
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 19 January 2023
Sec. Photosynthesis and Photobiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1050483
This article is part of the Research TopicEnhancement of Photosynthesis through Light Utilization in Plants and CropsView all 11 articles
 Daniel Caddell1,2
Daniel Caddell1,2 Noah J. Langenfeld3
Noah J. Langenfeld3 Madigan JH. Eckels3
Madigan JH. Eckels3 Shuyang Zhen4
Shuyang Zhen4 Rachel Klaras2
Rachel Klaras2 Laxmi Mishra2
Laxmi Mishra2 Bruce Bugbee3
Bruce Bugbee3 Devin Coleman-Derr1,2*
Devin Coleman-Derr1,2*Plants compete for light partly by over-producing chlorophyll in leaves. The resulting high light absorption is an effective strategy for out competing neighbors in mixed communities, but it prevents light transmission to lower leaves and limits photosynthesis in dense agricultural canopies. We used a CRISPR/Cas9-mediated approach to engineer rice plants with truncated light-harvesting antenna (TLA) via knockout mutations to individual antenna assembly component genes CpSRP43, CpSRP54a, and its paralog, CpSRP54b. We compared the photosynthetic contributions of these components in rice by studying the growth rates of whole plants, quantum yield of photosynthesis, chlorophyll density and distribution, and phenotypic abnormalities. Additionally, we investigated a Poales-specific duplication of CpSRP54. The Poales are an important family that includes staple crops such as rice, wheat, corn, millet, and sorghum. Mutations in any of these three genes involved in antenna assembly decreased chlorophyll content and light absorption and increased photosynthesis per photon absorbed (quantum yield). These results have significant implications for the improvement of high leaf-area-index crop monocultures.
In plants, pigments such as chlorophyll and carotenoids harvest light and transfer the energy toward reaction centers, where it is converted to chemical energy via photochemical charge separation, initiating photosynthesis. In full sunlight, plants absorb more light than can be used for photosynthesis. This overaccumulation enables a selective advantage in natural communities, whereby plants with large light-harvesting antenna complexes can outcompete neighboring plants for sunlight, which is a major growth-limiting factor in most environments. Prevention of weed establishment in agricultural settings is often accomplished through herbicidal or management practices, such as planting in dense monocultures. The absorption of excess photons by upper leaves reduces the amount of light within the canopy that is used for photosynthesis, and can negatively impact plant yields (Bugbee, 1992). Recent studies have suggested that mutations that reduce the size of light-harvesting antenna complexes, producing mutants commonly referred to as truncated light antenna (TLA) mutants, may improve quantum yield of PSII by increasing light penetration deeper into the canopy, particularly under high density, high light conditions (Gu et al., 2017a; Gu et al., 2017b; Kirst et al., 2017; Kirst et al., 2018).
Chloroplast light-harvesting antenna assembly relies on cytosolic synthesis of nuclear-encoded light-harvesting chlorophyll a/b binding proteins (LHCPs). To function, LHCPs must be imported into the chloroplast, transported, and ultimately assembled into the thylakoid membrane. This process is mediated by the chloroplast signal recognition particle (CpSRP) pathway (Ziehe et al., 2017). In the stroma, CpSRP43 and CpSRP54 form a heterodimer that binds LHCPs, forming a transit complex (DeLille et al., 2000; Tu et al., 2000; Jonas-Straube et al., 2001; Hermkes et al., 2006). This complex is important for preventing aggregation of hydrophobic LHCPs prior to insertion into the thylakoid membrane (Payan and Cline, 1991; Li et al., 1995; Schuenemann et al., 1998; Klimyuk et al., 1999; Tu et al., 1999). At the thylakoid membrane, the CpSRP receptor, CpFTSY, binds the transit complex (Kogata et al., 1999; Tu et al., 1999; Stengel et al., 2007; Chandrasekar et al., 2008) and positions it such that LHCPs can be integrated into the thylakoid membrane by the insertase ALBINO3 (ALB3) (Moore et al., 2000; Yuan et al., 2002; Moore et al., 2003).
Reducing light harvesting pigment concentration has been shown to improve photosynthesis in microalgae (Nakajima and Ueda, 1997; Nakajima et al., 2001) and cyanobacteria (Kirst et al., 2014). In plants, mutants of the CpSRP pathway (TLA mutants) have been identified in genetic screens by pale green leaf phenotypes that correspond to higher chlorophyll a/b ratios (Liu et al., 2016; Kirst et al., 2017; Kirst et al., 2018). Multiple studies have demonstrated trait alterations in TLA mutant plants with the potential for agronomic benefit, with particular focus on plants with mutations in CpSRP43 (TLA3 mutants) and CpSRP54 (TLA4 mutants). For example, the Arabidopsis TLA3 mutant chaos showed significantly higher tolerance to photooxidative stress in lab and field conditions (Klenell et al., 2005), and CpSRP43 knockdown in Nicotiana tobacum was recently demonstrated to enhance leaf-to-stem ratio and plant biomass (Kirst et al., 2018). Similarly, a TLA3 mutation in rice reduced plant height and tillering with no loss of grain mass (Lv et al., 2015), and three distinct TLA4 mutant alleles have been identified in rice that cause reduced tillering with no loss of grain weight (Zhang et al., 2013; Liu et al., 2016). In the Poales family, mutations to paralogous proteins CpSRP54a and CpSRP54b (creating TLA4 and TLA4L mutants, respectively) also caused plants to display a pale-green phenotype, although TLA4L mutants died before reaching maturity (Shi et al., 2020). In contrast to potential benefits associated with TLA3 and TLA4 mutants, plants lacking CpFTSY (TLA2 mutants) or ALB3 have been shown to suffer significant pleiotropic effects on growth and development. For example, Arabidopsis cpftsY and alb3 mutants (Durrett et al., 2006; Asakura et al., 2008), and a maize cpftsY null-mutant (Asakura et al., 2004) had pleiotropic defects associated with broad roles in thylakoid biogenesis in addition to a substantial reduction in chlorophyll.
Previous research has indirectly compared various molecular characteristics of TLA mutants in Arabidopsis (Amin et al., 1999; Asakura et al., 2008; Walter et al., 2015). However, direct comparisons of potential agronomic benefits between TLA mutants in crop plants have not been performed, in part because they have been independently identified and characterized in different genetic backgrounds, which can prevent the ability to distinguish phenotypic differences due to broad genotypic contributions from specific TLA mutations. Gene engineering mediated by CRISPR/Cas9 now provides the ability to compare gene function in near-isogenic backgrounds, allowing for a direct dissection of the contribution of each TLA mutation to photosynthesis and fitness phenotypes. In this study, we used CRISPR/Cas9 to create three independent mutant lines, each with a single null mutation in predicted components of the rice CpSRP pathway; CpSRP43 (TLA3), CpSRP54a (TLA4), and CpSRP54b (TLA4L), and assessed their potential agronomic benefits.
Assembly of CRISPR/Cas9 T-DNA vectors was performed as described previously (Lowder et al., 2015). In short, sgRNA oligo pairs targeting CpSRP43 (LOC_Os03g03990), CpSRP54a (LOC_Os11g05552), and CpSRP54b (LOC_Os11g05556) were designed and annealed (Table 1). Annealed oligos with a single target near the N-terminal of CpSRP54a or CpSRP54b were cloned into the gateway entry vector pYPQ141C (Lowder et al., 2015) to create 1-TLA4 and 1-TLA4L. Annealed oligos for golden gate assembly were cloned into the golden gate recipient vectors: pYPQ131C, pYPQ132C, or pYPQ133C (Lowder et al., 2015). Proper ligation of oligos was confirmed at all stages by Sanger sequencing with either primer M13-F (for pYPQ141C) or pTC14-F2 (for pYPQ131C, pYPQ132C, and pYPQ133C). See Table 2 for the sequences of primers used in this study. The following golden gate assemblies were also performed: assembly of two sgRNAs targeting the N- and C-terminal regions of CpSRP43, CpSRP54a, or CpSRP54b into pYPQ142 (Lowder et al., 2015) to create 2-TLA3, 2-TLA4, and 2-TLA4L, respectively, and assembly of three sgRNAs targeting near the N- and C-terminal of CpSRP54a and the C-terminal of CpSRP54b into pYPQ143 (Lowder et al., 2015) to create TLA4/TLA4L. Gateway assembly of the sgRNA entry vectors, the Cas9 entry vector pYPQ167 (Lowder et al., 2015), and the gateway-compatible destination vector Ubi-CAMBIA-1300 (Chern et al., 2001) were performed using LR clonase II enzyme (Thermo Fisher Scientific, Waltham, MA), to generate the final CRISPR/Cas9 T-DNA vectors Ubi-2-TLA3, Ubi-1-TLA4, Ubi-2-TLA4, Ubi-1-TLA4L, Ubi-2-TLA4L, and Ubi-TLA4/TLA4L. Proper assembly was verified by restriction digestion and sequencing with Ubi-pro-F and Cas9-seq-R primers (Table 2).
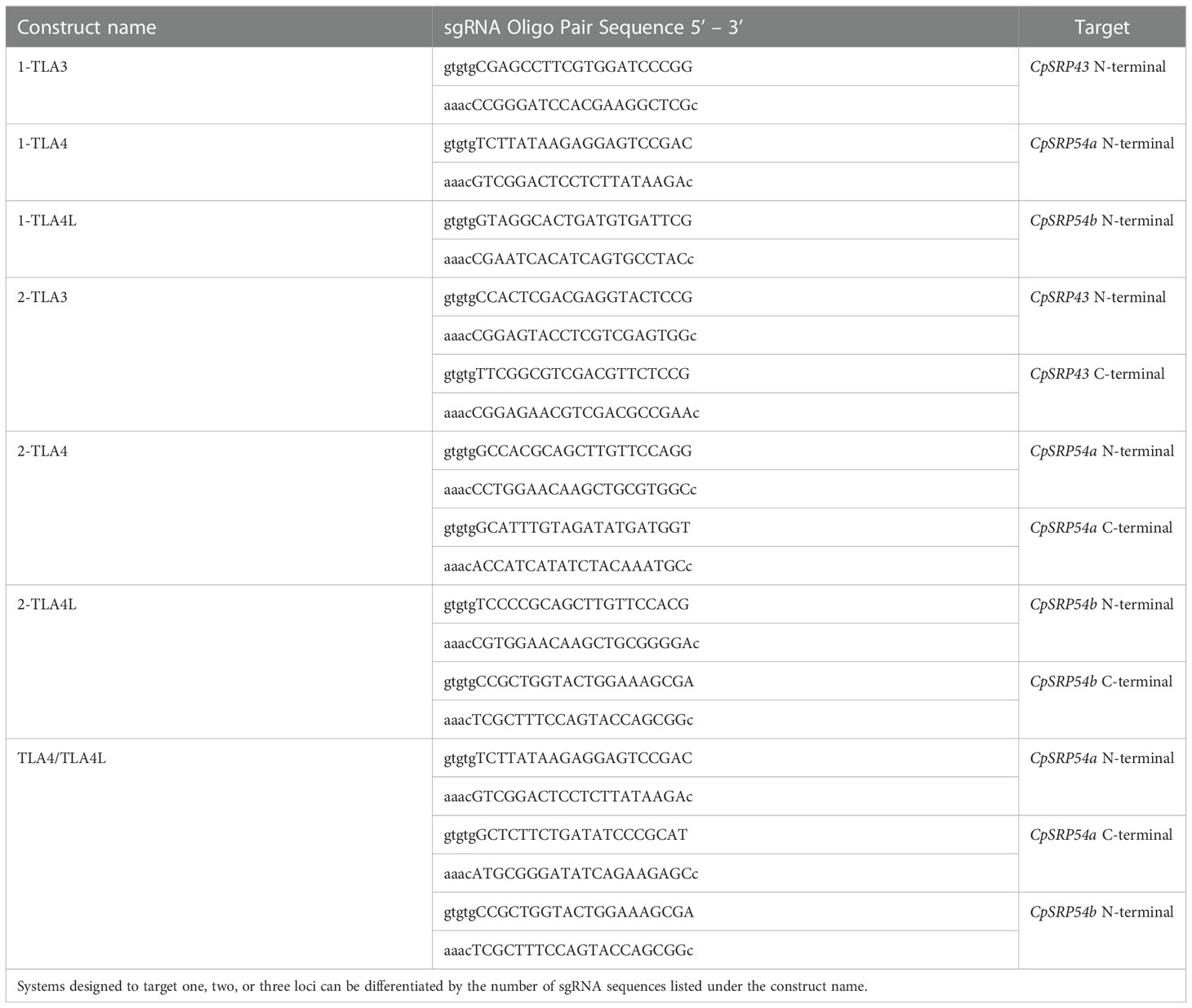
Table 1 sgRNA oligo pairs designed to produce CRISPR/Cas9-based knockouts of CpSRP43, CpSRP54a, and CpSRP54b.
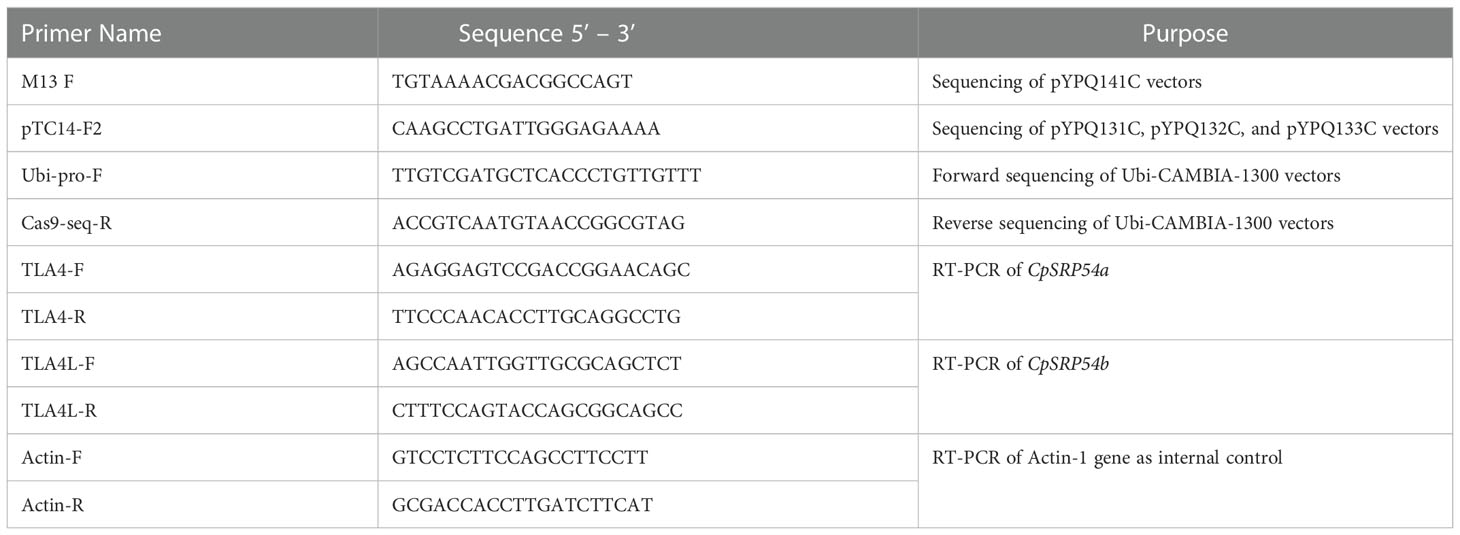
Table 2 Primers used in this study. Both sequencing and RT-PCR primers are listed with corresponding purposes.
Transformation of Agrobacterium tumefaciens strain EHA105 with plasmids Ubi-2-TLA3, Ubi-1-TLA4, Ubi-2-TLA4, Ubi-1-TLA4L, Ubi-2-TLA4L, or Ubi-TLA4/TLA4L was performed by electroporation using the Gene Pulser Xcell (Bio-Rad), per the manufacturer’s instructions. Rice transformation was performed at the University of California, Davis Rice Transformation Facility using A. tumefaciens strain EHA105 to infect rice calli (Oryza sativa ssp. japonica L., cv. Kitaake). Selection of transformants carrying the transgenes was performed using hygromycin as a selectable marker and later confirmed by PCR with transgene specific primers Ubi-pro-F and Cas9-seq-R (Table 2). Identification of successful CRISPR/Cas9-mediated indels was determined by PCR using primers flanking the target cut site. Indels were further validated by Sanger sequencing. All plants used in this study were homozygous TLA mutants unless otherwise specified as null mutants (n).
Rice plants were grown in the greenhouse with 16-h day and 8-h night cycle, 28°C day and 26°C night temperature, and 50-60% relative humidity. Plants were grown in a peat-based soilless media and watered daily with a complete nutrient solution (Zhen and Bugbee, 2020). The solution was supplemented with 20 ppm ethylenediamine-N,N′-bis(2-hydroxyphenylacetic acid) (EDDHA) chelated iron to minimize any possible iron chlorosis. Photosynthetic rate was measured using a portable photosynthesis system (model LI-6800, LICOR Biosciences, Lincoln, NE) with a fluorometer head attached (model LI-6800-01A, LICOR Biosciences). The chamber settings held constant across measurements were: 25°C air temperature, 150 µmol s-1 flow rate, 5000 rpm fan speed, 50% relative humidity, 90% red photon fraction, and 10% blue photon fraction. Light response curves (LRCs) were generated by measuring assimilation rates stepwise at 400, 100, 0, 50, 200, 400, 800, 1400, and 2000 µmol photons m-2 s-1 using the built-in light response curve program. A minimum chamber stabilization time of 10 minutes was set, and carbon dioxide and water infrared gas analyzers were matched following the logging of each measurement. LRCs were run first at 400 ppm CO2 and then at 1200 ppm CO2 without removing the leaf from the chamber. All photosynthetic rate measurements were collected on rice plants ranging from 33 to 35 d old.
Photon absorbance was calculated by subtraction from measurements of transmission and calculated reflectance. Photon transmission was measured after placing the leaf over quantum sensor (model LI-190R, LICOR Biosciences). The transmitted fraction of photons was multiplied by 2 to account for an equal number of reflected and transmitted photons (Jacquemoud and Ustin, 2019). Photosynthetic rates were then divided by the absorbed photon flux to calculate photosynthetic rate per photon absorbed. Chlorophyll content was measured using an optical chlorophyll meter (Model MC-100, Apogee Instruments, Logan, UT) using generic coefficients (Parry and Bugbee, 2017). Laboratory measurements were made on nine leaves and averaged. To confirm optical chlorophyll measurements, 0.05 g of tissue was cut from the newest fully expanded leaf of mature rice plants and ground to a fine powder in liquid N2. Photosynthetic pigment was extracted using 80% acetone and incubated in the dark at room temperature for 24-h. Absorbance measurements at 663.2 nm, 646.8 nm, and 470 nm were recorded using a Shimadzu UV-1280 spectrophotometer. Estimates of chlorophyll and carotenoid content were calculated with the equations previously described by Wellburn (1994).
Protein blast was performed using Arabidopsis CpSRP54 (At5g03940) as an input in phytozome v12.1 against other genomes, retrieving 188 hits. Because plants have both an SRP54 and CpSRP54 system, we used ChloroP v1.1 (Emanuelsson et al., 1999) to predict which proteins were chloroplast localized, reducing our protein lists to 60 hits. Of those 60, 32 genes that were predicted to encode the SRP receptor (CpFTSY) were removed from the list, leaving 28 genes predicted to encode CpSRP54 across 21 species. CpSRP54 proteins were aligned using the MUSCLE alignment algorithm, with 100 iterations. The phylogenetic tree was built using the Jukes-cantor neighbor joining method, with 1,000 bootstraps. The CpSRP pathway functional gene network predictions were made using RiceNet v2 (Lee et al., 2015) with default parameters.
Leaf tissue from the youngest fully expanded leaves of three independent TLA4 mutant lines and Kitaake WT was collected from four-week-old plants (five plants per line) and RNA was extracted using a Qiagen Plant RNA extraction kit. RNA was converted to cDNA using a cDNA synthesis kit, and quantitative RT-PCR was performed using primers specific to CpSRP54a (TLA4-F/R) or CpSRP54b (TLA4L-F/R) (Table 2). Actin-1 (LOC_Os03g50885) gene expression was amplified as a control using the primers Actin-F/R (Table 2).
Statistical tests were performed using a one-way analysis of variance (ANOVA) test. Statistically significant results were followed up with a Tukey’s honest significance post-hoc test using R (Version 3.6.1, R Core Team). Significance for all statistical tests was predetermined at p < 0.05, and significant differences are reported in figures using capital letters to differentiate between groups. Groups with the same letter are not significantly different from one another.
Rice TLA3 mutations have emerged as the cause of pale-green leaf phenotypes in multiple rice ethyl methane sulfonate (EMS) screens (Lv et al., 2015; Wang et al., 2016; Ye et al., 2018). Likewise, previous EMS and T-DNA screens have also identified putative TLA4 mutations in rice (Zhang et al., 2013; Liu et al., 2016). However, direct comparisons between TLA mutants in rice have been difficult, as these studies were performed in different cultivars, and as EMS-generated mutants may not be complete loss-of-function mutants or may carry additional genetic mutations that contribute to phenotypic outcomes (Wang et al., 2016). To gain a better understanding of the agronomic potential of these TLA mutants, we utilized CRISPR/Cas9 to generate targeted TLA knock-out mutants in the rice cultivar Kitaake. Rice CpSRP43 is encoded by a single gene copy with a single exon. Using CRISPR/Cas9, we generated near-complete gene knockouts of CpSRP43 in rice (Figure 1D) to create TLA3 mutants. In agreement with previous studies, TLA3 mutants had reduced plant height (Figure 1A, C). Likewise, these mutants displayed a pale green leaf phenotype and had reduced chlorophyll content (Figure 1B, C). These results demonstrate that CRISPR/Cas9 can be successfully implemented to generate targeted TLA-engineered rice plants with phenotypes consistent with the previous studies described above.
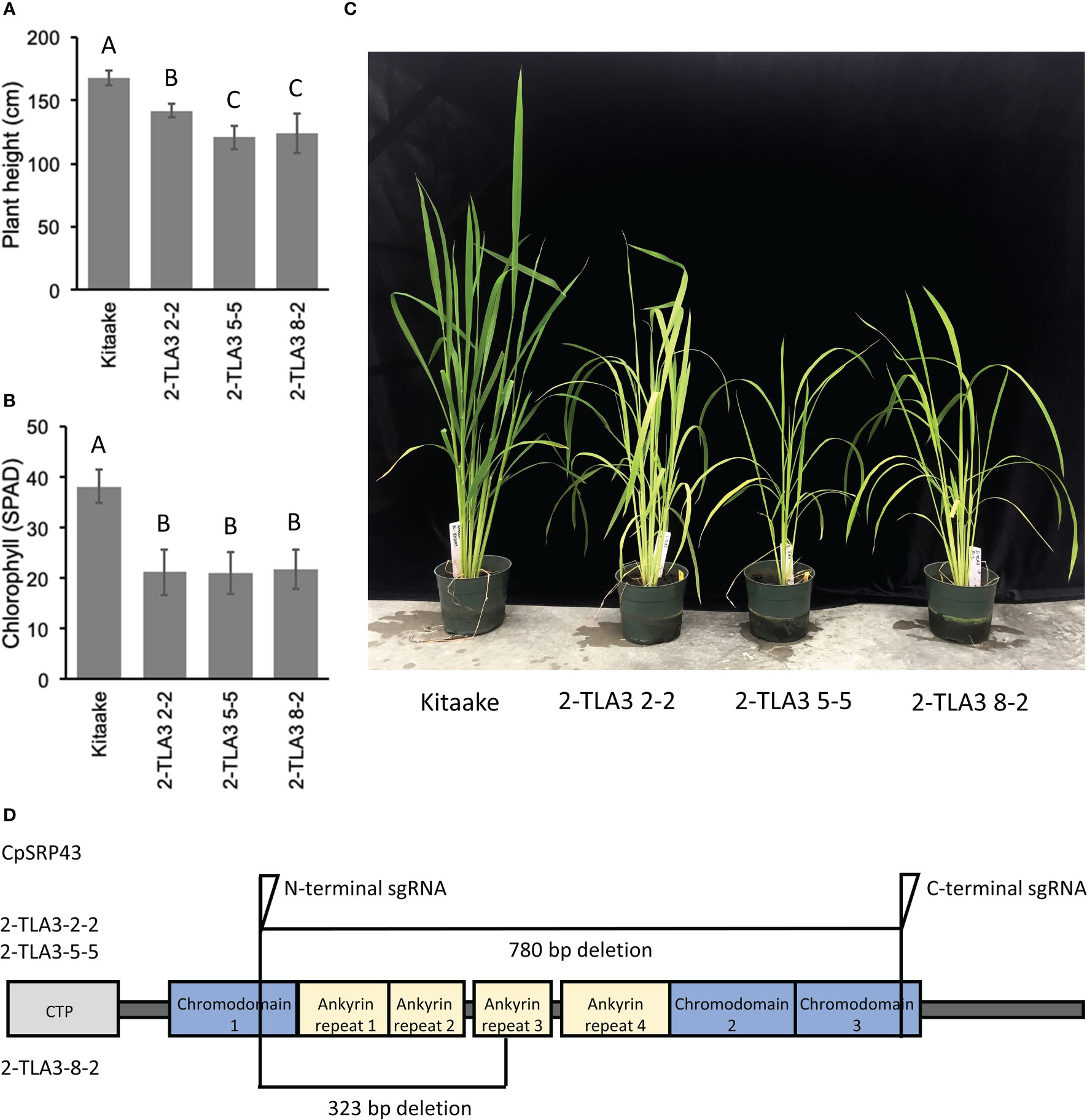
Figure 1 TLA3 mutants have reduced height and chlorophyll content. (A) Measurements of plant height for three independent CRISPR/Cas9-generated TLA3 mutant lines as compared to Kitaake wild type (WT). (B) Measurements of chlorophyll content for these three mutants, as measured using a SPAD meter. (C) Representative images of the homozygous parental line of each of the three mutant lineages characterized in (A) and (B), demonstrating reduced stature and a pale green leaf phenotype. (D) Protein domain model of validated CRISPR/Cas9-mediated TLA3 mutations generated using two sgRNAs indicating the relative position and size in base pairs of large deletions within CpSRP43. Frameshift mutations occurred in the middle of chromodomain 1 in each line. CTP, Chloroplast targeting peptide.
Two previous EMS and T-DNA screens have identified the putative CpSRP54 gene in rice encoded by LOC Os11g05552 (Zhang et al., 2013; Liu et al., 2016), here called CpSRP54a. The canonical CpSRP54a protein structure consists of conserved N, G, and M domains (Figure 2A). While CpSRP54 is predicted to be encoded by a single gene copy in algae, mosses, and dicots, a tandem duplication of CpSRP54a occurred in an ancestor of the Poales lineage of monocots (here referred to as CpSRP54b). This duplication is absent from non-Poales monocot species, such as duckweed and banana, and is present in all sequenced members of the Poales family (Figure 2B). To predict whether both genes are involved in the CpSRP pathway, we performed a computational functional gene network prediction using CpSRP43, CpSRP54a, and CpSRP54b as inputs using RiceNet v2 (Lee et al., 2015), which utilizes a large repository of published expression data for rice for its calculations. Network predictions demonstrated that the predicted rice CpFTSY was a node shared by all three genes, suggesting that they are all involved in the anticipated CpSRP pathway (Supplemental Figure 1). Likewise, CpSRP43 is predicted to functionally interact with the insertase ALB3 (Supplemental Figure 1), which has been previously characterized in Arabidopsis as part of the CpSRP pathway (Tzvetkova-Chevolleau et al., 2007). The most direct connections between inputs were shared between CpSRP54a and CpSRP54b, suggesting a strong level of functional overlap. Notably, CpSRP54b has more predicted connections with CpSRP43 than does CpSRP54a (Supplemental Figure 1), suggesting that it may play a more important role in CpSRP pathway function than its paralog. Collectively these data suggest that both putative CpSRP54 paralogs are expressed and are predicted to interact with other CpSRP pathway components.
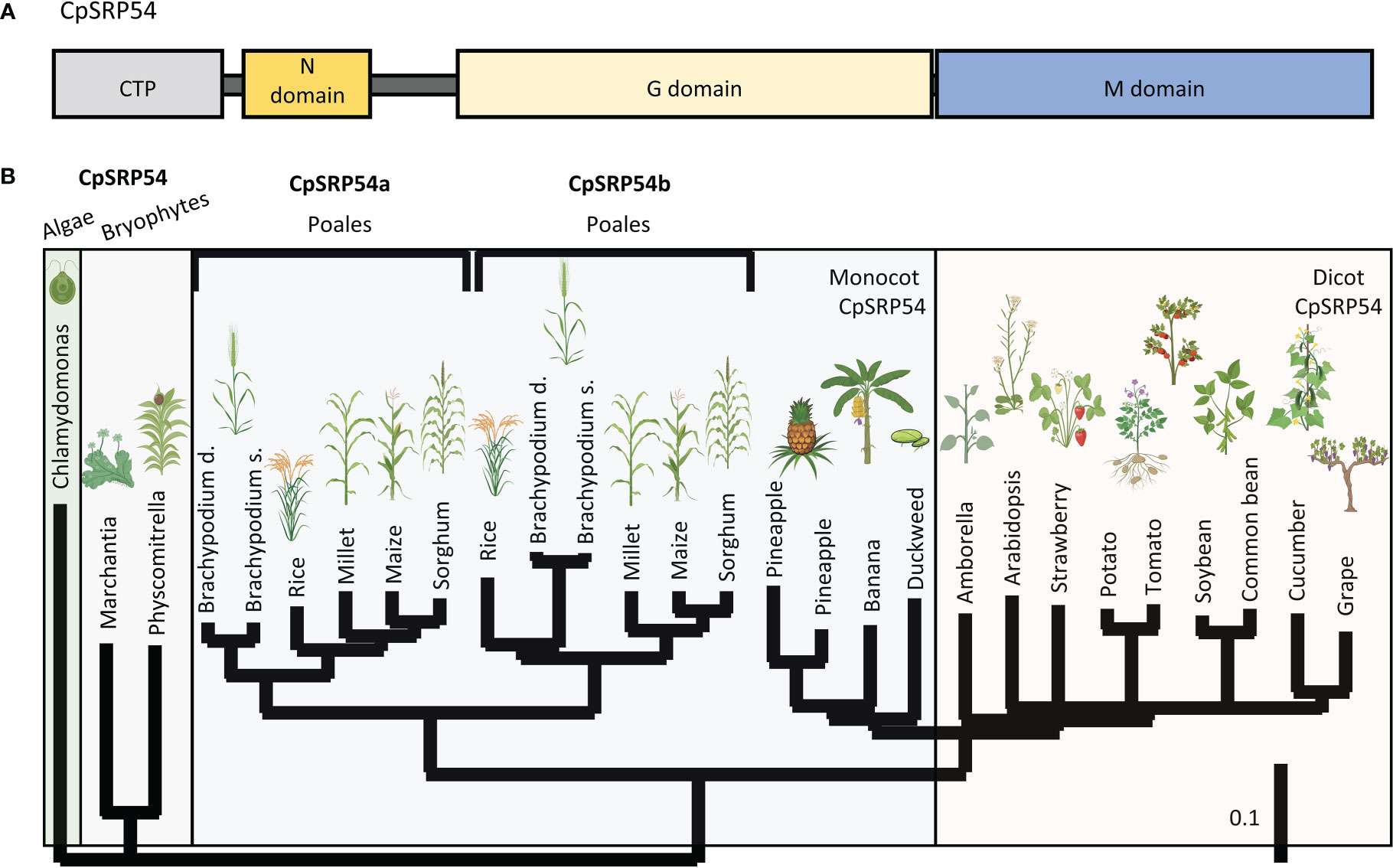
Figure 2 A duplication of CpSRP54 occurred in the Poales lineage. (A) Protein domain model of CpSRP54 with CTP (chloroplast targeting peptide). (B) Phylogenetic tree of CpSRP54 showing a duplication event in the Poales lineage to produce CpSRP54a and CpSRP54b Created with BioRender.com.
To allow for direct comparisons between mutations in CpSRP54a, CpSRP54b, and CpSRP43 in the context of photosynthetic and agronomic traits within the same genetic background, we used CRISPR/Cas9 to generate TLA4 and TLA4L single null mutants and TLA4/TLA4L double null mutants in the Kitaake cultivar (Figure 3F). In agreement with previous findings (Shi et al., 2020), we confirmed that TLA4 mutants had reduced plant height, and a demonstrable decrease in chlorophyll content (Figure 3A-E). Notably, unlike the findings of Shi et al. (2020), TLA4L rice plants were able to set seed and be propagated (Figure 3C). However, compared with the TLA4 mutants, the TLA4L mutants had a greater decrease in both plant height (Figure 3D) and chlorophyll content (Figure 3E), further suggesting that TLA4L and TLA4 do not perform identical roles within the CpSRP pathway. The inclusion of TLA4L null segregants with restored plant height and chlorophyll content demonstrate that observed phenotypes are due to specific mutations in the target gene and not the result of off-target cutting by CRISPR/Cas9.
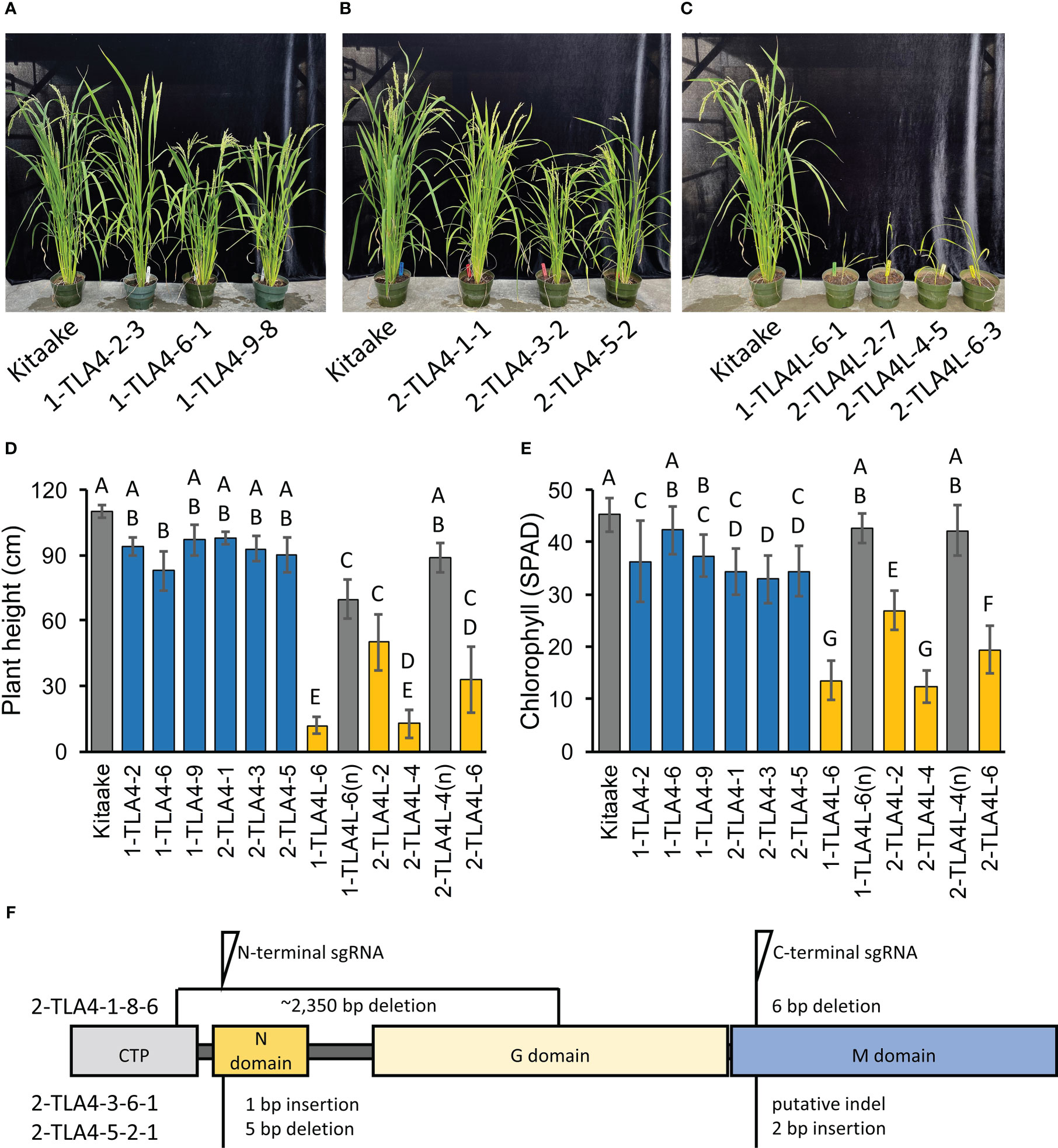
Figure 3 CpSRP54a and CpSRP54b both contribute to leaf chlorophyll accumulation. (A, B, C) Representative images of several mutant TLA4 (A, B) and TLA4L (C) lineages demonstrate reduced plant height and a pale green leaf phenotype as compared to WT. TLA4L mutants display a particularly severe phenotype. (D) Plant height of representative TLA4 and TLA4L mutant individuals from independently CRISPR/Cas9-generated lineages. Blue bars represent TLA4 mutants, yellow bars represent TLA4L mutants, and gray bars represent null-segregants (n). Null-segregants show restored WT phenotypes. (E) Measurements of chlorophyll content for TLA4, TLA4L, and TLA4L(n) individuals as measured using a SPAD meter. Blue bars represent TLA4 mutants, yellow bars represent TLA4L mutants, and gray bars represent null-segregants (n). (F) Protein domain model with CTP (Chloroplast targeting peptide) of validated CRISPR/Cas9-mediated TLA4 mutations generated using two sgRNAs indicating the relative position and size in base pairs of indels within CpSRP54a. All three mutant lines have multiple indels.
Due to the close proximity of CpSRP54a and CpSRP54b in the rice genome, and the fact that TLA4 mutations reside upstream of the CpSRP54b promoter, we hypothesized that the loss of function mutation in TLA4 may have led to altered expression of CpSRP54b, making the dissection of CpSRP54a and CpSRP54b contributions to phenotype difficult in TLA4 mutant lines. To test this, we used quantitative RT-PCR to first confirm the reduced expression of CpSRP54a in three independent TLA4 lines (2-TLA4-1-8-6, 2-TLA4-3-6, and 2-TLA4-5-2) compared with WT Kitaake (Supplemental Figure 2A). We observed significantly reduced CpSRP54a expression in TLA4 mutant lines 2-TLA4-3-6 and 2-TLA4-5-2, and a complete loss of CpSRP54a expression in 2-TLA4-1-8-6 (Supplemental Figure 2A). Contrary to our hypothesis, no significant reduction in CpSRP54b expression was observed in any of the three independent TLA4 mutant lines as compared with Kitaake (Supplemental Figure 2B). These results indicate that the phenotypes observed in TLA4 mutants are not due to reduced expression of CpSRP54b, but can be attributed to the knockout of CpSRP54a alone.
To evaluate the degree to which CpSRP54a and CpSRP54b perform similar functions in the CpSRP pathway, 30 rice plantlets were recovered from tissue culture containing the TLA4/TLA4L sgRNA designed to generate double knockout mutants. However, unlike either single mutant, all putative TLA4/TLA4L double mutant plantlets died shortly after transplanting to the greenhouse. While not definitive, this observation suggests that CpSRP54a and CpSRP54b may contribute additively to chlorophyll accumulation in rice. Combined with the observation that TLA4 mutants do not have reduced expression of CpSRP54b yet display significant phenotypes, these results suggest that CpSRP54a and CpSRP54b are both functional proteins with overlapping but not identical roles in rice.
To more precisely determine the relative contributions of CpSRP43 and CpSRP54a to photosynthesis and fitness, a series of experiments were performed to evaluate chlorophyll and carotenoid content, plant height, and photosynthetic rate across physiological development and a range of lighting conditions. TLA4L mutants were not included in this study due to the extremely severe phenotypes associated with the mutations, including dramatically smaller plant size.
First, the pigment content and plant height of three independent lines of TLA3 (2-TLA3-2-5, 2-TLA3-5-5-1-5, 2-TLA3-8-2-3) and TLA4 (2-TLA4-1-8-6, 2-TLA4-3-6, 2-TLA4-5-2) mutants were compared. At five weeks post germination, both the chlorophyll and carotenoid content were significantly reduced in TLA mutants compared to wild-type plants, with TLA3 having lower levels of each pigment than TLA4 (Supplemental Figure 3). Similarly, the plant heights of both TLA3 and TLA4 mutants were reduced two weeks after panicle emergence (Supplemental Figure 3A). In agreement with previous reports (Lv et al., 2015; Liu et al., 2016; Wang et al., 2016; Shi et al., 2020), chlorophyll a/b ratio was elevated and the ratio of chlorophyll to carotenoids was reduced in TLA mutants (Supplemental Figure 3F).
A further characterization of TLA phenotypes was performed using two independent mutant lines for both TLA3 (2-TLA3-5-5-1-5-1, 2-TLA3-8-2-3-4) and TLA4 (2-TLA4-1-8-6-1, 2-TLA4-3-6-1).We observed that while chlorophyll concentration increased over time for all lines, rates of accumulation differed between WT and TLA mutant lines (Figure 4A). Specifically, while the WT reached a steady chlorophyll concentration after 40 days, all the genetically altered lines continued to increase until heading at 78 days. As expected, plant height steadily increased over time for all lines, but the height of the mutant lines was less than the WT until 60 days after planting (Figure 4B). Additionally, we saw that optically measured chlorophyll content was positively correlated with leaf photon absorbance fraction across all lines (Figure 5). Collectively, these results confirm earlier observations of decreased height and chlorophyll content during early and mid-development.
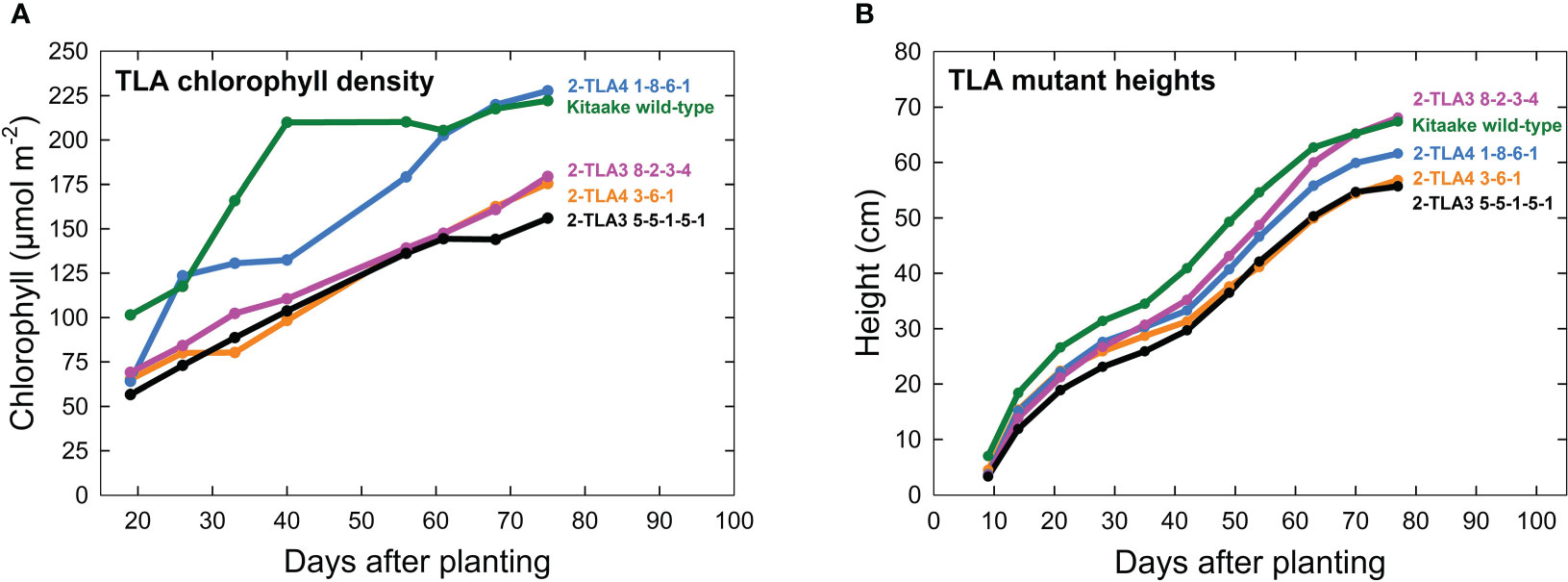
Figure 4 TLA mutants have reduced chlorophyll content and height. (A) Plots of chlorophyll content, (y-axis) across time (x-axis) for WT and all mutant lines demonstrate that chlorophyll content increases over time for all lines. (B) Most mutants are reduced in height compared to the wild-type.
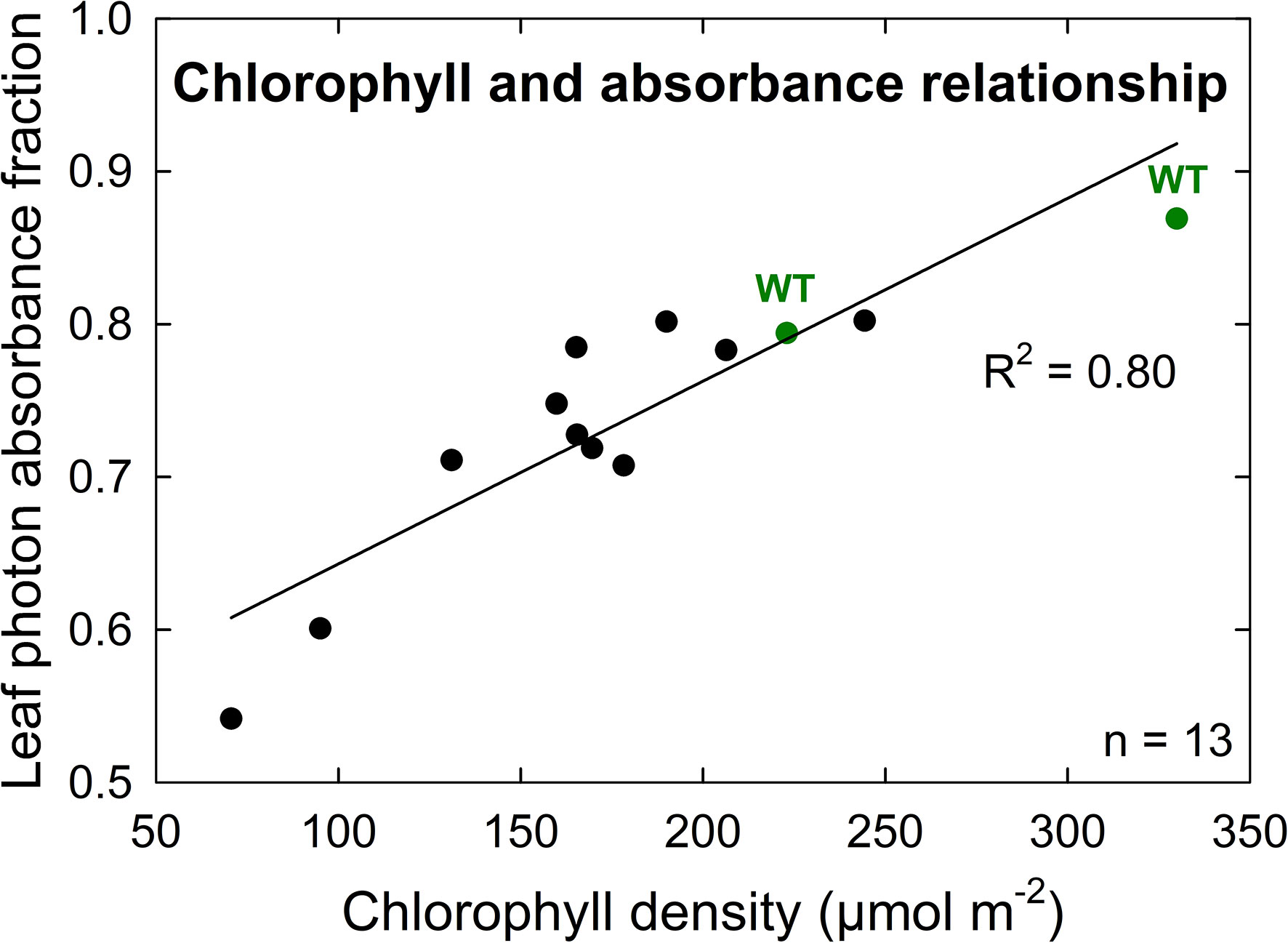
Figure 5 Chlorophyll density is positively correlated with leaf photon absorbance fraction among both wild-type (WT, green) and TLA mutant (black) lines. The high correlation (R2 = 0.80) of this relationship implies similar chlorophyll structures and orientations between TLA mutant and WT lines.
As hypothesized, we also observed that mutant lines exhibited only minimally reduced photosynthetic rate compared to the wild type under standard conditions (Figure 6A). When considering the reduced chlorophyll content of all mutant lines, this formally demonstrates that photosynthetic rates per photons absorbed increased in most TLA mutants compared to WT (Figure 6B). Furthermore, we demonstrated that the photosynthetic rate across all lines increased at elevated CO2, and there was no interaction between increased CO2 and mutant lines (Figure 6C, D). Significant increases in photosynthesis per photon absorbed in TLA mutants are consistent with the theory that plants overproduce chlorophyll.
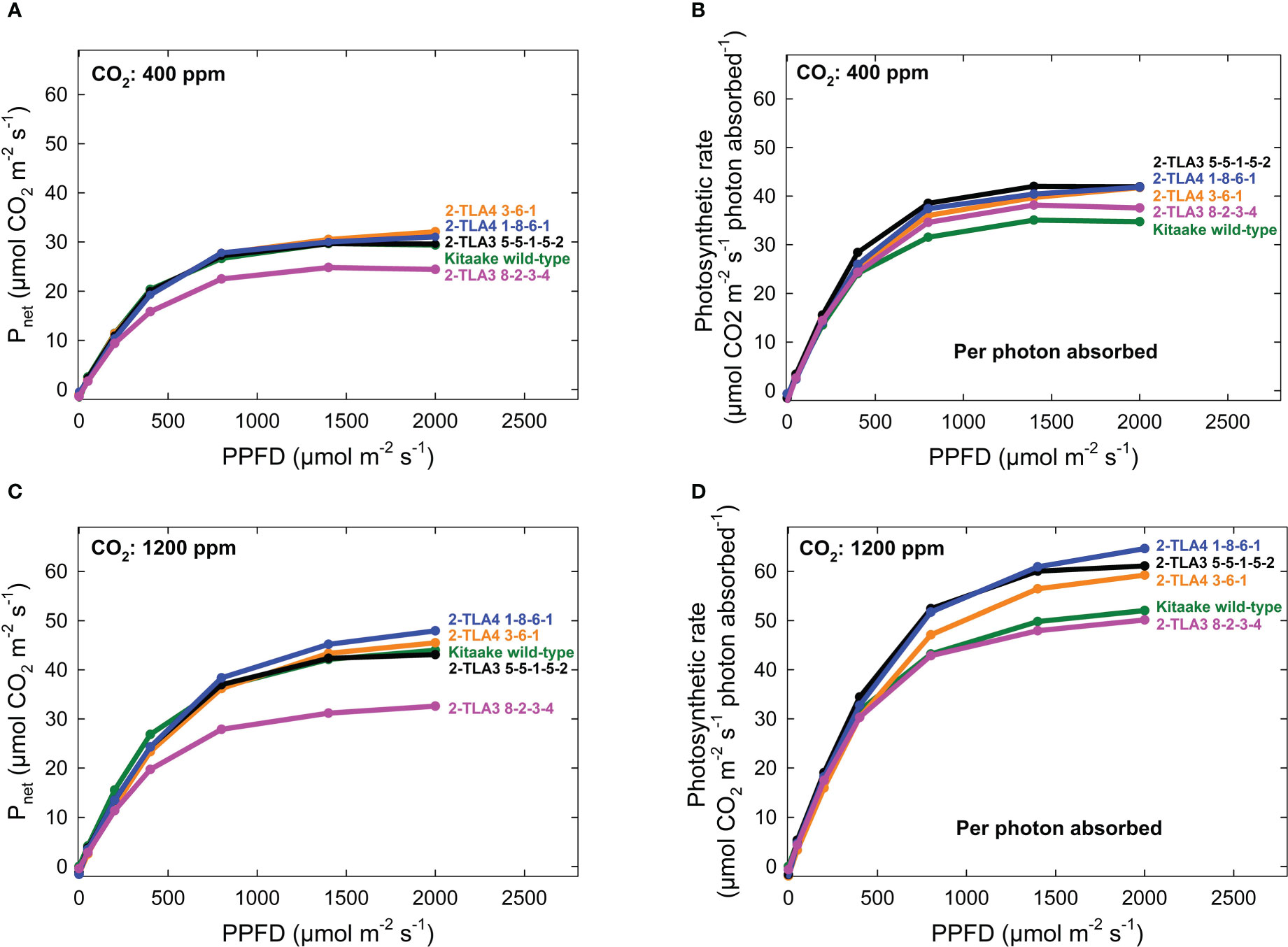
Figure 6 Photosynthetic rates in TLA mutants. The photosynthetic rates for mutant lines are increased compared to the Kitaake wild-type (WT) when accounting for the photosynthetic rate per photon absorbed at 400 ppm CO2 (A, B). The relationship is the same at 1200 ppm CO2 (C, D). The photosynthetic rate of 2-TLA3 8-2-3-4 (pink) is often less than the photosynthetic rate Kitaake WT (green).
Organisms need to safeguard the ability to capture light, which varies diurnally, changes with weather conditions, and can be blocked by shade produced by other plants (Kirst and Melis, 2018). As a genetically encoded strategy to ensure access to light even under suboptimal conditions, photosynthetic organisms often overproduce light-harvesting machinery. The net result is an overproduction of chlorophyll in leaves (Zhao et al., 2020), and at high light intensities there is wasteful absorption of photons by upper leaves, as they far exceed the capacity of the thylakoid membrane for electron transport and the biochemical rate of the carbon reactions of photosynthesis (Kirst et al., 2017; Kirst and Melis, 2018). The excess photons absorbed by upper leaves are used inefficiently for photosynthesis, with an increased fraction of the absorbed light being dissipated as heat (Demmig-Adams et al., 1996; Baker, 2008). By engineering TLA3 and TLA4 mutants with reduced light-harvesting antenna size and chlorophyll content (resulting in more uniform light distribution within the plant canopy) and higher quantum yield compared to WT lines, we show that reduced chlorophyll content is a valid method to improve photosynthetic solar energy conversion efficiency on a whole-plant level. This reduction in chlorophyll content helps to reduce the excess absorption of light by the upper canopy, which increases light transmission to lower leaves, improves photochemical efficiency, and reduces the ensuing wasteful non-photochemical dissipation of excitation energy (Kirst et al., 2018).
Reduced chlorophyll content in leaves can lead to an overall reduction in heat dissipation processes, such as nonphotochemical quenching (NPQ), by limiting the amount of excess energy that is received by the photosynthetic reaction center. A failure in these photoprotective processes can lead to overaccumulation of energy, generation of reactive oxygen species (ROS), and damage to the photosynthetic machinery. However, plants that are too deficient in chlorophyll will capture suboptimal amounts of light energy and suffer a growth penalty as a result. In the case of TLA3 and TLA4 mutants, we observed no detrimental effects in plant growth or appearance throughout the lifecycle of the rice, indicating that this potential reduction had a minimal impact on plant health. We also observed no signs of photoinhibition among the lines and therefore did not measure ROS. However, it is possible that the initial decrease in height observed in TLA3 and TLA4 mutants may be due in part to a decrease in NPQ and associated ROS, which has been shown to decrease leaf elongation in maize (Rodríguez et al., 2002).
Previous research indicates that the CpSRP pathway may behave differently among plant species. In Arabidopsis, single mutants in either CpSRP43 (chaos) or CpSRP54 (ffc) do not drastically reduce chlorophyll accumulation. The CpSRP54 mutant ffc first produces yellow leaves, but these eventually recover and produce green leaves by maturity (Amin et al., 1999). A double mutant chaos/ffc was required to see a significant and lasting reduction of LCHPs (Hutin et al., 2002). In rice, we observed that TLA4 mutants follow this trend, but TLA4L mutants do not. We also observed that although the engineered TLA3 and TLA4 plants increased in chlorophyll content over time, levels did remain lower than WT until plants were 60 days old.
The potential redundancy between components of this pathway may allow for plants to compensate for the loss of individual components, and is alluded to by the apparent increase in CpSRP54b expression in plants with mutated CpSRP54a (Shi et al., 2020). This putatively-observed compensation may allow plants with reduced chlorophyll synthesis to reach WT levels given sufficient time, and may indicate that the greatest advantages of these mutants in terms of photosynthetic efficiency and light penetration would occur during early growth, when the most significant reductions in chlorophyll content are observed. Future work is needed to further characterize the change in CpSRP pathway function across developmental stages, which remains largely unexplained.
This study, and other recently published work, also suggests that a Poales-specific duplication of CpSRP54 has allowed for further evolution of this ancient and essential pathway. In bacteria, the SRP pathway requires the conserved SRP54 protein and an SRP RNA component (Ziehe et al., 2017). In higher plants, the CpSRP pathway maintains the CpSRP54 protein, lacks bacterially-required RNA, and contains a novel, chloroplast-specific CpSRP43 (Ziehe et al., 2017). A recent study in rice found that CpSRP54a and CpSRP54b both independently interact with CpSRP43, though their specific functions in this regard are not yet known (Shi et al., 2020). Specifically, this research has shown that while both CpSRP54a and CpSRP54b appear to colocalize with CpSRP43 in the chloroplast, the phenotypes of the two mutants TLA4 and TLA4L and their impact on gene expression of the remaining members of the pathway are distinct. These observed differences in phenotype for TLA4 and TLA4L mutants suggest that the functions of the two proteins do not strictly overlap. Indeed, it has recently been proposed that these two closely related paralogs may be responsible for transporting different chloroplast proteins into the thylakoid (Shi et al., 2020). Our study confirms that mutations in these two proteins have distinct phenotypes when compared within a common genetic background. While initial heights of TLA4 mutants were reduced compared to WT, they approached normal heights during the grain-fill stage, which suggests that impact on grain yield should be minimal. However, TLA4L mutants showed a more drastic reduction in plant height and chlorophyll content than did TLA4 mutants and never recovered to the stature observed for WT individuals. The difference in severity of observed phenotypes is also consistent with the findings of our CpSRP network analysis, which shows a greater number of predicted connections between CpSRP54b and CpSRP43 than between CpSRP54a and CpSRP43, indicating a potentially more integral role for the former. Previous work also corroborates these findings to suggest that CpSRP54b might be more essential for chloroplast development, as single knockout mutants are significantly smaller and were previously thought to be seedling lethal (Shi et al., 2020). The TLA4L plants generated in this study, however, survived beyond the seedling stage, although they remained severely stunted, developmentally delayed, and had poor seed set.
The prior study conducted by Shi et al. (2020) compared an EMS mutation of TLA4 in the cultivar IR64 and a CRISPR mutation in TLA4L in the cultivar Kitaake, preventing a comparison of TLA4 and TLA4L function in an isogenic background, and making the development and analysis of a double mutant challenging. Finally, it is worth noting that our study utilizing CRISPR/Cas9 enabled a direct test of the viability of a TLA4/TLA4L double mutant, which is not feasible by genetic crossing, as the two genes are located at the same genetic locus. In our study, we failed to produce any double TLA4/TLA4L mutants, suggesting that CpSRP54a and CpSRP54b collectively contribute to an indispensable part of plant development. The large number of overlapping functions between these proteins as shown in our CpSRP network analysis substantiates that our lack of double mutants was likely not due to CRISPR/Cas9 methodological challenges but is instead indicative of plant lethality in double mutants. Collectively, the phenotypic differences among TLA mutants in both the present and prior studies suggest that CpSRP genes likely vary in suitability as targets for plant engineering, and that care must be taken in choosing the target to avoid detrimental phenotypes.
In the wild, plants maximize light absorption through synthesis of large light-harvesting antenna, which makes individual plants more competitive by capturing more photons than neighboring plants (Kirst and Melis, 2018). However, in monoculture agriculture this normally beneficial trait leads to decreased light use efficiency and reduced yield potential because the top leaves absorb about 95% of the photons and the remaining leaves are light-starved. In theory, increased light transmission to lower leaves means more equal light distribution among leaves, especially in plants with a planophile (horizontal) leaf architecture, where excessive light absorption by the top-most leaves results in non-uniform light distribution. More uniform distribution of light among leaves should lead to increased canopy photosynthesis and yield. This study demonstrates the potential for this novel yet counterintuitive approach toward crop improvement via the knockout of essential genes and paves the way for work in other crop species. Future investigations involve expanding this work to larger scale field studies under multiple planting densities to show community-level improvements.
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Conceptualization, DC-D, BB and DC; methodology, DC, NL, SZ, LM; formal analysis, DC, NL; data curation, RK; writing—original draft preparation, DC, DC-D, BB, NL, ME, RK; supervision, DC-D, BB; project administration, DC-D. All authors contributed to the article and approved the submitted version.
This research was funded by NASA, grant number NNX17AJ31G.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1050483/full#supplementary-material
Amin, P., Sy, D. A. C., Pilgrim, M. L., Parry, D. H., Nussaume, L., Hoffman, N. E. (1999). Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant Physiol. 121, 61–70. doi: 10.1104/pp.121.1.61
Asakura, Y., Hirohashi, T., Kikuchi, S., Belcher, S., Osborne, E., Yano, S., et al. (2004). Maize mutants lacking chloroplast FtsY exhibit pleiotropic defects in the biogenesis of thylakoid membranes. Plant Cell 16, 201–214. doi: 10.1105/tpc.014787
Asakura, Y., Kikuchi, S., Nakai, M. (2008). Non-identical contributions of two membrane-bound cpSRP components, cpFtsY and Alb3, to thylakoid biogenesis. Plant J. 56, 1007–1017. doi: 10.1111/j.1365-313X.2008.03659.x
Baker, N. R. (2008). Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59, 89–113. doi: 10.1146/annurev.arplant.59.032607.092759
Bugbee, B. (1992). Determining the potential productivity of food crops in controlled environments. Adv. Space Res. 12, 85–95. doi: 10.1016/0273-1177(92)90014-O
Chern, M. S., Fitzgerald, H. A., Yadav, R. C., Canlas, P. E., Dong, X., Roland, P. C. (2001). Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J. 27(2), 101–113. doi: 10.1046/j.1365-313x.2001.01070.x
Chandrasekar, S., Chartron, J., Jaru-Ampornpan, P., Shan, S. (2008). Structure of the chloroplast signal recognition particle (SRP) receptor: Domain arrangement modulates SRP–receptor interaction. J. Mol. Biol. 375, 425–436. doi: 10.1016/j.jmb.2007.09.061
DeLille, J., Peterson, E. C., Johnson, T., Moore, M., Kight, A., Henry, R. (2000). A novel precursor recognition element facilitates posttranslational binding to the signal recognition particle in chloroplasts. Proc. Natl. Acad. Sci. 97, 1926–1931. doi: 10.1073/pnas.030395197
Demmig-Adams, B., Adams, W. W., III, Barker, D. H., Logan, B. A., Bowling, D. R., Verhoeven, A. S. (1996). Using chlorophyll fluorescence to assess the fraction of absorbed light allocated to thermal dissipation of excess excitation. Physiol. Plant 98, 253–264. doi: 10.1034/j.1399-3054.1996.980206.x
Durrett, T. P., Connolly, E. L., Rogers, E. E. (2006). Arabidopsis cpFtsY mutants exhibit pleiotropic defects including an inability to increase iron deficiency-inducible root Fe(III) chelate reductase activity. Plant J. 47, 467–479. doi: 10.1111/j.1365-313X.2006.02803.x
Emanuelsson, O., Nielsen, H., Heijne, G. V. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. doi: 10.1110/ps.8.5.978
Gu, J., Zhou, Z., Li, Z., Chen, Y., Wang, Z., Zhang, H. (2017b). Rice ( oryza sativa l.) with reduced chlorophyll content exhibit higher photosynthetic rate and efficiency, improved canopy light distribution, and greater yields than normally pigmented plants. Field Crops Res. 200, 58–70. doi: 10.1016/j.fcr.2016.10.008
Gu, J., Zhou, Z., Li, Z., Chen, Y., Wang, Z., Zhang, H., et al. (2017a). Photosynthetic properties and potentials for improvement of photosynthesis in pale green leaf rice under high light conditions. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01082
Hermkes, R., Funke, S., Richter, C., Kuhlmann, J., Schünemann, D. (2006). The α-helix of the second chromodomain of the 43 kDa subunit of the chloroplast signal recognition particle facilitates binding to the 54 kDa subunit. FEBS Lett. 580, 3107–3111. doi: 10.1016/j.febslet.2006.04.055
Hutin, C., Havaux, M., Carde, J., Kloppstech, K., Meiherhoff, K., Hoffman, N., et al. (2002). Double mutation cpSRP43 – /cpSRP54 – is necessary to abolish the cpSRP pathway required for thylakoid targeting of the light-harvesting chlorophyll proteins. Plant J. 29, 531–543. doi: 10.1046/j.0960-7412.2001.01211.x
Jacquemoud, S., Ustin, S. (2019). Leaf optical properties. 1st ed (Cambridge, United Kingdom:Cambridge University Press). . doi: 10.1017/9781108686457
Jonas-Straube, E., Hutin, C., Hoffman, N. E., Schünemann, D. (2001). Functional analysis of the protein-interacting domains of chloroplast SRP43. J. Biol. Chem. 276, 24654–24660. doi: 10.1074/jbc.M100153200
Kirst, H., Formighieri, C., Melis, A. (2014). Maximizing photosynthetic efficiency and culture productivity in cyanobacteria upon minimizing the phycobilisome light-harvesting antenna size. Biochim. Biophys. Acta BBA - Bioenerg. 1837, 1653–1664. doi: 10.1016/j.bbabio.2014.07.009
Kirst, H., Gabilly, S. T., Niyogi, K. K., Lemaux, P. G., Melis, A. (2017). Photosynthetic antenna engineering to improve crop yields. Planta 245, 1009–1020. doi: 10.1007/s00425-017-2659-y
Kirst, H., Melis, A. (2018). “Improving photosynthetic solar energy conversion efficiency: the truncated light-harvesting antenna (TLA) concept,” in Comprehensive series in photochemical & photobiological sciences. Eds. Seibert, M., Torzillo, G. (Cambridge: Royal Society of Chemistry), 335–354. doi: 10.1039/9781849737128-00335
Kirst, H., Shen, Y., Vamvaka, E., Betterle, N., Xu, D., Warek, U., et al. (2018). Downregulation of the CpSRP43 gene expression confers a truncated light-harvesting antenna (TLA) and enhances biomass and leaf-to-stem ratio in Nicotiana tabacum canopies. Planta 248, 139–154. doi: 10.1007/s00425-018-2889-7
Klenell, M., Morita, S., Tiemblo-Olmo, M., Mühlenbock, P., Karpinski, S., Karpinska, B. (2005). Involvement of the chloroplast signal recognition particle cpSRP43 in acclimation to conditions promoting photooxidative stress in arabidopsis. Plant Cell Physiol. 46, 118–129. doi: 10.1093/pcp/pci010
Klimyuk, V. I., Persello-Cartieaux, F., Havaux, M., Contard-David, P., Schuenemann, D., Meiherhoff, K., et al. (1999). A chromodomain protein encoded by the arabidopsis CAO gene is a plant-specific component of the chloroplast signal recognition particle pathway that is involved in LHCP targeting. Plant Cell 11, 87–99. doi: 10.1105/tpc.11.1.87
Kogata, N., Nishio, K., Hirohashi, T., Kikuchi, S., Nakai, M. (1999). Involvement of a chloroplast homologue of the signal recognition particle receptor protein, FtsY, in protein targeting to thylakoids. FEBS Lett. 447, 329–333. doi: 10.1016/s0014-5793(99)00305-1
Lee, T., Oh, T., Yang, S., Shin, J., Hwang, S., Kim, C. Y., et al. (2015). RiceNet v2: An improved network prioritization server for rice genes. Nucleic Acids Res. 43, W122–W127. doi: 10.1093/nar/gkv253
Li, X., Henry, R., Yuan, J., Cline, K., Hoffman, N. E. (1995). A chloroplast homologue of the signal recognition particle subunit SRP54 is involved in the posttranslational integration of a protein into thylakoid membranes. Proc. Natl. Acad. Sci. U. S. A. 92, 3789–3793. doi: 10.1073/pnas.92.9.3789
Liu, S., Deng, L., Fu, Y., Hu, G., Liu, W., Zhao, X. (2016). Identification and characterization of the yls mutation in rice (Oryza sativa l.) with lower photosynthetic pigment content. Czech J. Genet. Plant Breed. 52, 101–107. doi: 10.17221/42/2016-CJGPB
Lowder, L. G., Zhang, D., Baltes, N. J., Paul, J. W., Tang, X., Zheng, X., et al. (2015). A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 169, 971–985. doi: 10.1104/pp.15.00636
Lv, X., Shi, Y., Xu, X., Wei, Y., Wang, H., Zhang, X., et al. (2015). Oryza sativa chloroplast signal recognition particle 43 (OscpSRP43) is required for chloroplast development and photosynthesis. PloS One 10, e0143249. doi: 10.1371/journal.pone.0143249
Moore, M., Goforth, R. L., Mori, H., Henry, R. (2003). Functional interaction of chloroplast SRP/FtsY with the ALB3 translocase in thylakoids. J. Cell Biol. 162, 1245–1254. doi: 10.1083/jcb.200307067
Moore, M., Harrison, M. S., Peterson, E. C., Henry, R. (2000). Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J. Biol. Chem. 275, 1529–1532. doi: 10.1074/jbc.275.3.1529
Nakajima, Y., Tsuzuki, M., Ueda, R. (2001). Improved productivity by reduction of the content of light-harvesting pigment in Chlamydomonas perigranulata. J. Appl. Phycol. 13, 95–101. doi: 10.1023/A:1011192832502
Nakajima, Y., Ueda, R. (1997). Improvement of photosynthesis in dense microalgal suspension by reduction of light harvesting pigments. Hydrobiologia 9, 503–510. doi: 10.1023/A:1007920025419
Parry, C., Bugbee, B. (2017). Reduced root-zone phosphorus concentration decreases iron chlorosis in maize in soilless substrates. HortTechnology 27, 490–493. doi: 10.21273/HORTTECH03735-17
Payan, L. A., Cline, K. (1991). A stromal protein factor maintains the solubility and insertion competence of an imported thylakoid membrane protein. J. Cell Biol. 112, 603–613. doi: 10.1083/jcb.112.4.603
Rodríguez, A. A., Grunberg, K. A., Taleisnik, E. L. (2002). Reactive oxygen species in the elongation zone of maize leaves are necessary for leaf extension. Plant Physiol. 129, 1627–1632. doi: 10.1104/pp.001222
Schuenemann, D., Gupta, S., Persello-Cartieaux, F., Klimyuk, V. I., Jones, J. D., Nussaume, L., et al. (1998). A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes. Proc. Natl. Acad. Sci. U. S. A. 95, 10312–10316. doi: 10.1073/pnas.95.17.10312
Shi, Y., He, Y., Lv, X., Wei, Y., Zhang, X., Xu, X., et al. (2020). Chloroplast SRP54s are essential for chloroplast development in rice. Rice 13, 54. doi: 10.1186/s12284-020-00415-2
Stengel, K. F., Holdermann, I., Wild, K., Sinning, I. (2007). The structure of the chloroplast signal recognition particle (SRP) receptor reveals mechanistic details of SRP GTPase activation and a conserved membrane targeting site. FEBS Lett. 581, 5671–5676. doi: 10.1016/j.febslet.2007.11.024
Tu, C. J., Peterson, E. C., Henry, R., Hoffman, N. E. (2000). The L18 domain of light-harvesting chlorophyll proteins binds to chloroplast signal recognition particle 43. J. Biol. Chem. 275, 13187–13190. doi: 10.1074/jbc.c000108200
Tu, C.-J., Schuenemann, D., Hoffman, N. E. (1999). Chloroplast FtsY, chloroplast signal recognition particle, and GTP are required to reconstitute the soluble phase of light-harvesting chlorophyll protein transport into thylakoid membranes. J. Biol. Chem. 274, 27219–27224. doi: 10.1074/jbc.274.38.27219
Tzvetkova-Chevolleau, T., Hutin, C., Noël, L. D., Goforth, R., Carde, J.-P., Caffarri, S., et al. (2007). Canonical signal recognition particle components can be bypassed for posttranslational protein targeting in chloroplasts. Plant Cell 19, 1635–1648. doi: 10.1105/tpc.106.048959
Walter, B., Pieta, T., Schünemann, D. (2015). Arabidopsis thaliana mutants lacking cpFtsY or cpSRP54 exhibit different defects in photosystem II repair. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00250
Wang, Z., Zhang, T., Xing, Y., Zeng, X., Wang, L., Liu, Z., et al. (2016). YGL9, encoding the putative chloroplast signal recognition particle 43 kDa protein in rice, is involved in chloroplast development. J. Integr. Agric. 15, 944–953. doi: 10.1016/S2095-3119(15)61310-7
Wellburn, A. R. (1994). The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 144, 307–313. doi: 10.1016/S0176-1617(11)81192-2
Ye, J., Yang, Y., Wei, X., Niu, X., Wang, S., Xu, Q., et al. (2018). PGL3 is required for chlorophyll synthesis and impacts leaf senescence in rice. J. Zhejiang Univ.-Sci. B 19, 263–273. doi: 10.1631/jzus.B1700337
Yuan, J., Kight, A., Goforth, R. L., Moore, M., Peterson, E. C., Sakon, J., et al. (2002). ATP stimulates signal recognition particle (SRP)/FtsY-supported protein integration in chloroplasts. J. Biol. Chem. 277, 32400–32404. doi: 10.1074/jbc.M206192200
Zhang, F., Luo, X., Hu, B., Wan, Y., Xie, J. (2013). YGL138(t), encoding a putative signal recognition particle 54 kDa protein, is involved in chloroplast development of rice. Rice 6, 7. doi: 10.1186/1939-8433-6-7
Zhao, X., Jia, T., Hu, X. (2020). HCAR is a limitation factor for chlorophyll cycle and chlorophyll b degradation in chlorophyll-b-overproducing plants. Biomolecules 10, 1639. doi: 10.3390/biom10121639
Zhen, S., Bugbee, B. (2020). Substituting far-red for traditionally defined photosynthetic photons results in equal canopy quantum yield for CO2 fixation and increased photon capture during long-term studies: Implications for re-defining PAR. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.581156
Keywords: truncated light antenna, photosynthesis, chlorophyll, CRISPR/Cas9, rice
Citation: Caddell D, Langenfeld NJ, Eckels MJH, Zhen S, Klaras R, Mishra L, Bugbee B and Coleman-Derr D (2023) Photosynthesis in rice is increased by CRISPR/Cas9-mediated transformation of two truncated light-harvesting antenna. Front. Plant Sci. 14:1050483. doi: 10.3389/fpls.2023.1050483
Received: 21 September 2022; Accepted: 09 January 2023;
Published: 19 January 2023.
Edited by:
Agepati S Raghavendra, University of Hyderabad, IndiaReviewed by:
Zhanguo Xin, Agricultural Research Service, United States Department of Agriculture (USDA), United StatesCopyright © 2023 Caddell, Langenfeld, Eckels, Zhen, Klaras, Mishra, Bugbee and Coleman-Derr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Devin Coleman-Derr, ZGV2aW4uY29sZW1hbi1kZXJyQHVzZGEuZ292
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.