- 1State Key Laboratory of Desert and Oasis Ecology, Key Laboratory of Ecological Safety and Sustainable Development in Arid Lands, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences, Urumqi, China
- 2College of Resources and Environment, University of Chinese Academy of Sciences, Beijing, China
- 3Xinjiang Key Laboratory of Conservation and Utilization of Plant Gene Resources, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences, Urumqi, China
- 4Turpan Eremophytes Botanical Garden, Chinese Academy of Sciences, Turpan, China
Among the most important transcription factors in plants, the v-myb avian myeloblastosis viral oncogene homolog (MYB) regulates the expression network of response genes under stresses such as fungal infection. In China, the canker disease Valsa mali threatens the survival of Malus sieversii, an ancestor of cultivated apples. Using the M. sieversii genome, we identified 457 MsMYB and 128 R2R3-MsMYB genes that were randomly distributed across 17 chromosomes. Based on protein sequence and structure, the R2R3-MsMYB genes were phylogenetically divided into 29 categories, and 26 conserved motifs were identified. We further predicted cis-elements in the 2000-kb promoter region of R2R3-MsMYBs based on the genome. Transcriptome analysis of M. sieversii under V. mali infection showed that 27 R2R3-MsMYBs were significantly differentially expressed, indicating their key role in the response to V. mali infection. Using transient transformation, MsMYB14, MsMYB24, MsMYB39, MsMYB78, and MsMYB108, which were strongly induced by V. mali infection, were functionally identified. Among the five MsMYBs, MsMYB14 and MsMYB78 were both important in enhancing resistance to diseases, whereas MsMYB24 inhibited resistance. Based on the results of this study, we gained a better understanding of the MsMYB transcription factor family and laid the foundation for a future research program on disease prevention strategies in M. sieversii.
1 Introduction
Plants rely on a wide range of transcription factors (TFs), including the v-myb avian myeloblastosis viral oncogene homolog (MYB) TFs, to respond to biotic stress and other environmental factors (Romero et al., 1998; Riechmann et al., 2000). In 1987, the first MYB gene, COLORED1 (C1), was identified in plants (Paz-Ares et al., 1987). The MYB repeats (R) that form the MYB DNA-binding domain (DBD) comprise four typical subfamilies and atypical MYB-like subfamilies (Mmadi et al., 2017).
MYB TFs have a modular structure, with an N-terminal domain called the MYB domain and a C-terminal that could either be activator, repressor, or both (Stracke et al., 2001). Several encoded proteins have highly conserved domains composed of 1-4 imperfect amino acid sequence repeats (R) of 50–53 amino acids arranged in a helix-turn-helix structure (HTH) (Ogata et al., 1996; Rosinski and Atchley, 1998; Dubos et al., 2010; Hurtado-Gaitán et al., 2021). In the three-helical structure, there is a hydrophobic core formed by three tryptophans positioned periodically (Ogata et al., 1992; Dubos et al., 2010; Zhang et al., 2012). In accordance with the number of repeats, four main subfamilies were classified: R1 or R2-MYB (1R), R2R3-MYB (2R), R1R2R3-MYB (3R), and R4-MYB (4R), with one, two, three, and four conservative MYB repeats, respectively (Kanei-Ishii et al., 1990). The R2R3-MYB subfamily is considered the largest of the MYB family (Sun et al., 2019; Chen X. et al., 2021). Furthermore, R1R2R3-MYB has been reported to exist predominantly in land plants (Kranz et al., 2000). The most remarkable is the 4R-MYB group, which includes four R1/R2-like repeats (Dubos et al., 2010). Because of these DBD characteristics, the R2R3-MYB family was also divided into 25 subgroups (SGs) in Arabidopsis (Stracke et al., 2001). In recent years, genome-wide sequencing has been performed in various species of plants, and MYB genes were discovered in many species (Pucker et al., 2020; Chen G. et al., 2021; Chen L. et al., 2021; Song et al., 2021). Moreover, MYB genes have been found to function in several regulatory mechanisms and simultaneously perform multiple functions. MYB genes were also found to be expressed in different organs and tissues by expression profiling, and several genes were classified as being induced by external stress conditions (Li X. et al., 2019; Xing et al., 2021; Yuan et al., 2021). Moreover, MYB genes are also responsible for disease resistance in many plants, including sugarcane (Saccharum officinarum) (Yuan et al., 2021). In addition to improving product quality and yield, screening MYB genes can increase bioenergy resources in biotechnology (Cardenas-Hernandez et al., 2021).
Apples are very significant fruit crops (Dubos et al., 2010) and prominent economic trees. Infection of apple tree trunks, scaffolds, branches, and leaves by diseases such as Valsa canker and Botryosphaeria canker causes huge destruction to production (Li et al., 2021; Liang et al., 2022). In particular, the ascomycete V. mali has been reported to greatly reduce apple production (Lee et al., 2006; Wang et al., 2014). Recent research suggests that MYB plays a significant role in controlling apple canker. MYB TFs are known to play a crucial role in stress perception and signal transduction, especially in the stress response in plants (Yanhui et al., 2006; Lu et al., 2009; Li J. et al., 2019). The balance between homeostasis and the environment is maintained by several genes involved in hormone regulation (Chen X. et al., 2021). In recent years, many differentially expressed genes that participate in biotic and abiotic stress responses have been identified in plants by transcriptomic analysis. In recent years, R2R3-MYB proteins perform a variety of functions under biotic and abiotic stress, including secondary metabolic regulation and the cell cycle, growth, and development (Paz-Ares et al., 1987; IH and EP, 1999; Stracke et al., 2001; Dubos et al., 2010; Blanco et al., 2018; Chen et al., 2019; Cao et al., 2020; Pucker et al., 2020). Several studies have shown that R2R3-MYB proteins regulate anthocyanin and lignin biosynthesis in plants (Tuan et al., 2015; Arce-Rodriguez et al., 2021; Song et al., 2021; Zhou et al., 2021). As a result of R2R3-MYB regulation, anthocyanin biosynthesis is upregulated through hypomethylation of DNA in their promoter regions (Zhou et al., 2021). A certain level of canker resistance can be indirectly enhanced by the growth of anthocyanins and lignin (Faize et al., 2020; Zhang Q. et al., 2021). Overexpression of R2R3-MYB genes as a method for preventing phytopathogenic fungi such as V. mali and B. kuwatsukai is effective for protecting plants from phytopathogenic fungi (Chen L. et al., 2021).
Herbaceous and woody plants have been studied for the majority of the functions and structural characteristics of the MYB gene family. Several studies have shown that MYB TFs are involved in regulating plant growth and development in wild apples (Malus sieversii) and in modulating cold tolerance (An et al., 2018; Xie et al., 2018). Many R2R3-MYB proteins in M. sieversii cannot be accurately identified as redundant in their respective functions, but they probably have overlapped functions (Jin and Martin, 1999). In this study, the resistance functions of five R2R3-MYB TF in Valsa canker were identified. As a first step, this research collected genome-wide information about M. sieversii and snapped all MYB gene sequences from A. thaliana. These analyses were conducted to predict protein physicochemical properties, build a phylogenetic tree, analyze conserved motifs and chromosomal positions, and forecast promoter cis-elements. Five MsMYB transcripts (MsMYB14, MsMYB24, MsMYB39, MsMYB78, and MsMYB108) were cloned to determine their resistance to Valsa canker by analyzing the transcriptome data of the MYB gene. This study helps to understand the mechanisms of MYB genes and to identify V. mali-resistant genes.
2 Materials and methods
2.1 Identification and classification of the MYB gene family in Malus sieversii
In this study, the Hidden Markov model (HMM) was used to identify MsMYB based on the M. sieversii genome, which was retrieved from the Pfam database (PF00249) (http://pfam.xfam.org/). Additionally, AtMYB protein sequences were retrieved from TAIR (http://www.arabidopsis.org/) (Stracke et al., 2001) and used for searching more MYB transcription factor candidates using BlastP and E-value < 1e-5.
To predict the molecular weight (Da) and isoelectric point (PI) of potential proteins, the ProtParam tool was used (https://web.expasy.org/protparam/). TBtools were used to measure length of the MsMYB gene and MsMYB protein sequences (Chen et al., 2020) (Supplementary Table S1).
2.2 Phylogenetic and conserved motif analysis of the MYB gene family
To identify and classify 1R, 2R, 3R, and 4R subfamilies of MYB and thoroughly classify the R2R3-MYB subfamily, AtMYB and MsMYB were used to analyze multiple sequence alignments and construct a phylogenetic tree with MEGA 10 using the neighbor-joining (NJ) model and 1000 replicate bootstraps. Finally, a phylogenetic tree was constructed using itol (https://itol.embl.de/).
To evaluate the classification results of MsMYB, MEME, an online analysis tool (https://meme-suite.org/meme/doc/meme.html), was used to analyze and show the MYB domain structure. The parameters included motif occurrence distribution, zero or one motif per sequence, the maximum number of motifs (26), and the optimal width of motifs between 6 and 10 residues.
2.3 Analysis of gene collinearity and cis-elements of promoter
The Malus domestica Borkh genome was downloaded from the GDR database (https://www.rosaceae.org/species/malus/all) and used for comparative analysis (Daccord et al., 2017). To explore the collinearity relationship of the MsMYB gene family, the McScan module in TBtools was used to analyze collinearity of intragenome and intergenome, and Advance Circos and Multiple Synteny Plot were used to perform the collinearity analysis and duplication type, including the whole genome duplication or segmental (WGD or segmental), tandem, Dispersed and Proximal. Based on the genome, the chromosomal distribution of R2R3-MsMYB was also shown using TBtools. To determine the type and number of cis-elements of the R2R3-MsMYB gene, the promoter region, upstream 2000 bp of the gene was extracted from the genome for analysis. We submitted the promoter sequences to the PlantCARE database (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) for excavating cis-elements.
2.4 Expression profiling of MsMYB genes in response to Valsa mali
A set of transcriptions of M. sieversii under V. mali infection data was downloaded from NCBI for analysis of R2R3-MsMYB gene expression patterns (https://www.ncbi.nlm.nih.gov/) (Liu et al., 2021). A heat map of R2R3-MsMYB gene expression was created using the TBtools software. Additionally, a gene with a differential expression scale of Log2|foldchange| ≥ 1 and p-value < 0.05, i.e., a differentially expressed gene (DEG) under infection, was compared to the control (Supplementary Table S3).
2.5 Experimental materials and gene cloning
Malus sieversii seedlings were grown under controlled conditions in a greenhouse with soil (nutrient soil: vermiculite: perlite = 3:1:1). They were kept growing until they were 2–3 months old, at 24–26°C, 70%–75% relative humidity, and sufficient watering.
The intact coding sequence (CDS) of the examined TF was fused in-frame with the C-terminus of a 3 × Flag tag under the control of the CaMV 35S promoter in the p1307-Flag plant expression vector. All primers used for construction are listed in Supplementary Table S4. DNA sequencing was performed on all constructs before transferring them to Agrobacterium EHA105.
2.6 Identification of the resistance of MsMYB transcription factors in response to Valsa mali
Transient transformation was performed as described by Wen et al. (2020). The colonies of Agrobacterium tumefaciens EHA105 harboring the studied constructs were cultured in LB medium to OD 08-1.0, and harvested by centrifugation at 3500 rpm. Agrobacterium tumefaciens EHA105 colonies harboring the studied constructs were adjusted to an OD600 of 50 mL of transformation solution [ 5% (w/v) sucrose + 150 μM acetosyringone + 5 mM CaCl2 + 0.015% DTT (w/v) + 20 μM 5-AZA + Tween 20 (0.01%, v/v), pH 5.8]. Malus sieversii leaves were soaked in the transformation solution at room temperature while shaking at 100 rpm. Three hours after the leaves were removed from the transformation solution, excess water was quickly wiped off with sterile filter paper. The leaves were rinsed twice with sterile water. Control plants (Con) were transformed with empty P1307-Flag.
Aseptically, mycelial plugs (diameter of 0.5 mm) were removed from the edge of the three-day-cultured isolate V. mali on PDA medium. Following transient expression, the leaves were wounded by tips and inoculated with mycelial plugs for 24 h. For three days, the inoculated leaves were placed on dishes tapped with parafilm to maintain moisture. Incidence ratios (%) were calculated using photographs. We measured the lesion areas using ImageJ software. Three biological replicates were used for each experiment. Using the LSD method, the mean and standard error (SD) of the data were analyzed using one-way ANOVA (p < 0.05). For the measurement of fungal biomass, we used the following equation:6.02 × 10 ^ 23 × (ng/μl × 10 ^ −9) × (DNA length × 660) × volume of extraction liquid ÷ fresh weight of sample; for measuring H2O2 and MDA content, we used the H2O2 content detection kit (Jiancheng, Nanjing) and the MDA content detection kit (Jiancheng, Nanjing), respectively.
2.7 Real-time PCR analysis
Malus sieversii RNA was isolated using the CTAB method, and DNA contamination was removed with DNase I. Using the PrimeScript TMRT reagent kit (Transgen, Beijing, China) with oligo (dT) primers, 1 µg of total RNA from each sample was reverse-transcribed into cDNA. The resulting cDNA product was diluted 10-fold before use in the PCR. To normalize the number of templates generated, EF1αgene (Elongation factor 1-α) was used as the internal reference.
A CFX96 Real-Time PCR Detection System (Bio-Rad, CFX96, USA) was used to perform real-time PCR under the following conditions: 94°C for 60 s; 45 cycles at 94°C for 10 s, 59°C for 20 s, 72°C for 30 s, and 80°C for 1 s for plate reading. In a 20-L volume, the reaction mixture included 10 L of SYBR Green Real-time PCR Master Mix (Transgenic, Beijing), 0.5 L of forward and reverse primers, and 2 L of cDNA template. A melting curve was generated to ensure the purity of the amplified products. Three biological replicates were used in all experiments, and relative expression levels were assessed using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Detailed primer information is provided in Supplementary Table S4.
3 Results
3.1 Genome-wide identification of the MYB gene family in Malus sieversii
The Hemmer search that based PF00249 model and BlastP that use ATMYB protein sequences as query were used to identify MsMYB TFs in M. sieversii. When 458 proteins were analyzed for R1-4 motifs, one protein without any conserved motifs was found. Finally, the 457 proteins were identified. Based on conserved motifs and phylogenetic analysis, we identified 323, 128, 5, and 1 member in the 1R, 2R(R2R3), 3R, and 4R subfamilies, respectively. The 1R-MsMYB subfamily was divided into 52 groups. The isoelectric point (PI) of R1-MsMYB proteins ranged from 4.38 for MS12G07430.1 to 10.24 for MS04G02770.1. The 2R-MsMYB subfamily was divided into 29 groups. The PI of R2R3-MsMYB proteins ranged from 4.83 for MS03G17360.1 to 10.43 for MS13G03930.1, and 56% of the proteins had isoelectric points between 5 and 7. The average PI of the 2R-MsMYB subfamily was 7.05 (STD = 0.13). The maximum and minimum molecular weights were 259.85 kDa (MS03G19210.1) and 6.76 kDa (MS10G15090.1), respectively. The average gene length of each subfamily was arranged in the following order: 2R (992 bp), 1R (1303 bp), 3R (2030 bp), and 4R (2928 bp) (Supplementary Table S1).
3.2 Phylogenetic and motif analysis of R2R3-MsMYB transcription factors
The 2R subfamily was further classified and studied for diverse and important biological functions. The 94 R2R3-AtMYB and 128 R2R3-MsMYB that obtained the R2R3 domain were used to construct a phylogenetic tree and analyze conserved motifs. Finally, R2R3-MsMYBs were classified into 29 groups and named I-XXIX, and MsMYBs belonging to the same cluster had the same motif pattern (Figure 1).
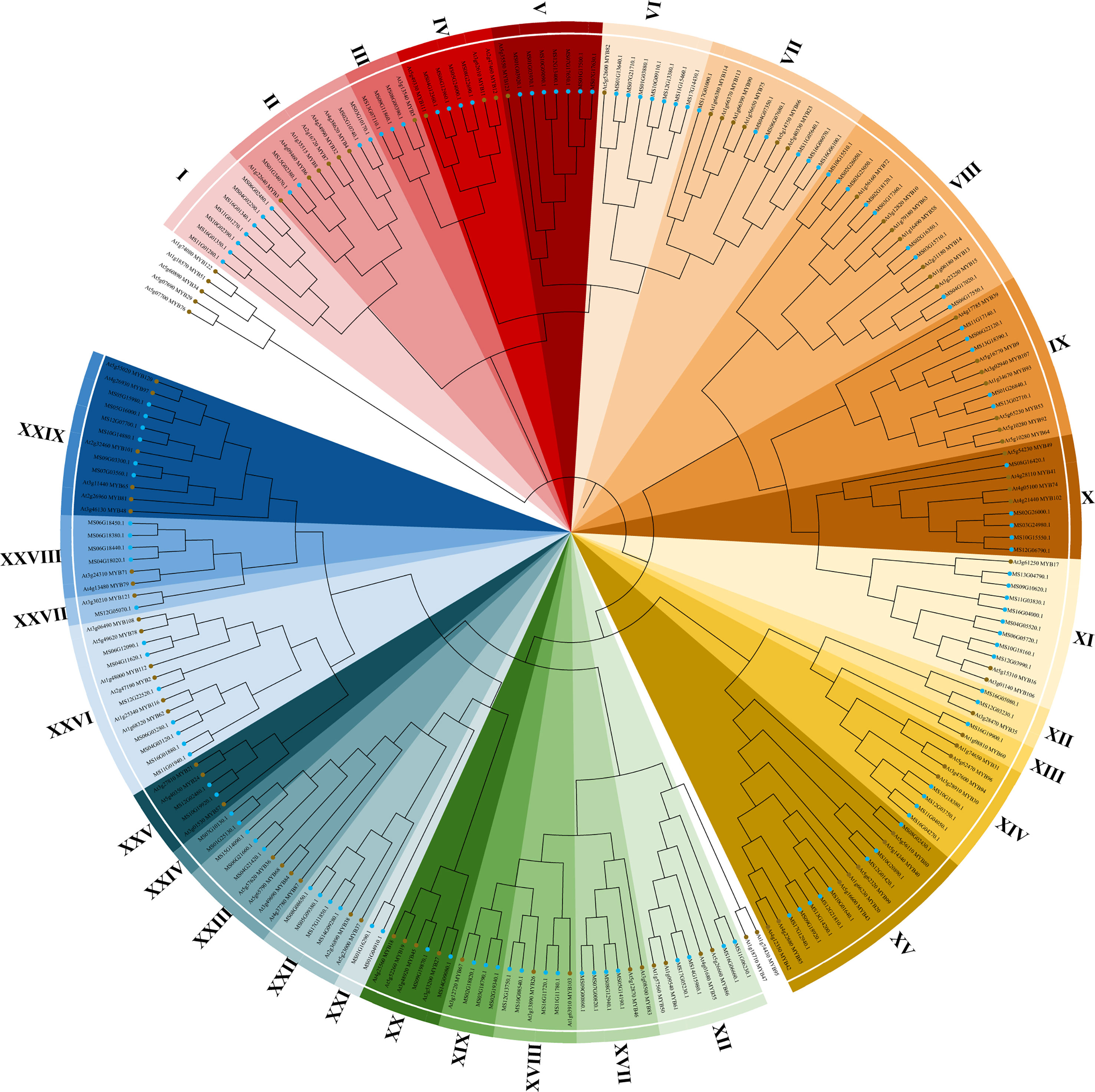
Figure 1 Phylogenetic analysis of R2R3-MYB transcription factors from Malus sieversii and Arabidopsis thaliana. Neighbor-joining tree of 128 M. sieversii (MsMYBs) and 94 A. thaliana (AtMYBs) R2R3-MYB TFs. The bootstrap test used 1000 replicates. The R2R3-MYB transcription factors of M. sieversii and A. thaliana are marked in blue and brown, respectively. The tree shows 29 clades (I–XXIX) with a high bootstrap value (with a different color for each clade). The uncolored region represents A. thaliana (AtMYBs).
To further verify the 128 R2R3-MsMYB classification results by phylogenetic analysis, the MEME website was used to study the MYB domain structure. A total of 26 conserved motifs were identified in 128 R2R3-MsMYB proteins. Nine motifs, i.e., 1, 2, 3, 4, 5, 6, 7, 8, and 9, were highly conserved in the subfamily and constructed a basal MYB domain of 50–53 amino acids (Figure 2). Among them, motifs 4, 8, 6, 1, 3, and the first half of motif 7 formed a complete R2 domain, whereas the second half of motif 7 and motifs 5, 2, and 9 formed a complete R3 domain. In addition, MsR2R3-MYB members within the same clade shared similar motifs and highly conserved MYB domains. Among these, motif 17 formed a common element in the V group. Groups I and II had a common motif 15; the X group had unique motifs, including motifs 23 and 24 in R2R3-MYB TF; motifs 12 and 16 were unique to the XI group; motif 19 was unique to the XIV group; and motif 18 was unique to XXVI. Specifically, motif 21 was repeated twice in MS10G01640.1 and MS12G21810.1, but repeated once in MS02G16350.1, MS03G15710.1, MS04G17020.1, and MS06G17550.1. MS16G1880.1 (MsMYB24), MS06G12090.1 (MsMYB78), and MS06G12090.1 (MsMYB108) were clustered in group XXVI (Figure 2). A phylogenetic tree is generally built based on protein structures and conservation domains that are characteristic of members of the subfamily. Phylogenetic trees can be verified for their accuracy and authenticity based on these characteristics.
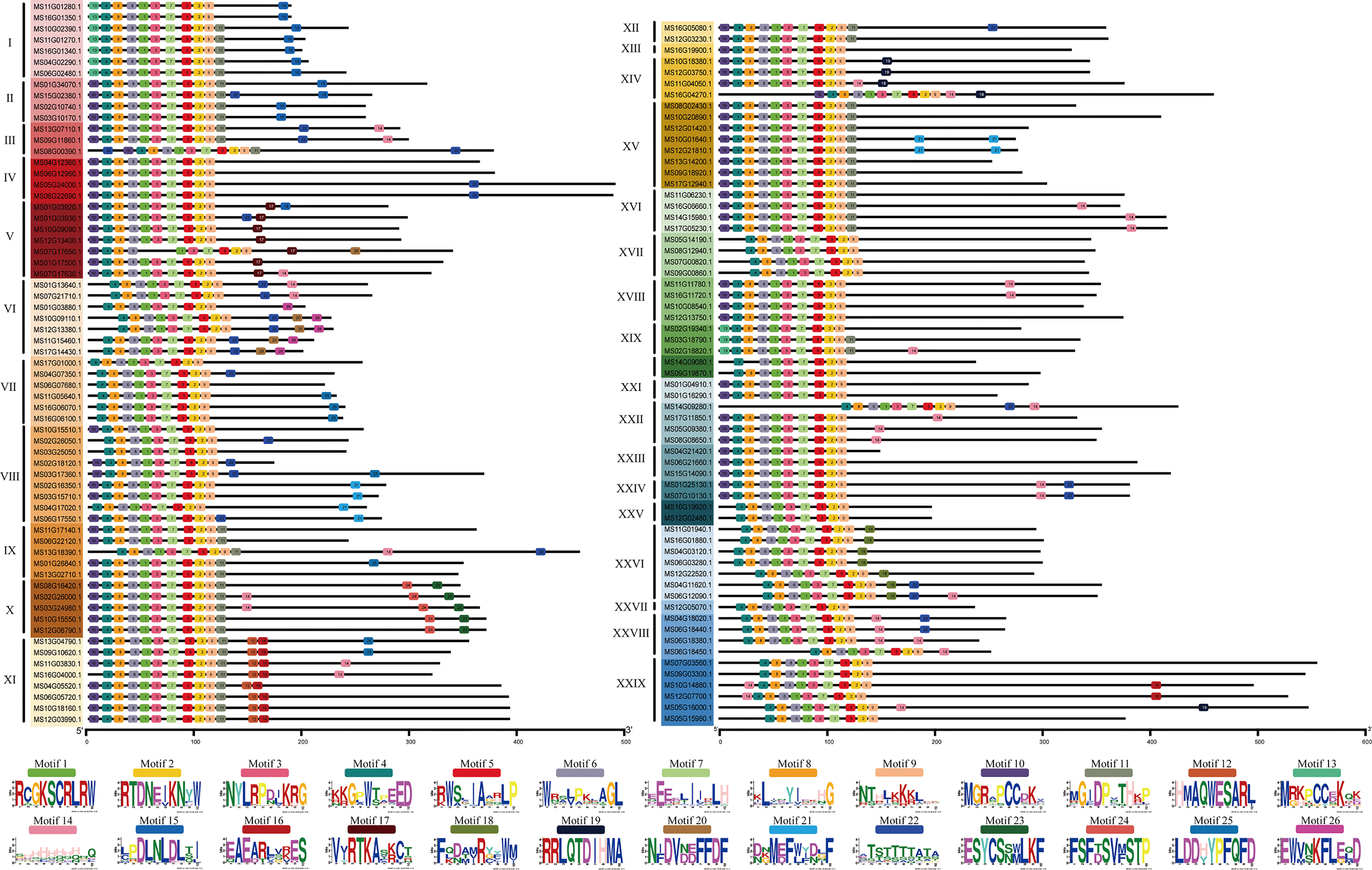
Figure 2 Motif composition of R2R3-MYBs in Malus sieversii. Roman numbers and different colors indicate different groups. Each solid line length indicate the different protein sequences length. The 9 Colored boxes indicate different motifs, which composes the MYB conserved domain region.
3.3 Chromosomal distribution and gene collinearity of MsMYB genes
To clearly show the distribution and better understand the origin of the MsMYB gene family, McScan was used to analyze collinearity relationships in self-MsMYB genes and between M. sieversii and M. domestica. The 457 MsMYB genes distributed across 17 chromosomes did not cluster as a subfamily. Based on the R2R3-MsMYB, a total of 13 MYB genes were found on chromosome 12, which contained the most genes, followed by 12 genes on chromosome 10 and 12 genes on chromosome 6. In contrast, only two genes were identified on chromosome 15: MS15G02380 and MS14G15980. The 1R-MsMYB subfamily with the largest genes and R2R3-MsMYB uneven are present on each chromosome. Five genes were identified in the 3R subfamily. In addition, two 3R-MsMYB genes were distributed on the same chromosome, whereas the other three were distributed on different chromosomes. Meanwhile, 452 link gene pairs were produced, including 323 1R-MYB genes, 128 R2R3-MYB genes, and one 3R-MYB gene. Among 1R-MYB genes, they mainly matched 1R-MYB (88.75%), whereas 11.25% 1R-MYBs were matched with the R2R3-MYB genes. In R2R3-MYB subfamily, there were 84.72% self-matches and 15.28% others-matches, including 13.89% 1R-MYB subfamily and 1.39% 3R-MYB subfamily (Figure 3). The collinearity result showed that for 457 MsMYB genes, 457 duplication events occurred, including 392 WGD or segmental, 45 dispersed, 10 tandem, seven proximal, and three singleton events. The results indicated that WGD or segmental was the main amplification driving force in MsMYB gene family formation process (Supplementary Table S2).
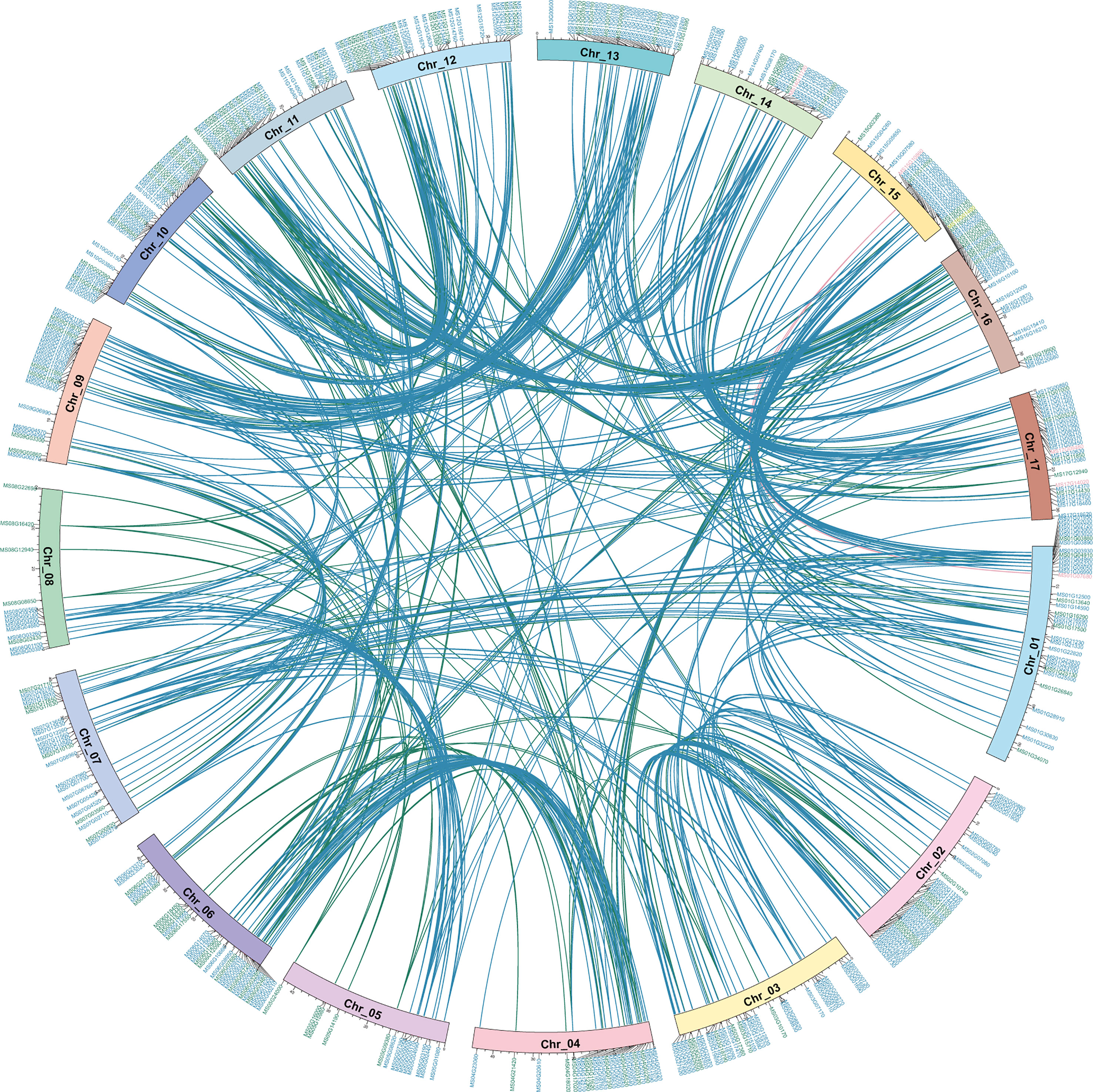
Figure 3 Self-Collinearity analysis and distribution of MsMYB genes in the Malus sieversii genome. Circle plot shows collinearity and distribution of the MYB gene family, and the colored lines highlightly link two syntenic MYB genes. Colored lines indicate collinearity relationship of 1R-MsMYB (blue), R2R3-MsMYB (green), and 3R-MsMYB (pink). Location of MYB genes on apple chromosomes (Chr_01~17). Colored IDs indicate 1R-MsMYB (blue), R2R3-MsMYB (green), 3R-MsMYB (pink), and 4R-MsMYB (yellow), respectively.
To better reveal the evolution and function of the MYB gene, we performed a collinearity analysis between M. sieversii and M. domestica. In total, 413 MYB collinearity gene pairs were obtained between the genomes of M. sieversii and M. domestica. The MsMYB gene family contained 286 1R-MYB, 122 R2R3-MYB, 4 3R-MYB, and 1 4R-MYB collinear gene pairs. In addition, 44 MsMYB genes did not correspond to M. domestica genes, including 37 1R-MsMYBs and seven R2R3-MsMYBs, which indicates that those MsMYB genes were lost during evolution (Figure 4).
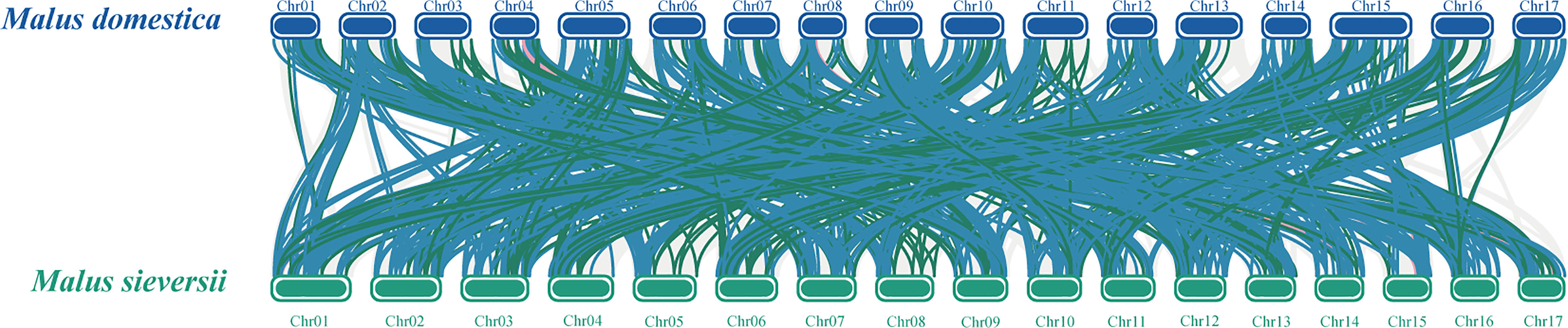
Figure 4 Collinearity analysis of MYB genes between Malus domestica and Malus sieversii. Syntenic regions between Malus domestica and Malus sieversii chromosomes are connected by different color lines. Gray lines and colored lines represent the collinear blocks and syntenic MYB gene pairs, respectively, including 1R-MYB (blue), R2R3-MYB (green), and 3R-MYB (pink).
3.4 Identifying cis-elements in R2R3-MsMYB promoters
The cis-elements in the promoter region of 128 R2R3-MsMYB genes were analyzed to better understand the functions of MsR2R3-MYBs in wild apple plants. A total of 59 cis-acting elements were identified, including 13 abiotic stress elements, five biotic stress elements, six growth development-related elements, 20 light-response elements, and 15 hormone response elements (Figure 5). Among them, the maximum number of light-response elements, especially Box 4 (114/128, 89%) and G-box elements (111/128, 86%), was followed by 87 GT1-motifs (68%), 85 TCT-motifs (66%), among others. Among the growth- and development-related elements, only one was greater than 50%, namely the O2-site (65/128, 50%), followed by 54 CAT-boxes (42%), 26 CCGTCC-boxes (20%), 25 GCN4_motif (19%), 24 RY-elements (18%), and 8 HD-Zip1 (6.2%). Among the hormone response elements, ABRE (113/128, 88%), GARE-motif (96/128, 75%), TGACG-motif (96/128, 75%), and MYB1 (81/128, 63%) were related to abscisic acid (ABA) and methylated jasmonic acid (MeJA), respectively. Among the biotic response elements, only one was greater than 80%, namely STRE (89/128, 83%), while the others were less than 50%. Among the abiotic stress elements, several higher numbers of cis-elements were found: ARE (110/128, 86%), AAGAA-motif (94/128, 73%), GT1-motif (87/128, 68%), and MYB (78/128, 61%). Promoter analysis showed that 55 of the 128 selected MsMYB promoters harbored the same number of CGTCA-motif (75%) and TGACG-motif (75%). It also showed that 55 of the 128 selected MsMYB genes harbored the same number of ABRE3a (43%) and ABRE4 (43%) cis-elements. Compared to MsMYB14 and MsMYB39, MsMYB24, MsMYB78, and MsMYB108 had the same number of cis-elements, ABRE3a and ABRE4, and they were in group XXVI. According to the distribution of biological stress response cis-elements, each gene had a common biotic stress response cis-elements STRE in group IV, and MS06G12960.1 had the highest number of STRE, which was seven. Ultimately, MsMYB24, MsMYB14, MsMYB39, MsMYB78, and MsMYB108 had one, seven, four, three and five biotic stress elements, respectively.
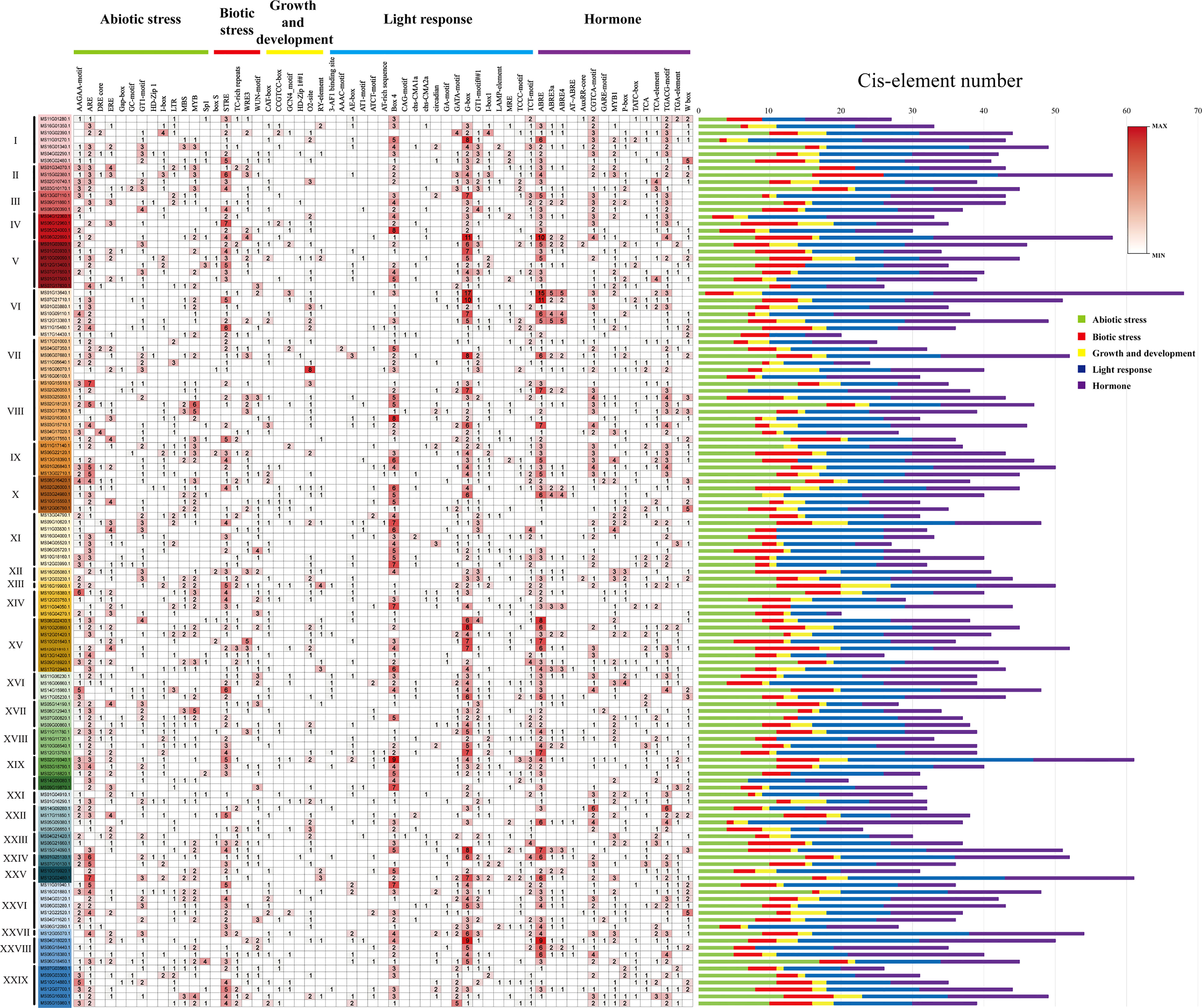
Figure 5 Analysis of the cis-acting elements in the promoter region of the R2R3-MsMYB genes. Top: different color bars indicate the classification of different cis-elements. I–XXIX show the subfamily of R2R3-MsMYB genes.
3.5 Expression patterns of R2R3-MsMYB genes upon the Valsa mali infection
We analyzed the transcript levels of MsR2R3-MYB genes in wild apple leaves at different time points (0, 1, 2, and 5 dpi). According to the expression pattern after infection with V. mali and GO annotation analysis, the expression pattern was divided into several modules (Figure 6A). Figure 6A (a) and Figure 6A (b) show high and low expression of these genes, respectively. A total of 27 MsMYB genes showed the highest changes in transcription levels, with significant differences. Compared with these MsMYB genes on day 0, five MsMYB genes showed up-regulated transcriptional levels and seven MsMYB genes showed down-regulated transcriptional levels on day 1. In addition, 11 MsMYB genes were down-regulated and five MsMYB genes were up-regulated on day 2. Furthermore, 11 MsMYB genes were down-regulated and 13 MsMYB genes were up-regulated on day 5. On days 1, 2, and 5, there were four sustained up-regulated genes, namely MS06G02480.1, MS01G34070.1, MS12G03750.1, and MS01G16290.1. At all time points, there were five sustained down-regulated genes: MS16G01340.1, MS03G10170.1, MS12G21810.1, MS01G03930.1, and MS01G13640.1. Of the remaining genes, MS04G11620.1 (MsMYB108) was upregulated on day 2. MS06G17550.1 (MsMYB14) and MS06G12090.1 (MsMYB78) only showed up-regulated expression on day 5. MS06G17550.1 (MsMYB14) only showed up-regulated expression on day 1.
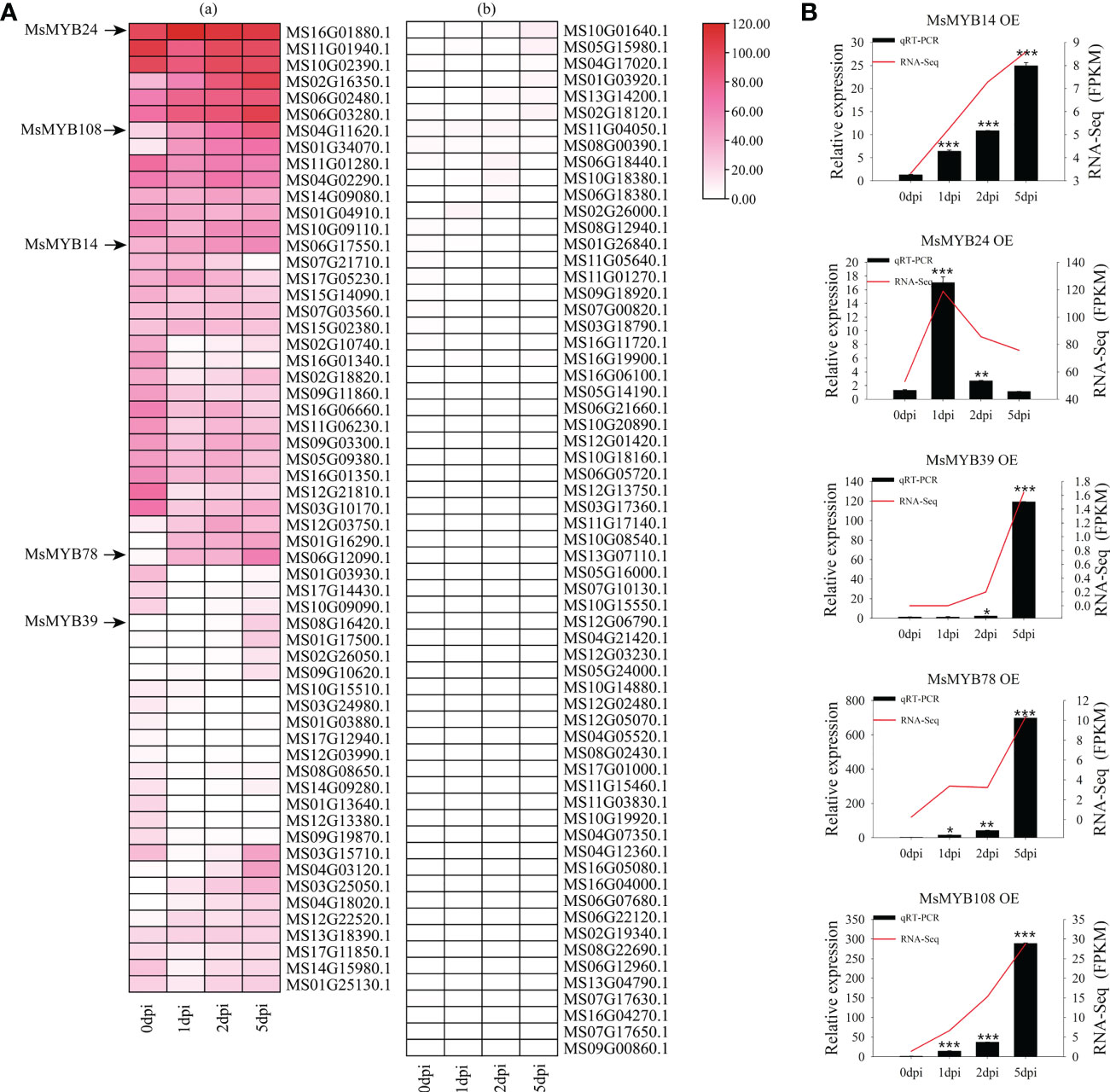
Figure 6 Expression profiles of the 128 R2R3-MsMYB genes during response to infection by Valsa mali. (A) Heatmaps with clusters represent the expression profiles of R2R3-MsMYB genes at 0, 1, 2, and 5 dpi. Color scale represents the normalized FPKM values. Red, pink, and white indicate high expression, low expression, and no expression, respectively. The cut-off of the differentially expressed R2R3-MsMYB transcripts islog2 (fold change)≥ 1 (Q-value < 0.05). (B) Comparison of RNA-seq data (red line) with qRT-PCR data (black column). The FPKM values from the RNA-seq are shown on the right y-axis. The relative expression levels are shown on the left y-axis. One asterisk indicates a significant difference between treatment and control plants (p ≤ 0.05). Two asterisks indicate an extremely significant difference between treatment and control plants (p ≤ 0.01). Three asterisks indicate an extremely significant difference between treatment and control plants (p ≤ 0.001).
Figure 6B shows that the expression profiles were accurate. The RNA-Seq (FPKM) results were consistent with the qRT-PCR results. According to the results of qRT-PCR, MsMYB14 continued to be highly expressed at 0, 1, 2, and 5 days. Compared with day 0, MsMYB14 expression levels were 3.9 times higher on day 1, 7.3 times higher on day 2, and 18.1 times higher on day 5. On day 1, MsMYB24 expression levels were 12-fold higher compared with day 0. As shown in Figure 6B, it was up-regulated and subsequently down-regulated. MsMYB39 was upregulated in the late stage. MsMYB78 gene expression was up-regulated 10-fold and 31-fold on days 1 and 2, respectively, compared to day 0, and was then up-regulated 536-fold. MsMYB108 expression levels were 10.1 times higher on day 1, 28.3 times higher on day 2, and 222.1 times higher on day 5.
3.6 Identification of R2R3-MsMYB members in Malus sieversii in response to Valsa mali infection
Five MsMYBs were cloned, and their expression levels in transiently transformed leaves were quantified by qRT-PCR. Figure 7A shows the lowest and highest relative expression level (7.45, MsMYB108; and 19.9, MsMYB78, respectively). This indicated that we successfully overexpressed MsMYB14, MsMYB24, MsMYB39, MsMYB78, and MsMYB108 genes in wild apple leaves. The incidence of leaves overexpressing MsMYB78 was 7.8% and 25.9% lower than that of the control at 48 h and 72 h, respectively. The leaves overexpressing MsMYB78 reduced leaf incidence by 18.1% at 72 h compared to 48 h. This indicated that MsMYB78 has a certain resistance phenotype. In contrast to the control plants, the remaining overexpressed leaves showed no significant difference in leaf incidence (Figure 7B). The leaves overexpressing the MsMYB14 gene decreased the area of lesion leaves by 29%, which was statistically significant after 48 h. Additionally, it was decreased by 19% at 72 h, which was extremely significant. The lesion area in the leaves overexpressing the MsMYB24 gene was increased by 26% and 13%, which were extremely significant and significant at 48 h and 72 h, respectively. When compared with the control plants, the lesion area of leaves overexpressing MsMYB108 had been reduced by approximately 17% at 48 h. This indicates that MsMYB14 was resistant, MsMYB24 was susceptible, and MsMYB108 had a slightly resistant phenotype (Figure 7C). Figure 7D shows a significant increase in fungal biomass in the MsMYB24 overexpressed leaves when compared to the control leaves by approximately 600%, which was statistically significant. Compared to the control, the fungal biomass of MsMYB14 and MsMYB78 overexpressed leaves was reduced by 46% and 24%, respectively. The relative fungal biomass of the control was similar to that of MsMYB39 and MsMYB108 overexpressed leaves. Figure 7D supports these results and shows that MsMYB78 has a disease resistant phenotype. Compared to the control, the leaves overexpressing MsMYB24 had a 2.2-fold increase in H2O2 content, and the H2O2 content of the leaves overexpressing MsMYB14 and MsMYB78 decreased by 21% and 25%, respectively (Figure 7E). Moreover, compared with the control, the MDA content of the leaves overexpressing MsMYB24 increased by 2.4 times and decreased by 27% and 39% in the leaves overexpressing MsMYB14 and MsMYB78, respectively (Figure 7F). As shown in Figures 7E and 7F, MsMYB14 and MsMYB78 were resistant and MsMYB24 was sensitive to V. mali. Figure 7G shows increased lesion area under V. mali infection at 48 h and 72 h on the leaves of wild apples. In summary, combined with the phenotypic traits shown in Figure 7G, we found that MsMYB24 was sensitive to V. mali, MsMYB14 and MsMYB78 were resistance genes, and MsMYB39 and MsMYB108 had no function in response to V. mali.
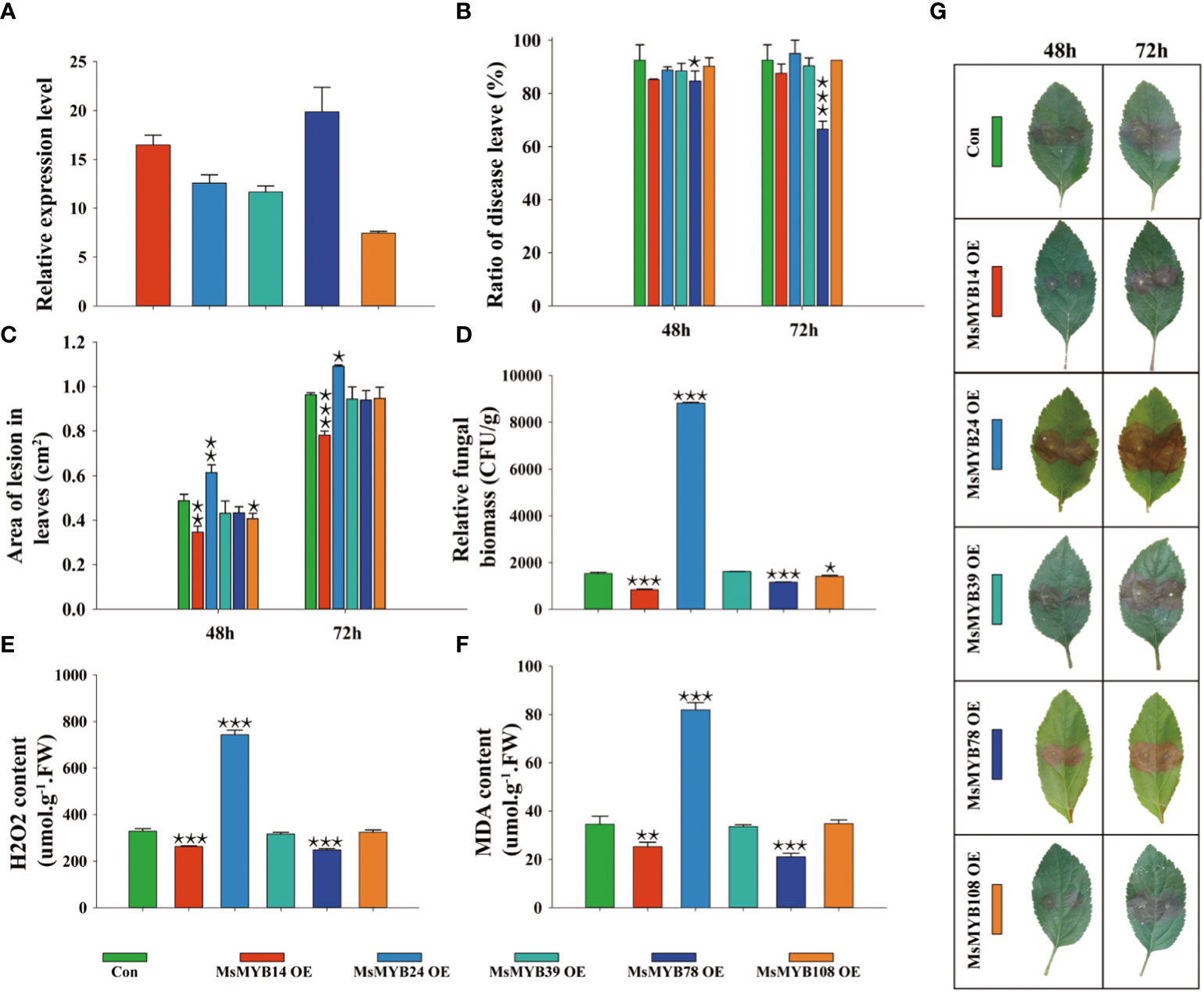
Figure 7 Functional identification of MsMYB family members in Malus sieversii in response to infection by Valsa mali. (A) Relative expression levels of MsMYBs in transiently transformed leaves determined by qRT-PCR at 72 h after infection by V. mali. The Valsa canker disease resistance of five MsMYBs is compared to that of the control. The lesion ratio (B) and area (C) were measured from detached leaves at 48 h and 72 h Each sample contained three or more independent biological replicates. One-way ANOVA was used for significances comparison with the control leaves (Con). (D) Relative fungal biomass of transiently transformed leaves of wild apple after 72 h of infection by V. mali. (E) H2O2 content in transiently transformed leaves of wild apple after 72 h of infection by V. mali. (F) MDA content in transiently transformed leaves of wild apple after 72 h of infection by V. mali. (G) The phenotype of transiently transformed leaves in wild apple changes following V. mali infection. One asterisk indicates a significant difference between treatment and control plants (p ≤ 0.05). Two asterisks indicate an extremely significant difference between treatment and control plants (p ≤ 0.01). Three asterisks indicate an extremely significant difference between treatment and control plants (p ≤ 0.001).
4 Discussion
Recent research has revealed that wild apples (Malus sieversii) are the most primitive and chief ancestors of domesticated apples in the Tianshan Mountains of Central Asia and Western Europe (Cornille et al., 2014). The abundance of germplasm and genetic resources is conducive to molecular breeding of domesticated apples (Duan et al., 2016). There has been a drastic decrease in M. sieversii populations in Xinjiang as a result of apple canker disease caused by V. mali, a necrotrophic pathogen that has become widespread across Asia (Li X. et al., 2019). Therefore, it is imperative to explore the resistance gene resources in M. sieversii upon infection with V. mali to provide a basis for molecular breeding of apple trees and develop more effective strategies for preventing disease infection.
Our study focused on the MYB transcription factor family by conducting a genome-wide screening of the M. sieversii genome. Initially, 457 MsMYB transcription factor genes were identified in the genome of M. sieversii, including 128 genes of the R2R3 type. Following V. mali infection, 27 R2R3-type MsMYB genes were found to be associated with the response to V. mali infection based on M. sieversii transcriptome data. To identify the genes involved in the response to infection with V. mali, we combined the transcriptome with the transient transformation method. In the present study, we found that wild apples can resist V. mali infection with MsMYB14 and MsMYB78. In contrast, MsMYB24 may play a negative role in the immunoregulation of wild apples.
According to the results of the present study, infection with V. mali significantly induced MsMYB14 expression (Figures 6A, B). Compared with the control plants, leaves transiently overexpressing MsMYB14 showed reduced lesion areas (Figure 7C), whereas the ratio of diseased leaves did not seem to differ. This suggests that MsMYB14 enhances wild apple resistance by reducing V. mali propagation in leaves instead of invading the plant. According to Hurtado-Gaitán et al. (2021), MYB14 enhanced the synthesis of resveratrol by down-regulating phosphoenolpyruvate carboxylase kinase in grapevines. Additionally, it may be directly associated with stilbene synthases (Holl et al., 2013; Fang et al., 2014; Wang et al., 2015) and stilbenoid pathways (Orduna et al., 2022), which are responsible for the production of resveratrol in grapevines. Plants produce stilbenes and resveratrol under biotic and abiotic stress as a defense mechanism. Duan et al. (2016) found that after a series of upstream signals (e.g., RboH-dependent oxidative burst, calcium influx, MAPK cascade, and jasmonate), FLG22 induces the MYB14 promoter, thus influencing resveratrol accumulation and resistance to pathogenic bacteria in plants. A recent study found that VqMAPKKK38, a member of the MAPK cascade, activates transcription factor MYB14 to positively regulate stilbene synthase transcription. It has been shown that the promoter of VqMYB14 (pVqMYB14) is triggered by the elicitors flg22 and harpin, respectively, and participates in both PAMP-triggered immunity and effector-triggered immunity (Luo et al., 2020). Using structure-activity relationships, Wang et al. (2020) showed that VqWRKY53 directly interacts with VqMYB14 and VqMYB15, enhancing stilbene synthesis. In addition, MYB14 plays a role in the accumulation of flavonoids in Marchantia polymorpha (Hamashima et al., 2019) and conifers (Bedon et al., 2010), which were found to be defense phytoalexins in response to stress and infection. In contrast, no studies have been conducted to determine whether MYB14 is involved in the response to pathogenic fungi, specifically V. mali. Based on these findings, it is likely that V. mali infection induces MsMYB14 expression, which in turn may result in increased biotic stress resistance through stilbenoid, resveratrol, or flavonoid accumulation.
However, leaves transiently overexpressing MsMYB78 showed a reduced ratio of diseased leaves (Figure 7B), whereas no obvious differences were observed in the lesion area compared to the control plants (Figure 7C). This suggests that MsMYB78 may improve wild apple resistance by reducing the incidence of V. mali invasion rather than by slowing the propagation of pathogens in leaves. CaMYB78 has been demonstrated to negatively regulate the anthocyanin biosynthetic pathway in chickpeas, culminating in increased resistance to Fusarium oxysporum (Shriti et al., 2022). It is likely that MYB78 is a broad-spectrum resistance gene that is responsive to pathogenic fungi such as V. mali and F. oxysporum.
Interestingly, MsMYB24 played a negative role in the response to V. mali infection. The leaf lesion area (Figure 7C) and fungal biomass (Figure 7D) were significantly increased. Interaction between MYB24 and DELLA regulates filament elongation. Activation of MYB24 encodes flavonol biosynthesis during Pollen Coat Patterning by regulating FLS1 gene expression (Zhang X. et al., 2021) and phenylpropanoid biosynthesis during anther/pollen development by interacting with JAZ1/2 (Li et al., 2013). MYB24 is an influential regulator of jasmonate-mediated stamen development by interacting with bHLH TF (Song et al., 2011; Qi et al., 2015; Chen et al., 2016; Huang et al., 2017). Moreover, MYB24 plays a role in the growth of gynoecium, development of nectaries, and production of volatile sesquiterpenes, which may attract insects and/or repel pathogens (Reeves et al., 2012). As a result, MYB24 promotes plant reproduction by disrupting the balance between development and the stress response.
A synergistic effect of MYB24 and MYB108 on jasmonate-mediated stamen maturation has been reported in Arabidopsis (Mandaokar and Browse, 2009). The MYB108 gene has been implicated in defense against Verticillium dahliae infection (Cheng et al., 2016) and has been shown to significantly increase anthocyanin biosynthesis (Khan et al., 2022) and regulate ABA-dependent wound-induced spreading cell death (Cui et al., 2013). Using MsMYB108 overexpressing leaves, we found significant reductions in lesion areas after 48 h, but no difference in fungal biomass was observed. MYB108 appears to participate in the disease response only at an early stage.
We identified 128 MsMYB genes of the R2R3 type within the genome of M. sieversii. Based on the RNA-Seq results, we selected five TFs that may play an important role in the response to V. mali. In response to infection with V. mali, we characterized the functions of these five TFs in response to V. mali infection. We successfully identified two resistance genes and one sensitivity gene using RNA-Seq coupled with a molecular and physiological assay based on a transient genetic transformation platform. The methods used to identify resistance genes in response to biotic stress in the present study can be applied to a wide range of situations.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
XW and DZ designed the research. YD, QY, and XW performed the research. QY, MZ, and XW analyzed the data and discussed the results. and QY, XW, and AW wrote the paper. All authors contributed to the article and approved the submitted version. The first two authors contributed equally.
Funding
This work was supported by the NSFC-XJ key project (No. U1903206), the National Natural Science Foundation of China (No. 32101539) and China Postdoctoral Science Foundation (No. 2021M693379).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1112681/full#supplementary-material
References
An, J. P., Li, R., Qu, F. J., You, C. X., Wang, X. F., Hao, Y. J. (2018). R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 96 (3), 562–577. doi: 10.1111/tpj.14050
Arce-Rodriguez, M. L., Martinez, O., Ochoa-Alejo, N. (2021). Genome-wide identification and analysis of the MYB transcription factor gene family in chili pepper (Capsicum spp.). Int. J. Mol. Sci. 22 (5), 2229. doi: 10.3390/ijms22052229
Bedon, F., Bomal, C., Caron, S., Levasseur, C., Boyle, B., Mansfield, S. D., et al. (2010). Subgroup 4 R2R3-MYBs in conifer trees: gene family expansion and contribution to the isoprenoid- and flavonoid-oriented responses. J. Exp. Bot. 61 (14), 3847–3864. doi: 10.1093/jxb/erq196
Blanco, E., Sabetta, W., Danzi, D., Negro, D., Passeri, V., Lisi, A., et al. (2018). Isolation and characterization of the flavonol regulator CcMYB12 from the globe artichoke [Cynara cardunculus var. scolymus (L.) fiori]. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00941
Cao, Y., Li, K., Li, Y., Zhao, X., Wang, L. (2020). MYB transcription factors as regulators of secondary metabolism in plants. Biol. (Basel) 9 (3), 61. doi: 10.3390/biology9030061
Cardenas-Hernandez, H., Titaux-Delgado, G. A., Castaneda-Ortiz, E. J., Torres-Larios, A., Brieba, L. G., Del Rio-Portilla, F., et al. (2021). Genome-wide and structural analysis of the Myb-SHAQKYF family in entamoeba histolytica. Biochim. Biophys. Acta Proteins Proteomics 1869 (4), 140601. doi: 10.1016/j.bbapap.2021.140601
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, G., He, W., Guo, X., Pan, J. (2021). Genome-wide identification, classification and expression analysis of the MYB transcription factor family in Petunia. Int. J. Mol. Sci. 22 (9), 4838. doi: 10.3390/ijms22094838
Chen, L., Hu, B., Qin, Y., Hu, G., Zhao, J. (2019). Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Biochem. 136, 178–187. doi: 10.1016/j.plaphy.2019.01.024
Chen, X., Huang, H., Qi, T., Liu, B., Song, S. (2016). New perspective of the bHLH-MYB complex in jasmonate-regulated plant fertility in Arabidopsis. Plant Signal. Behav. 11 (2), e1135280. doi: 10.1080/15592324.2015.1135280
Chen, X., Wang, P., Gu, M., Lin, X., Hou, B., Zheng, Y., et al. (2021). R2R3-MYB transcription factor family in tea plant (Camellia sinensis): Genome-wide characterization, phylogeny, chromosome location, structure and expression patterns. Genomics 113 (3), 1565–1578. doi: 10.1016/j.ygeno.2021.03.033
Chen, L., Yang, H., Fang, Y., Guo, W., Chen, H., Zhang, X., et al. (2021). Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol. J. 19 (4), 702–716. doi: 10.1111/pbi.1349615
Cheng, H. Q., Han, L. B., Yang, C. L., Wu, X. M., Zhong, N. Q., Wu, J. H., et al. (2016). The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection. J. Exp. Bot. 67 (6), 1935–1950. doi: 10.1093/jxb/erw016
Cornille, A., Giraud, T., Smulders, M. J., Roldan-Ruiz, I., Gladieux, P. (2014). The domestication and evolutionary ecology of apples. Trends Genet. 30 (2), 57–65. doi: 10.1016/j.tig.2013.10.002
Cui, F., Brosche, M., Sipari, N., Tang, S., Overmyer, K. (2013). Regulation of ABA dependent wound induced spreading cell death by MYB108. New Phytol. 200 (3), 634–640. doi: 10.1111/nph.12456
Daccord, N., Celton, J. M., Linsmith, G., Becker, C., Choisne, N., Schijlen, E., et al. (2017). High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 49 (7), 1099–1106. doi: 10.1038/ng.3886
Duan, D., Fischer, S., Merz, P., Bogs, J., Riemann, M., Nick, P. (2016). An ancestral allele of grapevine transcription factor MYB14 promotes plant defence. J. Exp. Bot. 67 (6), 1795–1804. doi: 10.1093/jxb/erv569
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15 (10), 573–581. doi: 10.1016/j.tplants.2010.06.005
Faize, M., Faize, L., Alburquerque, N., Venisse, J. S., Burgos, L. (2020). Hydrogen peroxide generated by over-expression of cytosolic superoxide dismutase in transgenic plums enhances bacterial canker resistance and modulates plant defence responses. Mol. Biol. Rep. 47 (8), 5889–5901. doi: 10.1007/s11033-020-05660-8
Fang, L., Hou, Y., Wang, L., Xin, H., Wang, N., Li, S. (2014). Myb14, a direct activator of STS, is associated with resveratrol content variation in berry skin in two grape cultivars. Plant Cell Rep. 33 (10), 1629–1640. doi: 10.1007/s00299-014-1642-3
Hamashima, N., Xie, X., Hikawa, M., Suzuki, T., Kodama, Y. (2019). A gain-of-function T-DNA insertion mutant of marchantia polymorpha hyper-accumulates flavonoid riccionidin a. Plant Biotechnol. (Tokyo) 36 (3), 201–204. doi: 10.5511/plantbiotechnology.19.0722a
Holl, J., Vannozzi, A., Czemmel, S., D’Onofrio, C., Walker, A. R., Rausch, T., et al. (2013). The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 25 (10), 4135–4149. doi: 10.1105/tpc.113.117127
Huang, H., Gao, H., Liu, B., Qi, T., Tong, J., Xiao, L., et al. (2017). Arabidopsis MYB24 regulates jasmonate-mediated stamen development. Front. Plant Sci. 8, 1525. doi: 10.3389/fpls.2017.01525
Hurtado-Gaitán, E., Sellés-Marchart, S., Hartwell, J., Martínez-Esteso, M. J., Bru-Martínez, R. (2021). Down-regulation of phosphoenolpyruvate carboxylase kinase in grapevine cell cultures and leaves is linked to enhanced resveratrol biosynthesis. Biomolecules 11 (11), 1641. doi: 10.3390/biom11111641
IH, O., EP, R. (1999). The myb gene family in cell growth, differentiation and apoptosis. Oncogene 118 (19), 3017–3033. doi: 10.1038/sj.onc.1202839
Jin, H., Martin, C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41 (5), 577–585. doi: 10.1023/a:1006319732410
Kanei-Ishii, C., Sarai, A., Sawazaki, T., Nakagoshi, H., He, D. N., Ogata, K., et al. (1990). The tryptophan cluster: a hypothetical structure of the DNA-binding domain of the myb protooncogene product. J. Biol. Chem. 265 (32), 19990–19995. doi: 10.1016/s0021-9258(17)45472-x
Khan, I. A., Cao, K., Guo, J., Li, Y., Wang, Q., Yang, X., et al. (2022). Identification of key gene networks controlling anthocyanin biosynthesis in peach flower. Plant Sci. 316, 111151. doi: 10.1016/j.plantsci.2021.111151
Kranz, H., Scholz, K., Weisshaar, B. (2000). C-MYB oncogene-like genes encoding three MYB repeats occur in all major plant lineages. Plant J. 21 (2), 231–235. doi: 10.1046/j.1365-313x.2000.00666.x
Lee, D. H., Lee, S. W., Choi, K. H., Kim, D. A., Uhm, J. Y. (2006). Survey on the occurrence of apple diseases in Korea from 1992 to 2000. Plant Pathol. J. 22 (4), 375–380. doi: 10.5423/ppj.2006.22.4.375
Li, X., Guo, C., Ahmad, S., Wang, Q., Yu, J., Liu, C., et al. (2019). Systematic analysis of MYB family genes in potato and their multiple roles in development and stress responses. Biomolecules 9 (8), 317. doi: 10.3390/biom9080317
Li, J., Han, G., Sun, C., Sui, N. (2019). Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 14 (8), 1613131. doi: 10.1080/15592324.2019.1613131
Li, Y., Jiang, J., Du, M. L., Li, L., Wang, X. L., Li, X. B. (2013). A cotton gene encoding MYB-like transcription factor is specifically expressed in pollen and is involved in regulation of late anther/pollen development. Plant Cell Physiol. 54 (6), 893–906. doi: 10.1093/pcp/pct038
Li, L., Zhang, S., Wang, B. (2021). Apple leaf disease identification with a small and imbalanced dataset based on lightweight convolutional networks. Sens. (Basel) 22 (1), 173. doi: 10.3390/s22010173
Liang, X., Zhang, R., Gleason, M. L., Sun, G. (2022). Sustainable apple disease management in China: Challenges and future directions for a transforming industry. Plant Dis. 106 (3), 786–799. doi: 10.1094/PDIS-06-21-1190-FE
Liu, X., Li, X., Wen, X., Zhang, Y., Ding, Y., Zhang, Y., et al. (2021). PacBio full-length transcriptome of wild apple (Malus sieversii) provides insights into canker disease dynamic response. BMC Genom. 22 (1), 52. doi: 10.1186/s12864-021-07366-y
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Lu, S. X., Knowles, S. M., Andronis, C., Ong, M. S., Tobin, E. M. (2009). CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 150 (2), 834–843. doi: 10.1104/pp.108.133272
Luo, Y., Wang, Q., Bai, R., Li, R., Chen, L., Xu, Y., et al. (2020). The effect of transcription factor MYB14 on defense mechanisms in Vitis quinquangularis-pingyi. Int. J. Mol. Sci. 21 (3), 706. doi: 10.3390/ijms21030706
Mandaokar, A., Browse, J. (2009). MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in arabidopsis. Plant Physiol. 149 (2), 851–862. doi: 10.1104/pp.108.132597
Mmadi, M. A., Dossa, K., Wang, L., Zhou, R., Wang, Y., Cisse, N., et al. (2017). Functional characterization of the versatile MYB gene family uncovered their important roles in plant development and responses to drought and waterlogging in Sesame. Genes (Basel) 8 (12), 362. doi: 10.3390/genes8120362
Ogata, K., Hojo, H., Aimoto, S., Nakai, T., Nakamura, H., Sarai, A., et al. (1992). Solution structure of a DNA-binding unit of myb: A helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc. Natl. Acad. Sci. 89 (14), 6428–6432. doi: 10.1073/pnas.89.14.6428
Ogata, K., Kanei-Ishii, C., Sasaki, M., Hatanaka, H., Nagadoi, A., Enari, M., et al. (1996). The cavity in the hydrophobic core of myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Biol. 3 (2), 178–187. doi: 10.1038/nsb0296-178
Orduna, L., Li, M., Navarro-Paya, D., Zhang, C., Santiago, A., Romero, P., et al. (2022). Direct regulation of shikimate, early phenylpropanoid, and stilbenoid pathways by subgroup 2 R2R3-MYBs in grapevine. Plant J. 110 (2), 529–547. doi: 10.1111/tpj.15686
Paz-Ares, J., Ghosal, D., Wienand, U., A.Petersont, P., Saedler, H. (1987). The regulatory cl locus of zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6 (12), 3553–3558. doi: 10.1002/j.1460-2075
Pucker, B., Pandey, A., Weisshaar, B., Stracke, R. (2020). The R2R3-MYB gene family in banana (Musa acuminata): Genome-wide identification, classification and expression patterns. PloS One 15 (10), e0239275. doi: 10.1371/journal.pone.0239275
Qi, T., Huang, H., Song, S., Xie, D. (2015). Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 27 (6), 1620–1633. doi: 10.1105/tpc.15.00116
Reeves, P. H., Ellis, C. M., Ploense, S. E., Wu, M. F., Yadav, V., Tholl, D., et al. (2012). A regulatory network for coordinated flower maturation. PLoS Genet. 8 (2), e1002506. doi: 10.1371/journal.pgen.1002506
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290 (5499), 2105–2110. doi: 10.1126/science.290.5499.2105
Romero, I., Fuertes, A., Benito, M. J., Malpica, J. M., Leyva, A., Paz-Ares, J. (1998). More than 80R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 14 (3), 273–283. doi: 10.1046/j.1365-313x.1998.00113.x
Rosinski, J. A., Atchley, W. R. (1998). Molecular evolution of the myb family of transcription factors: Evidence for polyphyletic origin. J. Mol. Evol. 46 (1), 74–83. doi: 10.1007/pl00006285
Shriti, S., Paul, S., Das, S. (2022). Overexpression of CaMYB78 transcription factor enhances resistance response in chickpea against fusarium oxysporum and negatively regulates anthocyanin biosynthetic pathway. Protoplasma 260 (2), 589–605. doi: 10.1007/s00709-022-01797-4
Song, S., Qi, T., Huang, H., Ren, Q., Wu, D., Chang, C., et al. (2011). The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23 (3), 1000–1013. doi: 10.1105/tpc.111.083089
Song, X., Yang, Q., Liu, Y., Li, J., Chang, X., Xian, L., et al. (2021). Genome-wide identification of pistacia R2R3-MYB gene family and function characterization of PcMYB113 during autumn leaf coloration in Pistacia chinensis. Int. J. Biol. Macromol. 192, 16–27. doi: 10.1016/j.ijbiomac.2021.09.092
Stracke, R., Werber, M., Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 (5), 447–456. doi: 10.1016/s1369-5266(00)00199-0
Sun, W., Ma, Z., Chen, H., Liu, M. (2019). MYB gene family in potato (Solanum tuberosum l.): Genome-wide identification of hormone-responsive reveals their potential functions in growth and development. Int. J. Mol. Sci. 20 (19), 4847. doi: 10.3390/ijms20194847
Tuan, P. A., Bai, S., Yaegaki, H., Tamura, T., Hihara, S., Moriguchi, T., et al. (2015). The crucial role of PpMYB10.1 in anthocyanin accumulation in peach and relationships between its allelic type and skin color phenotype. BMC Plant Biol. 15, 280. doi: 10.1186/s12870-015-0664-5
Wang, D., Jiang, C., Liu, W., Wang, Y. (2020). The WRKY53 transcription factor enhances stilbene synthesis and disease resistance by interacting with MYB14 and MYB15 in Chinese wild grape. J. Exp. Bot. 71 (10), 3211–3226. doi: 10.1093/jxb/eraa097
Wang, J. F., Ma, L., Xi, H. F., Wang, L. J., Li, S. H. (2015). Resveratrol synthesis under natural conditions and after UV-c irradiation in berry skin is associated with berry development stages in ‘Beihong’ (V. vinifera×V. amurensis). Food Chem. 168, 430–438. doi: 10.1016/j.foodchem.2014.07.025
Wang, X., Zang, R., Yin, Z., Kang, Z., Huang, L. (2014). Delimiting cryptic pathogen species causing apple valsa canker with multilocus data. Ecol. Evol. 4 (8), 1369–1380. doi: 10.1002/ece3.1030
Wen, X., Ding, Y., Tan, Z., Wang, J., Zhang, D., Wang, Y. (2020). Identification and characterization of cadmium stress-related LncRNAs from Betula platyphylla. Plant Sci. 299, 110601. doi: 10.1016/j.plantsci.2020.110601
Xie, Y., Chen, P., Yan, Y., Bao, C., Li, X., Wang, L., et al. (2018). An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol. 218 (1), 201–218. doi: 10.1111/nph.14952
Xing, G., Li, J., Li, W., Lam, S. M., Yuan, H., Shui, G., et al. (2021). AP2/ERF and R2R3-MYB family transcription factors: potential associations between temperature stress and lipid metabolism in Auxenochlorella protothecoides. Biotechnol. Biofuels 14 (1), 22. doi: 10.1186/s13068-021-01881-6
Yanhui, C., Xiaoyuan, Y., Kun, H., Meihua, L., Jigang, L., Zhaofeng, G., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60 (1), 107–124. doi: 10.1007/s11103-005-2910-y
Yuan, Y., Yang, X., Feng, M., Ding, H., Khan, M. T., Zhang, J., et al. (2021). Genome-wide analysis of R2R3-MYB transcription factors family in the autopolyploid saccharum spontaneum: an exploration of dominance expression and stress response. BMC Genom. 22 (1), 622. doi: 10.1186/s12864-021-07689-w
Zhang, X., He, Y., Li, L., Liu, H., Hong, G. (2021). Involvement of the R2R3-MYB transcription factor MYB21 and its homologs in regulating flavonol accumulation in Arabidopsis stamen. J. Exp. Bot. 72 (12), 4319–4332. doi: 10.1093/jxb/erab156
Zhang, Q., Wang, L., Wang, Z., Zhang, R., Liu, P., Liu, M., et al. (2021). The regulation of cell wall lignification and lignin biosynthesis during pigmentation of winter jujube. Hortic. Res. 8 (1), 238. doi: 10.1038/s41438-021-00670-4
Zhang, L., Zhao, G., Jia, J., Liu, X., Kong, X. (2012). Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J. Exp. Bot. 63 (1), 203–214. doi: 10.1093/jxb/err264
Keywords: MsMYB transcription factor, Malus sieversii, Valsa canker, fungal resistance, function identification
Citation: Ding Y, Yang Q, Waheed A, Zhao M, Liu X, Kahar G, Haxim Y, Wen X and Zhang D (2023) Genome-wide characterization and functional identification of MYB genes in Malus sieversii infected by Valsa mali. Front. Plant Sci. 14:1112681. doi: 10.3389/fpls.2023.1112681
Received: 30 November 2022; Accepted: 22 March 2023;
Published: 05 April 2023.
Edited by:
Ting Wu, China Agricultural University, ChinaReviewed by:
Jianwei Gao, Shandong Academy of Agricultural Sciences, ChinaKourosh Vahdati, University of Tehran, Iran
Copyright © 2023 Ding, Yang, Waheed, Zhao, Liu, Kahar, Haxim, Wen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daoyuan Zhang, zhangdy@ms.xjb.ac.cn; Xuejing Wen, wenxj@ms.xjb.ac.cn
†These authors have contributed equally to this work
 Yu Ding1,2†
Yu Ding1,2†