Abstract
Plant parasitic nematodes (PPNs) cause an important class of diseases that occur in almost all types of crops, seriously affecting yield and quality and causing great economic losses. Accurate and rapid diagnosis of nematodes is the basis for their control. PPNs often have interspecific overlays and large intraspecific variations in morphology, therefore identification is difficult based on morphological characters alone. Instead, molecular approaches have been developed to complement morphology-based approaches and/or avoid these issues with various degrees of achievement. A large number of PPNs species have been successfully detected by biochemical and molecular techniques. Newly developed isothermal amplification technologies and remote sensing methods have been recently introduced to diagnose PPNs directly in the field. These methods have been useful because they are fast, accurate, and cost-effective, but the use of integrative diagnosis, which combines remote sensing and molecular methods, is more appropriate in the field. In this paper, we review the latest research advances and the status of diagnostic approaches and techniques for PPNs, with the goal of improving PPNs identification and detection.
1 Introduction
The phylum Nematoda is one of the largest in the animal kingdom, including many species and a wide variety of lifestyles. More than 25 000 nematode species are currently known (Zhang, 2013). Of these, 50% are marine salt water and 25% dwell in soil and freshwater. (Hassan et al., 2015). Over 4100 species of PPNs have been described to date (Decraemer and Hunt, 2006) representing an important constraint on global food security. They parasitize a wide range of plant species, including monocots and dicots, and are one of the most severe limiting factors for major crops, causing an estimated annual crop loss of at least 80$ billion worldwide (Nicol et al., 2011). Nematode diseases are difficult to control because their symptoms could be largely inapparent, hence, they are often overlooked. Nematode identification and differentiation can allow accurate decisions for the control of these plant parasites and the conservation of non‐parasitic nematodes.
The challenge in differentiating nematodes is not only the selection of the most accurate and suitable methods, but also due to other factors possibly effects the performance of the identification assays, such as the small size of the nematode, the high number of nematodes found in the samples, and/or the lack of particular morphological characteristics. (Floyd et al., 2002; Chitwood, 2003). The traditional classification of PPNs is based on morphological characteristics combined with morphometric values. The variations in some of these morphological and morphometric features are often conjectural, subtle, and have overlapping characteristics or show intraspecific variation that compromises accurate identification or may result in mistaken identification of species (Oliveira et al., 2011). Moreover, morphological identification is complex and time-consuming, requiring specialized and experienced researchers to be accurate (Carneiro et al., 2017).
The recent rapid development of Polymerase Chain Reaction (PCR)-based methods has facilitated their wide use for the detection and identification of PPNs. Since its invention, PCR has been one of the most prevalent and essential molecular biology methods. Currently, the PCR detection techniques applied to PPNs mainly include DNA barcoding, restriction fragment length polymorphism of the internal transcribed spacer region of ribosomal DNA (ITS-RFLP), sequence characterized amplified regions (SCAR), random amplified polymorphic DNA (RAPD), and Real-time Quantitative Polymerase Chain Reaction (RT-qPCR). Many target genes for PCR methods have been used to identify PPNs using the universal primers (Table 1),such as rDNA -ITS, rDNA - intergenic spacer region (IGS) (Wishart et al., 2002), 28S D2-D3 (Vallejo et al., 2021), heat shock proteins (Green et al., 2019), 18S (small subunit; SSU) (Floyd et al., 2002), and mitochondrial DNA (mtDNA) (Stanton et al., 1997). These molecular approaches compensate for the failings of traditional morphological identification to a certain extent. One or more nematode species can be detected in a mixed sample by a PCR assay, reducing the time and cost of diagnosis (Keçici et al., 2022).
Table 1
| Primercombination and code(direction) | Primersequence(5′-3′) | Amplified region | References |
|---|---|---|---|
| G18SU(f) | GCTTGCCTCAAAGATTAAGCC | 18SrRNA | (Blaxter et al., 1998) |
| R18Tyl1(r) | GGTCCAAGAATTTCACCTCTC | (Chizhov et al., 2006) | |
| F18Tyl2(f) | CAGCCGCGGTAATTCCAGC | 18SrRNA | (Chizhov et al., 2006) |
| R18Tyl2(r) | CGGTGTGTACAAAGGGCAGG | ||
| 988F(f) | CTCAAAGATTAAGCCATGC | 18SrRNA | (Holterman et al., 2006) |
| 1912R(r | TTTACGGTCAGAACTAGGG | ||
| 1096F(f) | GGTAATTCTGGAGCTAATAC | 18SrRNA | (Holterman et al., 2006) |
| 1912R(r) | TTTACGGTCAGAACTAGGG | ||
| 1813F(f) | CTGCGTGAGAGGTGAAAT | 18SrRNA | (Holterman et al., 2006) |
| 2646R(r) | GCTACCTTGTTACGACTTTT | ||
| SSU_F_04 | GCTTGTCTCAAAGATTAAGCC | 18SrRNA | (Blaxter et al., 1998) |
| SSU_R_09 | AGCTGGAATTACCGCGGCTG | ||
| SSU_F_22 | TCCAAGGAAGGCAGCAGGC | 18SrRNA | (Blaxter et al., 1998) |
| SSU_R_13 | GGGCATCACAGACCTGTTA | ||
| SSU_F_23 | ATTCCGATAACGAGCGAGA | 18SrRNA | (Blaxter et al., 1998) |
| SSU_R_81 | TGATCCWKCYGCAGGTTCAC | ||
| designated Nem_18S_F |
CGCGAATRGCTCATTACAACAGC | 18SrRNA | (Floyd et al., 2005) |
| Nem_18S_R | GGGCGGTATCTGATCGCC | ||
| 18S-CL-F3 | CTTGTCTCAAAGATTAAGCCATGCAT | 18SrRNA+ ITS1-5.8S- ITS2rRNA+ 28SrRNA |
(Carta and Li, 2018; Carta and Li, 2019) |
| 28S-CL-R | CAGCTACTAGATGGTTCGATTAGTC | ||
| 18S(f) | TTGATTACGTCCCTGCCCTTT | ITS1-rRNA | (Vrain et al., 1992) |
| rDNA1.58S(r) | ACGAGCCGAGTGATCCACCG | (Szalanski et al., 1997) | |
| TW81(f) | GTTTCCGTAGGTGAACCTGC | ITS1-rRNA | (Curran et al., 1994) |
| 5.8SM5(r) | GGCGCAATGTGCATTCGA | (Zheng et al., 2000) | |
| 18S(f) | TTGATTACGTCCCTGCCCTTT | ITS1-5.8S- ITS2rRNA |
(Vrain et al., 1992) |
| 26S(r) | TTTCACTCGCCGTTACTAAGG | ||
| F194(f) | CGTAACAAGGTAGCTGTAG | ITS1-5.8S- ITS2rRNA |
(Ferris et al., 1993) |
| F195(r) | TCCTCCGCTAAATGATATG | ||
| TW81(f) | GTTTCCGTAGGTGAACCTGC | ITS1-5.8S- ITS2rRNA |
(Curran et al., 1994) |
| AB21(r) | ATATGCTTAAGTTCAGCGGGT | ||
| D2A(f) | ACAAGTACCGTGAGGGAAAGTTG | D2-D3of28S rRNA |
(Nunn, 1992) |
| D3B(r) | TCGGAAGGAACCAGCTACTA | ||
| D2Tyl(f) | GAGAGAGTTAAANAGBACGTGA | D2-D3of28S | (Chizhov et al., 2012) |
| D3B(r) | TCGGAAGGAACCAGCTACTA | rRNA | (Nunn, 1992) |
| D2A(f) | ACAAGTACCGTGAGGGAAAGTTG | D2of 28S rRNA |
(Nunn, 1992) |
| D2A(r) | GACCCGTCTTGAAACACGGA |
Some universal primer combinations used for amplification of ribosomal RNA genes of PPNs.
The advent of detection techniques for isothermal amplification, including loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA), provides additional options for the identification of PPNs. These technologies are characterized by high specificity and sensitivity. These two methods combined with the Lateral Flow Dipstick (LFD) allow for the clear visualization of the amplification products in the field, which can be identified by the naked eye (Yao et al., 2021). They also work well for field or point-of-service-based nematode detection and diagnosis. The combination of CRISPR Cas12a with RPA and LAMP methods has a detection sensitivity at the attomolar level. The specificity is enhanced by the isothermal detection technique (Gootenberg et al., 2017). CRISPR/Cas12a-based nucleic acid detection technology has been successfully used to test Heterodera schachtii (Yao et al., 2021), H. avenae, and H. filipjevi (Shao et al., 2022). Additionally, the development of remote sensing technology has brought new opportunities for extensive field monitoring and management of nematodes.
This article is a review of common methods for the identification of PPNs, which focuses on new isothermal amplification technologies and remote sensing methods capable of revolutionizing the approach for PPNs detection in the field.
2 Biochemical detection methods for PPNs
2.1 Isozymes analyses
Enzyme phenotyping methods, also named multifocal enzyme electrophoresis (MEE), were determined by the transport modes of isozymes, as variations in charges, molecular volumes and conformations arise from slight changes in their amino acid composition (Bogale et al., 2020). This method has the advantages of high stability, high polymorphism, and accuracy (Carneiro et al., 2017). It was first applied in the early 1970s for the identification of several common root-knot nematodes (Meloidogyne spp.) (Dickson et al., 1970). Many root-knot nematodes including Meloidogyne javanica, M. incognita, M. arenaria, M. exigua, and M. paranaensis have been identified using isozyme techniques (Carneiro et al., 2004; Carneiro et al., 2008; Muniz et al., 2008). Although this technique has been studied for other nematodes such as H. glycines, Ditylenchus triformic, and Aphelenchus avenae (Dickson et al., 1970), it has been best applied only for root-knot nematodes. The main reason is that certain proteins are only expressed at specific stages of the nematode life cycle, hence the isozyme extraction has strict requirements vis a vis the worm’s state (Esbenshade and Triantaphyllou, 1985; Esbenshade and Triantaphyllou, 1990). Generally, only young females can be used. Except for root-knot nematodes, young females of other plant nematode species are relatively difficult to obtain.
2.2 Mass spectral analyses
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) has been widely used as a diagnostic technology in laboratories for the analysis of complex molecules, by producing protein fingerprint signatures from protein extracts of organisms (Bizzini et al., 2010). MALDI-TOF MS is a highly sensitive, rapid, and reliable diagnosis method (Seng et al., 2009; Sandrin et al., 2013). Recently, researchers have discovered that MALDI biotechnology can be used for viruses, protozoa, and arthropods in addition to bacteria, mycobacteria, and fungi (Sjöholm et al., 2008; Yssouf et al., 2016; Angeletti, 2017; Vega-Rúa et al., 2018). Today, MALDI-TOF MS has also been applied for the identification of the PPNs Anguina tritici, A. funesta, M. javanica and M. incognita (Perera et al., 2005; Ahmed et al., 2011). With the increasing development of MALDI-TOF MS technology, the reduction of instrument cost, and the improvement of related databases, the technique will become a powerful tool for PPNs identification soon.
3 Molecular diagnosis of PPNs
3.1 Traditional PCR methods
3.1.1 RFLP
RFLP uses restriction enzymes to either digest genomic DNA or amplified fragments, producing DNA banding patterns based on sequence divergence (Brown, 1981). The RFLP technique has the characteristics of high sensitivity, a requirement for a low amount of DNA, rapidity, and accuracy (Jarcho, 2001). The technique was first applied to the identification of nematode species by Curran et al. (1985). This method has been applied successively to identify root-knot nematodes and their physiological subspecies (Curran et al., 1986; Powers and Sandall, 1988; Zijlstra et al., 1995), Xiphinema aameracanum (Vrain, 1993), Diylenchus spp. (Wendt et al., 1993; Mahmoudi et al., 2020), Bursaphelenchus spp. (Aikawa et al., 2013), and Heterodera spp. (Zheng et al., 2003; Ou et al., 2008b; Baklawa et al., 2015). Although this method is valid in differentiating nematode isolates, it is less frequently used today owing to the complicated nature of its technique and the need for significant numbers of target DNA, usually requiring pre-culture of nematode populations (Currie et al., 2000).
3.1.2 RAPD and SCAR
The RAPD method was invented by Williams et al. (1991) and is a novel genetic marker. The method involves PCR amplification of target DNA using a random sequence of 9–10 nucleotides as a primer. Polymorphism can occur due to a difference of one base in the DNA sequence from the complementary oligonucleotide primer. The use of RAPD markers for PPNs identification has the benefits of rapidity, ease, and sensitivity. Caswell-Chen et al. (1992) distinguished H. curicifrae from H. schachtii by RAPD and detected differences among six geographic populations of H. schachtii. Subsequently, this method was studied on both root-knot nematodes and cyst nematodes (Cenis, 1993). Because the RAPD assay is performed at a low temperature, creating a lower degree of severity for primer reductions, and replicability, in particular between laboratories. It also imposes a restriction, making it impossible to use in the field.
To compensate for the shortcomings of RAPD, it can be converted into a SCAR marker technique as proposed and applied by Paran and Michelmore in 1993. This technique not only has the characteristics of high specificity and sensitivity of the RAPD method, but has the advantages of good stability and reproducibility (Li et al., 2022). This method solves the problem of long primers and a high annealing temperature for RAPD. Fullaondo et al. (1999) transformed RAPD markers into SCAR markers to differentiate between Globodera rostochiensis and G. pallida. Subsequently, SCAR markers have been successfully used to identify Meloidogyne spp. (Lecouls et al., 1999; Zijlstra et al., 2000; Randig et al., 2002), Heterodera spp. (Ou et al., 2008a; Qi et al., 2012; Liu et al., 2014; Jiang et al., 2021), and Bursaphelenchus spp. (Chen et al., 2011; Feng et al., 2011).
3.1.3 DNA barcoding
The DNA barcoding technique was first proposed by Hebert et al. (2003), and it uses a universal barcode to build a barcode database and analyze DNA data based on sample information to achieve identification. The advantages of this method are high primer versatility, a stable amplification system, a convenient fragment size, and low DNA sample quality requirements (Ahmed et al., 2015). DNA barcoding techniques have recently been used to study the species and phylogenetic relationships of nematodes including Meloidogyne spp. (Rashidifard et al., 2019), Heterodera spp. (Subbotin et al., 2019), and Bursaphelenchus spp. (Wang et al., 2015). Metabarcoding is a combination of barcoding and high-throughput sequencing (NGS). Metabarcoding was described by Taberlet et al. (2012) as the automatic identification of multiple species from a single bulk sample including several different taxa. Waite et al. (2003) used this method for community analysis of nematodes using 18S rDNA. Palomares-Rius et al. (2017) applied barcoding methods using mtDNA and rDNA regions to the phylogenetic analysis of PPNs from Longidoridae (Nematoda, Enoplea). There are several difficulties in the analysis of DNA metabarcoding of environmental DNA (eDNA). The eDNA is susceptible to contamination during sampling, extraction, and storage; the availability of species-specific DNA barcodes relies on the mass of the available databases. The identification of PPNs species is difficult due to the lack of available data for DNA barcoding of most known plant nematodes (Sikder et al., 2020). DNA barcoding is a tool with much potential for taxonomy. Currently, the metabarcoding technique is little utilized for PPNs detection and can be more developed in the future for PPNs identification.
3.1.4 Quantitative real-time PCR (qPCR)
The fluorescent qPCR technique adds fluorescent moieties to a PCR reaction system and monitors the entire PCR process in real-time by the accumulation of the fluorescent signal. The qPCR method allows continuous monitoring of the sample during PCR using fluorescence probes or double-stranded dyes such as SYBR Green I. The method is used to quantify the unknown template by means of a standard curve. The qPCR method has the advantages of sensitivity, reliability, safety, and allowing high throughput (Smith and Osborn, 2009). A quantitative PCR technique has been developed for targeting PPNs, containing M. enterolobii (Kiewnick et al., 2015), M. javanica, Xiphinema elongatum, and Pratylenchus zeae (Berry et al., 2008), P. penetrans (Sato et al., 2007), H. avenae and H. latipons (Toumi et al., 2013), H. schachtii (Madani et al., 2005), and H. glycines (Goto et al., 2009; Baidoo et al., 2017). Specific technologies for PPNs identification and quantification directly from the soil or plant tissues before DNA extraction and amplification have been recently explored (Goto et al., 2009; Lopez-Nicora et al., 2012; Li et al., 2014; Jian et al., 2022). These molecular detection methods can reduce the time and labor required for identification since they eliminate the need to extract nematodes from the soil and microscopy. Although qPCR is a sensitive method for detecting low concentrations of target DNA, its use for the identification of PPNs is hampered by its cost and dependence on expensive equipment.
3.1.5 Droplet digital PCR (ddPCR) technology
The concept of digital PCR was first described in 1992 by Sykes et al. (1992). It quantifies DNA molecules using a combination of the Poisson distribution and the dilution of templates to the single molecule level (Espy et al., 2006). The principle of ddPCR is to reduce a traditional PCR reaction mixture, which is like the Taqman assay, into a smaller reaction system either by diluting it in microwell plates, oil emulsion, or capillaries (Rougemont et al., 2004). It has the advantage of being very accurate at very low concentrations, with less contamination, and may be easier to sample for some diseases that are difficult to diagnose accurately (Li et al., 2018). Compared to qPCR, the ddPCR system could be used for the absolute quantitation of DNA copy numbers. The ddPCR method has high sensitivity and does not depend on a pre-enrichment for templates in extremely low concentrations. The ddPCR method has been successfully introduced into the clinic for the diagnosis of infectious diseases. Also the ddPCR has been utilized for the identification of a variety of plant pathogens including fungi, bacteria, and viruses. (Rani et al., 2019). Currently, this method has been applied to M. enterolobii (Chen et al., 2022).
3.2 Isothermal amplification technologies
3.2.1 LAMP
LAMP is designed on based on automated cycling and high DNA strand replacement activity mediated by Bst polymerase. It uses 4-6 oligonucleotide primers to produce a significant amount of amplicons within 10-20min (Notomi et al., 2000). LAMP is becoming a popular assay for the detection of PPNs, because it is rapid, sensitive and easy to use in a point-of-service environment (Ahuja, 2020). The inclusion of a fluorescent dye in a positive LAMP reaction generated a color difference that enabled observation by the naked eye. (He et al., 2013). Moreover, it has also been advanced by using Lateral flow devices (LFDs) to confirm visually the existence of amplicons (Kiatpathomchai et al., 2008; Ding et al., 2010). The LAMP-LFD method allows both nucleic acid amplification and amplicon visualization to be conducted without any complex or costly equipment, which promises to enhance usability for field investigations and common field monitoring. However, the disadvantage of LAMP is that once the tube is opened, aerosol contamination can easily form, causing more serious problem of false positives. In combination with a real-time turbidimeter, LAMP results can be measured accurately and contamination can be avoided (Mori et al., 2004). With these benefits, LAMP technology has been packaged in commercially available assay kits for the testing of a diversity of pathogens which include viruses, fungi and bacteria. (Mori et al., 2001). The use of this method on PPNs has been very popular in recent years. In particular, LAMP technology has been developed for diagnosing many species of PPNs including Bursaphelenchus spp., Meloidogyne spp., Anguina spp., Radopholus spp., Ditylenchus spp., and Tylenchulus spp. (Table 2).
Table 2
| Genus name | Nematode species | Target region | Host | References |
|---|---|---|---|---|
| Bursaphelenchus | B. xylophilus | ITS-rDNA | Pine | (Kikuchi et al., 2009) |
| B. xylophilus | ITS-rDNA | Pinus armandii var. | (Kanetani et al., 2011) | |
| B. xylophilus | Pectate lyase-3 | Pine | (Kang et al., 2015) | |
| B. cocophilus | D2-D3 of rDNA | coconut and oil palm trees | (Ide et al., 2017) | |
| Meloidogyne | Meloidogyne incognita, M. arenaria, M. javanica, M. hapla | ITS of rDNA | tomato | (Niu et al., 2011) |
| M. enterolobii | 5S rDNA-IGS2 | tomato | (Niu et al., 2012) | |
| M. mali | ITS-5.8S rDNA | tomato | (Zhou et al., 2017) | |
| M. chitwoodi and M. fallax | IGS2-18S | tomato | (Zhang and Gleason, 2019) | |
| M. partityla | ITS-5.8S rDNA | mature pecan trees | (Waliullah et al., 2020) | |
| Anguina | A. wevelli | ITS rDNA | — | (Yu et al., 2018) |
| A. agrostis | ITS rDNA | — | (Yu et al., 2020) | |
| Radopholus | R. similis | D2-D3 of rDNA | Anthurium | (Peng et al., 2012) |
| Ditylenchus | D. destructor | 28S rRNA | patato | (Deng et al., 2019) |
| Tylenchulus | T. semipenetrans | ITS-rDNA | cirus orchards | (Lin et al., 2016) |
| T. semipenetrans | ITS1 | citrus rhizosphere soil | (Song et al., 2017) |
The application of the LAMP technique to plant parasitic nematodes.
3.2.2 RPA
RPA is a novel, highly sensitive, isothermal DNA amplification and detection assay (Piepenburg et al., 2006). The technique is performed at 37-42°C and only requires a minimum number of DNA samples to amplify 1-10 target copies of DNA within 20 minutes (Sabate del Rio et al., 2017). RPA products can be detected by using fluorescent probes in real-time or by agarose gel electrophoresis or a lateral flow assay (Lobato and O'sullivan, 2018). The main advantages of RPA technology over other PCR detection technologies are that it is quick, sensitive, simple, and easy to use in the field. Compared to the LAMP, which needs 6-8 primers for amplification, RPA technology is simpler and requires only one pair of primers to finish amplification. RPA has been successfully applied to different species of target organisms including viruses, fungi, bacteria, animals and plants. (Lobato and O'sullivan, 2018). It has recently been reported to be highly effective in testing for a large range of PPNs including M. javanica (Chi et al., 2020), M. enterolobii (Subbotin, 2019), M. hapla (Subbotin and Burbridge, 2021), B. xylophilus, M. incognita, M. javanica, and M. arenaria (Ju et al., 2019) (Table 3). Although RPA has been described as highly specific, it has been reported that RPA depends on the number and distribution of mismatches in sequences of closely related DNA molecules. If one or more bases are mismatched, nematode populations cannot be differentiated based on their distribution.
Table 3
| Nematode species | Target | Time (min) | Temp (°C) | Sensitivity | References |
|---|---|---|---|---|---|
| M. enterolobii | IGS rRNA | 20 | 37 | 1/10 of a second-stage juvenile(J2) | (Subbotin, 2019) |
| M. javanica | SCAR marker | 40 | 39 | 1 pg purified genomic DNA, or 0.01 adult female, or 0.1 J2 | (Chi et al., 2020) |
| M. hapla | IGS rRNA | 20 | 39 | 1/100 of a J2 and 1/1000 of a female | (Subbotin and Burbridge, 2021) |
| M. enterolobii, M. incognita, M. javanica and M. arenaria | SCAR marker | 20 | 38 | 10−2, 10−2, 10−1, and 10−1 dilutions of DNA from a single J2 | (Ju et al., 2019) |
| H. schachtii | RAPD marker | 15-60 | 37 | 10−4 single cysts and single females, 4−3 single second-stage juveniles, and a 0.001 ng genomic DNA | (Yao et al., 2021) |
| H. avenae and H. filipjevi | SCAR marker | 15 | 35 | 10-4 single second-stage juvenile (J2), 10-5 single cyst, and 0.001 ng of genomic DNA | (Shao et al., 2022) |
| B. xylophilus | ITS2 | 25 | 37 | 308 ± 51 of B. xylophilus per 10 g of pinewood |
Information about reported studies of RPA in nematodes.
Coupling RPA with CRISPR (Cas) systems identifies stable differences in individual bases. Cas12a and CRISPR form ribonucleoprotein, which recognizes the protospacer adjacent motif (PAM) site on the target nucleic acid and then guides the effector Cas protein to shear the target sequence. The Cas12a enzyme can non-specifically be a shear single-stranded DNA reporter-labeled fluorophore and quencher (Chen et al., 2018). It has been concluded that RPA- CRISPR/Cas12a is more sensitive and specific than RPA alone. The PPNs H. schachtii (Yao et al., 2021), H. avenae, and H. filipjevi (Shao et al., 2022) have been detected using RPA-CRISPR/Cas12a technology. The combination of LAMP and CRISPR/Cas12a can also be used for pathogen detection. The use of Cas12a is a powerful method for virus detection (Broughton et al., 2020) set up a DETECTOR platform that combined RT-LAMP and CRISPR/Cas12a for SARS-CoV-2 diagnosis. In the future, this technique could also be applied to the detection of PPNs.
4 Direct detection of PPNs in the field
In order to truly implement field testing, the feasibility of field operation encompassing the entire detection process must be considered, including sample handling, the amplification process, and visualization of the results. In a previous study, isolating nematodes from a Baermann funnel or directly picking nematodes from plant root galls was time-consuming and required specialized techniques. DNA could be extracted directly from plant root nodules using the Flinders Technology Associates (FTA) technique, reducing the cost and time for diagnosis by simplifying sample storage, transport, and extraction. All of the steps of FTA-based archiving and DNA preparation are carried out at room temperature, which significantly reduces the expense and is environmentally friendly (Marek et al., 2014). FTA technology has been used for the DNA extraction of D. dipsaci, H. schachtii, and M. hapla (Marek et al., 2014; Peng et al., 2017). The drawback of this method is that it is limited to the extraction of pathogenic DNA from plant tissues and cannot be utilized for the extraction of DNA from soil or other media. Commercial kits for direct extraction of nematode soil DNA have now been developed and used successfully in several laboratories. The use of these kits also saves the time consumed by nematode isolation and the cost of instruments. Using only a small amount of DNA in this template, the target nematode can be detected. Soil DNA, including that from Pratylenchus neglectus, P. thornei, M. incognita, R. similis, and H. schachtii, was extracted using soil kits for the successful detection of these nematodes (Yan et al., 2008; Hu et al., 2011; Min et al., 2011; Peng et al., 2012; Jiang et al., 2021; Yao et al., 2021). Although this method has several advantages such as time-saving, simplicity, and efficiency, the soil kit can extract no more than 10 g of soil at a time. The uneven distribution of nematodes makes it difficult to extract DNA containing the target nematodes. This problem might be solved by repeating the assay multiple times to improve the detection rate for nematodes. In the amplification stage, using RPA and LAMP techniques or these two methods combined with CRISPR/Cas12a allows DNA amplification in 15-60 min without thermal cycling and expensive instruments (i.e., PCR instruments or fluorescence PCR instruments) compared to conventional PCR. It offers the possibility of field application for PPNs detection. The results of a combination of LFD technology and these methods are visible to the naked eye. RPA combined with the CRISPR/Cas12 assay has been applied to the detection of the PPN H. schachtii in the field (Yao et al., 2021), H. avenae, and H. filipjevi (Shao et al., 2022). Therefore, the combination of the FTA technique, the kit method for soil sample extraction, and a combination of LFD technology and RPA/LAMP-CRISPR/Cas12 can fully and truly realize the field detection of nematodes.
5 The remote sensing method for PPNs
Remote sensing is a method of observing and acquiring information about the properties of the studied entity without physically coming in contact with it (Kundu et al., 2022). The method could determine the presence of a nematode species by the change of symptoms in the above-ground parts of a plant. It avoids damage to the host and saves time and cost of diagnosis. Remote sensing is a fast, non-invasive, and highly effective process of acquiring information that has a wide coverage. Various spectroscopic and imaging approaches have been performed for the detection of PPNs, such as visible, multiband, infrared, and fluorescence spectroscopy, fluorescence imaging, multispectral and hyperspectral imaging, thermography, and nuclear magnetic resonance spectroscopy. Norman and Fritz (1965) were the first to use infrared sensors for pre-sign detection of R. similis in citrus trees. Subsequently, R. reniformis was detected by Heald et al. (1972) using airborne infrared imaging methods in cotton fields. Heath et al. (2000) predicted the amount of the nematodes G. rostochiensis and G. pallida on potatoes based on non-destructive hyperspectral measurements with a combination of GIS and RS technologies. Remote sensing coupled with GIS technologies was employed to identify and quantify an H. glycines population (Nutter et al., 2002). Three data preprocessing approaches were tested to evaluate their suitability for detecting H. schachtii and R. solanii (Hillnhutter et al., 2012). Pine wood nematode disease was discovered by Pan et al. (2014) based on hyperspectral remote sensing technology. Three methods, visible light imaging, thermometry and spectroscopy, were compared for their ability to detect H. schachtii in two sugar beet varieties (Joalland et al., 2017). Currently, remote sensing techniques have accuracy issues, as some nematodes are misdiagnosed due to similar symptoms and a lack of sufficient survey data for nematode surveillance modeling.
6 Machine learning for PPNs identification
Machine Learning or Artificial intelligence (AI) is a novel technology for nematode identification and quantitation based on image analysis (Bogale et al., 2020). It is an effective method for processing a large number of samples and identifying unique and minute items such as nematodes eggs and cysts in a complex background (Akintayo et al., 2018). Biological image datasets for multiple genera of PPNs were established and used to identify them based on the deep convolutional neural networks (CNNs) method (Lu et al., 2021). A convolutional CNNs model for identification of nematodes in soybean crop was developed by Abade et al. (2022).AI or Deep learning combined with hyperspectral image analysis is more popular because of the advantages this assay presents over direct soil methods (Arjoune et al., 2022). A combination of infrared spectra analysis and AI assay was used to detect rootknot nematode M. enterorlobii at the early stage of infection (San-Blas et al., 2020). AI, a relatively new technology, is gradually being applied to the field of PPNs detection. Though the technique could overcome the drawbacks of reduced specialist and subjective judgment, the generation of sufficient data may become a bottleneck in the development of AI.
7 Conclusion and future perspectives
In this article, we reviewed various existing methods for the detection and diagnosis of PPNs, such as morphological and biochemical methods, traditional PCR, isothermal amplification technologies, and remote sensing techniques. Practically, no single method or technique exists for diagnosing PPNs. Each approach has its strengths and weaknesses, therefore, we concluded the characteristics of each method (Table 4). Morphology-based classification forms the foundation of taxonomy, but morphological and morphometric characters are subtle and subjective, which may lead to inaccurate identification of a species (Floyd et al., 2002; Chitwood, 2003). In the future, the Protein-based approach play an important role in studies of species identification. However, the complexity of protein expression patterns and the ease of degradation of extracted proteins may affect the accuracy of the assessment; this restriction is the major challenge in the use of this technique. PCR molecular marker technologies have been widely used for PPNs detection, which compensate for the lack of morphological identification. The representative PCR, ddPCR, and qPCR technologies use dynamics of denaturation that drive replication events in control, and show excellent testing capability (Agüero et al., 2003; Mika et al., 2020; Wang et al., 2020). However, the requirements for expensive equipment and lack of trained scientist lead to their restriction for use in the laboratory and field detection. The isothermal amplification method is suitable for field testing because it does not require a device for temperature loop control (Niemz et al., 2011). The isothermal amplification technique takes much less time and cost than conventional PCR amplification. Among isothermal amplification techniques, LAMP-LFD and RPA-LFD are quickly evolving in the field of identification, because of their obvious specificity, efficiency, and visualization (Figure 1). The previously mentioned techniques are only useful for identifying small-scale samples, but remote sensing techniques could be quickly applied to detect large infected areas in the field employing various instruments such as drones, spectrometers, and satellite imagers. Remote sensing technology has contributed greatly to the prediction of damage caused by PPNs in the field.
Table 4
| Category | Technology | Advantages | Disadvantages | Site | References |
|---|---|---|---|---|---|
| Morphology | Morphological methods | Intuitive, low cost | Difficult to judge accurately; complex to operate and requires specialized technicians | In the lab | (Oliveira et al., 2011) |
| Biochemical Methods | Isozymes | It can reflect phylogenetic relationships; High sensitivity | Mainly used only for root-knot nematodes; Time-consuming | (Dickson et al., 1970) | |
| Mass spectral analyses | fast, reliable, high sensitivity | Time- consuming, requires specialized skills | (Rivero et al., 2022) | ||
| PCR Methods | DNA barcoding | Accuracy | Time-consuming | (Hebert et al., 2003) | |
| Droplet digital PCR | High sensitivity and low amount of template DNA | Expensive reagents and instruments | (Rougemont et al., 2004) | ||
| Gene chip technology | Fast, accuracy | Expensive equipment, immature technology | (Fodor, 1997) | ||
| RFLPs | Reliable and reproducible | Complex operations, requiring large amounts of DNA | Blok and Powers, 2009 | ||
| RAPD | Generates a large amount of information | Lacks repeatability, requires strict experimental reaction | (Feng et al., 2005) | ||
| SCAR | High sensitivity and specificity | Time-consuming | (Chen et al., 2011) | ||
| RT-qPCR | Sensitive, reliable | Time- consuming, equipment relatively expensive | (Berry et al., 2008) | ||
| ddPCR | High sensitivity, Simple, convenient | Expensive instruments | Chen et al., 2022) | ||
| Isothermal Amplification Technology | LAMP | Low cost, simple operation, low equipment demand | False-positive results | Outdoors and in the field | (Ahuja and Somvanshi, 2021) |
| RPA | Fast, high sensitivity and specificity, Low cost, simple operation, low equipment demand, visualization of results | False-positive results; Required to design specific primers, probes, and gRNA | (Babu et al., 2018) | ||
| LRPA- CRISPR/Cas12a | |||||
| Spectral techniques | Remote sensing systems | Fast, large-area detection, dynamic monitoring | Requires technical personnel expertise, difficult to capture detailed changes | (Tao et al., 2020) | |
| Machine Learning | Artificial intelligence | fast, accurate and eliminate human errors | Lack of professional classification experts and a sufficient number of databases | In the lab | (Almalki, 2022) |
Comparison of different plant nematode detection methods.
Figure 1
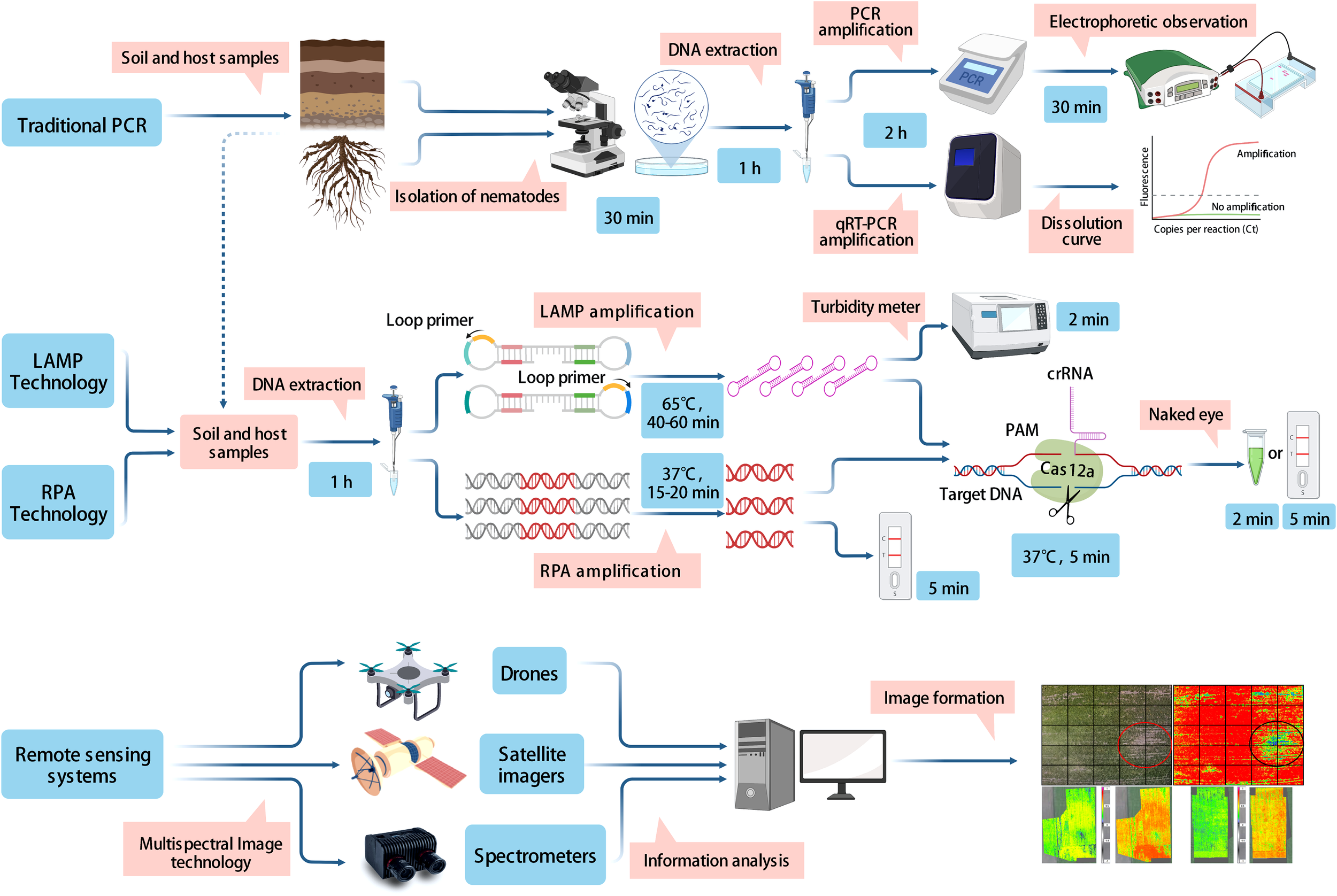
Demonstration of the principles and working processes involved in traditional PCR methods, isothermal amplification techniques, and remote sensing techniques.
In the field, rapid and accurate early diagnosis of PPNs is essential to control nematode damage. PPNs mainly damage the root tissues of plants, symptoms on aboveground parts are often not apparent, and are difficult to differentiate with the naked eye unless the damage is particularly serious. At the early stage of a nematode infection, no obvious changes are evident in the aboveground parts of the plant. However, hyperspectral methods can find significant differences in the leaf area index, absorptivity, photosynthetically active radiation, or canopy depression. This may be due to changes in the chlorophyll content of the above-ground parts of the plant, causing a change in the spectrum of the host plant (Din et al., 2017). Thus, the first use of remote sensing technology would be a prediction of the presence of the location of nematode infestation in the field. While remote sensing techniques face the problem that many nematode symptoms (i.e., wavelength, lutein, and chlorophyll, etc.) are similar, it is difficult to capture these changes in detail, leading to misjudgment. To solve this problem, RPA/LAMP-LFD or RPA/LAMP-CRISPR/Cas and other detection methods can be used to accurately survey the samples in a potential occurrence area. Time and economic losses caused by the blind application and ineffective use of nematicide can be avoided. It is noteworthy that in the field environment, every step from sampling and nucleic acid extraction to obtaining test results is exposed to the risk of contamination. Therefore, the integration of sample pretreatment, target identification and signal acquisition into a single device to establish an integrated nucleic acid detection system is a major development trend for future pathogenic nematode detection.
Statements
Author contributions
Conceptualization, HP, DP, and EL; Article framework, L-aK, WH, and CL; software, HS and PZ; Literature collection, HS, CL, and PZ; writing—original draft preparation, HS and PZ; writing—review and editing, HP and EL; project administration, DP; funding support, HP. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Key R&D Program of China (2021YFD1400100), the National Natural Science Foundation of China (31972247), the Norwegian Ministry of Foreign Affairs (SINOGRAIN II, CHN-17/0019), the Open Fund of Key Laboratory of Integrated Pest Management on Crop in Northwestern Oasis, Ministry of Agriculture and Rural Affairs (KFJJ202101), and the Science and Technology Innovation Project of the Chinese Academy of Agricultural Sciences (ASTIP-2016-IPP-15).
Acknowledgments
The authors are truly grateful to Dr. Sulaiman Abdulsalam of the Ahmadu Bello University for English editing of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Abade A. Porto L. Ferreira P. Vidal F. (2022). NemaNet : A convolutional neural network model for identification of soybean nematodes. Biosyst. Eng.213, 39–62. doi: 10.1016/j.biosystemseng.2021.11.016
2
Agüero M. Fernández J. Romero L. Sánchez Mascaraque C. Arias M. Sánchez-Vizcaíno J. M. (2003). Highly sensitive PCR assay for routine diagnosis of African swine fever virus in clinical samples. J. Clin. Microbiol.41, 4431–4434. doi: 10.1128/jcm.41.9.4431-4434.2003
3
Ahmed H. A. Macleod E. T. Hide G. Welburn S. C. Picozzi K. (2011). The best practice for preparation of samples from FTA®cards for diagnosis of blood borne infections using African trypanosomes as a model system. Parasit. Vectors4, 68. doi: 10.1186/1756-3305-4-68
4
Ahmed M. Sapp M. Prior T. Karssen G. Back M. (2015). Nematode taxonomy: from morphology to metabarcoding. Soil Discussions2, 1175–1220. doi: 10.5194/soild-2-1175-2015
5
Ahuja A. (2020). Daignosis of plant parasitic nematodes using loop mediated isothermal amplification (LAMP): A review. Crop Prot.147, 105459. doi: 10.1016/j.cropro.2020.105459
6
Ahuja A. Somvanshi V. S. (2021). Diagnosis of plant-parasitic nematodes using loop-mediated isothermal amplification (LAMP). A Rev. Crop Prot.147, 105459. doi: 10.1016/j.cropro.2020.105459
7
Aikawa T. Kanzaki N. Maehara N. (2013). ITS-RFLP pattern of bursaphelenchus xylophilus (Nematoda: Aphelenchoididae) does not reflect nematode virulence. J. For. Res.18, 384–388. doi: 10.1007/s10310-012-0354-1
8
Akintayo A. Tylka G. L. Singh A. K. Ganapathysubramanian B. Singh A. Sarkar S. (2018). A deep learning framework to discern and count microscopic nematode eggs. Sci. Rep.8, 9145. doi: 10.1038/s41598-018-27272-w
9
Almalki W. (2022). Advances in nematode identification: A journey from fundamentals to evolutionary aspects. Diversity14, 536. doi: 10.3390/d14070536
10
Angeletti S. (2017). Matrix assisted laser desorption time of flight mass spectrometry (MALDI-TOF MS) in clinical microbiology. J. Microbiol. Methods138, 20–29. doi: 10.1016/j.mimet.2016.09.003
11
Arjoune Y. Sugunaraj N. Peri S. Nair S. V. Skurdal A. Ranganathan P. et al . (2022). Soybean cyst nematode detection and management: a review. Plant Methods18, 110. doi: 10.1186/s13007-022-00933-8
12
Babu B. Ochoa-Corona F. M. Paret M. L. (2018). Recombinase polymerase amplification applied to plant virus detection and potential implications. Analytical Biochem.546, 72–77. doi: 10.1016/j.ab.2018.01.021
13
Baidoo R. Yan G. P. Nelson B. Skantar A. M. Chen S. Y. (2017). Use of chemical flocculation and nested PCR for heterodera glycines detection in DNA extracts from field soils with low population densities. Plant Dis.101, 1153–1161. doi: 10.1094/pdis-08-16-1163-re
14
Baklawa M. Niere B. Heuer H. Massoud S. (2015). Characterisation of cereal cyst nematodes in Egypt based on morphometrics. RFLP rDNA-ITS Sequence Analyses Nematol.17, 103–115. doi: 10.1163/15685411-00002855
15
Berry S. D. Fargette M. Spaull V. W. Morand S. Cadet P. (2008). Detection and quantification of root-knot nematode (Meloidogyne javanica), lesion nematode (Pratylenchus zeae) and dagger nematode (Xiphinema elongatum) parasites of sugarcane using real-time PCR. Mol. Cell Probes22, 168–176. doi: 10.1016/j.mcp.2008.01.003
16
Bizzini A. Durussel C. Bille J. Greub G. Prod'hom G. (2010). Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol.48, 1549–1554. doi: 10.1128/jcm.01794-09
17
Blaxter M. L. De Ley P. Garey J. R. Liu L. X. Scheldeman P. Vierstraete A. et al . (1998). A molecular evolutionary framework for the phylum Nematoda. Nature392, 71–75. doi: 10.1038/32160
18
Blok V. Powers T. (2009). Biochemical and Molecular Identification. Root-knot Nematodes. 3, 21–32. doi: 10.1079/9781845934927.0098
19
Bogale M. Baniya A. Digennaro P. (2020). Nematode identification techniques and recent advances. Plants-Basel9, 1260. doi: 10.3390/plants9101260
20
Broughton J. P. Deng X. Yu G. Fasching C. L. Servellita V. Singh J. et al . (2020). CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol.38, 870–874. doi: 10.1038/s41587-020-0513-4
21
Brown W. M. (1981). Mechanisms of evolution in animal mitochondrial DNA. Ann. N Y Acad. Sci.361, 119–134. doi: 10.1111/j.1749-6632.1981.tb46515.x
22
Carneiro R. Lima F. Correa V. (2017). Methods and tools currently used for the identification of plant parasitic nematodes. Nematology6, 45–60. doi: 10.5772/intechopen.69403
23
Carneiro R. Santos M. Almeida M. Mota F. Gomes A. Tigano M. (2008). Diversity of meloidogyne arenaria using morphological, cytological and molecular approaches. Nematology10, 819–834. doi: 10.1163/156854108786161526
24
Carneiro R. Tigano M. Randig O. Almeida M. R. . Sarah J. L. . (2004). Identification and genetic diversity of meloidogyne spp. (Tylenchida: Meloidogynidae) on coffee from Brazil, central America and Hawaii. Nematology6, 287–298. doi: 10.1163/1568541041217942
25
Carta L. K. Li S. (2018). Improved 18S small subunit rDNA primers for problematic nematode amplification. J. Nematol50, 533–542. doi: 10.21307/jofnem-2018-051
26
Carta L. K. Li S. (2019). PCR amplification of a long rDNA segment with one primer pair in agriculturally important nematodes. J. Nematol.51, 1–8. doi: 10.21307/jofnem-2019-026
27
Caswell-Chen E. P. Williamson V. M. Wu F. F. (1992). Random amplified polymorphic DNA analysis of heterodera cruciferae and h. schachtii populations. J. Nematol24, 343–351. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2619296/
28
Cenis J. L. (1993). Identification of four major meloidogyne spp. by random amplified polymorphic DNA (RAPD-PCR). phytopathology. Phytopathology83, 76–80. doi: 10.1094/phyto-83-76
29
Chen Y. Long H. B. Feng T. Z. Pei Y. L. Sun Y. F. Zhang X. C. (2022). Development of a novel Primer–TaqMan probe set for diagnosis and quantification of meloidogyne enterolobii in soil using qPCR and droplet digital PCR assays. Int. J. Mol. Sci.23, 11185. doi: 10.3390/ijms231911185
30
Chen F. M. Negi S. Ye J. R. (2011). A SCAR molecular marker to distinguish bursaphelenchus mucronatus from the pinewood nematode, b. xylophilus. For. Pathol.41, 376–381. doi: 10.1111/j.1439-0329.2010.00693.x
31
Chen J. S. Ma E. Harrington L. B. Da Costa M. Tian X. Palefsky J. M. et al . (2018). CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science360, 436–439. doi: 10.1126/science.aar6245
32
Chitwood D. J. (2003). Research on plant-parasitic nematode biology conducted by the united states department of agriculture-agricultural research service. Pest Manag Sci.59, 748–753. doi: 10.1002/ps.684
33
Chi Y. K. Zhao W. Ye M. D. Ali F. Wang T. Qi R. D. (2020). Evaluation of recombinase polymerase amplification assay for detecting meloidogyne javanica. Plant Dis.104, 801–807. doi: 10.1094/pdis-07-19-1473-re
34
Chizhov V. Butorina N. Subbotin S. (2012). Entomoparasitic nematodes of the genus skarbilovinema: S. laumondi and s. lyoni (Nematoda: Tylenchida), parasites of the flies of the family syrphidae (Diptera), with phylogeny of the suborder hexatylina. Russian J. Nematol.20, 141–155. doi: 10.1111/j.1744-7410.2012.00275.x
35
Chizhov V. N. Chumakova O. A. Subbotin S. A. Baldwin J. G. (2006). Morphological and molecular characterization of foliar nematodes of the genus aphelenchoides: A. fragariae and a. ritzemabosi (Nematoda: Aphelenchoididae) from the main botanical garden of the Russian academy of sciences, Moscow. Russian J. Nematol.14, 179–184. doi: 10.1016/j.dsr2.2007.12.007
36
Curran J. M. Baillie D. L. Webster J. M. J. P. (1985). Use of genomic DNA restriction fragment length differences to identify nematode species. Parasitology90, 137–144. doi: 10.1017/S0031182000049088
37
Curran J. Driver F. Ballard J. W. O. Milner R. J. (1994). Phylogeny of metarhizium: analysis of ribosomal DNA sequence data. Mycological Res.98, 547–552. doi: 10.1016/S0953-7562(09)80478-4
38
Curran J. Mcclure M. A. Webster J. M. (1986). Genotypic differentiation of meloidogyne populations by detection of restriction fragment length difference in total DNA. J. Nematol18, 83–86. https://eurekamag.com/research/001/375/001375845.php
39
Currie V. Hold G. Pryde S. Rehbein H. Quinteiro J. Rey-Méndez M. et al . (2000). Use of restriction fragment length polymorphism to distinguish between salmon species. J. Agric. Food Chem.48, 2184–2188. doi: 10.1021/jf991213e
40
Decraemer W. Hunt D. J. (2006). Structure and classification (Wallingford, UK; Cambridge, MA, USA: CABI), 3–32.
41
Deng M. H. Zhong L. Y. Kamolnetr O. Limpanont Y. Lv Z. Y. (2019). Detection of helminths by loop-mediated isothermal amplification assay: a review of updated technology and future outlook. Infect. Dis. Poverty8, 20–29. doi: 10.1186/s40249-019-0530-z
42
Dickson D. W. Sasser J. N. Huisingh D. M. (1970). Comparative disc-electrophoretic protein analyses of selected meloidogyne, ditylenchus, heterodera and aphelenchus spp. J. Nematol.2, 286–293. doi: 10.1080/09505431.2011.605921
43
Ding W. C. Chen J. Shi Y. H. Lu X. J. Li M. Y. (2010). Rapid and sensitive detection of infectious spleen and kidney necrosis virus by loop-mediated isothermal amplification combined with a lateral flow dipstick. Arch. Virol.155, 385–389. doi: 10.1007/s00705-010-0593-4
44
Din M. Zheng W. Rashid M. Wang S. Shi Z. (2017). Evaluating hyperspectral vegetation indices for leaf area index estimation of oryza sativa l. at diverse phenological stages. Front. Plant Sci.8. doi: 10.3389/fpls.2017.00820
45
Esbenshade P. R. Triantaphyllou A. C. (1985). Use of enzyme phenotypes for identification of meloidogyne species. J. Nematol17, 6–20. http://europepmc.org/backend/ptpmcrender.fcgi?accid=PMC2618420&blobtype=pdf
46
Esbenshade P. R. Triantaphyllou A. C. (1990). Isozyme phenotypes for the identification of meloidogyne species. J. Nematol22, 10–15. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2619005/
47
Espy M. J. Uhl J. R. Sloan L. M. Buckwalter S. P. Jones M. F. Vetter E. A. et al . (2006). Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin. Microbiol. Rev.19, 165–256. doi: 10.1128/cmr.19.1.165-256.2006
48
Feng M. C. Jian R. Y. Jian T. (2005). A study on detection technique of bursaphelenchus xylophilus and b. mucronatus by RAPD. J. Nanjing Forestry Univ.4, 25–28. doi: 10.1360/biodiv.050121
49
Feng M. C. Negi S. Ye J. R. (2011). A SCAR molecular marker to distinguish bursaphelenchus mucronatus from the pinewood nematode, b. xylophilus. For. Pathol.41, 13–18. doi: 10.1111/j.1439-0329.2010.00693.x
50
Ferris V. R. Ferris J. M. Faghihi J. (1993). Variation in spacer ribosomal DNA in some cyst-forming species of plant parasitic nematodes. Fundam. Appl. Nematol.16, 177–184. https://horizon.documentation.ird.fr/exl-doc/pleins_textes/fan/40289.pdf
51
Floyd R. Abebe E. Papert A. Blaxter M. (2002). Molecular barcodes for soil nematode identification. Mol. Ecol.11, 839–850. doi: 10.1046/j.1365-294x.2002.01485.x
52
Floyd R. M. Rogers A. D. Lambshead P. J. D. Smith C. R. (2005). Nematode-specific PCR primers for the 18S small subunit rRNA gene. Mol. Ecol. Notes5, 611–612. doi: 10.1111/j.1471-8286.2005.01009.x
53
Fodor S. P. A. (1997). DNA Sequencing - massively parallel genomics. Science277, 393–398. doi: 10.1126/science.277.5324.393
54
Fullaondo A. Barrena E. Viribay M. Barrena I. Salazar A. Ritter E. (1999). Identification of potato cyst nematode species Globodera rostochiensis and G. pallida by PCR using specific primer combinations. Nematology1, 157–163. doi: 10.1163/156854199508126
55
Gootenberg J. S. Abudayyeh O. O. Lee J. W. Essletzbichler P. Dy A. J. Joung J. et al . (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science356, 438–442. doi: 10.1126/science.aam9321
56
Goto K. Sato E. Toyota K. (2009). A novel detection method for the soybean cyst nematode heterodera glycines ichinohe using soil compaction and real-time PCR. Japanese J. Nematol.39, 1–7. doi: 10.3725/jjn.39.1
57
Green M. Rott M. Leal I. Humble L. Allen E. (2019). Application of a real-time PCR method for the detection of pine wood nematode, bursaphelenchus xylophilus, in wood samples from lodgepole pine. Nematol. Int. J. Fundam. Appl. nematological Res.9, 351–362. doi: 10.1163/156854107781352098
58
Hassan T. Martinelli H. T. Pr P. (2015). Impact of phytonematode on agriculture economy. Bio. age. phytonematodes.chapter1, 3–4. doi: 10.1079/9781780643755.0000
59
Heald C. M. Thames W. H. Wiegand C. L. (1972). Detection of totylenchulus reniformis infestations by aerial infrared photography. J. Nematol4, 298–300. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2619957/
60
Heath W. L. Haydock P. P. J. Wilcox A. C. Evans K. (2000). The potential use of spectral reflectance from the potato crop for remote sensing of infection by potato cyst nematodes. Aspects Appl. Biol.60, 185–188. https://eurekamag.com/research/003/592/003592696.php
61
Hebert P. D. Cywinska A. Ball S. L. Dewaard J. R. (2003). Biological identifications through DNA barcodes. Proc. Biol. Sci.270, 313–321. doi: 10.1098/rspb.2002.2218
62
He X. F. Das P. Peng H. Ding Z. He W. T. Huang W. K. et al . (2013). Loop-mediated isothermal amplification assay for rapid diagnosis of meloidogyne enterolobii directly. Chin. Agric. Sci.46, 534–544. doi: 10.1016/j.plantsci.2022.111376
63
Hillnhutter C. Mahlein A. K. Sikora R. A. Oerke E. C. (2012). Use of imaging spectroscopy to discriminate symptoms caused by heterodera schachtii and rhizoctonia solani on sugar beet. Precis. Agric.13, 17–32. doi: 10.1007/s11119-011-9237-2
64
Holterman M. van der Wurff A. Van Den Elsen S. Van Megen H. Bongers T. Holovachov O. et al . (2006). Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol. Biol. Evol.23, 1792–1800. doi: 10.1093/molbev/msl044
65
Hu M. X. Zhuo K. Liao J. L. (2011). Multiplex PCR for the simultaneous identification and detection of meloidogyne incognita, m. enterolobii and m. javanica using DNA extracted directly from individual galls. Phytopathology101, 1270–1277. doi: 10.1094/phyto-04-11-0095
66
Ide T. Kanzaki N. Giraldo P. P. P. Giblin-Davis R. M. (2017). Loop-mediated isothermal amplification (LAMP) for detection of the red ring nematode, bursaphelenchus cocophilus. Nematology19, 559–565. doi: 10.1163/15685411-00003069
67
Jarcho J. (2001). Restriction fragment length polymorphism analysis. Curr. Protoc. Hum. Genet. Chapter 2, Unit 2.7. 1–15. doi: 10.1002/0471142905.hg0207s01
68
Jiang C. Zhang Y. D. Yao K. Abdulsalam S. Li G. K. Gao H. F. et al . (2021). Development of a species-specific SCAR-PCR assay for direct detection of sugar beet cyst nematode (Heterodera schachtii) from infected roots and soil samples. Life-Basel11, 1358. doi: 10.3390/life11121358
69
Jian J. Z. Huang W. K. Kong L. A. Jian H. Abdulsalam S. Peng D. L. et al . (2022). Molecular diagnosis and direct quantification of cereal cyst nematode (Heterodera filipjevi) from field soil using TaqMan real-time PCR1. J. Integr. Agriculture. doi: 10.1016/j.jia.2022.09.016
70
Joalland S. Screpanti C. Liebisch F. Varella H. V. Gaume A. Walter A. (2017). Comparison of visible imaging, thermography and spectrometry methods to evaluate the effect of heterodera schachtii inoculation on sugar beets. Plant Methods13, 73. doi: 10.1186/s13007-017-0223-1
71
Ju Y. Lin Y. Yang G. Wu H. Pan Y. (2019). Development of recombinase polymerase amplification assay for rapid detection of meloidogyne incognita, m. javanica, m. arenaria and m. enterolobii. Eur. J. Plant Pathol.155, 1155–1163. doi: 10.1007/s10658-019-01844-6
72
Kanetani S. Kikuchi T. Akiba M. Nakamura K. Ikegame H. Tetsuka K. (2011). Detection of bursaphelenchus xylophilus from old discs of dead pinus armandii var. amamiana trees using a new detection kit. For. Pathol.41, 387–391. doi: 10.1111/j.1439-0329.2010.00695.x
73
Kang J. S. Kim A. Y. Han H. R. Moon Y. S. Koh Y. H. (2015). Development of two alternative loop-mediated isothermal amplification tools for detecting pathogenic pine wood nematodes. For. Pathol.45, 127–133. doi: 10.1111/efp.12147
74
Keçici A. Bozbuğa R. Öcal A. Yüksel E. Özer G. Yildiz Ş. et al . (2022). Diversity and identification of plant-parasitic nematodes in wheat-growing ecosystems. Microorganisms10, 1534. doi: 10.3390/microorganisms10081534
75
Kiatpathomchai W. Jaroenram W. Arunrut N. Jitrapakdee S. Flegel T. W. (2008). Shrimp taura syndrome virus detection by reverse transcription loop-mediated isothermal amplification combined with a lateral flow dipstick. J. Virol. Methods153, 214–217. doi: 10.1016/j.jviromet.2008.06.025
76
Kiewnick S. Frey J. E. Braun-Kiewnick A. (2015). Development and validation of LNA-based quantitative real-time PCR assays for detection and identification of the root-knot nematode meloidogyne enterolobii in complex DNA backgrounds. Phytopathology105, 1245–1249. doi: 10.1094/phyto-12-14-0364-r
77
Kikuchi T. Aikawa T. Oeda Y. Karim N. Kanzaki N. (2009). A rapid and precise diagnostic method for detecting the pinewood nematode bursaphelenchus xylophilus by loop-mediated isothermal amplification. Phytopathology99, 1365–1369. doi: 10.1094/phyto-99-12-1365
78
Kundu A. Chakraborty R. Sarkar S. (2022). Remote sensing in nematode detection and forecasting. Agriculture & Food : e-Newsletter4, 74–77. doi: 10.1117/12.614376
79
Lecouls A. C. Rubio-Cabetas M. J. Minot J. C. Voisin R. Bonnet A. Salesses G. et al . (1999). RAPD and SCAR markers linked to the Ma1 root-knot nematode resistance gene in myrobalan plum (Prunus cerasifera ehr.). Theor. Appl. Genet.99, 328–335. doi: 10.1007/s001220051240
80
Li H. Bai R. Zhao Z. Tao L. Ma M. Ji Z. et al . (2018). Application of droplet digital PCR to detect the pathogens of infectious diseases. Biosci. Rep.38, BSR20181170. doi: 10.1042/BSR20181170
81
Li Y. Lawrence G. W. Lu S. E. Balbalian C. Klink V. P. (2014). Quantitative field testing heterodera glycines from metagenomic DNA samples isolated directly from soil under agronomic production. PloS One9, e89887. doi: 10.1371/journal.pone.0089887
82
Li M. Li H. Ding X. Wang L. Wang X. Chen F. (2022). The detection of pine wilt disease: A literature review. Int. J. Mol. Sci.23, 10797. doi: 10.3390/ijms231810797
83
Lin B. Wang H. Zhuo K. Liao J. (2016). Loop-mediated isothermal amplification for the detection of tylenchulus semipenetrans in soil. Plant Dis.100, 877–883. doi: 10.1094/pdis-07-15-0801-re
84
Lobato I. M. O'sullivan C. K. (2018). Recombinase polymerase amplification: Basics, applications and recent advances. Trends Analyt Chem.98, 19–35. doi: 10.1016/j.trac.2017.10.015
85
Lopez-Nicora H. D. Craig J. P. Gao X. B. Lambert K. N. Niblack T. L. (2012). Evaluation of cultivar resistance to soybean cyst nematode with a quantitative polymerase chain reaction assay. Plant Dis.96, 1556–1563. doi: 10.1094/pdis-12-11-1083-re
86
Lu X. Wang Y. Fung S. Xue Q. (2021). A Biological Image Dataset for Nematode Recognition. I-Nema. 4, 1–13. doi: 10.48550/arXiv.2103.08335
87
Madani M. Subbotin S. A. Moens M. (2005). Quantitative detection of the potato cyst nematode, globodera pallida, and the beet cyst nematode, heterodera schachtii, using real-time PCR with SYBR green I dye. Mol. Cell. Probes19, 81–86. doi: 10.1016/j.mcp.2004.09.006
88
Mahmoudi N. Pakina E. N. Limantceva L. A. Ivanov A.V.J.R.J.O.A. Industries A. (2020). Diagnosis of potato rot nematode ditylenchus destructor using PCR-RFLP. Plant Prot.3, 353–362. doi: 10.22363/2312-797X-2020-15-4-353-362
89
Marek M. Zouhar M. Douda O. Maňasová M. Ryšánek P. (2014). Exploitation of FTA cartridges for the sampling, long-term storage, and DNA-based analyses of plant-parasitic nematodes. Phytopathology104, 306–312. doi: 10.1094/phyto-03-13-0067-r
90
Mika T. Maghnouj A. Klein-Scory S. Ladigan-Badura S. Baraniskin A. Thomson J. et al . (2020). Digital-droplet PCR for quantification of CD19-directed CAR T-cells. Front. Mol. Biosci.7. doi: 10.3389/fmolb.2020.00084
91
Min Y. Y. Toyota K. Goto K. Sato E. Mizuguchi S. Abe N. et al . (2011). Development of a direct quantitative detection method for meloidogyne incognita in sandy soils and its application to sweet potato cultivated fields in tokushima prefecture, Japan. Nematology13, 95–102. doi: 10.1163/138855410x504916
92
Mori Y. Kitao M. Tomita N. Notomi T. (2004). Real-time turbidimetry of LAMP reaction for quantifying template DNA. J. Biochem. Biophys. Methods59, 145–157. doi: 10.1016/j.jbbm.2003.12.005
93
Mori Y. Nagamine K. Tomita N. Notomi T. (2001). Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun.289, 150–154. doi: 10.1006/bbrc.2001.5921
94
Muniz M. Campos V. Castagnone-Sereno P. Castro J. Almeida M. Carneiro R. (2008). Diversity of meloidogyne exigua (Tylenchida: Meloidogynidae) populations from coffee and rubber tree. Nematology10, 897–910. doi: 10.1163/156854108786161418
95
Nicol J. M. Turner S. J. Coyne D. L. Nijs L. D. Hockland S. Maafi Z. T. (2011). “Current nematode threats to world agriculture,” in Genomics and molecular genetics of plant-nematode interactions. Eds. JonesJ.GheysenG.FenollC. (Dordrecht: Springer Netherlands), 21–43.
96
Niemz A. Ferguson T. M. Boyle D. S. (2011). Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol.29, 240–250. doi: 10.1016/j.tibtech.2011.01.007
97
Niu J. H. Guo Q. X. Jian H. Chen C. L. Yang D. Liu Q. et al . (2011). Rapid detection of meloidogyne spp. by LAMP assay in soil and roots. Crop Prot.30, 1063–1069. doi: 10.1016/j.cropro.2011.03.028
98
Niu J. H. Jian H. Guo Q. X. Chen C. L. Wang X. Y. Liu Q. et al . (2012). Evaluation of loop-mediated isothermal amplification (LAMP) assays based on 5S rDNA-IGS2 regions for detecting meloidogyne enterolobii. Plant Pathol.61, 809–819. doi: 10.1111/j.1365-3059.2011.02562.x
99
Norman G. G. Fritz N. L. (1965). Infrared photography as an indicator of disease and decline in citrus trees. Proc. Fla. State Horticult. Soc. 12, 31–41. https://www.adobe.com/creativecloud/photography/discover/infrared-photography.html Powers and Sandall 1988: URL:https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2618847/
100
Notomi T. Okayama H. Masubuchi H. Yonekawa T. Watanabe K. Amino N. et al . (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res.28, E63. doi: 10.1093/nar/28.12.e63
101
Nunn G. B. (1992). Nematode Molecular Evolution. Ph.D. Thesis. University of Nottingham, Nottingham, UK. Available at: https://ethos.bl.uk/OrderDetails.do?uin=uk.bl.ethos.334855
102
Nutter F. W. Tylka G. L. Guan J. Moreira A. J. Marett C. C. Rosburg T. R. et al . (2002). Use of remote sensing to detect soybean cyst nematode-induced plant stress. J. Nematol34, 222–231. doi: 10.1016/S0022-2011(02)00111-8
103
Oliveira C. Monteiroi A. Blok V. (2011). Morphological and molecular diagnostics for plant-parasitic nematodes: Working together to get the identification done. Trop. Plant Pathol.36, 65–76. doi: 10.1590/S1982-56762011000200001
104
Ou S. Q. Peng D. L. Liu X. M. Li Y. Moens M. (2008a). Identification of heterodera glycines using PCR with sequence characterised amplified region (SCAR) primers. Nematology10, 397–403. doi: 10.1163/156854108783900212
105
Ou S. Q. Peng D. L. Yu L. L. (2008b). Restriction fragment length polymorphism and sequences analysis of rDNA-ITS region of cereal cyst nematode(Heterodera avenae) on wheat from zhengzhou. Acta Phytopathologica Sin.4, 407–413. doi: 10.13926/j.cnki.apps.2008.04.008
106
Palomares-Rius J. E. Cantalapiedra-Navarrete C. Archidona-Yuste A. Subbotin S. A. Castillo P. (2017). The utility of mtDNA and rDNA for barcoding and phylogeny of plant-parasitic nematodes from longidoridae (Nematoda, enoplea). Sci. Rep.7, 10905. doi: 10.1038/s41598-017-11085-4
107
Pan J. Ju Y. Zhang H. Wang X. (2014). Detection of bursaphelenchus xylophilus infection in pinus massoniana from hyperspectral data. Nematology16, 1197–1207. doi: 10.1163/15685411-00002846
108
Peng H. Long H. Huang W. Liu J. Cui J. Kong L. et al . (2017). Rapid, simple and direct detection of meloidogyne hapla from infected root galls using loop-mediated isothermal amplification combined with FTA technology. Sci. Rep.7, 44853. doi: 10.1038/srep44853
109
Peng H. Peng D. L. Hu X. Q. He X. F. Wang Q. Huang W. K. et al . (2012). Loop-mediated isothermal amplification for rapid and precise detection of the burrowing nematode, radopholus similis, directly from diseased plant tissues. Nematology14, 977–986. doi: 10.1163/156854112x638415
110
Perera M. R. Vanstone V. A. Jones M. G. K. (2005). A novel approach to identify plant parasitic nematodes using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrometry19, 1454–1460. doi: 10.1002/rcm.1943
111
Piepenburg O. Williams C. H. Stemple D. L. Armes N. A. (2006). DNA Detection using recombination proteins. PloS Biol.4, e204. doi: 10.1371/journal.pbio.0040204
112
Powers T. O. Sandall L. J. (1988). Estimation of genetic divergence in meloidogyne mitochondrial DNA. J. Nematol20, 505–511.
113
Qi X. L. Peng D. Peng H. Long H. Huang W. He W. T. (2012). Rapid molecular diagnosis based on SCAR marker system for cereal cyst nematode. Sci. Agric. Sin.45, 4388–4395. doi: 10.3864/j.issn.0578-1752.2012.21.007
114
Randig O. Bongiovanni M. Carneiro R. M. D. G. Castagnone-Sereno P. (2002). Genetic diversity of root-knot nematodes from Brazil and development of SCAR markers specific for the coffee-damaging species. Genome45 5, 862–870. doi: 10.1111/ppa.12108
115
Rani A. Donovan N. Mantri N. (2019). Review: The future of plant pathogen diagnostics in a nursery production system. Biosens Bioelectron145, 111631. doi: 10.1016/j.bios.2019.111631
116
Rashidifard M. Marais M. Daneel M. S. Mienie C. M. S. Fourie H. (2019). Molecular characterisation of meloidogyne enterolobii and other meloidogyne spp. from south Africa. Trop. Plant Pathol.44, 213–224. doi: 10.1007/s40858-019-00281-4
117
Rivero J. Zurita A. Cutillas C. Callejon R. (2022). The use of MALDI-TOF MS as a diagnostic tool for adult trichuris species. Front. Veterinary Sci.9. doi: 10.3389/fvets.2022.867919
118
Rougemont M. Van Saanen M. Sahli R. Hinrikson H. P. Bille J. Jaton K. (2004). Detection of four plasmodium species in blood from humans by 18S rRNA gene subunit-based and species-specific real-time PCR assays. J. Clin. Microbiol.42, 5636–5643. doi: 10.1128/jcm.42.12.5636-5643.2004
119
Sabate del Rio J. Magrina Lobato I. Mayboroda O. Katakis I. O'sullivan C. K. (2017). Enhanced solid-phase recombinase polymerase amplification and electrochemical detection. Analytical Bioanalytical Chem.409, 3261–3269. doi: 10.1007/s00216-017-0269-y
120
San-Blas E. Paba G. Cubillan N. Portillo E. Casassa-Padrón A. González-González C. et al . (2020). The use of infrared spectroscopy and machine learning tools for detection of meloidogyne infestations. Plant Pathol.69, 1589–1600. doi: 10.1111/ppa.13246
121
Sandrin T. R. Goldstein J. E. Schumaker S. (2013). MALDI TOF MS profiling of bacteria at the strain level: a review. Mass Spectrom Rev.32, 188–217. doi: 10.1002/mas.21359
122
Sato E. Min Y. Y. Shirakashi T. Wada S. Toyota K. (2007). Detection of the root-lesion nematode, Pratylenchus penetrans (Cobb), in a nematode community using real-time PCR. Nematological Research37, 87–92. doi: 10.3725/jjn.37.87
123
Seng P. Drancourt M. Gouriet F. La Scola B. Fournier P. E. Rolain J. M. et al . (2009). Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis.49, 543–551. doi: 10.1086/600885
124
Shao H. Jian J. Peng D. Yao K. Abdulsalam S. Huang W. et al . (2022). Recombinase polymerase amplification coupled with CRISPR-Cas12a technology for rapid and highly sensitive detection of heterodera avenae and heterodera filipjevi. Plant Dis. doi: 10.1094/pdis-02-22-0386-re
125
Sikder M. M. Vestergård M. Sapkota R. Kyndt T. Nicolaisen M. (2020). A novel metabarcoding strategy for studying nematode communities (Cold Spring Harbor Laboratory).
126
Sjöholm M. I. Dillner J. Carlson J. (2008). Multiplex detection of human herpesviruses from archival specimens by using matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol.46, 540–545. doi: 10.1128/jcm.01565-07
127
Smith C. J. Osborn A. M. (2009). Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol. Ecol.67, 6–20. doi: 10.1111/j.1574-6941.2008.00629.x
128
Song Z. Q. Cheng J. E. Cheng F. X. Zhang D. Y. Liu Y. (2017). Development and evaluation of loop-mediated isothermal amplification assay for rapid detection of tylenchulus semipenetrans using DNA extracted from soil. Plant Pathol. J.33, 184–192. doi: 10.5423/ppj.Oa.10.2016.0224
129
Stanton J. Hugall A. F. Moritz C. C. (1997). Nucleotide polymorphisms and an improved PCR-based mtDNA diagnostic for parthenogenetic root-knot nematodes (Meloidogyne spp.). Fundam. Appl. Nematol.20, 261–268. https://horizon.documentation.ird.fr/exl-doc/pleins_textes/fan/010011563.pdf
130
Subbotin S. A. (2019). Recombinase polymerase amplification assay for rapid detection of the root-knot nematode meloidogyne enterolobii. Nematology21, 243–251. doi: 10.1163/15685411-00003210
131
Subbotin S. A. Burbridge J. (2021). Sensitive, accurate and rapid detection of the northern root-knot nematode, meloidogyne hapla, using recombinase polymerase amplification assays. Plants-Basel10, 336. doi: 10.3390/plants10020336
132
Subbotin S. Franco J. Knoetze R. Roubtsova T. Bostock R. Cid I. et al . (2019). DNA Barcoding, phylogeny and phylogeography of the cyst nematode species from the genus globodera (Tylenchida: Heteroderidae). Nematology22, 1–29. doi: 10.1163/15685411-00003305
133
Sykes P. J. Neoh S. H. Brisco M. J. Hughes E. Condon J. Morley A. A. (1992). Quantitation of targets for PCR by use of limiting dilution. Biotechniques13, 444–449. https://pubmed.ncbi.nlm.nih.gov/1389177/
134
Szalanski A. L. Sui D. D. Harris T. S. Powers T. O. (1997). Identification of cyst nematodes of agronomic and regulatory concern with PCR-RFLP of ITS1. J. Nematol.29, 255–267. doi: 10.1006/jipa.1997.4682
135
Taberlet P. Coissac E. Pompanon F. Brochmann C. Willerslev E. (2012). Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol.21, 2045–2050. doi: 10.1111/j.1365-294X.2012.05470.x
136
Tao H. Li C. Cheng C. Jiang L. Hu H. (2020). Progress in remote sensing monitoring for pine wilt disease induced tree mortality: a review. For. Res.3, 172–183. doi: 10.13275/j.cnki.lykxyj.2020
137
Toumi F. Waeyenberge L. Viaene N. Dababat A. Nicol J. M. Ogbonnaya F. et al . (2013). Development of two species-specific primer sets to detect the cereal cyst nematodes heterodera avenae and heterodera filipjevi. Eur. J. Plant Pathol.136, 613–624. doi: 10.1007/s10658-013-0192-9
138
Vallejo D. Rojas D. A. Martinez J. A. Marchant S. Holguin C. M. Pérez O. Y. (2021). Occurrence and molecular characterization of cyst nematode species (Globodera spp.) associated with potato crops in Colombia. PloS One16, e0241256. doi: 10.1371/journal.pone.0241256
139
Vega-Rúa A. Pagès N. Fontaine A. Nuccio C. Hery L. Goindin D. et al . (2018). Improvement of mosquito identification by MALDI-TOF MS biotyping using protein signatures from two body parts. Parasit. Vectors11, 574. doi: 10.1186/s13071-018-3157-1
140
Vrain T. C. (1993). Restriction fragment length polymorphism separates species of the xiphinema americanum group. J. Nematol25, 361–364. doi: 10.1006/jipa.1993.1100
141
Vrain T. C. Wakarchuk D. A. Lévesque A. Hamilton R. I. (1992). Intraspecific rDNA restriction fragment length polymorphism in the xiphinema americanum group. Fundam. Appl. Nematol.15, 563–573. doi: 10.1159/000156634
142
Waite I. S. O'donnell A. G. Harrison A. Davies J. T. Colvan S. R. Ekschmitt K. et al . (2003). Design and evaluation of nematode 18S rDNA primers for PCR and denaturing gradient gel electrophoresis (DGGE) of soil community DNA. Soil Biol. Biochem.35, 1165–1173. doi: 10.1016/s0038-0717(03)00177-9
143
Waliullah S. Bell J. Jagdale G. Stackhouse T. Hajihassani A. Brenneman T. et al . (2020). Rapid detection of pecan root-knot nematode, meloidogyne partityla, in laboratory and field conditions using loop-mediated isothermal amplification. PloS One15, e0228123. doi: 10.1371/journal.pone.0228123
144
Wang X. Wang L. Wang Y. Huang Y. Ding Z. Zhou J. et al . (2015). Identification and genetic analysis of the pinewood nematode bursaphelenchus xylophilus from pinus yunnanensis. For. Pathol.45, 388–399. doi: 10.1111/efp.12181
145
Wang X. Xiong E. Tian T. Cheng M. Lin W. Wang H. et al . (2020). Clustered regularly interspaced short palindromic Repeats/Cas9-mediated lateral flow nucleic acid assay. ACS Nano14, 2497–2508. doi: 10.1021/acsnano.0c00022
146
Wendt K. R. Vrain T. C. Webster J. M. (1993). Separation of three species of ditylenchus and some host races of d. dipsaci by restriction fragment length polymorphism. J. Nematol25, 555–563. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2619421/
147
Williams J. Kubelik A. R. Rafalski J. A. Tingey S. V. (1991). Genetic analysis with RAPD markers. More Gene Manipulations Fungi81, 431–439. doi: 10.1016/B978-0-12-088642-5.50026-0
148
Wishart J. Phillips M. S. Blok V. C. (2002). Ribosomal intergenic spacer: A polymerase chain reaction diagnostic for meloidogyne chitwoodi, m. fallax and m. hapla. Phytopathology92, 884–892. doi: 10.1094/phyto.2002.92.8.884
149
Yan G. Smiley R. W. Okubara P. A. Skantar A. Easley S. A. Sheedy J. G. et al . (2008). Detection and discrimination of pratylenchus neglectus and p. thornei in DNA extracts from soil. Plant Dis.92, 1480–1487. doi: 10.1094/pdis-92-11-1480
150
Yao K. Peng D. Jiang C. Zhao W. Li G. Huang W. et al . (2021). Rapid and visual detection of heterodera schachtii using recombinase polymerase amplification combined with Cas12a-mediated technology. Int. J. Mol. Sci.22, 12577. doi: 10.3390/ijms222212577
151
Yssouf A. Almeras L. Raoult D. Parola P. (2016). Emerging tools for identification of arthropod vectors. Future Microbiol.11, 549–566. doi: 10.2217/fmb.16.5
152
Yu L. Z. Song S. Y. Yu C. Jiao B. B. Tian Y. M. Li Y. J. (2020). Rapid detection of anguina agrostis by loop-mediated isothermal amplification. Eur. J. Plant Pathol.156, 819–825. doi: 10.1007/s10658-020-01932-y
153
Yu L. Z. Song S. Y. Yu C. Qi L. J. Yu Z. X. Jiao B. B. et al . (2018). A loop mediated isothermal amplification (LAMP) assay for rapid and reliable detection of anguina wevelli, a grass parasitic nematode. Eur. J. Plant Pathol.150, 725–734. doi: 10.1007/s10658-017-1320-8
154
Zhang Z. Q. (2013). Animal biodiversity: An update of classification and diversity in 2013. Zootaxa3703, 5–11. doi: 10.11646/zootaxa.3703.1.3
155
Zhang L. Gleason C. (2019). Loop-mediated isothermal amplification for the diagnostic detection of meloidogyne chitwoodi and m. fallax. Plant Dis.103, 12–18. doi: 10.1094/pdis-01-18-0093-re
156
Zheng J. W. Subbotin S. A. He S. S. Gu J. F. Moens M. (2003). Molecular characterisation of some Asian isolates of bursaphelenchus xylophilus and b. mucronatus using PCR-RFLPs and sequences of ribosomal DNA. Russian J. Nematol.11, 17–22. doi: 10.1051/rnd:2003010
157
Zheng J.-W. Subbotin S. Waeyenberge L. Moens M. (2000). Molecular characterisation of Chinese heterodera glycines and h. avenae populations based on RFLPs and sequences of rDNA-ITS regions. Russian J. Nematol.8, 109–113. doi: 10.1071/RD99037
158
Zhou Q. J. Cai Y. Gu J. F. Wang X. Chen J. (2017). Rapid and sensitive detection of meloidogyne mali by loop-mediated isothermal amplification combined with a lateral flow dipstick. Eur. J. Plant Pathol.148, 755–769. doi: 10.1007/s10658-016-1130-4
159
Zijlstra C. Donkers-Venne D. Fargette M. (2000). Identification of meloidogyne incognita, m-javanica and m-arenaria using sequence characterised amplified region (SCAR) based PCR assays. Nematology2, 847–853. doi: 10.1163/156854100750112798
160
Zijlstra C. Lever A. M. L. Uenk B. J. Silfhout C.H.V.J.P. (1995). Differences between ITS regions of isolates of the root-knot nematodes meloidogyne hapla and m. chitwoodi. Phytopathology85, 1231–1237. https://research.wur.nl/en/publications/differences-between-its-regions-of-isolates-of-the-root-knot-nema
Summary
Keywords
plant parasitic nematodes, diagnosis, PCR, Isothermal amplification, remote sensing, field detection
Citation
Shao H, Zhang P, Peng D, Huang W, Kong L-a, Li C, Liu E and Peng H (2023) Current advances in the identification of plant nematode diseases: From lab assays to in-field diagnostics. Front. Plant Sci. 14:1106784. doi: 10.3389/fpls.2023.1106784
Received
24 November 2022
Accepted
10 January 2023
Published
24 January 2023
Volume
14 - 2023
Edited by
Rahul Kumar Tiwari, Indian Council of Agricultural Research (ICAR), India
Reviewed by
Ashish Kumar Singh, Vivekananda Parvatiya Krishi Anusandhan Sansthan (ICAR), India; Lee Robertson, National Institute of Agricultural and Food Research and Technology, Spain
Updates
Copyright
© 2023 Shao, Zhang, Peng, Huang, Kong, Li, Liu and Peng.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huan Peng, penghuan@caas.cn; Enliang Liu, liuenliang@cau.edu.cn
†These authors have contributed equally to this work and share first authorship
This article was submitted to Plant Pathogen Interactions, a section of the journal Frontiers in Plant Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.