- Guangdong Key Laboratory of Plant Adaptation and Molecular Design, Guangzhou Key Laboratory of Crop Gene Editing, Innovative Center of Molecular Genetic and Evolution, School of Life Science, Guangzhou University, Guangzhou, China
Flowering time influences reproductive success in plants and has a significant impact on yield in grain crops. Flowering time is regulated by a variety of environmental factors, with daylength often playing an important role. Crops can be categorized into different types according to their photoperiod requirements for flowering. For instance, long-day crops include wheat (Triticum aestivum), barley (Hordeum vulgare), and pea (Pisum sativum), while short-day crops include rice (Oryza sativa), soybean (Glycine max), and maize (Zea mays). Understanding the molecular regulation of flowering and genotypic variation therein is important for molecular breeding and crop improvement. This paper reviews the regulation of flowering in different crop species with a particular focus on how photoperiod-related genes facilitate adaptation to local environments.
Flowering is a central developmental process in the life cycle of plants. Plants must integrate internal factors and external cues to determine the optimal time to flower. This process is crucial for successful reproduction in all flowering plants but has added importance in crop species because of its major effect on yield. Daylength is one of the critical environmental cues that influence flowering time. Based on their flowering responses to daylength, plants can be categorized into three major types: long-day (LD) plants, short-day (SD) plants, and day-neutral (DN) plants. LD plants flower when the daylength is longer than a critical threshold, while SD plants flower when the daylength is shorter than a critical threshold. The flowering of DN plants is not affected by day length (Garner and Allard, 1920). LD plants, such as wheat (Triticum aestivum), barley (Hordeum vulgare), and pea (Pisum sativum), generally originated at higher latitudes, and tend to flower in late spring or early summer when periods of uninterrupted light extend past a certain threshold. SD plants, such as rice (Oryza sativa), soybean (Glycine max), and maize (Zea mays), originated at lower latitudes and tend to flower after they perceive a certain period of uninterrupted darkness (Brambilla et al., 2017). Here, we summarize the centers of domestication for major crop species and track their dissemination around the world. We also compare the different flowering regulation strategies in LD and SD crops, and discuss genotypic variation that arose during their dispersal.
1 Historical dissemination routes of major crops
1.1 LD crops
LD crops include wheat, barley, and pea, which were domesticated at a relatively high latitude in the Fertile Crescent, a narrow range that extends from 30°N to 40°N, and then were spread into different parts of world via different routes (Figure 1) (Zohary et al., 2012). The domestication of hexaploid bread wheat (Triticum aestivum L. ssp. aestivum) involved two hybridization events. The initial hybridization event took place between Triticum urartu (AA) and Aegilops speltoides (BB), resulting in emmer wheat, Triticum durum ssp. dicoccum (BBAA). The hexaploid Triticum aestivum ssp. aestivum (BBAADD) subsequently arose from a hybridization between domesticated emmer wheat (BBAA) and Aegilops tauschii (goat grass; DD) (Marcussen et al., 2014). Cultivated barley (Hordeum vulgare L. ssp. vulgare), however, was domesticated directly from a single wild progenitor, Hordeum vulgare ssp. spontaneum (Haas et al., 2019).
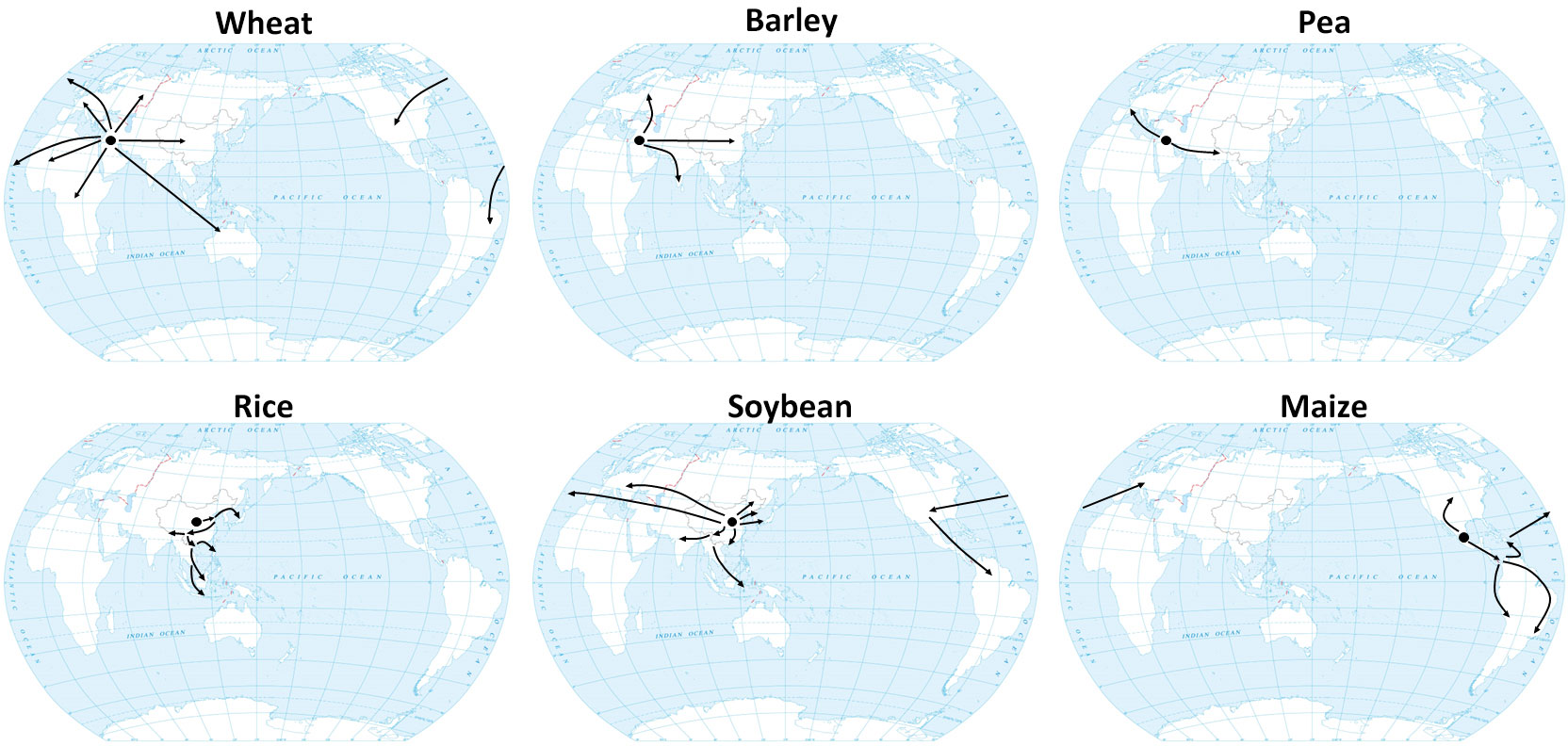
Figure 1 Domestication centers and hypothetical dissemination routes of major crops. Black arrows indicate the expansion scenario of wheat (Gohar et al., 2022), barley (Lister et al., 2018), pea (Jing et al., 2010), rice (Gutaker et al., 2020), soybean (Kihara, 1969; Hymowitz and Shurtleff, 2005; Stacey, 2008; Sedivy et al., 2017) and maize (Wang et al., 2021).
Pea (Pisum sativum) is also a LD crop. Its wild progenitor, P. sativum subsp. elatius, was initially domesticated in the Fertile Crescent as well. After domestication, P. sativum subsp. elatius began to be dispersed in two different directions. Eastward expansion into the Indian subcontinent and the Himalayan region gave rise to the Afghanistan germplasm group. The more prominent western expansion into Mediterranean Europe gave rise to the European domestic pea (P. sativum ssp. sativum) germplasm group, which eventually was developed into modern elite varieties (Figure 1) (Jing et al., 2010).
1.2 SD crops
SD crops include rice, soybean, and maize, which were domesticated at relatively low latitudes. There are two cultivated species of rice, Asian rice (O. sativa L.) and African rice (O. glaberrima). The archaeological record suggests that Asian rice was first domesticated in the middle and lower Yangtze River corridor in southern China as early as 9,000 years ago (Figure 1) (Doebley et al., 2006; Fuller et al., 2007; Wang et al., 2018). Two conflicting models exist to account for the domestication history of Asian rice: 1) the single domestication with introgression model and 2) the multiple independent domestications model. The first model suggests that wild rice can be divided into three major subpopulations: Or-I, Or-II, and Or-III. Population genetic analysis indicates that early-cultivated Asian rice, belonging to the japonica subspecies, was first domesticated from the Or-III-type O. rufipogon in southern China before spreading to other parts of Asia. Another subspecies of Asian rice, indica, was domesticated later by crossing japonica rice with local Or-I and Or-II-type wild rice, also belonging to O. rufipogon (Huang et al., 2012). By contrast, the multiple independent domestication model speculates that japonica was domesticated in China while indica was domesticated independently from local wild rice in China and India (Civáň et al., 2015).
Compared to Asian cultivated rice, African cultivated rice was grown in a limited area and, because of its low yield, has gradually been replaced by Asian rice (van Andel et al., 2016; Chen et al., 2019). African cultivated rice (O. glaberrima) was domesticated from its wild progenitor O. barthii around 3,000 years ago (Sweeney and McCouch, 2007; Wang et al., 2014; Huang et al., 2015). Some studies have proposed that African rice was domesticated in the Inner Niger Delta, while other studies suggest the domestication of African rice was multiregional (Huang et al., 2015; Snodgrass and Hufford, 2018; Choi et al., 2019; Veltman et al., 2019).
Soybean (Glycine max) was most likely domesticated from wild soybean (Glycine soja) in the Huang-Huai Valley of Central China (around 30–45°N) 5,000 years ago (Sedivy et al., 2017). Cultivated soybean then expanded to Korea, Japan, and other parts of Asia about 2,000 years ago (Kihara, 1969). In the 18th century, soybean was disseminated to Europe and North America, and was introduced to Central and South America in the first half of the 20th century (Figure 1) (Hymowitz and Shurtleff, 2005; Stacey, 2008).
Maize (Z. mays ssp. mays) was domesticated from its wild ancestor teosinte (Z. mays subsp. parviglumis) about 9,000 years ago in the Balsas region of southwest Mexico (Matsuoka et al., 2002; Piperno et al., 2009). From there, maize spread south and north to the rest of the Western Hemisphere around 1500 years ago. After Columbus arrived in the New World and brought maize to Europe, it rapidly spread around the world (Figure 1) (Brandolini and Brandolini, 2009; Nunn and Qian, 2010).
Through the broad dissemination of these major crops over their long history, the cultivation of both LD and SD crops has expanded from their original sites of domestication to the rest of the world. However, these species have a variety of distinct or partially overlapping strategies for regulating flowering time. Thus, a comparison of the molecular mechanisms underlying flowering time regulation is critical for identifying conserved and divergent mechanisms among these species.
2 Regulation of flowering time in LD plants
Temperate crops such as wheat, barley, and pea are LD plants, and as such their flowering is primarily controlled by daylength. For wheat and barley, flowering time under long day conditions is regulated by the PHOTOPERIOD1 (PPD1) gene (Table 1, Figure 2) (Turner et al., 2005; Seki et al., 2011). PPD1 encodes an ortholog of the Arabidopsis (Arabidopsis thaliana) PSEUDO-RESPONSE REGULATOR (PRR) protein, which is characterized by a pseudo-receiver and a CCT (CONSTANTS, CONSTANTS-like and TOC1) domain. PPD1 expression is repressed by circadian clock genes EARLY FLOWERING 3 (ELF3) and LUX ARRHYTHMO (LUX) in wheat and barley (Faure et al., 2012; Mizuno et al., 2012; Zakhrabekova et al., 2012; Campoli et al., 2013; Alvarez et al., 2016). In addition, PHYTOCHROME family, PHYB and PHYC mediate activation of PPD1 expression in the acceleration of wheat and barley flowering under LD conditions (Chen et al., 2014; Pankin et al., 2014; Pearce et al., 2016). LD conditions induce PPD1 and upregulate VERNALIZATION3 (VRN3), a homologue of the Arabidopsis gene FLOWERING LOCUS T (FT), by controlling CONSTANS activity to promote flowering (Turner et al., 2005; Chen et al., 2014). In Arabidopsis, FT protein moves from leaves to the shoot apical meristem (SAM) through the phloem (Turck et al., 2008). In the SAM, FT interacts with the bZIP transcription factor (TF) FLOWERING LOCUS D (FD) and binds to the promoters of APETALA1 (AP1) and FRUITFULL (FUL) to induce the switch from vegetative to reproductive growth (Abe et al., 2005; Wigge et al., 2005). Similarly, in wheat, VRN3 interacts with an FD-like protein (TaFDL2) and subsequently binds to the promoter of TaVRN1, which is the ortholog of Arabidopsis AP1 and FUL (Li and Dubcovsky, 2008). Under SD conditions, VRN3 transcript levels are low (Yan et al., 2006). However, many varieties of wheat and barley can also flower under SD conditions, although flowering is delayed. Studies have identified a gene, PPD2, which is a paralog of FT referred to as FT3, that confers the ability to flower under SD conditions in barley and barley (Laurie et al., 1995; Faure et al., 2007; Kikuchi et al., 2009; Casao et al., 2011; Halliwell et al., 2016). However, how PPD2 affects flowering under SD conditions and how PPD2 regulates downstream genes remain unknown.
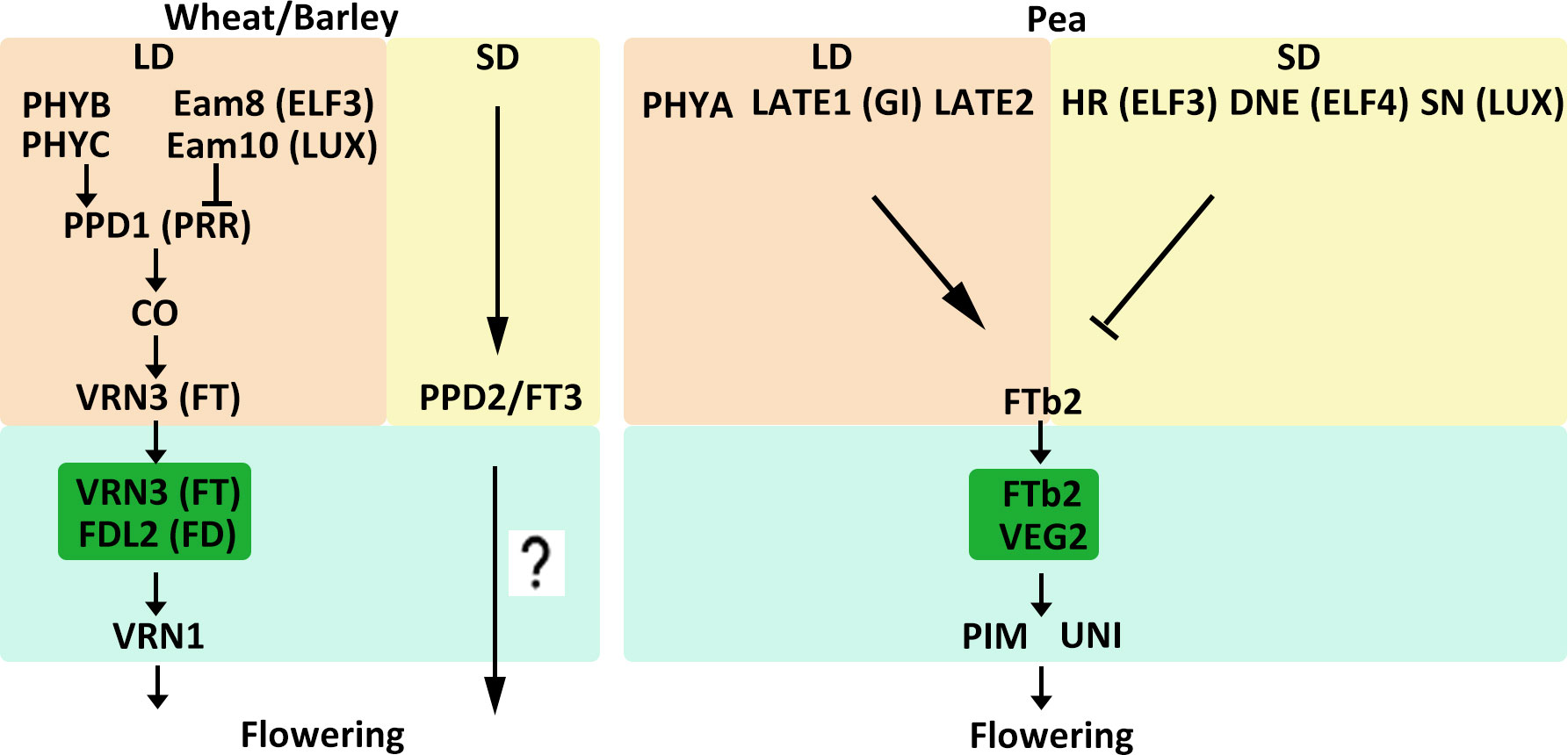
Figure 2 Model of flowering time control pathways in major LD crops. The different external and internal cues are highlighted in different colors. Positive and negative regulatory actions are indicated by arrows and lines with bares, respectively.
Pea (Pisum sativum) is another crop grown in temperate environments. A number of loci related to flowering time have recently been identified in pea. Several of these are known to promote flowering in LD conditions. For instance, phyA, late1, and late2 mutants all displayed a late-flowering phenotype under LD conditions (Weller et al., 1997; Hecht et al., 2007). LATE2 has not yet been characterized, but LATE1 is an ortholog of the Arabidopsis circadian clock-related gene GIGANTEA (GI) (Hecht et al., 2007). Loci that delay flowering under SD conditions have also been identified. Recessive alleles of HIGH RESPONSE (HR), DIE NEUTRALIS (DNE), and STERILE NODES (SN) can cause early flowering in SD conditions (Table 1, Figure 2) (Weller et al., 2009). HR, DNE, and SN have been identified as homologs of the circadian clock genes ELF3, ELF4, and LUX, respectively. (Weller et al., 2012; Liew et al., 2014). In Arabidopsis, ELF3, ELF4, and LUX work together to form the evening complex (EC) and participate in the evening loop of the circadian clock (Nagel and Kay, 2012). This mechanism might explain why hr, dne, and sn mutants have similar phenotypes. The legume FT-like genes are divided into three subclasses: FTa, FTb, and FTc (Surkova and Samsonova, 2022). Five FT-like genes have been identified in pea: FTa1, FTa2, FTb1, FTb2, and FTc (Hecht et al., 2011). FTa and FTb are expressed in leaves and are probably involved in vernalization and photoperiod responses, respectively. FTc, however, is expressed in the SAM and might be involved in the integration of signals from leaf-expressed FT genes (Hecht et al., 2011). FTb2 is expressed in leaves, and grafting experiments suggest that it might generate a flowering stimulus that travels from leaves to the SAM and promotes flowering (Beveridge and Murfet, 1996; Hecht et al., 2011). FTb2 interacts with VEGETATIVE2 (VEG2), which is an ortholog of the Arabidopsis FD protein, and may participate in a florigen activation complex (FAC) to activate downstream floral meristem-identity genes (Sussmilch et al., 2015). Flowering regulation-related genes in pea also include PROLIFERATING INFLORESCENCE MERISTEM (PIM) and UNIFOLIATA (UNI), which are orthologs of Arabidopsis AP1 and LEAFY (LFY), respectively (Hofer et al., 1997; Taylor et al., 2002).
3 Regulation of flowering time in SD plants
Rice is a SD crop, and its heading date is primarily determined by photoperiod sensitivity. Here, we summarize the current understanding of core molecular regulatory networks involved in rice flowering in both LD and SD conditions (Table 1, Figure 3). In LD conditions, rice has the GI-CO-FT pathway. Heading date 1 (Hd1), an ortholog of the Arabidopsis CO, is expressed from nightfall to dawn. The diurnal expression of Hd1 is regulated by OsGI (Hayama et al., 2003). Heading date 3a (Hd3a) is the ortholog of the Arabidopsis FT gene (Komiya et al., 2009). Different from Arabidopsis, rice Hd1 negatively regulates Hd3a expression under LD conditions but positively regulate Hd3a expression under SD conditions (Hayama et al., 2003). The switch of Hd1 function is mediated by DAYS TO HEADING 8 (DTH8) which encodes a CCAAT-box-binding TF. The DTH8-Hd1 complex increases H3K27 trimethylation at the Hd3a locus and represses Hd3a expression in LD conditions (Du et al., 2017). Hd6 encodes one subunit of the protein kinase CASEIN KINASE 2 (CK2), which indirectly promotes Hd1-induced repression of Hd3a expression under LD conditions (Ogiso et al., 2010).
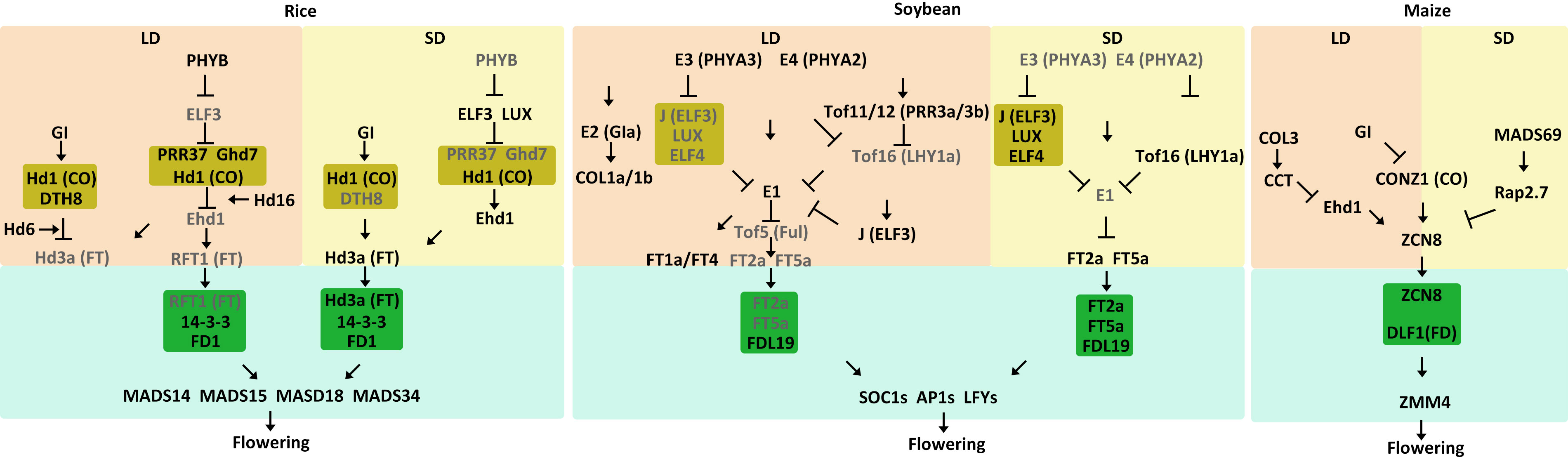
Figure 3 Model of flowering time control pathways in major SD crops. The different external and internal cues are highlighted in different colors. Positive and negative regulatory actions are indicated by arrows and lines with bares, respectively.
Aside from the conserved GI-CO-FT pathway, rice also has the PHYB-ELF3-GHD7/OSPRR37-Ehd1-Hd3a/RFT1 regulatory pathway, which includes Grain number, plant height, and heading date 7 (Ghd7), Early heading date 1 (Ehd1) and RICE FLOWERING LOCUS T1 (RFT1). Recently, it has been found that PHYB is activated under LD conditions and promotes the degradation of ELF3, thereby releasing the EC-mediated repression of Ghd7 and OsPRR37 (Andrade et al., 2022). Ghd7, which encodes a CCT-domain TF, is significantly induced by LD conditions (Xue et al., 2008). Ghd7 directly interacts with Hd1 to repress Ehd1, leading to suppression of the downstream gene Hd3a and RFT1 to delay flowering (Xue et al., 2008; Weng et al., 2014). Hd16, which encodes CASEIN KINASE 1, enhances the repressive function of Ghd7 on Ehd1 expression (Hori et al., 2013). Similarly, OsPRR37 and Hd1 together form a transcriptional repressor complex that downregulates Ehd1 to suppress flowering under LD conditions (Goretti et al., 2017; Zhang et al., 2019).Both RFT1 and Hd3a are orthologs of the Arabidopsis FT gene and function as florigens. However, RFT1 is a major LD activator whereas Hd3a is a major SD activator for rice. (Komiya et al., 2009). RFT1 protein, which is produced in leaves, moves to the SAM and forms a FAC with 14-3-3 and OsFD1, subsequently activating the expression of three AP1/FUL-like genes OsMADS14, OsMADS15, and OsMADS18 (Peng et al., 2021).
Under SD conditions, ELF3 activity increases during periods of darkness, enabling the EC to reduce expression of key floral repressors such as OsPRR37 and Ghd7 in SD conditions. This converts Hd1 from a suppressor to an activator, promoting the expression of Hd3a at night (Andrade et al., 2022). DTH8 also interacts with Hd1, but Hd1 continues to act as an activator of Hd3a expression to promote flowering (Du et al., 2017). SD activator Hd3a protein is generated in leaves and moves to the SAM, where it interacts with 14-3-3 (Taoka et al., 2011). The Hd3a-14-3-3 complex enters the nucleus and forms an FAC with OsFD1, activating OsMADS14, OsMADS15 and OsMADS18 to promote floral transition (Komiya et al., 2008; Taoka et al., 2011; Tsuji et al., 2011; Yin et al., 2019).
Soybean is a typical SD plant that is very sensitive to photoperiod. Many important genes that control soybean flowering have been identified, including E1-E11, J, and quantitative trait loci including Time of Flowering 5 (Tof5), Tof11, Tof12, Tof16, and Tof18 (Bernard, 1971; Buzzell, 1971; Buzzell and Voldeng, 1980; McBlain and Bernard, 1987; Ray et al., 1995; Bonato and Vello, 1999; Cober and Voldeng, 2001; Cober et al., 2010; Kong et al., 2014; Zhai et al., 2014; Wang et al., 2019; Lu et al., 2020; Dong et al., 2021; Dong et al., 2022; Kou et al., 2022). In the past decade, scientists have uncovered the molecular mechanisms of the key genes involved in soybean flowering regulation, namely the E3/E4-E1-GmFTs regulatory module (Table 1, Figure 3). E3 and E4 are the phytochrome genes GmPHYA3 and GmPHYA2, respectively (Cober et al., 1996; Watanabe et al., 2009). E1 is a soybean-specific TF that is a core regulator of the flowering pathway (Xia et al., 2012). Under LD conditions, E3 and E4 function as photoreceptors that perceive light signals to influence downstream genes (Liu et al., 2008; Watanabe et al., 2009; Tsubokura et al., 2013). E3 and E4 physically interact with E1 to stabilize the E1 protein. In addition, E3 and E4 interact with LUXs and promote their degradation, releasing the suppression of EC on E1 expression (Lin et al., 2022). E1 upregulates expression of the floral inhibitors GmFT1a and GmFT4, which further suppresses the expression of several downstream floral genes (Zhai et al., 2014; Liu et al., 2018). Although GmFT1a and GmFT4 are highly expressed in leaves and are expressed in the SAM at low levels, there is no direct evidence to support GmFT1a and GmFT4 being transported from leaves to the SAM to inhibit flowering in soybean. (Zhai et al., 2014; Liu et al., 2018). Additionally, E1 represses transcription of Tof5, an ortholog of Arabidopsis FUL, by binding to its promoter. E9 and E10 encode GmFT2a and GmFT4, respectively, which are orthologs of Arabidopsis FT (Kong et al., 2014; Zhao et al., 2016; Samanfar et al., 2017). Tof5 physically associates with the promoters of the floral activators GmFT2a and GmFT5a to induce their expression (Kong et al., 2010; Dong et al., 2022). The downregulation of GmFT2a and GmFT5a ultimately represses the induction of SUPPRESSOR OF OVEREXPRESSION OF CONSTANTS 1 (GmSOC1), GmAP1, and GmLFY (Nan et al., 2014). Thus, the E3/E4-EC-E1-GmFTs regulatory module delays flowering in LD conditions. In addition, Tof11 and Tof12, two PRR homologs, directly bind to the promoter of LATE ELONGATED HYPOCOTYL (GmLHY) to repress its expression. This prevents its transcriptional repression of E1, resulting in the E3/E4-Tof11/Tof12-Tof16-E1-GmFTs module (Li et al., 2020; Lu et al., 2020). Moreover, E2 (GIa) encodes an ortholog of the Arabidopsis circadian clock component GI. The recessive e2 allele leads to an early flowering phenotype by activating the expression of GmFT2a (Watanabe et al., 2011). Soybean has two additional GI orthologs, GIb and GIc, but their functions remain unknown (Wang et al., 2016). CONSTANS-LIKE 1a (GmCOL1a) and GmCOL1b are homologs of the Arabidopsis CO, and they suppress flowering in soybean under LD conditions (Cao et al., 2015; Cao et al., 2016). Compared to the conserved GI-CO-FT pathway in Arabidopsis, however, the functions of E2 and GmCOLs in soybean remain relatively unknown.
Under SD conditions, the functions of E3 and E4 become repressed (Xia et al., 2012). Functional reduction of E3 and E4 releases the suppression of Tof16, which encodes GmLHY1a, and J, which encodes an ortholog of the Arabidopsis EC component ELF3 (Lu et al., 2017; Dong et al., 2021). Tof16 and J control soybean flowering both additively and independently. Tof16 directly binds to the E1 promoter to suppress its expression (Dong et al., 2021). J combines with LUX and ELF4 to also inhibit the expression of E1 (Lu et al., 2017). Decreased expression of E1 releases the transcriptional suppression of GmFT2a and GmFT5a, further promoting flowering. In general, two regulatory modules are formed under SD conditions: EC-E1-GmFTs and Tof16-E1-GmFTs. Both GmFT2a and GmFT5a move from the leaves to the SAM and interact with the soybean ortholog of Arabidopsis FD, GmFDL19, to upregulate several downstream genes that promote flowering, like GmSOC1, GmAP1, and GmLFY (Nan et al., 2014).
Maize (Zea mays ssp. mays) was domesticated from its wild progenitor teosinte (Zea mays ssp. parviglumis) (Doebley et al., 2006). Modern maize and teosinte are, however, quite different from each other. Teosinte grows in tropical regions and requires SD photoperiods to induce flowering, while maize is grown at higher latitudes and is primarily photoperiod insensitive, with some varieties being DN plants (Minow et al., 2018). When early Native American farmers migrated to higher latitudes, they selected maize lines that were less dependent on SD photoperiods to flower. A CCT domain-containing gene ZmCOL3 has been identified as an inhibitor of flowering, itself being inhibited in SD conditions but activated in LD conditions. In LD conditions, ZmCOL3 becomes activated and directly induces ZmCCT transcription (Table 1, Figure 3) (Ducrocq et al., 2009; Hung et al., 2012; Yang et al., 2013; Huang et al., 2018). The ZmCCT protein binds to the promoter of ZmEhd1, which is homologous to the rice OsEhd1 gene, repressing its transcription. Downregulation of ZmEhd1 reduces expression of the florigen gene ZEA CENTRORADIALIS 8 (ZmZCN8) (Zhong et al., 2021). Thus, this ZmCOL3-ZmCCT-ZmEhd1-ZmZCN8 module regulates flowering in LD conditions. The gi mutation in maize, however, leads to an early flowering phenotype in LD conditions. Maize contains two homologs of the Arabidopsis GI, ZmGI1 and ZmGI2 (Mendoza et al., 2012). Transcription analysis has demonstrated that ZmGI1 represses expression of ZmZCN8 and ZmCONZ1, which is the homolog of Arabidopsis CO gene (Bendix et al., 2013). Although there is no evidence that ZmCONZ1 activates ZmZCN8 expression, the data suggest that ZmCONZ1 is downstream of GI1 but possibly upstream of ZmZCN8, acting as a positive regulator (Miller et al., 2008). Thus, ZmGI might repress flowering in LD condition via a ZmGI-ZmCONZ1-ZmZCN8 regulatory module.
In maize, an important flowering time QTL has been identified, VEGETATIVE TO GENERATIVE TRANSITION 1 (Vlăduţu et al., 1999). VGT1 corresponds to a noncoding regulator of the AP2-like TF, ZmRAP2.7. ZmRAP2.7 functions as a negative regulator of maize flowering (Salvi et al., 2007). A MADS-box TF, ZmMADS69 was identified as the causal gene at the VGT3 QTL, functioning as a constitutive activator of flowering (Castelletti et al., 2020). Maize also contains the ZmMADS69-ZmRAP2.7-ZmZCN8 regulatory module that functions in both LD and SD conditions. ZmMADS69 functions as a flowering activator by suppressing expression of the flowering repressor ZmRAP2.7, thereby relieving its transcriptional repression of the florigen gene ZmZCN8 to induce early flowering (Liang et al., 2019). ZmZCN8 is transcribed and translated in the leaf vasculature, then moves through the phloem to the SAM where it interacts with DELAYED FLOWERING 1 (DLF1), a homolog of the Arabidopsis FD protein, to activate downstream floral organ identity genes like ZEA MAYS MADS-BOX 4 (ZmZMM4) (Muszynski et al., 2006; Danilevskaya et al., 2008; Lazakis et al., 2011; Meng et al., 2011).
4 Genotypic variation helps crops adapt to different latitudes
When LD and SD crops move out of their native ranges and adapt to new locations, they must alter their flowering behavior and become less sensitive to photoperiods to ensure reproductive success in their non-native zones. During this process, natural and artificial selection act on genotypic variation to produce individuals that harbor suitable alleles and flower optimally, becoming locally adapted.
In the LD crops barley and wheat, major regulators conferring photoperiod sensitivity are encoded by PPD1 genes. PPD1 was identified as the core determinant of photoperiod responses in barley (Turner et al., 2005). The recessive ppd1 allele was selected for in barley from northern Europe, conferring delayed flowering and maturity in LD conditions. The dominant allele PPD1, however, was selected for in barley from southern Europe, promoting flowering in response to longer days (Turner et al., 2005; Hemming et al., 2008; Jones et al., 2008). Similar to barley, photoperiod-sensitive wheat is stimulated to flower only after exposure to long days, and flowering is delayed during short days. Photoperiod-insensitive wheat flowers independently of day length and can be grown at lower latitudes (Worland and Snape, 2001). In wheat, dominant PPD1 greatly reduce sensitivity to photoperiod and confer early flowering phenotype under both LD and SD conditions, resulting yield benefits in Europe (Cockram et al., 2007). Another gene involved in adaptation of LD crops to different latitudes is PPD2. In barley, the dominant PPD2 allele was selected for in spring cultivars at low latitudes to promote flowering in SD conditions, while the recessive ppd2 allele was selected for in winter cultivars grown at higher latitudes (Halliwell et al., 2016).
Wild peas display a typical winter habit, which consists of germination in autumn, vegetative growth during winter, and flowering in response to long days in spring (Abbo et al., 2003). However, the majority of cultivated peas can flower in SD and are grown as a spring crop, suggesting this ability has been an important factor for the expansion of pea cultivation (Lejeune-Hénaut et al., 1999). Four flowering loci HR, SN, LATE FLOWERING (LF), and EARLY (E) have been found to contribute to this variation (Foucher et al., 2003; Weller et al., 2012; Liew et al., 2014). The recessive hr allele causes early flowering in SD and decreases photoperiod response (Weller et al., 2012). sn mutants flowered early in SD conditions and eliminated PS (Liew et al., 2014). lf mutants displayed an extremely early, photoperiod-insensitive initiation of flowering (Foucher et al., 2003). The E locus can promote flowering without influencing the general PS of the plant, but the mechanism is not well understood at the molecular level (Lejeune-Hénaut et al., 2008; Weller et al., 2012). Various allelic combinations of HR, SN, LF, and E confer a wide range of flowering times in various conditions. The lf sn allelic combination, for instance, contributes to extremely early flowering and complete photoperiod insensitivity. The LF SN HR e allelic combination, however, contributes to late flowering in LD conditions and completely prevents flowering in SD conditions (Murfet, 1985; Weller et al., 2012). Most spring flowering (hr) pea cultivars carry at least one additional mutation of sn or lf alleles, with many also carrying mutations at the E locus (Weller and Ortega, 2015).
In rice, Hd1, Ghd7, DTH8, and OsPRR37 are core genes regulating flowering, and different combinations of these genes determine the photoperiod response and latitudinal adaptability of rice (Zhang et al., 2015; Zhang et al., 2019; Zong et al., 2021). The wild rice O. rufipogon, which is grown in tropical and subtropical regions of Asia, has strong PS and contains functional Hd1, Ghd7, DTH8, and OsPRR37 alleles. As wild rice evolved into modern varieties with different levels of PS, various allelic combinations of these genes were selected for to adapt rice to different latitudes. The sixteen possible allelic combinations of these four genes can be divided into three main groups with different PS. The first group exhibits strong PS and contains either four functional alleles (HGDP), three functional alleles (HGDp, HgDP, HGdP, hGDP), or functional alleles of only Hd1 and Ghd7 (HGdp). These combinations lead to a long vegetative growth phase and plants carrying them are mainly cultivated in tropical and subtropical regions of China. Rice cultivars containing combination of Hd1, Ghd7 and Hd1, Ghd7, DTH8 have LD repression and SD promotion effects, resulting sufficient vegetative growth for maxima photoassimilation and higher yield under LD conditions (Sun et al., 2022). The second group exhibits no PS and contains only a functional Hd1 allele (Hgdp) or four non-functional alleles (hgdp). These combinations lead to early heading dates and are generally cultivated in the northern part of China. The third group exhibits moderate photoperiod sensitivity contains the other eight allelic combinations, and is suitable for planting in the middle latitudes of China (Zhang et al., 2015; Zong et al., 2021; Chen et al., 2022). In summary, non-functional alleles of Hd1, Ghd7, DTH8, and OsPRR37 allow rice to be grown at higher latitudes, while functional alleles facilitate adaptation to lower latitudes. In addition, variation in other flowering-related genes also helps rice to adapt to different latitudes. For example, as rice began to be cultivated at higher latitudes, a functional early-heading RFT1 allele was selected for, while the late-heading non-functional rft1 allele was retained in wild or cultivated rice grown at lower latitudes (Ogiso-Tanaka et al., 2013; Naranjo et al., 2014; Zhao et al., 2015). In rice breading, the late-heading allele could be utilized for increasing yield when growth duration is not limited. The early-heading allele is preferred when the constraint comes in multiple season-cropping systems and in the northernmost region of rice cultivation (Zhu et al., 2017). Non-functional alleles of Hd6 and Hd16 also contributed to the expansion of rice cultivation to higher latitudes (Kwon et al., 2014; Nemoto et al., 2018).
Soybean, as a SD crop, became acclimated to LD conditions at higher latitudes in Asia and North America by accumulating early-flowering alleles to reduce or completely eliminate its photoperiod sensitivity. Tof11 and Tof12 have played essential roles in soybean domestication for growth at high latitudes. The tof12 mutation has been selected for in cultivated soybean, resulting in earlier flowering and maturity. The tof11 mutation, which occurred after that of tof12, further accelerated flowering and maturity, also contributing to adaptation to higher latitudes (Li et al., 2019; Li et al., 2020; Lu et al., 2020). As plants acclimated to higher latitudes, different combinations of E1/E1lb, E3, and E4 alleles were selected for, such as e3e4, e1e3e4, e1e3, e1e4, e1-ase3 and e1-ase1lbe3 (Xu et al., 2013; Zhu et al., 2019). e2, which leads to an early flowering phenotype, is prevalent in soybean cultivated in northern China (Langewisch et al., 2014; Wang et al., 2016). Recent research found that the Tof5H1 allele was artificially selected for in cultivated soybean and promotes adaptation to higher latitudes. Moreover, the early flowering allele Tof18G promotes adaptation to high latitudes in both cultivated and wild soybean (Kou et al., 2022).
Conversely, for soybean to acclimate to the SD conditions at lower latitudes in Brazil, required a long juvenile (LJ) trait to delay flowering time and improve yield. When soybean was first imported to Brazil from North America, it could only be cultivated farther south than 22°S. This barrier remained until the LJ trait was identified and introduced into soybean cultivars in central-western Brazil in 1970, allowing soybean production to expand to lower latitude regions and even to the equator (Lin et al., 2021b). J is the main locus regulating the LJ trait. Loss-of-function mutations in J can increase soybean yield by 30–50% by prolonging the flowering phase (Lu et al., 2017; Fang et al., 2021). A recent study identified a novel locus, Tof16, that delays flowering time and improves yield at low latitudes. Mutations in Tof16 and J were gradually selected for as soybean acclimated to tropical regions. When soybean was initially disseminated to lower latitudes, weak tof16 and j mutants were selected for, leading to delayed flowering. However, these weak mutations in tof16 and j did not lead to complete adaptation to tropical regions. Thus, null alleles of tof16 and j were selected for based on the two earlier weak alleles, which prolonged the flowering period even further and improved soybean yield in tropical regions (Dong et al., 2021). In addition, the ft2aft5a double mutant could overcome the genetic compensation effect and showed an enhanced LJ phenotype and high yield at low latitudes (Li et al., 2021). Moreover, the lux1lux2 double mutant of soybean completely lost photoperiod sensitivity, resulting in extremely late flowering. This phenotype was similar to the famous photoperiod insensitive tobacco (Nicotiana tabacum) mutant Maryland Mammoth. Thus, lux1lux2 was named the Guangzhou Mammoth (Bu et al., 2021). All these alleles and varieties provided important genetic resources for improving soybean yield in tropical areas.
In maize, many genes controlling flowering time have been found to play important roles in the expansion of cultivation from tropical and subtropical regions to higher latitudes. ZmCOL3 is a repressor of flowering that functions under LD conditions. The loss of one cytosine in the 3’UTR of ZmCOL3 and the presence of a 551bp fragment in the promoter region have been found to reduce transcription of ZmCOL3 and help maize adapt to temperate regions (Jin et al., 2018). Temperate maize exhibits higher ZmMADS69 expression than tropical maize in both the apex and leaf tip tissues, which indicates ZmMADS69 might have been selected for as maize adapted to temperate regions (Liang et al., 2019). Defective alleles of ZmCCT9 and ZmCCT10 were selected for in maize cultivars that are grown in North and South America; these alleles result in the activation of florigen ZmZCN8 and consequently accelerated flowering in LD conditions (Yang et al., 2013; Guo et al., 2018; Huang et al., 2018). A miniature transposon (MITE) inserted 70kb upstream of ZmRAP2.7 was another major target of selection and contributed to adaptation of maize to temperate regions (Ducrocq et al., 2008). Two genotypes, SNP-1245A and Indel-2339 in the promoter of ZmZCN8, have also been identified. The early flowering SNP-1245A allele was initially selected for during the early domestication of maize. The Indel-2339 allele was later introgressed into the SNP-1235A haplotype and was subsequently selected for as maize cultivation expanded from its tropical origin to more temperate regions (Guo et al., 2018). In summary, the ZmMADS69-ZmRAP2.7-ZmZCN8 regulatory module has been targeted by selection and contributed to the expansion of maize cultivation to higher latitudes.
5 Future perspectives
In this review, we discussed recent studies in the field of flowering time regulation in LD crops like wheat, barley, and pea as well as SD crops like rice, soybean, and maize. In these major crops, flowering time is regulated by genetic networks that respond to day length. All LD crops and SD crops have similar regulatory modules that control flowering time (Figure 4).
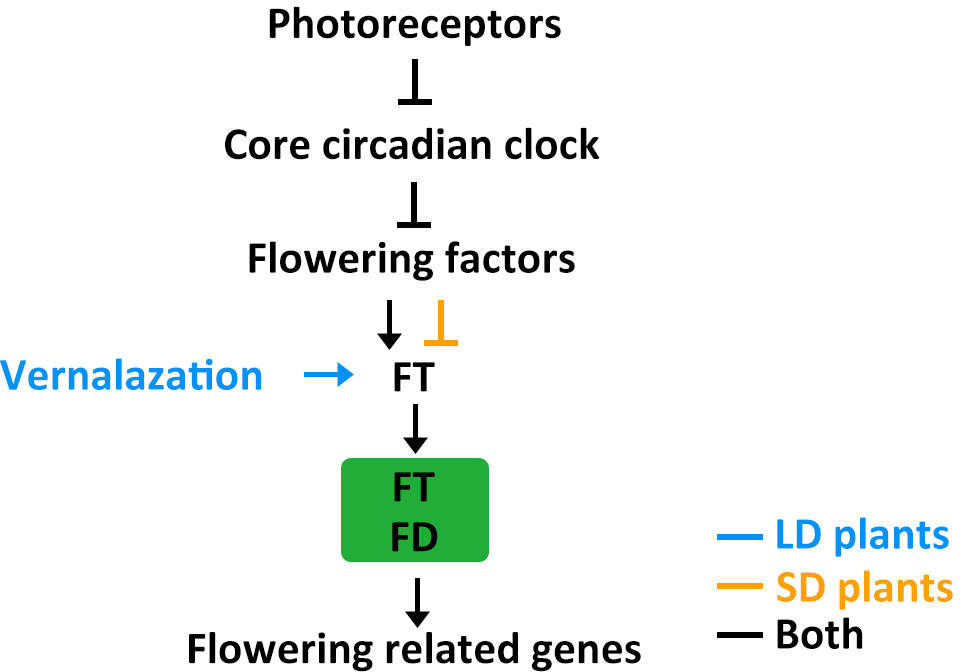
Figure 4 The putative model of flowering regulatory module in LD crops and SD crops. Positive and negative regulatory actions are indicated by arrows and lines with bares, respectively.
Photoreceptors detect light signals and transmit this information to the circadian clock through several different signalling mechanisms. The circadian clock then integrates the light cues and regulates flowering factors. Flowering factors modulate expression of the florigen gene FT. FT moves from leaves to the SAM and interacts with FD to activate flowering related genes, such as AP1 and FUL. The core circadian clock genes ELF3, ELF4, LUX, and PRRs are conserved in crops and play important roles in natural adaptation to different latitudes. The FT-FD complex is also conserved in crops and further regulates downstream flowering related genes, such as AP1/FUL-like genes like VRN1 in wheat, VEG1, PIM, and UNI in pea, OsMADS14, OsMADS15, and OsMADS18 in rice, GmAP1 in soybean, and ZmZMM4 in maize (Hofer et al., 1997; Taylor et al., 2002; Danilevskaya et al., 2008; Li and Dubcovsky, 2008; Berbel et al., 2012; Kobayashi et al., 2012; Nan et al., 2014; Yin et al., 2019; Dong et al., 2022). Usually, the flowering factor is a homolog of Arabidopsis CO, such as CO in wheat and barley, Hd1 in rice, and CONZ1 in maize (Griffiths et al., 2003; Miller et al., 2008). However, in addition to the conserved CO-FT module, rice has specifically evolved the (Hd1/Ghd7/DTH8)-Ehd1-Hd3a/RFT1 pathway (Zong et al., 2021). The interplay between SD-promotion and LD-repression pathways determines the differential effects of daylength on rice heading, highlighting the genetic diversity of flowering control mechanisms in crops. Soybean, in addition to having Arabidopsis CO homologs, also possesses the unique central flowering factor E1. Further research is needed, however, to fully understand why this specific central flowering factor evolved in soybean (Xu et al., 2015). For example, it is unknown whether E1 also appeared in other crops but disappeared during evolutionary history or was selected against during domestication and improvement.
Both LD and SD crops require changes in their flowering behavior when they move away from their native ranges and adapt to a new locality. In general, they become less sensitive to photoperiods to ensure reproductive success in non-native zones. However, different crops have evolved different strategies for adjusting their flowering time. For example, crops differ in the role played by other agronomic traits, such as temperature tolerance, during adaptation. Natural variation in flowering time regulation in SD crops often involves just photoperiod-related genes, but in many LD crops it may also involve low temperature-related genes (Figure 4) (Lin et al., 2021a). These differences may reflect where these lineages evolved: SD crops evolved in equatorial regions characterized by stable temperatures and daylengths throughout the year, while LD crops evolved in temperate regions with fluctuating temperatures and changing daylengths. SD crops mainly developed photoperiodic control of flowering, while LD crops acquired additional vernalization requirements as an adaptation to the cold. Further studies are needed to systematically evaluate natural variation in other agronomic traits that influence flowering time. This information would help clarify the evolution of flowering regulatory pathways and possibly help generate new cultivars with improved yields.
As major crops were disseminated to new continents, landraces were developed by farmers as the outcome of artificial selection that facilitated adaptation to the new environments. Subsequently, breeders used introduction, selection, and cross breeding to introduce desirable agronomic traits into crops and generate commercial cultivars. Different allelic combinations, such as combinations of Hd1, Ghd7, DTH8, and OsPRR37 in rice, could help crops withstand a range of ecological and climatic conditions (Zhang et al., 2015; Zong et al., 2021; Chen et al., 2022). Thus, by investigating the great genetic diversity in crop populations, reintroducing useful genetic resources into crops, and exploiting appropriate gene combinations during breeding, farmers and breeders may be able to improve yields and increase the agricultural and geographic flexibility of crops in the future.
In summary, a comprehensive understanding of the molecular networks regulating flowering time in different crops is needed to maximize production. Additionally, it is important for breeders to investigate variation in flowering time regulation, including how key genes are retained or lost through the process of crop domestication, in order to reveal the crops’ histories, select better alleles, and develop improved cultivars for future breeding applications.
Author contributions
FW drafted the manuscript. SLi and FK revised the manuscript. XL and SLu supervised this work and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Natural Science Foundation of China (32201866) to FW, the open competition program of the top ten critical priorities of Agricultural Science and Technology Innovations for the 14th Five-Year Plan of Guangdong Province (2022SDZG05) and Major Program of Guangdong Basic and Applied Research (Grant 2019B030302006) to FK, and City-School (Institute) Joint Funding Program for Basic and Applied Basic Research (202201020130) to SL.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbo, S., Shtienberg, D., Lichtenzveig, J., Lev-Yadun, S., Gopher, A. (2003). The chickpea, summer cropping, and a new model for pulse domestication in the ancient near east. Q. Rev. Biol. 78, 435–448. doi: 10.1086/378927
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi: 10.1126/science.1115983
Alvarez, M. A., Tranquilli, G., Lewis, S., Kippes, N., Dubcovsky, J. (2016). Genetic and physical mapping of the earliness per se locus eps-a (m) 1 in triticum monococcum identifies EARLY FLOWERING 3 (ELF3) as a candidate gene. Funct. Integr. Genomics 16, 365–382. doi: 10.1007/s10142-016-0490-3
Andrade, L., Lu, Y., Cordeiro, A., Costa, J. M. F., Wigge, P. A., Saibo, N. J. M., et al. (2022). The evening complex integrates photoperiod signals to control flowering in rice. Proc. Natl. Acad. Sci. U. S. A. 119, e2122582119. doi: 10.1073/pnas.2122582119
Bendix, C., Mendoza, J. M., Stanley, D. N., Meeley, R., Harmon, F. G. (2013). The circadian clock-associated gene gigantea1 affects maize developmental transitions. Plant Cell Environ. 36, 1379–1390. doi: 10.1111/pce.12067
Berbel, A., Ferrándiz, C., Hecht, V., Dalmais, M., Lund, O. S., Sussmilch, F. C., et al. (2012). VEGETATIVE1 is essential for development of the compound inflorescence in pea. Nat. Commun. 3, 797. doi: 10.1038/ncomms1801
Bernard, R. L. (1971). Two major genes for time of flowering and maturity in soybeans. Crop Sci. 11, 242–244. doi: 10.2135/cropsci1971.0011183X001100020022x
Beveridge, C. A., Murfet, I. C. (1996). The gigas mutant in pea is deficient in the floral stimulus. Physiol. Plant 96, 637–645. doi: 10.1111/J.1399-3054.1996.TB00237.X
Bonato, E. R., Vello, N. A. (1999). E6, a dominant gene conditioning early flowering and maturity in soybeans. Genet. Mol. Biol. 22, 229–232. doi: 10.1590/s1415-47571999000200016
Brambilla, V., Gomez-Ariza, J., Cerise, M., Fornara, F. (2017). The importance of being on time: Regulatory networks controlling photoperiodic flowering in cereals. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00665
Brandolini, A., Brandolini, A. (2009). Maize introduction, evolution and diffusion in Italy. Maydica 54, 233–242.
Bu, T., Lu, S., Wang, K., Dong, L., Li, S., Xie, Q., et al. (2021). A critical role of the soybean evening complex in the control of photoperiod sensitivity and adaptation. Proc. Natl. Acad. Sci. 118, e2010241118. doi: 10.1073/pnas.2010241118
Buzzell, R. I. (1971). Inheritance of a soybean flowering response to fluorescent daylength conditions. Can. J. Genet. Cytol. 13, 703–707. doi: 10.1139/g71-100
Buzzell, R., Voldeng, H. (1980). Inheritance of insensitivity to long daylength. Soybean Genet. Newsl. 7, 26–29.
Campoli, C., Pankin, A., Drosse, B., Casao, C. M., Davis, S. J., Korff, M. (2013). HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: Phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol. 199, 1045–1059. doi: 10.1111/nph.12346
Cao, D., Li, Y., Lu, S., Wang, J., Nan, H., Li, X., et al. (2015). GmCOL1a and GmCOL1b function as flowering repressors in soybean under long-day conditions. Plant Cell Physiol. 56, 2409–2422. doi: 10.1093/pcp/pcv152
Cao, D., Takeshima, R., Zhao, C., Liu, B., Jun, A., Kong, F. (2016). Molecular mechanisms of flowering under long days and stem growth habit in soybean. J. Exp. Bot. 68, 1873–1884. doi: 10.1093/jxb/erw394
Casao, M. C., Karsai, I., Igartua, E., Gracia, M. P., Veisz, O., Casas, A. M. (2011). Adaptation of barley to mild winters: A role for PPDH2. BMC Plant Biol. 11, 164. doi: 10.1186/1471-2229-11-164
Castelletti, S., Coupel-Ledru, A., Granato, I., Palaffre, C., Cabrera-Bosquet, L., Tonelli, C., et al. (2020). Maize adaptation across temperate climates was obtained via expression of two florigen genes. PLoS Genet. 16, e1008882. doi: 10.1371/journal.pgen.1008882
Chen, A., Li, C., Hu, W., Lau, M. Y., Lin, H., Rockwell, N. C., et al. (2014). Phytochrome c plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proc. Natl. Acad. Sci. U. S. A. 111, 10037–10044. doi: 10.1073/pnas.1409795111
Chen, E., Huang, X., Tian, Z., Wing, R. A., Han, B. (2019). The genomics of oryza species provides insights into rice domestication and heterosis. Annu. Rev. Plant Biol. 70, 639–665. doi: 10.1146/annurev-arplant-050718-100320
Chen, R., Deng, Y., Ding, Y., Guo, J., Qiu, J., Wang, B., et al. (2022). Rice functional genomics: Decades’ efforts and roads ahead. Sci. China Life Sci. 65, 33–92. doi: 10.1007/s11427-021-2024-0
Choi, J. Y., Zaidem, M., Gutaker, R., Dorph, K., Singh, R. K., Purugganan, M. D. (2019). The complex geography of domestication of the African rice oryza glaberrima. PLoS Genet. 15, e1007414. doi: 10.1371/journal.pgen.1007414
Civáň, P., Craig, H., Cox, C. J., Brown, T. A. (2015). Three geographically separate domestications of Asian rice. Nat. Plants 1, 15164. doi: 10.1038/nplants.2015.164
Cober, E. R., Molnar, S. J., Charette, M., Voldeng, H. D. (2010). A new locus for early maturity in soybean. Crop Sci. 50, 524–527. doi: 10.2135/cropsci2009.04.0174
Cober, E. R., Tanner, J. W., Voldeng, H. D. (1996). Genetic control of photoperiod response in early-maturing, near-isogenic soybean lines. Crop Sci. 36, 601–605. doi: 10.2135/cropsci1996.0011183X003600030013x
Cober, E. R., Voldeng, H. D. (2001). A new soybean maturity and photoperiod-sensitivity locus linked to E1 and T. Crop Sci. 41, 698–701. doi: 10.2135/CROPSCI2001.413698X
Cockram, J., Jones, H., Leigh, F. J., O’Sullivan, D., Powell, W., Laurie, D. A., et al. (2007). Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. J. Exp. Bot. 58, 1231–1244. doi: 10.1093/jxb/erm042
Danilevskaya, O. N., Meng, X., Selinger, D. A., Deschamps, S., Hermon, P., Vansant, G., et al. (2008). Involvement of the MADS-box gene ZMM4 in floral induction and inflorescence development in maize. Plant Physiol. 147, 2054–2069. doi: 10.1104/pp.107.115261
Doebley, J. F., Gaut, B. S., Smith, B. D. (2006). The molecular genetics of crop domestication. Cell 127, 1309–1321. doi: 10.1016/j.cell.2006.12.006
Dong, L., Cheng, Q., Fang, C., Kong, L., Yang, H., Hou, Z., et al. (2022). Parallel selection of distinct Tof5 alleles drove the adaptation of cultivated and wild soybean to high latitudes. Mol. Plant 15, 308–321. doi: 10.1016/j.molp.2021.10.004
Dong, L., Fang, C., Cheng, Q., Su, T., Kou, K., Kong, L., et al. (2021). Genetic basis and adaptation trajectory of soybean from its temperate origin to tropics. Nat. Commun. 12, 5445. doi: 10.1038/s41467-021-25800-3
Du, A., Tian, W., Wei, M., Yan, W., He, H., Zhou, D., et al. (2017). The DTH8-Hd1 module mediates day-length-dependent regulation of rice flowering. Mol. Plant 10, 948–961. doi: 10.1016/j.molp.2017.05.006
Ducrocq, S., Giauffret, C., Madur, D., Combes, V., Dumas, F., Jouanne, S., et al. (2009). Fine mapping and haplotype structure analysis of a major flowering time quantitative trait locus on maize chromosome 10. Genetics 183, 1555–1563. doi: 10.1534/genetics.109.106922
Ducrocq, S., Madur, D., Veyrieras, J.-B., Camus-Kulandaivelu, L., Kloiber-Maitz, M., Presterl, T., et al. (2008). Key impact of Vgt1 on flowering time adaptation in maize: Evidence from association mapping and ecogeographical information. Genetics 178, 2433–2437. doi: 10.1534/genetics.107.084830
Fang, C., Liu, J., Zhang, T., Su, T., Li, S., Cheng, Q., et al. (2021). A recent retrotransposon insertion of J caused E6 locus facilitating soybean adaptation into low latitude. J. Integr. Plant Biol. 63, 995–1003. doi: 10.1111/jipb.13034
Faure, S., Higgins, J., Turner, A., Laurie, D. A. (2007). The flowering locus T -like gene family in barley (Hordeum vulgare). Genetics 176, 599–609. doi: 10.1534/genetics.106.069500
Faure, S., Turner, A. S., Gruszka, D., Christodoulou, V., Davis, S. J., von Korff, M., et al. (2012). Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc. Natl. Acad. Sci. 109, 8328–8333. doi: 10.1073/pnas.1120496109
Foucher, F., Morin, J., Courtiade, J., Cadioux, S., Ellis, N., Banfield, M. J., et al. (2003). DETERMINATE and LATE FLOWERING are two TERMINAL FLOWER1/CENTRORADIALIS homologs that control two distinct phases of flowering initiation and development in pea. Plant Cell 15, 2742–2754. doi: 10.1105/tpc.015701
Fuller, D. Q., Harvey, E., Qin, L. (2007). Presumed domestication? evidence for wild rice cultivation and domestication in the fifth millennium BC of the lower Yangtze region. Antiquity 81, 316–331. doi: 10.1017/S0003598X0009520X
Garner, W. W., Allard, H. A. (1920). Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. Mon. Weather Rev. 48, 415. doi: 10.1175/1520-0493(1920)48<415
Gohar, S., Sajjad, M., Zulfiqar, S., Liu, J., Wu, J., Rahman, M. (2022). Domestication of newly evolved hexaploid wheat–a journey of wild grass to cultivated wheat. Front. Genet. 13. doi: 10.3389/fgene.2022.1022931
Goretti, D., Martignago, D., Landini, M., Brambilla, V., Gómez-Ariza, J., Gnesutta, N., et al. (2017). Transcriptional and post-transcriptional mechanisms limit heading date 1 (Hd1) function to adapt rice to high latitudes. PLoS Genet. 13, e1006530. doi: 10.1371/journal.pgen.1006530
Griffiths, S., Dunford, R. P., Coupland, G., Laurie, D. A. (2003). The evolution of CONSTANS -like gene families in barley, rice, and arabidopsis. Plant Physiol. 131, 1855–1867. doi: 10.1104/pp.102.016188
Guo, L., Wang, X., Zhao, M., Huang, C., Li, C., Li, D., et al. (2018). Stepwise cis-regulatory changes in ZCN8 contribute to maize flowering-time adaptation. Curr. Biol. 28, 3005–3015.e4. doi: 10.1016/j.cub.2018.07.029
Gutaker, R. M., Groen, S. C., Bellis, E. S., Choi, J. Y., Pires, I. S., Bocinsky, R. K., et al. (2020). Genomic history and ecology of the geographic spread of rice. Nat. Plants 6, 492–502. doi: 10.1038/s41477-020-0659-6
Haas, M., Schreiber, M., Mascher, M. (2019). Domestication and crop evolution of wheat and barley: Genes, genomics, and future directions. J. Integr. Plant Biol. 61, 204–225. doi: 10.1111/jipb.12737
Halliwell, J., Borrill, P., Gordon, A., Kowalczyk, R., Pagano, M. L., Saccomanno, B., et al. (2016). Systematic investigation of FLOWERING LOCUS T-like poaceae gene families identifies the short-day expressed flowering pathway gene, TaFT3 in wheat (Triticum aestivum l.). Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00857
Hayama, R., Yokoi, S., Tamaki, S., Yano, M., Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
Hecht, V., Knowles, C. L., Vander Schoor, J. K., Liew, L. C., Jones, S. E., Lambert, M. J. M., et al. (2007). Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 144, 648–661. doi: 10.1104/pp.107.096818
Hecht, V., Laurie, R. E., Vander Schoor, J. K., Ridge, S., Knowles, C. L., Liew, L. C., et al. (2011). The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 23, 147–161. doi: 10.1105/tpc.110.081042
Hemming, M. N., Peacock, W. J., Dennis, E. S., Trevaskis, B. (2008). Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiol. 147, 355–366. doi: 10.1104/pp.108.116418
Hofer, J., Turner, L., Hellens, R., Ambrose, M., Matthews, P., Michael, A., et al. (1997). UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr. Biol. 7, 581–587. doi: 10.1016/S0960-9822(06)00257-0
Hori, K., Ogiso-Tanaka, E., Matsubara, K., Yamanouchi, U., Ebana, K., Yano, M. (2013). Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J. 76, 36–46. doi: 10.1111/tpj.12268
Huang, C., Sun, H., Xu, D., Chen, Q., Liang, Y., Wang, X., et al. (2018). ZmCCT9 enhances maize adaptation to higher latitudes. Proc. Natl. Acad. Sci. 115, 334–341. doi: 10.1073/pnas.1718058115
Huang, X., Kurata, N., Wei, X., Wang, Z.-X., Wang, A., Zhao, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. doi: 10.1038/nature11532
Huang, X., Zhao, Q., Han, B. (2015). Comparative population genomics reveals strong divergence and infrequent introgression between Asian and African rice. Mol. Plant 8, 958–960. doi: 10.1016/j.molp.2015.01.010
Hung, H.-Y., Shannon, L. M., Tian, F., Bradbury, P. J., Chen, C., Flint-Garcia, S. A., et al. (2012). ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc. Natl. Acad. Sci. U. S. A. 109, E1913–E1921. doi: 10.1073/pnas.1203189109
Hymowitz, T., Shurtleff, W. R. (2005). Debunking soybean myths and legends in the historical and popular literature. Crop Sci. 45, 473–476. doi: 10.2135/cropsci2005.0473
Jin, M., Liu, X., Jia, W., Liu, H., Li, W., Peng, Y., et al. (2018). ZmCOL3, a CCT gene represses flowering in maize by interfering with the circadian clock and activating expression of ZmCCT. J. Integr. Plant Biol. 60, 465–480. doi: 10.1111/jipb.12632
Jing, R., Vershinin, A., Grzebyta, J., Shaw, P., Smýkal, P., Marshall, D., et al. (2010). The genetic diversity and evolution of field pea (Pisum) studied by high throughput retrotransposon based insertion polymorphism (RBIP) marker analysis. BMC Evol. Biol. 10, 44. doi: 10.1186/1471-2148-10-44
Jones, H., Leigh, F. J., Mackay, I., Bower, M. A., Smith, L. M. J., Charles, M. P., et al. (2008). Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the fertile crescent. Mol. Biol. Evol. 25, 2211–2219. doi: 10.1093/molbev/msn167
Kihara, H. (1969). History of biology and other sciences in Japan in retrospect. Proc. XII Intern. Congr Genet. 3, 49–70.
Kikuchi, R., Kawahigashi, H., Ando, T., Tonooka, T., Handa, H. (2009). Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 149, 1341–1353. doi: 10.1104/pp.108.132134
Kobayashi, K., Yasuno, N., Sato, Y., Yoda, M., Yamazaki, R., Kimizu, M., et al. (2012). Inflorescence meristem identity in rice is specified by overlapping functions of three AP1 / FUL -like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24, 1848–1859. doi: 10.1105/tpc.112.097105
Komiya, R., Ikegami, A., Tamaki, S., Yokoi, S., Shimamoto, K. (2008). Hd3a and RFT1 are essential for flowering in rice. Development 135, 767–774. doi: 10.1242/dev.008631
Komiya, R., Yokoi, S., Shimamoto, K. (2009). A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136, 3443–3450. doi: 10.1242/dev.040170
Kong, F., Liu, B., Xia, Z., Sato, S., Kim, B. M., Watanabe, S., et al. (2010). Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 154, 1220–1231. doi: 10.1104/pp.110.160796
Kong, F., Nan, H., Cao, D., Li, Y., Wu, F., Wang, J., et al. (2014). A new dominant gene E9 conditions early flowering and maturity in soybean. Crop Sci. 54, 2529–2535. doi: 10.2135/cropsci2014.03.0228
Kou, K., Yang, H., Li, H., Fang, C., Chen, L., Yue, L., et al. (2022). A functionally divergent SOC1 homolog improves soybean yield and latitudinal adaptation. Curr. Biol. 32, 1–15. doi: 10.1016/j.cub.2022.02.046
Kwon, C.-T., Yoo, S.-C., Koo, B.-H., Cho, S.-H., Park, J.-W., Zhang, Z., et al. (2014). Natural variation in early flowering1 contributes to early flowering in japonica rice under long days. Plant Cell Environ. 37, 101–112. doi: 10.1111/pce.12134
Langewisch, T., Zhang, H., Vincent, R., Joshi, T., Xu, D., Bilyeu, K. (2014). Major soybean maturity gene haplotypes revealed by SNPViz analysis of 72 sequenced soybean genomes. PLoS One 9, e94150. doi: 10.1371/journal.pone.0094150
Laurie, D. A., Pratchett, N., Snape, J. W., Bezant, J. H. (1995). RFLP mapping of five major genes and eight quantitative trait loci controlling flowering time in a winter × spring barley (Hordeum vulgare l.) cross. Genome 38, 575–585. doi: 10.1139/g95-074
Lazakis, C. M., Coneva, V., Colasanti, J. (2011). ZCN8 encodes a potential orthologue of arabidopsis FT florigen that integrates both endogenous and photoperiod flowering signals in maize. J. Exp. Bot. 62, 4833–4842. doi: 10.1093/jxb/err129
Lejeune-Hénaut, I., Bourion, V., Etévé, G., Cunot, E., Delhaye, K., Desmyter, C. (1999). Floral initiation in field-grown forage peas is delayed to a greater extent by short photoperiods, than in other types of European varieties. Euphytica 109, 201–211. doi: 10.1023/A:1003727324475
Lejeune-Hénaut, I., Hanocq, E., Béthencourt, L., Fontaine, V., Delbreil, B., Morin, J., et al. (2008). The flowering locus hr colocalizes with a major QTL affecting winter frost tolerance in pisum sativum l. Theor. Appl. Genet. 116, 1105–1116. doi: 10.1007/s00122-008-0739-x
Li, C., Dubcovsky, J. (2008). Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 55, 543–554. doi: 10.1111/j.1365-313X.2008.03526.x
Li, C., Li, Y., Li, Y., Lu, H., Hong, H., Tian, Y., et al. (2020). A domestication-associated gene GmPRR3b regulates the circadian clock and flowering time in soybean. Mol. Plant 13, 745–759. doi: 10.1016/j.molp.2020.01.014
Li, M.-W., Liu, W., Lam, H.-M., Gendron, J. M. (2019). Characterization of two growth period QTLs reveals modification of PRR3 genes during soybean domestication. Plant Cell Physiol. 60, 407–420. doi: 10.1093/pcp/pcy215
Li, X., Fang, C., Yang, Y., Lv, T., Su, T., Chen, L., et al. (2021). Overcoming the genetic compensation response of soybean florigens to improve adaptation and yield at low latitudes. Curr. Biol. 31, 3755–3767. doi: 10.1016/j.cub.2021.06.037
Liang, Y., Liu, Q., Wang, X., Huang, C., Xu, G., Hey, S., et al. (2019). ZmMADS69 functions as a flowering activator through the ZmRap2.7-ZCN8 regulatory module and contributes to maize flowering time adaptation. New Phytol. 221, 2335–2347. doi: 10.1111/nph.15512
Liew, L. C., Hecht, V., Sussmilch, F. C., Weller, J. L. (2014). The pea photoperiod response gene STERILE NODES is an ortholog of LUX ARRHYTHMO. Plant Physiol. 165, 648–657. doi: 10.1104/pp.114.237008
Lin, X., Dong, L., Tang, Y., Li, H., Cheng, Q., Li, H., et al. (2022). Novel and multifaceted regulations of photoperiodic flowering by phytochrome a in soybean. Proc. Natl. Acad. Sci. U. S. A. 119, e2208708119. doi: 10.1073/pnas.2208708119
Lin, X., Fang, C., Liu, B., Kong, F. (2021a). Natural variation and artificial selection of photoperiodic flowering genes and their applications in crop adaptation. Abiotech 2, 156–169. doi: 10.1007/s42994-021-00039-0
Lin, X., Liu, B., Weller, J. L., Abe, J., Kong, F. (2021b). Molecular mechanisms for the photoperiodic regulation of flowering in soybean. J. Integr. Plant Biol. 63, 981–994. doi: 10.1111/jipb.13021
Lister, D. L., Jones, H., Oliveira, H. R., Petrie, C. A., Liu, X., Cockram, J., et al. (2018). Barley heads east: Genetic analyses reveal routes of spread through diverse Eurasian landscapes. PLoS One 13, e0196652. doi: 10.1371/journal.pone.0196652
Liu, W., Jiang, B., Ma, L., Zhang, S., Zhai, H., Xu, X., et al. (2018). Functional diversification of flowering locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. New Phytol. 217, 1335–1345. doi: 10.1111/nph.14884
Liu, B., Kanazawa, A., Matsumura, H., Takahashi, R., Harada, K., Abe, J. (2008). Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome a gene. Genetics 180, 995–1007. doi: 10.1534/genetics.108.092742
Lu, S., Dong, L., Fang, C., Liu, S., Kong, L., Cheng, Q., et al. (2020). Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 52, 428–436. doi: 10.1038/s41588-020-0604-7
Lu, S., Zhao, X., Hu, Y., Liu, S., Nan, H., Li, X., et al. (2017). Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 49, 773–779. doi: 10.1038/ng.3819
Marcussen, T., Sandve, S. R., Heier, L., Spannagl, M., Pfeifer, M., Jakobsen, K. S., et al. (2014). Ancient hybridizations among the ancestral genomes of bread wheat. Science 345, 1250092. doi: 10.1126/science.1250092
Matsuoka, Y., Vigouroux, Y., Goodman, M. M., Sanchez G., J., Buckler, E., Doebley, J. (2002). A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. 99, 6080–6084. doi: 10.1073/pnas.052125199
McBlain, B. A., Bernard, R. L. (1987). A new gene affecting the time of flowering and maturity in soybeans. J. Hered. 78, 160–162. doi: 10.1093/oxfordjournals.jhered.a110349
Mendoza, J. M., Bendix, C., Meeley, R., Harmon, F. G. (2012). The homeologous zea mays gigantea genes: Characterization of expression and novel mutant alleles. Maydica 57, 252–259.
Meng, X., Muszynski, M. G., Danilevskaya, O. N. (2011). The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 23, 942–960. doi: 10.1105/tpc.110.081406
Miller, T. A., Muslin, E. H., Dorweiler, J. E. (2008). A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta 227, 1377–1388. doi: 10.1007/s00425-008-0709-1
Minow, M. A. A., Ávila, L. M., Turner, K., Ponzoni, E., Mascheretti, I., Dussault, F. M., et al. (2018). Distinct gene networks modulate floral induction of autonomous maize and photoperiod-dependent teosinte. J. Exp. Bot. 69, 2937–2952. doi: 10.1093/jxb/ery110
Mizuno, N., Nitta, M., Sato, K., Nasuda, S. (2012). A wheat homologue of PHYTOCLOCK 1 is a candidate gene conferring the early heading phenotype to einkorn wheat. Genes Genet. Syst. 87, 357–367. doi: 10.1266/ggs.87.357
Murfet, I. C. (1985). “Pisum sativum,” in CRC Handbook of flowering, vol. 97–126 . Ed. Halevy, A. H.. (Boca Raton: CRC Press).
Muszynski, M. G., Dam, T., Li, B., Shirbroun, D. M., Hou, Z., Bruggemann, E., et al. (2006). Delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 142, 1523–1536. doi: 10.1104/pp.106.088815
Nagel, D. H., Kay, S. A. (2012). Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 22, R648–R657. doi: 10.1016/j.cub.2012.07.025
Nan, H., Cao, D., Zhang, D., Li, Y., Lu, S., Tang, L., et al. (2014). GmFT2a and GmFT5a redundantly and differentially regulate flowering through interaction with and upregulation of the bZIP transcription factor GmFDL19 in soybean. PLoS One 9, e97669. doi: 10.1371/journal.pone.0097669
Naranjo, L., Talón, M., Domingo, C. (2014). Diversity of floral regulatory genes of japonica rice cultivated at northern latitudes. BMC Genomics 15, 101. doi: 10.1186/1471-2164-15-101
Nemoto, Y., Hori, K., Izawa, T. (2018). Fine-tuning of the setting of critical day length by two casein kinases in rice photoperiodic flowering. J. Exp. Bot. 69, 553–565. doi: 10.1093/jxb/erx412
Nunn, N., Qian, N. (2010). The columbian exchange: a history of disease, food, and ideas. J. Econ. Perspect. 24, 163–188. doi: 10.1257/jep.24.2.163
Ogiso, E., Takahashi, Y., Sasaki, T., Yano, M., Izawa, T. (2010). The role of casein kinase II in flowering time regulation has diversified during evolution. Plant Physiol. 152, 808–820. doi: 10.1104/pp.109.148908
Ogiso-Tanaka, E., Matsubara, K., Yamamoto, S., Nonoue, Y., Wu, J., Fujisawa, H., et al. (2013). Natural variation of the RICE FLOWERING LOCUS T 1 contributes to flowering time divergence in rice. PLoS One 8, e75959. doi: 10.1371/journal.pone.0075959
Pankin, A., Campoli, C., Dong, X., Kilian, B., Sharma, R., Himmelbach, A., et al. (2014). Mapping-by-sequencing identifies HvPHYTOCHROME c as a candidate gene for the early maturity 5 locus modulating the circadian clock and photoperiodic flowering in barley. Genetics 198, 383–396. doi: 10.1534/genetics.114.165613
Pearce, S., Kippes, N., Chen, A., Debernardi, J. M., Dubcovsky, J. (2016). RNA-Seq studies using wheat PHYTOCHROME b and PHYTOCHROME c mutants reveal shared and specific functions in the regulation of flowering and shade-avoidance pathways. BMC Plant Biol. 16, 141. doi: 10.1186/s12870-016-0831-3
Peng, L.-T., Shi, Z.-Y., Li, L., Shen, G.-Z., Zhang, J.-L. (2007). Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter. Biochem. Biophys. Res. Commun. 360, 251–256. doi: 10.1016/j.bbrc.2007.06.041
Peng, Q., Zhu, C., Liu, T., Zhang, S., Feng, S., Wu, C. (2021). Phosphorylation of OsFD1 by OsCIPK3 promotes the formation of RFT1-containing florigen activation complex for long-day flowering in rice. Mol. Plant 14, 1135–1148. doi: 10.1016/j.molp.2021.04.003
Piperno, D. R., Ranere, A. J., Holst, I., Iriarte, J., Dickau, R. (2009). Starch grain and phytolith evidence for early ninth millennium B.P. maize from the central balsas river valley, Mexico. Proc. Natl. Acad. Sci. 106, 5019–5024. doi: 10.1073/pnas.0812525106
Ray, J. D., Hinson, K., Mankono, J. E. B., Malo, M. F. (1995). Genetic control of a long-juvenile trait in soybean. Crop Sci. 35, 1001–1006. doi: 10.2135/cropsci1995.0011183X003500040012x
Salvi, S., Sponza, G., Morgante, M., Tomes, D., Niu, X., Fengler, K. A., et al. (2007). Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. U. S. A. 104, 11376–11381. doi: 10.1073/pnas.0704145104
Samanfar, B., Molnar, S. J., Charette, M., Schoenrock, A., Dehne, F., Golshani, A., et al. (2017). Mapping and identification of a potential candidate gene for a novel maturity locus, E10, in soybean. Theor. Appl. Genet. 130, 377–390. doi: 10.1007/s00122-016-2819-7
Scarth, R., Law, C. N. (1983). The location of the photoperiod gene, Ppd2 and an additional genetic factor for ear-emergence time on chromosome 2B of wheat. Heredity 51, 607–619. doi: 10.1038/hdy.1983.73
Sedivy, E. J., Wu, F., Hanzawa, Y. (2017). Soybean domestication: the origin, genetic architecture and molecular bases. New Phytol. 214, 539–553. doi: 10.1111/nph.14418
Seki, M., Chono, M., Matsunaka, H., Fujita, M., Oda, S., Kubo, K., et al. (2011). Distribution of photoperiod-insensitive alleles ppd-B1a and ppd-D1a and their effect on heading time in Japanese wheat cultivars. Breed. Sci. 61, 405–412. doi: 10.1270/jsbbs.61.405
Snodgrass, S. J., Hufford, M. B. (2018). Domestication genomics: untangling the complex history of African rice. Curr. Biol. 28, R786–R788. doi: 10.1016/j.cub.2018.05.072
Stacey, G. (2008). Genetics and genomics of soybean (New York, NY: Springer New York). doi: 10.1007/978-0-387-72299-3
Sun, K., Huang, M., Zong, W., Xiao, D., Lei, C., Luo, Y., et al. (2022). Hd1, Ghd7, and DTH8 synergistically determine the rice heading date and yield-related agronomic traits. J. Genet. Genomics 49, 437–447. doi: 10.1016/j.jgg.2022.02.018
Surkova, S. Y., Samsonova, M. G. (2022). Mechanisms of vernalization-induced flowering in legumes. Int. J. Mol. Sci. 23, 9889. doi: 10.3390/ijms23179889
Sussmilch, F. C., Berbel, A., Hecht, V., Vander Schoor, J. K., Ferrándiz, C., Madueño, F., et al. (2015). Pea VEGETATIVE2 is an FD homolog that is essential for flowering and compound inflorescence development. Plant Cell 27, 1046–1060. doi: 10.1105/tpc.115.136150
Sweeney, M., McCouch, S. (2007). The complex history of the domestication of rice. Ann. Bot. 100, 951–957. doi: 10.1093/aob/mcm128
Taoka, K., Ohki, I., Tsuji, H., Furuita, K., Hayashi, K., Yanase, T., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335. doi: 10.1038/nature10272
Taylor, S. A., Hofer, J. M. I., Murfet, I. C., Sollinger, J. D., Singer, S. R., Knox, M. R., et al. (2002). PROLIFERATING INFLORESCENCE MERISTEM, a MADS-box gene that regulates floral meristem identity in pea. Plant Physiol. 129, 1150–1159. doi: 10.1104/pp.001677
Tsubokura, Y., Matsumura, H., Xu, M., Liu, B., Nakashima, H., Anai, T., et al. (2013). Genetic variation in soybean at the maturity locus E4 is involved in adaptation to long days at high latitudes. Agronomy 3, 117–134. doi: 10.3390/agronomy3010117
Tsuji, H., Taoka, K., Shimamoto, K. (2011). Regulation of flowering in rice: Two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 14, 45–52. doi: 10.1016/j.pbi.2010.08.016
Turck, F., Fornara, F., Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594. doi: 10.1146/annurev.arplant.59.032607.092755
Turner, A., Beales, J., Faure, S., Dunford, R. P., Laurie, D. A. (2005). The pseudo-response regulator ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034. doi: 10.1126/science.1117619
van Andel, T. R., Meyer, R. S., Aflitos, S. A., Carney, J. A., Veltman, M. A., Copetti, D., et al. (2016). Tracing ancestor rice of Suriname maroons back to its African origin. Nat. Plants 2, 16149. doi: 10.1038/nplants.2016.149
Veltman, M. A., Flowers, J. M., van Andel, T. R., Schranz, M. E. (2019). Origins and geographic diversification of African rice (Oryza glaberrima). PLoS One 14, e0203508. doi: 10.1371/journal.pone.0203508
Vlăduţu, C., McLaughlin, J., Phillips, R. L. (1999). Fine mapping and characterization of linked quantitative trait loci involved in the transition of the maize apical meristem from vegetative to generative structures. Genetics 153, 993–1007. doi: 10.1093/genetics/153.2.993
Wang, F., Nan, H., Chen, L., Fang, C., Zhang, H., Su, T., et al. (2019). A new dominant locus, E11, controls early flowering time and maturity in soybean. Mol. Breed. 39, 70. doi: 10.1007/s11032-019-0978-3
Wang, L., Josephs, E. B., Lee, K. M., Roberts, L. M., Rellán-Álvarez, R., Ross-Ibarra, J., et al. (2021). Molecular parallelism underlies convergent highland adaptation of maize landraces. Mol. Biol. Evol. 38, 3567–3580. doi: 10.1093/molbev/msab119
Wang, M., Yu, Y., Haberer, G., Marri, P. R., Fan, C., Goicoechea, J. L., et al. (2014). The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat. Genet. 46, 982–988. doi: 10.1038/ng.3044
Wang, Y., Gu, Y., Gao, H., Qiu, L., Chang, R., Chen, S., et al. (2016). Molecular and geographic evolutionary support for the essential role of GIGANTEAa in soybean domestication of flowering time. BMC Evol. Biol. 16, 79. doi: 10.1186/s12862-016-0653-9
Watanabe, S., Hideshima, R., Xia, Z., Tsubokura, Y., Sato, S., Nakamoto, Y., et al. (2009). Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 182, 1251–1262. doi: 10.1534/genetics.108.098772
Wang, W., Mauleon, R., Hu, Z., Chebotarov, D., Tai, S., Wu, Z., et al. (2018). Genomic variation in 3,010 diverse accessions of Asian cultivated rice[J]. Nature 557 (7703), 43–49.
Watanabe, S., Xia, Z., Hideshima, R., Tsubokura, Y., Sato, S., Yamanaka, N., et al. (2011). A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188, 395–407. doi: 10.1534/genetics.110.125062
Weller, J. L., Hecht, V., Liew, L. C., Sussmilch, F. C., Wenden, B., Knowles, C. L., et al. (2009). Update on the genetic control of flowering in garden pea. J. Exp. Bot. 60, 2493–2499. doi: 10.1093/jxb/erp120
Weller, J. L., Liew, L. C., Hecht, V. F. G., Rajandran, V., Laurie, R. E., Ridge, S., et al. (2012). A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc. Natl. Acad. Sci. U. S. A. 109, 21158–21163. doi: 10.1073/pnas.1207943110
Weller, J. L., Murfet, I. C., Reid, J. B. (1997). Pea mutants with reduced sensitivity to far-red light define an important role for phytochrome a in day-length detection. Plant Physiol. 114, 1225–1236. doi: 10.1104/pp.114.4.1225
Weller, J. L., Ortega, R. (2015). Genetic control of flowering time in legumes. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00207
Weng, X., Wang, L., Wang, J., Hu, Y., Du, H., Xu, C., et al. (2014). Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol. 164, 735–747. doi: 10.1104/pp.113.231308
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., et al. (2005). Integration of spatial and temporal information during floral induction in arabidopsis. Science 309, 1056–1059. doi: 10.1126/science.1114358
Worland, T., Snape, J. W. (2001). “Genetic basis of worldwide wheat varietal improvement,” in The world wheat book: a history of wheat breeding. Eds. Bonjean, A. P., Angus, W. J. (Paris: Lavoisier Publishing), 55–100.
Xia, Z., Watanabe, S., Yamada, T., Tsubokura, Y., Nakashima, H., Zhai, H., et al. (2012). Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. 109, E2155–E2164. doi: 10.1073/pnas.1117982109
Xu, M., Yamagishi, N., Zhao, C., Takeshima, R., Kasai, M., Watanabe, S., et al. (2015). The soybean-specific maturity gene E1 family of floral repressors controls night-break responses through down-regulation of FLOWERING LOCUS T orthologs. Plant Physiol. 168, 1735–1746. doi: 10.1104/pp.15.00763
Xu, M., Xu, Z., Liu, B., Kong, F., Tsubokura, Y., Watanabe, S., et al. (2013). Genetic variation in four maturity genes affects photoperiod insensitivity and PHYA-regulated post-flowering responses of soybean. BMC Plant Biol. 13, 91. doi: 10.1186/1471-2229-13-91
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yan, L., Fu, D., Li, C., Blechl, A., Tranquilli, G., Bonafede, M., et al. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. 103, 19581–19586. doi: 10.1073/pnas.0607142103
Yang, Q., Li, Z., Li, W., Ku, L., Wang, C., Ye, J., et al. (2013). CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc. Natl. Acad. Sci. U. S. A. 110, 16969–16974. doi: 10.1073/pnas.1310949110
Yin, X., Liu, X., Xu, B., Lu, P., Dong, T., Yang, D., et al. (2019). OsMADS18, a membrane-bound MADS-box transcription factor, modulates plant architecture and the abscisic acid response in rice. J. Exp. Bot. 70, 3895–3909. doi: 10.1093/jxb/erz198
Zakhrabekova, S., Gough, S. P., Braumann, I., Müller, A. H., Lundqvist, J., Ahmann, K., et al. (2012). Induced mutations in circadian clock regulator mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc. Natl. Acad. Sci. 109, 4326–4331. doi: 10.1073/pnas.1113009109
Zhai, H., Lü, S., Liang, S., Wu, H., Zhang, X., Liu, B., et al. (2014). GmFT4, a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. PLoS One 9, e89030. doi: 10.1371/journal.pone.0089030
Zhang, B., Liu, H., Qi, F., Zhang, Z., Li, Q., Han, Z., et al. (2019). Genetic interactions among Ghd7, Ghd8, OsPRR37 and Hd1 contribute to large variation in heading date in rice. Rice 12, 48. doi: 10.1186/s12284-019-0314-x
Zhang, J., Zhou, X., Yan, W., Zhang, Z., Lu, L., Han, Z., et al. (2015). Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol. 208, 1056–1066. doi: 10.1111/nph.13538
Zhao, J., Chen, H., Ren, D., Tang, H., Qiu, R., Feng, J., et al. (2015). Genetic interactions between diverged alleles of early heading date 1 (Ehd1) and heading date 3a (Hd3a) RICE FLOWERING LOCUS T1 (RFT 1) control differential heading and contribute to regional adaptation in rice (Oryza sativa). New Phytol. 208, 936–948. doi: 10.1111/nph.13503
Zhao, C., Takeshima, R., Zhu, J., Xu, M., Sato, M., Watanabe, S., et al. (2016). A recessive allele for delayed flowering at the soybean maturity locus E9 is a leaky allele of FT2a, a FLOWERING LOCUS T ortholog. BMC Plant Biol. 16, 20. doi: 10.1186/s12870-016-0704-9
Zhong, S., Liu, H., Li, Y., Lin, Z. (2021). Opposite response of maize ZmCCT to photoperiod due to transposon jumping. Theor. Appl. Genet. 134, 2841–2855. doi: 10.1007/s00122-021-03862-7
Zhu, J., Takeshima, R., Harigai, K., Xu, M., Kong, F., Liu, B., et al. (2019). Loss of function of the E1-like-b gene associates with early flowering under long-day conditions in soybean. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01867
Zhu, Y.-J., Fan, Y.-Y., Wang, K., Huang, D.-R., Liu, W.-Z., Ying, J.-Z., et al. (2017). Rice flowering locus T 1 plays an important role in heading date influencing yield traits in rice. Sci. Rep. 7, 4918. doi: 10.1038/s41598-017-05302-3
Zohary, D., Hopf, M., Weiss, E. (2012). Domestication of plants in the old world: The origin and spread of domesticated plants in southwest Asia, Europe, and the Mediterranean basin (Oxford, United Kingdom: Oxford University Press). doi: 10.1093/acprof:osobl/9780199549061.001.0001
Keywords: flowering, regional adaptation, crops, long day, short day
Citation: Wang F, Li S, Kong F, Lin X and Lu S (2023) Altered regulation of flowering expands growth ranges and maximizes yields in major crops. Front. Plant Sci. 14:1094411. doi: 10.3389/fpls.2023.1094411
Received: 10 November 2022; Accepted: 04 January 2023;
Published: 19 January 2023.
Edited by:
Alexios Polidoros, Aristotle University of Thessaloniki, GreeceReviewed by:
Changhui Sun, Sichuan Agricultural University, ChinaYong Guo, Institute of Crop Science, CAAS, China
Copyright © 2023 Wang, Li, Kong, Lin and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoya Lin, eGlhb3lhbGluQGd6aHUuZWR1LmNu; Sijia Lu, bHVzaWppYUBnemh1LmVkdS5jbg==
 Fan Wang
Fan Wang Fanjiang Kong
Fanjiang Kong Xiaoya Lin
Xiaoya Lin Sijia Lu
Sijia Lu