
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 March 2023
Sec. Crop and Product Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1110366
This article is part of the Research Topic Woody Oil Crops: Key Trait Formation and Regulation View all 14 articles
Camellia oleifera Abel is a highly valued woody edible oil tree, which is endemic to China. It has great economic value because C. oleifera seed oil contains a high proportion of polyunsaturated fatty acids. C. oleifera anthracnose caused by Colletotrichum fructicola, poses a serious threat to C. oleifera growth and yield and causes the benefit of the C. oleifera industry to suffer directly. The WRKY transcription factor family members have been widely characterized as vital regulators in plant response to pathogen infection. Until now, the number, type and biological function of C. oleifera WRKY genes are remains unknown. Here, we identified 90 C. oleifera WRKY members, which were distributed across 15 chromosomes. C. oleifera WRKY gene expansion was mainly attributed to segmental duplication. We performed transcriptomic analyses to verify the expression patterns of CoWRKYs between anthracnose-resistant and -susceptible cultivars of C. oleifera. These results demonstrated that multiple candidate CoWRKYs can be induced by anthracnose and provide useful clues for their functional studies. CoWRKY78, an anthracnose-induced WRKY gene, was isolated from C. oleifera. It was significantly down-regulated in anthracnose-resistant cultivars. Overexpression of CoWRKY78 in tobacco markedly reduced resistance to anthracnose than WT plants, as evidenced by more cell death, higher malonaldehyde content and reactive oxygen species (ROS), but lower activities of superoxide dismutase (SOD), peroxidase (POD), as well as phenylalanine ammonia-lyase (PAL). Furthermore, the expression of multiple stress-related genes, which are associated with ROS-homeostasis (NtSOD and NtPOD), pathogen challenge (NtPAL), and pathogen defense (NtPR1, NtNPR1, and NtPDF1.2) were altered in the CoWRKY78-overexpressing plants. These findings increase our understanding of the CoWRKY genes and lay the foundation for the exploration of anthracnose resistance mechanisms and expedite the breeding of anthracnose-resistant C. oleifera cultivars.
Camellia oleifera Abel is an evergreen small tree or shrub in the family Theaceae. Together with coconut (Cocos nucifera), olive (Canarium album), and palm (Trachycarpus fortunei), they are called the world’s four major woody oil tree species (Zhang et al., 2021). As one kind of oil-rich seed tree, C. oleifera has great economic value, nutritional and medicinal value because its seed oil is rich in unsaturated fatty acids and natural bioactive ingredients (Ye et al., 2021). In the past several years, most C. oleifera studies have focused mainly on oil extraction technology (Zhang et al., 2019a), self-incompatibility (Zhou et al., 2020), seed oil biosynthesis (Zhang et al., 2021), seed development (Wu et al., 2022), and fruit development (He et al., 2022a). Specifically, with ecological value, C. oleifera can not only live in the cold climate (Wu et al., 2020), but also grow well in drought and barren soil (He et al., 2022b). However, C. oleifera is vulnerable to a number of fungal and bacterial infections, which seriously threaten the healthy and sustainable development of C. oleifera industry. C. oleifera anthracnose, which is caused by Colletotrichum fructicola, is the primary disease of C. oleifera, and seriously affects yield and tea-oil quality (Zhang et al., 2019b). Chemical pesticides can prevent C. oleifera anthracnose, but this may induce problems such as chemical residues on the tree, fungicide resistance, and environmental pollution. Therefore, selecting resistance genes to develop resistant cultivars would be an effective method to manage diseases. Nevertheless, limited knowledge exists regarding the molecular mechanisms underlying anthracnose resistance.
Plants have evolved sophisticated defense mechanism to defend themselves from various pathogenic diseases (Kang et al., 2018). This process requires different types of transcription factors (TFs), which play essential roles in transcriptional regulation (Amorim et al., 2017; Yan et al., 2022). WRKY proteins constitute one of the largest TF families in land plants, and each member has one or two conserved WRKY domains at the N-terminus region, followed closely by a Cys2HisCys-type or a Cys2His2-type zinc-finger domain at the C-terminus region (Lim et al., 2022). WRKY TFs can specifically recognize the W-box, with sequence TTGAC/T within the target genes’ promoter regions (Ciolkowski et al., 2008). WRKY TF families can comprise 3 groups (Du et al., 2022). The members in the group I have two WRKY structural domains with Cys2His2-type motifs. Furthermore, members in group II or group III that have one WRKY structural domain with Cys2His2-type or Cys2HisCys-type motif. Lots of evidence showing that WRKY TFs plays a central role involved in disease resistance in the plant through a variety of pathways. In Arabidopsis thaliana, overexpression of AtWRKY75 enhanced plant resistance to Sclerotinia sclerotiorum and increased expression of PDF1.2 (Chen et al., 2013). Meanwhile, transgenic A. thaliana overexpressing AtWRKY70 showed resistance against Pseudomonas syringae and increased the PR gene expression level (Li et al., 2004). In Oryza sativa, overexpression of OsWRKY30 (Peng et al., 2012) or OsWRKY45 (Huangfu et al., 2016) led to increased resistance against Magnaporthe grisea. Moreover, ShWRKY41 (Lian et al., 2022), CmWRKY15-1 (Bi et al., 2021), PlWRKY65 (Wang et al., 2020), and FaWRKY25 (Jia et al., 2020) have been shown to function as negative or positive regulators involved in the plant defense response to various pathogen infection. In woody plants, overexpression of WRKY60 in Populus tomentosa led to enhanced resistance to Dothiorella gregaria and increased PR gene expression (Ye et al., 2014). As well, transgenic poplars overexpressing PtrWRKY18 or PtrWRKY35 showed enhanced resistance against Melampsora (Jiang et al., 2017). In addition, RcWRKY41, was inferred as a candidate regulator in response to Botrytis cinerea infection in roses (Liu et al., 2019). So far, 72 and 109 WRKY TFs have already been identified in Arabidopsis or rice (Eulgem and Somssich, 2007; Ross et al., 2007). Furthermore, 104 and 80 WRKY members were identified in the poplar and grape genome (He et al., 2012; Zhang and Feng, 2014). However, systematic information on WRKYs in C. oleifera was unclear.
WRKY TFs are of great importance in plant-pathogen interactions (Li et al., 2015). However, there have not been any reports investigating the expression pattern and function of WRKY genes directly involved in C. oleifera-C. fructicola interaction. The genome sequence of C. oleifera has recently been completed (Lin et al., 2022). Key metabolites involved in C. oleifera against anthracnose have been investigated through integrated transcriptome and metabolome analysis (Yang et al., 2022a). Overexpression of CoDFR in Nicotiana tabacum L. increased salicylic acid content as well as promoted the accumulation of flavonoids and thereby increased resistance to anthracnose (Yang et al., 2022b). Nonetheless, there is still a lack of deep and systematic research on the C. oleifera anthracnose-resistance mechanisms at the molecular level. Relatively, we still know little about the information of WRKY members in C. oleifera. A systematic investigation of CoWRKYs is needed.
Here, we identified WRKY TFs from C. oleifera genome, and analysis of their sequence features, conserved motifs, chromosome location, evolutionary relationship, and gene duplication events. Furthermore, the expression patterns of CoWRKYs after infection with C. fructicola between anthracnose-susceptible and -resistant C. oleifera cultivars were also determined. We found CoWRKY78 showed high expression in leaf and peel, and the expression level of CoWRKY78 in the anthracnose-resistant C. oleifera cultivars showed the greatest decline after inoculation. To investigate its function in anthracnose resistance, we overexpression of CoWRKY78 in tobacco, and further analysis of physiological changes and the difference in the expression of stress-related genes in WT and CoWRKY78-overexpressing lines after inoculation with Collettrichum nicotianae. Our study provides valuable guiding information for a deeper investigation of the functional properties and anthracnose defense mechanisms of C. oleifera WRKYs.
Genomic data and annotation information were downloaded from https://github.com/Hengfu-Yin/CON_genome_data. All the WRKY amino acid sequences of A. thaliana and Populus trichocarpa were obtained from Phytozome ver11 (https://phytozome-next.jgi.doe.gov/). The HMM profile of the WRKY domain was obtained from the Pfam database (PF03106) (El-Gebali et al., 2019), and then used to explore potential WRKYs in C. oleifera. We confirm the authenticity of the obtained WRKY sequences by the CDD and SMART (Marchler-Bauer et al., 2017; Letunic and Bork, 2018). According to their positions in the chromosomes of C. oleifera, we named these CoWRKY genes.
Protein properties of CoWRKY were determined using the online software ProtParam (https://web.expasy.org/protparam/). Furthermore, signal peptide was predicted using SignalP (http://www.cbs.dtu.dk/services/SignalP/) (Duvaud et al., 2021). Subcellular localization was predicted using Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) and experiments were performed according to our previous research (Li et al., 2022). Alignment was performed by using the program MUSCLE. Maximum likelihood phylogenetic trees were constructed using the MEGA 7 software with the JTT+G model, and then was visualized using iTOL (Letunic and Bork, 2019).
We performed conserved motif analysis using MEME version 5.5.0 (parameters: -nmotifs 10 -minw 6 -maxw 50) (Bailey et al., 2009), and gene structures were constructed via TBtools software (Chen et al., 2020). Moreover, the PlantCARE was performed to predict and analyze the promoter elements of all C. oleifera WRKY genes (Liu et al., 2022). Subsequently, the predicted cis-acting elements were visualized using TBtools.
Based on the C. oleifera genome database, the chromosomal locations of CoWRKYs were physically mapped on the 15 chromosomes of C. oleifera. To analyze the duplication events of WRKY genes, MCScanX was run with default parameters except -s (the number of colinear genes to claim a syntenic block) set to 5. Non-synonymous (Ka) and synonymous (Ks) substitutions of identified gene pairs were also calculated using TBtools (Zhao et al., 2020). The synteny of WRKYs between C. oleifera and the other two species (A. thaliana and P. trichocarpa) were determined by using Dual Systeny Plotter software and visualized via TBtools software.
Two C. oleifera cultivars, MY53 (anthracnose-resistant cultivar) and MY01 (anthracnose-susceptible cultivar) were grown at the C. oleifera orchard in Yuping Dong Autonomous County, China (N27°17′, E108°54′). The annual mean temperature and precipitation were 16.4°C (61.52°F) and 1174.1 mm (Wu et al., 2022). These two cultivars had similar genetic backgrounds, but different resistance to anthracnose.
The pathogenic C. fructicola was cultured on PDA medium under the dark condition at 28 °C for 1 week. C. oleifera young fruits were inoculation with C. fructicola. A sterile needle was used to puncture the peels (four wounds per fruit), 10 μL of C. fructicola zoospores suspension (1×106 zoospores/mL) was applied to each wound. After the inoculation, plastic tents were constructed to cover the plants and maintain almost 100% humidity by an automatic sprinkler system that switched on every 1 h to facilitate spore germination and infection. The fruits were harvested at 0, 2, 4, and 6 days after inoculation (Figure S1) and the peel of the fruits were used for the experiment.
Using the transcriptome data obtained in our laboratory (NCBI accession number: PRJNA898339), we examined the expression patterns of C. oleifera WRKYs after inoculation with C. fructicola. A heatmap of CoWRKYs was generated by TBtools and the gene expression was estimated by FPKM value (Li et al., 2019). Tissues from the root, stem, leaf, and peel were collected from C. oleifera to analyze the tissue-specific expression. As previously described, RNA isolation, cDNA synthesis followed by quantitative real-time RT-PCR (qRT-PCR) analysis were conducted (Wu et al., 2022). Gene expression level was determined via the 2−ΔΔCt method, and EF1α and GAPDH were used as the housekeeping gene. The sequences of all primers are listed in Table S1.
The coding sequence of CoWRKY78 was cloned by PCR with primers containing BamH I and Sac I restriction enzyme sites. The amplification products were ligated into the pBI121 vector. The recombinant plasmid was introduced into the Agrobacterium strain GV3101 and then transformed into the tobacco plants (N. tabacum L.cv. NC89) as previously described (Xiong et al., 2020). Firstly, kanamycin-resistant seedlings were analyzed by PCR. We examined the expression level of CoWRKY78 in transgenic tobacco plants via qRT-PCR. The tobacco actin and L25 were used for the normalization of the qRT-PCR analysis.
The C. nicotimiae strain was grown and maintained on the PDA media at 28 °C in the dark for 2 weeks before sporangia collection. The suspension was then incubated at 4°C for 1 h to stimulate the release of zoospores whose concentration was adjusted to 1×106 zoospores/mL. Five-leaf stage seedlings of CoWRKY78-overexpressing and WT tobacco plants were treated with C. nicotimiae by spraying a zoospores suspension. Inoculated tobacco plants were placed under dark and high humidity conditions for 1 day, then moved to a culture room at 28°C with 16-h/8-h photoperiod cycle. The inoculated leaves were collected for physiological parameter monitoring at 7 days after infection. The lesion areas were quantified via ImageJ software.
Based on the lesion area, disease grades were categorized: grade 0, no symptoms; grade 1, few lesions (less than 5%) shown on leaves; grade 2, about 6-10% of leaves area are infected; grade 3, about 11-20% of leaves area are infected; grade 4, about 21-40% of leaves area are infected; grade 5, larger than 41% of leaves area are infected. Disease index (DI) was calculated by the formula: DI (%) = [Σ (rating number × number of plants in the rating)/(the highest rating × total number of plants)] × 100%. Each experiment was performed at least thrice.
Cell death was determined using a trypan blue staining assay (Li et al., 2015). H2O2 or -accumulation was detected by 3, 3′-diaminobenzidine (DAB) or nitroblue tetrazolium (NBT) staining (Xiong et al., 2020). The malondialdehyde (MDA) content and the activities of superoxide dismutase (SOD), peroxidase (POD) as well as phenylalanine ammonia-lyase (PAL) were measured by the previously reported method (Li et al., 2015).
QRT-PCR was used to quantify the expression of NtSOD, NtPOD, NtPAL, NtNPR1, NtPR1, and NtPDF1.2. Total RNA was isolated from the leaves of CoWRKY78-overexpressing and WT tobacco plants before and after inoculation with C. nicotianae and then was converted into cDNA for qRT-PCR analysis. PLACE database was employed to identify W-box in their promoter regions.
Results were shown as means ± standard deviations (SD) from three independent experiments replicates. Statistically significant differences (p-values below 0.05) were determined via one-way ANOVA followed by Tukey’s multiple-comparison test.
In the C. oleifera genome, 91 C. oleifera WRKY genes were identified, and then renamed according to their chromosome distribution (Table S2). The longest CoWRKY is CoWRKY42, containing 758 amino acids, while the shortest CoWRKY63 has 133 amino acids (Table S2). Moreover, the molecular weights and isoelectric points of CoWRKYs range from 15.04 kDa (CoWRKY63) to 81.99 kDa (CoWRKY42) and 5.01 (CoWRKY86) to 10.17 (CoWRKY50), respectively. The grand average of hydropathicity of the CoWRKYs ranged from -1.216 (CoWRKY70) to -0.391 (CoWRKY17), suggesting that they are all hydrophilic proteins. Furthermore, we found that all CoWRKYs were localized in the nucleus and showed no signal peptide (Table S2).
We also investigate the evolutionary relationships of WRKYs in C. oleifera and A. thaliana. Ultimately, 91 CoWRKYs were assigned to 3 groups (Figure 1). Group I contained 19 CoWRKY members. According to the phylogenetic tree, group II can be divided into 5 subgroups, 6, 10, 21, 10, and 11 CoWRKY members belonged to the respective group. Moreover, 14 CoWRKY members were assigned to group III.
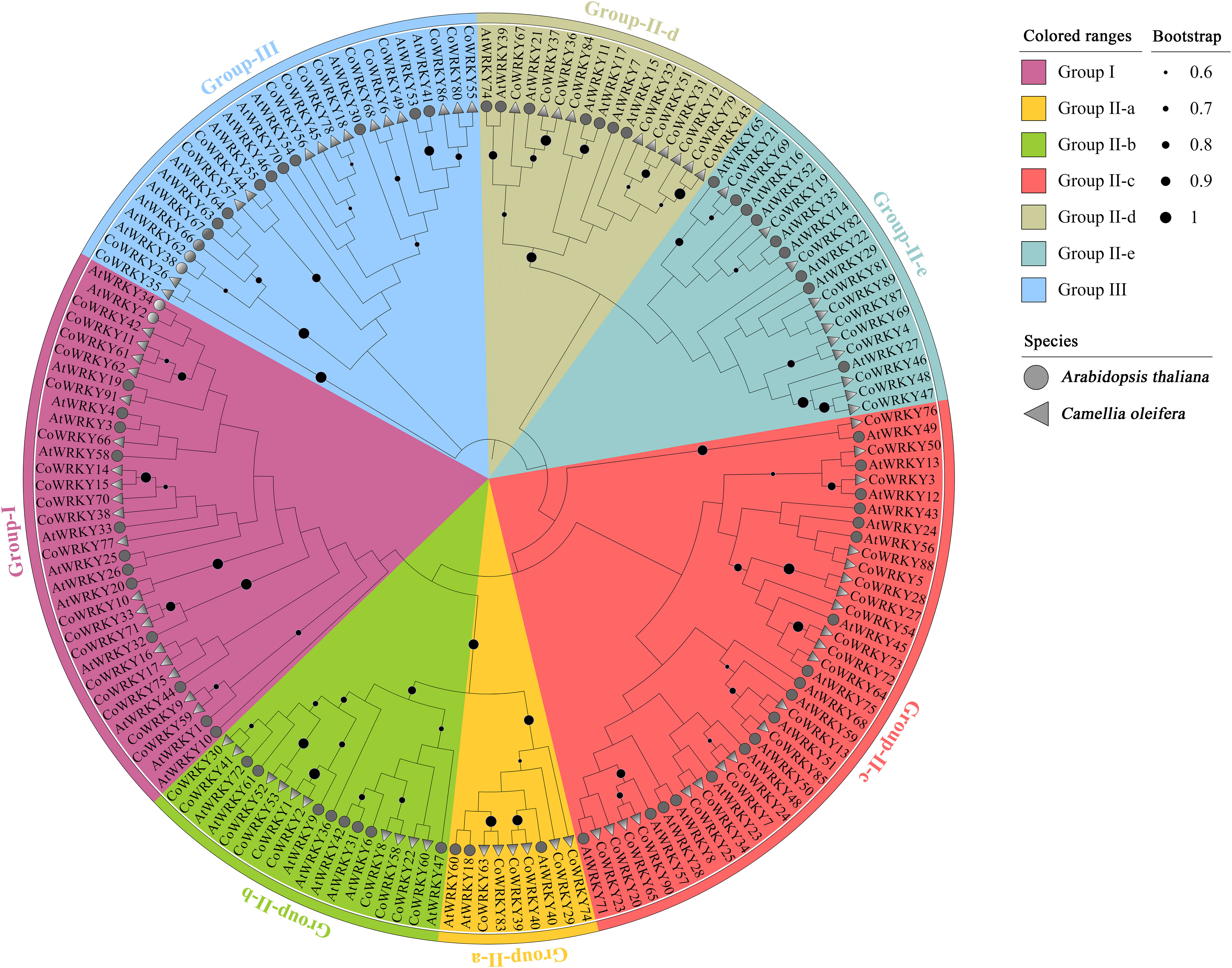
Figure 1 Phylogenetic tree of WRKY TFs from C. oleifera and A. thaliana. The clades representing different groups were filled with different colors.
The analysis of conserved motifs of C. oleifera WRKYs showed that there were 10 conserved motifs in 91 CoWRKYs (Figure S2), and the length of these motifs varied from 8 to 50 amino acids (Table S3). CoWRKYs in the same group had similar motif compositions (Figure 2A). Among the 10 identified motifs, motifs 1 and 3, characterized as WRKY domains. Meanwhile, all CoWRKY members in C. oleifera possessed motif 1 (Figure 2A). Regarding the gene structure of the CoWRKYs, the intron number of the 91 CoWRKYs ranged from 1 to 8. CoWRKY42 had the greatest number of introns (8) (Figure 2B).
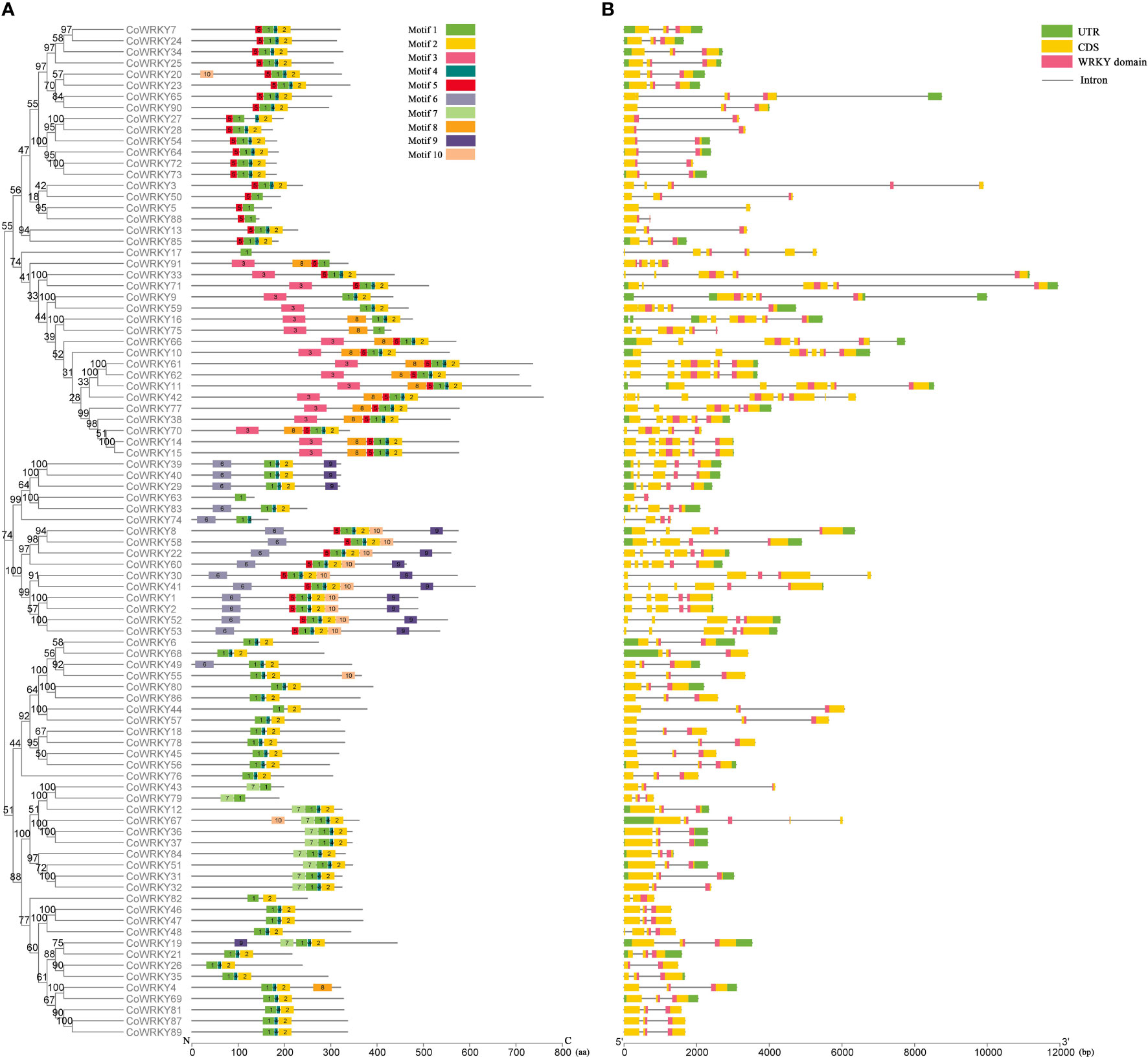
Figure 2 Conserved motifs and gene structure of CoWRKYs in C. oleifera. (A) Conserved motifs analysis of CoWRKYs. The conserved motifs were displayed in different colors. (B) Gene structures analysis of CoWRKYs. Different domains were shown in different colors.
The cis-elements of each CoWRKY gene in the 2 kb promoter region were identified using PlantCARE. Three stress response elements were widely distributed in these gene promoters (Figure S3). Meanwhile, a variety of plant hormone response elements were also found in their promoters. The results suggested that C. oleifera WRKY family may plays an important role in stress and hormone pathways.
There are 90 of the 91 CoWRKY genes are unevenly distributed on the 15 C. oleifera chromosomes (Figure 3A). Among these genes, 13 CoWRKY genes distributed on chromosome 10, followed by chromosome 12, which had 9 CoWRKY genes (Figure S4). Chromosomes 11 and 13 had 8 CoWRKY genes, and chromosomes 3 and 7 had 7 CoWRKY genes. Two chromosomes (chromosome 1 and 14) contained 6 CoWRKY genes each, and four chromosomes (chromosome 2, 4, 8, and 15) harbored 5 CoWRKY genes each. In addition, 3 CoWRKY genes on chromosome 9, 2 CoWRKY genes on chromosome 6, and 1 CoWRKY gene on chromosome 5.
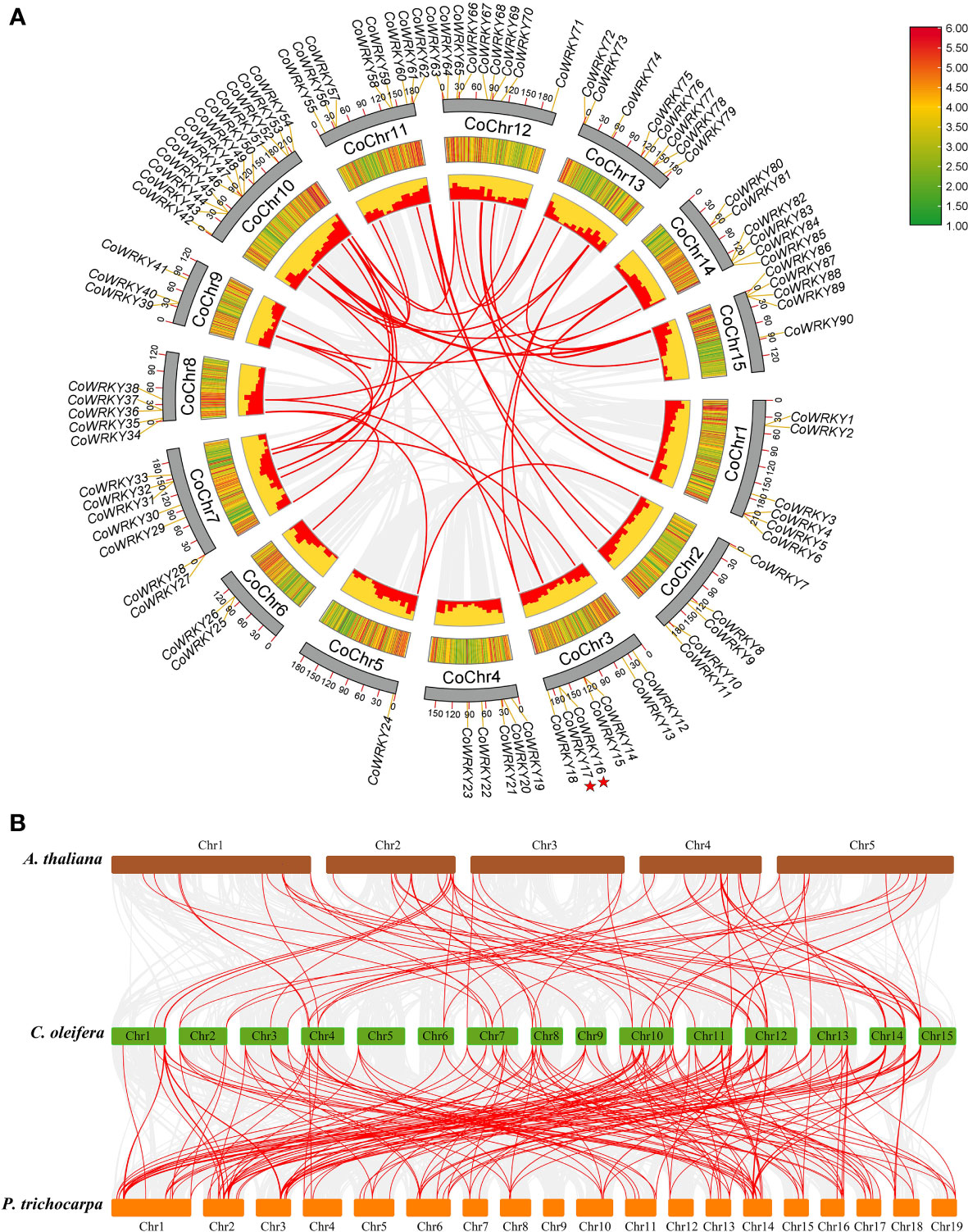
Figure 3 Chromosomal distribution and synteny analysis of CoWRKYs in C. oleifera. (A) A total of 90 WRKY genes are located in 15 chromosomes. Tandem duplication genes are marked with red stars. Gene pairs of WGD/segmental duplication in C. oleifera are linked using red lines. (B) Synteny relationship of WRKY genes from A. thaliana, P. trichocarpa, and C. oleifera genomes. The red lines highlight gene pairs with a collinear relationship. Gray lines in the background indicate the collinear blocks within two genomes.
In this study, we investigated gene duplication events in A. thaliana, P. trichocarpa and C. oleifera genomes. The numbers of whole-genome duplications (WGD)/segmental, tandem, proximal and dispersed duplication events in A. thaliana were 30 (41.7%), 2 (2.8%), 2 (2.8%) and 38 (52.7%), respectively (Table S4). In P. trichocarpa, 90 (92.8%) of the WRKY genes originated from WGD/segmental, which indicates WGD/segmental duplication made a valuable contribution to the evolution of the PtWRKY family (Table S4). Of the genes of the C. oleifera WRKY family, 57 (62.6%) originated from WGD/segmental duplication, 2 (2.2%) appeared to have been created through tandem duplication, 6 (6.6%) were proximal duplicated genes and 26 (28.6%) were dispersed duplicated genes (Table S4). These results indicated that WGD/segmental duplication explained the majority of gene duplication events in the CoWRKY family.
We further studied the collinear relationship between C. oleifera and two dicotyledons (A. thaliana and P. trichocarpa) (Figure 3B). The 74 orthologous gene pairs were identified between C. oleifera and A. thaliana (Table S5). In comparison to C. oleifera and P. trichocarpa genomes, 196 gene pairs were observed (Table S6). Significantly, among these gene pairs, 48 C. oleifera WRKYs have collinear relationships with A. thaliana and P. trichocarpa. Nine C. oleifera WRKY genes, including CoWRKY 7, 20, 22, 34, 54, 68, 69, 80, and 86, were associated with at least 6 syntenic gene pairs, indicating that they might have played a crucial role in C. oleifera WRKYs evolution. Moreover, the synonymous substitution rates (Ka/Ks) of the gene pairs were calculated to identify the evolutionary forces. All of the 243 orthologous gene pairs had Ka/Ks< 1, suggesting that purifying selection may be the dominant force driving the evolution of CoWRKY genes.
To screen the potential CoWRKYs in response to anthracnose infection, we created a heatmap to compare the profile of expression of CoWRKY genes in peels of two C. oleifera cultivars after C. fructicola inoculation. In group I, the CoWRKY genes in the anthracnose-resistant cultivars of C. oleifera have a higher expression level than that in the anthracnose-susceptible cultivars of C. oleifera after inoculation with C. fructicola. In another group, the expression level of CoWRKYs in the susceptible cultivar occurred earlier and stronger while being induced later in the resistant cultivar (Figure 4).
We selected 6 CoWRKY members from 2 groups for qRT-PCR analysis (Figure S5). CoWRKY4, CoWRKY28, and CoWRKY82 were from group I, which showed higher expression level in the anthracnose-resistant C. oleifera cultivars than that in the anthracnose-susceptible C. oleifera cultivars after inoculation with C. fructicola. In contrast, after inoculation, CoWRKY36, CoWRKY74, and CoWRKY78 in group II had lower expression levels in the anthracnose-resistant C. oleifera cultivars than in the anthracnose-susceptible C. oleifera cultivars (Figure S5). These expression patterns were consistent with the results from transcriptome sequencing. It is noteworthy that, CoWRKY78 in the resistant cultivar showed the greatest decline in expression level at 2 d after inoculation. Furthermore, CoWRKY78 showed high expression levels in the leaf and peel (Figure S6A). Therefore, CoWRKY78 was selected to explore its function in response to anthracnose infection.
Under confocal microscope, the green fluorescence signal was observed mainly in the nucleus of tobacco leaf epidermal cells, suggesting that CoWRKY78 is located in the nucleus (Figure S6B). To investigate CoWRKY78 function in anthracnose resistance, the pBI121-CoWRKY78 overexpression construct was transformed into tobacco. Firstly, kanamycin-resistant seedlings were verified by PCR. The result showed that the transgenic lines (lines 1, 2, 3, 5, and 6) exhibited expected bands (Figure S7A). We further verified the expression level of CoWRKY78 using qRT-PCR analysis. The line 1 showed 3.1-fold higher expression, while the line 5 showed a 2.87-fold higher level relative to line 6. Hence, these lines were selected for further experiments (Figure S7B).
After inoculation, CoWRKY78-overexpressing tobacco plants developed more severe disease symptoms on the leaves compared with WT tobacco plants (Figure 5A). Moreover, transgenic plants showed a higher ratio of lesion area to whole leaf area than WT tobacco plants after inoculation (Figure 5B). Furthermore, CoWRKY78-overexpressing tobacco plants had higher DI than WT plants (Figure 5C), suggesting that the areas of anthracnose lesions were more prominent in transgenic tobacco plants. After inoculation, we found that cell death was more prominent in the CoWRKY78-overexpressing tobacco plants than in the WT tobacco plants according to trypan blue staining (Figure 6A, upper panel). These results suggested that overexpression of CoWRKY78 led to decreased resistance to anthracnose in tobacco.
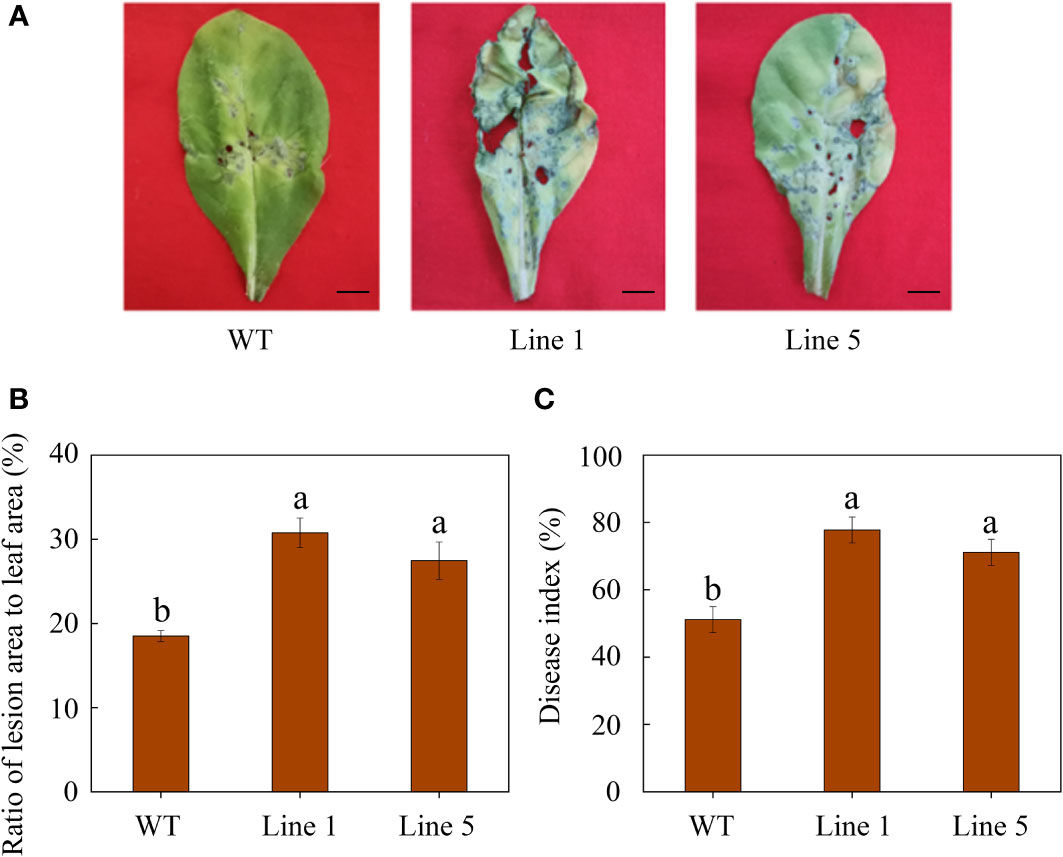
Figure 5 Overexpression of CoWRKY78 decreased anthracnose resistance in transgenic tobacco. (A) Disease symptoms on the leaves from transgenic and WT plants at 7 days after C. nicotianae inoculation. Scale bar indicates 1 cm. (B) The ratio of lesion area to leaf area in inoculated tobacco leaves. (C) Measurement of DI at 7 days after C. nicotianae inoculation. Data represent the means ± SD and bars denoted by a different letter are significantly different (p-values below 0.05).
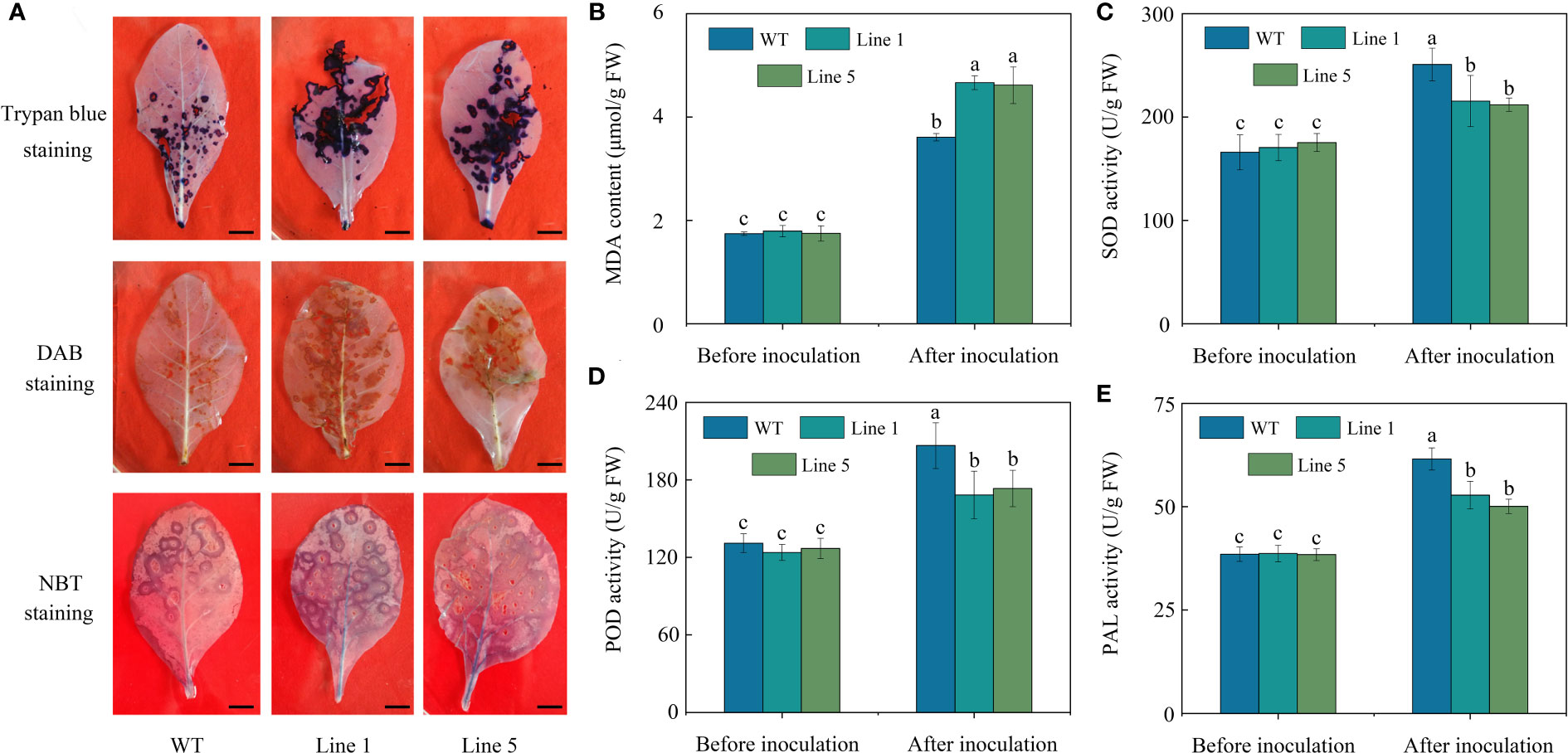
Figure 6 Physiological changes in transgenic and WT tobacco plants before and after C. nicotianae inoculation. (A) Histochemical staining via trypan blue, DAB, and NBT. Scale bar indicates 1 cm. (B) MDA content. (C) SOD activity. (D) POD activity. (E) PAL activity. Data represent the means ± SD and bars denoted by a different letter are significantly different (p-values below 0.05).
ROS and MDA often accumulate in plants after pathogen infection (Li et al., 2015). After C. nicotianae inoculation, more intense brown coloration (Figure 6A, middle panel), blue coloration (Figure 6A, lower panel), and higher MDA content (Figure 6B) in the transgenic tobacco plants than that in WT plants. Antioxidant enzymes like SOD or POD play an important role in removing excess ROS. PAL activity is often used as one of the important indicators for plant resistance evaluation (Li et al., 2015). The activities of SOD (Figure 6C), POD (Figure 6D), and PAL (Figure 6E) in the transgenic tobacco plants were significantly lower compared with those in the WT plants after inoculation with C. nicotianae. These data suggested that overexpression of CoWRKY78 in tobacco resulted in decreased activities of defense-related enzymes, which lead to increased sensitivity to anthracnose.
To gain further insight into the regulated role of CoWRKY78, we further analysed the expression profiles of several stress-related genes which are related to ROS-scavenging (NtSOD and NtPOD), pathogen challenge (NtPAL), and pathogen defense (NtPR1, NtNPR1, and NtPDF1.2). No significant differences were observed in the expression of NtSOD (Figure 7A), NtPOD (Figure 7B), NtPAL (Figure 7C), and NtNPR1 (Figure 7E) between CoWRKY78-overexpressing and WT tobacco plants before inoculation. Interestingly, the transgenic lines exhibited higher expression of NtPR1 (Figure 7D), but lower expression of NtPDF1.2 (Figure 7F) than the WT plants. After inoculation, all of these genes were upregulated. Compared with WT tobacco plants, the expression levels of NtSOD, NtPOD, NtPAL, and NtPDF1.2 were lower in transgenic tobacco plants, but the expression of NtNPR1 and NtPR1 was significantly higher in transgenic plants. Furthermore, we observed several W-box in their promoter regions (Table S7), suggesting CoWRKY78 may participate in anthracnose resistance by regulating these genes expression.
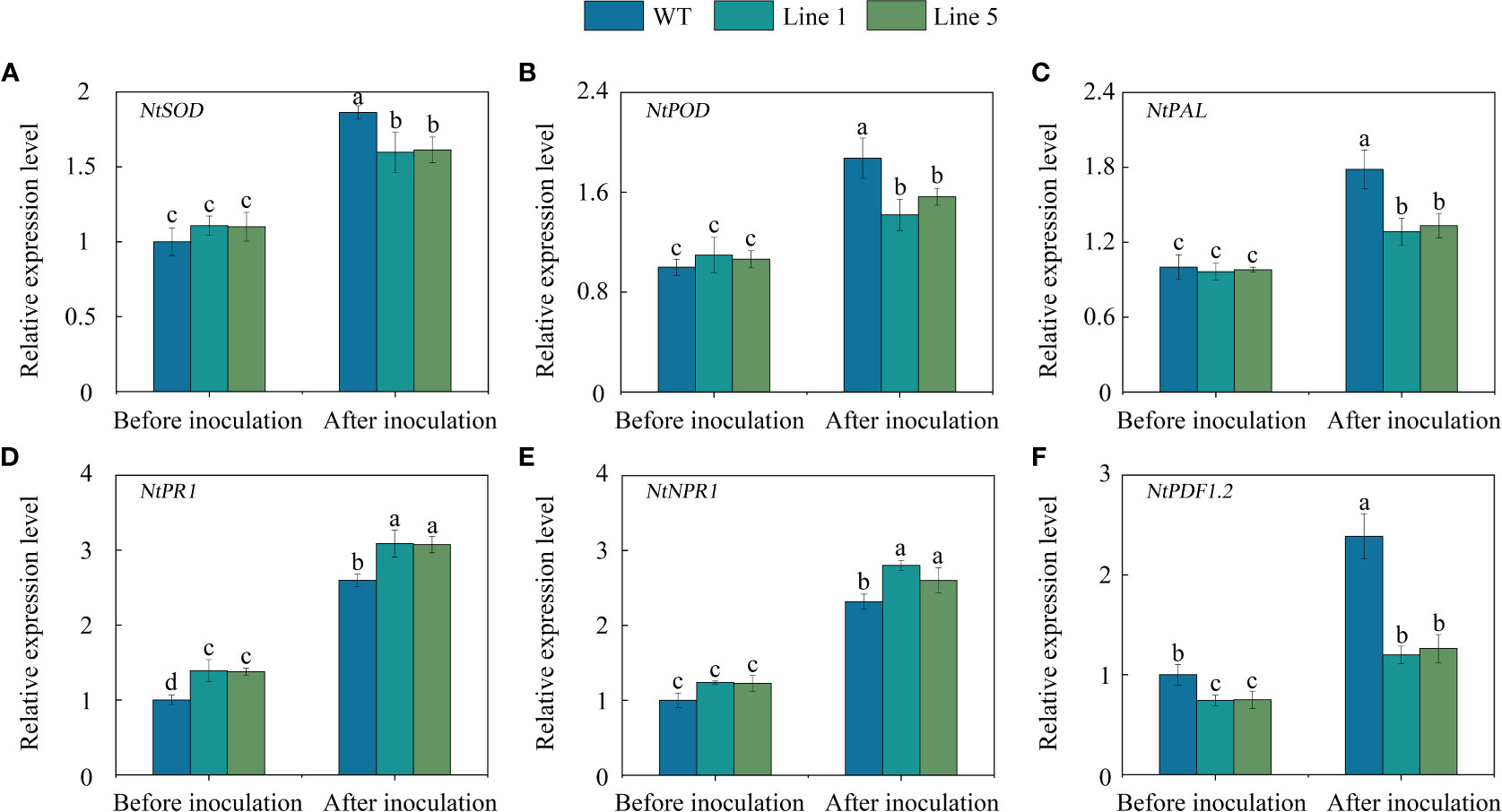
Figure 7 The expression of some stress-related genes in the transgenic and WT tobacco plants. (A-F) Analysis of expression of NtSOD, NtPOD, NtPAL, NtPR1, NtNPR1, and NtPDF1.2 in the leaves of transgenic and WT tobacco plants before and after C. nicotianae inoculation. Data represent the means ± SD and bars denoted by a different letter are significantly different (p-values below 0.05).
WRKY TFs have been reported in many woody plants, including 104 WRKYs in poplar (He et al., 2012), 80 WRKYs in grape (Zhang and Feng, 2014), 132 WRKYs in Musa balbisiana (Goel et al., 2016), 56 WRKYs in tea (Wang et al., 2019), and 103 WRKYs in common walnut (Hao et al., 2021). As an important woody oil plant, not much is known about the exact number of WRKYs in C. oleifera, so we began our investigations.
We identified 91 WRKY TFs in C. oleifera, which can be subdivided into 3 groups (Figure 1). There were 19, 58, and 14 members that were included in group I, II, and III, respectively. Understanding the exon and intron organization of these genes may help us to obtain more information about their evolutionary history (Liu et al., 2022). Regarding the structure of WRKY family in C. oleifera, the intron number of the 91 CoWRKYs ranged from 1 to 8 (Figure 2). Obvious differences in gene structures exist in CoWRKYs, but members clustered in the same group exhibited similar structures, implying the important roles of these features have led to functional divergence.
The duplication of genes plays an important role in gene family evolution (Yan et al., 2022). Previous research has established that the three WGD in A. thaliana have been directly responsible for over 90% of the increase in TFs, signal transducers, and developmental genes in the last 350 million years (Maere et al., 2005). We investigated gene duplication events in C. oleifera genomes, 57 (62.6%) of the WRKY genes originated from WGD/segmental duplication, which indicates WGD/segmental duplication made a valuable contribution to the evolution of the C. oleifera WRKY family. Our finding that is aligns with the previous findings (Waqas et al., 2019; Yan et al., 2022). Moreover, orthologous relationships of WRKY genes among C. oleifera, A. thaliana, and P. trichocarpa genomes were detected, including C. oleifera-A. thaliana (74 pairs) and C. oleifera-P. trichocarpa (196 pairs) (Figure 3B). The synonymous substitution rates (Ka/Ks) of the gene pairs were calculated to identify the evolutionary forces. All of the 243 orthologous gene pairs had Ka/Ks< 1, suggesting that purifying selection may be the dominant force driving the evolution of CoWRKY genes.
Anthracnose, caused by C. fructicola, is an extremely destructive disease of C. oleifera (Zhang et al., 2019b). The pathogen attacks many C. oleifera organs, including buds, fruits, and leaves. WRKY TFs are of great importance in plant-pathogen interactions. However, there have not been any reports investigating the expression pattern of WRKY genes directly involved in C. oleifera-C. fructicola interaction. A systematic investigation of CoWRKYs is needed.
In this study, the peels of two C. oleifera cultivars showing different resistance to anthracnose were analyzed by transcriptomics. We created a heatmap to analyze the profile of expression of CoWRKY genes in peels of two C. oleifera cultivars after C. fructicola inoculation. In group I, the expression level of CoWRKYs in the resistant C. oleifera cultivar occurred stronger and earlier, while CoWRKY genes in group II have a lower expression level in the anthracnose-resistant C. oleifera cultivars than that in the anthracnose- susceptible C. oleifera cultivars (Figure 4). Our analysis provides a unique opportunity to understand the candidate WRKY genes involved in the anthracnose resistance.
A majority of the WRKY genes have been shown to respond to stress and phytohormone treatments (Xiong et al., 2020; Lim et al., 2022). Consistent with previous results, we found that abundant cis-acting regulatory elements in CoWRKY promoters were related to abiotic stress (drought and cold) and hormones (ABA, MeJA, and SA). A total of 33 (35.2%) CoWRKY genes have MBS, implying their important roles in drought stress. In addition, 70 (76.9%) CoWRKY genes have ABA-responsive element, implying they also plays a critical role in ABA signaling pathways. With ecological value, C. oleifera can grow well in drought and barren soil (He et al., 2022b). Our finding show that CoWRKY genes were important and necessary for the responses to drought stress and further exploration of the potential biological functions of CoWRKY genes is needed.
CoWRKY78 showed high expression in leaf and peel, and its expression in the resistant cultivar showed the greatest decline at 2 d after inoculation. Therefore, CoWRKY78 was selected to explore its function in response to anthracnose infection. Overexpression of CoWRKY78 decreased anthracnose resistance in transgenic tobacco plants, which was demonstrated by a higher ratio of lesion area to leaf area, higher DI, and more severe cell death than WT plants (Figures 5, 6A).
Pathogen infection can cause oxidative stress by increasing the production of ROS in plants (Bloem et al., 2015; Li et al., 2015). However, late massive ROS generations may lead to damage to cellular membranes. MDA, SOD, and POD activities are common indicators for assessing plant resistance to diseases (Li et al., 2015; Prabhukarthikeyan et al., 2018). Previous studies showed that the accumulation of MDA, SOD, and POD in both disease-resistant and susceptible C. oleifera lines increased by anthracnose infection. However, disease resistant lines exhibited lower MDA, but higher SOD and POD activities compared to susceptible lines (Yang et al., 2022b). To investigate the physiological differences between WT and CoWRKY78-overexpression tobacco plants before and after anthracnose infection, these important physiological indices were measured. After inoculation, the transgenic tobacco plants accumulated more ROS and MDA than WT plants (Figures 6A, B). Moreover, the activities of SOD and POD in the transgenic tobacco plants were lower than those in the WT plants (Figures 6C, D). Furthermore, overexpression of CoWRKY78 decreased the expression of NtSOD and NtPOD (Figures 7A, B), which was consistent with the enzyme activity. These suggested that CoWRKY78 negatively regulates the resistance of anthracnose via impairing the antioxidant abilities, which in turn excess accumulation of ROS.
We identified 91 WRKY TFs in C. oleifera, which can be divided into 3 groups. Segmental duplications were the main contributor to the expansion of C. oleifera WRKY TF family. We mined multiple anthracnose-responsive CoWRKY genes. Overexpression of CoWRKY78 in tobacco resulted in increased sensitivity to anthracnose. The transgenic tobacco plants had higher ROS and lower activity of defense-related enzymes than WT tobacco plants. Furthermore, the expression of stress-related genes involved in ROS-homeostasis was also reduced in the CoWRKY78-overexpressing plants. These findings increase our understanding of the C. oleifera WRKYs and provide further evidence for exploration of anthracnose resistance mechanisms in C. oleifera.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA898339.
JL and CR conceived and designed the research. JL and CX conducted the bioinformatics analysis and performed the experiments. DR, WD, and HL analyzed data. JL wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the Science and Technology Support Project of Guizhou Province (Qiankehezhicheng [2022] Zhongdian 017, Qinkehezhicheng [2019] 2404), the Special Forestry Industry Science and Technology R&D project for Guizhou Rural Industrial Revolution (Telinyan2020-07), the Agricultural Special Projects of Tongren City (Tongshikeyan [2021]), and the Guizhou Provincial Science and Technology Planning Project of China (Qinkehejichu [2018] 1159, [2020] 1Y131).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1110366/full#supplementary-material
Supplementary Figure 1 | Phenotype of MY53 and MY01 after inoculation with C. fructicola. Scale bar indicates 1 cm.
Supplementary Figure 2 | Sequences of conserved motifs.
Supplementary Figure 3 | The cis-acting regulatory elements in the promoters of C. oleifera WRKY genes.
Supplementary Figure 4 | Chromosome distribution of C. oleifera WRKY genes.
Supplementary Figure 5 | Expression analysis of selected CoWRKY genes using qRT-PCR. (A-F) The expression profiles of 6 C. oleifera WRKY genes after inoculation with C. fructicola in MY53 and MY01 peels. Data represent the means ± SD and bars denoted by a different letter are significantly different (p-values below 0.05).
Supplementary Figure 6 | Characterization of CoWRKY78. (A) Tissue-specific expression patterns analysis of CoWRKY78. Data represent the means ± SD and bars denoted by a different letter are significantly different (p-values below 0.05). (B) Subcellular localization of CoWRKY78. Scale bar indicates 20 μm in all images.
Supplementary Figure 7 | Molecular analysis of putatively transgenic tobacco plants. (A) Genomic DNA-PCR analysis of transgenic plants. H: blank control (The amplified PCR products using the H2O as templates); P: positive control (The amplified PCR products using the pBI121-CoWRKY78 plasmid DNA as templates); M: DL2000 DNA Marker. (B) QRT-PCR analysis of the selected transgenic lines for determination of the expression level of CoWRKY78. Data represent the means ± SD and bars denoted by a different letter are significantly different (p-values below 0.05).
Amorim, L. L. B., da Fonseca Dos Santos, R., Neto, J. P. B., Guida-Santos, M., Crovella, S., Benko-Iseppon, A. M. (2017). Transcription factors involved in plant resistance to pathogens. Curr. Protein Pept. Sci. 18, 335–351. doi: 10.2174/1389203717666160619185308
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bi, M., Li, X., Yan, X., Liu, D., Gao, G., Zhu, P., et al. (2021). Chrysanthemum WRKY15-1 promotes resistance to Puccinia horiana henn. via the salicylic acid signaling pathway. Hortic. Res. 8, 6. doi: 10.1038/s41438-020-00436-4
Bloem, E., Haneklaus, S., Schnug, E. (2015). Milestones in plant sulfur research on sulfur-induced-resistance (SIR) in Europe. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00779
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, X., Liu, J., Lin, G., Wang, A., Wang, Z., Lu, G. (2013). Overexpression of AtWRKY28 and AtWRKY75 in arabidopsis enhances resistance to oxalic acid and Sclerotinia sclerotiorum. Plant Cell Rep. 32, 1589–1599. doi: 10.1007/s00299-013-1469-3
Ciolkowski, I., Wanke, D., Birkenbihl, R. P., Somssich, I. E. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68, 81–92. doi: 10.1007/s11103-008-9353-1
Du, P., Wu, Q., Liu, Y., Cao, X., Yi, W., Jiao, T., et al. (2022). WRKY transcription factor family in lettuce plant (Lactuca sativa): Genome-wide characterization, chromosome location, phylogeny structures, and expression patterns. Peer J. 10, e14136. doi: 10.7717/peerj.14136
Duvaud, S., Gabella, C., Lisacek, F., Stockinger, H., Ioannidis V. and Durinx, C. (2021). Expasy, the Swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Res. 49 (W1), W216–W227. doi: 10.1093/nar/gkab225
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Eulgem, T., Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Goel, R., Pandey, A., Trivedi, P. K. (2016). Genome-wide analysis of the musa WRKY gene family: Evolution and differential expression during development and stress. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00299
Hao, F., Yang, G., Zhou, H., Yao, J., Liu, D., Zhao, P., et al. (2021). Genome-wide identification and transcriptional expression profiles of transcription factor WRKY in common walnut (Juglans regia l.). Genes (Basel) 12, 1444. doi: 10.3390/genes12091444
He, Y., Chen, R., Yang, Y., Liang, G., Zhang, H., Deng, X., et al. (2022a). Sugar metabolism and transcriptome analysis reveal key sugar transporters during Camellia oleifera fruit development. Int. J. Mol. Sci. 23, 822. doi: 10.3390/ijms23020822
He, H., Dong, Q., Shao, Y., Jiang, H., Zhu, S., Cheng, B., et al. (2012). Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 31, 1199–1217. doi: 10.1007/s00299-012-1241-0
He, Z., Liu, C., Zhang, Z., Wang, R., Chen, Y. (2022b). Integration of mRNA and miRNA analysis reveals the differentially regulatory network in two different Camellia oleifera cultivars under drought stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022
Huangfu, J., Li, J., Li, R., Ye, M., Kuai, P., Zhang, T., et al. (2016). The transcription factor OsWRKY45 negatively modulates the resistance of rice to the brown planthopper Nilaparvata lugens. Int. J. Mol. Sci. 17, 697. doi: 10.3390/ijms17060697
Jia, S., Wang, Y., Zhang, G., Yan, Z., Cai, Q. (2020). Strawberry FaWRKY25 transcription factor negatively regulated the resistance of strawberry fruits to Botrytis cinerea. Genes (Basel) 12, 56. doi: 10.3390/genes12010056
Jiang, Y., Guo, L., Ma, X., Zhao, X., Jiao, B., Li, C., et al. (2017). The WRKY transcription factors PtrWRKY18 and PtrWRKY35 promote melampsora resistance in populus. Tree Physiol. 37, 665–675. doi: 10.1093/treephys/tpx008
Kang, Z. W., Liu, F. H., Zhang, Z. F., Tian, H. G., Liu, T. X. (2018). Volatile β-ocimene can regulate developmental performance of peach aphid Myzus persicae through activation of defense responses in chinese cabbage brassica pekinensis. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00708
Letunic, I., Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Letunic, I., Bork, P. (2019). Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, J., Brader, G., Palva, E. T. (2004). The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331. doi: 10.1105/tpc.016980
Li, J. B., Jian, D., Yu, X., Li, H., Ruan, C. J. (2019). Identification and expression analysis of critical microRNA-transcription factor regulatory modules related to seed development and oil accumulation in developing Hippophae rhamnoides seeds. Ind. Crop Prod. 137, 33–42. doi: 10.1016/j.indcrop.2019.05.011
Li, J. B., Luan, Y. S., Liu, Z. (2015). SpWRKY1 mediates resistance to Phytophthora infestans and tolerance to salt and drought stress by modulating reactive oxygen species homeostasis and expression of defense-related genes in tomato. Plant Cell Tiss. Organ Cult. 123, 67–81. doi: 10.1007/s11240-015-0815-2
Li, J. B., Zhou, H., Xiong, C. W., Peng, Z. J., Du, W., Li, H., et al. (2022). Genome-wide analysis R2R3-MYB transcription factors in Xanthoceras sorbifolium bunge and functional analysis of XsMYB30 in drought and salt stresses tolerance. Ind. Crop Prod. 178, 114597. doi: 10.1016/j.indcrop.2022.114597
Lian, Q., He, X., Zhang, B., Wang, Y., Ma, Q. (2022). Identification and characterization of wrky41, a gene conferring resistance to powdery mildew in wild tomato (Solanum habrochaites) LA1777. Int. J. Mol. Sci. 23, 1267. doi: 10.3390/ijms23031267
Lim, C., Kang, K., Shim, Y., Yoo, S. C., Paek, N. C. (2022). Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways. Plant Physiol. 188, 1900–1916. doi: 10.1093/plphys/kiab492
Lin, P., Wang, K., Wang, Y., Hu, Z., Yan, C., Huang, H., et al. (2022). The genome of oil-camellia and population genomics analysis provide insights into seed oil domestication. Genome Biol. 23, 14. doi: 10.1186/s13059-021-02599-2
Liu, X., Li, D., Zhang, S., Xu, Y., Zhang, Z. (2019). Genome-wide characterization of the rose (Rosa chinensis) WRKY family and role of RcWRKY41 in gray mold resistance. BMC Plant Biol. 19, 522. doi: 10.1186/s12870-019-2139-6
Liu, Z., Zhang, Y., Altaf, M. A., Hao, Y., Zhou, G., Li, X., et al. (2022). Genome-wide identification of myeloblastosis gene family and its response to cadmium stress in ipomoea aquatica. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.979988
Maere, S., De Bodt, S., Raes, J., Casneuf, T., Van Montagu, M., Kuiper, M., et al. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 5454–5459. doi: 10.1073/pnas.0501102102
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Peng, X., Hu, Y., Tang, X., Zhou, P., Deng, X., Wang, H., et al. (2012). Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 236, 1485–1498. doi: 10.1007/s00425-012-1698-7
Prabhukarthikeyan, S., Keerthana, U., Raguchander, T. (2018). Antibioticproducing pseudomonas fluorescens mediates rhizome rot disease resistance and promotes plant growth in turmeric plants. Microbiol. Res. 210, 65–73. doi: 10.1016/j.micres.2018.03.009
Ross, C. A., Liu, Y., Shen, Q. J. (2007). The WRKY gene family in rice (Oryza sativa). J. Integr. Plant Biol. 49, 827–842. doi: 10.1111/j.1672-9072.2007.00504.x
Wang, X., Li, J., Guo, J., Qiao, Q., Guo, X., Ma, Y. (2020). The WRKY transcription factor PlWRKY65 enhances the resistance of Paeonia lactiflora (herbaceous peony) to Alternaria tenuissima. Hortic. Res. 7, 57. doi: 10.1038/s41438-020-0267-7
Wang, P. J., Yue, C., Chen, D. (2019). Genome-wide identification of WRKY family genes and their response to abiotic stresses in tea plant (Camellia sinensis). Genes Genom. 41, 17–33. doi: 10.1007/s13258-018-0734-9
Waqas, M., Azhar, M. T., Rana, I. A., Azeem, F., Ali, M. A., Nawaz, M. A., et al. (2019). Genome-wide identification and expression analyses of WRKY transcription factor family members from chickpea (Cicer arietinum l.) reveal their role in abiotic stress-responses. Genes Genomics 41, 467–481. doi: 10.1007/s13258-018-00780-9
Wu, L., Li, J., Li, Z., Zhang, F., Tan, X. (2020). Transcriptomic analyses of Camellia oleifera 'Huaxin' leaf reveal candidate genes related to long-term cold stress. Int. J. Mol. Sci. 21, 846. doi: 10.3390/ijms21030846
Wu, B., Ruan, C., Shah, A. H., Li, D., Li, H., Ding, J., et al. (2022). Identification of miRNA-mRNA regulatory modules involved in lipid metabolism and seed development in a woody oil tree (Camellia oleifera). Cells 11, 71. doi: 10.3390/cells11010071
Xiong, C., Zhao, S., Yu, X., Sun, Y., Li, H., Ruan, C., et al. (2020). Yellowhorn drought-induced transcription factor XsWRKY20 acts as a positive regulator in drought stress through ROS homeostasis and ABA signaling pathway. Plant Physiol. Biochem. 155, 187–195. doi: 10.1016/j.plaphy.2020.06.037
Yan, L., Jin, H., Raza, A., Huang, Y., Gu, D., Zou, X. (2022). WRKY genes provide novel insights into their role against ralstonia solanacearum infection in cultivated peanut (Arachis hypogaea l.). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.986673
Yang, C., Wu, P., Cao, Y., Yang, B., Liu, L., Chen, J., et al. (2022b). Overexpression of dihydroflavonol 4-reductase (CoDFR) boosts flavonoid production involved in the anthracnose resistance. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1038467
Yang, C., Wu, P., Yao, X., Sheng, Y., Zhang, C., Lin, P., et al. (2022a). Integrated transcriptome and metabolome analysis reveals key metabolites involved in Camellia oleifera defense against anthracnose. Int. J. Mol. Sci. 23, 536. doi: 10.3390/ijms23010536
Ye, S., Jiang, Y., Duan, Y., Karim, A., Fan, D., Yang, L., et al. (2014). Constitutive expression of the poplar WRKY transcription factor PtoWRKY60 enhances resistance to Dothiorella gregaria sacc. in transgenic plants. Tree Physiol. 34, 1118–1129. doi: 10.1093/treephys/tpu079
Ye, Z., Yu, J., Yan, W., Zhang, J., Yang, D., Yao., G., et al. (2021). Integrative iTRAQ-based proteomic and transcriptomic analysis reveals the accumulation patterns of key metabolites associated with oil quality during seed ripening of Camellia oleifera. Hortic. Res. 8, 157. doi: 10.1038/s41438-021-00591-2
Zhang, Y., Feng, J. C. (2014). Identification and characterization of the grape WRKY family. BioMed. Res. Int. 2014, 787680. doi: 10.1155/2014/787680
Zhang, S., Guo, Y., Li, S., Zhou, G., Liu, J., Xu, J., et al. (2019b). Functional analysis of CfSnf1 in the development and pathogenicity of anthracnose fungus Colletotrichum fructicola on tea-oil tree. BMC Genet. 20, 94. doi: 10.1186/s12863-019-0796-y
Zhang, F., Li, Z., Zhou, J., Gu, Y., Tan, X. (2021). Comparative study on fruit development and oil synthesis in two cultivars of Camellia oleifera. BMC Plant Biol. 21, 348. doi: 10.1186/s12870-021-03114-2
Zhang, S., Pan, Y. G., Zheng, L., Yang, Y., Zheng, X., Ai, B., et al. (2019a). Application of steam explosion in oil extraction of camellia seed (Camellia oleifera abel.) and evaluation of its physicochemical properties, fatty acid, and antioxidant activities. Food Sci. Nutr. 7, 1004–1016. doi: 10.1002/fsn3.924
Zhao, K., Cheng, Z., Guo, Q., Yao, W., Liu, H., Zhou, B., et al. (2020). Characterization of the poplar R2R3-MYB gene family and over-expression of PsnMYB108 confers salt tolerance in transgenic tobacco. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.571881
Keywords: Camellia oleifera, anthracnose, CoWRKY genes, gene expression patterns, ROS homeostasis
Citation: Li J, Xiong C, Ruan D, Du W, Li H and Ruan C (2023) Identification of Camellia oleifera WRKY transcription factor genes and functional characterization of CoWRKY78. Front. Plant Sci. 14:1110366. doi: 10.3389/fpls.2023.1110366
Received: 28 November 2022; Accepted: 23 February 2023;
Published: 09 March 2023.
Edited by:
Deyi Yuan, Central South University Forestry and Technology, ChinaReviewed by:
Dianyun Hou, Henan University of Science and Technology, ChinaCopyright © 2023 Li, Xiong, Ruan, Du, Li and Ruan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengjiang Ruan, cnVhbkBkbG51LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.