- 1National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan, China
- 2Hongshan Laboratory, Wuhan, China
HOX32, a member of the HD-ZIP III family, functions in the leaf morphogenesis and plant photosynthesis. However, the regulatory mechanism of HOX32 in maize has not been studied and the regulatory relationship in photosynthesis is unclear. We conducted a comprehensive study, including phylogenetic analysis, expression profiling at both transcriptome and translatome levels, subcellular localization, tsCUT&Tag, co-expression analysis, and association analysis with agronomic traits on HOX32 for the dissection of the functional roles of HOX32. ZmHOX32 shows conservation in plants. As expected, maize HOX32 protein is specifically expressed in the nucleus. ZmHOX32 showed constitutively expression at both transcriptome and translatome levels. We uncovered the downstream target genes of ZmHOX32 by tsCUT&Tag and constructed a cascaded regulatory network combining the co-expression networks. Both direct and indirect targets of ZmHOX32 showed significant gene ontology enrichment in terms of photosynthesis in maize. The association study suggested that ZmHOX32 plays an important role in regulation of plant architecture. Our results illustrate a complex regulatory network of HOX32 involving in photosynthesis and plant architecture, which deepens our understanding of the phenotypic variation in plants.
Introduction
It is predicted that the world population will reach up to 9 billion in 2050. According to the current food production, only 70-90% of human can be fed, which largely lags behind our needs for food in future. Increasing food production is the only option (Salas Fernandez et al., 2009).
Maize (Zea mays L.), a most widely planted crop in the world, is particularly important in ensuring food security (Assefa et al., 2018). Photosynthesis enables the synthesis of carbohydrates, which plays an important role in guaranteeing yield of crops as well as maize. Similar to other C4 plants, the carbon reaction of maize is also carried out in mesophyll cells and vascular bundle sheath cells (Dai et al., 2022). The C4 photosynthetic pathway is very complicated, where numerous genes are involved in the regulation. However, the regulatory relationships of photosynthesis related genes are still limited.
Leaf is the main place of photosynthesis, respiration, and transpiration in plants. Plant architecture traits such as leaf size, shape, thickness, and angle can affect the photosynthesis rate through regulating the utilization rate of light energy, thereby affecting the accumulation of carbohydrates. Therefore, the improvement of plant architecture is also one of the main goals of crops breeding. Plant leaves start from the flanks of the stem apical meristem, begin to develop asymmetrically, and establish three-dimensional spatial polarity along three directions: the base-apex axis, the middle-edge axis, and the adaxial-distal axis. Among them, the paraxial maintenance-distal axial patterning plays a crucial role in leaf morphogenesis, which is caused by antagonism between specialized adaxial and abaxial tissue-specific genes (Moon and Hake, 2011).
Many studies have reported that HD-ZIP III is involved in the regulation of leaf morphogenesis. HD-ZIP protein genes (homeodomain leucine zipper), a class of plant-specific transcription factors, belong to the homeobox family because of the containment of highly conserved homeodomain (HD) and leucine zipper structures domain (leucine zipper, LZ). Besides the basic HD and LZ domains, HD-ZIP III family proteins also contain a START domain that is able to bind to steroid ligands, which is highly conserved in evolution (Ponting and Aravind, 1999; Schrick et al., 2004). Moreover, several HD-ZIP III family proteins also contain a MEKHLA (Met-Glu-Lys-His-Leu-Ala) domain that consists of 6 conserved amino acids. This domain is involved in the signal transduction pathways mediated by chemical and physical stimulation, and play a potential role in affecting plant photosynthesis (Mukherjee and Bürglin, 2006). Hox32 is a member of HD-ZIP III family genes. In rice, overexpression of OsHOX32 caused narrow adaxial curling leaves, reduced leaf angle, erect plant type, dwarf plants, and reduced chlorophyll levels, thus repressing the photosynthesis efficiency (Li et al., 2016). In Arabidopsis, Hox32 homologs IFL1, ATHB-9 and ATHB-14 are involved in the development of the apical meristem, vascular bundle and the paraxial region of the lateral tissue, regulating the embryonic development process in the formation of the root tip (Palena et al., 2001). However, this gene has not been studied in maize at present and the regulatory network of Hox32 in maize is largely unknown.
To the end, we explore the function of ZmHOX32 using a cutting-edge molecular technique tsCUT&Tag and dissect the functional roles of ZmHOX32 in plant architecture by association mapping on a global diverse association panel. The cascading regulatory network in photosynthesis and association signals with plant architecture of ZmHOX32 may lay a foundation for the improvement of plant architecture and photosynthesis in maize.
Experimental process
Bioinformatics analysis for ZmHOX32
The cis-acting elements of the ZmHOX32 gene promoter were predicted using Plantcare (Rombauts et al., 1999)1. The phylogenetic tree of ZmHXO32 and other plant homologous proteins was constructed using the NJ method in MEGA software (Kumar et al., 1994). The conserved structural domains of ZmHOX32 and other homologous proteins were detected using the NCBI Conserved Domain Database (CDD) search tool (Marchler-Bauer, 2002)2.
Vector construction
The pM999-GFP vector was digested using Xba1 under condition 37 °C for 2-3 h. The CDS of the ZmHOX32 with removement of stop codon was inserted into the pM999-GFP vector upstream the GFP sequence, the amplification primers are shown in Supplementary Table 1. PCR fragment and linearized vector were recombined using ClonExpress® II One Step Cloning Kit (Vazyme C113-02), and the resultant was transformed into DH5α. The transformed bacterial solution was evenly coated on LB medium plates that contain Ampicillin and incubating for 12 h-16 h at 37 °C. Positive colonies were filtered by PCR and sanger sequencing. After expanding the cultivation of positive clone, the plasmid was extracted using the Endo-Free Plasmid DNA Maxi Kit (OMEGA D6926-03).
Isolation and transformation of protoplasts
Protoplasts were isolated from yellowing seedlings grown in dark culture for about 9-11 days at the nutritional growth V3 stage. The plasmids were transformed into protoplasts according to the described method (Yoo et al., 2007). The GFP fluorescence signal was observed under a confocal microscope (Leica) with a 485 nm laser. The protoplasts with successful transformation were subsequently subjected to subcellular localization and CUT&Tag.
tsCUT&Tag experimental procedure
A new cutting-edge technique tsCUT&Tag was employed for the dissection of regulatory network of ZmHOX32 (Wu et al., 2022). The Hyperactive In-Situ ChIP Library Prep Kit for Illumina (pG-Tn5) kit (Vazyme TD901) was used for the operation. Transformed protoplasts were observed by fluorescence microscopy to measure transformation efficiency. The samples with transformation efficiency no less than 60% were selected for subsequent CUT&Tag experiments. Two biological replicates were set for the constructed vector containing CDS of ZmHOX32. Cells were collected by low-speed centrifugation at 100 r/min for 2 min, and resuspended using 100 μl resuspension solution. After treating the resultant with ConA beads, incubation was performed with GFP antibody and corresponding secondary antibody. pG-Tn5 Transposon was used to fragment the DNAs and insert adaptors. Finally, the fragmentated DNA was extracted for library construction. After quantifying by Qubit, the constructed libraries were sequenced with pair-end 150 bp in Illumina Hiseq X-Ten platform. The transformed protoplasts with pM999-GFP vector were as the control group.
Data analysis for tsCUT&Tag
The reads were mapped into B73 reference genome (AGPv4) using Bowtie2 (Langmead and Salzberg, 2012) with the parameters “-p 10 –phred33 -I 0 -X 1000 –no-discordant –no-mixed”. PCR duplicates and reads with low quality (mapping quality score < 30) were removed. The searching for high confidence peaks (peaks p < 1×10-5) was performed using MACS (Feng et al., 2012) with the parameters “callpeak -g 2.2e+9 -s 150 -B -p 1e-5 -f BAMPE”. The distribution of peaks over the whole genome was analyzed using the ChIPseeker (Yu et al., 2015) in R. If the peak is located within the range of 3 kb upstream to 3 kb downstream of the gene, we will assume that this gene is the target of the protein. The performance was finished by the intersect function of the BEDtools software (Quinlan and Hall, 2010). GO enrichment analysis of target genes was performed using the AgriGOv2 (Tian et al., 2017)3 and the enriched GO terms were visualized using R. The generated downstream target genes were presented in Supplementary Table 2.
Association analysis
An association analysis was performed using genotypes and phenotypes of 690 inbred lines of maize with the general linear model (Abe et al., 2012; Gui et al., 2020). The phenotypes contained Tassel main axis length, Tassel branch number, Silking time, Pollen shed, Plant height, Leaf number above ear, Kerner number per row, Kernel width, Kernel thickness, Kernel length, Heading date, Ear length, Ear leaf width, Ear leaf length, Ear height, Ear diameter, Cob weight, Cob diameter, 100 Grain weight. All 19 phenotypes were distinguished by two categories of plant architecture and yield. Among them, Ear length, ear diameter, and cob diameter are contained in both of two types. The two types of phenotypic data were dimensionally reduced using R function “prcomp” and the setting “retx=T,scale=T,center=T”. The first two PCs were corrected for normal distribution by the R function “qnorm”. After the above processing, we obtained four phenotype data: Yield-pc1, Yield-pc2, Plant architecture-pc1, Plant architecture-pc2. In order to retain high-confidence SNPs, a total of 4069278 SNPs were retained through the filtering of VCFtools. (–maf 0.05 –minDP 5 –remove-indels –max-missing 0.9 –min-alleles 2 –max-alleles 2) (Danecek et al., 2011). Tassel software was used for SNP sorting and hapmap format conversion (Bradbury et al., 2007). Finally, the processed hapmap files and phenotype data were used as input for GWAS analysis by GAPIT software (Lipka et al., 2012). The threshold for significant SNPs is -Log10(1/4069278), which is 6.609517.
Results
Phylogenetic tree of ZmHOX32 demonstrates the potential functional conservation of HOX32 proteins across different plants
HD-ZIP III gene family is one of the important transcription factor family. Rice OsHOX32 has been reported to function in leaf morphogenesis. Comparative genomics has identified that Zm00001d033246 in maize is homologous to OsHOX32 in rice and so named as ZmHOX32. ZmHOX32 contains 18 exons, with 3,259 bp in genomic length and 2,570 bp of CDS, which encodes a HOX protein of 856 amino acids. To sense the potential function of ZmHOX32, we constructed a phylogenetic tree of HOX32 in plants and found that the closest evolutionary homologs are XP_002464180.1 in Sorghum bicolor and CAD6205937.1 in Miscanthus lutarioriparius, both of which are C4 crops. Further analysis of the conservation of HOX32 proteins showed that HOX32 is conserved across different plants with nearly identical functional domains, suggestive that HOX32 genes are likely to have conserved function in plants (Figure 1).

Figure 1 Phylogenetic tree and conserved domain analysis of ZmHOX32 with different plants. (A) Construction of phylogenetic tree using MEGA7. (B) Conserved domains of HOX32 proteins in different plants.
Expression pattern of ZmHOX32
To clarify the location of HOX32 protein in cells, we constructed a vector with the coding sequence of ZmHOX32 that fused to the upstream of green fluorescent protein (GFP) sequence and performed a subcellular localization assay. Successfully fused plasmid and empty vector were delivered into maize protoplasts to generate transient expression by infiltration. The GFP signal in empty vector (as control) was detected throughout the whole cell. In contrary, the ZmHOX32-GFP signal is specifically detected in the nucleus of maize protoplast cells (Figure 2). These results suggested that ZmHOX32 protein tends to function in nucleus, which is compatible with the intrinsic functional role as a transcription factor.
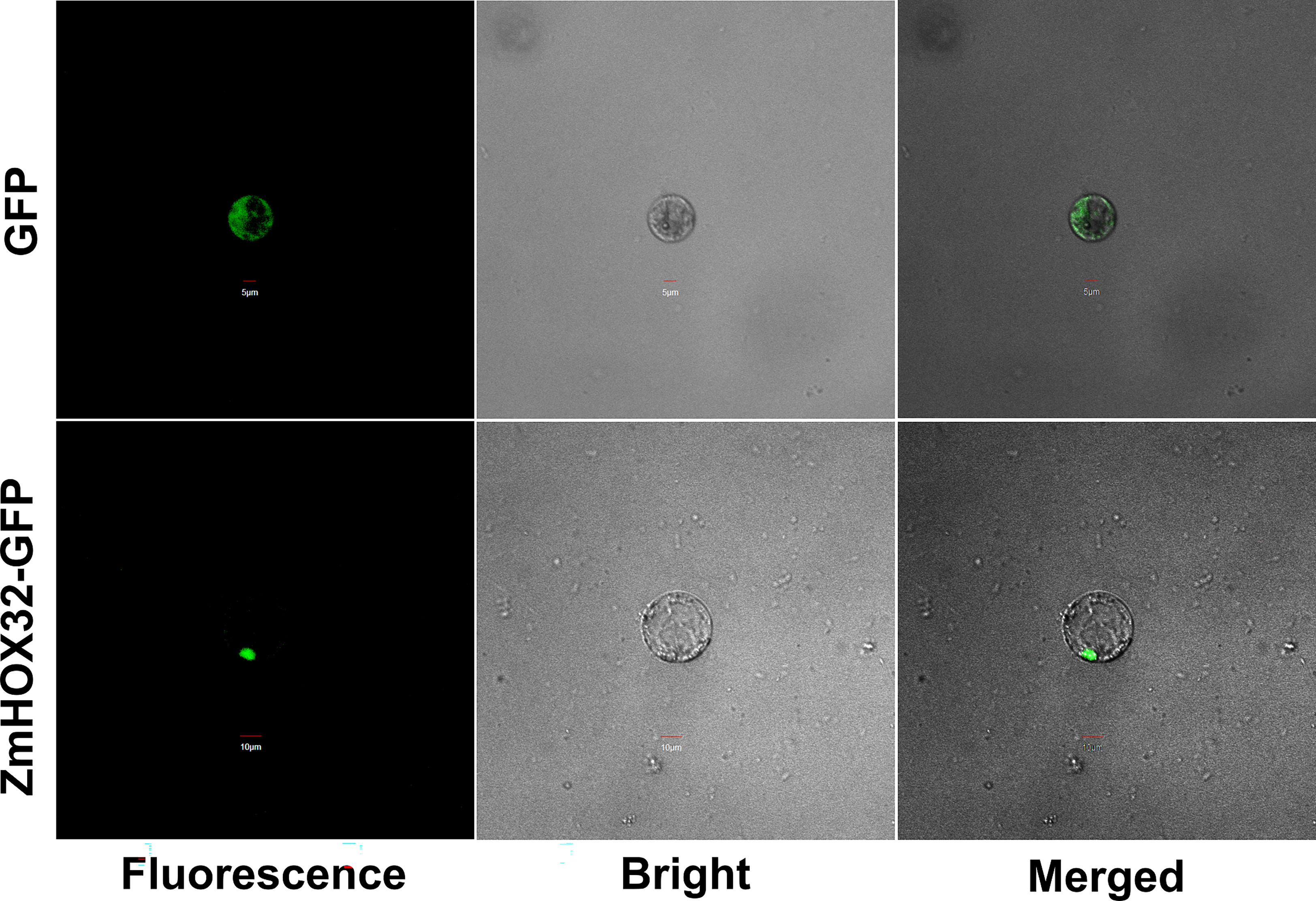
Figure 2 Subcellular localization of the HOX32 protein in maize. Subcellular localization of 35S::GFP and 35S::ZmHOX32-GFP in maize protoplasts; maize protoplast cells were used for taking images of green fluorescence, chloroplast autofluorescence, visible light, and merged visible light.
To profile the expression pattern of ZmHOX32 across maize development, we extracted a comprehensive transcriptome and translatome data from 33 different tissues or stages of maize different development from a previous study (Han et al., 2023). The expression abundance of ZmHOX32 was detected in roots, stems, leaves, and other tissues (Figures 3A, B). Interestingly, we noticed that ZmHOX32 was preferentially expressed in SAM and tassel etc, suggesting a potential function in plant development.
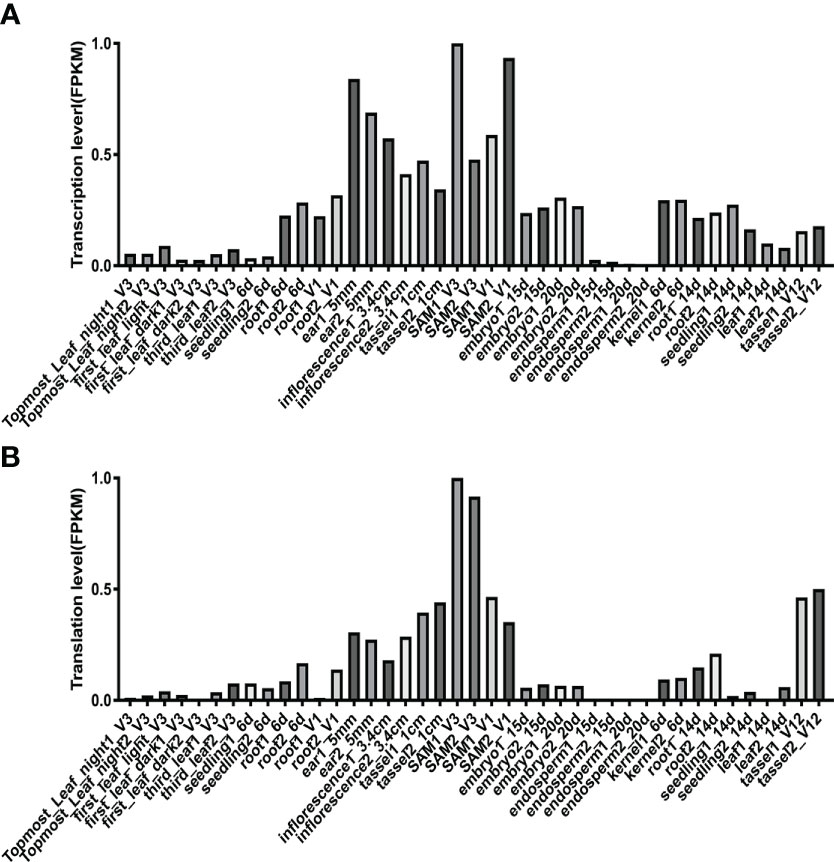
Figure 3 Expression of ZmHOX32 gene in maize tissues. (A) Transcription of ZmHOX32 gene. (B) Translation of the ZmHOX32 gene.
Potential upstream regulators of ZmHOX32 are associated to light responsive
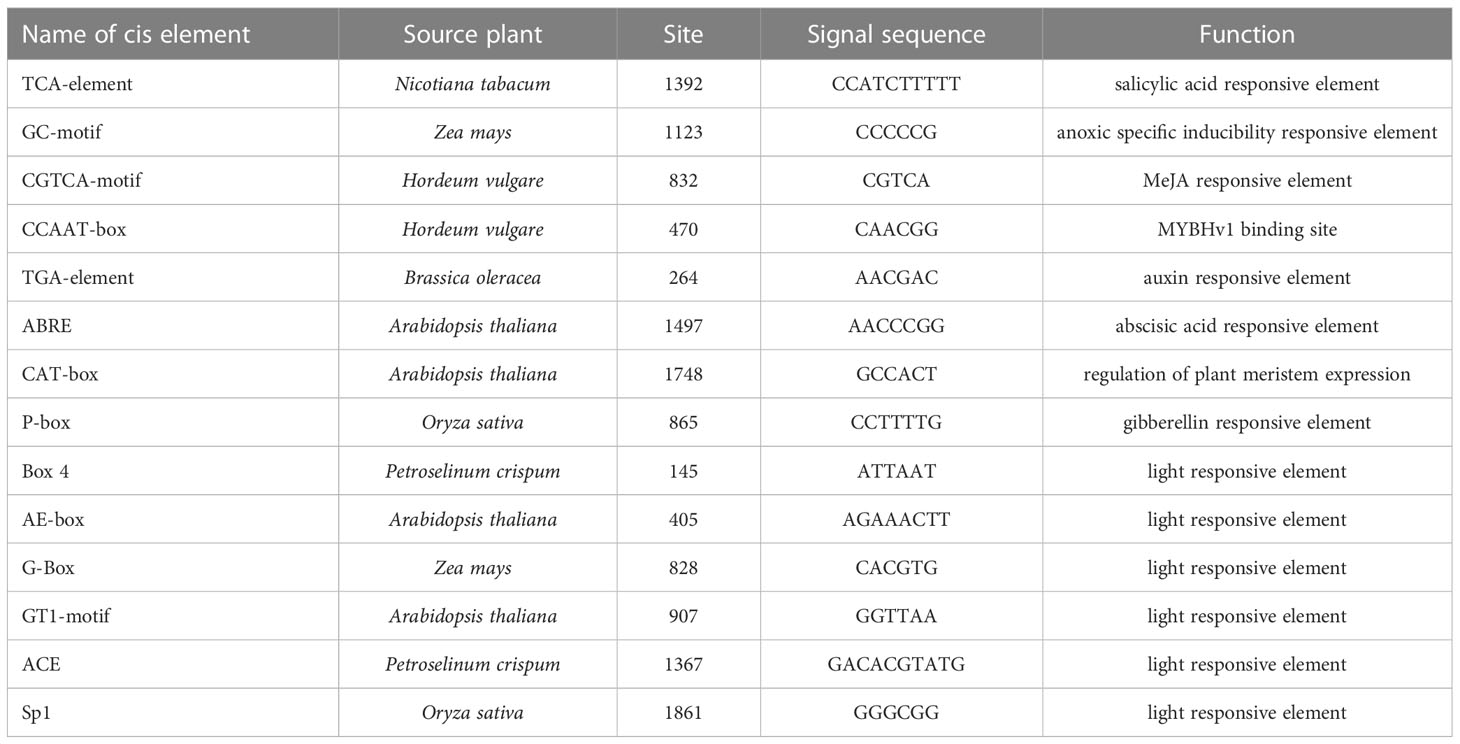
To identify and characterize the upstream regulators of ZmHOX32, the PlantCARE (Rombauts et al., 1999) database was used to analyze the promoter sequence of ZmHOX32, which suggested that ZmHOX32 gene contains a variety of functional response elements (Table 1). Besides the TATA-box and CAAT-box that belonged to the basic core element of the promoter, there were several specific motifs with potential different functions through the binding of the upstream regulators. It has been reported that TCA-element is involved in salicylic acid signal transduction pathway and GC-motif plays an important role in antioxidant response (Zhou et al., 2019). Some motifs are associated with hormone signal or response, such as CGTCA-motif, TGA-element, ABRE and P-box. Furthermore, we identified six different motifs that all related to light responsive. These cis-acting elements include Box 4, AE-box, G-Box, GT1-motif, ACE and Sp1. These results suggested that ZmHOX32 was likely to be regulated by different genes that associated to photosynthesis.
Interaction proteins of ZmHOX32 are associated with photosynthesis
To explore the interaction proteins of ZmHOX32, we analyzed the data generated by RLL-Y2H-seq in maize (Yang et al., 2018; Han et al., 2023). We found that ZmHOX32 was likely to interacted to the proteins encoded by the genes of NAC, AP2/ERF, and MYB families (Figure 4A). GO enrichment analysis of these genes showed that they are correlated to hormone signaling, leaf development, and response to light stimulation (Figure 4B). These results implied complicated function of ZmHOX32, which may be related to photosynthesis.
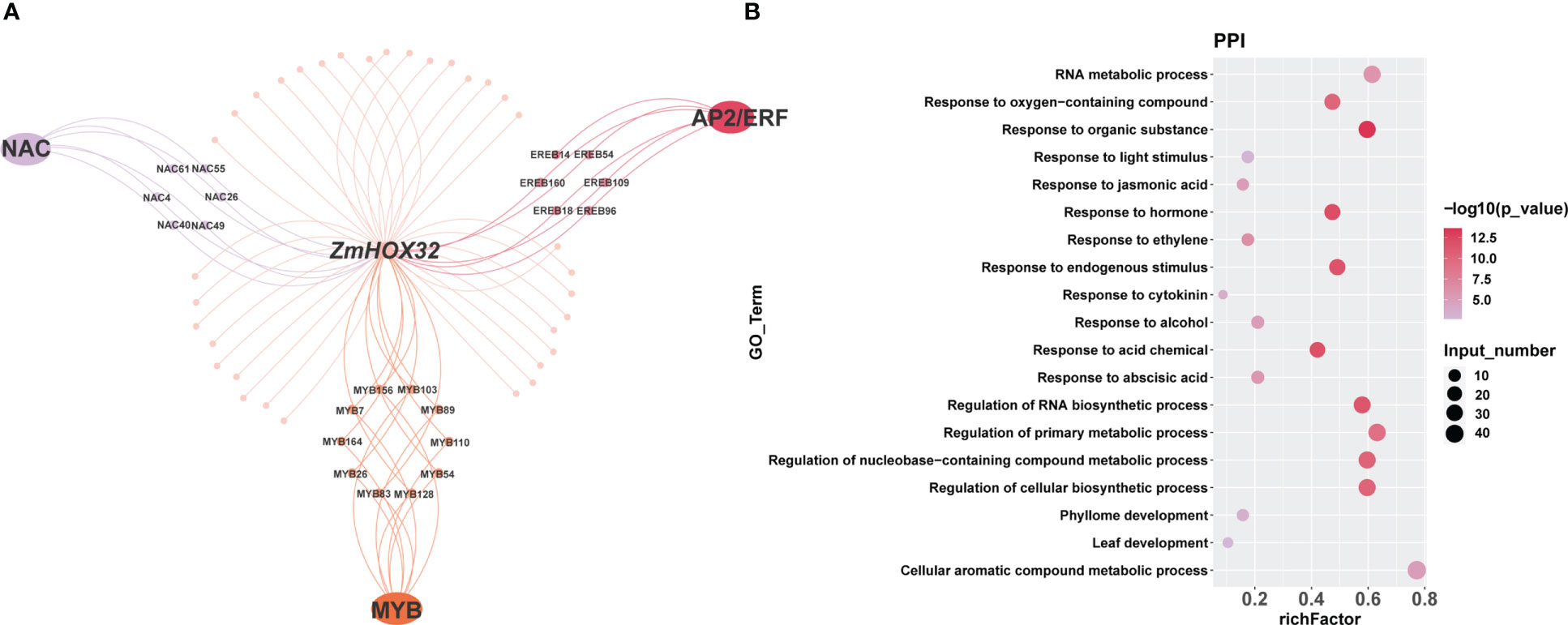
Figure 4 Enrichment of intercalating proteins of ZmHOX32. (A) Regulatory network of the intercalating proteins of ZmHOX32. (B) Degree of enrichment of intercalating proteins of ZmHOX32.
A regulatory network showed the complicated function of ZmHOX32 in participating photosynthesis
To uncover the downstream target genes of ZmHOX32, we performed a tsCUT&Tag assay to investigate the binding sites of ZmHOX32 protein in maize B73 genome. The library and sequences showed good quality. The library fragments ranged from 200 to 650 bp, without primer dimer contamination (Supplementary Figure 1A). Sequencing results showed that the quality values of most bases were above 30, suggesting the good quality of this data (Supplementary Figure 1B). We also performed correlation analyses for the two replicates, which showed high correlations (Pearson Correlation Coefficient = 1, Spearman Correlation Coefficient = 0.98) (Supplementary Figures 1C, D). These results indicate the effective of tsCUT&Tag experiment. Two biological tsCUT&Tag replicates detected 2,262 and 1,903 target genes, respectively. Of these targets, 1,473 target genes were detected by both replicates and considered to be high-confidence targets (Figure 5A). Transcription factors usually bind to cis-acting elements in promoter of targets to determine the transcription of downstream genes (Lambert et al., 2018). So, we scanned the binding sites of ZmHOX32 across the whole genome and uncovered that they were mainly located in promoter 3kb regions of target genes, which accounted for 70.25% and 69.99% of all binding sites in two replicates (Figure 5B). The signal heat map confirmed that the reads of tsCUT&Tag were significantly enriched near the transcription start site (TSS) (Figure 5C). These results demonstrate a genome-wide binding landscape of ZmHOX32, which provides us an unprecedented resource to dissect the function of ZmHOX32 in maize.
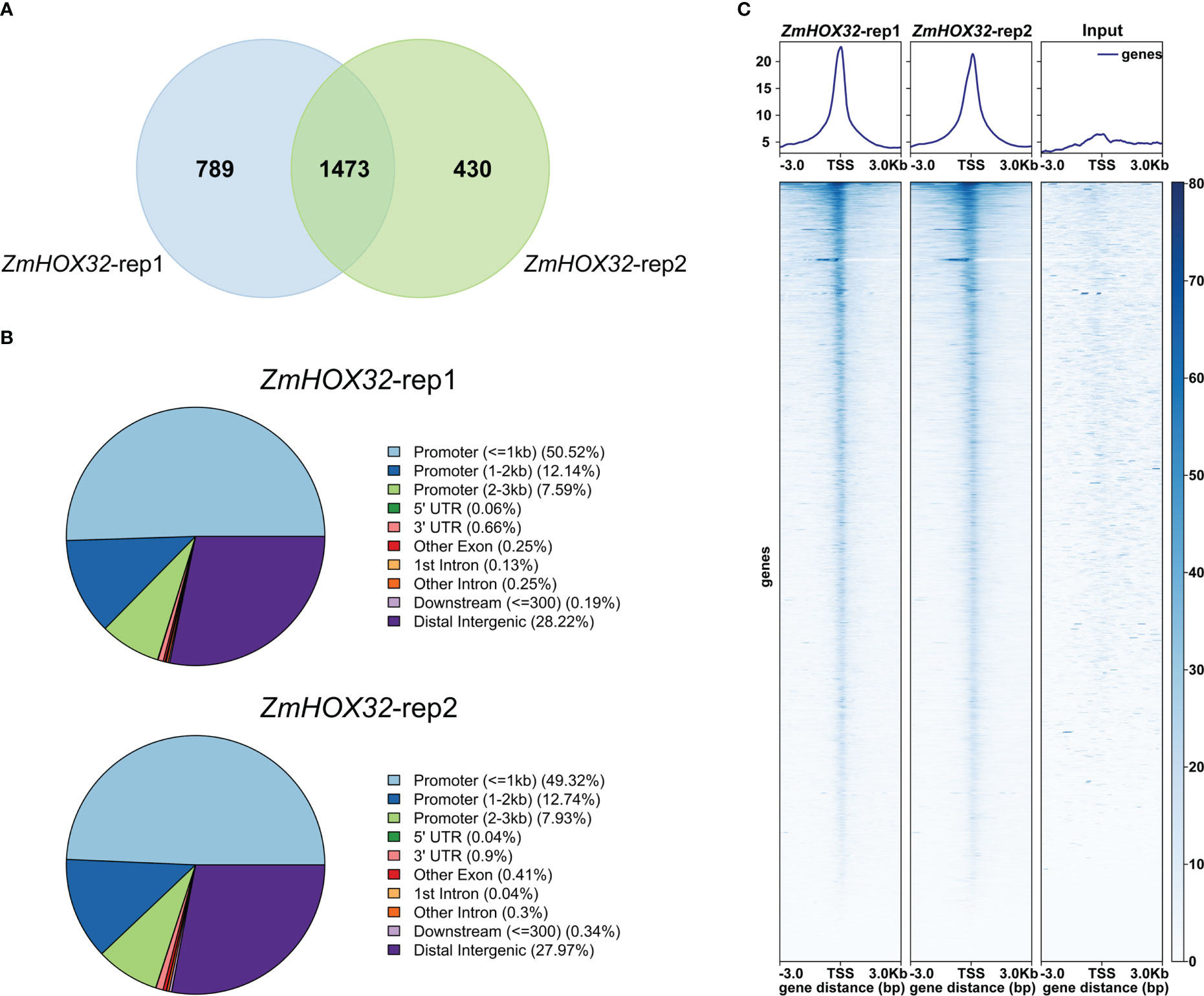
Figure 5 tsCUT&Tag reveals the genome-wide binding position of ZmHOX32. (A) The overlap between the downstream target genes of the two replicates of ZmHOX32. (B) Genome-wide binding site map of ZmHOX32 two repeats. (C) Distribution of ZmHOX32 tsCUT&Tag peaks at 3 Kb upstream and downstream of the gene.
The target genes of ZmHOX32 were significantly enriched in multiple gene families, such as WRKY, AUXIN, AP2/ERF, MYB and so on (Figure 6A), which are mainly related to plant growth, development and signal transduction, as well as photosynthesis in different plants (Chuck et al., 2008; De Boer et al., 2011; Zheng et al., 2018; Dickinson et al., 2020; Feng et al., 2020; Zhang et al., 2021). GO enrichment for these genes also revealed that most of them were involved in hormone signal transduction, development, and response to light stimulation (Figure 6B). These results are in line with the expectation and suggest the complicated functions of ZmHOX32.
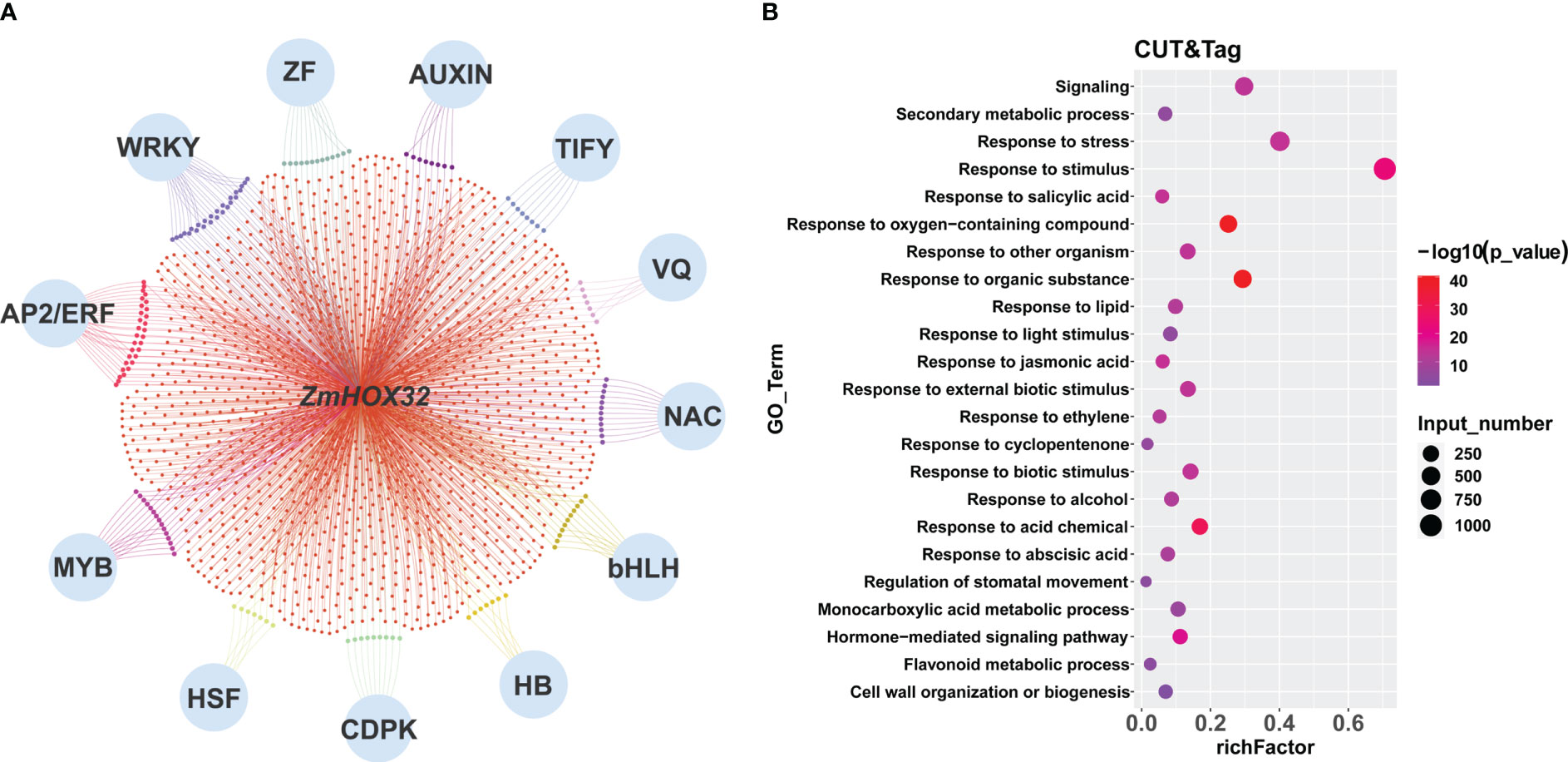
Figure 6 Enrichment of downstream target genes of ZmHOX32 protein. (A) Regulatory network of downstream target genes of HOX32 protein. (B) Go enrichment for downstream target genes of HOX32 protein.
In previous study, the ChIP-seq of 104 TFs were performed in maize protoplasts to explore their potential target genes (Tu et al., 2020). These TFs are specifically expressed in leaf tissues, implying their possible role in light harvesting. Interestingly, we found 20 TFs of these 104 TFs could be targeted by HOX32 protein (Figure 7A), which is significantly higher than random (P value =8.13e-4). All the TFs belonged to the classical families associated with metabolism, signaling, transport, hormone, and cell wall (Tu et al., 2020) (Supplementary Figure 2). Of the targets of these 20 downstream TFs of ZmHOX32, 66 genes are annotated as photosynthesis-related genes that participated in light reaction (Thimm et al., 2004) (Figure 7B; Supplementary Table 3), suggesting a potential function of ZmHOX32 in photosynthesis. In addition, we found three of the 66 photosynthesis-related genes were also directly targeted by the HOX32 protein. These three genes Zm00001d012293 (FD4), Zm00001d011826 (NDHO1) and Zm00001d035185 (CDB1) have been evidenced to play important roles in photosynthesis (Hanke et al., 2005; Chen et al., 2022; Su et al., 2022). Furthermore, three leaf-related transcription factors - Zm00001d050816 (ALF7), Zm00001d033267 (bHLH43), and Zm00001d031044 (bHLH163) were evidenced to directly target to ZmHOX32 in a previous study (Tu et al., 2020) (Figure 7C). It has been shown that the transcription factors bHLH43 and bHLH163 play important roles in the light regulation mechanism of carotenoids and tricarboxylic acid cycle by light, respectively (Eprintsev et al., 2022; Xiang et al., 2022). These results demonstrate a regulatory module of ZmHOX32 that affect photosynthesis in maize.
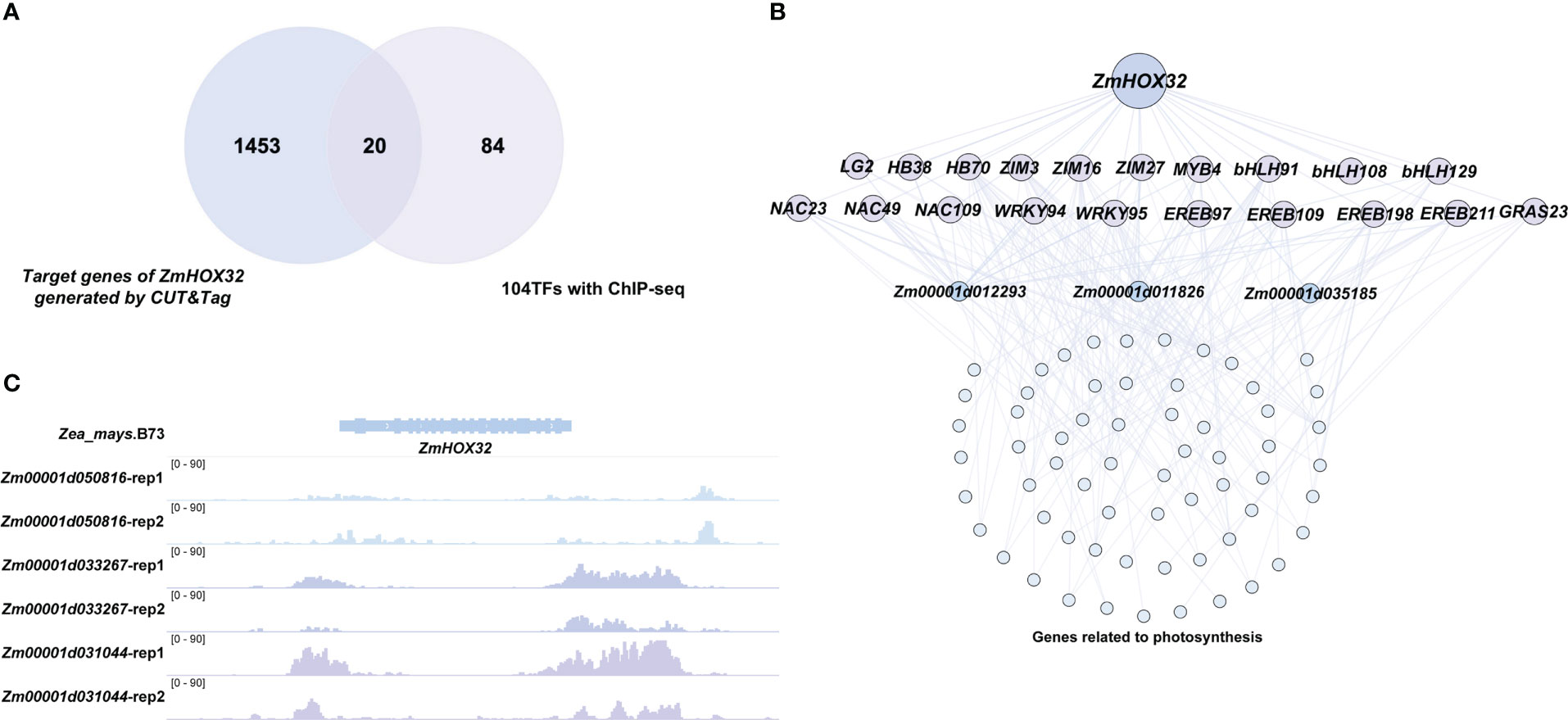
Figure 7 Characterization of the regulatory network of ZmHOX32 using the ChIP-seq data of 104 transcription factors. (A) Overlap of ZmHOX32 downstream target genes with 104 transcription factors. (B) Preliminary construction of the regulatory network of ZmHOX32. (C) Transcriptional binding sites of three ZmHOX32 upstream transcription factors in the promoter region of ZmHOX32.
Association analysis suggest ZmHOX32 is also likely to function in plant architecture of maize
To clarify the functions of ZmHOX32 in regulation of the agronomic traits of maize given the tight relationship between photosynthesis and agronomic traits, we performed the genome-wide association study (GWAS) with general linear model in a diverse association mapping panel of 690 maize inbred lines (Abe et al., 2012; Gui et al., 2020). Multiple agronomic traits of the maize association population had been investigated before. These traits could be classified into two categories with Yield and Plant architecture based on their effects on the maize phenotype. The Yield is composed of 9 traits that related to the kernel size and ear size, while the category Plant architecture contained the traits that associated to tassel branch number, leaf, and flowering time (Figures 8A, B). To reduce dimensionality of these complex traits, the Principal Component Analysis (PCA) were performed for the two main traits categories. The PC1 explained almost of the variation (95.8% and 99.1%) for categories Yield and Plant architecture, respectively (Figures 8A, B), which suggested that the Yield traits and Plant architecture were likely to be determined or represent by the PC1. The values of PC1 and PC2 of two categories all showed the normal distribution in the association population (shapiro.test, P = 1). Therefore, we used the PC1 and PC2 as phenotypic data, and performed a panel of genome-wide association studies (GWAS) by combining 4,069,278 SNPs. The four phenotypic data (Yield-PC1, Yield-PC2, Plant architecture-PC1, Plant architecture-PC2) identified 28, 824, 2671, 452 significant SNPs at the genome-wide level, respectively (Supplementary Figures 3, 4). For ZmHox32, we found three SNPs located on the promoter region were significantly associated to the PC1 of Plant architecture (Figure 8C). The PC2 of two trait types all showed no association with the sequence variation in the population (Figure 8C), which may be associated to their low explained rate of variation (PCA) for the Yield and Plant architecture. The three SNPs that significantly associated to PC1 of Plant architecture could divide the population into three Haplotypes (Haps). The plants of Hap1 (CCC) showed significant higher plant height, higher ear height, larger ear leaf width, larger ear leaf length, larger tassel main axis length, more leaf number above ear, larger ear length, longer silking time, longer pollen shed time, and longer heading date than the plants of Hap3 (GTG) (Supplementary Figure 5). Tassel branch number, ear diameter and cob diameter showed no significant difference among Hap1, Hap2 (CTC) and Hap3 (Supplementary Figure 5). The Hap2 showed highest in most traits in category of Plant architecture, except in ear length, ear diameter and cob diameter. In summary, ZmHOX32 likely to play an important role in regulation of plant architecture.
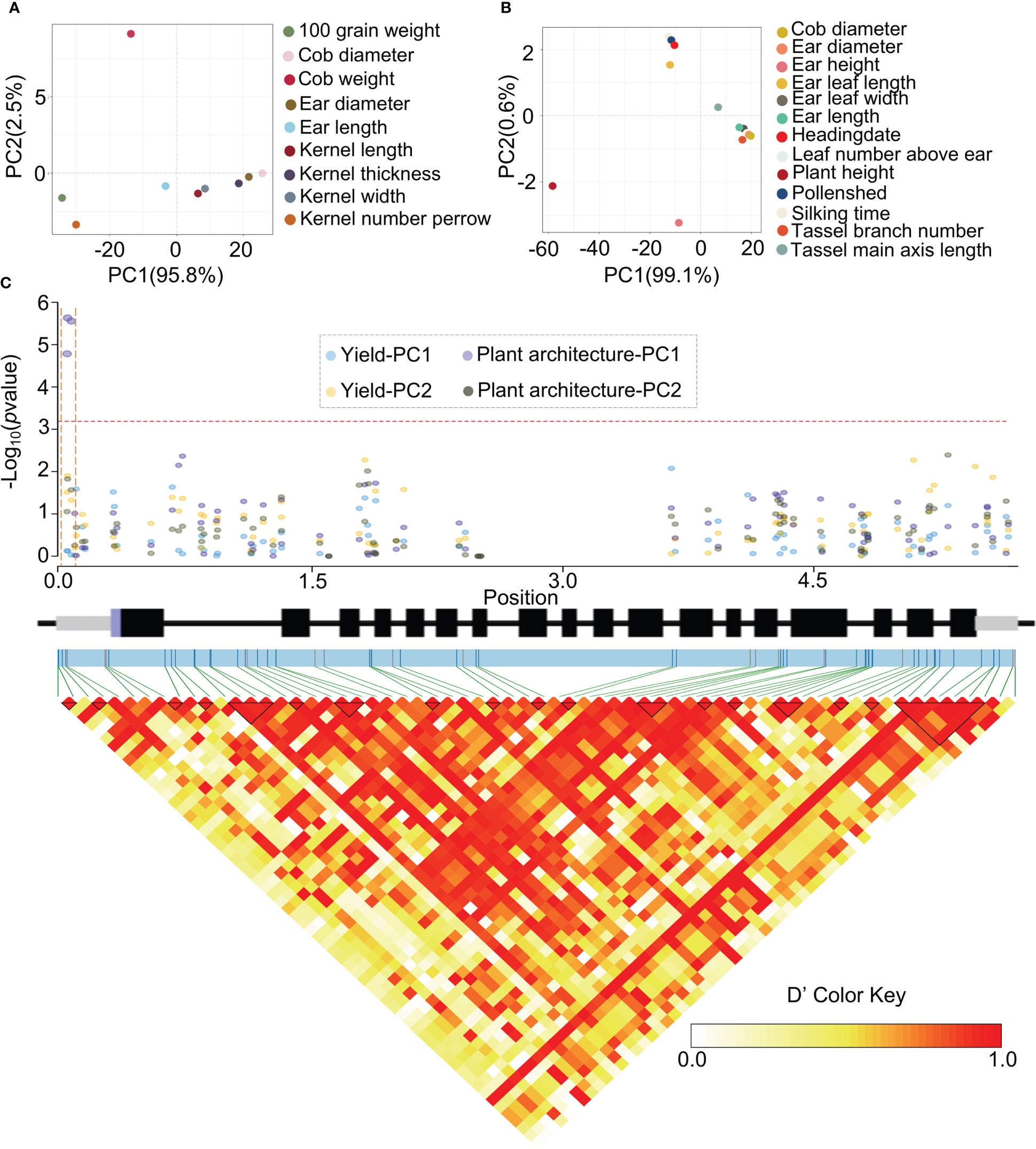
Figure 8 ZmHOX32 association analysis. (A) PCA of maize yield. (B) PCA of plant architecture. (C) Association analysis of Hox32 in four phenotypic data (Yield-pc1, Yield-pc2, Plant architecture-pc1, Plant architecture-pc2).
Discussion
The HD-Zip III family exists in both C3 and C4 crops, plays an important role in regulating various cell differentiation processes in plants, including embryo morphogenesis, meristem formation, lateral organogenesis, lateral organ polarity establishment, and vascular system development, etc. (Prigge et al., 2005). Within this family, the regulatory mechanism of HOX32 influencing leaf morphogenesis has been studied in the C3 crops (rice and Arabidopsis) (Emery et al., 2003; Li et al., 2016). However, the function of HOX32 in C4 crop maize has not been studied. In present study, we focused on ZmHOX32 in maize and found its high conservation in plants. As expected, ZmHOX32 as one TF showed specific expression in nucleus, and constitutive expression in several tissues or organs, including the leaves at different growth stages or different leaf parts. Multiple cis-acting elements related to light-responsive were identified in promoter of ZmHOX32, suggesting a potential role in photosynthesis. The functions and the regulatory networks constructed for the interacting proteins and target genes of HOX32 also suggest a potential role in photosynthesis.
It has been studied that HOX32 is involved in the regulation of leaf morphogenesis and thus affects photosynthesis. In Arabidopsis, the homologous gene PHV was expressed in the abaxial side of cotyledons and center of the protovascular bundle (McConnell et al., 2001; Emery et al., 2003). Mutation of PHV caused changes in leaf polarity and the development of shoot and root apical meristems. Alteration in leaf morphology may indirectly affect photosynthesis efficiency. Overexpression of OsHOX32 resulted in a variety of phenotype changes, including narrow adaxially curled leaves, reduced leaf angle, erect plant type, dwarf plants, and reduced chlorophyll levels, which all affected photosynthetic efficiency (Li et al., 2016). OsHOX32 is the main target of microRNA166, the regulatory module may be involved in regulation of cell wall formation and vascular tissue development (Zhang et al., 2018b). The OsHOX32 protein could directly bind to the promoters of cinnamyl alcohol dehydrogenase (CAD) gene and cellulose synthase (CESA) gene, repressing their expression level and affecting leaf shape (Chen et al., 2021). In this study, to explore the regulatory pathway of ZmHOX32 functioned on photosynthesis, tsCUT&Tag, an upgrade of ChIP-seq, was employed to investigate the downstream genes of ZmHOX32. The downstream genes of ZmHOX32 were enriched in multiple gene families, such as WRKY, AUXIN, AP2/ERF, MYB, NAC, HB and VQ, which are related to leaf structure, signal transduction, and photosynthesis directly or indirectly. For examples, the inhibition of OsSWNs (encoding one NAC domain protein) expression in rice results in leaf drooping and reduced plant height (Yoshida et al., 2013). The mutation of WUSCHEL-related homeobox1 (WOX1) caused significant developmental defects in mid-lateral axis polarity and narrowed leaf width during leaf morphogenesis (Wang et al., 2020). In Arabidopsis, AtTIFY4a and AtTIFY4b regulate leaf development by affecting leaf size and leaf edge curvature (White, 2006). The drought tolerance of JAZ7 can be induced by modulating photosynthesis, redox, amino acids, phytohormones, and defense metabolites in plants (Meng et al., 2019). We also focused on the genes upstream of ZmHOX32, and identified three transcription factors that function in the leaf. Previous studies showed that the transcription factor bHLH43 was related to the light regulation mechanism of carotenoids, and bHLH163 participated in the regulation of the tricarboxylic acid cycle by light (Eprintsev et al., 2022; Xiang et al., 2022). These potential effects on leaf development of the targets suggesting potential function of ZmHOX32 in photosynthesis.
Protein-protein interaction study demonstrated that NAC, AP2/ERF, and MYB genes would interact with ZmHOX32 as co-factors. Current studies have found that the genes of the NAC family play important roles in different physiological developmental processes in plants, including stem tip meristem formation, leaf senescence, secondary cell wall formation, and hormone signaling (Grant et al., 2010; Yoshida et al., 2013; Ren et al., 2018; Zhang et al., 2018a). AP2/ERF family transcription factors regulate a variety of biological processes such as growth, development and differentiation, hormone signaling, and metabolic synthesis in different tissues (Feng et al., 2020). In the study of MYB family, it has been shown that AtMYB76 regulates photosynthesis (Duan et al., 2017).
Here, we explored the upstream regulators, co-factors, and downstream targets of ZmHOX32, which for the first time construct the cascading regulatory network of ZmHOX32 in maize. Based on such regulatory network, we uncovered functional roles of ZmHOX32 in photosynthesis and plant architecture of maize.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject: PRJNA909077 Accession: SRR22580376· SRR22580377· SRR22580378· SRR22580379.
Author contributions
LL, XM, and WZ designed the project. XM, WZ, QJ, and ZS performed the experiments, data analysis, and collection of electronic resources. LL supported the work financially and participated in its planning. XM and WZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The research was funded by the National Natural Science Foundation of China (grant number: 32261143463), Hainan Yazhou Bay Seed Lab (grant number: B21HJ8102), the major Program of Hubei Hongshan laboratory (grant number: 2021hszd008), Huazhong Agricultural University Scientific & Technological Self-innovation Foundation (grant number: 2021ZKPY001), HZAU-AGIS Cooperation Fund (grant number: SZYJY2021006), National Natural Science Foundation of China (grant number: 32272158) , which were applied for from Dr. Lin Li. This research was also funded by the China Postdoctoral Science Foundation (grant number: 2022M721282), which was applied for from Dr. Zhu Wanchao.
Acknowledgments
The authors wish to thank the High-throughput Computing Platform of National Key Laboratory of Crop Genetic Improvement.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1119678/full#supplementary-material
Footnotes
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ http://www.ncbi.nlm.nih.gov/cdd/
- ^ http://systemsbiology.cau.edu.cn/agriGOv2/
References
Abe, A., Kosugi, S., Yoshida, K., Natsume, S., Takagi, H., Kanzaki, H., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30 (2), 174–178. doi: 10.1038/nbt.2095
Assefa, Y., Carter, P., Hinds, M., Bhalla, G., Schon, R., Jeschke, M., et al. (2018). Analysis of long term study indicates both agronomic optimal plant density and increase maize yield per plant contributed to yield gain. Sci. Rep. 8 (1)4937. doi: 10.1038/s41598-018-23362-x
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23 (19), 2633–2635. doi: 10.1093/bioinformatics/btm308
Chen, H., Fang, R., Deng, R., Li, J. (2021). The OsmiRNA166b-OsHox32 pair regulates mechanical strength of rice plants by modulating cell wall biosynthesis. Plant Biotechnol. J. 19 (7), 1468–1480. doi: 10.1111/pbi.13565
Chen, W., Huang, J., Chen, S., Zhang, L., Rochaix, J.-D., Peng, L., et al. (2022). Stromal protein chloroplast development and Biogenesis1 is essential for chloroplast development and biogenesis in arabidopsis thaliana. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.815859
Chuck, G., Meeley, R., Hake, S. (2008). Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135 (18), 3013–3019. doi: 10.1242/dev.024273
Dai, X., Tu, X., Du, B., Dong, P., Sun, S., Wang, X., et al. (2022). Chromatin and regulatory differentiation between bundle sheath and mesophyll cells in maize. PlJ 109 (3), 675–692. doi: 10.1111/tpj.15586
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27 (15), 2156–2158. doi: 10.1093/bioinformatics/btr330
De Boer, K., Tilleman, S., Pauwels, L., Vanden Bossche, R., De Sutter, V., Vanderhaeghen, R., et al. (2011). APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis: ERFs and bHLHs control nicotine biosynthesis. PlJ 66 (6), 1053–1065. doi: 10.1111/j.1365-313X.2011.04566.x
Dickinson, P. J., Kneřová, J., Szecówka, M., Stevenson, S. R., Burgess, S. J., Mulvey, H., et al. (2020). A bipartite transcription factor module controlling expression in the bundle sheath of arabidopsis thaliana. Nat. Plants 6 (12), 1468–1479. doi: 10.1038/s41477-020-00805-w
Duan, S., Jin, C., Li, D., Gao, C., Qi, S., Liu, K., et al. (2017). MYB76 inhibits seed fatty acid accumulation in arabidopsis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00226
Emery, J. F., Floyd, S. K., Alvarez, J., Eshed, Y., Hawker, N. P., Izhaki, A., et al. (2003). Radial patterning of arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13 (20), 1768–1774. doi: 10.1016/j.cub.2003.09.035
Eprintsev, A. T., Fedorin, D. N., Igamberdiev, A. U. (2022). Light dependent changes in adenylate methylation of the promoter of the mitochondrial citrate synthase gene in maize (Zea mays l.) leaves. Int. J. Mol. Sci. 23 (21), 13495. doi: 10.3390/ijms232113495
Feng, K., Hou, X.-L., Xing, G.-M., Liu, J.-X., Duan, A.-Q., Xu, Z.-S., et al. (2020). Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 40 (6), 750–776. doi: 10.1080/07388551.2020.1768509
Feng, J., Liu, T., Qin, B., Zhang, Y., Liu, X. S. (2012). Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7 (9), 1728–1740. doi: 10.1038/nprot.2012.101
Grant, E. H., Fujino, T., Beers, E. P., Brunner, A. M. (2010). Characterization of NAC domain transcription factors implicated in control of vascular cell differentiation in arabidopsis and populus. Planta 232 (2), 337–352. doi: 10.1007/s00425-010-1181-2
Gui, S., Yang, L., Li, J., Luo, J., Xu, X., Yuan, J., et al. (2020). ZEAMAP, a comprehensive database adapted to the maize multi-omics era. iScience 23 (6), 101241. doi: 10.1016/j.isci.2020.101241
Han, L., Zhong, W., Qian, J., Jin, M., Tian, P., Zhu, W., et al. (2023). A multi-omics integrative network map of maize. Nat. Genet. 55 (1), 144–153. doi: 10.1038/s41588-022-01262-1
Hanke, G. T., Okutani, S., Satomi, Y., Takao, T., Suzuki, A., Hase, T. (2005). Multiple iso-proteins of FNR in arabidopsis: evidence for different contributions to chloroplast function and nitrogen assimilation. Plant Cell Environ. 28 (9), 1146–1157. doi: 10.1111/j.1365-3040.2005.01352.x
Kumar, S., Tamura, K., Nei, M. (1994). MEGA: Molecular evolutionary genetics analysis software for microcomputers. Bioinformatics 10 (2), 189–191. doi: 10.1093/bioinformatics/10.2.189
Lambert, S. A., Jolma, A., Campitelli, L. F., Das, P. K., Yin, Y., Albu, M., et al. (2018). The human transcription factors. Cell 172 (4), 650–665. doi: 10.1016/j.cell.2018.01.029
Langmead, B., Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9 (4), 357–359. doi: 10.1038/nmeth.1923
Li, Y.-y., Shen, A., Xiong, W., Sun, Q.-L., Luo, Q., Song, T., et al. (2016). Overexpression of OsHox32 results in pleiotropic effects on plant type architecture and leaf development in rice. Rice 9 (1), 46. doi: 10.1186/s12284-016-0118-1
Lipka, A. E., Tian, F., Wang, Q., Peiffer, J., Li, M., Bradbury, P. J., et al. (2012). GAPIT: genome association and prediction integrated tool. Bioinformatics 28 (18), 2397–2399. doi: 10.1093/bioinformatics/bts444
Marchler-Bauer, A. (2002). CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 30 (1), 281–283. doi: 10.1093/nar/30.1.281
McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J., Barton, M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 (6838), 709–713. doi: 10.1038/35079635
Meng, L., Zhang, T., Geng, S., Scott, P. B., Li, H., Chen, S. (2019). Comparative proteomics and metabolomics of JAZ7-mediated drought tolerance in arabidopsis. J. Proteomics 196, 81–91. doi: 10.1016/j.jprot.2019.02.001
Moon, J., Hake, S. (2011). How a leaf gets its shape. Curr. Opin. Plant Biol. 14 (1), 24–30. doi: 10.1016/j.pbi.2010.08.012
Mukherjee, K., Bürglin, T. R. (2006). MEKHLA, a novel domain with similarity to PAS domains, is fused to plant homeodomain-leucine zipper III proteins. Plant Physiol. 140 (4), 1142–1150. doi: 10.1104/pp.105.073833
Palena, C. M., Tron, A. E., Bertoncini, C. W., Gonzalez, D. H., Chan, R. L. (2001). Positively charged residues at the n-terminal arm of the homeodomain are required for efficient DNA binding by homeodomain-leucine zipper proteins11Edited by m. yaniv. J. Mol. Biol. 308 (1), 39–47. doi: 10.1006/jmbi.2001.4563
Ponting, C. P., Aravind, L. (1999). START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem. Sci. 24 (4), 130–132. doi: 10.1016/S0968-0004(99)01362-6
Prigge, M. J., Otsuga, D., Alonso, J. M., Ecker, J. R., Drews, G. N., Clark, S. E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in arabidopsis development. Plant Cell 17 (1), 61–76. doi: 10.1105/tpc.104.026161
Quinlan, A. R., Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26 (6), 841–842. doi: 10.1093/bioinformatics/btq033
Ren, T., Wang, J., Zhao, M., Gong, X., Wang, S., Wang, G., et al. (2018). Involvement of NAC transcription factor SiNAC1 in a positive feedback loop via ABA biosynthesis and leaf senescence in foxtail millet. Planta 247 (1), 53-68. doi: 10.1007/s00425-017-2770-0
Rombauts, S., Dehais, P., Van Montagu, M., Rouze, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27 (1), 295–296. doi: 10.1093/nar/27.1.295
Salas Fernandez, M. G., Becraft, P. W., Yin, Y., Lübberstedt, T. (2009). From dwarves to giants? plant height manipulation for biomass yield. Trends Plant Sci. 14 (8), 454–461. doi: 10.1016/j.tplants.2009.06.005
Schrick, K., Nguyen, D., Karlowski, W. M., Mayer, K. F. (2004). START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 5 (6), R41. doi: 10.1186/gb-2004-5-6-r41
Su, X., Cao, D., Pan, X., Shi, L., Liu, Z., Dall’Osto, L., et al. (2022). Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from arabidopsis thaliana. Mol. Plant 15 (3), 454–467. doi: 10.1016/j.molp.2022.01.020
Thimm, O., Bläsing, O., Gibon, Y., Nagel, A., Meyer, S., Krüger, P., et al. (2004). Mapman: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. PlJ 37 (6), 914–939. doi: 10.1111/j.1365-313X.2004.02016.x
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., et al. (2017). agriGO v2.0: a GO analysis toolkit for the agricultural communitUpdate. Nucleic Acids Res. 45 (W1), W122–W129. doi: 10.1093/nar/gkx382
Tu, X., Mejía-Guerra, M. K., Valdes Franco, J. A., Tzeng, D., Chu, P.-Y., Shen, W., et al. (2020). Reconstructing the maize leaf regulatory network using ChIP-seq data of 104 transcription factors. Nat. Commun. 11 (1), 5089. doi: 10.1038/s41467-020-18832-8
Wang, H., Niu, H., Li, C., Shen, G., Liu, X., Weng, Y., et al. (2020). WUSCHEL-related homeobox1 (WOX1) regulates vein patterning and leaf size in cucumis sativus. Horticul. Res. 7 (1), 182. doi: 10.1038/s41438-020-00404-y
White, D. W. R. (2006). PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. 103 (35), 13238–13243. doi: 10.1073/pnas.0604349103
Wu, L., Luo, Z., Shi, Y., Jiang, Y., Li, R., Miao, X., et al. (2022). A cost-effective tsCUT&Tag method for profiling transcription factor binding landscape. J. Integr. Plant Biol. 64 (11), 2033–2038. doi: 10.1111/jipb.13354
Xiang, N., Zhao, Y., Wang, S., Guo, X. (2022). The modulation of light quality on carotenoids in maize (Zea mays l.) sprouts. Food Chem: Mol. Sci. 5, 100128. doi: 10.1016/j.fochms.2022.100128
Yang, F., Lei, Y., Zhou, M., Yao, Q., Han, Y., Wu, X., et al. (2018). Development and application of a recombination-based library versus library high- throughput yeast two-hybrid (RLL-Y2H) screening system. Nucleic Acids Res. 46 (3), e17–e17. doi: 10.1093/nar/gkx1173
Yoo, S.-D., Cho, Y.-H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2 (7), 1565–1572. doi: 10.1038/nprot.2007.199
Yoshida, K., Sakamoto, S., Kawai, T., Kobayashi, Y., Sato, K., Ichinose, Y., et al. (2013). Engineering the oryza sativa cell wall with rice NAC transcription factors regulating secondary wall formation. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00383
Yu, G., Wang, L.-G., He, Q.-Y. (2015). ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31 (14), 2382–2383. doi: 10.1093/bioinformatics/btv145
Zhang, J., Huang, G., Zou, D., Yan, J., Li, Y., Hu, S., et al. (2018a). The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol. 217 (2), 625–640. doi: 10.1111/nph.14864
Zhang, H., Yang, X., Lu, J., Song, F., Sun, J., Wang, C., et al. (2021). OsIAA20, an Aux/IAA protein, mediates abiotic stress tolerance in rice through an ABA pathway. Plant Sci. 308, 110903. doi: 10.1016/j.plantsci.2021.110903
Zhang, J., Zhang, H., Srivastava, A. K., Pan, Y., Bai, J., Fang, J., et al. (2018b). Knockdown of rice MicroRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development. Plant Physiol. 176 (3), 2082–2094. doi: 10.1104/pp.17.01432
Zheng, X., Zhao, Y., Shan, D., Shi, K., Wang, L., Li, Q., et al. (2018). MdWRKY9 overexpression confers intensive dwarfing in the M26 rootstock of apple by directly inhibiting brassinosteroid synthetase MdDWF4 expression. New Phytol. 217 (3), 1086–1098. doi: 10.1111/nph.14891
Zhou, C., Zhu, C., Fu, H., Li, X., Chen, L., Lin, Y., et al. (2019). Genome-wide investigation of superoxide dismutase (SOD) gene family and their regulatory miRNAs reveal the involvement in abiotic stress and hormone response in tea plant (Camellia sinensis). PloS One 14 (10), e0223609. doi: 10.1371/journal.pone.0223609
Keywords: maize, ZmHOX32, leaf, photosynthesis, tsCUT&Tag
Citation: Miao X, Zhu W, Jin Q, Song Z and Li L (2023) ZmHOX32 is related to photosynthesis and likely functions in plant architecture of maize. Front. Plant Sci. 14:1119678. doi: 10.3389/fpls.2023.1119678
Received: 09 December 2022; Accepted: 09 March 2023;
Published: 22 March 2023.
Edited by:
Hao Tong, University of Potsdam, GermanyReviewed by:
Haiming Zhao, China Agricultural University, ChinaChuang Ma, Northwest A&F University, China
Copyright © 2023 Miao, Zhu, Jin, Song and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Li, aHphdWxpbGluQG1haWwuaHphdS5lZHUuY24=
†These authors have contributed equally to this work
 Xinxin Miao
Xinxin Miao Wanchao Zhu
Wanchao Zhu Qixiao Jin
Qixiao Jin Zemeng Song1,2
Zemeng Song1,2 Lin Li
Lin Li