- 1Rice Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 2Guangdong Key Laboratory of New Technology in Rice Breeding, Guangzhou, China
- 3Guangdong Rice Engineering Laboratory, Guangzhou, China
- 4College of Agriculture and Biology, Zhongkai University of Engineering, Zhongkai, China
- 5Guangdong Key Laboratory of New Technology in Plant Protection, Plant Protection Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China
High-salinity and blast disease are two major stresses that cause dramatic yield loss in rice production. GF14 (14-3-3) genes have been reported to play important role in biotic and abiotic stresses in plants. However, the roles of OsGF14C remain unknown. To understand the functions and regulatory mechanisms of OsGF14C in regulating salinity tolerance and blast resistance in rice, we have conducted OsGF14C-overexpressing transgenic experiments in the present study. Our results showed that overexpression of OsGF14C enhanced salinity tolerance but reduced blast resistance in rice. The enhanced salinity tolerance is related to the reduction of methylglyoxal and Na+ uptake instead of exclusion or compartmentation and the negative role of OsGF14C in blast resistance is associated with the suppression of OsGF14E, OsGF14F and PR genes. Our results together with the results from the previous studies suggest that the lipoxygenase gene LOX2 which is regulated by OsGF14C may play roles in coordinating salinity tolerance and blast resistance in rice. The current study for the first time revealed the possible roles of OsGF14C in regulating salinity tolerance and blast resistance in rice, and laid down a foundation for further functional study and crosstalk regulation between salinity and blast resistance in rice.
Introduction
Rice (Oryza sativa L.) is the staple food for over half of the world’s population. Therefore, rice production plays a crucial role in world food security. However, rice production is frequently subjected to biotic and abiotic stresses. High-salinity and blast disease are two major stresses that cause dramatic yield loss in rice production (Jacobs and Wang, 2021; Yin et al., 2021; Liu et al., 2022).
It has been reported that more than 955 million hectares of land (approximately 6.5% of the world’s total land area), 20% of the world’s cultivated land area and nearly 50% of the world’s irrigated land area are affected by salinity (Tufail et al., 2018), and this problem continues to worsen. In addition, the marginal lands such as coastal regions which are saline soils and unsuitable for crop cultivation need to be utilized for mitigating the issues of food demands.
Salinity tolerance in rice is a quantitative trait which is regulated by a host of genes in plants. It is imperative to understand the molecular mechanism of salinity tolerance in rice to minimize salinity related yield penalty. So far, a number of salinity tolerance genes involving in transcription regulation, signal transduction, ion transportation and metabolic homeostasis have been identified in plants (Apse et al., 1999; Shi et al., 2000; Singla-Pareek et al., 2003; Conti et al., 2008; Munns et al., 2012; Kesten et al., 2019; Ma et al., 2019; Zhao et al., 2019; Song et al., 2021; Ganapati et al., 2022). However, the molecular mechanisms underlying salinity tolerance are still not well understood. Particularly, the crosstalk between salinity and other stresses remain largely unknown (Gong et al., 2020; Zelm et al., 2020; Ganapati et al., 2022).
Rice blast disease caused by Magnaporthe oryzae is one of the most destructive rice diseases leading to severe yield losses in rice production worldwide (Yin et al., 2021). Rice blast resistance can be divided into qualitative resistance conferred by major genes (R) and quantitative (partial) resistance contributed by multiple genes (Kou and Wang, 2010; Fu et al., 2011). Despite the powerful and universally used of R genes, quantitative resistance is presumably non race specific and is generally considered to be broader spectrum and more durable in natural conditions (Kou and Wang, 2010).
The GF14 (14-3-3) genes are ubiquitous in eukaryotic organisms. Plant GF14 proteins interact with a broad range of phosphorylated proteins and function in signaling, transcription activation or defense (Roberts, 2003), which endow GF14 proteins different roles in plant development, metabolism, environmental stress responses and may establish a platform for crosstalk in diverse pathways (Oecking and Jaspert, 2009). Many GF14 genes have been reported to be involved in biotic stresses in various crop plants. For example, the barley GF14 proteins have been demonstrated to interact with plasma membrane H+-ATPases in epidermal cells as they respond to infection by the powdery mildew fungus (Finni et al., 2002). The Arabidopsis GF14-λ confers resistance to the fungal pathogen Golovonomyces spp. through interaction with the RPW8.2 protein (Yang et al., 2009). The tomato GF14 protein 7 directly interacts with mitogen-activated protein kinase kinase kinase (MAPKKKa) to positively regulate immunity-associated programmed cell death (Oh et al., 2010). There are eight GF14 members in rice (GF14A - GF14H) (Chen et al., 2006), among which, OsGF14B may interact with mitogen-induced MAP kinase 1 (BIMK1) which was induced by rice blast infection and participates in systemic acquired disease resistance (Cooper et al., 2003). OsGF14E was the first GF14 gene confirmed to play a negative role in bacterial Xoo and fungal pathogen Rhizoctonia solani disease resistance (Manosalva et al., 2011). In previous study, we also found that OsGF14E positively regulates panicle blast resistance in rice (Liu et al., 2016b), while OsGF14B positively regulates panicle blast resistance but negatively regulates leaf blast resistance in rice (Liu et al., 2016a). In addition to the roles in biotic stresses, GF14 genes have been also reported to function in abiotic stresses. Overexpression of the Arabidopsis GF14λ in cotton leads to a “stay-green” phenotype and improves stress resistance under moderate drought conditions (Yan et al., 2004). However, GF14λ and GF14κ were reported to negatively regulate salinity tolerance in Arabidopsis (Zhou et al., 2014). Co-expression of D-myo-inositol-3-phosphate synthase (IPS) and 14-3-3-like protein GF14 is responsible for the interaction between maize and Rhizophagus intraradices under drought stress, and potentially induces the synergistic actions of the symbiotic partners in enhancing plant drought resistance (Li et al., 2016). Ectopic expression of TaGF14b in tobacco led to longer root, better growth status, and higher relative water content, survival rate, photosynthetic rate, and water use efficiency in transgenic tobaccos compared to those in control plants under drought and salinity stresses (Zhang et al., 2018a). Studies also showed that the binding of GF14ω to Glutamate Receptor-Like Protein GLR3.7 inhibited primary root length under salinity stress in Arabidopsis thaliana (Wang et al., 2019). In addition to its function in blast, the OsGF14B was also found to regulate rice response to drought stress. The OsGF14B-overexpression lines exhibited enhanced sensitivity to drought and osmotic stress (Liu et al., 2019).
OsGF14C has been reported to be responsive to different biotic and abiotic stresses in rice (Chen et al., 2006). However, its actual functions in different stresses in rice have not been fully characterized yet. In the present study, we found that overexpression of OsGF14C showed opposite roles in response to salinity and blast stress in rice. The overexpression of OsGF14C positively regulates salinity tolerance by reducing the level of methylglyoxal and Na+ uptake instead of exclusion or compartmentation, while negatively regulates blast resistance by inhibiting the expression of GF14E, GF14F and PR genes. Furthermore, our results and the previous studies together implied that the lipoxygenase gene LOX2 which is regulated by OsGF14C may play roles in both salinity tolerance and blast resistance in rice. Thus, the present study for the first time revealed the possible roles of OsGF14C in regulating salinity tolerance and blast resistance in rice, and provided insights into the crosstalk regulation between salt and blast resistance in rice.
Materials and methods
Vector construction and genetic transformation
For OsGF14C overexpression vector construction, the coding region sequence of OsGF14C was amplified from the blast-resistant line BC10 using the primers in Supplemental Table 1. The resulting product (843bp) was cloned into pEASY-T1 (TransGen) vector and verified by sequencing. The entry clone for overexpressing plants was then inserted into PHQSN (modified from pCAMBIA1390) which harbors a CaMV35S promoter. The positive plasmid was electroporated into Agrobacterium tumefaciens EHA105 and then introduced into calli of the cultivar Lijiangxintuanheigu (LTH) via Agrobacterium-mediated genetic transformation.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted with Trizol reagent (Invitrogen) and purified with NucleoSpin RNA Clean-up (MACHEREYNAGEL) according to the manufacturers’ instructions. RNA quality and quantity were assessed by formaldehyde denaturing agarose gel electrophoresis and spectrophotometry (Nanodrop-1000), respectively. The purified total RNA was reverse-transcribed using the Primescript™ RT reagent kit (Takara) to generate cDNA and real-time PCR was carried out using SYBR ExTaq™ (Takara). Actin gene was chosen as a reference gene. Gene expression was quantified by the comparative CT method. Experiments were performed in triplicate, and the results were presented by their means ± standard derivation (SD). Gene-specific primers used were listed in Supplemental Table 1.
Salinity treatment and evaluation of salinity tolerance
We got OsGF14C over expression plants by vector construction and rice transformation. The relative expression level of OsGF14C in every line was measured by quantitative real-time PCR. The seeds of T0, T1, and T2 segregating progeny were used for salinity treatment. After incubation at 49°C for 96 h to break dormancy, seeds were soaked in water for 36 h and then incubated at 30°C for 48 h for pre-germination. The seeds that germinated uniformly were sown in cultural boxes with kimura nutrient mixture and grown in the tissue culture incubator for ~10 d. The kimura nutrient mixture was changed into new kimura nutrient mixture containing 120 mmol/L Nacl. After 7 d treatment, the culture solution was change back to normal kimura nutrient mixture without Nacl. After ~10 d recovery, the survival rate was calculated. The treatment was repeated three times.
Measurement of electrolyte leakage
Electrolyte leakage was measured using the method reported in our previous study (Dong et al., 2019). Briefly, at 0 h, 2 d, 3 d, 4 d before and after Nacl treatment, 0.1 g of leaf material was collected and washed several times in ultrapure water and then immersed in 20 mL ultrapure water for 24 h. The electrical conductivity of the resulting solution was measured and defined as R1. This solution was placed in boiling water for 20-30 min. After cooling, the electrical conductivity was measured and defined as R2. Relative electrolyte leakage (%) = (R1/R2)×100%. The experiment were repeated three times.
Blast treatment and evaluation of disease resistance
We got OsGF14C over expression plants by vector construction and rice transformation. Wild-type LTH plants and 15 lines of T1 segregating progeny germinated from T0 transgenic seeds were grown in soil in greenhouse. The relative OsGF14C transcription level was measured by quantitative real-time PCR. M. oryzae isolate GD08-T13 inoculum was used for blast resistance evaluation as described in our previous study (Dong et al., 2021). Briefly, when the plants grow to tillering stage, the leaves of the plants were inoculated with GD08-T13 using the punch method. The inoculated plants were sprayed with water for 2-3 min every 2 h to maintain the humidity. After 10 days inoculation, the lesion size of each leaf was measured. The correlation between the disease area and relative OsGF14C transcription was analysed in excel. The treatment was repeated three times.
Sub-cellular localization analysis
We amplified the protein coding region of OsGF14C from BC10 using the primers in Supplemental Table 1 and cloned it into the pUC18 vector with the coding sequence for enhanced green fluorescent protein (eGFP) under the control of the Cauliflower mosaic virus (CaMV) 35S promoter provided by Yang et al. (2014). Dehulled rice seeds of LTH were surface-sterilized using 1.5% sodium hypochlorite for 40 min, then germinated and cultured on half-strength Murashige and Skoog medium in the tissue culture room for ~10 d. Seedlings were grown hydroponically under natural light for 3 d. The protoplast isolation and transformation were performed following the method of Yang et al. (2014). After 24 h incubation at 28°C without light, the rice protoplasts were observed and photographed under a laser confocal microscopy (Zeiss LSM710, Germany).
Measurement of methylglyoxal content
The samples were ground in liquid nitrogen to powder. 1 g of sample was dissolved in 5 mL of water, and placed in an ice water bath for ultrasonic extraction for 1 h. After centrifugation at 4°C, 12000 rpm for 5 min, 1 mL supernatant extract was mixed with 1 mL aqueous anthalenediamine solution (6 g/L), derivatization reaction was performed for 8 h at room temperature and dark conditions, filtered at 0.22 um, and then conducted chromatograph analysis. Chromatographic conditions are as follows: chromatographic column: Kromasil; Anti-phase chromatography column (150 mm × 4.6 mm, 5 um); Mobile phase: 0.1%(V/V) acid aqueous solution (A) and methyl alcohol (B); gradient elution program: 0~5 min: B= 30%; 5~10 min: B up from 30% gradient to 90%; 10~15 min: B= 90%; 15~16 min: B gradually decreased from 90% to 30%; 16~20 min: B= 30%; strength: 1.0 ml/min. Detection wavelength: 318nm; Column temperature: 30°C; Sample volume: 10uL; Retention time: 10.3min. Methylglyoxal content= Detection concentration*Sample extraction volume (dilute volume)/the weight of sample.
Results
OsGF14C is induced by salinity stress but repressed by blast infection
To investigate the biological functions of OsGF14C in salinity and blast resistance in rice, we first analyzed the expression patterns of OsGF14C under salinity stress and blast infection. The experiments were conducted at 0h, 12h, 24h and 48h after stress treatments. The results indicated that the expression levels of OsGF14C significantly increased at 24h and 48h after salinity treatment, while significantly decreased at 24h after blast inoculation (Figure 1).
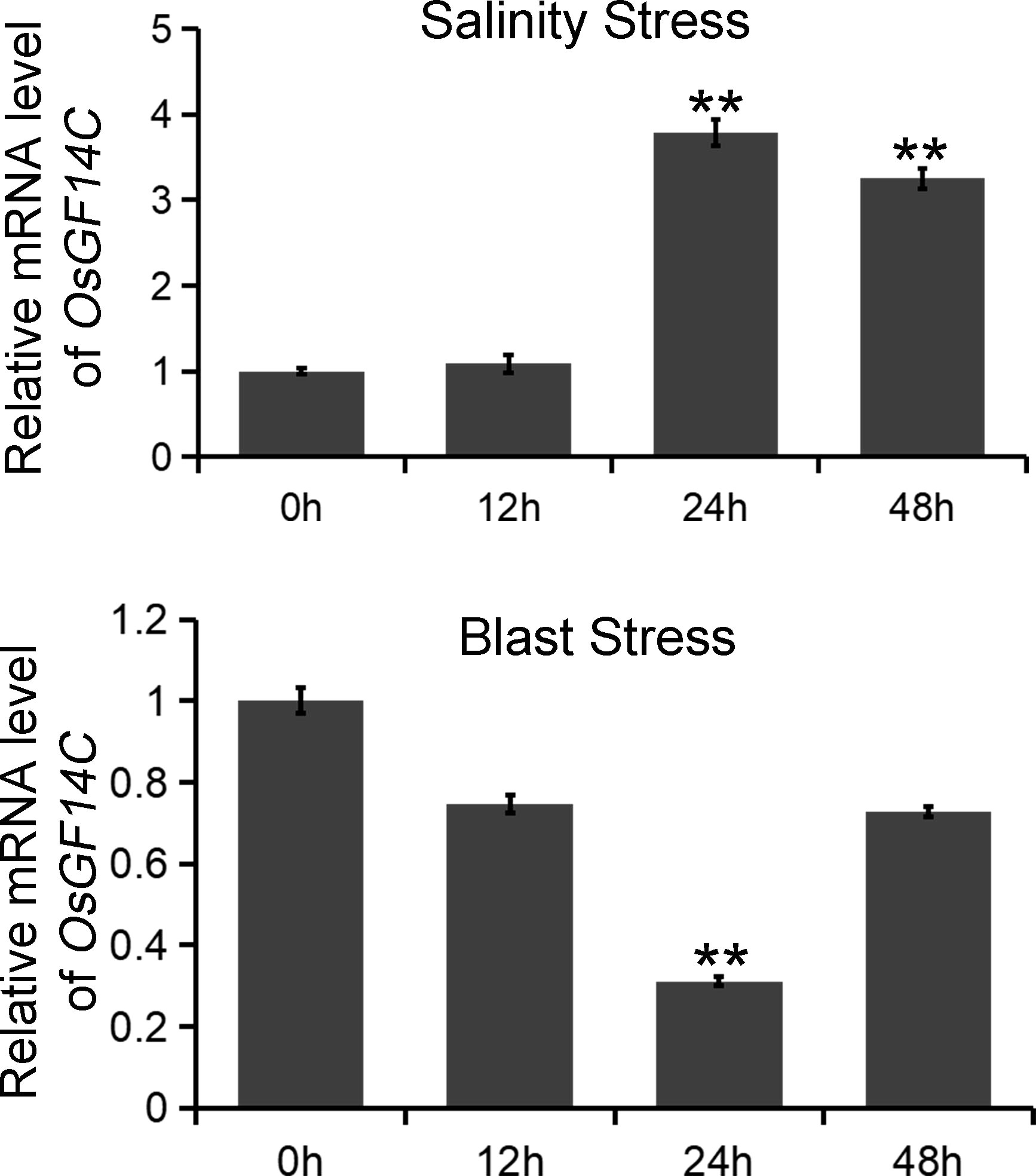
Figure 1 The expression patterns of OsGF14C in responses to salinity stress and blast stress, respectively. The expression of OsGF14C during salinity stress and blast stress as assessed by quantitative real-time PCR at 0 h before treatment and 12 h, 24 h, 48 h after treatment. Error bars indicate the standard deviation (SD) from three biological replicates and ** indicates a statistically significant difference compared with 0 h treatment (t test, p < 0.01).
Overexpression of OsGF14C enhances tolerance to salinity stress but reduces resistance to blast in rice
To confirm the functions of OsGF14C in salinity and blast resistance in rice, we produced the OsGF14C overexpressing plants (OsGF14C-OX) using the variety Lijiangxintuanheigu (LTH) as material, which is susceptible to blast. Fifteen independent transgenic lines were generated and the OX plants showed no differences in morphological phenotypes compared to wild type plants.
Four independent homozygous lines (OX-4, OX-9, OX-12 and OX-15) with different high transcription levels of OsGF14C were selected for evaluation of salinity tolerance (Figure 2A). The results showed that the relative electrolyte leakages, which is an indicator of membrane injury under salinity stress, of all the OsGF14C-OX lines were significantly lower than the wild-type plants on the fourth day after high salinity stress treatment (Figure 2B). After salinity stress treatment for seven days and then recovery for ten days, the survival rates of the OsGF14C-OX lines were significantly higher than their wild-type plants (Figures 2C, D). We also measured the transcription level of OsGF14C in the fifteen OX lines and conducted blast resistance evaluation using M. oryzae isolate GD08-T13 by measuring the infected lesion areas (Figures 3A–C). Correlation analysis demonstrated a positive correlation between the expression level of OsGF14C and infected leaf area (R2 = 0.643; Figure 3D), suggesting that increased OsGF14C expression may reduce the blast resistance in rice.
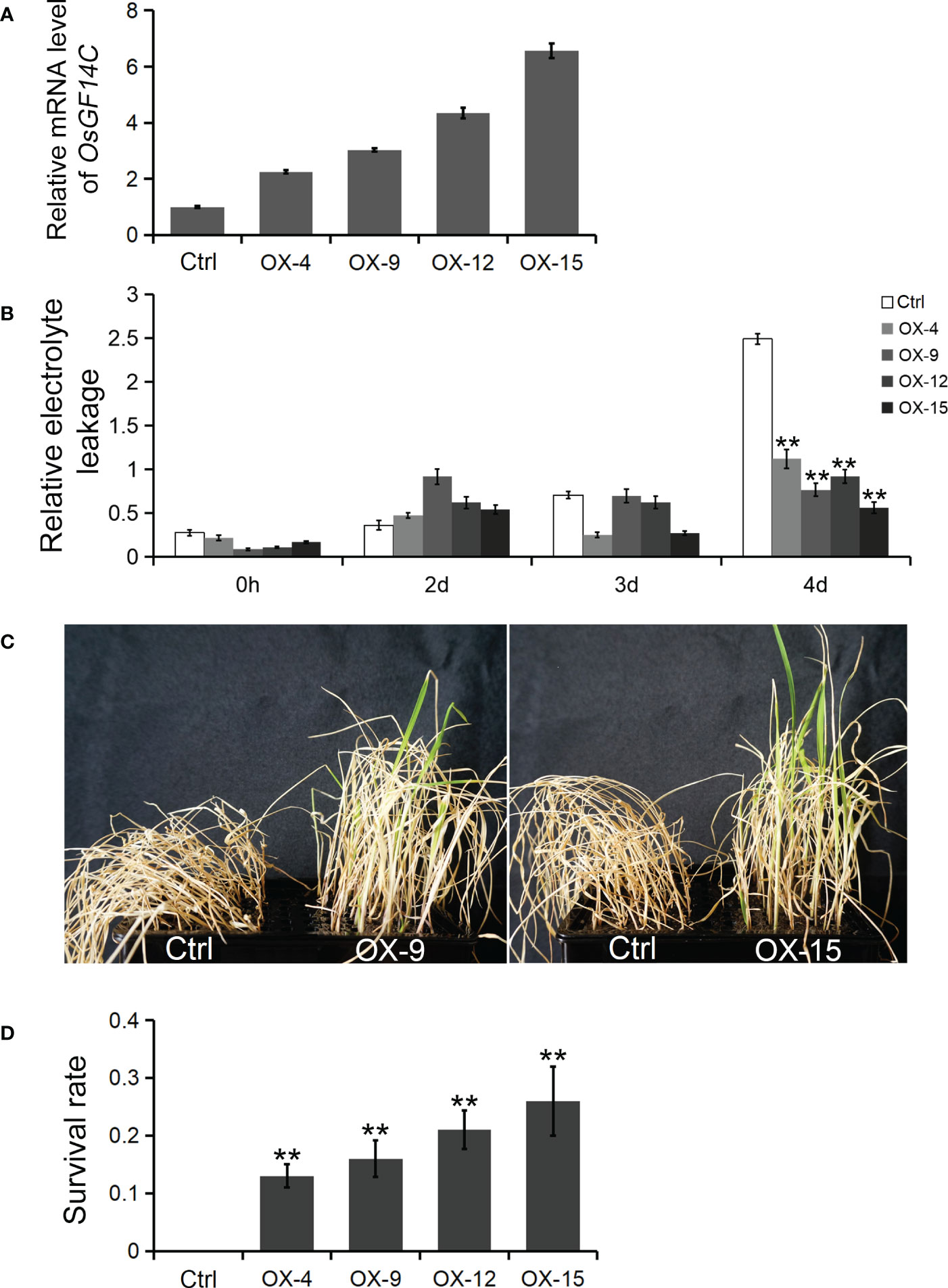
Figure 2 The role of OsGF14C in resistance to salinity stress in rice. (A), the relative OsGF14C transcription level of four independent homozygous lines (OX-4, OX-9, OX-12 and OX-15) used in salinity tolerance evaluation experiment; (B), the relative electrolyte leakages of Ctrl, OX-9, and OX-15 on the forth day after salinity treatment; (C), the growth state of Ctrl, OX-9, and OX-15 after seven days of salinity treatment and ten days of recovery; (D), the survival rate of Ctrl, OX-4, OX-9, OX-12 and OX-15 after seven days of salinity treatment and ten days of recovery. Error bars indicate the standard deviation (SD) from three biological replicates and ** indicates a statistically significant difference compared with 0 h treatment (t test, p < 0.01).
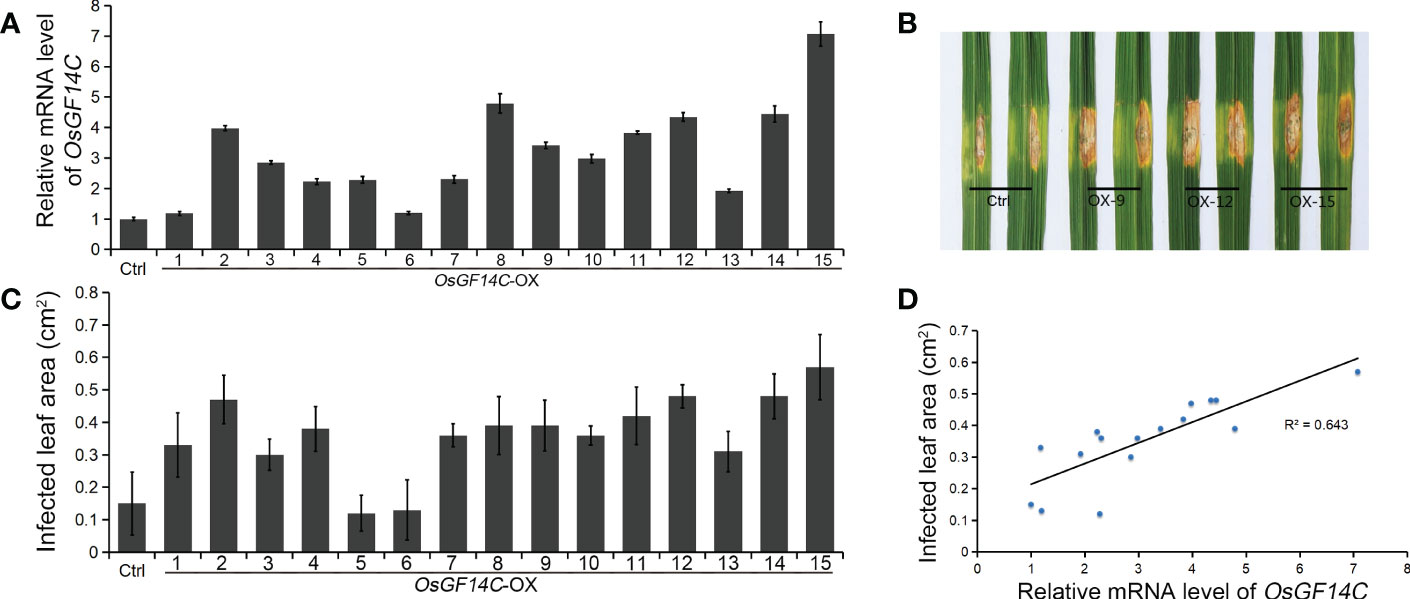
Figure 3 The role of OsGF14C in resistance to blast stress in rice. (A), the relative OsGF14C transcription of independent 15 OsGF14C-OX lines; (B), the infected lesions areas of Ctrl and OsGF14C-OX lines after inoculation using punch method; (C), the infected lesions areas on leaves of the 15 OsGF14C-OX lines; (D), the correlation between (A, C).
OsGF14C is ubiquitously expressed in different tissues and localized in cytoplasm
In order to gain further insights into the function of OsGF14C in rice response to stresses, the temporal and spatial expression patterns of OsGF14C in root, stem and leaf at different stages were analyzed. As shown in Figure 4A, the expression of OsGF14C can be detected in all rice tissues examined in the present study and relative higher expression level was observed in leaf at seedling stage.
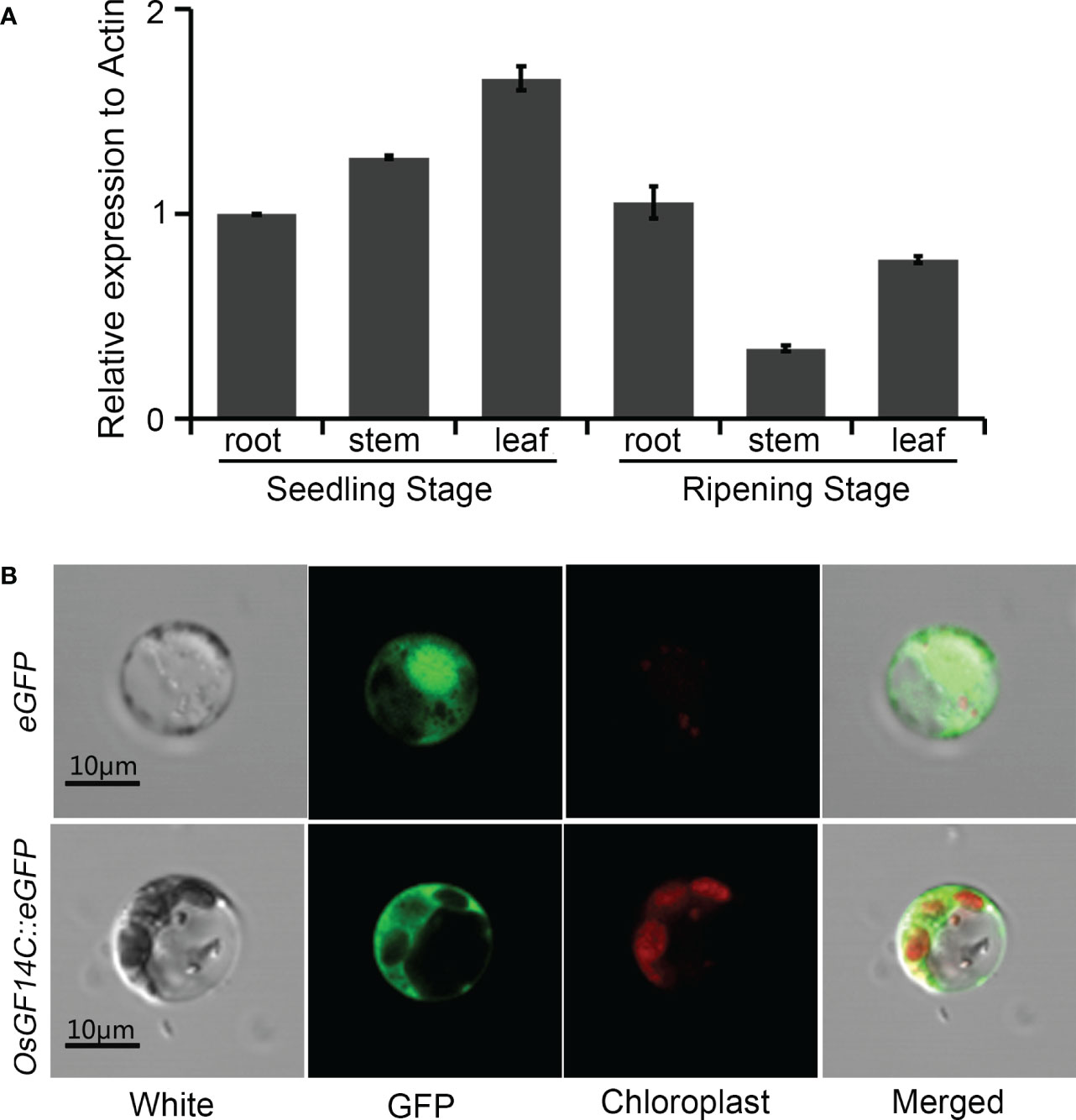
Figure 4 The temporal and spatial expression patterns of OsGF14C and its subcellular localizations. (A) Expression analysis of OsGF14C in different rice tissues by quantitative real-time PCR. Error bars indicate the standard deviation (SD) from three biological replicates; (B) Sub-cellular localization of OsGF14C in rice protoplasts. White indicates the protoplast under visible light, GFP indicates the eGFP fluorescence, Chloroplast indicates the chloroplast fluorescence, Merge indicates the overlapping situation of White, GFP, and Chloroplast.
To investigate the sub-cellular localization of OsGF14C, the coding domain sequence of OsGF14C was fused with the green fluorescence protein (eGFP) under the control of the cauliflower mosaic virus 35S promoter. The OsGF14C::eGFP fusion vector and the control vector (empty eGFP vector) were transiently expressed in rice protoplasts. The laser confocal microscope showed that the green fluorescent signal in the protoplasts expressing eGFP control vector was distributed both in the nucleus and the cytoplasm, whereas the green fluorescent signal was distributed in the cytoplasm of the rice protoplasts transfected with the OsGF14C::eGFP vector (Figure 4B). The results demonstrated OsGF14C was localized in cytoplasm.
OsGF14C modulate the expression of defense-related genes and Na+ transport-related genes
According to the previous studies, the defense-related genes play important roles in blast resistance in rice, and the accumulation of toxic ions, such as Na+ is the main reason for ion toxicity in rice under salinity stresses (Yan et al., 2022). To elucidate the regulatory mechanisms underlying OsGF14C in response to salinity and blast stresses in rice, we detected the expression changes of defense-related genes and Na+ transport-related genes in transgenic and wild type plants.
The results indicated that the expression of the lipoxygenase gene LOX2 was significantly induced in OsGF14C-OX lines compared to the wild type plants. In contrast, the expression levels of three pathogenesis-related genes (PR1, PR5, PR10a) and a chitinase gene (CHT1, also named PR-3 chitinase 1), were significantly lower in OsGF14C-OX lines than that in wild type plants (Figure 5).
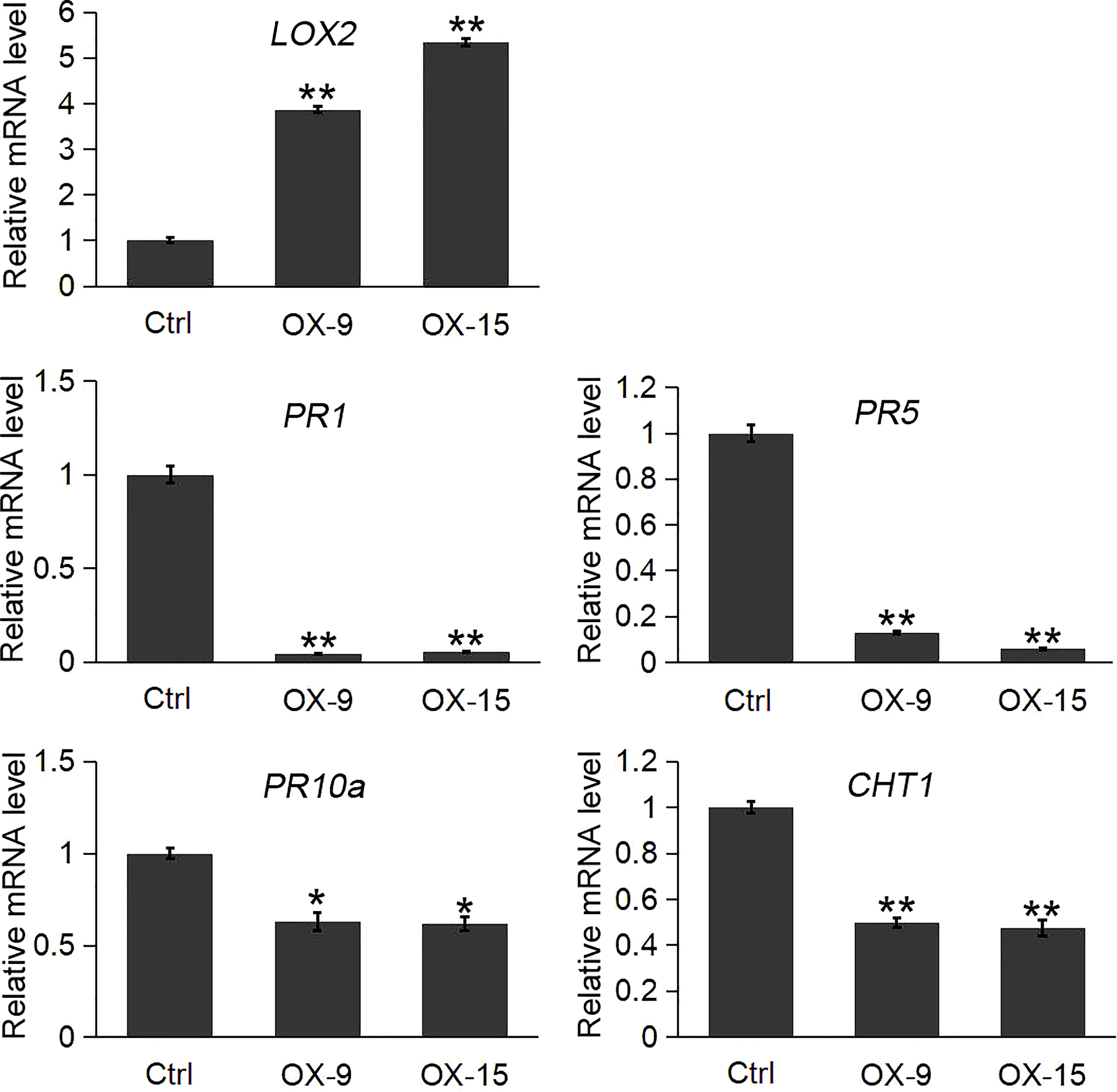
Figure 5 Expression analysis of LOX2 and pathogenesis-related (PR) genes in the OsGF14C-OX transgenic plants and control plants (Ctrl) by quantitative real-time PCR. Error bars indicate the SD from three biological replicates and asterisks indicate statistically significant differences compared to the control plants (t test, **P < 0.01; *P < 0.05).
We also evaluated the expression pattern of the HKT, NHX and SOS1 genes, which are the important player in regulating the ion homeostasis in rice under salinity stress. Our results showed that the expression levels of the three HKT genes (HKT1, HKT4, HKT6) were significantly lower in OsGF14C-OX lines than that in wild type plants both before and after salinity stress treatment (Figure 6). However, there was no significant difference in the expression levels of NHX1, NHX5, and SOS1 between the OsGF14C-OX lines and the wild type plants (Supplemental Figure 1).
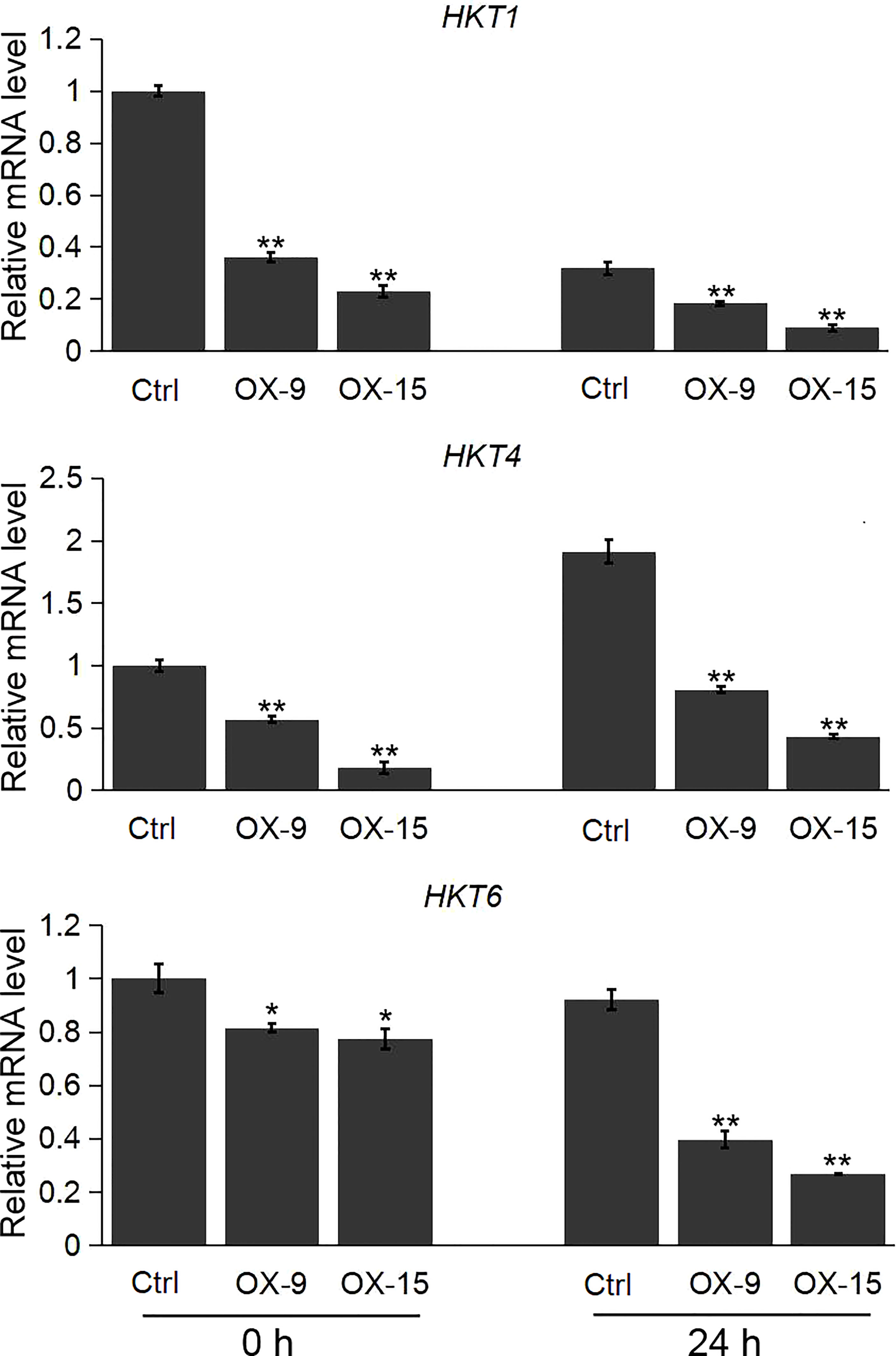
Figure 6 Expression analysis of HKT genes in the OsGF14C-OX transgenic plants and control plants (Ctrl) by quantitative real-time PCR. Error bars indicate the SD from three biological replicates and asterisks indicate statistically significant differences compared to the control plants (t test, **P < 0.01; *P < 0.05).
The expression levels of the client proteins were differently regulated by OsGF14C
Plant GF14 proteins usually interact with a broad range of proteins to exert their functions. In the previous study, Zhang et al. (2017) reported 29 (exclude OsGF14C itself) client proteins of OsGF14C in rice root by affinity chromatography. Among the 29 client protein genes, 22 of them were found to be expressed in our OsGF14C-OX RNA-seq results. Among these 22 genes, only 6 of them had significant expression changes in OsGF14C-OX plants compared to wild-type plants (Table S2). In order to confirm these results, we further evaluated the expression levels of these gene by qRT-PCR. The results demonstrated that the expression levels of two other OsGF14 genes (GF14E and GF14F) and a sucrose synthase gene SUS1 were significantly lower in OsGF14C-OX lines than that in the wild type plants. In contrast, the expression levels of two self-defense mechanism related genes (GLYI-11 and GSTF2), as well as a cell structure and growth-related gene TUBC3 were significantly higher in OsGF14C-OX lines than that in the wild type plants (Figure 7).
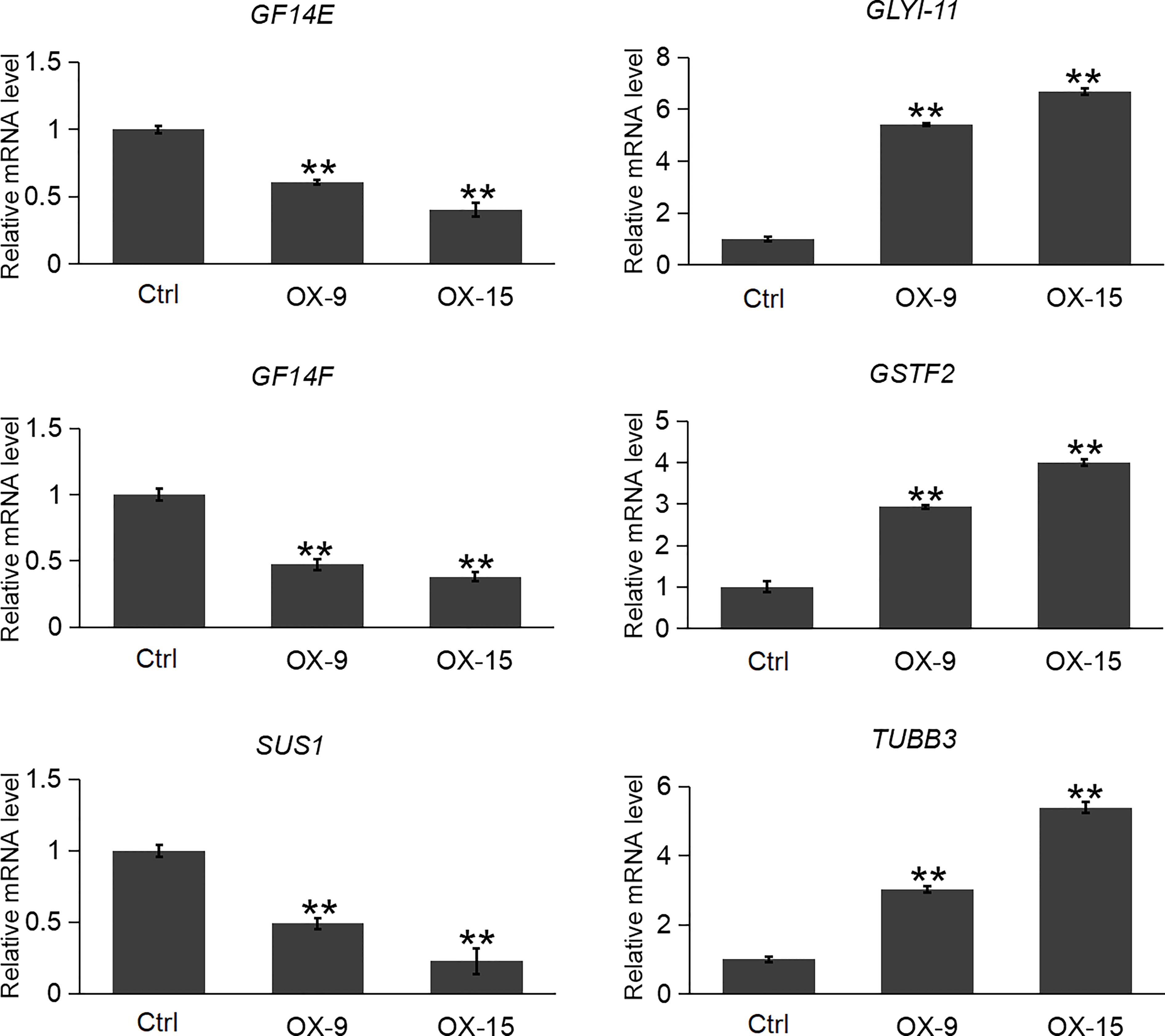
Figure 7 Expression analysis of several clients genes in the OsGF14C-OX transgenic plants and control plants (Ctrl) by quantitative real-time PCR. Error bars indicate the SD from three biological replicates and asterisks indicate statistically significant differences compared to the control plants (t test, **P < 0.01).
Disscussion
Overexpression of OsGF14C positively regulates salinity tolerance while negatively regulates blast resistance in rice
It has been reported that GF (14-3-3) played very important roles in plant growth and development, as well as in response to various stresses. There are eight GF genes in rice, named GF14A-GF14H. The other studies and our previous studies have confirmed that GF14B and GF14E functioned in regulating disease resistance and drought tolerance in rice (Manosalva et al., 2011; Liu et al., 2016a; Liu et al., 2016b). Although it was reported that OsGF14C responded differently in various biotic and abiotic stresses in rice (Chen et al., 2006), the molecular functions of OsGF14C involved in biotic and abiotic stresses resistances in rice remain to be elucidated. In present study, the sub-cellular localization of OsGF14C in rice protoplasts revealed that OsGF14C accumulated only in the cytoplasm, which is consistent with the observations in rice callus and onion epidermal cells (Figure 4B). Differently, GF14B, GF14E and GF14F were localized in both nuclei and cytoplasm (Chen et al., 2006). Furthermore, the temporal and spatial expression analysis indicated that OsGF14C exhibited higher expression levels in all the tissues examined in the present study at the seedling stage compared to those at the ripening stage (Figure 4A). The different sub-cellular localization, temporal and spatial expression patterns imply that OsGF14C might have different functions in rice. Our transgenic experiments further confirmed this speculation.
Our results showed that the expression of OsGF14C was induced by salinity stress while suppressed by blast infection (Figure 1). The relative electrolyte leakages, which is an indicator of salinity resistance, of the OsGF14C-OX lines were significantly lower than that of the wild-type plants after salinity stress treatment (Figure 2B). In consistence with these results, after salinity treatment for seven days and then recovery for ten days, the survival rates of the OsGF14C-OX lines were significantly higher than that of their wild type plants (Figures 2C, D). On the contrary, for the blast disease resistance, the infected lesion areas of OsGF14C-OX lines were larger than the wild type plants and the expression levels of OsGF14C was positively correlated with infected leaf areas in OsGF14C-OX lines (Figure 3), indicating negative effect of OsGF14C in regulating blast resistance in rice. Taken together, our results suggest that overexpression of OsGF14C can improve salinity tolerance but reduce blast resistance in rice.
The negative role of OsGF14C overexpression in blast resistance is associated with the suppression of GF14E, GF14F and PR genes
It is well known that PR genes play important roles in plant defense responses (Kaur et al., 2017). Our previous study and other studies also suggest that the PR genes are involved in blast resistance in rice (Tezuka et al., 2019; Dong et al., 2021). CHT1, which is also named as pathogenesis related (PR)-3 chitinase 1, is another typical defense response gene. Previous studies showed that CHT1 played important roles in disease resistances, such as sheath blight disease and blast disease in rice (Hao et al., 2012). To investigate whether OsGF14C mediated blast resistance is associated with PR genes, we analyzed the expression of the PR genes in the present study. The results showed that the expression levels of PR1, PR5, PR10a and CHT1 in OsGF14C overexpressing lines were all significantly lower than that in wild type plants, suggesting that OsGF14C negatively regulates blast resistance in rice by suppression of PR genes.
In plants, GF proteins are known to be involved in regulating a large number of biological processes via their interaction with numerous client proteins in a phosphorylation-dependent manner. Recently, 87 client proteins were identified through affinity chromatography assay using four GF proteins in rice, including the GF14C (Zhang et al., 2017). Twenty-nine (exclude OsGF14C itself) out of the 87 client proteins can interact with OsGF14C. Among these 29 client proteins, the expression levels of OsGF14E and OsGF14F, were significantly lower in OsGF14C-OX plants than that in wild type plants. In our previous study, we have functionally confirmed that OsGF14E and OsGF14F could positively regulate blast resistance in rice (Liu et al., 2016b; Ma et al., 2022). These results suggest that OsGF14C might negatively regulate blast resistance by suppressing the expression of OsGF14E and OsGF14F. However, further studies are needed to elucidate whether OsGF14C regulates blast resistance by an indirect way or by both indirect and direct ways.
OsGF14C positively regulates salinity tolerance by reducing the level of methylglyoxal and Na+ uptake
GFP (Glyoxalase family protein, also named GLY or GLO) and GSTF2 (Glutathione S-transferase) are another two client proteins of OsGF14C reported by Zhang et al. (2017). GLYI catalyzes methylglyoxal (MG) metabolism in the presence of GSH and produces an intermediate compound S-lactoylglutathione. According to previous studies, MG is a potent cytotoxic compound produced from glycolysis. The rate of glycolysis increases under stresses, and lead to MG accumulation in plants. MG accumulation is toxic to cells as it inhibits cell proliferation and results in a number of adverse effects, such as increasing the degradation of proteins and inactivating the antioxidant defense system (Hossain et al., 2009). Previous studies showed that overexpression of GLY I+II together in tobacco led to enhancement of salinity and zinc tolerance (Singla-Pareek et al., 2003; Singla-Pareek et al., 2006). Heterologous expression of OsGLYI-11.2 in Escherichia coli and tobacco also resulted in improved adaptation to various abiotic stresses caused by increased scavenging of MG, lowering the Na(+)/K(+) ratio and maintenance of reduced glutathione levels (Mustafiz et al., 2014). In the present study, the expression levels of GLYI-11 and GSTF2, and the contents of MG were significantly lower in OsGF14C-OX lines than that in wild type plants (Figures 7, 8). Taken together, these results suggest that the positive role of OsGF14C in regulating salinity tolerance might be partially attributed to its interactions with GLYI-11 and GSTF2, and leading to the reduction of MG.
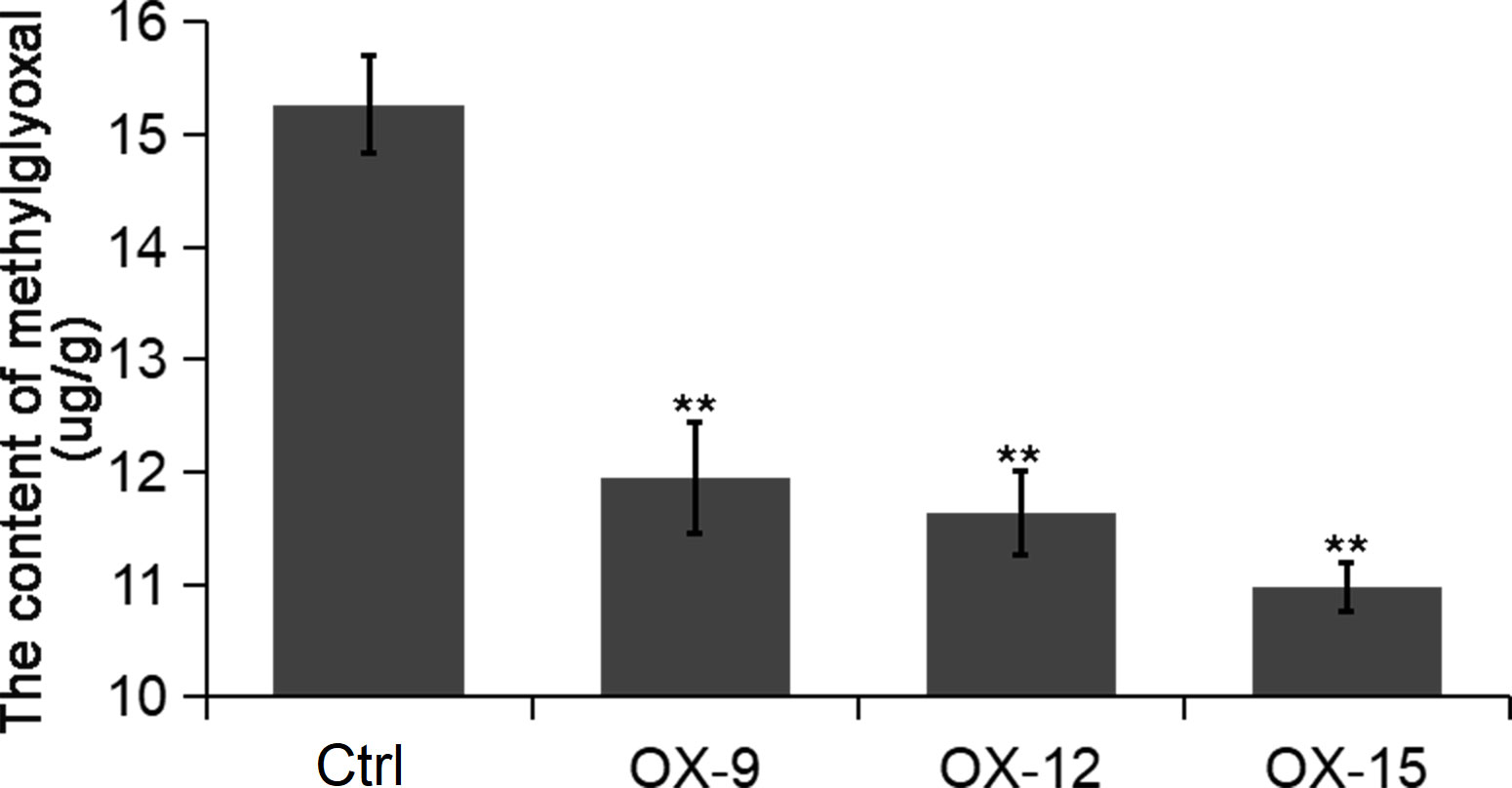
Figure 8 The content of methylglyoxal in the OsGF14C-OX transgenic plants and control plants (Ctrl). Error bars indicate the SD from three biological replicates and asterisks indicate statistically significant differences compared to the control plants (t test, **P < 0.01).
Plants have developed intricate mechanisms for Na+ uptake, export and compartmentation to reduce the accumulation of Na+ (Wu, 2018). So far, the salt-overly-sensitive (SOS) signaling pathway has been considered an importance pathway in salt resistance (Gong et al., 2020; Zelm et al., 2020). The SOS1 encodes a plasma membrane Na+/H+ antiporter, which uses the H+ gradient to drive Na+ efflux and thus reducing cytosolic Na+ concentration (Shi et al., 2003). However, in the present study, there was no significant difference in the expression levels of SOS1 between OsGF14C-OX plants and wild type plants, either before or after salinity treatment (Supplemental Figure 1). In addition, the central vacuole is an ideal location for Na+ sequestration to reduce the Na+ accumulation in the cytoplasm. NHX-type transporters play important roles in Na+ translocation from cytoplasm to vacuole through a Na+/H+ exchanger (Blumwald and Poole, 1985; Gong et al., 2020). Nevertheless, our results showed that the expression levels of NHX1 and NHX5 in OsGF14C-OX lines were similar to that in wild type plants, both with and without salinity treatment (Supplemental Figure 1). In contrast, the expression levels of HKT-type transporters including HKT1, HKT4, and HKT6, which mediate the translocation of Na+ from extracellular to cytoplasm in OsGF14C-OX plants were significantly lower than that in wild type plants, both before and after salinity treatment (Figure 6). Taken together, these results suggest that overexpression of OsGF14C may positively regulate salinity tolerance by lowering the expression of HKT-type transporter genes and inhibiting the Na+ uptake instead of exclusion or compartmentation.
LOX2 may play important roles in coordinating salinity tolerance and blast resistance in rice
In natural environment, plants are often subjected to both abiotic (such as drought, salt, cold) and biotic (necrotrophic and biotrophic pathogens) stresses simultaneously, which likely demands cross-talk between stress-response pathways to minimize the fitness costs. It is essential to find out the genes which could play roles in different stresses resistance and understand how the various stress response pathways interact with one another. In the present study, overexpression of OsGF14C enhanced the resistance to salinity stress but reduced the resistance to blast in rice. The opposite roles of OsGF14C overexpression played in salinity stress and blast disease are consistent with the previous findings that plants exposed to abiotic stresses such as high salinity and drought often display reduced immune activity (Bostock et al., 2014). It is understandable for this conflict as stress responses are costly, plants likely coordinate these responses to minimize fitness costs.
According to previous studies, LOXs, the key enzymes of JA synthesis, have been reported to involve in various physiological processes, including responses to biotic and abiotic stresses (Mostofa et al., 2015). Chu et al. (2019) showed that the Arabidopsis plants inoculated with Pseudomonas PS01 could survive under 225 mM NaCl, while the non-inoculated plants were dead above 200 mM NaCl. The gene expression analysis of the genes related to stress resistance indicated that only LOX2 was up-regulated in the inoculated plants in comparison to the non-inoculated controls (Chu et al., 2019). Furthermore, Zhang et al. (2018b) found that LOX2 and LOX5 were specifically suppressed by Guy11 infection, which are the key JA synthesis genes hijacked by M. oryzae strain Guy11 to subvert host immunity and facilitate pathogenicity. They also proposed that induced expression of OsLOX2/5 may improve resistance to the blast disease in rice (Zhang et al., 2018b). In the present study, our results showed that the expression levels of LOX2 were significantly higher in the OsGF14C-OX plants than that in wild type plants. Given that LOX genes play important roles in regulating both biotic and abiotic stresses in plants, the increased expressions of LOX2 in OsGF14C-OX plants implied LOX2 may partially lead to the differences responses of transgenetic plants to salinity and blast resistance. However, despite that OsGF14C-OX plants showed increased expression of LOX2, the transgenetic plants were more sensitive to rice blast disease than their wild type plants, which seems to be conflict with previous studies. But we also detected reduced expression of some PR genes in OsGF14C-OX plants, which may be the principal reason for the reduction of blast resistance. This also indicated the complexity between different stress response pathways and the importance for understanding the crosstalk between different stress pathways. According to the results from this study, LOX2 may play important roles in coordinating salinity tolerance and blast resistance in rice. However, it need more studies to confirm if LOX2 is the switch gene and how it regulates the salinity tolerance and blast resistance in rice. It is of great significance to study the regulation mechanisms of LOX2 underlying different stresses in crops.
Conclusion
In conclusion, in the present study, we have functionally confirmed that overexpression of OsGF14C enhances salinity tolerance but reduces blast resistance in rice. The negative role of overexpression of OsGF14C plays in blast resistance may be associated with the suppression of OsGF14E, OsGF14F and PR genes. The improvement of salinity tolerance by overexpression of OsGF14C may be resulted from reducing the level of methylglyoxal and Na+ uptake instead of exclusion or compartmentation. Our results and the previous studies together suggest that the lipoxygenase gene LOX2 may an important gene which play roles in both abiotic and biotic stresses. Finally, a molecular model of OsGF14C-mediated salinity and blast resistance pathways in rice was proposed (Figure 9).
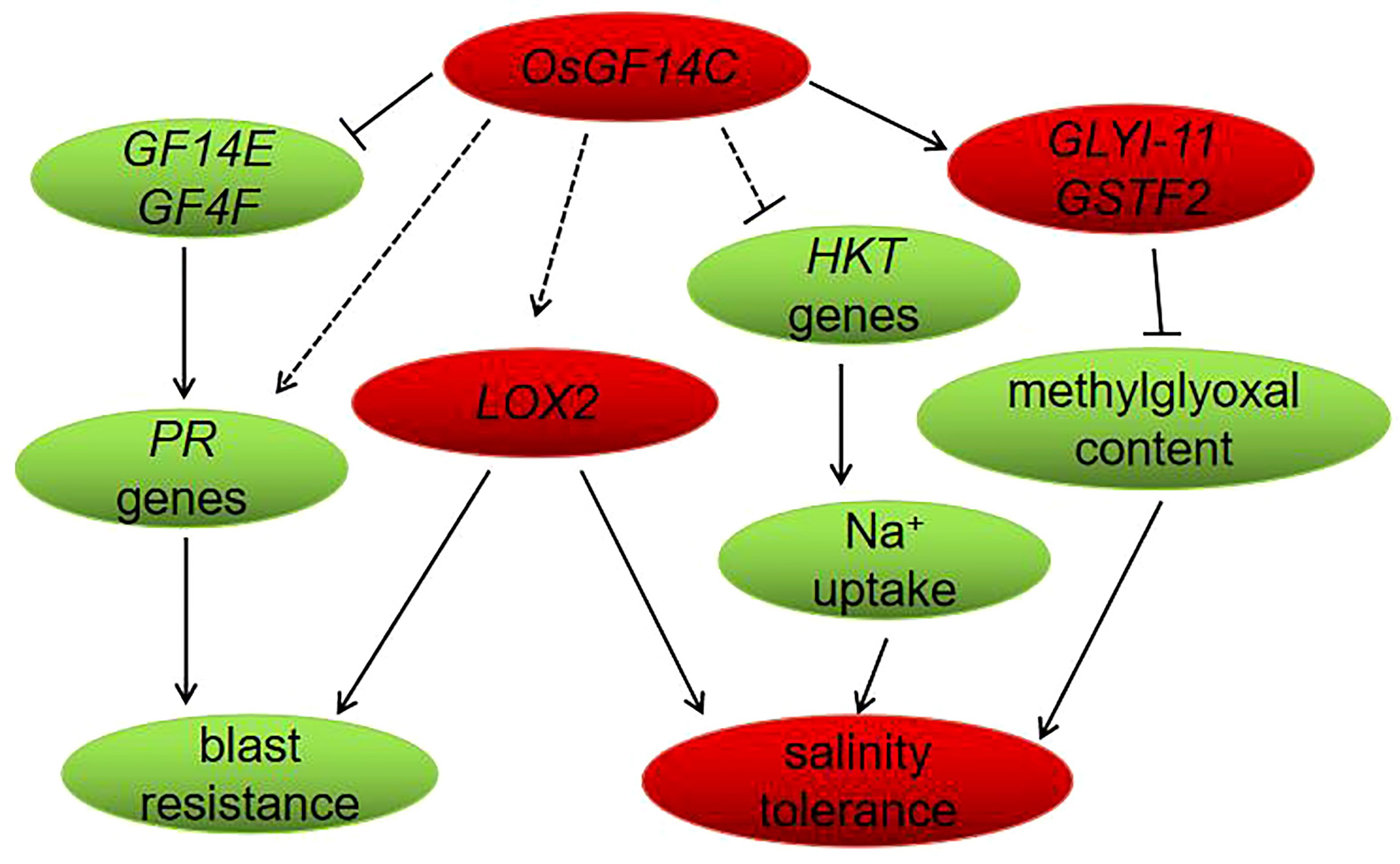
Figure 9 Proposed model of OsGF14C in salinity tolerance and blast resistance in rice. When the expression level of OsGF14C is up regulated, the expression of GF14E, GF14F, PR genes, and HKT genes were repressed, while the expression of GLYI-11, GSTF2, and LOX2 were induced.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
JD conducted the experiments, performed data analysis and wrote the manuscript. XL participated in writing editing and format modification, JY and XZ provided the strain of rice fungus. YM, JC, WY, LiaZ, and JW participated in material development, sample preparation and data analysis. TY, SZ, QL and LinZ provided technical assistance. BL and JZ conceived and designed the experiment, and writing - review & editing. All authors contributed to the article and approved the submitted version.
Funding
This work was jointly supported by grants from the Natural Science Foundation of Guangzhou City (202102021006), the Excellent Valley program of Rice Research Institute, Guangdong Academy of Agricultural Sciences (2021YG10, 2021YG001), the Natural Science Foundation of Guangdong Province (2019A1515010003), the Key Areas Research Projects of Guangdong Province (2020B0202090003), the Innovation Team Project of Guangdong Modern Agricultural Industrial System (2022KJ106), and the foundation of Guangdong Key Laboratory of New Technology in Rice Breeding (2020B1212060047).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1098855/full#supplementary-material
Supplementary Figure 1 | Expression analysis of SOS1, NHX1, and NHX5 genes in the GF14C transgenic plants (OX-9, OX-15) and control plants by quantitative real-time PCR before (0 h) and after (24 h) salinity treatment. Error bars indicate the SD from three biological replicates.
References
Apse, M. P., Aharon, G. S., Snedden, W. A., Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in arabidopsis. Science 285 (5431), 1256–1258. doi: 10.1126/science.285.5431.1256
Blumwald, E., Poole, R. J. (1985). Na+ /H+ antiport in isolated tonoplast vesicles from storage tissue of beta vulgaris. Plant Physiol. 78 (1), 163–167. doi: 10.1104/pp.78.1.163
Bostock, R. M., Pye, M. F., Roubtsova, T. V. (2014). Predisposition in plant disease: Exploiting the nexus in abiotic and biotic stress perception and response. Annu. Rev. Phytopathol. 52 (1), 517–549. doi: 10.1146/annurev-phyto-081211-172902
Chen, F., Li, Q., Sun, L. X., He, Z. H. (2006). The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res. 13 (2), 53–63. doi: 10.1093/dnares/dsl001
Chu, T. N., Tran, B. T. H., Bui, L. V., Hoang, M. T. T. (2019). Plant growth-promoting rhizobacterium pseudomonas PS01 induces salinity tolerance in arabidopsis thaliana. BMC Res. Notes 12 (1), 11. doi: 10.1186/s13104-019-4046-1
Conti, L., Price, G., O'Donnell, E., Schwessinger, B., Dominy, P., Sadanandom, A. (2008). Small ubiquitinlike modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in arabidopsis. Plant Cell 20 (10), 2894–2908. doi: 10.1105/tpc.108.058669
Cooper, B., Clarke, J. D., Budworth, P., Kreps, J., Hutchison, D., Park, S., et al. (2003). A network of rice genes associated with stress response and seed development. Proc. Natl. Acad. Sci. 100 (8), 4945–4950. doi: 10.1073/pnas.0737574100
Dong, J. F., Zhao, J., Zhang, S. H., Yang, T. F., Liu, Q., Mao, X. X., et al. (2019). Physiological and genome-wide gene expression analyses of cold-induced leaf rolling at the seedling stage in rice (Oryza sativa l.). Crop J. 13 (4), 431–443. doi: 10.1016/j.cj.2019.01.003
Dong, J. F., Zhou, L., Feng, A. Q., Zhang, S. H., Fu, H., Chen, L., et al. (2021). The OsOXO2, OsOXO3 and OsOXO4 positively regulate panicle blast resistance in rice. Rice 14 (1), 51. doi: 10.1186/s12284-021-00494-9
Finni, C., Andersen, C. H., Borch, J., Gjetting, S., Christensen, A. B., de Boer, A. H., et al. (2002). Do 14-3-3 proteins and plasma membrane h+-AtPases interact in the barley epidermis in response to the barley powdery mildew fungus? Plant Mol. Biol. 49 (2), 137–147. doi: 10.1023/A:1014938417267
Fu, J., Liu, H. B., Li, Y., Yu, H. H., Li, X. H., Xiao, J. H., et al. (2011). Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol. 155 (1), 589–602. doi: 10.1104/pp.110.163774
Ganapati, R. K., Naveed, S. A., Zafar, S., Wang, W. S., Xu, J. L. (2022). Saline-alkali tolerance in rice: Physiological response, molecular mechanism, and QTL identification and application to breeding. Rice Sci. 29 (5), 412–434. doi: 10.1016/j.rsci.2022.05.002
Gong, Z. Z., Xiong, L. M., Shi, H. Z., Yang, S. H., Herrera-Estrella, L. R., Xu, G. H., et al. (2020). Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 63 (5), 635–674. doi: 10.1007/s11427-020-1683-x
Hao, Z. N., Wang, L. P., Huang, F. D., Tao, R. X. (2012). Expression patterns of defense genes in resistance of the panicles exserted from the caulis and from the tillers to neck blast in rice. Plant Physiol. Biochem. 60, 150–156. doi: 10.1016/j.plaphy.2012.08.006
Hossain, M. A., Hossain, M. Z., Masayuki, F. (2009). Stress-induced changes of methyglyoxal level and glyoxalase I activity in pumpkin seedlings and cDNA cloning of glyoxalase I gene. Aust. J. Crop Sci. 3 (2), 53–64. doi: 10.2134/agronj2008.0083x
Jacobs, J., Wang, G. L. (2021). Next generation rice disease research. Rice 14, 84. doi: 10.1186/s12284-021-00523-7
Kaur, A., Pati, P. K., Pati, A. M., Nagpal, A. K. (2017). In-silico analysis of cis-acting regulatory elements of pathogenesis-related proteins of arabidopsis thaliana and oryza sativa. PLos One 12 (9), e0184523. doi: 10.1371/journal.pone.0184523
Kesten, C., Wallmann, A., Schneider, R., McFarlane, H. E., Diehl, A., Khan, G. A., et al. (2019). The companion of cellulose synthase 1 confers salt tolerance through a tau-like mechanism in plants. Nat. Commun. 10 (1), 857. doi: 10.1038/s41467-019-08780-3
Kou, Y. J., Wang, S. P. (2010). Broad-spectrum and durability: understanding of quantitative disease resistance. Curr. Opin. Plant Biol. 13 (2), 181–185. doi: 10.1016/j.pbi.2009.12.010
Li, T., Sun, Y. Q., Ruan, Y., Xu, L. J., Hu, Y. J., Hao, Z. P., et al. (2016). Potential role of d-myo-inositol-3-phosphate synthase and 14-3-3 genes in the crosstalk between zea mays and rhizophagus intraradices under drought stress. Mycorrhiza 26 (8), 879–893. doi: 10.1007/s00572-016-0723-2
Liu, C. T., Mao, B. G., Yuan, D. Y., Chu, C. C., Duan, M. J. (2022). Salt tolerance in rice:physiological responses and molecular mechanisms. Crop J. 10 (1), 13–25. doi: 10.1016/j.cj.2021.02.010
Liu, J. P., Sun, X. J., Liao, W. C., Zhang, J. H., Liang, J. S., Xu, W. F. (2019). Involvement of OsGF14b adaptation in the drought resistance of rice plants. Rice 12 (1), 82. doi: 10.1186/s12284-019-0346-2
Liu, Q., Yang, J. Y., Zhang, S. H., Zhao, J. L., Feng, A. Q., Yang, T. F., et al. (2016a). OsGF14b positively regulates panicle blast resistance but negatively regulates leaf blast resistance in rice. Mol. Plant Microbe Interact. 29 (1), 46–56. doi: 10.1094/MPMI-03-15-0047
Liu, Q., Yang, J. Y., Zhang, S. H., Zhao, J. L., Feng, A. Q., Yang, T. F., et al. (2016b). OsGF14e positively regulates panicle blast resistance in rice. Biochem. Biophys. Res. Commun. 471 (1), 247–252. doi: 10.1016/j.bbrc.2016.02.005
Ma, Y. M., Yang, J. Y., Dong, J. F., Zhang, S. H., Yang, W., Zhao, J. L., et al. (2022). Overexpression of OsGF14fEnhances quantitative leaf blast and bacterial blight resistance in rice. Int. J. Mol. Sci. 23 (13), 7440. doi: 10.3390/ijms23137440
Ma, L., Ye, J. M., Yang, Y. Q., Lin, H. X., Yue, L. L., Luo, J., et al. (2019). The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress. Dev. Cell 48 (5), 697–709. doi: 10.1016/j.devcel.2019.02.010
Manosalva, P. M., Bruce, M., Leach, J. E. (2011). Rice 14-3-3 protein (GF14e) negatively affects cell death and disease resistance. Plant J. 68 (5), 777–787. doi: 10.1111/j.1365-313X.2011.04728.x
Mostofa, M. G., Hossain, M. A., Fujita, M. (2015). Trehalose pretreatment induces salinity tolerance in rice (Oryza sativa l.) seedlings: Oxidative damage and co-induction of antioxidant defense and glyoxalase systems. Protoplasma 252 (2), 461–475. doi: 10.1007/s00709-014-0691-3
Munns, R., James, R. A., Xu, B., Athman, A., Conn, S. J., Jordans, C., et al. (2012). Wheat grain yield on saline soils is improved by an ancestral na+ transporter gene. Nat. Biotechnol. 30 (4), 360–364. doi: 10.1038/nbt.2120
Mustafiz, A., Ghosh, A., Tripathi, A. K., Kaur, C., Ganguly, A. K., Bhavesh, N. S., et al. (2014). A unique Ni2+ -dependent and methylglyoxal-inducible rice glyoxalase I possesses a single active site and functions in abiotic stress response. Plant J. 78 (6), 951–963. doi: 10.1111/tpj.12521
Oecking, C., Jaspert, N. (2009). Plant 14-3-3 proteins catch up with their mammalian orthologs. Curr. Opin. Plant Biol. 12 (6), 760–765. doi: 10.1016/j.pbi.2009.08.003
Oh, C. S., Pedley, K. F., Martin, G. B. (2010). Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKK {alpha}. Plant Cell 22 (1), 260–272. doi: 10.1105/tpc.109.070664
Roberts, M. R. (2003). 14-3-3 proteins find new partners in plant cell signaling. Trends Plant Sci. 8 (5), 218–223. doi: 10.1016/S1360-1385(03)00056-6
Shi, H., Ishitani, M., Kim, C., Zhu, J. K. (2000). The arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. 97 (12), 6896–6901. doi: 10.1073/pnas.120170197
Shi, H. Z., Lee, B. H., Wu, S. J., Zhu, J. K. (2003). Overexpression of a plasma membrane na+ /H+ antiporter gene improves salt tolerance in arabidopsis thaliana. Nat. Biotechnol. 21 (1), 81–85. doi: 10.1038/nbt766
Singla-Pareek, S. L., Reddy, M. K., Sopory, S. K. (2003). Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc. Natl. Acad. Sci. 100 (25), 14672–14677. doi: 10.1073/pnas.2034667100
Singla-Pareek, S. L., Yadav, S. K., Pareek, A., Reddy, M. K., Sopory, S. K. (2006). Transgenic tobacco overexpressing glyoxalase pathway enzymes grow and set viable seeds in zinc-spiked soils. Plant Physiol. 140 (2), 613–623. doi: 10.1104/pp.105.073734
Song, T. Z., Shi, Y. Y., Shen, L., Cao, C. J., Shen, Y., Jing, W., et al. (2021). An endoplasmic reticulum-localized cytochrome b5 regulates high-affinity k+ transport in response to salt stress in rice. Proc. Natl. Acad. Sci. 118 (50), e2114347118. doi: 10.1073/pnas.2114347118
Tezuka, D., Kawamata, A., Kato, H., Saburi, W., Mori, H., Imai, R. (2019). The rice ethylene response factor OsERF83 positively regulates disease resistance to magnaporthe oryzae. Plant Physiol. Biochem. 135, 263–271. doi: 10.1016/j.plaphy.2018.12.017
Tufail, A., Li, H., Naeem, A., Li, T. X. (2018). Leaf cell membrane stability-based mechanisms of zinc nutrition in mitigating salinity stress in rice. Plant Biol. 20 (2), 338–345. doi: 10.1111/plb.12665
Wang, P. H., Lee, C. E., Lin, Y. S., Lee, M. H., Chen, P. Y., Chang, H. C., et al. (2019). The glutamate receptor-like protein GLR3.7 interacts with 14-3-3ω and participates in salinity stress response in arabidopsis thaliana. Front. Plant Sci. 10, 1169. doi: 10.3389/fpls.2019.01169
Wu, H. H. (2018). Plant salt tolerance and na~+ sensing and transport. Crop J. 6 (03), 215–225. doi: 10.1016/j.cj.2018.01.003
Yan, J. Q., He, C. X., Wang, J., Mao, Z. H., Holaday, S. A., Allen, R. D., et al. (2004). Overexpression of the arabidopsis 14-3-3 protein GF14 lambda in cotton leads to a "stay-green" phenotype and improves stress resistance under moderate drought conditions. Plant Cell Physiol. 45 (8), 1007–1014. doi: 10.1093/pcp/pch115
Yang, J. W., Fu, J. X., Li, J., Cheng, X. L., Li, F., Dong, J. F., et al. (2014). A novel co-immunoprecipitation protocol based on protoplast transient gene expression for studying protein–protein interactions in rice. Plant Mol. Biol. Rep. 32 (1), 153–161. doi: 10.1007/s11105-013-0633-9
Yan, J. W., Yang, L., Liu, Y., Zhao, Y. D., Han, T., Miao, X. F., et al. (2022). Calcineurin b-like protein 5 (SiCBL5) in setaria italica enhances salt tolerance by regulating na+ homeostasis. Crop J. 10 (1), 234–242. doi: 10.1016/j.cj.2021.06.006
Yang, X. H., Wang, W. M., Coleman, M., Orgil, U., Feng, J. Y., Ma, X. F., et al. (2009). Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. Plant J. 60 (3), 539–550. doi: 10.1111/j.1365-313X.2009.03978.x
Yin, J. J., Zou, L. J., Zhu, X. B., Cao, Y. Y., He, M., Chen, X. W. (2021). Fighting the enemy: How rice survives the blast pathogen’s attack. Crop J. 9 (3), 543–552. doi: 10.1016/j.cj.2021.03.009
Zelm, E. V., Zhang, Y. X., Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71 (1), 403–433. doi: 10.1146/annurev-arplant-050718-100005
Zhang, Z. X., Zhang, Y. P., Zhao, H., Huang, F. L., Zhang, Z. F., Lin, W. X. (2017). The important functionality of 14-3-3 isoforms in rice roots revealed by affinity chromatography. J. Proteomics 158, 20–30. doi: 10.1016/j.jprot.2017.02.008
Zhang, Y., Zhao, H. Y., Zhou, S. Y., He, Y., Luo, Q. C., Zhang, F., et al. (2018a). Expression of TaGF14b, a 14-3-3 adaptor protein gene from wheat, enhances drought and salinity tolerance in transgenic tobacco. Planta 248 (1), 117–137. doi: 10.1007/s00425-018-2887-9
Zhang, X., Bao, Y. L., Shan, D. Q., Wang, Z. H., Song, X. N., Wang, Z. Y., et al. (2018b). Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiol. 177 (1), 352–368. doi: 10.1104/pp.17.01665
Zhao, J. L., Zhang, L. Q., Liu, N., Xu, S. L., Yue, Z. L., Zhang, L. L., et al. (2019). Mutual regulation of receptor-like kinase SIT1 and b’κ-PP2A shapes the early response of rice to salt stress. Plant Cell 31 (9), 2131–2151. doi: 10.1105/tpc.18.00706
Keywords: OsGF14C, salinity, blast, Na+ Uptake, Lox2, rice
Citation: Dong J, Li X, Ma Y, Yang J, Chen J, Yang W, Zhou L, Wang J, Yang T, Zhang S, Zhao J, Liu Q, Zhou L, Zhu X and Liu B (2023) Overexpression of OsGF14C enhances salinity tolerance but reduces blast resistance in rice. Front. Plant Sci. 14:1098855. doi: 10.3389/fpls.2023.1098855
Received: 15 November 2022; Accepted: 27 January 2023;
Published: 10 February 2023.
Edited by:
Mehdi Rahimi, Graduate University of Advanced Technology, IranReviewed by:
Muhammad Sajjad, COMSATS University, Islamabad Campus, PakistanLiqun Du, Hangzhou Normal University, China
Copyright © 2023 Dong, Li, Ma, Yang, Chen, Yang, Zhou, Wang, Yang, Zhang, Zhao, Liu, Zhou, Zhu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junliang Zhao, emhhb19qdW5saWFuZ0BnZGFhcy5jbg==; Bin Liu, bGJnejEwMDlAMTYzLmNvbQ==
 Jingfang Dong
Jingfang Dong Xuezhong Li1,2,3,4
Xuezhong Li1,2,3,4 Jianyuan Yang
Jianyuan Yang Wu Yang
Wu Yang Jian Wang
Jian Wang TiFeng Yang
TiFeng Yang Shaohong Zhang
Shaohong Zhang Junliang Zhao
Junliang Zhao Qing Liu
Qing Liu Lingyan Zhou
Lingyan Zhou Bin Liu
Bin Liu