
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 19 January 2023
Sec. Plant Pathogen Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1116147
This article is part of the Research TopicPlant Resistance to Soil-Borne DiseasesView all 8 articles
 Long Lin1
Long Lin1 Zixiang Yang1
Zixiang Yang1 Min Tao1
Min Tao1 Danyu Shen1
Danyu Shen1 Chuanbin Cui2
Chuanbin Cui2 Pingping Wang2
Pingping Wang2 Limin Wang1
Limin Wang1 Maofeng Jing1
Maofeng Jing1 Guoliang Qian1*
Guoliang Qian1* Xiaolong Shao1*
Xiaolong Shao1*The Phytophthora pathogen causes enormous damage to important agricultural plants. This group of filamentous pathogens is phylogenetically distant from fungi, making them difficult to control by most chemical fungicides. Lysobacter enzymogenes OH11 (OH11) is a biocontrol bacterium that secretes HSAF (Heat-Stable Antifungal Factor) as a broad-spectrum antifungal weapon. Here, we showed that OH11 could also control a variety of plant Phytophthora diseases caused by three major oomycetes (P. sojae, P. capsici and P. infestans). We provided abundant evidence to prove that OH11 protected host plants from Phytophthora pathogen infection by inhibiting mycelial growth, digesting cysts, suppressing cyst germination, and eliciting plant immune responses. Interestingly, the former two processes required the presence of HSAF, while the latter two did not. This suggested that L. enzymogenes could prevent Phytophthora infection via multiple previously unknown mechanisms. Therefore, this study showed that L. enzymogenes could serve as a promising alternative resource for promoting plant resistance to multiple Phytophthora pathogens.
Phytophthora belonging to the phylum Oomycota includes a large number of plant pathogens that cause devastating diseases of important agricultural crops (Kamoun et al., 2015). For examples, Potato late blight caused by Phytophthora infestans led to the Irish famine in the 1840s and still threatens potato production worldwide to date (Fry, 2008). Soybean root rot caused by Phytophthora sojae is one of the most destructive diseases in soybean production (Tyler, 2007). Phytophthora capsici is an extremely destructive pathogen with a broad host range that attacks hosts in the Solanaceous, Fabaceae, and most Cucurbitaceae (Lamour et al., 2012). Phytophthora shares similar morphology and habitat with fungi, but it is evolutionarily distant from fungi, belonging to the kingdom Stramenopiles (Baldauf et al., 2000; Kamoun et al., 2015). The differences between Phytophthora and fungi in terms of genome structure, metabolic pattern, and pathogenic mechanism make Phytophthora difficult to control by most fungicides (Latijnhouwers et al., 2003). In this context, environment-friendly biocontrol bacteria are emerging as an important microbial resource for effective control of Phytophthora diseases (Sang et al., 2011; de Andrade Lourenco et al., 2022). Members of plant-beneficial genera Bacillus and Pseudomonas are representatives of such biocontrol bacteria (Caulier et al., 2018). For example, Bacillus amyloliquefaciens and Bacillus subtilis have been reported to inhibit mycelial growth, cyst germination, and zoospore motility of P. sojae (Liu et al., 2019). Bacillus velezensis FZB42 antagonizes the growth and virulence of P. sojae by producing bacilysin (Han et al., 2021). Pseudomonas aurantiaca ST-TJ4 inhibits the growth of Phytophthora cinnamomi through production of antimicrobial phenazine compounds and volatile organic compounds (VOCs) (Zhang et al., 2022).
The genus Lysobacter comprises numerous environmentally ubiquitous biocontrol bacteria with unique ability to prey on other microorganisms by secreting abundant antibiotics, antimicrobial compounds, and lytic enzymes (Puopolo et al., 2018). These powerful weapons enable certain members of Lysobacter to efficiently kill the Phytophthora pathogens in the laboratory or in the field (Puopolo et al., 2018; Lin et al., 2021). For instance, Lysobacter antibioticus HS124 produces 4-hydroxyphenylacetic acid and several lytic enzymes to against P. capsici mycelial growth (Ko et al., 2009). Lysobacter capsici AZ78 generates cyclo (L-Pro-L-Tyr), a 2,5-diketopiperazine that inhibits sporangia development and virulence in P. infestans (Puopolo et al., 2014). VOCs from L. capsici have been reported to suppress mycelial growth of various fungi and oomycete (Vlassi et al., 2020a; Vlassi et al., 2020b).
Lysobacter enzymogenes is the most studied species in the genus Lysobacter (Puopolo et al., 2018; Lin et al., 2021). As the name suggested, L. enzymogenes can secrete abundant extracellular lytic enzymes (i.e protease and β-1,3-glucanase) that degrade the cell walls of filamentous fungi (Qian et al., 2013). Notably, this bacterium is primarily considered an antifungal agent, because it produces a well-characterized antibiotic metabolite called Heat-Stable Antifungal Factor (HSAF), a novel macrocyclic-lactam compound (Yu et al., 2007). HSAF in L. enzymogenes is synthesized by a gene cluster called the HSAF biosynthetic gene operon, in which lafB encodes a hybrid polyketide synthase and a nonribosomal peptide synthetase (Li et al., 2014; Wang et al., 2017). HSAF targets filamentous fungi with a unique action mode that disrupts the biosynthesis of fungal cell membrane-associated sphingolipids (Li et al., 2006). Due to its broad-spectrum antifungal activity, HSAF is found to disrupt mycelial growth and spore germination of various fungi such as Aspergillus nidulans and Alternaria alternata (Li et al., 2006; He et al., 2018). In the field, HSAF shows significant biocontrol activity in controlling wheat Fusarium head blight caused by Fusarium graminearum (Zhao et al., 2019). Although we recently reported that seed coatings assembled with HSAF inhibited mycelial growth of the oomycete pathogen Pythium graminerum (Ren et al., 2020), biocontrol potential of L. enzymogenes and HSAF in controlling Phytophthora diseases have not been systematically investigated.
Here, we used L. enzymogenes OH11 (herein refers to OH11) as a working model to verify the above viewpoint. We found that OH11 effectively controlled P. sojae, P. capsici and P. infestans on host plants via multiple HSAF-dependent and HSAF-independent mechanisms, suggesting this strain may be considered as a promising alternative biocontrol agent against Phytophthora diseases in important agricultural crops.
To study the biocontrol ability of L. enzymogenes OH11 (OH11) against Phytophthora infection, we cultivated OH11 in LB media overnight and transferred the culture to 1/10 TSB (herein refers to as TSB) media for 24 hours to induce HSAF production, as previously described (Qian et al., 2013). The cultures were normalized to OD600 = 1.0 with fresh TSB for further use. Phytophthora zoospores were prepared and normalized to 100 zoospores/μL. Afterwards, 200 zoospores of P. sojae strain P6497 were mixed with the prepared OH11 culture and inoculated on soybean (Glycine max) etiolated seedlings, where both TSB and an irrelevant E. coli strain Top10 were used as negative controls. Two days after inoculation, P. sojae treated by TSB or E. coli caused obvious lesions on soybean seedlings, while OH11 significantly inhibited the virulence of P. sojae (Figure 1A). We analysed infected soybean seedlings in more detail at the microscopic level with GFP-labelled P. sojae. At 6 hpi (hours post inoculation), both TSB- and E. coli-treated zoospores had germinated and started to infect soybean, whereas no cyst or hyphae was observed in OH11-treated zoospores. After 24 hpi, the TSB- or E. coli-treated P. sojae invaded into soybean and grew in soybean cells, while OH11-treated P. sojae showed no invasive hyphae (Figure 1B), suggesting that OH11 completely inhibited the infection ability of P. sojae to soybean cells. Using a similar method, we found that OH11 also inhibited the virulence of P. capsici (LT263) on N. benthamiana leaves and P. infestans (88069) on potato (Solanum tuberosum) leaves (Figure 1A). These results collectively indicated that L. enzymogenes OH11 could effectively prevent infection of its host plants by various Phytophthora pathogens.
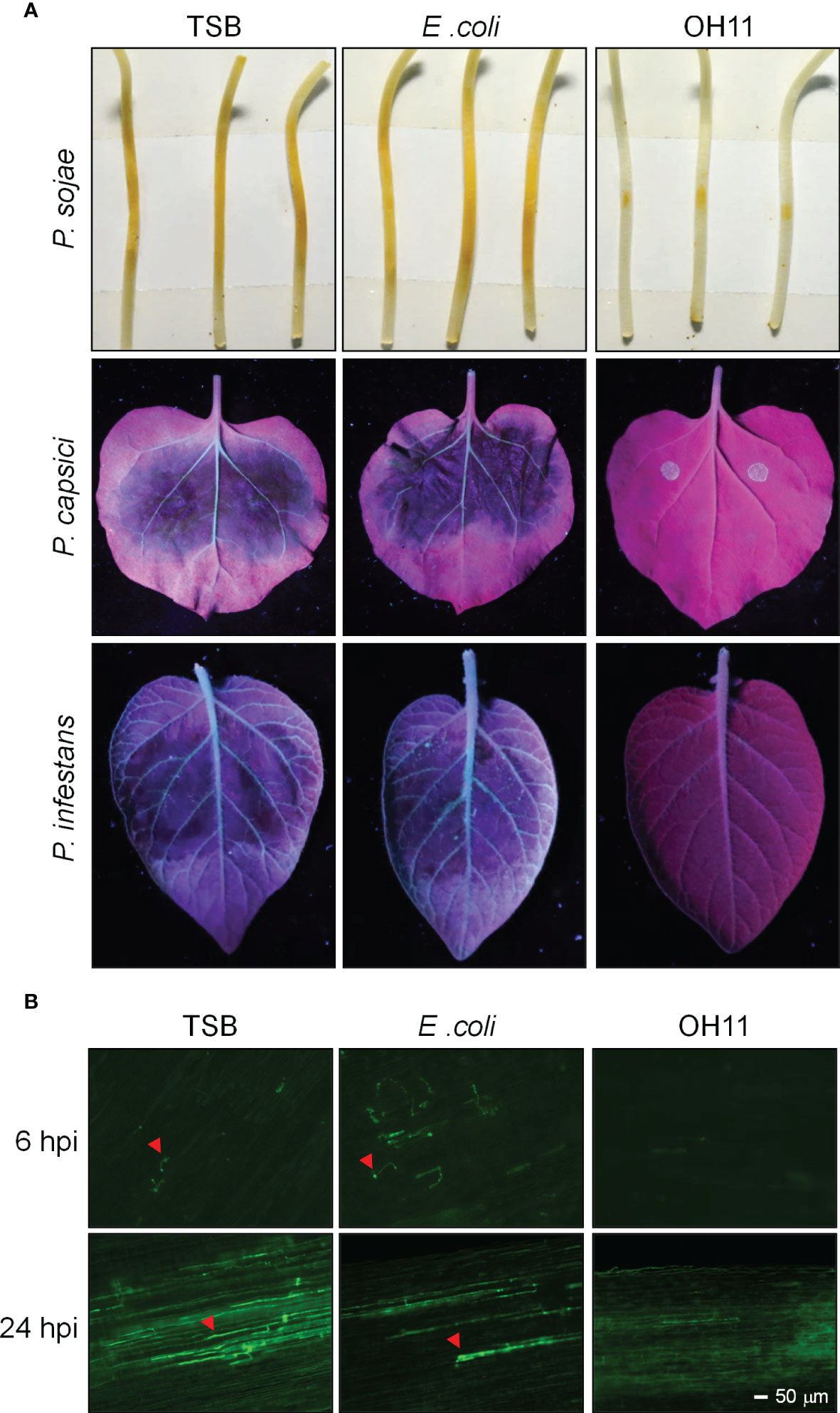
Figure 1 L. enzymogenes OH11 inhibited the virulence of Phytophthora. (A) OH11 inhibited the virulence of P. sojae, P. capsici and P. infestans. A total of 200 Phytophthora zoospores were mixed with an equal volume of TSB media, bacterial cultures of E coli and OH11, individually. The mixtures were inoculated onto host plants (soybean, N. benthamiana, or potato). TSB- and E. coli-treated Phytophthora had obvious lesions on host plants, while OH11- treated Phytophthora revealed no lesion. (B) OH11 inhibited infection of GFP-labeled P. sojae zoospores. At 6 hpi, TSB- and E coli-treated P. sojae showed germinated cysts that could efficiently infect soybean cells, whereas OH11-treated P. sojae was not able to infect plants. At 24 hpi, TSB- or E coli-treated P. sojae infected soybean cells, whereas OH11-treated GFP-labeled P. sojae showed no invasive hyphae in soybean cells. Red arrows indicate the invasive hyphae of P. sojae.
To understand how OH11 targets Phytophthora pathogens to suppress their infection, we first tested whether the antifungal metabolite HSAF is a potentially key factor (Yu et al., 2007). For this purpose, we carried out a plate-based anti- Phytophthora assay in which 5 μL cultures of wild-type OH11 or the HSAF-deficient mutant ΔlafB generated in a previous study (Wang et al., 2017) were spotted on the surface edge of V8 or RSA agar plates followed by inoculation of three Phytophthora pathogens (P. sojae P6497, P. capsici LT263 and P. infestans 88069) in the center of the plate. After incubation for 3 days, we found that wild-type OH11 produced a distinct zone of inhibition around the tested Phytophthora colonies, whereas HSAF-deficient mutants almost lost this antagonistic activity (Figure 2A).
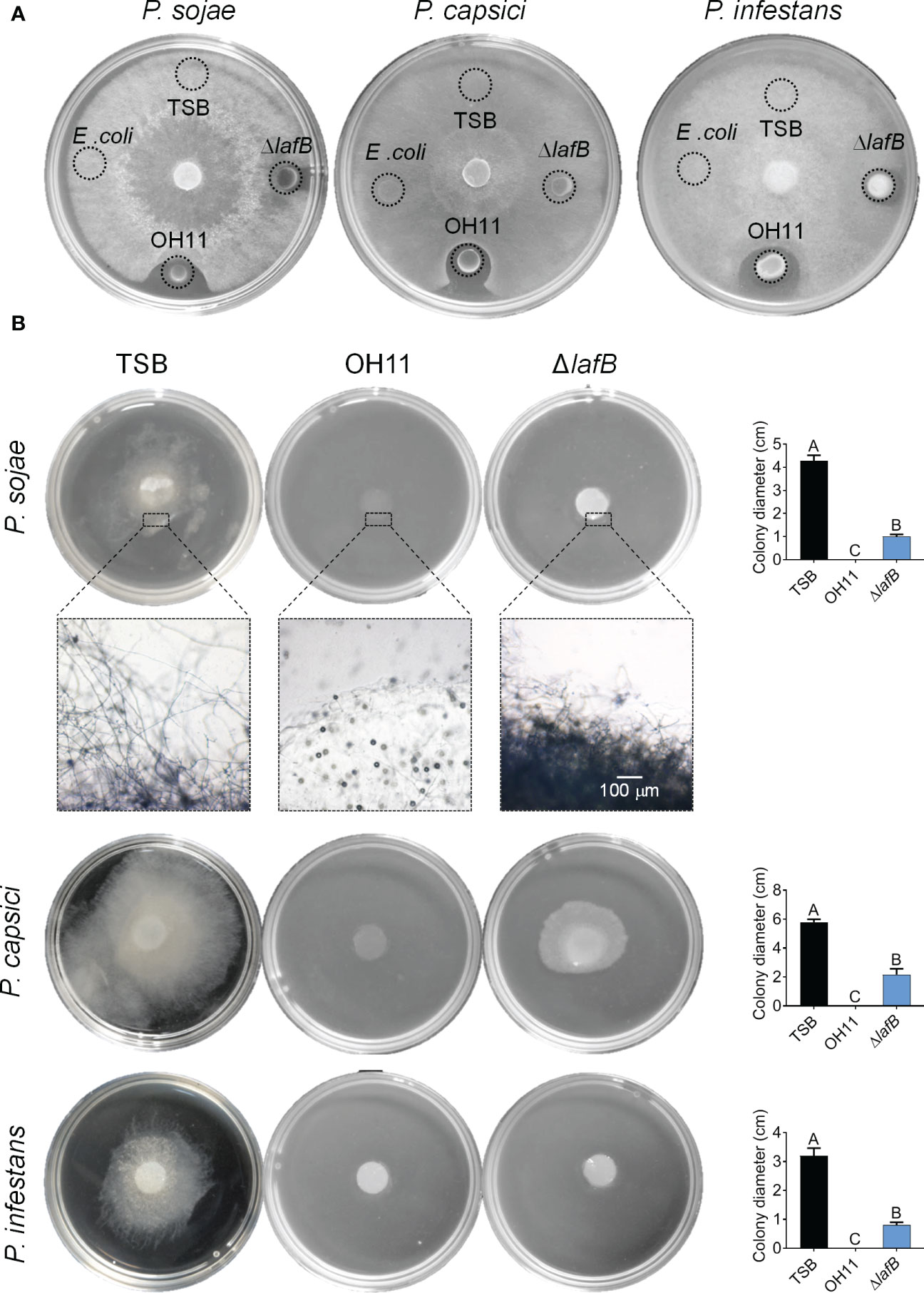
Figure 2 L. enzymogenes OH11 inhibited mycelial growth of the Phytophthora pathogens. (A) Compared with wild-type OH11, the HSAF-deficient mutant ΔlafB had less antagonistic activity against Phytophthora on solid media. lafB is a key gene for HSAF biosynthesis. (B) Wild-type OH11 degraded mycelia of Phytophthora. Phytophthora hyphal plugs were cultured in liquid media mixed with TSB, OH11, and ΔlafB, respectively. OH11 completely inhibited Phytophthora mycelial growth and degraded the mycelia in hyphal plugs, while ΔlafB inhibited mycelial growth of Phytophthora. Three replicates of each sample were analyzed using a one-way ANOVA test.
To confirm the above findings, we carried out similar assays in liquid media in which hyphal plugs of P. sojae P6497, P. capsici LT263 or P. infestans 88069 were mixed with empty V8 broth (negative control) or L. enzymogenes cultures, respectively. After 3-day co-inoculation, we found that wild-type OH11 significantly inhibited mycelial growth of the three Phytophthora pathogens mentioned above compared with the negative control (Figure 2B). Under similar testing conditions, we were surprised to find that the HSAF-deficient mutant ΔlafB also showed significant inhibition of mycelial growth compared to the negative control (Figure 2B). Via microscopic observation represented by P. sojae, we further observed that wild-type OH11 could completely degrade mycelia, while HSAF-deficient mutants and negative control (TSB) could not, although ΔlafB also effectively inhibited hyphal growth in liquid medium (Figure 2B). These results suggested that, while HSAF is important during the antagonistic interaction between L. enzymogenes and Phytophthora pathogens, other HSAF-independent factors were also involved in this process.
Besides mycelial growth, cyst germination is another key step preceding Phytophthora infection (Judelson and Blanco, 2005). We therefore investigated whether L. enzymogenes OH11 could inhibit cyst germination of Phytophthora pathogens in an HSAF-dependent and/or HSAF-independent manner. We carried out a corresponding assay to address this issue using GFP-labeled P. sojae as a representative. We mixed P. sojae cysts with cultures of wild-type OH11 or HSAF-deficient mutant ΔlafB in liquid broth, followed by microscopic observation. As shown in Figure 3A, approximately 80% of P. sojae cysts were degraded by OH11 after 6 hours of co-incubation compared to the negative control (TSB), and almost all remaining cysts showed undetectable GFP signals, indicating that L. enzymogenes was able to kill P. sojae cysts instantly. This killing effect appeared to be specific, as another negative control (E. coli) failed to do so. Under similar testing conditions, although ΔlafB failed to directly kill P. sojae cysts, this mutant also significantly reduced germination rate and germ tube length of P. sojae (Figure 3A). After 24 hours of co-incubation, wild-type OH11 completely digested cysts, whereas ΔlafB only inhibited cyst germination (Supplemental Figure S1). Similar findings were also observed when P. capsici and P. infestans cysts were applied (Figures 3B, C). Together, these results indicated that L. enzymogenes OH11 had the ability to directly degrade Phytophthora cysts, and that HSAF was involved in this process. In the absence of HSAF, L. enzymogenes might also inhibit cyst germination through other uncharacterized factors/pathways.
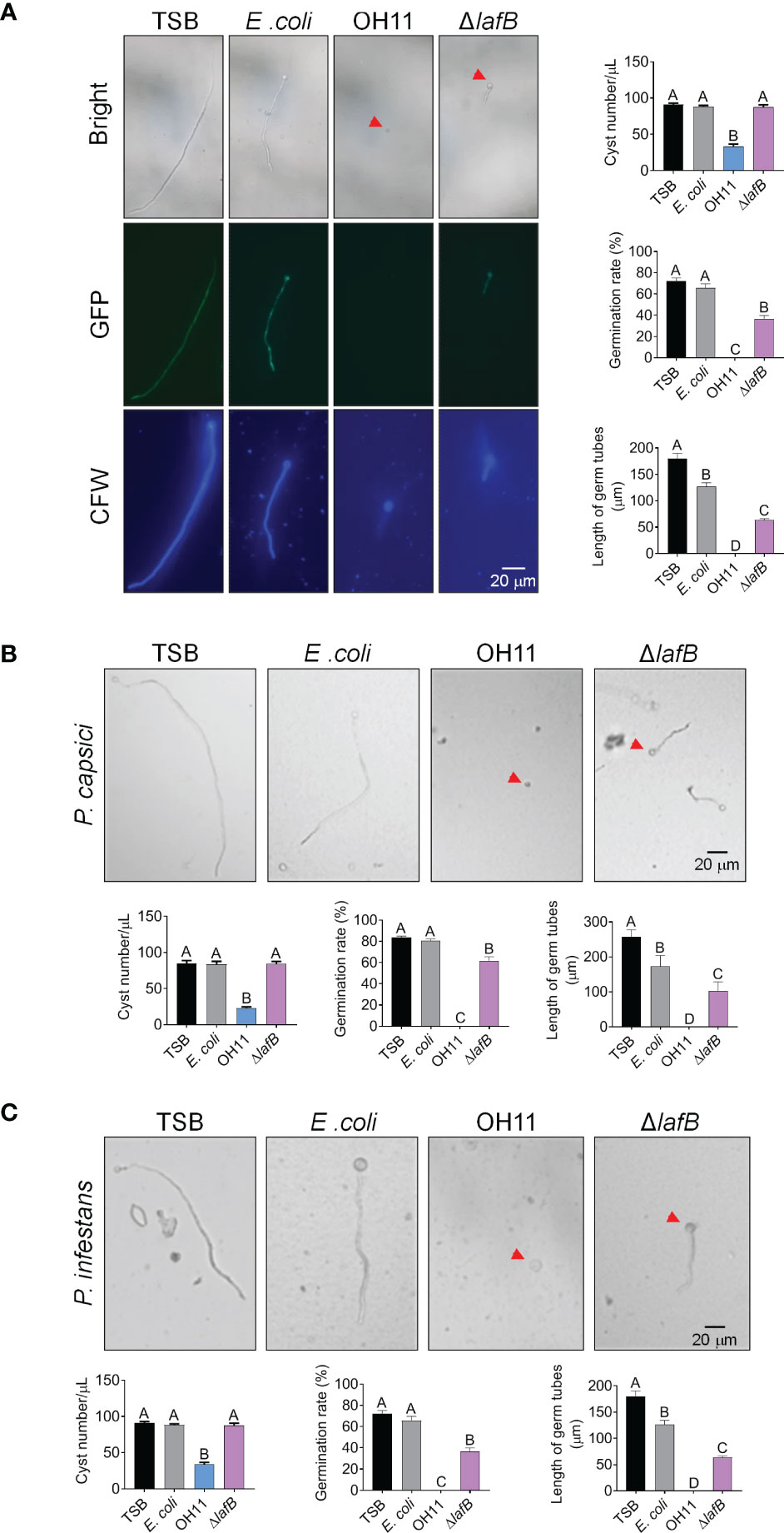
Figure 3 L. enzymogenes OH11 inhibited cyst germination of Phytophthora. (A) Wild-type OH11 degraded P. sojae cysts, whereas the HSAF-deficient mutant ΔlafB did not, but inhibited cyst germination. GFP-labeled P. sojae cysts were mixed with TSB, E coli, OH11, or ΔlafB, and cultured at 25°C for 6 hours. Compared to TSB control, OH11 treatment completely inhibited cyst germination in P. sojae and the number of cysts after OH11 treatment was significantly reduced. At the same time, both the germination rate and the germinated tube length of P. sojae were significantly reduced after ΔlafB treatment. Three replicates of each sample were analyzed using a one-way ANOVA test. Red arrows indicate P. sojae cysts. WT-treated cysts showed a lower GFP signal, indicating that WT-treatment resulted in cell death of P. sojae cysts. Cysts were stained with chitin-binding CFW to make them clearer to observe. (B, C) WT degraded P. capsici (B) and P. infestans (C) cysts, whereas ΔlafB only inhibited germination of the corresponding cysts. P. capsici and P. infestans cysts were mixed with TSB, E coli, OH11, and ΔlafB, respectively, and cultured at 25°C (P. capsici) or 18°C (P. infestans) for 6 hours (P. capsici) or 12 hours (P. infestans). Three replicates of each sample were analyzed with a one-way ANOVA test. Red arrows indicate P. capsici or P. infestans cysts.
The interesting findings above prompted us to explore the underlying mechanisms at the molecular level. To this end, we carried out an RNA-seq assay to identify P. sojae cyst genes whose transcription could be significantly altered by L. enzymogenes OH11 treatment. To facilitate this assay and considering that wild-type OH11 can degrade the cysts in liquid medium, we therefore depleted OH11 cells by filter and treated P. sojae cysts with the cell-free supernatant that exhibited resistance to P. sojae activity similar to total OH11 culture (Supplemental Figure S2). The results of RNA-seq assay showed that among the 13014 expressed P. sojae genes, 1741 genes were up-regulated and 2176 genes were down-regulated in the OH11-treated P. sojae compared with the negative control (TSB-treated) (Figure 4A). Details of differentially expressed genes (DEGs) were listed in Supplemental Table S1. The functions of DEGs were enriched in basal metabolism, including translation initiation, RNA binding, and amino acid metabolism (Supplemental Figure S3). Meanwhile, a large number of genes involved in signal transduction (46 in total number), stimulus response (95 in total number), antioxidation (20 in total number), and transporting (210 in total number) were also differentially expressed (Figure 4B). Considering that Phytophthora can secrete effectors to subvert plant immunity and promote pathogen infection (Wang and Wang, 2018), and several transcription factors (TFs) of bZIP, MYB and HSF families are key transcriptional regulators of during Phytophthora cyst germination (Zhang et al., 2012; Ye et al., 2013; Sheng et al., 2015), we also investigated whether these genes were included in the RNA-seq data. As shown in Figure 4C, we found that the expression levels of 27 RxLR effector genes and 21 CRN effector genes as well as 15 bZIP TFs, 5 HSF TFs and 20 MYB TF genes were significantly changed when P. sojae interacted with L. enzymogenes OH11. In addition, the expression levels of PsAvh457 (RxLR effector), PsCRN70 (CRN effector), PsBZP4 (bZIP TF), Ps138282 (MYB TF), Ps156741 (homolog of apoptosis-inducing centromere protein) and Ps131084 (cell wall endo-beta-1, 3-glucanase) were verified by quantitative real-time PCR (qRT-PCR) (Figure 4D). These results provided molecular evidence supporting the conclusion that L. enzymogenes prevented P. sojae infection by attacking the cysts.
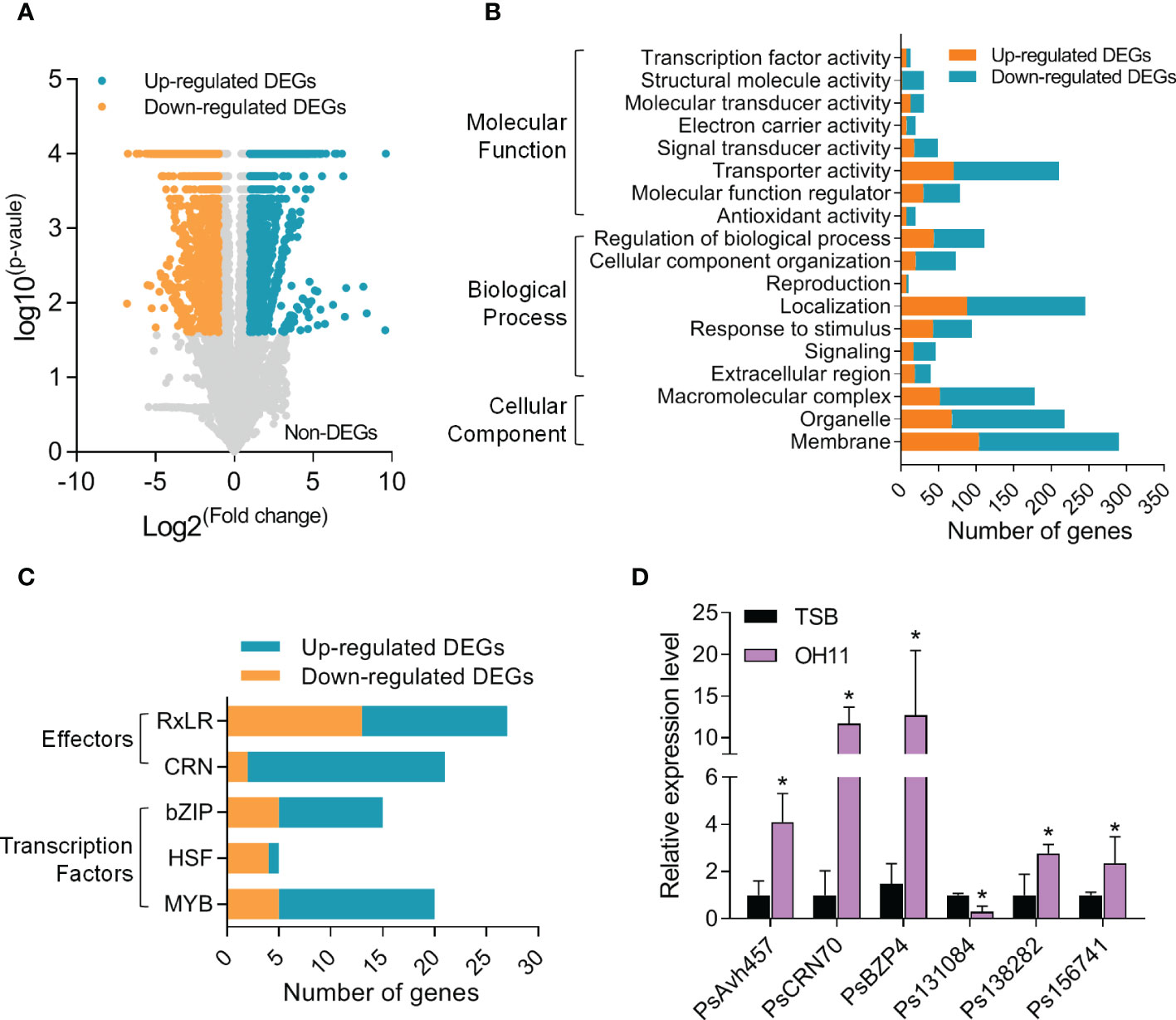
Figure 4 L. enzymogenes OH11 treatment resulted in global changes in P. sojae cyst gene expression. (A) Volcano plot of OH11-treated P. sojae DEGs (differentially expressed genes) compared to medium (TSB) control. Orange spots indicate the 1741 up-regulated genes; blue spots indicate the 2176 down-regulated genes; grey spots list those non-differentially expressed genes. (B) Number of DEGs categorized into different GO terms. (C) Number of DEGs categorized into different effector and transcription factor families. (D) Expression levels of representative DEGs verified by qRT-PCR. Three replicates of each sample were analyzed with a t-test. Asterisks indicate significant differences (P < 0.01).
In addition to directly targeting pathogens, eliciting host plant immune response is another common biocontrol strategy shared by plant-beneficial Pseudomonas and Bacillus species (Yu et al., 2022). We were therefore also interested in finding out whether L. enzymogenes could use this similar strategy to prevent Phytophthora infection. To address this question, we selected P. capsici-N. benthamiana interaction system as a working model. We first treated leaves of N. benthamiana with TWEEN-20 to promote bacterial adhesion to plant surfaces (Paul et al., 2012), and then sprayed cultures of wild-type OH11 and ΔlafB on the leaves. Meanwhile, empty TSB broth was used as a negative control. After 12 hours and 24 hours of treatment, we examined the expression levels of those well-characterized plant defence-related genes in N. benthamiana that serve as molecular indicators of plant immune responses, as previously described (Yang et al., 2021). The result showed that both wild-type OH11 and ΔlafB induced the expression of NbPR1b (Pathogenesis-related protein 1b), NbPR2, NbPR2b, NbPR4 and NbLOX (Lipoxygenase) at 24h. At 12 h, OH11 induced higher levels of NbPR2, NbPR4 and NbLOX gene expression compared with ΔlafB (Figure 5A). Among them, NbPR1b, NbPR2 and NbPR2b are known maker genes of the salicylic acid (SA) signaling pathway, while NbPR4 and NbLOX are well characterized to participate in the jasmonic acid (JA) signaling pathway (Zhang et al., 2015; Yu et al., 2022). These results indicated that OH11 could elicit plant immune response in a HSAF-independent manner by activating the expression of defense genes involved in the SA and JA signaling pathways. To validate this point, we conducted another plant-based biocontrol assay. We inoculated P. capsici zoospores on N. benthamiana leaves at 24 hours after L. enzymogenes treatment and found that both wild-type OH11 and ΔlafB effectively inhibited the infection of N. benthamiana by P. capsici (Figure 5B).
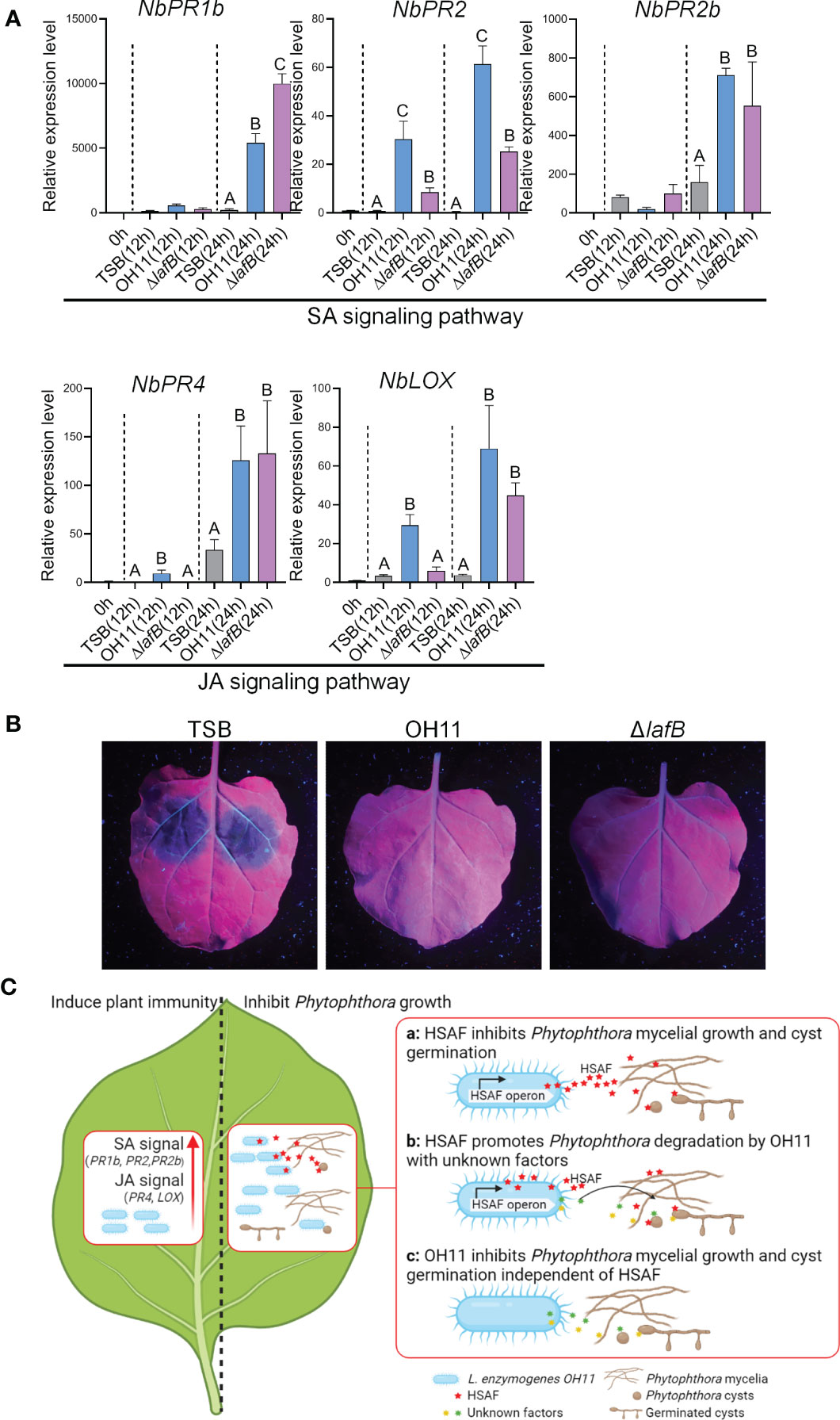
Figure 5 L. enzymogenes OH11 treatment elicited immunity responses in N. benthamiana. (A) Treatment of wild-type OH11 and HSAF-deficient mutant ΔlafB up-regulated expression levels of defense-related genes in N. benthamiana leaves. Gene expression levels were measured 12 and 24 hours after treatment, respectively. The relative expression level of untreated leaves (0h) was set to 1.0. Three replicates of each sample were analyzed using a one-way ANOVA test. (B) Pretreatment with WT and ΔlafB prevented infection of N. benthamiana leaves from P. capsici. P. capsici zoospores were inoculated onto N. benthamiana leaves 24 hours after pretreatment. TSB-treated N. benthamiana leaves showed obvious lesions, while OH11- and ΔlafB-treated leaves did not show any lesion. (C) A working model showing how L. enzymogenes prevented Phytophthora infection. When L. enzymogenes colonized plant surfaces through biofilms, it induced an immune response (left panel) to protect plants from pathogen infection by activating genes involved in SA and JA signaling. On the right, L. enzymogenes could directly target and inhibit the growth of Phytophthora. HSAF is a key weapon of OH11 that inhibited Phytophthora mycelial growth and cyst germination (a). HSAF also enhanced the degradation activity of OH11 on Phytophthora mycelia and cysts by unknown factors (b). When HSAF was blocked by environmental factors, OH11 could also inhibit Phytophthora mycelial growth and cyst germination through other unknown factors (c). The symbol ABCs indicate significant differences (P < 0.01).
Oomycetes represented by Phytophthora are phylogenetically distant from fungi, and are difficult to control by most antifungal agents (Latijnhouwers et al., 2003). On the other hand, most Phytophthora species have sizeable, repeat-rich genomes. The variability of the genome leads to fast adaptive evolution, which is conducive to the rapid evasion of host resistance genes of Phytophthora and the development of resistance to chemical fungicides (Leesutthiphonchai et al., 2018). Biological control is a sustainable and environmentally friendly solution for managing Phytophthora diseases. Most biocontrol activity against the bacterial Phytophthora is measured by antagonistic activity against the mycelium. Previously, the studies on the interaction between Lysobacter and Phytophthora were mainly limited to the inhibition of mycelial growth, while zoospores are the main carrier of Phytophthora dispersal and infection (Judelson and Blanco, 2005). In the field, zoospores can sense chemical signals released by host plant and swim chemotactically to the host. After attaching to the host, zoospores will lose their flagella and form walled cyst. The cyst will then germinate and extend a germ tube to start infection (Judelson and Blanco, 2005). In addition to inhibiting mycelium growth, this study also reported that L. enzymogenes OH11 could degrade cysts and inhibited infection by various Phytophthora, making OH11 a promising agent for field control of Phytophthora pathogens.
HSAF is a polycyclic tetramate macrolactam synthesized and secreted by L. enzymogenes with broad-spectrum inhibitory activity against filamentous fungi (Lou et al., 2011). Compared with wild-type OH11, the HSAF-deficient mutant ΔlafB showed defective activity against Phytophthora, consistent with the earlier finding that HSAF has a direct inhibitory effect on Phytophthora mycelium growth (Li et al., 2008). This was also consistent with earlier reports that L. enzymogenes C3 mutants unable to release HSAF shows reduced activity in inhibiting fungal mycelial growth and spore germination (Yu et al., 2007; Li et al., 2008). Interestingly, unlike wild-type OH11, ΔlafB failed to degrade mycelia and cysts of Phytophthora, indicating that HSAF had an additional previously unknown role beyond its direct anti-Phytophthora effect.
HSAF targets fungal sphingolipids biosynthesis by blocking ceramide synthase in A. nidulans, resulting in arrest of hyphal tip elongation (Li et al., 2006). HSAF also induces apoptosis in Candida albicans by triggering the generation of reactive oxygen species and binding to β-tubulin (Ding et al., 2016). Besides, HSAF treatment disrupts multiple signaling networks and fundamental cellular metabolisms in A. alternata (He et al., 2018). Our RNA-seq data showed that OH11 treatment resulted in differential expression of 20 antioxidant-related genes and one apoptotic-inducing centromere protein homolog in Phytophthora. These Phytophthora gene expression changes might be due to HSAF-induced apoptosis and ROS (reactive oxygen species) burst, providing clues for future investigations on the mode of action of HSAF against the Phytophthora pathogens.
It is interesting to observe the direct digestion of Phytophthora mycelia and cysts by L. enzymongenes OH11. Although we do not have a definitive answer to explain how this occurs, abundant cell wall-targeting cleavage enzymes (i.e. protease and β-1,3-glucanase) secreted by L. enzymogenes might be involved in this process. In agreement, previous reports have shown that HSAF-deficient mutants produce some unidentified thermostable antifungal factors that degrade fungal mycelia (Odhiambo et al., 2017). Therefore, these unidentified factors might also have activity against Phytophthora.
Interactions between biocontrol bacteria and phytopathogenic fungi/oomycetes are complex. Bacteria attack fungi/oomycetes with various weapons, and fungi/oomycetes defend bacteria attack through different patterns (Tomada et al., 2017; Wang et al., 2020). 15 bZIP TFs, 5 HSF TFs and 20 MYB TFs of P. sojae showed differential expression levels during interacting with L. enzymogenes. Disorder of gene expression regulation may involve in degradation of Phytophythora cysts. On the other hand, differentially expression of TFs may be a feedback response to defend the OH11 inhibition. Phytophthora pathogens cohabited with other microbes and showed anti-bacteria activity to gain ecological adaptation in the field. Effectors secreted by Phytophthora not only subvert plant immunity and promote pathogen infection, but also interfere bacteria physiology to compete with bacteria in the same habitat and defend attack from biocontrol bacteria (Wang and Wang, 2018; Wang et al., 2020). It is noteworthy that expression of 14 RxLR effectors and 19 CRN effectors were induced when P. sojae interacting with L. enzymogenes. These effectors may involve in inhibiting or defending the OH11 inhibition.
Many biocontrol bacteria, represented by Bacillus and Pseudomonas, can stimulate plant immune responses and help plants acquire broad-spectrum resistance to pathogens (Yu et al., 2022). Cellular components of these bacteria, such as flagellin and lipopolysaccharides (LPS), can be recognized by plant cell surface receptors to generate pattern-triggered immunity (PTI) (Zamioudis and Pieterse, 2012). In addition to cellular components, some secondary metabolites produced by beneficial bacteria also induce plant defense responses (Yu et al., 2022). For example, pyocyanin produced by Pseudomonas activates plant defense responses by inducing the accumulation of H2O2 (De Vleesschauwer et al., 2006). L. enzymogenes C3 were reported to induce common resistance of tall fescue and wheat to fungi pathogens, and this immune induction was independent of the strain’s antifungal ability (Kilic-Ekici and Yuen, 2003). We found that L. enzymogenes OH11 could induce the expression levels of defence-related genes involved in SA and JA signalling pathways, which appeared to be independent of HSAF, but seemed to enable L. enzymogenes to protect host plants from pathogen infection under natural conditions where HSAF generation was hindered by environmental factors. It is also noteworthy that L. enzymogenes is a non-flagellated bacterium that does not produce flagellin (Xia et al., 2018). Instead, it displays twitching motility driven by type IV pili (T4P) on solid surface and is essential for the bacterium to form biofilms that facilitate its colonization on plants (Xia et al., 2018). Using T4P, L. enzymogenes can secrete pili that are the first to contact the plant surface. Therefore, it is possible that non-flagellated L. enzymogenes used type IV pilin as the initial signal to elicit plant immune response, a hypothesis we are now testing in our laboratory.
In conclusion, we reported that L. enzymogenes OH11 could protect plants against Phytophthora infection by inducing plant immune responses through activation of SA and JA signaling pathways. OH11 also protected plants by directly targeting Phytophthora, which not only secreted HSAF to inhibit Phytophthora mycelium growth as previously described (Li et al., 2008), but also directly degraded Phytophthora mycelia and cysts in a HSAF-dependent manner. Furthermore, OH11 could also inhibit Phytophthora mycelial growth and cyst germination in the absence of HSAF through unknown factors (Figure 5C). These multiple mechanisms made L. enzymogenes OH11 an ideal bacterium for the control of various Phytophthora diseases.
The strains used in this study are listed in Supplementary Table S2. P. sojae strain P6497 and P. capsici strain LT263 were routinely cultured on 10% V8 media at 25°C, while P. infestans strain 88069 was routinely cultured on RSA media at 18°C. Soybean cultivar Hefeng47 was grown at 25°C in the dark for 4 days in environmentally controlled growth chamber, and etiolated soybean seedlings were used for infection assays. N. benthamiana was grown at 25°C in a growth chamber with a 16 h light and 8 h dark photoperiod, and 4-week-old leaves were selected for infection assay. Potato cultivar Desiree was grown at 22°C in a growth chamber with a 16 h light and 8 h dark photoperiod, and 4-week-old leaves were selected for infection assay. Bacteria strains were grown overnight in LB media at 28°C and then transferred to 10% TSB media for 24 h at 28°C to induce HSAF production.
For mycelial growth assays on solid media, 5 × 5-mm hyphal plugs of P. sojae or P. capsici were inoculated onto 10% V8 medium plates at 25°C, while P. infestans hyphal plugs were inoculated onto RSA medium plates at 18°C. When the fresh mycelia grew to a suitable size (3 cm in diameter), 5 μL of bacterial culture (diluted to an OD600 = 1.0) was inoculated to the edge of the plates. Antagonistic activity was measured by an inhibition zone around the colony. For mycelial growth assays in liquid media, 5 × 5-mm hyphal plugs of P. sojae or P. capsici were inoculated onto the plates containing 4 mL 10% V8 liquid medium at 25°C, while P. infestans hyphal plugs were inoculated onto plates containing 4 mL PEA liquid medium at 18°C. 1 mL of bacterial culture (diluted to an OD600 = 1.0) was inoculated onto the plates. Antagonistic activity was measured by the size of Phytophthora colony. Mycelia colonies were observed with a microscope (Zeiss Axio Observer 3).
To induce zoospore production, mycelia of P. sojae or P. capsici were cultured in 10% liquid V8 medium for 3 days, washed with sterile water, and incubated at 25°C until zoospores were produced. To induce zoospore production of P. infestans, mycelia was grown on RSA medium plate at 18°C for 14 days. The sporangia were scraped into sterile water and incubated at 4°C until zoospores were released. Zoospores were diluted to 100 zoospores/μL and mixed with an equal volume of bacterial cultures (diluted to an OD600 = 1.0). A final mixture containing 200 zoospores were inoculated onto the host plants. Infected soybean and N. benthamiana were incubated in the dark at 25°C for 2 days before observation. Infected potatoes were incubated in the dark at 18°C for 3 days before observation. Lesions of plant leaves were observed under UV light. Infection details of GFP-labelled P. sojae zoospores were observed with a microscope (Zeiss Axio Observer 3).
To examine the immune response of N. benthamiana, leaves of N. benthamiana were quickly soaked into 0.5% (v/v) TWEEN-20 and washed with H2O to remove TWEEN-20. The TSB, OH11 and ΔlafB cultures were then sprayed on the leaves individually. Treated leaves were incubated at 25°C in the dark before sampling. To detect immune gene expression, leaf samples were collected 12 and 24 hours after treatment. For the infection assay, 200 P. capsici zoospores were inoculated onto N. benthamiana leaves 24 hours after treatment. Infected leaves were incubated at 25°C in the dark for 2 days before observation.
Phytophthora zoospores were induced and harvested into 1.5 mL tubes. Tubes containing 100 zoospores/μL in a 500 μL suspension were vortexed for 90 s to induce cyst formation. 500 μL cyst suspension were mixed with 500 μL 10% V8 medium and 500 μL bacterial culture (diluted to an OD600 = 1.0), and then incubated at 25°C to observe the germination of P. sojae or P. capsici. 500 μL P. infestans cyst suspension were mixed with 500 μL PEA medium and 500 μL bacterial culture (diluted to an OD600 = 1.0), and then incubated at 18°C to observe the germination, which was observed with a microscope (Zeiss Axio Observer 3). GFP-labeled P. sojae cysts were stained with 1 μg/mL chitin-binding CFW (Caleofluor White) for 5 min to make cysts clearer to observe.
For RNA-seq sampling, bacterial culture of L. enzymogenes OH11 were collected and diluted to a density of 1.0 at 600 nm. The bacteria culture was then centrifuged and filtered through a 0.22-μm filter to remove OH11 cells. P. sojae cysts were diluted to 100 cyst/µL with sterile ddH2O. Bacterial suspension and cysts were mixed in a 1:1 ratio and cultured for 6 hours at 25°C prior to sampling. TSB media-treated cysts were used as negative controls. Three replicates of each sample were collected and sent to BGI Genomics Corporation for RNA-seq. P. sojae RNA was extracted using the TRIzol® method following the manufacturer’s protocol. RNA-seq was conducted using the DNBseq platform and 100-bp paired-end modules.
Raw reads were filtered for subsequent analysis by removing reads containing adapter, poly-N, and low-quality reads. Clean reads were mapped to the genome of P. sojae (v1.1 for isolate P6497) using Tophat with up to two mismatches. Mapped reads were quantified using the Cufflinks program, and transcript levels for each gene were quantified as RPKM (reads per kilobase transcript length per million reads mapped). Differentially expressed genes were identified using featureCounts software, and log2 fold change (log2FC) values and adjusted P-values were calculated using DESeq2 software. Genes with adjusted P-value < 0.05 and absolute log2FC ≥1 were considered differentially expressed.
For qRT-PCR assays, cDNA was synthesized with PrimeScript First Strand cDNA Synthesis Kit (TaKaRa Bio Inc.) following the manufacturer’s protocol. qRT-PCR was performed in a reaction mixture of 20 μL of SYBR Premix ExTaq (TaKaRa Bio Inc.) following the manufacturer’s protocol. PCR was performed on an ABI Prism 7500 Fast Real-Time PCR System (Applied Biosystems Inc.) The P. sojae actin gene (Psactin, Ps108986) was used as a constitutively expressed endogenous control for P. sojae. The N. benthamiana elongation factor 1-alpha gene (NbEF1α) was used as endogenous control for N. benthamiana. Primers used in this study are listed in Supplementary Table S3.
The RNA-seq data presented in the study are deposited in the NCBI database, accession number PRJNA909446.
GQ and XS conceived the project and designed experiments. LL, ZY, MT, CC and PW carried out experiments. LL, DS, LW, GQ and XS analyzed data and prepared figures and tables. LL, MJ, GQ and XS wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was funded by Science and Technology Project of Shaanxi Branch of China National Tobacco Corporation (KJ-2022-04 to GQ), the National Natural Science Foundation of China (32272619 to XS, U22A20486 and 32072470 to GQ, 32001955 to LL), and the project of dominant discipline in Jiangsu province (80900246 to XS).
We thank Prof. Yuanchao Wang (Nanjing Agricultural University) and Prof. Suomeng Dong (Nanjing Agricultural University) for providing Phytophthora and plant strains used in this study.
The author CC and PW are employed by Shaanxi Provincial Tobacco Corporation of CNTC. They contributed to the pathogenicity test of P. capsici to N. benthamiana.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1116147/full#supplementary-material
Supplementary Figure 1 | L. enzymogenes OH11 completely digested the cysts of P. sojae 24 hours after treatment.
Supplementary Figure 2 | L. enzymogenes OH11 supernatant inhibited cyst germination of P. sojae 6 hours after treatment.
Supplementary Figure 3 | GO enrichment of Phytophthora cyst DEGs treated with OH11.
Baldauf, S. L., Roger, A. J., Wenk-Siefert, I., Doolittle, W. F. (2000). A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 290, 972–977. doi: 10.1126/science.290.5493.972
Caulier, S., Gillis, A., Colau, G., Licciardi, F., Liepin, M., Desoignies, N., et al. (2018). Versatile antagonistic activities of soil-borne bacillus spp. and Pseudomonas spp. against Phytophthora infestans and other potato pathogens. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00143
de Andrade Lourenco, D., Branco, I., Choupina, A. (2022). A systematic review about biological control of phytopathogenic Phytophthora cinnamomi. Mol. Biol. Rep. 49 (10), 9947–9962. doi: 10.1007/s11033-022-07547-2
De Vleesschauwer, D., Cornelis, P., Hofte, M. (2006). Redox-active pyocyanin secreted by Pseudomonas aeruginosa 7NSK2 triggers systemic resistance to Magnaporthe grisea but enhances Rhizoctonia solani susceptibility in rice. Mol. Plant Microbe Interact. 19, 1406–1419. doi: 10.1094/MPMI-19-1406
Ding, Y., Li, Z., Li, Y., Lu, C., Wang, H., Shen, Y., et al. (2016). HSAF-induced antifungal effects in Candida albicans through ROS-mediated apoptosis. Rsc. Adv. 6, 30895–30904. doi: 10.1039/C5RA26092B
Fry, W. (2008). Phytophthora infestans: the plant (and r gene) destroyer. Mol. Plant Pathol. 9, 385–402. doi: 10.1111/j.1364-3703.2007.00465.x
Han, X., Shen, D., Xiong, Q., Bao, B., Zhang, W., Dai, T., et al. (2021). The plant-beneficial rhizobacterium Bacillus velezensis FZB42 controls the soybean pathogen Phytophthora sojae due to bacilysin production. Appl. Environ. Microbiol. 87 (23), e0160121. doi: 10.1128/AEM.01601-21
He, F., Li, B., Ai, G., Kange, A., Zhao, Y., Zhang, X., et al. (2018). Transcriptomics analysis of the Chinese pear pathotype of Alternaria alternata gives insights into novel mechanisms of HSAF antifungal activities. Int. J. Mol. Sci. 19 (7), 1841. doi: 10.3390/ijms19071841
Judelson, H. S., Blanco, F. A. (2005). The spores of Phytophthora: weapons of the plant destroyer. Nat. Rev. Microbiol. 3, 47–58. doi: 10.1038/nrmicro1064
Kamoun, S., Furzer, O., Jones, J. D., Judelson, H. S., Ali, G. S., Dalio, R. J., et al. (2015). The top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 16, 413–434. doi: 10.1111/mpp.12190
Kilic-Ekici, O., Yuen, G. Y. (2003). Induced resistance as a mechanism of biological control by Lysobacter enzymogenes strain C3. Phytopathology. 93, 1103–1110. doi: 10.1094/PHYTO.2003.93.9.1103
Ko, H. S., Jin, R. D., Krishnan, H. B., Lee, S. B., Kim, K. Y. (2009). Biocontrol ability of Lysobacter antibioticus HS124 against Phytophthora blight is mediated by the production of 4-hydroxyphenylacetic acid and several lytic enzymes. Curr. Microbiol. 59, 608–615. doi: 10.1007/s00284-009-9481-0
Lamour, K. H., Stam, R., Jupe, J., Huitema, E. (2012). The oomycete broad-host-range pathogen Phytophthora capsici. Mol. Plant Pathol. 13, 329–337. doi: 10.1111/j.1364-3703.2011.00754.x
Latijnhouwers, M., de Wit, P. J., Govers, F. (2003). Oomycetes and fungi: similar weaponry to attack plants. Trends. Microbiol. 11, 462–469. doi: 10.1016/j.tim.2003.08.002
Leesutthiphonchai, W., Vu, A. L., Ah-Fong, A. M. V., Judelson, H. S. (2018). How does Phytophthora infestans evade control efforts? modern insight into the late blight disease. Phytopathology .108 (8), 916–924. doi: 10.1094/PHYTO-04-18-0130-IA
Li, Y., Chen, H., Ding, Y., Xie, Y., Wang, H., Cerny, R. L., et al. (2014). Iterative assembly of two separate polyketide chains by the same single-module bacterial polyketide synthase in the biosynthesis of HSAF. Angew. Chem. Int. Ed. Engl. 53, 7524–7530. doi: 10.1002/anie.201403500
Li, S., Du, L., Yuen, G., Harris, S. D. (2006). Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell. 17, 1218–1227. doi: 10.1091/mbc.e05-06-0533
Li, S., Jochum, C. C., Yu, F., Zaleta-Rivera, K., Du, L., Harris, S. D., et al. (2008). An antibiotic complex from Lysobacter enzymogenes strain C3: antimicrobial activity and role in plant disease control. Phytopathology. 98, 695–701. doi: 10.1094/PHYTO-98-6-0695
Lin, L., Xu, K., Shen, D., Chou, S. H., Gomelsky, M., Qian, G. (2021). Antifungal weapons of Lysobacter, a mighty biocontrol agent. Environ. Microbiol. 23, 5704–5715. doi: 10.1111/1462-2920.15674
Liu, D., Li, K., Hu, J., Wang, W., Liu, X., Gao, Z. (2019). Biocontrol and action mechanism of Bacillus amyloliquefaciens and Bacillus subtilis in soybean Phytophthora blight. Int. J. Mol. Sci. 20 (12), 2908. doi: 10.3390/ijms20122908
Lou, L., Qian, G., Xie, Y., Hang, J., Chen, H., Zaleta-Rivera, K., et al. (2011). Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J. Am. Chem. Soc 133, 643–645. doi: 10.1021/ja105732c
Odhiambo, B. O., Xu, G., Qian, G., Liu, F. (2017). Evidence of an unidentified extracellular heat-stable factor produced by Lysobacter enzymogenes (OH11) that degrade Fusarium graminearum PH1 hyphae. Curr. Microbiol. 74, 437–448. doi: 10.1007/s00284-017-1206-1
Paul, A., Bakshi, S., Sahoo, D. P., Kalita, M. C., Sahoo, L. (2012). Agrobacterium-mediated genetic transformation of Pogostemon cablin (Blanco) benth. using leaf explants: bactericidal effect of leaf extracts and counteracting strategies. Appl. Biochem. Biotechnol. 166, 1871–1895. doi: 10.1007/s12010-012-9612-0
Puopolo, G., Cimmino, A., Palmieri, M. C., Giovannini, O., Evidente, A., Pertot, I. (2014). Lysobacter capsici AZ78 produces cyclo(L-Pro-L-Tyr), a 2,5-diketopiperazine with toxic activity against sporangia of Phytophthora infestans and Plasmopara viticola. J. Appl. Microbiol. 117, 1168–1180. doi: 10.1111/jam.12611
Puopolo, G., Tomada, S., Pertot, I. (2018). The impact of the omics era on the knowledge and use of Lysobacter species to control phytopathogenic micro-organisms. J. Appl. Microbiol. 124, 15–27. doi: 10.1111/jam.13607
Qian, G., Wang, Y., Liu, Y., Xu, F., He, Y., Du, L., et al. (2013). Lysobacter enzymogenes uses two distinct cell-cell signaling systems for differential regulation of secondary-metabolite biosynthesis and colony morphology. Appl. Environ. Microbiol. 79, 6604–6616. doi: 10.1128/AEM.01841-13
Ren, X., Ren, S., Xu, G., Dou, W., Chou, S. H., Chen, Y., et al. (2020). Knockout of diguanylate cyclase genes in Lysobacter enzymogenes to improve production of antifungal factor and increase its application in seed coating. Curr. Microbiol. 77, 1006–1015. doi: 10.1007/s00284-020-01902-x
Sang, M. K., Kim, J. D., Kim, B. S., Kim, K. D. (2011). Root treatment with rhizobacteria antagonistic to Phytophthora blight affects anthracnose occurrence, ripening, and yield of pepper fruit in the plastic house and field. Phytopathology. 101, 666–678
Sheng, Y., Wang, Y., Meijer, H. J., Yang, X., Hua, C., Ye, W., et al. (2015). The heat shock transcription factor PsHSF1 of Phytophthora sojae is required for oxidative stress tolerance and detoxifying the plant oxidative burst. Environ. Microbiol. 17, 1351–1364. doi: 10.1111/1462-2920.12609
Tomada, S., Sonego, P., Moretto, M., Engelen, K., Pertot, I., Perazzolli, M., et al. (2017). Dual RNA-seq of Lysobacter capsici AZ78 - Phytophthora infestans interaction shows the implementation of attack strategies by the bacterium and unsuccessful oomycete defense responses. Environ. Microbiol. 19, 4113–4125. doi: 10.1111/1462-2920.13861
Tyler, B. M. (2007). Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 8, 1–8. doi: 10.1111/j.1364-3703.2006.00373.x
Vlassi, A., Nesler, A., Parich, A., Puopolo, G., Schuhmacher, R. (2020a). Volatile-mediated inhibitory activity of rhizobacteria as a result of multiple factors interaction: the case of Lysobacter capsici AZ78. Microorganisms. 8 (11), 1761. doi: 10.3390/microorganisms8111761
Vlassi, A., Nesler, A., Perazzolli, M., Lazazzara, V., Buschl, C., Parich, A., et al. (2020b). Volatile organic compounds from Lysobacter capsici AZ78 as potential candidates for biological control of soilborne plant pathogens. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01748
Wang, P., Chen, H., Qian, G., Liu, F. (2017). LetR is a TetR family transcription factor from Lysobacter controlling antifungal antibiotic biosynthesis. Appl. Microbiol. Biotechnol. 101, 3273–3282. doi: 10.1007/s00253-017-8117-8
Wang, J., Shen, D., Ge, C., Du, Y., Lin, L., Liu, J., et al. (2020). Filamentous Phytophthora pathogens deploy effectors to interfere with bacterial growth and motility. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.581511
Wang, Y., Wang, Y. (2018). Phytophthora sojae effectors orchestrate warfare with host immunity. Curr. Opin. Microbiol. 46, 7–13. doi: 10.1016/j.mib.2018.01.008
Xia, J., Chen, J., Chen, Y., Qian, G., Liu, F. (2018). Type IV pilus biogenesis genes and their roles in biofilm formation in the biological control agent Lysobacter enzymogenes OH11. Appl. Microbiol. Biotechnol. 102, 833–846. doi: 10.1007/s00253-017-8619-4
Yang, K., Dong, X., Li, J., Wang, Y., Cheng, Y., Zhai, Y., et al. (2021). Type 2 Nep1-like proteins from the biocontrol oomycete Pythium oligandrum suppress Phytophthora capsici infection in solanaceous plants. J. Fungi. (Basel). 7 (7), 496. doi: 10.3390/jof7070496
Ye, W., Wang, Y., Dong, S., Tyler, B. M., Wang, Y. (2013). Phylogenetic and transcriptional analysis of an expanded bZIP transcription factor family in Phytophthora sojae. BMC. Genomics 14, 839. doi: 10.1186/1471-2164-14-839
Yu, Y., Gui, Y., Li, Z., Jiang, C., Guo, J., Niu, D. (2022). Induced systemic resistance for improving plant immunity by beneficial microbes. Plants (Basel). 11, 386. doi: 10.3390/plants11030386
Yu, F., Zaleta-Rivera, K., Zhu, X., Huffman, J., Millet, J. C., Harris, S. D., et al. (2007). Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob. Agents. Chemother. 51, 64–72. doi: 10.1128/AAC.00931-06
Zamioudis, C., Pieterse, C. M. (2012). Modulation of host immunity by beneficial microbes. Mol. Plant Microbe Interact. 25, 139–150. doi: 10.1094/MPMI-06-11-0179
Zhang, M., Ahmed Rajput, N., Shen, D., Sun, P., Zeng, W., Liu, T., et al. (2015). A Phytophthora sojae cytoplasmic effector mediates disease resistance and abiotic stress tolerance in Nicotiana benthamiana. Sci. Rep. 5, 10837. doi: 10.1038/srep10837
Zhang, Y., Kong, W. L., Wu, X. Q., Li, P. S. (2022). Inhibitory effects of phenazine compounds and VOCs produced by Pseudomonas aurantiaca ST-TJ4 against Phytophthora cinnamomi. Phytopathology 112 (9), 1867–1876. doi: 10.1094/PHYTO-10-21-0442-R
Zhang, M., Lu, J., Tao, K., Ye, W., Li, A., Liu, X., et al. (2012). A myb transcription factor of Phytophthora sojae, regulated by MAP kinase PsSAK1, is required for zoospore development. PLoS. One 7, e40246. doi: 10.1371/journal.pone.0040246
Keywords: Lysobacter, Phytophthora, plant immunity, HSAF, biocontrol
Citation: Lin L, Yang Z, Tao M, Shen D, Cui C, Wang P, Wang L, Jing M, Qian G and Shao X (2023) Lysobacter enzymogenes prevents Phytophthora infection by inhibiting pathogen growth and eliciting plant immune responses. Front. Plant Sci. 14:1116147. doi: 10.3389/fpls.2023.1116147
Received: 07 December 2022; Accepted: 02 January 2023;
Published: 19 January 2023.
Edited by:
Meixiang Zhang, Shaanxi Normal University, ChinaReviewed by:
Xiao-Ren Chen, Yangzhou University, ChinaCopyright © 2023 Lin, Yang, Tao, Shen, Cui, Wang, Wang, Jing, Qian and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolong Shao, eGxzaGFvQG5qYXUuZWR1LmNu; Guoliang Qian, Z2xxaWFuQG5qYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.