- 1Key Laboratory of Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu, China
- 2College of Grassland, Resources and Environment, Inner Mongolia Agricultural University, Hohhot, China
Long-lived tree species need to cope with changing environments and pathogens during their lifetime. Fungal diseases cause damage to trees growth and forest nurseries. As model system for woody plants, poplars are also hosts of a large variety of fungus. The defense strategies to fungus are generally associated with the type of fungus, therefore, the defense strategies of poplar against necrotrophic and biotrophic fungus are different. Poplars initiate constitutive defenses and induced defenses based on recognition of the fungus, hormone signaling network cascades, activation of defense-related genes and transcription factors and production of phytochemicals. The means of sensing fungus invasion in poplars are similar with herbs, both of which are mediated by receptor proteins and resistance (R) proteins, leading to pattern-triggered immunity (PTI) and effector-triggered immunity (ETI), but poplars have evolved some unique defense mechanisms compared with Arabidopsis due to their longevity. In this paper, current researches on poplar defensive responses to necrotrophic and biotrophic fungus, which mainly include the physiological and genetic aspects, and the role of noncoding RNA (ncRNA) in fungal resistance are reviewed. This review also provides strategies to enhance poplar disease resistance and some new insights into future research directions.
Introduction
Forest trees play crucial roles in mitigating effects of climate change and increasing industrial demand, and have considerable economic and ecological value (Kovalchuk et al., 2013). Poplars are predominantly distributed worldwide as model species of woody plants due to their rapid growth and stress tolerance. Populus is also hosts of a large variety of fungus (Miranda et al., 2007; Wang et al., 2017a; Wang et al., 2022). Fungal diseases not only affect the growth, but also cause large numbers of tree deaths and ecosystem degradation (Eyles et al., 2010; Kovalchuk et al., 2013). This makes it ecologically and economically important to deepen the knowledge of poplar defense mechanisms against fungus (Kovalchuk et al., 2013). Plant pathogens can be broadly divided into biotrophic (feeding on living plant tissue), necrotrophic (feeding on dead plant tissue) and hemibiotrophic (infect living plant tissues to first establish infection before switching to necrotrophy) (McCombe et al., 2022). Biotrophs infecting poplars like leaf rust, caused by obligate parasitic fungus Melampsora spp., powdery mildews caused by Phyllactinia spp. or Uncinula spp., while necrotrophs or hemibiotrophs, like leaf blight (caused by Septoria spp.), leaf spot (caused by Marssonina spp., Venturia spp., Coryneum spp.), canker (Septoria spp), and so on (Steenackers et al., 1996; Weiland et al., 2003; Feau et al., 2010). In order to improve the resistance of trees to disease, it is necessary to understand the defense mechanisms.
Similar with herbaceous plants, poplars defense mechanism categorized as constitutive defenses and induced defenses. Induced defenses only expressed when plants suffered external stimulus, and constitutive defenses are always expressed in the plants (Eyles et al., 2010). Constitutive defense is the first line of defense contributing to non-host resistance, including inherent physical structures and phytochemicals, which provide basic defense against pathogens (Alkan and Fortes, 2015). To reduce morbidity during long life cycle of poplar, they enhance the physical and phytochemical defenses. Furthermore, plants enhance defensive capacity through a series of complex regulations when infected by pathogens, which is called induced defenses. According to the expression range, induced resistance can be divided into local induced resistance and systemic induced resistance (De Kesel et al., 2021). Trees evolve induced defenses due to the lower resource allocation costs than constitutive defenses (Eyles et al., 2010). Regardless of the lifestyle of the attacking pathogens, plants have developed several means in protection against fungal pathogens: pathogen-associated molecular pattern PAMP-triggered immunity (PTI), effector-triggered immunity (ETI) and noncoding RNA (ncRNA)-mediated defense (De Kesel et al., 2021) (Figure 1). To against fungal pathogens, plants need to distinguish different fungal life cycles, for example, programmed cell-death (PCD) around the infected sites is an effective way against biotrophs but not an appropriate response to some necrotrophy.
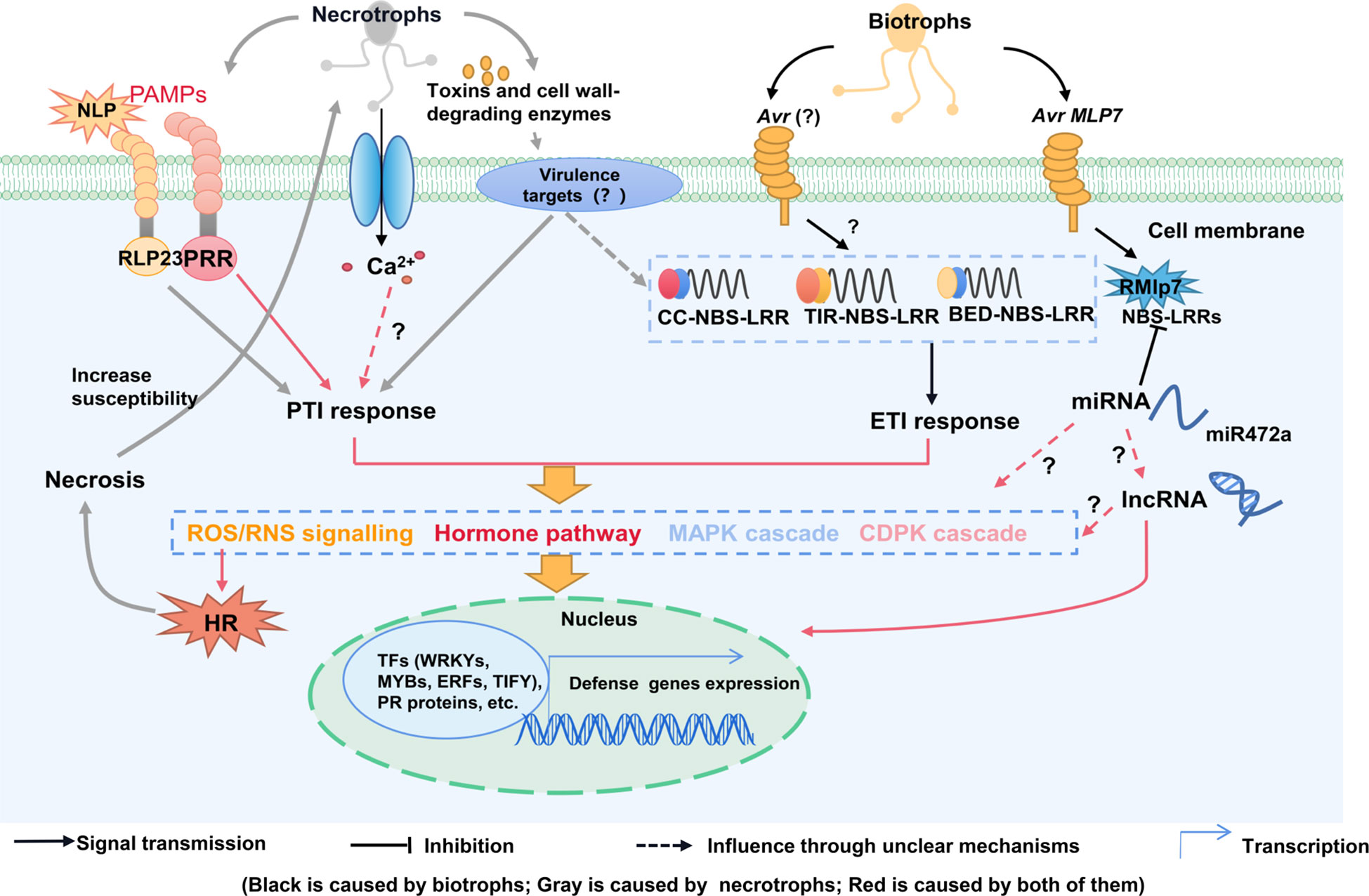
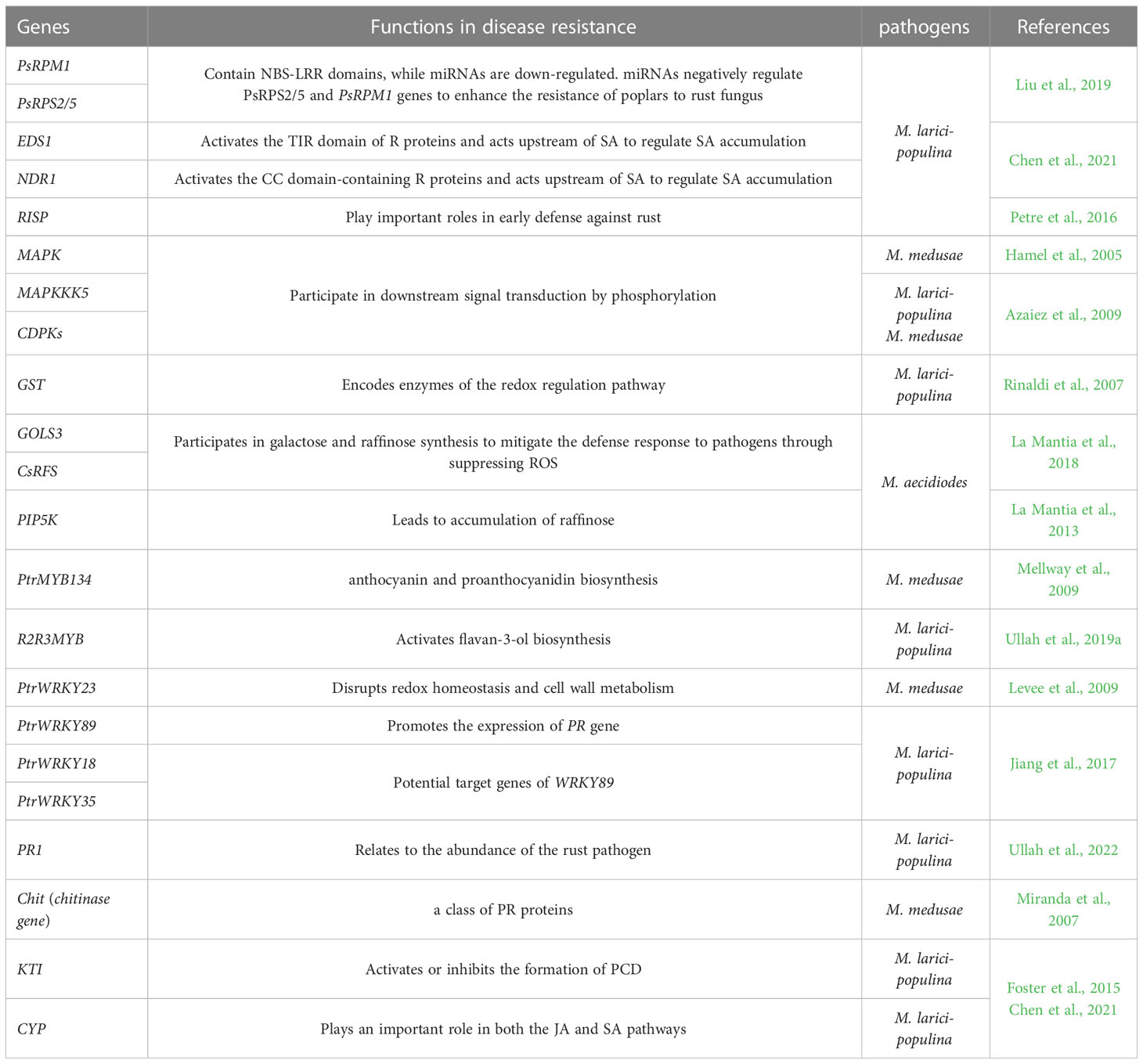
Figure 1 A model of poplars immunity to fungal pathogens. When plants attacked by biotrophic pathogens, PRRs recognize extracellular pathogenic characteristics and activate PTI, R proteins (NBS-LRRs) recognize intracellular effector proteins from pathogens and activate ETI. They induce a series of defense responses including the production of ROS, RNS, phytohormone, CDPKs and MAPK signals. But necrotrophic fungus may only recognized by PRRs. Many defensive genes and transcription factors are key players, including PR, MYB, WRKY, TIFY, and ERF, lncRNAs and miRNAs also participate in defense responses against fungal pathogens. miRNAs also play a role in the regulation of NBS-LRR. Avr, avirulence gene; CDPK, calcium-dependent protein kinase; ETI, effector-triggered immunity; HR, hypersensitive response; lncRNAs, long ncRNAs; MAPK, mitogen-activated protein kinase; miRNAs, microRNAs; PCD, programmed cell death; PR, pathogenesis-related proteins; PRRs, pattern recognition receptors; PTI, pattern-triggered immunity; ROS, reactive oxygen species.
The completion of the whole genome of P. trichocarpa marks that the study of poplar disease has entered the genomic era (Tuskan et al., 2006). However, it is difficult to study the disease resistance of poplars using genetic methods due to the long generation time. To date, research on poplar-fungus interactions at the molecular level has mostly focused on genes related to host defense. Therefore, this paper mainly summarizes the research on physical and physiological mechanisms in poplars against fungus with different life cycles (Figure 2). Also, we have focused on the molecular mechanism especially on defense genes, transcription factors and non-coding RNAs in poplars against biotrophic and necrotrophic fungus (Tables 1, 2).
Figure 2 A model for the interaction between poplar physical and biochemical defenses. Constitutive defense responses mainly include physical barriers and phytochemicals defenses, when attacked by pathogens. Defense-related metabolites include peroxide, phenolics and phytohormones. The black arrows represent responses caused by biotrophic fungus, grey arrows represent responses caused by necrotrophic fungus, red arrows represent responses caused by all fungus. ABA, abscisic acid; APX, ascorbate peroxidase; C4H, cinnamate 4-hydroxylase; CAT, catalase; 4CL, 4-coumarate-CoA ligase; DIR, dirigent; ET, ethylene; GST, glutathione sulfur transferase; JA, jasmonic acid; JAZ, jasmonate-zim domain; PAL, phenylalanine ammonia lyase; PAs, proanthocyanidins; POD, peroxidase; ROS, reactive oxygen species; SA, salicylic acid; SOD, superoxide dismutase; SAR, systemic acquired resistance.
Defense responses of poplars to fungal pathogens
Constitutive defense responses
Biotrophic fungal pathogens such as rust and powdery mildew produce invasive filaments through appressorium, and penetrate the cuticle, and form haustoria in the epidermal cells to absorb nutrients from the hosts, and produce cell wall degrading enzymes and sporulate without killing the host cells (Maupetit et al., 2018). Whereas most necrotrophic fungi kill their hosts by secreting cell wall-degrading enzymes or toxins to impair cuticles and cell walls to facilitate infection (Shi et al., 2016). Biotrophic fungal pathogens spend most of their life cycle on living plant tissues, therefore, their fitness appeared to be more influenced by constitutive defense (Maupetit et al., 2018). Constitutive defense is the first line of protection, including physical structures and phytochemicals. Woody plants have many mechanical barriers to against pathogen invasion, such as the leaf cuticle, the pectin and lignin of cell walls. Cuticle and cell wall are the first line of defense in plants, the components of cell wall are cellulose, hemicellulose, pectin, and lignin (Ziv et al., 2018). Many biotrophic fungal species use their appressoria to penetrate the cuticular layer and then infect internal cells. The cuticle is a polyester that is partly covered with waxes (epicuticular and intracuticular) (Serrano et al., 2014). A study found PtoMYB142 could directly regulate the transcriptional activity of wax biosynthesis genes, e.g., fatty acid hydroxylase (CER4) and 3-ketoacyl CoA synthase (KCS6), to adapt drought conditions for poplars (Song et al., 2022). But the contribution of wax biosynthesis on poplar disease resistance is poorly studied. However, many rust fungi failed to penetrate the cuticle and thus had to invade the mesophyll cells through the stomata by germ tubes, hyphae or appressorium (Rinaldi et al., 2007), but the cuticle was also found to contribute to non-host resistance to leaf rust by impeding the germination and growth of urediniospores from M. larici-populina (Yu et al., 2019). Lignin also acts as physical barrier during pathogen infection, preventing water and nutrients transferring from host cells to pathogens (Ellinger et al., 2013; Miedes et al., 2014). Genes involved in lignin biosynthesis are critical for plant cell walls in immunity, e.g., phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate-CoA ligase (4CL), cinnamyl alcohol dehydrogenase (CAD), cinnamoyl-CoA reductase (CCR) and hydroxycinnamoyl transferase (HCT), were highly expressed after attack by fungi pathogens (Azaiez et al., 2009; Polle et al., 2013; Huang et al., 2022). Interestingly, in incompatible interaction, localized lignin formation was observed, while there was an accumulation in compatible interaction when poplars infected with Melampsora (Boyle et al., 2010). In the incompatible interactions, abundant lignin was deposited around leaf vessels (Rinaldi et al., 2007). Regulation of the lignification pathway may be critical for improved poplar tolerance (Polle et al., 2013). Dirigent (DIR) proteins have been identified in many plants, which are involved in process of plant lignin synthesis. Furthermore, overexpression of PtDIR11 in poplars could improve lignin biosynthesis and enhance poplar resistance to Septotis populiperda (Li et al., 2022). These results imply that lignins are effective antifungal chemical defenses against pathogens infection.
Some necrotrophic fungus prefer to plants with cell walls are rich in pectins because they possess a strong pectin decomposition machinery. Pectin methylesterase (PME), a class of pectin modification enzyme, plays an important role in cell wall modification, pectins function not only in primary cell walls, but also in secondary cell walls. It is an effective way in restricting necrotrophic fungal pathogens to inhibit PME (Pelloux et al., 2007). It has been reported PtoPME35 involved in stomatal closure, and the overexpression of its homologous gene AtPME35 in Arabidopsis leading to plant lodging, but overexpression of PtoPME35 do not influence poplar growth, indicating that woody plants have more complex defensive networks than annual herbs (Yang et al., 2020). However, cell wall-related genes showed differential expression patterns when two susceptible poplars infected by hemibiotrophic pathogen of M. brunnea. For example, pectin methylesterase inhibitors were enriched in P. deltoids but they were mostly reduced in P. alba. Transcriptomic data showed that P. deltoids differentially expressed genes were most responsive at the initial biotrophic stage, while P. alba was mainly responsive to Marssonina brunnea at the necrotrophic phase (Zhang et al., 2018). Additionally, when susceptible poplars suffered two specific forms of M. brunnea, the number of DEGs expressed among three stages of infection were changed in a significantly different pattern. The results showed there were more differentially expressed genes in the necrotrophic stage than biotrophic stage when poplars infected with M. brunnea. In particular, pectinlyase was significantly induced in multi-strain infections during the initial invasion phase, but not expressed in monoculture infections (Ren et al., 2020).
Induced defense responses
Innate immunity against fungal pathogens
Similar with herbs, woody plants sense pathogen invasion through resistant (R) proteins or receptor proteins (Kovalchuk et al., 2013). When fungal pathogens penetrate plant physical barriers, PRRs may recognize pathogen-associated molecular patterns (PAMPs) from pathogens and activate PTI. As shown in Figure 1, PTI acts as a basic defense system and triggers a set of plant defense responses, including activation of signaling molecules, like calcium-dependent protein kinase (CDPK) and mitogen-activated protein kinase (MAPK) cascades. CDPKs and MAPK are involved in the regulation of downstream immune responses (Meng and Zhang, 2013). In poplars, MAPK has been reported to be an important component in biotic and abiotic stresses, and functioned as common points of cross-talk between pathogen defense and oxidant stress signaling cascades (Hamel et al., 2005). A study indicated that genes encoding mitogen-activated protein kinase-kinase-kinase 5 (MAPKKK5) and CDPKs were upregulated after infection by rust (Azaiez et al., 2009). When two susceptible poplars infected with M. brunnea (Marssonina) that causes leaf spot disease, MPK3 and MPKK9 were detected in both poplars (Zhang et al., 2018), and the MPKK9-MPK3 modules have been identified to participated in ethylene (ET) signaling (Yoo and Sheen, 2008). However, the specific functional mechanisms of MAPK and CDPK regulating poplars defense against pathogens remains to be further studied. The PAMP flg22 is a highly conserved 22-amino-acid peptide of the N-terminal of bacterial flagellin, which can induce PTI responses in many plants (Zhao and Cheng, 2022). The activation of MAPK cascades after flg22 treatment was found in P. davidiana × P. bolleana, but necrosis and ethylene-inducing peptide 1-like proteins (NLPs), which mainly expressed in hemibiotrophs or necrotrophs, cannot generate PTI responses in general. Heterologously expressed receptor-like protein (AtRLP23) in poplar recognizes NLP24s and generates PTI responses and ROS bursts after M. brunnea and Elsinoë australis infections, thereby enhancing broad-spectrum disease resistance to the fungus (Zhao and Cheng, 2022) (Figure 1). Pathogens have evolved effectors that promote their growth by suppressing PTI, which results in effector-triggered susceptibility (ETS). However, plants will evolve R gene to sense the effectors and trigger ETI, which is associated with the hypersensitive response (HR) (Chisholm et al., 2006). PTI and ETI reinforcing each other to enhance plant defensive responses against pathogen infections by inducing downstream signal transduction (Chang et al., 2022). However, the crosstalk of PTI and ETI in poplar still needs to be studied.
Plants recognition events are mostly mediated by a class of receptor proteins containing nucleotide-binding (NB) and leucine-rich repeat proteins (LRR) domains (Dodds and Rathjen, 2010). The NBS-LRR class is the most abundant R protein and is responsible for pathogen identification of intracellular effectors (Dodds and Rathjen, 2010). R-mediated defense response seem not to function in necrotrophs, while PRRs like receptor-like kinases (RLKs) involved in the perception of necrotrophs (Wang et al., 2014), but R-mediated plant cell necrosis increased susceptibility to necrotrophs. Therefore, R protein acts indirectly on necrotrophic fungus (Su et al., 2018) (Figure 1). In dicotyledons, there are two main classes of NBS-LRRs: TIR-NBS-LRRs and CC-NBS-LRRs, which have Toll-interleukin-1 receptor (TIR) and amino-terminal coiled-coil (CC) domains, respectively (Bresson et al., 2011). TIR-NBS-LRRs are entirely missing from the monocotyledon’s genome like rice. In poplars, a third class of NBS-LRR genes, called BED-NB-LRRs, which containing a BED domain, has been reported. BED-NB-LRR family comprising 32 members, seems to be unique to poplars (Kohler et al., 2008; Germain and Seguin, 2011). Poplar possesses 400 NBS-LRRs nearly twofold that of Arabidopsis, indicating that woody plants may have developed more intracellular receptors compared with herbaceous plants due to their longevity, which may result in greater disease resistance ability (Duplessis et al., 2009). There were 34 NBS-LRRs differentially expressed after rust fungi infected poplars (Kohler et al., 2008). It has been reported disease resistance proteins (CC-NBS-LRR class) and LRR proteins were induced by biotrophic fungus in hybrid poplars (Azaiez et al., 2009). In addition, enhanced disease susceptibility 1 (EDS1) and nonrace-specific disease resistance 1 (NDR1) are required for activation of R protein-mediated resistance. EDS1 regulates defense signaling by activating R proteins with TIR domains, while NDR1 is required for the activation of CC domain-containing R proteins (Lang et al., 2022). EDS1 and NDR1 were found to regulate SA accumulation in poplars in response to leaf rust disease (Chen et al., 2021). Rust-induced secreted protein (RISP) is a small (82 amino acids), cysteine-rich protein and was highly inducible in poplar leaves after infection with rust, and RISP was found near the LRR-RLP gene (Petre et al., 2014). The two genes have similar promoter regions and expression profiles in response to rust infection. RISP inhibits M. larici-populina growth on poplar leaves by binding to M. larici-populina urediniospores and inhibiting germination and germ tube elongation. Thus, RISP plays a role in early defense against fungal pathogens (Petre et al., 2016) (Table 1).
In the plant-pathogen interaction system, pathogen-induced host-specific resistance mainly depends on pathogen avirulence (Avr) genes and R genes to activate downstream defense cascades, forming a HR at the infected site (Gururani et al., 2012). However, as Avr evolves rapidly, it allows pathogens to break down R-mediated plant immunity. For example, the AvrL567, a virulence gene, was found in M. lini haustoria and induced by HR (Dodds et al., 2004). The transcriptome data indicated that six known Avr genes in M. lini (AvrM, AvrM14, AvrL2, AvrL567, AvrP123 and AvrP4) showed similar patterns of early expression during infection (Wu et al., 2019). However, the R genes that sense these AVR genes in poplars are still unknown. Another study showed that the candidate locus AvrMlp7 drives rust fungus adaptation to poplar RMlp7-mediated immunity (Figure 1) (Louet et al., 2021).
HR and ROS inhibit fungus infection
HR is the most typical response after infection and causes programmed cell death (PCD). HR has been extensively described with relation to biotrophic fungi, but promote necrotrophic pathogens infection (Mayer et al., 2001). A study found that several genes in poplars that cause HR were upregulated at 96 hpi (necrotrophic phase) when response to M. brunnea, indicating that necrotrophic fungus may induce HR in poplars to increase their susceptibility (Zhang et al., 2018). HR varies among different poplar-pathogen interaction types. In incompatible Populus-Melampsora interactions, HR appears the day after infection, and poplars then show PCD characteristics at 7 dpi, however, there is no HR performance in the compatible interaction (Rinaldi et al., 2007). One of the most striking features accompanying HR is the burst of ROS after pathogen infection. ROS burst is one of the earliest responses of plants to fungus, leading to necrosis of host tissue, which develop an effective defense against biotrophic fungi but may increase susceptibility to necrotrophs (Mayer et al., 2001). Therefore, ROS homeostasis regulation is important for regulating plant resistance to different pathogens.
Plants have initiated a series of enzymatic and nonenzymatic oxidation systems to reduce ROS accumulation and to avoid oxidative damage in cells. Several transcripts encoding ascorbate peroxidase (APX), superoxide dismutase (SOD), glutathione sulfur transferase (GST) and peroxidase (POD) were upregulated (Figure 2), which were involved in the oxidative burst and even reached approximately 20-fold higher levels in M. larici-populina and M. medusae infected poplars (Miranda et al., 2007; Azaiez et al., 2009). However, the accumulation of ROS is varied from different poplar species. A study on male and female P. cathayana infected with rust showed that the production of was higher in males than in females, while the H2O2 content was higher in females than in males. The results indicated that male poplars showed higher antioxidant activities and less H2O2 accumulation than females after being infected by leaf rust; therefore, rust disease was more severe in female poplars (Zhang et al., 2010). In addition, when poplars are attacked by pathogens, guard cells produce H2O2 to form a rapid defense, but it is weaker in the compatible interactions (Boyle et al., 2010). Moreover, second messengers, e.g., Ca2+, hydrogen sulfide (H2S), inositol triphosphate (IP3) and NO, are produced within seconds to enhance plant responses (Agurla et al., 2018). For example, NO-activated antioxidant enzymes can decrease ROS and reactive nitrogen species (RNS) toxicity to improve poplar tolerance to environment stress (Cheng et al., 2016). However, how these second messengers regulate biotic stress in poplars remains unclear. Additionally, inositol, galactitol and raffinose are important regulators of ROS homeostasis. Inositol negatively impacted SA, while galactitol enhances systemic resistance to necrotic pathogens induced by JA (Figure 2). The overexpression of galactose synthase gene (GOLS3) and raffinose synthase gene (CsRFS) mitigated the defensive responses to poplar leaf rust by suppressing ROS and attenuating calcium and phosphatidic acid signaling events. The accumulation of galactinol could constitutively repress defense signaling events upstream of SA biosynthesis (La Mantia et al., 2018). Phosphatidylinositol 4-phosphate 5-kinase (PIP5K) encoding phosphatidylinositol 4-phosphate 5-kinase led to the accumulation of raffinose. In P. trichocarpa × deltoides, the expression of PIP5K decreased after 48 hpi inoculation with rust (La Mantia et al., 2013). WRKY-TF also plays an important role in multiple defense responses (Spoel and Loake, 2011). For example, WRKY23 affects poplar resistance to fungi infection by disrupting redox homeostasis and cell wall metabolism. WRKY23 can increase poplar susceptibility to rust disease (Levee et al., 2009). Thus, precise regulation of the transcription factors and genes related to ROS production could improve the resistance of poplars to pathogens (Figure 2).
SA-mediated signaling pathway against pathogens
Phytohormones play essential roles in plant resistance to pathogens and plant immune responses (Pieterse et al., 2012; Chanclud and Morel, 2016). SA dominates in the execution of host defense response against biotrophic, while JA and ET are key players to facilitate host defense response against necrotrophic (Li et al., 2019). In plants, SA is synthesized through two routes. One route is the chloroplast-localized isochorismate synthase (ICS) pathway, and the other is PAL-mediated pathway. ICS pathway is believed to be responsible for the most of SA synthesized during the activation of pathogens in Arabidopsis (Yuan et al., 2009). While Populus is primarily dependent on the PAL pathway, therefore, PAL genes are critical for SA synthesis. SA might be converted to MeSA at infected sites and transported as a signaling molecule to uninfected sites, induced SAR (Li et al., 2018). SA signaling positively regulates plant defense against biotrophic pathogens by enhancing secondary metabolites and inducing pathogenesis-related genes (PRs), which are necessary for the establishment of SAR (Shah, 2003; Thaler et al., 2012). SA was proved to activate flavan-3-ol biosynthesis against biotrophic fungus M. larici-populina in poplars (Ullah et al., 2019a). In addition, it was indicated that the accumulation of flavanols was negatively regulated by cytokinin (CK) in poplars (Ullah et al., 2019b).
High SA levels induce PR gene expression in Arabidopsis, which may lead to high metabolic costs. However, for perennial woody plants, it may have better balance of resource allocation, therefore, PR gene expression is not necessarily with SA levels, but positively correlate with the degree of disease susceptibility in poplars (Ullah et al., 2022). Evidence has shown that an increase of PR gene expression after pathogens invasion, is probably mediated by SA and JA (Irigoyen et al., 2020). PR proteins have been classified into 17 families. Among them, PR-15 and PR-16 families were only found in monocots. Although long-living trees would suffer more diverse set of fungal pathogens, the number of defense-related genes in poplars showed no different from Arabidopsis and rice, only PR-encoding chitinases and kunitz-type protease inhibitors are found to be more abundant in poplars (Kovalchuk et al., 2013). There are abundant PR genes in poplars, which are key for SA-mediated defense (Wei et al., 2020c). Several transcriptomic studies have found that PR-1 expression is increased 100-fold higher than controls (Miranda et al., 2007; Azaiez et al., 2009; Boyle et al., 2010). In addition, the expression levels of PR-2 (3-glucanase), PR-3, PR-4 (chitinase), PR-5 (thaumatin-like protein), PR-6 (protease inhibitor), PR-7 (l-aspartic acid protease) and PR-9 (lignin peroxidase) are also increased after infection with pathogens (Azaiez et al., 2009; Duplessis et al., 2009). Interestingly, PR-1 and PR-2 genes induction are much larger in the susceptible genotype compared with the rust-resistant genotypes, indicating that the abundance of the rust pathogen determines the degree of PR gene induction in poplars (Ullah et al., 2022). Additionally, chitinase genes were upregulated in poplars after inoculation with M. medusae (Miranda et al., 2007) or S. musiva (Liang et al., 2014). Most of chitinase genes showed high transcript levels in early stage when infection with Alternaria alternata (Huang et al., 2022) (Tables 1, 2). A chitinase gene (Bbchit1) from Beauveria bassiana was overexpressed in white poplar and enhanced resistance to a pathogenic fungus C. chrysosperma (Jia et al., 2010). Overexpression of TLP gene that belongs to PR-5 family, could inhibit the growth of pathogens in poplars and enhance resistance to spots disease (Table 2) (Sun et al., 2020).
JA/ET-mediated signaling pathway against pathogens
Meanwhile, the JA/ET pathway is generally required for the predominant defense response against necrotrophic pathogens and herbivores (Yang et al., 2015). Genes involved in JA synthesis including lipoxygenase (LOX), allene oxide synthase (AOS), allene oxide cyclase (AOC), 12-oxophytodienoate reductase (OPR) and acyl-coenzyme oxidase (ACX) were induced during biotrophic or necrotrophic pathogens infection (Azaiez et al., 2009; Huang et al., 2022). Interestingly, LOX can regulate the ROS accumulation in poplars to against pathogens (Huang et al., 2022). Genes involved in JA signal transduction pathways were also significantly induced, such as the MYC proteins, which are positive regulators in the JA signaling pathway, were upregulated during pathogen infection. While JAZ, which contains TIFY domains, a negative regulator of JA signal transduction (Pauwels and Goossens, 2011), was mainly downregulated during pathogen infection. The transcription factor TIFY has also been found to be a key element that contributes to phytohormone or stress responses. TIFY genes can be divided into four subfamilies, TIFY, JAZ, ZML and PPD (Xia et al., 2017). It has been reported that there are 24 TIFY genes in poplars (Xia et al., 2017). A study showed that most of the TIFY genes could respond to M. larici-populina infection, while genes expression patterns were different at different time points (Xia et al., 2017).
Furthermore, ET synthesis genes including 1-aminocyclopropane-1-carboxylate synthase (ACS), and 1-aminocyclopropane-1-carboxylate oxidase (ACO) were also induced after pathogens infection (Azaiez et al., 2009; Huang et al., 2022). ET activated PtoRbohD/RbohF expressions, which encode NADPH oxidases to induce H2O2 production in poplars to Dothiorella gregaria fungi (Liu et al., 2022). ET response factors (ERFs) bind to the ethylene-responsive element GCC-box, and the target genes are related to wounding and pathogen infection (Meng et al., 2013). A recent study showed that the transcript levels of 21 ERF genes were strikingly upregulated and 72 genes were downregulated in P. nigra × P. deltoides under M. larici-populina infection at 4 dpi, and the inactivation of ERF genes and disease resistance-related ERF target genes might result in poplar susceptibility to rust disease (Chen et al., 2019). Therefore, JA/ET pathway not only enhances necrotrophic fungus resistance on poplars, but also regulates the susceptibility of poplars to biotrophic fungus, which relate to the timing of the infection.
Crosstalk of SA and JA in poplar disease resistance
It is generally assumed that SA and JA are antagonistic in disease resistance in Arabidopsis, rice and tomato. Plants have to balance the costs and potential benefits of investing in defense to external stimulus (Pieterse et al., 2012; Thaler et al., 2012), but this antagonism is not obvious in poplar against rust disease. A study reported that both SA and JA contents were increased upon rust infection in black poplars, and transgenic poplar lines with high SA levels increased JA and flavonoid contents, and enhanced rust resistance (Ullah et al., 2019a; Ullah et al., 2022). Therefore, SA and JA pathways interact positively in poplars to decrease biotrophic pathogen growth (Figure 2). This difference may attribute to perennial woody plants can store large reserves of resources for defense, might have evolved SA- and JA-mediated co-defense systems without antagonism, while annual plants lack the defensive resources.
WRKY-TFs and NONEXPRESSOR OF PR1 (NPR1) are known to be involved in modulating between SA- and JA-dependent responses in plants. There are 65 and 64 PtrWRKY gene promoters involved in SA and MeJA responses in poplars, respectively (Jiang et al., 2014). WRKY70 activates the expression of NPR1 and thus enhances PR expression, leading to the resistance of Arabidopsis to both biotrophic and hemibiotrophic pathogens but increasing plant susceptibility to necrotizing vegetative fungal pathogens (Li et al., 2004; Li et al., 2006; Shim et al., 2013). In poplars, PsnWRKY70 enhance the resistance to A. alternata by activating genes involved in MAPK cascade and Ca2+ signaling, other members of WRKYs, and LRR domain proteins (Wang et al., 2022). Arabidopsis homolog transcripts WRKY70, WRKY51 and WRKY40 were strongly increased (more than 10-fold) in poplars after infection by Melampsora (Miranda et al., 2007; Azaiez et al., 2009). After exogenous SA treatment, nine genes in PtrWRKYIII were upregulated (WRKY 89, WRKY 62, WRKY 64, WRKY63, WRKY41, WRKY55, WRKY-53, WRKY-54, WRKY-30), while one gene (PtrWRKY90) was significantly downregulated in poplar (Wang et al., 2015). Particularly, PtrWRKY89 induced by SA plays an important role in rust resistance by upregulating PR gene expression. PtrWRKY18 and PtrWRKY35 are potential target genes of PtrWRKY89, and they can increase resistance to M. larici-populina fungus (Jiang et al., 2017). Additionally, PtrWRKY73 induced by SA in P. tomentosa could increase plant resistance to biotrophic pathogens but enhance sensitivity to the necrotrophic fungal pathogen (Duan et al., 2015). However, PtrWRKY40 is similar with AtWRKY40, AtWRKY18 and AtWRKY60, which had a side effect on the resistance of poplars to hemibiotrophic fungus (D. gregaria) by negatively regulating SA-related genes expression (Karim et al., 2015). To date, the role of WRKY genes has been explored extensively in poplars, which are associated with stress responses and phytohormones (Jiang et al., 2014).
Additionally, poplars can activate or inhibit the SA or JA pathway by proteins, such as kunitz-type serine endopeptidase inhibitor (KTI) regulation, which can restrict fungus growth by PCD (Chen et al., 2021). KTI controlled by the cytochrome P450 family (CYP). Thus, CYP genes have great effects on the JA and SA pathways (Xu et al., 2015a; Chen et al., 2021), while KTI and CYP genes were significantly induced by biotrophic fungus Melampsora (Miranda et al., 2007; Azaiez et al., 2009) and necrotrophic fungus Sphaerulina (Foster et al., 2015). In addition, plant defensins are antimicrobial peptides that represent a major barrier to invasion by pathogens. A study found overexpression of PtDefensin in poplars may change the crosstalk between the SA and JA signal pathways to increase the resistance to S. populiperda infection at the early stages (Wei et al., 2020a) (Figure 2). Similarly, PtDefensin overexpression transgenic poplars enhanced resistance to S. populiperda may be due to the upregulation of PR1-1 and MYC2-1 and downregulation of JAZ1, COI1-1 and COI1-2, leading to activation of SA and JA signaling pathways. Host defense peptides (HDPs) are known as cationic antimicrobial peptides and almost found in all living organisms. MsrA2 peptide is proved to have the best antimicrobial potential among all HDPs, and overexpression of MsrA2 in poplar leaves inhibited S. musiva growth (Yevtushenko and Misra, 2019). These findings suggest that antimicrobial peptides could be used for genetic engineering on woody plants to enhance disease resistance.
The role of ABA in fungal resistance
The role of abscisic acid (ABA) in disease resistance remains complex. ABA play positive role in plant immunity through stomatal closure and callose deposition during early stages of pathogen invasion, but may suppresses SA- or JA-dependent immunity in late disease resistance (Ton et al., 2009). ABA has been proven to regulate stomatal closure and rapidly accumulate ROS in stomatal cells (Kohler et al., 2003). It was indicated that exogenous ABA treatment increased poplar resistance against rust. Ullah et al. (2019a) subjected black poplars to drought stress followed by rust inoculation and found that endogenous ABA increased approximately 3-fold, and the growth of M. larici-populina significantly decreased by 10- and 6-fold at 4 and 8 dpi under drought stress, respectively. Due to the different invasion ways of biotrophic and necrotrophic fungus, ABA exerts greater influence on biotrophic fungi like rust which invaded through stomata (Figure 2). However, the mechanisms of ABA regulating poplars against pathogens in later stages of infection need to be explored.
Phytochemicals involved in defense responses
Several antimicrobial compounds are directly involved in the plant defense response to pathogens, including plant antibiotics, which are present in plants prior to infection, and plant antitoxins, which are produced in response to plant attack by pathogens (Silva et al., 2018). Many plants secondary metabolites are thought to serve as phytoalexins, such as flavan-3-ol (catechin, epicatechin) and phenolic acids that involved in phenylpropanoid pathway (Ullah et al., 2017; Ullah et al., 2019b). Phenols and flavonoids are vital products of the phenylpropanoid pathway, which plays important roles in plant disease resistance (Dixon et al., 2005; Syvertsen and Garcia-Sanchez, 2014; Dong and Lin, 2021). In poplars, it was reported that the accumulation of proanthocyanidin (PA) and flavan-3-ol in poplar leaves could inhibit rust hyphal growth and reduce rust colonization. The contents of catechin and PAs were strongly increased at 7 dpi, and their accumulation was significantly induced by SA (Ullah et al., 2017; Ullah et al., 2019a). In addition, moderately resistant poplars accumulate higher amounts of flavan-3-ols at the site of rust infection than susceptible poplars (Ullah et al., 2017). Additionally, the genes were significantly enriched in the flavonoid biosynthesis pathway in poplars after being infected by M. brunnea (Marssonina) (Zhang et al., 2018). The transcriptional responses of P. trichocarpa × P. deltoides to M. medusae showed that genes encoding enzymes of proanthocyanidin and flavonoid were strongly induced, such as flavanone 3-hydroxylase (F3H), PAL, 4CL, dihydroflavonol reductase (DFR), anthocyanidin reductase (ANR) and leucoanthocyanidin reductase (LAR) (Miranda et al., 2007). Similarly, these genes were changed at different time points in poplars when attacked by A. alternata, but they were up-regulated at 2 dpi (Huang et al., 2022).
The MYB family is involved in the regulation of various physiological processes in plants. Flavonoid biosynthesis is regulated by the MYB family at the transcriptional level (Xu et al., 2015b). The activation of anthocyanin and PAs is different from that of other flavonoid branches because they need coactivators, e.g., basic-helix-loop-helix (bHLH) and WD40 (WDR), to interact with MYB to form MYB-bHLH-WD40 (MBW) (Ma and Constabel, 2019). SA can stimulate the expression of MBW that positively regulating the biosynthesis of anthocyanins and proanthocyanidins in poplars to reduce rust proliferation (Ullah et al., 2019a). In addition, MYB134 (Mellway et al., 2009), MYB119 (Cho et al., 2016) and MYB115 (Wang et al., 2017a) are positive regulators to enhance resistance to fungus, while MYB182 (Yoshida et al., 2015), MYB57 (Wan et al., 2017), MYB165 and MYB192 (Ma et al., 2018) are negative regulators of proanthocyanins synthesis in poplars. However, MYB6 can promote the biosynthesis of anthocyanins and proanthocyanins but suppress the formation of secondary cell walls in P. euphratica (Wang et al., 2019). MYB118 (Wang et al., 2020), MYB120 (Kim et al., 2021) and MYB117 (Ma et al., 2021) are involved in anthocyanin and lignin biosynthesis, respectively. Additionally, MYB is widely involved in the phenylpropanoid pathway at the transcriptional level and can potentially precisely regulate lignin and flavonoid synthesis genes, which can enhance poplar disease resistance.
Non-coding RNAs on poplars response to fungal pathogens
Most studies have focused on the function of protein-coding genes like PR in biotic stresses. However, large proportions of eukaryotic genomes are transcribed into RNAs that do not encode proteins. These transcripts are called noncoding RNAs (ncRNAs) and can be directly involved in the regulation of disease resistant genes (Li et al., 2021). ncRNAs are mainly classified into microRNAs (miRNAs), long ncRNAs (lncRNAs) and circular RNAs (circRNAs). Among them, miRNAs play important roles in disease resistance by cleaving target genes or repressing the translation of target mRNAs (Li et al., 2016). Li et al. (2016) studied the susceptibility of P. nigra × P. deltoides to M. larici-populina, they found that miRNAs could act directly or indirectly on disease-related genes or proteins. For example, CC-NBS-LRR class protein family, TIR-NBS-LRR class protein family, cellulose synthase genes and stress-inducible protein genes. However, none of them was responding to rust infection. Conversely, miRNAs related to PAMPs and PTIs were responsive to rust infection. Moreover, the miRNA-mediated posttranscriptional regulation of defense signaling genes was inactivated at the ETI and HR stages by infection with M. larici-populina (Li et al., 2016). After P. trichocarpa induced with canker pathogen (Botryosphaeria dothidea), 12 miRNAs were upregulated. Especially, miR156 responded to biotic and abiotic stresses in Populus, and miRNA-TF interaction networks in poplar canker were revealed. For example, miR159, miR164, and miR319 targeted MYB factors and MYB involved in their activation or repression, miR160-ARF (Auxin receptor factor) and miR167-ARF interaction were related to disease resistance (Zhao et al., 2012). Additionally, miRNAs also play crucial roles in the regulation of NBS-LRR and host defense responses (Figure 1). Liu et al. (2019) found PsRPM1 and PsRPS2/5, which containing NBS-LRR domains, were significantly increased at later infection stages with rust, while miRNAs were down-regulated. These results indicated miRNAs were negatively regulated the expression of their target genes to enhance the resistance of poplars to rust fungus. Similarly, when poplars exposed to the hemibiotrophic fungus C. gloeosporioides, miR472a was down-regulated and NBS-LRRs were up-regulated, leading to a ROS burst and HR to against hemibiotrophic fungus. But when poplars are exposed to the necrotrophic fungus Cytospora chrysosperma miR472a negatively regulated NBS-LRRs, leading to PCD and resulting in necrotrophic fungus susceptibility (Su et al., 2018). Furthermore, plant lncRNAs might be a target of miRNAs and decrease the interaction between mRNAs and miRNAs by binding specific miRNAs. Moreover, lncRNAs that were located closed to protein-coding genes, were differentially expressed during pathogen infection (Wang et al., 2017b). Therefore, these results indicate that sRNAs play important roles in plant-pathogen interactions in poplars.
Conclusion and prospects
Currently, the formation mechanism of plants induced disease resistance has been preliminarily revealed in some model species. However, the mechanisms of plants induced disease resistance are complex and may correspond with plant and pathogen species. Woody plants have more receptors and R proteins than herbs, providing them with better defenses strategies (Duplessis et al., 2009). When receptor proteins and R proteins sense the pathogen, then activate downstream signals. Different with Arabidopsis, SA and JA cooperated to against both biotrophic and necrotrophic fungus in poplar and the genes regulated SA signaling may be different from Arabidopsis (Ullah et al., 2019a; Ullah et al., 2022). Because perennial woody plants have higher resource utilization efficiency than annual plants, they generally use more resources for defense.
Although development and application of omics technologies have provided broadened insights into poplars defensive responses against fungal pathogens, there are still many gaps in our understanding of poplar defense against fungus. For instance, the mechanisms of diverse R proteins and receptors in poplar that sense different fungus are still unclear. Many plant-pathogen interaction candidate genes, such as PRs, WRKYs, ERFs, TIFY, NDR1, EDS1 and RISP, which play important roles in poplar disease resistance, still need to be explored. In addition, plants defensive responses are complex, it is urgent to explore diverse crosstalk between different types of defensive responses in woody plants. The mechanism by which phytohormones interact with signaling molecules, e.g., ROS and NO, to influence poplar defense against pathogens also need to be investigated further. Further studies on the functions of ncRNAs in disease resistance and in regulating transcription and RNA silencing are of great significance.
Author contributions
YZ collected the data and wrote the manuscript. HS, LX, and LY gave some advice. SZ designed the framework and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Second Tibetan Plateau Scientific Expedition and Research Program (2019QZKK0404), the National Natural Science Foundation of China (32271830) and the Fundamental Research Funds for the Central Universities.
Acknowledgments
Because of space limitations, we apologize to our colleagues whose important works were not cited in this review.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
References
Agurla, S., Gahir, S., Munemasa, S., Murata, Y., Raghavendra, A. S. (2018). Mechanism of Stomatal Closure in Plants Exposed to Drought and Cold Stress. In: Iwaya-Inoue, M., Sakurai, M., Uemura, M. (eds) Survival Strategies in Extreme Cold and Desiccation. Advances in Experimental Medicine and Biology, vol 1081. Springer, Singapore. doi: 10.1007/978-981-13-1244-1_12
Alkan, N., Fortes, A. M. (2015). Insights into molecular and metabolic events associated with fruit response to post-harvest fungal pathogens. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00889
Azaiez, A., Boyle, B., Levee, V., Seguin, A. (2009). Transcriptome profiling in hybrid poplar following interactions with Melampsora rust fungi. Mol. Plant Microbe Interact. 22, 190–200. doi: 10.1094/MPMI-22-2-0190
Boyle, B., Levee, V., Hamel, L. P., Nicole, M. C., Seguin, A. (2010). Molecular and histochemical characterisation of two distinct poplar Melampsora leaf rust pathosystems. Plant Biol. 12, 364–376. doi: 10.1111/j.1438-8677.2009.00310.x
Bresson, A., Jorge, V., Dowkiw, A., Guerin, V., Bourgait, I., Tuskan, G. A., et al. (2011). Qualitative and quantitative resistances to leaf rust finely mapped within two nucleotide-binding site leucine-rich repeat (NBS-LRR)- rich genomic regions of chromosome 19 in poplar. New Phytol. 192, 151–163. doi: 10.1111/j.1469-8137.2011.03786.x
Chanclud, E., Morel, J. B. (2016). Plant hormones: a fungal point of view. Mol. Plant Pathol. 17, 1289–1297. doi: 10.1111/mpp.12393
Chang, M., Chen, H., Liu, F., Fu, Z. Q. (2022). PTI and ETI: convergent pathways with diverse elicitors. Trends Plant Sci. 27, 113–115. doi: 10.1016/j.tplants.2021.11.013
Chen, Q., Wang, Z., Li, D., Wang, F., Zhang, R., Wang, J. (2019). Molecular characterization of the ERF family in susceptible poplar infected by virulent Melampsora larici-populina. Physiol. Mol. Plant 108, 101437. doi: 10.1016/j.pmpp.2019.101437
Chen, Q., Zhang, R., Li, D., Wang, F. (2021). Integrating transcriptome and coexpression network analyses to characterize salicylic acid- and jasmonic acid-related genes in tolerant poplars infected with rust. Int. J. Mol. Sci. 22, 5001. doi: 10.3390/ijms22095001
Cheng, T., Chen, J., Ef, A. A., Wang, P., Wang, G., Hu, X., et al. (2016). Quantitative proteomics analysis reveals that s-nitrosoglutathione reductase (GSNOR) and nitric oxide signaling enhance poplar defense against chilling stress. Planta 243, 1081–1081. doi: 10.1007/s00425-016-2494-6
Chisholm, S. T., Coaker, G., Day, B., Staskawicz, B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814. doi: 10.1016/j.cell.2006.02.008
Cho, J. S., Nguyen, V. P., Jeon, H. W., Kim, M. H., Eom, S. H., Lim, Y. J., et al. (2016). Overexpression of PtrMYB119, a R2R3-MYB transcription factor from Populus trichocarpa, promotes anthocyanin production in hybrid poplar. Tree Physiol. 36, 162–1176. doi: 10.1093/treephys/tpw046
De Kesel, J., Conrath, U., Flors, V., Luna, E., Mageroy, M. H., Mauch-Mani, B., et al. (2021). The induced resistance lexicon: do’s and don’ts. Trends Plant Sci. 26, 685–691. doi: 10.1016/j.tplants.2021.01.001
Dixon, R. A., Xie, D. Y., Sharma, S. B. (2005). Proanthocyanidins - a final frontier in flavonoid research? New Phytol. 165, 9–28. doi: 10.1111/j.1469-8137.2004.01217.x
Dodds, P. N., Lawrence, G. J., Catanzariti, A. M., Ayliffe, M. A., Ellis, J. G. (2004). The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16, 755–768. doi: 10.1105/tpc.020040
Dodds, P. N., Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Dong, N. Q., Lin, H. X. (2021). Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 63, 180–209. doi: 10.1111/jipb.13054
Duan, Y., Jiang, Y., Ye, S., Karim, A., Ling, Z., He, Y., et al. (2015). PtrWRKY73, a salicylic acid-inducible poplar WRKY transcription factor, is involved in disease resistance in Arabidopsis thaliana. Plant Cell Rep. 34, 831–841. doi: 10.1007/s00299-015-1745-5
Duplessis, S., Major, I., Martin, F., Seguin, A. (2009). Poplar and pathogen interactions: insights from Populus genome-wide analyses of resistance and defense gene families and gene expression profiling. Crit. Rev. Plant Sci. 28, 309–334. doi: 10.1080/07352680903241063
Ellinger, D., Naumann, M., Falter, C., Zwikowics, C., Jamrow, T., Manisseri, C., et al. (2013). Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol. 161, 1433–1444. doi: 10.1104/pp.112.211011
Eyles, A., Bonello, P., Ganley, R., Mohammed, C. (2010). Induced resistance to pests and pathogens in trees. New Phytol. 185, 893–908. doi: 10.1111/j.1469-8137.2009.03127.x
Feau, N., Mottet, M. J., Périnet, P., Hamelin, R. C., Bernier, L. (2010). Recent advances related to poplar leaf spot and canker caused by Septoria musiva. can. J. Plant Pathol. 32, 122–134. doi: 10.1080/07060661003740009
Foster, A. J., Pelletier, G., Tanguay, P., Séguin, A. (2015). Transcriptome analysis of poplar during leaf spot infection with sphaerulina spp. PloS One 10, e0138162. doi: 10.1371/journal.pone.0138162
Germain, H., Seguin, A. (2011). Innate immunity: has poplar made its BED? New Phytol. 189, 678–687. doi: 10.1111/j.1469-8137.2010.03544.x
Gururani, M. A., Venkatesh, J., Upadhyaya, C. P., Nookaraju, A., Pandey, S. K., Park, S. W. (2012). Plant disease resistance genes: Current status and future directions. Physiol. Mol. Plant 78, 51–65. doi: 10.1016/j.pmpp.2012.01.002
Hamel, L. P., Miles, G. P., Samuel, M. A., Ellis, B. E., Séguin, A., Beaudoin, N. (2005). Activation of stress-responsive mitogen-activated protein kinase pathways in hybrid poplar (Populus trichocarpa x Populus deltoides). Tree Physiol. 25, 277–288. doi: 10.1093/treephys/25.3.277
Huang, Y., Ma, H., Yue, Y., Zhou, T., Zhu, Z., Wang, C. (2022). Integrated transcriptomic and transgenic analyses reveal potential mechanisms of poplar resistance to Alternaria alternata infection. BMC Plant Biol. 22, 413. doi: 10.1186/s12870-022-03793-5
Irigoyen, M. L., Garceau, D. C., Bohorquez-Chaux, A., Lopez-Lavalle, L. A. B., Perez-Fons, L., Fraser, P. D., et al. (2020). Genome-wide analyses of cassava pathogenesis-related (PR) gene families reveal core transcriptome responses to whitefly infestation, salicylic acid and jasmonic acid. BMC Genomics 21, 93. doi: 10.1186/s12864-019-6443-1
Jia, Z., Sun, Y., Yuan, L., Tian, Q., Luo, K. (2010). The chitinase gene (Bbchit1) from Beauveria bassiana enhances resistance to Cytospora chrysosperma in Populus tomentosa Carr. Biotechnol. Lett. 32, 1325–1332. doi: 10.1007/s10529-010-0297-6
Jiang, Y., Duan, Y., Yin, J., Ye, S., Zhu, J., Zhang, F., et al. (2014). Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J. Exp. Bot. 65, 6629–6644. doi: 10.1093/jxb/eru381
Jiang, Y., Guo, L., Ma, X., Zhao, X., Jiao, B., Li, C., et al. (2017). The WRKY transcription factors PtrWRKY18 and PtrWRKY35 promote Melampsora resistance in Populus. Tree Physiol. 37, 665–675. doi: 10.1093/treephys/tpx008
Karim, A., Jiang, Y., Guo, L., Ling, Z., Ye, S., Duan, Y., et al. (2015). Isolation and characterization of a subgroup iia WRKY transcription factor PtrWRKY40 from Populus trichocarpa. Tree Physiol. 35, 1129–1139. doi: 10.1093/treephys/tpv084
Kim, M. H., Cho, J. S., Bae, E. K., Choi, Y. I., Eom, S. H., Lim, Y. J., et al. (2021). PtrMYB120 functions as a positive regulator of both anthocyanin and lignin biosynthetic pathway in a hybrid poplar. Tree Physiol. 41, 2409–2423. doi: 10.1093/treephys/tpab082
Kohler, B., Hills, A., Blatt, M. R. (2003). Control of guard cell ion channels by hydrogen peroxide and abscisic acid indicates their action through alternate signaling pathways. Plant Physiol. 131, 385–388. doi: 10.1104/pp.016014
Kohler, A., Rinaldi, C., Duplessis, S., Baucher, M., Geelen, D., Duchaussoy, F., et al. (2008). Genome-wide identification of NBS resistance genes in Populus trichocarpa. Plant Mol. Biol. 66, 619–636. doi: 10.1007/s11103-008-9293-9
Kovalchuk, A., Keriö, S., Oghenekaro, A. O., Jaber, E., Raffaello, T., Asiegbu, F. O. (2013). Antimicrobial defenses and resistance in forest trees: challenges and perspectives in a genomic era. Annu. Rev. Phytopathol. 51, 221–244. doi: 10.1146/annurev-phyto-082712-102307
La Mantia, J., Klapste, J., El-Kassaby, Y. A., Azam, S., Guy, R. D., Douglas, C. J., et al. (2013). Association analysis identifies Melampsora×columbiana poplar leaf rust resistance SNPs. PloS One 8, e78423. doi: 10.1371/journal.pone.0078423
La Mantia, J., Unda, F., Douglas, C. J., Mansfield, S. D., Hamelin, R. (2018). Overexpression of AtGolS3 and CsRFS in poplar enhances ROS tolerance and represses defense response to leaf rust disease. Tree Physiol. 38, 457–470. doi: 10.1093/treephys/tpx100
Lang, J., Genot, B., Bigeard, J., Colcombet, J. (2022). MAP kinases 3 and 6 control salicylic acid signaling by upregulating NLR receptors during pattern- and effector-triggered immunity. J. Exp. Bot. 73, 2190–2205. doi: 10.1093/jxb/erab544
Levee, V., Major, I., Levasseur, C., Tremblay, L., MacKay, J., Seguin, A. (2009). Expression profiling and functional analysis of Populus WRKY23 reveals a regulatory role in defense. New Phytol. 184, 48–70. doi: 10.1111/j.1469-8137.2009.02955.x
Li, J., Brader, G., Kariola, T., Palva, E. T. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46, 477–491. doi: 10.1111/j.1365-313X.2006.02712.x
Li, J., Brader, G., Palva, E. T. (2004). The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331. doi: 10.1105/tpc.016980
Li, S., Fang, Y., Chen, L., Wang, J., Chen, X. W. (2021). Role of non-coding RNAs in plant immunity. Plant Commun. 2, 100180. doi: 10.1016/j.xplc.2021.100180
Li, N., Han, X., Feng, D., Yuan, D., Huang, L. J. (2019). Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: do we understand what they are whispering? Int. J. Mol. Sci. 20, 671. doi: 10.3390/ijms20030671
Li, L., Sun, W., Wang, P., Li, H., Rehman, S., Li, D., et al. (2022). Characterization, expression, and functional analysis of the pathogenesis-related gene PtDIR11 in transgenic poplar. Int. J. Biol. Macromol. 210, 182–195. doi: 10.1016/j.ijbiomac.2022.05.012
Li, D. L., Wang, F., Wang, C., Zou, L., Wang, Z. Y., Chen, Q. L., et al. (2016). MicroRNA-mediated susceptible poplar gene expression regulation associated with the infection of virulent Melampsora larici-populina. BMC Genomics 17, 59. doi: 10.1186/s12864-015-2286-6
Li, Y. X., Zhang, W., Dong, H. X., Liu, Z. Y., Ma, J., Zhang, X. Y. (2018). Salicylic acid in Populus tomentosa is a remote signalling molecule induced by Botryosphaeria dothidea infection. Sci. Rep. 8, 14059. doi: 10.1038/s41598-018-32204-9
Liang, H., Staton, M., Xu, Y., Xu, T., Leboldus, J. (2014). Comparative expression analysis of resistant and susceptible Populus clones inoculated with Septoria musiva. Plant Sci. 223, 69–78. doi: 10.1016/j.plantsci.2014.03.004
Liu, X., Chen, M., Zhou, X., Cao, Z. (2019). Identification of novel miRNAs and their target genes from Populus szechuanica infected with Melampsora larici-populina. Mol. Biol. Rep. 46, 3083–3092. doi: 10.1007/s11033-019-04746-2
Liu, H., Zhang, H., Yang, F., Chai, S., Wang, L., de, Dios, V. R., et al. (2022). Ethylene activates poplar defense against Dothiorella gregaria sacc by regulating reactive oxygen species accumulation. Physiol. Plant 174, e13726. doi: 10.1111/ppl.13726
Louet, C., Saubin, M., Andrieux, A., Persoons, A., Gorse, M., Petrowski, J., et al. (2021). A point mutation and large deletion at the candidate avirulence locus AvrMlp7 in the poplar rust fungus correlate with poplar RMlp7 resistance breakdown. Mol. Ecol 00, 1–12. doi: 10.1111/mec.16294
Ma, D., Constabel, C. P. (2019). MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 24, 275–289. doi: 10.1016/j.tplants.2018.12.003
Ma, D., Reichelt, M., Yoshida, K., Gershenzon, J., Constabel, C. P. (2018). Two R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar. Plant J. 96, 949–965. doi: 10.1111/tpj.14081
Ma, D., Tang, H., Reichelt, M., Piirtola, E. M., Salminen, J. P., Gershenzon, J., et al. (2021). Poplar MYB117 promotes anthocyanin synthesis and enhances flavonoid b-ring hydroxylation by up-regulating the flavonoid 3’, 5’-hydroxylase gene. J. Exp. Bot. 72, 3864–3880. doi: 10.1093/jxb/erab116
Maupetit, A., Larbat, R., Pernaci, M., Andrieux, A., Guinet, C., Boutigny, A. L., et al. (2018). Defense compounds rather than nutrient availability shape aggressiveness trait variation along a leaf maturity gradient in a biotrophic plant pathogen. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01396
Mayer, A. M., Staples, R. C., Gil-ad, N. L. (2001). Mechanisms of survival of necrotrophic fungal plant pathogens in hosts expressing the hypersensitive response. Phytochemistry 58, 33–41. doi: 10.1016/s0031-9422(01)00187-x
McCombe, C. L., Greenwood, J. R., Solomon, P. S., Williams, S. J. (2022). Molecular plant immunity against biotrophic, hemibiotrophic, and necrotrophic fungi. Essays Biochem. 66, 581–593. doi: 10.1042/EBC20210073
Mellway, R. D., Tran, L. T., Prouse, M. B., Campbell, M. M., Constabel, C. P. (2009). The wound-, pathogen-, and ultraviolet b-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiol. 150, 924–941. doi: 10.1104/pp.109.139071
Meng, X. Z., Xu, J., He, Y. X., Yang, K. Y., Mordorski, B., Liu, Y. D., et al. (2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25, 1126–1142. doi: 10.1105/tpc.112.109074
Meng, X., Zhang, S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266. doi: 10.1146/annurev-phyto-082712-102314
Miedes, E., Vanholme, R., Boerjan, W., Molina, A. (2014). The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00358
Miranda, M., Ralph, S. G., Mellway, R., White, R., Heath, M. C., Bohlmann, J., et al. (2007). The transcriptional response of hybrid poplar (Populus trichocarpa x p. deltoides) to infection by Melampsora medusae leaf rust involves induction of flavonoid pathway genes leading to the accumulation of proanthocyanidins. Mol. Plant Microbe Interact. 20, 816–831. doi: 10.1094/MPMI-20-7-0816
Pauwels, L., Goossens, A. (2011). The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23, 3089–3100. doi: 10.1105/tpc.111.089300
Pelloux, J., Rustérucci, C., Mellerowicz, E. J. (2007). New insights into pectin methylesterase structure and function. Trends Plant Sci. 12, 267–277. doi: 10.1016/j.tplants.2007.04.001
Petre, B., Hacquard, S., Duplessis, S., Rouhier, N. (2014). Genome analysis of poplar LRR-RLP gene clusters reveals RISP, a defense-related gene coding a candidate endogenous peptide elicitor. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00111
Petre, B., Hecker, A., Germain, H., Tsan, P., Sklenar, J., Pelletier, G., et al. (2016). The poplar rust-induced secreted protein (RISP) inhibits the growth of the leaf rust pathogen Melampsora larici-populina and triggers cell culture alkalinisation. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00097
Pieterse, C. M., Van, der. Does, D., Zamioudis., C., Leon-Reyes, A., Van, Wees, S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Polle, A., Janz, D., Teichmann, T., Lipka, V. (2013). Poplar genetic engineering: promoting desirable wood characteristics and pest resistance. Appl. Microbiol. Biotechnol. 97, 5669–5679. doi: 10.1007/s00253-013-4940-8
Ren, F., Yan, D. H., Wu, G., Sun, X., Song, X., Li, R. (2020). Distinctive gene expression profiles and effectors consistent with host specificity in two formae speciales of Marssonina brunnea. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00276
Rinaldi, C., Kohler, A., Frey, P., Duchaussoy, F., Ningre, N., Couloux, A., et al. (2007). Transcript profiling of poplar leaves upon infection with compatible and incompatible strains of the foliar rust Melampsora larici-populina. Plant Physiol. 144, 347–366. doi: 10.1104/pp.106.094987
Serrano, M., Coluccia, F., Torres, M., L'Haridon, F., Métraux, J. P. (2014). The cuticle and plant defense to pathogens. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00274
Shah, J. (2003). The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 6, 365–371. doi: 10.1016/s1369-5266(03)00058-x
Shi, G., Zhang, Z., Friesen, T. L., Raats, D., Fahima, T., Brueggeman, R. S., et al. (2016). The hijacking of a receptor kinase-driven pathway by a wheat fungal pathogen leads to disease. Sci. Adv. 2, e1600822. doi: 10.1126/sciadv.1600822
Shim, J. S., Jung, C., Lee, S., Min, K., Lee, Y. W., Choi, Y., et al. (2013). AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 73, 483–495. doi: 10.1111/tpj.12051
Silva, M. S., Arraes, F. B. M., Campos, M. A., Grossi-de-Sa, M., Fernandez, D., Cândido, E. S., et al. (2018). Review: Potential biotechnological assets related to plant immunity modulation applicable in engineering disease-resistant crops. Plant Sci. 270, 72–84. doi: 10.1016/j.plantsci.2018.02.013
Song, Q., Kong, L., Yang, X., Jiao, B., Hu, J., Zhang, Z., et al. (2022). PtoMYB142, a poplar R2R3-MYB transcription factor, contributes to drought tolerance by regulating wax biosynthesis. Tree Physiol. 42, 2133–2147. doi: 10.1093/treephys/tpac060
Spoel, S. H., Loake, G. J. (2011). Redox-based protein modifications: the missing link in plant immune signalling. 40rabidops Plant Biol. 14, 358–364. doi: 10.1016/j.pbi.2011.03.007
Steenackers, J., Steenackers, M., Steenackers, V., Stevens, M. (1996). Poplar diseases, consequences on growth and wood quality. Biomass Bioenerg. 10, 267–274. doi: 10.1016/0961-9534(95)00121-2
Su, Y., Li, H. G., Wang, Y., Li, S., Wang, H. L., Yu, L., et al. (2018). Poplar miR472a targeting NBS-LRRs is involved in effective defence against the necrotrophic fungus Cytospora chrysosperma. J. Exp. Bot. 69, 5519–5530. doi: 10.1093/jxb/ery304
Sun, W., Zhou, Y., Movahedi, A., Wei, H., Zhu, Q. (2020). Thaumatin-like protein (Pe-TLP) acts as a positive factor in transgenic poplars enhanced resistance to spots disease. Physiol. Mol. Plant 2020, 101512. doi: 10.1016/j.pmpp.2020.101512
Syvertsen, J. P., Garcia-Sanchez, F. (2014). Multiple abiotic stresses occurring with salinity stress in citrus. Environ. Exp. Bot. 103, 128–137. doi: 10.1016/j.envexpbot.2013.09.015
Thaler, J. S., Humphrey, P. T., Whiteman, N. K. (2012). Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17, 260–270. doi: 10.1016/j.tplants.2012.02.010
Ton, J., Flors, V., Mauch-Mani, B. (2009). The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14, 310–317. doi: 10.1016/j.tplants.2009.03.006
Tuskan, G. A., DiFazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Ullah, C., Schmidt, A., Reichelt, M., Tsai, C.-J., Gershenzon, J. (2022). Lack of antagonism between salicylic acid and jasmonate signalling pathways in poplar. New Phytol. 235, 701–717. doi: 10.1111/nph.18148
Ullah, C., Tsai, C. J., Unsicker, S. B., Xue, L., Reichelt, M., Gershenzon, J., et al. (2019a). Salicylic acid activates poplar defense against the biotrophic rust fungus Melampsora larici-populina via increased biosynthesis of catechin and proanthocyanidins. New Phytol. 221, 960–975. doi: 10.1111/nph.15396
Ullah, C., Unsicker, S. B., Fellenberg, C., Constabel, C. P., Schmidt, A., Gershenzon, J., et al. (2017). Flavan-3-ols are an effective chemical defense against rust infection. Plant Physiol. 175, 1560–1578. doi: 10.1104/pp.17.00842
Ullah, C., Unsicker, S. B., Reichelt, M., Gershenzon, J., Hammerbacher, A. (2019b). Accumulation of catechin and proanthocyanidins in black poplar stems after infection by plectosphaerella populi: hormonal regulation, biosynthesis and antifungal activity. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01441
Wan, S. Z., Li, C. F., Ma, X. D., Luo, K. M. (2017). PtrMYB57 contributes to the negative regulation of anthocyanin and proanthocyanidin biosynthesis in poplar. Plant Cell Rep. 36, 1263–1276. doi: 10.1007/s00299-017-2151-y
Wang, W., Bai, X. D., Chen, K., Gu, C. R., Yu, Q. B., Jiang, J., et al. (2022). Role of PsnWRKY70 in regulatory network response to infection with Alternaria alternata (Fr.) keissl in Populus. Int. J. Mol. Sci. 23, 7537. doi: 10.3390/ijms23147537
Wang, N. A., Cao, P., Xia, W. X., Fang, L. C., Yu, H. Y. (2017b). Identification and characterization of long non-coding RNAs in response to early infection by Melampsora larici-populina using genome-wide high-throughput RNA sequencing. Tree Genet. Genomes 13, 34. doi: 10.1007/s11295-017-1116-1
Wang, Y., Feng, L., Zhu, Y., Li, Y., Yan, H., Xiang, Y. (2015). Comparative genomic analysis of the WRKY III gene family in populus, grape, 44rabidopsis and rice. Biol. Direct 10, 48. doi: 10.1186/s13062-015-0076-3
Wang, X., Jiang, N., Liu, J., Liu, W., Wang, G. L. (2014). The role of effectors and host immunity in plant-necrotrophic fungal interactions. Virulence 5, 722–732. doi: 10.4161/viru.29798
Wang, L., Lu, W., Ran, L., Dou, L., Yao, S., Hu, J., et al. (2019). R2R3-MYB transcription factor MYB6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J. 99, 733–751. doi: 10.1111/tpj.14364
Wang, L. J., Ran, L. Y., Hou, Y. S., Tian, Q. Y., Li, C. F., Liu, R., et al. (2017a). The transcription factor MYB115 contributes to the regulation of proanthocyanidin biosynthesis and enhances fungal resistance in poplar. New Phytol. 215, 351–367. doi: 10.1111/nph.14569
Wang, H., Wang, X., Yu, C., Wang, C., Jin, Y., Zhang, H. (2020). MYB transcription factor PdMYB118 directly interacts with bHLH transcription factor PdTT8 to regulate wound-induced anthocyanin biosynthesis in poplar. BMC Plant Biol. 20, 173. doi: 10.1186/s12870-020-02389-1
Wei, H., Movahedi, A., Xu, C., Sun, W., Wang, X., Li, D., et al. (2020a). Overexpression of PtDefensin enhances resistance to Septotis populiperda in transgenic poplar. Plant Sci. 292, 110379. doi: 10.1016/j.plantsci.2019.110379
Wei, H., Movahedi, A., Xu, C., Sun, W., Wang, P., Li, D., et al. (2020b). Characterization, expression profiling, and functional analysis of PtDef, a defensin-encoding gene from Populus trichocarpa. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00106
Wei, S., Wu, H., Li, X., Chen, Y., Yang, Y., Dai, M., et al. (2020c). Identification of genes underlying the resistance to Melampsora larici-populina in an R gene supercluster of the populus deltoides genome. Plant Dis. 104, 1133–1143. doi: 10.1094/PDIS-08-19-1699-RE
Weiland, J. E., Stanosz, J. C., Stanosz, G. R. (2003). Prediction of long-term canker disease damage from the responses of juvenile poplar clones to inoculation with Septoria musiva. Plant Dis. 87, 1507–1514. doi: 10.1094/PDIS.2003.87.12.1507
Wu, W., Nemri, A., Blackman, L. M., Catanzariti, A. M., Sperschneider., J., Lawrence, G. J., et al. (2019). Flax rust infection transcriptomics reveals a transcriptional profile that may be indicative for rust Avr genes. PloS One 14, e0226106. doi: 10.1371/journal.pone.0226106
Xia, W., Yu, H., Cao, P., Luo, J., Wang, N. (2017). Identification of TIFY family genes and analysis of their expression profiles in response to phytohormone treatments and Melampsora larici-populina infection in poplar. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00493
Xu, W. J., Dubos, C., Lepiniec, L. (2015b). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20, 176–185. doi: 10.1016/j.tplants.2014.12.001
Xu, J., Wang, X.-y., Guo, W.-z. (2015a). The cytochrome P450 superfamily: Key players in plant development and defense. J. Integr. Agr. 14, 1673–1686. doi: 10.1016/s2095-3119(14)60980-1
Yang, Y. X., Ahammed, G. J., Wu, C., Fan, S. Y., Zhou, Y. H. (2015). Crosstalk among jasmonate, salicylate and ethylene signaling pathways in plant disease and immune responses. Curr. Protein Pept. Sci. 16, 450–461. doi: 10.2174/1389203716666150330141638
Yang, W., Ruan, M., Xiang, M., Deng, A., Du, J., Xiao, C. (2020). Overexpression of a pectin methylesterase gene PtoPME35 from Populus tomentosa influences stomatal function and drought tolerance in Arabidopsis thaliana. biochem. Biophys. Res. Commun. 523, 416–422. doi: 10.1016/j.bbrc.2019.12.073
Yevtushenko, D. P., Misra, S. (2019). Enhancing disease resistance in poplar through modification of its natural defense pathway. Plant Mol. Biol. 100, 481–494. doi: 10.1007/s11103-019-00874-2
Yoo, S. D., Sheen, J. (2008). MAPK signaling in plant hormone ethylene signal transduction. Plant Signal Behav. 3, 848–849. doi: 10.4161/psb.3.10.5995
Yoshida, K., Ma, D. W., Constabel, C. P. (2015). The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol. 167, 693–69+. doi: 10.1104/pp.114.253674
Yu, Z. D., Kuo, C. S., Newcombe, G., Jun, F. F., Qian, W. C. (2019). Leaf cuticle can contribute to non-host resistance to poplar leaf rust. Forests 10, 870. doi: 10.3390/f10100870\
Yuan, Y., Chung, J. D., Fu, X., Johnson, V. E., Ranjan, P., Booth, S. L., et al. (2009). Alternative splicing and gene duplication differentially shaped the regulation of isochorismate synthase in Populus and Arabidopsis. proc. Natl. Acad. Sci. U.S.A. 106, 22020–22025. doi: 10.1073/pnas.0906869106
Zhang, S., Lu, S., Xu, X., Korpelainen, H., Li, C. (2010). Changes in antioxidant enzyme activities and isozyme profiles in leaves of male and female Populus cathayana infected with Melampsora larici-populina. Tree Physiol. 30, 116–128. doi: 10.1093/treephys/tpp094
Zhang, Y., Tian, L., Yan, D. H., He, W. (2018). Genome-wide transcriptome analysis reveals the comprehensive response of two susceptible poplar sections to Marssonina brunnea infection. Genes (Basel) 9, 154. doi: 10.3390/genes9030154
Zhao, L., Cheng, Q. (2022). Heterologous expression of Arabidopsis pattern recognition receptor RLP23 increases broad-spectrum resistance in poplar to fungal pathogens. Mol. Plant Pathol. 24, 80–86. doi: 10.1111/mpp.13275
Zhao, J. P., Jiang, X. L., Zhang, B. Y., Su, X. H. (2012). Involvement of microRNA-mediated gene expression regulation in the pathological development of stem canker disease in Populus trichocarpa. PloS One 7, e44968. doi: 10.1371/journal.pone.0044968
Keywords: poplar, fungus, physiology, molecular mechanism, defense
Citation: Zeng Y, Song H, Xia L, Yang L and Zhang S (2023) The responses of poplars to fungal pathogens: A review of the defensive pathway. Front. Plant Sci. 14:1107583. doi: 10.3389/fpls.2023.1107583
Received: 25 November 2022; Accepted: 06 February 2023;
Published: 16 February 2023.
Edited by:
Hongjie Feng, National Key Laboratory of Cotton Biology (CAAS), ChinaReviewed by:
Jonathan Gershenzon, Max Planck Institute for Chemical Ecology, GermanyQingxue Guo, Hangzhou Normal University, China
Fan Yang, Hainan University, China
Copyright © 2023 Zeng, Song, Xia, Yang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheng Zhang, c2hlbmd6aGFuZ0BzY3UuZWR1LmNu
 Yi Zeng
Yi Zeng Haifeng Song2
Haifeng Song2 Linchao Xia
Linchao Xia Le Yang
Le Yang Sheng Zhang
Sheng Zhang