mainly assigned to carbon and sugar metabolisms (Figure S5).
To gain insight into the molecular mechanisms involved in AM symbiosis in cassava, we conducted transcriptome analyses of CK, Ri-3w, and Ri-6w root samples. The summary of the transcriptome sequencing is presented in Table 1. The raw data and clean data ranged from 5.78 to 7.17 Gb and 5.71 to 7.07 Gb, respectively (Table 1). The Q20 varied from 97.45 to 97.76%, while the Q30 ranged from 92.95 to 93.68%. The clean reads were further mapped into the cassava reference genome, and the unique and total mapped reads varied from 74.18 to 90.99% and 75.71 to 92.86%, respectively (Table 1). These results show the high quality of the RNA-seq data. To examine the influence of AMF inoculation on cassava transcriptome, we conducted correlation and principal component analysis (PCA) of samples (Figure S6). The results revealed that the transcriptomes of CK and Ri-3w were not too much different. Samples of Ri-6w clustered separately and could be discriminated by PC1 (72.2%), suggesting a dynamic transcriptome change in inoculated cassava along with root development (Figure S6B).
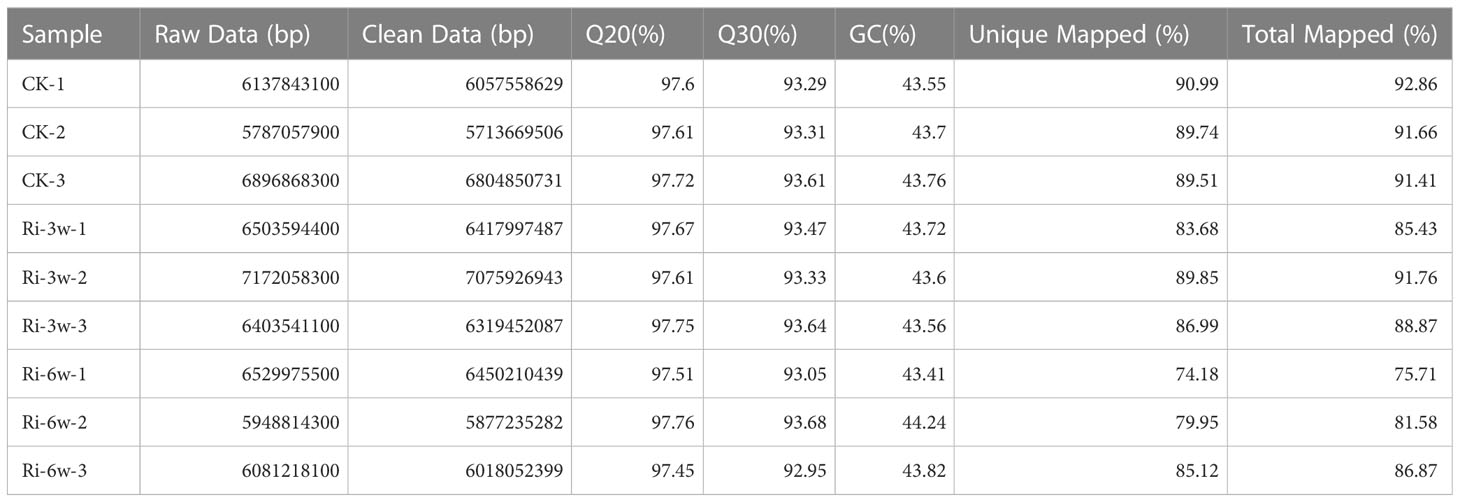
Table 1 Summary of throughput and quality Illumina-based transcriptome sequencing of cassava roots during AM symbiosis.
To unveil key differentially regulated pathways, we first screened out DEGs in pairwise comparison between groups. The volcano plots of the DEGs in the comparison of CK against Ri-3w and Ri-6w are shown in Figure S7. We detected 693 DEGs, including 659 up-regulated and 34 down-regulated between CK and Ri-3w (Figure 4A; Table S6). Between CK and Ri-6w, there were 6,481 DEGs, of which 2,637 and 3,844 were up- and down-regulated, respectively (Figure 4A; Table S7). Of them, 88 DEGs were commonly identified in the pairwise comparison between all groups (Figure 4B).
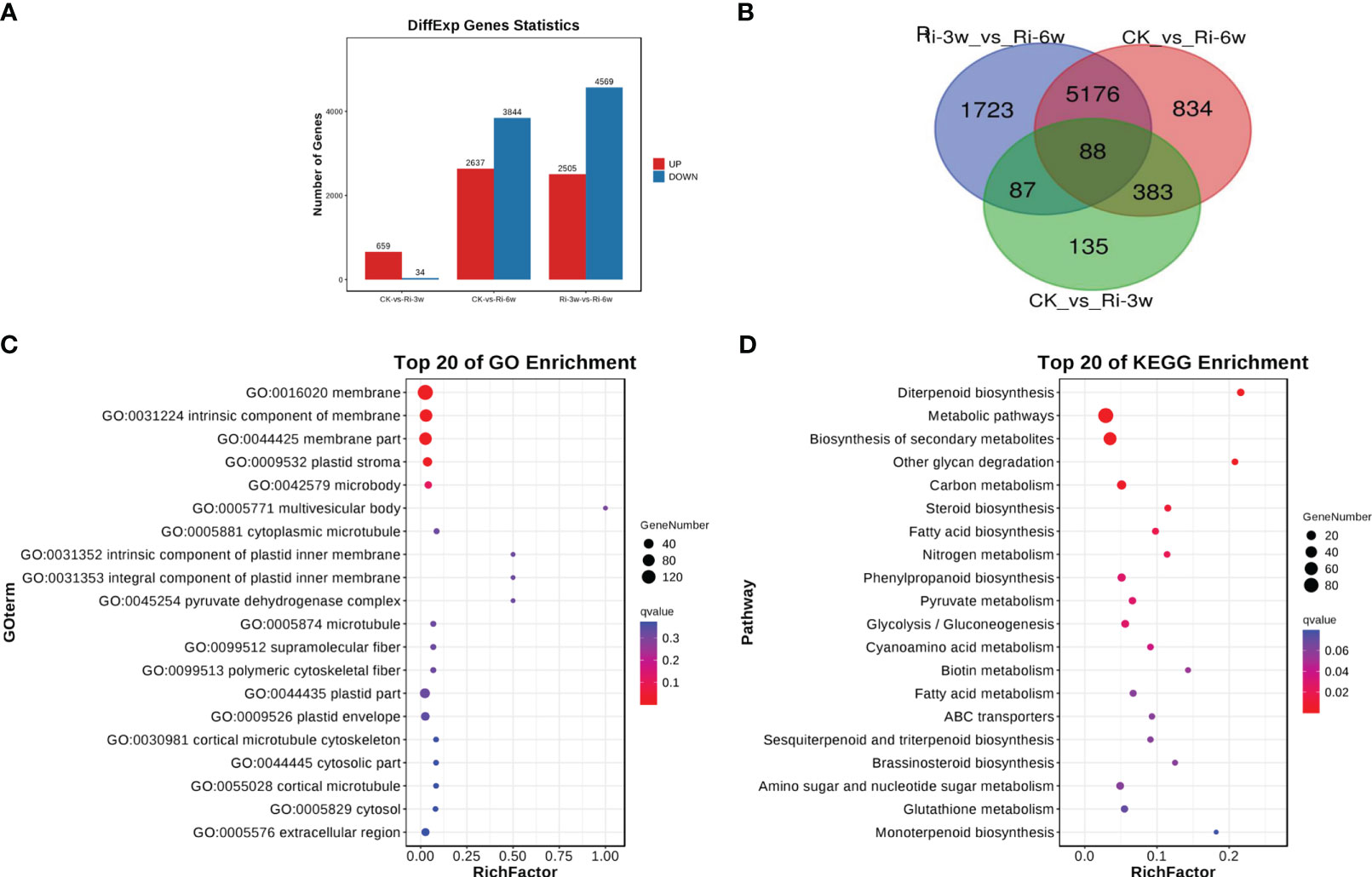
Figure 4 Differentially expressed genes (DEGs) and functional analyses. (A) Number of up- and down-regulated DEGs in pairwise comparison between groups. (B) Venn diagram showing the number of common DEGs shared by groups. (C, D) GO and KEGG annotation and enrichment analyses of DEGs between CK and Ri-3w, respectively.
We selected the DEGs in the pairwise comparison between CK and the other two groups for functional annotation and enrichment analyses. The most enriched GO terms that involve DEGs between CK and Ri-3w were related to membrane and plastid (Figure 4C). Meanwhile, DEGs between CK and Ri-6w were mainly associated with membranes, cell walls and junctions, and extracellular regions (Figure S8A). KEGG annotation and enrichment analysis of DEGs between CK and Ri-3w indicated that they are primarily involved in secondary metabolites biosynthesis, carbon metabolism, steroid and diterpenoid biosynthesis, phenylpropanoid biosynthesis, pyruvate metabolism, glycolysis/gluconeogenesis, fatty acid metabolism, and nitrogen metabolism (Figure 4D). The DEGs between CK and Ri-6w were mainly assigned to the biosynthesis of secondary metabolites, plant hormone signal transduction, phenylpropanoid biosynthesis, MAPK signaling pathway, plant-pathogen interaction, and metabolism of diverse sugars (Figure S8B). The lists of all identified pathways are shown in Tables S8 and S9.
As we above reported, the 6w samples were taken in order to examine the dynamic changes in the expression levels of DEGs identified at 3w. Therefore, we mainly focused on DEGs between CK and Ri-3w for identifying key candidate genes underlying AM symbiosis in cassava.
AM symbiosis promotes phosphorus and nitrogen uptake and metabolisms. We identified six inorganic phosphate transporters (PHT) and 20 nitrogen transporters related DEGs (Figure 5A; Table S10A). Of the PHT-related DEGs, Manes_15G190400 and Manes_03G164700 were the most significantly induced (Figure 5A). The identified nitrogen transporters included 5 and 15 ammonium (AMT) and nitrate (NRT) transporters, respectively. Both five AMTs were up-regulated, with Manes_05G082500 and Manes_16G119600 being the most highly induced (Figure 5A). Eleven of the NRTs were up-regulated, among which Manes_16G113300, Manes_01G191900, Manes_17G061600, Manes_08G106700, and Manes_18G018900 were the most significantly induced (Figure 5A).
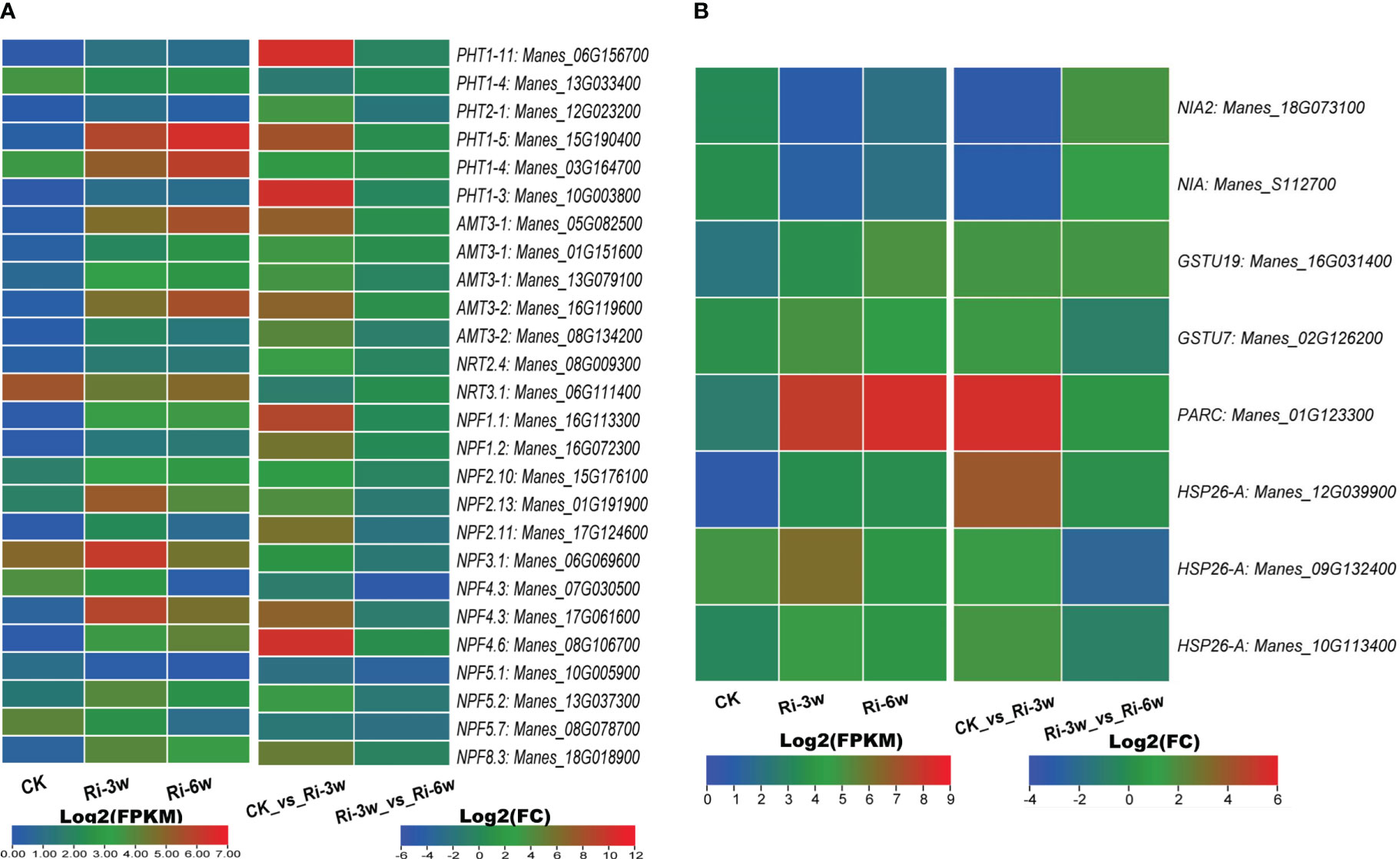
Figure 5 Transcription levels of DEGs related to nitrogen and phosphorus metabolisms. (A) Phosphorus and nitrogen transporters. (B) Other DEGs related to nitrogen metabolism. FPKM, Fragments Per Kilobase of transcript per Million mapped reads. PHT, inorganic phosphate transporter; AMT, ammonium transporter; NRT, high-affinity nitrate transporter; NPF, protein NRT1/PTR family-like. Genes’ annotation is presented in Tables S9A, B.
We also identified eight nitrogen metabolism related-DEGs, including two nitrate reductase (Manes_18G073100 and Manes_S112700) and seven glutathione S-transferase family genes (Figure 5B; Table S10B). Both two nitrate reductase genes were down-regulated, while the glutathione S-transferase genes were up-regulated. Of them, Manes_01G123300 (Log2(FC) = 5.5) and Manes_12G039900 (Log2(FC) = 3.9) were the most significantly induced (Figure 5B; Table S10B).
AM symbiosis influence sugar metabolism and phenylpropanoid biosynthesis. We identified 40 sugar metabolism and seven phenylpropanoid pathway-related DEGs (Figure 6; Tables S10C, D). It is noteworthily that all these DEGs were up-regulated in Ri at 3w (Figures 6A, B). Of the sugar metabolism-related DEGs, one beta-glucosidase (Manes_S042900) and six UDP-glycosyltransferases (Manes_S049100, Manes_01G018000, Manes_01G018100, Manes_01G017900, Manes_07G123300, and Manes_02G125300) were more than 8.5 folds significantly induced. The gene Manes_S091700 encoding a 4-coumarate-CoA ligase-like was the most significantly induced (Log2(FC) = 7.72) in the phenylpropanoid pathway (Figure 6B).
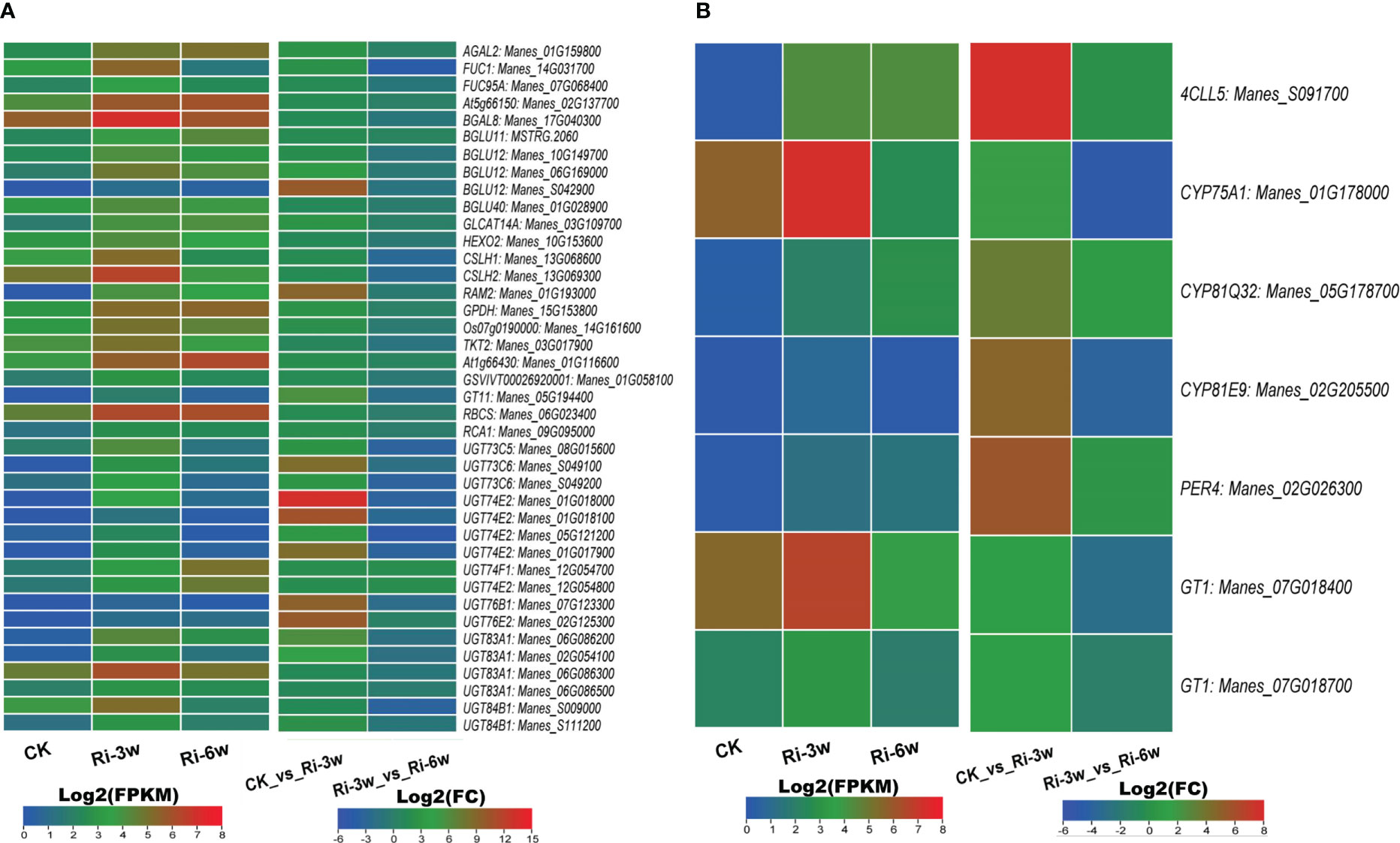
Figure 6 Transcription levels of DEGs related to sugar metabolism (A) and phenylpropanoid pathway (B). FPKM, Fragments Per Kilobase of transcript per Million mapped reads. Genes’ annotation is presented in Tables S9C, D.
Besides, 49 (four down-regulated) diverse other transporter family-related DEGs were also filtered out (Table S10E), and their expression patterns are shown in Figure S9. Of them, three ABC transporters (Manes_11G015600, Manes_13G104000, and Manes_11G148900), two acyl carrier proteins (Manes_03G055600 and Manes_16G087900), one oligopeptide transporter (Manes_03G142900), one ascorbate-specific transmembrane electron transporter (Manes_17G086900), one transmembrane protein (Manes_15G046500), four wall-associated receptors (Manes_02G171700, Manes_11G070400, Manes_11G070300, and Manes_11G070200), and two lipid transporters (Manes_14G109400 and Manes_S095100) were more than 6.8 folds up-regulated in Ri at 3w.
TFs and phytohormones play essential roles in AM symbiosis. Therefore, we screened out their respective DEGs and examined their expression profiles (Figure 7; Table S10F). The main differentially regulated TFs were DELLA (9 DEGs), EP2/ERF (13 DEGs), and MYBs (5 DEGs) (Figure 7A; Table S10F). As shown in Figure 7A, the genes Manes_17G055300 (MYB), Manes_05G040000 and Manes_13G048400 (EP2), Manes_13G041400 (ERF), Manes_11G083500 and Manes_14G054900 (DELLA), were the most induced TFs by the AMF inoculation.
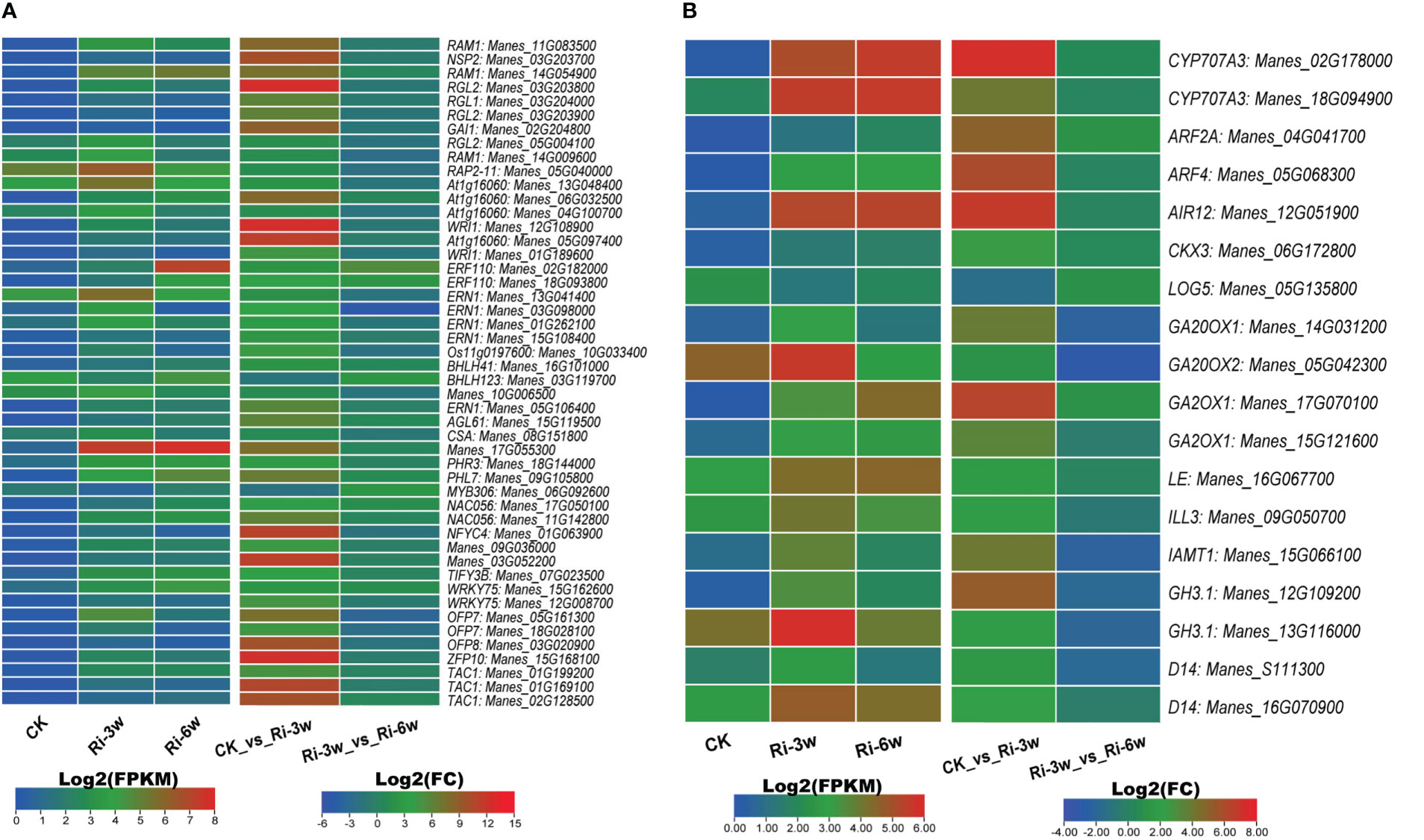
Figure 7 Transcription levels of DEGs belonging to transcription factor (A) and phytohormones (B) families. ABA, abscisic acid; AUX, auxin; GA, gibberellic acid; SL, strigolactones. FPKM, Fragments Per Kilobase of transcript per Million mapped reads. Genes’ annotation is presented in Tables S9F, G.
We identified 18 phytohormone-related DEGs, including 2, 7, 2, 5, and 2 ABA (abscisic acid), auxins (AUXs), cytokinin (CYTs), gibberellic acid (GAs), and strigolactones (STs), respectively (Figure 7B; Table S10G). Most of them were highly induced, and only Manes_05G135800 (CYT) was down-regulated in Ri at 3w (Figure 7B).
To unveil key symbiosis (SYM) signaling pathway-related candidate genes in cassava, we performed a Blast search against Medicago truncatula SYM pathway genes. In total, we identified 41 genes homologous to M. truncatula SYM pathway genes, with a similarity ranging from 37.42 to 81.99% (Table S11). Thereafter, we examined their expression profiles and found that 28 of them were differentially regulated at least at one-time point. Accordingly, they were selected as potential candidate genes for further study on the SYM signaling pathway in cassava (Table 2). Among them, MeSTR2, MeRAM1, MeKinG2, MeEXO70, MeABCB12, MeRAD1, MeHYP4, MeKinG1, and MeCYTB561 were more than 7 folds significantly induced in Ri at 3w.
To confirm the reliability of the transcriptome data, twelve random DEGs were selected for quantitative real-time PCR analysis. The results showed that the expression patterns of these genes through the RNA-seq and qRT-PCR were consistent (R2 = 0.80; Figure 8), supporting the high confidence level of our findings.
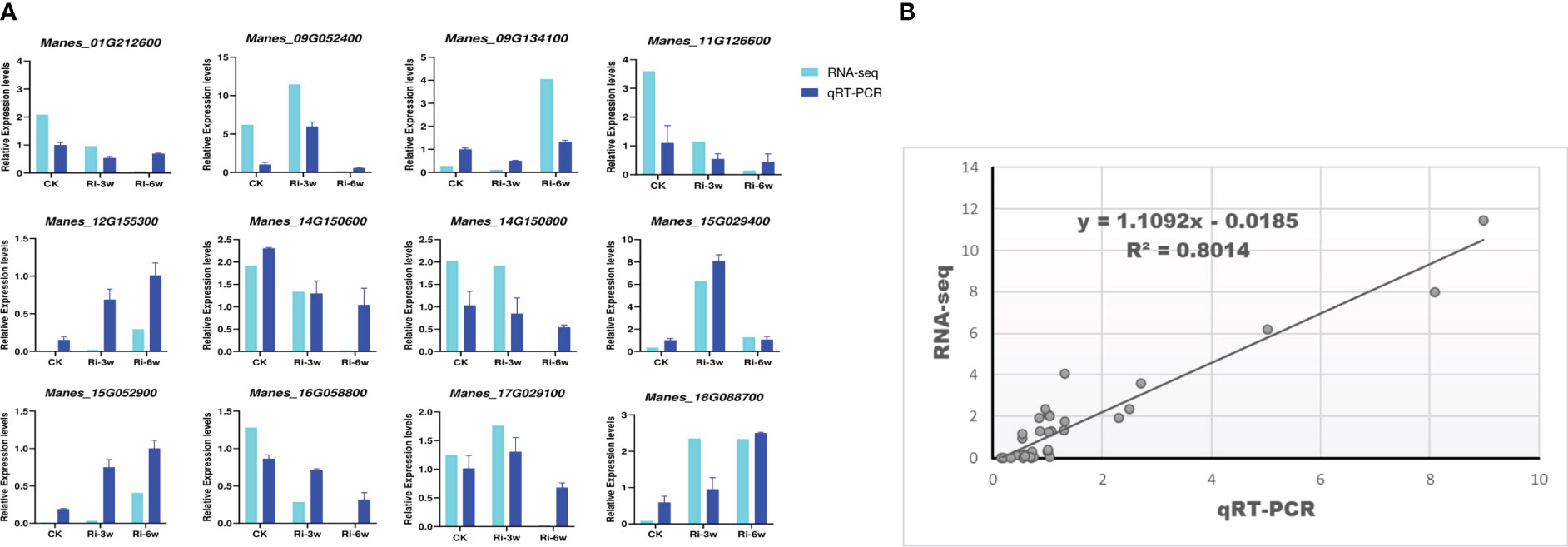
Figure 8 qRT-PCR validation of the RNA-seq data. Twelve DEGs were randomly selected for the qRT-PCR analysis. (A) Expression patterns of selected genes by RNA-seq and qRT-PCR. (B) Linear regression analysis of RNA-seq and qRT-PCR data. Relative expression indicates FPKM values for RNA-seq and the quantified expression levels by the 2¯ΔΔCT method for qRT-PCR.
With regard to their huge beneficial effects on plant growth, productivity, adaptation, and resistance capability, there is an increasing interest in using AMF as bio-fertilizers to achieve sustainable agriculture and food security. A deep understanding of the molecular mechanisms and regulation of AM symbiosis is a prerequisite to attending this goal. Accordingly, this study combined metabolome and transcriptome analyses and explored the biochemical and molecular changes that occur during AM symbiosis in cassava.
The inoculation of cassava with AMF improved the plants’ physiological performances. For instance, plant height and root length were significantly increased in inoculated cassava plants after three weeks. These results are consistent with previous reports in cassava and other crops, indicating that AM improves plant biomass, growth, yield, and quality (Ceballos et al., 2013; Chen et al., 2017; Aliyu et al., 2018; Gao et al., 2020; Felföldi et al., 2022; Yan et al., 2022). The positive effects of AM on plants’ performances are due to the fact that AM improves water and mineral nutrient (primarily phosphorus and nitrogen) uptake and metabolism, which in turn stimulates primary and secondary metabolisms (Kaur and Suseela, 2020; Püschel et al., 2020; Fu et al., 2021; Saboor et al., 2021; Kaur et al., 2022; Rui et al., 2022). DAMs analysis revealed that AMF inoculation induced primary metabolism in cassava roots. The major DAMs were amino acids and derivatives, nucleotides and derivatives, lipids, and flavonoids. It is reported that lipid metabolism is stimulated during AM symbiosis as plants supply fungi with carbon, mainly in the form of photosynthate and lipids (Wewer et al., 2014). Moreover, it is found that fatty acid pathway is the key pathway that modulates the currency of exchange between AMF and cassava roots during symbiosis (Savary et al., 2020). Fourteen (14/18) flavonoids were downregulated in inoculated plants after six weeks, indicating that AM altered flavonoid biosynthesis in developing roots. This result may also suggest the catabolism of flavonoids via signaling or defense mechanisms. Indeed, flavonoids play important roles in mediating selective cross-talk between beneficial soil microbiomes and plants (Bag et al., 2022). The most DAMs were 5-aminolevulinic acid, L-glutamic acid, and lysoPC 18:2. Glutamic acid is essential for plant growth and development. It has emerged as a signaling molecule involved in various developmental processes, such as root architecture, seed germination, pollen germination, tolerance to abiotic stresses, resistance to pathogens, etc. (Qiu et al., 2020; Kim et al., 2021). 5-aminolevulinic acid modulates mineral nutrient uptake and enhances plant abiotic stress tolerance (Rhaman et al., 2021; Zhang et al., 2022). These findings infer that biofertilization with AMF may confer cassava plants abiotic and biotic stresses tolerance and improve production. Moreover, these key metabolites may represent key markers for discriminating efficient AM symbiosis in cassava.
Transcriptome analysis identified 693 and 6,481 DEGs in Ri at 3w and 6w, respectively. These DEGs and DAMs represent valuable resources for further study to better understand cassava biology, specifically its interactions with AMF. Functional enrichment analyses of DAMs and DEGs uncovered transport, amino acids and sugar metabolisms, biosynthesis of secondary metabolites, plant hormone signal transduction, phenylpropanoid biosynthesis, and plant-pathogen interactions as being the most significant differentially regulated pathways. Supportively, many transporter and phytohormone family genes and sugar metabolism-related genes were significantly up-regulated in inoculated plants. Previous studies have demonstrated that AM symbiosis significantly induces the expression of plant transporter genes (Porcel et al., 2016; Kameoka et al., 2019; Banasiak et al., 2021; Rui et al., 2022). Savary et al. have reported the significant up-regulation of the fatty acid biosynthesis pathway during AM symbiosis in cassava (Savary et al., 2020). Sugars and amino acids (primary metabolites) are precursors of diverse secondary metabolites and sources of energy. The identified phytohormone-related DEGs belong to ABA, AUXs, CYTs, GAs, and STs. These phytohormones are reported to play essential roles during AM symbiosis (Garrido et al., 2010; Gutjahr, 2014; Lanfranco et al., 2018; Faizan et al., 2020; Ho-Plágaro and García-Garrido, 2022a). STs are key signaling molecules used by plants to attract AMF through interactions with other phytohormones (Lanfranco et al., 2018; Faizan et al., 2020). GAs specific roles in AMF colonization and AM symbiosis are still unclear and confusing (Takeda et al., 2015; Tominaga et al., 2020; Tominaga et al., 2021; Tominaga et al., 2022). According to these studies, GA may promote or alter AMF penetration and proliferation in plants’ roots depending on the type of fungi. Two gibberellin-20-oxidase genes and three gibberellin 2-beta-dioxygenase genes were significantly up-regulated, suggesting they might modulate cassava plants and AMF symbiosis by regulating the expression of DELLA and other GRAS TFs. Gibberellin-2-oxidase was previously identified as a potential candidate gene for promoting AMF symbiosis in cassava (Savary et al., 2020). Therefore, functional characterization of these phytohormone-related genes is required to understand their regulatory effects on AM symbiosis in cassava.
It is worth noting that AM symbiosis improves phosphorus and nitrogen uptake and metabolism (Aliyu et al., 2018; Fu et al., 2021; Rui et al., 2022). We identified significantly up-regulated inorganic phosphate transporters (6), ammonium transporters (5), nitrate transporters (11), and glutathione S-transferases (6), and two significantly down-regulated nitrate reductase genes. The up-regulation of nitrogen and phosphorus transporters, coupled with the down-regulation of nitrate reductase in roots, indicate an improved uptake and a coordinated transport of these essential mineral nutrients from roots to the aboveground organs. The up-regulation of glutathione-related genes supports that AM symbiosis may improve cassava abiotic stress tolerance capability. Tremendous studies have demonstrated that AM enhances various biotic and abiotic stress tolerance abilities of plants (Sabra et al., 2018; Gao et al., 2020; Zhang et al., 2020; Jajoo, 2021; Lin et al., 2021; Jumrani et al., 2022).
The establishment of mutualistic interactions between plant roots and beneficial microorganisms is governed by a common pathway known as the SYM signaling pathway (Bonfante and Genre, 2010; Genre and Russo, 2016). This pathway has been well-studied in M. truncatula and rice (Harrison, 2005; Bonfante and Genre, 2010; Genre and Russo, 2016). It is mainly regulated by DELLA proteins, members of the GRAS TF family (Ho-Plágaro and García-Garrido, 2022a). Our analysis revealed that DELLA (9 DEGs), EP2/ERF (13 DEGs), and MYBs (5 DEGs) were the major differentially regulated TFs. These TFs might play critical roles in regulating AM symbiosis in cassava (Diédhiou and Diouf, 2018). The regulation of AM symbiosis in cassava involves complex mechanisms and is governed mainly by genetic variations in the genome of the two symbionts and genotypes × genotypes specifications (Mateus et al., 2019; Savary et al., 2020). Through blast analysis, we identified 28 significantly up-regulated SYM signaling pathway homologous DEGs in cassava. Twelve other significantly up-regulated SYM pathway ortholog genes have been identified in cassava (Mateus et al., 2019; Savary et al., 2020). These genes represent key resources to dissecting AM symbiosis regulatory network in cassava. Therefore, functional validation of all identified potential candidate genes is required to understand the regulation of AM symbiosis and uncover key markers and genes for genomics-assisted improvement of cassava. Particular attention should be given to the gene MeRAM1 that encodes a DELLA protein GAI-like. RAM1 was unveiled as the dominant gene in regulating the fatty acid pathway during AM symbiosis in cassava (Savary et al., 2020). Moreover, previous studies in other crops have shown that RAM1 play central roles during AM symbiosis (Hohnjec et al., 2015; Rich et al., 2017; Hartmann et al., 2019; Ho-Plágaro and García-Garrido, 2022a).
Overall, this study provided a comprehensive data set by integrating metabolomics and transcriptomics analyses and enabled a global view of the complex biochemical and molecular changes that occur during AMF and cassava root symbiosis. DAMs and DEGs were identified, and Key differentially regulated pathways were revealed as being transport, amino acids and sugar metabolisms, biosynthesis of secondary metabolites, plant hormone signal transduction, phenylpropanoid biosynthesis, and plant-pathogen interactions. The AM symbiosis significantly stimulated nitrogen, phosphorus, and sugar metabolisms. In contrast, it altered flavonoid biosynthesis. GRAS (DELLA) TFs and some phytohormone family genes might be the key regulators of the two symbionts’ interactions. Potential candidate genes were uncovered for future functional studies. Our findings offer fundamental resources to dissect the regulatory network of the SYM signaling pathway in cassava and the efficient use of AMF in improving the crop production.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Conceptualization, YC and SC. Methodology, YG and ZP. Software, YG and SH. Validation, YG, YW. Formal analysis, YG. Investigation, YG, YW. and SZ. Resources, HL and JZ. Data curation, YG. Writing—original draft preparation, YG. Writing—review and editing, ZP. Visualization, WW. Supervision, YC and SC. Project administration, YC. Funding acquisition, YC. All authors contributed to the article and approved the submitted version.
This research was supported by the National Key Research and Development Program of China (2018YFD1000500), and Earmarked fund for China Agriculture Research System (CARS-11-HNCYH), and the Hainan Provincial Innovative Research Projects of Postgraduates (no. Hyb2019-04).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1130924/full#supplementary-material
Abdallah, C., Valot, B., Guillier, C., Mounier, A., Balliau, T., Zivy, M., et al. (2014). The membrane proteome of medicago truncatula roots displays qualitative and quantitative changes in response to arbuscular mycorrhizal symbiosis. J. Proteomics 108, 354–368. doi: 10.1016/j.jprot.2014.05.028. Elsevier B.V.
Aliyu, I. A., Yusuf, A. A., Uyovbisere, E. O., Masso, C., Sanders, I. R. (2018). Effect of co-application of phosphorus fertilizer and in vitro-produced mycorrhizal fungal inoculants on yield and leaf nutrient concentration of cassava. PLos One 14, 1–19. doi: 10.1371/journal.pone.0218969
Bag, S., Mondal, A., Majumder, A., Mondal, S. K., Banik, A. (2022). Flavonoid mediated selective cross-talk between plants and beneficial soil microbiome. Phytochem. Rev. 21, 1739–1760. doi: 10.1007/s11101-022-09806-3
Banasiak, J., Jamruszka, T., Murray, J. D., Jasinski, M. (2021). A roadmap of plant membrane transporters in arbuscular mycorrhizal and legume-rhizobium symbioses. Plant Physiol. 187, 2071–2091. doi: 10.1093/plphys/kiab280
Bapaume, L., Reinhardt, D. (2012). How membranes shape plant symbioses: Signaling and transport in nodulation and arbuscular mycorrhiza. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00223
Begum, N., Qin, C., Ahanger, M. A., Raza, S., Khan, M. I., Ashraf, M., et al. (2019). Role of arbuscular mycorrhizal fungi in plant growth regulation: Implications in abiotic stress tolerance. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01068
Blagbrough, I. S., Bayoumi, S. A. L., Rowan, M. G., Beeching, J. R. (2010). Phytochemistry Cassava : An appraisal of its phytochemistry and its biotechnological prospects. Phytochemistry 71, 1940–1951. doi: 10.1016/j.phytochem.2010.09.001
Bonfante, P., Genre, A. (2010). Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosiss. Nat. Commun. 1, 48. doi: 10.1038/ncomms1046
Bredeson, J. V., Lyons, J. B., Prochnik, S. E., Wu, G. A., Ha, C. M., Edsinger-Gonzales, E., et al. (2016). Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nat. Biotechnol. 34, 562–570. doi: 10.1038/nbt.3535
Ceballos, I., Ruiz, M., Fernández, C., Peña, R., Rodríguez, A., Sanders, I. R. (2013). The In vitro mass-produced model mycorrhizal fungus, rhizophagus irregularis, significantly increases yields of the globally important food security crop cassava. PLos One 8, e70633. doi: 10.1371/journal.pone.0070633
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, W., Gong, L., Guo, Z., Wang, W., Zhang, H., Liu, X., et al. (2013). A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 6, 1769–1780. doi: 10.1093/mp/sst080
Chen, W., Li, J., Zhu, H., Xu, P., Chen, J., Yao, Q. (2017). Arbuscular mycorrhizal fungus enhances lateral root formation in poncirus trifoliata (L.) as revealed by RNA-seq analysis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02039
Chen, X., Sun, M., Chong, S., Si, J., Wu, L. (2022). Transcriptomic and metabolomic approaches deepen our knowledge of plant–endophyte interactions. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.700200
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., Robles, M. (2005). Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Diédhiou, I., Diouf, D. (2018). Transcription factors network in root endosymbiosis establishment and development. World J. Microbiol. Biotechnol. 0, doi: 10.1007/s11274-018-2418-7
Dossou, S. S. K., Xu, F., Cui, X., Sheng, C., Zhou, R., You, J., et al. (2021). Comparative metabolomics analysis of different sesame (Sesamum indicum l.) tissues reveals a tissue-specific accumulation of metabolites. BMC Plant Biol. 21, 1–14. doi: 10.1186/s12870-021-03132-0
Dossou, S. S. K., Xu, F., You, J., Zhou, R., Li, D., Wang, L. (2022). Widely targeted metabolome profiling of different colored sesame (Sesamum indicum l.) seeds provides new insight into their antioxidant activities. Food Res. Int. 151, 110850. doi: 10.1016/j.foodres.2021.110850
Faizan, M., Faraz, A., Sami, F., Siddiqui, H., Yusuf, M., Gruszka, D., et al. (2020). Role of strigolactones: Signalling and crosstalk with other phytohormones. Open Life Sci. 15, 217–228. doi: 10.1515/biol-2020-0022
Farag, M. A., Elsebai, M. F., Khattab, A. R. (2018). Metabolome based classification of artichoke leaf: A prospect for phyto-equivalency of its different leaf origins and commercial preparations. J. Pharm. Biomed. Anal. 158, 151–159. doi: 10.1016/j.jpba.2018.05.046
Felföldi, Z., Vidican, R., Stoian, V., Roman, I. A., Sestras, A. F., Rusu, T., et al. (2022). Arbuscular mycorrhizal fungi and fertilization influence yield, growth and root colonization of different tomato genotype. Plants 11, 1743. doi: 10.3390/plants11131743
Floss, D. S., Levy, J. G., Lévesque-Tremblay, V., Pumplin, N., Harrison, M. J. (2013). DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. 110, 5025–5034. doi: 10.1073/pnas.1308973110
Fu, D., Rui, Y., Zevenbergen, C., Prasad, R. (2021). Nitrogen absorption efficiency and mechanism in arbuscular mycorrhizal fungi - canna indica symbiosis. Chemosphere 282, 130708. doi: 10.1016/j.chemosphere.2021.130708
Gao, X., Guo, H., Zhang, Q., Guo, H., Zhang, L., Zhang, C. (2020). Arbuscular mycorrhizal fungi ( AMF ) enhanced the growth , yield , fiber quality and phosphorus regulation in upland cotton (gossypium hirsutum l.). Sci. Rep 10. 2084. doi: 10.1038/s41598-020-59180-3
Garrido, J. M. G., Morcillo, R. J. L., Rodríguez, J.Á.M., Bote, J. A. O. (2010). Variations in the mycorrhization characteristics in roots of wild-type and ABA-deficient tomato are accompanied by specific transcriptomic alterations. MPMI 23, 651–664. doi: 10.1094/MPMI-23-5-0651
Genre, A., Russo, G. (2016). Does a common pathway transduce symbiotic signals in plant–microbe interactions? Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00096
Gutjahr, C. (2014). Phytohormone signaling in arbuscular mycorhiza development. Curr. Opin. Plant Biol. 20, 26–34. doi: 10.1016/j.pbi.2014.04.003
Harrison, M. J. (2005). SIGNALING IN THE ARBUSCULAR MYCORRHIZAL SYMBIOSIS. Annu. Rev. Microbiol. 59, 19–42. doi: 10.1146/annurev.micro.58.030603.123749
Hartmann, R. M., Schaepe, S., Nübel, D., Petersen, A. C., Bertolini, M., Vasilev, J., et al. (2019). Insights into the complex role of GRAS transcription factors in the arbuscular mycorrhiza symbiosis. Sci. Rep. 9, 3360. doi: 10.1038/s41598-019-40214-4
Hohnjec, N., Czaja-hasse, L. F., Hogekamp, C., Küster, H. (2015). Pre-announcement of symbiotic guests : transcriptional reprogramming by mycorrhizal lipochitooligosaccharides shows a strict co-dependency on the GRAS transcription factors NSP1 and RAM1. BMC Genomics 16, 994. doi: 10.1186/s12864-015-2224-7
Ho-Plágaro, T., García-Garrido, J. M. (2022a). Molecular regulation of arbuscular mycorrhizal symbiosis. Int. J. Mol. Sci. 23, 5960. doi: 10.3390/ijms23115960
Ho-Plágaro, T., García-Garrido, J. M. (2022b). Multifarious and interactive roles of GRAS transcription factors during arbuscular mycorrhiza development. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.836213
Ho-Plágaro, T., Molinero-Rosales, N., Fariña Flores, D., Villena Díaz, M., García-Garrido, J. M. (2019). Identification and expression analysis of GRAS transcription factor genes involved in the control of arbuscular mycorrhizal development in tomato. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00268
Hu, W., Ji, C., Liang, Z., Ye, J., Ou, W., Ding, Z., et al. (2021). Resequencing of 388 cassava accessions identifies valuable loci and selection for variation in heterozygosity. Genome Biol. 22, 316. doi: 10.1186/s13059-021-02524-7
Igiehon, N. O., Babalola, O. O. (2017). Biofertilizers and sustainable agriculture: exploring arbuscular mycorrhizal fungi. Appl. Microbiol. Biotechnol. 101, 4871–4881. doi: 10.1007/s00253-017-8344-z
Jajoo, A. (2021). Role of arbuscular mycorrhizal fungi as an underground saviuor for protecting plants from abiotic stresses. Physiol. Mol. Biol. Plants 27, 2589–2603. doi: 10.1007/s12298-021-01091-2
Jiang, Y., Xie, Q., Wang, W., Yang, J., Zhang, X., Yu, N., et al. (2018). Medicago AP2-domain transcription factor WRI5a is a master regulator of lipid biosynthesis and transfer during mycorrhizal symbiosis. Mol. Plant 11 (11), 1344–1359. doi: 10.1016/j.molp.2018.09.006
Jumrani, K., Bhatia, V. S., Kataria, S., Alamri, S. A., Siddiqui, M. H., Rastogi, A. (2022). Inoculation with arbuscular mycorrhizal fungi alleviates the adverse effects of high temperature in soybean. Plants 11, 2210. doi: 10.3390/plants11172210
Kameoka, H., Maeda, T., Okuma, N., Kawaguchi, M. (2019). Structure-specific regulation of nutrient transport and metabolism in arbuscular mycorrhizal fungi. Plant Cell Physiol. 60, 2272–2281. doi: 10.1093/pcp/pcz122
Kanehisa, M., Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kaur, S., Campbell, B. J., Suseela, V. (2022). Root metabolome of plant–arbuscular mycorrhizal symbiosis mirrors the mutualistic or parasitic mycorrhizal phenotype. New Phytol. 234, 672–687. doi: 10.1111/nph.17994
Kaur, S., Suseela, V. (2020). Unraveling arbuscular mycorrhiza-induced changes in plant primary and secondary metabolome. Metabolites 10, 335. doi: 10.3390/metabo10080335
Kim, D. R., Jeon, C. W., Cho, G., Thomashow, L. S., Weller, D. M., Paik, M. J., et al. (2021). Glutamic acid reshapes the plant microbiota to protect plants against pathogens. Microbiome 9, 1–18. doi: 10.1186/s40168-021-01186-8
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., Salzberg, S. L. (2013). TopHat2 : accurate alignment of transcriptomes in the presence of insertions , deletions and gene fusions. Genome Biol. 14 (4), R36. doi: 10.1186/gb-2013-14-4-r36
Lanfranco, L., Fiorilli, V., Venice, F., Bonfante, P. (2018). Strigolactones cross the kingdoms: plants, fungi, and bacteria in the arbuscular mycorrhizal symbiosis. J. Exp. Bot. 69, 2175–2188. doi: 10.1093/jxb/erx432
Li, S., Cui, Y., Zhou, Y., Luo, Z., Liu, J., Zhao, M. (2017). The industrial applications of cassava: current status, opportunities and prospects. J. Sci. Food Agric. 97, 2282–2290. doi: 10.1002/jsfa.8287
Lin, P., Zhang, M., Wang, M., Li, Y., Liu, J., Chen, Y. (2021). Inoculation with arbuscular mycorrhizal fungus modulates defense-related genes expression in banana seedlings susceptible to wilt disease. Plant Signal. Behav. 16, 1884782. doi: 10.1080/15592324.2021.1884782
Liu, F., Xu, Y., Wang, H., Zhou, Y., Cheng, B., Li, X. (2020). APETALA 2 transcription factor CBX1 is a regulator of mycorrhizal symbiosis and growth of lotus japonicus. Plant Cell Rep 39 (4), 445–445. doi: 10.1007/s00299-019-02501-2
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Ma, J., Wang, W., Yang, J., Qin, S., Yang, Y., Sun, C., et al. (2022). Mycorrhizal symbiosis promotes the nutrient content accumulation and affects the root exudates in maize. BMC Plant Biol. 22, 64. doi: 10.1186/s12870-021-03370-2
Malik, A. I., Kongsil, P., Nguyễn, V. A., Ou, W., Sholihin, Srean, P., et al. (2020). Cassava breeding and agronomy in Asia: 50 years of history and future directions. Breed. Sci. 70, 145–166. doi: 10.1270/jsbbs.18180
Mateus, I. D., Masclaux, F. G., Aletti, C., Rojas, E. C., Savary, R., Dupuis, C., et al. (2019). Dual RNA-seq reveals large-scale non-conserved genotype × genotype-specific genetic reprograming and molecular crosstalk in the mycorrhizal symbiosis. ISME J. 13, 1226–1238. doi: 10.1038/s41396-018-0342-3
Mishra, S., Priyanka, Sharma, S. (2022). Metabolomic insights into endophyte-derived bioactive compounds. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.835931
Mitra, D., Valizadeh, K., Chaudhary, P., Ruparelia, J., Smruthi, M., Boutaj, H., et al. (2021). Current research in microbial sciences involvement of strigolactone hormone in root development , influence and interaction with mycorrhizal fungi in plant : Mini-review. Curr. Res. Microb. Sci. 2, 100026. doi: 10.1016/j.crmicr.2021.100026
Mombo, S., Dumat, C., Shahid, M., Schreck, E. (2016). A socio-scientific analysis of the environmental and health benefits as well as potential risks of cassava production and consumption. Environ. Sci. pollut. Res 24 (6), 5207–5221. doi: 10.1007/s11356-016-8190-z
Müller, L. M., Harrison, M. J. (2019). Phytohormones, miRNAs, and peptide signals integrate plant phosphorus status with arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 50, 132–139. doi: 10.1016/j.pbi.2019.05.004
Nagae, M., Takeda, N., Kawaguchi, M. (2014). Common symbiosis genes CERBERUS and NSP1 provide additional insight into the establishment of arbuscular mycorrhizal and root nodule symbioses in lotus japonicus. Plant Signal. Behav. 9, e28544. doi: 10.4161/psb.28544
Parniske, M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6, 763–775. doi: 10.1038/nrmicro1987
Pimprikar, P., Gutjahr, C. (2018). Transcriptional regulation of arbuscular mycorrhiza development. Plant Cell Physiol. 59, 678–695. doi: 10.1093/pcp/pcy024
Porcel, R., Aroca, R., Azcon, R., Ruiz-lozano, J. M., Ruiz-lozano, J. M. (2016). Regulation of cation transporter genes by the arbuscular mycorrhizal symbiosis in rice plants subjected to salinity suggests improved salt tolerance due to reduced Na + root-to-shoot distribution. Mycorrhiza 26 (7), 673–684. doi: 10.1007/s00572-016-0704-5
Püschel, D., Bitterlich, M., Rydlová, J., Jansa, J. (2020). Facilitation of plant water uptake by an arbuscular mycorrhizal fungus: a Gordian knot of roots and hyphae. Mycorrhiza 30, 299–313. doi: 10.1007/s00572-020-00949-9
Qiu, X. M., Sun, Y. Y., Ye, X. Y., Li, Z. G. (2020). Signaling role of glutamate in plants. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01743
Ren, C., Kong, C., Yan, K., Xie, Z. (2019). Transcriptome analysis reveals the impact of arbuscular mycorrhizal symbiosis on sesbania cannabina expose to high salinity. Sci. Rep. 9, 2780. doi: 10.1038/s41598-019-39463-0
Rhaman, M. S., Imran, S., Karim, M. M., Chakrobortty, J., Mahamud, M. A., Sarker, P., et al. (2021). 5-aminolevulinic acid-mediated plant adaptive responses to abiotic stress. Plant Cell Rep. 40, 1451–1469. doi: 10.1007/s00299-021-02690-9
Rich, M. K., Courty, P., Roux, C., Reinhardt, D. (2017). Role of the GRAS transcription factor ATA/RAM1 in the transcriptional reprogramming of arbuscular mycorrhiza in petunia hybrida. BMC Genomics 18, 589. doi: 10.1186/s12864-017-3988-8
Rui, W., Mao, Z., Li, Z. (2022). The roles of phosphorus and nitrogen nutrient transporters in the arbuscular mycorrhizal symbiosis. Int. J. Mol. Sci. 23, 11027. doi: 10.3390/ijms231911027
Saboor, A., Arif, M., Hussain, S., El, H. A., Hussain, S., Ahmed, N., et al. (2021). Zinc nutrition and arbuscular mycorrhizal symbiosis effects on maize (zea mays l.) growth and productivity. Saudi J. Biol. Sci 28 (11), 6339–6351. doi: 10.1016/j.sjbs.2021.06.096
Sabra, M., Aboulnasr, A., Franken, P., Perreca, E., Wright, L. P., Camehl, I. (2018). Beneficial root endophytic fungi increase growth and quality parameters of sweet basil in heavy metal contaminated soil. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01726
Sakamoto, K., Ogiwara, N., Kaji, T., Sugimoto, Y., Ueno, M. (2019). Transcriptome analysis of soybean ( glycine max ) root genes differentially expressed in rhizobial , arbuscular mycorrhizal , and dual symbiosis. J. Plant Res. 132, 541–568. doi: 10.1007/s10265-019-01117-7
Savary, R., Dupuis, C., Masclaux, F. G., Mateus, I. D., Rojas, E. C., Sanders, I. R. (2020). Genetic variation and evolutionary history of a mycorrhizal fungus regulate the currency of exchange in symbiosis with the food security crop cassava. ISME J. 14, 1333–1344. doi: 10.1038/s41396-020-0606-6
Scalbert, A., Andres-Lacueva, C., Arita, M., Kroon, P., Manach, C., Urpi-Sarda, M., et al. (2011). Databases on food phytochemicals and their health-promoting effects. J. Agric. Food Chem. 59, 4331–4348. doi: 10.1021/jf200591d
Shtark, O., Puzanskiy, R., Avdeeva, G., Yemelyanov, V., Shavarda, A., Romanyuk, D., et al. (2021). Metabolic alterations in pisum sativum roots during plant growth and arbuscular mycorrhiza development. Plants 10, 1033. doi: 10.3390/plants10061033
Song, S., You, J., Shi, L., Sheng, C., Zhou, W., Dossou, S. S. K., et al. (2021). Genome-wide analysis of nsltp gene family and identification of siltps contributing to high oil accumulation in sesame (Sesamum indicum l.). Int. J. Mol. Sci. 22, 5291. doi: 10.3390/ijms22105291
Sonnewald, U., Fernie, A. R., Gruissem, W., Schläpfer, P., Anjanappa, R. B., Chang, S., et al. (2020). The cassava source–sink project: opportunities and challenges for crop improvement by metabolic engineering. Plant J. 103, 1655–1665. doi: 10.1111/tpj.14865
Takeda, N., Handa, Y., Tsuzuki, S., Kojima, M., Sakakibara, H., Kawaguchi, M. (2015). Gibberellins interfere with symbiosis signaling and gene expression and alter colonization by arbuscular mycorrhizal fungi in lotus japonicus. Plant Physiol. 167, 545–557. doi: 10.1104/pp.114.247700
Tominaga, T., Miura, C., Sumigawa, Y., Hirose, Y., Yamaguchi, K., Shigenobu, S., et al. (2021). Conservation and diversity in gibberellin-mediated transcriptional responses among host plants forming distinct arbuscular mycorrhizal morphotypes. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.795695
Tominaga, T., Miura, C., Takeda, N., Kanno, Y., Takemura, Y., Seo, M., et al. (2020). Gibberellin promotes fungal entry and colonization during Paris-type arbuscular mycorrhizal symbiosis in eustoma grandiflorum. Plant Cell Physiol. 61, 565–575. doi: 10.1093/pcp/pcz222
Tominaga, T., Yao, L., Saito, H., Kaminaka, H. (2022). Conserved and diverse transcriptional reprogramming triggered by the establishment of symbioses in tomato roots forming arum-type and Paris-type arbuscular mycorrhizae. Plants 11, 747. doi: 10.3390/plants11060747
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Tsiokanos, E., Cartabia, A., Tsafantakis, N., Lalaymia, I., Termentzi, A., Miguel, M., et al. (2022). The metabolic profile of anchusa officinalis l. differs according to its associated arbuscular mycorrhizal fungi. Metabolites 12, 573. doi: 10.3390/metabo12070573
Wang, L., Dossou, S. S. K., Wei, X., Zhang, Y., Li, D., Yu, J., et al. (2020). Transcriptome dynamics during black and white sesame (Sesamum indicum l.) seed development and identification of candidate genes associated with black pigmentation. Genes (Basel). 11, 1–14. doi: 10.3390/genes11121399
Wang, W., Feng, B., Xiao, J., Xia, Z., Zhou, X., Li, P., et al. (2014). Cassava genome from a wild ancestor to cultivated varieties. Nat. Commun. 5, 5110. doi: 10.1038/ncomms6110
Wewer, V., Brands, M., Dörmann, P. (2014). Fatty acid synthesis and lipid metabolism in the obligate biotrophic fungus rhizophagus irregularis during mycorrhization of lotus japonicus. Plant J. 79, 398–412. doi: 10.1111/tpj.12566
Yan, Q., Li, X., Xiao, X., Chen, J., Liu, J., Lin, C., et al. (2022). Arbuscular mycorrhizal fungi improve the growth and drought tolerance of cinnamomum migao by enhancing physio-biochemical responses. Ecol. Evol. 12, 1–18. doi: 10.1002/ece3.9091
Zai, X. M., Fan, J. J., Hao, Z. P., Liu, X. M., Zhang, W. X. (2021). Effect of co − inoculation with arbuscular mycorrhizal fungi and phosphate solubilizing fungi on nutrient uptake and photosynthesis of beach palm under salt stress environment. Sci. Rep. 11, 5761. doi: 10.1038/s41598-021-84284-9
Zhang, X., Bai, L., Sun, H., Yang, C., Cai, B. (2020). Transcriptomic and proteomic analysis revealed the effect of funneliformis mosseae in soybean roots differential expression genes and proteins. J. Proteome Res. 19, 3631–3643. doi: 10.1021/acs.jproteome.0c00017
Zhang, Z., Yuan, L., Ma, Y., Kang, Z., Zhou, F., Gao, Y., et al. (2022). Exogenous 5-aminolevulinic acid alleviates low-temperature damage by modulating the xanthophyll cycle and nutrient uptake in tomato seedlings. Plant Physiol. Biochem. 189, 83–93. doi: 10.1016/j.plaphy.2022.08.013
Keywords: cassava, symbiosis, arbuscular mycorrhiza, transcriptome, metabolome, candidate gene, fungi
Citation: Gao Y, Huang S, Wang Y, Lin H, Pan Z, Zhang S, Zhang J, Wang W, Cheng S and Chen Y (2023) Analysis of the molecular and biochemical mechanisms involved in the symbiotic relationship between Arbuscular mycorrhiza fungi and Manihot esculenta Crantz. Front. Plant Sci. 14:1130924. doi: 10.3389/fpls.2023.1130924
Received: 23 December 2022; Accepted: 22 February 2023;
Published: 07 March 2023.
Edited by:
Katsuharu Saito, Shinshu University, JapanReviewed by:
Wei Wang, Chinese Academy of Tropical Agricultural Sciences, ChinaCopyright © 2023 Gao, Huang, Wang, Lin, Pan, Zhang, Zhang, Wang, Cheng and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinhua Chen, eWhjaGVuQGhhaW5hbnUuZWR1LmNu; Shanhan Cheng, OTkwODY1QGhhaW5hbnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.