Corrigendum: Editing of StSR4 by Cas9-RNPs confers resistance to Phytophthora infestans in potato
- 1Plant Systems Engineering Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon, Republic of Korea
- 2Department of Biosystems and Bioengineering, KRIBB School of Biotechnology, University of Science and Technology, Daejeon, Republic of Korea
- 3Department of Functional Genomics, KRIBB School of Bioscience, University of Science and Technology, Daejeon, Republic of Korea
- 4Center for Eco-Friendly New Materials, Korea Research Institute of Chemical Technology, Daejeon, Republic of Korea
- 5Department of Biological Sciences, Korea Advanced Institute for Science and Technology, Daejeon, Republic of Korea
- 6Genome Editing Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon, Republic of Korea
- 7GenKOre, Daejeon, Republic of Korea
- 8Department of Biological Sciences, Chungnam National University, Daejeon, Republic of Korea
Potato (Solanum tuberosum L.) cultivation is threatened by various environmental stresses, especially disease. Genome editing technologies are effective tools for generating pathogen-resistant potatoes. Here, we established an efficient RNP-mediated CRISPR/Cas9 genome editing protocol in potato to develop Phytophthora infestans resistant mutants by targeting the susceptibility gene, Signal Responsive 4 (SR4), in protoplasts. Mutations in StSR4 were efficiently introduced into the regenerated potato plants, with a maximum efficiency of 34%. High co-expression of StEDS1 and StPAD4 in stsr4 mutants induced the accumulation of salicylic acid (SA), and enhanced the expression of the pathogen resistance marker StPR1. In addition, increased SA content in the stsr4 mutant enhanced its resistance to P. infestans more than that in wild type. However, the growth of stsr4_3-19 and stsr4_3-698 mutants with significantly high SA was strongly inhibited, and a dwarf phenotype was induced. Therefore, it is important to adequate SA accumulation in order to overcome StSR4 editing-triggered growth inhibition and take full advantages of the improved pathogen resistance of stsr4 mutants. This RNP-mediated CRISPR/Cas9-based potato genome editing protocol will accelerate the development of pathogen-resistant Solanaceae crops via molecular breeding.
Introduction
Plants evolve innate immune systems to defend against pathogens. However, most plant pathogens that escape or suppress plant immunity establish a supply system in the host plant in cooperation with plant cells to derive nutrition, thus ensuring their survival and maintenance. The host plant genes that promote pathogen infection and aid in its survival and maintenance are called susceptibility (S) genes (van Schie and Takken, 2014). Loss of susceptibility function by S gene modification could be a practical and alternative approach for improving the pathogen tolerance of the plant because S genes no longer facilitate infection and support compatibility of the pathogen (Jørgensen, 1992; Vogel et al., 2002; van Schie and Takken, 2014). Many studies use genome editing tools for developing pathogen-resistant plants by targeting the S genes. For example, MLO and eIF4 confer resistance to powdery mildew (Malnoy et al., 2016; Nekrasov et al., 2017) and potyvirus (Pyott et al., 2016), respectively. Furthermore, SWEET and DMR6 confer resistance to Xanthomonas bacteria (Oliva et al., 2019) and a broad spectrum of bacterial and fungal pathogens (de Toledo Thomazella et al., 2021), respectively. In particular, DND1, CHL1, and DMR6 are known as S genes that confer resistance to P. infestans in potato (Kieu et al., 2021).
The Signal Response 1 (SR1) protein, also known as calmodulin (CaM)-binding transcription activator 3 (CAMTA3), mediates the inhibition of effector-triggered immunity, and plays an essential role in hypersensitive response signaling by binding to the promoters of enhanced disease susceptibility 1 (EDS1) and nonrace-specific disease resistance 1 (NDR1) (Du et al., 2009; Nie et al., 2012). Mutated Arabidopsis thaliana (Atsr1) exhibit enhanced disease resistance against several pathogens, such as Pseudomonas syringae pv. tomato DC3000, Golovinomyces cichoracearum, and Botrytis cinerea (Galon et al., 2008; Du et al., 2009; Nie et al., 2012) by elevating salicylic acid (SA) levels. However, side effects caused by mutations in S genes must be considered because S gene-mediated resistance is often accompanied by a decrease in growth, yield, and fertility (Zaidi et al., 2018). Phylogenetic analysis of data available from the potato genome database reveals that StSR4 (PGSC0003DMP400026345, Sotub01g012330) is the CAMTA3 gene, and the deduced amino acid sequence of StSR4 is most similar to that of A. thaliana SR1 (Sun et al., 2016). Sun et al. (2016) report that RNAi silencing of StSR4 enhances late blight resistance in potato. Although StSR4-silenced plants enhanced resistance to the P. infestans, they are designated as genetically-modified organisms (GMOs). Therefore, regulatory restrictions for GMOs limit their use in commercial agricultural settings.
Late blight is caused by the pathogen P. infestans. It is one of the most devastating bacterial diseases worldwide, resulting in yield losses of 40% in susceptible potato cultivars (Arévalo-Marín et al., 2021). One of the main strategies for managing late blight is to select resistant cultivars using traditional breeding methods that rely on cross-breeding or artificial mutagenesis. However, these methods are time-consuming and laborious, and are limited by the availability of resistant germplasm. Moreover, developing disease-resistant potato cultivars through traditional breeding is particularly challenging because of high heterogeneity and tetraploid inheritance (Muthoni et al., 2015). Therefore, new breeding methods to overcome these hurdles are required.
Genome editing through CRISPR-Cas9 is now a major component of new crop breeding technologies for increasing productivity or disease resistance. In particular, inducing mutations using pre-assembled Cas9 protein-synthesized guideRNA (RNP) transfection into protoplasts is highly advantageous when foreign gene-free plants are required. The lower off-target effects of Cas9 protein-synthesized RNP approaches than that of plasmid-based genome editing is also a positive aspect. Overall, RNP-mediated DNA-free genome editing provides a promising strategy for the rapid commercialization of improved crop cultivars given the stringent policies for handling GMOs. To date, RNP-based CRISPR/Cas9 technology is widely used in maize (Zea mays) (Svitashev et al., 2016), bread wheat (Triticum aestivum) (Liang et al., 2017), grapevine (Vitis vinifera) (Malnoy et al., 2016), apple (Malus domestica) (Malnoy et al., 2016), and more recently potato (Andersson et al., 2018; Kieu et al., 2021; Zhao et al., 2021).
In this study, a CRISPR/Cas9 system was used to generate gene edited knock-out potato mutants to the putative S gene, StSR4, as a strategy to confer resistance to P. infestans. Two guide RNAs (gRNAs) targeting the StSR4 locus were each introduced into potato protoplasts in association with the Cas9 nuclease protein in the form of pre-assembled RNPs to analyze insertion and deletion (indel) frequencies in regenerated plants. Editing of StSR4 in potato increased the expression of StEDS1, StPAD4, and StPR1, and triggered an elevation in SA content. Although resistance to late blight improved, increased StEDS1, StPAD4, and StPR1 expression coupled with high SA levels had a significant inhibitory effect on plant growth.
Materials and methods
Plant materials and Cas9 protein
Potato (S. tuberosum cv. Desiree) was used in this study. Plants were grown in vitro at a constant temperature of 24°C under 16 h light/8 h dark photoperiod and 80 μE m−2 s−1 light intensity. Leaves of 2-week-old in vitro-grown plants were used as the explant source for protoplast isolation. To conduct protoplast isolation and regeneration, plants were grown in plant containers containing basal Murashige and Skoog (MS; pH 5.8) medium composed of MS salts and vitamins, 3% (w/v) sucrose, and 0.8% (w/v) Plant agar (Duchefa Biochime B.V.). Streptococcus pyogenes Cas9 (SpCas9) protein (Thermo Fisher Scientific, Waltham, MA, USA), containing the nuclear localization signal, was used to enhance the rate of genomic DNA cleavage. A CRISPR RNA (crRNA), which contains a specific sequence for guiding the Cas9 protein to the target site in the potato genome, and a trans-acting CRISPR RNA (tracrRNA), which hybridizes to the crRNA to activate the Cas9 enzyme, formed the crRNA: tracrRNA complex.
Protoplast isolation and purification
Mesophyll protoplasts were isolated from the leaves of in vitro-grown potato plants, as described previously (Moon et al., 2021). Briefly, partially wounded leaves (~1 g) were soaked in 30 mL extraction solution, containing 20 mM MES, 1% Cellulase Onozuka R10 (Yakult Pharmaceutical, Inc. Co., Tokyo, Ja Hsu), 0.5% Macerozyme (Yakult Pharmaceutical, Inc. Co., Tokyo, Japan), 0.5 M Mannitol, 20 mM KCl, 10 mM CaCl2, and 0.1% BSA (pH 5.7) in the dark at 24°C on a shaker (50 rpm) for 6 h, and then purified using a sucrose cushion to measure their density. The protoplasts were washed twice by resuspending the pellet in 3-fold volume of wash solution (0.24 M NaCl, 2 mg/L NAA, 0.5 mg/L BA in basal medium, pH 5.6) and centrifuged at 50 × g for 5 min. Finally, protoplast pellets were resuspended in MMG solution (4 mM MES [pH 5.7], 400 mM mannitol, and 15 mM MgCl2) at a final concentration of 1 × 106 protoplasts/mL. To confirm the protoplast density, the number of protoplasts in a 10 µL aliquot of the protoplast suspension was measured using a Fuchs-Rosenthal Hemocytometer chamber under a microscope.
Target site selection and gRNA design
Three specific gRNAs targeting the StSR4 gene were designed using the CRISPR RGEN Cas-Designer software (Park et al., 2015). The synthetic crRNAs were purchased from Bioneer Co. (Daejeon, South Korea), and tracrRNA was purchased from Integrated DNA Technologies Co. (Coralville, IA, USA).
Genotyping
To characterize the StSR4 exon1 region, the genomic DNA of cv. Desiree was amplified with StSR4_F1(446) and SR4_R-T7 primers (Table S1) using 2X pfu PCR Master Mix (BioFACT, Daejeon, South Korea). The amplified PCR products were purified and sequenced by Sanger sequencing at Bioneer Co. (Daejeon, South Korea). As a result, a SNP of C/T was observed at the +39bp region of exon1 (Figure S1). Therefore, the target site was selected within the sequence except for the site with the SNP.
RNP assembly
The RNP complex was formed using the Cas9 protein and synthetic crRNA and tracrRNA. First, to generate the gRNA complex, a mixture of crRNA and tracrRNA (1:1 molar ratio) was incubated at room temperature for 10 min. Then, 10 µg each of the Cas9 protein and gRNA complex (1:10 molar ratio) were mixed and incubated at room temperature for 10 min.
Protoplast transfection
The RNP was introduced into potato protoplasts by polyethylene glycol (PEG)-mediated transformation, as described previously (Nicolia et al., 2015; Woo et al., 2015). Briefly, resuspended protoplasts (3 × 105) were mixed with the pre-assembled RNP and incubated at room temperature for 10 min. Then, an equal volume of freshly prepared 40% (w/v) PEG solution was added and the mixture was incubated at 25°C for 10 min in the dark. After stopping the reaction, the transfected protoplasts were collected by centrifugation at 50 × g for 5 min and then gently resuspended in differentiation (D) medium (Nicolia et al., 2015; Moon et al., 2021). Then, an equal volume of alginate solution (2.8% w/v alginic acid-sodium salt and 0.4 M sorbitol) was added to the protoplast suspension, and mixed thoroughly by inverting. After approximately 2 h of incubation in the dark at room temperature, alginate droplets solidified on solid agar (0.4 M sorbitol, 50 mM CaCl2·2H2O, 2 g/L gelrite, and 6 g/L plant agar) were cultured for plant regeneration.
T7E1 assay
One week after transfection, targeted mutagenesis in potato protoplasts was detected by the T7E1 assay. Total genomic DNA was isolated from the protoplast population or calli using the HiYield Genomic DNA Mini Kit (Real Biotech Corporation, Taiwan), according to the manufacturer’s instructions. Then, the target StSR4 site (289 bp) was amplified from the genomic DNA using StSR4_F1 (446) and StSR4_R-T7 primers (Table S1), and the PCR product was used for the T7E1 assay as described previously (Kim et al., 2009). Single band PCR products were denatured at 95°C for 5 min and then re-annealed by decreasing the temperature by 1°C per min until reaching 4°C. The hetero-complexed PCR product (15 µL) was incubated with 5 U of the T7E1 enzyme (New England Bio Labs, Ipswich, MA, USA) at 37°C for 30 min. The mismatched products were analyzed by electrophoresis.
Callus formation and plant regeneration
A method for regenerating plants from micro callus was optimized for our experimental conditions, based on previously reported methods (Moon et al., 2021). Briefly, the transfected protoplasts immobilized in alginate lens were incubated on callus induction medium in the dark at 25°C for 4 weeks, with subculture at weekly intervals, to produce micro callus. Micro calli were transferred to the proliferation medium for mini calli formation, which were subsequently transferred to greening medium. After 6 weeks, greened calli were transferred to regeneration medium and subcultured every 2 weeks with special care to avoid breaking the callus cluster. Regenerated plants from green callus were transplanted into medium and soil by inducing roots. These regenerated plants were obtained by inducing single micro callus and mini callus from protoplast fixed to the alginate lens, and since the callus from which on plant was regenerated was discarded, each regenerated plant can be seen as an individual plant.
Sanger sequencing assay
PCR-Sanger sequencing analysis can identify mutations or indel efficiencies by sequencing the genomic region surrounding the amplified target site and analyzing the sequence difference from the wild-type sequence through sequence alignment soft (Gaillochet et al., 2021).
To identify CRISPR/Cas9-induced mutations in the regenerated plants, the region flanking the StSR4 target site was amplified directly from plant tissue with HelixAmpTM Direct PCR [3G] (NANOHELIX, Daejeon, South Korea), according to the manufacturer’s instructions. The 289 bp amplicon was purified using the Expin PCR SV Kit (GeneAll, Seoul, South Korea) and subjected to Sanger sequencing (BioFACT, Daejeon, South Korea). The insertion/deletion mutation (indel) at the target site was analyzed based on the sequencing chromatogram. The frequency of mutagenesis was defined as the number of mutants divided by the total number of regenerated plants (Figure S2).
Next generation sequencing-based targeted deep sequencing and mutation analysis
Genomic DNA was isolated from protoplasts at 3–4 days after transfection and from regenerated potato plants. Libraries were constructed using a two-step PCR protocol. A primary PCR was performed using 2X pfu PCR Master Mix (BioFACT, Daejeon, South Korea) to attach index and sequencing adaptors to both ends of the 289-bp StSR4 target site containing sequences complementary to gRNA_1 and gRNA_3. To create adaptor overhangs at both ends of the amplicon by primary PCR, the 5’-ILMN preadapter-sequencing primer and sequence-specific locus primer-3’ were designed. Primers SR4_F (adaptor) and SR4_R (adaptor) were purchased from Bioneer. The amplicon was purified using the Expin PCR SV Kit (GeneAll, Seoul, South Korea), according to the manufacturer’s instructions, and was used as the DNA template for the secondary PCR, which was performed using the Nextera XT Index Kit (Illumina, San Diego, CA USA) to attach the index adaptors. Targeted deep sequencing was performed using the Illumina MiSeq platform at Macrogen (Seoul, South Korea). Raw reads for all accessions are available from stsr4_1 mutants (SRA accession No. SRR20664905-SRR20664984) and stsr4_3 mutants (SRA accession No. SRR20664783-SRR20664862) in GenBank Sequence Read Archive (SRA) under BioProject number (PRJNA862654).
Morphological analysis of stsr4 mutants
The morphology of at least five plants of each stsr4 mutant line was examined. Single nodes were cut from cultured potato plantlets and cultured in basal MS medium at 24°C, 16 h light/8 h dark photoperiod, and 50% humidity. All plants were photographed daily with a high-resolution camera (Nikon D750 with AF-S Micro Nikkor 105 mm 1:2.8G ED lens; Nikon, Inc., Japan) for up to 30 days. Phenotypic traits including number of branches per plant, number of leaves per branch, stem length, and midrip length were measured using four plants excluding maximum and minimum values from six plants with ImageJ software (https://imagej.nih.gov/ij). Midrip and petiole lengths were measured with an average of three leaves from the first leaf excluding the shoot-tip in each plant. However, the early stage phenotype (0 day-5days) was measured with an average of two leaves in each plant due to the small size plant.
P. infestans resistance assay
To perform the P. infestans resistance assay, fully expanded leaves of stsr4 mutants (selected based on targeted deep sequencing data) and wild-type Desiree plants, grown in a controlled room at 25°C and 16 h light/8 h dark photoperiod, were inoculated with P. infestans strain 88069 zoospore suspension (3 × 104 sporangia/mL), as described previously (Ha et al., 2017). The inoculated plants were incubated at 20°C under high humidity (95–100%), and disease symptoms were examined at 5 days after inoculation (DAI). Five days after inoculation, the area of necrotic symptoms to the total leaf area in the whole plant was investigated, and the average ratio (%) of symptom was quantified from high-resolution camera (Nikon D750 with AF-S Micro Nikkor 105 mm 1:2.8G ED lens; Nikon, Inc., Japan) and ImageJ software. All experiments were repeated five times.
Expression analysis of pathogen related genes
Total RNA was extracted from the leaf tissue using TRIzol Reagent (Invitrogen, Waltham, MA, USA) and then treated with DNase I (Qiagen), according to the manufacturer’s instructions. The total RNA isolated from three biological replicates per treatment was pooled, and the quality of the pooled samples was evaluated using the NanoDrop ND-2000 Spectrophotometer (Thermo Fisher Scientific). Then, first-strand cDNA was synthesized from 1 μg total RNA using the AccuPower® CycleScript™ RT PreMix (Bioneer), according to the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was performed on the CFX Connect Real-Time PCR System (Bio-Rad, Hercules, CA, USA) using the SYBR Green Master Mix (Enzynomics Co., Daejeon, South Korea) and StEDS1, StPAD4, and StPR1 specific primers (Table S1). The EF1a gene was used as an internal control for data normalization. Three biological replicates were performed for each experiment.
Quantification of SA
Five biological replicates, each containing 10 plants, were performed for each stsr4 mutant line. The sample used for SA accumulation analysis was harvested from leaves of plants cultured in vitro for 20 days in basal MS medium at 24°C, 16 h light/8 h dark photoperiod, and 50% humidity. Leaf tissues were frozen in liquid nitrogen, and then ground with a mortar and pestle. One millilitre of ethylacetate containing 1 µL/mL SA (as an internal standard) was added to approximately 100 mg frozen ground plant tissue in 2 mL EP tubes. The supernatant was evaporated to complete dryness at 30°C for 1 h using a vacuum concentrator. The dried pellet was resuspended in 100 µL of 70% methanol and subjected to liquid chromatography-mass spectrometry (LC-MS) analysis for measuring the SA of the leaves.
Analysis of candidate off-target editing in mutants
To validate the accuracy of Cas9-RNP-mediated genome editing, eight candidate off-target sites containing 2–4 nt mismatches were identified using the Cas-OFFinder program (http://rgenome.net/cas-offinder/) (Bae et al., 2014). The sequence-specific primer sets (Table S1) were designed to PCR amplify the region flanking the eight putative off-target sites from genomic DNA isolated from wild-type and stsr4 mutants. The amplified PCR products were subjected to NGS-based targeted deep sequencing using the Illumina MiSeq platform (Macrogen, Seoul, South Korea). The raw reads of targeted deep sequencing are available from off-target data (BioSample accession No. SAMN30672769-SAMN30672792) in BioProject number (PRJNA862654).
Statistical analysis
The statistical significance of the difference between two average values was analyzed using Student’s t‐test at p < 0.05. Quantitative data are displayed as mean ± standard deviation (SD).
Results
Generation of stsr4 knockout mutants
Two guide RNAs (SR4_1 and 3) complementary to the exon 1 sequence were designed to knockout the StSR4 gene (Sun et al., 2016) in the tetraploid potato cultivar ‘Desiree’. The SR4_1 and SR4_3 gRNAs without SNPs in the target sequences were used for subsequent experiments. These targets were confirmed that there appears to be no allelic variation (Figure 1A).
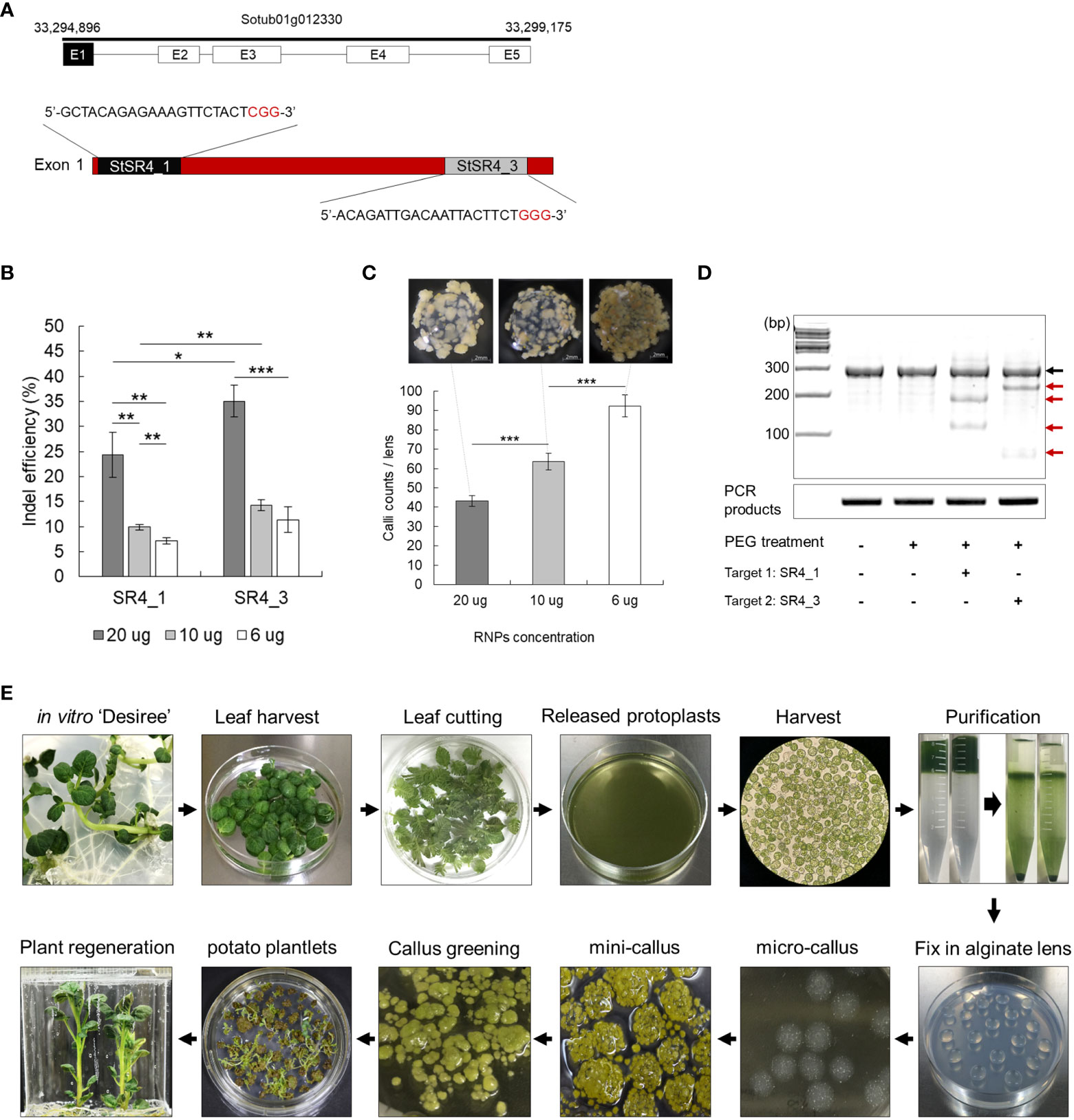
Figure 1 Ribonucleoprotein (RNP)-mediated CRISPR/Cas9 genome editing in potato. (A) Identification of target susceptibility (S) genes. Selection and design of two crRNA target sites in exon 1 of StSR4 in the potato cultivar Desiree. Black sequence, crRNA target sequence; red sequence, PAM site. (B) Indel efficiency at two target sites, depending on the concentration of the Cas9 protein and guide RNA (gRNA) in protoplasts. Values represent the mean ± SE of three replicated experiments. Student’s t-test; *p < 0.05, **p < 0.01, ***p < 0.001. (C) Efficiency of mini callus induction, depending on the concentration of RNPs in protoplasts. Callus growth as a function of RNP concentration: 20 µg, 10 µg, and 6 µg. (D) The T7E1 assay of mutations in protoplasts independently treated with SR4_1 and SR4_3 RNPs. Black arrow indicates the wild-type fragment; red arrow indicates indel-carrying fragments. (E) Schematic diagram showing the process of inducing plant regeneration from potato protoplast.
Overcoming the low editing efficiency of the RNP-mediated CRISPR-Cas9 system remains a challenge. In an attempt to mitigate this problem, the effect of injecting different concentrations of RNP on editing efficiency was examined. Increasing the amount of RNP to 20 µg improved indel efficiency (Figure 1B), but simultaneously decreased micro calli formation (Figure 1C). The transfection of 20 µg RNP into protoplasts yielded fewer, but larger, light-yellow-colored calli than those of 6 µg RNP, which induced the formation of micro calli to approximately 92.4 calli/6 mm alginate lens mini brown-calli. PEG-mediated transfection of 20 µg RNP into protoplasts resulted in indel frequencies of 24.3 ± 4.5% and 35.1 ± 3.2% for SR4_1 and SR4_3, respectively, as assessed by the T7E1 assay (Figures 1B, D). Regenerated plants were successfully induced from RNP-infected protoplasts using the established protoplast-derived regeneration system (Figures 1E; S3), and 170 and 240 mutants of 24.6% and 33.9% were obtained by Sanger sequencing, respectively (Figure 2A; Table S2). In addition, mutations were reconfirmed by T7E1 assay in 30 randomly selected mutants (13 stsr4_1 and 17 stsr4_3 mutants) (Figure S2).
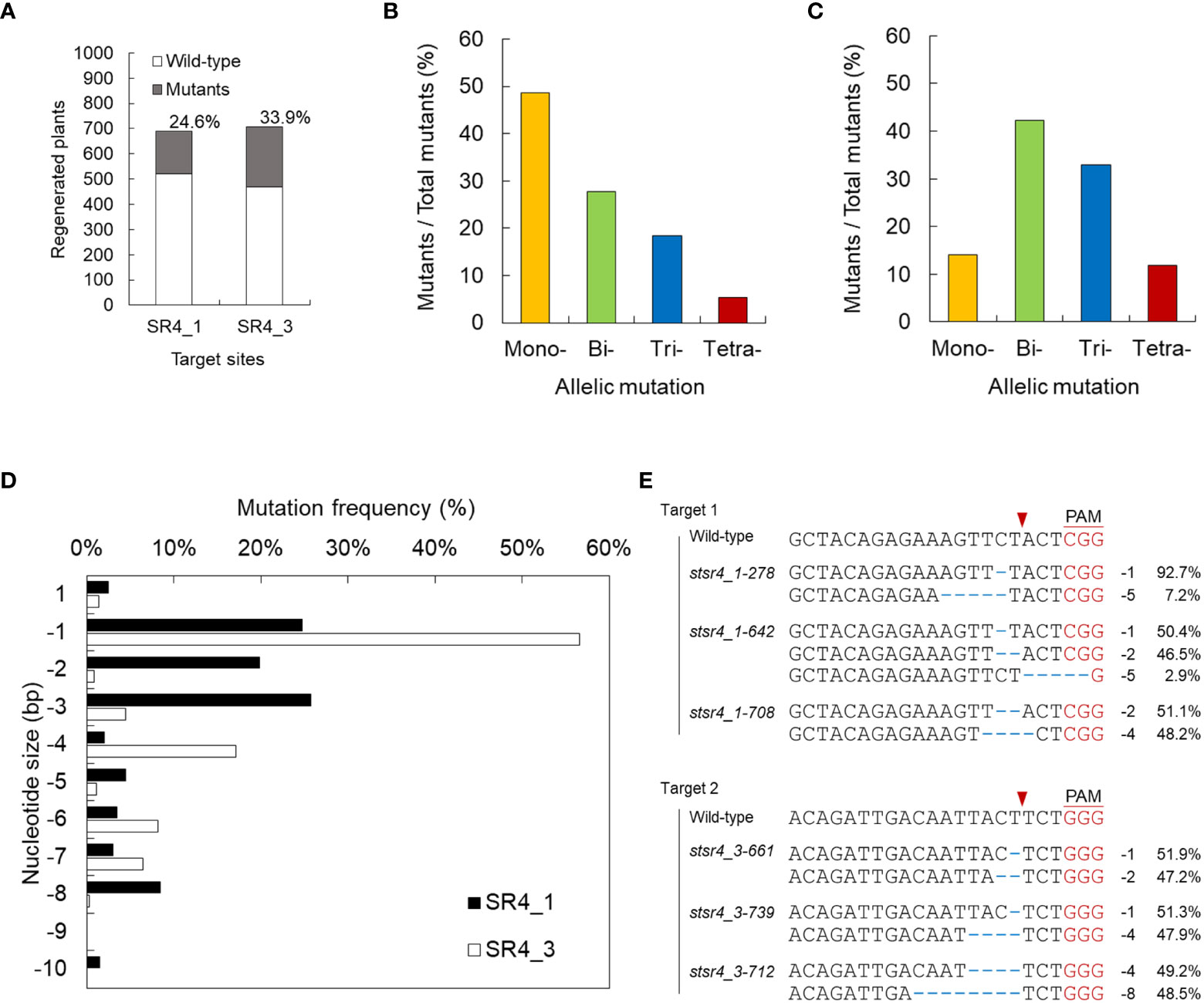
Figure 2 Screening for mutants induced by StSR4 gene editing of regenerated plants. (A) Efficiency of stsr4 knockout mutant regeneration from protoplasts transfected with SR4_1 and SR4_3 RNPs. A total of 170 and 240 mutants were identified by PCR, followed by Sanger sequencing, among 691 and 708 SR4_1 and SR4_3 transfected protoplast-derived regenerated potato plants, respectively. (B, C) Mutation type of mutants induced by the RNPs of SR4_1 (B) and SR4_3 (C). Mono-, only one allele is mutated; Bi-, the two alleles are mutated; Tri-, the three alleles are mutated; Tetra-, the four alleles are fully mutated. (D) Targeted deep sequencing-based analysis of indel patterns in the regenerated mutants. (E) Sequence analysis by targeted deep sequencing of 4 KO stsr4 mutants.
We performed targeted deep sequencing with randomly selected 76 individual mutant lines edited by SR1_1_RNP and 78 by SR4_3. Mutants were obtained in all alleles with frequencies of 3.9% (4 mutants) and 11.8% (9 mutants) in both SR4_1 and SR4_3 target sequences, respectively. Mutations induced in 1, 2, and 3 alleles showed efficiencies of 48.7%, 27.6%, and 18.4% in SR4_1 mutants, respectively, and 14.1%, 42.3%, and 32.9% in SR4_3 mutants, respectively (Figure 2B; Table S2). In regard to the indel type of each mutant, various deletions of 1, 2, and 3 nt were detected in SR4_1, whereas in the target sequence of SR4_3, 1 nt was deleted with more than 50% frequency, followed by deletion of 4 nt (Figure 2C). Finally, six plants carried a frameshift mutation in 90% or more NGS reads obtained from each target site (Figure 2D).
StSR4 acts as a negative regulator of SA-related gene expression
In Arabidopsis, AtSR1 not only inhibits the expression of several SA immunity pathway genes, but also directly controls the expression of AtEDS1, which encodes a putative lipase-like protein that promotes the accumulation of SA through a poorly understood mechanism (Kim et al., 2017) (Figure 3A). To determine the effect of StSR4 gene suppression in potato, transcript levels of the SA pathway-related gene, StEDS1, was analyzed by qRT-PCR. StEDS1 expression was evaluated on randomly selected representative mutants for two allele-types (two allele knockouts, 2KO: stsr4_3-55; three allele knockouts, 3KO: stsr4_3-19 and stsr4_3-698), and four allele knockouts (4KO:srst4_1-278, stsr4_1-642, stsr4_1-708, and stsr4_3-739) (Figures 2D; Figure S4). On the other hand, single allele knockout (1KO) mutants were excluded from the analysis because it was expected that their growth would not be significantly different from that of wild type. The results showed that relative StEDS1 transcript levels increased at least 2-fold in the six mutants except for stsr4_1-642 (Figure 3B). In particular, the stsr4_3-698 mutants showed about 7.51 ± 0.57-fold higher relative StEDS1 transcript levels. These results indicate that the StSR4 acts as a negative regulator of StEDS1 expression.
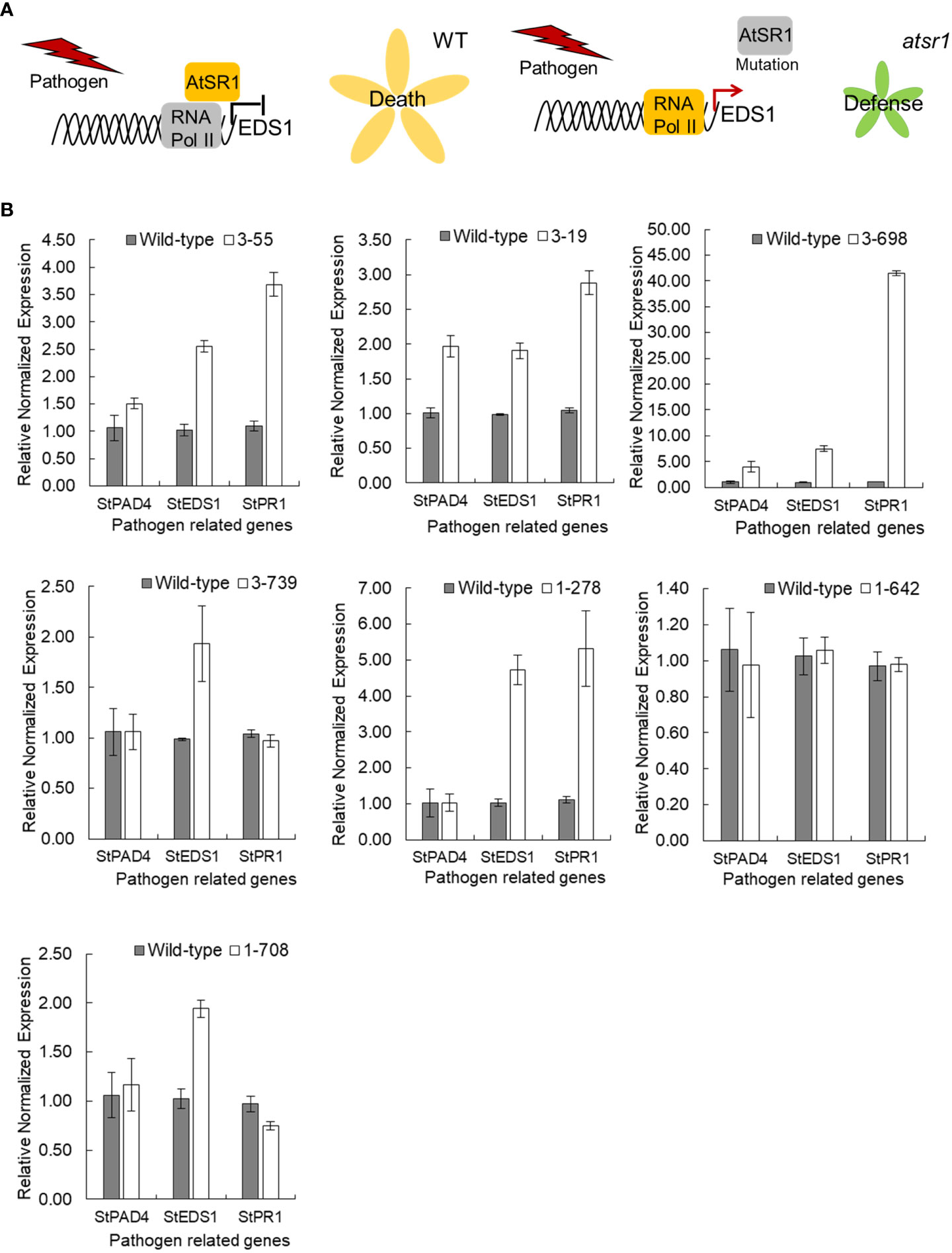
Figure 3 Transcript levels of StEDS1, StPAD4 and StPR1 in stsr4 mutants. (A) A model demonstrating the function of AtSR1 as a negative regulator of pathogen resistance. (B) Analysis of mRNA expression of SA-related genes, StEDS1, StPAD4, and StPR1, in stsr4 mutants.
Because an increase in EDS1 expression causes a rise in SA levels, the expression of the SA marker gene, pathogenesis-related gene 1 (PR1), was analyzed in six StSR4-edited mutants with elevated StEDS1 expression (Figure 3B). Results showed that relative StPR1 transcript levels increased in the four mutants, stsr4_3-19, stsr4_3-55, stsr4_1-278, and stsr4_3-698. StPR1 expression increased by 41.55 ± 0.48 fold in the stsr4_3-698 mutant, which was the highest among the mutants with elevated StEDS1. StPAD4 transcript levels were quantified to determine whether the increased StPR1 expression is caused by SA accumulation resulting from the interaction between PAD4 and EDS1. Results revealed that relative StPAD4 expression was significantly higher in stsr4_3-19 and stsr4_3-698 mutants, and slightly increased in stsr4_3-55 mutants. By contrast, StPAD4 expression did not change in stsr4_1-278. Mutating SR1 in A. thaliana leads to a dwarf phenotype because of elevated SA (Du et al., 2009). In addition, the EDS1 and PAD4 protein complex promotes SA biosynthesis (Liang et al., 2017). The stsr4_3-55 and stsr4_1-278 mutants with slightly elevated StPAD4 expression and no change in StPAD4 transcript levels, respectively, showed a phenotype almost similar to that of wild type except for petiole length (Figure 4). The stsr4_3-19 and stsr4_3-698 mutants, which showed enhanced expression of StPAD4 and StEDS1, had dwarf phenotypes. The dwarf phenotypes of stsr4_3-19 and stsr4_3-698 were expected given that these mutants had elevated SA. The stsr4_3-19 mutant showed a typical dwarf phenotype, with increased branch numbers and leaves per plant, as well as reduced midrib and stem lengths (Figure 4). The stsr4_3-698 mutant, which had the highest levels of StPR1 transcripts among the mutants, displayed inhibited stem and leaf growth, and had almost no growth for 20 days in vitro. Because of the severely inhibited growth phenotype of stsr4_3-698, it was difficult to obtain samples to further evaluate this mutant. As such, subsequent experiments excluded analysis of the stsr4_3-698 mutant.
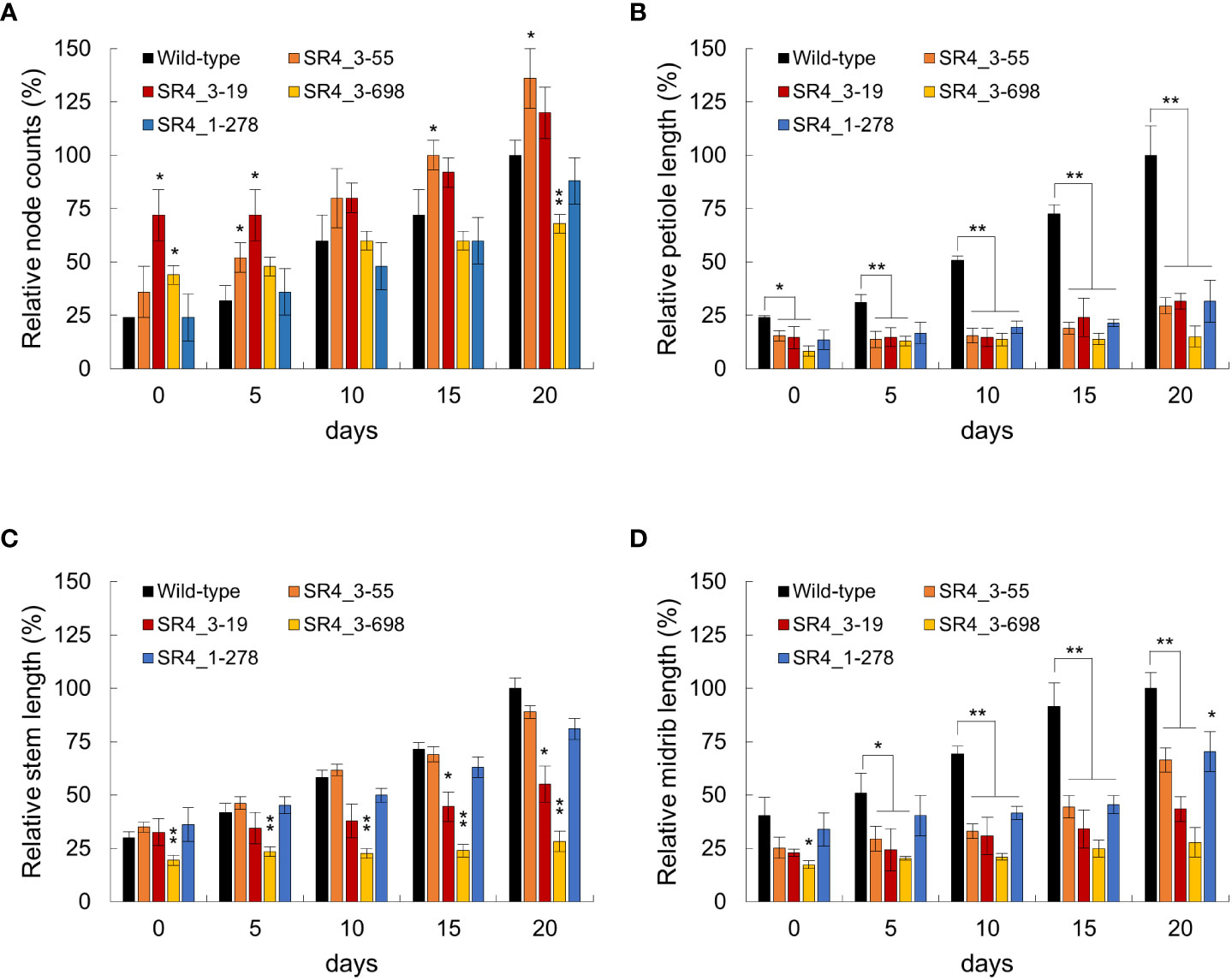
Figure 4 Phenotypic characterization of wild type and stsr4 mutants (stsr4_3-55, stsr4_3-19, stsr4_3-278, and stsr4_3-698). Plants were grown in vitro for 20 days at 16 h light/8 h dark. Representative whole plant phenotypes in wild type and stsr4 mutants. The typical phenotype with node counts (A), petiole length (B), stem length (C), and midrib length (D) of each genotype was observed and photographed every 5 days. Node counts (number), petiole, stem, and midrip length (mm) of 20 days old wild-type plants were 8.3 ± 0.57, 10.73 ± 1.47, 38.37 ± 1.85, and 11.63 ± 0.85, respectively. Values represent the mean ± SE of six replicated experiments (each mean value represents the measured phenotype of four plants). Bars indicate SE. Student’s t-test; *p < 0.05, **p < 0.01.
High SA content in stsr4 mutants leads to enhanced pathogen resistance
Results thus far showed that StSR4 editing triggered increased StEDS1 and StPR1 expression. Endogenous SA content in three stsr4 mutants, namely, stsr4_3-19, stsr4_3-55, and stsr4_1-278, was measured to determine whether increased StPR1 expression and the dwarf phenotypes were caused by elevated SA through interactions with StPAD4. The stsr4_3-19 mutant, which had increased expression of StPAD4 and StEDS1, accumulated the highest amount of SA of 1,153 ng/g FW. This increase in SA level in stsr4_3-19 was significantly higher than that of the wild type by 11.45-fold. The stsr4_3-55 mutant had SA levels of 366 ng/g FW, while the stsr4_1-278 mutant, which had the lowest StPAD4 expression, showed an SA content of 266 ng/g FW. Endogenous SA content in stsr4_1-278 was 2.6-fold higher than that of wild type (Figure 5A). It is worth noting that stsr4_3-19, which had higher SA levels than those of stsr4_3-55 and stsr4_1-278, also had the most severe dwarf phenotype among the mutants analyzed (Figures 5A, B).
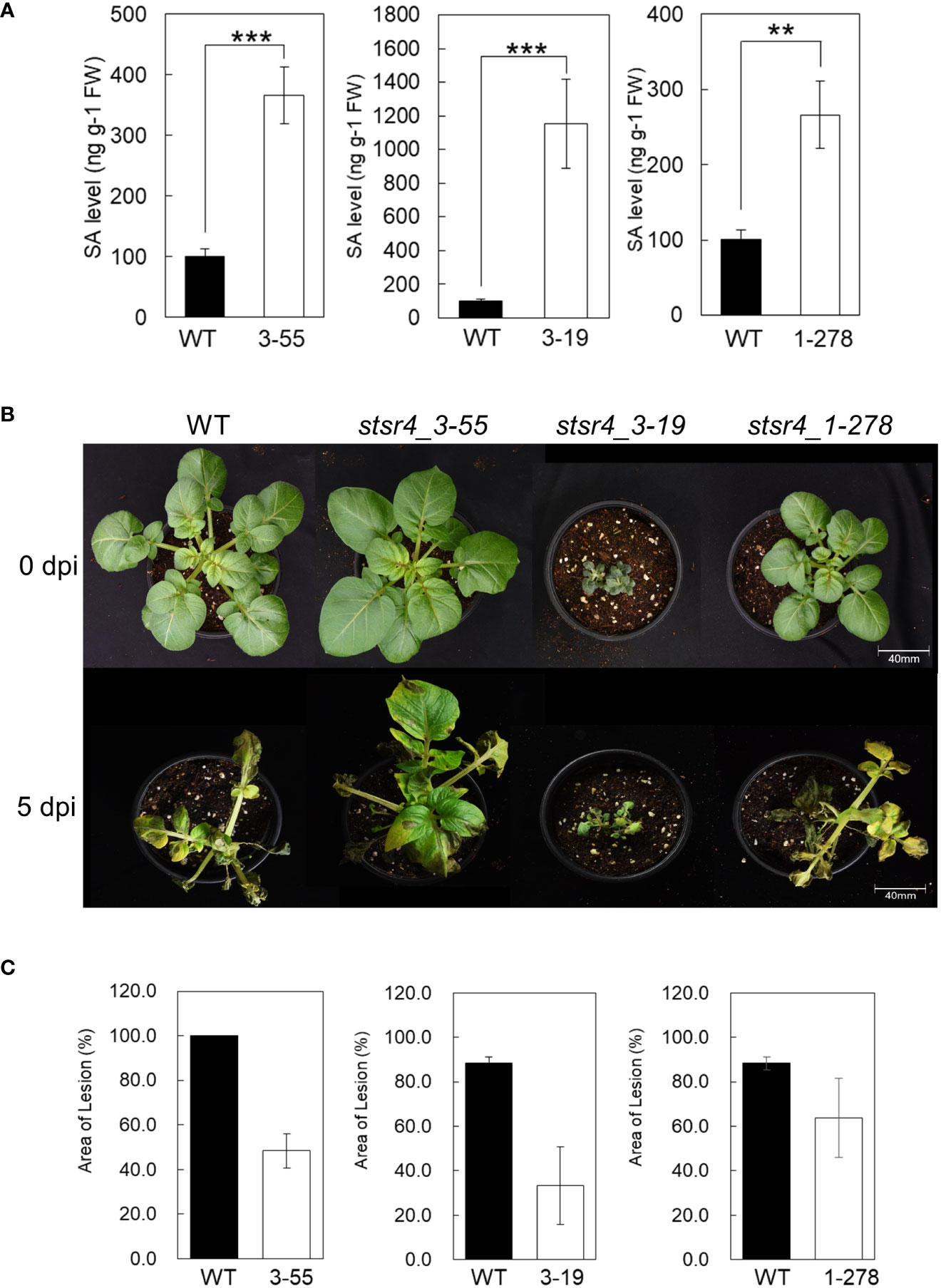
Figure 5 Plant immune response through the suppression of StSR4 expression is regulated by SA. (A) Quantification of free SA in three stsr4 mutants (stsr4_3-55, stsr4_3-19, and stsr4_1-278). Data represent the mean ± SE of five biological replicates. Asterisks indicate significant differences (Student’s t-test; **p < 0.01, ***p < 0.001). (B) Growth at 25°C for 3 weeks in pots of three mutants before pathogen resistance experiment. (C) Disease symptoms observed on the leaves of wild type and stsr4_3-55 and stsr4_3-19 mutant plants at 0 days post-inoculation (dpi) and 5 dpi with Phytophthora infestans. Leaves of wild type and stsr4 mutant plants grown at 25°C for 3 weeks were sprayed with a zoospore suspension of P. infestans strain 88069. At least three biological replicates were performed.
High SA accumulation enhances the plant innate immunity system, and therefore improves plant resistance to pathogens (Ryals et al., 1996). P. infestans, which is the causal organism of potato late blight disease, was inoculated into leaves of three stsr4 mutants to determine whether increased SA content affects resistance to pathogens. Results revealed that at 5 DAI, most tissues of wild-type were necrotic. By contrast, tissues of stsr4 mutants had fewer necrotic lesions than those in wild type. For example, the leaf area of the stsr4_3-55 mutant was only 48.3% necrotic. Furthermore, leaf area of the stsr4_3-19 mutant, which had the highest SA content, was only 38% necrotic, and therefore exhibited the highest resistance to P. infestans among the stsr4 mutants (Figure 5C). However, stsr4_1-278 mutant, which showed the lowest SA levels, had weak resistance with 72% of its leaf area exhibiting necrosis. These results suggest that SA accumulation in potato improves resistance to pathogens, and that increased StEDS1 expression resulting from StSR4 editing increases SA content through its interaction with StPAD4.
As noted earlier, the stsr4_3-55 mutant had a wild-type phenotype, whereas the stsr4_3-19 mutant exhibited a dwarf phenotype when grown on MS medium. Two stsr4 mutants were grown for 1 month in soil to determine whether the stsr4_3-19 dwarf phenotype was still observed under a different growth environment (Figure 6A). Similar to results obtained from MS medium, the stsr4_3-55 mutant showed a wild-type phenotype, whereas the dwarf phenotype was maintained in the stsr4_3-19 mutant. These results suggest that the dwarf phenotype of stsr4_3-19 mutant was caused by StSR4–triggered SA accumulation, rather than an environmental effect. The results with potato described here are consistent with a previous report showing that inhibition of SR1 induces a dwarf phenotype in A. thaliana (Du et al., 2009).
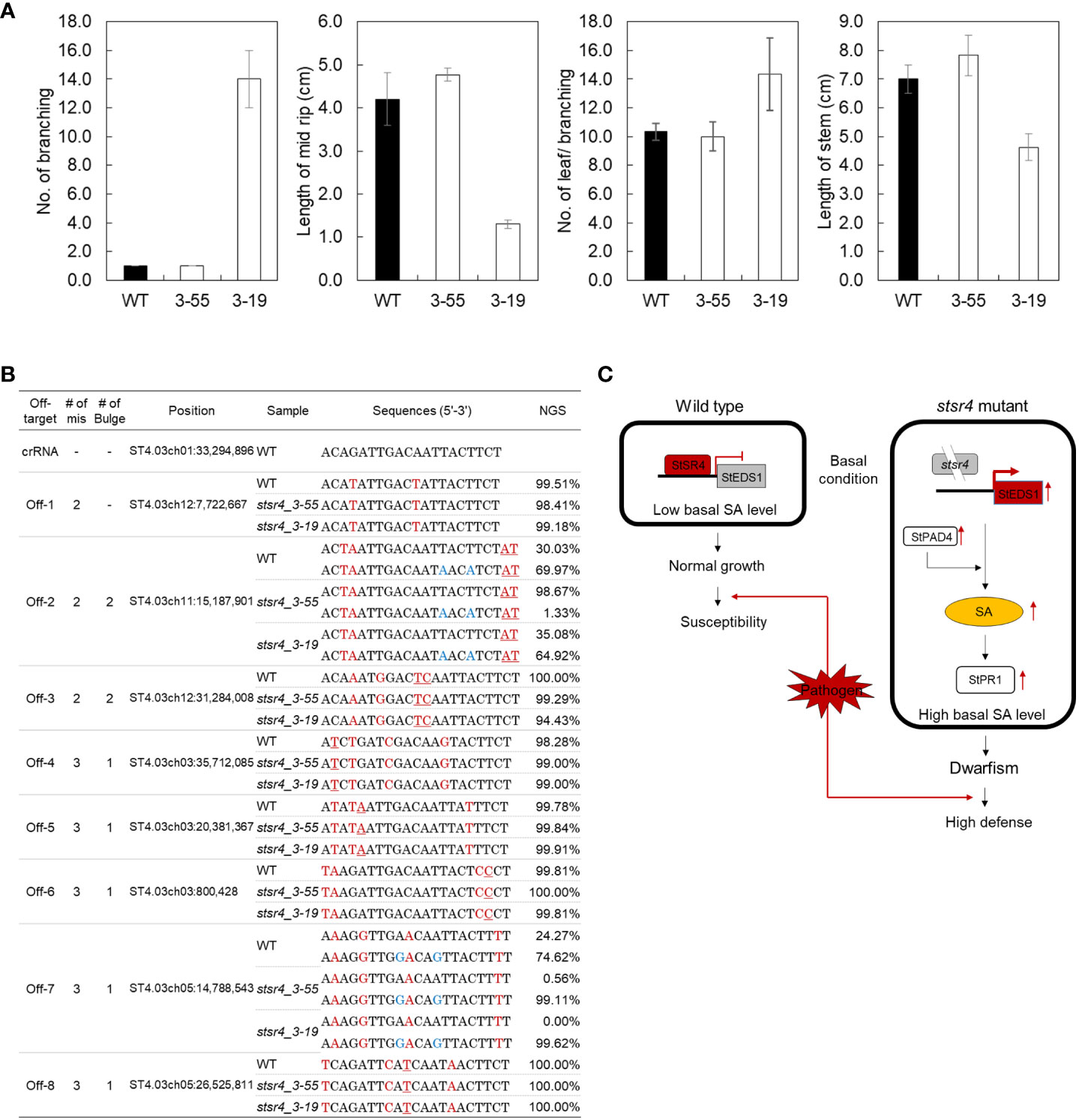
Figure 6 In vivo phenotype assays and off-target analysis of two mutants showing improved pathogen resistance. (A) Measurement of branch number per plant, midrib length, leaf number per branch, and stem length in wild type and stsr4_3-55 and stsr4_3-19 mutants. The plants were grown in a greenhouse under 16 h light/8 h dark photoperiod for 1 month. Values represent mean ± SE of four biological replicates. Asterisks indicate significant differences (Student’s t-test; *p < 0.05). (B) Analysis of off-target editing in two stsr4 mutants. Eight candidate off-target sequences were analyzed by PCR, followed by targeted deep sequencing (Read counts > 100). The mismatched nucleotides between the on- and off-target sequences are indicated in red, and the bulges are underlined red. Blue sequences represent SNPs. (C) Model explaining the basis of increased SA levels and pathogen resistance in potato, based on the suppression of StSR4 expression, and consequently increased expression of StEDS1 and StPAD4.
Off-target editing in two stsr4 mutants
The CRISPR-Cas9 technology has the potential to cause off-target mutations at sites similar to the target genomic sequences (Peng et al., 2016). To determine whether genome editing using RNPs can cause off-target editing, we validated mutations in eight candidate off-target sites including 2–4 mismatches with 1–2 bulges in stsr4_3_55 and stsr4_3_19 mutants (Figure 6B). No indels were observed at the eight candidate off-target sites in the two mutants, based on the analysis of Sanger sequencing-generated chromatograms and Targeted deep sequencing.
Discussion
CRISPR/Cas9-mediated genome editing is a promising approach for enhancing specific agricultural traits of plants, while preserving their existing elite characteristics through targeted gene mutation. To improve pathogen resistance of the potato cultivar ‘Desiree’, an S gene (SR4) was edited in potato protoplasts using RNP-mediated genome editing. A first step was to improve gene editing efficiency by considering RNP concentration. Optimizing the concentration of gRNA and Cas9, which are components of the RNP, is important for gene editing. According to previous studies, increasing the molar concentration of gRNA, rather than using the 1:1 molar ratio of Cas9:gRNA, increases the indel efficiency (Min et al., 2019; Wei et al., 2020). The editing efficiency of RNP-mediated genome editing using the 1:10 molar ratio of Cas9:gRNA has been reported, but not applied, in plants (Min et al., 2019). Accordingly, we compared the editing efficiency obtained using different concentrations of RNPs, based on the 1:10 molar ratio of Cas9:gRNA. At high RNP concentrations, the editing efficiency was improved but the micro calli induction rate was decreased, whereas low RNP concentrations decreased the editing efficiency and increased the micro calli induction rate (Figure 1C). Excessive induction of callus within a confined space of the alginate lens hinders callus growth (Moon et al., 2021). This indicates that callus size is an important factor affecting plant regeneration. Therefore, 20 µg RNPs, which showed the highest mutation efficiency and lowest callus induction rate, was determined to be an appropriate concentration for genome editing in this study. This implies that mutation efficiency is more important than callus induction rate.
The gRNA could be used as a pre-assembled RNP complex of synthetic crRNA and tracrRNA or as an in vitro transcribed RNA (IVT-RNP) derived from a DNA template. Synthetic gRNA might provide possible benefits of increasing the specificity of gRNA, though IVT-RNP can lead to the insertion of the residual DNA template (Andersson et al., 2018). Here, using a high-efficiency protoplast regeneration system as described (Moon et al., 2021) along with two different StSR4-specific gRNAs, low mutation frequency of RNPs, the higher mutation frequency of 25% (170 mutants from 691 regenerated plants) and 34% (240 mutants from 708 regenerated plants) stsr4 mutants were obtained. These results show that the indel efficiency observed in protoplasts extend to regenerated plants. In summary, in order to improve the efficiency of DNA-free genome editing, a high-efficiency protoplast-derived plant regeneration system, appropriate RNP infection concentration, and gRNA target site selection should be considered as factors. In addition, it is necessary for stable mutant supply to ensure that protoplast-level mutation efficiency can be maintained until the regeneration process (Figure S3).
Furthermore, we investigated the reason for the difference in indel efficiency at the two target sites of StSR4. Although Cas9 activity is affected by the sequence specificity of gRNA (Doench et al., 2014), there was no difference within the sequence of the StSR4 two target sites; therefore, indel efficiency at the two target sites is unlikely to be affected by Cas9 activity. Rather, the existence of mononucleotide (T) repeats at +1 and +4 bp, where frequent deletions were observed in the SR4_3 target sequence (Figure 2C), would have been more effective in creating a frame shift and is considered to have a higher mutation rate than SR4_1 (Figure 2B). As such, the targeting specificity of Cas9 is tightly controlled by the 20 nt sequence of gRNA and the presence of PAM, but potential off-target effects can still occur in the genome (Zhang et al., 2015). On the other hand, the RNP-mediated CRISPR/Cas9 system cleaves the target DNA site and is rapidly degraded by endogenous proteases, thereby limiting off-target effects (Foster et al., 2018; Wang et al., 2018). Additionally, we found no evidence of off-target mutations at the eight off-target candidate sites in 16 mutants, supporting the finding that RNP-mediated genome editing is highly specific in plants.
Suppression of the S gene, a negative regulator of plant defense, improves resistance against various diseases (Pessina, 2016; Kieu et al., 2021). In this study, improved late blight disease resistance was expected through the suppression of the S gene, StSR4, in potato. The stsr4 mutants exhibited improved resistance to P. infestans (Figure 5C). StSR4 editing in potato triggered increased expression of StEDS1, which led to higher transcript levels of the pathogen resistance marker gene, StPR1 (Figure 3B). These results are consistent with reports of increased SA accumulation and pathogen resistance following the suppression of SR1 expression in Arabidopsis (Du et al., 2009). However, SA levels did not increase in all stsr4 mutants with high StPR1 expression. SA accumulation in the three mutants with high StPR1 expression was proportional to StPAD4 transcript levels. This observation suggests that StPAD4 expression is related to StEDS1-induced SA accumulation. These results support a previous report showing that EDS1 and PAD4 play a parallel role in SA synthesis in A. thaliana basal- and effector-triggered immunity (Liang et al., 2017).
An increase in SA accumulation provides improved immunity to plants but also inhibits plant growth, as supported by the suppression of plant growth because of SA accumulation (Galon et al., 2008; Du et al., 2009; Nie et al., 2012). In this study, the stsr4_3-19 mutant, in which StSR4 was edited in three alleles by CRISPR/Cas9, showed a significantly high level of SA, and improved resistance to P. infestans. However, growth of this mutant was also severely inhibited as manifested by a dwarf phenotype (Figure 4). Furthermore, the stsr4_3-698 mutant with StSR4 edited in three alleles did not survive because growth was strongly inhibited along with significantly high StEDS1, StPAD4, and StPR1 transcript levels, which were associated with high SA. By contrast, the stsr4_3-55 mutant with StSR4 edited in two alleles, showed a slight increase in SA and a wild-type phenotype, except for short petioles. Therefore, high SA content triggered by StSR4 editing in potato not only improves resistance to pathogens, but also induces a dwarf phenotype. Here, we did not obtain mutants with improved P. infestans resistance in the full allele mutants. In the stsr4_1-278 mutant, the expression of StEDS1 and StPR1 increased (Figure 3B), which indicated that StSR4 was strongly suppressed by the allele mutation. Nevertheless, the stsr4_1-278 mutant showed low StPAD4 expression and SA levels, which resulted in weaker resistance to P. infestans than that in stsr4_3-55 or stsr4_3-19 mutants. In addition, with low SA content, the plant recovered from dwarf, unlike the stsr4_3-19 mutant, and was phenotypically similar to the wild type. Previously, in the atsr1 mutant, overexpression of the NahG gene, which encodes a SA degrading enzyme, reduced SA accumulation and restored the wild-type phenotype (Du et al., 2009). However, NahG overexpression led to the suppression of AtPR1 expression, unlike the results of the current study, which showed high StPR1 expression in the stsr4_1-278 mutant. Next, we investigated whether the function of StSR4 in the stsr4 mutants could be complemented by its functionally redundant homologs. It has been previously reported that the loss of function of the eIF4E1 gene by CRISPR/Cas9-based editing is compensated by eIF4E2, a homolog of eIF4E1 (Bastet et al., 2017). We analyzed the expression of StSR4 gene homologs (accession no.: XM_006337904, XM_006352111, XM_006355330, and XM_006349770) to investigate the effect of gene redundancy in the stsr4_1-278 mutant; however, we could not confirm any association between the knockout mutation of StSR4 and the expression of StSR4 gene homologs (Figure S5). Because the accumulation of SA is regulated by various factors, such as the phenylalanine ammonia-lyase pathway and isochorismate pathway, the recovery of SA expression in the obtained full allele mutants is expected to be influenced by factors other than SA degrading enzymes or gene redundancy. In summary, the SR4 full allele mutation induces a greater increase in SA, which strongly inhibits plant growth, making it difficult to obtain a full knockout mutant, which is considered to be the cause of low regeneration efficiency. However, in some of the obtained full knockout mutants, the expression of SA-related genes was suppressed due to unknown factors, resulting in lower resistance to P. infestans and recovering normal plant growth (Figure S6).
In conclusion, the genome editing strategy utilized in this study enables the development of disease-resistant genotypes by editing the S gene via the delivery of synthetic RNPs. We show that RNP concentration affects gene editing efficiency and plant regeneration (Figure 1E). This indicates the importance of selecting an appropriate RNP concentration for the alginate method. Both targets of the StSR4 gene were efficiently edited, and plant regeneration was successfully induced, without off-target mutations in potato protoplasts. Increased expression of StEDS1 and StPAD4 in stsr4 mutants induced SA accumulation, and improved resistance to P. infestans (Figure 6C). This implies that the function of StSR4 in potato is similar to that of AtSR1 in A. thaliana. For the first time to our knowledge, we developed P. infestans-resistant potato through RNP-mediated genome editing method of the susceptibility gene ‘StSR4’ and investigated the mechanism by which the resistance works. This is expected to contribute to encouraging the development of foreign gene-free potato cultivars through genome editing in the future.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
H-SK and Y-iP, conceptualized and supervised the study; K-BM performed the experiments, analyzed the results, and wrote the first draft of the manuscript; S-JP, J-SP, SS, and J-HJ performed experiments involving protoplast isolation, callus induction, shoot regeneration, and plant cultivation; SL and GC performed P. infestans inoculation experiments; S-GK measured the SA content; HC, Y-SK, and H-JL analyzed the data and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by grants from the National Research Foundation (NRF-2019R1A2C1003798), New Breeding Technologies Development Program (Project No. PJ01653001) of the Rural Development Administration, and the KRIBB Research Initiative Program funded by the government of the Republic of Korea.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.997888/full#supplementary-material
References
Andersson, M., Turesson, H., Olsson, N., Fält, A. S., Ohlsson, P., Gonzalez, M. N., et al. (2018). Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiologia Plantarum 164, 378–384. doi: 10.1111/ppl.12731
Arévalo-Marín, D. F., Briceño-Robles, D. M., Mosquera, T., Melgarejo, L. M., Sarmiento, F. (2021). Jasmonic acid priming of potato uses hypersensitive response-dependent defense and delays necrotrophic phase change against phytophthora infestans. Physiol. Mol. Plant Pathol., 115, 101680. doi: 10.1016/j.pmpp.2021.101680
Bae, S., Park, J., Kim, J.-S. (2014). Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475. doi: 10.1093/bioinformatics/btu048
Bastet, A., Robaglia, C., Gallois, J.-L. (2017). eIF4E resistance: natural variation should guide gene editing. Trends Plant Sci. 22, 411–419. doi: 10.1016/j.tplants.2017.01.008
de Toledo Thomazella, D. P., Seong, K., Mackelprang, R., Dahlbeck, D., Geng, Y., Gill, U. S., et al. (2021). Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. 118(27), e2026152118. doi: 10.1073/pnas.2026152118
Doench, J. G., Hartenian, E., Graham, D. B., Tothova, Z., Hegde, M., Smith, I., et al. (2014). Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267. doi: 10.1038/nbt.3026
Du, L., Ali, G. S., Simons, K. A., Hou, J., Yang, T., Reddy, A., et al. (2009). Ca 2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457, 1154–1158. doi: 10.1038/nature07612
Foster, A. J., Martin-Urdiroz, M., Yan, X., Wright, H. S., Soanes, D. M., Talbot, N. J. (2018). CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus. Sci. Rep. 8, 1–12. doi: 10.1038/s41598-018-32702-w
Gaillochet, C., Develtere, W., Jacobs, T. B. (2021). CRISPR screens in plants: approaches, guidelines, and future prospects. Plant Cell 33, 794–813. doi: 10.1093/plcell/koab099
Galon, Y., Nave, R., Boyce, J. M., Nachmias, D., Knight, M. R., Fromm, H. (2008). Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in arabidopsis. FEBS Lett. 582, 943–948. doi: 10.1016/j.febslet.2008.02.037
Ha, J. H., Jang, H. A., Moon, K.-B., Baek, K. H., Choi, G. J., Choi, D., et al. (2017). Nicotiana benthamiana matrix metalloprotease 1 (NMMP1) gene confers disease resistance to phytophthora infestans in tobacco and potato plants. J. Plant Physiol. 218, 189–195. doi: 10.1016/j.jplph.2017.08.010
Jørgensen, I. H. (1992). Discovery, characterization and exploitation of mlo powdery mildew resistance in barley. Euphytica 63, 141–152. doi: 10.1007/BF00023919
Kieu, N. P., Lenman, M., Wang, E. S., Petersen, B. L., Andreasson, E. (2021). Mutations introduced in susceptibility genes through CRISPR/Cas9 genome editing confer increased late blight resistance in potatoes. Sci. Rep. 11, 1–12. doi: 10.1038/s41598-021-83972-w
Kim, Y. S., An, C., Park, S., Gilmour, S. J., Wang, L., Renna, L., et al. (2017). CAMTA-mediated regulation of salicylic acid immunity pathway genes in arabidopsis exposed to low temperature and pathogen infection. Plant Cell 29, 2465–2477. doi: 10.1105/tpc.16.00865
Kim, H. J., Lee, H. J., Kim, H., Cho, S. W., Kim, J.-S. (2009). Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 19, 1279–1288. doi: 10.1101/gr.089417.108
Liang, Z., Chen, K., Li, T., Zhang, Y., Wang, Y., Zhao, Q., et al. (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8, 1–5. doi: 10.1038/ncomms14261
Malnoy, M., Viola, R., Jung, M.-H., Koo, O.-J., Kim, S., Kim, J.-S., et al. (2016). DNA-Free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7, 1904. doi: 10.3389/fpls.2016.01904
Min, Y.-L., Li, H., Rodriguez-Caycedo, C., Mireault, A. A., Huang, J., Shelton, J. M., et al. (2019). CRISPR-Cas9 corrects duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci. Adv. 5, eaav4324. doi: 10.1126/sciadv.aav4324
Moon, K.-B., Park, J.-S., Park, S.-J., Lee, H.-J., Cho, H.-S., Min, S.-R., et al. (2021). A more accessible, time-saving, and efficient method for In vitro plant regeneration from potato protoplasts. Plants 10, 781. doi: 10.3390/plants10040781
Muthoni, J., Kabira, J., Shimelis, H., Melis, R. (2015). Tetrasomic inheritance in cultivated potato and implications in conventional breeding. Aust. J. Crop Sci. 9, 185.
Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7, 1–6. doi: 10.1038/s41598-017-00578-x
Nicolia, A., Proux-Wéra, E., Åhman, I., Onkokesung, N., Andersson, M., Andreasson, E., et al. (2015). Targeted gene mutation in tetraploid potato through transient TALEN expression in protoplasts. J. Biotechnol. 204, 17–24. doi: 10.1016/j.jbiotec.2015.03.021
Nie, H., Zhao, C., Wu, G., Wu, Y., Chen, Y., Tang, D. (2012). SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol. 158, 1847–1859. doi: 10.1104/pp.111.192310
Oliva, R., Ji, C., Atienza-Grande, G., Huguet-Tapia, J. C., Perez-Quintero, A., Li, T., et al. (2019). Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350. doi: 10.1038/s41587-019-0267-z
Park, J., Bae, S., Kim, J.-S. (2015). Cas-designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 31, 4014–4016. doi: 10.1093/bioinformatics/btv537
Peng, R., Lin, G., Li, J. (2016). Potential pitfalls of CRISPR/Cas9-mediated genome editing. FEBS J. 283, 1218–1231. doi: 10.1111/febs.13586
Pessina, S. (2016). Role of MLO genes in susceptibility to powdery mildew in apple and grapevine. Wageningen NL: Wageningen University. Available at: http://library.wur.nl/WebQuery/wurpubs/fulltext/367864
Pyott, D. E., Sheehan, E., Molnar, A. (2016). Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free arabidopsis plants. Mol. Plant Pathol. 17, 1276–1288. doi: 10.1111/mpp.12417
Ryals, J. A., Neuenschwander, U. H., Willits, M. G., Molina, A., Steiner, H.-Y., Hunt, M. D. (1996). Systemic acquired resistance. Plant Cell 8, 1809. doi: 10.1105/tpc.8.10.1809
Sun, K., Wolters, A.-M. A., Vossen, J. H., Rouwet, M. E., Loonen, A. E., Jacobsen, E., et al. (2016). Silencing of six susceptibility genes results in potato late blight resistance. Transgenic Res. 25, 731–742. doi: 10.1007/s11248-016-9964-2
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K., Cigan, A. M.. (2016). Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nature communications 7, 1–7.
van Schie, C. C., Takken, F. L. (2014). Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52, 551–581. doi: 10.1146/annurev-phyto-102313-045854
Vogel, J. P., Raab, T. K., Schiff, C., Somerville, S. C. (2002). PMR6, a pectate lyase–like gene required for powdery mildew susceptibility in arabidopsis. Plant Cell 14, 2095–2106. doi: 10.1105/tpc.003509
Wang, W., Pan, Q., He, F., Akhunova, A., Chao, S., Trick, H., et al. (2018). Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. CRISPR J. 1, 65–74. doi: 10.1089/crispr.2017.0010
Wei, T., Cheng, Q., Min, Y.-L., Olson, E. N., Siegwart, D. J. (2020). Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun. 11, 1–12. doi: 10.1038/s41467-020-17029-3
Woo, J. W., Kim, J., Kwon, S. I., Corvalán, C., Cho, S. W., Kim, H., et al. (2015). DNA-Free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi: 10.1038/nbt.3389
Zaidi, S.-e., Mukhtar, M. S., Mansoor, S. (2018). Genome editing: targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 36, 898–906. doi: 10.1016/j.tibtech.2018.04.005
Zhang, X.-H., Tee, L. Y., Wang, X.-G., Huang, Q.-S., Yang, S.-H. (2015). Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Therapy-Nucleic Acids 4, e264. doi: 10.1038/mtna.2015.37
Zhao, X., Jayarathna, S., Turesson, H., Fält, A.-S., Nestor, G., González, M. N., et al (2021). Amylose starch with no detectable branching developed through DNA-free CRISPR-Cas9 mediated mutagenesis of two starch branching enzymes in potato. Scientific reports 11, 1-13. doi: 10.1038/s41598-021-83462-z
Keywords: RNPs, protoplast, genome editing, susceptibility gene, late blight
Citation: Moon K-B, Park S-J, Park J-S, Lee H-J, Shin SY, Lee SM, Choi GJ, Kim S-G, Cho HS, Jeon J-H, Kim Y-S, Park Y-I and Kim H-S (2022) Editing of StSR4 by Cas9-RNPs confers resistance to Phytophthora infestans in potato. Front. Plant Sci. 13:997888. doi: 10.3389/fpls.2022.997888
Received: 19 July 2022; Accepted: 06 September 2022;
Published: 23 September 2022.
Edited by:
Jong-Seong Jeon, Kyung Hee University, Republic of KoreaReviewed by:
Sang-Tae Kim, Catholic University of Korea, Republic of KoreaKaushik Ghose, Texas Tech University, United States
Copyright © 2022 Moon, Park, Park, Lee, Shin, Lee, Choi, Kim, Cho, Jeon, Kim, Park and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youn-Il Park, eWlwYXJrQGNudS5hYy5rcg==; Hyun-Soon Kim, aHl1bnNAa3JpYmIucmUua3I=
 Ki-Beom Moon
Ki-Beom Moon Su-Jin Park1,2
Su-Jin Park1,2 Ji-Sun Park
Ji-Sun Park Hyo-Jun Lee
Hyo-Jun Lee Gyung Ja Choi
Gyung Ja Choi Sang-Gyu Kim
Sang-Gyu Kim Hye Sun Cho
Hye Sun Cho Yong-Sam Kim
Yong-Sam Kim Youn-Il Park
Youn-Il Park Hyun-Soon Kim
Hyun-Soon Kim