
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 25 January 2023
Sec. Plant Pathogen Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1082395
This article is part of the Research Topic Plant Defense Mechanisms in Plant-pathogen Interactions View all 20 articles
Introduction: The clubroot disease caused by Plasmodiophora brassicae (P. brassicae) poses a serious threat to the economic value of cruciferous crops, which is a serious problem to be solved worldwide. Some resistance genes to clubroot disease in Brassica rapa L. ssp pekinensis cause by P. brassicae have been located on different chromosomes. Among them, Rcr1 and Rcr2 were mapped to the common candidate gene Bra019410, but its resistance mechanism is not clear yet.
Methods: In this experiment, the differences of BrRPP1 between the resistant and susceptible material of Chinese cabbage were analyzed by gene cloning and qRT-PCR. The gene function was verified by Arabidopsis homologous mutants. The expression site of BrRPP1 gene in cells was analyzed by subcellular localization. Finally, the candidate interaction protein of BrRPP1 was screened by yeast two-hybrid library.
Results: The results showed that the cDNA sequence, upstream promoter sequence and expression level of BrRPP1 were quite different between the resistant and susceptible material. The resistance investigation found that the Arabidopsis mutant rpp1 was more susceptible to clubroot disease than the wild type, which suggested that the deletion of rpp1 reduces resistance of plant to clubroot disease. Subcellular location analysis confirmed that BrRPP1 was located in the nucleus. The interaction proteins of BrRPP1 screened from cDNA Yeast Library by yeast two-hybrid are mainly related to photosynthesis, cell wall modification, jasmonic acid signal transduction and programmed cell death.
Discussion: BrRPP1 gene contains TIR-NBS-LRR domain and belongs to R gene. The cDNA and promoter sequence of BrRPP1 in resistant varieties was different from that in susceptible varieties led to the significant difference of the gene expression of BrRPP1 between the resistant varieties and the susceptible varieties. The high expression of BrRPP1 gene in resistant varieties enhanced the resistance of Chinese cabbage to P. brassicae, and the interaction proteins of BrRPP1 are mainly related to photosynthesis, cell wall modification, jasmonic acid signal transduction and programmed cell death. These results provide important clues for understanding the mechanism of BrRPP1 in the resistance of B. rapa to P. brassicae.
Chinese cabbage is a cruciferous Brassica crop with both nutritional value and economic value. However, Chinese cabbage is often harmed by P. brassicae, which leads to a soil-borne disease-clubroot disease, and brings huge losses to yield and economy (Ludwig-Müller and Schuller, 2008). The infection cycle of P. brassicae was mainly divided into two stages, infection stage (root hair infected) and disease stage (hypocotyls, cortex and stele infected) (Hirai, 2006). At the later stage of the disease, the overground part of the host plant shows the phenomenon of yellowing and wilting leaves, and the underground part forms swollen nodules, which will affect the root function and reduce the host’s the absorption of water and nutrients (Wang et al., 2016).
So far, there is no effective strategy to control clubroot disease of Chinese cabbage (Diederichsen et al., 2009). Therefore, it has become an important research direction to find more disease resistance genes and reveal their resistant mechanism. In nature, plants are often invaded by various pathogens. Cell wall is the first cell barrier against pressure, which is composed of polysaccharide skeleton, protein and polymer (Lampugnani et al., 2018). Plants can resist a large number of pathogens through the waxy layer and stratum corneum of the cell wall and outer epidermis (Houston et al., 2016). In plants, the key steps of photosynthesis and synthesis of defense related hormones occur in chloroplasts (Lu et al., 2018). In addition, chloroplast is the main production site of reactive oxygen species and nitric oxide, and the site of calcium signal transduction. Photosynthesis can also be affected by defense related hormones and signaling molecules (Fischer et al., 1986; Seemann and Sharkey, 1987; Popova et al., 1996).
When infected by pathogens, the biological stress induced by pathogens will trigger complex signal cascade responses regulated by hormones. The stress and defense related hormone jasmonic acid (JA) is considered as one of the basic components of the pathogen induced response (Kazan and Lyons, 2014). JA defense response is necessary for defense against biotic stress and abiotic stress, and works cooperatively with other hormones (Wasternack, 2007; Wasternack and Hause, 2013). Hormone dependent pathways lead to the expression of defense related genes and the production of antimicrobial secondary metabolites (Glazebrook, 2001).
In recent years, many resistance genes to clubroot disease have been located in Chinese cabbage including Crr1, Crr2 (Suwabe et al., 2003), Crr3 (Saito et al., 2006), Crr4 (Suwabe, 2006), CRa (Matsumoto et al., 1998), CRb (Kato et al., 2013), CRc (Sakamoto et al., 2008), CRk (Suwabe et al., 2003), Rcr1 (Chu et al., 2014), Rcr2 (Huang et al., 2017), PbBa3.1, PbBa3.2 (Chen et al., 2013). It is worth noting that Bra019410 has also been mapped as a candidate resistant (R) gene for many times in the research of Chinese cabbage resistance to P. brassicae infection. For example, Yu et al. (2016) mapped Bra019409 and Bra019410 as candidate genes for the target region of Rcr1 by the combined technology of BSA and KASP. Rcr2 was also fine-located between two SNP sites by the combined technology of BSA and KASP, and finally Bra019410 and Bra019413 were located as the candidate genes of Rcr2 (Huang et al., 2017). In summary, Rcr1 and Rcr2 share a common candidate gene Bra019410. This suggested that Bra019410 should be a key resistant gene in Brassica rapa resisted to clubroot disease.
Our research found Bra019410 is homologous to the Arabidopsis RPP1 (Recognition of Peronospora parasitica1), which is a R gene and encodes the TIR-NBS-LRR protein, so we named it as BrRPP1. RPP1 is also considered as a candidate gene when crops respond to pathogens. For example, RPP1 plays a key role in the resistance of Brassica napus to blackleg disease (caused by the fungal pathogen Leptosphaeria maculans) (Larkan et al., 2014) and Brassica juncea resistance to Sclerotinia sclerotiorum (Rana et al., 2019).
Pathogen ‘recognition’ is considered as the first step of disease resistance, as a signal of general defense response (Botella, 1998). Most R genes in plants belong to NBS-LRR (Nucleotide binding sites rich in leucine repeats) family genes. The multi-domain structure of the NBS-LRR family gene enables it to have multiple functions and is responsible for pathogen identification and signal transduction (Takken et al., 2006). As a R gene, RPP1, participates in host-pathogen interaction and is confirmed to be a downy mildew resistant gene (Holub and Beynon, 1997; Botella, 1998). RPP1 can bind to ATR1 and form a tetramer through the oligomeric interface of the NBS domain. This tetramer has enzymatic activity and can catalyze the hydrolysis of NAD+ and activate downstream EDS1 proteins such as (enhance disease susceptibility 1) and NRG1 (N requirement gene 1) can regulate the death response of cells (Wan et al., 2019).
Although BrRPP1 has been mapped as a candidate gene for resistance to clubroot disease in Chinese cabbage, its resistance function and its resistance mechanism to clubroot is still not clear. In this study, the sequence difference of BrRPP1 between resistant and susceptible material of Chinese cabbage was found, and its interaction proteins were obtained and analyzed, which can provide important information for the study of mechanism of BrRPP1 in the resistance of Chinese cabbage to P. brassicae.
The materials used in the experiment were resistant variety ‘SN205’ and susceptible variety ‘SN742’ of Chinese cabbage, physiological race no.4 of P. brassicae, Colombian wild-type ‘WT’ of Arabidopsis thaliana and tobacco ‘Nicotiana Benthamiana’. All the above experimental materials were preserved by the Vegetable Genetics and Breeding Laboratory of Shenyang Agricultural University. The Arabidopsis mutant rpp1 (‘SALK_065253’) was purchased from ABRC (https://abrc.osu.edu/).
The reference sequence of BrRPP1 was obtained by searching from Brassica database (BRAD (brassicadb.cn)). Then the sequence validity was checked by Nucleic acid BLAST (http://www.ncbi.nlm.nih.gov/blast/) in the National Center of Biotechnology Information (NCBI). The gene conservative structure domain was predicted by NCBI protein conserved domain database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
To obtain accurate cDNA sequences of BrRPP1 from the ‘SN205’ and ‘SN742’, three segmented primers (BrRPP1-1, BrRPP1-2 and BrRPP1-3) with Eco3II sites were designed using Primer 5 software (Table 1). The three corrected amplified fragments from the cDNA of ‘SN205’ or ‘SN742’ were digested by Eco3II (Beijing NEB, China) respectively, and then ligated to pBWA(V)BS vector linearized by Eco3II using T4 ligase (Tiangen, Beijing, China). The connecting products pBWA(V)BS-SN205-BrRPP1 and pBWA(V)BS-SN742-BrRPP1 were transformed into the competent cells of Trans10 E. coli respectively. The correctness of recombinant clones were detected with primers (F: cagtGGTCCacacatgatttcgatcgattttttgaaaaaaaaa and R: cagtgGTCTCatacataacatgaggaggaggaggtttc) and the colony with the correct band position was sequenced (Sangon Inc, Shanghai, China). The three-dimensional structure of BrRPP1 protein in ‘SN205’ and ‘SN742’ was predicted using SWISS-MODEL online tool (https://swissmodel.expasy.org/interactive).
Promoter primers (BrRPP1pro-F: CTGTTCTCGTATCCTTCAC and BrRPP1pro-R: CTCATCTGCTTTCTCTTTTTC) were designed according the upstream 2 kb sequence of BrRPP1 from Brassica database (brassicadb.cn). The DNA of ‘SN205’ and ‘SN742’ were used as templates respectively to amplify the promoter sequences of BrRPP1. Promoter elements were analyzed using online software PlantCARE (http://bioinformatics.psb.ugent.be/webtools/PlantCARE/htmL/).
cDNA was obtained from ‘SN742’ roots on the 14th day after inoculation of P. brassicae (at this time, P. brassicae spores were found in root hairs under the compound microscope) and on the 42nd day after inoculation (at this time, obvious swelling appeared in the roots), respectively. In addition, the roots cDNA of uninoculated ‘SN742’ and ‘SN205’ at above same stage was used as control and then to analyze the difference of BrRPP1 expression between resistance and susceptible materials. Using Quantstudio6 (ThermoFisher, Waltham, MA, USA) and the fluorescence quantification kit UltraSYBR Mixture (Low ROX, Cwbio, Jiaosu, China), we analyzed the expression pattern of BrRPP1 in the roots of Chinese cabbage at two sampling stages after infection by P. brassicae by qRT-PCR. The specific primers were designed, and the BrActin gene (Gao et al., 2020) was used as the internal control (primers are shown in Table S1). The relative expression of BrRPP1 was analyzed using the 2−ΔΔCt method (Livak and Schmittgen, 2001) and SigmaPlot 12.5 (Systat Software, Inc., San Jose, CA, USA). Three replications were performed for each different treatment. The difference significance test was analyzed using the program SPSS v.26 (IBM, Armonk, NY).
Seeds of Arabidopsis mutant rpp1 and wild-type ‘WT’ were vernalized at 4 °C low-temperature for 3-5 days and then evenly sowed in nutritious bowl. When seedlings grew to 10 true leaves, DNAs of rpp1 and ‘WT’ leaves were extracted by modified CTAB method (Springer, 2010). The ‘three-primer’ PCR was performed to select the homozygous mutants. The detection primers (LP: ACCATTCATTGTTCCTTGCAG; RP: ATGATAATGAAGGACCCCTCC; LB: ATTTTGCCGATTTCGGAAC) were obtained from the SIGnAL website (http://signal.salk.edu/tdnaprimers.2.htmL). The homozygous mutants were further cultured until offsprings were obtain.
The P. brassicae suspension with a concentration of 107/mL was prepared according to Lv et al. (2021). The seeds of the rpp1 and ‘WT’ were sterilized and seeded in a petri dish containing moist filter paper to accelerate germination at 25 °C. After about 24 hours in darkness, the culture dishes were transfered into a light incubator at 25 °C, with 16 hours of light and 60% humidity. When two cotyledons were extended, P. brassicae suspension were sprayed on the roots of mutant rpp1 and ‘WT’ for inoculation. Five randomly selected plants were observed at 24 h intervals. The roots of seedlings were cut into 1 cm sections, dyed with 1% toluidine blue for 15 min, and decolorized with sterile water for 5 min, and then observed under a composite microscope (Eclipse 80i; Nikon, Tokyo, Japan).
To construct the 35S::pGPTVII.GFP-BrRPP1 expression vector, the BrRPP1 coding sequence with the termination codon removed was amplified from pBWA(V)BS-SN205-BrRPP1 using BrRPP1-GFP-F:CGCCACTAGTGGATCCatgagatttcgatcgtttttgaaaga; BrRPP1-GFP -R:GAGCGGTACCCTCGAGacatgagggagccaaggtttcc, and inserted into the pGPTVII.GFP vector using NovoRec homologous recombinase (Novprotein, Shanghai, China).
The plasmid of 35S::pGPTVII.GFP-BrRPP1 and 35S::pGPTVII.GFP (as a control) was introduced into Agrobacterium strain GV3101 respectively. The strains were injected into 4-week-old tobacco leaves as described previously (Sainsbury et al., 2009). After 48 hours in darkness, the tobacco leaves were stained with 4,6-diamidino-2-phenylindole (DAPI, Coolaber, Beijing, China), and observed under a Confocal laser microscope (TCS SP8-SE, Leica, Wetzlar, Germany).
To construct pGBKT7-BrRPP1 bait vector, the BrRPP1 coding sequence with the termination codon removed was amplified from pBWA(V)BS-SN205-BrRPP1 using BrRPP1-BD-F: CGCACTAGTGGATCCatgagatttcgatcgtttttgaaaga and BrRPP1-BD-R: AAGGAAAAAAGCGGCCGCacatgagggagccaaggtttcc, and the products and PGBKT7-T7 were respectively digested with BamHI and NotI (NEB, Beijing, China) and connected. The obtained pGBKT7-BrRPP1 bait vector was transferred into the freshly prepared competent cells of Y2H yeast. The self-activation and toxicity of positive colonies and yeast library screening was carried out by mating hybridization according to Xu et al. (2020). Functional annotation of candidate interaction proteins of BrRPP1 obtained from the screening library were analyzed by NCBI and Brassica database, and the expression pattern of those proteins with high detection rate was analyzed by qRT-PCR (primers are shown in Table S1).
The reference sequence of BrRPP1 gene (Bra019410) obtained from Brassica database contains an open reading frame (ORF) with a length of 3471 bp, which is located on chromosome A03. The accession number of Bra019410 in the NCBI database is XM_009138983.3. The conservative structure domain prediction of BrRPP1 showed that there are a TIR conserved domain in the region of 85-259 aa, and a LRR conserved domain in the region of 690-929 aa (Figure 1A). Therefore, BrRPP1 is a TIR-NBS-LRR-like protein.
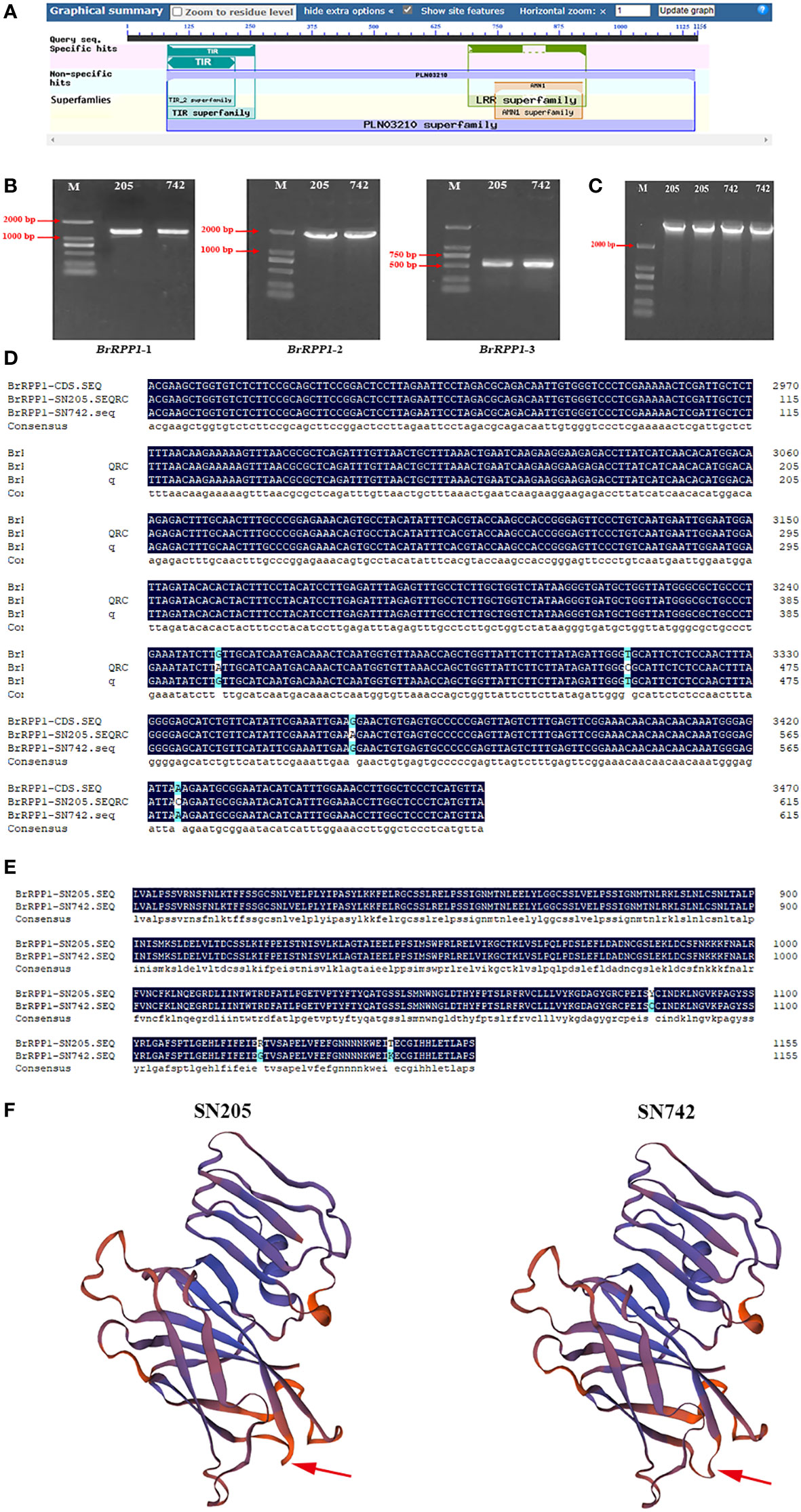
Figure 1 Acquisition and analysis of BrRPP1 gene sequence. (A) Conservative domain prediction of BrRPP1. (B) BrRPP1 fragment amplified from ‘SN205’ and ‘SN742’ with three segmented primers BrRPP1-1, BrRPP1-2 and BrRPP1-3. (C) The cDNA full-length of BrRPP1 in ‘SN205’ and ‘SN742’. (D) cDNA sequence alignment of BrRPP1 in different regions between ‘SN205’ and ‘SN742’. (E) Amino acid sequences alignment of BrRPP1 in different regions between ‘SN205’ and ‘SN742’. (F) Protein structure prediction of BrRPP1 in ‘SN205’ and ‘SN742’. The arrow points to the different positions.
The lengths of products amplified by BrRPP1-1, BrRPP1-2 and BrRPP1-3 were 1141 bp, 1654 bp and 617 bp, respectively, and the results were consistent in ‘SN205’ and ‘SN742’ (Figure 1B). The ligation products of the three cDNA fragments were both 3471 bp in ‘SN205’ and ‘SN742’ (Figure 1C). Comparing the cDNA sequences of BrRPP1 from ‘SN205’ and ‘SN742’ with the reference genome, it was found that the sequence in ‘SN742’ is consistent with the reference sequence. However, it is cDNA different between ‘SN205’ and reference sequence, with four single nucleotide polymorphisms (SNPs) (G to A, T to C, G to A and A to C) in the 3450-3471 bp region (Figure 1D). The change of base leads to the change of amino acid sequence and three-dimensional structure of BrRPP1 protein (Figures 1E, F).
The upstream 1167 bp promoter region of coding sequence of BrRPP1 were cloned from ‘SN205’ and ‘SN742’. Sequences alignment found that there are two additional insert fragments (IF-1, IF-2) and multiple SNPs in the promoter region of BrRPP1 in the ‘SN205’ compared with in the ‘SN742’. The prediction of promoter elements found that the BrRPP1 promoter region amplified form ‘SN742’ or ‘SN205’ contains basic elements, light-responsive elements and corresponding elements of biotic and abiotic stress (Table 2). However, the number of cis-acting elements was different in the two materials. Analyzing the difference of biotic and abiotic stress elements in BrRPP1 promoter between ‘SN205’ and ‘SN742’, it was found that there was a MYC transcription factor binding site in one of inserted segments (IF-1) in BrRPP1 promoter of ‘SN205’, and a MYB binding site was truncated by the inserted segment IF-1. In addition, compared with BrRPP1 promoter of ‘SN742’, an ARE element was missing and an ARBE element was adding in that of ‘SN205’ (Figure 2).
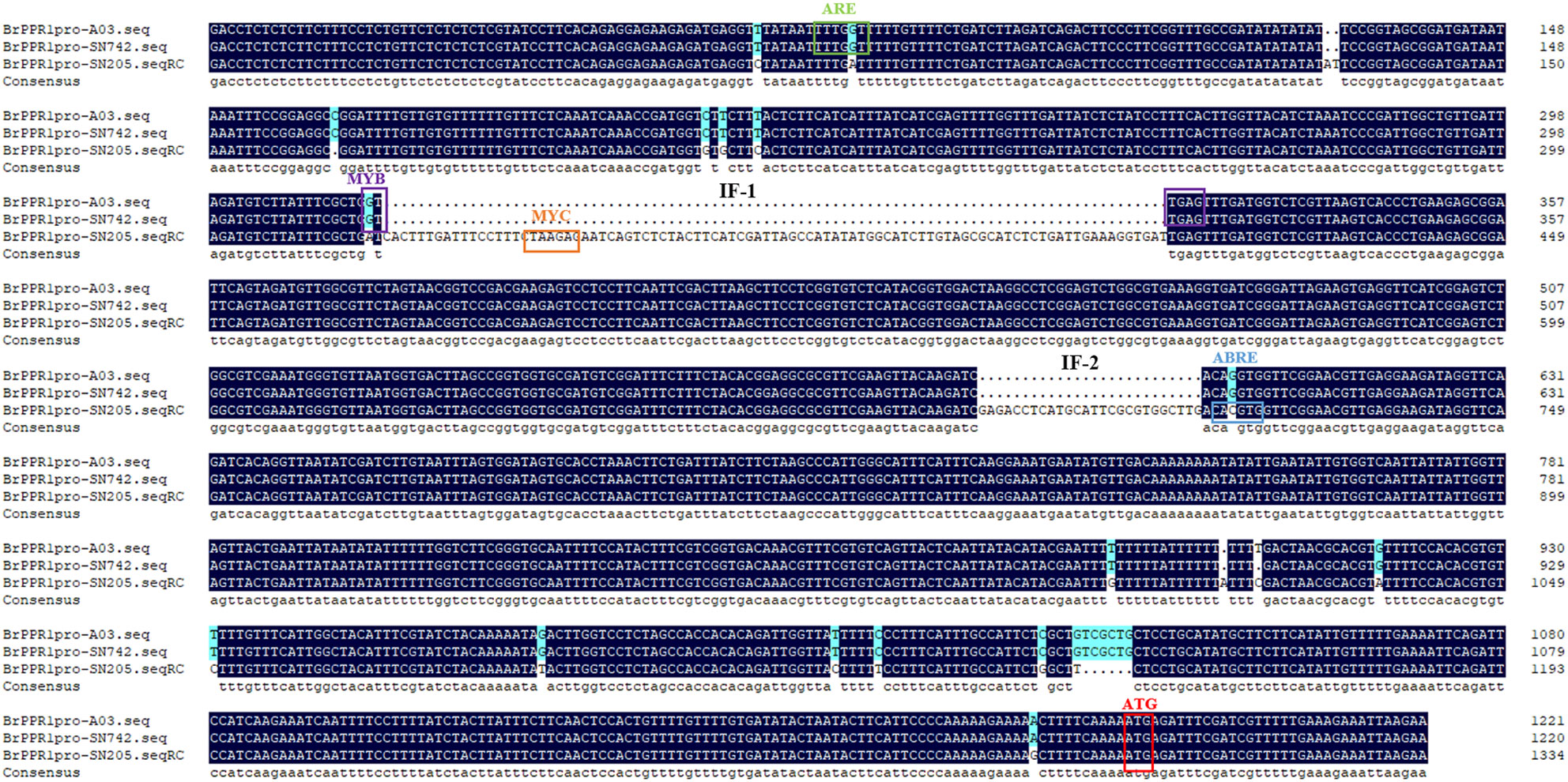
Figure 2 Promoter sequence and element analysis of BrRPP1 in resistance and susceptible materials of Chinese cabbage.
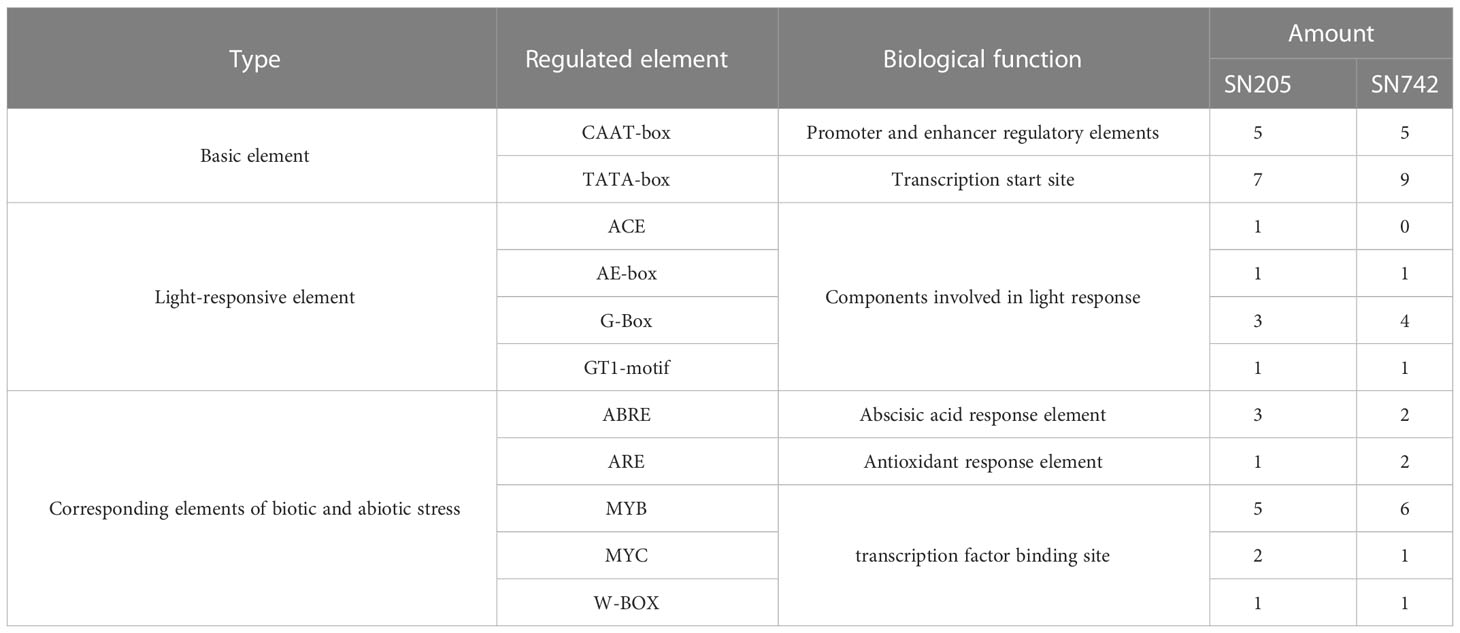
Table 2 Promoter element analysis of BrRPP1 in resistance and susceptible materials of Chinese cabbage.
In order to analyze the expression pattern of BrRPP1 gene, qRT-PCR showed that there was no significant difference between the expression level of BrRPP1 in ‘SN742’ roots of 14th and 42nd days after inoculation of P. brassicae and that in uninfected roots of Chinese cabbage (Figure 3A). However, the comparative analysis of the expression of BrRPP1 between ‘SN742’ and ‘SN205’ showed that it was significantly higher in ‘SN205’ than in ‘SN742’ at both sampling dates (Figure 3B).
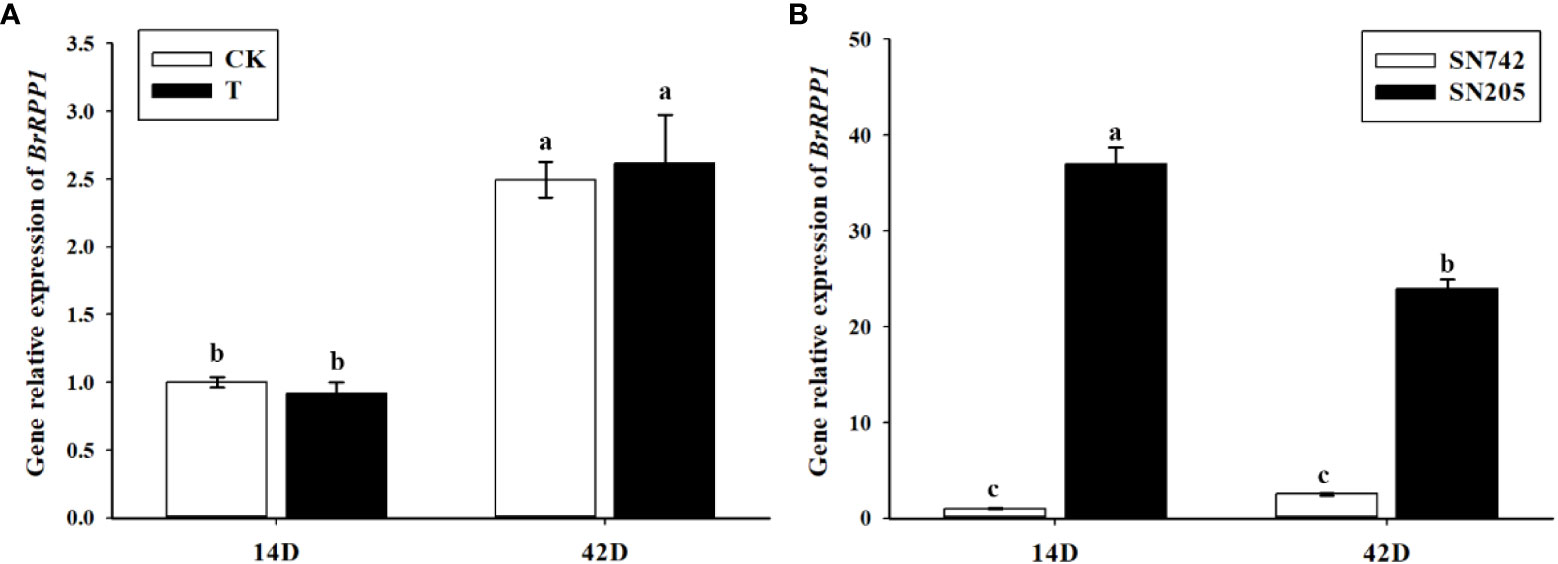
Figure 3 The analysis of relative expression of BrRPP1 gene. (A) The expression difference of BrRPP1 between uninoculated and inoculated roots with P. brassicae. 14D, the 14th day after inoculation of P. brassicae. 42D, the 42th day after inoculation of P. brassicae. CK, uninoculated control roots. T, roots treated with P. brassicae. (B) The expression of BrRPP1 in roots of ‘SN742’ and ‘SN205’ uninoculated with P. brassicae. Data represent the mean ± standard deviation (n = 3); letters indicate significant differences at p ≤ 0.05, according to Duncan’s multiple range test.
Identification of Arabidopsis mutants by ‘three-primers’ showed that a fragment of about 1280 bp was amplified in No.1 and No.3 by LP+RP, and a fragment of about 720 bp was amplified in No.2, 3, 4, 5 and 6 by LB + RP (Figure 4A). In theory, LP and RP are located on both ends of T-DNA insertion site, and LB is located at T-DNA insertion site, the length of LP + RP amplification products should be 1280 bp, and the length of LB + RP amplification products should be 594-894 bp.
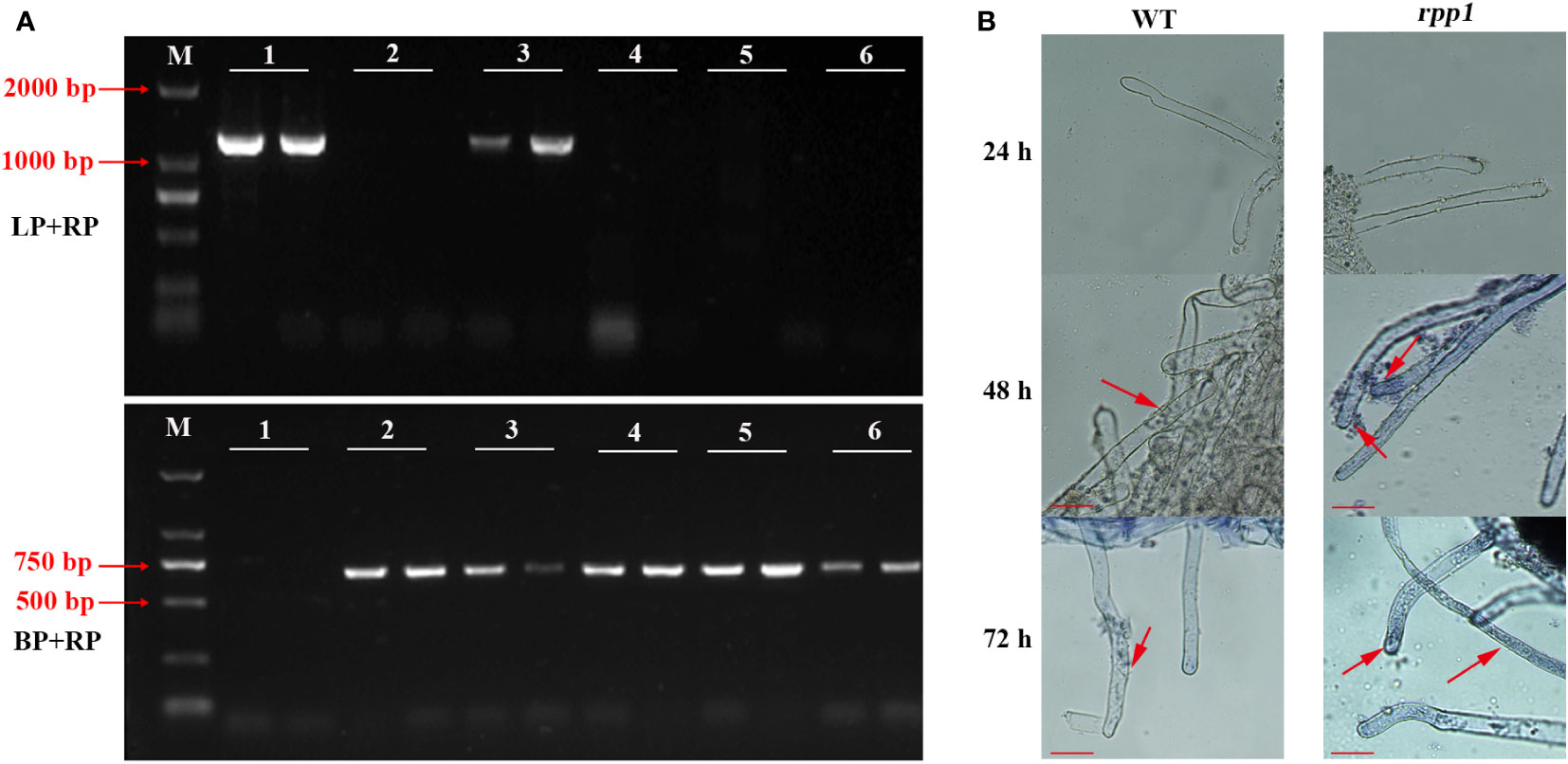
Figure 4 Disease resistance identification to P. brassicae in Arabidopsis mutant rpp1. (A) The identification results of Arabidopsis mutant rpp1 by ‘three-primers’. M, D2000 bp marker; 1, wild type of Arabidopsis with two replicates; 2-6, different plants of Arabidopsis mutant rpp1 with two replicates. (B) P. brassicae infection observation for wild-type (WT) and Arabidopsis mutant rpp1. The red arrow, the position of the spore or sporangium. Scale bar=50 μm.
There was only a band of 1280 bp in No. 1, so it was a wild-type mutant without T-DNA insertion, while two bands of 1280 bp and 720 bp was amplified in No.3, so it was a heterozygous mutant with unilateral T-DNA insertion. There was only a band of 720 bp in No. 2, 4 and 6, so they were homozygous mutants.
After infection with P. brassicae, the roots of ‘rpp1’ and ‘WT’ were both not be infected within 24 h. After 48 h, primary spores appeared in the root hairs of the ‘rpp1’ mutants, while zoospores only attached to the surface of root hair of ‘WT’. After 72 h, the root hair of ‘rpp1’ was more severely infected by spores, and the primary spores appeared in the root hairs of ‘WT’ (Figure 4B). The results indicated that the absence of RPP1 could accelerate the infection process of P. brassicae.
After A. tumefaciens infection solution containing pGPTVII.GFP-BrRPP1 plasmid was injected into tobacco leaves, observation under Confocal laser microscope found that pGPTVII.GFP-BrRPP1 carrier only produced fluorescence signal in the nucleus, and the fluorescence signal overlapped with DAPI nuclear dye (Figure 5).
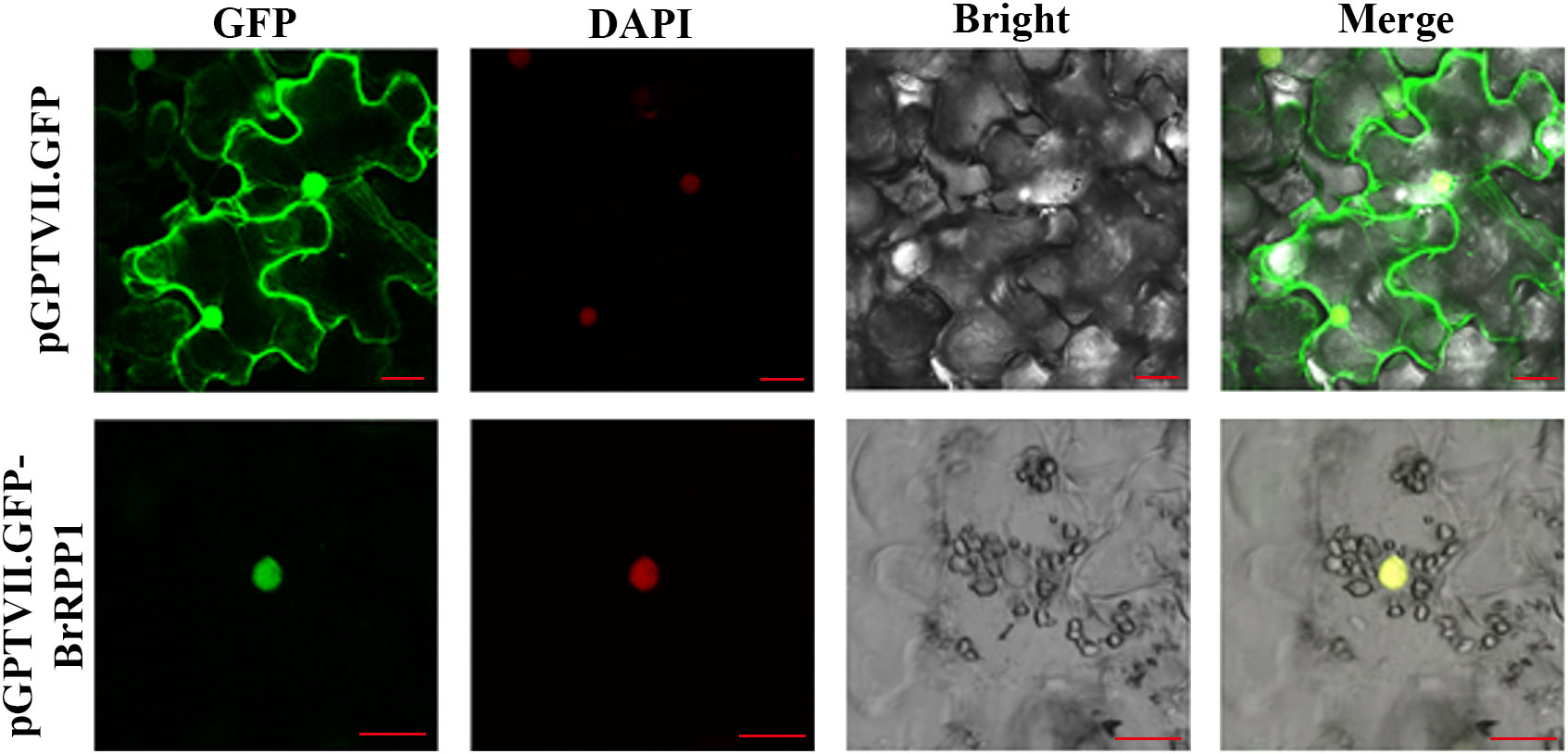
Figure 5 Subcellular localization of BrRPP1. GFP, GFP fluorescence. DAPI, DAPI fluorescence. Bright, brightfield image. Merge, merged pictures of GFP, bright, and DAPI fluorescence. Three views were selected for electron microscope observation. Scale bar=20 μm.
The toxicity test results of pGBKT7-BrRPP1 showed that the yeast strains containing pGBKT7-BrRPP1 could grow normally on SD/-Trp medium plate, and there was no significant difference in colony size and number between the strains containing pGBKT7-BrRPP1 and those containing pGBKT7-T, which indicated that BrRPP1 is not toxic to yeast growth (Figure 6A).
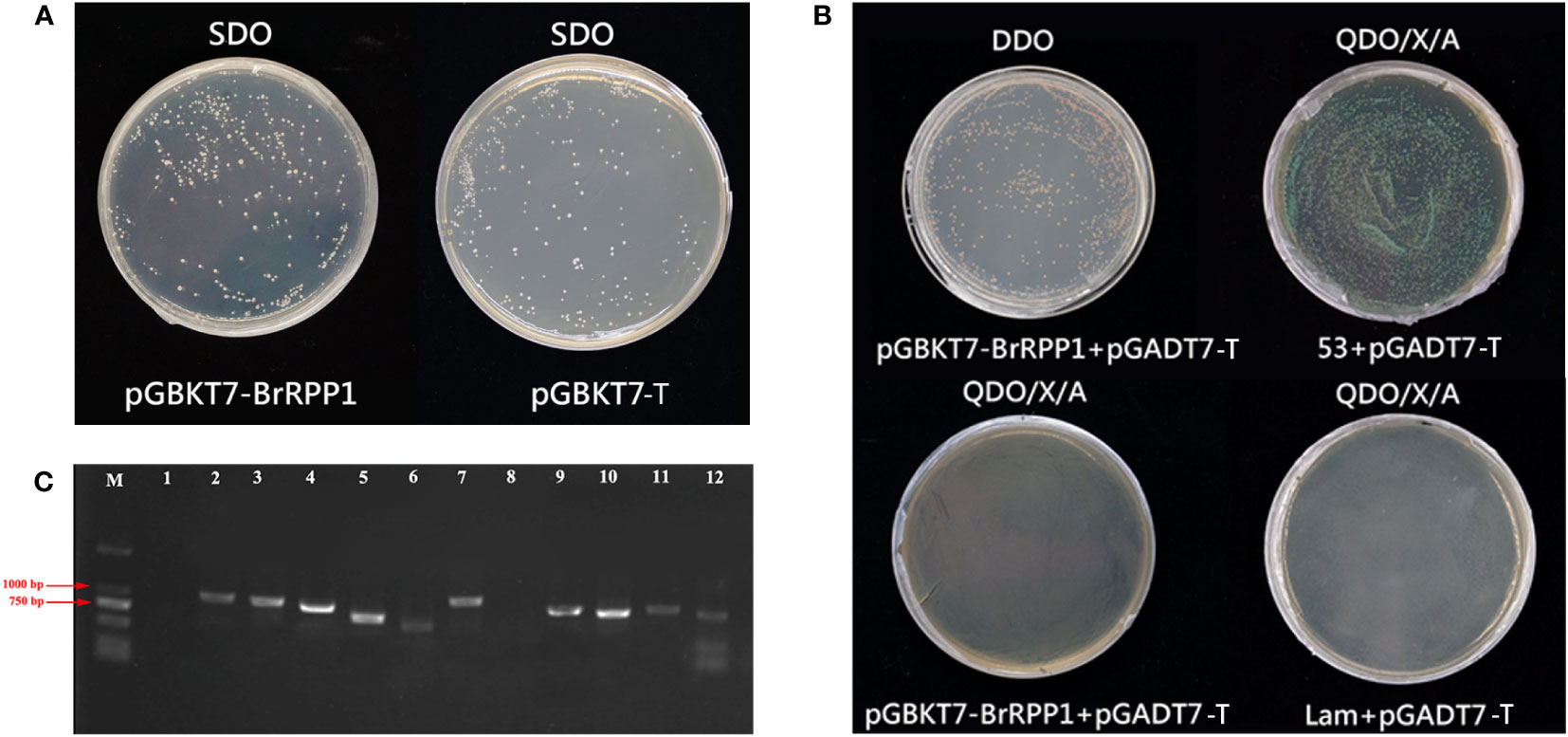
Figure 6 Interaction proteins screening of pGBKT7-BrRPP1. (A) Toxicity test of pGBKT7- BrRPP1. (B) Self-activation detection of pGBKT7- BrRPP1. (C) PCR identification of positive interaction proteins of BrRPP1 (part).
To avoid the false positive interaction proteins caused by the self-activation of pGBKT7-BrRPP1, self-activation analysis found that the yeast colonies co-transfected with pGBKT7-BrRPP1 and pGADT7-T could grow normally on DDO deficient medium, indicating that pGBKT7-BrRPP1 and pGADT7-T were successfully co-transfected into yeast cells. On the QDO/X/A medium, the positive control group (pGBKT7-53 + pGADT7-T) grew normally, activated the reporter genes and showed blue colonies. However, the negative control group (pGBKT7-Lam + pGADT7-T) could not grow on the QDO/X/A medium. The experimental group (pGBKT7-BrRPP1 + pGADT7-T) also could not grow on the QDO/X/A medium, indicating that the bait vector pGBKT7-BrRPP1 had no self-activation effect and could be used for yeast two-hybrid library screening (Figure 6B). Through Y2H library screening, 35 positive colonies were screened on QDO/X/A medium, which may interact with BrRPP1. The length of the inserted fragments of these clones ranged from 750-1000 bp (Figure 6C).
Functional analysis of these candidate interacting proteins shows that they participate in six pathways, including programmed cell death, JA pathway, plant cell wall synthesis, plant photosynthesis, nitrogen metabolism and transcriptional regulation. XM_009125371.3 (cytochrome c6, Cytc) involved in programmed cell death, the detection rate was 17.14%; XM_009112750.3 (Phospholipase A1, PLA1) involved in the synthesis of jasmonic acid (JA), the detection rate was 14.28%; XM_013801607.2 (galacturonosyltransferase 8-like, GT8) involved in cell wall modification, the detection rate was 11.42%; XM_009151660.3 (HNH endonuclease, HNH), XM_018654018.2 (phosphoribulokinase, PRK) and XM_009132313.1 (oxygen enhancing protein, OEE) involved in photosynthesis, the detection rate were 17.14%, 14.28% and 11.42% respectively. The detection rate of other candidate proteins is lower than 3%. In addition, there were two other unknown functional proteins (Table 3).
In order to further analyze whether the expression of these candidate interacting proteins were influenced by P. brassicae infection, qRT-PCR results showed that the gene expression of all of these proteins were significantly changed after infection with P. brassicae. After inoculation treatment by P. brassicae, XM_009132313.1 and XM_018654018.2 were only significantly down-regulated expressed at the early stage of infection, and the expression of XM_009125371.3 and XM_009151660.3 were significantly up-regulated at both sampling dates, and XM_013801607.2 was significantly down-regulated at both sampling dates, while the expression of XM_009112750.3 was significantly down-regulated on the 14th day, and significantly up-regulated on the 42nd day (Figure 7A). In addition, expression differences of these genes were also happened between ‘SN205’ and ‘SN742’ (Figure 7B).
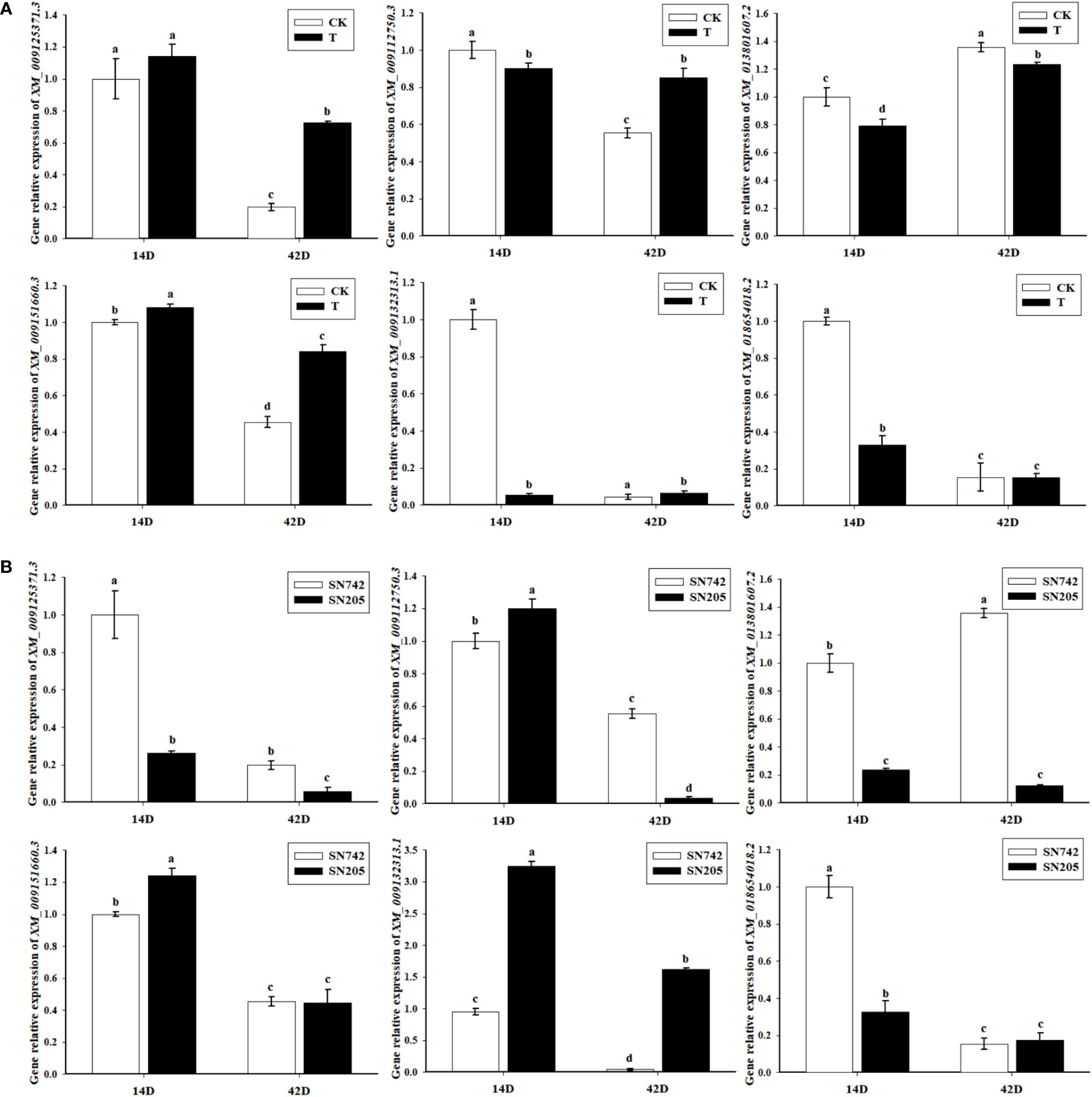
Figure 7 Gene expression pattern analysis of the candidate interacting protein of BrRPP1. (A) The expression difference of the candidate interacting proteins of BrRPP1 between uninoculated and inoculated roots with P. brassicae. 14D, the 14th day after inoculation of P. brassicae. 42D, the 42th day after inoculation of P. brassicae. CK, uninoculated control roots. T, roots treated with P. brassicae. (B) The expression of the candidate interacting proteins of BrRPP1 in roots of ‘SN742’ and ‘SN205’ uninoculated with P. brassicae. Data represent the mean ± standard deviation (n = 3); letters indicate significant differences at p ≤ 0.05, according to Duncan’s multiple range test.
The clubroot disease caused by the P. brassicae has seriously affected the economic value of cruciferous crops. At present, many R genes to clubroot disease have been mapped in Chinese cabbage. As a common candidate gene for Rcr1 and Rcr2 (Chu et al., 2014; Yu et al., 2016; Huang et al., 2017), Bra019410 was a highly homology protein with RPP1 in Arabidopsis, so it was named BrRPP1. Through the analysis of the conservative domain, we found that the protein contains TIR and LRR and other conservative domains, belonging to TIR-NBS-LRR proteins (Figure 1A). The LRR domain of TIR-NBS-LRR has the function of identifying pathogens and can trigger the ETI resistance of plants (Ghelder and Esmenjaud, 2016). Previous studies have confirmed the LRR domain of RPP1-NdA (allele of RPP1) of hyaloperospora Arabidopsidis can recognize the effector protein ATR1 of the oomycete pathogen, but the LRR domain alone cannot activate the hypersensitivity of plants, which indicates that other domains of RPP1 have signal transduction functions (Steinbrenner, 2015).
After cloning and sequencing of cDNA sequence of BrRPP1 in ‘SN205’ and ‘SN742’, it was found that the cDNA sequence was different between ‘SN205’ and ‘SN742’, and the difference mainly occured after the LRR conservative domain (Figure 1). In addition, sequence and elements number of BrRPP1 promoter were significantly differences between resistant and susceptible materials (Figure 2). The analysis of gene expression pattern showed that the expression of BrRPP1 was not affected by the infection of P. brassicae, while the expression of BrRPP1 in ‘SN205’ was significantly higher than that in ‘SN742’ (Figure 3). Therefore, it can be inferred that the sequence and the expression difference of BrRPP1 leads to resistance difference between resistant and susceptible materials.
Arabidopsis thaliana can also be infected by P. brassicae, so it can be used as a useful model host for studying the clubroot disease (Koch et al., 1991; Mithen and Magrath, 1992; Ludwig-Müller et al., 2009). In order to confirm the disease resistance of BrRPP1, the disease resistance of the Arabidopsis mutant rpp1 was identified. The results showed that the deficiency of RPP1 accelerated the infection of P. brassicae, which indicated that BrRPP1 should play a role in disease resistance to P. brassicae (Figure 4).
In order to analyze the expression site of BrRPP1 in cells, we constructed the expression vector pGPTVII.GFP-BrRPP1 for subcellular localization analysis. In previous subcellular localization studies, the RPP1-WsA fragment was located in the endoplasmic reticulum and Golgi apparatus, RPP1-WsB fragments are located in the plasma membrane, and RPP4 and RPP8 fragments are located in the nucleus (Takemoto et al., 2012). However, the localization result of the N-terminal fragment alone cannot completely determine the expression pattern of the full-length protein. GhTIR-NBS-LRR1 containing the structure of TIR-NBS-LRR was a homologous protein of BrRPP1, which localized in the nucleus (Li et al., 2019). This result provided a reference for our prediction results. In this experiment, Confocal laser microscope observation showed that pGPTVII.GFP-BrRPP1 appeared fluorescence signal in the nucleus, which confirmed BrRPP1 was expressed in the nucleus (Figure 5). Therefore, yeast two-hybrid screening can be carried out using nuclear library.
The RPP1 (Recognition of Peronospora parasitica1) was first found in Arabidopsis ‘Wassilewskija’ (Botella, 1998). The current research about RPP1 is mostly in its interaction with effector proteins, intramolecular interactions and the process of inducing plant cell death. The Arabidopsis RPP1 is a typical TIR-NBS-LRR gene, RPP1 has been shown to be able to directly recognize pathogens in Arabidopsis thaliana, and it also has an indirect recognition function by recognizing the target proteins of pathogenic effector proteins (Renier et al., 2008). TIR domain in RPP1 has been confirmed to have a self-binding effect, which plays an important role in inducing cell death (Wan et al., 2019). In this study, 35 interacting proteins of BrRPP1 were screened by yeast-two hybrid. Among them, Cytochrome C (Cytc) is widely present in the mitochondria of plants and organisms, and is considered to be related to the programmed cell death. The release of Cytc from mitochondria is common in the process of programmed cell death in plant cells (Vacca et al., 2006; Qi et al., 2018; Matilla, 2021). Wang et al. (2016) also found that rice NBS-LRR disease-resistant protein Pik-h interacts with Cytochrome c oxidase assembly protein (COX11). In our study, it was found that the expression of Cytc was significantly up-regulated after infection by P. brassicae, and its expression was significantly different between resistant and susceptible materials (Figure 7). Therefore, we can speculate that BrRPP1 as a TIR-NBS-LRR gene may regulate the programmed death of plant cells by promoting the release of cytochromes.
Jasmonic acid (JA) defense response is necessary for defense against biotic stress (Wasternack, 2007). AtPLAs family is involved in the transduction of auxin and pathogen signals (Holk et al., 2002). PLA2 can be strongly induced when tobacco is infected by bacterium Erwinia carovora or the fungus Botrytis cinerea, and accumulate large amounts of JA and 12-oxo-phytodienoic acid (Dhondt et al., 2010). We also found that a lipoyl hydrolase phospholipase A1 (PLA1) interacts with BrRPP1, which is homologous to Arabidopsis AtPLAIIα (Table 3).
There is a conflict between the understanding of relationship of JA pathway and the infection of P. brassicae. Eight genes related to JA biosynthesis and signaling were down-regulated in Brassica rapa ‘CR BJN3-2’ at early defense response induced by P. brassicae infection (Chen et al., 2016). Jubault et al. (2013) also reported the occurrence of a suppressed JA/ET signaling pathway in Arabidopsis on the 7th day after infection by P. brassicae. However, in the transcriptome experiment of Luo et al. (2018), the JA pathway was significantly up-regulated after infection by P. brassicae. Lemarié et al. (2015) demonstrated that both the SA and JA pathways contribute to resistance against P. brassicae in Arabidopsis.
In our research, the relative expression of PLA1 was significantly down-regulated on the 14th day after infection by P. brassicae, while the expression was significantly up-regulated on the 42nd day after infection, and its expression was significantly different between resistant and susceptible materials (Figure 7). Therefore, we speculate that BrRPP1 may interact with PLA1 to mediate the regulation of JA, but the process of JA pathway is complex in response to infection of P. brassicae. The confliction of JA pathway responsing to infection of P. brassicae should be from the different detection stage after infecting by P. brassicae.
In addition, one of the screened interacting proteins of BrRPP1, XM_013801607.2, belongs to the glycosyltransferase family 8 (GT8), which has high homology with Arabidopsis AtGAUT8 (At3g25140). It has been proved that alpha-galacturonosyltransferase (GAUT) can synthesize pectin with UDP-GalA as substrate to participate in the synthesis of plant cell wall (Sterling et al., 2006). Plant cell walls provide a structural framework to support plant growth and act as the first line of defense when plants encounter pathogens (Houston et al., 2016). The changes of cell wall components affect the downstream functions of cells as storage units, structural networks and solute transporters. In many cases, it also affects the ability of cells to respond to stress caused by pathogens and the environment (Tucker et al., 2018). Genes encoding enzymes that can synthesize or hydrolyze plant cell wall components show different expressions under different pressures, indicating that they may promote stress tolerance by changing cell wall components (Seki et al., 2002). The arabinogalactan protein (Unigenes40439), a cell wall component, was changed significantly after infection by P. brassicae (Luo et al., 2018). In this study, the relative expression of GT8 was significantly down-regulated after infection by P. brassicae, and its expression was significantly different between resistant and susceptible materials (Figure 7). Therefore, it can be speculated that BrRPP1 may modify the plant cell wall to respond to the infection of P. brassicae by regulating glycosyltransferase.
Among these interacting proteins, some proteins related to plant photosynthesis have been found, including oxygen-evolving enhancer protein (OEE), phosphoribulokinase (PRK) and His-Asn-His endonuclease (HNH) (Table 3).
OEE is the peripheral protein of photosystem II and plays an important role in the oxygen releasing activity of photosystem II (Seidler et al., 1996). Studies have found that Arabidopsis oxygen enhanced protein AtOEE2 may participate in the generation of reactive oxygen species in the ETI reaction of plants by acting as the downstream signal of WAK1 (Yang et al., 2003). Oxygen enhancing protein has been proved to be involved in the transduction of disease resistance signals. The Phytophthora capsici Effect Factor RxLR19781 can regulate its zoospore infection by combining with the oxygen enhancing NbOEE2, which is the target protein of the effector protein of pathogenic bacteria (Liang et al., 2019). In this study, it was found that the expression of OEE was significantly down-regulated on the 14th day after inoculation of P. brassicae, and its expression level in resistant and susceptible materials was significantly different (Figure 7), so we speculated that OEE, as a peripheral protein of photosystem II, may be the target protein of effector protein of P. brassicae, and participate in the indirect recognition of BrRPP1 to P. brassicae.
The PRK is a part of the Calvin cycle of plant dark reaction, which exists in chloroplasts, and is responsible for the fixation of CO2 in photosynthetic organisms by catalyzing ribulose 5-phosphate to form the CO2 receptor ribulose 1, 5-diphosphate (Hariharan and Cattolico, 1998). Although people usually focus on the role of PRK in carbon assimilation and in the light environment, PRK also plays a role in biological and abiotic stresses. For example, OsPRK gene may participate in the induced defense response of rice to pests (Chen et al., 2020), and PRK in wheat responded to the infection of Puccina striiformis (Liang et al., 2007). In this study, it was found that the expression of PRK was significantly down-regulated after inoculation with P. brassicae, and its expression was significantly different between resistant and susceptible materials (Figure 7). This suggests that PRK, as a part of the Calvin cycle of plant dark reaction, may respond to the infection of P. brassicae.by interacting with BrRPP1,
The HNH nuclease domain contains three of the most conserved His and Asn amino acid residues and has DNA cleaving activity (Zhang et al., 2016). The role of proteins containing HNH nuclease motif has been rarely reported in plants. A recent study showed that a White Stripe Leaf 9 (WSL9) protein containing HNH domain plays a crucial role in the early development of chloroplasts (Zhu et al., 2020). There is no report on the role of HNH in disease resistance. In this study, it was found that HNH interacted with BrRPP1, and its expression was significantly down-regulated after being infected by P. brassicae. The expression of HNN was significantly different between resistant and susceptible materials (Figure 7). This result suggests that HNH plays a crucial role in the early development of chloroplasts, and may respond to the infection of P. brassicae.
Above all, OEE, PRK and HNH all participate in plant photosynthesis, and respond to the infection of P. brassicae by interacting with BrRPP1.
BrRPP1 has been located as a candidate R gene to clubroot disease for many times. In this study, we found that BrRPP1 contains typical TIR-NBS-LRR domain of R gene. Although the gene expression of BrRPP1 was not affected by the infection of P. brassicae, the sequence difference of its cDNA and promoter between resistant and susceptible materials leads to the change of protein structure, and the significant difference of BrRPP1 gene expression between resistant materials and susceptible materials. Arabidopsis homologous deletion mutant of BrRPP1 reduced the resistance to clubroot disease. Moreover, the interaction proteins of BrRPP1 with high detection rate were involved in the pathways of programmed cell death, JA signal, cell wall synthesis and photosynthetic, and their expressions were significantly changed by infection with P. brassicae. So it was speculate that BrRPP1 participated in the resistance of Chinese cabbage to P. brassicae by inducing the above pathways (Figure 8).
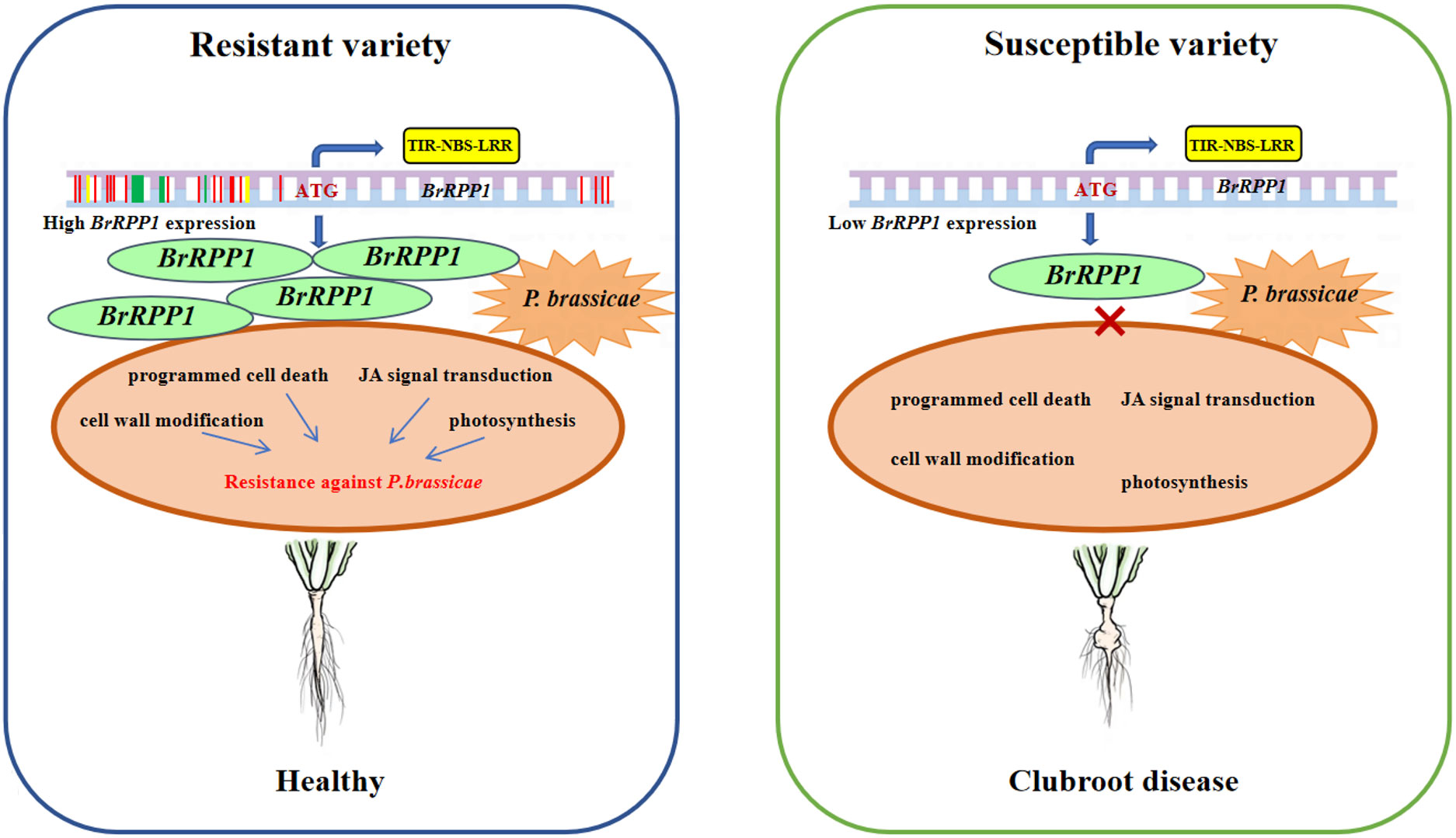
Figure 8 Role pattern of BrRPP1 in Chinese cabbage response to Plasmodiophora brassicae. BrRPP1 gene contains TIR-NBS-LRR domain and belongs to R gene. The gene sequence of BrRPP1 in resistant varieties was different from that in susceptible varieties. Green represents base insertion, yellow represents base deletion, and red represents base mutation. The difference of BrRPP1 promoter sequence leads to the significant difference of gene expression between resistant varieties and susceptible varieties.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
WG, ML and RJ defined the research theme and wrote the manuscript. HF, XW and BZ designed methods and experiments, carried out the laboratory experiments, analyzed the data, and interpreted the results. KL, JZh, and JZo co-designed the experiments and discussed the analyses and interpretation. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China [grant number 31972412; 32272717] and the Science and Technology Mission Project of Shenyang Science and Technology Council [grant number 22-319-2-05].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1082395/full#supplementary-material
Botella, M. A. (1998). Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct peronospora parasitica avirulence determinants. Plant Cell. 10 (11), 1847–1860. doi: 10.1105/tpc.10.11.1847
Chen, J., Jing, J., Zhan, Z., Zhang, T., Zhang, C., Piao, Z. (2013). Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa. PloS One 8 (12), e85307. doi: 10.1371/journal.pone.0085307
Chen, L., Lv, J., Li, C. Y., Zhou, S. X., Lou, Y. G. (2020). Cloning, subcellular localization and expression patterns of the phosphoribulokinase gene OsPRK in the rice plant. J. Plant Protection. 47 (2), 283–291. doi: 10.13802/j.cnki.zwbhxb.2020.2019113
Chen, J., Pang, W., Chen, B., Zhang, C., Piao, Z. (2016). Transcriptome analysis of Brassica rapa near-isogenic lines carrying clubroot-resistant and-susceptible alleles in response to Plasmodiophora brassicae during early infection. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01183
Chu, M., Song, T., Falk, K. C., Zhang, X., Liu, X., Chang, A., et al. (2014). Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genomics 15 (1), 1166. doi: 10.1186/1471-2164-15-1166
Dhondt, S., Gouzerh, G., Müller, A., Legrand, M., Heitz, T. (2010). Spatio-temporal expression of patatin-like lipid acyl hydrolases and accumulation of jasmonates in elicitor-treated tobacco leaves are not affected by endogenous levels of salicylic acid. Plant J. 32 (5), 749–762. doi: 10.1046/j.1365-313x.2002.01465.x
Diederichsen, E., Frauen, M., Linders, E., Hatakeyama, K., Hirai, M. (2009). Status and perspectives of clubroot resistance breeding in crucifer crops. J. Plant Growth Regul. 28 (3), 265–281. doi: 10.1007/s00344-009-9100-0
Fischer, E., Raschke, K., Stitt, M. (1986). Effects of abscisic acid on photosynthesis in whole leaves: changes in CO2 assimilation, levels of carbon-reduction-cycle intermediates, and activity of ribulose-1,5-bisphosphate carboxylase. Planta. 169 (4), 536–545. doi: 10.1007/BF00392104
Gao, Y., Huang, S., Qu, G., Fu, W., Zhang, M., Liu, Z., et al. (2020). The mutation of ent-kaurene synthase, a key enzyme involved in gibberellin biosynthesis, confers a non-heading phenotype to Chinese cabbage (Brassica rapa l. ssp. pekinensis). Hortic. Res. 7 (1), 178. doi: 10.1038/s41438-020-00399-6
Ghelder, C. V., Esmenjaud, D. (2016). TNL genes in peach: insights into the post-LRR domain. BMC Genom. 17 (1), 317. doi: 10.1186/s12864-016-2635-0
Glazebrook, J. (2001). Genes controlling expression of defense responses in Arabidopsis–2001 status. Curr. Opin. Plant Biol. 4 (4), 301–308. doi: 10.1016/s1369-5266(00)00177-1
Hariharan, T., Cattolico, J. R. A. (1998). Purification and characterization of phosphoribulokinase from the marine chromophytic alga Heterosigma carterae. Plant Physiol. 117 (1), 321. doi: 10.1104/pp.117.1.321
Hirai, M. (2006). Genetic analysis of clubroot resistance in Brassica crops. Breed Sci. 56 (3), 223–229. doi: 10.1270/jsbbs.56.223
Holk, A., Rietz, S., Zahn, M., Quader, H., Scherer, G. F. (2002). Molecular identification of cytosolic, patatin-related phospholipases a from Arabidopsis with potential functions in plant signal transduction. Plant Physiol. 130 (1), 90–101. doi: 10.1104/pp.006288
Holub, E. B., Beynon, J. L. (1997). Symbiology of mouse-ear cress (Arabidopsis thaliana) and oomycetes. Adv. Bot. Res. 24, 227–273. doi: 10.1016/S0065-2296(08)60075-0
Houston, K., Tucker, M. R., Chowdhury, J., Shirley, N., Little, A. (2016). The plant cell wall: a complex and dynamic structure as revealed by the responses of genes under stress conditions. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00984
Huang, Z., Peng, G., Liu, X., Deora, A., Falk, K. C., Gossen, B. D., et al. (2017). Fine mapping of a clubroot resistance gene in Chinese cabbage using SNP markers identified from bulked segregant RNA sequencing. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01448
Jubault, M., Lariagon, C., Taconnat, L., Renou, J. P., Gravot, A., Delourme, R., et al. (2013). Partial resistance to clubroot in Arabidopsis is based on changes in the host primary metabolism and targeted cell division and expansion capacity. Funct. Integr. Genomics 13 (2), 191–205. doi: 10.1007/s10142-013-0312-9
Kato, T., Hatakeyama, K., Fukino, N., Matsumoto, S. (2013). Fine mapping of the clubroot resistance gene CRb and development of a useful selectable marker in Brassica rapa. Breed Sci. 63 (1), 116–124. doi: 10.1270/jsbbs.63.116
Kazan, K., Lyons, R. (2014). Intervention of phytohormone pathways by pathogen effectors. Plant Cell. 26 (6), 2285–2309. doi: 10.1105/tpc.114.125419
Koch, E., Cox, R., Williams, P. H. (1991). Infection of Arabidopsis thaliana by plasmodiophora brassicae. J. Phytopathol. 132 (2), 99–104. doi: 10.1111/j.1439-0434.1991.tb00100.x
Lampugnani, E. R., Khan, G. A., Somssich, M., Persson, S. (2018). Building a plant cell wall at a glance. J. Cell Sci. 131 (2), jcs207373. doi: 10.1242/jcs.207373
Larkan, N. J., Lydiate, D. J., Yu, F., Rimmer, S. R., Borhan, M. H. (2014). Co-Localisation of the blackleg resistance genes Rlm2 and LepR3 on Brassica napus chromosome A10. BMC Plant Biol. 14, 387. doi: 10.1186/s12870-014-0387-z
Lemarié, S., Robert-Seilaniantz, A., Lariagon, C., Lemoine, J., Marnet, N., Jubault, M., et al. (2015). Both the jasmonic acid and the salicylic acid pathways contribute to resistance to the biotrophic clubroot agent Plasmodiophora brassicae in arabidopsis. Plant Cell Physiol. 56 (11), 2158–2168. doi: 10.1093/pcp/pcv127
Liang, G., Ji, H., Zhang, Z., Wei, H., Kang, Z., Peng, Y., et al. (2007). Proteome analysis of slow-rusting variety Chuanmai 107 inoculated by wheat stripe rust (Puccina striiformis). J. Triticeae Crops. 27 (2), 335–340. doi: 10.7606/j.issn.1009-1041.2007.02.083
Liang, T., Wang, L., Zhu, T., Ma, Y., Jiang, W., Ji, R. (2019). Effect of silencing oxygen-enhancing protein (OEE2) on immune function of Phytophthora capsici effect factor RxLR1978. J. Shandong Agric. University. 50 (1), 40–43. doi: 10.3969/j.issn.1000-2324
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods. 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Li, T., Wang, B., Yin, C., Zhang, D., Wang, D., Song, J., et al. (2019). The gossypium hirsutum TIR-NBS-LRR gene GhDSC1 mediates resistance against Verticillium wilt. Mol. Plant Pathol. 20 (6), 857–876. doi: 10.1111/mpp.12797
Ludwig-Müller, J., Prinsen, E., Rolfe, S. A., Schoies, J. D. (2009). Metabolism and plant hormone action during clubroot disease. J. Plant Growth Regul. 28, 229–244. doi: 10.1007/s00344-009-9089-4
Ludwig-Müller, J., Schuller, A. (2008). What can we learn from clubroots: Alterations in host roots and hormone homeostasis caused by Plasmodiophora brassicae. Eur. J. Plant Pathol. 121, 291–302. doi: 10.1007/s10658-007-9237-2
Luo, Y., Dong, D., Su, Y., Wang, X., Peng, Y., Peng, J. (2018). Transcriptome analysis of Brassica juncea var. tumida tsen responses to Plasmodiophora brassicae primed by the biocontrol strain zhihengliuella aestuarii. Funct. Integr. Genomics 18 (3), 301–314. doi: 10.1007/s10142-018-0593-0
Lu, Y., Yao, J. (2018). Chloroplasts at the crossroad of photosynthesis, pathogen infection and plant defense. Int. J. Mol. Sci. 19 (12), 3900-3936. doi: 10.3390/ijms19123900
Lv, M., Liu, Y., Wu, Y., Zhang, J., Liu, X., Ji, R., et al. (2021). An improved technique for isolation and characterization of single-spore isolates of Plasmodiophora brassicae. Plant Dis. 105 (12), 3932–3938. doi: 10.1094/PDIS-03-21-0480-RE
Matilla, A. (2021). Cellular oxidative stress in programmed cell death: focusing on chloroplastic 1O2 and mitochondrial cytochrome-c release. J. Plant Res. 134 (2), 179–194. doi: 10.1007/s10265-021-01259-7
Matsumoto, E., Yasui, C., Ohi, M., Tsukada, M. (1998). Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage (Brassica rapa ssp. pekinensis). Euphytica. 104 (2), 79–86. doi: 10.1023/A:1018370418201
Mithen, R., Magrath, R. (1992). A contribution to the life history of Plasmodiophora brassicae: secondary plasmodia development in root galls of Arabidopsis thaliana. Mycol Res. 96 (10), 877–885. doi: 10.1016/S0953-7562(09)81035-6
Popova, L. P., Tsonev, T. D., Lazova, G. N., Stoinova, Z. G. (1996). Drought- and ABA-induced changes in photosynthesis of barley plants. Physiol. Plant 96 (4), 623–629. doi: 10.1111/j.1399-3054.1996.tb00235.x
Qi, Y., Mao, F., Zhou, Z., Liu, D., Min, Y., Deng, X., et al. (2018). The release of cytochrome c and the regulation of the programmed cell death progress in the endosperm of winter wheat (Triticum aestivum l.) under waterlogging. Protoplasma. 255 (6), 1651–1665. doi: 10.1007/s00709-018-1256-7
Rana, K., Atri, C., Akhatar, J., Kaur, R., Goyal, A., Singh, M. P., et al. (2019). Detection of first marker trait associations for resistance against Sclerotinia sclerotiorum in Brassica juncea-erucastrum cardaminoides introgression lines. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01015
Renier, D., Liu, Y., Yang, K. Y., Han, L., Mao, G., Glazebrook, J., et al. (2008). A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105 (14), 5638–5643. doi: 10.1073/pnas.0711301105
Sainsbury, F., Thuenemann, E. C., Lomonossoff, G. P. (2009). pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7 (7), 682–693. doi: 10.1111/j.1467-7652.2009.00434.x
Saito, M., Kubo, N., Matsumoto, S., Suwabe, K., Tsukada, M., Hirai, M. (2006). Fine mapping of the clubroot resistance gene, Crr3, in Brassica rapa. Theor. Appl. Genet. 114 (1), 81–91. doi: 10.1007/s00122-006-0412-1
Sakamoto, K., Saito, A., Hayashida, N., Taguchi, G., Matsumoto, E. (2008). Mapping of isolate-specific QTLs for clubroot resistance in Chinese cabbage (Brassica rapa l. ssp. pekinensis). Theor. Appl. Genet. 117 (5), 759–767. doi: 10.1007/s00122-008-0817-0
Seemann, J. R., Sharkey, T. D. (1987). The effect of abscisic acid and other inhibitors on photosynthetic capacity and the biochemistry of CO(2) assimilation. Plant Physiol. 84 (3), 3900–3936. doi: 10.1104/pp.84.3.696
Seidler, A., Rutherford, A. W., Michel, H. (1996). On the role of the n-terminus of the extrinsic 33 kDa protein of photosystem II. Plant Mol. Biol. 31 (1), 183–188. doi: 10.1007/BF00020619
Seki, M., Narusaka, M., Ishida, J., Tokihiko, N., Fujita, M., Oono, Y., et al (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high‐Salinity stresses using a full-length cDNA microarray. Plant J 31 (3), 279–292. doi: 10.1046/j.1365-313X.2002.01359.x
Springer, N. M. (2010). Isolation of plant DNA for PCR and genotyping using organic extraction and CTAB. Cold Spring Harb. Protoc. 1, 2010(11):5515. doi: 10.1101/pdb.prot5515
Steinbrenner, A. D. (2015). Recognition, activation, and signaling functions of the Arabidopsis NLR receptor RPP1. Doctoral thesis. [United States (IL)]: Univ. California.
Sterling, J. D., Atmodjo, M. A., Inwood, S. E., Kumar Kolli, V. S., Quigley, H. F., Hahn, M. G., et al. (2006). Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 103 (13), 5236–5241. doi: 10.1073/pnas.0600120103
Suwabe, K. (2006). Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics. 173 (1), 309–319. doi: 10.1534/genetics.104.038968
Suwabe, K., Tsukazaki, H., Iketani, H., Hatakeyama, K., Fujimura, M., Nunome, T., et al. (2003). Identification of two loci for resistance to clubroot (Plasmodiophora brassicae woronin) in Brassica rapa l. Theor. Appl. Genet. 107 (6), 997–1002. doi: 10.1007/s00122-003-1309-x
Takemoto, D., Rafiqi, M., Hurley, U., Lawrence, G. J., Bernoux, M., Hardham, A. R., et al. (2012). N-terminal motifs in some plant disease resistance proteins function in membrane attachment and contribute to disease resistance. Mol. Plant Microbe Interact. 25 (3), 379–392. doi: 10.1094/MPMI-11-10-0272
Takken, F. L., Albrecht, M., Tameling, W. I. (2006). Resistance proteins: molecular switches of plant defence. Curr. Opin. Plant Biol. 9 (4), 383–390. doi: 10.1016/j.pbi.2006.05.009
Tucker, M. R., Lou, H., Aubert, M. K., Wilkinson, L. G., Little, A., Houston, K., et al. (2018). Exploring the role of cell wall-related genes and polysaccharides during plant development. Plants 7 (2), 42. doi: 10.3390/plants7020042
Vacca, R. A., Valenti, D., Bobba, A., Merafina, R. S., Passarella, S., Marra, E. (2006). Cytochrome c is released in a reactive oxygen species-dependent manner and is degraded via caspase-like proteases in tobacco bright-yellow 2 cells en route to heat shock-induced cell death. Plant Physiol. 141 (1), 208–219. doi: 10.1104/pp.106.078683
Wan, L., Essuman, K., Anderson, R. G., Sasaki, Y., Monteiro, F., Chung, E. H., et al. (2019). TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science. 365 (6455), 799–803. doi: 10.1126/science.aax1771
Wang, J., Liu, H., Wang, H., Chen, Z. (2016). Screening of interaction proteins of RICE blast resistance PROTEIN PIK-h from NBS-LRR family. Scientia Agricultura Sinica. 49 (03), 482–490. doi: 10.3864/j.issn.0578-1752.2016.03.007
Wasternack, C. (2007). Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100 (4), 681–697. doi: 10.1093/aob/mcm079
Wasternack, C., Hause, B. (2013). Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. an update to the 2007 review in annals of botany. Ann. Bot. 111 (6), 1021–1058. doi: 10.1093/aob/mct067
Xu, Y., Zhou, J., Liu, Q., Li, K., Zhou, Y. (2020). Construction and characterization of a high-quality cDNA library of Cymbidium faberi suitable for yeast one-and two-hybrid assays. BMC Biotechnol. 20 (1), 1–9. doi: 10.1186/s12896-020-0599-2
Yang, E., Oh, Y. A., Lee, E. S., Park, A. R., Cho, S. K., Yoo, Y. J., et al. (2003). Oxygen-evolving enhancer protein 2 is phosphorylated by glycine-rich protein 3/wall-associated kinase 1 in Arabidopsis. Biochem. Biophys. Res. Commun. 305 (4), 862–868. doi: 10.1016/s0006-291x(03)00851-9
Yu, F., Zhang, X., Huang, Z., Chu, M., Song, T., Falk, K. C., et al. (2016). Identification of genome-wide variants and discovery of variants associated with Brassica rapa clubroot resistance gene Rcr1 through bulked segregant RNA sequencing. PloS One 11 (4), e0153218. doi: 10.1371/journal.pone.0153218
Zhang, L., Huang, Y., Xu, D., Yang, L., Qian, K., Chang, G., et al. (2016). Biochemical characterization of a thermostable HNH endonuclease from deep-sea thermophilic bacteriophage GVE2. Appl. Microbiol. Biotechnol. 100 (18), 8003–8012. doi: 10.1007/s00253-016-7568-7
Keywords: clubroot disease, Chinese cabbage, BrRPP1, Plasmodiophora brassicae, resistance gene
Citation: Ge W, Lv M, Feng H, Wang X, Zhang B, Li K, Zhang J, Zou J and Ji R (2023) Analysis of the role of BrRPP1 gene in Chinese cabbage infected by Plasmodiophora brassicae. Front. Plant Sci. 14:1082395. doi: 10.3389/fpls.2023.1082395
Received: 28 October 2022; Accepted: 12 January 2023;
Published: 25 January 2023.
Edited by:
Huan Peng, Institute of Plant Protection (CAAS), ChinaReviewed by:
Zhansheng Li, Insititute of Vegetables and Flowers (CAAS), ChinaCopyright © 2023 Ge, Lv, Feng, Wang, Zhang, Li, Zhang, Zou and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruiqin Ji, cnVpcWluamlAc3lhdS5lZHUuY24=
†Present address: Ken Li, State Key Laboratory of Vegetable Germplasm Innovation, Vegetable Research Institute, Tianjin, China
Tianjin Key Laboratory of Vegetable Genetics and Breeding/Tianjin Kernel, Vegetable Research Institute, Tianjin, China
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.