
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 20 January 2023
Sec. Plant Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1044918
Plant biological processes, such as growth and metabolism, hormone signal transduction, and stress responses, are affected by gene transcriptional regulation. As gene expression regulators, transcription factors activate or inhibit target gene transcription by directly binding to downstream promoter elements. DOF (DNA binding with One Finger) is a classic transcription factor family exclusive to plants that is characterized by its single zinc finger structure. With breakthroughs in taxonomic studies of different species in recent years, many DOF members have been reported to play vital roles throughout the plant life cycle. They are not only involved in regulating hormone signals and various biotic or abiotic stress responses but are also reported to regulate many plant biological processes, such as dormancy, tissue differentiation, carbon and nitrogen assimilation, and carbohydrate metabolism. Nevertheless, some outstanding issues remain. This article mainly reviews the origin and evolution, protein structure, and functions of DOF members reported in studies published in many fields to clarify the direction for future research on DOF transcription factors.
Regulation of gene transcription is fundamental to plant germination, growth, and tissue differentiation, and numerous transcription factors (TFs) are essential for these processes (Xian et al., 2020). Transcription factors, also referred to as trans-acting factors, are DNA-binding proteins that selectively bind to cis-acting elements in the promoter regions of target genes to promote or repress their transcription (Feng et al., 2016). Therefore, the structure and function of TFs have aroused great interest from scholars and are trending topics in the field of plant molecular biology. According to their conserved domains, TFs are divided into different families, including the WRKY, NAC, bZIP, AP2/ERF, bHLH, and Cys2-His2 zinc finger (ZF) families. Different types of TFs have evolved to regulate the expression of various plant-specific genes or signals. DOF (DNA binding with One Finger) is a classic protein in the ZF superfamily (Yang et al., 2022; Zhang et al., 2022). An increasing number of members of the DOF TF family have been reported to be involved in regulating plant germination (Chen et al., 2017), growth and development (Liu et al., 2022), and responses to abiotic stress (Sun et al., 2021). A recent perspective article reported that DOFs are involved in multiple hormonal pathways during abiotic stresses (Wang et al., 2022). However, an in-depth understanding of the DOF family reveals that DOFs also play an important role in other processes in plants. To direct follow-up studies of DOF TFs, this article mainly reviews the origin and evolution, protein structure, and biological functions of DOF in various aspects of plant life.
DOF proteins were originally found in maize (Yanagisawa and Izui, 1993) but not in yeast or Drosophila. These proteins are members of a plant-specific TF family that has not been detected in other eukaryotes (Riechmann et al., 2000). Compared to other TFs, fewer DOF members are present in the plant genome, and significant variation has been observed across different species. Although Chlamydomonas reinhardtii has only one DOF transcription factor, studies have shown that DOFs seem to have originated from this copy (Moreno-Risueno et al., 2007b). Since no identifiable DOF gene was identified in the genomes of red algae (Cyanidioschyzon merolae) or diatoms (Thalassiosira pseudonana), researchers have proposed that the origin of DOF TFs predates the isolation of green algae and terrestrial plant ancestors (Shigyo et al., 2007). The multiplication of DOF family genes seems to be related to the emergence of various transcriptional regulatory mechanisms in plant evolution. Some DOF TFs may play conserved roles in both vascular and non-vascular plants (Sugiyama et al., 2012). Earlier studies identified 37 DOF members in Arabidopsis thaliana, one of which is a pseudogene (Yanagisawa, 2002), while 30 DOF members were identified in rice (Oryza sativa) (Lijavetzky et al., 2003). With the analysis of the whole genomes of many species, an increasing number of DOF members have been identified in different species (Table 1), including 26 DOF genes in birch (Betula platyphylla) (Sun et al., 2021), 36 in watermelon (Citrullus lanatus) (Zhou et al., 2020), 35 in foxtail millet (Setaria italica) (Zhang et al., 2017), 22 in spinach (Spinacia oleracea) (Yu et al., 2021a) and 24 in rose (Rosa chinensis) (Nan et al., 2021). Despite the species differences, the functions of DOF members in the monocot model plant rice are still questioned. In particular, the issue of functional differences or redundancy is highly controversial. Some scientists believe that the functions of DOF proteins are highly redundant (Noguero et al., 2013; Huang et al., 2020), while others argue that large differences exist (Yanagisawa, 2016; Renau-Morata et al., 2020). Huang et al. (2020) suggested that the functional redundancy of DOF genes in rice strain ZH11 might mask the phenotypes of individual mutants. In contrast, Yu et al. (2021b) confirm that knockout lines of all OsDOF family members have significant phenotypic defects in the japonica rice strain TP309, indicating that the OsDOF family has low redundancy. The structural diversity of DOF TFs may be closely related to complex physiological regulatory networks established during plant evolution. However, the functions of many DOFs are still unknown. Consequently, elucidating the function of each DOF protein in plants is crucial for understanding the connection between the functional diversity of plant-specific TFs and plant species evolution.
DOF proteins consist of 200-400 amino acids, and a conserved DOF domain with a zinc finger (ZF) structure containing 50-52 amino acid residues is located at its N-terminal region, along with a transcriptional regulatory domain in the C-terminal region (Figure 1). Unlike other ZF proteins, DOF TFs contain only one Cys2/Cys2 ZF, and the ZF structure specifically recognizes the upstream core sequence 5’-(T/A)/AAAG-3’ of the target gene (Umemura et al., 2004; Kim et al., 2010). More specifically, the pumpkin DOF protein AOBP recognizes the AGTA motif and plays a role in facilitating binding of this protein to DNA (Kisu et al., 1998). The positions of four cysteines that comprise the ZF structure are necessary to achieve loop stability. The tryptophan in the C-terminal region of the ZF protein is important for DNA binding. In steroid hormone receptors, the tryptophan residue seems to play a role in stabilizing the structure (Noguero et al., 2013). However, the affinity of the conserved ZF domain in the DOF protein for AAAG motifs of downstream target genes is unknown. Moghaddas Sani et al. (2018) determined the binding affinity between the DOF-ZF domain and target oligonucleotides in Arabidopsis thaliana using microscale thermophoresis. The authors proposed that when two binding sites are present, the affinity of the DOF domain for an oligonucleotide is 100 times higher than that for the single binding site. This result explains why many repeated AAAG motifs exist in the promoters of downstream target genes of DOF TFs. Compared to other TF families, DOF proteins recognize relatively shorter motifs (Umemura et al., 2004), which has led to the discovery of many putative DOF binding sites in the promoter regions of many genes. However, most sites may be non-functional in vivo, and accumulating evidence suggests that the location of the AAAG motif limits DOF protein binding to DNA (Cavalar et al., 2003). Thus, DOF proteins may need to interact with other TFs to ensure the precise targeting and binding of DNA and subsequently promote transcription.
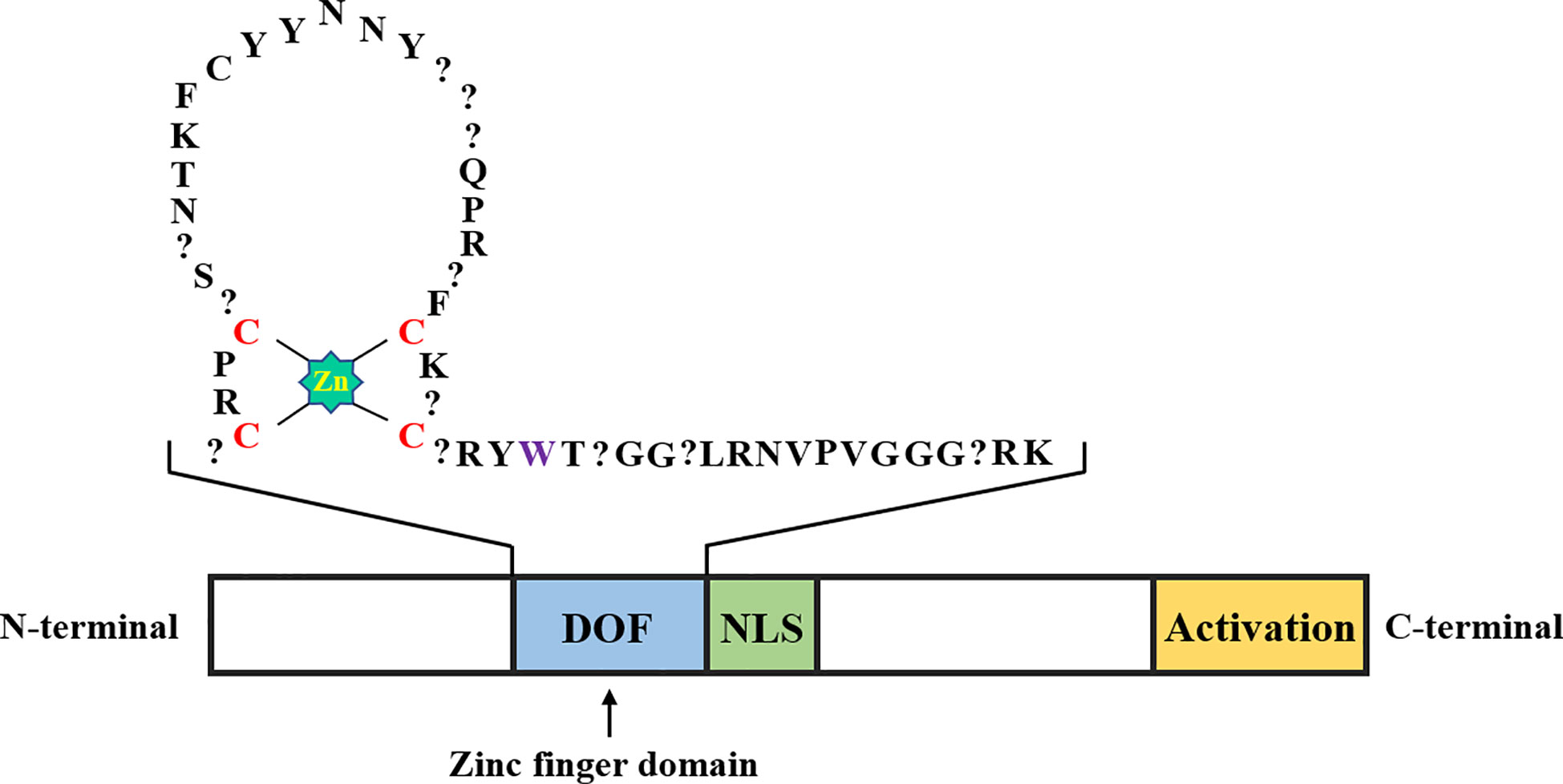
Figure 1 Schematic representation of the DOF transcription factor structure. The DOF zinc finger domain, nuclear localization signal, and transcriptional activation domain are shown in blue, green, and yellow, respectively. The amino acid sequence in the zinc finger domain of DOF is represented by abbreviations, the zinc coordination is shown in bright green, the position of the four cysteines in the zinc finger structure is shown in red, the question mark represents any amino acid, and the tryptophan residue mentioned in the second part is shown in purple.
The presence of two distinct domains in the N-terminal and C-terminal regions indicates that the DOF proteins are bifunctional (Noguero et al., 2013; Zou et al., 2019). The C-terminal region of DOF proteins contains a nonconserved domain that may interact with various TFs, including WRKY, bZIP, and MYB proteins, and thus function at different levels of expression (Zhang et al., 1995; Isabel-LaMoneda et al., 2003; Zou et al., 2008). The Arabidopsis DOF protein OBP1 was shown for the first time to specifically enhance the binding of the bZIP transcription factor OBF to its DNA target OSC element through a protein-protein interaction (Zhang et al., 1995). Subsequently, the ZmDOF2 protein in maize was shown to interact with HMG (High Mobility Group) proteins and affect the binding efficiency to downstream target sites (Yanagisawa, 1997). The barley DOF protein BPBF has been reported to interact with the MYB transcription factor GAMYB to activate the promoter of the target gene (Diaz et al., 2002). In addition, a binary nuclear localization signal (NLS) is located between the two domains of DOF proteins, which may partially overlap with them (Krebs et al., 2010).
Dormancy and germination are complex multistage processes in the plant life cycle that are tightly regulated by the coordinated expression of various genes in different tissues. As key TFs regulating gene expression, studies of DOF proteins are crucial for analyzing the molecular mechanism and regulatory network of plant dormancy and germination. In monocotyledons, the barley DOF proteins HvDOF17 and HvDOF19 were shown to regulate the expression of aleurone hydrolase genes and induce seed germination (Diaz et al., 2005; Moreno-Risueno et al., 2007a). An earlier study reported that OsDOF3 may be a mediator of GA signaling during rice germination (Washio, 2001). Some DOF TFs have been reported to regulate seed germination in response to exogenous GA3 in barley (Mena et al., 2002) and rice (Washio, 2003), but the regulatory mechanism between DOF and GA signaling remains unknown. The DOF protein DAG1, which affects germination, was detected in the dicotyledonous model plant A. thaliana and shown to play a key role in the establishment and maintenance of seed dormancy by controlling the dynamic balance of GA and ABA (Gabriele et al., 2010). However, the molecular mechanism by which DOF regulates seed germination has not been fully resolved. Subsequently, DAG1 was reported to interact with the DELLA protein GAI to inhibit the expression of the GA biosynthesis gene AtGA3ox1 and maintain seed dormancy (Boccaccini et al., 2014). The stability of the DAG1 protein was also controlled by GA (Boccaccini et al., 2016). Unlike DAG1, Arabidopsis DAG2 plays an active role in seed germination but is negatively regulated by DAG1 (Santopolo et al., 2015). As more DOF members were identified, AtDOF6 was shown to interact with AtTCP14 and inhibit seed germination (Rueda-Romero et al., 2012). Likewise, AtDOF6 interacted with the DELLA protein RGL2 to form the RGL2-DOF6 complex, which bound to the downstream AtGATA12 promoter and promoted seed dormancy (Ravindran et al., 2017). In addition to seed dormancy, DOF has also been reported to regulate bud dormancy in woody plants. For example, through transcriptome analysis of European beech (Fagus sylvatica) (Lesur et al., 2015) and whole-genome analysis of peach (Chen et al., 2017), researchers found that the DOF family may be involved in regulating plant bud dormancy. However, the molecular mechanism by which DOF members regulate bud dormancy and the potential ability of DOF proteins to regulate geophyte dormancy still remain to be determined.
DOF members are involved in regulating the growth and developmental stages of many plant tissues, such as root growth, hypocotyl elongation, plant morphogenesis, leaf development, and floral organ development. The tobacco (Nicotiana tabacum) DOF protein NtBBF1 binds to the rolB promoter in an auxin-induced manner, thus regulating root growth (Baumann et al., 1999). Arabidopsis cycling DOF factor 4 (AtCDF4) can promote the differentiation of root columella stem cells, but this effect is inhibited by WOX5 (WUSCHEL HOMEOBOX 5) via recruitment of TPL/TPR co-inhibitors and histone deacetylase HDA19 (Pi et al., 2015). Between the cotyledon and the root, the part near the root is called the hypocotyl. The Arabidopsis DOF TF COG1 (COGWHEEL1) regulates BR biosynthesis and ultimately promotes hypocotyl growth by binding to the PIF4 and PIF5 promoters and inducing their expression (Wei et al., 2017). Arabidopsis DAG1 can directly bind to the promoters of the downstream ethylene-related gene ERF2 (ETHYLENE RESPONSE FACTOR2), auxin-responsive gene SAUR67 (SMALL AUXIN UP RNA 67), and WRKY18 gene, which are involved in ABA signaling to promote hypocotyl elongation (Lorrai et al., 2018). Arabidopsis AtCDF1 and AtCDF5 promote hypocotyl elongation under short-day conditions (Martín et al., 2020). A recent study showed that Arabidopsis CDF2 and PIF4 (PHYTOCHROME-INTERACTING FACTOR 4) form a complex that co-regulates the downstream target gene YUCCA8, thereby promoting hypocotyl elongation (Gao et al., 2022). Plants activate phytochromes or phototropin and cryptochrome light receptors in vivo following exposure to light to promote growth and development. StCDF1 was active during potato tuber development under short-day (SD) conditions (Kondhare et al., 2019). Arabidopsis AtCOG1 depends on phytochromes upon light induction but plays a negative regulatory role in the phytochrome signaling pathway (Park et al., 2003). Interestingly, the antagonistic effect between phytochromes and COG1 may be explained in terms of the inhibition and activation of gibberellin, respectively. Phytochromes inhibit GA biosynthesis (Hedden and Thomas, 2012), while COG1 overexpression increases endogenous GA1 levels in Arabidopsis siliques by promoting the expression of the gibberellin biosynthetic gene GA3ox3, which is ultimately involved in seed coat development (Bueso et al., 2016).
Studies examining plant structural regulation have shown that overexpression of AtDOF5.4/OBP4 resulted in dwarfing of Arabidopsis plants by reducing cell size and number (Xu et al., 2016). Similarly, overexpression of OsDOF12 may lead to structural changes in rice, such as a reduced plant height, erect leaves, smaller leaves, and shortened panicles, by inhibiting BR signaling (Wu et al., 2015). As shown in the study by Nilsen et al. (2020), wheat TdDOF may regulate stem structure by directly or indirectly downregulating the NAC and CEP genes involved in programmed cell death (PCD) and may also affect responses to oxidative stress, metal ion transport, cell wall modification, and cation transport.
According to a previous study, maize DOF1 was involved in regulating leaf development in a light-dependent manner (Yanagisawa and Sheen, 1998). As an important component of the plant foliar epidermis, guard cells control the opening or closing of stomata. The DOF transcription factor SCAP1 (STOMATAL CARPENTER 1) was shown to play a major role in the critical period of stomatal guard cell differentiation and directly contribute to the maturation of guard cells in Arabidopsis (Negi et al., 2013). Subsequent results indicated that SCAP1 overexpression increased the stomatal density and index in Arabidopsis (Castorina et al., 2016). The functions of DOF members in the development of leaf veins have also attracted much attention. Arabidopsis VDOF1 (VASCULAR-RELATED DOF1) and VDOF2 (VASCULAR-RELATED DOF2) may inhibit cotyledon vein formation and lignin deposition by regulating brassinosteroid (BR) signaling and lignin-related gene transcription in inflorescence stems (Ramachandran et al., 2020). Overexpression of AtDOF5.8 has been found to inhibit the formation of higher-order veins in cotyledons and leaves (Konishi and Yanagisawa, 2015). Using single-cell sequencing, Liu et al. (2022) identified cell types in 10 cell clusters. They elucidated the potential functions of AtCDF5 and the DELLA protein RGA in early cotyledon vein development through the TF network. Furthermore, by obtaining the cdf5 mutant and analyzing the DAP-seq results, the authors inferred that CDF5 might control the expression of the downstream target genes BZIP9, SWEET11, SWEET12, and SULTR2;1 to regulate leaf vein function. Overexpression of AtCDF4 has been reported to impair the vein development of 3-day-old cotyledons, indicating that CDF4 is also involved in regulating cotyledon vein development (Liu et al., 2022). Additionally, DOFs have also been shown to participate in JA-induced leaf senescence in monocotyledons and dicotyledons. For example, in rice, OsDOF24 was found to delay leaf senescence by suppressing the activity of the OsAOS gene related to JA biosynthesis (Shim et al., 2019). However, Arabidopsis AtDOF2.1 has been reported to play an active role in JA-induced leaf senescence through the MYC2-DOF2.1-MYC2 feed-forward transcription loop (Zhuo et al., 2020).
Auxin can control the expression of AtDOF2.1, AtDOF4.6, AtDOF5.3, and AtDOF5.8 in Arabidopsis, which are crucial for vascular development (Gardiner et al., 2010; Konishi et al., 2015). Interestingly, both auxin and other hormones regulate DOFs. The expression of the AtDOF2.4/PEAR1 (PHLOEM EARLY DOF1), AtDOF5.1/PEAR2 (PHLOEM EARLY DOF2), AtDOF3.2/DOF6, AtDOF5.3/TMO6, AtDOF1.1/OBP2 (OBF BINDING PROTEIN2) and AtDOF5.6/HCA2 (HIGH CAMBIAL ACTIVITY2) genes was reported to be regulated by auxin and cytokinin in the sieve tube of the primary phloem (Miyashima et al., 2019; Smetana et al., 2019). Downregulation of tomato SlDOF10 triggered parthenocarpic development while suppressing vascular tissue development (Rojas-Gracia et al., 2019). MdDOF15, MdDOF18, and MdDOF48 were significantly upregulated in apple during pollen tube growth and development (Yang et al., 2018b). Moreover, DOF members also play vital roles in regulating floral organ development. LOW (LOVE ON WINGS) is a legume DOF gene that controls floral organ differentiation by regulating the floral vasculature pattern and petal asymmetry of mung beans (Guo et al., 2019).
Floral transition is one of the most important biological processes, ensuring reproductive success by integrating internal and external signals in plants. Overexpression of the oilseed rape BnCDF1 gene in Arabidopsis reduced the expression of the signal integration factors CO (CONSTANS) and FT (FLOWERING LOCUS T) in the photoperiod flowering pathway, thus delaying the flowering time (Xu and Dai, 2016). Similarly, overexpression of pear PbDOF9.2 in Arabidopsis also resulted in a delayed flowering time. PbDOF9.2 increased the activity of the PbTFL1a and PbTFL1b promoters by inhibiting FT expression and inhibiting flowering (Liu et al., 2020b). Overexpression of Medicago MtCDFd1-1 (Zhang et al., 2019) and tomato SlCDF3 (Xu et al., 2021) also resulted in delayed flowering. In contrast, overexpression of moso bamboo PheDOF12-1 caused Arabidopsis to flower earlier under long-day (LD) conditions. PheDOF12-1 regulated the flowering time by combining with the downstream PheCOL4 promoter (Liu et al., 2019). Similarly, OsDOF4 also promoted rice flowering under LD conditions (Wu et al., 2017a). In contrast, studies in pea (Pisum sativum) revealed that a CDF homolog LATE2 (LATE BLOOMER2), which functions downstream of light signaling and the biological clock, repressed the expression of FTb2, an FT gene regulated by the pea photoperiod, ultimately caused a late-flowering phenotype (Ridge et al., 2016). TaDOF5, TaDOF16, and TaDOF19 were found to be homologous to Arabidopsis CDF in wheat, and diurnal changes in their expression during the light-dark cycle were similar to the pattern of CDF, presumably indicating that these genes have potential roles in the photoperiodic regulation of flowering (Shaw et al., 2009). AtDOF4.7 caused flower organ abscission defects by inhibiting the expression of cell wall hydrolase genes (Wei et al., 2010). Furthermore, AtDOF4.7 might be affected by ethylene and short peptide IDA (INFLORESCENCE DEFICIENT IN ABSCISSION) to negatively regulate floral organ abscission (Wang et al., 2016b). Conversely, AtCDF4 was reported to contribute to floral organ abscission in Arabidopsis (Xu et al., 2020).
Exogenous GA3-induced expression of DOFs has been proven to be a key factor in the initiation of tomato fruit formation (Rojas-Gracia et al., 2019). Furthermore, overexpression of SlCDF4 induced high expression of GA20ox and GA3ox and increased tomato fruit size by regulating endogenous GA4 biosynthesis (Renau-Morata et al., 2020). The high expression of DzDOF2.2 in durian increased the level of auxin and upregulated ethylene biosynthesis through the transcriptional activation of the ACC synthase gene, thus initiating auxin-ethylene crosstalk in advance and ultimately promoting early fruit ripening (Khaksar et al., 2019). Similarly, banana MaDOF23 interacted with MaERF9 to regulate fruit ripening (Feng et al., 2016). Although ethylene induced MaDOF23 expression, MaDOF23 in turn inhibited the expression of maturity-related genes. Based on these findings, MaDOF23 acted as a repressor to fine-tune ethylene biosynthesis during fruit ripening, possibly by balancing the induction of transcriptional activators such as MaERF9. Another study showed that the downregulation of StCDF1 affected potato propagation and the internal water balance by activating downstream StFLORE expression (Ramírez et al., 2021). In addition, overexpression of AtDOF4.2 or its homologous gene AtDOF4.4 increased the seed number and silique size (Zou et al., 2013). DASH (DOF Acting in Seed embryogenesis and Hormone accumulation) is a DOF member unique to the endosperm. Auxin transport in the dash mutant is impaired, resulting in embryonic defects and affecting seed size in Medicago truncatula (Noguero et al., 2015). It can be seen that some DOFs can also affect fruit size, quantity, and quality to a certain extent, and DOF functions must be clarified to improve crop yields and promote the development of the fruit and vegetable industry.
Several results suggest that DOF TFs respond to biotic stresses by improving the ability of plants to defend against pathogens. Transient expression of the DOF genes BBF2 and BBF3 in tobacco increased plant resistance to pathogens (Sasaki et al., 2015). Overexpression of grape VvDOF3 can improve plant resistance to the powdery mildew pathogen Golovinomyces cichoracearum (Yu et al., 2019). However, more studies have suggested that DOF proteins resist various abiotic stresses in plants (Figure 2). ThDOF1.4 improved tolerance to salt and osmotic stress by increasing the proline level and improving the ROS scavenging capability of Tamarix hispida (Zang et al., 2017). Likewise, ThDOF14 can bind specifically to the DOF motif in the downstream TheIF1A promoter and may participate in plant salt stress and osmotic stress responses by regulating or interacting with TheIF1A (Yang et al., 2017). Tomato SlDOF22 affected ascorbic acid (AsA) accumulation and improved salt tolerance in plants (Cai et al., 2016). The wheat DOF protein TaZNF increased Na+ excretion by controlling the expression of many downstream genes and ultimately significantly improved salt tolerance (Ma et al., 2016). Arabidopsis AtDOF5.8 regulated the expression of ANAC069 and played a role in the salt signaling pathway (He et al., 2015). Similarly, watermelon ClDOF29 was the main regulator of the salt stress response (Zhou et al., 2020). In banana, many MaDOF genes were affected by salt and drought stresses, resulting in downregulated expression levels (Dong et al., 2016). In contrast, the alfalfa MsDOF10, MsDOF35, and MsDOF39 genes were significantly upregulated under the same drought and salt stress conditions (Cao et al., 2020). As a DOF transcription factor, CDF has been reported to be widely involved in the responses to various abiotic stresses in plants. The mutant cdf3-1 gene rendered Arabidopsis sensitive to drought and cold stresses, while overexpression of this gene unexpectedly increased plant resistance to osmotic stress (Corrales et al., 2017). Likewise, overexpression of tomato SlCDF1 or SlCDF3 also improved the salt tolerance and drought resistance of plants (Corrales et al., 2014). Moreover, DOF family genes also play prominent roles in improving the drought resistance of woody plants. Overexpression of apple MdDOF54 resulted in a higher photosynthetic rate and stronger branch water transport capacity under long-term drought conditions than the wild-type lines but substantially increased the survival rate under short-term drought conditions (Chen et al., 2020). Undoubtedly, MdDOF54 could improve the drought tolerance of plants. Similarly, overexpression of the birch BpDOF4, BpDOF11, and BpDOF17 genes improved drought tolerance by enhancing the ROS scavenging ability (Sun et al., 2021). Notably, DOF proteins can also improve plant survivability in extreme temperature environments. In Brassica, BnCDF1 expression was induced at low temperatures, while overexpression of BnCDF1 improved plant cold tolerance (Xu and Dai, 2016). Similarly, overexpression of cotton GhDOF1 (Su et al., 2017), grape VaDOF17d (Wang et al., 2021), and rice OsDOF1 (Liu et al., 2021) significantly enhanced plant cold tolerance. One study has shown that walnut JrDOF3 directly regulates the transcription of its target gene JrGRAS2, effectively regulates the expression of heat shock protein genes (HSPs), and ultimately improves heat tolerance (Yang et al., 2018a). In Chinese cabbage (Ma et al., 2015), chrysanthemum (Song et al., 2016), and spinach (Yu et al., 2021a), the expression of DOFs has also been found to be regulated by salt stress, cold stress, and heat stress.
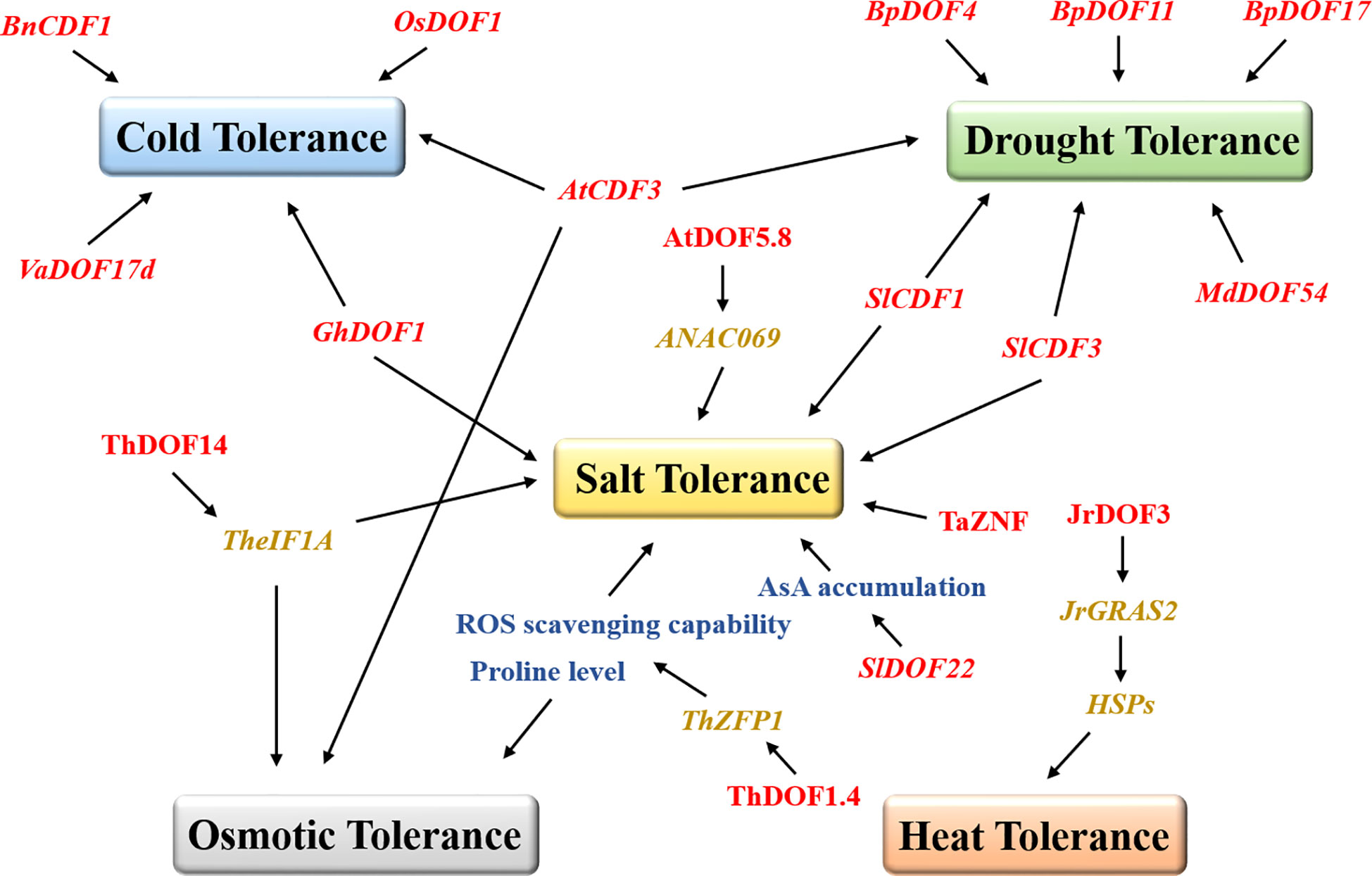
Figure 2 DOFs promote plant tolerance under different abiotic stresses. The DOF family members are represented in red, and the box marks resistance to different stresses. The blue, green, yellow, grey, and orange boxes represent cold tolerance, drought tolerance, salt tolerance, osmotic tolerance, and heat tolerance, respectively. The black arrows indicate facilitated relationships. AsA, ascorbic acid.
In addition, some DOF proteins are widely involved in plant stress responses by responding to some plant hormone signals, such as ABA and SA (Figure 3). Many RcDOFs in castor were expressed at two different levels in response to ABA treatment (Jin et al., 2014). Similarly, researchers have also proposed that chrysanthemum CmDOFs may be involved in the response to ABA and SA, thus resulting in different expression patterns, in which CmDOF12 and CmDOF20 are significantly upregulated by exogenous ABA, while the expression levels of CmDOF2, CmDOF5, CmDOF6, CmDOF10, and CmDOF12 are upregulated by SA (Song et al., 2016). CmDOF12 is a homolog of Arabidopsis OBP3 (AtDOF3.6), consistent with the previous result that Arabidopsis OBP3 expression was upregulated by SA induction (Kang and Singh, 2000). DOF proteins may improve the submergence tolerance of rice at the early stage of germination by regulating GA and other plant hormones (Mohanty, 2021). OsDOF15 contributes to the inhibition of rice primary root elongation under salt stress by mediating ethylene biosynthesis (Qin et al., 2019). Clearly, DOF family genes play vital roles in plant abiotic stresses, and many members are involved in regulating multiple stress pathways simultaneously. An in-depth exploration of the DOF regulatory network will help to reveal the mechanism of the plant abiotic stress response.
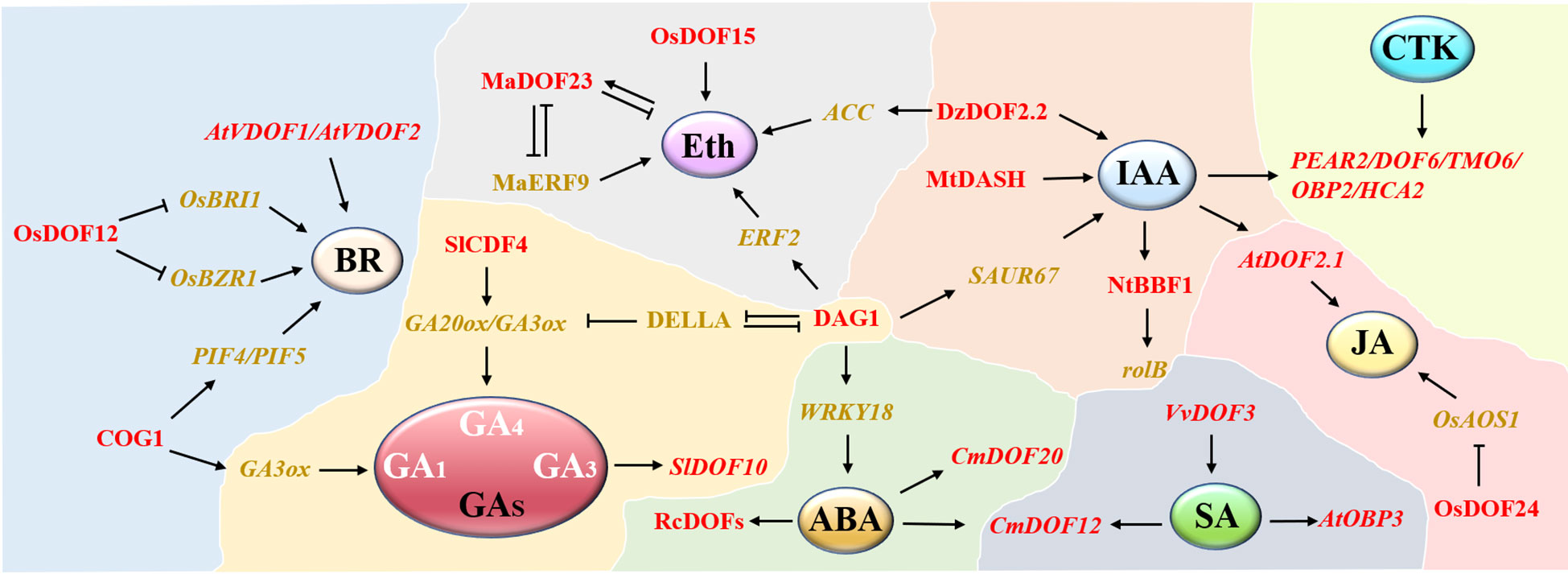
Figure 3 A network model of DOF members participating in different phytohormone pathways. A total of 8 hormone modules are distributed. DOF members are shown in red. Positive and negative regulatory relationships are represented by black arrows and blunted lines, respectively.
Plant metabolic regulation includes primary metabolic processes that supply the immediate needs for plant growth and secondary metabolic processes that regulate the interaction between plants and the environment. DOF members have been reported to be involved in plant metabolic regulatory networks (Figure 4). The metabolism of non-structural carbohydrates in plants affects growth, development, and responses to environmental factors to a great extent. Overexpression of the DOF gene SRF1 in sweet potato promoted the formation of dry matter in storage roots, led to the decreases in the glucose and fructose contents, and ultimately regulated carbohydrate metabolism by inhibiting the expression of the vacuolar invertase gene (Tanaka et al., 2009). Likewise, overexpression of PpDOF5 in transgenic hybrid poplars significantly increased carbohydrate accumulation (Rueda-López et al., 2017). A study of maize found that ZmDOF3 knockdown inhibited the expression of genes involved in starch synthesis (Qi et al., 2017). Similarly, overexpression of ZmDOF36 decreased the contents of reduced sugars and soluble sugars in maize seed endosperm, and the presence of higher soluble sugar levels led to starch synthesis. ZmDOF36 positively regulated the expression of different starch synthesis-related genes, such as ZmAGPS1a, ZmAGPL1, ZmISA1, ZmISA3, ZmGBSSI, and ZmSSIIa, by binding to specific motifs in downstream promoters (Wu et al., 2019). In contrast, kiwifruit (Actinidia deliciosa) AdDOF3 affected fruit flavor by transactivating the starch degradation gene AdBAM3 L upon binding to downstream AAAG/CTTT elements (Zhang et al., 2018b). Furthermore, studies in Arabidopsis showed that AtDOF4.6/VDOF1 and AtDOF1.8/VDOF2 modulate secondary cell wall (SCW) properties related to glycation efficiency (Ohtani et al., 2017). Rice OsDOF11 promoted sucrose transport by directly regulating the transcription of the downstream sucrose transport-related genes OsSUT1, OsSWEET11, and OsSWEET14 (Wu et al., 2018). In addition, the only DOF protein present in C. reinhardtii was involved in regulating fatty acid metabolism (Ibáñez-Salazar et al., 2014). The results reported by Jia et al. (2019) confirmed that overexpression of Chlamydomonas CrDOF could regulate the expression of crucial genes related to lipid metabolism and promote intracellular lipid accumulation. Overexpression of soybean GmDOF4 and GmDOF11 increased the seed lipid content (Wang et al., 2007). Similarly, overexpression of GmDOF4 in Chlorella ellipsoidea increased the cellular lipid content by approximately 50% (Zhang et al., 2014). These results provide new methods for the future development of the food processing and biofuel industries.
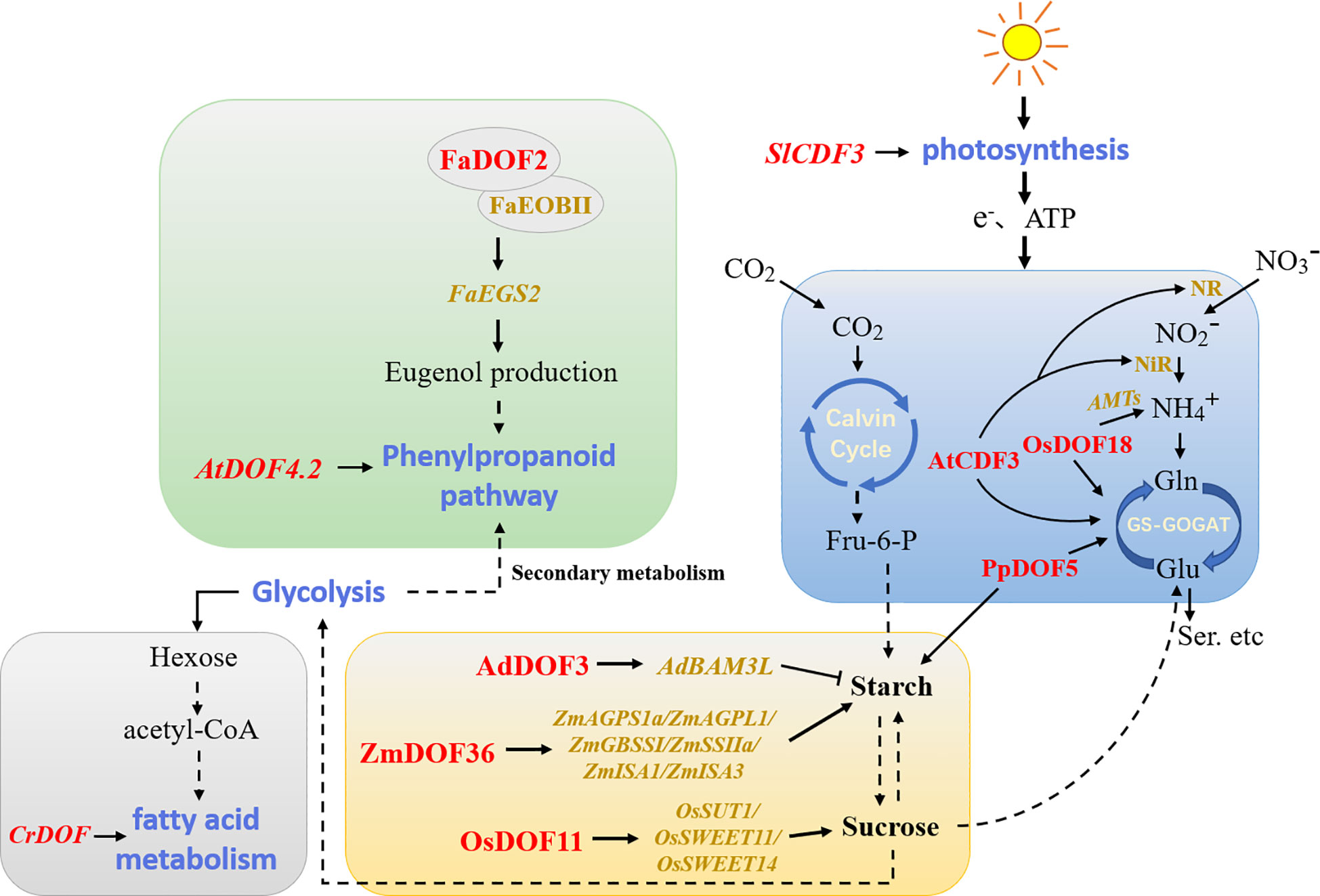
Figure 4 Model of DOFs involved in the regulation of plant metabolism. The blue, yellow, grey, and green modules represent carbon and nitrogen assimilation, carbohydrate metabolism, fatty acid metabolism, and phenylpropanoid metabolism, respectively. Red indicates DOF members in different plants. Dotted lines indicate that some processes have been omitted. NR/NiR, nitrate/nitrite reductases; AMTs, ammonium transporters; GS, glutamine synthetase; GOGAT, glutamate synthase; Fru-6-P, fructose-6-phosphate.
The antagonistic relationship between carbon and nitrogen also regulates various developmental processes in plants. A recent study has shown that OsDOF11 regulates nitrogen metabolism by affecting sugar transport and distribution, thereby coordinating the balance of carbon and nitrogen to maintain plant life processes (Huang et al., 2021). OsDOF18 was reported to regulate ammonium transport by inducing the expression of ammonium transporter genes, thus affecting the efficiency of nitrogen assimilation (Wu et al., 2017b). Maritime pine (Pinus pinaster) PpDOF5 was also crucial in controlling conifer ammonium assimilation for glutamine production (Rueda-López et al., 2008). Arabidopsis AtCDF3 promoted root development by regulating the expression of genes related to carbon and nitrogen assimilation (Domínguez-Figueroa et al., 2020). To some extent, improving the nitrogen use efficiency or photosynthetic rate of plants is the key factor in increasing crop yields. Overexpression of SlCDF3 in tomato increased the photosynthetic rate, which also resulted in higher sucrose utilization and changes in plant primary metabolism while enhancing nitrogen assimilation, promoting an increase in biomass, and ultimately increasing yields (Renau-Morata et al., 2017). Moreover, some DOF members are also involved in secondary plant metabolism. Arabidopsis AtDOF4.2 regulated phenylpropane metabolism in different tissues and under various stresses (Skirycz et al., 2007). FaDOF2 regulated the volatile phenylpropanoid pathway in ripe strawberry (Fragaria × ananassa) receptacles by controlling the expression of the FaEGS2 and FaEOBII genes, which are involved in eugenol synthesis (Molina-Hidalgo et al., 2017). This regulation of volatile organic compounds may become a breakthrough in analyzing the mechanism of plant aroma formation.
The large number of DOF functional studies summarized here indicates that many unanswered questions about DOF TFs still remain. Although the functional diversity of DOF has attracted increasing attention in recent years, research on the molecular mechanisms by which these proteins regulate certain biological processes, such as plant dormancy and germination, has been limited to some model plants and thus further studies are needed. Some DOFs coordinate internal factors such as phytochromes and plant hormones to affect plant growth and development upon light induction, which also provides an important reference for the analysis of light signaling pathways in non-model plants. Accumulating evidence suggests that DOFs respond to a variety of plant hormone signals and participate in regulating plant growth and development, stress response, metabolic senescence, and other life stages. However, further studies are needed to explore the roles of DOFs in some phytohormone signaling pathways and determine which members are central to integrating the crosstalk between different hormones. Based on species specificity, substantial differences in bioactive GA types regulated by various DOFs exist, which provides a new direction for traditional gibberellin pathway research. The establishment of a more comprehensive DOF-GA regulatory network is an urgent need.
To date, despite our keen interest in DOF genes, only half of the DOF members have been characterized in model plants, and thus, our knowledge of DOF transcription factors is limited. Although some variation in the number of DOFs has been observed in different species, the potential functional redundancy in these DOF family genes still must be further confirmed by obtaining multiple gene knockout lines using CRISPR and other techniques. Some DOF members may become potential factors to improve crop yields in the future and have quite wide application prospects for the development of the food processing and biofuel industries. Accordingly, in-depth research on DOF TFs must be conducted in many fields to reveal their possible novel functions that will help us understand the intricate life processes of plants.
XZ and HS conceived the structure and content of the article, and finished the manuscript. All authors contributed to the article and approved the submitted version.
This work was financed by LiaoNing Revitalization Talents Program (XLYC2002052), Shenyang Innovation Program of Seed Industry (21-110-3-12), the earmarked fund for CARS (CARS-23), and the National Key R&D Program of China (2018YFD1000407).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Baumann, K., De Paolis, A., Costantino, P., Gualberti, G. (1999). The DNA binding site of the dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell. 11 (3), 323–334. doi: 10.1105/tpc.11.3.323
Boccaccini, A., Lorrai, R., Ruta, V., Frey, A., Mercey-Boutet, S., Marion-Poll, A., et al. (2016). The DAG1 transcription factor negatively regulates the seed-to-seedling transition in Arabidopsis acting on ABA and GA levels. BMC Plant Biol. 16 (1), 198. doi: 10.1186/s12870-016-0890-5
Boccaccini, A., Santopolo, S., Capauto, D., Lorrai, R., Minutello, E., Serino, G., et al. (2014). The DOF protein DAG1 and the DELLA protein GAI cooperate in negatively regulating the AtGA3ox1 gene. Mol. Plant 7 (9), 1486–1489. doi: 10.1093/mp/ssu046
Bueso, E., Muñoz-Bertomeu, J., Campos, F., Martínez, C., Tello, C., Martínez-Almonacid, I., et al. (2016). Arabidopsis COGWHEEL1 links light perception and gibberellins with seed tolerance to deterioration. Plant J. 87 (6), 583–596. doi: 10.1111/tpj.13220
Cai, X., Zhang, C., Shu, W., Ye, Z., Li, H., Zhang, Y. (2016). The transcription factor SlDof22 involved in ascorbate accumulation and salinity stress in tomato. Biochem. Biophys. Res. Commun. 474 (4), 736–741. doi: 10.1016/j.bbrc.2016.04.148
Cai, X., Zhang, Y., Zhang, C., Zhang, T., Hu, T., Ye, J., et al. (2013). Genome-wide analysis of plant-specific dof transcription factor family in tomato. J. Integr. Plant Biol. 55 (6), 552–566. doi: 10.1111/jipb.12043
Cao, B., Cui, Y., Lou, K., Luo, D., Liu, Z., Zhou, Q. (2020). Genome-wide identification and expression analysis of the Dof gene family in Medicago sativa l. under various abiotic stresses. DNA Cell Biol. 39 (11), 1976–1989. doi: 10.1089/dna.2020.5652
Castorina, G., Fox, S., Tonelli, C., Galbiati, M., Conti, L. (2016). A novel role for STOMATAL CARPENTER 1 in stomata patterning. BMC Plant Biol. 16 (1), 172. doi: 10.1186/s12870-016-0851-z
Cavalar, M., Möller, C., Offermann, S., Krohn, N. M., Grasser, K. D., Peterhänsel, C. (2003). The interaction of DOF transcription factors with nucleosomes depends on the positioning of the binding site and is facilitated by maize HMGB5. Biochemistry. 42 (7), 2149–2157. doi: 10.1021/bi026761r
Chattha, W. S., Atif, R. M., Iqbal, M., Shafqat, W., Farooq, M. A., Shakeel, A. (2020). Genome-wide identification and evolution of dof transcription factor family in cultivated and ancestral cotton species. Genomics. 112 (6), 4155–4170. doi: 10.1016/j.ygeno.2020.07.006
Chen, M., Liu, X., Huan, L., Sun, M. Y., Liu, L., Chen, X. D., et al. (2017). Genome-wide analysis of dof family genes and their expression during bud dormancy in peach (Prunus persica). Sci. Hortic. 214, 18–26. doi: 10.1016/j.scienta.2016.11.014
Chen, P., Yan, M., Li, L., He, J., Zhou, S., Li, Z., et al. (2020). The apple DNA-binding one zinc-finger protein MdDof54 promotes drought resistance. Hortic. Res. 7 (1), 195. doi: 10.1038/s41438-020-00419-5
Corrales, A. R., Carrillo, L., Lasierra, P., Nebauer, S. G., Dominguez-Figueroa, J., Renau-Morata, B., et al. (2017). Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 40 (5), 748–764. doi: 10.1111/pce.12894
Corrales, A. R., Nebauer, S. G., Carrillo, L., Fernández-Nohales, P., Marqués, J., Renau-Morata, B., et al. (2014). Characterization of tomato cycling dof factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 65 (4), 995–1012. doi: 10.1093/jxb/ert451
da Silva, D. C., da Silveira Falavigna, V., Fasoli, M., Buffon, V., Porto, D. D., Pappas, G. J., Jr., et al. (2016). Transcriptome analyses of the Dof-like gene family in grapevine reveal its involvement in berry, flower and seed development. Hortic. Res. 3, 16042. doi: 10.1038/hortres.2016.42
Diaz, I., Martinez, M., Isabel-LaMoneda, I., Rubio-Somoza, I., Carbonero, P. (2005). The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant J. 42 (5), 652–662. doi: 10.1111/j.1365-313X.2005.02402.x
Diaz, I., Vicente-Carbajosa, J., Abraham, Z., Martínez, M., Isabel-La Moneda, I., Carbonero, P. (2002). The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J. 29 (4), 453–464. doi: 10.1046/j.0960-7412.2001.01230.x
Domínguez-Figueroa, J., Carrillo, L., Renau-Morata, B., Yang, L., Molina, R. V., Marino, D., et al. (2020). The Arabidopsis transcription factor CDF3 is involved in nitrogen responses and improves nitrogen use efficiency in tomato. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.601558
Dong, C., Hu, H., Xie, J. (2016). Genome-wide analysis of the DNA-binding with one zinc finger (Dof) transcription factor family in bananas. Genome. 59 (12), 1085–1100. doi: 10.1139/gen-2016-0081
Feng, B. H., Han, Y. C., Xiao, Y. Y., Kuang, J. F., Fan, Z. Q., Chen, J. Y., et al. (2016). The banana fruit dof transcription factor MaDof23 acts as a repressor and interacts with MaERF9 in regulating ripening-related genes. J. Exp. Bot. 67 (8), 2263–2275. doi: 10.1093/jxb/erw032
Gabriele, S., Rizza, A., Martone, J., Circelli, P., Costantino, P., Vittorioso, P. (2010). The dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 61 (2), 312–323. doi: 10.1111/j.1365-313X.2009.04055.x
Gao, H., Song, W., Severing, E., Vayssières, A., Huettel, B., Franzen, R., et al. (2022). PIF4 enhances DNA binding of CDF2 to co-regulate target gene expression and promote arabidopsis hypocotyl cell elongation. Nat. Plants. 8 (9), 1082–1093. doi: 10.1038/s41477-022-01213-y
Gardiner, J., Sherr, I., Scarpella, E. (2010). Expression of DOF genes identifies early stages of vascular development in Arabidopsis leaves. Int. J. Dev. Biol. 54 (8-9), 1389–1396. doi: 10.1387/ijdb.093006jg
Guo, W., Zhang, X., Peng, Q., Luo, D., Jiao, K., Su, S. (2019). Love on wings, a dof family protein regulates floral vasculature in Vigna radiata. BMC Plant Biol. 19 (1), 495. doi: 10.1186/s12870-019-2099-x
Gupta, S., Kushwaha, H., Singh, V. K., Bisht, N. C., Sarangi, B. K., Yadav, D. (2014). Genome wide in silico characterization of dof transcription factor gene family of sugarcane and its comparative phylogenetic analysis with Arabidopsis, rice and sorghum. Sugar Tech. 16, 372–384. doi: 10.1007/s12355-013-0288-8
Hedden, P., Thomas, S. G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444 (1), 11–25. doi: 10.1042/BJ20120245
Hernando-Amado, S., González-Calle, V., Carbonero, P., Barrero-Sicilia, C. (2012). The family of DOF transcription factors in Brachypodium distachyon: phylogenetic comparison with rice and barley DOFs and expression profiling. BMC Plant Biol. 12, 202. doi: 10.1186/1471-2229-12-202
He, L., Su, C., Wang, Y., Wei, Z. (2015). ATDOF5.8 protein is the upstream regulator of ANAC069 and is responsive to abiotic stress. Biochimie. 110, 17–24. doi: 10.1016/j.biochi.2014.12.017
Huang, Y., Han, Z., Cheng, N., Luo, M., Bai, X., Xing, Y. (2020). Minor effects of 11 Dof family genes contribute to the missing heritability of heading date in rice (Oryza sativa l.). Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01739
Huang, W., Huang, Y., Li, M. Y., Wang, F., Xu, Z. S., Xiong, A. S. (2016). Dof transcription factors in carrot: genome-wide analysis and their response to abiotic stress. Biotechnol. Lett. 38 (1), 145–155. doi: 10.1007/s10529-015-1966-2
Huang, X., Zhang, Y., Wang, L., Dong, X., Hu, W., Jiang, M., et al. (2021). OsDOF11 affects nitrogen metabolism by sucrose transport signaling in rice (Oryza sativa l.). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.703034
Ibáñez-Salazar, A., Rosales-Mendoza, S., Rocha-Uribe, A., Ramírez-Alonso, J. I., Lara-Hernández, I., Hernández-Torres, A., et al. (2014). Over-expression of dof-type transcription factor increases lipid production in Chlamydomonas reinhardtii. J. Biotechnol. 184, 27–38. doi: 10.1016/j.jbiotec.2014.05.003
Isabel-LaMoneda, I., Diaz, I., Martinez, M., Mena, M., Carbonero, P. (2003). SAD: a new DOF protein from barley that activates transcription of a cathepsin b-like thiol protease gene in the aleurone of germinating seeds. Plant J. 33 (2), 329–340. doi: 10.1046/j.1365-313x.2003.01628.x
Jiang, Y., Zeng, B., Zhao, H., Zhang, M., Xie, S., Lai, J. (2012). Genome-wide transcription factor gene prediction and their expressional tissue-specificities in maize. J. Integr. Plant Biol. 54 (9), 616–630. doi: 10.1111/j.1744-7909.2012.01149.x
Jia, B., Xie, X., Wu, M., Lin, Z., Yin, J., Lou, S., et al. (2019). Understanding the functions of endogenous DOF transcript factor in Chlamydomonas reinhardtii. Biotechnol. Biofuels. 12, 67. doi: 10.1186/s13068-019-1403-1
Jin, Z., Chandrasekaran, U., Liu, A. (2014). Genome-wide analysis of the dof transcription factors in castor bean (Ricinus communis l.). Genes Genomics 36 (4), 527–537. doi: 10.1007/s13258-014-0189-6
Kang, H. G., Singh, K. B. (2000). Characterization of salicylic acid-responsive, Arabidopsis dof domain proteins: overexpression of OBP3 leads to growth defects. Plant J. 21 (4), 329–339. doi: 10.1046/j.1365-313x.2000.00678.x
Khaksar, G., Sangchay, W., Pinsorn, P., Sangpong, L., Sirikantaramas, S. (2019). Genome-wide analysis of the Dof gene family in durian reveals fruit ripening-associated and cultivar-dependent dof transcription factors. Sci. Rep. 9 (1), 12109. doi: 10.1038/s41598-019-48601-7
Kim, H. S., Kim, S. J., Abbasi, N., Bressan, R. A., Yun, D. J., Yoo, S. D., et al. (2010). The DOF transcription factor Dof5.1 influences leaf axial patterning by promoting Revoluta transcription in Arabidopsis. Plant J. 64 (3), 524–535. doi: 10.1111/j.1365-313X.2010.04346.x
Kisu, Y., Ono, T., Shimofurutani, N., Suzuki, M., Esaka, M. (1998). Characterization and expression of a new class of zinc finger protein that binds to silencer region of ascorbate oxidase gene. Plant Cell Physiol. 39 (10), 1054–1064. doi: 10.1093/oxfordjournals.pcp.a029302
Kondhare, K. R., Vetal, P. V., Kalsi, H. S., Banerjee, A. K. (2019). BEL1-like protein (StBEL5) regulates CYCLING DOF FACTOR1 (StCDF1) through tandem TGAC core motifs in potato. J. Plant Physiol. 241, 153014. doi: 10.1016/j.jplph.2019.153014
Konishi, M., Donner, T. J., Scarpella, E., Yanagisawa, S. (2015). MONOPTEROS directly activates the auxin-inducible promoter of the Dof5.8 transcription factor gene in Arabidopsis thaliana leaf provascular cells. J. Exp. Bot. 66 (1), 283–291. doi: 10.1093/jxb/eru418
Konishi, M., Yanagisawa, S. (2015). Transcriptional repression caused by Dof5.8 is involved in proper vein network formation in Arabidopsis thaliana leaves. J. Plant Res. 128 (4), 643–652. doi: 10.1007/s10265-015-0712-0
Krebs, J., Mueller-Roeber, B., Ruzicic, S. (2010). A novel bipartite nuclear localization signal with an atypically long linker in DOF transcription factors. J. Plant Physiol. 167 (7), 583–586. doi: 10.1016/j.jplph.2009.11.016
Kushwaha, H., Gupta, S., Singh, V. K., Rastogi, S., Yadav, D. (2011). Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and arabidopsis. Mol. Biol. Rep. 38 (8), 5037–5053. doi: 10.1007/s11033-010-0650-9
Lesur, I., Bechade, A., Lalanne, C., Klopp, C., Noirot, C., Leplé, J. C., et al. (2015). A unigene set for european beech (Fagus sylvatica l.) and its use to decipher the molecular mechanisms involved in dormancy regulation. Mol. Ecol. Resour. 15, 1192–1204. doi: 10.1111/1755-0998.12373
Lijavetzky, D., Carbonero, P., Vicente-Carbajosa, J. (2003). Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis dof gene families. BMC Evol. Biol. 3, 17. doi: 10.1186/1471-2148-3-17
Liu, J., Cheng, Z. C., Xie, L. H., Li, X. Y., Gao, J. (2019). Multifaceted role of PheDof12-1 in the regulation of flowering time and abiotic stress responses in moso bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 20 (2), 424. doi: 10.3390/ijms20020424
Liu, Y., Liu, N., Deng, X., Liu, D., Li, M., Cui, D., et al. (2020a). Genome-wide analysis of wheat DNA-binding with one finger (Dof) transcription factor genes: evolutionary characteristics and diverse abiotic stress responses. BMC Genomics 21 (1), 276. doi: 10.1186/s12864-020-6691-0
Liu, X., Liu, Z., Hao, Z., Chen, G., Qi, K., Zhang, H., et al. (2020b). Characterization of dof family in Pyrus bretschneideri and role of PbDof9.2 in flowering time regulation. Genomics. 112 (1), 712–720. doi: 10.1016/j.ygeno.2019.05.005
Liu, J., Meng, Q., Xiang, H., Shi, F., Ma, L., Li, Y., et al. (2021). Genome-wide analysis of Dof transcription factors and their response to cold stress in rice (Oryza sativa l.). BMC Genomics 22 (1), 800. doi: 10.1186/s12864-021-08104-0
Liu, Z., Wang, J., Zhou, Y., Zhang, Y., Qin, A., Yu, X., et al. (2022). Identification of novel regulators required for early development of vein pattern in the cotyledons by single-cell RNA-sequencing. Plant J. 110 (1), 7–22. doi: 10.1111/tpj.15719
Lorrai, R., Gandolfi, F., Boccaccini, A., Ruta, V., Possenti, M., Tramontano, A., et al. (2018). Genome-wide RNA-seq analysis indicates that the DAG1 transcription factor promotes hypocotyl elongation acting on ABA, ethylene and auxin signaling. Sci. Rep. 8 (1), 15895. doi: 10.1038/s41598-018-34256-3
Lucas-Reina, E., Romero-Campero, F. J., Romero, J. M., Valverde, F. (2015). An evolutionarily conserved DOF-CONSTANS module controls plant photoperiodic signaling. Plant Physiol. 168 (2), 561–574. doi: 10.1104/pp.15.00321
Ma, X., Liang, W., Gu, P., Huang, Z. (2016). Salt tolerance function of the novel C2H2-type zinc finger protein TaZNF in wheat. Plant Physiol. Biochem. 106, 129–140. doi: 10.1016/j.plaphy.2016.04.033
Ma, J., Li, M. Y., Wang, F., Tang, J., Xiong, A. S. (2015). Genome-wide analysis of dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics 16 (1), 33. doi: 10.1186/s12864-015-1242-9
Martín, G., Veciana, N., Boix, M., Rovira, A., Henriques, R., Monte, E. (2020). The photoperiodic response of hypocotyl elongation involves regulation of CDF1 and CDF5 activity. Physiol. Plant 169 (3), 480–490. doi: 10.1111/ppl.13119
Mena, M., Cejudo, F. J., Isabel-Lamoneda, I., Carbonero, P. (2002). A role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley aleurone. Plant Physiol. 130 (1), 111–119. doi: 10.1104/pp.005561
Miyashima, S., Roszak, P., Sevilem, I., Toyokura, K., Blob, B., Heo, J. O., et al. (2019). Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature. 565 (7740), 490–494. doi: 10.1038/s41586-018-0839-y
Moghaddas Sani, H., Hamzeh-Mivehroud, M., Silva, A. P., Walshe, J. L., Mohammadi, S. A., Rahbar-Shahrouziasl, M., et al. (2018). Expression, purification and DNA-binding properties of zinc finger domains of DOF proteins from Arabidopsis thaliana. Bioimpacts 8 (3), 167–176. doi: 10.15171/bi.2018.19
Mohanty, B. (2021). Promoter architecture and transcriptional regulation of genes upregulated in germination and coleoptile elongation of diverse rice genotypes tolerant to submergence. Front. Genet. 12. doi: 10.3389/fgene.2021.639654
Molina-Hidalgo, F. J., Medina-Puche, L., Cañete-Gómez, C., Franco-Zorrilla, J. M., López-Vidriero, I., Solano, R., et al. (2017). The fruit-specific transcription factor FaDOF2 regulates the production of eugenol in ripe fruit receptacles. J. Exp. Bot. 68 (16), 4529–4543. doi: 10.1093/jxb/erx257
Moreno-Risueno, M. A., Díaz, I., Carrillo, L., Fuentes, R., Carbonero, P. (2007a). The HvDOF19 transcription factor mediates the abscisic acid-dependent repression of hydrolase genes in germinating barley aleurone. Plant J. 51 (3), 352–365. doi: 10.1111/j.1365-313X.2007.03146.x
Moreno-Risueno, M. A., Martínez, M., Vicente-Carbajosa, J., Carbonero, P. (2007b). The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol. Genet. Genom. 277 (4), 379–390. doi: 10.1007/s00438-006-0186-9
Nan, H., Ludlow, R. A., Lu, M., An, H. (2021). Genome-wide analysis of Dof genes and their response to abiotic stress in rose (Rosa chinensis). Front. Genet. 12. doi: 10.3389/fgene.2021.538733
Negi, J., Moriwaki, K., Konishi, M., Yokoyama, R., Nakano, T., Kusumi, K., et al. (2013). A dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr. Biol. 23 (6), 479–484. doi: 10.1016/j.cub.2013.02.001
Nilsen, K. T., Walkowiak, S., Xiang, D., Gao, P., Quilichini, T. D., Willick, I. R., et al. (2020). Copy number variation of TdDof controls solid-stemmed architecture in wheat. Proc. Natl. Acad. Sci. U. S. A. 117 (46), 28708–28718. doi: 10.1073/pnas.2009418117
Noguero, M., Atif, R. M., Ochatt, S., Thompson, R. D. (2013). The role of the DNA-binding one zinc finger (DOF) transcription factor family in plants. Plant Sci. 209, 32–45. doi: 10.1016/j.plantsci.2013.03.016
Noguero, M., Le Signor, C., Vernoud, V., Bandyopadhyay, K., Sanchez, M., Fu, C., et al. (2015). DASH transcription factor impacts Medicago truncatula seed size by its action on embryo morphogenesis and auxin homeostasis. Plant J. 81 (3), 453–466. doi: 10.1111/tpj.12742
Ohtani, M., Ramachandran, V., Tokumoto, T., Takebayashi, A., Ihara, A., Matsumoto, T., et al. (2017). Identification of novel factors that increase enzymatic saccharification efficiency in Arabidopsis wood cells. Plant Biotechnol. (Tokyo). 34 (4), 203–206. doi: 10.5511/plantbiotechnology.17.1107a
Park, D. H., Lim, P. O., Kim, J. S., Cho, D. S., Hong, S. H., Nam, H. G. (2003). The Arabidopsis COG1 gene encodes a dof domain transcription factor and negatively regulates phytochrome signaling. Plant J. 34 (2), 161–171. doi: 10.1046/j.1365-313x.2003.01710.x
Pi, L., Aichinger, E., van der Graaff, E., Llavata-Peris, C. I., Weijers, D., Hennig, L., et al. (2015). Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev. Cell. 33 (5), 576–588. doi: 10.1016/j.devcel.2015.04.024
Qi, X., Li, S., Zhu, Y., Zhao, Q., Zhu, D., Yu, J. (2017). ZmDof3, a maize endosperm-specific dof protein gene, regulates starch accumulation and aleurone development in maize endosperm. Plant Mol. Biol. 93 (1-2), 7–20. doi: 10.1007/s11103-016-0543-y
Qin, H., Wang, J., Chen, X., Wang, F., Peng, P., Zhou, Y., et al. (2019). Rice OsDOF15 contributes to ethylene-inhibited primary root elongation under salt stress. New Phytol. 223 (2), 798–813. doi: 10.1111/nph.15824
Ramachandran, V., Tobimatsu, Y., Masaomi, Y., Sano, R., Umezawa, T., Demura, T., et al. (2020). Plant-specific dof transcription factors VASCULAR-RELATED DOF1 and VASCULAR-RELATED DOF2 regulate vascular cell differentiation and lignin biosynthesis in Arabidopsis. Plant Mol. Biol. 104 (3), 263–281. doi: 10.1007/s11103-020-01040-9
Ramírez, G. L., Shi, L., Bergonzi, S. B., Oortwijn, M., Franco-Zorrilla, J. M., Solano-Tavira, R., et al. (2021). Potato CYCLING DOF FACTOR 1 and its lncRNA counterpart StFLORE link tuber development and drought response. Plant J. 105 (4), 855–869. doi: 10.1111/tpj.15093
Ravindran, P., Verma, V., Stamm, P., Kumar, P. P. (2017). A novel RGL2-DOF6 complex contributes to primary seed dormancy in Arabidopsis thaliana by regulating a GATA transcription factor. Mol. Plant 10 (10), 1307–1320. doi: 10.1016/j.molp.2017.09.004
Renau-Morata, B., Carrillo, L., Cebolla-Cornejo, J., Molina, R. V., Martí, R., Domínguez-Figueroa, J., et al. (2020). The targeted overexpression of SlCDF4 in the fruit enhances tomato size and yield involving gibberellin signalling. Sci. Rep. 10 (1), 10645. doi: 10.1038/s41598-020-67537-x
Renau-Morata, B., Molina, R. V., Carrillo, L., Cebolla-Cornejo, J., Sánchez-Perales, M., Pollmann, S., et al. (2017). Ectopic expression of CDF3 genes in tomato enhances biomass production and yield under salinity stress conditions. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00660
Ridge, S., Sussmilch, F. C., Hecht, V., Vander, Schoor, J. K., Lee, R., Aubert, G., et al. (2016). Identification of LATE BLOOMER2 as a CYCLING DOF FACTOR homolog reveals conserved and divergent features of the flowering response to photoperiod in pea. Plant Cell. 28 (10), 2545–2559. doi: 10.1105/tpc.15.01011
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 290 (5499), 2105–2110. doi: 10.1126/science.290.5499.2105
Rojas-Gracia, P., Roque, E., Medina, M., López-Martín, M. J., Cañas, L. A., Beltrán, J. P., et al. (2019). The DOF transcription factor SlDOF10 regulates vascular tissue formation during ovary development in tomato. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00216
Rueda-López, M., Crespillo, R., Cánovas, F. M., Avila, C. (2008). Differential regulation of two glutamine synthetase genes by a single dof transcription factor. Plant J. 56 (1), 73–85. doi: 10.1111/j.1365-313X.2008.03573.x
Rueda-López, M., Pascual, M. B., Pallero, M., Henao, L. M., Lasa, B., Jauregui, I., et al. (2017). Overexpression of a pine dof transcription factor in hybrid poplars: A comparative study in trees growing under controlled and natural conditions. PloS One 12 (4), e0174748. doi: 10.1371/journal.pone.0174748
Rueda-Romero, P., Barrero-Sicilia, C., Gómez-Cadenas, A., Carbonero, P., Oñate-Sánchez, L. (2012). Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J. Exp. Bot. 63 (5), 1937–1949. doi: 10.1093/jxb/err388
Santopolo, S., Boccaccini, A., Lorrai, R., Ruta, V., Capauto, D., Minutello, E., et al. (2015). DOF AFFECTING GERMINATION 2 is a positive regulator of light-mediated seed germination and is repressed by DOF AFFECTING GERMINATION 1. BMC Plant Biol. 15, 72. doi: 10.1186/s12870-015-0453-1
Sasaki, N., Matsumaru, M., Odaira, S., Nakata, A., Nakata, K., Nakayama, I., et al. (2015). Transient expression of tobacco BBF1-related dof proteins, BBF2 and BBF3, upregulates genes involved in virus resistance and pathogen defense. Physiol. Mol. Plant Pathol. 89, 70–77. doi: 10.1016/j.pmpp.2014.12.005
Shaw, L. M., McIntyre, C. L., Gresshoff, P. M., Xue, G. P. (2009). Members of the dof transcription factor family in Triticum aestivum are associated with light-mediated gene regulation. Funct. Integr. Genomics 9 (4), 485–498. doi: 10.1007/s10142-009-0130-2
Shigyo, M., Tabei, N., Yoneyama, T., Yanagisawa, S. (2007). Evolutionary processes during the formation of the plant-specific dof transcription factor family. Plant Cell Physiol. 48 (1), 179–185. doi: 10.1093/pcp/pcl044
Shim, Y., Kang, K., An, G., Paek, N. C. (2019). Rice DNA-binding one zinc finger 24 (OsDOF24) delays leaf senescence in a jasmonate-mediated pathway. Plant Cell Physiol. 60 (9), 2065–2076. doi: 10.1093/pcp/pcz105
Skirycz, A., Jozefczuk, S., Stobiecki, M., Muth, D., Zanor, M. I., Witt, I., et al. (2007). Transcription factor AtDOF4;2 affects phenylpropanoid metabolism in Arabidopsis thaliana. New Phytol. 175 (3), 425–438. doi: 10.1111/j.1469-8137.2007.02129.x
Smetana, O., Mäkilä, R., Lyu, M., Amiryousefi, A., Sánchez Rodríguez, F., Wu, M. F., et al. (2019). High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature. 565 (7740), 485–489. doi: 10.1038/s41586-018-0837-0
Song, A., Gao, T., Li, P., Chen, S., Guan, Z., Wu, D., et al. (2016). Transcriptome-wide identification and expression profiling of the DOF transcription factor gene family in Chrysanthemum morifolium. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00199
Sugiyama, T., Ishida, T., Tabei, N., Shigyo, M., Konishi, M., Yoneyama, T., et al. (2012). Involvement of PpDof1 transcriptional repressor in the nutrient condition-dependent growth control of protonemal filaments in Physcomitrella patens. J. Exp. Bot. 63 (8), 3185–3197. doi: 10.1093/jxb/ers042
Su, Y., Liang, W., Liu, Z., Wang, Y., Zhao, Y., Ijaz, B., et al. (2017). Overexpression of GhDof1 improved salt and cold tolerance and seed oil content in Gossypium hirsutum. J. Plant Physiol. 218, 222–234. doi: 10.1016/j.jplph.2017.07.017
Sun, S., Wang, B., Jiang, Q., Li, Z., Jia, S., Wang, Y., et al. (2021). Genome-wide analysis of BpDof genes and the tolerance to drought stress in birch (Betula platyphylla). PeerJ. 9, e11938. doi: 10.7717/peerj.11938
Tanaka, M., Takahata, Y., Nakayama, H., Nakatani, M., Tahara, M. (2009). Altered carbohydrate metabolism in the storage roots of sweet potato plants overexpressing the SRF1 gene, which encodes a dof zinc finger transcription factor. Planta. 230 (4), 737–746. doi: 10.1007/s00425-009-0979-2
Umemura, Y., Ishiduka, T., Yamamoto, R., Esaka, M. (2004). The dof domain, a zinc finger DNA-binding domain conserved only in higher plants, truly functions as a Cys2/Cys2 zn finger domain. Plant J. 37 (5), 741–749. doi: 10.1111/j.1365-313x.2003.01997.x
Venkatesh, J., Park, S. W. (2015). Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiol. Biochem. 94, 73–85. doi: 10.1016/j.plaphy.2015.05.010
Wang, P., Li, J., Gao, X., Zhang, D., Li, A., Liu, C. (2018). Genome-wide screening and characterization of the Dof gene family in physic nut (Jatropha curcas l.). Int. J. Mol. Sci. 19 (6), 1598. doi: 10.3390/ijms19061598
Wang, Z., Wang, Y., Tong, Q., Xu, G., Xu, M., Li, H., et al. (2021). Transcriptomic analysis of grapevine Dof transcription factor gene family in response to cold stress and functional analyses of the VaDof17d gene. Planta. 253 (2), 55. doi: 10.1007/s00425-021-03574-8
Wang, G. Q., Wei, P. C., Tan, F., Yu, M., Zhang, X. Y., Chen, Q. J., et al. (2016b). The transcription factor AtDOF4.7 is involved in ethylene- and IDA-mediated organ abscission in Arabidopsis. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00863
Wang, Z., Wong, D. C. J., Chen, Z., Bai, W., Si, H., Jin, X. (2022). Emerging roles of plant DNA-binding with one finger transcription factors in various hormone and stress signaling pathways. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.844201
Wang, T., Yue, J. J., Wang, X. J., Xu, L., Li, L. B., Gu, X. P. (2016a). Genome-wide identification and characterization of the dof gene family in moso bamboo (Phyllostachys heterocycla var. pubescens). Genes Genomics 38 (8), 733–745. doi: 10.1007/s13258-016-0418-2
Wang, H. W., Zhang, B., Hao, Y. J., Huang, J., Tian, A. G., Liao, Y., et al. (2007). The soybean dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J. 52 (4), 716–729. doi: 10.1111/j.1365-313X.2007.03268.x
Washio, K. (2001). Identification of dof proteins with implication in the gibberellin-regulated expression of a peptidase gene following the germination of rice grains. Biochim. Biophys. Acta 1520 (1), 54–62. doi: 10.1016/s0167-4781(01)00251-2
Washio, K. (2003). Functional dissections between GAMYB and dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol. 133 (2), 850–863. doi: 10.1104/pp.103.027334
Wei, P. C., Tan, F., Gao, X. Q., Zhang, X. Q., Wang, G. Q., Xu, H., et al. (2010). Overexpression of AtDOF4.7, an Arabidopsis DOF family transcription factor, induces floral organ abscission deficiency in Arabidopsis. Plant Physiol. 153 (3), 1031–1045. doi: 10.1104/pp.110.153247
Wei, Q., Wang, W., Hu, T., Hu, H., Mao, W., Zhu, Q., et al. (2018). Genome-wide identification and characterization of dof transcription factors in eggplant (Solanum melongena l.). PeerJ. 6, e4481. doi: 10.7717/peerj.4481
Wei, Z., Yuan, T., Tarkowská, D., Kim, J., Nam, H. G., Novák, O., et al. (2017). Brassinosteroid biosynthesis is modulated via a transcription factor cascade of COG1, PIF4, and PIF5. Plant Physiol. 174 (2), 1260–1273. doi: 10.1104/pp.16.01778
Wen, C. L., Cheng, Q., Zhao, L., Mao, A., Yang, J., Yu, S., et al. (2016). Identification and characterisation of dof transcription factors in the cucumber genome. Sci. Rep. 6, 23072. doi: 10.1038/srep23072
Wu, J., Chen, L., Chen, M., Zhou, W., Dong, Q., Jiang, H., et al. (2019). The DOF-domain transcription factor ZmDOF36 positively regulates starch synthesis in transgenic maize. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00465
Wu, Z., Cheng, J., Cui, J., Xu, X., Liang, G., Luo, X., et al. (2016). Genome-wide identification and expression profile of dof transcription factor gene family in pepper (Capsicum annuum l.). Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00574
Wu, Y., Lee, S. K., Yoo, Y., Wei, J., Kwon, S. Y., Lee, S. W., et al. (2018). Rice transcription factor OsDOF11 modulates sugar transport by promoting expression of Sucrose transporter and SWEET genes. Mol. Plant 11 (6), 833–845. doi: 10.1016/j.molp.2018.04.002
Wu, Q., Li, D., Li, D., Liu, X., Zhao, X., Li, X., et al. (2015). Overexpression of OsDof12 affects plant architecture in rice (Oryza sativa l.). Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00833
Wu, Q., Liu, X., Yin, D., Yuan, H., Xie, Q., Zhao, X., et al. (2017a). Constitutive expression of OsDof4, encoding a C2-C2 zinc finger transcription factor, confesses its distinct flowering effects under long- and short-day photoperiods in rice (Oryza sativa l.). BMC Plant Biol. 17 (1), 166. doi: 10.1186/s12870-017-1109-0
Wu, Y., Yang, W., Wei, J., Yoon, H., An, G. (2017b). Transcription factor OsDOF18 controls ammonium uptake by inducing ammonium transporters in rice roots. Mol. Cells 40 (3), 178–185. doi: 10.14348/molcells.2017.2261
Xian, J., Wang, Y., Niu, K., Ma, H., Ma, X. (2020). Transcriptional regulation and expression network responding to cadmium stress in a cd-tolerant perennial grass Poa pratensis. Chemosphere. 250, 126158. doi: 10.1016/j.chemosphere.2020.126158
Xu, P. P., Chen, H. Y., Cai, W. M. (2020). Transcription factor CDF4 promotes leaf senescence and floral organ abscission by regulating abscisic acid and reactive oxygen species pathways in Arabidopsis. EMBO Rep. 21 (7), e48967. doi: 10.15252/embr.201948967
Xu, P., Chen, H., Ying, L., Cai, W. (2016). AtDOF5.4/OBP4, a DOF transcription factor gene that negatively regulates cell cycle progression and cell expansion in Arabidopsis thaliana. Sci. Rep. 6, 27705. doi: 10.1038/srep27705
Xu, J., Dai, H. (2016). Brassica napus cycling dof Factor1 (BnCDF1) is involved in flowering time and freezing tolerance. Plant Growth Regul. 80, 315–322. doi: 10.1007/s10725-016-0168-9
Xu, D., Li, X., Wu, X., Meng, L., Zou, Z., Bao, E., et al. (2021). Tomato SlCDF3 delays flowering time by regulating different FT-like genes under long-day and short-day conditions. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.650068
Yanagisawa, S. (1997). Dof DNA-binding domains of plant transcription factors contribute to multiple protein-protein interactions. Eur. J. Biochem. 250 (2), 403–410. doi: 10.1111/j.1432-1033.1997.0403a.x
Yanagisawa, S. (2002). The dof family of plant transcription factors. Trends Plant Sci. 7 (12), 555–560. doi: 10.1016/s1360-1385(02)02362-2
Yanagisawa, S. (2016). “Structure, function, and evolution of the dof transcription factor family,” in Plant transcription factors. Ed. Gonzalez, D. H. (Boston: Academic Press), 183–197. doi: 10.1016/B978-0-12-800854-6.00012-9
Yanagisawa, S., Izui, K. (1993). Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J. Biol. Chem. 268 (21), 16028–16036. doi: 10.1080/10425170400002439
Yanagisawa, S., Sheen, J. (1998). Involvement of maize dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell. 10 (1), 75–89. doi: 10.1105/tpc.10.1.75
Yang, Q., Chen, Q., Zhu, Y., Li, T. (2018b). Identification of MdDof genes in apple and analysis of their response to biotic or abiotic stress. Funct. Plant Biol. 45 (5), 528–541. doi: 10.1071/FP17288
Yang, G., Gao, X., Ma, K., Li, D., Jia, C., Zhai, M., et al. (2018a). The walnut transcription factor JrGRAS2 contributes to high temperature stress tolerance involving in dof transcriptional regulation and HSP protein expression. BMC Plant Biol. 18 (1), 367. doi: 10.1186/s12870-018-1568-y
Yang, Y., He, Z., Bing, Q., Duan, X., Chen, S., Zeng, M., et al. (2022). Two dof transcription factors promote flavonoid synthesis in kumquat fruit by activating c-glucosyltransferase. Plant Sci. 318, 111234. doi: 10.1016/j.plantsci.2022.111234
Yang, X., Tuskan, G. A., Cheng, M. Z. (2006). Divergence of the dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol. 142 (3), 820–830. doi: 10.1104/pp.106.083642
Yang, G., Yu, L., Wang, Y., Wang, C., Gao, C. (2017). The translation initiation factor 1A (TheIF1A) from Tamarix hispida is regulated by a dof transcription factor and increased abiotic stress tolerance. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00513
Yu, Y. H., Bian, L., Wan., Y. T., Jiao, Z. L., Yu, K. K., Zhang, G. H., et al. (2019). Grape (Vitis vinifera) VvDOF3 functions as a transcription activator and enhances powdery mildew resistance. Plant Physiol. Biochem. 143, 183–189. doi: 10.1016/j.plaphy.2019.09.010
Yu, Q., Li, C., Zhang, J., Tian, Y., Wang, H., Zhang, Y., et al. (2020). Genome-wide identification and expression analysis of the Dof gene family under drought stress in tea (Camellia sinensis). PeerJ. 8, e9269. doi: 10.7717/peerj.9269
Yu, H., Ma, Y., Lu, Y., Yue, J., Ming, R. (2021a). Expression profiling of the dof gene family under abiotic stresses in spinach. Sci. Rep. 11 (1), 14429. doi: 10.1038/s41598-021-93383-6
Yu, L., Ma, S., Zhang, X., Tian, D., Yang, S., Jia, X., et al. (2021b). Ancient rapid functional differentiation and fixation of the duplicated members in rice Dof genes after whole genome duplication. Plant J. 108 (5), 1365–1381. doi: 10.1111/tpj.15516
Zang, D., Wang, L., Zhang, Y., Zhao, H., Wang, Y. (2017). ThDof1.4 and ThZFP1 constitute a transcriptional regulatory cascade involved in salt or osmotic stress in Tamarix hispida. Plant Mol. Biol. 94 (4-5), 495–507. doi: 10.1007/s11103-017-0620-x
Zhang, B., Chen, W., Foley, R. C., Büttner, M., Singh, K. B. (1995). Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell. 7 (12), 2241–2252. doi: 10.1105/tpc.7.12.2241
Zhang, J., Hao, Q., Bai, L., Xu, J., Yin, W., Song, L., et al. (2014). Overexpression of the soybean transcription factor GmDof4 significantly enhances the lipid content of Chlorella ellipsoidea. Biotechnol. Biofuels. 7 (1), 128. doi: 10.1186/s13068-014-0128-4
Zhang, L. L., Jiang, A., Thomson, G., Kerr-Phillips, M., Phan, C., Krueger, T., et al. (2019). Overexpression of medicago MtCDFd1_1 causes delayed flowering in medicago via repression of MtFTa1 but not MtCO-like genes. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01148
Zhang, L., Liu, B., Zheng, G., Zhang, A., Li, R. (2017). Genome-wide characterization of the SiDof gene family in foxtail millet (Setaria italica). Biosystems. 151, 27–33. doi: 10.1016/j.biosystems.2016.11.007
Zhang, A., Matsuoka, K., Kareem, A., Robert, M., Roszak, P., Blob, B., et al. (2022). Cell-wall damage activates DOF transcription factors to promote wound healing and tissue regeneration in Arabidopsis thaliana. Curr. Biol. 32 (9), 1883–1894.e7. doi: 10.1016/j.cub.2022.02.069
Zhang, A. D., Wang, W. Q., Tong, Y., Li, M. J., Grierson, D., Ferguson, I., et al. (2018b). Transcriptome analysis identifies a zinc finger protein regulating starch degradation in kiwifruit. Plant Physiol. 178 (2), 850–863. doi: 10.1104/pp.18.00427
Zhang, Z., Yuan, L., Liu, X., Chen, X., Wang, X. (2018a). Evolution analysis of dof transcription factor family and their expression in response to multiple abiotic stresses in Malus domestica. Gene. 639, 137–148. doi: 10.1016/j.gene.2017.09.039
Zhou, Y., Cheng, Y., Wan, C., Li, J., Yang, Y., Chen, J. (2020). Genome-wide characterization and expression analysis of the Dof gene family related to abiotic stress in watermelon. PeerJ. 8, e8358. doi: 10.7717/peerj.8358
Zhuo, M., Sakuraba, Y., Yanagisawa, S. (2020). A jasmonate-activated MYC2-Dof2.1-MYC2 transcriptional loop promotes leaf senescence in Arabidopsis. Plant Cell. 32 (1), 242–262. doi: 10.1105/tpc.19.00297
Zou, X., Neuman, D., Shen, Q. J. (2008). Interactions of two transcriptional repressors and two transcriptional activators in modulating gibberellin signaling in aleurone cells. Plant Physiol. 148 (1), 176–186. doi: 10.1104/pp.108.123653
Zou, Z., Zhang, X. (2019). Genome-wide identification and comparative evolutionary analysis of the dof transcription factor family in physic nut and castor bean. PeerJ. 7, e6354. doi: 10.7717/peerj.6354
Zou, H. F., Zhang, Y. Q., Wei, W., Chen, H. W., Song, Q. X., Liu, Y. F., et al. (2013). The transcription factor AtDOF4.2 regulates shoot branching and seed coat formation in Arabidopsis. Biochem. J. 449 (2), 373–388. doi: 10.1042/BJ20110060
Keywords: transcription factor, DOF, zinc finger, plant hormones, metabolic regulation
Citation: Zou X and Sun H (2023) DOF transcription factors: Specific regulators of plant biological processes. Front. Plant Sci. 14:1044918. doi: 10.3389/fpls.2023.1044918
Received: 15 September 2022; Accepted: 03 January 2023;
Published: 20 January 2023.
Edited by:
Magda Pál, Centre for Agricultural Research (ELKH), HungaryReviewed by:
Paola Vittorioso, Sapienza University of Rome, ItalyCopyright © 2023 Zou and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongmei Sun, aG1iaEBzaW5hLmNvbQ==, c3VuaG1Ac3lhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.