- 1Zhejiang Provincial Key Laboratory of Crop Germplasm, Institute of Crop Science, Zhejiang University, Hangzhou, Zhejiang, China
- 2Hainan Institute of Zhejiang University, Sanya, Hainan, China
- 3Seed Management Station of Zhejiang Province, Hangzhou, Zhejiang, China
The HD-Zip transcription factors play a crucial role in plant development, secondary metabolism, and abiotic stress responses, but little is known about HD-Zip I genes in soybean. Here, a homeodomain-leucine zipper gene designated GmHdz4 was isolated. Chimeric soybean plants, GmHdz4 overexpressing (GmHdz4-oe), and gene-editing via CRISPR/Cas9 (gmhdz4) in hairy roots, were generated to examine the GmHdz4 gene response to polyethylene glycol (PEG)-simulated drought stress. Bioinformatic analysis showed GmHdz4 belonged to clade δ, and was closely related to other drought tolerance-related HD-Zip I family genes such as AtHB12, Oshox12, and Gshdz4. The GmHdz4 was located in the plant nucleus and showed transcriptional activation activity by yeast hybrid assay. Quantitative real-time PCR analysis revealed that GmHdz4 expression varied in tissues and was induced by PEG-simulated drought stress. The gmhdz4 showed promoted growth of aboveground parts, and its root system architecture, including the total root length, the root superficial area, and the number of root tips were significantly higher than those of GmHdz4-oe even the non-transgenic line (NT) on root tips number. The better maintenance of turgor pressure by osmolyte accumulation, and the higher activity of antioxidant enzymes to scavenge reactive oxygen species, ultimately suppressed the accumulation of hydrogen peroxide (H2O2), superoxide anion (O2−), and malondialdehyde (MDA), conferring higher drought tolerance in gmhdz4 compared with both GmHdz4-oe and NT. Together, our results provide new insights for future research on the mechanisms by which GmHdz4 gene-editing via CRISPR/Cas9 system could promote drought stress and provide a potential target for molecular breeding in soybean.
Introduction
Soybean (Glycine max [L.] Merr.) is an important oilseed crop, used as both food and feed due to its high protein content. China is a traditional soybean-producing country, and the people have long consumed soybeans and processed soybean products. However, a 100 million tons of soybeans need to be imported each year to meet domestic soybean consumption (Sun, 2020), accounting for about 60% of the global soybean trade volume, and external dependence is as high as 80% (Niu et al., 2021). In addition, due to the increase in extreme weather events, the impact of abiotic stresses such as drought, salinity, and high temperature on crop yields and economic losses has increased (Lesk et al., 2016). Soybean is sensitive to water shortage compared with many other crops, requiring about 450–700 mm of water throughout the growth period (Dogan et al., 2007). Especially at podding and filling stages, drought stress can seriously restrict soybean yield, resulting in a loss of about 40% of yield. During the soybean growth period, most areas of China experience different degrees of drought (You et al., 2021). Drought in Northeast China begins in mid-March, and develops into severe and extreme drought conditions from late April to May, which is the growing season for soybean. In addition to water-saving irrigation and raising irrigation efficiency, an effective way to improve soybean yield in water deficit areas is to identify the mechanism of drought tolerance and explore key genes to breed drought-tolerant soybean germplasm.
Homeodomain-leucine zipper (HD-Zip) protein is a class of transcription factor specific to higher plants (Schena and Davis, 1992). Its conserved domain is composed of a homeodomain (HD) and a closely linked leucine zipper (LZ). The HD-Zip proteins can be divided into four subfamilies: I, II, III, and IV. Studies on Arabidopsis thaliana (Peterson et al., 2013), rice (Oryza sativa) (Zhang et al., 2012), and sesame (Sesamum indicum) (Wei et al., 2019) have shown that HD-Zip I is mainly responsible for the regulation of plant growth and development, morphogenesis, and abiotic stress responses (Harris et al., 2011; Gong et al., 2019). For example, genes AtHB6 and AtHB7 were strongly induced under drought stress or exogenous abscisic acid (ABA) treatment and stomata were subsequently closed to adapt to these stresses (Perotti et al., 2017). Genes AtHB7 and AtHB12 acted as negative feedback regulation factors in the ABA signaling pathway, which influenced lateral root development under water-deficient conditions (Himmelbach et al., 2002; Romani et al., 2016). Expression of MeHDZ14 was also upregulated under drought and exogenous ABA stress in cassava leaves and roots (Yu et al., 2017). Agalou et al. (2008) found that the expression of Oshox22 was induced by polyethylene glycol (PEG)-simulated drought but that of Oshox4, which was specifically expressed in vascular bundles, was inhibited. In addition, Oshox4 overexpression led to accelerated gibberellin (GA) metabolism, shortened internode, and prolonged the vegetative growth period in rice. This suggested that Oshox4 was involved in the negative regulation of GA under drought stress (Zhou et al., 2015b). Overexpression of the Helianthus annuus HD-Zip homolog HaHB4 in wheat increased the number of spikelets, tillers, and fertile florets, and improved grain yield under drought stress (González et al., 2019). Overexpression of HaHB4 also improved drought tolerance in soybean by increasing xylem area and heat shock protein expression (Ribichich et al., 2020). The mechanisms of drought tolerance were found to be independent of traditional candidate genes such as RD19 and DREB1a (Liu et al., 1998).
There are 36 members of the HD-Zip I transcription factor family in soybean (Chen et al., 2014), among which GmHB13 was especially induced by water deficit in a drought-tolerant variety (EMBRAPA 48), while expression of GmHB6 was inhibited in a drought-sensitive variety BR16 (Belamkar et al., 2014). Limited HD-Zip I genes were identified in previous studies. Gene Gshdz4 (accession number: KHN 42008.1) was reported to participate in drought and salt tolerance regulation (Cao et al., 2017), while GmHdz4 (Glyma11g06940) was a homologous gene of Gshdz4 which had a 98.0% amino acid sequence similarity. Therefore, we speculate that GmHdz4 may also be involved in drought stress response in soybean. The aims of this study were to (1) clone the coding sequence (CDS) of GmHdz4, analyze its phylogenetic relationship and the spatio-temporal expression pattern in response to drought stress, (2) analyze the subcellular localization and transcription activity of GmHdz4, (3) construct overexpression and CRISPR/cas9 vectors of GmHdz4 to generate overexpression and knockout chimeric lines to study the function of GmHdz4, and (4) reveal the physiological and biochemical mechanisms of drought stress in overexpressing and gene-edited plants. These results may lay a theoretical foundation for further drought-tolerant soybean germplasm innovation.
Materials and methods
Plant materials and growth conditions
Soybean (Glycine max [L.] Merr.) cv. Tianlong No. 1 was used as the transgenic receptor in this experiment. The wild type and transgenic chimeric plants were grown in a temperature/light-controlled greenhouse at Zhejiang University, Zijingang Campus. All plants were grown under short-day conditions (light/dark of 10/14 h at 26/22°C). The roots, stems, unifoliolate leaves, flowers, and tender pods were taken at V1, R1, and R3 stages, for analysis of tissue-specific expression of GmHdz4. The PEG-simulated drought stress was imposed as follows: soybean seeds were germinated in a moist sand bed for 3 weeks. Uniform seedlings were selected and grown using half-strength Hoagland (Xu et al., 2017) solution with 15% PEG6000 (osmotic potential, Ψπ = −0.32 MPa, pH 5.8) to simulate drought stress. The drought-stressed roots were sampled at 0, 3, 6, and 12 h; 1, 2, and 4 days to measure the relative expression of GmHdz4. Three biological replicates were used for each analysis. All samples were immediately frozen in liquid nitrogen and stored at −80°C for further analysis.
Sequence alignment and phylogenetic analysis
The sequences of HD-Zip I family members from Arabidopsis, rice, maize, tomato, sunflower, wild soybean, and soybean were screened using BLAST in Phytozome.1 Multiple amino acid sequences were aligned using ClustalX 2.0 (Thompson et al., 1994) with default parameters. The phylogenetic tree was produced using MEGA 5.0 software (Tamura et al., 2011) with the neighbor-joining (NJ) method, and the bootstrap value was 1,000 times for each node to test branch reliability.
Total RNA extraction and cDNA synthesis
The CDS and genome sequence of GmHdz4 (Glyma11g06940) were obtained and aligned from NCBI,2 TAIR,3 and Phytozome databases. Total RNA was extracted according to the modified Trizol method (Bekesiova et al., 1999), and RNA integrity was verified by 2% agarose gel electrophoresis. First-strand cDNA was synthesized using a reverse transcription kit (Vazyme, Nanjing, China). Three independent biological replicates were used for RNA extraction and subsequent cDNA synthesis. The CDS of GmHdz4 was amplified by gene-specific primers GmHdz4_CDS_F (5′-ATGAATCATCGACCACCTTTCC-3′) and GmHdz4_CDS_R (5′-CAGATTAATCCATTCCATGCCG-3′) using Taq Master Mix (Vazyme, Nanjing, China). The target PCR products were cut and recycled with a gel extraction kit (Sangon Biotech, Shanghai, China) after separation by 2% agarose gel electrophoresis. The product was cloned into pUCI-T vector (Sangon Biotech, Shanghai, China) then transformed to Escherichia coli DH5α competent cells by the freeze–thaw method (Knopf, 1976). The selected single colonies were sequenced and stored at −20°C for subsequent experiments.
Real-time fluorescent quantitative PCR (qRT-PCR) analysis
The qRT-PCR was performed using ChamQ SYBR qPCR Master Mix (Vazyme, Nanjing, China), according to the manufacturer’s instruction. Specific primers designed for GmHdz4 were GmHdz4Q_F (5′-GACCACCTTTCCAAGACCACA-3′) and GmHdz4Q_R (5′-AGCTCCATTGCCAGCCTATC-3′). The normalizing reference gene was Actine11 (Xue et al., 2012). Abnormal CT values (> 0.2 + CTmedian) were eliminated, and relative expression levels were calculated according to the 2−ΔΔCT method (Livak and Schmittgen, 2001). All samples were selected randomly under the same greenhouse conditions. Three technical replicates for each biological replicate were used in qRT-PCR analysis.
Plasmid construction
For overexpression plant transformation, the full-length CDS of GmHdz4 isolated from soybean root tissue was amplified by PCR using premiers: GmHdz4-plus-F (5′-CAGTGAATTCCTGGACGTCCGTACGTTCGA-3′) and GmHdz4-plus-R (5′-CGATGAATTCCGGCGCAAAAATCACCAGTC-3′). The amplification products were inserted into the pTF102 vector EcoRI site to generate pTF102–GmHdz4 vector, containing a bar gene under the control of the CaMV35S promoter for the herbicide-based plant selection (Supplementary Figure S2A). For gene-editing plant transformation, the target sequence of GmHdz4 (Supplementary Figure S2C) was predicted by CRISPR-P4 (Lei et al., 2014) and the sgRNA sequence was 5′-GTCCGAAAGAAAGGATAGGC-3′. Specific designed oligo probes were 5′-GGGTTGGTCCGAAAGAAAGGATAGGC-3′ and 5′-AAACGTCCGAAAGAAAGGATAGGCCA-3′. The amplified dimer was inserted into the pBGK041 vector according to the manufacturer’s instructions to generate gene-editing pBGK041–GmHdz4 vector. The pBGK041 vector (Biogle, Jiangsu) contained Cas9 cassettes, driven by S35, and the inserted sgRNA was driven by GmU6 promoters (Supplementary Figure S2D). The plasmids of pTF102–GmHdz4 recombinant vector and pBGK041–GmHdz4 gene-editing vector were transformed into Agrobacterium rhizogenes K599 by the freeze–thaw method (Knopf, 1976), respectively.
Subcellular localization and transcriptional activation analysis
For subcellular localization, the recombinant vector pCAMBIA1302–GmHdz4 expressing the GmHdz4–GFP fusion protein and the positive marker vector pBWD-NLSmKAT expressing a nuclear-localized red fluorescent protein (Zoonbio, Nanjing, China) were transferred into Agrobacterium tumefaciens GV3101 by electroporation, respectively. After incubating at 30°C for 12 h, single colonies were picked out and cultured at 170 rpm/min for 1 h. A needle-free syringe was used to inject the resuspended bacterial solution under the epidermis of Nicotiana tabacum leaves, and this was marked. The GmHdz4–GFP fusion protein was examined 2 days after culture under weak light using a confocal laser scanning microscope (FV1000; Olympus, Japan). This experiment was repeated three times independently.
For transcriptional activation analysis, the experiment was based on the GAL4 yeast two-hybrid (Y2H) system (Chien et al., 1991), and the GmHdz4 gene was introduced into pGBKT7 vector. Plasmids pGBKT7-Lam and pGADT7-T were co-transformed into Y2HGold yeast as a negative control, and plasmids pGBKT7-53 and pGADT7-T were used as a positive control. Transcription activation activity was analyzed following the user manual of Takara Bio USA, Inc.
Hairy roots system generation and identification
Healthy and vigorous seeds of Tianlong No. 1 were selected, sterilized by chlorine gas, and germinated in sterile vermiculite for 5 days. After the hypocotyl grew about 2–3 cm length, 200 μl resuspended A. rhizogenes solution (OD600 ≈ 0.6) carrying recombinant plasmid was injected into each hypocotyl about 2 mm below the cotyledon node according to Kereszt et al. (2007). The A. rhizogenes without the target gene was used as a control. The photoperiod was controlled for 6 h of light and18 h of dark, temperature was maintained at 28°C, and relative humidity was about 70%. The hypocotyl and primary roots were excised when enough hairy roots were induced 1 week after injection. The chimeric seedlings were transplanted into half-strength Hoagland nutrient solution. The chimeric seedlings containing pTF102–GmHdz4 and pBGK041–GmHdz4 vectors were labeled GmHdz4-oe and gmhdz4, respectively, and seedlings without the target gene plasmids were labeled non-transgenic (NT).
The hairy roots of GmHdz4-oe chimeric seedlings were verified by PCR amplification using specific primers GmHdz4_CDS, which were mentioned above. The hairy roots of gmhdz4 chimeric seedlings were verified by PCR using Cas9 gene specific primers (5′-ACAAGGTGAGCGTTGTTTAT-3′ and 5′- CATGATGGTGGTATGTTTCG-3′). And the target site on the GmHdz4 CDS of each gmhdz4 hairy root was sequenced. The hairy roots from the independent chimera seedling line were collected and mixed to detect the expression of target sequence on GmHdz4 CDS. Three independent transgenic lines as biological replicates were performed.
Identification of GmHdz4 function by PEG-simulated drought stress
The positive overexpressed and gene-editing chimeric soybean seedlings were transplanted into half-strength Hoagland solution containing 15% PEG6000 (pH 5.8) for drought stress treatment. The phenotypic traits and physiological indexes were determined at 0, 6, and 12 h; 1, 2, 4, and 8 days after PEG treatment. Fresh root samples were immediately frozen in liquid nitrogen and stored at −80°C for further experiments. The following experiments had three biological replicates at least.
Biomass determination
After 8 days of PEG treatment, the fresh shoots and roots were dried for 3 h at 105°C and then for another 24 h at 80 ± 1.5°C to obtain a constant dry weight for the measurement for dry biomass. The ratios of root mass to shoot mass were then calculated.
Root system architecture analysis
The chimeric seedlings were cleaned in nutrient solution 8 days after PEG treatment. The hairy roots were scanned by Epson Perfection V850 Pro. The root system architecture was analyzed using WinRHIZO software (Win et al., 2016).
Hairy root activity determination
The root activity of each sample was measured by improved 2,3,5-triphenyl tetrazolium chloride (TTC) method (Higa et al., 2010). A microplate reader was used to detect the OD value of formazan at a wavelength of 485 nm. The TTC-reducing activity was converted to represent the root activity.
Osmoprotectant estimation
The content of free proline was determined according to Bates et al. (1973) and of soluble sugar according to Bailey (1958). The absorbance was measured using a microplate reader (Synergy™, BioTek).
Determination of hydrogen peroxide (H2O2), superoxide anion radical (O2−), and malondialdehyde (MDA) contents and antioxidant enzyme activities
The H2O2 and O2− contents in hairy roots were measured according to Alexieva et al. (2001) using reactive oxygen species (ROS) content reagent kits (Solarbio, Beijing). The enzyme activities of superoxide dismutase (SOD, EC1.15.1.1), peroxidase (POD, EC 1.11.1.7), and catalase (CAT, EC 1.11.1.6) were measured by NBT illumination, guaiacol reaction, and oxidation–reduction methods, respectively (Wang et al., 2021). The MDA was measured according to Heath and Packer (1968) and Tang et al. (2013).
Statistical analysis
All data are presented as the mean values with the standard deviation of three replications in figures. Statistical analysis was performed using EXCEL 2019 (Microsoft Inc., Redmond, WA, United States) and SPSS 20 (SPSS Inc., Chicago, IL, United States). Significant differences were determined using one-way ANOVA and Duncan’s multiple range tests. The differences between treatments were considered significant at p < 0.05. Prism7 (GraphPad Inc., San Diego, CA, United States) was used to visualize the data.
Results
GmHdz4 cloning and phylogenetic analysis
The CDS sequence of GmHdz4 (Glyma11g06940) was amplified by high-fidelity PCR using cDNA from total RNA of soybean cultivar Tianlong No. 1 as a template. The PCR product was cloned into pUCI-T vector to obtain pUCI–GmHd4 (Supplementary Figure S1) and sequenced. The sequencing result was completely consistent with the NCBI database. Gene GmHdz4 contained a 648-bp open reading frame encoding a protein of 215 amino acids; the molecular weight of GmHdz4 protein was 25.11 kDa, and the isoelectric point was 6.48. Sequence analyses revealed that most of the GmHdz4 amino acid sequence formed hydrophilic regions except for the aa10–24, aa64–72, and aa88–102, and nine out of 12 sites that may undergo phosphorylation modification were located in these regions. The results of multiple sequence alignment showed that GmHdz4 had a highly conserved HD compared with other HD-Zip I transcription factors, which was related with drought stress in Arabidopsis, rice, maize, tomato, wild soybean, and sunflower. The LZ domain was also relatively conserved, with only individual leucine sites mutated to threonine or valine (Figure 1A). The 3D model of GmHdz4 protein predicted by SWISS MODEL Workspace showed that the HD domain folded into three α-helices. This spatial conformation provided the conditions to recognize and bind the promoter elements of the target genes, and is consistent with the typical structural features of HD-Zip I transcription factors.

Figure 1. Sequence alignment and phylogenetic analysis. (A) Multiple sequence alignment for GmHdz4 and other drought stress-related HD-Zip I proteins. HD and LZ domains are underlined by black and gray, respectively. The corresponding IDs of Oshox22, Oshox4, AtHB7, AtHB12, AtHB6, Gshdz4, GmHdz4, HAHB4, Zmhdz10, SlHZ48, and SlHZ5, are Q7XUJ5.2, Q6K498.1, P46897.2, Q9M276.1, P46668.1, AOG74801, Glyma11g06940, XP_022022563, AFT92045, XP_004245456, and XP_004230017, respectively. (B) Phylogenetic analysis of HD-Zip I transcription factors from Arabidopsis, rice, maize, tomato, soybean, and wild soybean. The phylogenetic tree was built using NJ method in MEGA. Bootstrap support is indicated on the branches. Genes from each of the species are marked with different symbols: Arabidopsis (○), rice (■), maize (□), soybean (▼), wild soybean (▽), and tomato (●).
Phylogenetic analysis was performed among HD-Zip I subfamily members in Arabidopsis, rice, maize, tomato, wild soybean, and soybean. The clades in the tree were labeled according to previous studies in Arabidopsis and rice. The HD-Zip I subfamily members were clustered into eight subclasses: α, β1, ζ, γ, ɛ, β2, δ, and φ (Figure 1B). The distribution showed an obvious species preference. Six species contained HD-Zip I protein clades α, β1, β2, γ, and δ while only dicotyledons such as Arabidopsis, tomato, and soybean included clades ζ, ɛ, and φ. The results suggested that the main characteristics of this HD-Zip I subfamily were generated before the dicot–monocot split and differentiated continually after the split. Gene GmHdz4 was located in clade δ, and was closely related to drought stress-related HD-Zip I genes such as AtHB12, Oshox12, and Gshdz4.
GmHdz4 spatio-temporal and tissue-specific expression
The relative expressions of GmHdz4 in roots, stems, leaves, flowers, and pods significantly differed with the main expression in flowers and roots (Figure 2A). Expression of GmHdz4 in roots was about 60-fold higher than that in leaves, and 1.3-fold higher than that in flowers. The relative expression of GmHdz4 in roots showed a stepwise significant decrease within 12 h after 15% PEG6000 (Ψπ = −0.32 MPa) simulated drought stress (Figure 2B). The relative expression of GmHdz4 was reduced by 47.0% at 6 h after drought stress, and dropped to about 18% by 12 h after drought stress. Later, GmHdz4 expression decreased slightly and basically remained at 16% before treatment. The results suggested that GmHdz4 expression was inhibited by drought in soybean roots.
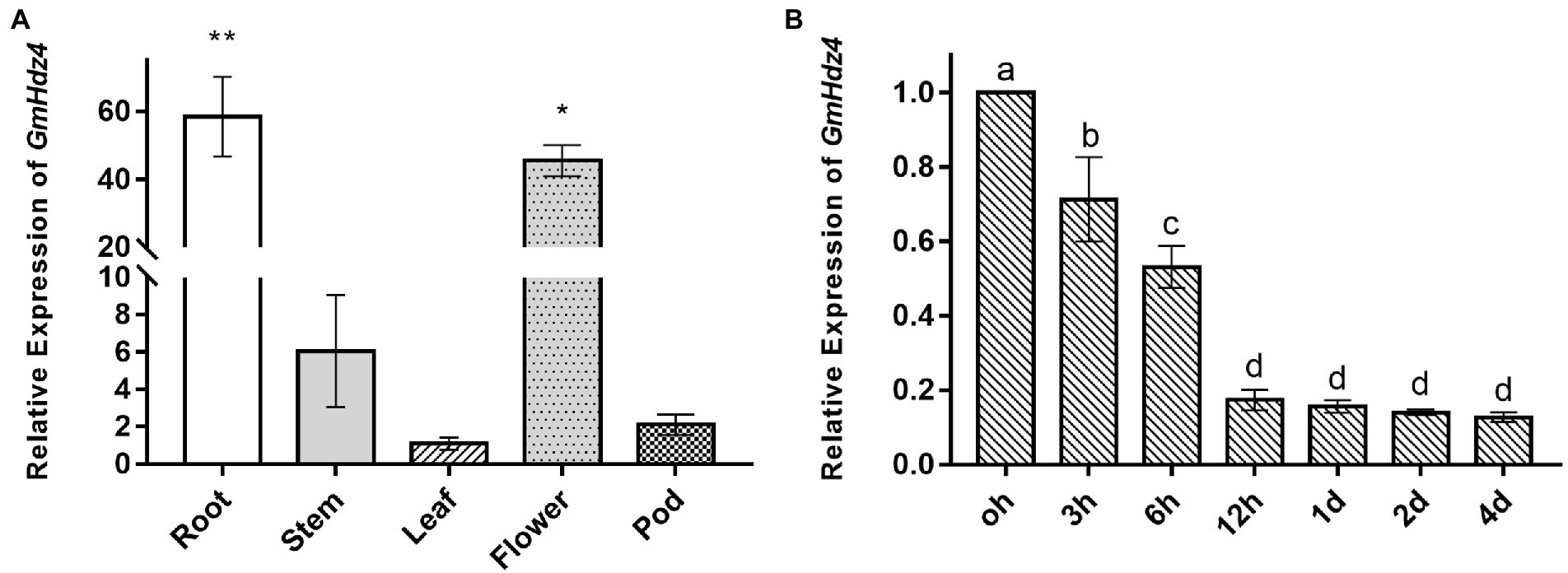
Figure 2. Tissue-specific and spatio-temporal expression of GmHdz4. Expression pattern of GmHdz4 in various tissues in soybean (A) and PEG-induced expression pattern of GmHdz4 in root (B). Significant differences are indicated by * and ** for p < 0.05 and p < 0.01, respectively, according to Duncan’s test in panel A; values with different letters (a–d) significantly differ at p < 0.05 according to Duncan’s test in panel B.
GmHdz4 subcellular localization and transcriptional activity
Gene GmHdz4 might target the nucleus according to its amino acid sequence, which contained an NLS sequence at the region of aa35–60. The fluorescence signal of GmHdz4 was only detected in the nucleus when tobacco cells were injected with a 35 s::GmHdz4–mGFP fusion construct and the NLS Red Marker (Figure 3B). However, the GFP signal was found throughout the cell using 35 s::mGFP as a control (Figure 3A).
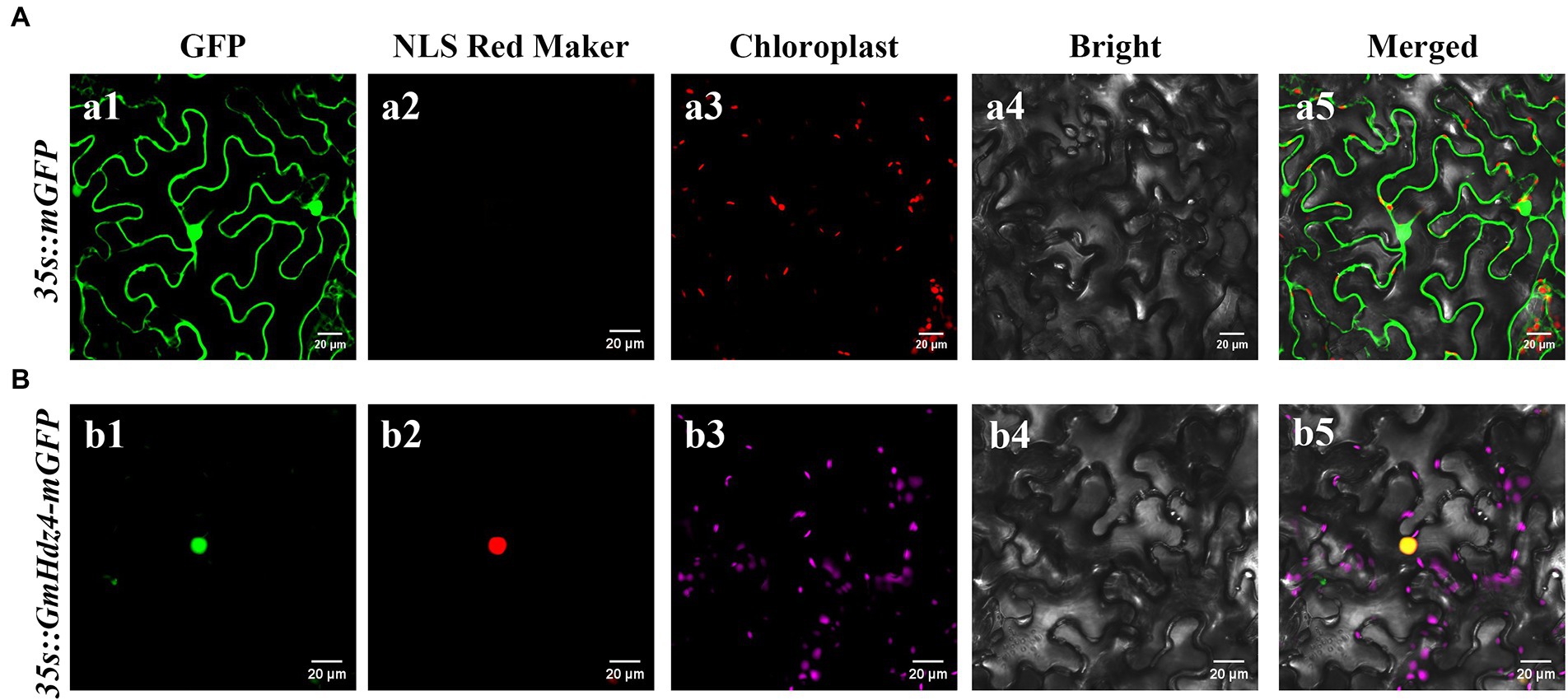
Figure 3. Subcellular localization analysis of GmHdz4. The fusion plasmid (35S::GmHdz4–GFP), the negative control plasmid (35S::mGFP), and pBWD-NLSmKAT expressing an NLS Red Marker protein were transiently transformed into tobacco epidermal cells. Images were taken in the dark field for green fluorescence (a1, b1), red fluorescence (a2, b2), and chloroplast fluorescence (a3, b3), while the outline of cells (a4, b4) and merged image (a5, b5) were photographed in a bright field. Microscopy detections of 35S::mGFP were displayed in penal (A), and the detections of 35S::GmHdz4–mGFP were displayed in penal (B). Bars represent 20 μm.
The pGBKT7–Gmhdz4 recombination vector was delivered into yeast strain Y2HGold to identify the transcriptional activation activity of Gmhdz4 (Figure 4A). The interaction between pGBKT7-Lam and pGADT7-T was the negative control, while the interaction between pGBKT7-p53 and pGADT7-T was the positive control. Yeast cells carrying the pGBKT7 empty plasmid and the negative control could not grow on SD/Trp-/His−/Ade- medium (Figure 4B); however, yeast cells carrying pGBKT7–GmHdz4 and the positive control grew well. This showed that the fusion expression of GmHdz4 and GAL4 DNA-binding domain could activate reporter genes ADE2 and HIS3. Meanwhile, pGBKT7–GmHdz4 also showed high galactosidase activity in the LacZ assay. The results indicated that GmHdz4 regulated transcription activities in the nucleus, consistent with two basic characteristics of transcription factors.
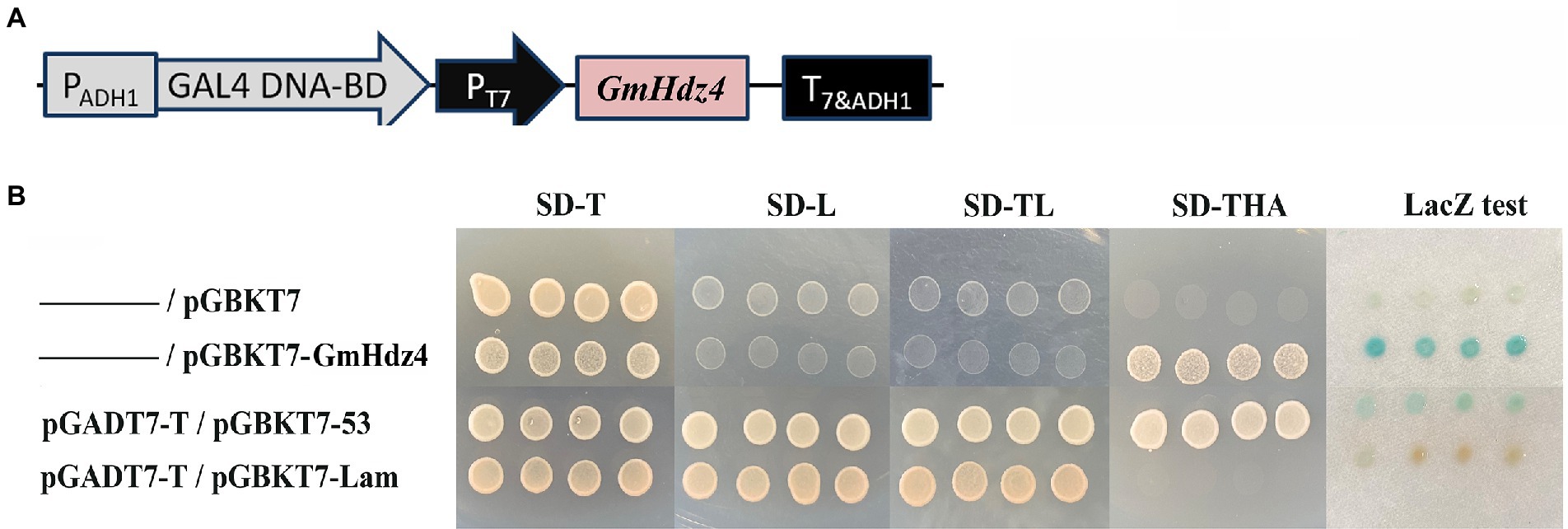
Figure 4. Transcription activity analysis of GmHdz4. (A) The construct of pGBKT7–GmHdz4; (B) ADE2 and HIS3 reporter assay, and galactosidase (LacZ) assay. pGADT7-T/pGBKT7-53 and pGADT7-T/pGBKT7-Lam vector groups were used as positive and negative controls, respectively.
The gmhdz4 mutant were more tolerant to drought stress
The A. rhizogenes-mediated transformation system was used to generate GmHdz4 overexpression and gene-editing chimeric soybean in order to explore GmHdz4 function. The fragments of GmHdz4 CDS with correct size could be amplified from most hairy roots of the three random selected GmHdz4-oe independent lines (Supplementary Figure S2B), that meant most individual hairy roots of GmHdz4-oe lines were successfully transformed. Similarly, majority hairy roots of gmhdz4 lines have Cas9 gene sequence (Supplementary Figure S2E), and there were three mutation types including a single base deletion on the 256 bp site, a single base insertion between the 256 bp site and the 257 bp site, and a four bases deletion of 254 bp to 257 bp (Supplementary Figure S2F). However, the target site of some transformed hairy roots were not edited successfully. In addition, the relative expression of GmHdz4 in GmHdz4-oe hairy roots was 5.1–8.8-fold higher than that for NT, whereas GmHdz4 expression in gmhdz4 hairy roots was 2.7–5.3-fold lower than that of NT (Supplementary Figure S2G).
The phenotypes obviously differed among NT, GmHdz4-oe, and gmhdz4 at 8 days after 15% PEG6000 simulated drought stress. The leaves were green and vigorous in NT (Figure 5A1) and gmhdz4 (Figure 5B1) before PEG treatment, and hairy roots developed normally (Figures 5A2,B2). However, leaves in GmHdz4-oe (Figures 5C1,C2) were etiolated and short, and hairy roots were stunted. Eight days after PEG treatment, most leaves in GmHdz4-oe were wilted and yellow, and the first trifoliolate leaf was dried and shed (Figures 5F1,F2). The dry weight of aboveground parts showed no significant difference between gmhdz4 and NT after PEG treatment, while that of GmHdz4-oe was relatively low (Figure 6A). Eight days after PEG treatment, gmhdz4 had the greatest root dry weight among the three chimeric lines with an average of 2.52 g/plant, which increased 411.2% before treatment. Emphatically, root dry weight in GmHdz4-oe was clearly the lowest, with less than one-half of that of gmhdz4 and NT. Additionally, the root–shoot ratio in descending order was gmhdz4, NT, and GmHdz4-oe (Figure 6B).
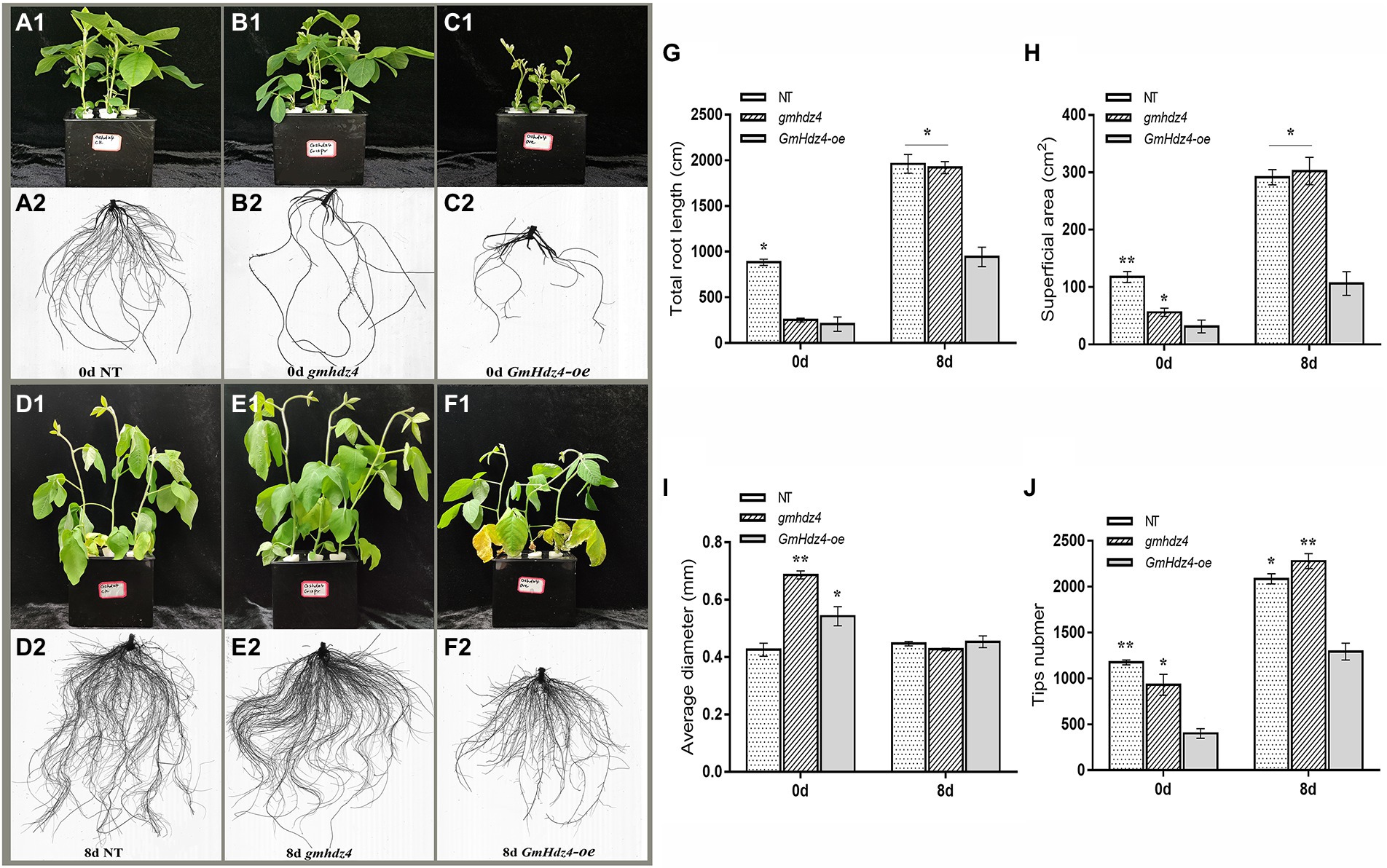
Figure 5. The phenotype and root system architecture among NT, gmhdz4 chimeric lines, and GmHdz4-oe chimeric lines. Phenotypic comparison of transgenic hairy roots chimera and NT soybeans before and after PEG treatment (A1–F2). The root system architecture analysis of total root length (G), superficial area (H), average diameter (I), and tip numbers (J). NT, non-transgenic soybeans; gmhdz4, gene-editing chimeric line; GmHdz4-oe, overexpression GmHdz4 chimeric line. At least three biological replicates were performed, and the data before (0d) and after (8 days) treatment were statistically analyzed, respectively. Significant differences are indicated by * and ** for p < 0.05 and p < 0.01, respectively, according to Duncan’s test.
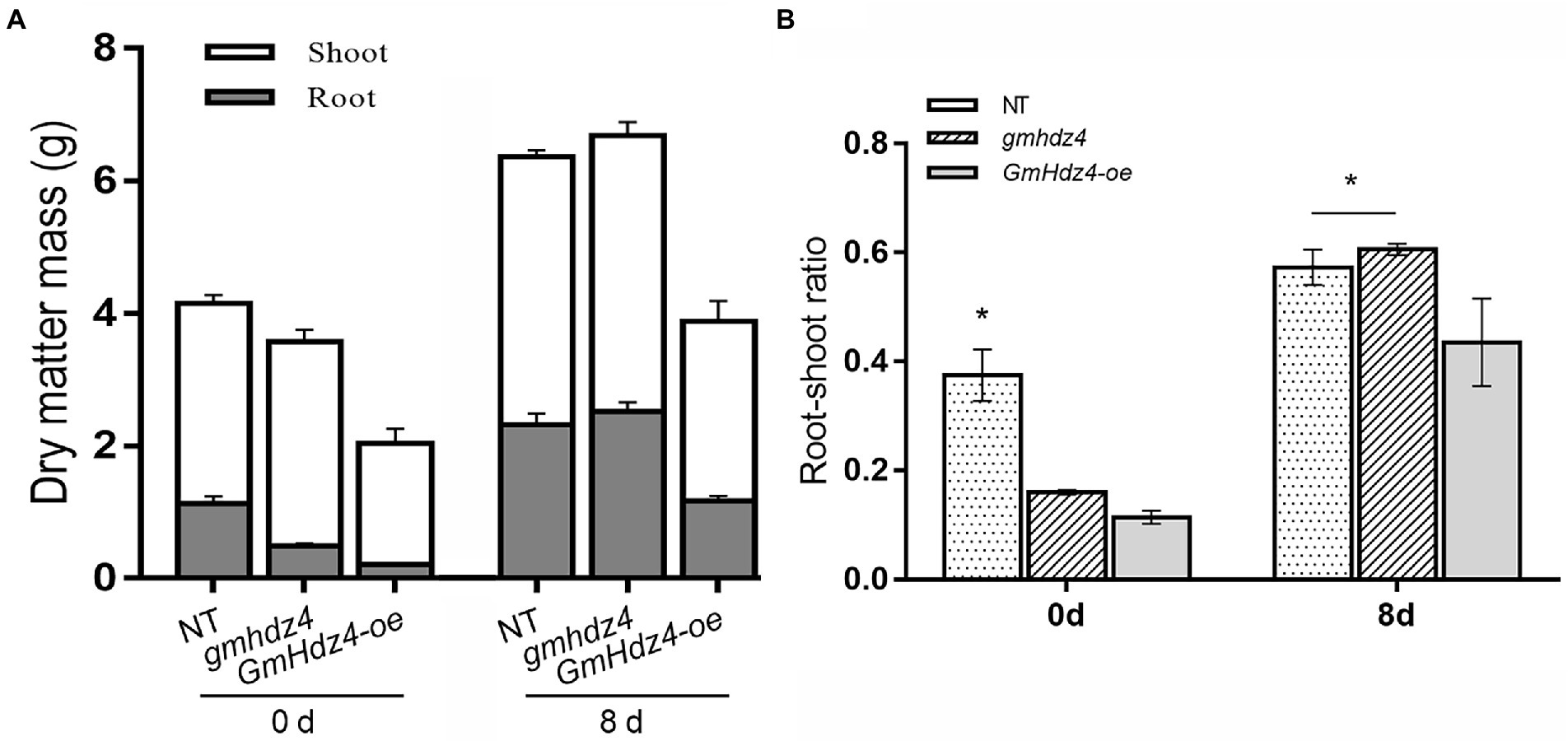
Figure 6. The dry matter mass (A) and root–shoot ratio (B) of each line before and after PEG treatment. Significant differences are indicated by * for p < 0.05, according to Duncan’s test.
The hairy root system in GmHdz4-oe was much weaker than that of NT (Figures 5D1,D2), while the root system in gmhdz4 developed well (Figures 5E1,E2). Analysis of root system architecture showed that the total root length (Figure 5G), superficial area (Figure 5H), and tip number (Figure 5J) in gmhdz4 were significantly higher than that of GmHdz4-oe, but not average diameter (Figure 5I) after PEG treatment. The total root length in gmhdz4 increased from 240 cm (before PEG treatment) to 2,100 cm (after PEG treatment), an increase of 775%, which was 5.8 times that of NT. The root surface area in gmhdz4 also increased from 55.97 to 302.33 cm2 and the number of root tips in gmhdz4 increased from about 900 to 2,300, with a significantly higher increase than that of NT and GmHdz4-oe. These results suggested that gene editing of GmHdz4 promoted the root morphogenesis under PEG treatment and enhanced drought tolerance in soybeans.
Accumulation of osmoprotectants, and hairy root activity enhanced in gmhdz4 mutant
There was no significant difference in soluble sugar and free-proline contents among hairy roots of GmHdz4-oe, gmhdz4, and NT lines before PEG treatment (Figure 7). With the prolongation of PEG-simulated drought stress, the soluble sugar and free-proline contents increased gradually among the three different lines (Figures 7A,B). However, the osmoprotectants in gmhdz4 increased significantly more than in GmHdz4-oe and NT. Eight days after PEG treatment, the soluble sugar content in gmhdz4 was 1.76 and 1.84 times higher than that of NT and GmHdz4-oe, respectively (Figure 7A); correspondingly, the proline content was 1.48 and 1.63 times higher (Figure 7B). This suggested that gmhdz4 obtained stronger osmotic accumulation ability under drought stress. Additionally, the root activity of gmhdz4 and NT showed no significant difference, while that of GmHdz4-oe significantly decreased after PEG treatment. The results suggested that gene editing of GmHdz4 maintained a high osmotic potential in hairy roots to ensure lower cellular water potential under drought stress, so that roots of gmhdz4 maintained normal physiological activity to cope with drought stress.
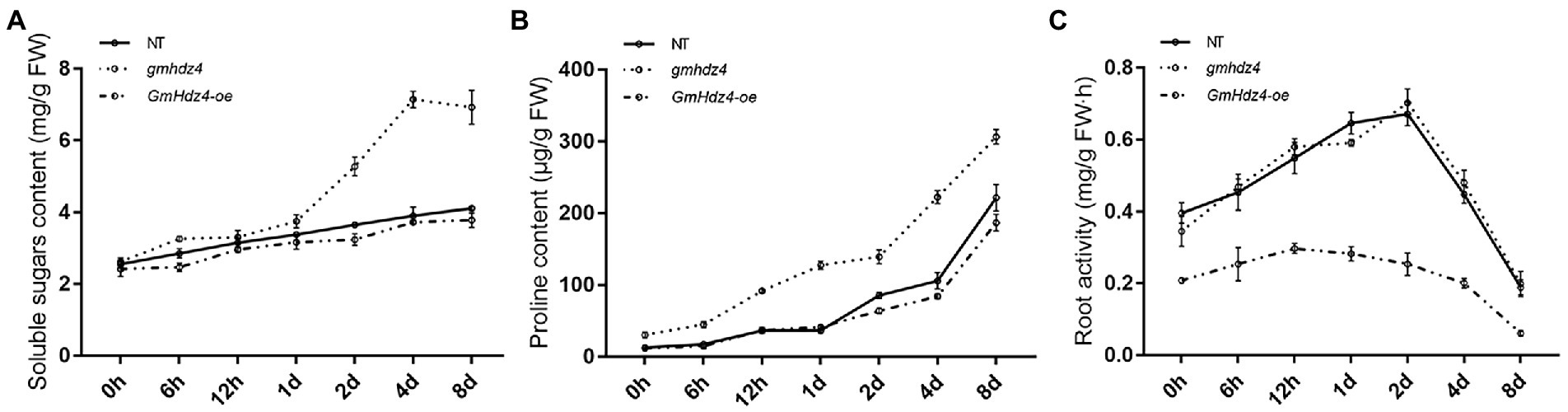
Figure 7. Soluble sugar content (A), proline content (B), and root activity (C) after PEG treatment. The straight line, dotted line, and dash-dotted line represent non-transgenic soybeans (NT), gmhdz4 chimeric line, and GmHdz4-oe line, respectively.
The gmhdz4 mutant promotes ROS scavenging capacity under drought stress
Accumulation of ROS increased gradually with continuing PEG treatment in soybean hairy roots. There was no significant difference in H2O2 contents in the hairy roots of NT and gmhdz4 in the first 3 days after PEG treatment. However, the O2− content in GmHdz4-oe was significantly higher than that of NT and gmhdz4. Subsequently, H2O2 accumulated rapidly in GmHdz4-oe chimeric lines (Figure 8A). The differences in O2− contents between GmHdz4-oe and the other two lines reached a maximum of about 2.1 times at 4 days after PEG treatment (Figure 8B). Eight days after PEG treatment, H2O2 content in gmhdz4 was only 12.0 μmol/g and was the lowest, compared with GmHdz4-oe and NT. Correspondingly, the MDA content was also significantly lower in gmhdz4 chimeric lines (Figure 8C).
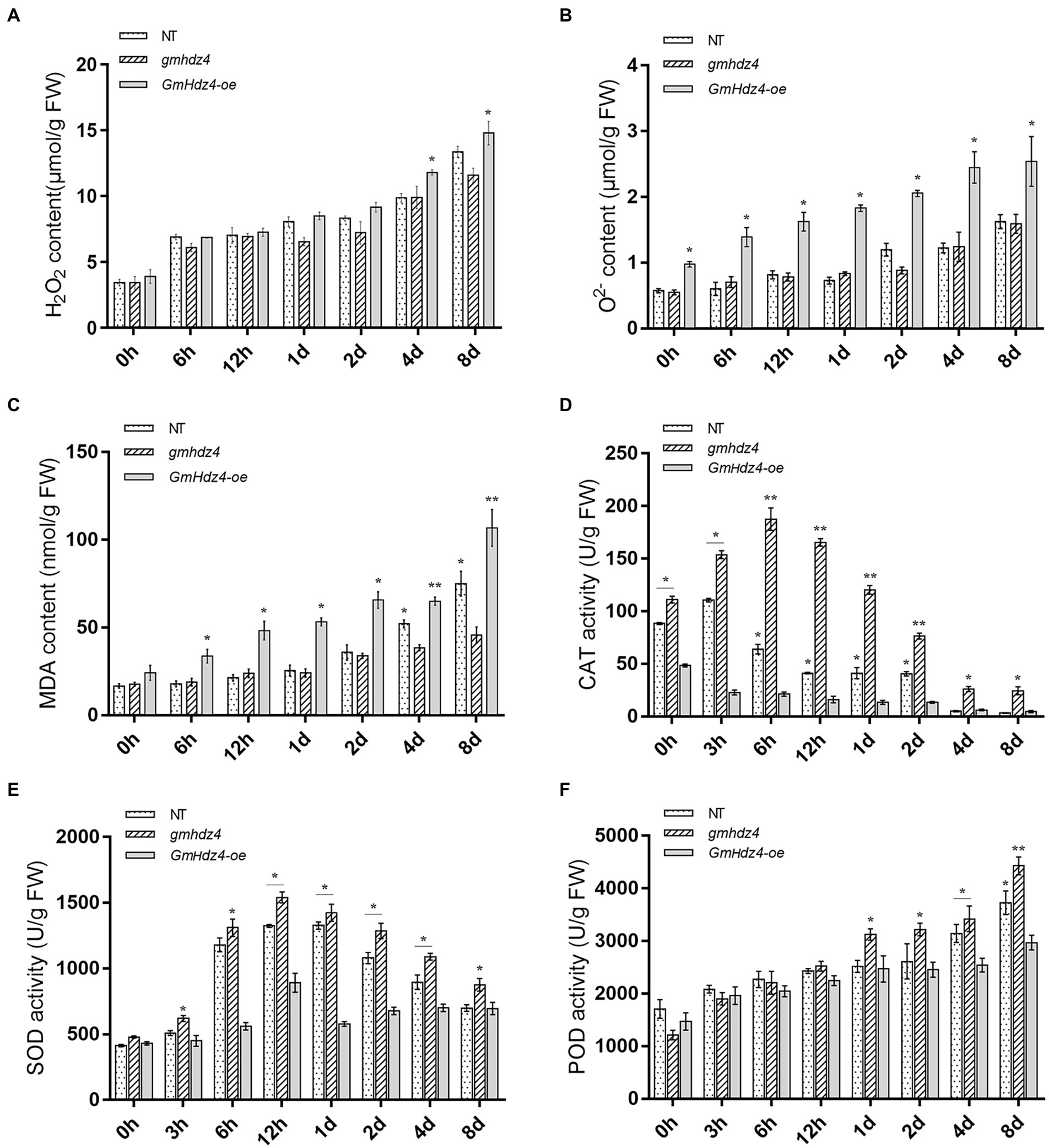
Figure 8. Knockout of GmHdz4 promotes ROS scavenging in response to drought stress. Contents of H2O2 (A), O2− (B), and malondialdehyde (MDA) (C), and activities of catalase (CAT) (D), superoxide dismutase (SOD) (E), and peroxidase (POD) (F) were tested to assess the antioxidation systems under drought stress. Significant differences are indicated by * and ** for p < 0.05 and p < 0.01, respectively, according to Duncan’s test.
The antioxidase activities of hairy roots after PEG treatment are shown in Figures 8D–F. The CAT and SOD enzyme activities initially increased and then declined within 8 days after PEG treatment (Figures 8D,E). The CAT activity in gmhdz4 and NT reached a maximum at 6 and 3 h after PEG treatment, respectively. The CAT activity in gmhdz4 was higher than that of NT, while GmHdz4-oe maintained the lowest CAT activity during the PEG treatment (Figure 8D). The SOD activity in NT and gmhdz4 was significantly higher than that of GmHdz4-oe from 6 h to 4 days after PEG treatment, with a peak at 12 h (Figure 8E). In addition, POD activity increased with extension of PEG treatment. The POD activity in gmhdz4 was significantly higher than that of NT and GmHdz4-oe from 1 days after PEG treatment, and POD activity in NT was also higher than that of GmHdz4-oe during 4–8 days after PEG treatment (Figure 8F). The results suggested that gmhdz4 created by CRISPR/Cas9 could stimulate the activities of antioxidases in soybean hairy roots under drought stress, and was beneficial to maintain the redox balance of the membrane system.
Discussion
The regulation of abiotic stress response by transcription factors is a hot topic in plant research. Most of the reported transcription factors associated with drought resistance in soybean are concentrated in the MYB, WRKY, bZIP, bHLH, and NAC families, accounting for about 63% of the total (Chai et al., 2015; Khan et al., 2018). In recent years, studies on Arabidopsis, rice, maize, and tomato showed that a plant-specific transcription factor, HD-Zip, was also involved in the response to abiotic stress. It was documented that 75% of HD-Zip genes in sesame were associated with drought and salt stress (Wei et al., 2019), but few studies in soybean were reported. In the present study, a novel HD-Zip I subfamily gene, GmHdz4, was cloned from the cDNA library of soybean cultivar Tianlong No. 1. It encodes a typical HD-Zip I protein which contains a conserved HD and a closely linked LZ domain (Ariel et al., 2007). The subcellular localization (Figure 3) and yeast hybridization assay (Figure 4) showed that GmHdz4 had the function of nuclear localization and transcription activation. Phylogenetic analysis (Figure 1B) showed that GmHdz4 was clustered in clade δ with other drought-tolerant HD-Zip I genes such as AtHB7, SlHZ48, and SlHZ5 (Jiang, 2017). Compared with rice, maize, and other gramineous crops, soybean HD-Zip I members were more numerous and most of them had paralogous genes. Our study confirmed that soybean underwent two duplication events and genome expansion in a species-specific manner after mono–dicot differentiation (Kong et al., 2007; Jin et al., 2017). This approach is one of the main evolutionary mechanisms for generating new genes that help plants adapt to environmental stresses such as drought (Skirycz and Inze, 2010).
We found that GmHdz4 was expressed in various organs in soybean, and notably its expression level was significantly higher in roots and flowers (Figure 2A). Whereas, Belamkar et al. (2014) found that GmHdz4 homologs, GmHdz5 and GmHdz10, were only expressed in flowers. Transcriptome studies showed that GmHdz38 was highly expressed in the whole plant; GmHdz24, GmHdz60, and GmHdz84 were mainly expressed in roots and flowers; and GmHdz66 was only expressed in roots (Libault et al., 2010a, 2010b; Belamkar et al., 2014). The complexity role of expression in the HD-Zip I family determines their diversity of gene functions. Expressions of GmHdz19 and GmHdz27 increased under drought stress, while those of GmHdz24, GmHdz84, and GmHdz66 were significantly downregulated (Chen et al., 2014). Overexpression of Zmhdz10 improved drought and salt tolerance in transgenic maize by regulating the ABA signaling pathway (Zhao et al., 2014). Furthermore, expression levels of ATHB7 and ATHB12 in Arabidopsis seedlings were upregulated 12–25 times after hydropenia or exogenous ABA treatment (Söderman et al., 1996; Lee et al., 2001; Valdes et al., 2012). Expressions of ATHB40, ATHB21, and ATHB53, which are orthologs of GmHdz4, were also upregulated by at least twice (Henriksson et al., 2005). We found that GmHdz4 expression decreased by about 82% at 12 h after PEG-simulated drought stress (Figure 2B), with a similar response pattern to root-specifically expressed gene GmHdz66 (Chen et al., 2014). The root system is the most important organ of plants in response to drought signals. Expression of GmHdz4 was suppressed in roots by drought, suggesting that this transcription factor gene may function in soybean roots in response to drought stress.
The plant root system architecture is plastic and dynamic. Plants make their root systems stronger and more developed in order to absorb water. The more lateral roots formed and the wider they are distributed, the more drought resistant is the plant (Battisti and Sentelhas, 2017). In general, the root system is the first organ to sense changes in soil water content, when plants suffer water deficit and produce a large number of root signals for transport to the aboveground parts. Thus, plants can promote root growth and development by regulating the distribution of assimilated substance to avoid hydropenia in the early stage of drought stress (Gilbert et al., 2011; Hu and Xiong, 2014). However, severe drought can inhibit growth of the root system and affect the formation of final yield (Huang et al., 2014; Ye et al., 2018). In our study, gmhdz4 chimeric plants generated by CRISPR/Cas9 promoted hairy root growth to some extent, while overexpression of GmHdz4 chimeric plants inhibited the development of hairy roots under drought stress (Figures 5A1–F2). The root system architecture results suggested that GmHdz4 negatively regulated the development of lateral roots, since the root surface area and root tip numbers of gmhdz4 increased significantly (Figures 5G–J).
Among the HD-Zip I subfamily in Arabidopsis, only AtHB7 and AtHB12 were found to be related to lateral root development under water deficit conditions, and played a negative feedback regulation role in the ABA pathway (Romani et al., 2016). The roots of the athb7/athb12 double mutant were shorter than the wild type (Ré et al., 2014). The AtHB53 gene, a GmHdz4 homolog expressed in the root apical meristem, was induced by hyperthermia and osmotic stress, and played an important role in auxin/cytokinin signaling pathways in roots (Son et al., 2005). A recent study by Perotti et al. (2021) showed that most of the HD-Zip members related to root development belonged to subfamilies III and IV. Gene AtHDG11 was involved in the auxin signaling pathway, which regulated drought resistance, and root biomass increased significantly in overexpressing plants. Expression of IAA28 in mutant hdg11 was downregulated and exhibited a drought-sensitive root phenotype (Rogg et al., 2001; Nakamura et al., 2006). In addition, AtHB10 acted downstream of MYBs and was a negative regulator of root hair formation (Wang et al., 2010). Smetana et al. (2019) found downregulated expression of AtHB8, AtHB9, and AtHB15 after drought stress, and their functions for differentiation and recognition of xylem were observed in the apical mature zone of roots. Gene MtHB1 was strongly expressed in root tips and regulated lateral root formation (Ariel et al., 2010). Wild soybean GsHdz4 was highly similar to GmHdz4, and regulated alkaline and osmotic stresses through different pathways. This positively regulated HCO3−-stress tolerance, but sensitivity to osmotic stress was aggravated (Cao et al., 2017). This is consistent with the root phenotype of the soybean line overexpressing GmHdz4 in the present study. Overexpression of GmHdz4 affected aboveground growth and biomass accumulation by suppressing differentiation and elongation of lateral roots (Figure 6), resulting in plants with diminished drought tolerance. However, the molecular basis of GmHdz4 regulating root morphology needs further study.
In addition to affecting root system architecture, the physiological mechanism of GmHdz4 reducing the drought tolerance of soybean was further explored. Overexpression of GmHdz4 resulted in decreased osmotic regulation capacity, accumulation of proline and soluble sugar were blocked (Figures 7A,B), and oxidative damage in hairy roots was aggravated under drought stress (Figures 8A–C). Conversely, gene-editing GmHdz4 alleviated these damages to some extent. Free proline and soluble sugars are the most common osmoregulation substances in plants (Liu and Zhu, 1997). The content of free proline in plants is very low under normal circumstances, but it accumulates rapidly under abiotic stress such as drought and is positively correlated with plant tolerance to stress (Gilmour et al., 2000; Du et al., 2020). Varieties with stronger osmotic regulation ability had less yield reduction under drought stress (Blum, 2017). The soybean osmotic regulation inhibited by GmHdz4 was similar to that by MtHB2, which reduced proline synthesis and accumulation by inducing expression of a proline dehydrogenase gene ProDH, which led to adverse effects on drought tolerance (Song et al., 2012). In addition, proline acts as a molecular chaperone to stabilize protein structure and reduce oxidative damage to cells (Székely et al., 2008). This oxidative damage is a complication of plants suffering from drought stress, which is manifest in significant accumulation of ROS and plasma membrane oxidative damage marker, MDA (Mittler, 2002; Verslues et al., 2007; Sun et al., 2018). We found that the activities of antioxidases (e.g., CAT, SOD, and POD) in gmhdz4 increased rapidly and rhythmically under drought stress (Figures 8D–F). It is crucial to reduce the content of H2O2 and O2− and relieve the oxidative senescence of roots. In contrast, these antioxidases in hairy roots overexpressing GmHdz4 were relatively sluggish in response to drought, and the ROS could not be scavenged promptly, leading to continuous accumulation of the end-product of membrane lipid oxidation. Stability of plasma membrane and maintenance of redox balance is critical to plant drought resistance. Transgenic tomato plants created by SlHB2-RNAi had significantly higher expression levels of CAT1 and APX1 than NT plants, and showed less membrane damage (Hu et al., 2017). This indicated that SlHB2 played a negative regulatory role in response to drought and high salt stress. Our findings in this experiment were consistent with those of Hu et al. (2017). However, in plants with high expression of HD-Zip genes such as ATHB12-like, Zmhdz10, EsHdzip1, and MdHB-7, the root system was more developed and activity of antioxidant enzymes significantly higher compared to the wild type under drought or salt stress (Zhao et al., 2014, 2020; Zhou et al., 2015a; Wu et al., 2016). This also illustrates the complexity of HD-Zip transcription factors in regulating plant response to drought.
Soybean HD-Zip transcription factor GmHdz4 was isolated and identified for the first time in this study. The A. rhizogenes transient transformation system was used to preliminarily verify GmHdz4 function. The gene negatively regulated drought tolerance in soybean hairy roots by inhibiting lateral root differentiation and weakening osmotic regulation ability and ROS scavenging ability. Our study provides some theoretical basis for further studies on HD-Zip I genes involved in soybean drought tolerance, and indicates the feasibility of breeding new germplasm for drought-tolerant soybean by gene editing. Due to the complex and variable regulation of transcription factors on abiotic stress, the signaling pathway of GmHdz4 involved in drought response and the specific regulatory mechanism related to root development need to be studied in the future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
XZ and GT proposed cloning of and investigating GmHdz4 gene function under drought conditions. XZ cloned this gene, implemented the molecular and hydroponic work, analyzed the data, and wrote the manuscript. GT, FI, and WZ contributed substantially to the manuscript writing and revisions. WH, YS, JL, and LL participated in the phenotyping identification and root scanning. XC provided plant materials and contributed to paper revisions. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Key Research Foundation of Science and Technology Department of Zhejiang Province (2021C02064-5-5), Hainan Provincial Joint Project of Sanya Yazhou Bay Science and Technology City (320LH033), Key R&D Projects of Zhejiang Province (2021C02057), and Collaborative Innovation Center for Modern Crop Production co-sponsored by Province and Ministry (CIC-MCP).
Acknowledgments
We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.988505/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | PCR amplification of GmHdz4 M: DNA DL2000 marker; −, negative control; 1 and 2 represent amplification products using soybean total DNA and pUCI-GmHd4 as templates, respectively.
SUPPLEMENTARY FIGURE S2 | Overexpression and targeted modification of the GmHdz4 gene in the soybean hairy roots. (A) Schematic diagram of the pTF102–GmHdz4 vector. The GmHdz4 gene and bar gene were driven by the CaMV 35S promoter. (B) The GmHdz4-oe hairy root lines were identified by PCR amplification of the GmHdz4 CDS. The length of the PCR product was 648 bp. (C) Schematic illustration of the sgRNA target sequence in the GmHdz4 gene. The black rectangles represent exons, the black line represents the introns, and the red vertical bars represent the locations of the target sequence. (D) Schematic diagram of the pBGK041–GmHdz4 vector. The Cas9 expression cassette was driven by the CaMV 35S promoter, and the sgRNA cassette was driven by the GmU6 promoter. (E) The gmhdz4 hairy root lines were identified by PCR amplification of Cas9 gene. The length of the PCR product was 502 bp. (F) Mutations induced by sgRNA in GmHdz4. The green letter represents the nucleotide insertion and the green dashes represent the deletions, and the labels on the left (gmhdz4 L1–R1 ~ gmhdz4 L3-R8) corresponds to each individual hairy root in panel E. M represents the DNA marker DL2000. Each individual hairy root on the chimera GmHdz4-oe Line 1 ~ 3 and gmhdz4 Line 1 ~ 3 were examined. -, non-transgenic plant; +, positive control. (G) Relative expression of GmHdz4 among transformed GmHdz4-oe chimeric lines, gmhdz4 chimeric lines, and NT hairy roots. Significant differences are indicated by * and ** for p < 0.05 and p < 0.01, respectively, according to Duncan’s test.
Footnotes
1. ^http://www.phytozome.net/index.php/
2. ^http://www.ncbi.nlm.nih.gov/
References
Agalou, A., Purwantomo, S., Overnas, E., Johannesson, H., Zhu, X., Estiati, A., et al. (2008). A genome-wide survey of HD-zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 66, 87–103. doi: 10.1007/s11103-007-9255-7
Alexieva, V., Sergiev, I., Mapelli, S., and Karanov, E. (2001). The effect of drought and ultraviolet radiation on growth and stress markers in pea and wheat. Plant Cell Envi. 24, 1337–1344. doi: 10.1046/j.1365-3040.2001.00778.x
Ariel, F. D., Diet, A., Crespi, M., and Chan, R. L. (2010). The LOB-like transcription factor MtLBD1 controls Medicago truncatula root architecture under salt stress. Plant Signal. Behav. 5, 1666–1668. doi: 10.4161/psb.5.12.14020
Ariel, F. D., Manavella, P. A., Dezar, C. A., and Chan, R. L. (2007). The true story of the HD-zip family. Trends Plant Sci. 12, 419–426. doi: 10.1016/j.tplants.2007.08.003
Bailey, R. W. (1958). The reaction of pentoses with anthrone. Biochem. J. 68, 669–672. doi: 10.1042/bj0680669
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Battisti, R., and Sentelhas, P. C. (2017). Improvement of soybean resilience to drought through deep root system in Brazil. Agron. J. 109, 1612–1622. doi: 10.2134/agronj2017.01.0023
Bekesiova, I., Nap, J. P., and Mlynarova, L. (1999). Isolation of high quality DNA and RNA from leaves of the carnivorous plant Drosera rotundifolia. Plant Mol. Biol. Rep. 17, 269–277. doi: 10.1023/A:1007627509824
Belamkar, V., Weeks, N. T., Bharti, A. K., Farmer, A. D., Graham, M. A., and Cannon, S. B. (2014). Comprehensive characterization and RNA-Seq profiling of the HD-zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genom. 15:950. doi: 10.1186/1471-2164-15-950
Blum, A. (2017). Osmotic adjustment is a prime drought stress adaptive engine in support of plant production. Plant Cell Envi. 40, 4–10. doi: 10.1111/pce.12800
Cao, L., Yu, Y., Ding, X., Zhu, D., Yang, F., Liu, B., et al. (2017). The Glycine soja NAC transcription factor GsNAC019 mediates the regulation of plant alkaline tolerance and ABA sensitivity. Plant Mol. Biol. 95, 253–268. doi: 10.1007/s11103-017-0643-3
Chai, C., Wang, Y., Joshi, T., Valliyodan, B., Prince, S., Michel, L., et al. (2015). Soybean transcription factor ORFeome associated with drought resistance: a valuable resource to accelerate research on abiotic stress resistance. BMC Genom. 16:6. doi: 10.1186/s12864-015-1743-6
Chen, X., Chen, Z., Zhao, H., Zhao, Y., Cheng, B., and Xiang, Y. (2014). Genome-wide analysis of soybean HD-zip gene family and expression profiling under salinity and drought treatments. PLoS ONE 9:e87156. doi: 10.1371/journal.pone.0087156
Chien, C. T., Bartel, P. L., Sternglanz, R., and Fields, S. (1991). The 2-hybrid system - A method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA 88, 9578–9582. doi: 10.1073/pnas.88.21.9578
Dogan, E., Kirnak, H., and Copur, O. (2007). Deficit irrigations during soybean reproductive stages and CROPGRO-soybean simulations under semi-arid climatic conditions. Field Crops Res. 103:9. doi: 10.1016/j.fcr.2007.05.009
Du, Y., Zhao, Q., Chen, L., Yao, X., Zhang, W., Zhang, B., et al. (2020). Effect of drought stress on sugar metabolism in leaves and roots of soybean seedlings. Plant Physiol. Biochem. 146, 1–12. doi: 10.1016/j.plaphy.2019.11.003
Gilbert, M. E., Zwieniecki, M. A., and Holbrook, N. M. (2011). Independent variation in photosynthetic capacity and stomatal conductance leads to differences in intrinsic water use efficiency in 11 soybean genotypes before and during mild drought. J. Exp. Bot. 62, 2875–2887. doi: 10.1093/jxb/erq461
Gilmour, S. J., Sebolt, A. M., Salazar, M. P., Everard, J. D., and Thomashow, M. F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124, 1854–1865. doi: 10.1104/pp.124.4.1854
Gong, S., Ding, Y., Hu, S., Ding, L., Chen, Z., and Zhu, C. (2019). The role of HD-zip class I transcription factors in plant response to abiotic stresses. Physiol. Plantarum 167, 516–525. doi: 10.1111/ppl.12965
González, F. G., Capella, M., Ribichich, K. F., Curín, F., Giacomelli, J. I., Ayala, F., et al. (2019). Field-grown transgenic wheat expressing the sunflower gene HaHB4 significantly outyields the wild type. J. Exp. Bot. 70, 1669–1681. doi: 10.1093/jxb/erz037
Harris, J. C., Hrmova, M., Lopato, S., and Langridge, P. (2011). Modulation of plant growth by HD-zip class I and II transcription factors in response to environmental stimuli. New Phytol. 190, 823–837. doi: 10.1111/j.1469-8137.2011.03733.x
Heath, R. L., and Packer, L. (1968). Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198. doi: 10.1016/0003-9861(68)90654-1
Henriksson, E., Olsson, A. S., Johannesson, H., Johansson, H., Hanson, J., Engstrom, P., et al. (2005). Homeodomain leucine zipper class I genes in Arabidopsis: expression patterns and phylogenetic relationships. Plant Physiol. 139, 509–518. doi: 10.1104/pp.105.063461
Higa, A., Mori, Y., and Kitamura, Y. (2010). Iron deficiency induces changes in riboflavin secretion and the mitochondrial electron transport chain in hairy roots of Hyoscyamus albus. J. Plant Physiol. 167, 870–878. doi: 10.1016/j.jplph.2010.01.011
Himmelbach, A., Hoffmann, T., Leube, M., Hohener, B., and Grill, E. (2002). Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 21, 3029–3038. doi: 10.1093/emboj/cdf316
Hu, J., Chen, G., Yin, W., Cui, B., Yu, X., Lu, Y., et al. (2017). Silencing of SlHB2 improves drought, salt stress tolerance, and induces stress-related gene expression in tomato. J. Plant Growth Regulation 36, 578–589. doi: 10.1007/s00344-017-9664-z
Hu, H., and Xiong, L. (2014). Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Biol. 65, 715–741. doi: 10.1146/annurev-arplant-050213-040000
Huang, X., Chaparro, J. M., Reardon, K. F., Zhang, R., Shen, Q., and Vivanco, J. M. (2014). Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany 92:75. doi: 10.1139/cjb-2013-0225
Jiang, X. (2017). Bioinformatics Analysis of HD-Zip Gene Family and Identification of HD-Zip I Genes Related to Stress Resistance in Tomato (in Chinese). Northeast Agricultural University.
Jin, J., Tian, F., Yang, D., Meng, Y., Kong, L., Luo, J., et al. (2017). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045. doi: 10.1093/nar/gkw982
Kereszt, A., Li, D., Indrasumunar, A., Nguyen, C. D., Nontachaiyapoom, S., Kinkema, M., et al. (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protocols 2, 948–952. doi: 10.1038/nprot.2007.141
Khan, S. A., Li, M. Z., Wang, S. M., and Yin, H. J. (2018). Revisiting the role of plant transcription factors in the Battle against abiotic stress. International Journal of Molecular Sciences 19, 1634–1662. doi: 10.3390/ijms19061634
Knopf, U. C. (1976). Studies on the bacteriophage PS8 of agrobacterium tumefaciens (smith and Townsend) conn: physico-chemical properties of its DNA. Microbios 17, 231–237.
Kong, H., Landherr, L. L., Frohlich, M. W., Leebens-Mack, J., Ma, H., and DePamphilis, C. W. (2007). Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 50, 873–885. doi: 10.1111/j.1365-313X.2007.03097.x
Lee, Y. H., Oh, H. S., Cheon, C. I., Hwang, I. T., Kim, Y. J., and Chun, J. Y. (2001). Structure and expression of the Arabidopsis thaliana homeobox gene Athb-12. Biochem. Biophys. Res. Commun. 284, 133–141. doi: 10.1006/bbrc.2001.4904
Lei, Y., Lu, L., Liu, H. Y., Li, S., Xing, F., and Chen, L. L. (2014). CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7, 1494–1496. doi: 10.1093/mp/ssu044
Lesk, C., Rowhani, P., and Ramankutty, N. (2016). Influence of extreme weather disasters on global crop production. Nature 529, 84–87. doi: 10.1038/nature16467
Libault, M., Farmer, A., Brechenmacher, L., Drnevich, J., Langley, R. J., Bilgin, D. D., et al. (2010a). Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 152, 541–552. doi: 10.1104/pp.109.148379
Libault, M., Farmer, A., Joshi, T., Takahashi, K., Langley, R. J., Franklin, L. D., et al. (2010b). An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 63, 86–99. doi: 10.1111/j.1365-313X.2010.04222.x
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. doi: 10.1105/tpc.10.8.1391
Liu, J. P., and Zhu, J. K. (1997). Proline accumulation and salt-stress-induced gene expression in a salt-hypersensitive mutant of Arabidopsis. Plant Physiol. 114, 591–596. doi: 10.1104/pp.114.2.591
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends in Plant Science 7, 405–410. doi: 10.1016/s1360-1385(02)02312-9
Nakamura, M., Katsumata, H., Abe, M., Yabe, N., Komeda, Y., Yamamoto, K. T., et al. (2006). Characterization of the class IV homeodomain-Leucine zipper gene family in Arabidopsis. Plant Physiol. 141, 1363–1375. doi: 10.1104/pp.106.077388
Niu, Y., Xie, G., Xiao, Y., Liu, J., Wang, Y., Luo, Q., et al. (2021). Spatiotemporal patterns and determinants of grain self-sufficiency in China. Foods 10:747. doi: 10.3390/foods10040747
Perotti, M. F., Arce, A. L., and Chan, R. L. (2021). The underground life of homeodomain-leucine zipper transcription factors. J. Exp. Bot. 72, 4005–4021. doi: 10.1093/jxb/erab112
Perotti, M. F., Ribone, P. A., and Chan, R. L. (2017). Plant transcription factors from the homeodomain-leucine zipper family I. role in development and stress responses. IUBMB Life 69, 280–289. doi: 10.1002/iub.1619
Peterson, K. M., Shyu, C., Burr, C. A., Horst, R. J., Kanaoka, M. M., Omae, M., et al. (2013). Arabidopsis homeodomain-leucine zipper IV proteins promote stomatal development and ectopically induce stomata beyond the epidermis. Development 140, 1924–1935. doi: 10.1242/dev.090209
Ré, D. A., Capella, M., Bonaventure, G., and Chan, R. L. (2014). Arabidopsis AtHB7 and AtHB12 evolved divergently to fine tune processes associated with growth and responses to water stress. BMC Plant Biol. 14:150. doi: 10.1186/1471-2229-14-150
Ribichich, K. F., Chiozza, M., Ávalos-Britez, S., Cabello, J. V., Arce, A. L., Watson, G., et al. (2020). Successful field performance in dry-warm environments of soybean expressing the sunflower transcription factor HaHB4. J. Exp. Bot. 71, 3142–3156. doi: 10.1101/2019.12.21.885798
Rogg, L. E., Lasswell, J., and Bartel, B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13, 465–480. doi: 10.1105/tpc.13.3.465
Romani, F., Ribone, P. A., Capella, M., Miguel, V. N., and Chan, R. L. (2016). A matter of quantity: common features in the drought response of transgenic plants overexpressing HD-zip I transcription factors. Plant Sci. 251, 139–154. doi: 10.1016/j.plantsci.2016.03.004
Schena, M., and Davis, R. W. (1992). HD-zip proteins: members of an Arabidopsis homeodomain protein superfamily. Proc. Natl. Acad. Sci. USA 89, 3894–3898. doi: 10.1073/pnas.89.9.3894
Skirycz, A., and Inze, D. (2010). More from less: plant growth under limited water. Curr. Opin. Biotechnol. 21, 197–203. doi: 10.1016/j.copbio.2010.03.002
Smetana, O., Makila, R., Lyu, M., Amiryousefi, A., Sanchez, R. F., Wu, M. F., et al. (2019). High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature 565, 485–489. doi: 10.1038/s41586-018-0837-0
Söderman, E., Mattsson, J., and Engstrom, P. (1996). The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J. 10, 375–381. doi: 10.1046/j.1365-313x.1996.10020375.x
Son, O., Cho, H. Y., Kim, M. R., Lee, H., Lee, M. S., Song, E., et al. (2005). Induction of a homeodomain-leucine zipper gene by auxin is inhibited by cytokinin in Arabidopsis roots. Biochem. Biophys. Res. Commun. 326, 203–209. doi: 10.1016/j.bbrc.2004.11.014
Song, S., Chen, Y., Zhao, M., and Zhang, W. (2012). A novel Medicago truncatula HD-zip gene, MtHB2, is involved in abiotic stress responses. Envi. Exp. Bot. 80, 1–9. doi: 10.1016/j.envexpbot.2012.02.001
Sun, L. (2020). Analysis on the strategy of developing soybean science and technology and revitalizing soybean industry under the background of new era of China. Soybean Science & Technology 4, 20–31.
Sun, X., Wang, P., Jia, X., Huo, L., Che, R., and Ma, F. (2018). Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol. J. 16, 545–557. doi: 10.1111/pbi.12794
Székely, G., Abraham, E., Cseplo, A., Rigo, G., Zsigmond, L., Csiszar, J., et al. (2008). Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 53, 11–28. doi: 10.1111/j.1365-313X.2007.03318.x
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tang, L., Cai, H., Ji, W., Luo, X., Wang, Z., Wu, J., et al. (2013). Overexpression of GsZFP1 enhances salt and drought tolerance in transgenic alfalfa (Medicago sativa L.). Plant Physiology and Biochemistry 71, 22–30. doi: 10.1016/j.plaphy.2013.06.024
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Valdes, A. E., Overnas, E., Johansson, H., Rada-Iglesias, A., and Engstrom, P. (2012). The homeodomain-leucine zipper (HD-zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Mol. Biol. 80, 405–418. doi: 10.1007/s11103-012-9956-4
Verslues, P. E., Batelli, G., Grillo, S., Agius, F., Mm, Y., Zhu, J., et al. (2007). Interaction of SOS2 with nucleoside diphosphate kinase 2 and catalases reveals a point of connection between salt stress and H2O2 signaling in Arabidopsis thaliana. Mol. Cell. Biol. 27, 7771–7780. doi: 10.1128/MCB.00429-07
Wang, S., Barron, C., Schiefelbein, J., and Chen, J. G. (2010). Distinct relationships between GLABRA2 and single-repeat R3 MYB transcription factors in the regulation of trichome and root hair patterning in Arabidopsis. New Phytol. 185, 387–400. doi: 10.1111/j.1469-8137.2009.03067.x
Wang, R., Zhang, Y., Wang, C., Wang, Y. C., and Wang, L. Q. (2021). ThNAC12 from Tamarix hispida directly regulates ThPIP2;5 to enhance salt tolerance by modulating reactive oxygen species. Plant Physiol. Biochem. 163, 27–35. doi: 10.1016/j.plaphy.2021.03.042
Wei, M., Liu, A., Zhang, Y., Zhou, Y., Li, D., Dossa, K., et al. (2019). Genome-wide characterization and expression analysis of the HD-zip gene family in response to drought and salinity stresses in sesame. BMC Genom. 20:748. doi: 10.1186/s12864-019-6091-5
Win, K. T., Oo, A. Z., Ookawa, T., Kanekatsu, M., and Hirasawa, T. (2016). Changes in hydraulic conductance cause the difference in growth response to short-term salt stress between salt-tolerant and -sensitive black gram (Vigna mungo) varieties. J. Plant Physiol. 193, 71–78. doi: 10.1016/j.jplph.2016.02.013
Wu, L., Yao, L., Ma, R., Zhu, X., Yang, J., Zhang, N., et al. (2016). Cloning and functional identification of the ATHB12 gene of HD-zip I family in potato (Solanum tuberosum L.). Acta Agron. Sin. 42, 1112–1121. doi: 10.3724/SP.J.1006.2016.01112
Xu, L., Islam, F., Ali, B., Pei, Z., Li, J., Ghani, M. A., et al. (2017). Silicon and water-deficit stress differentially modulate physiology and ultrastructure in wheat (Triticum aestivum L.). 3 Biotech 7:273. doi: 10.1007/s13205-017-0904-5
Xue, Z., Zhang, X., Lei, C., Chen, X., and Fu, Y. (2012). Molecular cloning and functional analysis of one ZEITLUPE homolog GmZTL3 in soybean. Mol. Biol. Rep. 39, 1411–1418. doi: 10.1007/s11033-011-0875-2
Ye, H., Roorkiwal, M., Valliyodan, B., Zhou, L., Chen, P., Varshney, R. K., et al. (2018). Genetic diversity of root system architecture in response to drought stress in grain legumes. J. Exp. Bot. 69, 3267–3277. doi: 10.1093/jxb/ery082
You, N., Dong, J., Huang, J., Du, G., Zhang, G., He, Y., et al. (2021). The 10-m crop type maps in Northeast China during 2017–2019. Sci. Data 8:9. doi: 10.1038/s41597-021-00827-9
Yu, X., Ruan, M., Wang, B., Yang, Y., Wang, S., and PENG, M. (2017). Cloning and analysis of the transcription factor gene MeHDZ14 in cassava. Acta Agron. Sin. 43, 1181–1189. doi: 10.3724/SP.J.1006.2017.01181
Zhang, S., Haider, I., Kohlen, W., Jiang, L., Bouwmeester, H., Meijer, A. H., et al. (2012). Function of the HD-zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice. Plant Mol. Biol. 80, 571–585. doi: 10.1007/s11103-012-9967-1
Zhao, S., Gao, H., Jia, X., Wang, H., Ke, M., and Ma, F. (2020). The HD-zip I transcription factor MdHB-7 regulates drought tolerance in transgenic apple (Malus domestica). Environ. Exp. Bot. 180:46. doi: 10.1016/j.envexpbot.2020.104246
Zhao, Y., Ma, Q., Jin, X., Peng, X., Liu, J., Deng, L., et al. (2014). A novel maize homeodomain-leucine zipper (HD-zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and Arabidopsis. Plant Cell Physiol. 55, 1142–1156. doi: 10.1093/pcp/pcu054
Zhou, W., Malabanan, P. B., and Abrigo, E. (2015b). OsHox4 regulates GA signaling by interacting with DELLA-like genes and GA oxidase genes in rice. Euphytica 201, 97–107. doi: 10.1007/s10681-014-1191-4
Keywords: Glycine max, HD-ZIP, GmHdz4, drought stress, root system architecture
Citation: Zhong X, Hong W, Shu Y, Li J, Liu L, Chen X, Islam F, Zhou W and Tang G (2022) CRISPR/Cas9 mediated gene-editing of GmHdz4 transcription factor enhances drought tolerance in soybean (Glycine max [L.] Merr.). Front. Plant Sci. 13:988505. doi: 10.3389/fpls.2022.988505
Edited by:
Jiban Shrestha, Nepal Agricultural Research Council, NepalReviewed by:
Linhui Yu, Northwest A&F University, ChinaParviz Heidari, Shahrood University of Technology, Iran
Copyright © 2022 Zhong, Hong, Shu, Li, Liu, Chen, Islam, Zhou and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guixiang Tang, dGFuZ2d4QHpqdS5lZHUuY24=
 Xuanbo Zhong
Xuanbo Zhong Wei Hong1
Wei Hong1 Faisal Islam
Faisal Islam Weijun Zhou
Weijun Zhou Guixiang Tang
Guixiang Tang