
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 15 February 2023
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1109941
This article is part of the Research Topic Omics technology in agriculture: molecular breeding for sustainable crop production View all 7 articles
Male sterility is classified as either cytoplasmic male sterility (CMS) or genic male sterility (GMS). Generally, CMS involves mitochondrial genomes interacting with the nuclear genome, while GMS is caused by nuclear genes alone. Male sterility is regulated by multilevel mechanisms in which non-coding RNAs (ncRNAs), including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and phased small interfering RNAs (phasiRNAs), which have been proven to be critical elements. The development of high-throughput sequencing technology offers new opportunities to evaluate the genetic mechanism of ncRNAs in plant male sterility. In this review, we summarize the critical ncRNAs that regulate gene expression in ways dependent on or independent of hormones, which involve the differentiation of the stamen primordia, degradation of the tapetum, formation of microspores, and the release of pollen. In addition, the key mechanisms of the miRNA–lncRNA–mRNA interaction networks mediating male sterility in plants are elaborated. We present a different perspective on exploring the ncRNA-mediated regulatory pathways that control CMS in plants and create male-sterile lines through hormones or genome editing. A refined understanding of the ncRNA regulatory mechanisms in plant male sterility for the development of new sterile lines would be conducive to improve hybridization breeding.
Heterosis has been utilized to increase productivity in many crops. The male sterility system provides crucial breeding tools to harness heterosis due to its biosafety and high efficiency and because it is less labor-intensive and time-consuming (Parodi and Gaju, 2009; Chen and Liu, 2014). Generally, the male reproductive development in plants comprises a series of stages of specification of the stamen primordia, generation of sporogenous cells, development of the tapetum and microspore mother cells, meiosis, formation of free haploid microspores, degeneration of the tapetum, and the release of mature pollen grains. The arrest of any of these steps could result in male sterility (Huang et al., 2014; Bohra et al., 2021; Han et al., 2021; Zhang et al., 2021). Plant male sterility refers to plants’ normal male organs failing to produce functional microspores, pollen, or anthers, but their pistils still having adequate flowering and crossing abilities with other plants (Parodi and Gaju, 2009; Brownfield, 2021). Male sterility, which was first observed by the German botanist Joseph Gottlieb Kolreuter in 1763 and has now been reported in hundreds of plant species (Laser and Lersten, 1972; Mayr, 1986; Kaul, 1988), can be classified as either cytoplasmic male sterility (CMS) or genic male sterility (GMS) (Zhu et al., 2020). Generally, CMS is ascribed to the uncoordinated inheritance between the organellar (mitochondrial, MT) and nuclear genomes, while GMS is caused by nuclear genes independently (Bohra et al., 2016).
Non-coding RNAs (ncRNAs) are important regulators of gene expression involved in various metabolisms (Xia et al., 2019; Chao et al., 2022) that are classified into three types: small ncRNAs [sncRNAs; 20–50 nucleotides (nt)], intermediate-sized ncRNAs (50–200 nt), and long ncRNAs (lncRNAs; >200 nt) (Hajieghrari and Farrokhi, 2022). MicroRNAs (miRNAs) comprise a well-studied subset of sncRNAs (Reinhart et al., 2002) whose biogenesis begins with the MIR genes (genes encoding miRNAs) inside the nucleus (Figure 1) (Guleria et al., 2011). Subsequently, these MIR genes are processed to form primary miRNAs (pri-miRNAs) and precursor miRNAs (pre-miRNAs) mediated by RNA polymerase II (RNA Pol II), ribonuclease III-like enzyme, and DICER-LIKE 1 (DCL1) (Guleria et al., 2011). Pre-miRNAs are small hairpin-shaped RNAs that are further processed by DCL1 to produce a mature miRNA duplex. This miRNA duplex contains miRNA and miRNA* strands. After transportation into the cytoplasm, the mature miRNA strand is loaded into the AGO (Argonaute) protein and forms a functional RNA-induced silencing complex (RISC) (Figure 1) (Wahid et al., 2010; Zhang et al., 2022a). Based on the degree of the miRNA–messenger RNA (mRNA) complementarity, miRNAs primarily mediate gene regulation in three different ways, namely, mRNA cleavage, translation inhibition, and expression silencing mechanism (Figure 1) (Zhang et al., 2022b). Another type of ncRNAs includes the lncRNAs, which are longer than 200 nt in length and lack a coding sequence or an open reading frame (ORF) (Wang et al., 2015; Rai et al., 2019). They are classified into three types based on their genomic location: intronic lncRNAs, antisense lncRNAs, and long intergenic ncRNAs (Figure 2A) (Ma et al., 2013; Fan et al., 2015). LncRNAs regulate plant development via affecting protein–protein interactions, interacting with miRNAs, and encoding small peptides, among others (Figures 2B–I) (Sun and Lin, 2019; Fan et al., 2020; Wierzbicki et al., 2021). Phased small interfering RNAs (phasiRNAs) are 21- or 24-nt sncRNAs that are produced in a phased pattern from the PHAS locus and are typically triggered by miRNAs (Fei et al., 2013). Intriguingly, phasiRNAs are specifically expressed in the reproductive organs and are associated with anther development and male fertility in plants, but their mechanism of function is not very clear (Xia et al., 2019).
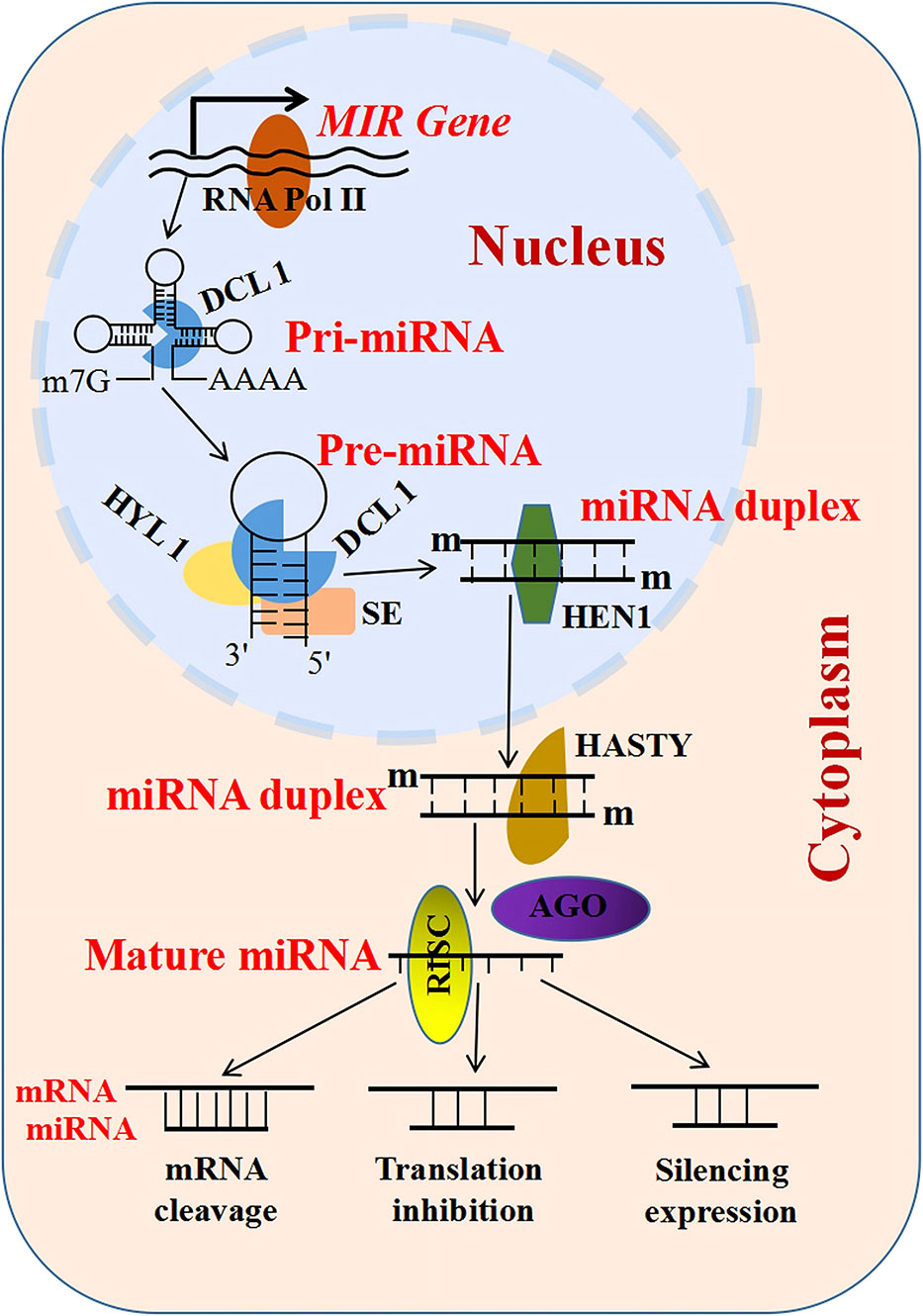
Figure 1 Characteristic features of microRNAs (miRNAs) and long non-coding RNAs (lncRNAs). Biosynthesis and mode of action of miRNAs: primary miRNAs (pri-miRNAs) self-fold and are complementarily paired to form one or more stable stem–loop structures, and are then processed by the ribonuclease III-like enzyme and DICER-LIKE 1 (DCL1) to generate a small hairpin-shaped RNA (precursor miRNA, pre-miRNA). The pre-miRNAs are further processed by DCL1 to produce a mature miRNA duplex. In this process, two proteins, HYPONASTIC LEAVES 1 (HYL1) and SERRATE (SE), also play important roles in the association with DCL1. HYL1 is a nuclear double-stranded RNA-binding protein, acting in the DCL1 complex to regulate the first step of the pri-miRNA process. SE encodes a C2H2 zinc finger protein, which is located in the nucleus and physically interacts with HYL1. RISC, RNA-induced silencing complex.
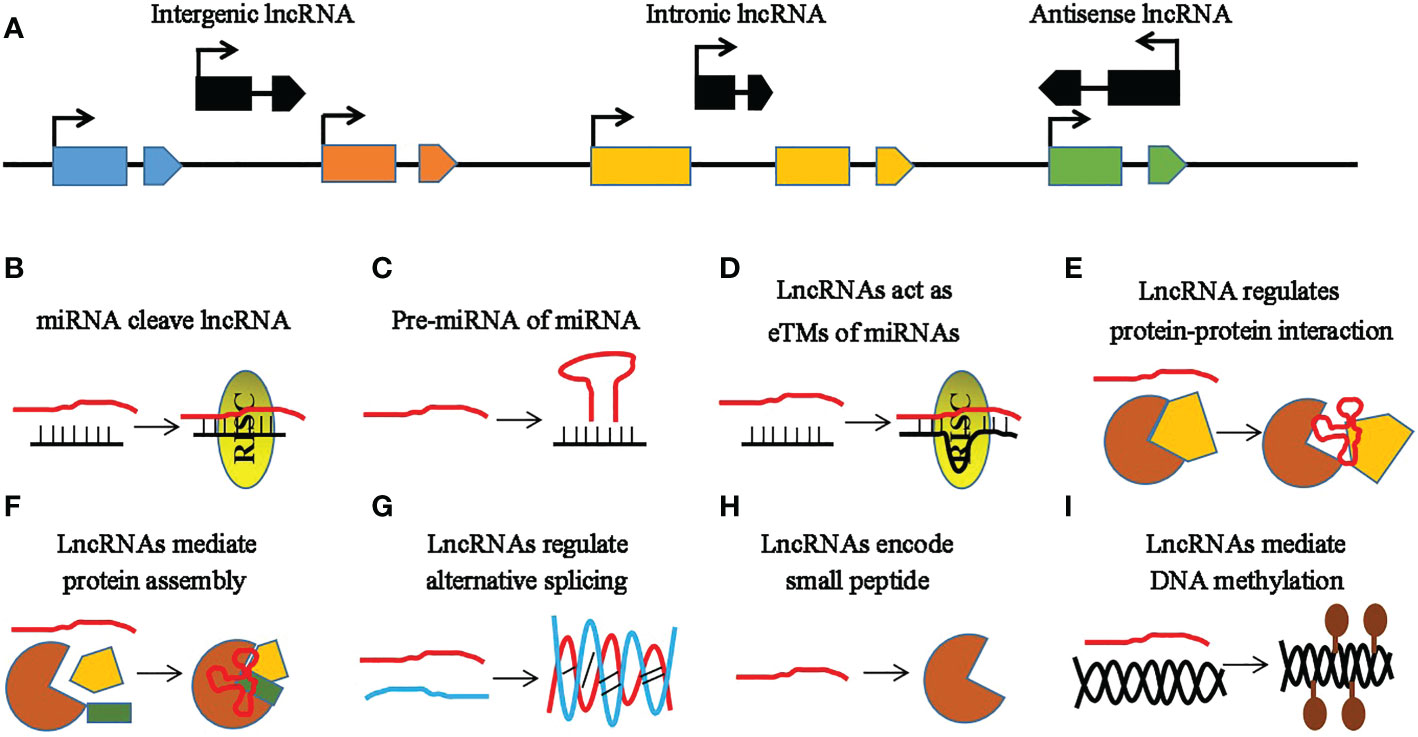
Figure 2 Characteristic features of long non-coding RNAs (lncRNAs). (A) Categories of lncRNAs based on their locations. The black line represents the chromosome, while the rectangles and polygons with similar colors represent the different exons of the protein-coding genes or lncRNAs. The blue, brown, yellow, and green rectangles and polygons represent different genes, while the black rectangles and polygons represent different lncRNAs. Intergenic, intronic, and antisense lncRNAs are the lncRNAs located outside the protein-coding genes, in the intron of a protein-coding gene, and those located in the antisense orientation to a protein-coding gene, respectively. Arrows indicate the direction of the gene or lncRNA. (B–I) Functional mechanisms of lncRNAs. The red line indicates the lncRNA, the black line represents the microRNA (miRNA), and the brown semicircle and yellow diagram indicate the proteins. (B) LncRNAs act as targets of miRNAs. (C) LncRNAs as precursor miRNAs (pre-miRNAs) of miRNAs. (D) LncRNAs as target mimics of miRNAs. (E) LncRNAs regulate protein–protein interactions. (F) LncRNAs as scaffolds regulating the assembly of protein complex subunits. (G) LncRNAs paired with their targeted RNAs. (H) LncRNAs encode small peptides. (I) LncRNAs contribute to epigenetic silencing via RNA-directed DNA methylation (RdDM).
A large number of ncRNAs have been elucidated as regulators of the transcription factors (TFs) or protein-coding genes that participate in the formation of male sterility (Mishra and Bohra, 2018; Dong et al., 2022; Yan and Ham, 2022). Their mechanisms of action are either dependent on or independent of plant hormones, which ultimately affect the degeneration of the tapetum, formation of microspores, and the release of mature pollen grains (Wei et al., 2015; Lei and Liu, 2020; Bohra et al., 2021; Han et al., 2021; Li et al., 2021). How do ncRNAs regulate the male-sterile phenotypes in plants? The functions of the new ncRNAs related to male sterility are summarized in this review.
Plant hormones are critical regulators of reproductive development, and inhibition of their synthesis and signal transduction will cause male sterility. Auxin was the first plant hormone to be recognized. The auxin signaling F-box (AFB) proteins, auxin response factors (ARFs), heat shock protein 90 (HSP90), and co-chaperone SGT1 (suppressor of the G2/M allele of skp1) are important moderators of auxin signaling transduction (Tian et al., 2017). ARF6 and ARF8 regulate the expression patterns of the genes that respond to auxin through binding to their promoter elements (Nagpal et al., 2005). The arf6 and arf8 double loss-of-function mutants showed flower arrest, stamen shortening, and anther indehiscence to release pollen grains (Nagpal et al., 2005). ARF17 is essential for pollen wall patterning in Arabidopsis through modulating primexine formation. The arf17 mutant did not have primexine within tetrads and exhibited a male-sterile phenotype (Yang et al., 2013). miRNAs, which target ARF, AP2, AFB2, HD-ZIP, and TIR, among others, regulate hormone metabolism and cause the expression of the pollen development genes to change, ultimately leading to male sterility (Figure 3) (Nie et al., 2018). Expression and function analysis revealed that the miR160/ARF16, miR393a/TIR1, and miR3444a/F-box interaction events participate in auxin signaling transduction and are important to CMS formation in radish (Zhang et al., 2016a). The same results were also found in male-sterile lines in cotton (Wei et al., 2013; Nie et al., 2018). Comparison of the small RNA expression profiles of the CMS line UPAS 120A and the isogenic fertile line UPAS 120B revealed 316 miRNAs and 961 target protein-coding genes, which include a variety of TFs, play key roles in plant reproduction, such as auxin and ethylene response factors. This result indicates that the signaling pathways activated by abscisic acid (ABA), auxin, etc., are important to the formation of male sterility (Bohra et al., 2021).
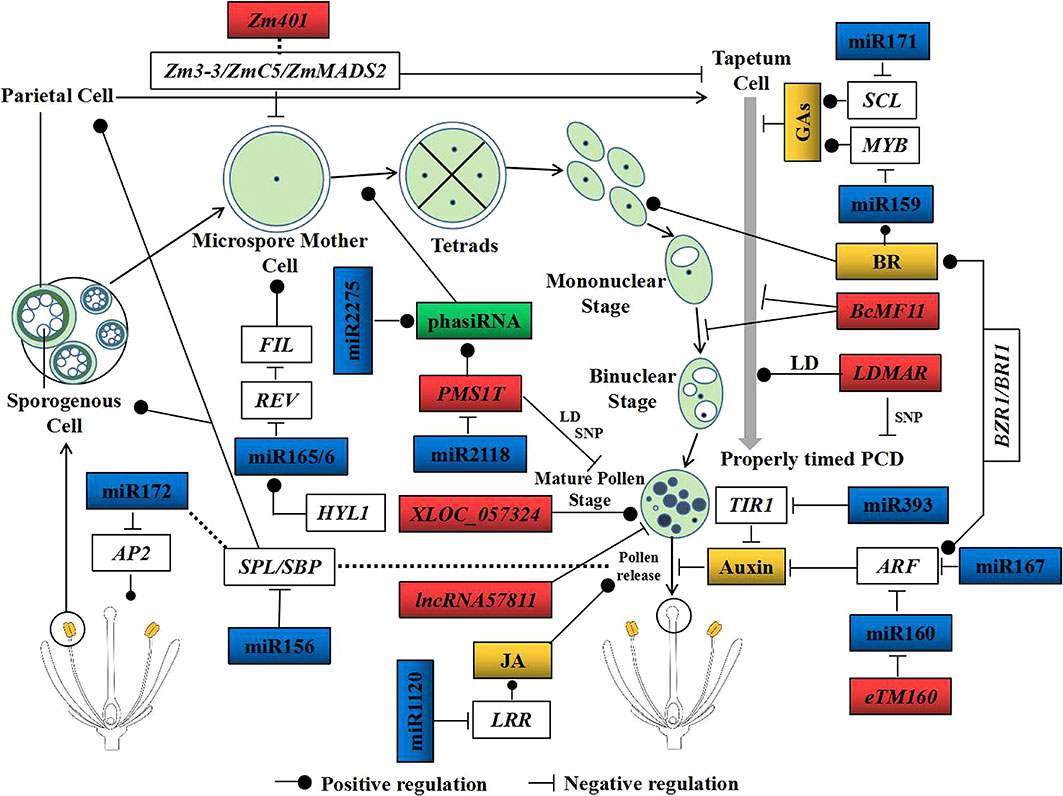
Figure 3 Critical non-coding RNAs (ncRNAs) participate in male organ development. Male organ development consists of six stages: the sporogenous cell stage, microspore cell stage, tetrad stage, mononuclear stage, binuclear stage, and the mature pollen stage. The blue rectangles represent the miRNAs that participate in male sterility, the red rectangles indicate the long non-coding RNAs (lncRNAs) related to pollen development in crops, the green rectangles indicate the phased small interfering RNAs (phasiRNAs) found in crops, and the yellow rectangles indicate the plant hormones involved in male organ development. NcRNAs participate in the dependence or non-dependence of male sterility on hormones. Among them, miR160, miR167, miR393, miR171, miR159, miR1120, and eTM160 regulate pollen development mediated by auxin, BRs, GAs, and JA. Others function by regulating the transcription factors or methylation levels. The dashed line denotes indirect regulation and control. MYB, MYB domain protein; SCL, scarecrow-like protein; ARF, auxin response factor; TIR, transport inhibitor response protein; SPL, SQUAMOSA promoter binding protein-like; FIL, filamentous flower; HYL1, hyponastic leaves 1; LD, long-day condition; SNP, single nucleotide polymorphism; BRs, brassinosteroids; JA, jasmonate; GAs, gibberellins.
Both miR160 and miR167 interact with ARFs and participate in the auxin signaling pathway (Wu et al., 2006; Ding et al., 2017). In high temperature (HT)-insensitive (84021) and HT-sensitive (H05) cotton cultivars, miR160 targeted ARF10, ARF16, and ARF17 (Ding et al., 2017). Overexpression of miR160 in cotton (e.g., in the miR160 OE2 line) resulted in higher miR160 and lower ARF10 and ARF17 expression, and the miR160 OE2 line was fertile, similar to the null control under normal temperature condition (Ding et al., 2017). However, after HT treatment, only the miR160 OE2 line showed anther indehiscence. Therefore, the overexpression of miR160 in cotton inhibited the expression levels of ARF10 and ARF17 and then activated auxin signaling, ultimately leading to male sterility under HT stress (Ding et al., 2017). Interestingly, the biosynthesis of jasmonic acid (JA) was suppressed in the anthers of the miR160 OE2 line under HT stress (Ding et al., 2017). In contrast to miR160, the overexpression of miR157 in cotton disrupted the signal transduction of auxin in ovules and young buds, generating indehiscent anthers and sterile pollen grains in the HT condition (Liu et al., 2017). miR167 has also been found to regulate the ARF TFs and cleave the ARF6 transcripts in vivo (Wu et al., 2006). mARF6 was obtained from the introduction of eight mutations into the miR167 target sites in the coding sequence of ARF6. When mARF6 was overexpressed in Arabidopsis, transgenic plants with higher ARF6 protein abundance and their flowers were sterile. Transforming the ARF6 overexpression vector into the wild-type Arabidopsis, although the ARF6 transgenic plants had ARF6 levels similar to or higher than those of mARF6 transgenic plants, the ARF6 transgenic plants showed fertile flowers (Wu et al., 2006). These results indicate that the loss of miR167/ARF6 interactions caused abortive pollen and that miR167 is essential for normal fertility in Arabidopsis. TamiR167a was upregulated in anthers during the pollen development processes in the photo-thermosensitive genic male-sterile (PTGMS) wheat line BS366, while its target TaARF8 was downregulated under sterile conditions (Wang et al., 2019a). The overexpression of miR167a in Arabidopsis resulted in the downregulation of both AtARF6 and AtARF8. Transgenic Arabidopsis showed fewer contents of endogenous JA and indole acetic acid (IAA), and their flowers were male sterile (Wang et al., 2019a).
The brassinosteroids (BRs) are a group of steroid hormones isolated from rape pollen that have high biological activity (Li and He, 2020; Nolan et al., 2020). BRASSINOSTEROID INSENSITIVE 1 (BRI1) is the receptor of BRs, and BES1/BZR1 are essential TFs in the BR signaling pathway (Yan et al., 2020). Although auxin and BR are two different hormones, their signaling transduction pathways interact and together regulate a variety of developmental processes in plants (Zakharova et al., 2022). BZR1 interacts with ARF proteins to directly target multiple auxin signaling components. In addition, ARFs could bind to the promoter of BRI1 and then activate BR signaling (Figure 3) (Tian et al., 2017). The BR biosynthesis mutant cpd, the responsive mutant bril-116, and the signal transduction mutants bin2-1 and qui-1 all presented male-sterile phenotypes (Ye et al., 2010; Chen et al., 2019). Leucine-rich repeat receptor-like kinase (LRR-RLK) interacts with BRI1 and modulates BR signaling. It plays an important role in plant developmental and physiological processes (Zhao et al., 2002). Some researchers have proven that LRR-RLKs participate in the differentiation of microsporocytes and tapetum cells, which eventually led to male sterility (Zhao et al., 2002). In the wheat thermosensitive cytoplasmic male-sterile line YS3038, TaeRPK (a LRR-RLK gene) was targeted by unconservative_chr3B_part2_17629 (belonging to the MIR9657 family) and was upregulated under sterile conditions compared to the fertile conditions. Silencing of TaeRPK using the VIGS (virus-induced gene silencing) method in YS3038 resulted in the seed setting rates being significantly lower than those of the negative control plants (Han et al., 2021). In general, gibberellins (GAs) and BRs play similar roles in plant developmental processes (Barro-Trastoy et al., 2020). BRs regulate the expression of miR159d and its target gene OsGAMYBL2 in rice (Gao et al., 2018). Furthermore, OsGAMYBL2 coordinates BR signaling and GA biosynthesis by regulating the expression of the BR signaling gene BU1 and two GA biosynthetic genes, CPS1 and GA3ox2 (Gao et al., 2018). GAs also participate in the formation of male sterility. The overexpression of gai (a GA signal transduction gene) using the reproductive tissue-specific promoter in Nicotiana tabacum and Arabidopsis resulted in the abortion of the male genital organ (Huang et al., 2003).
In addition, miRNAs and their targets mediate a complex regulatory network associated with the metabolism of auxin, cytokinin, ABA, and GAs and play important roles in cell division and flower induction in apple. Among them, miR396, miR160, and miR393 and their target genes are related to auxin, while miR159, miR319, and miR164 and their targets are involved in ABA and GA biosynthesis and signal transduction, eventually regulating bud growth and flower bud formation (Xing et al., 2016). The interaction events of miR172/AP2, miR156/SPL, and miR171/SCL participate in the GA/ABA and GA/auxin signaling pathways, further regulating the flowering time. Moreover, miR825 and miR1120 interact with their respective target genes (CaBP and LRR) to modulate pollen development via the CRY/PHY and JA signaling pathways (Bai et al., 2017).
Several miRNAs appear to be universally expressed and conserved among diverse angiosperms, while a small number is also conserved in bryophytes, lycopods, and gymnosperms (Axtell and Bowman, 2008). Conservative miRNAs are known to function in diverse development processes, and a proportion of them has been reported to participate in the development of male sterility. Research studies in cotton CMS have demonstrated that miR399, miR172, and miR393 were differentially expressed between wild-type and male-sterile anthers (Wei et al., 2013; Nie et al., 2018). Similar results have also been found in the soybean CMS line NJCMS1A and the wild-type line NJCMS1B (Ding et al., 2016b). To date, at least nine conserved miRNA families have been reported to regulate the male organ development, namely, miR156, miR159, miR160, miR164, miR166/165, miR167, miR169, miR172, and miR319 (Luo et al., 2013b; Spanudakis and Jackson, 2014; Sun et al., 2021). These miRNAs target various protein-coding genes and are involved in flower development processes independently of plant hormones (Figure 3).
miR156 is conserved in all angiosperms studied thus far, and its mature sequences from 40 species showed 75% similarity, such as in Arabidopsis, Gossypium hirsutum, N. tabacum, Oryza sativa, Triticum aestivum, and Zea mays (Sunkar and Jagadeeswaran, 2008). miR156 regulated 142 protein-coding genes, including three ATPase family AAA domain-containing proteins (e.g., ATD1A), four SQUAMOSA promoter binding protein-like (SPL), two calcium-transporting ATPases (ACA1 and ACA2), two cyclin-dependent kinases (e.g., CKB22), and a floral homeotic protein (MADS2) (Qu et al., 2021). Research in the soybean CMS line NJCMS1A and its restorer line NJCMS1C provided proof that the expression of miR156b is more than 1.5-fold greater in NJCMS1A, and the expression patterns of miR156b and SPL9 showed opposing trends in early-stage flower buds (Figure 3) (Ding et al., 2019). SPL interacted with FT (FLOWERING LOCUS T) and affected the flowering metabolic pathway (Schmid et al., 2003). In Arabidopsis seedlings, a higher miR156 and a lower SPL expression prevented precocious flowering. Subsequently, miR156 was downregulated and SPL was upregulated, which provided a permissive environment for flowering (Wang et al., 2009). miR156 targets all SPL genes, except for SPL8, and silencing the SPL8 gene caused a semi-sterile phenotype in Arabidopsis. Furthermore, the overexpression of miR156 in the spl8 mutant resulted in fully sterile plants, while overexpression of the miR156-resistant SPL could restore fertility of the spl8 mutant (Xing et al., 2011). In addition, miR172 was regulated by miR156 through SPL9, and the overexpression of miR172 in Arabidopsis promoted plant flowering (Figure 3) (Chen, 2004; Jung et al., 2007).
Members of the miR159 family have been found in Arabidopsis, O. sativa, T. aestivum, Z. mays, and other angiosperms with more than 80% sequence similarity. Their target genes are also conserved across different plant families (Luo et al., 2013b; Guo et al., 2021). In rice, miR159 and its putative targets (OsGAMYB and OsGAMYBL1) were co-expressed in reproductive organs, and the miR159-mediated OsGAMYB and OsGAMYBL1 mRNAs were degraded in anthers (Figure 3) (Tsuji et al., 2006). GAMYB has been proven to participate in flower development in many plants (Millar and Gubler, 2005). Overexpression of miR159a in rice inhibited the expression patterns of OsGAMYB and OsGAMYBL1, which finally led to abnormal flowers and a male-sterile phenotype (Tsuji et al., 2006). Moreover, GAMYB-like was also regulated by miR159 in Arabidopsis, wherein its overexpression led to the downregulation of AtMYB33 and caused anther defects and male sterility (Millar and Gubler, 2005).
Members of the miR164 family are completely identical among 28 species, and they are potentially capable of targeting the NAC TFs, including CUP-SHAPED COTYLEDON1 (CUC1) and CUC2 in Arabidopsis and EgNAM1 and PdNAM1 in palms (Luo et al., 2013b). miR164 was upregulated in pollen at the later developmental stages in sterile wheat plants, while CUC/NAM was downregulated in the sterile group, particularly in the later anther development stages (Bai et al., 2017). Osa-miR164d had higher expression in the meiosis stage of the photo-thermosensitive genic male-sterile (PTGMS) rice line WXS and showed negative correlation with its target, NAM. The pollen meiosis stage is important for the fertility transition of the WXS line, and the upregulation of osa-miR164d in this stage indicated that miR164d is critical to pollen development (Zhang et al., 2016b).
miR165/6 are involved in sporangium, microspore mother cell, and microspore development. Their mature sequences have 90% similarity among 36 species, and their target genes [Class III HOEMEODOMAIN LEUCINE ZIPPER (HD-ZIP III) TFs] are highly conserved in land plants (Luo et al., 2013b). REVOLUTA (REV) is a member of HD-ZIP III that has shown negative correlation with FILAMENTOUS FLOWER (FIL). REV regulates the establishment of anther polarity, while FIL is critical to the development of sporangium. HYPONASTIC LEAVES1 (HYL1) participates in miRNA biogenesis and plays a critical role in establishing the stamen architecture in Arabidopsis. HYL1 deficiency was shown to reduce the accumulation of miR165/6 and to increase the expression level of REV in anthers (Wu et al., 2007; Lian et al., 2013), further resulting in a decreased sporangium and even male infertility (Figure 3) (Lian et al., 2013).
Plant lncRNAs act as endogenous target mimics (eTMs) of miRNAs (Figures 2D and 3). Based on their incomplete binding to the miRNA seed regions, lncRNAs could block the interaction between the miRNAs and their authentic targets (Yoon et al., 2014; Meng et al., 2021). The lncRNA eTM160 acted as the eTM of miR160 and was specifically expressed in rice anthers during the meiotic and young microspore stages (Wang et al., 2017). miR160 targets ARF18, a gene that encodes an ARF and, as a transcriptional regulator, mediates auxin responses (Liu et al., 2015). When eTM160 (OE-eTM160) and the eTM160 mutant (OE-eTM160M) were overexpressed in rice, it was found that miR160 was downregulated, but ARF18 was upregulated, in OE-eTM160 plants; however, this phenomenon did not exist in OE-eTM160M plants. These results indicate that eTM160 blocked the binding of miR160 to ARF18 and overexpressed eTM160 in rice, resulting in the upregulation of ARF18. It is worth noting that the lemma and palea of OE-eTM160 failed to open and showed abnormal fertility during the reproductive development stage; a similar phenomenon also occurred in rice with overexpressed ARF18 (Wang et al., 2017). In B. rapa, the lncRNA bra-eTM160-2, which acted as the target mimic of miR160, was specifically and predominantly expressed in inflorescence. The overexpression of bra-eTM160-2 in B. rapa (OE-bra-eTM160-2) led to bra-miR160-5p being significantly decreased in inflorescence, while the target genes of miR160, including BrARF10, BrARF16-1, BrARF16-2, BrARF17-1, and BrARF17-2, were significantly increased. OE-bra-eTM160-2 plants possessed small and curled rosette leaves and showed symptoms of auxin deficiency. Although the floral organs of OE-bra-eTM160-2 appeared normal, the anthers started aborting from the uninucleate microspore stage and the microspores were hollow and collapsed, which finally caused the pollen grains to be small, shrunken, and without viability (Huang et al., 2018).
Based on transcriptome sequencing during anther development in the CMS-D2 line A, maintainer line B, and restorer-of-fertility line R in cotton, 328 and 431 differentially expressed lncRNAs were identified as unique in the A–B and A–R comparisons, respectively. TCONS_00807084, which was predicted to be a putative precursor of miR160b, showed higher abundance in the A line than in both the B and R lines. miR160b targeted GhARF17; therefore, it was speculated that miR160b and TCONS_00807084 might regulate anther development together through influencing the auxin metabolic pathway (Figure 3). In addition, based on the analysis of the putative miRNA–lncRNA–mRNA regulatory network, it was found that the interaction events, such as csi-miR160 negatively regulating Gh_D06G0360 and TCONS_01087328 and TCONS_00675896 positively regulating Gh_D02G0666, were important to CMS and fertility restoration through their involvement in auxin and GA metabolic processes in cotton (Zhang et al., 2019).
In summary, the lncRNA–miR160–ARF interaction networks might participate in pollen development by participating in auxin regulation in many plants.
Most plant miRNAs are generated from long non-coding precursors (Bielewicz et al., 2013), with similar results also found in animals (Raini et al., 2007). Generally, miRNAs are produced by the introns of lncRNAs, which suggests that the donor lncRNAs not only act as pre-miRNAs but also have additional roles mediated by exons (Bielewicz et al., 2013). In the rice photoperiod-sensitive male-sterile line Nongken 58S, LDMAR (long-day-specific male fertility-associated RNA) was important in pollen abortion. It could be processed into a 136-nt intermediate precursor firstly, then produce a 21-nt small RNA for function (Zhu and Deng, 2012). In addition, sufficient abundance of complete LDMAR transcripts is also important for pollen to develop normally under long-day conditions. Elevated methylation levels inhibited the transcription of LDMAR, specifically under long-day conditions, then led to abnormal degeneration of the tapetum due to premature programmed cell death (PCD), eventually causing photoperiod-sensitive male sterility (Ding et al., 2012). Analysis of the lncRNA expression profiles in the cotton CMS and the restorer (Rf) and maintainer line flower buds found that 1,531 lncRNAs and parts of them showed significant differential expression patterns between these three lines. Among them, 94 lncRNAs were identified as putative precursors of 49 miRNAs, indicating that the miRNA/lncRNA interaction networks probably function as a complex regulatory pathway during pollen development (Hamid et al., 2020).
Aside from interacting with miRNAs, lncRNAs regulate plant development through multifarious ways. In recent years, more and more reproduction organ-specific lncRNAs have been identified, and the number has surpassed that of protein-coding transcripts (Yang et al., 2020). These findings indicate the great potential of plant lncRNAs to function specifically in reproductive organs.
FSNR is a unique lncRNA identified from the MT genomes in the CMS line. It presented much higher expression and more edited sites in females than in hermaphrodites (Stone et al., 2017). Short open reading frame mRNA (sORF-mRNA) refers to the mRNA containing only short ORFs and encoding less than 100 amino acids. It is usually expressed in specific development stages (MacIntosh et al., 2001). Zm401 is a sORF-mRNA in maize that is specifically expressed in tapetal cells and in the microspores of pollen grains (Ma et al., 2008). The protein-coding genes Zm3-3, ZmC5, and ZmMADS2 have been proven to be important in pollen development. Silencing Zm401 in maize altered the expression patterns of ZmMADS2, Zm3-3, and ZmC5 and caused abnormal microspore and tapetum development (Figure 3) (Ma et al., 2008). BcMF11, a novel pollen-specific lncRNA from Chinese cabbage, delayed the degradation of the tapetum, caused asynchronous separation of the microspore, and aborted the development of pollen grains (Figure 3) (Song et al., 2013). Through strand-specific RNA sequencing (ssRNA-seq), Zhang et al. (2014) found that the lncRNA XLOC_057324 was specifically expressed in young panicles and pistils. Transfer DNA (T-DNA) insertion decreased the abundance of XLOC_057324 in rice, and the mutants were flowering earlier and were male sterile (Figure 3) (Zhang et al., 2014). A total of 444 differentially expressed lncRNAs were detected in rice anthers. Among them, TCONS_00057811 showed anther specificity and had the highest abundance during the meiosis stage (Figure 3) (Li et al., 2020). The overexpression of TCONS_00057811 in rice altered the expression pattern of LOC_Os12g41350 (Meiotic Asynaptic Mutant 1), a meiosis-related gene, which resulted in lower pollen fertility (29.70%) and seed setting (33%) than in control plants (Li et al., 2020). Interestingly, all of the aforementioned lncRNAs (Figure 3), which participated in the formation of male sterility through interacting with miRNAs, also presented tissue-specific expression. Apart from the well-studied lncRNAs, some lncRNAs also participate in male sterility, but with unknown mechanisms (Table 1).
Apart from being processed into miRNAs, lncRNAs have been identified as putative targets of miRNAs. After cleavage, lncRNAs form small intermediate RNAs to function. Similar to LDMAR, PMS1T (photoperiod-sensitive genic male sterility 1) was also specifically more highly expressed in young panicles in the rice photoperiod-sensitive male-sterile line (Fan et al., 2016). miR2118 is conserved in plants and is critical for inducing 21-nt phasiRNAs through the cleavage of lncRNAs. PMS1T is a target of miR2118 and could produce 21-nt-dominated phasiRNAs after cleavage. A sufficient number of phasiRNAs in panicles is necessary for the formation of male sterility under long-day conditions, but the mechanism is not well understood. A single nucleotide polymorphism (SNP) nearby the miR2118 target sites of PMS1T inhibited the action of miR2118 and reduced the abundance of phasiRNAs. A lower number of phasiRNAs eventually restored the male fertility in rice under long-day conditions (Fan et al., 2016).
In addition to the aforementioned miR2118–PMS1T–phasiRNA–target regulatory cascades, loss of miR2118 in rice was also demonstrated by Araki et al. (2020) to cause severe male and female sterility since the number of miR2118-dependent 21-nt phasiRNAs significantly decreased in the anther wall from stage 1 to stage 6. Through sncRNA and degradome sequencing of the developing flowers in the male-sterile cybrid pummelo (G1+HBP) and its fertile parent Hirado Buntan pummelo (HBP), a total of 2,235 unique 21-nt phasiRNAs were identified, with 88 of them showing significant expression differences in developing flowers between G1+HBP and HBP. Of these 88 phasiRNAs, 34 were generated from the protein-coding genes, while the others were produced from lncRNAs (Fang et al., 2020). In a derivative line of the male-sterile outer cell layer 4 (ocl4) mutant of maize, the biogenesis of the 21-nt phasiRNAs was largely dependent on Ocl4, and it was mediated by miR2118 (Yadava et al., 2020). Further research found that the accumulation levels and the fractions of phasiRNAs were the highest in the wild type, as expected, but were also significantly higher in the conditionally fertile ocl4 compared to the ocl4 sterile samples, suggesting partial restoration of the phasiRNAs in the conditionally fertile plants (Yadava et al., 2020). On the other hand, miR2118, miR390, miR472, miR3954, miR158, and miR482 could trigger the production of 21-nt phasiRNAs (Fang et al., 2020; Tripathi et al., 2021). miR2275 is the unique miRNA known to trigger the production of 24-nt-dominated phasiRNAs, and all of these trigger events preferentially occurred in the tapetum and meiocytes. miR2275 is specifically expressed in the tapetum during the early meiosis stage, and its expression is quickly reduced in other stages of male gametogenesis in both rice and maize (Tamim et al., 2018). In maize, the tapetum is the primary site of the 24-PHAS precursor and Dcl5 transcripts, as well as the resulting 24-nt phasiRNAs, and then the phasiRNAs are mobile from the tapetum to meiocytes and to other somatic cells (Zhou et al., 2022). Generally, maize male-sterile mutants with subepidermal defects lack 24-nt phasiRNAs (Zhai et al., 2015). Male fertility in maize requires all four basic helix–loop–helix genes, including Ms23, Ms32, basic helix–loop–helix 122 (bHLH122), and bHLH51. Interestingly, the male-sterile ms23, ms32, and bhlh122-1 mutants lack 24-nt phasiRNAs and the precursor transcripts from the corresponding 24-PHAS loci, while the bhlh51-1 mutant has wild-type levels of both precursors and small RNA products, suggesting that bHLH TFs regulate male sterility by directly binding to their target promoters or through their indirect regulation of the downstream TFs dependent on phasiRNAs (Nan et al., 2022).
Although ncRNAs play critical roles in male sterility formation, most of them are related to GMS. CMS is generally caused by the uncoordinated interactions between the MT and nuclear genomes (Bohra et al., 2016; Nie et al., 2020; Takatsuka et al., 2022). Plant MT genomes are dynamic genomes, the sequences of which frequently undergo rearrangement, recombination, structural variation, and exogenous or internal sequence migration (Chen et al., 2017). Previous studies have shown that CMS-associated genes were produced by dynamic changes in the MT genomes (Chen et al., 2017; Kim et al., 2019). To date, more than 30 CMS-related MT genes have been identified in different crop species (Kim and Zhang, 2018), including orfH79 in CMS-HL (Wang et al., 2013; Ding et al., 2016a), orf352 in CMS-RT102 (Okazaki et al., 2013), and orf138 in CMS-O (Bellaoui et al., 1998). Most of these genes have the following characteristics: 1) are located nearby the MT functional genes; 2) are associated and co-transcribed with the essential MT genes; 3) have chimeric ORFs; and 4) possess transmembrane domains (Chen et al., 2017; Li et al., 2018). Orf79 encodes a protein that includes 79 amino acids and contains an N-terminus with similarity to COX1 in cytoplasmic male-sterile rice. It is also co-transcribed with the upstream gene atp6 (Kazama et al., 2016). In CMS-T maize, compelling evidence has indicated that the MT gene T-urf13 (ORF 13 unique to the T cytoplasm of maize) is critical to CMS formation. T-urf13 is a unique chimeric sequence that comprises part of atp6 and encodes a nonessential amino acid (URF13) with an inner MT membrane domain (Dewey et al., 1987). In petunia CMS, pcf is located close to two MT genes, nad3 and rps72, and contains part of the coding region of atp9 and part of each exon of cox11, the same as T-urf13 (Young and Hanson, 1987).
Generally, four critical models of MT genes mediate CMS (Figure 4), and each CMS gene causes male sterility possibly through multiple mechanisms (Chen et al., 2017). Pollen development is a highly energy-consuming process, and a large number of research studies have indicated that CMS may be triggered by abnormal mitochondria that failed to provide enough energy for rapid cell differentiation (Chen and Liu, 2014). In the Honglian CMS system of rice, the MT protein ORFH79 interacted with the complex III QCR10 subunit homologous protein P61 and inhibited the activity of complex III, then causing male sterility due to the decrease in energy and the increase in ROS content (Wang et al., 2013). A properly timed PCD of the tapetum provides nutrients for pollen maturation; otherwise, a premature or delayed PCD leads to pollen abortion (Twell, 2011; Xie et al., 2022). In the wild-abortive CMS (CMS-WA) system of rice, WA352 interacted with the nuclear-encoded MT protein COX11, destroyed the function of COX11 in peroxide metabolism, and triggered premature tapetal PCD, consequently leading to pollen abortion (Luo et al., 2013a). The MT gene atp6c conferred male sterility in CMS-C maize, and it more strongly interacted with ATP8 and ATP9 than with ATP6 during the assembly of the F1Fo-ATP synthase (F-type ATP synthase, ATPase), thereby causing a reduction of the quantity and activity of F1Fo-ATP synthase. The reduced F1Fo-ATP synthase activity caused the accumulation of excess protons in the inner membrane space of the mitochondria, triggering a burst of ROS, premature PCD of the tapetal cells, and the abortion of pollen (Yang et al., 2022). Confirmed evidence from transgenic experiments has provided proof that the maize MT gene urf13 is toxic to cell viability (Levings, 1993). Retrograde regulation refers to the phenomenon of nuclear genes being regulated by organelle genes (Fujii et al., 2007). In the rice CMS-CW (rice hybrids obtained reciprocally between cultivated and wild varieties) system, MT genes acted as regulators of some nuclear genes that are preferentially expressed in male-sterile lines and related to alternative oxidase; the upregulation of these nuclear genes in anthers resulted in male sterility (Fujii et al., 2007). ZmDREB1.7, which showed higher expression in the sterile microspores in maize, activated the CMS-related MT gene orf355 and caused male sterility. Interestingly, orf355 also upregulated the expression pattern of ZmDREB1.7 through activating MT retrograde signaling. These results indicate that the retrograde regulation pattern between the nuclear regulator and the mitochondrial CMS gene plays a critical role in the formation of male sterility in maize (Xiao et al., 2020).
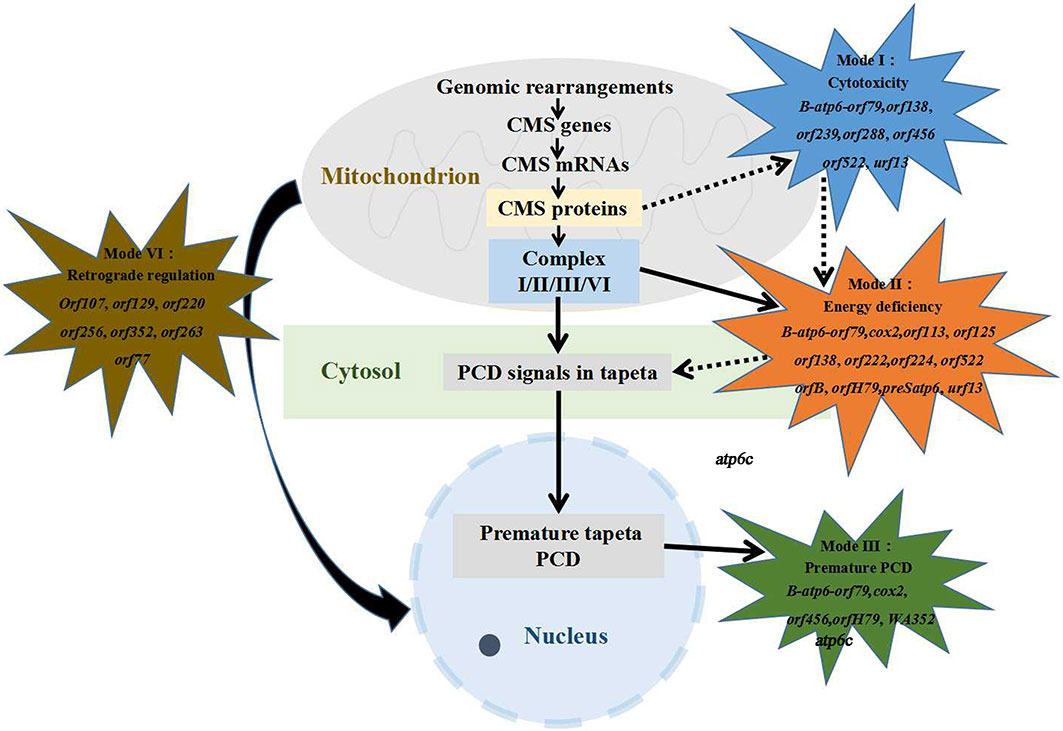
Figure 4 Four function mechanisms of cytoplasmic male sterility (CMS) genes. More than 30 mitochondrial genes have been found to participate in CMS formation, with several genes possibly possessing multiple mechanisms of action affecting male sterility. Some of the genes involved in CMS formation have unknown mechanisms. Dotted arrows represent the potential co-action between two modes.
Male sterility is a crucial attribute in the production of plant hybrids. To date, many ncRNAs have been proven to function in the formation of male sterility; however, most of them are related to GMS. To explore the mechanism of CMS, it is absolutely imperative to determine the large-scale ncRNAs encoded by the cytoplasmic genome and to reveal their roles in chimeric cytoplasmic genes. NcRNAs target hormone-responsive TFs and regulate plant male organ development by affecting hormone homeostasis, such as auxin, BRs, GAs, ABA, and JA (Xing et al., 2016; Bai et al., 2017). However, the development of male-sterile lines using exogenous hormone analogues is both an opportunity and a challenge. The rapid development of sequencing techniques has made it easier to discover ncRNAs (Li, 2020), and the CRISPR/Cas9 editing system has been used in the development of male sterility in plants (Jiang et al., 2021). Advancements in technology have offered new opportunities to apply the CRISPR/Cas9 system to reveal the functions of ncRNA mutants (Goyal et al., 2017; Wang et al., 2019b), which will be beneficial for exploring the mechanisms of ncRNAs in male sterility and will be conducive to creating new sterile lines using the transgenic method.
All authors listed have made direct contributions to the work and approved it for publication. JH, HN, CC, and HL wrote parts of the manuscript. JH, HN, and JK contributed to language revision and figure design. All authors contributed to the article and approved the submitted version.
This work was supported in part by the National Natural Science Foundation of China (31671741) and the National Key R&D Program for Crop Breeding (2016YFD0101407) to JH.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Araki, S., Le, N. T., Koizumi, K., Villar-Briones, A., Nonomura, K., Endo, M., et al. (2020). miR2118-dependent U-rich phasiRNA production in rice anther wall development. Nat. Commun. 11, 3115–3128. doi: 10.1038/s41467-020-16637-3
Axtell, M. J., Bowman, J. L. (2008). Evolution of plant microRNAs and their targets. Trends Plant Sci. 13, 343–349. doi: 10.1016/j.tplants.2008.03.009
Bai, J. F., Wang, Y. K., Wang, P., Duan, W. J., Yuan, S. H., Sun, H., et al. (2017). Uncovering male fertility transition responsive miRNA in a wheat photo-thermosensitive genic male sterile line by deep sequencing and degradome analysis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01370
Barro-Trastoy, D., Carrera, E., Baos, J., Palauodríguez, J., Miguel, A. (2020). Regulation of ovule initiation by gibberellins and brassinosteroids in tomato and Arabidopsis, two plant species., two molecular mechanisms. Plant J. 102, 1026–1041. doi: 10.1111/tpj.14684
Bellaoui, M., Martin-Canadell, A., Pelletier, G., Budar, F. (1998). Low copy number molecules are produced by recombination., actively maintained and can be amplified in the mitochondrial genome of Brassicaceae. relationship to reversion of the male sterile phenotype in some cybrids. Mol. Genet. Genomics 257, 177–185. doi: 10.1007/s004380050637
Bielewicz, D., Kalak, M., Kalyna, M., Windels, D., Barta, A., Vazquez, F., et al. (2013). Introns of plant pri-miRNAs enhance miRNA biogenesis. EMBO Rep. 14, 622–628. doi: 10.1038/embor.2013.62
Bohra, A., Gandham, P., Rathore, A., Thakur, V., Saxena, R. K., Satheesh, Naik, S. J., et al. (2021). Identifcation of microRNAs and their gene targets in cytoplasmic male sterile and fertile maintainer lines of pigeonpea. Planta 253, 59–73. doi: 10.1007/s00425-021-03568-6
Bohra, A., Jha, U. C., Adhimoolam, P., Bisht, D., Singh, N. (2016). Cytoplasmic male sterility (CMS) in hybrid breeding in field crops. Plant Cell Rep. 35, 967–993. doi: 10.1007/s00299-016-1949-3
Brownfield, L. (2021). Plant breeding: Revealing the secrets of cytoplasmic male sterility in wheat. Curr. Biol. 31, 724–726. doi: 10.1016/j.cub.2021.04.026
Chao, H., Hu, Y., Zhao, L., Xin, S., Ni, Q., Zhang, P., et al. (2022). Biogenesis, functions, interactions, and resources of non-coding RNAs in plants. Int. J. Mol. Sci. 23, 3695–3718. doi: 10.3390/ijms23073695
Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. doi: 10.1126/science.1088060
Chen, L., Liu, Y. G. (2014). Male Sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 65, 579–606. doi: 10.1146/annurev-arplant-050213-040119
Chen, W., Lv, M., Wang, Y., Wang, P. A., Cui, Y. W., Li, M. Z., et al. (2019). BES1 is activated by EMS1-TPD1-SERK1/2-mediated signaling to control tapetum development in Arabidopsis thaliana. Nat. Commun. 10, 1–12. doi: 10.1038/s41467-019-12118-4
Chen, Z., Zhao, N., Li, S., Grover, C. E., Nie, H., Wendel, J. F., et al. (2017). Plant mitochondrial genome evolution and cytoplasmic male sterility. Crit. Rev. Plant Sci. 36, 54–69. doi: 10.1080/07352689.2017.1327762
Cho, J., Koo, D., Nam, Y., Han, C., Lim, H., Bang, J., et al (2006). Isolation and characterization of cDNA clones expressed under male sex expression conditions in a monoecious cucumber plant (Cucumis sativus L. cv. Winter Long). Euphytica 146 (3), 271–281.
Csorba, T., Questa, J. I., Sun, Q., Dean, C. (2014). Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. P Natl. Acad. Sci. U.S.A. 111, 16160–16165. doi: 10.1073/pnas.1419030111
Dewey, R. E., Timothy, D. H., Levings, C. S. (1987). A mitochondrial protein associated with cytoplasmic male sterility in the T cytoplasm of maize. P Natl. Acad. Sci. U.S.A. 84, 534–5378. doi: 10.1073/pnas.84.15.5374
Ding, X., Chen, Q., Bao, C., Ai, A., Zhou, Y., Li, S., et al. (2016a). Expression of a mitochondrial gene orfH79 from CMS honglian rice inhibits Escherichia coli growth via deficient oxygen consumption. SpringerPlus 5, 1125–1131. doi: 10.1186/s40064-016-2822-0
Ding, X. L., Li, J. J., Zhang, H., He, T. T., Han, S. H., Li, Y. W., et al. (2016b). Identification of miRNAs and their targets by high-throughput sequencing and degradome analysis in cytoplasmic male-sterile line NJCMS1A and its maintainer NJCMS1B of soybean. BMC Genomics 17, 24–39. doi: 10.1186/s12864-015-2352-0
Ding, J., Lua, Q., Ouyanga, Y., Mao, H., Zhang, P., Yao, J., et al. (2012). A long noncoding RNA regulates photoperiod-sensitive male sterility., an essential component of hybrid rice. P Natl. Acad. Sci. U.S.A. 109, 2654–2659. doi: 10.1073/pnas.1121374109
Ding, Y., Ma, Y., Liu, N., Xu, J., Hu, Q., Li, Y., et al. (2017). microRNAs involved in auxin signaling modulate male sterility under high-temperature stress in cotton (Gossypium hirsutum). Plant J. 91, 977–994. doi: 10.1111/tpj.13620
Ding, X., Zhang, H., Ruan, H., Li, Y., Chen, L., Wang, T., et al. (2019). Exploration of miRNA-mediated fertility regulation network of cytoplasmic male sterility during flower bud development in soybean. 3 Biotech. 9, 22–35. doi: 10.1007/s13205-018-1543-1
Dong, Q., Hu, B., Zhang, C. (2022). microRNAs and their roles in plant development. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.824240
Fan, C., Hao, Z., Yan, J., Li, G. (2015). Genome-wide identification and functional analysis of lincRNAs acting as miRNA targets or decoys in maize. BMC Genomics 16, 793–811. doi: 10.1186/s12864-015-2024-0
Fan, Y., Yang, J., Mathioni, S. M., Yu, J., Shen, J., Yang, X., et al. (2016). PMS1T, producing phased small-interfering RNAs., regulates photoperiod-sensitive male sterility in rice. P Natl. Acad. Sci. U.S.A. 113, 15144–15149. doi: 10.1073/pnas.1619159114
Fan, T., Zhang, Q., Hu, Y., Wang, Z., Huang, Y. (2020). Genome-wide identification of lncRNAs during hickory (Carya cathayensis) flowering. Funct. Integr. Genomic. 20, 591–607. doi: 10.1007/s10142-020-00737-w
Fang, Y. N., Yang, X. M., Jiang, N., Wu, X. M., Guo, W. W. (2020). Genome-wide identification and expression profiles of phased siRNAs in a male-sterile somatic cybrid of pummelo (Citrus grandis). Tree Genet. Genomes. 16, 46–60. doi: 10.1007/s11295-020-01437-z
Fei, Q., Xia, R., Meyers, B. C. (2013). Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25, 2400–2415. doi: 10.1105/tpc.113.114652
Fujii, S., Komatsu, S., Toriyama, K. (2007). Retrograde regulation of nuclear gene expression in CW-CMS of rice. Plant Mol. Biol. 63, 405–417. doi: 10.1007/s11103-006-9097-8
Gao, J., Chen, H., Yang, H., He, Y., Tian, Z., Li, J. (2018). A brassinosteroid responsive miRNA target module regulates gibberellin biosynthesis and plant development. New Phytol. 220, 488–501. doi: 10.1111/nph.15331
Goyal, A., Myacheva, K., Groß, M., Klingenberg, M., Arqué, B., Diederichs, S. (2017). Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 45, 12–24. doi: 10.1093/nar/gkw883
Guleria, P., Mahajan, M., Bhardwaj, J., Yadav, S. K. (2011). Plant small RNAs, biogenesis, mode of action and their roles in abiotic stresses. Genom Proteom Bioinf. 9, 183–199. doi: 10.1016/S1672-0229(11)60022-3
Guo, C. K., Jiang, Y. Q., Shi, M., Wu, X., Wu, G. (2021). ABI5 acts downstream of miR159 to delay vegetative phase change in Arabidopsis. New Phytol. 231, 339–350. doi: 10.1111/nph.17371
Hajieghrari, B., Farrokhi, N. (2022). Plant RNA-mediated gene regulatory network. Genomics 114, 409–442. doi: 10.1016/j.ygeno.2021.12.020
Hamid, R., Jacob, F., Marashi, H., Rathod, V., Tomar, R. S. (2020). Uncloaking lncRNA-meditated gene expression as a potential regulator of CMS in cotton (Gossypium hirsutum l.). Genomics 112, 3354–3364. doi: 10.1016/j.ygeno.2020.06.027
Han, Y. C., Zhang, Y. Y., Cao, G. N., Shao, L. L., Ding, Q., Ma, J. (2021). Dynamic expression of miRNAs and functional analysis of target genes involved in the response to male sterility of the wheat line YS3038. Plant Physiol. Bioch. 162, 363–377. doi: 10.1016/j.plaphy.2021.02.047
Heo, J. B., Sung, S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79. doi: 10.1126/science.1197349
Huang, S., Cerny, R. E., Qi, Y. L., Bhat, D., Aydt, C. M., Hanson, D. D., et al. (2003). Transgenic studies on the involvement of cytokinin and gibberellin in male development. Plant Physiol. 131, 1270–1282. doi: 10.1104/pp.102.018598
Huang, L., Dong, H., Zhou, D., Li, M., Liu, Y., Zhang, F., et al. (2018). Systematic identification of long non-coding RNAs during pollen development and fertilization in Brassica rapa. Plant J. 96, 203–222. doi: 10.1111/tpj.14016
Huang, J. Z., E, Z. G., Zhang, H. L., Shu, Q. Y. (2014). Workable male sterility systems for hybrid rice, genetics, biochemistry, molecular biology, and utilization. Rice 7, 13–26. doi: 10.1186/s12284-014-0013-6
Jiang, Y. L., An, X. L., Li, Z. W., Yan, T. W., Zhu, T. T., Xie, K., et al. (2021). CRISPR/Cas9-based discovery of maize transcription factors regulating male sterility and their functional conservation in plants. Plant Biotechnol. J. 19, 1769–1784. doi: 10.1111/pbi.13590
Jung, J. H., Seo, Y. H., Seo, P. J., Reyes, J. L., Yun, J., Chua, N., et al. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19, 2736–2748. doi: 10.1105/tpc.107.054528
Kazama, T., Itabashi, E., Fujii, S., Nakamura, T., Toriyama, K. (2016). Mitochondrial ORF79 levels determine pollen abortion in cytoplasmic male sterile rice. Plant J. 85, 707–716. doi: 10.1111/tpj.13135
Kim, B., Yang, T. J., Kim, S. (2019). Identification of a gene responsible for cytoplasmic male-sterility in onions (Allium cepa l.) using comparative analysis of mitochondrial genome sequences of two recently diverged cytoplasms. Theor. Appl. Genet. 132, 313–322. doi: 10.1007/s00122-018-3218-z
Kim, Y. J., Zhang, D. (2018). Molecular control of male fertility for crop hybrid breeding. Trends Plant Sci. 23, 53–65. doi: 10.1016/j.tplants.2017.10.001
Laser, K. D., Lersten, N. R. (1972). Anatomy and cytology of microsporogenesis in cytoplasmic male sterile angiosperms. Bot. Rev. 38, 425–454. doi: 10.1007/BF02860010
Lei, X., Liu, B. (2020). Tapetum-dependent male meiosis progression in plants, increasing evidence emerges. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01667
Levings, C. S. (1993). Thoughts on cytoplasmic male sterility in CMS-T maize. Plant Cell 5, 1285–1290. doi: 10.1105/tpc.5.10.1285
Li, F. (2020). Towards complete deconstruction of cotton transcriptional landscape. J. Cotton Res. 3, 8–10. doi: 10.1186/s42397-020-00050-8
Li, M., Chen, L., Khan, A., Kong, X., Khan, M. R., Rao, M. J., et al. (2021). Transcriptome and miRNAomics analyses identify genes associated with cytoplasmic male sterility in cotton (Gossypium hirsutum l.). Int. J. Mol. Sci. 22, 4684–4697. doi: 10.3390/ijms22094684
Li, S., Chen, Z., Zhao, N., Wang, Y., Nie, H., Hua, J. (2018). The comparison of four mitochondrial genomes reveals cytoplasmic male sterility candidate genes in cotton. BMC Genomics 19, 775–789. doi: 10.1186/s12864-018-5122-y
Li, Z., He, Y. (2020). Roles of brassinosteroids in plant reproduction. Int. J. Mol. Sci. 21, 872–888. doi: 10.3390/ijms21030872
Li, X., Shahid, M. Q., Wen, M., Chen, S., Yu, H., Jiao, Y., et al. (2020). Global identification and analysis revealed differentially expressed lncRNAs associated with meiosis and low fertility in autotetraploid rice. BMC Plant Biol. 20, 82–101. doi: 10.1186/s12870-020-2290-0
Lian, H., Li, X., Liu, Z., He, Y. (2013). HYL1 is required for establishment of stamen architecture with four microsporangia in Arabidopsis. J. Exp. Bot. 64, 3397–3410. doi: 10.1093/jxb/ert178
Liu, J., Hua, W., Hua, Z., Yang, H., Zhang, L., Li, R., et al. (2015). Natural variation in ARF18 gene simultaneously affects seed weight and silique length in polyploid rapeseed. P Natl. Acad. Sci. U.S.A. 112, 5123–5132. doi: 10.1073/pnas.1502160112
Liu, N., Tu, L., Wang, L., Hu, H., Xu, J., Zhang, X. (2017). MicroRNA 157-targeted SPL genes regulate floral organ size and ovule production in cotton. BMC Plant Biol. 17, 7–22. doi: 10.1186/s12870-016-0969-z
Luo, Y., Guo, Z., Li, L. (2013b). Evolutionary conservation of microRNA regulatory programs in plant flower development. Dev. Biol. 380, 133–144. doi: 10.1016/j.ydbio.2013.05.009
Luo, D., Xu, H., Liu, Z., Guo, J., Li, H., Chen, L., et al. (2013a). A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat. Genet. 45, 573–577. doi: 10.1038/ng.2570
Ma, L., Bajic, V. B., Zhang, Z. (2013). On the classification of long non-coding RNAs. RNA Biol. 10, 925–933. doi: 10.4161/rna.24604
Ma, J., Yan, B., Qu, Y., Qin, F., Yang, Y., Hao, X., et al. (2008). Zm401, a short-open reading-frame mRNA or noncoding RNA, is essential for tapetum and microspore development and can regulate the floret formation in maize. J. Cell. Biochem. 105, 136–146. doi: 10.1002/jcb.21807
MacIntosh, G. C., Wilkerson, C., Green, P. J. (2001). Identification and analysis of Arabidopsis expressed sequence tags characteristic of non-coding RNAs. Plant Physiol. 127, 765–776. doi: 10.2307/4280139
Mayr, E. (1986). Joseph Gottlieb kolreuter’s contributions to biology. Osiris 2, 135–176. doi: 10.1086/368655
Meng, X., Li, A., Yu, B., Li, S. (2021). Interplay between miRNAs and lncRNAs: Mode of action and biological roles in plant development and stress adaptation. Compu Struct. Biotec. 19, 2567–2574. doi: 10.1016/j.csbj.2021.04.062
Millar, A. A., Gubler, F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17, 705–721. doi: 10.1105/tpc.104.027920
Mishra, A., Bohra, A. (2018). Non-coding RNAs and plant male sterility, current knowledge and future prospects. Plant Cell Rep. 37, 177–191. doi: 10.1007/s00299-018-2248-y
Nagpal, P., Ellis, C. M., Weber, H., Ploense, S. E., Barkawi, L. S., Guilfoyle, T. J., et al. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132, 4107–4118. doi: 10.1242/dev.01955
Nan, G. L., Teng, C., Fernandes, J., O'Connor, L., Meyers, B. C., Walbot, V. (2022). A cascade of bhlh-regulated pathways programs maize anther development. Plant Cell 34, 1207–1225. doi: 10.1093/plcell/koac007
Nie, H. S., Cheng, C., Hua, J. P. (2020). Mitochondrial proteomic analysis reveals that proteins relate to oxidoreductase activity play a central role in pollen fertility in cotton. J. Proteomics. 225, 103861–103875. doi: 10.1016/j.jprot.2020.103861
Nie, H. S., Wang, Y. M., Su, Y., Hua, J. P. (2018). Exploration of miRNAs and target genes of cytoplasmic male sterility line in cotton during flower bud development. Funct. Integr. Genomic. 18, 457–476. doi: 10.1007/s10142-018-0606-z
Nolan, T. M., Vukašinović, N., Liu, D., Russinova, E., Yin, Y. (2020). Brassinosteroids, multidimensional regulators of plant growth, development, and stress responses. Plant Cell 32, 295–318. doi: 10.1105/tpc.19.00335
Okazaki, M., Kazama, T., Murata, H., Motomura, K., Toriyama, K. (2013). Whole mitochondrial genome sequencing and transcriptional analysis to uncover an RT102-type cytoplasmic male sterility-associated candidate gene derived from Oryza rufipogon. Plant Cell Physiol. 54, 1560–1568. doi: 10.1093/pcp/pct102
Parodi, P. C., Gaju, M. D. (2009). Male Sterility induced by the chemical hybridizing agent clofencet on wheat., Triticum aestivum and T. turgidum var. durum. Cien Investigación Agraria 36, 267–276. doi: 10.4067/S0718-16202009000200011
Qu, H., Liu, Y., Jiang, H., Liu, Y., Chen, L. (2021). Identification and characterization of miRNAs associated with sterile flower buds in the tea plant based on small RNA sequencing. Hereditas 158, 26–44. doi: 10.1186/s41065-021-00188-8
Rai, M. I., Alam, M., Lightfoot, D. A., Gurha, P., Afzal, A. J. (2019). Classification and experimental identification of plant long non-coding RNAs. Genomics 111, 997–1005. doi: 10.1016/j.ygeno.2018.04.014
Raini, H. K., Griffiths-Jones, S., Enright, A. J. (2007). Genomic analysis of human microRNA transcripts. P Natl. Acad. Sci. U.S.A. 104, 17719–17724. doi: 10.1073/pnas.0703890104
Reinhart, B. J., Weinstein, E. G., Rhoades, M. W., Bartel, B., Bartel, D. P. (2002). MicroRNAs in plants. Gene Dev. 16, 1616–1626. doi: 10.1101/gad.1004402
Schmid, M., Uhlenhaut, N. H., Godard, F., Demar, M., Bressan, R., Weigel, D., et al. (2003). Dissection of floral induction pathways using global expression analysis. Development 130, 6001–6012. doi: 10.1242/dev.00842
Song, J., Cao, J., Wang, C. (2013). BcMF11, a novel non-coding RNA gene from Brassica campestris, is required for pollen development and male fertility. Plant Cell Rep. 32, 21–30. doi: 10.1007/s00299-012-1337-6
Spanudakis, E., Jackson, S. (2014). The role of microRNAs in the control of flowering time. J. Exp. Bot. 65, 365–380. doi: 10.1093/jxb/ert453
Stone, J. D., Koloušková, P., Sloan, D. B., Štorchová, H. (2017). Non-coding RNA may be associated with cytoplasmic malesterility in Silene vulgaris. J. Exp. Bot. 68, 1599–1612. doi: 10.1093/jxb/erx057
Sun, L., Lin, J. D. (2019). Function and mechanism of long noncoding RNAs in adipocyte biology. Diabetes 68, 887–896. doi: 10.2337/dbi18-0009
Sun, Y., Xiong, X., Wang, Q., Zhu, L., Wang, L., He, Y., et al. (2021). Integrated analysis of small RNA, transcriptome, and degradome sequencing reveals the miR156, miR5488 and miR399 are involved in the regulation of male sterility in PTGMS rice. Int. J. Mol. Sci. 22, 2260–2287. doi: 10.3390/ijms22052260
Sunkar, R., Jagadeeswaran, G. (2008). In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol. 8, 37–50. doi: 10.1186/1471-2229-8-37
Takatsuka, A., Kazama, T., Arimura, S., Toriyama, K. (2022). TALEN-mediated depletion of the mitochondrial gene orf312 proves that it is a tadukan-type cytoplasmic male sterilitycausative gene in rice. Plant J. 110, 994–1004. doi: 10.1111/tpj.15715
Tamim, S., Cai, Z., Mathioni, S., Zhai, J., Meyers, B. C.. (2018). Cis-directed cleavage and nonstoichiometric abundances of 21-nt reproductive phasiRNAs in grasses. New phytol 220 (3), 865–877. doi: 10.1101/243907
Tian, H., Lv, B., Ding, T., Bai, M., Ding, Z. (2017). Auxin-BR interaction regulates plant growth and development. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02256
Tripathi, M., Singh, R., Singh, A., Verma, A., Mishra, P., Narayan, S., et al. (2021). Abolished miR158 activity leads to 21-nucleotide tertiary phasiRNA biogenesis that targets NHX2 in arabidopsis thaliana (Cold Spring Harbor Laboratory). bioRxiv. doi: 10.1101/2021.01.27.428373
Tsuji, H., Aya, K., Ueguchi-Tanaka, M., Shimada, Y., Nakazono, M., Watanabe, R., et al. (2006). GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J. 47, 427–444. doi: 10.1111/j.1365-313X.2006.02795.x
Twell, D. (2011). Male Gametogenesis and germline specification in flowering plants. Sex Plant Reprod. 24, 149–160. doi: 10.1007/s00497-010-0157-5
Wahid, F., Shehzad, A., Khan, T., Kim, Y. (2010). MicroRNAs, synthesis, mechanism, function, and recent clinical trials. BBA-Mol Cell Res. 1803, 1231–1243. doi: 10.1016/j.bbamcr.2010.06.013
Wang, J., Czech, B., Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. doi: 10.1016/j.cell.2009.06.014
Wang, Y., Duan, W., Bai, J., Wang, P., Yuan, S., Zhao, C., et al. (2019a). Constitutive expression of a wheat microRNA, TaemiR167a, confers male sterility in transgenic Arabidopsis. Plant Growth Regul. 88, 227–239. doi: 10.1007/s10725-019-00503-4
Wang, K., Gao, F., Ji, Y., Liu, Y., Dan, Z., Yang, P., et al. (2013). ORFH79 impairs mitochondrial function via interaction with a subunit of electron transport chain complex III in honglian cytoplasmic male sterile rice. New Phytol. 198, 408–418. doi: 10.1111/nph.12180
Wang, C., Lu, T., Emanuel, G., Babcock, H. P., Zhuang, X. (2019b). Imaging-based pooled CRISPR screening reveals regulators of lncRNA localization. P Natl. Acad. Sci. U.S.A. 116, 10842–10851. doi: 10.1073/pnas.1903808116
Wang, M., Wu, H., Fang, J., Chu, C., Wang, X. (2017). A long noncoding RNA involved in rice reproductive development by negatively regulating osa-miR160. Sci. Bull. 62, 470–475. CNKI:SUN:JXTW.0.2017-07-006. doi: 10.1016/j.scib.2017.03.013
Wang, M., Yuan, D., Tu, L., Gao, W., He, Y., Hu, H., et al. (2015). Long noncoding RNAs and their proposed functions in fibre development of cotton (Gossypium spp.). New Phytol. 207, 1181–1197. doi: 10.1111/nph.13429
Wei, M., Wei, H., Wu, M., Song, M., Zhang, J., Yu, J., et al. (2013). Comparative expression profiling of miRNA during anther development in genetic male sterile and wild type cotton. BMC Plant Biol. 13, 66–79. doi: 10.1186/1471-2229-13-66
Wei, X., Zhang, X., Yao, Q., Yuan, Y., Li, X., We, F., et al. (2015). The miRNAs and their regulatory networks responsible for pollen abortion in ogura-CMS Chinese cabbage revealed by high-throughput sequencing of miRNAs, degradomes, and transcriptomes. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00894
Wierzbicki, A., Blevins, T., Swiezewski, S. (2021). Long noncoding RNAs in plants. Annu. Rev. Plant Biol. 72, 245–271. doi: 10.1146/annurev-arplant-093020-035446
Wu, M. F., Tian, Q., Reed, J. W. (2006). Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211–4218. doi: 10.1242/dev.02602
Wu, F., Yu, L., Cao, W., Mao, Y., Liu, Z., He, Y. (2007). The n-terminal double stranded RNA binding domains of Arabidopsis HYPONASTIC LEAVES1 are sufficient for pre-microRNA processing. Plant Cell 19, 914–925. doi: 10.1105/tpc.106.048637
Xia, R., Chen, C., Pokhrel, S., Ma, W., Huang, K., Patel, P., et al. (2019). 24-nt reproductive phasiRNAs are broadly present in angiosperms. Nat. Commun. 10, 627–634. doi: 10.1038/s41467-019-08543-0
Xiao, S., Zang, J., Pei, Y., Liu, J., Chen, H. (2020). Activation of mitochondrial orf355 gene expression by a nuclear-encoded DREB transcription factor causes cytoplasmic male sterility in maize. Mol. Plant 13, 1–14. doi: 10.1016/j.molp.2020.07.002
Xie, F., Vahldick, H., Lin, Z., Nowack, M. (2022). Killing me softly-programmed cell death in plant reproduction from sporogenesis to fertilization. Curr. Opin. Plant Biol. 69, 102271. doi: 10.1016/j.pbi.2022.102271
Xing, S., Salinas, M., Höhmann, S., Berndtgen, R., Huijser, P. (2011). miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22, 3935–3950. doi: 10.1105/tpc.110.079343
Xing, L., Zhang, D., Zhao, C., Li, Y., Ma, J., An, N., et al. (2016). Shoot bending promotes flower bud formation by miRNA-mediated regulation in apple (Malus domestica borkh.). Plant Biotechnol. J. 14, 749–770. doi: 10.1111/pbi.12425
Yadava, P., Tamim, S., Zhang, H., Teng, C., Zhou, X., Meyers, B., et al. (2020). Environment-conditioned male fertility of HD-ZIP IV transcription factor mutant ocl4: impact on 21-nt phasiRNA accumulation in pre-meiotic maize anthers (Cold Spring Harbor Laboratory). Plant reprod. 34 (2), 117–129. doi: . doi: 10.1007/s00497-021-00406-3
Yan, Y., Ham, B. K. (2022). The mobile small RNAs: Important messengers for long-distance communication in plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.928729
Yan, M., Xie, D., Cao, J., Xia, X., Shi, K., Zhou, Y., et al. (2020). Brassinosteroid-mediated reactive oxygen species are essential for tapetum degradation and pollen fertility in tomato. Plant J. 102, 931–947. doi: 10.1111/tpj.14672
Yang, J., Tian, L., Sun, M. X., Huang, X. Y., Zhu, J., Guan, Y. F., et al. (2013). Auxin response factor 17 is essential for pollen wall pattern formation in Arabidopsis. Plant Physiol. 162, 720–731. doi: 10.1104/pp.113.214940
Yang, H., Xue, Y., Li, B., Lin, Y., Li, H., Guo, Z., et al. (2022). The chimeric gene atp6c confers cytoplasmic male sterility in maize by impairing the assembly of the mitochondrial ATP synthase complex. Mol. Plant 15, 872–886. doi: 10.1016/j.molp.2022.03.002
Yang, Z., Yang, Z., Yang, C., Wang, Z., Chen, D., Xie, Y., et al. (2020). Identification and genetic analysis of alternative splicing of long non-coding RNAs in tomato initial flowering stage. Genomics 112, 897–907. doi: 10.1016/j.ygeno.2019.06.005
Ye, Q., Zhu, W., Li, L., Zhang, S., Yin, Y., Ma, H., et al. (2010). Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. P Natl. Acad. Sci. U.S.A. 107, 6100–6105. doi: 10.1073/pnas.0912333107
Yoon, J. H., Abdelmohsen, K., Gorospe, M. (2014). Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 34, 9–14. doi: 10.1016/j.semcdb.2014.05.015
Young, E. G., Hanson, M. R. (1987). A fused mitochondrial gene associated with cytoplasmic male sterility is developmentally regulated. Cell 50, 41–49. doi: 10.1016/0092-8674(87)90660-x
Zakharova, E., Khaliluev, M., Kovaleva, L. (2022). Hormonal signaling in the progamic phase of fertilization in plants. Horticulturae 8, 365. doi: 10.3390/horticulturae8050365
Zhai, J., Zhang, H., Arikit, S., Huang, K., Nan, G. L., Walbot, V., et al. (2015). Spatiotemporally dynamic., cell-type–dependent premeiotic and meiotic phasiRNAs in maize anthers. P Natl. Acad. Sci. U.S.A. 112, 3146–3151. doi: 10.1073/pnas.1418918112
Zhang, H., Hu, J., Qian, Q., Chen, H., Jin, J., Ding, Y. (2016b). Small RNA profiles of the rice PTGMS line wuxiang s reveal miRNAs involved in fertility transition. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00514
Zhang, Y. C., Liao, J. Y., Li, Z. Y., Yu, Y., Zhang, J. P., Li, Q. F., et al. (2014). Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol. 15, 512–527. doi: 10.1186/s13059-014-0512-1
Zhang, S. M., Wu, S. W., Niu, C. F., Liu, D. C., Yan, T. W., Tian, Y. H., et al. (2021). ZmMs25 is critical for anther and pollen development in maize. J. Exp. Bot. 72, 4298–4318. doi: 10.1093/jxb/erab142
Zhang, L., Xiang, Y., Chen, S., Shi, M., Jiang, X., He, Z., et al. (2022a). Mechanisms of microRNA biogenesis and stability control in plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.844149
Zhang, W., Xie, Y., Xu, L., Wang, Y., Zhu, X., Wang, R., et al. (2016a). Identification of microRNAs and their target genes explores miRNA-mediated regulatory network of cytoplasmic male sterility occurrence during anther development in radish (Raphanus sativus l.). Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01054
Zhang, F., Yang, J., Zhang, N., Wu, J., Si, H. (2022b). Roles of microRNAs in abiotic stress response and characteristics regulation of plant. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.919243
Zhang, B. B., Zhang, X. X., Zhang, M., Guo, L. P., Qi, T. X., Wang, H. L., et al. (2019). Transcriptome analysis implicates involvement of long noncoding RNAs in cytoplasmic male sterility and fertility restoration in cotton. Int. J. Mol. Sci. 220, 5530–5548. doi: 10.3390/ijms20225530
Zhao, Y., Li, H., Fang, S. S., Kang, Y., Wu, W., Hao, Y. J., et al. (2016). NONCODE 2016, an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 44, 203–208. doi: 10.1093/nar/gkv1252
Zhao, D. Z., Wang, G. F., Speal, B., Ma, H. (2002). The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16, 2021–2031. doi: 10.1101/gad.997902
Zhou, X., Huang, K., Teng, C., Abdelgawad, A., Batish, M., Meyers, B. C., et al. (2022). 24-nt phasirnas move from tapetal to meiotic cells in maize anthers. New Phytol. 235, 488–501. doi: 10.1101/2021.08.24.457464
Zhu, D., Deng, X. W. (2012). A non-coding RNA locus mediates environment-conditioned male sterility in rice. Cell Res. 22, 791–792. doi: 10.1038/cr.2012.43
Keywords: microRNA (miRNA), long non-coding RNA (lncRNA), plant hormone, mitochondrial gene, male sterility
Citation: Nie H, Cheng C, Kong J, Li H and Hua J (2023) Plant non-coding RNAs function in pollen development and male sterility. Front. Plant Sci. 14:1109941. doi: 10.3389/fpls.2023.1109941
Received: 28 November 2022; Accepted: 23 January 2023;
Published: 15 February 2023.
Edited by:
Zhiwen Chen, Hainan Yazhou Bay Seed Laboratory, ChinaReviewed by:
Lei Gong, Northeast Normal University, ChinaCopyright © 2023 Nie, Cheng, Kong, Li and Hua. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinping Hua, amlucGluZ19odWFAY2F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.