
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 February 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1119162
This article is part of the Research Topic Advanced Breeding for Abiotic Stress Tolerance in Crops View all 21 articles
A correction has been applied to this article in:
Frontiers Corrigendum Template for Authors
Drought is a major abiotic stress that reduces crop yields and quality worldwide. Although some genes involved in the response to drought stress have been identified, a more in-depth understanding of the mechanisms underlying wheat tolerance to drought is needed for the control of drought tolerance. Here, we evaluated the drought tolerance of 15 wheat cultivars and measured their physiological–biochemical parameters. Our data showed that the drought tolerance of the resistant wheat cultivars was significantly higher than that of drought-sensitive cultivars, which was associated with a greater antioxidant capacity of the former. Transcriptomic analysis revealed that different mechanisms of drought tolerance exist between the wheat cultivars Ziyou 5 and Liangxing 66. Transcriptomic analysis also revealed a large number of DEGs, including those involved in flavonoid biosynthesis, phytohormone signalling, phenolamides and antioxidants. qRT–PCR was performed, and the results showed that the expression levels of TaPRX-2A were significantly different among the various wheat cultivars under drought stress. Further study revealed that overexpression of TaPRX-2A enhanced tolerance to drought stress through the maintenance of increased antioxidase activities and reductions in ROS contents. Overexpression of TaPRX-2A also increased the expression levels of stress-related genes and ABA-related genes. Taken together, our findings show that flavonoids, phytohormones, phenolamides and antioxidants are involved in the plant response to drought stress and that TaPRX-2A is a positive regulator of this response. Our study provides insights into tolerance mechanisms and highlights the potential of TaPRX-2A overexpression in enhancing drought tolerance in crop improvement programmes.
Environmental stresses severely impact plant growth, development and productivity. Drought is a major abiotic stress that reduces crop yields and quality worldwide and is occurring more frequently due to climate change and water scarcity (Mahajan and Tuteja, 2005; Zampieri et al., 2017). When persistently exposed to drought stress, plants can experience multiple types of damage (including oxidative injury) (Joshi et al., 2016). Reactive oxygen species (ROS), including superoxide radicals (), hydrogen peroxide (H2O2), and hydroxyl radicals (OH−), constitute the primary cause of oxidative damage, which damages cells by destroying membrane lipids (McCord, 2000). To counter environmental stresses, plants have evolved sophisticated molecular mechanisms, such as ROS scavenging, phytohormone signalling pathway (those of abscisic acid (ABA)) activation, and secondary metabolism (including that involving flavonoids, melatonin, and secoisolariciresinol) (Campo et al., 2014; Zhu, 2016; Meng et al., 2021; Feng et al., 2022; Song et al., 2022). Moreover, the antioxidant system plays important roles in reducing ROS levels by the action of several endogenous antioxidant enzymes (catalase (CAT), ascorbate peroxidase (APX), and superoxide dismutase (SOD)) (Mittler, 2002). In addition, by catalysing redox reactions, peroxidases (PRXs) protect cells from ROS damage (Wood et al., 2003).
PRXs are antioxidant enzymes that are widely distributed in living organisms, including plants, microorganisms, and animals. The members of the PRX superfamily are categorized into haem (animal and nonanimal) and nonheme PRXs based on their structure (Welinder, 1992; Taurog, 1999). In plants, PRXs can be divided into three classes (class I, class II, and class III) based on their functions. Class I PRXs include APXs, class II subgroups include lignin PRXs, and class III subgroups (guaiacol PRXs, EC1.11.17) include secretory PRXs that are plant specific (Bhatt and Tripathi, 2011). Currently, a large number of class III PRX families have been identified as multigenic families in plants. For example, there are 155 members of the class III PRX gene family in rice. Arabidopsis thaliana class III PRXs comprise 75 isoenzymes, and Brachypodium distachyon class III PRXs comprise 151 isoenzymes (Fawal et al., 2013). Maize class III PRXs comprise 119 members (Wang et al., 2015). In addition, 374, 159 and 169 PRX members have been identified in Triticum aestivum, Triticum urartu and Aegilops tauschii, respectively (Yan et al., 2019).
Class III subgroups comprise large multigene families in the plant kingdom, and these members are involved in various biophysiological processes, such as lignin biosynthesis, cell wall hardening, defence against pathogens, H2O2 removal, and tolerance to abiotic stresses (Cosio and Dunand, 2009; Wang et al., 2015). In plants, class III PRXs can oxidize phenolic compounds, thereby reducing the level of H2O2, which makes these enzymes efficient components of the antioxidant system against stresses. The function of members of the class III PRX gene family has been widely studied and demonstrated. In A. thaliana, AtPrx71 is involved in cell growth, cell wall damage, and inhibition of cell expansion through accumulation of H2O2 (Raggi et al., 2015). At low temperature, the class III PRXs PRX62 and PRX69 promote root hair growth in Arabidopsis by modulating ROS homeostasis (Pacheco et al., 2022). It was reported that wheat PRX-1 is a specific wheat allergen that causes grass pollen-related wheat allergies (Ogino et al., 2021). Additionally, some class III PRXs respond to biotic and abiotic stresses. For example, T. aestivum PRX111, PRX112 and PRX113 respond to nematode infection (Simonetti et al., 2009). The French bean (Phaseolus vulgaris) PRX gene (FBP1) can cause oxidative burst impairments to increase Arabidopsis susceptibility to fungi and bacteria through the downregulation of AtPrx33 and AtPrx34 transcription (Daudi et al., 2012). In tomato, APX is phosphorylated by the protein kinase CPK28, which enhances thermotolerance (Hu et al., 2021). OsPrx30 transcription is modulated by the AT-hook protein OsATH1, and its overexpression enhances rice susceptibility to rice bacterial blight through the maintenance of increased PRX activity and reductions of H2O2 contents (Liu et al., 2021). Overexpression of AtPrx64 was shown to improve tolerance to aluminium stress through a reduction in ROS accumulation and increase in lignin content (Wu et al., 2017), and by mediating the IbBBX24-IbTOE3-IbPRX17 module, IbPRX17 overexpression was shown to enhance abiotic stress tolerance (Zhang et al., 2022). Some studies have demonstrated that wheat glutathione peroxidase (GPXs) can improve salt tolerance in transgenic Arabidopsis (Zhai et al., 2013). Our previous study showed that, by scavenging ROS and regulating stress-related genes, TaPRX-2A can improve wheat salt tolerance (Su et al., 2020).
Wheat, an important commercial crop species, yields are often restricted by abiotic stresses. In this study, we evaluated the drought tolerance of various wheat cultivars and performed a transcriptomic analysis between Ziyou 5 (a drought-resistant line) and Liangxing 66 (a drought-sensitive line). Our results showed that drought stress altered several metabolic pathways, including those involving flavonoids, phytohormones, phenolamides and antioxidants. Furthermore, we functionally characterized the class III PRX gene TaPRX-2A. Overexpression of TaPRX-2A enhanced the tolerance to drought stress of the transgenic lines, which occurred through the scavenging of ROS and increase in stress-responsive gene transcription. This work provides researchers with new insights into the class III PRXs molecular mechanisms underlying drought stress tolerance of wheat.
“Nonglin20”, “Shanrong 3”, “Jimai 22”, “Liangxing 66”, “Xiaoyan 6”, “Haomai 1”, “Ailiduo”, “Pubing 51”, “Qunzhong 10”, “Liangmai 2”, “LS117TL”, “Ziyou 5”, “Baokemai 1330”, “NSA98-1018” and “Pubing 9946” wheat (T. aestivum) seedlings were used in this study and obtained from Shandong Agricultural University. The seedlings were grown at 25°C under a photoperiod of 16/8 hours (h). The seedlings of the various wheat (T. aestivum) cultivars were subjected to drought stress when the plants produced one leaf and one heart. Concerning the drought stress treatment, the seedlings were not watered for 21 days (d) and then rewatered for 7 days.
Ziyou 5 and Liangxing 66 wheat (T. aestivum) seedlings were treated with 20% PEG6000, and the leaves were harvested at 48 h after treatment. The RNA of the samples was extracted using an RNAprep Pure Plant Kit (TIANGEN) and sent to Metware Corporation (Wuhan) for RNA sequencing (RNA-seq), which was performed using an Illumina HiSeq™ 2000 instrument. The clean RNA-seq reads were subsequently mapped to the wheat reference genome (http://plants.ensembl.org/Triticum_aestivum/Info/Index). Then, the transcripts were assembled, and the differentially expressed genes (DEGs) were analyzed using the DEGseq R package with the default parameters (|log2(fold-change)| >= 1, false discovery rate (FDR) < 0.05) (Mortazavi et al., 2008).
We cloned the TaPRX-2A gene from the wheat cultivar Sumai 3. The wheat leaves were harvested from three-week-old plants and the RNA was extracted with TRIzol reagent (TransGen). The open reading frame (ORF) of TaPRX-2A (Ensembl ID: TraesCS2A02G573900) was obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/). Specific primers were designed according to the TaPRX-2A sequence, and the primer sequences are listed in Table S3. Subsequently, the TaPRX-2A clone was ligated into the overexpression (OE) vector PC186 (pUbi::GWOE::Nos), after which the vector was transformed into the wheat KN199 background using particle gun-mediated gene transformation (Yao et al., 2006).
The leaves of various wheat cultivars were collected under control and drought stress treatment after 21 days. The all samples were used to measure physiological parameters related to drought stress, such as relative water content (RWC), malondialdehyde (MDA), ROS, and antioxidant enzymes. The following formula was used to measure the leaf RWC: RWC = (FW − DW)/(TW − DW) × 100% (FW, fresh weight; DW, dry weight; TW, turgid fresh weight) (Zhou et al., 2014). We used the thiobarbituric acid method to measure the MDA content (Heath and Packer, 1968), and we used the ninhydrin reaction method to measure the proline content and the anthrone method to measure the total soluble sugar content (Spiro, 1966; Bates et al., 1973). The ROS (H2O2, ) levels were visualized using 3,3’-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) staining (Gay et al., 1999; Tian et al., 2013). The activity of antioxidant enzymes (SOD, CAT, PRX) was measured using a previously described method (Chance and Maehly, 1955; Dhindsa et al., 1981; Aebi, 1984).
The RNA of leaves of various wheat cultivars, namely, “Shanrong 3”, “Liangxing 66”, “Xiaoyan 6”, “Ailiduo”, “Ziyou 5”, “Baokemai 1330”, “Shannong 41”, “JM22”, “Guomai 115”, “Taimai 198”, “Huaimai 33”, “NSA98-1018”, and “Pubing 9946”, TaPRX-2A transgenic lines and wild-type (WT) plants was extracted with TRIzol reagent (TransGen). Then, the mRNAs were reverse transcribed into first-strand cDNAs. The expression patterns of stress-related genes were determined using qRT–PCR (Roche LightCycler® 480 system). The 18S rRNA gene of wheat was used as an endogenous control, and the relative expression of the stress-related genes was calculated using the 2–ΔΔCT method. All the qRT–PCR primers used in this study are listed in Table S3.
To comprehensively evaluate the drought tolerance of the different wheat cultivars, we assessed the drought tolerance of 15 different wheat cultivars. The seedlings of all the different wheat cultivars were subjected to drought stress for 21 days. Then, we measured their physiological–biochemical indices. The results showed that different wheat cultivars had different drought tolerances. Some wheat cultivars (Liangmai 2, LS117TL, Ziyou 5, Baokemai 1330, and NSA98-1018) were drought-tolerant and had high survival rates, while others (Nonglin 20, Shanrong 3, Jimai 22, Liangxing 66, Xiaoyan 6, Haomai 1 and Ailiduo) were drought-sensitive and had low survival rates. Similarly, several wheat cultivars (Qunzhong 10, Pubing 51, and Pubing 9946) were moderately drought tolerant. After drought stress for 21 days, the leaves of the drought-tolerant cultivars were slightly wilted compared with those of the drought-sensitive cultivars. The phenotypic changes and survival rates of different the wheat cultivars under both the control and the drought stress treatments were recorded (Figure S1).
ROS constitute the main cause of oxidative damage, which is associated with the physiological response to abiotic stresses in plants. To gain further insight into the differences in drought tolerance between different wheat cultivars, we measured several important physiological–biochemical properties associated with oxidation resistance. First, we measured the levels of ROS accumulation and the activities of antioxidant enzymes under drought stress. Under drought stress, we observed that the O2- and H2O2 contents in the drought-tolerant wheat cultivars (Qunzhong 10, Liangmai 2, LS117TL, and Ziyou 5) were significantly lower than those in the drought-sensitive cultivars (Nonglin 20, Shanrong 3, Jimai 22, Liangxing 66, and Ailiduo) (Figures 1A, B). The activities of antioxidant enzymes (PRX, CAT) in the different wheat cultivars were subsequently measured. The results showed that the activities of PRX and CAT were higher in the drought-tolerant and drought-sensitive wheat cultivars under drought stress than in the controls. Specifically, the PRX activity in the drought-tolerant wheat cultivar Baokemai 1330 was significantly higher than that in the drought-sensitive cultivars (Figures 1C, D).
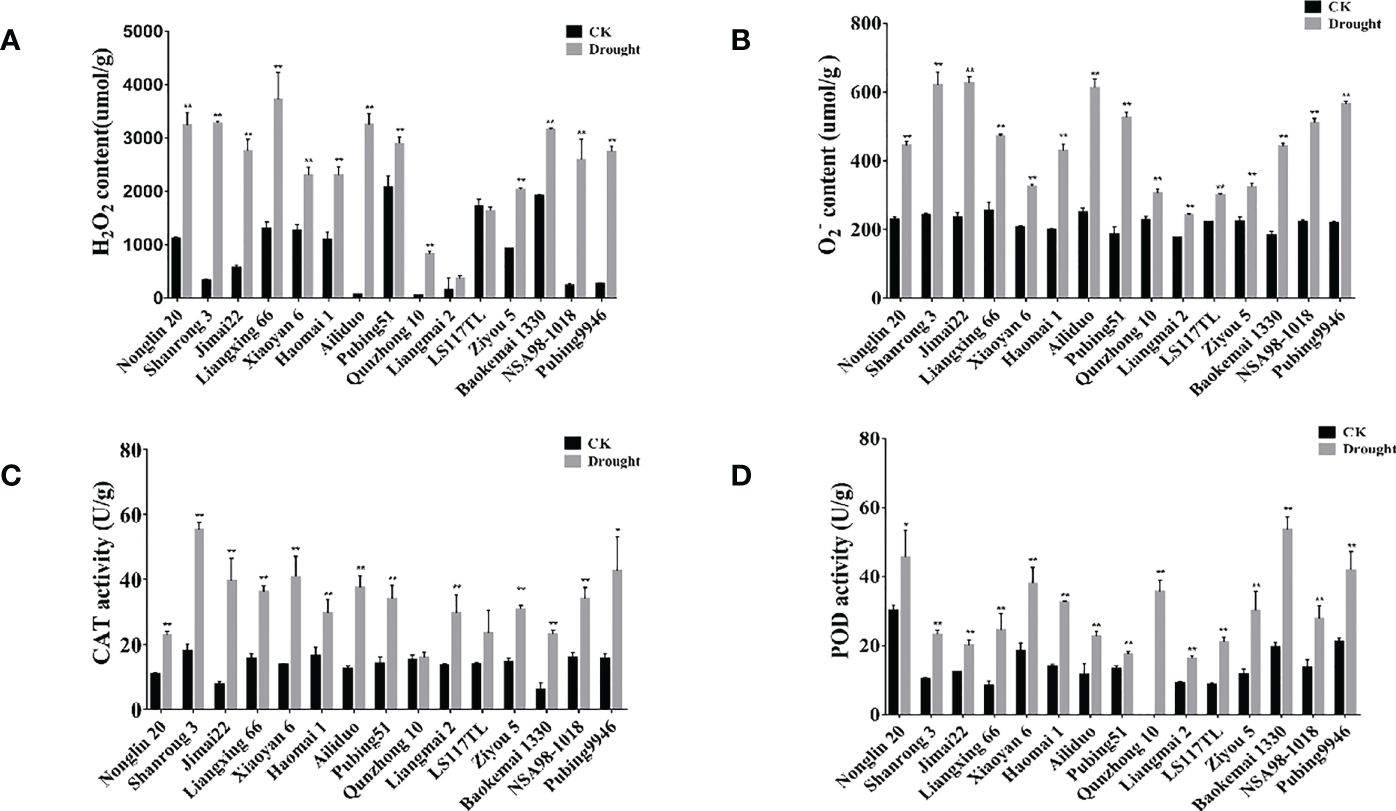
Figure 1. Analysis of ROS scavenging capacity and antioxidant enzymes activity. (A) H2O2 content for different wheat cultivars under control and drought treatment. (B) content for different wheat cultivars under control and drought treatment. (C) Detection of CAT activity for different wheat cultivars under control and drought treatment. (D) Detection of POD activity for different wheat cultivars under control and drought treatment. All experiments included three replicates and the data present the mean ± SD. *P < 0.05 and **P < 0.01 indicate a significant difference compared with WT.
To study the mechanism of drought tolerance between different wheat cultivars, we sequenced the transcriptome of a drought-tolerant wheat cultivar (Ziyou 5) and a drought-sensitive cultivar (Liangxing 66). The genes that were differentially between the control and treated samples were analysed. The results showed that 20663 (down, 8635; up, 12028) and 23775 (down, 12170; up, 11605) genes were differentially expressed in Liangxing 66 and Ziyou 5 (control vs. drought), respectively (Figure S2A–C and Table S1). According to our constructed Venn diagram, we focused on the 734 DEGs that cover 4 different combinations and found that these genes were mainly involved in the flavonoid, plant hormone, MAPK signalling, phenolamide and antioxidant pathways (Figure S2C; Table S1). Based on the DEG expression levels, principal component analysis (PCA) was performed on the samples (three biological replicates) (Figure S2D). According to the PCA score plot, the first principal components (PCs) accounted for 41.36% and the second PCs accounted for 18.47% of the total variance among the four sample groups. The four sample groups (i.e., Liangxing 66-CK, Liangxing 66-D, Ziyou 5-CK, Ziyou 5-D) could be easily distinguished from each other, since they were clustered into four distinct areas in the PCA score plot, indicating that each of the sample groups exhibited distinct expression profiles.
To determine the potential functions, all the DEGs were annotated by Gene Ontology (GO) enrichment analysis. The results showed that most DEGs were enriched in biological processes, including cellular processes (9319, 10610), metabolic processes (7824, 8884), responses to stimulus (5026, 5363), biological regulation (3623, 4040), and developmental processes (1930, 2099) in Liangxing 66 and Ziyou 5, respectively (control vs. drought). In the cellular component category, the top two categories were cellular anatomical entity (13490, 15364) and protein-containing complex (1239, 1603). In the molecular function category, the top four terms were binding (8968, 10076), catalytic activity (8639, 9664), transporter activity (1200, 1335), and transcription regulator activity (850, 917) (Figures S3A, B; Table S1).
To better understand the interactions of the DEGs, we performed a Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.genome.jp/kegg) functional enrichment analysis of the DEGs (control vs. drought). The results showed that the related KEGG pathways were mostly enriched in metabolic pathways (3615, 4066), biosynthesis of secondary metabolites (2271, 2557), plant−pathogen interactions (962, 942), the MAPK signalling pathway (507, 517), plant hormone signal transduction (541, 570), starch and sucrose metabolism (346, 361), phenylpropanoid biosynthesis (319, 351), oxidative phosphorylation (164, 193), flavonoid biosynthesis (161, 179), glutathione metabolism (157, 189), peroxisomes (155, 182), tryptophan metabolism (106, 136), arginine and proline metabolism (92, 114), and flavone and flavonol biosynthesis (40, 55) in Liangxing 66 and Ziyou 5, respectively (control vs. drought) (Figures S3C, D; Table S1).
To better understand the influence of drought stress on the expression profiles of genes involved in the flavonoid biosynthesis pathway, we analysed the transcriptomic data of the drought-stressed wheat cultivars (Ziyou 5 and Liangxing 66). Based on the transcript levels, we constructed a network involving all the relationships of genes involved in the flavonoid biosynthesis pathway (Figure 2). The analysis showed that the expression levels of 372 (Liangxing 66-CK vs. Liangxing 66-D) and 405 (Ziyou 5-CK vs. Ziyou 5-D) genes in the flavonoid biosynthesis pathway changed. When combining data obtained from each group, we found that most expression levels of these genes increased between the two groups, as was the case for phenylalanine ammonia lyase (PAL), phenylalanine/tyrosine ammonia-lyase (PTAL), trans-cinnamate 4-monooxygenase (CYP73A), ferulate-5-hydroxylase (F5H), caffeoyl-CoA O-methyltransferase (CMT), 5-O-(4-coumaroyl)-D-quinate 3’-monooxygenase (CYP98A), flavanone 4-reductase (DFR), flavonoid 3’-monooxygenase (CYP75B1), and flavanone 7-O-glucoside 2’-O-beta-L-rhamnosyltransferase (C12RT1). However, the expression level of flavonoid 3’,5’-hydroxylase (CYP75A) decreased. In addition, the expression levels of 4-coumarate-CoA ligase (4CL), shikimate O-hydroxycinnamoyl transferase (HCT), cinnamoyl-CoA reductase (CCR), chalcone synthase (CHS), chalcone isomerase (CHI), anthocyanidin reductase (ANR), anthocyanidin synthase (ANS), PRX, and flavonol synthase (FLS) were different, with some increasing and some decreasing (Table S2). Considering that data are from the two different individual groups, the results of our transcriptomic analysis revealed specific increases in naringenin 3-dioxygenase (F3H) expression (Ziyou 5-CK vs. Ziyou 5-D) (Figure 2, Table S2). These results suggested that the flavonoid biosynthesis pathway in wheat plays a major role in the response to drought stress. Moreover, these results highlight that treatment of distinct wheat cultivars can consistently elicit discrete profiles of specific genes. In addition, the RNA-seq-based transcript levels of these genes involved in the flavonoid biosynthesis pathway were also verified via qRT‐PCR. The expression trends according to the qRT‐PCR data were consistent with the trends according to the transcriptomic data (Figure S4).
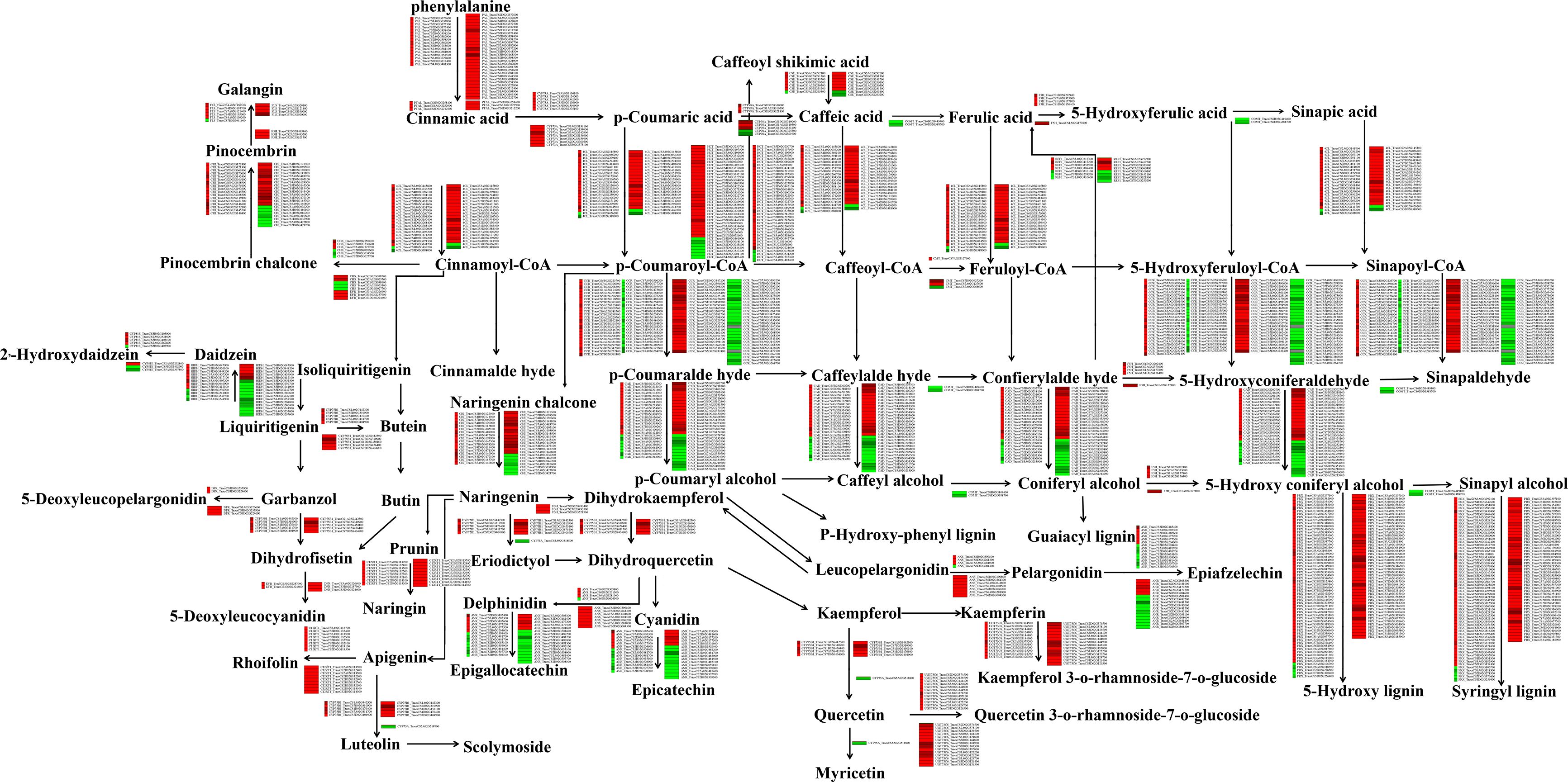
Figure 2. Effects of drought stress on flavonoid biosynthesis‐related genes. Regulatory network for the flavonoid metabolites, selected integrated expression levels of known genes involved in flavonoid biosynthetic pathway for wheat leaves treated with drought stress. The green and red represent doen-regulated and up-regulated gene expression under drought stress. PAL, phenylalanine ammonia-lyase; PTAL, phenylalanine/tyrosine ammonia-lyase; CYP73A, trans-cinnamate 4-monooxygenase; COMT, caffeic acid 3-O-methyltransferase; CMT, caffeoyl-CoA O-methyltransferase; HCT, shikimate O-hydroxycinnamoyltransferase; CYP98A, 5-O-(4-coumaroyl)-D-quinate 3’-monooxygenase; CSE, caffeoylshikimate esterase; REF1, coniferyl-aldehyde dehydrogenase; DFR, flavanone 4-reductase; CYP75A, flavonoid 3’,5’-hydroxylase; PRX, peroxidase; 4CL, 4-coumarate–CoA ligase; CCR, cinnamoyl-CoA reductase; HCT, shikimate O-hydroxycinnamoyl transferase; CYP98A, 5-O-(4-coumaroyl)-D-quinate 3’-monooxygenase; F5H, ferulate-5-hydroxylase; CAD, cinnamyl-alcohol dehydrogenase; CHS, chalcone synthase; CHI, chalcone isomerase; FLS, flavonol synthase; CYP75B1, flavonoid 3’-monooxygenase; ANS, anthocyanidin synthase; ANR, anthocyanidin reductase; HIDH, 2-hydroxyisoflavanone dehydratase; UGT73C6, flavonol-3-O-L-rhamnoside-7-O-glucosyltransferase; CYP81E, 4’-methoxyisoflavone 2’-hydroxylase; F3H, naringenin 3-dioxygenase; C12RT1, flavanone 7-O-glucoside 2’’-O-beta-L-rhamnosyltransferase.
Our transcriptome analysis also indicated that, in addition to the flavonoid biosynthesis pathway, the ABA (a plant hormone) pathway is involved in the response to drought stress. Therefore, we generated a network to explore the relationships between drought stress and ABA signalling pathways. Under drought stress, 148 (Liangxing 66-CK vs. Liangxing 66-D) and 166 (Ziyou 5-CK vs. Ziyou 5-D) ABA-related genes were enriched in plant hormone pathways. The expression levels of beta-carotene 3-hydroxylase (crtZ), xanthoxin dehydrogenase (ABA2), and abscisic-aldehyde oxidase (AAO3) increased, while those of prolycopene isomerase (crtISO), violaxanthin de-epoxidase (VDE), and PYR/PYL decreased. Moreover, we found that carotenoid epsilon hydroxylase (CYP97C1) was specifically expressed in the samples (Ziyou 5-CK vs. Ziyou 5-D) (Figure 3; Table S2). Similarly, the expression trends of genes involved in the ABA pathway according to the qRT‐PCR data were consistent with the trends according to the transcriptomic data (Figure S5).
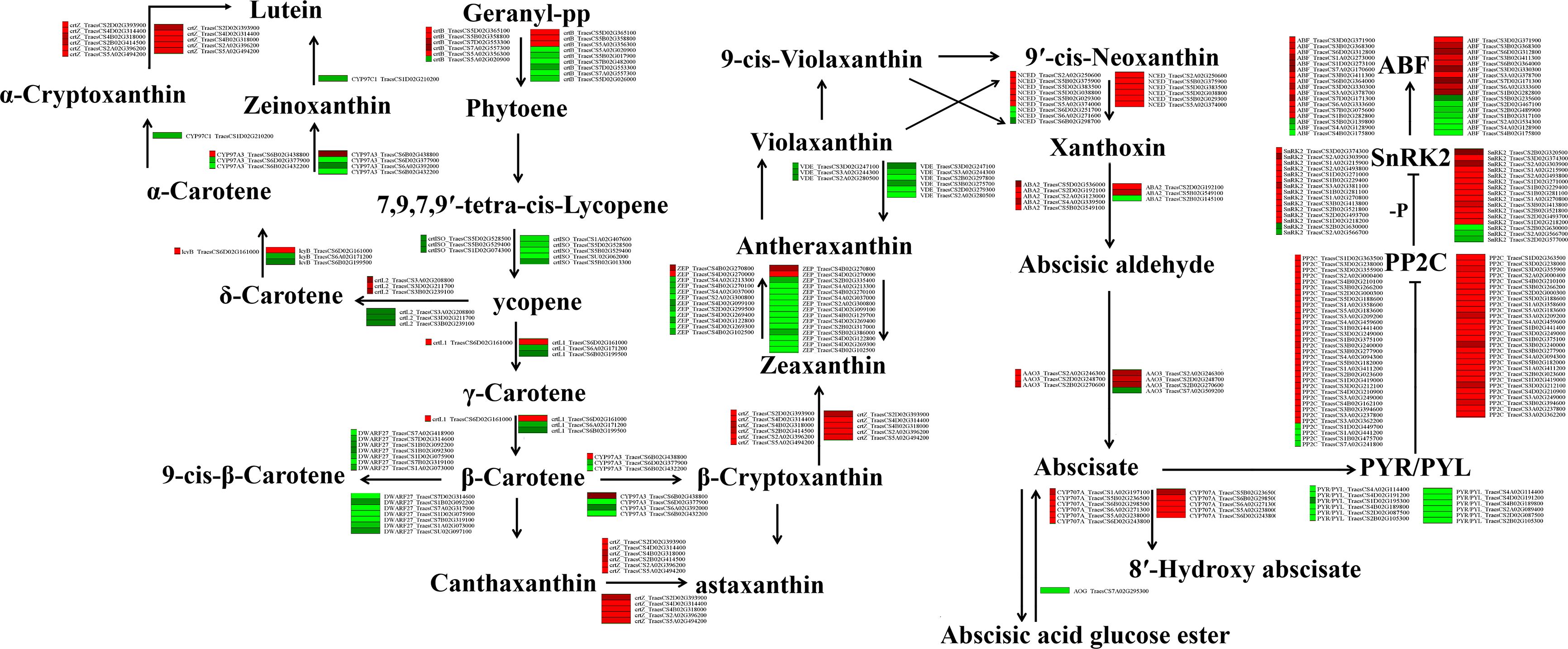
Figure 3. Effects of drought stress on ABA biosynthesis and signal transduction in wheat. Overview of ABA biosynthesis and signal transduction. The green and red represent down-regulated and up-regulated gene expression under drought stress. ctrB, 15-cis-phytoene synthase; ctrZ, beta-carotene 3-hydroxylase; ZEP, zeaxanthin epoxidase; crtISO, prolycopene isomerase; crtL2, lycopene epsilon-cyclase; lcyB, lycopene beta-cyclase; CYP97A3, beta-ring hydroxylase; crtZ, beta-carotene 3-hydroxylase; CYP97C1, carotenoid epsilon hydroxylase; crtL1, lycopene beta-cyclase; DWARF27, beta-carotene isomerase; VDE, violaxanthin de-epoxidase; NCED, 9-cis-epoxycarotenoid dioxygenase; ABA2, xanthoxin dehydrogenase; AAO3, abscisic-aldehyde oxidase; AOG, abscisate beta-glucosyltransferase; PP2C, protein phosphatase 2C; CYP707A, (+)-abscisic acid 8’-hydroxylase. ABF, ABA responsive element binding factor; PYL, abscisic acid receptor PYR/PYL family; SnRK2, serine/threonine-protein kinase SRK2.
We also generated networks about auxin, SA and JA pathways. (1) auxin: Under drought stress, 171 (Liangxing 66-CK vs Liangxing 66-D) and 210 (Ziyou 5-CK vs Ziyou 5-D) auxin-related genes were enriched in plant hormone pathways. Among them, the expression levels of 3-phosphoshikimate 1-carboxyvinyltransferase (AROA), chorismate synthase (AROC), anthranilate phosphoribosyl transferase (TRPD), aromatic aminotransferase (ISS1), and transport inhibitor response 1 (TIR1), exhibited upregulation, while arylalkylamine N-acetyltransferase (AANAT), indole-3-acetaldehyde oxidase (AAO) exhibited downregulation. Indole-3-pyruvate monooxygenase (YUCCA), amidase (AMI), L-tryptophan—pyruvate aminotransferase (TAA1), auxin-responsive protein (AUX/IAA), auxin response factor (ARF), auxin responsive GH3 gene family (GH3), SAUR family protein (SAUR) were different expression trends (upregulations, or downregulations). (2) SA: Under drought stress, 46 (Liangxing 66-CK vs Liangxing 66-D) and 59 (Ziyou 5-CK vs Ziyou 5-D) genes in SA pathways were enriched in plant hormone pathways. Among them, the expression levels of PAL, PTAL, and PR1 were significantly upregulated, whereas ICS and NPR1 were significantly downregulated. (3) JA: Under drought stress, 179 (Liangxing 66-CK vs Liangxing 66-D) and 168 (Ziyou 5-CK vs Ziyou 5-D) genes in JA pathway were enriched in plant hormone pathways. Among them, the expression levels of hydroperoxide dehydratase (AOS), allene oxide cyclase (AOC), and acetyl-CoA acyltransferase (FAD) were upregulated, whereas hydroperoxide lyase (HPL) was significantly downregulated. Fatty acid alpha-dioxygenase (DOX) was significantly upregulated in “Liangxing 66” (control vs. drought treated), while DOX was significantly downregulated in “Ziyou 5” (control vs. drought treated). OPC-8:0 CoA ligase 1 (OPCL1) and jasmonate O-methyltransferase (JAM) specially expressed in “Liangxing 66” (control vs. drought treated), while COI1 specially expressed in “Ziyou 5” (control vs. drought treated) (Figure S6, Table S2). In addition, qRT-PCR analysis confirmed the significantly differential gene expression trends detected in transcriptome data (Figure S7).
We found that drought stress modulates the phenolamide and antioxidant pathways. The transcriptomic data revealed significant differences in the expression of many phenolamide-related and antioxidant pathway-related genes. Among them, arginase (ARG), agmatine coumaroyl transferase (ACT), glutamate 5-kinase (proB), pyrroline-5-carboxylate reductase (proC), ornithine–oxo-acid transaminase (ROCD), prolyl 4-hydroxylase (P4HA), aspartate aminotransferase (GOT1), ornithine decarboxylase (ODC1), spermidine synthase (SPE), polyamine oxidase (PAO), phosphomannomutase (PMM), and mannose-1-phosphate guanylyltransferase (GMPP) were upregulated in response to drought stress. In contrast, ribonucleoside-diphosphate reductase subunit M1 (RRM1) was significantly downregulated under drought stress. Moreover, the expression trends of genes involved in the antioxidant pathway, such as GPX, L-APX, peroxiredoxin (PRDX6), glutathione reductase (GSR), gamma-glutamyltranspeptidase (GGT), glutathione S-transferase (GST), L-ascorbate oxidase (AOX), monodehydroascorbate reductase (NADH), (S)-2-hydroxy-acid oxidase (HAO), and CAT, were different, with the expression of some increasing and some decreasing, in the Liangxing 66-CK vs. Liangxing 66-D and Ziyou 5-CK vs. Ziyou 5-D comparison groups. GDP-D-mannose 3’,5’-epimerase (GME) was expressed specifically in Ziyou 5 (control vs. drought stressed) (Figure 4; Table S2). We also compared the expression levels of the genes involved in the phenolamide and the antioxidant pathways between the qRT‐PCR data and the transcriptomic data, and the we found that the expression trends were also the same (Figure S8).
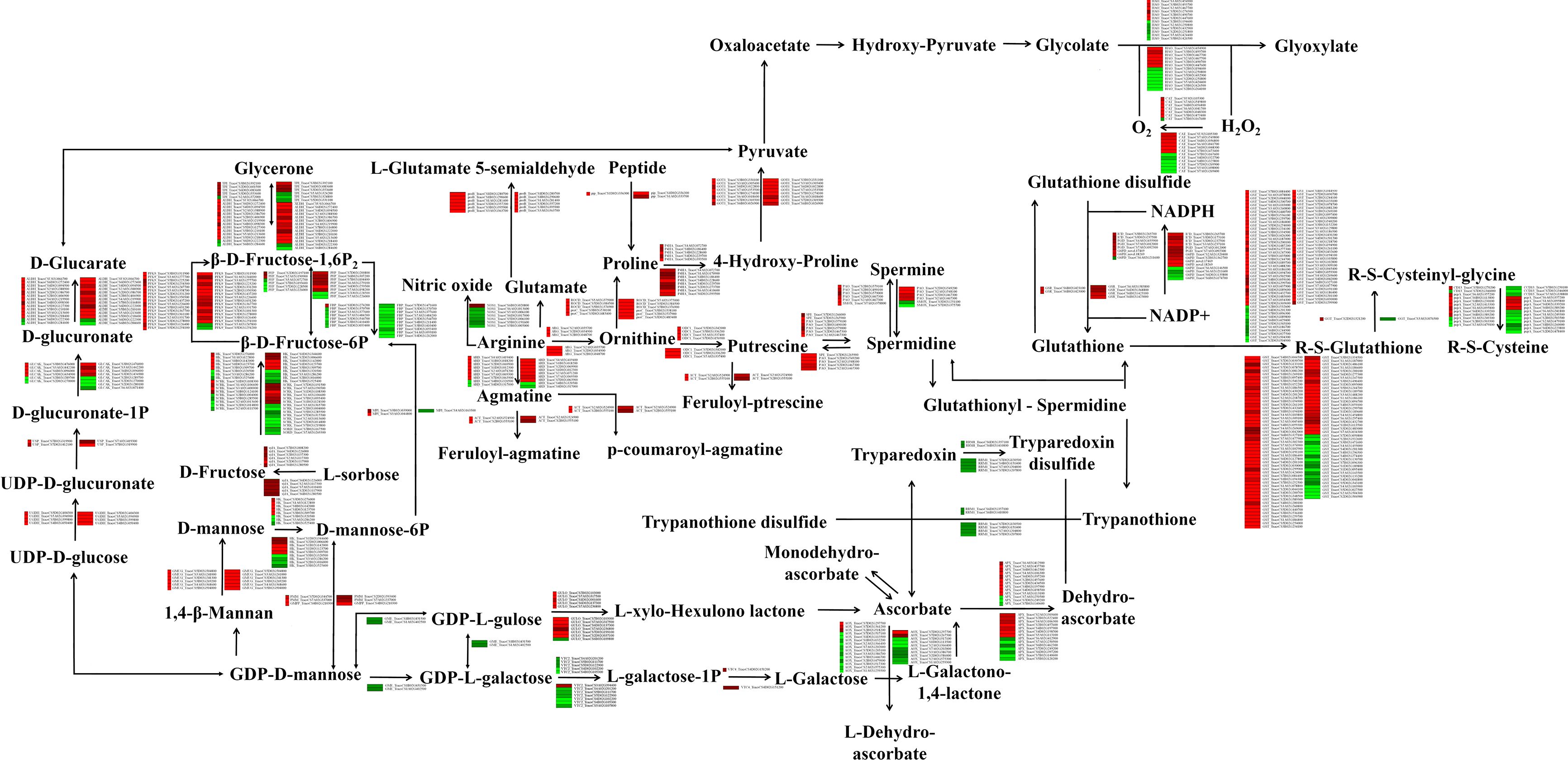
Figure 4. Effects of drought stress on phenolamides and antioxidant biosynthesis-related genes in wheat. Overview of phenolamides and antioxidant biosynthesis. The green and red represent doen-regulated and up-regulated gene expression under drought stress. ALDH, aldehyde dehydrogenase (NAD+); SMOX, spermine oxidase; ROCD, ornithine–oxo-acid transaminase; ACT, agmatine coumaroyltransferase; PRDX6, peroxiredoxin 6; G6PD, glucose-6-phosphate 1-dehydrogenase; GPX, glutathione peroxidase; APX, L-ascorbate peroxidase; RRM1, ribonucleoside-diphosphate reductase subunit M1; NOS1, nitric-oxide synthase; proB, glutamate 5-kinase; proC, pyrroline-5-carboxylate reductase; pip, proline iminopeptidase; P4HA, prolyl 4-hydroxylase; GOT1, aspartate aminotransferas; ODC1, ornithine decarboxylase; GLDH, L-galactono-1,4-lactone dehydrogenase; SPE, spermidine synthase; GSR, glutathione reductase (NADPH); PGD, 6-phosphogluconate dehydrogenase; ICD, isocitrate dehydrogenase; GGT, gamma-glutamyltranspeptidase/glutathione hydrolase; UGDH, UDP glucose 6-dehydrogenase; GMPP, mannose-1-phosphate guanylyltransferase; SORD, L-iditol 2-dehydrogenase; xylA, xylose isomerase; RRM1, ribonucleoside-diphosphate reductase subunit M1; GST, glutathione S-transferase; CD13, aminopeptidase N; pepA, leucyl aminopeptidase; GME, GDP-D-mannose 3’, 5’-epimerase; VTC2, GDP-L-galactose phosphorylase; VTC4, inositol-phosphate phosphatase; GULO, L-gulonolactone oxidase; AOX, L-ascorbate oxidase; NADH, monodehydroascorbate reductase; HK, hexokinase; scrK, fructokinase; PFP, diphosphate-dependent phosphofructokinase; PFK9, 6-phosphofructokinase; FBP, fructose-1,6-bisphosphatase I; ALDO, fructose-bisphosphate aldolase; TPI, triosephosphate isomerase; PMM, phosphomannomutase; GMUG, mannan endo-1,4-beta-mannosidase; MPI, mannose-6-phosphate isomerase; USP, UDP-sugar pyrophosphorylase; GLCAK, glucuronokinase; HAO, (S)-2-hydroxy-acid oxidase; CAT, catalase.
To explore the regulatory mechanisms of genes involved in different metabolic pathways, we performed a coexpression cluster analysis of these genes involved in different metabolic pathways and all differentially expressed transcription factor (TF)-encoding genes. We found that these DEGs could be divided into 9 clusters: subclass 1-subclass 9 (Figure 5A). We performed an additional analysis to explore which types of TFs have regulatory functions with respect to these genes among various metabolic pathways and counted the number of TF-encoding genes in the 9 clusters. The results showed that bHLH, bZIP, NAC, C2H2, MYB and AP2/ERF TFs were the most abundant (Figure 5B). We analysed the TF-binding sites of these gene promoters in the 2000 bp upstream sequences, and the results showed that the binding sites of bHLH, bZIP, Tify, MYB, ERF, and NF-Y TFs throughout the promoter regions of these genes were the most abundant, including in the ALDH, GST, JAZ, OPR, AOS, SnRK2, PP2C, PYL, ABA2, NCED, ANR, ANS, F3H, CHS, 4CL and PRX genes (Figures 5C, D). Taken together, these results suggested that the regulation of TFs and pathway genes affects the response to drought stress in wheat.
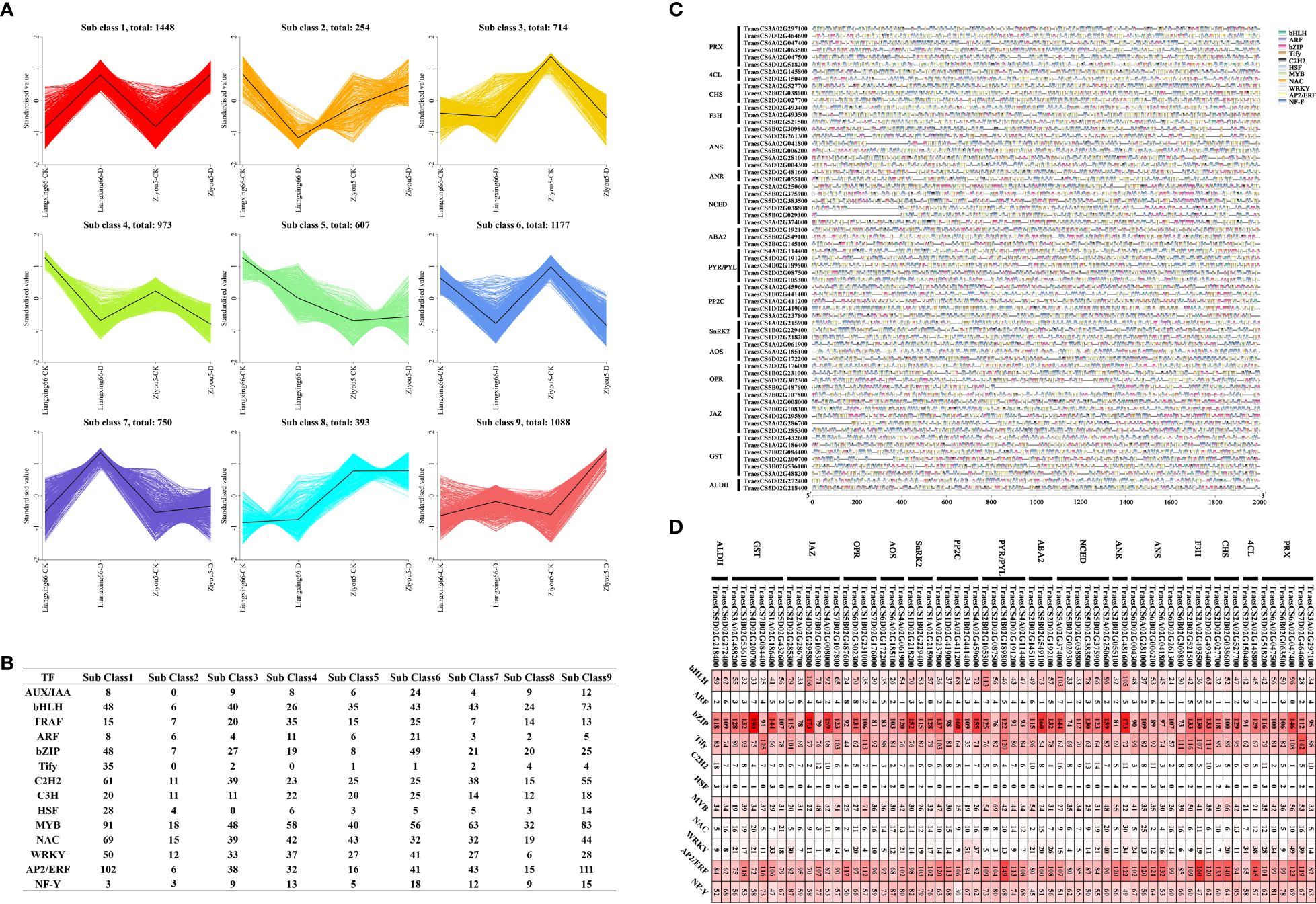
Figure 5. (A) Cluster analysis of co-expression patterns of genes associated with flavonoid, phytohormone, phenolamides and antioxidant metabolic pathways and all differentially expressed transcription factors. (B) Quantity statistics of the transcription factors in different sub clusters. (C) The promoter binding sites analysis of genes associated with different pathways. (D) The cis-elements distribution. We analyzed transcription factor binding site based on the 2000 bp DNA sequence upstream of the gene.
PRXs have been shown to be involved in plant tolerance to abiotic stresses. Our RNA‐seq data indicated that the expression levels of many PRXs changed after drought stress treatment. In our previous study, we confirmed that a PRX gene, TaPRX-2A, responded to various abiotic stresses (salt and drought), and TaPRX-2A overexpression was shown to enhance the salt tolerance of wheat significantly (Su et al., 2020). However, what’s interesting is that this gene TaPRX-2A is not found in our transcriptome. To further explore the detailed temporal and spatial expression patterns of TaPRX-2A at different time after drought treatment, we measured the expression levels of TaPRX-2A in the wheat cultivars with varying degrees of drought tolerance at 24h and 48h after drought treatment. Compared with that of “Liangxing 66”, the expression profile of TaPRX-2A was significantly different in several wheat cultivars with different drought tolerances, such as “Ziyou 5”, “Xiaoyan 6”, “Guomai 115”, and “Baokemai 1330”, at 24 h after treatment (Figure S9). Thus, we grew three independent TaPRX-2A overexpression transgenic lines (TaOE1, TaOE2, and TaOE3) and WT wheat plants under natural drought-stress conditions for 21 days, followed by rewatering for 7 days. Under nonstress conditions, we observed no significant differences in phenotypic characteristics between the TaPRX-2A transgenic lines and WT wheat plants. However, the TaPRX-2A transgenic lines grew more robustly than did the WT plants. The WT plants were more withered after drought stress treatment for 21 days. After being rewatered for 7 days, the TaPRX-2A transgenic lines presented higher survival percentages (65%, 65.2, 69.3%) than did the WT plants (38.7%) (Figures 6A, B). Then, the shoot length, RWC, and root length of the TaOE and WT plants were compared between the normal and drought-stress conditions. Compared with the WT plants, the TaOE plants had longer shoots and a higher RWC (Figures 6C, D). Interestingly, the lengths of the roots of the TaOE plants were not significantly different from those of the WT plants (Figure 6E). Taken together, these results indicated that TaPRX-2A overexpression drastically enhanced wheat drought tolerance.
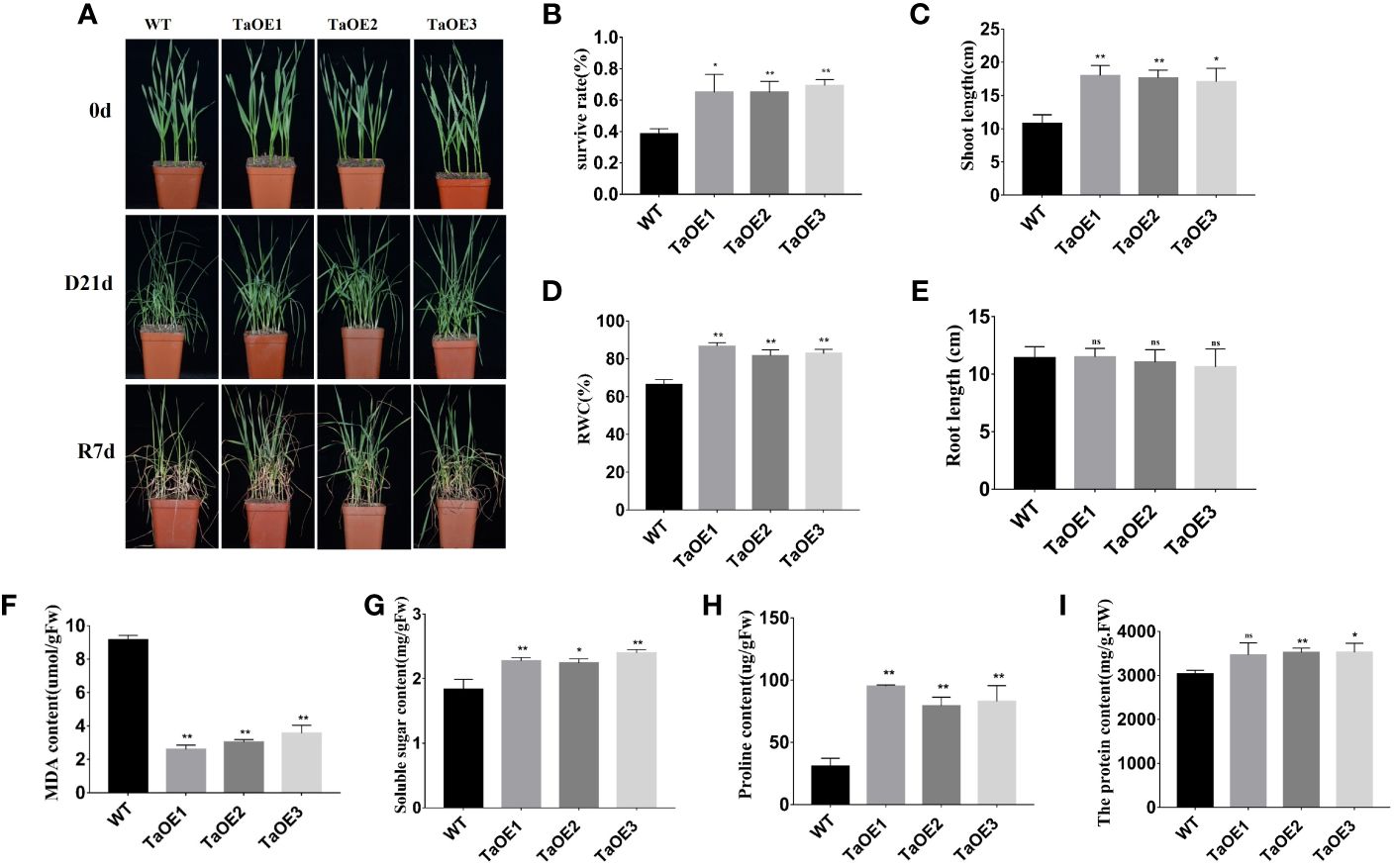
Figure 6. TaPRX-2A overexpression increased the drought tolerance. (A) Phenotype of TaPRX-2A-overexpressing transgenic and WT wheat (the cultivar “KN199”) with drought treatment. (B) Survival rates of TaPRX-2A-overexpressing transgenic lines and WT wheat. (C) shoot length of TaPRX-2A-overexpressing transgenic lines and WT wheat. (D) Relative water content (RWC), and (E) root length. (F) MDA content of TaPRX-2A-overexpressing transgenic lines and WT wheat. (G) soluble sugar content of TaPRX-2A-overexpressing transgenic lines and WT wheat. (H) proline content, and (I) soluble protein content of TaPRX-2A-overexpressing and WT plants. All experiments included three replicates and the data present the mean ± SD. *P < 0.05 and **P < 0.01 indicate a significant difference compared with WT. The “ns” presents “no differences”.
We further measured physiological–biochemical indices, including MDA, soluble sugar, proline, and soluble protein contents, to explore the mechanism of TaPRX-2A-mediated drought resistance. The results showed that the MDA, soluble sugar, proline, and soluble protein contents in the WT were not significantly different from those of the TaOE lines under normal conditions. However, the MDA content in the WT was significantly higher than that in the TaOE lines under drought stress (Figure 6F). The soluble sugar, proline, and soluble protein contents in the WT were significantly lower than those in the TaOE lines (Figures 6G–I). These results demonstrated that TaPRX-2A overexpression improved transgenic plant tolerance to drought stress through changes to metabolite (MDA, soluble sugar, proline, and soluble protein) contents (Figure 6).
PRXs can scavenge ROS to maintain steady-state levels of ROS when plants experience stress conditions. Therefore, the ROS levels of the TaOE and WT lines under drought stress were assessed. The levels of and H2O2 were major indicators of the ROS level. Therefore, the accumulation of and H2O2 was measured in both the TaOE and the WT lines. The accumulation of and H2O2 was determined by NBT staining and DAB staining. The results showed that the levels of (stained blue with NBT) and H2O2 (stained brown by DAB) were significantly higher in the WT plants than in the transgenic lines under drought stress (Figures 7A–D). In addition, we measured the activities of antioxidant enzymes (SOD, PRX, and CAT). The results showed that the SOD, CAT, and PRX activities in the transgenic plants were significantly higher than those in the WT plants (Figures 7E–G).
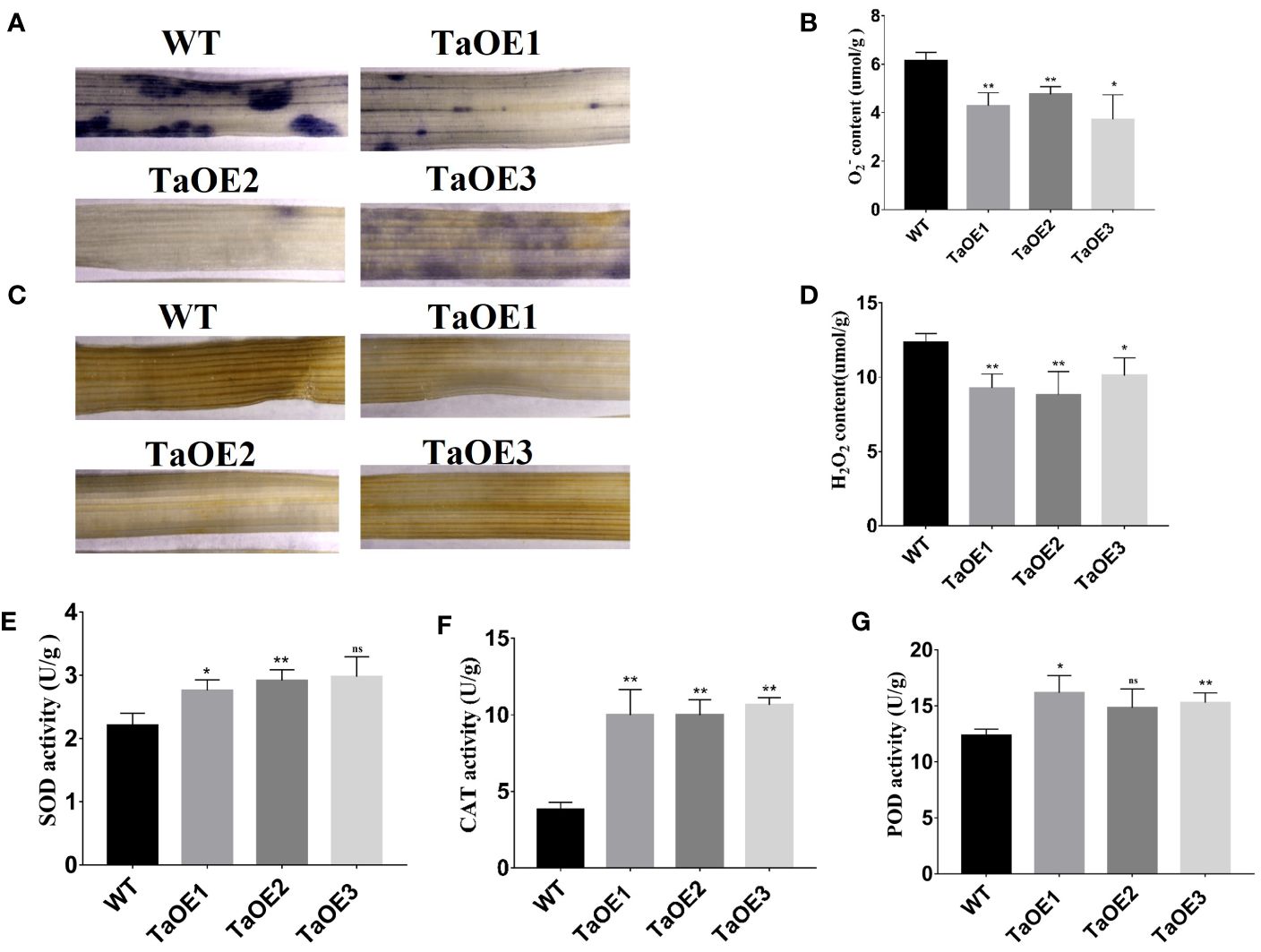
Figure 7. Analysis of ROS scavenging capacity andantioxidant enzymes activity in transgenic wheat lines. (A) Detection of generation by NBT staining and content (B). (C) Detection of H2O2 accumulation by DAB staining and H2O2 content (D). (E) Detection of SOD activity in TaPRX-2A-overexpressing transgenic lines and WT wheat. (F) Detection of CAT activity in TaPRX-2A-overexpressing transgenic lines and WT wheat. (G) Detection of POD activity in TaPRX-2A-overexpressing transgenic lines and WT wheat. All experiments included three replicates and the data present the mean ± SD. *P < 0.05 and **P < 0.01 indicate a significant difference compared with WT. The “ns” presents “no differences”.
To determine whether TaPRX-2A affects the expression of stress-related genes that contribute to drought tolerance, the expression patterns of various stress-related genes were determined in the transgenic lines and WT plants (Figure S10). These stress-related genes were selected on the basis of their purported involvement in the response to various abiotic stresses. These genes include TLP4, which encodes a thaumatin-like protein; DE6, which encodes dehydrin 6; RD22, which encodes a dehydration-responsive protein; ABA-related genes including CRTB, ZEP, PYR, PP2c, and SnRK2; ABAI, which encodes an ABA-inducing protein; GLP4, which encodes a germin-like protein; GST22, which encodes glutathione S-transferase; and FeSOD, CuSOD, APX, and CAT, which encode ROS-scavenging enzymes. We found that several of these genes (RD22, ABAI, CAT and APX) were highly expressed in the transgenic lines and in the WT plants under both nonstress and drought-stress conditions. In addition, the expression of several stress-related genes (TLP4, GLP4, GST22, FeSOD, CuSOD) was higher in the transgenic lines than in the WT plants under drought stress. Nevertheless, the expression of the DE6 gene was not significantly different between the transgenic lines and the WT plants (Figure S10A). In addition, we identified ABA-related genes, including CRTB, ZEP, PYR, PP2c, and SnRK2, in the transgenic lines and WT plants, and the results showed that the expression levels of these ABA-related genes were higher in the transgenic lines than in the WT plants under drought stress. Other genes (ZEP, PYR) were expressed as low levels in both the transgenic lines and in the WT plants under drought stress (Figure S10B). Taken together, these results suggested that the overexpression of TaPRX-2A improved drought tolerance by the control of stress-responsive and ABA-related gene expression.
Drought stress is a widespread abiotic stress that causes major crop yield and economic losses (Mahajan and Tuteja, 2005; Zampieri et al., 2017). The molecular mechanisms through which plants respond to drought conditions have been extensively reported. Plants increase their tolerance to drought stress by activating signalling pathways that drive biochemical and physiological responses. Secondary metabolism, including the production of flavonoids, melatonin, secoisolariciresinol and plant hormones (ABA), is a major signalling event (Campo et al., 2014; Zhu, 2016; Meng et al., 2021; Feng et al., 2022; Song et al., 2022). Class III PRXs compose a large gene family with multiple genes in higher plants. It has been reported that class III PRXs respond to abiotic stresses. However, information regarding class III PRX resistance mechanisms used by wheat to combat drought stress is limited. Here, we revealed that, by scavenging ROS and enhancing stress-responsive gene expression, TaPRX-2A enhanced wheat tolerance to drought conditions.
Many studies have revealed that integrated analysis of transcriptomic data can yield functional insights into various biological processes (Hoefgen and Nikiforova, 2008). It is also known that flavonoid biosynthesis, plant hormonal signalling, and peroxidases play an important role in plant against abiotic stresses (Ma et al., 2014; Guo et al., 2020). Like these studies, our study revealed changes in flavonoids, plant hormones, phenolamides and activity of antioxidant signalling pathways, thereby expanding the scope of known drought tolerance‐related DEGs. The previous study showed that PRXs enhanced wheat tolerance against salt stress through ABA signalling (Su et al., 2020). As components with antioxidant properties, the flavonoids and peroxidases has been widely reported to be involved in plant resistance to abiotic stress, respectively. But, little information has been showed about the regulatory relationship between flavonoid biosynthesis and peroxidases in plant resistance to abiotic stress. Therefore, the regulatory relationship in wheat against abiotic stresses between different metabolic pathways and peroxidases still needs further study in future.
When plants experience drought for an extended period, a large number of ROS are induced. ROS exist in many forms, such as , H2O2, and OH− in plants (McCord, 2000; Joshi et al., 2016). The class III PRXs catalyse H2O2 reduction in the peroxidative cycle through the transfer of electrons from different donors (Hiraga et al., 2001; Gill and Tuteja, 2010). Some studies have revealed that class III PRXs play an important role in enhancing tolerance to stresses through the regulation of ROS balance in plants. For example, the Oryza sativa class III PRX gene OsPRX38 activates the antioxidant system and scavenges H2O2 to enhance arsenic (As) tolerance (Maria et al., 2019). Furthermore, plants have evolved a complex antioxidant system (involving SOD, CAT, and PRX) to protect cells from damage by maintaining the balance of ROS levels (Dat et al., 2000; Gill and Tuteja, 2010). Our results showed that TaPRX-2A enhanced transgenic plant tolerance to drought stress by increasing antioxidant enzyme activity, thereby reducing and H2O2 levels. Moreover, TaPRX-2A also increased the expression of antioxidant-related genes, such as CuSOD, FeSOD, CAT, and APX. Therefore, these results showed that TaPRX-2A enhanced drought tolerance by regulating the expression of antioxidant genes that affect enzyme activity. In rice, the class III PRX gene OsPrx30 mediates rice bacterial blight (Xanthomonas oryzae pv. oryzae)-induced ROS accumulation by the AT-hook protein OsATH1 (Liu et al., 2021). Do wheat class III PRXs also respond to environmental stresses in this way? Future research will explore the mechanisms through which transcription factors regulate TaPRX-2A and regulate other antioxidant-encoding genes.
The signalling pathway of the plant hormone ABA is central to abiotic stress responses, and ABA is involved in a variety of cross-regulatory networks in response to stresses. For example, the transcription of the Arabidopsis dehydration-responsive gene RD22 may be induced by ABA (Shinozaki et al., 2003). In addition, it was reported that ABA can induce the expression of dehydrin (DE)- and thaumatin-like protein (TLP)-encoding genes, which are essential for tolerance to abiotic stresses (Jung et al., 2005; Seo and Park, 2016). It has also been reported that ABA is involved in the PRX-mediated stress response. For example, the ABA signalling pathway controls the expression of class III PRX genes in Tamarix hispida (Gao et al., 2012). We previously identified that TaPRX-2A enhanced wheat tolerance to salt stress through activation of the ABA signalling pathway and expression of stress-related genes. Similar to the function of TaPRX-2A in response to salt stress, TaPRX-2A also improved transgenic wheat drought tolerance by altering the expression of genes involved in the ABA signalling pathway and stress-related genes, including RD22, TLP4, GLP4, and GST22, in our work. Further study is needed to explore the cross-talk regulatory mechanisms through which TaPRX-2A regulates the ABA signalling pathway and stress-related genes under various stresses.
Finally, we also preliminarily explore the SNP distributions of TaPRX-2A in different wheat cultivars and found 23 SNPs (16 in the promoter, 7 in the coding DNA sequence (CDS), and 3 in the 5’-untranslated region (UTR)) in the TaPRX-2A sequences. Among the 7 SNPs in the CDS, we diagrammed the four SNPs that cause changes in amino acids. The 16 SNPs in promoter influenced only a few cis-acting elements. Therefore, it will be interesting to explore the question about what kind of variations caused the different expression patterns of TaPRX-2A in different cultivars merits further consideration in subsequent investigations. In the future, TaPRX-2A can be used to develop SNP markers or GWAS for screening drought-resistant wheat varieties. This work will have strong application value for the cultivation of the drought-tolerant wheat varieties in the future, which is especially relevant given the anticipated hydrodynamics changes associated with ongoing climate change.
In this study, we evaluated drought tolerance of various wheat cultivars and identified a class III PRXs gene TaPRX-2A in response to drought stress by RNA-Seq analysis in wheat. The overexpression of TaPRX-2A enhanced transgenic wheat tolerance against drought stress through improving antioxidant enzymes activity, activating ABA signaling pathway, and regulating stress-related genes expression, resulting in lower ROS accumulation. This work and its findings have strong future application value in the cultivation of drought-tolerant wheat varieties, which is especially relevant given the anticipated crop losses associated with the future impacts of climate change.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA914511. The accession number of the raw data is PRJNA914511.
PS and SG conceived and designed the experiments. PS performed most experiments. JY performed the bioinformatics analysis of TaPRX-2A, and revised the manuscript. CS,YN, SW, FS, and JL performed the transcriptome sequencing analysis, physiological-biochemical indices measures, and expression analysis. PS wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by Natural Science Foundation of Shandong Province (ZR2022QC129); Doctoral research start-up funds, Liaocheng University (318052018); State Key Laboratory of Crop Biology, Shandong Agricultural University (2021KF03); Industrial Promotion Project of Shandong Agricultural Science and Technology Park (2019YQ035), and Shandong Province Higher Educational Program for Introduction and Cultivation of Young Innovative Talents in 2021.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1119162/full#supplementary-material
Supplementary Figure 1 | The comparison of drought tolerance in 15 different wheat cultivars. (A) The phenotype of drought tolerance in different wheat cultivars was observed at 21 days after drought stress treatments. (B) The survive rate. Each treatment included at least 15 plants.
Supplementary Figure 2 | Overview of transcriptome changes in wheat treated with drought stress. (A) Hierarchical clustering of DEGs in drought‐treated and control wheat. FPKM expression values were used to create the heatmap. (B) Number of DEGs of Liangxing 66-CK vs Liangxing 66-D, Liangxing 66-CK vs Ziyou 5-CK, Ziyou 5-CK vs Ziyou 5-D and Liangxing 66-D vs Ziyou 5-D between control and drought treatment, respectively. (C) Venn diagram among Liangxing 66-CK vs Liangxing 66-D, Liangxing 66-CK vs Ziyou 5-CK, Ziyou 5-CK vs Ziyou 5-D and Liangxing 66-D vs Ziyou 5-D. (D) Principal component analysis (PCA) of transcriptomedata obtained from drought‐treated and control wheat. Each sample included three biological replicates.
Supplementary Figure 3 | Transcriptomic analysis reveals the effect of drought stress in wheat. (A) GO classification diagram of DEGs of Liangxing 66-CK vs Liangxing 66-D in biological process (BP), cellular component (CC) and molecular function (MF). (B) GO classification diagram of DEGs of Ziyou 5-CK vs Ziyou 5-D in BP, CC and MF. (C) KEGG enrichment dotplot of DEGs of Liangxing 66-CK vs Liangxing 66-D. (D) KEGG enrichment dotplot of DEGs of Ziyou 5-CK vs Ziyou 5-D.
Supplementary Figure 4 | The relative expression of flavonoid biosynthesis genes. 18SrRNA was as an endogenous control. The gene relative expression was calculated by the cycle threshold (Ct) values using formula 2–ΔΔCT. All experiments included three replicates and the data present the mean ± SD.
Supplementary Figure 5 | Expression levels of ABA biosynthesis and signal genes. 18SrRNA was as an endogenous control. The gene relative expression was calculated by the cycle threshold (Ct) values using formula 2–ΔΔCT. All experiments included three replicates and the data present the mean ± SD.
Supplementary Figure 6 | Effects of drought stress on SA, JA, and IAA biosynthesis and signal transduction in wheat. Overview of SA, JA, and IAA biosynthesis and signal transduction. The green and red represent doen-regulated and up-regulated gene expression under drought stress. AROK, shikimate kinase; AROA, 3-phosphoshikimate 1-carboxyvinyltransferase; AROC, chorismate synthase; TPR3, anthranilate synthase; TRPA, tryptophan synthase alpha chain; TRPB, tryptophan synthase beta chain; DDC, aromatic-L-amino-acid; AAO, indole-3-acetaldehyde oxidase; YUCCA, indole-3-pyruvate monooxygenase; trpD, anthranilate phosphoribosyltransferase; TRP1, anthranilate synthase; DDC, L-tryptophan decarboxylase; TAA1, L-tryptophan—pyruvate aminotransferase; ISS1, aromatic aminotransferase; ALDH, aldehyde dehydrogenase (NAD+); AANAT, arylalkylamine N-acetyltransferase; ASMT, acetylserotonin O-methyltransferase, AMI, amidase; PLA2G16, HRAS-like suppressor 3; ISS1, aromatic aminotransferase; LOX, lipoxygenase; AOS, hydroperoxide dehydratase; AOC, allene oxide cyclase; OPR, 12-oxophytodienoic acid reductase; PLA2G, secretory phospholipase A2; HPL, hydroperoxide lyase; DOX, fatty acid alpha-dioxygenase; OPCL1, OPC-8:0 CoA ligase 1; ACOX, acyl-CoA oxidaseL; MFP2, enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase; FAD, acetyl-CoA acyltransferase; JAM, jasmonate O-methyltransferase; JAZ, jasmonate ZIM domain-containing protein; aroC, chorismate synthase; aroA, 3-phosphoshikimate 1-carboxyvinyltransferase; AUX1, auxin influx carrier; TIR1, transport inhibitor response 1; AUX/IAA, auxin-responsive protein; ARF, auxin response factor; GH3, auxin responsive GH3 gene family; SAUR, SAUR family protein; PR1, pathogenesis-related protein 1.
Supplementary Figure 7 | The relative expression of genes involved in phytohormones (SA, IAA, and JA) pathways. 18SrRNA was as an endogenous control. The gene relative expression was calculated by the cycle threshold (Ct) values using formula 2–ΔΔCT. All experiments included three replicates and the data present the mean ± SD.
Supplementary Figure 8 | The relative expression of genes involved in phenolamides and antioxidant pathway. 18SrRNA was as an endogenous control. The gene relative expression was calculated by the cycle threshold (Ct) values using formula 2–ΔΔCT. All experiments included three replicates and the data present the mean ± SD.
Supplementary Figure 9 | The expression profile of TaPRX-2A among wheat cultivars with different levels of drought tolerance. 18SrRNA was as an endogenous control. The gene relative expression was calculated by the cycle threshold (Ct) values using formula 2–ΔΔCT. All experiments included three replicates and the data present the mean ± SD.
Supplementary Figure 10 | The expression profile of stress-related genes in TaPRX-2A-overexpressing transgenic lines and WT plants under drought stress. (A) The expression levels of stress-related genes in transgenic lines and WT plants under drought stress. (B) The expression levels of ABA-related genes in transgenic lines and WT plants. 18SrRNA was as an endogenous control. The gene relative expression was calculated by the cycle threshold (Ct) values using formula 2–ΔΔCT. All experiments included three replicates and the data present the mean ± SD.
Supplementary Table 1 | EXECL file with RNA-Seq data.
Supplementary Table 2 | DEGs involved in flavonoid biosynthesis pathway, phytohormones, and phenolamides and antioxidant biosynthesis pathways.
Supplementary Table 3 | Primers used for analysis.
Aebi, H. (1984). Catalase in vitro. Methods Enzymol. 105, 121–126. doi: 10.1016/S0076-6879(84)05016-3
Bates, L. S., Waldren, P. R., Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil. 39, 205–207. doi: 10.1007/BF00018060
Bhatt, I., Tripathi, B. N. (2011). Plant peroxiredoxins: catalytic mechanisms, functional significance and future perspectives. Biotechnol. Adv. 29, 850–859. doi: 10.1016/j.biotechadv.2011.07.002
Campo, S., Baldrich, P., Messeguer, J., Lalanne, E., Coca, M., San, S. B. (2014). Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol. 165, 688–704. doi: 10.1104/pp.113.230268
Chance, B., Maehly, A. (1955). Assay of catalases and peroxidases. Methods Enzymol. 2, 764–775. doi: 10.1016/S0076-6879(55)02300-8
Cosio, C., Dunand, C. (2009). Specific functions of individual class III peroxidase genes. J. Exp. Bot. 60, 391–408. doi: 10.1093/jxb/ern318
Dat, J., Vandenabeele, S., Vranová, E., Van, M. M., Inzé, D., Van, B. F. (2000). Dual action of the active oxygen species during plant stress responses. Cell Mol. Life Sci. 57, 779–795. doi: 10.1007/s000180050041
Daudi, A., Cheng, Z., O'Brien, J. A., Mammarella, N., Khan, S., Ausubel, F. M., et al. (2012). The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell. 24, 275–287. doi: 10.1105/tpc.111.093039
Dhindsa, R. A., Plumb-Dhindsa, P., Thorpe, T. A. (1981). Leaf senescence: correlated with increased permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J. Exp. Bot. 126, 93–101. doi: 10.1093/jxb/32.1.93
Fawal, N., Li, Q., Savelli, B., Brette, M., Passaia, G., Fabre, M., et al. (2013). PeroxiBase: a database for large-scale evolutionary analysis of peroxidases. Nucleic Acids Res. 41, D441–D444. doi: 10.1093/nar/gks1083
Feng, X., Peng, F., Yin, Z., Wang, J., Zhang, Y., Zhang, H., et al. (2022). Secondary metabolite pathway of SDG (secoisolariciresinol) was observed to trigger ROS scavenging system in response to Ca2+ stress in cotton. Genomics 114 (4), 110398. doi: 10.1016/j.ygeno.2022.110398
Gao, C., Zhang, K., Yang, G., Wang, Y. (2012). Expression analysis of four peroxiredoxin genes from Tamarix hispida in response to different abiotic stresses and Exogenous Abscisic Acid (ABA). Int J Mol Sci. 13, 3751–3764. doi: 10.3390/ijms13033751.
Gay, C., Collins, J., Gebicki, J. M. (1999). Hydroperoxide assay with the ferric-xylenol orange complex. Anal. Biochem. 273, 149–155. doi: 10.1006/abio.1999.4208
Gill, S. S., Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Guo, X., Xin, Z., Yang, T., Ma, X., Zhang, Y., Wang, Z., et al. (2020). Metabolomics response for drought stress tolerance in chinese wheat genotypes (Triticum aestivum). Plants (Basel) 9 (4), 520. doi: 10.3390/plants9040520
Heath, R., Packer, L. (1968). Photoperoxidation in isolated chloroplasts: I. kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198. doi: 10.1016/0003-9861(68)90654-1
Hiraga, S., Sasaki, K., Ito, H., Ohashi, Y., Matsui, H. (2001). A large family of class III plant peroxidases. Plant Cell Physiol. 42, 462–468. doi: 10.1093/pcp/pce061
Hoefgen, R., Nikiforova, V. J. (2008). Metabolomics integrated with transcriptomics:Assessing systems response to sulfur-deficiency stress. Physiol. Plant 132, 190–198. doi: 10.1111/j.1399-3054.2007.01012.x
Hu, Z., Li, J., Ding, S., Cheng, F., Li, X., Jiang, Y., et al. (2021). The protein kinase CPK28 phosphorylates ascorbate peroxidase and enhances thermotolerance in tomato. Plant Physiol. 186 (2), 1302–1317. doi: 10.1093/plphys/kiab120
Joshi, R., Wani, S. H., Singh, B., Bohra, A., Dar, Z. A., Lone, A. A., et al. (2016). Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 7 1029. doi: 10.3389/fpls.2016.01029
Jung, Y. C., Lee, H. J., Yum, S. S., Soh, W. Y., Cho, D. Y., Auh, C. K., et al. (2005). Drought-inducible-but ABA-independent-thaumatin-like protein from carrot (Daucus carota l.). Plant Cell Rep. 24, 366–373. doi: 10.1007/s00299-005-0944-x
Liu, H., Dong, S. Y., Li, M., Gu, F. W., Yang, G. L., Guo, T., et al. (2021). The class III peroxidase gene OsPrx30, transcriptionally modulated by the AT-hook protein OsATH1, mediates rice bacterial blight-induced ROS accumulation. J. Integr. Plant Biol. 63 (2), 393–408. doi: 10.1111/jipb.13040
Ma, D., Sun, D., Wang, C., Li, Y., Guo, T. (2014). Expression of flavonoid biosynthesis genes and accumulation of flavonoid in wheat leaves in response to drought stress. Plant Physiol. Biochem. 80, 60–66. doi: 10.1016/j.plaphy.2014.03.024
Mahajan, S., Tuteja, N. (2005). Cold, salinity and drought stresses: an overview. Arch. Biochem. Biophys. 444, 139–158. doi: 10.1016/j.abb.2005.10.018
Maria, K., Yogeshwar, V. D., Neelam, G., Madhu, T., Iffat, Z. A., Mehar, H. A., et al. (2019). Oryza sativa class III peroxidase (Osprx38) overexpression in Arabidopsis thaliana reduces arsenic accumulation due to apoplastic lignification. J. Hazard Mater. 362, 383–393. doi: 10.1016/j.jhazmat.2018.09.029
McCord, J. M. (2000). The evolution of free radicals and oxidative stress. Am. J. Med. 108, 652–659. doi: 10.1016/S0002-9343(00)00412-5
Meng, D., Dong, B., Niu, L., Song, Z., Wang, L., Amin, R., et al. (2021). The pigeon pea CcCIPK14-CcCBL1 pair positively modulates drought tolerance by enhancing flavonoid biosynthesis. Plant J. 106 (5), 1278–1297. doi: 10.1111/tpj.15234
Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. doi: 10.1016/S1360-1385(02)02312-9
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., Wold, B. (2008). Mappingand quantifying mammalian transcriptomes by RNA-seq. NatMethods 5, 621–628. doi: 10.1038/nmeth.1226
Ogino, R., Chinuki, Y., Yokooji, T., Takizawa, D., Matsuo, H., Morita, E. (2021). Identification of peroxidase-1 and beta-glucosidase as cross-reactive wheat allergens in grass pollen-related wheat allergy. Allergol. Int. 70 (2), 215–222. doi: 10.1016/j.alit.2020.09.005
Pacheco, J. M., Ranocha, P., Kasulin, L., Fusari, C. M., Servi, L., Aptekmann, A. A., et al. (2022). Apoplastic class III peroxidases PRX62 and PRX69 promote Arabidopsis root hair growth at low temperature. Nat. Commun. 13 (1), 1310. doi: 10.1038/s41467-022-28833-4
Raggi, S., Ferrarini, A., Delledonne, M., Dunand, C., Ranocha, P., De Lorenzo, G., et al. (2015). The Arabidopsis class III peroxidase AtPRX71 negatively regulates growth under physiological conditions and in response to cell wall damage. Plant Physiol. 169, 2513–2525. doi: 10.1104/pp.15.01464
Seo, P. J., Park, C. M. (2016). MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 186, 471–483. doi: 10.1111/j.1469-8137.2010.03183.x
Shinozaki, K., Yamaguchi-Shinozaki, K., Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. doi: 10.1016/S1369-5266(03)00092-X
Simonetti, E., Veronico, P., Melillo, M. T., Delibes, Á., Andrés, M. F., López-Braña, I. (2009). Analysis of class III peroxidase genes expressed in roots of resistant and susceptible wheat lines infected by heterodera avenae. Mol. Plant-Microbe Interact. 22, 1081–1092. doi: 10.1094/MPMI-22-9-1081
Song, Z., Yang, Q., Dong, B., Li, N., Wang, M., Du, T. (2022). Melatonin enhances plant stress tolerance by promoting flavonoid enrichment, focusing on luteolin for salt stress. J. Exp. Bot. 73, 5992–6008. doi: 10.1093/jxb/erac276
Spiro, R. G. (1966). Analysis of sugars found in glycoprotein. Method enzymol. 8, 3–26. doi: 10.1016/0076-6879(66)08005-4
Su, P. S., Yan, J., Li, W., Wang, L., Zhao, J. X., Ma, X., et al. (2020). A member of wheat class III peroxidase gene family, TaPRX-2A, enhanced the tolerance of salt stress. BMC Plant Biol. 20, 392. doi: 10.1186/s12870-020-02602-1
Taurog, A. (1999). Molecular evolution of thyroid peroxidase. Biochimie 81, 557–562. doi: 10.1016/S0300-9084(99)80110-2
Tian, F., Gong, J., Zhang, J., Zhang, M., Wang, G., Li, A., et al. (2013). Enhanced stability of thylakoid membrane proteins and antioxidant competence contribute to drought stress resistance in the tasg1 wheat stay-green mutant. J. Exp. Bot. 64, 1509–1520. doi: 10.1093/jxb/ert004
Wang, Y., Wang, Q., Zhao, Y., Han, G., Zhu, S. (2015). Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene 566, 95–108. doi: 10.1016/j.gene.2015.04.041
Welinder, K. G. (1992). Superfamily of plant, fungal and bacterial peroxidases. Curr. Opin. Struct. Biol. 2, 388–393. doi: 10.1016/0959-440X(92)90230-5
Wood, Z. A., Schröder, E., Robin Harris, J., Poole, L. B. (2003). Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 28, 32–40. doi: 10.1016/S0968-0004(02)00003-8
Wu, Y., Yang, Z., How, J., Xu, H., Chen, L., Li, K. (2017). Overexpression ofa peroxidase gene (AtPrx64) of arabidopsis thaliana in tobaccoimproves plant’s tolerance to aluminum stress. Plant Molec. Biol. 95, 157–168. doi: 10.1007/s11103-017-0644-2
Yan, J., Su, P. S., Li, W., Xiao, G. L., Zhao, Y., Ma, X., et al. (2019). Genome-wide and evolutionary analysis of the class III peroxidase gene family in wheat and aegilops tauschii reveals that some members are involved in stress responses. BMC Genomics 20, 666. doi: 10.1186/s12864-019-6006-5
Yao, Q., Cong, L., Chang, J. L., Li, K. X., Yang, G. X., He, G. Y. (2006). Low copy number gene transfer and stable expression in a commercial wheat cultivar via particle bombardment. J. Exp. Bot. 57, 3737–3746. doi: 10.1093/jxb/erl145
Zampieri, M., Ceglar, A., Dentener, F., Toreti, A. (2017). Wheat yield loss attributable to heat waves, drought and water excess at the global, national and subnational scales. Environ. Res. Lett. 12, 064008. doi: 10.1088/1748-9326/aa723b
Zhai, C. Z., Zhao, L., Yin, L. J., Chen, M., Wang, Q. Y., Li, L. C., et al. (2013). Two wheat glutathione peroxidase genes whose products are located in chloroplasts improve salt and H2O2tolerances in Arabidopsis. PloS One 8, e73989. doi: 10.1371/journal.pone.0073989
Zhang, H., Wang, Z., Li, X., Gao, X., Dai, Z., Cui, Y., et al. (2022). The IbBBX24-IbTOE3-IbPRX17 module enhances abiotic stress tolerance by scavenging reactive oxygen species in sweet potato. New Phytol. 233 (3), 1133–1152. doi: 10.1111/nph.17860
Zhou, S. M., Sun, X. D., Yin, S. H., Kong, X. Z., Zhou, S., Xu, Y., et al. (2014). The role of the f-box gene TaFBA1 from wheat (Triticum aestivum l.) in drought tolerance. Plant Physiol. Biochem. 84, 213–223. doi: 10.1016/j.plaphy.2014.09.017
Keywords: transcriptomics, drought tolerance, class III peroxidase, TaPRX-2A, ROS
Citation: Su P, Sui C, Niu Y, Li J, Wang S, Sun F, Yan J and Guo S (2023) Comparative transcriptomic analysis and functional characterization reveals that the class III peroxidase gene TaPRX-2A regulates drought stress tolerance in transgenic wheat. Front. Plant Sci. 14:1119162. doi: 10.3389/fpls.2023.1119162
Received: 08 December 2022; Accepted: 30 January 2023;
Published: 15 February 2023.
Edited by:
Xinyang Wu, China Jiliang University, ChinaReviewed by:
Xuan Chen, Xi’an University of Technology, ChinaCopyright © 2023 Su, Sui, Niu, Li, Wang, Sun, Yan and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peisen Su, cHNzdTIwMTRAMTYzLmNvbQ==; Shangjing Guo, Z3Vvc2hhbmdqaW5nQGxjdS5lZHUuY24=; Jun Yan, eGluc2luaWFuMjAwNkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.