- 1The New Zealand Institute for Plant and Food Research Ltd, Auckland, New Zealand
- 2University of Auckland, School of Biological Sciences, Auckland, New Zealand
- 3The New Zealand Institute for Plant and Food Research Ltd, Hamilton, New Zealand
- 4The New Zealand Institute for Plant and Food Research Limited, Palmerston North, New Zealand
Anthocyanins are a major group of red to blue spectrum plant pigments with many consumer health benefits. Anthocyanins are derived from the flavonoid pathway and diversified by glycosylation and methylation, involving the concerted action of specific enzymes. Blueberry and bilberry (Vaccinium spp.) are regarded as ‘superfruits’ owing to their high content of flavonoids, especially anthocyanins. While ripening-related anthocyanin production in bilberry (V. myrtillus) and blueberry (V. corymbosum) is regulated by the transcriptional activator MYBA1, the role of specific structural genes in determining the concentration and composition of anthocyanins has not been functionally elucidated. We isolated three candidate genes, CHALCONE SYNTHASE (VmCHS1), ANTHOCYANIDIN SYNTHASE (VmANS) and UDP-GLUCOSE : FLAVONOID-3-O-GLYCOSYLTRANSFERASE (VcUFGT2), from Vaccinium, which were predominantly expressed in pigmented fruit skin tissue and showed high homology between bilberry and blueberry. Agrobacterium-mediated transient expression of Nicotiana benthamiana showed that overexpression of VcMYBA1 in combination with VmANS significantly increased anthocyanin concentration (3-fold). Overexpression of VmCHS1 showed no effect above that induced by VcMYBA1, while VcUFGT2 modulated anthocyanin composition to produce delphinidin-3-galactosylrhamnoside, not naturally produced in tobacco. In strawberry (Fragaria × ananassa), combined transient overexpression of VcUFGT2 with a FLAVONOID 3´,5´-HYDROXYLASE from kiwifruit (Actinidia melanandra) modulated the anthocyanin profile to include galactosides and arabinosides of delphinidin and cyanidin, major anthocyanins in blueberry and bilberry. These findings provide insight into the role of the final steps of biosynthesis in modulating anthocyanin production in Vaccinium and may contribute to the targeted breeding of new cultivars with improved nutritional properties.
Introduction
Anthocyanins are a diverse group of natural plant pigments, providing a full spectrum of visible colours ranging from bright orange, pink and red to purple and blue (Chen, 2015). In plants, the colours provided by anthocyanins are known for their roles in attracting pollinators and seed dispersers, especially in reproductive organs such as flowers and fruits (Jimenez-Garcia et al., 2013). In many plant species anthocyanins only accumulate during fruit ripening, particularly in fruit skin, which makes these pigments visual cues for ripeness (Jimenez-Garcia et al., 2013).
As polyphenolic pigments, anthocyanins form part of the flavonoid phytochemical group and also act as antioxidants, photoprotectants, and metal-chelating agents (Stintzing and Carle, 2004; Landi et al., 2015). Anthocyanins are composite molecules, consisting of an anthocyanidin moiety which is linked with a glycoside at the C3-position of the heterocyclic C-ring structure (Castañeda-Ovando et al., 2009).
There are over 600 anthocyanins identified in nature, diversified primarily by different glycosylation patterns and other chemical modifications such as methylations and acylations (Smeriglio et al., 2016). Based on hydroxylation of the B-ring, there are three main groups of anthocyanidins: pelargonidin, cyanidin and delphinidin. Subsequent methylation of cyanidin generates peonidin, while methylation delphinidin produces petunidin and malvidin (Castañeda-Ovando et al., 2009).
The regulation of anthocyanin production in plants is highly conserved (Zhang et al., 2014). Activation of anthocyanin biosynthesis is transcriptionally controlled by the MBW complex, which is formed by subgroup 6 of the R2R3-MYB (R2R3-MYB) transcription factor (TF) family, the bHLH TF family and the WD Repeat (WDR) proteins (Zhang et al., 2014). To regulate the production of anthocyanins, R2R3-MYB TFs directly interact with promoters of structural genes from the anthocyanin biosynthesis pathway through their highly conserved DNA binding domain. It is the overall activity of the R2R3-MYB TFs that is thought to be the main determinant for the amount of anthocyanin produced (Schwinn et al., 2006; Allan et al., 2008). Furthermore, meta-analysis shows that overexpression of MYB TFs significantly impact the regulation of a set of structural genes together with increased anthocyanin production (Liu et al., 2021b). However, the manipulation of wide-acting MYB TFs can potentially lead to collateral consequences. For example, a red-fleshed transgenic apple line with high anthocyanin accumulation in fruit flesh was generated by constitutive expression of MdMYB10 (Espley et al., 2013), but early browning of fruit flesh was commonly observed (Espley et al., 2019). Therefore, altering the expression of key structural genes of the flavonoid pathway is an alternative strategy to provide more specific targets for modulating anthocyanin production in fruits.
The anthocyanin biosynthesis pathway in plants has been well researched across different species (Liu et al., 2021a) and can be subdivided into three sections: the general phenylpropanoid pathway, the flavonoid pathway and the specific biosynthesis of anthocyanins (Zhang et al., 2014). The general phenylpropanoid pathway first utilises phenylalanine to synthesise p-coumaroyl CoA. Chalcone synthase (CHS) then directs the carbon flux into the flavonoid pathway via the conversion of p-coumaroyl CoA into naringenin chalcone. Subsequent activities of chalcone isomerase (CHI) and flavanone 3-hydroxylase (F3H) then produce the structural flavonoid backbone, which is further modified and converted to leucoanthocyanidins by dihydroflavonol 4-reductase (DFR). As part of the anthocyanin-specific biosynthesis pathway, anthocyanidin synthase (ANS) converts leucoanthocyanidins into coloured anthocyanidins, which are then stabilised by esterification with sugar molecules by glycosyltransferases such as UDP-glucose:flavonoid-3-O-glycosyltransferase (UFGT) (Liu et al., 2018). The positive relationship between anthocyanin accumulation and the expression of structural genes in the pathway has been documented in various plant species. For example, significantly reduced anthocyanin production was found in kiwifruit when CHS or UFGT expression were silenced using RNAi (Montefiori et al., 2011; Sun et al., 2014), while individual overexpression of CHS and ANS promoted anthocyanin production in Arabidopsis and red sage (Salvia miltiorrhiza), respectively (Zhang et al., 2018; Li et al., 2019). The combined overexpression of key anthocyanin structural genes affects anthocyanin accumulation in flower species, e.g. rose (Katsumoto et al., 2007) and orchids (Zhou et al., 2021).
Dietary anthocyanins are well regarded for their health benefits, such as providing neuroprotection, lowering cardiovascular disease risks, and having anti-carcinogenic and anti-diabetic properties (Khoo et al., 2017). Blueberry (Vaccinium spp.) are known to be one of the best sources of dietary bioactive compounds, with their especially high anthocyanin content purported to confer a wide range of health-promoting benefits (Skrovankova et al., 2015; Ma et al., 2018; Kalt et al., 2020) through their antioxidant and anti-inflammation properties (Riso et al., 2013; Shi et al., 2017; Ma et al., 2018). It has been reported that the type of glycoside moiety conjugated to the anthocyanidin aglycone affects anthocyanin bioavailability (Zhao et al., 2014). However, findings on the effect of each type of glycosylation are inconsistent and this suggests that the sugar/anthocyanidin combination might play a determining role (McGhie et al., 2003; Ichiyanagi et al., 2006; Milbury et al., 2010). Bilberry (V. myrtillus) accumulates up to a 4-fold higher concentration of anthocyanins compared with blueberry (Kalt et al., 1999), and is therefore highly valued by the pharmaceutical and dietary supplement industries for their superior health benefits and potential applications due to their high content of bioactive compounds (Camire, 2002; Chu et al., 2011).
Bilberry and the blueberries Northern Highbush (V. corymbosum) and Rabbiteye (V. virgatum) contain 15 types of anthocyanins distributed over five anthocyanidin groups (cyanidin, peonidin, delphinidin, malvidin and petunidin), with malvidins and delphinidins being the most abundant in blueberry, and cyanidins and delphinidins in bilberry (Kader et al., 1996; Kalt et al., 1999; Dare et al., 2022). While blueberry and bilberry anthocyanidins are commonly linked with glucose, galactose and arabinose, the ratios of the respective glycosides are reported to be species-specific for blueberries (Fang, 2015; Günther et al., 2020; Mengist et al., 2020) and ecotype-specific for bilberry (Dare et al., 2022). Across geographically distinct bilberry populations, the glycosylation pattern varies depending on the region of origin. Such distinct variation in glycosylation pattern has been suggested as a feature for geographical authentication (Primetta et al., 2013).
Despite being one of the richest sources of anthocyanins in common fruits (Kalt et al., 2020), cultivated blueberry species such as Northern Highbush and Rabbiteye blueberry produce anthocyanins only in the fruit skin but not the fruit flesh. In contrast, the undomesticated European bilberry species (V. myrtillus L.) has a deep blue colour and pigmented flesh (Riihinen et al., 2008).
Based on the integration of metabolomics and transcriptomics data in blueberry skin and flesh during fruit development, Günther et al. (2020) showed the co-correlation between transcript abundance of blueberry genes VcCHS1, VcANS and VcUFGT2 with the accumulation of anthocyanin during blueberry fruit development. In particular, there was a significant decrease in CHS and ANS expression (between 5- and 15-fold lower) in non-anthocyanic fruit flesh of ripe blueberry fruit when compared with pigmented fruit skin (Günther et al., 2020). A study on wild albino bilberry with highly reduced anthocyanin accumulation found that gene expression of CHS, ANS and UFGT was 7.6, 11 and 33-fold lower, respectively, than in non-albino berry (Zorenc et al., 2017). Similarly, a study on a pink bilberry mutant with lowered anthocyanin accumulation also found a 100-fold decrease in transcript levels of CHS, ANS and UFGT compared with wild-type (Die et al., 2020). Such findings suggest that these three genes may act as bottlenecks in the pathway. These genes, however, have not been functionally characterised in Vaccinium before. We hypothesise that there is more than one rate-limiting step in the flavonoid pathway, affecting anthocyanin concentration. A combined effect from a set of these structural genes may be the key to determine the accumulation of anthocyanins. To investigate their roles driving anthocyanin composition and concentration, CHS, ANS and UFGT were cloned from bilberry and blueberry. We then employed the rapid in vivo transient expression system using different combinations of these genes in two different model systems (N. benthamiana and strawberry) to assess changes in anthocyanin production.
Methods
Gene expression analysis
Fruit material
Blueberry fruit were harvested from cultivated collections of tetraploid V. corymbosum ‘Nui’ (Plant & Food Research, Motueka, New Zealand) (Günther et al., 2020). Three different developmental stages, S5 to S7, were sampled (Zifkin et al., 2012). Bilberry fruit were harvested from wild V. myrtillus L. plants in Kvaløya, Norway (Dare et al., 2022). Fruit from three different developmental stages, S3 to S5 (Karppinen et al., 2016), were included in the sampling process. Collected berries were snap-frozen using liquid nitrogen. Frozen berry skins were separated as much as possible from the flesh using a scalpel on dry ice to allow skin-enriched samples. Special care was taken to avoid contaminating fruit flesh samples with anthocyanins from fruit skin. Separated tissues were homogenized in liquid nitrogen to a fine powder using a grinder (IKA A11 basic mill) and kept at −80°C.
RNA extraction and cDNA synthesis
RNA was extracted from pulverised fruit tissues (150 mg) using the Spectrum™ Plant Total RNA extraction Kit (Sigma Alrich). RNA concentration was quantified using the DeNovix DS-11 FX Spectrophotometer/Fluorometers (Denovix®). Gel electrophoresis was performed to confirm the integrity of the extracted RNA. cDNA synthesis was carried out on 1 μg of extracted RNA using the Quantitect® Reverse Transcription Kit (QIAGEN) with inbuilt gDNA removal step. Control samples without added RT enzyme (-RT) were included for each tissue type to check for possible gDNA contamination.
Quantitative real-time PCR
Quantitative real-time reverse transcription PCR (qRT-PCR) was performed on cDNA (1:20 dilution) from blueberry and bilberry fruit skin and flesh tissues. Reactions were performed in four technical replicates to a total volume of 10 μL (5 μL LightCycler SYBR Green I Master, 0.5 μL of 10 μM forward and reverse primer mix, 2.5 μL diluted cDNA (1:20) and 2 μL nuclease free water). The LightCycler®480 system and software (version 1.5, Roche) was used for amplification at the following conditions: 95°C for 5 min, followed by 50 cycles consisting of 95°C for 10 s, 60°C for 10 s and 72°C for 20 s, then continuous fluorescence data acquisition from 65 to 95°C for melting curve detection. The expression of six anthocyanin biosynthetic genes CHALCONE SYNTHASE (CHS1 and CHS2), DIHYDROFAVONOL REDUCTASE (DFR), FLAVANONE 3-HYDROXYLASE 1 (F3H1), ANTHOCYANIDIN SYNTHASE (ANS) and UDP-GLYCOSE: FLAVONOID-3-O-GLYCOSYLTRANSFERASE 2 (UFGT2) as well as the anthocyanin-related TF MYBA1 was analysed. For both blueberry and bilberry, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (GAPDH) and SAND FAMILY PROTEIN (SAND) were used as reference/house-keeping genes. Primer sequences (Supplementary Table 1) were designed based on RNA-Seq data from Günther et al. (2020) and primer efficiency was determined using the LinReqPCR software (Tuomi et al., 2010). A melting curve analysis was performed using the LightCycler®480 software to verify the amplification of only a single reaction product. Relative gene expression compared to housekeeping genes was calculated based on method described by André et al. (2009).
Gene identification and phylogenetic analysis
The open reading frame (ORF) sequences for VcCHS1 (maker-VaccDscaff35-augustus-gene-1.21-mRNA-1), VcANS (maker-VaccDscaff46-augustus-gene-11.23-mRNA-1) and VcUFGT2 (maker-VaccDscaff6-augustus-gene-420.36) originated from the tetraploid V. corymbosum genome (Colle et al., 2019) as described in (Günther et al., 2020). Bilberry homologues were identified from the V. myrtillus reference genome (Wu et al., 2021) using the Basic Local Alignment Search Tool (BLAST) algorithm.
Protein sequences encoded by these candidate genes from blueberry, were used to search for orthologues from publicly available proteome data from Arabidopsis (Berardini et al., 2015) and five other red fruits – Fragaria vesca (strawberry) V4.0 a1 genome database (Edger et al., 2018), Actinidia chinensis (kiwifruit) Red5 genome database (Pilkington et al., 2018), V. macrocarpon (cranberry) (Polashock et al., 2014), Vitis vinifera (grape) (Enrico, 2007), Rubus occidentalis (raspberry) (VanBuren et al., 2018), and Solanum lycopersicum (tomato) (Tomato Genome Consortium, 2012). Additional protein sequences were retrieved from other species using protein BLAST from non-redundant protein sequences (nr) database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple nucleotide and amino acid sequence alignments and phylogeny trees were created using Geneious software version 10.2.5 (Kearse et al., 2012). The CHS phylogenetic tree was constructed with 54 amino acid sequences from 39 plant species. The ANS phylogeny consisted of 51 sequences from 41 species. LDOX sequences from grape, tobacco, apple, kiwifruit and blueberry were also included in this analysis because of their high similarity in sequence and function of LDOX and ANS (Jun et al., 2018; Lafferty et al., 2022a). There were 62 sequences from 52 species included in the UFGT phylogenetic tree. Consensus phylogenetic trees were constructed using the Neighbor-Joining method with bootstrap at 10,000 replicates. Phylogenetic tree visualisation was generated using iTOL v6 (Letunic and Bork, 2021).
Gene cloning
For VmCHS1 (GenBank accession OP966665) and VmANS (GenBank accession OP966664), total RNA was extracted from bilberry leaf and used for cDNA synthesis (RNA extraction and cDNA synthesis were as described above). Gene-specific primers were designed to match the sequence of both bilberry and blueberry (Supplementary Table 2). High-Fidelity DNA Polymerase (iProof, BioRad) was used for ORF amplification, introducing HindIII and BamHI restriction sites at the 3’ and 5’ end, respectively, at optimised PCR conditions (98°C for 30 s, 30 cycles at 98°C for 10 s, 61°C for 15 s and 72°C for 1.5 min, followed by a final extension at 72°C for 10 min). The Zymoclean Gel DNA Recovery Kit (Zymo Research) was used for purifying VmCHS1 and VmANS fragments, respectively, which were then cloned into the pJET1.2/blunt cloning (pJet) vector and transformed into Escherichia coli DH5α competent cells through heatshock. E. coli cells were grown on Luria-Bertani (LB) media containing ampicillin (50 μg/mL) to amplify the plasmid for Sanger-sequencing (Macrogen). Sequence identity was confirmed against the reference sequence of the V. myrtillus reference genome (Wu et al., 2021). The ORFs of VmCHS1 and VmANS were subsequently cut using restriction enzymes (BamHI and HindIII, NEB) and cloned into the vector pGreen0029-62-SK (pGreen) (Hellens et al., 2000) using T4 DNA ligase (Thermo Scientific). After amplification in E. coli DH5α cells on LB-media with kanamycin (50 μg/mL), rifampicin (50 μg/mL) and gentamycin (10 μg/mL), the Zyppy™ Plasmid Miniprep Kit (Zymo Research) was used for plasmid purification and the correct orientation of the gene confirmed by Sanger sequencing. Recombinant pGreen vectors were transformed into Agrobacterium tumefaciens (Agrobacterium) strain GV3101 (carrying the pSoup helper vector) using electroporation (2.5 kV, capacitance 25 μFd, resistance: 400 Ohms with pulse time of 7-9 ms). Agrobacterium were recovered in Super optimal broth with catabolite repression (SOC) media after transformation and used for subsequent transient expression assays.
For VcUFGT2, double-stranded gblocks™ DNA fragments (Integrated DNA Technologies) were synthesised for the entire VcUFGT2 (GenBank accession OP966666, maker-VaccDscaff6-augustus-gene-420.36, Günther et al., 2020) ORF (Integrated DNA Technologies) and used as template (1.5 ng/µl) for amplification using High-Fidelity DNA Polymerase (iProof, BioRad) with gene-specific primers, introducing attB-attachment sites for subsequent Gateway® cloning. As Forward Gateway primer 3’ VcUFGT2 and as Reverse Gateway primer, 5’ VcUFGT2 was used (Supplementary Table 2). PCR conditions were: 98°C for 30 s, 10 cycles at 98°C for 10 s, 71°C for 15 s and 72°C for 30 sec, followed by a final extension at 72°C for 5 min. The gel purified (Zymoclean Gel DNA Recovery Kit, Zymo Research) PCR product (50-100 ng) was then cloned overnight (20°C) into pDONR221™ vector (150 ng) using BP Clonase™ II enzyme mix in a 10 µL reaction volume. The pHex destination vector (150 ng) and 2µL LR Clonase™ II enzyme mix was then added to and aliquot of the reaction mixture to a final volume of 10 µL and incubated overnight (20°C). OneShot™ Top10 chemically competent E. coli cells (Invitrogen™) were transformed with 2 μL of the reaction and plated onto selective LB-agar containing spectinomycin (25 μg/mL). Sanger-sequencing (Macrogen) was used to verify the VcUFGT2 sequence from eight random colonies after amplification of the plasmid and subsequent purification.
Agrobacterium-mediated transient transformation in tobacco and strawberry
Plant materials
Tobacco plants (N. benthamiana, ecotype ‘Northern Territory’, kindly provided by Prof. Peter Waterhouse at Queensland University of Technology) were grown from seed. Mature strawberry plants (Fragaria × ananassa ‘Camarosa’) were purchased from a garden centre in Auckland, New Zealand. These plant materials were grown in a containment glasshouse at Plant and Food Research (Auckland, New Zealand) at standard glasshouse conditions (20-24°C, 12 hr day lighting condition) for transient gene expression assays. N. benthamiana plants were used at 5.5 weeks old and the two youngest leaves chosen for at least six infiltration patches. Mature strawberry fruit (with fully developed seeds) were infiltrated at the initial reddening stage as described by (Jia et al., 2020) (Supplementary Figure 1A).
Agrobacterium-mediated transient transformation
VcMYBA1 (GenBank accession MH105054.1; Plunkett et al., 2018), MdCHS2 (GenBank accession EB120544.1; Dare et al., 2013), AmF3´5´H (GenBank accession OP963713; Peng et al., 2019) and the reporter gene GUS (Peng et al., 2020) as control have been previously reported. Agrobacterium were grown on LB-agar containing rifampicin (50 μg/mL), gentamycin (10 μg/mL) and kanamycin (50 μg/mL, for VmCHS1 and VmANS) or spectinomycin (25 μg/mL, for VcMYBA1, VcUFGT2, MdCHS2, AmF3´5´H and GUS) at 28°C overnight to achieve exponential growth phase. Using a 10 μL inoculation loop, Agrobacterium cells were then suspended in infiltration buffer (10 mM magnesium chloride, 10 μM acetosyringone) to appropriate OD600 levels and incubated at 28°C for 2 h before infiltration. Different combinations of MdCHS2, VmCHS1, VmANS and VcUFGT2 (OD600 = 0.3 each) were mixed in excess with minimal amounts of the TF VcMYBA1 (OD600 = 0.03) to activate the expression of the anthocyanin biosynthesis pathway in tobacco leaves. GUS was used as a negative control in both tobacco and strawberry, and to ensure equal final OD600 = 1.2 and 35S promoter activity of all mixtures. Each N. benthamiana plant was considered a separate biological replicate and infiltrated individually to avoid contamination between treatments. At least three biological replicates were harvested per treatment. Gene expression of some infiltrated genes in N. benthamiana leaves was confirmed using end-point PCR (Supplementary Table 3; Supplementary Figure 2).
AmF3´5´H was functionally tested using transient overexpression in N. tabacum leaves as described by Peng et al. (2019). Co-infiltration of VcUFGT2 with both VcMYBA (Plunkett et al., 2018) and AmF3´5´H was used in strawberry to produce delphinidin-based anthocyanin, which is not naturally occurring in strawberry. Strawberry fruit were infiltrated by injections of approximately 1 mL mixed culture into the fruit flesh. Infiltrated fruits were harvested when fully ripened after five to seven days (Supplementary Figure 1B). Only visibly transformed fruit tissues (showing dark colour development) were sampled. Each treatment was harvested in at least four biological replicates with each replicate comprising a pool of three to four berries. All infiltrated tissues were snap-frozen in liquid nitrogen and stored at -80°C.
Anthocyanin extraction and quantification from plant tissues
High-Performance-Liquid chromatography
Freeze-dried plant tissue (5 to 15 mg) was extracted in methanol containing 0.1% (v/v) HCl (500 μL) for 3 h in the dark at room temperature with hourly vortexing and then centrifuged at 15,000 rpm for 10 min. The supernatant was transferred, evaporated to dryness using a CentriVap® concentrator (Labconco, CA, USA) and resuspended in 20% (v/v) methanol (400 µL). The solutions were vortexed and centrifuged for 10-30 seconds at 15,000 rpm. Supernatant was filtered into a new Eppendorf tube using 1 mL syringe and RC membrane, pore size 0.45 μm, 15 mm syringe filter (Phenomenex®) prior to analysis.
The HPLC system used was a Dionex UltiMate 3000 UHPLC coupled to a Photodiode Array (PDA) detector (Dionex, Sunnyvale, CA, USA). Anthocyanins were separated on an Acclaim PolarAdvantage II (PA2), 4.6 × 150 mm, 5 µm, analytical LC column (Thermo Fisher Scientific), maintained at 35°C. The solvents were (A) 95:5 water:formic acid v/v and (B) acetonitrile (flow rate, 0.35 mL/min). The initial mobile phase, 100% A, was ramped up linearly to 93% A at 5 min, then to 88% A at 10 min, and 65% A at 25 min, followed by a column flush at 0% A before resetting to the original conditions. The sample injection volume was 5 µL and PDA detection was from 200 to 600 nm. Anthocyanin concentrations were quantified as cyanindin-3-O-glucoside equivalents using standard curves constructed from serial dilutions of cyanindin-3-O-glucoside (concentration range from 15 to 150 μg/μL) with PDA detection at 520 nm. Total anthocyanin concentration was normalised to μg total anthocyanin/g dry weight (DW).
Liquid chromatography-mass spectrometry (LC-MS)
Freeze-dried plant tissue (15 mg) was extracted in methanol+1% formic acid (1 mL) for 3 h at room temperature and vortexed every hour. Cyanidin rutinoside (4 µg/mL) was added as internal standard. The supernatant was transferred after centrifugation (15 min; 15,000 rpm), dried under nitrogen and re-suspended in 5% acetonitrile (100 µL) for analysis.
The samples were analysed by LC-MS using an LTQ linear ion trap mass spectrometer fitted with an ESI interface (electrospray voltage: 5 kV, source temperature: 275°C) (ThermoFisher Scientific, San Jose, CA, USA) coupled to a Dionex Ultimate 3000 UHPLC and PDA detector (Dionex, Sunnyvale, CA, USA). Anthocyanins were separated on an Accucore 2.6 µm C18 150 × 2.1 mm column (Thermo Scientific, CA, USA). The flow rate was 0.4 mL/minute, column temperature 40°C and injection volume was 2 µL. The solvents were (A) 90:10 water:formic acid v/v and (B) 22.5:22.5:45 methanol:acetonitrile:water + 10% formic acid v/v/v. The initial mobile phase, 97% A was held for 0.5 min, then ramped up linearly to 70% A at 22.5 min, and 50% A at 25.5 min, followed by a column flush at 30% A before resetting to the original conditions. The MS data were acquired in the positive mode. The Xcalibur software was used for peak integration and concentrations were calculated in equivalence to cyanidin-3-O-glycoside, based on a standard curve of serial dilution.
Statistical analysis
All statistical analyses were conducted using R 4.0.3 version “Bunny-Wunnies Freak Out” (R Core Team, 2020). The Shapiro-Wilk test and histograms plots were used to inspect distribution of data populations using the ‘MVN’ package (Korkmaz et al., 2014). Where normality was rejected, Box-Cox transformation of the data was performed. Standard parametric analysis was performed, namely Student’s t-test (Student, 1908) for qRT-PCR data and for transient expression data one-way analysis of variance (ANOVA) followed by Tukey-Kramer Honest significant difference (Yandell, 2017) post-hoc test was used. All analyses were conducted at α = 0.05 using the Benjamini-Hochberg correction (Benjamini and Yekutieli, 2001) for multiple comparisons.
Results
RT-qPCR analysis shows that ANS, CHS1 and UFGT2 expression correspond with anthocyanin production in both blueberry and bilberry
Structural genes were previously identified as relevant for anthocyanin biosynthesis in blueberry and bilberry using RNA-Seq (Günther et al., 2020; Lafferty et al., 2022b). To validate their expression before cloning, RT-qPCR was performed on fruit skin and flesh tissues from both blueberry and bilberry at three distinct stages during fruit ripening. Transcript abundances of the three candidate genes CHS1, ANS and UFGT2 (Figure 1A) was analysed in comparison with the main anthocyanin activator MYBA1 and additional non-candidate genes from the flavonoid pathway (CHS2, F3H1 and DFR). In both Vaccinium species, identical primers were used and candidate genes were successfully quantified from pigmented as well as non-pigmented tissues of unripe, veraison (start of skin colour change) and ripe fruit (Figure 1B).
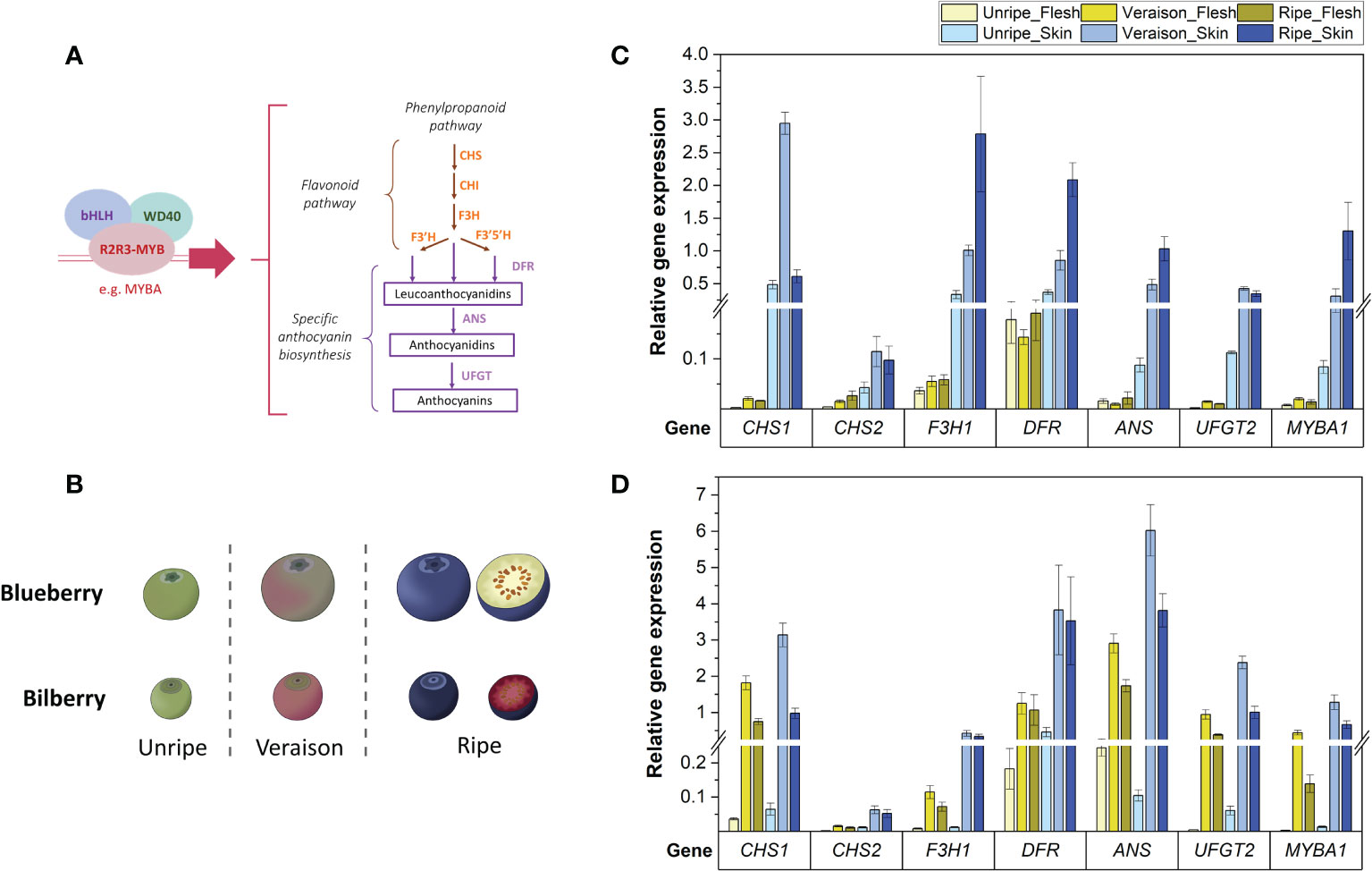
Figure 1 (A) The anthocyanin biosynthesis pathway showing structural genes and the MBW complex controlling the pathway. (B) Developmental stages used for RT-qPCR analysis of blueberry and bilberry fruit. Fruit stages unripe-ripe correspond to blueberry stages S5-7 (Zifkin et al., 2012) and bilberry stages S3-5 (Karppinen et al., 2016). (C, D) RT-qPCR analysis of the TF MYBA1 and structural anthocyanin genes in the flesh and skin of blueberry (C) and bilberry (D) at three fruit developmental stages (unripe, veraison and ripe). Mean relative expression from at least three technical replicates is shown. Error bars represent ± standard deviation.
In blueberry, the relative expression of all tested genes was significantly higher in fruit skin compared with fruit flesh across all fruit developmental stages (p-value < 2.5 x 10-7) and was highest in the skin of pigmented (veraison and ripe) fruit (Figure 1C). Transcript abundances of MYBA1, F3H1, DFR and ANS were highest in ripe fruit skin (p-value < 0.009). The expression level of UFGT2 remained at similar levels between both veraison and ripe stages (p-value = 0.53). Meanwhile, transcript abundance of CHS1 was approximately six-times higher in fruit skin at the veraison stage compared with both unripe and ripe fruit (p-value < 6.4 x 10-4). In bilberry, gene expression of tested flavonoid genes was also highest in fruit skin of ripening (veraison and ripe) fruit (Figure 1D) and reflecting MYBA1 expression levels.
The expression level of CHS1, ANS, UFGT2 and MYBA1 were considerably higher in blueberry fruit skin (between 30- to 103-fold) than in flesh tissue (Figure 1C). In contrast to blueberry, bilberry produced anthocyanins in fruit flesh and transcript abundance of the three candidate genes and MYBA1 was on average only 2.3-fold higher in fruit skin when compared with flesh (Figure 1D). Such changes in the fruit skin to flesh expression ratio in the two Vaccinium species confirmed the association between candidate gene expression and anthocyanin production.
In contrast, the additional CHS isoform (CHS2) and DFR only showed an approximately 3.4-fold increase in expression in fruit skin compared with fruit flesh for both blueberry and bilberry (Figure 1C, D). DFR also showed high abundance in the non-anthocyanic blueberry fruit flesh, which was on average 44-fold higher at the unripe fruit, and 10-fold higher at the veraison and ripe fruit stage than the other tested genes. CHS2, conversely, showed relatively low expression in the fruit skin and flesh of both bilberry and blueberry (Figure 1C, D). F3H1 had a high fruit skin/flesh ratio similar to the three candidate genes and MYBA1 (27-fold) in blueberry (Figure 1C). However, in bilberry, the expression of F3H1 was on average 8-fold lower than CHS1, ANS and UFGT2 in both the pigmented fruit flesh and skin (veraison and ripe stages) (Figure 1D).
In summary, CHS1 was the most highly expressed isoform for anthocyanin production in the fruit of both Vaccinium species. ANS and UFGT2 had the most similar expression pattern to the TF MYBA1 regarding the difference between fruit skin/flesh in blueberry and between blueberry/bilberry fruit flesh. All candidate genes were highly expressed in the fruit skin of berries of both species.
Based on the gene expression analyses, CHS, ANS and UFGT2 were confirmed as suitable gene candidates modulating anthocyanin production in both blueberry and bilberry. Bioinformatic analysis was performed for each of these three genes to further study their sequence similarities across different plant species in detail. First, nucleotide sequences of CHS, ANS and UFGT2 from both bilberry and blueberry were identified to investigate the similarity between the two species. Two putative VmCHS versions, one VmANS and four VmUFGTs were retrieved from the bilberry genome (Wu et al., 2021) using the nucleotide sequences from blueberry (VcCHS1, VcANS and VcUFGT2) as previously identified by Günther et al. (2020). For CHS and UFGT from bilberry, the sequences with the highest similarity with VcCHS1 and VcUFGT2 (VmCHS1 and VmUFGT2) were chosen for further experimental characterisation. The previous annotation as anthocyanidin-3-O-glycosyl transferase 2 (A3GT2) has not been experimentally validated, therefore, UFGT was used as a general term. For all three genes, the bilberry and blueberry sequences were near identical with identities ranging from 95% to 99% for both nucleotide and deduced amino acid sequences.
To evaluate the relevance of CHS1, ANS and UFGT2 in the anthocyanin biosynthesis pathway, the amino acid sequences from both blueberry and bilberry were aligned with homologous sequences from other plant species, including Arabidopsis and anthocyanin-producing fruits. For UFGT, seven anthocyanin-producing species with functionally characterised flavonoid-3-glycosyltransferes activity were included (Furtek et al., 1988; Miller et al., 1999; Lunkenbein et al., 2006; Offen et al., 2006; Kubo et al., 2007; Montefiori et al., 2011). Alignment results of CHS amino acid sequences showed that four CHS active site residues, Cys164, Phe215, His303 and Asn336 (Jez et al., 2001), were fully conserved and identical in all sequences considered (Figure 2A). For ANS, all eight active site residues identified in AtANS (Wilmouth et al., 2002) were fully conserved in blueberry, bilberry and seven other species (Figure 2B). Similarly, the PSPG (plant secondary product glycosyltransferases) motif (Hughes and Hughes, 1994) of UFGT was fully conserved between the considered sequences (Figure 2C), except for the last residue of the motif. VcUFGT2 and VmUFGT2 have a histidine residue in this position which was previously found to be highly conserved among galactosyltransferases, such as the kiwifruit AcF3GT (Montefiori et al., 2011). This is in contrast with glucosyltransferases such as the grape VvUFGT (Offen et al., 2006), which has a conserved glutamine residue at the end of the PSPG motif.
The phylogenetic relationship between bilberry and blueberry amino acid sequences with other plant species was investigated for each candidate gene. CHS and ANS were highly conserved across the different plant species considered. UFGT was less conserved, with the majority of sequences showing 40-50% identity to each other. For all CHS, ANS and UFGT, most sequences from the same taxonomic order clustered well with each other (Figure 2D-F). Most sequences from the order Ericales clustered together (purple clusters) with the exception of VcUFGT3, VcUFGT5 and VcCHS2. Within the Ericales subcluster, Vaccinium species showed the highest homology. Especially, blueberry sequences (highlighted blue) were most closely related to their corresponding homolog in bilberry (highlighted purple) (e.g. VcCHS1 and VmCHS1, VcUFGT2 and VmUFGT2) (Figure 2D, F). Overall, the high similarity and close relationship between the blueberry and bilberry candidate genes validated the use of cloned genes from either species for testing their function in Vaccinium species using transient expression.
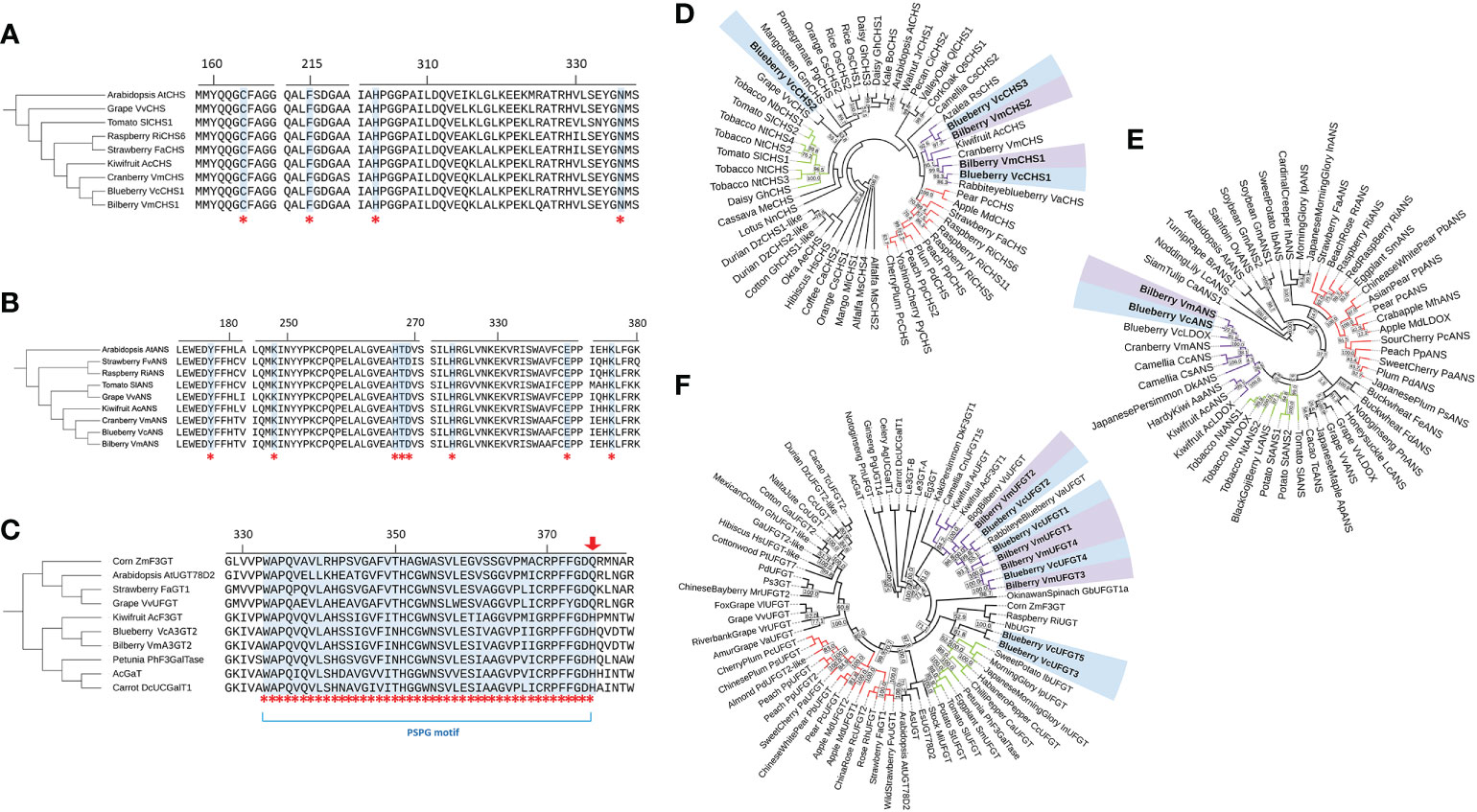
Figure 2 Active site alignment and phylogenetic tree of the protein sequence of CHS (A, D), ANS (B, E) & UFGT2 (C, F). (A-C) Active site residues are highlighted and indicated by asterisks and are referred to in the text. Arrow indicates conserved residue for glucosyltransferases/galactosyltransferases. Residue numbering is based on blueberry sequence. (B-F): Blueberry and bilberry sequences are in bold and highlighted (blueberry – blue; bilberry – purple). Clusters of the three most represented taxonomic orders are indicated by branch colour (Rosales – red; Solanales – green; Ericales – purple; others - black). Phylogenetic trees were constructed using the Neighbour-Joining algorithm with 0.5% topology support threshold. Bootstrap analysis was performed at 10,000 replicates, values > 50% are shown on the tree. Sequence accession numbers are indicated in Supplementary Tables 2, 3 and 4.
Overexpression of VmANS increases anthocyanin concentration
Agrobacterium-mediated infiltration assays in N. benthamiana (Hellens et al., 2005) were used to characterise the function of VmCHS1, VmANS and VcUFGT2 in modulating anthocyanin biosynthesis in vivo. Co-infiltration with VcMYBA at an OD600 ratio of 1:10 to other genes was found to be suitable for further assaying activities by providing a low but visible and quantifiable background of anthocyanin production in leaves.
First, effects on anthocyanin production by single candidate gene overexpression assays were assessed. VcMYBA1-only samples produced anthocyanin concentration of 1.81 ± 0.88 mg/g DW (Figure 3). Compared to the infiltration of VcMYBA1 alone, co-infiltration with VmCHS1 did not result in a significant change in anthocyanin production (p-value = 0.985). This was observed both visually, in similar pigmentation intensity of the infiltrated sites (Figure 3A), and in the quantification of anthocyanin concentration using HPLC with PDA detector (Figure 3B). An additional experiment was conducted using co-infiltration of VcMYBA1 and the apple MdCHS2, which has been previously functionally characterised (Dare et al., 2013). However, transient expression of MdCHS2 did not significantly change anthocyanin production either compared with the VcMYBA1 control (p-values = 0.969, Figure 3C). This indicated that neither CHS led to increased anthocyanin production compared with the VcMYBA1 control in N. benthamiana. Similarly, VcUFGT2 + VcMYBA1 infiltration resulted in a 1.3 times higher mean anthocyanin concentration (2.43 ± 0.28 mg/g DW, Figure 3B) compared to with the VcMYBA1 control. However, this was not a significant increase (p-value = 0.906). In contrast, infiltration of VmANS + VcMYBA1 resulted in a significant 3.57-fold increase in total anthocyanin compared with the VcMYBA1 control samples with 6.08 ± 1.20 of mg total anthocyanin/g DW produced (p-value < 10-4).
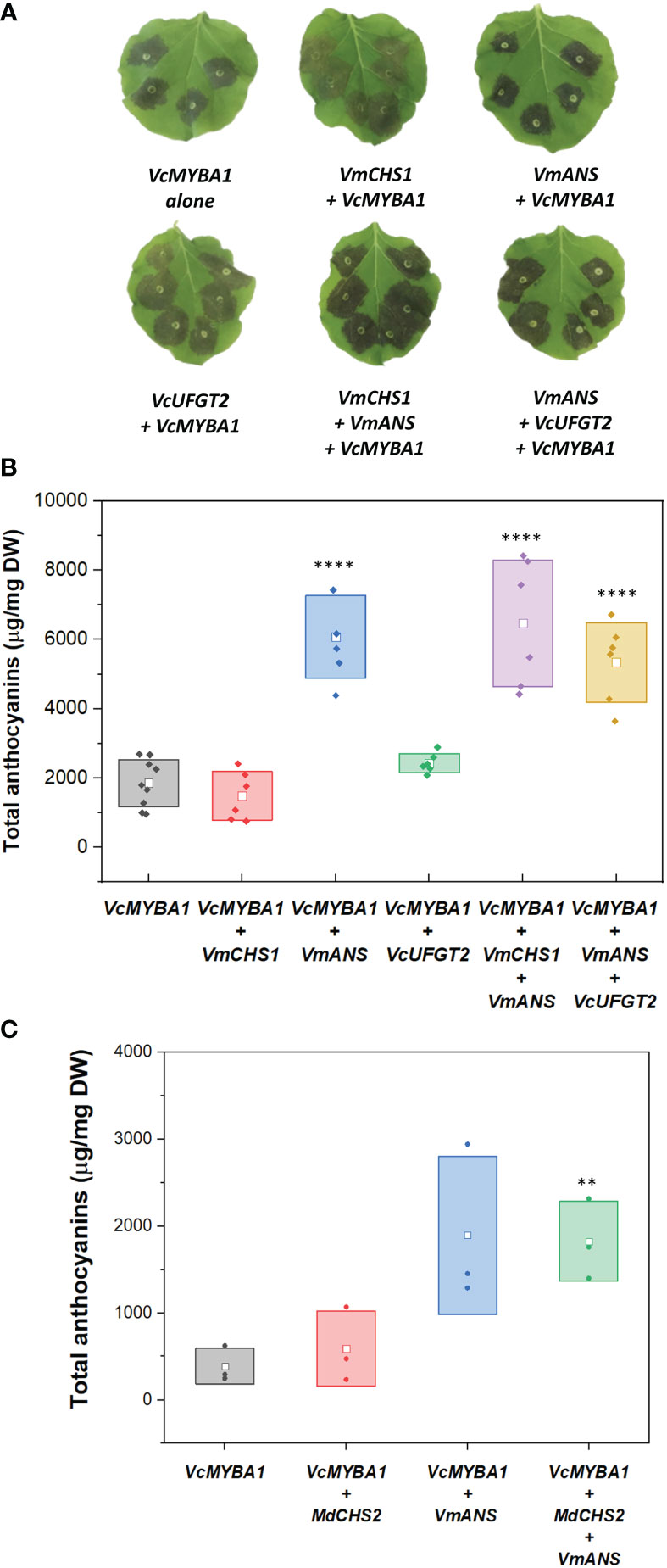
Figure 3 (A) Images of N. benthamiana leaves infiltrated with different combinations of VcMYBA1, VmCHS1, VmANS, and VcUFGT2. The combinations of genes used are indicated below each leaf image. Images were taken at 5 days post-infiltration. (B) Total anthocyanins from leaves infiltrated with different gene combinations. (C) Total anthocyanins from leaves infiltrated with MdCHS2. Filled dots represent data points from each biological replicate. Empty squares represent the mean concentration value of each treatment. Boxes represent the mean ± one SD from at least three biological replicates. Statistical analysis from pairwise comparison with VcMYBA1 control samples (Student’s t-test, N ≥ 3, α = 0.05), p-value < 0.005 is indicated by **; p-value < 10-4 is indicated by ****.
Co-infiltrations of different candidate genes with VmANS were tested to evaluate possible combination effects on anthocyanin production. When VmANS was co-infiltrated with either VmCHS1 or VmUFGT2, there was a significant increase (p-value < 10-4) in anthocyanin production when compared with the VcMYBA control (Figure 3B). In particular, combinations with VmANS (i.e. VcMYBA1 + VmCHS1 + VmANS and VcMYBA1 + VmANS + VcUFGT2) resulted in anthocyanin concentrations of 5.34 ± 1.13 mg/g DW and 6.47 ± 1.20 mg/g DW, respectively. However, there was no significant difference between the two combinations (p-value = 0.46). Therefore, the same trend as single candidate gene infiltrations regarding changes in anthocyanin concentration was observed when different gene combinations were tested. This suggested that VmANS was the only candidate gene with a strong effect in increasing anthocyanin concentration in this assay. These results reflected the visual evaluation of infiltration sites, which were darker in colour compared to that of the VcMYBA control and VmCHS1/VcUFGT2 infiltrations (Figure 3A). Co-infiltration of VmCHS1, VmANS and VcUFGT2 with other candidate genes without VcMYBA was also performed, but no anthocyanin was produced (Supplementary Figure 3).
Interestingly, while overexpression of VcMYBA1, VmCHS1 or VmANS resulted in the biosynthesis of a single type of anthocyanin (delphinidin-3-O-rutinoside/delphinidin-3-O-glucorhamnoside), overexpression of VcMYBA1 with VcUFGT2 or VcUFGT2 + VmANS resulted in an additional anthocyanin peak, which was identified as delphinidin-3-O-galactorhamnoside using LC-MS (Figure 4). This supports a primary function for UFGT2 in modulating anthocyanin composition, instead of concentration, in N. benthamiana and suggests a high affinity for utilising UDP-galactose as substrate.
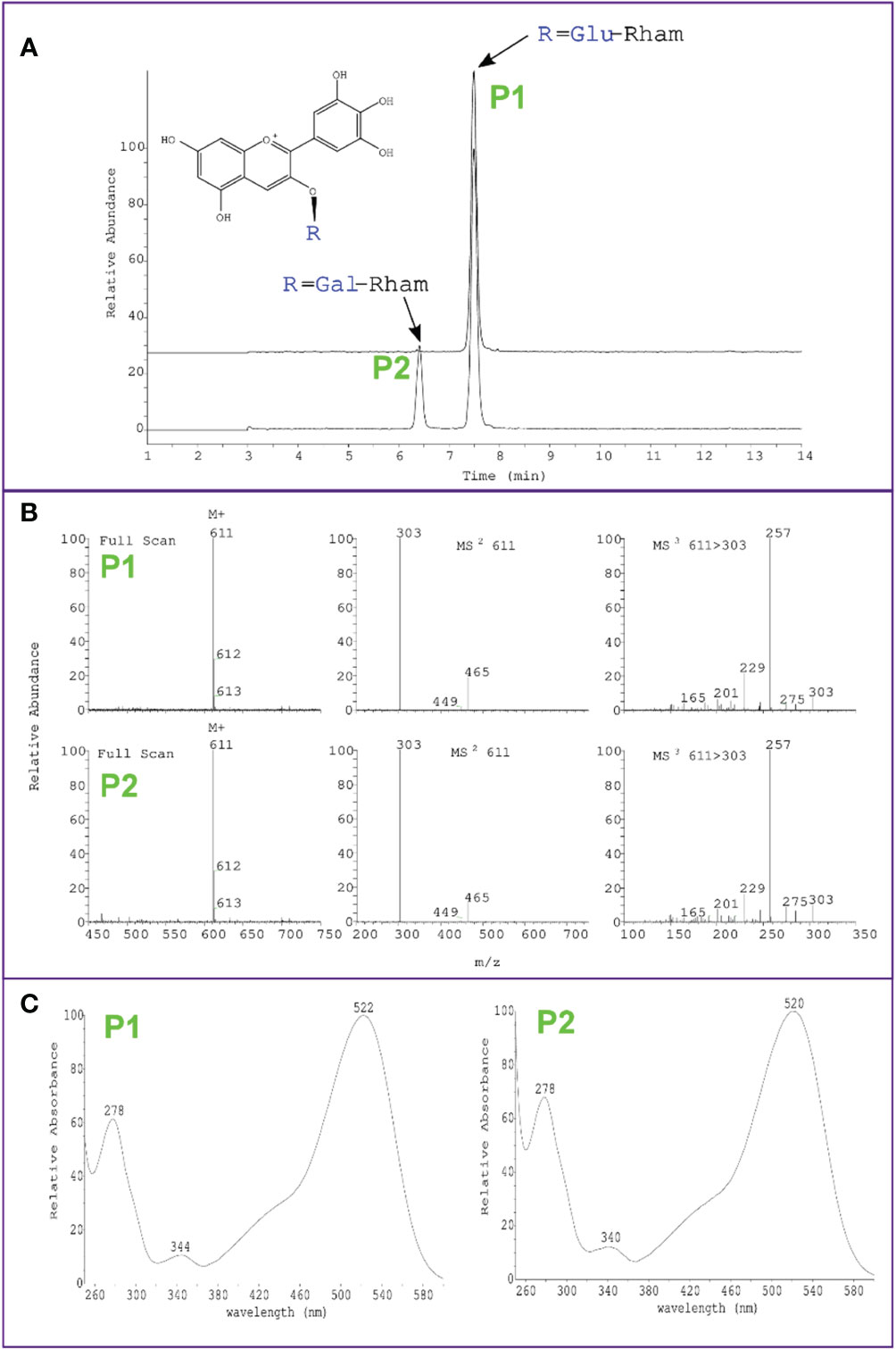
Figure 4 Identification of delphinidin-3-O-galactorhamnoside production in N. benthamiana leaves using Agrobacterium-mediated transient overexpression of VcMYBA1 + VcUFGT2. (A) Chromatogram showing peak detection at 530 nm and earlier retention time of delphinidin-3-O-galactorhamnoside (P2) compared with delphinidin-3-O-rutinoside (P1) of VcMYBA1 + VcUFGT2 (bottom trace) in comparison to VcMYBA1 + VmANS as control (top trace). (B) Mass spectra of positive ion mode full scan MS, MS2 and MS3 fragmentation of the respective selected main ion, showing loss of rhamnose and hexose in MS2 and fragmentation of the delphinidin aglycone in MS3. Molecular weight and fragmentation spectra are identical for P1 and P2, indicating that both anthocyanins have the same chemical formula and are therefore stereoisomers. (C) Full UV-Vis spectral scan, showing typical absorption spectrum of delphinidin for both compounds.
UFGT2 alters the composition of anthocyanin produced
As observed in Figure 4, overexpression of VcUFGT2 in N. benthamiana resulted in the production of an additional anthocyanin linked with galactorhamnoside, which is not naturally occurring in either Nicotiana or Vaccinium. The types of anthocyanins produced are likely dependent on both the affinity of the glycosyltransferase and the availability of the respective anthocyanidin and UDP-sugar substrates (Montefiori et al., 2011). To further investigate the role of VcUFGT2 in inducing the anthocyanin profile common to blueberry and bilberry in a fruit system, ‘Camarosa’ strawberry was chosen as a fruit transient overexpression system. Ripe ‘Camarosa’ strawberry naturally produces only two types of anthocyanins, pelargonidin-3-O-glucoside as the major compound (95%) and cyanidin-3-O-glucoside as the minor anthocyanin (5%). Preliminary experiments showed that overexpression of VcMYBA1 in strawberry fruit significantly increased the amount of cyanidin-3-O-glucoside to up to 35% total anthocyanins (Supplemental Figure 5).
In contrast to blueberries and N. benthamiana, strawberries do not produce delphinidin-based (blue) pigments due to the absence of FLAVONOID 3´5´ HYDROXYLASE (Song et al., 2015). To functionally test the substrate specificity of VcUFGT2 with respect to the anthocyanidin and UDP-sugar used for biosynthesis in fruit, we aimed to modify the chemical environment in strawberry fruit to include trihydroxylated anthocyanidins. This was achieved by the transient overexpression of VcMYBA1 with an active kiwifruit (A. melanandra) AmF3´5´H (Peng et al., 2019) (Supplementary Figure 5) resulting in the production of delphinidin-3-O-glucoside, which is not naturally present in strawberry due to the absence of this enzyme. Additionally, modified strawberry produced dark-pigmented patches as a result of delphinidin being produced (Figure 5A), thus, providing a visual cue for sampling transformed tissues.
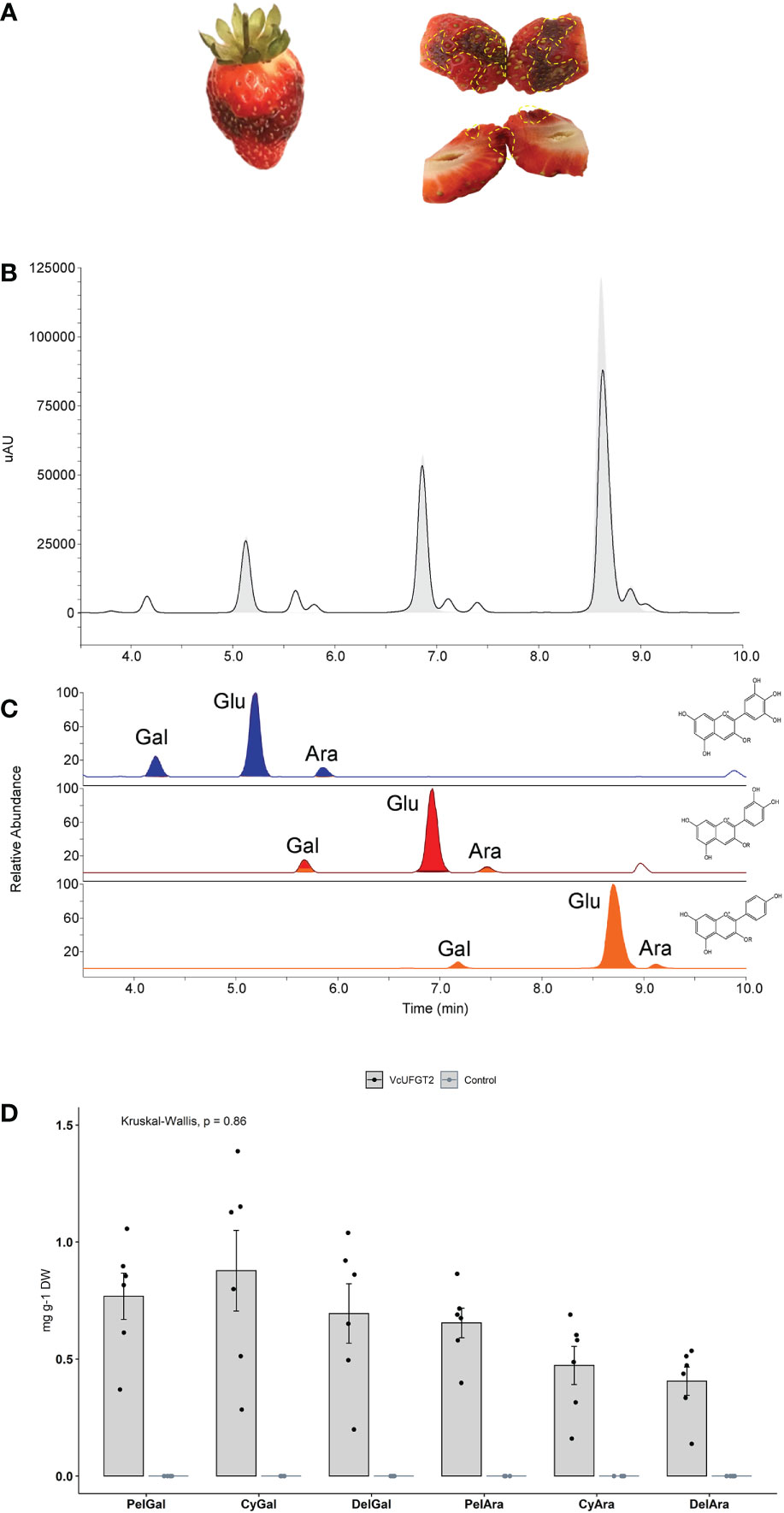
Figure 5 Anthocyanin profile of strawberry fruit overexpressing VcUFGT2 in a VcMYBA1 and AmF3´5´H–co-expression background. (A) Representative images of whole (left) and bisected (right) strawberry fruit infiltrated with VcMYBA1 + AmF3´5´H. Dotted yellow lines highlight transformed tissue showing darken colour that was sampled for further analysis. (B) Chromatogram indicating retention times of compounds from control (grey fill) and VcUFGT2-infiltrated (not filled) fruit at 530 nm. (C) Retention times of MS transitions for delphinidin- (blue; 465, 435), cyanidin- (red; 449, 419) and pelargonidin-derived (orange; 433, 403) anthocyanins. Gal: Galactoside; Glu: Glucoside; Ara: Arabinoside. (D) Bargraph visualising mean concentration ± standard deviation (N=6) of anthocyanidin-3-O-galactosides and -arabinosides following strawberry transient transformation 5 days after infiltration. While these anthocyanins were not detected in control samples, concentrations of individual anthocyanins were not significantly different in VcUFGT2-infiltrated fruit. Pel, Pelargonidin; Cy, Cyanidin; Del, Delphinidin.
The total amount of anthocyanins and that of the primary anthocyanidin-3-O-glucosides did not change significantly in response to VcUFGT2 overexpression in a VcMYBA1 and AmF3´5´H–co-expression background (Supplementary Figure 6). However, six additional anthocyanins were produced, linking each of the respective anthocyanidins to either galactoside or arabinoside (Figure 5). Thus, VcUFGT2 was confirmed in a fruit system to utilise UDP-galactose and UDP-arabinose as substrates to stabilise a range of anthocyanidins as precursors, including pelargonidin, which is not naturally present in blueberry.
Discussion
In this study, we characterised three genes, CHS1, ANS and UFGT2, from the Vaccinium flavonoid pathway leading to anthocyanin biosynthesis, and assessed their role in modulating anthocyanin production both individually and in combination. qRT-PCR was performed for the fruit skin and flesh tissue of blueberry and bilberry, which showed highest expression of CHS1, ANS, UFGT2 and the TF MYBA1 in anthocyanin-rich skin of ripe fruit. The expression of these genes also corresponded to anthocyanin accumulation in both Vaccinium species as there was very low expression in blueberry flesh, but substantially higher in bilberry flesh. Additional non-candidate structural genes from the flavonoid pathway (CHS2, F3H1 and DFR) included in the analysis, did not exhibit such pattern. Markedly, despite being an isoform of Vaccinium CHS1, CHS2 expression was low in both tissue types throughout fruit development in blueberry and bilberry. DFR expression was high even in the non-pigmented blueberry fruit flesh, likely associated with proanthocyanidin biosynthesis (Lafferty et al., 2022b), while F3H1 was less expressed in bilberry fruit skin and flesh compared with the candidate genes and VcMYBA1. This result confirmed the expression patterns previously reported from RNA-Seq (Günther et al., 2020), which correlated transcript abundance of the three genes to anthocyanin accumulation over fruit development in blueberry fruit skin and flesh. Analysis of the expression of some flavonoid genes had previously been performed on whole bilberry fruit as well as fruit skin and flesh showing their correlation with anthocyanin accumulation (Jaakola et al., 2002; Lafferty et al., 2022b). Our experiments using qRT-PCR further confirmed the connection between the expression of CHS1, ANS, UFGT2 and anthocyanin accumulation in Vaccinium fruit. Moreover, the exact same primers were used for both bilberry and blueberry, showing that conserved genes were amplified between the two species.
Phylogenetic analysis showed that CHS1, ANS and UFGT2 predicted protein sequences are highly conserved between blueberry and bilberry, and clustered well with related protein sequences from other plant species, in particular with fruit from the same taxonomic order (Ericales). For all three genes, bilberry and blueberry nucleotide and amino acid sequences showed high sequence similarity (95–99%), which validated the use of the same candidate gene sequences for functional characterisation from both Vaccinium species. The active sites of all three genes are highly conserved among anthocyanin-rich fruit species, supporting their relevance in anthocyanin production.
Overexpression of Vaccinium R2R3 MYBA-type TFs in N. benthamiana leaves induced the expression of endogenous anthocyanin biosynthetic genes which led to anthocyanin production (Karppinen et al., 2021; Lafferty et al., 2022b). Hence, the infiltration of low concentration of Agrobacterium carrying VcMYBA1 in N. benthamiana leaves was successfully employed in our experiments to induce a base level of anthocyanin production, which can be elevated through co-infiltration of candidate structural genes.
Transient overexpression in N. benthamiana leaves using both the bilberry VmCHS1 and apple MdCHS2 did not enhance anthocyanin production above endogenous tobacco CHS that might be induced by VcMYBA, suggesting that CHS is not a rate-limiting step in in this testing system. However, it is well-known that expression of CHS is critical for the production of anthocyanins and flavonoids in general. Downregulation/silencing of CHS has resulted in abolished or decreased anthocyanin accumulation in many plant species, including tobacco (Wang et al., 2006), kiwifruit (Sun et al., 2014), strawberry (Hoffmann et al., 2006), gentian (Nakatsuka et al., 2008; Ohta et al., 2022) and dahlia (Ohno et al., 2011). Especially, MdCHS2-KO transgenic apple lines had no detectable anthocyanins, together with reduced flavonoid accumulation and other changes in plant development (Dare et al., 2013). In our assay, co-infiltration with low amounts of VcMYBA1 was required to initiate anthocyanin production. VcMYBA1 has previously been found to control the activation of anthocyanin structural genes, such as DFR, ANS and UFGT (Plunkett et al., 2018; Lafferty et al., 2022a).
In contrast to VmCHS1, VmANS was shown to significantly increase anthocyanin production by 3 to 3.5 times using in vivo expression in N. benthamiana. This demonstrated a key role of ANS as a rate-limiting step for anthocyanin accumulation in N. benthamiana. ANS had previously been identified as a key enzyme for anthocyanin production in potato tuber (Zhang et al., 2020), Salvia miltiorrhiza (Li et al., 2019), apple (Szankowski et al., 2009), Medicago (Pang et al., 2007) and cotton (Long et al., 2018). Here, we showed that, of the tested candidate gene combinations, only ANS had a significant effect on anthocyanin production and this is in contrast to a number of previous studies showing that it was the combined expression of ANS with other structural genes, including UFGT and CHS, that correlated with increased anthocyanin production. For instance, in pink-flowered strawberry (Fragaria × Potentila), FpANS and two glycosyltransferases (FpBZ1 and FpUGT75C1) were found to be key factors in modulating anthocyanin accumulation in flower petals (Xue et al., 2019). Similarly, upregulation of both VvANS and VvUFGT in grape were found to correlate with increased anthocyanin accumulation (Yang et al., 2021). In the case of our study, the emphasis on the function of an individual gene, namely VmANS, to achieve a significantly higher anthocyanin concentration might be due to the co-expression to a low degree of VcMYBA1. Overexpression of bilberry VmMYBA1 has been shown to induce all endogenous N. benthamiana anthocyanin biosynthetic genes (Karppinen et al., 2021). Similarly, the low expression of VcMYBA1 in our optimised N. benthamiana transient assay allowed for the production of anthocyanins via the expression of native structural anthocyanin genes, for example, the stabilisation of anthocyanins by NbUFGT. Hence, substrates for anthocyanin production were not competed with by other pathways such as the proanthocyanin one. Indeed, when ANR, an proanthocyanin structural gene, was silenced in grape and strawberry, expression of ANS and anthocyanin accumulation increased (Fischer et al., 2014; Yang et al., 2021). Nevertheless, ANS expression was found to associate with anthocyanin accumulation in all aforementioned studies, which further supports the finding that ANS plays a key role in anthocyanin accumulation in combination with MYBA1. Intriguingly, both our qRT-PCR results and previous RNA-Seq data (Lafferty et al., 2022b) showed low, but not total absence of VcMYBA1 and VcANS expression in blueberry flesh despite the tissue not producing anthocyanins and similar observations have been reported for red-skinned but anthocyanin-deficient Muscat Hamburg grape (Xie et al., 2015). As overexpression of VmANS was able to elevate anthocyanin production in a low VcMYBA1 expression background in N. benthamiana leaves, it will be insightful to study fruit flesh anthocyanin production in stable ANS- as well as MYBA1- overexpressing blueberry plants to see if flesh colour can be induced. Evidently both genes are good candidates to evaluate as molecular markers for flesh colour in Vaccinium spp.
Our infiltration system showed that VcUFGT2 is important for modulating anthocyanin composition by modifying the sugar moiety of cyanidin, delphinidin and pelargonidin. Therefore, VcUFGT2 can utilise anthocyanidins as substrate and can be referred to as an anthocyanindin 3-O glycosyltransferase. Kubo et al. (2004) suggested that preference for UDP-glucose and UDP-galactose can be ascribed to the last amino acid residue of the PSPG motif, dependent on whether this is a glutamine or a histidine, respectively. Various characterised glucosyltransferases and galactosyltransferases follow this pattern such as residue Q382 of the Arabidopsis glucosyltransferase AtUGT78D2 (Kubo et al., 2007) and residue H375 of the kiwifruit galactosyltransferase AcF3GT1 (Montefiori et al., 2011). As both VmUFGT2 and VcUFGT2 have a histidine at this position (H376 and H378, respectively), it is likely that they can utilise UDP-galactose as a sugar donor. This is consistent with transient assay results where the production of additional, not naturally occurring galactosylated anthocyanins were observed following overexpression of VcUFGT2 in N. benthamiana leaves and strawberry fruit. Phylogenetic analysis of the UFGT sequences from other plant species showed that most UFGT amino acid sequences from blueberry and bilberry cluster well with other UFGT sequences from the order Ericales, except for VcUFGT3 and VcUFGT5. Interestingly, both the latter two have a glutamine at the conserved last amino acid of the PSPG motif. This suggests that these two enzymes may have a higher affinity for UDP-glucose as their sugar donor, unlike Vc/VmUFGT2. This amino acid residue, however, may not fully determine the sugar donor specificity of a glycosyltransferase as suggested in studies on freesia Fh3GT1, grape VvGT1 and kiwifruit AcF3GT1 (Offen et al., 2006; Montefiori et al., 2011; Sun et al., 2016). For instance, though VvGT1 was shown to accept both glucose and galactose as sugar donor, 200-fold reduced catalytic activity was seen when galactose was used as substrate (Offen et al., 2006). Indeed, the presence of both additional galactosylated and arabinosylated anthocyanins without trade-off in content of glucosylated anthocyanins in strawberry fruit suggested that galactose is not the sole sugar donor accepted by VcUFGT2. Glycosyltransferases can also catalyse the transfer of UDP-sugars to the sugar moiety of anthocyanidin glycosides (GGTs). Such enzymes have previously been characterised in tobacco and kiwifruit (Montefiori et al., 2011; Yonekura‐Sakakibara et al., 2012). Considering our transient assay results and the anthocyanin profile of blueberry, it is unlikely for VcUFGT2 to have GGT activity. Anthocyanidin-3-O-glucosides are a major glycoside group in blueberry, although proportions were shown to greatly differ between species with galactosides representing the primary sugar moiety in anthocyanins from Rabbiteye (Günther et al., 2020; Mengist et al., 2020). Hence, it is likely that VcUFGT2 can also accept glucose as sugar donor, although different isoforms, such as VcUFGT3 and VcUFGT5, may also be responsible for glycosylation of glucose. Because the in vivo assays used to characterise VcUFGT2 in this study was not suitable for assessing VcUFGT2 affinity to UDP-glucose, future work employing in vitro enzyme assays or an in vivo system using UFGT-KO strawberry would be needed for this. Regardless, our development of the strawberry transient system overexpressing both VcMYBA and AmF3´5´H was shown to be a fast and high throughput system useful for in vivo qualitative functional test.
A Vaccinium fruit transient assay system such as the non-anthocyanic blueberry flesh, would be ideal for verifying the functionality of Vaccinium anthocyanin structural genes, particularly for ANS. However, this has been proven difficult due to stress-induced anthocyanin biosynthesis when blueberry fruit is injected with Agrobacterium during the transient expression process (Günther et al., 2022). Nonetheless, our experimental findings for the function of VmANS and especially, VcUFGT2 in modulating anthocyanin concentrations in heterologous transient expression systems (N. benthamiana leaf and strawberry fruit) suggested fundamental key roles for these enzymes in the biosynthesis pathway. Further research on these target enzymes may provide an alternate strategy for the production of specific anthocyanin products. Potentially, in addition to important anthocyanin-related MYB TFs (e.g. MYBA1), ANS and UFGT may also be co-targeted to fine-tune high-quality blueberry cultivars with superior anthocyanin profiles.
In conclusion, we have shown that VmANS activity is a rate-limiting step within the flavonoid pathway, modulating the anthocyanin concentration in plantae. Co-expression with either VmCHS or VcUFGT2 did not further increase anthocyanin content. VcUFGT2 played a critical role in modulating anthocyanin composition via the modification of the sugar moiety. VcUFGT2 also demonstrated affinity for both galactose and arabinose as sugar donor. Our study provided evidence for the key roles of these two final steps in the Vaccinium anthocyanin biosynthesis pathway and contributed a new breeding target for novel blueberry cultivars.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
HN performed the experimental work, data analysis and wrote the manuscript. CG cloned VcUFGT2, contributed to data analysis and writing of the manuscript. YP cloned and performed functional characterisation of AmF3´5´H. CG and JC performed LTQ LC-MS experiments and data analysis. ES assisted with the cloning and bioinformatic analysis of VcUFGT2. BP and AD assisted with cloning of VmCHS1 and VmANS. NA contributed in figure generation and editing. CG conceptualised the study and supervised all aspects with RE and JP. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the New Zealand Ministry of Business, Innovation, and Employment contract C11X1704 “Filling the Void: boosting the nutritional content of New Zealand fruit.”
Acknowledgments
We thank Ross Atkinson and Andrew Allan for constructive feedback on the manuscript. We appreciate Niels Nieuwenhuizen, Simona Nardozza, Rubina Jibran and Jibran Tahir for the help with experiments; Yar-khing Yauk for providing Agrobacterium stock carrying MdCHS2. We appreciate Heather Jenkins for consultation of the statistical analyses. We further thank Monica Dragulescu and her team for maintenance of plant materials in the glasshouse.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1082246/full#supplementary-material
References
Allan, A. C., Hellens, R. P., Laing, W. A. (2008). MYB transcription factors that colour our fruit. Trends Plant Sci. 13, 99–102. doi: 10.1016/j.tplants.2007.11.012
André, C. M., Schafleitner, R., Legay, S., Lefèvre, I., Aliaga, C., Nomberto, G., et al. (2009). Gene expression changes related to the production of phenolic compounds in potato tubers grown under drought stress. Phytochemistry 70, 1107–1116. doi: 10.1016/j.phytochem.2009.07.008
Benjamini, Y., Yekutieli, D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Stat 29(4):1165–1188. doi: 10.1214/aos/1013699998
Berardini, T. Z., Reiser, L., Li, D., Mezheritsky, Y., Muller, R., Strait, E., et al. (2015). The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 53, 474–485. doi: 10.1002/dvg.22877
Camire, M. E. (2002). Phytochemicals in the Vaccinium family: Bilberries, blueberries, and cranberries. In: Meskin, M. S., Bidlack, W. R., Davies, A. J., Omaye, S. T., Phytochemicals in Nutrition and Health. Boca Raton, FL: CRC Press 19–40. doi: 10.1201/9781420031690.ch3
Castañeda-Ovando, A., Pacheco-Hernández, M. D. L., Páez-Hernández, M. E., Rodríguez, J. A., Galán-Vidal, C. A. (2009). Chemical studies of anthocyanins: A review. Food Chem. 113, 859–871. doi: 10.1016/j.foodchem.2008.09.001
Chen, C. (2015). “Overview of plant pigments,” in Pigments in fruits and vegetables (New York, NY: Springer), 1–7.
Chu, W.-K., Cheung, S. C. M., Lau, R., Benzie, I. F. F. (2011). Bilberry (Vaccinium myrtillus l.). Herbal Med. 20115386, 55–71. doi: 10.1201/b10787-5
Colle, M., Leisner, C. P., Wai, C. M., Ou, S., Bird, K. A., Wang, J., et al. (2019). Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry. GigaScience 8, giz012. doi: 10.1093/gigascience/giz012
Dare, A. P., Günther, C. S., Grey, A. C., Guo, G., Demarais, N. J., Cordiner, S., et al. (2022). Resolving the developmental distribution patterns of polyphenols and related primary metabolites in bilberry (Vaccinium myrtillus) fruit. Food Chem. 374, 131703. doi: 10.1016/j.foodchem.2021.131703
Dare, A. P., Tomes, S., Jones, M., Mcghie, T. K., Stevenson, D. E., Johnson, R. A., et al. (2013). Phenotypic changes associated with RNA interference silencing of chalcone synthase in apple (Malus × domestica). Plant J. 74, 398–410. doi: 10.1111/tpj.12140
Die, J. V., Jones, R. W., Ogden, E. L., Ehlenfeldt, M. K., Rowland, L. J. (2020). Characterization and analysis of anthocyanin-related genes in wild-type blueberry and the pink-fruited mutant cultivar ‘Pink lemonade’: New insights into anthocyanin biosynthesis. Agronomy 10(9), 1296–1314. doi: 10.3390/agronomy10091296
Edger, P. P., Vanburen, R., Colle, M., Poorten, T. J., Wai, C. M., Niederhuth, C. E., et al. (2018). Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. Gigascience 7, gix124. doi: 10.1093/gigascience/gix124
Enrico, E. (2007). The French–Italian public consortium for grapevine genome characterization the grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467.
Espley, R. V., Bovy, A., Bava, C., Jaeger, S. R., Tomes, S., Norling, C., et al. (2013). Analysis of genetically modified red-fleshed apples reveals effects on growth and consumer attributes. Plant Biotechnol. J. 11, 408–419. doi: 10.1111/pbi.12017
Espley, R. V., Leif, D., Plunkett, B., Mcghie, T., Henry-Kirk, R., Hall, M., et al. (2019). Red to brown: An elevated anthocyanic response in apple drives ethylene to advance maturity and fruit flesh browning. Front. Plant Science. 10, 1248. doi: 10.3389/fpls.2019.01248
Fang, J. (2015). Classification of fruits based on anthocyanin types and relevance to their health effects. Nutrition 31, 1301–1306. doi: 10.1016/j.nut.2015.04.015
Fischer, T. C., Mirbeth, B., Rentsch, J., Sutter, C., Ring, L., Flachowsky, H., et al. (2014). Premature and ectopic anthocyanin formation by silencing of anthocyanidin reductase in strawberry (Fragaria × ananassa). New Phytol. 201, 440–451. doi: 10.1111/nph.12528
Furtek, D., Schiefelbein, J. W., Johnston, F., Nelson, O. E. (1988). Sequence comparisons of three wild-type Bronze-1 alleles from Zea mays. Plant Mol. Biol. 11, 473–481. doi: 10.1007/BF00039028
Günther, C. S., Dare, A. P., Mcghie, T. K., Deng, C., Lafferty, D. J., Plunkett, B. J., et al. (2020). Spatiotemporal modulation of flavonoid metabolism in blueberries. Front. Plant Science. 11, 545. doi: 10.3389/fpls.2020.00545
Günther, C. S., Plunkett, B. J., Cooney, J., Jensen, D., Trower, T., Elborough, C., et al. (2022). Biotic stress-induced and ripening-related anthocyanin biosynthesis are regulated by alternate phytohormone signals in blueberries. Environ. Exp. Bot. 105065.
Hellens, R. P., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 1–14. doi: 10.1186/1746-4811-1-13
Hellens, R. P., Edwards, E. A., Leyland, N. R., Bean, S., Mullineaux, P. M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. doi: 10.1023/A:1006496308160
Hoffmann, T., Kalinowski, G., Schwab, W. (2006). RNAi-induced silencing of gene expression in strawberry fruit (Fragaria × ananassa) by agroinfiltration: A rapid assay for gene function analysis. Plant J. 48, 818–826. doi: 10.1111/j.1365-313X.2006.02913.x
Hughes, J., Hughes, M. A. (1994). Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta crantz) cotyledons. DNA sequence 5, 41–49. doi: 10.3109/10425179409039703
Ichiyanagi, T., Shida, Y., Rahman, M. M., Hatano, Y., Konishi, T. (2006). Bioavailability and tissue distribution of anthocyanins in bilberry (Vaccinium myrtillus l.) extract in rats. J. Agric. Food Chem. 54, 6578–6587. doi: 10.1021/jf0602370
Jaakola, L., Määttä, K., Pirttilä, A. M., Törrönen, R., Kärenlampi, S., Hohtola, A. (2002). Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol. 130, 729–739. doi: 10.1104/pp.006957
Jez, J. M., Ferrer, J. L., Bowman, M. E., Austin, M. B., Schröder, J., Dixon, R. A., et al. (2001). Structure and mechanism of chalcone synthase-like polyketide synthases. J. Ind. Microbiol. Biotechnol. 27, 393–398. doi: 10.1038/sj.jim.7000188
Jia, K., Zhang, Q., Xing, Y., Yan, J., Liu, L., Nie, K. (2020). A development-associated decrease in osmotic potential contributes to fruit ripening initiation in strawberry (Fragaria ananassa). Front. Plant Science. 11, 1035. doi: 10.3389/fpls.2020.01035
Jimenez-Garcia, S. N., Guevara-Gonzalez, R. G., Miranda-Lopez, R., Feregrino-Perez, A. A., Torres-Pacheco, I., Vazquez-Cruz, M. A. (2013). Functional properties and quality characteristics of bioactive compounds in berries: Biochemistry, biotechnology, and genomics. Food Res. Int. 54, 1195–1207. doi: 10.1016/j.foodres.2012.11.004
Jun, J. H., Xiao, X., Rao, X., Dixon, R. A. (2018). Proanthocyanidin subunit composition determined by functionally diverged dioxygenases. Nat. Plants 4, 1034–1043. doi: 10.1038/s41477-018-0292-9
Kader, F., Rovel, B., Girardin, M., Metche, M. (1996). Fractionation and identification of the phenolic compounds of highbush blueberries (Vaccinium corymbosum, l.). Food Chem. 55, 35–40. doi: 10.1016/0308-8146(95)00068-2
Kalt, W., Cassidy, A., Howard, L. R., Krikorian, R., Stull, A. J., Tremblay, F., et al. (2020). Recent research on the health benefits of blueberries and their anthocyanins. Adv. Nutr. 11, 224–236. doi: 10.1093/advances/nmz065
Kalt, W., Mcdonald, J. E., Ricker, R. D., Lu, X. (1999). Anthocyanin content and profile within and among blueberry species. Can. J. Plant Sci. 79, 617–623. doi: 10.4141/P99-009
Karppinen, K., Lafferty, D. J., Albert, N. W., Mikkola, N., Mcghie, T., Allan, A. C., et al. (2021). MYBA and MYBPA transcription factors co-regulate anthocyanin biosynthesis in blue-coloured berries. New Phytol. 232, 1350–1367. doi: 10.1111/nph.17669
Karppinen, K., Zoratti, L., Sarala, M., Carvalho, E., Hirsimäki, J., Mentula, H., et al. (2016). Carotenoid metabolism during bilberry (Vaccinium myrtillus l.) fruit development under different light conditions is regulated by biosynthesis and degradation. BMC Plant Biol. 16, 1–16. doi: 10.1186/s12870-016-0785-5
Katsumoto, Y., Fukuchi-Mizutani, M., Fukui, Y., Brugliera, F., Holton, T. A., Karan, M., et al. (2007). Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 48, 1589–1600. doi: 10.1093/pcp/pcm131
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Khoo, H. E., Azlan, A., Tang, S. T., Lim, S. M. (2017). Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 61, 1–21. doi: 10.1080/16546628.2017.1361779
Korkmaz, S., Göksülük, D., Zararsiz, G. (2014). MVN: An r package for assessing multivariate normality. R Journal. 6 (2), 151–162. doi: 10.32614/RJ-2014-031
Kubo, A., Arai, Y., Nagashima, S., Yoshikawa, T. (2004). Alteration of sugar donor specificities of plant glycosyltransferases by a single point mutation. Arch. Biochem. Biophysics 429, 198–203. doi: 10.1016/j.abb.2004.06.021
Kubo, H., Nawa, N., Lupsea, S. A. (2007). Anthocyaninless1 gene of Arabidopsis thaliana encodes a UDP-glucose: flavonoid-3-O-glucosyltransferase. J. Plant Res. 120, 445–449. doi: 10.1007/s10265-006-0067-7
Lafferty, D. J., Espley, R. V., Deng, C. H., Dare, A. P., Günther, C. S., Jaakola, L., et al. (2022a). The coordinated action of MYB activators and repressors controls proanthocyanidin and anthocyanin biosynthesis in Vaccinium. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.910155
Lafferty, D. J., Espley, R. V., Deng, C. H., Günther, C. S., Plunkett, B., Turner, J. L., et al. (2022b). Hierarchical regulation of MYBPA1 by anthocyanin-and proanthocyanidin-related MYB proteins is conserved in Vaccinium species. J. Exp. Bot. 73, 1344–1356. doi: 10.1093/jxb/erab460
Landi, M., Tattini, M., Gould, K. S. (2015). Multiple functional roles of anthocyanins in plant-environment interactions. Environ. Exp. Bot. 119, 4–17. doi: 10.1016/j.envexpbot.2015.05.012
Letunic, I., Bork, P. (2021). Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, H., Liu, J., Pei, T., Bai, Z., Han, R., Liang, Z., et al. (2019). Overexpression of SmANS enhances anthocyanin accumulation and alters phenolic acids content in Salvia miltiorrhiza and Salvia miltiorrhiza alba plantlets. Int. J. Mol. Sci. 20, 2225. doi: 10.3390/ijms20092225
Liu, W., Feng, Y., Yu, S., Fan, Z., Li, X., Li, J., et al. (2021a). The flavonoid biosynthesis network in plants. Int. J. Mol. Sci. 22, 12824. doi: 10.3390/ijms222312824
Liu, Y., Tikunov, Y., Schouten, R. E., Marcelis, L. F. M., Visser, R. G. F., Bovy, A. (2018). Anthocyanin biosynthesis and degradation mechanisms in solanaceous vegetables: A review. Front. Chem. 6, 52. doi: 10.3389/fchem.2018.00052
Liu, W., Zheng, T., Yang, Y., Li, P., Qiu, L., Li, L., et al. (2021b). Meta-analysis of the effect of overexpression of MYB transcription factors on the regulatory mechanisms of anthocyanin biosynthesis. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.781343
Long, L., Zhao, J.-R., Xu, F.-C., Yang, W.-W., Liao, P., Gao, Y., et al. (2018). Silencing of GbANS reduces cotton resistance to Verticillium dahliae through decreased ROS scavenging during the pathogen invasion process. Plant Cell Tissue Organ Culture (PCTOC) 135, 213–221. doi: 10.1007/s11240-018-1457-y
Lunkenbein, S., Bellido, M., Aharoni, A., Salentijn, E. M., Kaldenhoff, R., Coiner, H. A., et al. (2006). Cinnamate metabolism in ripening fruit. characterization of a UDP-glucose: cinnamate glucosyltransferase from strawberry. Plant Physiol. 140, 1047–1058. doi: 10.1104/pp.105.074955
Ma, L., Sun, Z., Zeng, Y., Luo, M., Yang, J. (2018). Molecular mechanism and health role of functional ingredients in blueberry for chronic disease in human beings. Int. J. Mol. Sci. 19, 2785. doi: 10.3390/ijms19092785
Mcghie, T. K., Ainge, G. D., Barnett, L. E., Cooney, J. M., Jensen, D. J. (2003). Anthocyanin glycosides from berry fruit are absorbed and excreted unmetabolized by both humans and rats. J. Agric. Food Chem. 51, 4539–4548. doi: 10.1021/jf026206w
Mengist, M. F., Grace, M. H., Xiong, J., Kay, C. D., Bassil, N., Hummer, K., et al. (2020). Diversity in metabolites and fruit quality traits in blueberry enables ploidy and species differentiation and establishes a strategy for future genetic studies. Front. Plant Sci. 11, 370. doi: 10.3389/fpls.2020.00370
Milbury, P. E., Vita, J. A., Blumberg, J. B. (2010). Anthocyanins are bioavailable in humans following an acute dose of cranberry juice. J. Nutr. 140, 1099–1104. doi: 10.3945/jn.109.117168
Miller, K. D., Guyon, V., Evans, J. N., Shuttleworth, W. A., Taylor, L. P. (1999). Purification, cloning, and heterologous expression of a catalytically efficient flavonol 3-o-galactosyltransferase expressed in the male gametophyte of Petunia hybrida. J. Biol. Chem. 274, 34011–34019. doi: 10.1074/jbc.274.48.34011
Montefiori, M., Espley, R. V., Stevenson, D., Cooney, J., Datson, P. M., Saiz, A., et al. (2011). Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis). Plant J. 65, 106–118. doi: 10.1111/j.1365-313X.2010.04409.x
Nakatsuka, T., Mishiba, K.-I., Abe, Y., Kubota, A., Kakizaki, Y., Yamamura, S., et al. (2008). Flower color modification of gentian plants by RNAi-mediated gene silencing. Plant Biotechnol. 25, 61–68. doi: 10.5511/plantbiotechnology.25.61
Offen, W., Martinez-Fleites, C., Yang, M., Kiat-Lim, E., Davis, B. G., Tarling, C. A., et al. (2006). Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 25, 1396–1405. doi: 10.1038/sj.emboj.7600970
Ohno, S., Hosokawa, M., Kojima, M., Kitamura, Y., Hoshino, A., Tatsuzawa, F., et al. (2011). Simultaneous post-transcriptional gene silencing of two different chalcone synthase genes resulting in pure white flowers in the octoploid dahlia. Planta 234, 945–958. doi: 10.1007/s00425-011-1456-2
Ohta, Y., Atsumi, G., Yoshida, C., Takahashi, S., Shimizu, M., Nishihara, M., et al. (2022). Post-transcriptional gene silencing of the chalcone synthase gene CHS causes corolla lobe-specific whiting of Japanese gentian. Planta 255, 1–12. doi: 10.1007/s00425-021-03815-w
Pang, Y., Peel, G. J., Wright, E., Wang, Z., Dixon, R. A. (2007). Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol. 145, 601–615. doi: 10.1104/pp.107.107326
Peng, Y., Lin-Wang, K., Cooney, J. M., Wang, T., Espley, R. V., Allan, A. C. (2019). Differential regulation of the anthocyanin profile in purple kiwifruit (Actinidia species). Horticulture Res. 6, 1–16. doi: 10.1038/s41438-018-0076-4
Peng, Y., Thrimawithana, A. H., Cooney, J. M., Jensen, D. J., Espley, R. V., Allan, A. C. (2020). The proanthocyanin-related transcription factors MYBC1 and WRKY44 regulate branch points in the kiwifruit anthocyanin pathway. Sci. Rep. 10, 1–19. doi: 10.1038/s41598-020-70977-0
Pilkington, S. M., Crowhurst, R., Hilario, E., Nardozza, S., Fraser, L., Peng, Y., et al. (2018). A manually annotated Actinidia chinensis var. chinensis (kiwifruit) genome highlights the challenges associated with draft genomes and gene prediction in plants. BMC Genomics 19, 1–19. doi: 10.1186/s12864-018-4656-3
Plunkett, B. J., Espley, R. V., Dare, A. P., et al. (2018). Warren, bMYBA from blueberry (Vaccinium section Cyanococcus) is a subgroup 6 type R2R3MYB transcription factor that activates anthocyanin production. a.W Front. Plant Science. 9, 1300. doi: 10.3389/fpls.2018.01300
Polashock, J., Zelzion, E., Fajardo, D., Zalapa, J., Georgi, L., Bhattacharya, D., et al. (2014). The American cranberry: first insights into the whole genome of a species adapted to bog habitat. BMC Plant Biol. 14, 1–18. doi: 10.1186/1471-2229-14-165
Primetta, A. K., Jaakola, L., Ayaz, F. A., Inceer, H., Riihinen, K. R. (2013). Anthocyanin fingerprinting for authenticity studies of bilberry (Vaccinium myrtillus l.). Food Control 30, 662–667. doi: 10.1016/j.foodcont.2012.09.009
R Core Team (2020). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Riihinen, K., Jaakola, L., Kärenlampi, S., Hohtola, A. (2008). Organ-specific distribution of phenolic compounds in bilberry (Vaccinium myrtillus) and ‘northblue’ blueberry (Vaccinium corymbosum x V. angustifolium). Food Chem. 110, 156–160. doi: 10.1016/j.foodchem.2008.01.057
Riso, P., Klimis-Zacas, D., Del Bo, C., Martini, D., Campolo, J., Vendrame, S., et al. (2013). Effect of a wild blueberry (Vaccinium angustifolium) drink intervention on markers of oxidative stress, inflammation and endothelial function in humans with cardiovascular risk factors. Eur. J. Nutr. 52, 949–961. doi: 10.1007/s00394-012-0402-9
Schwinn, K., Venail, J., Shang, Y., Mackay, S., Alm, V., Butelli, E., et al. (2006). A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18, 831–851. doi: 10.1105/tpc.105.039255
Shi, M., Loftus, H., Mcainch, A. J., Su, X. Q. (2017). Blueberry as a source of bioactive compounds for the treatment of obesity, type 2 diabetes and chronic inflammation. J. Funct. Foods 30, 16–29. doi: 10.1016/j.jff.2016.12.036
Skrovankova, S., Sumczynski, D., Mlcek, J., Jurikova, T., Sochor, J. (2015). Bioactive compounds and antioxidant activity in different types of berries. Int. J. Mol. Sci. 16, 24673–24706. doi: 10.3390/ijms161024673
Smeriglio, A., Barreca, D., Bellocco, E., Trombetta, D. (2016). Chemistry, pharmacology and health benefits of anthocyanins. Phytotherapy Res. 30, 1265–1286. doi: 10.1002/ptr.5642
Song, J., Du, L., Li, L., Kalt, W., Palmer, L. C., Fillmore, S., et al. (2015). Quantitative changes in proteins responsible for flavonoid and anthocyanin biosynthesis in strawberry fruit at different ripening stages: a targeted quantitative proteomic investigation employing multiple reaction monitoring. J. Proteomics 122, 1–10. doi: 10.1016/j.jprot.2015.03.017
Stintzing, F. C., Carle, R. (2004). Functional properties of anthocyanins and betalains in plants, food, and in human nutrition. Trends Food Sci. Technol. 15, 19–38. doi: 10.1016/j.tifs.2003.07.004
Sun, W., Liang, L. J., Meng, X. Y., Li, Y. Q., Gao, F. Z., Liu, X. X., et al. (2016). Biochemical and molecular characterization of a flavonoid 3-o-glycosyltransferase responsible for anthocyanins and flavonols biosynthesis in Freesia hybrida. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00410
Sun, J., Tang, W., Liu, Y. (2014). Transient expression of CHS-RNAi effectively influences the accumulation of anthocyanin in fruit of kiwifruit (Actinidia chinensis). Chin. J. Appl. Environ. Biol. 20(5), 929–933. doi: 10.3724/SP.J.1145.2014.03021
Szankowski, I., Flachowsky, H., Li, H., Halbwirth, H., Treutter, D., Regos, I., et al. (2009). Shift in polyphenol profile and sublethal phenotype caused by silencing of anthocyanidin synthase in apple (Malus sp.). Planta 229, 681–692. doi: 10.1007/s00425-008-0864-4
The French–Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. doi: 10.1038/nature06148
Tomato Genome Consortium, X. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635. doi: 10.1038/nature11119
Tuomi, J. M., Voorbraak, F., Jones, D. L., Ruijter, J. M. (2010). Bias in the cq value observed with hydrolysis probe based quantitative PCR can be corrected with the estimated PCR efficiency value. Methods 50, 313–322. doi: 10.1016/j.ymeth.2010.02.003
Vanburen, R., Wai, C. M., Colle, M., Wang, J., Sullivan, S., Bushakra, J. M., et al. (2018). A near complete, chromosome-scale assembly of the black raspberry (Rubus occidentalis) genome. Gigascience 7, giy094. doi: 10.1093/gigascience/giy094
Wang, C.-K., Chen, P.-Y., Wang, H.-M., To, K.-Y. (2006). Cosuppression of tobacco chalcone synthase using Petunia chalcone synthase construct results in white flowers. Botanical Stud. 47 (1), 71–82. https://ejournal.sinica.edu.tw/bbas/content/2006/1/Bot471-08.pdf
Wilmouth, R. C., Turnbull, J. J., Welford, R. W. D., Clifton, I. J., Prescott, A. G., Schofield, C. J. (2002). Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure 10, 93–103. doi: 10.1016/S0969-2126(01)00695-5
Wu, C., Deng, C., Hilario, E., Albert, N. W., Lafferty, D., Grierson, E. R. P., et al (2021). A chromosome-scale assembly of the bilberry genome identifies a complex locus controlling berry anthocyanin composition. Mol. Ecol. Resources. 22(1), 345–360. doi: 10.1111/1755-0998.13467.
Xie, S., Song, C., Wang, X., Liu, X., Zhang, Z., Xi, Z. (2015). Tissue-specific expression analysis of anthocyanin biosynthesis genes in white-and red-fleshed grape cultivars. Molecules 20, 22767. doi: 10.3390/molecules201219883
Xue, L., Wang, J., Zhao, J., Zheng, Y., Wang, H.-F., Wu, X., et al. (2019). Study on cyanidin metabolism in petals of pink-flowered strawberry based on transcriptome sequencing and metabolite analysis. BMC Plant Biol. 19, 1–16. doi: 10.1186/s12870-019-2048-8
Yang, B., Wei, Y., Liang, C., Guo, J., Niu, T., Zhang, P., et al. (2021). VvANR silencing promotes expression of VvANS and accumulation of anthocyanin in grape berries. Protoplasma 259(3), 743–753. doi: 10.1007/s00709-021-01698-y
Yonekura-Sakakibara, K., Fukushima, A., Nakabayashi, R., Hanada, K., Matsuda, F., Sugawara, S., et al. (2012). Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J. 69, 154–167. doi: 10.1111/j.1365-313X.2011.04779.x
Zhang, Y., Butelli, E., Martin, C. (2014). Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 19, 81–90. doi: 10.1016/j.pbi.2014.05.011
Zhang, H., Zhao, X., Zhang, J., Yang, B., Yu, Y., Liu, T., et al. (2020). Functional analysis of an anthocyanin synthase gene StANS in potato. Scientia Hortic. 272, 109569. doi: 10.1016/j.scienta.2020.109569
Zhang, X.-H., Zheng, X.-T., Sun, B.-Y., Peng, C.-L., Chow, W. S. (2018). Over-expression of the CHS gene enhances resistance of Arabidopsis leaves to high light. Environ. Exp. Bot. 154, 33–43. doi: 10.1016/j.envexpbot.2017.12.011
Zhao, C. L., Chen, Z. J., Bai, X. S., Ding, C., Long, T. J., Wei, F. G., et al. (2014). Structure–activity relationships of anthocyanidin glycosylation. Mol. Diversity 18, 687–700. doi: 10.1007/s11030-014-9520-z
Zhou, Z., Ying, Z., Wu, Z., Yang, Y., Fu, S., Xu, W., et al. (2021). Anthocyanin genes involved in the flower coloration mechanisms of Cymbidium kanran. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.737815
Zifkin, M., Jin, A., Ozga, J. A., Zaharia, L. I., Schernthaner, J. P., Gesell, A., et al. (2012). Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol. 158, 200–224. doi: 10.1104/pp.111.180950
Zorenc, Z., Veberic, R., Slatnar, A., Koron, D., Miosic, S., Chen, M.-H., et al. (2017). A wild a’lbino’bilberry (Vaccinium myrtillus L.) from Slovenia shows three bottlenecks in the anthocyanin pathway and significant differences in the expression of several regulatory genes compared to the common blue berry type. PloS One 12, e0190246. doi: 10.1371/journal.pone.0190246
Keywords: anthocyanins, chalcone synthase, anthocyanidin synthase, UDP-glucose:flavonoid 3-O-glucosyltransferase, blueberry, bilberry, vaccinium
Citation: Nguyen HM, Putterill J, Dare AP, Plunkett BJ, Cooney J, Peng Y, Souleyre EJF, Albert NW, Espley RV and Günther CS (2023) Two genes, ANS and UFGT2, from Vaccinium spp. are key steps for modulating anthocyanin production. Front. Plant Sci. 14:1082246. doi: 10.3389/fpls.2023.1082246
Received: 28 October 2022; Accepted: 11 January 2023;
Published: 02 February 2023.
Edited by:
Francesco Emanuelli, Berrytech, ItalyCopyright © 2023 Nguyen, Putterill, Dare, Plunkett, Cooney, Peng, Souleyre, Albert, Espley and Günther. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Han M. Nguyen, Alex.Nguyen@plantandfood.co.nz; Catrin S. Günther, Catrin.Guenther@plantandfood.co.nz
 Han M. Nguyen
Han M. Nguyen Joanna Putterill
Joanna Putterill Andrew P. Dare1
Andrew P. Dare1 Blue J. Plunkett
Blue J. Plunkett Janine Cooney
Janine Cooney Yongyan Peng
Yongyan Peng Edwige J. F. Souleyre
Edwige J. F. Souleyre Nick W. Albert
Nick W. Albert Richard V. Espley
Richard V. Espley Catrin S. Günther
Catrin S. Günther