
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 November 2022
Sec. Plant Development and EvoDevo
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.995734
This article is part of the Research Topic Orchid Genomics and Developmental Biology, Volume II View all 15 articles
 Qian-Qian Wang1
Qian-Qian Wang1 Yuan-Yuan Li1
Yuan-Yuan Li1 Jiating Chen1
Jiating Chen1 Meng-Jia Zhu2
Meng-Jia Zhu2 Xuedie Liu1
Xuedie Liu1 Zhuang Zhou1,3
Zhuang Zhou1,3 Diyang Zhang1
Diyang Zhang1 Zhong-Jian Liu1,2,3*
Zhong-Jian Liu1,2,3* Siren Lan1,2*
Siren Lan1,2*Members of the YABBY gene family play significant roles in lamina development in cotyledons, floral organs, and other lateral organs. The Orchidaceae family is one of the largest angiosperm groups. Some YABBYs have been reported in Orchidaceae. However, the function of YABBY genes in Cymbidium is currently unknown. In this study, 24 YABBY genes were identified in Cymbidium ensifolium, C. goeringii, and C. sinense. We analyzed the conserved domains and motifs, the phylogenetic relationships, chromosome distribution, collinear correlation, and cis-elements of these three species. We also analyzed expression patterns of C. ensifolium and C. goeringii. Phylogenetic relationships analysis indicated that 24 YABBY genes were clustered in four groups, INO, CRC/DL, YAB2, and YAB3/FIL. For most YABBY genes, the zinc finger domain was located near the N-terminus and the helix-loop-helix domain (YABBY domain) near the C-terminus. Chromosomal location analysis results suggested that only C. goeringii YABBY has tandem repeat genes. Almost all the YABBY genes displayed corresponding one-to-one relationships in the syntenic relationships analysis. Cis-elements analysis indicated that most elements were clustered in light-responsive elements, followed by MeJA-responsive elements. Expression patterns showed that YAB2 genes have high expression in floral organs. RT-qPCR analysis showed high expression of CeYAB3 in lip, petal, and in the gynostemium. CeCRC and CeYAB2.2 were highly expressed in gynostemium. These findings provide valuable information of YABBY genes in Cymbidium species and the function in Orchidaceae.
The seed plant-specific YABBY gene family, belonging to the zinc-finger superfamily, plays significant roles in lamina development in cotyledons, floral organs, and outer ovule integuments (Finet et al., 2016). YABBY genes encode transcription factors which contain two domains: a zinc finger domain located near the N-terminus and a helix-loop-helix domain (YABBY domain) located near the C-terminus (Bowman and Smyth, 1999). Six genes have been identified in Arabidopsis thaliana, and were clustered into five subfamilies: FIL/YAB3, CRC, INO, YAB2, and YAB5 (Siegfried et al., 1999). FIL, YAB2, YAB3, and YAB5 are expressed in leaf and floral organs and have been termed ‘vegetative YABBYs’. CRC and INO are essential in developing carpels and ovules, respectively, and have been termed ‘reproductive YABBYs’ (Bowman and Smyth, 1999; Siegfried et al., 1999; Villanueva et al., 1999; Bartholmes et al., 2012; Soundararajan et al., 2019).
According to previous studies from expression characterization in Arabidopsis YABBY genes, FIL, YAB2 and YAB3 play essential roles in lateral organ development (Siegfried et al., 1999; Rudall and Bateman, 2002; Lora et al., 2011). CRC is restricted to carpels and nectaries in angiosperms (Siegfried et al., 1999). INO functions in the development of the outer integument of the ovule to the seed coat in Arabidopsis, and INO expresses in eudicots, eumagnoliids, and some basal angiosperms (Bowman, 2000; Yamada et al., 2003; McAbee et al., 2005; Lora et al., 2011; Yamada et al., 2011).
The genome-wide YABBY gene family has been identified in Averrhoa carambola (star fruit), Cucumis sativus (cucumber), Lycopersicon esculentum (tomato), Oryza sativa (rice), Triticum aestivum (wheat) and Vitis vinifera (grape) (Toriba et al., 2007; Han et al., 2015; Zhang et al., 2019; Hao et al., 2022; Li et al., 2022; Yin et al., 2022). In monocot plants, YABBY genes show functional divergence and are crucial for vegetative and reproductive development. For example, the YAB3 clade genes ZYB9 and ZYB14 play essential roles in flower development and regulate lateral outgrowth (Juarez et al., 2004). OsDL, a member of the CRC subfamily in O. sativa, is necessary for the development of the leaf midrib and the flower carpel specification (Nagasawa et al., 2003; Yamaguchi et al., 2004; Ohmori et al., 2008; Zhang et al., 2020). OsYAB1, belonging to the YAB2 clade, is mainly expressed in the primordia of the carpel and stamen (Jang et al., 2004). The OsYAB3 gene may be necessary for the development of lateral organs and the growth and differentiation of leaf cells (Jang et al., 2004).
With an estimated > 28000 species, the Orchidaceae family is one of the largest angiosperm groups (Christenhusz and Byng, 2016). There are five subfamilies of Orchidaceae: Apostasioideae, Cypripedioideae, Vanilloideae, Orchidoideae, and Epidendroideae (Chase et al., 2003). The Orchidaceae show considerable diversity in epiphytic and terrestrial life forms and show unique flower morphologies and reproductive biology (Hsiao et al., 2011). Orchidaceae flowers show a variety of reliable floral morphological synapomorphies, such as a gynostemium (a fused structure of the pistils and stamens), a highly evolved petal termed labellum, and flowers with pollinia (Chase et al., 2003; Tsai et al., 2004). In the Orchidaceae family, genome-wide identification and expression patterns of YABBY genes were analyzed in Apostasia shenzhenica (Apostasioideae), Dendrobium catenatum (Epidendroideae), Gastrodia elata (Epidendroideae), and Phalaenopsis equestris (Epidendroideae) (Chen et al., 2020). However, studies of YABBY genes in the orchid tribe Cymbideae are still limited. Cymbidium is one of the most significant orchid genera for ornamental value because of its beautiful flowers (Ramya et al., 2019). Given the considerable role of YABBY genes in both vegetative and reproductive development, the identification of Cymbidium ensifolium, C. goeringii, and C. sinense will be employed, and the expression patterns of C. ensifolium will be analyzed in this study. This study provides new insights into the roles of YABBY genes and their contribution to the development of flower morphologies in Cymbidium subfamily of Orchidaceae.
The YABBY domain (PF04690) from PFAM was used as a query to search the protein database (El-Gebali et al., 2019). The genomes from Cymbidium ensifolium, C. goeringii, and C. sinense can be downloaded from their whole-genome sequencing data (Sun et al., 2021; Yang et al., 2021; Ai et al., 2021). HMM analysis (built in Tbtools) was used at an e value of 10-5 (Chen et al., 2018). BLASTP (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was also used to search the protein database using A. thaliana’s YABBY sequences, which can be downloaded in the TAIR database (https://www.arabidopsis.org). Then, the CDD website (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) was used to confirm the retrieved putative sequences. The aliphatic index (AI), grand average of hydrophobicity (GRAVY), instability index (II), and isoelectric points (pI) of the YABBY proteins were predicted using the ExPASy website (https://www.expasy.org/; Artimo et al., 2012). AtSubP (http://bioinfo3.noble.org/AtSubP/) was used to predict the subcellular localization of YABBY genes (Kaundal et al., 2010). The secondary structure was predicted using the SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html) program (Rozewicki et al., 2019).
The TAIR database (https://www.arabidopsis.org/) was used to download the protein sequences of Arabidopsis thaliana. The sequences of Oryza sativa, Phalaenopsis equestris, V. vinifera, and Zea mays were downloaded from the NCBI website (https://www.ncbi.nlm.nih.gov/genbank/). The protein sequences of YABBY genes from C. ensifolium, C. goeringii, and C. sinense can be downloaded from their whole-genome sequencing data (Ai et al., 2021; Sun et al., 2021; Yang et al., 2021). Multiple alignments were carried out using the program MAFFT (Rozewicki et al., 2019). Maximum likelihood (ML) tree inference was carried out using RAxML (RAxML-HPC2 on XSEDE; Miller et al., 2011), and was under a GTRGAMMA substitution model with 1,000 bootstraps. The EVOLVIEW website (https://evolgenius.info/) was used for layouting the phylogenetic tree (He et al., 2016).
Conserved domains of YABBY genes were analyzed using the CDD website (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi), and motifs were analyzed using the default parameters of the MEME website (http://meme-suite.org/) (Artimo et al., 2012). Fifteen motifs were identified in this study. To investigate the YABBY domains and C2C2 zinc-finger domain, the WEBLOGO tool (built in Tbtools) was employed. Multiple sequence alignments were carried out using MAFFT (Rozewicki et al., 2019).
To analyze the chromosomal location of YABBY genes in three Cymbidium species, the Tbtools software was used to create gene distribution maps by uploading the YABBY sequence (Chen et al., 2018). To analyze syntenic relationships, one step MCScanx (built in Tbtools) was used to analyze YABBY genes of C. ensifolium, C. goeringii, and C. sinense (Chen et al., 2018).
The 2000 bp regions upstream of the YABBY genes in C. ensifolium, C. goeringii, and C. sinense were extracted by TBTOOLS (Chen et al., 2018). Then, the cis-acting elements were identified by the PlantCare website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; Zhang et al., 2018).
Flower organs (petal, lip, and gynostemium) and leaves of C. ensifolium were collected, frozen in liquid nitrogen, and stored at 80°C until use. Total RNA was extracted using the Biospin Plant Total RNA Extraction Kit (Bioer Technology, Hangzhou, China). TransScript® All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (TransGen Biotech, Beijing, China) was used to create first-strand DNA and remove genomic DNA. The reaction conditions were 30 s at 94 °C and 45 cycles of 5 s at 94°C and 30 s at 60°C. Primers for the RT–qPCR analysis were designed by Primer Premier 5 software. GAPDH (JL008987) was used for normalization. Three biological replicates were performed in this study, and the expression data were quantified via the 2-△△CT method (Livak and Schmittgen, 2001).
Seven YABBY genes were found in C. ensifolium, nine in C. goeringii, and eight in C. sinense. The deduced protein length of YABBY genes ranged from 63 to 243 amino acids. The theoretical isoelectric point (pI) ranged from 6.11 to 10.75, and instability index (II) ranged from 32.78 to 57.09. The deducted grand average of hydrophilic values (GRAVY) of YABBY genes ranged from -1.155 to -0.232, and we found all the YABBY proteins were hydrophilic. The molecular weight (Mw) ranged from 7744.05 to 27185.43, and the aliphatic index (AI) ranged from 52.92 to 83.98 (Table 1). Subcellular localization results showed that all the YABBY genes were located in the nucleus, indicating that the nucleus may be where the YABBY genes function (Supplementary Table S1; Kaundal et al., 2010). The results of secondary structure prediction revealed that the average of α-helices, extended strands, β-turns, and random coils comprised 27.61, 14.13, 5.65, and 52.6% of the structure, respectively (Supplementary Table S2; Geourjon and Deléage, 1995).
To analyze the evolution patterns of YABBY genes in Cymbidium species, a phylogenetic tree was created by using the ML (maximum likelihood) method. Protein sequences from C. ensifolium, C. goeringii, C. sinense, A. thaliana O. sativa, P. equestris, V. vinifera, and Z. mays were used. The IDs of these species are listed in Supplementary Table S3. The results indicated that all Cymbidium species except C. goeringii have one member in the INO cluster. The number of YAB2 genes ranged from 3-6 (C. ensifolium: 3; C. goeringii: 6; C. sinense: 3). C. goeringii and C. sinense have two genes in the CRC subfamily, but C. ensifolium has only one. With the exception of C. goeringii, all Cymbidium species have two YAB3 genes (Figure 1).
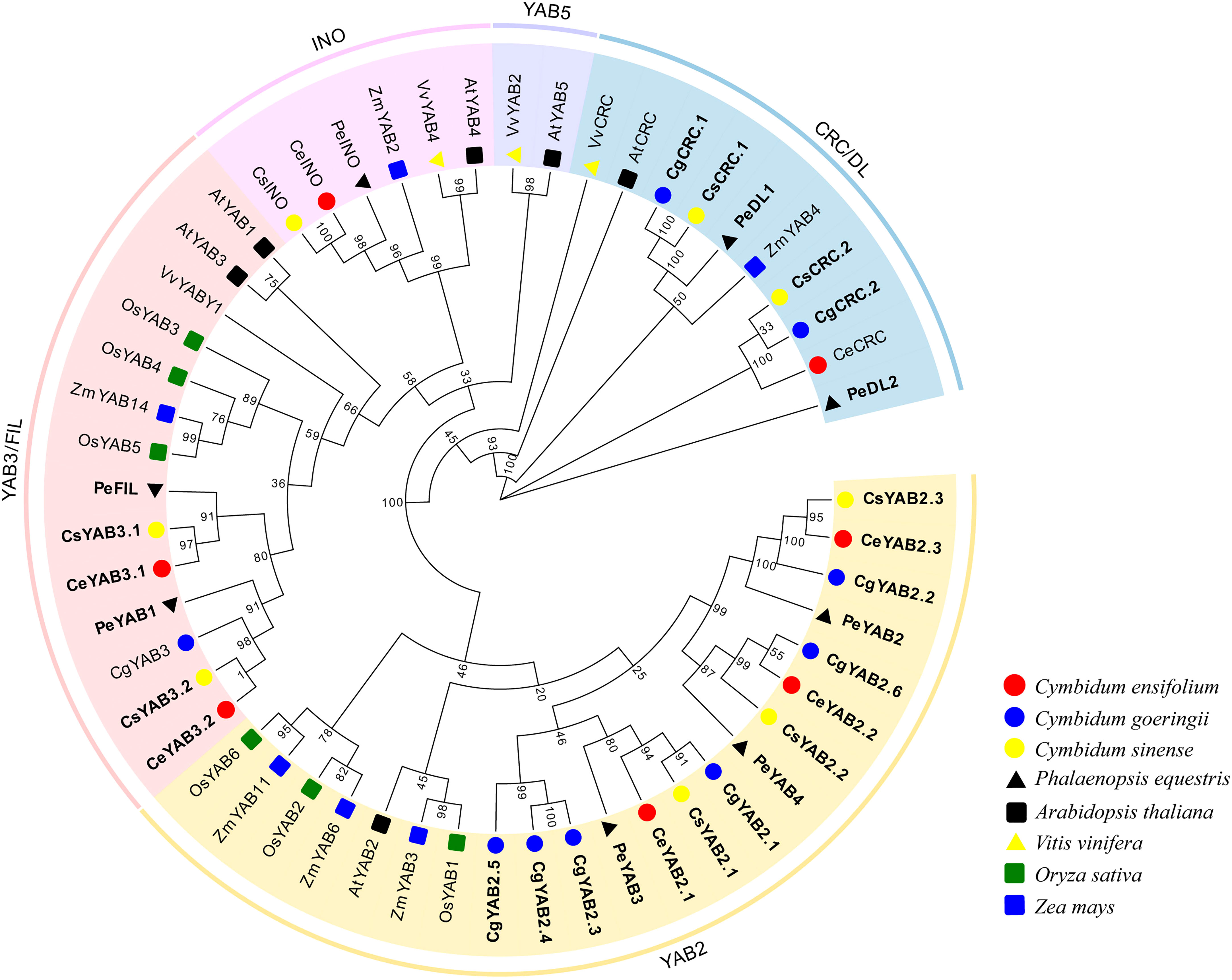
Figure 1 Phylogenetic tree of YABBY genes in eight plant species. The phylogenetic tree was created with the maximum-likelihood (ML) method using RAxML on the CIPRES Science Gateway web server (RAxML-HPC2 on XSEDE; Miller et al., 2011). Bootstrap values based on 1000 replicates are shown along the branches. Ce, C. ensifolium; Cg, C. goeringii; Cs, C. sinensis; At, A. thaliana; Os, O. sativa; Pe, P. equestris; Vv, V. vinifera; Zm, Z. mays; The duplicated genes are shown in bold.
To analyze the conserved domains of YABBY genes, the sequence logo of YABBY domains and c2c2 zinc-finger domains from three Cymbidium species and A. thaliana was generated. The multiple sequence alignment was also generated. The results showed that Cymbidum species and A. thaliana have highly conserved c2c2 zinc-finger domains and YABBY domains. However, the YABBY domain is more conserved than the c2c2 domain in Cymbidium species (Figure 2). Additionally, the motifs, domains, and phylogenetic tree of three Cymbidium species were analyzed (Figure 2). Fifteen motifs were analyzed by MEME software (Supplementary Table S4; Bailey et al., 2009). The results indicated that all the Cymbidium species have YABBY domains, and most YABBY genes of Cymbidium have motif 2 and motif 4. The findings also revealed that the conserved motifs of YABBY genes in the same clusters are similar.
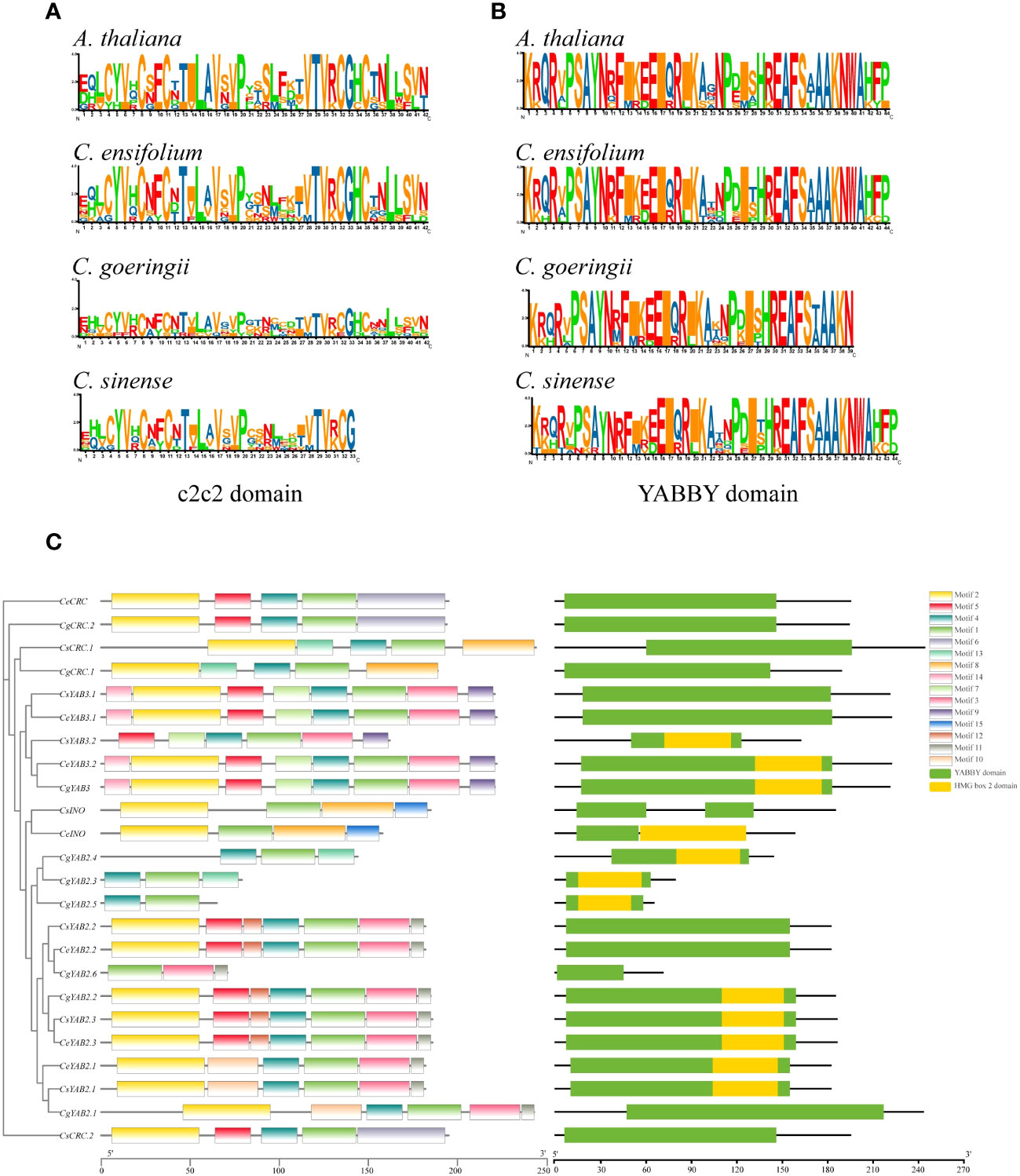
Figure 2 Conserved domains from three Cymbidium species and A. thaliana. (A) Sequence logo of the zinc-finger domain in the N-terminus. (B) Sequence logo of the YABBY domain in the C-terminus. (C) Motifs and conserved domains in the YABBY protein amino acid sequences in Cymbidium species.
To analyze the chromosome distribution of YABBY genes in three Cymbidium species, we create gene distribution maps. The results suggest that YABBY genes were distributed in seven chromosomes in C. ensifolium, C. goeringii, and C. sinense (Figure 3). In addition, YABBY genes were located in different chromosomes in C. ensifolium and C. sinense. Nevertheless, in C. goeringii, CgYAB2.3, CgYAB2.4, and CgYAB2.5 were located on same chromosome (chr17). We also analyzed the syntenic relationships of YABBY genes in three Cymbidium species. There are seven, nine, and eight YABBY genes in C. ensifolium, C. goeringii, and C. sinense (Figure 4). The results indicated that almost all the YABBY genes displayed corresponding one-to-one relationships in these three Cymbidium species.
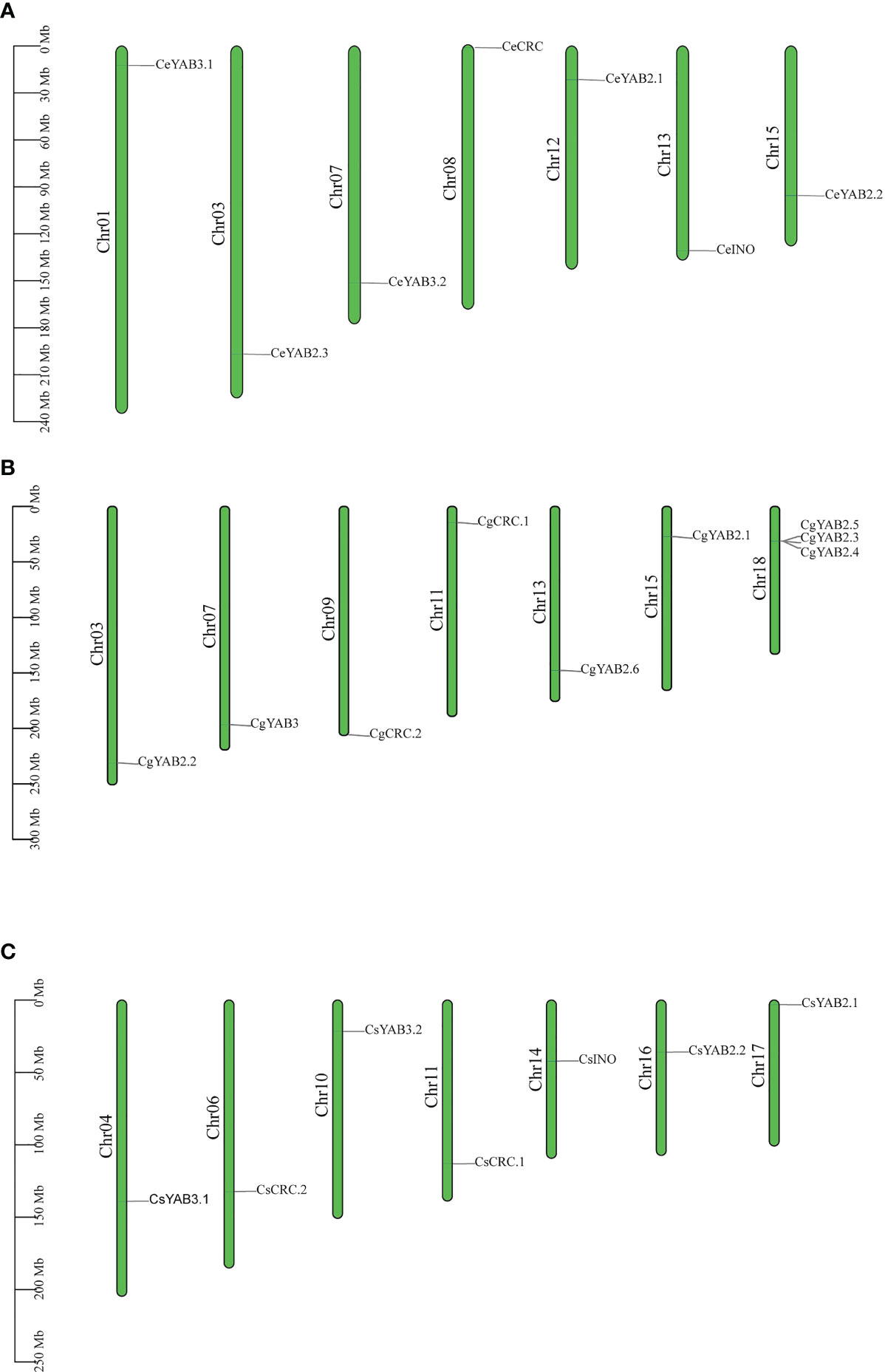
Figure 3 Chromosome distribution in three Cymbidium species. (A) Chromosome distribution in C. ensifolium. (B) Chromosome distribution in C. goeringii. (C) Chromosome distribution in C. sinense.
To predict the regulatory function of YABBY genes, we retrieved a 2000-bp region upstream of 24 YABBY genes and analyze them in C. ensifolium, C. goeringii, and C. sinense. We identified 12 types of cis-elements: abscisic acid responsiveness element, anaerobic induction element, auxin responsiveness element, circadian control element, defense and stress responsiveness element, endosperm expression element, light responsive element, low-temperature responsiveness element, MeJA-responsiveness element, meristem expression element, salicylic acid responsiveness element, and zein metabolism regulation element. In total, we found 412 cis-elements in three Cymbidium species, and C. sinense has most of the cis-elements (192/412), followed by C. goeringii (120/412), and C. ensifolium (100/412). The results also indicated that most of the elements were clustered in light-responsive elements (199/412), followed by MeJA-responsive elements (64/412), anaerobic induction element (27/412), and abscisic acid responsiveness element (24/412). All YABBY genes have light-responsive elements, and CsYAB3.1 contains the most (35/199). In addition, only CeYAB2.1, CeYAB3.2, and CgYAB2.1 have circadian control elements (Figure S1).
To analyze the expression patterns of YABBY genes, we sampled vegetative and floral organs from C. ensifolium and C. goeringii. The results suggested that in C. ensifolium, CeCRC showed high expression in pseudobulbs and pedicel, CeYAB2.1 and CeYAB 2.2 showed high expression in leaf and gynostemium, and CeYAB3.2 showed high expression in bud. CeYAB2.1, CeYAB2.2, CeYAB3.1, and CeYAB3.2 had expression in both vegetative and floral organs (Figure 5). In C. goeringii, CgCRC.1 showed high expression in gynostemium, and CgCRC.2 showed high expression in pseudobulbs and gynostemium. CgYAB3 showed high expression in pseudobulbs, leaves, and petals. CgCRC.2, CgYAB2.1, CgYAB2.2, CgYAB2.6, and CgYAB3 had expression in both vegetative and floral organs.
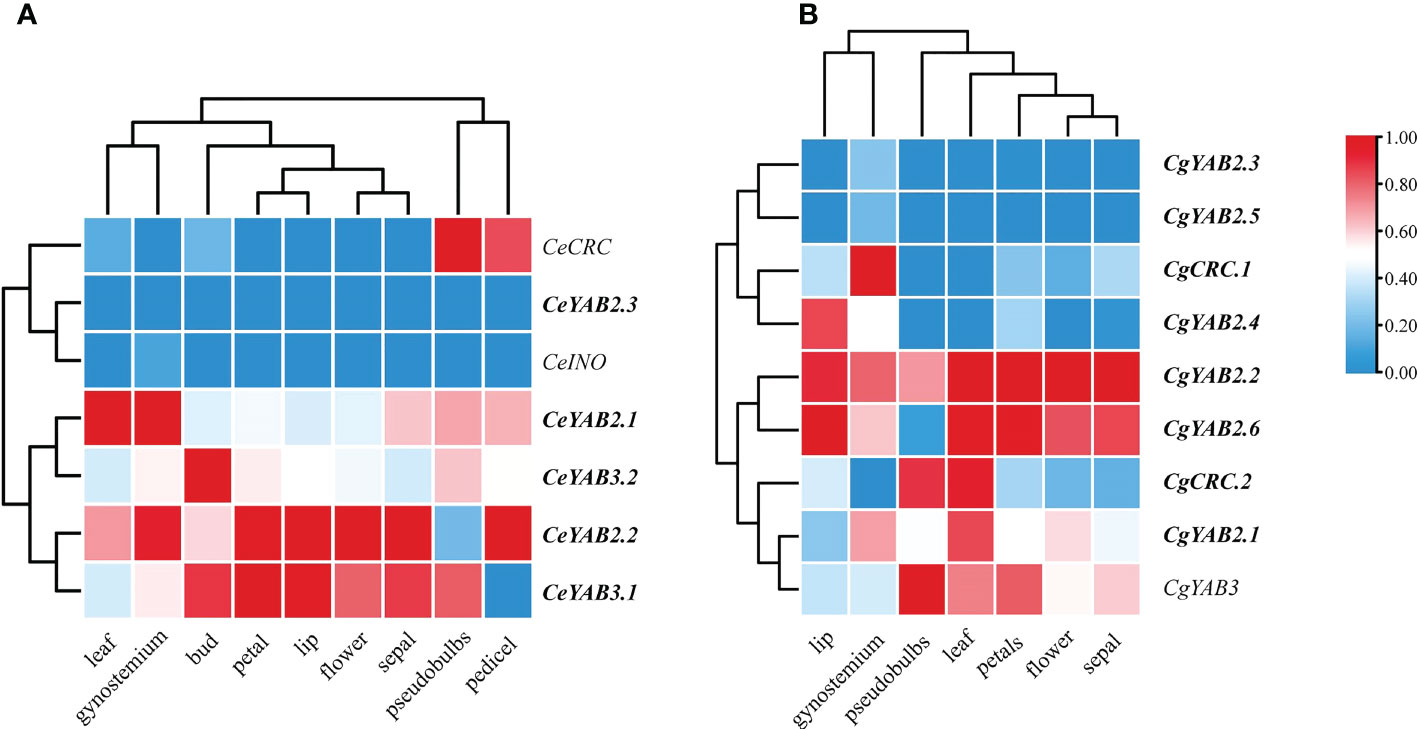
Figure 5 Expression patterns of YABBY genes in different organs from three Cymbidium species. (A, B) show the expression patterns of YABBY genes in different organs in C. ensifolium and C. goeringii. The heatmap was produced in Tbtools (Chen et al., 2018). The fragments per kilobase of transcript per million fragments (FPKM) values can be found in Supplementary Table S5. The duplicated genes are shown in bold.
To analyze the expression patterns of YABBY genes, we collected three floral organs (petal, lip, and gynostemium) and leaves from C. ensifolium. Four YABBY genes, CeCRC, CeINO, CeYAB2.2, and CeYAB3.1 were chosen for RT–qPCR analysis. The results showed that CeYAB3 showed high expression in the lip, petal, and gynostemium. CeCRC and CeYAB2.2 showed high expression in gynostemium. CeCRC, CeYAB2.2, and CeYAB3.1 had higher expression levels in floral organs than in leaves. However, the expression levels in leaves were higher than those in floral organs from CeINO (Figure 6).
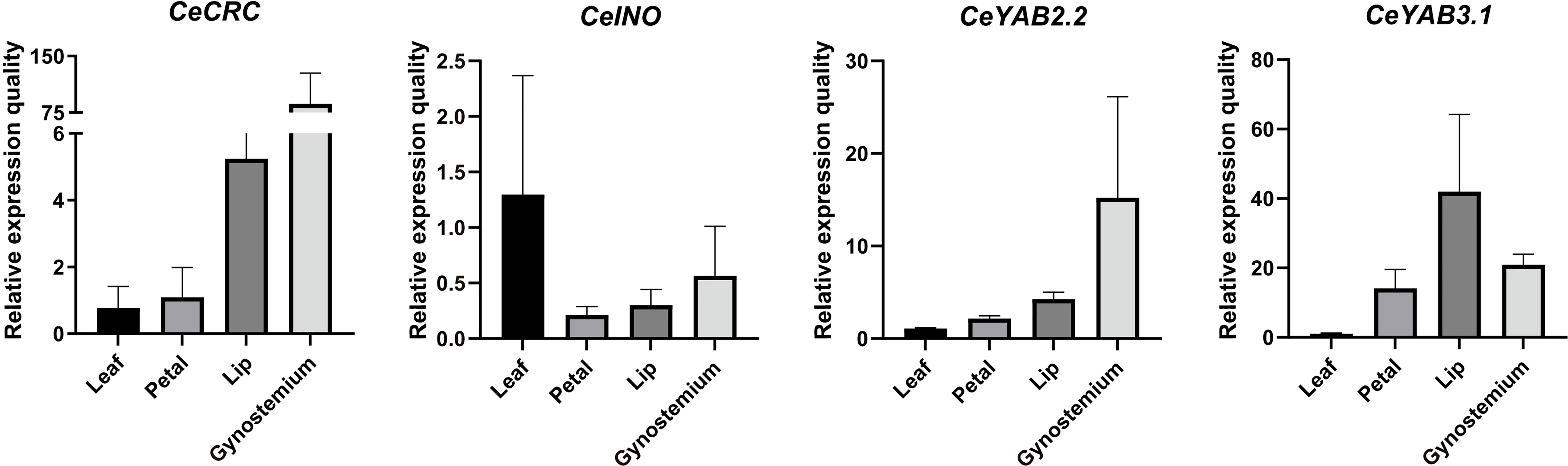
Figure 6 Relative expression patterns of YABBY genes in C. ensifolium. Raw data are listed in Supplementary Tables S6 and S7.
YABBY genes, which include a zinc finger domain near the N-terminus and a helix-loop-helix domain (YABBY domain) near the C-terminus, play important roles in lamina development in cotyledons, floral organs, and outer ovule integuments (Finet et al., 2016). In monocots, eight genes have been identified in O. sativa; in core eudicots, six YABBY genes have been found in A. thaliana (Bowman and Smyth, 1999; Sawa et al., 1999; Villanueva et al., 1999). Orchidaceae, belonging to monocots, is one of the largest angiosperm families and show unique flower morphologies and reproductive biology (Hsiao et al., 2011; Christenhusz and Byng, 2016). Recent studies have indicated that six YABBY genes were identified in A. shenzhenica, eight in D. catenatum, five in G. elata, and eight in P. equestris (Chen et al., 2020). However, studies of YABBY genes in Cymbidium are still limited. In this study, YABBY genes were identified in three Cymbidium species and the number of YABBY genes ranged from 7-9 (C. ensifolium: 7; C. goeringii: 9; C. sinense: 8). These results indicated that the number of YABBY genes in Cymbidum orchids were comparable to those in monocot and dicot species. However, the absence of YABBY genes in YAB 5 subfamily in orchids and other monocots is an exception.
The phylogenetic analysis indicated that YABBY genes in Cymbidium species are clustered into four subfamilies: YAB2, CRC, YAB3, and INO. There were no YABBY genes that clustered in the YAB5 subfamily. The results were consistent with some monocot species, such as A. shenzhenica, D. catenatum, G. elata, pineapple, and rice (Toriba et al., 2007; Li et al., 2019; Chen et al., 2020). However, seven species of magnoliids and A. thaliana have YABBY genes clustered in the YAB 5 clade (Siegfried et al., 1999; Liu et al., 2021). Early in the evolution of angiosperms, the lineages of basal flowering plants diverged, and then the magnoliids, eudicots, and monocots underwent rapid diversification (Tang et al., 2014; Chen et al., 2019). Magnoliids have two cotyledons and pollen with a single pore, and they are not monocots or eudicots (Tang et al., 2014). Recent reports also studied the comparative development of the androecial form in the Zingiberales and found one YAB2 gene, which was less homologous to YAB5 (De Almeida et al., 2014). Based on this, they suggested that after the divergence of monocots and eudicots, duplication led to separate YAB2 and YAB5 gene lineages (De Almeida et al., 2014). The YAB5 clade was exclusively composed of basal angiosperms and eudicot in recent studies (Chen et al., 2017; Liu et al., 2021). These results suggested that YAB5 gene clade might have been lost in monocot plants.
INO are restricted to the development of the outer ovule integument (Villanueva et al., 1999). Interestingly, we found C. ensifolium and C. sinense only has one number in the INO clade. These results were consistent with A. shenzhenica, A. thaliana, D. catenatum, G. elata, P. equestris, and V. vinifera, and indicated INO clade genes might be conserved in angiosperm plants and play essential roles in the outer integument (Siegfried et al., 1999; Zhang et al., 2019; Chen et al., 2020).
YABBY genes include a zinc finger domain near the N-terminus and a helix-loop-helix domain (YABBY domain) near the C-terminus. The results showed that the YABBY domain is more conserved than the c2c2 domain in three Cymbidium species. Fifteen motifs were analyzed in three Cymbidum species, and most YABBY genes of Cymbidium have motif 4 and motif 2. These findings revealed that the gene structure of YABBY genes are conserved during evolution. In the evolution of gene families, two main methods are tandem duplication and fragment duplication (Cannon et al., 2004). Chromosomal location analysis results suggested that YABBY genes were located in different chromosomes in C. ensifolium and C. sinense. But in C. goeringii, CgYAB2.3, CgYAB2.4, and CgYAB2.5 were located on same chromosome (chr17). The results indicated those genes might be tandem repeat genes. The syntenic relationships analysis indicated that almost every YABBY gene displayed corresponding one-to-one relationships in these three Cymbidium species.
Cis-elements were found in promoter areas in YABBY genes. The results indicated that most of the elements were clustered in light-responsive elements (199/412), followed by MeJA-responsive elements (64/412), anaerobic induction elements (27/412), and abscisic acid responsiveness element (24/412). The MeJA (methyl jasmonate) is a phytohormone involved in defense signaling of plants (Howe, 2004). The results indicated YABBY genes might play essential roles in plant growth and stress.
The growth of lateral organs in A. thaliana is thought to be redundantly controlled by the genes YAB2 and FIL, which are expressed in the leaves, cotyledons, and floral organs (Siegfried et al., 1999; Rudall and Bateman, 2002). FIL gene orthologues have similarly acted in flower development in Oryza (Tanaka et al., 2017). Our study indicated that three Cymbidium species contained one or two FIL genes and had high expression in the floral organs of C. ensifolium and C. goeringii. The results suggested that FIL may play important roles in the development of floral organ in Cymbidium species. CRC showed high expression in pseudobulbs in C. ensifolium and C. goeringii, and CRC showed high expression in pedicels in C. ensifolium. CRC also showed high expression in gynostemium in C. goeringii. The results suggested that CRC in different Cymbidum had different expression patterns. INO expressed in the gynostemium and pedicel in C. ensifolium. It may play important roles in the development of gynostemium and pedicel. YAB2 genes (CeYAB2.1, CeYAB2.2, CgYAB2.1, CgYAB2.2, and CgYAB2.6) showed high expression in all organs in Cymbidium species, indicating that the YAB2 clade may have functions in both reproductive and vegetative organs.
The results indicated that YABBY genes in Cymbidium species showed higher expression in reproductive tissues than in vegetative tissues. The results were consistent with the expression patterns reported in A. shenzhenica, D. catanum, and P. equestris (Chen et al., 2020). RT–qPCR analysis showed that CeCRC, CeYAB2.2, and CeYAB3.1 have higher expression levels in floral organs than in leaves. However, the expression levels in leaves were slightly higher than those in floral organs in CeINO. These findings indicated that YABBY genes play important roles in floral organ development in orchids. Orchids display unique flower morphologies, and their flowers possess several reliable floral morphological synapomorphies, including a gynostemium (a fused structure of the pistils and stamens) (Chase et al., 2003; Tsai et al., 2004). The results of this study indicated that CeCRC might play essential roles in floral organs, especially in gynostemium.
The data presented in the study are deposited in the National Centre for Biotechnology Information (NCBI) and National Genomics Data Center (NGDC). The raw data can be found under the following accession numbers: SAMN20059972 (NCBI), PRJNA749652 (NCBI) and PRJCA005355 (NGDC).
SL, Z-JL, and DZ contributed to conceptualization and validation. Q-QW, Y-YL, and ZZ prepared the original draft. Q-QW, JC, and M-JZ analyzed data, Q-QW and XL make the images. All authors contributed to the article and approved the submitted version.
This research was funded by Forestry Peak Discipline Construction Project of Fujian Agriculture and Forestry University (72202200205), The National Natural Science Foundation of China (no. 31870199), The Key Laboratory of National Forestry and Grassland Administration for Orchid Conservation and Utilization Construction Funds (Grant 115/118990050, 115/KJG18016A), Natural Science Foundation of Zhejiang Province (Grant nos. LY20C160005, LY19C150003), Key Research and Development Program of Zhejiang Province (Grant no. 2021C02043), and Wenzhou Agricultural New Variety Breeding Cooperative Group Project (Grant no. 2019ZX004-3).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.995734/full#supplementary-material
Ai, Y., Li, Z., Sun, W. H., Chen, J., Zhang, D., Ma, L., et al. (2021). The cymbidium genome reveals the evolution of unique morphological traits. Horticult. Res. 8, 255. doi: 10.1038/s41438-021-00683-z
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., De Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, 597–603. doi: 10.1093/nar/gks400
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME suite: Tools for motif discovery and searching. Nucleic Acids Res. 37, 1–7. doi: 10.1093/nar/gkp335
Bartholmes, C., Hidalgo, O., Gleissberg, S. (2012). Evolution of the YABBY gene family with emphasis on the basal eudicot eschscholzia californica (Papaveraceae). Plant Biol. 14, 11–23. doi: 10.1111/j.1438-8677.2011.00486.x
Bowman, J. L. (2000). The YABBY gene family and abaxial cell fate. Curr. Opin. Plant Biol. 3, 17–22. doi: 10.1016/S1369-5266(99)00035-7
Bowman, J. L., Smyth, D. R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126, 2387–2396. doi: 10.1242/dev.126.11.2387
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in arabidopsis thaliana. BMC Plant Biol. 4, 1–21. doi: 10.1186/1471-2229-4-10
Chase, M., Cameron, K., Barrett, R., Freudenstein, J. V. (2003). DNA Data and orchidaceae systematics: A new phylogenetic classification. Orchid Conserv. 69 (89), 32. Available at: https://www.researchgate.net/publication/234814296.
Chen, C., Chen, H., He, Y., Xia, R. (2018). TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. bioRxiv. 289660 (10.1101), 289660. doi: 10.1101/289660
Chen, J., Hao, Z., Guang, X., Zhao, C., Wang, P., Xue, L., et al. (2019). Liriodendron genome sheds light on angiosperm phylogeny and species–pair differentiation. Nat. Plants 5 (1), 18–25. doi: 10.1038/s41477-018-0323-6
Chen, Y. Y., Hsiao, Y. Y., Chang, S. B., Zhang, D., Lan, S. R., Liu, Z. J., et al. (2020). Genome-wide identification of yabby genes in orchidaceae and their expression patterns in phalaenopsis orchid. Genes 11, 1–17. doi: 10.3390/genes11090955
Chen, F., Liu, X., Yu, C., Chen, Y., Tang, H., Zhang, L. (2017). Water lilies as emerging models for darwin’s abominable mystery. Horticult. Res. 4, 17051. doi: 10.1038/hortres.2017.51
Christenhusz, M. J. M., Byng, J. W. (2016). The number of known plants species in the world and its annual increase. Phytotaxa 261, 201–217. doi: 10.11646/phytotaxa.261.3.1
De Almeida, A. M. R., Yockteng, R., Schnable, J., Alvarez-Buylla, E. R., Freeling, M., Specht, C. D. (2014). Co-Option of the polarity gene network shapes filament morphology in angiosperms. Sci. Rep. 4, 1–9. doi: 10.1038/srep06194
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Finet, C., Floyd, S. K., Conway, S. J., Zhong, B., Scutt, C. P., Bowman, J. L. (2016). Evolution of the YABBY gene family in seed plants. Evol. Dev. 18, 116–126. doi: 10.1111/ede.12173
Geourjon, C., Deléage, G. (1995). Sopma: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 11, 681–684. doi: 10.1093/bioinformatics/11.6.681
Han, H. Q., Liu, Y., Jiang, M. M., Ge, H. Y., Chen, H. Y. (2015). Identification and expression analysis of YABBY family genes associated with fruit shape in tomato (Solanum lycopersicum l.). Genet. Mol. Res. 14, 7079–7091. doi: 10.4238/2015.June.29.1
Hao, L., Zhang, J., Shi, S., Li, P., Li, D., Zhang, T., et al. (2022). Identification and expression profiles of the YABBY transcription factors in wheat. PeerJ 10, 1–15. doi: 10.7717/peerj.12855
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W. H., Hu, S. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44, W236–W241. doi: 10.1093/nar/gkw370
Howe, G. A. (2004). Jasmonates as signals in the wound response. J. Plant Growth Regul. 23 (3), 223–237. doi: 10.1007/s00344-004-0030-6
Hsiao, Y. Y., Pan, Z. J., Hsu, C. C., Yang, Y. P., Hsu, Y. C., Chuang, Y. C., et al. (2011). Research on orchid biology and biotechnology. Plant Cell Physiol. 52, 1467–1486. doi: 10.1093/pcp/pcr100
Jang, S., Hur, J., Kim, S. J., Han, M. J., Kim, S. R., An, G. (2004). Ectopic expression of OsYAB1 causes extra stamens and carpels in rice. Plant Mol. Biol. 56, 133–143. doi: 10.1007/s11103-004-2648-y
Juarez, M. T., Twigg, R. W., Timmermans, M. C. P. (2004). Specification of adaxial cell fate maize leaf development. Development 131, 4533–4544. doi: 10.1242/dev.01328
Kaundal, R., Saini, R., Zhao, P. X. (2010). Combining machine learning and homology-based approaches to accurately predict subcellular localization in arabidopsis. Plant Physiol. 154, 36–54. doi: 10.1104/pp.110.156851
Li, C., Dong, N., Shen, L., Lu, M., Zhai, J., Zhao, Y., et al. (2022). Genome-wide identification and expression profile of YABBY genes in averrhoa carambola. PeerJ 9, 1–22. doi: 10.7717/peerj.12558
Li, Z., Li, G., Cai, M., Priyadarshani, S. V. G. N., Aslam, M., Zhou, Q., et al. (2019). Genome-wide analysis of the YABBY transcription factor family in pineapple and functional identification of AcYABBY4 involvement in salt stress. Int. J. Mol. Sci. 20, 1–17. doi: 10.3390/ijms20235863
Liu, X., Liao, X. Y., Zheng, Y., Zhu, M. J., Yu, X., Jiang, Y. T., et al. (2021). Genome-wide identification of the YABBY gene family in seven species of magnoliids and expression analysis in litsea. Plants 10, 1–18. doi: 10.3390/plants10010021
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lora, J., Hormaza, J. I., Herrero, M., Gasser, C. S. (2011). Seedless fruits and the disruption of a conserved genetic pathway in angiosperm ovule development. Proc. Natl. Acad. Sci. United States America 108, 5461–5465. doi: 10.1073/pnas.1014514108
McAbee, J. M., Kuzoff, R. K., Gasser, C. S. (2005). Mechanisms of derived unitegmy among impatiens species. Plant Cell 17, 1674–1684. doi: 10.1105/tpc.104.029207
Miller, M. A., Pfeiffer, W., Schwartz, T. (2011). The CIPRES science gateway: A community resource for phylogenetic analyses. Proceedings of the TeraGrid 2011 Conference: Extreme Digital Discovery, TG’11. doi: 10.1145/2016741.2016785
Nagasawa, N., Miyoshi, M., Sano, Y., Satoh, H., Hirano, H., Sakai, H., et al. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130, 705–718. doi: 10.1242/dev.00294
Ohmori, Y., Abiko, M., Horibata, A., Hirano, H. Y. (2008). A transposon, ping, is integrated into intron 4 of the DROOPING LEAF gene of rice, weakly reducing its expression and causing a mild drooping leaf phenotype. Plant Cell Physiol. 49, 1176–1184. doi: 10.1093/pcp/pcn093
Ramya, M., Park, P. H., Chuang, Y. C., Kwon, O. K., An, H. R., Park, P. M., et al. (2019). RNA Sequencing analysis of cymbidium goeringii identifies floral scent biosynthesis related genes. BMC Plant Biol. 19, 1–14. doi: 10.1186/s12870-019-1940-6
Rozewicki, J., Li, S., Amada, K. M., Standley, D. M., Katoh, K. (2019). MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 47, W5–W10. doi: 10.1093/nar/gkz342
Rudall, P., Bateman, R. (2002). Roles of synorganisation, zygomorphy and heterotopy in floral evolution: The gynostemium and labellum of orchids and other lilioid monocots. Biol. Rev. 77 (3), 403–441. doi: 10.1017/S1464793102005936
Sawa, S., Ito, T., Shimura, Y., Okada, K. (1999). FILAMENTOUS FLOWER controls the formation and development of arabidopsis inflorescences and floral meristems. Plant Cell 11, 69–86. doi: 10.1105/tpc.11.1.69
Siegfried, K. R., Eshed, Y., Baum, S. F., Otsuga, D., Drews, G. N., Bowman, J. L. (1999). Members of the YABBY gene family specify abaxial cell fate in arabidopsis. Development 126 (18), 4117–4128. doi: 10.1242/dev.126.18.4117
Soundararajan, P., Won, S. Y., Park, D. S., Lee, Y. H., Sun Kim, J. (2019). Comparative analysis of the YABBY gene family of bienertia sinuspersici, a single-cell c4 plant. Plants 8. doi: 10.3390/plants8120536
Sun, Y., Chen, G., Huang, J., Liu, D., Xue, F., Chen, X., et al. (2021). The cymbidium goeringii genome provides insight into organ development and adaptive evolution in orchids. Ornamental Plant Research 1 (1), 1–13. doi: 10.48130/OPR-2021-0010
Tanaka, W., Toriba, T., Hirano, H. Y. (2017). Three TOB1-related YABBY genes are required to maintain proper function of the spikelet and branch meristems in rice. New Phytol. 215, 825–839. doi: 10.1111/nph.14617
Tang, H., Lyons, E., Schnable, J. C. (2014). “Earlay history of the angiosperms,” in Advances in botanical research, (Academic Press) 69, 195–222. doi: 10.1016/B978-0-12-417163-3.00008-1
Toriba, T., Harada, K., Takamura, A., Nakamura, H., Ichikawa, H., Suzaki, T., et al. (2007). Molecular characterization the YABBY gene family in oryza sativa and expression analysis of OsYABBY1. Mol. Genet. Genomics 277, 457–468. doi: 10.1007/s00438-006-0202-0
Tsai, W. C., Kuoh, C. S., Chuang, M. H., Chen, W. H., Chen, H. H. (2004). Four DEF-like MADS box genes displayed distinct floral morphogenetic roles in phalaenopsis orchid. Plant Cell Physiol. 45, 831–844. doi: 10.1093/pcp/pch095
Villanueva, J. M., Broadhvest, J., Hauser, B. A., Meister, R. J., Schneitz, K., Gasser, C. S. (1999). INNER NO OUTER regulates abaxial-adaxial patterning in arabidopsis ovules. Genes Dev. 13, 3160–3169. doi: 10.1101/gad.13.23.3160
Yamada, T., Ito, M., Kato, M. (2003). Expression pattern of INNER NO OUTER homologue in nymphaea (water lily family, nymphaeaceae). Dev. Genes Evol. 213, 510–513. doi: 10.1007/s00427-003-0350-8
Yamada, T., Yokota, S., Hirayama, Y., Imaichi, R., Kato, M., Gasser, C. S. (2011). Ancestral expression patterns and evolutionary diversification of YABBY genes in angiosperms. Plant J. 67, 26–36. doi: 10.1111/j.1365-313X.2011.04570.x
Yamaguchi, T., Nagasawa, N., Kawasaki, S., Matsuoka, M., Nagato, Y., Hirano, H. Y. (2004). The yabby gene drooping leaf regulates carpel specification and midrib development in oryza sativa. Plant Cell 16, 500–509. doi: 10.1105/tpc.018044
Yang, F., Gao, J., Wei, Y., Ren, R., Zhang, G., Lu, C., et al. (2021). The genome of cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 19 (12), 2501. doi: 10.1111/pbi.13676
Yin, S., Li, S., Gao, Y., Bartholomew, E. S., Wang, R., Yang, H., et al. (2022). Genome-wide identification of YABBY gene family in cucurbitaceae and expression analysis in cucumber (Cucumis sativus l.). Genes 13 (3), 467. doi: 10.3390/genes13030467
Zhang, T., Li, C., Li, D., Liu, Y., Yang, X. (2020). Roles of YABBY transcription factors in the modulation of morphogenesis, development, and phytohormone and stress responses in plants. J. Plant Res. 133, 751–763. doi: 10.1007/s10265-020-01227-7
Zhang, J., Li, Y., Liu, B., Wang, L., Zhang, L., Hu, J., et al. (2018). Characterization of the populus rab family genes and the function of PtRabE1b in salt tolerance. BMC Plant Biol. 18, 1–15. doi: 10.1186/s12870-018-1342-1
Keywords: YABBY genes, Orchidaceae, Cymbidium, expression pattern, genome-wide
Citation: Wang Q-Q, Li Y-Y, Chen J, Zhu M-J, Liu X, Zhou Z, Zhang D, Liu Z-J and Lan S (2022) Genome-wide identification of YABBY genes in three Cymbidium species and expression patterns in C. ensifolium (Orchidaceae). Front. Plant Sci. 13:995734. doi: 10.3389/fpls.2022.995734
Received: 16 July 2022; Accepted: 07 November 2022;
Published: 24 November 2022.
Edited by:
Katharina Nargar, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaCopyright © 2022 Wang, Li, Chen, Zhu, Liu, Zhou, Zhang, Liu and Lan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhong-Jian Liu, empsaXVAZmFmdS5lZHUuY24=; Siren Lan, bGt6eEBmYWZ1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.