- 1Guangdong Provincial Key Laboratory of Plant Molecular Breeding, State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, South China Agricultural University, Guangzhou, China
- 2Guangdong Laboratory for Lingnan Modern Agriculture, South China Agricultural University, Guangzhou, China
Stigma exsertion rate (SER) is an index of outcrossing ability in rice and is a key trait of male sterile lines (MSLs) in hybrid rice. In this study, it was found that the maintainer lines carrying gs3 and gs3/gw8 showed higher SER. Single-segment substitution lines (SSSLs) carrying gs3, gw5, GW7 or gw8 genes for grain shape and gene pyramiding lines were used to reveal the relationship between grain shape and SER. The results showed that the grain shape regulatory genes had pleiotropic effects on SER. The SERs were affected by grain shapes including grain length, grain width and the ratio of length to width (RLW) not only in low SER background, but also in high SER background. The coefficients of determination (R2) between grain length and SER, grain width and SER, and grain RLW and SER were 0.78, 0.72, and 0.91 respectively. The grain RLW was the most important parameter affecting SER, and a larger grain RLW was beneficial to stigma exsertion. The pyramiding line PL-gs3/GW7/gw8 showed the largest grain RLW and the highest SER, which will be a fine breeding resource. Further research showed that the grain shape regulatory genes had pleiotropic effects on stigma shape, although the R2 values between grain shape and stigma shape, and stigma shape and SER were lower. Our results demonstrate that grain shape is a factor affecting SER in rice, in part by affecting stigma shape. This finding will be helpful for breeding MSLs with high SER in hybrid rice.
Introduction
Hybrid rice has made a great contribution to maintaining the world’s major food production and food security (Yuan, 2017). The commercialization of hybrid rice depends on large-scale hybrid seed production (Qian et al., 2016). Cultivated rice is a self-pollination crop (Virmani and Athwal, 1973). The yield of hybrid seeds mainly depends on the outcrossing ability of male sterile lines (MSLs) (Virmani et al., 1982; Guo et al., 2017). The stigma exsertion in MSLs can capture more pollens from male parents, thus improving their ability of outcrossing (Marathi and Jena, 2015). Therefore, the stigma exsertion rate (SER) is an important trait for outcrossing ability in MSLs.
In the past decades, dozens of quantitative trait loci (QTLs) responsible for the SER have been identified from rice germplasm resources (Marathi and Jena, 2015; Guo et al., 2022; Liu et al., 2022). Recently, eighteen QTLs for SER from O. sativa, O. glaberrima, and O. glumaepatula were detected in the single-segment substitution lines (SSSLs) with the Huajingxian74 (HJX74) genetic background (Tan et al., 2020; Tan et al., 2021; Tan et al., 2022a). Eleven of the QTLs were used to develop the pyramiding lines with 2- to 6-QTLs in the HJX74 genetic background. The results showed that the SER can be improved with increasing QTLs in pyramiding lines. The pyramiding lines carrying 5-6 QTLs showed as high SER as wild rice (Tan et al., 2022b). The results indicate that SER is a complex trait controlled by a series of QTLs, and the high SER trait can be reconstructed by pyramiding of the QTLs in rice. However, no QTL for SER has been cloned, so the mechanism of stigma exsertion is still unclear.
SER is the result of phenotypic balance between stigma and other parts of rice spikelet (Zhou et al., 2017; Jiang et al., 2021). The relationship between SER and grain shape has become an interesting focus for researchers. Among detected QTLs, major QTLs qES3 and qSER8 were demonstrated to act pleiotropic effects on grain length by controlling the longitudinal axis direction of grains (Bakti and Tanaka, 2019). On the other hand, it is documented that the GS3 gene not only determines grain length (Fan et al., 2006), but also acts a pleiotropic effect on stigma size (Takano-Kai et al., 2011; Zhou et al., 2017; Dang et al., 2020) and stigma exsertion (Miyata et al., 2007; Li et al., 2014; Zhou et al., 2017; Xu et al., 2019; Liu et al., 2022). Two cloned genes for grain width, GW5 and GW2, were also reported to extend their effects on SER (Zhou et al., 2017). However, it was also reported that the gs3 gene for long grain didn’t always result in stigma exsertion, some rice accessions with very low SER also possessed the gs3 gene (Zhou et al., 2017; Xu et al., 2019). These results indicate that the influence of grain shape on SER is very complex, and the relationship between grain shape and SER is still not very clear.
During the process of domestication, cultivated rice has already lost the ability of natural outcrossing (Parmar et al., 1979). Wild rice has long and large stigma and long floret opening period, which provides a biological basis for outcrossing (Marathi et al., 2015). Therefore, stigma trait may be another important factor influencing SER. Some QTLs for stigma size including stigma length and stigma width were mapped (Dang et al., 2016; Zhou et al., 2017; Dang et al., 2020). Among of the QTLs, some controlled stigma size and SER (Jiang et al., 2021), while others only controlled stigma size and didn’t influence SER (Uga et al., 2003; Yan et al., 2009; Zhou et al., 2017). In addition, it was found that glume opening angle was positively correlated with SER (Kato and Namai, 1987; Uga et al., 2003; Mahalingam et al., 2013). Therefore, the relationship between stigma shape and SER is still ambiguous.
In present study, we found that the maintainer lines with long grains showed higher SERs in rice. To reveal the relationship between grain shape and SER, SSSLs carrying grain shape regulatory genes and pyramiding lines with different gene combinations were used to analyze the relationship. We show that grain shape regulatory genes have pleiotropic effects on stigma shape and SER. The grain shape is a factor affecting SER, in part by affecting stigma shape. This finding reveals the contribution of grain shape to SER, which is helpful to rebuild the outcrossing ability of MSLs in hybrid rice.
Materials and methods
Plant materials
The maintainer line H121B carrying four substitution segments was previously developed in the HJX74 genetic background with the rf3 and rf4 genes from XieqingzaoB. The maintainer line H131B was previously bred in the HJX74 genetic background with the rf3 and rf4 genes from XieqingzaoB and the OsMADS50, gs3 and Wxt genes from SSSLs in the HJX74 genetic background (Dai et al., 2015). Seven SSSLs, W17-46-40-10-07-05, W23-07-06-05-02-02, W12-11-22-03-03-163, W02-08-08-08-01, W07-07-02-03-02, W23-19-06-07-19-03 and W09-38-60-07-18-04 carrying respectively fgr, qBLAST-11, gs3, gs3, gw5, GW7 and gw8 were selected from the HJX74-SSSL library (Wang et al., 2012; Zhang, 2021). The pyramiding lines carrying 2-4 QTLs for SER, 2QL-1, 2QL-5, 3QL-3, 3QL-10, 4QL-1 and 4QL-3, were previously constructed by pyramiding QTLs for SER from HJX74-SSSLs through maker assisted selection (MAS) (Tan et al., 2022b).
Field experiment
All plant materials were planted in the experimental station, South China Agricultural University, Guangzhou (23°07′N, 113°15′E). The materials were planted in 2015-2020, two cropping seasons per year. The first cropping season (FCS) was from late February to middle July and the second cropping season (SCS) was from late July to middle November. The seeds were sown on seedbeds and the seedlings were transplanted to the paddy field as single seedlings. Field management and controlling of diseases and insect pests followed normal agricultural practices.
Genotyping
Molecular markers were applied to detect the substitution segments from SSSLs. The length of substitution segments was measured by the method described previously (Tan et al., 2020). The target genes in substitution segments were identified using closely linked markers. The target genes were genotyped by linkage markers, functional markers, and phenotypic analysis. Genomic DNA was extracted from fresh leaf using a modified CTAB method (Murray et al., 1980). The PCR products were separated on the 6% denatured PAGE gel, and banded by the silver staining.
Phenotyping and statistical analysis
For stigma investigation, five panicles were collected from each line during the flourishing florescence. Six mature spikelets at the upper part of each panicle were selected to carefully separate pistils from glumes and then took photos under a stereomicroscope (Leica M205FA). Rice stigma was divided into two parts, brush-shaped part (BSP) and non-brush-shaped part (NBSP). The stigma length was the sum of BSP and NBSP lengths (Takano-Kai et al., 2011). The stigma width was the maximum width of BSP (Zhou et al., 2017). Stigma length and stigma width were measured by using the software of ImageJ (https://imagej.nih.gov/ij/) described by Zhou et al. (2017). The SER was investigated following the previous method (Tan et al., 2020; Tan et al., 2021; Tan et al., 2022a). Grain traits were measured by the yield traits scorer (YTS), a rice phenotypic facility (Yang et al., 2014).
For statistical analysis, percentage data was converted to the arcsine square root. The least significance range (LSR) was used for multiple range test among multiple groups (Duncan, 1955). Student’s t test was used to detect the difference between two groups. The correlation between traits was analyzed by regression correlation. MapChart2.3 (https://www.wur.nl/en/show/Mapchart.htm) and OriginPro 9.0 (https://www.originlab.com) were used to make figures.
Results
Long grain maintainer lines showed higher SER
The maintainer line H131B previously developed in the HJX74 genetic background was improved by pyramiding two target genes on the substitution segments of HJX74-SSSLs. The new maintainer line H211B carrying seven target genes on substitution segments in the HJX74 genetic background was developed (Supplementary Figure 1 and Supplementary Table 1). Phenotype investigation showed that the traits controlled by target genes have been significantly improved, while other traits had no significant difference (Supplementary Table 2). Both H131B and H211B carried the gs3 gene. Compared with HJX74, H131B and H211B showed significant differences in grain length, but no significant differences in grain width. The grain length of HJX74 was 8.36 mm, while that of H131B and H211B increased to 9.50 mm and 9.51 mm respectively (Figures 1A, B, D, E and Supplementary Table 2). Interestingly, the SERs of H131B and H211B were also significantly different from HJX74. The SER of HJX74 was 27.8%, while that of H131B and H211B increased to 43.9% and 43.4% respectively (Figures 1C, F and Supplementary Table 2). These results showed that the long grain trait controlled by gs3 increased the SERs of maintainer lines H131B and H211B.
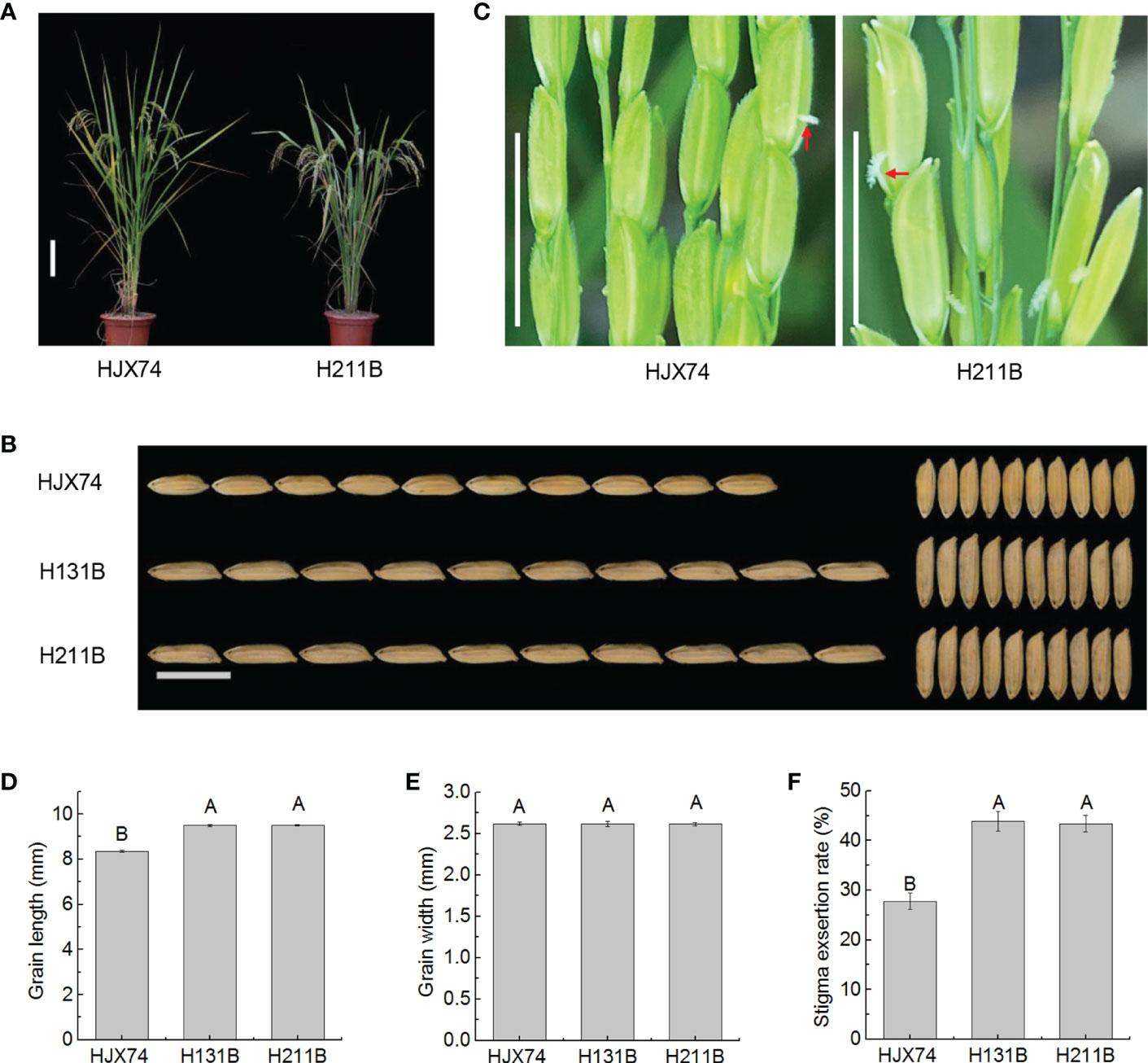
Figure 1 Grain shape and stigma exsertion rate (SER) in maintainer line H211B. (A), Plant type of HJX74 and H211B. Scale bar, 15 cm. (B), Grain shape in HJX74, H131B and H211B. Scale bar, 1 cm. (C), SER in HJX74 and H211B. Scale bar, 1 cm. The arrows point the exserted stigmas. Grain length (D), grain width (E), and SER (F) in HJX74, H131B and H211B. Data are shown as mean ± S.E. of two cropping seasons. Capital letters indicate significant differences at the 0.01 level.
The maintainer line H121B previously developed in the HJX74 genetic background was improved by pyramiding gs3 and gw8 genes on substitution segments of HJX74-SSSLs. The new maintainer line H212B carrying gs3and gw8 genes in the HJX74 genetic background was developed (Supplementary Figure 2 and Supplementary Table 3). Phenotype investigation showed that the traits controlled by target genes have been significantly improved, while other traits had no significant differences (Supplementary Table 4). Compared with HJX74 and H121B, H212B carrying gs3 and gw8 genes had significant differences in grain length and grain width. In grain length, HJX74 and H121B were 8.36 mm and 8.42 mm respectively, while H212B increased to 9.31 mm. In grain width, HJX74 and H121B were both 2.62 mm, while H212B reduced to 2.43 mm (Figures 2A, B, D, E and Supplementary Table 4). Interestingly, the SER of H212B was also significantly different from that of HJX74 and H121B. SERs of HJX74 and H121B were 27.8% and 27.4% respectively, while SER of H212B increased to 55.0% (Figures 2C, F and Supplementary Table 4). Compared with H131B and H211B carrying gs3 (Figure 1 and Supplementary Table 2), H212B with gs3 and gw8 had slender grain and high SER.
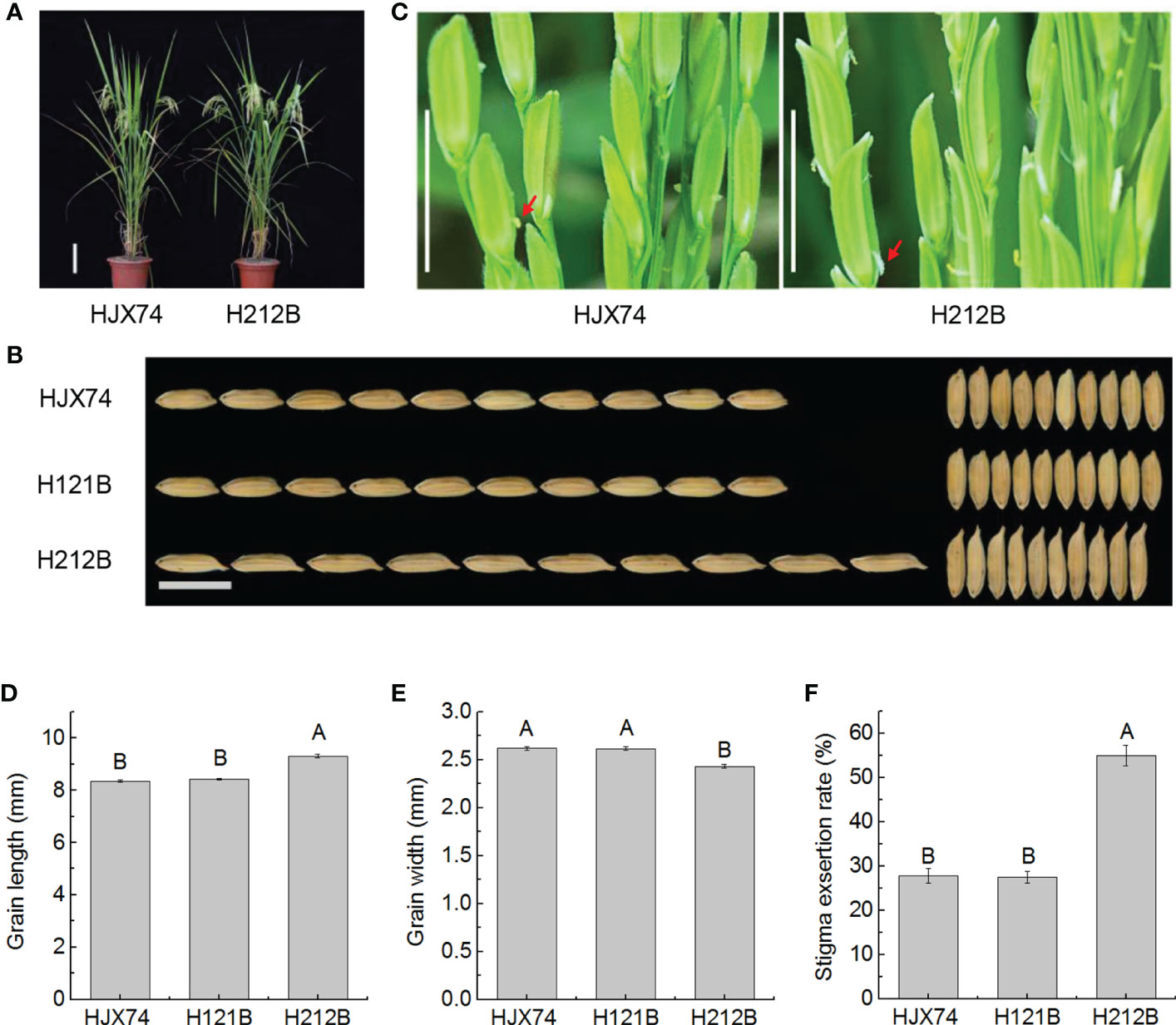
Figure 2 Grain shape and stigma exsertion rate (SER) in maintainer line H212B. (A), Plant type of HJX74 and H212B. Scale bar, 15 cm. (B), Grain shape in HJX74, H121B and H212B. Scale bar, 1 cm. (C), SER in HJX74 and H212B. Scale bar, 1 cm. The arrows point the exserted stigmas. Grain length (D), grain width (E), and SER (F) in HJX74, H121B and H212B. Data are shown as mean ± S.E. of two cropping seasons. Capital letters indicate significant differences at the 0.01 level.
Pleiotropic effects on SER of the genes controlling grain shape in SSSLs
In order to confirm the finding that the trait of slender grain led to higher SER, four SSSLs carrying a grain shape regulatory gene, gs3, gw5, GW7 or gw8, on the substitution segments in the HJX74 genetic background were used to analyze the relationship between grain shape and SER (Figure 3A and Supplementary Table 5). The grain of HJX74 was 8.38 mm long and 2.55 mm wide. SSSL-gs3, SSSL-gw5, SSSL-GW7 and SSSL-gw8 had grain lengths of 9.77 mm, 7.79 mm, 9.29 mm and 8.79 mm, and grain widths of 2.55 mm, 2.88 mm, 2.47 mm and 2.40 mm, respectively. In the ratio of length to width (RLW) in grains, HJX74 was 3.29, while SSSL-gs3, SSSL-GW7 and SSSL-gw8 increased to 3.83, 3.77 and 3.65 respectively, and SSSL-gw5 decreased to 2.71 (Figures 3B–E and Supplementary Table 6). Correspondingly, the SER of HJX74 was 28.3%, while that of SSSL-gs3, SSSL-GW7 and SSSL-gw8 increased to 47.4%, 45.6% and 40.8% respectively, and that of SSSL-gw5 decreased to 22.8% (Figures 3E, F and Supplementary Table 6). These results showed that the genes of gs3, gw5, GW7 and gw8 had significant effects on grain shape and SER in the SSSLs, indicating that the genes controlling grain shape had significant pleiotropic effects on SER.
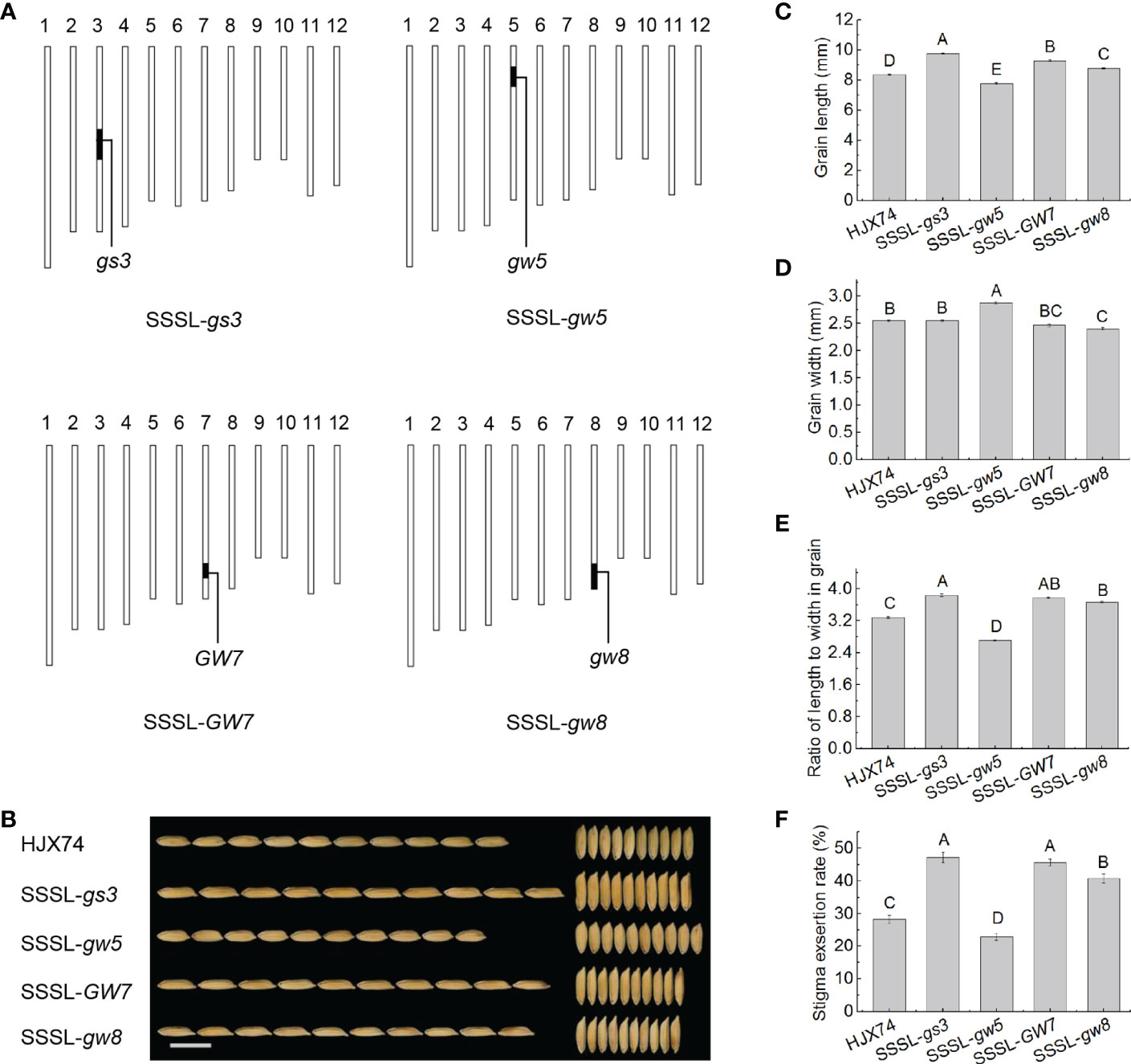
Figure 3 Grain shape and SER in the SSSLs carrying a gene for grain shape. (A), Graphical genotypes of the SSSLs. The vertical bars are a graphical representation of chromosomes. The black bars represent substitution segments containing target genes for grain shape, and the white regions represent the HJX74 genetic background. (B), Appearance of the grains in HJX74 and SSSLs. Scale bar, 1 cm. Grain length (C), grain width (D), grain RLW (E), and SER (F) in HJX74 and SSSLs. Data are shown as mean ± S.E. of two cropping seasons. Capital letters indicate significant differences at the 0.01 level. SSSL, single-segment substitution line. SER, stigma exsertion rate. RLW, ratio of length to width.
Effects of the grain shape controlled by different gene combinations of gs3, gw5, GW7 and gw8 on SER in pyramiding lines
To further evaluate the effect of grain shape on SER, four SSSLs, SSSL-gs3, SSSL-gw5, SSSL-GW7 and SSSL-gw8, were used to develop a series of pyramiding lines. A total of 11 pyramiding lines were developed through MAS, including six 2-gene pyramiding lines, four 3-gene pyramiding lines, and one 4-gene pyramiding line (Supplementary Table 7).
In six 2-gene pyramiding lines, PL-gs3/gw5, PL-gs3/GW7, PL-gs3/gw8, PL-gw5/GW7, PL-gw5/gw8 and PL-GW7/gw8, grain lengths were 9.04 mm, 10.17 mm, 9.57 mm, 7.88 mm, 8.29 mm and 8.85 mm, and grain widths were 2.90 mm, 2.41 mm, 2.39 mm, 2.86 mm, 2.52 mm and 2.41 mm, respectively, which differed from 8.38 mm long and 2.55 mm wide of the HJX74 grain (Figures 4A, B and Supplementary Table 8). Compared with the grain RLW of HJX74 3.29, grain RLWs of PL-gs3/gw5, PL-gs3/GW7, PL-gs3/gw8, PL-gw5/GW7, PL-gw5/gw8 and PL-GW7/gw8 were 3.12, 4.22, 4.01, 2.75, 3.29 and 3.67 respectively. Correspondingly, SERs of PL-gs3/gw5, PL-gs3/GW7, PL-gs3/gw8, PL-gw5/GW7, PL-gw5/gw8 and PL-GW7/gw8 were 20.0%, 63.1%, 60.3%, 19.0%, 25.0% and 44.5% respectively, with 28.3% SER of HJX74 as control (Figures 4C, D and Supplementary Table 8). In five pyramiding lines with multiple genes for grain shape, PL-gs3/gw5/GW7, PL-gs3/gw5/gw8, PL-gs3/GW7/gw8, PL-gw5/GW7/gw8 and PL-gs3/gw5/GW7/gw8, grain lengths were 9.43 mm, 9.49 mm, 10.93 mm, 8.22 mm and 10.59 mm, grain widths were 2.65 mm, 2.67 mm, 2.10 mm, 2.62 mm and 2.51 mm, and grain RLWs were 3.55, 3.55, 5.21, 3.14 and 4.21, respectively. SERs of the five pyramiding lines were 34.6%, 38.5%, 74.6%, 25.0% and 59.5% respectively (Figures 4A–D and Supplementary Table 8). These results showed that larger grain RLWs led to higher SERs in the pyramiding lines. Four pyramiding lines, PL-gs3/GW7, PL-gs3/gw8, PL-gs3/GW7/gw8, and PL-gs3/gw5/GW7/gw8, had larger than 4.0 of grain RLWs and higher than 50.0% of SERs. The pyramiding line PL-gs3/GW7/gw8 showed the largest grain RLW and the highest SER.
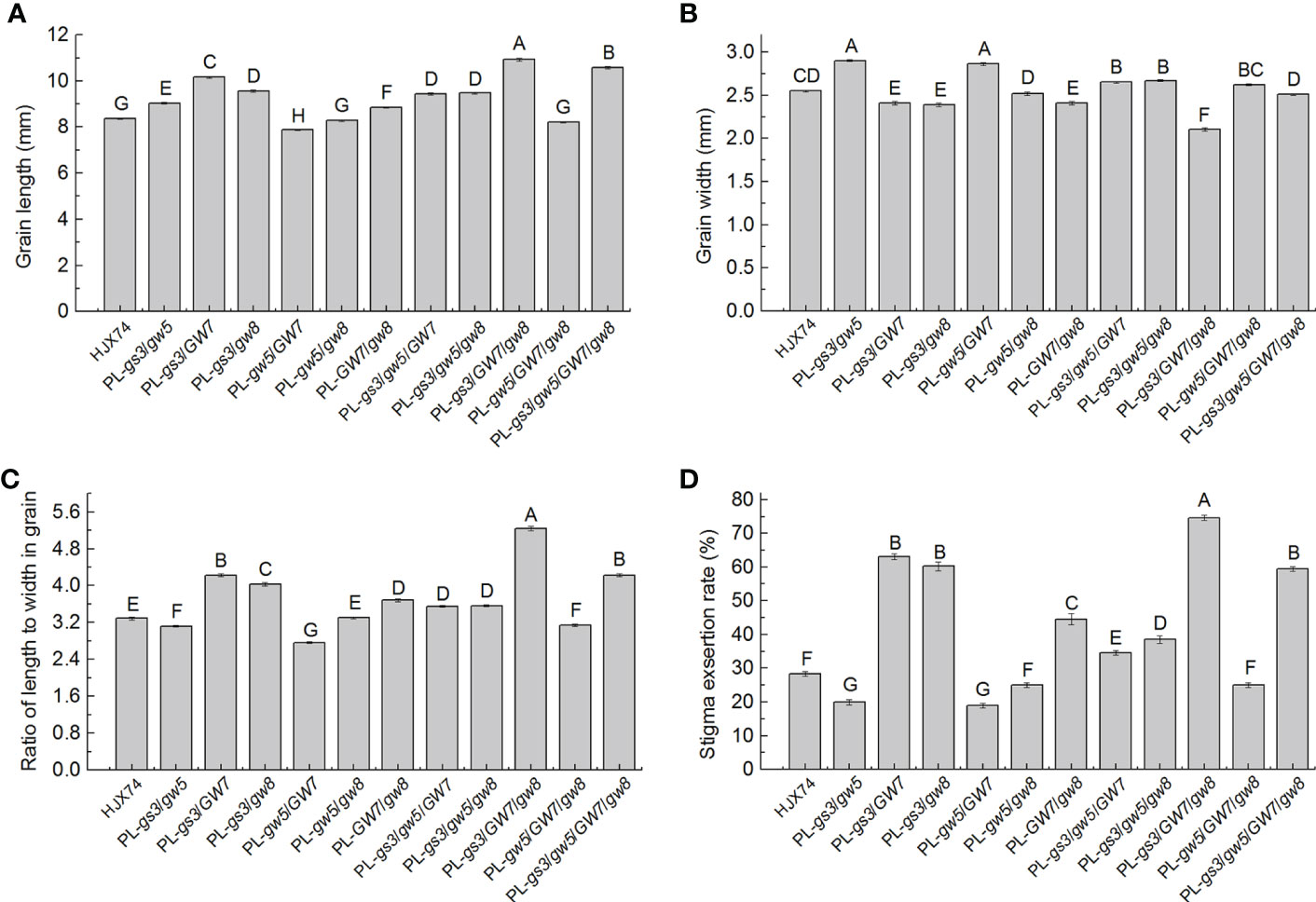
Figure 4 Grain shape and SER in pyramiding lines carrying genes for grain shape. Grain length (A), grain width (B), grain RLW (C), and SER (D) in HJX74 and pyramiding lines carrying genes for grain shape. Data are shown as mean ± S.E. of two cropping seasons. Capital letters indicate significant differences at the 0.01 level. SER, stigma exsertion rate. PL, pyramiding line. RLW, ratio of length to width.
Effects of grain shape on SER in high SER background
To investigate the effect of grain shape on SER at high SER level, the gs3 gene was introduced into the pyramiding lines carrying multiple SER-QTLs, 2-QTL pyramiding lines (2QLs), 3-QTL pyramiding lines (3QLs) and 4-QTL pyramiding lines (4QLs) developed previously. Three new pyramiding lines, 2QL-1/gs3, 3QL-3/gs3 and 4QL-3/gs3, were developed in the HJX74 genetic background (Supplementary Figure 3). In the same way, gs3 and gw8 genes were introduced into the pyramiding lines carrying multiple SER-QTLs, 2QLs, 3QLs and 4QLs developed previously. Three new pyramiding lines, 2QL-5/gs3/gw8, 3QL-10/gs3/gw8 and 4QL-1/gs3/gw8, were developed in the HJX74 genetic background (Supplementary Figure 4).
Compared with the pyramiding lines without gs3, the grain lengths of the pyramiding lines with gs3, 2QL-1/gs3, 3QL-3/gs3 and 4QL-3/gs3, were significantly longer, but the grain width had no significant difference. The grains of pyramiding lines with gs3 and gw8, 2QL-5/gs3/gw8, 3QL-10/gs3/gw8 and 4QL-1/gs3/gw8, were significantly longer and narrower than those without gs3 and gw8 (Figures 5A–D). In grain RLW, the pyramiding lines with gs3 and gs3/gw8 were larger than those without gs3 and gs3/gw8, while the pyramiding lines with gs3/gw8 were larger than those with only gs3 (Figures 5E, F).
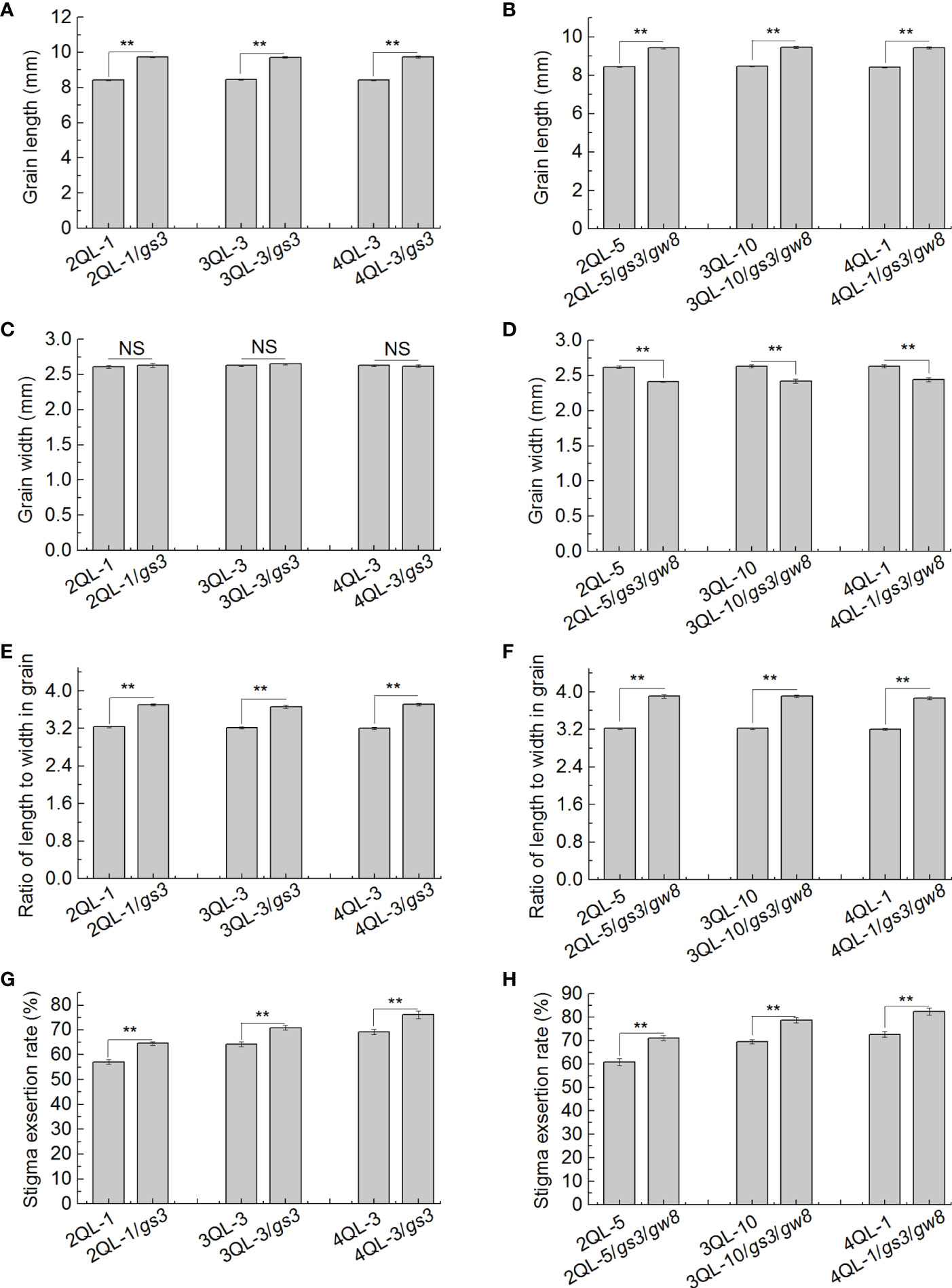
Figure 5 Grain shape and SER in pairs of pyramiding lines. Grain length (A, B), grain width (C, D), grain RLW (E, F), and SER (G, H) in pairs of pyramiding lines with and without grain shape gene gs3 and pyramiding lines with and without grain shape genes gs3 and gw8. **P ≤ 0.01, Student’s t test. SER, stigma exsertion rate. RLW, ratio of length to width.
SERs of three pairs of pyramiding lines with and without gs3 and three pairs of pyramiding lines with and without gs3 and gw8 were compared in pairs. In three pairs of pyramiding lines with and without gs3, SERs of 2QL-1, 3QL-3 and 4QL-3 were 57.0%, 64.2% and 69.0% respectively, while those of 2QL-1/gs3, 3QL-3/gs3 and 4QL-3/gs3 increased to 64.5%, 70.8% and 76.2% respectively. The SERs of pyramiding lines with gs3 were significantly higher than those of the pyramiding lines without gs3 (Figure 5G). In three pairs of pyramiding lines with and without gs3 and gw8, SERs of 2QL-5, 3QL-10 and 4QL-1 were 60.7%, 69.5% and 72.6% respectively, while those of 2QL-5/gs3/gw8, 3QL-10/gs3/gw8, 4QL-1/gs3/gw8 increased to 71.1%, 78.7% and 82.4% respectively. SERs of pyramiding lines with gs3 and gw8 were significantly higher than those of the pyramiding lines without gs3 and gw8 (Figure 5H). These results indicated that the effect of grain shape on SER occurred not only in low SER background, but also in high SER background.
Correlation between grain shape and stigma shape
To reveal the relationship between grain shape and stigma shape, stigma length and stigma width were measured in HJX74, 4 SSSLs, SSSL-gs3, SSSL-gw5, SSSL-GW7 and SSSL-gw8, and 11 pyramiding lines, PL-gs3/gw5, PL-gs3/GW7, PL-gs3/gw8, PL-gw5/GW7, PL-gw5/gw8, PL-GW7/gw8, PL-gs3/gw5/GW7, PL-gs3/gw5/gw8, PL-gs3/GW7/gw8, PL-gw5/GW7/gw8 and PL-gs3/gw5/GW7/gw8. Stigma RLWs were then calculated by the values of stigma length and stigma width in each line.
In stigma lengths, HJX74 was 1.92 mm, while 4 SSSLs were 1.77 mm to 2.27 mm, and 11 pyramiding lines were 1.81 mm to 2.41 mm (Figures 6A, B and Supplementary Table 9). In grain lengths, HJX74 was 8.38 mm, while 4 SSSLs were 7.79 mm to 9.77 mm, and 11 pyramiding lines were 7.88 mm to 10.93 mm (Figures 3C, 4A; Supplementary Tables 6, 8). Regression correlation between stigma length and grain length was a positive with the coefficient of determination (R2) of 0.58 (Figure 6E). In stigma widths, HJX74 was 0.44 mm, while 4 SSSLs were 0.36 mm to 0.46 mm, and 11 pyramiding lines were 0.37 mm to 0.46 mm (Figures 6A, C and Supplementary Table 9). In grain widths, HJX74 was 2.55 mm, while 4 SSSLs were 2.40 mm to 2.88 mm, and 11 pyramiding lines were 2.10 mm to 2.90 mm (Figures 3D, 4B; Supplementary Tables 6, 8). Regression correlation between stigma width and grain width was positive with R2 value of 0.46 (Figure 6F). In stigma RLWs, HJX74 was 4.40 mm, while 4 SSSLs were 3.83 mm to 6.35 mm, and 11 pyramiding lines were 4.39 mm to 6.26 mm (Figures 6A, D and Supplementary Table 9). In grain RLWs, HJX74 was 3.29 mm, while 4 SSSLs were 2.71 mm to 3.83 mm, and 11 pyramiding lines were 2.75 mm to 5.21 mm (Figures 3E, 4C; Supplementary Tables 6, 8). The stigma RLW was positively correlated with the grain RLW, and the R2 value was 0.53 (Figure 6G).
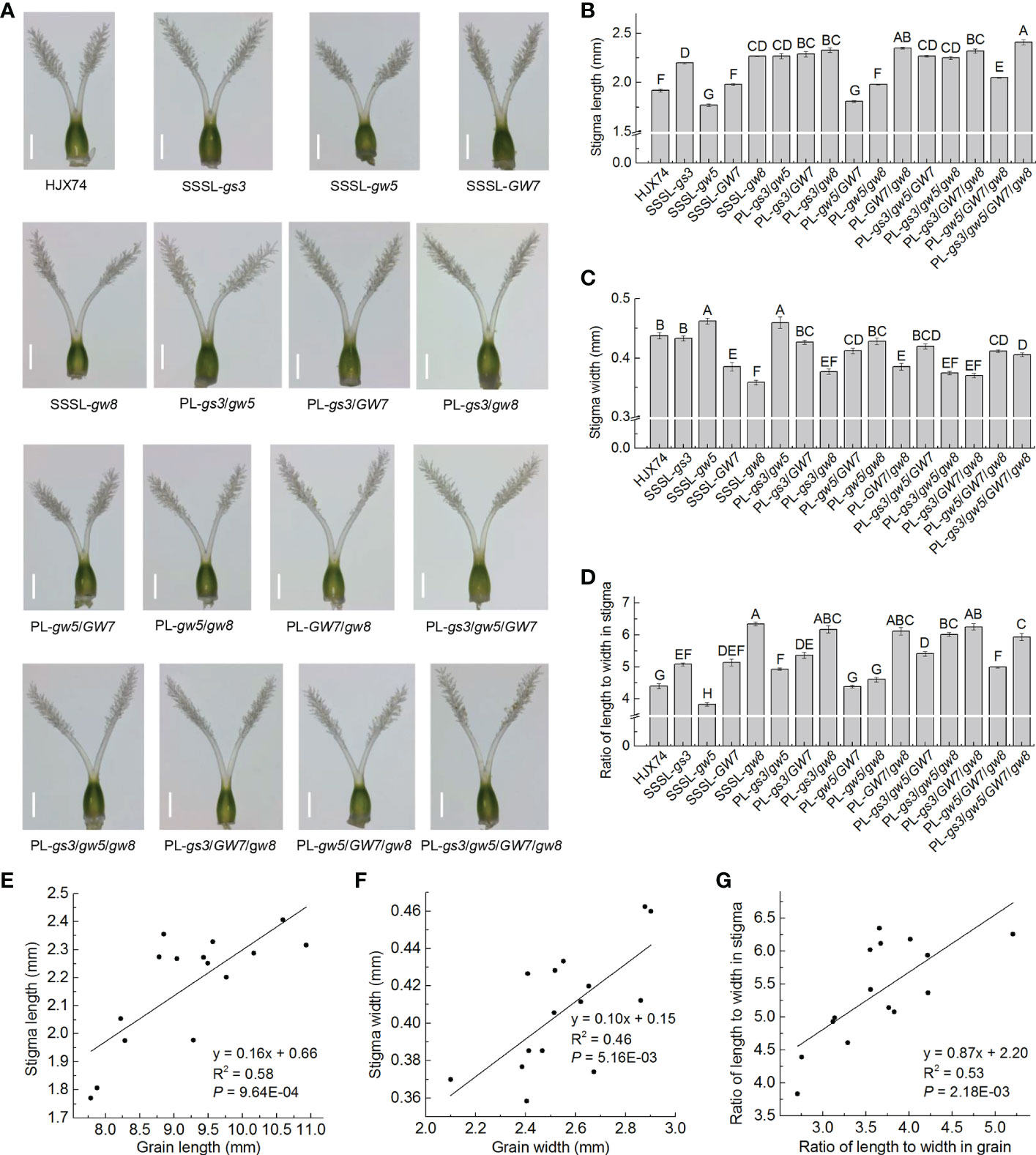
Figure 6 Stigma shape and its correlations with grain shape. (A), Stigma shape of the SSSLs and PLs. Scale bar, 0.5 mm. (B-D), Stigma length (B), stigma width (C), and stigma RLW (D) in the SSSLs and PLs. Data are shown as mean ± S.E. of two cropping seasons. Capital letters indicate significant differences at the 0.01 level. (E-G), Correlations between grain length and stigma length (E), between grain width and stigma width (F), and between grain RLW and stigma RLW (G). The average value of each line in two cropping seasons is used to perform correlation analysis. R2 represents the percentage of x contribution to y phenotype variation. SSSL, single-segment substitution line. PL, pyramiding line. RLW, ratio of length to width.
Taken together, stigma shape, including stigma length, stigma width and stigma RLW, was positively correlated with grain shape. The R2 values between stigma length and grain length, stigma width and grain width, and stigma RLW and grain RLW were close to 0.5. These results demonstrate that the genes gs3, gw5, GW7 and gw8 controlling grain shape have pleiotropic effects on stigma shape, and the stigma shape is partially affected by grain shape in rice.
Correlation between grain shape and SER
Regression correlation analysis was used to detect the relationships between stigma shape and SER, and grain shape and SER in the lines containing different genotypes of grain shape in the HJX74 genetic background. In stigma shape, stigma length was positively correlated with SER (R2 = 0.43) (Figure 7A), stigma width was negatively correlated with SER (R2 = 0.28) (Figure 7B), and stigma RLW was positively correlated with SER (R2 = 0.50) (Figure 7C). In grain shape, grain length was positively correlated with SER (R2 = 0.78) (Figure 7D), grain width was negatively correlated with SER (R2 = 0.72) (Figure 7E), and grain RLW was positively correlated with SER (R2 = 0.91) (Figure 7F). These results showed that the effect of grain shape on SER was greater than that of stigma shape. In grain shape, the grain RLW had a greater effect on SER than grain length and grain width. Therefore, the grain RLW was the most important factor affecting SER.
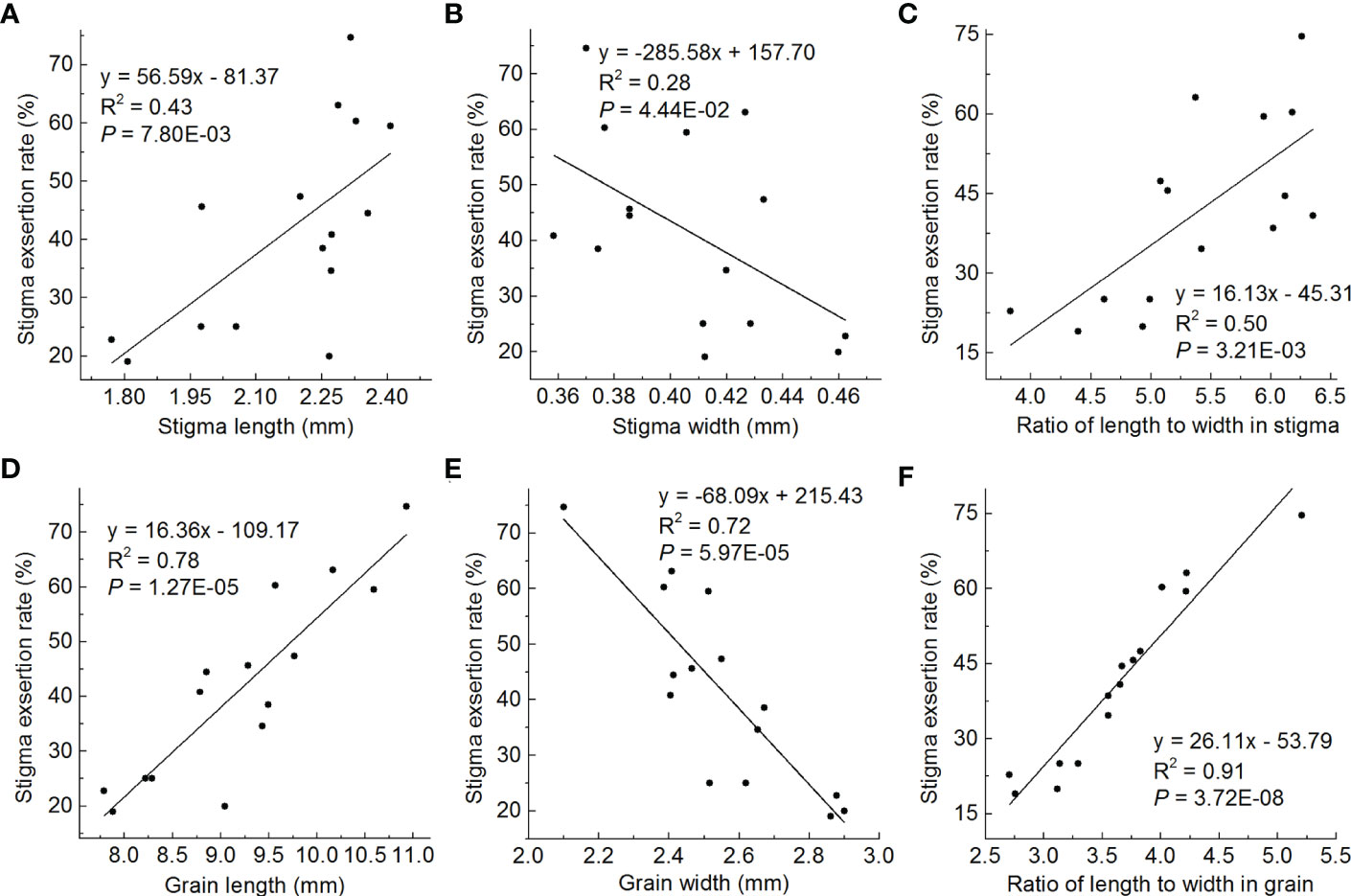
Figure 7 Regression correlations between stigma shape and SER, and grain shape and SER. (A-C), Correlations between stigma length and SER (A), stigma width and SER (B), and stigma RLW and SER (C). (D-F), Correlations between grain length and SER (D), grain width and SER (E), and grain RLW and SER (F). The average value of each line in two cropping seasons is used to perform statistical analysis. R2 represents the percentage of x contribution to y phenotype variation. SER, stigma exsertion rate. RLW, ratio of length to width.
Discussion
The grain shape has a partial effect on SER
In Asian cultivated rice, indica and japonica are two subspecies, in which indica tended to has slender grain and japonica mostly has a short and wide grain (Zhang et al., 2021). It is well known that the SER of indica rice is generally higher than that of japonica rice, which leads to generally obtain higher hybrid seed yield using indica MSLs than using japonica MSLs (Zhou et al., 2017; Dang et al., 2020; Jiang et al., 2021). It was found that the GS3 gene controlling grain length had a positive effect on SER (Miyata et al., 2007; Li et al., 2014; Zhou et al., 2017; Xu et al., 2019; Liu et al., 2022) and GW5 and GW2 genes controlling grain width had a negative effect on SER (Zhou et al., 2017). However, the pleiotropic effect of the genes controlling grain shape were not always obvious (Zhou et al., 2017; Xu et al., 2019). In this study, we found that although grain length and grain width had significant correlated with SER, the grain RLW had the greatest effect on SER, with R2 value as high as 0.91 (Figures 7D–F). This finding can explain the fact that the SER of japonica rice is generally lower than that of indica rice. On the other hand, dozens of QTLs responsible for the SER have been identified in rice genome, many of them are independent of grain shape (Marathi and Jena, 2015; Liu et al., 2019; Tan et al., 2020; Tan et al., 2021; Tan et al., 2022a). Therefore, SER is only partially affected by the grain shape.
Relationships of stigma shape with grain shape and SER
Stigma is a part of spikelet, located above ovary, surrounded by palea and lemma. At flowering, palea and lemma open, and the BSP of stigma may extend outside the glume. After flowering, palea and lemma are closed, and the BSP of stigma may exsert outside the glume (Khumto et al., 2018). Therefore, the stigma shape should be closely related to the grain shape and SER (Dang et al., 2016; Zhou et al., 2017). Wild rice has long and large stigma and strong outcrossing ability (Marathi et al., 2015). During the domestication process of cultivated rice, the stigma trait and natural outcrossing ability have already degenerated (Parmar et al., 1979; Marathi et al., 2015). Only few QTLs for stigma size including stigma length and stigma width were mapped. Some QTLs controlled stigma size and SER (Jiang et al., 2021), while others only controlled stigma size and didn’t influence SER (Uga et al., 2003; Yan et al., 2009; Zhou et al., 2017). In this study, we found that the genes controlling grain shape had pleiotropic effects on stigma shape. Correlation analysis showed that stigma length, stigma width and stigma RLW were weakly positively correlated with grain length, grain width and grain RLW respectively, with R2 values of about 0.50 (Figures 6E–G). Similarly, stigma length, stigma width and stigma RLW were also weakly correlated with SER, with R2 values of 0.50 or less (Figures 7A–C). These results showed that stigma shape was partly affected by grain shape, and has a partial effect on SER. The influence of grain shape on SER was partly caused by the influence on stigma shape. Recently, cellular examination and transcriptomic analyses revealed that three grain shape regulatory genes, GS3, GW8 and GS9, cooperatively regulate cell division during pistil development (Zhu et al., 2023). This may be the molecular mechanism of the pleiotropy of grain shape regulatory genes on stigma shape and SER.
The strategy for improving SER in rice
Cultivated rice lost the outcrossing ability during domestication (Parmar et al., 1979; Marathi et al., 2015). MSLs of hybrid rice need to restore the outcrossing ability to meet the needs of hybrid seed production (Taillebois et al., 2017). The stigma exsertion of rice breaks through the closure of palea and lemma, prolongs the pollination period, compensates for the asynchronous flowering time of male and female parents, and thus improves the outcrossing ability of MSLs (Rahman et al., 2017a; Zou et al., 2020). Therefore, improving SER is the key goal of MSL breeding. In recent decades, a large number of QTLs for SER have been located in rice genome, laying a foundation for improving SER (Marathi and Jena, 2015; Rahman et al., 2017b; Guo et al., 2022; Liu et al., 2022). Recently, eighteen QTLs for SER from O. sativa, O. glaberrima, and O. glumaepatula were mapped in the SSSLs of the HJX74 genetic background (Tan et al., 2020; Tan et al., 2021; Tan et al., 2022a). Eleven of the QTLs were used to develop pyramiding lines in the HJX74 genetic background. The pyramiding lines carrying 5-6 QTLs showed as high SER as wild rice. The phenotypic analysis showed that the increase of SER did not lead to the change of grain shape in most pyramiding lines. The results indicated that the high-SER trait can be reconstructed by pyramiding of the QTLs in rice (Tan et al., 2022b). In this study, we found that grain shape is another factor affecting rice SER, which affects SER by partially affecting stigma shape (Figures 6 and 7). Grain shape regulatory genes had pleiotropic effect on SER, and genes controlling long grain or narrow grain, such as gs3, GW7 and gw8, could improve SER (Figure 3). The pyramiding line PL-gs3/GW7/gw8 showed the largest grain RLW and the highest SER (Figure 4), which will be a fine breeding resource. Our results demonstrate that long grain rice is beneficial to the reconstruction of high SER trait. Therefore, the MSLs with high SER can be developed by pyramiding of the QTLs for SER and the genes controlling slender grain. Our finding implies that the breeding of MSLs with high SER is more difficult in japonica rice with short and wide grain than in indica rice with slender grain.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
GZ and SW designed the experiments and supervised the research works. QT performed most of the experiments. SC, ZG, QL, ZY, GC, SL, WY, JZ, YB, SB, and ZL performed a part of experiments. HZ and GL participated in material development. GZ and QT analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Major Program of Transgenic New Variety Breeding of China (2014ZX08009-037B), from the National Natural Science Foundation of China (91435207, 32201841), and from the Major Science and Technology Research Projects of Guangdong Laboratory for Lingnan Modern Agriculture (NT2021001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1087285/full#supplementary-material
References
Bakti, C., Tanaka, J. (2019). Detection of dominant QTLs for stigma exsertion ratio in rice derived from Oryza rufipogon accession ‘W0120’. Breed. Sci. 69, 143–150. doi: 10.1270/jsbbs.18139
Dai, Z., Lu, Q., Luan, X., Cai, J., Zhu, H., Liu, Z., et al. (2015). Development of a platform for breeding by design of CMS lines based on an SSSL library in rice (Oryza sativa L.). Euphytica 205, 63–72. doi: 10.1007/s10681-015-1384-5
Dang, X., Liu, E., Liang, Y., Liu, Q., Breria, C. M., Hong, D. (2016). QTL detection and elite alleles mining for stigma traits in Oryza sativa by association mapping. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01188
Dang, X., Yang, Y., Zhang, Y., Chen, X., Fan, Z., Liu, Q., et al. (2020). OsSYL2AA, an allele identified by gene-based association, increases style length in rice (Oryza sativa L.). Plant J. 104, 1491–1503. doi: 10.1111/tpj.15013
Duncan, D. B. (1955). Multiple range and multiple F tests. Biometrics 11, 1–42. doi: 10.2307/3001478
Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain with and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. doi: 10.1007/s00122-006-0218-1
Guo, L., Qiu, F., Gandhi, H., Kadaru, S., De Asis, E. J., Zhuang, J., et al. (2017). Genome-wide association study of outcrossing in cytoplasmic male sterile lines of rice. Sci. Rep. 7, 3223. doi: 10.1038/s41598-017-03358-9
Guo, N., Wang, Y., Chen, W., Tang, S., An, R., Wei, X., et al. (2022). Fine mapping and target gene identification of qSE4, a QTL for stigma exsertion rate in rice (Oryza sativa L.). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.959859
Jiang, J., Xu, L., Xiao, M., Hu, C., Zhang, Y., Wang, D., et al. (2021). Genetic analysis and QTLs identification of stigma traits in japonica rice (Oryza sativa L.). Euphytica 217, 82. doi: 10.1007/s10681-021-02813-z
Kato, H., Namai, H. (1987). Floral characteristics and environmental factors for increasing natural outcrossing rate for F1 hybrid seed production of rice Oryza sativa L. Breed. Sci. 37, 318–330. doi: 10.1270/jsbbs1951.37.318
Khumto, S., Sreethong, T., Pusadee, T., Rerkasem, B., Jamjod, S. (2018). Variation of floral traits in Thai rice germplasm (Oryza sativa). Genet. Resour. Crop Ev. 65, 1123–1132. doi: 10.1007/s10722-017-0600-7
Li, P., Feng, F., Zhang, Q., Chao, Y., Gao, G., He, Y. (2014). Genetic mapping and validation of quantitative trait loci for stigma exsertion rate in rice. Mol. Breed. 34, 2131–2138. doi: 10.1007/s11032-014-0168-2
Liu, Y., Fu, D., Kong, D., Ma, X., Zhang, A., Wang, F., et al. (2022). Linkage mapping and association analysis to identify a reliable QTL for stigma exsertion rate in rice. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.982240
Liu, Y., Zhang, A., Wang, F., Kong, D., Li, M., Bi, J., et al. (2019). Fine mapping a quantitative trait locus, qSER-7, that controls stigma exsertion rate in rice (Oryza sativa L.). Rice 12, 46. doi: 10.1186/s12284-019-0304-z
Mahalingam, A., Saraswathi, R., Ramalingam, J., Jayaraj, T. (2013). Genetics of floral traits in cytoplasmic male sterile (CMS) and restorer lines of hybrid rice (Oryza sativa L.). Pak. J. Bot. 45, 1897–1904.
Marathi, B., Jena, K. K. (2015). Floral traits to enhance outcrossing for higher hybrid seed production in rice: present status and future prospects. Euphytica 201, 1–14. doi: 10.1007/s10681-014-1251-9
Marathi, B., Ramos, J., Hechanova, S. L., Oane, R. H., Jena, K. K. (2015). SNP genotyping and characterization of pistil traits revealing a distinct phylogenetic relationship among the species of Oryza. Euphytica 201, 131–148. doi: 10.1007/s10681-014-1213-2
Miyata, M., Yamamoto, T., Komori, T., Nitta, N. (2007). Marker-assisted selection and evaluation of the QTL for stigma exsertion under japonica rice genetic background. Theor. Appl. Genet. 114, 539–548. doi: 10.1007/s00122-006-0454-4
Murray, M. G., Thompson, C. L., Wendel, J. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 432–4326. doi: 10.1093/nar/8.19.4321
Parmar, K. S., Siddiq, E. A., Swaminathan, M. S. (1979). Variation in components of flowering behaviour of rice. Indian J. Genet. Plant Breed. 39, 542–550.
Qian, Q., Guo, L., Smith, S. M., Li, J. (2016). Breeding high-yield superior quality hybrid super rice by rational design. Natl. Sci. Rev. 3, 283–294. doi: 10.1093/nsr/nww006
Rahman, M. H., Zhang, Y., Sun, L., Zhang, K., Rahman, M. S., Wu, W., et al. (2017b). Genetic mapping of quantitative trait loci for the stigma exsertion rate in rice (Oryza sativa L.). J. Integr. Agr. 16, 1423–1431. doi: 10.1016/S2095-3119(16)61540-X
Rahman, M. H., Zhang, Y., Zhang, K., Rahman, M. S., Barman, H. N., Riaz, A., et al. (2017a). Genetic dissection of the major quantitative trait locus (qSE11), and its validation as the major influence on the rate of stigma exsertion in rice (Oryza sativa L.). Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01818
Taillebois, J., Dosmann, J., Cronemberger, H., Paredes, H., CaO, T., Neves, P., et al. (2017). Breeding for outcrossing ability in rice, to enhance seed production for hybrid rice cropping. Rice Research-Open Access 5, 3. doi: 10.4172/2375-4338.1000184
Takano-Kai, N., Doi, K., Yoshimura, A. (2011). GS3 participates in stigma exsertion as well as seed length in rice. Breed. Sci. 61, 244–250. doi: 10.1270/jsbbs.61.244
Tan, Q., Bu, S., Chen, G., Yan, Z., Chang, Z., Zhu, H., et al. (2022b). Reconstruction of the high stigma exsertion rate trait in rice by pyramiding multiple QTLs. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.921700
Tan, Q., Wang, C., Luan, X., Zheng, L., Ni, Y., Yang, W., et al. (2021). Dissection of closely linked QTLs controlling stigma exsertion rate in rice by substitution mapping. Theor. Appl. Genet. 134, 1253–1262. doi: 10.1007/s00122-021-03771-9
Tan, Q., Zhu, H., Liu, H., Ni, Y., Wu, S., Luan, X., et al. (2022a). Fine mapping of QTLs for stigma exsertion rate from Oryza glaberrima by chromosome segment substitution. Rice Sci. 29, 55–66. doi: 10.1016/j.rsci.2021.12.005
Tan, Q., Zou, T., Zheng, M., Ni, Y., Luan, X., Li, X., et al. (2020). Substitution mapping of the major quantitative trait loci controlling stigma exsertion rate from Oryza glumaepatula. Rice 13, 37. doi: 10.1186/s12284-020-00397-1
Uga, Y., Fukuta, Y., Cai, H. W., Iwata, H., Ohsawa, R., Morishima, H., et al. (2003). Mapping QTLs influencing rice floral morphology using recombinant inbred lines derived from a cross between Oryza sativa L. and Oryza rufipogon Griff. Theor. Appl. Genet. 107, 218–226. doi: 10.1007/s00122-003-1227-y
Virmani, S. S., Aquino, R. C., Khush, G. S. (1982). Heterosis breeding in rice (Oryza sativa L.). Theor. Appl. Genet. 63, 373–380. doi: 10.1007/BF00303911
Virmani, S. S., Athwal, D. S. (1973). Genetic variability in floral characteristics influencing outcrossing in Oryza sativa L. Crop Sci. 13, 66–67. doi: 10.2135/cropsci1973.0011183X001300010019x
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. doi: 10.1038/ng.2327
Xu, S., Zheng, Y., Liu, Y., Guo, X., Tan, Y., Qian, Q., et al. (2019). Identification of a major quantitative trait locus and its candidate underlying genetic variation for rice stigma exsertion rate. Crop J. 7, 350–359. doi: 10.1016/j.cj.2018.11.006
Yang, W., Guo, Z., Huang, C., Duan, L., Chen, G., Jiang, N., et al. (2014). Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat. Commun. 5, 5087. doi: 10.1038/ncomms6087
Yan, W., Li, Y., Agrama, H. A., Luo, D., Gao, F., Lu, X., et al. (2009). Association mapping of stigma and spikelet characteristics in rice (Oryza sativa L.). Mol. Breed. 24, 277–292. doi: 10.1007/s11032-009-9290-y
Yuan, L. (2017). Progress in super-hybrid rice breeding. Crop J. 5, 100–102. doi: 10.1016/j.cj.2017.02.001
Zhang, G. (2021). Target chromosome-segment substitution: a way to breeding by design in rice. Crop J. 9, 658–668. doi: 10.1016/j.cj.2021.03.001
Zhang, X., Yang, C., Lin, H., Wang, J., Xue, H. (2021). Rice SPL12 coevolved with GW5 to determine grain shape. Sci. Bull. 66, 2353–2357. doi: 10.1016/j.scib.2021.05.005
Zhou, H., Li, P., Xie, W., Hussain, S., Li, Y., Xia, D., et al. (2017). Genome-wide association analyses reveal the genetic basis of stigma exsertion in rice. Mol. Plant 10, 634–644. doi: 10.1016/j.molp.2017.01.001
Zhu, X., Gou, Y., Heng, Y., Ding, W., Li, Y., Zhou, D., et al. (2023). Targeted manipulation of grain shape genes effectively improves outcrossing rate and hybrid seed production in rice. Plant Biotechnol. J. doi: 10.1111/pbi.13959
Keywords: grain shape, stigma shape, stigma exsertion, pleiotropic effect, outcrossing ability, rice
Citation: Tan Q, Chen S, Gan Z, Lu Q, Yan Z, Chen G, Lin S, Yang W, Zhao J, Ba Y, Zhu H, Bu S, Liu G, Liu Z, Wang S and Zhang G (2023) Grain shape is a factor affecting the stigma exsertion rate in rice. Front. Plant Sci. 14:1087285. doi: 10.3389/fpls.2023.1087285
Received: 02 November 2022; Accepted: 16 January 2023;
Published: 31 January 2023.
Edited by:
Hongwei Wang, Shandong Agricultural University, Taian, ChinaReviewed by:
Xiaoding Ma, Chinese Academy of Agricultural Sciences (CAAS), ChinaJian Sun, Shenyang Agricultural University, China
Copyright © 2023 Tan, Chen, Gan, Lu, Yan, Chen, Lin, Yang, Zhao, Ba, Zhu, Bu, Liu, Liu, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guiquan Zhang, Z3F6aGFuZ0BzY2F1LmVkdS5jbg==; Shaokui Wang, c2hhb2t1aXdhbmdAc2NhdS5lZHUuY24=
 Quanya Tan
Quanya Tan Songliang Chen1,2
Songliang Chen1,2 Qimiao Lu
Qimiao Lu Weifeng Yang
Weifeng Yang Suhong Bu
Suhong Bu Guifu Liu
Guifu Liu Zupei Liu
Zupei Liu Shaokui Wang
Shaokui Wang Guiquan Zhang
Guiquan Zhang