
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 27 February 2023
Sec. Plant Cell Biology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1123059
This article is part of the Research TopicCooperation of Gene Regulatory Networks And Phytohormones in Cell Development and MorphogenesisView all 6 articles
The key phytohormone auxin is involved in practically every aspect of plant growth and development. Auxin regulates these processes by controlling gene expression through functionally distinct AUXIN RESPONSE FACTORs (ARFs). As a noncanonical ARF, ARF3/ETTIN (ETT) mediates auxin responses to orchestrate multiple developmental processes during the reproductive phase. The arf3 mutation has pleiotropic effects on reproductive development, causing abnormalities in meristem homeostasis, floral determinacy, phyllotaxy, floral organ patterning, gynoecium morphogenesis, ovule development, and self-incompatibility. The importance of ARF3 is also reflected in its precise regulation at the transcriptional, posttranscriptional, translational, and epigenetic levels. Recent studies have shown that ARF3 controls dynamic shoot apical meristem (SAM) maintenance in a non-cell autonomous manner. Here, we summarize the hierarchical regulatory mechanisms by which ARF3 is regulated and the diverse roles of ARF3 regulating developmental processes during the reproductive phase.
Classically, auxin regulates gene expression by controlling the activity of AUXIN RESPONSE FACTORs (ARFs) through the Aux/IAA pathway. The Arabidopsis (Arabidopsis thaliana) genome encodes 23 ARFs that can be divided into three subclasses: A, B, and C (Guilfoyle and Hagen, 2007; Finet et al., 2013; Roosjen et al., 2018). ARF3/ETTIN (ETT) belongs to class B and is an important regulator of many developmental processes. Located on chromosome 2, ARF3 contains 10 exons and 9 introns, has a total length of 3170 bp, and encodes a protein that is 608 amino acids long. Most ARFs possess three major domains: an N-terminal DNA-binding domain (DBD), with higher affinity for the DNA-binding motifs TGTCGG and TGTCTC; a middle region (MR), which can be used as a reference for determining activators and repressors in the ARF family; and a C-terminal Phox and Bem1 domain (PB1 or domain III/IV). The PB1 domain allows ARFs to form homodimers and heterodimers with other ARFs (Boer et al., 2014; Weijers and Wagner, 2016; Roosjen et al., 2018). Canonically, ARF activity is inhibited by Aux/IAA proteins when auxin levels are low, but this repression is released when auxin concentrations increase (Vernoux et al., 2011; Calderon Villalobos et al., 2012). ARF3, ARF13, ARF17, and ARF23 are distinct in that they lack the PB1 domain (Guilfoyle and Hagen, 2007; Guilfoyle, 2015; Li et al., 2016b), raising the possibility that auxin may not regulate ARF3 activity through the ARF-Aux/IAA pathway (Chandler, 2016). However, a unique C-terminal ETT-specific domain (ES domain) in ARF3 can sense auxin signals (Simonini et al., 2016), through binding auxin directly, and that this interaction determines the expression of ARF3 target genes (Kuhn et al., 2020).
A flowering plant goes through four stages in its life: embryonic development, vegetative growth, reproductive growth, and senescence. The reproductive growth phase is a critical period for plant fitness and is a focal point for biologists and breeders alike (Roeder and Yanofsky, 2006; Alvarez-Buylla et al., 2010). In Arabidopsis, reproduction begins with the transition to flowering, in which the shoot apical meristem (SAM) is transformed into an inflorescence meristem (IM) that continuously forms an orderly arrangement of flower primordia around a central axis (Traas, 2013; Li and He, 2020). IM (or SAM) homeostasis is the basis of indeterminate growth in plants and is modulated by phytohormones, specific genes, and the environment (Lee et al., 2019; Ma et al., 2019). Floral primordia differentiate into a specific number of floral organs at specific locations (Sessions et al., 1997; Thomson and Wellmer, 2019). Floral meristem (FM) determinacy occurs after the initiation of carpel primordia, which is required for normal gynoecium development (Cao et al., 2015; Sun and Ito, 2015). Many mutants with FM determinacy defects exhibit abnormal gynoecia and thus abnormal ovule development (Liu et al., 2011; Liu et al., 2014; Li et al., 2016a). The formation of male and female gametes as well as successful fertilization are crucial for the development of fruit (Drews and Koltunow, 2011; Verma, 2019). Genetic and phylogenetic analyses have confirmed that there is widespread functional redundancy among ARF family members; most arf single mutants display no obvious phenotypes (Remington et al., 2004; Tian et al., 2004; Okushima et al., 2005). However, arf3 mutations have pleiotropic effects on reproductive development, causing abnormalities in meristem homeostasis, floral determinacy, patterning formation, gynoecium morphogenesis, ovule development, and self-incompatibility (Sessions et al., 1997; Tantikanjana and Nasrallah, 2012; Liu et al., 2014; Chandler, 2016; Su et al., 2017). These observations suggest that ARF3 has a special role in auxin signaling and developmental responses.
In a regulatory network, the critical aspects tend to be more precisely and intricately regulated. In addition to mediating auxin signaling using a noncanonical auxin-sensing mechanism, ARF3 transcription is regulated by changes in DNA methylation as well as changes in the abundance of many transcription factors, such as ASYMMETRIC LEAVES1 (AS1)-AS2, APETALA2 (AP2), and AGAMOUS (AG). ARF3 transcript abundance is further fine-tuned by microRNAs (miRNAs) and trans-acting short-interfering RNA–auxin response factors (tasiR-ARFs) at the posttranscriptional level (Marin et al., 2010; Liu et al., 2014; Machida et al., 2015; Simonini et al., 2016). ARF3 translation is controlled by upstream open reading frames (uORFs)-mediated translation reinitiation, followed by intercellular migration that leads to a precise distribution of the protein (Nishimura et al., 2005; Liu et al., 2014). Furthermore, ARF3 has a number of interacting factors that allow it to execute complex and diverse biological functions (Bao et al., 2010; Kelley et al., 2012).
ARF3 expression and activity of the encoded protein are controlled at multiple levels, including transcriptional, posttranscriptional, translational, and epigenetic levels. Furthermore, ARF3 regulates plant development in both cell- and non-cell-autonomous manners. In this section, the modulation and mechanisms of ARF3 at various levels are summarized.
At the transcriptional level, in addition to being induced by auxin, ARF3 is directly repressed by the AS1–AS2 complex, which is involved in the establishment of leaf polarity (Iwakawa et al., 2020). GIANT KILLER (GIK), containing an AT-hook DNA binding motif, is a target of AG that can bind to the ARF3 promoter and inhibit its transcription, indicating that GIK mediates the transcriptional activation of ARF3 depending on AG activity (Ng et al., 2009; Zhang et al., 2018b). The AP2 domain-containing protein APETALA2 (AP2) also directly represses ARF3 transcription to mediate its own role in floral determinacy (Liu et al., 2014; Figure 1; details below).
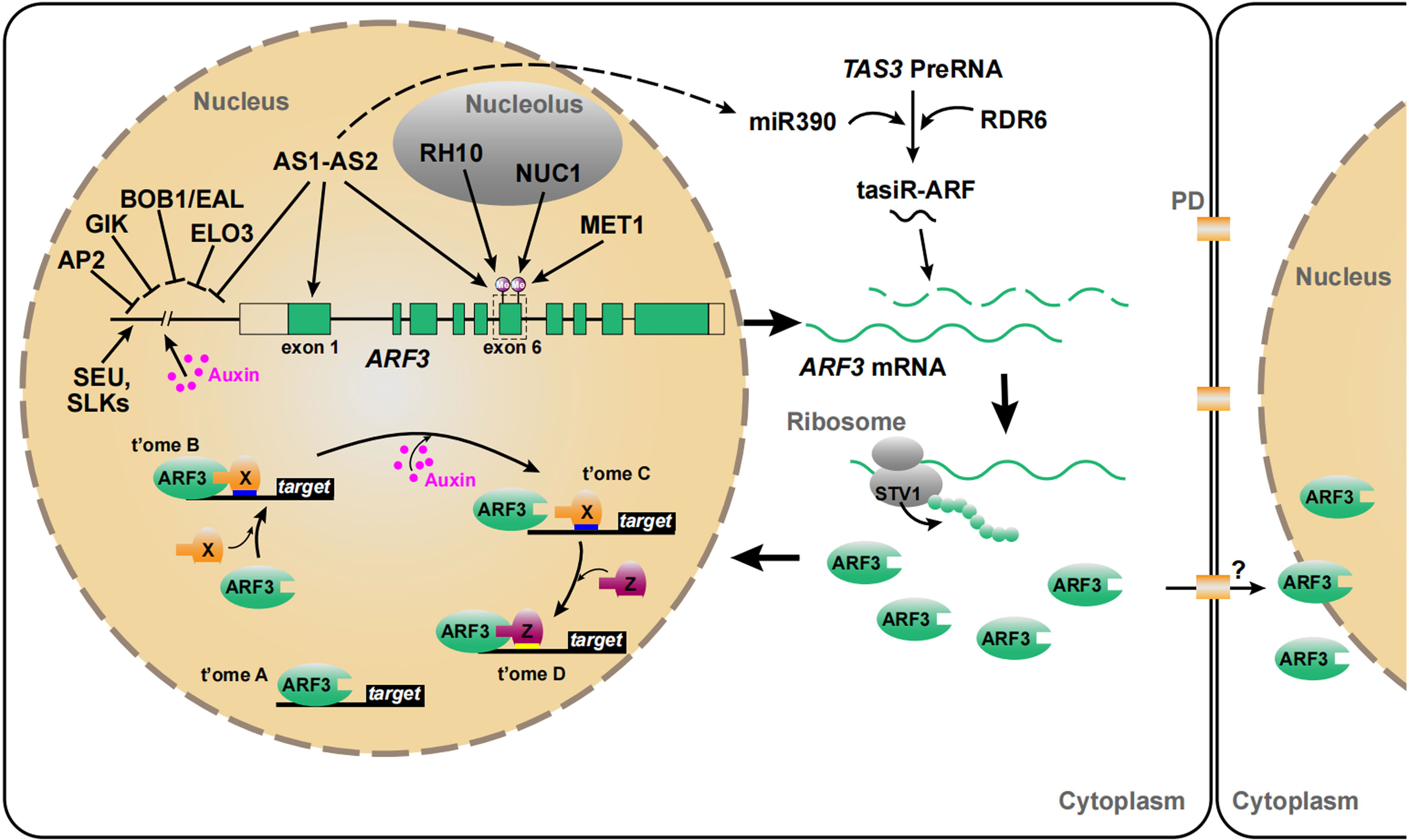
Figure 1 Regulation of ARF3 at multiple levels. ARF3 is affected by transcriptional regulation, tasiR-ARF, translation reinitiation, and DNA methylation (Nishimura et al., 2005; Vial-Pradel et al., 2018; Iwakawa et al., 2020). ARF3 regulates its target genes in two ways, either dependent on auxin or independent of auxin (Simonini et al., 2017). ARF3 can also migrate intercellularly (Zhang et al., 2022). The ARF3 gene, mRNA, and protein are in green. Auxin is in magenta. t’ome A/B/C/D, transcriptome A/B/C/D. “X” and “Z”, the partners of ARF3. PD, plasmodesmata (Whether ARF3 travels from cell to cell via PD remains to be confirmed).
ARF3 is also regulated at the post-transcriptional level by endogenous tasiR-ARF, encoded by the trans-acting siRNA3 (TAS3) locus, which shares a 21- and 22-nt region of sequence similarity with ARF3 and ARF4, respectively (Williams et al., 2005; Hunter et al., 2006b; Figure 1). The biogenesis of tasiR-ARF shares similarities with both the siRNA and miRNA pathways. tasiR-ARF production begins with specific microRNA 390 (miR390)-mediated cap and polyadenosine cleavage of the primary TAS3 transcripts (pri-TAS3) (Vaucheret, 2005; Allen and Howell, 2010; Yoshikawa, 2013). Subsequently, the 5′ cleavage fragments are transformed into double-stranded RNAs (dsRNAs) by RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) and processed by DICER-LIKE 4 to form tasiRNA3 (Vaucheret, 2005; Yoshikawa, 2013). Trans-acting siRNA-mediated repression of ARF3 is involved in the regulation of juvenile development (Peragine et al., 2004), heteroblasty (Hunter et al., 2006), morphology and patterning of leaves and floral organs (Fahlgren et al., 2006; Liu et al., 2014), ovule development (Su et al., 2017), self-incompatibility (Tantikanjana and Nasrallah, 2012), and lateral root growth (Marin et al., 2010).
uORFs have a negative effect on gene translation, but this can be mitigated by ribosomal translation reinitiation (von Arnim et al., 2014). Translation reinitiation is a regulatory mechanism that controls the expression of specific genes involved in developmental programs and responses to environmental signals (Nishimura et al., 2005; Rahmani et al., 2009; Ivanov et al., 2010). ARF3 is predicted to contain two uORFs in the 5′ untranslated region (Nishimura et al., 2005). SHORT VALVE1 (STV1), encoding the large ribosomal subunit protein Ribosomal protein L24B (RPL24B), affects ARF3 translation by mediating translation reinitiation and is associated with apical–basal patterning in gynoecia (Nishimura et al., 2005). STV1 transcripts are distributed throughout shoot apices and in young developing flowers, but are most abundant in vigorously dividing cells such as IMs, flower meristems (FMs), and flower organ primordia. The gynoecia of stv1 mutants display a similar phenotype to ett-2 mutants, which contain a weak allele of ett. stv1-1 ett-2 double mutants display even more serious defects in their gynoecia, including fewer ovaries and morphological abnormalities (Nishimura et al., 2005). In addition, ARF3 uORFs are partially responsible for auxin-related defects of root development in rpl4d and rpl5a (Rosado et al., 2012; Figure 1). Auxin might efficiently use ribosomes to fulfill global and specialized regulation during plant development. (Rosado et al., 2012; Figure 1)
DNA methylation is an epigenetic modification involved in gene regulation, genome stability, and plant development (Bartels et al., 2018; Zhang et al., 2018a). The cytosine residues at CG sites in exons 6 and 10 of ARF3 are often strongly methylated in wild-type plants, suggesting that ARF3 transcription is regulated by epigenetic modification (Zhang et al., 2006; Cokus et al., 2008). AS1–AS2, a nuclear protein complex, plays an important role in maintaining DNA methylation within the ARF3 coding region to regulate adaxial–abaxial partitioning in leaves (Takahashi et al., 2013; Machida et al., 2015; Iwakawa et al., 2020). Genetic analysis revealed that ARF3 and ARF4 are regulated by the modifiers BOBBER1 (BOB1) and ELONGATA3 (ELO3) along with AS1–AS2 (Takahashi et al., 2013). In as1 and as2 mutants, the cytosine residues in CG pairs have a lower methylation level in exon 6 of ARF3, and ARF3 transcript levels increased correspondingly. An AS2-eoe/as2-1, ectopic overexpression line, however, had similar levels of CG methylation as the wild type (Iwasaki et al., 2013). NUCLEOLIN1 (NUC1) and RNA HELICASE10 (RH10), encoding nucleolus-localized proteins, are also involved in the epigenetic repression of ARF3 via gene body DNA methylation through several independent pathways (Vial-Pradel et al., 2018). METHYL TRANSFERASE1 (MET1), encoding a cytosine methyltransferase, can also repress ARF3 transcription in shoot apices by maintaining the CG methylation status of ARF3 (Ronemus et al., 1996; Iwasaki et al., 2013; Figure 1).
Non-cell-autonomous transcription factors can function as cell-to-cell communication signals and play pivotal roles in most processes related to the formation and development of plant organs (Han et al., 2014). ARF3 protein has a distribution pattern distinct from that of its transcript in the meristem, indicating that it is capable of intercellular migration (Liu et al., 2014). This notion has been verified by comparing the distribution patterns of ARF3:ARF3–GFP and ARF3:ARF3-nls-GFP (in which GFP contains a nuclear localization tag, or nls) in IMs. Longitudinal sections imaged with confocal microscopy exhibit ARF3-GFP signals throughout the IM, but ARF3-nls-GFP signals could only be detected in the peripheral zone (PZ) and central zone (CZ) of meristems (Zhang et al., 2022), suggesting that ARF3 functions in a non-cell-autonomous manner (see section 2 for details).
ARF3 does not mediate auxin signaling through the canonical ARF–Aux/IAA pathway since it lacks the PB1 domain (Guilfoyle and Hagen, 2007; Vernoux et al., 2011; Calderon Villalobos et al., 2012; Chandler, 2016). Nonetheless, exogenous auxin was shown to up-regulate ARF3 expression during floral development and de novo organ regeneration processes (Cheng et al., 2013; Zhang et al., 2018b). ARF3 mRNA has been detected in the meristem periphery and in organ primordia, which corresponds with auxin maxima and suggests that ARF3 expression depends on auxin activity (Sessions et al., 1997; Vernoux et al., 2011).
Recently, a noncanonical auxin-sensing mechanism was proposed to explain how the auxin-dependent modulation of ARF3 activity regulates target gene expression (Simonini et al., 2016; Simonini et al., 2017; Simonini et al., 2018; Kuhn et al., 2020). The ETT-ES domain directly binds to auxin to perceive auxin and affects the expression of ARF3 target genes. (Kuhn et al., 2020). Simonini et al. found that ARF3 physically interacts with INDEHISCENT (IND) in an auxin-sensitive manner to control polarity at the gynoecium apex (Simonini et al., 2016). Furthermore, Kuhn et al., showed under low auxin conditions an ARF3- TOPLESS (TPL)- HISTONE DEACETYLASE 19 (HDA19) complex binds to the promoter of PINOID (PID) and HECATE1 (HEC1) keeping their chromatin environments repressed, through de-acetylation, while high nuclear auxin concentrations abolish the ARF3-TPL-HDA19 complex through direct ARF3-auxin interaction (Kuhn et al., 2020). Simonini et al. proposed that auxin affects ARF3 action by controlling its dimerization and transcription. For example, ARF3 regulates target gene expression alone (transcriptome A, t’ome A), and ARF3 function by coupling with process-specific protein partners (“x”) (t’ome B); the dimerization of ARF3 with partners is reversed as auxin concentrations increase, leading to a different transcriptional outcome (t’ome C); and the released ARF3 may interact with new partners (“Z”) (t’ome D) (Simonini et al., 2017; Figure 1).
Plant growth and development are based on the precise control of meristem fate. For example, the SAM needs to maintain homeostasis to continuously produce lateral organs, while FM activity requires appropriate termination, known as FM determinacy, to form the correct number of whorls and floral organs (Lee et al., 2019; Chang et al., 2020). Many regulatory factors, including plant hormones, transcription factors, secreted peptides, and environmental signals, work together to construct a complex regulatory network that controls meristem fate (Cao et al., 2015; Sun and Ito, 2015). Recent studies revealed that ARF3-mediated auxin signaling contributes to the fine-tuning of meristem homeostasis and FM determinacy (Liu et al., 2014; Zhang et al., 2018b; Zhang et al., 2022).
Meristems possess the ability to self-renew and continuously produce new tissues and organs (Zhang and Yu, 2014). To maintain meristem homeostasis, dynamic signals and gene expression need to be precisely regulated both spatially and temporally (Lee et al., 2019). Auxin plays a critical role in specifying the fate of organ primordia in the SAM (Vernoux et al., 2011; Janocha and Lohmann, 2018). High levels of auxin in the peripheral regions of the SAM mediate primordial differentiation, but low levels are required in the organizing center (OC) and CZ (Schaller et al., 2015; Shi et al., 2018; Ma et al., 2019). WUSCHEL (WUS), a homeodomain transcription factor, mediates stem cells’ resistance to auxin but allows low levels to persist in the CZ to ensure stem cell maintenance (Ma et al., 2019). Auxin also interacts synergistically with cytokinin (CK) signals to regulate WUS expression in the SAM and FMs (Schaller et al., 2015; Zhang et al., 2018b).
However, how auxin, which is concentrated in the PZ, regulates cytokinin signaling and WUS expression in the OC remains additional research. Recent observations that ARF3 migrates between cells suggest that ARF3 may act as a messenger in the auxin-mediated regulation of WUS and cytokinin signaling (Lanctot, 2022; Zhang et al., 2022). ARF3 transcript patterns coincide with auxin maxima in regions of primordia initiation, but ARF3 protein has been detected throughout the meristem (Liu et al., 2014; Simonini et al., 2017). Additionally, the transgenic expression of ARF3:ARF3-nls-GFP effectively prevents ARF3-nls-GFP from translocating into the OC from adjacent cells in the SAM (Zhang et al., 2022). Statistical analysis demonstrated that arf3-29 mutants have larger SAMs than the wild type (Figures 2A, B), and ARF3:ARF3-nls-GFP can only partially rescue this mutant phenotype, suggesting that ARF3 protein migration is required for SAM activity. Furthermore, ARF3 intercellular migration is necessary for its inhibition of cytokinin signaling and WUS expression. Compared with ARF3:ARF3-GFP arf3-29, ARF3-nls-GFP enrichment at the ARABIDOPSIS HISTIDINE KINASE4 (AHK4) and WUS loci was significantly reduced, while the transcript levels of AHK4 and WUS-DsRed increased. These findings reveal a network of ARF3-mediated crosstalk between phytohormones and gene expression that maintains meristem homeostasis (Lanctot, 2022; Zhang et al., 2022; Figure 2E).
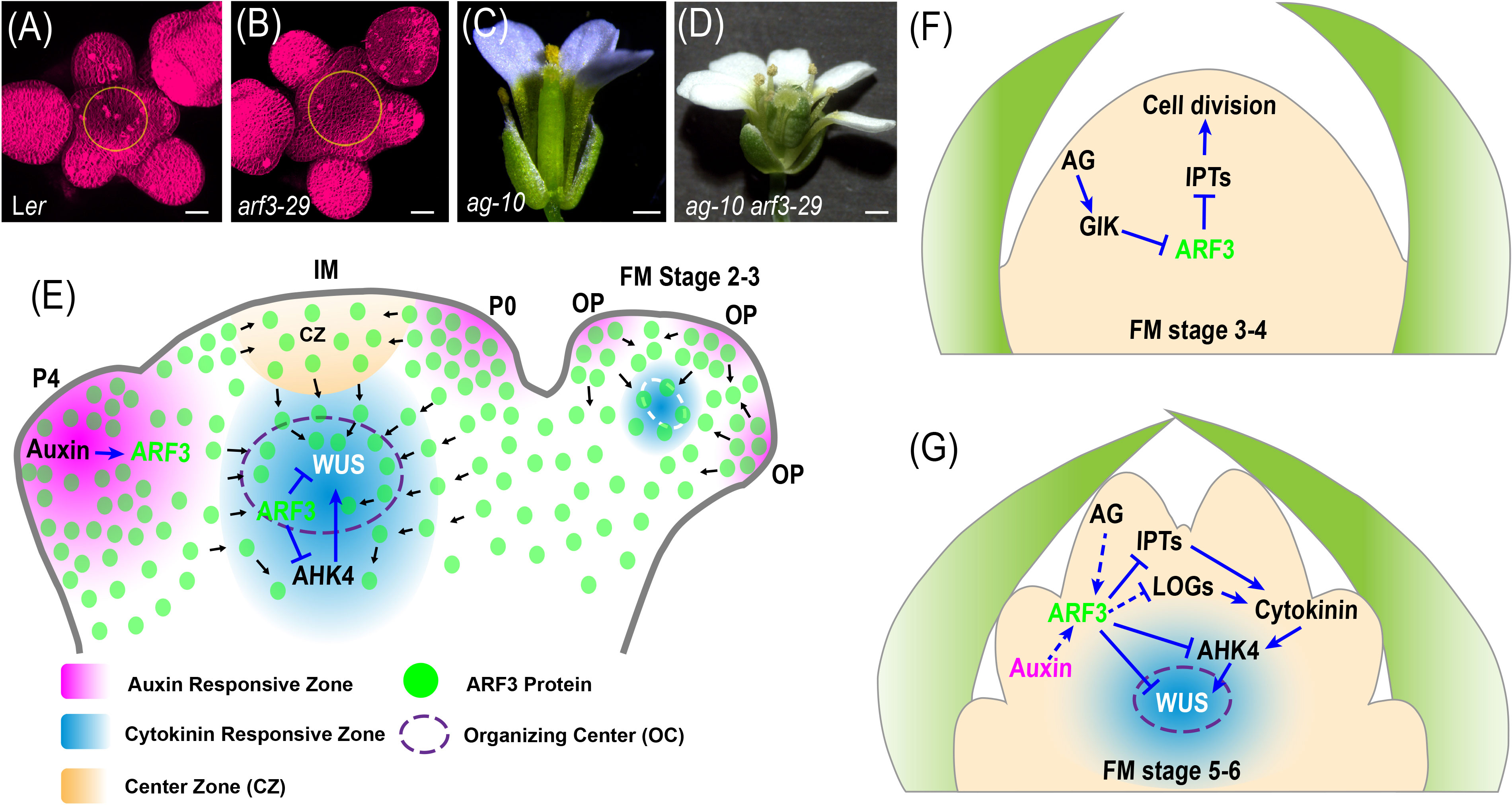
Figure 2 Models of SAM and FM activity control by ARF3. (A, B), Representative images of SAM for the indicated genotypes (from Zhang et al., 2022, provided by Dr. Ke Zhang). (C, D), Representative images of flower for the indicated genotypes. (E), In the IM, auxin promotes ARF3, which migrates from the peripheral zone (PZ) to the organizing center (OC) where it directly represses AHK4 and WUS expression to control meristem activity in a non-cell-autonomous manner. Figures adapted from (Zhang et al., 2022). P0 and P4, primordia of flower buds at different developmental stages; OP, flower organ primordia. (F), At stages 3-4 of floral development, AG indirectly represses ARF3 expression through GIK, and consequently enhancing IPTs to promote cell division and maintain the meristem cell population, thereby favoring floral meristem (FM) maintenance. Figures adapted from (Zhang et al., 2018b) (G), At stages 5-6 of floral development, auxin and AG promote ARF3 expression, which in turn directly represses IPTs, and AHK4 and indirectly represses LOG genes expression. The resulting inhibition of cytokinin activity is required for proper temporal termination of WUS expression during FM determinacy. Figures adapted from (Zhang et al., 2018b). Blue solid and dashed arrows indicate direct and indirect positive regulation, respectively. Blue solid and dashed block lines indicate direct and indirect negative regulation, respectively. Black arrows indicate the direction of protein movement.
Unlike stem cells in the SAM, floral stem cells cease to be stem cells after generating carpel primordia (Liu et al., 2011). At stages 3–4 of floral development, AG indirectly represses ARF3 expression through GIK (Ng et al., 2009) and other factors, consequently enhancing ISOPENTENYLTRANSFERASE (IPT) and cell cycle gene expression to promote cell division and maintain the meristem cell population, thereby favoring FM maintenance (Figure 2F) (Zhang et al., 2018b). However, some mutations uncouple the formation of carpel primordia from the loss of stem cell identity, suggesting that the termination of floral stem cell maintenance is not synonymous with the differentiation of stem cells into carpel cells (Prunet et al., 2008; Sun et al., 2009; Ji et al., 2011). The orchestration of multiple regulators ensures precise timing of stem cell termination (Sun and Ito, 2010; Xu et al., 2019). In spite of FM determinacy being normal in arf3-29 mutants, this mutation can enhance FM determinacy defects in ag-10 plants, which harbor a weak ag allele (Liu et al., 2014). arf3-29 ag-10 double mutants have bulged and unfused carpels with hyperplasia tissue on the inside, although most of the siliques in ag-10 single mutant plants are morphologically similar to wild-type siliques (Figures 2C, D). WUS is a key regulator of stem cell maintenance in both the SAM and FM (Laux et al., 1996; Cao et al., 2015). WUS expression was undetectable at stage 6 of floral development in wild-type flowers and 90% of ag-10 flowers have normal WUS expression patterns (Yumul et al., 2013; Liu et al., 2014). All ag-10 arf3-29 double mutant flowers, however, had prolonged WUS expression until or beyond stage 7 (Liu et al., 2014). ARF3 integrates the functions of AP2 and AG to repress WUS expression in floral meristem determinacy (Liu et al., 2014; Shang et al., 2019; Thomson and Wellmer, 2019). AG acts independently of AP2 to terminate the FM (Lenhard et al., 2001; Zhao et al., 2007). AG promotes the binding of ARF3 to the WUS locus in vivo and ARF3 is a direct target of AP2 (Liu et al., 2014).
At stages 5–6 of floral development, ARF3 mediates AG to promote FM determinacy by repressing cytokinin biosynthesis and signaling (Zhang et al., 2018b; Lee et al., 2019; Xu et al., 2019) (Figure 2C). Exogenous cytokinin treatment enhances the FM determinacy defect of ag-10 mutants by modulating the temporal termination of WUS expression and cell division. ARF3 directly represses the expression of IPT3, 5, and 7, cytokinin biosynthetic genes, and indirectly represses LONELY GUYs (LOGs) to fine-tune cytokinin levels and activity (Cheng et al., 2013; Zhang et al., 2018b; Figure 2G). Furthermore, ARF3 directly inhibits the expression of cytokinin receptor genes like AHK4 to regulate the perception of cytokinin in OC regions of the FM (Schaller et al., 2015; Zhang et al., 2018b; Figure 2G). Additionally, ARF3 regulates FM determinacy in a manner that depends on its ability to move between cells. The non-mobile ARF3 (ARF3-nls-GFP) only partially rescues the ag-10 arf3-29 double mutant phenotype (Zhang et al., 2022), but ARF3:ARF3-GFP ag-10 arf3-29 displayed full complementation (Liu et al., 2014; Zhang et al., 2018b). However, the function of ARF3 in FM determinacy is irrelevant to its function in organ patterning. Considering that ag-10 kanadi1 (kan1) kan2 triple mutant flowers have no FM determinacy defects, the KANs-ARF3 complex is not responsible for FM determinacy (Zheng et al., 2018).
The regularity of plant architecture and morphology is a striking and mysterious phenomenon. The lateral organs, such as leaves, flowers, and floral organs, are arranged in regular patterns around a central axis (Reinhardt, 2005; Traas, 2013; Bartlett and Thompson, 2014). Leaves and flowers arise from the meristem in stereotypical patterns, known as phyllotaxis, which are related to the divergence angle between successive organs, most commonly approximating 137.5° (Smith et al., 2006; Okabe, 2012). The floral organs also exhibit a characteristically symmetrical arrangement, with their number, position, and shape being precisely regulated (Reinhardt, 2005; Thomson and Wellmer, 2019). arf3 mutants exhibit obvious abnormalities in phyllotactic patterning, floral organ patterning, floral organ number, the patterning of abaxial tissues, and the establishment of apical and basal boundaries in primordia (Figures 3A, B, G–J; see section 4 for details).
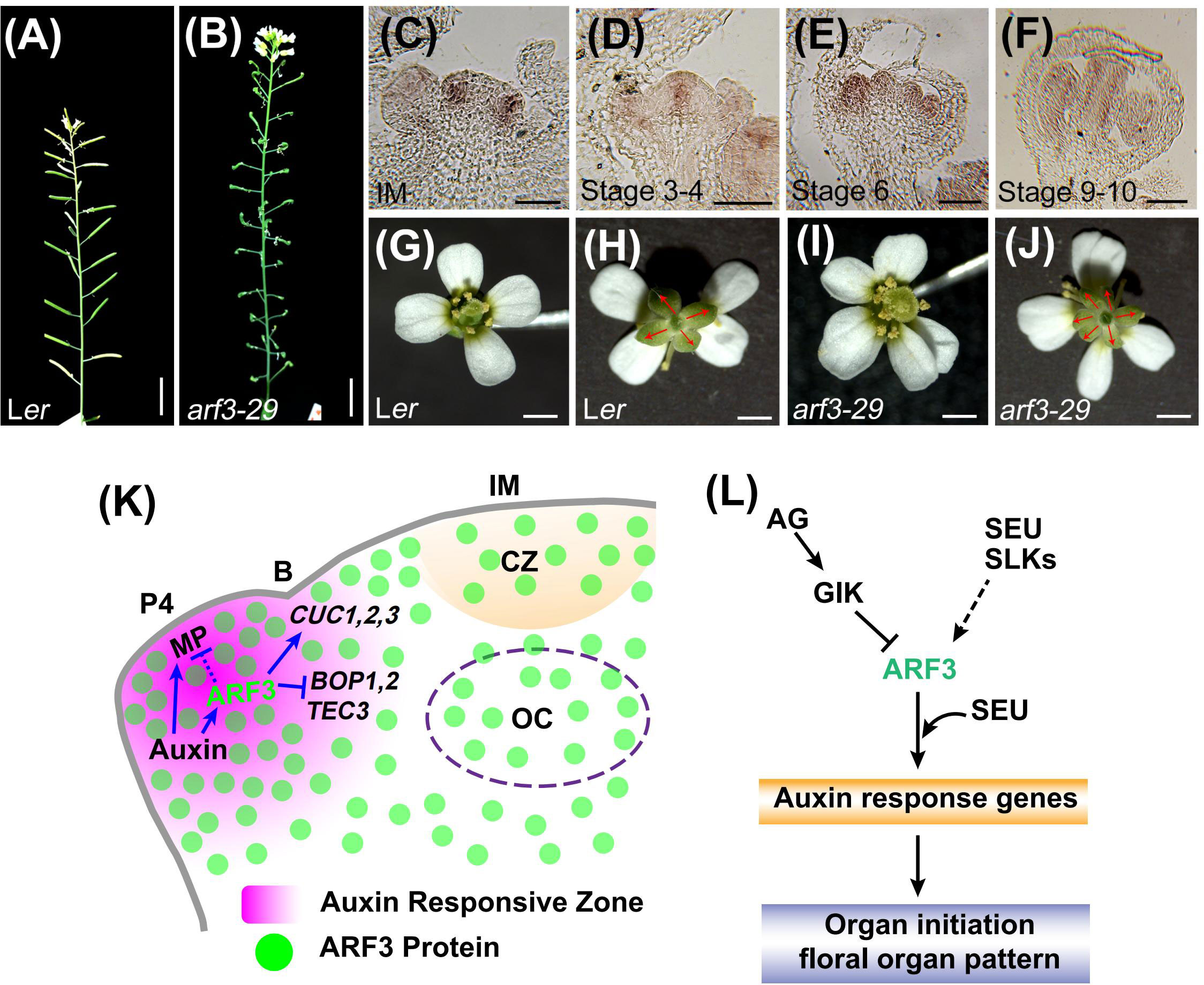
Figure 3 ARF3 controls phyllotaxy and floral organ patterning cell autonomously. (A, B), Representative images of inflorescence stems for the indicated genotypes (from Zhang et al., 2022, provided by Dr. Ke Zhang). Scale bars = 1 cm. (C–F), ARF3 expression in IM (C) and FM at stage 3 (D), stage 6 (E) and stage 9–10 (F) examined by in situ hybridization (from Zhang et al., 2022, provided by Dr. Ke Zhang). Bars = 50 μm. (G–J), The number of floral organs in the indicated genotypes. Compared with Ler (G, H), arf3 flowers have more sepals, petals and stamens (I, J). The sepals are marked by red arrows. Bars = 1 mm. (K), (A) model of phyllotactic patterning control by ARF3. Auxin promotes ARF3 in floral primordia, where it regulates meristem–organ boundary-specific genes (CUC1–3, BOP1–2, and TEC3) and MP in a cell-autonomous manner. Figures adapted from (Zhang et al., 2022). P4, primordia of flower buds at different developmental stages; (B), meristem–organ boundary. Blue solid arrows indicate direct and indirect positive regulation. Blue solid and dashed block lines indicate direct and indirect negative regulation, respectively. (L), A model of floral organ patterning control by ARF3. ARF3 cooperates with SEU to control organ initiation and floral organ pattern by regulating auxin response genes, and ARF3 is inhibited by GIK directly and promoted by SEU and SLKs. Solid and dashed arrow indicates positive regulation. Block lines indicate negative regulation.
Although many mathematical, physical, and chemical models have been proposed to explain the mechanism of phyllotactic pattern formation, the regular initiation of organs and their spatial and temporal positioning is the basis for the formation of phyllotactic patterns from a developmental biology perspective (Reinhardt, 2005; Smith et al., 2006; Traas, 2013; Bergeron and Reutenauer, 2019). The phytohormone auxin plays a critical role in establishing phyllotactic patterns by promoting organ initiation (Prasad et al., 2011; Bhatia et al., 2016). Loss-of-function mutations in boundary genes such as CUP-SHAPED COTYLEDON2 (CUC2), REPLUMLESS (RPL), and BELLRINGER (BLR), which are involved in the formation of meristem-to-organ boundaries, often cause altered phyllotactic patterning (Byrne et al., 2003; Peaucelle et al., 2007; Bencivenga et al., 2016). arf3 mutant plants display dramatically altered phyllotaxis. ETTC2S, an auxin-insensitive ett line, has a phyllotaxy defect, suggesting that ARF3 functions in an auxin-dependent manner (Simonini et al., 2017). Recently, evidence has emerged that ARF3 shares a common target gene, TARGETS UNDER ETTIN CONTROL3 (TEC3), with RPL that regulates phyllotactic patterning (Simonini et al., 2017). Furthermore, ARF3 can directly regulate the expression of boundary-specific genes, including CUC3, BLADE ON PETIOLE (BOP1), and BOP2, to control phyllotaxy in a cell-autonomous manner (Figure 3K). Both ARF3-GFP and ARF3-nls-GFP can occupy the CUC3, BOP1, BOP2, and TEC3 loci with similar enrichment levels in the arf3 background. In arf3-29, CUC1, CUC2, and CUC3 expression were lower than in the corresponding wild-type plants, while the transcript levels of BOP1, BOP2, and TEC3 were higher (Zhang et al., 2022). Therefore, ARF3 regulates boundary specific genes to determine the location of primordium initiation and thus influence phyllotactic patterning.
During flower development, FM and organ identity genes LEAFY (LFY) and APETALA1 (AP1)/CAULIFLOWER (CAL) play key roles in the patterning of FMs by setting the spatial limits of expression of floral organ identity genes, such as AP1, AP3, PISTILLATA (PI), AG, and SEPALLATA (SEP) (John L. Bowman et al., 1989; Lee et al., 1997; Lenhard et al., 2001; Lohmann et al., 2001; Krizek and Fletcher, 2005; Chae et al., 2008; Alvarez-Buylla et al., 2010;). An “ABC” model, later extended to an “ABCDE” model, was formulated that proposes floral organ identity genes function combinatorially (Coen and Meyerowitz, 1991; Krizek and Fletcher, 2005; Thomson and Wellmer, 2019).
ARF3 displays a complex expression pattern during early-stage floral development (Figures 3C–F). For example, ARF3 mRNA is detected throughout the FM during stages 1-2. In stages 3-4, it is expressed in incipient stamen and gynoecium primordia. In stage 5-7, it concentrated in the abaxial side of petal, stamen, and gynoecium primordia (Sessions et al., 1997; Zhang et al., 2022). In line with its expression patterns, ARF3 regulates the FM and floral organ initiation, and loss of function in ARF3 leads to abnormal floral patterning. For example, the sepals and petals numbers are increased, while the number of stamens decreases (Sessions et al., 1997; Zhang et al., 2022). SEUSS (SEU) encodes a protein that interacts with ARF3 to affect auxin responses and facilitate floral organ patterning and growth (Pfluger and Zambryski, 2004; Figure 3L). In addition, three SEU-LIKE genes (SLK1, 2, and 3) have a degree of functional redundancy with SEU, and the expression level of ARF3 was reduced in seu slk1 inflorescences (Bao et al., 2010; Figure 3L).
ARF3 plays a prominent role in gynoecium patterning and identity (Simonini and Ostergaard, 2019). Wild-type Arabidopsis gynoecia are composed of a stigma-and-style-capped bilocular ovary on an unelongated internode. The ovary has two medial furrows and two lateral valves, and is the largest region of the gynoecium (Sessions et al., 1997; Roeder and Yanofsky, 2006). In arf3 (ett) mutants, the valve tissue disappears basally, being replaced by structures intermediate between the abaxial style and internode. These aberrant phenotypes indicate ARF3 participates in apical/basal axis patterning of the gynoecium. (Sessions and Zambryski, 1995; Sessions et al., 1997). However, abaxial/adaxial polarity is also affected in ett gynoecia where an everted transmitting tract tissue develops in outgrowths on the exterior of the gynoecium (Nemhauser et al., 2000; Roeder and Yanofsky, 2006).
ARF3 mRNA is expressed in a ring around the FM preceding the formation of gynoecia at stage 5. The upper and bottom edges of the ring are required for proper specification of the apical and basal boundaries, respectively (Roeder and Yanofsky, 2006). Previous studies have shown auxin gradients play a key role in establishing the apical/basal axis of a gynoecium. (Nemhauser et al., 2000; Hawkins and Liu, 2014; Sehra and Franks, 2015; Marsch-Martinez and de Folter, 2016). Treatment with N-1-naphthylphthalamic acid (NPA), an inhibitor of polar auxin transport, promotes ett-like phenotypes. ARF3 affects the auxin gradient by negatively regulating PIN-FORMED1 (PIN1) and PIN3 auxin efflux transporters (Simonini et al., 2017), and two basic helix-loop-helix (bHLH) transcription factors, SPATULA (SPT) and (HEC1) (Heisler et al., 2001; Moubayidin and Ostergaard, 2014; Schuster et al., 2015; Figure 4). HEC1 is able to interact with SPT in yeast, and they act together to buffer auxin and cytokinin signals during gynoecium development (Schuster et al., 2014; Schuster et al., 2015; Figure 4). HEC1 can stimulate auxin biosynthesis and directly promotes the expression of PIN1and PIN3 auxin efflux transporters. It also inhibits CK signals by promoting the expression of type-A ARRs. TEOSINTE BRANCHED1-CYCLOIDEA-PCF 15 (TCP15), a class I TCP transcription factor, can also modulate cytokinin and auxin responses during gynoecium development. TCP15 can not only affect auxin homeostasis by inhibiting auxin synthesis-related YUC genes, but it can also be induced by cytokinin and modulates the expression of cytokinin-responsive genes (Steiner et al., 2012; Lucero et al., 2015; Figure 4). The SPT- IND complex, however, directly represses the expression of PID, which modulates PIN polarization (Girin et al., 2011; Schuster et al., 2015). Recently, Sara Simonini et al., showed ARF3 and IND interact to form a complex that mediates gynoecium patterning via PID transcriptional control, and auxin affects the activity of this complex (Simonini et al., 2016). Furthermore, Andre´ Kuhn et al., showed under low auxin conditions an ARF3-TPL-HDA19 complex binds to the promoter of PID and HEC1 keeping their chromatin environments repressed, through de-acetylation, while high nuclear auxin concentrations abolish the ETT-TPL-HDA19 complex through direct ARF3-auxin interaction (Kuhn et al., 2020). TOUSLED (TSL), a nuclear serine/threonine protein kinase, plays a role in apical tissue formation (Roe et al., 1993; Roe et al., 1997). In ett mutants, TSL is ectopically expressed in the stylar medial tissue and valves, which indicates ARF3 limits TSL expression to the apex of gynoecia to maintain apical boundary identity (Roe et al., 1997; Sehra and Franks, 2015). STYLISH1 (STY1) and STY2 are members of the SHI/STY-family, and sty1-1 sty2-1 double mutants show a reduced production of stylar xylem and a basalized point of medial vein bifurcation in the gynoecium (Kuusk et al., 2002). STY1 synergistically interacts with ARF3 during apical-basal patterning and apical fusion of gynoecia by controlling YUCCA-mediated auxin biosynthesis and polar auxin transport (PAT) (Sohlberg et al., 2006; Figure 4). GIK, as a direct target of AG, negatively regulates ARF3 expression and plants overexpressing GIK displays stigmatic tissue outgrowth, short valves, and bipartite stigmas with ectopic ovules, closely resembling ett mutants (Ng et al., 2009; Figure 4).
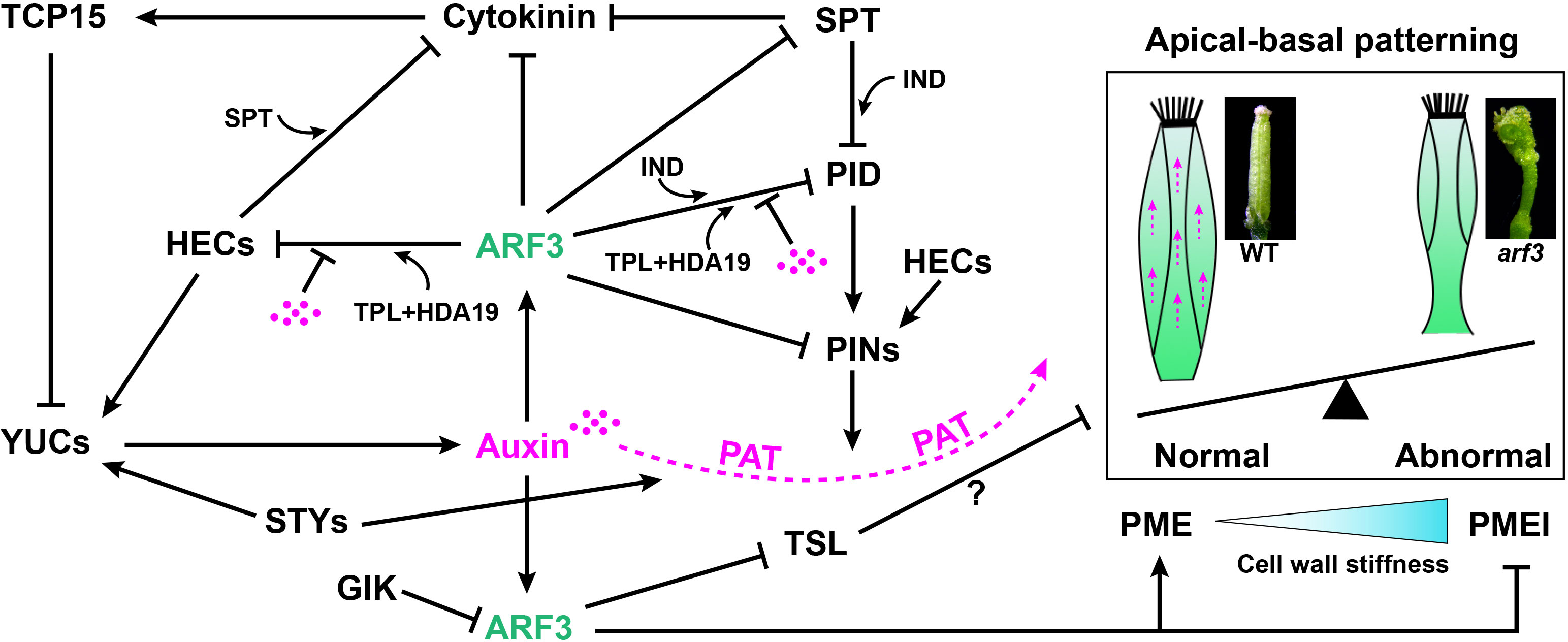
Figure 4 A model of the ARF3-mediated regulatory network in the apical/basal axis formation of gynoecia. Solid arrow indicates positive regulation. Block lines indicate negative regulation. Dashed arrow indicates polar auxin transport (PAT). The magenta dots indicate auxin.
Recently, evidence has shown ARF3 modulates gynoecium morphogenesis via cell wall dynamics (Andres-Robin et al., 2018). Amélie Andres-Robin et al., proposed ARF3 functions by reducing cell wall stiffness through the covalent modification of pectin. ARF3 acts to increase pectin methylesterase (PME) activity by stimulating PME genes and repressing pectin methylesterase inhibitor (PMEI) gene expression in the valves (Pelloux et al., 2007; Andres-Robin et al., 2018; Wormit and Usadel, 2018). It is possible that ARF3 affects the activity of PME/PMEI indirectly, and it is in fact a consequence of other processes affected by ARF3 such as auxin dynamics (Nemhauser et al., 2000; Andres-Robin et al., 2018; Figure 4).
In leaf and gynoecium primordia, ARF3 transcription is restricted to the abaxial cells during stages 5-8 and this is essential in determining adaxial-abaxial polarity. (Sessions et al., 1997; Iwasaki et al., 2013; Liu et al., 2014; Husbands et al., 2015). Previous reports revealed transcription of ARF3 and its homolog ARF4 is restricted to the abaxial side of the leaf by TAS3-derived trans-acting small interfering RNAs (tasiRNAs) known as tasiR-ARFs (Williams et al., 2005; Hunter et al., 2006a). The tasiR-ARFs are distributed on the adaxial side, forming a gradient towards the abaxial side that limits ARF3 and ARF4 to the abaxial side (Garcia et al., 2006; Chitwood et al., 2009). Recently, ARF3:ARF3m-GFP, a TAS3 tasiRNA-insensitive transgene, led to a ubiquitous distribution of ARF3 mRNA in gynoecium primordia and ovules, suggesting the expression pattern of ARF3 is regulated by tasiR-ARFs during gynoecium development (Liu et al., 2014; Su et al., 2017).
ARF3, cooperating with ARF4, regulates the adaxial/abaxial polarity of valves in conjunction with KANADI (KAN) (Pekker et al., 2005; Figure 5). The GARP transcription factor KAN1 specifies abaxial polarity in both the gynoecium and the leaves (Eshed et al., 2001; Kerstetter et al., 2001). ARF3 interacts physically with KAN1 in yeast, and the activity of KAN1 is required for ARF3 (Pekker et al., 2005; Kelley et al., 2012). YABBY genes are expressed on the abaxial side of all lateral organ primordia and are capable of inducing the differentiation of abaxial cell types when expressed adaxially (Eshed et al., 1999; Sawa et al., 1999; Siegfried et al., 1999). FILAMENTOUS FLOWER (FIL), a member of the YABBY family, is a target of ARF3, and both KAN and ARF3 promote the expression of FIL (Eshed et al., 2001; Garcia et al., 2006; Figure 5).
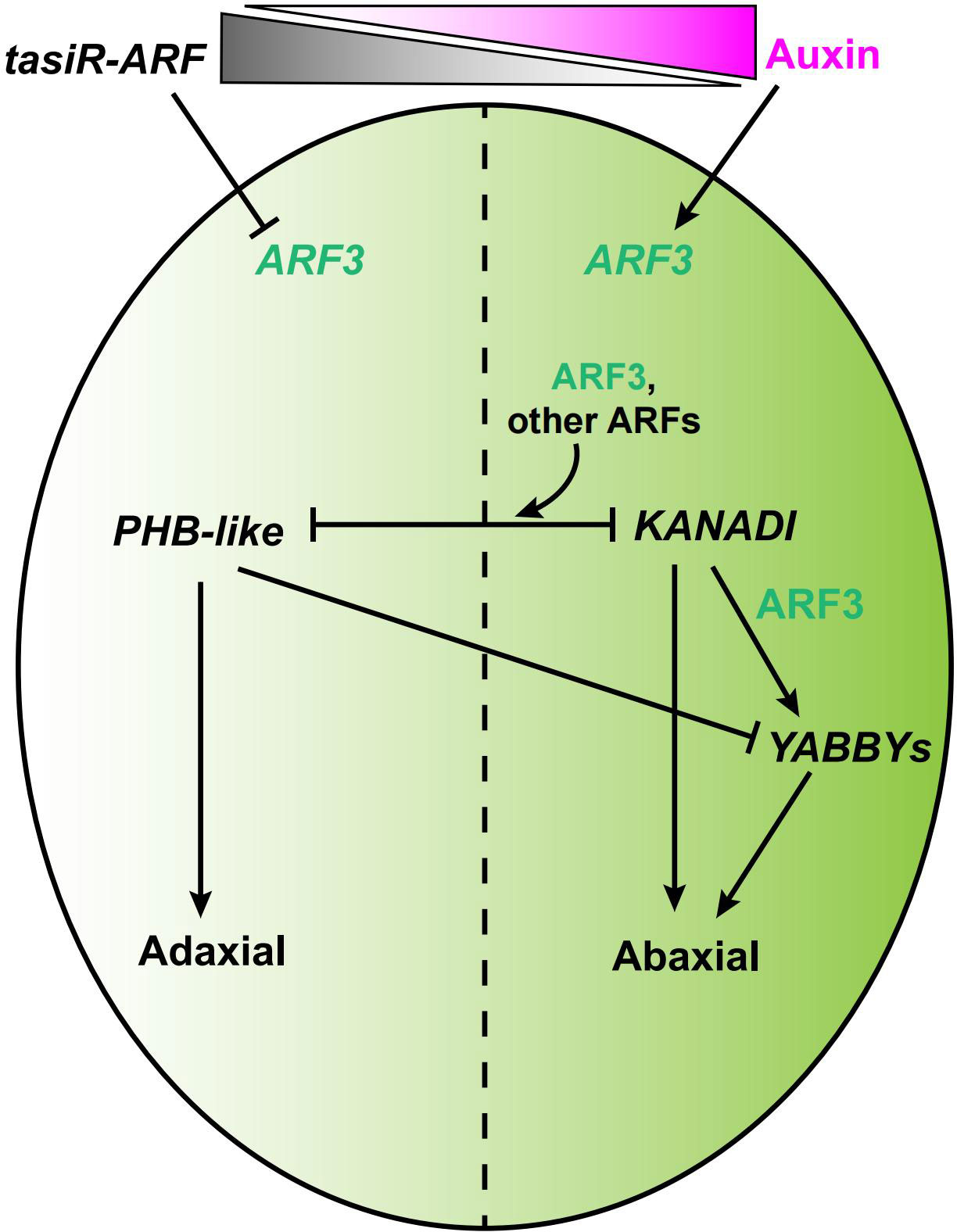
Figure 5 A model of the ARF3-mediated regulatory network in the abaxial/adaxial axis formation of gynoecia. ARF3 is indicated in green, auxin in magenta, and tasiR-ARF3 in gray. Solid arrow indicates positive regulation. Block lines indicate negative regulation.
Carpel fusion creates a protected cavity that houses the ovules and the placenta. Ovules develop from a ridge of tissue that originates on the adaxial side of the gynoecium at stages 8-10 (Bowman et al., 1999). The ovule primordia arise as finger-like projections from the placenta, and the megaspore mother cell (MeMC) is derived from a sub-epidermal cell at the distal end of the ovule primordium, which is committed to generating the haploid, multicellular female gametophyte (Drews and Koltunow, 2011). Recent studies have shown ARF3 is involved in ovule development and mediates the inhibition of TAS3 during MeMC formation (Su et al., 2017; Pinto et al., 2019). The THO/TREX complex plays a conserved role in processing and transporting long RNA molecules during small interfering RNA (siRNA) biosynthesis (Chavez et al., 2000; Strasser et al., 2002; Yelina et al., 2010). Mutations in THO complex members, such as TEX, HPR1, and THO6, can inhibit megaspore mother cell fate by accelerating the biogenesis of tasiRNAs that repress ARF3 expression. Loss of function in TEX1, RDR6, and TAS3 leads to supernumerary MeMC formation and the ectopic expression of ARF3 can phenocopy these MeMC defects (Su et al., 2017; Lora et al., 2019).
ARF3 also directly interacts with KAN4/ABERRANT TESTA SHAPE (ATS) to define the boundary between integument primordia (Kelley et al., 2012; Shirley et al., 2019; Figure 6). The expression pattern of ARF3 overlaps with ATS, being restricted to the inner integument during ovule development (Kelley et al., 2012; Lora et al., 2015). The ats mutant forms aberrant ovules, in which the inner and outer integument cell layers grow as a single fused structure (McAbee et al., 2006; Kelley et al., 2012). The ovule phenotype of ett resembles ats, and the ovules of ats ett double mutant plants showed no phenotypic differences to either single mutant. Thus, loss of either ATS or ETT is sufficient to disrupt a common regulatory pathway that is mediated by both TFs. (Kelley et al., 2012). Dior R. Kelley et al. proposed the ATS-ARF3 module might be directly linked to auxin signaling by restricting PIN activity and, thus, auxin flow (Benkova et al., 2003; Bencivenga et al., 2012; Kelley et al., 2012; Figure 6).
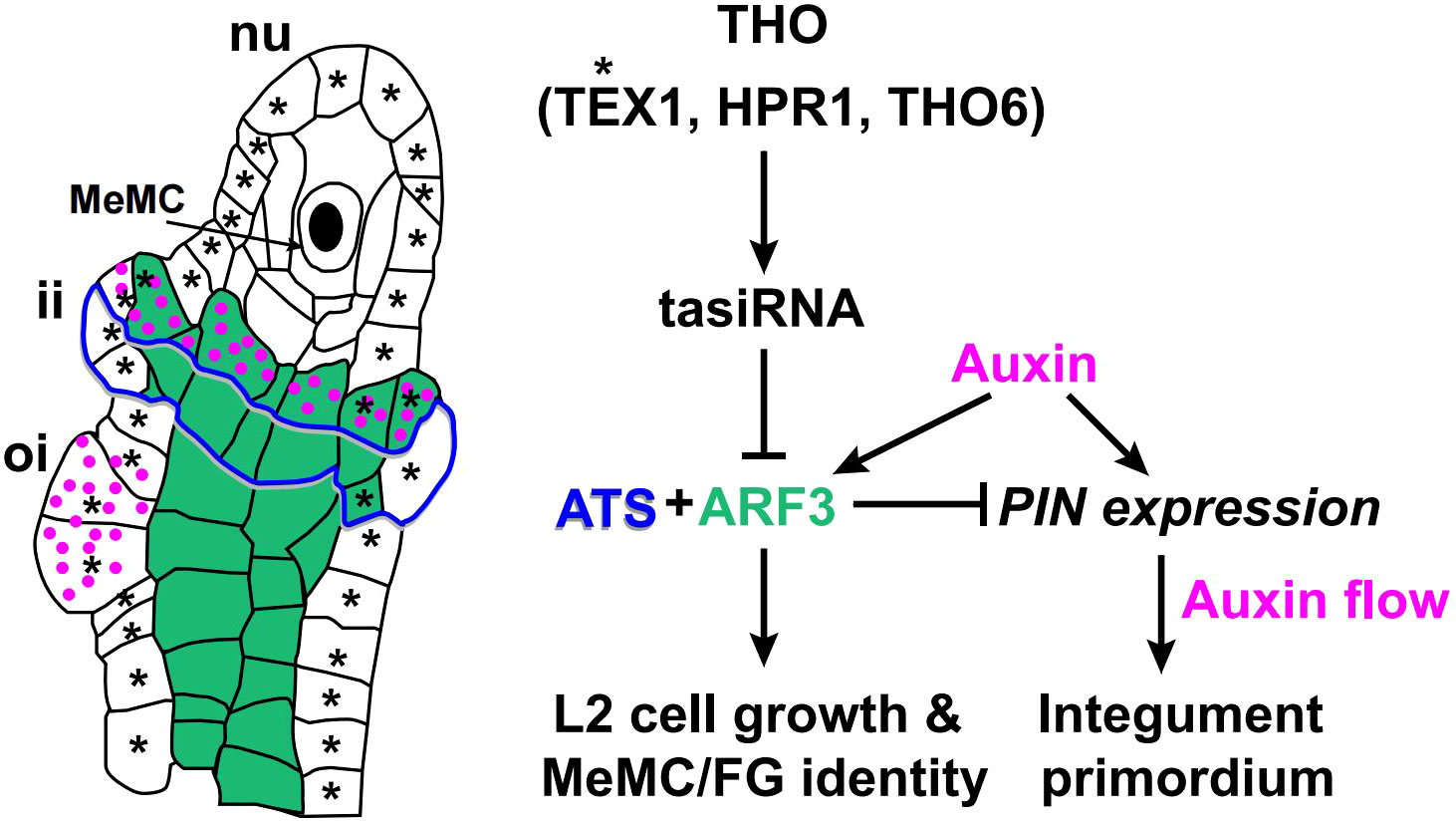
Figure 6 A model of the ARF3-mediated regulatory network during ovule development. ARF3 is indicated in green, auxin in magenta, tasiR-ARF in gray, and ATS in blue with shades of gray (Kelley et al., 2012). Green area indicates the distribution of ARF3 (Su et al., 2017; Lora et al., 2019). The magenta dots indicate the distribution of auxin. Blue lines with shades of gray indicate the distribution region of ATS. Asterisks indicate the distribution of TEX1. Solid arrows indicate positive regulation. Block lines indicate negative regulation. nu, nucellus; ii, inner integument; oi, outer integument.
Auxin gradients also modulate ovule and integument development by controlling the activity of ARF proteins, such as MONOPTEROS (MP)/ARF5 and ARF3 (Galbiati et al., 2013; Sehra and Franks, 2015; Marsch-Martinez and de Folter, 2016; Simonini et al., 2016). ARF3 regulates ovule integument development in an auxin-sensitive fashion. Previous research has shown ETT : ETT-GFP can complement the integument defect of ett, but ETT : ETT2C-S, an auxin-insensitive variant, was not able to rescue this defect (Simonini et al., 2016).
Self-incompatibility is one of the most common mechanisms by which angiosperms prevent self-fertilization and enforce outcrossing. A genetic self-incompatibility (SI) allows cells in the pistil to recognize and specifically prevent “self” pollen from affecting fertilization (Nasrallah, 2019). However, Arabidopsis is highly self-fertile due to it lacking functional alleles of two genes, the S-locus receptor kinase (SRK) and the S-locus cysteine-rich protein (SCR), which function in SI in the Brassicaceae (Tantikanjana and Nasrallah, 2012; Nasrallah, 2019). Tantikanjana and Nasrallah demonstrated that ARF3 acts non-cell-autonomously as a regulator of SI by mediating TAS3 tasiRNA to enhance SI. ARF3:ARF3mut, however, a tasiRNA-insensitive variant, enhances SI whereas loss-of-function ett mutations abolish SI (Tantikanjana and Nasrallah, 2012).
ARF3 is a noncanonical auxin response factor with a special structure, is regulated at multiple levels, and performs a wide range of functions. Because ARF3 lacks the C-terminal domain (PB1 domain), its response to auxin does not depend on the AUX/IAA pathway. In addition to inducing the transcription of ARF3, auxin affects its regulation of target genes. Studies suggest that ARF3 binds auxin and regulates its target genes in either an auxin-dependent or -independent manner. Aside from transcriptional regulation, ARF3 is also regulated by tasiR-ARF, translation reinitiation, and DNA methylation. Recent reports have shown that ARF3 can also modulate SAM maintenance through intercellular migration. Sophisticated regulation often implies complex functionality. ARF3 functions in both the regulation of patterning formation and meristem activity. ARF3 can affect phyllotactic patterning by changing the divergence angle between successive floral primordia, can control floral organ patterning by changing the number and arrangement of floral organs, and plays an important role in the regulation of apical/basal and abaxial/adaxial organ polarity. ARF3-mediated auxin signaling contributes to the fine-tuning of meristem activity, and is involved in SAM homeostasis and FM determinacy. Future research into the biological function and regulatory mechanisms of ARF3 may (1) further clarify and verify the auxin-response mechanism of ARF3, for example, by uncovering the mechanism by which auxin regulates ARF3 transcription, and the mechanism by which auxin affects the interaction between ARF3 and cooperators to modulate target gene expression; (2) explore the molecular mechanism that regulates ARF3 intercellular migration, screening for the domains that affect the spatial patterning of ARF3 protein; and (3) analyze the molecular mechanisms by which ARF3 regulates meristem homeostasis to better understand this regulatory network.
KZ and XL conceived and designed the project. YF and HZ wrote the paper. YM drew the model. CL provided some suggestions for the paper. All authors contributed to the article and approved the submitted version.
The authors apologize to the colleagues whose work or original publications could not be cited because of space limitations. This work was supported by research grants from National Science Foundation of China (31900623); Independent research project from State Key Laboratory of North China Crop Improvement and Regulation; Hebei Natural Science Foundation (C2021205013) and Full-time introduction of high-end talent research project (2020HBQZYC004).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Allen, E., Howell, M. D. (2010). miRNAs in the biogenesis of trans-acting siRNAs in higher plants. Semin. Cell Dev. Biol. 21, 798–804. doi: 10.1016/j.semcdb.2010.03.008
Alvarez-Buylla, E. R., Benitez, M., Corvera-Poire, A., Chaos Cador, A., de Folter, S., Gamboa de Buen, A., et al. (2010). “Flower development,” in Arabidopsis book. 8, e0127. doi: 10.1199/tab.0127
Andres-Robin, A., Reymond, M. C., Dupire, A., Battu, V., Dubrulle, N., Mouille, G., et al. (2018). Evidence for the regulation of gynoecium morphogenesis by ETTIN via cell wall dynamics. Plant Physiol. 178, 1222–1232. doi: 10.1104/pp.18.00745
Bao, F., Azhakanandam, S., Franks, R. G. (2010). SEUSS and SEUSS-LIKE transcriptional adaptors regulate floral and embryonic development in arabidopsis. Plant Physiol. 152, 821–836. doi: 10.1104/pp.109.146183
Bartels, A., Han, Q., Nair, P., Stacey, L., Gaynier, H., Mosley, M., et al. (2018). Dynamic DNA methylation in plant growth and development. Int. J. Mol. Sci. 19, 2144. doi: 10.3390/ijms19072144
Bartlett, M. E., Thompson, B. (2014). Meristem identity and phyllotaxis in inflorescence development. Front. Plant Sci. 5, 508. doi: 10.3389/fpls.2014.00508
Bencivenga, S., Serrano-Mislata, A., Bush, M., Fox, S., Sablowski, R. (2016). Control of oriented tissue growth through repression of organ boundary genes promotes stem morphogenesis. Dev. Cell 39, 198–208. doi: 10.1016/j.devcel.2016.08.013
Bencivenga, S., Simonini, S., Benkova, E., Colombo, L. (2012). The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in arabidopsis. Plant Cell 24, 2886–2897. doi: 10.1105/tpc.112.100164
Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G., et al. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. doi: 10.1016/S0092-8674(03)00924-3
Bergeron, F., Reutenauer, C. (2019). Golden ratio and phyllotaxis, a clear mathematical link. J. Math. Biol. 78, 1–19. doi: 10.1007/s00285-018-1265-3
Bhatia, N., Bozorg, B., Larsson, A., Ohno, C., Jönsson, H., Heisler, M. G. (2016). Auxin acts through MONOPTEROS to regulate plant cell polarity and pattern phyllotaxis. Curr. Biol. 26, 3202–3208. doi: 10.1016/j.cub.2016.09.044
Boer, D. R., Freire-Rios, A., van den Berg, W. A., Saaki, T., Manfield, I. W., Kepinski, S., et al. (2014). Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156, 577–589. doi: 10.1016/j.cell.2013.12.027
Bowman, J. L., Baum, S. F., Eshed, Y., Putterill, J., Alvarez, J. (1999). Molecular genetics of gynoecium development in arabidopsis. Curr. Top. Dev. Biol. 45, 155–205. doi: 10.1016/S0070-2153(08)60316-6
Bowman, J. L., Smith, D. R., Meyerowitz, E. M. (1989). Genes directing flower development in arabidopsis. Plant Cell 1, 37–52. doi: 10.1105/tpc.1.1.37
Byrne, M. E., Groover, A. T., Fontana, J. R., Martienssen, R. A. (2003). Phyllotactic pattern and stem cell fate are determined by the arabidopsis homeobox gene BELLRINGER. Development 130, 3941–3950. doi: 10.1242/dev.00620
Calderon Villalobos, L. I., Lee, S., De Oliveira, C., Ivetac, A., Brandt, W., Armitage, L., et al. (2012). A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8, 477–485. doi: 10.1038/nchembio.926
Cao, X., He, Z., Guo, L., Liu, X. (2015). Epigenetic mechanisms are critical for the regulation of WUSCHEL expression in floral meristems. Plant Physiol. 168, 1189–1196. doi: 10.1104/pp.15.00230
Chae, E., Tan, Q. K., Hill, T. A., Irish, V. F. (2008). An arabidopsis f-box protein acts as a transcriptional co-factor to regulate floral development. Development 135, 1235–1245. doi: 10.1242/dev.015842
Chandler, J. W. (2016). Auxin response factors. Plant Cell Environ. 39, 1014–1028. doi: 10.1111/pce.12662
Chang, W., Guo, Y., Zhang, H., Liu, X., Guo, L. (2020). Same actor in different stages: Genes in shoot apical meristem maintenance and floral meristem determinacy in arabidopsis. Front. Ecol. Evol. 8, 89. doi: 10.3389/fevo.2020.00089
Chavez, S., Beilharz, T., Rondon, A. G., Erdjument-Bromage, H., Tempst, P., Svejstrup, J. Q., et al. (2000). A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in saccharomyces cerevisiae. EMBO J. 19, 5824–5834. doi: 10.1093/emboj/19.21.5824
Cheng, Z. J., Wang, L., Sun, W., Zhang, Y., Zhou, C., Su, Y. H., et al. (2013). Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol. 161, 240–251. doi: 10.1104/pp.112.203166
Chitwood, D. H., Nogueira, F. T., Howell, M. D., Montgomery, T. A., Carrington, J. C., Timmermans, M. C. (2009). Pattern formation via small RNA mobility. Genes Dev. 23, 549–554. doi: 10.1101/gad.1770009
Coen, E. S., Meyerowitz, E. M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Cokus, S. J., Feng, S., Zhang, X., Chen, Z., Merriman, B., Haudenschild, C. D., et al. (2008). Shotgun bisulphite sequencing of the arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219. doi: 10.1038/nature06745
Drews, G. N., Koltunow, A. M. (2011). “The female gametophyte”, in Arabidopsis book. 9, e0155. doi: 10.1199/tab.0155
Eshed, Y., Baum, S. F., Bowman, J. L. (1999). Distinct mechanisms promote polarity establishment in carpels of arabidopsis. Cell 99, 199–209. doi: 10.1016/S0092-8674(00)81651-7
Eshed, Y., Baum, S. F., Perea, J. V., Bowman, J. L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11, 1251–1260. doi: 10.1016/S0960-9822(01)00392-X
Fahlgren, N., Montgomery, T. A., Howell, M. D., Allen, E., Dvorak, S. K., Alexander, A. L., et al. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in arabidopsis. Curr. Biol. 16, 939–944. doi: 10.1016/j.cub.2006.03.065
Finet, C., Berne-Dedieu, A., Scutt, C. P., Marlétaz, F. (2013). Evolution of the ARF gene family in land plants: Old domains, new tricks. Mol. Biol. Evol. 30, 45–56. doi: 10.1093/molbev/mss220
Galbiati, F., Sinha Roy, D., Simonini, S., Cucinotta, M., Ceccato, L., Cuesta, C., et al. (2013). An integrative model of the control of ovule primordia formation. Plant journal: For Cell Mol. Biol. 76, 446–455. doi: 10.1111/tpj.12309
Garcia, D., Collier, S. A., Byrne, M. E., Martienssen, R. A. (2006). Specification of leaf polarity in arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16, 933–938. doi: 10.1016/j.cub.2006.03.064
Girin, T., Paicu, T., Stephenson, P., Fuentes, S., Korner, E., O'Brien, M., et al. (2011). INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in arabidopsis. Plant Cell 23, 3641–3653. doi: 10.1105/tpc.111.090944
Guilfoyle, T. J. (2015). The PB1 domain in auxin response factor and Aux/IAA proteins: A versatile protein interaction module in the auxin response. Plant Cell 27, 33–43. doi: 10.1105/tpc.114.132753
Guilfoyle, T. J., Hagen, G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460. doi: 10.1016/j.pbi.2007.08.014
Han, X., Kumar, D., Chen, H., Wu, S., Kim, J. Y. (2014). Transcription factor-mediated cell-to-cell signalling in plants. J. Exp. Bot. 65, 1737–1749. doi: 10.1093/jxb/ert422
Hawkins, C., Liu, Z. (2014). A model for an early role of auxin in arabidopsis gynoecium morphogenesis. Front. Plant Sci. 5, 327. doi: 10.3389/fpls.2014.00327
Heisler, M. G., Atkinson, A., Bylstra, Y. H., Walsh, R., Smyth, D. R. (2001). SPATULA, a gene that controls development of carpel margin tissues in arabidopsis, encodes a bHLH protein. Development 128, 1089–1098. doi: 10.1242/dev.128.7.1089
Hunter, C., Willmann, M. R., Wu, G., Yoshikawa, M., de la Luz Gutierrez-Nava, M., Poethig, S. R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in arabidopsis. Development 133, 2973–2981. doi: 10.1242/dev.02491
Husbands, A. Y., Benkovics, A. H., Nogueira, F. T., Lodha, M., Timmermans, M. C. (2015). The ASYMMETRIC LEAVES complex employs multiple modes of regulation to affect adaxial-abaxial patterning and leaf complexity. Plant Cell 27, 3321–3335. doi: 10.1105/tpc.15.00454
Ivanov, I. P., Atkins, J. F., Michael, A. J. (2010). A profusion of upstream open reading frame mechanisms in polyamine-responsive translational regulation. Nucleic Acids Res. 38, 353–359. doi: 10.1093/nar/gkp1037
Iwakawa, H., Takahashi, H., Machida, Y., Machida, C. (2020). Roles of ASYMMETRIC LEAVES2 (AS2) and nucleolar proteins in the adaxial-abaxial polarity specification at the perinucleolar region in arabidopsis. Int. J. Mol. Sci. 21, 7314. doi: 10.3390/ijms21197314
Iwasaki, M., Takahashi, H., Iwakawa, H., Nakagawa, A., Ishikawa, T., Tanaka, H., et al. (2013). Dual regulation of ETTIN (ARF3) gene expression by AS1-AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial-abaxial partitioning in arabidopsis. Development 140, 1958–1969. doi: 10.1242/dev.085365
Janocha, D., Lohmann, J. U. (2018). From signals to stem cells and back again. Curr. Opin. Plant Biol. 45, 136–142. doi: 10.1016/j.pbi.2018.06.005
Ji, L., Liu, X., Yan, J., Wang, W., Yumul, R. E., Kim, Y. J., et al. (2011). ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in arabidopsis. PLoS Genet. 7, e1001358. doi: 10.1371/journal.pgen.1001358
Kelley, D. R., Arreola, A., Gallagher, T. L., Gasser, C. S. (2012). ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in arabidopsis. Development 139, 1105–1109. doi: 10.1242/dev.067918
Kerstetter, R. A., Bollman, K., Taylor, R. A., Bomblies, K., Poethig, R. S. (2001). KANADI regulates organ polarity in arabidopsis. Nature 411, 706–709. doi: 10.1038/35079629
Krizek, B. A., Fletcher, J. C. (2005). Molecular mechanisms of flower development: An armchair guide. Nat. Rev. Genet. 6, 688–698. doi: 10.1038/nrg1675
Kuhn, A., Ramans Harborough, S., McLaughlin, H. M., Natarajan, B., Verstraeten, I., Friml, J., et al. (2020). Direct ETTIN-auxin interaction controls chromatin states in gynoecium development. Elife 9, e51787. doi: 10.7554/eLife.51787
Kuusk, S., Sohlberg, J. J., Long, J. A., Fridborg, I., Sundberg, E. (2002). STY1 and STY2 promote the formation of apical tissues during arabidopsis gynoecium development. Development 129, 4707–4717. doi: 10.1242/dev.129.20.4707
Lanctot, A. (2022). At Home and away: A mobile transcription factor regulates meristem development in discrete spatial domains. Plant Physiol. 190, 2066–2068. doi: 10.1093/plphys/kiac453
Laux, T., Mayer, K. F., Berger, J., Jürgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in arabidopsis. Development 122, 87–96. doi: 10.1242/dev.122.1.87
Lee, I., Wolfe, D. S., Nilsson, O., Weigel, D. (1997). A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr. Biol. 7, 95–104. doi: 10.1016/S0960-9822(06)00053-4
Lee, Z. H., Hirakawa, T., Yamaguchi, N., Ito, T. (2019). The roles of plant hormones and their interactions with regulatory genes in determining meristem activity. Int. J. Mol. Sci. 20, 4065. doi: 10.3390/ijms20164065
Lenhard, M., Bohnert, A., Jurgens, G., Laux, T. (2001). Termination of stem cell maintenance in arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105, 805–814. doi: 10.1016/S0092-8674(01)00390-7
Li, D., Fu, X., Guo, L., Huang, Z., Li, Y., Liu, Y., et al. (2016a). FAR-RED ELONGATED HYPOCOTYL3 activates SEPALLATA2 but inhibits CLAVATA3 to regulate meristem determinacy and maintenance in arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, 9375–9380. doi: 10.1073/pnas.1602960113
Li, S. B., Xie, Z. Z., Hu, C. G., Zhang, J. Z. (2016b). A review of auxin response factors (ARFs) in plants. Front. Plant Sci. 7, 47. doi: 10.3389/fpls.2016.00047
Li, Z., He, Y. (2020). Roles of brassinosteroids in plant reproduction. Int. J. Mol. Sci. 21, 872. doi: 10.3390/ijms21030872
Liu, X., Dinh, T. T., Li, D., Shi, B., Li, Y., Cao, X., et al. (2014). AUXIN RESPONSE FACTOR 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. Plant journal: For Cell Mol. Biol. 80, 629–641. doi: 10.1111/tpj.12658
Liu, X., Kim, Y. J., Muller, R., Yumul, R. E., Liu, C., Pan, Y., et al. (2011). AGAMOUS terminates floral stem cell maintenance in arabidopsis by directly repressing WUSCHEL through recruitment of polycomb group proteins. Plant Cell 23, 3654–3670. doi: 10.1105/tpc.111.091538
Lohmann, J. U., Hong, R. L., Hobe, M., Busch, M. A., Parcy, F., Simon, R., et al. (2001). A molecular link between stem cell regulation and floral patterning in arabidopsis. Cell 105, 793–803. doi: 10.1016/S0092-8674(01)00384-1
Lora, J., Hormaza, J. I., Herrero, M. (2015). Transition from two to one integument in prunus species: Expression pattern of INNER NO OUTER (INO), ABERRANT TESTA SHAPE (ATS) and ETTIN (ETT). New Phytol. 208, 584–595. doi: 10.1111/nph.13460
Lora, J., Yang, X., Tucker, M. R. (2019). Establishing a framework for female germline initiation in the plant ovule. J. Exp. Bot. 70, 2937–2949. doi: 10.1093/jxb/erz212
Lucero, L. E., Uberti-Manassero, N. G., Arce, A. L., Colombatti, F., Alemano, S. G., Gonzalez, D. H. (2015). TCP15 modulates cytokinin and auxin responses during gynoecium development in arabidopsis. Plant journal: Cell Mol. Biol. 84, 267–282. doi: 10.1111/tpj.12992
Ma, Y., Miotk, A., Sutikovic, Z., Ermakova, O., Wenzl, C., Medzihradszky, A., et al. (2019). WUSCHEL acts as an auxin response rheostat to maintain apical stem cells in arabidopsis. Nat. Commun. 10, 5093. doi: 10.1038/s41467-019-13074-9
Machida, C., Nakagawa, A., Kojima, S., Takahashi, H., Machida, Y. (2015). The complex of ASYMMETRIC LEAVES (AS) proteins plays a central role in antagonistic interactions of genes for leaf polarity specification in arabidopsis. Wiley Interdiscip. Rev. Dev. Biol. 4, 655–671. doi: 10.1002/wdev.196
Marin, E., Jouannet, V., Herz, A., Lokerse, A. S., Weijers, D., Vaucheret, H., et al. (2010). miR390, arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22, 1104–1117. doi: 10.1105/tpc.109.072553
Marsch-Martinez, N., de Folter, S. (2016). Hormonal control of the development of the gynoecium. Curr. Opin. Plant Biol. 29, 104–114. doi: 10.1016/j.pbi.2015.12.006
McAbee, J. M., Hill, T. A., Skinner, D. J., Izhaki, A., Hauser, B. A., Meister, R. J., et al. (2006). ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of arabidopsis ovule integuments. Plant J. 46, 522–531. doi: 10.1111/j.1365-313X.2006.02717.x
Moubayidin, L., Ostergaard, L. (2014). Dynamic control of auxin distribution imposes a bilateral-to-radial symmetry switch during gynoecium development. Curr. Biol. 24, 2743–2748. doi: 10.1016/j.cub.2014.09.080
Nasrallah, J. B. (2019). Self-incompatibility in the brassicaceae: Regulation and mechanism of self-recognition. Curr. Top. Dev. Biol. 131, 435–452. doi: 10.1016/bs.ctdb.2018.10.002
Nemhauser, J. L., Feldman, L. J., Zambryski, P. C. (2000). Auxin and ETTIN in arabidopsis gynoecium morphogenesis. Development 127, 3877–3888. doi: 10.1242/dev.127.18.3877
Ng, K. H., Yu, H., Ito, T. (2009). AGAMOUS controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PloS Biol. 7, e1000251. doi: 10.1371/journal.pbio.1000251
Nishimura, T., Wada, T., Yamamoto, K. T., Okada, K. (2005). The arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 17, 2940–2953. doi: 10.1105/tpc.105.036533
Okabe, T. (2012). Systematic variations in divergence angle. J. Theor. Biol. 313, 20–41. doi: 10.1016/j.jtbi.2012.08.007
Okushima, Y., Overvoorde, P. J., Arima, K., Alonso, J. M., Chan, A., Chang, C., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463. doi: 10.1105/tpc.104.028316
Peaucelle, A., Morin, H., Traas, J., Laufs, P. (2007). Plants expressing a miR164-resistant CUC2 gene reveal the importance of post-meristematic maintenance of phyllotaxy in arabidopsis. Development 134, 1045–1050. doi: 10.1242/dev.02774
Pekker, I., Alvarez, J. P., Eshed, Y. (2005). Auxin response factors mediate arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17, 2899–2910. doi: 10.1105/tpc.105.034876
Pelloux, J., Rusterucci, C., Mellerowicz, E. J. (2007). New insights into pectin methylesterase structure and function. Trends Plant Sci. 12, 267–277. doi: 10.1016/j.tplants.2007.04.001
Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H. L., Poethig, R. S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in arabidopsis. Genes Dev. 18, 2368–2379. doi: 10.1101/gad.1231804
Pfluger, J., Zambryski, P. (2004). The role of SEUSS in auxin response and floral organ patterning. Development 131, 4697–4707. doi: 10.1242/dev.01306
Pinto, S. C., Mendes, M. A., Coimbra, S., Tucker, M. R. (2019). Revisiting the female germline and its expanding toolbox. Trends Plant Sci. 24, 455–467. doi: 10.1016/j.tplants.2019.02.003
Prasad, K., Grigg, S. P., Barkoulas, M., Yadav, R. K., Sanchez-Perez, G. F., Pinon, V., et al. (2011). Arabidopsis PLETHORA transcription factors control phyllotaxis. Curr. Biol. 21, 1123–1128. doi: 10.1016/j.cub.2011.05.009
Prunet, N., Morel, P., Thierry, A. M., Eshed, Y., Bowman, J. L., Negrutiu, I., et al. (2008). REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in arabidopsis thaliana. Plant Cell 20, 901–919. doi: 10.1105/tpc.107.053306
Rahmani, F., Hummel, M., Schuurmans, J., Wiese-Klinkenberg, A., Smeekens, S., Hanson, J. (2009). Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol. 150, 1356–1367. doi: 10.1104/pp.109.136036
Reinhardt, D. (2005). Regulation of phyllotaxis. Int. J. Dev. Biol. 49, 539–546. doi: 10.1387/ijdb.041922dr
Remington, D. L., Vision, T. J., Guilfoyle, T. J., Reed, J. W. (2004). Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 135, 1738–1752. doi: 10.1104/pp.104.039669
Roe, J. L., Nemhauser, J. L., Zambryski, P. C. (1997). TOUSLED participates in apical tissue formation during gynoecium development in arabidopsis. Plant Cell 9, 335–353. doi: 10.1105/tpc.9.3.335
Roe, J. L., Rivin, C. J., Sessions, R. A., Feldmann, K. A., Zambryski, P. C. (1993). The tousled gene in a. thaliana encodes a protein kinase homolog that is required for leaf and flower development. Cell 75, 939–950. doi: 10.1016/0092-8674(93)90537-Z
Roeder, A. H., Yanofsky, M. F. (2006). “Fruit development in arabidopsis,” in Abidopsis book. 4, e0075. doi: 10.1199/tab.0075
Ronemus, M. J., Galbiati, M., Ticknor, C., Chen, J., Dellaporta, S. L. (1996). Demethylation-induced developmental pleiotropy in arabidopsis. Science 273, 654–657. doi: 10.1126/science.273.5275.654
Roosjen, M., Paque, S., Weijers, D. (2018). Auxin response factors: Output control in auxin biology. J. Exp. Bot. 69, 179–188. doi: 10.1093/jxb/erx237
Rosado, A., Li, R., van de Ven, W., Hsu, E., Raikhel, N. V. (2012). Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc. Natl. Acad. Sci. U.S.A. 109, 19537–19544. doi: 10.1073/pnas.1214774109
Sawa, S., Watanabe, K., Goto, K., Liu, Y. G., Shibata, D., Kanaya, E., et al. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13, 1079–1088. doi: 10.1101/gad.13.9.1079
Schaller, G. E., Bishopp, A., Kieber, J. J. (2015). The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 27, 44–63. doi: 10.1105/tpc.114.133595
Schuster, C., Gaillochet, C., Lohmann, J. U. (2015). Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development 142, 3343–3350. doi: 10.1242/dev.120444
Schuster, C., Gaillochet, C., Medzihradszky, A., Busch, W., Daum, G., Krebs, M., et al. (2014). A regulatory framework for shoot stem cell control integrating metabolic, transcriptional, and phytohormone signals. Dev. Cell 28, 438–449. doi: 10.1016/j.devcel.2014.01.013
Sehra, B., Franks, R. G. (2015). Auxin and cytokinin act during gynoecial patterning and the development of ovules from the meristematic medial domain. Wiley Interdiscip. Rev. Dev. Biol. 4, 555–571. doi: 10.1002/wdev.193
Sessions, A., Nemhauser, J. L., McColl, A., Roe, J. L., Feldmann, K. A., Zambryski, P. C. (1997). ETTIN patterns the arabidopsis floral meristem and reproductive organs. Development 124, 4481–4491. doi: 10.1242/dev.124.22.4481
Sessions, R. A., Zambryski, P. C. (1995). Arabidopsis gynoecium structure in the wild and in ettin mutants. Development 121, 1519–1532. doi: 10.1242/dev.121.5.1519
Shang, E., Ito, T., Sun, B. (2019). Control of floral stem cell activity in arabidopsis. Plant Signaling Behav. 14, 1659706. doi: 10.1080/15592324.2019.1659706
Shi, B., Guo, X., Wang, Y., Xiong, Y., Wang, J., Hayashi, K. I., et al. (2018). Feedback from lateral organs controls shoot apical meristem growth by modulating auxin transport. Dev. Cell 44, 204–216.e206. doi: 10.1016/j.devcel.2017.12.021
Shirley, N. J., Aubert, M. K., Wilkinson, L. G., Bird, D. C., Lora, J., Yang, X., et al. (2019). Translating auxin responses into ovules, seeds and yield: Insight from arabidopsis and the cereals. J. Integr. Plant Biol. 61, 310–336. doi: 10.1111/jipb.12747
Siegfried, K. R., Eshed, Y., Baum, S. F., Otsuga, D., Drews, G. N., Bowman, J. L. (1999). Members of the YABBY gene family specify abaxial cell fate in arabidopsis. Development 126, 4117–4128. doi: 10.1242/dev.126.18.4117
Simonini, S., Ostergaard, L. (2019). Female reproductive organ formation: A multitasking endeavor. Curr. Top. Dev. Biol. 131, 337–371. doi: 10.1016/bs.ctdb.2018.10.004
Simonini, S., Bencivenga, S., Trick, M., Ostergaard, L. (2017). Auxin-induced modulation of ETTIN activity orchestrates gene expression in arabidopsis. Plant Cell 29, 1864–1882. doi: 10.1105/tpc.17.00389
Simonini, S., Deb, J., Moubayidin, L., Stephenson, P., Valluru, M., Freire-Rios, A., et al. (2016). A noncanonical auxin-sensing mechanism is required for organ morphogenesis in arabidopsis. Genes Dev. 30, 2286–2296. doi: 10.1101/gad.285361.116
Simonini, S., Mas, P. J., Mas, C., Ostergaard, L., Hart, D. J. (2018). Auxin sensing is a property of an unstructured domain in the auxin response factor ETTIN of arabidopsis thaliana. Sci. Rep. 8, 13563. doi: 10.1038/s41598-018-31634-9
Smith, R. S., Guyomarc'h, S., Mandel, T., Reinhardt, D., Kuhlemeier, C., Prusinkiewicz, P. (2006). A plausible model of phyllotaxis. Proc. Natl. Acad. Sci. U.S.A. 103, 1301–1306. doi: 10.1073/pnas.0510457103
Sohlberg, J. J., Myrenas, M., Kuusk, S., Lagercrantz, U., Kowalczyk, M., Sandberg, G., et al. (2006). STY1 regulates auxin homeostasis and affects apical-basal patterning of the arabidopsis gynoecium. Plant journal: For Cell Mol. Biol. 47, 112–123. doi: 10.1111/j.1365-313X.2006.02775.x
Steiner, E., Efroni, I., Gopalraj, M., Saathoff, K., Tseng, T. S., Kieffer, M., et al. (2012). The arabidopsis O-linked n-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24, 96–108. doi: 10.1105/tpc.111.093518
Strasser, K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., et al. (2002). TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417, 304–308. doi: 10.1038/nature746
Su, Z., Zhao, L., Zhao, Y., Li, S., Won, S., Cai, H., et al. (2017). The THO complex non-Cell-Autonomously represses female germline specification through the TAS3-ARF3 module. Curr. Biol. 27, 1597–1609.e1592. doi: 10.1016/j.cub.2017.05.021
Sun, B., Ito, T. (2010). Floral stem cells: From dynamic balance towards termination. Biochem. Soc. Trans. 38, 613–616. doi: 10.1042/BST0380613
Sun, B., Ito, T. (2015). Regulation of floral stem cell termination in arabidopsis. Front. Plant Sci. 6, 17. doi: 10.3389/fpls.2015.00017
Sun, B., Xu, Y., Ng, K. H., Ito, T. (2009). A timing mechanism for stem cell maintenance and differentiation in the arabidopsis floral meristem. Genes Dev. 23, 1791–1804. doi: 10.1101/gad.1800409
Takahashi, H., Iwakawa, H., Ishibashi, N., Kojima, S., Matsumura, Y., Prananingrum, P., et al. (2013). Meta-analyses of microarrays of arabidopsis asymmetric leaves1 (as1), as2 and their modifying mutants reveal a critical role for the ETT pathway in stabilization of adaxial-abaxial patterning and cell division during leaf development. Plant Cell Physiol. 54, 418–431. doi: 10.1093/pcp/pct027
Tantikanjana, T., Nasrallah, J. B. (2012). Non-cell-autonomous regulation of crucifer self-incompatibility by auxin response factor ARF3. proceedings of the national academy of sciences of the united states of America. Proc. Natl. Acad. Sci. U.S.A. 109, 19468–19473. doi: 10.1073/pnas.1217343109
Thomson, B., Wellmer, F. (2019). Molecular regulation of flower development. Curr. Top. Dev. Biol. 131, 185–210. doi: 10.1016/bs.ctdb.2018.11.007
Tian, C. E., Muto, H., Higuchi, K., Matamura, T., Tatematsu, K., Koshiba, T., et al. (2004). Disruption and overexpression of auxin response factor 8 gene of arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant journal: For Cell Mol. Biol. 40, 333–343. doi: 10.1111/j.1365-313X.2004.02220.x
Vaucheret, H. (2005). MicroRNA-dependent trans-acting siRNA production. Sci STKE5 2005, pe43. doi: 10.1126/stke.3002005pe43
Verma, N. (2019). Transcriptional regulation of anther development in arabidopsis. Gene 689, 202–209. doi: 10.1016/j.gene.2018.12.022
Vernoux, T., Brunoud, G., Farcot, E., Morin, V., Van den Daele, H., Legrand, J., et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7, 508. doi: 10.1038/msb.2011.39
Vial-Pradel, S., Keta, S., Nomoto, M., Luo, L., Takahashi, H., Suzuki, M., et al. (2018). Arabidopsis zinc-Finger-Like protein ASYMMETRIC LEAVES2 (AS2) and two nucleolar proteins maintain gene body DNA methylation in the leaf polarity gene ETTIN (ARF3). Plant Cell Physiol. 59, 1385–1397. doi: 10.1093/pcp/pcy031
von Arnim, A. G., Jia, Q., Vaughn, J. N. (2014). Regulation of plant translation by upstream open reading frames. Plant science: Int. J. Exp. Plant Biol. 214, 1–12. doi: 10.1016/j.plantsci.2013.09.006
Weijers, D., Wagner, D. (2016). Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol. 67, 539–574. doi: 10.1146/annurev-arplant-043015-112122
Williams, L., Carles, C. C., Osmont, K. S., Fletcher, J. C. (2005). A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl. Acad. Sci. U.S.A. 102, 9703–9708. doi: 10.1073/pnas.0504029102
Wormit, A., Usadel, B. (2018). The multifaceted role of pectin methylesterase inhibitors (PMEIs). Int. J. Mol. Sci. 19, 2878. doi: 10.3390/ijms19102878
Xu, Y., Yamaguchi, N., Gan, E. S., Ito, T. (2019). When to stop: An update on molecular mechanisms of floral meristem termination. J. Exp. Bot. 70, 1711–1718. doi: 10.1093/jxb/erz048
Yelina, N. E., Smith, L. M., Jones, A. M., Patel, K., Kelly, K. A., Baulcombe, D. C. (2010). Putative arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 13948–13953. doi: 10.1073/pnas.0911341107
Yoshikawa, M. (2013). Biogenesis of trans-acting siRNAs, endogenous secondary siRNAs in plants. Genes Genet. Syst. 88, 77–84. doi: 10.1266/ggs.88.77
Yumul, R. E., Kim, Y. J., Liu, X., Wang, R., Ding, J., Xiao, L., et al. (2013). POWERDRESS and diversified expression of the MIR172 gene family bolster the floral stem cell network. PloS Genet. 9, e1003218. doi: 10.1371/journal.pgen.1003218
Zhang, H., Lang, Z., Zhu, J. K. (2018a). Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489–506.
Zhang, K., Wang, R., Zi, H., Li, Y., Cao, X., Li, D., et al. (2018b). AUXIN RESPONSE FACTOR3 regulates floral meristem determinacy by repressing cytokinin biosynthesis and signaling. Plant Cell 30, 324–346. doi: 10.1105/tpc.17.00705
Zhang, K., Zhang, H., Pan, Y., Niu, Y., Guo, L., Ma, Y., et al. (2022). Cell- and non-cell-autonomous AUXIN RESPONSE FACTOR3 controls meristem proliferation and phyllotactic patterns. Plant Physiol. 190, 2335–2349. doi: 10.1093/plphys/kiac370
Zhang, X., Yazaki, J., Sundaresan, A., Cokus, S., Chan, S. W., Chen, H., et al. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126, 1189–1201. doi: 10.1016/j.cell.2006.08.003
Zhang, W., Yu, R. (2014). Molecule mechanism of stem cells in arabidopsis thaliana. Pharmacognosy Rev. 8, 105–112. doi: 10.4103/0973-7847.134243
Zhao, L., Kim, Y., Dinh, T. T., Chen, X. (2007). miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in arabidopsis floral meristems. Plant journal: Cell Mol. Biol. 51, 840–849.
Keywords: Arabidopsis thaliana, auxin, AUXIN RESPONSE FACTOR3, reproductive phase, meristem homeostasis, patterning formation
Citation: Fu Y, Zhang H, Ma Y, Li C, Zhang K and Liu X (2023) A model worker: Multifaceted modulation of AUXIN RESPONSE FACTOR3 orchestrates plant reproductive phases. Front. Plant Sci. 14:1123059. doi: 10.3389/fpls.2023.1123059
Received: 13 December 2022; Accepted: 16 February 2023;
Published: 27 February 2023.
Edited by:
Yongwei Sun, Inner Mongolia University, ChinaReviewed by:
Weibing Yang, Institute of Plant Physiology and Ecology (CAS), ChinaCopyright © 2023 Fu, Zhang, Ma, Li, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ke Zhang, emhhbmdrZTAxMjZAMTYzLmNvbQ==; Xigang Liu, eGdsaXVAaGVidHUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.