
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 29 September 2022
Sec. Functional Plant Ecology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.999990
This article is part of the Research Topic Physiological Ecology of Trees under Environmental Stresses View all 14 articles
 Ying Yang1,2†
Ying Yang1,2† Jiaqi Mei1,2†
Jiaqi Mei1,2† Juanjuan Chen1,2,3†
Juanjuan Chen1,2,3† Ying Yang1,2
Ying Yang1,2 Yujie Gu1,2
Yujie Gu1,2 Xiaoyu Tang1,2
Xiaoyu Tang1,2 Huijie Lu1,2
Huijie Lu1,2 Kangbiao Yang1,2
Kangbiao Yang1,2 Anket Sharma1,2
Anket Sharma1,2 Xiaofei Wang1,2
Xiaofei Wang1,2 Daoliang Yan1,2
Daoliang Yan1,2 Rongling Wu1,2
Rongling Wu1,2 Bingsong Zheng1,2*
Bingsong Zheng1,2* Huwei Yuan1,2*
Huwei Yuan1,2*Grafting is an effective way to improve Chinese hickory while salt stress has caused great damage to the Chinese hickory industry. Grafting and salt stress have been regarded as the main abiotic stress types for Chinese hickory. However, how Chinese hickory responds to grafting and salt stress is less studied. Auxin has been proved to play an essential role in the stress response through its re-distribution regulation mediated by polar auxin transporters, including PIN-formed (PIN) proteins. In this study, the PIN gene family in Chinese hickory (CcPINs) was identified and structurally characterized for the first time. The expression profiles of the genes in response to grafting and salt stress were determined. A total of 11 CcPINs with the open reading frames (ORFs) of 1,026–1,983 bp were identified. Transient transformation in tobacco leaves demonstrated that CcPIN1a, CcPIN3, and CcPIN4 were localized in the plasma membrane. There were varying phylogenetic relationships between CcPINs and homologous genes in different species, but the closest relationships were with those in Carya illinoinensis and Juglans regia. Conserved N- and C-terminal transmembrane regions as well as sites controlling the functions of CcPINs were detected in CcPINs. Five types of cis-acting elements, including hormone- and stress-responsive elements, were detected on the promoters of CcPINs. CcPINs exhibited different expression profiles in different tissues, indicating their varied roles during growth and development. The 11 CcPINs responded differently to grafting and salt stress treatment. CcPIN1a might be involved in the regulation of the grafting process, while CcPIN1a and CcPIN8a were related to the regulation of salt stress in Chinese hickory. Our results will lay the foundation for understanding the potential regulatory functions of CcPIN genes during grafting and under salt stress treatment in Chinese hickory.
Auxin is the first discovered phytohormone (Went, 1935) and plays an important role during different growth stages of plants (Yu et al., 2022), including gametogenesis, seed germination, root elongation, vascular patterning, and blossoming (Zhao, 2010). In addition, an important function of auxin is its regulation of the responses of plant species to different biotic and abiotic stresses (Kapazoglou et al., 2020; Dastborhan et al., 2021; Mearaji et al., 2021; Zhang et al., 2022). The functioning of auxin is closely related to its homeostatic regulation, which involves biosynthesis, conjugate formation, transport, and degradation (Zhang and Peer, 2017). Following biosynthesis, transport is the key step for auxin functioning (Gomes and Scortecci, 2021) because the site of action is usually different from that of synthesis.
Plants have evolved two methods for transporting auxin: the long-distance network (transporting auxin through phloem) and the cell-to-cell transport network (also named polar auxin transport) (Muday and DeLong, 2001; Robert and Friml, 2009). In plants, polar auxin transport is mainly mediated by AUXIN/LIKE AUXIN proteins (AUX/LAX, responsible for auxin influx), PIN-formed proteins (PIN, responsible for auxin efflux), PIN-LIKES (PILS, responsible for auxin transport between the cytosol and endoplasmic reticulum), and ATP-binding cassette subfamily B proteins (ABCB, responsible for auxin influx/efflux) (Swarup and Peret, 2012; Mohanta et al., 2018; Sauer and Kleine-Vehn, 2019). PIN proteins play major roles in auxin efflux because their polar localizations are always consistent with the directionality of auxin (Zhou and Luo, 2018).
PIN is a gene family composed of multiple genes with similar or different structures and functions. AtPIN1 was the first discovered PIN protein in Arabidopsis. The mutant of PIN1, pin1, has no leaves or flowers in cauline inflorescence (Kuhlemeier and Reinhardt, 2001) and resembles pins, which is the reason why PIN1 and the subsequent proteins in this family were named PIN-formed proteins (PINs). PIN family proteins contain conserved N- and C-terminal transmembrane domains and intracellular central hydrophilic loops. There are two types of PIN proteins categorized according to the differences in structures of the central hydrophilic loop: canonical PIN proteins with a long central hydrophilic loop and non-canonical PIN proteins with a short hydrophilic loop. In Arabidopsis, canonical PIN proteins (AtPIN1, 2, 3, 4, and 7) are localized to the plasma membrane (PM) and mediate auxin efflux to the extracellular space (Bennett et al., 2014). Non-canonical PINs (AtPIN5 and 8) are localized to the endoplasmic reticulum (ER) and mediate intracellular auxin transport between the cytoplasm and ER (Mravec et al., 2009; Sauer and Kleine-Vehn, 2019). AtPIN6 possesses a long hydrophilic loop and is located on the PM and ER, and thus is classified as a non-canonical PIN (Bennett et al., 2014).
The functions of PIN proteins have been well studied in Arabidopsis. AtPIN1 regulates the shoot apical meristem, root elongation, and the development of xylem (Blilou et al., 2005; Alabdallah et al., 2017). AtPIN2 influences root gravitropism and responds to salt stress (Wang et al., 2019; Gibson et al., 2020). AtPIN3 is mainly expressed in root and vascular tissues. It regulates auxin distribution in roots, participates in lateral root growth, and responds to light and gravity (Blilou et al., 2005; Zhai et al., 2021; Li et al., 2022). Similar to AtPIN3, AtPIN4 is related to light and gravity responses. In addition, AtPIN4 participates in auxin flow to the quiescent center (Bureau et al., 2010). AtPIN5 mediates not only the transport of auxin from the cytoplasm to the ER, but also regulates auxin homeostasis and metabolism (Mravec et al., 2009). AtPIN6 regulates lateral root formation (Sauer and Kleine-Vehn, 2019). The functions of AtPIN7 are similar to those of AtPIN3 and AtPIN4, and it participates in the gravimetric response of roots (Lewis et al., 2011). The function of AtPIN8 is the opposite of that of AtPIN5, transporting auxin from the ER to the cytoplasm (Ding et al., 2012). In addition, there is functional redundancy for some PIN proteins (Blilou et al., 2005; Wang et al., 2021). For example, the mutant genes pin1 and pin2 exhibit functional redundancy regarding the regulation of root meristem and root length (Vieten et al., 2005).
In addition to developmental functions, PIN family genes also participate in the regulation of abiotic stress (Yu et al., 2017). For example, in pepper (Capsicum annuum) and Sorghum bicolor, the expression profiles of PIN genes were changed under high salt treatment (Shen et al., 2010; Zhang et al., 2018). The process of grafting creates wounding stress, which also influences the expression of PIN genes (Melnyk, 2017; Sharma and Zheng, 2019). The overexpression of AtPINs-GUS in Arabidopsis shows that AtPINs are expressed in the grafted union, indicating their involvement in grafting regulation (Wang et al., 2014). Transcriptome sequencing of Arabidopsis grafting also supports this conclusion (Melnyk et al., 2018).
Chinese hickory (Carya cathayensis Sarg.), belonging to the Juglandaceae family, is one of the most important trees economically in the Zhejiang and Anhui Provinces of China. Nuts from Chinese hickory trees are rich in nutritional compounds, including polyunsaturated fatty acids, phenolics, and flavonoids (Huang et al., 2022). However, a long juvenile phase and narrow distribution have restricted the development of the Chinese hickory industry. Grafting is an effective way to solve these problems. In addition, soil salinization has limited the development of the Chinese hickory industry.
To date, members of the PIN gene family have not been identified, and their potential roles during grafting and salt stress treatment in Chinese hickory are still unknown. In this study, PIN family genes in Chinese hickory were cloned and structurally analyzed, and their expression profiles in response to grafting and salt stress treatment were determined. Our results will lay the foundation for revealing the molecular mechanism of PIN family genes in regulating grafting and salt stress in Chinese hickory.
Candidate gene sequences of PIN in Chinese hickory (CcPINs) were identified from the published genome (Huang et al., 2019). The process used to identify genes was the same as that used in a previous study (Yang et al., 2021). Hidden Markov model (HMM) profiles of the PIN domain (PF03547) were used for analysis. TBtools (Toolbox for Biologists, v1.098745) was used for the preliminary screening of PIN genes (Chen et al., 2020). The identified CcPINs were named according to their phylogenetic relationships with the homologs in Arabidopsis (Supplementary Figure 1).
In addition to predicting the functions of CcPINs in Chinese hickory, the PIN proteins in Chinese hickory and 14 other species were chosen to undergo phylogenetic relationship analysis. The PIN proteins of 11 species were obtained from previous studies. PINs of the other three species (including Juglans regia, Carya illinoinensis, and Nymphaea tetragona) were analyzed based on the genome in Phytozome v13.1 The identified method was the same as that used for Chinese hickory, as stated above. The PIN protein information for 15 species is shown in Supplementary Table 1.
The phylogenetic tree between PIN proteins in the 15 species was constructed by MEGAX using the neighbor-joining method and a bootstrap of 1,000 replicates. ClustalW was used for multiple sequence alignment analysis of CcPINs. The motif distribution of CcPIN proteins was predicted by MEME Suite.2 The motif number was set to 8, and the maximum and minimum numbers of amino acids were set to 50 and 6, respectively. The online tool ExPASy was used to analyze biochemical information (length, molecular weight, isoelectric point, and size) of CcPIN genes. The transmembrane regions of CcPIN proteins were predicted by the TMHMM2 tool (TMHMM--2.0).3 ‘‘Search for CARE’’ on the PlantCare website4 was used to identify cis-acting regulatory elements on the promoters of CcPIN genes.
One-year-old Chinese hickory seedlings were used for tissue-specific expression analysis, while 2-year-old Chinese hickory seedlings were used as rootstocks for grafting and materials for salt stress treatment. Seedlings were planted in seedling pots with a diameter of 17 cm and height of 22 cm and cultivated in the greenhouse of Zhejiang A&F University. One seedling was planted in each pot with 5 L of soil. The soil formula included 50% peat, 13% pastoral soil, 15% organic fertilizer, 5% exocarp, 10% agricultural bran, 5% perlite, and 2% release fertilizer. The seedlings were grown under the following conditions: temperature of 25 ± 3°C, humidity of 60–70%, and photoperiod of 12-h light/12-h dark. The seedlings were watered every 5 days. Tobacco (Nicotiana benthamiana) was cultivated in the culture room at the temperature of 22°C, humidity of 60–70%, and photoperiod of 16-h light/8-h dark. For subcellular localization analysis, 45-day-old tobacco leaves were used.
For the grafting experiment, 2-year-old seedlings with similar phenotypes were used for rootstocks, and the grafting operation was conducted on April 20th, 2018. One-year-old branches (7–8 cm in length) with a new bud collection on the fruit-bearing trees were chosen as scions. For salt stress treatment, the 2-year-old seedlings were treated with 150 mm NaCl (Shanghai Hushi Laboratorial Equipment Co., Ltd., China) solution three times every 3 days, following the same method as a previous study (Chen et al., 2015). The equivalent water treatment was used for the control check (CK).
Graft unions were collected at 0, 3, 7, and 14 days after grafting, representing the initiation stage, isolation layer formation stage, callus formation and isolating layer disappearance stage, and vascular bridge formation and linkage stage during Chinese hickory grafting, respectively, as detected by cytological observation (Liu et al., 2009). In each treatment, three graft unions were mixed together and regarded as a single sample. For salt stress treatment, samples from different tissues, including roots, stems, and leaves, were collected after 0, 1, 3, and 10 days of treatment, representing the initial-, short-, medium- and long-term treatment stages as reported in similar studies of other woody plants (Chen et al., 2015; Zhao et al., 2015; Hang et al., 2020; Huang et al., 2020). The day on which the seedlings were treated with NaCl solution for the first time was regarded as day 0. For each tissue, collections from five different seedlings were mixed together and regarded as a single sample. For tissue-specific expression analysis, different tissues, including roots, stems, leaves, and shoots, of 1-year-old Chinese hickory seedlings were collected in early spring (on April 16, 2022), representing the fast-growing stage of the seedlings. For each tissue, collections from five different seedlings were mixed together and regarded as a single sample. All the samples were wrapped in aluminum foil, immediately immersed in liquid nitrogen, and stored in an ultra-low temperature refrigerator (−80°C) for future use.
The total RNAs of Chinese hickory were extracted using a MiniBEST Universal RNA Extraction Kit [Code No. 9767, Takara Biomedical Technology (Beijing) Co., Ltd., China]. For the grafting experiment, the rootstocks and scions in a single sample were separated from each other, and the cells at the conjunction surfaces of the rootstocks and scions were scraped using blades, respectively. Total RNAs of the rootstocks and scions were extracted separately. For the salt stress experiment, the total RNAs of the roots, stems, and leaves collected at different time points were extracted separately. For the tissue-specific experiment, the total RNAs of the roots, stems, leaves, and shoots of 1-year-old seedlings were extracted separately. The cDNA used for cloning was synthesized using the PrimeScript™ IV 1st strand cDNA Synthesis Mix [Code No. 6215A, Takara Biomedical Technology (Beijing) Co., Ltd., China]. PrimeSTAR Max DNA Polymerase [Code No. R045Q, Takara Biomedical Technology (Beijing) Co., Ltd., China] was used for PIN gene cloning. The cDNA used for Real-Time Quantitative PCR analysis (qRT-PCR) was obtained using the PrimeScript™ RT Master Mix [Code No. RR036A, Takara Biomedical Technology (Beijing) Co., Ltd., China].
Real-Time Quantitative PCR analysis primers were designed on the National Center for Biotechnology Information (NCBI) primer prediction website.5 The CcActin gene was used as the internal standard for normalization. The primers for qRT-PCR are listed in Supplementary Table 2. The methods used for qRT-PCR were followed according to a previously published study (Yuan et al., 2018). The qRT-PCR analysis was replicated at least three times.
The coding sequence of CcPINs was cloned from the cDNA library of Chinese hickory and inserted into the modified pCAMBIA1300-GFP vector. Then, the obtained 35S:CcPINs-GFP was translated into Escherichia coli Trans1-T1 (TransGen Biotech Co., Ltd., China) and sequenced by Sangon Biotech (Shanghai) Co., Ltd., China. Plasmids from the identified strains were extracted and transformed into Agrobacterium strain GV3101 competent cells (AC1001, Shanghai Weidi Biotechnology Co., Ltd., China). Transient transformation of 45-day-old tobacco leaves was performed as previously described (Yamaji et al., 2009). The pm-rk (plasma membrane marker with red fluorescence, Nelson et al., 2007) was transiently co-transformed into tobacco leaves with CcPINs. After culturing for 48 h, the fluorescence signal in the transformed tobacco leaves was detected using a confocal microscope (LSM 880, Zeiss, Germany) with 488 and 594 nm argon lasers.
To calculate the relative expression levels of CcPIN genes, the 2–ΔΔCT method, introduced by Schmittgen and Livak (2008), was used. A one-way analysis of variance (ANOVA) and multiple comparisons [the least significant difference (LSD) method] were conducted using SPSS Statistics (version 17.0) to analyze the expression differences of CcPIN genes among different tissues and treatments.
The histogram and line figures were drawn using Microsoft Excel 2019. The phylogenetic tree was drawn using Mega X and embellished using Fig. Tree (v1.4.4). The figures illustrating motifs and gene structures were drawn using TBtools (Toolbox for Biologists, v1.098745). Finally, the figures were merged by Adobe Photoshop (version 2020).
Eleven CcPIN genes were identified, and the corresponding information is displayed in Table 1. The ORF length of the CcPINs ranged from 1,026 bp (CcPIN8a) to 1,983 bp (CcPIN3), encoding proteins with between 341 and 660 amino acids. The molecular weights of CcPINs ranged from 37,338.75 Da (CcPIN8a) to 72,046.05 Da (CcPIN3), and the predicted isoelectric points (pI) varied from 6.42 (CcPIN5) to 9.39 (CcPIN8a). The number of exons for CcPIN genes ranged from 5 to 7. To explore the potential locations where CcPINs perform their functions, several CcPINs were detected by subcellular localization. Transient transformation of CcPINs to tobacco leaves showed that CcPIN1a, CcPIN3, and CcPIN4 were expressed on the PM (Figure 1).
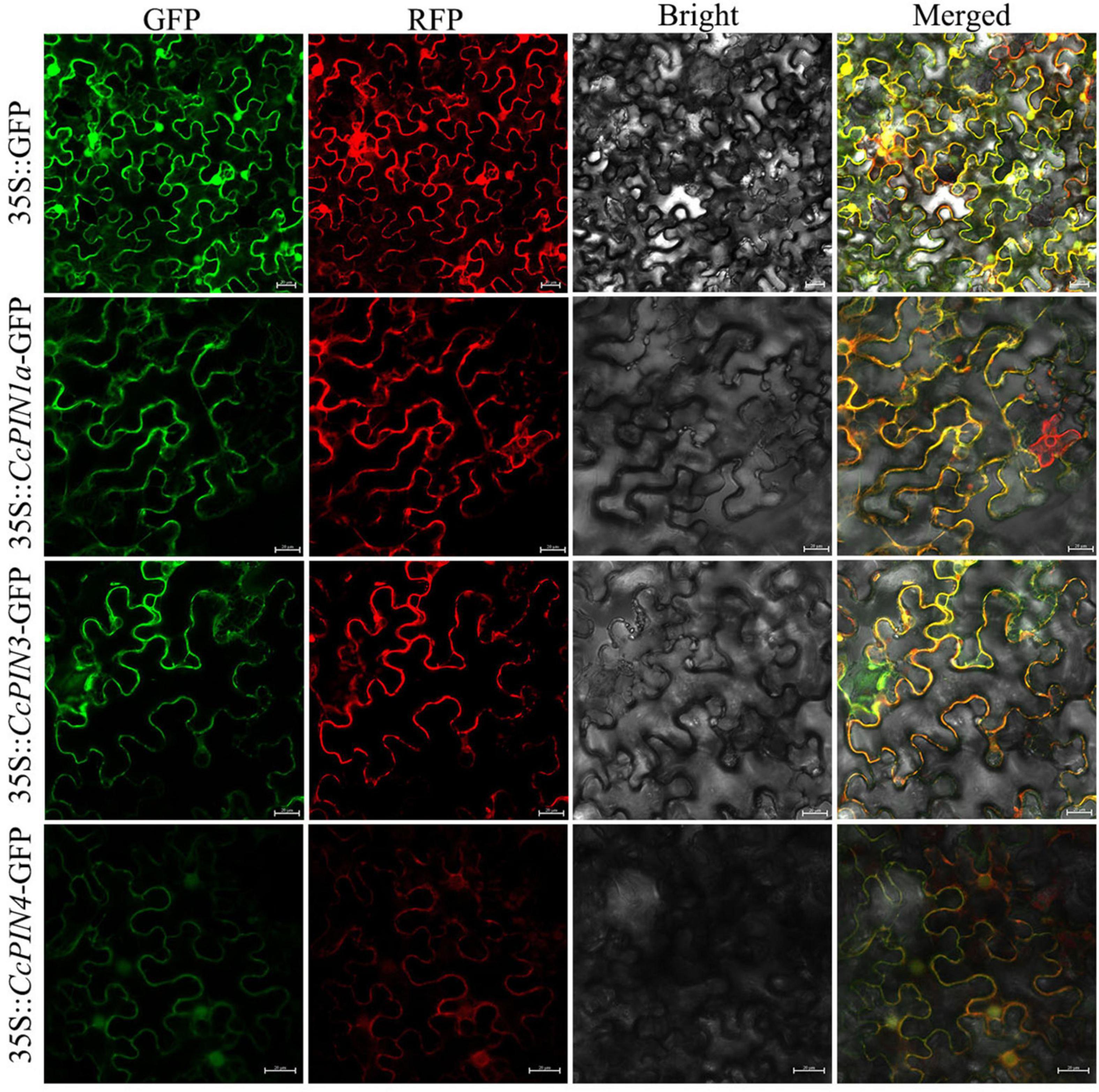
Figure 1. The subcellular localization of CcPIN1a, CcPIN3, and CcPIN4. The fluorescence images were captured in a dark field for green and red fluorescence, in a white field for the morphology of the cell, and in combination. GFP, green fluorescent protein fluorescence; RFP, red fluorescent protein fluorescence; Bright, bright field; Merged, GFP/RFP/bright field overlay. Bar = 20 μm.
To predict the potential functions of PIN family genes in Chinese hickory, a phylogenetic tree was constructed to explore the phylogenetic relationships between the 11 PIN proteins in Chinese hickory and 121 PIN proteins from 14 other plant species. The 15 plant species used to construct the phylogenetic tree belonged to 5 types of charophytes, bryophytes, lycophytes, monilophytes, gymnosperms, and angiosperms according to the consensus phylogeny reported by Doyle (2018). Detailed information on the 132 PIN proteins is shown in Supplementary Table 1. A total of eight groups (I, II, III, IV, V, VI, VII, and VIII) were identified (Figure 2A). There was significant variation in the number of PIN proteins in the different groups. There was only one protein in Group I, three proteins in Group II, seven proteins in Group VI, and 11 proteins in Group VII. The number of proteins in Groups III, IV, and V was 22, 19, and 23, respectively, and the highest number of proteins was in Group VIII, with 46 (Figure 2B).
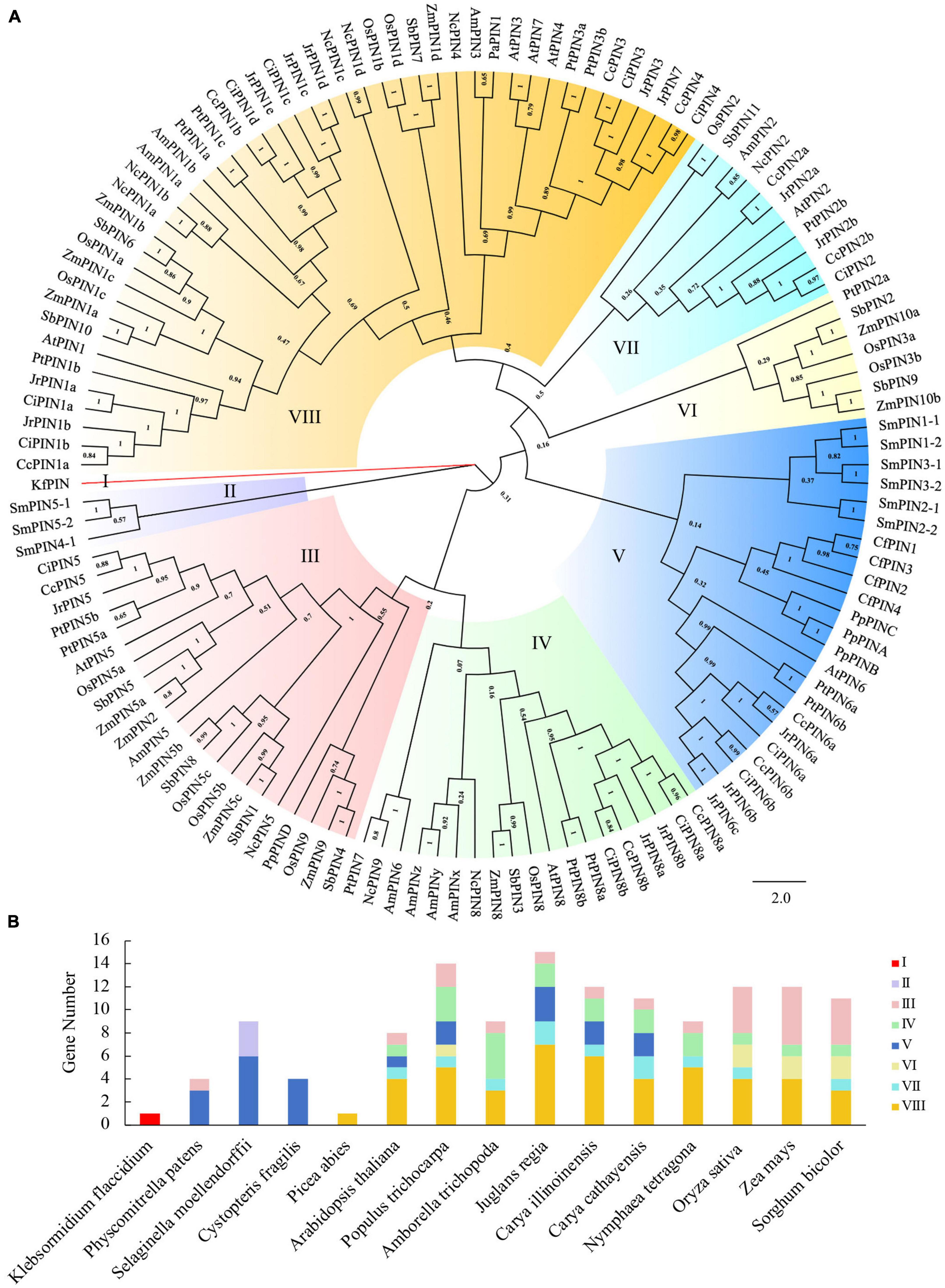
Figure 2. Phylogenetic analysis of PIN proteins from 15 species. (A) The phylogenetic tree. (B) The number and distribution of PINs in different species. Kf, Klebsormidium flaccidium; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii; Cf, Cystopteris fragilis; Pa, Picea abies; At, Arabidopsis thaliana; Pt, Populus trichocarpa; Am, Amborella trichopoda; Jr, Juglans regia; Ci, Carya illinoinensis; Cc, Carya cathayensis; Nc, Nymphaea tetragona; Os, Oryza sativa; Zm, Zea mays; Sb, Sorghum bicolor.
The number of PIN proteins varied in different species. There was only one PIN protein in Klebsormidium flaccidium and Picea abies, while the number of PIN proteins in the other 13 species changed from 4 to 15 (Figure 2B). In addition, there were more PINs (8–15) in angiosperm species than those in the other types (Figure 2B). Similar to C. illinoinensis and J. regia, the PINs in C. cathayensis were distributed in Groups III, IV, V, VII, and VIII, with the largest number of PINs in Group VIII. Most PINs in Oryza sativa, Zea mays, and S. bicolor were distributed in Groups III, IV, VI, VII, and VIII (Figures 2A,B).
Multiple sequence alignment of CcPIN proteins is shown in Figure 3, which displays the high level of conservation in the N-terminal and C-terminal regions. There were short sequences in the middle region for CcPIN5, CcPIN8a, and CcPIN8b, with long sequences for the other CcPINs. In addition, “F165,” the site controlling the distribution of PIN proteins on PM, the phosphorylation site “TPRXS,” and the conserved “NPXXY” site were detected (Figure 3). Motif analysis of the eleven CcPIN proteins was performed using the MEME Suite, with the eight conversed motifs set. Motif 6 and Motif 8 were not detected in short CcPINs (CcPIN5 and CcPIN8) (Figures 4A,B). The conserved element “NPXXY” was found in motif 5 (Figure 4D), and the phosphorylated site “TPRXS” of MPK 4/6 kinases was found in Motif 8 (Figure 4D). Transmembrane region prediction using TMHMM showed that there were N-terminal or C-terminal transmembrane regions for all CcPINs, with the total number ranging from 6 to 10 (Supplementary Figure 2). The coding sequences of CcPINs were compared with their corresponding DNA sequences on the genome. The numbers of exons in CcPINs ranged from five to eight (Figure 4C).
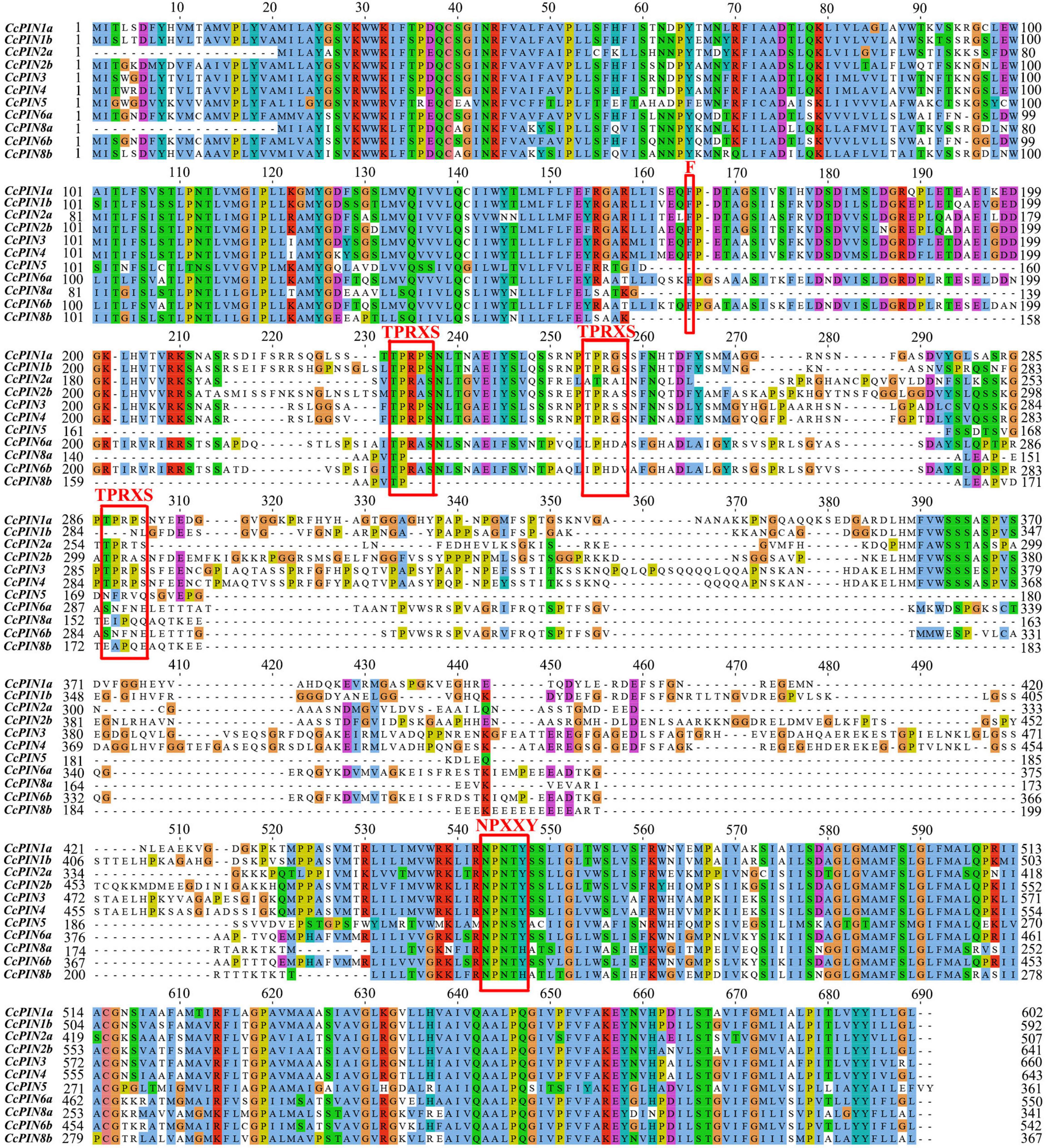
Figure 3. Multiple alignments of CcPINs by ClustalW. The possible functional sites are circled with red boxes.
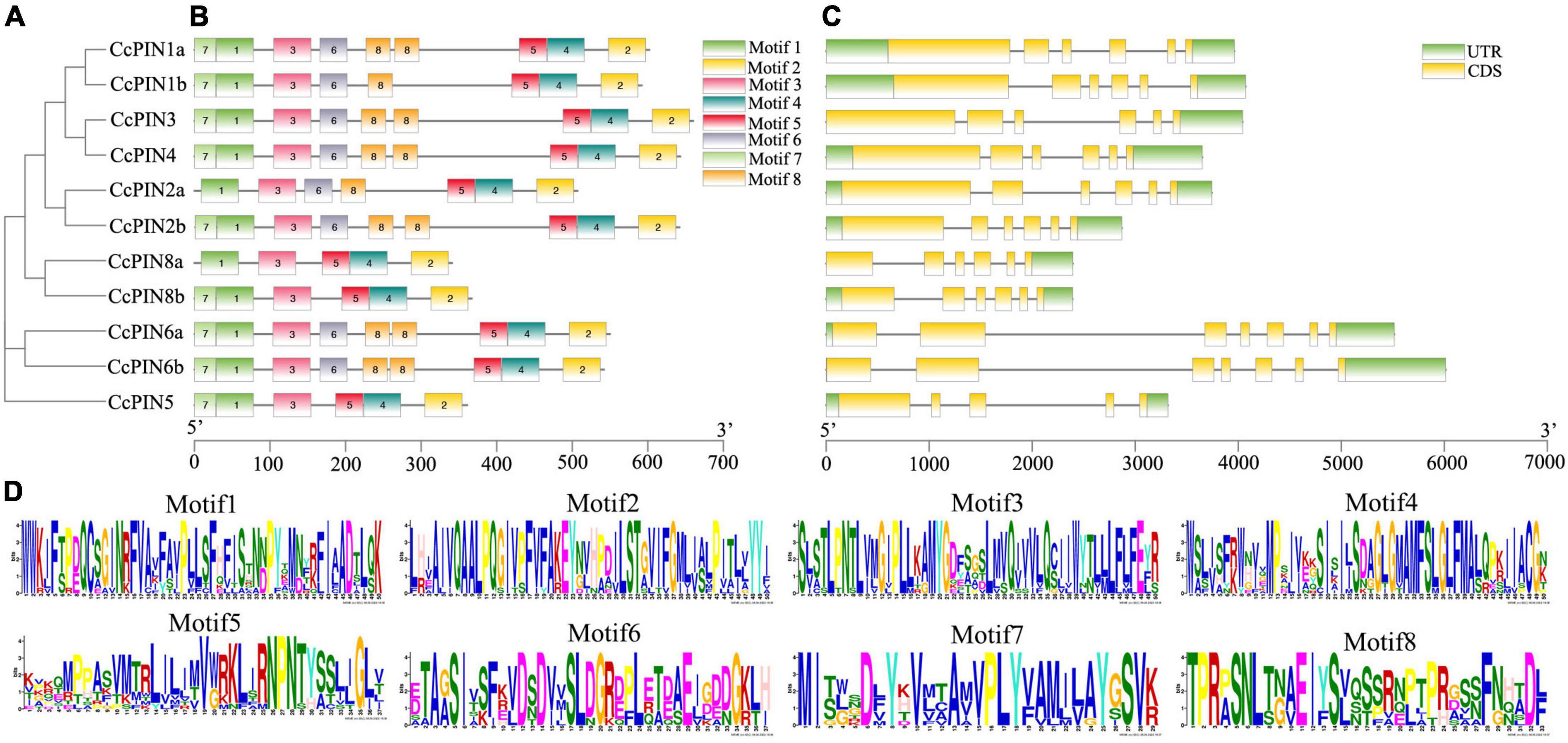
Figure 4. Motif and gene structure analysis of CcPINs. (A) Phylogenetic relationships. (B) Motif distribution. (C) Exon-intron structures. (D) Motif information.
The PPI network was predicted between the homologs of CcPINs in Arabidopsis and other proteins in Arabidopsis by the String database, with the minimum required interaction score set to a high confidence score of 0.700, and the maximum number of interactors was limited to no more than 10. The PPI network consisted of 17 nodes and 39 edges (Figure 5A). Seven nodes were representative of PIN proteins. The corresponding information is shown in Supplementary Table 3. Different line colors represent protein-protein associations.

Figure 5. (A) QQProtein–protein interaction (PPI) network of PIN-formed (PINs) and (B) gene ontology (GO) annotation of proteins in the PPI.
The nodes were annotated using Gene Ontology (GO) (Figure 5B and Supplementary Table 4). The results showed that most of the predicted proteins contributed to biological processes, such as “Cellular response to stimulus” (GO:0051716), “Auxin-activated signaling pathway” (GO:0009734), “Tissue development” (GO:0009888), and “Xylem and phloem pattern formation” (GO:0010051). The proteins also played a role in multiple cellular components including the “Plasma membrane” (GO:0005886), “Auxin polar transport” (GO:0009926), and “Cytoplasm” (GO:0005737). In addition, the molecular function of the proteins included “Auxin transmembrane transporter activity” (GO:0080161), “Auxin efflux transmembrane transporter activity” (GO:0010329), “Auxin influx transmembrane transporter activity” (GO:0010328), and “Auxin binding” (GO:0010011).
Regulatory elements that are cis-acting on the promoters of genes were the sites for expression regulation of genes. To explore how the expression of CcPINs was regulated by the different transcriptional factors, the section 2,000 bp upstream of the start codon (ATG) of CcPINs was used to query for cis-acting element detection using PlantCare. Five types of cis-acting elements, including a binding site element, hormone responsiveness, light responsiveness, plant development, and stress responsiveness, were detected, with numbers of 9, 71, 147, 28, and 40, respectively (Figure 6).
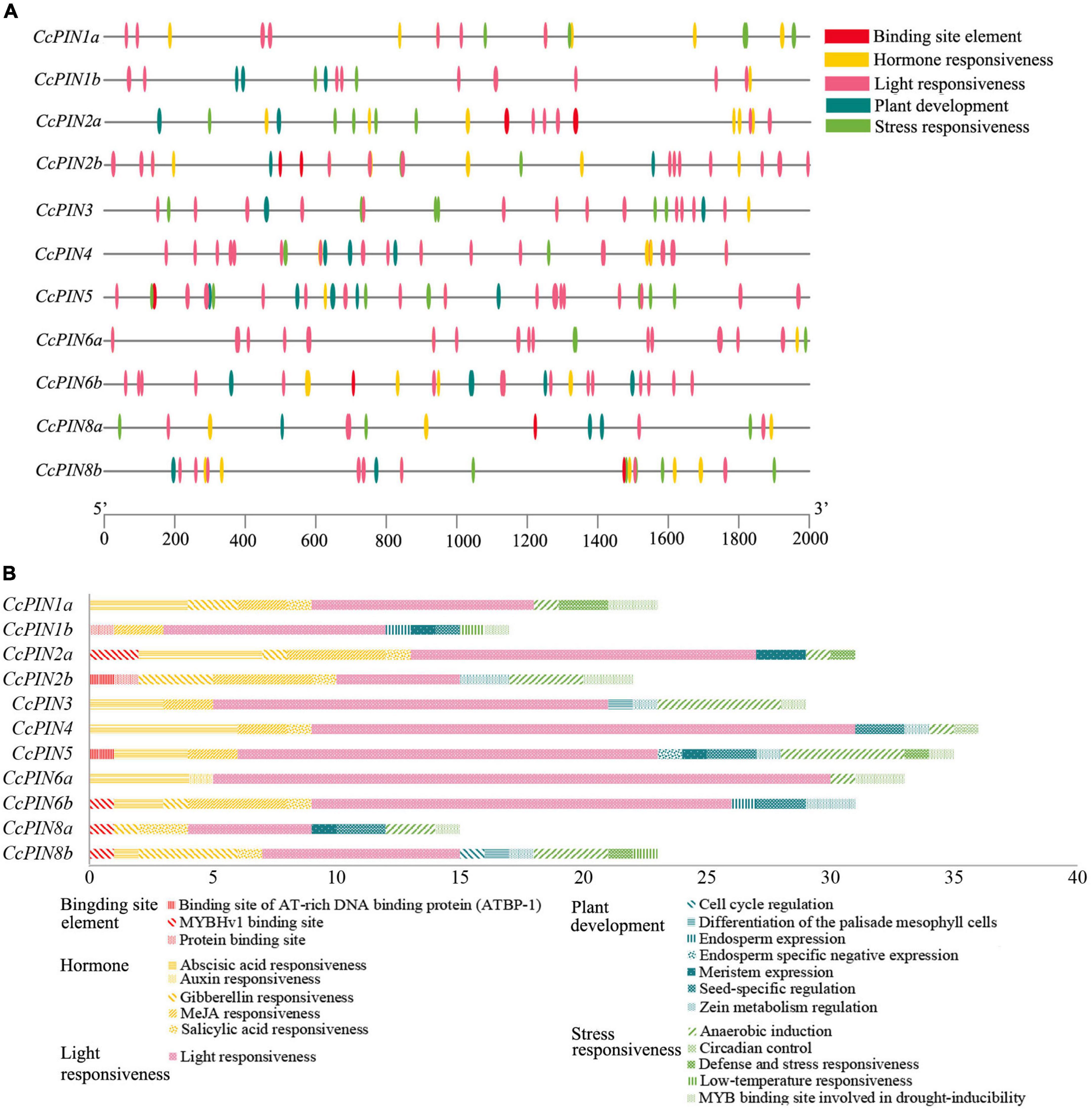
Figure 6. Cis-acting elements on the promoters of CcPIN family genes. (A) The distribution of cis-acting elements. (B) The number of cis-acting elements.
Within each type, specific cis-acting elements were detected. For example, five types of hormone-responsive elements, including gibberellin-responsive elements (GARE-motif, P-box, TATC-box, and TGA-element), abscisic acid-responsive elements (ABRE), MeJA-responsive elements (CGTCA-motif, TGACG-motif), and salicylic acid-responsive elements (TCA-element), were detected. The number of cis-acting elements for a certain CcPIN gene ranged from 15 (CcPIN8a) to 36 (CcPIN4), where light-responsive elements were the most abundant. In addition, five types of stress-responsive elements were detected on the promoters of CcPINs.
To explore the relative expression of PIN genes in different tissues, tissue-specific expression profiles of CcPIN genes in roots, stems, leaves, and shoots of Chinese hickory were obtained by qRT-PCR. The results showed that CcPIN1a, CcPIN2a, CcPIN5, and CcPIN8a were highly expressed in stems, CcPIN4 and CcPIN6a showed the highest expression levels in shoots, and the highest expression of CcPIN3 was in leaves (Figure 7). The tissue-specific expression patterns of CcPIN genes showed that they might play different roles in different tissues in Chinese hickory.
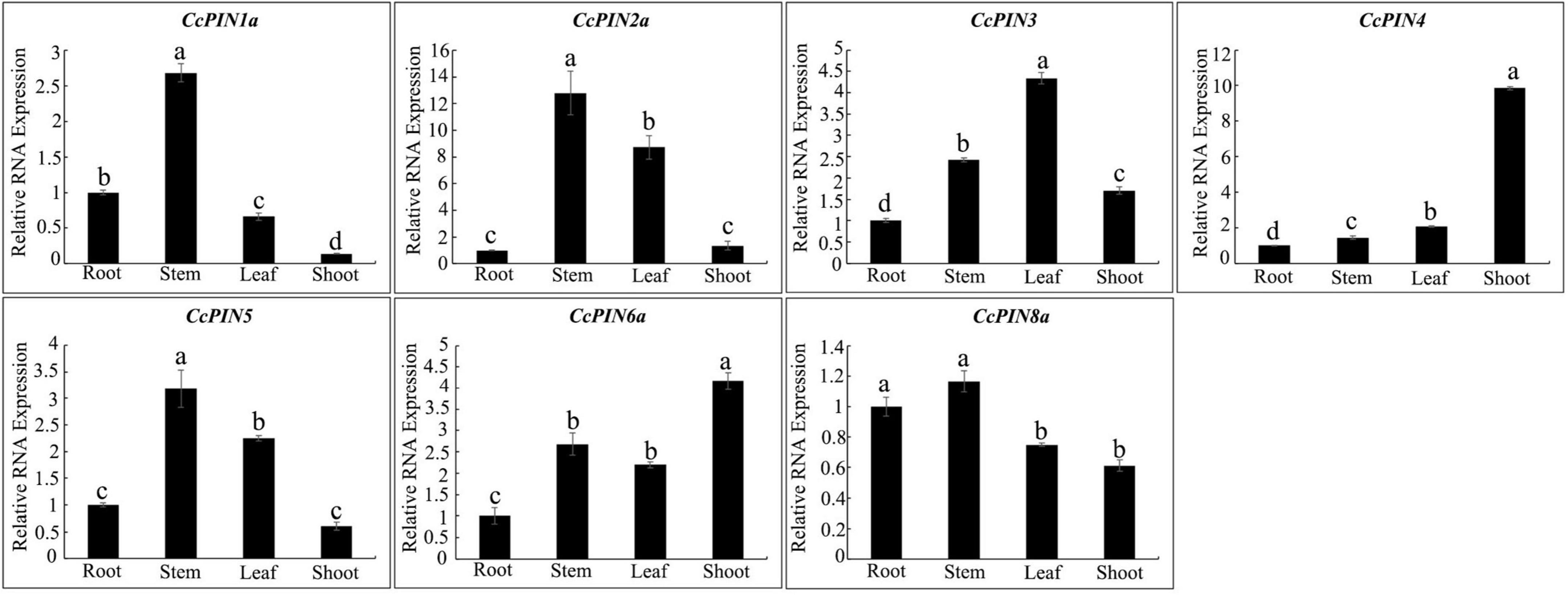
Figure 7. Tissue-specific expression profiles of CcPIN genes. Total RNA was extracted from roots, stems, leaves, and shoots of 1-year-old Chinese hickory. The relative expression levels of each CcPIN gene in roots were standardized as one. Different letters above the columns denote a significant difference in different tissues (P < 0.05).
To investigate whether CcPINs regulated grafting in Chinese hickory, the expression levels of CcPINs were measured at different stages of grafting (Figure 8). In scions, compared with 0 days after grafting, the expression level of CcPIN1a was significantly upregulated over sevenfold and eightfold at 3 and 7 days after grafting, respectively. However, at 14 days after grafting, the expression was decreased but still significantly higher than at 0 days after grafting. The expression of CcPIN3 and CcPIN6a was downregulated in scions. The expression levels of CcPIN4 and CcPIN8a significantly changed in rootstocks after grafting. The highest expression for CcPIN4 was at 14 days after grafting in rootstocks, while CcPIN2a exhibited the highest expression at 3 days after grafting. CcPIN5 showed stable expression during Chinese hickory grafting.
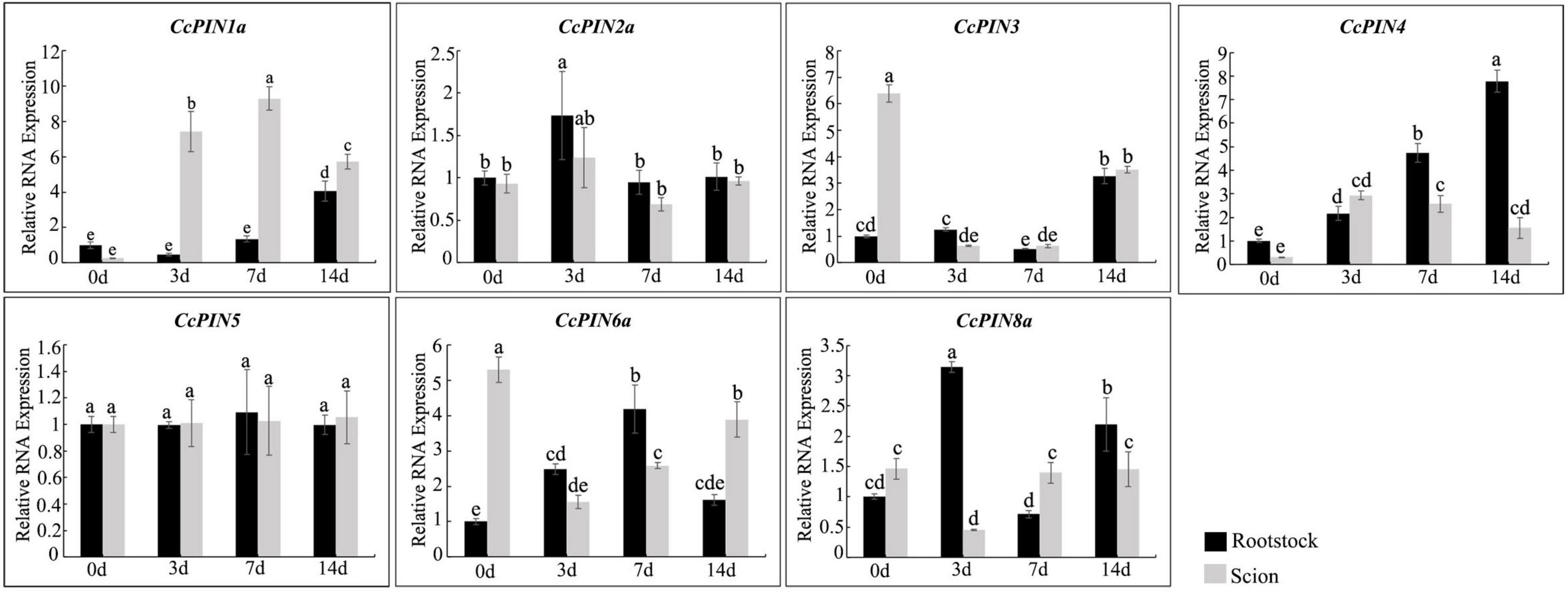
Figure 8. Relative RNA expression of CcPIN genes during the grafting of Chinese hickory. 0, 3, 7, and 14 d denote 0, 3, 7, and 14 days after grafting, respectively. The rootstocks collected at 0 day were regarded as the control sample. Different letters above the columns denote a significant difference in different treatments (P < 0.05).
To investigate how CcPINs respond to salt stress, the expression of CcPINs in the roots, stems, and leaves of Chinese hickory was determined at different time points in the salt stress treatment. CcPINs showed different expression profiles in different tissues (Figure 9). In roots, CcPIN1a was significantly upregulated after the salt stress treatment; CcPIN2a, CcPIN3, and CcPIN4 were downregulated at 3 days after the salt stress treatment; and CcPIN8a was downregulated at 1 day after the salt stress treatment. In stems, CcPIN1a, CcPIN3, and CcPIN4 were upregulated at 1 day after the salt stress treatment, while CcPIN3 and CcPIN4 were downregulated at 3 days after the salt stress treatment. The expression levels of CcPIN5, CcPIN6a, and CcPIN8a were similar to those after the CK treatment. In the leaves, most of the CcPIN genes were downregulated at 3 days after the salt stress treatment, while the expression of CcPIN8a was upregulated. The different expression profiles of CcPINs indicated that some of them might take part in the regulation of salt stress in Chinese hickory.
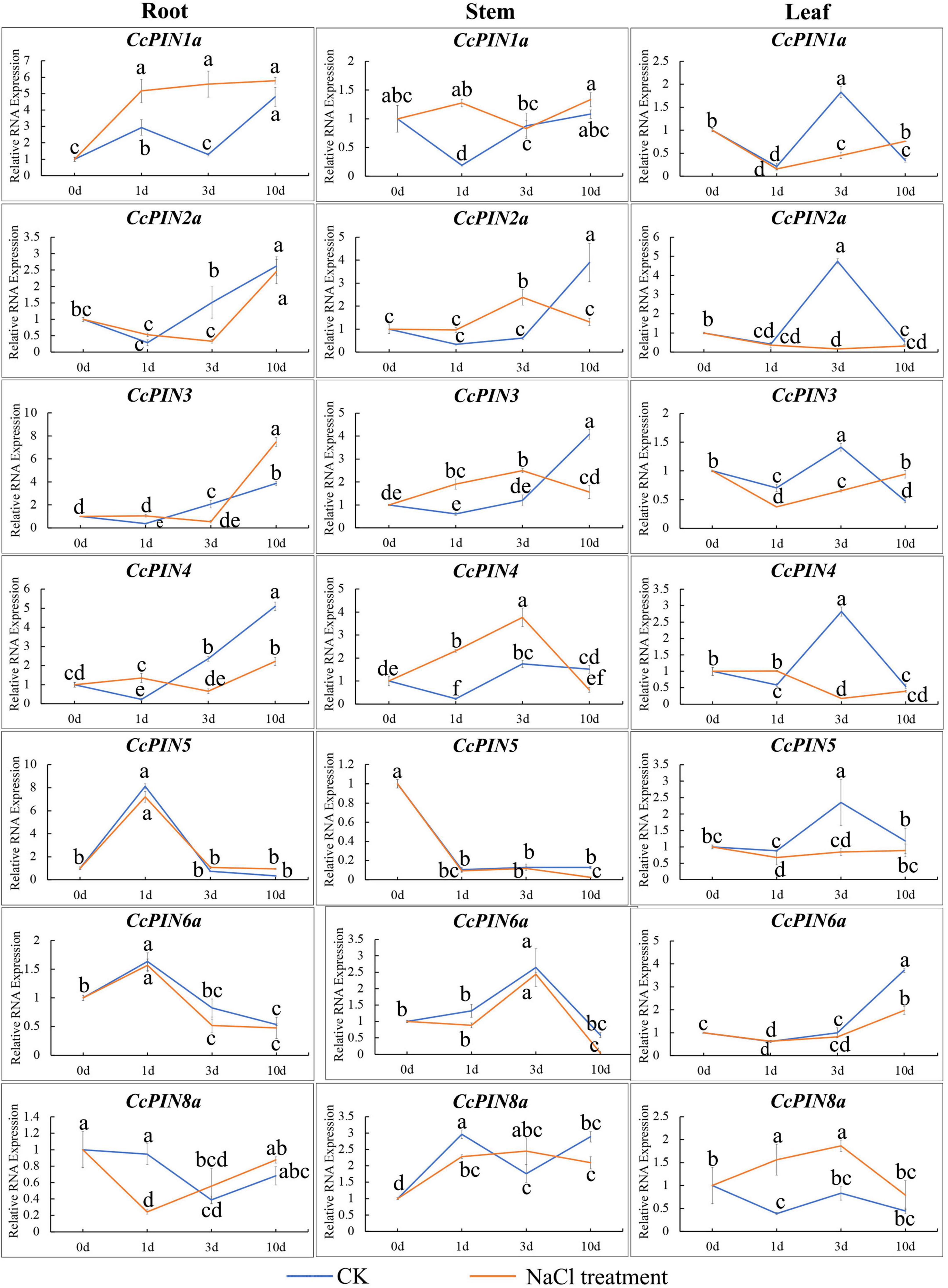
Figure 9. Relative RNA expression of CcPIN genes under CK (H2O) and salt stress treatments. 0, 1, 3, and 10 d represent 0, 1, 3, and 10 days after treatment, respectively. The sample from the CK group collected 0 days after treatment was regarded as the control sample. Different letters near the points indicate a significant difference in different treatments (P < 0.05).
Chinese hickory is a native nut tree in China that is widely cultivated in Zhejiang and Anhui provinces. The nuts of Chinese hickory are not only rich in nutrition such as saturated and unsaturated fatty acids, proteins, essential amino acids for humans, and trace elements (Peng et al., 2013; Huang et al., 2022) but also have an important role in the medicinal field (Gao et al., 2020). High nutritional values have brought huge economic benefits for farmers. In 2021, the total output value of Chinese hickory in Lin’an distinct (Hangzhou, China) was 845 million RMB (∼123.3 million dollars) (Gu, 2022). However, the slow vegetative growth and narrow growing range have limited the development of Chinese hickory. Grafting is a technique that is commonly used in horticulture and has been applied to Chinese hickory to solve the difficulties in the Chinese hickory industry. In addition, it has been predicted that the salt in soil will be increased with increasing global temperatures (Dhankher and Foyer, 2018). Therefore, the study of grafting and salt stress in Chinese hickory is essential. PIN is an auxin efflux protein that participates in the regulation of plant grafting and salt stress (Wang et al., 2014; Chen et al., 2015). In the present study, we identified eleven PIN genes in Chinese hickory according to the published genome. The expression profiles in grafting and salt stress were determined to elucidate the function of CcPINs.
The number of identified CcPIN genes (11) was more than that in Arabidopsis (8). Phylogenetic analysis of 132 PIN proteins in 15 plant species was carried out to compare with CcPINs. On the phylogenetic tree, CcPINs were close to CilPINs in C. illinoinensis, which indicated that they might come from a common ancestor. To further study the similarity and diversity of CcPIN genes, an analysis of motif and intron/exon structure was executed. The exons of CcPINs were similar, and the various intron lengths were the main reason for different gene lengths, with similar results in other species (Wang et al., 2009; Hu et al., 2021).
PIN proteins play important roles in cellular auxin transport, and their function relies on corresponding sites in conserved regions (Zhou and Luo, 2018). In contrast, functional sites distributed in non-conserved regions might be unique in different species, and the phenomenon is related to sequence evolution (Zwiewka et al., 2019). In Arabidopsis, PIN genes consist of two conserved hydrophobic loops at the N- and C-terminus and a variable hydrophilic loop in the middle (Zhou and Luo, 2018). The analysis of multiple sequence alignments and motifs showed that there were similar motifs for CcPINs, and the motifs were distributed in a gene-conserved area. It was determined that F165 in AtPIN1 controls the polar distribution of the PIN protein (Sancho-Andrés et al., 2016). Functional F165 sites were also found in CcPIN proteins, indicating that they might perform the same function.
TPRXS, a phosphorylation site, appeared in motif 8 (Figure 3), and control of the polar distribution of PIN proteins was performed by phosphorylase kinase (Bassukas et al., 2022). The NPNXY site on the conserved domain is also related to PIN transport. Mutants of NPNXY in AtPIN1 contributed to the accumulation of the PIN1 protein on the endoplasmic reticulum (Mravec et al., 2009). The multiple sequence alignment results show that all the CcPINs contained NPXXY in the conserved region, which suggests that the NPXXY motif performed the functional equivalent in Chinese hickory.
The plant hormone auxin regulates various developmental processes through its asymmetrical distribution on the PM (Vanneste and Friml, 2009). The different auxin distribution is executed by PINs (Wisniewska et al., 2006), and PIN-mediated auxin polar transport is crucial for auxin homeostasis. The PPI network predicted the multiple protein interactions with CcPINs through their homologous AtPINs genes. GO annotation showed that the predicted proteins were involved in many processes, and they participated in the regulation of auxin homeostasis together.
PINs are auxin efflux proteins that play important roles in plant life (Yu et al., 2022). To further explore the function of CcPINs, the expression levels from different tissues, including roots, stems, leaves, and shoots in Chinese hickory, were analyzed by qRT-PCR. The results showed that there was a high tissue-specific expression in the leaves, stems, and shoots of Chinese hickory, and the results were in accordance with those from previous studies. As previously reported, D6 PROTEIN KINASE and PINOID (PID)/WAG kinases activated auxin efflux transport PINs and influenced stem development (Wulf et al., 2019). In the woody plant Populus, PtaPIN1 regulated stem growth, with high expression in the developing xylem (Carraro et al., 2012). OsPIN5a and OSPIN5c in rice were highly expressed in leaves. PbPIN1-3, PbPIN2-2, PbPIN5-1, and PbPIN6 in pear trees were highly expressed in shoots (Qi et al., 2020). The tissue-specific transcriptional levels indicate that CcPINs participate in the regulation of hickory growth and development. Therefore, it is worthy of further research to clarify the detailed regulation process in Chinese hickory.
In the successful grafting of horticultural plants, the adhesion of different stems from rootstock and scion, namely the formation of the graft union, is necessary for grafting survival. During the process, callus formation and differentiation, and the reconnection of vascular tissues, are prominent (Melnyk et al., 2018; Sharma and Zheng, 2019). Auxin contributes to callus formation after a plant is wounded (Wulf et al., 2019). The imposed exogenous auxin also accelerates vascular reconnection (Wulf et al., 2019). In Chinese hickory grafting studies, morphological changes indicated that 3, 7, and 14 days after grafting were the crucial time points for isolation layer formation, callus formation, and vascular reconnection (Liu et al., 2009). Auxin-related genes, including CcIAAs and CcGH3, exhibited different expression levels at 0, 3, 7, and 14 days after grafting.
During Arabidopsis grafting, it has been proved that AtPIN1 participates in grafting with a high expression on the scion (Melnyk et al., 2018). To explore if auxin efflux proteins coding for CcPIN genes participated in the grafting process, the relative RNA expression of CcPINs was measured in rootstocks and scions at different time points by qRT-PCR. During the obtained results, the relative RNA expression level of CcPIN1a was upregulated 6–9 fold at the time points of 0, 3, 7, and 14 days after grafting. CcPIN1a is the ortholog of AtPIN1, indicating they might perform a similar function. Further study is necessary to elucidate the additional functions of CcPIN1a with respect to grafts. In addition to CcPIN1a, other PIN genes exhibited varied expression levels in grafting except for CcPIN5. The various expression profiles of CcPIN genes in different sections at the key stages of hickory grafting indicate their potential functions in grafting regulation.
When faced with salt stress, plants activate various mechanisms to resist the adverse environment. Auxin homeostasis and dynamic rearrangement are one of the regulatory pathways. A previous study showed that phospholipase D-derived phosphatidic acid combined with PID (PINOID/WAVY ROOT GROWTH) kinases enhanced the phosphorylation of PIN2, promoted auxin polar efflux ability, and contributed to the salt tolerance of Arabidopsis (Wang et al., 2019). PIN1 and PIN3 also participated in the process (Wang et al., 2019). During salt stress in watermelon, ClPINs performed functions of resisting salt with varying expressions (Yu et al., 2017). In the present study, samples of roots, stems, and leaves under salt stress were collected to explore the CcPINs’ expression profiles of anti-salt ability in Chinese hickory. As shown in Figure 9, the expression of CcPIN2a in roots and leaves was significantly downregulated, with variations similar to those in the previous report (Wang et al., 2019).
To investigate the potential roles of auxin efflux transporters during stress treatments, eleven CcPIN genes with an ORF of 1,026–1,983 bp were identified in Chinese hickory for the first time, and their structural characteristics and expression responses to grafting and salt stress were detected by qRT-PCR. CcPINs were localized on the PM and had the closest relationships with homologous genes in C. illinoinensis and J. regia. Different hormone- and stress-responsive elements were detected on the promoter of CcPINs, indicating their expression regulation by stress treatments. CcPINs performed different functions during grafting and salt treatment. CcPIN1a has the potential to respond to grafting, while CcPIN1a and CcPIN8a might be involved in salt-stress regulation. In addition, CcPINs displayed different expression levels in different tissues, suggesting their varying roles during growth and development. Further investigations might be conducted on the mechanism of CcPINs for grafting and salt stress regulation.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
HY, BZ, XW, and DY conceived and designed the concept of the manuscript. YY (1st author), JM, JC, YY (4th author), and KY performed the experiments. YG, XT, HL, and HY analyzed the data. YY (1st author), JM, YG, and JC performed the formal analysis. YY (1st author), JM, and HY drafted the manuscript. AS, XW, DY, RW, BZ, and HY revised and finalized the manuscript. All authors endorsed the final version of the manuscript.
This study was supported by the National Natural Science Foundation of China (32071807, 31901346, and 31971695); Key Project of Zhejiang Provincial Natural Science Foundation (LZ18C160001); National Key Research and Development Program of China (2018YFD1000604); Independent Research Project of State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University (ZY20180208 and ZY20180308); Open Foundation of State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University (KF201903, KF202007, and KF201708); The Talent Foundation of Zhejiang A&F University (2022LFR001); Overseas Expertise Introduction Project for Discipline Innovation (111 Project D18008); Key Research and Development Program of Zhejiang Province (2018C02004); National High Technology Research and Development Program of China (863 Program) (2013AA102605); Fruit Innovation Team Project of Zhejiang Province (2016C02052-12); Key Agricultural New Varieties Breeding Projects founded by the Zhejiang Province Science and Technology Department (2021C02066-12 and 2016C02052-13); Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholar (LR13C160001); Open Foundation of First-Class Discipline of Forestry, Zhejiang Province (201703); and the First-Class General Financial Grant from the China Postdoctoral Science Foundation (2017M610377).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.999990/full#supplementary-material
Alabdallah, O., Ahou, A., Mancuso, N., Pompili, V., Macone, A., Pashkoulov, D., et al. (2017). The Arabidopsis polyamine oxidase/dehydrogenase 5 interferes with cytokinin and auxin signaling pathways to control xylem differentiation. J. Exp. Bot. 68, 997–1012. doi: 10.1093/jxb/erw510
Bassukas, A. E. L., Xiao, Y., and Schwechheimer, C. (2022). Phosphorylation control of PIN auxin transporters. Curr. Opin. Plant Biol. 65:102146. doi: 10.1016/j.pbi.2021.102146
Bennett, T., Brockington, S., Rothfels, C., Graham, S., Stevenson, D., Kutchan, T., et al. (2014). Paralogous radiations of PIN proteins with multiple origins of noncanonical PIN structure. Mol. Biol. Evol. 31, 2042–2060. doi: 10.1093/molbev/msu147
Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., et al. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. doi: 10.1038/nature03184
Bureau, M., Rast, M., Illmer, J., and Simon, R. (2010). JAGGED LATERAL ORGAN (JLO) controls auxin dependent patterning during development of the Arabidopsis embryo and root. Plant Mol. Biol. 74, 479–491. doi: 10.1007/s11103-010-9688-2
Carraro, N., Tisdale-Orr, T. E., Clouse, R. M., Knoller, A. S., and Spicer, R. (2012). Diversification and expression of the PIN, AUX/LAX, and ABCB families of putative auxin transporters in Populus. Front. Plant Sci. 3:17. doi: 10.3389/fpls.2012.00017
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Y., Cai, J., Yang, F. X., Zhou, B., and Zhou, L. R. (2015). Ascorbate peroxidase from Jatropha curcas enhances salt tolerance in transgenic Arabidopsis. Genet. Mol. Res. 14, 4879–4889. doi: 10.4238/2015.May.11.20
Dastborhan, S., Kalisz, A., Kordi, S., Lajayer, B. A., and Pessarakli, M. (2021). “Physiological and biochemical responses of plants to drought and oxidative stresses,” in Handbook of plant and crop physiology, ed. M. Pessarakli (Boca Raton, FL: CRC Press), 517–541.
Dhankher, O. P., and Foyer, C. H. (2018). Climate resilient crops for improving global food security and safety. Plant Cell Environ. 41, 877–884.
Ding, Z., Wang, B., Moreno, I., Dupláková, N., Simon, S., Carraro, N., et al. (2012). ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 3:941. doi: 10.1038/ncomms1941
Doyle, J. A. (2018). Phylogenetic analyses and morphological innovations in land plants. Annu. Plant Rev. 45, 1–50. doi: 10.1002/9781119312994.apr0486
Gao, F., Wu, J., Zhou, Y., Huang, J., Lu, J., and Qian, Y. (2020). An appropriate ratio of unsaturated fatty acids is the constituent of hickory nut extract for neurite outgrowth in human SH-SY5Y cells. Food Sci. Nutr. 8, 6346–6356. doi: 10.1002/fsn3.1623
Gibson, C. L., Isley, J. W., Falbel, T. G., Mattox, C. T., Lewis, D. R., Metcalf, K. E., et al. (2020). A conditional mutation in SCD1 reveals linkage between PIN protein trafficking, auxin transport, gravitropism, and lateral root initiation. Front. Plant Sci. 11:910. doi: 10.3389/fpls.2020.00910
Gomes, G. L. B., and Scortecci, K. C. (2021). Auxin and its role in plant development: Structure, signalling, regulation and response mechanisms. Plant Biol. 23, 894–904. doi: 10.1111/plb.13303
Gu, X. B. (2022). Small Chinese hickory nuts has brought economic benefits for industry development in Lin’an. Zhejiang Forest. 5, 10–11.
Hang, L. T., Mori, K., Tanaka, Y., Morikawa, M., and Toyama, T. (2020). Enhanced lipid productivity of Chlamydomonas reinhardtii with combination of NaCl and CaCl2 stresses. Bioprocess Biosyst. Eng. 43, 971–980. doi: 10.1007/s00449-020-02293-w
Hu, L., Wang, P., Long, X., Wu, W., Zhang, J., Pan, Y., et al. (2021). The PIN gene family in relic plant L. chinense: Genome-wide identification and gene expression profiling in different organizations and abiotic stress responses. Plant Physiol. Biochem. 162, 634–646. doi: 10.1016/j.plaphy.2021.03.030
Huang, C., Li, Y., Wang, K., Xi, J., Xu, Y., Si, X., et al. (2022). Analysis of lipidomics profile of Carya cathayensis nuts and lipid dynamic changes during embryonic development. Food Chem. 370, 130975. doi: 10.1016/j.foodchem.2021.130975
Huang, J., Wang, S., Wang, X., Fan, Y., and Han, Y. (2020). Structure and expression analysis of seven salt-related ERF genes of Populus. PeerJ. 20:e10206. doi: 10.7717/peerj.10206
Huang, Y., Xiao, L., Zhang, Z., Zhang, R., Wang, Z., Huang, C., et al. (2019). The genomes of pecan and Chinese hickory provide insights into Carya evolution and nut nutrition. GigaScience 8, giz036. doi: 10.1093/gigascience/giz036
Kapazoglou, A., Tani, E., Avramidou, E. V., Abraham, E. M., Gerakari, M., Megariti, S., et al. (2020). Epigenetic changes and transcriptional reprogramming upon woody plant grafting for crop sustainability in a changing environment. Front. Plant Sci. 11:613004. doi: 10.3389/fpls.2020.613004
Kuhlemeier, C., and Reinhardt, D. (2001). Auxin and phyllotaxis. Trends Plant Sci. 6, 187–189. doi: 10.1016/s1360-1385(01)01894-5
Lewis, D., Negi, S., Sukumar, P., and Muday, G. (2011). Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development 138, 3485–3495. doi: 10.1242/dev.065102
Li, R., Pan, Y., Hu, L., Yang, D., Yuan, M., Hao, Z., et al. (2022). PIN3 from liriodendron may function in inflorescence development and root elongation. Forests 13:568. doi: 10.3390/f13040568
Liu, C., Hong, J., Xia, G., and Huang, J. (2009). Cytological observation on healing responses in grafting of Carya cathayensis. Sci. Silvae Sin. 45, 34–38.
Mearaji, H. S., Ansari, A., Igdelou, N. K. M., Lajayer, B. A., and Pessarakli, M. (2021). “Phytohormones and abiotic stresses: Roles of phytohormones in plants under abiotic stresses,” in Handbook of plant and crop physiology, ed. M. Pessarakli (Boca Raton, FL: CRC Press), 175–213.
Melnyk, C. W. (2017). Plant grafting: Insights into tissue regeneration. Regeneration 4, 3–14. doi: 10.1002/reg2.71
Melnyk, C. W., Gabel, A., Hardcastle, T. J., Robinson, S., Miyashima, S., Grosse, I., et al. (2018). Transcriptome dynamics at Arabidopsis graft junctions reveal an intertissue recognition mechanism that activates vascular regeneration. Proc. Natl. Acad. Sci. U.S.A. 115, E2447–E2456. doi: 10.1073/pnas.1718263115
Mohanta, T. K., Bashir, T., Hashem, A., Abd_Allah, E. F., Khan, A. L., and Al-Harrasi, A. S. (2018). Molecular players of auxin transport systems: Advances in genomic and molecular events. J. Plant Interact. 13, 483–495. doi: 10.1080/17429145.2018.1523476
Mravec, J., Skupa, P., Bailly, A., Hoyerova, K., Krecek, P., Bielach, A., et al. (2009). Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459, 1136–1140. doi: 10.1038/nature08066
Muday, G., and DeLong, A. (2001). Polar auxin transport: Controlling where and how much. Trends Plant Sci. 6, 535–542. doi: 10.1016/s1360-1385(01)02101-x
Nelson, B. K., Cai, X., and Nebenühr, A. (2007). A multicolor set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x
Peng, Q., Bian, W., Wang, J. L., Wu, J., Ge, L. Y., Wang, F., et al. (2013). Effect of production techniques on nutrient content in Li’an pecans. Sci. Technol. Food Ind. 34, 173–175. doi: 10.13386/j.issn1002-0306.2013.20.056
Qi, L., Chen, L., Wang, C., Zhang, S., Yang, Y., Liu, J., et al. (2020). Characterization of the auxin efflux transporter PIN proteins in pear. Plants (Basel) 9:349. doi: 10.3390/plants9030349
Robert, H., and Friml, J. (2009). Auxin and other signals on the move in plants. Nat. Chem. Biol. 5, 325–332. doi: 10.1038/nchembio.170
Sancho-Andrés, G., Soriano-Ortega, E., Gao, C., Bernabé-Orts, J. M., Narasimhan, M., Müller, A. O., et al. (2016). Sorting motifs involved in the trafficking and localization of the PIN1 auxin efflux carrier. Plant Physiol. 171, 1965–1982. doi: 10.1104/pp.16.00373
Sauer, M., and Kleine-Vehn, J. (2019). PIN-FORMED and PIN-LIKES auxin transport facilitators. Development 146:dev168088. doi: 10.1242/dev.168088
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108.
Sharma, A., and Zheng, B. (2019). Molecular responses during plant grafting and its regulation by auxins, cytokinins, and gibberellins. Biomolecules 9:397. doi: 10.3390/biom9090397
Shen, C., Bai, Y., Wang, S., Zhang, S., Wu, Y., Chen, M., et al. (2010). Expression profile of PIN, AUX/LAX and PGP auxin transporter gene families in Sorghum bicolor under phytohormone and abiotic stress. FEBS J. 277, 2954–2969. doi: 10.1111/j.1742-4658.2010.07706.x
Swarup, R., and Peret, B. (2012). AUX/LAX family of auxin influx carriers-an overview. Front. Plant Sci. 3:225. doi: 10.3389/fpls.2012.00225
Vanneste, S., and Friml, J. (2009). Auxin: A trigger for change in plant development. Cell 136, 1005–1016. doi: 10.1016/j.cell.2009.03.001
Vieten, A., Vanneste, S., Wisìniewska, J., Benkovaì, E., Benjamins, R., Beeckman, T., et al. (2005). Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132, 4521–4531. doi: 10.1242/dev.02027
Wang, J., Guo, X., Xiao, Q., Zhu, J., Cheung, A. Y., Yuan, L., et al. (2021). Auxin efflux controls orderly nucellar degeneration and expansion of the female gametophyte in Arabidopsis. New Phytol. 230, 2261–2274. doi: 10.1111/nph.17152
Wang, J., Jin, Z., Yin, H., Yan, B., Ren, Z. Z., Xu, J., et al. (2014). Auxin redistribution and shifts in PIN gene expression during Arabidopsis grafting. Russ. J. Plant Physiol. 61, 688–696. doi: 10.1134/s102144371405015x
Wang, J.-R., Hu, H., Wang, G.-H., Li, J., Chen, J.-Y., and Wu, P. (2009). Expression of PIN genes in rice (Oryza sativa L.): Tissue specificity and regulation by hormones. Mol. Plant. 2, 823–831. doi: 10.1093/mp/ssp023
Wang, P., Shen, L., Guo, J., Jing, W., Qu, Y., Li, W., et al. (2019). Phosphatidic acid directly regulates PINOID-dependent phosphorylation and activation of the PIN-FORMED2 auxin efflux transporter in response to salt stress. Plant Cell 31, 250–271. doi: 10.1105/tpc.18.00528
Wisniewska, J., Xu, J., Seifertová, D., Brewer, P. B., Ruzicka, K., Blilou, I., et al. (2006). Polar PIN localization directs auxin flow in plants. Science 312, 883–883. doi: 10.1126/science.1121356
Wulf, K. E., Reid, J. B., and Foo, E. (2019). Auxin transport and stem vascular reconnection–has our thinking become canalized? Ann. Bot. 123, 429–439. doi: 10.1093/aob/mcy180
Yamaji, N., Huang, C. F., Nagao, S., Yano, M., Sato, Y., Nagamura, Y., et al. (2009). A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 21, 3339–3349. doi: 10.1105/tpc.109.070771
Yang, Y., Huang, Q., Wang, X., Mei, J., Sharma, A., Tripathi, D. K., et al. (2021). Genome-wide identification and expression profiles of ABCB gene family in Chinese hickory (Carya cathayensis Sarg.) during grafting. Plant Physiol. Biochem. 168, 477–487. doi: 10.1016/j.plaphy.2021.10.029
Yu, C., Dong, W., Zhan, Y., Huang, Z. A., Li, Z., Kim, I. S., et al. (2017). Genome-wide identification and expression analysis of ClLAX, ClPIN and ClABCB genes families in Citrullus lanatus under various abiotic stresses and grafting. BMC Genet. 18:33. doi: 10.1186/s12863-017-0500-z
Yu, Z., Zhang, F., and Ding, Z. (2022). Auxin signaling: Research advances over the past 30 years. J. Integr. Plant Biol. 2, 371–392. doi: 10.1111/jipb.13225
Yuan, H., Zhao, L., Chen, J., Yang, Y., Xu, D., Tao, S., et al. (2018). Identification and expression profiling of the Aux/IAA gene family in Chinese hickory (Carya cathayensis Sarg.) during the grafting process. Plant Physiol. Biochem. 127, 55–63. doi: 10.1016/j.plaphy.2018.03.010
Zhai, S., Cai, W., Xiang, Z. X., Chen, C., Lu, Y. T., and Yuan, T. T. (2021). PIN3-mediated auxin transport contributes to blue light-induced adventitious root formation in Arabidopsis. Plant Sci. 312:111044. doi: 10.1016/j.plantsci.2021.111044
Zhang, C., Dong, W., Huang, Z. A., Cho, M., Yu, Q., Wu, C., et al. (2018). Genome-wide identification and expression analysis of the CaLAX and CaPIN gene families in pepper (Capsicum annuum L.) under various abiotic stresses and hormone treatments. Genome 61, 121–130. doi: 10.1139/gen-2017-0163
Zhang, H., Zhu, J., Gong, Z., and Zhu, J.-K. (2022). Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119. doi: 10.1038/s41576-021-00413-0
Zhang, J., and Peer, W. A. (2017). Auxin homeostasis: The DAO of catabolism. J. Exp. Bot. 68, 3145–3154. doi: 10.1093/jxb/erx221
Zhao, R., Sun, H., Zhao, N., Jing, X., Shen, X., and Chen, S. (2015). The Arabidopsis Ca2+-dependent protein kinase CPK27 is required for plant response to salt-stress. Gene 563, 203–214. doi: 10.1016/j.gene.2015.03.024
Zhao, Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61, 49–64. doi: 10.1146/annurev-arplant-042809-112308
Zhou, J. J., and Luo, J. (2018). The PIN-FORMED auxin efflux carriers in plants. Int. J. Mol. Sci. 19, 2759. doi: 10.3390/ijms19092759
Keywords: Carya cathayensis, PIN, transport, auxin, salt, grafting
Citation: Yang Y, Mei J, Chen J, Yang Y, Gu Y, Tang X, Lu H, Yang K, Sharma A, Wang X, Yan D, Wu R, Zheng B and Yuan H (2022) Expression analysis of PIN family genes in Chinese hickory reveals their potential roles during grafting and salt stress. Front. Plant Sci. 13:999990. doi: 10.3389/fpls.2022.999990
Received: 21 July 2022; Accepted: 29 August 2022;
Published: 29 September 2022.
Edited by:
Tongli Wang, University of British Columbia, CanadaReviewed by:
Behnam Asgari Lajayer, University of Tabriz, IranCopyright © 2022 Yang, Mei, Chen, Yang, Gu, Tang, Lu, Yang, Sharma, Wang, Yan, Wu, Zheng and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huwei Yuan, aHd5dWFuQHphZnUuZWR1LmNu; Bingsong Zheng, YnN6aGVuZ0B6YWZ1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.