
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 January 2023
Sec. Plant Pathogen Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1078100
This article is part of the Research Topic Systemic Resistance and Defense Priming Against Pathogens View all 9 articles
 Piao Yang1
Piao Yang1 Zhenzhen Zhao1
Zhenzhen Zhao1 Jiangbo Fan2
Jiangbo Fan2 Yinping Liang3
Yinping Liang3 Matthew C. Bernier4
Matthew C. Bernier4 Yu Gao5,6
Yu Gao5,6 Lijing Zhao1
Lijing Zhao1 Stephen Obol Opiyo1
Stephen Obol Opiyo1 Ye Xia1*
Ye Xia1*Pseudomonas syringae and Botrytis cinerea cause destructive bacterial speck and grey mold diseases in many plant species, leading to substantial economic losses in agricultural production. Our study discovered that the application of Bacillus proteolyticus strain OSUB18 as a root-drench enhanced the resistance of Arabidopsis plants against P. syringae and B. cinerea through activating Induced Systemic Resistance (ISR). The underlying mechanisms by which OSUB18 activates ISR were studied. Our results revealed that the Arabidopsis plants with OSUB18 root-drench showed the enhanced callose deposition and ROS production when inoculated with Pseudomonas syringae and Botrytis cinerea pathogens, respectively. Also, the increased salicylic acid (SA) levels were detected in the OSUB18 root-drenched plants compared with the water root-drenched plants after the P. syringae infection. In contrast, the OSUB18 root-drenched plants produced significantly higher levels of jasmonyl isoleucine (JA-Ile) than the water root-drenched control after the B. cinerea infection. The qRT-PCR analyses indicated that the ISR-responsive gene MYC2 and the ROS-responsive gene RBOHD were significantly upregulated in OSUB18 root-drenched plants upon both pathogen infections compared with the controls. Also, twenty-four hours after the bacterial or fungal inoculation, the OSUB18 root-drenched plants showed the upregulated expression levels of SA-related genes (PR1, PR2, PR5, EDS5, and SID2) or JA-related genes (PDF1.2, LOX3, JAR1 and COI1), respectively, which were consistent with the related hormone levels upon these two different pathogen infections. Moreover, OSUB18 can trigger ISR in jar1 or sid2 mutants but not in myc2 or npr1 mutants, depending on the pathogen’s lifestyles. In addition, OSUB18 prompted the production of acetoin, which was reported as a novel rhizobacterial ISR elicitor. In summary, our studies discover that OSUB18 is a novel ISR inducer that primes plants’ resistance against bacterial and fungal pathogens by enhancing the callose deposition and ROS accumulation, increasing the production of specific phytohormones and other metabolites involved in plant defense, and elevating the expression levels of multiple defense genes.
Food crops are the primary nutrient source of vitamins, minerals, and health-promoting antioxidants worldwide (Giovannoni et al., 2017). Diverse biotic and abiotic stresses could significantly reduce the yield and quality of crops. For instance, under favorable conditions of rainfall and humidity, up to 80% of the fruit crops’ flowers can be lost due to fungal pathogen infections if the fungicides were not used (Ries, 1995). Like the other major food crops, the yield, quality, and security of fruit crops are vastly reduced upon the pathogen infections at the household, national, and global levels (Savary et al., 2019). For example, the fungal pathogen Botrytis cinerea leads to gray mold diseases, which could cause significant economic losses to the tomato and strawberry industries (Petrasch et al., 2019). The total annual economic losses of crops caused by B. cinerea are over $10 billion globally (Petrasch et al., 2019). Similarly, the bacterial pathogen Pseudomonas syringae pathovars caused severe bacterial speck disease in more than 200 plant species, such as the tomato speck disease (pathovar. tomato; Shenge et al., 2007) and the bleeding canker of horse-chestnut (pathovar. aesculi; Green et al., 2010). Under wet conditions, these disease outbreaks were shown to be devastating (Murillo et al., 2010). For instance, a study reported that bacterial speck disease caused 75% yield losses in field plants infected at their early growth stage (Yunis et al., 1980).
Synthetic chemicals have long been used in the agricultural system to manage plant diseases in order to increase crop yield and improve crop quality (Tronsmo et al., 2020). However, chemical residues pose health risks to humans and wild animals (Fernández-Vizcaíno et al., 2022). More and more restrictions have been placed on the use of synthetic chemicals in crops (Provost and Pedneault, 2016). In addition, overuse of synthetic chemicals could cause pathogen resistance. For instance, copper-based treatments for bacterial pathogens, such as P. syringae, are becoming less and less effective due to resistance in bacterial populations (Cazorla et al., 2002). Thus, safer, sustainable, and effective alternatives to synthetic chemicals are urgently needed. Plant growth-promoting rhizobacteria (PGPR) have emerged in agriculture as suitable alternatives to synthetic chemicals for the environmentally safer control of plant diseases (Bhattacharyya and Jha, 2012; Maheshwari et al., 2015). For instance, biological control uses the natural enemies of the pathogens against them, which influences the balance of the plant-microbe interaction. Therefore, the pathogens are eliminated or at least decreased to below economic thresholds (Leppla and LeBeck, 2021). Many Bacillus species are popular biological control agents in the agricultural practice (Kumar et al., 2011).
Bacillus species are Gram-positive bacteria with rod shapes and are under the phylum of Firmicutes (Turnbull, 1996). Many Bacillus species can tolerate different stresses and are potential biological control agents in the agricultural practice (Kumar et al., 2011). For instance, B. thuringiensis has been applied as a bio-insecticide because it can produce toxins to kill insects that attach to cotton plants (May, 2011). B. siamensis can produce antimicrobial metabolites to inhibit plant pathogens, such as B. cinerea and Rhizoctonia solani, and it can also release volatiles to promote plant growth (Jeong et al., 2012). Some Bacillus species, such as B. amyliliquefaciens, can not only promote plant growth (Yang et al., 2022) but also secrete certain enzymes as the source of natural antibiotic proteins, including barnase, alpha-amylase, and the BamH1 restriction enzyme (Molohon et al., 2011). Bacillus simplex strain WY10 was reported to be able to uptake DNA by transformation (Keen et al., 2017).
Beyond the direct biological control effects, some Bacillus spp. have been reported to indirectly benefit plants by triggering an enhanced resistance known as Induced Systemic Resistance (ISR) (Yu et al., 2022). ISR is mediated by beneficial bacteria and fungi living in the rhizosphere. These beneficial microbes impact plant growth and boost plant defenses against phytopathogens and pests (Romera et al., 2019). MYC2 is a transcription factor constitutively expressed in rhizobacteria-induced systemic resistance (Pozo et al., 2008). MYC2 is required for beneficial microbe-triggered ISR and regulates the crosstalk between different phytohormones such as JA, SA, and IAA (Kazan and Manners, 2013). The signaling pathways mediating ISR can differ depending on the plant and microbial species (Ryu et al., 2003; Alizadeh et al., 2013). For example, Bacillus cereus C1L activated ISR in tobacco against B. cinerea infection via the production of volatile dimethyl disulfide (Huang et al., 2012). Bacillus amyloliquefaciens FZB42 conferred ISR in Nicotiana benthamiana against B. cinerea infection via inducing stomatal closure and activating SA-, JA/ET-Signaling Pathways (Wu et al., 2018).
Bacillus proteolyticus strain OSUB18 was initially isolated from the switchgrass plants through the previously reported approach (Xia et al., 2013). The objectives of this study were to elucidate the mechanisms of OSUB18 on activating the ISR of Arabidopsis plants against the pathogens of P. syringae and B. cinerea by integrated physiological, molecular, and biochemical approaches. Our studies provide new insights into how Bacillus proteolyticus could suppress pathogenic bacteria and fungi with different lifestyles by activating the host Arabidopsis plant’s systemic resistance through similar but also different defense responses.
All wild-type A. thaliana (Col-0) and mutant (sid2, npr1, jar1, and myc2) seeds were obtained from the Arabidopsis Biological Resource Center (ABRC) in Columbus, Ohio. The homozygous lines were identified based on the information from a published report (Jarret and McCluskey, 2019) and salk website (http://signal.salk.edu/). A. thaliana plants were grown in a potting mix (Lambert LM-111) in a walk-in growth chamber (Winnipeg, MB, Canada) at 22 °C with 12-hour light, 12-hour darkness and ~60% relative humidity (Yoo et al., 2022). The light intensity was adjusted as ~120 μmol/m2 s (Zhao et al., 2020). Tobacco and tomato seeds were purchased from Park Seed Company at U.S.A. Plants were grown in a professional potting mix (Lambert LM-111) in a greenhouse compartment with air temperatures set at 28 °C, day/night cycles with 16-hour light and 8-hour darkness, and relative humidity at ~45%. Plants were fertilized with Osmocote’s general fertilizer (14-14-4) (Calhoun et al., 2021) to support the needed nutrients.
OSUB18 was cultured on Tryptic Soy Agar (TSA) plates at 28 °C for 2 days before the bacteria were transferred tosterile 15 mL tubes. 2 mL sterile water was used to wash out all bacterial residues from each TSA plate. TSA plates without OSUB18 were also washed with 2ml sterile water and used as the controls. The 15 mL tubes with OSUB18 cells or controls were homogenized with pipette tips and then transferred to new sterile 2 mL tubes and centrifuged at 12000 rpm for 10 minutes. The above clear cell-free supernatant (CFS) was defined as 1x BEE and stored at -80°C until use. 10x BEE was obtained by concentrating the 1xBEE from 2 mL to 200 µL with a lyophilizer machine. The 5x BEE was diluted two times from the 10x BEE with sterile water.
OSUB18 was cultured on Tryptic Soy Agar (TSA) plates at 28 °C. Phytopathogenic P. syringae pv. tomato DC3000 (Pst DC3000) was cultured on King’s B Agar (KBA) plates containing 50 mg/L rifampicin at 28 °C. The grey mold fungus B. cinerea was cultured on Potato Dextrose Agar (PDA) plates at 22 °C. Antagonism of OSUB18 against B. cinerea was conducted on Potato Soy Agar (PSA, made of PDA and TSA at 1:1 ratio) plates by the dual-culture test. In brief, one agar disc of B. cinerea was first placed on one side of the PSA plate. Then, OSUB18 cells (10ul per drop at 10^8 CFU/ml) were spotted on the opposite side away from the fungal plugs in the plates. The plates were then incubated for five days at 22 °C before the fungal growth was measured. The control set was conducted simultaneously by replacing the OSUB18 cells with sterile water. A similar dual-culture assay was carried out to test the direct inhibition effect of OSUB18 against Pst DC3000. In brief, Pst DC3000 cells (100ul, 10^8 CFU/ml) were evenly distributed on KBA by sterile glass beads (Eyler, 2013). OSUB18 cells (20ul, 10^8 CFU/ml) were spotted to the same plate ~3 cm apart from the plate center. The plates were then incubated for two days at 28 °C before measuring the inhibition zone. The OSUB18/Pst DC3000 cells were collected freshly and washed 3 times with sterile water to remove the nutrient residues before use. The above methods were modified from previous reports (Hu et al., 2014; Chen et al., 2018). All experiments were repeated three times with four plates as the replicates for each treatment.
The bacterial 16S ribosomal DNA (16S rDNA) sequence of OSUB18 was amplified by polymerase chain reaction (PCR) from its genomic DNA extracted by the Quick-DNA Fungal/Bacterial Micro prep Kit (Zymo Research, Irvine, CA, USA) with the primer pair 799F/1193R (Chen et al., 2020). The purified 16S rDNA sequence was subjected to the Basic Local Alignment Search Tool (BLAST) provided by the National Center for Biotechnology Information (NCBI). We used the Molecular Evolutionary Genetics Analysis (MEGA) software (Tamura et al., 2021) to construct the phylogenetic tree with the Gram positive bacterium Deinococcus radiophilus DSM 20551T as an out-cluster control.
The leaves of the 4~6-week-old Arabidopsis plants were syringe-injected with Pst DC3000 at 1 x 106 CFU/mL, dipping inoculated, or spray inoculated with Pst DC3000 at 5 x 108 CFU/mL containing 0.05% Silwet L-77 based on the experimental needs. Three days after inoculation, the bacterial growth was quantified by measuring Pst DC3000 growth in infected leaves using the serial dilution technique (Katagiri et al., 2002; Yao et al., 2013).
The leaves of the 4~6-week-old Arabidopsis plants were inoculated with B. cinerea spores (10ul-drop, 5 x 105 spores/mL in half-strength V8 juice) and covered with a plastic dome to maintain high humidity to facilitate disease development at 22 °C. Three days later, the fungal disease symptom was assessed by measuring the lesion size using the ImageJ software (Weiberg et al., 2013).
The A. thaliana seedlings were transplanted into pots (3.5” x 3.5” x 2”). They were then subjected to a weekly treatment of OSUB18 (107 CFU/mL, 50ml per pot) or water (Ctrl) as the root drench for three consecutive weeks. A week after the last root drench, the treated A. thalianaplants were used for bacterial or fungal pathogen inoculation to check ISR responses.
Superoxide anion (•O2−) detection by nitroblue tetrazolium (NBT) staining was performed as described by Jambunathan (Jambunathan, 2010). Hydrogen peroxide (H2O2) detection by diaminobenzidine tetrahydrochloride (DAB) staining was performed based on a previously published method (Nie et al., 2017). Callose deposition was measured following a previous method (Wang et al., 2017). Because the bacterial pathogen was infiltrated into the plant leaves for the infection assay and the fungal pathogen was drop-inoculated on the plant leaves, different patterns of callose deposition were induced. Therefore, they were quantified as callose/mm2 (Jin and Mackey, 2017) in the bacterial infection assay and relative callose (Nie et al., 2017) in the fungal infection assay with Image J software (https://imagej.nih.gov/ij/ ). ROS burst assay was performed following a published protocol (Sang and Macho, 2017).
A. thaliana (Col-0) plants were root-drenched with OSUB18 or water (Ctrl) for bacterial or fungal phytopathogen infection assay. At 0 or 24 hours post-infection, the infected leaves were sampled for plant defense-related hormone extraction. SA, SA glucoside (SAG), JA, and JA isoleucine (JA-IIe) were extracted and quantified from ~0.1g fresh weight leaf tissues, as described previously (Zhao et al., 2022b). In brief, phytohormones were extracted from leaf samples with a methanol buffer with the addition of acetic acid and isotope-labeled internal standards (2H4-SA and 2H2-JA, CDN Isotopes) on ice. The supernatant was collected after being centrifued at 4 °C and analyzed by UPLC-MS on the Thermo Fisher Ultimate 3000 system (Thermo Fisher Scientific) equipped with a C18 (100 mm × 2.0 mm) column (Waters company, Milford, MA, USA). The solvent gradient used was 100%A (99.9% H2O: 0.1% CHOOH) to 100%B (99.9% CH3CN: 0.1% CHOOH) at 200 ul/min. The endogenous and isotope-labeled SA and JA were detected using the following mass transitions: SA 137 > 93, 2H4 SA 141 > 97, SAG 299 > 93, JA 209 > 59, 2H2-JA 211 > 61, JA-IIe 247 > 97.
The total RNA of A. thaliana (Col-0) leaves with 0 or 24 hours of the bacterial or fungal pathogen infection was extracted with TRIzol Reagent (Invitrogen, Carlsbad, MA, USA). First-strand cDNA was synthesized from 1 µg total RNA with a reverse transcription kit (Applied Biosystem, Waltham, MA, USA). The qRT-PCR was performed on the CFX96 real-time PCR machine (Bio-Rad, Hercules, CA, USA) with UBIQUITIN10 (UBQ10; At4g05320) as the internal reference gene. Table S1 lists the gene-specific primers. A standard curve was made by determining the threshold cycle (Ct) values for each primer pair’s dilution series of the cDNA product. The gene expression was calculated using the ΔCT method using the UBQ10 as the reference gene, following the Bio-Rad Real-Time Application Guide. In summary, the relative quantity of each gene is expressed in comparison to UBQ10 using the formula 2^ (CtUBQ10-CtGENE), where 2 represents the perfect PCR efficiency. For each qRT-PCR reaction, the Ct was determined by setting the threshold within the logarithmic amplification phase. The ΔCT method is a variation of the Livak (2-ΔΔCT) method. The ΔCT method is simpler to perform and gives essentially the same results as the Livak (2-ΔΔCT) method (BIO-RAD Real-Time PCR Applications Guide).
The O-CAS assay was conducted to detect the bacterial siderophore (Pérez-Miranda et al., 2007). The bacterial exopolysaccharide (EPS) was detected as previously described (Mu’minah et al., 2015). The bacterial acetoin and diacetyl were detected according to the previous report (Khalaf and Raizada, 2018). The bacterial hydrogen cyanide (HCN) was detected according to the previous study (Castric and Castric, 1983). The ammonia production was tested according to the previous report (Vyas et al., 2018).The production of indole acetic acid (IAA) and phosphate solubilization assays were performed according to the previous studies (Maheshwari et al., 2015; Batista et al., 2021).The organic acid was detected on Bromocresol purple Agar (BCPA) with 5′,5″-dibromo-o-cresolsulfophthalein as a pH indicator. The catalase activity test was performed according to the previous report (Odds, 1981).All the related experiments had at least 4 replicates and had been repeated 3 times with consistent results.
All statistical analyses were performed using the SPSS software. Data were summarized as mean ± s.e.m unless otherwise stated. Graph bars with different letters or * indicate the p-value < 0.05 by a Student t-test (to compare two groups) or analysis of variance (ANOVA, to compare three or more groups). Data from at least three biological replicates were presented. All experiments were repeated three times to ensure consistency.
OSUB18 was initially isolated from switchgrass plants (Panicum virgatum L.) in Ohio by the previous method (Xia et al., 2013). We investigated the potential capability of OSUB18 in plant pathogen inhibition and disease control. We noticed that one-day-old OSUB18 cultured on Tryptic Soy Agar (TSA) plates exhibited colonies with undulate margins (Figure 1A, upper). The representative crinkle colony was confirmed under a light microscope (Figure 1A, bottom). OSUB18 in vitro plate assay showed its ability to inhibit the growth of Pst DC3000 (Figures 1B, C), which is the causing agent of bacterial speck disease and the top first plant pathogenic bacterium with a hemibiotrophic lifestyle in molecular plant pathology study (Mansfield et al., 2012). Additionally, we examined the capacity of OSUB18 to control B. cinerea, a prevalent fungal pathogen with a necrotrophic lifestyle that causes the destructive grey mold disease on more than 200 plant species (Dean et al., 2012). We found that in vitro, OSUB18 significantly inhibited the growth of B. cinerea by releasing diffusible functional compounds to the agar media (Figures 1D, E). We also found that OSUB18 could increase Arabidopsis plants’ shoot biomass and seed yield by the root-drench treatment (Figure S1). To characterize OSUB18 at the molecular level, we amplified a PCR fragment from its genome DNA with the internal transcribed sequences (ITS) primers 799F/1193R (Chen et al., 2020) and conducted the Sanger sequencing for the purified PCR fragment. Based on the 16S ribosomal RNA sequences of OSUB18 and the other various sequenced Bacillus spp. from the NCBI database, we built a phylogenetic tree for OSUB18. The phylogenetic analysis indicated that OSUB18 belongs most closely to Bacillus proteolyticus at the species level (Figure S2).
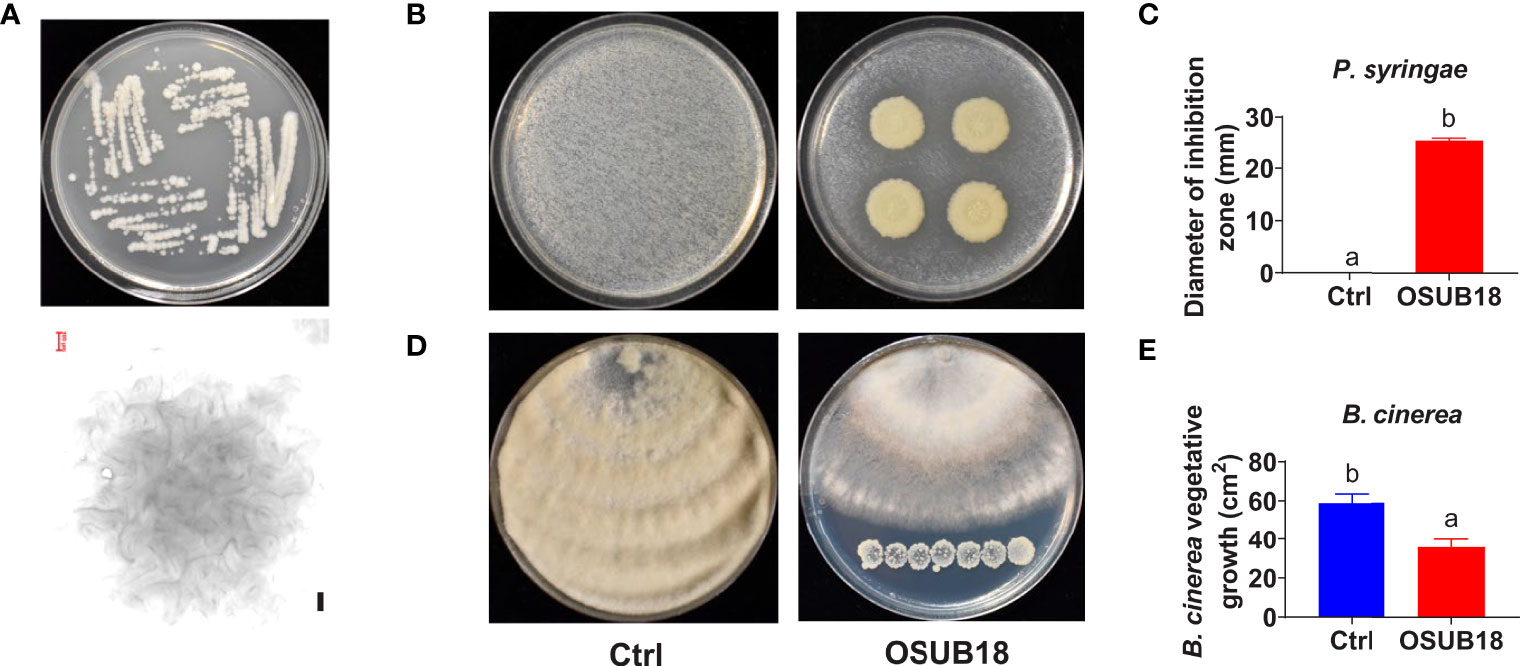
Figure 1 Isolation and identification of OSUB18 for bacterial speck disease and fungal gray mold disease control. (A) Colony morphology of OSUB18 on Tryptic Soy Agar (TSA) after 24 hours of incubation at 28°C. The bottom panel is the microscopic image of one representative colony from the upper panel. Scale bar = 100 µm. (B) OSUB18 antagonized the bacterial speck disease-causing agent P. syringae. (C) Quantification of the inhibition zone in (B). (D) OSUB18 antagonized the gray mold disease-causing agent B cinerea. Note that there was no physical contact between the bacterium and the fungus. (E) Quantification of the fungal growth in (D). Data present mean ± SE of three biological replicates. Data with different letters indicate a p-value < 0.05 on Student’s t-test.
To further study the efficacy of OSUB18 in antagonizing phytopathogens in plants, we mixed OSUB18 cells or sterile water (control) into the pathogen inoculum and conducted the pathogen infection assay in plants, which is a much more complicated and natural system than the in vitro co-culture agar plate assay. As expected, the mixing of OSUB18 in the pathogen inoculum significantly inhibited the growth of Pst DC3000 in A. thaliana Col-0 leaves (Figures 2A–C). In addition to the model plant A. thaliana, tobacco leaves co-syringe injected with Pst DC3000 and OSUB18 showed much weaker disease symptoms (Figure 2D) than the water control. In addition, similar results were obtained from 4 different cultivars of tomato plants (Figures 2E, F), validating the roles of OSUB18 against the bacterial phytopathogen in different plant species. Likewise, we mingled OSUB18 into the fungal pathogen inoculum of B. cinerea for its infection assay. Consistently, the presence of OSUB18 significantly reduced the virulence of B. cinerea on Arabidopsis Col-0 plants (Figures 3A, B), tobacco plants (Figures 3C, D), and tomato plants (Figures 3E, F). Our results indicate that OSUB18 has great potential in controlling bacterial and fungal diseases with a broad spectrum in different plant species.
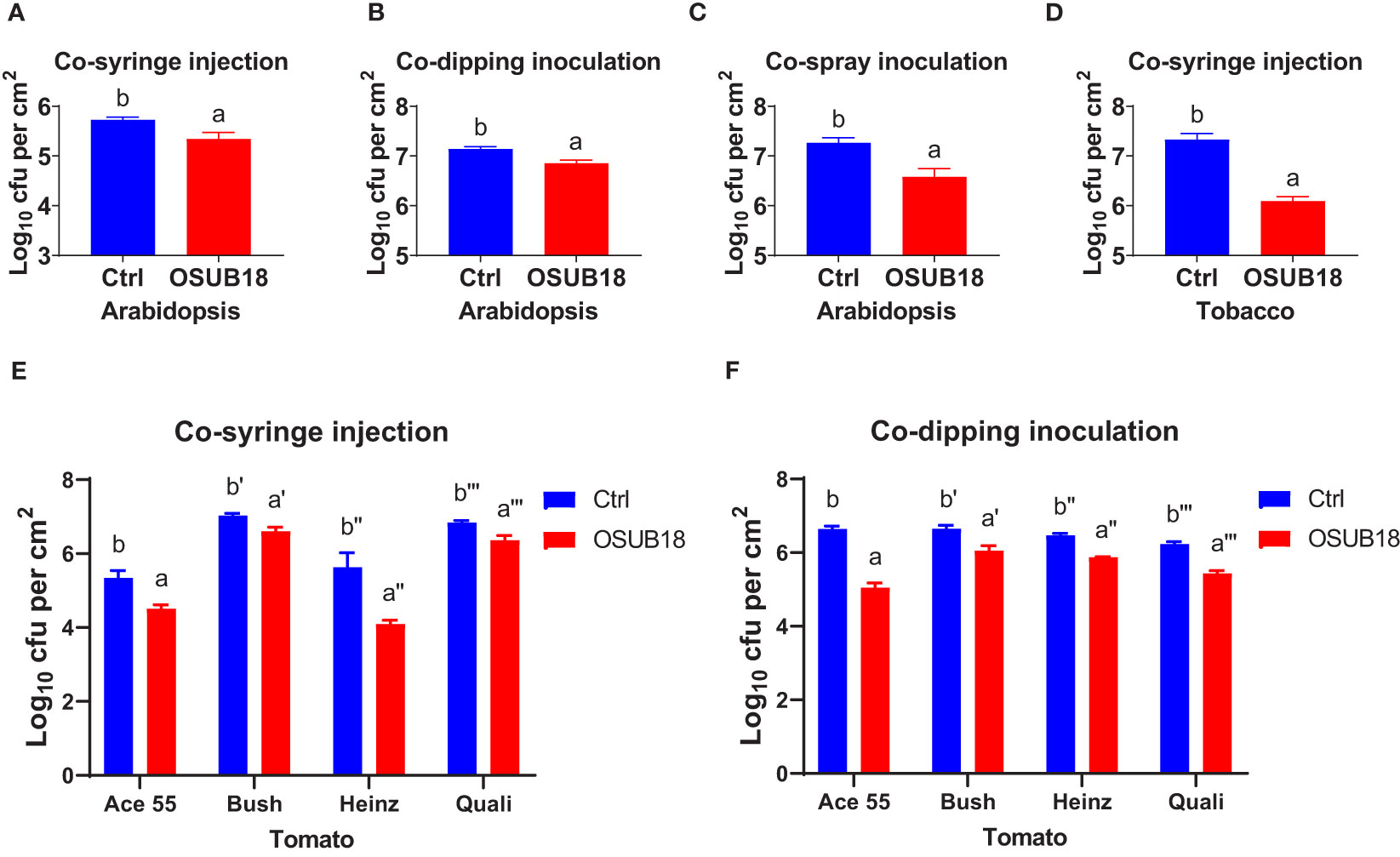
Figure 2 OSUB18 co-inoculation protected plants of different species from the bacterial pathogen Pst DC3000 infections. (A–C) OSUB18 inhibited Pst DC3000 growth in A thaliana Col-0 leaves after co-syringe injection (A), co-dipping inoculation (B), or co-spray inoculation (C). (D) OSUB18 inhibited Pst DC3000 growth in tobacco leaves after co-syringe injection. (E, F) OSUB18 inhibited Pst DC3000 growth in tomato leaves of diverse cultivars after co-syringe injection (E) or co-dipping inoculation (F). An equal volume of water (Ctrl) or OSUB18 was mixed into the Pst DC3000 (cell) inoculum for the phytopathogenic infection assay. Data present mean ± SE of three biological replicates. Data with different letters indicate a p-value < 0.05 on Student’s t-test.
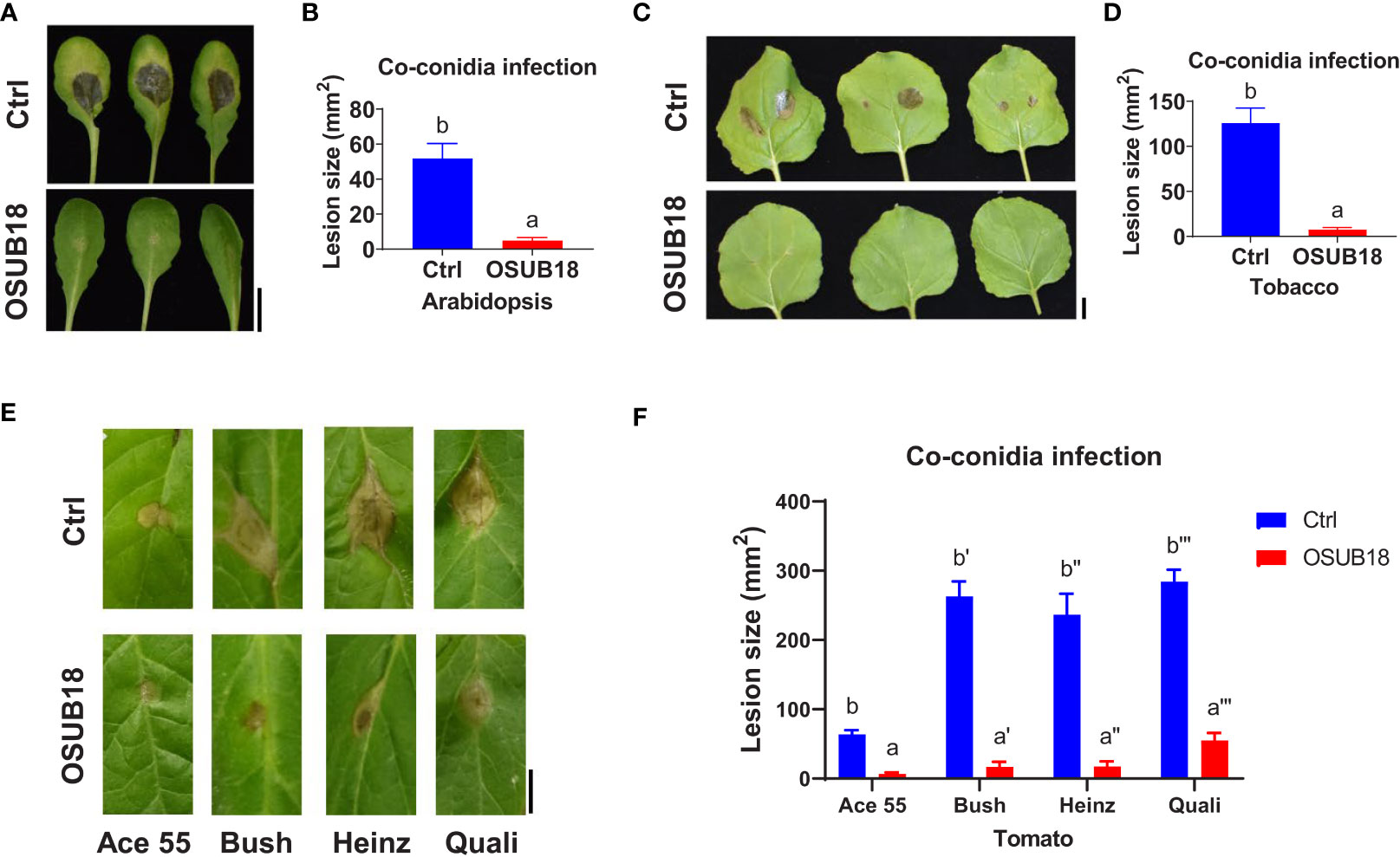
Figure 3 OSUB18 co-inoculation protected plants of different species from the B cinerea infections. (A) OSUB18 inhibited B cinerea growth in A thaliana Col-0 leaves after co-inoculation. Scale bar = 10 mm. (B) Quantification of the fungal disease symptoms in (A). (C) OSUB18 inhibited B cinerea growth in tobacco leaves after co-inoculation. Scale bar = 10 mm. (D) Quantification of the fungal disease symptoms in (C). (E) OSUB18 inhibited B cinerea growth in different tomato leaves after co-inoculation. Scale bar = 10 mm. (F) Quantification of the fungal disease symptoms in (E). We mixed an equal volume of water (Ctrl) or OSUB18 cells into the B cinerea inoculum for the phytopathogenic infection assay. Ace55, Bush, Heinz, and Quali are four popular tomato cultivars in the U.S. market. Data present mean ± SE of three biological replicates. Data with different letters indicate a p-value < 0.05 on Student’s t-test.
To further investigate the mechanisms of OSUB18 in activating the ISR of host plants against pathogens, we root-drenched A. thaliana Col-0 plants with OSUB18 and then challenged their above-ground leaves with the pathogenic bacterium Pst DC3000. Both syringe injection and dipping inoculation assays exhibited significantly higher ISR response in Arabidopsis against Pst DC3000 infection (Figure 4A). The leaves were collected 24 hours after the Pst DC3000 infection to proceed with assays assessing the callose deposition and ISR-responsive gene expression. Compared with control plants (Ctrl, pre-drenched with water), Col-0 plants pre-drenched with OSUB18 accumulated much more callose (Figures 4B, C) and significantly higher expression levels of the ISR-responsive gene MYC2 (Figure 4D) upon the Pst DC3000 inoculation. Unlike the hemibiotrophic phytopathogens such as Pst DC3000, B. cinerea has a necrotrophic lifestyle. As a result that OSUB18 antagonized both Pst DC3000 and B. cinerea (Figures 1–3), we were interested in the ISR efficacy of OSUB18 against phytopathogens with different lifestyles. We thus infected the pre-drenched OSUB18 and water control A. thaliana Col-0 plants with B. cinerea. We found that Col-0 plants pre-drenched with OSUB18 showed significantly higher resistance against B. cinerea infection (Figures 5A, B). The Col-0 plants pre-drenched with OSUB18 also exhibited a more robust callose deposition (Figures 5C, D) and significantly higher expression of the ISR-responsive gene MYC2 (Figure 5E) against B. cinerea. Consistent with previous reports that MYC2 was a transcription factor constitutively expressed in rhizobacteria-induced systemic resistance (Pozo et al., 2008), our data showed that OSUB18-drenched plants had a remarkably higher expression level of MYC2 compared to the control plants (Figures 5E, 6E). In agreement with the above results, Arabidopsis myc2 mutant plants were defective in OSUB18-triggered ISR response against both the bacterial pathogen Pst DC3000 (Figures S5) and the fungal pathogen B. cinerea (Figure S6). The result suggested that OSUB18 could successfully activate ISR induction against phytopathogens with different lifestyles.
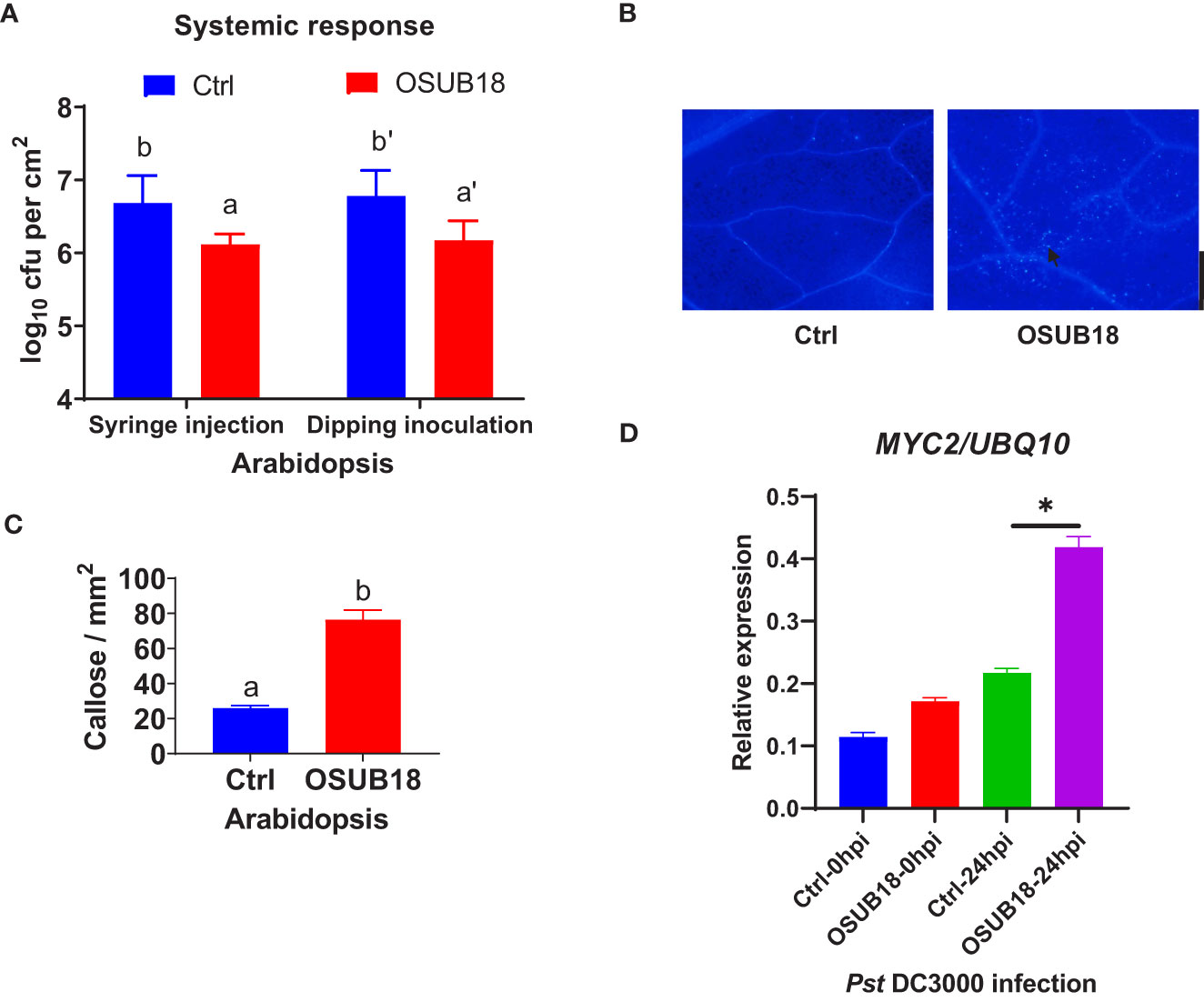
Figure 4 OSUB18 root-drench treatment triggered strong ISR in A thaliana against the bacterial pathogen Pst DC3000 attack via significantly more callose deposition and a significantly higher level of the MYC2 gene expression. (A) OSUB18 root-drench treatment increased the host plant defense of Col-0 plants against Pst DC3000 infection. (B) Col-0 plants drenched with OSUB18 deposited significantly more callose upon the Pst DC3000 infection (24hpi), as illustrated by the aniline blue staining assay. The black arrow indicates a representative callose. Scale bar = 500 µm. (C) Quantification of the callose deposition in (B). (D) The relative expression level of the ISR-responsive gene MYC2. Water or OSUB18-drenched Col-0 plants were infected with Pst DC3000 by syringe injection. 0 or 24 hours later, the injected leaves were collected for the qRT-PCR assay. The UBQ10 gene was used as an internal reference in the qRT-PCR assay. Data present mean ± SE of three biological replicates. Data with different letters or * indicate a p-value < 0.05 on Student’s t-test.
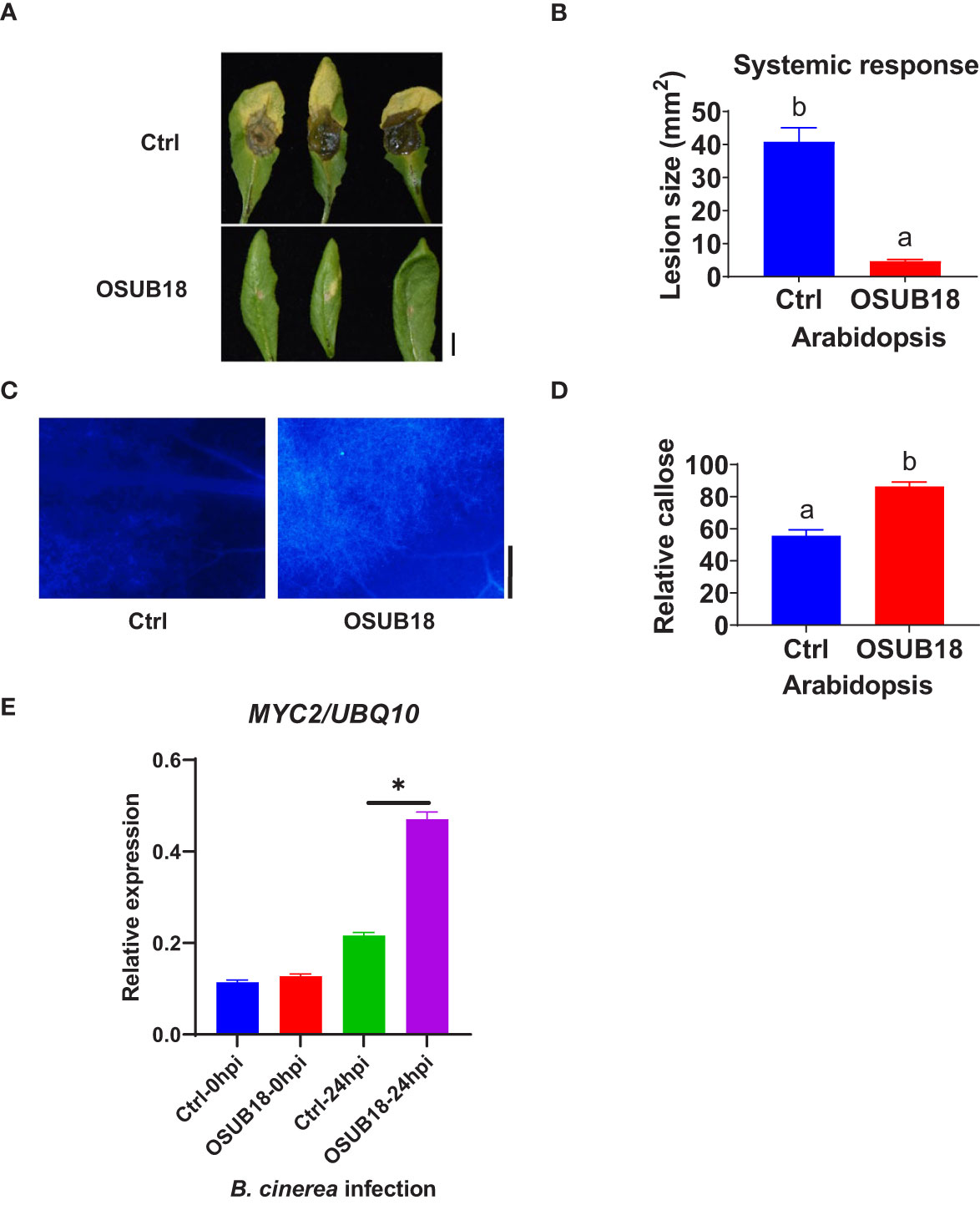
Figure 5 OSUB18 root drench treatment triggered strong ISR in A thaliana against the attack of the fungal pathogen B cinerea via significantly more callose deposition and higher MYC2 gene expression. (A) OSUB18 root drench treatment increased host plant defense of Col-0 plants against B cinerea infection. Scale bar = 5 mm. (B) Quantification of the disease symptoms in (A). (C) Col-0 plants drenched with OSUB18 deposited significantly more callose upon the B cinerea infection (24hpi), as illustrated by the aniline blue-staining assay. Scale bar = 500 µm. (D) Quantification of the callose deposition in (C). (E) The relative expression level of the ISR-responsive gene MYC2. Water or OSUB18-drenched plants were inoculated with B cinerea spores. 0 or 24 hours later, the inoculated leaves were collected for the qRT-PCR assay. The UBQ10 gene was used as an internal reference in the qRT-PCR assay. Data present mean ± SE of three biological replicates. Data with different letters or * indicate a p-value < 0.05 on Student’s t-test.
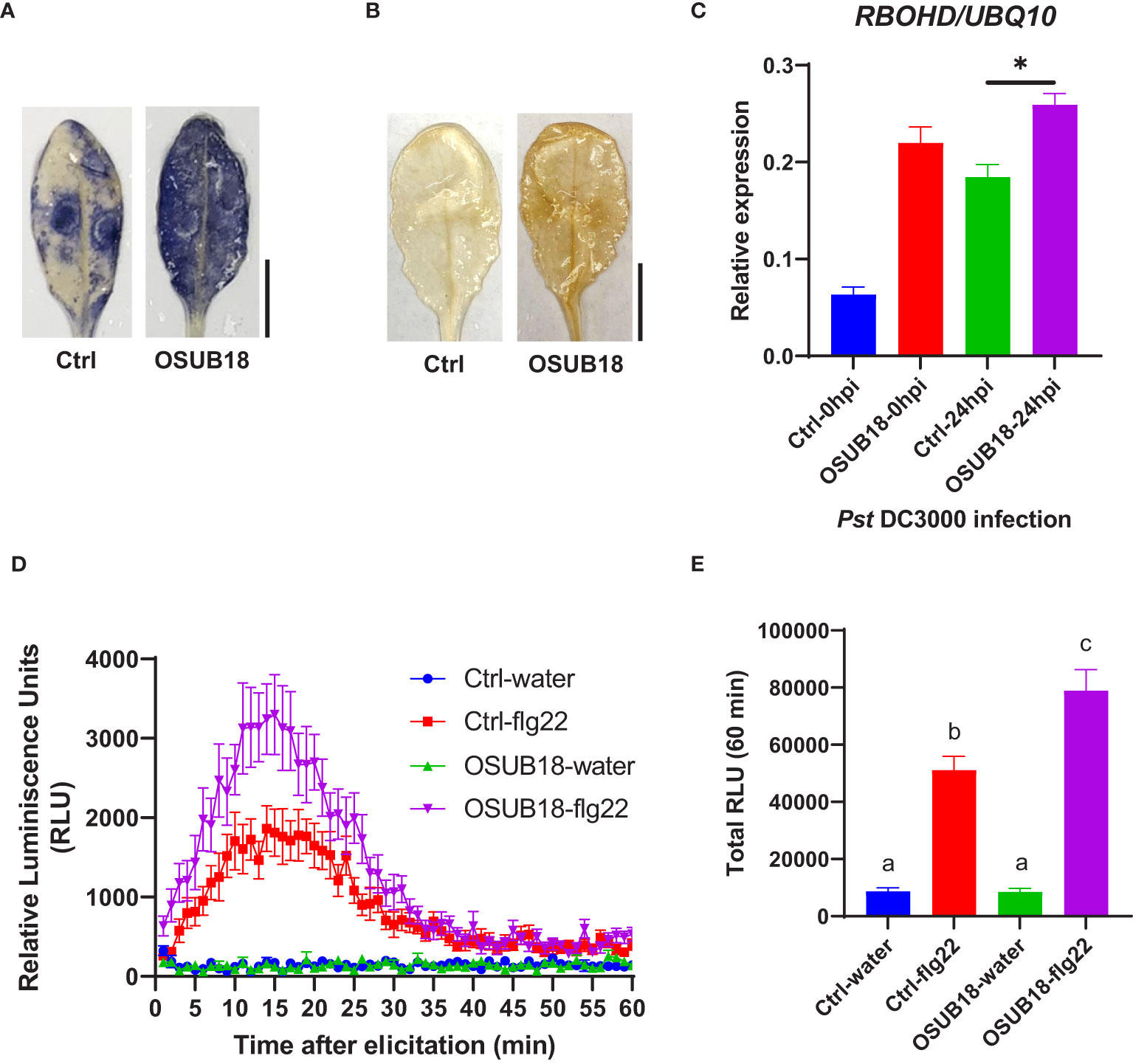
Figure 6 OSUB18 root drench treatment triggered strong ISR in A thaliana against the bacterial pathogen Pst DC3000 via significantly stronger ROS production and higher RBOHD gene expression. (A) Col-0 plants drenched with OSUB18 produced more intense superoxide anion upon the Pst DC3000 infection (24hpi), as illustrated by the nitroblue tetrazolium (NBT) staining assay. Scale bar = 10 mm. (B) Col-0 plants drenched with OSUB18 produced more intense hydrogen peroxide upon the Pst DC3000 infection (24hpi), as illustrated by the 3,3-diaminobenzidine (DAB) staining assay. Scale bar = 10 mm. (C) Relative gene expression of the ROS-responsive gene RBOHD. Water or OSUB18-drenched plants were infected with Pst DC3000 by syringe injection. 0 or 24 hours later, the injected leaves were collected for the qRT-PCR assay. The UBQ10 gene was used as an internal reference in the qRT-PCR assay. (D) Col-0 plants pre-drenched with OSUB18 showed a more vigorous ROS burst elicited by the bacterial PAMP flg22. (E) Quantification of the ROS burst in (D). Data present mean ± SE of three biological replicates. Data with different letters or * indicate a p-value < 0.05 on Student’s t-test or ANOVA.
Upon the Pst DC3000 infection, Arabidopsis leaves were collected to proceed with assays assessing the superoxide anion production, hydrogen peroxide production, and ROS-responsive gene expression. Compared with the control plants (Ctrl, pre-drenched with water), Col-0 plants pre-drenched with OSUB18 accumulated much more superoxide anion (Figure 6A), hydrogen peroxide (Figure 6B), and significantly higher expression of the ROS-responsive gene RBOHD (Figure 6C), which is essential for the accumulation of ROS in the apoplast (Torres et al., 2002; Zhao et al., 2022a). To verify whether a similar mechanism pattern is present in OSUB18-induced ISR against fungal pathogens, we also infected the pre-drenched OSUB18 A. thaliana Col-0 plants with B. cinerea. After B. cinerea infection, Arabidopsis leaves were collected to proceed with assays to assess the superoxide anion production, hydrogen peroxide production, and ROS-responsive gene expression. Indeed, the above-ground leaves of Col-0 plants pre-drenched with OSUB18 also exhibited more robust superoxide anion (Figure 7A), hydrogen peroxide (Figure 7B), and significantly higher expression level of the ROS-responsive gene RBOHD (Figure 7C) against the B. cinerea infection. To further validate the positive role of ROS production in OSUB18-activated ISR against pathogens in A. thaliana, we performed the PAMP-triggered ROS burst assay with leaf discs collected from OSUB18-drenched and water-drenched Col-0 plants. As expected, A. thaliana Col-0 plants pre-drenched with OSUB18 showed a significantly stronger ROS burst elicited by the bacterial PAMP flg22 (Figures 6D, E) and the fungal PAMP chitin (Figures 7D, E) than control plants.
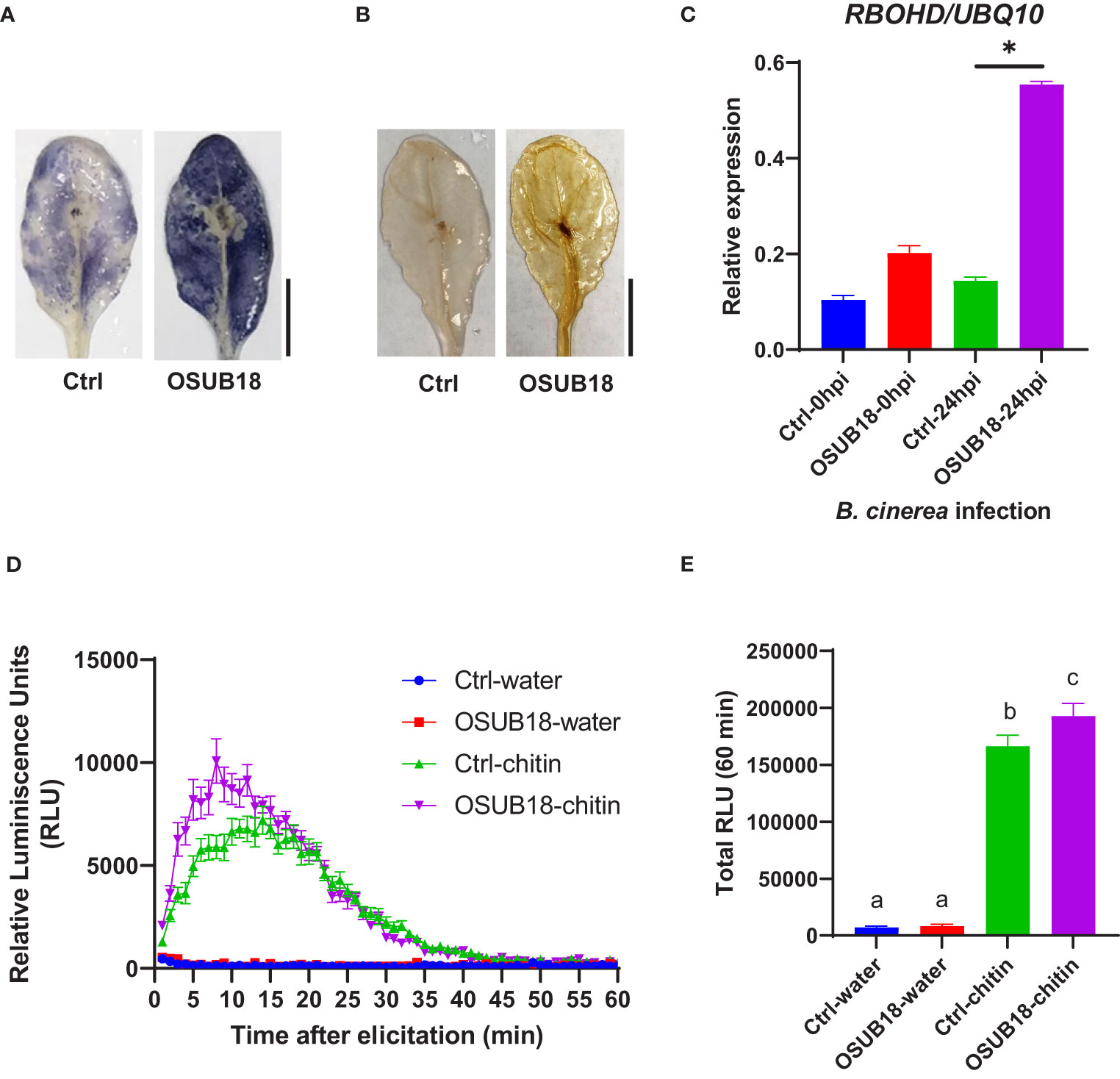
Figure 7 OSUB18 root drench treatment triggered strong ISR in A thaliana against the fungal pathogen B cinerea via significantly stronger ROS production and higher RBOHD gene expression. (A) Col-0 plants drenched with OSUB18 produced more intense superoxide anion upon the B cinerea infection (24hpi), as illustrated by the nitroblue tetrazolium (NBT) staining assay. Scale bar = 10 mm. (B) Col-0 plants drenched with OSUB18 produced more intense hydrogen peroxide upon the B cinerea infection (24hpi), as illustrated by the 3,3-diaminobenzidine (DAB) staining assay. Scale bar = 10 mm. (C) Relative gene expression of the ROS-responsive gene RBOHD. Water or OSUB18-drenched plants were infected with B cinerea spores. 0 or 24 hours later, the inoculated leaves were collected for the qRT-PCR assay. The UBQ10 gene was used as an internal reference in the qRT-PCR assay. (D) Col-0 plants pre-drenched with OSUB18 showed a more vigorous ROS burst elicited by the fungal PAMP chitin. (E) Quantification of the ROS burst in (D). Data present mean ± SE of three biological replicates. Data with different letters or * indicate a p-value < 0.05 on Student’s t-test or ANOVA.
To further decipher the underlying mechanism of OSUB18-triggered ISR against the hemibiotrophic phytopathogen Pst DC3000, we quantified the levels of SA and SAG in the water- and OSUB18-drenched A. thaliana Col-0 plants at 0 hpi and 24 hpi of the Pst DC3000 inoculation. We found that the free SA and conjugated SA (SAG) levels were significantly increased in OSUB18-drenched plants 24 hours after the Pst DC3000 inoculation compared with the control plants (Figures 8A, B). In line with the SA phytohormone production, the representative genes in the SA-signaling pathway (Wang et al., 2019) (PR1, PR2, PR5, EDS5, NPR1 and SID2) were significantly upregulated in OSUB18-drenched plants at 24 hours after the Pst DC3000 inoculation, compared with the control plants (Figures 8, S3). There were no significant differences in the JA or JA-lle levels in OSUB18-drenched plants at 24 hours after the Pst DC3000 inoculation, compared with the control plants (Figures 8E, F), as supported by the normal gene expression levels of LOX3 and JAR1 (Figures S3). Interestly, the plant JA responsive gene PDF 1.2 was significantly upregulated after the Pst DC3000 inoculation. One of the reasons could be that the PDF1.2 gene is also regulated by the other upstream genes, such as NPR1 (Nie et al., 2017). Indeed, we found that the NPR1 gene was significantly upregulated in the process of OSUB18-induced priming against Pst DC3000 (Figure S3D), which may lead to the increased expression of PDF 1.2.In agreement with the above results, Arabidopsis sid2 and npr1 mutant plants were defective in OSUB18-triggered ISR response against the bacterial pathogen Pst DC3000 but not jar1 mutant plants (Figures S5).
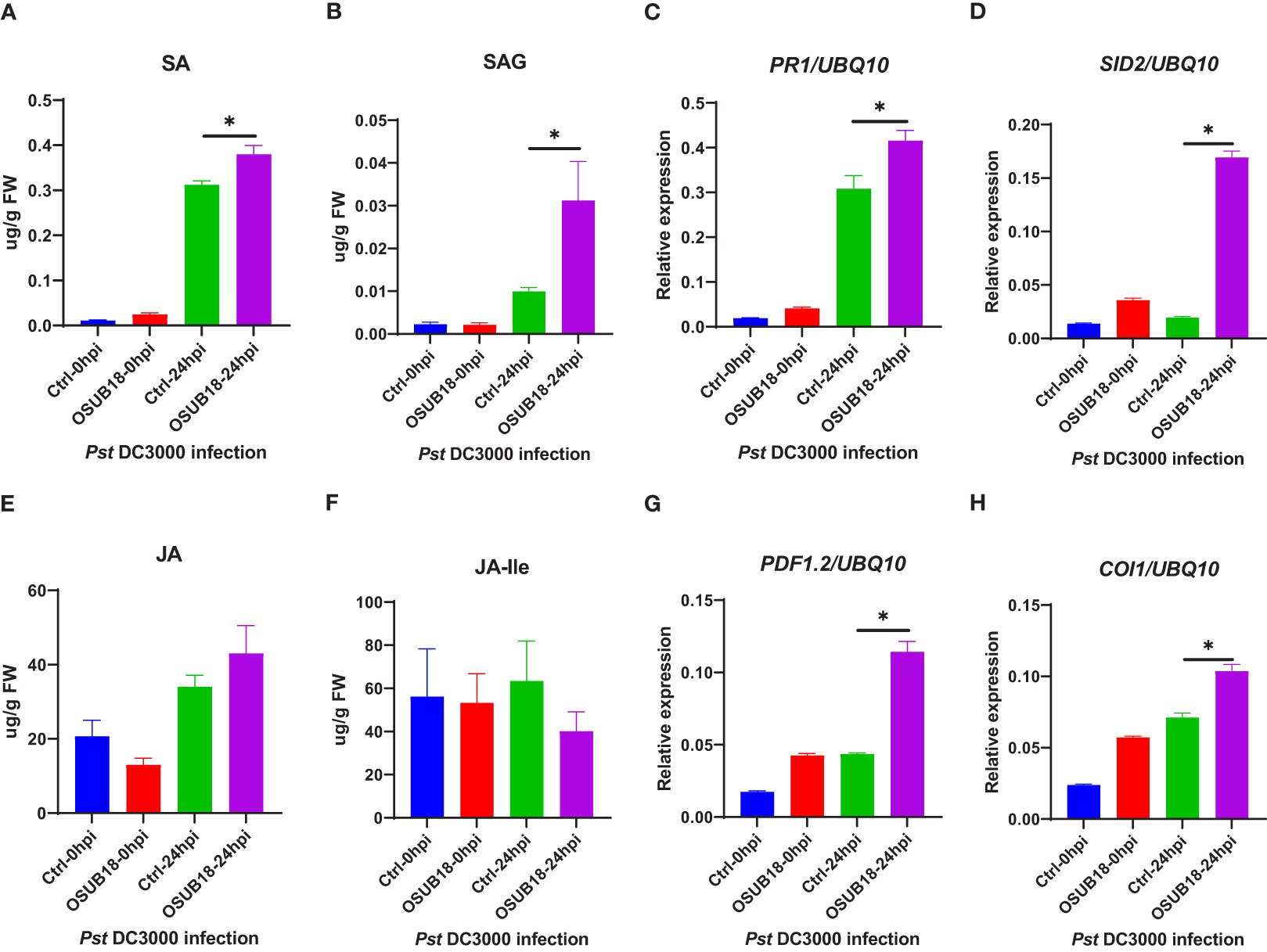
Figure 8 OSUB18 root drench treatment increased the SA and SA-related gene expression levels in A thaliana after the bacterial pathogen Pst DC3000 infection. (A) Free SA level. (B) Conjugated SA(SAG) level. (C) Relative gene expression of PR1. (D) Relative gene expression of SID2. (E) Free JA level. (F) Conjugated JA level. (G) Relative gene expression of PDF1.2. (H) Relative gene expression of COI1. Water or OSUB18-drenched plants were infected with Pst DC3000 by syringe injection. 0 or 24 hours later, the injected leaves were collected for phytohormone quantification and qRT-PCR assay. The UBQ10 gene was used as an internal reference in the qRT-PCR assay. Data present mean ± SE of three biological replicates. Data with * indicate a p-value < 0.05 on Student’s t-test.
To further examine the underlying mechanism of OSUB18-triggered ISR against the necrotrophic phytopathogen, we quantified the levels of JA and JA-lle in the water- and OSUB18-drenched A. thaliana Col-0 plants at 0hpi and 24hpi of the B. cinerea inoculation. Higher JA levels (Figures 9E) and significantly higher levels of JA-lle (the bioactive form of jasmonate) were detected in OSUB18-drenched plants at 24 hours after the B. cinerea inoculation, compared with the control plants (Figures 9F). Consistent with the phytohormone quantification results, the representative genes in the JA-signaling pathway (PDF1.2, COI1, LOX3, and JAR1) and the main ISR regulator NPR1 were significantly upregulated in OSUB18-drenched plants at 24 hours after the B. cinerea inoculation, compared with the control plants (Figures 9, S4). In agreement with the above results, Arabidopsis jar1 and npr1 mutant plants were defective in OSUB18-triggered ISR response against the fungal pathogen B. cinerea but not sid2 mutant plants (Figures S6). Though the representative genes in the SA-pathway (PR1, PR2, PR5, SID2) were also upregulated (Figures 9C, D, S3), there were no significant differences in the SA or SAG levels in OSUB18-drenched plants 24 hours after the B. cinerea inoculation, compared with the control plants (Figures 9A, B). Possibly, it is due to the downregulated expression of EDS5 (Figure S4), which is responsible for the transport of the SA precursor isochorismate (IC) from chloroplast to cytosol for the SA biosynthesis in cytosol.
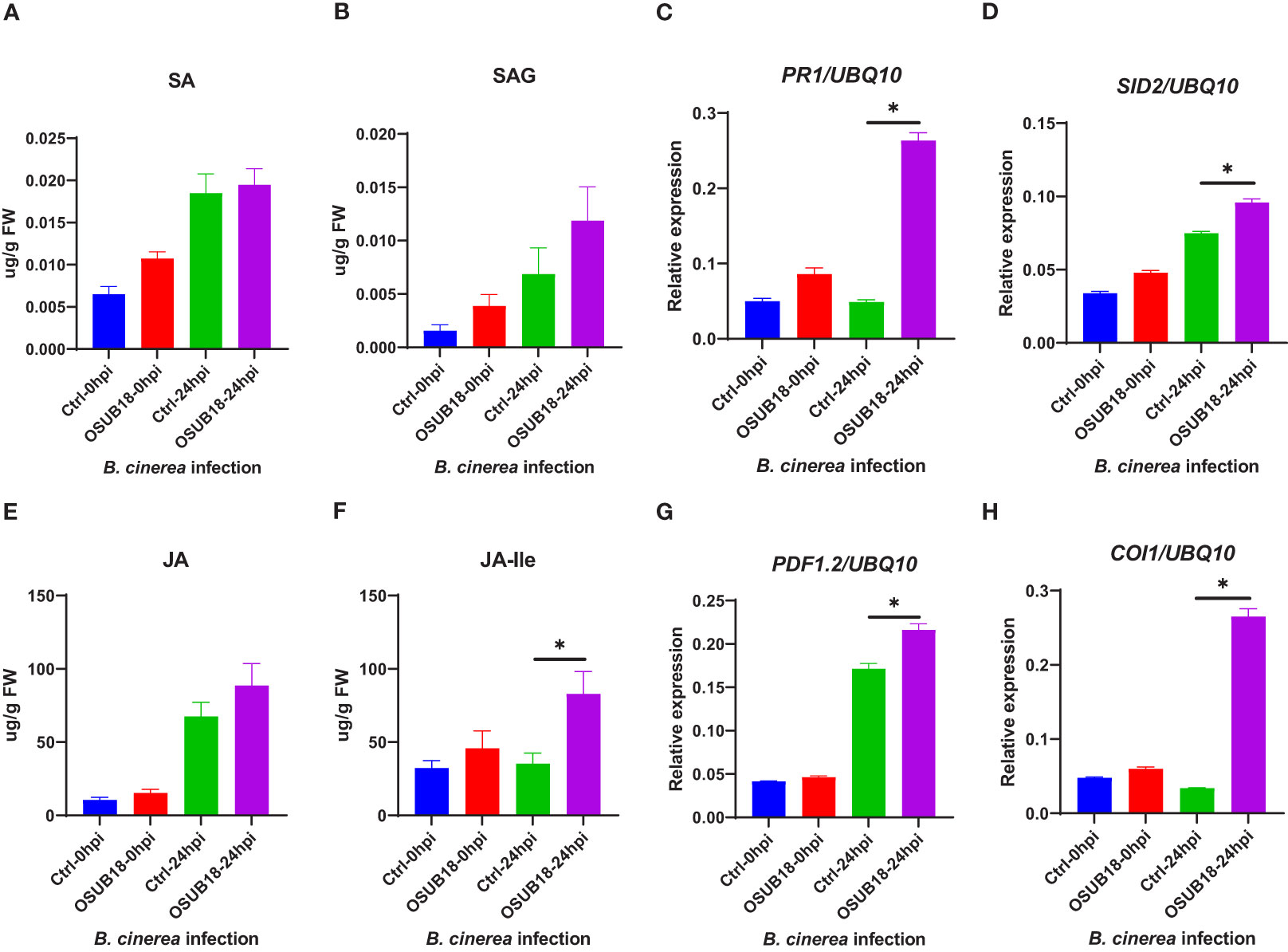
Figure 9 OSUB18 root drench treatment increased the JA and JA-related gene expression levels in A thaliana after the fungal pathogen B cinerea infection. (A) Free SA level. (B) Conjugated SA (SAG) level. (C) Relative gene expression of PR1. (D) Relative gene expression of SID2. (E) Free JA level. (F) Conjugated JA level. (G) Relative gene expression of PDF1.2. (H) Relative gene expression of COI1. Water or OSUB18-drenched plants were infected with B cinerea by spore inoculation. 0 or 24 hours later, the injected leaves were collected for phytohormone quantification and qRT-PCR assay. The UBQ10 gene was used as an internal reference in the qRT-PCR assay. Data present mean ± SE of three biological replicates. Data with * indicate a p-value < 0.05 on Student’s t-test.
Some chemical compounds, such as siderophores, polysaccharides, and volatile acetoin/diacetyl, are critical in ISR elicitation (Berendsen et al., 2015; Jiang et al., 2016). For our study, we found that OSUB18 was positive in producing siderophores (Figure 10A), exopolysaccharides (Figure 10B), and acetoin/diacetyl (Figure 10C). These data aligned with OSUB18’s efficacy in activating ISR against both bacterial and fungal phytopathogens (Figures 4, 5). OSUB18 was positive in producing acetoin/diacetyl (Figure 10C) but not HCN (Figure 10D), suggesting acetoin/diacetyl-related compounds can be the functional VOCs, in contrast to Pseudomonas fluorescens strain Pf5, one well-known bacterial strain involved in ISR. The production of ammonia by OSUB18 (Figure 8E) may also provide host plants with an additional nitrogen source for growth and defense. The phytohormone auxin could promote plant shoots’ and lateral roots’ growth (Woodward & Bartel, 2005). The enhanced shoot growth of OSUB18-inoculated plants (Figures S1C, D) is consistent with the fact that OSUB18 could produce ample amounts of IAA (Figure 10F). However, OSUB18 did not solubilize phosphate (Figure 10G) nor produce organic acid (Figure 10H), suggesting its plant growth-promoting role might be independent of these two factors. The catalase activity of OSUB18 (Figure 10I) may also play a positive role in protecting host plants by increasing oxygen availability in potting mix/soil and facilitating the gas exchange of plants with the environment. In agreement with the above beneficial traits, OSUB18 metabolites (cell-free crude extracts) inhibited the growth of Pst DC3000 (Figure S7A) and prevented the spore germination and hypha development of B. cinerea (Figure S7B).
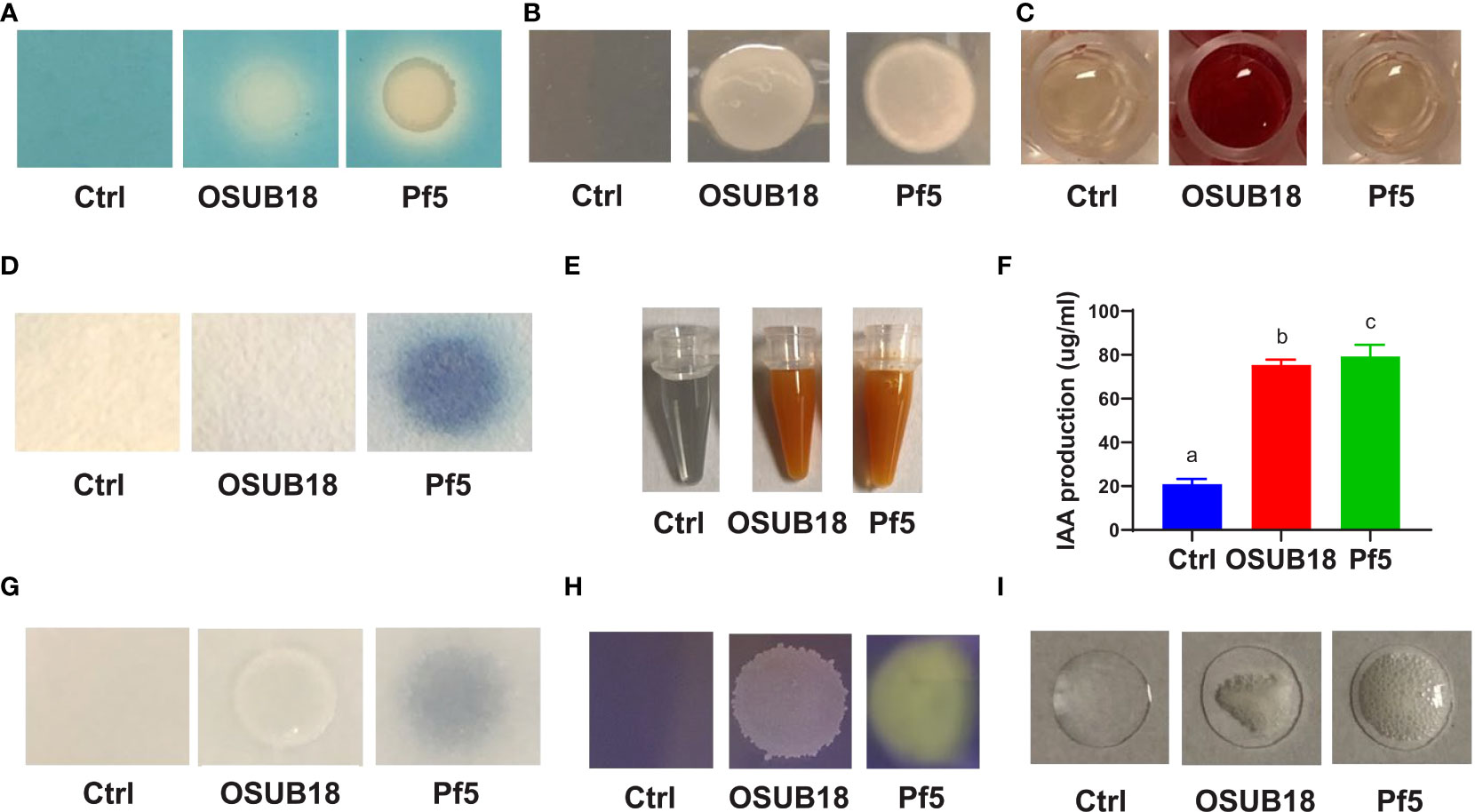
Figure 10 OSUB18 produced beneficial ISR-inducing metabolites suppressing plant pathogens and diseases and promoting plant growth. (A) Siderophore production. (B) Exopolysaccharide production. (C) Acetoin and diacetyl production. (D) HCN production. (E) Ammonia production. (F) IAA production. (G) Phosphate solubilization. (H) Organic acid production. (I) Catalase activity. The plant commensal bacterial Pseudomonas fluorescens strain Pf-5 was used as a control. Data present mean ± SE of three biological replicates. Data with different letters indicate a p-value < 0.05 on ANOVA.
OSUB18 was isolated from switchgrass and further characterized due to its capacity to promote plant growth (Figure S1). In this study, we investigated the roles and related mechanisms of OSUB18 in activating the ISR of Arabidopsis plants against diverse pathogens, including Pst DC3000 and B. cinerea. We discovered that OSUB18 antagonized bacterial and fungal phytopathogens both in-vitro plate assay (Figure 1) and in planta (Figures 2, 3) in Arabidopsis, tobacco, and tomato plants. Additionally, OSUB18 root treatment primed vigorous ISR activities of Arabidopsis plants against the bacterial phytopathogen Pst DC3000 (Figure 4) and the fungal phytopathogen B. cinerea (Figure 5). These data suggest that OSUB18 could be an excellent biological control agent with a broad-spectrum efficacy against bacterial and fungal phytopathogens via both direct antagonism and indirect ISR mechanisms in different plant species.
Plant leaves use cuticles and cell walls as the first barrier to avoid pathogen attacks (Malinovsky et al., 2014). For example, plants evolved to deposit polymers, such as callose, phenolic compounds, and antimicrobial toxins at the cuticle and cell walls to defend against pathogen infections (Hückelhoven, 2007; Zhao et al., 2020; Ziv et al., 2018). PAMPs, such as flg22 and chitin, can induce callose deposition, which was thus widely used to evaluate the plant PTI (Luna et al., 2010) by aniline blue staining (Jin and Mackey, 2017). In this study, we found that the callose deposition was significantly higher in OSUB18 root-treated plants than in water-treated plants upon the bacterial (Figures 4B, C) or fungal (Figures 5C, D) pathogen infections, indicating that cell wall reinforcement contributed to the OSUB18 associated ISR. Additionally, we found that the gene expression of transcription factor MYC2 was significantly higher in OSUB18-treated plants than that in the water-treated plants, consistent with the previous report showing that MYC2 could be involved in priming plant defense in the rhizobacterial-activated ISR of Arabidopsis (Pozo et al., 2008).
Reactive oxygen species (ROS) play central roles in plant signaling and immunity via diverse cellular processes (Qi et al., 2017). Plants evolved to harness toxic and signaling properties of ROS to defend themselves against invading pathogens (O’Brien et al., 2012). For example, plant plasma membrane-localized respiratory burst oxidase homologs (RBOHs) produce apoplastic ROS in response to pathogen infections. RBOHD is the most well studied member of the 10 RBOHs in Arabidopsis that plays essential roles in plant immunity by producing hydrogen peroxide (H2O2) (Kadota et al., 2015). In this study, we found that during the defense priming, OSUB18-treated plants exhibited significantly higher ROS production and gene expression of RBOHD than those in water-treated plants for both hemibiotrophic and necrotrophic pathogen infections (Figures 6, 7). These results indicate that ROS contributed to OSUB18-activated ISR.
Phytohormones play vital roles in plant development and defense (Bari and Jones, 2009; Santner et al., 2009). However, whether phytohormone pathways contribute to the OSUB18-activated ISR remains unclear. In this research study, we revealed that under the attacks of the hemibiotrophic bacterial pathogen Pst DC3000, SA levels were significantly higher in OSUB18-treated plants than in the water-treated plants at 24 hpi (Figure 8A). On the other hand, upon the attacks of the necrotrophic fungal pathogen B. cinerea, JA-lle (the bioactive form of JA) levels were significantly higher in OSUB18-treated plants than those in the water-treated plants at 24 hpi (Figure 9F). In line with the phytohormone quantification results, we detected the related gene expression by qRT-PCR and found that SA pathway-related genes, such as PR1, PR2, PR5, EDS5, NPR1, and SID2, were significantly induced in OSUB18-treated plants against the hemibiotrophic bacterial pathogen Pst DC3000 (Figures 8, S3).However, JA pathway-related genes, such as PDF1.2, LOX3, JAR1, and COI1 were significantly induced in OSUB18-treated plants against the necrotrophic fungal pathogen B. cinerea (Figures 9, S4), compared with water-treated plants. These data indicate that SA and JA contributed to the OSUB18-activated ISR against various pathogen infections, which depends on the pathogen’s lifestyles.
In addition, the representative genes in the SA pathway (PR1, PR2, PR5, SID2) were also upregulated, but no significant differences in the SA or SAG levels were found in OSUB18-drenched plants after the B. cinerea infection (Figure 9). One of the reasons could be that many genes are involved in the final outcome of the SA or SAG levels. For instance, among those genes involved in SA biosynthesis and metabolism, EDS5 controls the outport of SA precursor isochorismate (IC) from chloroplast to cytosol for the SA biosynthesis (Ding and Ding, 2020). We found that the experession level of EDS5 was significantly downregulated compared with the control in the case of B. cinerea infection (Figure S4C), suggesting a decreased IC pool in the cytosol and, thus leading to a none significant increase in the final SA or SAG levels. The other possible reason is that PR genes can be regulated by the other upstream genes. For instance, NPR1 is a crucial defense gene downstream of SA and JA but upstream of PRs and PDF1.2 in ISR against both biotrophic and necrotrophic phytopathogens (Nie et al., 2017). Indeed, we found that the NPR1 gene and its downstream PR genes (PR1, PR2, PR5) were significantly upregulated in OSUB18-drenched plants after the B. cinerea infection compared with the control plants (Figure S4). In agreement with the above results, Arabidopsis jar1 and npr1 mutant plants were defective in OSUB18-triggered ISR response against the fungal pathogen B. cinerea but not sid2 mutant plants (Figures S6).
We also noticed that the level of JA showed no significant change, but the expression of JA-related genes, such as PDF 1.2, were significantly changed in the process of OSUB18-induced priming against Pst DC3000 (Figure 8). One of the reasons could be that the PDF1.2 gene is also regulated by the other upstream genes, such as NPR1. As mentioned above NPR1 is a crucial defense gene upstream of PRs and PDF1.2 in ISR against plant pathogens with different lifestyles (Nie et al., 2017). Indeed, we found that the NPR1 gene was significantly upregulated in the process of OSUB18-induced priming against Pst DC3000 (Figure S3D). The other possible reason is that many genes are involved in the final outcome of the JA level. Among those genes involved in JA biosynthesis and metabolism, LOX3 converts 18:3 to the JA precursor OPDA in the chloroplast, and JAR1 converts JA to JA-lle, the bioactive form of JA in the cytoplasm (Ruan et al., 2019; Habash et al., 2020). We found that the expression leves of LOX3 and JAR1 did not significantly change in the process of OSUB18-induced priming against Pst DC3000 (Figure S3), suggesting a normal JA pool. In agreement with the above results, Arabidopsis sid2 and npr1 mutant plants were defective in OSUB18-triggered ISR response against the bacterial pathogen Pst DC3000 (Figure S5) but not jar1 mutant plants. Indeed, increasingly studies confirmed that the favorable signal transduction pathways during ISR not only depend on the host plants and beneficial microbes but also the pathogen lifestyles (Shoresh et al., 2010; Pieterse et al., 2014; Yu et al., 2022).
Microbial secondary metabolites function differently and diversely in biological activities, such as (Leong, 1986; Makras and De Vuyst, 2006; Kai et al., 2007; Mota et al., 2017; Anand et al., 2020; Abdalla et al., 2021), plant growth (Hayat et al., 2010; Etesami et al., 2015; Rijavec and Lapanje, 2016; Pahari et al., 2017; Alori et al., 2017; Naseem et al., 2018; Sharifi and Ryu, 2018; Macias-Benitez et al., 2020; Lopes et al., 2021), and insecticidal activities (Ullah et al., 2020). Interestingly, many traditional microbial secondary metabolites have recently been reported to play novel roles in (Berendsen et al., 2015; Jiang et al., 2016; Peng et al., 2019). For instance, Bacillus velezenisis was recently engineered to produce higher acetoin to prime a strong ISR in Arabidopsis (Peng et al., 2019), though acetoin was well-known for its volatile and antagonistic properties (Kai et al., 2007). Our study found that OSUB18 produced several secondary metabolites (siderophore, exopolysaccharide, and acetoin) reported to be ISR inducers (Table S2), partially explaining the promising activities of OSUB18 in ISR. OSUB18 was also positive in producing secondary metabolites, such as ammonia, with reported roles in pathogen inhibition (Mota et al., 2017). This might partially explain why OSUUB18 inhibited bacterial and fungal pathogen growth in vitro (Figure 1) and planta (Figures 2, 3). In addition, OSUB18 produced growth hormone IAA (Figure 10) with the reported roles in plant growth promotion (Table S2), which may contribute to plant growth activation. Although our work extends the plant protection properties of OSUB18 from switchgrass to A. thaliana, tobacco, and tomato (Figures 2, 3), little is known about the modes of action of OSUB18 in promoting plant growth. Recently, VOCs produced by PGPRs have been reported to induce ISR against phytopathogens and promote plant growth (Lee et al., 2012; Pérez-Flores et al., 2017). For example, the volatile 2,3-butanediol and 3-hydroxy-2-butanone (acetoin) were found in multiple beneficial bacteria, including Bacillus spp. (Effantin et al., 2011; Yi et al., 2016). Applying these ISR-eliciting VOCs to tobacco plants has been reported to increase host resistance against the fungal pathogen Colletotrichum orbiculare (D’alessandro et al., 2014).
In summary, our study discovered that OSUB18 is a novel strain that can activate plant ISR against diverse pathogens by enhancing the callose deposition, ROS accumulation, SA or JA-lle production, and the other ISR related signaling pathways in Arabidopsis plants. Further studies need to be conducted to reveal the underlying mechanisms of OSUB18-activated ISR in other plant species. In addition, investigating the persistence and propagation of OSUB18 in plants grown in potting mix or soils under different environmental conditions can yield critical information for its applications in agricultural practice. The results from these studies could deepen our understanding of the underlying mechanisms of plant ISR activated by the beneficial microbes, facilitate the application of these beneficial microbes in agriculture practice to protect plants against pathogens, increase crop yields, and improve food quality while lowering pesticide input.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
PY and YX conceived and designed the experiments. PY performed the experiments. YG helped with the drafting and editing of the manuscript. All authors developed experimental approaches or analyzed the data. PY and YX wrote the manuscripts with help from all the authors. All authors approved the final version of the manuscript.
This work was supported by Ohio Agricultural Research and Development Center seed grant funds (project numbers OHOA1591 and OHOA1615).
We thank Arabidopsis Biological Resource Center at Ohio State for contributing Arabidopsis seeds. We thank the Genomics Shared Resource at The Ohio State University Comprehensive Cancer Center, Columbus, OH, for Sanger Sequencing and Genotyping. This work was supported by Ohio Agricultural Research and Development Center Seed Grant Funds (project numbers OHOA1591 and OHOA1615). We extend special thanks to Nate Heiden (Ohio State University) and Dr. Lianhu Zhang (Jiangxi Agricultural University, China) for their helpful discussion.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1078100/full#supplementary-material
Abdalla, A. K., Ayyash, M. M., Olaimat, A. N., Osaili, T. M., Al-Nabulsi, A. A., Shah, N. P., et al. (2021). Exopolysaccharides as antimicrobial agents: Mechanism and spectrum of activity. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.664395
Alizadeh, H., Behboudi, K., Ahmadzadeh, M., Javan-Nikkhah, M., Zamioudis, C., Pieterse, C. M. J., et al. (2013). Induced systemic resistance in cucumber and arabidopsis thaliana by the combination of trichoderma harzianum Tr6 and pseudomonas sp. Ps14. Biol. Control 65, 14–23. doi: 10.1016/j.biocontrol.2013.01.009
Alori, E. T., Glick, B. R., Babalola, O. O. (2017). Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00971
Anand, A., Chinchilla, D., Tan, C., Mène-Saffrané, L., L’Haridon, F., Weisskopf, L. (2020). Contribution of hydrogen cyanide to the antagonistic activity of pseudomonas strains against phytophthora infestans. Microorganisms 8 (8), 1144. doi: 10.3390/microorganisms8081144
Bari, R., Jones, J. D. G. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Batista, B. D., Bonatelli, M. L., Quecine, M. C. (2021). “Fast screening of bacteria for plant growth promoting traits,” in The plant microbiome: Methods and protocols methods in molecular biology. Eds. Carvalhais, L. C., Dennis, P. G. (New York, NY: Springer US), 61–75. doi: 10.1007/978-1-0716-1040-4_7
Berendsen, R. L., van Verk, M. C., Stringlis, I. A., Zamioudis, C., Tommassen, J., Pieterse, C. M. J., et al. (2015). Unearthing the genomes of plant-beneficial pseudomonas model strains WCS358, WCS374 and WCS417. BMC Genomics 16, 539. doi: 10.1186/s12864-015-1632-z
Bhattacharyya, P. N., Jha, D. K. (2012). Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J. Microbiol. Biotechnol. 28, 1327–1350. doi: 10.1007/s11274-011-0979-9
Calhoun, C., Crist, D., Knee, E., Miller, J., Nagy, E., Somers, D. E. (2021). “Handling arabidopsis and other brassicaceae: Growth, preservation of seeds, transformation, and genetic crosses,” in Arabidopsis protocols methods in molecular biology. Eds. Sanchez-Serrano, J. J., Salinas, J. (New York, NY, Springer US: Humana), 3–23. doi: 10.1007/978-1-0716-0880-7_1
Castric, K. F., Castric, P. A. (1983). Method for rapid detection of cyanogenic bacteria. Appl. Environ. Microbiol. 45, 701–702. doi: 10.1128/aem.45.2.701-702.1983
Cazorla, F. M., Arrebola, E., Sesma, A., Pérez-García, A., Codina, J. C., Murillo, J., et al. (2002). Copper resistance in Pseudomonas syringae strains isolated from mango is encoded mainly by plasmids. Phytopathology® 92, 909–916. doi: 10.1094/PHYTO.2002.92.8.909
Chen, T., Nomura, K., Wang, X., Sohrabi, R., Xu, J., Yao, L., et al. (2020). A plant genetic network for preventing dysbiosis in the phyllosphere. Nature 580, 653–657. doi: 10.1038/s41586-020-2185-0
Chen, Y., Wang, J., Yang, N., Wen, Z., Sun, X., Chai, Y., et al. (2018). Wheat microbiome bacteria can reduce virulence of a plant pathogenic fungus by altering histone acetylation. Nat. Commun. 9, 3429. doi: 10.1038/s41467-018-05683-7
D’alessandro, M., Erb, M., Ton, J., Brandenburg, A., Karlen, D., Zopfi, J., et al. (2014). Volatiles produced by soil-borne endophytic bacteria increase plant pathogen resistance and affect tritrophic interactions. Plant Cell Environ. 37, 813–826. doi: 10.1111/pce.12220
Dean, R., Kan, J.a. L.V., Pretorius, Z. A., Hammond-Kosack, K. E., Pietro, A. D., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Ding, P., Ding, Y. (2020). Stories of salicylic acid: A plant defense hormone. Trends Plant Sci. 25, 549–565. doi: 10.1016/j.tplants.2020.01.004
Effantin, G., Rivasseau, C., Gromova, M., Bligny, R., Hugouvieux-Cotte-Pattat, N. (2011). Massive production of butanediol during plant infection by phytopathogenic bacteria of the genera dickeya and pectobacterium. Mol. Microbiol. 82, 988–997. doi: 10.1111/j.1365-2958.2011.07881.x
Etesami, H., Alikhani, H. A., Hosseini, H. M. (2015). Indole-3-acetic acid (IAA) production trait, a useful screening to select endophytic and rhizosphere competent bacteria for rice growth promoting agents. MethodsX 2, 72–78. doi: 10.1016/j.mex.2015.02.008
Eyler, E. (2013). “Chapter one - pouring agar plates and streaking or spreading to isolate individual colonies,” in Methods in enzymology laboratory methods in enzymology: Cell, lipid and carbohydrate. Ed. Lorsch, J. (Amsterdam, Netherlands: Academic Press), 3–14. doi: 10.1016/B978-0-12-420067-8.00001-5
Fernández-Vizcaíno, E., Ortiz-Santaliestra, M. E., Fernández-Tizón, M., Mateo, R., Camarero, P. R., Mougeot, F. (2022). Bird exposure to fungicides through the consumption of treated seeds: A study of wild red-legged partridges in central Spain. Environ. pollut. 292, 118335. doi: 10.1016/j.envpol.2021.118335
Giovannoni, J., Nguyen, C., Ampofo, B., Zhong, S., Fei, Z. (2017). The epigenome and transcriptional dynamics of fruit ripening. Annu. Rev. Plant Biol. 68, 61–84. doi: 10.1146/annurev-arplant-042916-040906
Green, S., Studholme, D. J., Laue, B. E., Dorati, F., Lovell, H., Arnold, D., et al. (2010). Comparative genome analysis provides insights into the evolution and adaptation of pseudomonas syringae pv. aesculi on aesculus hippocastanum. PloS One 5, e10224. doi: 10.1371/journal.pone.0010224
Habash, S. S., Könen, P. P., Loeschcke, A., Wüst, M., Jaeger, K.-E., Drepper, T., et al. (2020). The plant sesquiterpene nootkatone efficiently reduces heterodera schachtii parasitism by activating plant defense. Int. J. Mol. Sci. 21 9627. doi: 10.3390/ijms21249627
Hayat, R., Ali, S., Amara, U., Khalid, R., Ahmed, I. (2010). Soil beneficial bacteria and their role in plant growth promotion: a review. Ann. Microbiol. 60, 579–598. doi: 10.1007/s13213-010-0117-1
Huang, C.-J., Tsay, J.-F., Chang, S.-Y., Yang, H.-P., Wu, W.-S., Chen, C.-Y. (2012). Dimethyl disulfide is an induced systemic resistance elicitor produced by bacillus cereus C1L. Pest Manage. Sci. 68, 1306–1310. doi: 10.1002/ps.3301
Hückelhoven, R. (2007). Cell wall–associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 45, 101–127. doi: 10.1146/annurev.phyto.45.062806.094325
Hu, W., Gao, Q., Hamada, M. S., Dawood, D. H., Zheng, J., Chen, Y., et al. (2014). Potential of pseudomonas chlororaphis subsp. aurantiaca strain Pcho10 as a biocontrol agent against fusarium graminearum. Phytopathology 104, 1289–1297. doi: 10.1094/PHYTO-02-14-0049-R
Jambunathan, N. (2010). “Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants,” in Plant stress tolerance: Methods and protocols methods in molecular biology. Ed. Sunkar, R. (Totowa, NJ: Humana Press), 291–297. doi: 10.1007/978-1-60761-702-0_18
Jarret, R. L., McCluskey, K. (2019). The biological resources of model organisms (Boca Raton, Florida: CRC Press).
Jeong, H., Jeong, D.-E., Kim, S. H., Song, G. C., Park, S.-Y., Ryu, C.-M., et al. (2012). Draft genome sequence of the plant growth-promoting bacterium bacillus siamensis KCTC 13613T. J. Bacteriol. 194, 4148–4149. doi: 10.1128/JB.00805-12
Jiang, C.-H., Fan, Z.-H., Xie, P., Guo, J.-H. (2016). Bacillus cereus AR156 extracellular polysaccharides served as a novel micro-associated molecular pattern to induced systemic immunity to pst DC3000 in arabidopsis. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00664
Jin, L., Mackey, D. M. (2017). “Measuring callose deposition, an indicator of cell wall reinforcement, during bacterial infection in arabidopsis,” in Plant pattern recognition receptors. Eds. Shan, L., He, P. (New York, NY: Springer New York), 195–205. doi: 10.1007/978-1-4939-6859-6_16
Kadota, Y., Shirasu, K., Zipfel, C. (2015). Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 56, 1472–1480. doi: 10.1093/pcp/pcv063
Kai, M., Effmert, U., Berg, G., Piechulla, B. (2007). Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen rhizoctonia solani. Arch. Microbiol. 187, 351–360. doi: 10.1007/s00203-006-0199-0
Katagiri, F., Thilmony, R., He, S. Y. (2002). The arabidopsis thaliana-pseudomonas syringae interaction. Arab. Book Am. Soc Plant Biol. 1. doi: 10.1199/tab.0039
Kazan, K., Manners, J. M. (2013). MYC2: The master in action. Mol. Plant 6, 686–703. doi: 10.1093/mp/sss128
Keen, E. C., Bliskovsky, V. V., Adhya, S. L., Dantas, G. (2017). Draft genome sequence of the naturally competent bacillus simplex strain WY10. Genome Announc. 5, e01295–e01217. doi: 10.1128/genomeA.01295-17
Khalaf, E. M., Raizada, M. N. (2018). Bacterial seed endophytes of domesticated cucurbits antagonize fungal and oomycete pathogens including powdery mildew. Front. Microbiol. 0. doi: 10.3389/fmicb.2018.00042
Kumar, A., Prakash, A., Johri, B. N. (2011). “Bacillus as PGPR in crop ecosystem,” in Bacteria in agrobiology: Crop ecosystems. Ed. Maheshwari, D. K. (Berlin, Heidelberg: Springer), 37–59. doi: 10.1007/978-3-642-18357-7_2
Lee, B., Farag, M. A., Park, H. B., Kloepper, J. W., Lee, S. H., Ryu, C.-M. (2012). Induced resistance by a long-chain bacterial volatile: Elicitation of plant systemic defense by a C13 volatile produced by paenibacillus polymyxa. PloS One 7, e48744. doi: 10.1371/journal.pone.0048744
Leong, J. (1986). Siderophores: Their biochemistry and possible role in the biocontrol of plant pathogens. Annu. Rev. Phytopathol. 24, 187–209. doi: 10.1146/annurev.py.24.090186.001155
Leppla, N., LeBeck, L. M. (2021). 2021 guidelines for purchasing and using commercial natural enemies and biopesticides in north America. EDIS 2021, 14. doi: 10.32473/edis-in849-2021
Lopes, M. J., dos, S., Dias-Filho, M. B., Gurgel, E. S. C. (2021). Successful plant growth-promoting microbes: Inoculation methods and abiotic factors. Front. Sustain. Food Syst. 5. doi: 10.3389/fsufs.2021.606454
Luna, E., Pastor, V., Robert, J., Flors, V., Mauch-Mani, B., Ton, J. (2010). Callose deposition: A multifaceted plant defense response. Mol. Plant-Microbe Interactions® 24, 183–193. doi: 10.1094/MPMI-07-10-0149
Macias-Benitez, S., Garcia-Martinez, A. M., Caballero Jimenez, P., Gonzalez, J. M., Tejada Moral, M., Parrado Rubio, J. (2020). Rhizospheric organic acids as biostimulants: Monitoring feedbacks on soil microorganisms and biochemical properties. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00633
Maheshwari, D. K., Dheeman, S., Agarwal, M. (2015). “Phytohormone-producing PGPR for sustainable agriculture,” in Bacterial metabolites in sustainable agroecosystem sustainable development and biodiversity. Ed. Maheshwari, D. K. (Cham: Springer International Publishing), 159–182. doi: 10.1007/978-3-319-24654-3_7
Makras, L., De Vuyst, L. (2006). The in vitro inhibition of gram-negative pathogenic bacteria by bifidobacteria is caused by the production of organic acids. Int. Dairy J. 16, 1049–1057. doi: 10.1016/j.idairyj.2005.09.006
Malinovsky, F. G., Fangel, J. U., Willats, W. G. T. (2014). The role of the cell wall in plant immunity. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00178
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
May, B. (2011). A review of microbiology: An evolving science, second edition. J. Microbiol. Biol. Educ. JMBE 12, 216. doi: 10.1128/jmbe.v12i2.339
Molohon, K. J., Melby, J. O., Lee, J., Evans, B. S., Dunbar, K. L., Bumpus, S. B., et al. (2011). Structure determination and interception of biosynthetic intermediates for the plantazolicin class of highly discriminating antibiotics. ACS Chem. Biol. 6, 1307–1313. doi: 10.1021/cb200339d
Mota, M. S., Gomes, C. B., Souza Júnior, I. T., Moura, A. B. (2017). Bacterial selection for biological control of plant disease: criterion determination and validation. Braz. J. Microbiol. 48, 62–70. doi: 10.1016/j.bjm.2016.09.003
Mu’minah, B., Subair, H., Fahruddin (2015). Isolation and screening bacterial exopolysaccharide (EPS) from potato rhizosphere in highland and the potential as a producer indole acetic acid (IAA). Proc. Food Sci. 3, 74–81. doi: 10.1016/j.profoo.2015.01.007
Murillo, J., Bardaji, L., Führer, M. E. (2010)La grasa de las judías, causada por la bacteria pseudomonas syringea pv. phaseolicoda. In: Phytoma esp. rev. Prof. sanid. veg. Available at: https://dialnet.unirioja.es/servlet/articulo?codigo=3767170 (Accessed January 2, 2022).
Naseem, H., Ahsan, M., Shahid, M. A., Khan, N. (2018). Exopolysaccharides producing rhizobacteria and their role in plant growth and drought tolerance. J. Basic Microbiol. 58, 1009–1022. doi: 10.1002/jobm.201800309
Nie, P., Li, X., Wang, S., Guo, J., Zhao, H., Niu, D. (2017). Induced systemic resistance against botrytis cinerea by bacillus cereus AR156 through a JA/ET- and NPR1-dependent signaling pathway and activates PAMP-triggered immunity in arabidopsis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00238
O’Brien, J. A., Daudi, A., Butt, V. S., Paul Bolwell, G. (2012). Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta 236, 765–779. doi: 10.1007/s00425-012-1696-9
Odds, F. (1981). Biochemical tests for identification of medical bacteria. J. Clin. Pathol. 34, 572.
Pahari, A., Pradhan, A., Nayak, S. K., Mishra, B. B. (2017). “Bacterial siderophore as a plant growth promoter,” in Microbial biotechnology: Volume 1. applications in agriculture and environment. Eds. Patra, J. K., Vishnuprasad, C. N., Das, G. (Singapore: Springer), 163–180. doi: 10.1007/978-981-10-6847-8_7
Peng, G., Zhao, X., Li, Y., Wang, R., Huang, Y., Qi, G. (2019). Engineering bacillus velezensis with high production of acetoin primes strong induced systemic resistance in arabidopsis thaliana. Microbiol. Res. 227, 126297. doi: 10.1016/j.micres.2019.126297
Pérez-Flores, P., Valencia-Cantero, E., Altamirano-Hernández, J., Pelagio-Flores, R., López-Bucio, J., García-Juárez, P., et al. (2017). Bacillus methylotrophicus M4-96 isolated from maize (Zea mays) rhizoplane increases growth and auxin content in arabidopsis thaliana via emission of volatiles. Protoplasma 254, 2201–2213. doi: 10.1007/s00709-017-1109-9
Pérez-Miranda, S., Cabirol, N., George-Téllez, R., Zamudio-Rivera, L. S., Fernández, F. J. (2007). O-CAS, a fast and universal method for siderophore detection. J. Microbiol. Methods 70, 127–131. doi: 10.1016/j.mimet.2007.03.023
Petrasch, S., Knapp, S. J., Kan, J.A.L.v., Blanco-Ulate, B. (2019). Grey mould of strawberry, a devastating disease caused by the ubiquitous necrotrophic fungal pathogen botrytis cinerea. Mol. Plant Pathol. 20, 877–892. doi: 10.1111/mpp.12794
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C. M., Bakker, P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Pozo, M. J., Ent, S. V. D., Loon, L. C. V., Pieterse, C. M. J. (2008). Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in arabidopsis thaliana. New Phytol. 180, 511–523. doi: 10.1111/j.1469-8137.2008.02578.x
Provost, C., Pedneault, K. (2016). The organic vineyard as a balanced ecosystem: Improved organic grape management and impacts on wine quality. Sci. Hortic. 208, 43–56. doi: 10.1016/j.scienta.2016.04.024
Qi, J., Wang, J., Gong, Z., Zhou, J.-M. (2017). Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 38, 92–100. doi: 10.1016/j.pbi.2017.04.022
Ries (1995) IPM : Reports on plant Diseases : Gray mold of strawberry. Available at: http://ipm.illinois.edu/diseases/series700/rpd704/ (Accessed January 2, 2022).
Rijavec, T., Lapanje, A. (2016). Hydrogen cyanide in the rhizosphere: Not suppressing plant pathogens, but rather regulating availability of phosphate. Front. Microbiol. 0. doi: 10.3389/fmicb.2016.01785
Romera, F. J., García, M. J., Lucena, C., Martínez-Medina, A., Aparicio, M. A., Ramos, J., et al. (2019). Induced systemic resistance (ISR) and fe deficiency responses in dicot plants. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00287
Ruan, J., Zhou, Y., Zhou, M., Yan, J., Khurshid, M., Weng, W., et al. (2019). Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 20. 2479 doi: 10.3390/ijms20102479
Ryu, C.-M., Hu, C.-H., Reddy, M. S., Kloepper, J. W. (2003). Different signaling pathways of induced resistance by rhizobacteria in arabidopsis thaliana against two pathovars of pseudomonas syringae. New Phytol. 160, 413–420. doi: 10.1046/j.1469-8137.2003.00883.x
Sang, Y., Macho, A. P. (2017). “Analysis of PAMP-triggered ROS burst in plant immunity,” in Plant pattern recognition receptors: Methods and protocols methods in molecular biology. Eds. Shan, L., He, P. (New York, NY: Springer), 143–153. doi: 10.1007/978-1-4939-6859-6_11
Santner, A., Calderon-Villalobos, L. I. A., Estelle, M. (2009). Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5, 301–307. doi: 10.1038/nchembio.165
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Sharifi, R., Ryu, C.-M. (2018). Revisiting bacterial volatile-mediated plant growth promotion: lessons from the past and objectives for the future. Ann. Bot. 122, 349–358. doi: 10.1093/aob/mcy108
Shenge, K. C., Mabagala, R. B., Mortensen, C. N., Stephan, D., Wydra, K. (2007). First report of bacterial speck of tomato caused by pseudomonas syringae pv. tomato in Tanzania. Plant Dis. 91, 462–462. doi: 10.1094/PDIS-91-4-0462C
Shoresh, M., Harman, G. E., Mastouri, F. (2010). Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 48, 21–43. doi: 10.1146/annurev-phyto-073009-114450
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Torres, M. A., Dangl, J. L., Jones, J. D. G. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. 99, 517–522. doi: 10.1073/pnas.012452499
Tronsmo, A. M., Collinge, D. B., Djurle, A., Munk, L., Yuen, J., Tronsmo, A. (2020). “Plant pathology and plant diseases,” in CABI. (Boston, MA: CAB International).
Turnbull, P. C. B. (1996)Chapter 15 bacillus. In: Medical microbiology. 4th edition. Available at: http://www.ncbi.nlm.nih.gov/books/NBK7699/ (Accessed December 18, 2018).
Ullah, A., Bano, A., Janjua, H. T. (2020). “Chapter 3 - microbial secondary metabolites and defense of plant stress,” in Microbial services in restoration ecology. Eds. Singh, J. S., Vimal, S. R. (Amsterdam, Netherlands: Elsevier), 37–46. doi: 10.1016/B978-0-12-819978-7.00003-8
Vyas, P., Kumar, D., Dubey, A., Kumar, A. (2018). Screening and characterization of achromobacter xylosoxidans isolated from rhizosphere of jatropha curcas l. (Energy crop) for plant-growth-promoting traits. J. Adv. Res. Biotechnol. 3, 1–8. doi: 10.15226/2475-4714/3/1/00134
Wang, C., Huang, X., Li, Q., Zhang, Y., Li, J.-L., Mou, Z. (2019). Extracellular pyridine nucleotides trigger plant systemic immunity through a lectin receptor kinase/BAK1 complex. Nat. Commun. 10, 1–16. doi: 10.1038/s41467-019-12781-7
Wang, S., Zheng, Y., Gu, C., He, C., Yang, M., Zhang, X., et al. (2017). Bacillus cereus AR156 activates defense responses to pseudomonas syringae pv. tomato in arabidopsis thaliana similarly to flg22. Mol. Plant Microbe Interact. 31, 311–322. doi: 10.1094/MPMI-10-17-0240-R
Weiberg, A., Wang, M., Lin, F.-M., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Woodward, A. W., Bartel, B. (2005). Auxin: Regulation, action, and interaction. Ann. Bot. 95, 707–735. doi: 10.1093/aob/mci083
Wu, L., Huang, Z., Li, X., Ma, L., Gu, Q., Wu, H., et al. (2018). Stomatal closure and SA-, JA/ET-signaling pathways are essential for bacillus amyloliquefaciens FZB42 to restrict leaf disease caused by phytophthora nicotianae in nicotiana benthamiana. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00847
Xia, Y., Greissworth, E., Mucci, C., Williams, M. A., Bolt, S. D. (2013). Characterization of culturable bacterial endophytes of switchgrass (Panicum virgatum l.) and their capacity to influence plant growth. GCB Bioenergy 5, 674–682. doi: 10.1111/j.1757-1707.2012.01208.x
Yang, P., Bokros, N., Debolt, S., Zhao, Z., Xia, Y. (2022). Genome sequence source of bacillus amyloliquefaciens strain GD4a, a bacterial endophyte associated with switchgrass plants. Phytobiomes J. 6:4, 354–357. doi: 10.1094/PBIOMES-09-21-0054-A
Yao, J., Withers, J., He, S. Y. (2013). “Pseudomonas syringae infection assays in arabidopsis,” in Jasmonate signaling: Methods and protocols methods in molecular biology. Eds. Goossens, A., Pauwels, L. (Totowa, NJ: Humana Press), 63–81. doi: 10.1007/978-1-62703-414-2_6
Yi, H.-S., Ahn, Y.-R., Song, G. C., Ghim, S.-Y., Lee, S., Lee, G., et al. (2016). Impact of a bacterial volatile 2,3-butanediol on bacillus subtilis rhizosphere robustness. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00993
Yoo, S.-J., Choi, H. J., Noh, S. W., Cecchini, N. M., Greenberg, J. T., Jung, H. W. (2022). Genetic requirements for infection-specific responses in conferring disease resistance in arabidopsis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1068438
Yu, Y., Gui, Y., Li, Z., Jiang, C., Guo, J., Niu, D. (2022). Induced systemic resistance for improving plant immunity by beneficial microbes. Plants 11, 386. doi: 10.3390/plants11030386
Yunis, H., Bashan, Y., Okon, Y. (1980). Weather dependence, yield losses, and control of bacterial speck of tomato caused by Pseudomonas tomato. Plant Dis. 64, 937. doi: 10.1094/PD-64-937
Zhao, Z., Fan, J., Gao, Y. G., Wang, Z., Yang, P., Liang, Y., et al. (2022a). Arabidopsis plasma membrane ATPase AHA5 is negatively involved in PAMP-triggered immunity. Int. J. Mol. Sci. 23, 3857. doi: 10.3390/ijms23073857
Zhao, Z., Fan, J., Yang, P., Wang, Z., Stephen, O., Mackey, D., et al. (2022b). Involvement of arabidopsis acyl carrier protein 1 in PAMP-triggered immunity. Mol. Plant-Microbe Interactions®. 35 (8), 681–693. doi: 10.1094/MPMI-02-22-0049-R
Zhao, Z., Yang, X., Lü, S., Fan, J., Opiyo, S., Yang, P., et al. (2020). Deciphering the novel role of AtMIN7 in cuticle formation and defense against the bacterial pathogen infection. Int. J. Mol. Sci. 21, 5547. doi: 10.3390/ijms21155547
Keywords: Bacillus proteolyticus, Botrytis cinerea, Pseudomonas syringae, Arabidopsis thaliana, induced systemic resistance (ISR), callose, reactive oxygen species (ROS), defense priming
Citation: Yang P, Zhao Z, Fan J, Liang Y, Bernier MC, Gao Y, Zhao L, Opiyo SO and Xia Y (2023) Bacillus proteolyticus OSUB18 triggers induced systemic resistance against bacterial and fungal pathogens in Arabidopsis. Front. Plant Sci. 14:1078100. doi: 10.3389/fpls.2023.1078100
Received: 24 October 2022; Accepted: 04 January 2023;
Published: 23 January 2023.
Edited by:
Ho Won Jung, Dong-A University, Republic of KoreaReviewed by:
Shuishan Song, Hebei Academy of Sciences, ChinaCopyright © 2023 Yang, Zhao, Fan, Liang, Bernier, Gao, Zhao, Opiyo and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ye Xia, eGlhLjM3NEBvc3UuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.