- 1State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Chengdu, China
- 2Triticeae Research Institute, Sichuan Agricultural University, Chengdu, China
Fusarium crown rot (FCR), caused by various Fusarium species, is a primary fungal disease in most wheat-growing regions worldwide. A. tauschii, the diploid wild progenitor of the D-genome of common wheat, is a reservoir of genetic diversity for improving bread wheat biotic and abiotic resistance/tolerance. A worldwide collection of 286 A. tauschii accessions was used to evaluate FCR resistance. Population structure analysis revealed that 115 belonged to the A. tauschii ssp. strangulata subspecies, and 171 belonged to the A. tauschii ssp. tauschii subspecies. Five accessions with disease index values lower than 20 showed moderate resistance to FCR. These five originated from Afghanistan, China, Iran, Uzbekistan, and Turkey, all belonging to the tauschii subspecies. Genome-wide association mapping using 6,739 single nucleotide polymorphisms (SNPs) revealed that two SNPs on chromosome 2D and four SNPs on chromosome 7D were significantly associated with FCR resistance. Almost all FCR resistance alleles were presented in accessions from the tauschii subspecies, and only 4, 11, and 19 resistance alleles were presented in accessions from the strangulata subspecies. Combining phenotypic correlation analysis and genome-wide association mapping confirmed that FCR resistance loci were independent of flowering time, heading date, and plant height in this association panel. Six genes encoding disease resistance-related proteins were selected as candidates for further validation. The identified resistant A. tauschii accessions will provide robust resistance gene sources for breeding FCR-resistant cultivars. The associated loci/genes will accelerate and improve FCR in breeding programs by deploying marker-assisted selection.
Introduction
Fusarium crown rot (FCR), caused by various Fusarium species, is a severe fungal disease that affects the whole growth period of the plant in cereal crops, such as wheat, barley, sorghum, and so on (Li et al., 2012; Xu et al., 2017; Cuevas et al., 2019). It poses a considerable threat to wheat and barley production in regions with arid and semi-arid cropping globally, including major wheat-producing countries, such as Australia, the USA, Canada, New Zealand, China, and many others (Kazan and Gardiner, 2018). For example, yield loss estimates have indicated that FCR can cause a 35% reduction in wheat grain yield under natural inoculum levels in the Pacific Northwest of the USA (Smiley et al., 2005). Besides, FCR is estimated to routinely cause a 10% yield loss in cereals in Australia (Murray and Brennan, 2009). In China, FCR has become one of the major diseases that have caused wheat and barley yield losses in the past decade. It has generated more than 70% yield losses at individual sites with an average annual reduction of 9–35% from 2008 to 2019 (Li et al., 2012; Xu et al., 2016, 2017; Luan et al., 2022). In addition, cereal crop grains infected by Fusarium species will accumulate mycotoxins, which are harmful to humans and livestock. Developing resistant cereal crop cultivars is the most practical and sustainable to control FCR.
Marker-assisted selection is an important approach for breeding, which can improve the efficiency of breeding. FCR resistance genes/loci have been identified in chromosomes 1A, 1B, 1D, 2A, 2B, 2D, 3A, 3B, 3D, 4B, 4D, 5D, 6A, 6B, and 6D in wheat (Liu C. et al., 2015; Su et al., 2021; Yang et al., 2021) and 1H, 2H, 3H, 4H, 5H, and 6H in Barley (Chen et al., 2013; Liu C. et al., 2015; Gao et al., 2019). In wheat, 12 single nucleotide polymorphisms (SNPs) within a 0.5-Mb genomic region were identified by a genome-wide association study (GWAS) using the Wheat 660 K SNP array in an association panel containing 435 introgression lines (Yang et al., 2021). A candidate gene, TraesCS4B02G385500 (also known as TaDIR-B1: encoding dirigent protein), was cloned and confirmed to improve resistance FCR resistance by virus-induced gene silencing and EMS-mutagenized wheat lines (Yang et al., 2021). Using a set of 358 Chinese germplasms, a novel 13.78 Mb region targeted by five SNPs on chromosome arm 5DL was identified by GWAS using the wheat 55 K SNP array (Jin et al., 2020). Several candidate genes within this 5DL region encode TIR-NBS-LRR proteins associated with FCR resistance (Jin et al., 2020). Although these studies provided some important markers associated with FCR resistance, more major and stable genetic loci controlling the resistance to FCR are needed to deal with this severe fungal disease in wheat.
Aegilops tauschii (2n = 2 × = 14, DD) is the D-genome progenitor of common wheat. The diverse A. tauschii D-genome offers a valuable gene pool for biotic and abiotic resistance/tolerance, such as phosphorus-deficient tolerance (Liu et al., 2015a), drought resistance (Qin et al., 2016), stripe rust resistance (Zhang et al., 2019b), powdery mildew resistance (Xue et al., 2022), and so on. Although the hexaploid Triticum aestivum (AABBDD) was formed by hybridization with Triticum turgidum (AABB) and A. tauschii (DD), only a tiny fraction of the D- genome subpopulation participated in the hybridization (Giles and Brown, 2006; Zhou et al., 2020). Harnessing genetic diversity from wild wheat progenitor A. tauschii to improve hexaploid wheat has been proven fast and efficient (Zhang et al., 2019b; Awan et al., 2022; Gaurav et al., 2022). Here, using 286 A. tauschii accessions, we aim to (1) identify FCR resistant materials in A. tauschii; (2) identify loci significantly associated with FCR resistance using GWAS based on SNP array; (3) select candidate genes for loci of FCR resistance.
Materials and methods
Plant materials
A total of 286 A. tauschii accessions were used in the present study. Based on the morphological classification, 115 and 171 belong to A. tauschii ssp. strangulata and A. tauschii ssp. tauschii, respectively (Supplementary Table 1).
Filed experiment design and phenotypic evaluation
All accessions were planted for three straight years during the sowing seasons in October from 2017 to 2019. Five seeds of each accession were planted in three rows as previously described (Liu et al., 2015b; Wang et al., 2021). The row length was 1.5 m, and the space between rows was 0.6 m. The heading date and flowering time were recorded for each accession in Wenjiang from 2018 to 2020 and in Chongzhou in 2020. Five plants of each accession were used to evaluate plant height. At physiological maturity in 2020, spikes of each accession were harvested, and hand-threshed seeds were used to identify FCR resistance further.
Identification of Fusarium crown rot resistance
A highly aggressive Fusarium pseudograminearum isolate (Fp.322) was used in FCR inoculation. The inoculum preparation, injection, and FCR resistance identification were based on the method described by a previous study (Li et al., 2008) with some modifications. Briefly, the Fp.322 was incubated on half-strength potato dextrose agar plates at 25°C for 7 days. When the white mycelium grew over the leaves and showed peach-red to dark-red pigmentation, a piece of culture medium with mycelium was taken out from the margin of the plate and then transferred into a conical flask containing 100 ml carboxy-methyl-cellulose sodium liquid medium. The conical flask was put into a shaking incubator at 180 rpm at 28°C for approximately 6 days. The concentration of spore suspension was adjusted to 1 × 106 spores per ml. The Tween-20 was added to the spore suspension to a final concentration of 0.1% volume ratio before use for injection.
The seedling disease assessment contained three replicates. Twenty unified seeds of each accession were used for each replicate. Seeds were washed with 10% NaClO for 20 min and with sterile water three times. Seeds were soaked in water at 45°C for 5 min to break dormancy. Then, seeds were put in Petri dishes on three layers of filter paper saturated with water and germinated at 25°C in an incubator. Ten seedlings with the shoot length at 0.5–1.0 cm for each accession were immersed in the spore suspension for 1 min and were sown in 5-cm square seeding trays containing autoclaved potting mix. The seedlings were grown in the greenhouse at Sichuan Agricultural University with a 16 h photoperiod at 25/22 (±1) °C day/night temperature and 65/85% day/night relative humidity. Seedlings were watered only when wilt symptoms promoted FCR development, as in a previous study (Jin et al., 2020).
When the whole plants of “Janz” were severe to completely necrotic, the FCR severity of each accession was recorded using a 0–5 scale (Li et al., 2008). The FCR resistance was identified using the disease index (DI). The DI value of each replication was calculated according to the formula:
DI = (∑nX/5N) × 100
where X is the scale value of each plant, n is the number of plants in the category, and N is the total number of plants assessed for each line (Jin et al., 2020). The DI value represented FCR resistance was used for further analysis.
Phenotypic statistical analysis
The mean value, minima, maxima, coefficient of variation (CV), and Person’s correlation coefficient (r) were calculated using IBM SPSS Statistics 20 (IBM Corp., Armonk, NY, USA). The analysis of variance was calculated using the “GLM” procedure in SAS 9.4 (SAS Institute Inc., Cary, NC, USA). The broad-sense heritability was estimated using the formula:
H2 = Vg/(Vg + Vge + Ve)
where Vg, Vge, and Ve are the estimates of genetic variance, the genotype × environment interaction, and environmental variance, respectively (Smith et al., 1998). The best linear unbiased prediction (BLUP) values of each trait were calculated in SAS 9.4 (SAS Institute Inc., Cary, NC, USA) to reduce environmental effects.
Genotyping, population structure, and kinship analysis
DNA samples of each genotype were extracted from young leaves using the CTAB method (Murray and Thompson, 1980). All 286 accessions were genotyped using the Illumina 10 K SNP arrays. The physical location of SNPs was based on A. tauschii reference genome v4.0 (Aet v4.0) (Luo et al., 2017). The gathered SNPs with minor allele frequency (MAF) <0.05, heterozygosis >0.20, or missing data >0.20 were rejected for further analysis. Finally, 6,739 SNPs were used for population structure analysis STRUCTURE 2.3.4 based on the linkage ancestry model (Pritchard et al., 2000). The K value was set from 1 to 10, with five runs per K. A total of 10,000 burn-in iterations and 10,000 Markov Chain Monte Carlo iterations were developed for each K. The best K was determined using the Evanno method (Evanno et al., 2005) calculated by web-based STRUCTURE HARVESTER (Earl and VonHoldt, 2012). The population structure matrix was gathered using the CLUMPP among five repetitions (Jakobsson and Rosenberg, 2007). A neighbor-joining tree was created using TASSEL 5.0 (Bradbury et al., 2007) and visualized using the iTOL website (Letunic and Bork, 2021).
Genome-wide association analysis and candidate gene prediction
The mixed linear model adjusted by population structure and kinship was used to identify marker-trait associations in TASSEL 5.0 (Yu et al., 2006; Bradbury et al., 2007). The significance threshold was set at P < 0.001 correspondingly −log10(P) = 3.00. Manhattan plots of GWAS results were plotted in R 3.6.3 (R Core Team, 2014) using the package “CMplot.” Flanking sequences of significantly associated SNPs and reported QTL were used to obtain chromosomal information and physical distances from Chinese Spring Reference Sequence v2.1 (IWGSC RefSeq V2.1) (Zhu et al., 2021). The physical map of IWGSC RefSeq V2.1 was used as the reference genome to determine whether significantly associated loci in A. tauschii and significant QTL in the D-genome of wheat were overlapped.
According to previously reported linkage disequilibrium decay distances in A. tauschii (Wang et al., 2021; Gaurav et al., 2022), conservative genomic regions (500 kb upstream and downstream of the significant SNPs) were used to select candidate genes. The predicted high confidence (HC) genes in these genomic regions of reference genome Aet v4.0 were selected to annotate using arabidopsis as background species by KOBAS 3.0 (Bu et al., 2021). Candidate genes were identified based on annotated function information. Candidate genes were identified based on annotated function information.
Results
Single nucleotide polymorphisms distribution and population structure of the Aegilops tauschii natural population
A total of 6,739 high polymorphic SNPs (MAF ≥0.05, both missing and heterozygous ≤0.20) were gathered for further analysis (Supplementary Figure 1 and Table 1). These SNPs are evenly distributed over seven chromosomes corresponding to a total map length of 4,022.29 Mb. The number of SNPs ranged from 707 for chromosome 6D to 1,237 for chromosome 2D. For marker density, the average distance between two SNPs ranged from 0.53 Mb on chromosome 2D to 0.70 Mb on chromosome 6D, with an average value of 0.60 for all chromosomes. The average polymorphism information content was 0.43 for 6,739 SNPs showing a polymorphism.
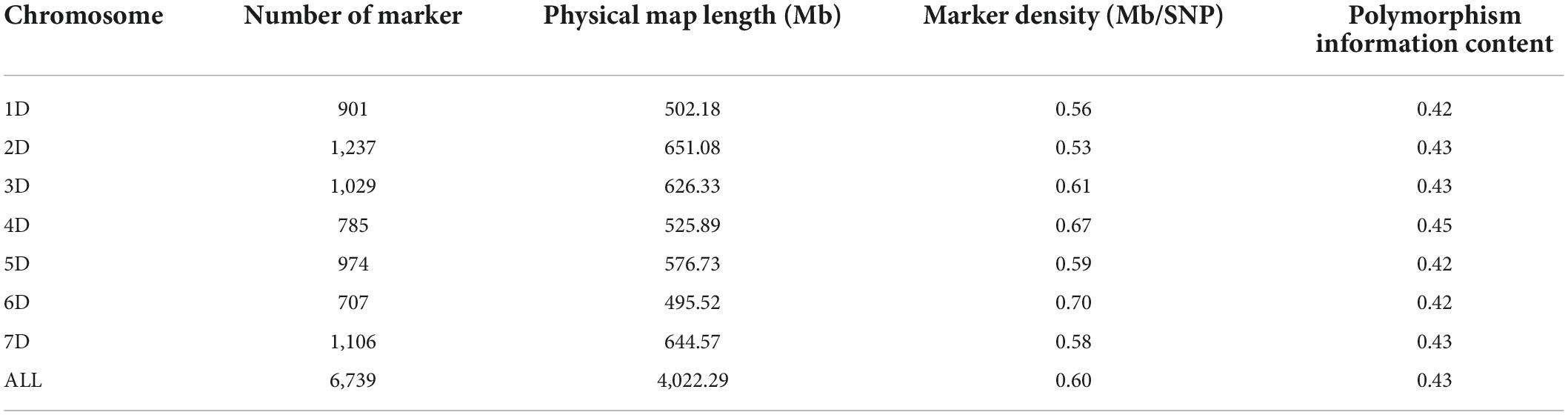
Table 1. Distributions of single nucleotide polymorphisms, density, and polymorphism information content of 286 Aegilops tauschii accessions.
Population structure analysis showed that Delta K had the highest value while K = 2 (Supplementary Figure 2). Thus, 286 accessions were divided into two groups. The first group included 114 A. tauschii ssp. strangulata accessions and one A. tauschii ssp. tauschii accessions, and the second group had 169 A. tauschii ssp. tauschii accessions and two A. tauschii ssp. strangulata accessions. Almost all strangulata accessions were divided into the first group (S-group), and tauschii accessions were divided into the second group (T-group). The cluster analysis by neighbor-joining tree also divided 286 accessions into two groups confirming the result revealed by population structure (Figure 1).
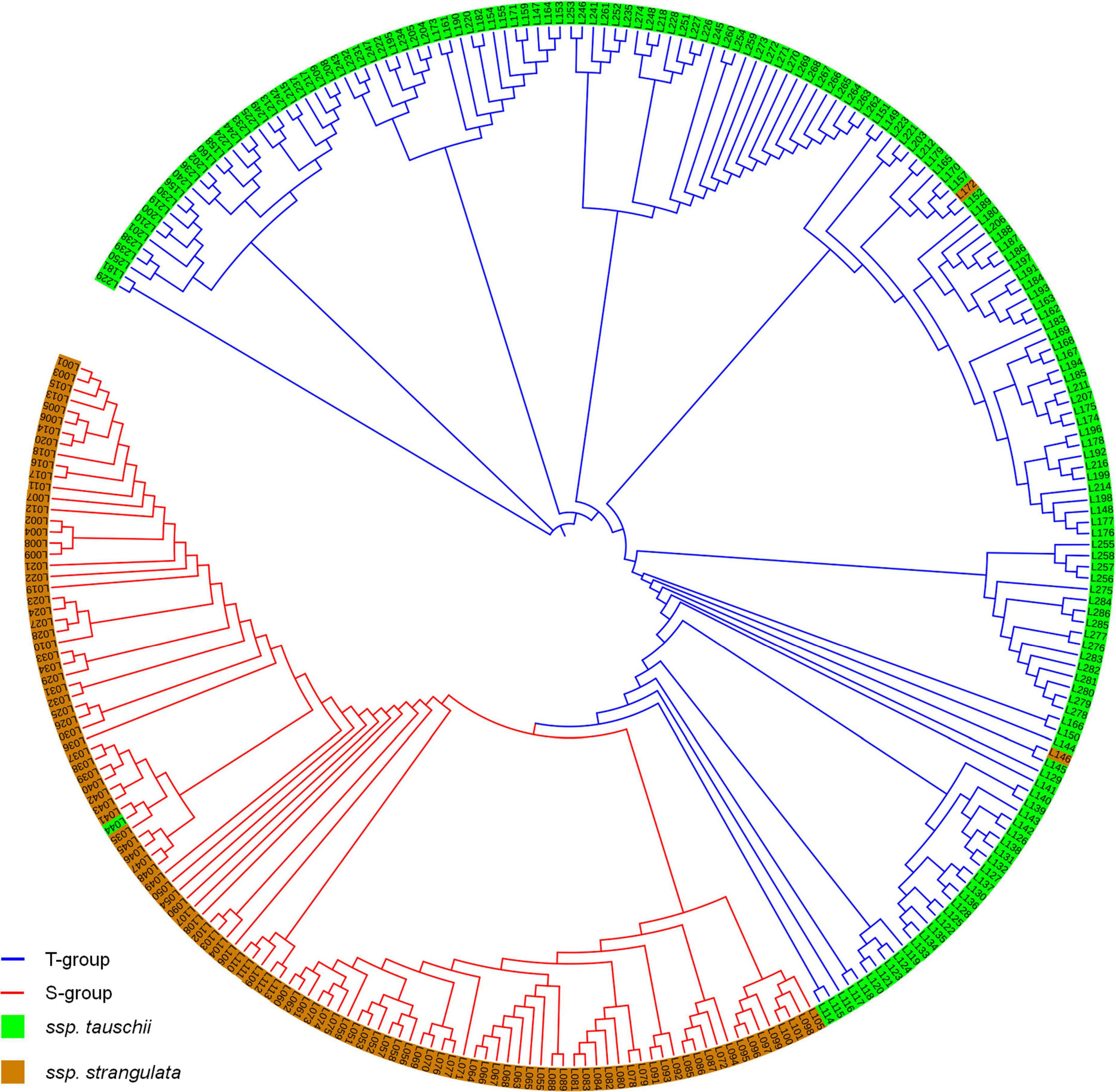
Figure 1. Population structures of 286 Aegilops tauschii revealed by STRUCTURE 2.3.4 and neighbor-joining tree.
Genetic variation of Fusarium crown rot resistance in Aegilops tauschii and correlations with flowering time, heading date, and plant height
In this panel, FCR severity ranged from 0.80 to 2.88, with a mean value of 1.82. The CV of FCR severity was 22.94% (Table 2). The DI values ranged from 15.81 to 57.66, with a mean value of 36.34 (Table 2). Frequency distributions of DI were continuous and normally (Figure 2). The ANOVA revealed significant (P < 0.01) genotypic and environmental effects of FCR resistance on the A. tauschii natural population. The broad-sense heritability of FCR resistance was 0.65, indicating it was major controlled by genetic effects. Among 286 A. tauschii, five accessions had DI lower than 20, indicating these five were moderate resistance to FCR. This five originated from Afghanistan, China, Iran, Turkey, and Uzbekistan (Supplementary Table 1). The five accessions all belonged to the T-group (Supplementary Table 1). The remaining 60 and 221 accessions were moderate and high susceptibility, respectively.
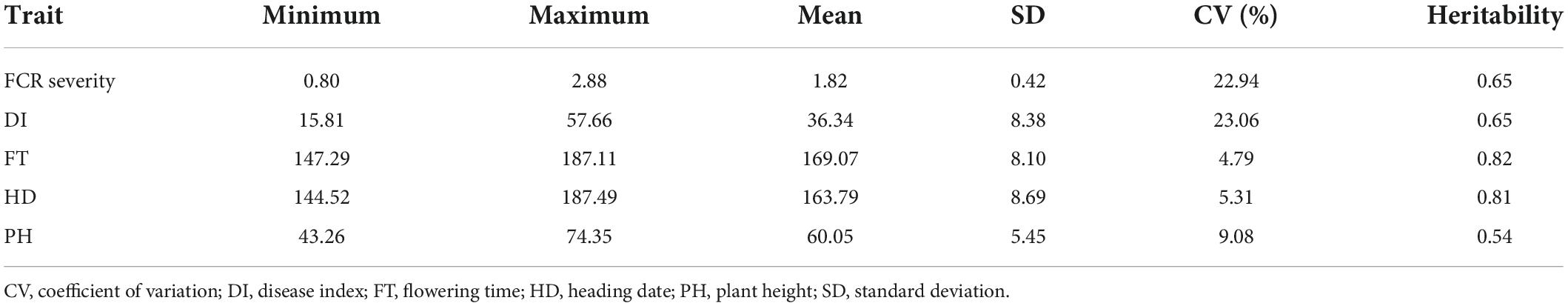
Table 2. Phenotypic variations and heritability of disease index, flowering time, heading date, and plant height.
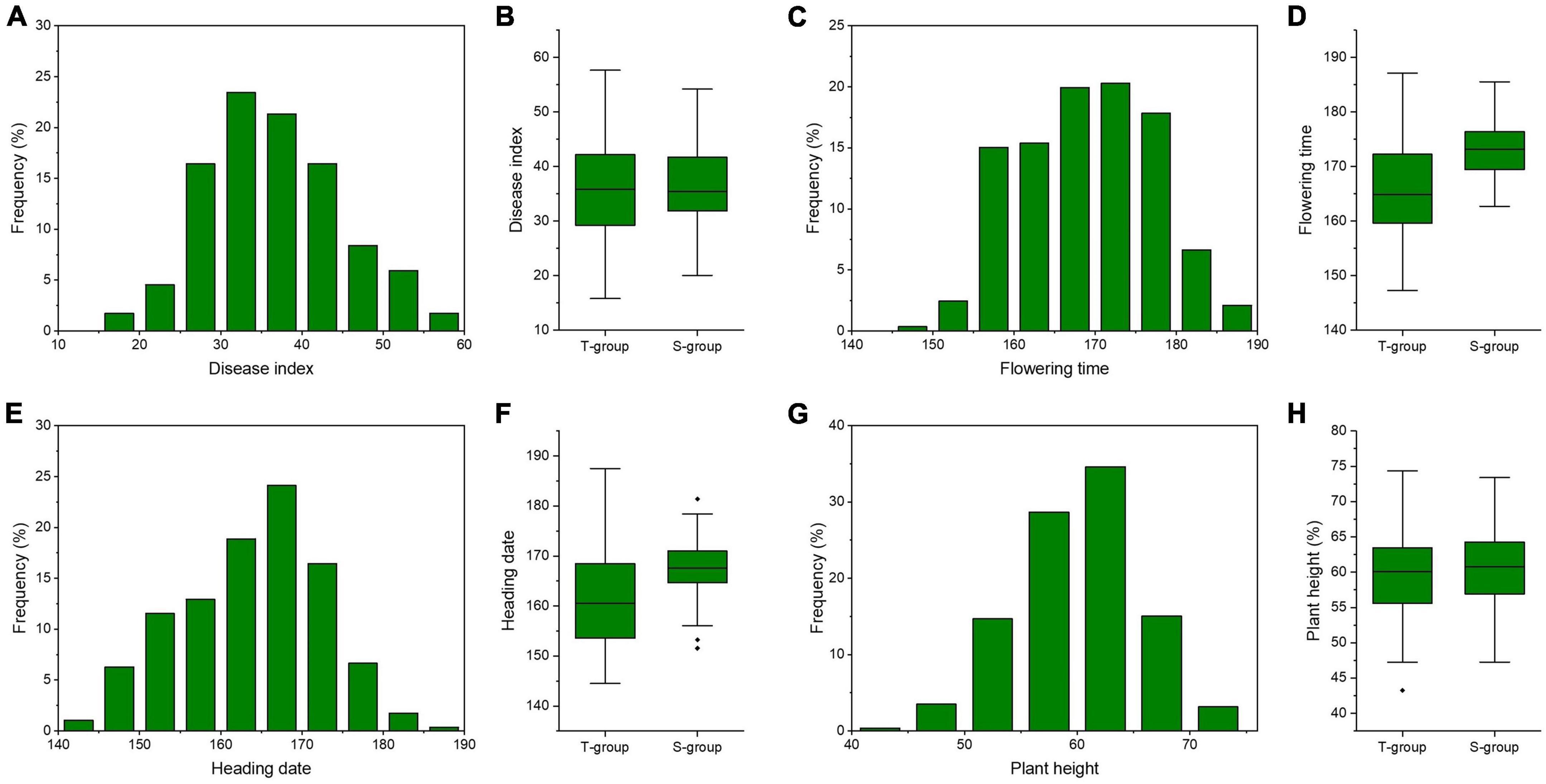
Figure 2. Phenotypic distributions of phenotypic traits among the whole population and subgroups. Frequency distributions of disease index (A), flowering time (C), heading time (E), and plant height (G) among 286 Aegilops tauschii accessions. Box plots of disease index (B), flowering time (D), heading time (F), and plant height (H) among two subgroups. S-group and T-group represent the Aegilops tauschii ssp. strangulata group and the Aegilops tauschii ssp. tauschii group, respectively.
To resolve relationships between FCR resistance and possible agronomic traits, BLUP values across four environments of flowering time (FT), heading date (HD) and plant height (PH) were gathered. The broad-sense heritability of FT, HD, and PH was 0.82, 0.81, and 0.54 (Table 2). It indicated FT and HD were highly inheritable characters, while PH was affected by genetic and environmental effects (Table 2). Frequency distributions of FT, HD, and PH showed continuous and normal distributions (Figure 2). Based on Person’s correlation analysis, correlation coefficients between DI and FT, HD, and PH were 0.12, 0.15, and 0.02, respectively. It indicated that FCR resistance was weakly correlated with FT and HD and not PH.
Fusarium crown rot resistance loci revealed by genome-wide association study in Aegilops tauschii and their associations with flowering time, heading date, and plant height
Based on the threshold value P < 0.001 correspondingly −log10(P) = 3.00, six SNPs were identified for FCR resistance by GWAS in the 286 A. tauschii natural population. These loci were located on chromosomes 2D and 7D, explaining 3.90–5.35% phenotypic variations (Figure 3 and Table 3). Among these, two were located on the short arm of chromosome 2D at 177.65 Mb and the long arm of chromosome 2D at 577.73 Mb, respectively. Four SNPs were located on chromosome 7D at 579.21, 612.48, 612.48, and 637.80 Mb, respectively. To resolve relationships between FCR resistance and possible-related agronomic traits, SNPs associated with FT, HD, and PH were also detected by genome-wide association mapping. The results showed that 19 SNPs were significantly associated with FT, HD, and PH (Figure 3 and Table 3). The SNP GCE8AKX02GFUOC on chromosome 7D at 67.90 Mb explained the highest phenotypic variation. Eleven SNPs were significantly associated with FT with PVE ranging from 3.80 to 6.90%. These SNPs are located on all seven chromosomes of A. tauschii. The SNP GCE8AKX02GFUOC on chromosome 7D at 67.90 Mb explained the highest phenotypic variation. For HD, 11 SNPs were identified with PVE ranging from 4.05 to 7.28%. These 11 SNPs were located on chromosomes 1D, 3D, 4D, and 7D, respectively. The SNP GCE8AKX02GFUOC associated with HD on chromosome 7D at 67.90 Mb explained the highest phenotypic variation. Three SNPs were significantly associated with PH. Two SNPs were located on chromosome 2D at 531.86 and 637.24 Mb, and one SNP was located on chromosome 5D at 405.67 Mb with PVE ranging from 4.09 to 6.54%. These loci, associated with FT, HD, and PH, were not overlapped loci associated with FCR resistance.
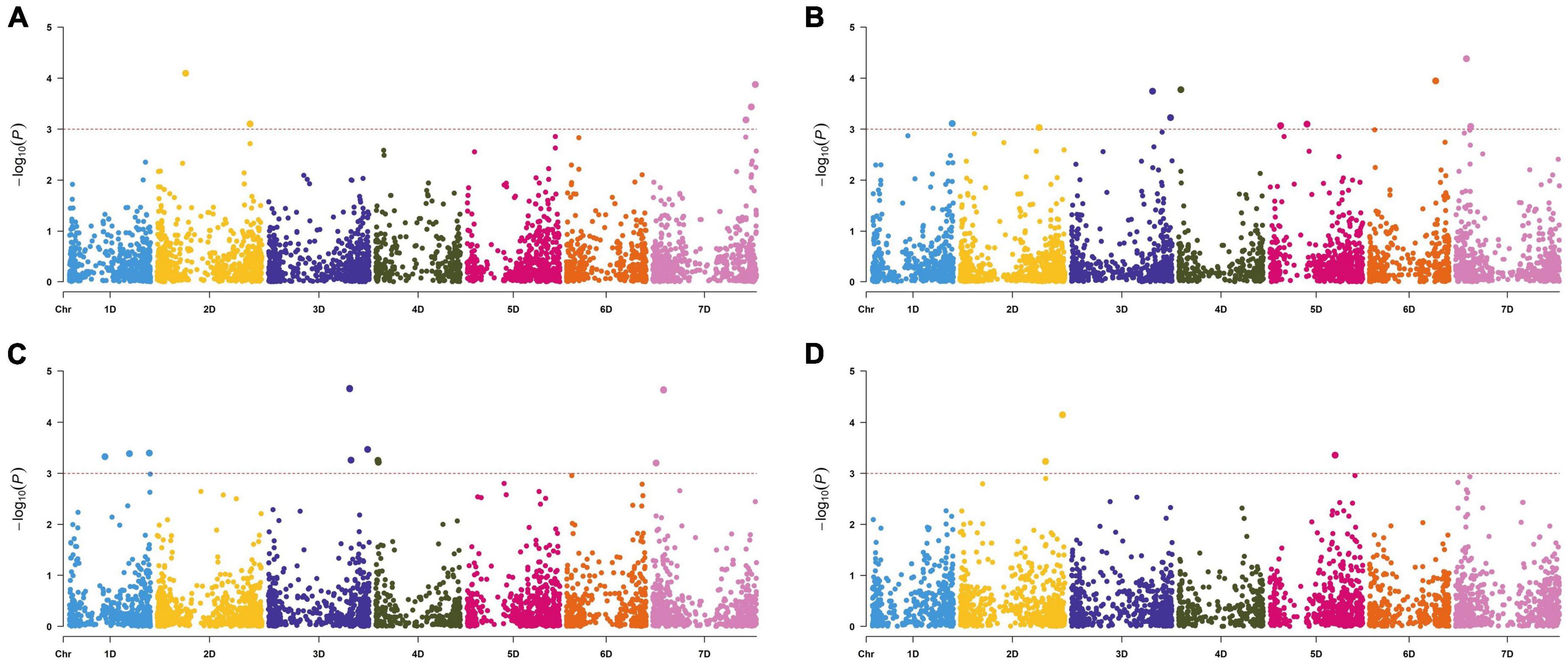
Figure 3. Manhattan plots of marker-trait associations detected by the mixed linear model using 6,739 single nucleotide polymorphisms in 286 Aegilops tauschii accessions. Panels (A–D) represents marker-trait associations of disease index (A), flowering time (B), heading time (C), and plant height (D). The horizontal dashed line represents the significant threshold –log10(P) = 3.00.
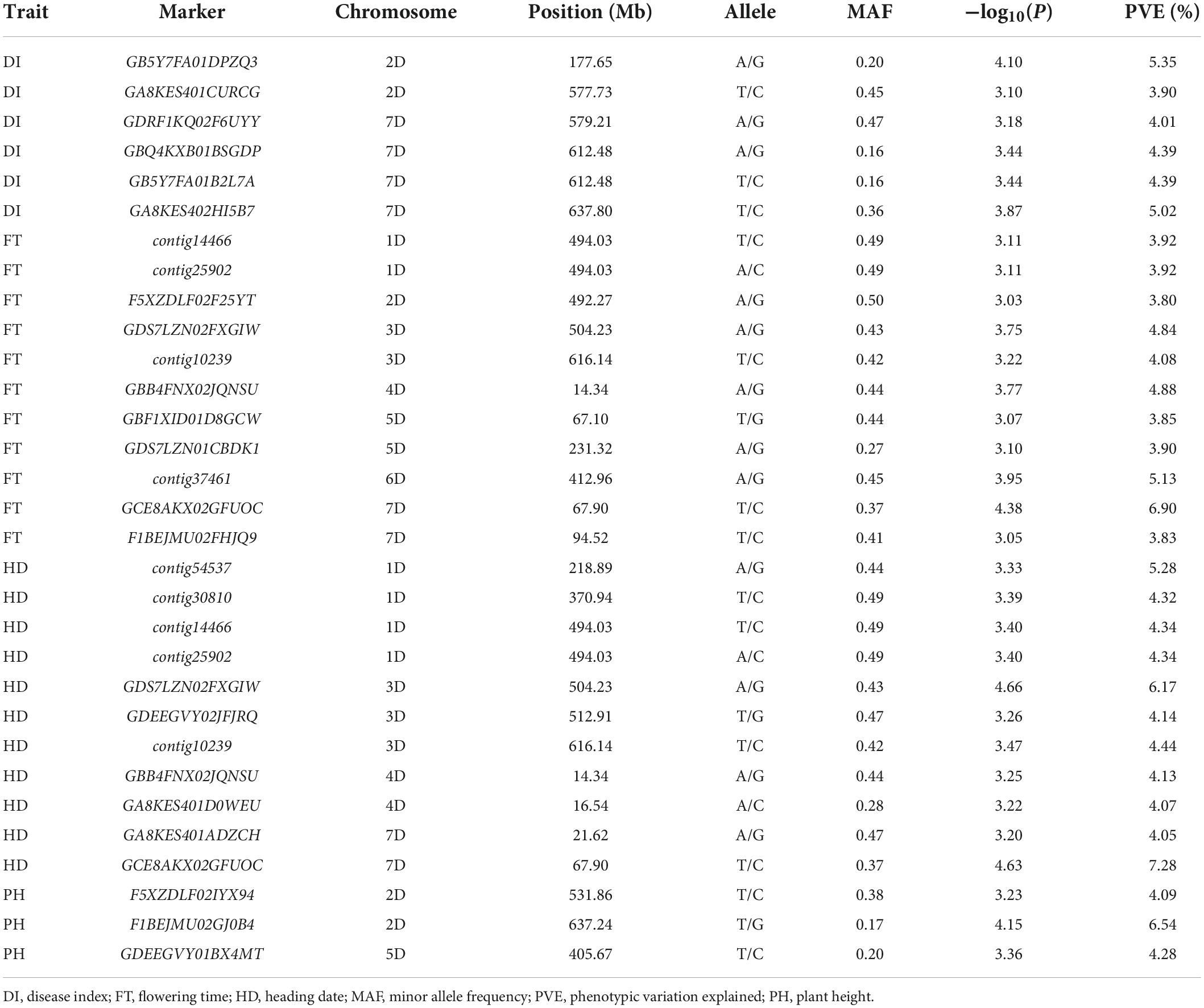
Table 3. Significant markers associated with disease index, flowering time, heading date, and plant height.
Distributions of Fusarium crown rot resistance alleles in T-group and S-group
In the whole panel, A. tauschii accessions with resistance alleles showed lower DI than accessions with susceptibility alleles (Figure 4 and Table 4), indicating A. tauschii accessions with resistance alleles were more resistant to FCR. A. tauschii accessions with resistance alleles could decrease by 9.16 to 15.21% DI values, corresponding to increased FCR resistance. Interestingly, most A. tauschii accessions with resistance alleles belong to the T-group. Resistance alleles of loci GB5Y7FA01DPZQ3, GBQ4KXB01BSGDP, and GB5Y7FA01B2L7A were only present in T-group accessions (100%) and all absent in S-group accessions (Figures 4A,D,E and Table 4). Resistance alleles were present in 129, 135, and 104 accessions for the other three loci. But only a few accessions with resistance alleles were in S-group, while most were in the T-group. Resistance alleles were only present in 11, 19, and 4 accessions of the S-group but in 118, 116, and 100 accessions of the T-group (Figures 4B,C,F and Table 4). Besides, accessions with 3–6 superior alleles all belonged to the T-group (Supplementary Table 1). These results suggested that A. tauschii accessions in the T-group (subspecies tauschii) harbored abundant resistance alleles to FCR.
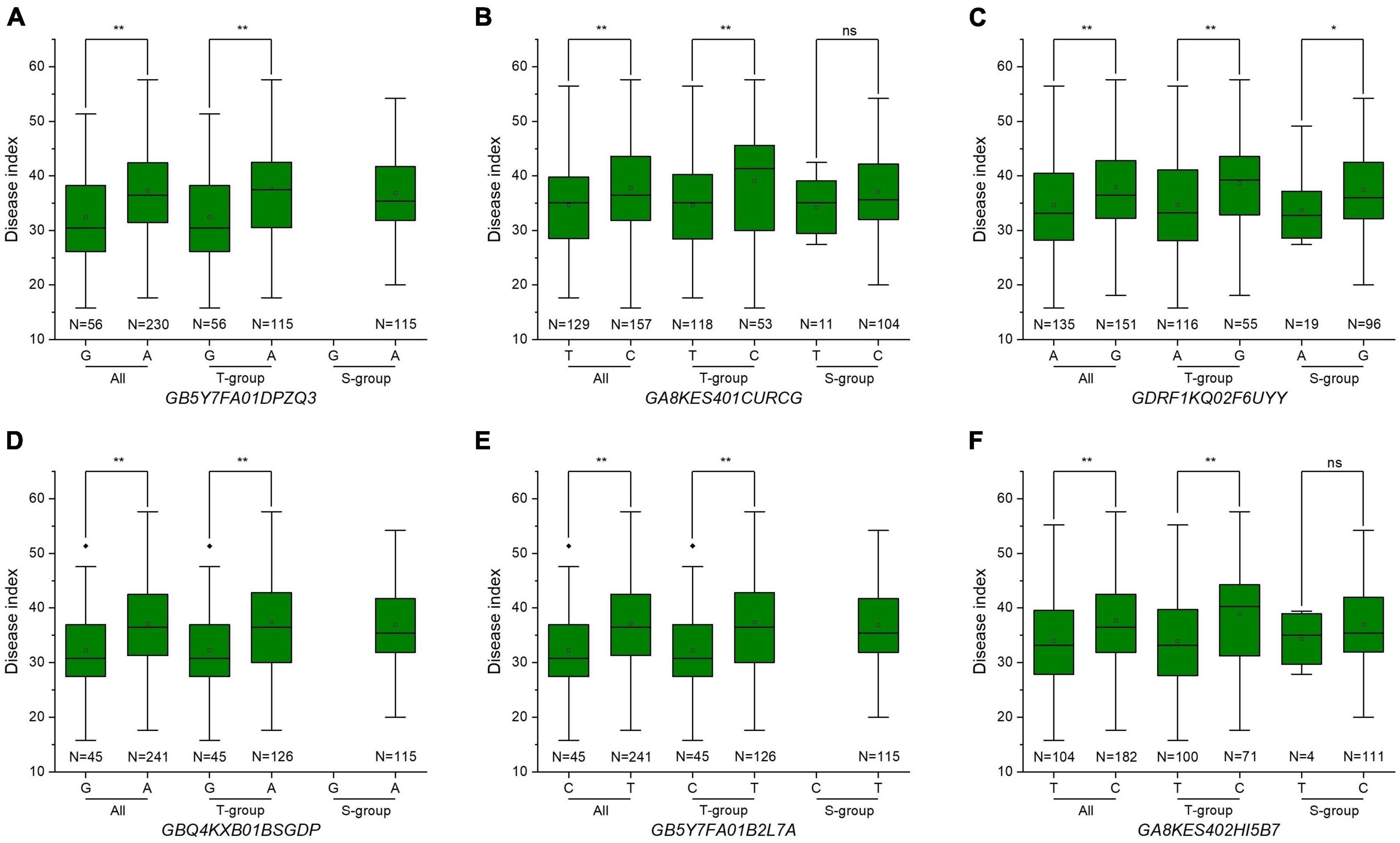
Figure 4. Allele distributions of significant single nucleotide polymorphisms (SNPs) significantly associated with Fusarium crown rot resistance. (A–F) Allele distributions of the six significantly associated SNPs GB5Y7FA01DPZQ3, GA8KES401CURCG, GDRF1KQ02F6UYY, GBQ4KXB01BSGDP, GB5Y7FA01B2L7A, and GA8KES402HI5B7 among the whole population and subgroups. S-group and T-group represent the Aegilops tauschii ssp. strangulata group and the Aegilops tauschii ssp. tauschii group, respectively. N represents the number of alleles.
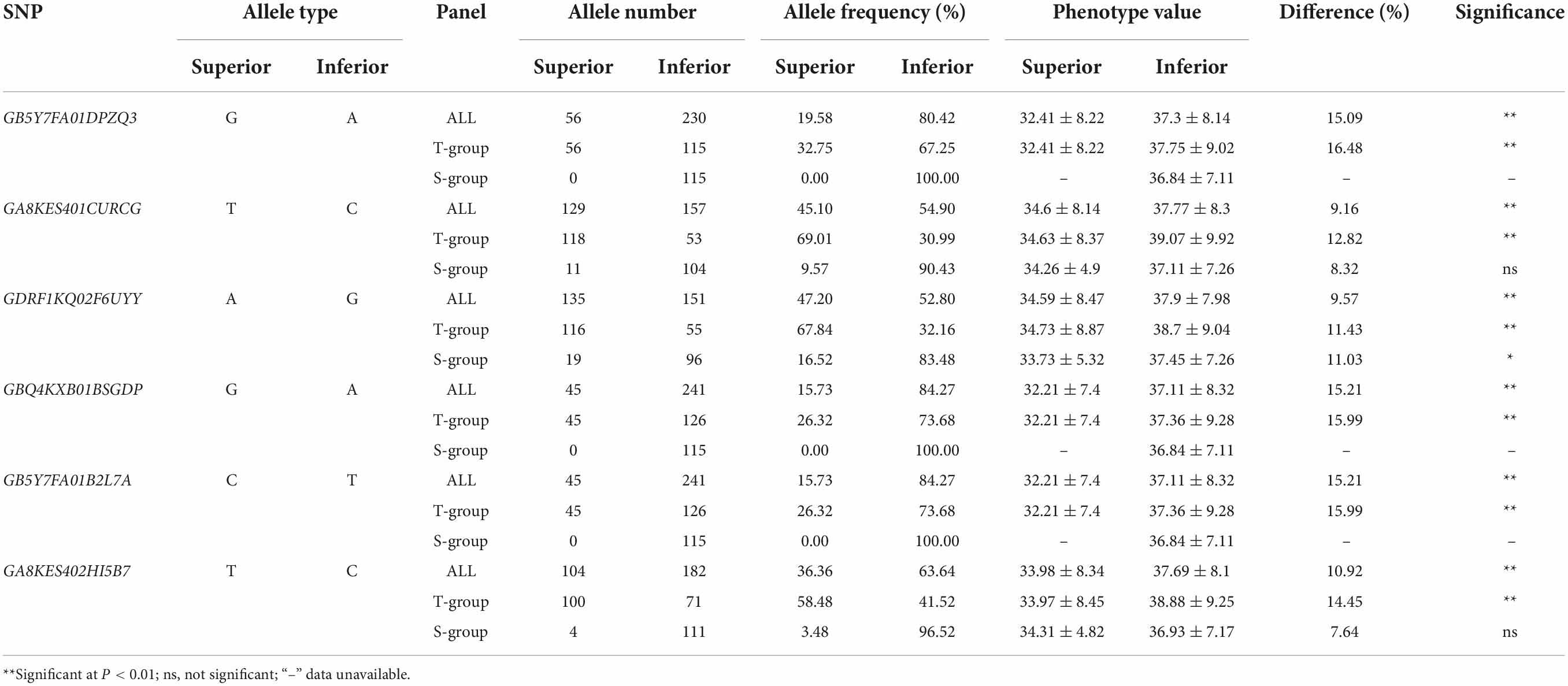
Table 4. Distributions of superior and inferior alleles for the six significant single nucleotide polymorphisms associated with Fusarium crown rot resistance.
Haplotypes based on these six SNP were also analyzed to detect the additive effect. Seven haplotypes that included more than ten accessions were used to compare the difference between groups. It showed that accessions with six superior alleles (the GTAGCT haplotype) were significantly (P < 0.05) more resistant than those with 0–3 superior alleles (Figure 5 and Supplementary Table 2). Accessions with 3–4 superior alleles were significantly (P < 0.05) more resistant than those without any superior allele. It indicated that pyramiding superior alleles could effectively increase FCR resistance of A. tauschii accessions.
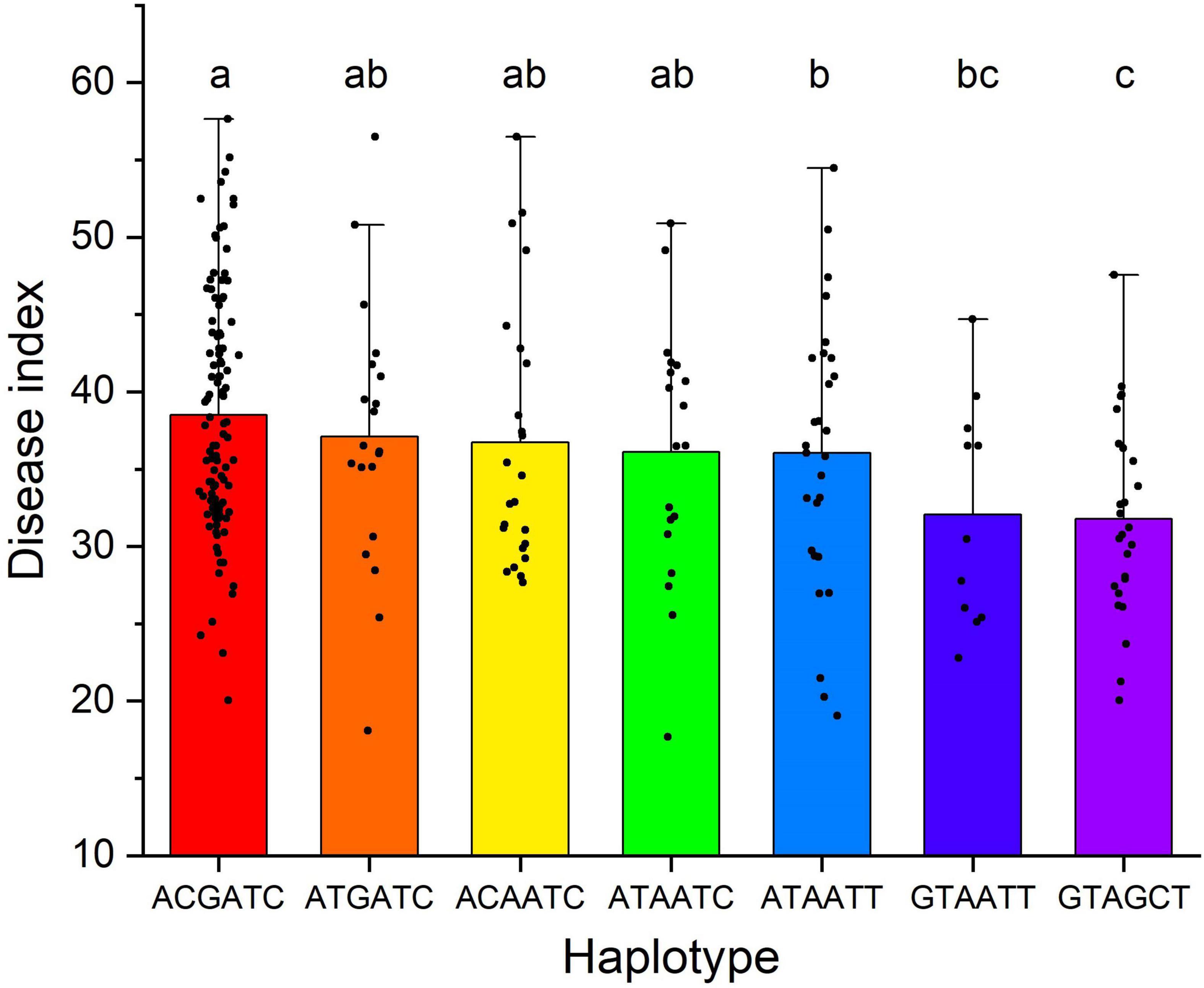
Figure 5. Haplotype analysis based on single nucleotide polymorphisms significantly associated with Fusarium crown rot (FCR) resistance. The different lowercase letters between haplotypes denote FCR resistance were significant (P < 0.05) differences. The seven haplotypes from left to right contained 0, 1, 1, 2, 3, 4, and 6 resistance alleles and presented 107, 20, 24, 19, 28, 11, and 26 accessions, respectively.
Prediction of candidate genes associated with Fusarium crown rot resistance
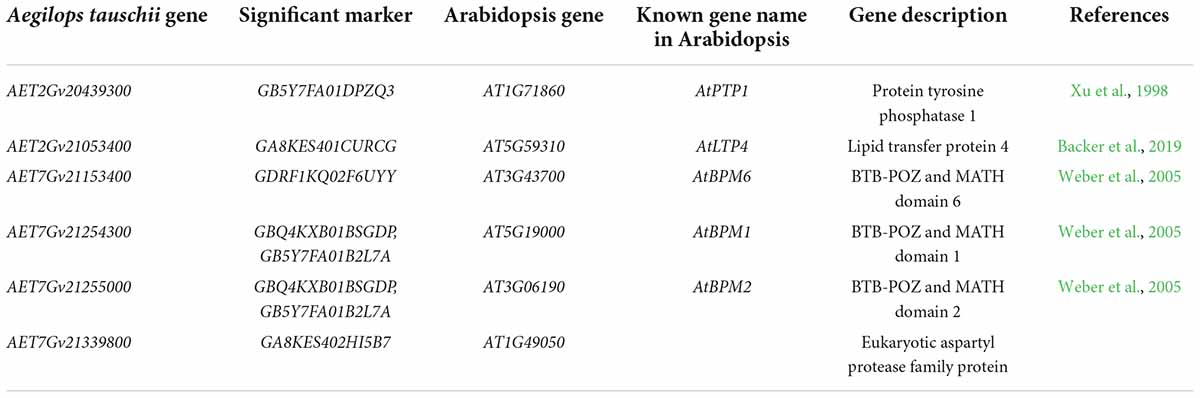
The A. tauschii genome reference sequence (Aet 4.0) was used to identify the candidate genes possibly associated with the significant FCR resistance loci. Seventy-three HC genes were around the six significant SNP for FCR resistance (Supplementary Table 3). The functions of HC genes were annotated using KOBAS 3.0. Fifty-six HC genes were successfully annotated, and 22 were homologous to the known function genes in arabidopsis. Six were selected as candidate genes based on the gene annotation information (Table 5). The first gene, AET2Gv20439300, was around at significant SNP GB5Y7FA01DPZQ3. It encoded protein tyrosine phosphatase 1 and was homologous to AtPTP1 (Xu et al., 1998). AET2Gv21053400 encoding lipid transfer protein 4 was considered the candidate gene for the locus on the long arm of chromosome 2D. It was homologous to the arabidopsis gene AtLTP4, a member of the pathogenesis-related (PR) protein family (Backer et al., 2019). Candidate genes AET7Gv21153400, AET7Gv21254300, and AET7Gv21255000, included BTB-POZ and MATH domain and were homologous to AtBPM6, AtBPM1, and AtBPM2, respectively (Weber et al., 2005). The last gene, AET7Gv21339800, could encode eukaryotic aspartyl protease family protein.
Discussion
Fusarium crown rot resistance alleles is abundant in Aegilops tauschii ssp. tauschii but rarely in Aegilops tauschii ssp. strangulata
Aegilops tauschii, the diploid wild progenitor of the D-genome of common wheat, is a reservoir of genetic diversity for improving bread wheat biotic and abiotic resistance/tolerance. Previous studies have identified resistance/tolerance genes/loci response to phosphorus deficiency, drought, stripe rust, and powdery mildew resistance (Liu et al., 2015a; Qin et al., 2016; Zhang et al., 2019b; Xue et al., 2022). It has proved that superior genes identified from A. tauschii could be quickly and efficiently used to improve hexaploid wheat (Zhang et al., 2019b; Awan et al., 2022; Gaurav et al., 2022). For example, the stripe rust resistance allele YrAS2388R on chromosome 4DS was cloned from A. tauschii. It was only present in A. tauschii (100 and 19% present in subspecies strangulata and tauschii, respectively) and the A. tauschii -derived synthetic wheat and absent in 100% of 461 tested common wheat lines (Zhang et al., 2019b). The synthetic hexaploid wheat method and transgenic test have proved that the allele YrAS2388R can be transferred into hexaploid wheat and improve the resistance to stripe rust (Zhang et al., 2019b). Recently, the first wheat cultivar Shumai1675 carrying YrAS2388R with high resistance to stripe rust was released in China. We identified five moderate resistance accessions and six SNPs associated with FCR resistance in 286 worldwide A. tauschii accessions in the present study. Interestingly, these five accessions with moderate resistance originated from five countries (Afghanistan, China, Iran, Turkey, and Uzbekistan), but all belonged to the subspecies tauschii (Supplementary Table 1). These results suggested subspecies tauschii may be richer in genetic resources of FCR resistance than subspecies strangulata. It was also found that resistance alleles were primarily present in the T-group and rarely present in S-group (Table 4). For three loci, resistance alleles were only present in accession of the T-group and 100% absent in accessions of the S-group. For the rest three, only 11, 19, and 4 resistance alleles were from accessions of the S-group. Besides, A. tauschii ssp. strangulata was known to be the D-genome donor of wheat. A study has proven that D-genome consensus carried only 68 (2.7%) alleles from subspecies tauschii and 2,406 (97.3%) alleles from subspecies strangulata (Singh et al., 2019). It indicated that subspecies tauschii contains more private alleles that were not transferred into wheat than subspecies strangulata. These results suggested that A. tauschii ssp. tauschii is the focus material for mining FCR resistance gene/loci in our further work.
Loci associated with Fusarium crown rot resistance revealed by genome-wide association study in the Aegilops tauschii natural population
QTL for FCR resistance have been identified in barley and wheat. To date, QTL of FCR resistance have been detected on chromosomes 1H, 2H, 3H, 4H, 5H, and 6H in barley (Chen et al., 2013; Liu C. et al., 2015; Gao et al., 2019). For example, two QTL conferring FCR resistance were detected on chromosomes 1H and 3H using a recombinant inbred line population derived from Baudin and AWCS079 (Chen et al., 2013). Using another recombinant inbred line population derived from Fleet and AWCS799, QTL were detected on chromosomes 1H, 2H, 3H, 4H, 5H, and 6H, but only Qcrs.caf-6H was detected in all four experiments (Gao et al., 2019). In wheat, QTL for FCR resistance have been detected on chromosomes 1A, 1B, 1D, 2A, 2B, 2D, 3A, 3B, 3D, 4B, 4D, 5D, 6A, 6B, and 6D (Liu C. et al., 2015; Su et al., 2021; Yang et al., 2021). These QTL are distributed on all D group chromosomes except chromosome 7D. Using one recombinant inbred line population derived from EGA Wylie and Sumai3, a major locus Qcrs.cpi-5D was detected in explained up to 31.1% of the phenotypic variance (Zheng et al., 2014). Besides, a major and stable QTL Qcrs.cpi-2D was detected in three replicated trials with PVE up to 20.2%. The simple repeat sequence (SSR) marker cfd73 linked closely to Qcrs.cpi-2D was used to genotype in two additional recombinant inbred line populations, indicating that Qcrs.cpi-2D could increase FCR resistance (Zheng et al., 2014). Based on these closely linked markers, Qcrs.cpi-2D was mapped on the long arm of chromosome 2D at 556.62–602.73 Mb of the Chinese Spring Reference Sequence physical map v2.1 (IWGSC RefSeq v2.1). In the present study, the SNP GA8KES401CURCG on the long arm of chromosome 2D was significantly associated with FCR resistance. Based on the SNP flanking sequence, GA8KES401CURCG was mapped on chromosome 2D at 582.53 Mb, overlapped with the genomic region of Qcrs.cpi-2D. These results indicated that these two loci might be the same. The resistance allele of this locus was present in both the T-group and S-group. It is well known that A. tauschii ssp. strangulata was the D-genome donor (Singh et al., 2019). Thus, this locus may be originated from A. tauschii and transferred into hexaploid wheat due to the natural hybridization of tetraploid wheat and A. tauschii about 8,000–10,000 years ago. However, more evidence is needed to prove it. In another previous study, the QTL flanking with SSR markers gwm484 and gwm102 on the short arm of chromosome 2D was significantly associated with FCR in field trials in two doubled haploid populations (Martin et al., 2015). This QTL was located on chromosome 2D at 50.63 Mb. In the present study, an SNP GB5Y7FA01DPZQ3 was significantly associated with FCR resistance on the short arm of chromosome 2D at 178.40 Mb. These two loci were far from each other. Thus, the locus identified in this study may be novel. Besides, four SNPs were significantly associated with FCR resistance on the long arm of chromosome 7D. Before this study, no loci/gene for FCR resistance was identified on chromosome 7D in A. tauschii or wheat. These results showed that A. tauschii is a valuable source of novel FCR resistance loci for wheat improvement.
Fusarium crown rot resistance loci are not associated with flowering time, heading date, and plant height in Aegilops tauschii
Heading date and PH are crucial agronomic traits in plants that determine many crop productivity. Previous studies have found that PH and HD affected FCR resistance in wheat (Liu et al., 2010; Liu C. et al., 2015; Zheng et al., 2014). Co-located QTL for resistance and HD and PH, and these loci impeded developing closely linked markers associated with FCR resistance (Liu C. et al., 2015). The present study also analyzed to detect possible relationships between FCR resistance and traits of agronomic importance in A. tauschii, FT, HD, and PH. In the present study, FCR resistance was weakly correlated with FT and HD and was not correlated with PH. A genome-wide association analysis was performed for these three traits to understand their relationship further. Eleven, 11, and 3 SNP were significantly associated with FT, HD, and PH, respectively (Table 4). The results showed no loci between FCR resistance and FT, HD, and PH were overlapped. It indicated that FCR resistance was independent of FT, HD, and PH in the A. tauschii natural population. Thus, these loci that did not influence these possible plant development traits are better used in breeding programs.
Potential candidate genes associated with Fusarium crown rot resistance loci
Using GWAS in a wheat population, the FCR resistance gene TaDIR-B1 encoding dirigent protein has been found and validated. In the present study, six candidate genes were identified for these candidate regions and will further validate their function response to FCR (Table 5). The SNP GB5Y7FA01DPZQ3 on chromosome 2D was significantly associated with FCR resistance. AET2Gv20439300 is flanking this locus and could encode protein tyrosine phosphatase 1. It was homologous to AtPTP1 (Xu et al., 1998). AtPTP1 is a member of phosphotyrosine-specific protein phosphatases in arabidopsis (Xu et al., 1998). The expression of AtPTP1 was altered in response to different environmental stresses. It was also confirmed that AtPTP1 repressed salicylic acid biosynthesis in the autoimmune-like response caused by toll interleukin one receptor/nucleotide-binding/leucine-rich repeat (TIR-NB-LRR) receptor-like resistance gene (Bartels et al., 2009). The SNP GB5Y7FA01DPZQ3 was significantly associated with FCR resistance and located on the long arm of chromosome 2D. AET2Gv21053400 could encode lipid transfer protein four and be homologous to AtLTP4. AtLTP4 is a member of the PR protein family, which plays a fundamental role in a plant’s response to pathogen challenges (Backer et al., 2019). In wheat, Fusarium species infection caused the induction of several PR proteins during the development of seedlings (Caruso et al., 1999). In garlic (Allium sativum L.), it was also found that PR genes, which encoded AsPR1, AsPR2, AsPR4, and AsPR5 proteins, could mediate protection against some Fusarium species infection (Anisimova et al., 2021). Thus, AET2Gv21053400 was suggested as a candidate gene for the locus on the long arm of chromosome 2D. Besides, three genes, AET7Gv21153400, AET7Gv21254300, and AET7Gv21255000, contained BTB-POZ and MATH domains and were homologous to AtBPM6, AtBPM1, and AtBPM2, respectively (Weber et al., 2005). Increasing evidence has found that BTB-POZ and MATH protein family plays a vital role in plant defense regulation (Xu et al., 2003; Zavaliev et al., 2020). In soybean, GmBTB/POZ encoding BTB/POZ domain-containing nuclear protein could positively regulate the response to fungal infection (Zhang et al., 2019a). It was also reported that NbBTB encoding a BTB/POZ domain-containing protein could regulate plant immunity in Nicotiana benthamiana (Zhao et al., 2022). Besides, MATH domain protein is associated with resistance to fungal disease in arabidopsis (Cosson et al., 2010) and apricot (Prunus armeniaca L.) (Mariette et al., 2016). The predicted gene Sobic.010G222400 containing BTB/POZ and MATH domains, was identified as a candidate for resistance to sorghum grain mold caused by Fusarium species (Cuevas et al., 2019). Thus, AET7Gv21153400, AET7Gv21254300, and AET7Gv21255000 may regulate FCR resistance in A. tauschii. The last candidate gene was AET7Gv21339800, which could encode an aspartyl protease family protein. The aspartyl protease plays a vital role in plant-pathogen interactions (Figueiredo et al., 2021). It has been demonstrated that AtAPCB1 encoding aspartyl protease is necessary for fungal resistance in Bcl-2-associated athanogene processing (Li et al., 2016). OsAP47 encoding aspartyl protease is associated with resistance to black-streaked dwarf virus disease and southern rice black-streaked dwarf virus disease (Wang et al., 2022). These six candidate genes were identified for FCR resistance, and further analysis, such as qRT-PCR and the transgenic test, is needed to confirm the function.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
YuL and QW drafted and revised the manuscript and contributed to data analysis. HC and NY performed the phenotypic evaluation and helped with data analysis. FW, ZW, and CL performed part of the statistical analyses. YaL designed and coordinated the study and revised the manuscript. All authors have read and approved the final manuscript for publication.
Funding
This study was funded by the Key Research and Development Program of Sichuan Province (2021YFN0034 and 2021YFG0028) and the National Natural Science Foundation of China (31771794).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.998622/full#supplementary-material
Supplementary Figure 1 | Distributions of 6,739 single nucleotide polymorphisms on Aegilops tauschii chromosomes. Color represents the number of single nucleotide polymorphisms within a 1 Mb window size, from 0 (white) to 10 (red).
Supplementary Figure 2 | Delta K over ten repetitions estimated by population structure analysis.
Supplementary Table 1 | Detail information and phenotypic data of 286 Aegilops tauschii accessions used in this study.
Supplementary Table 2 | Haplotype analysis based on single nucleotide polymorphisms significantly associated with Fusarium crown rot (FCR) resistance.
Supplementary Table 3 | Detail information of predicted genes flanking significant markers associated with Fusarium crown rot resistance.
References
Anisimova, O. K., Shchennikova, A. V., Kochieva, E. Z., and Filyushin, M. A. (2021). Pathogenesis-related genes of PR1, PR2, PR4, and PR5 families are involved in the response to Fusarium infection in garlic (Allium Sativum L.). Int. J. Mol. Sci. 22:6688. doi: 10.3390/IJMS22136688
Awan, M. J. A., Rasheed, A., Saeed, N. A., and Mansoor, S. (2022). Aegilops tauschii presents a genetic roadmap for hexaploid wheat improvement. Trends Genet. 38, 307–309. doi: 10.1016/j.tig.2022.01.008
Backer, R., Naidoo, S., and Van den Berg, N. (2019). The nonexpressor of pathogenesis-related genes 1 (NPR1) and related family: mechanistic insights in plant disease resistance. Front. Plant Sci. 10:102. doi: 10.3389/fpls.2019.00102
Bartels, S., Anderson, J. C., González Besteiro, M. A., Carreri, A., Hirt, H., Buchala, A., et al. (2009). Map kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell. 21, 2884–2897. doi: 10.1105/tpc.109.067678
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). Tassel: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Bu, D., Luo, H., Huo, P., Wang, Z., Zhang, S., He, Z., et al. (2021). KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 49, W317–W325. doi: 10.1093/nar/gkab447
Caruso, C., Chilosi, G., Caporale, C., Leonardi, L., Bertini, L., Magro, P., et al. (1999). Induction of pathogenesis-related proteins in germinating wheat seeds infected with Fusarium culmorum. Plant Sci. 140, 87–97. doi: 10.1016/S0168-9452(98)00199-X
Chen, G. D., Liu, Y. X., Wei, Y. M., McIntyre, C. L., Zhou, M. X., Zheng, Y. L., et al. (2013). Major QTL for Fusarium crown rot resistance in a barley landrace. Theor. Appl. Genet. 126, 2511–2520. doi: 10.1007/s00122-013-2151-4
Cosson, P., Sofer, L., Hien Le, Q., Léger, V., Schurdi-Levraud, V., Whitham, S. A., et al. (2010). RTM3, which controls long-distance movement of Potyviruses, is a member of a new plant gene family encoding a meprin and traf homology domain-containing protein. Plant Physiol. 154, 222–232. doi: 10.4161/psb.5.10.13244
Cuevas, H. E., Fermin Pérez, R. A., Prom, L. K., Cooper, E. A., Bean, S., and Rooney, W. L. (2019). Genome-wide association mapping of grain mold resistance in the US sorghum association panel. Plant Genome 12:180070. doi: 10.3835/plantgenome2018.09.0070
Earl, D. A., and VonHoldt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing structure output and implementing the evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Figueiredo, L., Santos, R. B., and Figueiredo, A. (2021). Defense and offense strategies: the role of aspartic proteases in plant–pathogen interactions. Biology 10:75. doi: 10.3390/biology10020075
Gao, S., Zheng, Z., Hu, H., Shi, H., Ma, J., Wei, Y., et al. (2019). A novel QTL conferring Fusarium crown rot resistance located on chromosome arm 6HL in barley. Front. Plant Sci. 10:1206. doi: 10.3389/fpls.2019.01206
Gaurav, K., Arora, S., Silva, P., Sánchez-Martín, J., Horsnell, R., Gao, L., et al. (2022). Population genomic analysis of Aegilops tauschii identifies targets for bread wheat improvement. Nat. Biotechnol. 40, 422–431. doi: 10.1038/s41587-021-01058-4
Giles, R. J., and Brown, T. A. (2006). Gludy allele variations in Aegilops tauschii and Triticum aestivum: implications for the origins of hexaploid wheats. Theor. Appl. Genet. 112, 1563–1572. doi: 10.1007/s00122-006-0259-5
Jakobsson, M., and Rosenberg, N. A. (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806. doi: 10.1093/bioinformatics/btm233
Jin, J., Duan, S., Qi, Y., Yan, S., Li, W., Li, B., et al. (2020). Identification of a novel genomic region associated with resistance to Fusarium crown rot in wheat. Theor. Appl. Genet. 133, 2063–2073. doi: 10.1007/s00122-020-03577-
Kazan, K., and Gardiner, D. M. (2018). Fusarium crown rot caused by Fusarium pseudograminearum in cereal crops: recent progress and future prospects. Mol. Plant Pathol. 19, 1547–1562. doi: 10.1111/mpp.12639
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, H. L., Yuan, H. X., Fu, B., Xing, X. P., Sun, B. J., and Tang, W. H. (2012). First report of Fusarium pseudograminearum causing crown rot of wheat in Henan. China. Plant Dis. 96, 1065–1065. doi: 10.1094/PDIS-01-12-0007-PDN
Li, X., Liu, C., Chakraborty, S., Manners, J. M., and Kazan, K. (2008). A simple method for the assessment of crown rot disease severity in wheat seedlings inoculated with Fusarium pseudograminearum. J. Phytopathol. 156, 751–754.
Li, Y., Kabbage, M., Liu, W., and Dickman, M. B. (2016). Aspartyl protease-mediated cleavage of BAG6 is necessary for autophagy and fungal resistance in plants. Plant Cell. 28, 233–247. doi: 10.1105/tpc.15.00626
Liu, C., Ogbonnaya, F. C., Bürstmayr, H., and Bürstmayr, H. (2015). Resistance to Fusarium crown rot in wheat and barley: a review. Plant Breed. 134, 365–372. doi: 10.1111/pbr.12274
Liu, Y., Wang, L., Deng, M., Li, Z., Lu, Y., Wang, J., et al. (2015a). Genome-wide association study of phosphorus-deficiency-tolerance traits in Aegilops tauschii. Theor. Appl. Genet. 128, 2203–2212. doi: 10.1007/s00122-015-2578-x
Liu, Y., Wang, L., Mao, S., Liu, K., Lu, Y., Wang, J., et al. (2015b). Genome-wide association study of 29 morphological traits in Aegilops tauschii. Sci. Rep. 5:15562. doi: 10.1038/srep15562
Liu, Y., Yang, X., Ma, J., Wei, Y., Zheng, Y., Ma, H., et al. (2010). Plant height affects Fusarium crown rot severity in wheat. Phytopathology 100, 1276–1281. doi: 10.1094/PHYTO-05-10-0142
Luan, D., Jia, J., Wang, G., and Zhang, J. (2022). Occurrence status and control strategies of wheat crown rot in China (in Chinese). Mai Lei Zuo Wu Xue Bao. 42, 512–520. doi: 10.7606/j.issn.1009-1041.2022.04.15
Luo, M., Gu, Y. Q., Puiu, D., Wang, H., Twardziok, S. O., Deal, K. R., et al. (2017). Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551, 498–502. doi: 10.1038/nature24486
Mariette, S., Wong Jun Tai, F., Roch, G., Barre, A., Chague, A., Decroocq, S., et al. (2016). Genome-wide association links candidate genes to resistance to plum pox virus in apricot (Prunus armeniaca). New Phytol. 209, 773–784. doi: 10.1111/nph.13627
Martin, A., Bovill, W. D., Percy, C. D., Herde, D., Fletcher, S., Kelly, A., et al. (2015). Markers for seedling and adult plant crown rot resistance in four partially resistant bread wheat sources. Theor. Appl. Genet. 128, 377–385. doi: 10.1007/s00122-014-2437-1
Murray, G. M., and Brennan, J. P. (2009). Estimating disease losses to the Australian wheat industry. Australas. Plant Pathol. 38, 558–570. doi: 10.1071/AP09053
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.3410/f.1015548.197423
Qin, P., Lin, Y., Hu, Y., Liu, K., Mao, S., Li, Z., et al. (2016). Genome-wide association study of drought-related resistance traits in Aegilops tauschii. Genet. Mol. Biol. 39, 398–407. doi: 10.1590/1678-4685-GMB-2015-0232
R Core Team (2014). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Singh, N., Wu, S., Tiwari, V., Sehgal, S., Raupp, J., Wilson, D., et al. (2019). Genomic analysis confirms population structure and identifies inter-lineage hybrids in Aegilops tauschii. Front. Plant Sci. 10:9. doi: 10.3389/fpls.2019.00009
Smiley, R. W., Gourlie, J. A., Easley, S. A., Patterson, L., and Whittaker, R. G. (2005). Crop damage estimates for crown rot of wheat and barley in the Pacific Northwest. Plant Dis. 89, 595–604. doi: 10.1094/PD-89-0595
Smith, S. E., Kuehl, R. O., Ray, I. M., Hui, R., and Soleri, D. (1998). Evaluation of simple methods for estimating broad-sense heritability in stands of randomly planted genotypes. Crop. Sci. 38, 1125–1129.
Su, J., Zhao, J., Zhao, S., Li, M., Pang, S., Kang, Z., et al. (2021). Genetics of resistance to common root rot (spot blotch), Fusarium crown rot, and sharp eyespot in wheat. Front. Genet. 12:699342. doi: 10.3389/fgene.2021.699342
Wang, Q., Yan, N., Chen, H., Li, S., Hu, H., Lin, Y., et al. (2021). Genome-wide association study of kernel traits in Aegilops tauschii. Front. Genet. 12:651785. doi: 10.3389/fgene.2021.651785
Wang, Z., Zhou, L., Lan, Y., Li, X., Wang, J., Dong, J., et al. (2022). An aspartic protease 47 causes quantitative recessive resistance to rice black-streaked dwarf virus disease and southern rice black-streaked dwarf virus disease. New Phytol. 233, 2520–2533. doi: 10.1111/nph.17961
Weber, H., Bernhardt, A., Dieterle, M., Hano, P., Mutlu, A., Estelle, M., et al. (2005). Arabidopsis ATCUL3a and ATCUL3b form complexes with members of the BTB/POZ-MATH protein family. Plant Physiol. 137, 83–93. doi: 10.1104/pp.104.052654
Xu, F., Song, Y. L., Wang, J. M., Liu, L. L., and Zhao, K. (2017). Occurrence of Fusarium crown rot caused by Fusarium pseudograminearum on barley in China. Plant Dis. 101, 837–837. doi: 10.1094/PDIS-10-16-1436-PDN
Xu, F., Song, Y. L., Zhou, Y. L., Zhang, H., Wang, J. M., and Li, Y. H. (2016). Occurrence dynamics and characteristics of Fusarium root and crown rot of wheat in Henan Province during 2013-2016 (in Chinese). Plant Prot. 42, 126–132.
Xu, L., Wei, Y., Reboul, J., Vaglio, P., Shin, T., Vidal, M., et al. (2003). BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425, 316–321. doi: 10.1038/nature01985
Xu, Q., Fu, H., Gupta, R., and Luan, S. (1998). Molecular characterization of a tyrosine-specific protein phosphatase encoded by a stress-responsive gene in Arabidopsis. Plant Cell 10, 849–857. doi: 10.1105/tpc.10.5.849
Xue, S., Hu, S., Chen, X., Ma, Y., Lu, M., Bai, S., et al. (2022). Fine mapping of Pm58 from Aegilops tauschii conferring powdery mildew resistance. Theor. Appl. Genet. 135, 1657–1669. doi: 10.1007/s00122-022-04061-8
Yang, X., Zhong, S., Zhang, Q., Ren, Y., Sun, C., and Chen, F. (2021). A loss-of-function of the dirigent gene TaDIR-B1 improves resistance to Fusarium crown rot in wheat. Plant Biotechnol. J. 19, 866–868. doi: 10.1111/pbi.13554
Yu, J., Pressoir, G., Briggs, W. H., Vroh Bi, I, Yamasaki, M., Doebley, J. F., et al. (2006). A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet 38, 203–208. doi: 10.1038/ng1702
Zavaliev, R., Mohan, R., Chen, T., and Dong, X. (2020). Formation of NPR1 condensates promotes cell survival during the plant immune response. Cell 182, 1093–1108. doi: 10.1016/j.cell.2020.07.016
Zhang, C., Huang, L., Zhang, H., Hao, Q., Lyu, B., and Wang, M. (2019b). An ancestral NB-LRR with duplicated 3’ UTRs confers stripe rust resistance in wheat and barley. Nat. Commun. 10, 1–12. doi: 10.1038/s41467-019-11872-9
Zhang, C., Gao, H., Li, R., Han, D., Wang, L., Wu, J., et al. (2019a). GmBTB/POZ, a novel BTB/POZ domain-containing nuclear protein, positively regulates the response of soybean to Phytophthora sojae infection. Mol. Plant Pathol. 20, 78–91. doi: 10.1111/mpp.12741
Zhao, M., Ge, Y., Xu, Z., Ouyang, X., Jia, Y., Liu, J., et al. (2022). A BTB/POZ domain-containing protein negatively regulates plant immunity in Nicotiana benthamiana. Biochem. Biophys. Res. Commun. 600, 54–59. doi: 10.1016/j.bbrc.2022.02.050
Zheng, Z., Kilian, A., Yan, G., and Liu, C. (2014). QTL conferring Fusarium crown rot resistance in the elite bread wheat variety EGA Wylie. PLoS One 9:e96011. doi: 10.1371/journal.pone.0096011
Zhou, Y., Zhao, X., Li, Y., Xu, J., Bi, A., Kang, L., et al. (2020). Triticum population sequencing provides insights into wheat adaptation. Nat. Genet. 52, 1412–1422. doi: 10.1038/s41588-020-00722-w
Keywords: Aegilops tauschii ssp. tauschii, Aegilops tauschii ssp. strangulata, candidate gene, FCR resistance, GWAS, QTL, resistance allele, SNP
Citation: Lin Y, Wang Q, Chen H, Yan N, Wu F, Wang Z, Li C and Liu Y (2022) Genome-wide association mapping of Fusarium crown rot resistance in Aegilops tauschii. Front. Plant Sci. 13:998622. doi: 10.3389/fpls.2022.998622
Received: 20 July 2022; Accepted: 29 August 2022;
Published: 30 September 2022.
Edited by:
Vijay Kumar Tiwari, University of Maryland, College Park, United StatesReviewed by:
Penghao Wang, Murdoch University, AustraliaYong Jia, Murdoch University, Australia
Junyan Feng, Institute of Biotechnology and Nuclear Technology, Sichuan Academy of Agricultural Sciences, China
Zehou Liu, Crop Research Institute, Sichuan Academy of Agricultural Sciences, China
Copyright © 2022 Lin, Wang, Chen, Yan, Wu, Wang, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaxi Liu, eWF4aS5saXVAb3V0bG9vay5jb20=; bGl1eWF4aUBzaWNhdS5lZHUuY24=
†These authors have contributed equally to this work
 Yu Lin
Yu Lin Qing Wang1,2†
Qing Wang1,2† Yaxi Liu
Yaxi Liu