- 1College of Life Science and Agronomy, Zhoukou Normal University, Henan, Zhoukou, China
- 2School of Journalism and Communication, Zhoukou Normal University, Henan, Zhoukou, China
HD-Zip is a plant-specific transcription factor that plays an important regulatory role in plant growth and stress response. However, there have been few reports on the functions of members of the physic nut HD-Zip gene family. In this study, we cloned a HD-Zip I family gene from physic nut by RT-PCR, and named JcHDZ21. Expression pattern analysis showed that JcHDZ21 gene had the highest expression in physic nut seeds, and salt stress inhibited the expression of JcHDZ21 gene. Subcellular localization and transcriptional activity analysis showed that JcHDZ21 protein is localized in the nucleus and has transcriptional activation activity. Salt stress results indicated that JcHDZ21 transgenic plants were smaller and had more severe leaf yellowing compared to those of the wild type. Physiological indicators showed that transgenic plants had higher electrical conductivity and MDA content, and lower proline and betaine content compared with wild-type plants under salt stress. In addition, the expression of abiotic stress-related genes in JcHDZ21 transgenic plants was significantly lower than that in wild type under salt stress. Our results showed that ectopic expression of JcHDZ21 increased the sensitivity of transgenic Arabidopsis to salt stress. This study provides a theoretical basis for the future application of JcHDZ21 gene in the breeding of physic nut stress-tolerant varieties.
Introduction
Salinity is one of the main abiotic stress factors that limit crop yields. Plants can initiate a series of corresponding stress tolerance mechanisms to resist environmental stress under abiotic stress conditions. During this process, almost all plants showed changes in the expression levels of various functional genes and regulatory protein genes. Among these genes, transcription factors can improve the tolerance of plants to extreme environmental stress by activating the expression of many downstream stress-resistant genes. The transcription factors related to abiotic stress discovered so far are mainly concentrated in the following gene family members, such as DREB, MYB, HD-Zip, WRKY and NAC (Dubos et al., 2010; Rushton et al., 2010; Harris et al., 2011; Javelle et al., 2011; Thirumalaikumar et al., 2018; Feng et al., 2020).
The HD-Zip family is a type of plant-specific transcription factor that contains a highly conserved homeodomain (HD) and a leucine zipper (LZ) domain (Johannesson et al., 2001). The HD-Zip protein forms a dimer through the LZ domain, and then acts as a transcription factor by combining its HD with a specific DNA sequence (Deng et al., 2002). According to the conservative and structural characteristics of the amino acid sequence, the family can be divided into 4 subfamilies, named HD-Zip I-IV (Henriksson et al., 2005). Most of the HD-Zip proteins related to plant stress response belong to HD-Zip I subfamily, and the expression of this family member is mainly affected by drought, salt, cold and osmotic pressure, and increasing the expression of these genes can change the tolerance of transgenic plants to extreme environmental stress (Harris et al., 2011). For example, overexpression of ATHB-6 increases drought stress tolerance in transgenic maize by activating the expression of reactive oxygen species-related genes (Jiao et al., 2022), PsnHDZ63 transgenic plants showed better phenotypic and physiological indicators under salt stress, indicating that increasing the expression of the PsnHDZ63 gene altered the tolerance of transgenic plants to salt stress (Guo et al., 2021). Overexpression of maize Zmhdz4 and Zmhdz10 genes in rice increases the tolerance of transgenic plants to drought and salt stress (Zhang et al., 2012; Zhao et al., 2014). In addition to participating in abiotic stress regulation, HD-Zip family genes also play important roles in plant growth and development (Turchi et al., 2013; Preciado et al., 2022). ABIG1 plays a role in leaf laminar growth as well as in adaxial-abaxial polarity establishment (Preciado et al., 2022). Overexpression of ATHB13 affects cotyledon shape by inhibiting lateral expansion of epidermal cells in sugar-treated seedlings (Hanson et al., 2001). OsHOX1 and OsHOX28 regulate the local distribution of auxin by reducing endogenous auxin content, thereby regulating the size of the tiller angle (Hu et al., 2020). Although many HD-Zip genes have been cloned from different plants, the physiological and molecular mechanisms by which HD-Zip family genes regulate plant responses to salt stress, especially in salt-tolerant Euphorbiaceae species, need to be further studied.
Physic nut (Jatropha curcas), a Euphorbiaceae species, has high salinity tolerance, and rich in oils suitable for production of biodiesel, which is one of the most promising biomass energy plants (Montes and Melchinger, 2016). Although physic nut has strong salt tolerance, the molecular mechanism of how physic nut responds to salt stress tolerance is still unclear. In previous study, we notice that a HD-Zip I family transcription factor, we named JcHDZ21, which is strongly repressed expression by salinity stress (Tang et al., 2019). On this basis, we cloned the JcHDZ21 gene and investigated its function in Arabidopsis. Up-regulation of JcHDZ21 gene expression increases the sensitivity of transgenic Arabidopsis to salt stress, indicating that JcHDZ21 acts as a negative regulator in the regulation of plant responses to salt stress. The results provide foundations for exploring roles of JcHDZ21 gene in responses to salinity stress in physic nut.
Material and methods
Plant material and stress treatment
After the physic nut seeds germinated, the three-week-old physic nut seedlings with uniform growth were divided into three groups for abiotic stress, and each group contained twenty seedlings. Then, the physic nut seedlings in the first and second groups were used to irrigate 1/2 MS nutrient solution containing 150 mM NaCl for salinity stress treatment. After the stress treatment, the fourth leaves of five treatment time points (0 h, 2 h, 4 h, 12 h and 24 h) were selected for RNA extraction and gene expression detection. In addition, the roots, cortex stem, flowers, and leaves of the 3-week-old physic nut seedlings without stress treatment were taken for tissue expression analysis. The material was quickly frozen in liquid nitrogen and stored in a refrigerator at -80°C. The growth conditions of physic nut under normal growth and stress treatment were as follows: 16 h light/8 h dark, and the temperature was controlled at 28°C.
JcHDZ21 protein characterization and sequence alignment analysis
Download the amino acid sequence of Arabidopsis proteins from TAIR database (https://www.arabidopsis.org/). Download the amino acid sequence of Oryza sativa L. and Zea mays L. proteins from NCBI database (https://www.ncbi.nlm.nih.gov/). SMART online software and NCBI were used to detect the conserved domain of JcHDZ21 protein. We selected HD-Zip I group proteins with positive regulator function in abiotic stress for multiple sequence alignment analysis. Amino acid multiple sequence alignment analysis was performed using the software DNAMAN XL.
Subcellular localization of JcHDZ21 protein
Using physic nut leaf cDNA as a template, the full-length CDS sequence of the JcHDZ21 gene (with the stop codon removed) was amplified by RT-PCR and verified by sequencing. Then connect the correct gene sequence to the subcellular localization vector to construct pBWA(V)HS-JcHDZ21-GLosgfp fusion expression vector. Finally, the constructed pBWA(V)HS-JcHDZ21-GLosgfp fusion expression vector and empty vector (control vector) were co-transformed into Arabidopsis protoplasts by PEG-mediated method. The isolation and extraction of Arabidopsis mesophyll protoplasts and the PEG-mediated transient transformation were carried out according to the method in the previously reported paper (Yoo et al., 2007). Add 100 μL of plasmid DNA to the 100 μL of protoplast suspension. Add 200 μL of PEG4000 solution to the protoplast suspension, store the tube on room temperature for 30 min. Finally, add 1 mL of W5 solution to the mixture, and culture in the dark at 23°C for 16 h, and then the fluorescence was observed under a laser confocal microscope (LSM800, Carl Zeiss).
Transcriptional activity assay
The cDNA sequence of JcHDZ21 was combined with the GAL4 DNA binding domain on the pGBKT7 expression vector to construct the pGBKT7-JcHDZ21 recombinant expression vector. The recombinant expression vector was transferred into Y2HGold yeast competent cells by PEG/LiAc method, and cultured on SD/-Trp solid medium for 3-5 d. Then pick a single colony and inoculate it in SD/-Trp liquid medium. Taking 2.5 uL of pGBKT7-GAL4, pGBKT7, pGBKT7-JcHDZ21 bacterial solution dropwise on SD/-Trp, SD/-Trp/-His/X-a-gal and SD/-Trp/-His/-Ade/X-a-gal plates. Finally, verify transcriptional activity based on colony growth and whether it turns blue.
Construction of plant expression vector and selection of transgenic plants
Using physic nut root cDNA as a template, the full-length cDNA sequence of JcHDZ21 was amplified by RT-PCR and verified by sequencing. Subsequently, the target gene and plant expression vector were digested by Kpn I and Xba I. After electrophoresis, gel excision, and recovery of DNA fragments, the target gene was ligated to the binary transformation vector pCAMBIA1301 by T4 DNA ligase, and the target gene was driven by the CaMV 35S promoter. The vector was transformed into Agrobacterium strain GV3101, and then we transformed the constructed vector into Arabidopsis thaliana (Columbia ecotype) by Agrobacterium-mediated transformation of Arabidopsis using the floral dip method (Zhang et al., 2006). Homozygous plants of the T3 generation were selected in hygromycin-resistant medium. The transgenic Arabidopsis was then identified by GUS staining and RT-PCR techniques. Finally, the effective three transgenic lines were used for subsequent functional analysis.
Analysis of salt tolerance of transgenic Arabidopsis
To avoid mechanical root damage of 4-day-old Arabidopsis seedlings during transferring for salinity treatment, sterile seeds were sown directly into 1/2 MS solid medium containing 100 mM NaCl in petri dishes for salinity stress. Then put them in the growing room to grow at 22 ± 2°C under a 16 h light/8 h dark photoperiod. After 25 days, observing the salt tolerance of wild-type and transgenic plants. Salt stress treatment experiments contained three biological replicates.
Determination of physiological indicators of wild-type and transgenic plants
Seeds of wild-type and transgenic plants were spotted into 1/2 MS solid medium containing 0 and 100 mM NaCl in petri dishes. The leaves of wild-type and transgenic plants growth in control and salt stress conditions for 15 days were taken, washed twice with deionized water, and then the surface water of the leaves was blotted with filter paper. Accurately weigh 0.1 g and cut into filaments and place in a test tube containing 10 mL of deionized water. The test tube was then placed in a vacuum pump for 30 min. After standing at room temperature for 1 h, we used a HANNA 546808 (Italy) conductivity meter to measure the initial conductivity value and named R1. Immediately after heating the test tube in boiling water for 30 min, the running water was cooled to room temperature and shaken well, the conductance value R2 was measured. Calculate the relative conductivity using the following formula: R1/R2×100%. The betaine content was detected by ELISA kit, the specific operation method is shown in the kit instructions. The detection methods of proline and MDA content refer to the papers published by the previous research group (Tang et al., 2019).
RNA isolation and qRT-PCR analysis
Seeds of wild-type and transgenic plants were spotted into 1/2 MS solid medium containing 0 and 100 mM NaCl in petri dishes. Then culture vessels containing the seeds were incubated at 22 ± 2°C under 16/8 h (light/dark) conditions. The leaves of wild-type and transgenic plants growth in control and salt-stressed conditions for 15 days were collected and used for qRT-PCR analysis. Plant RNA extraction kit (Magen, China) was used to extract the total RNA of each tissue in this study, and then the extracted RNA was synthesized into cDNA by TAKARA reverse transcription kit. TB Green® Fast qPCR Mix (TAKARA, Beijing) was used for real-time fluorescence quantitative PCR detection, and the BIO-RAD Mini Opticon fluorescence quantitative analyzer was used for qRT-PCR reaction and analysis. Each reaction contains 100 ng cDNA, primers (10 μmol/L) 0.4 μL, SuperMix 10 μL, PassiveDye 0.4 μL, add ddH2O to 20 μL. The amplification program was initial denaturation at 94°C for 30 s, denaturation at 94°C for 5 s, annealing at 60°C for 15 s, extension at 72°C for 34 s, 40 cycles. All the above specific operating methods were carried out according to the kit instructions. Physic nut JcActin (GenBank Accession JQ806331) and Arabidopsis Actin2 (GenBank Accession AT3G18780) were used as internal reference genes. Primer Premier 5 was used for the design of the gene-specific qRT-PCR primers. Primer sequences were shown in Supplementary Table 1. The specificity of qRT-PCR amplification was confirmed by melting curve analysis. Then using the 2–ΔΔCt method to calculate the relative expression of genes in different samples based on the Ct value of each sample at a specific fluorescence threshold.
Statistical analysis
The SPSS 25.0 software was used for data analysis. Significance analysis was performed using One-Way ANOVA for LSD testing of SPSS 25.0 data processing system (* P<0.05; ** P<0.01).
Results
JcHDZ21 encodes a HD-Zip protein belonging to HD-Zip I subfamily
In order to study the biological function of JcHDZ21 gene, we cloned the gene by RT-PCR using physic nut leaf cDNA as template. Sequence analysis showed that the full-length open reading frame of JcHDZ21 was 969 bp, encoding 322 amino acids. SMART analysis revealed that JcHDZ21 contained a highly conserved HD domain and an LZ domain.
We further analyzed the homology between JcHDZ21 and HD-Zip proteins of other species by DNAMAN XL software. Sequence alignment analysis showed that, like the reported HD-Zip proteins, JcHDZ21 also contained the same characteristic regions named homeodomain and leucine zipper domains (Figure 1). Results also showed that many amino acids residues distributed in the N-terminal and C-terminal of HD-Zip protein were variable. Taken together, these results indicated that the different functions of HD-Zip proteins in different species may be attributed to these highly variable amino acid residues.
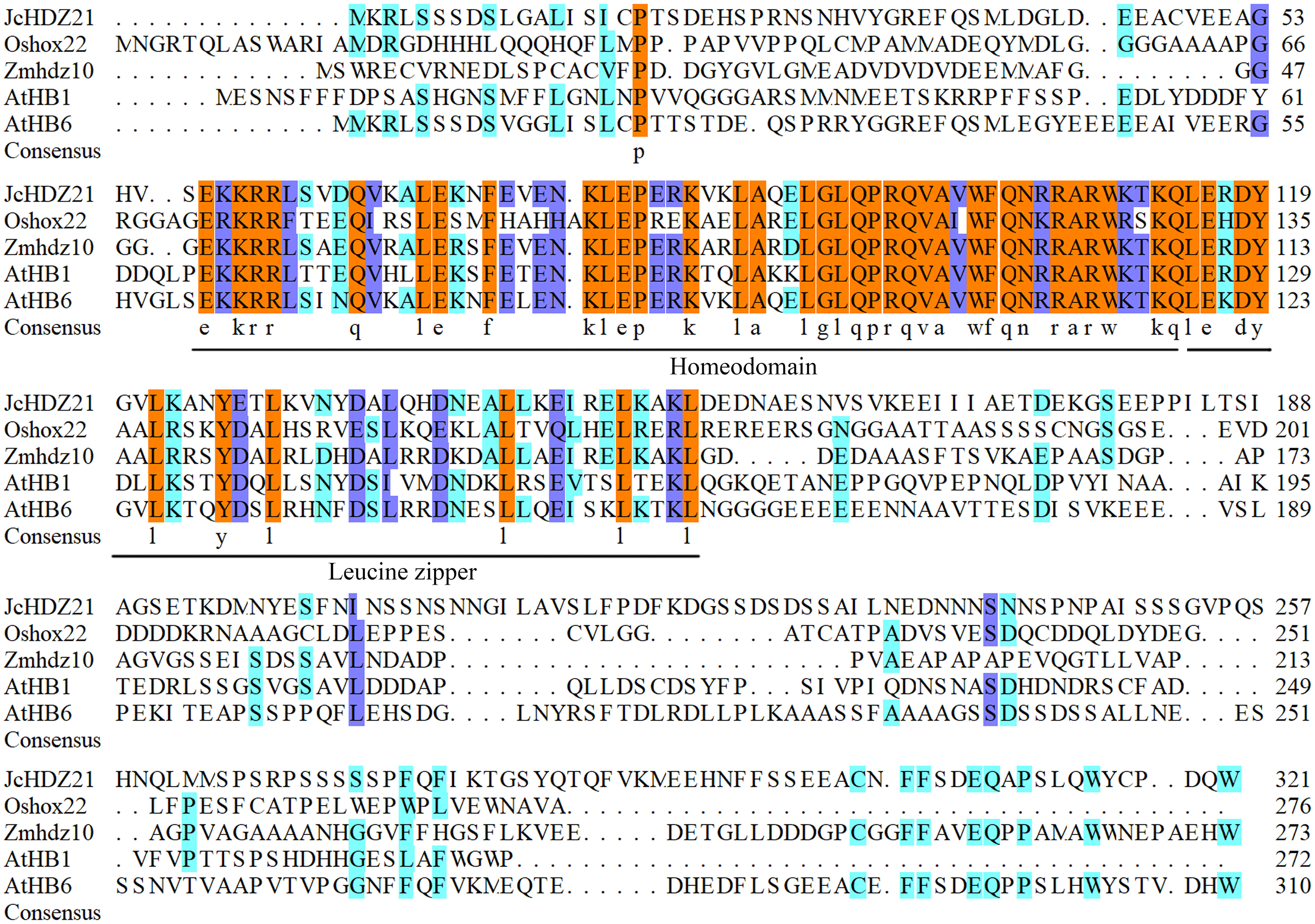
Figure 1 Sequence alignment analysis of JcHDZ21 and HD-Zip I family proteins that have been functionally studied. The DNAMAN XL software was used to perform the sequence alignment analysis.
Expression profile of JcHDZ21 gene
To study the expression profile of JcHDZ21 gene, we analyzed the expression of JcHDZ21 gene in roots, stems and leaves of physic nut at six-leaf stage, flowers and seeds 35 days after pollination by qRT-PCR. The results showed that the expression of JcHDZ21 gene could be detected in all tissues of physic nut, with the highest expression in seeds, followed by flowers and roots, and the lowest expression in stems (Figure 2A).
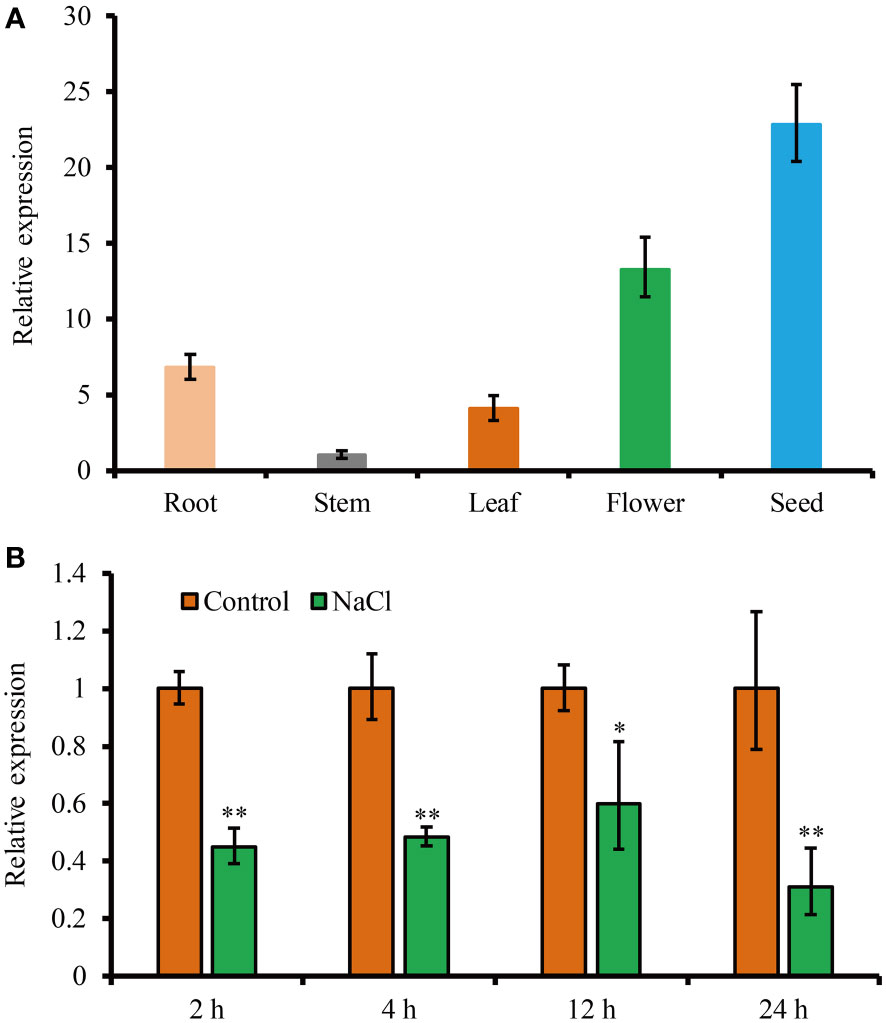
Figure 2 Expression level of JcHDZ21 gene in physic nut growth in control and salt stress conditions. (A) Expression levels of JcHDZ21 gene in different tissues of physic nut under normal growth conditions. (B) Expression level of JcHDZ21 gene in physic nut leaves treated with 150 mM NaCl. Data are from three independent biological replicates, asterisks represent significant differences (p<0.01).
In order to analyze the expression characteristics of JcHDZ21 gene under salt stress, we further detected the expression level of JcHDZ21 gene under salt stress. The results showed that the expression of JcHDZ21 under salt stress was significantly lower than that in the control group (Figure 2B). The above results suggest that the JcHDZ21 gene can be inhibited by salt stress, indicating that the JcHDZ21 gene may be involved in the regulation of plants to salt stress.
Subcellular localization of JcHDZ21 protein
To clarify the subcellular localization of JcHDZ21 protein, we transformed p16318hGFP-JcHDZ21 fusion expression vector and GFP empty vector plasmid into Arabidopsis protoplasts, and observed the subcellular localization of JcHDZ21-GFP fusion protein under laser confocal microscope. The result showed that the GFP empty control protein was expressed in all cells, while the JcHDZ21-GFP fusion protein suggested green fluorescence only in the nucleus (Figure 3), indicating that the JcHDZ21 protein is localized in the nucleus.
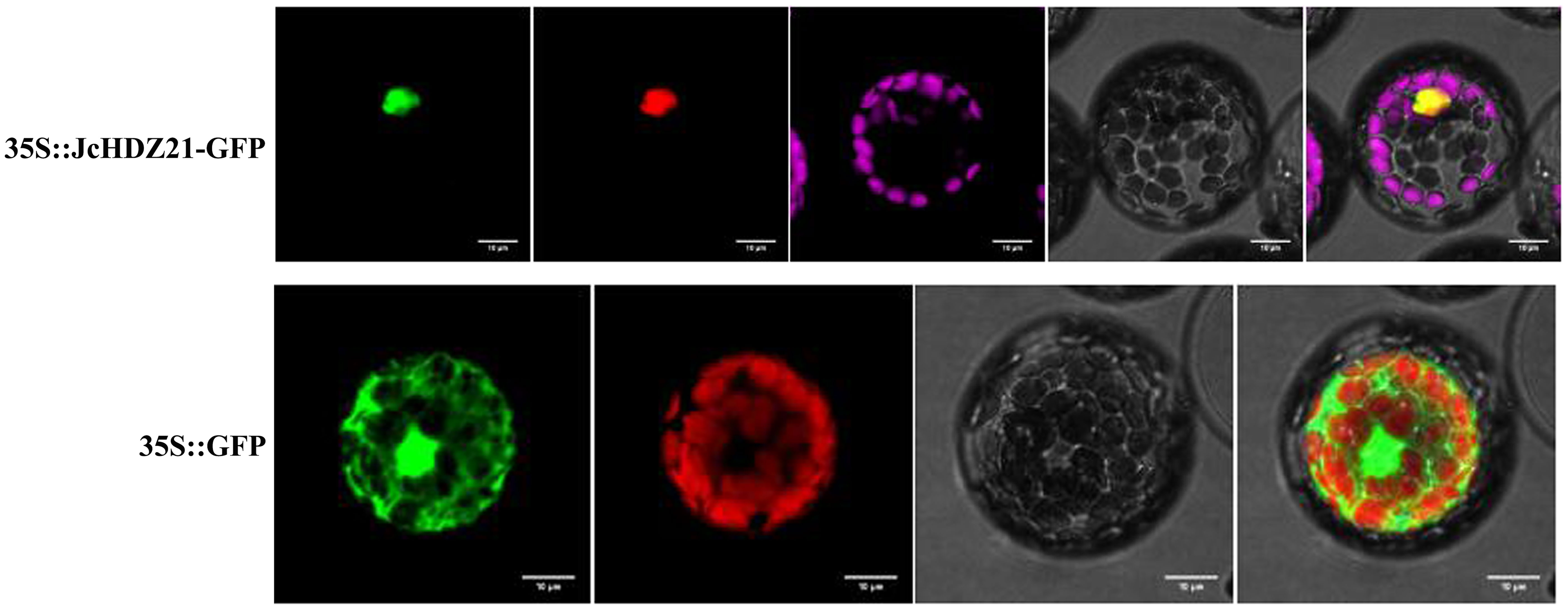
Figure 3 Subcellular localization of JcHDZ21 protein. 35S::JcHDZ21-GFP shows GFP, NLS-Cherry, Chlorophy II, Bright and Merge from left to right, whereas 35S::GFP shows GFP, Chlorophy II, Bright and Merge from left to right. Bars shows 10 μm.
Analysis of JcHDZ21 protein transcriptional activity
In order to further explore the transcriptional activity of JcHDZ21 protein, we used the yeast two-hybrid system to detect the transcriptional activity of JcHDZ21 in yeast. The results showed that all the combined vectors could grow on SD/-Trp medium. On SD/-Trp/-His/X-a-gal medium, the negative control grew weakly and did not turn blue, but pGBKT7-JcHDZ21 recombinant expression vector and the pGBKT7-GAL4 plaques showed blue. However, on SD/-Trp/-His/-Ade/X-a-gal medium, the pGBKT7 vector could not grow, but the pGBKT7-JcHDZ21 recombinant expression vector and the pGBKT7-GAL4 could grow well and the plaques were blue (Figure 4). These results indicate that JcHDZ21 acts as a transcriptional activator.
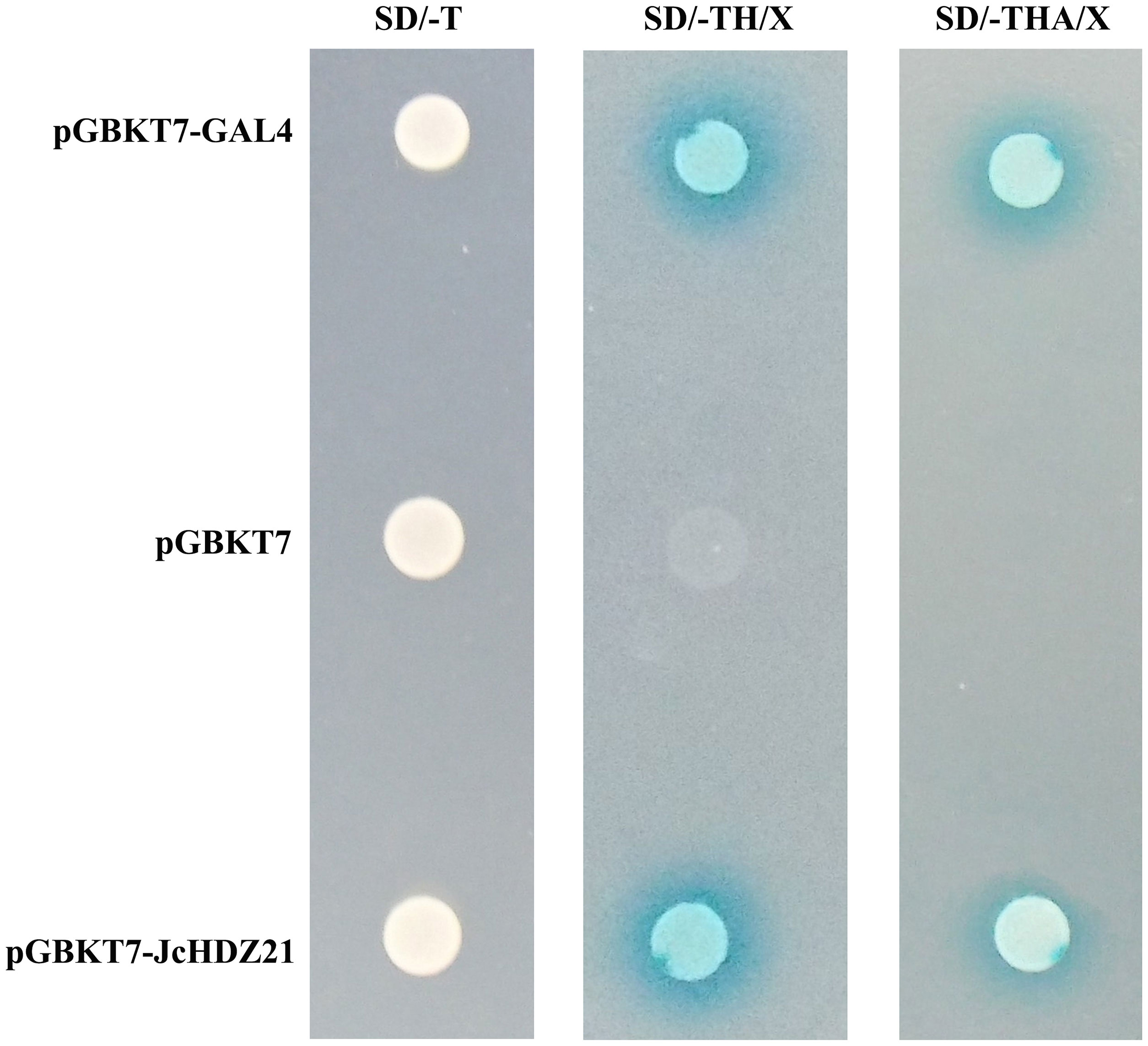
Figure 4 JcHDZ21 protein transcriptional activity. The left panel shows pGBKT7-GAL4, pGBKT7 and pGBKT7-JcHDZ21 grown on SD/-Trp plates for 4d, and the middle panel shows pGBKT7-GAL4, pGBKT7 and pGBKT7-JcHDZ21 grown on SD/-Trp/-His/X-a-gal plates for 4d, and the right panel shows pGBKT7-GAL4, pGBKT7 and pGBKT7-JcHDZ21 grown on SD/-Trp/-His/-Ade/X-a-gal plates for 4d.
Phenotypic analysis of JcHDZ21 transgenic plants
To verify the function of the JcHDZ21 gene, we constructed a JcHDZ21 overexpression vector and obtained JcHDZ21 overexpressing transgenic Arabidopsis plants. Homozygous plants of the T3 generation were selected in hygromycin-resistant medium for further study. We further detected the expression of JcHDZ21 gene in wild-type and transgenic plants by RT-PCR. The results showed that high expression levels of JcHDZ21 were detected in transgenic plants, but no expression was detected in wild-type plants (Figure 5A). We selected 4-week wild-type and transgenic plants for phenotype observation. Phenotypic analysis showed that there was no significant difference in the growth of JcHDZ21 transgenic plants and wild-type plants (Figures 5B, C). Statistical data showed that JcHDZ21 transgenic plants had no significant difference in root length compared with wild-type plants (Figure 5D). We further analyzed the flowering time and yield per plant of JcHDZ21 transgenic plants. The results showed that the flowering time and yield per plant of JcHDZ21 transgenic plants were not significantly different from those of wild-type plants (Figures 5E, F), indicating that ectopic expression of JcHDZ21 gene did not affect the growth and development of transgenic Arabidopsis.
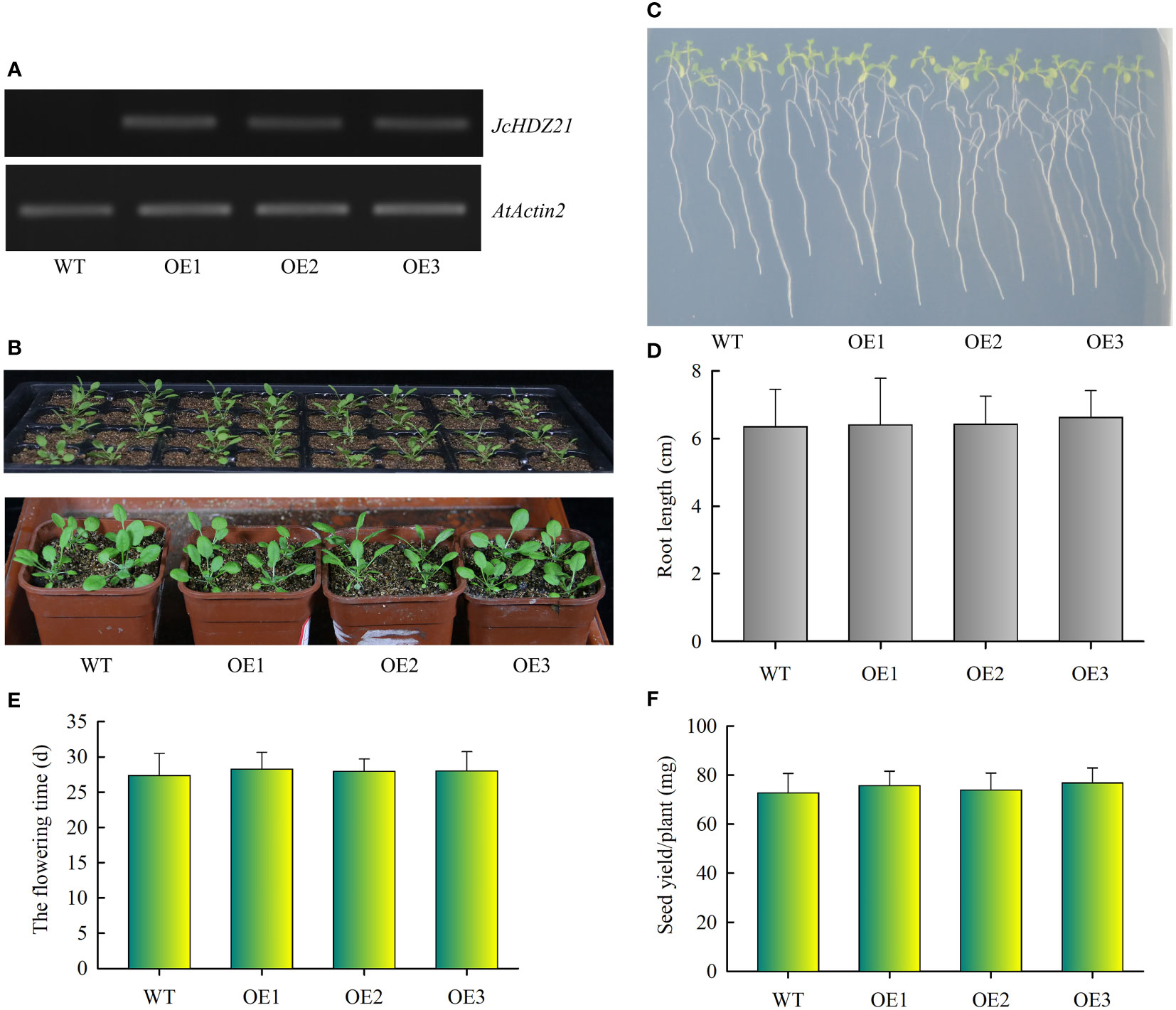
Figure 5 Phenotype of JcHDZ21 transgenic Arabidopsis plants under normal growth condition. (A) Expression levels of JcHDZ21 in wild-type and transgenic plants. (B) Growth status of wild-type and JcHDZ21 transgenic plants at 25 days. (C) Root morphology of wild-type and JcHDZ21 transgenic plants at day 10. The seeds of the sterilized wild-type and JcHDZ21 transgenic plants were sown on 1/2 MS medium for vertical cultivation for 4 days, and the plants with consistent growth were selected and transferred to 1/2 MS medium for vertical growth for 6 days. (D) Root length of wild-type and JcHDZ21 transgenic plants at day 10. Data in (D): means of n = 20 ± SD from three independent experiments. (E) The flowering time of wild-type and JcHDZ21 transgenic plants. (F) Seed yield of wild-type and JcHDZ21 transgenic plants under normal growth conditions. Values represent means of n = 30 ± SD from three independent experiments.
JcHDZ21 negatively regulates the tolerance of transgenic Arabidopsis to salt stress
Salt stress can repress the expression of JcHDZ21 gene, indicating that JcHDZ21 might be involved in plant response to salt stress. To prove this hypothesis, we directly spread the sterilized transgenic and wild type Arabidopsis seeds onto 1/2 MS medium containing 100 mM NaCl for 25 days. The results showed that the leaves of transgenic and wild-type Arabidopsis showed different degrees of yellow and smaller symptoms, but the degree of yellow of wild-type plants leaves was significantly lower than that of transgenic Arabidopsis plants, and the leaves of transgenic Arabidopsis were smaller than that of wild-type plants (Figure 6A). The growth status and survival rate of transgenic Arabidopsis plants were significantly lower than those of wild-type plants. The survival rates of JcHDZ21 transgenic plants were 22.58%, 21.24% and 19.27%, respectively, while the survival rate of wild-type plants reached 76.21%. The above results indicate that the heterologous expression of physic nut JcHDZ21 gene significantly enhances the sensitivity of transgenic Arabidopsis to salt stress.
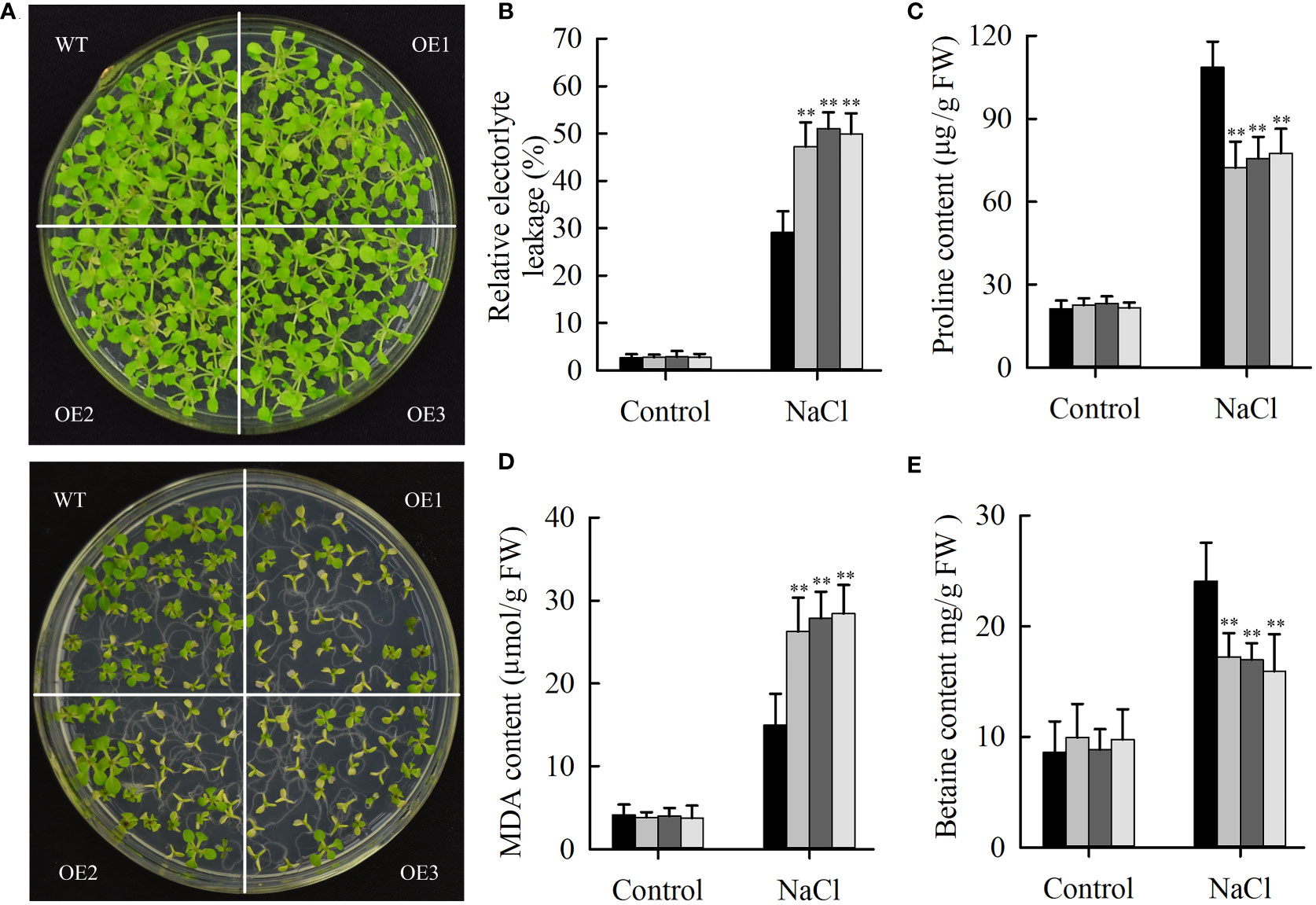
Figure 6 Phenotypic and physiological indicators analysis of wild-type and JcHDZ21 transgenic plants growth in control and salt stress conditions. (A) Phenotypes of wild-type and JcHDZ21 transgenic plants at 25 days growth in control and salt stress conditions. (B) Relative electrolyte leakage of leaves of wild-type and overexpressing plants after stress treatment and under non-stress conditions. (C) Proline content of leaves of wild-type and overexpressing plants after stress treatment and under non-stress conditions. (D) MDA content of leaves of wild-type and overexpressing plants after stress treatment and under non-stress conditions. (E) Betaine content of leaves of wild-type and overexpressing plants after stress treatment and under non-stress conditions. Data in (B–E): means of n = 21 ± SD from three independent experiments, asterisks above the bars indicate significant differences from wild-type controls at p < 0.01according to Duncan’s multiple range test.
Some physiological indicators were also tested in transgenic and wild-type plants under normal growth and salt stress conditions. The results showed that salinity stress increased MDA, proline and betaine content, and relative electrolyte leakage, whereas wild-type plants had higher proline and betaine content, lower relative electrolyte leakage and MDA content compared with transgenic plants under salinity stress (Figures 6B–E). In contrast, no significant differences in these stress response indicators between wild-type and transgenic plants were observed under non-stress conditions. The above results show that overexpression of JcHDZ21 increases the sensitivity of transgenic Arabidopsis to salt stress, which may be partly caused by changing these physiological indicators. Taken together, these results strongly suggest that JcHDZ21 acts as a negative regulator in responses to salt stress.
JcHDZ21 alters the expression of abiotic stress genes under salt stress
To clarify the molecular mechanism of JcHDZ21 regulating Arabidopsis in response to salt stress, we further detected the expression levels of abiotic stress-related genes by qRT-PCR method, such as AtP5CS1, AtNHX1, AtBADH and AtHKT1;1. The results showed that plants growth in control condition, these abiotic stress-related genes had no significant differences among different plants. However, the expression of these abiotic stress-related genes was significantly downregulated in JcHDZ21 transgenic plants compared to wild type when plants were exposed to salt stress (Figure 7).
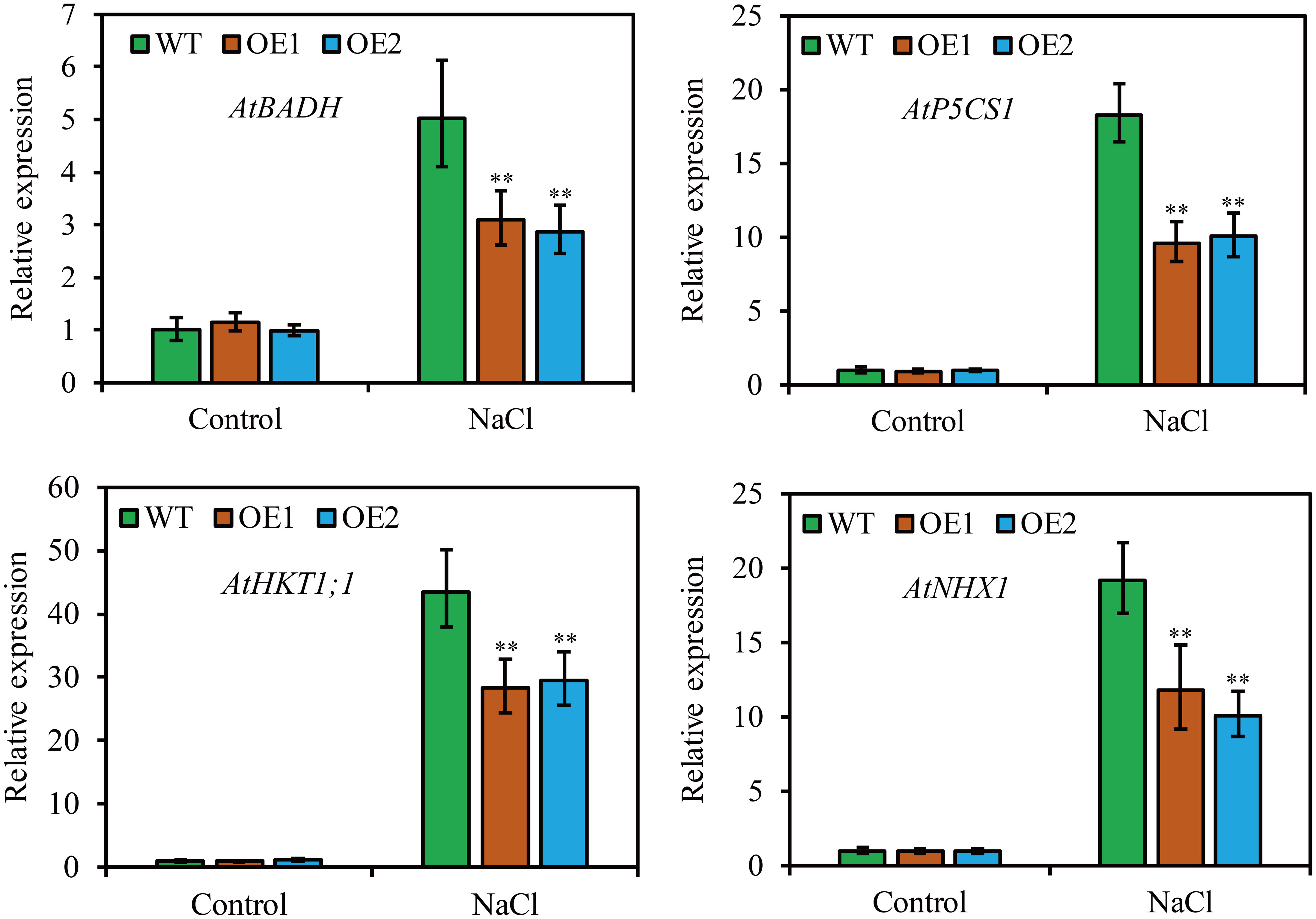
Figure 7 Expression levels of abiotic stress-related genes in wild-type and overexpressing plants under salt stress and non-stress conditions. Each experiment contains three biological replicates, and each with two technical replicates (means of n = 6 ± SD, asterisks above the bars indicate significant differences from wild-type controls at p < 0.01).
Discussion
HD-Zip group I transcription factors, as the main type in the HD-Zip transcription factor family, can respond to abiotic stresses by participating in various biological processes (Gong et al., 2019; Zhao et al.,2021). The properties of HD-Zip transcription factors closely related to abiotic stress have been widely reported in the model plant Arabidopsis and other plants (Perotti et al., 2021). For example, ectopic expression of the Arabidopsis AtHB6 gene increases drought stress tolerance in transgenic maize (Jiao et al., 2022); the expression of the maize HD-Zip transcription factor gene Zmhdz10 and the cotton transcription factor gene MdHB7-like enhanced the salt tolerance of the transgenic plants (Zhao et al., 2014; Zhao et al., 2021). After the physic nut genome sequencing was completed, a total of 32 HD-Zip family genes were identified in physic nut (Wu et al., 2015), but little is known about the regulatory function of HD-Zip transcription factors in physic nut. To date, only a few genes related to HD-Zip transcription factors have been identified in physic nut, such as JcHDZ16 (Tang et al., 2019), which both alter the ability of transgenic plants to tolerate abiotic stress. Therefore, we cloned a salt stress-responsive gene JcHDZ21 in physic nut and analyzed the function of this gene in Arabidopsis. In future research, we will use CRISPR/Cas9 genome editing technology to construct jchdz21 mutant physic nut plants to obtain more salt-tolerant physic nut species. Thus, our results may provide foundations to further promote application of JcHDZ21 gene in physic nut breeding.
When plants are under salt stress, some substances that regulate osmotic pressure (such as proline and betaine) will be induced to synthesize to adapt to the stress. The accumulation of proline and betaine in plants under salt stress is an important physiological phenomenon that is beneficial to the growth of plants under stress (Ganjavi et al., 2021; Hosseinifard et al., 2022). Their contents are positively correlated with plant salt tolerance and can protect plant cell membrane systems from abiotic stress (Ganjavi et al., 2021; Hosseinifard et al., 2022). Maize transcription factor ZmWRKY114 enhances the sensitivity of transgenic Arabidopsis to salt stress by regulating proline accumulation (Bo et al., 2020). GmNAC06 alters the salt tolerance of transgenic soybean by promoting the accumulation of proline and betaine (Li et al., 2021). Similarly, our study found that increasing the expression of the JcHDZ21 gene increased the sensitivity of transgenic Arabidopsis to salt stress, and the accumulation of proline and betaine in JcHDZ21 transgenic plants induced by salt stress was significantly lower than that in wild-type plants (Figure 6), indicating that proline and betaine may be the factors that confer lower salt stress tolerance in JcHDZ21 transgenic plants. Increases in malondialdehyde (MDA) and relative electrical conductivity caused by abiotic stress are also important indicators of plant oxidative damage (Xie et al., 2019). In our study, compared with wild type, the relative conductivity and MDA content of JcHDZ21 transgenic plants induced by salt stress were significantly increased (Figures 6B–E). Therefore, we speculate that the increase in relative conductivity and MDA content induced by salt stress may be one of the reasons for the weaker salt stress tolerance of JcHDZ21 transgenic plants. JcHDZ21 protein has transcriptional activation activity, suggesting that JcHDZ21 may interact with other transcriptional repressors to regulate the process of plant response to salt stress.
In addition to physiological indicators that can reflect the parameters of plants responding to adversity stress, some abiotic stress genes also play an important role in plant responses to abiotic stresses when plants encounter adversity stress. AGL16 negatively regulates salt stress progression by altering the expression of abiotic stress-related genes (Zhao et al., 2021). Oshox22 alters the tolerance of transgenic plants to salt stress by promoting the expression of abiotic stress-related genes (Zhang et al., 2012). Similarly, our study found that the expression of abiotic stress-related genes (AtP5CS1, AtNHX1, AtBADH and AtHKT1;1) in JcHDZ21 transgenic plants was significantly lower than that in wild type under salt stress conditions (Figure 7). P5CS1 has been identified as the major contributor to stress-induced proline accumulation (Feng et al., 2016), and ectopic expression of the Arabidopsis P5CS1 gene increases salt stress tolerance in transgenic tobacco by regulating proline accumulation (Ibragimova et al., 2015). AtHKT1;1 gene plays an important role in regulating the process of plant response to salt stress, and increasing the expression of this gene increases the tolerance of transgenic plants to salt stress (Plett et al., 2010). Increased expression of the AtNHX1 gene alters the tolerance of transgenic plants to salt stress (Asif et al., 2011). AtBADH increases salt stress tolerance in transgenic plants by altering betaine content (Zhou et al., 2008). Taken together, JcHDZ21 may affect the biosynthesis of proline and betaine by regulating the expression of proline and betaine metabolism genes under salt stress, thereby increasing the sensitivity of transgenic plants to salt stress. Another possibility is that JcHDZ21 may alter the tolerance of transgenic plants to salt stress by regulating the expression of abiotic stress-related genes.
Conclusion
JcHDZ21 is a member of the HD-Zip I family, which is highly expressed in seeds and responds to salt stress. JcHDZ21 protein is localized to the nucleus and has transcriptional activity. Under the condition of salt stress, the damage degree of salt stress on JcHDZ21 transgenic Arabidopsis was significantly higher than that of wild type. This study provides a reference for further research on the functions of the HD-Zip family genes in physic nut and other species, and also provides important candidate genes for the genetic breeding of physic nut.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
YHT conceived and designed the experiments; JP, JL, MZ, YT, SC, and YS performed experiments; QW and MZ analyzed the data; XB provided research advice; YHT wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Innovation and Entrepreneurship Training Program for College Students of Henan Province (grant no. 202210478018, 202210478019), Zhoukou Normal University, and the Research and Innovation Fund for College Students of Zhoukou Normal University (grant no. ZKNUD2022014).
Acknowledgments
We thank the reviewers and editor-in-chief for their valuable comments on our paper.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1097265/full#supplementary-material
References
Asif, M. A., Zafar, Y., Iqbal, J., Iqbal, M. M., Rashid, U., Ali, G. M., et al. (2011). Enhanced expression of AtNHX1, in transgenic groundnut (Arachis hypogaea l.) improves salt and drought tolerence. Mol. Biotechnol. 49, 250–256. doi: 10.1007/s12033-011-9399-1
Bo, C., Chen, H., Luo, G., Li, W., Zhang, X., Ma, Q., et al. (2020). Maize WRKY114 gene negatively regulates salt-stress tolerance in transgenic rice. Plant Cell Rep. 39, 135–148. doi: 10.1007/s00299-019-02481-3
Deng, X., Phillips, J., Meijer, A. H., Salamini, F., Bartels, D. (2002). Characterization of five novel dehydration-responsive homeodomain leucine zipper genes from the resurrection plant craterostigma plantagineum. Plant Mol. Biol. 49, 601–610. doi: 10.1023/a:1015501205303
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L. (2010). MYB transcription factors in arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Feng, K., Hou, X. L., Xing, G. M., Liu, J. X., Duan, A. Q., Xu, Z. S., et al. (2020). Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 40, 750–776. doi: 10.1080/07388551.2020.1768509
Feng, X. J., Li, J. R., Qi, S. L., Lin, Q. F., Jin, J. B., Hua, X. J. (2016). Light affects salt stress-induced transcriptional memory of P5CS1 in arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, 8335–8343. doi: 10.1073/pnas.1610670114
Ganjavi, A. S., Oraei, M., Gohari, G., Akbari, A., Faramarzi, A. (2021). Glycine betaine functionalized graphene oxide as a new engineering nanoparticle lessens salt stress impacts in sweet basil (Ocimum basilicum l.). Plant Physiol. Biochem. 162, 14–26. doi: 10.1016/j.plaphy.2021.02.028
Gong, S., Ding, Y., Hu, S., Ding, L., Chen, Z., Zhu, C. (2019). The role of HD-Zip class I transcription factors in plant response to abiotic stresses. Physiol Plant. 167, 516–525. doi: 10.1111/ppl.12965
Guo, Q., Jiang, J., Yao, W., Li, L., Zhao, K., Cheng, Z., et al. (2021). Genome-wide analysis of poplar HD-zip family and over-expression of PsnHDZ63 confers salt tolerance in transgenic populus simonii × p.nigra. Plant Sci. 311, 111021. doi: 10.1016/j.plantsci.2021.111021
Hanson, J., Johannesson, H., Engström, P. (2001). Sugar-dependent alterations in cotyledon and leaf development in transgenic plants expressing the HDZhdip gene ATHB13. Plant Mol. Biol. 45, 247–262. doi: 10.1023/a:1006464907710
Harris, J. C., Hrmova, M., Lopato, S., Langridge, P. (2011). Modulation of plant growth by HD-zip class I and II transcription factors in response to environmental stimuli. New Phytol. 190, 823–837. doi: 10.1111/j.1469-8137.2011.03733.x
Henriksson, E., Olsson, A. S., Johannesson, H., Johansson, H., Hanson, J., Engström, P., et al. (2005). Homeodomain leucine zipper class I genes in arabidopsis. expression patterns and phylogenetic relationships. Plant Physiol. 139, 509–518. doi: 10.1104/pp.105.063461
Hosseinifard, M., Stefaniak, S., Ghorbani Javid, M., Soltani, E., Wojtyla, Ł., Garnczarska, M. (2022). Contribution of exogenous proline to abiotic stresses tolerance in plants: A review. Int. J. Mol. Sci. 23, 5186. doi: 10.3390/ijms23095186
Hu, Y., Li, S., Fan, X., Song, S., Zhou, X., Weng, X., et al. (2020). OsHOX1 and OsHOX28 redundantly shape rice tiller angle by reducing HSFA2D expression and auxin content. Plant Physiol. 184, 1424–1437. doi: 10.1104/pp.20.00536
Ibragimova, S. M., Trifonova, E. A., Filipenko, E. A., Shymny, V. K. (2015). Evaluation of salt tolerance of transgenic tobacco plants bearing with P5CS1 gene of Arabidopsis thaliana. Genetika 51, 1368–1375. doi: 10.1134/S1022795415120078
Javelle, M., Klein-Cosson, C., Vernoud, V., Boltz, V., Maher, C., Timmermans, M., et al. (2011). Genome-wide characterization of the HD-ZIP IV transcription factor family in maize: Preferential expression in the epidermis. Plant Physiol. 157, 790–803. doi: 10.1104/pp.111.182147
Jiao, P., Jiang, Z., Wei, X., Liu, S., Qu, J., Guan, S., et al. (2022). Overexpression of the homeobox-leucine zipper protein ATHB-6 improves the drought tolerance of maize (Zea mays l.). Plant Sci. 316, 111159. doi: 10.1016/j.plantsci.2021.111159
Johannesson, H., Wang, Y., Engström, P. (2001). DNA-Binding and dimerization preferences of arabidopsis homeodomain-leucine zipper transcription factors in vitro. Plant Mol. Biol. 45, 63–73. doi: 10.1023/a:1006423324025
Li, M., Chen, R., Jiang, Q., Sun, X., Zhang, H., Hu, Z. (2021). GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 105, 333–345. doi: 10.1007/s11103-020-01091-y
Montes, J. M., Melchinger, A. E. (2016). Domestication and breeding of Jatropha curcas L. Trends Plant Sci 21, 1045–1057. doi: 10.1016/j.tplants.2016.08.008
Perotti, M. F., Arce, A. L., Chan, R. L. (2021). The underground life of homeodomain-leucine zipper transcription factors. J. Exp. Bot. 72, 4005–4021. doi: 10.1093/jxb/erab112
Plett, D., Safwat, G., Gilliham, M., Skrumsager Møller, I., Roy, S., Shirley, N., et al. (2010). Improved salinity tolerance of rice through cell type-specific expression of AtHKT1;1. PloS One 5, e12571. doi: 10.1371/journal.pone.0012571
Preciado, J., Begcy, K., Liu, T. (2022). The arabidopsis HDZIP class II transcription factor ABA INSENSITIVE TO GROWTH 1 functions in leaf development. J. Exp. Bot. 73, 1978–1991. doi: 10.1093/jxb/erab523
Rushton, P. J., Somssich, I. E., Ringler, P., Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Tang, Y., Wang, J., Bao, X., Liang, M., Lou, H., Zhao, J., et al. (2019). Genome-wide identification and expression profile of HD-ZIP genes in physic nut and functional analysis of the JcHDZ16 gene in transgenic rice. BMC Plant Biol. 19, 298. doi: 10.1186/s12870-019-1920-x
Thirumalaikumar, V. P., Devkar, V., Mehterov, N., Ali, S., Ozgur, R., Turkan, I., et al. (2018). NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 16, 354–366. doi: 10.1111/pbi.12776
Turchi, L., Carabelli, M., Ruzza, V., Possenti, M., Sassi, M., Peñalosa, A., et al. (2013). Arabidopsis HD-zip II transcription factors control apical embryo development and meristem function. Development 140, 2118–2129. doi: 10.1242/dev.092833
Wu, P., Zhou, C., Cheng, S., Wu, Z., Lu, W., Han, J., et al (2015). Integrated genome sequence and linkage map of physic nut (Jatropha curcas L.), a biodiesel plant. Plant J. 81, 810–821. doi: 10.1111/tpj.12761
Xie, Y., Sun, X., Feng, Q., Luo, H., Wassie, M., Amee, M., et al. (2019). Comparative physiological and metabolomic analyses reveal mechanisms of Aspergillus aculeatus-mediated abiotic stress tolerance in tall fescue. Plant Physiol. Biochem. 142, 342–350. doi: 10.1016/j.plaphy.2019.07.022
Yoo, S. D., Cho, Y. H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, S., Haider, I., Kohlen, W., Jiang, L., Bouwmeester, H., Meijer, A. H., et al. (2012). Function of the HD-zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice. Plant Mol. Biol. 80, 571–585. doi: 10.1007/s11103-012-9967-1
Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W., Chua, N. H. (2006). Agrobacterium-mediated transformation of arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhao, P. X., Zhang, J., Chen, S. Y., Wu, J., Xia, J. Q., Sun, L. Q., et al. (2021). Arabidopsis MADS-box factor AGL16 is a negative regulator of plant response to salt stress by downregulating salt-responsive genes. New Phytol. 232, 2418–2439. doi: 10.1111/nph.17760
Zhao, S., Wang, H., Jia, X., Gao, H., Mao, K., Ma, F. (2021). The HD-zip I transcription factor MdHB7-like confers tolerance to salinity in transgenic apple (Malus domestica). Physiol. Plant 172, 1452–1464. doi: 10.1111/ppl.13330
Zhao, Y., Ma, Q., Jin, X., Peng, X., Liu, J., Deng, L., et al. (2014). A novel maize homeodomain-leucine zipper (HD-zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and arabidopsis. Plant Cell Physiol. 55, 1142–1156. doi: 10.1093/pcp/pcu054
Keywords: HD-Zip gene family, salt stress, JcHDZ21, physic nut, arabidopsis
Citation: Tang Y, Peng J, Lin J, Zhang M, Tian Y, Shang Y, Chen S, Bao X and Wang Q (2023) A HD-Zip I transcription factor from physic nut, JcHDZ21, confers sensitive to salinity in transgenic Arabidopsis. Front. Plant Sci. 14:1097265. doi: 10.3389/fpls.2023.1097265
Received: 13 November 2022; Accepted: 03 February 2023;
Published: 15 February 2023.
Edited by:
Diaa Abd El Moneim, Arish University, EgyptReviewed by:
Yuri Shavrukov, Flinders University, AustraliaFei Zhou, Huazhong Agricultural University, China
Copyright © 2023 Tang, Peng, Lin, Zhang, Tian, Shang, Chen, Bao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuehui Tang, eWh0YW5nMjAwNUAxNjMuY29t
 Yuehui Tang
Yuehui Tang Jingrui Peng1
Jingrui Peng1 Xinxin Bao
Xinxin Bao