- 1State Key Laboratory of Biocontrol and Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou, China
- 2State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, The Chinese Academy of Sciences, Beijing, China
- 3College of Life Sciences, University of Chinese Academy of Sciences, Beijing, China
Mitogenomes of most flowering plants evolve slowly in sequence, but rapidly in structure. The rearrangements in structure are mainly caused by repeat-mediated recombination. However, patterns of repeat-mediated recombination vary substantially among plants, and to provide a comprehensive picture, characterization of repeat-mediated recombination should extend to more plant species, including parasitic plants with a distinct heterotrophic lifestyle. Here we assembled the mitogenome of the holoparasitic plant Aeginetia indica (Orobanchaceae) using Illumina sequencing reads. The mitogenome was assembled into a circular chromosome of 420,362 bp, 18,734 bp longer than that of another individual of A. indica which was assembled before as a linear molecule. Synteny analysis between the two mitogenomes revealed numerous rearrangements, unique regions of each individual and 0.2% sequence divergence in their syntenic regions. The A. indica mitogenome contains a gene content typical of flowering plants (33 protein-coding, 3 rRNA, and 17 tRNA genes). Repetitive sequences >30 bp in size totals 57,060 bp, representing 13.6% of the mitogenome. We examined recombination mediated by repeats >100 bp in size and found highly active recombination for all the repeats, including a very large repeat of ~16 kb. Recombination between these repeats can form much smaller subgenomic circular chromosomes, which may lead to rapid replication of mitochondrial DNA and thus be advantageous for A. indica with a parasitic lifestyle. In addition, unlike some other parasitic plants, A. indica shows no evidence for horizontal gene transfer of protein-coding genes in its mitogenome.
1 Introduction
The mitogenomes of most flowering plants evolve very slowly in sequence while very rapidly in genome rearrangement (Palmer and Herbon, 1988; Drouin et al., 2008; Gualberto and Newton, 2017; Sloan et al., 2017). The frequent rearrangement in the mitogenomes is thought to stem from recombination between repeated sequences, which are common in the mitogenomes of flowering plants (Alverson et al., 2011; Cole et al., 2018; Kozik et al., 2019). For a circular chromosome, isomeric forms of the circle would form when a repeat pair is present in reverse orientation; alternatively, two small circular chromosomes (subgenomes) would be expected when a repeat pair is present in direct orientation (Kubo and Newton, 2008). As expected, the presence of both direct and reverse repeat pairs would greatly enrich the mitochondrial DNA conformations. Repeats of intermediate length (100-1,000 bp) are thought to be subject to not-infrequent recombination (Maréchal and Brisson, 2010; Woloszynska, 2010; Kozik et al., 2019), while large repeats (>1,000 bp) are expected to generate equimolar or nearly equimolar recombined molecules in some plant species (Maréchal and Brisson, 2010), such as Ginkgo (Guo et al., 2016) and monkeyflower (Mower et al., 2012a). However, this is not the case for Nymphaea colorata, which have some large repeats but with very low recombination frequency (Dong et al., 2018). At the same time, moderate to considerable recombination frequencies were also found for small repeats (< 100 bp) in some plant species, for example 14.7% for a 55-bp repeat pair of Picea abies (Sullivan et al., 2020) and 24.1% for a 75-bp repeat pair of Viscum scurruloideum (Skippington et al., 2015). In the mitogenome of V. scurruloideum, the length of short repeats (30-100 bp) was positively correlated with recombination rate (Skippington et al., 2015). In contrast, little or no evidence for ongoing recombination mediated by repeats ranging from 100 to 800 bp in the mitogenomes of two Monsonia species (Cole et al., 2018).
Recombination mediated by small or medium repeats is usually of low frequency and symmetrical stoichiometry between different conformations is rare (Arrieta-Montiel et al., 2009). Being less well studied, the biological significance of this stoichiometric variation in different conformations is unclear. The degree of stoichiometry equality may potentially affect the rate of DNA replication, gene order, gene expression patterns, and further environmental adaptability. So far, the relationships between repeats and repeat-mediated recombination in the mitogenomes of plants remain elusive. Therefore, mitogenome sequencing from more plants are needed to generalize patterns of their mitogenome evolution, including the recombination activity of repeats.
Parasitic plants consist of ~1% flowering plants and have independently evolved at least 12 or 13 times (Westwood et al., 2010; Nickrent, 2020). They partially or completely obtain water and nutrients from their hosts via haustoria due to partial or complete loss of the capability of photosynthesis (Petersen et al., 2020). Plastid genomes of parasitic plants usually exhibit reduced size and gene content, and increased AT content and evolutionary rate (Wicke and Naumann, 2018). Parasitic plants also show deeply altered nuclear genome architecture, such as genome size expansion (>100 Gb in Viscum) (Marie and Brown, 1993), substantial gene loss (Cuscuta and Sapria) (Sun et al., 2018; Cai et al., 2021) and frequent horizontal gene transfer (HGT) from their hosts (Kado and Innan, 2018). Although rarely characterized, mitogenomes of some parasitic plants also show unusual features, such as minicircular chromosomes (Yu et al., 2022), extreme size reduction, gene loss and high substitution rate (Skippington et al., 2015); extreme heteroplasmy (Yu et al., 2022), and rampant HGT (Petersen et al., 2020). However, these unusual features are not universal to parasitic plants, for example, the hemi-parasitic plant Castilleja paramensis appears to have a mitogenome typical of flowering plants (Fan et al., 2016), and mitochondrial substitution rates of parasitic plants are not always higher than those of non-parasitic plants (Zervas et al., 2019).
Aeginetia is a holoparasitic genus of Orobanchaceae, mostly distributed in tropical Asia (Kuijt, 1969; POWO, 2022). This genus comprises four species, of which A. indica is the most widely distributed species. The hosts of A. indica are usually plants from Poaceae like Miscanthus and Saccharum (sugar-cane) (Kuijt, 1969). Previous studies showed that A. indica had lost almost all photosynthesis-related genes in its plastome (Chen et al., 2020) and 84 nuclear genes of A. indica had been obtained via horizontal gene transfer (HGT) from its hosts (Kado and Innan, 2018). A recent study attempted to assemble the mitogenome of A. indica, but failed to get a complete assembly (Choi and Park, 2021). Thus, its mitogenome structure remains unknown.
In this study, we successfully assembled a complete circular-mapping mitogenome of A. indica using Illumina sequencing reads. We found highly active repeat-mediated recombination in its mitogenome, implying numerous alternative genomic and subgenomic conformations in this holoparasitic plant, which may be advantageous for rapid replication of its mitochondrial DNA. In addition, unlike some other parasitic plants, A. indica shows no evidence for HGT of protein-coding genes in its mitogenome.
2 Results and discussion
2.1 The mitogenome of A. indica contains a gene content typical of flowering plants
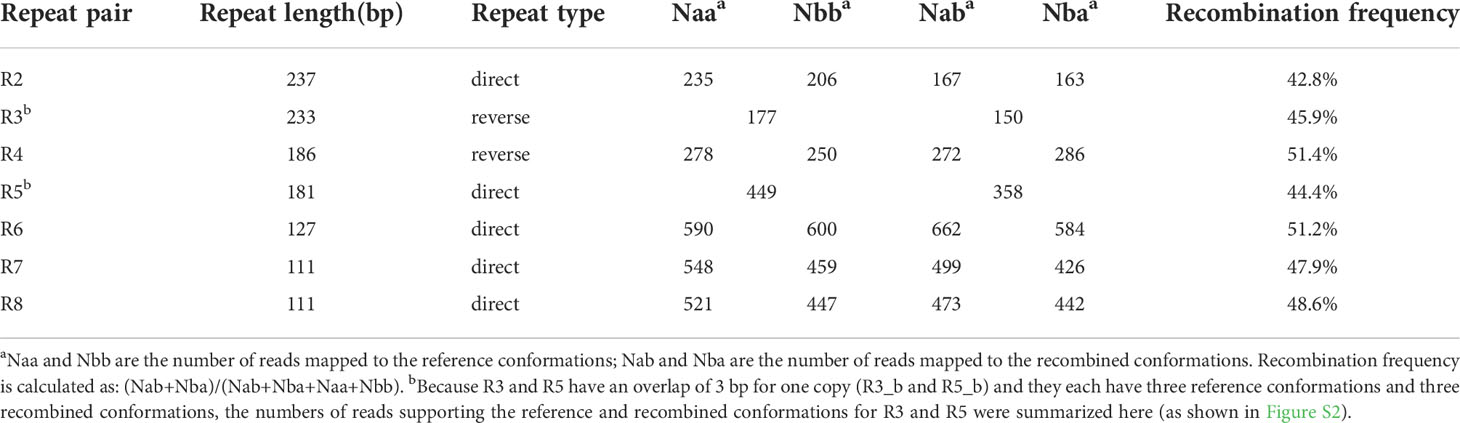
Genome assembly using Illumina reads generated 614,229 contigs > 127 bp in length for A. indica. We obtained 17 putative mitochondrial contigs by filtering out the contigs with very high (putative plastid contigs) and low (putative nuclear contigs) sequening depth. When visualized in Bandage, these putative mitochondrial contigs are connected to a network, and several contigs with four links to other contigs likely represent repeats in the A. indica mitogenome (Figure 1A). Many different conformations of the A. indica mitogenome can be inferred from the contig network. For the illustrative purpose, we selected for display a full-genome conformation consisting of all these contigs (Figure 1B). The mitogenome size is 420,362 bp, and its overall GC content of is 43.5%, similar to that found in most other angiosperms (Mower et al., 2012b; Skippington et al., 2015). The sequencing depth across the mitogenome is relatively even (Figure S1), with an average depth of 357 ×, validating the accuracy of our assembly.
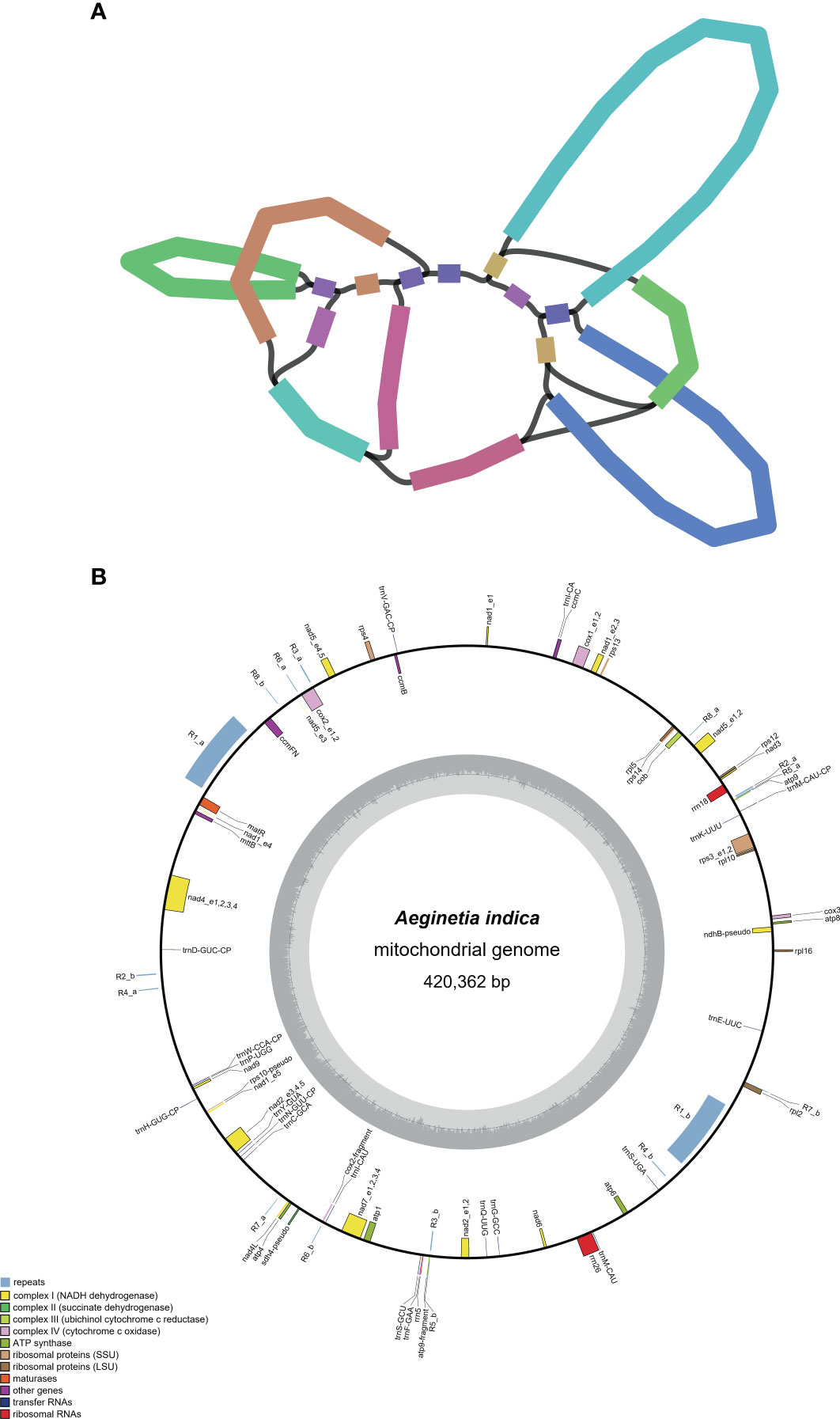
Figure 1 The mitogenome of Aeginetia indica. (A), A network of the mitochondrial contigs of Aeginetia indica visualized in Bandage. (B), Gene map. Genes shown outside the outer circle are transcribed counterclockwise, whereas those inside the outer circle are transcribed clockwise. The darker and light shading inside the inner circle indicates GC and AT content, respectively. The eight largest repeats (R1-R8) are also shown in blue in this figure.
The A. indica mitogenome contains 33 protein-coding, 3 rRNA, and 17 tRNA genes (Table S1). The 33 protein-coding genes includes 24 core genes usually present in seed plants (Adams et al., 2002) and nine genes variably present in seed plants. The protein-coding genes contain 16 cis-spliced and six trans-spliced introns. As shown in Table S2, the mitogenomes of all eight Orobanchaceae species (including the autotrophic plant Lindenbergia philippensis) have highly similar gene contents, with occasional loss and/or psedogenization of several genes variably present in seed plants. Compared with its close relative Castilleja paramensis, A. indica has lost one gene sdh3. In addition, rps10 is intact in C. paramensis while pseudogenized in A. indica, and rpl2 is intact in A. indica while pseudogenized in C. paramensis. In addition to the three introns (cox2-i1, nad7-i3, and rpl2-i1), which were also missing in seven other Orobanchaceae species (Fan et al., 2016), one more intron (rps10-i1) was lost in A. indica than Castilleja paramensis due to pseudogenization of rps10.
Available complete plant mitogenomes suggests no evolutionary correlation between the type of parasitic lifestyle and mitogenome size and structure (Petersen et al., 2020). Also, the effects of the parasitic lifestyle on mitogenome gene content of plants remain unclear, because there are so few well-annotated mitogenomes of parasitic plants. Although the mitochondrial genomes of the mistletoes Viscum (Petersen et al., 2015; Skippington et al., 2015) exhibit substantial gene loss, other parasitic plants do not show significant gene loss, including seven Orobanchaceae species (Fan et al., 2016), Cynomorium coccineum (Bellot et al., 2016), Rafflesia lagascae (Molina et al., 2014), Tolypanthus maclurei (Yu et al., 2021), Cuscuta (Lin et al., 2022), Rhopalocnemis phalloides (Yu et al., 2022), and A. indica in this study. This suggests mitochondrial gene loss may not be directly associated with the parasitic lifestyle in most parasitic plants.
2.2 Intraspecific variation in the mitogenomes of A. indica
Compared with the mitogenome of another individual of A. indica, which was collected from Jeju Island, Korea and comprised a linear molecule with a total length of 401,628 bp (Choi and Park, 2021), the mitogenome in this study is 18,734 bp larger in size. The syntenic analysis of the two A. indica mitogenomes revealed numerous rearrangements (Figure 2). 389,843 bp out of 420,362 bp (92.7% of the mitogenome in this study) can be aligned and the alignable regions had an identity greater than 99%. The alignable regions between the two individuals contains 864 nucleotide substitutions (0.2% divergence). The unalignable sequences are all in the intergenic regions. There are 33 and 25 unalignable regions with a total length of 49,642 bp and 30,512 bp, respectively, in the mitogenomes of the two individuals. The largest repeat, R1, (see section 2.3 below) in the mitogenome of this study is divided into three parts in the mitogenome of the other individual (Figure 2).
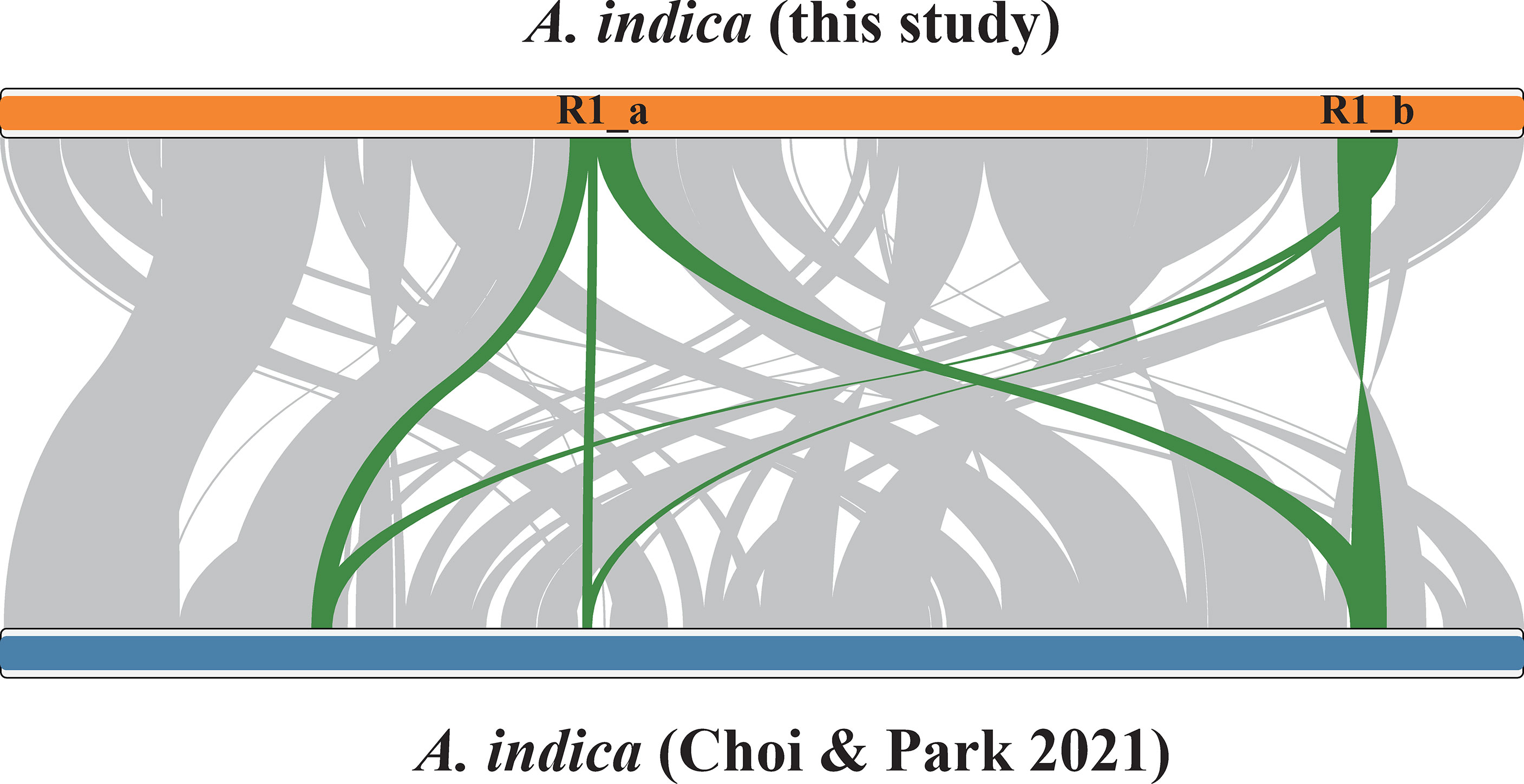
Figure 2 Numerous rearrangements between the two Aeginetia indica mitogenomes. The green curve corresponds to the ~16 kb repeat pair (R1) in the A. indica mitogenome of this study.
The A. indica mitogenome reported by Choi and Park (2021) annotated two more protein-coding genes (rps10 and atpI) than that in this study. rps10 was annotated as a pseudogene in this study due to substantial truncation, and careful sequence inspection for the mitogenome sequence reported by Choi and Park (2021) found an opening reading frame (ORF) of only 165 bp for this gene, much shorter than ORF length of this gene in other angiosperms (~360 bp). So it must be a pseudogene as well. atpI was suggested to be acquired by horizontal gene transfer (Choi and Park, 2021), but in fact it is of plastid origin and an only 216 bp gene fragment (~750 bp in other angiosperms) was found in both this study and Choi and Park (2021). Therefore, the mitochondrial protein-coding gene content is actually the same for the two individuals of A. indica.
We annotated 17 tRNAs and three rRNAs in the A. indica mitogenome (Table S1), two more tRNAs than those annotated in Choi and Park (2021). We reannotated the tRNAs in the A. indica mitogeome reported by Choi and Park (2021) using the same method. We found the mitogenome reported by Choi and Park (2021) had the same 17 tRNAs, plus one more private tRNA (trnS-GGA) in an unalignable region.
Previous studies, albeit very few, have revealed the existence of intraspecific variation in plant mitogenomes, including structural rearrangements in Beta vulgaris (Darracq et al., 2011), copy number variation in Arabidopsis thaliana (Wu et al., 2020), chromosome presence/absence in Silene noctiflora (Wu et al., 2015; Wu and Sloan, 2019), and nucleotide variations in Oryza rufipogon and O. sativa (He et al., 2020). The numerous rearrangements and abundant sequence variations between the mitogenomes of Choi and Park (2021) and this study suggests that there is a considerable intraspecific mitogenomic variation in A. indica.
2.3 Highly active repeat-mediated recombination
The A. indica mitogenome contains 377 repeat units ≥ 30 bp in size, which form 846 repeat copies and a total of 57,060 bp repetitive sequences, covering 13.6% of the mitogenome (Supplementary Excel file 2). All repeat units >100 bp in size, including one large repeat unit (R1, 16,366 bp) and seven relatively small repeat units (R2-R8, 111-237 bp), have two copies (Table 1). These eight repeat pairs are all located in the intergenic regions (Figure 1B). We analyzed recombination activity of the eight repeat pairs. The largest repeat pair can mediate recombination, as verified by the results of PCR amplification (Figure 3). Recombination mediated by seven other repeat pairs were verified by mapping Illumina reads to the reference and potentially recombined conformations.
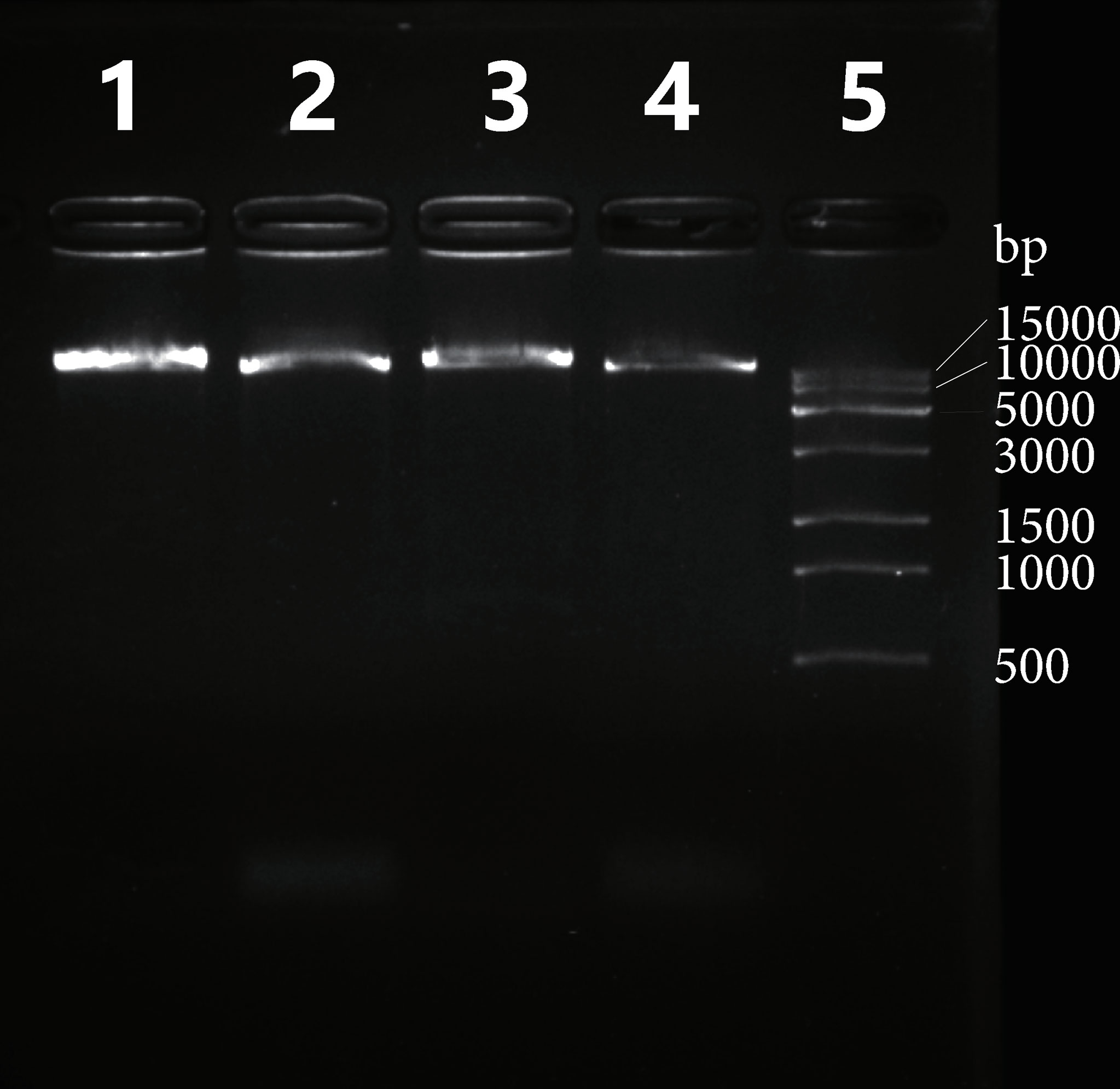
Figure 3 PCR evaluation of recombination mediated by a ~16 kp repeat pair (R1) in the Aeginetia indica mitogenome. Shown are results of DNA gel electrophoresis of amplified PCR products. Lane 1: PCR amplification of R1_a and flanking regions (primer: a_F and a_R); Lane 2: PCR amplification of R1_b and flanking regions (primer: b_F and b_R); Lane 3: PCR amplification for the recombined conformation 1 (primer: a_F and b_F); Lane 4: PCR amplification for the recombined conformation 2 (primer: a_R and b_R); Lane 5: DNA ladder. Expected sizes for these PCR products are 17734, 17589, 17809 and 17514 bp for Lane 1, 2, 3 and 4, respectively.
Although high-frequency recombinations is often mediated by large repeats in many plant species, this is not necessarily the case in A. indica. For all seven small repeat pairs of A. indica, we observed very high recombination rates, ranging from 42.8% to 51.4% (Table 1), in nearly equimolar amounts (50.0%). Even though recombination rates are not widely characterized in plant species, they are higher than many of the reported fractions of recombination. For example, Lactuca sativa and L. serriola have a very low fraction of recombinations (~1%-10%) mediated by their short repeats (Kozik et al., 2019), and Nymphaea colorata possesses 0.2% and 8.2% recombination frequencies for its two largest repeats (Dong et al., 2018). Evidence of recombination was detected in Vigna angularis mitogenome for twelve repeat units of 62-1,215 bp length, with recombination rates ranging from 5% to 33% (Naito et al., 2013). In Picea abies, most of the detected repeat pairs showed no to little evidence of repeat-mediated recombination, but a few ranging from 50 bp to 948 bp were found to have highly active recombination, reaching a maximum of ~32% for a 186 bp repeat pair (Sullivan et al., 2020). In addition, four repeat pairs (387-593 bp in length) in the mitogenome of a parasitic mistletoe Viscum scurruloideum have 39-58% recombination frequencies, which was considered to be the smallest repeats at recombinational equilibrium in plant mitogenomes (Skippington et al., 2015). The repeats at recombinational equilibrium are even smaller in the mitogenome of A. indica. This implies highly active repeat-mediated recombination occurring in the mitogenome of A. indica, which is consistent with the numerous rearrangements shown in the collinear analysis (Figure 2).
The A. indica mitogenome can form multiple alternative conformations through repeat-mediated recombination. All alternative conformations resulting from repeat-mediated recombination contain identical genomic content. These conformations differ only in the order and orientation of non-repeat regions and repeat regions separating them, and in some cases, also include differences in chromosome number (dividing one big circular chromosome into two or more small circular chromosomes). Here we show one of the possible recombination scenarios (Figure 4). The circular chromosome can shift into two small subgenomic circular chromosomes by R2 mediated recombination, and further into four smaller subgenomic circular chromosomes by R5 and R8 mediated recombination. A special case was found for R3 and R5, of which one copy (R3_b and R5_b) has an overlap of 3 bp. Unlike other repeats, R3 and R5 each have three predicted reference conformations and three predicted recombined conformations, as illustrated in Figure S2. Through repeat-mediated recombinations, the A. indica mitogenome can exist in various conformations, including many small subgenomic circular chromosomes. At the same DNA replication speed, the smaller chromosomes will be expected to replicate faster. Rapid replication of mitochondrial DNA may be beneficial for holoparasitic plants like A. indica to cope with a harsh heterotrophic lifestyle.
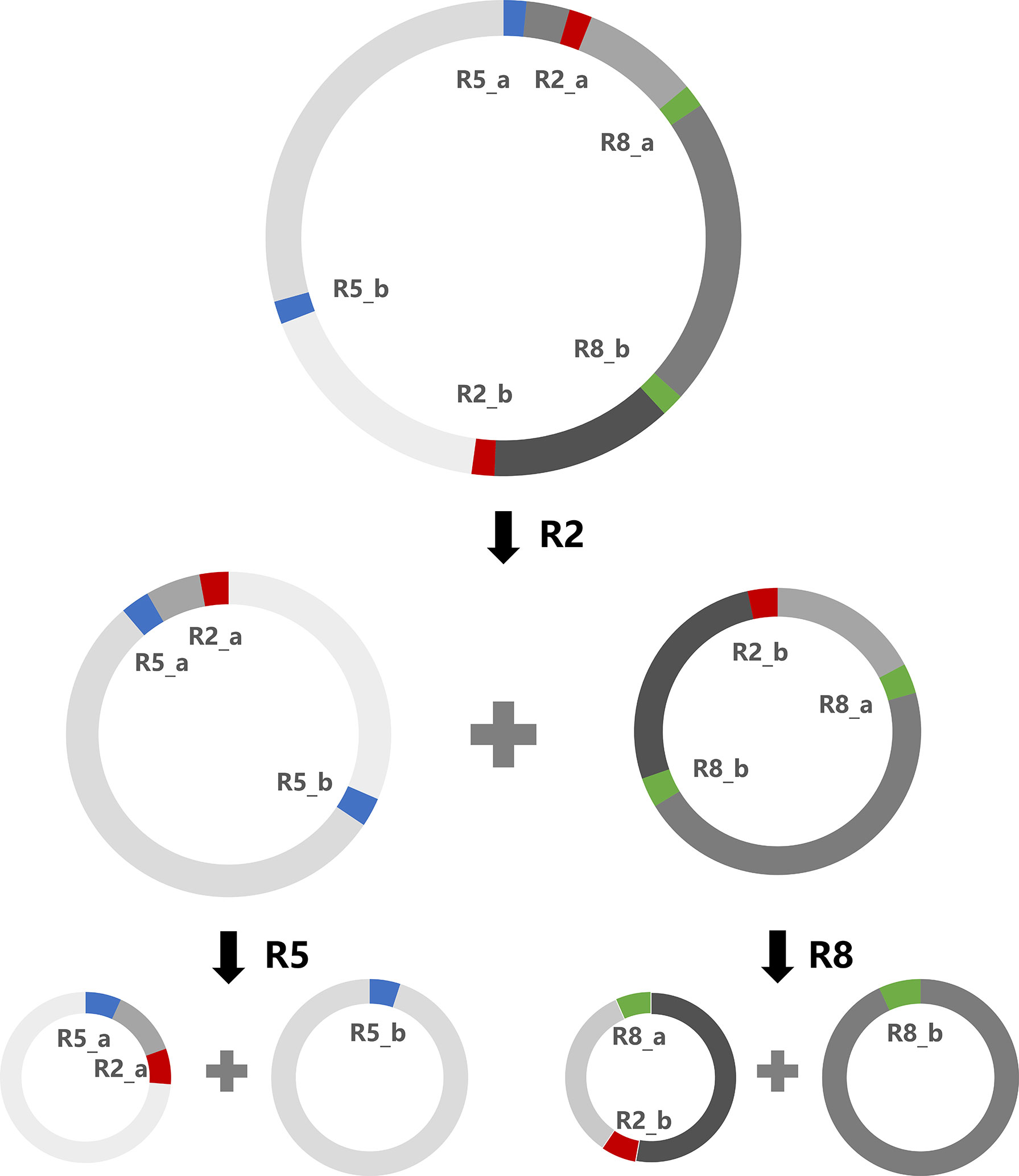
Figure 4 A schematic diagram of multiple alternative mitogenomic conformations formed by repeat-mediated recombination. This diagram shows only one of the possible ways of repeat-mediated recombination. These three repeat pairs (R2, R5 and R8) in the A. indica mitogenome are direct repeats.
2.4 No to little horizontal and intracellular gene transfer in the A. indica mitogenome
Phylogenetic analysis for each of the 33 mitochondrial protein-coding genes revealed that A. indica always clustered with other species of Lamiales with high bootstrap support, and that monocots, its potential hosts, formed a distant monophyletic clade (Supplementary Figure 1). This is also the case for another parasitic plant from the same family, Castilleja paramensis. Therefore, no evidence for HGT was found in the mitogenomes of the two parasitic plants in Orobanchaceae. Some parasitic plants show rampant HGT in their mitogenomes from their hosts (Sanchez-Puerta et al., 2019) and a plausible explanation is that parasitic plants establish intimate contact with their hosts via haustoria (Goyet et al., 2019). However, no evidence for HGT in the holoparasitic plant A. indica, the hemiparasitic plant Castilleja paramensis and two Cuscuta species (Anderson et al., 2021) suggests that physical contact between parasitic plants and their hosts is not always associated with host-to-parasitic-plants mitochondrial HGT. On the other hand, the A. indica nuclear genome indeed possesses some genes derived from its hosts (Kado and Innan, 2018), which implies that patterns of HGT from hosts to parasitic plants might differ between mitogenomes and nuclear genomes. Why the extent of HGT differ substantially between mitogenomes of parasitic plants, and between the mitogenome and nuclear genome of a parasitic plant deserves further investigation.
In terms of intracellular gene transfer (IGT) between the mitogenome and the plastome of A. indica, we found two regions (220 bp and 251 bp) in the mitogenome with hits to the psuedogene ψndhB in its plastome. BLASTN search of the A. indica mitogenome against the plastome of L. philippensis revealed a 611-bp hit to the ndhB gene of L. philippensis (gene length = 2,212 bp). Therefore, we suggest that the pseudogene ψndhB in the A. indica mitogenome was transferred from its plastome and that little IGT has occurred between the mitogenome and the plastome of A. indica.
3 Materials and methods
3.1 Plant sampling, Illumina sequencing and mitogenome assembly
Plant collection and Illumina sequencing of an individual of Aeginetia indica were performed as described in (Chen et al., 2020). The individual was collected from Shimentai Forest Park, Yingde, Guangdong, China. 21.4 Gb of 150 bp paired-end reads with an insert size of 300 bp were generated. Raw reads were filtered by Trimmomatic v 0.39 (Bolger et al., 2014) with default parameters. The mitogenome was assembled using Illumina reads in GetOrganelle v1.7 (Jin et al., 2020) with the parameters: -F embplant_mt, -k 57,77,97,117,127, and the mitogenomes of Liriodendron tulipifera and Castilleja paramensis were used as references. We identified putative mitochondrial contigs when they have a depth of coverage > 100× (based on the depths of several contigs containing the mitochondrial genes), BLASTN (evalue set to 1e-5) hits to the plant mitochondrial databases, or with direct or indirect links (based on the GFA output) to the contigs with mitochondrial hits. By visualizing these potential mitochondrial contigs in Bandage v0.8.1 (Wick et al., 2015), the complete mitogenome could be manually connected into a circular chromosome, but with many alternative conformations of the same genome size. We selected for display one full-genome conformation for subsequent analysis. All Illumina reads were mapped back to the mitogenome using BWA-mem (Li, 2013) with default parameters except -T set to 100 and the depth of coverage across the chromosome was calculated using Samtools v1.9 (Li et al., 2009). To avoid the influence of plastid-derived reads on the sequencing depth of the mitogenome, the plastid genome of A. indica (GenBank accession number MN529629) was also used as reference during mapping of Illumina reads. To avoid the influence of nuclear-derived reads, the mapped alignment shorter than 100 bp were filtered.
3.2 Mitogenome annotation
The mitochondrial protein-coding genes and rRNAs of A. indica were annotated with Geseq (Tillich et al., 2017) and BLASTN with default settings (Camacho et al., 2009). Available mitogenomes of Lamiales species were used as references and manual adjustment was conducted when necessary. Genes that contain one or more premature stop codons or frameshift mutations were considered as pseudogenes. Transfer RNAs (tRNAs) were identified using tRNAscan-SE v2.0.7 (Lowe and Chan, 2016) with the organelle option. Mitochondrial gene maps (including eight repeats >100 bp in size) were plotted by OGDRAW (Greiner et al., 2019).
3.3 Collinear analysis with another individual of A. indica and comparative analysis with other species in Orobanchaceae
The mitogenome of another individual of A. indica characterized by Choi and Park (2021) were used for comparison. This individual was collected from Jeju Island, Korea, which is more than 1,700 km away from the collection site of the individual we studied here. Collinear analysis between the two mitogenomes were conducted in Mummer (Kurtz et al., 2004). Collinear regions were identified using the nucmer module with 85% identity as the threshold with many-to-many alignment mode. Nucleotide substitutions in the collinear regions were analyzed using the dnadiff module. The collinear results were visualized using RIdeogram in the R package (Hao et al., 2020).
Comparative analysis of mitochondrial protein-coding genes and introns was also performed among the two A. indica individuals and seven other species of Orobanchaceae, including an autotrophic plant (Lindenbergia philippensis), three hemiparasitic plants (Bartsia pedicularioides, Castilleja paramensis, and Schwalbea americana), and three holoparasitic plants (Orobanche crenata, O. gracilis and Phelipanche ramosa), whose mitogenomes were characterized before (Fan et al., 2016).
3.4 Repeat identification and repeat-mediated recombination analysis
We used a Python script ROUSFinder2.0.py (Wynn and Christensen, 2019) to identify repeats (≥30 bp) in the A. indica mitogenome. Repeat units > 100 bp in size were used to assess repeat-mediated recombination. For all the repeats but the largest one (~16 kb), Illumina reads were mapped to sequences of the reference (the mitogenome we assembled) and potentially recombined conformations (repeat pair itself and flanking 300 bp single-copy sequences at both ends), and paired reads spanning these conformations were counted for calculating the recombination rate (the frequency of recombined conformations) for each repeat pair. The influence of plastid- and nuclear-derived reads was minimized by the same method in calculating the depth of coverage mentioned in section 3.1. We found one copy each of R3 and R5 (R3_b and R5_b) has an overlap of 3 bp, resulting in three predicted reference conformations and three predicted recombined conformations for each repeat pair (Figure S2), and we calculated the numbers of reads supporting each of these conformations and summarized in Table 1.
For the ~16 kb repeat pair, Illumina reads are too short to be used for assessing its recombination rate. We then designed the primers (Table S3) anchoring the regions flanking the repeat for the reference and potentially recombined conformations using Primer3 (Koressaar and Remm, 2007), performed long-fragment PCR amplification using KOD One™ PCR Master Mix (TOYOBO, Japan), and run agarose gel electrophoresis to verify the presence of repeat-mediated recombination.
3.5 Horizontal and intracellular gene transfer analysis for mitochondrial protein coding genes
For each of the 33 mitochondrial protein-coding genes of A. indica, we did phylogenetic analysis to infer its origin. Coding region sequences of as many as 18 other species, including species from the same family Orobanchaceae and its potential host family Poaceae, were downloaded from GenBank (https://www.ncbi.nlm.nih.gov/) (source details: Supplementary Excel file 1). Sequence alignment for each gene was performed with ClustalW (Thompson et al., 1997) and further adjusted manually. Phylogenetic trees were generated by RAxML v.8.2.11 using maximum likelihood (ML) method under the GTRGAMMAI model opting for one thousand bootstrap replicates. Either Amborella trichopoda or Liriodendron tulipifera was used as an outgroup in the phylogenetic analysis. Horizontal gene transfer (HGT) was inferred based on phylogenetic position of A. indica. HGT was identified if A. indica was grouped with non-Orobanchaceae species with high bootstrap support (>80%).
To detect the presence of intracellular gene transfer from the plastome to the mitogenome, the mitogenome of A. indica was searched against the A. indica plastome using BLASTN with the parameters of -evalue set to 1e-6. Due to massive gene loss in the A. indica plastome, the same BLASTN analysis was performed between the A. indica mitogenome and the plastome of the autotrophic plant L. philippensi (GenBank accession number NC_022859). Hits >100 bp in length and > 80% in identity were considered as potential intracellular gene transfer.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank under the accession number TPA: BK061406.
Author contributions
Conceptualization, RZ; methodology, YZ, RY, and JC; formal analysis, YZ and RY; data curation, YZ and JC; software, YZ; visualization, YZ; writing – original draft preparation, YZ and RZ; writing — review and editing, YZ, RY, and RZ; project administration, RZ and YL; funding acquisition, RZ. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported financially by the National Natural Science Foundation of China (31811530297).
Acknowledgments
We thank Choi Kyoung-Su and Park Seonjoo from Yeungnam University for sharing their mitogenome sequence of Aeginetia indica.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.988368/full#supplementary-material
References
Adams, K. L., Qiu, Y.-L., Stoutemyer, M., Palmer, J. D. (2002). Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci. United. States America 99, 9905–9912. doi: 10.1073/pnas.042694899
Alverson, A. J., Zhuo, S., Rice, D. W., Sloan, D. B., Palmer, J. D. (2011). The mitochondrial genome of the legume vigna radiata and the analysis of recombination across short mitochondrial repeats. PloS One 6, e16404. doi: 10.1371/journal.pone.0016404
Anderson, B. M., Krause, K., Petersen, G. (2021). Mitochondrial genomes of two parasitic cuscuta species lack clear evidence of horizontal gene transfer and retain unusually fragmented ccmFC genes. BMC Genomics 22, 816. doi: 10.1186/s12864-021-08105-z
Arrieta-Montiel, M. P., Shedge, V., Davila, J., Christensen, A. C., Mackenzie, S. A. (2009). Diversity of the arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics 183, 1261–1268. doi: 10.1534/genetics.109.108514
Bellot, S., Cusimano, N., Luo, S., Sun, G., Zarre, S., Gröger, A., et al. (2016). Assembled plastid and mitochondrial genomes, as well as nuclear genes, place the parasite family cynomoriaceae in the saxifragales. Genome Biol. Evol. 8, 2214–2230. doi: 10.1093/gbe/evw147
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Cai, L., Arnold, B. J., Xi, Z., Khost, D. E., Patel, N., Hartmann, C. B., et al. (2021). Deeply altered genome architecture in the endoparasitic flowering plant Sapria himalayana griff. (Rafflesiaceae). Curr. Biol. 31, 1002–1011.e1009. doi: 10.1016/j.cub.2020.12.045
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10, 1–9. doi: 10.1186/1471-2105-10-421
Chen, J., Yu, R., Dai, J., Liu, Y., Zhou, R. (2020). The loss of photosynthesis pathway and genomic locations of the lost plastid genes in a holoparasitic plant Aeginetia indica. BMC Plant Biol. 20, 199. doi: 10.1186/s12870-020-02415-2
Choi, K.-S., Park, S. (2021). Complete plastid and mitochondrial genomes of aeginetia indica reveal intracellular gene transfer (IGT), horizontal gene transfer (HGT), and cytoplasmic Male sterility (CMS). Int. J. Mol. Sci. 22, 6143. doi: 10.3390/ijms22116143
Cole, L. W., Guo, W., Mower, J. P., Palmer, J. D. (2018). High and variable rates of repeat-mediated mitochondrial genome rearrangement in a genus of plants. Mol. Biol. Evol. 35, 2773–2785. doi: 10.1093/molbev/msy176
Darracq, A., Varré, J. S., Maréchal-Drouard, L., Courseaux, A., Castric, V., Saumitou-Laprade, P., et al. (2011). Structural and content diversity of mitochondrial genome in beet: A comparative genomic analysis. Genome Biol. Evol. 3, 723–736. doi: 10.1093/gbe/evr042
Dong, S., Zhao, C., Chen, F., Liu, Y., Zhang, S., Wu, H., et al. (2018). The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genomics 19, 614. doi: 10.1186/s12864-018-4991-4
Drouin, G., Daoud, H., Xia, J. (2008). Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 49, 827–831. doi: 10.1016/j.ympev.2008.09.009
Fan, W., Zhu, A., Kozaczek, M., Shah, N., Pabón-Mora, N., González, F., et al. (2016). Limited mitogenomic degradation in response to a parasitic lifestyle in Orobanchaceae. Sci. Rep. 6, 36285. doi: 10.1038/srep36285
Goyet, V., Wada, S., Cui, S., Wakatake, T., Shirasu, K., Montiel, G., Simier, P., et al, et al. (2019). Haustorium inducing factors for parasitic orobanchaceae. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01056
Greiner, S., Lehwark, P., Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Gualberto, J. M., Newton, K. J. (2017). Plant mitochondrial genomes: dynamics and mechanisms of mutation. Annu. Rev. Plant Biol. 68, 225–252. doi: 10.1146/annurev-arplant-043015-112232
Guo, W., Grewe, F., Fan, W., Young, G.J., Knoop, V., Palmer, J.D., et al. (2016). Ginkgo and Welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution. Mol. Biol. Evol. 33, 1448–1460. doi: 10.1093/molbev/msw024
Hao, Z., Lv, D., Ge, Y., Shi, J., Weijers, D., Yu, G., et al. (2020). RIdeogram: drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ. Comput. Sci. 6, e251. doi: 10.7717/peerj-cs.251
He, W., Chen, C., Adedze, Y.M.N., Dong, X., Xi, K., Sun, Y., et al. (2020). Multicentric origin and diversification of atp6-orf79-like structures reveal mitochondrial gene flows in Oryza rufipogon and Oryza sativa. Evol. Appl. 13, 2284–2299. doi: 10.1111/eva.13022
Jin, J.-J., Yu, W. B., Yang, J. B., Song, Y., dePamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 241. doi: 10.1186/s13059-020-02154-5
Kado, T., Innan, H. (2018). Horizontal gene transfer in five parasite plant species in orobanchaceae. Genome Biol. Evol. 10, 3196–3210. doi: 10.1093/gbe/evy219
Koressaar, T., Remm, M. (2007). Enhancements and modifications of primer design program Primer3. Bioinformatics 23, 1289–1291. doi: 10.1093/bioinformatics/btm091
Kozik, A., Phillippy, A., Delcher, A. L., Smoot, M., Shumway, M., Antonescu, C., et al. (2019). The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PloS Genet. 15, e1008373. doi: 10.1371/journal.pgen.1008373
Kubo, T., Newton, K. J. (2008). Angiosperm mitochondrial genomes and mutations. Mitochondrion 8, 5–14. doi: 10.1016/j.mito.2007.10.006
Kuijt, J. (1969). The biology of parasitic flowering plants (Berkeley: University of California Press).
Kurtz, S., Phillippy, A., Delcher, A. L., Smoot, M., Shumway, M., Antonescu, M. C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5, R12. doi: 10.1186/gb-2004-5-2-r12
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 1–3. doi: 10.48550/arXiv.1303.3997
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lin, Q., Banerjee, A., Stefanović, S. (2022). Mitochondrial phylogenomics of Cuscuta (Convolvulaceae) reveals a potentially functional horizontal gene transfer from the host. Genome Biol. Evol. 14, 1–11. doi: 10.1093/gbe/evac091
Lowe, T. M., Chan, P. P. (2016). tRNAscan-SE on-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. doi: 10.1093/nar/gkw413
Maréchal, A., Brisson, N. (2010). Recombination and the maintenance of plant organelle genome stability. New Phytol. 186, 299–317. doi: 10.1111/j.1469-8137.2010.03195.x
Marie, D., Brown, S. C. (1993). A cytometric exercise in plant DNA histograms, with 2C values for 70 species. Biol. Cell 78, 41–51. doi: 10.1016/0248-4900(93)90113-S
Molina, J., Hazzouri, K. M., Nickrent, D., Geisler, M., Meyer, R. S., Pentony, M. M., et al. (2014). Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae). Mol. Biol. Evol. 31, 793–803. doi: 10.1093/molbev/msu051
Mower, J. P., Case, A. L., Floro, E. R., Willis, J. H. (2012a). Evidence against equimolarity of large repeat arrangements and a predominant master circle structure of the mitochondrial genome from a monkeyflower (Mimulus guttatus) lineage with cryptic CMS. Genome Biol. Evol. 4, 670–686. doi: 10.1093/gbe/evs042
Mower, J. P., Sloan, D. B., Alverson, A. J. (2012b). “Plant mitochondrial genome diversity: the genomics revolution,” in Plant genome diversity volume 1: plant genomes, their residents, and their evolutionary dynamics. Eds. Wendel, J. F., Greilhuber, J., Dolezel, J., Leitch, I. J. (New York: Springer), 123–144.
Naito, K., Kaga, A., Tomooka, N., Kawase, M. (2013). De novo assembly of the complete organelle genome sequences of azuki bean (Vigna angularis) using next-generation sequencers. Breed. Sci. 63, 176–182. doi: 10.1270/jsbbs.63.176
Nickrent, D. L. (2020). Parasitic angiosperms: How often and how many? TAXON 69, 5–27. doi: 10.1002/tax.12195
Palmer, J. D., Herbon, L. A. (1988). Plant mitochondrial DNA evolved rapidly in structure, but slowly in sequence. J. Mol. Evol. 28, 87–97. doi: 10.1007/BF02143500
Petersen, G., Anderson, B., Braun, H. P., Meyer, E. H., Moller, I. M. (2020). Mitochondria in parasitic plants. Mitochondrion 52, 173–182. doi: 10.1016/j.mito.2020.03.008
Petersen, G., Cuenca, A., Møller, I. M., Seberg, O. (2015). Massive gene loss in mistletoe (Viscum, viscaceae) mitochondria. Sci. Rep. 5, 17588. doi: 10.1038/srep17588
POWO (2022). Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew, Published on the Internet; http://www.plantsoftheworldonline.org / Retrieved 13 June 2022.
Sanchez-Puerta, M. V., Edera, A., Gandini, C. L., Williams, A. V., Howell, K. A., Nevill, P. G., et al. (2019). Genome-scale transfer of mitochondrial DNA from legume hosts to the holoparasite Lophophytum mirabile (Balanophoraceae). Mol. Phylogenet. Evol. 132, 243–250. doi: 10.1016/j.ympev.2018.12.006
Skippington, E., Barkman, T. J., Rice, D. W., Palmer, J. D. (2015). Miniaturized mitogenome of the parasitic plant viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc. Natl. Acad. Sci. United. States America 112, E3515–E3524. doi: 10.1073/pnas.1504491112
Sloan, D. B., Havird, J. C., Sharbrough, J. (2017). The on-again, off-again relationship between mitochondrial genomes and species boundaries. Mol. Ecol. 26, 2212–2236. doi: 10.1111/mec.13959
Sullivan, A. R., Eldfjell, Y., Schiffthaler, B., Delhomme, N., Asp, T., Hebelstrup, K. H., et al. (2020). The mitogenome of Norway spruce and a reappraisal of mitochondrial recombination in plants. Genome Biol. Evol. 12, 3586–3598. doi: 10.1093/gbe/evz263
Sun, G., Xu, Y., Liu, H., Sun, T., Zhang, J., Hettenhausen, C., et al. (2018). Large-Scale gene losses underlie the genome evolution of parasitic plant Cuscuta australis. Nat. Commun. 9, 1–8. doi: 10.1038/s41467-018-04721-8
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876. doi: 10.1093/nar/25.24.4876
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E.S., Fischer, A., Bock, R., et al. (2017). GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11. doi: 10.1093/nar/gkx391
Westwood, J. H., Yoder, J. I., Timko, M. P., dePamphilis, C. W. (2010). The evolution of parasitism in plants. Trends Plant Sci. 15, 227–235. doi: 10.1016/j.tplants.2010.01.004
Wicke, S., Naumann, J. (2018). Molecular evolution of plastid genomes in parasitic flowering plants. Adv. Bot. Res. 85, 315–347. doi: 10.1016/bs.abr.2017.11.014
Wick, R. R., Schultz, M. B., Zobel, J., Holt, K. E. (2015). Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. doi: 10.1093/bioinformatics/btv383
Woloszynska, M. (2010). Heteroplasmy and stoichiometric complexity of plant mitochondrial genomes–though this be madness, yet there’s method in’t. J. Exp. Bot. 61, 657–671. doi: 10.1093/jxb/erp361
Wu, Z., Cuthbert, J. M., Taylor, D. R., Sloan, D. B. (2015). The massive mitochondrial genome of the angiosperm silene noctiflora is evolving by gain or loss of entire chromosomes. Proc. Natl. Acad. Sci. United. States America 112, 10185–10191. doi: 10.1073/pnas.1421397112
Wu, Z., Sloan, D. B. (2019). Recombination and intraspecific polymorphism for the presence and absence of entire chromosomes in mitochondrial genomes. Heredity 122, 647–659. doi: 10.1038/s41437-018-0153-3
Wu, Z., Waneka, G., Sloan, D. B. (2020). The tempo and mode of angiosperm mitochondrial genome divergence inferred from intraspecific variation in Arabidopsis thaliana. G3 Genes|Genomes|Genetics 10, 1077–1086. doi: 10.1534/g3.119.401023
Wynn, E. L., Christensen, A. C. (2019). Repeats of unusual size in plant mitochondrial genomes: identification, incidence and evolution. G3.: Genes|Genomes|Genetics 9, 549–559. doi: 10.1534/g3.118.200948
Yu, R., Sun, C., Zhong, Y., Liu, Y., Sanchez-Puerta, M.V., Mower, J.P., et al. (2022). The minicircular and extremely heteroplasmic mitogenome of the holoparasitic plant Rhopalocnemis phalloides. Curr. Biol. 32, 470–479. doi: 10.1016/j.cub.2021.11.053
Yu, R., Sun, C., Liu, Y., Zhou, R. (2021). Shifts from cis-to trans-splicing of five mitochondrial introns in Tolypanthus maclurei. PeerJ 9, e12260. doi: 10.7717/peerj.12260
Keywords: Aeginetia indica, mitochondrial genome, repeat-mediated recombination, intraspecific variation, HGT
Citation: Zhong Y, Yu R, Chen J, Liu Y and Zhou R (2022) Highly active repeat-mediated recombination in the mitogenome of the holoparasitic plant Aeginetia indica. Front. Plant Sci. 13:988368. doi: 10.3389/fpls.2022.988368
Received: 07 July 2022; Accepted: 07 September 2022;
Published: 21 September 2022.
Edited by:
Daniel Potter, University of California, Davis, United StatesReviewed by:
Helena Storchova, Academy of Sciences of the Czech Republic, CzechiaZhiqiang Wu, Agricultural Genomics Institute at Shenzhen, (CAAS), China
Copyright © 2022 Zhong, Yu, Chen, Liu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Renchao Zhou, emhyZW5jaEBtYWlsLnN5c3UuZWR1LmNu
 Yan Zhong
Yan Zhong Runxian Yu
Runxian Yu Jingfang Chen1
Jingfang Chen1 Ying Liu
Ying Liu Renchao Zhou
Renchao Zhou