
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 18 October 2022
Sec. Plant Symbiotic Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.997037
This article is part of the Research TopicMetabolic Adjustments and Gene Expression Reprogramming for Symbiotic Nitrogen Fixation in Legume Nodules, Volume IIView all 6 articles
Symbiotic nitrogen fixation is an important component in the nitrogen cycle and is a potential solution for sustainable agriculture. It is the result of the interactions between the plant host, mostly restricted to legume species, and the rhizobial symbiont. From the first encounter between the host and the symbiont to eventual successful nitrogen fixation, there are delicate processes involved, such as nodule organogenesis, rhizobial infection thread progression, differentiation of the bacteroid, deregulation of the host defense systems, and reallocation of resources. All these processes are tightly regulated at different levels. Recent evidence revealed that non-coding RNAs (ncRNAs), including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), participate in these processes by controlling the transcription and translation of effector genes. In general, ncRNAs are functional transcripts without translation potential and are important gene regulators. MiRNAs, negative gene regulators, bind to the target mRNAs and repress protein production by causing the cleavage of mRNA and translational silencing. LncRNAs affect the formation of chromosomal loops, DNA methylation, histone modification, and alternative splicing to modulate gene expression. Both lncRNAs and circRNAs could serve as target mimics of miRNA to inhibit miRNA functions. In this review, we summarized and discussed the current understanding of the roles of ncRNAs in legume nodulation and nitrogen fixation in the root nodule, mainly focusing on their regulation of hormone signal transduction, the autoregulation of nodulation (AON) pathway and nutrient homeostasis in nodules. Unraveling the mediation of legume nodulation by ncRNAs will give us new insights into designing higher-performance leguminous crops for sustainable agriculture.
Legumes are important crops that benefit the world in diverse ways. Chief among them is the ability to form a specific organ called the root nodule to convert atmospheric nitrogen into bioavailable nutrients. Nodulation and nitrogen fixation are complicated processes that are regulated at different levels to maximize the benefits to the host and the microsymbiont. The interaction between the legume host and rhizobia is normally initiated with the release of flavonoids from the plant root to attract the rhizobium (Cohn et al., 1998), which is then entrapped in the root hair cells. The rhizobium multiplies to form infection foci, from which infection threads are developed toward the cortical cells to facilitate rhizobial entry (Oldroyd and Downie, 2008). Alternatively, the rhizobium can invade the cortical cells through cracks on the epidermis (Oldroyd and Downie, 2008), and produce nodulation factors (Nod factors [NFs]), which are recognized by the host to start nodule formation (Cohn et al., 1998). Nod factor perception and transduction at the epidermis trigger root cell swelling, Ca2+ spiking (Ehrhardt et al., 1996) and the activation of early nodulation genes to advance the early events of nodulation (Oldroyd and Downie, 2008). Several kinases, transcription factors (Oldroyd and Downie, 2008) and other genes (Stacey et al., 2006) are involved in the Nod factor signaling pathway. Upon sensing epidermal responses, the cortical cells divide and form the nodule primordium where nodule organogenesis and development take place. The rhizobium then enters the cortical cells and is encapsulated within the plant cell membrane. The cell membrane and rhizobium differentiate together to form a symbiosome (Oldroyd and Downie, 2008). A nodule then develops, within which the rhizobium performs nitrogen fixation (Oldroyd and Downie, 2008). Several phytohormones are essential for maintaining highly effective nitrogen fixation (Oldroyd and Downie, 2008).
Determinate and indeterminate nodules are developed in different species of legumes. For instance, soybean forms determinate nodules and alfalfa forms indeterminate ones (Mao et al., 2013). Determinate nodules originate from the middle or outer cortex of the plant with a transient nodule meristem that is absent in distant zonation while indeterminate nodules develop from the inner cortex or pericycle with a persistent nodule meristem that is spatially separated into five zones (Kohlen et al., 2018). These two kinds of nodules are suggested to respond differently to auxin or other diffusible signals during nodule development (Kohlen et al., 2018).
Biological nitrogen fixation (BNF) is crucial for maintaining nitrogen cycling in the biosphere. It converts atmospheric nitrogen into ammonia (Long, 1989), an ultimate goal of legume-rhizobium symbiosis. This symbiosis fixes 21 metric tons of nitrogen annually (Herridge et al., 2008) and increases bioavailable nitrogen, bringing about agricultural, natural and socio-economic benefits. Considering the high energy cost of producing synthetic fertilizers, the substitution of synthetic fertilizer with biological nitrogen fixation has become a crucial element of sustainable agriculture (Foyer et al., 2016).
Non-coding RNAs (ncRNAs) are functional transcripts without translation potential (Mattick and Makunin, 2006). Major groups of regulatory ncRNAs in plants are small RNAs (20-24 nt), which include microRNAs (miRNAs) and small interfering RNAs (siRNAs) (Borges and Martienssen, 2015), long non-coding RNAs (lncRNAs) (>200 nt) (Wang T. et al., 2017), and circular RNAs (circRNAs) (Ye et al., 2015).
MiRNAs are transcribed by RNA polymerase II (Pol II) in the nucleus to produce single-stranded primary miRNAs, which are then processed by DICER-LIKE (DCL) 1 protein to form the mature miRNA duplex. MiRNAs form a silencing effector complex with specific ARGONAUTE (AGO) proteins (Borges and Martienssen, 2015). The complementary sequences of miRNAs to their target mRNAs guide the complex to perform post-transcriptional gene-silencing by target mRNA cleavage or translational repression (Borges and Martienssen, 2015). For siRNAs, an endogenous gene is transcribed by Pol II or Pol IV and might be processed by RNA-DEPENDENT RNA POLYMERASEs (RDRs). The transcripts are then cleaved by DCLs to form siRNAs or secondary siRNAs (Borges and Martienssen, 2015). MiRNA-mediated gene-silencing might also produce secondary siRNAs (Manavella et al., 2012). SiRNAs are categorized into hairpin-derived siRNAs (hp-siRNAs), natural antisense trans- or cis-acting siRNAs (natsiRNAs), heterochromatic siRNAs (hetsiRNAs) and secondary siRNAs consisting of trans-acting siRNAs (ta-siRNA), phased siRNAs (phasiRNAs) and epigenetically activated siRNAs (easiRNA) (Borges and Martienssen, 2015). SiRNAs can silence their target genes by mRNA cleavage, transcriptional (Baulcombe, 2004) and translational repression, and DNA methylation (Borges and Martienssen, 2015).
Plant lncRNAs are classified into long intergenic ncRNAs (lincRNAs), intronic ncRNAs (incRNAs) and antisense RNAs and natural antisense transcripts (NATs) (Wang J. et al., 2017). LncRNAs are produced by Pol I, II, III and two plant-specific enzymes, Pol IV and V (Wang J. et al., 2017). LncRNAs can act in trans and in cis and they function by affecting the formation of chromosomal loops, DNA methylation, histone modification, and alternative splicing to modulate promoter and terminator activities and regulate gene expressions (Statello et al., 2020). LncRNAs can also act as miRNA sponges which served as the mimicry of the miRNA target to sponge up the miRNA, which help to control miRNA activities in gene regulation (Zhu and Wang, 2012).
CircRNAs are single-stranded RNAs with a phosphodiester linkage at the 3′ and 5′ ends that forms a stable circular molecule (Chen, 2016). CircRNAs are mainly produced by back-splicing and can be generated from exons (exonic circRNA), introns (intronic circRNA), intergenic regions (Ye et al., 2015) and transcripts of transposons (Chen et al., 2018). It can regulate other linear transcripts or genes by acting as target mimics of miRNAs to inhibit miRNA functions (Chen, 2016), modulating gene expression post-transcriptionally, or mediating alternative splicing (Chen, 2016).
Various ncRNAs are reported to play roles in plant development, plant immunity and responses to abiotic and biotic stress (Wang T. et al., 2017; Chand Jha et al., 2021), indicating the importance of ncRNAs in plants. There are also many studies illustrating that ncRNAs are critical regulators in the nodulation process.
Several legume species, such as soybean and peanut, are important crops worldwide as they are the major sources of proteins and carbohydrates. The symbiosis between legumes and rhizobia ensures the bioavailability of nitrogen, which is an essential part of the nitrogen cycle and contributes to sustainable agriculture and ecological balance (Salvagiotti et al., 2008). Up to now, many nodulation-related regulators have been identified. Among them, ncRNAs have been proven to play key roles in this process. In this review, we described the roles of miRNAs, lncRNAs and circRNAs in controlling nodulation through the regulation of hormone signal transduction, the AON pathway and nutrient transportation. With the widespread application of next-generation sequencing, more and more ncRNAs are identified to be involved in the interactions between legumes and rhizobia. MiRNAs play critical roles in nodule formation, nodule development and nitrogen fixation in mature nodules, while the functions of lncRNAs and circRNAs are still not well defined.
With the advances in sequencing methods, more and more research has been carried out to identify ncRNAs governing nodulation and nitrogen fixation. Up to now, miRNAs, siRNAs, lncRNAs and circRNAs have been identified in a number of legume species, especially in soybean, Medicago truncatula, common bean (Phaseolus vulgaris) and Lotus japonicus (Wang et al., 2020; Chand Jha et al., 2021).
MiRNA is the most studied class of ncRNAs, and is the most conserved type of ncRNAs among plant species (Li and Mao, 2007; De Lima et al., 2012). Most plant species usually have both conserved and species-specific miRNAs (De La Rosa et al., 2020). Even within the same legume species, different germplasms can harbor their own specific miRNAs. For example, cultivated soybean (Glycine max) and wild soybean (Glycine soja) have their own specific miRNAs (Zeng et al., 2012; Sun Y. et al., 2016). The functions of conserved miRNAs among plant species are quite similar (Paul et al., 2015; Li and Zhang, 2016), mostly involved in controlling development and stress responses (Rubio-Somoza and Weigel, 2013; Teotia and Tang, 2015; Fan et al., 2021). The expression profiles of conserved miRNAs vary a lot across plant species and organs (Tsikou et al., 2018). Since biological nitrogen fixation is a unique feature in legumes, (Salvagiotti et al., 2008), research on miRNAs in legumes is often focused on this symbiotic function. For example, 382 miRNA candidates, including 154 known mature miRNAs, 57 variants of known miRNAs and 171 novel miRNAs, were identified in soybean roots and nodules (Fan et al., 2021). About 68.1% of the miRNAs were differentially expressed in mature nodules (Fan et al., 2021) inferring their potential roles in nitrogen fixation and nodule maintenance. In M. truncatula, 73 miRNA belonging to 27 known miRNA families and 100 new miRNA candidates were identified (Lelandais-Brière et al., 2009). These miRNAs also showed differential expression profiles in roots and nodules (Lelandais-Brière et al., 2009). In chickpea nodules, 91 miRNAs were identified, including 84 conserved and 7 novel miRNAs (Tiwari et al., 2021). Among them, 28 miRNAs were legume-specific, of which 13 were chickpea-specific (Tiwari et al., 2021). These studies showed that the miRNA regulation on legume nodules is common among legume species, with diversities among the specific miRNAs involved.
Unlike miRNAs, lncRNAs are less conserved among plants (Deng et al., 2018). Compared to animals, plant lncRNAs are generally shorter, and have fewer exons than protein-coding genes (Deng et al., 2018). The abundance of lncRNAs is usually correlated with the number of mRNAs in plant species (Deng et al., 2018). Across species, lncRNAs exhibit a high divergence with conserved genomic positions, but distinct nucleotide sequences (Deng et al., 2018). However, those lncRNAs which are the precursors or targets of miRNAs normally show a certain percentage of sequence conservation among different plant species (Deng et al., 2018). Meanwhile, sequences of lncRNAs within individual species also show some degree of conservation (Deng et al., 2018). For instance, 48,752 and 35, 675 lncRNA gene loci were identified in cultivated soybeans and wild soybeans, respectively (Lin et al., 2020), with ~12,000 loci shared between cultivated and wild soybeans (Lin et al., 2020). However, most of the soybean lncRNAs cannot be aligned well with the genomes of other legume species, showing that lncRNAs are more diverged across species than other types of ncRNAs (Lin et al., 2020).
The biogenesis of circRNAs is conserved in both plants and animals, which depends on RNA polymerase II-mediated transcription and back-splicing reactions of the mRNA precursor (Sun X. et al., 2016; Zhao et al., 2019a). However, due to alternative circularization, circRNAs are rich in isoforms (Zhang et al., 2014; Zhao et al., 2019a). In soybean, 5,372 circRNAs were identified by high-throughput sequencing, with 80% of them being paralogous circRNAs formed from paralogous genes (Zhao et al., 2017). CircRNAs showed tissue-specific expression patterns and were potentially involved in many biological processes, such as multi-organism processes and developmental processes (Zhao et al., 2017). In common bean, 8,842 high-confidence circRNAs were identified, with 3,448 of them specifically expressed during symbiosis, peaking at the nitrogen-fixing stage (Wu et al., 2020). These findings provide evidence that circRNAs participate in legume nodulation.
Phytohormones play important roles in plant growth and development. Most of them were shown to be involved in nodulation and nitrogen fixation, especially auxins, cytokinins (CK), gibberellins (GA), ethylene (ET) and jasmonates (JA). Recently, miRNAs and siRNAs, both small ncRNAs, have been identified to control nodule formation and development by mediating phytohormone signal transduction. The specific ncRNAs involved in hormone signal transduction and the references are listed in Table 1.
Auxin signaling plays crucial roles in nodule formation and development (Kohlen et al., 2018). The alteration in auxin homeostasis upon rhizobial infection was shown to be a prerequisite for nodule formation (Mathesius and Mathesius, 2008). Specifically, the inhibition of polar auxin transport and down-regulation of the auxin-inducible promoter, GH3, are associated with the formation of the nodule primordium, where low levels of auxin are required for the initiation of nodule formation (Mathesius et al., 1998; Pacios-Bras et al., 2003; De Billy et al., 2001; Grunewald et al., 2009). During nodule development, auxin is responsible for controlling the cell cycle and nodule maturation (Kondorosi et al., 2005; Nizampatnam et al., 2015).
MiRNAs are important regulators in the auxin signaling pathway. MiRNAs, such as miR160, miR164, miR167, miR390 and miR393, were reported to control nodulation by affecting auxin signaling (Nizampatnam et al., 2015; Wang et al., 2015; Yan et al., 2015; Hobecker et al., 2017). MiR160, miR167 and miR390 target different auxin response factors (ARF) and perform different regulatory roles in nodulation (Nizampatnam et al., 2015; Wang et al., 2015; Hobecker et al., 2017). Auxin directly binds to TRANSPORT INHIBITOR RESPONSE1 (TIR1)-like F-box proteins to form part of an Skp, Cullin, F-box (SCF)-type ubiquitin ligase complex that regulates the degradation of auxin/indole-3-acetic acid (Aux/IAA) (Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Turner et al., 2013). Aux/IAAs are repressor proteins that form complexes with ARFs to repress the expression of auxin-responsive genes (Liscum and Reed, 2002; Remington et al., 2004). The degradation of Aux/IAAs caused by TIR1 in an auxin dose-dependent manner leads to the de-repression/expression of auxin-responsive genes (Gray et al., 2001; Schwechheimer et al., 2001). Overexpression of miR160 could inhibit the expressions of the auxin response repressing transcription factors, ARF10/16/17, to make the root hypersensitive to auxin and reduce sensitivity to cytokinin (Turner et al., 2013; Nizampatnam et al., 2015). Auxin hypersensitivity leads to a reduction in nodule primordium formation (Turner et al., 2013). Therefore, overexpression of miR160 reduces the nodule numbers, which can be restored by exogenous cytokinin application (Nizampatnam et al., 2015). Knockdown of miR160 results in reduced sensitivity to auxin and enhanced sensitivity to cytokinin to increase nodule formation and delay nodule maturation (Nizampatnam et al., 2015). Exogenous auxin rescues the proper nodule formation and maturation in miR160 knockdown roots (Nizampatnam et al., 2015). At the early stage of nodule formation in soybean, a low level of miR160 promotes nodulation, and high miR160 levels in nodule development could stimulate nodule maturation (Nizampatnam et al., 2015). In contrast to miR160, miR167c is induced by rhizobium inoculation and positively mediates nodulation efficiency under low microsymbiont density by targeting the negative regulators of nodulation, ARF8a and ARF8b, that impair nodule formation (Wang et al., 2015). The miR167c overexpression reduces the root sensitivity to auxin, while the knockdown of miR167c enhances the root sensitivity to auxin (Wang et al., 2015). MiR167c also positively controls the expression of a set of nodulation marker genes, like ENOD40 (Wang et al., 2015).
MiR390, an evolutionarily conserved miRNA, targets the Trans-Acting Short Interference RNA3 (TAS3) and causes the cleavage of TAS3 by ARGONAUTE7 at the 3’ ends most proximal site to produce ta-siRNA (Hobecker et al., 2017). TAS3-derived ta-siRNAs directly control the cleavage of complementary mRNAs encoding ARF2/3/4 (Hobecker et al., 2017; Kirolinko et al., 2021). Knockdown of ARF2/3/4 reduces the initiation and progression of infection events to inhibit nodule formation and downregulate the Nodulation Signaling Pathway 2 (NSP2) (Kirolinko et al., 2021). At the early stage of nodulation, the expression level of miR390 and tasiRNAs are at a low level, whereas TAS3 and ARF2/3/4 are substantially highly expressed to promote the nodule formation (Hobecker et al., 2017).
Other than regulating ARFs, nodulation can also be regulated by direct targeting of auxin-responsive genes by miRNAs to silence their expressions (Guo et al., 2005; D’haeseleer et al., 2011; Yan et al., 2015). In Medicago truncatula, miR164 causes a reduction in nodule numbers by causing the cleavage of the mRNA of the auxin-induced transcription factor, NAC1 (Guo et al., 2005; D’haeseleer et al., 2011). MiR393 also works as a negative regulator in nodule formation by directly targeting the transcripts of the auxin receptor, TIR1/AFB, to alter auxin signaling in roots (Cai et al., 2017; Kohlen et al., 2018). Overexpression of GmTIR1A and GmTIR1C, target genes of miR393, could increase the nodule numbers through modulation of cellular response to auxin (Cai et al., 2017).
CK plays a crucial role in the initiation of nodule organogenesis (Gamas et al., 2017). Upon rhizobial infection, nodulation factor (NF) induces the accumulation of CK (Van Zeijl et al., 2015). Exogenous application of a low concentration of CK to soybean could enhance nodule formation, while the application of a high concentration of CK did the opposite (Mens et al., 2018). Interestingly, CK plays an important role in the AON pathway (Sasaki et al., 2014). In Lotus japonicus, the CLE-RS1/2-HAR1 signaling pathway stimulates the transportation of shoot-derived CK to the root to suppress nodulation (Sasaki et al., 2014). CLE production and the whole AON pathway are regulated by miR156b and miR172c (Wang et al., 2014; Wang et al., 2019; Yun et al., 2022). The high expression of miR172c activates the production of ENOD40 and GmCLE through inhibiting its target gene, Nodule Number Control 1 (GmNNC1) (Wang et al., 2014). GmNNC1 is a transcription repressor of ENOD40 and GmCLE (Wang et al., 2014). Therefore, the shoot-derived CK regulation on nodulation is also controlled by miR156b and miR172c (Yun et al., 2022). The detailed information about ncRNAs regulation on the AON pathway will be introduced in section 4.
Exogenous application of GA demonstrated both positive and negative roles of GA in legume nodulation (Hayashi et al., 2014). Recently, gibberellin 20-oxidase 1a (GA20ox1a), GA3ox1a and GA2ox1a were shown to be induced by rhizobial infection in soybean roots and the induction of these three genes was required for nodule organogenesis (Chu et al., 2022). A reduced level of GA led to aberrant rhizobial infection and inhibited nodule formation (Chu et al., 2022). In soybean, miR166 targets a transcription factor ATHB14-LIKE, which directly represses the transcription of the GA biosynthesis genes GmGA1 and GmGA2, and activates the transcription of catabolic gene GIBBERLLIN 2 OXIDASE 2 (GmGA2ox2) (Zhao et al., 2022). Therefore, miR166 could regulate the level of bioactive gibberellic acid (GA3) (Zhao et al., 2022). In Medicago, miR166a targets several class-III homeodomain-leucine zipper (HD-ZIP III) genes: MtHB8, MtCNA1 and MtCNA2 (Boualem et al., 2008). These three HD-ZIP III genes show the strong expressions in nodule primordia followed by in the apical region and vascular bundles in the mature nodules, which suggests they are involved in the new meristem formation and vascular-bundle differentiation (Boualem et al., 2008). Overexpression of miR166a reduces the symbiotic nodule numbers and lateral roots and improves ectopic development of vascular bundles (Boualem et al., 2008). In alfalfa (Medicago sativa), transcriptomic analyses found that the overexpression of miR156 improved the expressions of GA receptors and the GA-regulated family of proteins, promoting nitrogen fixation activity (Aung et al., 2017). In peanut (Arachis hypogaea L.), an lncRNA, XR_001621694.1, was identified to target a GA receptor gene, GID1C-like, to regulate the GA signaling pathway, which may in turn regulate nodulation (Ma X. et al., 2020).
ET plays an important role in the nodulation process as a negative regulator (Guinel, 2015). Rhizobium-derived flagellin-like molecules (flg22) could trigger the production of ET (Nakagawa et al., 2011), activating the expression of ethylene response factor (ERF)-encoding genes, which in turn upregulate the expression of Pathogenesis-Related (PR) proteins to inhibit the progress of the infection thread (Breakspear et al., 2015; Guinel, 2015).
In chickpea, miR171b expression was negatively correlated with that of ERF (Ca_12975), and was shown to regulate nodule formation (De Luis et al., 2012; Garg et al., 2019). Another miRNA, miR5678 from soybean, was predicted to target an ethylene receptor 1-related gene (Glyma.12G241700) and its sequence is reverse-complementary to a circRNA, Gma-circ_0000352 (Wang et al., 2020), suggesting that Gma-circ_0000352 may affect nodulation through the interaction with an miRNA.
JAs serve either positive or negative roles in nodulation in a species-specific manner, by affecting the formation of infection threads and the expression of nodulation-related genes, such as ENOD11 (Sun et al., 2006; Ferguson and Mathesius, 2014). Studies on the relationship between the JA signaling pathway and ncRNAs are limited. One example is found in common bean, where miR319d indirectly suppresses the expression of a JA biosynthetic gene, lipoxygenase 2 (LOX2) through the direct inhibition of TEOSINTE BRANCHED/CYCLOIDEA/PCF10 (TCP10) (Martín-Rodríguez et al., 2018), thus exhibiting a role in regulating the JA content during nodulation. As a consequence, an increase in miR319d expression contributed to rhizobial infection but reduced nodule formation (Martín-Rodríguez et al., 2018). MiR319d expression level is induced after rhizobium infection in roots to promote nodule formation (Martín-Rodríguez et al., 2018).
Plant hormones could mediate stress responses when the plant encounter stresses like salinity, drought, heat, cold and heavy metals, and bacterial and viral infection. In recent studies, hormone-responsive miRNAs in symbiotic soybeans were also found to be important regulators for abiotic and biotic stresses in plants, which may contribute to the maintenance of nitrogen fixation and nodule formation under stresses. Auxin contributes to plant tolerance to salt, drought, heat stress, and microbe infection (Šečić et al., 2021; Waadt et al., 2022). As the main regulator of auxin signaling, miR160, miR167, miR390 and miR393 were differentially expressed under salt and drought stresses (Kumar, 2014; Jatan and Lata, 2019). MiR160 and miR167 can improve the level of auxin by targeting TIR1 or ARF to reset the development program, which in turn counters biomass damage due to salt stress (Yin et al., 2017). Thus, the alternation of the expression of these miRNAs under abiotic stress may affect the nodule functions. In chickpea, miR319 was upregulated by salt stress, while it was also the most down-regulated miRNA in soybean functional nodules upon salt stress (Dong et al., 2013). As mentioned above, a high level of miR319 could reduce the nodule number. Therefore, the downregulation of miR319 under salt stress may protect the nodule formation. Except for abiotic stress, miR160, miR167, miR390 and miR393 are also responsive to the pathogenic bacterium Pseudomonas syringae and the pathogenic fungus Verticillium dahlia (Šečić et al., 2021). Overexpression of miR160 leads to altered callose deposition and bacteria resistance (Li et al., 2010). MiR393 was induced by flag22 treatment to repress the auxin signaling to restrict P. syringae growth (Navarro et al., 2006). In soybean, miR393 was induced by heat-inactivated Phytophthora sojae hyphae, and the knockdown of miR393 reduces the defense against P. sojae and the production of isoflavonoids, one major group of soybean antimicrobial metabolites (Wong et al., 2014). MiR160, miR167 and miR393 are induced by soybean mosaic virus (SMV), and miR167 could reduce the harm of SMV by targeting ARF (Yin et al., 2013). Therefore, miRNAs regulation in both hormones and stresses is required for legume nodule formation and development.
Symbiotic nitrogen fixation in legume nodules is a highly energy-intensive process requiring a great amount of carbon in the form of sugars (Ferguson et al., 2019). Therefore, legumes have tight control over nodule numbers to balance the symbiotic relationship with their own development (Ferguson et al., 2019), by maintaining the optimal number of nodules through the AON pathway (Kosslak and Bohlool, 1984; Suzuki et al., 2008; Reid et al., 2011). MiRNAs and lncRNAs are the core regulators in this pathway (Ferguson et al., 2019).
During rhizobial infection, NFs are perceived by NF receptors (NFRs), such as NFR1a/5a, which then activate the expressions of downstream genes, such as nodule inception (NIN), miR172c and nodulation signaling pathway (NSP) (Madsen et al., 2003; Madsen et al., 2011). In soybean, besides being regulated by NFR1a/5a, miR172c is also under the control of the transcription factor, NINa and miR156b (Wang et al., 2019). NINa directly binds to the promoter of miR172c to stimulate its transcription (Wang et al., 2019), whereas miR172c represses its target gene, Nodule Number Control 1 (NNC1) (Wang et al., 2014). NNC1 encodes a protein that targets the promoter of the early nodulin gene, ENOD40 (Wang et al., 2014). The ENOD40 RNA is a special class of RNAs in plants, known as bifunctional RNAs, with both coding and non-coding functions (Röhrig et al., 2002; Ganguly et al., 2021). ENOD40 initiates the proliferation of root cortical cells and increases nodule numbers (Crespi et al., 1994; Charon et al., 1997; Campalans et al., 2004). In Pisum sativum, ENOD40 shows a high expression level in the nodule primordium and prefixing zones in mature nodules, suggesting that ENOD40 is involved in nodule development (Matvienko et al., 1994). Furthermore, ENOD40 encodes peptides that were found to bind to sucrose synthase and could potentially control the enzymatic activity to mediate sucrose use in soybean nodules (Röhrig et al., 2002).
MiR156b is a negative regulator of nodulation by targeting SQUAMOSA promoter-binding protein-like 9D (SPL9d). SPL9d improves nodulation through binding to the promoter of miR172c and stimulating its expression. SPL9d is co-expressed with nodulation marker genes, NINa and ENOD40-1, during nodule formation and development (Yun et al., 2022). Therefore, the miR156b-SPL9d module and NINa both act as upstream master regulators of soybean nodulation.
On the other hand, the inhibition of NNC1 by miR172c also upregulates the expression of Rhizobia-Induced CLEs (RICs), resulting in the activation of AON (Wang et al., 2019). Following processing, CLEs are transported to the shoot in the xylem, and perceived by a homodimeric or heterodimeric receptor complex in the parenchymal cells of the leaf vasculature, such as NARK (Nodule autoregulation receptor kinase) (reviewed by Ferguson et al., 2019). The perception of RICs activates a shoot-derived inhibitor that travels through the phloem to the root to suppress nodulation (Mortier et al., 2010; Sasaki et al., 2014). The precise identity of this shoot-derived inhibitor (SDI) is still unknown. So far, miR2111 is considered one possible candidate for SDI (Sasaki et al., 2014). When there is no rhizobial infection, miR2111 is maintained at a high level and is translocated from shoot to root to inhibit Too Much Love (TML), a negative regulator of nodulation, thus maintaining the legume at a default susceptible status (Tsikou et al., 2018). The CLE perception in the leaf suppresses miR2111 expression, leading to the accumulation of TML in roots and the subsequent inhibition of nodule organogenesis (Zhang et al., 2021). Taken together, both the upstream and downstream portions of the AON pathway are under the control of ncRNAs. A schematic of the regulation of AON by ncRNAs is shown in Figure 1.
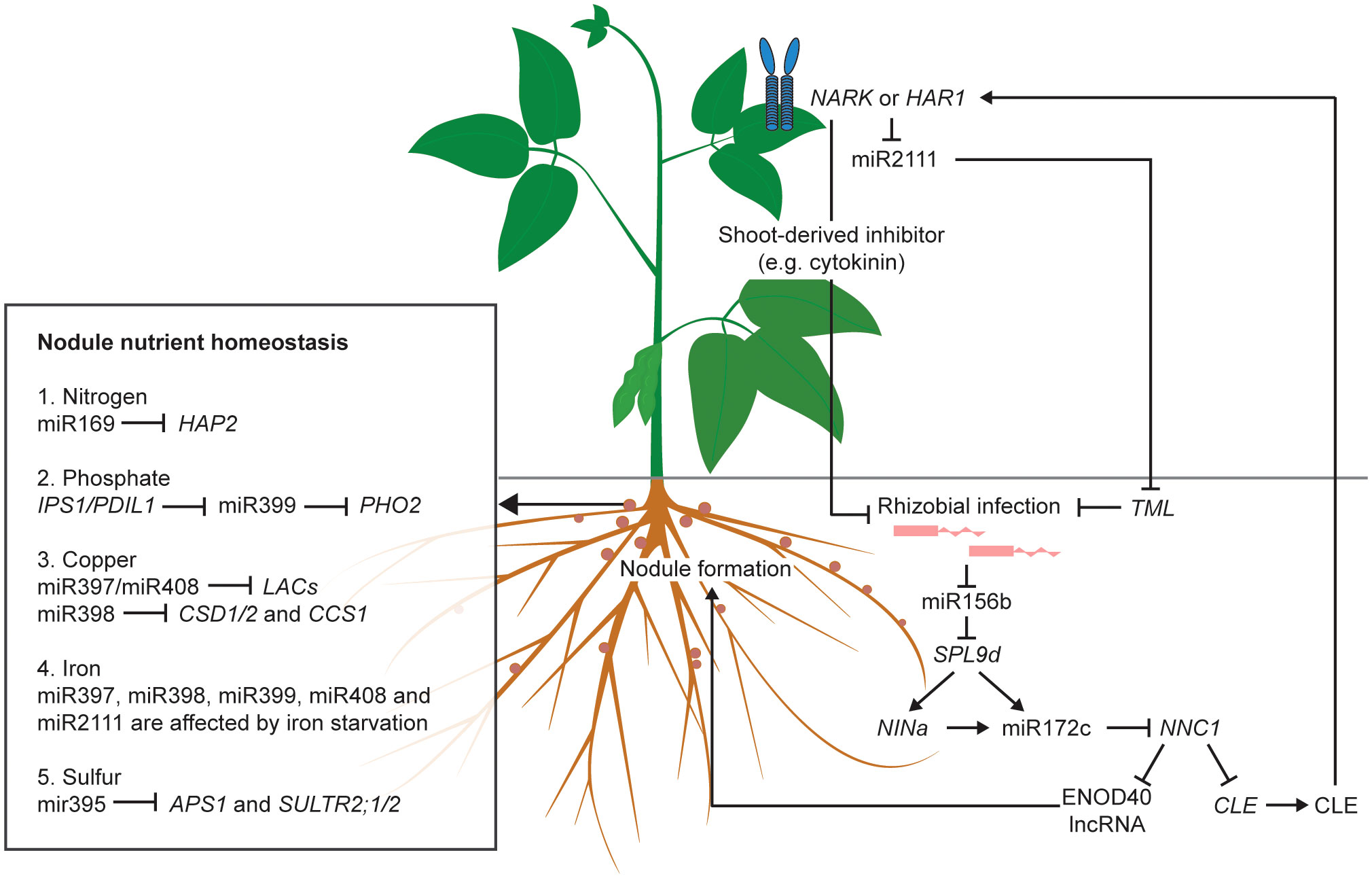
Figure 1 Roles of validated non-coding RNAs (ncRNAs) in AON pathway and nodule nutrient homeostasis of legumes. Without rhizobial infection, miR2111 is transported from shoot to root to inhibit the symbiosis suppressor TML, thus maintaining the legume host at an infection-susceptible default status. Rhizobial infection reduces the expression of miR156b to release its inhibition on the downstream symbiosis activators, such as miR172c and NINa. The activation of the long non-coding RNA (lncRNA) of ENOD40 directly induces nodule formation. The induction of CLE produces the CLE peptides which is transported from root to shoot and perceived by the NARK/HAR1 receptor to suppress nodule formation, either by activating the shoot-derived inhibitors, such as cytokinin, or by repressing miR2111, thereby allowing TML to inhibit rhizobial infection. In the nodule, miRNAs and lncRNAs regulate nutrient homeostasis, such as those of nitrogen, phosphate, copper, iron and sulfur. Arrows indicate activation. Blunt ended arrows indicate inhibition.
Nitrogen fixation is critical for legume growth. In soybean, nitrogen supplied by biological nitrogen fixation (BNF) is about 52% of the total nitrogen uptake (Salvagiotti et al., 2008). The legume host provides carbon and other nutrients, and the bacteroid supplies the legume with ammonium and amino acids in exchange (Udvardi and Day, 1997). Sufficient nutrient supply from the legume host to the bacteroid is critical for effective nitrogen fixation (Snowball et al., 1980). Many mineral nutrients, such as phosphate, nitrogen, sulfur, copper and iron, are required for high-efficiency nitrogen fixation, (Raei et al., 2013; González-Guerrero et al., 2014; Sulieman and Tran, 2015). NcRNAs, especially miRNAs, are key regulators of nutrient homeostasis in plants, by controlling various nutrient transporters (reviewed in Paul et al., 2015). The nutrient deficiency-responsive ncRNAs and their regulation on nutrient homeostasis are listed in Figure 1 and Table 2.
Nitrate negatively regulates all phases of nodulation, including rhizobial infection, nodule formation and development, and nitrogen fixation (Stephens and Neyra, 1983; Kanayama et al., 1990). Until now, many nitrogen starvation-responsive (NSR) miRNAs in plants were identified. For example, miR156, miR160, miR169, miR171, miR319, miR826 and miR846 were induced under low-nitrogen conditions, while miR167, miR172, miR395, miR399 and miR827 were suppressed (Liang et al., 2012; Paul et al., 2015). These NSR miRNAs were also differentially expressed in soybean upon nodulation. For instance, miR156, miR160, miR167, miR171, miR172, miR319, miR395 and miR399 were differentially expressed among uninfected roots, infected roots and nodules in soybean (Fan et al., 2021), suggesting that they also played roles in regulating the nitrogen status in nodules. In roots, miR169-NF-YA modules are important for the primary root growth and lateral root initiation (Sorin et al., 2014). In soybean, miR169c was induced by high nitrogen levels to target the transcription factor, GmNFYA-C (Xu et al., 2021). Overexpression of miR169c significantly reduced nodule number, whereas knockout miR169c promoted nodule formation (Xu et al., 2021). GmNFYA-C was proved to directly induce the expression of ENOD40 and attenuate the suppression of nodulation by nitrogen (Xu et al., 2021). In Medicago, miR169 was mainly expressed in nodules and targets MtNF-YA1, a central transcription regulator of symbiotic nodule development (Combier et al., 2006). MtNF-YA1 could control meristem persistence and potentially be involved in the symbiotic bacterial release (Combier et al., 2006). Overexpression of miR169 blocked the nodule development by targeting MtNF-YA1 (Combier et al., 2006). MiR169 exhibited a critical spatial and temporal restriction of MtNF-YA1 to the nodule meristematic zone to ensure the correct tissue identity and the transition from meristematic to differentiated cells (Combier et al., 2006). In Lotus japonicus, NF-YA1 is presumably under the regulation of NIN. NF-YA1 regulates the expression of the Short Internode/Stylish (STY) family transcript factors. In turn, STY regulates YUCCA1 and YUCCA11, two transcription factors that corresponding to the auxin biosynthesis (Shrestha et al., 2021). This regulatory circuit contributed to the auxin signaling important for the nodule differentiation (Shrestha et al., 2021). In Lotus japonicas, miR171c took part in bacterial infection in nodules by targeting the transcript of a transcription factor, NSP2 (De Luis et al., 2012). Accumulation of miR171c inhibited the formation of infection threads and symbiotically active nodules, whereas the miR171c-resistant NSP2 could restore the changes (De Luis et al., 2012). In soybean, miR171o and miR171g functioned as negative regulators for nodulation inhibiting the expression of NIN, ENOD40 and Ethylene Response Factor Required for Nodulation (ERN) by targeting SCL-6 and NSP2 (Montgomery et al., 2008).
There is a high demand for phosphate in the legume nodule during nitrogen fixation (Gunawardena et al., 1992). Phosphate is commonly incorporated into organic molecules, such as ATP, nucleic acids and enzymes. Deficiency in phosphate leads to shortage in ATP supply, which reduces the efficiency of energy-intensive nitrogen fixation in nodules (Vance et al., 2000; Thuynsma et al., 2014). Under phosphate-deficient conditions, 25 miRNAs are induced and 11 miRNAs are repressed in soybean (Xu et al., 2013). Among the 25 miRNAs, three miRNAs (miR399, miR2111 and miR159e-3p) are also important regulators in nodulation (Xu et al., 2013; Tsikou et al., 2018; Fan et al., 2021). For example, miR2111 is induced under phosphate stress and targets TML to regulate nodule formation (Xu et al., 2013; Tsikou et al., 2018). MiR399 was highly induced in soybean roots and nodules after inoculation with Sinorhizobium fredii, and could enhance the symbiotic soybean phosphate uptake by suppressing the phosphate over accumulator 2 (PHO2) (Fan et al., 2021). PHO2 is a negative regulator of phosphate transporters responsible for phosphate uptake (Dong et al., 1998; Branscheid et al., 2010; Du et al., 2018). Thus, the upregulation of miR399 upon rhizobium inoculation could relax the inhibition on phosphate uptake. This improvement in phosphate uptake by miR399 can translate into higher ureide production and better overall plant growth (Fan et al., 2021).
Apart from miRNAs, lncRNAs also play a part. An lncRNA, Induced by Phosphate Starvation 1 (IPS1), was also induced by phosphate starvation and functioned as an miR399 sponge (Franco-Zorrilla et al., 2007). IPS1 harbors a motif with a reverse-complementary sequence to miR399, but with a mismatched loop at the expected miR399 cutting site, which makes IPS1 not cleavable and instead serves to sequester miR399 (Franco-Zorrilla et al., 2007). Therefore, IPS1 competitively binds to miR399 to dampen the control by miR399 on PHO2 (Franco-Zorrilla et al., 2007; Xu et al., 2013). In soybean, four IPS1 members were found by blast search in the genome and showed a potential role in the regulation of miR399 (Xu et al., 2013).
In Medicago truncatula, 10,785 lncRNAs were identified in roots and leaves, and some of them were responsible for the phosphate starvation response (Wang T. et al., 2017). An lncRNA, Phosphate Deficiency-Induced lncRNA (PDIL) 1 also targets miR399, performing the same function as IPS1 (Wang T. et al., 2017). Another two lncRNAs, PDIL2 and PDIL3, directly target inorganic phosphate (Pi) transporters at the transcriptional level to maintain phosphate homeostasis (Wang T. et al., 2017).
Copper (Cu) is an indispensable element for nitrogen fixation. Sufficient copper supply increases nitrogen fixation efficiency per nodule and elevates the nitrogen content in plants. Copper is a cofactor of metabolic enzymes and protein complexes in the electron transport chain in Bradyrhizobium japonicum (Preisig et al., 1996). Copper is also a substrate of several superoxide dismutases involved in oxidative stress responses, which could affect the penetration of the rhizobial infection thread into the legume host (Rubio et al., 2007). In Arabidopsis, the expressions of miR397, miR398, and miR408 are responsive to copper starvation. However, the induction of these miRNAs is abolished in the spl7 mutant (Abdel-Ghany and Pilon, 2008; Pilon et al., 2009; Pilon, 2017). These three miRNAs could reduce the biosynthesis of non-essential copper-containing enzymes to conserve copper resources among various groups of copper-containing proteins in order to maintain copper homeostasis (Gifford et al., 2008). The expression of miR397 is increased in the nitrogen-fixing nodules but not in the inactive nodules, and miR397 is involved in the nitrogen fixation-related copper homeostasis (De Luis et al., 2012). MiR397 targets the transcripts of copper-containing laccases (LACs), such as LAC2, LAC4 and LAC17 (Abdel-Ghany and Pilon, 2008). Furthermore, LAC3, LAC12 and LAC13 are under the control of miR408 (Gifford et al., 2008), which has a higher expression level in soybean nodules than in roots (Fan et al., 2021). MiR398 is induced in common bean nodules under copper deficiency, and is responsible for copper ion availability by reducing the production of non-essential copper-containing enzymes (Naya et al., 2014). It targets Cu/Zn Superoxide Dismutase 1 and 2 (CSD1 and CSD2) and Cu chaperones for superoxide dismutase SOD1 (CCS1), and is also linked to plastocyanin (Sunkar et al., 2006; Beauclair et al., 2010; Juszczak and Baier, 2012; Naya et al., 2014). CSD1, CSD2, CCS1 and plastocyanin are the most abundant copper-binding proteins in chloroplasts (Abdel-Ghany, 2009; Juszczak and Baier, 2012).
Iron is critical for both the legume host and the rhizobium, and has a positive effect on nodulation. Iron deficiency leads to a reduction in nodule numbers in Lupinus angustifolius L, and affects nodule development in soybean and common bean (Phaseolus vulgaris L.) (Tang et al., 1990; Soerensen et al., 2008; Slatni et al., 2011). The iron concentration in the nodule is positively related to the rate of nitrogen fixation in common bean (Slatni et al., 2008). In Arabidopsis, Iron deficiency-responsive cis-Element 1 and 2 (IDE1/2) were identified in 24 miRNA gene promoter regions (Kong and Yang, 2010). Among the 24 miRNAs genes, 17 of them exhibited were differentially expressed under iron starvation (Kong and Yang, 2010). MiR397, miR398, miR399, miR408 and miR2111 were down-regulated by iron deficiency while up-regulated by copper deficiency (Waters et al., 2012). A possible explanation for this is that, under low copper availability, the function of Cu/Zn superoxide dismutases is replaced by iron superoxide dismutase (Waters et al., 2012; Paul et al., 2015). MiR159, miR164, miR172, miR173 and miR394 were also identified to be iron-responsive miRNAs, based on the small RNA sequencing result (Kong and Yang, 2010; Waters et al., 2012). As stated above, most of these iron-responsive miRNAs are important nodulation regulators.
Sulfur (S) is a critical nutrient for plant growth as a component of amino acids, cofactors, enzymes and secondary metabolites (reviewed by Davidian and Kopriva, 2010; Becana et al., 2018). Sulfur deficiency leads to a reduction in nitrogen fixation, lower nitrogenase activity and the slowing down of nodule metabolism (Anderson and Spencer, 1950; Scherer et al., 2006). Not surprisingly, sulfur starvation inhibits nodulation, such as reducing nodule numbers and nodule mass per unit root length (Varin et al., 2010). Sulfur starvation affects the expression of miR156, miR160, miR167, miR164, miR168, miR394 and miR395 in Brassica napus (Huang et al., 2010). MiR395 is the main miRNA responsible for sulfur uptake and translocation (Liang et al., 2010; Yang et al., 2022). It targets and inhibits the expression of the ATP sulfurylase gene, APS1, and the sulfate transporter genes, SULTR2;1 and SULTR2;2 (Liang et al., 2010). APS1 is responsible for sulfur assimilation, and SULTR2;1 and SULTR2;2 function in sulfur translocation (Liang et al., 2010; Matthewman et al., 2012; Yang et al., 2022). In Arabidopsis, the overexpression of miR395 resulted in the over-accumulation of sulfur in shoots, while a reduced expression of miR395 caused a decline in sulfate concentration (Ai et al., 2016). A high level of miR395 in soybean nodules may ensure higher sulfur accumulation in nodules (Fan et al., 2021). Hydrogen sulfide (H2S) in nodules improves the soybean-rhizobium symbiosis and enhances nitrogen fixation (Zou et al., 2019). In Phaseolus vulgaris, a circRNA (Chr01:7660079|7660536) potentially functions as a sponge to sequester miR-RH83, to prevent the interaction during symbiosis between miR-RH83 and its target gene, Phvul.008G170800, which encodes a sulfur transporter (Wu et al., 2020). Therefore, circRNA (Chr01:7660079|7660536) may play a role in sulfur accumulation in the nodule.
Non-coding-RNAs, especially miRNAs, are involved in many aspects of nodulation and nitrogen fixation in legumes. MiR160, miR167, miR390 and miR393 are critical regulators for auxin signaling during the legume nodulation process, responsive to nutrient starvations and required for the enhanced tolerance of nodules to many biotic and abiotic stresses. MiR156 works as an upstream master regulator of legume nodulation by affecting multiple hormone signaling pathways, maintaining nutrient homeostasis and regulating the AON pathway. For the AON pathway, miR172c showed an indispensable role in controlling the expression of both CLE and ENOD40. As the lack of functional studies of lncRNAs and circRNAs in legumes, only a few lncRNAs were well studied, like IPS1 and PDIL are controlling the Pi homeostasis. These ncRNAs act as key regulators for the nodulation process and are promising targets for legume crop improvement.
The regulation of nodulation and nitrogen fixation is complicated. Elimination of important regulator may lead to detrimental effects. However, a small change in the expression or specificity of some miRNAs, such as miR172c, may potentially help loosening the AON to improve nodule formation or improving nitrogen fixation. CRISPR/Cas9 system is a commonly used genome editing tool in biology research and is also applied in legume crops, such as soybean, cowpea and chickpea (Chang et al., 2016; Gao, 2018). Apart from complete knockout of target ncRNA, CRISPR/Cas9 system could also be adopted to knockdown ncRNA. Single-guide RNAs (sgRNAs) were designed to target biogenesis processing sites of selected miRNA, the cutting sites of Dicer-like RNase III endonucleases (DCLs) (Chang et al., 2016). The CRISPR/Cas9 construct with this kind of sgRNAs could reduce the expressions of selected miRNA up to 96% (Chang et al., 2016). On the other hand, CRISPR/Cas base editors can be used to alter the base of the miRNAs or their targets in order to fine-tuning the miRNA-target specificity. Furthermore, CRISPR activation system (Pan et al., 2021) may also be adopted to activate the expression of miRNAs such as miR395 and miR399 to improve the nutritional status of the nodules. Apart from CRISPR/Cas9, short tandem target mimic technology (STTM) is also a high-efficient technology to block small RNA functions in plants (Tang et al., 2012). In rice, a large collection of STTM lines silencing 35 miRNA families was established (Zhang et al., 2017). Blocking miR530 was proved to improve the rice grain yield (Sun et al., 2020). Based on the analysis result of the genetic variation, the miR530 locus was regarded as a potentially artificial selection site in rice breeding (Sun et al., 2020).
With the advance in sequencing technologies, the discovery of different classes of ncRNAs is no longer challenging. Nevertheless, investigation of the biological functions of ncRNAs is difficult, especially for lncRNAs. Unlike protein-coding genes, the sequences of lncRNAs show little homology across species. Therefore, predicting the functions of lncRNAs by homology is not feasible. Since there is no functional domain/motif on ncRNAs, functional annotation of ncRNA is also not practical. As the concept of frameshift mutation does not apply to ncRNAs, small INDELs (insertions/deletions) created by traditional genome editing methods are not sufficient to knockout the ncRNAs unless they hit upon some critical nucleotides. Large-fragment deletions or substitutions by more advanced genome editing protocols may be able to solve the problem, but they still require some optimization. Therefore, although hundreds and thousands of ncRNAs have been identified, knowledge of their functions is still limited. Since nodules and roots are buried under the soil, mutant phenotypes are less observable than above-ground phenotypes. Hence, studying the functions of ncRNAs in nodulation and nitrogen fixation requires extra effort.
Plants can take up exogenous miRNAs and siRNAs to activate the RNA interference (RNAi) machinery (Betti et al., 2021). For example, the exogenous application of miR399 and miR156 could trigger RNAi by a mechanism requiring both AGO1 and RDR6 to cause the cleavage of their target genes, PHO2 and SPL (Betti et al., 2021). MiRNAs produced by plants can also be secreted into the environment to affect the gene expressions of the nearby plants, acting as signaling molecules that enable communications between plants (Betti et al., 2021). Actually, several studies have demonstrated that exogenous double-stranded RNAs (dsRNAs), short interfering RNAs (siRNAs), or hairpin RNAs (hpRNAs) could trigger the plant RNAi machinery to protect themselves against plant pathogenic viruses (Gan et al., 2010; Konakalla et al., 2016; Das and Sherif, 2020; Vadlamudi et al., 2020). While the application of GMO is hotly debated, altering plant phenotypes through exogenous applications of double-stranded miRNAs (ds-miRNAs), siRNAs, dsRNAs and hpRNAs may be more acceptable by society and eco-friendlier and more sustainable. Another possibility was the use of rich genetic resources. Nowadays, the large volume of sequencing data allows the discovery of genetic variations of ncRNA in different accessions within the same species. The natural gain or loss of ncRNA loci or natural variation of ncRNA sequence may play role in the regulation of the nodule function. Introduction or elimination of these loci through breeding may also be a viable strategy to improve the nitrogen fixation capability of elite cultivars.
Recently, the miRNA, Pmic_miR-8, was found to be transported from a fungal symbiont, Pisolithus microcarpus, to the roots of its symbiotic host, Eucalyptus grandis (Wong-Bajracharya et al., 2022). Pmic_miR-8 may inhibit the host transcripts encoding an NB-ARC domain-containing disease resistance protein, to stabilize the symbiotic interaction (Wong-Bajracharya et al., 2022). In rhizobium, three transfer RNA (tRNA)-derived small RNA fragments (tRFs) target the host legume genes associated with the nodulation process by triggering the host RNAi machinery (Ren et al., 2019). These three tRFs, tRFs-dubbed Bj-tRF001, Bj-tRF002, and Bj-tRF003 are the predominant products come from three tRNAs: Val-1-tRNA (CAC), Gly-1-tRNA (UCC), and Gln-1-tRNA (CUG), respectively (Ren et al., 2019). Bj-tRF001, Bj-tRF002, and Bj-tRF003 were predicted to respectively target GmRHD3a/GmRHD3b, GmHAM4a/GmHAM4b and GmLRX5, and hijack the soybean GmAGO1b to cause the tRF-guided cleavage of targets genes (Ren et al., 2019). GmRHD3a/GmRHD3b, GmHAM4a/GmHAM4b and GmLRX5 are orthologs of the Arabidopsis ROOT HAIR DIRECTIVE 3 (RDH3), HAIRY MERISTEM 4 (HAM4), and LEUCINE-RICH REPEAT EXTENSION-LIKE 5 (LRX5), which are required for root hair and plant development in Arabidopsis (Ren et al., 2019). Gain and loss of function analyses proved that GmRHD3a/GmRHD3b, GmHAM4a/GmHAM4b and GmLRX5 negatively regulate the deformed and curled root hairs upon rhizobium infection and the nodule numbers (Ren et al., 2019). Therefore, rhizobial tRFs could promote rhizobial infection (Ren et al., 2019). These studies hinted that legume-rhizobium symbiosis may not be only controlled by the ncRNAs produced by the legume host, but also by the rhizobium-produced small ncRNAs. A more complicated cross-kingdom sRNA gene-silencing network could be involved in the symbiosis between legume and rhizobium, and this requires further exploration.
The picture of the functions of ncRNAs on legume-rhizobium interactions is far from complete. Further delineation of the functions of ncRNAs in the legume-rhizobium symbiosis is important for legume crop breeding and improvement. Artificial manipulations and exogenous applications of ncRNAs could be effective ways to make nodulation and nitrogen fixation more efficient, which in turn can provide a good solution for sustainable agriculture.
KF, M-WL, and H-ML conceptualized the manuscript. KF, C-CS wrote the first draft. KF, M-WL, and H-ML reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
The work was supported by Hong Kong Research Grants Council Area of Excellence Scheme [AoE/M-403/16] and Lo Kwee-Seong Biomedical Research Fund for H-ML.
Ms. Jee Yan Chu copy-edited the manuscript. Any opinions, findings, conclusions, or recommendations expressed in this publication do not reflect the views of the Government of the Hong Kong Special Administrative Region or the Innovation and Technology Commission.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdel-Ghany, S. E. (2009). Contribution of plastocyanin isoforms to photosynthesis and copper homeostasis in Arabidopsis thaliana grown at different copper regimes. Planta 229, 767–779. doi: 10.1007/S00425-008-0869-Z
Abdel-Ghany, S. E., Pilon, M. (2008). MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 283, 15932–15945. doi: 10.1074/JBC.M801406200
Ai, Q., Liang, G., Zhang, H., Yu, D. (2016). Control of sulfate concentration by miR395-targeted APS genes in Arabidopsis thaliana. Plant Diversity 38, 92–100. doi: 10.1016/J.PLD.2015.04.001
Anderson, A. J., Spencer, D. (1950). Sulphur in nitrogen metabolism of legumes and non-legumes. Aust. J. Biol. Sci. 3, 431–449. doi: 10.1071/BI9500431
Aung, B., Gao, R., Gruber, M. Y., Yuan, Z. C., Sumarah, M., Hannoufa, A. (2017). MsmiR156 affects global gene expression and promotes root regenerative capacity and nitrogen fixation activity in alfalfa. Transgenic Res. 26, 541–557. doi: 10.1007/s11248-017-0024-3
Beauclair, L., Yu, A., Bouché, N. (2010). MicroRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J. 62, 454–462. doi: 10.1111/J.1365-313X.2010.04162.X
Becana, M., Wienkoop, S., Matamoros, M. A. (2018). Sulfur transport and metabolism in legume root nodules. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01434
Betti, F., Ladera-Carmona, M. J., Weits, D. A., Ferri, G., Iacopino, S., Novi, G., et al. (2021). Exogenous miRNAs induce post-transcriptional gene silencing in plants. Nat. Plants 7, 1379–1388. doi: 10.1038/s41477-021-01005-w
Borges, F., Martienssen, R. A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16, 727–741. doi: 10.1038/nrm4085
Boualem, A., Laporte, P., Jovanovic, M., Laffont, C., Plet, J., Combier, J. P., et al. (2008). MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J. 54, 876–887. doi: 10.1111/J.1365-313X.2008.03448.X
Branscheid, A., Sieh, D., Datt Pant, B., May, P., Devers, E. A., Elkrog, A., et al. (2010). Expression pattern suggests a role of miR399 in the regulation of the cellular response to local pi increase during arbuscular mycorrhizal symbiosis. Mol. Plant-Microbe Interact. 23, 915–926. doi: 10.1094/MPMI-23-7-0915
Breakspear, A., Liu, C., Roy, S., Stacey, N., Rogers, C., Trick, M., et al. (2015). The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 26, 4680–4701. doi: 10.1105/TPC.114.133496
Bustos-Sanmamed, P., Mao, G., Deng, Y., Elouet, M., Khan, G. A., Bazin, J., et al. (2013). Overexpression of miR160 affects root growth and nitrogen-fixing nodule number in Medicago truncatula. Funct. Plant Biol. 40, 1208–1220. doi: 10.1071/FP13123
Cai, Z., Wang, Y., Zhu, L., Tian, Y., Chen, L., Sun, Z., et al. (2017). GmTIR1/GmAFB3-based auxin perception regulated by miR393 modulates soybean nodulation. New Phytol. 215, 672–686. doi: 10.1111/NPH.14632
Campalans, A., Kondorosi, A., Crespi, M. (2004). Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell 16, 1047–1059. doi: 10.1105/TPC.019406
Chand Jha, U., Nayyar, H., Mantri, N., Siddique, K. H. M. (2021). Non-coding RNAs in legumes: Their emerging roles in regulating biotic/abiotic stress responses and plant growth and development. Cells 10, 1674. doi: 10.3390/CELLS10071674
Chang, H., Yi, B., Ma, R., Zhang, X., Zhao, H., Xi, Y. (2016). CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci. Rep. 6, 1–12. doi: 10.1038/srep22312
Charon, C., Johansson, C., Kondorosi, E., Kondorosi, A., Crespi, M. (1997). Enod40 induces dedifferentiation and division of root cortical cells in legumes. Proc. Natl. Acad. Sci. 94, 8901–8906. doi: 10.1073/PNAS.94.16.8901
Chen, L. L. (2016). The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 17, 205–211. doi: 10.1038/nrm.2015.32
Chen, L., Zhang, P., Fan, Y., Lu, Q., Li, Q., Yan, J., et al. (2018). Circular RNAs mediated by transposons are associated with transcriptomic and phenotypic variation in maize. New Phytol. 217, 1292–1306. doi: 10.1111/NPH.14901
Chen, H., Yang, Q., Chen, K., Zhao, S., Zhang, C., Pan, R., et al. (2019). Integrated microRNA and transcriptome profiling reveals a miRNA-mediated regulatory network of embryo abortion under calcium deficiency in peanut (Arachis hypogaea L.). BMC Genomics 20, 1–17. doi: 10.1186/S12864-019-5770-6
Chu, X., Su, H., Hayashi, S., Gresshoff, P. M., Ferguson, B. J. (2022). Spatiotemporal changes in gibberellin content are required for soybean nodulation. New Phytol. 234, 479–493. doi: 10.1111/NPH.17902
Cohn, J., Bradley Day, R., Stacey, G. (1998). Legume nodule organogenesis. Trends Plant Sci. 3, 105–110. doi: 10.1016/S1360-1385(97)01185-0
Combier, J. P., Frugier, F., De Billy, F., Boualem, A., El-Yahyaoui, F., Moreau, S., et al. (2006). MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 20, 3084–3088. doi: 10.1101/GAD.402806
Crespi, M. D., Jurkevitch, E., Poiret, M., D’Aubenton-Carafa, Y., Petrovics, G., Kondorosi, E., et al. (1994). Enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 13, 5099. doi: 10.1002/J.1460-2075.1994.TB06839.X
Das, P. R., Sherif, S. M. (2020). Application of exogenous dsRNAs-induced RNAi in agriculture: Challenges and triumphs. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00946
Davidian, J. C., Kopriva, S. (2010). Regulation of sulfate uptake and assimilation–the same or not the same? Mol. Plant 3, 314–325. doi: 10.1093/MP/SSQ001
De Billy, F., Grosjean, C., May, S., Bennett, M., Cullimore, J. V. (2001). Expression studies on AUX1-like genes in Medicago truncatula suggest that auxin is required at two steps in early nodule development. Mol. Plant-Microbe Interact. 14, 267–277. doi: 10.1094/MPMI.2001.14.3.267
De La Rosa, C., Lozano, L., Castillo-Ramírez, S., Covarrubias, A. A., Reyes, J. L. (2020). Origin and evolutionary dynamics of the miR2119 and ADH1 regulatory module in legumes. Genome Biol. Evol. 12, 2355–2369. doi: 10.1093/GBE/EVAA205
De Lima, J. C., Loss-Morais, G., Margis, R. (2012). MicroRNAs play critical roles during plant development and in response to abiotic stresses. Genet. Mol. Biol. 35, 1069–1077. doi: 10.1590/S1415-47572012000600023
De Luis, A., Markmann, K., Cognat, V., Holt, D. B., Charpentier, M., Parniske, M., et al. (2012). Two microRNAs linked to nodule infection and nitrogen-fixing ability in the legume Lotus japonicus. Plant Physiol. 160, 2137–2154. doi: 10.1104/pp.112.204883
Deng, P., Liu, S., Nie, X., Weining, S., and Wu, L (2018). Conservation analysis of long non-coding RNAs in plants. Sci. China Life Sci. 2017 612 61, 190–198. doi: 10.1007/S11427-017-9174-9
D’haeseleer, K., Den Herder, G., Laffont, C., Plet, J., Mortier, V., Lelandais-Brière, C., et al. (2011). Transcriptional and post-transcriptional regulation of a NAC1 transcription factor in Medicago truncatula roots. New Phytol. 191, 647–661. doi: 10.1111/J.1469-8137.2011.03719.X
Dharmasiri, N., Dharmasiri, S., Weijers, D., Lechner, E., Yamada, M., Hobbie, L., et al. (2005). Plant development is regulated by a family of auxin receptor f box proteins. Dev. Cell 9, 109–119. doi: 10.1016/J.DEVCEL.2005.05.014
Dong, B., Rengel, Z., Delhaize, E. (1998). Uptake and translocation of phosphate by pho2 mutant and wild-type seedlings of Arabidopsis thaliana. Planta 205, 251–256. doi: 10.1007/s004250050318
Dong, Z., Shi, L., Wang, Y., Chen, L., Cai, Z., Wang, Y., et al. (2013). Identification and dynamic regulation of microRNAs involved in salt stress responses in functional soybean sodules by high-throughput sequencing. Int. J. Mol. Sci. 14, 2717–2738. doi: 10.3390/IJMS14022717
Du, Q., Wang, K., Zou, C., Xu, C., Li, W. X. (2018). The PILNCR1-miR399 regulatory module is important for low phosphate tolerance in maize. Plant Physiol. 177, 1743–1753. doi: 10.1104/pp.18.00034
Ehrhardt, D. W., Wais, R., Long, S. R. (1996). Calcium spiking in plant root hairs responding to rhizobium nodulation signals. Cell 85, 673–681. doi: 10.1016/S0092-8674(00)81234-9
Fan, K., Wong-Bajracharya, J., Lin, X., Ni, M., Ku, Y. S., Li, M. W., et al. (2021). Differentially expressed microRNAs that target functional genes in mature soybean nodules. Plant Genome 14, e20103. doi: 10.1002/TPG2.20103
Ferguson, B. J., Mathesius, U. (2014). Phytohormone regulation of legume-rhizobia interactions. J. Chem. Ecol. 40, 770–790. doi: 10.1007/s10886-014-0472-7
Ferguson, B. J., Mens, C., Hastwell, A. H., Zhang, M., Su, H., Jones, C. H., et al. (2019). Legume nodulation: The host controls the party. Plant Cell Environ. 42, 41–51. doi: 10.1111/PCE.13348
Feyissa, B. A., Amyot, L., Nasrollahi, V., Papadopoulos, Y., Kohalmi, S. E., Hannoufa, A. (2021). Involvement of the miR156/SPL module in flooding response in Medicago sativa. Sci. Rep. 11, 1–16. doi: 10.1038/s41598-021-82450-7
Foyer, C. H., Lam, H.-M., Nguyen, H. T., Siddique, K. H. M., Varshney, R. K., Colmer, T. D., et al. (2016). Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2, 1–10. doi: 10.1038/nplants.2016.112
Franco-Zorrilla, J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I., Rubio-Somoza, I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. doi: 10.1038/ng2079
Gamas, P., Brault, M., Jardinaud, M. F., Frugier, F. (2017). Cytokinins in symbiotic nodulation: When, where, what for? Trends Plant Sci. 22, 792–802. doi: 10.1016/J.TPLANTS.2017.06.012
Ganguly, P., Roy, D., Das, T., Kundu, A., Cartieaux, F., Ghosh, Z., et al. (2021). The natural antisense transcript DONE40 derived from the lncRNA ENOD40 locus interacts with SET domain protein ASHR3 during inception of symbiosis in Arachis hypogaea. Mol. Plant-Microbe Interact. 34, 1057–1070. doi: 10.1094/MPMI-12-20-0357-R
Gan, D., Zhang, J., Jiang, H., Jiang, T., Zhu, S., Cheng, B. (2010). Bacterially expressed dsRNA protects maize against SCMV infection. Plant Cell Rep. 29, 1261–1268. doi: 10.1007/S00299-010-0911-Z
Gao, C. (2018). The future of CRISPR technologies in agriculture. Nat. Rev. Mol. Cell Biol. 19, 275–276. doi: 10.1038/nrm.2018.2
Garg, V., Khan, A. W., Kudapa, H., Kale, S. M., Chitikineni, A., Qiwei, S., et al. (2019). Integrated transcriptome, small RNA and degradome sequencing approaches provide insights into ascochyta blight resistance in chickpea. Plant Biotechnol. J. 17, 914–931. doi: 10.1111/PBI.13026
Gifford, M. L., Dean, A., Gutierrez, R. A., Coruzzi, G. M., Birnbaum, K. D. (2008). Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. U. S. A. 105, 803–808. doi: 10.1073/PNAS.0709559105
González-Guerrero, M., Matthiadis, A., Sáez, Á., Long, T. A. (2014). Fixating on metals: New insights into the role of metals in nodulation and symbiotic nitrogen fixation. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00045
Gray, W. M., Kepinski, S., Rouse, D., Leyser, O., Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276. doi: 10.1038/35104500
Grunewald, W., Van Noorden, G., Van Lsterdael, G., Beeckman, T., Gheysen, G., Mathesius, U. (2009). Manipulation of auxin transport in plant roots during Rhizobium symbiosis and nematode parasitism. Plant Cell 21, 2553–2562. doi: 10.1105/TPC.109.069617
Guinel, F. C. (2015). Ethylene, a hormone at the center-stage of nodulation. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01121
Gunawardena, S. F. B. N., Danso, S. K. A., Zapata, F. (1992). Phosphorus requirements and nitrogen accumulation by three mungbean (Vigna radiata (L) welzek) cultivars. Plant Soil 147, 267–274. doi: 10.1007/BF00029078
Guo, H. S., Xie, Q., Fei, J. F., Chua, N. H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17, 1376–1386. doi: 10.1105/TPC.105.030841
Hayashi, S., Gresshoff, P. M., Ferguson, B. J. (2014). Mechanistic action of gibberellins in legume nodulation. J. Integrated Plant Biol. 56, 971–978. doi: 10.1111/JIPB.12201
Herridge, D. F., Peoples, M. B., Boddey, R. M. (2008). Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311, 1–18. doi: 10.1007/s11104-008-9668-3
Hobecker, K. V., Reynoso, M. A., Bustos-Sanmamed, P., Wen, J., Mysore, K. S., Crespi, M., et al. (2017). The microRNA390/TAS3 pathway mediates symbiotic nodulation and lateral root growth. Plant Physiol. 174, 2469–2486. doi: 10.1104/PP.17.00464
Huang, S. Q., Xiang, A. L., Che, L. L., Chen, S., Li, H., Song, J. B., et al. (2010). A set of miRNAs from Brassica napus in response to sulphate deficiency and cadmium stress. Plant Biotechnol. J. 8, 887–899. doi: 10.1111/J.1467-7652.2010.00517.X
Jatan, R., Lata, C. (2019). Role of MicroRNAs in abiotic and biotic stress resistance in plants. Proc. Indian Natl. Sci. Acad. 85, 553–567. doi: 10.16943/PTINSA/2019/49586
Juszczak, I., Baier, M. (2012). The strength of the miR398-Csd2-CCS1 regulon is subject to natural variation in Arabidopsis thaliana. FEBS Lett. 586, 3385–3390. doi: 10.1016/J.FEBSLET.2012.07.049
Kanayama, Y., Watanabe, I., Yamamoto, Y. (1990). Inhibition of nitrogen fixation in soybean plants supplied with nitrate i. nitrite accumulation and formation of nitrosylleghemoglobin in nodules. Plant Cell Physiol. 31, 341–346. doi: 10.1093/oxfordjournals.pcp.a077913
Kepinski, S., Leyser, O. (2005). The Arabidopsis f-box protein TIR1 is an auxin receptor. Nature 435, 446–451. doi: 10.1038/NATURE03542
Khoei, M. A., Karimi, M., Karamian, R., Amini, S., Soorni, A. (2021). Identification of the complex interplay between nematode-related lncRNAs and their target genes in Glycine max L. Front. Plant Sci., 12. doi: 10.3389/FPLS.2021.779597
Kirolinko, C., Hobecker, K., Wen, J., Mysore, K. S., Niebel, A., Blanco, F. A., et al. (2021). Auxin response factor 2 (ARF2), ARF3, and ARF4 mediate both lateral root and nitrogen fixing nodule development in Medicago truncatula. Front. Plant Sci. 12. doi: 10.3389/FPLS.2021.659061
Kohlen, W., Ng, J. L. P., Deinum, E. E., Mathesius, U. (2018). Auxin transport, metabolism, and signalling during nodule initiation: Indeterminate and determinate nodules. J. Exp. Botany 69, 229–244. doi: 10.1093/jxb/erx308
Konakalla, N. C., Kaldis, A., Berbati, M., Masarapu, H., Voloudakis, A. E. (2016). Exogenous application of double-stranded RNA molecules from TMV p126 and CP genes confers resistance against TMV in tobacco. Planta 244, 961–969. doi: 10.1007/S00425-016-2567-6
Kondorosi, E., Redondo-Nieto, M., Kondorosi, A. (2005). Ubiquitin-mediated proteolysis. to be in the right place at the right moment during nodule development. Plant Physiol. 137, 1197–1204. doi: 10.1104/PP.105.060004
Kong, W. W., Yang, Z. M. (2010). Identification of iron-deficiency responsive microRNA genes and cis-elements in Arabidopsis. Plant Physiol. Biochem. 48, 153–159. doi: 10.1016/J.PLAPHY.2009.12.008
Kosslak, R. M., Bohlool, B.B. (1984). Suppression of nodule development of one side of a split-root system of soybeans caused by prior inoculation of the other side. Plant Physiol. 75, 125–130. doi: 10.1104/pp.75.1.125
Kumar, R. (2014). Role of microRNAs in biotic and abiotic stress responses in crop plants. Appl. Biochem. Biotechnol. 174, 93–115. doi: 10.1007/S12010-014-0914-2
Kumar, N., Bharadwaj, C., Sahu, S., Shiv, A., Shrivastava, A. K., Reddy, S. P. P., et al. (2021). Genome-wide identification and functional prediction of salt- stress related long non-coding RNAs (lncRNAs) in chickpea (Cicer arietinum L.). Physiol. Mol. Biol. Plants 27, 2605–2619. doi: 10.1007/S12298-021-01093-0
Lelandais-Brière, C., Naya, L., Sallet, E., Calenge, F., Frugier, F., Hartmann, C., et al. (2009). Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell 21, 2780–2796. doi: 10.1105/TPC.109.068130
Li, X., Lei, M., Yan, Z., Wang, Q., Chen, A., Sun, J., et al. (2014). The REL3-mediated TAS3 ta-siRNA pathway integrates auxin and ethylene signaling to regulate nodulation in Lotus japonicus. New Phytol. 201, 531–544. doi: 10.1111/NPH.12550
Liang, G., He, H., Yu, D. (2012). Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PloS One 7, e48951. doi: 10.1371/journal.pone.0048951
Liang, G., Yang, F., Yu, D. (2010). MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 62, 1046–1057. doi: 10.1111/j.1365-313X.2010.04216.x
Li, A., Mao, L. (2007). Evolution of plant microRNA gene families. Cell Res. 17, 212–218. doi: 10.1038/sj.cr.7310113
Lin, X., Lin, W., Ku, Y. S., Wong, F. L., Li, M. W., Lam, H. M., et al. (2020). Analysis of soybean long non-coding RNAs reveals a subset of small peptide-coding transcripts. Plant Physiol. 182, 1359–1374. doi: 10.1104/PP.19.01324
Liscum, E., Reed, J. W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400. doi: 10.1023/A:1015255030047
Li, C., Zhang, B. (2016). MicroRNAs in control of plant development. J. Cell. Physiol. 231, 303–313. doi: 10.1002/jcp.25125
Li, Y., Zhang, Q. Q., Zhang, J., Wu, L., Qi, Y., Zhou, J. M. (2010). Identification of MicroRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 152, 2222–2231. doi: 10.1104/PP.109.151803
Li, Z., Xu, H., Li, Y., Wan, X., Ma, Z., Cao, J., et al. (2018). Analysis of physiological and miRNA responses to Pi deficiency in alfalfa (Medicago sativa L.). Plant Mol. Biol. 96, 473–492. doi: 10.1007/s11103-018-0711-3
Long, S. R. (1989). Rhizobium-legume nodulation: Life together in the underground. Cell 56, 203–214. doi: 10.1016/0092-8674(89)90893-3
Liu, X., Chu, S., Sun, C., Xu, H., Zhang, J., Jiao, Y., et al. (2020). Genome-wide identification of low phosphorus responsive microRNAs in two soybean genotypes by high-throughput sequencing. Funct. Integr. Genomics 20, 825–838. doi: 10.1007/S10142-020-00754-9
Madsen, E. B., Antolín-Llovera, M., Grossmann, C., Ye, J., Vieweg, S., Broghammer, A., et al. (2011). Autophosphorylation is essential for the in vivo function of the Lotus japonicus nod factor receptor 1 and receptor-mediated signaling in cooperation with nod factor receptor. Plant J. 65, 404–417. doi: 10.1111/J.1365-313X.2010.04431.X
Madsen, E. B., Madsen, L. H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., et al. (2003). A receptor kinase gene of the LysM type is involved in legumeperception of rhizobial signals. Nature 425, 637–640. doi: 10.1038/nature02045
Manavella, P. A., Koenig, D., Weigel, D. (2012). Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. U. S. A. 109, 2461–2466. doi: 10.1073/PNAS.1200169109
Mao, G., Turner, M., Yu, O., Subramanian, S. (2013). miR393 and miR164 influence indeterminate but not determinate nodule development. Plant Signaling Behav. 8, 1559–2316. doi: 10.4161/PSB.26753
Martín-Rodríguez, J., Leija, A., Formey, D., Hernández, G. (2018). The microRNA319d/TCP10 node regulates the common bean – rhizobia nitrogen-fixing symbiosis. Front. Plant Sci. 9. doi: 10.3389/FPLS.2018.01175
Mathesius, U., Mathesius, U. (2008). Auxin: at the root of nodule development? Funct. Plant Biol. 35, 651–668. doi: 10.1071/FP08177
Mathesius, U., Schlaman, H. R. M., Spaink, H. P., Sautter, C., Rolfe, B. G., Djordjevic, M. A. (1998). Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J. 14, 23–34. doi: 10.1046/J.1365-313X.1998.00090.X
Matthewman, C. A., Kawashima, C. G., Húska, D., Csorba, T., Dalmay, T., Kopriva, S. (2012). miR395 is a general component of the sulfate assimilation regulatory network in Arabidopsis. FEBS Lett. 586, 3242–3248. doi: 10.1016/J.FEBSLET.2012.06.044
Mattick, J. S., Makunin, I. V. (2006). Non-coding RNA. Hum. Mol. Genet. 15, R17–R29. doi: 10.1093/HMG/DDL046
Matvienko, M., Van De Sande, K., Yang, W. C., Van Kammen, A., Bisseling, T., Franssen, H. (1994). Comparison of soybean and pea ENOD40 cDNA clones representing genes expressed during both early and late stages of nodule development. Plant Mol. Biol. 26, 487–493. doi: 10.1007/BF00039559
Ma, J., Wang, Y., Li, J. (2020). Global identification and analysis of micrornas involved in salt stress responses in two alfalfa (Medicago sativa millennium) lines. Can. J. Plant Sci. 100, 445–455. doi: 10.1139/CJPS-2018-0327
Ma, X., Zhang, X., Traore, S. M., Xin, Z., Ning, L., Li, K., et al. (2020). Genome-wide identification and analysis of long noncoding RNAs (lncRNAs) during seed development in peanut (Arachis hypogaea l.). BMC Plant Biol. 20, 1–14. doi: 10.1186/S12870-020-02405-4
Mens, C., Li, D., Haaima, L. E., Gresshoff, P. M., Ferguson, B. J. (2018). Local and systemic effect of cytokinins on soybean nodulation and regulation of their Isopentenyl transferase (IPT) biosynthesis genes following rhizobia inoculation. Front. Plant Sci. 9. doi: 10.3389/FPLS.2018.01150
Montgomery, T. A., Howell, M. D., Cuperus, J. T., Li, D., Hansen, J. E., Alexander, A. L., et al. (2008). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133, 128–141. doi: 10.1016/J.CELL.2008.02.033
Mortier, V., den Herder, G., Whitford, R., van de Velde, W., Rombauts, S., D’haeseleer, K., et al. (2010). CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 153, 222–237. doi: 10.1104/pp.110.153718
Nakagawa, T., Kaku, H., Shimoda, Y., Sugiyama, A., Shimamura, M., Takanashi, K., et al. (2011). From defense to symbiosis: limited alterations in the kinase domain of LysM receptor-like kinases are crucial for evolution of legume–rhizobium symbiosis. Plant J. 65, 169–180. doi: 10.1111/J.1365-313X.2010.04411.X
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. doi: 10.1126/SCIENCE.1126088
Naya, L., Paul, S., Valdés-López, O., Mendoza-Soto, A. B., Nova-Franco, B., Sosa-Valencia, G., et al. (2014). Regulation of copper homeostasis and biotic interactions by microRNA398b in common bean. PloS One 9, e84416. doi: 10.1371/JOURNAL.PONE.0084416
Nizampatnam, N. R., Schreier, S. J., Damodaran, S., Adhikari, S., Subramanian, S. (2015). MicroRNA160 dictates stage-specific auxin and cytokinin sensitivities and directs soybean nodule development. Plant Journal 84, 140–153. doi: 10.1111/tpj.12965
Oldroyd, G. E. D., Downie, J. A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59, 519–546. doi: 10.1146/ANNUREV.ARPLANT.59.032607.092839
Pacios-Bras, C., Schlaman, H. R. M., Boot, K., Admiraal, P., Langerak, J. M., Stougaard, J., et al. (2003). Auxin distribution in Lotus japonicus during root nodule development. Plant Mol. Biol. 52, 1169–1180. doi: 10.1023/B:PLAN.0000004308.78057.F5
Pan, C., Wu, X., Markel, K., Malzahn, A. A., Kundagrami, N., Sretenovic, S., et al. (2021). CRISPR–Act3.0 for highly efficient multiplexed gene activation in plants. Nat. Plants 7, 942–953. doi: 10.1038/s41477-021-00953-7
Paul, S., Datta, S. K., Datta, K. (2015). MiRNA regulation of nutrient homeostasis in plants. Front. Plant Sci. 06. doi: 10.3389/fpls.2015.00232
Pilon, M., Cohu, C. M., Ravet, K., Abdel-Ghany, S. E., Gaymard, F. (2009). Essential transition metal homeostasis in plants. Curr. Opin. Plant Biol. 12, 347–357. doi: 10.1016/J.PBI.2009.04.011
Preisig, O., Zufferey, R., Thöny-Meyer, L., Appleby, C. A., Hennecke, H. (1996). A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J. Bacteriol. 178, 1532–1538. doi: 10.1128/JB.178.6.1532-1538.1996
Raei, Y., Weisany, W., Allahverdipoor, K. H. (2013). Role of some of mineral nutrients in biological nitrogen fixation. Bull. Environ. Pharmacol. Life Sci. 2, 77–84.
Reid, D. E., Ferguson, B. J., Hayashi, S., Lin, Y. H., Gresshoff, P. M. (2011). Molecular mechanisms controlling legume autoregulation of nodulation. Ann. Bot. 108, 789–795. doi: 10.1093/AOB/MCR205
Remington, D. L., Vision, T. J., Guilfoyle, T. J., Reed, J. W. (2004). Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 135, 1738–1752. doi: 10.1104/PP.104.039669
Ren, B., Wang, X., Duan, J., Ma, J. (2019). Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 365, 919–922. doi: 10.1126/SCIENCE.AAV8907
Röhrig, H., Schmidt, J., Miklashevichs, E., Schell, J., John, M. (2002). Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc. Natl. Acad. Sci. U. S. A. 99, 1915–1920. doi: 10.1073/PNAS.022664799
Rubio, M. C., Becana, M., Sato, S., James, E. K., Tabata, S., Spaink, H. P. (2007). Characterization of genomic clones and expression analysis of the three types of superoxide dismutases during nodule development in Lotus japonicus. Mol. Plant Microbe Interact. 20, 262–275. doi: 10.1094/MPMI-20-3-0262
Rubio-Somoza, I., Weigel, D. (2013). Coordination of flower maturation by a regulatory circuit of three microRNAs. PloS Genet. 9, e1003374. doi: 10.1371/journal.pgen.1003374
Salvagiotti, F., Cassman, K. G., Specht, J. E., Walters, D. T., Weiss, A., Dobermann, A. (2008). Nitrogen uptake, fixation and response to fertilizer n in soybeans: A review. F. Crop Res. 108, 1–13. doi: 10.1016/j.fcr.2008.03.001
Sasaki, T., Suzaki, T., Soyano, T., Kojima, M., Sakakibara, H., Kawaguchi, M. (2014). Shoot-derived cytokinins systemically regulate root nodulation. Nat. Commun. 5, 4983. doi: 10.1038/ncomms5983
Scherer, H. W., Pacyna, S., Manthey, N., Schulz, M. (2006). Sulphur supply to peas (Pisum sativum l.) influences symbiotic N2 fixation. Plant Soil Environ. 52, 72–77. doi: 10.17221/3348-PSE
Schwechheimer, C., Serino, G., Callis, J., Crosby, W. L., Lyapina, S., Deshaies, R. J., et al. (2001). Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science 292, 1379–1382. doi: 10.1126/SCIENCE.1059776
Šečić, E., Kogel, K. H., Ladera-Carmona, M. J. (2021). Biotic stress-associated microRNA families in plants. J. Plant Physiol. 263, 153451. doi: 10.1016/J.JPLPH.2021.153451
Shrestha, A., Zhong, S., Therrien, J., Huebert, T., Sato, S., Mun, T., et al. (2021). Lotus japonicus nuclear factor YA1, a nodule emergence stage-specific regulator of auxin signalling. New Phytol. 229, 1535–1552. doi: 10.1111/NPH.16950
Slatni, T., Krouma, A., Aydi, S., Chaiffi, C., Gouia, H., Abdelly, C. (2008). Growth, nitrogen fixation and ammonium assimilation in common bean (Phaseolus vulgaris l) subjected to iron deficiency. Plant Soil 312, 49–57. doi: 10.1007/S11104-007-9481-4
Slatni, T., Vigani, G., Salah, I., Kouas, S., Dell’Orto, M., Gouia, H., et al. (2011). Metabolic changes of iron uptake in N2-fixing common bean nodules during iron deficiency. Plant Sci. 181, 151–158. doi: 10.1016/J.PLANTSCI.2011.04.015
Snowball, K., Robson, A. D., Lonergan, J. F. (1980). The effect of copper on nitrogen fixation in subterranean clover (Trifolium subterraneum). New Phytol. 85, 63–72. doi: 10.1111/j.1469-8137.1980.tb04448.x
Soerensen, K. U., Terry, R. E., Jolley, V. D., Brown, J. C., Vargas, M. E. (2008). The interaction of iron-stress response and root nodules in iron efficient and inefficient soybeans. J. Plant Nutr. 11, 853–862. doi: 10.1080/01904168809363850
Sorin, C., Declerck, M., Christ, A., Blein, T., Ma, L., Lelandais-Brière, C., et al. (2014). A miR169 isoform regulates specific NF-YA targets and root architecture in arabidopsis. New Phytol. 202, 1197–1211. doi: 10.1111/NPH.12735
Stacey, G., Libault, M., Brechenmacher, L., Wan, J., May, G. D. (2006). Genetics and functional genomics of legume nodulation. Curr. Opin. In Plant Biol. 9, 110–121. doi: 10.1016/J.PBI.2006.01.005
Statello, L., Guo, C. J., Chen, L. L., Huarte, M. (2020). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118. doi: 10.1038/s41580-020-00315-9
Stephens, B. D., Neyra, C. A. (1983). Nitrate and nitrite reduction in relation to nitrogenase activity in soybean nodules and Rhizobium japonicum bacteroids. Plant Physiol. 71, 731–735. doi: 10.1104/pp.71.4.731
Sulieman, S., Tran, L. S. P. (2015). Phosphorus homeostasis in legume nodules as an adaptive strategy to phosphorus deficiency. Plant Sci. 239, 36–43. doi: 10.1016/j.plantsci.2015.06.018
Sun, J., Cardoza, V., Mitchell, D. M., Bright, L., Oldroyd, G., Harris, J. M. (2006). Crosstalk between jasmonic acid, ethylene and nod factor signaling allows integration of diverse inputs for regulation of nodulation. Plant J. 46, 961–970. doi: 10.1111/j.1365-313X.2006.02751.x
Sunkar, R., Kapoor, A., Zhu, J. K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18, 2051–2065. doi: 10.1105/tpc.106.041673
Sun, Y., Mui, Z., Liu, X., Kay-Yuen Yim, A., Qin, H., Wong, F. L., et al. (2016). Comparison of small RNA profiles of Glycine max and Glycine soja at early developmental stages. Int. J. Of Mol. Sci. 17, 2043. doi: 10.3390/IJMS17122043
Sun, X., Wang, L., Ding, J., Wang, Y., Wang, J., Zhang, X., et al. (2016). Integrative analysis of Arabidopsis thaliana transcriptomics reveals intuitive splicing mechanism for circular RNA. FEBS Lett. 590, 3510–3516. doi: 10.1002/1873-3468.12440
Sun, W., Xu, X. H., Li, Y., Xie, L., He, Y., Li, W., et al. (2020). OsmiR530 acts downstream of OsPIL15 to regulate grain yield in rice. New Phytol. 226, 823–837. doi: 10.1111/NPH.16399
Suzuki, A., Hara, H., Kinoue, T., Abe, M., Uchiumi, T., Kucho, K. I., et al. (2008). Split-root study of autoregulation of nodulation in the model legume Lotus japonicus. J. Plant Res. 121, 245–249. doi: 10.1007/S10265-007-0145-5
Tang, C., Roboson, A. D., DILWORTH, M. J. (1990). The role of iron in nodulation and nitrogen fixation in Lupinus angustifolius l. New Phytol. 114, 173–182. doi: 10.1111/J.1469-8137.1990.TB00388.X
Tang, G., Yan, J., Gu, Y., Qiao, M., Fan, R., Mao, Y., et al. (2012). Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 58, 118–125. doi: 10.1016/j.ymeth.2012.10.006
Teotia, S., Tang, G. (2015). To bloom or not to bloom: Role of microRNAs in plant flowering. Mol. Plant 8, 359–377. doi: 10.1016/j.molp.2014.12.018
Thuynsma, R., Valentine, A., Kleinert, A. (2014). Phosphorus deficiency affects the allocation of below-ground resources to combined cluster roots and nodules in Lupinus albus. J. Plant Physiol. 171, 285–291. doi: 10.1016/j.jplph.2013.09.001
Tiwari, M., Singh, B., Yadav, M., Pandey, V., Bhatia, S. (2021). High throughput identification of miRNAs reveal novel interacting targets regulating chickpea-rhizobia symbiosis. Environ. Exp. Bot. 186, 104469. doi: 10.1016/J.ENVEXPBOT.2021.104469
Tsikou, D., Yan, Z., Holt, D. B., Abel, N. B., Reid, D. E., Madsen, L. H., et al. (2018). Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 362, 233–236. doi: 10.1126/science.aat6907
Turner, M., Nizampatnam, N. R., Baron, M., Coppin, S., Damodaran, S., Adhikari, S., et al. (2013). Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 162, 2042–2055. doi: 10.1104/pp.113.220699
Udvardi, M. K., Day, D. A. (1997). Metabolite transport across symbiotic membranes of legume nodules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 493–523. doi: 10.1146/annurev.arplant.48.1.493
Vadlamudi, T., Patil, B. L., Kaldis, A., Sai Gopal, D. V. R., Mishra, R., Berbati, M., et al. (2020). DsRNA-mediated protection against two isolates of Papaya ringspot virus through topical application of dsRNA in papaya. J. Virol. Methods 275, 113750. doi: 10.1016/J.JVIROMET.2019.113750
Van Zeijl, A., Op Den Camp, R. H. M., Deinum, E. E., Charnikhova, T., Franssen, H., Op Den Camp, H. J. M., et al. (2015). Rhizobium lipo-chitooligosaccharide signaling triggers accumulation of cytokinins in Medicago truncatula roots. Mol. Plant 8, 1213–1226. doi: 10.1016/J.MOLP.2015.03.010
Vance, C. P., Graham, P. H., Allan, D. L.. (2000). “Biological nitrogen fixation: phosphorus - a critical future need?,” in Nitrogen Fixation: From Molecules to Crop Productivity (Dordrecht: Kluwer Academic Publishers), 509–514. doi: 10.1007/0-306-47615-0_291
Varin, S., Cliquet, J. B., Personeni, E., Avice, J. C., Lemauviel-Lavenant, S. (2010). How does sulphur availability modify n acquisition of white clover (Trifolium repens l.)? J. Of Exp. Bot. 61, 225–234. doi: 10.1093/JXB/ERP303
Waadt, R., Seller, C. A., Hsu, P. K., Takahashi, Y., Munemasa, S., Schroeder, J. I. (2022). Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 1–15. doi: 10.1016/J.GPB.2017.01.007
Wang, X., Chang, X., Jing, Y., Zhao, J., Fang, Q., Sun, M., et al. (2020). Identification and functional prediction of soybean CircRNAs involved in low-temperature responses. J. Plant Physiol. 250, 153188. doi: 10.1016/J.JPLPH.2020.153188
Wang, Y., Li, K., Hen, L., Zou, Y., Liu, H., Tian, Y., et al. (2015). Microrna167-directed regulation of the auxin response factors GmARF8a and GmARF8b is required for soybean nodulation and lateral root development. Plant Physiol. 168, 101–116. doi: 10.1104/pp.15.00265
Wang, J., Meng, X., Dobrovolskaya, O. B., Orlov, Y. L., Chen, M. (2017). Non-coding RNAs and their roles in stress response in plants. Genomics Proteomics Bioinf. 15, 301–312. doi: 10.1016/J.GPB.2017.01.007
Wang, L., Sun, Z., Su, C., Wang, Y., Yan, Q., Chen, J., et al. (2019). A GmNINa-miR172c-NNC1 regulatory network coordinates the nodulation and autoregulation of nodulation pathways in soybean. Mol. Plant 12, 1211–1226. doi: 10.1016/j.molp.2019.06.002
Wang, Y., Wang, L., Zou, Y., Cai, Z., Zhang, S., Zhang, S., et al. (2014). Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell 26, 4782–4801. doi: 10.1105/tpc.114.131607
Wang, T., Zhao, M., Zhang, X., Liu, M., Yang, C., Chen, Y., et al. (2017). Novel phosphate deficiency-responsive long non-coding RNAs in the legume model plant Medicago truncatula. J. Exp. Bot. 68, 5937–5948. doi: 10.1093/jxb/erx384
Waters, B. M., McInturf, S. A., Stein, R. J. (2012). Rosette iron deficiency transcript and microRNA profiling reveals links between copper and iron homeostasis in Arabidopsis thaliana. J. Exp. Bot. 63, 5903–5918. doi: 10.1093/JXB/ERS239
Wong-Bajracharya, J., Singan, V. R., Monti, R., Plett, K. L., Ng, V., Grigoriev, I. V., et al. (2022). The ectomycorrhizal fungus Pisolithus microcarpus encodes a microRNA involved in cross-kingdom gene silencing during symbiosis. Proc. Natl. Acad. Sci. U. S. A. 119, 1-9. doi: 10.1073/PNAS.2103527119
Wong, J., Gao, L., Yang, Y., Zhai, J., Arikit, S., Yu, Y., et al. (2014). Roles of small RNAs in soybean defense against Phytophthora sojae infection. Plant J. 79, 928–940. doi: 10.1111/TPJ.12590
Wu, Z., Huang, W., Qin, E., Liu, S., Liu, H., Grennan, A. K., et al. (2020). Comprehensive identification and expression profiling of circular RNAs during nodule development in Phaseolus vulgaris. Front. Plant Sci. 11. doi: 10.3389/FPLS.2020.587185
Xu, F., Liu, Q., Chen, L., Kuang, J., Walk, T., Wang, J., et al. (2013). Genome-wide identification of soybean microRNAs and their targets reveals their organ-specificity and responses to phosphate starvation. BMC Genomics 14, 66. doi: 10.1186/1471-2164-14-66
Xu, H., Li, Y., Zhang, K., Li, M., Fu, S., Tian, Y., et al. (2021). miR169c-NFYA-C-ENOD40 modulates nitrogen inhibitory effects in soybean nodulation. New Phytol. 229, 3377–3392. doi: 10.1111/NPH.17115
Yang, Z., Hui, S., Lv, Y., Zhang, M., Chen, D., Tian, J., et al. (2022). miR395-regulated sulfate metabolism exploits pathogen sensitivity to sulfate to boost immunity in rice. Mol. Plant 15, 671–688. doi: 10.1016/J.MOLP.2021.12.013
Yan, Z., Hossain, M. S., Arikit, S., Valdés-López, O., Zhai, J., Wang, J., et al. (2015). Identification of microRNAs and their mRNA targets during soybean nodule development: functional analysis of the role of miR393j-3p in soybean nodulation. New Phytol. 207, 748–759. doi: 10.1111/nph.13365
Ye, C. Y., Chen, L., Liu, C., Zhu, Q. H., Fan, L. (2015). Widespread noncoding circular RNAs in plants. New Phytol. 208, 88–95. doi: 10.1111/NPH.13585
Yin, Z., Han, X., Li, Y., Wang, J., Wang, D., Wang, S., et al. (2017). Comparative analysis of cotton small RNAs and their target genes in response to salt stress. Genes 8, 369. doi: 10.3390/GENES8120369
Yin, X., Wang, J., Cheng, H., Wang, X., Yu, D. (2013). Detection and evolutionary analysis of soybean miRNAs responsive to soybean mosaic virus. Planta 237, 1213–1225. doi: 10.1007/S00425-012-1835-3
Yun, J., Sun, Z., Jiang, Q., Wang, Y., Wang, C., Luo, Y., et al. (2022). The miR156b-GmSPL9d module modulates nodulation by targeting multiple core nodulation genes in soybean. New Phytol. 233, 1881–1899. doi: 10.1111/NPH.17899
Zeng, Q. Y., Yang, C. Y., Ma, Q., Li, X. P., Dong, W. W., Nian, H. (2012). Identification of wild soybean miRNAs and their target genes responsive to aluminum stress. BMC Plant Biol. 12, 1–16. doi: 10.1186/1471-2229-12-182
Zhang, H., Yang, Y., Sun, C., Liu, X., Lv, L., Hu, Z., et al. (2020). Up-regulating GmETO1 improves phosphorus uptake and use efficiency by promoting root growth in soybean. Plant Cell Environ. 43, 2080–2094. doi: 10.1111/PCE.13816
Zhang, H., Liu, X., Wang, X., Sun, M., Song, R., Mao, P., et al. (2021a). Genome-wide identification of GRAS gene family and their responses to abiotic stress in Medicago sativa. Int. J. Mol. Sci. 22. doi: 10.3390/IJMS22147729
Zhang, J., Xu, H., Yang, Y., Zhang, X., Huang, Z., Zhang, D. (2021b). Genome-wide analysis of long non-coding RNAs (lncRNAs) in two contrasting soybean genotypes subjected to phosphate starvation. BMC Genomics 22, 1–15. doi: 10.1186/S12864-021-07750-8
Zhang, M., Su, H., Gresshoff, P. M., Ferguson, B. J. (2021). Shoot-derived miR2111 controls legume root and nodule development. Plant Cell Environ. 44, 1627–1641. doi: 10.1111/PCE.13992
Zhang, X. O., Wang, H., Zhang, Y., Lu, X., Chen, L. L., Yang, L. (2014). Complementary sequence-mediated exon circularization. Cell 159, 134–147. doi: 10.1016/J.CELL.2014.09.001
Zhang, H., Zhang, J., Yan, J., Gou, F., Mao, Y., Tang, G., et al. (2017). Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. Proc. Natl. Acad. Sci. U. S. A. 114, 5277–5282. doi: 10.1073/PNAS.1703752114
Zhao, W., Cheng, Y., Zhang, C., You, Q., Shen, X., Guo, W., et al. (2017). Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci. Rep. 7, 1–11. doi: 10.1038/s41598-017-05922-9
Zhao, W., Chu, S., Jiao, Y. (2019a). Present scenario of circular RNAs (circRNAs) in plants. Front. Plant Sci. 10. doi: 10.3389/FPLS.2019.00379
Zhao, X., Gan, L., Yan, C., Li, C., Sun, Q., Wang, J., et al. (2019b). Genome-wide identification and characterization of long non-coding RNAs in peanut. Genes, 10. doi: 10.3390/GENES10070536
Zhao, C., Ma, J., Zhang, Y., Yang, S., Feng, X., Yan, J. (2022). The miR166 mediated regulatory module controls plant height by regulating gibberellic acid biosynthesis and catabolism in soybean. J. Integr. Plant Biol. 64, 995–1006. doi: 10.1111/JIPB.13253
Zhu, Q. H., Wang, M. B. (2012). Molecular functions of long non-coding RNAs in plants. Genes 3, 176–190. doi: 10.3390/GENES3010176
Keywords: nitrogen fixation, nodulation, non-coding RNA, legume, rhizobium, autoregulation of nodulation, nutrient homeostasis, phytohormone
Citation: Fan K, Sze C-C, Li M-W and Lam H-M (2022) Roles of non-coding RNAs in the hormonal and nutritional regulation in nodulation and nitrogen fixation. Front. Plant Sci. 13:997037. doi: 10.3389/fpls.2022.997037
Received: 18 July 2022; Accepted: 29 September 2022;
Published: 18 October 2022.
Edited by:
Attila Kereszt, Eötvös Lóránd Research Network, HungaryReviewed by:
Andres Cruz-Hernandez, Universidad de La Salle Bajío, MexicoCopyright © 2022 Fan, Sze, Li and Lam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hon-Ming Lam, aG9ubWluZ0BjdWhrLmVkdS5oaw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.