
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 18 August 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.992391
This article is part of the Research TopicThe Brassicaceae — Agri-Horticultural and Environmental Perspectives, Volume IIView all 19 articles
Male sterility is an ideal character for the female parent in commercial hybrid seed production in Chinese cabbages. We identified three allele male sterile mutants msm2-1/2/3 in progenies of ethyl methane sulfonate mutagenized Chinese cabbage. It was proved that their male sterilities were controlled by a same recessive nuclear gene. Cytological observation showed that the delayed tapetal programmed cell death (PCD) as well as the abnormal pollen exine and intine led to pollen abortion in these mutants. MutMap combined with KASP analyses showed that BraA10g019050.3C, a homologous gene of AtMS1 encoding a PHD-finger transcription factor and regulated pollen development, was the causal gene. A single-nucleotide mutation from G to A occurred at the 2443th base of BrMS1 in msm2-1 which results in premature termination of the PHD-finger protein translation; a single-nucleotide mutation from G to A existed at 1372th base in msm2-2 that makes for frameshift mutation; a single-nucleotide mutation from G to A distributed at 1887th base in msm2-3 which issues in the amino acid changed from Asp to Asn. The three allelic mutations in BrMS1 all led to the male sterile phenotype, which revealed its function in stamen development. Quantitative reverse transcription polymerase chain reaction analysis indicated that BrMS1 specially expressed in the anther at the early stage of pollen development and its expression level was higher in msm2-1/2/3 than that in the wild-type “FT.” BrMS1 was located at the nucleus and a length of 12 amino acid residues at the C-terminus had transcriptional activation activity. RNA-seq indicated that the mutation in BrMS1 affected the transcript level of genes related to the tapetum PCD and pollen wall formation, which brought out the pollen abortion. These male sterile mutants we developed provided a novel gene resource for hybrid breeding in Chinese cabbage.
Male sterility refers to the phenomenon in which stamens are degenerated or malformed, and viable male gametes are not produced, while female gametes are fertile in flowering plants. Male sterility has been widely used for hybrid seed production (Fan and Zhang, 2018). Up to now, many genic male sterility (GMS) genes have been identified in plants (Shi et al., 2015; Wan et al., 2019, 2020), and a great deal of effort has been made to develop male-sterility systems by using these GMS genes and their corresponding mutants to maintain and propagate GMS lines as female parents for hybrid seed production (Kim and Zhang, 2018). The male sterility system is also of immense significance in probing the molecular mechanism of pollen development. Spontaneous mutant male sterile mutants have been widely used in functional genomics (Liu et al., 2016; Liang et al., 2017; Zhou et al., 2017; Lin et al., 2020; Zhang et al., 2020). Due to its scarcity, artificial mutagenesis including physical mutagenesis, chemical mutagenesis, biological mutagenesis, and other methods to create male sterile mutants, has become an important measure for the study of gene regulatory networks for male gamete development (Ma et al., 2019; Tan et al., 2019; Huang et al., 2020).
Male gamete formation in anthers requires the coordinated participation of both sporophytic and gametophytic tissues (Hafidh et al., 2016). The tapetum, the innermost cell layer of the anther wall, plays a crucial role in regulating pollen development. The tapetum cells could provide the nutrients required for the meiosis of the microspore mother cell and the development of the microspore (Wu and Cheung, 2000; Parish and Li, 2010). During the tetrad period, the tapetum secrete sporopollenin precursor substances and they deposit continuously on the primexine to form a special T-shaped structure composed of the tectum, bacula, and foot layer (Shi et al., 2015). After the microspore mother cell completes meiosis, the tapetum synthesizes callose enzymes to degrade callose, which promotes the release of independent uninucleate microspores (Zhang et al., 2007; Parish and Li, 2010). Then, the tapetum undergoes programmed cell death (PCD), the pollen coat proteins (PCPs) and the tapetal cell debris produced by tapetum degradation, fill the pollen exine to form the pollen coat (Zhang et al., 2016). Therefore, abnormalities at any stage of the tapetum development could cause pollen aberrations.
The development of the tapetum is a precise and orderly process, which is jointly regulated by a variety of genes. To date, there are three major types of transcription factors (TFs) that regulate the development and function of the tapetum: bHLH TFs such as DYT1 and AMS, MYB TFs such as TDF1 and MS188, and PHD-fingers TFs such as MS1 and TIP3 (Zhu et al., 2008, 2011; Zhang and Yang, 2014; Wan et al., 2019; Yang et al., 2019a). These TFs cooperatively form the tapetal genetic pathway (DYT1-TDF1-AMS-MS188-MS1) and strictly control tapetum development (Zhu et al., 2011). Several orthologs of MS1 have been identified in different species. In Arabidopsis, AtMS1 directly regulated the expression of multiple PCPs such as GRP14, GRP19 and affected the expression of KCS7, KCS15, and KCS21 for pollen coat lipids (PCLs) synthesis (Lu et al., 2020; Zhang et al., 2021). In rice, OsMS1/OsPTC1 can interact with OsMADS15 and TIP2. It has been reported that TIP2 coordinated with TDR to modulate the expression of EAT1 and Cys protease gene (CP1) further regulated tapetal PCD and pollen exine formation in rice (Li et al., 2011; Yang et al., 2019b). In maize, ZmMs7 can interact with maize nuclear factor Y (NF-Y) subunits to form ZmMs7-NF-YA6-YB2-YC9/12/15 protein complexes that activate target gene ZmMT2C to regulate the tapetal development (An et al., 2020). However, the molecular mechanism by which the homologous gene of MS1 in Chinese cabbage affects pollen development remains unclear.
Chinese cabbage (Brassica rapa L. ssp. pekinensis) is one of the most important vegetable crops in eastern Asia, which is a typical cross-pollinated Brassica crops with obvious heterosis (Sharma et al., 2021). Breeding of male sterile lines is crucial for the commercialization of Chinese cabbage hybrid seed production (Singh et al., 2019). Mapping and cloning the male sterile genes are preconditions to the study of its molecular mechanism and applications. The genic male sterile lines could be created by artificial mutagenesis in Chinese cabbage. Tan et al. (2019) developed a male sterile mutant (ftms) from Chinese cabbage DH line “FT” by irradiating microspores with 60Co γ-rays while Bra010198 was a candidate gene for male sterile mutant (ftms), which encoded a putative β-(1,3)-galactosyltransferase that controls pollen exine development. Huang et al. (2020) obtained a stably inherited male sterile mutant (msm) by the identical methods and compared the differential expression genes of “FT” and msm flower buds using RNA-Seq technology.
Here, we identified three allele male sterile mutants named msm2-1/2/3 in an ethyl methane sulfonate (EMS)-mutagenized Chinese cabbage progenies. Cytological observation revealed delayed tapetum PCD, defective pollen exine and intine formation in msm2-1 mutant. We discovered by MutMap and KASP that the non-synonymous base-pair mutations in BrMS1 (BraA10g019050.3C) induced the pollen abortion in the allele mutants msm2-1-2/3. BrMS1 was a homologous gene of AtMS1, which encoded a PHD-finger TF and regulated the pollen development. The protein encoded by the BrMS1 was located at the nucleus in Chinese cabbage. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) performed that BrMS1 expressed specifically in the anther at the early stage of pollen development and its expression level was higher in msm2-1/2/3 versus in the wild-type “FT.” The analysis of RNA-seq exposed that the expression level of genes associated with the tapetum PCD and pollen wall synthesis has changed significantly in the mutant msm2-1. Our research is the first to clone an AtMS1 orthologs gene relevant to fertility regulation in Chinese cabbage, thereby providing basal information for applying this critical agronomic trait in hybrid breeding and studying the molecular mechanism of pollen development.
The Chinese cabbage doubled haploid (DH) line “FT,” obtained from a microspore culture of the commercial variety “Fukuda 50,” was used as the wild type. We treated “FT” seeds with 0.8% EMS, the live plants (M0) were self-pollinated, M1 seeds were harvested. After M1 self-pollination, we obtained stably inherited mutations in the M2 generation (Gao et al., 2022). Among them, nine male sterile mutants with stable inheritance and fertility not affected by environment were identified after multiple generations phenotypic identification (Supplementary Figure S1). The fertile and sterile plants in AB lines of the nine male sterile materials were performed the allelic test by mutual cross. The materials whose segregation ratio of the hybrid progeny conformed to 1:1 were allelic mutants of each other (Supplementary Table S1). We screened out two groups of allelic mutant materials in total and select one group (msm2-1, msm2-2, and msm2-3) as the experimental materials for this study. All plant materials were cultured in a greenhouse at 20–28°C in Shenyang Agricultural University (Shenyang, P. R. China).
For the genetic analysis, the wild-type “FT” (P1) was crossed with msm2-1/2/3 mutants (P2) to generate the F1 and F2 populations. Fertile plants and sterile plants from the F2 population were selected for crossing to generate AB line. Phenotypic characterization was obtained for each generation (P1, P2, F1, F2 and AB line), and the segregation ratio of the population was analyzed by the Chi-square test.
To observe the difference in floral organs between the wild-type “FT” and msm2-1/2/3, the five floral organs (sepals, petals, filament, stamens, pistils) were collected at the full-bloom stage and dissected under a stereomicroscope (Nikon SMZ800, Japan). For investigating the pollen vitality, the fresh anthers at the full-bloom stage were extruded and immersed in 0.1% 2,3,5-triphenyltetrazolium chloride dye solution on a carrier plate covered with a coverslip. Pollen viability was analyzed via applying an optical microscope (Nikon ECLIPSE 80i, Japan).
For understanding more about the period of pollen abortion, the flower buds of the wild-type “FT” and msm2-1/2/3 were fixed using FAA solution (50% ethanol, 5% glacial acetic, 10% formalin), then dyed using safranin and fast green, and observed under an optical microscope (Nikon ECLIPSE 80i; Nikon, Japan). The buds were divided into six grades according to the length of the buds (BUD1 < 1.5 mm, BUD2: 1.5–2.0 mm, BUD3: 2.0–2.5 mm, BUD4: 2.5–3.0 mm, BUD5: 3.0–3.5 mm, BUD6 > 3.5 mm.). The detailed paraffin sections operation steps were performed according to the method described by Zhou et al. (2017).
For scanning electron microscopy (SEM), to observe the morphology of anther and pollen exine, the fresh anthers of the wild-type “FT” and the msm2-1 mutant were collected at the full-bloom stage and fixed in 2.5% glutaraldehyde solution at 4°C for 48 h, processed according to the methods of Lin et al. (2014) and then examined under SEM (Hitachi TM3030, Japan).
For transmission electron microscopy (TEM), to further explore the morphology changes of pollen exine, the anthers are roughly divided into three periods: the early stage (BUDS < 2.0 mm), middle stage (2.0 mm < BUDS < 3.0 mm), and the late stage (BUDS > 3.0 mm) according to the pollen development process. Anthers at different pollen developmental stages from “FT” and msm2-1 plants were examined by TEM (Hitachi Ltd., Tokyo, Japan) after a series of treatments as previously described (Wang et al., 2020).
The candidate gene was identified by the modified MutMap method. Fifty plants with mutant phenotype were selected from the F2 plants, and a DNA pool was constructed by pooling equal amounts of plant leaf tissue from each plant. DNA was acquired from the wild-type “FT” and msm2-1 mutant, the DNA pool was extracted using a DNA-secure Plant Kit (Tiangen, Beijing, China) and was re-sequenced using a NovaSeq 6,000 System sequencer (Illumina, San Diego, United States). The analysis method was the same as described by Gao et al. (2020) and Wang et al. (2020).
KASP was applied for the genotypic assay to detect the co-segregation of each single nucleotide polymorphism (SNP) identified by MutMap and confirmed the candidate gene. Allele-specific primers bearing the FAM and HEX fluorescence probes and the common primer were designed by LGC (Laboratory of the Government Chemist, Shanghai, China) and used to determine the male sterile genotypes of 183 F2 plants (Supplementary Table S2). The primer mixed and PCRs were programmed as recommended by LGC. The KASP thermal cycling conditions were those described in Xi et al. (2018). Fluorescence was detected with a QuantStudio 6 instrument (Applied Biosystems, Foster City, CA, United States).
The full-length and CDS sequences of candidate genes were obtained by cloning and sequencing. Primers were designed according to the gene sequence information and are shown in Supplementary Tables S3, S4. We purified the PCR products with a Gel Extraction Kit (CWBIO, Beijing, China) and then introduced the purified products into a pGEM-T-Easy Vector (Promega, United States). The products were sequenced at GENEWIZ (Suzhou, China). We analyzed the sequences using DNAMAN V6 (Lynnon BioSoft, Canada).
The sequence and the information of BrMS1 were downloaded from the Brassica database. BrMS1 protein homologs in various plants were obtained from the GenBank database1 and aligned with ClustalW (Larkin et al., 2007). A phylogenetic tree was constructed with MEGA6.0 by the neighbor-joining (NJ) method based on a bootstrap test of 1,000 replicates (Tamura et al., 2013). The conserved domain of BrMS1 was analyzed online at NCBI.2
Total RNA was isolated from different tissues of the wild-type “FT” and msm2-1/2/3 at developmental stages with the TIANGEN RNA prep Pure Plant Kit. First-strand cDNA was synthesized using Fast Quant RT SuperMix (Tian gen). The cDNA was subsequently used as a template for qRT-PCR together with SYBR Green PCR Master Mix (TaKaRa, Dalian, China). ACTIN was used as the internal reference gene. Specific primers for each target gene are listed in Supplementary Table S5. Each sample had three biological replicates, each with three technical replicates.
To generate the BrMS1–GFP fusion protein, the full-length CDS of BrMS1 was amplified. The coding regions of the BrMS1 sequences were amplified without stop codons using the primers BrMS1-GFP-F and BrMS1-GFP-R. They were cloned into the C-terminal GFP fusion vector pBWA(V)HS driven by the 35S promoter. Vector-bearing, 35S-driven GFP was the negative control. Subcellular localization of the BrMS1 proteins was performed according to Yoo et al. (2007). The constructs were then transiently transformed into tobacco (Nicotiana benthamiana) leaves through Agrobacterium tumefaciens infiltration. After 48 h, the epidermis of the tobacco leaves was injected with DAPI and examined with a confocal laser-scanning microscope (Leica Microsystems, Wetzlar, Germany). The primer sequences used in this experiment are shown in Supplementary Table S6.
The transcriptional activity assay was performed using the Matchmaker GAL4 Yeast Two-Hybrid System 3 (Clontech). To generate the BD–BrMS1, BD–ΔN, BD–ΔC and BD–PHD constructs, the full-length CDS and various deletions of BrMS1 were PCR amplified and inserted into the pGBKT7 plasmid linearized. All recombinant plasmids and a pGBKT7 empty vector were separately transformed into the Y2H Gold Yeast strain. Then they were initially grown on the medium lacking Trp for 3 days at 30°C. Four of the resulting independent colonies were transferred onto the medium without Trp, His, and Ade for 3 days at 30°C to assay transcriptional activity. The primers used for vector construction are listed in Supplementary Table S7.
To identify transcripts involved in the regulation of pollen abortion in the Chinese cabbage, we performed an extensive transcriptomic analysis of entire anther. Total RNA was extracted from the wild-type “FT” and msm2-1 with the RNA prep Pure Plant Plus Kit (Tiangen, Beijing, China). Samples were collected with three biological replicates. Six cDNA libraries (F1, F2, F3, S1, S2, and S3) were constructed and sequenced using the Illumina HiSeq™ 2,500 sequencing platform at Beijing Genomics of Institute (BGI), Shenzhen, China. The detailed method and procedure have been described previously by Huang et al. (2017). In this study, different expression genes (DEGs) were defined as those genes with a false discovery rate (FDR) ≤ 0.01 and an absolute log2 ratio value ≥ 10. To further study the biological functions and metabolic pathways of the DEGs, we performed Gene Ontology (GO)3 functional analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of these genes (Ashburner et al., 2000; Kanehisa et al., 2007).
To elucidate the genetics of the male sterile phenotype, the wild-type “FT” (P1) and the mutants msm2-1/2/3 (P2) were employed as the male parent and the female parent to generate F1 and F2 populations. All F1 individuals displayed male fertile phenotype. As disclosed by the Chi-square test, the segregation rate of the F2 population corresponded to the expected Mendelian ratio of 3:1 (fertile:sterile; Table 1). Allelism test showed that the fertility separation ratio of msm2-1, msm2-2 and msm2-3 hybrid offspring was 1:1 (fertile:sterile), which implied that mutant msm2-1, msm2-2 and msm2-3 were controlled by an allelic gene (Table 2). Therefore, these results indicated that the male sterile trait of msm2-1/2/3 mutants was controlled by a single nuclear recessive gene. From the perspective of breeding, it also demonstrated that allele gene mutation result in male sterility in the mutants msm2-1/2/3.
At the full-bloom stage, the morphology of floral organs was investigated. The stamens of msm2-1/2/3 had developed normally yet no pollen attached to anthers (Figure 1A). Other flower organs in the msm2-1/2/3 mutants were found to be slightly smaller than those in the wild-type “FT” (Figures 1B–G). Pollen viability observations also illustrated that the wild-type “FT” could generate mature and active pollen grains, whereas msm2-1/2/3 could not produce pollen grains (Figures 1H–K). These three msm2-1/2/3 mutants were allelic as evidenced by allelic test and manifested similar phenotypes under the same growth conditions; thus, some of the subsequent analyses in this study were only performed on msm2-1.

Figure 1. Phenotypic analysis of the wild-type “FT” and msm2-1/2/3 mutants. Comparison of the floral organs of the wild-type “FT” and mutant msm2-1/2/3 (from left to right). (A) Inflorescence (B) buds, (C) petals, (D) sepals, (E) stamens (long), (F) stamens (short), (J) pistils. Scale bar 2 mm analysis of pollen viability of the wild-type “FT” (H), msm2-1 (I), msm2-2 (J), and msm2-3 (K). Scale bar 150 μm.
To explore the cytological characteristics of pollen abortion in msm2-1/2/3, we examined paraffin sections prepared at different anther development stages of the wild-type “FT” and the msm2-1/2/3. The paraffin section analysis showed there was no significant difference in msm2-1/2/3 compared with the wild-type “FT” at the early stage of the anther development (Figures 2A–H). At the middle stage of anthers development, the wild-type tapetum was concentrated with less vacuolation and darker staining. However, the tapetum cells became more vacuolated and less stained in msm2-1/2/3 (Figures 2I–L). Then, the wild-type tapetum cell appeared to undergo degeneration, and the cytoplasmic constituents remained densely stained. By contrast, we discovered that the tapetum PCD was retarded in the msm2-1/2/3 mutants and the tapetum cells had vacuolated (Figures 2M–T). At the late stage of anthers development, the tapetal cells degenerated and the pollen grains matured with a round shape in the wild-type “FT.” Nevertheless, cell debris of tapetal cells and immature pollen grains remained in the msm2-1/2/3 (Figures 2U–X). Paraffin sections of anther indicated that the degradation of tapetum cell delayed occurred in mutants msm2-1/2/3.
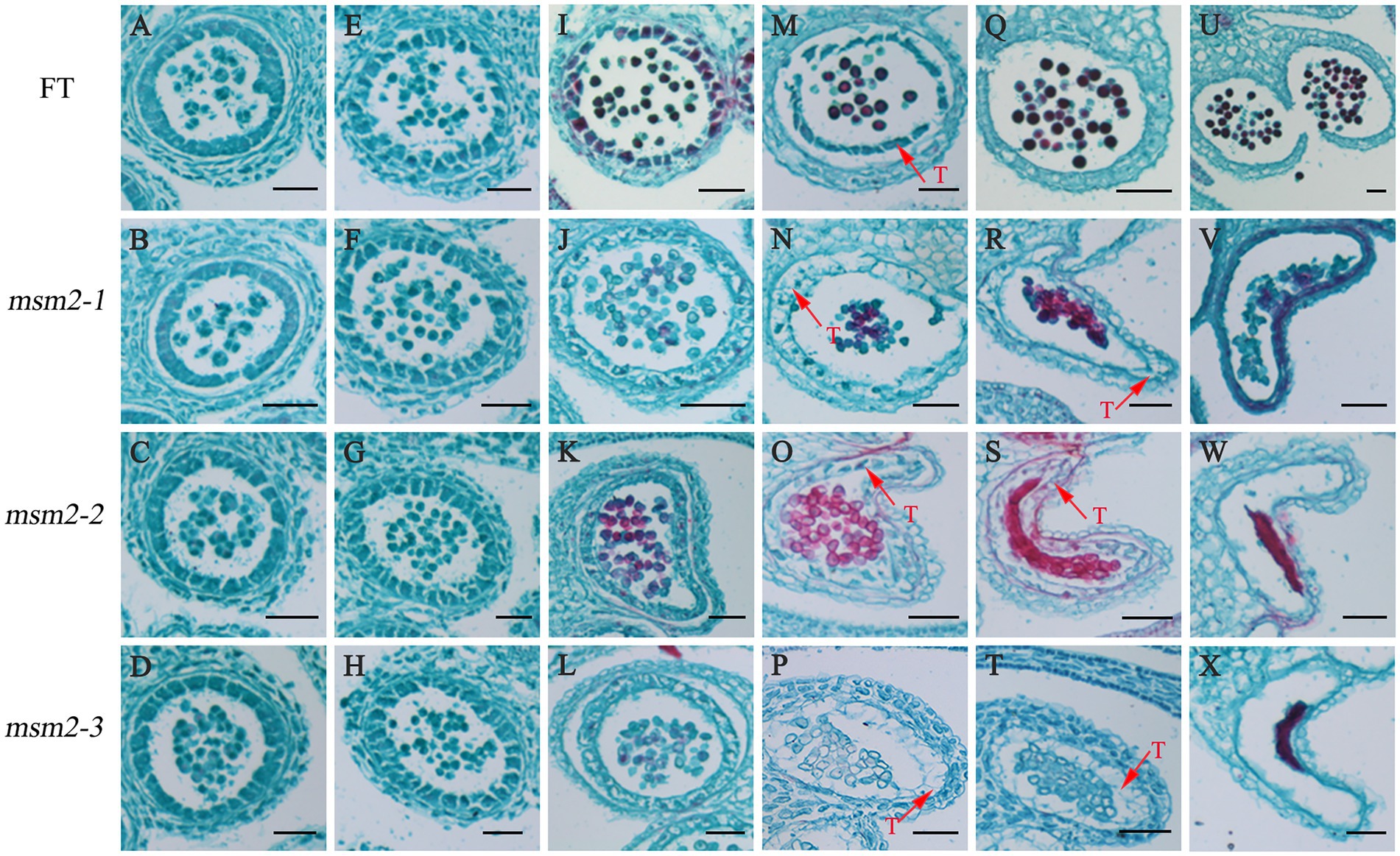
Figure 2. Anther paraffin section observation of the wild-type “FT” and msm2-1/2/3 mutants. (A–D) The stage of tapetal formation (i.e., the tetrad period), BUD1 < 1.5 mm; (E–H) The stage of the tapetum cell structure integrity and clearly demarcated (i.e., the uninucleate microspore stage), BUD2: 1.5–2.0 mm; (I–L) The stage of tapetal cytoplasm dense, BUD3: 2.0–2.5 mm; (M–P) The stage of tapetum cells begin the degradation, BUD4: 2.5–3.0 mm; (Q–T) The stage of tapetum cells complete the degradation, BUD5: 3.0–3.5 mm; (U–X) The stage of anthers dehiscence and mature pollen release, BUD6 > 3.5 mm. T tapetum. Scale bar, 50 μm.
To investigate the anther and pollen exine morphology, the fresh anthers of the wild-type “FT” and the msm2-1 were inspected by scanning electron microscopy. The SEM observation of the anther and pollen surface depicted the msm2-1 mutant had a slightly smaller anther versus the wild type at the anther maturation stage (Figures 3A,E). The cuticle ridges of the anther epidermis in wild-type “FT” are loosely distributed while the cuticles of msm2-1 are arranged relatively tightly (Figures 3B,F). The mature pollen grains of wild-type “FT” rounded surfaces having a reticular structure and the visible germinal furrow (Figures 3C,D), while pollen grain surface of mutant msm2-1 is smooth with neither reticular structure nor germination furrow, and finally degraded to pieces (Figures 3G,H).
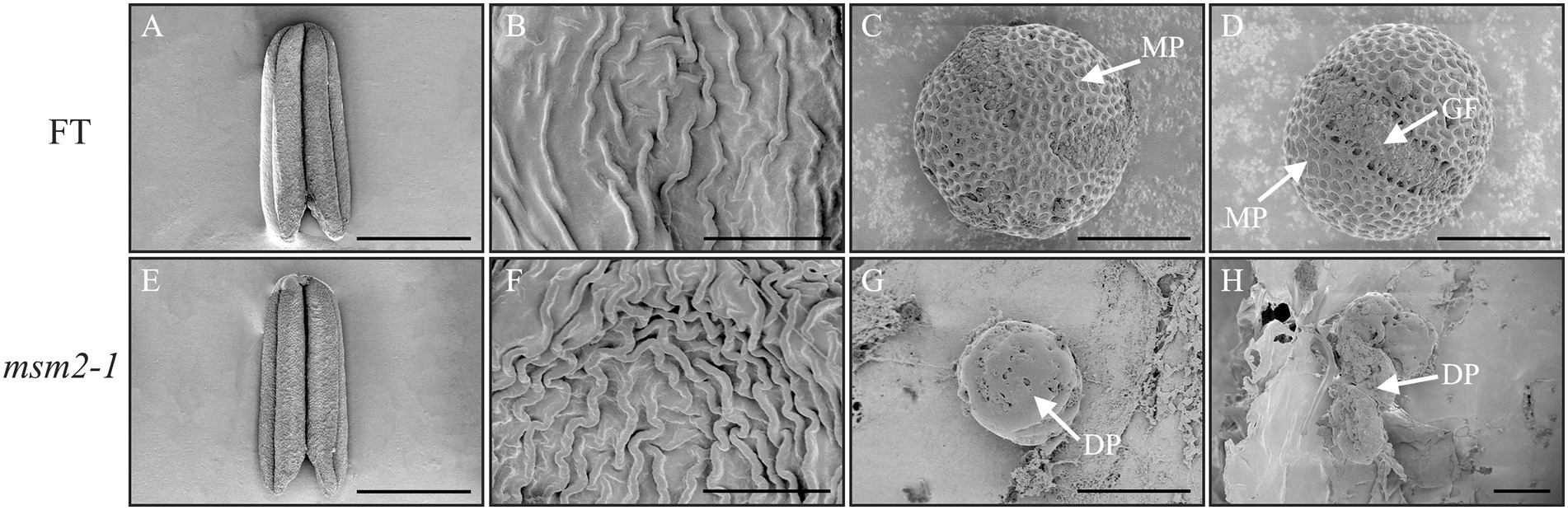
Figure 3. SEM analysis of the anthers and pollen grains in the wild-type “FT” and msm2-1 mutant. The anthers of the wild-type “FT” (A) and msm2-1 mutant (E). Scale bar, 1 mm. The anthers epidermis of the wild-type “FT” (B) and msm2-1 mutant (F), Scale bar. 5 μm. The surface on the pollen exine of the wild-type “FT” (C,D) and msm2-1 mutant (G,H). MP mature pollen grains. GF germinal furrow, DP degenerated pollen grains. Scale bar, 10 μm.
To reveal the msm2-1 pollen grain’s developmental defects in detail, TEM was conducted to observe the anther development. In the early stage of the anther development, there was no distinct difference between the wild-type “FT” and the msm2-1 mutant (Figures 4A,D,G,J). In the middle stage of the anther development, the tapetum cells of wild-type “FT” began to degrade and secrete lipid droplets and protein substances (Figure 4H). These substances were deposited on the wild-type pollen exine, which formed with the well-organized layers including the tectum, bacula, and foot layer (Figure 4B). The degradation of the tapetum was delayed in msm2-1 mutant (Figure 4K) and the skeletal structure of the pollen exine was thickened continuously (Figure 4E). At the late stage of the anther development, the pollen coat substances produced by tapetum degradation filled the sexine of the pollen exine to form the pollen coat in the wild-type “FT” (Figures 4C,I). However, the pollen of msm2-1 was surrounded by a thick abnormal pollen exine structure, while the filling of skeleton structure by pollen coats was not observed in msm2-1 (Figure 4F). In the wild type, a white pollen intine structure could be seen on the inner side of the pollen wall, while the pollen inner wall structure of the mutant msm2-1 was vague and amorphous (Figures 4C,F).
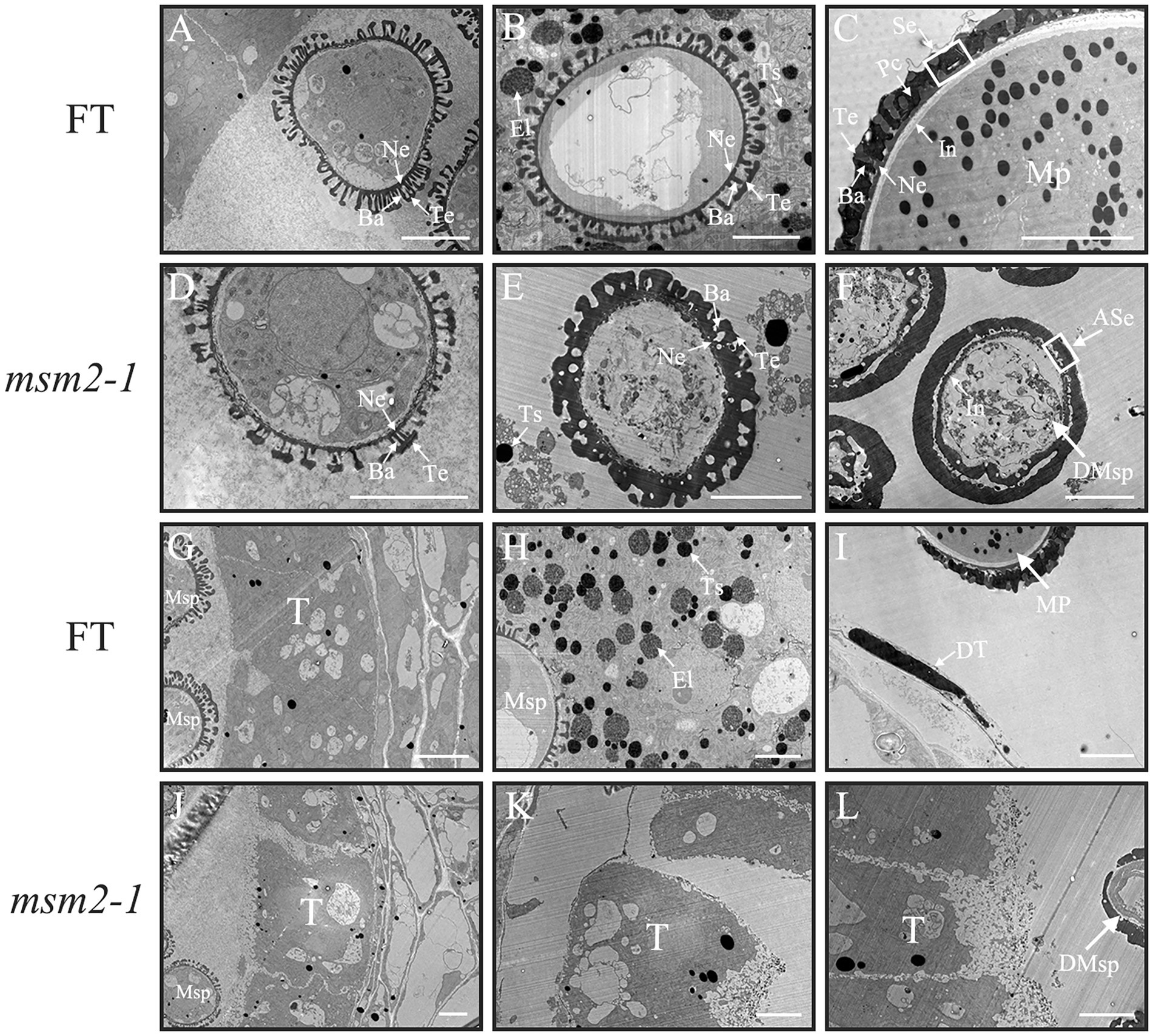
Figure 4. TEM analysis of pollen wall and tapetum development in the wild-type “FT” and msm2-1 mutant. The process of pollen exine development in wild-type “FT” and msm2-1 mutant at early (BUDS < 2.0 mm; A,D), middle (2.0 mm < BUDS <3.0 mm; B,E), late (BUDS > 3.0 mm; C,F) stage. The process of tapetum development in wild-type “FT” and msm2-1 mutant at early (BUDS <2.0 mm; G,J), middle (2.0 mm < BUDS <3.0 mm; H,K), late (BUDS >3.0 mm; I,L) stage; Scale bar, 5 μm. T, Tapetal layer; DT, Degraded tapetum; Msp, Microspores; DMsp, Degenerated microspores; MP, Mature pollen grains, Ba, Bacula; Te, Tectum; Se, Sexine; In, Intine; ASe, Abnormal sexine; El, Elaioplast; Ts, Tapetosome.
A modified MutMap method was applied to identify the mutant gene. By genome resequencing of the wild-type “FT,” msm2-1 mutant and the F2 mutant DNA pools, 93,975,658, 58,702,874 and 166,730,557 high-quality reads were obtained, respectively. Whereafter, these clean reads were aligned with the reference genome, resulting in 98.00%, 99.00% and 98.83% mapped to it, respectively. With the SNP index of 0.95 as the threshold, we mapped 1.08 Mb (14,034,596–15,116,841) candidate regions on chromosome A10 and detected three SNP mutations (SNP A10, 140,813,37; SNP A10, 143,901,24; SNP A10, 146,585,70), which were located at exons and resulted in nonsynonymous mutations (Figure 5A; Supplementary Table S8).
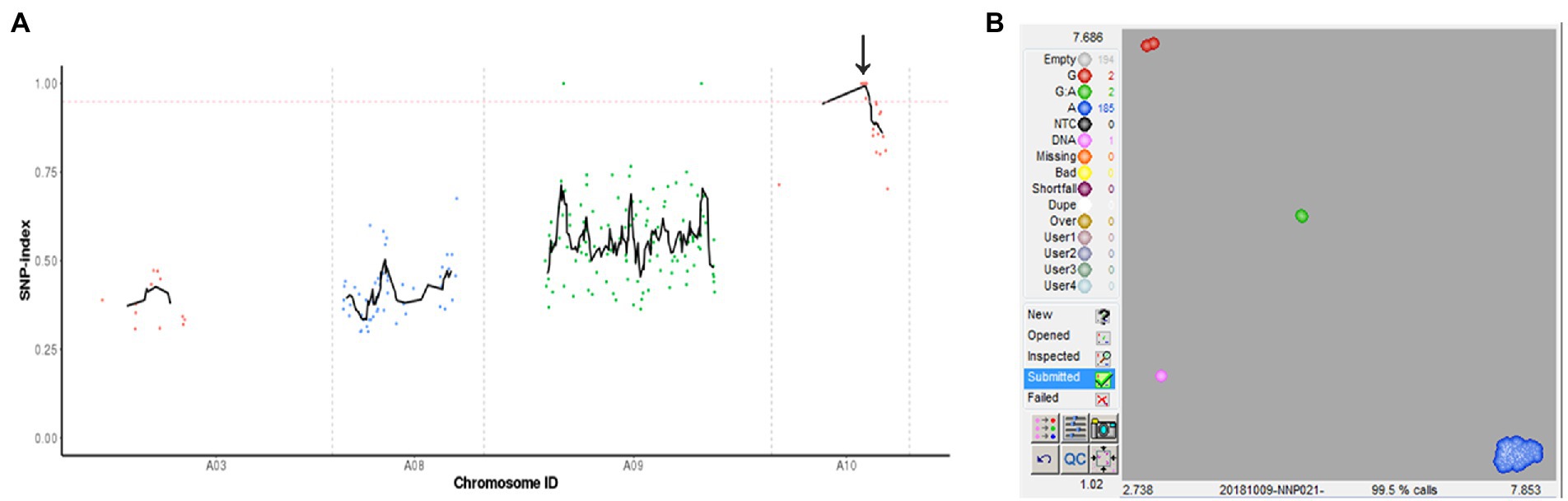
Figure 5. Identification of the msm2-1/2/3 candidate gene. (A) SNP index plot of ten chromosomes was produced by MutMap analysis. The x-axis represents the positions of the ten chromosomes and the y-axis represents the SNP index. The dotted pink line represents the index threshold (0.95). (B) The results of SNP genotyping arrays of BraA10g019050.3C.
For further determining the candidate genes, we adopted KASP technology to design primers for three candidate SNPs, and genotyping was performed in “FT,” mutant msm2-1, F1 and F2 mutant plants. The results showed that the genotypes of three SNPs in wild type, mutant and F1 generation were G: G, A: A and G: A, respectively. The gene phenotypes of BraA10g019050.3C SNP (A10:14,081,337) in F2 generation were all A: A (Figure 5B) and the SNPs of the other two genes (A10:14,390,124; A10:14,658,570) appeared six and nine heterozygous types (G: A) in the F2 generation sterile plants (Supplementary Figure S2). Therefore, BraA10g019050.3C was co-segregated with the mutant phenotype, which further confirmed that BraA10g019050.3C was the mutant gene of msm2-1. BraA10g019050.3C encodes a PHD-finger protein sharing 88% identity with the Arabidopsis MS1, which functions as a transcriptional activator that regulates tapetal development and pollen exine biosynthesis. Hence, BraA10g019050.3C was named BrMS1.
The mutants msm2-1, msm2-2 and msm2-3 are reciprocal allelic mutants, which means that the function of BrMS1 can be verified in the other two mutants. The results of the BraA10g019050.3C gene cloned from three mutants proved that at 2443 bp in exon 3 of the mutant msm2-1, the base sequence changed from G to A and the amino acid changed from tryptophan to terminator codon, leading to the termination of translation; At 1372 bp in the end of the second intron of mutant msm2-2, the base sequence changed from G to A, resulting in cutting off by mistake at the first base of third exon and giving rise to frameshift mutation; At 1887 bp in the third exon of msm2-3, the base sequence changed from G to A, and the amino acid changed from Asp to Asn (Figures 6A,B). The amino acid transformation attributed to the BrMS1 mutation likewise contributed to differences in the three-dimensional structure of the protein (Figures 6C–F). These results hinted that the mutation in BrMS1 led to the male sterile phenotype in msm2-1/2/3 and verified its function in stamen development.
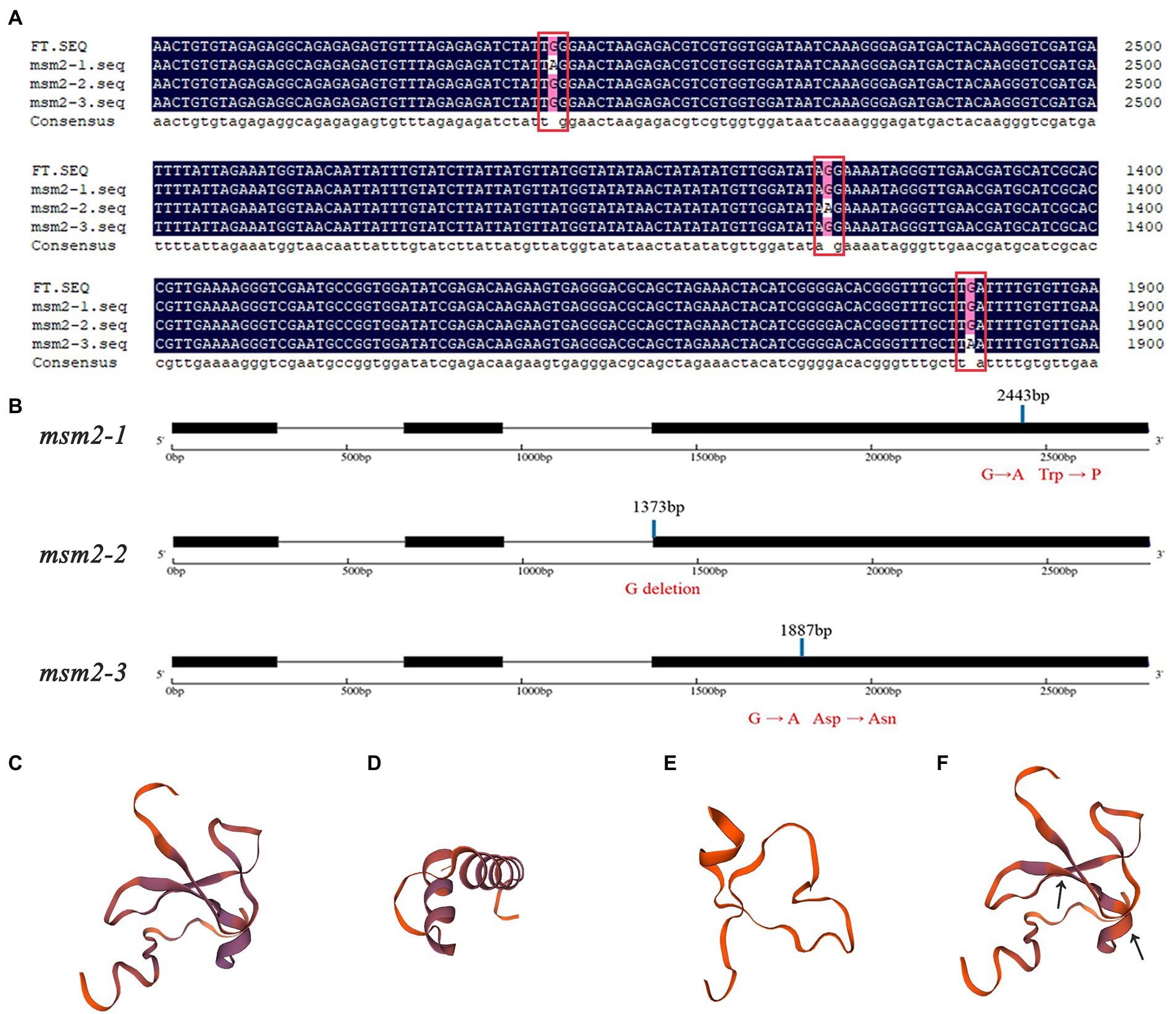
Figure 6. Sequence alignment and structure analysis of BrMS1 between the wild-type “FT” and msm2-1/2/3 mutants. Comparison of the BraA10g019050.3C nucleotide sequence (A) and structure (B) between the wild-type “FT” and msm2-1/2/3 mutants. Three-dimensional protein structure of BraA10g019050.3C in the wild-type “FT” (C), msm2-1 (D), msm2-2 (E) and msm2-3 (F).
To elucidate the evolutionary relationship between BrMS1 and its close homologs, an unrooted tree of BrMS1 and its 16 homologs from moss to flowering plants was constructed (Figure 7A). BrMS1 was clustered with many dicotyledons, and located in the same clade as AtMS1, BnMS1, BoMS1. BrMS1 has a zinc finger protein (PHD-finger) domain (Figure 7B). The PHD-finger domain is functionally conserved in a variety of plants. We compared the sequences of the PHD-finger structural regions among BrMS1 and homologous sequences in Arabidopsis, rice, maize, and wheat. The amino acid sequence of the PHD-finger domain sequence in Chinese cabbage was 90% consistent with that of AtMS1 (Figure 7C).
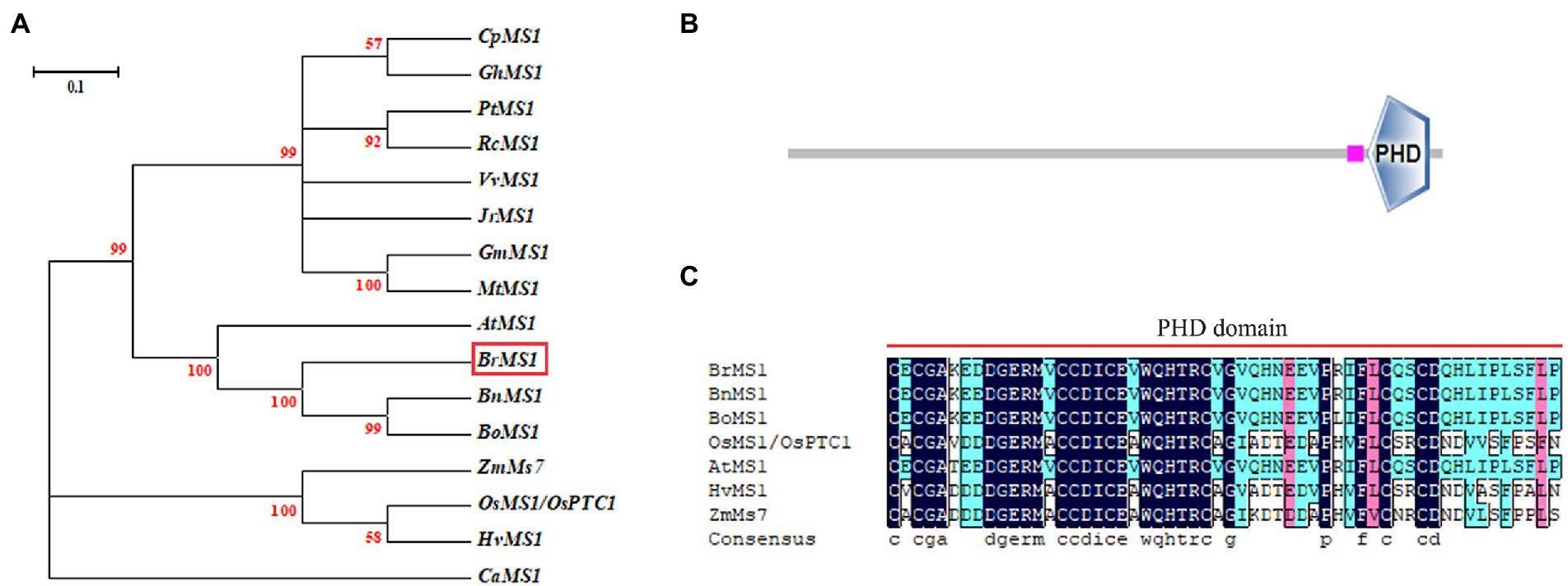
Figure 7. PHD-finger domain sequence alignment in different species and phylogenetic analysis of MS1. (A) Phylogenetic analysis of MS1 and its related proteins. An unrooted maximum likelihood tree was created by MEGA 3.1 using the MS1-related sequences from Carica papaya (Cp), Gossypium hirsutum (Gh), Populus trichocarpa (Pt), Ricinus communis (Rc), Vitis vinifera (Vv), Juglans regia (Jr), Glycine max (Gm), Medicago truncatula (Mt), Arabidopsis (At), Brassica rapa (Br), Brassica napu (Bn), Brassica oleracea (Bo), Zea mays (Zm), rice (Os), barley (Hv), Capsicum annuum (Ca). (B) Conserved domain structure of BrMS1. (C) PHD-finger domain sequence alignment. Residues with 100% similarity are shaded in black. Those with ≥75% similarity are shaded in pink. Those with ≥ 50% similarity are shaded in blue.
qRT-PCR analysis was conducted to recognize the spatial and temporal expression patterns of BrMS1 in wild-type “FT” and mutants msm2-1/2/3. The expression level of BrMS1 in the buds of the mutants msm2-1/2/3 was higher than in the wild-type “FT” line and the expression level of the BrMS1 gene in the mutant msm2-1/2 was significantly higher than that of the wild-type “FT,” while the expression level of the BrMS1 gene in msm2-3 was slightly higher than that of the wild-type “FT” (Figure 8A). The results were likewise made clear that BrMS1 was specifically expressed in stamens at stage I and stage II (i.e., the stage of tetrad to uninucleate microspore release). Afterwards, the expression level of BrMS1 declined rapidly and was almost undetectable in the mature anthers (Figures 8B,C). The expression patterns of BrMS1 in allelic mutants msm2-2/3 are same as that in mutant msm2-1 (Supplementary Figure S3).
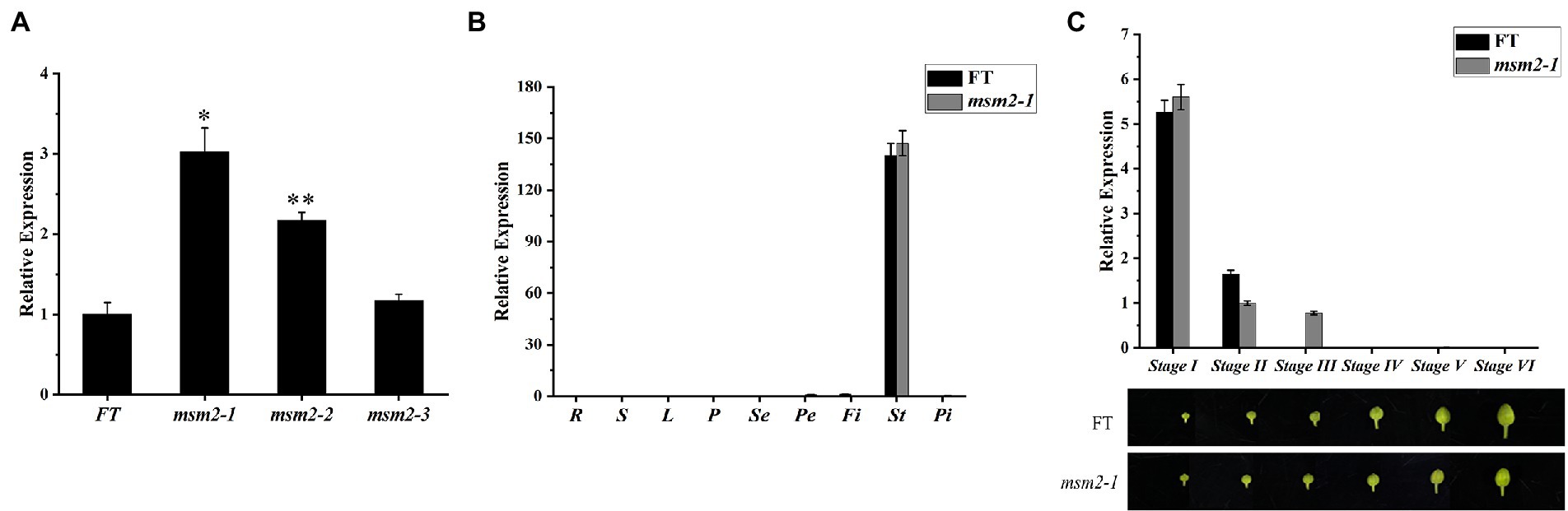
Figure 8. Expression pattern of BrMS1 between the wild-type “FT” and msm2-1/2/3 mutants. (A) Analysis of BrMS1 expression in the wild-type “FT” and the allelic mutants msm2-1/2/3. Asterisks indicate statistically significant differences determined using a t-test (*p < 0.05; **p < 0.01). (B) Expression of BrMS1 in the roots (R), stems (S), leaves (L), pods (P), sepal (Se), petal (Pe), filament (Fi), stamen (St) and pistil (Pi) of the wild-type “FT” and msm2-1 mutant. (C) Analysis of BrMS1 expression in different grades of buds in wild-type “FT” and msm2-1 mutant. The grading rules for flower buds are the same as those for paraffin sections.
Previous studies have shown that Arabidopsis MS1 functions as a transcriptional activator that regulates pollen and tapetum development (Ito and Shinozaki, 2002). Herein, to verify if BrMS1 functions as a TF in Chinese cabbage, a subcellular localization assay was initially performed. The transient expression assay showed that the GFP signal was only detected in the protoplast nucleus and indicated that BrMS1 encodes a PHD-finger protein localized in the nucleus, supporting that it functions as a TF (Figure 9A).
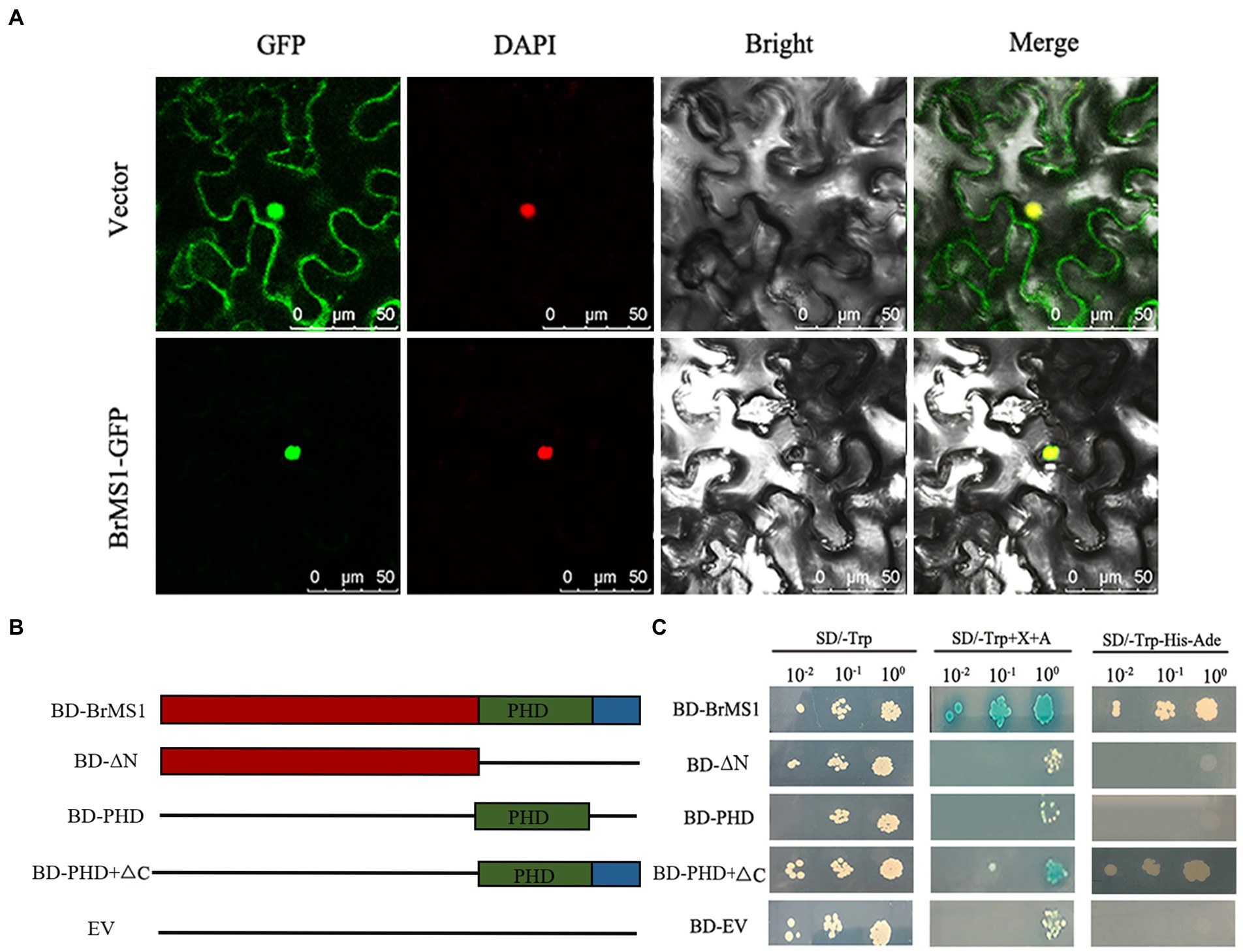
Figure 9. Subcellular localization and transcriptional activation assay of the BrMS1. (A) Subcellular localization of BrMS1 in Tobacco leaf. DAPI staining was used as a nuclear marker. Scale bar, 50 μm. (B) The schematic diagram displays various truncated constructs and pGBKT7 empty vector used for the transcriptional activation assay. (C) Transcriptional activation assay of BrMS1 in the Y2H yeast strain. BD GAL4–DNA binding domain, EV empty vector. The BD–EV used as a negative control.
For analyzing whether BrMS1 exhibits transcriptional activity activation, the full-length CDS and three truncated fragments of BrMS1 (ΔN, ΔC, PHD) were ligated into a pGBKT7 vector to generate BD-BrMS1, BD-ΔN, BD-ΔC, and BD-PHD fusion constructs (Figure 9B). The results showed all of vectors transformed yeast cells could grow in the control medium (SD/-Trp), but only BD-BrMS1 and BD-ΔC could grow in the selective medium (SD/-Trp/-His/-Ade). The findings demonstrated that BrMS1 exhibited transcriptional activation activity in yeast (Figure 9C). Subsequently, the characteristics of the deleted fragments were extensively compared. We discovered that a length of 12 C-terminal amino acid residues were required to activate activity. Taken together, these results suggested that BrMS1 was a nuclear-localized TF with a length of 12 C-terminal amino acid transcriptional activation activity.
As a TF, BrMS1 is expected to affect the anther development by regulating the expression of downstream genes. To identify genes that might be regulated by BrMS1, we performed a comparative transcriptome analysis of anther in wild-type “FT” and the msm2-1 using RNA-Seq. 4,779 DEGs were identified between the wild-type “FT” and the msm2-1, involving 1,475 upregulated genes and 3,304 downregulated genes in msm2-1 (fold change ≥ 10, false discovery rate, FDR < 0.01; Figure 10A). The KEGG analysis displayed that DEGs were enriched in multiple metabolic pathways associated with pollen development, including the pentose and glucuronate interconversions pathway (53 DEGs), phenylpropanoid metabolism pathway (52 DEGs), galactose metabolism pathway (21 DEGs) as well as cutin, suberine and wax biosynthesis pathway (15 DEGs; Figure 10B). In the pentose and glucuronate interconversions pathway, several genes related to pectin biosynthesis which affected the formation of pollen intines were identified, containing 10 pectinesterase inhibitor genes (PMEIs), 13 polygalacturonases genes (PGs) and 11 pectate lyasegenes (PLs; Figure 10C; Supplementary Table S9). Most of them were down-regulated in msm2-1 and some of these genes expressed specially in the wild-type “FT” such as PMEIs (BraA06g033760.3C and BraA08g011580.3C), PGs (BraA02g039370.3C, BraA02g039400.3C and BraA09g065620.3C), PLs (BraA09g059390.3C and BraA06g039690.3C). The phenylpropanoid pathway splits into two main branches leading to hydroxycinnamic acids and flavonoids, both of which are related to the synthesis of pollen exine (Preston et al., 2004). The expression levels of some genes involved in the synthesis of key enzymes e.g., cinnamyl alcohol dehydrogenase, feruloyl-CoA, caffeoyl-CoA were significantly changed in the mutant msm2-1, among which COMT (BraA02g024180.3C) was specifically expressed in the wild-type “FT” (Figure 10D; Supplementary Table S10). RNA-seq profiling suggested that the defect in BrMS1 affected the transcript levels of genes involved in pollen intine and exine biosynthesis metabolic pathways in msm2-1.
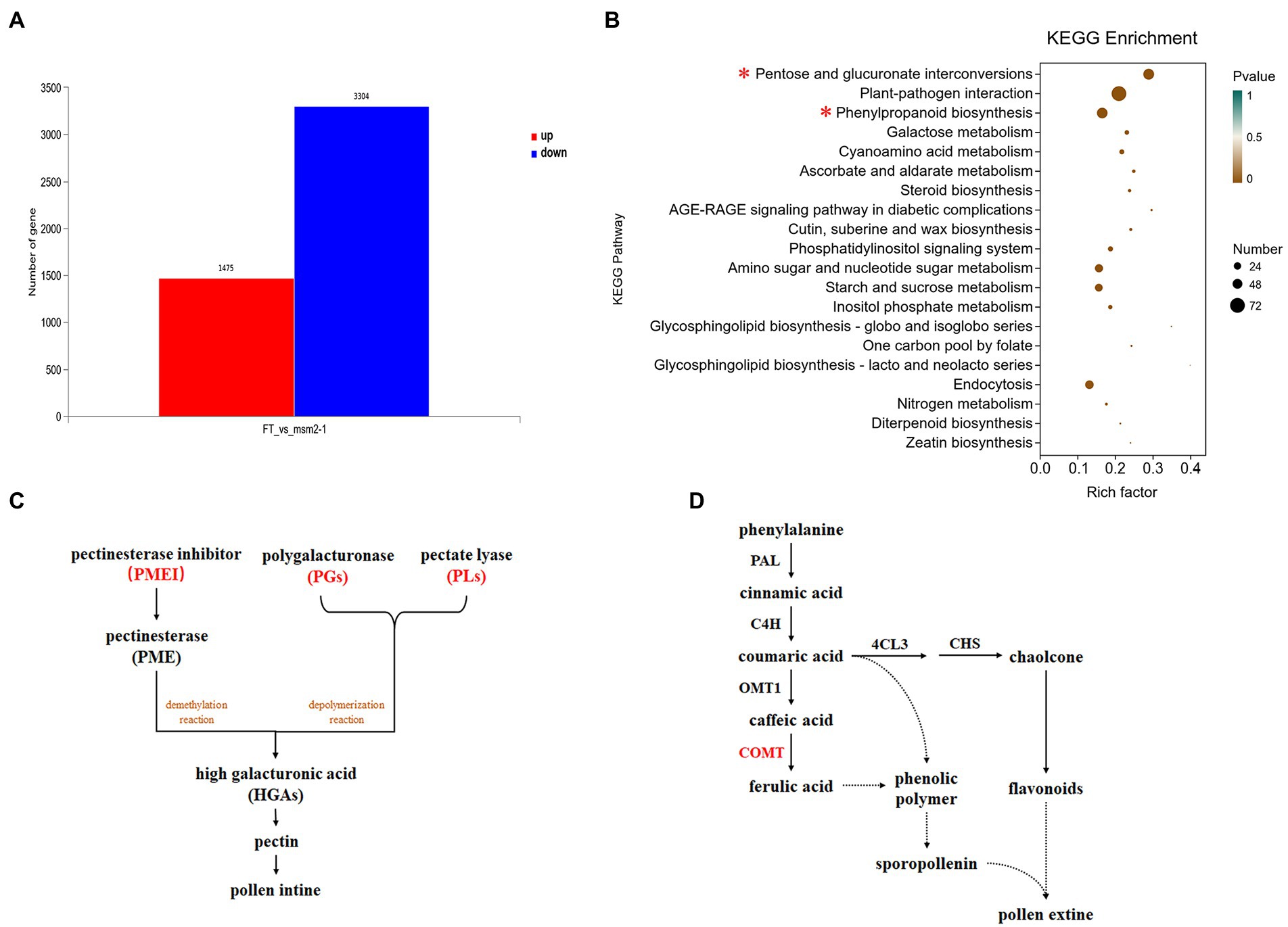
Figure 10. RNA-seq analysis between the wild-type “FT” and msm2-1 mutant. (A) The number of different expression genes (DEGs) in the anthers between the wild-type “FT” and msm2-1 mutant. (B) Kyoto Encyclopedia of Genes and Genomes enrichment results of the DEGs. (C) Schematic of pentose and glucuronate interconversions pathway related to pollen development. The red part represents the key enzymes enriched by the differentially expressed genes. (D) Schematic of phenylpropanoid metabolism pathway related to pollen development. The red part represents the key enzymes enriched by the differentially expressed genes.
In order to assess whether the defect in BrMS1 broadly affects other pollen development pathways, we compared gene expression associated with pollen development between the wild-type “FT” and the msm2-1. In Arabidopsis, AtMS1 regulates the expression of PCPs and PCLs genes, which participate in pollen coat formation (Lu et al., 2020; Zhang et al., 2021). In our study, several PCPs genes such as GRPs and EXLs exhibited a prominent reduction in the mutant msm2-1, but no significant changes were observed in the expression of PCL synthesis genes such as KCS7/5/11/21 (Supplementary Table S11). Excessive deposition of sporopollenin on the pollen primexine of the msm2-1 was observed by TEM. The results of the RNA-seq also showed that some crucial sporopollenin synthesis genes (SSGs) such as ACOS5, MS2, CYP703A2, PKSA were up-regulated in the msm2-1 (Supplementary Table S12). In the tapetum development regulatory network, MS188 is the upstream regulatory factor of MS1. The expression of MS188 genes (BraA10g013970.3C and BraA02g013010.3C) in the mutant msm2-1, was higher than in the wild-type “FT.” AMS (BraA07g004220.3C and BraA03g043400.3C), which was the upstream gene of MS188, also up-regulated expression in msm2-1 (Supplementary Table S13). The degradation of tapetum cells is regarded as a process of PCD, and papain-like cysteine proteases play a key control role in the timely degradation of tapetum cells (Liu et al., 2018; Wang et al., 2021). CEP1 is a crucial executor during tapetal PCD and that proper CEP1 expression is necessary for timely degeneration of tapetal cells and functional pollen formation in Arabidopsis thaliana (Zhang et al., 2014). BraA03g026210.3C, a homologous gene of CEP1, was up-regulated in msm2-1. We also observed the downregulation of the other two cysteine proteases genes CP1 (BraA10g004970.3C and BraA08g033980.3C) in msm2-1 compared with the wild-type “FT” (Supplementary Table S13). Overall, these findings suggested that BrMS1 might directly or indirectly regulate the expression levels of PCPs, SSGs and Cys proteases genes, further resulting in pollen abortion in msm2-1.
To test the reliability of RNA-seq data, we verified the transcript levels of four PCP genes and three cysteine protease genes between the wild-type “FT” and the mutant msm2-1 by qRT-PCR, and the analyses showed that their mRNA levels were all significantly reduced in msm2-1, apart from CEP1, which was up-regulated (Figure 11). These findings are consistent with the results from the RNA-seq analyses.
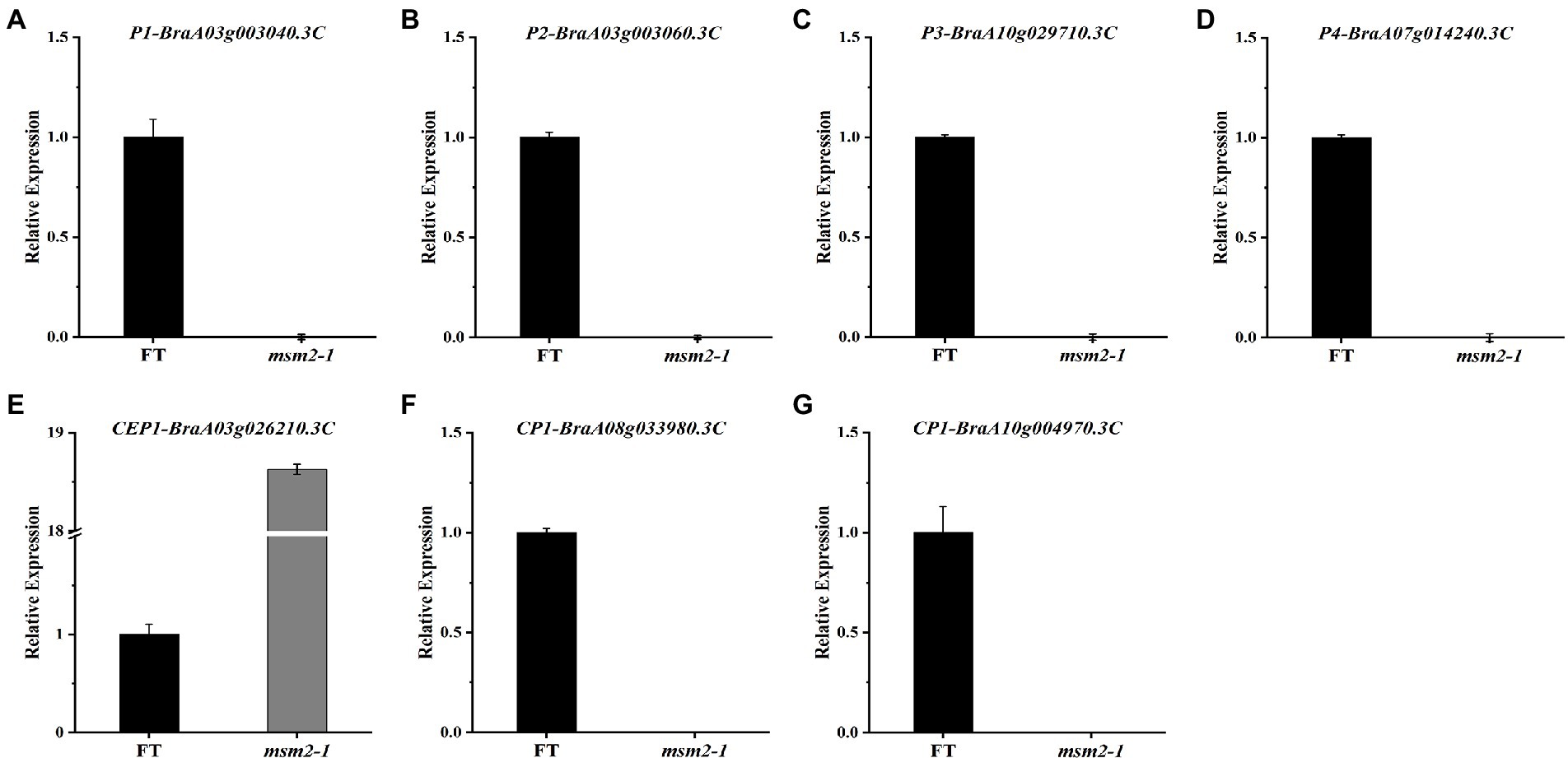
Figure 11. The quantitative reverse transcription polymerase chain reaction analysis of DEGs between the wild-type “FT” and msm2-1 mutant. (A–D) The expression level of pollen coat proteins in anthers between the wild-type “FT” and msm2-1 mutant. (E–G) The expression level of papain-like cysteine proteins in anthers between the wild-type “FT” and msm2-1 mutant.
The application of male sterility lines is an effective approach to commercial hybrid seed production and heterosis utilization in Chinese cabbage. Moreover, male sterility mutant is an important material to study pollen development and nuclear-cytoplasmic interactions. The creation of male sterile materials through artificial mutation has become a crucial mean of male sterility research. Discovery and research on male sterile mutants also had been a hot topic in research in various species. In this study, we cloned firstly the male sterility gene BrMS1 in Chinese cabbage by forwarding genetics strategy and revealed the critical role of BrMS1 in pollen development. In addition, the male sterile mutants we discovered have the characteristics of complete abortion, genetical stability and no concomitant adverse traits, which can be adopted as excellent sources of male sterility for Chinese cabbage hybrid seed production.
Tapetum timely degradation is essential for the pollen development in plants. The forms of abnormal tapetal development led to male sterile are diverse (Sanders et al., 1999; Zhang et al., 2011; Zhu et al., 2011; Zhang and Yang, 2014). Premature tapetum cell death caused the male sterile in rice osmyb80 and bm1 mutants (Pan et al., 2020; Xiang et al., 2021). Loss of RGAT1 function also induced the premature degeneration of tapetal cells and pollen abortion in Arabidopsis (Qian et al., 2021). In addition, delayed tapetum degradation brought out defective mature pollen in rice osmyb103 and sts1 mutants (Lei et al., 2022; Yuan et al., 2022). In the present study, delayed tapetum degeneration is a hallmark defect in msm2-1/2/3, but the tapetum cells enlargement was not observed in msm2-1/2/3 (Figures 2M–X). This phenotype was distinct from other tapetal PCD delayed mutants that have been described, such as gamyb and tdr mutants, which exhibited tapetal defects involving tapetum cell enlargement (Kaneko et al., 2004; Li et al., 2006), These findings suggested that the BrMS1 mutation in msm-1/2/3 mutants induced the delayed degradation of tapetum, but did not affect the morphology of tapetum cells.
Known tapetal PCD mutants also showed different types of pollen exine patterning. The manifestations of pollen exine abnormal in msm2-1 mutant were different from other MS1 homologous mutants. In the Arabidopsis ms1 mutant, exine formation is abnormal, with initial primexine deposition but with only limited sporopollenin deposition (Ito and Shinozaki, 2002; Ariizumi et al., 2003; Vizcay-Barrena and Wilson, 2006). In the rice, the osms1/ptc1 exines exhibited a two-layer structure, the sexine was abnormal because of the missing or decrease bacula (Yang et al., 2019b). In the maize, ubisch bodies were not enlarged in mutant ms7-6007 and exine was much thinner than WT (An et al., 2020). In this study, the msm2-1 mutant could form a complete pollen exine structure in the early stage of pollen development yet not observed the PCPs filled the pollen exine at the late stage. The excessive deposition of sporopollenin resulted in the pollen exine structure continuously thickened and formed a doughnut-like structure in the late stage of pollen development (Figures 4B,C,E,F). The SEM observation of pollen also exhibited a lack of elaborate patterning, giving rise to a smooth surface in msm2-1 mutant (Figures 3C,D,G,H). In addition, abnormal morphology of pollen inner wall was observed in msm2-1. Taken together, loss of function of BrMS1 results in delayed tapetal degeneration and pollen wall abnormal formation, and these developmental defects ultimately cause complete male sterility in the Chinese cabbage.
To decipher the molecular mechanism of the causal gene, MutMap and KASP analyses revealed that BrMS1 (BraA10g019050.3C) encoded a PHD-finger transcriptional factor, was the causal gene in mutant msm2-1 and confirmed function by BrMS1 cloning in the allelic mutants msm2-2/3 (Figures 5, 6). A previous study has shown that the PHD-finger motif was required for MS1 function in Arabidopsis (Ito et al., 2007). The mutation of lacking the PHD-finger motif in both OsMS1 and PTC1, caused male sterility in rice (Ito et al., 2007; Li et al., 2011; Yang et al., 2019b). Here, the base variations from G to A at 2443th of msm2-1, 1373th of msm2-2 and 1887th of msm2-3 made the PHD-finger domain abnormal in the allelic mutants and eventually led to pollen abortion (Figures 6A,B). Therefore, this demonstrates to some extent that the PHD-finger motif is indispensable for the development of anthers in Chinese cabbage.
Previous studies have implied MS1/PTC1 could down-regulate their own expression in wild-type plants, and conjectured that MS1 down-regulated its expression may due to conformational problem with the truncated mutant polypeptides and/or that the PHD-finger motif binding to its own promoter (Yang et al., 2007; Li et al., 2011). In this paper, qRT-PCR analysis revealed that BrMS1 transcripts were increased to varying degrees in the msm2-1/2/3 mutant, implying that self-regulatory based upon a normal gene functional could limit the expression of BrMS1 in Chinese cabbage (Figure 8A). The expression of BrMS1 in msm2-1/2 allele mutants showed a greater expression compared with in msm2-3, we speculated that this was on account of the smaller difference in the 3D structure of the BrMS1 gene in msm2-3 versus the wild type “FT” (Figures 6C–F).
To gain a better understanding of how BrMS1 affects pollen development, we produced transcriptomes of the wild-type “FT” and the msm2-1 mutant to identify potential genes and pathways involved in pollen development. The pollen coat rich in lipids and proteins plays an important role in the attachment and recognition of the pollen to the stigma (Kobayashi et al., 2021). The pollen coat is necessary for fertility, as pollen coat mutants in rice lead to the humidity-sensitive male sterility (Xue et al., 2018; Yu et al., 2019; Chen et al., 2020). In this study, RNA-seq profiling demonstrated that pollen coat proteins GRPs and EXLs significantly down-regulated in msm2-1 compared with the wild-type “FT” (Supplementary Table S11). Accumulating reports implicated that papain-like cysteine proteases play a role in tapetal PCD, our results indicated that cysteine proteases BrCEP1 (BraA03g026210.3C) was up-regulated and the other two cysteine proteases genes BrCP1 (BraA10g004970.3C and BraA08g033980.3C) were down-regulated in msm2-1 (Supplementary Table S13). These findings, to some extent, revealed a potential cause of the delayed degradation of tapetum in the msm2-1 mutant. KEGG pathway analysis indicated that differentially expressed genes were significantly enriched in the pentose and glucuronate interconversions pathway. In this pathway, most DEGs involved in pectin biosynthesis, the main component of the pollen intine. HGAs are the most abundant polymer of pectin (Wolf et al., 2009). The HGA-modifying process can be strictly regulated by multiple cell wall enzymes. It has been demonstrated that the demethylesterification of HGAs could be spatiotemporally controlled by pectin methylesterases (PMEs) and PMEs activity might be regulated by a specific proteinaceous PME inhibitors (PMEIs; Camardella et al., 2000). Polygalacturonases (PGs) and pectin lyases (PLs) can depolymerize HGAs (Wakabayashi et al., 2003). Most of PMEIs, PGs and PLs were down-regulated in msm2-1 (Figure 10C; Supplementary Table S9). Thus, we speculated that the defection of BrMS1 regulated the development of pollen intine by affecting the expression of genes relevant to pectin biosynthesis in msm2-1 mutant. However, the specific way in which BrMS1 regulates these fertility-related genes will require further investigation.
Sporopollenin is the main component of the pollen exine and that its biosynthesis is closely related to fatty acid and phenylpropanoid metabolism (Liu and Fan, 2013; Shi et al., 2015). In this study, DEGs was likewise enriched in the phenylpropanoid metabolism pathway. The expression of some key enzymes in this pathway were significantly changed in msm2-1 (Figure 10D; Supplementary Table S10). In addition, several SSGs were up-regulated expression in mutant msm2-1. This finding was consistent with the excessive deposition of sporopollenin of the msm2-1 genotype (Figure 4; Supplementary Table S12). As we know that MS188 directly regulates sporopollenin biosynthesis genes, and MS188 is an upstream regulator of MS1 in the tapetum regulatory network (DYT1-TDF1-AMS-MS188-MS1; Xiong et al., 2016; Wang et al., 2018). Herein, the expression level of MS188 was up-regulated (Supplementary Table S13). In addition, several reports have shown that MS1 could negatively regulate AMS expression, possibly through chromatin remodeling, or indirect protein degradation (Ferguson et al., 2017). This complex network of feedback loops is critical for the anther development. We suspected that mutation in BrMS1 broke the balance of this negative feedback mechanism, resulting in the up-regulation expression of AMS and its downstream target gene MS188. While the SSGs directly regulated by the MS188 were also up-regulated, giving rise to pollen exine abnormal in the mutant msm2-1. BrMS1 as a TF could not only regulate the expression of downstream potential pollen development-related genes, but also effect on the expression of upstream genes (Figure 12). Hence, BrMS1 plays an important role in maintaining the stable and timely expression of various TFs in the tapetum regulatory network.
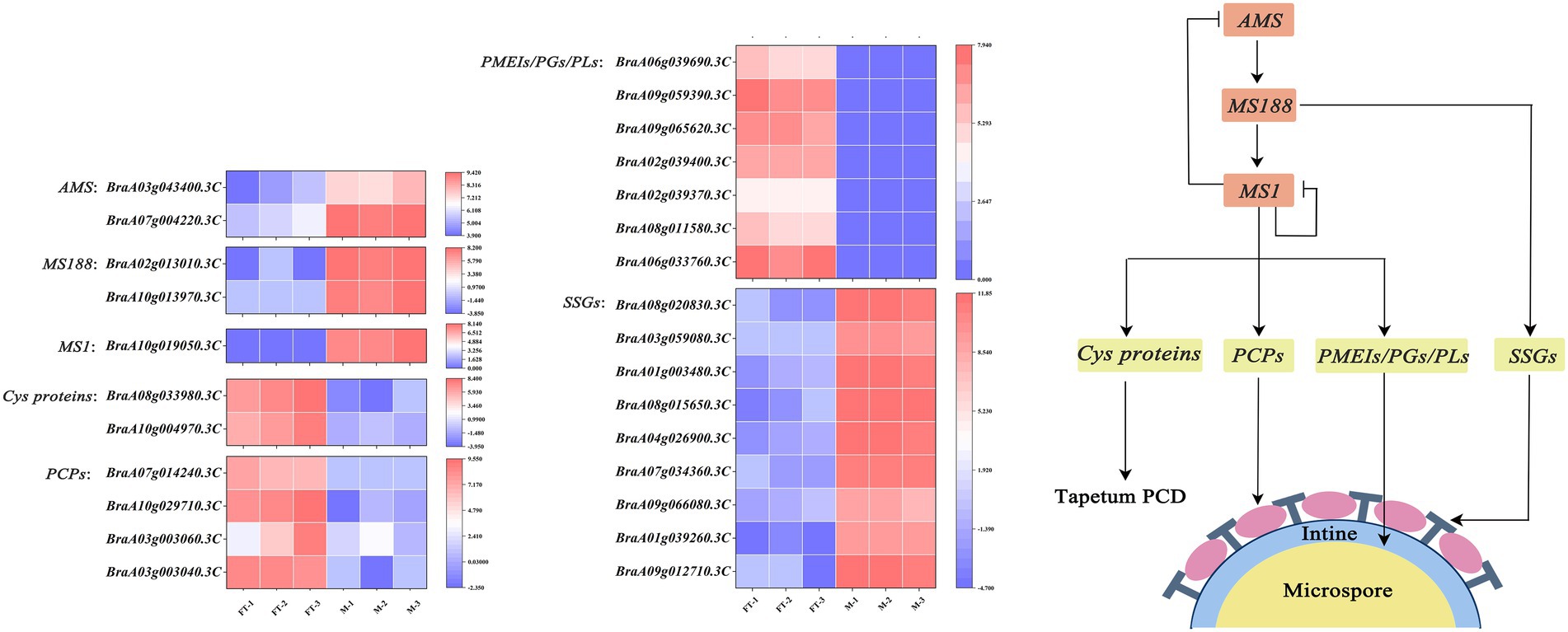
Figure 12. Proposed working model for BrMS1 regulation of male fertility in Chinese cabbage. MS1 which encodes a PHD-finger transcription factor could regulate the tapetum programmed cell death by influencing the transcription level of Cys proteins. MS1 functions in pollen coats and pollen intine formation by mediating the expression of related genes in anthers. In addition, MS1 could negatively regulate the transcription level of itself and the upstream regulator AMS, which in turn affects the expression of SSGs to control the formation of pollen exine. MS1 plays an important role as a downstream regulator in the male fertility regulatory network.
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/sra//PRJNA856064.
Data analysis and writing of the manuscript were performed by SD. Study conception and design were performed by SH and HF. JZ, YZ, FS, and GS carried out the experiments. BF took charge of English language editing. All authors contributed to the article and approved the submitted version.
The research was supported by the National Natural Science Foundation of China (grant no. 31730082).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.992391/full#supplementary-material
An, X. L., Ba, B., Duan, M. J., Dong, Z. Y., Liu, R. G., Yuan, D. Y., et al. (2020). Molecular regulation of ZmMs7 required for maize male fertility and development of a dominant male-sterility system in multiple species. Proc. Natl. Acad. Sci. U. S. A. 117, 23499–23509. doi: 10.1073/pnas.2010255117
Ariizumi, T., Hatakeyama, K., Hinata, K., Sato, S., Kato, T., Tabata, S., et al. (2003). A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol. Biol. 53, 107–116. doi: 10.1023/B:PLAN.0000009269.97773.70
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Camardella, L., Carratore, V., Ciardiello, M., Servillo, L., Balestrieri, C., and Giovane, A. (2000). Kiwi protein inhibitor of pectin methylesterase amino-acid sequence and structural importance of two disulfide bridges. Eur. J. Biochem. 267, 4561–4565. doi: 10.1046/j.1432-1327.2000.01510.x
Chen, H., Zhang, Z., Ni, E., Lin, J., Peng, G., Huang, J., et al. (2020). HMS1 interacts with HMS1I to regulate very-long-chain fatty acid biosynthesis and the humidity-sensitive genic male sterility in rice (Oryza sativa). New Phytol. 225, 2077–2093. doi: 10.1111/nph.16288
Fan, Y. R., and Zhang, Q. F. (2018). Genetic and molecular characterization of photoperiod and thermo-sensitive male sterility in rice. Plant Reprod. 31, 3–14. doi: 10.1007/s00497-017-0310-5
Ferguson, A. C., Pearce, S., Band, L. R., Yang, C. Y., Ferjentsikova, I., King, J., et al. (2017). Biphasic regulation of the transcription factor ABORTED MICROSPORES (AMS) is essential for tapetum and pollen development in Arabidopsis. New Phytol. 213, 778–790. doi: 10.1111/nph.14200
Gao, Y., Huang, S. N., Qu, G. Y., Fu, W., Zhang, M. D., Liu, Z. Y., et al. (2020). The mutation of ent-kaurene synthase, a key enzyme involved in gibberellin biosynthesis, confers a non-heading phenotype to Chinese cabbage (Brassica rapa L. ssp. pekinensis). Hortic Res. 7, 178. doi: 10.1038/s41438-020-00399-6
Gao, Y., Qu, G., Huang, S., Liu, Z., Zhang, M., Fu, W., et al. (2022). Comparison between germinated seed and isolated microspore EMS mutagenesis in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Horticulturae 8, 232. doi: 10.3390/horticulturae8030232
Hafidh, S., Fila, J., and Honys, D. (2016). Male gametophyte development and function in angiosperms: a general concept. Plant Reprod. 29, 31–51. doi: 10.1007/s00497-015-0272-4
Huang, S., Liu, Z., Li, C., Yao, R., Li, D., Hou, L., et al. (2017). Transcriptome analysis of a female-sterile mutant (fsm) in Chinese cabbage (Brassica campestris ssp. pekinensis). Front Plant Sci. 8, 546. doi: 10.3389/fpls.2017.00546
Huang, S., Peng, S., Liu, Z., Li, C., Tan, C., Yao, R., et al. (2020). Investigation of the genes associated with a male sterility mutant (msm) in Chinese cabbage (Brassica campestris ssp. pekinensis) using RNA-Seq. Mol. Gen. Genomics. 295, 233–249. doi: 10.1007/s00438-019-01618-z
Ito, T., Nagata, N., Yoshiba, Y., Ohme-Takagi, M., Ma, H., and Shinozaki, K. (2007). Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19, 3549–3562. doi: 10.1105/tpc.107.054536
Ito, T., and Shinozaki, K. (2002). The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol. 43, 1285–1292. doi: 10.1093/pcp/pcf154
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2007). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Kaneko, M., Inukai, Y., Ueguchi-Tanaka, M., Itoh, H., Izawa, T., Kobayashi, Y., et al. (2004). Loss-of-function mutations of the rice GAMYB gene impair α-amylase expression in aleurone and flower development. Plant Cell 16, 33–44. doi: 10.1105/tpc.017327
Kim, Y. J., and Zhang, D. B. (2018). Molecular control of male fertility for crop hybrid breeding. Trends Plant Sci. 23, 53–65. doi: 10.1016/j.tplants.2017.10.001
Kobayashi, K., Akita, K., Suzuki, M., Ohta, D., and Nagata, N. J. P. B. (2021). Fertile Arabidopsis cyp 704b1 mutant, defective in sporopollenin biosynthesis, has a normal pollen coat and lipidic organelles in the tapetum. Plant Biotechnol. 38, 109–116. doi: 10.5511/plantbiotechnology.20.1214b
Larkin, M., Blackshields, G., Brown, N., Chenna, R., McGettigan, P., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lei, T., Zhang, L. S., Feng, P., Liu, Y., Yin, W. Z., Shang, L. N., et al. (2022). OsMYB103 is essential for tapetum degradation in rice. Theor. Appl. Genet. 135, 929–945. doi: 10.1007/s00122-021-04007-6
Li, H., Yuan, Z., Vizcay-Barrena, G., Yang, C. Y., Liang, W. Q., Zong, J., et al. (2011). PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein That is required for Tapetal CELL death and pollen development in Rice. Plant Physiol. 156, 615–630. doi: 10.1104/pp.111.175760
Li, N., Zhang, D.-S., Liu, H.-S., Yin, C.-S., Li, X. X., Liang, W. Q., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18, 2999–3014. doi: 10.1105/tpc.106.044107
Liang, J., Ma, Y., Wu, J., Cheng, F., Liu, B., and Wang, X. (2017). Map-based cloning of the dominant genic male sterile Ms-cd1 gene in cabbage (Brassica oleracea). Theor. Appl. Genet. 130, 71–79. doi: 10.1007/s00122-016-2792-1
Lin, S., Dong, H., Zhang, F., Qiu, L., Wang, F. Z., Cao, J. S., et al. (2014). BcMF8, a putative arabinogalactan protein-encoding gene, contributes to pollen wall development, aperture formation and pollen tube growth in Brassica campestris. Ann. Bot. 113, 777–788. doi: 10.1093/aob/mct315
Lin, T.-K., Lin, Y.-P., and Lin, S.-F. (2020). Genetic analysis and fine mapping of a spontaneously mutated male sterility gene in Brassica rapa ssp. chinensis. G3 10, 1309–1318. doi: 10.1534/g3.120.401091
Liu, L., and Fan, X. D. (2013). Tapetum: regulation and role in sporopollenin biosynthesis in Arabidopsis. Plant Mol. Biol. 83, 165–175. doi: 10.1007/s11103-013-0085-5
Liu, H., Hu, M., Wang, Q., Cheng, L., and Zhang, Z. (2018). Role of papain-like cysteine proteases in plant development. Front. Plant Sci. 9:1717. doi: 10.3389/fpls.2018.01717
Liu, Q., Lan, Y., Wen, C., Zhao, H., Wang, J., and Wang, Y. (2016). Transcriptome sequencing analyses between the cytoplasmic male sterile line and its maintainer line in welsh onion (Allium fistulosum L.). Int. J. Mol. Sci. 17:1058. doi: 10.3390/ijms17071058
Lu, J. Y., Xiong, S. X., Yin, W. Z., Teng, X. D., Lou, Y., Zhu, J., et al. (2020). MS1, a direct target of MS188, regulates the expression of key sporophytic pollen coat protein genes in Arabidopsis. J. Exp. Bot. 71, 4877–4889. doi: 10.1093/jxb/eraa219
Ma, C. F., Zhu, C. Z., Zheng, M., Liu, M. C., Zhang, D. J., Liu, B. L., et al. (2019). CRISPR/Cas 9-mediated multiple gene editing in Brassica oleracea var. capitata using the endogenous tRNA-processing system. Hortic Res. 6, 20. doi: 10.1038/s41438-018-0107-1
Pan, X., Yan, W., Chang, Z., Xu, Y., Luo, M., Xu, C., et al. (2020). OsMYB80 regulates anther development and pollen fertility by targeting multiple biological pathways. Plant Cell Physiol. 61, 988–1004. doi: 10.1093/pcp/pcaa025
Parish, R. W., and Li, S. F. (2010). Death of a tapetum: a programme of developmental altruism. Plant Sci. 178, 73–89. doi: 10.1016/j.plantsci.2009.11.001
Preston, J., Wheeler, J., Heazlewood, J., Li, S. F., and Parish, R. W. J. T. P. J. (2004). AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 40, 979–995. doi: 10.1111/j.1365-313X.2004.02280.x
Qian, Q., Yang, Y., Zhang, W., Hu, Y., Li, Y., Yu, H., et al. (2021). A novel Arabidopsis gene RGAT1 is required for GA-mediated tapetum and pollen development. New Phytol. 231, 137–151. doi: 10.1111/nph.17314
Sanders, P. M., Bui, A. Q., Weterings, K., McIntire, K. N., Hsu, Y.-C., Lee, P. Y., et al. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11, 297–322. doi: 10.1007/s004970050158
Sharma, P., Kaur, M., Sharma, A., and Bhardwaj, N. (2021). Breeding vegetables for protected cultivation: A review. Himachal J. Agric. Res. 47, 1–17.
Shi, J., Cui, M., Yang, L., Kim, Y. J., and Zhang, D. (2015). Genetic and biochemical mechanisms of Pollen Wall development. Trends Plant Sci. 20, 741–753. doi: 10.1016/j.tplants.2015.07.010
Singh, S., Dey, S. S., Bhatia, R., Kumar, R., and Behera, T. K. (2019). Current understanding of male sterility systems in vegetable brassicas and their exploitation in hybrid breeding. Plant Reprod. 32, 231–256. doi: 10.1007/s00497-019-00371-y
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tan, C., Liu, Z., Huang, S., and Feng, H. (2019). Mapping of the male sterile mutant gene ftms in Brassica rapa L. ssp. pekinensis via BSR-Seq combined with whole-genome resequencing. Theor. Appl. Genet. 132, 355–370. doi: 10.1007/s00122-018-3223-2
Vizcay-Barrena, G., and Wilson, Z. (2006). Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J. Exp. Bot. 57, 2709–2717. doi: 10.1093/jxb/erl032
Wakabayashi, K., Hoson, T., and Huber, D. J. (2003). Methyl de-esterification as a major factor regulating the extent of pectin depolymerization during fruit ripening: a comparison of the action of avocado (Persea americana) and tomato (Lycopersicon esculentum) polygalacturonases. J. Plant Physiol. 160, 667–673. doi: 10.1078/0176-1617-00951
Wan, X. Y., Wu, S. W., Li, Z. W., An, X. L., and Tian, Y. H. (2020). Lipid metabolism: critical roles in male fertility and other aspects of reproductive development in plants. Mol. Plant 13, 955–983. doi: 10.1016/j.molp.2020.05.009
Wan, X. Y., Wu, S. W., Li, Z. W., Dong, Z. Y., An, X. L., Ma, B., et al. (2019). Maize genic male-sterility genes and their applications in hybrid breeding: progress and perspectives. Mol. Plant 12, 321–342. doi: 10.1016/j.molp.2019.01.014
Wang, K., Guo, Z. L., Zhou, W. T., Zhang, C., Zhang, Z. Y., Lou, Y., et al. (2018). The regulation of sporopollenin biosynthesis genes for rapid Pollen Wall formation. Plant Physiol. 178, 283–294. doi: 10.1104/pp.18.00219
Wang, Y., Ye, H., Bai, J., and Ren, F. (2021). The regulatory framework of developmentally programmed cell death in floral organs: a review. Plant Physiol. Biochem. 158, 103–112. doi: 10.1016/j.plaphy.2020.11.052
Wang, N., Zhang, Y., Huang, S., Liu, Z., Li, C., and Feng, H. (2020). Defect in Brnym1, a magnesium-dechelatase protein, causes a stay-green phenotype in an EMS-mutagenized Chinese cabbage (Brassica campestris L. ssp. pekinensis) line. Hortic Res. 7, 8. doi: 10.1038/s41438-019-0223-6
Wolf, S., Mouille, G., and Pelloux, J. (2009). Homogalacturonan methyl-esterification and plant development. Mol. Plant 2, 851–860. doi: 10.1093/mp/ssp066
Wu, H.-M., and Cheung, A. Y. (2000). Programmed cell death in plant reproduction. Plant Mol. Biol. 44, 267–281. doi: 10.1023/A:1026536324081
Xi, X., Wei, K. Y., Gao, B. Z., Liu, J. H., Liang, J. L., Cheng, F., et al. (2018). BrFLC5: a weak regulator of flowering time in Brassica rapa. Theor. Appl. Genet. 131, 2107–2116. doi: 10.1007/s00122-018-3139-x
Xiang, X., Sun, L., Yu, P., Yang, Z., Zhang, P., Zhang, Y., et al. (2021). The MYB transcription factor Baymax1 plays a critical role in rice male fertility. Theor. Appl. Genet. 134, 453–471. doi: 10.1007/s00122-020-03706-w
Xiong, S. X., Lu, J. Y., Lou, Y., Teng, X. D., Gu, J. N., Zhang, C., et al. (2016). The transcription factors MS 188 and AMS form a complex to activate the expression of CYP 703A2 for sporopollenin biosynthesis in Arabidopsis thaliana. Plant J. 88, 936–946. doi: 10.1111/tpj.13284
Xue, Z., Xu, X., Zhou, Y., Wang, X., Zhang, Y., Liu, D., et al. (2018). Deficiency of a triterpene pathway results in humidity-sensitive genic male sterility in rice. Nat. Commun. 9, 1–10. doi: 10.1038/s41467-018-03048-8
Yang, Z. F., Liu, L., Sun, L. P., Yu, P., Zhang, P. P., Abbas, A., et al. (2019a). OsMS1 functions as a transcriptional activator to regulate programmed tapetum development and pollen exine formation in rice. Plant Mol. Biol. 99, 175–191. doi: 10.1007/s11103-018-0811-0
Yang, Z., Sun, L., Zhang, P., Zhang, Y., Yu, P., Liu, L., et al. (2019b). TDR INTERACTING PROTEIN 3, encoding a PHD-finger transcription factor, regulates Ubisch bodies and pollen wall formation in rice. Plant J. 99, 844–861. doi: 10.1111/tpj.14365
Yang, C., Vizcay-Barrena, G., Conner, K., and Wilson, Z. A. (2007). MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19, 3530–3548. doi: 10.1105/tpc.107.054981
Yoo, S., Cho, Y., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Yu, B., Liu, L., and Wang, T. J. P. (2019). Deficiency of very long chain alkanes biosynthesis causes humidity-sensitive male sterility via affecting pollen adhesion and hydration in rice. Plant Cell Environ. 42, 3340–3354. doi: 10.1111/pce.13637
Yuan, G., Zou, T., He, Z., Xiao, Q., Li, G., Liu, S., et al. (2022). SWOLLEN TAPETUM AND STERILITY 1 is required for tapetum degeneration and pollen wall formation in rice. Plant Physiol. kiac307. doi: 10.1093/plphys/kiac307 [Epub ahead of print].
Zhang, X. J., Chen, H. Y., Zhang, Q., Zhang, Y. X., Xiao, Z. D., Guo, Y., et al. (2020). Cytological and genetic characterisation of dominant GMS line Shaan-GMS inBrassica napusL. J. Appl. Genet. 61, 477–488. doi: 10.1007/s13353-020-00570-8
Zhang, D., Liu, D., Lv, X., Wang, Y., Xun, Z., Liu, Z., et al. (2014). The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 26, 2939–2961. doi: 10.1105/tpc.114.127282
Zhang, D. B., Luo, X., and Zhu, L. (2011). Cytological analysis and genetic control of rice anther development. J. Genet. Genomics 38, 379–390. doi: 10.1016/j.jgg.2011.08.001
Zhang, D., Shi, J., and Yang, X. (2016). “Role of lipid metabolism in plant pollen Exine development,” in Lipids in Plant and Algae Development. eds. Y. Nakamura and Y. Li-Beisson (Cham: Springer International Publishing), 315–337.
Zhang, D., and Yang, L. (2014). Specification of tapetum and microsporocyte cells within the anther. Curr. Opin. Plant Biol. 17, 49–55. doi: 10.1016/j.pbi.2013.11.001
Zhang, Z. B., Zhan, H. D., Lu, J. Y., Xiong, S. X., Yang, N. Y., Yuan, H. Y., et al. (2021). Tapetal 3-ketoacyl-coenzyme a synthases are involved in pollen coat lipid accumulation for pollen-stigma interaction in Arabidopsis. Front. Plant Sci. 12:770311. doi: 10.3389/fpls.2021.770311
Zhang, Z., Zhu, J., Gao, J., Wang, C., Li, H., Li, H., et al. (2007). Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J. 52, 528–538. doi: 10.1111/j.1365-313X.2007.03254.x
Zhou, X., Liu, Z. Y., Ji, R. Q., and Feng, H. (2017). Comparative transcript profiling of fertile and sterile flower buds from multiple-allele-inherited male sterility in Chinese cabbage (Brassica campestris L. ssp pekinensis). Mol. Gen. Genomics. 292, 967–990. doi: 10.1007/s00438-017-1324-2
Zhu, J., Chen, H., Li, H., Gao, J.-F., Jiang, H., Wang, C., et al. (2008). Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 55, 266–277. doi: 10.1111/j.1365-313X.2008.03500.x
Keywords: Chinese cabbage, male sterility, allelic mutants, PHD-finger transcription factor, RNA-seq
Citation: Dong S, Zou J, Fang B, Zhao Y, Shi F, Song G, Huang S and Feng H (2022) Defect in BrMS1, a PHD-finger transcription factor, induces male sterility in ethyl methane sulfonate-mutagenized Chinese cabbage (Brassica rapa L. ssp. pekinensis). Front. Plant Sci. 13:992391. doi: 10.3389/fpls.2022.992391
Received: 12 July 2022; Accepted: 01 August 2022;
Published: 18 August 2022.
Edited by:
Ryo Fujimoto, Kobe University, JapanReviewed by:
Shengwu Hu, Northwest A&F University, ChinaCopyright © 2022 Dong, Zou, Fang, Zhao, Shi, Song, Huang and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Feng, ZmVuZ2h1aWFhYUBzeWF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.