- 1School of Biological Science and Technology, University of Jinan, Jinan, China
- 2Xilin Gol League Agricultural and Animal Product Quality and Safety Monitoring Center, Xilinhot, China
- 3Shandong Institute of Pomology, Tai’an, Shandong, China
The RAV (related to ABI3 and VP1) transcription factors are specific and exist in plants, which contain a B3 DNA binding domain and/or an APETALA2 (AP2) DNA binding domain. RAVs have been extensively studied in plants, and more and more evidences show that RAVs are involved in various aspects of plant growth and development, stress resistance and hormone signal transduction. However, the systematic analysis of RAV family in cucumber is rarely reported. In this study, eight CsRAV genes were identified in cucumber genome and we further comprehensively analyzed their protein physicochemical properties, conserved domains, gene structure and phylogenetic relationships. The synteny analysis and gene duplications of CsRAV genes were also analysed. Cis-element analysis revealed that the CsRAVs promoter contained several elements related to plant hormones and abiotic stress. Expression analysis showed that NaCl and ABA could significantly induce CsRAV genes expression. Subcellular localization revealed that all CsRAVs were localized in the nucleus. In addition, 35S:CsRAV1 transgenic Arabidopsis and cucumber seedlings enhanced NaCl and ABA tolerance, revealing CsRAV1 may be an important regulator of abiotic stress response. In conclusion, comprehensive analysis of CsRAVs would provide certain reference for understanding the evolution and function of the CsRAV genes.
Introduction
The RELATED TO ABI3/VP1 (RAV) family belongs to one of the plant-specific B3 superfamily, which also contains three other families encompassing the auxin response factor (ARF) family, leafy cotyledon2 (lec2)-abscisic acid insensitive3 (abi3)-val (LAV) family, and reproductive meristem arf (REM) family (Swaminathan et al., 2008; Wang et al., 2012a). All members of the B3 superfamily contain a region of about 110 amino acids called the B3 domain, which is a DNA binding domain named because it is the third basic domain in the maize gene VIVIPAROUS1 (VP1) (Kagaya et al., 1999). Most B3 genes in the ARF and LAV families have been extensively studied, but B3 genes in the RAV family are rarely unknown. The RAV proteins contain B3 domain and/or AP2 (APETALA2) domain (Lu et al., 2014). Therefore, RAV family members can reasonably be classified as B3 superfamily members or AP2/EREBP family members (Matías-Hernández et al., 2014). In Arabidopsis, there are 13 RAV genes, of which seven contain only the B3 domain and the other six contain both B3 and AP2 domains (Magnani et al., 2004; Romanel et al., 2009; Fu et al., 2014). Of these, AtRAV1 and AtRAV2, containing both B3 domain and AP2 domain, were the earliest discovered members of the RAV family (Kagaya et al., 1999).
Increasing evidence has shown that RAV transcription factors play key roles in a few aspects of plant growth and development. Arabidopsis RAV1 negatively regulates flowering time, hypocotyl elongation, and seed development (Hu et al., 2004; Woo et al., 2010; Shin and Nam, 2018). Similarly, ectopic expression of soybean RAV1 in tobacco results in slow development and delayed flowering time (Zhao et al., 2008; Lu et al., 2014). It was reported that members of RAV family TEMPRANILLO 1 and 2 (TEM1 and TEM2) negatively regulate flowering time by inhibiting FLOWERINGLOCUS T (FT) expression and the production of gibberellins in Arabidopsis (Castillejo and Pelaz, 2008; Marín-González et al., 2015; Kabir et al., 2021). The NGATHA genes (AtNGA1-AtNGA4), members of the RAV family in Arabidopsis thaliana, have been shown to play key roles in lateral organs development. Over-expression of AtNGA1 to AtNGA4 present small and narrow leaf and flower, whereas the nga1/nga2/nga3/nga4 quadruple mutant reveals large and wide lateral organs (Alvarez et al., 2009; Lee et al., 2015). Besides, overexpressing of Brassica rapa NGA1 (BrNGA1) in Arabidopsis thaliana gives rise to significantly smaller and narrower lateral organs such as roots, flowers, leaves and cotyledons due to a reduction in cell numbers compared to the wild type (Kwon et al., 2009). In rice, OsRAV9/OsTEM1 was identified as a negative regulator of floral transition. Moreover, the down-regulation of OsRAV11 gene was associated with ovary enlargement and seed weight increase (Osnato et al., 2020). The results of functional characterization of RAVs support the notion that RAVs are negative growth regulators in plant (Shin and Nam, 2018).
It is also reported that RAV transcription factors are involved in biotic and abiotic stress responses, such as salt, drought, plant pathogens and plant hormones. The ectopic expression of soybean GmRAV3 in Arabidopsis thaliana can improve the resistance of transgenic lines to high salt and drought, and lead to the insensitivity of transgenic plants to exogenous ABA (Lu et al., 2014; Zhao et al., 2017). The OsRAV2 expression can be significantly induced by salt in rice, indicating its important roles in salt response (Duan et al., 2016). Melon RAV1 (CmRAV1) can be induced by NaCl treatment and ectopic overexpression of CmRAV1 in Arabidopsis enhances salt tolerance at the seed germination and seedling stages (Zhao et al., 2019a). It has been reported that MeRAV1 and MeRAV2 play important roles in resistance to cassava bacterial blight through activation of melatonin biosynthesis genes in cassava (Wei et al., 2018). In addition, over-expression of tomato RAV2 (SlRAV2) gives rise to increased bacterial wilt (BW) tolerance, whereas knockdown SlRAV2 significantly decreases tomato resistance to BW (Li et al., 2011). Meanwhile, members of the RAV family can be induced by some plant hormones and have been shown to be involved in brassinosteroid and ethylene responses (Alonso et al., 2003; Hu et al., 2004). The expression of all soybean RAV genes increases dramatically under ABA treatment (Zhao et al., 2017). In cotton, GhRAV4, GhRAV9 and GhRAV20 genes are significantly induced by BL, JA and IAA hormones (Kabir et al., 2021). Therefore, it clearly shows that members of the RAV gene family play important roles in the development and stress response of different plant species.
Cucumber (Cucumis sativus L.) is one of the most important vegetable crops in the world (Huang et al., 2009). Despite the important roles of RAV genes in plant growth, development and stress tolerance, the functions of CsRAVs in cucumber are largely unknown. In this study, we identified eight CsRAV genes and classified them into four clades. The gene structures, conserved motifs, phylogenetic analysis, synteny analysis and gene duplications were further performed. In addition, the expression patterns of CsRAV genes under different abiotic stresses were also measured. Furthermore, overexpression of CsRAV1 increased salt tolerance and ABA resistance. These results lay a foundation for the evolutionary and functional characterization of RAVs in cucumber.
Materials and methods
Genome-wide identification of CsRAVs in cucumber
To identify the CsRAV genes from cucumber (Chinese Long) v3 genome database (http://cucurbitgenomics.org/organism/20), 13 AtRAV proteins were used as query sequences and Blastp was used to search for the predicted cucumber proteins. All candidate genes were further confirmed by the existence of B3 (PF02362.21) and/or AP2 (PF00847.20) domains using the Pfam and Simple Modular Architecture Research Tool (SMART) datebase (http://smart.embl-heidelberg.de). The RAV protein sequences in Arabidopsis and tomato were downloaded from the Arabidopsis Information Resource database (https://www.Arabidopsis.org) and the Solanaceae Genomics Network (https://solgenomics.net), respectively.
Phylogenetic analysis, conserved motif and gene structure analysis
A phylogenetic tree containing 115 RAV protein sequences from ten species and a phylogenetic tree with the full-length amino acid sequences of nine SlRAVs, 13 AtRAVs and eight CsRAVs were constructed using MEGA 7.0 respectively and the neighbour-joining (NJ) method was used with the following parameters: Poisson correction, pairwise deletion, and bootstrap (1000 replicates; random seed) (Li et al., 2022). The corresponding cDNA and DNA sequences of RAV genes were downloaded from corresponding genomes, and the gene structures were analyzed as described by Li et al. (2022). The conserved motifs in RAVs were identified using Multiple Expectation Maximization for Motif Elicitation (MEME) online program (http://meme-suite.org/index.html). CsRAV genes were classified according to its phylogenetic relationship with RAVs in other species. The visual evolutionary tree, conserved motif and gene structure maps were completed using TBtools (Li et al., 2020).
Chromosomal distribution and gene duplication
All CsRAVs were mapped to cucumber chromosomes based on physical location information from the database of cucumber genome using Circos (Krzywinski et al., 2009). Multiple Collinearity Scan toolkit (MCScanX) was used to analyze the gene duplication events, with the default parameters (Wang et al., 2012b). To show the synteny relationship of the orthologous RAVs in cucumber, Arabidopsis and tomato, we used TBtools to constructed the syntenic analysis maps (https://github.com/CJ-Chen/TBtools) (Chen et al., 2020).
Cis-element analysis on CsRAVs promoter in cucumber
The relevant data of cucumber genome were downloaded from cucumber genome database (Chinese Long 9930: http://cucurbitgenomics.org/), and we used TBtools to extract the 2 kb sequence of CsRAVs gene promoter. The cis-elements on the promoter regions of CsRAV genes were analysed using PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Li et al., 2020).
Vector construction, transient transformation of cucumber cotyledons and Arabidopsis transformation
The coding sequence of CsRAVs was recombined into pCAMBIA1300 vector with a GFP tag to obtain 35S:CsRAVs-GFP. The 35S:CsRAV1-GFP construct was transformed into Agrobacterium tumefaciens LBA4404, and then transferred into Arabidopsis (Col-0) or 8-d-old cucumber cotyledons (Li et al., 2020). The homozygous T3 transgenic Arabidopsis lines were screened and identified for subsequent experiments. The primers used are listed in Supplementary Table 5.
Subcellular localization of CsRAVs
Tobacco leaf epidermal cells were injected with the empty GFP vector and the 35S: CsRAVs-GFP recombinant plasmids, respectively. Then the injected tobaccos were grown under normal conditions for about 48 h, the subcellular localization of CsRAVs were determined by observing the fluorescence signal under a fluorescence microscope.
Analysis of CsRAV gene expression under different abiotic stresses
The cucumber inbred line (Xintaimici) was used as materials to undergo stress treatments and transient transformation. The leaves of vigorous two-week-old cucumber seedlings were sprayed with 100 mM NaCl and 100 μM ABA, respectively. Leaves were taken at 0, 0.5, 1, 3, 6, 9, 12 and 24h for quantitative analysis. The two-week-old cucumber seedlings were cultured in a 4°C incubator with 16 h light/8 h dark for low temperature treatment, and sampled at the same time point for storage. Each sample was taken from six plants and each treatment had three biological replicates.
Transient transformation of cucumber cotyledons
The coding sequence of CsRAV1 was recombined into the pCAMBIA1300 vector (universal vector: stored in our laboratory), which was then transformed into Agrobacterium tumefaciens LBA4404. The A. tumefaciens LBA4404 cells containing the recombinant vector was incubated in liquid medium overnight until the optical density (600 nm) was about 0.6-0.8. Then the Agrobacterium solution was centrifuged at 8000 × g for three min, and re-suspended with MES solution (consisting of 10 mM MES, 10 mM MgCl2 and 200 μM Acetosyringone) to OD600 value of 0.6-0.8. The suspensions containing the target gene were injected into the cotyledons of 6-7-d-old cucumber seedlings with a 1ml disposable syringe, and cultured in the dark at 20°C for one day, then the follow-up related treatment experiments were conducted.
Tolerance of transgenic plants to abiotic stress
The seeds of 35S:CsRAV1 T3-generation homozygous lines and WT were seeded in vermiculite soil and cultured at 22°C for 3 weeks under normal conditions. For salt treatment, the 3-week-old seedlings were irrigated with 200 mM NaCl solution every two days, and the growth of different lines was observed every three days. Under ABA treatment, the transgenic lines and WT were watered 100 μM ABA solution every two days, respectively, and phenotypic evaluation was performed every three days. To investigate the seed germination rate of transgenic and WT seeds under salt stress and ABA treatment, the seeds of transgenic lines and WT were sown on 1/2 MS medium containing 100 mM NaCl or 2 μM ABA, respectively, and cultured under normal conditions. Germination rate was measured after 7 days of culture. The cotyledons of 8-d-old transgenic cucumber seedlings with transient transformation of 35S and 35S:CsRAV1 were subjected to salt and ABA tolerance by hydroponic method. Transgenic cucumber seedlings with equivalent growth were selected and transferred to 2 L Hoagland nutrient solution for hydroponics. After two days of hydroponics, NaCl and ABA were added into the nutrient solution, and the final concentrations of NaCl and ABA in nutrient solution were 100 mM and 100 μM, respectively. To obtain accurate experimental results, the cucumber seedlings transfected with 35S and 35S:CsRAV1 were cultured in the same tank. The phenotypes of transgenic seedlings and control seedlings were observed at different periods (Li et al., 2020).
Results
Identification and analysis of RAV Genes in cucumber
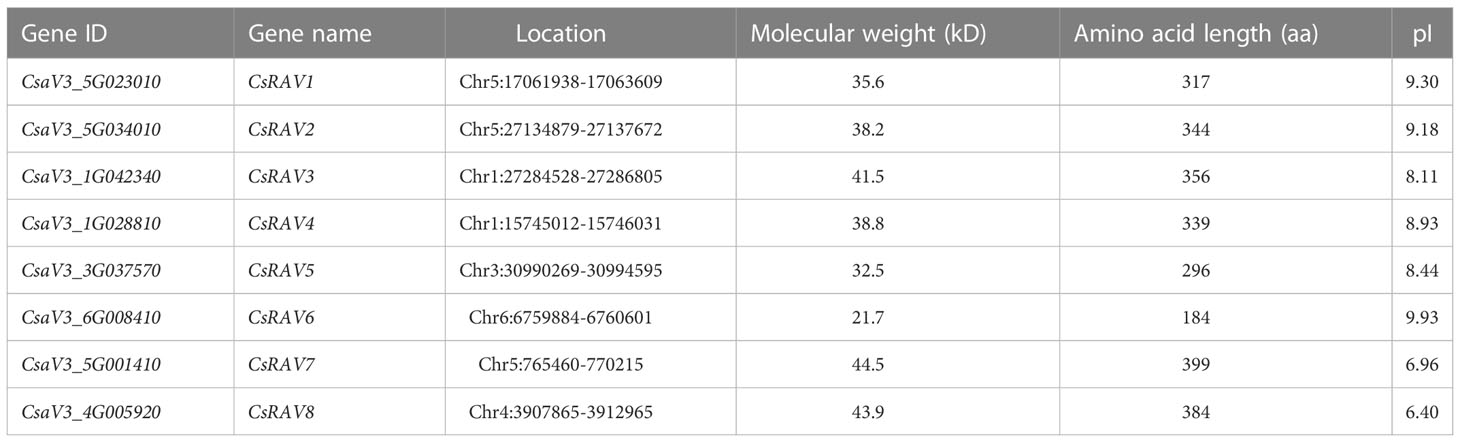
To identify the putative CsRAV family genes in cucumber genome, BlastP was used to search against cucumber genome database based on 13 Arabidopsis RAV proteins and consensus protein sequences of B3 (PF02362) and AP2 (PF00847). A total of eight CsRAVs were identified in the cucumber genome and the presence of the B3 and/or AP2 domains was also confirmed by Pfam and SMART. The identified CsRAVs were named CsRAV1 to CsRAV8 according to the similarities of CsRAVs with their characterized counterparts in Arabidopsis. The information of the CsRAVs, including the gene ID, gene name, location, molecular weight, amino acid length and isoelectric points (pI) was listed in Table 1. These eight CsRAV genes were distributed on chromosome 1, 3, 4, 5 and 6 of cucumber, respectively (Supplementary Figure 1). Except for CsRAV7 and CsRAV8, the pI of the other CsRAV proteins were all greater than 7, indicating that these proteins were basic proteins (Table 1).
Phylogenetic analysis, conserved motif and gene structure analysis of RAV gene family in cucumber, tomato and Arabidopsis
To explore the homology of RAV genes among different plant species, RAV genes of ten species were selected to construct a phylogenetic tree based on amino acid sequences (Figure 1). As could be seen from Figure 1, the 115 RAV proteins could be roughly divided into 23 groups, among which CsRAV genes were classified into different groups, such as CsRAV1 was classified in group 5, CsRAV2 were classified in group3, CsRAV3 and CsRAV4 were classified in group 22, CsRAV6 and CsRAV7 were classified in group 16, and CsRAV8 was classified in group 19 (Figure 1; Supplementary Table 1). To better assess the phylogenetic relationships of the cucumber RAV proteins, the phylogenetic tree, conserved motifs and gene structures of eight CsRAVs, 13 AtRAVs and nine SlRAVs were further analyzed in detail (Figure 2). As shown in Figure 2A, the phylogenetic tree divided these RAV proteins into four clades, which CsRAV1 and CsRAV2 were classified into Clade I, CsRAV5 belonged to Clade II, CsRAV6, CsRAV7 and CsRAV8 belonged to Clade III, and CsRAV3 and CsRAV4 were classified into Clade IV. Each clade contained Arabidopsis, cucumber, and tomato RAV genes, suggesting that proteins in the same clades might have similar functions. For example, CsRAV1 and CsRAV2 were simultaneously classified into Clade I with RAV1 and RAV2 of Arabidopsis and tomato, implying that CsRAV1 and CsRAV2 might regulate plant growth and improve plant tolerance (Figure 2A).
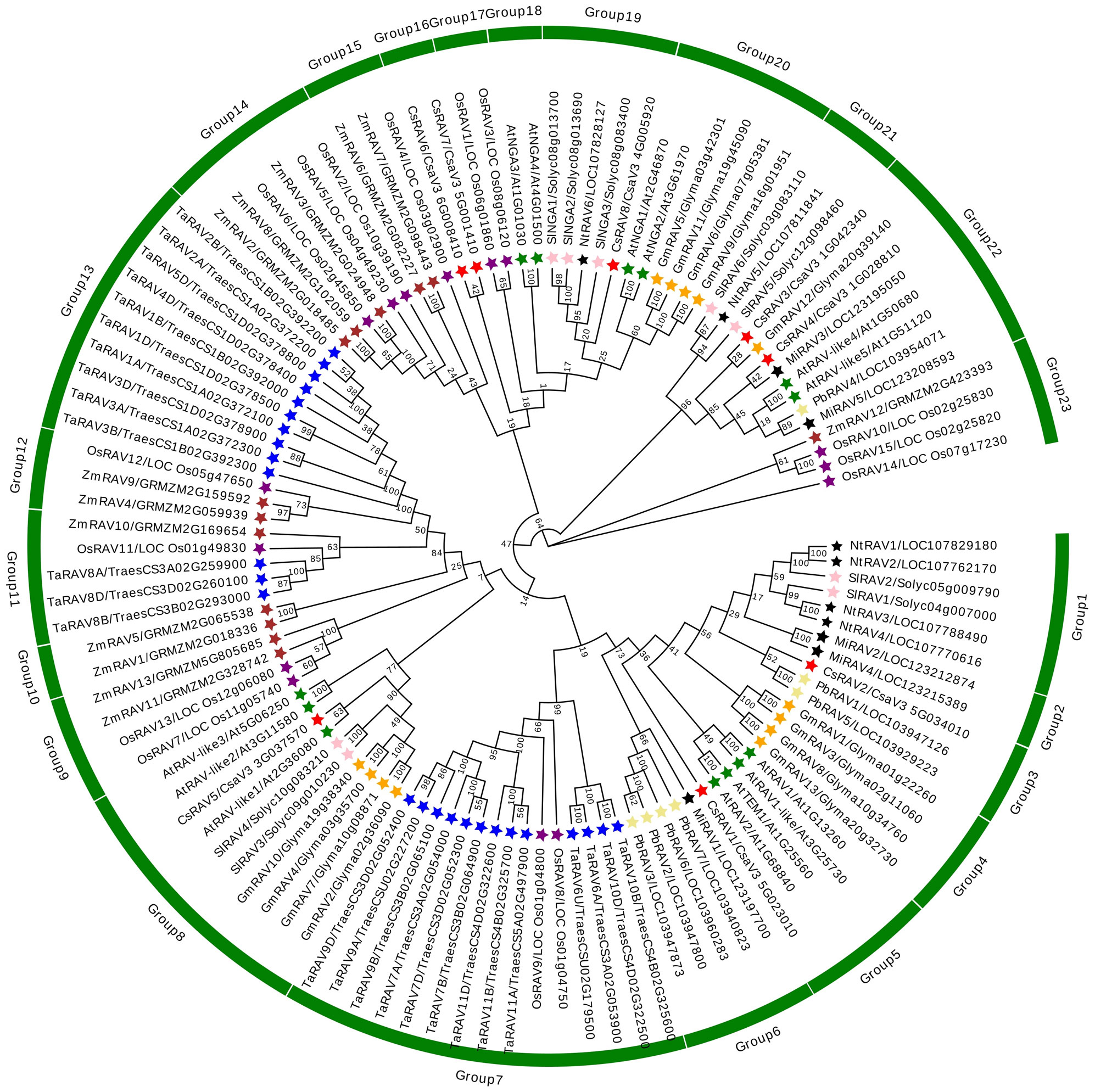
Figure 1 Phylogenetic relationships of 115 RAV proteins from Cucumis sativus, Arabidopsis thaliana, Solanum lycopersicum, Glycine max, Oryza sativa, Triticum aestivum, Zea mays, Mangifera indica, Pyrus bretschneideri and Nicotiana tabacum. The MEGA 7.0 software was used to construct the phylogenetic tree of 115 RAVs with complete amino acid sequences. The CsRAV proteins were marked with red stars.
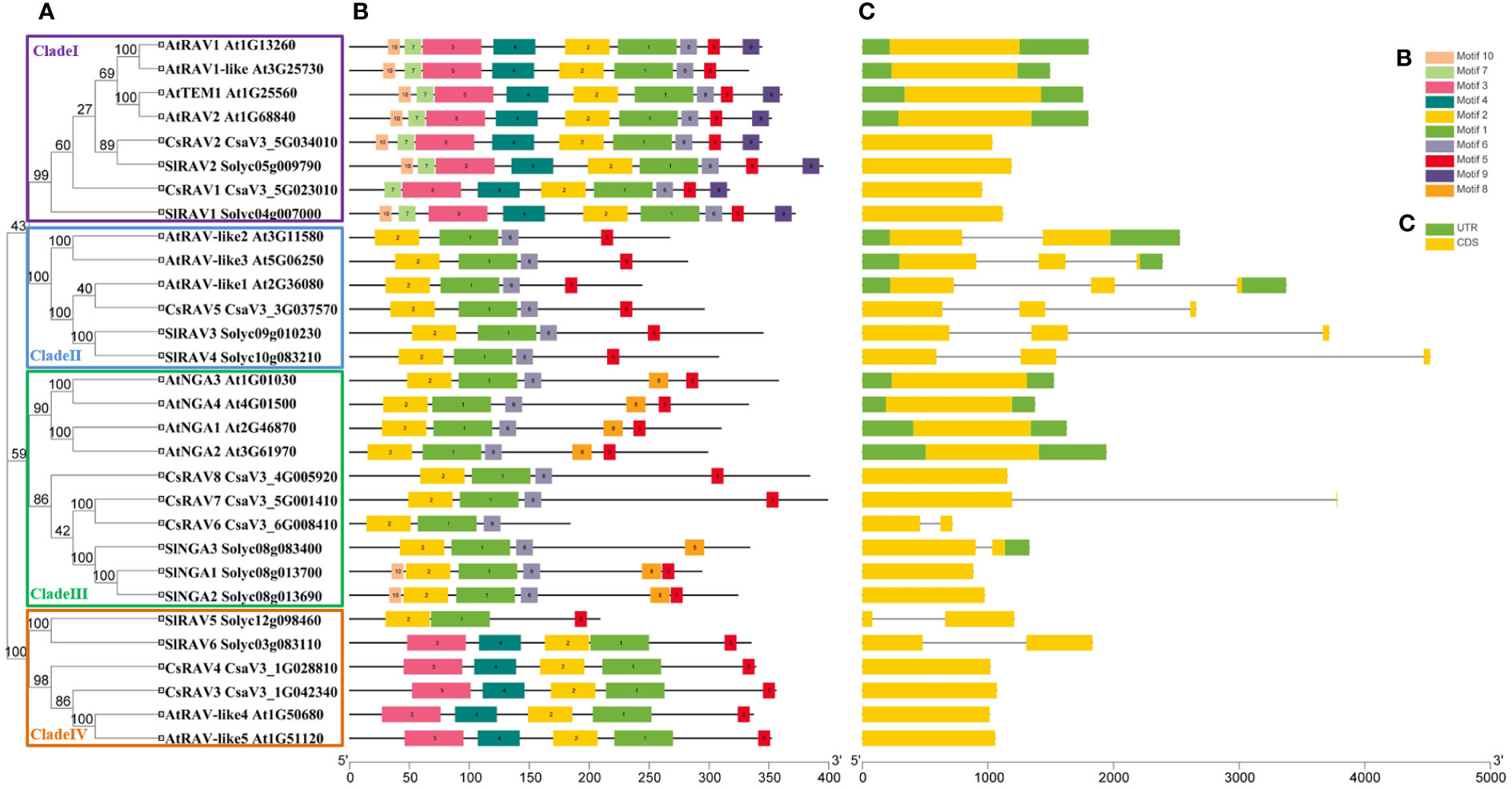
Figure 2 The analysis of phylogenetic relationships, conserved motifs and gene structures in RAV genes from cucumber, Arabidopsis and tomato. (A) A phylogenetic tree containing nine SlRAVs, 13 AtRAVs and eight CsRAVs was constructed by MEGA 7.0 software and divided into four clades named I, II, III and IV, respectively. (B) The conserved motifs in RAVs were identified using MEME online program. Ten conserved sequences were presented in Supplementary Figure S2. (C) Exon-intron structure of RAV genes.
In order to support the phylogenetic results, the gene structure of RAVs from tomato, Arabidopsis and cucumber were analyzed (Figure 2C). The number of exons in SlRAV, AtRAV and CsRAV genes was conserved, ranging from one to three exons. As shown in Figure 2C, the gene structures of RAVs in the same clade were highly conserved in all three species. For example, all RAVs in Clade I contained only one exon and all RAVs in clade II, except AtRAV-like2, contained three exons. In Clade III and Clade IV, the genes had one or two exons (Figure 2C).
To further analyze the structural diversity and predict the function of the RAV proteins, the number and composition of conservative motifs in the SlRAVs, AtRAVs and CsRAVs were analyzed by MEME (Figure 2B; Supplementary Figure 2). We analyzed 10 different motifs named Motif1-Motif10 (Supplementary Figure 2). Motifs 1 and 2, which is representative B3 domain (PF02362), were identified in all RAVs, and motifs 3 and 4, which are representative AP2 domain (PF00847), were identified in some RAV proteins, including all the RAVs in Clade I and Clade IV except SlRAV5. Some of the specific motifs were absent in specific clade (Figure 2B). For example, motif 6 was absent in all the member of the Clade IV subfamily, which further corroborates the accuracy of subfamily division. Motif 9 was only identified in Clade I (Figure 2B). Therefore, the functions of these motifs in relation to the functions of these proteins need to be investigated further.
In general, RAV genes with close evolutionary relationships in the phylogenetic tree contained similar conserved motifs and gene structures, suggesting that each subfamily in the three different species was evolutionarily conserved.
Synteny analysis of RAV genes in cucumber, tomato and Arabidpsis
Gene duplication is one of the main driving forces for the evolution of genomes and genetic systems (Das et al., 2020). Segmental duplication and tandem duplication are considered to be the two main reasons for the expansion of plant gene families (Cannon et al., 2004). To reveal the duplication of CsRAVs, the syntenic regions were analysed using MCscanX software. In cucumber genome, there were 231 segmental duplication blocks and 1468 tandem duplication gene pairs in all (Supplementary Table 2). The analysis revealed that there was one segmental duplication gene pair (CsRAV7 and CsRAV8) in cucumber RAV gene family, but no tandem duplication gene pairs (Figure 3A).
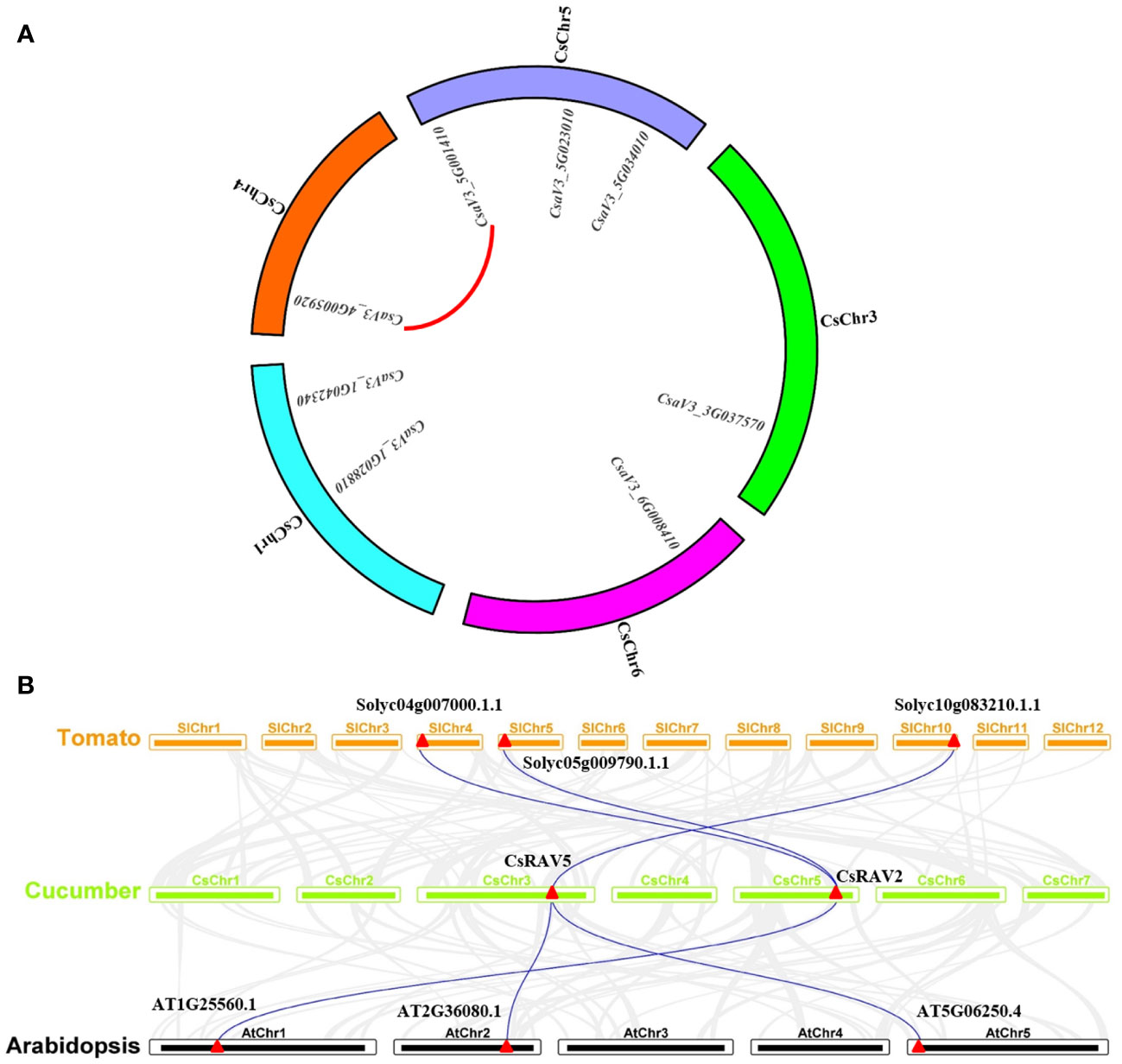
Figure 3 Gene duplication and synteny analysis of CsRAV genes. (A) Chromosome distribution and interchromosome relationships of CsRAV genes. The red line indicated segmental duplication gene pair. (B) Synteny analysis of RAV genes from cucumber, tomato and Arabidopsis. Gray lines represented the collinear blocks between cucumber and tomato or Arabidopsis; Blue lines represented the collinear blocks of RAV genes between cucumber and tomato or Arabidopsis.
To further elucidate the phylogenetic mechanism of the cucumber RAV family, a comparison of the syntenic map of cucumber connected with tomato and Arabidopsis, was constructed (Figure 3B; Supplementary Table 3). Interestingly, only two CsRAV genes in cucumber were collinear with RAV genes in tomato and Arabidopsis. As shown in Figure 3, CsRAV2 gene (CsaV3_5G034010) showed syntenic relationship with At1G25560.1 gene in Arabidopsis, Solyc04g007000.1.1 and Solyc05g009790.1.1 genes in tomato. Similarly, CsRAV5 gene also corresponded to syntenic gene pairs between tomato and Arabidopsis (Supplementary Table 3). These results indicated that these orthologous pairs might already exist before the ancestral divergence. In addition, we found the rest of the CsRAV genes (CsRAV1, CsRAV3, CsRAV4, CsRAV6, CsRAV7 and CsRAV8) were not associated with syntenic gene pairs in Arabidopsis or tomato, suggesting that they might be unique to cucumber during the evolutionary process.
Cis-element analysis of CsRAVs promoter in cucumber
Cis-acting elements on promoters are non-coding DNA sequences that play key roles in gene expression and mutation. To explore the potential function of CsRAV genes in cucumber, the putative cis-elements on the 2-kb promoter regions of CsRAV were analyzed by PlantCARE (Supplementary Table 4). As shown in Figure 4, the cis-elements associated with abiotic stress such as low temperature, drought, wound, defense and stress existed on the promoter of CsRAV genes. Moreover, cis-elements that respond to plant hormones were also found on the promoter regions of CsRAV genes, such as auxin, gibberellin (GA), abscisic acid (ABA), salicylic acid (SA) and jasmonic acid (MeJA). In addition, some cis-elements involved in regulating plant growth and development were present on promoters of certain CsRAV genes, for example, meristem expression, metabolism regulation and anaerobic induction (Figure 4). Furthermore, the core elements including transcription start site (TSS) and TATA-box of eight CsRAV genes were predicted using TSSPlant online software. The results showed that only four transcription start sites were predicted for CsRAV3 and no TATA-box, while three core elements were predicted for CsRAV5 and CsRAV7 and two core elements were predicted for all the other five genes (Supplementary Figure 3). In conclusion, the analyses of cis-elements suggested that CsRAVs might play vital functions in abiotic stress responses and multiple plant hormone signaling pathways.
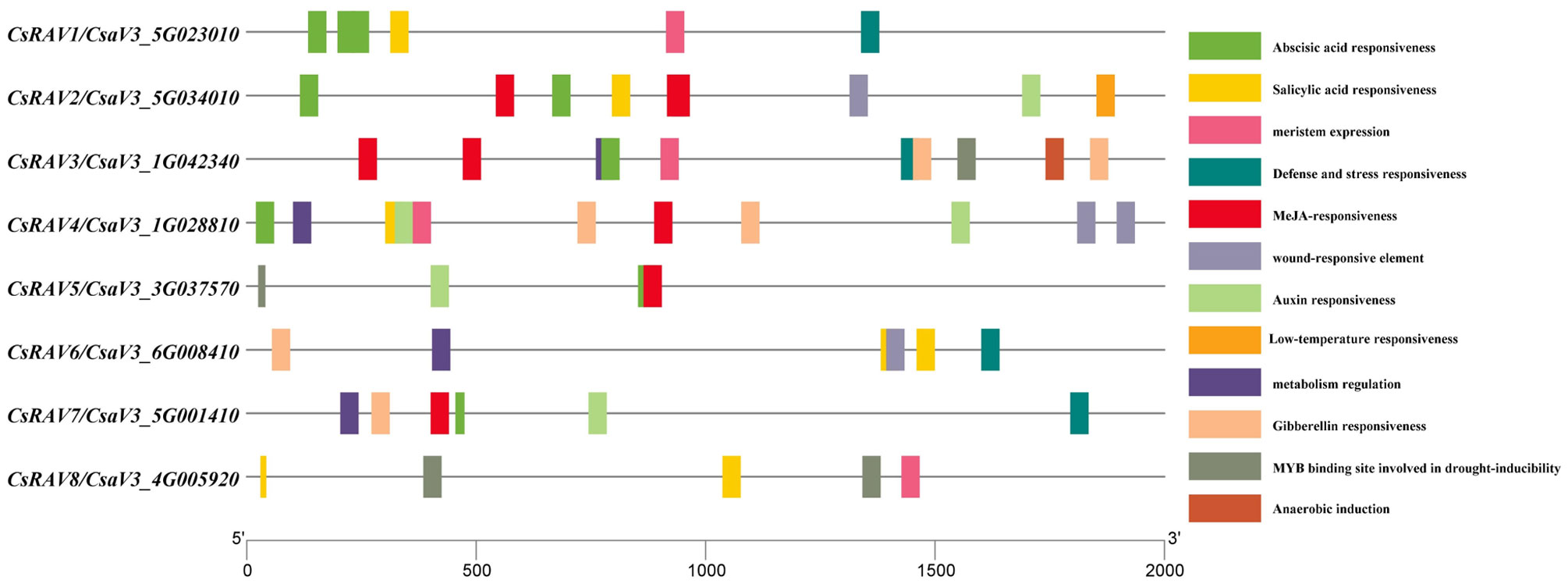
Figure 4 The promoter analysis of the CsRAV genes in cucumber. The potential cis-elements on the 2-kb promoter regions upstream of CsRAV genes were analysed using PlantCARE. Different colored boxes represented the corresponding cis-elements.
Responses of CsRAV genes to different abiotic stresses
Abiotic stresses are known to affect many physiological processes. To explore the response of CsRAV genes to different stresses, qRT-PCR was used to analyze the expression patterns of these genes under low temperature (4°C), ABA treatment (100 μΜ ABA) and salt stress (100 mM NaCl), respectively (Figure 5). Under salt stress, CsRAV1 and CsRAV2 genes were significantly up-regulated (>15-fold) after 0.5 h. Two genes, CsRAV6 and CsRAV8, were slightly upregulated, while others were obviously down-regulated (Figure 5A). Under ABA treatment, the expression levels of all CsRAV genes were markedly induced (>7-fold). For example, the expression levels of CsRAV5 and CsRAV8 were more than 30 times higher. In particular, the expression patterns of CsRAV1 and CsRAV2 genes showed a similar trend and the expression levels of these two genes were up-regulated 60 times after 0.5 h (Figure 5B). After low temperature treatment, all CsRAV genes were up-regulated except CsRAV4 and CsRAV6, and CsRAV1 and CsRAV2 were up-regulated most significantly. CsRAV1 reached maximum values at 1 h, while the expression levels of CsRAV2 reached its maximum at 6 h (Figure 5C). These results suggested that some CsRAV genes were involved in low temperature, salt and ABA responses.
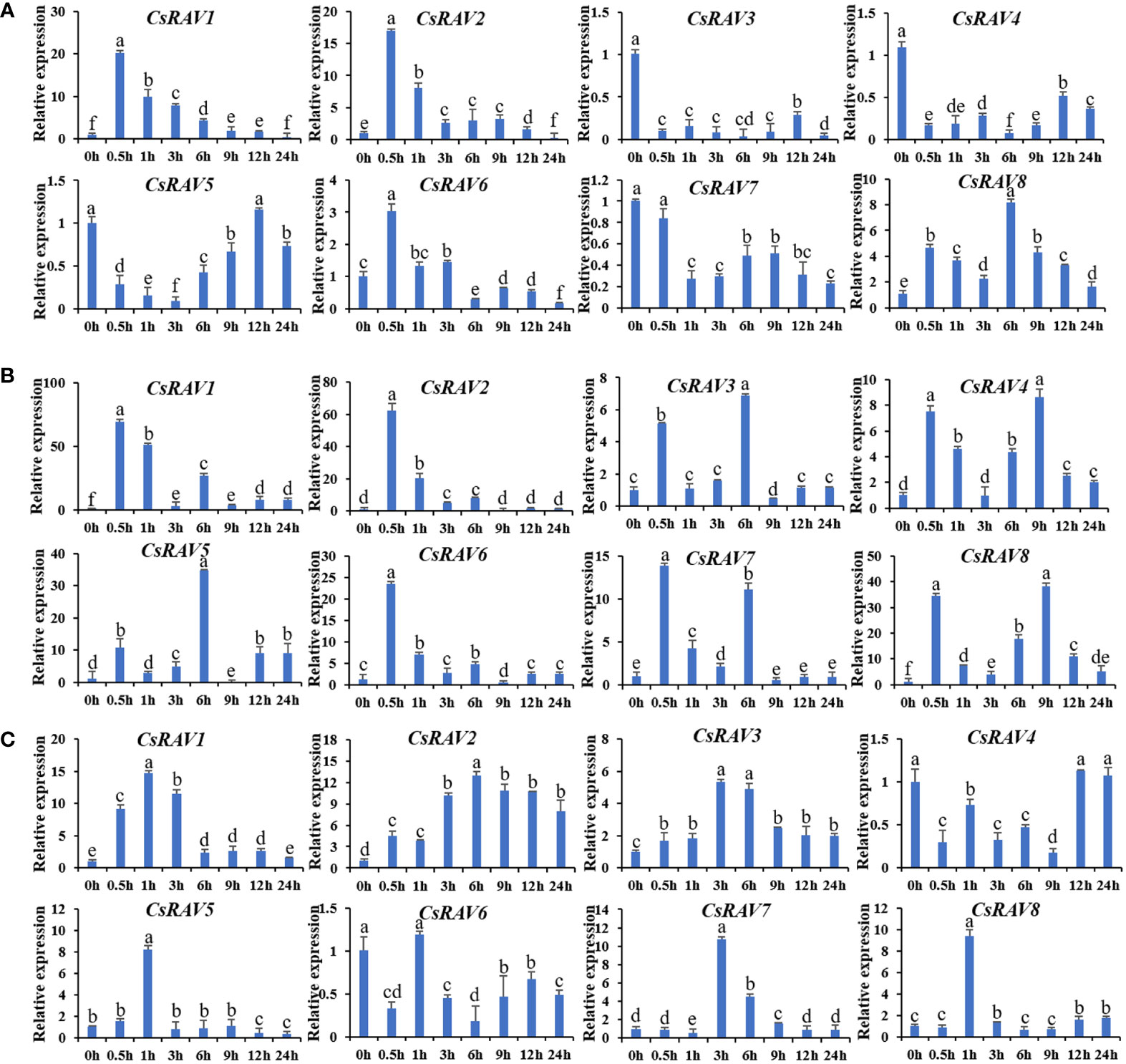
Figure 5 Relative expression of CsRAV genes in cucumber seedlings treated with NaCl, ABA and low temperature. QRT-PCR was used to analyze the expression levels of CsRAVs under NaCl (100 mM) (A), ABA (100 μM) (B) and 4°C (C). Using cucumber β-actin gene as an internal control, three biological replicates were used to analyze gene expression. Error bars were the standard errors (SE). Different lowercase letters represented significant differences (P < 0.05).
Subcellular localization of cucumber RAVs
To confirm the subcellular localization of CsRAV proteins, the GFP fusion protein vectors 35S:CsRAVs-GFP of eight CsRAV genes were constructed, respectively. The 35S:GFP vector was used as empty control. The green fluorescent signal of GFP expression in epidermal cells of tobacco leaves was observed after transformation. Under confocal laser scanning microscope, the different CsRAV fusion proteins were all localized in the nucleus, while 35S:GFP fluorescence signal was distributed throughout the whole cell (Figure 6). The data showed that all CsRAV proteins were nuclear localization proteins, suggesting that these CsRAVs might be involved in transcriptional regulation.
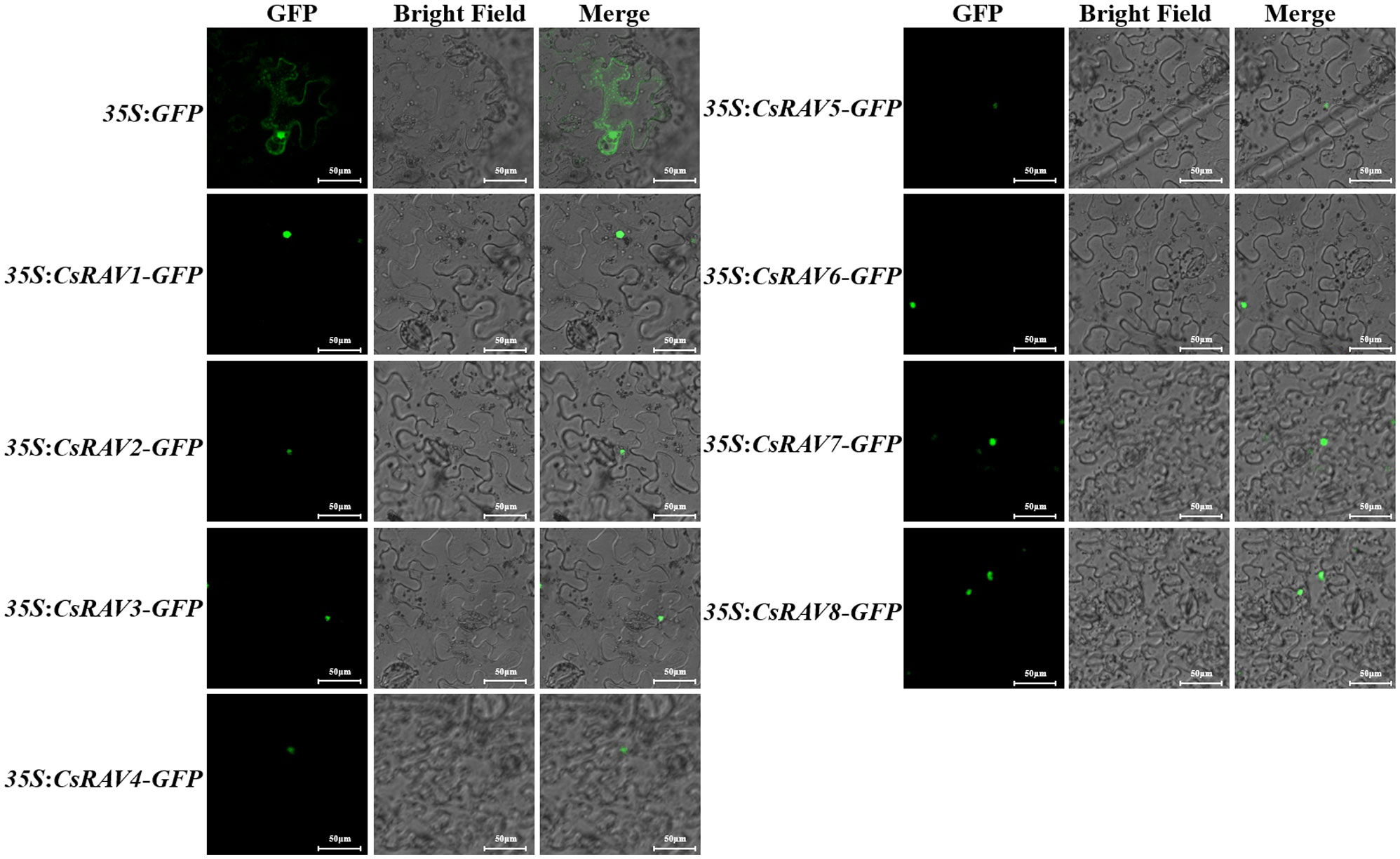
Figure 6 Subcellular localization of the cucumber RAV proteins in tobacco leaf. The green fluorescence signal was observed under confocal microscope 48 h after transformation.
Overexpression of CsRAV1 improved cucumber and transgenic Arabidopsis tolerance to NaCl and ABA
The expression analysis showed that CsRAV1 was significantly induced by NaCl and ABA in cucumber (Figures 5A, B). To determine the function of CsRAV1 in abiotic stress response, first, agrobacterium-mediated transient transformation of cucumber cotyledons was used to elucidate the tolerance of CsRAV1 to NaCl and ABA. In the preliminary experiments, the transient transfection of CsRAV1 was successfully verified by PCR amplification, and the expression levels of CsRAV1 in transgenic cucumber cotyledons were observably higher than that in 35S plants by qRT-PCR (Supplementary Figure 4). When cultured in a hydroponic solution containing 100 mM NaCl, the control seedlings (overexpressing 35S empty vector) showed significant wilting after 12 h compared with overexpressed CsRAV1 transgenic seedlings, and the difference in wilting degree was more obvious after 24 h (Figure 7A). After 24 h of NaCl treatment, the survival rate of transgenic seedlings was 44%, which was significantly higher than that of control, indicating that the overexpression of CsRAV1 produced significant salt tolerance (Figure 7C). Similarly, in the presence of exogenous ABA, the growth of both control and 35S:CsRAV1 seedlings was distinctly harmed, but the damage of control was much greater than that of transgenic seedlings. After 24 h of ABA treatment, about 46% of the transgenic seedlings remained green with extended cotyledons, while 89% of the control showed obvious injury symptoms, such as wilting and death (Figures 7B, C). To elucidate the underlying regulatory effect of CsRAV1 on tolerance to NaCl and ABA stress, the expression levels of four stress- and ABA- related marker genes were detected by qRT-PCR in 35S and 35S:CsRAV1 cucumber seedlings before and after NaCl and ABA treatments. As shown in Figures 7D–G, only CsCPK11 was slightly higher expressed in 35S:CsRAV1 transgenic plants before NaCl and ABA treatments. However, after NaCl and ABA treatments, all the four genes, CsSOS1 (Salt overly Sensitive 1), CsNHX1 (vacuolar sodium/proton antiporter), CsCPK11 (calcium-dependent protein kinase) and CsABI5 (ABA-insensitive factor 5) were strongly induced in 35S and 35S:CsRAV1 plants. Furthermore, the expression levels of the four genes were significantly higher in 35S:CsRAV1 transgenic plants than those in 35S plants after NaCl and ABA stress treatment. The enhanced expression of these genes in 35S:CsRAV1 transgenic plants might conduce to improve plant stress resistance, which also suggested that CsRAV1 may be involved in response to NaCl and ABA stress by influencing the transcription of multiple stress-related genes.
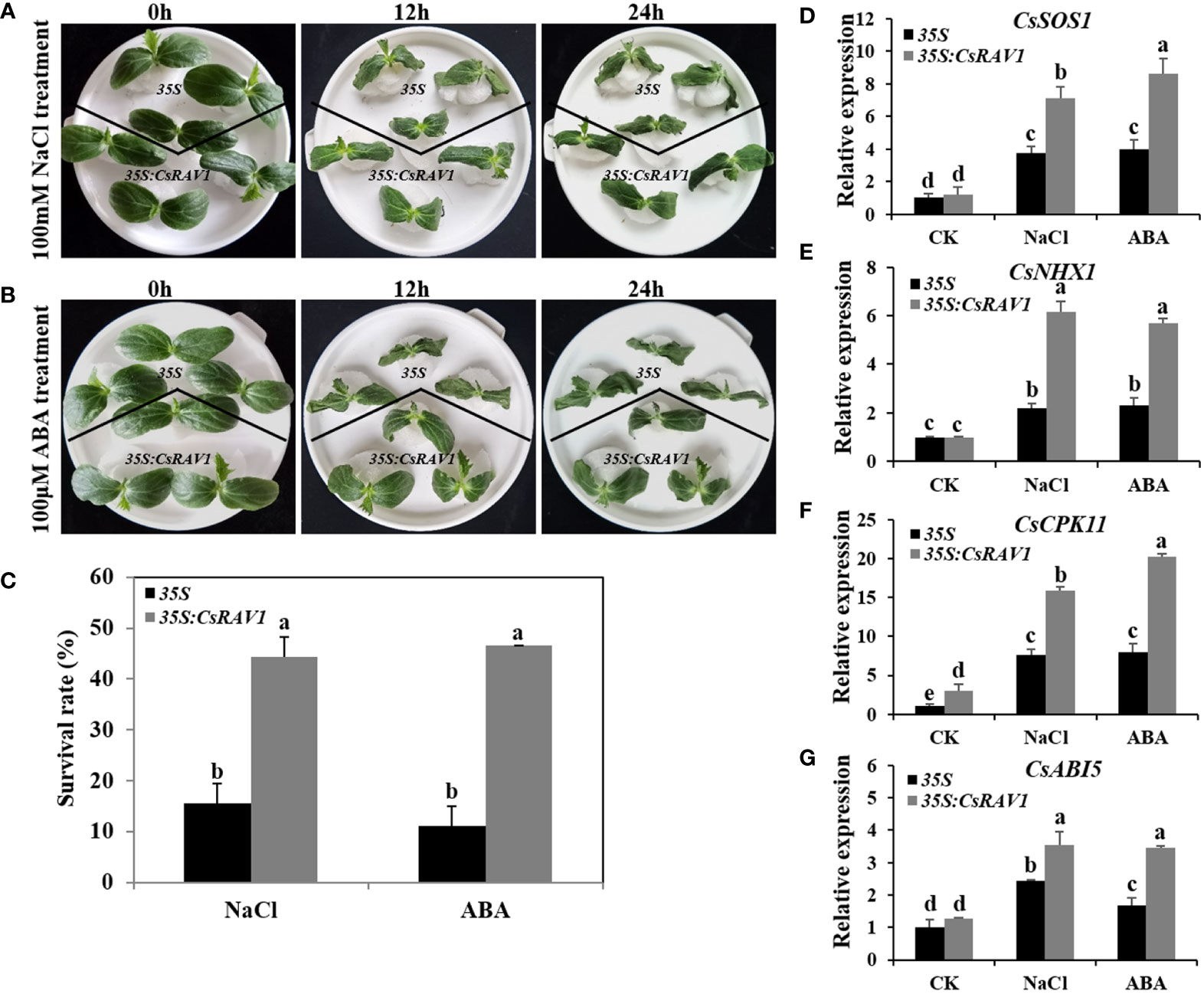
Figure 7 Overexpression of CsRAV1 improved NaCl and ABA tolerance in cucumber seedlings. (A, B) Phenotypes of 35S and 35S:CsRAV1 cucumber seedlings treated with NaCl and ABA under hydroponics. (C) Survival rate of 35S and 35S:CsRAV1 plants after 24 h NaCl and ABA treatments. The transcript levels of CsSOS1 (D), CsNHX1 (E), CsCPK11 (F), and CsABI5 (G) genes in 35S and 35S:CsRAV1 cucumber seedlings were analyzed with qRT-PCR under NaCl and ABA treatment for 6 h. The cucumber β-actin gene was used as internal control. Error bars were the standard errors (SE). Different letters indicated significant differences (P < 0.05).
To further study the function of CsRAV1 in plant resistance to abiotic stress, we obtained transgenic Arabidopsis plants overexpressing CsRAV1 and two highly expressed homozygous transgenic lines (L1 and L2) were selected for further analysis (Supplementary Figure 5). The NaCl and ABA tolerance of CsRAV1 transgenic plants was evaluated. For germination assays, the WT, L1 and L2 on 1/2 MS medium showed similar germination rate (Figure 8A). However, the germination of both WT and 35S:CsRAV1 transgenic seeds was significantly inhibited on 1/2 MS medium containing 100 mM NaCl or 2 μM ABA, but the inhibition of the WT was much higher than that of the transgenic seeds. With 100 mM NaCl or 2 μM ABA treatment, nearly 40-50% of CsRAV1 transgenic seeds were able to germinate, while only about 10-12% of the WT seeds could germinate (Figures 8A, B). The results showed that overexpressing CsRAV1 could remarkably improve seed germination rate under salt and ABA treatment. In addition, we determined the seedling growth of 3-week-old WT and CsRAV1 transgenic lines under 200 mM NaCl and 100μM ABA treatment, respectively. After four days of treatment with 200 mM NaCl or 100μM ABA, the leaves of CsRAV1 transgenic lines were still green, but the leaves of WT were severely yellowed (Figures 8C, D). After eight days, the CsRAV1 transgenic lines and WT had more obvious differences in NaCl and ABA resistance, indicating that CsRAV1 transgenic plants were more tolerant to NaCl and ABA stresses than WT. Following NaCl and ABA stress treatment, the expression levels of stress- and ABA- related marker genes were significantly up-regulated in transgenic lines (L1 and L2) and WT, but the induction levels of AtRD29A (response to desiccation 29A), AtSOS1, AtNHX1 and AtABI5 genes were markedly higher in L1 and L2 lines than in WT (Figures 8E–H). In conclusion, these results indicated that CsRAV1 might play key roles in ABA signaling and in plant response to high salinity during seed germination and seedling development.
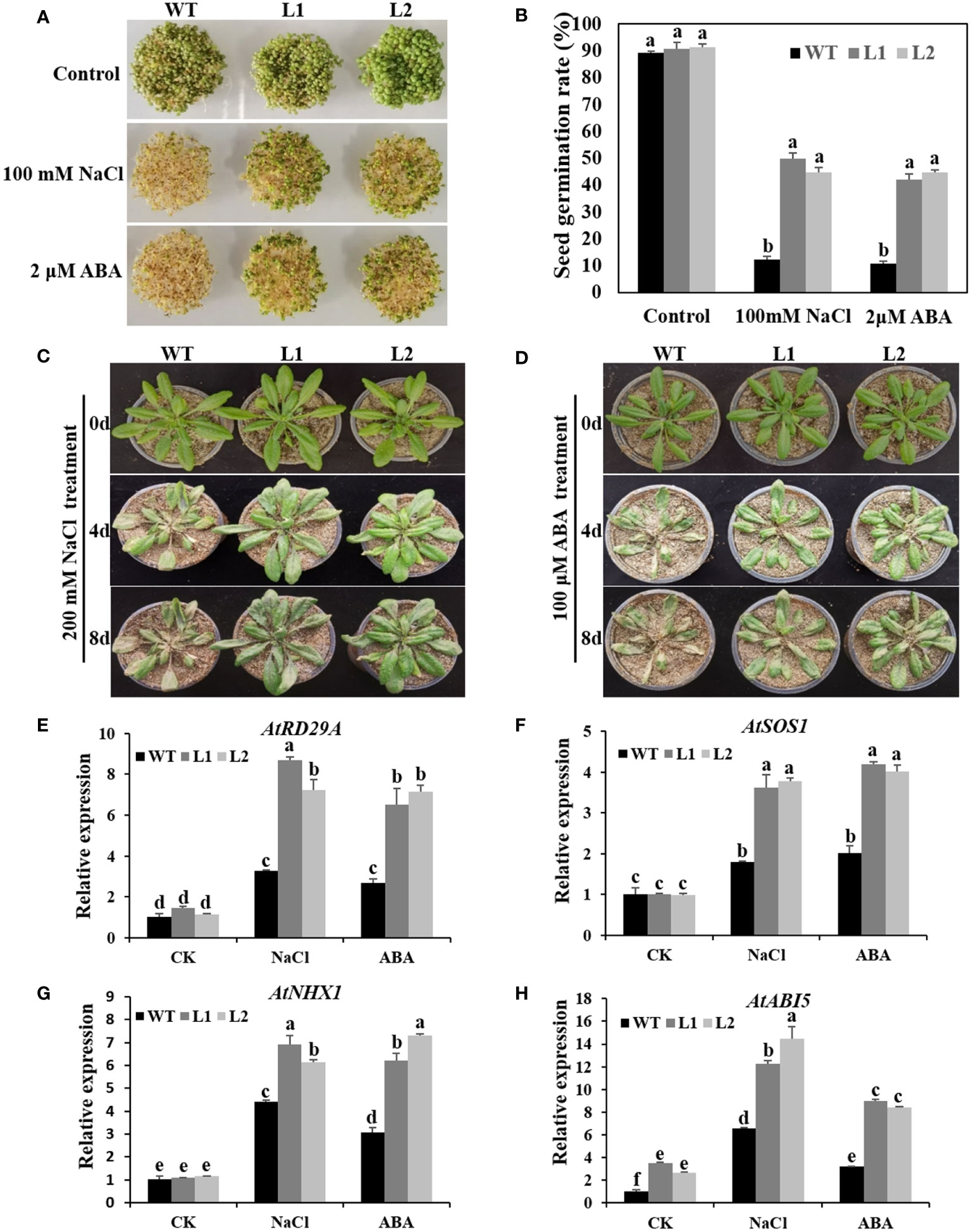
Figure 8 CsRAV1 enhanced NaCl and ABA tolerance in transgenic Arabidopsis. (A) Seed germination of WT and 35S:CsRAV1 plants (L1 and L2) after 7 d on 1/2 MS with NaCl and ABA. (B) Seed germination rate in (A). Different letters indicated significant differences (P < 0.05). (C, D) Phenotypes of WT and 35S:CsRAV1 transgenic plants treated with NaCl and ABA. After 6 h of NaCl and ABA stress treatment, the expression levels of stress- and ABA- related marker genes AtRD29A (E), AtSOS1 (F), AtNHX1 (G), and AtABI5 (H) were detected in WT and 35S:CsRAV1 plants (L1 and L2). The actin gene was used as internal control, with error bars of three biological replicates. Different letters indicated significant differences (P < 0.05).
Discussion
RAV gene family is widely distributed in higher plants and is one of the plant-specific regulatory gene families. As one of the B3 gene superfamilies in plants, the RAV family genes regulate many aspects of plant growth and development, including regulating flowering time and heading date (Hu et al., 2004; Woo et al., 2010; Duan et al., 2016; Shin and Nam, 2018). At present, RAV genes have been extensively studied in Arabidopsis, rice, soybean, cotton, pepper and other plants (Giraudat et al., 1992; Sohn et al., 2006; Zhao et al., 2008; Li et al., 2015; Osnato et al., 2020). However, little is known about the role of the RAVs in cucumber. In this study, we identified eight cucumber RAV genes through genome-wide analysis. The phylogenetic relationship, gene structure, conserved domains, gene duplication events and cis-acting elements on promoters of RAV family genes in cucumber were systematically analyzed.
To analyze the molecular evolutionary relationship between cucumber RAV proteins and RAVs among other plant species, a phylogenetic tree of 115 RAV proteins from ten species was constructed, and roughly divided into 23 groups, among which CsRAVs were classified into different groups (Figure 1). To further study the evolutionary relationship and diversity/conservativeness of RAV genes in tomato, Arabidopsis and cucumber, the phylogenetic tree, conserved motifs and gene structures of eight CsRAVs, 13 AtRAVs and nine SlRAVs were further analyzed in detail (Figure 2). The gene structure pattern and motif composition can give important insights for evolutionary relationships of multi-gene families (Boudet et al., 2001; Babenko et al., 2004). Tomato, Arabidopsis and cucumber are different in anatomy and physiology. Therefore, some branches may have different ways of expansion in the RAV family of tomato, Arabidopsis and cucumber. As shown in Figure 2, the RAVs within the same clade shared similar gene structure and motif composition. Genes with similar gene structures and conserved motifs usually have similar functions. The cucumber RAV proteins were clustered into some Arabidopsis functional clades, which will provide valuable information for studying the function of CsRAV genes.
Previous studies have investigated the role of RAV transcription factors in regulating plant responses to biotic and abiotic stresses such as plant hormones and pathogens (Chen et al., 2021). In our study, it was shown that the promoters of CsRAV genes contain cis-elements that respond to abiotic stresses and hormones such as drought, wound, defense and stress, low temperature, auxin, salicylic acid, abscisic acid, gibberellin and jasmonic acid (Figure 4). In addition, TATA box is one of the elements that constitute the eukaryotic promoters, and its sequence is TATA (A/T) A (A/T). It is generally about -30bp (-25~-32bp) upstream of the transcription start site of most eukaryotic genes. The TATA box is the selection that determines the initiation of gene transcription and is one of the binding sites for RNA polymerase, which can only be transcribed after the RNA polymerase is firmly bound to the TATA box (Pedersen et al., 1999). The transcription start site (TSS) is the location where transcription starts at the 5’-end of a gene sequence (Peng et al., 2006). Our data showed that all CsRAV gene promoters except CsRAV3 contained TSS and TATA-box (Supplementary Figure 3). The specific TSS and TATA-box on each CsRAV gene promoter need to be determined by robust analysis of 5’-transcript ends (5’-RATE) experiment. Previous studies indicated that AtRAV1, AtRAV2, BnaRAV-1, GhRAV1 and CARAV1 genes could be significantly induced after stress treatment (Sohn et al., 2006; Zhuang et al., 2011; Fu et al., 2014; Li et al., 2015). In this study, CsRAV genes were mainly up-regulated by salt, low temperature and ABA, and some of them were significantly affected in different degrees (Figure 5). These results suggested that the cucumber RAV genes, like the genes of other plants, played key roles in stress resistance. However, there were different response patterns under different stresses, indicating functional diversity among the genes.
The functions of RAV genes in Arabidopsis, soybean, cotton, pepper and rice have been widely reported (Hu et al., 2004; Zhao et al., 2017; Chen et al., 2021; Kabir et al., 2021). Overexpression of AtRAV1 and AtRAV2 can improve the drought resistance in cotton (Matías-Hernández et al., 2014). Overexpression of soybean GmRAV3 in Arabidopsis could significantly increase the resistance of transgenic lines to high salt and drought, and lead to the insensitivity of transgenic plants to exogenous ABA (Lu et al., 2014; Zhao et al., 2017). CsRAV1 was significantly induced by salt and ABA (Figures 5A, B), so CsRAV1 might play key roles in resistance to salt or ABA stress. To further determine the roles of CsRAV1 in abiotic stress response, we obtained the 35S:CsRAV1 transgenic Arabidopsis and transient transformed cucumber cotyledons. Under NaCl and ABA treatments, we found 35S:CsRAV1 cucumber seedlings indeed showed enhanced tolerance to salt and ABA compared with 35S plants (Figures 7A–C). Meanwhile, the transgenic Arabidopsis showed higher germination rates than WT, indicating that CsRAV1 played vital roles in seed germination and development (Figures 8A–D). The expression of CsRAV1 was also induced by low temperature (Figure 5C), but we found that there were no significant differences in phenotype between 35S:CsRAV1 transgenic seedlings and control treated at 4°C, which required further verification and discussion in the future.
Previous studies have shown that abiotic stress-related marker genes (such as RD29A, ABI5, SOS1, SOS2, NHX3, CPK1, RD22 and APX2) are involved in plant response and defense against environmental stress (Li et al., 2015; Zhang et al., 2020). Changes in the expression of these marker genes may contribute to plant resistance to abiotic stress. The transcriptional levels of stress- and ABA- related genes were regulated in transgenic plants under different stresses (Li et al., 2015; Zhang et al., 2020). Under NaCl treatment, the expressions of ABI1, RD29A and RAB18 were obvious enhanced in 35S:GhRAV1 transgenic plants compared with those in WT (Li et al., 2015). Compared with those in WT, the transcriptional levels AtSOS1, AtRD22 and AtRD29A were upregulated in 35S:TaRAV1 transgenic plants after NaCl treatment (Luo et al., 2022). The ABI5 gene is a key positive regulator of ABA signal transduction, and increased ABI5 gene expression enhances susceptibility to ABA during seed germination and early seedling development (Lopez-Molina et al., 2001). AtRAV1 is involved in ABA signaling by directly binding to the promoter of ABI5 and influencing its expression (Feng et al., 2014). MdRAV1 plays a key role in ABA signaling by directly binding to the promoters of MdABI3 and MdABI4 (Zhao et al., 2019b). Previous research showed the sodium/proton antiporter1 (NHX1) maintained Na+ homeostasis in plants and plays a vital role in plant salt tolerance (Zhang et al., 2020). In this study, qRT-PCR analysis indicated that the transcriptional levels of four stress-related genes, including CsSOS1, CsNHX1, CsCPK11 and CsABI5, in transgenic cucumber were higher than those in 35S plants after NaCl and ABA treatment (Figures 7D–G). These results indicated that CsRAV1 might directly regulate the expression of CsABI5, thereby participating in ABA signal pathway and enhancing ABA tolerance in cucumber. Furthermore, CsRAV1 might regulate the transcription of CsSOS1 and CsNHX1 genes to maintain ion homeostasis, thus enhancing salt tolerance. In addition, stress- and ABA- related marker genes (AtRD29A, AtSOS1, AtNHX1 and AtABI5) in transgenic Arabidopsis were significantly up-regulated compared with WT, suggesting that CsRAV1 might alleviate NaCl and ABA stress by regulating stress- related genes in Arabidopsis (Figures 8E–H). These results will provide a theoretical basis for further study on the function of RAV transcription factor in cucumber.
Conclusions
In this study, the cucumber RAV family genes were systematically analyzed. The expression patterns of CsRAV genes under different stress treatments were studied, and the potential functions of CsRAV1 were also analyzed using the transgenic method. This work provides a rich insight into the functions and regulatory mechanism of CsRAV genes in abiotic stress resistance of cucumber.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
JL, WH and ZW conceived and designed the experiments. JL, CS, HL, SW, LH and YY performed the experiments. JL analyzed the data and wrote the manuscript. WH and ZW revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by fundings from the National Natural Science Foundation of China (32202484, 31872415 and 32102310), the Natural Science Foundation of Shandong Province (ZR2022QC008), Scientific Research Leaders Studio of Jinan (2019GXRC052) and Doctoral Fund project of Jinan University (XBS2104). The funds played no role in study design, data analysis, and manuscript preparation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1115874/full#supplementary-material
References
Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 (5633), 653–657. doi: 10.1126/science.1086391
Alvarez, J. P., Goldshmidt, A., Efroni, I., Bowman, J. L., Eshed, Y. (2009). The NGATHA distal organ development genes are essential for style specification in Arabidopsis. Plant Cell 21 (5), 1373–1393. doi: 10.1105/tpc.109.065482
Babenko, V. N., Rogozin, I. B., Mekhedov, S. L., Koonin, E. V. (2004). Prevalence of intron gain over intron loss in the evolution of paralogous gene families. Nucleic Acids Res. 32 (12), 3724–3733. doi: 10.1093/nar/gkh686
Boudet, N., Aubourg, S., Toffano-Nioche, C., Kreis, M., Lecharny, A. (2001). Evolution of intron/exon structure of DEAD helicase family genes in Arabidopsis, Caenorhabditis, and Drosophila. Genome Res. 11 (12), 2101–2114. doi: 10.1101/gr.200801
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Castillejo, C., Pelaz, S. (2008). The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18 (17), 1338–1343. doi: 10.1016/j.cub.2008.07.075
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C., Li, Y., Zhang, H., Ma, Q., Wei, Z., Chen, J., et al. (2021). Genome-wide analysis of the RAV transcription factor genes in rice reveals their response patterns to hormones and virus infection. Viruses. 13 (5), 752. doi: 10.3390/v13050752
Das, L. S., Dutta, S., Schäffner, A. R., Das, M. (2020). Gene duplication and stress genomics in brassicas: Current understanding and future prospects. J. Plant Physiol. 255, 153293. doi: 10.1016/j.jplph.2020.153293
Duan, Y. B., Li, J., Qin, R. Y., Xu, R. F., Li, H., Yang, Y. C., et al. (2016). Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol. Biol. 90 (1-2), 49–62. doi: 10.1007/s11103-015-0393-z
Feng, C. Z., Chen, Y., Wang, C., Kong, Y. H., Wu, W. H., Chen, Y. F. (2014). Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 80 (4), 654–668. doi: 10.1111/tpj.12670
Fu, M., Kang, H. K., Son, S. H., Kim, S. K., Nam, K. H. (2014). A subset of Arabidopsis RAV transcription factors modulates drought and salt stress responses independent of ABA. Plant Cell Physiol. 55 (11), 1892–1904. doi: 10.1093/pcp/pcu118
Giraudat, J., Hauge, B. M., Valon, C., Smalle, J., Parcy, F., Goodman, H. M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4 (10), 1251–1261. doi: 10.1105/tpc.4.10.1251
Hu, Y. X., Wang, Y. X., Liu, X. F., Li, J. Y. (2004). Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res. 14 (1), 8–15. doi: 10.1038/sj.cr.7290197
Huang, S., Li, R., Zhang, Z., Li, L., Gu, X., Fan, W., et al. (2009). The genome of the cucumber, Cucumis sativus l. Nat. Genet. 41 (12), 1275–1281. doi: 10.1038/ng.475
Kabir, N., Lin, H., Kong, X., Liu, L., Qanmber, G., Wang, Y., et al. (2021). Identification, evolutionary analysis and functional diversification of RAV gene family in cotton (G. hirsutum l.). Planta. 255 (1), 14. doi: 10.1007/s00425-021-03782-2
Kagaya, Y., Ohmiya, K., Hattori, T. (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478. doi: 10.1093/nar/27.2.470
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19 (9), 1639–1645. doi: 10.1101/gr.092759.109
Kwon, S. H., Lee, B. H., Kim, E. Y., Seo, Y. S., Lee, S., Kim, W. T., et al. (2009). Overexpression of a Brassica rapa NGATHA gene in Arabidopsis thaliana negatively affects cell proliferation during lateral organ and root growth. Plant Cell Physiol. 50 (12), 2162–2173. doi: 10.1093/pcp/pcp150
Lee, B. H., Kwon, S. H., Lee, S. J., Park, S. K., Song, J. T., Lee, S., et al. (2015). The Arabidopsis thaliana NGATHA transcription factors negatively regulate cell proliferation of lateral organs. Plant Mol. Biol. 89 (4-5), 529–538. doi: 10.1007/s11103-015-0386-y
Li, C. W., Su, R. C., Cheng, C. P., Sanjaya, You, S. J., Hsieh, T. H., et al. (2011). Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway. Plant Physiol. 156 (1), 213–227. doi: 10.1104/pp.111.174268
Li, J., Li, H., Quan, X., Shan, Q., Wang, W., Yin, N., et al. (2022). Comprehensive analysis of cucumber c-repeat/dehydration-responsive element binding factor family genes and their potential roles in cold tolerance of cucumber. BMC Plant Biol. 22 (1), 270. doi: 10.1186/s12870-022-03664-z
Li, J., Wang, T., Han, J., Ren, Z. (2020). Genome-wide identification and characterization of cucumber bHLH family genes and the functional characterization of CsbHLH041 in NaCl and ABA tolerance in Arabidopsis and cucumber. BMC Plant Biol. 20 (1), 272. doi: 10.1186/s12870-020-02440-1
Li, X. J., Li, M., Zhou, Y., Hu, S., Hu, R., Chen, Y., et al. (2015). Overexpression of cotton RAV1 gene in Arabidopsis confers transgenic plants high salinity and drought sensitivity. PLoS One 10 (2), e0118056. doi: 10.1371/journal.pone.0118056
Lopez-Molina, L., Mongrand, S., Chua, N. H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 98, 8, 4782–4787. doi: 10.1073/pnas.081594298
Lu, Q., Zhao, L., Li, D., Hao, D., Zhan, Y., Li, W. (2014). A GmRAV ortholog is involved in photoperiod and sucrose control of flowering time in soybean. PLoS One 9 (2), e89145. doi: 10.1371/journal.pone.0089145
Luo, Y. X., Chen, S. K., Wang, P. D., Peng, D., Zhang, X., Li, H. F., et al. (2022). Genome-wide analysis of the RAV gene family in wheat and functional identification of TaRAV1 in salt stress. Int. J. Mol. Sci. 23 (16), 8834. doi: 10.3390/ijms23168834
Magnani, E., Sjölander, K., Hake, S. (2004). From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell. 16 (9), 2265–2277. doi: 10.1105/tpc.104.023135
Marín-González, E., Matías-Hernández, L., Aguilar-Jaramillo, A. E., Lee, J. H., Ahn, J. H., Suárez-López, P., et al. (2015). SHORT VEGETATIVE PHASE up-regulates TEMPRANILLO2 floral repressor at low ambient temperatures. Plant Physiol. 169 (2), 1214–1224. doi: 10.1104/pp.15.00570
Matías-Hernández, L., Aguilar-Jaramillo, A. E., Marín-González, E., Suárez-López, P., Pelaz, S. (2014). RAV genes: regulation of floral induction and beyond. Ann. Botany. 114 (7), 1459–1470. doi: 10.1093/aob/mcu069
Osnato, M., Matias-Hernandez, L., Aguilar-Jaramillo, A. E., Kater, M. M., Pelaz, S. (2020). Genes of the RAV family control heading date and carpel development in rice. Plant Physiol. 183 (4), 1663–1680. doi: 10.1104/pp.20.00562
Pedersen, A. G., Baldi, P., Chauvin, Y., Brunak, S. (1999). The biology of eukaryotic promoter prediction–a review. Comput. Chem. 23, 191–207. doi: 10.1016/S0097-8485(99)00015-7
Peng, Z. H., Chen, J., Cao, L. J., Gao, T. T. (2006). Identification of TSS in the human genome based on a RBF neural network. Int. J. Automation Computing. 3 (1), 35–40. doi: 10.1007/s11633-006-0035-7
Romanel, E. A., Schrago, C. G., Couñago, R. M., Russo, C. A., Alves-Ferreira, M. (2009). Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS One 4 (6), e5791. doi: 10.1371/journal.pone.0005791
Shin, H. Y., Nam, K. H. (2018). RAV1 negatively regulates seed development by directly repressing MINI3 and IKU2 in Arabidopsis. Molecules Cells 41 (12), 1072–1080. doi: 10.14348/molcells.2018.0259
Sohn, K. H., Lee, S. C., Jung, H. W., Hong, J. K., Hwang, B. K. (2006). Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol. Biol. 61 (6), 897–915. doi: 10.1007/s11103-006-0057-0
Swaminathan, K., Peterson, K., Jack, T. (2008). The plant B3 superfamily. Trends Plant Sci. 13 (12), 647–655. doi: 10.1016/j.tplants.2008.09.006
Wang, Y., Deng, D., Zhang, R., Wang, S., Bian, Y., Yin, Z. (2012a). Systematic analysis of plant-specific B3 domain-containing proteins based on the genome resources of 11 sequenced species. Mol. Biol. Rep. 39 (5), 6267–6282. doi: 10.1007/s11033-012-1448-8
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012b). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49. doi: 10.1093/nar/gkr1293
Wei, Y., Chang, Y., Zeng, H., Liu, G., He, C., Shi, H. (2018). RAV transcription factors are essential for disease resistance against cassava bacterial blight via activation of melatonin biosynthesis genes. J. Pineal Res. 64 (1). doi: 10.1111/jpi.12454
Woo, H. R., Kim, J. H., Kim, J., Kim, J., Lee, U., Song, I. J., et al. (2010). The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J. Exp. Bot. 61 (14), 3947–3957. doi: 10.1093/jxb/erq206
Zhang, X., Chen, L., Shi, Q., Ren, Z. (2020). SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato. Plant Sci. 291, 110356. doi: 10.1016/j.plantsci.2019.110356
Zhao, L., Luo, Q., Yang, C., Han, Y., Li, W. (2008). A RAV-like transcription factor controls photosynthesis and senescence in soybean. Planta 227 (6), 1389–1399. doi: 10.1007/s00425-008-0711-7
Zhao, L., Zhang, F., Liu, B., Yang, S., Xiong, X., Hassani, D., et al. (2019a). CmRAV1 shows differential expression in two melon (Cucumis melo l.) cultivars and enhances salt tolerance in transgenic Arabidopsis plants. Acta Biochim. Biophys. Sin. (Shanghai) 51 (11), 1123–1133. doi: 10.1093/abbs/gmz107
Zhao, S. P., Xu, Z. S., Zheng, W. J., Zhao, W., Wang, Y. X., Yu, T. F., et al. (2017). Genome-wide analysis of the RAV family in soybean and functional identification of GmRAV-03 involvement in salt and drought stresses and exogenous ABA treatment. Front. Plant Science. 8, 905. doi: 10.3389/fpls.2017.00905
Zhao, X. Y., Qi, C. H., Jiang, H., You, C. X., Guan, Q. M., Ma, F. W., et al. (2019b). The MdWRKY31 transcription factor binds to the MdRAV1 promoter to mediate ABA sensitivity. Horticulture Res. 6, 66. doi: 10.1038/s41438-019-0147-1
Keywords: abiotic stresses, cucumber, expression patterns, RAV family, salt tolerance, transcription factor
Citation: Li J, Song C, Li H, Wang S, Hu L, Yin Y, Wang Z and He W (2023) Comprehensive analysis of cucumber RAV family genes and functional characterization of CsRAV1 in salt and ABA tolerance in cucumber. Front. Plant Sci. 14:1115874. doi: 10.3389/fpls.2023.1115874
Received: 04 December 2022; Accepted: 25 January 2023;
Published: 02 February 2023.
Edited by:
Jihong Hu, Northwest A&F University, ChinaReviewed by:
Yongguang Li, Northeast Agricultural University, ChinaYong Zhou, Jiangxi Agricultural University, China
Copyright © 2023 Li, Song, Li, Wang, Hu, Yin, Wang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenxing He, MTYzLnhAMTYzLmNvbQ==; Zenghui Wang, Mzc0MTQxNjQ1QHFxLmNvbQ==
 Jialin Li1
Jialin Li1 Yanlei Yin
Yanlei Yin Wenxing He
Wenxing He