
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 29 September 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.994314
This article is part of the Research Topic Women in Plant Pathogen Interactions: 2022 View all 8 articles
Plant viruses are obligate intracellular pathogens, and most depend on insect vectors for transmission between plants. Viral infection causes various physiological and metabolic changes in host traits, which subsequently influence the behavior and fitness of the insect vectors. Cucumber mosaic virus (CMV), one of the most widespread pathogens in pepper (Capsicum annuum L.), is transmitted by aphid vectors in a non-persistent manner. Here, we examined whether CMV infection in pepper affects the behavior of aphid vectors (Myzus persicae and Aphis glycines) in pepper. Aphid preference test revealed that significantly more aphids were attracted to CMV-infected pepper plants than to healthy plants. Comparative transcriptome analysis revealed a significant activation of the ethylene biosynthesis pathway in CMV-infected pepper plants. Indeed, gas chromatography analysis demonstrated that ethylene emission was significantly increased by CMV infection in pepper plants. Elevated ethylene emission in ethephon-treated healthy pepper increased their attractiveness to aphids. In contrast, aphid preference decreased after chemical inhibition of ethylene biosynthesis in CMV-infected pepper plants. Our results suggest that the ethylene emitted by CMV infection is a volatile cue that regulates the attractiveness of pepper plants to M. persicae and A. glycines.
Infection by pathogens induces a variety of physiological and metabolic changes in host plants, and these changes can subsequently influence the behavior and fitness of herbivorous insects that feed on these plants. Accumulating evidence suggests that pathogens can induce the release of volatile organic compounds (VOCs) from infected plants in ways that can either attract or repel insect herbivores (Eigenbrode et al., 2002; De Vos and Jander, 2010; Mauck et al., 2010; Mauck et al., 2014; Tungadi et al., 2017; Bak et al., 2019). Consequently, these host responses can strongly affect the transmission rates of insect-borne pathogens, with significant implications for epidemiology and agricultural ecology of insect-vectored diseases (Mauck et al., 2010; Mauck et al., 2014; Bak et al., 2019; Safari et al., 2019; Carr et al., 2020). However, little is still known about the biochemical and molecular mechanisms that mediate these interactions among pathogens, host plants, and insect herbivores.
Plant viruses are obligate intracellular pathogens that cause dramatic reprogramming of the transcriptome of host cells. This not only induces defense responses, but also disrupts normal plant development, physiology, and metabolism (Seo et al., 2018; Han et al., 2021). In nature, most plant viruses are transmitted between plants by insect vectors. Therefore, altering host plant traits in a manner that causes plants to attract insect vectors may be beneficial to the virus. Indeed, physiological and metabolic changes in virus-infected host plants have been shown to influence plant-insect vector interactions in ways that can increase virus transmission (Mauck et al., 2014; Tungadi et al., 2017; Bak et al., 2019). For example, changes in plant nutritional quality for vectors can enhance their performance and population growth (Mauck et al., 2014; Shikano et al., 2017). Viral infection can also increase the emission of plant VOCs that mediate vector attraction to plants (Mauck et al., 2010; Bak et al., 2019).
Aphid-transmitted potyviruses have been shown to enhance aphid performance on infected plants (Casteel et al., 2014; Adachi et al., 2018). Aphid vectors prefer to settle on host plants infected with potyviruses such as turnip mosaic virus and potato virus Y (PVY) and reproduce in larger numbers on infected host plants than on uninfected plants (Casteel et al., 2014; Bak et al., 2019). Potential mechanisms suggested for the enhanced aphid performance on potyvirus-infected plants include improvement of plant nutritional quality for aphids and suppression of plant defenses against aphids (Casteel et al., 2014; Adachi et al., 2018). Recently, PVY infection-induced ethylene signaling has been shown to be associated with aphid attraction and settlement on potato plants (Bak et al., 2019).
Cucumber mosaic virus (CMV; the type member of the genus Cucumovirus) is one of the most successful RNA viruses in terms of dispersion (Roossinck, 2002; Heo et al., 2020). CMV is a tripartite icosahedral virus transmitted by more than 70 species of aphids (Palukaitis and Garcia-Arenal, 2003). Aphids are phloem-feeding insects that transmit CMV in a non-persistent manner; therefore, CMV particles are retained in the aphid stylet for relatively short periods (a few minutes to a few hours) (Ng and Falk, 2006). In infected plants, CMV causes severe physiological and metabolic changes with diverse symptoms, including mosaic, stunting, leaf malformation, and chlorosis. Interestingly, CMV-infected plants show elevated emission of the VOC blend, which increases their attractiveness to aphid vectors (Mauck et al., 2010; Mauck et al., 2014). However, the specific VOC(s) responsible for manipulating aphid behavior have not been identified.
Pepper (Capsicum annuum L.) is an economically important vegetable crop worldwide, and CMV is one of the most widespread pathogens in this plant species (Kim et al., 2014a; Heo et al., 2020). Molecular investigations of CMV-pepper-aphid interactions can improve our understanding of the transmission ecology of CMV, which is non-persistently vectored by aphids in pepper. In the present study, we performed comparative transcriptome analysis to gain a molecular understanding of the physiological and metabolic changes in pepper plants in response to infection with CMV. Based on this approach, we observed a significant activation of the ethylene biosynthesis pathway in CMV-infected pepper plants. Ethylene is a volatile hormone associated with the susceptibility of various plant species to aphid infestation (Mantelin et al., 2009; Louis et al., 2015). Therefore, in this study, we also examined whether ethylene functions as a volatile cue mediating the attraction of aphid vectors to CMV-infected pepper plants.
Nicotiana benthamiana, pepper (Capsicum annuum L. cv. Sinhong), and soybean (Glycine max cv. Lee 74) plants were grown in an insect-free growth chamber with a cycle of 16 h of light at 26°C and 8 h of darkness at 24°C. Full-length infectious cDNA clones of the CMV GTN strain (CMV-GTN) were used as viral sources (Heo et al., 2020). Infectious cDNA clones of CMV-GTN were inoculated into leaves of two-weeks-old N. benthamiana plants by agroinfiltration, as previously described (Seo et al., 2009; Heo et al., 2020). Crude sap prepared from the upper symptomatic leaves of N. benthamiana infected with CMV-GTN was used for mechanical inoculation of two-weeks-old pepper plants (Heo et al., 2020). Non-viruliferous clones of a pepper-adapted strain of the aphid Myzus persicae and a soybean-adapted strain of the aphid Aphis glycines were reared on pepper plants (cv. Sinhong) and soybean plants (cv. Lee 74), respectively, in a growth chamber with a cycle of 16 h of light at 26°C and 8 h of darkness at 24°C.
Aphid preference for plant volatile cues was tested using a glass Y-tube olfactometer (diameter, 50 mm; length, 36 cm; Figure 1A) as described previously (Da Costa et al., 2011) with minor modifications. In brief, incoming air was filtered through activated charcoal and passed through two sample-containing chambers: one chamber contained a control sample, and the other contained a test sample. From each chamber, the air flowed into the respective arms of the Y-tube, and through the main tube of the olfactometer. Airflow through the olfactometer was maintained at 3.5 l/min using a gas flowmeter (RMB- 50D-SSV, Dwyer, USA). For each pairwise test, 100 wingless morphs of M. persicae or A. glycines were placed at the end of the main tube of the olfactometer and allowed to choose between two pepper samples. Tests were performed in the dark to eliminate any visual cues. The number of aphids that made a choice was counted at 3, 4, and 5 h after they were released, and the average was determined for each treatment in each test. Pairwise tests were performed to evaluate aphid preference for pepper plants treated as follows: (I) healthy vs. healthy, (II) mock-inoculated (mechanically inoculated with inoculation buffer) vs. CMV-infected, (III) healthy and mock-treated (water-sprayed) vs. healthy and ethephon-treated, (IV) CMV-infected and mock-treated vs. CMV-infected and aminoethoxyvinylglycine (AVG)-treated. Each pairwise test was repeated eight times, and the data were averaged.
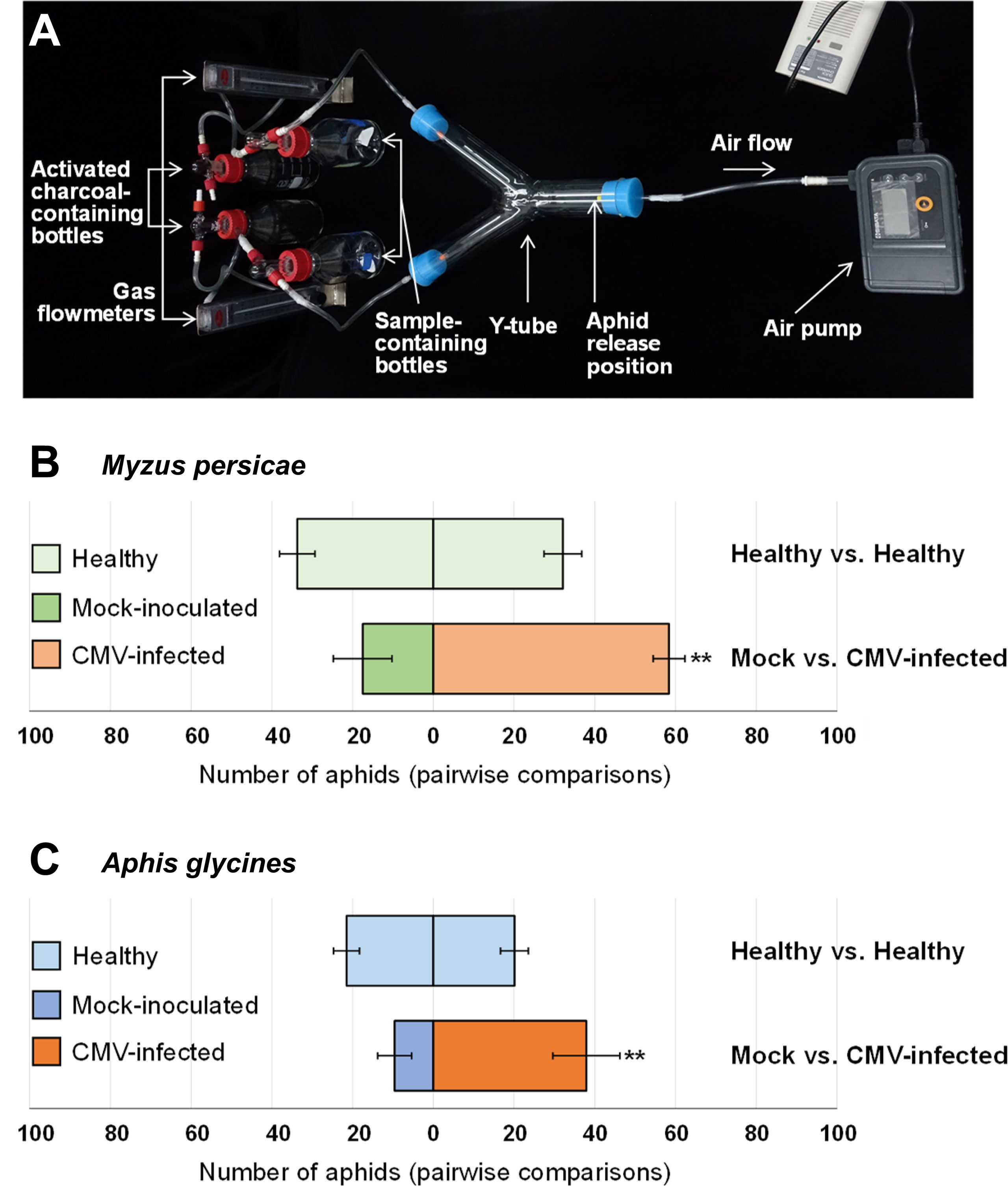
Figure 1 Aphid attraction to volatiles from healthy, mock-inoculated, or cucumber mosaic virus (CMV)-infected plants. (A) The glass Y-tube olfactometer used in this study for aphid preference tests. Pairwise preference tests for Myzus persicae (a species that colonizes pepper) (B) and Aphis glycines (a species that does not colonizes pepper) (C). In pairwise aphid preference tests, pepper plants were treated as follows: healthy vs. healthy; mock-inoculated vs. CMV-infected. Pairs involved in each preference test are shown horizontally. Each pairwise test was repeated eight times, and data were averaged and are presented as the mean ± SD. Significant differences were analyzed using paired Student’s t-test (**P < 0.01).
Fourteen days post-inoculation (dpi), symptomatic upper leaves were obtained from nine individual pepper plants infected with CMV-GTN. Leaf samples from three individual plants were pooled for RNA extraction. Thus, three biological replicates of RNA samples were obtained for each experimental group. Similarly, leaf samples from healthy plants were used as negative controls. Total RNA was isolated using PureLink® RNA Mini Kit (Ambion, USA) and used for library construction with the Illumina TruSeq RNA Sample Preparation Kit v2 (Illumina, Inc., USA). Nine libraries were generated and quantified with the KAPA library quantification kit (Kapa Biosystems, USA). Sequencing was performed using an Illumina HiSeq2000 sequencer (Illumina, Inc., USA) by TheragenEtex Inc. (South Korea).
RNA-seq data were analyzed as described previously (Han et al., 2021). Raw sequence reads were filtered using the Illumina pipeline and mapped to the reference transcripts of C. annuum cv. CM334 (Kim et al., 2014b) retrieved from Pepper Genome Platform (http://peppergenome.snu.ac.kr/). We used the RNA-seq mapping algorithm implemented in the bowtie2 software (v2.1.0) (Langmead et al., 2009) and allowed all alignments with a maximum of two mismatches. The raw data were deposited in the NCBI SRA database with BioProject accession numbers PRJNA751625 and PRJNA855293. To eliminate bias caused by variations in sequencing depth, the number of mapped clean reads for each gene was counted and normalized using the DESeq package in R software (Anders and Huber, 2010). Differentially expressed genes (DEGs) were identified based on by a ≥ two-fold change in read coverage and a false discovery rate (FDR) ≤ 0.01 in a binomial test. The FDR was applied to identify the threshold p-value for multiple tests and calculated using DESeq. Correlation analysis and hierarchical clustering were performed to categorize the genes according to their expression patterns using the AMAP library in R (Lucas, 2014).
Gene ontology (GO) analysis was used for functional annotation of the DEGs based on protein sequence similarity (e-value ≤ 1e-10) in the GO database (Ashburner et al., 2000). The in-house scripts of SEEDERS Inc. (Daejeon, South Korea) were used to count the number of DEGs assigned to each GO term. The PANTHER overrepresentation test was used to analyze GO term enrichment (Mi et al., 2013). The biological processes associated with enriched GO terms were visualized using the web-based REVIGO software (Supek et al., 2011), and the data were plotted in R. Functional enrichment analysis was performed to assign biological relevance to the gene network modules using agriGO v2.0 (Tian et al., 2017). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was based on sequence similarity (e-value ≤ 1e-10; identity ≥ 90) between pepper proteins in the KEGG database (Kanehisa and Goto, 2000).
To validate the RNA-seq results, the mRNA expression of the following genes was measured using qRT-PCR: ripening-related protein grip22 (CA.PGAv.1.6.scaffold631.48), glycine-rich protein (CA.PGAv.1.6.scaffold1405.6), AP2/ERF domain-containing transcription factor (CA.PGAv.1.6.scaffold291.8), basic-region leucine zipper transcription factor (CA.PGAv.1.6.scaffold588.80), n-methyltransferase 1-like (CA.PGAv.1.6.scaffold423.30), ACC oxidase-4-like (CA.PGAv.1.6.scaffold784.1), ACC oxidase-1-like (CA.PGAv.1.6.scaffold793.14), ACC synthase-2-like (CA.PGAv.1.6.scaffold630.30), ABC transporter B family member 11 (CA.PGAv.1.6.scaffold484.97), and glycine-rich protein 5-like (CA.PGAv.1.6.scaffold688.1). The ubiquitin2 gene (CA.PGAv.1.6.scaffold337.91) was used as an internal reference for the qRT-PCR experiments, as this gene showed stable expression pattern in the RNA sequencing results. The same RNA preparations (DNase-treated) used for RNA-seq were used for cDNA synthesis using a Superscript III kit (Invitrogen, USA) with oligo-dT primers. The resulting cDNA was analyzed using SYBR Green-based qRT-PCR with specific primers (listed in Supplementary Table S1) in an iCycler iQ5 qRT-PCR detection system (Bio-Rad, USA). Three biological and three technical replicates were used per sample. The qRT-PCR results were normalized with the reference gene ubiquitin2, expressed as log2 fold changes in CMV-infected samples relative to healthy samples, and compared with log2 fold change data acquired from the RNA-seq.
Gas chromatography (GC) analysis was performed to measure ethylene production by pepper plants as described previously (Hou et al., 2018; Heo et al., 2020), with minor modifications. Briefly, the upper leaves were collected from three plants, and samples weighing 5 g were placed in 400-ml glass jars for 5 h at 26°C in the dark. A 1 ml air sample of the jar headspace was collected and analyzed using a GC system equipped with a hydrogen flame ionization detector and an activated alumina column (Agilent Technologies, USA). The samples were compared with a standard of known concentration. Each experiment was repeated three times, and the data were averaged. Ethylene analysis was performed for four-weeks-old pepper plants treated as follows: (I) healthy, (II) CMV-infected, (III) healthy and mock-treated (water-sprayed), (IV) healthy and ethephon-treated, (V) CMV-infected and mock-treated, and (VI) CMV-infected and AVG-treated.
Pepper plants were treated with ethephon (Sigma-Aldrich Korea, Korea), which promotes ethylene release, or AVG (Sigma-Aldrich Korea, Korea), which inhibits ethylene biosynthesis. The treatments were performed as described previously (Bak et al., 2019), with minor modifications. To increase ethylene emission by plants, each four-weeks-old healthy pepper plant was sprayed homogeneously with 5 ml of 0.1 mg/l ethephon. Ethephon-treated plants were used in aphid preference tests and for ethylene analysis 2 h after the treatment. To inhibit ethylene release, 5 ml of 0.25 mM AVG was sprayed homogeneously on each four-weeks-old pepper plants inoculated with CMV-GTN 2 weeks previously. AVG-treated plants were used in aphid preference tests and for ethylene analysis 24 h after the treatment. Control plants (mock-treated) were sprayed with distilled water.
To examine whether CMV infection affects the attractiveness of host plants to aphid vectors, we performed pairwise preference tests using a Y-tube olfactometer. The tests were performed in the dark so that aphid vectors were exposed to volatile cues in the absence of visual cues. Significantly greater numbers of both M. persicae (a species that colonizes pepper) and A. glycines (a species that does not colonize pepper) preferred CMV-infected pepper plants instead of mock-inoculated plants (Figure 1). However, neither M. persicae nor A. glycines showed a preference between uninfected pepper plants (Figure 1). These results demonstrated that CMV infection in pepper plants resulted in increased release of volatile compounds to attract both colonizing and non-colonizing aphid vectors.
We next examined CMV-induced transcriptomic changes in pepper plants to identify host genes associated with the increased attractiveness to aphid vectors. Samples of total RNA isolated from healthy and CMV-infected pepper plants (Figure 2A) were used for cDNA library construction. Six cDNA libraries (three for each sample) were constructed and sequenced using an Illumina RNA-seq system. Each library yielded approximately 81–89 million clean paired-end reads (Supplementary Table S2) that were mapped to 35,884 reference transcripts of C. annuum cv. CM334 (Kim et al., 2014b). Approximately 92% of the nucleotides were mapped; the nucleotide coverage for healthy and CMV-infected samples was 175- and 192.33-fold, respectively (Supplementary Table S2).
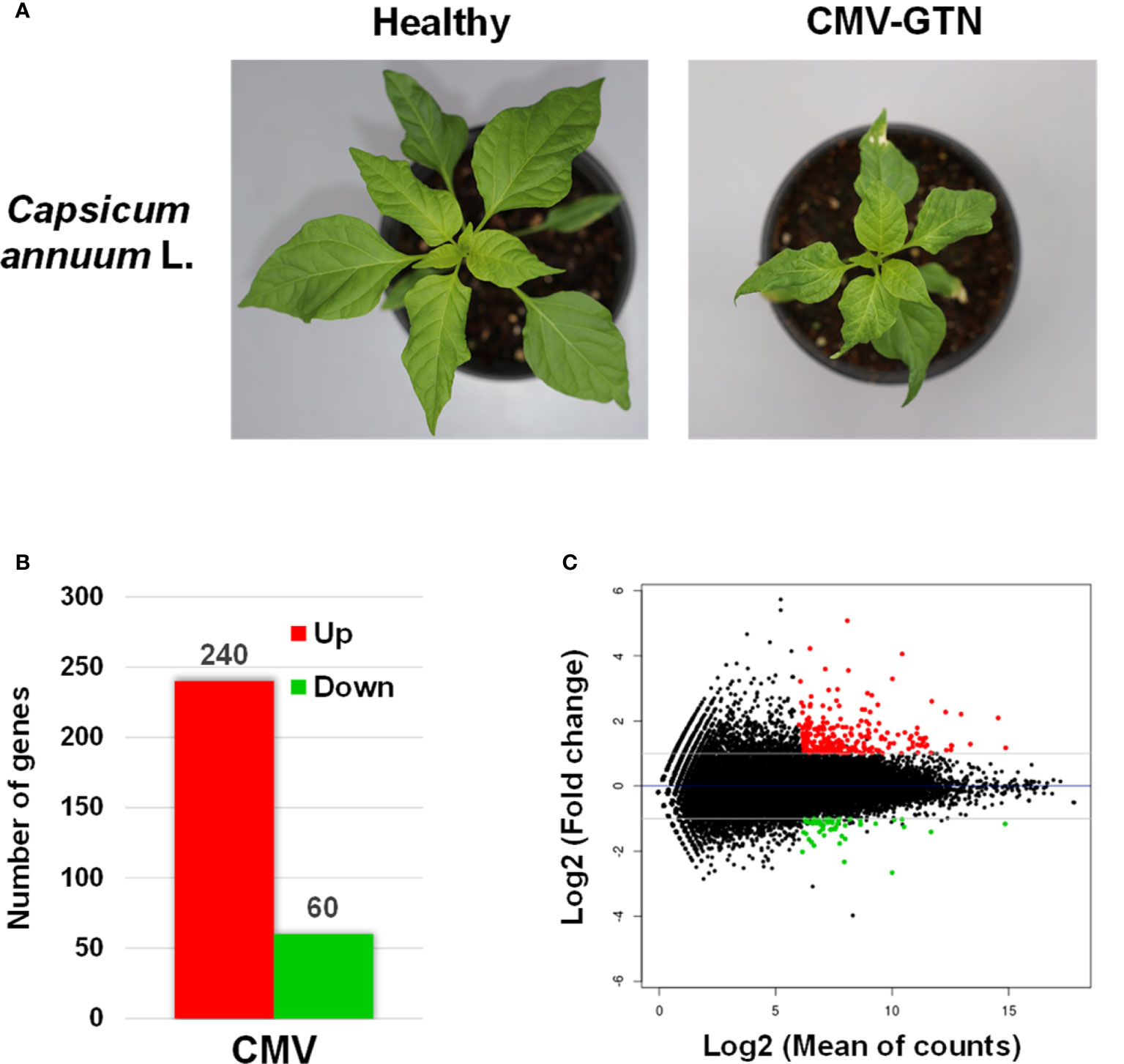
Figure 2 Transcriptomic reprogramming of pepper upon infection with CMV. (A) Phenotypes of healthy and CMV-infected pepper plants (Capsicum annuum L.) subjected to transcriptome analysis. Plants infected with CMV-GTN exhibited mosaic, severe leaf size reduction, and stunted growth. (B) Number of differentially expressed genes (DEGs) identified using RNA-seq of CMV-infected pepper plants. The DEGs were identified by comparing the transcriptomes of CMV-infected and healthy pepper plants (two-fold change in expression with a false discovery rate of ≤ 0.01). Red and green bar indicates the number of up- and downregulated DEGs, respectively. (C) MA-plot of RNA seq results. Each dot represents one gene. The normalized mean of read counts is shown on the x axis, and the log2 fold change of normalized counts is shown on the y axis. Up- and downregulated DEGs are represented by red and green dots, respectively.
To normalize the expression levels of the mapped genes, we used the fragments per kilobase of exon per million fragments mapped (FPKM) value. The FPKM values of the CMV-infected samples were compared with those of the healthy samples, and DEGs were identified based on a two-fold change in expression with FDR ≤ 0.01. We identified 300 DEGs (240 upregulated and 60 downregulated genes) in response to infection with CMV-GTN (Figure 2B, Tables 1, 2 and Supplementary Table S3). The top 30 DEGs upregulated upon infection with CMV included a few ethylene-responsive genes (e.g. CA.PGAv.1.6.scaffold631.48 and CA.PGAv.1.6.scaffold291.8) and various defense response-related genes (Table 1). The latter group included genes encoding proteins in the chitinase family (CA.PGAv.1.6.scaffold939.24, CA.PGAv.1.6.scaffold890.65, CA.PGAv.1.6.scaffold1306.1, CA.PGAv.1.6.scaffold13678.1, CA.PGAv.1.6.scaffold1611.1, CA.PGAv.1.6.scaffold1306.2, and CA.PGAv.1.6.scaffold1306.3) and receptor-like protein kinase genes (CA.PGAv.1.6.scaffold674.24, CA.PGAv.1.6.scaffold1110.29, and CA.PGAv.1.6.scaffold387.5) (Table 1). It was very Interesting that various chitinase family protein genes and ethylene-responsive genes were dramatically upregulated after infection with CMV (Table 1). To examine changes in global gene expression in CMV-infected samples, we visualized the magnitude distribution of DEG counts using an MA plot (Figure 2C). In general, fold changes were higher for upregulated DEGs than for down-regulated DEGs.
GO analysis was performed to examine how the up- and downregulated DEGs were associated with CMV infection-induced metabolic and physiological changes. A total of 98 GO terms (59 and 39 GO terms for up- and downregulated DEGs, respectively) were significantly enriched after infection with CMV (Supplementary Table S4). The enriched GO terms associated with the downregulated DEGs were related to regulation of cell division and included the following: cell cycle (GO:0007049), DNA replication (GO:0006260), cell division (GO:0051301), DNA conformation change (GO:0071103), and cell cycle process (GO:0022402) (Figures 3A, B). CMV infection severely reduced leaf size and caused stunted growth in pepper plants (Figure 2A). The morphology of plant organs is intricately regulated by cell division and expansion (Meyerowitz, 1997). Therefore, the symptoms induced by CMV may be directly associated with the suppression of genes that regulate cell division. The GO terms associated with the upregulated DEGs included those related to cellular responses to various stimuli: response to stimulus (GO:0050896), response to stress (GO:0006950), response to chemical (GO:0042221), and response to external stimulus (GO:0009605) (Figures 4A, B). Hierarchical GO enrichment analysis using agriGO v2.0 revealed that the enriched GO terms were highly correlated in a network context (Supplementary Figure S1).
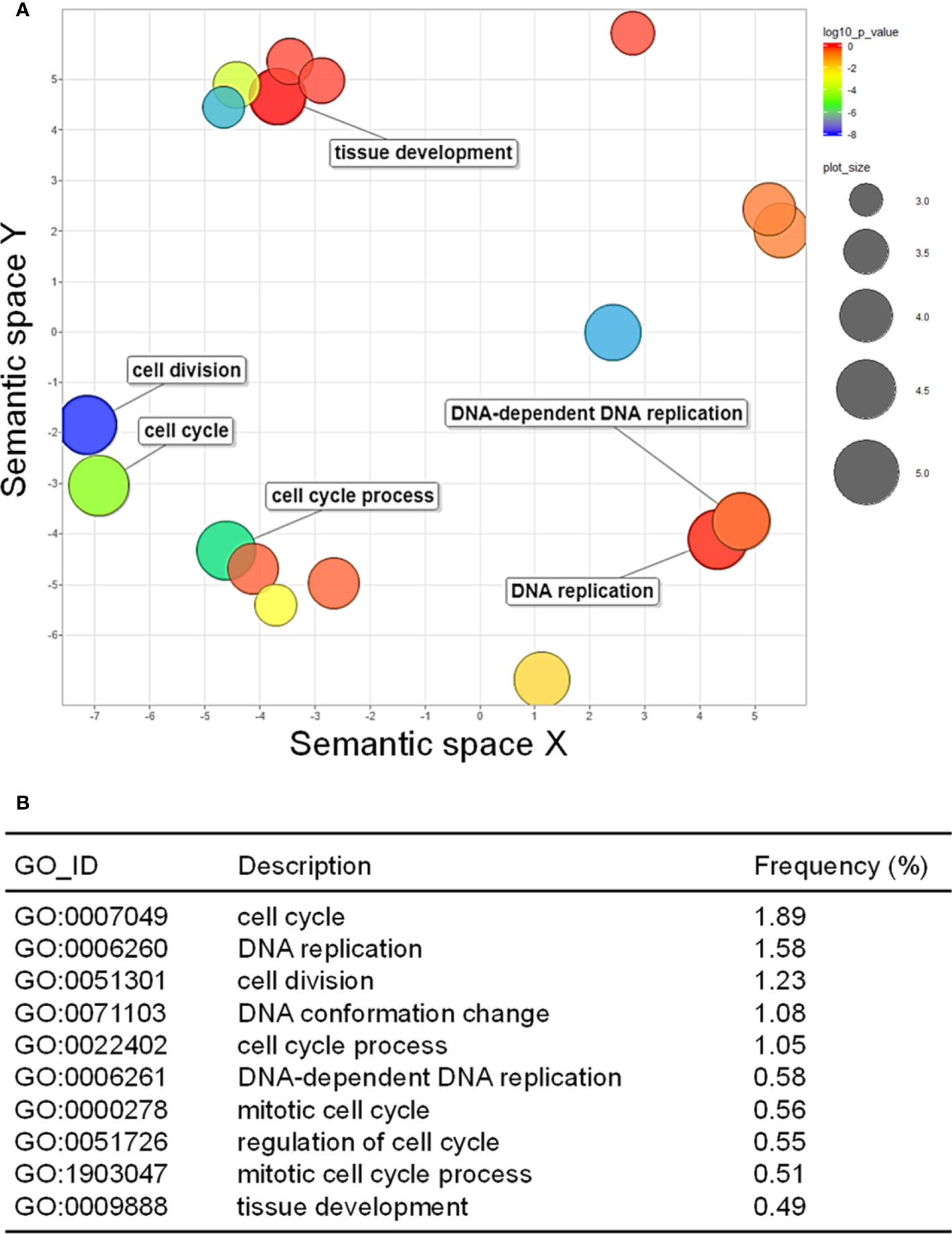
Figure 3 Gene ontology (GO) enrichment of downregulated differentially expressed genes (DEGs). (A) GO enrichment in the biological process category, as visualized using REVIGO. The scatter plot shows the significance of GO terms in a two-dimensional space. Multi-dimensional scaling was applied to a matrix of GO terms to calculate semantic similarities. Colors indicate the P-value for the false discovery rate, and circle sizes represent the frequency of the GO term. (B) Top 10 GO terms enriched for the downregulated DEGs in response to CMV infection.
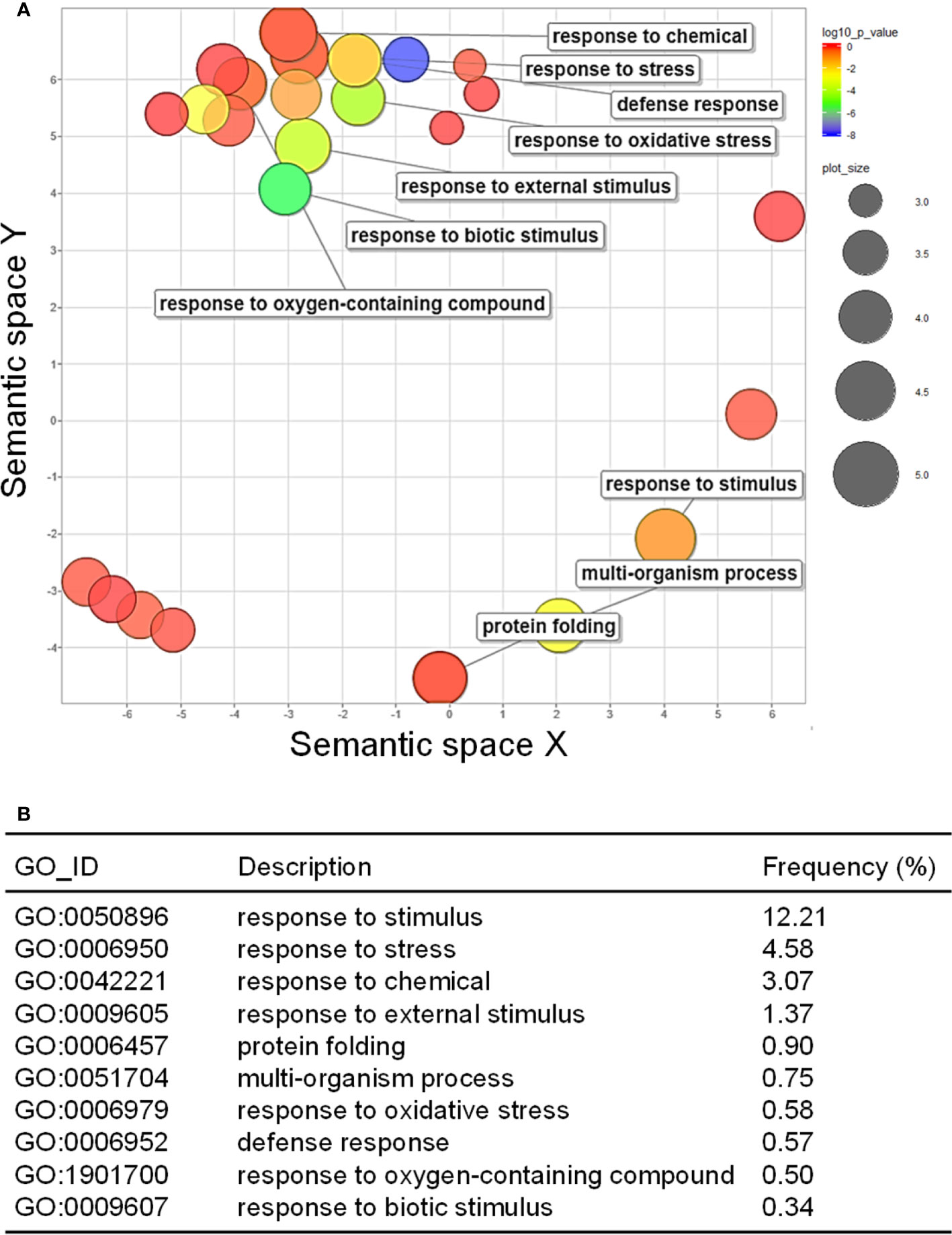
Figure 4 GO enrichment of upregulated DEGs. (A) GO enrichment in the biological process category, as visualized using REVIGO. The scatter plot shows the significance of GO terms in a two-dimensional space. Multi-dimensional scaling was applied to a matrix of GO terms to calculate semantic similarities. Colors indicate the P-value for the false discovery rate, and circle sizes represent the frequency of the GO term. (B) Top 10 GO terms enriched for the upregulated DEGs in response to CMV infection.
Next, KEGG pathway analysis was performed to examine the expression patterns of the DEGs in the associated pathways. KEGG analysis of the upregulated DEGs identified some highly ranked pathways, including plant hormone signal transduction (Pathway ID: cann04075), cysteine and methionine metabolism (Pathway ID: cann00270), and biosynthesis of secondary metabolites (Pathway ID: cann01110) (Table 3). It was interesting that CMV infection caused significant changes in important pathways related to metabolite biosynthesis, because we performed transcriptome analysis to identify the VOCs responsible for attracting aphids to plants. In particular, the cysteine and methionine metabolism pathway include the ethylene biosynthesis pathway, and we found that key genes involved in ethylene biosynthesis, including 1-aminocyclopropane-1-carboxylate (ACC) synthetase (ACS) and ACC oxidase (ACO), were significantly upregulated upon infection with CMV (Figure 5).
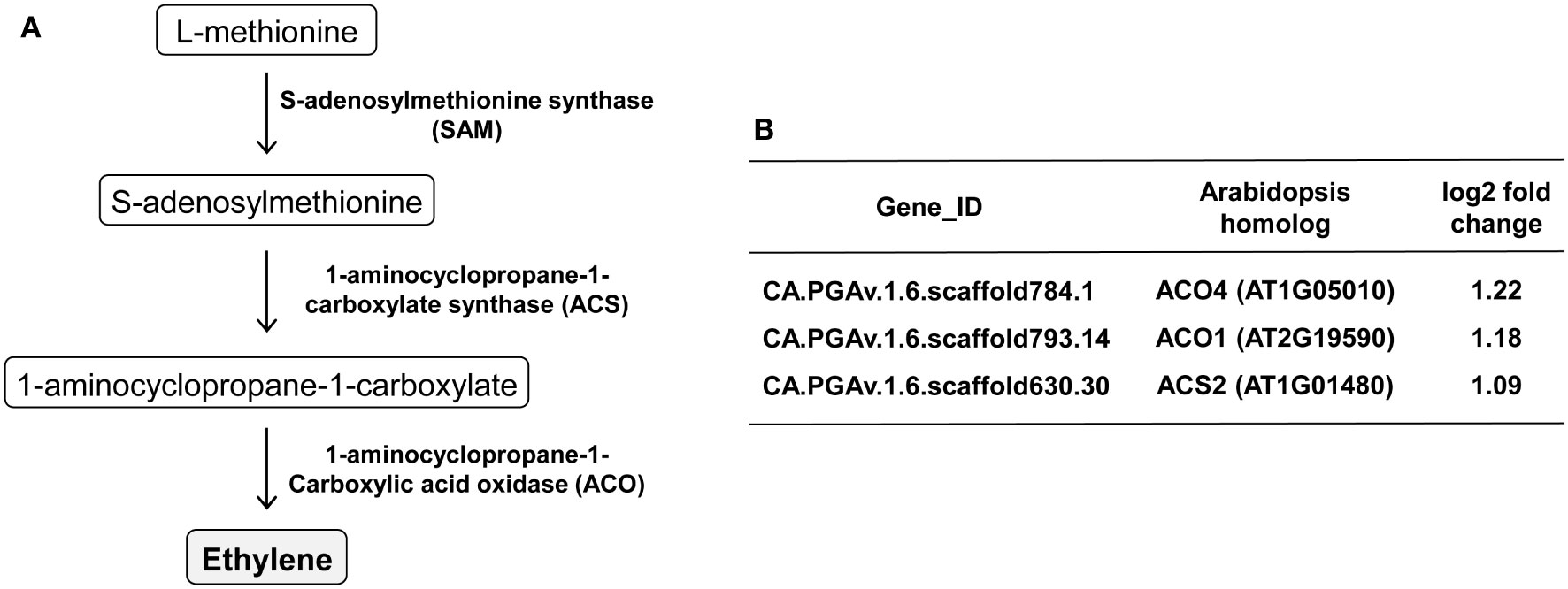
Figure 5 Regulation of genes involved in ethylene biosynthesis upon CMV infection in pepper. (A) The ethylene biosynthesis pathway and key enzymes. (B) Ethylene biosynthesis-associated genes upregulated in response to CMV infection. The changes in gene expression were calculated using a log scale of the FPKM data obtained using RNA-seq.
To validate the RNA-seq results, the expression of 10 representative DEGs was assessed by qRT-PCR using the same RNA preparations used for RNA-seq. The expression of these target genes was normalized to that of ubiquitin2 (CA.PGAv.1.6.scaffold337.91), which was used as an internal control. The relative expression of each gene was calculated and compared with the corresponding RNA-seq data. The qRT-PCR results of all evaluated genes were generally consistent with their respective RNA-seq results (Supplementary Table S5). This demonstrated the accuracy of gene expression data obtained using RNA-seq in this study.
Our transcriptomic analysis revealed that CMV infection activated the ethylene biosynthesis pathway in pepper. Therefore, we investigated whether ethylene production is indeed enhanced in CMV-infected pepper plants. GC analysis demonstrated that ethylene emission was considerably higher in CMV-infected leaves than in healthy leaves (Figure 6). This implied that CMV infection increased ethylene production in pepper, which was consistent with the transcriptional regulation of the ethylene biosynthesis pathway.
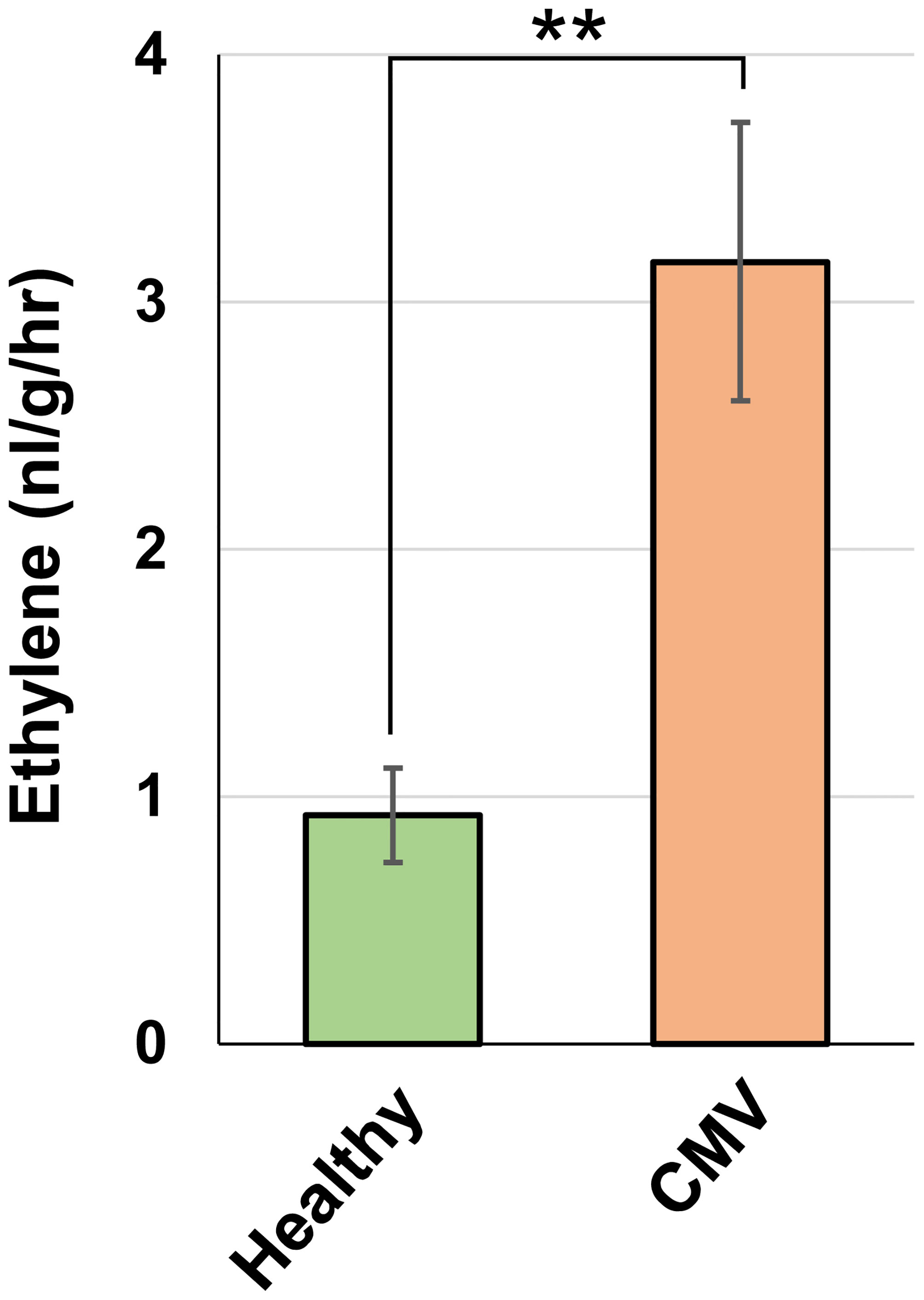
Figure 6 Effect of CMV infection on ethylene emission in pepper. Ethylene emission by upper uninoculated leaves of healthy and CMV-infected plants, as measured using gas chromatography. The mean ± SD values from three independent experiments are shown, and each column represents one group with nine plants. Significant differences were determined using paired Student’s t-test (**P < 0.01).
Ethylene is a volatile hormone, and its production was strongly upregulated in CMV-infected pepper plants. Therefore, we next investigated whether ethylene acts as a volatile cue to mediate aphid attraction to pepper plants. We utilized two chemical compounds: ethephon (a promoter of ethylene release) and AVG (an inhibitor of ethylene biosynthesis) (Bak et al., 2019). Ethephon treatment increased ethylene emission in healthy pepper plants (Figure 7A), and a greater number of M. persicae preferred ethephon-treated pepper plants over mock-treated plants (average 51.8 and 8.6, respectively; Figure 7C). Furthermore, AVG treatment inhibited ethylene emission in CMV-infected pepper plants (Figure 7B). The number of aphids that preferred AVG-treated pepper was less than half the number of those that preferred mock-treated pepper (average 20 and 46.3, respectively; Figure 7C). Therefore, our results suggested that ethylene emitted by pepper plants acts as a volatile cue responsible for attracting aphid vectors.
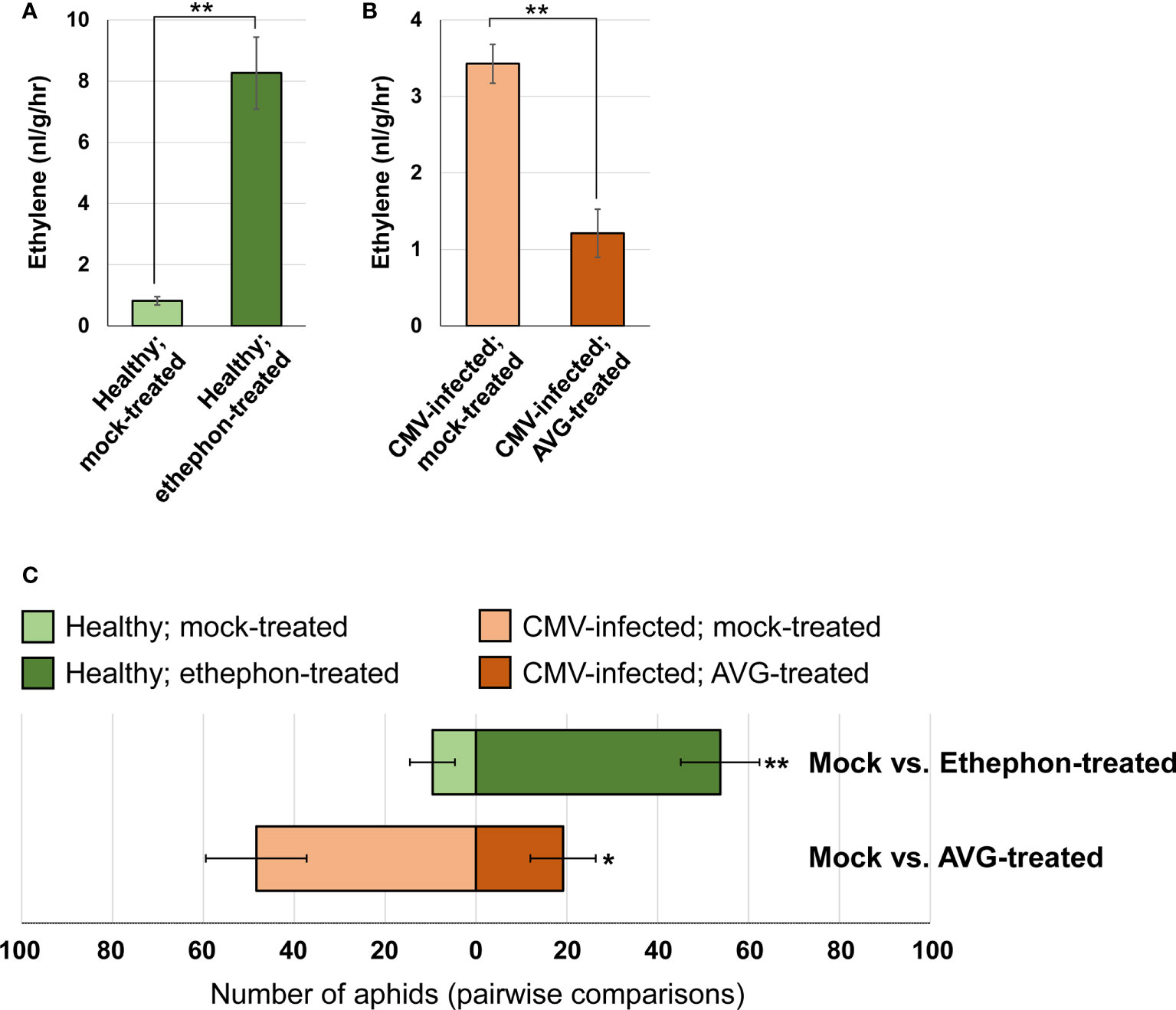
Figure 7 Effect of changes in ethylene emission on aphid attraction to pepper plants. (A) Ethylene emission by healthy pepper plants treated with mock or ethephon, as measured using gas chromatography. (B) Ethylene emission by CMV-infected pepper plants treated with mock or AVG, as measured using gas chromatography. The mean ± SD values from three independent experiments are shown, and each column represents one group with nine plants. Significant differences were determined using paired Student’s t-test (**P < 0.01). (C) Pairwise aphid preference tests between pepper plants treated as follows: healthy and mock-treated vs. healthy and ethephon-treated; CMV-infected and mock-treated vs. CMV-infected and AVG-treated. Pairs involved in each preference test are shown horizontally. Each pairwise test was repeated eight times, and data were averaged and are presented as the mean ± SD. Significant differences were determined using paired Student’s t-test (*P < 0.05; **P < 0.01).
Despite being a non-persistently transmitted virus, CMV has evolved into one of the most successful plant viruses in terms of worldwide dispersion (Palukaitis and Garcia-Arenal, 2003; Heo et al., 2020). The successful transmission of a virus requires that virus-infected plants attract insect vectors and stimulate them to acquire virus particles. Following this, the vectors must migrate to neighboring plants to transmit the virus within a short period. This pattern of transmission has been demonstrated in interactions between aphid vectors and squash (Cucurbita pepo) plants infected with CMV. In this system, the aphid vectors are preferentially attracted to CMV-infected squash plants but migrate to nearby plants soon after initial feeding (Mauck et al., 2010; Mauck et al., 2014). In this regard, previous studies found that CMV infection causes the following physiological and biochemical changes in host plants: (1) reduced plant nutritional quality for vectors; (2) induction of plant defense responses against aphid feeding; and (3) elevated emission of plant VOCs (Mauck et al., 2010; Mauck et al., 2014). These findings suggest that elucidating the molecular mechanisms underlying these virus-induced phenotypic and physiological changes in host plants is essential for better understanding of the interactions between host plants and insect vectors in relation to viral transmission.
Consistent with the aforementioned observations in squash plants (Mauck et al., 2010), we demonstrated that CMV infection increased the attractiveness of pepper plants to both colonizing and non-colonizing aphid vectors (Figure 1). We employed comparative transcriptome analysis to explore the molecular basis of physiological and metabolic changes in pepper plants in response to infection with CMV. Based on this approach, we identified several host genes associated with the increased attractiveness of CMV-infected plants to aphid vectors. Our analysis of DEGs and GO terms showed that many upregulated DEGs were associated with cellular responses to various stimuli, including defense responses (Figure 4; Supplementary Figure S1; Table 1). KEGG pathway analysis further highlighted that several DEGs were associated with biosynthesis of metabolites, especially ethylene (Figure 5; Table 3). The upregulation of key genes involved in ethylene biosynthesis, including ACS (CA.PGAv.1.6.scaffold630.30) and ACO(CA.PGAv.1.6.scaffold784.1 and CA.PGAv.1.6.scaffold793.14), resulted in elevated ethylene production in CMV-infected plants, indicating that CMV infection increases ethylene emission in pepper (Figure 6).
Ethylene is an important plant hormone that regulates many aspects of plant development, and plays a key role in modulating plant responses to biotic and abiotic stresses (Van Loon et al., 2006; Huang et al., 2016). Accumulating evidence suggests that virus-induced ethylene signaling is associated with the alteration of host plant susceptibility (Louis et al., 2015; Han et al., 2021). A recent study reported that in pepper plants infected with broad bean wilt virus 2 (BBWV2), elevated ethylene production enhances the severity of disease symptoms (Han et al., 2021). In rice, infection with rice dwarf virus (RDV) caused a dramatic increase in ethylene production, resulting in increased susceptibility of rice plants to RDV (Zhao et al., 2017). Interestingly, RDV infection increased the host preference of its insect vector, the green rice leafhopper (Wang et al., 2018), although the role of RDV-induced ethylene emission in causing this change has not been examined. A recent study demonstrated that PVY-induced ethylene mediates aphid attraction to infected potato plants, thus affecting virus spread (Bak et al., 2019). Several studies have also implicated virus-induced VOCs in vector attraction to infected plants (Eigenbrode et al., 2002; Ngumbi et al., 2007; Mauck et al., 2010; Mauck et al., 2014; Tungadi et al., 2017; Bak et al., 2019).
Ethylene is a volatile hormone, and its emission levels were higher in pepper plants infected with CMV (Figure 6). In this study, we showed that the elevated levels of ethylene emitted by ethephon-treated uninfected pepper plants increased their attractiveness to aphids (Figure 7). In contrast, the inhibition of ethylene biosynthesis in AVG-treated CMV-infected pepper plants reduced their attractiveness to aphids (Figure 7). Taken together, these results suggest that CMV-induced changes in ethylene production alters the attractiveness of pepper plants to aphid vectors. However, we cannot rule out the possibility that CMV-induced ethylene signaling affects the overall production of VOCs in pepper plants, which may be important for aphid attraction as well.
Rapid dispersal of insect vectors after initial feeding is important for successful transmission of non-persistently transmitted viruses (Mauck et al., 2010). Aphids show a strong tendency to leave CMV-infected squash plants after initial feeding (Mauck et al., 2010; Mauck et al., 2014), suggesting that CMV infection induces gustatory cues that stimulate aphids to disperse soon after feeding. A previous study in tobacco plants demonstrated that CMV infection induced plant defense signaling and H2O2 production. The authors suggested that CMV-induced H2O2 may function as a gustatory cue to modify aphid feeding behavior in ways that enhance its virus acquisition and aphid dispersal (Guo et al., 2019). Our transcriptome analysis revealed that several DEGs upregulated in response to CMV infection in pepper were associated with plant defense responses. These DEGs included gene encoding receptor-like protein kinases and pathogenesis-related proteins as well as genes involved in hormone signaling pathways (Table 1 and Figure 4). More interestingly, several genes encoding proteins in the chitinase family were also highly upregulated in CMV-infected pepper (Table 1). Chitin is a major component of the exoskeleton and alimentary canal of insects, whereas chitinases are hydrolytic enzymes that break down glycosidic bonds in chitin (Ding et al., 1998). Various insect chitinase genes have been used to generate transgenic plants resistant to insect pests (Ding et al., 1998; Schuler et al., 1998; Arakane and Muthukrishnan, 2010). Furthermore, the overexpression of plant chitinases enhanced the resistance of Arabidopsis and potato plants to aphids (Gatehouse et al., 1996; Zhong et al., 2021). Therefore, we suggest that the chitinase family proteins induced by CMV infection may affect plant resistance to aphids and act as a gustatory cue to stimulate post-feeding dispersal in insect vectors.
Vector-borne pathogens may have evolved to increase their transmission efficiency by altering host-vector-pathogen interactions to facilitate viral transmission (Mauck et al., 2016). Viral infection causes complex physiological and metabolic changes in host plants owing to transcriptomic reprogramming (Seo et al., 2018; Han et al., 2021). Our transcriptome analysis of CMV-pepper plant interactions revealed molecular genetic clues regarding the manipulated behaviors of aphid vectors feeding on CMV-infected pepper plants. CMV infection upregulated genes related to ethylene biosynthesis (Figure 5), and subsequent experiments (Figure 7) suggested that enhanced ethylene emission is responsible for aphid attraction in pepper. CMV infection also induced various genes encoding proteins in the chitinase family that may stimulate aphid dispersal (Table 1). Taken together, our findings improve our understanding of the molecular mechanisms underlying virus-induced manipulation of plant-insect interactions. Moreover, these results provide important insights into the evolutionary relationships among plant hosts, viruses, and insect vectors.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
S-JK and J-KS designed the experiments and supervised the project; S-JK, S-JH, M-HK, S-YJ, and J-SC performed the experiments; S-JK, S-JH, and J-KS analyzed the data; S-JK, S-JH, and J-KS wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported in part by grants from Basic Science Research Program (NRF-2020R1I1A1A01072564) funded by the National Research Foundation of Korea and Agenda Program (PJ015308) funded by the Rural Development Administration of Korea.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.994314/full#supplementary-material
Supplementary Table 1 | Primers used in this study for qRT-PCR.
Supplementary Table 2 | Summary statistics of sequences obtained from RNA-seq of pepper plants infected with CMV.
Supplementary Table 3 | DEGs in response to infection with CMV-GTN.
Supplementary Table 4 | GO terms for DEGs in response to infection with CMV-GTN.
Supplementary Table 5 | Validation of RNA sequencing data by qRT-PCR.
Supplementary Figure 1 | Hierarchical tree graph of overrepresented GO terms for upregulated DEGs in response to CMV infection.
Adachi, S., Honma, T., Yasaka, R., Ohshima, K., Tokuda, M. (2018). Effects of infection by turnip mosaic virus on the population growth of generalist and specialist aphid vectors on turnip plants. PloS One 13, e0200784. doi: 10.1371/journal.pone.0200784
Anders, S., Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. doi: 10.1186/gb-2010-11-10-r106
Arakane, Y., Muthukrishnan, S. (2010). Insect chitinase and chitinase-like proteins. Cell. Mol. Life Sci. 67, 201–216. doi: 10.1007/s00018-009-0161-9
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Bak, A., Patton, M. F., Perilla-Henao, L. M., Aegerter, B. J., Casteel, C. L. (2019). Ethylene signaling mediates potyvirus spread by aphid vectors. Oecologia 190, 139–148. doi: 10.1007/s00442-019-04405-0
Carr, J. P., Tungadi, T., Donnelly, R., Bravo-Cazar, A., Rhee, S. J., Watt, L. G., et al. (2020). Modelling and manipulation of aphid-mediated spread of non-persistently transmitted viruses. Virus Res. 277, 197845. doi: 10.1016/j.virusres.2019.197845
Casteel, C. L., Yang, C., Nanduri, A. C., De Jong, H. N., Whitham, S. A., Jander, G. (2014). The NIa-pro protein of turnip mosaic virus improves growth and reproduction of the aphid vector, myzus persicae (green peach aphid). Plant J. 77, 653–663. doi: 10.1111/tpj.12417
Da Costa, J. G., Pires, E. V., Riffel, A., Birkett, M. A., Bleicher, E., Sant’ana, A. E. G. (2011). Differential preference of capsicum spp. cultivars by aphis gossypii is conferred by variation in volatile semiochemistry. Euphytica 177, 299–307. doi: 10.1007/s10681-010-0250-8
De Vos, M., Jander, G. (2010). Volatile communication in plant-aphid interactions. Curr. Opin. Plant Biol. 13, 366–371. doi: 10.1016/j.pbi.2010.05.001
Ding, X., Gopalakrishnan, B., Johnson, L. B., White, F. F., Wang, X., Morgan, T. D., et al. (1998). Insect resistance of transgenic tobacco expressing an insect chitinase gene. Transgenic Res. 7, 77–84. doi: 10.1023/A:1008820507262
Eigenbrode, S. D., Ding, H., Shiel, P., Berger, P. H. (2002). Volatiles from potato plants infected with potato leafroll virus attract and arrest the virus vector, myzus persicae (Homoptera: Aphididae). Proc. Biol. Sci. 269, 455–460. doi: 10.1098/rspb.2001.1909
Gatehouse, A. M. R., Down, R. E., Powell, K. S., Sauvion, N., Rahbe, Y., Newell, C. A., et al. (1996). Transgenic potato plants with enhanced resistance to the peach-potato aphid myzus persicae. Entomol. Exp. Appl. 79, 295–307. doi: 10.1111/j.1570-7458.1996.tb00837.x
Guo, H., Gu, L., Liu, F., Chen, F., Ge, F., Sun, Y. (2019). Aphid-borne viral spread is enhanced by virus-induced accumulation of plant reactive oxygen species. Plant Physiol. 179, 143–155. doi: 10.1104/pp.18.00437
Han, S. J., Choi, B., Kim, M. H., Kwon, S. J., Kwak, H. R., Seo, J. K. (2021). Viral strain-specific activation of pathogen-associated molecular pattern-triggered immunity enhances symptom severity in broad bean wilt virus 2 infection. Front. Plant Sci. 12, 746543. doi: 10.3389/fpls.2021.746543
Heo, K. J., Kwon, S. J., Kim, M. K., Kwak, H. R., Han, S. J., Kwon, M. J., et al. (2020). Newly emerged resistance-breaking variants of cucumber mosaic virus represent ongoing host-interactive evolution of an RNA virus. Virus Evol. 6, veaa070. doi: 10.1093/ve/veaa070
Hou, B. Z., Li, C. L., Han, Y. Y., Shen, Y. Y. (2018). Characterization of the hot pepper (Capsicum frutescens) fruit ripening regulated by ethylene and ABA. BMC Plant Biol. 18, 162. doi: 10.1186/s12870-018-1377-3
Huang, P. Y., Catinot, J., Zimmerli, L. (2016). Ethylene response factors in arabidopsis immunity. J. Exp. Bot. 67, 1231–1241. doi: 10.1093/jxb/erv518
Kanehisa, M., Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kim, S., Park, M., Yeom, S. I., Kim, Y. M., Lee, J. M., Lee, H. A., et al. (2014b). Genome sequence of the hot pepper provides insights into the evolution of pungency in capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Kim, M. K., Seo, J. K., Kwak, H. R., Kim, J. S., Kim, K. H., Cha, B. J., et al. (2014a). Molecular genetic analysis of cucumber mosaic virus populations infecting pepper suggests unique patterns of evolution in Korea. Phytopathology 104, 993–1000. doi: 10.1094/PHYTO-10-13-0275-R
Langmead, B., Trapnell, C., Pop, M., Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. doi: 10.1186/gb-2009-10-3-r25
Louis, J., Basu, S., Varsani, S., Castano-Duque, L., Jiang, V., Williams, W. P., et al. (2015). Ethylene contributes to maize insect resistance1-mediated maize defense against the phloem sap-sucking corn leaf aphid. Plant Physiol. 169, 313–324. doi: 10.1104/pp.15.00958
Lucas, A. (2014) AMAP: Another multidimensional analysis package. Available at: http://CRAN.R-project.org/package=amap (Accessed 4 Apr 2016).
Mantelin, S., Bhattarai, K. K., Kaloshian, I. (2009). Ethylene contributes to potato aphid susceptibility in a compatible tomato host. New Phytol. 183, 444–456. doi: 10.1111/j.1469-8137.2009.02870.x
Mauck, K. E., De Moraes, C. M., Mescher, M. C. (2010). Deceptive chemical signals induced by a plant virus attract insect vectors to inferior hosts. Proc. Natl. Acad. Sci. U.S.A. 107, 3600–3605. doi: 10.1073/pnas.0907191107
Mauck, K. E., De Moraes, C. M., Mescher, M. C. (2014). Biochemical and physiological mechanisms underlying effects of cucumber mosaic virus on host-plant traits that mediate transmission by aphid vectors. Plant Cell Environ. 37, 1427–1439. doi: 10.1111/pce.12249
Mauck, K. E., De Moraes, C. M., Mescher, M. C. (2016). Effects of pathogens on sensory-mediated interactions between plants and insect vectors. Curr. Opin. Plant Biol. 32, 53–61. doi: 10.1016/j.pbi.2016.06.012
Meyerowitz, E. M. (1997). Genetic control of cell division patterns in developing plants. Cell 88, 299–308. doi: 10.1016/S0092-8674(00)81868-1
Mi, H., Muruganujan, A., Thomas, P. D. (2013). PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, 377–386. doi: 10.1093/nar/gks1118
Ng, J. C., Falk, B. W. (2006). Virus-vector interactions mediating nonpersistent and semipersistent transmission of plant viruses. Annu. Rev. Phytopathol. 44, 183–212. doi: 10.1146/annurev.phyto.44.070505.143325
Ngumbi, E., Eigenbrode, S. D., Bosque-Perez, N. A., Ding, H., Rodriguez, A. (2007). Myzus persicae is arrested more by blends than by individual compounds elevated in headspace of PLRV-infected potato. J. Chem. Ecol. 33, 1733–1747. doi: 10.1007/s10886-007-9340-z
Palukaitis, P., Garcia-Arenal, F. (2003). Cucumoviruses. Adv. Virus Res. 62, 241–323. doi: 10.1016/S0065-3527(03)62005-1
Roossinck, M. J. (2002). Evolutionary history of cucumber mosaic virus deduced by phylogenetic analyses. J. Virol. 76, 3382–3387. doi: 10.1128/JVI.76.7.3382-3387.2002
Safari, M., Ferrari, M. J., Roossinck, M. J. (2019). Manipulation of aphid behavior by a persistent plant virus. J. Virol. 93, e01781–e01718. doi: 10.1128/JVI.01781-18
Schuler, T. H., Poppy, G. M., Kerry, B. R., Denholm, I. (1998). Insect-resistant transgenic plants. Trends Biotechnol. 16, 168–175. doi: 10.1016/S0167-7799(97)01171-2
Seo, J. K., Kim, M. K., Kwak, H. R., Choi, H. S., Nam, M., Choe, J., et al. (2018). Molecular dissection of distinct symptoms induced by tomato chlorosis virus and tomato yellow leaf curl virus based on comparative transcriptome analysis. Virology 516, 1–20. doi: 10.1016/j.virol.2018.01.001
Seo, J. K., Kwon, S. J., Choi, H. S., Kim, K. H. (2009). Evidence for alternate states of cucumber mosaic virus replicase assembly in positive- and negative-strand RNA synthesis. Virology 383, 248–260. doi: 10.1016/j.virol.2008.10.033
Shikano, I., Rosa, C., Tan, C. W., Felton, G. W. (2017). Tritrophic interactions: Microbe-mediated plant effects on insect herbivores. Annu. Rev. Phytopathol. 55, 313–331. doi: 10.1146/annurev-phyto-080516-035319
Supek, F., Bosnjak, M., Skunca, N., Smuc, T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PloS One 6, e21800. doi: 10.1371/journal.pone.0021800
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., et al. (2017). agriGO v2.0: a GO analysis toolkit for the agricultural community 2017 Update. Nucleic Acids Res. 45, 122–129. doi: 10.1093/nar/gkx382
Tungadi, T., Groen, S. C., Murphy, A. M., Pate, A. E., Iqbal, J., Bruce, T. J. A., et al. (2017). Cucumber mosaic virus and its 2b protein alter emission of host volatile organic compounds but not aphid vector settling in tobacco. Virol. J. 14, 91. doi: 10.1186/s12985-017-0754-0
Van Loon, L. C., Geraats, B. P., Linthorst, H. J. (2006). Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 11, 184–191. doi: 10.1016/j.tplants.2006.02.005
Wang, Q., Li, J., Dang, C., Chang, X., Fang, Q., Stanley, D., et al. (2018). Rice dwarf virus infection alters green rice leafhopper host preference and feeding behavior. PloS One 13, e0203364. doi: 10.1371/journal.pone.0203364
Zhao, S., Hong, W., Wu, J., Wang, Y., Ji, S., Zhu, S., et al. (2017). A viral protein promotes host SAMS1 activity and ethylene production for the benefit of virus infection. Elife 6, e27529. doi: 10.7554/eLife.27529.036
Keywords: cucumber mosaic virus, vector attraction, volatile cue, virus transmission, Myzus persicae
Citation: Kwon S-J, Han S-J, Kim M-H, Jang S-Y, Choi J-S and Seo J-K (2022) Ethylene emitted by viral pathogen-infected pepper (Capsicum annuum L.) plants is a volatile chemical cue that attracts aphid vectors. Front. Plant Sci. 13:994314. doi: 10.3389/fpls.2022.994314
Received: 14 July 2022; Accepted: 07 September 2022;
Published: 29 September 2022.
Edited by:
Špela Baebler, National Institute of Biology (NIB), SloveniaReviewed by:
Xiaobin Shi, Hunan Academy of Agricultural Sciences, ChinaCopyright © 2022 Kwon, Han, Kim, Jang, Choi and Seo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jang-Kyun Seo, amFuZ3Nlb0BzbnUuYWMua3I=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.