
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 09 March 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1121809
This article is part of the Research TopicAbiotic Stress Alleviation in Plants: Morpho-Physiological and Molecular AspectsView all 31 articles
Foxtail millet (Setaria italica L.) is a vital cereal food crop with promising development and utilization potential because of its outstanding ability to resist drought stress. However, the molecular mechanisms underlying its drought stress resistance remain unclear. In this study, we aimed to elucidate the molecular function of a 9-cis-epoxycarotenoid dioxygenase gene, SiNCED1, in the drought stress response of foxtail millet. Expression pattern analysis showed that SiNCED1 expression was significantly induced by abscisic acid (ABA), osmotic stress, and salt stress. Furthermore, ectopic overexpression of SiNCED1 could enhance drought stress resistance by elevating endogenous ABA levels and promoting stomatal closure. Transcript analysis indicated that SiNCED1 modulated ABA-related stress responsive gene expression. In addition, we found that ectopic expression of SiNCED1 delayed seed germination under normal and abiotic stress conditions. Taken together, our results show that SiNCED1 plays a positive role in the drought tolerance and seed dormancy of foxtail millet by modulating ABA biosynthesis. In conclusion, this study revealed that SiNCED1 is an important candidate gene for the improvement of drought stress tolerance in foxtail millet and could be beneficial in the breeding and investigation of drought tolerance in other agronomic crops.
Foxtail millet (Setaria italica L.) is an important cereal food crop that has been cultivated for over 8000 years (Zhang et al., 2012). This species is an ideal model plant because of its short life cycle, self-pollination, small diploid genome, prolific seed production, and small adult stature (Doust et al., 2009). Moreover, foxtail millet is a promising crop for the study of drought stress because of its outstanding ability to resist it (Barton et al., 2009). However, the genes underlying the foxtail millet drought stress response have not been studied extensively to date.
Abscisic acid (ABA) is an essential phytohormone that regulates plant development and modulates abiotic stress tolerance. The main functions of ABA in plant development include dormancy, the delaying of seed germination, and induction of stomatal closure (Yang et al., 2022). For example, ABA plays a critical role in the protection of seed dormancy and inhibition of seed germination through ABA metabolism, which is modulated by regulatory factors, the environment, and natural variations of ABA metabolism signal pathway related genes (Sano and Marion-Poll, 2021). Moreover, ABA promotes stomatal closure in a dual way by increasing its concentration in guard cells and decreasing water permeability in leaf vascular tissues (Pantin et al., 2013). During exposure to abiotic stresses, such as drought, salinity, heat, and cold, ABA can act as a dominant hormone to regulate abiotic stress signaling pathways. For instance, ABA promotes desiccation-tolerance by controlling stomatal closure, enabling plants to adapt to water stress (Shang et al., 2020). In addition, ABA participates in plant development, and abiotic-stress tolerance depends on ABA metabolism (biosynthesis and catabolism), perception, the core signaling pathway, and factors that trigger ABA-mediated transcription (Nambara et al., 2010).
Endogenous ABA levels in plants are positively regulated by ABA biosynthesis (Ng et al., 2014). ABA biosynthesis occurs in two sites, starting in the plastids and ending in the cytosol (Sah et al., 2016). ABA biosynthesis has two pathways, but in higher plants, ABA is synthesized via an indirect pathway, also called the mevalonic acid-independent pathway. Several enzymes that originate from the catalysis of carotenoid precursors participate in this pathway (Xu et al., 2013). To date, almost all biosynthetic genes have been identified through the isolation of nutritionally deficient mutants or the creation of specific gene-editing mutants (Eiji and Annie, 2005; Miao et al., 2013), including zeaxanthin epoxidase (ZEP), 9-cis-epoxycarotenoid dioxygenase (NCED), and abscisic aldehyde oxidase (AAO) (Andrew and Roger, 1992; Finkelstein, 2013). The NCED enzyme regulates the key limiting step of ABA biosynthesis by catalyzing the cleavage of 9-cis-violaxanthin or 9-cis-neoxanthin to xanthoxin (C15) (Ye et al., 2012).
The first NCED gene, VP14, was cloned and identified in maize (Zea mays L.) (Schwartz et al., 1997). Subsequently, NCED genes have been isolated and studied from multiple plant species in both dicotyledons and monocotyledons, such as Arabidopsis (Arabidopsis thaliana (L.) Heynh.) (Sato et al., 2018), peaches (Prunus persica L.) (Wang et al., 2021), cotton (Gossypium hirsutum L.) (Li et al., 2021), Brassica napus L. (Xu and Cai, 2017), tomatoes (Solanum lycopersicum L.) (Sun et al., 2012), rice (Oryza sativa L.) (Ye et al., 2011), wheat (Triticum aestivum L.) (Lang et al., 2021), and Phaius tankervilliae (Banks ex L’Hér.) Blume (Lee et al., 2018). The rice NCED gene family consists of five members (Ye et al., 2011), with the following functions: OsNCED1, the overexpression of which can improve the heat tolerance of rice by enhancing its antioxidant capacity (Zhou et al., 2022); OsNCED3 and OsNCED5, which mediate multi-abiotic stress tolerance and leaf senescence by regulating endogenous ABA accumulation in rice (Huang et al., 2018; Huang et al., 2019); and OsNCED3, OsNCED4, and OsNCED5, whose ectopic expression in Arabidopsis can alter plant size and leaf morphology, delay seed germination, and advanced drought stress tolerance through the regulation of endogenous ABA content (Hwang et al., 2010; Hwang et al., 2018; Huang et al., 2019). In Arabidopsis, five NCED genes have been identified (Tan et al., 2003), which exhibit diverse expression patterns and share redundant functions. AtNCED6 cooperates with AtNCED9 to regulate seed germination and dormancy by controlling ABA levels in seeds. The transcription of AtNCED3 and AtNCED5 can be strongly induced by drought stress, which results in increased endogenous ABA levels and improved water stress tolerance (Hao et al., 2009; Frey et al., 2012). Two NCED isozymes, PpNCED1 and PpNCED5, which have been identified in peach fruits, control fruit ripening and senescence by regulating ABA biosynthesis (Wang et al., 2021).
The above studies have shown that increasing the transcript levels of NCED could promote ABA biosynthesis and enhance ABA accumulation in plants, which play important roles during the various phases of the plant life cycle, including seed development and dormancy, and in plant responses to variable environmental stress (Koshiba, 2002). However, the functions of ABA biosynthesis genes for NCED in foxtail millet are still unknown. In this study, we cloned four NCED genes from the Setaria italica cultivated variety Jigu 42. The expression pattern and ectopic expression of SiNCED1 in Arabidopsis was investigated. SiNCED1 was strongly induced by ABA, osmotic stress, and salt stress. Furthermore, SiNCED1 overexpression increased endogenous ABA contents to enhance drought stress tolerance and delay seed dormancy. Our findings indicate that SiNCED1 can be an important candidate gene to improve drought stress tolerance and keep seed dormancy in foxtail millet and other agronomic crops.
Foxtail millet (S. italica, cultivar Jigu 42) seeds were germinated in distilled water for 3 d at 28°C. The seedlings were grown in Hoagland’s culture solution (Li and Cheng, 2015) and cultured in growth chambers at 28°C under a 16 h light/8 h dark cycle. For abiotic stress, two-week-old seedlings were subjected to various stress treatments by placing them in a solution with a final concentration of 20% PEG6000, 150 mM NaCl, and 100 μM ABA. All treatments were repeated more than three times. The shoots and roots were harvested for RNA extraction.
A. thaliana (Columbia-0 ecotype; Col) was used in this study. The seeds were surface-sterilized with 75% (v/v) ethanol and sown on either a 1/2 MS medium or soil at 22°C under a 16 h light/8 h dark cycle (Lu et al., 2017). To analyze the seed-germination rate, sterilized seeds were sown on a 1/2 MS medium with 150 mM NaCl, 275 mM mannitol, and 6% glucose. The seed germination rate were calculated according to opened cotyledons. For drought stress treatment, the Arabidopsis transgenic lines and Col were grown in soil for 25 d under the same growth conditions mentioned above; after which, water was withheld for 30 d, followed by watering for 7 d.
The NCED protein sequences from four species, A. thaliana, Oryza sativa, Zea mays, and Setaria italica, were used for phylogenetic analysis. Phylogenetic tree analysis was performed using MEGA 7 software utilizing the maximum-likelihood method. Multiple sequence alignment was used to construct the phylogenetic tree in MEGA7 with the following parameter settings: Jones-Taylor-Thornton, pairwise deletion, and 1000 bootstrap replications.
The protein sequences of NCED genes used for multiple alignment were downloaded from the NCBI database (http://plants.ensembl.org/index.html). The multiple alignment was conducted using Bioedit software, applying ClustalW with the following parameters: full multiple alignment, number of bootstraps 1000, output clustal format with clustal consensus sequence generation.
The SiNCED1 open reading frame (ORF) was amplified through PCR using pHB-NCED1-F/R primers (Supplementary Table S1) and then inserted into the pHB binary vector with HindIII and XbaI restriction sites. The recombinant construct, 2×35S::SiNCED1, was transformed into Arabidopsis using the floral dip transformation method. The transgenic lines were planted on MS medium containing 50 μg hygromycin for 7 d. Homozygous lines with 100% resistance to hygromycin were selected and confirmed through semi-quantitative-PCR. Two genetically stable transgenic lines (OE3 and OE5) with high transcript levels of SiNCED1 were selected for further analysis.
Total RNA extraction was carried out for all samples using Trizol (Invitrogen, Shanghai, China) and was converted to cDNA using the Synthesis Kit (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. Subsequently, quantitative real-time PCR (RT-qPCR) was performed using the SYBR Premix Ex Taq kit (Takara, Dalian, China) on an Applied Biosystems Quant Studio 5 (Thermo Fisher Scientific). Relative changes in gene expression levels were quantitated based on three biological replicates using the 2-ΔΔCt method (Livak and Schmittgen, 2001). Each experiment included three technical replicates. The primer sequences used for RT-qPCR are listed in Supplementary Table S1.
The water loss rate was measured as described previously (Huang et al., 2018). The Arabidopsis rosette leaves of 25-day-old Col and SiNCED1-overexpressing lines were detached and placed in dishes with constant conditions. Fresh weight was measured at set times (0, 10, 20, 30, 40, 50, 60, 90,120, 150, and 180 min). The proportion of the initial fresh weight represented the water loss rate.
The proline content was measured according to previously described methods (Huang et al., 2018). Approximately 300 mg fresh leaf samples were homogenized in 3 mL of 3% aqueous sulfosalicylic acid and centrifuged at 4000 × g for 10 min. Then, 1 mL of supernatant was taken out and mixed with 1 mL of acid ninhydrin and 1 mL of glacial acetic acid in a new tube and then incubated in boiling water bath for 40 min. For every reaction, 2 mL toluene was added and oscillated for 30 min at room temperature. The proline content was measured using a spectrophotometer at 520 nm.
Relative electrolyte leakage was measured as previously described (Lou et al., 2017). The same number of detached leaf samples were placed in 20 mL of deionized water and then in a vacuum pump for 30 min. Samples were then sealed and placed in a 25 °C incubator for 6 h. The water conductance was measured using a conductivity meter.
ABA content was measured as previously described (Huang et al., 2019). Arabidopsis rosette leaves (300 mg) were collected from 25-day-old Col and SiNCED1-overexpressing lines and ground in liquid nitrogen. The internal standard was [2H6], and ABA content was quantified using UPLC–MS/MS with three independent biological replicates.
Stomatal apertures were analyzed using Arabidopsis leaf epidermis samples under normal or drought stress conditions. Stomata were observed and photographed under a fluorescence microscope (BX51, Olympus, Japan). The size of stomatal apertures was measured using Image J software and a digital ruler. Three independent experiments were performed, which obtained similar results. Each experiment included at least 30 stomata from each epidermal sample.
All experiments were repeated at least three times for each treatment. Data were analyzed using SPSS software (version 13.0). Differences were considered statistically significant at P < 0.05, as determined using ANOVA analysis and Tukey’s multiple comparisons test. All data are shown as mean ± SD from the three independent experiments.
Four 9-cis-epoxycarotenoid dioxygenase genes were found in the foxtail millet genome database (NCBI GenBank) and cloned (Supplementary Table S2). These genes were designated as SiNCED1 (LOC101783411), SiNCED3 (LOC101778945), SiNCED4 (LOC101766978), and SiNCED5 (LOC101770668). Among these, SiNCED1 was further investigated because of its promoter, which may have a drought related cis-element, as assessed through plantCARE prediction analysis (Supplementary Table S3). The full-length SiNCED1 CDS has a 1980 bp ORF, encoding 660 amino acids. To understand the phylogenetic relationship of SiNCED protein, a phylogenetic tree was constructed among SiNCED1, other SiNCEDs, and NCED proteins derived from Arabidopsis, rice, and maize. Based on the analysis, SiNCED proteins were clustered into two clades (Figure 1). SiNCED1 was separately clustered into a clade, while the other three SiNCEDs were aggregated into another clade. Alignment of the deduced protein sequences showed that SiNCED1 contains a large number of conserved amino acid residues in the NCED conserved domain RPE65, when compared with other NCED proteins (Figure 2). Therefore, SiNCED1 is homologous with NCED proteins from other species.
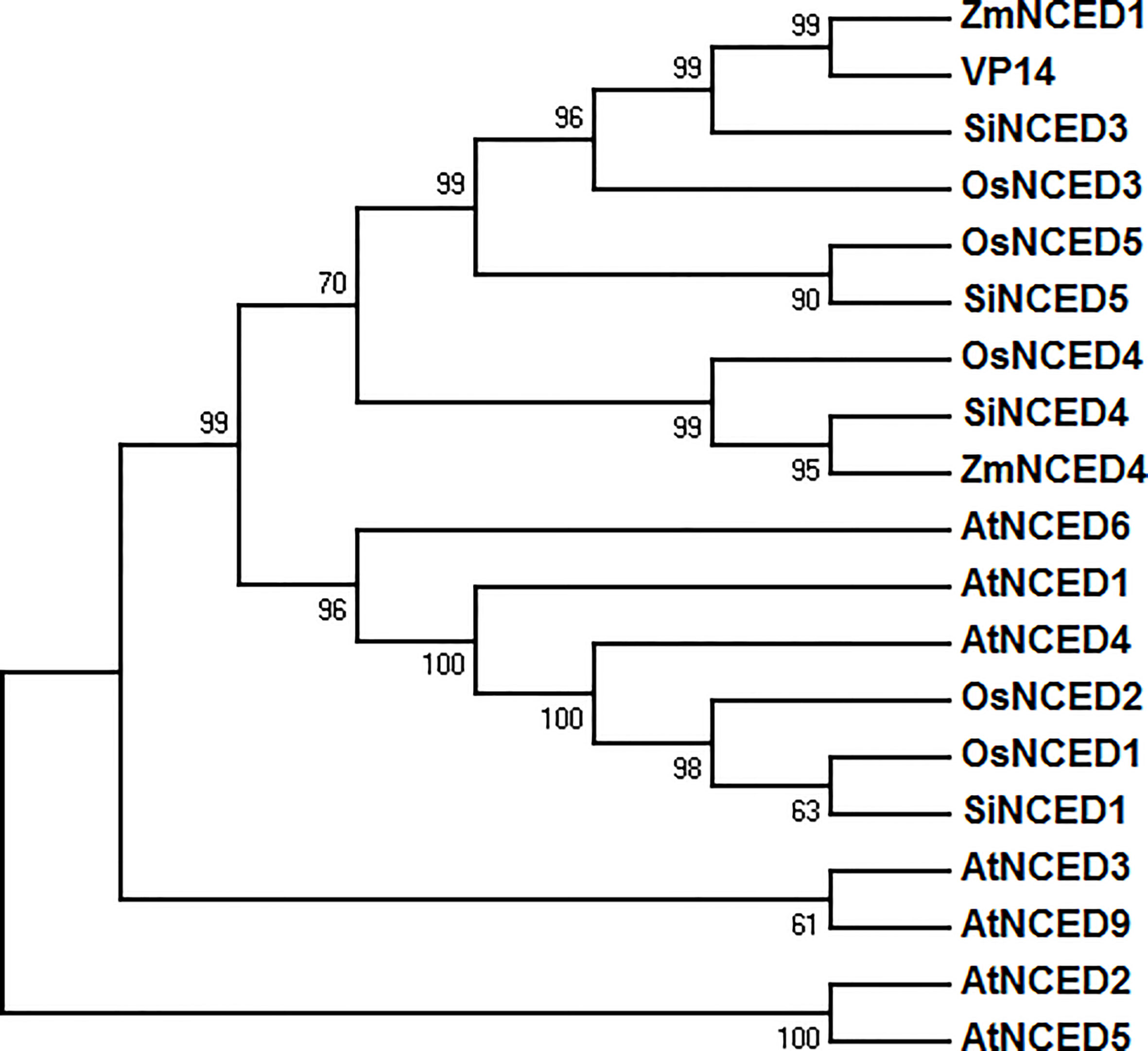
Figure 1 Phylogenetic tree analysis based on the amino acid sequences of SiNCED1 and other NCED from other plants. Nineteen amino acid sequences from four species were selected and analyzed by MEGA 7.

Figure 2 Multiple alignment of the deduced protein sequence of SiNCED1 and NCED proteins from other plants. Ten protein sequences from six species were selected and analyzed by Bioedit.
The temporal and spatial expression patterns of SiNCED1 were observed through RT-qPCR. SiNCED1 was constitutively expressed in the roots, young leaves, mature leaves, stems, and flowers but highly expressed in the roots, following by the stems, young leaves, mature leaves and flowers (Figure 3A).
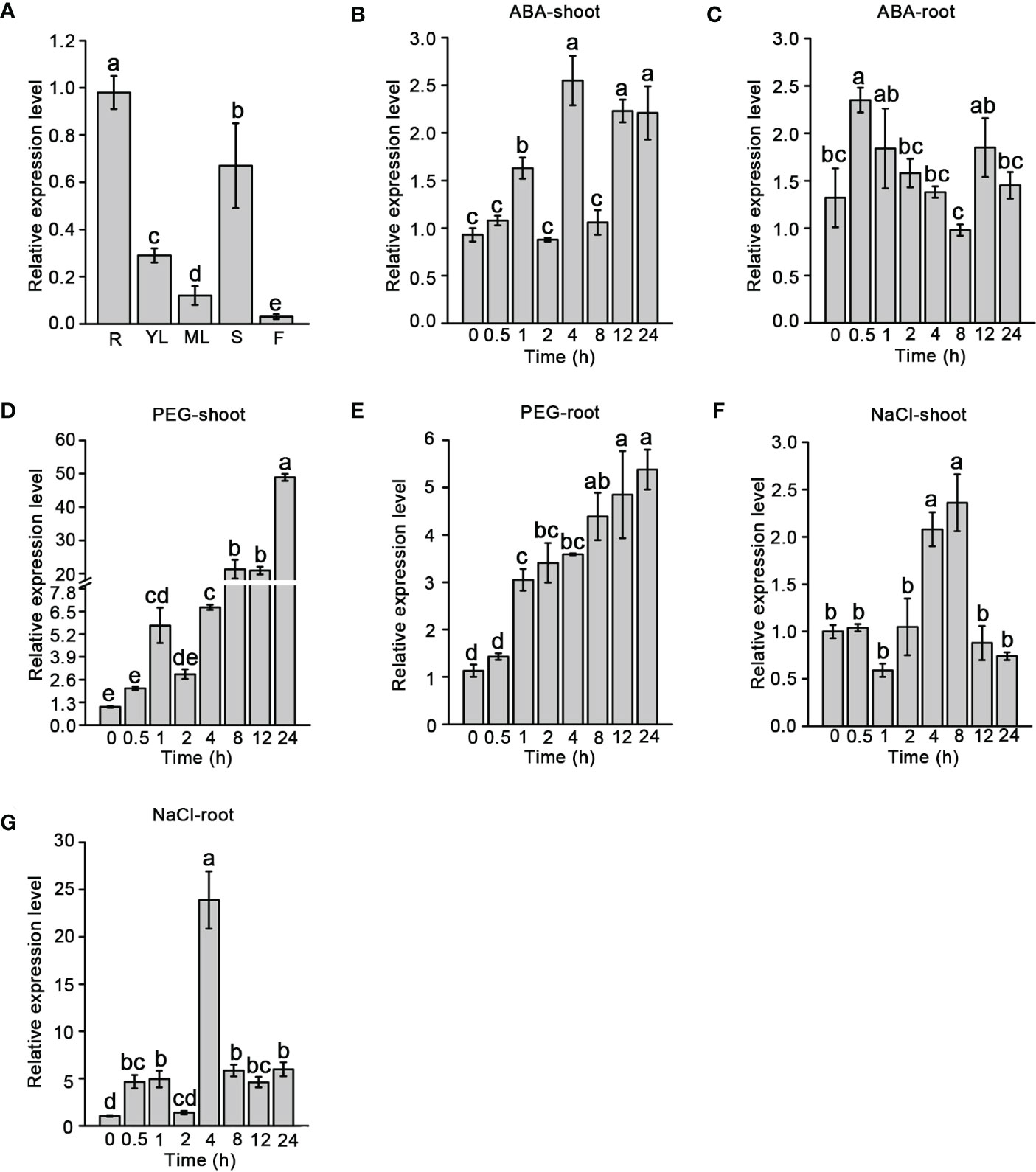
Figure 3 SiNCED1 gene expression pattern. (A) SiNCED1 gene expression in different organs. R(root), YL(young leaf), ML(mature leaf), S(stem), F(flower). (B–G) The three-leaf stage seedlings were submitted to ABA and 20% PEG, 150 mM NaCl treatment. (B, C) Expression of SiNCED1 under ABA treatment; (D, E) Expression of SiNCED1 under 20% PEG stress; (F, G) Expression of SiNCED1 under 150 mM NaCl stress. The shoot and root samples were collected for SiNCED1 expression analysis. Data are the mean ± SD for three replicates. Different letters represent significant differences at P<0.05 (ANOVA analysis, Tukey’s multiple comparisons test).
To investigate the response of SiNCED1 transcript levels under ABA, 20% PEG, and 150 mM NaCl treatments, RT-qPCR was performed to analyzed SiNCED1 expression in the shoots and roots of seedlings. Compared with the normal conditions, SiNCED1 was strongly induced in the shoots and roots of seedlings that underwent ABA treatment. We found that the expression levels of SiNCED1 reached their peak at 4 h in the shoots but 0.5 h in the roots (Figures 3B, C). In roots and shoots that had undergone PEG treatment, the expression of SiNCED1 was also induced, reaching their highest levels at 24 h in the shoots but 12 h in the roots (Figures 3D, E). Under high salt treatment, the level of SiNCED1 showed a trend of first increasing and then decreasing in the shoots and roots and reached its highest level in both at 4 h (Figures 3F, G). These results suggest that SiNCED1 plays a role in the tolerance of foxtail millet to multiple types of abiotic stress.
The heterogeneous expression of SiNCED1 in Arabidopsis was investigated to determine the function of SiNCED1. Semi-quantitative PCR analysis showed that SiNCED1 expression was only detected in SiNCED1-overexpressing lines (SiOE1-3 and SiOE1-5) but not in Col (Figure 4A). Subsequently, SiNCED1-overexpressing plants and Col were analyzed to identify the ability of drought stress tolerance. The SiNCED1-overexpressing plants and Col were grown in soil for 25 d under well-watered conditions, and water-stress conditions (water withheld) for 15 d and 30 d. When water was withheld for 30d, almost all Col plants withered, but SiNCED1-overexpressing lines did not show any wilted leaves (Figure 4B).
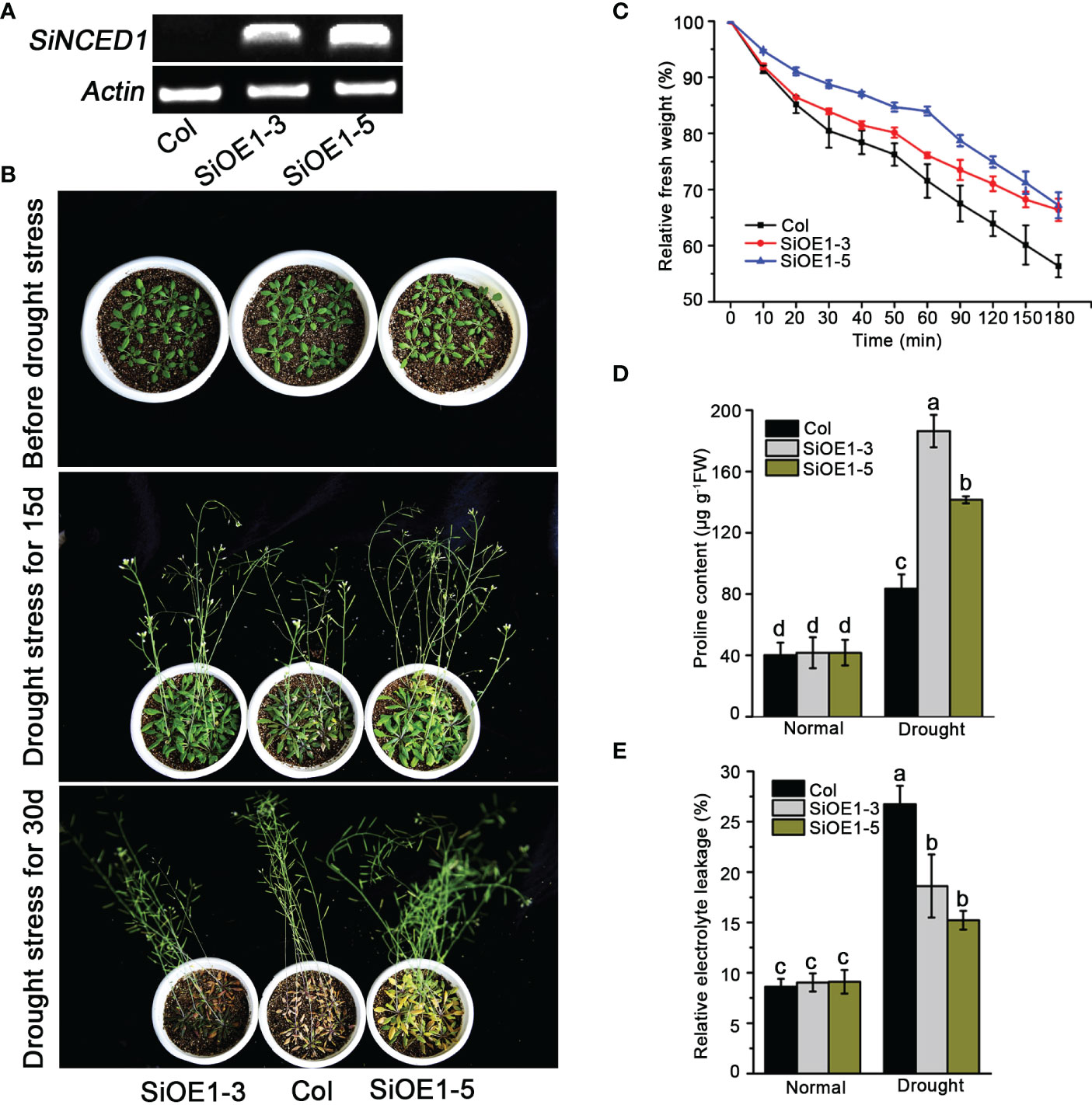
Figure 4 Drought stress tolerance of SiNCED1 overexpression plants. (A) Semi-quantitative RT-PCR detection of SiNCED1 expression in Col and SiOE1-3, SiOE1-5. (B) The phenotype of SiNCED1-overexpressing plants and Col seedlings under drought stress. (C) Measurement of water loss rate in Col and siNCED1-overexpressing seedlings. (D) Proline content determination after drought stress treatment (E) Relative electrolyte leakage determination after drought stress treatment. Data shown are mean ± SD from three independent replicates. Values are the mean ± SD (n = 6 plants). Similar results were obtained from three independent replicates. Different letters represent significant differences at P<0.05 (ANOVA analysis, Tukey’s multiple comparisons test).
Given that the altered physiological mechanism was possibly responsible for drought stress adaptation, we examined the water loss rate, proline content, and relative electrolyte leakage in detached Arabidopsis rosette leaves. We observed that Col rosette leaves lost water faster than those from the SiNCED1-overexpressing lines (Figure 4C). Meanwhile, the proline content and relative electrolyte leakage were not substantially different between Col plants and SiNCED1-overexpressing lines under normal conditions. However, SiNCED1-overexpressing lines had higher proline content and lower relative electrolyte leakage than Col plants under drought stress (Figures 4D, E). These results indicate that SiNCED1 could increase drought stress tolerance in foxtail millet.
We also investigated stomatal aperture to further assess the drought stress tolerance in Col and SiNCED1-overexpressing plants. Stomatal apertures were classified into three categories: completely open, partially open, and completely closed (Figure 5A). Under normal conditions, the percentage of completely open stomata in SiNCED1-overexpressing plants was slightly higher than that in Col plants, but the percentage of completely closed and partially open stomata showed no difference between SiNCED1-overexpressing and Col plants (Figure 5B). However, under drought stress conditions, 34% and 32.05% of stomata were completely closed in SiNCED1-overexpressing plants (SiOE1-3 and SiOE1-5, respectively), whereas only 20.97% were completely closed in Col plants. Furthermore, only 4.43% and 9.65% of stomata were completely open in SiNCED1-overexpressing plants (SiOE1-3 and SiOE1-5, respectively), whereas 14.92% were completely open in Col. Additionally, 64.11% of stomata were partially open in Col, and no marked differences were observed compared with 61.57% and 58.3% of partially open stomata in SiNCED1-overexpressing plants (SiOE1-3 and SiOE1-5, respectively) (Figure 5B). Consistent with these results, the stomatal apertures did not differ between Col and SiNCED1-overexpressing plants under normal conditions. However, the stomatal apertures of SiNCED1-overexpressing plants (SiOE1-3, SiOE1-5) were significantly smaller than those of Col plants under drought conditions (Figure 5C). These results indicate that SiNCED1 may reduce stomatal apertures to inhibit water loss under drought stress conditions.
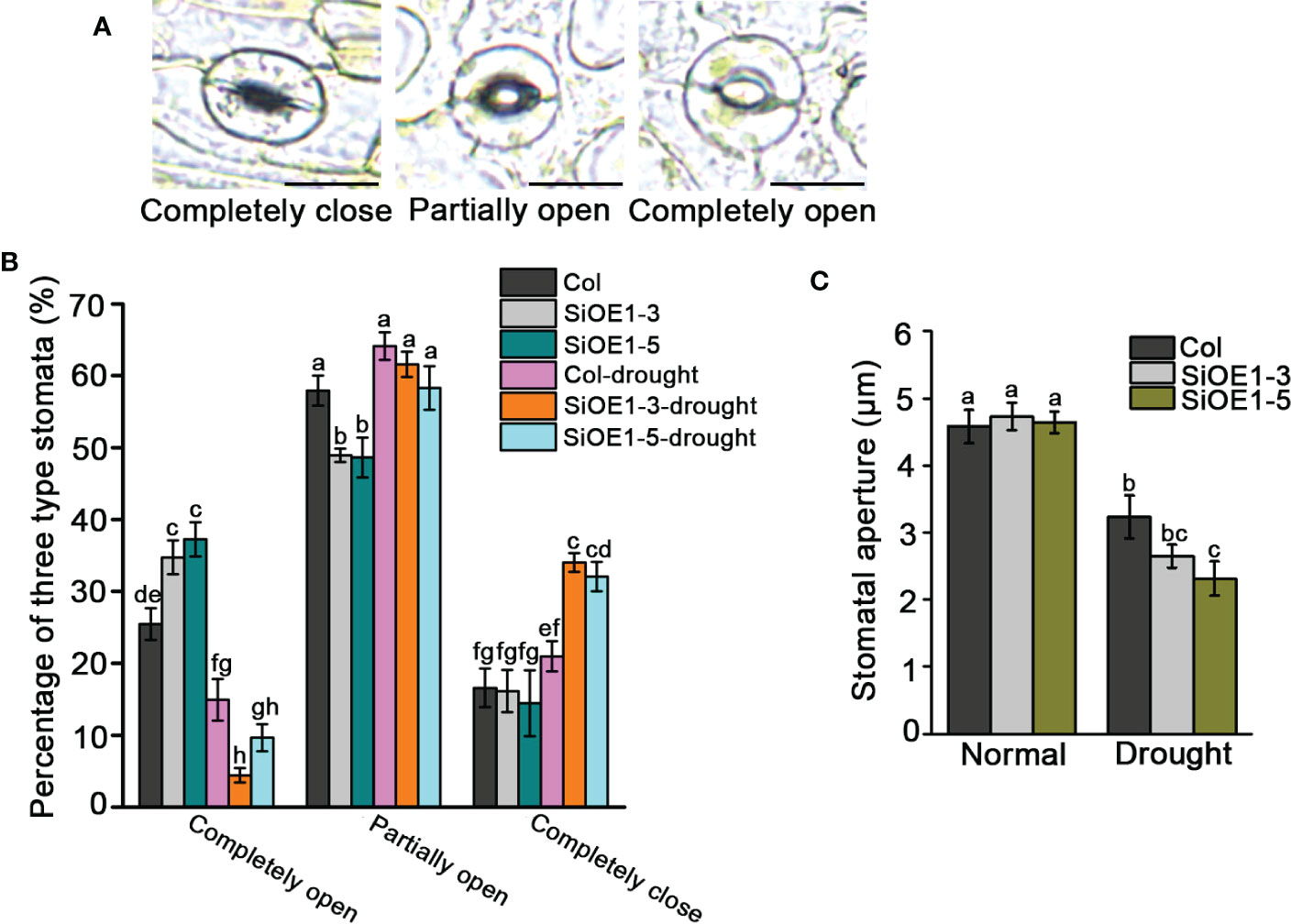
Figure 5 Stomata closure of SiNCED1-overexpressing plants under drought stress. (A) Images of three levels of stomatal apertures, Bars = 10 μm. (B) The percentage of three levels of stomatal apertures in SiNCED1-overexpressing plants and Col were calculated under normal and drought stress condition. (C) Comparison of stomatal apertures in SiNCED1-overexpressing plant and Col under normal and drought stress condition. Data shown are mean ± SD from there independent replicate. Each experiment includes at least 30 stomata from each epidermal peel. Different letters represent significant differences at P<0.05 (ANOVA analysis, Tukey’s multiple comparisons test).
Stomata movement acts as an important mechanism for plant resistance to drought stress through an ABA-dependent pathway. Moreover, NCED is a critical rate-limiting enzyme in ABA biosynthesis. To understand whether endogenous ABA is involved in altering drought stress tolerance, ABA levels were measured in Col and SiNCED1-overexpressing plants before and after drought stress treatment. We found that under normal conditions, ABA levels were not different between Col and SiNCED1-overexpressing plants (Figure 6A). However, ABA levels rapidly increased under drought stress condition in both Col and SiNCED1-overexpressing plants, and the ABA levels in SiNCED1-overexpressing plants (SiOE1-3, 44.2 ng/g; SiOE1-5, 39 ng/g) were much higher than those in Col plants (31.8 ng/g) (Figure 6A). Therefore, SiNCED1 increases drought stress tolerance by promoting endogenous ABA biosynthesis in plants.
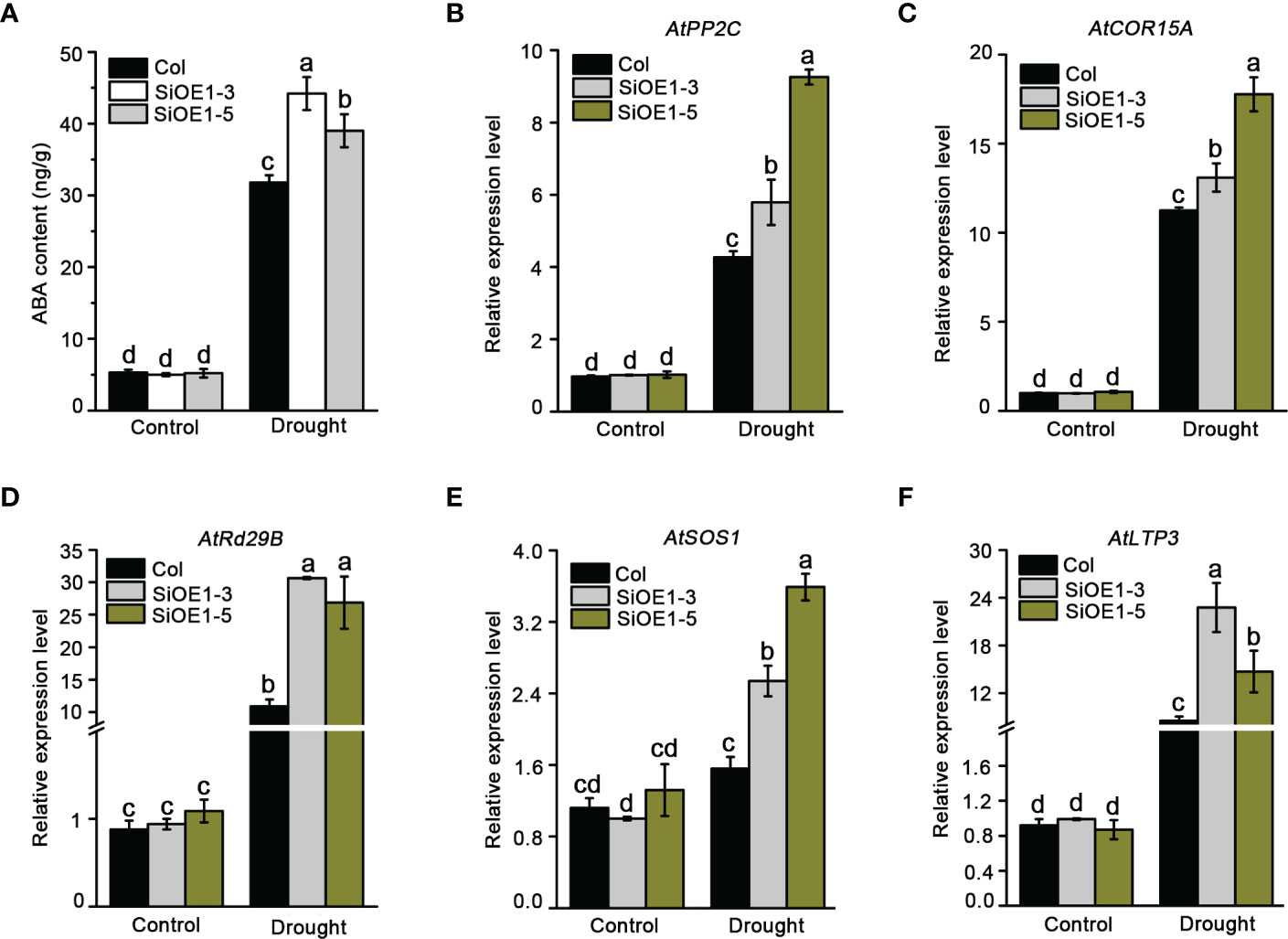
Figure 6 ABA content and abiotic stress related genes expression. (A) The ABA content in seedlings of Col and SiNCED1-overexpressing plants under drought stress. (B–F) The expression of ABA related stress responsive genes in Col and SiNCED1-overexpressing plants under drought stress for 7 d. Data shown are mean ± SD from there independent replicate. Different letters represent significant differences at P<0.05 (ANOVA analysis, Tukey’s multiple comparisons test).
We further determined the transcript levels of ABA-related stress-responsive genes under normal and drought stress conditions. Consistent with the changes in ABA content, the expression of AtPP2C, AtCOR15A, AtRd29B, AtSOS1, and AtLTP3 did not differ between Col and SiNCED1-overexpressing plants under normal conditions. However, the expression levels of these genes were noticeably higher in SiNCED1-overexpressing plants than in Col plants (Figures 6B–F). These results suggest that SiNCED1 up-regulated ABA-related stress-responsive genes under drought stress conditions.
The above results show that SiNCED1 regulates endogenous ABA biosynthesis. Since ABA plays pivotal roles in seed dormancy and germination, the seed germination rate of Col and SiNCED1-overexpressing lines were determined. The results showed that the seed germination rate of SiNCED1-overexpressing lines was slower and more inhibited than that of Col plants when the seeds were sown in normal MS medium (Figure 7A). Ectopic SiNCED1-overexpressing line seeds sown in MS medium containing high NaCl, mannitol, and glucose showed germination delay, which revealed that SiNCED1-overexpressing lines were more sensitive to high salt, high mannitol, and high glucose conditions than Col plants (Figures 7B–D). Furthermore, when we grew SiNCED1-overexpressing lines and Col seeds for 7 d in normal MS media, the Col seeds all germinated, while only 87.58% and 85.60% of the seeds of SiNCED1-overexpressing lines OE1-3 and OE1-5 germinated, respectively (Figures 7A, 8A). Sowing the seeds in the MS medium containing high NaCl, mannitol, and glucose for 7 d, the phenotype of SiNCED1-overexpressing lines OE1-3 and OE1-5 showed that the seeds could not germinate as well as those of Col (Figures 8B–D).
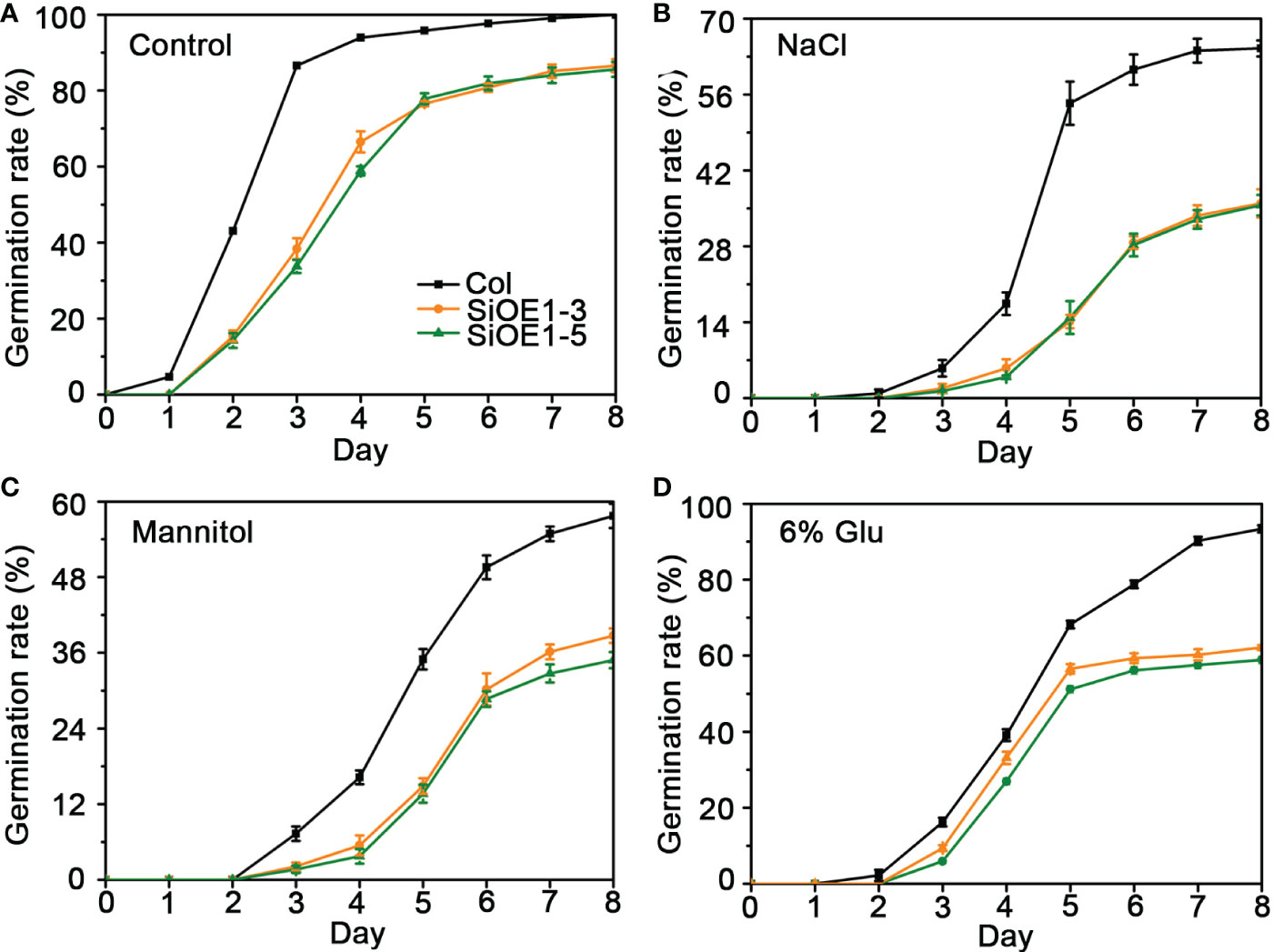
Figure 7 Germination rate of Col and SiNCED1 transgenic seeds grown in MS, Mannitol, NaCl and Glucose media. (A–D) The germination rate of Col and SiNCED1 transgenic seeds grown in MS media contain 150 mM NaCl, 275 mM Mannitol, or 6% Glucose. Data shown are mean ± SD from three independent replicates.
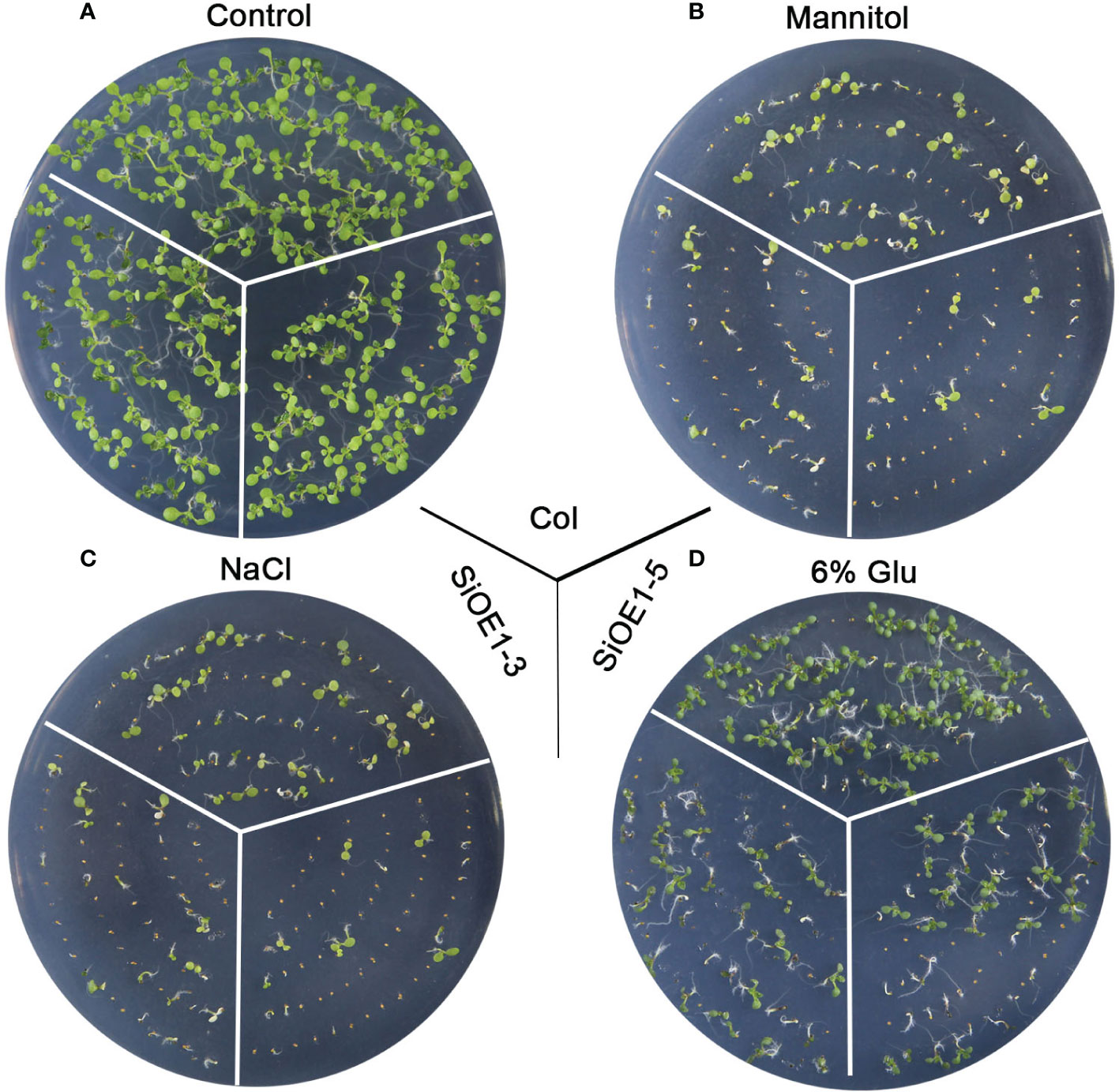
Figure 8 Phenotypic analysis of SiNCED1 transgenic plants under MS, mannitol, NaCl and Glucose. (A–D) The phenotype of Col and SiNCED1 transgenic seeds grown in MS (control), MS contain 275 mM Mannitol, 150 mM NaCl or 6% Glucose, respectively for 7 d.
ABA is a key hormone in plants that regulates plant development, including seed germination, seedling growth, stomatal aperture, flowering, and senescence. Additionally, it also elevates the ability of plants to withstand multiple stresses (Finkelstein, 2013; Kumar et al., 2022). NCED is the key rate-limiting enzyme in ABA biosynthesis that modulates endogenous ABA homeostasis by oxidatively cleaving 9-cis-violaxanthin or 9-cis-neoxanthin to produce xanthoxin (Priya and Siva, 2015). Currently, the NCED gene has been cloned in many plants, and its biological functions have been reported. However, the biological function of NCED in foxtail millet remains unclear. In this study, four SiNCED genes were found in the NCBI database and cloned, after which homologous comparison were performed. The following biological functions of SiNCED1 were identified: seed germination regulation, stomatal closure, and drought stress tolerance by modulating endogenous ABA synthesis.
NCED belongs to the CCD gene family, which ordinarily contains the conserved REP65 domain (Yue et al., 2022; Zhao et al., 2022). In our study, the phylogenetic tree and multiple alignment analysis showed that SiNCED1 was separately grouped into one cluster, while the other three SiNCEDs were aggregated into another cluster, but SiNCED1 was homologous with other NCED proteins and contains the conserved REP65 domain (Figures 1, 2). Members of the NCED subfamily are involved in the synthesis of ABA, which is involved in plant growth and development (Yang et al., 2022). OsNCED2 was constitutively expressed in various tissues in rice plants; osnced2 mutant seedlings with knockout of OsNCED2 showed growth inhibition (Copenhaver et al., 2021). OsNCED3 is expressed in multiple tissues, including embryo, coleoptile, root, leaf, culm, node, flower, stigma, and pollen, with especially high expression found in the flower and root. However, nced3 mutant seedlings with a mutation in the OsNCED3 gene showed a faster growth trend as well as longer roots and shoots compared with the wild type (Huang et al., 2018). Furthermore, osnced2 and nced3 mutants showed contrary seedling phenotypes. The regulation of endogenous ABA content was regarded as the main mechanism for the regulation of the growth and development of seedlings by OsNCED2 and OsNCED3, which also confirms the dual regulatory effect of endogenous ABA on plant growth and development (Cheng et al., 2002). Expression pattern analysis showed that SiNCED1 was expressed in multiple organs, especially the roots, which showed the highest expression (Figure 3A). However, the seedling growth and development of ectopic SiNCED1-overexpressing lines showed no difference from those of Col. Whether SiNCED1 modulates the growth and development of foxtail millet needs to be investigated by constructing nced1 mutants of foxtail millet. In addition, the transcript levels of SiNCED1 were significantly induced by ABA, osmotic stress, and high salt stress (Figures 3B–G). SiNCED1 is likely similar to other NCEDs in other vascular plants, such as AtNCED3 in Arabidopsis and OsNCED3 and OsNCED4 in rice, which are significantly induced by high salt and osmotic stress (Huang et al., 2018; Hwang et al., 2018).
ABA is an important plant stress hormone. Under water stress, ABA can rapidly synthesize and regulate ABA-dependent signaling pathways to resist water stress, which are in turn regulated by NCED genes (Ye et al., 2012; Kavi Kishor et al., 2022). In our study, the ectopic overexpression of SiNCED1 in Arabidopsis enhanced its ability to survive water stress (Figure 4B), and increased endogenous ABA levels under water stress (Figure 6A). NCED genes that participate in drought stress have also been reported in many plants. For instance, heterologous expression of CrNCED1 in tobacco has indicated that this gene can enhance drought stress tolerance by increasing ABA levels and decreasing the levels of reactive oxygen species (Xian et al., 2014). Constitutively overexpressing VaNCED1 in a drought-sensitive cultivar of V. vinifera increased ABA content and decreased stomatal density, thereby significantly strengthening drought resistance (He et al., 2018). GhNCED3a/3c participates in drought stress in cotton because GhirNAC2 directly binds to its promoter to modulate ABA biosynthesis and stomatal closure (Shang et al., 2020). AhNCED1, cloned from peanuts, can control ABA content, and the negative feedback loop regulated by the transcription factor complex AhAREB1 cooperates with AhNAC2 under water stress (Liu et al., 2016). AtNCED3 mediates drought stress by increasing endogenous ABA levels and promoting drought- and ABA-inducible genes expression. These signaling pathways are manipulated by NGATHA1, which binds directly to the AtNCED3 promoter of the NBE cis-element (Sato et al., 2018). OsNCED4 can also elevate ABA levels to increase drought tolerance through OsbZIP23, which is directly bound to its promoter cis-element, ABRE (Zong et al., 2016).
Under drought conditions, plants produce and accumulate ABA in their guard cells, which induces stomatal closure to reduce water loss (Lim et al., 2015; Wang et al., 2022). NCED genes have been identified as a major determinant of endogenous ABA levels by regulating ABA biosynthesis (Tan et al., 1997; Schwartz et al., 2003). Our results suggest that SiNCED1 overexpression in Arabidopsis can reduce water loss by increasing the percentage of completely closed stomata and reducing stomata apertures under water stress (Figure 5). This is consistent with the modulation of ABA biosynthesis and stomatal closure by GhNCED3a/3c under drought stress (Shang et al., 2020) and AtNCED3 expression induced by the small peptide CLE25 to enhance ABA levels and modulate stomatal movement to prevent water loss (Takahashi et al., 2018). In addition, proline content and electrolyte leakage are important physiological indices for the evaluation of plant resistance to water stress (Voetberg and Sharp, 1991; Jiang et al., 2016). The ectopic expression of BnNCED3 in Arabidopsis or overexpression of OsNCED5 in rice resulted in both exhibiting higher proline content and lower electrolyte leakage than the wild type following drought stress treatment (Xu and Cai, 2017; Huang et al., 2019). In the present study, SiNCED1-overexpressing transgenic lines had higher proline content and lower electrolyte leakage than Col under water stress (Figures 4D, F), which is reliable evidence for the role of SiNCED1 in resistance to drought stress. Furthermore, high expression levels of ABA-related stress-responsive marker genes, such as AtPP2C, AtCOR15A, AtRd29B, AtSOS1, and AtLTP3, enhance resistance to drought stress in Arabidopsis (Guo et al., 2013; Yang et al., 2018; Lu et al., 2019). We found that the expression levels of these stress-responsive genes increased substantially in SiNCED1 transgenic lines (Figures 6B–F), indicating that SiNCED1 can also help to maintain a high resistance to water stress.
ABA has an important effect on seed dormancy, which relies on its biosynthesis and catabolism (Bentsink and Koornneef, 2008). Numerous studies have showed that the overexpression of ABA biosynthesis pathway genes, including PtNCED1 (Lee et al., 2018), AtNCED9 (Seo et al., 2016), BdNCED1 (Barrero et al., 2012), TaNCED2 (Izydorczyk et al., 2018), and LeNCED1 (Tung et al., 2008), can promote seed dormancy by regulating endogenous ABA accumulation. In the present study, SiNCED1 overexpression in transgenic lines exhibited the inhibition of seed germination under normal and abiotic stress conditions (Figures 7 and 8), which showed a similar phenotype to that in the above-mentioned reports. However, the molecular mechanism underlying the regulation of dormancy and germination processes in foxtail millet seeds by SiNCED1 still requires further study.
In the present study, four NCED genes were identified and cloned into foxtail millet. The relative expression levels of SiNCED1 were strongly induced in the shoots and roots under osmotic stress. Overexpression of SiNCED confers drought stress tolerance to Arabidopsis. This function is supported by the finding that SiNCED1 regulates ABA biosynthesis to modulate stress-related physiological indices, stomatal closure, and ABA-related stress-responsive genes. Furthermore, SiNCED1 promotes seed dormancy by increasing ABA accumulation. Thus, SiNCED1 is likely crucial for improving the breeding of foxtail millet. The mechanisms by which SiNCED1 cooperates with other SiNCED members and how transcription factors regulate its expression and affect developmental processes and abiotic stress could be novel research topics for future studies.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Conceptualization, ML and LC. Formal analysis, SY and FW. Investigation, YH and YJ. Supervision, ML and LC. Writing—original draft, YH and SY. Writing—review and editing, ML and DM. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (No. 8226140781), the Basic Scientific Research Program from Guizhou Science and Technology Agency (No. Qian Ke He Ji Chu-ZK [2021]522).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1121809/full#supplementary-material
Andrew, D., Roger, H. (1992). Abscisic acid biosynthesis in root. Planta 187, 185–191. doi: 10.1007/BF00201936
Barrero, J. M., Jacobsen, J. V., Talbot, M. J., White, R. G., Swain, S. M., Garvin, D. F., et al. (2012). Grain dormancy and light quality effects on germination in the model grass brachypodium distachyon. New Phytol. 193 (2), 376–386. doi: 10.1111/j.1469-8137.2011.03938.x
Barton, L., Newsome, S., Chen, F., Wang, H., Guilderson, T., Bettinger, R. (2009). Agricultural origins and the isotopic identity of domestication in northern China. Proc. Natl. Acad. Sci. U.S.A. 106 (14), 5523–5528. doi: 10.1073/pnas.0809960106
Bentsink, L., Koornneef, L. (2008). “Seed dormancy and germination,” in The arabidopsis book 6, 1–18. doi: 10.1199/tab.0050
Cheng, W., Endo, A., Zhou, L., Penney, J., Chen, H., Arroyo, A., et al. (2002). A unique short-chain dehydrogenase/reductase in arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14 (11), 2723–2743. doi: 10.1105/tpc.006494
Copenhaver, G., Li, X., Yu, B., Wu, Q., Min, Q., Zeng, R., et al. (2021). OsMADS23 phosphorylated by SAPK9 confers drought and salt tolerance by regulating ABA biosynthesis in rice. PloS Genet. 17 (8), e1009699. doi: 10.1371/journal.pgen.1009699
Doust, A., Kellogg, E., Devos, K., Bennetzen, J. (2009). Foxtail millet: a sequence-driven grass model system. Plant Physiol. 149 (1), 137–141. doi: 10.1104/pp.108.129627
Eiji, N., Annie, M. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185. doi: 10.1146/annurev.arplant.56.032604.144046
Finkelstein, R. (2013). Abscisic acid synthesis and response. Am. Soc. Plant Biologists. 11. doi: 10.1199/tab.0166
Frey, A., Effroy, D., Lefebvre, V., Seo, M., Perreau, F., Berger, A., et al. (2012). Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 70 (3), 501–512. doi: 10.1111/j.1365-313X.2011.04887.x
Guo, L., Yang, H. B., Zhang, X. Y., Yang, S. H. (2013). Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in arabidopsis. J. Exp. Bot. 64 (6), 1755–1767. doi: 10.1093/jxb/ert040
Hao, G. P., Zhang, X. H., Wang, Y. Q., Wu, Z. Y., Huang, C. L. (2009). Nucleotide variation in the NCED3 region of Arabidopsis thaliana and its association study with abscisic acid content under drought stress. J. Integr. Plant Biol. 51 (2), 175–183. doi: 10.1111/j.1744-7909.2008.00786.x
He, R., Zhuang, Y., Cai, Y., Agüero, C. B., Liu, S., Wu, J., et al. (2018). Overexpression of 9-cis-epoxycarotenoid dioxygenase cisgene in grapevine increases drought tolerance and results in pleiotropic effects. Front. Plant Sci. 9 (970). doi: 10.3389/fpls.2018.00970
Huang, Y., Guo, Y., Liu, Y., Zhang, F., Wang, Z., Wang, H., et al. (2018). 9-cis-epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00162
Huang, Y., Jiao, Y., Xie, N., Guo, Y., Zhang, F., Xiang, Z., et al. (2019). OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci. 287, 110188. doi: 10.1016/j.plantsci.2019.110188
Hwang, S. G., Chen, H. C., Huang, W. Y., Chu, Y. C., Shii, C. T., Cheng, W. H. (2010). Ectopic expression of rice OsNCED3 in arabidopsis increases ABA level and alters leaf morphology. Plant Sci. 178 (1), 12–22. doi: 10.1016/j.plantsci.2009.09.014
Hwang, S. G., Lee, C. Y., Tseng, C. S. (2018). Heterologous expression of rice 9-cis-epoxycarotenoid dioxygenase 4 (OsNCED4) in arabidopsis confers sugar oversensitivity and drought tolerance. Botanical Stud. 59 (1). doi: 10.1186/s40529-018-0219-9
Izydorczyk, C., Nguyen, T. N., Jo, S., Son, S., Tuan, P. A., Ayele, B. T. (2018). Spatiotemporal modulation of abscisic acid and gibberellin metabolism and signalling mediates the effects of suboptimal and supraotimal temperatures on seed germination in wheat (Triticum aestivum l.). Plant Cell Environ. 41, 1022–1037. doi: 10.1111/pce.12949
Jiang, Y., Qiu, Y., Hu, Y., Yu, D. (2016). Heterologous expression of AtWRKY57 confers drought tolerance in Oryza sativa. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00145
Kavi Kishor, P. B., Tiozon, R. N., Fernie, A. R., Sreenivasulu, N. (2022). Abscisic acid and its role in the modulation of plant growth, development, and yield stability. Trends Plant Sci. 27 (12), 1283–1295. doi: 10.1016/j.tplants.2022.08.013
Koshiba, M. (2002). Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7 (1), 41–48. doi: 10.1016/s1360-1385(01)02187-2
Kumar, S., Shah, S. H., Vimala, Y., Jatav, H. S., Ahmad, P., Chen, Y., et al. (2022). Metabolism, transport, crosstalk with other plant growth regulators, and its role in heavy metal stress mitigation. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.972856
Lang, J., Fu, Y. X., Zhou, Y., Cheng, M. P., Deng, M., Li, M. L., et al. (2021). Myb10-d confers PHS-3D resistance to pre-harvest sprouting by regulating NCED in ABA. New Phytol. 230, 1940–1952. doi: 10.1111/NPH.17312
Lee, Y. I., Chen, M. C., Lin, L., Chung, M. C., Leu, W. M. (2018). Increased expression of 9- cis-epoxycarotenoid dioxygenase, PtNCED1, associated with inhibited seed germination in a terrestrial orchid, Phaius tankervilliae. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01043
Li, H. Z., Cheng, Z. H. (2015). Hoagland nutrient solution promotes the growth of cucumber seedlings under light-emitting diode lingt. Acta Agric. Scand. Sect. B Soil Plant Sci. 65, 74–82. doi: 10.1080/09064710.2014.967285
Li, Q. H., Yu, X. T., Chen, L., Zhao, G., Li, S. Z., Zhou, H., et al. (2021). Genome-wide identification and expression analysis of the NCED family in cotton (Gossypium hirsutum l.). PloS One 16 (2), e0246021. doi: 10.1371/journal.pone.0246021
Lim, C. W., Baek, W. H., Jung, J. H., Kim, J. H., Lee, S. C. (2015). Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 16, 15251–15270. doi: 10.3390/ijms160715251
Liu, S., Li, M. J., Su, L. C., Ge, K., Li, L. M., Li, X. Y., et al. (2016). Negative feedback regulation of ABA biosynthesis in peanut (Arachis hypogaea): a transcriptioin factor complex inhibits AhNCED1 expression during water stress. Sci. Rep. 6, 37943. doi: 10.1038/srep37943
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-DeltaDeltaC(T)). Method Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lou, D., Wang, H., Liang, G., Yu, D.. (2017). OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice. Front. Plant Sci. 8, 993. doi: 10.3389/fpls.2017.00993
Lu, C. Q., Yu, F., Tian, L. F., Huang, X. Y., Tan, H., Xie, Z. J., et al. (2017). RPS9M, a mitochondrial ribosomal protein, is essential for central cell maturation and endosperm development in Arabidopsis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02171
Lu, P., Magwanga, R. O., Kirungu, J. N., Hu, Y. G., Dong, Q., Cai, X. Y., et al. (2019). Overexpression of cotton a DTX/MATE gene enhances drought, salt, and cold stress tolerance in transgenic arabidopsis. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00299
Miao, J., Guo, D. S., Zhang, J. Z., Huang, Q. P., Qin, G. J., Zhang, X., et al. (2013). Targeted mutagenesis in rice using CRISPR-cas system. Cell Res. 23, 1233–1236. doi: 10.1038/cr.2013.123
Nambara, E., Okamoto, M., Tatematsu, K., Yano, R., Seo, M., Kamiya, Y. (2010). Abscisic acid and the control of seed dormancy and germination. Seed Sci. Res. 20 (2), 55–67. doi: 10.1017/s0960258510000012
Ng, L. M., Melcher, K., Teh, B. T., Xu, H. E. (2014). Abscisic acid perception and signaling: structural mechanisms and applications. Acta Pharmacologica Sin. 35 (5), 567–584. doi: 10.1038/aps.2014.5
Pantin, F., Monnet, F., Jannaud, D., Costa, J. M., Renaud, J., Muller, B., et al. (2013). The dual effect of abscisic acid on stomata. New Phytol. 197 (1), 65–72. doi: 10.1111/nph.12013
Priya, R., Siva, R. (2015). Analysis of phylogenetic and functional diverge in plant nine−cis epoxycarotenoid dioxygenase gene family. J. Plant Res. 128, 519–534. doi: 10.1007/s10265-015-0726-7
Sah, S. K., Reddy, K. R., Li, J. (2016). Abscisic ccid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00571
Sano, N., Marion-Poll, A. (2021). ABA metabolism and homeostasis in seed dormancy and germination. Int. J. Mol. Sci. 22 (10), 5069. doi: 10.3390/ijms22105069
Sato, H., Takasaki, H., Takahashi, F., Satoshi, L., Mitsuda, N., Ohme Takagi, M., et al. (2018). Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. PNAS 115, e11178–e11187. doi: 10.1073/pnas.1811491115
Schwartz, S. H., Qin, X., Zeevaart, J. A. D. (2003). Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 131, 1591–1601. doi: 10.1104/pp.102.017921
Schwartz, S. H., Tan, B., Gage, D. A., Zeevaart, J. A. D., McCarty, R. D. (1997). Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276, 1872–1874. doi: 10.1126/science.276.5320.1872
Seo, M., Kanno, Y., Frey, A., North, H. M., Marion-Poll, A. (2016). Dissection of arabidopsis NCED9 promoter regulatory regions reveals a role for ABA synthesized in embryos in the regulation of GA-dependent seed germination. Plant Sci. 246, 91–97. doi: 10.1016/j.plantsci.2016.02.013
Shang, X. G., Yu, Y. J., Zhu, L. J., Liu, H. Q., Chai, Q. C., Guo, W. Z. (2020). A cotton NAC transcription factor GhirNAC2 plays positive roles in drought tolerance via regulating ABA biosynthesis. Plant Sci. 296, 110498. doi: 10.1016/j.plantsci.2020.110498
Sun, L., Sun, Y. F., Zhang, M., Wang, L., Ren, J., Cui, M. M., et al. (2012). Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomato. Plant Physiol. 158, 283–298. doi: 10.1104/pp.111.186866
Takahashi, F., Suzuki, T., Osakabe, Y., Betsuyaku, S., Kondo, Y., Dohmae, N., et al. (2018). A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556 (7700), 235–238. doi: 10.1038/s41586-018-0009-2
Tan, B. T., Joseph, L. M., Deng, W. D., Liu, L., Li, Q., Cline, K., et al. (2003). Molecular characterization of the arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 33, 44–56. doi: 10.1046/j.1365-313x.2003.01786.x
Tan, B., Schwartz, S. H., Zeevaart, J. A. D., McCarty, D. R. (1997). Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. U.S.A. 94, 12235–12240. doi: 10.1073/pnas.94.22.12235
Tung, S. A., Smeeton, R., White, C. A., Black, C. R., Taylor, I. B., Hilton, H. W., et al. (2008). Over-expression of LeNCED1 in tomato (Solanum lycopersicum l.) with the rbcS3C promoter allows recovery of lines that accumulate very high levels of abscisic acid and exhibit severe phenotypes. Plant Cell Environ. 31, 968–981. doi: 10.1111/j.1365-3040.2008.01812.x
Voetberg, G. S., Sharp, R. E. (1991). Growth of the maize primary root at low water potentials III. role of increased proline deposition in osmotic adjustment. Plant Physiol. 96, 1125–1130. doi: 10.1104/pp.96.4.1125
Wang, P., Lu, S., Zhang, X., Hyden, B., Qin, L., Liu, L., et al. (2021). Double NCED isozymes control ABA biosynthesis for ripening and senescent regulation in peach fruits. Plant Sci. 304, 110739. doi: 10.1016/j.plantsci.2020.110739
Wang, P., Qi, S. J., Wang, X. H., Dou, L. R., Jia, M. A., Mao, T. L., et al. (2022). The OPEN STOMATA1-SPIRAL1 module regulates microtubule stability during abscisic acid-induced stomatal closure in arabidopsis. Plant Cell 35 (1), 260–278. doi: 10.1093/plcell/koac307
Xian, L., Sun, P., Hu, S., Wu, J., Liu, J. (2014). Molecular cloning and characterization of CrNCED1, a gene encoding 9−cis−epoxycarotenoid dioxygenase in Citrus reshni, with functions in tolerance to multiple abiotic stresses. Planta 239, 61–77. doi: 10.1007/s00425-013-1963-4
Xu, P., Cai, W. (2017). Functional characterization of the BnNCED3 gene in Brassica napus. Palnt Sci. 256, 16–24. doi: 10.1016/j.plantsci.2016.11.012
Xu, Z. Y., Kim, D. H., Hwang, I. (2013). ABA homeostasis and signaling involving multiple subcellular compartments and multiple receptors. Plant Cell Rep. 32 (6), 807–813. doi: 10.1007/s00299-013-1396-3
Yang, X. W., He, K., Chi, X. Y., Chai, G. H., Wang, Y. P., Jia, C. L., et al. (2018). Miscanthus NAC transcription factor MlNAC12 positively mediates abiotic stress tolerance in transgenic arabidopsis. Plant Sci. 277, 229–241. doi: 10.1016/j.plantsci.2018.09.013
Yang, X., Jia, Z., Pu, Q., Tian, Y., Zhu, F., Liu, Y. (2022). ABA mediates plant development and abiotic stress via alternative splicing. Int. J. Mol. Sci. 23 (7). doi: 10.3390/ijms23073796
Ye, N. H., Jia, L. H., Zhang, J. H. (2012). ABA signal in rice under stress conditions. Rice 5, 1. doi: 10.1186/1939-8433-5-1
Ye, N. H., Zhu, G. H., Liu, Y. G., Li, Y. X., Zhang, J. H. (2011). Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Phsiology 52 (4), 689–698. doi: 10.1093/pcp/pcp022
Yue, X. Q., Zhang, Y., Yang, C. K., Li, J. G., Rui, X., Ding, F., et al. (2022). Genome-wide identification and expression analysis of carotenoid cleavage oxygenase genes in litchi (Litchi chinensis sonn.). BMC Plant Biol. 22, 394. doi: 10.1186/s12870-022-03772-w
Zhang, G. Y., Liu, X., Quan, Z. W., Cheng, S. F., Xu, X., Pan, S. K., et al. (2012). Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 30 (6), 549–554. doi: 10.1038/nbt.2195
Zhao, X. L., Yang, Y. L., Xia, H. X., Li, Y. (2022). Genome-wide analysis of the carotenoid cleavage dioxygenases gene family in Forsythia suspensa: Expression profile and cold and drought stress responses. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.998911
Zhou, H., Wang, Y. F., Zhang, Y. J., Xiao, Y. H., Liu, X., Deng, H. B., et al. (2022). Comparative analysis of heat-tolerant and heat-susceptible rice highlights the role of OsNCED1 gene in heat stress tolerance. Plants 11 (8), 1062. doi: 10.3390/plants11081062
Keywords: foxtail millet, abscisic acid, SiNCED1, drought stress, seed germination
Citation: Huang Y, Jiao Y, Yang S, Mao D, Wang F, Chen L and Liang M (2023) SiNCED1, a 9-cis-epoxycarotenoid dioxygenase gene in Setaria italica, is involved in drought tolerance and seed germination in transgenic Arabidopsis. Front. Plant Sci. 14:1121809. doi: 10.3389/fpls.2023.1121809
Received: 12 December 2022; Accepted: 24 February 2023;
Published: 09 March 2023.
Edited by:
Mahmoud Magdy, Ain Shams University, EgyptReviewed by:
Agata Daszkowska-Golec, University of Silesia in Katowice, PolandCopyright © 2023 Huang, Jiao, Yang, Mao, Wang, Chen and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manzhong Liang, MTk2MmxpYW5nQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.