- 1Innovation Team of Stone Fruit Breeding and Cultivation, Shandong Institute of Pomology, Tai’an, Shandong, China
- 2College of Horticulture Science and Engineering, Shandong Agricultural University, Tai’an, China
Color is an essential appearance characteristic of sweet cherry (Prunus avium L.) fruits and mainly determined by anthocyanin. Temperature plays an important role in the regulation of anthocyanin accumulation. In this research, anthocyanin, sugar, plant hormone and related gene expression were analyzed using physiological and transcriptomic methods in order to reveal the effects of high temperature on fruit coloring and the related mechanism. The results showed that high temperature severely inhibited anthocyanin accumulation in fruit peel and slowed the coloring process. The total anthocyanin content in fruit peel increased by 455% and 84% after 4 days of normal temperature treatment (NT, 24°C day/14°C night) and high temperature treatment (HT, 34°C day/24°C night), respectively. Similarly, the contents of 8 anthocyanin monomers were significantly higher in NT than in HT. HT also affected the levels of sugars and plant hormones. The total soluble sugar content increased by 29.49% and 16.81% in NT and HT, respectively, after 4 days of treatment. The levels of ABA, IAA and GA20 also increased in both the two treatments but more slowly in HT. Conversely, the contents of cZ, cZR and JA decreased more rapidly in HT than in NT. The results of the correlation analysis showed that the ABA and GA20 contents were significantly correlated with the total anthocyanin contents. Further transcriptome analysis showed that HT inhibited the activation of structural genes in anthocyanin biosynthesis as well as the repression of CYP707A and AOG, which dominated the catabolism and inactivation of ABA. These results indicate that ABA may be a key regulator in the high-temperature-inhibited fruit coloring of sweet cherry. High temperature induces higher ABA catabolism and inactivation, leading to lower ABA levels and finally resulting in slow coloring.
1 Introduction
Sweet cherry (Prunus avium L.) is one of the earliest fresh fruits available in northern China and has high economic value. The fruit is deeply loved by people for its bright color and rich nutrition, conferred by a variety of carbohydrates, proteins, and vitamins as well as iron, calcium, potassium and other essential nutrients and minerals (Li et al., 2010; Prinsi et al., 2016; Gonçalves et al., 2021). The coloring of sweet cherry peel is a major factor affecting fruit quality and largely determines its market value. Anthocyanin is responsible for the red color of sweet cherry peel, and its content directly affects the color of fruits.
The anthocyanin biosynthetic pathway has been studied in many plants, and related genes have been identified (Petroni and Tonelli, 2011; Zhai et al., 2014; Dai et al., 2022). The regulation of anthocyanin biosynthesis mainly involves a number of structural genes such as phenylalanine ammonia-lyase (PAL), 4-coumarate-CoA ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone-3-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), UDP-glucose flavonoid 3-O-glucosyltransferase (UFGT) and leucoanthocyanidin dioxygenase (LDOX), as well as the MYB-bHLH-WD40 (MBW) transcription complex (Kim et al., 2006; Albert et al., 2014; Xue et al., 2016; Moglia et al., 2020). MYB10 promotes anthocyanin accumulation by activating the promoters of DFR and UGFT (Wang et al., 2013; Zhai et al., 2016). The coexpression of basic helix-loop-helix (bHLH) and MYB10 proteins activates the transcription of structural genes (CHS, DFR and UFGT) (Rahim et al., 2014). WD40 domain-containing protein (WD40) is associated with the stabilization of the MBW complex, and the interaction between TTG1 (a WD40 protein) and bHLH plays an important role in promoting anthocyanin biosynthesis (An et al., 2012).
Anthocyanin accumulation is affected by multiple biotic and abiotic factors, such as nutrition, temperature, light and wounding (Weiss, 2000; Lin-Wang et al., 2011; Günther et al., 2022). Low temperature can induce anthocyanin biosynthesis and the expression of related genes, while high temperature accelerates anthocyanin degradation (Rowan et al., 2009; Rehman et al., 2017) and is unfavorable for anthocyanin synthesis (Crifò et al., 2012; Ryu et al., 2020). In rose (Rosa hybrida), high temperature (39 °C/18 °C) decreases the anthocyanin content in flowers by inhibiting the transcriptional levels of CHS and DFR (Dela et al., 2003). In Arabidopsis thaliana, high temperature reduces anthocyanin accumulation by decreasing the expression of genes and transcription complexes related to anthocyanin synthesis and increasing the expression of the anthocyanin repressors AtMYB3, AtMYB6 and AtMYBL2 (Rowan et al., 2009).
Plant hormones are important regulators of anthocyanin biosynthesis. Exogenous application of abscisic acid and jasmonic acid promotes anthocyanin accumulation in multiple fruits (Shen et al., 2014; An et al., 2015; An et al., 2018; Wu et al., 2020; Gao et al., 2021). Gibberellins negatively regulate low-temperature-induced anthocyanin accumulation in Arabidopsis (Zhang et al., 2011). Cytokinin (Ji et al., 2015; Wang et al., 2019), auxin (Mori et al., 1994; Wang et al., 2020) and brassinolide (Zheng et al., 2018) are also effective in regulating anthocyanin biosynthesis in multiple fruits. Among these plant hormones, abscisic acid mediates anthocyanin biosynthesis in sweet cherry, and MYBA is involved in the process (Shen et al., 2014). Although the effect of abscisic acid on anthocyanin biosynthesis has been widely studied, the molecular mechanism by which the interaction between high temperature and abscisic acid regulates anthocyanin biosynthesis is still unknown.
Color changes occur as an adaptation to changes in the external environment during the development of plant tissue (Oren-Shamir, 2009). The coloring period of sweet cherry in northern China is from late April to early May. The weather in this period is atypical, and the temperature usually increases sharply, which may have a profound impact on fruit coloring. This phenomenon will be a common problem in sweet cherry production, which directly affects the color and quality of fruits under global warming. Although the effects of temperature on anthocyanin biosynthesis have been analyzed in fruit species such as apple (Fang et al., 2019), grape (Yang et al., 2020) and pear (Zhai et al., 2016), few related studies have been conducted on sweet cherry, and the associated mechanism remains unclear.
In the present study, we found that high-temperature-treated sweet cherry peels accumulated less anthocyanin than normal-temperature-treated sweet cherry peels. Further study revealed that ABA may play a key role in the high-temperature-inhibited fruit coloring of sweet cherry. This study provides a new approach for studying the effect of temperature on anthocyanin accumulation in sweet cherry peel and provides a theoretical basis for improving the quality and commercial value of sweet cherry.
2 Materials and methods
2.1 Plant materials and treatment
The branches of ‘Tieton’ sweet cherry (Prunus avium L. Tieton) were collected at the beginning of coloring from the experimental orchard of the Shandong Institute of Pomology (36°21′N, 117°12′E; Tai’an China). At 25 days after flowering, fruiting branches showing similar growth were selected, and equal numbers of leaves from each branch were brought back to the laboratory. The collected branches were inserted into a glass bottle containing sterile water and placed in an artificial climate chamber for different temperature treatments. The high temperature treatment (HT) was set at 24°C (night)/34°C (day), and the normal temperature treatment (NT) was set at 14°C (night)/24°C (day). The light intensity was set at 20,000 lx, and the light cycle was 8 hours dark/16 hours light. The relative humidity was approximately 70%. After 4 days of treatment, the peel of sweet cherry was cut, immediately frozen in liquid nitrogen and stored at -80°C until use. The abbreviations used for the samples are as follows: BT represents samples collected before treatment, NT represents the normal temperature treatment and HT represents those subjected to high temperature treatment. Finally, three biological replicates of three samples each (9 samples in total) were used for further analysis.
2.2 Measurement of total anthocyanin and anthocyanin components
The extraction of total anthocyanin was based on Harborne’s method. The sweet cherries collected at 25 days after flowering were placed in a phytotron (light intensity 20000 lx, humidity 70%) for four days. Samples of approximately 1 g of the HT (24°C night/34°C day)- and NT (14°C night/24°C day)-treated sweet cherries were incubated in 10 ml of 1% (v/v) HCl-95% ethanol at 24°C for 24 hours. The absorbance was measured at 530, 620, and 650 nm with a UV−vis spectrophotometer (UV-2600, SHIMADZU, Chengdu, China).
The analysis of anthocyanin components was conducted by MetWare Biotechnology Co., Ltd. (Wuhan, China) using a UPLC−MS/MS platform. The standard procedures from the separation of substances by chromatography to identification by mass spectrometry have been fully described by Yang et al. (Mori et al., 2007), Zhang, et al. (Zhang et al., 2019a) and Zhang, et al. (Zhang et al., 2019b). The mass spectrometry data were processed with Analyst 1.6.3 software (AB Sciex, Ontario, Canada). The total ion current (TIC) diagrams of the various quality normal (QC) samples were overlapped to determine the repeatability of the metabolic extraction and detection results. The metabolite content data were normalized according to unit variance scaling. A multivariate statistical analysis method with supervised pattern recognition, partial least square discriminant analysis (PLS-DA), was used to maximize the differentiation between groups. Based on the orthogonal projections to latent structures discriminant analysis (OPLS-DA) results, the differentially accumulated metabolites were preliminarily screened using variable importance in projection (VIP). Metabolites exhibiting a fold change of ≥2 or ≤0.5 and VIP≥1 were considered to present significant differences between groups. The KEGG database (Kanehisa et al., 2004) was used to annotate the differentially abundant metabolites.
2.3 Measurement of soluble sugar components, total acid and pH
The soluble sugar content was determined by high-performance liquid chromatography (Waters 510, PA, USA) with an RID-10 differential detector. The chromatographic column used for measuring fructose, glucose and sorbitol was a carbohydrate Ca2+ column (250 mm * 4.6 mm); the mobile phase was 1000 ml of water; the column temperature was 80°C; the flow rate was 0.4 ml/min; and the injection volume was 10 µl. The chromatographic column used for measuring sucrose and galactose was a Carbomi xH-NP column (300*7.8 mm), the mobile phase was 1000 ml of water and 136 µl of sulfuric acid, the column temperature was 55°C, the flow rate was 0.6 ml/min, and the injection volume was 10 µl.
The determination of the total acid content was performed with reference to the OIV method (OIV 2012). The pH was measured with a PH400 desktop pH meter (PH400, Alalis, USA).
2.4 Measurement of plant hormones
Approximately 50 mg of each sample was ground in liquid nitrogen and dissolved in 1 mL methanol/water/formic acid (15:4:1, V/V/V). Ten microliters of internal standard mixed solution (100 ng/mL) was added as an internal standard. The mixture was vortexed for 10 minutes and centrifuged for 5 min (12000 r/min, and 4°C). The supernatant was then transferred into clean microtubes, evaporated to dry, dissolved in 100 μL 80% methanol (V/V) and filtered through a 0.22 μm membrane filter for further analysis.
The analysis was performed using a UPLC−ESI−MS/MS system (UPLC, ExionLC™ AD, SCIEX, USA; MS, Applied Biosystems 6500 Triple Quadrupole, USA). The analytical conditions were as follows: LC: column, Waters ACQUITY UPLC HSS T3 C18 (100 mm×2.1 mm, 1.8 µm); solvent system: water with 0.04% acetic acid (A), acetonitrile with 0.04% acetic acid (B); gradient program: started at 5% B (0-1 min), increased to 95% B (1-8 min), 95% B (8-9 min), and finally decreased to 5% B (9.1-12 min); flow rate: 0.35 mL/min; temperature: 40°C; and injection volume: 2 μL.
Linear ion trap and triple quadrupole scans were acquired on a triple quadrupole–linear ion trap mass spectrometer (QTRAP), QTRAP® 6500+ LC−MS/MS System, controlled by Analyst 1.6.3 software (SCIEX, USA). The ESI source operation parameters were as follows: ion source, ESI+/-; source temperature 550°C; ion spray voltage (IS) 5500 V (positive), -4500 V (negative); curtain gas (CUR) was set at 35 psi. Plant hormones were analyzed using scheduled multiple reaction monitoring (MRM). Multiquant 3.0.3 software (Sciex) was used to quantify all metabolites. Mass spectrometer parameters, including the declustering potentials (DP) and collision energies (CE) for individual MRM transitions, were performed with further DP and CE optimization. A specific set of MRM transitions was monitored for each period according to the metabolites eluted within this period.
2.5 RNA extraction, identification and transcriptome analysis
Total RNA was extracted from 3 biological replicates of sweet cherry peel using the RNAprep Pure Plant Kit (for polysaccharide- and polyphenolic-rich plants) (DP441, TIANGEN, Beijing, China). The quality of the RNA was measured using 1% agarose gel electrophoresis (18S and 28S) to ensure that the RNA was not contaminated or degraded. Nanodrop (IMPLEN, CA, USA), Qubit 2.0 (Life Technologies, CA, USA) and Agilent 2100 (Agilent Technologies, CA, USA) systems were used to detect the purity, concentration and integrity of the RNA samples to ensure the use of qualified samples for transcriptome sequencing.
PCR enrichment was conducted to construct the cDNA library after the samples were qualified. Qubit 2.0 (Life Technologies, CA, USA) and Agilent 2100 (Agilent Technologies, CA, USA) systems were used to determine the library concentration and insert size, respectively. qRT−PCR was used to accurately quantify the effective concentration of the library. The library was then sequenced on the Illumina HiSeq platform, and 150-bp paired-end reads were obtained. Based on the use of sequencing-by-synthesis technology, large amounts of high-quality data were obtained, and most of the base-pair quality values reached or exceeded Q30. The transcriptome data were compared with the reference genome sequence (GenBank: JAAOZG010000000) by using the HISAT2 system (Kim et al., 2015). After the comparative analysis was completed, the mapped reads were assembled and quantified using StringTie.
BLAST software was used to compare the extracted new genes with the NR, Swiss-Prot, GO, COG, KOG, Pfam and KEGG databases. The KEGG orthology analysis of the new genes was performed by using KOBAS2.0 (Xie et al., 2011). Fragments per kilobase of transcript per million mapped reads (FPKM) (Florea et al., 2013) values were used as an indicator to measure the expression levels of the transcripts or genes. DESeq was used for differential expression analysis between different sample groups (Wang et al., 2010). Fold change (FC) ≥ 2 or ≤ 0.5 and a false discovery rate (FDR) < 0.01 were used as the criteria for the identification of differentially expressed genes (DEGs). The DEGs identified between samples were subjected to GO and KEGG analyses with the R package.
2.6 Quantitative real-time PCR (qRT−PCR) analysis
To verify the results of the transcriptome analysis, we selected 13 genes that were significantly enriched in the anthocyanin biosynthetic pathway for RT−PCR verification. First-strand cDNA was synthesized by using a cDNA reverse transcription kit (RR047A, TaKaRa, Dalian, China). The primers used in these steps are listed in Supplementary Material S1. qRT−PCR was performed on a CFX Connect™ real-time PCR system (Bio-Rad, Hercules, CA, USA) using SYBR® Premix Ex Taq™ (Tli RNaseH Plus) (RR420A, TaKaRa, Dalian, China). The reaction system included 12.5 μL of SYBR® Premix Ex Taq (2 ×), 1 μL of each forward and reverse primer, 1 μL of cDNA, and ddH2O to 25 μL. Three technical replicates were analyzed for each sample. The qRT−PCR conditions were as follows: 40 cycles of predenaturation at 95°C for 30 s, denaturation at 95°C for 5 s, and annealing at 60°C for 30 s. A fluorescence curve and a melting curve were obtained, and relative expression levels were calculated using the 2-△△CT method.
2.7 Statistical analysis
Statistical analysis was performed using the software Microsoft Excel 2010. Graphs were obtained from GraphPad Prism 6.01. The values in each figure were the mean ± SD of three replicates. Significant differences analysis was performed with SPSS 19.0 using Duncan’s test, and differences at p < 0.05 were labeled for the statistical tests. Pearson’s correlation analysis was conducted using SPSS 19.0 with a critical level of p < 0.05.
3 Results
3.1 High temperature inhibited anthocyanin accumulation in sweet cherry peel
HT inhibited anthocyanin accumulation in fruit peel and severely delayed the coloring process of sweet cherry fruits. The fruits turned bright red from green yellow after 4 days in NT, while the fruits in HT only colored slightly (Figures 1A–C). The total anthocyanin contents in fruit peel increased by 455% and 84% in NT and HT, respectively (Figure 1D). The final total anthocyanin level in HT was only approximately 1/3 of that in NT.
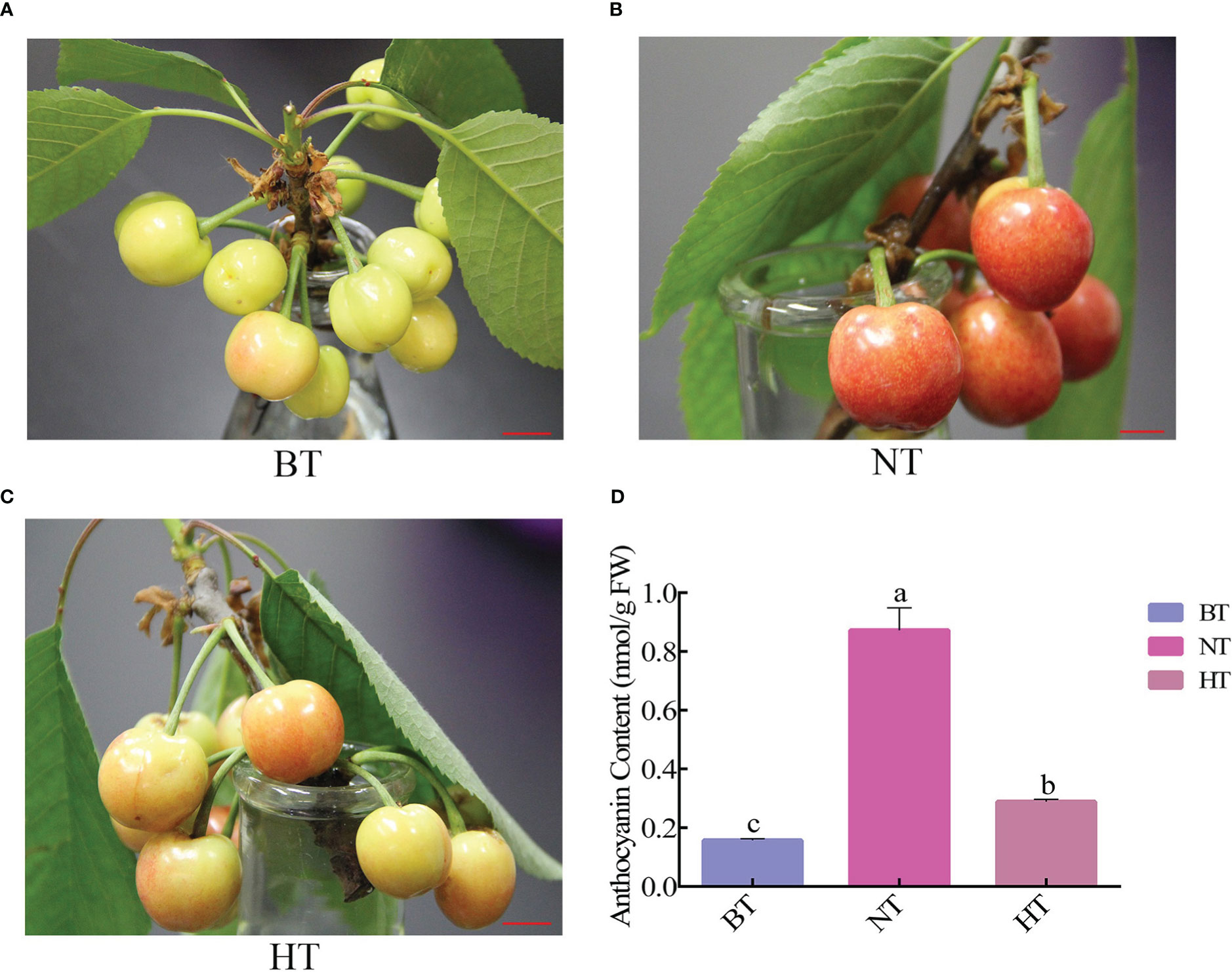
Figure 1 High temperature represses anthocyanin accumulation in sweet cherry peel. (A) Phenotype of sweet cherry before treatment (BT). (B) Phenotype of sweet cherry treated at normal temperature [NT, 14°C (night)/24°C (day)]. (C) Phenotype of sweet cherry treated with high temperature [HT, 24°C (night)/34°C (day)]. (D) The anthocyanin content in different temperature treatment groups. The different letters indicate significant differences among different treatments according to Duncan’s test (p < 0.05).
The variety and level of anthocyanin monomers were further analyzed using UPLC−MS/MS to reveal details of the anthocyanin change. A total of 511 metabolites, including 48 differentially accumulated metabolites (DAMs), were detected in NT and HT. The DAMs mainly included flavonoids, phenolic acids, lipids and alkaloids (Supplementary Material S2). In the DAMs, most lipids and flavonoids had higher abundance NT, while most phenolic acids and alkaloids had higher abundance in HT. Among the detected metabolites, 21 were identified to be anthocyanin monomers, and 8 of them were significantly less abundant in HT than in NT. The contents of the 8 anthocyanin monomers ranged from 18.01% to 49.06% of those in NT (Figure 2). The levels of the remaining 13 anthocyanin monomers showed no difference between HT and NT. The metabolic data are deposited in the figshare database (https://figshare.com/articles/dataset/metabolic_data_of_sweet_cherry/13571306).
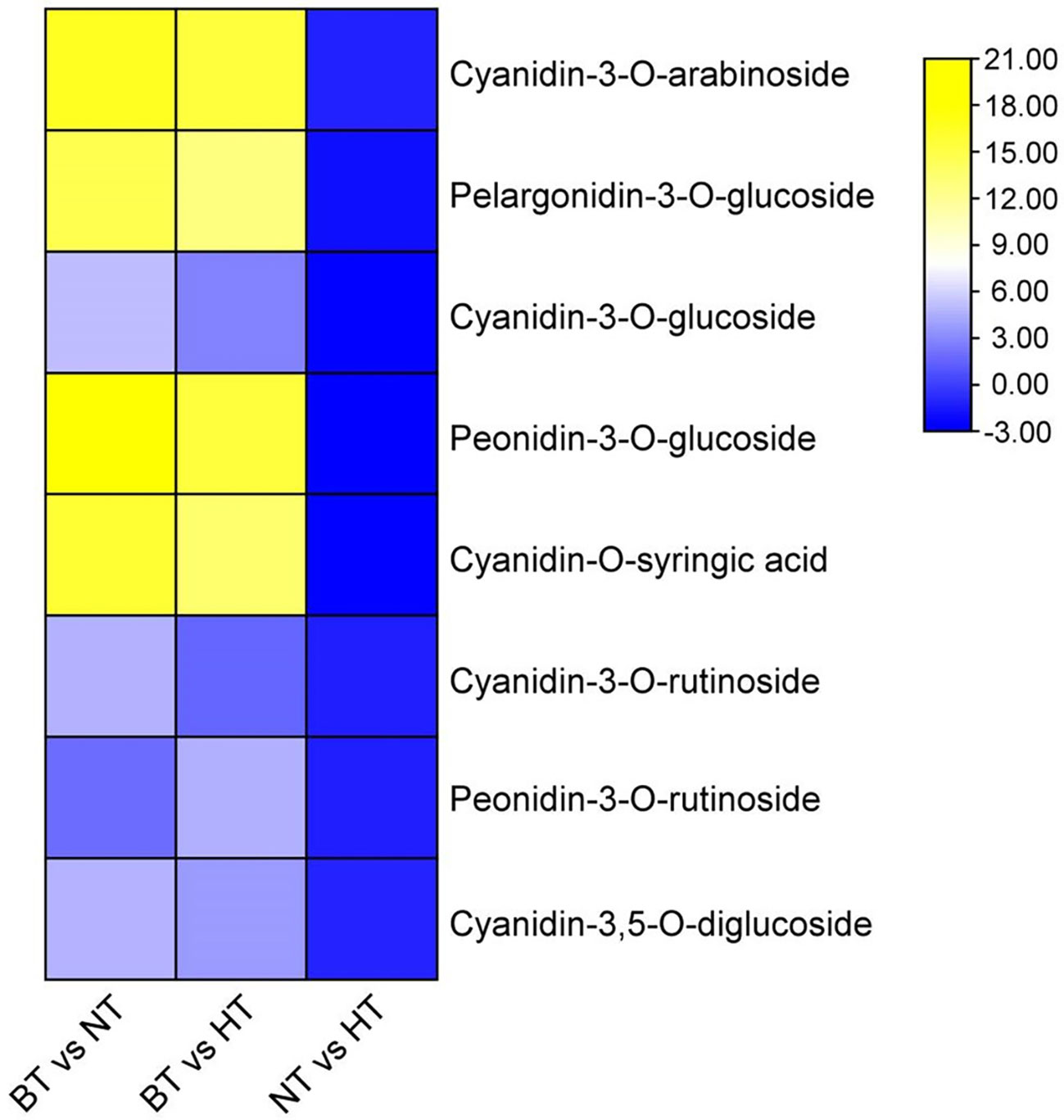
Figure 2 Differential accumulation of anthocyanin monomers in sweet cherry peel. Anthocyanin monomers were visualized as a heat map based on log2(fold change) obtained from comparisons among different treatments.
3.2 High temperature inhibited sugar accumulation in sweet cherry peel
In accordance with the change in anthocyanin, the accumulation of sugar in fruit peel in HT was less than that in NT (Figure 3). The total soluble sugar content increased by 29.49% and 16.81% in NT and HT, respectively, after 4 days of treatment. The total soluble sugar content of sweet cherry peel in the HT group was 9.79% lower than that in the NT group. Further analysis showed that the difference mainly came from glucose, fructose, sorbitol and galactose. The contents of the 4 sugars were 10.04%, 16.83% 6.45% and 3.40% lower in HT than in NT, respectively. Sucrose accounted for no more than 2% of the total soluble sugar and showed no significant difference in HT and NT. No significant difference in total acid content or pH was found between HT and NT (Figure 3).
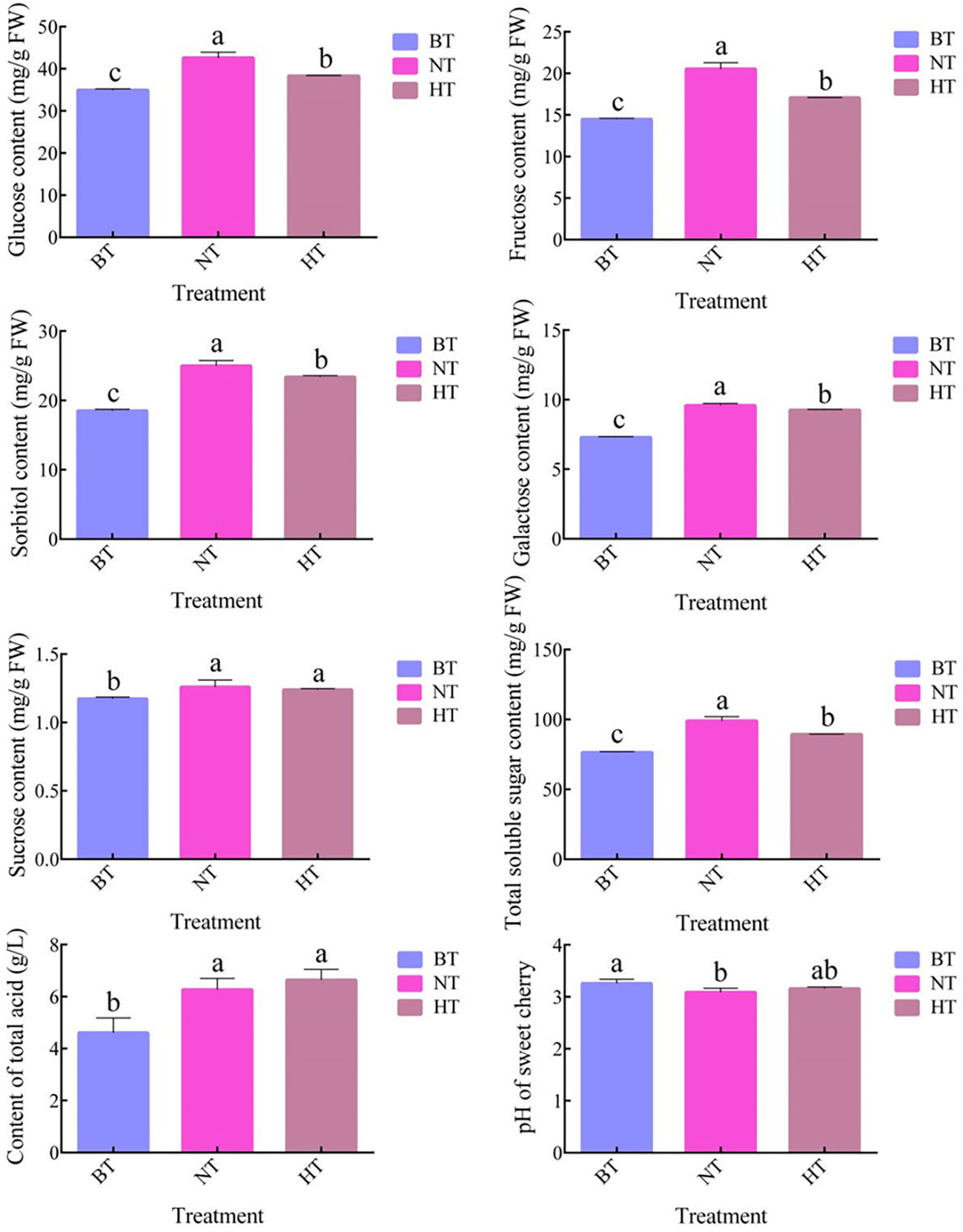
Figure 3 The soluble sugar content, total acid content and pH of sweet cherry peel in different treatments. The values represent the means ± SDs, n = 3. The different letters indicate significant differences among different treatments according to Duncan’s test (p < 0.05).
3.3 High temperature changed hormone levels in sweet cherry peel
Plant hormones are important regulators of anthocyanin biosynthesis in fruits. Our research showed that high temperature led to dramatic changes in plant hormone levels in sweet cherry fruit peel (Figure 4). The levels of abscisic acid (ABA), auxin (IAA) and gibberellic acid 20 (GA20) increased in both the NT and HT treatments, while the contents of cis-zeatin (cZ), cis-zeatin riboside (cZR) and jasmonic acid (JA) showed downward trends. Interestingly, the contents of the six compounds were all lower in HT than in NT. Notably, the level of ABA, an important regulator of anthocyanin biosynthesis in sweet cherry (Shen et al., 2014), was only 18.07% in HT compared with NT. The levels of GA1 and salicylic acid (SA) showed no significant difference among samples from BT, NT and HT. Some common hormones, including trans-zeatin (tZ), GA3, GA4 and GA7, were not detected in the samples.
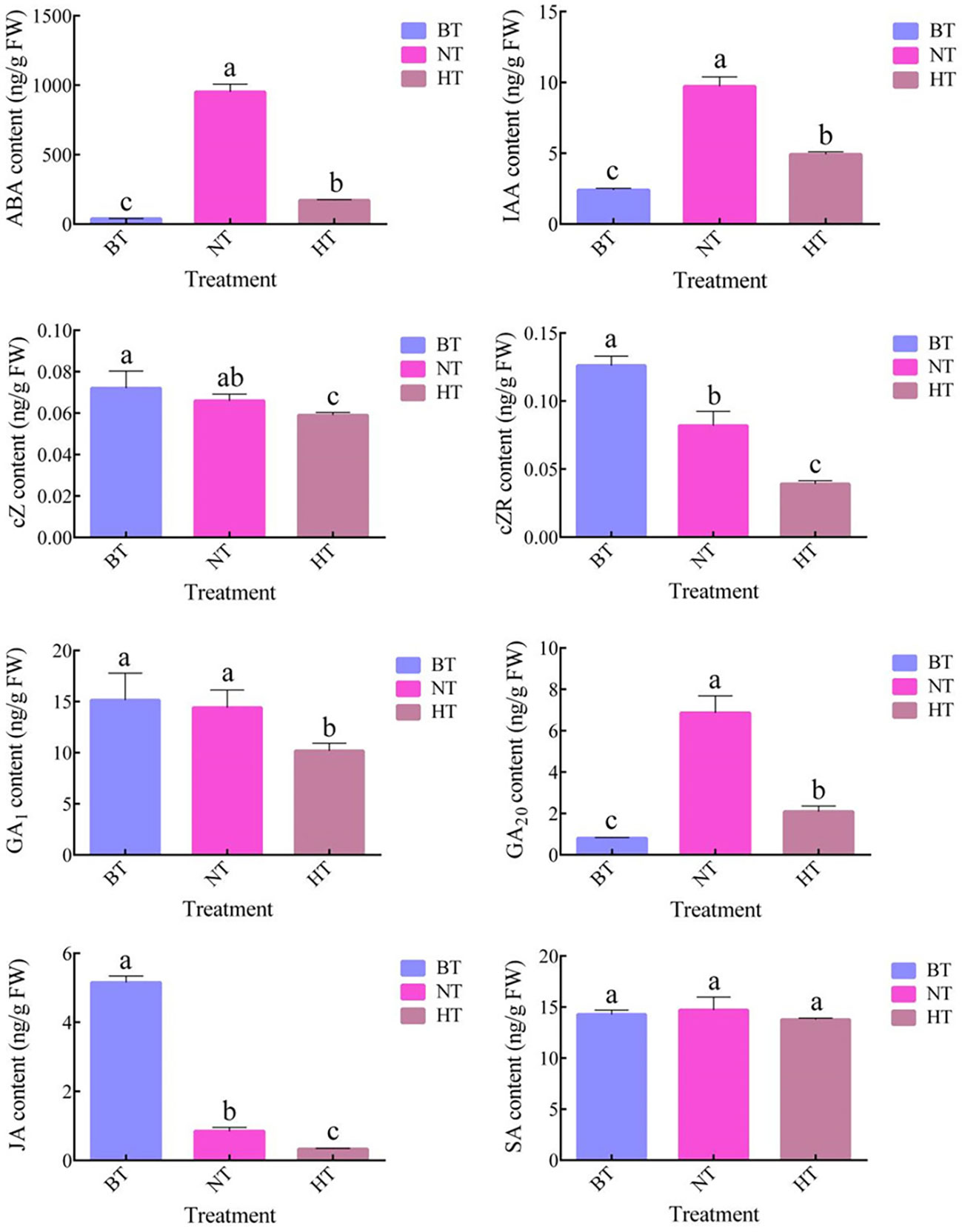
Figure 4 The contents of plant hormones in sweet cherry peel under different treatments. The values represent the means ± SDs, n = 3. The different letters indicate significant differences among different treatments according to Duncan’s test (p < 0.05).
3.4 Correlation analysis between anthocyanin content and other physiological parameters
Pearson’s correlation analysis was further conducted to screen potential physiological factors affecting anthocyanin biosynthesis in sweet cherry peels (Supplementary Material S3). The results showed that the anthocyanin level was significantly correlated with the ABA and GA20 contents. High correlation coefficients also existed between the anthocyanin levels and glucose, fructose, sorbitol, total sugar and IAA contents regardless of the significance analysis.
3.5 Comparative transcriptome analysis of sweet cherry peel under different treatments
Clean data (79.85 Gb) were generated after mRNA sequencing for 9 samples from BT, HT and NT (including 3 biological replicates). A total of 39,695 transcripts were obtained by processing the sequencing reads. The expression levels were calculated using the FPKM method (Trapnell et al., 2010). Pairwise Pearson’s correlation analysis showed high consistency of the biological replicates (Figure 5A). The RNAseq data generated for this study is available in the GenBank database (http://www.ncbi.nlm.nih.gov/bioproject/678548).
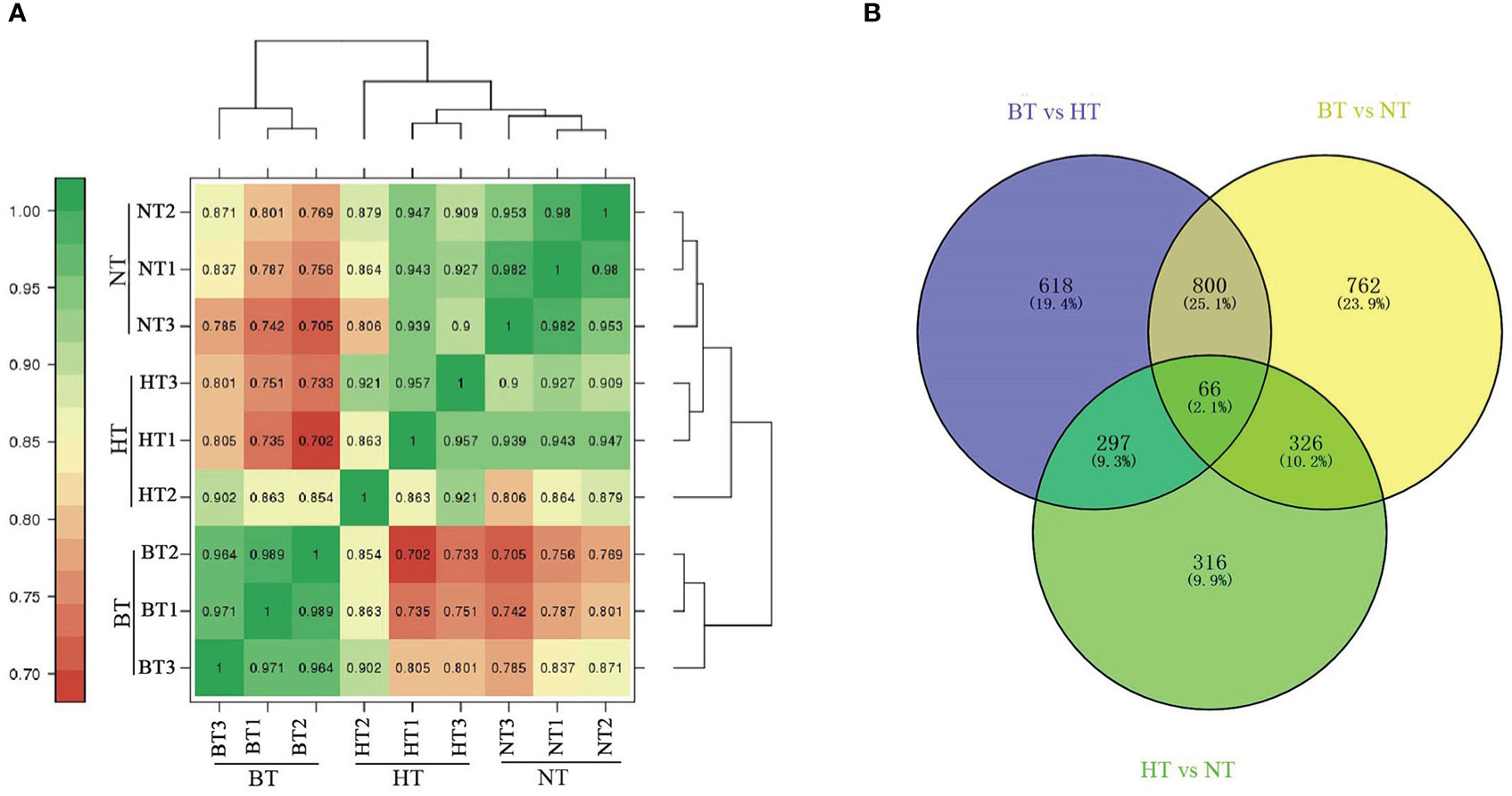
Figure 5 Analysis of DEGs in the transcriptome. (A) Heatmap of the correlation coefficients of different temperature treatment groups. (B) Venn diagram of DEGs.
Differentially expressed genes (DEGs) were further identified using the DESeq R package (1.18.0) with the criteria of |log2FoldChange| ≥ 1 and FDR < 0.01. A total of 1,781 and 1,954 DEGs were obtained from BT vs. HT and BT vs. NT, respectively, while only 1,005 DEGs were obtained from NT vs. HT. The Venn diagram showed that 866 DEGs were shared in BT vs. HT and BT vs. NT, accounting for more than 40% of the DEGs of each group (Figure 5B). However, the numbers in BT vs. HT and NT vs. HT (363 shared DEGs) or BT vs. NT and NT vs. HT (392 shared DEGs) were much lower. The statistical data of DEGs in each group are shown in Supplementary Material S4.
3.6 DEGs involved in anthocyanin biosynthesis
The DEGs involved in anthocyanin biosynthesis and its upstream regulation were further screened through KEGG analysis and manually. These DEGs mainly included structural genes involved in anthocyanin biosynthesis, genes encoding the MBW (MYB-bHLH-WD40) complex and genes participating in plant hormone metabolism and plant hormone signal transduction. Table 1 shows the expression of structural genes involved in anthocyanin biosynthesis. Compared with BT, the expression of most genes was significantly upregulated in NT, while only 4 of them were upregulated in HT. The expression of 2 PAL genes and 1 4CL genes was upregulated in both NT and HT, while UFGT was upregulated only in HT. The expression of CHS, CHI, DFR and ANS was significantly lower in HT than in NT.
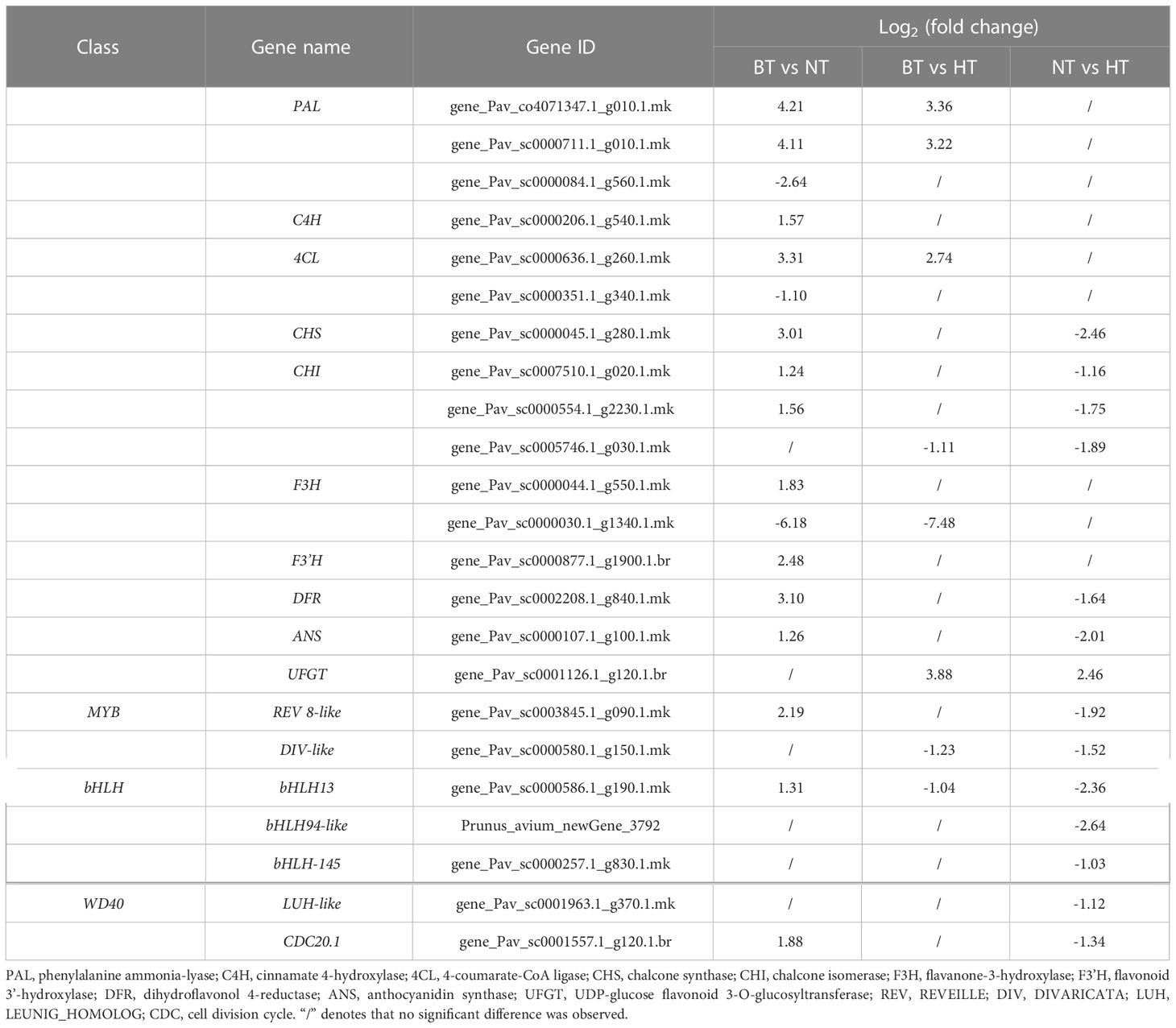
Table 1 Differentially expressed structural genes involved in anthocyanin biosynthesis and downregulated MYB, bHLH and WD40 genes in NT vs. HT.
The expression of anthocyanin biosynthesis-associated structural genes is regulated by the MYB-bHLH-WD40 complex comprising MYB transcription factor, bHLH transcription factor and WD40 protein (Jaakola, 2013). Considering the downregulation of these structural genes in HT compared with NT, the downregulated genes belonging to the MYB, bHLH and WD40 classes in NT vs. HT were screened to explore the upstream regulators. A total of 7 genes were obtained (Table 1), including 2 in the MYB class, 3 in the bHLH class and 2 in the WD40 class. The 4 genes belonging to the MYB and WD40 classes were not involved in anthocyanin biosynthesis according to the present literature. The expression levels of bHLH13, bHLH94-like and bHLH-145 were significantly lower in HT than in NT. Notably, the expression of bHLH13 was upregulated in NT but downregulated in HT.
3.7 DEGs involved in ABA metabolism and signal transduction
ABA has been proven to be an important regulator of anthocyanin biosynthesis in sweet cherry (Ren and Leng, 2010; Shen et al., 2014). The DEGs involved in ABA metabolism and signal transduction were further screened considering the significant correlation between ABA levels and anthocyanin levels in this research. A total of 4 DEGs were identified participating the biosynthesis and catabolism of ABA (Table 2). β-carotene 3-hydroxylase (BCH) and zeaxanthin epoxidase (ZEP) catalyzed the biosynthesis of violaxanthin (an intermediate products of ABA biosynthesis) from β-carotene (Hieber et al., 2000; Xia et al., 2022). Both BCH2 and ZEP showed lowed expression level in HT than in NT (Table 2). Cytochrome P450 707A (CYP707A), also known as ABA 8’-hydroxylase, is the key enzyme in ABA catabolism (Saito et al., 2004). Abscisate β-glucosyltransferase (AOG) catalyzes the glycosylation and inactivation of ABA (Xu et al., 2002). The expression levels of CYP707A and AOG were both lower in NT than in BT but higher in HT than in NT (Table 2). The expression levels of the 4 genes are consistent with the difference in ABA levels among the 3 treatments (Figure 4). Further analysis showed that the expression of most genes involved in ABA signal transduction showed little difference between NT and HT.
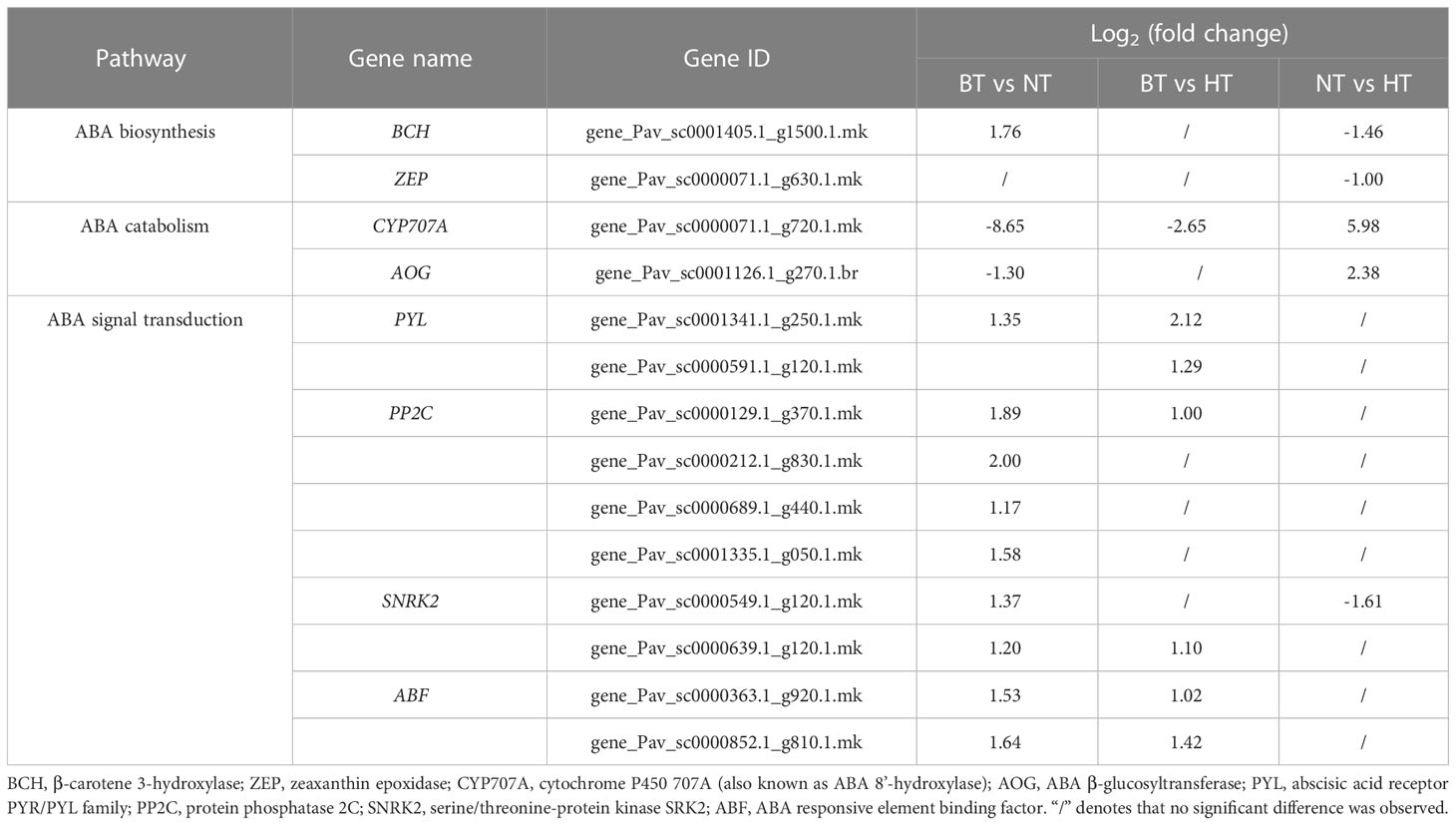
Table 2 Expression of DEGs involved in the biosynthesis, catabolism and signal transduction of plant hormones.
GA20 is an intermediate in GA biosynthesis with no hormonal activity (Hedden, 1999). The content of no other GAs varied among BT, NT and HT (Figure 4). Therefore, GAs were not considered regulators of anthocyanin biosynthesis, and related DEGs were not analyzed.
3.8 qRT−PCR validation of differentially expressed genes
To validate the reliability of the expression profiles obtained from RNA-Seq, 13 DEGs related to flavonoid biosynthesis and anthocyanin biosynthesis were selected for quantitative real-time PCR (qRT−PCR) (Figure 6). The results of the qRT−PCR analysis of genes in the anthocyanin biosynthetic pathway were consistent with the RNA-seq results. The expression levels of CHS, F3H and DFR, key genes in anthocyanin biosynthesis, were upregulated in NT but downregulated in HT. These results indicate that the RNA-seq data were reliable for assessing the transcriptomic changes induced by high temperature.
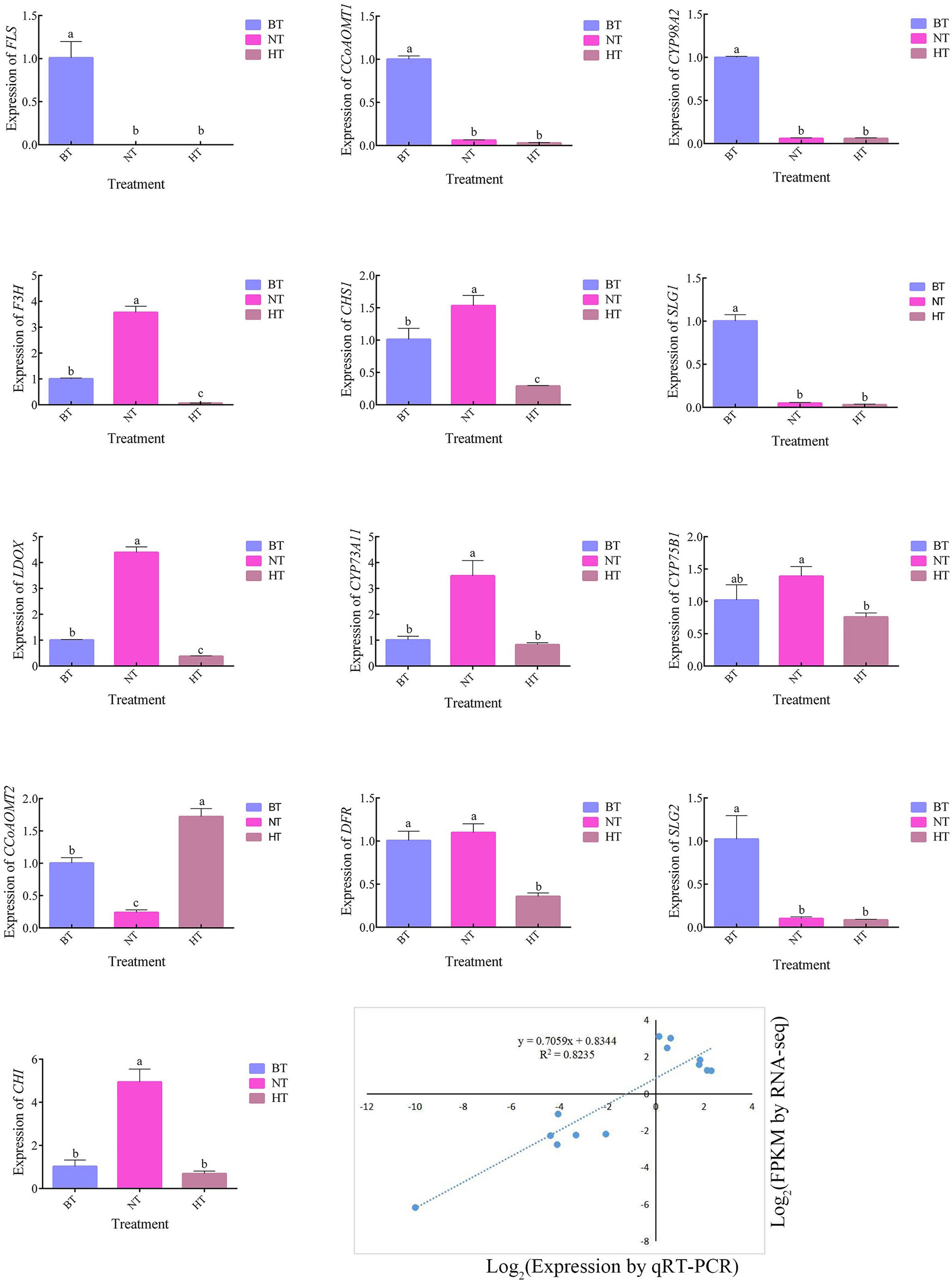
Figure 6 The genes of the anthocyanin and flavonoid synthesis pathways in the transcriptome were verified by qRT−PCR. BT was set as the reference group for each temperature treatment, in which the gene expression level was recorded as 1. The different letters indicate significant differences among different treatments according to Duncan’s test (p < 0.05).
4 Discussion
4.1 High temperature inhibits the activation of anthocyanin biosynthesis in sweet cherry peels
Sweet cherry presents high environmental requirements during coloring. An unfavorable environment leads to the inhibition of anthocyanin accumulation, which directly affects the appearance and commercial value of fruits (Esti et al., 2002). Anthocyanin biosynthesis is affected by various external factors, such as temperature and light. Low temperature promotes the expression of anthocyanin biosynthetic pathway genes and increases anthocyanin contents (Steyn, 2009; Ubi et al., 2006), while high temperature affects the activity of related enzymes (Mori et al., 2005) and inhibits the accumulation of anthocyanins and glycosides (Ban et al., 2007). Global warming and inconsistent temperature conditions are leading to serious implications for the coloring of sweet cherry fruits. Therefore, elucidating the molecular mechanism of temperature-regulated anthocyanin accumulation is important.
In this research, high temperature severely inhibited anthocyanin accumulation in sweet cherry peel and delayed the coloring process (Figure 1). This phenomenon is similar to that in apple (Lin-Wang et al., 2011), grape (Yamane et al., 2006) and eggplant (Lv et al., 2019). Transcriptome and qRT−PCR analyses showed that the general expression of structural genes involved in anthocyanin biosynthesis was significantly upregulated under normal temperature but changed little under high temperature (Table 1). The bHLH transcription factor is an essential component of the MYB-bHLH-WD40 complex that directly regulates the expression of anthocyanin biosynthesis-associated structural genes (Jaakola, 2013). The expression of 3 bHLH genes was also found to be downregulated under high temperature compared with normal temperature (Table 1). Among them, bHLH13 was annotated encoding the N-terminal and helix-loop-helix DNA-binding domains of bHLH-MYC and R2R3-MYB transcription factors in the Pfam database. These data indicated that high-temperature high temperature inhibited the activation of the anthocyanin biosynthesis pathway in sweet cherry peel during fruit development and finally led to slow coloring.
4.2 Roles of plant hormones in the coloring inhibition induced by high temperature
Plant hormones are essential regulators of anthocyanin biosynthesis in fruits. ABA plays an important role in the ripening and coloring of nonclimacteric fruits, such as strawberry, sweet cherry and grape (Setha et al., 2005; Jia et al., 2011; Wang et al., 2022). ABA treatment significantly induced anthocyanin accumulation in sweet cherry fruits, while treatment with an ABA biosynthesis inhibitor blocked anthocyanin production (Ren and Leng, 2010; Shen et al., 2014). In this research, high temperature severely inhibited the increase in ABA levels in sweet cherry peel (Figure 4). More genes involved in ABA signal transduction were upregulated in NT than in HT (Table 2). The ABA level showed a significant correlation with the anthocyanin level (Supplementary Material S3). This finding indicates that ABA is a key regulator in the high-temperature-inhibited anthocyanin biosynthesis in sweet cherry peel.
Further RNA-seq showed that HT changed the expression of several genes related with ABA metabolsim. BCH and ZEP encode enzymes, but not limiting enzymes, in ABA biosynthesis (Hieber et al., 2000; Xia et al., 2022). The expression levels of them were only slightly lower in HT than in NT with the log2(fold change) of no more than -1.5 (Table 2). Therefore, the downregulation of BCH and ZEP might have little effects on ABA accumulation. CYP707A and AOG encode key enzymes in the catabolism and inactivation of ABA, respectively (Xu et al., 2002; Saito et al., 2004). The expression of the two genes was much higher in HT than in NT and lower in NT than in BT (Table 2). This indicates that the expression of CYP707A and AOG decreased during fruit coloring, while high temperature strongly hindered the process. These data show that the regulation of ABA catabolism and inactivation rather than ABA biosynthesis was responsible for the high temperature-inhibited ABA accumulation. The study on detailed effects of high temperature on ABA metabolism in sweet cherry peel is still needed in subsequent studies.
Previous studies showed that GA mainly exerted a negative role in the ripening and coloring process of sweet cherry. GA3 application delays ABA accumulation, fruit size increase and ripening, reduces anthocyanin levels and produces fruits with less color (Kondo and Danjo, 2001; Usenik et al., 2005; Kuhn et al., 2021). GA4 is also significantly and inversely correlated with the ripening parameters of sweet cherry (Teribia et al., 2016). In this research, only the content of GA20 was observed to be significantly correlated with the anthocyanin level in sweet cherry peel (Supplementary Material S3). However, GA20 is an intermediate in GA biosynthesis and has no hormonal activity (Hedden, 1999). Therefore, GA could be inferred to be not related to high-temperature-inhibited anthocyanin biosynthesis.
Other plant hormones, such as IAA, cytokinins and JA, are also effective in regulating anthocyanin biosynthesis (Shan et al., 2009; An et al., 2015; Ji et al., 2015). However, related studies in sweet cherry are still scarce. In this research, although the levels of IAA, cZ, cZR, JA and SA in sweet cherry peel differed between NT and HT (Figure 4), they showed no significant correlation with the anthocyanin level (Supplementary Material S3), indicating their minor role in high-temperature-inhibited anthocyanin biosynthesis.
4.3 Roles of sugar and acid in the coloring inhibition induced by high temperature
Sugar is the precursor of anthocyanin (Smeekens, 2000). The metabolism and accumulation of sugar in fruits is affected by environmental and ecological factors (Kerepesi et al., 2000; Watanabe et al., 2000; Moustakas et al., 2011). In Arabidopsis, anthocyanin production in cotyledons or leaves increases when seedlings are grown in sugar-containing medium (Mita et al., 1997; Ohto et al., 2001). Sucrose is closely related to anthocyanin biosynthesis (Hara et al., 2004; Zhang et al., 2022). It acts as an endogenous trigger that modulates the expression of anthocyanin biosynthesis genes in grape berry skins (Boss et al., 1996). In Arabidopsis, exogenous sucrose increased the transcript levels of several structural genes in anthocyanin biosynthesis by several hundred-fold (Solfanelli et al., 2006) and showed a synergistic effect with ABA on anthocyanin accumulation (Loreti et al., 2008).
In this research, high temperature suppressed the accumulation of glucose, fructose, sorbitol and galactose in sweet cherry peel but not sucrose (Figure 3). Fruit sugars mainly come from the photosynthetic product of leaves. It was reported that 35°C/25°C inhibited the photosynthetic rate of ‘Satohnishiki’ sweet cherry compared to 25°C/15°C condition (Beppu et al., 2003). Therefore, the suppressed sugar accumulation could be resulted from the inhibition of photosynthesis by high temperature. Sorbitol is the major photosynthetic product as well as the major sugar transported to fruits in cherry (Roper et al., 1988). It is largely converted to fructose or glucose by sorbitol dehydrogenase (SDH) upon reaching fruits (Aguayo et al., 2013; Zhang et al., 2014). In this research, no DEGs were identified encode any sorbitol transporters, but the expression of two SDH-like genes were upregulated in HT than in BT and NT (Supplementary Material S5). Two genes respectively encoding glucan endo-1,3-β-glucosidase (GEBG) and β-glucosidase (β-Glu) were also upregulated in HT than in NT. GEBG and β-Glu participate the generation of glucose from glucan and cellulose, respectively. The upregulation of the 4 genes above are all related to the generation of fructose or glucose, and could be a response of sugar deficiency in fruits of HT. Conversely, the expression of a gene encoding invertase (INV) was down regulated in HT than in NT. INV catalyze the hydrolysis of sucrose to glucose and fructose. The inhibition of INV expression matches the relative stable sucrose level in HT in condition of lower total sugar (Figure 3).
Despite the significant difference of sugar levels no significant correlation was detected between any sugar and anthocyanin (Supplementary Material S3), indicating that lower sugar content is not a main cause of the high-temperature-inhibited anthocyanin biosynthesis. However, that sugar metabolism contributed to the process as a synergistic factor of ABA is still a possibility. Sucrose is a minor component of fruit sugar in sweet cherry, accounting for no more than 2% of the total soluble sugar. It may not be important in the regulation of anthocyanin biosynthesis.
Most studies exploring the relationship between acid and anthocyanin are focused on the effect of pH on anthocyanin stability and enzyme activity (Lou et al., 2019). A given pigment can exhibit different colors under different pH conditions (Fossen et al., 1998). The total acid content in sweet cherry peel increased and the pH decreased under both high temperature and normal temperature. Although the expression of a gene encoding NADP-malic enzyme (NADP-ME) was lower in HT than in NT (Supplementary Material S5), no significant difference of total acid and pH was detected between the two treatments (Figure 3). Therefore, acid metabolism should not be related to the high-temperature-induced coloring inhibition of sweet cherry fruits.
Overall, our results indicated that high temperature severely inhibits the coloring of sweet cherry. ABA, an important positive regulator of the ripening and coloring of nonclimacteric fruits, probably plays a key role in the process. High temperature delays the downregulation of CYP707A and AOG expression during fruit coloring and leads to higher ABA catabolism and ABA inactivation (Figure 7). This causes a lower ABA level in fruit peel and delays coloring.
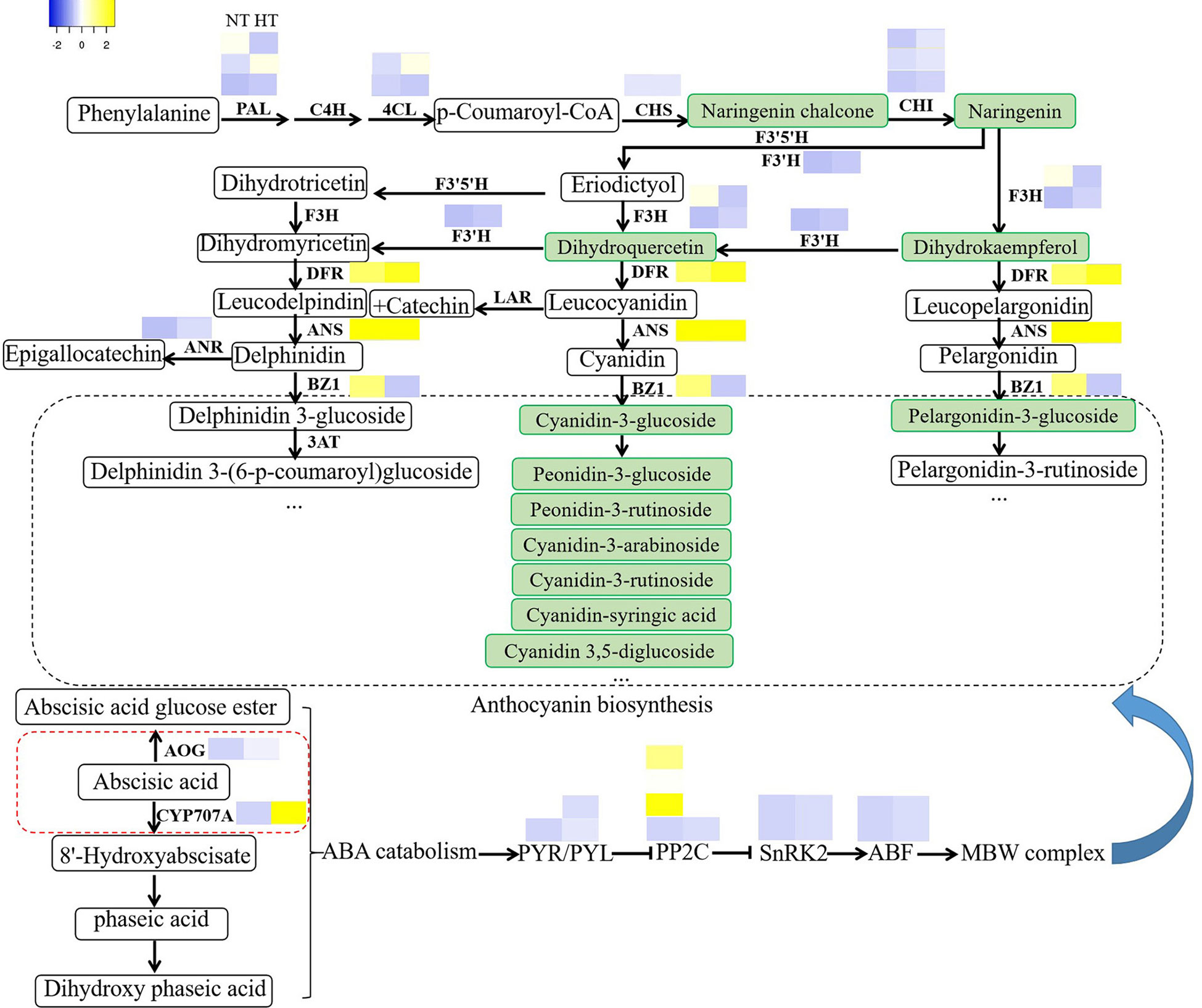
Figure 7 High temperature affects anthocyanin biosynthesis through the ABA catabolic pathway. DEGs from anthocyanin biosynthesis and ABA catabolism pathway genes and selected TFs were visualized as a heat map based on log2-fold changes obtained from high temperature treatments against the normal temperature treatment.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
HW conceived and designed the experiments. YT and BW conducted the experiments, collected the data and wrote the manuscript. LX and XZ collected the data and performed the statistical tests. GW, YS and HW read and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of Shandong Province, China (ZR2020MC133), Natural Science Foundation of Shandong Province, China (ZR2020MC119) and the earmarked fund for CARS (CARS-30).
Acknowledgments
We would like to thank Jiangang Jiang for the help in preparing plant materials.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The review X-FW declared a shared affiliation with the author BW to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1079292/full#supplementary-material
References
Aguayo, M. F., Ampuero, D., Mandujano, P., Parada, R., Muñoz, R., Gallart, M., et al. (2013). Sorbitol dehydrogenase is a cytosolic protein required for sorbitol metabolism in Arabidopsis thaliana. Plant Sci. 205-206, 63–75. doi: 10.1016/j.plantsci.2013.01.012
Albert, N. W., Davies, K. M., Lewis, D. H., Zhang, H., Montefiori, M., Brendolise, C., et al. (2014). A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26 (3), 962–980. doi: 10.1105/tpc.113.122069
An, J.-P., Yao, J.-F., Xu, R.-R., You, C.-X., Wang, X.-F., Hao, Y.-J. (2018). Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant. Cell Environ. 41 (11), 2678–2692. doi: 10.1111/pce.13393
An, X.-H., Tian, Y., Chen, K.-Q., Liu, X.-J., Liu, D.-D., Xie, X.-B., et al. (2015). MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 56 (4), 650–662. doi: 10.1093/pcp/pcu205
An, X.-H., Tian, Y., Chen, K.-Q., Wang, X.-F., Hao, Y.-J. (2012). The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 169 (7), 710–717. doi: 10.1016/j.jplph.2012.01.015
Ban, Y., Honda, C., Hatsuyama, Y., Igarashi, M., Bessho, H., Moriguchi, T. (2007). Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant and Cell Physiology 48(7), 958–970. doi: 10.1093/pcp/pcm066
Beppu, K., Suehara, T., Kataoka, I. (2003). High temperature and drought stress suppress the photosynthesis and carbohydrate accumulation in 'Satohnishiki' sweet cherry. Acta Hortic. 618, 371–377. doi: 10.17660/ActaHortic.2003.618.43
Boss, P. K., Davies, C., Robinson, S. P. (1996). Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol. Biol. 32 (3), 565–569. doi: 10.1007/BF00019111
Crifò, T., Petrone, G., Lo Cicero, L., Lo Piero, A. R. (2012). Short cold storage enhances the anthocyanin contents and level of transcripts related to their biosynthesis in blood oranges. J. Agric. Food Chem. 60 (1), 476–481. doi: 10.1021/jf203891e
Dai, X., Li, Q., Jiang, F., Song, Z., Tang, X., Su, S., et al. (2022). Transcriptome analysis of branches reveals candidate genes involved in anthocyanin biosynthesis of ‘Red bartlett’ pear (Pyrus communis l.). Sci. Hortic. 305, 111392. doi: 10.1016/j.scienta.2022.111392
Dela, G., Or, E., Ovadia, R., Nissim-Levi, A., Weiss, D., Oren-Shamir, M. (2003). Changes in anthocyanin concentration and composition in ‘Jaguar’ rose flowers due to transient high-temperature conditions. Plant Sci. 164 (3), 333–340. doi: 10.1016/S0168-9452(02)00417-X
Esti, M., Cinquanta, L., Sinesio, F., Moneta, E., Di Matteo, M. (2002). Physicochemical and sensory fruit characteristics of two sweet cherry cultivars after cool storage. Food Chem. 76 (4), 399–405. doi: 10.1016/S0308-8146(01)00231-X
Fang, H., Dong, Y., Yue, X., Hu, J., Jiang, S., Xu, H., et al. (2019). The b-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant. Cell Environ. 42 (7), 2090–2104. doi: 10.1111/pce.13552
Florea, L., Song, L., Salzberg, S. L. (2013). Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2, 188. doi: 10.12688/f1000research.2-188.v2
Fossen, T., Cabrita, L., Andersen, O.M. (1998). Colour and stability of pure anthocyanins influenced by pH including the alkaline region. Food Chem. 63 (4), 435–440. doi: 10.1016/S0308-8146(98)00065-X
Gao, H.-N., Jiang, H., Cui, J.-Y., You, C.-X., Li, Y.-Y. (2021). Review: The effects of hormones and environmental factors on anthocyanin biosynthesis in apple. Plant Sci. 312, 111024. doi: 10.1016/j.plantsci.2021.111024
Gonçalves, A. C., Campos, G., Alves, G., Garcia-Viguera, C., Moreno, D. A., Silva, L. R. (2021). Physical and phytochemical composition of 23 Portuguese sweet cherries as conditioned by variety (or genotype). Food Chem. 335, 127637. doi: 10.1016/j.foodchem.2020.127637
Günther, C. S., Plunkett, B. J., Cooney, J. M., Jensen, D. J. J., Trower, T. M., Elborough, C., et al. (2022). Biotic stress-induced and ripening-related anthocyanin biosynthesis are regulated by alternate phytohormone signals in blueberries. Environ. Exp. Bot. 203, 105065. doi: 10.1016/j.envexpbot.2022.105065
Hara, M., Oki, K., Hoshino, K., Kuboi, T. (2004). Effects of sucrose on anthocyanin production in hypocotyl of two radish (Raphanus sativus) varieties. Plant Biotechnol. 21 (5), 401–405. doi: 10.5511/plantbiotechnology.21.401
Hedden, P. (1999). Recent advances in gibberellin biosynthesis. J. Exp. Bot. 50 (334), 553–563. doi: 10.1093/jxb/50.334.553
Hieber, A. D., Bugos, R. C., Yamamoto, H. Y. (2000). Plant lipocalins: violaxanthin de-epoxidase and zeaxanthin epoxidase. Biochim. Biophys. Acta (BBA) - Protein Struct. Mol. Enzymol. 1482 (1), 84–91. doi: 10.1016/S0167-4838(00)00141-2
International Organisation of Vine and Wine. (2012). International Code of Oenological Practices. https://www.oiv.int/standards/international-code-of-oenological-practices.
Jaakola, L. (2013). New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 18 (9), 477–483. doi: 10.1016/j.tplants.2013.06.003
Jia, H.-F., Chai, Y.-M., Li, C.-L., Lu, D., Luo, J.-J., Qin, L., et al. (2011). Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 157 (1), 188–199. doi: 10.1104/pp.111.177311
Ji, X.-H., Wang, Y.-T., Zhang, R., Wu, S.-J., An, M.-M., Li, M., et al. (2015). Effect of auxin, cytokinin and nitrogen on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii f.niedzwetzkyana). Plant Cell Tissue Organ Cult. 120 (1), 325–337. doi: 10.1007/s11240-014-0609-y
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., Hattori, M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32 (Database issue), D277–D280. doi: 10.1093/nar/gkh063
Kerepesi, I., Galiba, G. (2000). Osmotic and salt stress-induced alteration in soluble carbohydrate content in wheat seedlings. Crop Science 40(2), 482–487. doi: 10.2135/cropsci2000.402482x
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12 (4), 357–360. doi: 10.1038/nmeth.3317
Kim, S.-H., Lee, J.-R., Kim, S.-R. (2006). Characterization of an apple anthocyanidin synthase gene in transgenic tobacco plants. J. Plant Biol. 49 (4), 326–330. doi: 10.1007/BF03031164
Kondo, S., Danjo, C. (2001). Cell wall polysaccharide metabolism during fruit development in sweet cherry 'Satohnishiki' as affected by gibberellic acid. J. Japanese Soc. Hortic. Sci. 70 (2), 178–184. doi: 10.2503/jjshs.70.178
Kuhn, N., Maldonado, J., Ponce, C., Arellano, M., Time, A., Multari, S., et al. (2021). RNAseq reveals different transcriptomic responses to GA3 in early and midseason varieties before ripening initiation in sweet cherry fruits. Sci. Rep. 11 (1), 13075. doi: 10.1038/s41598-021-92080-8
Li, B., Xie, Z., Zhang, A., Xu, W., Zhang, C., Liu, Q., et al. (2010). Tree growth characteristics and flower bud differentiation of sweet cherry (Prunus avium l.) under different climate conditions in China. Hortic. Sci. 31 (1), 6–13. doi: 10.17221/36/2009-HORTSCI
Lin-Wang, K. U. I., Micheletti, D., Palmer, J., Volz, R., Lozano, L., Espley, R., et al. (2011). High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant. Cell Environ. 34 (7), 1176–1190. doi: 10.1111/j.1365-3040.2011.02316.x
Loreti, E., Povero, G., Novi, G., Solfanelli, C., Alpi, A., Perata, P. (2008). Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol. 179 (4), 1004–1016. doi: 10.1111/j.1469-8137.2008.02511.x
Lou, L., Yue, Y., Yin, P., Chen, J., Ye, X., Liu, D. (2019). Study on the copigmentation effects of tannic acid and chlorogenic acid on the anthocyanins of bayberry. Food and Fermentation Industries 45 (4), 74–80. doi: 10.13995/j.cnki.11-1802/ts.017972
Lv, L. L., Feng, X. F., Li, W., Li, K. (2019). High temperature reduces peel color in eggplant (Solanum melongena) as revealed by RNA-seq analysis. Genome 62 (7), 503–512. doi: 10.1139/gen-2019-0021
Mita, S., Hirano, H., Nakamura, K. (1997). Negative regulation in the expression of a sugar-inducible gene in Arabidopsis thaliana. a recessive mutation causing enhanced expression of a gene for β-amylase. Plant Physiol. 114 (2), 575–582. doi: 10.1104/pp.114.2.575
Moglia, A., Florio, F. E., Iacopino, S., Guerrieri, A., Milani, A. M., Comino, C., et al. (2020). Identification of a new R3 MYB type repressor and functional characterization of the members of the MBW transcriptional complex involved in anthocyanin biosynthesis in eggplant (S. melongena l.). PloS One 15 (5), e0232986. doi: 10.1371/journal.pone.0232986
Mori, K., Sugaya, S., Gemma, H. (2005). Decreased anthocyanin biosynthesis in grape berries grown under elevated night temperature condition. Sci. Hortic. 105 (3), 319–330. doi: 10.1016/j.scienta.2005.01.032
Mori, T., Sakurai, M., Seki, M., Furusaki, S. (1994). Use of auxin and cytokinin to regulate anthocyanin production and composition in suspension cultures of strawberry cell. J. Sci. Food Agric. 65 (3), 271–276. doi: 10.1002/jsfa.2740650303
Mori, K., Goto-Yamamoto, N., Kitayama, M., Hashizume, K. (2007). Loss of anthocyanins in red-wine grape under high temperature. J. Exp. Bot. 58 (8), 1935–1945. doi: 10.1093/jxb/erm055.
Moustakas, M., Sperdouli, I., Kouna, T., Antonopoulou, C.-I., Therios, I. (2011). Exogenous proline induces soluble sugar accumulation and alleviates drought stress effects on photosystem II functioning of Arabidopsis thaliana leaves. Plant Growth Regulation 65(2), 315. doi: 10.1007/s10725-011-9604-z
Ohto, M., Onai, K., Furukawa, Y., Aoki, E., Araki, T., Nakamura, K. (2001). Effects of sugar on vegetative development and floral transition in arabidopsis. Plant Physiol. 127 (1), 252–261. doi: 10.1104/pp.127.1.252
Oren-Shamir, M. (2009). Does anthocyanin degradation play a significant role in determining pigment concentration in plants? Plant Sci. 177 (4), 310–316. doi: 10.1016/j.plantsci.2009.06.015
Petroni, K., Tonelli, C. (2011). Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 181 (3), 219–229. doi: 10.1016/j.plantsci.2011.05.009
Prinsi, B., Negri, A. S., Espen, L., Piagnani, M. C. (2016). Proteomic comparison of fruit ripening between ‘Hedelfinger’ sweet cherry (Prunus avium l.) and its somaclonal variant ‘HS’. J. Agric. Food Chem. 64 (20), 4171–4181. doi: 10.1021/acs.jafc.6b01039
Rahim, M. A., Busatto, N., Trainotti, L. (2014). Regulation of anthocyanin biosynthesis in peach fruits. Planta 240 (5), 913–929. doi: 10.1007/s00425-014-2078-2
Rehman, R. N. U., You, Y., Zhang, L., Goudia, B. D., Khan, A. R., Li, P., et al. (2017). High temperature induced anthocyanin inhibition and active degradation in Malus profusion. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01401
Ren, J., Leng, P. (2010). Role of abscisic acid and ethylene in fruit maturation of sweet cherry. Acta Hortic. Sin. 37 (2), 199–206. doi: 10.16420/j.issn.0513-353x.2010.02.007
Roper, T. R., Keller, J. D., Loescher, W. H., Rom, C. R. (1988). Photosynthesis and carbohydrate partitioning in sweet cherry: Fruiting effects. Physiol. Plant. 72 (1), 42–47. doi: 10.1111/j.1399-3054.1988.tb06620.x
Rowan, D. D., Cao, M., Lin-Wang, K., Cooney, J. M., Jensen, D. J., Austin, P. T., et al. (2009). Environmental regulation of leaf colour in red 35S:PAP1 arabidopsis thaliana. New Phytol. 182 (1), 102–115. doi: 10.1111/j.1469-8137.2008.02737.x
Ryu, S., Han, J. H., Cho, J. G., Jeong, J. H., Lee, S. K., Lee, H. J. (2020). High temperature at veraison inhibits anthocyanin biosynthesis in berry skins during ripening in ‘Kyoho’ grapevines. Plant Physiol. Biochem. 157, 219–228. doi: 10.1016/j.plaphy.2020.10.024
Saito, S., Hirai, N., Matsumoto, C., Ohigashi, H., Ohta, D., Sakata, K., et al. (2004). Arabidopsis CYP707As encode (+)-abscisic acid 8'-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 134 (4), 1439–1449. doi: 10.1104/pp.103.037614
Setha, S., Kondo, S., Hirai, N., Ohigashi, H. (2005). Quantification of ABA and its metabolites in sweet cherries usingdeuterium-labeled internal standards. Plant Growth Regul. 45 (3), 183–188. doi: 10.1007/s10725-005-3088-7
Shan, X., Zhang, Y., Peng, W., Wang, Z., Xie, D. (2009). Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 60 (13), 3849–3860. doi: 10.1093/jxb/erp223
Shen, X., Zhao, K., Liu, L., Zhang, K., Yuan, H., Liao, X., et al. (2014). A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong deng (Prunus avium l.). Plant Cell Physiol. 55 (5), 862–880. doi: 10.1093/pcp/pcu013
Smeekens, S. (2000). Sugar-induced signal transduction in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 49–81. doi: 10.1146/annurev.arplant.51.1.49
Solfanelli, C., Poggi, A., Loreti, E., Alpi, A., Perata, P. (2006). Sucrose-specific induction of the anthocyanin biosynthetic pathway in arabidopsis. Plant Physiol. 140 (2), 637–646. doi: 10.1104/pp.105.072579
Steyn, W. J., Wand, S. J. E., Jacobs, G., Rosecrance, R. C., Roberts, S. C. (2009). Evidence for a photoprotective function of low-temperature-induced anthocyanin accumulation in apple and pear peel. Physiologia Plantarum 136(4), 461–472. doi: 10.1111/j.1399-3054.2009.01246.x.
Teribia, N., Tijero, V., Munné-Bosch, S. (2016). Linking hormonal profiles with variations in sugar and anthocyanin contents during the natural development and ripening of sweet cherries. New Biotechnol. 33 (6), 824–833. doi: 10.1016/j.nbt.2016.07.015
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Ubi, B.E., Honda, C., Bessho, H., Kondo, S., Wada, M., Kobayashi, S., et al. (2006). Expression analysis of anthocyanin biosynthetic genes in apple skin: Effect of UV-B and temperature. Plant Science 170(3), 571–578. doi: 10.1016/j.plantsci.2005.10.009.
Usenik, V., Kastelec, D., Štampar, F. (2005). Physicochemical changes of sweet cherry fruits related to application of gibberellic acid. Food Chem. 90 (4), 663–671. doi: 10.1016/j.foodchem.2004.04.027
Wang, L., Brouard, E., Prodhomme, D., Hilbert, G., Renaud, C., Petit, J.-P., et al. (2022). Regulation of anthocyanin and sugar accumulation in grape berry through carbon limitation and exogenous ABA application. Food Res. Int. 160, 111478. doi: 10.1016/j.foodres.2022.111478
Wang, L., Feng, Z., Wang, X., Wang, X., Zhang, X. (2010). DEGseq: an r package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26 (1), 136–138. doi: 10.1093/bioinformatics/btp612
Wang, C.-K., Han, P.-L., Zhao, Y.-W., Ji, X.-L., Yu, J.-Q., You, C.-X., et al. (2020). Auxin regulates anthocyanin biosynthesis through the auxin repressor protein MdIAA26. Biochem. Biophys. Res. Commun. 533 (4), 717–722. doi: 10.1016/j.bbrc.2020.09.065
Wang, Z., Meng, D., Wang, A., Li, T., Jiang, S., Cong, P., et al. (2013). The methylation of the PcMYB10 promoter is associated with green-skinned sport in max red Bartlett pear. Plant Physiol. 162 (2), 885–896. doi: 10.1104/pp.113.214700
Wang, Y., Sun, J., Wang, N., Xu, H., Qu, C., Jiang, S., et al. (2019). MdMYBL2 helps regulate cytokinin-induced anthocyanin biosynthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana) callus. Funct. Plant Biol. 46 (2), 187–196. doi: 10.1071/fp17216
Watanabe, S., Kojima, K., Ide, Y., Sasaki, S. (2000). Effects of saline and osmotic stress on proline and sugar accumulation in Populus euphratica in vitro. Plant Cell, Tissue and Organ Culture 63(3), 199–206. doi: 10.1023/A:1010619503680
Weiss, D. (2000). Regulation of flower pigmentation and growth: Multiple signaling pathways control anthocyanin synthesis in expanding petals. Physiol. Plant. 110 (2), 152–157. doi: 10.1034/j.1399-3054.2000.110202.x
Wu, T., Liu, H.-T., Zhao, G.-P., Song, J.-X., Wang, X.-L., Yang, C.-Q., et al. (2020). Jasmonate and ethylene-regulated ethylene response factor 22 promotes lanolin-induced anthocyanin biosynthesis in 'Zaosu' pear (Pyrus bretschneideri rehd.) fruit. Biomolecules 10 (2), 278. doi: 10.3390/biom10020278
Xia, H., Zhou, Y., Lin, Z., Guo, Y., Liu, X., Wang, T., et al. (2022). Characterization and functional validation of β-carotene hydroxylase AcBCH genes in Actinidia chinensis. Horticult. Res. 9, uhac063. doi: 10.1093/hr/uhac063
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39 (Web Server issue), W316–W322. doi: 10.1093/nar/gkr483
Xu, Z. J., Nakajima, M., Suzuki, Y., Yamaguchi, I. (2002). Cloning and characterization of the abscisic acid-specific glucosyltransferase gene from adzuki bean seedlings. Plant Physiol. 129 (3), 1285–1295. doi: 10.1104/pp.001784
Xue, L., Wang, Z., Zhang, W., Li, Y., Wang, J., Lei, J. (2016). Flower pigment inheritance and anthocyanin characterization of hybrids from pink-flowered and white-flowered strawberry. Sci. Hortic. 200, 143–150. doi: 10.1016/j.scienta.2016.01.020
Yamane, T., Jeong, S. T., Goto-Yamamoto, N., Koshita, Y., Kobayashi, S. (2006). Effects of temperature on anthocyanin biosynthesis in grape berry skins. Am. J. enol. viticult. 57 (1), 54–59. doi: 10.5344/ajev.2006.57.1.54
Yang, B., He, S., Liu, Y., Liu, B., Ju, Y., Kang, D., et al. (2020). Transcriptomics integrated with metabolomics reveals the effect of regulated deficit irrigation on anthocyanin biosynthesis in Cabernet sauvignon grape berries. Food Chem. 314, 126170. doi: 10.1016/j.foodchem.2020.126170
Zhai, R., Liu, X.-T., Feng, W.-T., Chen, S.-S., Xu, L.-F., Wang, Z.-G., et al. (2014). Different biosynthesis patterns among flavonoid 3-glycosides with distinct effects on accumulation of other flavonoid metabolites in pears (Pyrus bretschneideri rehd.). PloS One 9 (3), e91945. doi: 10.1371/journal.pone.0091945
Zhai, R., Wang, Z., Zhang, S., Meng, G., Song, L., Wang, Z., et al. (2016). Two MYB transcription factors regulate flavonoid biosynthesis in pear fruit (Pyrus bretschneideri rehd.). J. Exp. Bot. 67 (5), 1275–1284. doi: 10.1093/jxb/erv524
Zhang, H. P., Wu, J. Y., Qin, G. H., Yao, G. F., Qi, K. J., Wang, L. F., et al. (2014). The role of sucrose-metabolizing enzymes in pear fruit that differ in sucrose accumulation. Acta Physiol. Plant. 36 (1), 71–77. doi: 10.1007/s11738-013-1387-6
Zhang, S., Ying, H., Pingcuo, G., Wang, S., Zhao, F., Cui, Y., et al. (2019a). Identification of potential metabolites mediating bird’s selective feeding on Prunus mira flowers. BioMed Research International 2019, 1395480. doi: 10.1155/2019/139548010
Zhang, X., Jiang, X., He, Y., Li, L., Xu, P., Sun, Z., et al. (2019b). AtHB2, a class II HD-ZIP protein, negatively regulates the expression of CsANS, which encodes a key enzyme in Camellia sinensis catechin biosynthesis. Physiologia Plantarum 166(4), 936–945. doi: 10.1111/ppl.12851.
Zhang, X., Li, B., Duan, R., Han, C., Wang, L., Yang, J., et al. (2022). Transcriptome analysis reveals roles of sucrose in anthocyanin accumulation in 'Kuerle xiangli' (Pyrus sinkiangensis yü). Genes 13 (6), 1064. doi: 10.3390/genes13061064
Zhang, Y., Liu, Z., Liu, R., Hao, H., Bi, Y. (2011). Gibberellins negatively regulate low temperature-induced anthocyanin accumulation in a HY5/HYH-dependent manner. Plant Signaling Behav. 6 (5), 632–634. doi: 10.4161/psb.6.5.14343
Keywords: high temperature, sweet cherry, coloring, anthocyanin, ABA
Citation: Tan Y, Wen B, Xu L, Zong X, Sun Y, Wei G and Wei H (2023) High temperature inhibited the accumulation of anthocyanin by promoting ABA catabolism in sweet cherry fruits. Front. Plant Sci. 14:1079292. doi: 10.3389/fpls.2023.1079292
Received: 25 October 2022; Accepted: 30 January 2023;
Published: 13 February 2023.
Edited by:
Yugang Zhang, Qingdao Agricultural University, ChinaReviewed by:
Jietang Zhao, South China Agricultural University, ChinaXiao-Fei Wang, Shandong Agricultural University, China
Shenghui Jiang, Qingdao Agricultural University, China
Copyright © 2023 Tan, Wen, Xu, Zong, Sun, Wei and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hairong Wei, aHJ3ZWkyMDA0QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Yue Tan
Yue Tan Binbin Wen
Binbin Wen Li Xu1
Li Xu1 Xiaojuan Zong
Xiaojuan Zong Hairong Wei
Hairong Wei