
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 20 March 2023
Sec. Plant Breeding
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1092992
This article is part of the Research TopicGenetics and Molecular Breeding in Cereal CropsView all 29 articles
Identifying genomic regions for root traits in bread wheat can help breeders develop climate-resilient and high-yielding wheat varieties with desirable root traits. This study used the recombinant inbred line (RIL) population of Synthetic W7984 × Opata M85 to identify quantitative trait loci (QTL) for different root traits such as rooting depth (RD), root dry mass (RM), total root length (RL), root diameter (Rdia) and root surface areas (RSA1 for coarse roots and RSA2 for fine roots) under controlled conditions in a semi-hydroponic system. We detected 14 QTL for eight root traits on nine wheat chromosomes; we discovered three QTL each for RD and RSA1, two QTL each for RM and RSA2, and one QTL each for RL, Rdia, specific root length and nodal root number per plant. The detected QTL were concentrated on chromosome groups 5, 6 and 7. The QTL for shallow RD (Q.rd.uwa.7BL: Xbarc50) and high RM (Q.rm.uwa.6AS: Xgwm334) were validated in two independent F2 populations of Synthetic W7984 × Chara and Opata M85 × Cascade, respectively. Genotypes containing negative alleles for Q.rd.uwa.7BL had 52% shallower RD than other Synthetic W7984 × Chara population lines. Genotypes with the positive alleles for Q.rm.uwa.6AS had 31.58% higher RM than other Opata M85 × Cascade population lines. Further, we identified 21 putative candidate genes for RD (Q.rd.uwa.7BL) and 13 for RM (Q.rm.uwa.6AS); TraesCS6A01G020400, TraesCS6A01G024400 and TraesCS6A01G021000 identified from Q.rm.uwa.6AS, and TraesCS7B01G404000, TraesCS7B01G254900 and TraesCS7B01G446200 identified from Q.rd.uwa.7BL encoded important proteins for root traits. We found germin-like protein encoding genes in both Q.rd.uwa.7BL and Q.rm.uwa.6AS regions. These genes may play an important role in RM and RD improvement. The identified QTL, especially the validated QTL and putative candidate genes are valuable genetic resources for future root trait improvement in wheat.
Wheat is the most important cereal crop exported in developing countries as a primary source of protein (20%) and calories (21%) (Singh, 2019; Vishwakarma et al., 2022). Climate change is stimulating multiple abiotic stresses affecting crop nutrient acquisition, grain yield and quality (Asif et al., 2019; Kumar et al., 2022), and thus threatening global crop productivity (Calleja-Cabrera et al., 2020). However, crop production needs to be at least double by 2050 to feed the future world population (Woo et al., 2021) including the current wheat production (775.6 million tons) (Alexandratos and Bruinsma, 2012; Singh, 2019; Halder et al., 2022).
Roots are pivotal for supplying water and nutrient to crops (Lynch, 2007) and for anchorage (Fitter, 2002), and thus directly affect grain yield (Lynch, 2007). However, due to the heterogeneous nature of soil environments—variations in soil texture, pH, water and nutrient (Li et al., 2021), root traits change according to environmental variations to capture edaphic resources (Grossman and Rice, 2012; Rogers and Benfey, 2015). For example, under well-watered and low moisture conditions, the shallow root system of durum wheat contributed more to yield than the deep root system while under water-limited conditions, deeper roots contributed to higher grain yields (El Hassouni et al., 2018). Furthermore, root traits vary genetically (Scheiner, 1993) and are highly heritable (Fitz Gerald et al., 2006). Therefore, genetic research on different root traits is essential for improving climate resilience and yield potential in crops (Zheng et al., 2019). Quantitative trait loci (QTL) identification is a popular approach for investigating genetic variation in quantitative traits (i.e., root traits) in many cereal crops including wheat. QTL identification requires molecular linkage maps coupled with precise phenotyping (Collard et al., 2006; Shukla et al., 2014; Soriano and Alvaro, 2019). However, obtaining reliable root data for identifying root trait associated QTL from a large number of genotypes grown in soil is challenging due to invasive nature of soil, labor intensity and is time-consuming approach (Atkinson et al., 2015; Ren et al., 2017; Chen et al., 2018). To overcome this limitation, hydroponic (Horn et al., 2016; Ren et al., 2017) and semi-hydroponic (Halder et al., 2021) systems have been used to study wheat root system, and are equally useful for QTL studies (Ren et al., 2017).
QTL studies have revealed the contribution of root trait QTL to grain yield, stress tolerance, and nutrient uptake at different growth stages in wheat (Cai et al., 2008; Bennett et al., 2012; Bai et al., 2013; Ayalew et al., 2017; Ren et al., 2017; Khalid et al., 2018; Alahmad et al., 2019; Salarpour et al., 2020), indicating the value of QTL identification in marker-assisted breeding (MAS) for root traits. For example, rooting depth (RD) and grain yield spike–1 were co-localized with the flanking marker D_GA8KES401CIKOJ–160-BS00067285_51 on chromosome 7D (Salarpour et al., 2020). Root trait QTL of wheat seedlings correlated with QTL at maturity (Bai et al., 2013; Atkinson et al., 2015). In a doubled haploid (DH) of Rialto × Savannah, a grain yield QTL was co-located with different root traits including RD and total root length (RL) of wheat seedling on chromosome 7D (Atkinson et al., 2015). Co-localized QTL for thousand grain weight and root traits, including RL, root surface area (RSA), and root dry mass (RM) were found on chromosomes 4D, 5A and 6A (Bai et al., 2013). Therefore, genetic studies of root traits at the seedling stage might play important role in wheat yield improvement. Under normal and drought conditions, Ayalew et al. (2017) reported a stable QTL for RM on chromosome 5AL of recombinant inbred lines (RILs) of Synthetic W7984 × Opata M85. Under 35°C heat stress, a QTL for RD was found on chromosome 4D in the same population (Lu et al., 2022). Ren et al. (2017) found three significant QTL for RM on chromosomes 2A, 2D and 3B under controlled conditions and two QTL each on chromosome 4B under both low N and phosphorus (P) conditions; they also reported two QTL for RD each on chromosome 2B under both P- and N- limited conditions. A QTL for RM, qRNAX.7A.3, showed salt stress tolerance in the F2 of WTSD91 × WN-64 (Hussain et al., 2017). As wheat is a polyploid with a large genome, many QTL for root traits in wheat remain unexplored. Additionally, only a few QTL for root traits have been validated including RD (Qrls.uwa.1AS and Qrls.uwa.3AL) on chromosome 3A (Ayalew et al., 2017), RL (QTrl.saw-2D.2) on chromosome 2D (Zheng et al., 2019), RM on chromosome 6B (AX-109558906–AX-110028322) and chromosome 7B (AX-95025477–AX-95121748) (Meng-jiao et al., 2020).
The availability of the wheat reference genome has improved the identification of traits controlling candidate genes in QTL regions of specific chromosome and the preciseness and usefulness of QTL mapping for MAS breeding (Appels et al., 2018). In the last decade, several candidate/putative candidate genes of wheat root traits have been reported in the identified QTL regions (Wu et al., 2017; Soriano and Alvaro, 2019; Zheng et al., 2019; Li et al., 2020; Yang et al., 2021b; Griffiths et al., 2022; Li et al., 2022). For example, Wu et al. (2017) identified five putative candidate genes from QTL for root diameter (Rdia) on chromosomes 1BL, 2BL, 3BL, 3DL and 7DS under P stress conditions. TraesCS2D02G594400 and TraesCS2D02G594700 candidate genes were reported for RL QTL on chromosome 2D under controlled conditions (Zheng et al., 2019). However, none of the genes were functionally validated.
The genetics of wheat root traits are complex (Griffiths et al., 2022) due to the large genome (17 Gb) and polyploidy nature of bread wheat (Borrill et al., 2019). Therefore, genetic studies that identify QTL and associated genes of multiple root traits will help to understand the molecular mechanism of wheat root systems (Zheng et al., 2019), ultimately helping to develop climate-resilient, high-yielding wheat genotypes. Therefore, this research aimed to identify QTL for different root traits from RILs developed from Synthetic W79804 and Opata M85, validate key QTL in two populations with different genetic backgrounds and identify candidate genes within the flanking markers of the validated QTL.
A population of 103 RILs developed from a cross between highly polymorphic parents Synthetic W7984 (T. turgidum cv. Altar 84/Aegilops tauschii Coss. line WPI 219) and Mexican spring wheat (Opata M85) accessed through the International Triticeae Mapping Initiative (Salem et al., 2007; Sorrells et al., 2011) was used for the genetic mapping study. In addition, F2 populations of Synthetic W7984 × Chara and Opata M85 × Cascade with different genetic backgrounds to RILs were developed to validate the phenotypic effect of two identified QTL for RD and RM, respectively.
Synthetic W7984, Opata M85, and the 103 RILs were grown in a semi-hydroponic system (Chen et al., 2011b) for 42 days in a randomized block design with four replicates for each genotype. The experimental conditions and trait measurements were the same as described by Halder et al. (2021). All plants were assessed at tiller onset [Zadoks 2.4; (Zadoks et al., 1974)], i.e. 42 days after transplanting. Briefly, the experiment was conducted in a temperature controlled (10–24°C) glasshouse at The University of Western Australia (UWA), Perth, from mid-June to late-August 2019. Wheat seedlings (4–5 cm long roots) grown in washed river sand were transplanted into bins for a semi-hydroponic system containing 35 L nutrient solution.
At harvest, the maximum depth of a plant root (RD) was measured with a ruler from its crown, and the number of nodal roots per plant (NNR) was counted manually. After capturing photographs of the root system using a portable photographing system, the root system were separated from the shoot. Root sections (≤ 20 cm) were scanned at 400 dpi using a desktop scanner (Epson Perfection V800/850) to determine other root traits—RL (sum of all root length types), Rdia, RSA and root diameter length (RDCL) of fine roots (root diameter< 0.25 mm) and coarse roots (root diameter > 0.25 mm)—were measured using WinRHIZO Pro software (v2009, Regent Instruments Inc., Montreal, QC, Canada). Specific root length (SRL) was calculated as the RL per unit of RM, and root length intensity (RLI) was the RL per unit of RD. Root growth rate is the RD per day. RM is the weight of the whole root system after air-forced oven drying (65°C for 72 h). Further, using the phenotypic data, broad-sense heritability (H2) of the root traits was calculated as:
H2 = , where is genotypic variance (mean sum of squares of a trait) and is environmental variance (residual mean sum of square) from the analysis of variance, and n is replication number per genotype (4) (Wu et al., 2013; Ben Sadok et al., 2015).
Molecular marker data and the linkage map of the Synthetic W7984 × Opata 85 RIL mapping population were accessed from the GrainGenes database (https://wheat.pw.usda.gov/cgi-bin/GG3/report.cgi?class=mapdata&name=Wheat%2C%20Synthetic%20x%20Opata%2C%20BARC). The linkage map comprised 1,476 simple-sequence repeats (SSR) and restriction fragment length polymorphism (RFLP) markers distributed across 21 linkage groups. Among the available markers, 1,018 with known chromosomal locations were used for QTL mapping of the target root traits. The genetic map spanned a length of about 500 cM with an average marker density of 1 cM after filtering the 20% missing values from the dataset.
The composite interval mapping method in Windows QTL Cartographer V2.5_011 was used to identify root traits QTL; the logarithm of odds (LOD) threshold value was set to ≥ 2.5 based on 500 and 1,000 permutations at the 5% significance level. LOD > 2.5 indicate the presence of significant QTL in a particular genomic region for an individual trait. The square of the partial correlation coefficient (R2) estimates the phenotypic variance of a single QTL (Balakrishnan et al., 2020). The sequences of the SSR and RFLP flanking markers (left-and right-hand sides closest to the QTL regions) were identified from GrainGenes database (https://wheat.pw.usda.gov/cgi-bin/GG3/browse.cgi?class=marker; accessed on 05 October 2022) and/or NCBI database (https://www.ncbi.nlm.nih.gov/; 05 October 2022), respectively. Further the sequences were blasted in JBrowse (https://urgi.versailles.inra.fr/blast/?dbgroup=wheat_iwgsc_refseq_v1_chromosomes&program=blastn) with the wheat reference genome RefSeq v1.0 to identify the physical position of the markers. The graphical representation of the QTL was drawn using MapChart 2.32 software.
One-third of an individual seed (excluding the embryo) was used to extract the genomic DNA of Synthetic W7984, Opata M85, Chara, Cascade, and the F2 of Synthetic W7984 × Chara and Opata M85 × Cascade. The remaining seed with embryo was preserved in the cold room for seed germination to validate the phenotypic effect of the targeted QTL. The one-third seed part was crushed manually using a small hammer, and then crushed further with a SPEX® SamplePrep 2010 GenoGrinder at 1,400 rpm for 2 minutes for DNA extractions following the cetyl trimethyl ammonium bromide (CTAB) method. The extracted DNA was suspended in 0.1× TE buffer (pH 8.0) for storage. DNA concentrations were measured by NanoDrop (NanoDrop-1000 spectrophotometer) and Qubit 2.0 fluorometer using the Qubit dsDNA BR (Broad-Range) Assay Kit. The primers (forward and reverse) for SSR marker Xbarc50 were obtained from Sigma-Aldrich (Sigma-Aldrich Pty Ltd., NSW, Australia). DNA primers of Xgwm334 (forward (5´) dyed with fluorescent PET) were obtained from Alpha ADN (225 Bridge CP 4023, Montreal, Quebec H3C 0J7, Canada: http://www.alphaadn.com/contact.html).
An EmeraldAmp®MAX HS PCR Master Mix reaction mixture (15 µL) containing 20 ng template DNA of Synthetic W7984 and Synthetic W7984 × Chara populations, 0.2 µM of each forward and reverse primers was amplified in a thermocycler (Eppendorf Mastercycler EP Gradient S) to validate the Q.rd.uwa.7BL with a flanking marker Xbarc50. The annealing temperature of the marker (53°C) was found in GrainGenes (https://wheat.pw.usda.gov/cgi-bin/GG3/report.cgi?class=marker&name=&id=86860). The PCR conditions were 98°C for 1 min, 35 cycles of denaturation at 98°C for 10 sec, annealing at 53°C for 30 sec, elongation at 72°C for 1 min kb–1 and final extension (Taq polymerase) at 72°C for 5 min. The PCR products were run on a 2.5% agarose gel electrophoresis using GelRed™ (1:10 ratio) at 120 V for 1 h 20 min. The experiment was conducted at the genetics and molecular genetics laboratories at the UWA School of Agriculture and Environment.
A DNA fragment analysis was undertaken using the Applied Biosystems Genetic Analyzer at Biodiversity Conservation Centre, Kings Park, WA, to validate Q.rm.uwa.6AS with a flanking marker Xgwm334. The annealing temperature of the marker was determined by a gradient PCR using RT-PCR. The master mix (1rxn) for gradient PCR was 5 µL SYBR Green, 1.5 µL of each forward and reverse primer and 2 µL template DNA of Opata M85 and Cascade. Using PCR conditions at 98°C for 2 min, 40 cycles denaturation at 98°C for 10 sec, a range of annealing at 52–67°C for 45 sec, elongation at 72°C for 30 sec kb–1 and final extension at 72°C for 5 min, the best annealing temperature for the marker was set at 58°C. Singleplex PCR of template DNAs (20 ng) from the Opata M85 × Cascade populations and both parents was done in the wheat genetics laboratory at UWA. The master mix (1×) for a singleplex PCR was 3.52 µL PCR grade water, 2 µL 5× buffer, 0.8 µL MgCl2 (25mM), 0.08 µL Taq polymerase (0.04 u µL-1), 0.8 µL of each fluorescent forward primer, and reverse primer, and 2 µL of template DNA (≥ 2 ng µL-1). Singleplex PCR conditions were 94°C for 5 min, 40 cycles denaturation at 94°C for 30 sec, annealing at 58°C for 1min, elongation at 72°C for 45 sec kb–1, final extension at 72°C for 7 min and hold at 10°C. A multiplex PCR was done using 1 µL PCR product mixed with 9 µL highly deionized (Hi-Di) formamide with LIZ Size Standard for fragment analysis in an ABI sequencer.
Further, 1 µL PCR product was mixed with 9 µL Hi-Di with LIZ Size Standard for fragment analysis using capillary electrophoresis in an Applied Biosystems 3500 series Genetic Analyzer at Biodiversity Conservation Centre, Kings Park, WA. The DNA fragment size were identified by analyzing the electrogram from SeqPartitioner, Geneious plugin.
Homozygous (AA from Synthetic W7984 or Opata M85, and BB from Cascade or Chara alleles) individuals were identified by comparing differences in band size in the agarose gel and the DNA fragment size of their respective parents. Selected individuals were grown in a semi-hydroponic system in a controlled environment (day/night 24°C, 14°C) as described above. The average RD and RM of the genotypes of two allelic combinations (AA and BB) were compared using a student’s t-test at 0.05% significance level.
Phenotypic data were analyzed using GenStat statistical software 19th edition, with the frequency analysis done in SPSS Version 28.0.0 (142) (https://www.ibm.com/support/pages/node/6525830).
Potential high confidence candidate genes for root traits were identified in the two QTL considered for validation. The physical position of the flanking markers of the QTL was found in the GrainGenes wheat database (https://wheat.pw.usda.gov/cgi-bin/GG3/browse.cgi?class=marker), and blasted in the JBrowse (http://www.wheatgenome.org/Tools-and-Resources/Sequences, accessed on 01 June 2022) wheat genome browser with RefSeq v1.0 to identify the candidate genes on the QTL region.
Further, the gene functions were identified in the International Wheat Genome Sequencing Consortium (IWGSC) RefSeq v1.0 website (https://wheaturgi.versailles.inra.fr/Seq-Repository/Annotation, accessed on 20 June 2022) (Appels et al., 2018). Genes involved in root growth and development from other studies were considered putative candidate genes. The biological functions of the individual genes were obtained from Uniprot (https://www.uniprot.org/?-id+2fYRW1ChXSa+-fun+Pagelibinfo+-info+TREMBL). WheatExp revealed expression of the candidate genes on root tissues in other wheat cultivars (Pearce et al., 2015) (http://www.wheat-expression.com/; accessed on 01 December 2022). Gene expression levels for the candidate genes in different wheat tissues, including roots were downloaded from WheatExp. Further, gene expression in root tissue was filtered, with the highest expression level considered for this study.
Root traits of the Synthetic W7984 × Opata M85 RILs and their parents varied considerably (Table 1). Opata M85 had higher RL, RD, RM, root surface area of fine roots (root diameter < 0.25 mm, cm2; RSA2), and total length of coarse roots (root diameter < 0.25 mm, cm; RDCL2) than Synthetic W7984, while Synthetic W7984 had higher root surface area of fine roots (root diameter > 0.25 mm, cm2; RSA1), SRL, and total length of coarse roots (root diameter > 0.25 mm, cm; RDCL1) than Opata M85 (Table 1). For the RILs, RL, RD, RM, Rdia, RSA1, RSA2, and SRL ranged from 173.10–12,783 cm, 8.00–158 cm, 0.03–0.44 g, 0.21–0.66 mm, 10.07–216.20 cm2, 1.16–55.17 cm2, and 2,150–74,013 cm g–1, respectively (Table 1). Transgressive segregation with approximately normal distribution for various root traits (RL, RD, RM, Rdia, RSA1, RSA2, and SRL) between the RILs and the parents was detected (Figure 1). H2 was high (84.5–92.1%) for all root traits except RLI (data not shown).
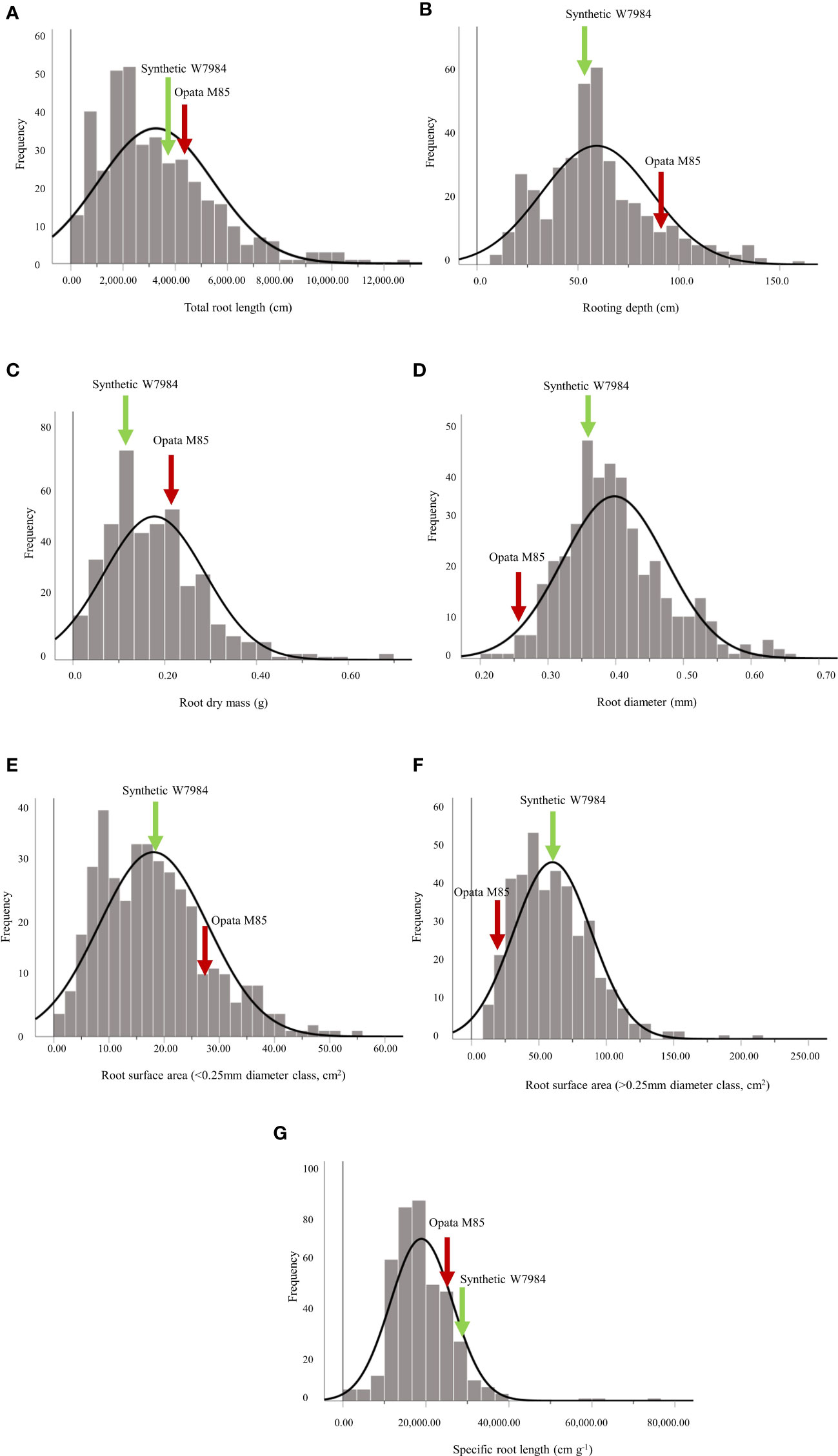
Figure 1 Distribution of (A) total root length (cm), (B) rooting depth (cm), (C) root dry mass (g), (D) root diameter (mm), (E) root surface area of fine roots (root diameter< 0.25 mm, cm2), (F) root surface area of coarse roots (root diameter > 0.25 mm, cm2), and (G) specific root length (cm g-1) of 103 recombinant inbred lines and their parents, Synthetic W7984 and Opata 85. The green and red arrows indicate the phenotypic performance of Synthetic W7984 and Opata M85, respectively.
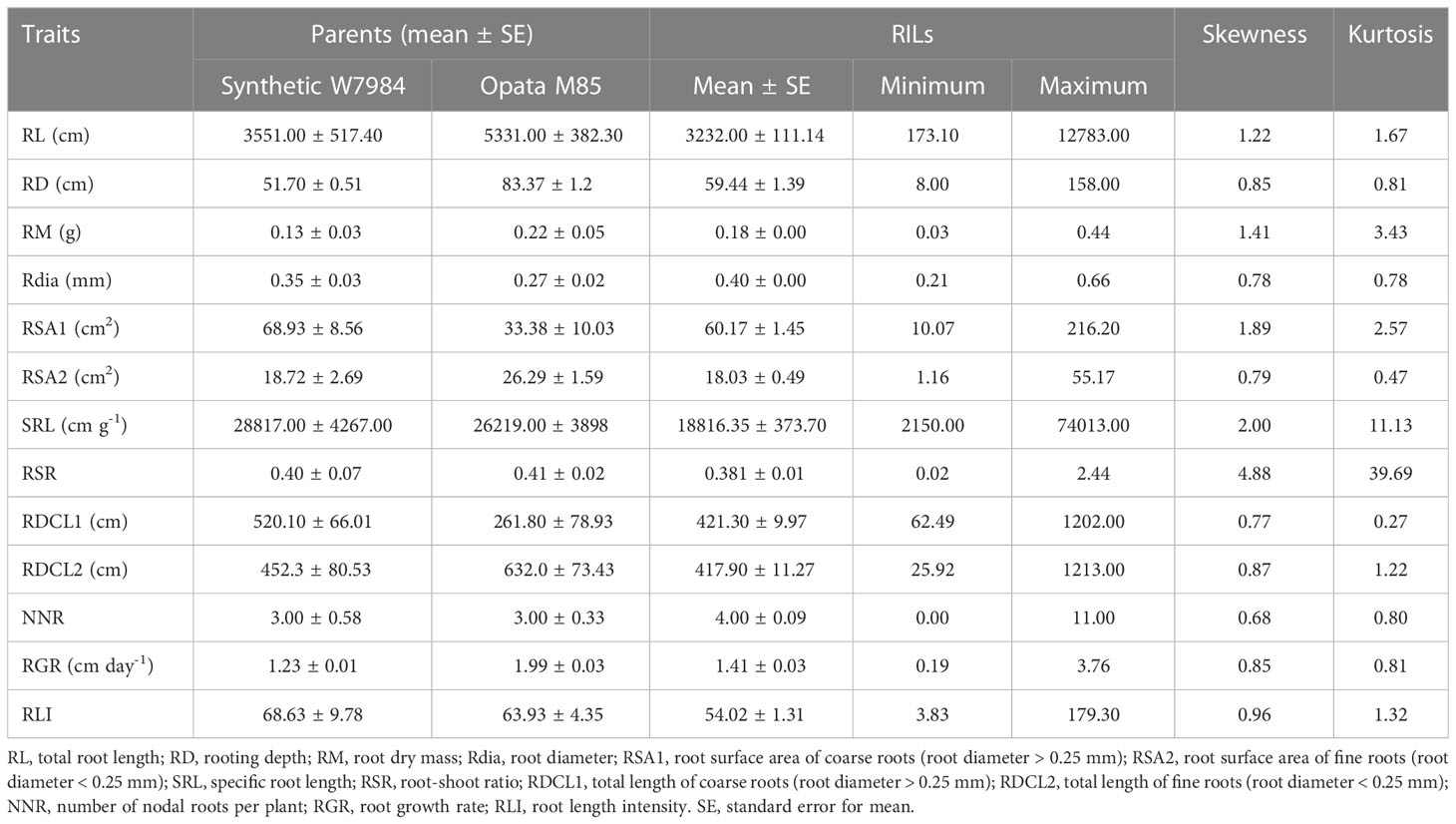
Table 1 Variations in root traits of recombinant inbred lines (RILs) and their parents, Synthetic W7984 and Opata M85.
The permutation tests identified 14 and nine QTL for eight root traits, with LOD scores ≥ 2.5 in CIM at 500 and 1,000 permutations, respectively. However, considering that root system architecture is complex and governed by many genes of small effect (Sharma et al., 2011), the study considered the QTL identified at 500 permutations. Most of the QTL were distributed on chromosome groups 5, 6 and 7, except 5B and 7D chromosomes. The QTL for RSA1 were found on chromosome 2A and 3B (Table 2). Opata M85 contributed alleles to all the QTL for RL, RM, RSA1 and NNR and a QTL for RD (Q.rd.uwa.5DL), and Synthetic W7984 contributed all other alleles.
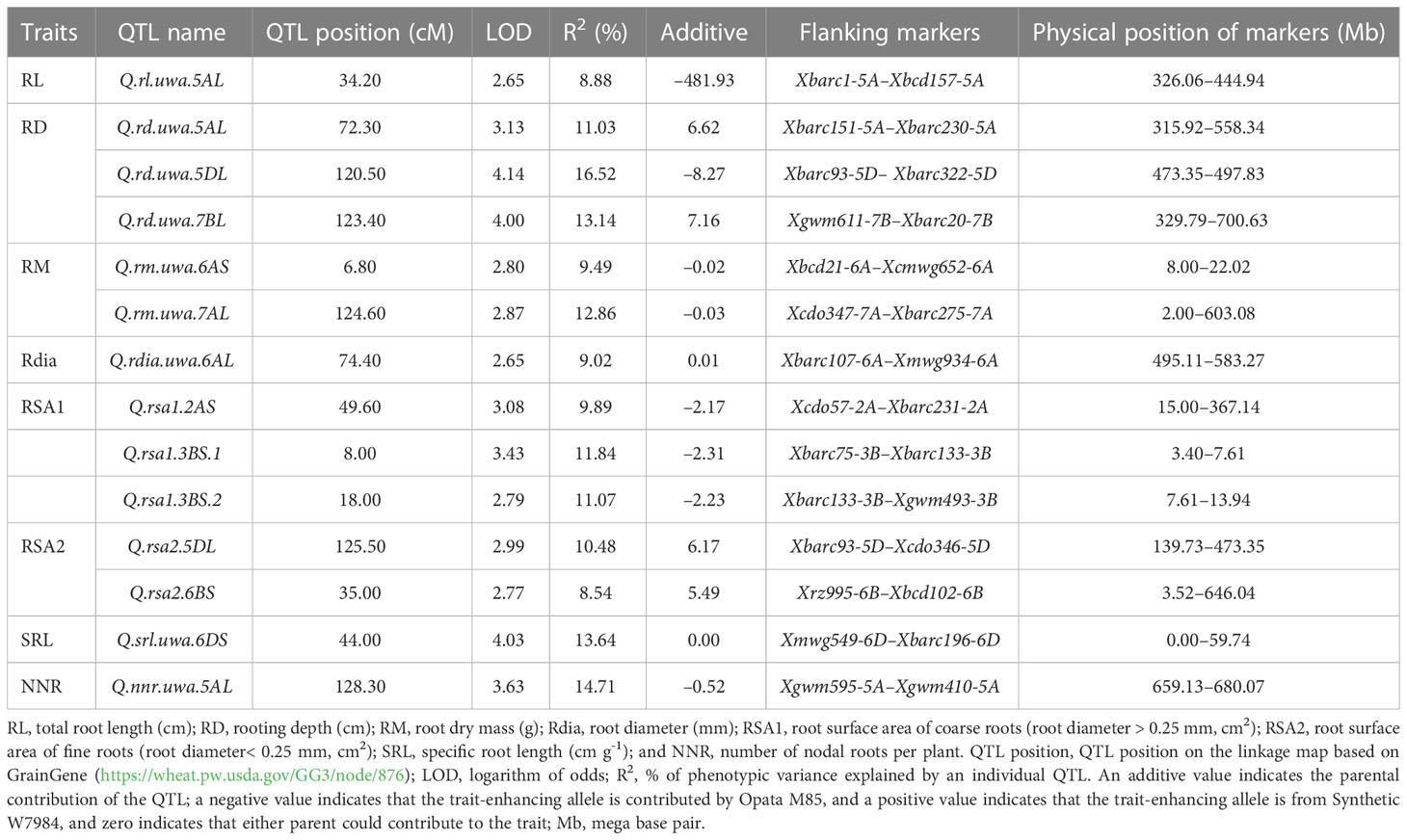
Table 2 QTL for eight root traits in recombinant inbred lines (RILs) of Synthetic W7984 × Opata M85 identified by composite interval mapping (CIM) at the logarithm of odds (LOD) threshold ≥ 2.5.
Seven of the QTL on chromosome groups 5, 6 and 7 explained more than 10% phenotypic variance (R2). Three QTL for RD (R2 = 11.03–16.52%; LOD 3.14–4.14) were identified and distributed on the long arms of chromosomes 5A, 5D and 7B (Figure 2). Synthetic W7984 contributed alleles to Q.rd.uwa.5AL and Q.rd.uwa.7BL, and Opata M85 contributed alleles to Q.rd.uwa.5DL. Among the 11 QTL, Q.rd.uwa.5DL had the highest LOD (4.14) and R2 (16.52) values. For the two QTL for RM (both contributed by Opata M85), one was distributed on the long arm of chromosome 7A (R2 = 12.86%; LOD = 2.87) and the other on the short arm of chromosome 6A (R2 = 9.49%; LOD = 2.80) (Figure 2C); the LOD and R2 were same for both 500 and 1,000 permutations. QTL for RL and NNR occurred on the long arm of chromosome 5A. Two QTL for RSA2 were detected on chromosomes 5DL (Figure 2B) and 6BS, and QTL for SRL was identified on chromosome 6DS.
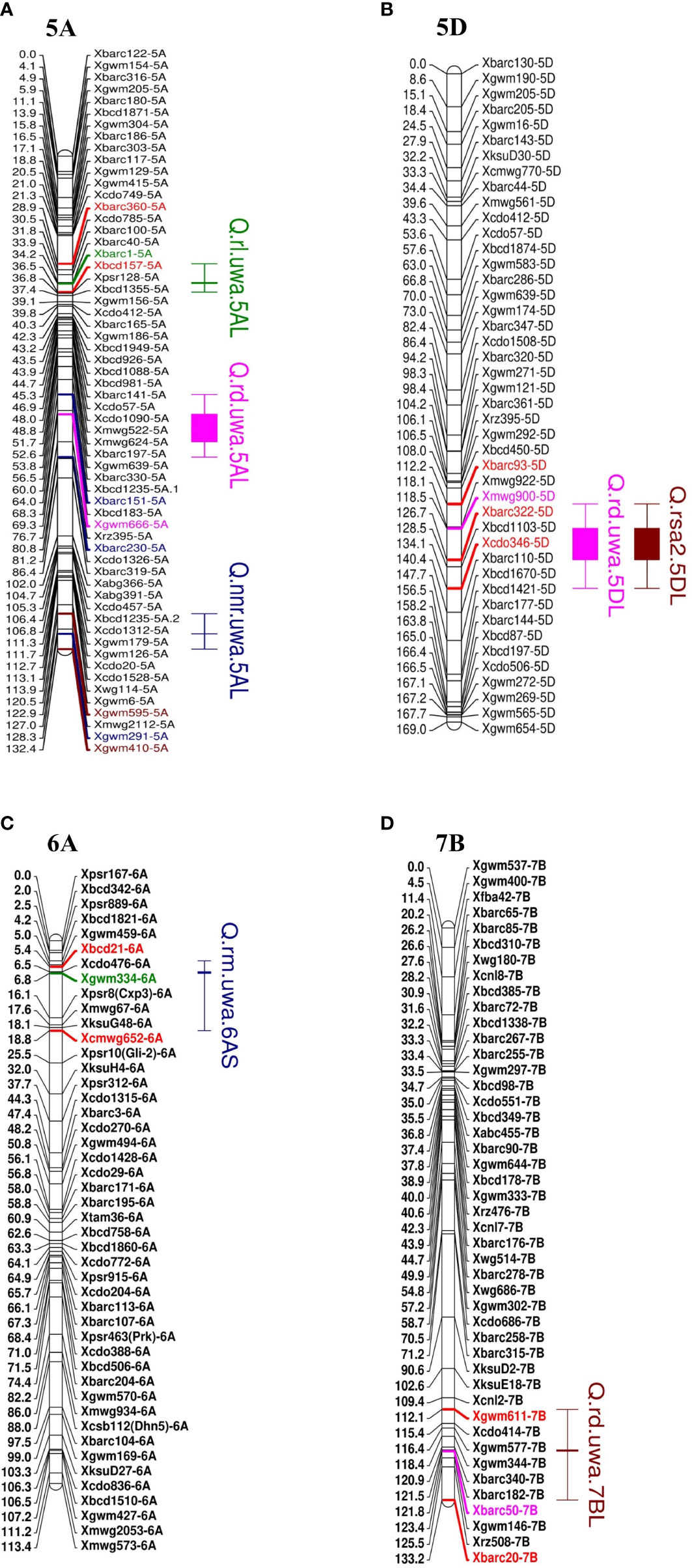
Figure 2 Mapping of important QTL for root traits in wheat in the Synthetic W7984 and Opata 85 RIL population (A) total root length (RL, cm), rooting depth (RD, cm), and nodal roots per plant (NNR) (B) rooting depth and root surface area of fine roots (root diameter < 0.25 mm, RSA2, cm2), (C) root dry mass (RM), and (D) rooting depth of recombinant inbred lines of Synthetic W7984 × Opata 85. Bars and caps indicate the QTL with LOD > 2.5. Red markers are flanking markers of different colours in individual chromosomes that represent the tightly linked marker of the respective QTL.
Co-location of Q.rd.uwa.5DL and Q.rsa2.5DL were identified at 120.50 cM (Figure 2B). The flanking marker interval was 112.2–126.7 cM for Q.rd.uwa.5DL and 112.2–134.1 cM for Q.rsa2.5DL.
Xbarc50, the closely linked marker of Q.rd.uwa.7BL, showed polymorphism between Synthetic W7984 and Chara in agarose gel electrophoresis. Xgwm334, the closely linked marker of Q.rm.uwa.6AS, showed polymorphism between Opata M85 and Cascade in DNA fragment analysis. The fragment size of the parents (Table 3) was used to score the randomly selected F2 populations of the two validation population lines—19 lines for Synthetic W7984 × Chara, and 13 lines for Opata M85 × Cascade. The Synthetic W7984 × Chara hybrids (F2) were divided into two groups (only homozygous lines). The group containing the negative allele from Synthetic W7984 (Q.rd.uwa.7BL) had a significantly (P < 0.01) shorter (52%) RD than Chara (Table 4). Similarly, in the Opata M85 × Cascade hybrids, the group containing the positive allele from Opata M85 (Q.rm.uwa.6AS) had a significantly (P < 0.01) higher (31.58%) RM than Cascade (Table 4).
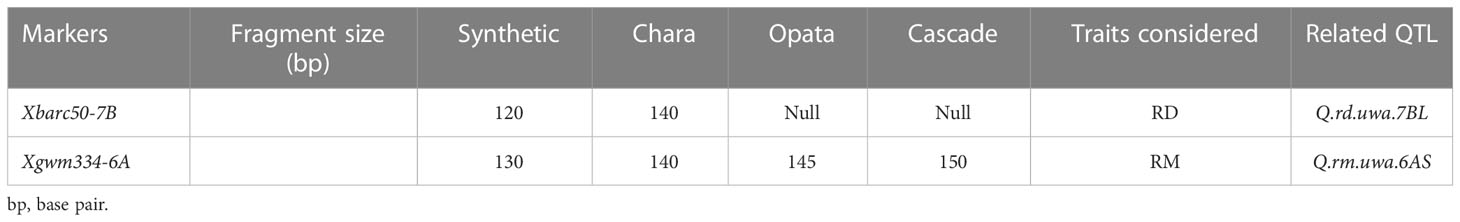
Table 3 Fragment size of two SSR markers, with polymorphism among parental lines (Synthetic W7984, Opata M85, Chara, and Cascade) of the validation population, related to QTL for rooting depth (RD) and root dry mass (RM).
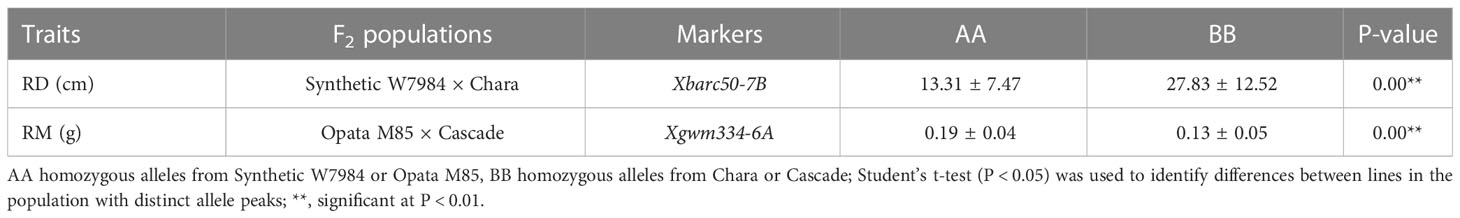
Table 4 Validation of quantitative trait loci (QTL) for rooting depth (RD) and root dry mass (RM) identified from Synthetic W7984 × Opata M85 recombinant inbred line (RIL) population in F2 populations of Synthetic W7984 × Chara and Opata M85 × Cascade.
The 329.79–700.63 Mb mapping interval of Q.rd.uwa.7BL contained 2,323 genes, with the functions of 215 genes associated with the wheat root system (Halder et al., 2022) (Supplementary Table S1). Twenty-one genes were putative candidate genes for root traits and abiotic and biotic stress tolerance in wheat, reported earlier in other crops and Arabidopsis (Table 5). The 8.00–22.02 Mb mapping interval of Q.rm.uwa.6AS contained 387 genes, with the functions of 34 genes reported in the wheat root system (Halder et al., 2022) (Supplementary Table S1) and 13 were putative candidate genes for root traits and abiotic and biotic stress tolerance in wheat (Table 5). The in-silico expression study identified, high expression levels of the candidate genes in wheat cultivars ‘Chinese Spring,’ ‘Nulliterea Chinese Spring,’ ‘Azhurnaya,’ and ‘N1DT1A’ (Supplementary Table S2). Among the putative candidate genes from both QTL, TraesCS7B01G374800, had the highest expression level (log2 of transcripts per million: 360.65) in the roots of ‘Chinese Spring’. In the roots of ‘Azhurnaya,’ TraesCS7B01G404000, TraesCS7B01G368400, and TraesCS6A01G026500 had high-expression levels.
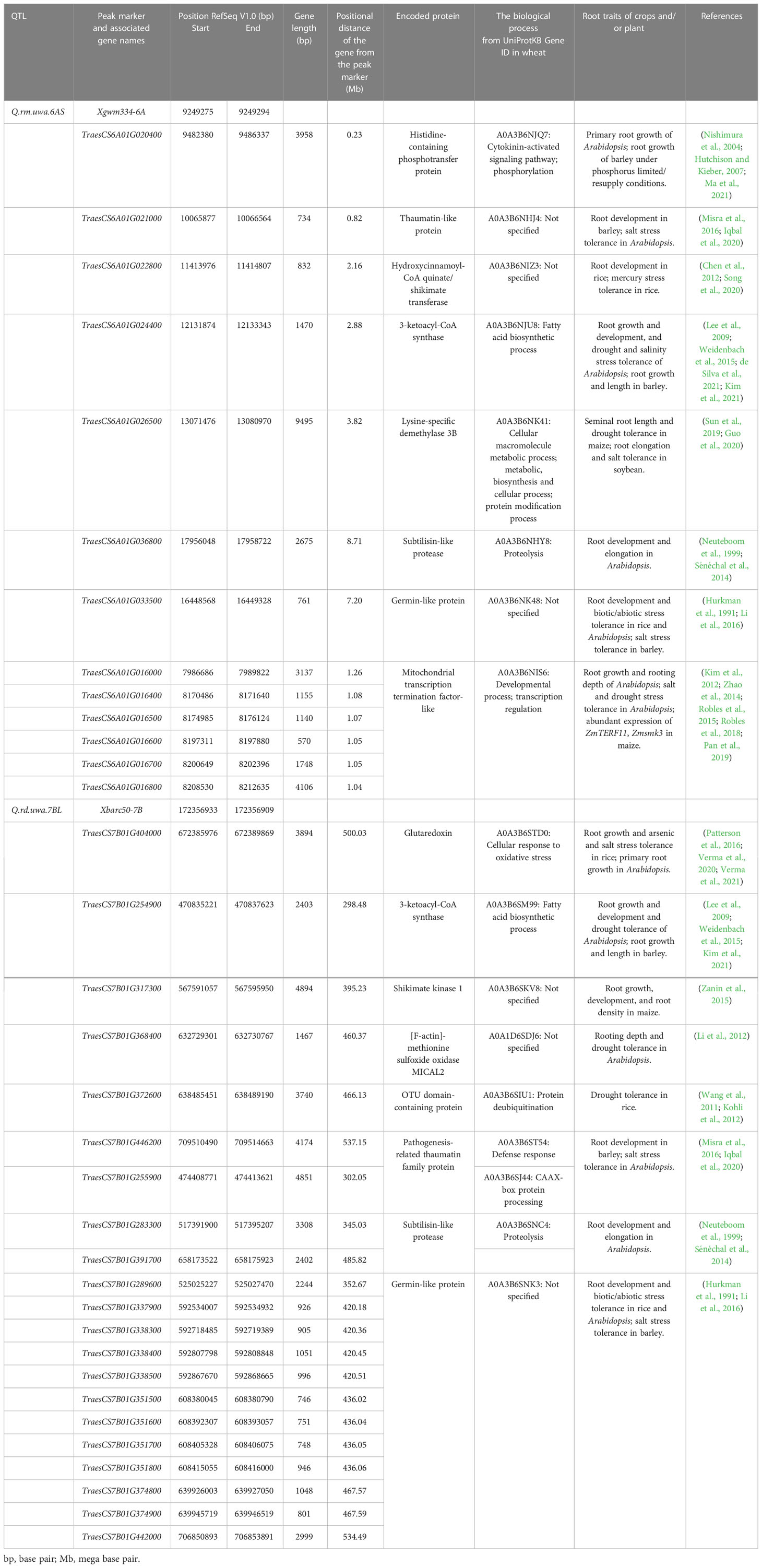
Table 5 Genes located within the two quantitative trait loci (QTL) and the encoded proteins related to root traits and abiotic stress tolerance in crops and Arabidopsis.
A semi-hydroponic phenotyping system was used for phenotyping a RIL mapping population of Synthetic W7984 × Opata M85 and to identify QTL for different root traits in wheat. A total of 14 QTL for eight root traits were detected on nine wheat chromosomes, with two important QTL validated in two independent F2 populations. The QTL identified were concentrated in wheat chromosome groups 5, 6 and 7. Several putative genes located in the QTL region were identified for the molecular breeding of root traits.
The semi-hydroponic system used in this study offered an excellent opportunity to acquire reliable root trait data with high accuracy and repeatability (Chen et al., 2020). In our recent study, root trait variability of 184 bread wheat genotypes originating from 37 countries was characterized in the same semi-hydroponic phenotyping system (Chen et al., 2020), followed by validation of genotypes with contrasting root systems in soil-filled rhizoboxes (Figueroa-Bustos et al., 2018). The consistent ranking of genotypes for some important root traits in the semi-hydroponic system and soil conditions indicates the reliability of the phenotyping study for root studies, as confirmed in other crop species, such as narrow-leafed lupin (Chen et al., 2011a; Chen et al., 2014), barley (Wang et al., 2021), and soybean (Liu et al., 2021; Salim et al., 2022). The wheat lines used in this study will be examined further under the field conditions. Significant phenotypic variation for all measured root traits in the biparental population indicates the successful identification of the polygenic trait (Table 1). Further, the continuous distribution of different root traits such as RL, RD, RM, Rdia, RSA1, RSA2 and SRL (Figure 1) indicates that the genetic architecture of individual trait has many genes responsible for the variation. Similarly, high broad-sense heritability (> 80%) for all root traits except RLI (data not shown) indicates the potential for selecting these traits in future wheat breeding. Earlier studies demonstrated significant phenotypic variation in root traits under drought stress (Ayalew et al., 2017), heat stress (Lu et al., 2022), and waterlogging stress (Yu and Chen, 2013) of the same population suggesting that the population in the current study is suitable for genetic mapping of root traits.
Understanding root trait variation is essential for manipulating the traits according to the soil and environmental conditions to improve stress tolerance and yield in wheat. For example, a large wheat root system (in terms of RL and RM) was beneficial for higher grain yield, capturing water and nutrient from sandy soil under well-watered conditions (Palta and Watt, 2009; Palta et al., 2011). However, large root systems reduced yield at terminal drought due to lower (59%) water use efficiency than shallow root systems (Figueroa-Bustos et al., 2020). RM is another important root trait positively correlated with grain yield under drought stress (Ehdaie et al., 2012). SRL (ratio of RL and RM) is an indicator of utilization in nutrient uptake. Wheat genotypes with large SRL and more fine roots take up nutrient and water from the subsoil and contribute to high yield (Chen et al., 2020). Similarly, Rdia significantly correlates with wheat yield and P acquisition (Atta et al., 2013; Nahar et al., 2022).
Several studies have reported the genetics of RL (Ibrahim et al., 2012; Atkinson et al., 2015; Danakumara et al., 2021; Yang et al., 2021a; Li et al., 2022), RD (Liu et al., 2013; Xu et al., 2013; Ayalew et al., 2017; Ren et al., 2017; Salarpour et al., 2020; Danakumara et al., 2021) and RM (Yu and Chen, 2013; Acuna et al., 2014; Meng-jiao et al., 2020; Danakumara et al., 2021) under different environmental conditions. In our study, we discovered 11 QTL on wheat chromosome groups 5, 6 and 7 responsible for root traits contributed by the two parents, Synthetic W7984 and Opata M85. Both parents could contribute favorable alleles for root traits (Bhoite et al., 2018) (Table 2).
Identification of putative QTL alone is insufficient for trait improvement using MAS. Therefore, validating QTL—testing the allelic effect in populations other than the original population—is essential to eliminate statistical error (Langridge et al., 2001). In this study, we validated two flanking markers for RD and RM QTL in populations different than the original population used for QTL identification.
Mapping QTL can also identify the relationship between the traits through the co-localization of QTL (Colombo et al., 2022) which is important for plant performance improvement. Q.rd.uwa.7BL (Xgwm611–Xbarc20) for RD had a LOD of 4.00 and phenotypic variation of 13.14% (Table 2; Figure 2D) for both 500 and 1,000 permutations. Q.rd.uwa.7BL were co-located with previously identified QTL including qMRL.CK-7B (Xbarc257.2–Xgwm46) under controlled conditions. Q.rd.uwa.7BL also co-located with qMRL.LP-7B and qMRL-7B1 (Xbarc1181–Xbarc1116) under low P (Ren et al., 2017) and well-watered conditions (Ren et al., 2012), respectively. Two other QTL under well-watered conditions—qLR-7B (Wms400–Wms573) (Ehdaie et al., 2016) and Q.RL-7BL (AX-94528392) (Danakumara et al., 2021) were co-located with Q.rd.uwa.7BL. Two drought-stress specific QTL for RD found from the RILs derived in Synthetic W7984 and Opata M85 (Ayalew et al., 2017) were co-located with Q.rd.uwa.7BL, suggesting that Q.rd.uwa.7BL may contribute to drought stress tolerance. Importantly, Q.rd.uwa.7BL was co-located with grain yield QTL (Xm43p78.14–Xm86p65.0) (Quarrie et al., 2005), kernel number per spike QTL, QKNPS-DH-7B-2.1 (Xbarc276.1–Xwmc396), and thousand-grain weight QTL, QTKW-DH-7B (Xgwm333–Xwmc10) (Zhang et al., 2016) on chromosome 7B. These co-location evidence strongly suggest that Q.rd.uwa.7BL contributes to improving RD and wheat yield. However, for the first time Q.rd.uwa.7BL was successfully validated for RD in other populations with different genetic backgrounds. The QTL may also contribute to biotic stress tolerance: Xgwm344, a closely linked marker of Q.rd.uwa.7BL previously validated for leaf rust resistance in wheat (Zhang et al., 2020). Comparing of Q.rd.uwa.7BL with other previously reported co-located QTL revealed that Q.rd.uwa.7BL is physically located in a larger interval (329.79–700.63 Mb) than the above-mentioned QTL, except qLR-7B (Figure 3).
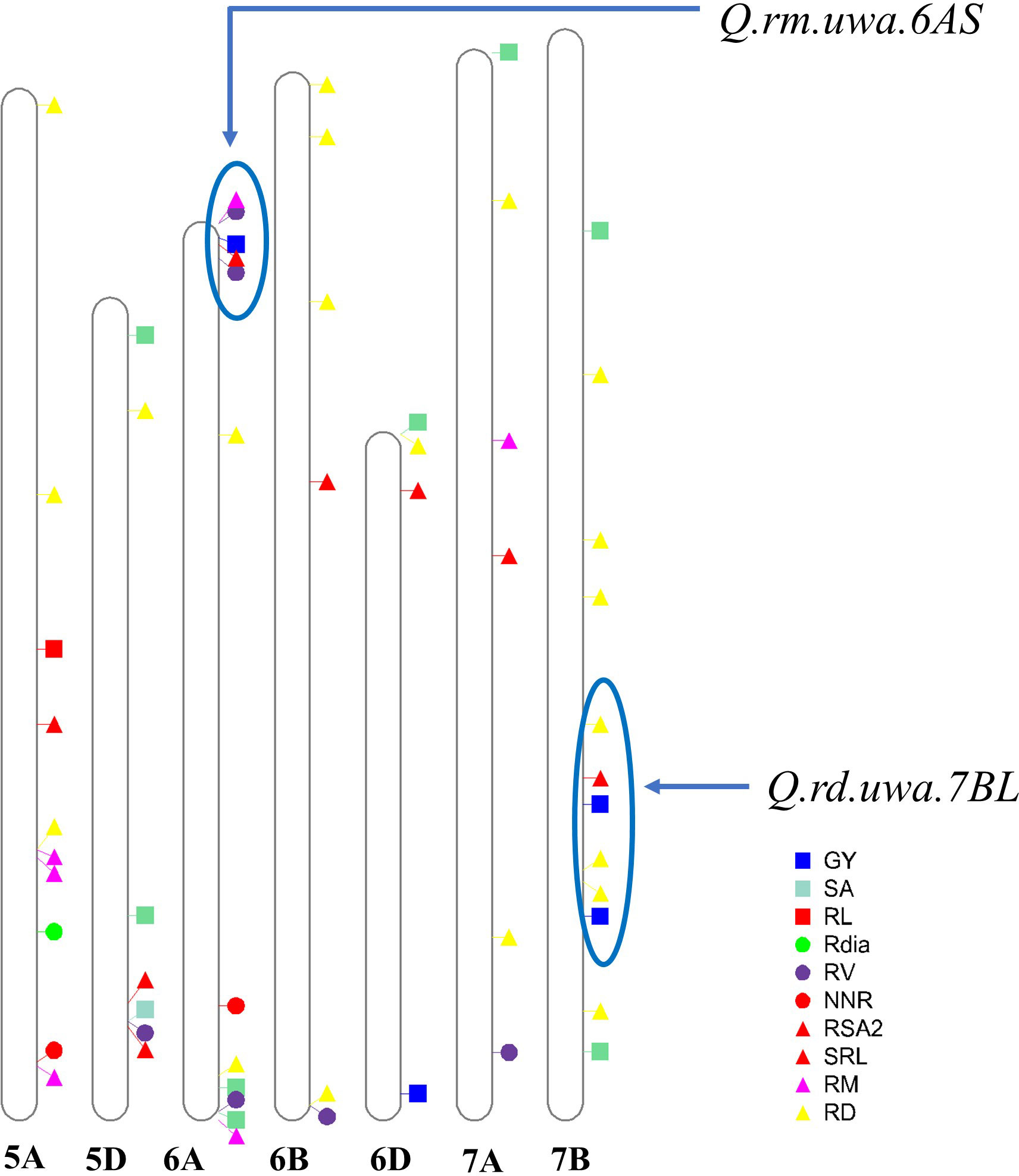
Figure 3 Quantitative trait loci (QTL) identified in this study and previous studies (Ibrahim et al., 2012; Ren et al., 2012; Liu et al., 2013; Xu et al., 2013; Atkinson et al., 2015; Ehdaie et al., 2016; Ayalew et al., 2017; Ren et al., 2017; Soriano and Alvaro, 2019; Li et al., 2020; Meng-jiao et al., 2020; Danakumara et al., 2021; Yang et al., 2021a) on chromosomes 5A, 5D, 6A, 6B, 6D, 7A and 7B. Previously discovered QTL for root traits and grain yield were depicted. The QTL identified in this study were labelled in red colour, and the previously studied QTL were represented by other colours (blue, yellow, green, green-cyan, magenta and blue-magenta). Blue circles represent the genomic regions of the validated QTL (Q.rm.uwa.6AS and Q.rd.uwa.7BL) positions from this study and their overlapping with other QTL from previous studies. Table 2 is referred to the detailed QTL identified in this study on different chromosomes; GY, grain yield; SA, surface area; RL, total root length; Rdia, root diameter; RV, root volume; NNR, number of nodal roots per plant; RSA2 root surface area of fine roots (root diameter < 0.25 mm); SRL, specific root length; RM, root dry mass; and RD, rooting depth.
A number of QTL for RD have been reported on chromosome 7B (Figure 3) (Ren et al., 2012; Liu et al., 2013; Xu et al., 2013; Ehdaie et al., 2016; Ren et al., 2017; Ayalew et al., 2018; Li et al., 2020; Danakumara et al., 2021) under different stress conditions, suggesting that chromosome 7B harbors important genes for RD to improve stress tolerance in wheat. However, none of the QTL has been validated in other populations. In this study, Q.rd.uwa.7BL with a peak marker of Xbarc50 was used to screen the validation population. Synthetic W7984 contributed to Q.rd.uwa.7BL for shallow RD; therefore, Xbarc50 was validated in Synthetic W7984 × Chara hybrids (F2). A significant reduction in RD in the validation population confirmed the reliability of the marker performance. However, the marker could be further tested in other genotypes for application in wheat breeding. Previous studies have reported the significance of shallow RD in high yield of wheat. Under well-watered conditions, genotypes with shallow RD in durum wheat (Bellario and Jabal2), contributed to high yield. Under irrigated conditions, the shallow-rooted genotypes contributed to 20–40% higher yield than deep-rooted genotypes (El Hassouni et al., 2018). The short root length gene, TaSRL1 (on chromosome 4A: 3.37 Mb) improved thousand grain weight as a pleiotropic effect (Zhuang et al., 2021). Importantly, it was revealed in an earlier study that despite taking up 20% less water under drought conditions, Synthetic W7984 (donor parent of Q.rd.uwa.7BL in our study) had higher grain numbers per spike than Opata M85 (Onyemaobi et al., 2018). Therefore, the validated marker for shallow RD from this study could be used to improve the grain yield and stress tolerance in wheat.
Q.rm.uwa.6AS (Xbcd21–Xcmwg652) identified in this study, was co-located with a previously identified grain yield QTL, QGY.cgb-6A (Xgwm334–WMC297) under both well-watered and water-stressed conditions (Liu et al., 2013) indicating its potential for drought tolerance and grain yield improvement. A meta-QTL for RM, Root_MQTL_67 (Soriano and Alvaro, 2019), overlapped with Q.rm.uwa.6AS (Xgwm334), but was not validated. In this study, Opata M85 contributed positive alleles to Q.rm.uwa.6AS for increased RM. Testing the allelic performance of Xgwm334, in Opata M85 × Cascade hybrids (F2), Opata M85 had significantly higher RM than Cascade confirming the functionality of the identified QTL. Therefore, using of Q.rm.uwa.6AS in future MAS or other advanced genetic approaches may help improve RM, yield, and stress tolerance in wheat. The QTL could be tested in wider populations for wheat breeding. On the other hand, Q.rm.uwa.7AL for RM identified in this study was co-located with previously identified QTrl.D84-7A (Xbarc275) for RL under both well-watered and drought-stress conditions in a back cross population of Devon × Syn084 (Ibrahim et al., 2012). Another grain yield QTL (Xpsp3094.1–Xm68p78.6) on chromosome 7A was co-located with Q.rm.uwa.7AL (Quarrie et al., 2005). As no similar QTL for root traits on chromosome 6AS and 7AL were reported, Q.rm.uwa.6AS and Q.rm.uwa.7AL were novel discoveries in this study. Validation of Q.rm.uwa.7AL is recommended in the future.
In this study, QTL for RD (Q.rd.uwa.5DL) and QTL for RSA2 (Q.rsa2.5DL) overlapped and shared the same marker, Xmwg900 (Figure 2B). These QTL also overlapped with a previously reported QTL for root volume (Ibrahim et al., 2012). Furthermore, the marker interval was found within a recently identified QTL (IWB61072–IWB49479) for grain yield (Li et al., 2018) suggesting the importance of Q.rd.uwa.5DL and Q.rsa2.5DL over other root traits and grain yield. However, there was no overlapping evidence for Q.rl.uwa.5AL, Q.rd.uwa.5AL, Q.rdia.uwa.6AL, Q.rm.uwa.7AL, or Q.srl.uwa.6DS discovered in this study with any previous root trait QTL (Figure 3) which suggesting that they are novel QTL for controlling root traits.
We identified 2,323 genes in the Q.rd.uwa.7BL regions and 387 genes in the Q.rm.uwa.6AS regions. Proteins encoded by 215 genes of Q.rd.uwa.7BL and 34 genes of Q.rm.uwa.6AS were associated with the wheat root traits (Supplementary Table S1). In a recent review article, Halder et al. (2022) listed the number of proteins associated with the wheat root system. However, among the identified genes, proteins encoded by 21 and 13 genes in Q.rd.uwa.7BL (329.79–700.63 Mb) and in Q.rm.uwa.6AS (8.00–22.02 Mb), respectively, had roles in controlling root traits in different crops such as rice, barley, maize and soybean, and Arabidopsis (Table 5).
Phytohormones such as cytokinin play important role in root development (Aloni et al., 2006), with histidine-containing phosphotransfer (HK) and glutaredoxin proteins regulating cytokinin signaling. Transgenic plants with reduced cytokinin had greater root growth and more lateral roots than those plants with high cytokinin (Nishimura et al., 2004). HK proteins regulate phosphorylation to control the root growth of Arabidopsis (Hutchison and Kieber, 2007) and barley (under limited/resupply of P) (Ma et al., 2021). We found TraesCS6A01G020400 encoded HK proteins which located very close (0.23 Mb) to the validated marker, Xgwm334, and TraesCS7B01G404000 encoded glutaredoxin protein which located on the QTL for shallow RD. In Arabidopsis, a genotypes AtGRXS3/4/5/8 with silenced glutaredoxin proteins had large primary roots (Patterson et al., 2016) indicating the negative role of glutaredoxin protein in RD. Glutaredoxin also play important role in stress tolerance through redox state of cell, redox dependent pathway regulation, and also improve nutrient uptake. In rice root, glutaredoxin improved arsenic (Verma et al., 2020) and salinity (Verma et al., 2021) stress tolerance. In Arabidopsis root, glutaredoxin improved nitrogen uptake and ammonium stress tolerance (Patterson et al., 2016).
Another protein, 3-ketoacyl-CoA synthase (KCS) condenses very-long-chain fatty acids essential for cuticular waxes and suberin production in roots (Lee et al., 2009; Kim et al., 2021). Suberin is critical role in drought and salinity stress tolerance in root (de Silva et al., 2021). For example, reduced suberin restricted root growth in Arabidopsis (Lee et al., 2009). A KCS6 barley mutant with reduced cuticular waxes had reduced seminal root length but increased lateral root length (Weidenbach et al., 2015). Therefore, future exploration of the KCS encoding genes (TraesCS6A01G024400 and TraesCS7B01G254900) identified in this study could be useful for improving stress tolerance in wheat through improved RM, and RD improvement. TraesCS6A01G026500, identified in our study, encoded lysine-specific demethylase (LSD). LSD belongs to histone demethylase (amine oxidase superfamily) which contributes to root elongation and abiotic stress tolerance through histone modification (Sun et al., 2019; Liu et al., 2022). In maize, the LSD encoding hub gene [Zm00001d002266: genes with top 10% correlation within a module (Liu et al., 2019)], controlled seminal root length under drought and controlled conditions (Guo et al., 2020). Transgenic Arabidopsis with overexpressed LSD encoding Glyma.17G022500 improved salinity stress tolerance (Sun et al., 2019).
Few candidate genes for RD and RM encode proteins with similar functions indicating their importance for future wheat breeding. Germin-like protein (GLP) was first identified in germinating wheat grains (Bernier and Berna, 2001). However, the role of GLP genes in wheat root trait control is unclear, except for an association between GLP and cell wall modification for improved aluminum stress tolerance in wheat (Delisle et al., 2001; Houde and Diallo, 2008). GLP genes Gs1 and Gs2 are highly expressed in barley roots and expressed salinity stress tolerance (Hurkman et al., 1991). GLP contributed to multiple stress (e.g. as drought, heat, cold, and oxidative stress) tolerance in Arabidopsis and rice (Li et al., 2016). Therefore, it would be interesting to explore the genes encoding GLP for RD and RM (Table 5). We also found six candidate genes encoding mitochondrial transcription termination factor (mTERF) in Q.rm.uwa.6AS, and thaumatin-like protein encoding genes—TraesCS6A01G021000 in Q.rm.uwa.6AS and TraesCS7B01 G446200 and TraesCS7B01G25590 in Q.rd.uwa.7BL. mTERF and thaumatin-like protein expressed in roots of different crops and Arabidopsis and play important role in abiotic and biotic stress tolerance. For example, mTERF encoding genes shot1 and mterf6-5 in an Arabidopsis mutant expressed heat tolerance (Kim et al., 2012) and salt tolerance (Robles et al., 2018), respectively. Thaumatin-like protein highly expressed in roots in barley (Iqbal et al., 2020) and Arabidopsis, and contributed to abiotic (e.g. drought and salt) and biotic (e.g. fungus) stress tolerance in Arabidopsis (Misra et al., 2016; de Jesús-Pires et al., 2020). In-silico studies of the putative candidate genes suggested that the genes expressed at different levels (0.02–360.65) in root tissues of different wheat cultivars. Important genes, TraesCS7B01G404000, TraesCS7B01G254900 and TraesCS7B01G446200, from Q.rd.uwa.7BL showed high (124.32) to low (0.89) gene expression while important genes from Q.rm.uwa.6AS, TraesCS6A01G024400, TraesCS6A01G021000 and TraesCS6A01G020400 showed low (0.20–0.11) gene expression in roots of previously studied wheat cultivars (Supplementary Table S2). However, the study conditions and the studied root traits of the previous studies may cause variation in gene expression, which could be confirmed by future gene expression approaches. Moreover, after further functional validation, the identified putative candidate genes may be useful for wheat breeding programs for root improvement.
Eleven QTL were identified on chromosomes 5A, 5D, 6A, 6B, 6D, 7A and 7B for seven root traits in bread wheat suggesting that wheat chromosome groups 5, 6 and 7 harbor major QTL/genes for root traits. Q.rd.uwa.7BL co-located with previously identified grain yield and biotic and abiotic stress tolerance markers. Q.rm.uwa.6AS, is a novel QTL for RM. Validation studies confirmed the functionality of Q.rd.uwa.7BL and Q.rm.uwa.6AS in two independent F2 populations. The putative candidate genes located in the validated QTL encode important proteins for root traits in other crops. Further gene validation is required to confirm their role in wheat breeding. The identified and validated QTL/markers and putative candidate genes in this study provide a genetic foundation for marker-assisted breeding of root traits in bread wheat.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Investigation, formal analysis, and preparation of the original draft: TH. Supervision: KS, GY, HL, and YC. Writing—review and editing: KS, GY, HL, and YC. All authors have read and agreed to the published version of the manuscript. All authors contributed to the article and approved the submitted version.
TH was supported by an Australian Government Research Training Program scholarships and Underwood PhD Completion Scholarship (UWA). The research was partly supported by the Global Innovation Linkage Program (GIL53853) from the Australian Department of Industry, Science and Resources, and The UWA Institute of Agriculture.
The authors thank Dr. Siegy Krauss and Dr. Janet Anthony for their assistance in DNA fragment analysis at the Biodiversity Conservation Centre, Kings Park, WA; Prof. Jacqueline Batley and Dr. Anita Severn-Ellis for their assistance in agarose gel electrophoresis; Mukesh Choudhary for help in glasshouse work; Dr. Asad Prodhan for constructive suggestions for analysis; Robert Creasy and Bill Piasini for the glasshouse facility setup.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1092992/full#supplementary-material
Acuna, T. B., Rebetzke, G., He, X., Maynol, E., Wade, L. (2014). Mapping quantitative trait loci associated with root penetration ability of wheat in contrasting environments. Mol. Breed. 34 (2), 631–642. doi: 10.1007/s11032-014-0063-x
Alahmad, S., El Hassouni, K., Bassi, F. M., Dinglasan, E., Youssef, C., Quarry, G., et al. (2019). A major root architecture QTL responding to water limitation in durum wheat. Front. Plant Sci. 10, 436. doi: 10.3389/fpls.2019.00436
Alexandratos, N., Bruinsma, J. (2012). World agriculture towards 2030/2050: The 2012 revision. Rome, FAO.
Aloni, R., Aloni, E., Langhans, M., Ullrich, C. (2006). Role of cytokinin and auxin in shaping root architecture: regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. 97 (5), 883–893. doi: 10.1093/aob/mcl027
Appels, R., Eversole, K., Stein, N., Feuillet, C., Keller, B., Rogers, J., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361 (6403), eaar7191. doi: 10.1126/science.aar7191
Asif, M., Tunc, C. E., Yazici, M. A., Tutus, Y., Rehman, R., Rehman, A., et al. (2019). Effect of predicted climate change on growth and yield performance of wheat under varied nitrogen and zinc supply. Plant Soil 434 (1), 231–244. doi: 10.1007/s11104-018-3808-1
Atkinson, J. A., Wingen, L. U., Griffiths, M., Pound, M. P., Gaju, O., Foulkes, M. J., et al. (2015). Phenotyping pipeline reveals major seedling root growth QTL in hexaploid wheat. J. Exp. Bot. 66 (8), 2283–2292. doi: 10.1093/jxb/erv006
Atta, M. B., Mahmood, T., Trethowan, M. (2013). Relationship between root morphology and grain yield of wheat in north-western NSW, Australia. Aust. J. Crop Sci. 13, 2108–2115.
Ayalew, H., Liu, H., Borner, A., Kobiljski, B., Liu, C., Yan, G. (2018). Genome-wide association mapping of major root length QTLs under PEG induced water stress in wheat. Front. Plant Sci. 9, 1759. doi: 10.3389/fpls.2018.01759
Ayalew, H., Liu, H., Yan, G. (2017). Identification and validation of root length QTLs for water stress resistance in hexaploid wheat (Triticum aestivum L.). Euphytica 213 (6), 126. doi: 10.1007/s10681-017-1914-4
Bai, C., Liang, Y., Hawkesford, M. J. (2013). Identification of QTLs associated with seedling root traits and their correlation with plant height in wheat. J. Exp. Bot. 64 (6), 1745–1753. doi: 10.1093/jxb/ert041
Balakrishnan, D., Surapaneni, M., Yadavalli, V. R., Addanki, K. R., Mesapogu, S., Beerelli, K., et al. (2020). Detecting CSSLs and yield QTLs with additive, epistatic and QTL x environment interaction effects from Oryza sativa x O. nivara IRGC81832 cross. Sci. Rep. 10 (1), 7766. doi: 10.1038/s41598-020-64300-0
Bennett, D., Reynolds, M., Mullan, D., Izanloo, A., Kuchel, H., Langridge, P., et al. (2012). Detection of two major grain yield QTL in bread wheat (Triticum aestivum L.) under heat, drought and high yield potential environments. Theor. Appl. Genet. 125 (7), 1473–1485. doi: 10.1007/s00122-012-1927-2
Ben Sadok, I., Martinez, S., Moutier, N., Garcia, G., Leon, L., Belaj, A., et al. (2015). Plasticity in vegetative growth over contrasted growing sites of an F1 olive tree progeny during its juvenile phase. PLoS One 10 (6), e0127539. doi: 10.1371/journal.pone.0127539
Bernier, F., Berna, A. (2001). Germins and germin-like proteins: Plant do-all proteins. but what do they do exactly? Plant Physiol. Biochem. 39 (7-8), 545–554.
Bhoite, R., Onyemaobi, I., Si, P., Siddique, K., Yan, G. (2018). Identification and validation of QTL and their associated genes for pre-emergent metribuzin tolerance in hexaploid wheat (Triticum aestivum L.). BMC Genet. 19 (1), 102. doi: 10.1186/s12863-018-0690-z
Borrill, P., Harrington, S. A., Uauy, C. (2019). Applying the latest advances in genomics and phenomics for trait discovery in polyploid wheat. Plant J. 97 (1), 56–72. doi: 10.1111/tpj.14150
Cai, S., Bai, G. H., Zhang, D. (2008). Quantitative trait loci for aluminum resistance in Chinese wheat landrace FSW. Theor. Appl. Genet. 117 (1), 49–56. doi: 10.1007/s00122-008-0751-1
Calleja-Cabrera, J., Boter, M., Oñate-Sánchez, L., Pernas, M. (2020). Root growth adaptation to climate change in crops. Front. Plant Sci. 11, 544. doi: 10.3389/fpls.2020.00544
Chen, Y.-A., Chi, W.-C., Huang, T.-L., Lin, C.-Y., Nguyeh, T. T. Q., Hsiung, Y.-C., et al. (2012). Mercury-induced biochemical and proteomic changes in rice roots. Plant Physiol. Biochem. 55, 23–32. doi: 10.1016/j.plaphy.2012.03.008
Chen, Y., Djalovic, I., Siddique, K. H. M. (2018). “Advances in understanding grain legume physiology: understanding root architecture, nutrient uptake and response to abiotic stress,” in Achieving sustainable cultivation of grain legumes Burleigh Dodds Science Publishing Limited. 1, 11–28.
Chen, Y. L., Dunbabin, V. M., Diggle, A. J., Siddique, K. H. M., Rengel, Z. (2011a). Assessing variability in root traits of wild Lupinus angustifolius germplasm: Basis for modelling root system structure. Plant Soil 354 (1-2), 141–155. doi: 10.1007/s11104-011-1050-1
Chen, Y. L., Dunbabin, V. M., Diggle, A. J., Siddique, K. H. M., Rengel, Z. (2011b). Development of a novel semi-hydroponic phenotyping system for studying root architecture. Funct. Plant Biol. 38 (5), 355–363. doi: 10.1071/FP10241
Chen, Y. L., Palta, J., Clements, J., Buirchell, B., Siddique, K. H. M., Rengel, Z. (2014). Root architecture alteration of narrow-leafed lupin and wheat in response to soil compaction. Field Crops Res. 165, 61–70. doi: 10.1016/j.fcr.2014.04.007
Chen, Y., Palta, J., Prasad, P. V. V., Siddique, K. H. M. (2020). Phenotypic variability in bread wheat root systems at the early vegetative stage. BMC Plant Biol. 20 (1), 185. doi: 10.1186/s12870-020-02390-8
Collard, B. C. Y., Jolley, R., Bovill, W. D., Grams, R. A., Wildermuth, G. B., Sutherland, M. W. (2006). Confirmation of QTL mapping and marker validation for partial seedling resistance to crown rot in wheat line ‘2-49’. Aust. J. Agric. Res. 57 (9), 967–973. doi: 10.1071/AR05419
Colombo, M., Roumet, P., Salon, C., Jeudy, C., Lamboeuf, M., Lafarge, S., et al. (2022). Genetic analysis of platform-phenotyped root system architecture of bread and durum wheat in relation to agronomic traits. Front. Plant Sci. 13, 853601. doi: 10.3389/fpls.2022.853601
Danakumara, T., Kumari, J., Singh, A. K., Sinha, S. K., Pradhan, A. K., Sharma, S., et al. (2021). Genetic dissection of seedling root system architectural traits in a diverse panel of hexaploid wheat through multi-locus genome-wide association mapping for improving drought tolerance. Int. J. Mol. Sci. 22 (13), 7188. doi: 10.3390/ijms22137188
de Jesús-Pires, C., Ferreira-Neto, J. R., Pacifico Bezerra-Neto, J., Kido, E. A., de Oliveira Silva, R. L., Pandolfi, V., et al. (2020). Plant thaumatin-like proteins: function, evolution and biotechnological applications. Curr. Protein Pept. Sci. 21 (1), 36–51. doi: 10.2174/1389203720666190318164905
Delisle, G., Champoux, M., Houde, M. (2001). Characterization of oxalate oxidase and cell death in Al-sensitive and tolerant wheat roots. Plant Cell Physiol. 42 (3), 324–333. doi: 10.1093/pcp/pce041
de Silva, N. D., Murmu, J., Chabot, D., Hubbard, K., Ryser, P., Molina, I., et al. (2021). Root suberin plays important roles in reducing water loss and sodium uptake in Arabidopsis thaliana. Metabolites 11 (11), 735. doi: 10.3390/metabo11110735
Ehdaie, B., Layne, A. P., Waines, J. G. (2012). Root system plasticity to drought influences grain yield in bread wheat. Euphytica 186 (1), 219–232. doi: 10.1007/s10681-011-0585-9
Ehdaie, B., Mohammadi, S. A., Nouraein, M. (2016). QTLs for root traits at mid-tillering and for root and shoot traits at maturity in a RIL population of spring bread wheat grown under well-watered conditions. Euphytica 211 (1), 17–38. doi: 10.1007/s10681-016-1670-x
El Hassouni, K., Alahmad, S., Belkadi, B., Filali-Maltouf, A., Hickey, L., Bassi, F. (2018). Root system architecture and its association with yield under different water regimes in durum wheat. Crop Sci. 58 (6), 2331–2346. doi: 10.2135/cropsci2018.01.0076
Figueroa-Bustos, V., Palta, J. A., Chen, Y. L., Siddique, K. H. (2018). Characterization of root and shoot traits in wheat cultivars with putative differences in root system size. Agronomy 8 (7), 109. doi: 10.3390/agronomy8070109
Figueroa-Bustos, V., Palta, J. A., Chen, Y., Stefanova, K., Siddique, K. H. M. (2020). Wheat cultivars with contrasting root system size responded differently to terminal drought. Front. Plant Sci. 11, 1285. doi: 10.3389/fpls.2020.01285
Fitter, A. ,. H. (2002). “Characteristics and functions of root systems,,” in Plant roots: The hidden half, 4th ed. Eds. Eshel, A., Beeckman, T. (New York: Dekker: Taylor & Francis Group), 15–32.
Fitz Gerald, J. N., Lehti-Shiu, M. D., Ingram, P. A., Deak, K. I., Biesiada, T., Malamy, J. E. (2006). Identification of quantitative trait loci that regulate Arabidopsis root system size and plasticity. Genetics 172 (1), 485–498. doi: 10.1534/genetics.105.047555
Griffiths, M. D., Atkinson, J. A., Gardiner, L.-J., Swarup, R., Pound, M. P., Wilson, M. H., et al. (2022). Identification of QTL and underlying genes for root system architecture associated with nitrate nutrition in hexaploid wheat. J. Integr. Agric. 21 (4), 591529. doi: 10.1016/S2095-3119(21)63700-0
Grossman, J. D., Rice, K. J. (2012). Evolution of root plasticity responses to variation in soil nutrient distribution and concentration. Evol. Appl. 5 (8), 850–857. doi: 10.1111/j.1752-4571.2012.00263.x
Guo, J., Li, C., Zhang, X., Li, Y., Zhang, D., Shi, Y., et al. (2020). Transcriptome and GWAS analyses reveal candidate gene for seminal root length of maize seedlings under drought stress. Plant Sci. 292, 110380. doi: 10.1016/j.plantsci.2019.110380
Halder, T., Choudhary, M., Liu, H., Chen, Y., Yan, G., Siddique, K. H. (2022). Wheat proteomics for abiotic stress tolerance and root system architecture: Current status and future prospects. Proteomes 10 (2), 17. doi: 10.3390/proteomes10020017
Halder, T., Liu, H., Chen, Y., Yan, G., Siddique, K. H. M. (2021). Identification of candidate genes for root traits using genotype–phenotype association analysis of near-isogenic lines in hexaploid wheat (Triticum aestivum L.). Int. J. Mol. Sci. 22 (7), 3579. doi: 10.3390/ijms22073579
Horn, R., Wingen, L. U., Snape, J. W., Dolan, L. (2016). Mapping of quantitative trait loci for root hair length in wheat identifies loci that co-locate with loci for yield components. J. Exp. Bot. 67 (15), 4535–4543. doi: 10.1093/jxb/erw228
Houde, M., Diallo, A. O. (2008). Identification of genes and pathways associated with aluminum stress and tolerance using transcriptome profiling of wheat near-isogenic lines. BMC Genome. 9, 400. doi: 10.1186/1471-2164-9-400
Hurkman, W. J., Tao, H. P., Tanaka, C. K. (1991). Germin-like polypeptides increase in barley roots during salt stress. Plant Physiol. 97 (1), 366–374. doi: 10.1104/pp.97.1.366
Hussain, B., Lucas, S. J., Ozturk, L., Budak, H. (2017). Mapping QTLs conferring salt tolerance and micronutrient concentrations at seedling stage in wheat. Sci. Rep. 7 (1), 1–14. doi: 10.1038/s41598-017-15726-6
Hutchison, C. E., Kieber, J. J. (2007). Signaling via histidine-containing phosphotransfer proteins in Arabidopsis. Plant Signal. Behav. 2 (4), 287–289. doi: 10.4161/psb.2.4.4039
Ibrahim, S. E., Schubert, A., Pillen, K., Léon, J. (2012). QTL analysis of drought tolerance for seedling root morphological traits in an advanced backcross population of spring wheat. Int. J. AgriScience 2 (7), 619–629.
Iqbal, I., Tripathi, R. K., Wilkins, O., Singh, J. (2020). Thaumatin-like protein (TLP) gene family in barley: Genome-wide exploration and expression analysis during germination. Genes 11 (9), 1080. doi: 10.3390/genes11091080
Khalid, M., Gul, A., Amir, R., Ali, M., Afzal, F., Quraishi, U. M., et al. (2018). QTL mapping for seedling morphology under drought stress in wheat cross synthetic (W7984)/Opata. Plant Genet. Res.: Characterization Util. 16 (4), 359–366. doi: 10.1017/S1479262118000023
Kim, M., Lee, U., Small, I., des Francs-Small, C. C., Vierling, E. (2012). Mutations in an Arabidopsis mitochondrial transcription termination factor–related protein enhance thermotolerance in the absence of the major molecular chaperone HSP101. Plant Cell 24 (8), 3349–3365. doi: 10.1105/tpc.112.101006
Kim, J., Lee, S. B., Suh, M. C. (2021). Arabidopsis 3-ketoacyl-CoA synthase 4 is essential for root and pollen tube growth. J. Plant Biol. 64 (2), 155–165. doi: 10.1007/s12374-020-09288-w
Kohli, A., Narciso, J. O., Miro, B., Raorane, M. (2012). Root proteases: Reinforced links between nitrogen uptake and mobilization and drought tolerance. Physiol. Plant. 145 (1), 165–179. doi: 10.1111/j.1399-3054.2012.01573.x
Kumar, P., Choudhary, M., Halder, T., Prakash, N. R., Singh, V., Sheoran, S., et al. (2022). Salinity stress tolerance and omics approaches: Revisiting the progress and achievements in major cereal crops. Heredity, 1–22. doi: 10.1038/s41437-022-00516-2
Langridge, P., Lagudah, E. S., Holton, T. A., Appels, R., Sharp, P. J., Chalmers, K. J. (2001). Trends in genetic and genome analyses in wheat: A review. Aust. J. Agric. Res. 52 (12), 1043–1077. doi: 10.1071/AR01082
Lee, S. B., Jung, S. J., Go, Y. S., Kim, H. U., Kim, J. K., Cho, H. J., et al. (2009). Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 60 (3), 462–475. doi: 10.1111/j.1365-313X.2009.03973.x
Li, C.-W., Lee, S.-H., Chieh, P.-S., Lin, C.-S., Wang, Y.-C., Chan, M.-T. (2012). Arabidopsis root-abundant cytosolic methionine sulfoxide reductase b genes MsrB7 and MsrB8 are involved in tolerance to oxidative stress. Plant Cell Physiol. 53 (10), 1707–1719. doi: 10.1093/pcp/pcs114
Li, L., Liu, Z., Wu, J. (2022). Genetic mapping of QTL for three root-related traits in wheat (Triticum aestivum). Biotechnol. Biotechnol. Equip. 36 (1), 506–512. doi: 10.1080/13102818.2022.2098817
Li, T., Ma, J., Zou, Y., Chen, G., Ding, P., Zhang, H., et al. (2020). Quantitative trait loci for seeding root traits and the relationships between root and agronomic traits in common wheat. Genome 63 (1), 27–36. doi: 10.1139/gen-2019-0116
Li, G., Wang, M., Ma, C., Tao, R., Hou, F., Liu, Y. (2021). Effects of soil heterogeneity and species on plant interactions. Front. Ecol. Evol. 734. doi: 10.3389/fevo.2021.756344
Li, F., Wen, W., He, Z., Liu, J., Jin, H., Cao, S., et al. (2018). Genome-wide linkage mapping of yield-related traits in three Chinese bread wheat populations using high-density SNP markers. Theor. Appl. Genet. 131 (9), 1903–1924. doi: 10.1007/s00122-018-3122-6
Li, L., Xu, X., Chen, C., Shen, Z. (2016). Genome-wide characterization and expression analysis of the germin-like protein family in rice and Arabidopsis. Int. J. Mol. Sci. 17 (10), 1622. doi: 10.3390/ijms17101622
Liu, S., Begum, N., An, T., Zhao, T., Xu, B., Zhang, S., et al. (2021). Characterization of root system architecture traits in diverse soybean genotypes using a semi-hydroponic system. Plants 10 (12), 2781. doi: 10.3390/plants10122781
Liu, Y., Gu, H.-Y., Zhu, J., Niu, Y.-M., Zhang, C., Guo, G.-L. (2019). Identification of hub genes and key pathways associated with bipolar disorder based on weighted gene co-expression network analysis. Front. Physiol. 10, 1081. doi: 10.3389/fphys.2019.01081
Liu, M., Jiang, J., Han, Y., Shi, M., Li, X., Wang, Y., et al. (2022). Functional characterization of the lysine-specific histone demethylases family in soybean. Plants 11 (11), 1398. doi: 10.3390/plants11111398
Liu, X., Li, R., Chang, X., Jing, R. (2013). Mapping QTLs for seedling root traits in a doubled haploid wheat population under different water regimes. Euphytica 189 (1), 51–66. doi: 10.1007/s10681-012-0690-4
Lu, L., Liu, H., Wu, Y., Yan, G. (2022). Identification and validation of a chromosome 4D quantitative trait locus hotspot conferring heat tolerance in common wheat (Triticum aestivum L.). Plants 11 (6), 729. doi: 10.3390/plants11060729
Lynch, J. P. (2007). Roots of the second green revolution. Aust. J. Bot. 55 (5), 493–512. doi: 10.1071/BT06118
Ma, Z., Wang, J., Li, C., Ren, P., Yao, L., Li, B., et al. (2021). Global profiling of phosphorylation reveals the barley roots response to phosphorus starvation and resupply. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.676432
Meng-jiao, Y., Cai-rong, W., Hassa, M. A., Wu, Y.-Y., Xian-chun, X., Shu-Bing, S., et al. (2020). QTL mapping of seedling biomass and root traits under different nitrogen conditions in bread wheat (Triticum aestivum L.). J. Integr. Agric. 19 (0), 2–14. doi: 10.1016/S2095-3119(20)63192-6
Misra, R. C., Kamthan, M., Kumar, S., Ghosh, S. (2016). A thaumatin-like protein of Ocimum basilicum confers tolerance to fungal pathogen and abiotic stress in transgenic Arabidopsis. Sci. Rep. 6 (1), 1–14. doi: 10.1038/srep25340
Nahar, K., Bovill, W., McDonald, G. (2022). Assessing the contribution of seedling root traits to phosphorus responsiveness in wheat. J. Plant Nutr. 1-19, 2170–2188. doi: 10.1080/01904167.2022.2046079
Neuteboom, L. W., Veth-Tello, L. M., Clijdesdale, O. R., Hooykaas, P. J., van der Zaal, B. J. (1999). A novel subtilisin-like protease gene from Arabidopsis thaliana is expressed at sites of lateral root emergence. DNA Res. 6 (1), 13–19. doi: 10.1093/dnares/6.1.13
Nishimura, C., Ohashi, Y., Sato, S., Kato, T., Tabata, S., Ueguchi, C. (2004). Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16 (6), 1365–1377. doi: 10.1105/tpc.021477
Onyemaobi, I., Ayalew, H., Liu, H., Siddique, K. H. M., Yan, G. (2018). Identification and validation of a major chromosome region for high grain number per spike under meiotic stage water stress in wheat (Triticum aestivum L.). PLoS One 13 (3), e0194075. doi: 10.1371/journal.pone.0194075
Palta, A. J., Chen, X., Milroy, P. S., Rebetzke, J. G., Dreccer, M. F., Watt, M. (2011). Large Root systems: Are they useful in adapting wheat to dry environments? Funct. Plant Biol. 38, 347–354. doi: 10.1071/FP11031
Palta, J., Watt, M. (2009). “Vigorous crop root systems: form and function for improving the capture of water and nutrients,” in Applied crop physiology: boundaries between genetic improvement and agronomy (Academic, San Diego: Elsevier), 309–325. doi: 10.1016/B978-0-12-374431-9.00013-X
Pan, Z., Ren, X., Zhao, H., Liu, L., Tan, Z., Qiu, F. (2019). A mitochondrial transcription termination factor, ZmSmk3, is required for nad1 intron4 and nad4 intron1 splicing and kernel development in maize. G3: Genes Genomes Genet. 9 (8), 2677–2686. doi: 10.1534/g3.119.400265
Patterson, K., Walters, L. A., Cooper, A. M., Olvera, J. G., Rosas, M. A., Rasmusson, A. G., et al. (2016). Nitrate-regulated glutaredoxins control Arabidopsis primary root growth. Plant Physiol. 170 (2), 989–999. doi: 10.1104/pp.15.01776
Pearce, S., Vazquez-Gross, H., Herin, S. Y., Hane, D., Wang, Y., Gu, Y. Q., et al. (2015). WheatExp: An RNA-seq expression database for polyploid wheat. BMC Plant Biol. 15, 299. doi: 10.1186/s12870-015-0692-1
Quarrie, S. A., Steed, A., Calestani, C., Semikhodskii, A., Lebreton, C., Chinoy, C., et al. (2005). A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese spring x SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor. Appl. Genet. 110 (5), 865–880. doi: 10.1007/s00122-004-1902-7
Ren, Y., He, X., Liu, D., Li, J., Zhao, X., Li, B., et al. (2012). Major quantitative trait loci for seminal root morphology of wheat seedlings. Mol. Breed. 30 (1), 139–148. doi: 10.1007/s11032-011-9605-7
Ren, Y., Qian, Y., Xu, Y., Zou, C., Liu, D., Zhao, X., et al. (2017). Characterization of QTLs for root traits of wheat grown under different nitrogen and phosphorus supply levels. Front. Plant Sci. 8, 2096. doi: 10.3389/fpls.2017.02096
Robles, P., Micol, J. L., Quesada, V. (2015). Mutations in the plant-conserved MTERF9 alter chloroplast gene expression, development and tolerance to abiotic stress in Arabidopsis thaliana. Physiol. Plant. 154 (2), 297–313. doi: 10.1111/ppl.12307
Robles, P., Navarro-Cartagena, S., Ferrández-Ayela, A., Núñez-Delegido, E., Quesada, V. (2018). The characterization of Arabidopsis mterf6 mutants reveals a new role for mTERF6 in tolerance to abiotic stress. Int. J. Mol. Sci. 19 (8), 2388. doi: 10.3390/ijms19082388
Rogers, E. D., Benfey, P. N. (2015). Regulation of plant root system architecture: Implications for crop advancement. Curr. Opin. Biotechnol. 32, 93–98. doi: 10.1016/j.copbio.2014.11.015
Salarpour, M., Pakniyat, H., Abdolshahi, R., Heidari, B., Razi, H., Afzali, R. (2020). Mapping QTL for agronomic and root traits in the Kukri/RAC875 wheat (Triticum aestivum L.) population under drought stress conditions. Euphytica 216, 105. doi: 10.1007/s10681-020-02627-5
Salem, K. F. M., Roder, M. S., Borner, A. (2007). Identification and mapping quantitative trait loci for stem reserve mobilisation in wheat (Triticum aestivum L.). Cereal Res. Commun. 35 (3), 1367–1374. doi: 10.1556/CRC.35.2007.3.1
Salim, M., Chen, Y., Ye, H., Nguyen, H. T., Solaiman, Z. M., Siddique, K. H. M. (2022). Screening of soybean genotypes based on root morphology and shoot traits using the semi-hydroponic phenotyping platform and rhizobox technique. Agronomy 12 (1), 56. doi: 10.3390/agronomy12010056
Scheiner, S. M. (1993). Genetics and evolution of phenotypic plasticity. Annu. Rev. Ecol. Systematics, 35–68. doi: 10.1146/annurev.es.24.110193.000343
Sénéchal, F., Graff, L., Surcouf, O., Marcelo, P., Rayon, C., Bouton, S., et al. (2014). Arabidopsis PECTIN METHYLESTERASE17 is co-expressed with and processed by SBT3. 5, a subtilisin-like serine protease. Ann. Bot. 114 (6), 1161–1175. doi: 10.1093/aob/mcu035
Sharma, S., Xu, S., Ehdaie, B., Hoops, A., Close, T. J., Lukaszewski, A. J., et al. (2011). Dissection of QTL effects for root traits using a chromosome arm-specific mapping population in bread wheat. Theor. Appl. Genet. 122 (4), 759–769. doi: 10.1007/s00122-010-1484-5
Shukla, S., Singh, K., Patil, R. V., Kadam, S., Bharti, S., Prasad, P., et al. (2014). Genomic regions associated with grain yield under drought stress in wheat (Triticum aestivum L.). Euphytica 203 (2), 449–467. doi: 10.1007/s10681-014-1314-y
Singh, R. (2019). “Wheat strategy delivers stronger grains,” in Annual performance report 2019: Synthetic-wheat-breeding-strategy (Consultative Group for International Agricultural Research (CGIAR)).
Song, T., Das, D., Yang, F., Chen, M., Tian, Y., Cheng, C., et al. (2020). Genome-wide transcriptome analysis of roots in two rice varieties in response to alternate wetting and drying irrigation. Crop J. 8 (4), 586–601. doi: 10.1016/j.cj.2020.01.007
Soriano, J. M., Alvaro, F. (2019). Discovering consensus genomic regions in wheat for root-related traits by QTL meta-analysis. Sci. Rep. 9 (1), 10537. doi: 10.1038/s41598-019-47038-2
Sorrells, M. E., Gustafson, J. P., Somers, D., Chao, S., Benscher, D., Guedira-Brown, G., et al. (2011). Reconstruction of the synthetic W7984 x opata M85 wheat reference population. Genome 54 (11), 875–882. doi: 10.1139/g11-054
Sun, L., Song, G., Guo, W., Wang, W., Zhao, H., Gao, T., et al. (2019). Dynamic changes in genome-wide histone3 lysine27 trimethylation and gene expression of soybean roots in response to salt stress. Front. Plant Sci. 10, 1031. doi: 10.3389/fpls.2019.01031
Verma, P. K., Verma, S., Tripathi, R. D., Chakrabarty, D. (2020). A rice glutaredoxin regulate the expression of aquaporin genes and modulate root responses to provide arsenic tolerance. Ecotoxicology Environ. Saf. 195, 110471. doi: 10.1016/j.ecoenv.2020.110471
Verma, P. K., Verma, S., Tripathi, R. D., Pandey, N., Chakrabarty, D. (2021). CC-type glutaredoxin, OsGrx_C7 plays a crucial role in enhancing protection against salt stress in rice. J. Biotechnol. 329, 192–203. doi: 10.1016/j.jbiotec.2021.02.008
Vishwakarma, S., Zhang, X., Lyubchich, V. (2022). Wheat trade tends to happen between countries with contrasting extreme weather stress and synchronous yield variation. Commun. Earth Environ. 3 (1), 261. doi: 10.1038/s43247-022-00591-7
Wang, J., Chen, Y., Zhang, Y., Zhang, Y., Ai, Y., Feng, Y., et al. (2021). Phenotyping and validation of root morphological traits in barley (Hordeum vulgare L.). Agronomy 11 (8), 1583. doi: 10.3390/agronomy11081583
Wang, D., Pan, Y., Zhao, X., Zhu, L., Fu, B., Li, Z. (2011). Genome-wide temporal-spatial gene expression profiling of drought responsiveness in rice. BMC Genomics 12 (1), 1–15. doi: 10.1186/1471-2164-12-149
Weidenbach, D., Jansen, M., Bodewein, T., Nagel, K. A., Schaffrath, U. (2015). Shoot and root phenotyping of the barley mutant kcs6 (3-ketoacyl-CoA synthase6) depleted in epicuticular waxes under water limitation. Plant Signal. Behav. 10 (4), e1003752. doi: 10.1080/15592324.2014.1003752
Woo, D., Riley, W., Paez-Garcia, A., Marklein, A., Mekonnen, Z., Liu, X., et al. (2021). Impoverishing roots will improve wheat yield and profitability through increased water and nitrogen use efficiencies. J. Geophys. Res. Biogeosciences. 126 (9), e2020JG005829. doi: 10.1029/2020JG005829
Wu, J., Cai, G., Tu, J., Li, L., Liu, S., Luo, X., et al. (2013). Identification of QTLs for resistance to sclerotinia stem rot and BnaC. IGMT5.a as a candidate gene of the major resistant QTL SRC6 in Brassica napus. PLoS One 8 (7), e67740.
Wu, F., Yang, X., Wang, Z., Deng, M., Ma, J., Chen, G., et al. (2017). Identification of major quantitative trait loci for root diameter in synthetic hexaploid wheat under phosphorus-deficient conditions. J. Appl. Genet. 58 (4), 437–447. doi: 10.1007/s13353-017-0406-5
Xu, Y., Li, S., Li, L., Zhang, X., Xu, H., An, D., et al. (2013). Mapping QTLs for salt tolerance with additive, epistatic and QTL × treatment interaction effects at seedling stage in wheat. Plant Breed. 132 (3), 276–283. doi: 10.1111/pbr.12048
Yang, M.-j., Wang, C.-R., Hassan, M. A., Wu, Y.-Y., Xia, X.-C., Shi, S.-B., et al. (2021a). QTL mapping of seedling biomass and root traits under different nitrogen conditions in bread wheat (Triticum aestivum L.). J. Integr. Agric. 20 (5), 1180–1192. doi: 10.1016/S2095-3119(20)63192-6
Yang, M., Wang, C., Hassan, M. A., Li, F., Xia, X., Shi, S., et al. (2021b). QTL mapping of root traits in wheat under different phosphorus levels using hydroponic culture. BMC Genomics 22 (1), 1–12. doi: 10.1186/s12864-021-07425-4
Yu, M., Chen, G.-Y. (2013). Conditional QTL mapping for waterlogging tolerance in two RILs populations of wheat. Springerplus 2 (1), 1–7. doi: 10.1186/2193-1801-2-245
Zadoks, J. C., Chang, T. T., Konzak, C. F. (1974). A decimal code for the growth stages of cereals. Weed Res. 14, 415–421. doi: 10.1111/j.1365-3180.1974.tb01084.x
Zanin, L., Zamboni, A., Monte, R., Tomasi, N., Varanini, Z., Cesco, S., et al. (2015). Transcriptomic analysis highlights reciprocal interactions of urea and nitrate for nitrogen acquisition by maize roots. Plant Cell Physiol. 56 (3), 532–548. doi: 10.1093/pcp/pcu202
Zhang, H., Chen, J., Li, R., Deng, Z., Zhang, K., Liu, B., et al. (2016). Conditional QTL mapping of three yield components in common wheat (Triticum aestivum L.). Crop J. 4 (3), 220–228. doi: 10.1016/j.cj.2016.01.007
Zhang, N., Zhao, L., Mawcha, K. T., Zhao, C., Yang, W., Liu, D. (2020). Evaluation of leaf rust resistance in the Chinese wheat cultivar ‘Een1’. PeerJ 8, e8993. doi: 10.7717/peerj.8993
Zhao, Y., Cai, M., Zhang, X., Li, Y., Zhang, J., Zhao, H., et al. (2014). Genome-wide identification, evolution and expression analysis of mTERF gene family in maize. PLoS One 9 (4), e94126. doi: 10.1371/journal.pone.0094126
Zheng, X., Wen, X., Qiao, L., Zhao, J., Zhang, X., Li, X., et al. (2019). A novel QTL QTrl.saw-2D.2 associated with the total root length identified by linkage and association analyses in wheat (Triticum aestivum L.). Planta 250 (1), 129–143. doi: 10.1007/s00425-019-03154-x
Keywords: QTL, markers, candidate genes, phenotyping, recombinant inbred lines, genomic region
Citation: Halder T, Liu H, Chen Y, Yan G and Siddique KHM (2023) Chromosome groups 5, 6 and 7 harbor major quantitative trait loci controlling root traits in bread wheat (Triticum aestivum L.). Front. Plant Sci. 14:1092992. doi: 10.3389/fpls.2023.1092992
Received: 16 November 2022; Accepted: 27 February 2023;
Published: 20 March 2023.
Edited by:
Colin W. Hiebert, Agriculture and Agri-Food Canada (AAFC), CanadaReviewed by:
Zhiqiang Zhou, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2023 Halder, Liu, Chen, Yan and Siddique. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tanushree Halder, dGFudXNocmVlLmhhbGRlckByZXNlYXJjaC51d2EuZWR1LmF1; Hui Liu, aHVpLmxpdUB1d2EuZWR1LmF1; Kadambot H. M. Siddique, a2FkYW1ib3Quc2lkZGlxdWVAdXdhLmVkdS5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.