
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 20 January 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1102200
This article is part of the Research TopicPeanut Genomics and Biotechnology in Breeding ApplicationsView all 11 articles
Abiotic stresses such as cold, drought and salinity are the key environmental factors that limit the yield and quality of oil crop peanut. Phospholipase Ds (PLDs) are crucial hydrolyzing enzymes involved in lipid mediated signaling and have valuable functions in plant growth, development and stress tolerance. Here, 22, 22 and 46 PLD genes were identified in Arachis duranensis, Arachis ipaensis and Arachis hypogaea, respectively, and divided into α, β, γ, δ, ε, ζ and φ isoforms. Phylogenetic relationships, structural domains and molecular evolution proved the conservation of PLDs between allotetraploid peanut and its diploid progenitors. Almost each A. hypogaea PLD except for AhPLDα6B had a corresponding homolog in A. duranensis and A. ipaensis genomes. The expansion of Arachis PLD gene families were mainly attributed to segmental and tandem duplications under strong purifying selection. Functionally, the most proteins interacting with AhPLDs were crucial components of lipid metabolic pathways, in which ahy-miR3510, ahy-miR3513-3p and ahy-miR3516 might be hub regulators. Furthermore, plenty of cis-regulatory elements involved in plant growth and development, hormones and stress responses were identified. The tissue-specific transcription profiling revealed the broad and unique expression patterns of AhPLDs in various developmental stages. The qRT-PCR analysis indicated that most AhPLDs could be induced by specific or multiple abiotic stresses. Especially, AhPLDα3A, AhPLDα5A, AhPLDβ1A, AhPLDβ2A and AhPLDδ4A were highly up-regulated under all three abiotic stresses, whereas AhPLDα9A was neither expressed in 22 peanut tissues nor induced by any abiotic stresses. This genome-wide study provides a systematic analysis of the Arachis PLD gene families and valuable information for further functional study of candidate AhPLDs in peanut growth and abiotic stress responses.
Cultivated peanut (Arachis hypogaea L.) is one of the most important grain legumes worldwide, ranking second in production among all grain legumes and fifth among oilseeds. China contributes the highest share and India ranks second by 36.48% and 13.97% in world production, respectively (United States Department of Agriculture (USDA) Foreign Agricultural Service, 2020). Peanut seeds, containing 40%-56% oil, 20%-30% protein and 10%-20% carbohydrates, have been primarily used to provide vegetable oil and proteins for human nutrition (Huang et al., 2015). In some Third World countries, peanut shows greater potential to reduce hunger and malnutrition as it is also a good source of quality fodder, calories, vitamins, minerals, and other antioxidant molecules (Krishna et al., 2015). However, majority of the world’s peanut is often grown on marginal soils with lesser inputs or intercropped with cereals in many developing countries. A huge gap is developed between its demand and supply due to the constrained quality and productivity resulting from various abiotic factors such as drought, salinity and temperature aberrations (Zhang et al., 2020; Shi et al., 2021). Thus, identification of key genes that can confer abiotic stress tolerance and can be utilized in biotechnological programs to generate improved varieties is an urgent requirement in peanut production (Raza et al., 2022a; Raza et al., 2022b).
One of the most crucial signaling networks for plants in response to multiple stimuli is mediated by lipid molecules (Rizwan et al., 2022a). Environmental cues can increase the activities of phospholipases and trigger the hydrolysis of membrane phospholipids, thus leading to the generation of different classes of lipids and lipid-derived signal messengers (Hou et al., 2016). Phospholipase D (PLD) represents a major family of membrane phospholipases in plants. It acts upon and cleaves the terminal phosphodiester bond of glycerophospholipids to produce phosphatidic acid (PA) and water-soluble free head group (Wang et al., 2012). PLD was first identified in plants as early as in 1940s (Hanahan and Chaikoff, 1947), but did not receive detailed attention until the 1980s (Bocckino et al., 1987). In the past two decades, the disclosure of numerous genomic resources facilitates the characterization of plant PLDs at a genome-wide level. The PLD gene families have been identified in Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), soybean (Glycine max), cotton (Gossypium spp.), rape (Brassica napus L.), chickpea (Cicer arietinum) and other plants successively (Qin and Wang, 2002; Li et al., 2007; Liu et al., 2010; Zhao et al., 2012; Tang et al., 2016a; Lu et al., 2019; Sagar et al., 2021). Commonly, all plant PLDs contain two conserved HKD domains (HxKxxxxD) responsible for hydrolysis activity and can be divided into three sub-classes based on the presence of different domains near the N-terminus: the PLDs with a calcium/phospholipid-binding C2 domain belong to C2-PLD subclass; the PLDs with phox homology (PX) or pleckstrin homology (PH) domains belong to PX/PH-PLD subclass; the PLDs possessing a signal peptide (SP) in place of the usual C2 or PX/PH domains belong to SP-PLD subclass (Tang et al., 2016b). According to sequence characteristics and biochemical properties, the PLD members can be further subdivided into different isoforms, including PLDαs, PLDβs, PLDγs, PLDδs, PLDεs, PLDζs, and/or PLDφs. The isoforms α, β, γ, δ and ε are part of C2-PLDs; ζ and φ isoforms are attached to PX/PH-PLDs and SP-PLDs, respectively (Yao et al., 2021).
The different PLD isoforms are found to have specific subcellular localizations, lipid selectivity and reaction requirements, which lead to their unique cytological and biological functions in particular signaling pathways (Wang, 2005). In Arabidopsis, the phenotypic changes caused by the absence of one PLD member cannot be offset by the other 11 PLDs (Hong et al., 2016). PLDα1 is the predominant PLD and has been proved to regulate drought and salt tolerance by stimulating the accumulation of abscisic acid (ABA) and jasmonic acid (JA) (Li et al., 2009). The repressed expression of PLDα1 may result in a reduced sensitivity to ABA and drought-induced stomatal closure (Sang et al., 2001). The PLDα1 knock-out mutants display increased sensitivities to salinity and water deficiency and tend to induce ABA-responsive genes more readily, whereas the overexpression of PLDα1 have decreased sensitivities (Bargmann et al., 2009). PLDδ is a signal enzyme that can connect microtubules with plasma membrane. The expression levels of PLDδ significantly increase under dehydration, salinity, and ABA treatments (Angelini et al., 2018). The knockout of PLDδ makes plants sensitive to freezing, heat and oxidative stresses (Li et al., 2004; Liu et al., 2021; Song et al., 2021). PLDζs are similar to animal PLDs in structure and have unique functions in root growth. PLDζ2-derived PA can promote root hair development under phosphorus deficiency by suppressing the vacuolar degradation of auxin efflux carrier PIN-FORMED2 (Lin et al., 2020). PLDζ1 shows crucial roles in both ionic and osmotic stress-induced auxin carrier dynamics during salt stress (Korver et al., 2020). Overall, most of the knowledge about PLDs and PLD-mediated lipid signaling has been revealed from studies on model plant Arabidopsis and some other crops. But the information about peanut PLDs and their roles in regulating developmental features and abiotic stresses yet need to be studied.
Cultivated peanut is a classic natural allotetraploid (AABB, 2n=4x=40). It arose from the interspecific hybridization and subsequent chromosome doubling of two diploid species Arachis duranensis (AA, 2n=2x=20) and Arachis ipaensis (BB, 2n=2x=20). Recently, the genomes of A. duranensis, A. ipaensis and A. hypogaea have been completely sequenced, which open a new chapter of Arachis genomic studies (Bertioli et al., 2016; Bertioli et al., 2019). In present study, we identified 90 PLD genes in these three Arachis species and evaluated their sequence characteristics, gene structures, conserved domains, phylogenetic relationships, expansion patterns, physiochemical properties of proteins, cis-regulatory elements, protein-protein interactions, miRNA-genes regulatory networks and expression profiles in different tissues and multiple abiotic stresses. These fundings may provide a comprehensive characterization of Arachis PLDs and lay a theoretical basis for further functional analysis of PLDs in regulating abiotic stress tolerance in peanut.
The genome data (version 1.0) of A. duranensis, A. ipaensis and A. hypogaea were downloaded from the PeanutBase database (https://www.peanutbase.org/peanut_genome). The protein and nucleotide sequences of PLDs in Arabidopsis (TAIR10), soybean (version 2.1) and cotton (version 2.0) were retrieved from the Ensemble database (http://plants.ensembl.org/index.html) and used as queries to perform BLASTP and BLASTN searches against the Arachis genome data. The Hidden Markov Model (HMM) profiles for PLDs (PF00614) were obtained from the Pfam (https://pfam.xfam.org/) and used to perform HMMER searches in the Arachis proteome database. The protein sequences identified by both above methods were integrated and parsed by manual editing to remove the redundant. The remaining were considered as candidate Arachis PLD proteins and finally submitted to the SMART (http://smart.embl-heidelberg.de/) and InterProScan (http://www.ebi.ac.uk/interpro/) to analyze the presence of characteristic and functional domains. The protein size (aa), molecular weight (Mw), theoretical isoelectric point (pI), instability index (II), aliphatic index (AI) and grand average of hydropathicity (GRAVY) of Arachis PLD proteins were calculated by the ExPASy (https://web.expasy.org/protparam/). The subcellular localization was predicted using the Plant-mPLoc server (http://www.csbio.sjtu.edu.cn/).
The multiple sequence alignment of non-redundant PLD protein sequences in A. duranensis, A. ipaensis, A. hypogaea, Arabidopsis, soybean and cotton was performed using the Clustal W with the default settings (Thompson et al., 1994). A phylogenetic tree was constructed using the neighbor joining (NJ) method in MEGA7 software with following parameters: P-distance, pairwise gap deletion and bootstrap with 1000 replicates (Kumar et al., 2016).
The information regarding the detailed positions of Arachis PLDs on chromosomes were obtained from the Arachis genome database and visualized using the MapChart 2.32 software (Voorrips, 2002). The nomenclature of Arachis PLD genes was based on the results of phylogenetic analysis and chromosomal localization. Identified PLD genes in A. duranensis, A. ipaensis and A. hypogaea were named as AdPLD, AiPLD and AhPLD followed by roman letters, numbers and capital letters corresponding to their respective orthologs and chromosomal coordinates.
The exon-intron organization of Arachis PLD genes was displayed using the Gene Structure Display Server (GSDS2.0) (http://gsds.cbi.pku.edu.cn/) by comparing their coding sequences (CDS) and corresponding genomic sequences. The conserved domains of Arachis PLD proteins were identify using the Pfam and SMART tools with the default cut off parameters. The conserved motifs of Arachis PLD proteins were investigated by the online tool Multiple Expectation maximization for Motif Elicitation (MEME) (https://meme-suite.org/meme/tools/meme) with following parameters: the maximum motif number was 30; the minimum motif width was 6; and the maximum motif width was 50. The identified protein motifs were further annotated with InterProScan.
The homologous PLD gene pairs were identified based on the results of chromosomal localization, multiple sequence alignments and phylogenetic analysis. The adopted criteria for gene duplication events were that the shared aligned sequence covered over 70% of the longer sequence and the minimum similarity of aligned regions was 70% (Tang et al., 2016b). The tandem duplications have been characterized as multiple members of one gene family occurring within the same or neighboring intergenic regions. The segmental duplications have been defined as homologous genes that result from large-scale events, such as whole genome duplications or large chromosomal region duplications. The duplication events and collinear relationships of Arachis PLDs were analyzed using the MCScanX and MCScanX-transposed toolkits (Wang et al., 2013) and visualized by the Circos software (Krzywinski et al., 2009).
The non-synonymous substitution rate (Ka) and synonymous substitution rate (Ks) of the duplicated gene pairs were calculated to assess the molecular selection effect using the KaKs_Calculator 2.0 software (Wang et al., 2010). The gene pairs with the Ka and Ks value of 0 as well as the Ks value more than 2 were discarded, as they might result from the sequence saturation or misalignment. The Ka/Ks ratio was then calculated to show the selection pressure for duplicated PLD genes. The Ka/Ks ratio >1, <1 or =1 represented positive, negative (purifying selection) and neutral evolution, respectively. The divergence time of AhPLD gene pairs was estimated by the formula T= Ks/2r, where r indicates the neutral substitution rate (r = 8.12 × 10-9 Ks yr−1) (Bertioli et al., 2016).
The 2000 base pair (bp) DNA sequences in upstream regions of AhPLDs were extracted from the peanut genome database and submitted to the PlantCare (http://bioinformatice.psb.ugent.be/webtools/plantcare/) for the identification of putative cis-regulatory elements. The results were finally visualized by the TBtools software (Chen et al., 2020).
The STRING database (https://string-db.org/) was used to analyze the interaction relationships between AhPLDs and other proteins with the confidence parameter set at the threshold of 0.4. The well-characterized plant Arabidopsis was as the query organism. The predicted protein-protein interaction (PPI) network was displayed by the Cytoscape 3.7.2 software (Shannon et al., 2003).
The targeting relationship between AhPLDs and microRNA (miRNA) were predicated using the psRNATarget Server (http://plantgrn.noble.org/psRNATarget/) with the expectation score of 3.5 (Rizwan et al., 2022b). The peanut miRNA sequences were obtained from the miRbase database (http://www.mirbase.org/). The cDNA sequences of AhPLDs were extracted as the candidate targets. The linkage of the predicted miRNAs and corresponding target genes were displayed by the Cytoscape 3.7.2 software (Shannon et al., 2003).
The RNA-sequencing (RNA-seq) data of 22 peanut tissues at vegetative, reproductive and seed development stages were retrieved from the National Center for Biotechnology Information BioProject (https://www.ncbi.nlm.nih.gov/bioproject/) with the accession number PRJNA291488. The fragments per kilobase of exon model per million mapped reads (FPKM) method in Cufflinks software (http://cufflnks.cbcb.umd.edu/) were used to calculate the transcript abundance. The log2FPKM values were displayed in the form of heatmaps by the HemI software (Deng et al., 2014).
The allotetraploid peanut (A. hypogaea cultivar ‘Nonghua 5’) planted in large areas of northeast China was used as experimental materials in this study. The seeds were surface sterilized with 3% sodium hypochlorite, and washed five times with distilled water, and kept in dark to germinate. The germinated seeds were sown in round plastic pots filled with clean sandy soil and grown in a climate chamber with a 16 h light (28 °C)/8 h dark (23 °C) cycle, a photosynthetic photon flux density of 400 µmol m−2 s−1, and a relative humidity of 70%. After 14 days, the three-leaf seedlings were transferred from sandy soil into hydroponic cultures and grown for 3 days to recover before initiating any stress treatments. For drought and salt stresses, seedlings were incubated in 20% (w/v) polyethylene glycol (PEG-6000) and 250mM NaCl solution, respectively. For cold stress, the temperature of the climate chamber was reduced to 6°C without changing other growth conditions. The second leaves were collected at 0, 6, 12, 24 and 48 h of each treatment with three biological replicates, and frozen in liquid nitrogen immediately and stored at -80°C.
The total RNA for each sample was extracted by TRIzol reagent (Carlsbad, CA, USA) according to the manufacturer’s protocol. The RNA concentration was tested using a micro-spectrophotometer (OD260/280). The RNA integrity was tested using the Agilent Bioanalyzer 2100 system. 1 μg of total RNA was used to synthesize first strand cDNA by Takara Reverse Transcription System (TaKaRa, Shuzo, Otsu, Japan). The expression profiles of AhPLDs under various abiotic stresses was detected by quantitative real-time PCR (qRT-PCR) using the specific primers as listed in Table S1. The real-time quantification was performed with SYBR Premix Ex Taq™ (TaKaRa, Shuzo, Otsu, Japan) according to the manufacturer’s instructions. PCR mixtures (10 µL) contained 1.0 µL cDNA, 0.3 µL each primer, 3.4 µL ddH2O and 5.0 µL SYBRfi Green Master Mix. The amplification conditions were as follows: 60 s denaturation at 95°C, followed by 40 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 60 s. Three biological replicates per sample and three technical replicates per biological replicate were used for the analysis. The AhACT11 was served as the internal reference gene, and the relative expression values were calculated using the 2-ΔΔct method (Chi et al., 2012).
Using the method as described above, a total of 22 AdPLDs, 22 AiPLDs and 46 AhPLDs were identified in A. duranensis, A. ipaensis and A. hypogaea genomes, respectively (Table S2). Chromosome localization found that the identified AdPLDs and AiPLDs were unevenly distributed in nine out of ten chromosomes of A. duranensis (A genome) and A. ipaensis (B genome), respectively (Figure 1A). Among them, A01 and A08 chromosomes possessed the most abundant PLDs with each containing five AdPLDs, followed by B01, B08 and B09 chromosomes with each containing four AiPLDs, but no PLDs was found on chromosomes A06 and B06. In A. hypogaea, 46 AhPLDs were located on 18 out of 20 chromosomes (Figure 1B), of which chromosomes 01, 08, 11 and 18 contained the most AhPLDs but chromosomes 06 and 16 had no AhPLD.
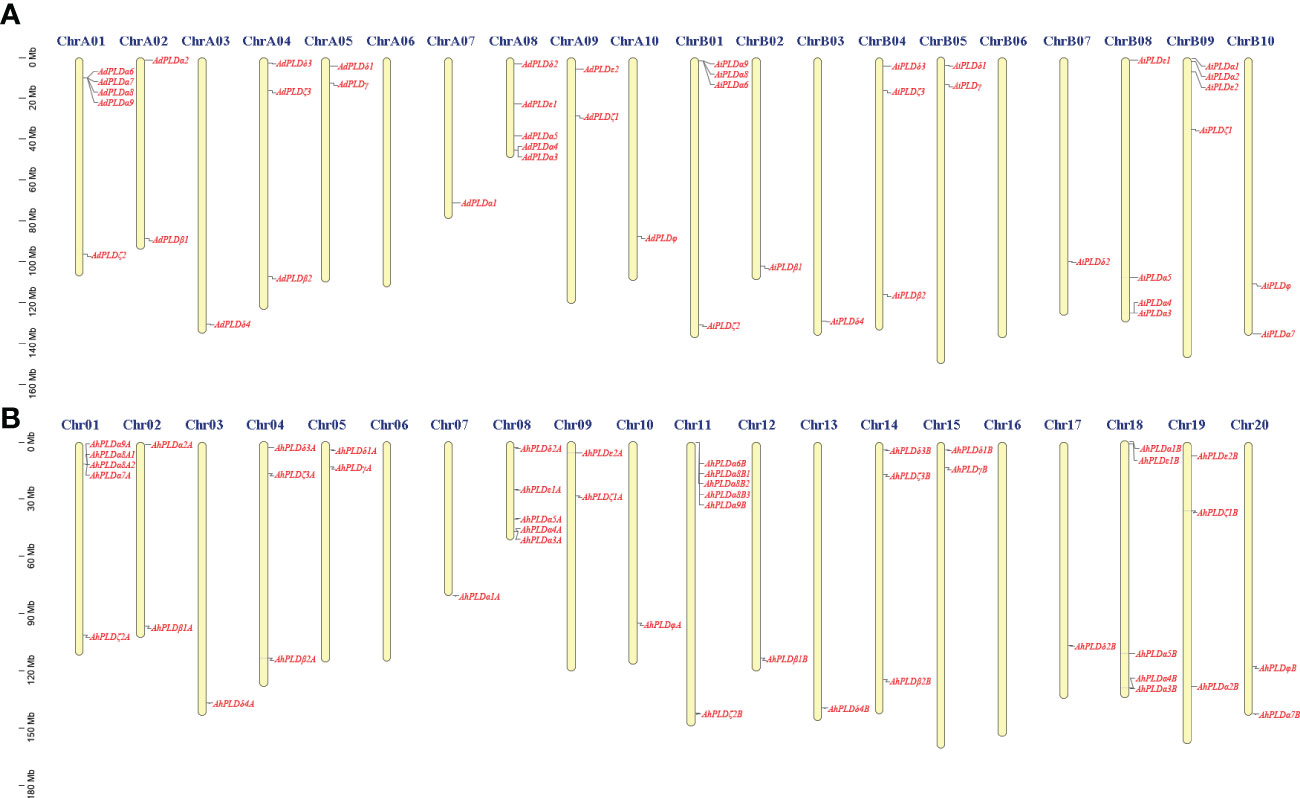
Figure 1 Chromosomal distribution of Arachis PLD genes. (A) Chromosomal distribution of PLD genes in A. duranensis and A. ipaensis. (B) Chromosomal distribution of PLD genes in A. hypogaea. Yellow color bars represent the chromosomes, and the location of PLD genes has been marked alongside.
Furthermore, the sequence characteristics of PLD proteins in three Arachis species were analyzed (Table 1). The opening reading frame (ORF) lengths of all the 90 PLDs ranged from 1305 bp to 3417 bp, which encoded polypeptides of 507 aa to 1138 aa with predicted MWs ranging from 57.9 kD to 126.94 kD and theoretical pIs ranging from 5.16 to 8.44. According to the II values, 49 Arachis PLDs were considered as the stable proteins (II<40), and 41 Arachis PLDs belonged to the unstable proteins (II>40). Besides, all Arachis PLD proteins had high AI values from 69.47 to 86.41 but minus GRAVY values from -0.179 to -0.756, indicating they were stable over a wide temperature range and were hydrophilic and highly soluble in water. Subcellular localization prediction revealed that most Arachis PLD proteins were in the cytoplasm, endoplasmic reticulum and vacuole, only a few were in the chloroplast, nucleus and plasma membrane.
To explore the phylogenetic relationships and evolutionary patterns of the PLDs in peanut and its two progenitors, a neighbor-joining tree was constructed using the full-length protein sequence alignments of 90 Arachis PLDs, 12 AtPLDs, 23 GmPLDs and 20 GrPLDs. All the PLDs from different plant species were clearly divided into seven well-supported sub-clades based on the similarity with cotton PLDs, comprising α, β, γ, δ, ε, ζ and φ isoforms (Figure 2). Among them, the α constituted the largest clade containing 38 Arachis PLDs (20 AhPLDαs, nine AdPLDαs and AiPLDαs), the δ formed the second largest clade containing 16 Arachis PLDs (eight AhPLDδs, four AdPLDδs and AiPLDδs), and the φ was the smallest subgroup containing only four Arachis PLDs (two AhPLDφs, one AdPLDφs and AiPLDφs). Besides, the β and γ showed closely together and were not explicitly separated from each other. Within the separate clades, isoforms ε and φ were absent in Arabidopsis, isoforms γ and φ were absent in soybean, but all seven isoforms were present in Arachis and cotton. Obviously, Arachis PLDs showed closer to GmPLDs but more distant from AtPLDs. According to the phylogenetic relationships with orthologs in soybean, cotton and Arabidopsis, as well as the physical location on chromosomes, 22 AdPLDs and 22 AiPLDs were renamed as Ad/AiPLDα1-9, Ad/AiPLDβ1, Ad/AiPLDβ2, Ad/AiPLDγ, Ad/AiPLDδ1-4, Ad/AiPLDε1, Ad/AiPLDε2, Ad/AiPLDζ1-3 and Ad/AiPLDφ. The 46 AhPLDs were renamed as AhPLDα1A/1B-5A/5B, AhPLDα6B, AhPLDα7A/7B, AhPLDα8A1/8A2/8B1/8B2/8B3, AhPLDα9A/9B, AhPLDβ1A/1B, AhPLDβ2A/2B, AhPLDγA/B, AhPLDδ1A/1B-4A/4B, AhPLDε1A/1B, AhPLDε2A/2B, AhPLDζ1A/1B- 3A/3B and AhPLDφA/B.
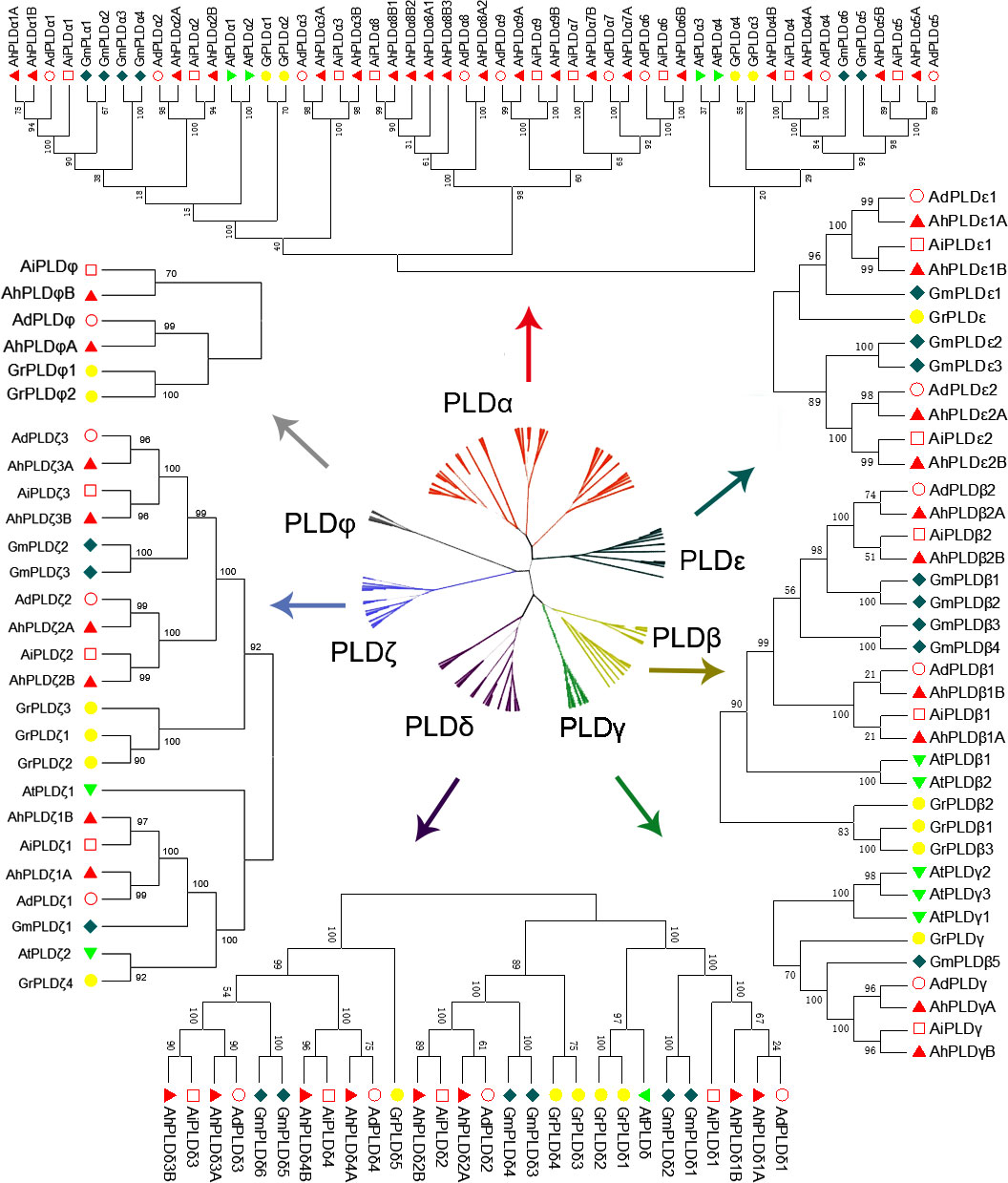
Figure 2 Phylogenetic tree of PLD gene family with bootstrap values among three Arachis species, Arabidopsis, soybean and cotton. The red, yellow, green, purple, dark green, blue and gray branches represent α, β, γ, δ, ε, ζ and ϕ isoforms, respectively. The red circles, red squares, red triangles, green triangles, dark green diamonds and yellow circles represent the proteins of A. duranensis (Ad), A. ipaensis (Ai), A. hypogaea (Ah), Arabidopsis (At), soybean (Gm) and cotton (Gr), respectively.
To confirm the authenticity and integrity of identified Arachis PLD genes, their functional domains were analyzed by searching the Pfam and SMART databases. As expected, all Arachis PLD proteins possessed two structurally conserved HKD domains at C-terminal and were divided into three subgroups (Figure 3A). Isoforms α, β, γ, δ and ε contained C2 domain at the N-terminal and belonged to the C2-PLDs. PLDζs contained PX and/or PH domain at the N-terminal and belonged to the PX/PH-PLDs. PLDφs contained signal peptide at the N-terminal instead of C2 or PX/PH domain and belonged to the SP-PLDs. Furthermore, the distribution of exons and introns within each PLD protein was also analyzed. As shown in Figure 3B, all the 90 Arachis PLDs were comprised of multiple exons and introns. The number of introns determined for Arachis PLDs ranged from one in AdPLDα6, AdPLDα9, AiPLDα9 and AhPLDα9B to 20 in AiPLDζ1. The members of all Arachis PLD isoforms showed similar exon-intron structure withing their respective subgroups. It was consistent with the phylogenetic classification depicted in the left panel of Figure 3A. For instance, both α and ε subgroups included members with one to five introns, the members in β/γ and δ subgroups possessed eight or nine introns (except AdPLDδ2 that had six introns), the members in ζ subgroup contained 15 to 20 introns, and all six members in φ subgroup had six introns.
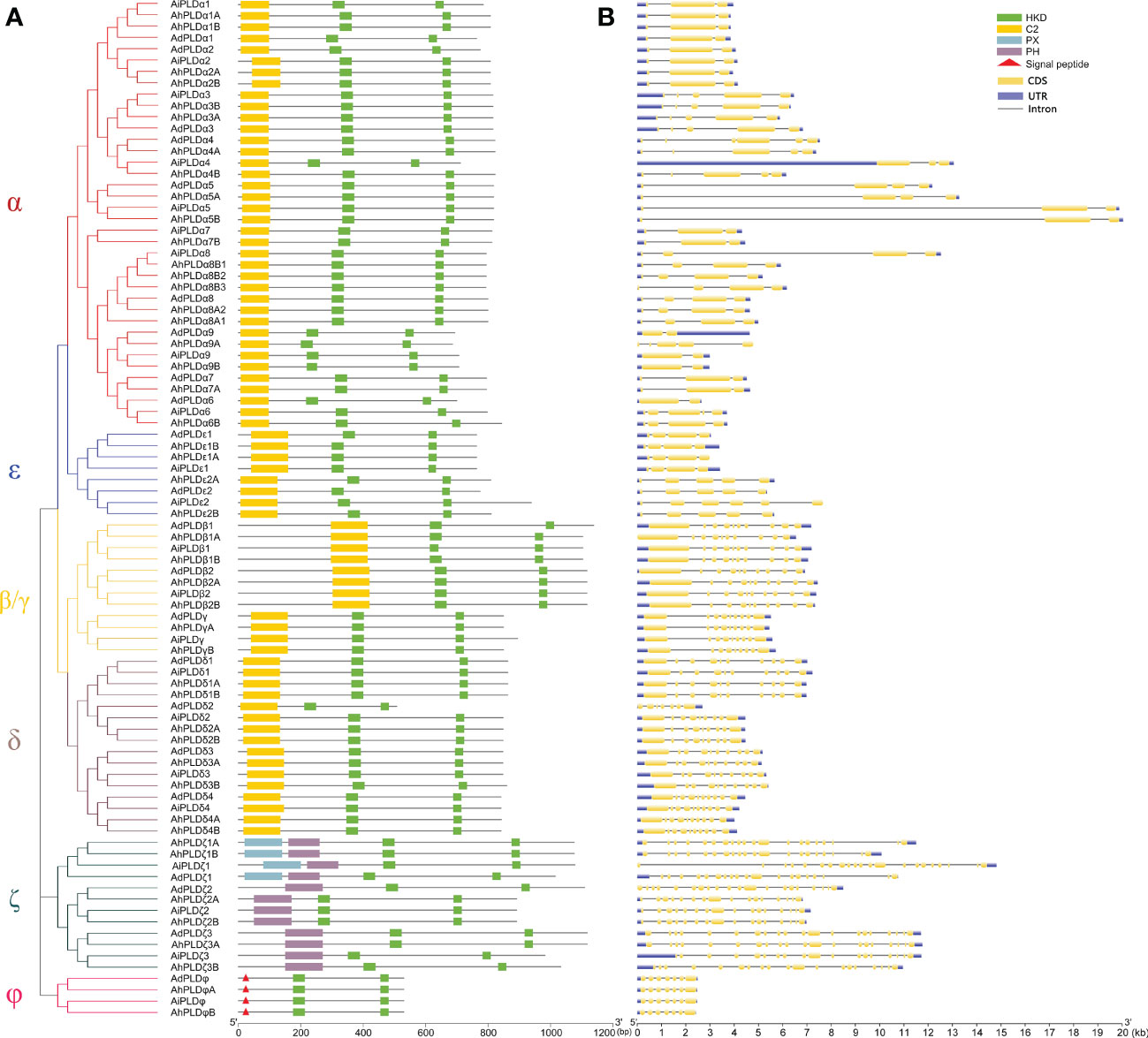
Figure 3 Phylogenetic relationship, conserved domains and gene structure of PLD genes in peanut and its two progenitors. (A) Schematic representation of the phylogenetic tree and conserved domain of PLD genes in three Arachis species. The red, blue, yellow, purple, dark green and pink branches on phylogenetic tree represent the α, ε, β/γ, δ, ζ and ϕ isoforms, respectively. The yellow, green, blue and purple rectangles represent the C2, HKD, PX and PH domains, respectively. The red triangles represent the signal peptides. (B) Exon-intron structure of PLD genes in three Arachis species. The blue boxes represent exons. The yellow boxes represent coding sequences. The black lines represent introns.
To reveal the typical domain characteristics of Arachis PLD subgroups, the conservation of amino acid residues in functional domains were analyzed based on the alignments of these PLDs. A total of 30 distinct motifs designated as Motif 1 to Motif 30 were identified (Figures 4, S1; Table S3). The members within the same subfamilies were usually shared similar motif composition, for example, C2-PLDs possessed 17 to 25 motifs, of which Motif 1-11, Motif 13, Motif 15 and Motif 25 were highly conserved; PX/PH-PLDs possessed 18 motifs, including Motif 1-9, Motif 13, Motif 15, Motif 19, Motif 24 and Motif 26-30; whereas SP-PLDs only contained five motifs, including Motif 1, Motif 4, Motif 15, Motif 28 and Motif 29. By comparison, the Motif 26 and Motif 27 were only present in the C-terminus of PX/PH-PLDs but absent in C2-PLDs and SP-PLDs, suggesting these two motifs were specific to the PX and PH domains. Moreover, almost all these motifs could be matched to the annotated motifs in InterProScan and were functionally associated with PLD activity. The Motif 1, Motif 6, Motif 14 and Motif 29 were annotated as the conserved HKD domains (HxKxxxxD), especially the Motif 1 was uniformly observed in all Arachis PLDs. The Motif 4, also observed in all PLD proteins, contained a core triplet of amino acids “ERF” followed by a highly conserved hydrophobic region “VYVVV”. The Motif 2 contained a regular-expression sequence “IYIENQ[F/Y]F”, of which the seventh amino acid, Phenylalanine (F), appeared in all PX/PH-PLDs but was often substituted by the Tyrosine (Y) in C2-PLDs. The Motif 3 contained the conserved amino acid”xxGPRxPWHDxHxxxxGPAxxDVLTNFExRWRKxGx”, which was considered as the binding sites of PIP2.
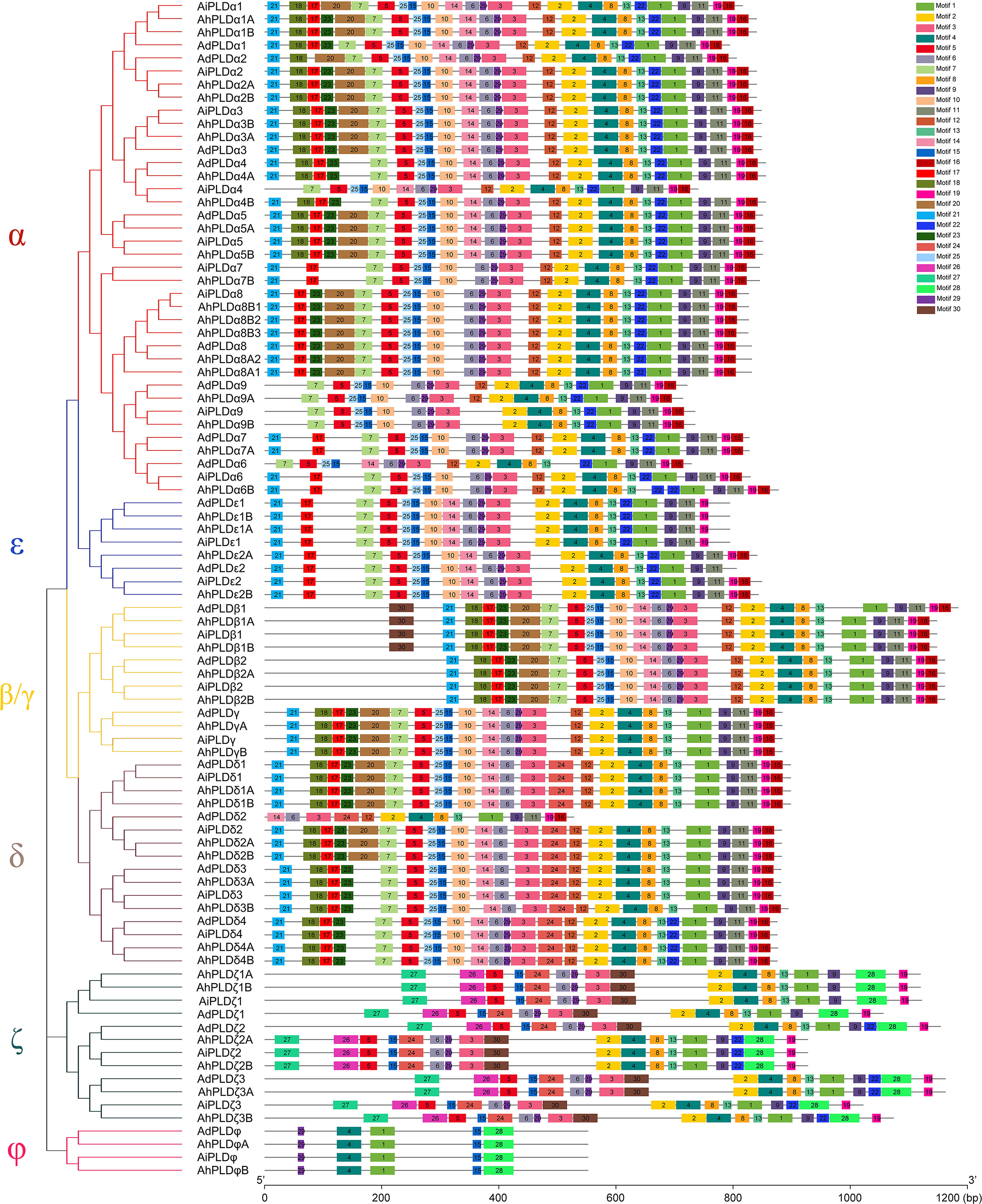
Figure 4 Motif composition of PLD proteins in peanut and its two progenitors. The red, blue, yellow, purple, dark green and pink branches on phylogenetic tree represent the α, ε, β/γ, δ, ζ and ϕ isoforms, respectively. The motifs, numbers 1-30, are displayed in different colored boxes.
To clarify the roles of gene duplications in the expansion of Arachis PLD gene families, the synteny analysis including tandem duplication and whole genome duplication (WGD)/segmental duplication was performed based on the multiple and pairwise alignments of Arachis PLDs. As a result, a total of 131 orthologous/paralogous gene pairs were identified, of which 54 pairs were predicted to form paralogous gene pairs within the A. duranensis, A. ipaensis and A. hypogaea genomes, and nine pairs underwent the tandem duplications (AdPLDα3-AdPLDα4, AdPLDα8-AdPLDα9, AiPLDα1-AiPLDα2, AiPLDα3-AiPLDα4, AiPLDα6-AiPLDα8-AiPLDα9, AhPLDα3A-AhPLDα4A, AhPLDα3B-AhPLDα4B, AhPLDα8A1-AhPLDα8A2-AhPLDα9A, AhPLDα6B-AhPLDα8B1-AhPLDα8B2-AhPLDα8B3-AhPLDα9B) (Figure 5A; Table S4). Besides, 25, 28 and 24 segmental duplications were found in groups AhPLDs-AdPLDs, AhPLDs-AiPLDs and AdPLDs-AiPLDs respectively, but one group only contained AiPLDα6 homoeologous gene (AhPLDα6B), suggesting the corresponding AhPLDα6A had lost during peanut evolution (Figure 5B; Table S4). Consequently, we presumed that the putative gene duplication events were main causes of Arachis PLD gene family expansion, and homoeologous gene pairs were generally raised from tandem or WGD/segmental duplication before polyploidization involved in evolution process.
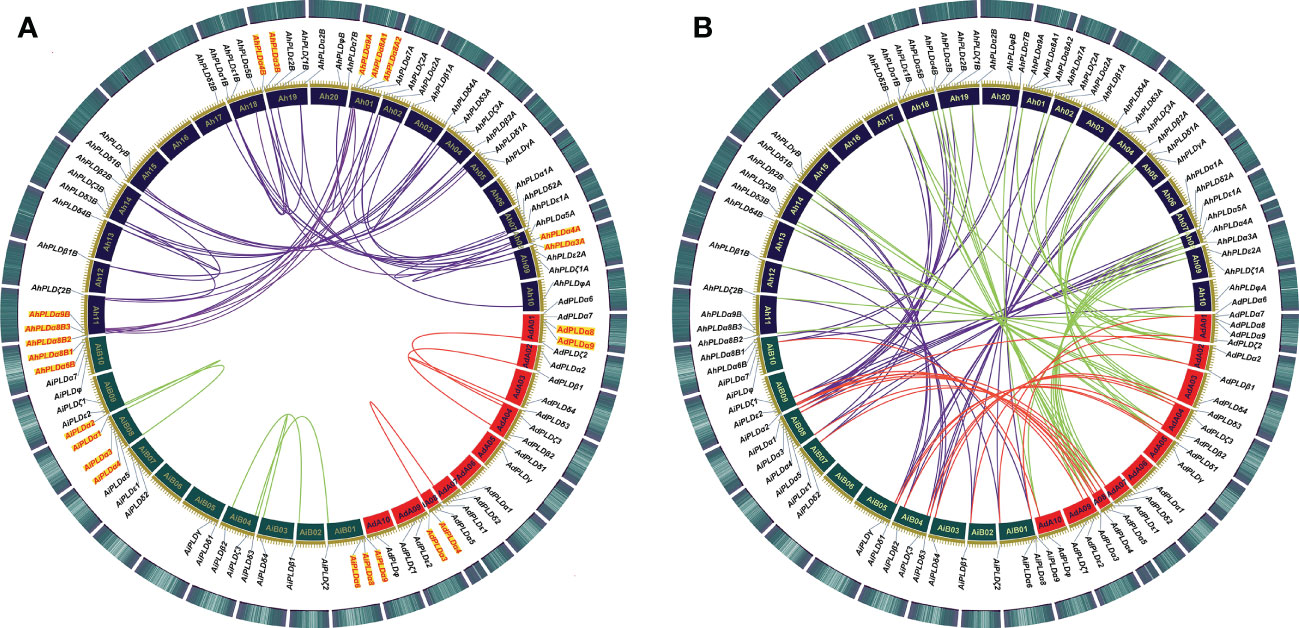
Figure 5 Syntenic relationships among PLD genes in peanut and its two progenitors. (A) Syntenic relationships of PLD genes within A. duranensis, A. ipaensis and A. hypogaea, respectively. (B) Syntenic relationships of PLD genes between A. duranensis, A. ipaensis and A. hypogaea. The chromosomes of A. duranensis, A. ipaensis and A. hypogaea were shown with red, dark green and dark purple colors, respectively. The putative homologous PLD genes are linked by different colored lines. The tandem duplicated PLD genes are marked with red font next to the chromosomes.
To investigate which type of selection pressure had been involved in PLD gene divergence after duplication, the Ka/Ks ratios of duplicated Arachis PLD gene pairs were calculated on the basis of coding sequences (Table S5). The resulting pairwise comparison data showed that most homoeologous gene pairs had Ka/Ks ratios of < 1, indicating these gene pairs might have undergone strong purifying selection pressure with limited functional divergence that occurred after tandem or WGD/segmental duplication. Only three gene pairs (AhPLDα8A2-AdPLDα8, AhPLDδ3B-AiPLDδ3 and AhPLDδ3A-AhPLDδ3B) had Ka/Ks ratios of > 1, suggesting these gene pairs might have experienced relatively rapid evolution following duplication. Meanwhile, according to Ks values, the divergence time of duplicated Arachis PLD gene pairs were estimated. As a result, these tandem and segmental duplications may occur in 3.02-70.62 and 0.10-119.83 Mya, respectively (Table S5).
To better understand the transcriptional regulation and potential functions of peanut PLD genes, the cis-regulatory elements present in promoters (2000 bp of 5’ upstream regions) of AhPLD genes were identified. Totally, 39 functional cis-elements related to growth and development, plant hormones and stress responses were obtained from 46 AhPLDs (Table S6). All AhPLD promoters had variable number of cis-regulatory elements and most of them were present in multiples (Figure 6). Among the 25 growth and development-related elements, light-responsive elements (87.59%), such as Box 4, G-box, GT1-motif, TCT-motif and GATA-motif, accounted for the most and widely distributed in all AhPLD promoters; others mainly included the circadian element (3.28%) related to circadian control, the O2-site (2.57%) related to zein metabolism regulation, the CAT-box (2.00%) related to meristem expression and the GCN4_motif (1.85%) related to endosperm expression.
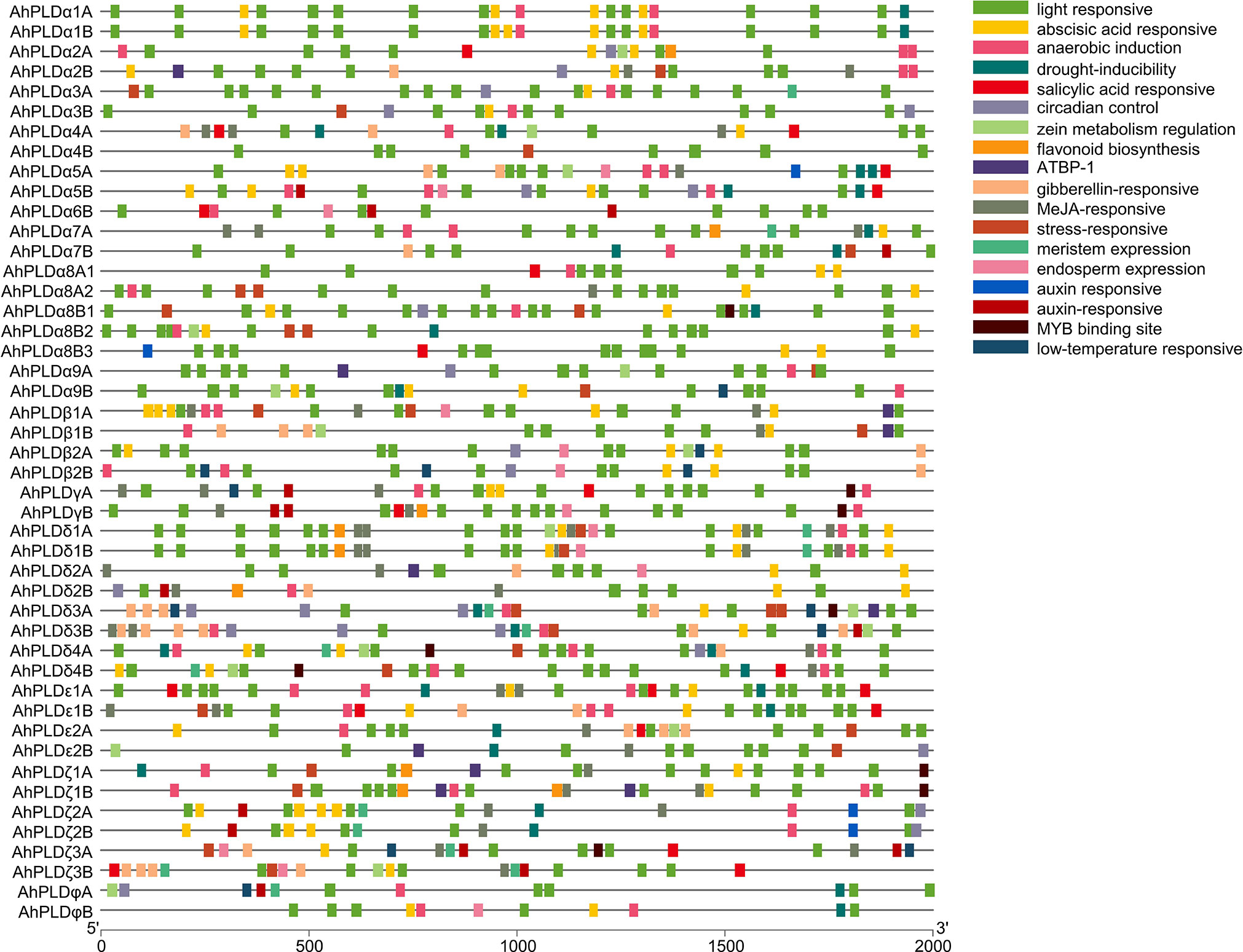
Figure 6 Cis-regulatory elements in the promoters of peanut PLD genes. Various cis-regulatory elements are displayed in different colored boxes.
The nine hormone-responsive cis-elements were related to five plant hormones. Among them, abscisic acid responsiveness element (35.84%, ABRE) was identified in 39 AhPLDs; MeJA-responsive elements (37.99%, CGTCA-motif/TGACG-motif) were identified in 27 AhPLDs; gibberellin-responsive elements (12.19%, GARE-motif/P-box/TATC-box) were identified in 17 AhPLDs; salicylic acid-responsive element (7.17%, TCA-element) was identified in 15 AhPLDs; auxin-responsive elements (6.81%, TGA-element/AuxRR-core) were identified in 14 AhPLDs (Figure 6; Table S6). There were three AhPLDs (AhPLDα5A, AhPLDζ3A and AhPLDζ3B) contained the five hormone-responsive elements above-mentioned, but AhPLDα9A did not contain any hormone-responsive elements. Besides, the promoter regions of eight AhPLDs (AhPLDα1A, AhPLDα1B, AhPLDα3A, AhPLDα3B, AhPLDα8B1, AhPLDα8B2, AhPLDα9B and AhPLDφB) only contained ABA-responsive elements, whereas AhPLDε2B and AhPLDφA only contained MeJA- and auxin- responsive elements, respectively. However, the ethylene-responsive cis-elements were not found in any AhPLD promoters.
Moreover, there were five cis-elements found to be responsive to various stresses, including anaerobic element (42.76%, ARE), MYB binding site involved in drought-inducibility (27.59%, MBS), low temperature-responsive (8.28%, LTR) element and stress-responsive element (21.38%, TC-rich repeats) (Figure 6; Table S6). Except for AhPLDα8B3 and AhPLDδ2A, almost all AhPLDs contained at least one stress-responsive element, indicating AhPLDs played an important role not only in peanut growth and development, but also in stress responses. The simultaneous appearance of stress and hormone related cis-elements in some AhPLDs suggesting that AhPLDs could mediate the cross-talk of stress and hormone signaling pathways and may have potential role in hormone mediated abiotic stress signaling in peanut.
To elucidate the metabolic regulation network mediated by PLDs in peanut and further understand the biological function of AhPLDs, the protein-protein interactions (PPIs) of AhPLDs were analyzed using ortholog-based method. Totally, 24 AhPLDs had orthologous relationships with 12 Arabidopsis PLDs and interacted with 55 functional proteins (Figure 7A; Table S7). As expected, most proteins were the functionally validated components of lipid biosynthetic and lipid metabolic processes, such as amino alcohol-phosphotransferase (AAPT), diacylglycerol O-acyltransferase 1 (DGAT1), diacylglycerol kinases (DGKs), lysophosphatidyl acyltransferase 2 (LPAT2) and triglyceride lipase (TGL4). Some others like phospholipase C1 (PLC1), phosphatidylinositol 3-kinase (PI3K), phosphatidylinositol 4-OH kinase 1 (PI4K1) and phosphatidylinositol-4-phosphate 5-kinase 1 (PIP5K1) mainly participated in the phosphatidylinositol-mediated signaling. Besides, several AhPLDs could interact with phospholipid/glycerol acyltransferase family protein ATS2, G protein alpha subunit 1 (GPA1), lipoxygenase 1 (LOX1) and plasma-membrane choline transporter-like protein CTL1. These proteins may regulate plant root development, pollen tube growth, post-embryonic development, seed development and other system development processes.
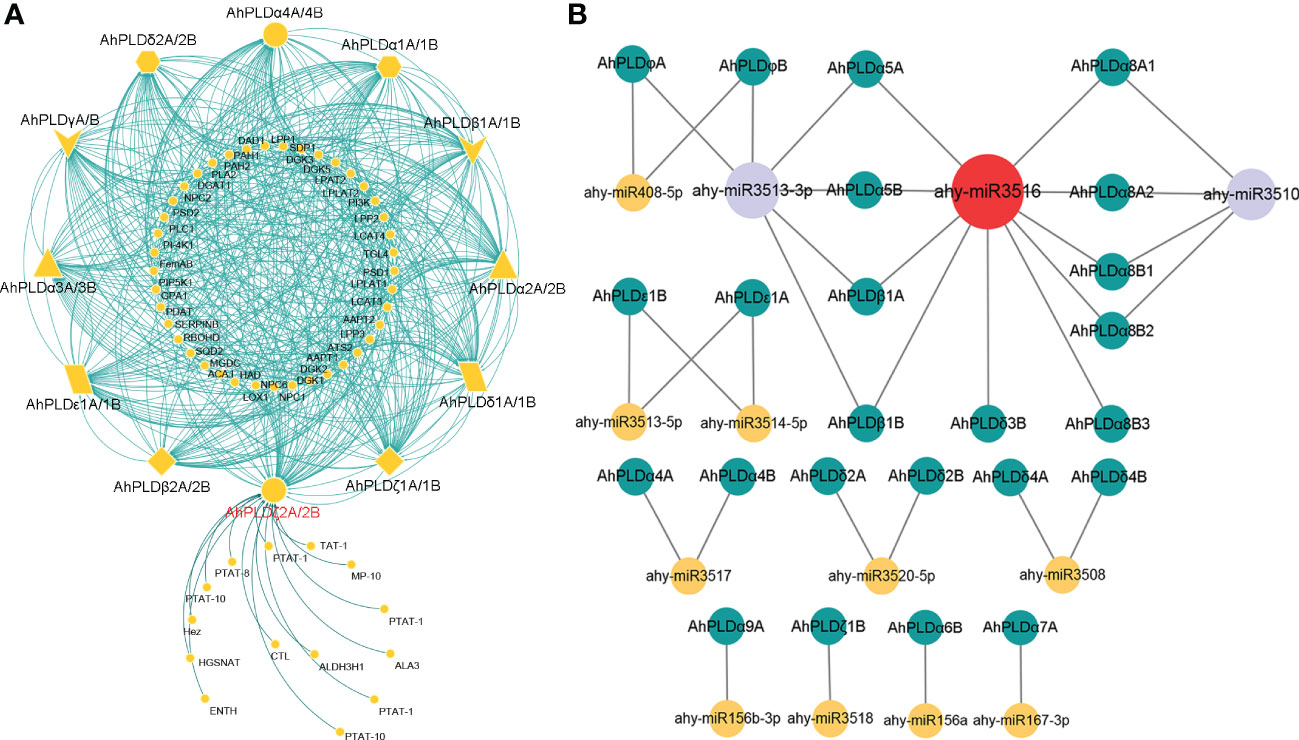
Figure 7 Interactions and regulations of peanut PLD genes with other proteins and miRNAs. (A) Protein-protein interaction network of peanut PLD proteins with other proteins. The yellow geometric figures represent peanut PLD proteins. The little yellow dots represent other proteins that interact with AhPLDs. The interaction relationships are shown by green lines. (B) Regulatory network of putative miRNAs and their targeted AhPLDs. The yellow, purple and red circles represented miRNAs. The green circles represent targeted AhPLDs. The putative regulatory relationships between miRNAs and their targeted AhPLDs are shown in grey lines.
Moreover, the pathways in response to stress or abiotic stimulus were also significantly enriched by abundant proteins that interacted with AhPLDs, for example, there were five proteins (DGAT1, DGK2, PLC1, respiratory burst oxidase homologue D (RBOHD) and phospholipid-transporting ATPase (PTAT-1)) involved in the response to temperature stimulus, four proteins (DGAT1, PI3K, PLC1 and aldehyde dehydrogenase 3H1 (ALDH3H1)) involved in the response to salt stress, four proteins (DGAT1, PI3K, PLC1 and RBOHD) involved in the response to osmotic stress, three proteins (phosphatidate phosphatase PAH1, monogalactosyldiacylglycerol synthase type C (MGDC) and sulfoquinovosyldiacylglycerol 2 (SQD2)) involved in the response to phosphate starvation, and six proteins (four DGKs, phosphatidic acid phosphatase 1 (LPP1) and RBOHD) involved in the defense response. Notably, there were 14 predicted proteins only interacted with AhPLDζ2A/2B and mainly responsible for lipid transport and phospholipid translocation.
Furthermore, the putative miRNA-targeted AhPLDs were predicted to obtain the potential association between lipid metabolic pathways and miRNA regulations. As shown in Figure 7B, a total of 24 AhPLDs were targeted by 13 miRNAs, of which 12 AhPLDs were targeted by only one miRNA, and 12 AhPLDs were targeted by two miRNAs. These miRNAs belonged to 12 different families. The comprehensive data of all miRNAs targeted sites/genes was given in Table S8. Most miRNAs only targeted one or two AhPLDs, but ahy-miR3516, ahy-miR3510 and ahy-miR3513-3p could target ten (AhPLDα5A/5B, AhPLDα8A1/8A2/8B1/8B2/8B3, AhPLDβ1A/1B and AhPLDδ3B), six (AhPLDα5A/5B, AhPLDβ1A/1B and AhPLDφA/B) and four AhPLDs (AhPLDα8A1/8A2/8B1/8B2), respectively, indicating these three miRNAs may be crucial in regulating the lipid metabolic processes.
The tissue-specific expression profiles of AhPLDs were investigated by using the RNA-Seq data from Clevenger et al. (2016). A total of 22 different tissues encompassing almost all peanut tissues and developmental stages were analyzed (Figure 8; Table S9). The results showed that the expression patterns of 46 AhPLDs in various tissues were quite different, though the expression levels of most homologous copies from the A genome and the B genome (such as AhPLDα1A and AhPLDα1B) were similar because of their extremely high similarity of mRNAs and transcript sizes. The AhPLDα1A/1B, AhPLDα7B, AhPLDβ2A/2B, AhPLDγA/B, AhPLDδ1A/1B, AhPLDδ3A/3B, AhPLDζ3A/3B and AhPLDφA/B were expressed in all 22 tissues, especially the AhPLDδ3A/3B exhibited higher expressions in all tissues. Conversely, AhPLDα9A/9B, the members of PLDα subgroup, had weak or even undetected expression in any tissues. Besides, almost all AhPLDs except for AhPLDα9A/9B were highly expressed in all above-ground tissues during the vegetative growth stage, indicating AhPLDs might play essential roles in the growth and development of peanut seedlings. The expression levels of AhPLDεs, AhPLDζs and AhPLDφs in pods and seeds were significantly lower than in leaves, shoots, roots and flowers, suggesting these AhPLDs might mainly involve in peanut seedling growth, root elongation and flowering.
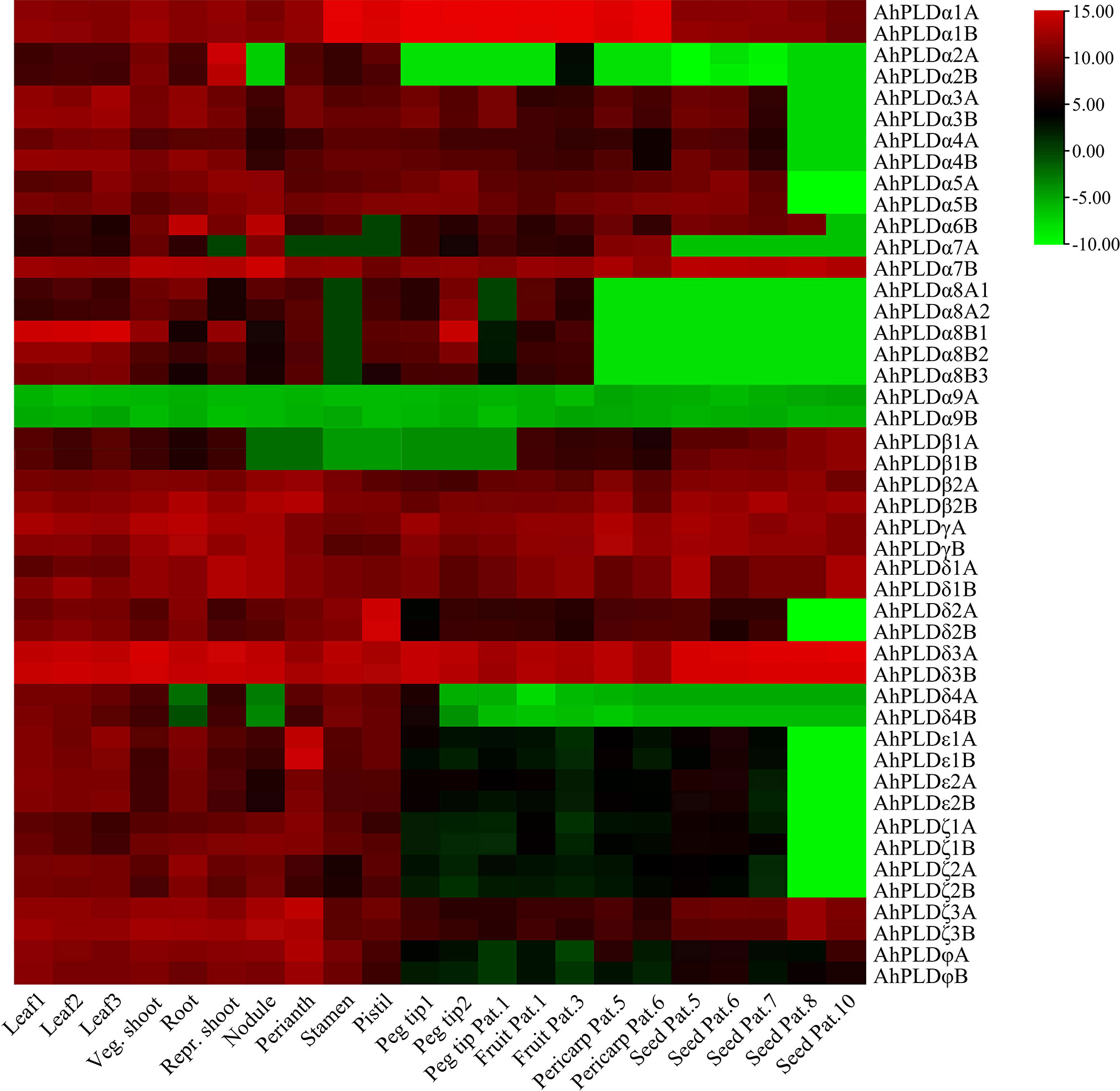
Figure 8 Expression profiles of 46 AhPLDs in 22 different tissues. The X axis represents various tissues. The Y axis represents 46 AhPLDs. The color scale represents Log2 expression values (FPKM).
Moreover, some AhPLDs displayed tissue-specific or preferential expression patterns, for example, the AhPLDα1A/1B showed preferential expressions in flowers and pod development; the AhPLDα2A/2B had higher expressions in shoot at the reproductive growth stage but hardly expressed in pod or seed developmental processes; the AhPLDα6B preferred to express in root and nodule; the AhPLDα7B maintained obvious expression levels in all 22 tissues, but its homologue AhPLDα7A had almost undetected expression in flowers and nearly mature seeds; the AhPLDδ2A/2B and AhPLDε1A/1B showed highest expression levels in pistil and perianth, respectively, indicating their significant roles in the floral organ development in peanut; but only a few AhPLDs were highly expressed in seed Pat. 8 and seed Pat. 10.
To explore the potential involvement of peanut PLD genes in abiotic stresses, their expression profiles under cold, drought and salt were investigated by qRT-PCR (Figure 9; Table S10). Since the paralogous pairs of AhPLDs displayed highly similar nucleotide sequence and expression pattern in 22 different tissues, only AhPLDs from the A subgenome were selected for analysis. Similarly, most AhPLDs within the same phylogenetic subgroup were also similar in nucleotide sequence and expression pattern, so one representative AhPLD in each subclade was chosen for analysis, such as AhPLDα8A1. Besides, given that AhPLDα7B showed distinct expression pattern in 22 different tissues compared with its homologue AhPLDα7A, it was also included. Finally, the expression profiles of 23 AhPLDs under three major abiotic stresses were analyzed. The results in Figure 9 showed that all AhPLDs except for AhPLDα9A could be induced by at least one abiotic stress, of which 13 AhPLDs (AhPLDα3A, AhPLDα5A-7A, AhPLDβ1A, AhPLDβ2A, AhPLDδ1A, AhPLDδ3A, AhPLDδ4A, AhPLDζ1A-3A and AhPLDφA) were found to be induced by all three abiotic stresses commonly, and seven AhPLDs (AhPLDα1A, AhPLDα2A, AhPLDα7B, AhPLDα8A1, AhPLDδ2A, AhPLDε1A and AhPLDε2A) could be induced by both cold and drought stresses, and one AhPLD gene (AhPLDα4A) could be induced by both cold and salt stresses, and AhPLDγA was only induced by cold stress. Surprisingly, almost all AhPLDs induced by cold and drought were up-regulated compared with controls.
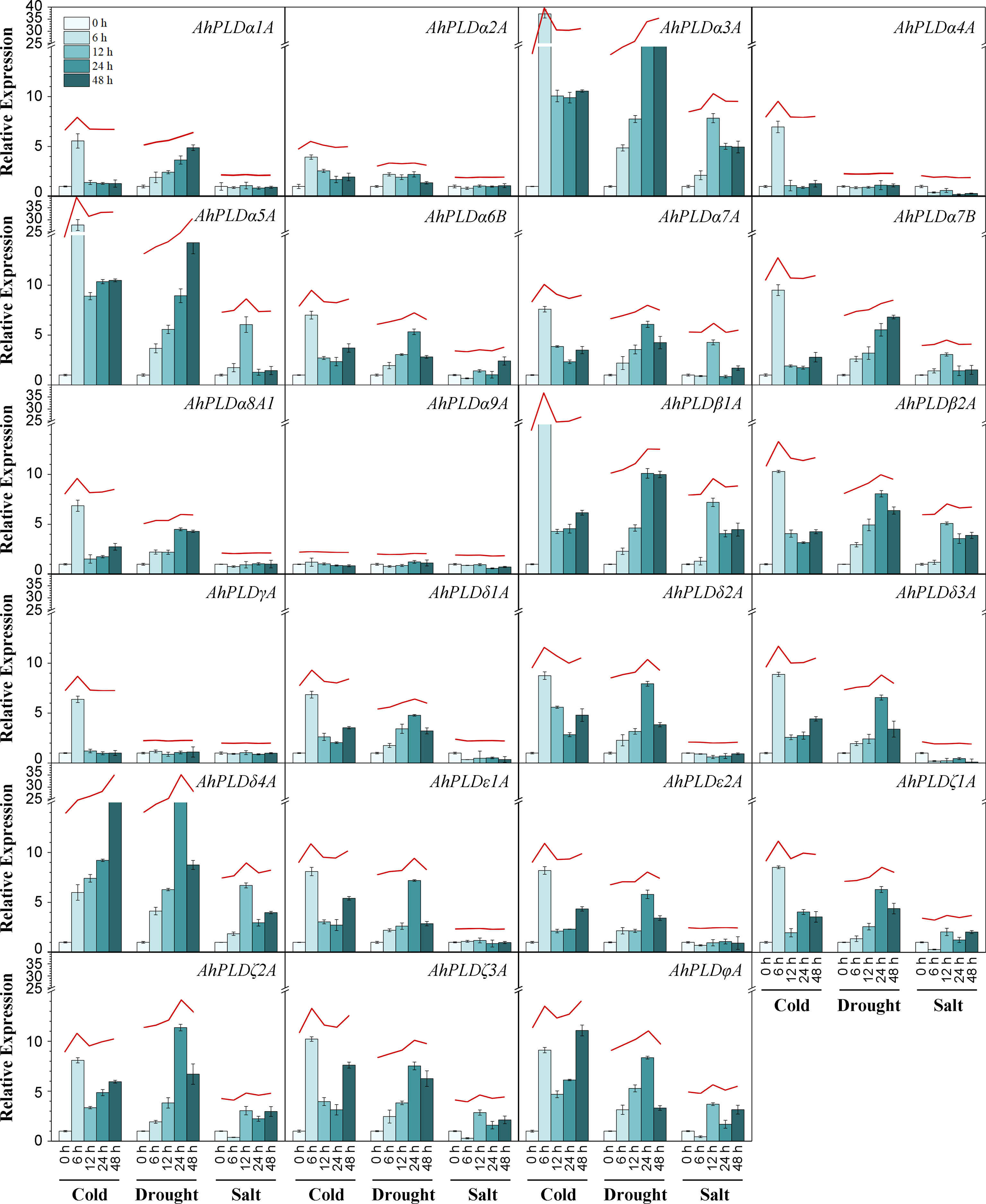
Figure 9 Expression profiles of AhPLDs under abiotic stresses. The expression levels of AhPLDs under cold, drought and salt were generated using qRT-PCR. The X axis represents different treatments. The Y axis represents relative expression levels of AhPLDs. Each bar represents average of three replicates. Standard error is indicated by error bars. The red lines above bars represent the overall trends of AhPLDs expressions under different abiotic stresses.
Under cold stress, the expressions of AhPLDα3A, AhPLDα5A, AhPLDβ1A, AhPLDδ4A, AhPLDζ3A and AhPLDφA were most significantly up-regulated. Among them, AhPLDδ4A showed a continuous increase with the prolongation of cold time, the fold change (FC) at 48 h of cold stress reached 23.48; AhPLDα3A, AhPLDα5A and AhPLDβ1A showed a trend of increasing (0-6 h) first and then decreasing (6-48 h), peaking at 6 h of cold stress with the FC values of 37.19, 27.86 and 16.11, respectively; similarly, AhPLDζ3A and AhPLDφA also showed a trend of increasing first and then decreasing, while their expression level was increased again after 48 h of cold stress. Under drought stress, the expressions of most AhPLDs were significantly up-regulated in the early stage (24 h) of stress condition, but their expressions did not continue to increase with the duration of stress, typically like AhPLDδ4A, AhPLDζ2A and AhPLDφA; while such AhPLDs as AhPLDα3A (19.36 FC), AhPLDα5A (14.25 FC) and AhPLDβ1A (9.99 FC) kept up-regulated expression to 48 h of drought stress. Compared with cold and drought, the transcript levels of AhPLDs under salt tress were lower, some of which were even not differentially expressed or down-regulated. For example, AhPLDα3A (7.84 FC), AhPLDα5A (6.05 FC), AhPLDβ1A (7.20 FC), AhPLDβ2A (5.09 FC) and AhPLDδ4A (6.70 FC) were found to be induced to highly express exclusively at 12 h of salt treatment, the expressions of AhPLDζ1A, AhPLDζ2A, AhPLDζ3A and AhPLDφA were down-regulated at 6 h but up-regulated with the duration of salt stress, whereas AhPLDα4A, AhPLDδ1A and AhPLDδ3A were continuously down-regulated with the log2FC values of -1.74, -1.47 and -3.32 at 48 h, respectively. Above fundings suggested that AhPLDs were potentially involved in signaling triggered by multiple abiotic stresses, and many of them may act as positive regulators of cold and drought stresses in peanut, especially such as AhPLDα3A, AhPLDα5A, AhPLDβ1A and AhPLDδ4A.
As one of the most representative phospholipases in plants, PLD can catalyze the hydrolysis of membrane lipids for lipid remodeling and mediate many physiological processes in plant growth, development and responses to abiotic stresses (Wang, 2005). The identification and functional validation of the PLDs may hold the promise to breed the improved crops with excellent agronomic traits and stress tolerances to combat the challenge of global climate change. In this study, we obtained 46 AhPLDs in allotetraploid peanut as well as 22 AdPLDs and 22 AiPLDs in its two diploid progenitors A. duranensis and A. ipaensis, respectively (Table 1). Obviously, the number of PLDs in A. hypogaea was greater than already reported plant species (Qin and Wang, 2002; Li et al., 2007; Liu et al., 2010; Zhao et al., 2012; Lu et al., 2019; Sagar et al., 2021), which might be due to the hybridization of A. duranensis and A. ipaensis with succeeding polyploidization. The similar results are also reported in cotton (Tang et al., 2016a; Tang et al., 2016b). Based on the conserved structural domains and sequence properties, these 90 Arachis PLD genes could be divided into seven subgroups with distinct biochemical, regulatory and catalytic properties, including α, β, γ, δ, ε, ζ and φ isoforms but excluding isoform κ (Figure 2). At present, PLDκ has only been identified in rice and encodes a C2-PLD (Li et al., 2007).
The specific patterns of retention or dispersion of family genes are vital clues to understand the homoeologous chromosome interaction and genetic evolution during plant allo-polyploidization. In this study, the integrated results of phylogeny, gene structure and gene duplication showed that most allotetraploid peanut PLDs had at least two homoeologous copies in A and B subgenomes as well as orthologous genes with its progenitors (Figures 3–5). It proved that a specific large-scale genome duplication event has occurred during the peanut origin, with the segmental duplication and tandem duplication jointly taking place at some locations (Figure 5; Table S4). But the orthologous gene of AhPLDα6B was not found in peanut genome, suggesting a AhPLDα6A loss event occurred during peanut evolution. Like Arabidopsis and other plant species, the small clade of PLDγs was closer to the clades of PLDβs and PLDδs on the phylogenetic tree (Figure 2), and they contained similar numbers of exons and introns (Figure 3). This suggested that the isoforms β, γ and δ might have originated as one group in plants but be separated into different functional isoforms during the evolution due to the possible gene duplication events. Similarly, α and ε isoforms might also originate from the common ancestor. But PLDζs and PLDφs had dissimilar intron numbers and belonged to the PX/PH-PLD and SP-PLD subfamilies, respectively, suggesting their convergent evolution via two independent evolutionary paths. Besides, the conservation of isoform φ could be also proved by the rate of molecular evolution, in which the Ka/Ks ratios of gene pairs in subgroup PLDφ were smaller than those in other subgroups (Table S5).
Based on the sequence similarity, structural conservation and close evolutionary relationships of ortholog genes among different species, a functional conservation of peanut PLDs might also be predicted. The PPI network of AhPLDs showed that most proteins that interacted with AhPLDs (like non-specific phospholipase C (NPC), DGAT1 and DGKs) were the major components of lipid biosynthesis, lipid metabolism and lipid signaling pathways (Figure 7A; Table S7), which proved the central and conservative functions of peanut PLDs in lipid-related biological processes. Currently, it has become increasingly difficult to find an area of cell biology in which lipids do not have important roles as signaling and regulatory molecules (Hou et al., 2016). For example, DGAT1, DGK2, DGK3 and DGK5 could enhance plant cold tolerance by balancing triacylglycerol (TAG) and PA production (Tan et al., 2018); NPC4 knockout plants displayed increased sensitivity to salt stress in root elongation, seedling biomass, and seed germination (Kocourkova et al., 2011); PLDζs could hydrolyze phosphatidylcholine to supply phosphorus for cell metabolism and DAG for galactolipid synthesis during phosphorus starvation (Lin et al., 2020). Besides, AhPLDs also interacted with other proteins such as protease inhibitors, GPA1 and RBOHD (Figure 7A, Table S7). These interactions can regulate PLD activity and intracellular locations, thus affecting cellular functions. For instance, PLDα1 could interact with the GPA1 through its DRY motif to mediate ABA signaling in Arabidopsis (Zhao and Wang, 2004). Furthermore, there were 24 AhPLDs targeted by 13 miRNAs, suggesting the complex regulation network of AhPLDs involved in and providing the clues to genetically engineer AhPLDs precisely through miRNA mediation.
Hormones are important regulators of plant growth and development. Many basic biological processes in peanut, such as seed germination, root hair growth, pollen tube elongation, blossom and leaf senescence, are known to be regulated by auxin, ABA, gibberellic acid (GA), JA and ethylene (Wang et al., 2018; Guo et al., 2021). Here, the AhPLDs expressed in various peanut tissues were found to contain at least one class of hormone-responsive cis-elements in their respective promoters (Figure 6; Table S6). AhPLDα1A/1B that contained ABA-responsive elements could be expressed in all peanut tissues and showed preferential expression in flowers and pod development. In Arabidopsis, PLDα1 can interact with a low-affinity nitrate transporter NRT1.2 to positively regulate ABA-mediated seed germination and seedling development (Li et al., 2020). It proved that the regulation of AhPLDα1A/1B on peanut growth and development may be mediated by ABA. Isoform PLDδ is the second abundant subfamily next only to isoform PLDα. It represents the majority of PLD isoforms expressed in male gametophyte throughout angiosperms evolution and has been found to be expressed higher in old leaves, stem, roots and flowers than in young leaves and siliques (Qin and Wang, 2002). Here, all AhPLDδs had high expression levels in perianths, stamens and pistil, suggesting their central roles in peanut floral organ development. AhPLDδ3A/B even showed high expressions in all 22 tissues. This might be supported by the abundant hormone-responsive elements in their promoters. In tobacco (Nicotiana tabacum L.), PLDδ3 is the most important member active in pollen tubes. Tightly controlled production of PA generated by PLDδ3 is crucial for maintaining the balance between various membrane trafficking processes that are vital for plant cell tip growth (Pejchar et al., 2020). However, AhPLDα9A did not contain any cis-regulatory elements that respond to hormones, which may be the major factor that caused its non-expression in all peanut tissues. There is also evidence that PLD genes can be induced by ethylene to regulate the programmed death of plant cells (Lanteri et al., 2008), but no ethylene-responsive element could be found in any peanut PLD promoters here.
Besides, many cis-elements in response to diverse environmental stimuli were also found in AhPLDs’ promoters, including ARE, MBS, LTR and T-rich (Figure 6; Table S6). It has been proved that abiotic stresses, such as cold, drought and salinity, could trigger high expressions of most PLDs low or weakly expressed under normal growth conditions, as a result of that a number of specific elements are located in their promoters (Wei et al., 2022). As expected, almost all AhPLDs (except for AhPLDα9A) could be induced by specific or multiple adversities (Figure 9), proving their potential roles in abiotic stress tolerances. At present, the functions of 14 AhPLDs’ Arabidopsis orthologs in stress resistances have been determined (Table 2). For example, AtPLDα1 (the ortholog of AhPLDα1A/B) has been found to participate in ABA signaling and responses to cold, drought and salt stress (Bargmann et al., 2009); AtPLDα3 (the ortholog of AhPLDα3A/B) knockout mutant plants exhibit high sensitivity towards salinity, dehydration and ABA, while gain-of-function of AtPLDα3 leads to reduced sensitivity in transgenic plants (Hong et al., 2008); AtPLDδ (the ortholog of AhPLDδ1A/1B) also has been proved to regulate stress resistances, such as freezing, severe dehydration, high salt, oxidative assault and ultra-violet irradiation (Zhang et al., 2003; Li et al., 2004; Liu et al., 2021); AtPLDδ and/or AtPLDα1 can form a regulatory feedback loop with MPK3 and MPK6 to regulate PLD stability and submergence-induced PA production (Zhou et al., 2022).

Table 2 Orthologous PLD genes in peanut and Arabidopsis with known abiotic stress tolerant functions.
In this study, there were five AhPLDs (AhPLDα3A, AhPLDα5A, AhPLDβ1A, AhPLDβ2A and AhPLDδ4A) found to be highly up-regulated under all three abiotic stresses commonly, suggesting these five AhPLDs might be involved in multiple regulatory pathways at the same time and lead to a wider range of stress resistances in peanut. But the up-regulated expressions of AhPLDα1A and several other AhPLDs were only induced by cold and drought rather than salinity. AhPLDα4A, AhPLDδ1A and AhPLDδ3A were even continuously down-regulated under salt stress. These results suggested that most AhPLDs were mainly involved in cold and drought tolerances but had little or even negative regulation on salt tolerance of peanut. Besides, AhPLDγA was significantly up-regulated only at the early stage (6 h) of cold stress, which may be caused by the low-temperature responsive element (LTR) in its promoter. AhPLDα9A could neither be expressed in any peanut tissues, nor be induced by any abiotic stresses in peanut. This deviation proved that AhPLDα9A might have functional roles in some processes other than peanut growth or stress signaling and its specific biological mechanism need to be further studied.
In conclusion, a total of 22, 22 and 46 PLD genes were identified in A. duranensis, A. ipaensis and A. hypogaea, respectively. Our comparative analyses provided valuable insight into the understanding of sequence characteristics, conserved domains, phylogenetic relationships and molecular evolution of PLD genes in allotetraploid peanut and its diploid progenitors. The predictive analytics of cis-regulatory elements, protein-protein interactions, putative miRNA expanded the view of transcriptional regulation and potential functions of AhPLD genes. Importantly, the expression patterns of tissue-specific and abiotic stress-responsive AhPLDs obtained from RNA-seq and qRT-PCR results offered useful information for further functional investigations. Several candidate AhPLDs, such as AhPLDα3A, AhPLDα5A, AhPLDβ1A, AhPLDβ2A and AhPLDδ4A, can be utilized for genetic manipulation of peanut and other legume crops for improved abiotic stress tolerance and productivity.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
HZ and HY designed the research study. YY, SW and CZ conducted the bioinformatics analysis. JY, XA, NZ and XL performed the experiments. HZ and XZ analyzed the data. HZ and HY wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the earmarked fund for CARS-13 and LiaoNing Revitalization Talents Program (XLYC1902002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1102200/full#supplementary-material
Angelini, J., Vosolsobě, S., Skůpa, P., Ho, A. Y. Y., Bellinvia, E., Valentová, O., et al. (2018). Phospholipase dδ assists to cortical microtubule recovery after salt stress. Protoplasma 255 (4), 1195–1204. doi: 10.1007/s00709-018-1204-6
Bargmann, B. O., Laxalt, A. M., ter Riet, B., van Schooten, B., Merquiol, E., Testerink, C., et al. (2009). Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol. 50 (1), 78–89. doi: 10.1093/pcp/pcn173
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K., et al. (2016). The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48 (4), 438–446. doi: 10.1038/ng.3517
Bertioli, D. J., Jenkins, J., Clevenger, J., Dudchenko, O., Gao, D., Seijo, G., et al. (2019). The genome sequence of segmental allotetraploid peanut arachis hypogaea. Nat. Genet. 51 (5), 877–884. doi: 10.1038/s41588-019-0405-z
Bocckino, S. B., Blackmore, P. F., Wilson, P. B., Exton, J. H. (1987). Phosphatidate accumulation in hormone-treated hepatocytes via a phospholipase d mechanism. J. Biol. Chem. 262 (31), 15309–15315. doi: 10.1016/S0021-9258(18)48176-8
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chi, X., Hu, R., Yang, Q., Zhang, X., Pan, L., Chen, N., et al. (2012). Validation of reference genes for gene expression studies in peanut by quantitative real-time RT-PCR. Mol. Genet. Genomics 287 (2), 167–176. doi: 10.1007/s00438-011-0665-5
Clevenger, J., Chu, Y., Scheffler, B., Ozias-Akins, P. (2016). A developmental transcriptome map for allotetraploid arachis hypogaea. Front. Plant Sci. 7, 1446. doi: 10.3389/fpls.2016.01446
Deng, W., Wang, Y., Liu, Z., Cheng, H., Xue, Y. (2014). HemI: a toolkit for illustrating heatmaps. PloS One 9 (11), e111988. doi: 10.1371/journal.pone.0111988
Guo, F., Hou, L., Ma, C., Li, G., Lin, R., Zhao, Y., et al. (2021). Comparative transcriptome analysis of the peanut semi-dwarf mutant 1 reveals regulatory mechanism involved in plant height. Gene 791, 145722. doi: 10.1016/j.gene.2021.145722
Hanahan, D. J., Chaikoff, I. L. (1947). A new phospholipide-splitting enzyme specific for the ester linkage between the nitrogenous base and the phosphoric acid grouping. J. Biol. Chem. 169 (3), 699–705. doi: 10.1016/S0021-9258(17)30887-6
Hong, Y., Pan, X., Welti, R., Wang, X. (2008). Phospholipase Dalpha3 is involved in the hyperosmotic response in arabidopsis. Plant Cell 20 (3), 803–816. doi: 10.1105/tpc.107.056390
Hong, Y., Zhao, J., Guo, L., Kim, S. C., Deng, X., Wang, G., et al. (2016). Plant phospholipases d and c and their diverse functions in stress responses. Prog. Lipid Res. 62, 55–74. doi: 10.1016/j.plipres.2016.01.002
Hou, Q., Ufer, G., Bartels, D. (2016). Lipid signalling in plant responses to abiotic stress. Plant Cell Environ. 39 (5), 1029–1048. doi: 10.1111/pce.12666
Huang, L., He, H., Chen, W., Ren, X., Chen, Y., Zhou, X., et al. (2015). Quantitative trait locus analysis of agronomic and quality-related traits in cultivated peanut (Arachis hypogaea l.). Theor. Appl. Genet. 128 (6), 1103–1115. doi: 10.1007/s00122-015-2493-1
Kocourkova, D., Krckova, Z., Pejchar, P., Kroumanova, K., Podmanicka, T., Danek, M., et al. (2020). Phospholipase Dα1 mediates the high-Mg2+ stress response partially through regulation of k+ homeostasis. Plant Cell Environ. 43 (10), 2460–2475. doi: 10.1111/pce.13831
Kocourkova, D., Krckova, Z., Pejchar, P., Veselkova, S., Valentova, O., Wimalasekera, R., et al. (2011). The phosphatidylcholine-hydrolysing phospholipase c NPC4 plays a role in response of Arabidopsis roots to salt stress. J. Exp. Bot. 62 (11), 3753–3763. doi: 10.1093/jxb/err039
Korver, R. A., van den Berg, T., Meyer, A. J., Galvan-Ampudia, C. S., Ten Tusscher, K. H. W. J., Testerink, C. (2020). Halotropism requires phospholipase Dζ1-mediated modulation of cellular polarity of auxin transport carriers. Plant Cell Environ. 43 (1), 143–158. doi: 10.1111/pce.13646
Krishna, G., Singh, B. K., Kim, E. K., Morya, V. K., Ramteke, P. W. (2015). Progress in genetic engineering of peanut (Arachis hypogaea l.) –a review. Plant Biotechnol. J. 13 (2), 147–162. doi: 10.1111/pbi.12339
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19 (9), 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Lanteri, M. L., Laxalt, A. M., Lamattina, L. (2008). Nitric oxide triggers phosphatidic acid accumulation via phospholipase d during auxin-induced adventitious root formation in cucumber. Plant Physiol. 147 (1), 188–198. doi: 10.1104/pp.107.111815
Li, M., Hong, Y., Wang, X. (2009). Phospholipase d- and phosphatidic acid-mediated signaling in plants. Biochim. Biophys. Acta 1791 (9), 927–935. doi: 10.1016/j.bbalip.2009.02.017
Li, G., Lin, F., Xue, H. W. (2007). Genome-wide analysis of the phospholipase d family in Oryza sativa and functional characterization of PLD beta 1 in seed germination. Cell Res. 17 (10), 881–894. doi: 10.1038/cr.2007.77
Li, W., Li, M., Zhang, W., Welti, R., Wang, X. (2004). The plasma membrane-bound phospholipase ddelta enhances freezing tolerance in arabidopsis thaliana. Nat. Biotechnol. 22 (4), 427–433. doi: 10.1038/nbt949
Li, M., Welti, R., Wang, X. (2006). Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. roles of phospholipases d zeta1 and d zeta2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol. 142 (2), 750–761. doi: 10.1104/pp.106.085647
Li, J., Zhao, C., Hu, S., Song, X., Lv, M., Yao, D., et al. (2020). Arabidopsis NRT1.2 interacts with the PHOSPHOLIPASE Dα1 (PLDα1) to positively regulate seed germination and seedling development in response to ABA treatment. biochem. biophys. Res. Commun. 533 (1), 104–109. doi: 10.1016/j.bbrc.2020.08.025
Lin, D. L., Yao, H. Y., Jia, L. H., Tan, J. F., Xu, Z. H., Zheng, W. M., et al. (2020). Phospholipase d-derived phosphatidic acid promotes root hair development under phosphorus deficiency by suppressing vacuolar degradation of PIN-FORMED2. New Phytol. 226 (1), 142–155. doi: 10.1111/nph.16330
Liu, Q., Zhang, C., Yang, Y., Hu, X. (2010). Genome-wide and molecular evolution analyses of the phospholipase d gene family in poplar and grape. BMC Plant Biol. 10, 117. doi: 10.1186/1471-2229-10-117
Liu, Q., Zhou, Y., Li, H., Liu, R., Wang, W., Wu, W., et al. (2021). Osmotic stress-triggered stomatal closure requires phospholipase dδ and hydrogen sulfide in arabidopsis thaliana. Biochem. Biophys. Res. Commun. 534, 914–920. doi: 10.1016/j.bbrc.2020.10.074
Lu, S., Fadlalla, T., Tang, S., Li, L., Ali, U., Li, Q., et al. (2019). Genome-wide analysis of phospholipase d gene family and profiling of phospholipids under abiotic stresses in brassica napus. Plant Cell Physiol. 60 (7), 1556–1566. doi: 10.1093/pcp/pcz071
Pejchar, P., Sekereš, J., Novotný, O., Žárský, V., Potocký, M. (2020). Functional analysis of phospholipase dδ family in tobacco pollen tubes. Plant J. 103 (1), 212–226. doi: 10.1111/tpj.14720
Qin, C., Wang, X. (2002). The arabidopsis phospholipase d family. Characterization of a calcium-independent and phosphatidylcholine-selective PLD zeta 1 with distinct regulatory domains. Plant Physiol. 128 (3), 1057–1068. doi: 10.1104/pp.010928
Raza, A., Mubarik, M. S., Sharif, R., Habib, M., Jabeen, W., Zhang, C., et al. (2022a). Developing drought-smart, ready-to-grow future crops. Plant Genome 10, e20279. doi: 10.1002/tpg2.20279
Raza, A., Tabassum, J., Fakhar, A. Z., Sharif, R., Chen, H., Zhang, C., et al. (2022b). Smart reprograming of plants against salinity stress using modern biotechnological tools. Crit. Rev. Biotechnol. 15, 1–28. doi: 10.1080/07388551.2022.2093695
Rizwan, H. M., Shaozhong, F., Li, X., Bilal Arshad, M., Yousef, A. F., Chenglong, Y., et al. (2022a). Genome-wide identification and expression profiling of KCS gene family in passion fruit (Passiflora edulis) under fusarium kyushuense and drought stress conditions. Front. Plant Sci. 13, 872263 doi: 10.3389/fpls.2022.872263.
Rizwan, H. M., Waheed, A., Ma, S., Li, J., Arshad, M. B., Irshad, M., et al. (2022b). Comprehensive genome-wide identification and expression profiling of eceriferum (CER) gene family in passion fruit (Passiflora edulis) under fusarium kyushuense and drought stress conditions. Front. Plant Sci. 13, 898307. doi: 10.3389/fpls.2022.898307
Sagar, S., Deepika, Biswas, D. K., Chandrasekar, R., Singh, A. (2021). Genome-wide identification, structure analysis and expression profiling of phospholipases d under hormone and abiotic stress treatment in chickpea (Cicer arietinum). Int. J. Biol. Macromol. 169, 264–273. doi: 10.1016/j.ijbiomac.2020.12.102
Sang, Y., Zheng, S., Li, W., Huang, B., Wang, X. (2001). Regulation of plant water loss by manipulating the expression of phospholipase dalpha. Plant J. 28 (2), 135–144. doi: 10.1046/j.1365-313x.2001.01138.x
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi: 10.1101/gr.1239303
Shi, X., Zhao, X., Ren, J., Dong, J., Zhang, H., Dong, Q., et al. (2021). Influence of peanut, sorghum, and soil salinity on microbial community composition in interspecific interaction zone. Front. Microbiol. 12, 678250. doi: 10.3389/fmicb.2021.678250
Song, P., Jia, Q., Xiao, X., Tang, Y., Liu, C., Li, W., et al. (2021). HSP70-3 interacts with phospholipase dδ and participates in heat stress defense. Plant Physiol. 185 (3), 1148–1165. doi: 10.1093/plphys/kiaa083
Su, Y., Li, M., Guo, L., Wang, X. (2018). Different effects of phospholipase Dζ2 and non-specific phospholipase C4 on lipid remodeling and root hair growth in arabidopsis response to phosphate deficiency. Plant J. 94 (2), 315–326. doi: 10.1111/tpj.13858
Tang, K., Dong, C. J., Liu, J. Y. (2016a). Genome-wide comparative analysis of the phospholipase d gene families among allotetraploid cotton and its diploid progenitors. PloS One 11 (5), e0156281. doi: 10.1371/journal.pone.0156281
Tang, K., Dong, C. J., Liu, J. Y. (2016b). Genome-wide analysis and expression profiling of the phospholipase d gene family in gossypium arboreum. Sci. China Life Sci. 59 (2), 130–141. doi: 10.1007/s11427-015-4916-2
Taniguchi, Y. Y., Taniguchi, M., Tsuge, T., Oka, A., Aoyama, T. (2010). Involvement of arabidopsis thaliana phospholipase Dzeta2 in root hydrotropism through the suppression of root gravitropism. Planta 231 (2), 491–497. doi: 10.1007/s00425-009-1052-x
Tan, W. J., Yang, Y. C., Zhou, Y., Huang, L. P., Xu, L., Chen, Q. F., et al. (2018). DIACYLGLYCEROL ACYLTRANSFERASE and DIACYLGLYCEROL KINASE modulate triacylglycerol and phosphatidic acid production in the plant response to freezing stress. Plant Physiol. 177 (3), 1303–1318. doi: 10.1104/pp.18.00402
Thompson, J. D., Higgins, D. G., Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 (22), 4673–4680. doi: 10.1093/nar/22.22.4673
United States Department of Agriculture (USDA) Foreign Agricultural Service (2020). Available at: www.fas.usda.gov.
Urrea Castellanos, R., Friedrich, T., Petrovic, N., Altmann, S., Brzezinka, K., Gorka, M., et al. (2020). FORGETTER2 protein phosphatase and phospholipase d modulate heat stress memory in arabidopsis. Plant J. 104 (1), 7–17. doi: 10.1111/tpj.14927
Voorrips, R. E. (2002). MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 93 (1), 77–78. doi: 10.1093/jhered/93.1.77
Wang, X. (2005). Regulatory functions of phospholipase d and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol. 139 (2), 566–573. doi: 10.1104/pp.105.068809
Wang, Y., Li, J., Paterson, A. H. (2013). MCScanX-transposed: detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 29 (11), 1458–1460. doi: 10.1093/bioinformatics/btt150
Wang, G., Ryu, S., Wang, X. (2012). Plant phospholipases: an overview. Methods Mol. Biol. 861, 123–137. doi: 10.1007/978-1-61779-600-5_8
Wang, P., Shi, S., Ma, J., Song, H., Zhang, Y., Gao, C., et al. (2018). Global methylome and gene expression analysis during early peanut pod development. BMC Plant Biol. 18 (1), 352. doi: 10.1186/s12870-018-1546-4
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinf. 8 (1), 77–80. doi: 10.1016/S1672-0229(10)60008-3
Wang, C., Zien, C. A., Afitlhile, M., Welti, R., Hildebrand, D. F., Wang, X. (2000). Involvement of phospholipase d in wound-induced accumulation of jasmonic acid in arabidopsis. Plant Cell 12 (11), 2237–2246. doi: 10.1105/tpc.12.11.2237
Wei, J., Shao, W., Liu, X., He, L., Zhao, C., Yu, G., et al. (2022). Genome-wide identification and expression analysis of phospholipase d gene in leaves of sorghum in response to abiotic stresses. Physiol. Mol. Biol. Plants 28 (6), 1261–1276. doi: 10.1007/s12298-022-01200-9
Yao, Y., Li, J., Lin, Y., Zhou, J., Zhang, P., Xu, Y. (2021). Structural insights into phospholipase d function. Prog. Lipid Res. 81, 101070. doi: 10.1016/j.plipres.2020.101070
Yao, S., Wang, G., Wang, X. (2022). Effects of phospholipase dε overexpression on soybean response to nitrogen and nodulation. Front. Plant Sci. 13, 852923. doi: 10.3389/fpls.2022.852923
Zhang, H., Jiang, C., Ren, J., Dong, J., Shi, X., Zhao, X., et al. (2020). An advanced lipid metabolism system revealed by transcriptomic and lipidomic analyses plays a central role in peanut cold tolerance. Front. Plant Sci. 11, 1110. doi: 10.3389/fpls.2020.01110
Zhang, W., Wang, C., Qin, C., Wood, T., Olafsdottir, G., Welti, R., et al. (2003). The oleate-stimulated phospholipase d, PLDdelta, and phosphatidic acid decrease H2O2-induced cell death in Arabidopsis. Plant Cell 15 (10), 2285–2295. doi: 10.1105/tpc.013961
Zhao, J., Wang, X. (2004). Arabidopsis phospholipase Dalpha1 interacts with the heterotrimeric G-protein alpha-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J. Biol. Chem. 279 (3), 1794–1800. doi: 10.1074/jbc.M309529200
Zhao, J., Zhou, D., Zhang, Q., Zhang, W. (2012). Genomic analysis of phospholipase d family and characterization of GmPLDαs in soybean (Glycine max). J. Plant Res. 125 (4), 569–578. doi: 10.1007/s10265-011-0468-0
Keywords: PLDs, comparative genomics, molecular evolution, lipid metabolic network, growth and development, stress tolerance, Arachis
Citation: Zhang H, Yu Y, Wang S, Yang J, Ai X, Zhang N, Zhao X, Liu X, Zhong C and Yu H (2023) Genome-wide characterization of phospholipase D family genes in allotetraploid peanut and its diploid progenitors revealed their crucial roles in growth and abiotic stress responses. Front. Plant Sci. 14:1102200. doi: 10.3389/fpls.2023.1102200
Received: 18 November 2022; Accepted: 09 January 2023;
Published: 20 January 2023.
Edited by:
Weijian Zhuang, Fujian Agriculture and Forestry University, ChinaReviewed by:
Bin Zhang, Beijing Vegetable Research Center, ChinaCopyright © 2023 Zhang, Yu, Wang, Yang, Ai, Zhang, Zhao, Liu, Zhong and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiqiu Yu, eXVoYWlxaXVAc3lhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.