- 1Institute of Plant Protection/Henan Key Laboratory of Crop Pest Control/Key Laboratory of Integrated Pest Management on Crops in Southern Region of North China, Henan Academy of Agricultural Sciences, Zhengzhou, China
- 2College of Bioscience and Resource Environment/Key Laboratory of Urban Agriculture (North China), Ministry of Agriculture and Rural Affairs of the People’s Republic of China, Beijing University of Agriculture, Beijing, China
- 3Institute of Plant Protection, Luohe Institute of Agricultural Sciences, Luohe, China
- 4Henan Yunfei Technology Development Co., Ltd, Zhengzhou, China
Sitobion miscanthi, Rhopalosiphum padi, and Schizaphis graminum are the three main pests in Chinese wheat-producing regions. In 2020, they are classified into the Chinese Class I list of agricultural diseases and pests, due to their severe harm to wheat plantings. S. miscanthi, R. padi, and S. graminum are migrant pests, and understanding their migration patterns and simulating their migration trajectories would improve forecasting and controlling them. Furthermore, the bacterial community of the migrant wheat aphid is also less known. In this study, we employed a suction trap to uncover the migration patterns of the three wheat aphid species in Yuanyang county, Henan province, during 2018 to 2020. And then the migration trajectories of S. miscanthi and R. padi were simulated using the NOAA HYSPLIT model. The interactions between wheat aphids and bacteria were further revealed by specific PCR and 16S rRNA amplicon sequencing. The results showed that the population dynamics of migrant wheat aphids was varied. Most of the trapped samples were identified to be R. padi, and S. graminum was the least collected sample. Typically, R. padi had two migration peaks in the 3 years, whereas S. miscanthi and S. graminum only exhibited one migration peak in 2018 and 2019. Moreover, the aphid migration trajectories varied over the years. Generally, the aphids originated from the south and migrated to the north. Herein, the infections of three main aphid facultative bacterial symbionts, Serratia symbiotica, Hamiltonella defensa, and Regiella insercticola, were detected in S. miscanthi and R. padi with specific PCR. Rickettsiella, Arsenophonus, Rickettsia, and Wolbachia were further identified with 16S rRNA amplicon sequencing. Biomarker searching indicated that Arsenophonus was significantly enriched in R. padi. Furthermore, diversity analyses showed that the bacterial community of R. padi had a higher richness and evenness than that of S. miscanthi. In conclusion, this study expands our knowledge about the migration patterns of aphids in the main wheat plant region of China and reveals the interactions between bacterial symbionts and migrant aphids.
Introduction
Wheat (Triticum aestivum L.) is the third major grain crop plant in China. The most common pest on wheat is the wheat aphid, which not only feeds on the grain but also spreads plant viruses like the barley yellow dwarf virus (BYDV). Studies have reported that approximately 10–15 million hectares of wheat fields in China are affected by aphid infestations, resulting in 10% yield losses annually (Hu et al., 2016; Gong et al., 2021). Indian grain aphid Sitobion miscanthi, Bird cherry-oat aphid Rhopalosiphum padi (Linnaeus), and greenbug Schizaphis graminum are three main wheat aphids in China. In 2020, due to their severe harm to wheat plantings, the three wheat aphids are classified into the Chinese Class I list of agricultural diseases and pests (http://www.zzys.moa.gov.cn/gzdt/202006/t20200604_6345940.htm). It is well known that wheat aphids can perform long-distance migration by air (Sun et al., 2022); hence, uncovering their seasonal migration patterns would improve forecasting and controlling them.
The insect marking–release–recapture technique (Hagler and Jackson, 2001) and trapping aerial insect samples using the sticky nets attached to the balloons and airplanes (Florio et al., 2020) have been used to track the migration of insects, but these strategies are labor-intensive. To date, the economical searchlight trap and high effective radar are widely employed to investigate insect migrations. However, the searchlight trap is mostly used to uncover the migrations of lepidopterous pests with large bodies (Guo et al., 2020; Zhou et al., 2021; Guo et al., 2022; Wang et al., 2022b), and the commonly used centimeter-wave vertical-looking radar is unsuitable for monitoring small insects, such as aphids, and a more expensive radar with a shorter wavelength is required (Chapman et al., 2003; Chapman et al., 2004; Feng et al., 2009). Suction traps have been used to monitor the aerial movement of aphids since 1964 (Macaulay et al., 1988), which probably filled the gap in the investigation of aphid migrations. Suction trap networks have been established in the United Kingdom and the United States to monitor seasonal distribution and abundance of diverse aphid species for a few decades (Bell et al., 2015; Lagos-Kutz et al., 2020). In Spain, suction traps are useful for monitoring the flight of damson-hop aphid (Phorodon humuli) at the start of spring (Pérez et al., 2006). In 2011, to investigate the migrations of soybean and wheat aphids, a Chinese aphid suction trap network was constructed (Qiao et al., 2011).
It is well known that aphids and bacteria share intimate relationships. In recent years, the 16S ribosomal RNA (rRNA) gene amplicon high-throughput sequencing has been applied to explore the aphid bacterial community (Xu et al., 2020; Xu et al., 2021a; Xu et al., 2021b). However, these bacterial surveys have mainly been performed in the field-collected samples; the interactions between migrant aphids and bacterial symbionts are less studied. In this study, which employed a suction trap, we uncovered the occurrence and migration patterns of the three main wheat aphids in Yuanyang county from 2018 to 2020 and simulated their migration trajectories. Furthermore, using specific PCR and 16S amplicon sequencing, we uncovered the bacterial composition in the migrant S. miscanthi and R. padi.
Materials and methods
Flying wheat aphid collection and identification
Henan province in China, which contributes a quarter of China’s annual wheat harvest, is the core wheat production area. Notably, the first suction trap in Henan was constructed in Yuanyang county, Xinxiang city (35.01 N, 113.69 E). The suction trap is 8.8 m tall and collected the tiny flying insects weekly from April to June 2018–2020. Morphological characteristics were used to identify the three main wheat aphids, S. miscanthi, R. padi, and S. graminum, among the trapped samples. All samples were deposited in the Institute of Plant Protection, Henan Academy of Agricultural Sciences, Zhengzhou, China. They were stored in 75% ethanol and maintained under −20°C until DNA extraction.
Wheat aphid migration pattern and trajectory analysis
The population dynamics of migration wheat aphids was summarized, and their migrating peaks were revealed. The HYSPLIT (HYbrid Single-Particle Lagrangian Integrated Trajectory) model is designed to simulate the trajectory of substances transported and dispersed through atmosphere using gridded meteorological data (Stein et al., 2015; Rolph et al., 2017). It is widely used to simulate the global dust distribution, volcanic ash dispersion, and pollutant transport (Stein et al., 2007; Stunder et al., 2007; Wang et al., 2011). Tiny insects like aphids have limited flight capacity. The flight speed of aphids is approximately 0.9 m s−1 (Robert, 1987), and the aphid flights are controlled by the wind when they move above at approximately 1 m from the ground (Parry, 2013). Hence, the HYSPLIT model is also suitable to calculate the migration trajectory of tiny insects with meteorological data.
In this study, the migration trajectories of S. miscanthi and R. padi were simulated with the online HYSPLIT model (https://www.ready.noaa.gov/HYSPLIT_traj.php). In these analyses, the meteorological data were obtained with the one-degree GDAS (Global Data Assimilation System) model, the model calculated star times were set on the 02:00 UTC time (10 a.m. in Beijing time) on the aphid migration peak days, and the flight heights were chosen as 10, 50, and 100 m AGL (above ground level). The aphid migration trajectories were simulated in 24 h.
DNA extraction, bacterial symbiont detections, and phylogenetic analysis
In this study, the infections of three main aphid bacterial symbionts, Serratia symbiotica, Hamiltonella defensa, Regiella insercticola, and one common insect bacterial symbiont, Wolbachia, were detected in S. miscanthi and R. padi with specific primers listed in Table S1. In these specific amplifications, aphid total DNA was extracted from a single aphid using an Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech, Shanghai), following the manufacturer’s recommendations. Before DNA extraction, every aphid was washed with 70% ethanol and sterile water several times to remove surface contamination. Aphid elongation factor-1α gene was used as a reference to evaluate the extracted DNA quality, and the low-quality ones were excluded in the bacterial symbiont detections. The 16S rRNA sequences of the known aphid bacterial symbiont strains were retrieved to uncover the phylogenetic positions of the identified bacterial symbiont strains of S. miscanthi and R. padi. The sequences were aligned using the MUSCLE program in MEGA 7.0 (Kumar et al., 2016). Phylogenetic analysis was performed in IQ-TREE 1.6.12 (Nguyen et al., 2015). The support for each node was assessed by resampling 5,000 ultrafast bootstraps (Hoang et al., 2018). The best substitution model was selected by Bayesian information criterion in ModelFinder (Kalyaanamoorthy et al., 2017). The phylogenetic tree was visualized in Figtree 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Bacterial 16S rRNA amplicon amplification and sequencing
Herein, the bacterial composition of S. miscanthi and R. padi was further uncovered by amplicon sequencing. We randomly selected 15 individuals of S. miscanthi and R. padi collected in the same year and divided them into five replicates. The genomic DNA of the pooled aphids in each replicate was extracted using the DNeasy Blood and Tissue Kit (QIAGEN, Germany) according to the manufacturer’s instructions. The possible surface contamination was removed as described above. An approximately 460-bp fragment of the bacterial 16S rRNA V3–V4 region was amplified with primers 341F: CCTACGGGNGGCWGCAG and 806R: GGACTACHVGGGTATCTAAT (Qin et al., 2021). The PCR reactions were performed in triplicate in a 50-μl mixture containing 5 μl of 10 × KOD Buffer, 5 μl of 2 mM dNTPs, 3 μl of 25 mM MgSO4, 1.5 μl of each primer (10 μM), 1 μl of KOD Polymerase, and 100 ng of template DNA. PCR amplifications were carried out with the following program: 94°C for 2 min, followed by 30 cycles at 98°C for 10 s, 62°C for 30 s, and 68°C for 30 s and a final extension at 68°C for 5 min. The amplicons were further purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to the manufacturer’s instructions. Subsequently, the purified amplicons were quantified using the ABI StepOnePlus Real-Time PCR System (Life Technologies, Foster City, USA), pooled in equimolar and generated the sequencing libraries. The next sequencing was performed by the Gene Denovo Biotechnology Co. (Guangzhou, China) on an Illumina HiSeq 2500 platform with a 2×250-bp paired-end method according to the standard protocols.
Bioinformatics analyses
The reads were trimmed using Btrim with the cutoff for average quality scores higher than 20 over a 5-bp window size and a minimum length of 180 bp (Kong, 2011). The reads ranging from 242 bp to 244 bp were employed in the following analyses with QIIME2 (version 2022.11) (Bolyen et al., 2019). Primers and adaptor sequences were removed by cutadapt plugin (Martin, 2011). ASVs (amplicon sequence variants) were obtained by merging the sequences and removing chimera with the DADA2 plugin (Callahan et al., 2016). The taxonomic assignment of representative sequences was carried out using the feature-classifier plugin against the SILVA database (silva-138-99). Four alpha diversity indexes, ACE (abundance-based coverage estimator), Chao1, Shannon, and Simpson, were calculated in QIIME2; Bray–Curtis distance matrix was calculated in QIIME2, and then PCoA (principal coordinates analysis) and NMDS (non-metric multi-dimensional scaling) were visualized with Bray–Curtis distances and plotted in the ggplot2 package. The relative abundance of the top 10 bacterial taxa among the samples and the cladogram of the differential species were analyzed and visualized by the MicrobiotaProcess package (https://github.com/YuLab-SMU/MicrobiotaProcess), with the feature table and taxonomy table obtained in QIIME2.
Results
Annual migration patterns of three wheat aphids
In this study, most of the migrant wheat aphids were trapped in 2018 (4,051 samples), and 2020 had the least number of trapped migrant wheat aphids (737 samples). In 2019, 1,271 migrant wheat aphids were trapped (Figure 1A). R. padi was predominant among the samples of trapped wheat aphids, whereas S. miscanthi and S. graminum were less common. However, among the three aphid species, the number of trapped aphids was not significantly divergent (ANOVA test, F = 2.334, p = 0.1778). In addition, different patterns of wheat aphid migration were discovered over the years (Figures 1B–D). The results indicated that the migrations of R. padi had two distinct peaks during 2018–2020. Specifically, in 2018 and 2019, the first R. padi migration peak occurred in the second week of May, and the second peak occurred in the first week of June. However, in 2020, the two R. padi migration peaks happened 1 week earlier than that of 2018 and 2019. In contrast, only one peak was observed in the migration of S. graminum and S. miscanthi in 2018 and 2019, which occurred in the first and third weeks of May, respectively. The S. miscanthi migration experienced two adjacent peaks in 2020, which occurred in the fourth week of April and the second week of May, respectively. However, no obvious peak was revealed in S. graminum migration in 2020, since at most three samples were trapped in the weeks and a total of 15 samples were collected.
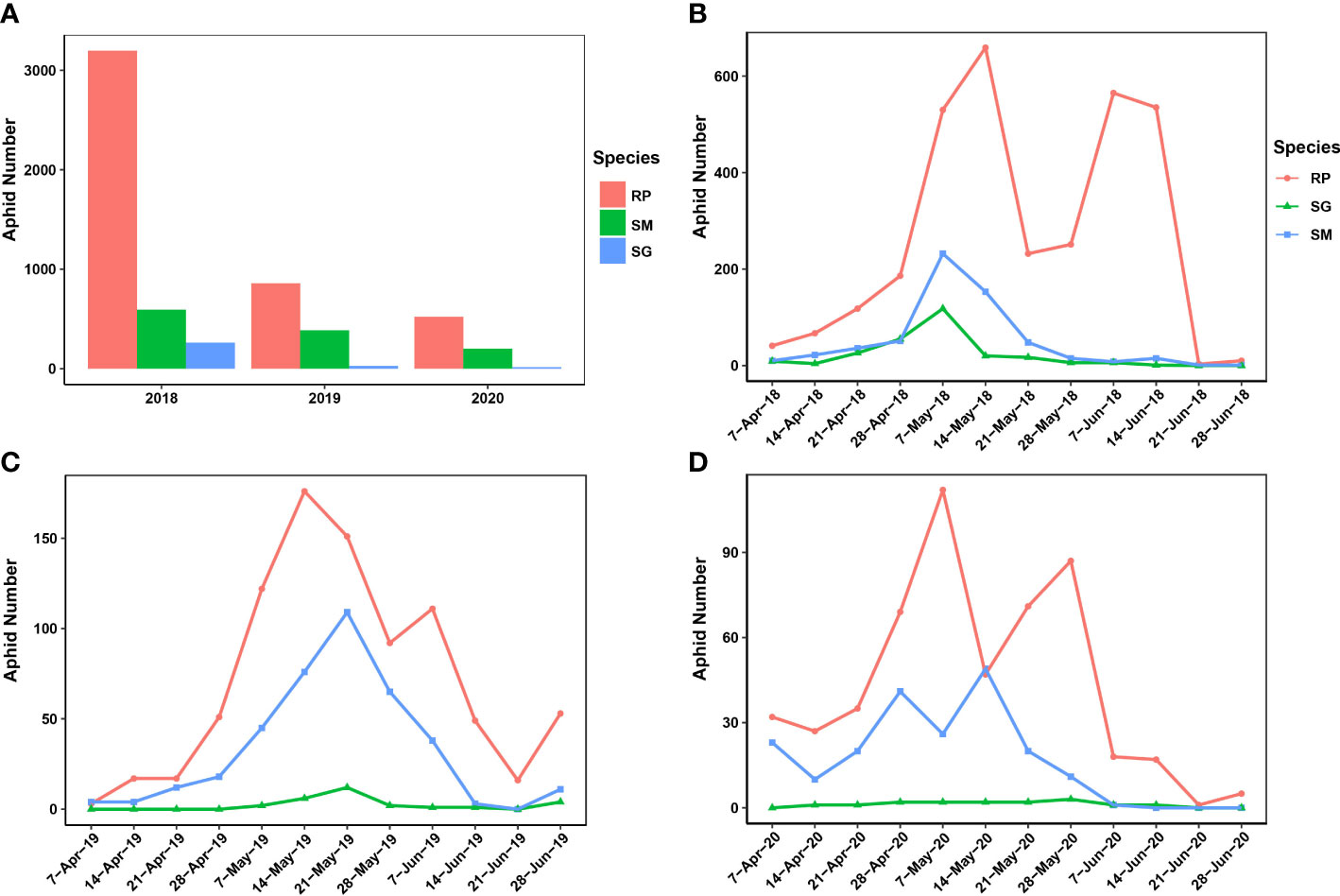
Figure 1 Population dynamics and migration patterns of the wheat aphids uncovered by a suction trap in Yuanyang country from 2018 to 2020. (A) The population dynamics of the three wheat aphids during 2018–2020. (B–D) The migration pattern of the three wheat aphids was revealed during 2018–2020. SM, Sitobion miscanthi; RR, Rhopalosiphum padi; SG, Schizaphis graminum.
Here, the meteorological data reflecting the local climate conditions near the suction trap (thereafter named Yuangyang), as well as those reflecting the climate conditions of a wider area, such as Xinxiang city, were involved in the statistical analyses to comprehensively uncover the underlying roles of temperature and humidity on the migrations of wheat aphids. The results indicated that there was no significant difference in the temperatures of April and May during 2018–2020, in either the Yuanyang or Xinxiang region (ANOVA test, p > 0.05) (Figures 2A, B). However, a significant difference in their relative humidity was observed (ANOVA test, p < 0.05) (Figures 2C, D). Furthermore, the multiple comparisons using Tukey’s test revealed that the relative humidity of April 2018 was significantly different with that of 2020 either in the Yuanyang or Xinxiang region. However, the relative humidity of May 2018 was significantly different from that of 2019 and 2020 in the Yuanyang region, and in the Xinxiang region, the relative humidity of May 2018 was significantly different from that of 2019.
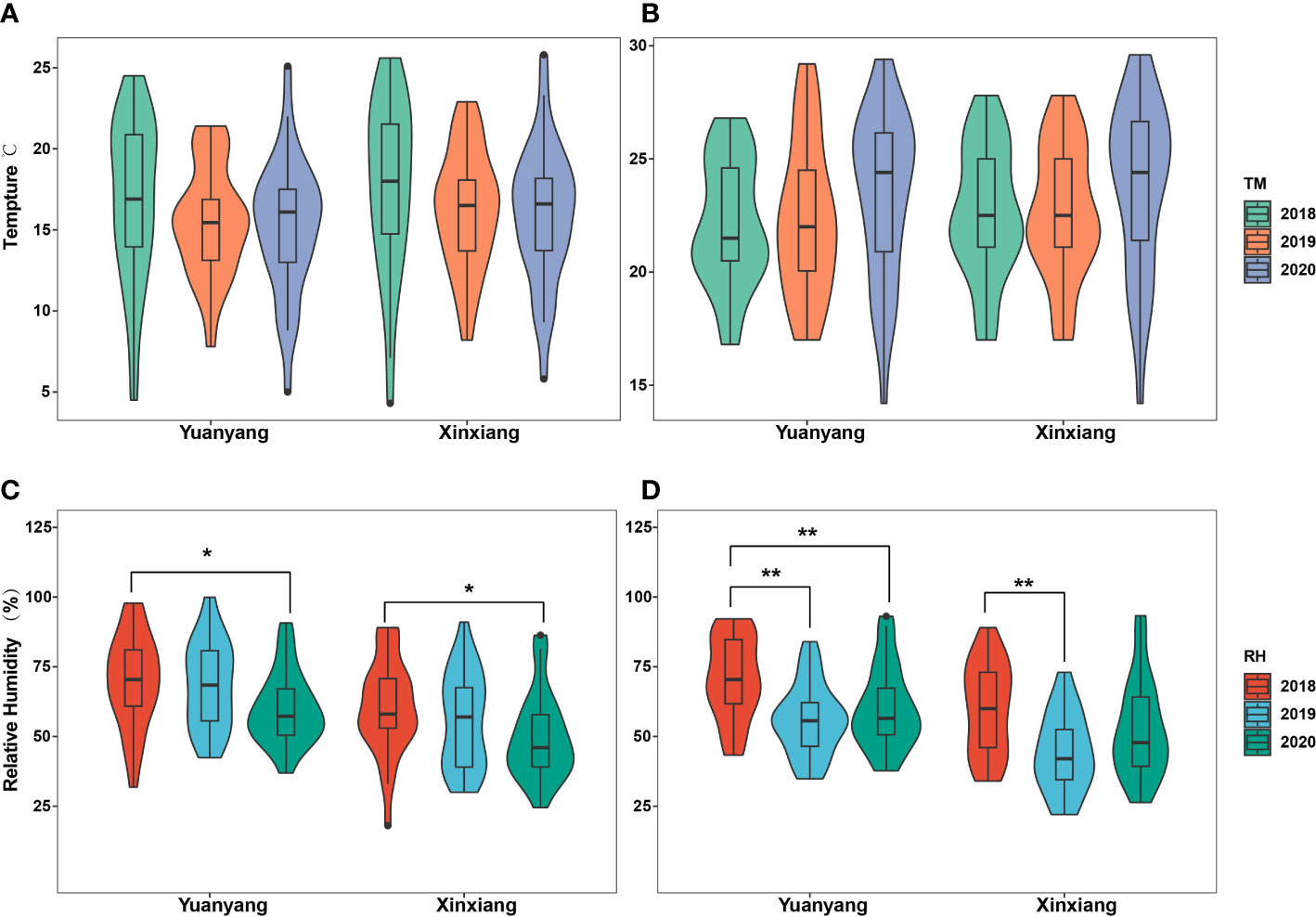
Figure 2 Statistical analysis of temperature and relative humidity among years in Yuanyang and Xinxiang regions. (A, B) Analysis with the April and May temperature obtained in Yuanyang and Xinxiang regions, respectively. (C, D) Analysis with the April and May relative humidity in Yuanyang and Xinxiang regions, respectively. *, indicating significant (p<0.05); **, indicating very significant (p<0.01).
Aphid migration trajectory analyses
To identify the origin of the migrating aphids, we simulated the backward migration trajectories of immigrations. On the other hand, the forward migration trajectories of emigrations were simulated to uncover the destinations of the flying aphids. The results demonstrated similar migration trajectories at the three layers: 10 m, 50 m, and 100 m AGL (Figures 3, 4). Most S. miscanthi migrations on 7 May 2018 and 28 April 2020 originated from the west and southeast of Yuanyang county, respectively (Figures 3A, B). In addition, on 21 May 2019 and 14 May 2020, S. miscanthi migrated to the northeast and northwest of Yuanyang county, respectively (Figures 3C, D). The migration trajectories of R. padi are summarized in Figure 4. The results indicated that most R. padi migrations on 14 May 2018 originated north of Yuanyang country (Figure 4A). However, most of the migrating R. padi on 14 May 2019 and 7 May 2020 originated northeast of Yuanyang country (Figures 4B, C). According to the simulation of forward migration trajectories, most R. padi would migrate to the south and east of Yuanyang county on 7 June 2018 (Figure 4D). On the contrary, most R. padi would migrate to the northeast and north of Yuanyang county on 7 June 2019 and 28 May 2020, respectively (Figures 4E, F).
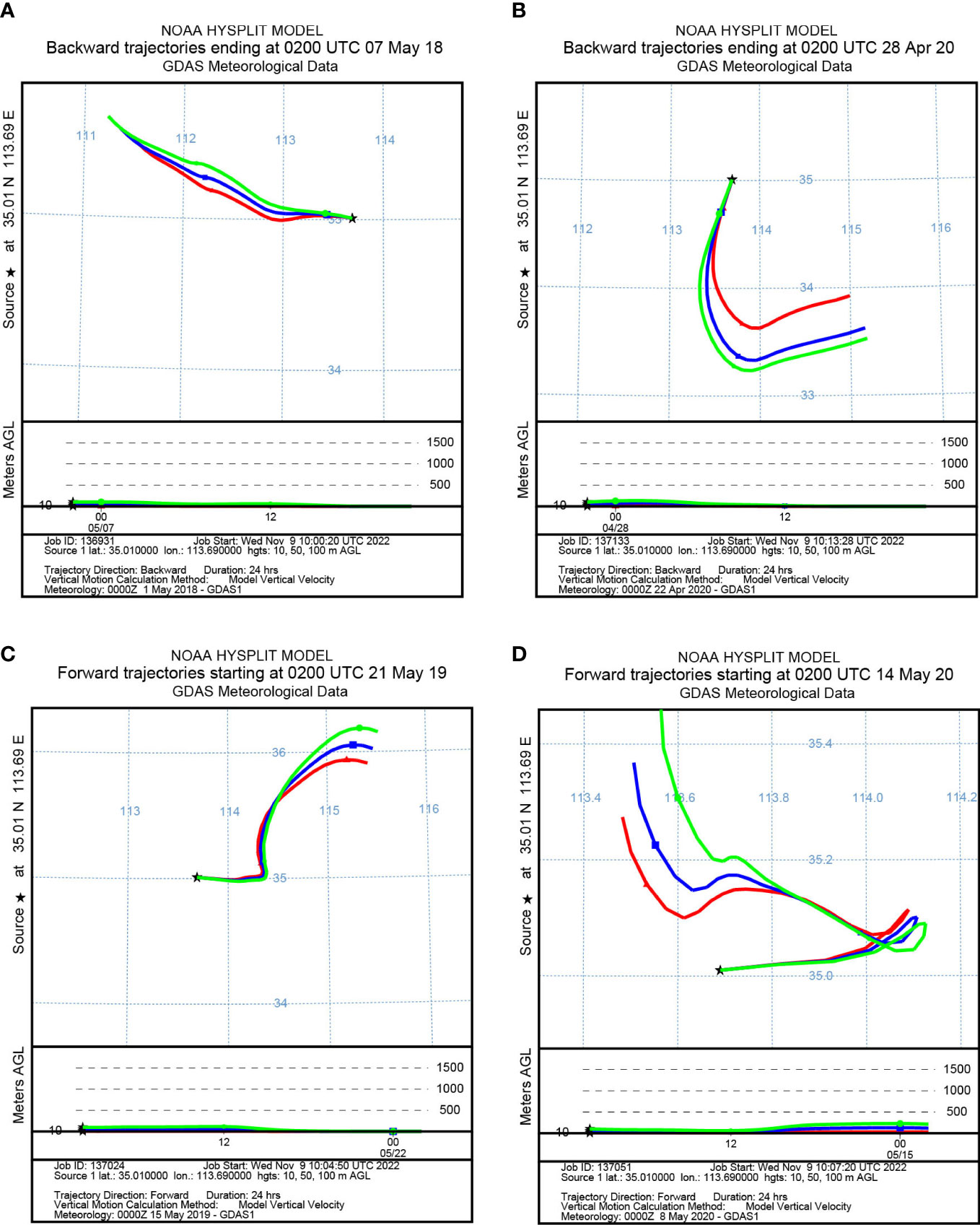
Figure 3 The migration trajectories of Sitobion miscanthi simulated by HYSPLIT. (A, B) Simulation of the backward trajectories on 7 May 2018 and 28 April 2020, respectively. (C, D) Simulation of the forward trajectories on 21 May 2019 and 14 May 2020, respectively.
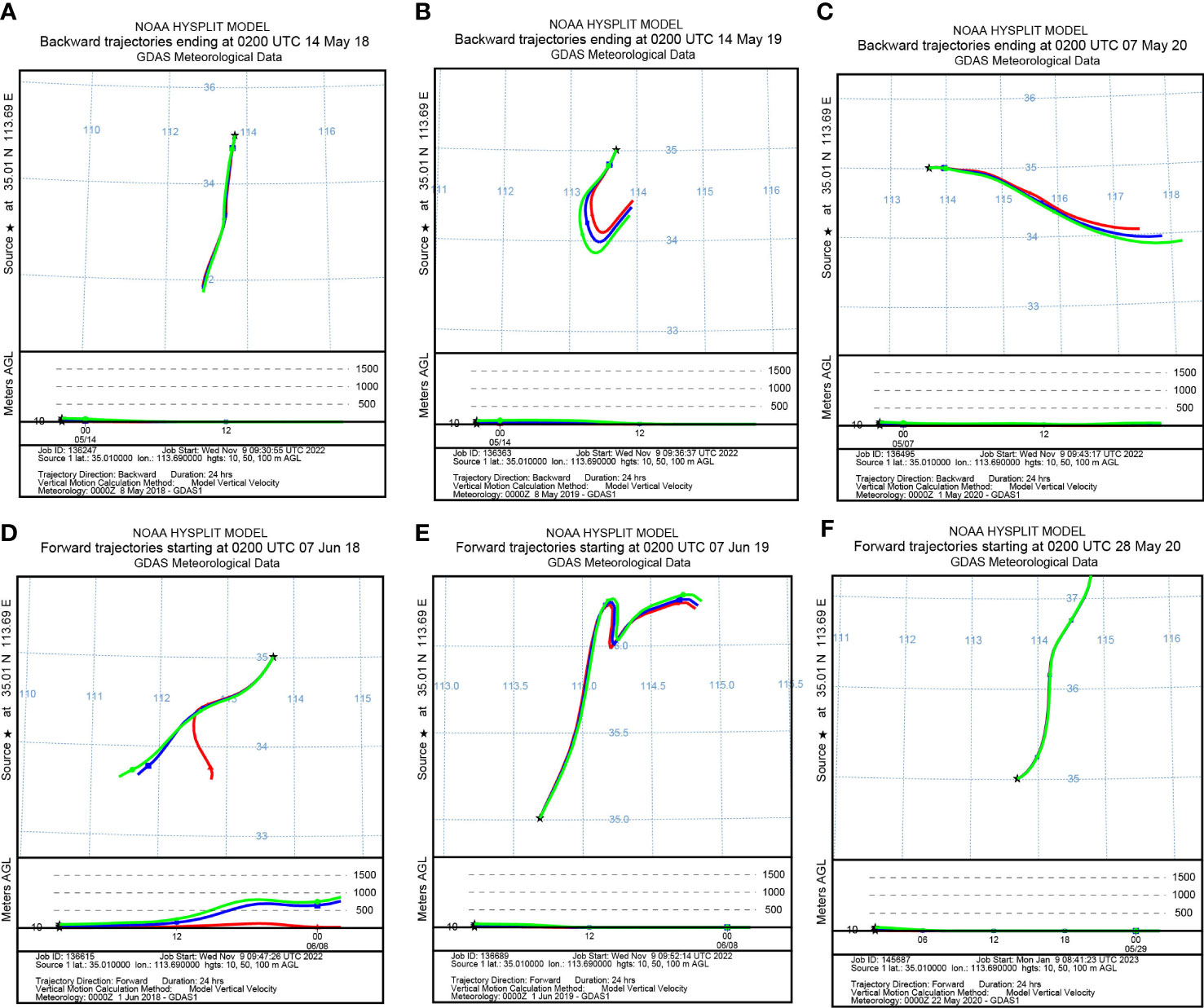
Figure 4 The migration trajectories of Rhopalosiphum padi simulated by HYSPLIT. (A, B) Simulation of the backward trajectories on 14 May 2018 and 14 May 2019, respectively. (C) Simulation of the backward trajectories on 7 May 2020. (D, E) Simulation of the forward trajectories on 7 June 2018 and 7 June 2019, respectively. (F) Simulation of the forward trajectories on 28 May 2020.
Specific aphid facultative symbiont detection
In this study, a total of 237 and 350 samples of S. miscanthi and R. padi were subjected to the specific bacterial symbiont detections, respectively (Table 1). S. symbiotica, H. defensa, and R. insercticola were detected in the aphids; however, no infection of Wolbachia was identified. In S. miscanthi, infection rates of S. symbiotica and H. defensa were 12.23% (29/237) and 1.27% (3/237), respectively. On the other side, in R. padi, infection rates of S. symbiotica and H. defensa were 7.71% (27/350) and 1.42% (5/350), respectively. The infection rates of R. insercticola in S. miscanthi and R. padi were 2.95% (7/237) and 4.29 (15/350), respectively. However, between S. miscanthi and R. padi, no significant difference was observed in the infections of S. symbiotica (χ2 = 2.28, p = 0.13), H. defensa (χ2 = 0.027, p = 0.86), and R. insercticola (χ2 = 0.69, p = 0.40). In the phylogenetic tree (Figure 5), the monophyly of S. symbiotica, H. defensa, and R. insercticola was robustly supported. In these symbiont clades, the strains identified in S. miscanthi and R. padi shared close relationships with that isolated in other aphid species, which further verified their infections.
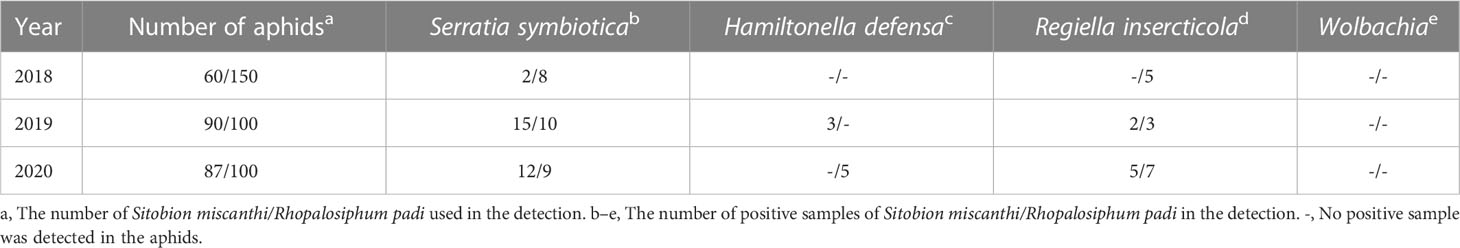
Table 1 Detection of bacterial symbionts in Sitobion miscanthi and Rhopalosiphum padi with specific primers.
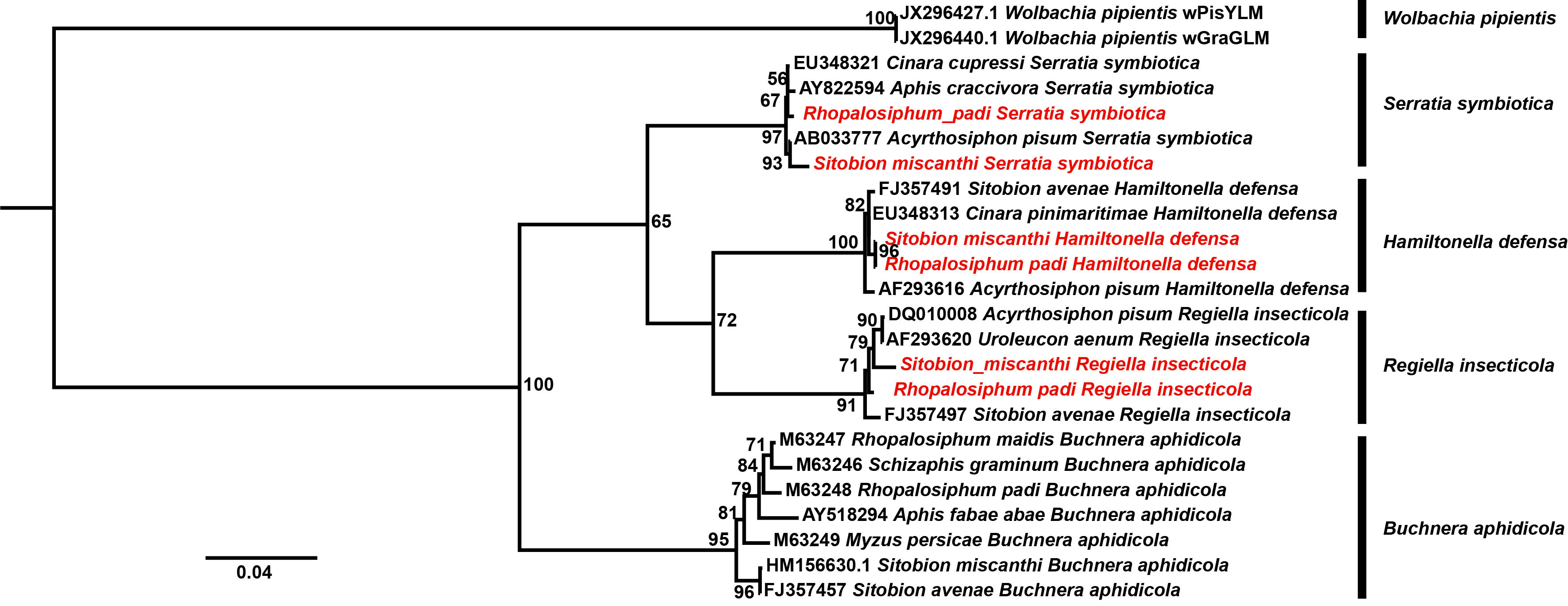
Figure 5 The phylogenetic analysis of aphid bacterial symbionts. The strains identified herein were highlighted with red.
Analyses of 16S rRNA amplicon sequencing
In this study, a total of 3,874,860 raw reads (average 129,162 reads per sample) were yielded in the 16S rRNA V3–V4 amplicon sequencing. After trimming, denoising, and chimera removal, 8,518 representative ASVs were obtained, and their taxonomic positions were identified against the SILVA database. The relative abundance of the top 10 bacterial taxa among the samples was revealed at four taxonomic levels (Figure 6). The results showed that compositions of bacterial community harbored in the S. miscanthi 2018 population was different from that of the remaining aphid populations. For instance, at the phylum level, in the 2018 S. miscanthi population, Firmicutes was dominant with 79.60% relative abundance and Proteobacteria was the second main taxon with 8.46% relative abundance. However, in the remaining aphid populations, Proteobacteria was dominant with a relative abundance ranging from 34.49% in the 2018 R. padi population to 78.47% in the 2019 R. padi population. A similar divergence was also observed at the order and family level. At the genus level, Buchnera was identified in all samples with an average relative abundance of 11.26%. Furthermore, two aphid facultative symbionts belonging to Serratia and Rickettsiella and three common insect bacterial symbionts, Arsenophonus, Rickettsia, and Wolbachia, were identified. In S. miscanthi, Serratia was the dominant aphid facultative symbiont with an average abundance of 8.41%. On the other side, Arsenophonus was the dominant aphid facultative symbiont in R. padi with an average abundance of 23.30%. In S. miscanthi, the average abundance of Rickettsiella was 10.11% much higher than that of R. padi, which had only 0.13% average abundance. Rickettsia was mainly identified in the 2019 S. miscanthi population with a 9.49% abundance. Wolbachia was only identified in the 2019 R. padi population with a 0.05% abundance. The results of biomarker discovery indicated that Arsenophonus was the most differentially abundant bacterial taxon in R. padi (Figure S1). Additionally, within S. miscanthi and R. padi, the abundance of Rickettsiella, Arsenophonus, and Serratia was varied among the years. Their lowest relative abundance was observed in the 2018 aphid populations (Figure S2). In S. miscanthi, the relative abundance of Rickettsiella in the 2018 and 2019 aphid populations was significantly different. A similar result was also observed in Serratia (Figure S2A). In R. padi, the relative abundance of Arsenophonus in the 2018 aphid population was significantly different from that in the 2019 and 2020 aphid populations (Figure S2B).
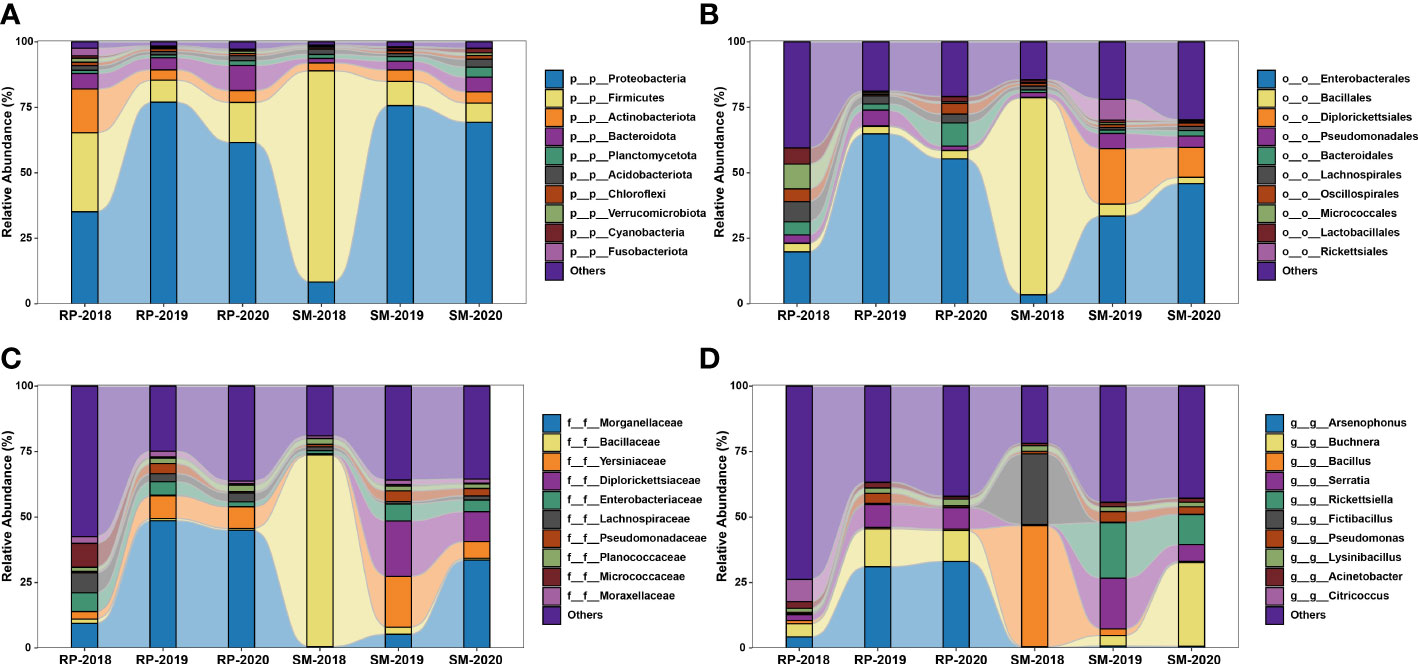
Figure 6 Stacked bar plot showing the relative abundance of the top 10 bacterial taxa identified in Sitobion miscanthi and Rhopalosiphum padi samples at four taxonomic levels. (A), phylum; (B), order; (C), family; and (D), genus.
In this study, the alpha diversity among the samples was varied (Figure S3A). Furthermore, it was found that R. padi had a relatively higher richness and evenness than S. miscanthi (Figure S3B). In beta diversity analyses, both PCoA and NMDS analyses indicated that the bacterial community of the 2018 S. miscanthi population was distinct from the other samples (Figure S4). The samples of S. miscanthi trapped in 2019 and 2020 had similar bacterial communities. In R. padi, the bacterial communities were separated by years in PCoA. However, in the NMDS analysis, bacterial communities of the R. padi samples trapped in 2018 and 2020 and that of S. miscanthi samples trapped in 2019 and 2020 were not well separated.
Discussion
Population dynamics of wheat migrant wheat aphids
In this study, the amount of migration wheat aphids varied among years. Previous studies indicated that temperature and humidity are the key environmental factors influencing aphid flights (Parry, 2013). Herein, we analyzed the underlying influences of temperature and humidity on the wheat aphid migrations with two types of meteorological data. The results showed that the difference in relative humidity among the years was significant, indicating that relative humidity probably influenced the wheat aphid flight. We also found that the migrating populations of the three wheat aphids varied from each other. R. padi was the most abundant among the identified migrating aphids, followed by S. miscanthi, and S. graminum was the least abundant. This result was consistent with the findings of our aphid surveying in the wheat field (data not shown). In Yuanyang country, R. padi was the dominant wheat aphid species, particularly in the late wheat development stages characterized by higher temperatures. Previous studies demonstrated that R. padi performed better in high-temperature habitats (Asin and Pons, 2001) and showed greater ecological plasticity when exposed to harsh environments (Zhu et al., 2021). We hypothesize that the R. padi population would proliferate faster with a temperature increase than other wheat aphids; thus, more winged R. padi would be induced due to the low host plant quality and crowding.
Migration patterns and simulated trajectories of wheat aphids
A previous study indicated that in Chinese northern winter wheat plant regions, aphids would probably migrate into wheat fields during the wheat heading to flowering stages, and then migrate out in the wheat milk-ripe stage (Li et al., 2014a). In the Yuanyang country, the periods of wheat heading to flowering stages ranged from around late April to early May, and the milk-ripe stage of wheat ranged from around late May to early June. Hence, in this study, we roughly used the half of May as the threshold to distinguish the types of aphid migrations; those that occurred earlier were categorized as immigrations, while those that occurred later were categorized as emigrations. Although alate aphids will undertake appetitive flight in short distances (Loxdale et al., 1993), long-distance migrations are also observed in various aphid species (Kring, 1972). This study found that R. padi and S. miscanthi had different migration patterns. Specifically, two migration peaks were observed in R. padi, identified as immigrations and emigrations. However, in S. miscanthi, the migration patterns varied over the years. Only immigrations were observed in 2018, whereas only emigrations were observed in 2019. In 2020, two adjacent migration peaks were uncovered. These results suggest that the field population of S. miscanthi in Yuanyang county in 2018 likely included the migrated aphids, but few S. miscanthi migrated out in late June. However, in 2019, most S. miscanthi field specimens might have been overwintering individuals that migrated out in late May.
Wind speed and direction are the key factors influencing the initiation, path, speed, distance, and duration of aphid flight (Parry, 2013). HYSPLIT is widely used to simulate the migration trajectories of insects, such as Helicoverpa armigera (Feng et al., 2009), Spodoptera frugiperda (Westbrook et al., 2016), Pantala flavescens (Cao et al., 2018), and Anopheles mosquitos (Huestis et al., 2019). Herein, HYSPLIT was used to simulate the trajectories of R. padi and S. miscanthi on their peak migration days. The results indicated that the immigrations typically originated from the south, whereas the north was the primary destination of the emigrations. These findings were consistent with the previous conclusions that wheat aphid migration trajectories in Chinese wheat fields were typically from south to north in line with the monsoons (Li et al., 2014a). However, there were also some exceptions. In 2018, it was predicted that the S. miscanthi would migrate from the west, whereas the predicted direction of the R. padi in emigrations was south.
Bacterial composition of wheat aphids
Previously, using specific PCR, H. defensa and R. insercticola have been detected in S. miscanthi (Li et al., 2014b). Recently, the bacterial communities of S. miscanthi are investigated with 16S amplicon sequencing (Wang et al., 2022a). However, the examined aphids in these studies are collected in wheat fields; the bacterial community of the migrant wheat aphids is less known. In this study, using specific PCR and phylogenetic analysis, the infections of S. symbiotica, H. defensa, and R. insercticola were identified in S. miscanthi and R. padi migrants. However, no significant difference was observed in their infection rates. Amplicon sequencing indicated that Proteobacteria and Firmicutes were the dominant bacterial taxa in the wheat aphid migrants. Moreover, we identified two aphid facultative symbionts (S. symbiotica and R. viridis), and three common insect bacterial symbionts (Arsenophonus, Rickettsia, and Wolbachia). Arsenophonus has been detected in diverse aphid groups (Jousselin et al., 2012), whereas R. viridis has been identified in pea aphid (Tsuchida et al., 2010). To our knowledge, it is the first report of them in S. miscanthi and R. padi. Furthermore, Arsenophonus was identified to be the most differentially abundant bacterial taxon in R. padi. Additionally, to uncover the potential interactions between the infections of bacterial symbionts and aphid flight, the relative abundance of Rickettsiella, Arsenophonus, and Serratia, was used in the statistical analyses. In both S. miscanthi and R. padi, the lowest relative abundance of the bacterial symbionts was observed in the 2018 aphid populations. Furthermore, the specific PCR results revealed that the 2018 populations of S. miscanthi and R. padi had the lowest rates of bacterial symbiont infection. Since most of the aphid migrants were trapped in 2018, it indicated that aphid bacterial symbionts did not probably promote the wheat aphid migrations. However, this hypothesis needs to be further verified by comparing the aphid flight capabilities of bacterial symbiont infected and uninfected populations.
In conclusion, in this study, we used the data obtained from the Yuanyang suction trap site, uncovered the population dynamics of migrant wheat aphids in the main wheat planting region in China, and simulated their migration trajectories. Furthermore, we revealed the potential roles of climate conditions on the aphid migrations. Additionally, the interactions between wheat aphid migrants and bacteria were investigated with specific PCR and amplicon sequencing.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA898692.
Author contributions
TL, YW, and CL conceived the study. TL, GY, QL, and YJ drafted the manuscript. DK, ZF, RL, and GZ collected and identified the aphid species.TL prepared the figures. TL, YW, and QL revised the manuscript. TL, YW, and CL designed the whole study and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the China Agriculture Research System of MOF and 342 MARA (CARS-03), the Henan Provincial Science and Technology R&D Program Joint Fund (grant no. 222301420107), the Special Fund for Agro-Scientific Research in the Public Interest of China (grant no. 201103022), and the Major Public Welfare Scientific Research Project of Henan Province (grant no. 201300111600).
Acknowledgments
We thank Gangqi Fang from CAS Key Laboratory of Insect Developmental and Evolutionary Biology, Chinese Academy of Sciences, Shanghai, China, for providing help in the data analyses.
Conflict of interest
Author GZ is employed by Henan Yunfei Technology Development Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor JC declared a past collaboration with the authors QL, YW.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1103236/full#supplementary-material
Supplementary Figure 1 | The cladogram and abundance of differential species. (A) A cladogram showing the significant differentially abundant taxa in Sitobion miscanthi and Rhopalosiphum padi. Significant differentially abundant taxa were identified by the Kruskal-Wallis test (p< 0.05). (B) The abundance and LDA effect size of differential taxa in Sitobion miscanthi and Rhopalosiphum padi.
Supplementary Figure 2 | Statistic analysis of the relative abundance of Rickettsiella, Arsenophonus, and Serratia among years. (A) Analysis performed in Sitobion miscanthi. (B) Analysis performed in Rhopalosiphum padi.
Supplementary Figure 3 | The boxplots of alpha diversity indexes. (A) The ACE, Chao1, Shannon and Simpson indexes of bacterial communities in the Sitobion miscanthi and Rhopalosiphum padi populations trapped during 2018-2020. (B) The ACE, Chao1, Shannon and Simpson indexes of bacterial communities of the entire Sitobion miscanthi and Rhopalosiphum padi samples. SM, Sitobion miscanthi; RR, Rhopalosiphum padi.
Supplementary Figure 4 | The results of beta diversity analyses. (A) The PCoA analyses among the Sitobion miscanthi and Rhopalosiphum padi samples. (B) The NMDS analyses among the Sitobion miscanthi and Rhopalosiphum padi samples.
References
Asin, L., Pons, X. (2001). Effect of High Temperature on the Growth and Reproduction of Corn Aphids (Homoptera: Aphididae) and Implications for Their Population Dynamics on the Northeastern Iberian Peninsula. Environ. Entomol. 30 (6), 1127–1134. doi: 10.1603/0046-225X-30.6.1127
Bell, J. R., Alderson, L., Izera, D., Kruger, T., Parker, S., Pickup, J., et al. (2015). Long-term phenological trends, species accumulation rates, aphid traits and climate: Five decades of change in migrating aphids. J. Anim. Ecol. 84 (1), 21–34. doi: 10.1111/1365-2656.12282
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using qiime 2. Nat. Biotechnol. 37 (8), 852–857. doi: 10.1038/s41587-019-0209-9
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., Holmes, S. P. (2016). Dada2: High-resolution sample inference from illumina amplicon data. Nat. Methods 13 (7), 581–583. doi: 10.1038/nmeth.3869
Cao, L. Z., Fu, X. W., Hu, C. X., Wu, K. M. (2018). Seasonal migration of Pantala flavescens across the bohai strait in northern China. Environ. Entomol. 47 (2), 264–270. doi: 10.1093/ee/nvy017
Chapman, J. W., Reynolds, D. R., Smith, A. D. (2003). Vertical-looking radar: A new tool for monitoring high-altitude insect migration. BioScience 53 (5), 503–511. doi: 10.1641/0006-3568(2003)053[0503:VRANTF]2.0.CO;2
Chapman, J., Reynolds, D., Smith, A. (2004). Migratory and foraging movements in beneficial insects: A review of radar monitoring and tracking methods. Int. J. Pest Manage. 50 (3), 225–232. doi: 10.1080/09670870410001731961
Feng, H., Wu, X., Wu, B., Wu, K. (2009). Seasonal migration of Helicoverpa armigera (Lepidoptera: Noctuidae) over the bohai Sea. J. Econ. Entomol. 102 (1), 95–104. doi: 10.1603/029.102.0114
Florio, J., Veru, L. M., Dao, A., Yaro, A. S., Diallo, M., Sanogo, Z. L., et al. (2020). Diversity, dynamics, direction, and magnitude of high-altitude migrating insects in the sahel. Sci. Rep. 10 (1), 20523. doi: 10.1038/s41598-020-77196-7
Gong, P., Li, X., Gao, H., Wang, C., Li, M., Zhang, Y., et al. (2021). Field evolved resistance to pyrethroids, neonicotinoids, organophosphates and macrolides in Rhopalosiphum padi (Linnaeus) and Sitobion avenae (Fabricius) from China. Chemosphere 269, 128747. doi: 10.1016/j.chemosphere.2020.128747
Guo, J. L., Fu, X. W., Zhao, S. Y., Shen, X. J., Wyckhuys, K. A. G., Wu, K. M. (2020). Long-term shifts in abundance of (Migratory) crop-feeding and beneficial insect species in northeastern Asia. J. Pest Sci. 93, 583–594. doi: 10.1007/s10340-019-01191-9
Guo, J., Yang, F., Zhang, H., Lin, P., Zhai, B., Lu, Z., et al. (2022). Reproduction does not impede the stopover departure to ensure a potent migration in Cnaphalocrocis medinalis moths. Insect Sci. 29 (6), 1672–1684. doi: 10.1111/1744-7917.13020
Hagler, J. R., Jackson, C. G. (2001). Methods for marking insects: Current techniques and future prospects. Annu. Rev. Entomol. 46, 511–543. doi: 10.1146/annurev.ento.46.1.511
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., Vinh, L. S. (2018). Ufboot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35 (2), 518–522. doi: 10.1093/molbev/msx281
Hu, X. S., Liu, Y. J., Wang, Y. H., Wang, Z., Yu, X. L., Wang, B., et al. (2016). Resistance of wheat accessions to the English grain aphid Sitobion avenae. PLoS One 11 (6), e0156158. doi: 10.1371/journal.pone.0156158
Huestis, D. L., Dao, A., Diallo, M., Sanogo, Z. L., Samake, D., Yaro, A. S., et al. (2019). Windborne long-distance migration of malaria mosquitoes in the sahel. Nature 574 (7778), 404–408. doi: 10.1038/s41586-019-1622-4
Jousselin, E., Coeur d'Acier, A., Vanlerberghe-Masutti, F., Duron, O. (2012). Evolution and diversity of Arsenophonus endosymbionts in aphids. Mol. Ecol. 22 (1), 260–270. doi: 10.1111/mec.12092
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., Jermiin, L. S. (2017). Modelfinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14 (6), 587–589. doi: 10.1038/nmeth.4285
Kong, Y. (2011). Btrim: A fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98 (2), 152–153. doi: 10.1016/j.ygeno.2011.05.009
Kring, J. B. (1972). Flight behaviour of aphids. Annu. Rev. Entomol. 17, 461–492. doi: 10.1146/annurev.en.17.010172.002333
Kumar, S., Stecher, G., Tamura, K. (2016). Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Lagos-Kutz, D., Voegtlin, D. J., Onstad, D., Hogg, D., Ragsdale, D., Tilmon, K., et al. (2020). The soybean aphid suction trap network: Sampling the aerobiological “Soup”. Am. Entomol. 66, 48–55. doi: 10.1093/ae/tmaa009
Li, K. B., Du, G. Q., Yin, J., Zhang, S., Cao, Y. Z. (2014a). Monitoring the migration of Sitobion avenae (Fabricius) bySuction trapping (in Chinese). Chin. J. Entomol. 51 (6), 1504–1515. doi: 10.7679/j.issn.2095–1353.2014.175
Li, T., Xiao, J. H., Wu, Y. Q., Huang, D. W. (2014b). Diversity of bacterial symbionts in populations of Sitobion miscanthi (Hemiptera: Aphididae) in China. Environ. Entomol. 43, 605–611. doi: 10.1603/EN13229
Loxdale, H. D., Hardie, J., Halbert, S., Foottit, R., Kidd, N. A. C., Carter, C. I. (1993). The relative importance of short- and long-range movement of flying aphids. Biol. Rev. 68, 291–311. doi: 10.1111/j.1469-185X.1993.tb00998.x
Macaulay, E. D. M., Tatchell, G. M., Taylor, L. R. (1988). The rothamsted insect survey '12-metre' suction trap. Bull. Ent. Res. 78, 121—129. doi: 10.1017/S0007485300016126
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 17 (1), 10–12. doi: 10.14806/ej.17.1.200
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., Minh, B. Q. (2015). Iq-tree: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi: 10.1093/molbev/msu300
Parry, H. R. (2013). Cereal aphid movement: General principles and simulation modelling. Mov. Ecol. 1 (1), 14. doi: 10.1186/2051-3933-1-14
Pérez, N., Seco, M. V., Valenciano, J. B., Lorenzana, A. (2006). Use of suction-trap and moericke traps for monitoring the migration of damson-hop aphid (Phorodon humuli) (Hemiptera, aphididae). New Zeal. J. Crop Hortic. 35, 455–461. doi: 10.1080/01140670709510214
Qiao, G. X., Qin, Q. L., Liang, H. B., Cao, Y. Z., Xu, G. Q., CAO, Z. L., et al. (2011). A new aphid-monitoring network system based on suction trapping and development of "Green techniques" for aphid management (in Chinese). Chin. J. Appl. Entomol. 48 (6), 1596–1601.
Qin, M., Chen, J., Xu, S., Jiang, L., Qiao, G. (2021). Microbiota associated with Mollitrichosiphum aphids (Hemiptera: Aphididae: Greenideinae): Diversity, host species specificity and phylosymbiosis. Environ. Microbiol. 23 (4), 2184–2198. doi: 10.1111/1462-2920.15391
Robert, Y. (1987). “Aphids and their environment: Dispersion and migration,” in Aphids: Their biology, natural enemies and control. Eds. Minks, A. K., Harrewijn, P. (Amsterdam: Elsevier), 299–313.
Rolph, G., Stein, A., Stunder, B. (2017). Real-time environmental applications and display system: Ready. Environ. Model. Software 95, 210–228. doi: 10.1016/j.envsoft.2017.06.025
Stein, A. F., Draxler, R. R., Rolph, G. D., Stunder, B. J. B., Cohen, M. D., Ngan, F. (2015). Noaa's hysplit atmospheric transport and dispersion modeling system. Bull. Amer. Meteor. Soc 96, 2059–2077. doi: 10.1175/BAMS-D-14-00110.1
Stein, A. F., Isakov, V., Godowitch, J., Draxler, R. R. (2007). A hybrid modeling approach to resolve pollutant concentrations in an urban area. Atmo. Environ. 41 (40), 9410–9426. doi: 10.1016/j.atmosenv.2007.09.004
Stunder, B. J. B., Heffter, J. L., Draxler, R. R. (2007). Airborne volcanic ash forecast area reliability. Weather Forecast 22 (5), 1132–1139. doi: 10.1175/WAF1042.1
Sun, J. J., Li, Q., Tan, X. L., Fan, J., Zhang, Y., Qin, Y. G., et al. (2022). Population genetic structure of Sitobion miscanthi in China. J. Integr. Agric. 21 (1), 178–187. doi: 10.1016/S2095-3119(20)63445-1
Tsuchida, T., Koga, R., Horikawa, M., Tsunoda, T., Maoka, T., Matsumoto, S., et al. (2010). Symbiotic bacterium modifies aphid body color. Science 330 (6007), 1102–1104. doi: 10.1126/science.1195463
Wang, C., Li, X., Jin, D., Gong, P., Li, Q., Zhang, Y., et al. (2022a). Implications of environmentally shaped microbial communities for insecticide resistance in Sitobion miscanthi. Environ. Res. 215 (Pt 2), 114409. doi: 10.1016/j.envres.2022.114409
Wang, X., Yang, X., Zhou, L., Wyckhuys, K. A., Jiang, S., Van Liem, N., et al. (2022b). Population genetics unveils Large-scale migration dynamics and population turnover of Spodoptera exigua. Pest. Manage. Sci. 78 (2), 612–625. doi: 10.1002/ps.6670
Wang, Y., Stein, A. F., Draxler, R. R., Rosa, J. D. D. L., Zhang, X. (2011). Global sand and dust storms in 2008: Observation and hysplit model verification. Atmo. Environ. 45 (35), 6368–6381. doi: 10.1016/j.atmosenv.2011.08.035
Westbrook, J. K., Nagoshi, R. N., Meagher, R. L., Fleischer, S. J., Jairam, S. (2016). Modeling seasonal migration of fall armyworm moths. Int. J. Biometeorol. 60 (2), 255–267. doi: 10.1007/s00484-015-1022-x
Xu, S., Jiang, L., Qiao, G., Chen, J. (2020). The bacterial flora associated with the polyphagous aphid Aphis gossypii glover (Hemiptera: Aphididae) is strongly affected by host plants. Microb. Ecol. 79 (4), 971–984. doi: 10.1007/s00248-019-01435-2
Xu, S., Jiang, L., Qiao, G., Chen, J. (2021a). Diversity of bacterial symbionts associated with Myzus persicae (Sulzer) (Hemiptera: Aphididae: Aphidinae) revealed by 16s rRNA illumina sequencing. Microb. Ecol. 81 (3), 784–794. doi: 10.1007/s00248-020-01622-6
Xu, T. T., Chen, J., Jiang, L. Y., Qiao, G. X. (2021b). Diversity of bacteria associated with hormaphidinae aphids (Hemiptera: Aphididae). Insect Sci. 28 (1), 165–179. doi: 10.1111/1744-7917.12746
Zhou, X. Y., Wu, Q. L., JIA, H. R., Wu, K. M. (2021). Searchlight trapping reveals seasonal cross-ocean migration of fall armyworm over the south China Sea. J. Integr. Agric. 20, 673–684. doi: 10.1016/S2095-3119(20)63588-2
Keywords: wheat aphid, aphid symbionts, insect migration, Sitobion miscanthi, Rhopalosiphum padi, Schizaphis graminum
Citation: Li T, Yang G, Li Q, Jiang Y, Kang D, Fan Z, Gong Z, Lu R, Zhou G, Wu Y and Lu C (2023) Population dynamics of migrant wheat aphids in China’s main wheat production region and their interactions with bacterial symbionts. Front. Plant Sci. 14:1103236. doi: 10.3389/fpls.2023.1103236
Received: 20 November 2022; Accepted: 27 January 2023;
Published: 09 February 2023.
Edited by:
Julian Chen, Institute of Plant Protection (CAAS), ChinaReviewed by:
Xiangfeng Jing, Northwest A&F University, ChinaXun Zhu, Institute of Plant Protection (CAAS), China
Copyright © 2023 Li, Yang, Li, Jiang, Kang, Fan, Gong, Lu, Zhou, Wu and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuqing Wu, eXVxaW5nd3UzNkBob3RtYWlsLmNvbQ==; Chuantao Lu, Y2h1YW50YW9sdUBxcS5jb20=
 Tong Li
Tong Li Gongqiang Yang1
Gongqiang Yang1 Qian Li
Qian Li Zhongjun Gong
Zhongjun Gong Guotao Zhou
Guotao Zhou