
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 28 September 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.998841
This article is part of the Research Topic Physiological, Biochemical and Molecular Approaches in Response to Abiotic Stresses in Plants View all 17 articles
 Jinhao Sun1,2†
Jinhao Sun1,2† Zhen Tian1†
Zhen Tian1† Xiaoxu Li2†
Xiaoxu Li2† Shaopeng Li1
Shaopeng Li1 Zhiyuan Li2
Zhiyuan Li2 Jinling Wang1
Jinling Wang1 Zongyu Hu1
Zongyu Hu1 Haiqing Chen1
Haiqing Chen1 Cun Guo2,3
Cun Guo2,3 Minmin Xie2*
Minmin Xie2* Ruyan Xu1*
Ruyan Xu1*The pectin methylesterases (PMEs) play multiple roles in regulating plant development and responses to various stresses. In our study, a total of 121 PME genes were identified in the tobacco genome, which were clustered into two groups based on phylogenetic analysis together with Arabidopsis members. The investigations of gene structure and conserved motif indicated that exon/intron and motif organizations were relatively conserved in each group. Additionally, several stress-related elements were identified in the promoter region of these genes. The survey of duplication events revealed that segmental duplications were critical to the expansion of the PME gene family in tobacco. The expression profiles analysis revealed that these genes were expressed in various tissues and could be induced by diverse abiotic stresses. Notably, NtPME029 and NtPME043, were identified as homologues with AtPME3 and AtPME31, respectively. Furthermore, NtPME029 was highly expressed in roots and the over-expression of the NtPME029 gene could promote the development of roots. While NtPME043 could be induced by salt and ABA treatments, and the over-expression of the NtPME043 gene could significantly enhance the salt-stress tolerance in tobacco. Overall, these findings may shed light on the biological and functional characterization of NtPME genes in tobacco.
The plant cell walls mainly contain structural proteins and three types of polysaccharides, including cellulose, hemicellulose, and pectins (Zhao et al., 2019). Pectins as highly heterogeneous polymers exist between the intercellular layer and the primary wall and it accounts for 30-40% of the cell wall of dicots and non-graminaceous monocots. The pectin modification leads to crucial changes in plant cell walls’ structure and function, affecting plant development, including fruit ripening and softening, organ abscission and senescence, pollen dehiscence, pollen tube growth, and stress resistance (Al-Qsous et al., 2004; Bosch et al., 2005; An et al., 2008; Zhang et al., 2010; Verma et al., 2014; Wallace and Williams, 2017). Pectins are synthesized with a highly methylesterified form and then are catalyzed to decrease methyl esterification by a large enzyme family of pectin methylesterases (PMEs) (EC. 3.1.1.11) that reside in the cell wall (Yapo et al., 2007; Wolf et al., 2009).
In plants, PMEs can be divided into two groups (Type I and Type II) according to whether it contains the PMEI (pectin methylesterase inhibitor domain) domain. Type I (PME type) contain the PME domain and the pro-region of its N-terminal, which is very short or missing. However, type II (ProPME type) contains simultaneously PME and PMEI domain and a long N-end pro-region (Udall et al., 2006). The pro-region was reported to fold the protein correctly and inhibit the PME activity (Micheli, 2001; Pelloux et al., 2007). Previous research has reported that there are five conserved structure regions, including Region I (GxYxE), Region II (QAVAxR), Region III (QDTL), Region IV (DFIFG), and Region V (YLGRxWx) in the secondary structure of PME proteins (Markovic and Janecek, 2004). The Region I is conserved in all PME proteins, while the remaining regions have diverse conservation in different plants.
A total of 66 PMEs have been identified in the Arabidopsis, 22 members falling into type I and 44 into type II, and the contributions of some PMEs to various developmental processes have been characterized (Tian et al., 2006; Levesque-Tremblay et al., 2015). For example, AtPME6 and AtPME58 express highly during mucilage secretion, which is involved in embryo development and mucilage extrusion, respectively (Levesque-Tremblay et al., 2015; Turbant et al., 2016). In addition, the knockout mutants of AtPME1 specifically exhibit curved, irregular morphology and are significantly stunted in the pollen tube compared that with wild type (Tian et al., 2006). AtPME48 specifically expressed in the male gametophyte and functioned to influence pollen grain germination (Leroux et al., 2015). Furthermore, AtPME5-overexpressing transgenic plants were reported to increase the cell walls elasticity in shoot apical meristem (Peaucelle et al., 2011). AtPME35 influences the mechanical strength of the stem by regulating the PME demethylesterifification (Hongo et al., 2012). AtPME17 is significantly expressed in vegetative tissues and could be processed by SBT3.5 (subtilisin-type serine protease 3.5) to modify PME activity which is involved in root growth (Sénéchal et al., 2014). Moreover, AtPME3-overexpressing plants showed longer roots compared to wild-type, whereas the RNAi plants showed the opposite phenotype (Hewezi et al., 2008).
Besides, many PMEs facilitate responses to multiple biotic and abiotic stresses in Arabidopsis. Over-expressed AtPME31 provides broad-spectrum insect resistance in transgenic tobacco plants (Dixit et al., 2013). Moreover, AtPME31 could be significantly induced under salt stress and positively modulate the expression levels of salt stress-induced genes to enhance the salt stress tolerance of plants (Yan et al., 2018). AtPME34 is proposed that responds to heat and salt stress through ABA (abscisic acid) pathway (Huang et al., 2017). It was verified that AtPME41 contributes to freezing tolerance by modulating the mechanical properties of cell walls through the BR (brassinosteroid) signaling (Qu et al., 2011).
Tobacco is one of the most significant economic crops and widely cultivated all over the world, while various abiotic stresses can severely threaten tobacco quality and yield. The PME genes have been reported to be involved in response to abiotic stresses and affect development in a variety of species. Although the PME family proteins have been characterized in several plants (Duan et al., 2016; Zega and D'Ovidio, 2016; Zhang et al., 2019; Li et al., 2020), limited information is available about PME family proteins in tobacco. In this study, a comprehensive analysis was performed, including phylogenetic analyses, gene structure, motif organization, cis-elements, chromosomal distributions, duplication events, and expression profiles. The results of the current study imply that the tobacco NtPME family proteins play multiple roles in various biological processes.
The full-length protein sequences for each AtPME in Arabidopsis were downloaded from TAIR (The Arabidopsis Information Resource) database (http://www.arabidopsis.org/), while the genomic data for tobacco (N. tabacumV1.0) were obtained from the SGN (Sol Genomics Network) database (https://solgenomics.net/). Then, the protein sequences of AtPME were used as queries to detect the potential NtPME proteins in tobacco by the BLASTp searches (E-value < 0.01) (Li et al., 2021). Subsequently, the potential sequences were examined using the SMART (Letunic et al., 2015) and PFAM (Finn et al., 2016) databases to determine the PME domains. The remaining NtPME numbers were renamed according to their physical locations on the chromosome or scaffold. The Mw (molecular weight) and theoretical pI (isoelectric points) of these finally identified NtPME numbers were predicted by ProtParam toolkits (Garg et al., 2016). The sequences of NtPME and AtPME proteins were aligned using Clustal X (Larkin et al., 2007), and the alignments were used to construct the neighbor-joining (NJ) tree by MEGA 6 with the default parameters method (Kumar et al., 2018). The subcellular localization of NtPME proteins was also predicted by Cell-PLoc 2.0 and WOLF POSRT II (https://www.genscript.com/psort.html) (Ren et al., 2019).
The chromosomal location image of NtPME genes was visualized by MG2C, according to the data obtained from the SGN database (Chao et al., 2015). The segmental duplication events were identified and visualized by the MCScanX program and Circos, respectively (Cao et al., 2016). The syntenic analysis of the orthologous genes obtained from tobacco and other four plant species was investigated by Synteny Plotter (Xie et al., 2018). Then, the synonymous substitution (Ks) and non-synonymous substitution (Ka) rates were calculated by DnaSP 5.0 software (Librado and Rozas, 2009).
The gene structure of NtPME genes was analyzed and visualized using GSDS (http://gsds.cbi.pku.edu.cn) according to their genomic and coding sequence (Hu et al., 2015). The Multiple Em for Motif Elicitation (MEME) online tool was adopted to identify and visualize conserved motifs of the NtPME full-length protein sequences (Bailey et al., 2015). The promoter regions, 2000-bp sequences upstream of tobacco NtPME genes, were obtained by TBtools software (Chen et al., 2020), and then the cis-regulatory elements of these promoters were analyzed by the PlantCARE tool (Lescot et al., 2002).
The RNA-seq data for each tissue of NtPME genes in tobacco K326 were obtained from the GEO database (accession number: GSE95717) (Edwards et al., 2017). The absolute transcript abundance values of three tissues (root, shoot, and apex) of the NtPME genes were normalized and illustrated using R.
The Cultivated tobacco K326 (Nicotiana tabacum L. Cv. K326) was used in the current study. The tobacco plants were cultivated in the growth conditions described in the previous report (Li et al., 2021). Different tissues, including the shoots, roots, leaves, and flowers were harvested to explore tissue-specific expression patterns and visualized in a heat map by TBtools (Wu et al., 2020). For abiotic stress (salt and ABA) treatments, the seedlings were treated with 50 µM Abscisic Acid (ABA) or 150 mM NaCl, and then harvested at 0, 1, 3, or 6 h after treatment (Sun et al., 2021). All the collected samples were frozen by the liquid nitrogen and then stored at -80 °C for RNA extraction.
Total RNA was extracted and 1 µg RNA was synthesized to the first-strand complementary DNA (cDNA) by the method previously reported (Li et al., 2019a; Ren et al., 2019). The qRT-PCR was carried out on an ABI7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, United States) with 2.5 µL template cDNA. The ribosomal protein gene L25 (GenBank No. L18908) of tobacco was adopted as the internal control. All reactions were performed with three biological replications and the resulting data were evaluated by the 2 −ΔΔCT method (Livak et al., 2001). The specific relative primer sequences were designed by Primer Premier 5.0 and the details were shown in Supplementary Table S1.
The coding regions of NtPME029 and NtPME043 were amplified by PCR with specific cloning primers and then generated into the pCHF3-GFP vector. The NtPME029-GFP and NtPME043-GFP fragments were driven by the CaMV-35S promoter, and the control was the GFP fragment driven by the CaMV-35S promoter. Subsequently, these constructs were injected into Nicotiana benthamiana leaves separately for transient expression as previously reported (Li et al., 2019b). After 3 days of growth in a light condition, the GFP fluorescence signals in these leaves were monitored by using the confocal microscope (TCS-SP8 Leica, Wetzlar, Germany) (Li et al., 2018).
The coding sequences of NtPME029 and NtPME043 were amplified and then generated into the expression vector (pCAMBIA1300). The NtPME029 and NtPME043 over-expression vectors were transformed into K326 tobacco plants by an Agrobacterium-mediated method and the empty vector as the control (Buschmann, 2016). The T0-generation seeds were screened in half-strength MS medium with 20 mg/L of hygromycin to obtain T1 seeds as the previous report (Li et al., 2021). The one-week-old T1 plants and wild-type tobacco plants were transferred to MS medium with or without 150 mM NaCl to grow for two weeks. Subsequently, the changes in root length were statistically analyzed.
Student’s t-tests in GraphPad Prism 8 revealed significant differences from the control. P values less than 0.01 were considered significant. All data were analyzed in three replicates.
To identify PME family genes in tobacco, we used the PME numbers of Arabidopsis as references to the BLASTP search. A total of 121 PME genes were identified in tobacco and named NtPME001 to NtPME121 based on their physical locations on chromosomes or scaffolds (Supplementary Table S2). Subsequently, the biochemical characteristics of PME genes were analyzed. As shown in Supplementary Table S2, the ORF (open reading frame) lengths ranged from 348 bp (NtPME037) to 3348 bp (NtPME84) and their protein weights (MWs) ranged from 12.63 (NtPME037) to 124.45 kDa (NtPME84). The isoelectric points (PIs) values of different NtPME proteins ranged from 4.30 (NtPME087) to 10.47 (NtPME034). The prediction of subcellular localization indicated that all NtPME proteins were located on the cell wall.
To better elucidate the evolutionary relationship of PME family members in tobacco, the MEGA7.0 software was employed to construct a neighbor-joining (NJ) tree consisting of tobacco (121 members) and Arabidopsis (66 members). The 121 NtPME proteins could be clustered into two groups(Group I and Group II) according to the PME or PMEI domain contained in these proteins with previous support (Li et al., 2020). Group II was further subdivided into three subclasses Group II-a, Group II-b, and Group II-c in this neighbor-joining (NJ) tree (Figure 1). Group I included 30 NtPME members with only PME domains, accounting for 24.8% of the total numbers. Seven of the members contained two PME domains, including NtPME20, NtPME23, NtPME27, NtPME60, NtPME63, NtPME99. However, the remaining others contained only one PME domain. Group II included 91 NtPME members with simultaneously PME and PMEI domains, accounting for 75.2% of the total numbers. To further understand the number and proportion of NtPME gene family members in plant genomes, six representative plant species were counted the members and the proportion of PME and PMEI, such as tobacco, Arabidopsis, rice, potato, Asian cotton, and physcomitrella patens (Table 1). As shown in Table 1, the number of PME proteins in Group I was generally less than that in Group II in plant species except for rice. The number of genes in Group I is approximately the same in different plant species. Notably, the 13 PME numbers in physcomitrella patens all belong to Group I. This result indicated that the PME genes of Group II may appear after the evolutionary differentiation of bryophytes plants. However, the number of PME proteins of Group II in tobacco far exceeded other species, which may be since tobacco is allopolytraploid.
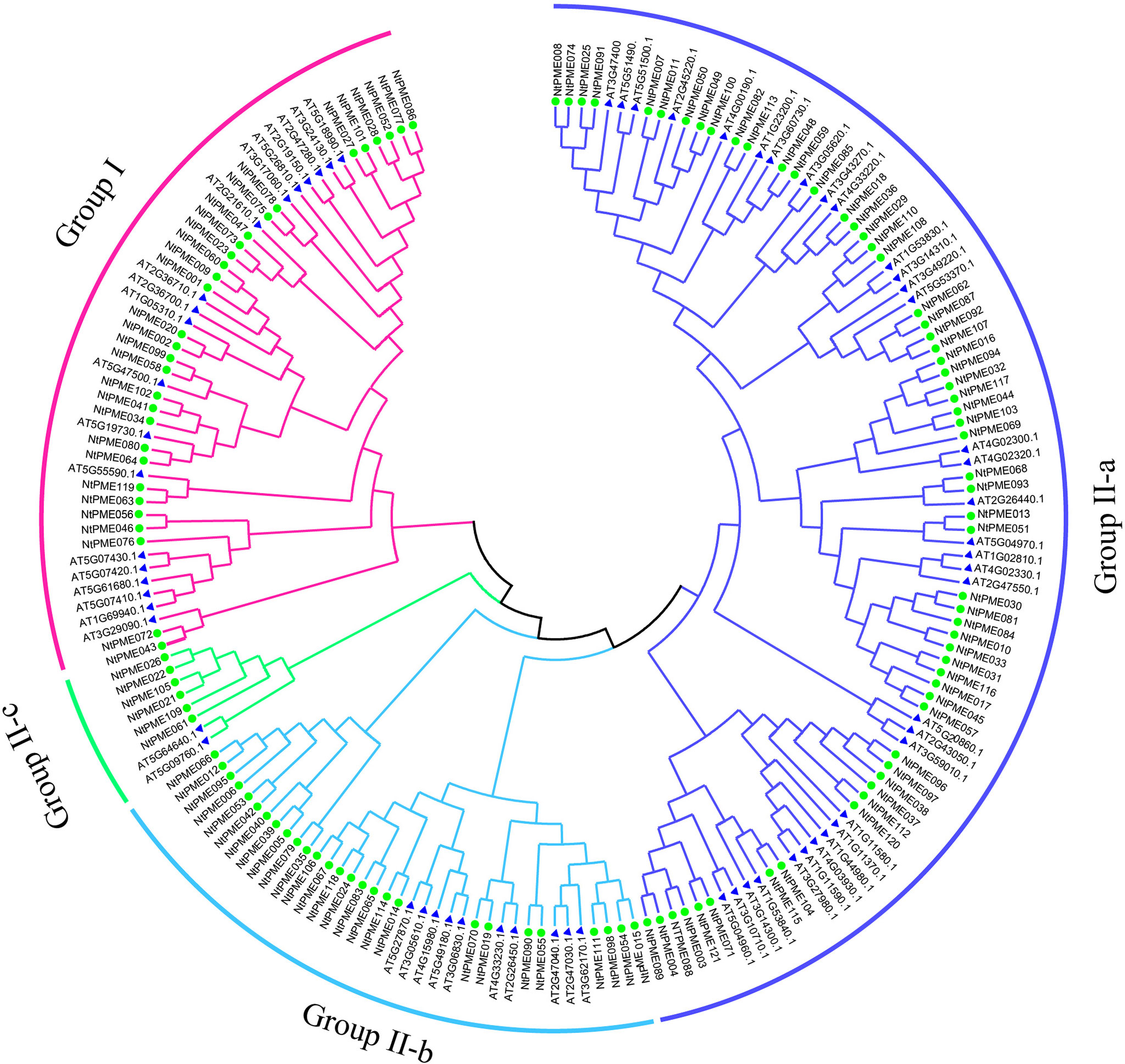
Figure 1 Phylogenetic relationship dendrogram of NtPME members in tobacco. The phylogenetic tree was generated by NtPME proteins of tobacco and Arabidopsis using the Neighbor-Joining method.
In this study, a total of 121 NtPMEs were identified in tobacco and the information on chromosomal distribution was shown in Figure 2. The results showed that only 60 NtPMEs were mapped on 22 chromosomes, and the remaining genes were located on scaffolds. Chromosome 17 contained the largest number of NtPMEs (11 NtPMEs), while the other remaining chromosomes harbored less than four NtPMEs, for instance, chromosome 2 and chromosome 12. In this study, tandem events were carefully screened in the tobacco genome and indicated that no many tandem events were found between these NtPME genes.
In addition, the segmental duplication analysis of NtPMEs was visualized using the MCScanX program (Cao et al., 2016). Notably, nine segmental duplication pairs in 17 NtPMEs were identified (Figure 3) and the details of these genes were listed in Supplementary Table S3. Four pairs of segmental duplication occurred in Group I (NtPME001/NtPME060, NtPME002/NtPME020, NtPME023/NtPME047 and NtPME028/NtPME052) and Group II-a (NtPME007/NtPME048, NtPME10/NtPME30, NtPME10/NtPME33 and NtPME031/NtPME045), respectively. However, only one pair of duplications (NtPME005/NtPME039) occurred in Group II-b. These results suggested that about 14% of the NtPMEs may be generated by segmental duplication events, which played a major role in the expansion of the NtPMEs family in tobacco. In addition, the ratio between non-synonymous and synonymous substitutions (Ka/Ks) can be used to estimate the purifying selection, neutral mutations, and beneficial mutations. In the present study, the Ka/Ks ratios of 9 segmental duplication gene pairs were calculated and the results showed that all the Ka/Ks ratios were less than 1, revealing that these NtPMEs may have undergone purifying selective pressure in the process of evolution.
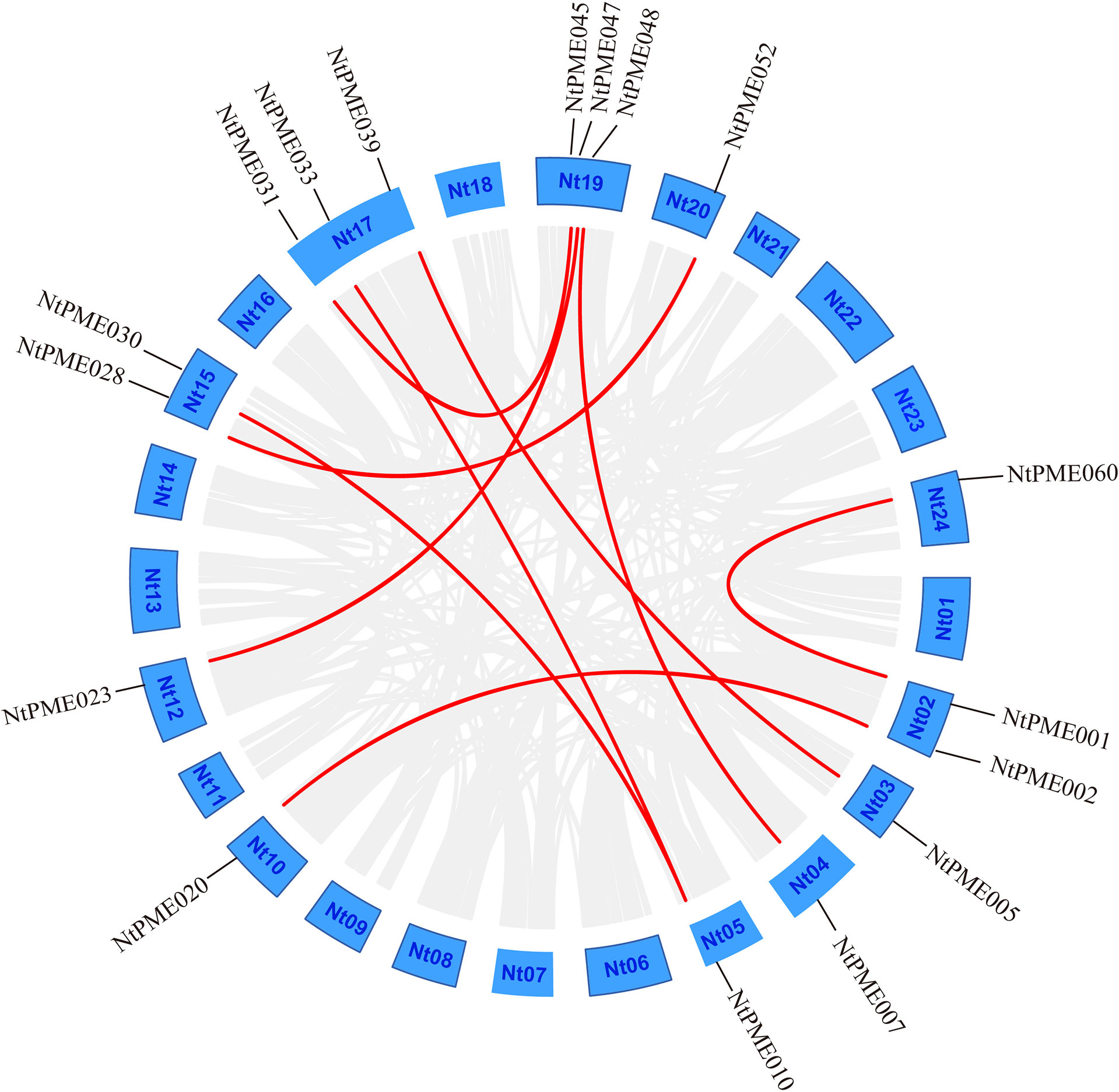
Figure 3 Segmental duplication analysis of the NtPME genes. The putative segmental duplication pairs are displayed by the gray line, whereas the NtPME segmental duplication pairs are revealed by the red line.
To further understand the genetic relationship of NtPMEs, the synthetic analysis of PMEs from dicotyledonous plants (Arabidopsis, tomato, and grape) and monocotyledonous plants (rice) were performed with NtPMEs (Figure 4A). The results showed that the numbers of predicted collinear pairs between tobacco and tomato were 46, followed by grape (28), Arabidopsis (16), and rice (7), respectively. The collinearity of tobacco with the other four plants indicated that a close genetic relationship existed between NtPMEs and tomato PME genes. Meanwhile, only four NtPMEs (NtPME32, NtPME44, NtPME45, NtPME58) formed collinear pairs with PME genes from all the other four plants, suggesting that these NtPMEs may have existed before the divergence of these plant species. (Figure 4B and Supplementary Table S4)
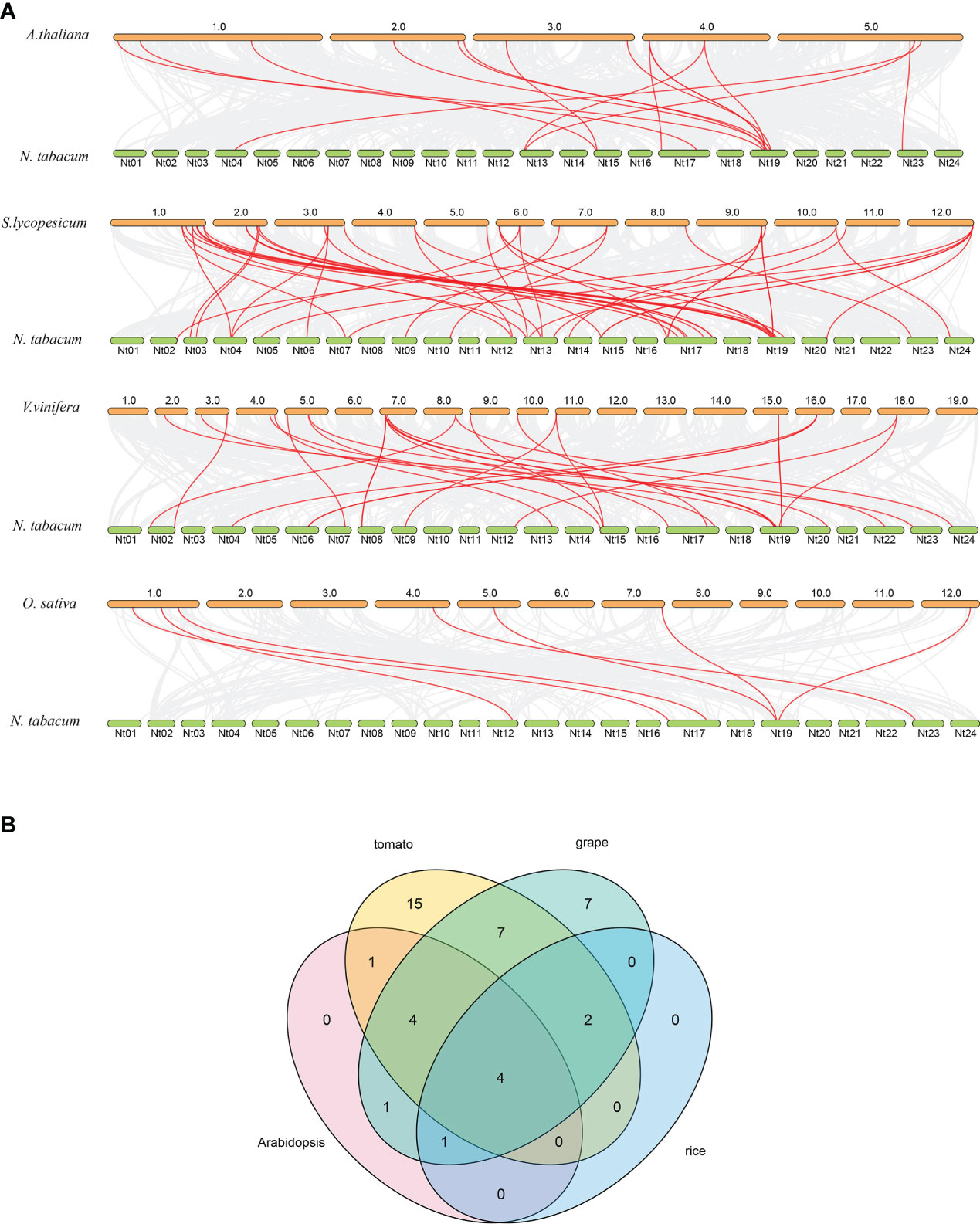
Figure 4 Syntenic relationship analysis dendrogram of PME genes among tobacco and four other plant species. (A) The collinear pairs between tobacco and four other plant species are displayed by the gray line, whereas the collinear PME gene pairs are revealed by the red color; (B) The PME genes formed the syntenic pairs between tobacco and all the other four plant species.
To further analyze the structural diversity of NtPMEs in tobacco, the neighbor-joining phylogenetic tree had been constructed with only NtPME members (Figure 5A). Each NtPME was clustered into different groups, consistent with its distribution in the evolutionary tree of Arabidopsis and tobacco, suggesting that the construction of the neighbor-joining phylogenetic tree is reasonable. In addition, the number and arrangement of exon-intron in NtPMEs were also identified to provide insights into the evolution of these genes in tobacco (Figure 5B). The results showed that these NtPMEs clustered into the same group generally have almost identical exon-intron structures. The coding sequences of the NtPMEs were interrupted by introns, and the number of exons varied from one to 11. Notably, NtPMEs in Group I generally have more introns than that NtPMEs in Group II. In Group I, only one NtPME gene (NtPME060) has two introns, and the remaining NtPMEs contained more than three introns, of which PME063 contained 10 introns. In Group II, there are 50 NtPMEs with only one intron (NtPME017, NtPME030, NtPME045, NtPME068, NtPME093, etc) and most of the remaining genes contain 2-3 introns (NtPME010, NtPME031, NtPME033, NtPME071, etc). These results indicate that the structure of NtPMEs in Group II is relatively simple compared to Group I. The effect of different intron numbers in NtPMEs on the function of these genes is a question worthy of further research.
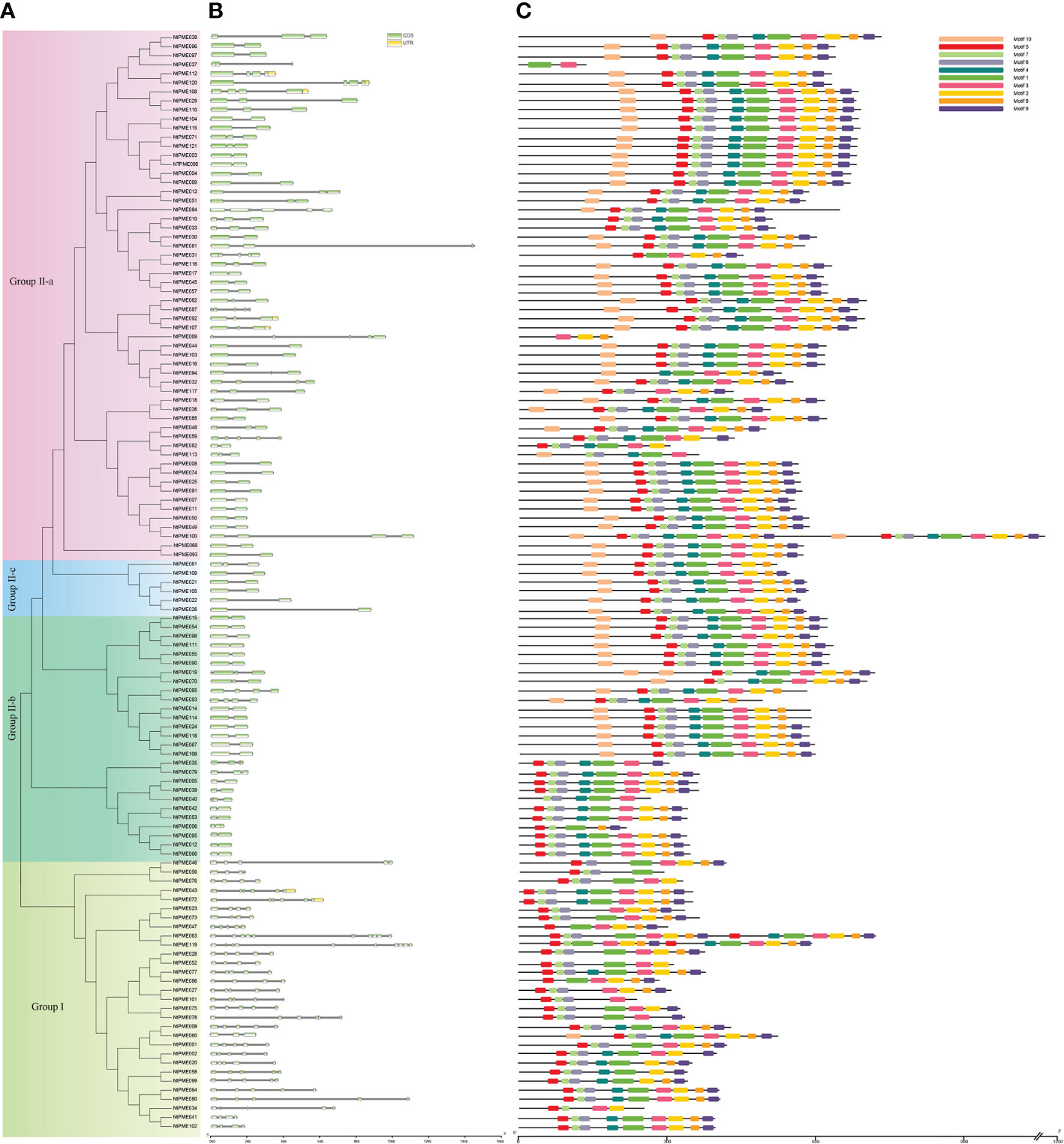
Figure 5 Gene structure and motif organizations analysis of NtPME members in tobacco. (A) The phylogenetic analysis tree of NtPME genes; (B) Gene structure of NtPME genes. Introns and exons are revealed by grey lines and light green boxes, respectively; (C) Distribution of conserved motifs in NtPME proteins. Different color boxes highlight different motifs.
The PME proteins generally have five conserved domains, including Region I (GxYxE), Region II (QAVAxR), Region III (QDTL), Region IV (DFIFG), and Region V (YLGRxWx). To further investigate the conserved domains of NtPME proteins in tobacco, 121 NtPME proteins were subjected to multiple sequence alignments of amino acid sequences, and the basic region was counted in Supplementary Table S5. As the results show that not all NtPME proteins contained all the conserved structural regions. There were 28 NtPME proteins contained less than five conserved structural regions, such as the NtPME006 protein sequence only contained four conserved structural regions. Moreover, there are ten conserved motifs (motif 1-motif 10) were analyzed by using MEME. The NtPME numbers in the same branch of the evolutionary tree usually have similar motif types and orders (Figure 5C). In addition, the motif 7, motif 4, and motif 2 correspond to Region I (GxYxE), Region II (QAVAxR), and V (YLGRxWx) conserved domains, respectively. Motif 1 contains Region III (QDTL) and Region IV (DFIFG) conserved domains.
In general, it was found that various cis-elements were identified in the tobacco NtPMEs promoters (Supplementary Table S6). Furthermore, 11 cis-elements involved in different hormone responses, developmental processes, and stress responses were selected for further analysis (Figure S1). The result showed that there were many cis-acting elements related to stress response in promoter regions of NtPME genes, for instance, ARE (anaerobic induction element), LTR (low-temperature-responsive element), MBS (MYB binding site), TC-rich repeats (stress-responsive element) and WUN-motif (wound-responsive element). The genes containing these cis-acting elements accounted for 88%, 49%, 50%, 45%, and 52% of all NtPMEs, respectively. In addition, the hormone-response elements were also identified in promoter regions, including ABRE, AuxRR-core, TCA-element, and CGTCA-motif. These cis-elements are potential responses to abscisic acid, auxin, ethylene, and methyl jasmonate, respectively.
To preliminarily investigate the significant roles of NtPMEs in tobacco growth and development, the transcript data of NtPMEs in three tissues (root, shoot, and shoot apex) were downloaded from the GEO database and analyzed (Figure S2). The results showed that 2/3 of 121 NtPME genes were detected to be expressed in at least one tested tissue, and seven NtPMEs (NtPME029, NtPME032, NtPME038, NtPME092, NtPME096, NtPME108 and NtPME116) were highly expressed in all three tested tissues. Furthermore, many NtPMEs were exclusively expressed in a tissue-specific manner. For instance, NtPME038, NtPME073, and NtPME096 genes were the most exclusively expressed in the roots. NtPME046 and NtPME103 were detected highly expressed in the shoot apex. In addition, high-level expressions of NtPME062, and NtPME093 were detected in the shoot and shoot apex.
To verify the reliability of NtPMEs RNA sequencing data analysis, we selected representative NtPME genes in different groups to carry out qRT-PCR analysis (Figure S3). NtPME058, NtPME062, and NtPME093 were detected highly expressed in the shoot apex and shoot, respectively, which were consistent with the RNA sequencing data. In Group I, NtPME072 was observed to be highly expressed in both leaves and flowers, while NtPME47 was highly expressed in both roots and flowers. In Group II-a, NtPME108 was detected highly expressed in the shoot, shoot apex, and flowers, whereas NtPME029 was found to be highly expressed in root and shoot apex. Furthermore, NtPME024 and NtPME106, classified into Group II-b, were found to exhibit similar expression patterns to NtPME082 (Group II-a) and NtPME056 (Group II-c), which were highly expressed in flowers. The results showed slight differences between qRT-PCR results and RNA sequencing data analysis, which might be due to the harvest of different sample methods and tissue development status.
In addition, to further investigate the expression patterns of NtPMEs under abiotic stress, the expression levels of 12 NtPME genes were determined under ABA (Figure 6A) and salt (Figure 6B) treatments. In Group I, NtPME043, NtPME047, NtPME056, and NtPME058 were induced and up-regulated by ABA treatments, whereas these four genes were detected to exhibit different expression patterns under salt stress, NtPME043 and NtPME047 were induced and up-regulated, and NtPME056 and NtPME058 were down-regulated. Interestingly, the expression patterns of NtPMEs in Group II-a were different under ABA and salt treatments. It was found that ABA-induced and continually up-regulated NtPME029 with the prolongation of treatment time, while the expression levels of NtPME062, NtPME082, and NtPME108 first increased and then decreased with the increase of treatment time. Furthermore, with increasing treatment time, NtPME029 and NtPME062 were down-regulated by salt, while NtPME082 and NtPME108 behaved oppositely. In Group II-b, NtPME005, NtPME039, and NtPME106 were found to induce by NaCl stress and down-regulated under ABA treatments. The expression of selected NtPME genes in NaCl and ABA treatment seemed to have opposite trend, suggesting that these genes may be involved in ABA pathway to response to NaCl abiotic stresses. How NtPME members including NtPME005 and NtPME039 involved in ABA and NaCl pathway will be the next questions to be addressed in this field.
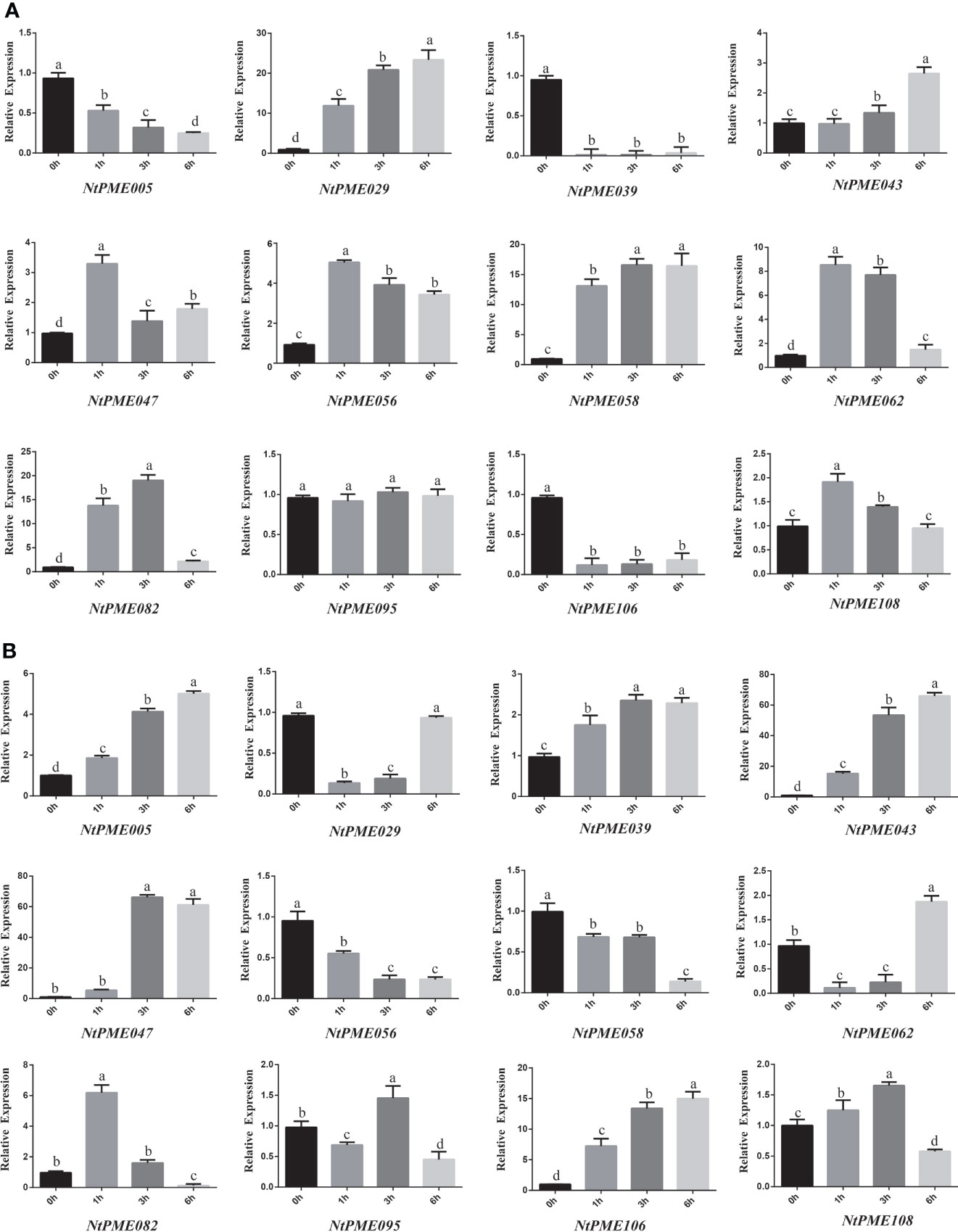
Figure 6 The expression patterns of NtPME genes under (A) Abscisic Acid (ABA) and (B) NaCl. For abiotic stress (salt and ABA) treatments, the seedlings were treated with 50 µM Abscisic Acid (ABA) or 150 mM NaCl, and then harvested at 0, 1, 3, or 6 h after treatment. Three biological replicates were performed. Values with superscript letters a, b, c, and d are significantly different across columns (P < 0.005).
To further investigate the potential functions of the NtPMEs, the NtPME029 and NtPME043 were selected for subcellular localization analysis (Figure 7). The p35S::NtPME029-GFP and p35S::NtPME043-GFP constructs were introduced into the leaves of Nicotiana benthamiana and the p35S:: GFP as a contrast. The fluorescence signals showed that NtPME029 and NtPME043 may be the cell wall localization proteins.
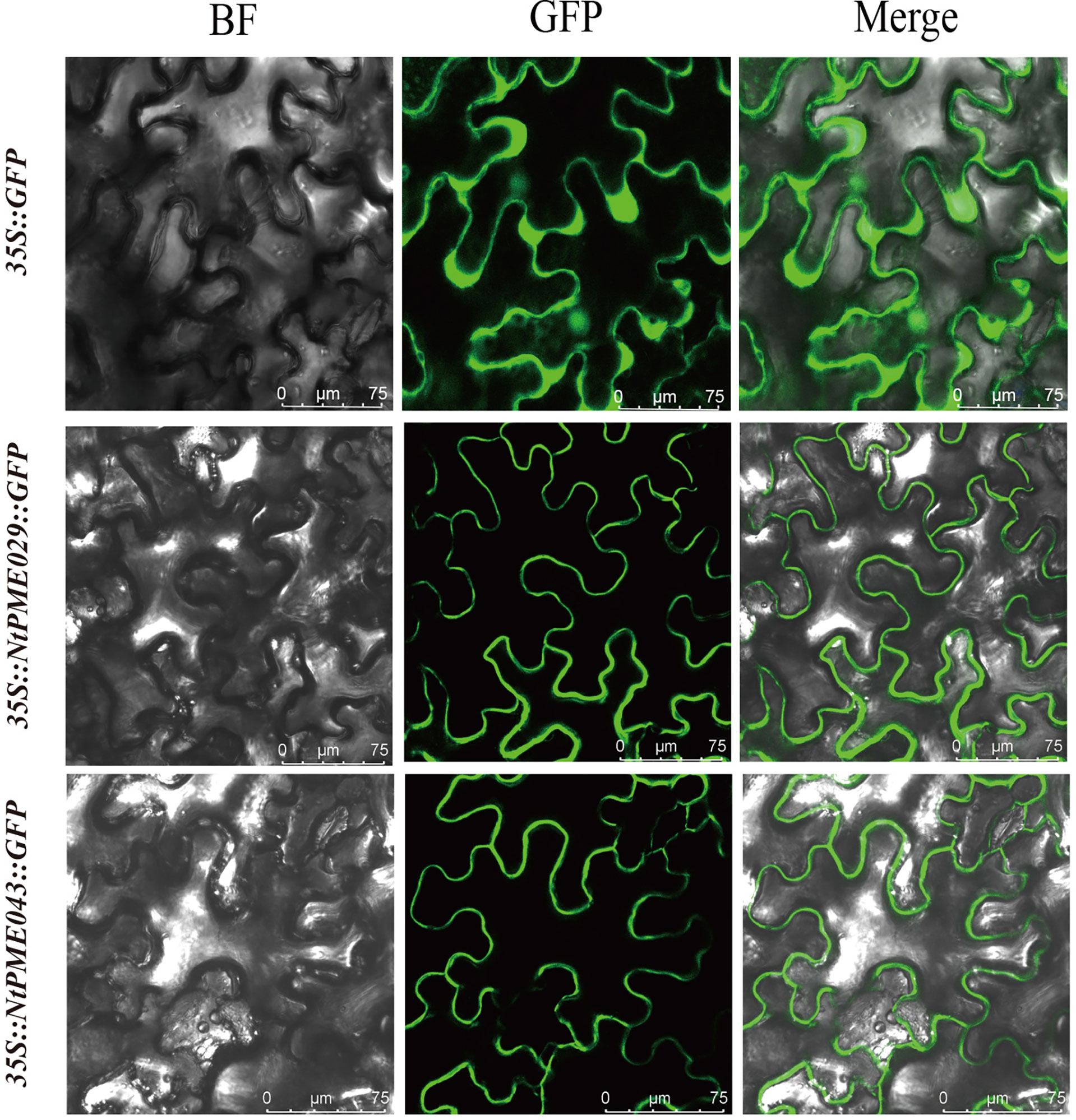
Figure 7 Subcellular localization of NtPME029 and NtPME043 proteins. The NtPME029-GFP, NtPME043-GFP fusion construct, and GFP gene all driven by the CaMV-35S promoter were transiently expressed in N.benthamiana leaves individually. BF, the signal of bright filed; GFP, the signal of GFP protein; Merge, the merge signal of GFP and BF signals. Scale bar = 75 µm.
In Arabidopsis, AtPME3-overexpressing plants showed longer root development compared to wild-type. In addition, AtPME31 could be significantly induced under salt stress and enhance the salt stress tolerance of plants. NtPME029 and NtPME043, were identified as homologues with AtPME3 and AtPME31, respectively. Furthermore, NtPME029 was highly expressed in roots, while NtPME043 could be induced by salt treatments.
To further explore the function of the NtPME029 gene in root development, the root elongation assay of wild-type (WT) and the NtPME029-overexpressed transgenic tobacco plants were assessed (Figures 8A–C). As result, the over-expressing NtPME029 displayed longer root lengths than WT after three weeks of growth under normal conditions, suggesting that the NtPME029 gene positively regulates the root development in tobacco.
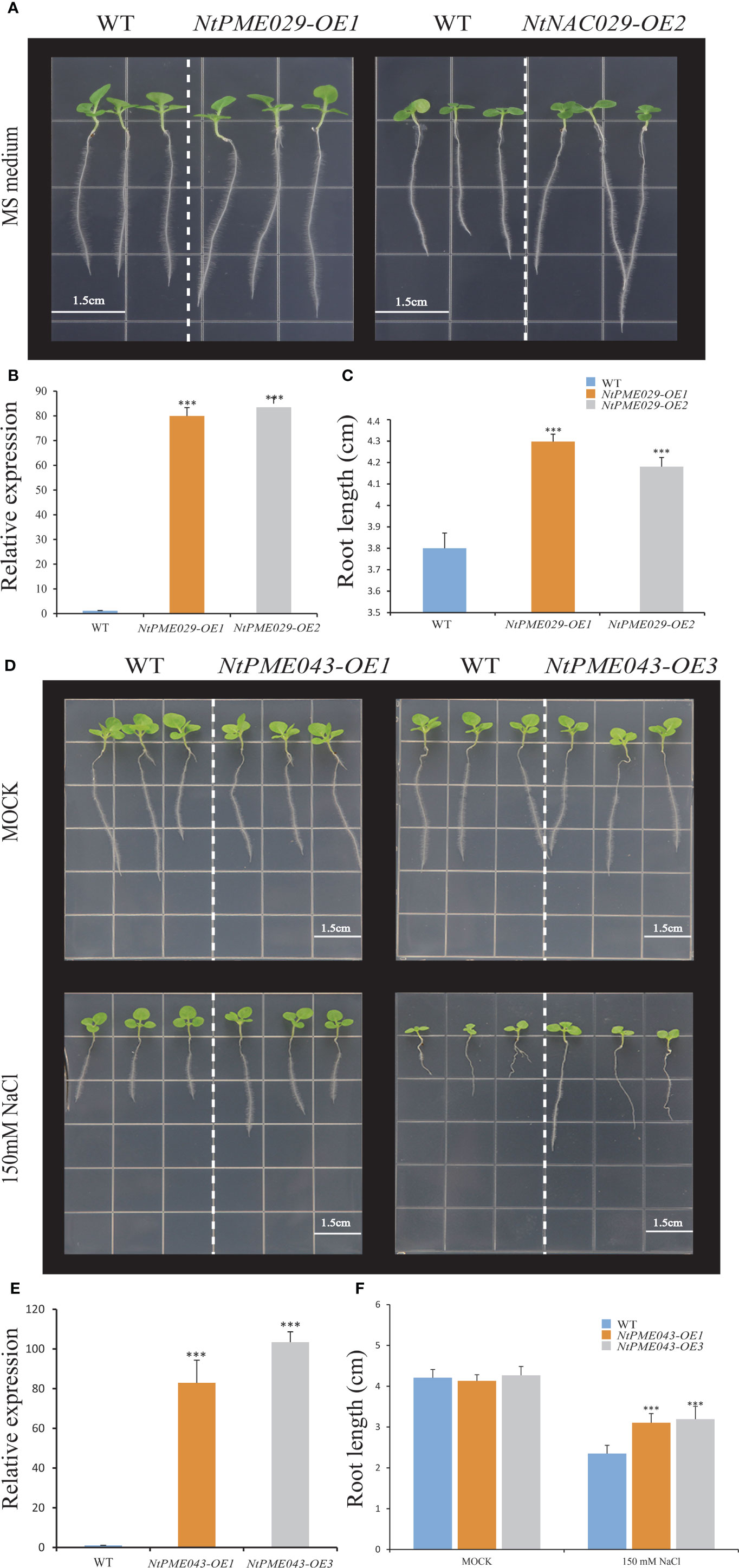
Figure 8 The over-expressing NtPME029 and over-expressing NtPME043 transgenic plants promote root growth and enhance salt stress, respectively. (A, D) Root growth of WT (wild type) and over-expressing NtPME029 and over-expressing NtPME043 lines under MS medium or MS contained 150 mM NaCl; (B, E) The expression level analysis of over-expressing NtPME029 and over-expressing NtPME043 tobacco plants, respectively; (C, F) Quantification and analysis of primary root length on media were retrieved from 27 plants of each genotype with three biological replicates. *** means P < 0.01 (Student’s t-test).
To further analyze the function of the NtPME043 gene in salt tolerance of tobacco, the root elongation assay of WT and the over-expressing NtPME043 transgenic tobacco plants were also assessed (Figures 8D–F). The NtPME043-overexpressed plants displayed no significant differences in root length grown normal nutrient medium. However, the transgenic tobacco plants produced longer root lengths than WT after three weeks of growing under 150 mM NaCl plates. It indicated that the NtPME043 gene could enhance salt tolerance when overexpressed in tobacco.
Previous studies reported that PMEs play critical roles in response to diverse abiotic stresses and plant development processes (Jeong et al., 2015; Duan et al., 2016; Zhang et al., 2019). In the present study, a total of 121 members of the NtPME genes were identified in the tobacco genome using BLASTP searches. Furthermore, the NtPMEs were analyzed using phylogeny, gene structure, motif organization, chromosomal distributions, duplication events, cis-elements, expression profiles, and potential functions.
The 121 NtPME members were divided into two groups according to the phylogeny analysis of Arabidopsis NtPME members. (Louvet et al., 2006). The NtPME members in the same group contained similar gene structure and motif organization consistent with the previous reports (Li et al., 2020), suggesting the evolutionary relationship and classification analysis of NtPME genes were reliable. Additionally, some NtPME genes of the same group contained similar cis-elements types, implying that they may reflect similar functions in plant development and abiotic stress responses. Interestingly, statistics on the number of PME genes in six plant species found the PME genes in tobacco significantly larger than in other plants (Table 1), probably due to tobacco being an allopolytraploid. Meanwhile, five higher plants all have Group I and Group II PME genes, while hyscomitrella patens only have Group I PME genes, implying that the PME genes of Group II appeared after the differentiation of moss and vascular plants, which is consistent with the study of Pelloux and Markovic et al., (Markovic and Janecek, 2004; Pelloux et al., 2007). The synthetic analysis of NtPMEs found four genes may have existed before the divergence of dicotyledonous and monocotyledonous plant species (Figure 4), suggesting that these genes were relatively conserved during evolution.
Previous studies suggested that the whole genome duplication events contribute to the expansion of gene families (Duan et al., 2016; Khan et al., 2019). In this study, the expansion of the tobacco NtPME gene family may be mainly attributable to segmental duplications, not tandem duplication. A total of nine segmental duplication pairs were identified in NtPMEs, with the Ka/Ks ratios of these duplication pairs being less than 1 (Figure 3 and Supplementary Table S3), suggesting that these NtPMEs might have undergone purifying selective pressure. Especially, NtPME005 and NtPME039 as duplicated gene pair was found to exhibit similar gene structure, conserved motif organization, and expression patterns (Figures 5, 6), which indicates the similarity of their functions.
According to previous reports, several PME members affect the plant development in Arabidopsis. Specifically, AtPME5 (AT5G47500) facilitates shoot development (Peaucelle et al., 2011), while NtPME058 (Group II-a) as its syntenic pair gene in tobacco was detected to be highly expressed in the shoots and shoot apex, indicating that NtPME058 may regulate the shoot development (Figure S3). In plants, floral development could affect seed formation, with implications for seed embryo development. In Group II-b, AtPME58 reportedly played a significant role in seed maturation (Turbant et al., 2016). As tobacco homologs, NtPME024 and NtPME106, are highly expressed in flowers. In Group I, AtPME48 (AT5G07410) is highly expressed in the flower and functions to influence pollen development (Leroux et al., 2015). NtPME043, NtPME056, and NtPME072 classified in the same group as AtPME48 were both highly expressed in flowers, implying that these genes may also modulate floral development. The evidence indicates that AtPME3 and AtPME17 specifically regulate the root growth and development of Arabidopsis (Hewezi et al., 2008; Sénéchal et al., 2014). Notably, the NtPME029 was highly expressed in the root and identified as homologues with AtPME3, suggesting that they might have similar biological functions in the root growth and development. The experiment of NtPME029-overexpressing also validated this idea (Figures 6, 8A, C). Additionally, the promoter regions analysis of NtPME029 was found to harbor ABRE and AuxRR-core elements, implying that this gene might participate in ABA and auxin pathways to mediate the root development. Whereas, in the same group, NtPME082 was highly expressed in the flowers, indicating possible functional divergence between these NtPME genes.
Previous studies have indicated that AtPME genes mediate responses to various abiotic stresses. In Group II-b, AtPME28 (AT5G27870) is induced by NaCl treatment to regulate salt stress (Qu et al., 2011). Consistent with AtPME28, the expression of NtPME005, NtPME039 and NtPME106 could also be induced under salt treatments, suggesting that these genes clustered in the same group may display similar functions in response to abiotic stress. NtPME047 embedded low-temperature-responsive element (LTR) was closely related to AtPME41 (AT4G02330), enhancing freezing tolerance (Qu et al., 2011), indicating that NtPME047 may have a similar function under chilling stress. In-plant tissues, PME genes with the ABA response element (ABRE) can be induced by ABA treatment. In Group II-a, AtPME34 (AT3G49220) is verified to respond to heat and salt stress through the signaling of the ABA (abscisic acid) pathway (Huang et al., 2017). NtPM0E62 and NtPME092, containing ABRE elements, were detected to induce by both ABA and NaCl treatment, implying that they may be involved in the ABA synthesis pathway in response to salt stress. In Group I, AtPME31 (AT3G29090) could be significantly induced under salt stress and positively modulate the expression levels of salt stress-induced genes to enhance the salt stress tolerance of plants (Yan et al., 2018). Its tobacco homolog, NtPME043, was also induced under salt stress and contained the stress-responsive element (TC-rich repeats) and MYB binding site (MBS) associated with stress response, implying that NtPME043 may contribute to the salt stress response of tobacco. Furthermore, NtPME043-overexpressing analyses demonstrated that the NtPME043 gene could enhance the salt tolerance of tobacco (Figures 8D, F). Additionally, the NtPME029-GFP and NtPME043-GFP fusion proteins located on the cell wall are consistent with previous evidence of PME proteins’ location (Yapo et al., 2007; Wolf et al., 2009).
In this study, 121 NtPME genes were identified in the tobacco genome. The comprehensive analysis indicated that the NtPME gene family may play multiple roles in various biological processes of tobacco. The PME homologous members between Arabidopsis and tobacco displayed the conserved function in plant development and stress responses. Notably, the NtPME029 was highly expressed in the root and NtPME029-overexpressing transgenic plants could promote root development. Furthermore, NtPME043 could be induced by salt and ABA treatments, and the NtPME043-overexpressing transgenic plants significantly enhanced the salt-stress tolerance. This study could provide insight into the further biological functional characterization of NtPME genes in tobacco.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
JS, ZT, and XL conducted the research and participated in drafting the manuscript. The other authors assisted in data collection and analysis. MX and RX conceived this research, designed the experiments and drafted the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Optimization of Quality Assurance Technology for China Tobacco special aroma 301 Variety and Its Application in Middle and High-end Cigarettes (Y02J202102); the Agricultural Science and Technology Innovation Program (ASTIP-TRIC02); the China Tobacco Genome Project [110202001029(JY-12)] and the China Tobacco Hunan Industrial Co., Ltd. Technology Project (KY2022YC0010).
JS, ZT, SL, JW, ZH, HC, and RX were employed by China Tobacco Jiangsu Industrial Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.998841/full#supplementary-material
Al-Qsous, S., Carpentier, E., Klein-Eude, D., Burel, C., Mareck, A., Dauchel, H., et al. (2004). Identification and isolation of a pectin methylesterase isoform that could be involved in flax cell wall stiffening. Planta. 219, 369–378. doi: 10.1007/s00425-004-1246-1
An, S. H., Sohn, K. H., Choi, H. W., Hwang, I. S., Lee, S. C., Hwang, B. K. (2008). Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance, and abiotic stress tolerance. Planta. 228, 61–78. doi: 10.1007/s00425-008-0719-z
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Bosch, M., Cheung, A. Y., Hepler, P. K. (2005). Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol. 138, 1334–1346. doi: 10.1104/pp.105.059865
Buschmann, H. (2016). Plant cell division analyzed by transient agrobacterium-mediated transformation of tobacco BY-2 cells. Methods Mol. Biol. 1370, 17–25. doi: 10.1007/978-1-4939-3142-2_2
Cao, Y., Han, Y., Li, D., Lin, Y., Cai, Y. (2016). MYB transcription factors in Chinese pear (Pyrus bretschneideri rehd.): Genome-wide identification, classification, and expression profiling during fruit development. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00577
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chao, J., Kong, Y., Wang, Q., Sun, Y., Liu, G. (2015). MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Hereditas 37, 91–97. doi: 10.16288/j.yczz.2015.01.013
Dixit, S., Upadhyay, S. K., Singh, H., Sidhu, O. P., Verma, P. C., Chandrashekar, C. (2013). Enhanced methanol production in plants provides broad spectrum insect resistance. PloS One 8, e79664. doi: 10.1371/journal.pone.0079664
Duan, W., Huang, Z., Song, X., Liu, T., Liu, H., Hou, X., et al. (2016). Comprehensive analysis of the polygalacturonase and pectin methylesterase genes in Brassica rapa shed light on their different evolutionary patterns. Sci. Rep. 6, 25107. doi: 10.1038/srep25107
Edwards, K. D., Fernandez-Pozo, N., Drake-Stowe, K., Humphry, M., Evans, A. D., Bombarely, A., et al. (2017). A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen efficiency. BMC Genom. 18, 448. doi: 10.1186/s12864-017-3791-6
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Garg, V. K., Avashthi, H., Tiwari, A. (2016). MFPPI–multi FASTA protparam interface. Bioinformation. 12, 74–77. doi: 10.6026/97320630012074
Hewezi, T., Howe, P., Maier, T. ,. R., Hussey, R. S., Mitchum, M. G., Davis, E. L., et al. (2008). Cellulose binding protein from the parasitic nematode heterodera schachtii interacts with Arabidopsis pectin methylesterase: cooperative cell wall modification during parasitism. Plant Cell. 20, 3080–3093. doi: 10.1105/tpc.108.063065
Hongo, S., Sato, K., Yokoyama, R., Nishitani, K. (2012). Demethylesterification of the primary wall by PECTIN METHYLESTERASE35 provides mechanical support to the Arabidopsis stem. Plant Cell. 24, 2624–2634. doi: 10.1105/tpc.112.099325
Huang, Y. C., Wu, H. C., Wang, Y. D., Liu, C. H., Lin, C. C., Luo, D. L., et al. (2017). PECTIN METHYLESTERASE34 contributes to heat tolerance through its role in promoting stomatal movement. Plant Physiol. 174, 748–763. doi: 10.1104/pp.17.00335
Hu, B., Jin, J. P., Guo, A. Y., Zhang, H., Luo, J. C., Gao, G. (2015). GSDS 2.0: An upgraded gene feature visualization server. Bioinf. 31(8), 1296–1297. doi: 10.1093/bioinformatics/btu817
Jenkins, J., Mayans, O., Smith, D., Worboys, K., Pickersgill, R. W. (2001). The third-dimensional structure of erwinia chrysanthemi pectin methylesterase reveals a novel esterase active site. J. Mol. Biol. 305, 951–960. doi: 10.1006/jmbi.2000.4324
Jeong, H. Y., Nguyen, H. P., Lee, C. (2015). Genome-wide identification and expression analysis of rice pectin methylesterases: Implication of functional roles of pectin modification in rice physiology. J. Plant Physiol. 183, 23. doi: 10.1016/j.jplph.2015.05.001
Jiangtao, C., Yingzhen, K., Qian, W., Yuhe, S., Guanshan, L. (2015). MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Hereditas. 37, 91. doi: 10.16288/j.yczz.2015.01.013
Khan, N., Fatima, F., Haider, M. S., Shazadee, H., Liu, Z., Zheng, T., et al. (2019). Genome-wide identification and expression profiling of the polygalacturonase (PG) and pectin methylesterase (PME) genes in grapevine (Vitisvinifera l.). Int. J. Mol. Sci. 20 (13), 3180. doi: 10.3390/ijms20133180
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics. 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Leroux, C., Bouton, S., Kiefer-Meyer, M. C., Fabrice, T. N., Mareck, A., Guénin, S., et al. (2015). PECTIN METHYLESTERASE48 is involved in Arabidopsis pollen grain germination. Plant Physiol. 167, 367–380. doi: 10.1104/pp.114.250928
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Doerks, T., Bork, P. (2015). SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Levesque-Tremblay, G., Müller, K., Mansfield, S. D., Haughn, G. W. (2015). Highly methyl esterified seeds is a pectin methyl esterase involved in embryo development. Plant Physiol. 167, 725–737. doi: 10.1104/pp.114.255604
Li, X., Hamyat, M., Liu, C., Ahmad, S., Gao, X., Guo, C., et al. (2018). Identification and characterization of the WOX family genes in five solanaceae species reveal their conserved roles in peptide signaling. Genes. 9, 260. doi: 10.3390/genes9050260
Li, X., Ahmad, S., Ali, A., Guo, C., Li, H., Yu, J., et al. (2019a). Characterization of somatic embryogenesis receptor-like kinase 4 as a negative regulator of leaf senescence in Arabidopsis. Cells. 8, 50. doi: 10.3390/cells8010050
Li, X., Guo, C., Ahmad, S., Wang, Q., Yu, J., Liu, C., et al. (2019b). Systematic analysis of MYB family genes in potato and their multiple roles in development and stress responses. Biomolecules. 9 (8), 317. doi: 10.3390/biom9080317
Li, Y., He, H., He, L. ,. F. (2020). Genome-wide analysis of the pectin methylesterase gene family in potato. Potato Res. 64, 1–19. doi: 10.1007/s11540-020-09453-1
Li, Z., Chao, J., Li, X., Li, G., Song, D., Guo, Y., et al. (2021). Systematic analysis of the bZIP family in tobacco and functional characterization of NtbZIP62 involvement in salt stress. Agronomy. 11, 148. doi: 10.3390/agronomy11010148
Librado, P., Rozas, J. (2009). DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-delta delta c (T)) method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Louvet, R., Cavel, E., Gutierrez, L., Guénin, S., Roger, D., Gillet, F., et al. (2006). Comprehensive expression profiling of the pectin methylesterase gene family during silique development in Arabidopsis thaliana. Planta. 224 (4), 782–791. doi: 10.1007/s00425-006-0261-9
Markovic, O., Janecek, S. (2004). Pectin methylesterases: sequence-structural features and phylogenetic relationships. Carbohydr Res. 339 (13), 2281–2295. doi: 10.1016/j.carres.2004.06.023
Micheli, F. (2001). Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Science. 6, 414–419. doi: 10.1016/s1360-1385(01)02045-3
Peaucelle, A., Braybrook, S. A., Le Guillou, L., Bron, E., Kuhlemeier, C., Höfte, H. (2011). Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol. 21, 1720–1726. doi: 10.1016/j.cub.2011.08.057
Pelloux, J., Rustérucci, C., Mellerowicz, E. J. (2007). New insights into pectin methylesterase structure and function. Trends Plant Sci. 12 (6), 267–277. doi: 10.1016/j.tplants.2007.04.001
Qu, T., Liu, R., Wang, W., An, L., Chen, T., Liu, G., et al. (2011). Brassinosteroids regulate pectin methylesterase activity and AtPME41 expression in Arabidopsis under chilling stress. Cryobiology. 63, 111–117. doi: 10.1016/j.cryobiol.2011.07.003
Ren, A., Ahmed, R. I., Chen, H., Han, L., Sun, J., Ding, A., et al. (2019). Genome-wide identification, characterization and expression patterns of the pectin methylesterase inhibitor genes in sorghum bicolor. Genes. 10 (10), 755. doi: 10.3390/genes10100755
Sénéchal, F., Graff, L., Surcouf, O., Marcelo, P., Rayon, C., Bouton, S., et al. (2014). Arabidopsis PECTIN METHYLESTERASE17 is co-expressed with and processed by SBT3.5, a subtilisin-like serine protease. Ann. Bot. 114, 1161–1175. doi: 10.1093/aob/mcu035
Sun, J., Xie, M., Li, X., Li, Z., Sun, Y. (2021). Systematic investigations of the ZF-HD gene family in tobacco reveal their multiple roles in abiotic stresses. Agronomy. 11, 406. doi: 10.3390/agronomy11030406
Tian, G. W., Chen, M. H., Zaltsman, A., Citovsky, V. (2006). Pollen-specific pectin methylesterase involved in pollen tube growth. Dev. Biol. 294, 83–91. doi: 10.1016/j.ydbio.2006.02.026
Turbant, A., Fournet, F., Lequart, M., Zabijak, L., Pageau, K., Bouton, S., et al. (2016). PME58 plays a role in pectin distribution during seed coat mucilage extrusion through homogalacturonan modification. J. Exp. Bot. 67, 2177–2190. doi: 10.1093/jxb/erw025
Udall, J. A., Swanson, J. M., Haller, K., Rapp, R. A., Sparks, M. E., Hatfield, J., et al. (2006). A global assembly of cotton ESTs. Genome Res. 16, 441–450. doi: 10.1101/gr.4602906
Verma, C., Singh, R. K., Mishra, S. (2014). Biochemical characterization of pectin methylesterase from musa acuminata referring to delayed ripening. IOSR-JPBS. 9, 42–47. doi: 10.9790/3008-49709134247
Wallace, S., Williams, J. H. (2017). Evolutionary origins of pectin methylesterase genes associated with novel aspects of angiosperm pollen tube walls. Biochem. Biophys. Res. Commun. 487, 509–516. doi: 10.1016/j.bbrc.2017.04.027
Wolf, S., Mouille, G., Pelloux, J. (2009). Homogalacturonan methyl-esterification and plant development. Mol. Plant 2, 851–860. doi: 10.1093/mp/ssp066
Wu, X., Lai, Y., Lv, L., Ji, M., Han, K., Yan, D., et al. (2020). Fasciclin-like arabinogalactan gene family in Nicotiana benthamiana: genome-wide identification, classification and expression in response to pathogens. BMC Plant Biol. 20, 305. doi: 10.1186/s12870-020-02501-5
Xie, T., Chen, C., Li, C., Liu, J., Liu, C., He, Y. (2018). Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 19, 490. doi: 10.1186/s12864-018-4880-x
Yan, J., He, H., Fang, L., Zhang, A. (2018). Pectin methylesterase31 positively regulates salt stress tolerance in. Arabidopsis. Biochem. Biophys. Res. Commun. 496, 497–501. doi: 10.1016/j.bbrc.2018.01.025
Yapo, B. M., Lerouge, P., Thibault, J. F., Ralet, M. C. (2007). Pectins from citrus peel cell walls contain homogalacturonans homogenous with respect to molar mass, rhamnogalacturonan I and rhamnogalacturonan II. Carbohydr. Polym. 69, 426–435. doi: 10.1016/j.carbpol.2006.12.024
Zega, A., D'Ovidio, R. (2016). Genome-wide characterization of pectin methyl esterase genes reveals members differentially expressed in tolerant and susceptible wheats in response to Fusarium graminearum. Plant Physiol. Biochem. 108, 1–11. doi: 10.1016/j.plaphy.2016.06.033
Zhang, G. Y., Feng, J., Wu, J., Wang, X. W. (2010). BoPMEI1, a pollen-specific pectin methylesterase inhibitor, has an essential role in pollen tube growth. Planta 231, 1323–1334. doi: 10.1007/s00425-010-1136-7
Zhang, P., Wang, H., Qin, X., Chen, K., Zhao, J., Zhao, Y., et al. (2019). Genome-wide identification, phylogeny and expression analysis of the PME and PMEI gene families in maize. Sci. Rep. 9, 19918. doi: 10.1038/s41598-019-56254-9
Keywords: tobacco, PME genes, genome-wide analysis, abiotic stresses, root development
Citation: Sun J, Tian Z, Li X, Li S, Li Z, Wang J, Hu Z, Chen H, Guo C, Xie M and Xu R (2022) Systematic analysis of the pectin methylesterase gene family in Nicotiana tabacum and reveal their multiple roles in plant development and abiotic stresses. Front. Plant Sci. 13:998841. doi: 10.3389/fpls.2022.998841
Received: 20 July 2022; Accepted: 05 September 2022;
Published: 28 September 2022.
Edited by:
Caroline Müller, Universidade Federal da Fronteira Sul, BrazilReviewed by:
Yunpeng Cao, Chinese Academy of Sciences (CAS), ChinaCopyright © 2022 Sun, Tian, Li, Li, Li, Wang, Hu, Chen, Guo, Xie and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruyan Xu, eHlfeHVyeUBqc3p5Z3MuY29t; Minmin Xie, eGllbWlubWluQGNhYXMuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.