- 1Wageningen University, Laboratory of Phytopathology, Wageningen, Netherlands
- 2Wageningen University, Laboratory of Biochemistry, Wageningen, Netherlands
Fungal plant pathogens secrete proteins that manipulate the host in order to facilitate colonization. Necrotrophs have evolved specialized proteins that actively induce plant cell death by co-opting the programmed cell death machinery of the host. Besides the broad host range pathogen Botrytis cinerea, most other species within the genus Botrytis are restricted to a single host species or a group of closely related hosts. Here, we focused on Botrytis squamosa and B. elliptica, host specific pathogens of onion (Allium cepa) and lily (Lilium spp.), respectively. Despite their occurrence on different hosts, the two fungal species are each other’s closest relatives. Therefore, we hypothesize that they share a considerable number of proteins to induce cell death on their respective hosts. In this study, we first confirmed the host-specificity of B. squamosa and B. elliptica. Then we sequenced and assembled high quality genomes. The alignment of these two genomes revealed a high level of synteny with few balanced structural chromosomal arrangements. To assess the cell death-inducing capacity of their secreted proteins, we produced culture filtrates of B. squamosa and B. elliptica that induced cell death responses upon infiltration in host leaves. Protein composition of the culture filtrate was analysed by mass spectrometry, and we identified orthologous proteins that were present in both samples. Subsequently, the expression of the corresponding genes during host infection was compared. RNAseq analysis showed that the majority of the orthogroups of the two sister species display similar expression patterns during infection of their respective host. The analysis of cell death-inducing proteins of B. squamosa and B. elliptica provides insights in the mechanisms used by these two Botrytis species to infect their respective hosts.
Introduction
In the battle for survival, plant pathogenic fungi have evolved different strategies to colonize their hosts. While biotrophic fungi obtain nutrients from living host cells via specialized feeding structures, necrotrophs kill the cells of their host to acquire nutrients. For a long time, fungi with a necrotrophic lifestyle have been considered to invade their hosts in a rather unsophisticated manner. Using lytic and degradative enzymes, the fungus first kills and subsequently grows into host cells and eventually colonizes host tissue. In the past decades however, evidence has been accumulating that the interaction between necrotrophs and their hosts is more subtle and sophisticated than previously appreciated. Necrotrophs actively induce plant cell death by co-opting the programmed cell death machinery of the host using specialized effector proteins, to a certain extent similar to biotrophs (Dickman et al., 2001; van Kan, 2006). The induction of plant cell death using proteinaceous effectors is vital for the success of infection (Tan et al., 2010; Stergiopoulos et al., 2013; Veloso and van Kan, 2018). For some necrotrophic effectors the corresponding plant receptor proteins required for effector recognition and triggering of the cell death response have been identified (Schürch et al., 2004; Abeysekara et al., 2009; Gao et al., 2015; Shi et al., 2016). The cell death response to effectors of necrotrophs is based on similar mechanisms as the recognition of effectors from biotrophic pathogens, but leads to disease susceptibility instead of disease resistance as is the case for biotrophs (Lorang et al., 2007; Tan et al., 2010).
The role of effectors of necrotrophs in plant-pathogen interactions is best studied in Septoria nodorum blotch in wheat, caused by the Ascomycete Parastagonospora nodorum. The first identified effector from P. nodorum was SnTox1, a small secreted protein that induced necrosis upon infiltration in wheat lines carrying the sensitivity gene Snn1 (Liu et al., 2004). SnTox1 knockout mutants lost virulence on Snn1-carrying wheat lines, and introduction of SnTox1 into an avirulent P. nodorum isolate conferred pathogenicity on wheat lines harbouring Snn1 (Liu et al., 2012). By genetic analysis, more P. nodorum effectors and corresponding wheat sensitivity genes were identified such as SnToxA-Tsn1, SnTox4-Snn4, and SnTox6-Snn6 (Abeysekara et al., 2009; Faris et al., 2010; Tan et al., 2012; Gao et al., 2015). To date, nine necrotrophic effectors and corresponding host sensitivity loci have been identified in the P. nodorum-wheat pathosystem (Duba et al., 2018; Cowger et al., 2020). The severity of infection depends in a quantitative manner on the number and identity of pathogen effectors and corresponding wheat sensitivity genes (Oliver et al., 2012; Phan et al., 2016; Ruud et al., 2017; Haugrud et al., 2019).
The broad host range model necrotroph Botrytis cinerea is also known to secrete proteins that induce cell death upon recognition by the host. B. cinerea endopolygalacturonases (PGs) induce cell death upon infiltration in Arabidopsis thaliana, but only in lines harbouring leucine-rich repeat (LRR) receptor-like protein RBPG1 (Zhang et al., 2014). Other proteinaceous virulence factors that function as cell death-inducing effectors in B. cinerea are BcXyl11a, BcXyl1 and BcXYG1, hemicellulases that induce necrosis in plant tissue independent from their catalytic activity (Noda et al., 2010; Zhu et al., 2017; Yang et al., 2018). Xylanases are recognized by the LRR receptor-like proteins LeEix1 and LeEix2 and the cell death response to BcXyl1 and BcXYG1 requires the LRR receptor-like kinases BAK1 and SOBIR1 (Ron and Avni, 2004; Zhu et al., 2017; Yang et al., 2018). In addition to the cell wall degrading enzymes with cell death-inducing activity, often considered as PAMPs or catalytic necrosis-inducing proteins (NIPs), B. cinerea also secretes cell death-inducing proteins that lack a known enzymatic domain and are referred to as effectors. Amongst others BcNep1, BcNep2, BcSpl1 and BcIEB1 have been heterologously produced and induced cell death upon infiltration in plant leaf tissue (Arenas et al., 2010; Frías et al., 2011; Frías et al., 2016).
The genus Botrytis comprises ~40 described species of which a few, including B. cinerea, are generalist plant pathogens with very broad host ranges (Hyde et al., 2014; Garfinkel et al., 2017). Most Botrytis species, however, are considered host-specific since they are pathogenic on a single host or a few taxonomically related hosts. An inventory by Mercier et al., 2019 of polyphagy indices of Botrytis species indicated that B. elliptica (commonly considered to be a pathogen specifically to lily and known as “lily fire blight”) was reported to infect as many as 21 host species from 10 distinct genera, and has a polyphagy index of 7.1. By contrast, its close relatives B. deweyae, B. sinoalli and B. squamosa, that cluster with B. elliptica in a subclade of the phylogenetic tree of the genus Botrytis (Valero-Jimenéz et al., 2020), have polyphagy indices below 1.5 (Mercier et al., 2019). In this study we focussed on B. elliptica and B. squamosa, two closely related sister species that are reported to cause disease on lily and onion, respectively. For both species, induction of programmed cell death is a key step in the infection process and it is achieved by means of secreted proteinaceous effectors (Van Baarlen et al., 2004; Malvestiti et al., 2021; Steentjes et al., 2022). We aimed to analyse whether the shared secreted proteins between these two closely related species can contribute to cell death induction in their respective host and non-host plant. We verified the host specificity and sequenced and assembled high quality genomes of B. squamosa and B. elliptica and analysed their synteny. We compared the cell death inducing capacity of the culture filtrates of B. squamosa and B. elliptica, and analysed the protein composition of these culture filtrates and the expression of the corresponding genes during host infection.
Materials and methods
Plant material and growth conditions
Bulbs of Lilium spp. cultivar “Asiatic” were planted in plastic crates containing potting soil and grown in a greenhouse under natural day light at a minimum night temperature between 12 and 15°C and maximum day temperature between 24 and 26°C. Mature leaves as described by Bar and Ori (2014) were harvested before the flowering stage and used for disease and infiltration assays.
Nicotiana benthamiana plants were grown in potting soil in a greenhouse with 16 h light at 24°C and 8 hours darkness at 22°C and 75% relative humidity. Both disease and infiltration assays were carried out on leaves that were still attached to the plant. Allium cepa (cv Ceresco F1) plants were grown from seeds in potting soil in a climate chamber with 12h light, 70% relative humidity, 18°C day temperature and 16°C night temperature. Fully grown leaves of 8 to 10 weeks-old onion plants were used for inoculation and infiltration assays. Disease assays were conducted on cut onion leaves, while for infiltration assays the leaves remained attached to the plant.
Fungal material and growth conditions
Botrytis squamosa isolate MUCL31421 (Steentjes et al., 2021), Botrytis elliptica isolate 9401 (Malvestiti et al., 2021) and Botrytis cinerea isolate B05.10 (Van Kan et al., 2017) used in this research were stored as conidia suspensions in 20% glycerol at -80°C. To obtain conidia for inoculation, B. elliptica and B. cinerea were grown on Malt Extract Agar (50 g/L, Oxoid), whereas B. squamosa was grown on autoclaved onion leaves on top of water agar (Oxoid). Sporulation was induced by illumination with UV-A lamps. After harvesting, the conidia were collected and washed in demineralized water, counted with the Bürker-Türk counting chamber, adjusted to a concentration of 106 conidia/mL and stored until use in darkness at 4°C.
Disease assays
Freshly harvested conidia of B. squamosa, B. elliptica and B. cinerea were inoculated in 12 g/L Potato Dextrose Broth (Difco) in 2 μL droplets at a spore concentration of 105 conidia/mL. Detached lily and onion leaves were placed on petri dishes in wet plastic boxes. Three lily leaves were abaxially inoculated with each fungus with four droplets per leaf avoiding leaf veins and leaf edges. Three detached onion leaves were first gently wiped with clean tissue paper to remove the epicuticular layer and then inoculated each with four droplets per fungus. Two N. benthamiana plants were kept in the pot and placed in boxes containing water. Three leaves per plant were adaxially inoculated with each fungus by applying four droplets on distinct sectors of the leaves, avoiding leaf veins and edges. Each plant-fungus combination was inoculated in a separate box. All boxes were closed with lids to maintain humid conditions and sealed with paper tape. The boxes were kept at room temperature under ambient light. After four days of incubation inoculated leaves were photographed and lesions were measured with a caliper. Lesion diameters were plotted using ggplot2 in R (Version 4.0.2).
DNA extraction, genome assembly and annotation
High molecular weight genomic DNA for Nanopore sequencing was extracted from in vitro grown mycelium as described in Valero-Jiménez et al. (2020). Both genomes of B. elliptica and B. squamosa were assembled with Oxford Nanopore reads. The adapters of the reads were firstly trimmed with porechop, version 0.2.4 (https://github.com/rrwick/Porechop). NECAT (Chen et al., 2021) was then launched for genome assembly, with genome sizes of 48 Mb and 55 Mb for B. elliptica and B. squamosa respectively and other parameters were set as default. After the assembly, we polished the B. elliptica genome with Illumina short reads using Pilon (Walker et al., 2014) and the B. squamosa genome by aligning with the previously published genome assembly (Valero-Jiménez et al., 2020). Subsequently, the two genomes were aligned with MUMer 3.07 (Kurtz et al., 2004) using PROmer (-l 150, -c 500, -g 100, -b 200). The genome of B. elliptica was annotated using Funannotate (https://funannotate.readthedocs.io). The previously predicted secretome of B. elliptica strain 9612 was subsequently mapped to the new genome assembly and the missing secretome genes in the new assembly were manually added. The previously published annotation of B. squamosa of the same strain was mapped to the new assembly.
Production of culture filtrate, ammonium sulfate precipitation and leaf infiltration
B. squamosa and B. elliptica Culture Filtrate (CF) samples were obtained by growing each fungus in a 250 mL flask containing 50 mL of liquid medium with 3 g/L Gamborg B5 salts (Duchefa, Haarlem, Netherlands), 10 mM potassium phosphate pH=6.0, 0.1% D-Glucose and 5mL of lily or onion leaf extract in demi water, respectively. The leaf extract was obtained by grinding 30g of fresh harvested leaf material in 250 mL demineralized water. The homogenate was centrifuged (3500 rpm, 20 min) and the supernatant was concentrated by freeze drying. The concentrate was redissolved in 10mL water, filter-sterilized and added to the liquid medium. The liquid culture was inoculated with a spore concentration of 105 conidia/mL medium. The cultures were incubated at 140 rpm, at 20°C in the dark. After 5 days of growth, the CF was passed through a layer of Miracloth (Calbiochem, San Diego, CA, United States), filter-sterilized (0.45 μm pore size, Millipore, Amsterdam, Netherlands) and kept on ice. A mock liquid medium was prepared without fungal conidia. Of the ca 40 mL harvested CF samples, 2mL were stored at 0°C for one night. To the remaining volume ammonium sulfate (LabChem, Tiel, Netherlands) was gradually added and mixed on a rolling bench at 4°C until the CFs were saturated with salt. Samples were centrifuged 30min at 4°C (4000rpm). After centrifugation a solid pellet of precipitated compounds was collected. The pellet was redissolved in 10mM potassium phosphate pH=6.0 and transferred to a prewetted dialysis membrane (Spectra/Por 3 Dialysis Membrane 3.5 kD MWCO, Repligen), and dialyzed overnight at 4°C in 5 L 10mM potassium phosphate pH=6.0. The dialyzed samples were collected and stored at 0°C until infiltration. The same plant genotypes used in the disease assays were tested for the infiltration assays. For lily, ca 100 μL of each sample (crude CFs, dialyzed pellets and mock liquid media) was infiltrated with a 1 mL syringe in two spots of two different detached leaves on their abaxial side. The leaves were placed in moist plastic boxes and incubated at 20°C under ambient light. The same volume of sample was infiltrated in two spots on two onion and N. benthamiana leaves whereby onion and N. benthamiana leaves were not detached from the plant. After 3 days incubation all leaves were photographed and the response was evaluated using red light imaging (Villanueva et al., 2021).
Proteomic analysis
B. elliptica and B. squamosa proteins that were analyzed with mass spectrometry were sampled from the liquid culture as described in the above section. Proteins were prepared for mass spectrometry as described in Supplementary Information 1. The samples were subjected to LC–MS/MS according to the parameters described in (Xiong et al., 2020). For protein identification and quantification, each run with all MS/MS spectra obtained was analyzed with Maxquant 2.0.3.0 with the Andromeda search engine. The B. squamosa and B. elliptica annotated genomes were used for protein mapping. A maximum of two missed cleavages and a mass deviation of 20 ppm for the fragment MS/MS peaks were allowed. The false discovery rate (FDR) was set to 1% on both peptide and protein level. The length of peptides was set to at least seven amino acids. Protein identification required minimally two distinct peptides of which at least one unique and at least one unmodified. Intensity based absolute quantification (iBAQ) algorithm was calculated as the sum of all peptide intensities divided by the number of theoretically observable tryptic peptides.
RNA extraction and gene expression analysis
For B. elliptica, RNAs used to create RNAseq library and to analyze gene expression profiles were isolated from 5 days old mycelial tissue of B. elliptica 9401 grown in vitro on MAE and from lily leaf tissue of cultivar “Asiatic” inoculated with conidia suspension of B. elliptica 9401 harvested at 16, 24 and 40 hours post inoculation (hpi), with three replicate samples per timepoint. RNA extraction was carried out from 100 mg freeze-dried, grinded material using the RNA extraction protocol of Maxwell® RSC Plant RNA Kit (Product AS 1500, Promega). For B. squamosa RNAs were extracted from 5 days old mycelial tissue grown on MAE supplemented with onion extract and from onion leaf tissue inoculated with B. squamosa isolate MUCL31421 at 16, 24 and 48 hpi with three replicate samples per timepoint. 100 mg of freeze-dried grinded material was incubated with Trizol (Ambion, Life Technology) and treated with chloroform. Technical EtOH (Sigma Aldrich) was added to the aqueous phase and the mixture was used as input for RNeasy Plant Mini Kit (Qiagen) for RNA extraction. RNAs of both Botrytis species were sequenced at Beijing Genomic Institute (BGI), Shenzhen, China. Mapping and quantifying gene transcripts from sequenced RNA-seq reads of both B. elliptica and B. squamosa were performed using Kallisto (Bray et al., 2016). The +1 log10 TPM value was calculated for plotting the expression heatmap.
Results
Host specificity of Botrytis spp.
B. squamosa and B. elliptica are reported to be host-specific pathogens of onion and lily, respectively. To assess this host specificity under laboratory conditions, we tested the virulence of B. squamosa and B. elliptica on their hosts and non-host plants and compared them to the broad host range pathogen B. cinerea (Figure 1). All three species were inoculated on onion, lily and N. benthamiana leaves and lesion development was monitored. The generalist B. cinerea was able to develop expanding lesions exclusively when inoculated on N. benthamiana leaves (Figure 1A) whereas the lesions on lily and onion leaves remained limited to the size of the inoculation droplet. Lesion expansion and disease progression were observed when B. squamosa was inoculated on onion leaves (Figure 1B) and when B. elliptica was inoculated on lily leaves (Figure 1C). In contrast, when B. squamosa and B. elliptica were cross-inoculated on lily and onion, respectively, the primary necrotic lesions did not expand over time and except for a few cases, they remained limited to the size of the inoculation droplet. In addition, inoculation of B. squamosa and B. elliptica on N. benthamiana leaves did not cause formation of expanding lesions. In conclusion, B. squamosa and B. elliptica caused expanding lesions on their reported natural host but not on the other tested plants.

Figure 1 Inoculation of B. cinerea (A), B. squamosa (B) and B. elliptica (C) on (N. benthamiana), onion and lily leaves confirms host-specificity. Disease symptoms were observed at 4dpi and box plot diagrams represent the lesion diameters at 4dpi upon inoculation of the different Botrytis species on the respective plants. Each box represents all compiled lesion diameters (in mm) for a given host plant.
Novel genome sequence assembly and annotation
High molecular weight genomic DNA of B. squamosa and B. elliptica was isolated from mycelial cultures and was used for whole genome sequencing using Nanopore technology. Assembly of the reads resulted in genomes with a size of 55 and 50 Mb for B. squamosa and B. elliptica, respectively (Table 1). The assembly sizes are in accordance to the available genomes that were sequenced using PacBio technology as described in Valero-Jiménez et al. (2020) (55 and 48 Mb), but the quality of the assemblies was significantly improved. The new assemblies consisted of 19 and 26 contigs for B. squamosa and B. elliptica, respectively (Table 1), as compared to 29 and 137 contigs in the previously published genomes (Valero-Jiménez et al., 2020). Many contigs resembled complete chromosomes. For B. squamosa we assembled 10 out of the 19 contigs with telomeric repeats at both ends, while the remaining 9 contigs contained telomeric sequences at one end. For the genome of B. elliptica, we obtained 8 complete chromosomes containing telomeres at both ends.
A whole genome synteny analysis of B. elliptica and B. squamosa was performed by pairwise alignment of the assemblies. The genomes displayed a high level of synteny, with perfect alignment along 6 of the 16 contigs (Figure 2). There was evidence for 3 structural rearrangements between the contigs of B. elliptica and B. squamosa. For instance contig 1 of B. elliptica aligned to contig 3, 4, and 16 of B. squamosa, suggesting chromosomal rearrangements between the species. Similarly, contig 10 from B. elliptica aligned with contig 4 and 16 from B. squamosa (Figure 2).
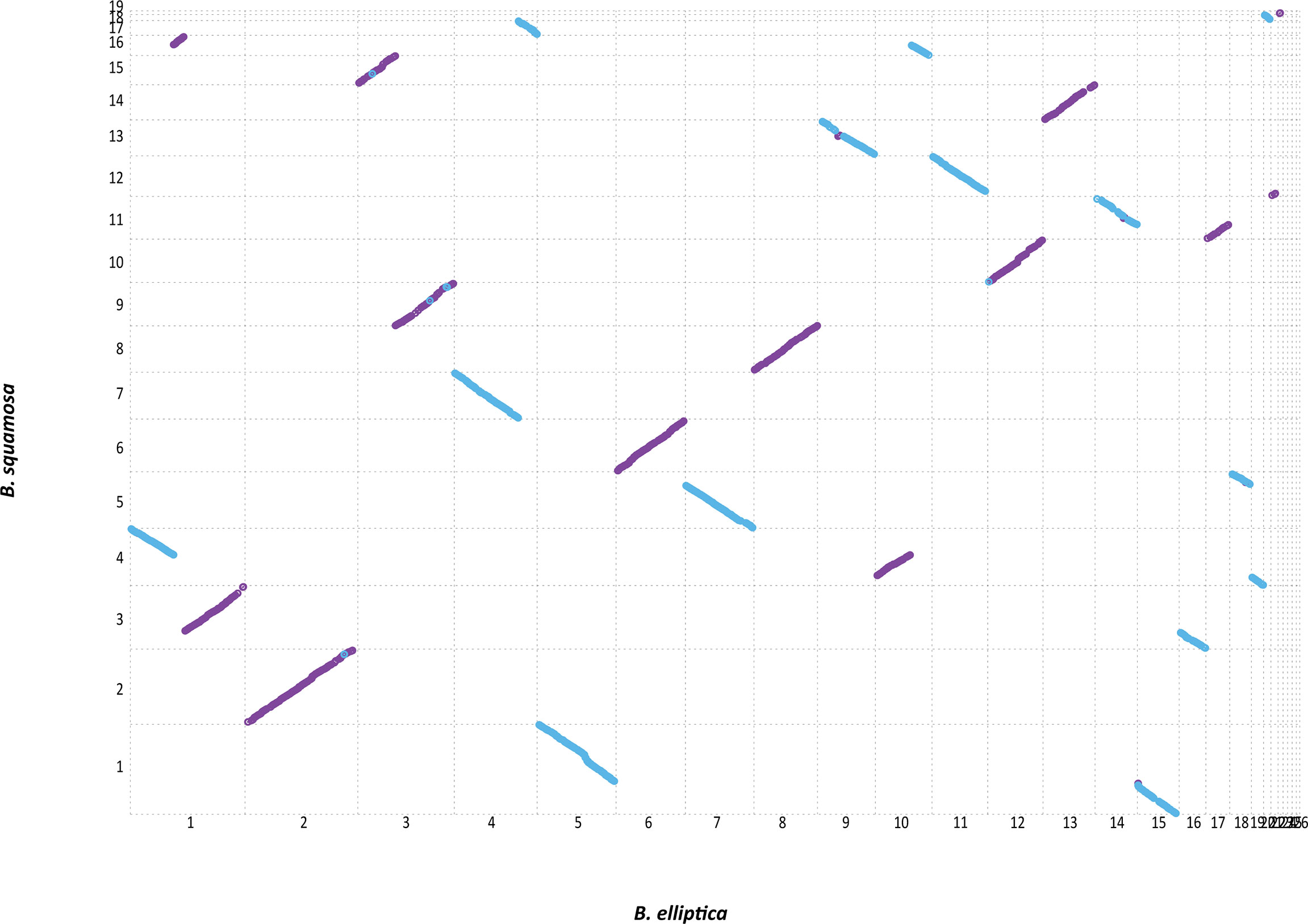
Figure 2 Synteny analysis of the genomes of B. elliptica and B. squamosa reveals a high level of synteny with few balanced structural rearrangements. Aligned contigs are color coded indicating the orientation of the synteny segments with purple indicating forward and blue indicating reverse orientation of the alignment.
The manually curated annotation of Valero-Jiménez et al. (2020) was used as input for annotation of the B. squamosa and B. elliptica genomes, together with RNAseq data from mycelial cultures and infected plant material. The genomes of B. squamosa and B. elliptica were predicted to contain 11,933 and 13,585 genes respectively, and we specifically examined and manually curated the gene models of the secretome (Table 1). The predicted secretome of B. squamosa consists of 897 genes (7.52%) while the predicted secretome of B. elliptica consists of 925 genes (6.81%) (Table 1). Based on the orthogroups described in Valero-Jiménez et al. (2020), we analyzed the predicted secretome for shared proteins between B. squamosa and B. elliptica and the more distantly related species B. cinerea which functions as the model species for the genus Botrytis (Pedro et al., 2019). The vast majority of the predicted secreted proteins could be assigned to 734 orthogroups that were shared between all three species (Supplementary Figure 1). A total of 91 orthogroups is shared only between B. squamosa and B. elliptica which reflects the recent divergence of these sister species as compared to B. cinerea. Only a small proportion of the predicted secretome is shared between B. cinerea and either B. elliptica or B. squamosa (31 and 20 orthogroups, respectively). Only 17, 26 and 55 predicted secreted proteins are unique for B. squamosa, B. elliptica and B. cinerea, respectively (Supplementary Figure 1; Supplementary Data 1).
The genome of B. elliptica isolate 9401 contains two mini-chromosomes (comprising both telomeres), of which the largest one is 199 Kb. These mini-chromosomes have no detectable synteny to the genome of B. squamosa (Figure 2). The largest mini-chromosome (contig 23, 199 kbp) contains 18 predicted genes, of which two appeared interesting. The gene BELL_9401_23g00040 encodes a predicted effector protein and has homologs in only B. tulipae (BTUL_0255g00010) and B. hyacinthi (BHYA_0337g00090) (Valero-Jiménez et al., 2019). The gene BELL_9401_23g00180 encodes a cysteine protease with potential ubiquitin-deconjugating activity, and it is orthologous to the gene Bcin18g00060 that is located on B. cinerea isolate B05.10 mini-chromosome Chr18. The second B. elliptica mini-chromosome (contig 25, 175 kbp) contains nine predicted genes, of which three have interesting features. BELL_9401_25g00050 has an ortholog in B. cinerea B05.10 on mini-chromosome Chr18 (Bcin18g00040, of unknown function), while BELL_9401_25g00060 encodes a putative Forkhead-domain protein kinase with a homolog in B. cinerea on mini-chromosome Chr17 (Bcin17g00060) and BELL_9401_25g00080 is orthologous to Bcin17g00070 (a putative ULP cysteine protease). All three genes appear to be expressed during in vitro growth and plant infection. The genome of B. squamosa includes a single mini-chromosome of 253 Kbp that contains 12 predicted genes. All of them have an ortholog in B. elliptica 9401 mini-chromosome (contig 22) but not in B. elliptica 9612 (Valero-Jiménez et al., 2020). Four of the 12 genes also have orthologs in B. cinerea B05.10, of which two are located on Chr9 and two on mini-chromosome Chr17. The orthologous B. cinerea genes on Chr9 encode a CAZyme GH43 protein and a secreted effector-like protein, whereas one of the genes on Chr17 (Bcin17g00060) encodes a Forkhead-domain protein kinase and is also orthologous to the gene in the B. elliptica mini-chromosome contig 25, as mentioned above.
Liquid cultures of Botrytis spp. contain cell-death inducing proteins
In order to analyze the cell death-inducing activity of proteins secreted by the different Botrytis spp., we collected culture filtrate (CF) samples from B. squamosa and B. elliptica grown in liquid cultures. The CFs were infiltrated into N. benthamiana, onion and lily to assess plant cell death responses. Plant responses were recorded at 3 dpi (Figure 3) and red light imaging (Villanueva et al., 2021) was used to visualize plant cell death responses (Supplementary Figure 2). B. elliptica grown in presence of lily leaf extract, caused the most severe response in all tested plant species. Upon infiltration of this sample (CF number 1 in Figure 3, left panel), we observed collapse of the tissue, drying and intense yellowing/browning at the infiltrated area. By contrast, leaf infiltration with compounds collected from B. elliptica grown in presence of onion leaf extract (CF number 2 in Figure 3 left panel) caused a milder response and only in lily leaves, where the infiltrated tissue showed a slight translucence. On the other hand, compounds collected from B. squamosa grown in presence of lily leaf extract (CF number 3 in Figure 3, left panel) caused a more severe response when compared to the compounds collected from B. squamosa grown in presence of onion leaf extract (CF number 4 in Figure 3, left panel). Both B. squamosa CF samples did not cause any clear response upon infiltration in lily and N. benthamiana leaves, although in the highly sensitive red light imaging system some cell death response could still be observed (Supplementary Figure 2). When CF samples from B. squamosa grown in the presence of lily leaf extract were infiltrated in onion leaves, tissue collapse, drying and intense yellowing was observed. Infiltration of the medium used for the fungal liquid culture caused no detectable response in any of the tested plants (Supplementary Figure 2). CF samples contain proteins and secondary metabolites secreted by the fungus, both of which potentially possess cell death-inducing activities. To distinguish the proteinaceous activity from the activity of other compounds, ammonium sulfate precipitation was performed to concentrate the proteins secreted by the fungi in the liquid medium. After dialysis of the protein pellet, the resuspended precipitates were infiltrated in N. benthamiana, onion and lily leaves. The ammonium sulfate precipitation of the proteins did not notably affect the cell death inducing activity of B. elliptica proteins, since the precipitated proteins derived from B. elliptica liquid culture grown in presence of lily leaf extract showed the strongest cell death inducing activity in all tested plants (sample A in Figure 3, right panel; Supplementary Figure 3). The symptoms appeared similar to those observed upon infiltration of the crude CF. In contrast, ammonium sulfate precipitation of the proteins contained in B. squamosa CF sample grown in presence of lily leaf extract substantially lost their cell death inducing activity when infiltrated in onion leaves (Figure 3, right panel sample C; Supplementary Figure 3). This result suggests that the cell death-inducing activity of the culture filtrate does not originate from proteins but from phytotoxic secondary metabolites such as botrydial, for which a functional biosynthetic gene cluster is present in B. squamosa (Colmenares et al., 2002; Valero-Jiménez et al., 2020). Infiltration of the dialysis buffer caused no visible response in any of the plants tested (Supplementary Figure 3).
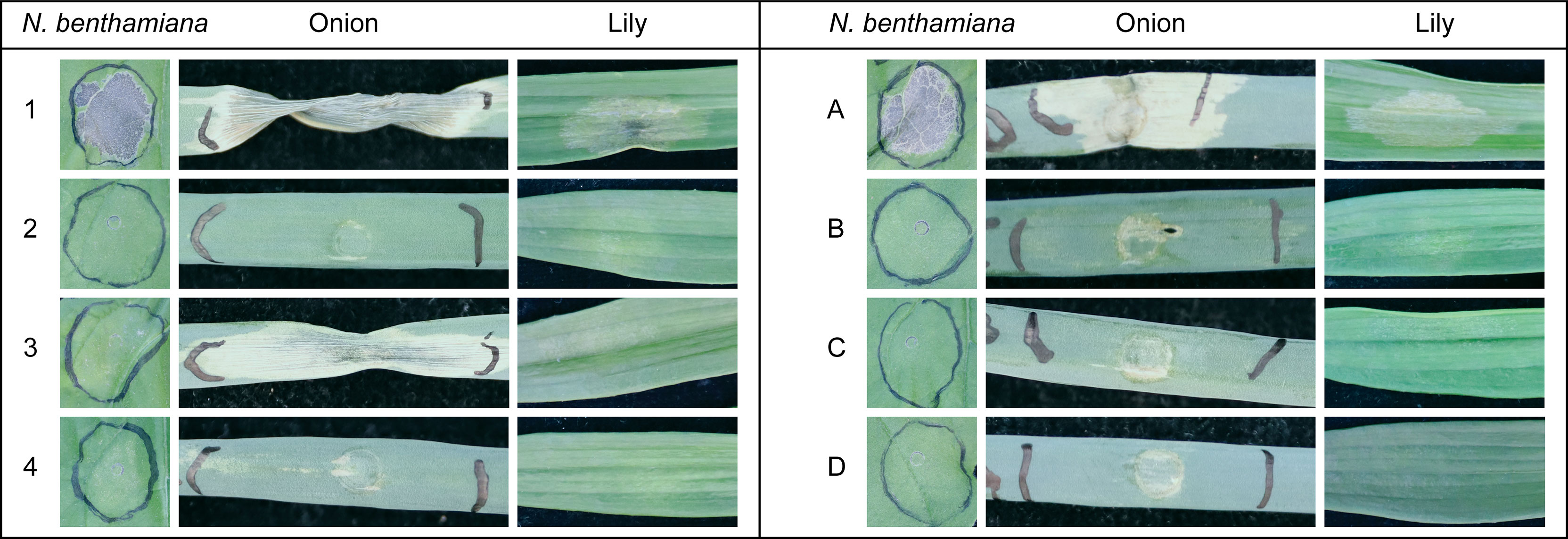
Figure 3 Leaf responses observed at 3dpi caused by infiltration of crude CF samples (left panel) and AS precipitated compounds (right panel) obtained from liquid cultures of B. squamosa and B. elliptica on leaves of N. benthamiana, onion and lily. 1 = B. elliptica grown in GB5_lily; 2 = B. elliptica grown in GB5_onion; 3 = B. squamosa grown in GB5_lily; 4 = B. squamosa grown in GB5_onion. (A) = AS precipitation of B. elliptica CF grown in GB5_lily; (B) = AS precipitation of B. elliptica CF grown in GB5_onion; (C) = AS precipitation of B. squamosa CF grown in GB5_lily; (D) = AS precipitation of B. squamosa CF grown in GB5_onion.
Protein composition of the cell death-inducing culture filtrates
The identity of proteins present in the CF samples that showed cell death-inducing activity was analyzed using mass spectrometry. The B. elliptica CF sample contained significant hits for a total of 165 proteins identified in the predicted secretome. More specifically, we detected 70 CAZymes (42% of total number), 15 putative effectors, 8 oxidoreductases and 24 proteases (14,5%). The remaining proteins represented enzymes related to lipid metabolism, different phosphatases, sugar and ion transporters, or proteins without a described domain. In the B. squamosa CF sample, 114 proteins with a predicted signal peptide could be identified. Compared to the CF of B. elliptica, the B. squamosa CF sample contains a considerably higher proportion of CAZymes (69 correspondig to 60% of the total number) but a lower proportion of proteases (7%) and only three putative effectors. In total 85 proteins are present both in B. elliptica and B. squamosa CF samples (Figure 4). Only three proteins present in both CF samples are predicted to be putative effectors: G17 (orthologs to B. cinerea Spl1, Frías et al., 2011) and G58, a small (16KDa) cysteine rich protein of unknown function. Both CFs contained a protein with a chorismate mutase domain that also carries a predicted chloroplast targeting peptide (G63). Interestingly, the orthologous region in B. cinerea appeared to be pseudogenized since it carries a point mutation in the start codon.
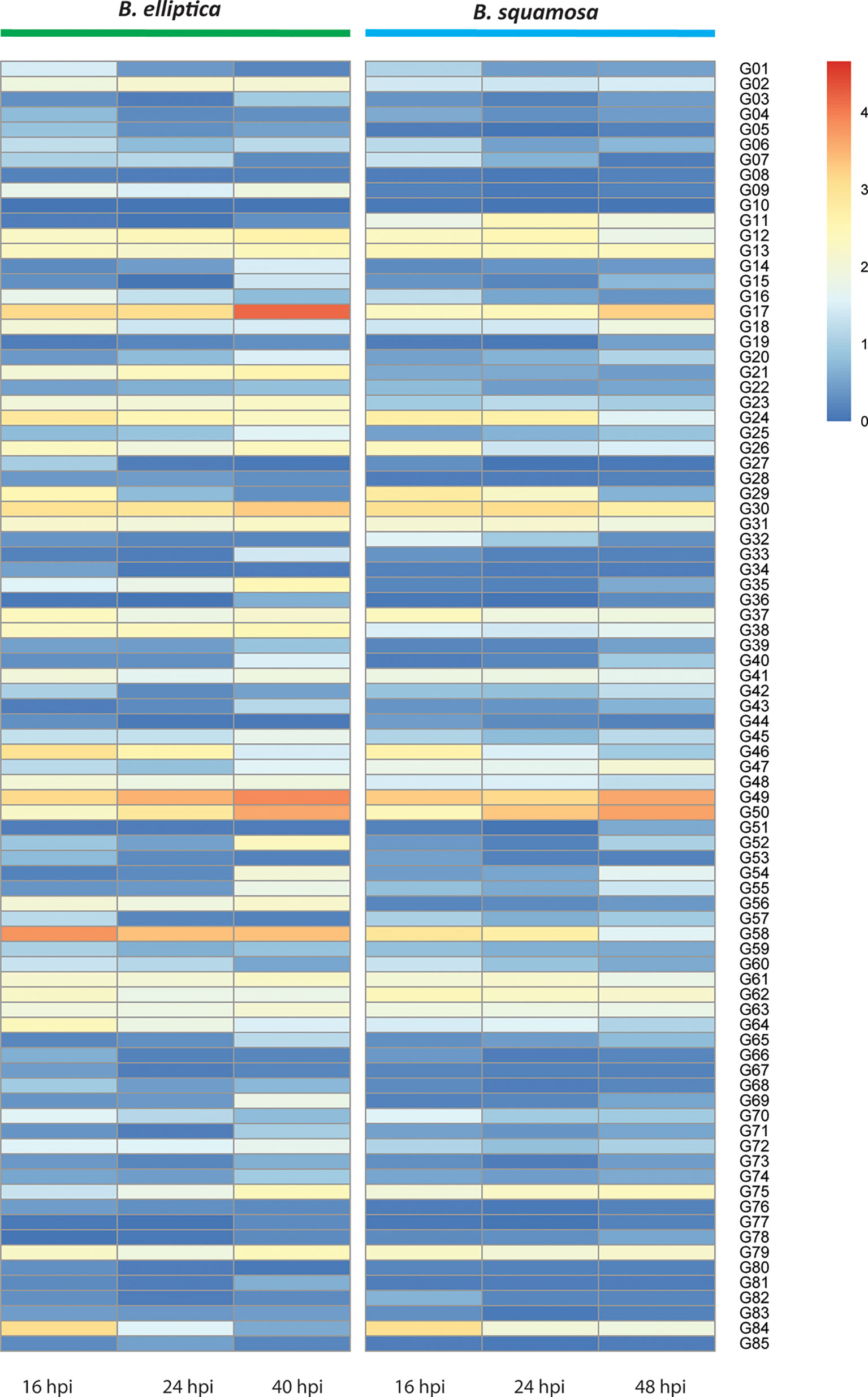
Figure 4 Heatmap showing expression profiles of genes of which the proteins were identified in the CF samples of B. elliptica and B. squamosa. Genes from both species that belong to the same orthogroups are aligned horizontally. Expression is based on the average of three replicate RNAseq samples during different timepoints of infection of B. elliptica and B. squamosa on their respective host plants.
To investigate the potential contribution to virulence of the 85 shared proteins detected in the CF samples, we analyzed the transcript levels of their corresponding genes at different timepoints during host infection (Supplementary Data 2). Figure 4 shows a heatmap of the gene expression profile of orthologous genes of B. squamosa and B. elliptica during infection. When examining the transcript levels in the two Botrytis spp., there are three orthogroups with relatively high expression at 40-48 hpi (G17, 49 and 50). For example,G17 which is orthologous to the B. cinerea cell death-inducing protein Spl1 (Frias et al., 2011) and G49 which encodes an aspartic protease, both peaked in expression in both species at 40-48 hpi. In contrast, some orthogroups had the highest relative expression at 16 hpi (G24, G29, G46, 58, 62 and 84) in both B. squamosa and B. elliptica. G24 encodes a putative endoglucanase (GH12) and G58 is predicted as a putative effector. Some orthologs showed different expression patterns between the two fungi, such as G09, G11, G18, G21, G23, G35, G38, G58. For instance, the orthogroup G21, which encodes a polygalacturonase also found in B. cinerea, was relatively highly expressed throughout infection in B. elliptica, but not in B. squamosa. spp.
Discussion
Previously the genomes of 16 Botrytis spp. with distinct host specificity were analyzed in order to identify unique genes that might encode virulence factors associated to host specificity (Valero-Jiménez et al., 2020). However, no specific elements were found, suggesting that host specificity may be determined by a combination of different (quantitative) traits rather than by presence or absence of unique virulence factors. The aim of this study was to analyse shared features of the secretome in B. elliptica and B. squamosa that contribute to cell death induction and virulence on their host and non-host plants. This was done by comparing their genomes and by testing the cell death-inducing activity of their secreted proteins.
Genome sequences were available for both species (Valero-Jiménez et al., 2020). However, we considered it useful to further improve the assembly of the B. squamosa reference genome and to newly sequence a different B. elliptica isolate 9401 (Malvestiti et al., 2021) that was much more virulent than the isolate B. elliptica 9612 previously sequenced by Valero-Jiménez et al. (2020). The new genome assemblies of B. elliptica 9401 and B. squamosa MUCL31421 indeed provided significant improvement over the previous assemblies and enabled us to perform a whole genome alignment and synteny analysis between B. squamosa and B. elliptica. They display a high degree of synteny, with evidence for only three balanced chromosomal rearrangements. A reconstruction of the ancestral genome of the genus Botrytis by Valero-Jiménez et al. (2020) had provided indications for the occurrence of very low numbers of balanced chromosomal rearrangements between the common ancestor of the genus Botrytis and the extant species, despite the estimate that they diverged ~5 Million years ago (Shen et al., 2020). The chromosome configurations of B. squamosa and B. elliptica provide further indications for the remarkable stability of genomes in the genus Botrytis, as proposed by Valero-Jiménez et al. (2020). Moreover, the manually curated annotations by Valero-Jiménez et al. (2020) could largely be transferred to the new assemblies of B. squamosa and B. elliptica, and provided a reliable insight into their proteome and especially secreted protein repertoires. The predicted secretomes of B. squamosa and B. elliptica were more similar to each other than either of the two species resembled the B. cinerea secretome, in concordance with their recent divergence and larger evolutionary distance to B. cinerea.
Interestingly, the genome assemblies yielded evidence for the occurrence in both species of mini-chromosomes with lengths of 175-253 kb. One mini-chromosome was identified in B. squamosa and two in B. elliptica. These mini-chromosomes contain few (predicted) genes, however, remarkably they contain orthologs of genes that are also present on the mini-chromosomes of B. cinerea, which is believed to have diverged from Botrytis species in clade 2 around 5 Million years ago (Shen et al., 2020). Intriguingly, homologs of several genes on B. elliptica and B. squamosa mini-chromosomes were detected in distant species (B. tulipae and B. hyacinthi) but none were detected in the close relatives B. deweyae and B. sinoalli, suggesting that certain more distant Botrytis species may possess homologous mini-chromosomes, while related species might not possess them. One gene that seems to be shared among mini-chromosomes in B. cinerea, B. elliptica and B. squamosa encodes a putative Forkhead-domain protein kinase, while other genes that are shared between two of the three species include genes encoding a putative effector, a glycosyl hydrolase (GH43) and a cysteine peptidase with potential ubiquitin-deconjugating activity. The presence of such genes on mini-chromosomes of three distinct species might explain their retention in the species and the population. The function of these genes in the biology and virulence of these species clearly deserves further analyses.
Besides genome comparisons, we tested the cell death-inducing activity of B. elliptica and B. squamosa proteins secreted during liquid culture. The CF sample of B. elliptica contained a much larger diversity of proteins than B. squamosa. Upon leaf infiltration, the B. elliptica CF sample caused cell death not only in lily, but also in onion and N. benthamiana (Figure 3). On the other hand, the B. squamosa CF samples only caused severe necrotic responses in onion leaves. These observations suggest that the larger diversity and the combined activity of B. elliptica secreted proteins might confer the capability of causing cell death not only in lily (Liliaceae), its natural host, but also in phylogenetically distant plant species such as N. benthamiana (Solanaceae) and onion (Amaryllidaceae). This hypothesis is supported by the fact that cell death induction and plant susceptibility in plant-necrotroph interactions often correlate in a quantitative manner. As shown for the wheat-P. nodorum interaction, the quantitative level of plant susceptibility is correlated to the number of cell death-inducing effectors that are produced by the fungus and the number of effector target loci present in the host (Tan et al., 2012; Haugrud et al., 2019). One should also consider that during fungal infection, cell death-inducing proteins are gradually expressed and secreted by the fungus and their levels must be sufficiently high to induce cell death responses that indeed facilitate fungal colonization. Experiments with CF samples can only partly mimic the events occurring during plant-necrotroph interactions.
CF samples that could induce host cell death on onion or lily contained subsets of (predicted) effectors as well as CAZymes. The samples of B. elliptica and B. squamosa contained proteins that could be grouped into 85 orthogroups shared between the species. Not all genes in these orthogroups were expressed during host infection, suggesting that they may have different roles in the biology of these pathogens. Based on transcriptome data of B. elliptica-infected lily and B. squamosa-infected onion, there were 15-20 genes in each of the species that appeared to display significant expression, including a subset of genes that displayed a transient expression peak in early phases of infection. Especially proteins from G58, encoding a putative effector, and G17 which encodes orthologs of the B. cinerea cell death-inducing protein Spl1 (Frías et al., 2011) are interesting candidates for analysis.
Both CF samples from B. squamosa and B. elliptica contained proteins that were uniquely present in that sample. Since the focus of this study is on the shared cell death inducing proteins between the two Botrytis spp., these unique proteins were not analysed. However, such unique proteins showing high gene expression during infection can be further investigated to explore their contribution to virulence in the respective host plants. This list still comprises a few dozen genes, and experiments are ongoing to purify CF samples through ion exchange chromatography and to test the activity of fractions for cell death-inducing activity. These efforts should result in reducing the number of candidate genes to a manageable number for functional analysis through CRISPR/Cas9-mediated deletion (Leisen et al., 2020; Leisen et al., 2022).
Data availability statement
The Whole Genome Shotgun project of B. elliptica 9401 has been deposited at DDBJ/ENA/GenBank under the accession JANCTG000000000. The version described in this paper is version JANCTG010000000. The Whole Genome Shotgun project of Botrytis squamosa MUCL31421 has been deposited at DDBJ/ENA/GenBank under the accession RCTC00000000. The version described in this paper is version RCTC02000000.
Author contributions
MM and MS contributed equally to this work. MM, MS, XS-K and JK designed the study. MM, MS, HB and SB performed the lab experiments and analysed the data. XS-K performed the genome assemblies and computational analyses. MM, MS, XS-K and JK wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The research of MCM is funded by NWO-Science domain (NWO-ENW), project GSGT.2018.008. The research of MBFS was financially supported by the Dutch Technology Foundation TTW, which is part of the Netherlands Organisation for Scientific Research (NWO) and which is partly funded by the Ministry of Economic Affairs (project 15003).
Acknowledgments
The plant material used was kindly provided by De Jong Lelies, Hobaho, Mak Breeding, Royal van Zanten, Vletter-den Haan, and World Breeding. The authors are grateful to Rinie Verwoert, Rohan van Genderen, and Wilfred Krijnen for their support in cultivation of the lilies.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.993325/full#supplementary-material
Supplementary Information 1 | Detailed procedure for protein preparation for Mass Spectrometry analysis.
Supplementary Data 1 | Detailed information of orthogroups, and Interproscan results of specific groups of proteins.
Supplementary Figure 1 | Secretome conservation of three Botrytis spp. Presence or absence of secreted protein genes among B. cinerea, B. elliptica and B. squamosa are indicated by dark and light blue, respectively.
Supplementary Figure 2 | Leaf responses observed at 3dpi caused by infiltration of crude CF samples obtained from liquid cultures of B. squamosa and B. elliptica on leaves of N. benthamiana, lily and onion visualized with normal light and with the red light imaging system. 1 = B. elliptica grown in GB5_lily; 2 = B. elliptica grown in GB5_onion; 3 = B. squamosa grown in GB5_lily; 4 = B. squamosa grown in GB5_onion; 5 = GB5_onion mock; 6 = GB5_lily mock.
Supplementary Figure 3 | Leaf responses observed at 3dpi caused by infiltration of AS precipitated compounds contained in the CF samples in N. benthamiana, lily and onion visualized with normal light and with the red light imaging system. A = AS precipitation of B. elliptica CF grown in GB5_lily; B = AS precipitation of B. elliptica CF grown in GB5_onion; C = AS precipitation of B. squamosa CF grown in GB5_lily; D = AS precipitation of B. squamosa CF grown in GB5_onion; E = mock dialysis buffer.
Supplementary Data 2 | Expression levels and functional annotation of the 85 shared proteins detected in the culture filtrates of B. squamosa and B. elliptica.
References
Abeysekara, N. S., Friesen, T. L., Keller, B., Faris, J. D. (2009). Identification and characterization of a novel host–toxin interaction in the wheat–Stagonospora nodorum pathosystem. Theor. Appl. Genet. 120 (1), 117–126. doi: 10.1007/s00122-009-1163-6
Arenas, Y. C., Kalkman, E. R., Schouten, A., Dieho, M., Vredenbregt, P., Uwumukiza, B., et al. (2010). Functional analysis and mode of action of phytotoxic Nep1-like proteins of Botrytis cinerea. Physiol. Mol. Plant Pathol. 74 (5-6), 376–386. doi: 10.1016/j.pmpp.2010.06.003
Bar, M., Ori, N. (2014). Leaf development and morphogenesis. Development 141 (22), 4219–4230. doi: 10.1242/dev.106195
Bray, N. L., Pimentel, H., Melsted, P., Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34 (5), 525–527. doi: 10.1038/nbt.3519
Chen, Y., Nie, F., Xie, S.-Q., Zheng, Y.-F., Dai, Q., Bray, T., et al. (2021). Efficient assembly of nanopore reads via highly accurate and intact error correction. Nat. Commun. 12 (1), 1–10. doi: 10.1038/s41467-020-20236-7
Colmenares, A., Aleu, J., Duran-Patron, R., Collado, I., Hernandez-Galan, R. (2002). The putative role of botrydial and related metabolites in the infection mechanism of Botrytis cinerea. J. Chem. Ecol. 28 (5), 997–1005. doi: 10.1023/A:1015209817830
Cowger, C., Ward, B., Brown-Guedira, G., Brown, J. (2020). Role of effector-sensitivity gene interactions and durability of quantitative resistance to Septoria nodorum blotch in Eastern US wheat. Front. Plant Sci. 11, 155. doi: 10.3389/fpls.2020.00155
Dickman, M., Park, Y., Oltersdorf, T., Li, W., Clemente, T., French, R. (2001). Abrogation of disease development in plants expressing animal antiapoptotic genes. Proc. Natl. Acad. Sci. 98 (12), 6957–6962. doi: 10.1073/pnas.091108998
Duba, A., Goriewa-Duba, K., Wachowska, U. (2018). A review of the interactions between wheat and wheat pathogens: Zymoseptoria tritici, Fusarium spp. and Parastagonospora nodorum. Int. J. Mol. Sci. 19 (4), 1138. doi: 10.3390/ijms19041138.
Faris, J. D., Zhang, Z., Lu, H., Lu, S., Reddy, L., Cloutier, S., et al. (2010). A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. 107 (30), 13544–13549. doi: 10.1073/pnas.1004090107
Frías, M., González, C., Brito, N. (2011). BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 192 (2), 483–495. doi: 10.1111/j.1469-8137.2011.03802.x
Frías, M., González, M., González, C., Brito, N. (2016). BcIEB1, a Botrytis cinerea secreted protein, elicits a defense response in plants. Plant Sci. 250, 115–124. doi: 10.1016/j.plantsci.2016.06.009
Gao, Y., Faris, J., Liu, Z., Kim, Y., Syme, R., Oliver, R., et al. (2015). Identification and characterization of the SnTox6-Snn6 interaction in the Parastagonospora nodorum–wheat pathosystem. Mol. Plant-Microbe Interact. 28 (5), 615–625. doi: 10.1094/MPMI-12-14-0396-R
Garfinkel, A. R., Lorenzini, M., Zapparoli, G., Chastagner, G. A. (2017). Botrytis euroamericana, a new species from peony and grape in North America and Europe. Mycologia 109 (3), 495–507. doi: 10.1080/00275514.2017.1354169
Haugrud, A. R. P., Zhang, Z., Richards, J. K., Friesen, T. L., Faris, J. D. (2019). Genetics of variable disease expression conferred by inverse gene-for-gene interactions in the wheat-Parastagonospora nodorum pathosystem. Plant Physiol. 180 (1), 420–434. doi: 10.1104/pp.19.00149
Hyde, K. D., Nilsson, R. H., Alias, S. A., Ariyawansa, H. A., Blair, J. E., Cai, L., et al. (2014). One stop shop: Backbones trees for important phytopathogenic genera). Fungal Diversity 67 (1), 21–125. doi: 10.1007/s13225-014-0298-1
Kurtz, S., Phillippy, A., Delcher, A. L., Smoot, M., Shumway, M., Antonescu, C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5 (2), 1–9. doi: 10.1186/gb-2004-5-2-r12.
Leisen, T., Bietz, F., Werner, J., Wegner, A., Schaffrath, U., Scheuring, D., et al. (2020). CRISPR/Cas with ribonucleoprotein complexes and transiently selected telomere vectors allows highly efficient marker-free and multiple genome editing in Botrytis cinerea. PloS Pathog. 16 (8), e1008326. doi: 10.1371/journal.ppat.1008326
Leisen, T., Werner, J., Pattar, P., Safari, N., Ymeri, E., Sommer, F., et al. (2022). Multiple knockout mutants reveal a high redundancy of phytotoxic compounds contributing to necrotrophic pathogenesis of Botrytis cinerea. PloS Pathog. 18 (3), e1010367. doi: 10.1371/journal.ppat.1010367
Liu, Z., Faris, J., Meinhardt, S., Ali, S., Rasmussen, J., Friesen, T. (2004). Genetic and physical mapping of a gene conditioning sensitivity in wheat to a partially purified host-selective toxin produced by stagonospora nodorum. Phytopathology 94 (10), 1056–1060. doi: 10.1094/PHYTO.2004.94.10.1056
Liu, Z., Zhang, Z., Faris, J. D., Oliver, R. P., Syme, R., McDonald, M. C., et al. (2012). The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. PloS Pathog. 8 (1), e1002467. doi: 10.1371/journal.ppat.1002467
Lorang, J. M., Sweat, T. A., Wolpert, T. J. (2007). Plant disease susceptibility conferred by a “resistance” gene. Proc. Natl. Acad. Sci. 104 (37), 14861–14866. doi: 10.1073/pnas.0702572104
Malvestiti, M. C., Immink, R. G., Arens, P., Quiroz Monnens, T., van Kan, J. A. L. (2021). Fire blight susceptibility in Lilium spp. correlates to sensitivity to Botrytis elliptica secreted cell death inducing compounds. Front. Plant Sci. 1270. doi: 10.3389/fpls.2021.660337.
Mercier, A., Carpentier, F., Duplaix, C., Auger, A., Pradier, J. M., Viaud, M., et al. (2019). The polyphagous plant pathogenic fungus Botrytis cinerea encompasses host‐specialized and generalist populations. Environmental microbiology 21 (12), 4808–21.
Noda, J., Brito, N., González, C. (2010). The Botrytis cinerea xylanase Xyn11A contributes to virulence with its necrotizing activity, not with its catalytic activity. BMC Plant Biol. 10 (1), 1–15. doi: 10.1186/1471-2229-10-38
Oliver, R. P., Friesen, T. L., Faris, J. D., Solomon, P. S. (2012). Stagonospora nodorum: From pathology to genomics and host resistance. Annu. Rev. Phytopathol. 50, 23–43. doi: 10.1146/annurev-phyto-081211-173019
Pedro, H., Yates, A. D., Kersey, P. J., De Silva, N. H. (2019). Collaborative annotation redefines gene sets for crucial phytopathogens. Front. Microbiol. 10, 2477. doi: 10.3389/fmicb.2019.02477
Phan, H. T., Rybak, K., Furuki, E., Breen, S., Solomon, P. S., Oliver, R. P., et al. (2016). Differential effector gene expression underpins epistasis in a plant fungal disease. Plant J. 87 (4), 343–354. doi: 10.1111/tpj.13203
Ron, M., Avni, A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16 (6), 1604–1615. doi: 10.1105/tpc.022475
Ruud, A. K., Windju, S., Belova, T., Friesen, T. L., Lillemo, M. (2017). Mapping of SnTox3–Snn3 as a major determinant of field susceptibility to Septoria nodorum leaf blotch in the SHA3/CBRD× naxos population. Theor. Appl. Genet. 130 (7), 1361–1374. doi: 10.1007/s00122-017-2893-5
Schürch, S., Linde, C. C., Knogge, W., Jackson, L. F., McDonald, B. A. (2004). Molecular population genetic analysis differentiates two virulence mechanisms of the fungal avirulence gene NIP1. Mol. Plant-Microbe Interact. 17 (10), 1114–1125. doi: 10.1094/MPMI.2004.17.10.1114
Shen, X.-X., Steenwyk, J. L., LaBella, A. L., Opulente, D. A., Zhou, X., Kominek, J., et al. (2020). Genome-scale phylogeny and contrasting modes of genome evolution in the fungal phylum ascomycota. Sci. Adv. 6 (45), eabd0079. doi: 10.1126/sciadv.abd0079.
Shi, G., Zhang, Z., Friesen, T. L., Raats, D., Fahima, T., Brueggeman, R. S., et al. (2016). The hijacking of a receptor kinase–driven pathway by a wheat fungal pathogen leads to disease. Sci. Adv. 2 (10), e1600822. doi: 10.1126/sciadv.1600822
Steentjes, M. B. F., Herrera Valderrama, A. L., Fouillen, L., Bahammou, D., Leisen, T., Albert, I., et al. (2022). Cytotoxic activity of Nep1-like proteins on monocots. New Phytol. 235, 690–700. doi: 10.1111/nph.18146
Steentjes, M. B. F., Tonn, S., Coolman, H., Langebeeke, S., Scholten, O. E., van Kan, J. A. L. (2021). Visualization of three Sclerotiniaceae species pathogenic on onion reveals distinct biology and infection strategies. Int. J. Mol. Sci. 22 (4), 1865. doi: 10.3390/ijms22041865
Stergiopoulos, I., Collemare, J., Mehrabi, R., De Wit, P. J. G. M. (2013). Phytotoxic secondary metabolites and peptides produced by plant pathogenic Dothideomycete fungi. FEMS Microbiol. Rev. 37 (1), 67–93. doi: 10.1111/j.1574-6976.2012.00349.x
Tan, K.-C., Ferguson-Hunt, M., Rybak, K., Waters, O. D., Stanley, W. A., Bond, C. S., et al. (2012). Quantitative variation in effector activity of ToxA isoforms from Stagonospora nodorum and Pyrenophora tritici-repentis. Mol. Plant-Microbe Interact. 25 (4), 515–522. doi: 10.1094/MPMI-10-11-0273
Tan, K.-C., Oliver, R. P., Solomon, P. S., Moffat, C. S. (2010). Proteinaceous necrotrophic effectors in fungal virulence. Funct. Plant Biol. 37 (10), 907–912. doi: 10.1071/FP10067
Valero-Jiménez, C. A., Steentjes, M. B., Slot, J. C., Shi-Kunne, X., Scholten, O. E., van Kan, J. A. L. (2020). Dynamics in secondary metabolite gene clusters in otherwise highly syntenic and stable genomes in the fungal genus Botrytis. Genome Biol. Evol. 12 (12), 2491–2507. doi: 10.1093/gbe/evaa218
Valero-Jiménez, C. A., Veloso, J., Staats, M., van Kan, J. A. L. (2019). Comparative genomics of plant pathogenic Botrytis species with distinct host specificity. BMC Genomics 20 (1), 1–12. doi: 10.1186/s12864-019-5580-x.
Van Baarlen, P., Staats, M., van Kan, J. A. L. (2004). Induction of programmed cell death in lily by the fungal pathogen Botrytis elliptica. Mol. Plant Pathol. 5 (6), 559–574. doi: 10.1111/j.1364-3703.2004.00253.x
van Kan, J. A. L. (2006). Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 11 (5), 247–253. doi: 10.1016/j.tplants.2006.03.005
Van Kan, J. A. L., Stassen, J. H., Mosbach, A., van der Lee, T. A., Faino, L., Farmer, A. D., et al. (2017). A gapless genome sequence of the fungus Botrytis cinerea. Mol. Plant Pathol. 18 (1), 75–89. doi: 10.1111/mpp.12384
Veloso, J., van Kan, J. A. L. (2018). Many shades of grey in Botrytis–host plant interactions. Trends Plant Sci. 23 (7), 613–622. doi: 10.1016/j.tplants.2018.03.016
Villanueva, S. L., Malvestiti, M. C., Joosten, M. H., van Ieperen, W., van Kan, J. A. L. (2021). Red-light imaging for programmed cell death visualization and quantification in plant-pathogen interactions. Mol. Plant Pathol. 22, 361–372. doi: 10.1111/mpp.13027
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS One 9 (11), e112963. doi: 10.1371/journal.pone.0112963
Xiong, L., Boeren, S., Vervoort, J., Hettinga, K. (2020). Dataset on proteomic changes of whey protein after different heat treatment. Data Brief 29, 105227. doi: 10.1016/j.dib.2020.105227
Yang, Y., Yang, X., Dong, Y., Qiu, D. (2018). The Botrytis cinerea xylanase BcXyl1 modulates plant immunity. Front. Microbiol. 9, 2535. doi: 10.3389/fmicb.2018.02535
Zhang, L., Kars, I., Essenstam, B., Liebrand, T. W., Wagemakers, L., Elberse, J., et al. (2014). Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 164 (1), 352–364. doi: 10.1104/pp.113.230698
Keywords: Botrytis spp., plant-necrotroph interaction, host specificity, cell death-inducing proteins, synteny
Citation: Malvestiti MC, Steentjes MBF, Beenen HG, Boeren S, van Kan JAL and Shi-Kunne X (2022) Analysis of plant cell death-inducing proteins of the necrotrophic fungal pathogens Botrytis squamosa and Botrytis elliptica. Front. Plant Sci. 13:993325. doi: 10.3389/fpls.2022.993325
Received: 13 July 2022; Accepted: 06 September 2022;
Published: 11 October 2022.
Edited by:
Jan Schirawski, Friedrich Schiller University Jena, GermanyReviewed by:
Martijn Rep, University of Amsterdam, NetherlandsInés Ponce De León, Instituto de Investigaciones Biológicas Clemente Estable (IIBCE), Uruguay
Copyright © 2022 Malvestiti, Steentjes, Beenen, Boeren, van Kan and Shi-Kunne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jan A. L. van Kan, amFuLnZhbmthbkB3dXIubmw=
†These authors have contributed equally to this work and share first authorship
 Michele C. Malvestiti
Michele C. Malvestiti Maikel B. F. Steentjes
Maikel B. F. Steentjes Henriek G. Beenen1
Henriek G. Beenen1 Sjef Boeren
Sjef Boeren Jan A. L. van Kan
Jan A. L. van Kan Xiaoqian Shi-Kunne
Xiaoqian Shi-Kunne