
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 April 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1116214
This article is part of the Research Topic Abiotic and Biotic Stress in Horticultural Crops: Insight into Recent Advances in the Underlying Tolerance Mechanism View all 46 articles
 Caixia Li1†
Caixia Li1† Shaoyun Dong1†
Shaoyun Dong1† Diane M. Beckles2
Diane M. Beckles2 Xiaoping Liu1
Xiaoping Liu1 Jiantao Guan1
Jiantao Guan1 Xingfang Gu1
Xingfang Gu1 Han Miao1*
Han Miao1* Shengping Zhang1*
Shengping Zhang1*Low temperatures (LTs) negatively affect the percentage and rate of cucumber (Cucumis sativus L.) seed germination, which has deleterious effects on yield. Here, a genome-wide association study (GWAS) was used to identify the genetic loci underlying low temperature germination (LTG) in 151 cucumber accessions that represented seven diverse ecotypes. Over two years, phenotypic data for LTG i.e., relative germination rate (RGR), relative germination energy (RGE), relative germination index (RGI) and relative radical length (RRL), were collected in two environments, and 17 of the 151 accessions were found to be highly cold tolerant using cluster analysis. A total of 1,522,847 significantly associated single-nucleotide polymorphism (SNP) were identified, and seven loci associated with LTG, on four chromosomes, were detected: gLTG1.1, gLTG1.2, gLTG1.3, gLTG4.1, gLTG5.1, gLTG5.2, and gLTG6.1 after resequencing of the accessions. Of the seven loci, three, i.e., gLTG1.2, gLTG4.1, and gLTG5.2, showed strong signals that were consistent over two years using the four germination indices, and are thus strong and stable for LTG. Eight candidate genes associated with abiotic stress were identified, and three of them were potentially causal to LTG: CsaV3_1G044080 (a pentatricopeptide repeat-containing protein) for gLTG1.2, CsaV3_4G013480 (a RING-type E3 ubiquitin transferase) for gLTG4.1, and CsaV3_5G029350 (a serine/threonine-protein kinase) for gLTG5.2. The function for CsPPR (CsaV3_1G044080) in regulating LTG was confirmed, as Arabidopsis lines ectopically expressing CsPPR showed higher germination and survival rates at 4°C compared to the wild-type, which preliminarily illustrates that CsPPR positively regulates cucumber cold tolerance at the germination stage. This study will provide insights into cucumber LT-tolerance mechanisms and further promote cucumber breeding development.
Cucumber (Cucumis sativus L.), as one of the most economically important vegetable crops, is widely cultivated on ~2.3 Mha globally and its production reached 91.26 M metric tons in 2020 (FAO, 2020). Cucumber, however, is a temperature-sensitive crop that is easily injured by low temperatures (LTs) (Cabrera et al., 1992). LT is one of the most common abiotic stresses; it can affect cucumber seed architecture and germination, slow the growth rate, delay seed maturation, decrease the rate of seed set, and even indirectly affect reproduction, which collectively threatens cucumber production and quality (Nienhuis et al., 1983; Røeggen, 1987). When cucumber seeds encounter LTs, either in the soil or in irrigation water, the percentage of seeds that germinate decreases, and more seriously, LTs can cause seed death (Smeets and Wehner, 1997). Clarifying the genetic basis of cucumber LT germinability, will enable molecular breeding vital to improving yield in cold regions, as well as extending the cultivated area from low- to high-altitudes.
Previous studies reported that low temperature germination (LTG) is a complex quantitative trait loci (QTL) commonly controlled by multiple genes (Kłosińska et al., 2013). Germination rate (GR), germination energy (GE), germination index (GI), and radicle length (RL) were the common used LTG evaluation indices (Song et al., 2018; Yagcioglu et al., 2019). To date, only three QTL analyses were performed in cucumber using these indices. First, Dong (2017) identified eight QTLs using F2:3 families. Next, three QTLs (qLTG1.1, qLTG2.1 and qLTG4.1) were detected using Recombinant Inbred Lines (RILs) in 2018 (Song et al., 2018; Li et al., 2022a). Subsequently, Yagcioglu et al. (2019) also identified three QTLs (qLTG1.2, qLTG2.1 and qLTG4.1) using RILs and F2:3 families. Collectively these studies provided important information on LTG-related genetics and accelerated the development of cucumber breeding programs.
A genome-wide association study (GWAS) is an effective way to identify complex quantitative traits, and it has been widely used in crops. In rice, Sales et al. (2017) showed that 31 markers were related to growth rate and germination rate at 25°C and 15°C using 200 diverse germplasms and 1672 single nucleotide polymorphisms (SNPs). Schläppi et al. (2017) evaluated 202 core germplasms and identified two new loci, i.e., qLTSS3-4 and qLTSS4-1, using seedling survival rate as an evaluation index under LT. A member of the 14-3-3 gene family (GF14h), was associated with changes in the germination rate of rice under optimal temperature conditions using GWAS (Yoshida et al., 2022). In sorghum, based on five evaluation indicators (stem weight, root weight, stem length, root length and anthocyanin content) at LT, transcription factors Sb06g024820 and Sb05g001215 were identified as candidate genes capable of regulating LT responses (Chopra et al., 2017). In maize, Zm00001d017932, a MADS-transcription factor 26 (MADS26), were verified as affecting seed germination via an association GWAS analysis of 300 lines and 24 transcriptome libraries analysis (Ma et al., 2022). In cotton, using GWAS mapping with 200 accessions, Gh_A01G1740 (GhSAD), which encodes a short-chain alcohol dehydrogenase, was found to confer cold tolerance (Ge et al., 2022). Further, Gh_D09G0189 (GhSAL1), a bifunctional protein, was identified as affecting cold tolerance in upland cotton at the seedling emergence stage, using 200 accessions from five ecological distributions (Shen et al., 2023). With the development of cucumber genome sequencing, GWAS has also been widely used to accelerate genes related to LTG. GWAS has also been successfully used to detect many QTLs in cucumber, for example, for biotic and abiotic stress resistance, and for fruit quality traits. For biotic stress resistance, novel powdery mildew (PM) resistance genes, i.e., Csa5G453160 and Csa5G471070, found on chromosome 5 were detected using a set of 264 from a core germplasm collection (Lee et al., 2020). Meanwhile, eight PM-resistance, five downy mildew resistance and four gummy stem blight resistance genes were putatively identified (Liu et al., 2020b; Liu et al., 2021; Han et al., 2022). For abiotic stress, Zhang et al. (2019) evaluated heat tolerance with a collection of 96 accessions and found two loci - GGI4.1 and GGI5.1 using GR, GE and GI. Wei et al. (2019) detected seven GWAS locus (gHII4.1, gHII4.2, gHII5.1, gHII5.2, gHII6.1, gHII6.2 and gHII7.1) using a heat injury index. Wang et al. (2019) detected five LT-tolerance loci (gLTS1.1, gLTS3.1, gLTS4.1 and gLTS5.1) on Chr.1, Chr.3, Chr.4 and Chr.5, respectively using low temperature injury indices. Seven loci (gST2.1, gST3.1, gST4.1, gST4.2, gST5.1, gST6.1) associated with salt tolerance in cucumber seedlings were repeatedly detected (Liu et al., 2022a). For quality traits, the ultra-high fruit spine density formation genes (fsd6.2) (Bo et al., 2019a) and the green flesh formation gene (qgf5.1 and qgf3.1) (Bo et al., 2019b) were identified using a group of 115 cucumber accessions from a core germplasm collection. However, there have been no reported studies investigating LTG tolerance using the GWAS approach.
In this study, we uncovered the genetic architecture of LTG in re-sequenced lines of a 151 core cucumber germplasm population which represented genotypes of diverse origins and ecotypes. Low temperature tolerance at the cucumber germination stage was assessed via four indices and 1,522,847 SNP markers. Three loci related to LTG were detected in two different environments by GWAS. Moreover, three novel candidate genes were discovered and assessed via functional annotation, sequence alignment and expression analysis. Finally, the CsaV3_1G044080 gene within the gLTG1.2 locus, encoded a pentatricopeptide repeat-containing protein (CsPPR) was further verified; When ectopically expressed in Arabidopsis, it engendered better LTG in the transgenic lines. The objectives of this study are to (i) enrich the diverse genetic variation and screen low temperature tolerance accession; (ii) provide insights into the genetic basis of cucumber LTG tolerance, as well as resources for the genetic improvement of cucumber; and (iii) identify the SNP markers and the causal gene affecting seed germination ability at LT in cucumber. This study will provide a basis for elucidating the regulatory mechanisms underlying LT-tolerance in cucumber at the germination stage, and accelerate the screening of LT-tolerant varieties.
A set of 151 accessions from different geographical origins (Table S1), was used for the GWAS analysis, and was provided by the cucumber research group at the Institute of Vegetables and Flowers, at the Chinese Academy of Agricultural Sciences in China. The 151 diverse plants genotypes were grown, each line includes 5 individuals and self-pollinated under natural field conditions in the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Science at Beijing in 2018 (39°90N, 116°30E; average day/night temperatures 26/14°C; day length ~14 h in May and June). The individuals of each line were collected, and mixed seeds were further used for phenotype identification.
Accessions of the 151 cucumber core germplasms were phenotyped in two different environments: a phytotron in Beijing in summer of 2019 (2019S) and an incubator in Shandong (36°51N, 118°50E) in Shandong province in summer of 2020 (2020S) (Table S1). Fifteen seeds per triplicate were placed at 13°C for 14 days in the dark and at 28 °C for 7 days as control (Kłosińska et al., 2013). For each experiment, three replicates were applied. Each replicate was randomly placed and the core germplasm collection was randomly placed in each replicate (block). Fifteen seeds per line were placed in a 90-mm-diameter Petri dish containing two layers of filter paper soaked 6 h with 30 ml 55°C water, then were exposed to 6 ml of distilled water. Seed germination (radicle ≥ 2 mm long) was recorded daily during the treatment stage (14 days for low temperature and 7 days for control) and radical length were measured at 14 day for low temperature and 7 day for control by Image J (Song et al., 2018). The relative value of four evaluated indices, including relative germination rate (RGR), relative germination energy (RGE), relative germination index (RGI) and relative radical length (RRL) were calculated between 13°C and 28°C (Song et al., 2018). The four indices, i.e., RGR, RGE, RGI and RRL were used for GWAS analysis.
Statistical analysis of the phenotypic data was implemented using SAS v.9.4 (SAS Institute Inc., Cary, NC, USA, 2012). Pearson’s liner correlation coefficients were found among the four indices, and were analyzed using R software (R Core Team, 2019). An analysis of variance (ANOVA), principal component analysis (PCA) analysis was also performed in SAS v.9.4 (SAS Institute Inc., Cary, NC, USA, 2012). Membership function and comprehensive evaluation values (D-value) were analyzed in Excel 2016 (Xie, 1983; He, 2006). Principal component Wj = Ij/∑Ij (j = 1, 2, …, n), where Ij represented the contribution rate of the jth principal component; Membership function value (Xj) = (Xj - Xmin)/(Xmax-Xmin) (j = 1, 2, …, n), where Xj represented the jth principal component value, Xmin and Xmax represented the minimum and maximum value of the jth principal component in different germplasms, respectively; Comprehensive evaluation values D = ∑ (Uj × Wj) (j = 1, 2, …, n). The D-value as a comprehensive evaluation index was used to represent the LT resistance of various germplasm. The D-value was used as an input for cluster and GWAS analysis. The sum of squares of dispersion (Ward) method was used for cluster analysis and the Euclidean distance method was used to measure in R software (R Core Team, 2019) based on the average D-value of each core germplasms in two experiments.
The re-sequenced genome data and SNPs of the 151 accessions of the core germplasm collection are available in the NCBI Database (PRJNA831637). A FaST-LMM model (Jiang et al., 2021) was used for association tests of LT tolerance with an estimated relatedness matrix as covariate. All of the 151 genotypes were included in the GWAS. P=1.0 × 10−6 was used as the significant threshold. The linkage disequilibrium (LD) decay coefficients (r2) between high-density SNPs was calculated using Plink software (Purcell et al., 2007), and was used to evaluate LD decay (Liu et al., 2020b). Association mapping of LTG traits using SNP genotyping, population structure, relative kinship and LD analysis were identified (unpublished). Manhattan plots and the LD heatmaps were drawn by qqman package (Turner, 2014) in the R environment.
The candidate locus was identified by repeatedly detected loci of various indices in two environments. The regions with significant trait-associated SNPs were described as candidate regions by unifying a set of various indices. The annotations of candidate genes in QTL regions were obtained based on “cucumber genome V3 (‘9930’ as reference genome) (http://cucurbitgenomics.org/organism/20)” (Li et al., 2019). The candidate genes were identified by the following steps: first, many genes related to abiotic stresses were predicted using Swiss-Prot (https://www.uniprot.org/)、TAIR (https://www.arabidopsis.org/index.jsp) and the gene ontology (GO) database (http://amigo.geneontology.org/amigo/landing). Secondly, SNPs in the exons or promoters of different genes, that differentiated the 47 LT-sensitive lines from the 21 LT-resistant lines were identified based on sequence alignment using MEGA X (Kumar et al., 2018). Finally, the expression level of these candidate gene was analyzed at varying time points, between germplasm differing in their LTG response, to further identify candidate genes.
Promoter (the 2.0 kb upstream of ATG) were used for searching regulatory elements of the CsPPR gene using PlantCARE online (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). And the result was visualized by TBtools (Chen et al., 2020). Cucumber orthologs of the Arabidopsis PPR proteins were identified using the Cucurbit (http://cucurbitgenomics.org/feature/gene/CsaV3_1G044080) and Arabidopsis databases (https://www.arabidopsis.org/servlets/TairObject?type=locus&name=AT4G04370). The functional interaction networks of proteins were identified by the online STRING database (https://cn.string-db.org/cgi/network?taskId=baqwil7ulR5D&sessionId=bZBjkvzPG61G).
A time-course expression analysis of the four sensitive lines (‘R13’, ‘R42’, ‘R77’, ‘R137’), together with the four resistant lines (‘R99’, ‘R152’, ‘R167’, ‘R174’) were examined during seed germination. Cucumber seeds exposed to 13°C in an incubator for 0 d,12 h, 1 d, 3 d and 7 days were collected. There were three biological replicates for each treatment. For Arabidopsis thaliana, Columbia-0 ecotype (wild-type) Col-0 and five T1 transgenic lines were grown 1/2 MS with hygromycin agar medium. Ten-days-old seedlings were collected for RNA extraction.
Total RNA was extracted, and the first-strand complementary DNA (cDNA) was synthesized using RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and a PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa, Kyoto, Japan), respectively, according to the manufacturer’s instructions. Quantitative reverse transcription PCR analysis (qRT-PCR) was performed using SYBR Premix Ex TaqTM II (TaKaRa, Kyoto, Japan). The PCR primers (Table S2) were designed with DNAMAN7.0 (Woffelman, 2004). Actin1 (Csa3G806800) was employed as the reference gene to normalize gene expression values (Xie et al., 2018). Each qRT-PCR experiment was performed with three biological replicates. The analysis of gene relative expression data was performed using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Significant differences between different materials were detected using two-tailed, two-sample Student’s t-test (p<0.05 or p<0.01) in Excel (2016).
Seeds of A.thaliana Col-0 ecotype was obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/). To obtain transgenic lines in which CsPPR was driven by the 35S promoter, the coding sequence of CsPPR was amplified from cucumber and subsequently cloned to the BamHI and PmI I sites of the pCAMBIA1305.4 vector to generate the 35S::CsPPR recombinant plasmid. Agrobacterium tumefaciens strain GV3101 carrying 35S::CsPPR plasmid was used to transform A.thaliana ecotype Columbia (Col-0) plants using the floral dip method (Clough and Bent, 1998). Transgenic seedlings were selected for their resistance to 40 mg/mL hygromycin. The resistant T1 transgenic seedlings were selected and propagated and T2 homozygotes selected for further analysis. The expression of CsPPR were compared between the transgenic plants with Col-0 using qRT-PCR.
Seeds of the Col-0 and transgenic plants were harvested at the same time from plants grown under the same conditions. For LT treatment, 50~100 seeds of each genotype were planted on the same 1/2MS medium and exposed to 4°C for 40 days under a 16 h-light/8-h dark cycle. At 4°C for 20 days, the number of seedlings that germinated was recorded, and the germination rate calculated. An obvious emergence of the radicle through the seed coat was used to indicate seed germination. At 4°C for 40 days, the number of surviving individuals was recorded, and the survival rate calculated. For the non-chilling control, treatment and analyses was identical to that described for the chilling treatment, except that the seeds were stored at 22°C for 5 days.
Phenotypic data related to the germination of 151 cucumber accessions of the core germplasm collection were obtained from two environments daily. From these data, four indices, i.e., RGR, RGE, RGI and RRL, were calculated (Table S3). Seedlings were tested in phytotrons in the summer of 2019 and in 2020, i.e., 2019S and 2020S, in Beijing and Shandong province respectively. The phenotypic data showed diverse variation among germplasms (Figure 1). RGR, RGE, RGI and RRL exhibited values of 0-100%, 0-96.59%, 0-75.20% and 0-1.37%, with average values of 29.01%, 21.07%, 16.24%, 0.35%, respectively (Figures 2A–D; Table 1). There was a clear bimodal distribution of RGR, RGE, RGI and RRL among the accessions, indicating that these traits were quantitative in nature, and were controlled by multiple genes (Figures 2E–H). In total, seven cucumber ecotypes were evaluated, including the Indian (n = 3 lines), Japanese (n = 6 lines), American (n = 13 lines), European (n = 17 lines), South China (n = 21 lines), Eurasian (n = 21 lines), and the North China ecotypes (n = 70 lines) (Table S3). Three market-types have higher LT resistant in the two environments, e.g., the North China, the Eurasian, and the South China types (Figures 2I–L). The coefficients of variation (CVs) of RGR, RGE, RGI and RRL were 112.05, 130.03, 119.89, and 117.12%, respectively (Table 1). Significant (P < 0.001) positive correlations were observed between each pair of the four traits, i.e., RGR, RGE, RGI, and RRL, when tested in both environments, with the correlation coefficients ranging from 0.56 (RGE_2020S and RRL_2019S) to 0.97 (RGR_2019S and RGE_2020S) (Figure S1). RGR, RGE, RGI in 2019S and 2020S showed high positive correlations, ranging from 0.62-0.97 (Figure S1). These findings indicate that the four traits probably exert a synergistic effect on cucumber seed germination at LT. Meanwhile, data collected from each of the two years were highly correlated and consistent, providing a solid foundation for GWAS analysis.
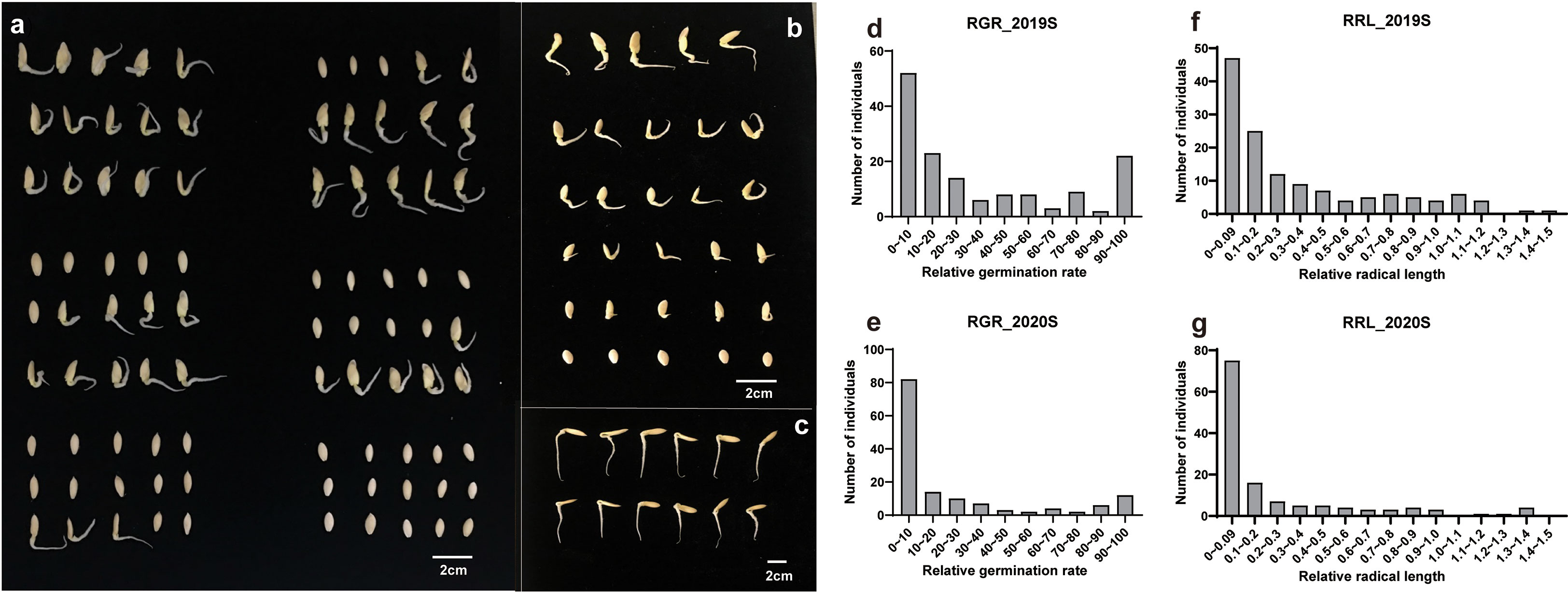
Figure 1 Differences in germination rate at low temperature stress among accessions from the core germplasm. (A) Germination rate and (B) radical lengths of accessions from the core germplasms incubated at 13°C. (C) Germination under control, i.e., 28°C, temperatures. Bars = 2 cm. (D, E) Relative germination rate and (F, G) relative radical length in 2019S and 2020S.
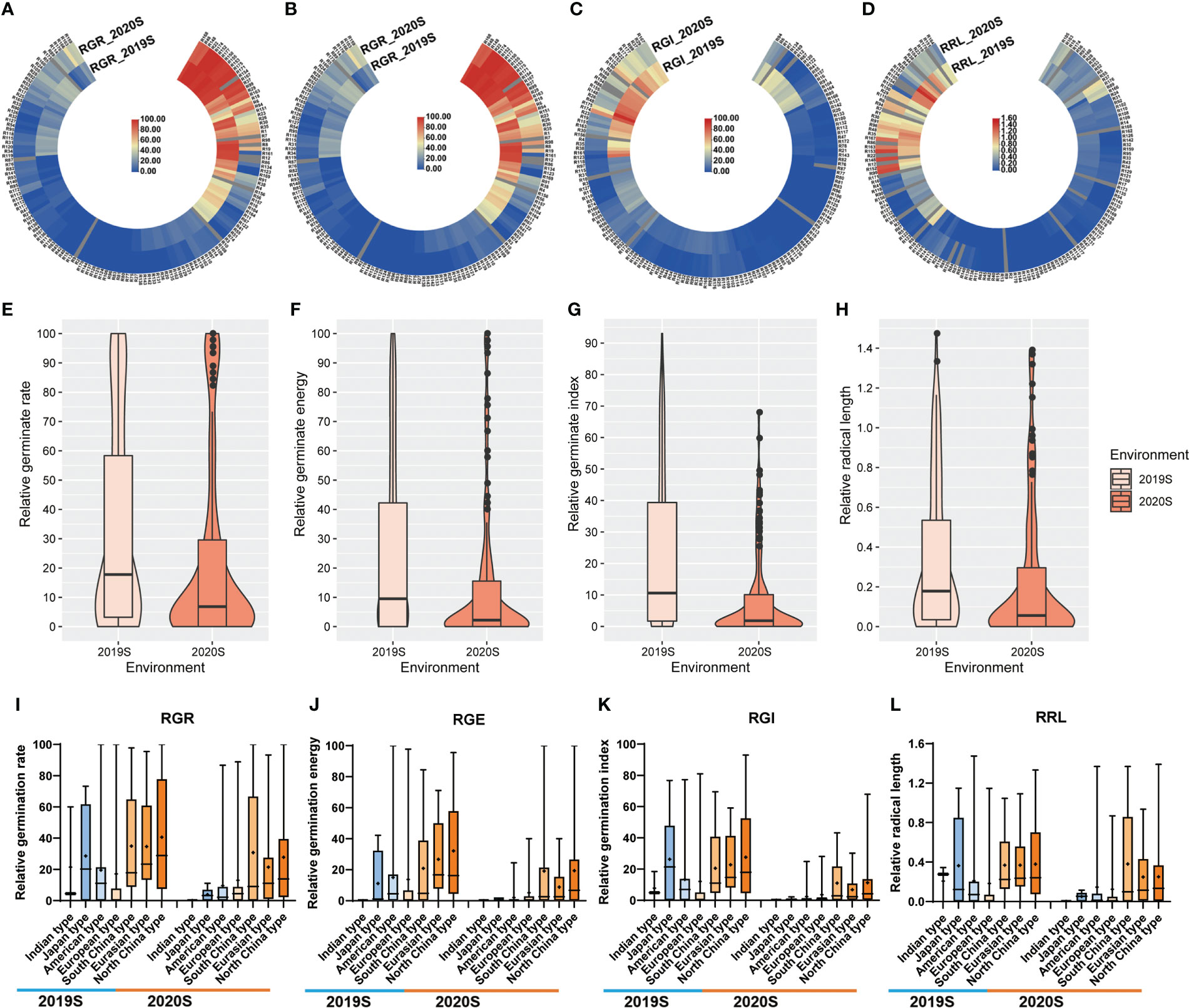
Figure 2 Heatmap, violin and box plots showing the phenotypic distribution of RGR, RGE, RGI and RRL among cucumber accessions grown under two environments. (A–D) Heatmap depicting the repeatability of the four germination indices, i.e., RGR, RGE, RGI and RRL. Blue and red bars represent accessions sensitive and tolerant to LTs, respectively, with the color intensity indicating the level of sensitivity or tolerance. (E–H) Violin and box plots depicting the distribution of the germination indices in two experiments. (I–L) Box plots depicting the distribution of the germination indices in seven ecotypes. Key: RGR, relative germination rate; RGE, relative germination energy; RGI, relative germination energy; RRL, relative radical length.
To evaluate accurately the LT tolerance ability of core germplasms, four principal (prin) components were identified by principal component analysis. Prin1 and prin2 represented 97.59% in 2019S and 98.07% in 2020S of the four indices (Table S4). Weight of prin1 was 95.98%, while prin2 was 4.02% by membership function (Table S4). Finally, a comprehensive evaluation (D-value) of the germination ability of the core germplasm, that encompassed all four germination indices, was calculated. The line with bigger D-value was more resistant for LT. Based on the average D-value per line of two environments, these core germplasms were divided into 5 clusters (I, II, III, IV, V) by the Ward method, when Euclidean distance was 13.5 (Figure 3A). Cluster I consisted of highly sensitive accessions, those in Cluster II were sensitive, while Clusters III, IV and V consisted of accessions that were intermediate in tolerance, tolerant, and highly-tolerant respectively (Table S5; Figure 3A). Each cluster contained different ecotypes (Figure 3C). The higher the D-value, the higher the LT resistance of the germplasms (Figure 3B). Significant differences were found among various groups (Figure 3B). In general, most accessions in the Indian, Japanese and European groups were sensitive to LT, while those in the Chinese group showed higher tolerance (Figure 3C). However, the LTG of some germplasms differed greatly among experiments, e.g., the ‘R161’ line grouped in the highly resistant cluster in 2019S, but showed susceptibility to LTs in 2020S (Figures 2A–D). Thus, we combined all the phenotypic data from the two experiments to differentiate the susceptible from the resistant lines. Of the 151 tested accessions, we could define 47 as sensitive, i.e., belonging to the highly sensitive group: ‘R13’, ‘R42’, ‘R77’, and 21 as resistant, i.e., belonging to the highly tolerant group: ‘R99’, ‘R152’, ‘R167’.
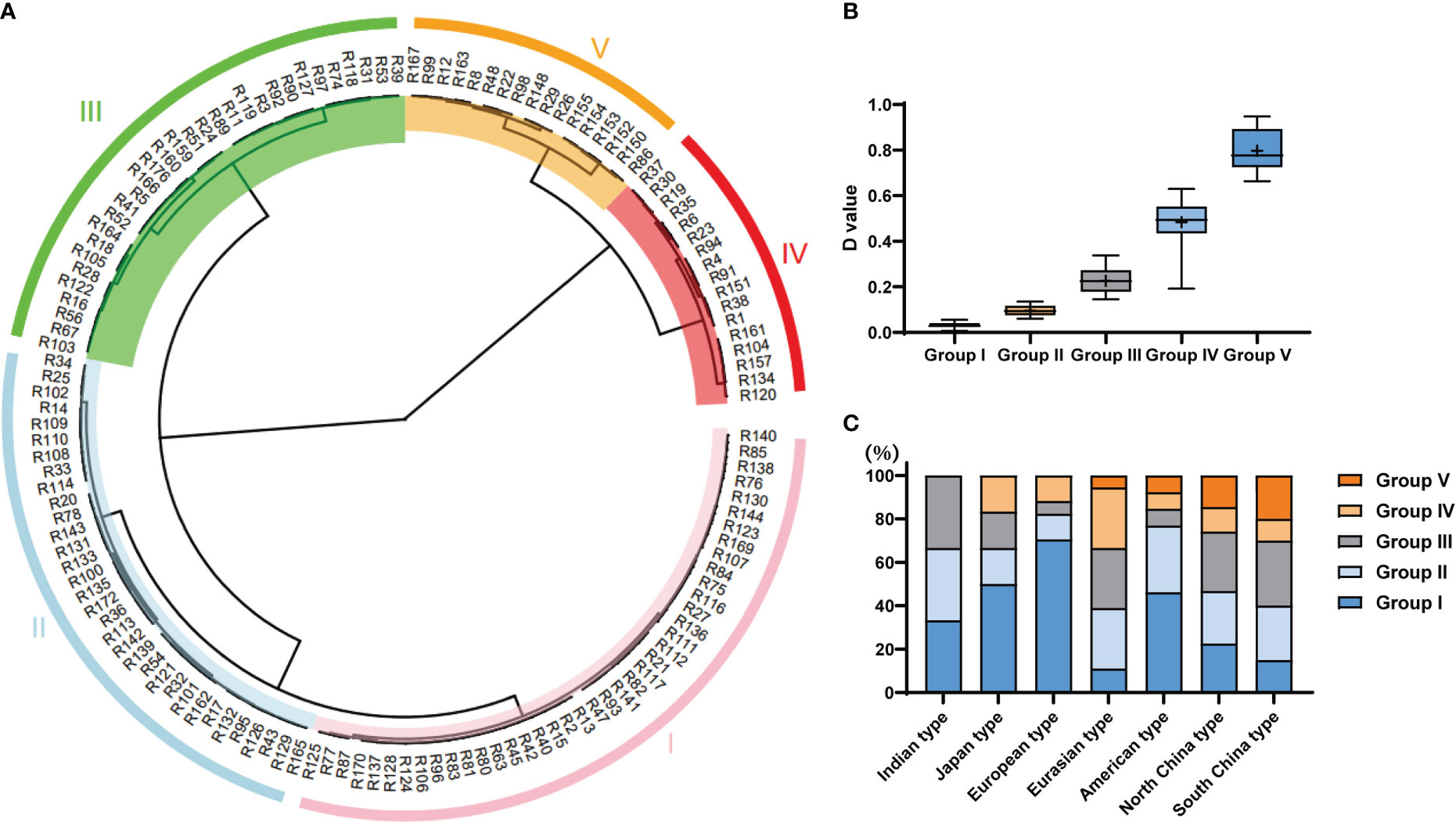
Figure 3 Analysis of the cucumber accessions based on their tolerance to germination at low temperatures. (A) Clustering of accessions of the core germplasm collection using the comprehensive D-value to indicate germination at low temperature. The I, II, III, IV and V indicate group I, group II, group III, group IV, and group V, respectively. (B) The range of D-values from Group I to Group V. (C) The proportion of Group I to Group V of D-values found within the different ecotypes.
The various index (D-value, RGR, RGE, RGR and RRL) were used to repeatedly detect loci at the threshold of 5.0 across all 7 chromosomes with well-fitted quantile-quantile (Q-Q) plots in two environments. SNP loci distributed on chromosomes 1, 4, 5 and 6 were detected, and were named gLTG1.1, gLTG1.2, gLTG1.3, gLTG4.1, gLTG5.1, gLTG5.2 and gLTG6.1, respectively (Figure 4, Figure S2, Table 2).
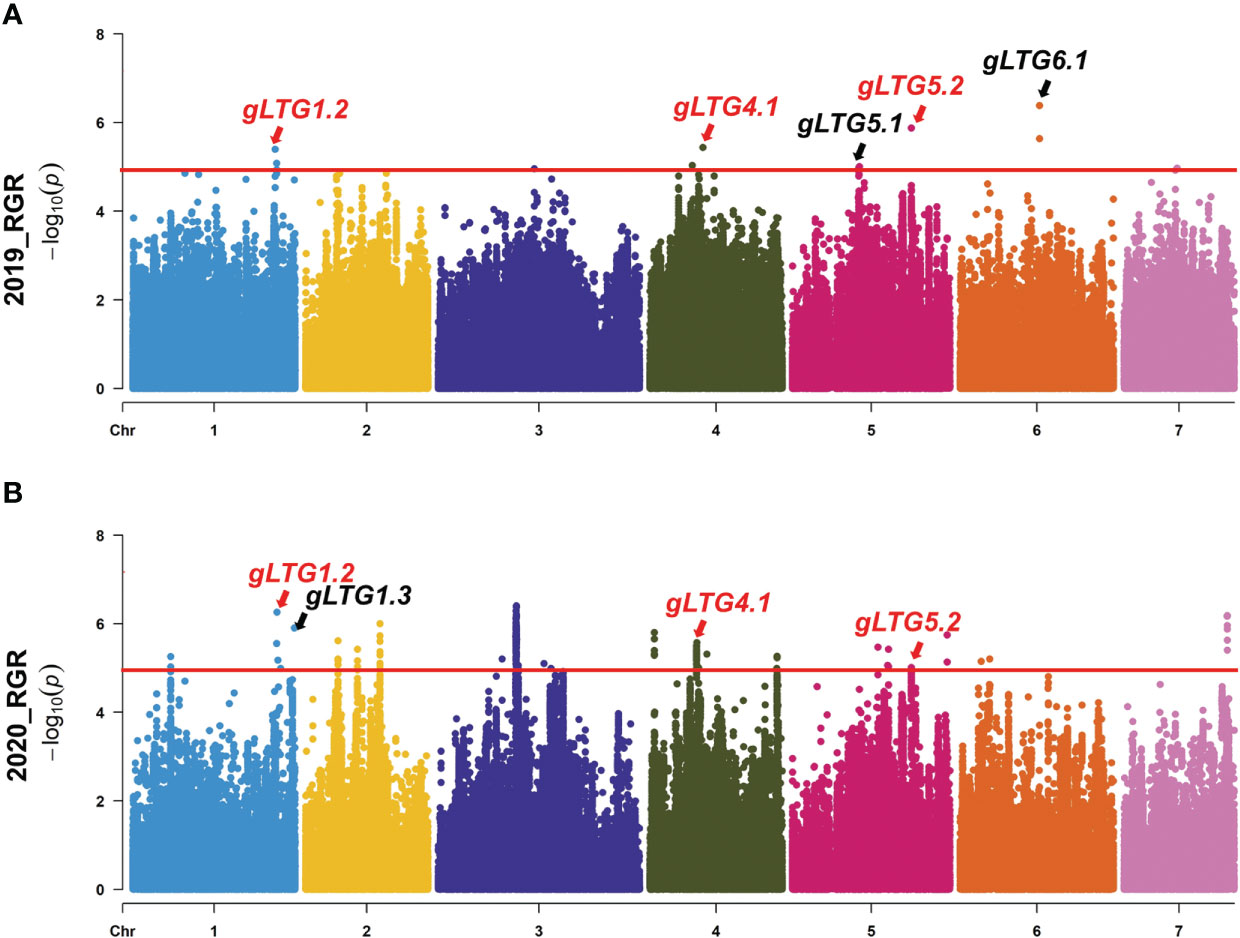
Figure 4 Genome-wide association analysis (GWAS) Manhattan plots of (A) RGR in 2019 (B), and RGR in 2020. The red horizontal lines indicate the significance threshold (P < 10−6). Arrow heads indicate the position of strong peaks.
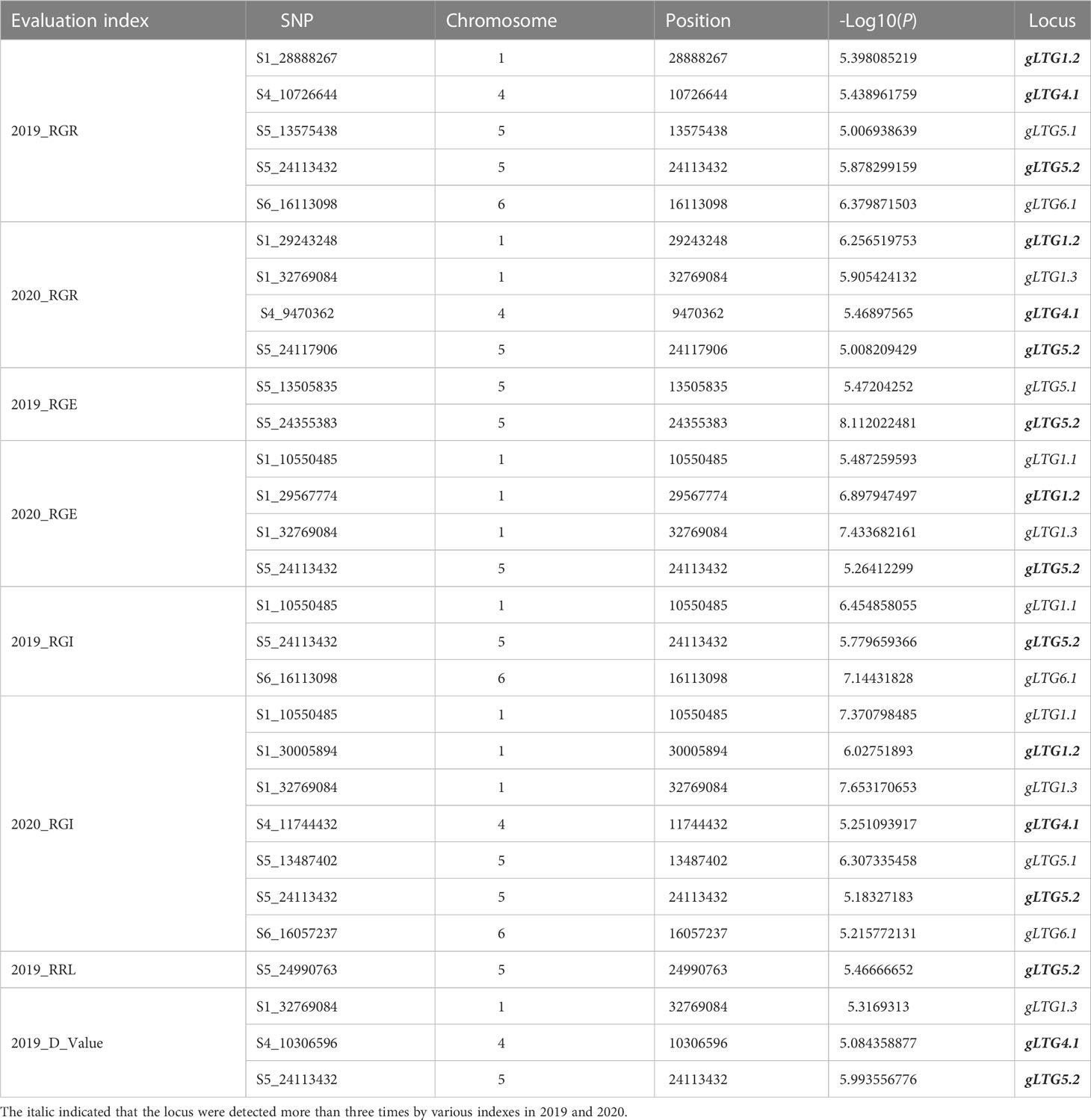
Table 2 The highly significant SNPs by GWAS in gLTG1.1, gLTG1.2, gLTG1.3, gLTG4.1, gLTG5.1, gLTG5.2 and gLTG6.1 in two environments.
Of these seven loci, gLTG5.2 was detected eight times, and two loci (gLTG1.2 and gLTG4.1) were detected four times, using the four indices in each of the two experiments, respectively. These loci were thus considered to have a stable effect on LTG (Table S6). For the rest, four loci (gLTG1.1, gLTG1.3, gLTG5.1, and gLTG6.1) were detected in three times. By comparing the SNP (single nucleotide polymorphism) positions of these 7 loci with QTLs reported in previous studies (Table 2), it was evident that one (gLTG6.1) of the 7 loci in this study colocalized with the known ones (Figure 5), which indicating the locus were reliable via GWAS.
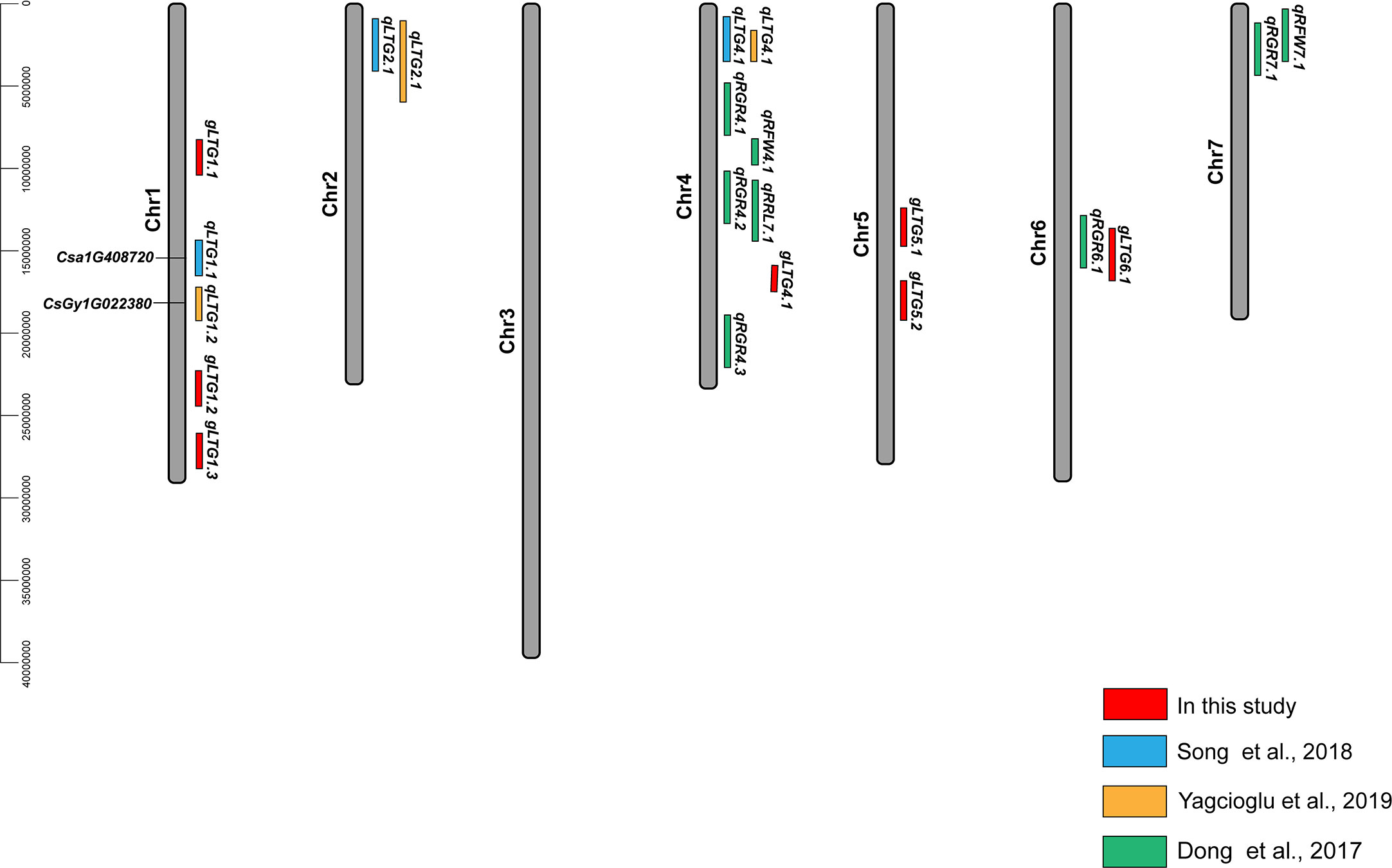
Figure 5 Locations of QTLs for LTG in cucumber reported in this study and previous studies (2015–2022). The red font indicates the loci of this study and their exact SNP locations can be found in (Table S5). The black font indicates candidate gene identified in previous studies (Li et al., 2022a).
The novel and stable loci on chromosomes 1 (gLTG1.2), 4 (gLTG4.1) and 5 (gLTG5.2) that exhibited significantly association with LT germinability (Figure 4 S2; Table S6). So, their candidate regions were further analyzed using LD decay around the peak SNPs, haplotype analysis, functional annotation of the orthologs in Arabidopsis and qRT-PCR. Given that all of them were detected in the GWAS for RGR, therefore RGR is the best choice to further analyze coincidence rate between genotype and loci SNP. Based on RGR in two environments, 21 resistant lines (Group V and IV) and 47 sensitive lines (Group I and II) were used for haplotype analysis (Table S7; Figure S3). And ‘R13’, ‘R42’, ‘R77’, ‘R137’ of LT sensitive group (Group I) and ‘R99’, ‘R152’, ‘R167’, ‘R174’ of LT tolerance group (Group V) were used for expression analysis (Figure 3A).
For the novel and stable gLTG4.1 locus, the candidate region on Chr.4 was analyzed by pairwise LD correlations leading to a region from 9,703,054 bp to 9,849,931 bp by LD block (Figure 6A). Ten annotated genes are located in this region, four of which are associated with different abiotic stresses (Figure 6B, Table S8). CsaV3_4G013370, encodes an enzyme with ATPase and ADPase activity (an apyrase), that are involved in stress-responses in Arabidopsis (Kim et al., 2009); CsaV3_4G013420, encodes a NAC-domain containing protein, which has a role in seed germination (Dekkers et al., 2013) and plant abiotic stress responses (Nakashima et al., 2012); CsaV3_4G013430, encodes a pentatricopeptide repeat-containing (PPR) protein; while CsaV3_4G013480, encodes a RING-type E3 ubiquitin transferase, and takes part in ABA-mediated stress responses (Nakashima et al., 2012). Of these candidate genes, only CsaV3_4G013480 was of interest since five SNPs – three in the exon and two in the intron, were found. Two of the three SNPs are predicted to lead to non-synonymous substitutions (Figure 6C), i.e., a Ser →Pro and a Gln→ Glu respectively (Figure 6C). We also found that 62% of the 47 sensitive lines carried the Hap1 “CCGCT” sequence, while 86% of the 21 resistant lines had the alternate Hap2 “GGAGA” motif (Figure 6D). Interestingly, CsaV3_4G013480 expression was highly induced 1 d after LT treatment in the susceptible lines ‘R13’ and ‘R42’ while in contrast, its expression was lower in the resistant lines ‘R99’ and ‘R152’ (Figure 6E). Moreover, after 3 d at 13°C, CsaV3_4G013480 expression in the susceptible lines (‘R13’ ‘R42’ ‘R77’ ‘R137’) was significantly higher than that in the resistant lines (‘R99’ ‘R152’ ‘R167’ ‘R174’) (Figure 6F). It is possible that haplotype and the expression of this gene might play a role in cucumber LTG response.
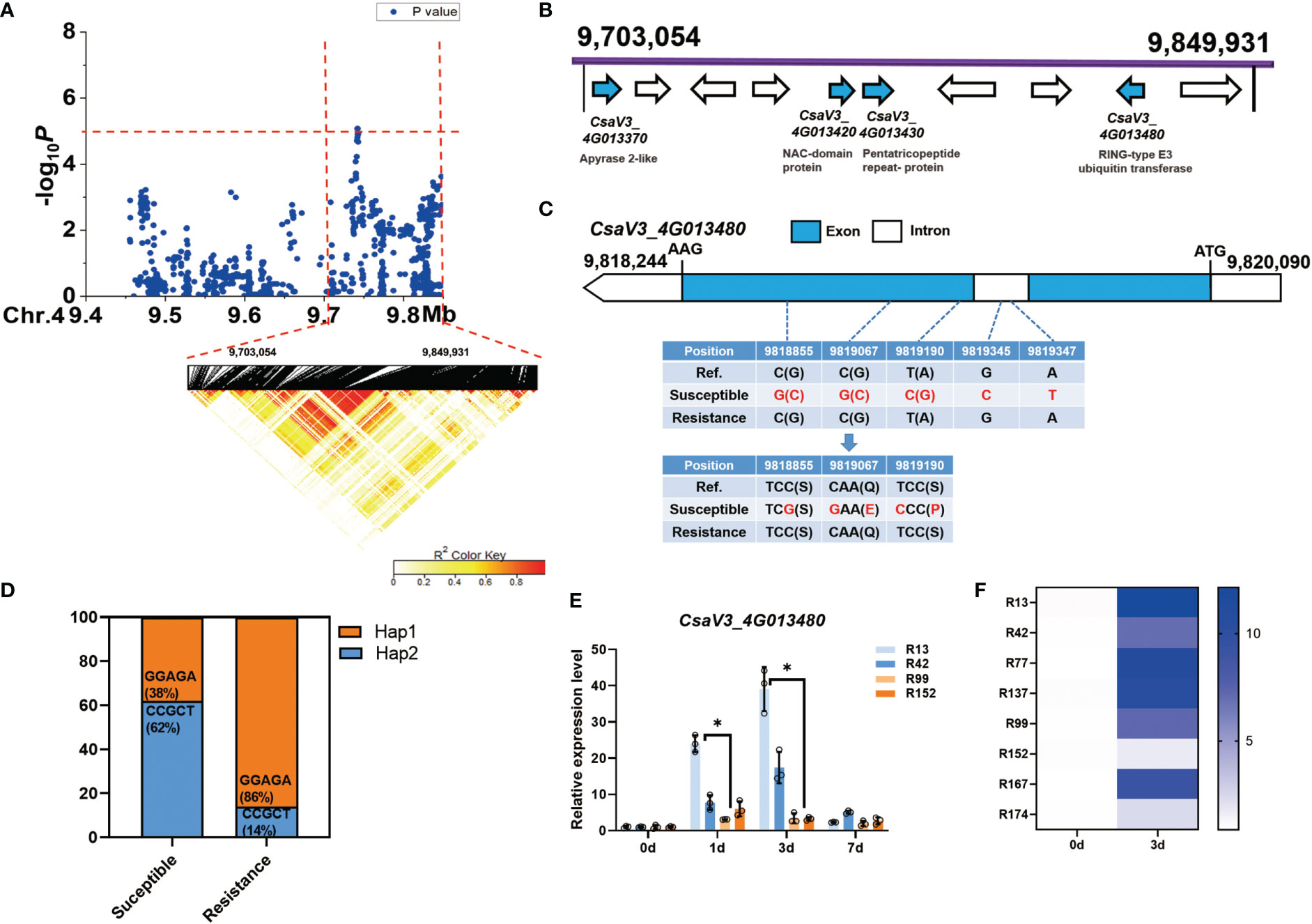
Figure 6 Identification of cucumber low-temperature germination trait-associated gene CsaV3_4G013480. (A) Local Manhattan plot (top) and LD heatmap (bottom) surrounding the peak on chromosome 4. The dashed line indicates the significance threshold (P < 10−6). Dashed lines indicate the candidate region (~ 146.8 kb) for the peak. (B) Ten functionally annotated genes were predicted in the gLTG4.1 candidate region. The arrows represent the direction of the genes. The genes shown as blue arrows are related to abiotic stress. Gene annotation is listed below. (C) Gene structure of CsaV3_4G013480. Blue and white rectangles indicate exons and introns, respectively. (D) Orange represents the proportion of accessions that are Hap1 (GGAGA) while blue represents the proportion that are Hap2 (CCGCT) among the susceptible and resistant lines. (E) The expression level of CsaV3_4G013480 in the LT-sensitive lines (‘R13’, ‘R42’) and LT-resistant lines (‘R99’, ‘R152’) after LT treatment (0, 1, 3 and 7 d), was tested through qRT–PCR. Actin was used an internal control. Data are represented as average values with the SD of three independent biological replicates. (F) The heatmap indicates the relative expression level of CsaV3_4G013480 in LT-sensitive lines (‘R13’, ‘R42’, ‘R77’, ‘R137’) and LT-resistant lines (‘R99’, ‘R152’, ‘R167’, ‘R174’) after LT treatment (0 d and 3 d), tested through qRT–PCR. Actin was used an internal control. *Significant difference (P <0.05).
For the novel and stable gLTG5.2 locus, the common peak SNPs were further analyzed and a candidate region from 24,329,867 bp to 24,478,603 bp on Chr.5 was estimated using pairwise LD correlations (r2 ≥ 0.6) (Figure 7A). In this region, 17 annotated genes were found, and five candidate genes related to abiotic stress identified (Figure 7B, Table S8), they include CsaV3_5G029350 (a serine/threonine-protein kinase), CsaV3_5G029450 (a zinc finger protein), CsaV3_5G029520 (A type I inositol polyphosphate 5-phosphatase), CsaV3_5G029540 (a cysteine proteinase inhibitor), CsaV3_5G029550 (a cytochrome P450). Nevertheless, 84 SNPs significantly associated with LTG in the gene CsaV3_5G029350 (Figure 7D). Two SNPs in the third exon caused a base change from A-to-T and from A-to-G, which resulted in amino acid change from Thr→Ser and Ile→Val, respectively (Figure 7D). Two SNPs located in the fourth exon, led to a synonymous mutation. All 47 sensitive lines were (Hap1), carried the haplotype “AAGG” while 24% of the resistant lines had the “TGAA” (Hap2) (Figure 7E). However, the SNP variation in the resistant lines were not consistent with the phenotype, indicating that cucumber LT germinability could be controlled by multiple genes (such as gLTG1.2, gLTG4.1, and gLTG5.2). The relative expression of CsaV3_5G029350 was higher than that of the susceptible lines ‘R13’, ‘R42’ at 1 d at 13°C (Figure 7F). In addition, the expression level of CsaV3_5G029350 in the susceptible lines (‘R99’ ‘R152’ ‘R167’) was significantly higher than in the susceptible resistant lines (‘R42’ ‘R77’ ‘R137’) after 3 d at 13°C (Figure 7G).
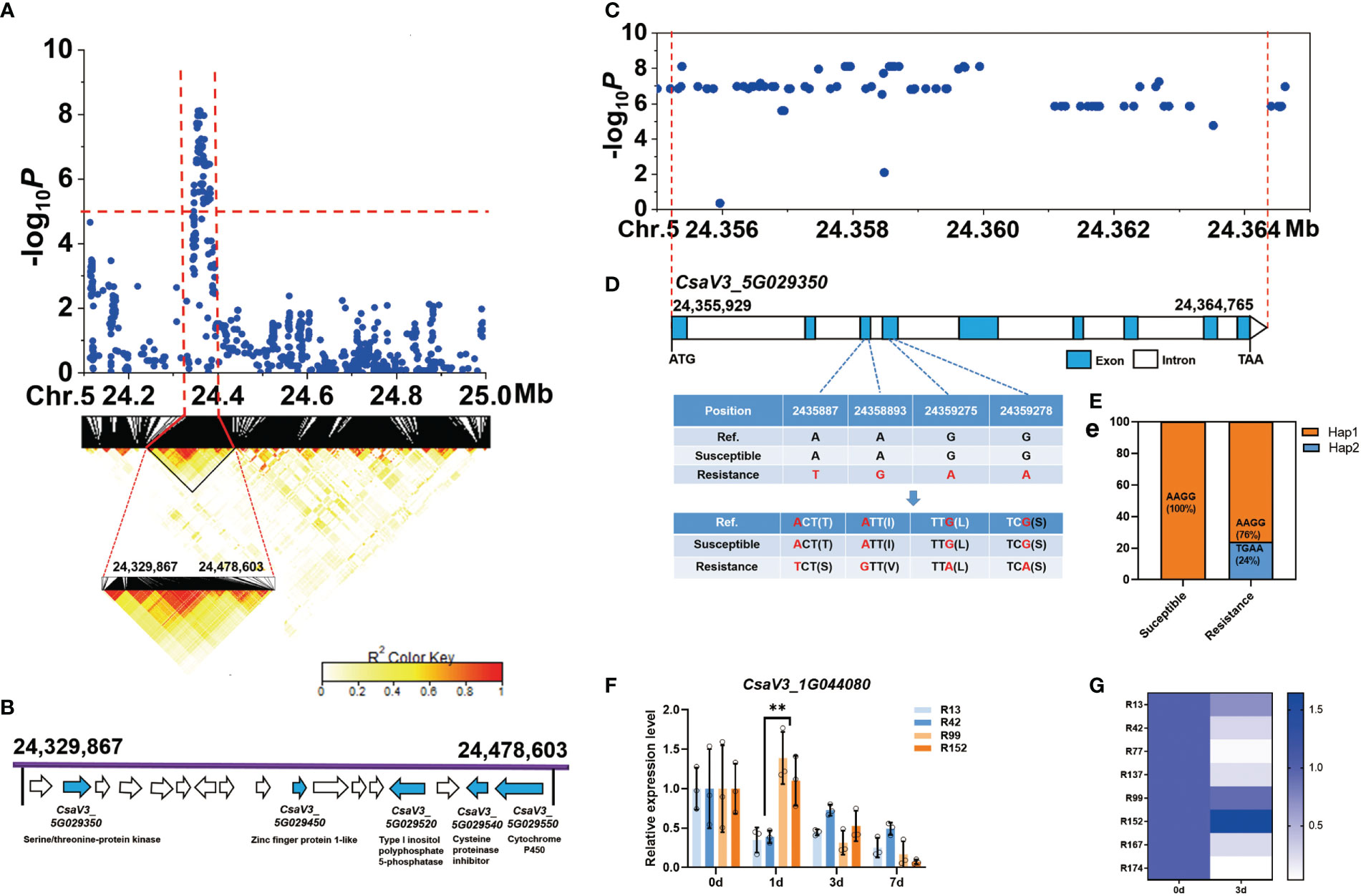
Figure 7 Identification of cucumber low-temperature germination trait-associated gene CsaV3_5G029350. (A) Local Manhattan plot (top) and LD heatmap (bottom) surrounding the peak on chromosome 5. The dashed line indicates the significance threshold (P < 10−6). Dashed lines indicate the candidate region (~ 148.7 kb) for the peak. (B) Seventeen functionally annotated genes were predicted in the gLTG5.2 candidate region. The arrows represent the direction of the genes. The genes shown as blue arrows are related to abiotic stress and their annotation is indicated. (C) The corresponding DNA polymorphisms of CsaV3_5G029350 with significant associations. (D) Gene structure of CsaV3_5G029350. Blue and white rectangles indicate exons and introns, respectively. (E) Bar chart showing the proportion of haplotypes - orange represents the proportion of accessions that are Hap1 (TGAA) while blue represents the proportion that are Hap2 (AAGG) among the susceptible and resistant lines, (F) The relative expression level of CsaV3_5G029350 in low-temperature sensitive lines (‘R13’, ‘R42’) and low-temperature resistant materials (‘R99’, ‘R152’) after low temperature treatment (0, 1, 3 and 7 d), tested through qRT–PCR. Actin was used an internal control. Data are represented as average values with SD of three independent biological replicates. (G) The heatmap indicates relative expression level of CsaV3_4G013480 in LT-sensitive lines (‘R13’, ‘R42’, ‘R77’, ‘R137’) and LT-resistant lines (‘R99’, ‘R152’, ‘R167’, ‘R174’) after low temperature treatment (0 and 3 d), tested through qRT–PCR. Actin was used an internal control. Data are represented as the average values with the SD of three independent biological replicates. **Significant difference (P <0.01).
For the gLTG1.2 locus, the signals were detected that consistently exceeded a significant threshold (−log10P ≥ 5.0) in 2019_RGR, 2020_RGR, 2020_RGE, 2020_RGI. The haplotype blocks were estimated using Plink tool (Purcell et al., 2007) in the gLTG1.2 locus; this effort further narrowed this region into only one haplotype block harboring significantly associated SNPs (Chr1: 29,185,530—29,524,806 bp) (Figure 8A). Fourteen annotated genes were located in this haplotype block (Figure 8A, Table S8). And only one gene (CsaV3_1G044080), encoding pentatricopeptide repeat-containing (PPR) protein (namely CsPPR), was reported to be related to abiotic stress. In Arabidopsis, AtPPR played a central role in abiotic stress during seed imbibition and seed development stages (Liu et al., 2016). AtCsPPR might interact with AtOTP (Figure S2) (Lee et al., 2009). To determine if the candidate gene CsPPR could function in a LTG response in cucumber, the haplotypes of 21 resistant and 47 sensitive cucumber lines were compared (Figure S3). Six variants of the significantly associated SNPs were identified in the 68 lines (Figure 8B). For 21 resistant cucumber lines and 47 sensitive cucumber accessions, 21 significantly associated SNPs located at the upstream (< 2 kb) and genic region of CsPRR were classified into 2 haplotypes (Figure 8B). Approximate 50% sensitive lines had the haplotype2 (Hap2; “ACTTGT”), while 86% resistant lines carried the Hap1 (“TTCAAC”) (Figure 8C). By further analysis of the accessions with different haplotypes, the “ACTTGT” (Hap2) haplotype was only existed in European type, American type, Indian type, South China type and Eurasian type (Figure 8D).

Figure 8 Identification of cucumber LT germination trait-associated gene CsaV3_1G044080 (CsPPR). (A) Local Manhattan plot (top) and LD heatmap (bottom) surrounding the peak on chromosome 1. The dashed line indicates the significance threshold (-log10(P)=5.0). Dashed lines indicate the candidate region (~ 339 kb) for the peak. Fourteen functionally annotated genes were predicted in the gLTG1.2 candidate region. The arrows represent the direction of the genes. Function of red arrow gene is relative to abiotic stress. (B) Schematic representation of two haplotypes of CsPPR. Orange and blue box represent promoter elements. (C) Orange represents the proportion of accessions that are Hap1 (TTCAAC) while blue represents the proportion that are Hap2 (ACTTGT) among the susceptible and resistant lines genotype. Orange represents the genotype Hap1 proportion in susceptible or resistant lines. Blue represents the genotype Hap2 proportion in susceptible or resistant lines. (D) The percentage of Hap1 and Hap2 genotypes within each ecotype. (E) Relative expression level of CsaV3_1G044080 in LT sensitive lines (‘R13’, ‘R42’) and LT resistant lines (‘R99’, ‘R152’) after LT treatment (0 d, 12 h, 1 d, 2 d, 3 d and 7 d), tested through qRT–PCR. Actin was used an internal control. Data are represented as average values with SD of three independent biological replicates. (F) The heatmap indicates relative expression level of CsaV3_1G044080 in LT sensitive lines (‘R13’, ‘R42’, ‘R77’, ‘R137’) and LT resistant lines (‘R99’, ‘R152’, ‘R167’, ‘R174’) after LT treatment (0 d and 3 d), tested through qRT–PCR. Actin was used an internal control. **Significant difference (P <0.01).
Of these significantly associated SNPs, in CDS region, one located at nucleotide 29,278,310 resulted in a C-to-T conversion, but led to no amino acid changes (Figure 8B); many cis-elements in CsPPR promoter were related to LT (Figure S4), eg. ABRE (ABA-responsive element), MYB (cold responsive element), ERE (ethylene-responsive element), CGTCA-motif (MeJA-responsive element), TGACG-motif (MeJA-responsive element). Of these, 5 motifs in promoter were altered by 5 SNP. SNP_29,275,643, SNP_29,277,326, SNP_29,277,544 caused change in TATA-box, SNP_29,276,402 bp caused change in TATA-box, and SNP_29276532 caused changes in MYB recognition site, respectively (Figure 8B). But the remaining SNPs has no change function of gene. The expression level of this gene in resistant lines (‘R99’ ‘R152’) was significantly higher than that in sensitive lines at 1 d after LT treatment (Figure 8E). In a larger panel of 8 randomly selected accessions, a general trend was observed that the expression level of resistant lines (‘R99’ ‘R152’ ‘R167’) were significantly higher than that of susceptible lines (‘R42’ ‘R77’ ‘R137’) at 3 d at 13°C (Figure 8F).
To further determine the reliability of the GWAS result, we determined if candidate gene CsPPR, identified at the gLTG1.2 locus, could have a role in seed germination under LT stress. A recombinant plasmid 35S::CsPPR was introduced into A. thaliana by A. tumefaciens-mediated floral dip method (Figure 9A). Five homozygous T2 transgenic lines (OE1-5) were obtained. The transgenic lines (OE-4, OE-5) were randomly selected for further experiments.
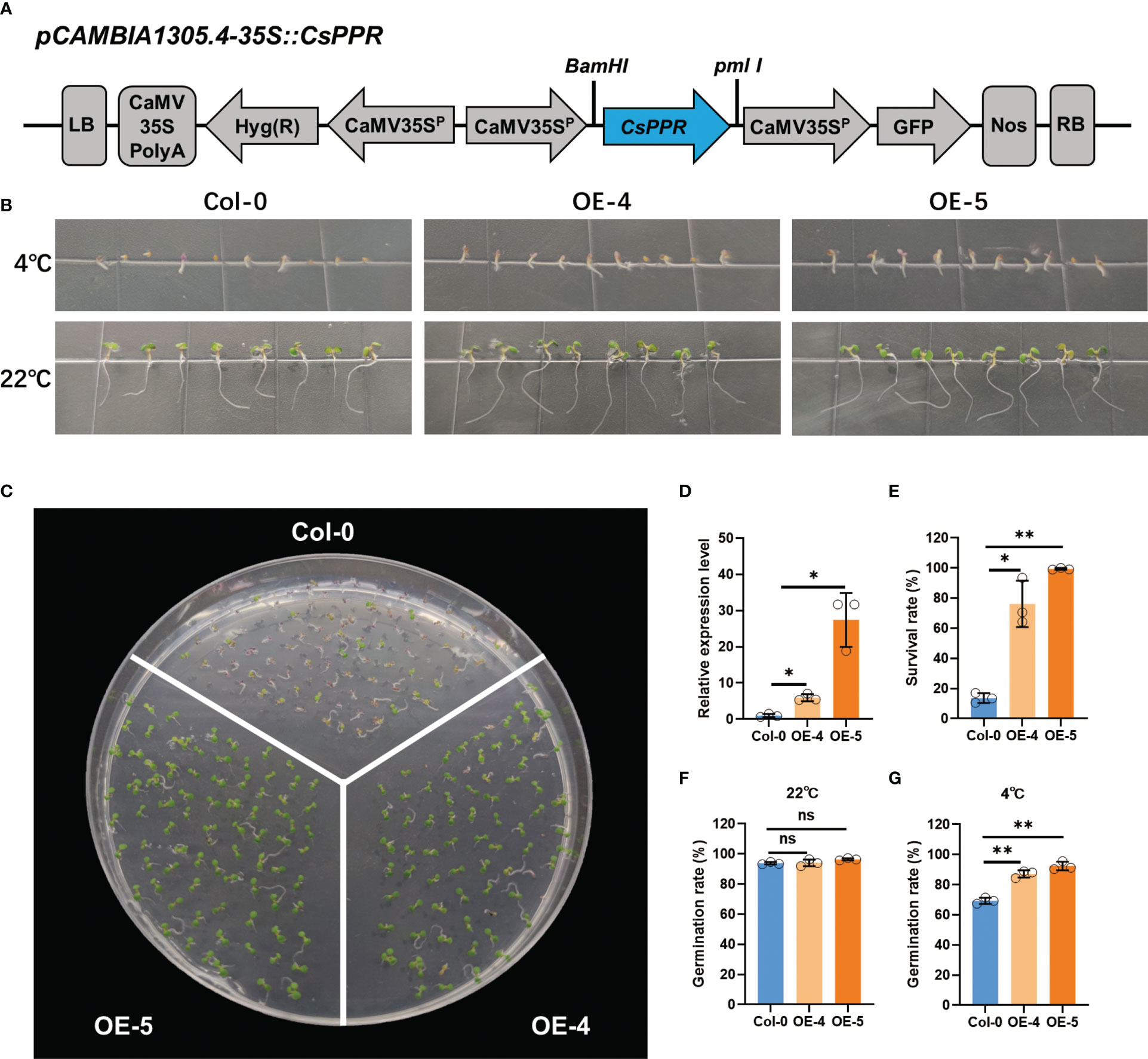
Figure 9 Low temperature tolerance of transgenic A.thalina ectopically expressing CsPPR. (A) Schematic diagram of the CsPPR construct for A.thaliana transformation. LB: Left border; CaMV35SPolyA: untranslated region of the CaMV 35S gene; hyg (R): Hygromycin phosphotransferase gene that confers hygromycin resistance; CaMV35Sp: CaMV 35S promoter; CsPPR: full-length of cucumber CsPPR; GFP: green fluorescent protein; NOS: transcriptional terminator sequence of the nopaline synthase gene; RB: right border. (B) Phenotype of the Col-0, CsPPR-expressing plants exposed to 4°C for 20 days and 22°C for 5 days. WT, wild-type (C) Phenotypes of acclimated WT, OE-4 and OE-5 plants exposed to 4°C for 40 days. At least three plates were used per treatment. (D) qRT-PCR showing the relative expression level of CsPPR in three independent CsPPR-ectopically expressing transgenic lines OE-4 and OE-5. Error bars represent the standard deviation of three replicates. (E) Survival rates after 4°C for 40 days. The data are the mean ± SD values of three independent experiments. Germination rates of the Col-0, OE-4 and OE-5 seeds germinated on 1/2 MS medium after (F) 5 days at 22°C, and (G) 20 days at 4°C. For (F, G) the data shown is the mean ± SD (n=50~100) of three replicates, with the error bars representing the standard deviation. The asterisks indicate the statistically significant differences of expression between control and transgenic lines. “**” represents “p<0.01”, “*” represents “p<0.05”.
The expression level of wild-type (Col-0), the lines ectopically expressing CsPPR, i.e., OE-4 and OE-5, were detected by qRT-PCR. OE-4 and OE-5 had 6-fold and 27-fold higher PPR transcripts compared to those in Col-0 (Figure 9D). Between 50-100 seeds of Col-0, OE-4 and OE-5 were germinated at 22°C for 5 days or 4°C for 40 days. Col-0 generally showed germination inhibition, and the germination rate was significantly lower than that of the CsPPR transgenic seeds (OE-4 and OE-5) at 4°C for 20 days (Figures 9B, G). In contrast, the germination rate of Col-0, OE-4 and OE-5 were same when exposed to a non-chilling temperature, i.e., 22°C for 5 days (Figures 9B, F). Further, the survival rates of the transgenic lines (OE-4 and OE-5) were higher than that of the Col-0 genotypes at 4°C for 40 days (Figures 9C, E). Interestingly, CsPPR expression showed a dosage effect for survival rate. For example, the expression of CsPPR on OE-5 had 5-fold higher transcripts than OE-4; Meanwhile, the survival rate of OE-4 was higher than Col-0 by over 63%, and the survival rate of OE-5 was 86%, higher than that of Col-0. These data were consistent between cucumber and Arabidopsis. Viewed collectively, these results support that CsPPR is very likely the causal gene to regulate LTG.
LT stress is one of the major abiotic stresses seriously affecting cucumber growth, development, and quality. The ability of seeds to germinate at LT is also a key factor affecting cucumber production at early stage of cucumber lifecycle. Therefore, it is urgent to identify LT-tolerant accessions, and develop cucumber varieties with LT-tolerance at the germination stage. GWAS was an efficient and effective way to excavate novel LT-tolerant genes, active during germination in cucumber.
In this study, cucumber LTG assessment was analyzed by four traits: RGR, RGE, RGI and RRL. In the association analysis, the phenotypic values of each of these traits exhibited high variation, with the CVs ranging from 112% to 130% (Table 1). The results indicated that the collected accessions represented a range of genetic backgrounds, and that they showed wide variation in the ability to germinate under LT, and were thus suitable for studying the genetic basis of LTG via GWAS.
Accurate phenotypic data is critical for successful gene mapping. In this study, phenotypic data were collected from two sites (Beijing or Shandong), two environments (phytotron or incubator) and over two years (2019 or 2020), which better ensured the reliability of the data and reduces spurious environmental effects.
Many studies including this one, have reported that the cucumber LTG trait is polygenic (Gu et al., 2002; Kłosińska et al., 2013), and have identified major or minor effect QTLs (Song et al., 2018; Yagcioglu et al., 2019). In the past five years, QTLs associated with LTG (GR, GE, GI, RL and RFW) have been identified using RILs or F2:3 lines. In this GWAS, QTLs for LTG tolerance were found on four chromosomes (Figure 4, Table 2), suggesting a complex genetic architecture underlying LTG in cucumber (Figure 2, Table 1), consistent with a previous study (Song et al., 2018). Among the 17 QTLs detected in the previous study, two of the three signals contributed to QTL intervals identified in this present study. QTLs qLTG1.1 and qLTG1.2 were close to the GWAS locus gLTG1.2. In addition, the GWAS locus gLTG4.1 was located between QTLs qLTG4.1 and qRGR4.1. Similar loci were detected by GWAS and QTL analysis, in genotypes of different genetic background, different segregating populations and modes of mapping, suggesting that the GWAS results were reliable. Simultaneously, more associated loci broaden the potential for breeding development.
The regulatory mechanisms of LT response are complex and dynamic processes (Ding et al., 2019; Ding and Yang, 2022; Kidokoro et al., 2022), involving the perception of signals at the cytomembrane, transduction of the signal from the cytomembrane to the cell nucleus, and the activation of regulatory mechanisms in the cell nucleus. Correspondingly, multiple factors participate in the LT regulatory response, including, transcription factors PPR (Xing et al., 2018), NAC (Zhuo et al., 2018; Liu et al., 2020a), GRAS (Wang et al., 2021; Li et al., 2022a), bZIP (Li et al., 2022b) and WRKY (Liu et al., 2022b; Sun et al., 2022a; Zhang et al., 2022; Zhao et al., 2022), phytohormones ABA (Zhu, 2016; Song et al., 2022; Zhang et al., 2023), JA (Li et al., 2021; Sun et al., 2022b) and GA (Magome et al., 2004; Achard et al., 2008) and various metabolic pathways especially those producing compatible solutes. In this study, ten genes in the GWAS regions, possibly related to abiotic stress were identified (Table S8). Three candidate genes were predicted as potential causal genes underlying the stable and novel loci, including CsaV3_1G044080 for gLTG1.2, CsaV3_4G013480 for gLTG4.1 and CsaV3_5G029350 for gLTG5.2 (Figures 6–8). These candidate genes either have significantly associated nonsynonymous SNPs which resulted in amino acid changes, or motif variations in the promoter cis-elements important for stress response.
(a) The A.thaliana orthologue of CsaV3_4G013480, encodes a plasma-membrane-localized RING E3 ubiquitin ligase, that regulates tolerance to cold stress (Kim and Kim, 2013). For example, Arabidopsis RING E3 ubiquitin ligase AtATL7 improves cold tolerance, while AtATL80 negatively regulate the cold tolerance response (Suh and Kim, 2015).
(b) For gLTG5.2, the A. thaliana s orthologue of CsaV3_5G029350 encodes a serine/threonine-protein kinase. Ectopic expression of a SNF1- serine/threonine-protein kinase TaSnRK2.4 in A.thaliana improved tolerance to freezing stress compared to the non-transformed, wild-type plants (Mao et al., 2010). Based on these collective data, we speculate that these three genes, i.e., CsaV3_1G044080, CsaV3_4G013480 and CsaV3_5G029350 might be part of the cucumber seed response to LT at germination. However, the specific role, and mode of action of the products of these genes in a signal transduction response for regulating LTG requires further study.
(c) The CsaV3_1G044080 gene, encoding a PPR family protein, is homologous to the A.thaliana gene At4G04370. In rice, the PPR protein OsV4 plays an important role in tolerance to cold stress (Gong et al., 2014; Tan et al., 2014). Arabidopsis transgenic lines, overexpressing a gene for a PPR protein called SOAR1, were able to tolerate various stresses, including cold stress (Jiang et al., 2015). SOAR1 positively regulated plant response to cold stress through the CBF signaling pathway, and ABA-dependent, and -independent signaling pathways (Jiang et al., 2015). MYB, an essential cold regulatory transcriptional factor, has a recognition cis-acting element at 29,277,326 bp in the promoter of CsaV3_1G044080 (An et al., 2018). There are also additional stress-responsive cis-acting elements (e.g. TGACG motif) in CsaV3_1G044080 that could be activated in response to LT and which are important in to cold resistance (Baker et al., 1994).
The sequence variation in the promoter and transcribed region of the gene(s) identified at a QTL, are crucial for linking DNA polymorphisms to a phenotypic effect. In this study, the CsPPR association study identified sequence variation that were significantly correlated with variation in RGR, RGE and RGI among the population studied. Of the 21 SNPs identified, SNP_29277326, within a MYB recognition site in the CsPPR promoter, might be the most significant. The expression levels could be affected by the genetic loci variation in promoter (Yu et al., 2019; Li et al., 2020; Zhang et al., 2020). qRT-PCR and contrasting haplotypes further identified its expression level by the effect of the promoter. The findings indicated that the promoter of resistant lines (haplotype “TTCAAC”) supported significantly higher transcriptional activity than that in the sensitive lines (Figures 8E, F), which was consistent with previous data that the favorable haplotypes of candidate genes were closely associated with the phenotypic performances of traits. Hence, the favorable haplotype of CDS (CsPPR’9930’) was transformed into A.thaliana and the seed germination was investigated. The seed germination rate and survival rate in transgenic A.thaliana were significantly higher than those in the wild-type Col-0 under LT, indicating that CsPPR is a causal factor in regulating seed germination in transgenic A.thaliana under LT. The molecular mechanism of G protein α subunit (CsGPA1) and heat-shock transcription factor (CsHSFA1d) were studied in cucumber (Qi et al., 2022; Yan et al., 2022). Thus, to verify whether, and how CsPPR controls cucumber seed germination and response to LT, the generation of overexpression or knockout in cucumber should be given priority.
In our study, we identified novel genes related to LTG and demonstrated that CsPPR contributes to cucumber LTG ability. Our results will help to facilitate future studies on the mechanism underlying LT tolerance in germinating cucumber. Meanwhile, the newly-discovered molecular markers and LT-tolerant germplasm will aid future marker-assisted breeding of cucumber.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
CL drafted the manuscript. CL and SD conducted the experiments and analyzed the data. DB helped analyzed the data and drafted the manuscript. XL helped collected the data, HM helped analyzed the data. SZ and XG designed the experiments. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (No.31902028), Key-Area Research and Development Program of Guangdong Province (2020B020220001), Beijing Joint Research Program for Germplasm Innovation and New Variety Breeding (G20220628003-03). The Earmarked Fund for Modern Agro-industry Technology Research System (CARS-23), Science and Technology Innovation Program of the Chinese Academy of Agricultural Science (CAAS-ASTIP-IVFCAAS), Central Public-interest Scientific Institution Basal Research Fund (Y2017PT52), and the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P.R.China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1116214/full#supplementary-material
Achard, P., Gong, F., Cheminant, S., Alioua, M., Hedden, P., Genschik, P. (2008). The cold-inducible CBF1 factor–dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20, 2117–2129. doi: 10.1105/tpc.108.058941
An, J. P., Li, R., Qu, F. J., You, C. X., Wang, X. F., Hao, Y. J. (2018). R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 96, 562–577. doi: 10.1111/tpj.14050
Baker, S. S., Wilhelm, K. S., Thomashow, M. F. (1994). The 5′-region of arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought-and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. doi: 10.1007/BF00029852
Bo, K., Miao, H., Wang, M., Xie, X., Song, Z., Xie, Q., et al. (2019a). Novel loci fsd6.1 and Csgl3 regulate ultra-high fruit spine density in cucumber. Theor. Appl. Genet. 132, 27–40. doi: 10.1007/s00122-018-3191-6
Bo, K., Wei, S., Wang, W., Miao, H., Gu, X. (2019b). QTL mapping and genome-wide association study reveal two novel loci associated with green flesh color in cucumber. BMC Plant Biol. 19, 243. doi: 10.1186/s12870-019-1835-6
Cabrera, R. M., Saltveit, M. E., Owens, K. (1992). Cucumber cultivars differ in their response to chilling temperatures. J. Am. Soc. Hortic. Sci. 117, 802–807. doi: 10.21273/JASHS.117.5.802
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Xia, R. (2020). TBtools-an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, G., Zou, Y., Hu, J., Ding, Y. (2018). Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments. BMC Genomics 19, 720. doi: 10.1186/s12864-018-5088-9
Chopra, R., Burow, G., Burke, J. J., Gladman, N., Xin, Z. (2017). Genome-wide association analysis of seedling traits in diverse sorghum germplasm under thermal stress. BMC Plant Biol. 17, 1–15. doi: 10.1186/s12870-016-0966-2
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dekkers, B. J., Pearce, S., Van Bolderen-Veldkamp, R., Marshall, A., Widera, P., Gilbert, J., et al. (2013). Transcriptional dynamics of two seed compartments with opposing roles in arabidopsis seed germination. Plant Physiol. 163, 205–215. doi: 10.1104/pp.113.223511
Ding, Y., Shi, Y., Yang, S. (2019). Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 222, 1690–1704. doi: 10.1111/nph.15696
Ding, Y., Yang, S. (2022). Surviving and thriving: How plants perceive and respond to temperature stress. Dev. Cell 57, 947–958. doi: 10.1016/j.devcel.2022.03.010
Dong, H. (2017). Mining genes with cold tolerance of elite germplasm in cucumber. Chin. Acad. Agric. Sci.
FAO (2020). FAOSTA. https://www.fao.org/faostat/en/#home.
Ge, C., Wang, L., Yang, Y., Liu, R., Liu, S., Chen, J., et al. (2022). Genome-wide association study identifies variants of GhSAD1 conferring cold tolerance in cotton. J. Exp. Bot. 73, 2222–2237. doi: 10.1093/jxb/erab555
Gong, X., Su, Q., Lin, D., Jiang, Q., Xu, J., Zhang, J., et al. (2014). The rice OsV4 encoding a novel pentatricopeptide repeat protein is required for chloroplast development during the early leaf stage under cold stress. J. Integr. Plant Biol. 56, 400–410. doi: 10.1111/jipb.12138
Gu, X., Feng, L., Zhang, C., Fang, X., Zhang, T. (2002). Genetic analysis of the cucumber germinating ability in low temperature. China Vegetables 2 (3), 5–7.
Han, J., Dong, S., Shi, Y., Dai, Z., Miao, H., Li, B., et al. (2022). Genome-wide association study reveals candidate genes for gummy stem blight resistance in cucumber. Hortic. Plant J. 1-12. doi: 10.1016/j.hpj.2022.06.004
He, Y. (2006). Some usual kinds of methods of fuzzy mathematics in agriculture research (Changchun: Jilin University).
Jiang, S., Mei, C., Liang, S., Yu, Y., Lu, K., Wu, Z., et al. (2015). Crucial roles of the pentatricopeptide repeat protein SOAR1 in arabidopsis response to drought, salt and cold stresses. Plant Mol. Biol. 88, 369–385. doi: 10.1007/s11103-015-0327-9
Jiang, L., Zheng, Z., Fang, H., Yang, J. (2021). A generalized linear mixed model association tool for biobank-scale data. Nat. Genet. 53, 1616–1621. doi: 10.1038/s41588-021-00954-4
Kidokoro, S., Shinozaki, K., Yamaguchi-Shinozaki, K. (2022). Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 27, 922–935. doi: 10.1016/j.tplants.2022.01.008
Kim, S. J., Kim, W. T. (2013). Suppression of arabidopsis RING E3 ubiquitin ligase AtATL78 increases tolerance to cold stress and decreases tolerance to drought stress. FEBS Lett. 587, 2584–2590. doi: 10.1016/j.febslet.2013.06.038
Kim, S., Yang, S., Kim, T., Han, J., Suh, J. (2009). Hypertonic stress increased extracellular ATP levels and the expression of stress-responsive genes in arabidopsis thaliana seedlings. Biosci. biotech. Biochem. 73, 1252–1256. doi: 10.1271/bbb.80660
Kłosińska, U., Kozik, E. U., Nowicki, M., Wehner, T. C. (2013). Low temperature seed germination of cucumber: genetic basis of the tolerance trait. J. Hortic. Res. 21, 125–130. doi: 10.2478/johr-2013-0031
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547. doi: 10.1093/molbev/msy096
Lee, H., Kim, J., Kang, B., Song, K. (2020). Assessment of the genetic diversity of the breeding lines and a genome wide association study of three horticultural traits using worldwide cucumber (Cucumis spp.) germplasm collection. Agronomy 10, 1736. doi: 10.3390/agronomy10111736
Lee, J., Lee, S., Jang, H., Lee, Y., Park, J., Park, S., et al. (2009). Heat-shock dependent oligomeric status alters the function of a plant-specific thioredoxin-like protein, AtTDX. Proc. Natl. Acad. Sci. 106, 5978–5983. doi: 10.1073/pnas.0811231106
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, C., Dong, S., Beckles, D. M., Miao, H., Sun, J., Liu, X., et al. (2022a). The qLTG1.1 candidate gene CsGAI regulates low temperature seed germination in cucumber. Theor. Appl. Genet. 135, 2593–2607. doi: 10.1007/s00122-022-04097-w
Li, H., Guo, Y., Lan, Z., Xu, K., Chang, J., Ahammed, G. J., et al. (2021). Methyl jasmonate mediates melatonin-induced cold tolerance of grafted watermelon plants. Hortic. Res. 8, 57. doi: 10.1038/s41438-021-00496-0
Li, Q., Li, H., Huang, W., Xu, Y., Zhou, Q., Wang, S., et al. (2019). A chromosome-scale genome assembly of cucumber (Cucumis sativus L.). GigaScience 8, 1–10. doi: 10.1093/gigascience/giz072
Li, Z., Liu, P., Zhang, X., Zhang, Y., Ma, L., Liu, M., et al. (2020). Genome-wide association studies and QTL mapping uncover the genetic architecture of ear tip-barrenness in maize. Physiologia plantarum 170, 27–39. doi: 10.1111/ppl.13087
Li, P., Zheng, T., Li, L., Wang, J., Cheng, T., Zhang, Q. (2022b). Genome-wide investigation of the bZIP transcription factor gene family in prunus mume: Classification, evolution, expression profile and low-temperature stress responses. Hortic. Plant J. 8, 230–242. doi: 10.1016/j.hpj.2021.01.009
Liu, D., Dong, S., Miao, H., Liu, X., Li, C., Han, J., et al. (2022a). A Large-scale genomic association analysis identifies the candidate genes regulating salt tolerance in cucumber (Cucumis sativus L.) seedlings. Int. J. Mol. Sci. 23, 8260. doi: 10.1007/s00122-022-04117-9
Liu, X., Gu, X., Lu, H., Liu, P., Miao, H., Bai, Y., et al. (2021). Identification of novel loci and candidate genes for resistance to powdery mildew in a resequenced cucumber germplasm. Genes (Basel) 12, 584. doi: 10.3390/genes12040584
Liu, X., Lu, H., Liu, P., Miao, H., Bai, Y., Gu, X., et al. (2020b). Identification of novel loci and candidate genes forcucumber downy mildew resistance using GWAS. Plants (Basel) 9, 1659. doi: 10.3390/plants9121659
Liu, Q., Sun, C., Han, J., Li, L., Zhang, M. (2020a). Identification, characterization and functional differentiation of the NAC gene family and its roles in response to cold stress in ginseng, panax ginseng C.A. Meyer. PLoS One 15, e0234423. doi: 10.1371/journal.pone.0234423
Liu, H., Yang, L., Xu, S., Lyu, M. J., Wang, J., Wang, H., et al. (2022b). OsWRKY115 on qCT7 links to cold tolerance in rice. Theor. Appl. Genet. 135, 2353–2367. doi: 10.1007/s00122-022-04117-9
Liu, J., Zhao, J. Y., Lu, P., Chen, M., Guo, C., Xu, Z., et al. (2016). The e-subgroup pentatricopeptide repeat protein family in arabidopsis thaliana and confirmation of the responsiveness PPR96 to abiotic stresses. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01825
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, L., Wang, C., Hu, Y., Dai, W., Liang, Z., Zou, C., et al. (2022). GWAS and transcriptome analysis reveal MADS26 involved in seed germination ability in maize. Theor. Appl. Genet. 135, 1717–1730. doi: 10.1007/s00122-022-04065-4
Magome, H., Yamaguchi, S., Hanada, A., Kamiya, Y., Oda, K. (2004). Dwarf and delayed-flowering 1, a novel arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 37, 720–729. doi: 10.1111/j.1365-313x.2003.01998.x
Mao, X., Zhang, H., Tian, S., Chang, X., Jing, R. (2010). TaSnRK2. 4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in arabidopsis. J. Exp. Bot. 61, 683–696. doi: 10.1093/jxb/erp331
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nienhuis, J., Lower, R., Staub, J. (1983). Selection for improved low temperature germination in cucumber. Journal-American Soc. Hortic. Sci. (USA) 108 (6),1040–1043. doi: 10.21273/JASHS.108.6.1040
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Qi, C., Dong, D., Li, Y., Wang, X., Guo, L., Liu, L., et al. (2022). Heat shock-induced cold acclimation in cucumber through CsHSFA1d-activated JA biosynthesis and signaling. Plant J. 111, 85–102. doi: 10.1111/tpj.15780
Røeggen, O. (1987). Variation in minimum germination temperature for cultivars of bean (Phaseolus vulgaris L.), cucumber (Cucumis sativus L.) and tomato (Lycopersicon esculentum mill.). Scientia Hortic. 33, 57–65. doi: 10.1016/0304-4238(87)90032-X
R Core Team (2019). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Sales, E., Viruel, J., Domingo, C., Marqués, L. (2017). Genome wide association analysis of cold tolerance at germination in temperate japonica rice (Oryza sativa L.) varieties. PLoS One 12, e0183416. doi: 10.1371/journal.pone.0183416
SAS Institute (2012). SAS/OR 9.3 user’s guide: Mathematical programming examples (Cary, NC, USA: SAS Institute Inc).
Schläppi, M. R., Jackson, A. K., Eizenga, G. C., Wang, A., Chu, C., Shi, Y., et al. (2017). Assessment of five chilling tolerance traits and GWAS mapping in rice using the USDA mini-core collection. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00957
Shen, Q., Zhang, S., Ge, C., Liu, S., Chen, J., Liu, R., et al. (2023). Genome-wide association study identifies GhSAL1 affects cold tolerance at the seedling emergence stage in upland cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 136, 4. doi: 10.1007/s00122-023-04317-x
Smeets, L., Wehner, T. C. (1997). Environmental effects on genetic variation of chilling resistance in cucumber. Euphytica 97, 217–225. doi: 10.1023/a:1003084821178
Song, Z., Lai, X., Chen, H., Wang, L., Pang, X., Hao, Y., et al. (2022). Role of MaABI5-like in abscisic acid-induced cold tolerance of ‘Fenjiao’ banana fruit. Horticult. Res. 9, uhac130. doi: 10.1093/hr/uhac130
Song, Z., Wang, W., Shi, L., Zhang, S., Xie, Q., Wei, S., et al. (2018). Identification of QTL s controlling low-temperature tolerance during the germination stage in cucumber (Cucumis sativus L.). Plant Breed. 137, 629–637. doi: 10.1111/pbr.12601
Suh, J. Y., Kim, W. T. (2015). Arabidopsis RING E3 ubiquitin ligase AtATL80 is negatively involved in phosphate mobilization and cold stress response in sufficient phosphate growth conditions. Biochem. Biophys. Res. Commun. 463, 793–799. doi: 10.1016/j.bbrc.2015.06.015
Sun, J., Chen, J., Si, X., Liu, W., Yuan, M., Guo, S., et al. (2022a). WRKY41/WRKY46-miR396b-5p-TPR module mediates abscisic acid-induced cold tolerance of grafted cucumber seedlings. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1012439
Sun, M., Shen, Y., Chen, Y., Wang, Y., Cai, X., Yang, J., et al. (2022b). Osa-miR1320 targets the ERF transcription factor OsERF096 to regulate cold tolerance via JA-mediated signaling. Plant Physiol. 189, 2500–2516. doi: 10.1093/plphys/kiac208
Tan, J., Tan, Z., Wu, F., Sheng, P., Heng, Y., Wang, X., et al. (2014). A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol. Plant 7, 1329–1349. doi: 10.1093/mp/ssu054
Turner, S. D. (2014). Qqman: an r package for visualizing GWAS results using q-q and manhattan plots. Biorxiv 3 (25),1731. doi: 10.21105/joss.00731
Wang, W., Song, Z., Bo, K., Dong, S., Wei, S., Miao, H., et al. (2019). Evaluation and genome-wide Association Study (GWAS) of low-temperature tolerance at seedling stage in cucumber core germplasm. Journal of Plant Genetic Resources 20 (6), 1606–1612. doi: 10.13430/j.cnki.jpgr.20190225001
Wang, Z., Wong, D. C. J., Wang, Y., Xu, G., Ren, C., Liu, Y., et al. (2021). GRAS-domain transcription factor PAT1 regulates jasmonic acid biosynthesis in grape cold stress response. Plant Physiol. 186, 1660–1678. doi: 10.1093/plphys/kiab142
Wei, S., Zhang, S., Bo, K., Wang, Y., Miao, H., Dong, S., et al. (2019). Evaluation and genome-wide Association Study (GWAS) of low-temperature tolerance at seedling stage in cucumber core germplasm. Journal of Plant Genetic Resources 20 (5), 1223–1231. doi: 10.13430/j.cnki.jpgr.20181226001
Woffelman, C. (2004). DNAMAN for windows (Lynon Biosoft, Institute of Molecular Plant Sciences, Leiden University).
Xie, Z. (1983). Fuzzy mathematics method in agricultural science (Wuhan: Huazhong University of Science and Technology Press), 99–193.
Xie, Q., Liu, P., Shi, L., Miao, H., Bo, K., Wang, Y., et al. (2018). Combined fine mapping, genetic diversity, and transcriptome profiling reveals that the auxin transporter gene ns plays an important role in cucumber fruit spine development. Theor. Appl. Genet. 131, 1239–1252. doi: 10.1007/s00122-018-3074-x
Xing, H., Fu, X., Yang, C., Tang, X., Guo, L., Li, C., et al. (2018). Genome-wide investigation of pentatricopeptide repeat gene family in poplar and their expression analysis in response to biotic and abiotic stresses. entific Rep. 8, 2817. doi: 10.1038/s41598-018-21269-1
Yagcioglu, M., Jiang, B., Wang, P., Wang, Y., Ellialtioglu, S. S., Weng, Y. (2019). QTL mapping of low temperature germination ability in cucumber. Euphytica 215, 84. doi: 10.1007/s10681-019-2408-3
Yan, Y., M, S., Si, M., Qian, F., Wang, Y., Qinghua, D., et al. (2022). Mechanism of CsGPA1 in regulating cold tolerance of cucumber. Hortic. Res. 9, uhac109. doi: 10.1093/hr/uhac109
Yoshida, H., Hirano, K., Yano, K., Wang, F., Mori, M., Kawamura, M., et al. (2022). Genome-wide association study identifies a gene responsible for temperature-dependent rice germination. Nat. Commun. 13, 5665. doi: 10.1038/s41467-022-33318-5
Yu, F., Liang, K., Fang, T., Zhao, H., Han, X., Cai, M., et al. (2019). A group VII ethylene response factor gene, ZmEREB180, coordinates waterlogging tolerance in maize seedlings. Plant Biotechnol. J. 17, 2286–2298. doi: 10.1111/pbi.13140
Zhang, S., Miao, H., Song, Z., Bo, K., Wang, Y., Gu, X., et al. (2019). Evaluation and genome-wide association study (GWAS) of thermotolerance for cucumber at the germination Stage. Journal of Plant Genetic Resources 20 (2), 335–346. doi: 10.13430/j.cnki.jpgr.20180624001
Zhang, Y., Hu, Y., Guan, Z., Liu, P., He, Y., Zou, C., et al. (2020). Combined linkage mapping and association analysis reveals genetic control of maize kernel moisture content. Physiologia plantarum 170, 508–518. doi: 10.1111/ppl.13180
Zhang, Y., Peng, Y., Liu, J., Yan, J., Zhu, K., Sun, X., et al. (2023). Tetratricopeptide repeat protein SlREC2 positively regulates cold tolerance in tomato. Plant Physiol 00, 1–18. doi: 10.1093/plphys/kiad085
Zhang, M., Zhao, R., Huang, K., Huang, S., Wang, H., Wei, Z., et al. (2022). The OsWRKY63–OsWRKY76–OsDREB1B module regulates chilling tolerance in rice. Plant J. 112, 383–398. doi: 10.1111/tpj.15950
Zhao, H., Mallano, A. I., Li, F., Li, P., Wu, Q., Wang, Y., et al. (2022). Characterization of CsWRKY29 and CsWRKY37 transcription factors and their functional roles in cold tolerance of tea plant. Beverage Plant Res. 2, 1–13. doi: 10.1016/j.bbrc.2015.07.085
Zhu, J. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029
Keywords: cucumber, low temperature germination, GWAS, polygenic, CsPPR
Citation: Li C, Dong S, Beckles DM, Liu X, Guan J, Gu X, Miao H and Zhang S (2023) GWAS reveals novel loci and identifies a pentatricopeptide repeat-containing protein (CsPPR) that improves low temperature germination in cucumber. Front. Plant Sci. 14:1116214. doi: 10.3389/fpls.2023.1116214
Received: 05 December 2022; Accepted: 24 March 2023;
Published: 26 April 2023.
Edited by:
Ravinder Kumar, Central Potato Research Institute (ICAR), IndiaReviewed by:
Mazahar Moin, University of Hyderabad, IndiaCopyright © 2023 Li, Dong, Beckles, Liu, Guan, Gu, Miao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengping Zhang, emhhbmdzaGVuZ3BpbmdAY2Fhcy5jbg==; Han Miao, bWlhb2hhbkBjYWFzLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.