
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 23 November 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.989406
This article is part of the Research TopicUnderstanding Plant Cell Wall Recalcitrance for Efficient Lignocellulose ProcessingView all 8 articles
 Baojian Guo1,2,3†
Baojian Guo1,2,3† Xinyu Huang1,2,3†
Xinyu Huang1,2,3† Jiang Qi1,2,3†
Jiang Qi1,2,3† Hongwei Sun1,2,3
Hongwei Sun1,2,3 Chao Lv1,2,3
Chao Lv1,2,3 Feifei Wang1,2,3
Feifei Wang1,2,3 Juan Zhu1,2,3
Juan Zhu1,2,3 Rugen Xu1,2,3*
Rugen Xu1,2,3*The cell wall plays an important role in plant mechanical strength. Cellulose is the major component of plant cell walls and provides the most abundant renewable biomass resource for biofuels on earth. Mutational analysis showed that cellulose synthase (CESA) genes are critical in cell wall biosynthesis in cereal crops like rice. However, their role has not been fully elucidated in barley. In this study, we isolated a brittle culm mutant brittle culm 3 (bc3) derived from Yangnongpi 5 ethyl methanesulfonate (EMS) mutagenesis in barley. The bc3 mutants exhibited reduced mechanical strength of the culms due to impaired thickening of the sclerenchyma cell wall and reduced cellulose and hemicellulose content in the culms. Genetic analysis and map-based cloning revealed that the bc3 mutant was controlled by a single recessive gene and harbored a point mutation in the HvCESA5 gene, generating a premature stop codon near the N-terminal of the protein. Quantitative real-time PCR (qRT-PCR) analysis showed that the HvCESA5 gene is predominantly expressed in the culms and co-expressed with HvCESA4 and HvCESA8, consistent with the brittle culm phenotype of the bc3 mutant. These results indicate that the truncated HvCESA5 affects cell wall biosynthesis leading to a brittle culm phenotype. Our findings provide evidence for the important role of HvCESA5 in cell wall biosynthesis pathway and could be a potential target to modify cell wall in barley.
Stem mechanical strength is an important agronomic trait for improving crop lodging resistance and yield in cereal crops, which is mainly determined by plant cell walls (Ma et al., 2021). Plant cell walls are dynamic structures and supply 80% of the plant biomass on earth, which largely consists of cellulose, hemicellulose, and lignin (Cosgrove, 2005). Plant cell walls contain primary cell walls (PCW) and secondary cell walls (SCW). Cellulose and hemicellulose exist mainly in PCW and SCW while lignin exists only in SCW (Taylor et al., 2000). Therefore, cell walls differ in structure and their composition affecting the mechanical strength of stems were important for crop production (Zhang and Zhou, 2011).
In plants, cellulose synthesis defects often lead to a reduction in cellulose content, termed a brittle culm or fragile stem phenotype (Kokubo et al., 1989). Cellulose, the most abundant polysaccharide in plant cell walls, is chemically a long chain of sugar and β−D−glucose linked by glycosidic linkages (Bhat et al., 2019). In higher plants, cellulose is synthesized at the plasma membrane by cellulose synthases (CESA) complexes assembled in the Golgi apparatus using uridine diphosphate-glucose as substrates (Somerville, 2006; Taylor, 2008; Polko and Kieber, 2019). In rice, recessive mutations in OsCESA4, OsCESA7 and OsCESA9 resulted in reduced mechanical strength and proportion of cellulose as well as increased hemicellulose content in the culms (Tanaka et al., 2003; Yan et al., 2007; Zhang et al., 2009; Kotake et al., 2011; Wang et al., 2012; Wang et al., 2016). However, rice Bc19 harboring a dominant brittle mutation in OsCESA4 leads to a decrease in cellulose, hemicellulose and lignin in the culms, but no differences were observed in plant growth and yield (Ma et al., 2021). A fs2 mutant in the HvCesA4 gene, which is an orthologue of rice OsCESA7, leads to a reduction of cellulose content compared with its parental lines in barley (Burton et al., 2010). Therefore, CESA genes have the conserved function in regulating mechanical strength through changing cell wall composition.
The COBRA-like (COBL) genes exhibit typical structural features of a glycosylphosphatidylinositol-anchored protein, which plays a vital role in cellulose biosynthesis in primary and secondary cell walls (Roudier et al., 2002; Li et al., 2003). Mutation of Brittle Culm1/CWA1 encoding COBL gene which shows brittle culms and reduced cellulose content in culms, and also exhibits a little disruption in vascular bundle (Li et al., 2003; Sato et al., 2010). A series of studies suggest that COBL genes play critical roles in regulating cellulose biosynthesis in cereal crops, such as maize Brittle stalk 2 (Ching et al., 2006; Sindhu et al., 2007), rice OsBC1L4 (Dai et al., 2011), Sorghum SbBC1 (Li et al., 2018) and wheat TmBr1 (Deng et al., 2019). BC10 encodes a Golgi-located type II membrane protein, and bc10 mutant plants exhibit whole plant brittleness and growth delay along with reduced cellulose content, suggesting that BC10 is required for cell-wall biosynthesis in rice (Zhou et al., 2009). The bc14 gene harbors a mutation in Golgi-localized Nucleotide Sugar Transporter1 (OsNST1), which shows a deficiency in the synthesis of glucagon-conjugated polysaccharides. Further analysis revealed that OsNST1 supplies the glucosyl substrate for the formation of matrix polysaccharides and affects cell wall biosynthesis and plant growth (Zhang et al., 2011). BC12, encoding a dual-targeting kinesin-4 protein, controls cell-cycle progression and wall properties in rice. bc12 mutants displayed dwarfism resulting from a significant reduction in cell number and brittleness due to an alteration in cellulose microfibril orientation and wall composition (Zhang et al., 2010). In addition, OsBC3 and BC15/OsCTL1 encode a classic dynamin-related gene in rice and membrane-associated chitinase-like protein is involved in cellulose biosynthesis through reducing cellulose content and mechanical strength (Hirano et al., 2010; Wu et al., 2012). Therefore, the underlying mechanisms involved in regulating cellulose synthesis were still elusive.
In this study, we characterized a barley mutant, brittle culm 3 (bc3), developed from an ethyl methanesulfonate (EMS) mutagenized library. The mutant exhibited a brittle culm phenotype and abnormal growth. Genetic analysis showed that the mutant phenotype was controlled by a single recessive gene. Map-based cloning identified a point mutation in the 4th exon of the HvCESA5 gene, which introduced a premature stop codon in the bc3 mutant. Our findings reveal that a missense mutation led to reduced cellulose levels in the bc3 mutant, and HvCESA5 is required for cell wall biosynthesis in barley.
The mutant bc3 was derived from Yangnongpi 5 (wild type, WT) by EMS mutagenesis (Caldwell et al., 2004), which displayed stable and inherited brittle culm. F2 and F2:3 populations were generated by crossing the bc3 mutant to Morex. The parents and mapping populations were planted at Yangzhou University farm. At the heading stage, the F2 and F2:3 individuals with brittle culm phenotype were collected and used for gene mapping. The mutant-type and wild-type plants were recorded in the segregating F2 and F2:3 generations, and the segregation ratio was tested by Chi square test.
The main culms were used for measuring agronomic traits at mature stage, including plant height, tiller number, spikelet length, and grain number per spike. Ten plants were harvested from each of the three replicates. At the heading stage, the third internodes of wild type and mutant plants were cut into segments of 5 cm and used for assays. The breaking force was measured by a digital force/length tester (5848 Microtester; Instron, USA) (Zhou et al., 2009). Student’s t-test were used to determine statistical significance. P values < 0.05 were considered statistically significant. Values were presented as mean ± standard deviation of the mean (SD).
At the heading stage, the third internodes were cut into thin pieces and segments, then fixed in the fixative solution (75% ethanol, 5% acetic acid, 5% glycerol, and 5% formaldehyde) for at least 24 h, and then dehydrated through an ethanol series. For scanning electron microscopy, the samples were critical point-dried, sputter-coated with gold and observed with Gemini SEM300 scanning electron microscope (ZEISS, Germany). For lignin staining, the third internode of fresh barley stem was sliced by freehand, and then one to two drops of 5% phloroglucinol ethanol solution was applied to the material, followed by one drop of concentrated hydrochloric acid to soak the material, which was immediately observed under a light microscope and photographed.
Cell wall components, including cellulose content, hemicellulose content, and lignin content were measured as previously described (Li et al., 2019). Briefly, at the heading stage, the third internodes were dried at 105°C for 1 h and then dried at 50°C until a constant weight was reached. Next, the samples were ground to fine powder and were assayed for cellulose content with the anthrone/H2SO4 with Whatman 3MM paper as the standard. Total hemicellulose contents were calculated subjective to total pentoses in the hemicellulose fraction. Total pentoses were detected using the HCl/3, 5-dinitrosalicylic acid reagent. To measure lignin content, the powder was extracted with methanol. After drying, lignin content was determined by the two-step acid hydrolysis method as described previously (Li et al., 2014). The lignin monomer contents were quantified by using HPLC-MS analysis (Dos Santos et al., 2008). Three biological replicates were set in the present study.
A total of 1878 F2 individuals were used for gene mapping. Genomic DNA of parents and each F2 plants were extracted by CTAB method (Clarke, 2009). InDel markers on the barley genome showing polymorphism between bc3 and Morex were designed for rough gene mapping (Guo et al., 2022) (Table S1). In the primary mapping, two DNA pools composed of 30 mutant and wild-type phenotype individuals and two parents DNA were used for the bulked segregation analysis (Michelmore et al., 1991). For the fine mapping, new Indel markers were designed by utilizing information of genomic sequences from Morex and Yangnongpi 5 (http://wheat.cau.edu.cn/Wheat_SnpHub_Portal/collaboration_GBJ_191007/) (Wang et al., 2020). PCR was carried out in a final reaction volume of 10 μL containing 50 ng DNA template, 1.0 μL forward and reverse primer (2 μM), 5 μL 2×Taq Master Mix (Vazyme Biotech Co., Ltd, China). The reaction was performed with an initial cycle of 5 min at 95°C, 32 cycles of 30 s at 94°C, 30 s at 58-60°C, and 30 s at 72°C; and a final 10 min at 72°C. PCR products were separated on 8% polyacrylamide gel.
According to the reference genome of the barley cultivar Morex V3 (Mascher et al., 2021), genomic DNA sequences of the candidate gene were amplified from both WT and bc3 mutants using the primer pairs shown in Table S1. The PCR program included 5 min at 95°C, followed by 32 cycles of 98°C for 15 s, 55°C for 5 s, and 72°C for 3.5 min, and a final extension at 72°C for 10 min. PCR products were separated by 1.0% agarose gel electrophoresis. DNA fragments were cut from the gel and purified with the GeneJET Gel Extraction kit (Thermo Scientific, Waltham, USA). The fragments were connected to the pEASY-T1 cloning vector and sequenced. Sequence analysis was performed using DNAMAN software version 10 (https://www.lynnon.com/dnaman.html).
The rice and Arabidopsis CESA proteins were used as queries (Tan et al., 2015). Domain searches were predictions by PredictProtein (https://predictprotein.org/). Multiple sequence alignments were performed using the CLUSTAL X program (Thompson et al., 1997; Finn et al., 2015). Based on protein alignment results, the phylogenetic tree was constructed using the neighbor-joining method in MEGA version 6 with default parameters, and the phylogeny test was performed by bootstrap method with 1000 replications (Tamura et al., 2013).
At the heading stage, the roots, leaf blades, leaf sheaths, culms and spikes were collected from WT and bc3 mutant. The RNA extraction, reverse transcription, and qRT-PCR assay were carried out using RNA extraction kit (TRIzol reagent, Invitrogen, USA), M-MLV reverse transcriptase (TaKaRa, Japan) and SYBR Premix Ex Taq2 kit (TaKaRa, Japan) based on the manufacturer’s recommendations. Specific primers were designed for qRT-PCR analysis by using Primer3 (https://bioinfo.ut.ee/primer3-0.4.0/) (Table S1). The HvActin was used as an internal control (Table S1). Relative expression levels were calculated using 2-ΔΔCt method with CFX Manager 3.1 software (Bio-Rad, USA) (Livak and Schmittgen, 2001). Experiments were carried out with three biological replicates.
The bc3 mutant exhibited brittle culms and leaves that could be easily broken (Figures 1A, B). In addition, the bc3 mutant also displayed leaf tip wilting (Figure 1C). We quantitatively compared the breaking forces of the second upper internodes and leaves between bc3 and WT plants at the heading stage. The breaking strength of the bc3 culms and leaves were significantly reduced by 82.5% and 18.7% compared to the WT (Figures 1D, E). In addition, the bc3 mutants also displayed dwarfism compared with WT plants (Figure 2A). At the maturation stage, plant height, spike length and grain number per spike were significantly decreased in the bc3 mutant, but the tiller number was unchanged (Figures 2B-E). Therefore, the bc3 mutation caused not only defective mechanical strength but also changes in plant morphology.
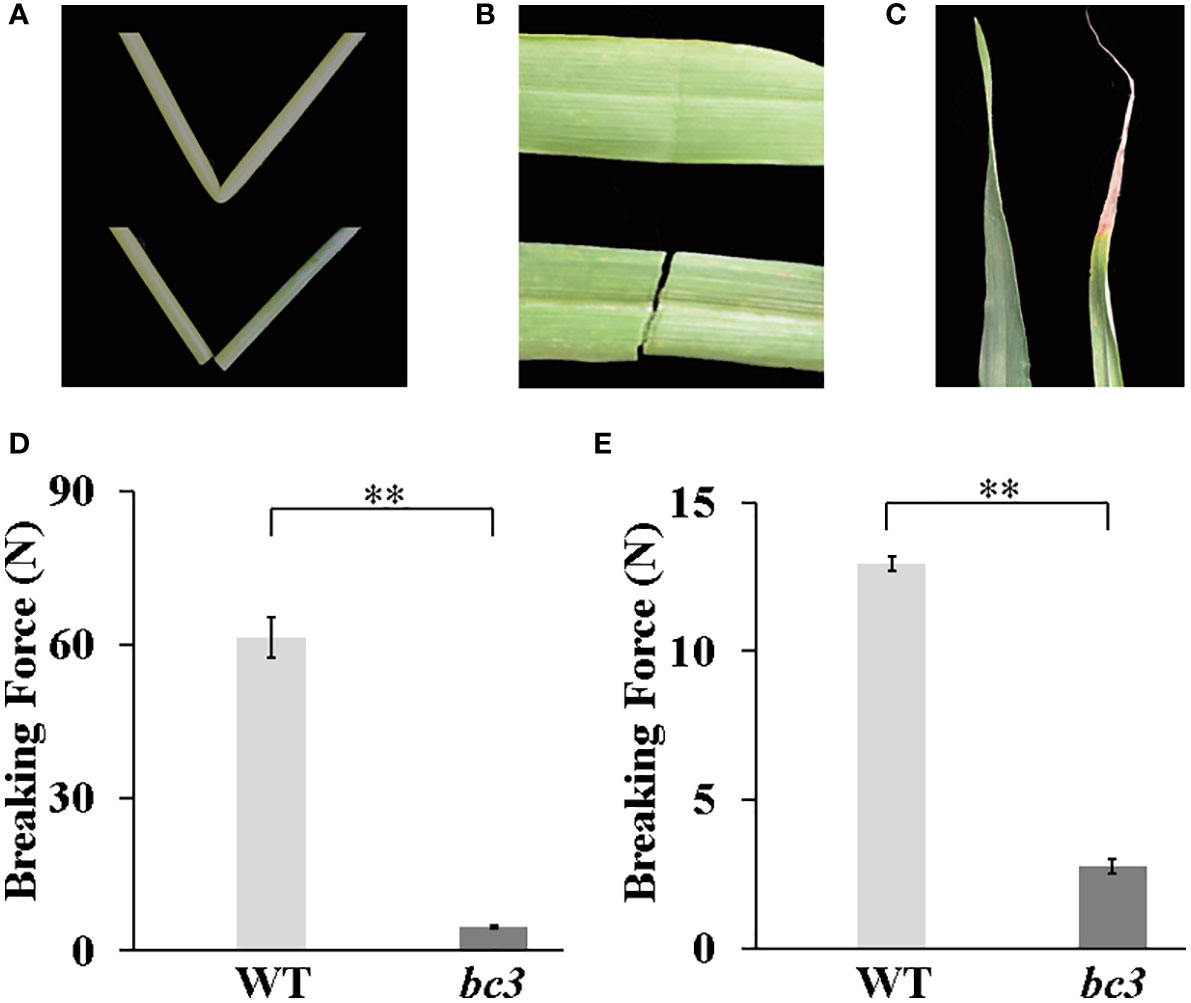
Figure 1 Physical properties of the bc3 mutant. (A) An easily broken culm of bc3 (lower) compared with a wild type in second upper internode (upper). (B) An easily broken leave of bc3 (upper) compared with a wild type (lower) in top second flag leaf (C) Leaf tip wilting of bc3 (right) compared with a wild type in top second flag (left). (D, E) The force required to break the third internodes and leaves, respectively. The bars represent standard errors, ** represent t-test at P < 0.01.
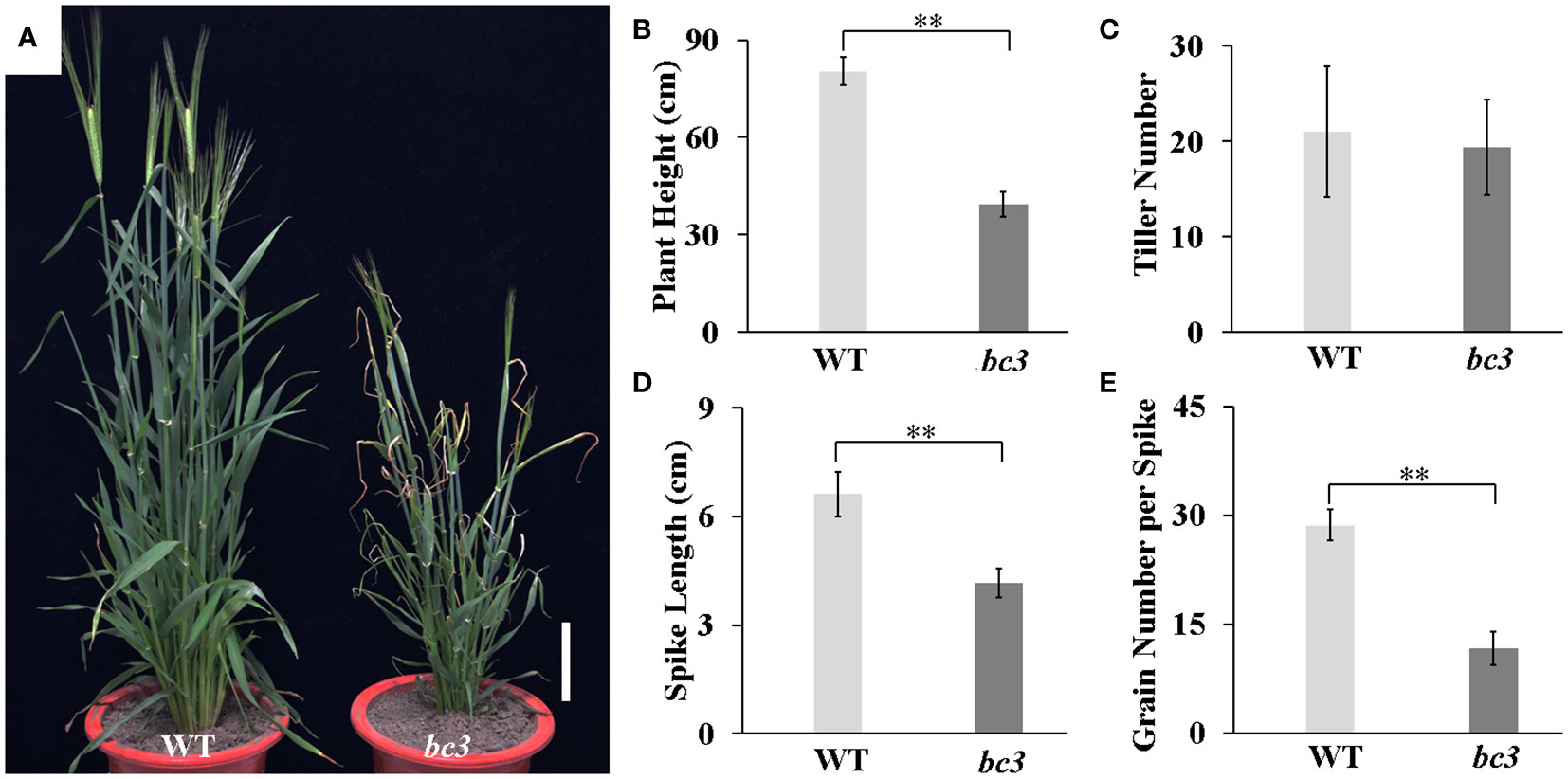
Figure 2 Phenotypes of the bc3 mutant compare with wide type. (A) Plant appearance at the heading stage. Scale bar is 10 cm. (B–D) The plant height (B), tiller number (C) and spike length (D) at the maturity stage. Error bars represent standard errors, ** represents P < 0.01.
A change in cell morphology usually leads to a reduction of mechanical strength in brittle culm mutants. Thus, the anatomical features of cells were compared in the culms between WT and bc3 mutants (Figures 3A–F). Scanning electron microscopy showed that the cell walls of sclerenchyma tissues were much thinner in bc3 mutants than those of WT plants (Figure 3G), while smaller sclerenchyma cell size was observed in bc3 mutants (Figure 3H). On the contrary, no obvious differences were observed in parenchyma cell wall and vascular bundles between bc3 mutant plants and WT (Figure S1). These results suggested that the reduction in mechanical strength was due to the thinning in sclerenchyma cell wall in the bc3 mutant.
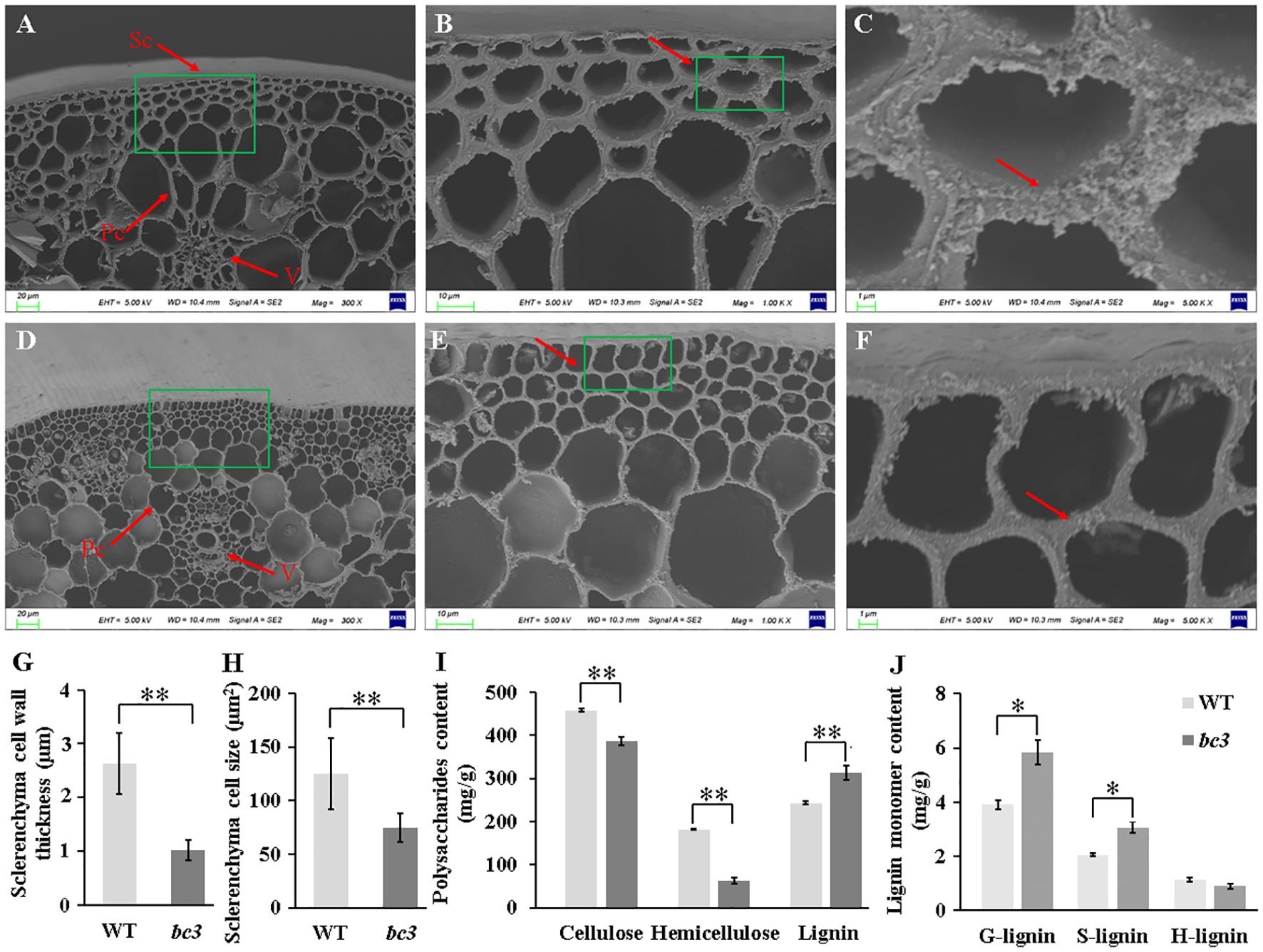
Figure 3 Histology of stem tissues. (A–F) Scanning electron microscopy showing the differences between wild type (A–C) and bc3 mutant (D–F). Sc, sclerenchyma cell; Pc, parenchyma cell; V, vascular bundles. Different magnifications (300, 1000 and 5000×) were shown. (G, H) Quantitative comparison of the wall thickness and size of sclerenchyma. Polysaccharides content (I) and lignin monomers content (J) of cell wall in the second upper internode at heading stage. Values shown are averages of 10 plants, and the bars represent standard errors. * represent t-test P < 0.05, ** represent t-test P < 0.01.
To characterize the underlying cell wall defects in the bc3 mutant, we measured the amount of cellulose, hemicellulose and lignin content between WT and bc3 mutant. In culms of bc3 mutants, cellulose and hemicellulose contents decreased by 15.2% and 65.4%, respectively (Figure 3I). Moreover, the lignin content of bc3 culms increased 29.8% compared with that of WT (Figure 3I). G- and S-lignin, the major lignin monomers, increased by 49.36% and 49.55% in the bc3 mutant, while H-lignin displayed no significant difference between the bc3 mutant and WT (Figure 3J). In addition, optical microscopy analysis showed that the sclerenchyma and vascular bundles were significantly lighter stained in the bc3 mutant than in WT (Figures S1E–H), suggesting a higher lignin content in the bc3 mutant. These results indicate that the bc3 mutant is deficient in cell wall biosynthesis.
To analyze whether the brittle culm phenotype was controlled by a single gene, the bc3 mutant was crossed to Morex. F1 plants displayed no brittle culm, indicating recessiveness of the mutant gene. Within the F2 population, 1878 plants showed the wild type phenotype and 574 showed brittle culm phenotype, indicating segregation at a single locus (χ2 = 3.308<χ2(0.05, 1)=3.84). These findings indicated that the brittle culm phenotype was controlled by a single recessive gene. The map-based cloning approach was used to isolate the bc3 mutant gene. Bulked segregation analysis showed that Indel marker 3H-20 on chromosome 3H was closely linked to the mutant phenotype. Fifteen Indel markers distributed on the primary mapping interval were designed to detect polymorphisms between bc3 and Morex. Eight out of the 15 markers exhibited polymorphism between bc3 and Morex and were used to genotype 60 F2 mutant individuals (Table S1). We mapped bc3 to a 25.478 Mb interval between the molecular markers 3H-25 and 3H-31 (Figure 4A). Next, six new polymorphic Indel markers and 574 F2 individuals with the brittle culm phenotype were used for fine mapping. The gene was localized within a 1.666 Mb interval region between Indel markers 3H-52 and 3H-61 (Figure 4B). According to the barley reference sequence Morex V3 (Mascher et al., 2021), ten high-confidence genes were present in the 1.666 Mb interval (Figure 5C). Microsynteny analysis of barley genomic region (485.821 Mb-487.487 Mb) with rice was performed (Figure 4C). A cellulose synthase A subunit 5 (HvCESA5) (HORVU.MOREX.r3.3HG0288960) as an orthologous gene of OsCESA4 was found in this region and prioritized for further study.
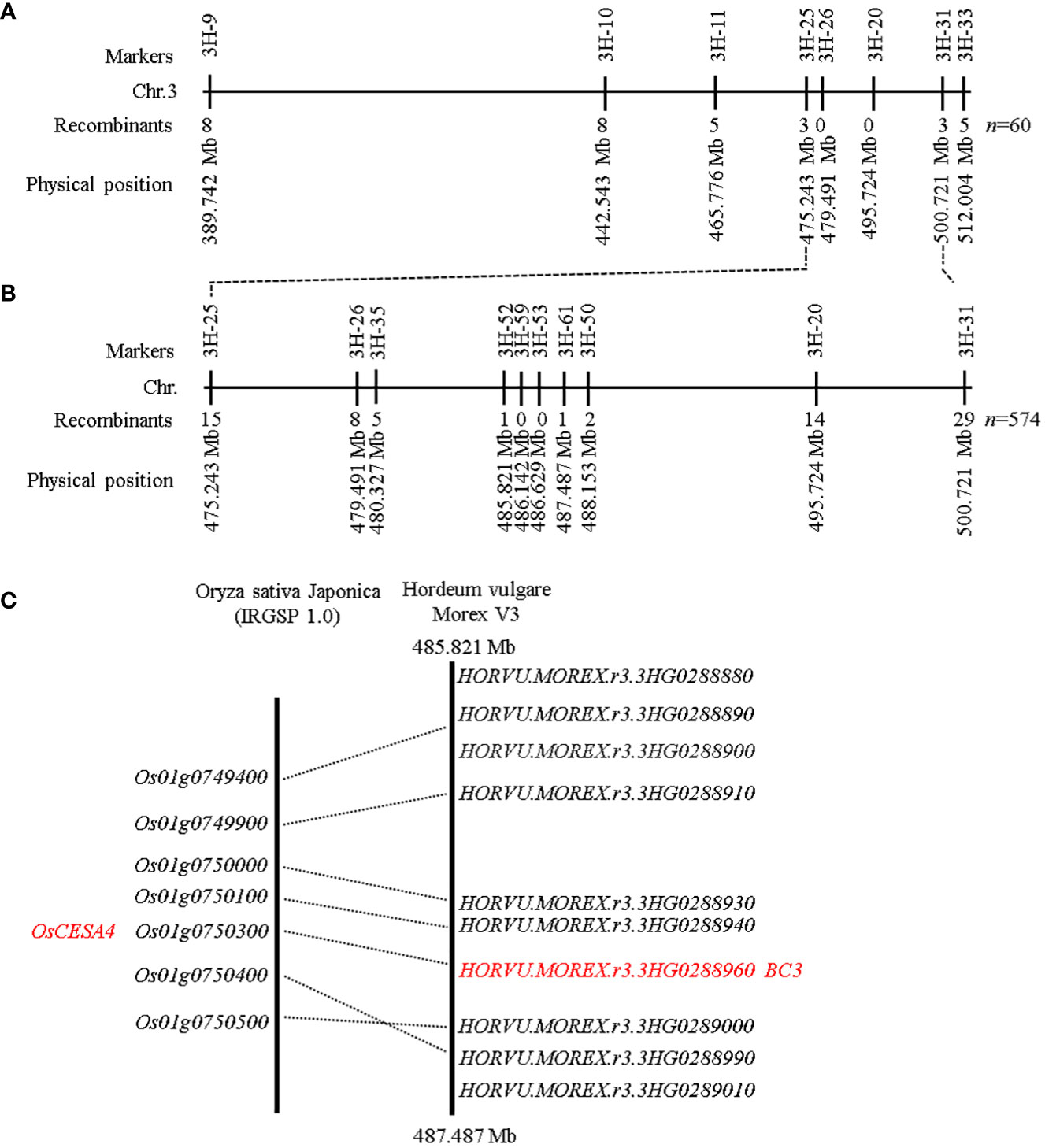
Figure 4 Map-based cloning of the bc3 gene in barley. (A) Physical positions of DNA markers (according to the Morex V3 gene model) used for primary mapping of bc3 using 60 F2 plants homozygous for the bc3 mutant phenotype. (B) Physical positions of DNA markers used for fine mapping of bc3 using 574 F2:3 plants homozygous with the bc3 mutant phenotype. (C) Micro-synteny analysis of the candidate region in barley and rice.
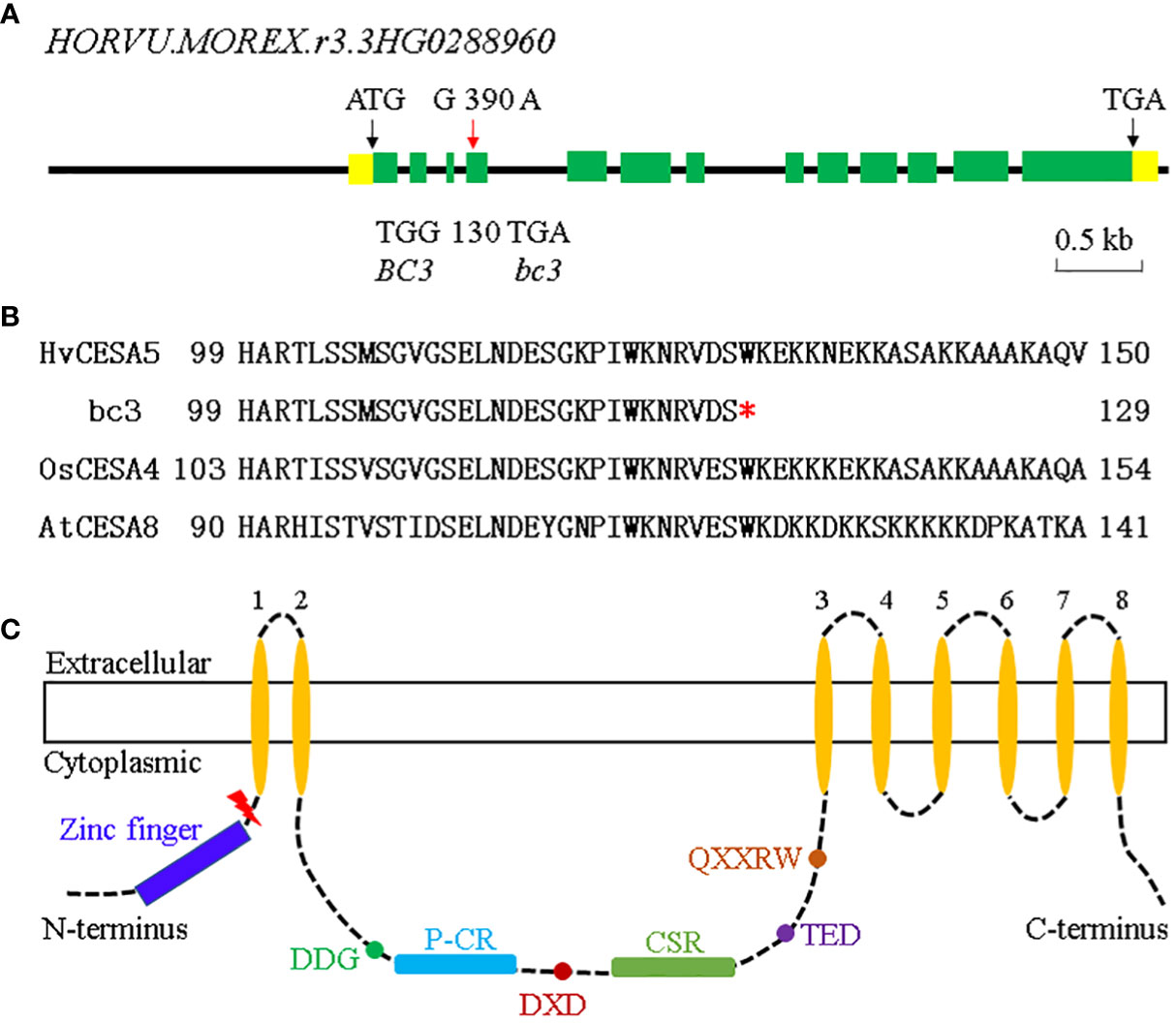
Figure 5 Gene structure of BC3. (A) Gene structure of BC3 and the mutation sites in bc3 mutant (red arrow). Yellow boxes indicate 5’ and 3’ UTR, green boxes indicate exons and black lines indicate introns. (B) The conservation for the mutation site among barley, rice, and Arabidopsis. The red star indicated the premature stop codon in the bc3 mutant. (C) Schematic representations of the structure of a CESA protein. Key motifs, such as the zinc finger, plant-conserved region (P-CR), class-specific region (CSR), and transmembrane regions (TMRs) are showed by different colors. The premature stop codon in the bc3 mutant site is highlighted with red lightning.
Bioinformatics analyses revealed that the HvCESA5 gene contained 13 exons and 12 introns with 2955 bp CDS, encoding a 984 amino-acid protein (Figure 5A). Sequencing of the HORVU.MOREX.r3.3HG0288960 gene identified a G-to-A substitution at position 2412 in the bc3 mutant (Figure 5A), resulting in a change of the coding amino acid from conserved Tryptophan (W) to a premature stop codon at the 130th amino acid position (Figure 5B). The truncated protein with 129 amino acids only contains zinc finger domain but lacks a central cytoplasmic domain which consists of some key motifs, including the transmembrane regions (TMRs), plant-conserved region (P-CR), class-specific region (CSR), DDG, DXD, TED, and QXXRW motif (Huang et al., 2020) (Figure 5C).
To investigate whether the premature stop codon affects HvCESA5 gene expression, we performed qRT-PCR at the heading stage and found that HvCESA5 was predominantly expressed in the culms, and the abundance of HvCESA5 transcripts in the culms was greatly reduced in the mutant plants compared with that of WT plants (Figure 6A; Figure S2). To investigate whether the expression of other CESA genes is affected in bc3 mutant plants, the expression levels of nine genes were monitored by qRT-PCR (Table S2). Remarkably, all HvCESA genes except HvCESA9 showed decreased expressions in the culms of the bc3 mutant (Figure 6B), and this was consistent with the reduced cellulose levels (Figure 1F). All of these results suggested that the mutation in HvCESA5 leads to the brittle culm phenotype in barley and that HvCESA5 is the candidate gene for BC3.
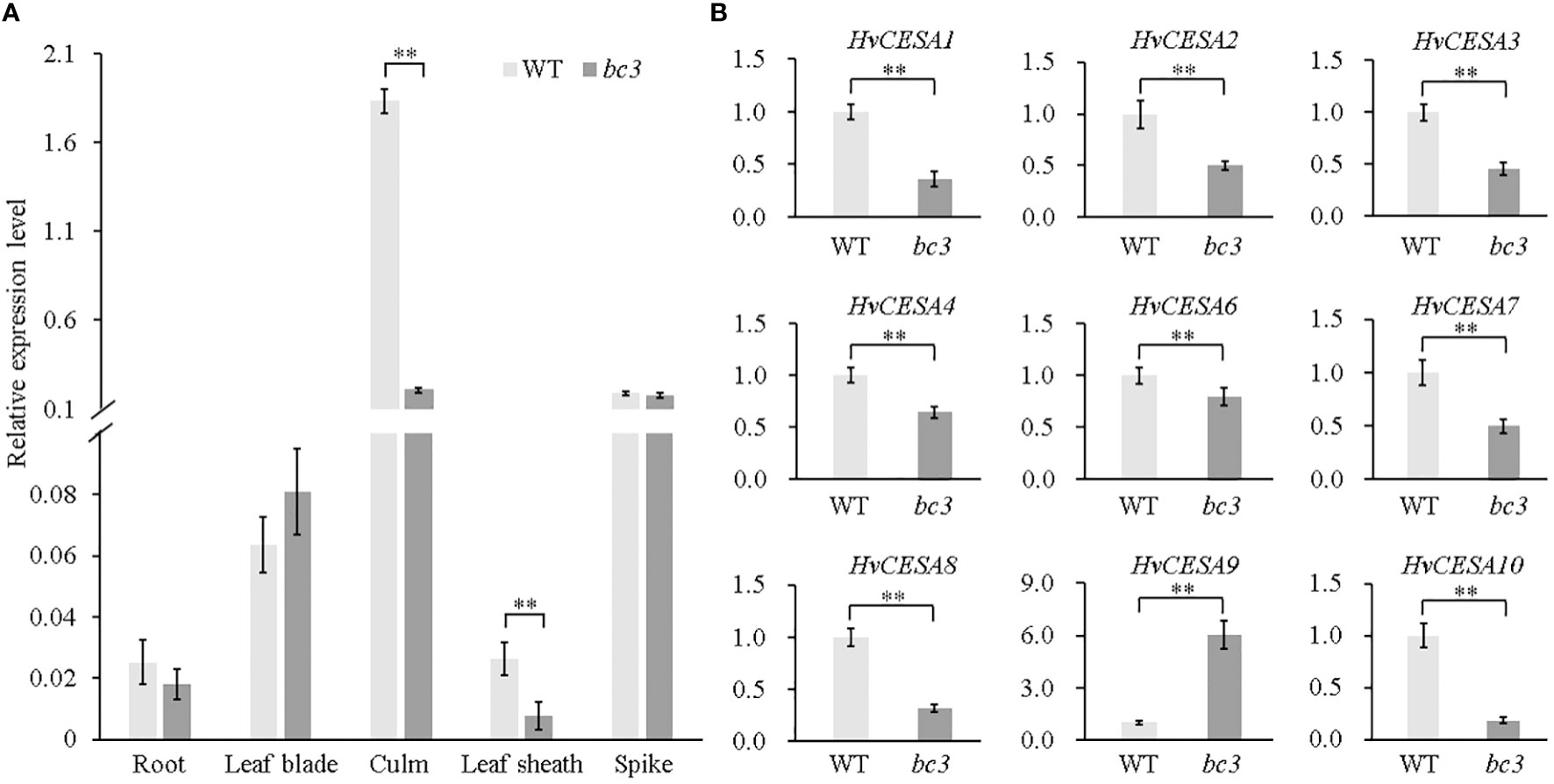
Figure 6 HvCESA genes expression analysis at the heading stage. (A) HvCESA5 gene expression analysis by qRT-PCR in root, leaf blade, leaf sheath, culm and spike between WT and bc3 mutant. (B) Expression analysis of HvCESA genes in culms between WT and bc3 mutant. HvActin gene as endogenous control. WT data is normalized as 1. ** indicates significant difference at P < 0.01.
The plant cell wall plays an important role in providing mechanical support for plant growth. Cellulose is the main chemical component in plant cell walls, and the defect of cell wall structure and components affects stem mechanical strength (Taylor, 2008). The barley brittle culm (bc) mutants were first described based on the physical properties of the culms, which had only minimal cellulose content, great loss of mechanical strength as well as fewer numbers of cellulose molecules compared with those of wild type plants in the cell walls (Kokubo et al., 1989; Kokubo et al., 1991). Mutational analysis of brittle culms has supplied us with a deeper understanding of cell wall synthesis. In barley, the fs2 mutant identified in the HvCESA4 gene might be responsible for the brittle stem phenotype together with decreased cellulose content and stem strength compared with the parental line (Burton et al., 2010). In the present study, the bc3 recessive mutant exhibited a similar brittle culm phenotype as fs2 and rice brittle culm mutants. The reduction in mechanical strength and change in thickening of the sclerenchyma cell wall demonstrated that the bc3 mutant has defective cell wall biosynthesis (Burton et al., 2010). Similar to rice bc5 and brittle sheath1 (bsh1) which is involved in brittle node and brittle sheath in rice (Aohara et al., 2009; Wang et al., 2016), decreased hemicellulose content was also observed in the bc3 mutant. The reduced cellulose and hemicellulose production in the bc3 mutant could be compensated by the increased level of lignin content which was observed in bc1, bc11, and bc15 mutants in rice (Li et al., 2003; Zhang et al., 2009; Wu et al., 2012).
Cellulose synthase, a major component of CESA complexes, catalyzes the chain elongation step in glucan polymerization (Kurek et al., 2002; Somerville, 2006). The CESAs contain the classic RING-type zinc finger with a cysteine-rich domain at N-terminal region and can interact with other CESAs to assemble a large CESA complex (Kurek et al., 2002). The CESAs have eight putative transmembrane domains, two of them near the zinc finger domain, a cluster of six transmembrane domains near the carboxy terminus, and a large central cytoplasmic domain (Somerville, 2006). In rice, S1-24 (C40Y) is the first mutant in the zinc finger domain of OsCESA7gene. Two allelic mutants, NC0259 and ND8759, harboring an altered central cytoplasmic domain exhibit more severely impaired plant growth than S1-24 (Tanaka et al., 2003; Wang et al., 2016). Rice bc11 (G858R) and S1-60 (G905 D) mutants carrying missense mutations in the transmembrane domain of OsCESA4 and OsCESA9 genes show reduced cellulose content, dwarfism as well as partial sterility (Zhang et al., 2009; Wang et al., 2012). Moreover, fc16 (W481C, P482S) and fc17 (F426S) mutants harboring the amino acid substitution at the plant-conserved region (P-CR) of OsCESA9 and OsCESA4 protein exhibit slightly affected plant growth and increased lodging resistance and biomass in rice. P-CR region is a potential target for lodging resistance breeding in rice (Li et al., 2017; Li et al., 2018). Furthermore, bc3 mutants have a truncated protein which might affect the function of HvCESA5 in CSCs. Taken together, the conserved domain and motif are critical for CESA catalytic activity and different mutation sites lead to diverse phenotypes in plants.
In plants, AtCESA4/7/8 and OsCESA4/7/9 are highly co-expressed and required for SCW cellulose synthesis, whereas AtCESA1/3/6 and OsCESA1/3/8 are implicated in forming PCW CSCs (Taylor et al., 1999; Taylor et al., 2000; Tanaka et al., 2003; Burton et al., 2004; Persson et al., 2005; Persson et al., 2007). Phylogenetic tree analysis revealed that the HvCESA5 gene clustered with OsCESA4 and AtCESA8, HvCESA4 clustered with OsCESA7 and AtCESA4, HvCESA8 clustered with OsCESA9 and AtCESA7 (Figure S3). In consideration of HvCESA4, HvCESA5 and HvCESA8 appeared to be coordinately expressed, especially in culm tissue (Figure 6B; Figure S4), we speculate that the combination of all three genes is required for cellulose synthesis during secondary cell wall deposition in barley (Brown et al., 2005; Burton et al., 2010). In addition. HvCESA1, HvCESA2, and HvCESA6 are also coexpressed and comprise CSCs for PCWs (Burton et al., 2004); their downregulation was consistent with the decrease of hemicellulose content in the bc3 mutant. Therefore, we speculate that the HvCESA5 gene plays a critical role in cell wall synthesis in barley.
In the present study, a barley brittle culm mutant with a point mutation in HvCESA5 was identified. The mutant protein loses its catalytic activity due to lacking eight transmembrane domains and the central cytoplasmic domain, resulting in impaired cell wall biosynthesis and mechanical strength, brittle culm phenotype and defective plant growth. Our work demonstrates that HvCESA5 is a potential target gene for improving mechanical strength of stem and lodging resistance in barley breeding.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
BG and RX designed and supervised the project. BG and XH performed map-based cloning of the bc3 gene. JQ performed cell wall composition analysis. HS and CL performed scanning electron microscopy. FW performed phylogenetic analysis. JZ performed qRT-PCR analysis. BG and RX wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Jiangsu Agriculture Science and Technology Innovation Fund (CX(20)2036), Natural Science Foundation of the Jiangsu Higher Education Institutions of China (19KJA560005), National Barley and Highland Barley Industrial Technology Specially Constructive Foundation of China (CARS-05), and a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.989406/full#supplementary-material
Supplementary Figure 1 | Histology observation of WT and bc3 mutant plants. (A–D) Scanning electron microscopy analysis of parenchyma cell wall and vascular bundles of WT (A, B) and bc3 mutant plants (C, D). Pc, parenchyma cell; V, vascular bundles. Different magnifications (300, 1000×) were shown. (E–H) The cross sections of the third internodes of WT (E, F) and bc3 mutant (G, H) were stained with phloroglucinol. (E, G), bar=200μm; (F, H), bar=100μm. Sc, sclerenchyma cell; Pc, parenchyma cell; V, vascular bundles.
Supplementary Figure 2 | Expression analysis of HvCESA5 genes between WT and bc3 mutant in different internodes. 1st, 2nd, 3rd, 4th, 5th represent the internode from the top. ** represents t-test P< 0.01, respectively.
Supplementary Figure 3 | Phylogenetic tree of CESAs protein among rice, Arabidopsis, and barley. Amino acids were used for the phylogenetic analysis (Tanaka et al., 2003).
Supplementary Figure 4 | Expression of the CESAs gene based on publicly available expression data across different tissues (Mascher et al., 2017). Transcript level is given as fragments per kilo base of exon per million read mapped (FPKM).
Aohara, T., Kotake, T., Kaneko, Y., Takatsuji, H., Tsumuraya, Y., Kawasaki, S. (2009). Rice BRITTLE CULM 5 (BRITTLE NODE) is involved in secondary cell wall formation in the sclerenchyma tissue of nodes. Plant Cell Physiol. 50, 1886–1897. doi: 10.1093/pcp/pcp133
Bhat, A. H., Khan, I., Usmani, M. A., Umapathi, R., Al-King, S. M. Z. (2019). Cellulose an ageless renewable green nanomaterial for medical applications: An overview of ionic liquids in extraction, separation, and dissolution of cellulose. Int. J. Biol. Macromol. 129, 750–777. doi: 10.1016/j.ijbiomac.2018.12.190
Brown, D. M., Zeef, L. A., Ellis, J., Goodacre, R., Turner, S. R. (2005). Identification of novel genes in arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell. 17, 2281–2295. doi: 10.1105/tpc.105.031542
Burton, R. A., Ma, G., Baumann, U., Harvey, A. J., Shirley, N., Taylor, J., et al. (2010). A customized gene expression microarray reveals that the brittle stem phenotype fs2 of barley is attributable to a retroelement in the HvCesA4 cellulose synthase gene. Plant Physiol. 153, 1716–1728. doi: 10.1104/pp.110.158329
Burton, R. A., Shirley, N. J., King, B. J., Harvey, A. J., Fincher, G. B. (2004). The CesA gene family of barley. quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol. 134, 224–236. doi: 10.1104/pp.103.032904
Caldwell, D. G., McCallum, N., Shaw, P., Muehlbauer, G. J., Marshall, D. F., Waugh, R. (2004). A structured mutant population for forward and reverse genetics in barley (Hordeum vulgare l.). Plant J. 40, 143–150. doi: 10.1111/j.1365-313X.2004.02190.x
Ching, A., Dhugga, K. S., Appenzeller, L., Meeley, R., Bourett, T. M., Howard, R. J., et al. (2006). Brittle stalk 2 encodes a putative glycosylphosphatidylinositol-anchored protein that affects mechanical strength of maize tissues by altering the composition and structure of secondary cell walls. Planta 224, 1174–1184. doi: 10.1007/s00425-006-0299-8
Clarke, J. D. (2009). “Cold spring harbor protocols,” in Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Eds. Weigel, D., Glazebrook, J. (New York, NY, USA: Cold Spring Harbor Laboratory Press), 1–2.
Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell. Bio. 6, 850–861. doi: 10.1038/nrm1746
Dai, X., You, C., Chen, G., Li, H., Zhang, Q., Wu, C. (2011). OsBC1L4 encodes a COBRA-like protein that affects cellulose synthesis in rice. Plant Mol. Biol. 75, 333–345. doi: 10.1007/s11103-011-9730-z
Deng, Q., Kong, Z., Wu, X., Ma, S., Yuan, Y., Jia, H., et al. (2019). Cloning of a COBL gene determining brittleness in diploid wheat using a MapRseq approach. Plant Sci. 285, 141–150. doi: 10.1016/j.plantsci.2019.05.011
Dos Santos, W. D., Ferrarese, M. L. L., Nakamura., C. V., Mourão, K. S. M., Mangolin, C. A., Ferrarese-Filho, O. (2008). Soybean (Glycine max) root lignification induced by ferulic acid. the possible mode of action. J. Chem. Ecol. 34, 1230–1241. doi: 10.1007/s10886-008-9522-3
Finn, R. D., Clements, J., Arndt, W., Miller, B. L., Wheeler, T. J., Schreiber, F., et al. (2015). HMMER web server: 2015 update. Nucleic Acids Res. 43, W30–W38. doi: 10.1093/nar/gkv397
Guo, B. J., Qi, J., Li, D. F., Sun, H. W., Lv, C., Wang, F. F., et al. (2022). Genetic analysis and gene mapping of a dwarf and liguleless mutation in barley. Crop J 10, 1094–1102. doi: 10.1016/j.cj.2022.01.006
Hirano, K., Kotake, T., Kamihara, K., Tsuna, K., Kawasaki, S. (2010). Rice BRITTLE CULM 3 (BC3) encodes a classical dynamin OsDRP2B essential for proper secondary cell wall synthesis. Planta 232, 95–108. doi: 10.1007/s00425-010-1145-6
Huang, L., Li, X., Zhang, W., Ung, N., Liu, N., Yin, X., et al. (2020). Endosidin20 targets the cellulose synthase catalytic domain to inhibit cellulose biosynthesis. Plant Cell. 32, 2141–2157. doi: 10.1105/tpc.20.00202
Kokubo, A., Kuraishi, S., Sakurai, N. (1989). Culm strength of barley: correlation among maximum bending stress, cell wall dimensions, and cellulose content. Plant Physiol. 91, 876–882. doi: 10.1104/pp.91.3.876
Kokubo, A., Sakurai, N., Kuraishi, S., Takeda, K. (1991). Culm brittleness of barley (Hordeum vulgare l.) mutants is caused by smaller number of cellulose molecules in cell wall. Plant Physiol. 97, 509–514. doi: 10.1104/pp.97.2.509
Kotake, T., Aohara, T., Hirano, K., Sato, A. (2011). Rice brittle culm 6 encodes a dominant negative form of CesA protein that perturbs cellulose synthesis in secondary cell walls. J. Exp. Bot. 62, 2053–2062. doi: 10.1093/jxb/erq395
Kurek, I., Kawagoe, Y., Jacob-Wilk, D., Doblin, M., Delmer, D. (2002). Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc. Natl. Acad. Sci. U.S.A 99, 11109–11114. doi: 10.1073/pnas.162077099
Li, P., Liu, Y., Tan, W., Jun, C., Zhu, M., Lv, Y., et al. (2019). Brittle culm 1 encodes a COBRA-like protein involved in secondary cell wall cellulose biosynthesis in sorghum. Plant Cell Physiol. 60, 788–801. doi: 10.1093/pcp/pcy246
Li, F., Liu, S., Xu, H., Xu, Q. (2018). A novel FC17/CESA4 mutation causes increased biomass saccharification and lodging resistance by remodeling cell wall in rice. Biotechnol. Biofuels. 11, 1–13. doi: 10.1186/s13068-018-1298-2
Li, Y., Qian, Q., Zhou, Y., Yan, M., Sun, L., Zhang, M., et al. (2003). BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell. 15, 2020. doi: 10.1105/tpc.011775
Li, M., Si, S., Hao, B., Zha, Y., Wan, C., Hong, S., et al. (2014). Mild alkali-pretreatment efectively extracts guaiacyl-rich lignin for high lignocellulose digestibility coupled with largely diminishing yeast fermentation inhibitors in miscanthus. Bioresour. Technol. 169, 447–454. doi: 10.1016/j.biortech.2014.07.017
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real time quantitative PCR and the 2ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Li, F., Xie, G., Huang, J., Zhang, R., Li, Y., Zhang, M. (2017). OsCESA9 conserved-site mutation leads to largely enhanced plant lodging resistance and biomass enzymatic saccharification by reducing cellulose DP and crystallinity in rice. Plant Biotechnol. J. 15, 1093–1104. doi: 10.1111/pbi.12700
Ma, X., Li, C., Huang, R., Zhang, K., Wang, Q., Fu, C., et al. (2021). Rice Brittle Culm19 encoding cellulose synthase subunit CESA4 causes dominant brittle phenotype but has no distinct influence on growth and grain yield. Rice 14, 1–12. doi: 10.1105/tpc.011775
Mascher, M., Gundlach, H., Himmelbach, A., Beier, S., Twardziok, S. O., Wicker, T., et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433. doi: 10.1038/nature22043
Mascher, M., Wicker, T., Jenkins, J., Plott, C., Lux, T., Koh, C. S., et al (2021). Long-read sequence assembly: a technical evaluation in barley. Plant Cell 33, 1888–1906. doi: 10.1093/plcell/koab077
Michelmore, R. W., Paran, I., Kesseli, R. V. (1991). Identification of markers linked to disease resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Persson, S., Paredez, A., Carroll, A., Palsdottir, H., Doblin, M., Poindexter, P., et al. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 15566–15571. doi: 10.1073/pnas.0706592104
Persson, S., Wei, H., Milne, J., Page, G. P., Somerville, C. R. (2005). Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. U.S.A. 102, 8633–8638. doi: 10.1073/pnas.0503392102
Polko, J. K., Kieber, J. J. (2019). The regulation of cellulose biosynthesis in plants. Plant Cell. 31, 282–296. doi: 10.1105/tpc.18.00760
Roudier, F., Schindelman, G., DeSalle, R., Benfey, P. N. (2002). The COBRA family of putative GPI-anchored proteins in Arabidopsis, a new fellowship in expansion. Plant Physiol. 130, 538–548. doi: 10.1104/pp.007468
Sato, K., Suzuki, R., Nishikubo, N., Takenouchi, S., Ito, S., Nakano, Y., et al. (2010). Isolation of a novel cell wall architecture mutant of rice with defective Arabidopsis COBL4 ortholog BC1 required for regulated deposition of secondary cell wall components. Planta 232, 257–270. doi: 10.1007/s00425-010-1171-4
Sindhu, A., Langewisch, T., Olek, A., Multani, D. S., McCann, M. C., Vermerris, W., et al. (2007). Maize brittle stalk2 encodes a COBRA-like protein expressed in early organ development but required for tissue flexibility at maturity. Plant Physiol. 145, 1444–1459. doi: 10.1104/pp.107.102582
Somerville, C. (2006). Cellulose synthesis in higher plants. Annu. Rev. Cell. Dev. Biol. 22, 53–78. doi: 10.1146/annurev.cellbio.22.022206.160206
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tanaka, K., Murata, K., Yamazaki, M., Onosato, K., Hirochika, H. (2003). Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol. 133, 73–83. doi: 10.1104/pp.103.022442
Tan, H. T., Shirley, N. J., Singh, R. R., Henderson, M., Dhugga, K. S., Mayo, G. M., et al. (2015). Powerful regulatory systems and post-transcriptional gene silencing resist increases in cellulose content in cell walls of barley. BMC. Plant Biol. 15, 1–16. doi: 10.1186/s12870-015-0448-y
Taylor, N. G. (2008). Cellulose biosynthesis and deposition in higher plants. New Phytol. 178, 239–252. doi: 10.1111/j.1469-8137.2008.02385.x
Taylor, N. G., Laurie, S., Turner, S. R. (2000). Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell. 12, 2529–2539. doi: 10.1105/tpc.12.12.2529
Taylor, N. G., Scheible, W. R., Cutler, S., Somerville, C. R., Turner, S. R. (1999). The irregular xylem3 locus of arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell. 11, 769–779. doi: 10.2307/3870813
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., Higgins, D. G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Wang, X., Cheng, Z., Zhao, Z., Gan, L., Qin, R. Z., Zhou, K. N., et al. (2016). BRITTLE SHEATH1 encoding OsCYP96B4 is involved in secondary cell wall formation in rice. Plant Cell Rep. 35, 745–755. doi: 10.1007/s00299-015-1916-4
Wang, D. F., Qin, Y. L., Fang, J. J., Yuan, S. J., Peng, L. X., Zhao, J. F., et al. (2016). A missense mutation in the zinc finger domain of OsCESA7 deleteriously affects cellulose biosynthesis and plant growth in rice. PloS One 11, e0153993. doi: 10.1371/journal.pone.0153993
Wang, W. X., Wang, Z. H., Li, X. T., Ni, Z. F., Hu, Z. R., Xin, M. M., et al. (2020). SnpHub: an easy-to-set-up web server framework for exploring large-scale genomic variation data in the post-genomic era with applications in wheat. GigaScience 9, 1–8. doi: 10.1093/gigascience/giaa060
Wang, D. F., Yuan, S., Yin, L., Zhao, J. F., Guo, B. T., Lan, J. H., et al. (2012). A missense mutation in the transmembrane domain of CESA9 affects cell wall biosynthesis and plant growth in rice. Plant Sci. 196, 117–124. doi: 10.1016/j.plantsci.2012.08.002
Wu, B., Zhang, B., Dai, Y., Zhang, L., Guan, K. S., Peng, Y. G., et al. (2012). Brittle culm15 encodes a membrane-associated chitinase-like protein required for cellulose biosynthesis in rice. Plant Physiol. 159, 1440–1452. doi: 10.1104/pp.112.195529
Yan, C., Yan, S., Zeng, X., Zhang, Z., Gu, M. (2007). Fine mapping and isolation of Bc7 (t), allelic to OsCesA4. J. Genet. Genomics 34, 1019–1027. doi: 10.1016/S1673-8527(07)60115-5
Zhang, B., Deng, L., Qian, Q., Xiong, G., Zeng, D., Li, R., et al. (2009). A missense mutation in the transmembrane domain of CESA4 affects protein abundance in the plasma membrane and results in abnormal cell wall biosynthesis in rice. Plant Mol. Biol. 71, 509–524. doi: 10.1007/s11103-009-9536-4
Zhang, B., Liu, X., Qian, Q., Liu, L., Dong, G., Xiong, G., et al. (2011). Golgi nucleotide sugar transporter modulates cell wall biosynthesis and plant growth in rice. Proc. Natl. Acad. Sci. U.S.A. 108, 5110–5115. doi: 10.1073/pnas.1016144108
Zhang, M., Zhang, B., Qian, Q., Yu, Y., Li, R., Zhang, J., et al. (2010). Brittle culm 12, a dual-targeting kinesin-4 protein, controls cell-cycle progression and wall properties in rice. Plant J. 63, 312–328. doi: 10.1111/j.1365-313X.2010.04238.x
Zhang, B., Zhou, Y. (2011). Rice brittleness mutants: a way to open the 'black box' of monocot cell wall biosynthesis. J. Integr. Plant Biol. 53, 136–142. doi: 10.1111/j.1744-7909.2010.01011.x
Keywords: barley, cell wall, CesA, cellulose synthesis, brittle culm
Citation: Guo B, Huang X, Qi J, Sun H, Lv C, Wang F, Zhu J and Xu R (2022) Brittle culm 3, encoding a cellulose synthase subunit 5, is required for cell wall biosynthesis in barley (Hordeum vulgare L.). Front. Plant Sci. 13:989406. doi: 10.3389/fpls.2022.989406
Received: 08 July 2022; Accepted: 28 October 2022;
Published: 23 November 2022.
Edited by:
Seonghee Lee, University of Florida, United StatesReviewed by:
Chaofeng Li, The University of Tokyo, JapanCopyright © 2022 Guo, Huang, Qi, Sun, Lv, Wang, Zhu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rugen Xu, cmd4dUB5enUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.