
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 07 February 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1105521
This article is part of the Research Topic Identification of Genes Involved in Environmental Adaption and Yield Traits in Forage and Other Ecologically Important Plants View all 10 articles
 Yuanyuan Cheng1†
Yuanyuan Cheng1† Chenglin Liang2†
Chenglin Liang2† Zhiyun Qiu1
Zhiyun Qiu1 Siqi Zhou1
Siqi Zhou1 Jianlong Liu1
Jianlong Liu1 Yingjie Yang1
Yingjie Yang1 Ran Wang1
Ran Wang1 Jie Yin1
Jie Yin1 Chunhui Ma1
Chunhui Ma1 Zhenhua Cui1
Zhenhua Cui1 Jiankun Song1
Jiankun Song1 Dingli Li1*
Dingli Li1*The quality of seedlings is an important factor for development of the pear industry. A strong seedling with few branches and suitable internodes is ideal material as a rootstock for grafting and breeding. Several branching mutants of pear rootstocks were identified previously. In the present study, ‘QAU-D03’ (Pyrus communis L.) and it’s mutants were used to explore the mechanism that affects branch formation by conducting phenotypic trait assessment, hormone content analysis, and transcriptome analysis. The mutant plant (MP) showed fewer branches, shorter 1-year-old shoots, and longer petiole length, compared to original plants (OP), i.e., wild type. Endogenous hormone analysis revealed that auxin, cytokinin, and jasmonic acid contents in the stem tips of MP were significantly higher than those of the original plants. In particular, the jasmonic acid content of the MP was 1.8 times higher than that of the original plants. Transcriptome analysis revealed that PcCOI1, which is a transcriptional regulatory gene downstream of the jasmonic acid signaling pathway, was expressed more highly in the MP than in the original plants, whereas the expression levels of PcJAZ and PcMYC were reduced in the MP compared with that of the original plants. In response to treatment with exogenous methyl jasmonate, the original plants phenotype was consistent with that of the MP in developing less branches. These results indicate that jasmonic acid negatively regulates branch growth of pear trees and that jasmonic acid downstream regulatory genes play a crucial role in regulating branching.
Grafting is a common method for asexual propagation of plants and for raising fruit tree seedlings. Pear rootstock with a strong stem, few branches, and suitable internodes is ideal material for grafting and breeding. Branch development and growth are regulated by a complex network of factors involving plant growth and development, hormone regulation, transcriptional regulation, and other factors.
Bud mutation is an important means of causing changes in branching through plant somatic variation (Zhao et al., 2021). The meristem cells of buds undergo genetic changes during cell division, leading to the development of lateral branches (Leng et al., 2021). The shoot apical meristem (SAM) is also crucial to plant development and is responsible for the development of leaves, stems, and flowers (Han et al., 2019). The SAM gives rise to leaf primordia during the vegetative growth stage and an inflorescence meristem in the transitional stage from vegetative growth to reproductive growth. An axillary meristem (AM) gives rise to a lateral bud primordium, which develops into a lateral bud and ultimately forms lateral branches (Grbić and Bleecker, 2000; Tanaka et al., 2013; ). The development of plant meristems mainly depends on the population of multifunctional stem cells in plants, and is affected by hormones and environmental factors.
Plant growth and development adapt to environmental changes through hormone signals, which provide a biochemical connection between the environment and cellular responses (Mur et al., 2006; Clarke et al., 2009). The auxin and cytokinin regulatory pathways are the main factors that regulate meristem formation and differentiation (Depuydt and Hardtke, 2011). Auxin combines with brassinolide (BR) to induce SMALL AUXIN UPREGULATED RNA 10 (SAUR10), thus participating in the regulation of branching angle (Bemer et al., 2017). Cytokinin is considered to be a second messenger that transmits auxin signals to lateral buds (Leyser, 2003). Cytokinin plays an important role in regulating leaf tip dominance and axillary bud growth. The expression level of PsIPT, a critical enzyme involved in cytokinin biosynthesis in pea, increases at the node after decapitation. Bud growth after decapitation is caused by the local accumulation of cytokinin (Tanaka et al., 2006). In poplar, overexpression of GA2ox leads to an increase in the number of tillers or branches, which indicates that gibberellin may play a negative role in the control of poplar bud branching (Zawaski and Busov, 2014). RAX1 promotes early formation of the AM, negatively regulates the gibberellin content of the stem tip, and affects the timing of AM development (Fambrini et al., 2017; Nie et al., 2018). Jasmonic acid is involved in regulating responses to biological and abiotic stresses, as well as growth and development of plants (Wasternack and Hause, 2013; Campos et al., 2014). In the jasmonic acid signaling pathway, COI1, as a downstream regulatory element of jasmonic acid, participates in almost all JAs regulatory processes (Ye et al., 2012). The COI1 gene encodes a F-box protein, which is a component of E3 ubiquitin ligase (Xie et al., 1998). The enzyme is associated with meristem arrest and apical dominance (Zhai et al., 2015). The coi1 mutant shows strong apical dominance and enhanced meristem longevity (Kim et al., 2013). Jasmonate ZIM-domain (JAZ) proteins play an inhibitory role in the JA signaling pathway (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). Overexpression of JAZ can cause increase in the number of lateral branches in tomato (Yu et al., 2018).
The regulation of transcription is important in the control of eukaryotic gene expression. Transcription factors are involved in all aspects of plant growth and development. The SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 13 (SPL13) gene encodes a SBP transcription factor, which is mainly expressed in meristems and is critical to regulating the branching and vegetative growth of alfalfa plants (Gao et al., 2018). Overexpression of SPL13 inhibits the growth of axillary buds and reduces the number of lateral branches. MsMYB112 RNA interference promotes branching, which indicates that MYB112 inhibits the growth of lateral branches in alfalfa (Stracke et al., 2001; Gao et al., 2018; ). In addition, TCP transcription factors are involved in lateral meristem growth, cell proliferation, and regulatory hormone effects in many cases (Aguilar-Martínez et al., 2007; Martin-Trillo and Cubas, 2010). PpTCP18 controls peach branching by positive feedback regulation of SL biosynthesis (Wang et al., 2022). In apple, MdWUS2 can regulate branching by inhibiting MdTCP12 expression (Li et al., 2021). The TB1 transcription factor, also known as FINECULM 1 (FC1), is a member of the TCP family that negatively regulates rice tillering and inhibits the subsequent growth of axillary buds (Takeda et al., 2003). The TB1 gene is expressed at the base of the axillary bud and SAM. Its overexpression leads to a significant reduction in number of tillers, whereas in the tb1 mutant an increased number of tillers develop (Wai and An, 2017). The transcription factor ERF BUD ENHANCE (EBE) affects cell proliferation, axillary bud growth, and branching of Arabidopsis thaliana. This gene encodes an AP2/ERF transcription factor and is highly expressed in proliferating cells (Mehrnia et al., 2013). Genes involved in plant hormone biosynthesis, transduction, and SAM formation are also associated with branching (Kurakawa et al., 2007; Ligerot et al., 2017; Zhang et al., 2018). These studies indicate that branching development is a complex process in plants.
The growth characteristics of pear rootstock have important influence on grafting effect. Sturdy seedlings and few branches make ideal stock material. In this study, the mechanism affecting pear branching formation were explored through phenotypic trait assessment, hormone content analysis and transcriptomic analysis, by using pear ‘QAU-D03’ (Pyrus communis L.) and less-branching mutants as materials. The results provide novel insights to improve understanding of the molecular mechanism of branch development in pear, which is of considerable importance for rootstock breeding.
The original plant (OP) with more branches was a seedling progeny of ‘QAU-D03’ (Pyrus communis L.). The shoots of OP were exposed to gamma radiation at 50 Gy (60Co source, 10 Gy min-1) and then grafted onto one-year-old rootstocks of Pyrus betulifolia Bunge. Among the resulting trees, one less-branching mutant was found and named MP. One-year-old shoots of OP and MP plants were collected from the Jiaozhou Demonstration Park in Qingdao, Shandong Province, China, for tissue culture to obtain tissue-cultured plantlets. The plants were grown on MS medium with 1.0 mg·L-1 6-BA, 0.1 mg·L-1 IBA, 30 g·L-1 sucrose, and 7 g·L-1 agar in a tissue culture room. Tissue-cultured shoots with 20 days of uniform growth were selected for rooting. Once the shoots had produced 3-4 roots of length 2-3 cm, the plantlets were transplanted to a mixture of perlite:vermiculite:peat (1:1:1, v/v/v). Samples were collected at the shoot dormancy in mid-March and bud expansion stages at the end of April, with three biological replicates per sample, for transcriptome sequencing analysis.
Annual branches from the periphery of crown of OP and MP plants, and mature leaves were randomly selected to observe leaf characteristics. Each treatment contains at least three biological replicates. The following quantitative characters were determined, including annual branch length, annual branch thickness, internode length, leaf length, leaf width and petiole length. The OP and MP of phenotypic difference was evaluated based on characteristics of pear plant.
Buds and stems of MP and OP plants were sampled for conventional paraffin-embedded sectioning. The samples were fixed in formaldehyde–alcohol–acetic acid for 1 d and then dehydrated in an ethanol series (50%, 60%, 70%, 80%, and 95%) for 60 min at each step. Samples were placed in 100% ethanol and left overnight. After decoloration with dimethylbenzene, the samples were embedded in paraffin. Sections (8 μm) were cut using a rotary microtome (HI1220, Leica, Nussloch, Germany), then dewaxed, rehydrated, cleaned, stained with toluidine blue, and the coverslip mounted with neutral balata. Sections were observed and photographed using an optical microscope (RM2235, Leica, Nussloch, Germany). In addition, buds were observed by scanning electron microscopy. After manual removal of the bud scales, the buds were fixed, washed, and dehydrated as described. The material was dried by carbon dioxide critical point drying and sputter-coated with gold. The material was observed and photographed with a scanning electron microscope (JSM-7500F, JEOL, China).
Fresh plant stem tips material (0.2-1.0 g) was ground in an ice-cooled mortar in 10 mL of 80% (v/v) methanol extraction medium containing 1 mM butylated hydroxytoluene as an antioxidant. The extract was incubated at 4°C for 4 h and then centrifuged at 4000 rpm for 15 min at 4°C. The supernatant was filtered through Chromosep C18 columns (C18 Sep-Park Cartridge, Waters Corp., Milford, MA, USA), and prewashed with 10 mL of 100% (w/v) and 5 mL of 80% (v/v) methanol. The hormone fractions eluted with 10 mL of 100% (v/v) methanol and 10 mL ether from the columns were dried under N2 gas, dissolved in 2 mL phosphate buffer saline containing 0.1% (v/v) Tween 20 and 0.1% (w/v) gelatin (pH 7.5) for analysis by an enzyme linked immunosorbent assay (ELISA) (Bollmark et al., 1988). The ELISA was performed in a 96-well microtitration plate. Each well was coated with 100 μL coating buffer (1.5 g·L−1 Na2CO3, 2.93g·L−1 NaHCO3, and 0.02g·L−1 NaN3, pH 9.6) containing 0.25 μg·mL−1 antigens against the hormones. The coated plates were incubated for 4 h at 37°C and then kept at room temperature for 30–40 min. The plate was incubated for 3 h at 28°C for measurement of dihydrozeatin riboside (DHZR), zeatin riboside (ZR), brassinolide (BR), methyl jasmonate (JA-Me), indole-3-acetic acid (IAA), indolepyruvic acid (IPA), gibberellins (GAs), and overnight at 4°C for IAA, and then washed as described above. Color development in each well was detected using an ELISA Reader (EL310, BioTek, Winooski, VT, USA) at an optical density of A490. The contents of DHZR, ZR, BR, JA-Me, IAA, IPA, GAs, and ABA were calculated following the method of Weiler et al. (1981).
Plant total RNA isolation kit (TaKaRa, Beijing, China) was used to extract total RNA from samples, following the manufacturer’s instructions, and different samples were subjected to three biological replicates. After qualification using a bioanalyzer (2,100, Agilent, United States), 1 μg of each sample was used for cDNA library construction. Four RNA sequencing libraries were constructed, in which MP1 and OP1 were the dormant shoot tips, MP2 and OP2 were the buds in expansion period. For library construction, 1 μg of RNA per sample was used as the input material. Library quality was assessed with a 2100 Bioanalyzer system (Agilent, Santa Clara, CA, USA). The clean dataset was obtained by removing reads containing the adapter sequence, poly-N, and low-quality reads from the raw data. The Q20, Q30, and GC content of the clean data were calculated. All downstream analyses used the high-quality clean data. The reference genome (Pyrus communis Bartlett DH Genome v2.0) and gene model annotation files were downloaded from GDR database (https://www.rosaceae.org/species/pyrus/pyrus_communis/genome_v2.0). An index of the reference genome was generated using HISAT2 v2.0.5 and paired-end clean reads were aligned to the reference genome using HISAT2 v2.0.5. FeatureCounts v1.5.0-p3 was used to count the read numbers mapped to each gene. The fragments per kilobase of exon per million mapped fragments (FPKM) value of each gene was calculated based on the length of the gene and number of reads mapped to the gene. Differential expression analysis of two conditions/groups (two biological replicates per condition) was performed using the DESeq2 R package (v1.20.0). Gene ontology (GO) enrichment analysis of differentially expressed genes (DEGs) was implemented with the clusterProfiler R package for which gene length bias was corrected. Statistical enrichment of DEGs in KEGG pathways was detected with the clusterProfiler R package.
Total RNA was extracted from pear using the RNAprep Pure Plant Kit (Tiangen Biotech Co., Beijing, China). The cDNA was synthesized using the HiScript II 1st Strand cDNA Synthesis Kit (Vazyme Biotech Co., Nanjing, China). The Lightcycler® 480 II System (Roche, Basel, Switzerland) and the ChamQ SYBR Color qPCR Master Kit (Vazyme Biotech Co.) were used to estimate relative gene expression levels under the different treatments. The reaction system (20 μL total volume) consisted of 2 μL template cDNA, 1 μL each forward and reverse primer, 10 μL Supermix, and 6 μL RNA-free water. The reaction protocol was as follows: 95°C for 5 min, then 45 cycles at 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. The Actin gene was used as an internal control. The relative expression level for each gene was calculated with the 2−ΔΔCt method. Each sample analysis was repeated three times. The primers used are listed in Supplementary Table S1.
The seedlings of ‘OP’ and ‘MP’ clones were selected and subcultured with Murashige and Skoog (MS) medium (Coolaber, Coolaber Science & Technology Co.,Ltd., China). The seedlings were divided into different treatments: (i) ‘OP’ and ‘MP’ control: continued use of the MS nutrient solution; (ii) ‘OP’ and ‘MP’ treated with JA-Me: MS with 100 μmol L-1 Methyl jasmonate (Macklin, Shanghai Macklin Biochemical Technology Co., Ltd., China). After JA-Me treatment, observations were carried out after 10 days.
Statistical analysis was performed using IBM SPSS Statistics 23.0 (IBM Corporation, Armonk, NY, USA). Values are presented as the mean ± SD of at least three independent biological replicates. The significance of differences between means was analyzed with Duncan’s multiple range test or Student’s t‐test. The probability level p < 0.05 was considered to be significant.
The wild type (OP) plants had more branches, whereas the mutant (MP) plants developed fewer branches and the leaves were larger (Figure 1A). The leaf lamina base of MP was broadly wedge-shaped or rounded, and the tip was tapered or blunt, whereas the OP leaf lamina base was mostly wedge-shaped or broadly wedge-shaped, and the tip was acute (Figure 1B). The mutant (MP) developed fewer branches, shorter 1-year-old shoots, and longer petiole, compared to OP, whereas no significant difference in leaf length, leaf width, leaf shape index, and annual branch thickness between OP and MP were observed (Table 1). With further development, the axillary meristem of OP developed into a spine-tipped spur shoot, whereas in MP axillary bud outgrowth did not occur (Figure 1C). All the above results confirmed that MP has the morphological characteristics of large leaves, short branches, few branches and no spines. Compared with OP, there were fewer branches in the early stage and no thorns in the leaf axils in the late stage, both of which were related to the activity of axillary meristem.
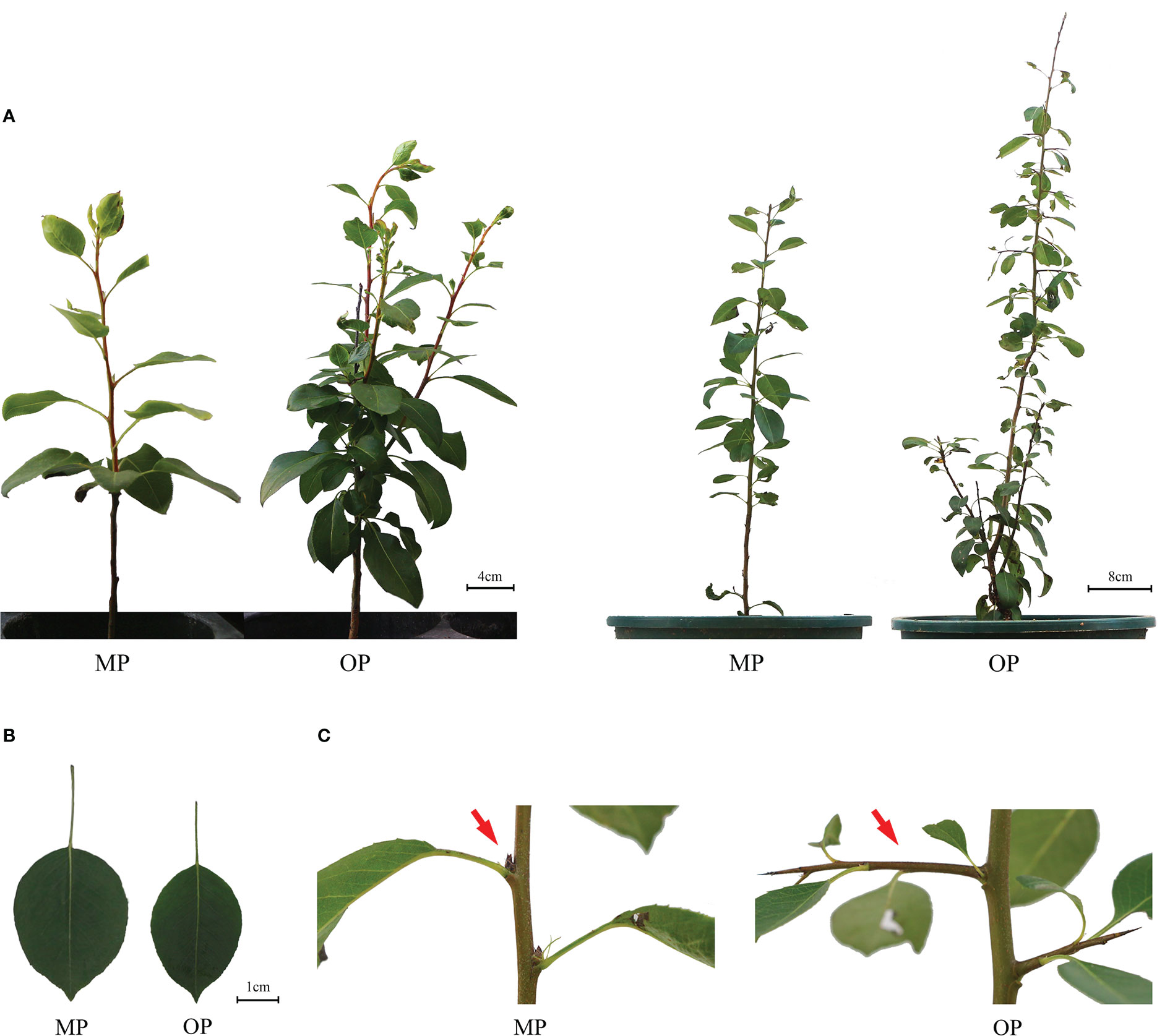
Figure 1 Phenotypes of the wild type (OP) and mutant (MP). (A) Plant phenotype of the same plant after growth for 9 months (left plant) and 12 months (right plant). (B) Leaf phenotype of MP and OP plants. (C) Axillary buds of MP and OP. The red arrow indicates transformation of the axillary meristem into a spur shoot (thorn) in OP and a dormant bud containing an undifferentiated axillary meristem in MP.
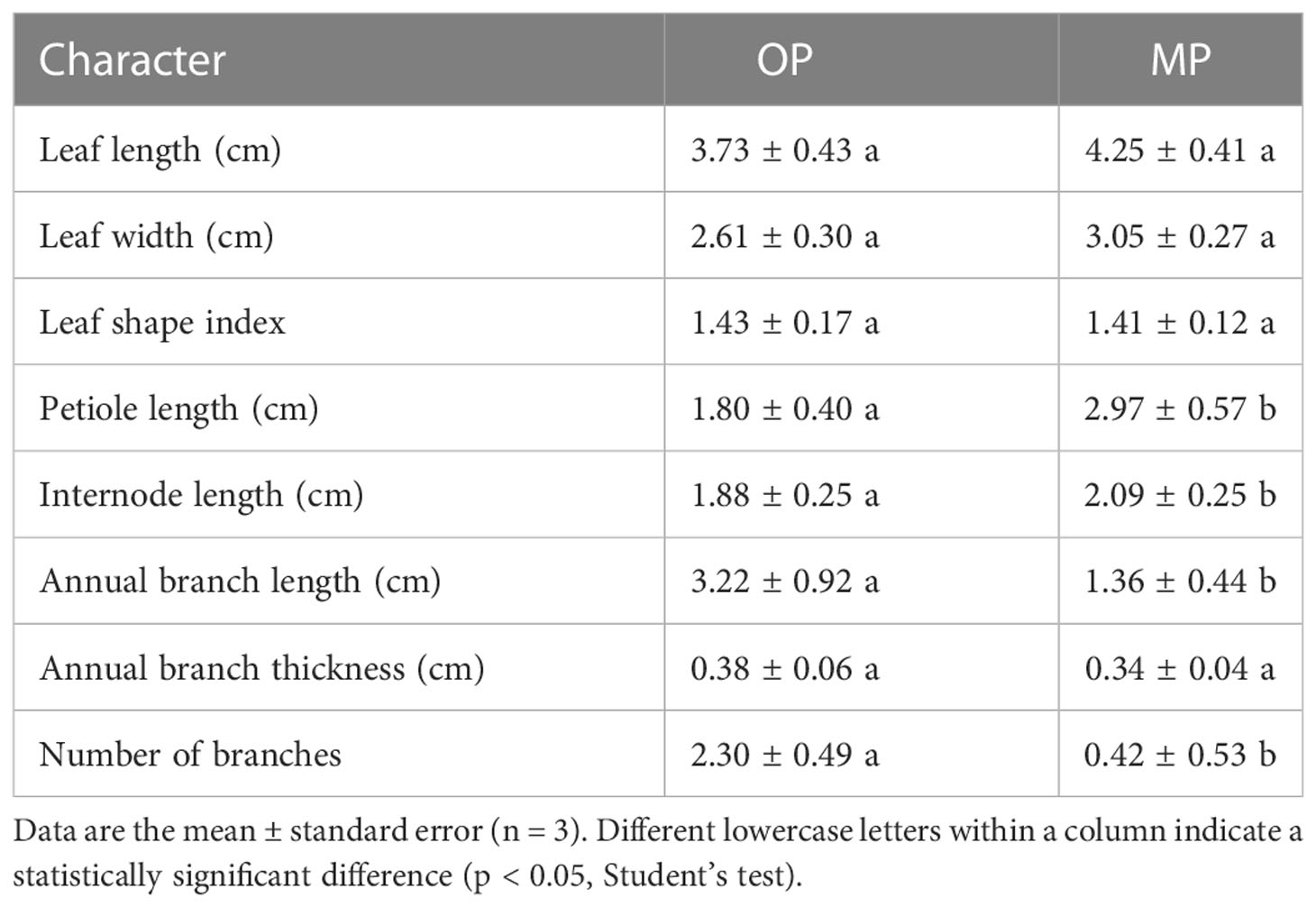
Table 1 Analysis of quantitative morphological characters between the mutant (MP) and wild type (OP).
The MP vascular bundle was larger than that of OP in the leaf, stem, and axillary buds at different development stages (Figure 2A; Table 2). The pith radius, xylem width and phloem width in the stem of new shoots of OP increased with stem development, and the phloem width changed most obviously (Figure 2B; Table 3). Axillary buds were divided into four developmental stages, namely bud primordium initiation preparation stage (I), bud primordium initiation stage (II), bud primordium formation stage (III), and bud primordium maturity stage (IV). Stage I was observed in the axillary buds of MP and OP; a group of darkly stained cells were present above the bud base, in the leaf axils of the new shoot, which represented the bud primordium (Figure 2C). With the further development of the bud primordium, the bud primordium entered the initiation stage (II). The growth point differentiated scale primordia from the outer to the inner, and the outermost is the scale developed from the scale primordium (LP). At the tip, the bud primordium formed the SAM and gradually differentiated into the scale leaf and bud primordium. When OP buds were in stage II, the MP buds had already entered stage III. In stage III, the bud base began to form the bud primordium, which was round and spherical, and gradually developed into a triangular form with time. In stage IV, the bud base formed a branch primordium and bracts one by one. When MP were in stage IV, the OP buds were still in stage III (Figure 2C). Observation of dormant axillary buds by scanning electron microscopy revealed that branch primordia and bract primordia were present in both MP and OP (Figure 2D). The above results showed that the bud structure of MP and OP were in different developmental stages at the same time, and the bud development of MP was faster than that of OP.

Figure 2 Anatomy of the vegetative organs of the wild type (OP) and mutant (MP). (A) Transverse sections of the leaf. EL, epithelial layer; PC, palisade mesophyll; SM, spongy mesophyll; VB, vascular bundle; LE, abaxial epidermis. (B) Transverse sections of the stem of the new shoots. Sections from leaf to right are from the shoot tip, mid-stem, and the shoot base, respectively. PE, periderm; C, cortex; PH, phloem; X, xylem; PI, pith. (C) Longitudinal sections of bud primordia at different stages of development, namely initiation preparation stage (I), initiation stage (II), formation stage (III), and maturation stage (IV). SAM, shoot apical meristem; LP, leaf primordium; P, bud primordium. (D) Scanning electron micrographs of dormant axillary buds.
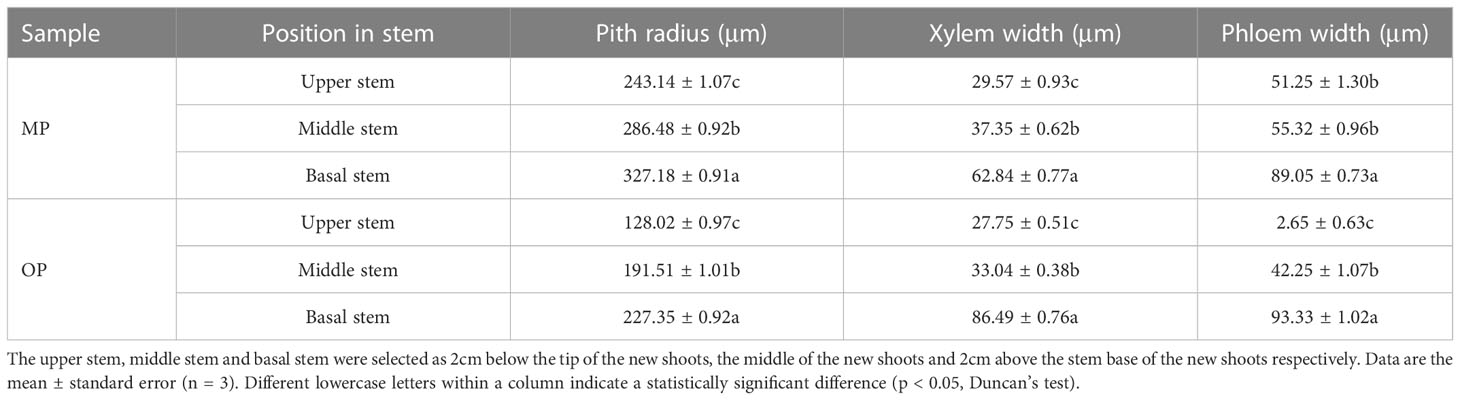
Table 3 Anatomical characters of the stem in transverse section for the mutant (MP) and wild type (OP).
Endogenous hormone analysis revealed that the content of GA4 in OP was significantly higher than that in MP, whereas the contents of auxin (IAA), methyl jasmonate (JA-Me), indolepro pionic acid (IPA), trans-zeatin-riboside (ZR), brassinolide (BR), and dihydrozeatin-riboside (DHZR) in MP were higher than those in OP. The difference in JA-Me content between OP and MP was most marked (Figure 3). Based on these results and the following transcriptome analysis, we speculate that jasmonic acid may play a role in regulating pear branching.
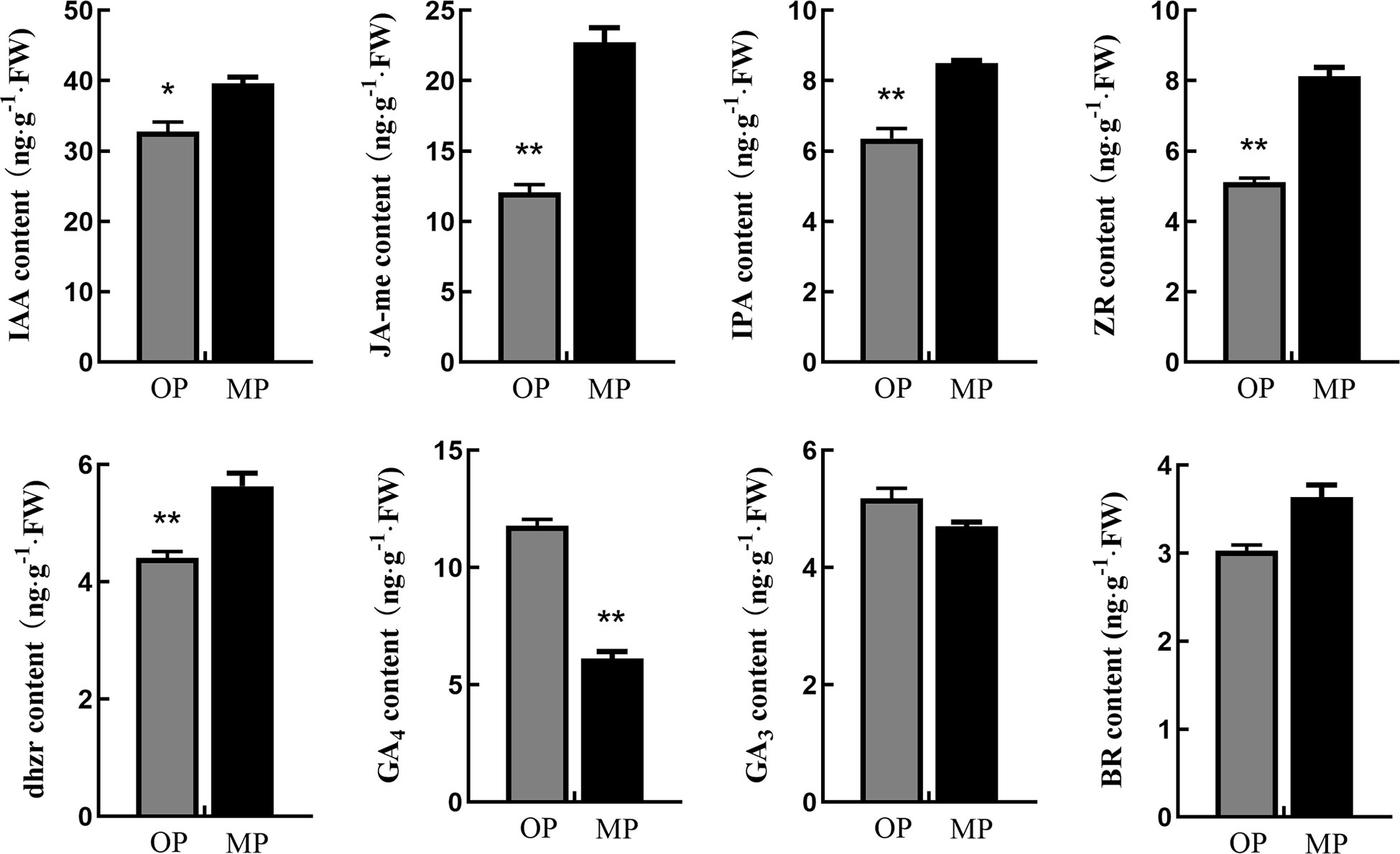
Figure 3 Hormone contents in stem tips of the wild type (OP) and mutant (MP). BR, brassinolide; DHZR, dihydrozeatin riboside; GA3 and GA4, gibberellins; IAA, indole-3-acetic acid; IPA, indolepyruvic acid; JA-Me, methyl jasmonate; ZR, zeatin riboside. Error bars indicate the standard error. *p < 0.05, **p < 0.01 (Duncan’s test).
The MP and OP shoot tips were sampled for transcriptome analysis. A total of 334 DEGs were common to the MP1 vs. OP1 and MP2 vs. OP2 comparison groups. Enrichment analysis of the GO terms indicated that the DEGs were mainly enriched in the apoplast in the biological process category, extracellular regions in the cellular component category, and xyloglucosyl transferase activity in the molecular function category (Figure 4A). The enrichment of KEGG pathways indicated that the DEGs were mainly enriched in the pathways of plant-pathogen interaction, fatty acid elongation, and nitrogen metabolism (Figure 4B). Thirty-eight of the 334 DEGs were associated with plant growth, cell division and differentiation, and SAM activity. Among these 38 DEGs, 23 genes encoded transcription factors, including members of the AP2/ERF, P450/CYP, MYB, WRKY, TCP, and NAC families (Figure 4C).
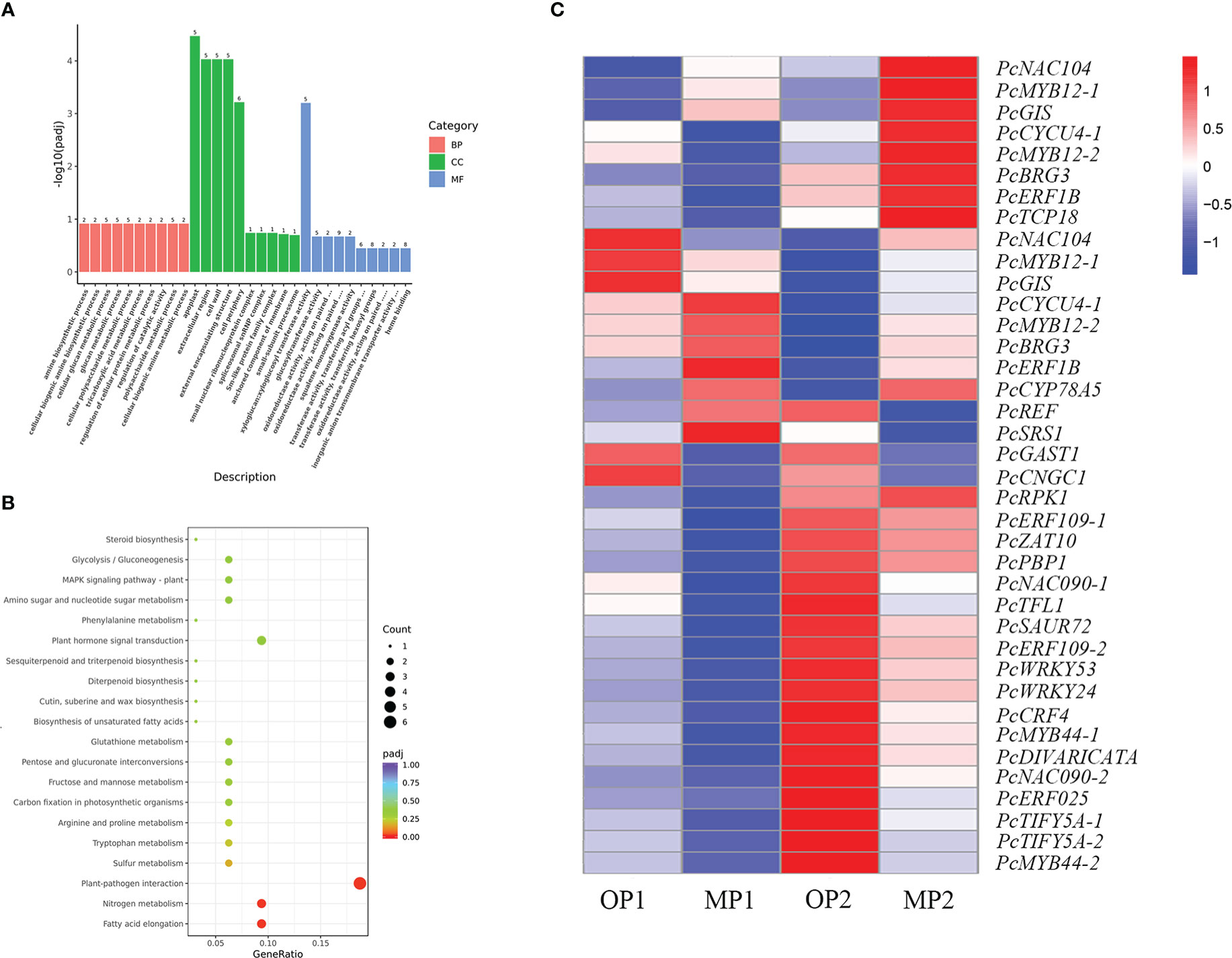
Figure 4 Functional annotation of differentially expressed genes (DEGs) in the mutant (MP) and wild type (OP). (A) DEGs enriched among the top 30 genes ontology terms. The annotated items are divided into three categories: cellular components(CC), molecular functions(MF), and biological processes(BP). (B) DEGs enriched among the top 20 KEGG pathways. (C) DEGs involved in cell division, differentiation, and shoot apical meristem activity, in which MP1 and OP1 were the shoot dormancy, MP2 and OP2 were the bud expanding stage. FPKM values are log2-based.
The hormone-related DEGs were screened and the changes in their expression in the two control groups were analyzed. Among these DEGs, most genes were upregulated in OP2, including IAA, JA-Me, Ethylene, ABA, CTK. Genes involved in the gibberellin signaling pathway were highly expressed in OP1 (Figure 5A). With regard to the JA biosynthesis and regulation pathways, 35 detected genes were associated with the octadecane synthesis pathway starting from α-linolenic acid, including genes encoding plant dienoic acid reductase (OPR3), acetyl coenzyme A oxidase (ACX), peroxisome fatty acid oxidized multifunctional protein (MFP2), and ketoethyl coenzyme A thiolase (3-KAT2), which are all involved in JA biosynthesis in the peroxisome. COI1-related genes were highly expressed in MP, whereas JAZ protein-related genes and MYC-related transcription factors were highly expressed in OP (Figure 5B). In OP and MP, the expressions of COI1-related genes and JAZ protein-related genes showed regular changes. We speculated that the phenomenon of plant branching might be related to COI1 and JAZ.
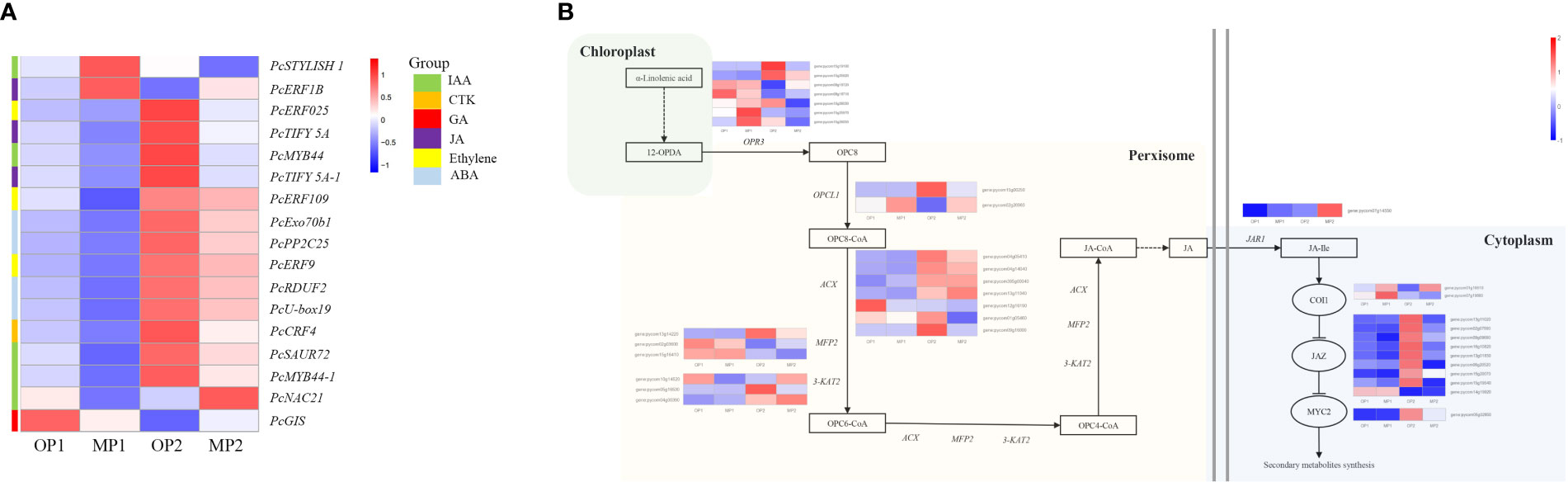
Figure 5 Differentially expressed genes involved in plant hormone signaling pathways and in jasmonate biosynthetic and signal transduction pathways. (A) Hormone-related DEGs, in which MP1 and OP1 were the shoot dormancy, MP2 and OP2 were the bud expanding stage. (B) Pathway constructed based on KEGG pathways and the literature. The enzymes and intermediates are indicated as follows: OPR3, OPDA reductase; ACX, acyl-coenzyme A oxidase; MFP2, peroxisomal fatty acid beta-oxidation multifunctional protein; 3-KAT2, 3-ketoacyl-CoA thiolase 2; JAR1, jasmonate resistant 1; OPC8, 8-(3-oxo-2-(pent-2-enyl)cyclopentyl) octanoic acid; JA, jasmonic acid; JA-Ile, jasmonoyl-L-isoleucine. FPKM values are log2-based.
Exogenous application of 100 μmol·L−1 JA-Me did not notably affect the branching of MP plants, whereas the branching of OP plants was visibly inhibited, in contrast to the control but consistent with the phenotype of MP plants. Expression analysis showed that the expression of PcOPR3 was significantly up-regulated after JA-Me treatment. In OP plants, PcCOI1 was not significantly increased in response to JA-Me treatment, whereas PcCOI1 was significantly upregulated in MP. PcJAZ1 was significantly upregulated in OP and MP in response to JA-Me treatment, but the extent of upregulation in OP was less than that in MP (Figure 6).
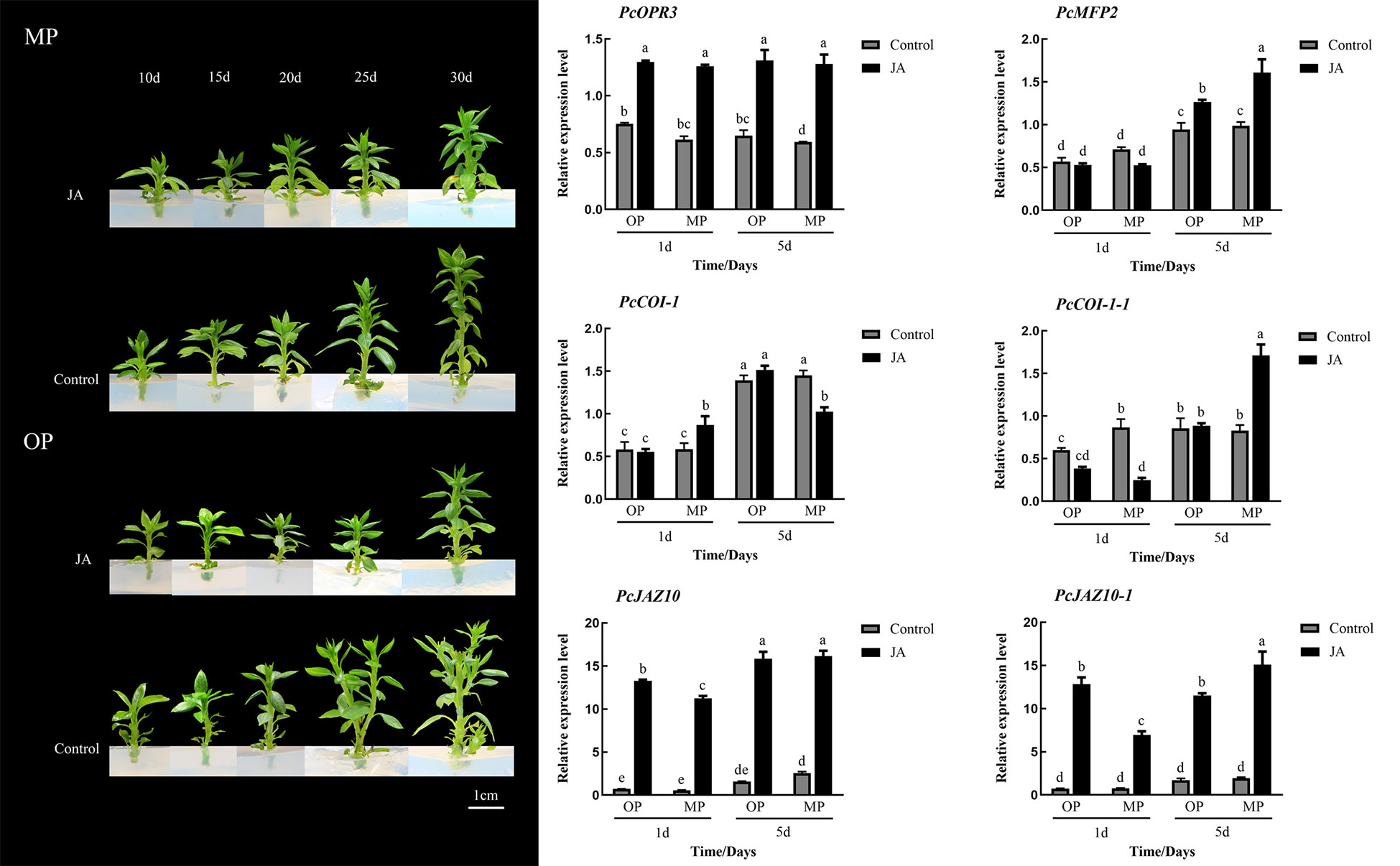
Figure 6 Effects of exogenous methyl jasmonate (JA) on plant phenotype and relative expression of six genes associated with the jasmonate signaling pathway at different stages of shoot development. Different lowercase letters above bars within a graph indicate a statistically significant difference (p < 0.05, Duncan’s multiple range test).
Pear is an economically important deciduous fruit tree. However, pear has a long growth cycle and complex genetic background, which is usually propagated by grafting. The growth characteristics of rootstocks play an important role in the outcome of grafting. Strong seedlings with few branches are ideal rootstock materials. In the present study, compared with OP, MP developed fewer branches, but the regulatory mechanism for these phenotypic changes remains unclear. Exploration of the regulation of pear branching through comparative analysis is important for the breeding of seedlings suitable for rootstocks.
Morphological assessment is the most common and direct method of identifying bud mutations. In this study, the morphology of OP and MP differed significantly. The MP plants had the phenotypic characteristics of few branches and large leaves, whereas OP had the contrasting traits of many branches and small leaves. Compared with MP, OP developed more branches at an early stage and the leaf axillary buds produced spiny spur shoots. Similar results have been reported in other species, such as birch and larch (Han et al., 2019; Cai et al., 2021). In addition, the vascular bundles and abaxial epidermal cells of MP leaves were larger than those of OP, and the areal proportion of the pith, xylem, and phloem in new shoots of MP from the base to the tip was greater than that of OP, and almost no conducting tissue had differentiated at the shoot tip of OP. These results indicated that MP had a greater capacity for nutrient and water transport than OP, which may be one reason why MP had stronger main branches and fewer branches. The bud structure and development period of MP and OP were different; bud development was more rapid in MP than in OP, thus the bud growth patterns were indicated to differ between OP and MP.
Cell division, expansion, and differentiation affect the basic processes of plant organ growth and development, and ultimately affect plant phenotype (Sugiyama, 2005; Yang et al., 2015). Plant morphogenesis is closely associated with genes involved in cell division, expansion, and differentiation (Lopez-Hernandez et al., 2020). The diversity of plant morphology is largely affected by SAM activity (Sussex and Kerk, 2001). In the current study, 38 DEGs associated with cell division, differentiation, and SAM activity were identified, including 23 transcription factors belonging to the AP2/ERF, P450/CYP, TCP, and NAC families. Transcription factors play a vital role in plant development and regulation of gene expression, forming a complex gene regulatory network (Muiño et al., 2016; Yang et al., 2018). Most of the 23 transcription factors were highly expressed in OP and participate in the regulation of mutant branching traits. The TCP transcription factors are involved in the growth of lateral meristems, cell proliferation, and regulation of hormones (Aguilar-Martínez et al., 2007; Martin-Trillo and Cubas, 2010). In citrus, the TCP transcription factors THORN IDENTITY 1 (TI1) and TI2 are essential to preclude the proliferation of meristems and simultaneously produce thorns (Zhang et al., 2020; Zhang et al., 2021). In the present study, the expression of PcTCP18 in OP was higher than that in MP at an early stage of development, whereas expression was higher in MP than in OP at an advanced stage of development. The gene PcTCP18 has high homology with the citrus TI1 gene. In peach, PpTCP18 can reduce secondary branches through brassinolide pathway (Wang et al., 2022). Based on these results, we speculated that PcTCP18 may be a candidate gene responsible for reducing the branching of MP at an early stage of development.
Plant hormones can regulate plant development processes, such as cell division, bud development, branch branching, and senescence (Heyl et al., 2007). Genes associated with hormone metabolism and signal transduction play an important role in regulating plant and organ size (Guo et al., 2010). In the present study, MP and OP were revealed to differ significantly in hormone contents. The IAA and cytokinin contents in MP were significantly higher than those in OP. Furthermore, the expression level cytokinin-related genes in OP were also higher than that in MP, indicating that cytokinins played an important role in plant branching. This result was consistent with a previous report that cytokinin promotes plant branching, tillering development, and lateral bud growth (Foo et al., 2007). Gibberellins play an important regulatory role in plant growth and development, and can control vertical growth and branching (Arend et al., 2009). In the current study, the expression of DEGs associated with GAs was higher in OP. Similarly, the GA4 content in OP was higher than that in MP. Gibberellin had been proved to have the effect of inhibiting branching, especially the development of axillary buds was closely related to the content of gibberellin (Martinez-Bello et al., 2015; Tan et al., 2018). This also showed that gibberellin played an important role in the branch development of pear trees. In addition, the JA content in MP was significantly higher than that in OP. Jasmonic acid has diverse roles in regulating developmental processes such as seed germination, root development, and senescence (Jin and Zhu, 2017). Exogenous JA-Me treatment and JAZ mutants all can cause the phenotype of branch reduction (Hong et al., 2020). Auxin can also cause changes in plant branching through jasmonic acid (Wang et al., 2013). In our study, 35 genes related to jasmonate biosynthesis and signal transduction were differentially expressed. Among them, in the JA signaling pathway, PcCOI1-related genes were highly expressed in both stages of shoot development in MP, whereas PcJAZs were expressed at a lower level in MP, indicating that Jasmonate signal transduction is involved in the formation of branching. Application of exogenous JA-Me did not notably change the MP branching phenotype, whereas OP treated with exogenous JA-Me showed a decrease in frequency of lateral branching, resulting in a branching phenotype similar to that of MP. The expression of PcOPR3 was significantly up-regulated after JA-Me treatment. The downstream JA regulatory gene PcCOI1 was not highly expressed in OP, but was significantly upregulated in MP plants, in response to JA-Me treatment. JAZ negatively regulates the downstream regulatory pathway of JA. In response to JA-Me treatment, the expression level of PcJAZ was significantly increased. So far, there were few reports about the effect of jasmonic acid on the branches, but previous studies had confirmed that JAZ could promote growth and inhibit aging and COI1 also participated in plant growth and development (Huang et al., 2017; Oblessuc et al., 2020; ). Therefore, we concluded that JA caused the decrease of plant branch development due to the inhibition of jasmonate on growth, and the jasmonate regulatory gene in OP was not sensitive to jasmonate.
In conclusion, through observation of the phenotypic characters of MP and OP, the mutation in MP results in reduced branching, large leaves, and slower development of bud primordia. The contents of JA, IAA, IPA, and ZR were significantly higher in MP than in OP, especially that of JA. The wild type showed a little-branching phenotype after treatment with exogenous JA-Me. The expression levels of JA synthesis regulatory genes in MP were increased and these genes were responsive to application of exogenous JA-Me.
The data presented in the study are deposited in the National Center for Biotechnology Information (NCBI) BioProject database repository, accession number PRJNA916958.
YC and CL conducted the experiment, and YC prepared the manuscript. ZQ, SZ, and JL provided important technical help in transcriptional analysis, YY, RW, and CM gave useful guidance and valuable discussion. JY, ZC, and JS provided the plant materials and important technical help in histological Observation. DL conceived the idea and provided financial support. All authors contributed to the article and approved the submitted version.
This research was supported by the National Key Research and Development Program of China (2019YFD1001404), the earmarked fund for CARS (CARS-28-07), the Agricultural Variety Improvement Project of Shandong Province (2022LZGC011), Qingdao Agricultural University Doctoral Start-Up Fund (2019).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1105521/full#supplementary-material
Aguilar-Martinez, J. A., Poza-Carrión., C., Cubas, P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell. 19, 458–472. doi: 10.1105/tpc.106.048934
Arend, M., Schnitzler, J. P., Ehlting, B., Hänsch, R., Lange, T., Rennenberg, H., et al. (2009). Expression of the Arabidopsis mutant ABI1 gene alters abscisic acid sensitivity, stomatal development, and growth morphology in gray poplars. Plant Physiol. 151, 2110–2119. doi: 10.1104/pp.109.144956
Bemer, M., van Mourik, H., Muiño, J. M., Ferrándiz, C., Kaufmann, K., Angenent, G. C. (2017). FRUITFULL controls SAUR10 expression and regulates Arabidopsis growth and architecture. J. Exp. Bot. 68, 3391–3403. doi: 10.1093/jxb/erx184
Bollmark, M., Kubát, B., Eliasson, L. (1988). Variations in endogenous cytokinin content during adventitious root formation in pea cuttings. J. Plant Physiol. 132, 262–265. doi: 10.1016/S0176-1617(88)80102-0
Cai, K. W., Zhou, X. Y., Li, X., Kang, Y., Yang, X. M., Cui, Y. H., et al. (2021). Insight into the multiple branches traits of a mutant in larix olgensis by morphological, cytological, and transcriptional analyses. Front. Plant sci. 12. doi: 10.3389/fpls.2021.787661
Campos, M. L., Kang, J. H., Howe, G. A. (2014). Jasmonate-triggered plant immunity. J. Chem. Ecol. 40, 657–675. doi: 10.1007/s10886-014-0468-3
Chini, A., Fonseca, S., Fernández, G., Adie, B., Chico, J., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Clarke, S. M., Cristescu, S. M., Miersch, O., Harren, F. J., Wasternack, C., Mur, L. A. (2009). Jasmonates act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytol. 182, 175–187. doi: 10.1111/j.1469-8137.2008.02735.x
Depuydt, S., Hardtke, C. S. (2011). Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 21, R365–R373. doi: 10.1016/j.cub.2011.03.013
Fambrini, M., Salvini, M., Pugliesi, C. (2017). Molecular cloning, phylogenetic analysis, and expression patterns of LATERAL SUPPRESSOR-LIKE and REGULATOR OF AXILLARY MERISTEM FORMATION-LIKE genes in sunflower (Helianthus annuus l.). Dev. Genes Evol. 227, 159–170. doi: 10.1007/s00427-016-0571-2
Foo, E., Morris, S. E., Parmenter, K., Young, N., Wang, H., Jones, A., et al. (2007). Feedback regulation of xylem cytokinin content is conserved in pea and Arabidopsis. Plant Physiol. 143, 1418–1428. doi: 10.1104/pp.106.093708
Gao, R., Gruber, M. Y., Amyot, L., Hannoufa, A. (2018). SPL13 regulates shoot branching and flowering time in medicago sativa. Plant Mol. Biol. 96, 119–133. doi: 10.1007/s11103-017-0683-8
Grbić, V., Bleecker, A. B. (2000). Axillary meristem development in Arabidopsis thaliana. Plant J. 21, 215–223. doi: 10.1046/j.1365-313x.2000.00670.x
Guo, M., Rupe, M. A., Dieter, J. A., Zou, J. J., Spielbauer, D., Duncan, K. E., et al. (2010). Cell number Regulator1 affects plant and organ size in maize: implications for crop yield enhancement and heterosis. Plant Cell. 22, 1057–1073. doi: 10.1105/tpc.109.073676
Han, R., Wang, S., Liu, C. Y., Xu, W. D., Bian, X. Y., Liu, G. F., et al. (2019). Transcriptome analysis of a multiple-branches mutant terminal buds in betula platyphylla×B. pendula. Forests 10, 374. doi: 10.3390/f10050374
Heyl, A., Wulfetange, K., Pils, B., Nielsen, N., Romanov, G. A., Schmülling, T. (2007). Evolutionary proteomics identifies amino acids essential for ligand binding of the cytokinin receptor CHASE domain. BMC Evol. Biol. 7, 62. doi: 10.1186/1471-2148-7-62
Hong, S. Y., Sun, B., Straub, D., Blaakmeer, A., Mineri, L., Koch, J., et al. (2020). Heterologous microProtein expression identifies LITTLE NINJA, a dominant regulator of jasmonic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 117, 26197–26205. doi: 10.1073/pnas.2005198117
Huang, H., Liu, B., Liu, L. Y., Song, S. S. (2017). Jasmonate action in plant growth and development. J. Exp. Bot. 6, 1349–1359. doi: 10.1093/jxb/erw495
Jin, H. H., Zhu, Z. Q. (2017). Temporal and spatial view of jasmonate signaling. Trends Plant Sci. 22, 451–454. doi: 10.1016/j.tplants.2017.04.001
Kim, J., Dotson, B., Rey, C., Lindsey, J., Bleecker, A. B., Binder, B. M., et al. (2013). New clothes for the jasmonic acid receptor COI1: delayed abscission, meristem arrest and apical dominance. PloS One 8 (4), e60505. doi: 10.1371/journal.pone.0060505
Kurakawa, T., Ueda, N., Maekawa, M., Kobayashi, K., Kojima, M., Nagato, Y., et al. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655. doi: 10.1038/nature05504
Leng, F., Ye, Y. L., Zhu, X. H., Zhang, Y., Zhang, Z. Y., Shi, J. Y., et al. (2021). Comparative transcriptomic analysis between ‘Summer black’ and its bud sport ‘Nantaihutezao’ during developmental stages. Planta 253, 23. doi: 10.1007/s00425-020-03543-7
Leyser, O. (2003). Regulation of shoot branching by auxin. Trends Plant Sci. 8, 541–545. doi: 10.1016/j.tplants.2003.09.008
Ligerot, Y., de Saint Germain, A., Waldie, T., Troadec, C., Citerne, S., Kadakia, N., et al. (2017). The pea branching RMS2 gene encodes the PsAFB4/5 auxin receptor and is involved in an auxin-strigolactone regulation loop. PloS Genet. 13 (12), e1007089. doi: 10.1371/journal.pgen.1007089
Li, G. F., Tan, M., Ma, J. J., Cheng, F., Li, K., Liu, X. J., et al. (2021). Molecular mechanism of MdWUS2-MdTCP12 interaction in mediating cytokinin signaling to control axillary bud outgrowth. J. Exp. Bot. 72, 4822–4838. doi: 10.1093/jxb/erab163
Lopez-Hernandez, F., Tryfona, T., Rizza, A., Yu, X. L., Harris, M. O. B., Webb, A. A. R., et al. (2020). Calcium binding by arabinogalactan polysaccharides is important for normal plant development. Plant Cell. 32, 3346–3369. doi: 10.1105/tpc.20.00027
Martinez-Bello, L., Moritz, T., López-Díaz, I. (2015). Silencing c-19-GA 2-oxidases induces parthenocarpic development and inhibits lateral branching in tomato plants. J. Exp. Bot. 19, 5897–5910. doi: 10.1093/jxb/erv300
Martin-Trillo, M., Cubas, P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. doi: 10.1016/j.tplants.2009.11.003
Mehrnia, M., Balazadeh, S., Zanor, M. I., Mueller-Roeber, B. (2013). EBE, an AP2/ERF transcription factor highly expressed in proliferating cells, affects shoot architecture in Arabidopsis. Plant Physiol. 162, 842–857. doi: 10.1104/pp.113.214049
Muiño, J. M., de Bruijn, S., Pajoro, A., Geuten, K., Vingron, M., Angenent, G. C., et al. (2016). Evolution of DNA-binding sites of a floral master regulatory transcription factor. Mol. Biol. Evol. 33, 185–200. doi: 10.1093/molbev/msv210
Mur, L. A., Kenton, P., Atzorn, R., Miersch, O., Wasternack, C. (2006). The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262. doi: 10.1104/pp.105.072348
Nie, J., Wen, C., Xi, L., Lv, S. H., Zhao, Q. C., Kou, Y. P., et al. (2018). The AP2/ERF transcription factor CmERF053 of chrysanthemum positively regulates shoot branching, lateral root, and drought tolerance. Plant Cell Rep. 37, 1049–1060. doi: 10.1007/s00299-018-2290-9
Oblessuc, P. R., Obulareddy, N., DeMott, L., Matiolli, C. C., Thompson, B. K., Melotto, M. (2020). JAZ4 is involved in plant defense, growth, and development in Arabidopsis. Plant J. 101, 371–383. doi: 10.1111/tpj.14548
Stracke, R., Werber, M., Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/S1369-5266(00)00199-0
Sugiyama, S. I. (2005). Polyploidy and cellular mechanisms changing leaf size: comparison of diploid and autotetraploid populations in two species of lolium. Ann. Bot. 96, 931–938. doi: 10.1093/aob/mci245
Sussex, I. M., Kerk, N. M. (2001). The evolution of plant architecture. Curr. Opin. Plant Biol. 4, 33–37. doi: 10.1016/S1369-5266(00)00132-1
Takeda, T., Suwa, Y., Suzuki, M., Kitano, H., Ueguchi-Tanaka, M., Ashikari, M., et al. (2003). The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33, 513–520. doi: 10.1046/j.1365-313X.2003.01648.x
Tanaka, W., Pautler, M., Jackson, D., Hirano, H. Y. (2013). Grass meristems II: Inflorescence architecture, flower development and meristem fate. Plant Cell Physiol. 54, 313–324. doi: 10.1093/pcp/pct016
Tanaka, M., Takei, K., Kojima, M., Sakakibara, H., Mori, H. (2006). Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 45, 1028–1036. doi: 10.1111/j.1365-313X.2006.02656.x
Tan, M., Li, G. F., Liu, X. J., Cheng, F., Ma, J. J., Zhao, C. P., et al. (2018). Exogenous application of GA(3) inactively regulates axillary bud outgrowth by influencing of branching-inhibitors and bud-regulating hormones in apple (Malus domestica borkh.). Mol. Genet. Genomics 293, 1547–1563. doi: 10.1007/s00438-018-1481-y
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G. H., et al. (2007). JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signaling. Nature 448, 661–665. doi: 10.1038/nature05960
Wai, A. H., An, G. (2017). Axillary meristem initiation and bud growth in rice. J. Plant Biol. 60, 440–451. doi: 10.1007/s12374-017-0088-x
Wang, X. B., Wang, Q. P., Yan, L. X., Hao, Y. H., Lian, X. D., Zhang, H. P., et al. (2022). PpTCP18 is upregulated by lncRNA5 and controls branch number in peach (Prunus persica) through positive feedback regulation of strigolactone biosynthesis. Horticul. Res. 10, uhac224. doi: 10.1093/hr/uhac224
Wang, J., Yan, D. W., Yuan, T. T., Gao, X., Lu, Y. T. (2013). A gain-of-function mutation in IAA8 alters Arabidopsis floral organ development by change of jasmonic acid level. Plant Mol. Biol. 82, 71–83. doi: 10.1007/s11103-013-0039-y
Wasternack, C., Hause, B. (2013). Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. an update to the 2007 review in annals of botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Weiler, E. W., Jordan, P. S., Conrad, W. (1981). Levels of indole 3-acetic acid in intact and decapitated coleoptiles as determined by a specific and highly sensitive solid-phase enzyme immunoassay. Planta 153, 561–571. doi: 10.1007/BF00385542
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M., Turner, J. G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. doi: 10.1126/science.280.5366.1091
Yang, Y., Bao, S. Y., Zhou, X. H., Liu, J., Zhuang, Y. (2018). The key genes and pathways related to male sterility of eggplant revealed by comparative transcriptome analysis. BMC Plant Biol. 18, 209. doi: 10.1186/s12870-018-1430-2
Yang, C. X., Gao, Y. N., Gao, S. H., Yu, G., Xiong, C., Chang, J., et al. (2015). Transcriptome profile analysis of cell proliferation molecular processes during multicellular trichome formation induced by tomato Wov gene in tobacco. BMC Genomics 16, 868. doi: 10.1186/s12864-015-2099-7
Yan, Y. X., Stolz, S., Chételat, A., Reymond, P., Pagni, M., Dubugnon, L., et al. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19, 2470–2483. doi: 10.1105/tpc.107.050708
Ye, M., Luo, S. M., Xie, J. F., Li, Y. F., Xu, T., Liu, Y., et al. (2012). Silencing COI1 in rice increases susceptibility to chewing insects and impairs inducible defense. PloS One 7 (4), e36214. doi: 10.1371/journal.pone.0036214
Yu, X., Chen, G. P., Tang, B. Y., Zhang, J. L., Zhou, S. G., Hu, Z. L. (2018). The jasmonate ZIM-domain protein gene SlJAZ2 regulates plant morphology and accelerates flower initiation in solanum lycopersicum plants. Plant Sci. 267, 65–73. doi: 10.1016/j.plantsci.2017.11.008
Zawaski, C., Busov, V. B. (2014). Roles of gibberellin catabolism and signaling in growth and physiological response to drought and short day photoperiods in Populus trees. PloS One 9, e86217. doi: 10.1371/journal.pone.0086217
Zhai, Q. Z., Zhang, X., Wu, F. M., Feng, H. L., Deng, L., Xu, L., et al. (2015). Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell. 27, 2814–2828. doi: 10.1105/tpc.15.00619
Zhang, B., Liu, J., Yang, Z. E., Chen, E. Y., Zhang, C. J., Zhang, X. Y., et al. (2018). Genome-wide analysis of GRAS transcription factor gene family in gossypium hirsutum l. BMC Genomics 19, 348. doi: 10.1186/s12864-018-4722-x
Zhang, F., Rossignol, P., Huang, T., Wang, Y. W., May, A., Dupont, C., et al. (2020). Reprogramming of stem cell activity to convert thorns into branches. Curr. Biol. 30, 2951–2961.e5. doi: 10.1016/j.cub.2020.05.068
Zhang, F., Wang, Y. W., Irish, V. F. (2021). CENTRORADIALIS maintains shoot meristem indeterminacy by antagonizing THORN IDENTITY1 in citrus. Curr. Biol. 31, 2237–2242.e4. doi: 10.1016/j.cub.2021.02.051
Keywords: pear, branch, JA, transcriptional regulation, less branching mutant plant
Citation: Cheng Y, Liang C, Qiu Z, Zhou S, Liu J, Yang Y, Wang R, Yin J, Ma C, Cui Z, Song J and Li D (2023) Jasmonic acid negatively regulates branch growth in pear. Front. Plant Sci. 14:1105521. doi: 10.3389/fpls.2023.1105521
Received: 22 November 2022; Accepted: 16 January 2023;
Published: 07 February 2023.
Edited by:
Shangang Jia, China Agricultural University, ChinaReviewed by:
Pu Liu, Anhui Agricultural University, ChinaCopyright © 2023 Cheng, Liang, Qiu, Zhou, Liu, Yang, Wang, Yin, Ma, Cui, Song and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dingli Li, cWF1bGRsQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.