
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci., 21 March 2023
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1086847
This article is part of the Research TopicCRISPR-based genome editing for seed oil improvements in Brassica napus L.View all 6 articles
Rapeseed not only provide considerable amount of edible oil with high nutritional properties but can also be used as a raw material for biofuel production in many industries. It is therefore in high demand to bring genetic changes in order to fulfill the need of human and of industries. Though traditional breeding techniques such as hybridization and mutagenesis remained the top methods for long time to create improved varieties in oilseed rape. Clustered regularly interspaced short palindromic repeats (CRISPR) is becoming one of the most valuable gene editing technologies that allow precise genome engineering, and open new ways for research in plant functional genomics. Though CRISPR has been used in many other crops for genetic improvement it is expected to be an effective tool for genome editing and molecular design in oilseed rape for seed oil improvement. This mini review will discuss and summarize the past and ongoing research and development in rapeseed in terms of seed oil improvement and fatty acid composition using CRISPR technology. In addition, the factors that hinder the efficiency of this tool and how to eliminate those factors will be briefly summarized. The improvement of CRISPR technology for getting better results in oilseed rape will also be considered here. This minireview will open new windows for researchers in Brassica napus oil improvement research and genetic improvement using CRISPR technology.
CRISPR/CRISPR-associated (Cas) systems, which were first discovered to give bacteria and archaea adaptive protection against viral and plasmid infection (Barrangou et al., 2007), have been widely used in recent years to modify the genomic DNA in many organisms. Since their inception, the CRISPR-mediated genome editing systems have promised to significantly improve crop genetics with exceptional features (Zhu et al., 2020). CRISPR-based genome editing technique has unmatched advantages over other genome editing systems such ZFN- zinc finger nuclease system and TALEN- transcription activator-like effector nuclease system (Zhang et al., 2017). The quick genetic studies of intricate regulatory circuits by allowing several sgRNAs encoded in a single CRISPR array to delete different target sites is one of those advantages. In addition to deleting genes, it may also be used to insert specific DNA fragments into target sites and specifically change the transcriptional activity of genes (Li et al., 2013; Lowder et al., 2015). Since its initial application in 2013 for plant gene editing, CRISPR/Cas9 technology has been used extensively in food crop species, including rice, wheat, tomato, etc.; industrial crops, including cichorium intybus, coffee, dandelion, etc.; ornamental crop species, including lily, lotus, rose, etc.; oil crop species, including rapeseed, soybean, sunflower, etc.; fiber crop (cotton); and feed crop (alfalfa) (Ricroch et al., 2017; Liu et al., 2021). These findings demonstrate the critical role CRISPR/Cas9 is playing in crop improvement and plant functional genomics research.
Brassica napus, also known as canola in North America and oilseed rape (OSR) in Europe, the third largest vegetable oil source in the world is grown primarily for its oil-rich seed. The meal produced as a byproduct is utilized as animal feed, primarily in poultry and dairy industries (Wallenhammar et al., 2022). China alone contributes about 20% of the total world production. It is also cultivated in other countries like India, Canada, and the European Union (Hu et al., 2017). For centuries rapeseed farmers have relied on traditional breeding methods in an effort to develop better varieties. Contrarily, B. napus is an allotetraploid species with a number of repetitive sequences. Gene redundancy makes it very inefficient to alter traits using traditional genetic methods like hybridization and random mutation. However, gene editing technologies may assist to speed up the process. A CRISPR-Cas9 strategy was successfully used in B. oleracea for genome editing, indicating that a transfer to oilseed rape might also be possible (Lawrenson et al., 2015). To prevent seed loss by boosting shatter resistance, the CRISPR/Cas9 technology was initially used in rapeseed in 2017 to target two BnALC homologous genes for site-directed mutagenesis (Braatz et al., 2017). The last five years have seen significant agronomical trait advancements in rapeseed using CRISPR/Cas9-mediated genome editing. As of 2022, numerous studies on the effective use of CRISPR/Cas9 technology in Brassica napus breeding have been published (Zhang et al., 2019; Yan et al., 2021; Liu Y. et al., 2022; Tan et al., 2022). Indicating the application of this genome editing technology in rapeseed is becoming even more mature and has been extensively used in the creation of germplasm resources and genetic improvement of oilseed rape. CRISPR/Cas9 technology has become an important tool to analyze gene function and molecular mechanisms in rapeseed. We aim to provide pioneer research and will indicate the path for further application of this technology in seed oil improvement in this mini-review.
The mechanism of CRISPR/Cas9 technology is briefly highlighted in Figure 1. Brassica napus is an allotetraploid species, and the majority of its genes have redundant functions and multiple copies (Chalhoub et al., 2014). It is exceedingly inefficient to alter features by random mutations because of gene redundancy. To enhance one feature, it is frequently required to modify several genes. The Cas9 protein can be directed to specific regions by a variety of single-guide RNAs (sgRNAs), which presents an opportunity to carry out multiple gene editing by expressing Cas9 along with the various sgRNAs (Cong et al., 2013; Lawrenson et al., 2015). Because numerous mutations can be caused simultaneously in polyploid rapeseed, CRISPR/Cas9 technology provides clear advantages. There are currently a growing number of research using CRISPR/Cas9 to knock out multiple copies of genes in B. napus, demonstrating how promising this technology is for understanding of polyploidy in this organism. Whether using a single or multiplex genome editing strategy, the CRISPR/Cas9 system is a useful tool and is frequently employed for crop improvement (Lohani et al., 2020). Multiple gene mutants are typically needed when analyzing the functions of homologous genes and members of gene families with high sequence similarity. Due to their ability to effectively produce mutants with numerous gene mutations, distinct sgRNAs connected in tandem exhibit a significant advantage in multiple gene editing (Xiong et al., 2019). It is also of prime concern that how specific CRISPR/Cas9 technology is when used for targeted gene editing in plants. Typically, any off-target locations should be examined to guarantee that mutation is as specific as desired.
Rapeseed breeding has historically focused on improving the fatty acid (FA) composition and increased oil content. Since the introduction of CRISPR/Cas9 in 2017 in rapeseed, the research on genome editing in this species has expanded (Braatz et al., 2017). After that, a significant number of rapeseed genes were modified, creating a useful germplasm resource for studying the fundamental biology of rapeseed and cultivating novel varieties (Table 1; Figure 2).
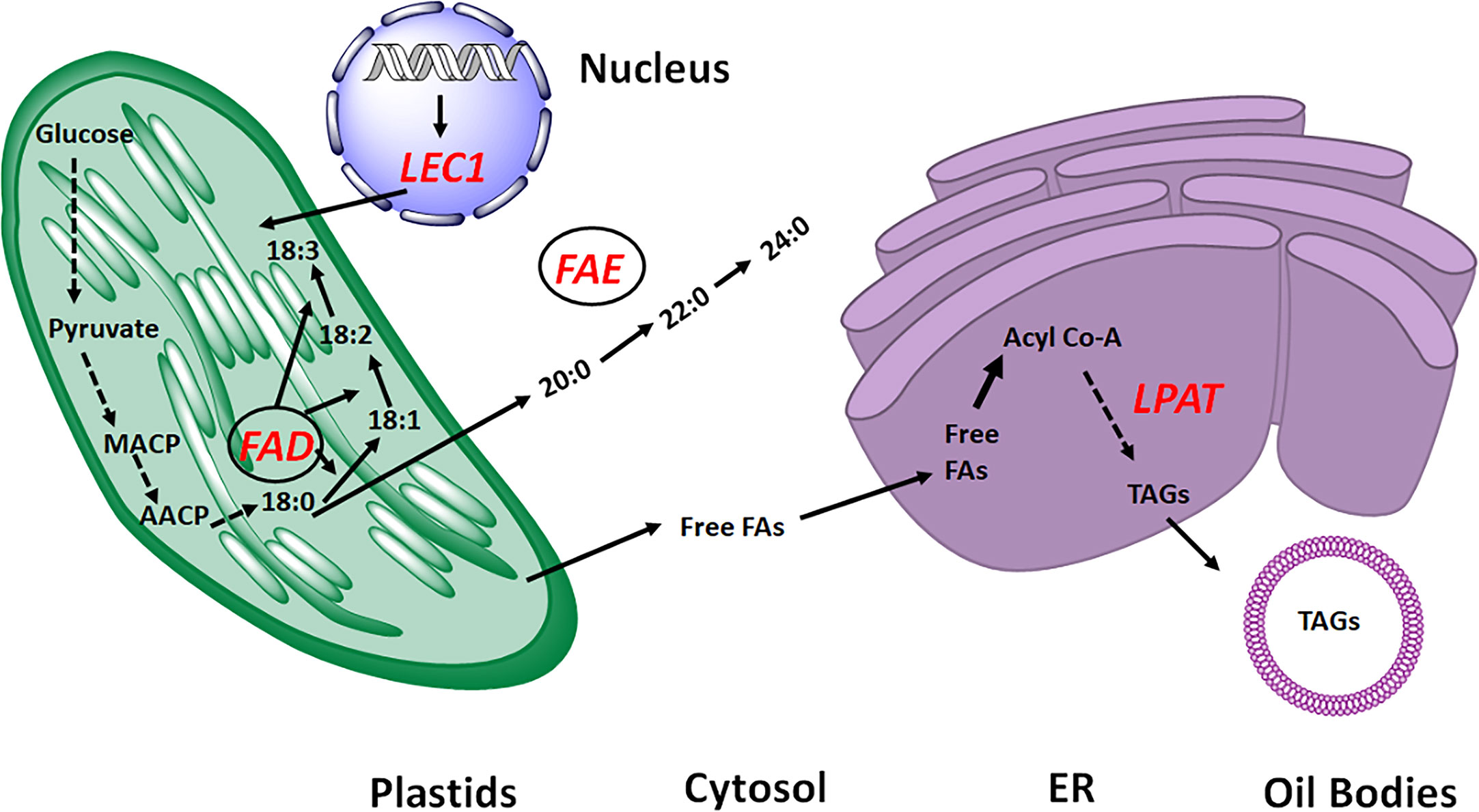
Figure 2 Overview of Fatty acid biosynthesis pathway. The genes targeted for seed oil improvement in Brassica napus by CRISPR technology are highlighted as red. FFAs, free fatty acids; MACP, Malonyle ACP; AACP, Acyle ACP; LPAT, Lysophosphatidic acid acyltransferase; FAE, Fatty acid elongase; LEC1, Leafy cotyledon; FAD, Fatty acid desaturase; ER, Endoplasmic Reticulum.
The Kennedy pathway’s key enzyme, lysophosphatidic acid acyltransferase (LPAT), catalyzes the conversion of FAs into triacylglycerol (TAG), which encourages more oil production (Chapman and Ohlrogge, 2012). Using CRISPR/Cas9, all seven BnLPAT2 homologous copies and four BnLPAT5 homologous copies were eliminated to examine their respective functions in the rapeseed oil biosynthesis. The oil content was reduced to diverse degrees in the knockout mutants of the various copies of BnLPAT2 and BnLPAT5, with a 32% decrease in Bnlpat2 lines, a 29% decrease in Bnlpat5 lines, and a 39% decrease in Bnlpat2/Bnlpat5 double mutant lines. This study confirmed that all BnLPAT copies could change the oil content, and that oil bodies with knockouts of all copies were larger and contained less oil (Zhang et al., 2019).
For instance, by knocking out the Brassica napus Transparent Testa-8 (BnTT8) gene by using CRISPR/Cas9 technology, yellow-seeded mutants with increased seed oil and protein content as well as altered fatty acid (FA) composition were created. Transparent testa 8 (TT8), according to mounting evidence, controls the accumulation of flavonoids in a variety of crops including Medicago truncatula, Lotus species (Li et al., 2016; Escaray et al., 2017). It has been determined that the B. napus transparent testa 2 (BnTT2) allelic variation in the C genome (BnaC) plays a role in rapeseed FA production and seed color development (Zhou et al., 2016). Yellow-seeded mutants were produced using the CRISPR/Cas9 technique by knocking out BnTT8 and BnTT2 homologs (Xie et al., 2020; Zhai et al., 2020), which displayed enhanced seed oil and changed FA composition. Only two yellow-seeded mutants, BnTT8 and BnTT2, have been produced so far. It is highly encouraging to produce additional yellow-seeded mutants by selectively knocking out essential flavonoid pathway genes.
One of the main priorities for genetic manipulation in oilseed rape, is fatty acid content, which influences the nutritional and manufacturing quality of vegetable oil. In oilseed rape, oleic (18:1), linoleic (18:2), and linolenic acid (18:3), synthesis is influenced by the fatty acid desaturase 2 gene (FAD2). The allotetraploid rapeseed contains four copies of BnFAD2. All the copies of BnFAD2 were altered using CRISPR/Cas9-based genome editing technology to produce novel allelic variants in oleic acid and other fatty acid levels. Two targeting sites produced a large number of mutants, and the phenotypic diversity of the mutants was extensively assessed. Compared to the wild type’s 66%, the oleic acid content of the mutant seeds grew dramatically, reaching a maximum of 80%, while the levels of linoleic and linolenic acids declined. Compared to BnFAD2 mutations, BnFAD2.A5 mutations led to more significant changes in the fatty acid profile. Additionally, mixing various BnFAD2 mutant alleles can even drastically widen the variety. It was discovered that the impact of various mutation types at BnFAD2 alleles on oleic acid levels varied, indicating a potential for fatty acid level manipulation by precise mutation at specific region of a gene (Huang et al., 2020). A recent study on BnFAD2 showed that the CRISPR/Cas9-mediated genome editing of the BnFAD2’s double loci (A5 and C5) can precisely edit the targeted genes, increasing the seed’s oleic acid content to a much greater extent than can a single locus-editing technique (Liu H. et al., 2022). Recent findings also reveal that induced mutations in the BnFAD2 genes by CRISPR/Cas9-mediated editing has produce high oleic acid germplasm from the CY2 cultivar (Shi et al., 2022).
Leafy cotyledon1 (LEC1) controls more than 50% of the genes in Arabidopsis involved in plastid FA production (Mu et al., 2008), demonstrating its importance as a transcriptional activator for controlling seed oil content. In Brassica napus, though some signal has also seen in the cytoplasm, BnLEC1 is primarily confined to the nucleus. The two BnLEC1 homologs have comparable expression patterns, with expression rising during seed development and peaking at 23 DAP. Additionally, the high-oil content line had higher BnLEC1 gene expression than the low-oil content. The decreasing and increasing pattern of seed oil content were caused by overexpressing and knocking out BnLEC1, respectively, demonstrating that LEC1 homologs have conserved functions in positively regulating seed oil content (Yan et al., 2021).
Vegetable oils’ processing and edible qualities are influenced by the fatty acid composition, and erucic acid is the fatty acid that may be harmful to the human health. Low erucic acid has therefore always been a breeding characteristic of B. napus. In the synthesis of erucic acid, fatty acid elongase 1 (FAE1) is essential. Three B. napus germplasms with high erucic acid (>30%) and high oil (>50%), the CRISPR/Cas9 technology was employed to produce targeted mutations on two homologous copies of BnFAE1. The double mutation of BnA08.FAE1 and BnC03.FAE1 (a08c03), resulted in nearly zero erucic acid in all BnFAE1-edited germplasms, and the level of oleic acid was increased. This demonstrate that the erucic acid content was significantly reduced by more than ten percent (10%) in the mutant of BnC03.FAE1 (c03). Furthermore, elimination of BnA08.FAE1 or/and BnC03.FAE1 had no appreciable impact on other agronomic parameters but marginally reduced seed oil content. These results showed that crisper based technology can be successfully use in producing low erucic acid germplasms of B. napus, which offers a workable method for upcoming low erucic acid breeding (Liu Y. et al., 2022). In another study, CRISPR/Cas9 mediated genome editing was used to generate novel knockout plants of BnFAE1 genes in order to improve the nutritional quality of rapeseed cultivar CY2. As a result a significant increase in oleic acid (70–80%) content was shown in new lines (Shi et al., 2022).
Off-target effects can result in unwanted outcomes even though they are not as serious for CRISPR/Cas applications in plants, still removing off-target effects is always necessary for genome editing. Off-target effects can be greatly reduced by direct delivery of the protein or RNA of Cas enzymes and sgRNAs, such as by RNP transformation. This is due to the fact that these functional molecules are not integrated into plant genomes; as a result, they can swiftly decay after performing their roles and only have a short life in plant cells, which reduces their ability to target additional areas (Zhang et al., 2021). In addition, utilizing the whole twenty three nucleotide (protospacer plus PAM) sequence as a query in the Brassica genome using BLAST searches in conjunction with other CRISPR/Cas9 online resources can enable the selection of guide RNA with no or a low number of projected off-targets (Lawrenson et al., 2019). According to recent research, precision sgRNA design can be used to combat off-target mutations (Jacinto et al., 2020). To prevent unexpected results, it is crucial to design sgRNAs with a minimum amount of off-target activity. Currently, a number of free online prediction tools, like CRISPOR (http://crispor.org) and CCTop (http://crispr.cos.uni-heidelberg.de) (Stemmer et al., 2017; Concordet and Haeussler, 2018), have been utilized to assist researchers in designing sgRNA in an effort to minimize off-target effects. Additionally, a number of machine learning-based techniques have been created and applied to the prediction of sgRNA activity in agronomy to discover sgRNAs with high on-target activity (Hesami et al., 2021; Niu et al., 2021). The creation of these techniques aids in determining the activity of intended sgRNA in crops and helps prevent unanticipated outcomes. Numerous studies have mentioned that they did not find any off-target activity in possible off-target locations. The off-target editing is insignificant and much less common when gRNAs are carefully generated than naturally occurring variability in plants.
In order to create rapeseed variants with less regulatory restrictions, it is also necessary to develop transgene-free genome editing techniques in this plant. A recent approach for efficiently delivering CRISPR complexes to rapeseed protoplasts using transient transfection has been discovered by researchers (Li et al., 2021). Other rapeseed researchers will benefit greatly from the advice provided by this improved protoplast regeneration methodology. Additionally, by infecting pollen with magnetic nanoparticles and then using that pollen to pollinate plants, transgenic cotton was effectively produced. Other nanoparticles, such as DNA origami and DNA nanostructures, as well as carbon nanotubes have also been successfully tested for the unaided transport of exogenous DNA (Zhao et al., 2017; Demirer et al., 2019; Kwak et al., 2019). If CRISPR/Cas9 for rapeseed genome editing could effectively use delivery by nanomaterials without tissue culture, it would be a quick way to create non-GM rapeseed while evading stringent GM laws. The CRISPR/Cas9 technology will produce additional top-notch rapeseed germplasm resources and provide enormous economic value with future advancement and the progressive resolution of remaining difficulties.
In order to effectively utilize this technology for seed oil improvement in Brassica napus, we seek to present a concise description of the developments in CRISPR-mediated genome editing technology, as well as its applications and potential future prospects. The further advancements in CRISPR/Cas technology, coupled with other breakthroughs, will significantly benefit in the genetic improvement and molecular breeding of Brassica crops in terms of seed oil improvement and other agronomic traits. More gene targets for CRISPR/Cas9-based crop improvement will be discovered with the aid of current multiple omics, such as whole genome sequencing, RNA-seq, and small RNA-seq. The advent of straightforward, precise, and genome-editing tools using CRISPR/Cas9 has revolutionized crop breeding and plant biology research. In addition, new applications for CRISPR/Cas technologies will be created, with the potential to change the tempo and focus of brassica research. Improvements in Brassica napus basic genetic research, the development of innovative delivery methods, boosting public confidence in the security of CRISPR-modified crops, and the development of supporting regulatory frameworks are just a few of the many more successes that are needed in the future. In order to promote functional genomics and aid in the breeding of new Brassica varieties with improved seed oil characteristics, we believe that CRISPR/Cas technologies will be extensively used in this crop species.
With an emphasis on the most recent discoveries in precise genome editing, we discuss current developments in the use of CRISPR/Cas technology in this mini-review. We also provide an outline of how Brassica napus traits for seed oil and fatty acid composition were altered using CRISPR/Cas technology. Finally, we discuss the challenges and future opportunities for the broad use of these technologies in Brassica napus. It will bolster the confidence of researchers studying rapeseed gene function and genetic improvement using genome editing technology to talk about the current state of application for novel, precise genome editing technology in plants as well as a number of potential limitations and technical hurdles.
EA: presented the idea and design the project, and write the manuscript KZ: Revised it critically, Figure creation. All authors contributed to the article and approved the submitted version.
This study was supported by grant from the NATURAL SCIENCE FOUNDATION OF Zhejiang province. Grant number LZ23C020001 to KZ.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor NH declared a past collaboration with the author EA.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315 (5819), 1709–1712. doi: 10.1126/science.1138140
Braatz, J., Harloff, H.-J., Mascher, M., Stein, N., Himmelbach, A., Jung, C. (2017). CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 174 (2), 935–942. doi: 10.1104/pp.17.00426
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-neolithic brassica napus oilseed genome. science 345 (6199), 950–953. doi: 10.1126/science.1253435
Chapman, K. D., Ohlrogge, J. B. (2012). Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 287 (4), 2288–2294. doi: 10.1074/jbc.R111.290072
Concordet, J.-P., Haeussler, M. (2018). CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46 (W1), W242–W2W5. doi: 10.1093/nar/gky354
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339 (6121), 819–823. doi: 10.1126/science.1231143
Demirer, G. S., Zhang, H., Matos, J. L., Goh, N. S., Cunningham, F. J., Sung, Y., et al. (2019). High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat. Nanotechnol 14 (5), 456–464. doi: 10.1038/s41565-019-0382-5
Escaray, F. J., Passeri, V., Perea-García, A., Antonelli, C. J., Damiani, F., Ruiz, O. A., et al. (2017). The R2R3-MYB TT2b and the bHLH TT8 genes are the major regulators of proanthocyanidin biosynthesis in the leaves of lotus species. Planta 246 (2), 243–261. doi: 10.1007/s00425-017-2696-6
Hesami, M., Yoosefzadeh Najafabadi, M., Adamek, K., Torkamaneh, D., Jones, A. M. P. (2021). Synergizing off-target predictions for in silico insights of CENH3 knockout in cannabis through CRISPR/CAS. Molecules 26 (7), 2053. doi: 10.3390/molecules26072053
Hu, Q., Hua, W., Yin, Y., Zhang, X., Liu, L., Shi, J., et al. (2017). Rapeseed research and production in China. Crop J. 5, 127–135. doi: 10.1016/j.cj.2016.06.005
Huang, H., Cui, T., Zhang, L., Yang, Q., Yang, Y., Xie, K., et al. (2020). Modifications of fatty acid profile through targeted mutation at BnaFAD2 gene with CRISPR/Cas9-mediated gene editing in brassica napus. Theor. Appl. Genet. 133 (8), 2401–2411. doi: 10.1007/s00122-020-03607-y
Jacinto, F. V., Link, W., Ferreira, B. I. (2020). CRISPR/Cas9-mediated genome editing: From basic research to translational medicine. J. Cell. Mol. Med. 24 (7), 3766–3778. doi: 10.1111/jcmm.14916
Kwak, S.-Y., Lew, T. T. S., Sweeney, C. J., Koman, V. B., Wong, M. H., Bohmert-Tatarev, K., et al. (2019). Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat. Nanotechnol 14 (5), 447–455. doi: 10.1038/s41565-019-0375-4
Lawrenson, T., Hundleby, P., Harwood, W. (2019). Creating targeted gene knockouts in brassica oleracea using CRISPR/Cas9. Plant Genome Editing CRISPR Syst. 1917, 155–170. doi: 10.1007/978-1-4939-8991-1_12
Lawrenson, T., Shorinola, O., Stacey, N., Li, C., Østergaard, L., Patron, N., et al. (2015). Induction of targeted, heritable mutations in barley and brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 16 (1), 1–13. doi: 10.1186/s13059-015-0826-7
Li, P., Chen, B., Zhang, G., Chen, L., Dong, Q., Wen, J., et al. (2016). Regulation of anthocyanin and proanthocyanidin biosynthesis by m edicago truncatula b HLH transcription factor m t TT 8. New Phytol 210 (3), 905–921. doi: 10.1111/nph.13816
Li, J.-F., Norville, J. E., Aach, J., McCormack, M., Zhang, D., Bush, J., et al. (2013). Multiplex and homologous recombination–mediated genome editing in arabidopsis and nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31 (8), 688–691. doi: 10.1038/nbt.2654
Li, X., Sandgrind, S., Moss, O., Guan, R., Ivarson, E., Wang, E. S., et al. (2021). Efficient protoplast regeneration protocol and CRISPR/Cas9-mediated editing of glucosinolate transporter (GTR) genes in rapeseed (Brassica napus l.). Front. Plant Sci. 1303. doi: 10.3389/fpls.2021.680859
Liu, Y., Du, Z., Lin, S., Li, H., Lu, S., Guo, L., et al. (2022). CRISPR/Cas9-targeted mutagenesis of BnaFAE1 genes confers low-erucic acid in brassica napus. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.848723
Liu, H., Lin, B., Ren, Y., Hao, P., Huang, L., Xue, B., et al. (2022). CRISPR/Cas9-mediated editing of double loci of BnFAD2 increased the seed oleic acid content of rapeseed (Brassica napus l.). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1034215
Liu, Q., Yang, F., Zhang, J., Liu, H., Rahman, S., Islam, S., et al. (2021). Application of CRISPR/Cas9 in crop quality improvement. Int. J. Mol. Sci. 22 (8), 4206. doi: 10.3390/ijms22084206
Lohani, N., Jain, D., Singh, M. B., Bhalla, P. L. (2020). Engineering multiple abiotic stress tolerance in canola, brassica napus. Front. Plant Sci 11, 3. doi: 10.3389/fpls.2020.00003
Lowder, L. G., Zhang, D., Baltes, N. J., Paul, J. W., III, Tang, X., Zheng, X., et al. (2015). A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 169 (2), 971–985. doi: 10.1104/pp.15.00636
Mu, J., Tan, H., Zheng, Q., Fu, F., Liang, Y., Zhang, J., et al. (2008). LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in arabidopsis. Plant Physiol. 148 (2), 1042–1054. doi: 10.1104/pp.108.126342
Niu, M., Lin, Y., Zou, Q. (2021). sgRNACNN: identifying sgRNA on-target activity in four crops using ensembles of convolutional neural networks. Plant Mol. Biol. 105 (4), 483–495. doi: 10.1007/s11103-020-01102-y
Okuzaki, P. A., Ogawa, T, Koizuka, C, Kaneko, K, Inaba, M, Imamura, J, et al. (2018). CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol Biochem 131, 63–69. doi: 10.1016/j.plaphy.2018.04.025
Ricroch, A., Clairand, P., Harwood, W. (2017). Use of CRISPR systems in plant genome editing: toward new opportunities in agriculture. Emerg. Top. Life Sci. 1, 169–182. doi: 10.1042/ETLS20170085
Shi, J., Ni, X., Huang, J., Fu, Y., Wang, T., Yu, H., et al. (2022). CRISPR/Cas9-mediated gene editing of BnFAD2 and BnFAE1 modifies fatty acid profiles in brassica napus. Genes 13 (10), 1681. doi: 10.3390/genes13101681
Stemmer, M., Thumberger, T., del Sol Keyer, M., Wittbrodt, J., Mateo, J. L. (2017). Correction: CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PloS One 12 (4), e0176619. doi: 10.1371/journal.pone.0176619
Tan, Z., Xie, Z., Dai, L., Zhang, Y., Zhao, H., Tang, S., et al. (2022). Genome-and transcriptome-wide association studies reveal the genetic basis and the breeding history of seed glucosinolate content in brassica napus. Plant Biotechnol. J. 20 (1), 211–225. doi: 10.1111/pbi.13707
Wallenhammar, A.-C., Vilvert, E., Bergqvist, S., Olson, Å, Berlin, A. (2022). Scientific evidence of sustainable plant disease protection strategies for oilseed rape (Brassica napus) in Sweden: a systematic map. Environ. Evidence 11 (1), 22. doi: 10.1186/s13750-022-00277-9
Xie, T., Chen, X., Guo, T., Rong, H., Chen, Z., Sun, Q., et al. (2020). Targeted knockout of BnTT2 homologues for yellow-seeded brassica napus with reduced flavonoids and improved fatty acid composition. J. Agric. Food Chem. 68 (20), 5676–5690. doi: 10.1021/acs.jafc.0c01126
Xiong, X., Liu, W., Jiang, J., Xu, L., Huang, L., Cao, J. (2019). Efficient genome editing of brassica campestris based on the CRISPR/Cas9 system. Mol. Genet. Genomics 294 (5), 1251–1261. doi: 10.1007/s00438-019-01564-w
Yan, G., Yu, P., Tian, X., Guo, L., Tu, J., Shen, J., et al. (2021). DELLA proteins BnaA6. RGA and BnaC7. RGA negatively regulate fatty acid biosynthesis by interacting with BnaLEC1s in brassica napus. Plant Biotechnol. J. 19 (10), 2011–2026. doi: 10.1111/pbi.13628
Zhai, Y., Yu, K., Cai, S., Hu, L., Amoo, O., Xu, L., et al. (2020). Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in brassica napus l. Plant Biotechnol. J. 18 (5), 1153–1168. doi: 10.1111/pbi.13281
Zhang, K., Nie, L., Cheng, Q., Yin, Y., Chen, K., Qi, F., et al. (2019). Effective editing for lysophosphatidic acid acyltransferase 2/5 in allotetraploid rapeseed (Brassica napus l.) using CRISPR-Cas9 system. Biotechnol. Biofuels 12 (1), 1–18. doi: 10.1186/s13068-019-1567-8
Zhang, K., Raboanatahiry, N., Zhu, B., Li, M. (2017). Progress in genome editing technology and its application in plants. Front. Plant Sci 8, 177. doi: 10.3389/fpls.2017.00177
Zhang, D., Zhang, Z., Unver, T., Zhang, B. (2021). CRISPR/Cas: A powerful tool for gene function study and crop improvement. J. Adv Res. 29, 207–221. doi: 10.1016/j.jare.2020.10.003
Zhao, X., Meng, Z., Wang, Y., Chen, W., Sun, C., Cui, B., et al. (2017). Pollen magnetofection for genetic modification with magnetic nanoparticles as gene carriers. Nat. Plants 3 (12), 956–964. doi: 10.1038/s41477-017-0063-z
Zhou, L., Li, Y., Hussain, N., Li, Z., Wu, D., Jiang, L. (2016). Allelic variation of BnaC. TT2. a and its association with seed coat color and fatty acids in rapeseed (Brassica napus l.). PloS One 11 (1), e0146661. doi: 10.1371/journal.pone.0146661
Keywords: CRISPR, Brassica napus, seed oil improvement, SgRNA, off-target effects
Citation: Ali E and Zhang K (2023) CRISPR-mediated technology for seed oil improvement in rapeseed: Challenges and future perspectives. Front. Plant Sci. 14:1086847. doi: 10.3389/fpls.2023.1086847
Received: 01 November 2022; Accepted: 10 March 2023;
Published: 21 March 2023.
Edited by:
Nazim Hussain, Independent Researcher, Sharjah, United Arab EmiratesReviewed by:
Kaushik Ghose, Texas Tech University, United StatesCopyright © 2023 Ali and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kewei Zhang, a3d6aGFuZ0B6am51LmVkdS5jbg==; Essa Ali, ZXNzYWFsaUB1cHIuZWR1LnBr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.